Abstract
Vitamin D can modulate the innate and adaptive immune system. Vitamin D deficiency has been associated with various autoimmune diseases. Th9 cells are implicated in the pathogenesis of numerous autoimmune diseases. Thus, we investigated the role of calcitriol (active metabolite of vitamin D) in the regulation of Th9 cell differentiation. In this study, we have unraveled the molecular mechanisms of calcitriol-mediated regulation of Th9 cell differentiation. Calcitriol significantly diminished IL-9 secretion from murine Th9 cells, associated with downregulated expression of the Th9-associated transcription factor, PU.1. Ectopic expression of VDR in Th9 cells attenuated the percentage of IL-9-secreting cells. VDR associated with PU.1 in Th9 cells. Using a series of mutations, we were able to dissect the VDR domain involved in the regulation of Il9 gene. The VDR-PU.1 interaction prevented the accessibility of PU.1 to the Il9 gene promoter thereby restricting its expression. However, the expression of Foxp3, Treg-specific transcription factor, was enhanced in the presence of calcitriol in Th9 cells. When Th9 cells are treated with both calcitriol and TSA (histone deacetylase inhibitor), the level of IL-9 reached to the level of wild-type untreated Th9 cells. Calcitriol attenuated specific histone acetylation at the Il9 gene. In contrast, calcitriol enhanced the recruitment of the histone modifier, HDAC1 at the Il9 gene promoter. In summary, we have identified that calcitriol blocked the access of PU.1 to Il9 gene by reducing its expression and associating with it as well as regulated the chromatin of Il9 gene to regulate expression.
Keywords: Vitamin D, Calcitriol, VDR, IL-9, HDAC1, PU.1, IL-10
Introduction:
The fat soluble vitamin D plays crucial function in immune system regulation apart from its conventional function in maintaining calcium and phosphorous homeostasis (1, 2). The biologically active form of vitamin D, designated as vitamin D3 or calcitriol, is a secosteroid hormone that is predominantly synthesized in the skin from 7-dehydrocholesterol by UV-B rays (2). Calcitriol acts via vitamin D receptor (VDR), which belongs to the nuclear receptor superfamily (2, 3). Binding of calcitriol brings about a change in conformation of VDR, followed by heterodimerization with an orphan receptor, retinoid X receptor (RXR). This heterodimer of VDR-RXR moves to the nucleus, binds to the vitamin D response elements (VDRE) to regulate the transcription of more than 200 genes (2, 3). VDR is expressed by numerous tissues such as brain, breast, kidney, intestine, liver, lung (4). Interestingly, VDR is also known to be expressed by a wide-range of immune cells such as monocytes, macrophages, NK cells, dendritic cells, B cells and T cells, highlighting the function of calcitriol in the immune system (2, 5). Active vitamin D promotes the antimicrobial mechanisms induced by macrophages and neutrophils. Calcitriol prevents the maturation and differentiation of the dendritic cells and thereby impairs T cell activation and differentiation, which upon dysregulation leads to the initiation and perpetuation of autoimmune diseases (2, 6, 7). Globally, there has been increased prevalence of autoimmune diseases including rheumatoid arthritis, multiple sclerosis, Crohn’s disease, type-I diabetes mellitus and other immune-mediated diseases (8). Vitamin D3 deficiency is associated with increased risk of several autoimmune diseases (2, 4). Several studies suggest that calcitriol supplementation attenuates the disease activity score and reduces disease exacerbation of various autoimmune diseases (9). Thus a strong correlation between low vitamin D levels and the development of autoimmune diseases exists and calcitriol supplementation might play a protective role in the pathophysiology of autoimmune diseases.
The principal immunological mechanism employed by calcitriol in providing protection against autoimmune diseases involves modulating the differentiation of various T helper (Th) cell subsets (6, 7, 10). Increased frequencies of Th1 and Th17 cells are known to be responsible for the pathogenesis of multiple sclerosis, rheumatoid arthritis, and inflammatory bowel disease (11). Interestingly, calcitriol treatment resulted in reduced frequency of pathogenic Th1 and Th17 cells in various autoimmune disease models and patients suffering from autoimmune diseases (10–12). This affirms the protective role of calcitriol in autoimmune diseases via modulating the development of pathogenic T helper cells. In contrast, calcitriol enhances the polarization of Th2 and Treg cells. The impact of calcitriol in the differentiation of IL-9-secreting Th9 cells is however not clearly understood. Th9 cells were discovered more than a decade ago and are known to impart protection against helminth infections as well as exhibit anti-tumor immune responses (13). Conversely, Th9 cells are also involved in the pathophysiology of several autoimmune diseases (14). Hence, modulating Th9 cell development might be an alternate strategy for restraining the perpetuation and progression of autoimmune diseases. Thus, owing to the pathological role of Th9 cells and the protective effects of vitamin D supplementation in the pathophysiology of autoimmune diseases, we have investigated the role and the molecular mechanisms employed by calcitriol in regulating Th9 cell development.
In this study, we have observed that calcitriol attenuated the secretion of IL-9 in murine Th9 cells associated with impaired level of Th9-specific transcription factors (PU.1, IRF4 and Batf). Ectopic expression of VDR resulted in decreased IL-9 secretion from murine Th9 cells. Mechanistically, VDR associated with PU.1 in Th9 cells that impaired the binding of PU.1 to the Il9 gene culminating in impaired IL-9 expression and secretion by Th9 cells. Using a series of mutations of the ligand binding and DNA binding domains of VDR, we have identified specific domains of VDR that interacted with PU.1. Furthermore, Th9 cells treated with calcitriol upregulated the secretion of IL-10, an anti-inflammatory cytokine and enhanced the expression and recruitment of Foxp3 at Il9 gene. Calcitriol also regulated Il9 gene expression epigenetically. The inhibitory effect of calcitriol on Th9 cell development was rescued by trichostatin A, HDAC inhibitor. We also observed increased recruitment of HDAC1 at the Il9 gene in the presence of calcitriol concomitant with decrease in the specific histone modifications essential for the permissive chromatin. Thus, our results suggest multiple novel mechanisms that are employed by calcitriol for the regulation of Th9 cell development and their potential therapeutic role to treat autoimmune diseases.
Materials and Methods
Mice
Female C57BL/6 mice were procured from National Institute of Nutrition (Hyderabad, India). They were housed and bred at small animal facility of IIT Kharagpur in pathogen-free conditions. All the studies were performed according to the guidelines laid out by Institutional Animal Ethics Committee of IIT Kharagpur.
Murine T helper cell differentiation
Naïve CD4+ T cells were isolated from the spleen of 6–8 w/o female mice by magnetic cell sorting (Biolegend, CA, USA). Magnetically sorted naïve CD4+ T cells were grown in RPMI 1640 medium supplemented with 10% FBS and 1% antibiotic-antimycotic solution at 37°C in an incubator with 5% CO2. Plate-bound anti-CD3 (2 μg/mL, Biolegend) and soluble anti-CD28 (1 μg/mL, Biolegend) were used to activate naïve CD4+ T cells, followed by differentiation into Th1 (IL-12 [5 ng/mL; Peprotech, NJ, USA], IL-2 [(50 U/mL); Biolegend] and anti-IL-4 [10 μg/mL; Peprotech]); Th2 (IL-4 [20 ng/mL; Biolegend] and anti-IFN-γ [10 μg/mL; Biolegend]), Th9 (IL-4 [20 ng/mL], TGF-β [2 ng/mL; Biolegend] and anti-IFN-γ [10 μg/mL]), Th17 (TGF-β [2 ng/mL], IL-6 [100 ng/mL; Peprotech], IL-1β [10 ng/mL; Peprotech], IL-23 [10 ng/mL; Peprotech], anti-IFN-γ [10 μg/mL] and anti-IL-4 [10 μg/mL]) and Treg cell differentiating conditions (TGF-β [2 ng/mL], anti-IL-4 [10 μg/mL; Peprotech] for 3 days. The cultures were expanded for further 2 days by adding three times of fresh media for all the culture conditions with IL-4 and TGF-β for Th9, half the concentration of IL-6, IL-1β, IL-23 for Th17 and IL-2 for Treg conditions. Calcitriol used in the experiments was procured from Sigma Aldrich (MO, USA).
Intracellular cytokine staining and flow cytometry
Day 5 differentiated Th9 cells were stimulated with phorbol 12-myristate 13-acetate (PMA) and ionomycin (Sigma Aldrich) for 6 hrs. Monensin was added for the last 2 hrs of stimulation. The cells were surface stained using PE/Cy7 anti-CD4 antibody (Biolegend). The cells were then fixed using paraformaldehyde followed by permeabilization and staining using fluorochrome-conjugated anti-mouse IL-9, anti-mouse IL-4 and anti-mouse IL-10 (Biolegend). The flow cytomter FACS Calibur (BD Biosciences, CA, USA) was used to analyse the stained cells. The data was analysed by FlowJo software (Tree Star, OR, USA)
Quantitative RT-PCR and cytokine analysis
On day 5, differentiated Th cells were re-stimulated using anti-CD3 for 6 hrs and total mRNA was isolated by TRIzol (Thermo Fisher Scientific, MA, USA). Reverse transcription of RNA into cDNA was performed using the verso cDNA synthesis kit and analysed using PowerUp SYBR™ Green PCR master mix in QuantStudio 5 real-time PCR system (Thermo Fisher Scientific). The change in gene expression was normalized using β-actin as an endogenous control. The relative fold change in the gene expression was estimated using the formula 2(−ΔΔCT). The primer sequences for real-time PCR have been depicted in the Table S1. Cytokine analysis was performed using ELISA by re-stimulating 5-day differentiated T helper cells with anti-CD3 for 24 hrs and detecting the amount of IL-9 in cell-free supernatant by using anti-IL-9 capture and biotin-conjugated anti-IL-9 detection antibodies (Biolegend).
Immunoblot
Th0, Th1, Th2, Th9, Th17 and Treg cells were differentiated for 5 days. Upon differentiation, total nuclear lysates were prepared from the indicated T helper cell subsets using CelLytic M (Merck, NJ, USA). The cell lysates were resuspended in Laemmli buffer, boiled at 100°C for 5 mins, run on SDS-PAGE gel, transferred on PVDF membrane and probed with antibodies against PU.1, VDR (Cell Signaling Technology, MA, USA), IRF4 and BATF (Santa Cruz Biotechnology, TX, USA) and β-Actin (Biorbyt, Cambridge, UK).
Plasmids
The MIEG-hCD4 retroviral plasmid was a generous gift from Prof. Mark H. Kaplan, Indiana University and pCDNA3.1 was a kind gift from Dr Arindam Mondal (School of Bioscience, IIT Kharagpur). The murine VDR open reading frame, pGEM-mVDR was purchased from Sino Biological Inc. (Beijing, China). The mVDR was cloned into MIEG-hCD4 vector using EcoRI and XhoI restriction sites. The DNA binding domain deletion mutant of VDR (VDRΔDBD) was made from wild type VDR using stitch PCR. The primer pairs 1, 2 and 3, 4 were used to generate individual fragments. The final stitching of both fragments was carried out using equimolar concentrations of each fragment. The mouse VDR lacking the amino acids from 403 to 422 (VDRΔ403), from 304 to 422 (VDRΔ304) and from 276 to 422 (VDRΔ276) were generated using forward primer 5 and different reverse primers 6, 7 and 8, respectively and cloned into pCDNA3.1. The primer sequences of primers 1–8 have been mentioned in Table S2.
Transient transfection and retroviral transduction
HEK293T cells were cultured in Dulbecco’s Modified Eagle’s medium supplemented with 10% FBS and 1% antibiotic-antimycotic solution. HEK293T cells were transfected with MIEG-mVDR-hCD4 using lipofectamine 3000 (Thermo Fisher Scientific) along with viral envelope constructs gag-pol and Env (provided by Prof. Mark H. Kaplan). The retroviral supernatant was collected after 24 hrs of transfection and was filtered using 0.45 μm filter. Naïve CD4+ T cells differentiated for 2 days under Th9 differentiating conditions were transduced with the retroviral supernatant using polybrene (Merck). On day 5, the transduced cells were analysed for the secretion of IL-9 by flow cytometry.
Immunoprecipitation
Nuclear extracts were prepared from T helper cells differentiating in Th9 developing conditions and HEK293T cells transfected with pCDNA3.1-mVDR, MIEG-PU.1-GFP (gift from Prof. Mark H. Kaplan) as well as different deletion constructs lacking mVDR DNA and ligand binding domains. 0.5 mg of nuclear lysate was immunoprecipitated with 2 μg of anti-VDR and anti-PU.1 antibody at 4°C overnight using protein G agarose beads (Cell Signaling Technology). The conjugate was eluted from the beads by boiling at 100°C for 5 minutes, followed by centrifugation at 10000g for 10 minutes. The eluted conjugate was run on SDS-PAGE, transferred onto PVDF membrane and probed against VDR/PU.1 antibodies.
Chromatin immunoprecipitation
Th9 cells were cross-linked with formaldehyde and the protein-chromatin conjugate was immunoprecipitated with protein-G agarose beads overnight at 4°C. The nuclear lysates were immunoprecipitated with antibodies against VDR, PU.1, Foxp3, H4K5Ac, H3K9Ac, H3K14Ac, HDAC1 and rabbit IgG (Cell Signaling Technology). Protein-chromatin cross-links were reversed by incubation at 65°C, followed by DNA elution by phenol-chloroform extraction and ethanol precipitation. The binding of the above mentioned proteins at the three previously identified conserved regions of Il9 (CNS1, CNS0 and CNS-25) was determined using real-time PCR analysis. The list of primers for all the three conserved non-coding sequences in Il9 gene has been mentioned in Table S1.
Statistics
One-way analysis of variance followed by post-hoc Tukey multiple comparison test was used to determine statistical significance among all the data sets using GraphPad Prism 6.0 software (CA, USA). p<0.05 and p<0.01 were considered to be statistically significant for all the experiments.
Results
Vitamin D receptor is expressed by Th9 cells and treatment with calcitriol leads to enhanced recruitment of VDR at vitamin D response element on Il9 gene
Calcitriol, is known to modulate the development of different T helper cell subsets (15). Calcitriol binds to its receptor VDR that leads to modulation of immune responses (2). VDR expression is usually associated with dampened Th1 and Th17 cell development and promotion of Th2 and Treg cell polarization (15). However, the role of calcitriol has remained understudied in Th9 cell differentiation. To begin with we assessed the level of VDR expression across different T helper subsets. VDR was expressed by activated Th cells and by differentiated Th cell subsets at various levels (Fig 1a). VDR was abundantly expressed by Th9 cells when compared with Th1, Th2, Th17 and Treg cells observed by qPCR and immunoblot (Fig 1a). This prompted us to investigate whether Th9 cells respond to calcitriol by altering VDR expression or by regulating signal transduction via VDR (16, 17). The addition of calcitriol (ligand) did not significantly alter VDR (receptor) expression in Th9 cells as evident from the qPCR and immunoblot experiments (Fig 1b). Subsequently, we wanted to determine if there was enhanced recruitment of VDR to the putative VDRE in the presence of calcitriol (150 bp downstream of the Il9 gene promoter) using chromatin immunoprecipitation (ChIP) (Fig 1c). We demonstrated that calcitriol treatment led to enhanced recruitment of VDR at the putative VDRE, suggesting that calcitriol might alter the activation status of VDR and thereby stimulate VDR signaling in order to regulate Th9 cell differentiation (Fig 1c). Collectively, our preliminary results support calcitriol-mediated regulation of Th9 cells.
Figure 1: Calcitriol regulates Th9 cell development via vitamin D receptor.
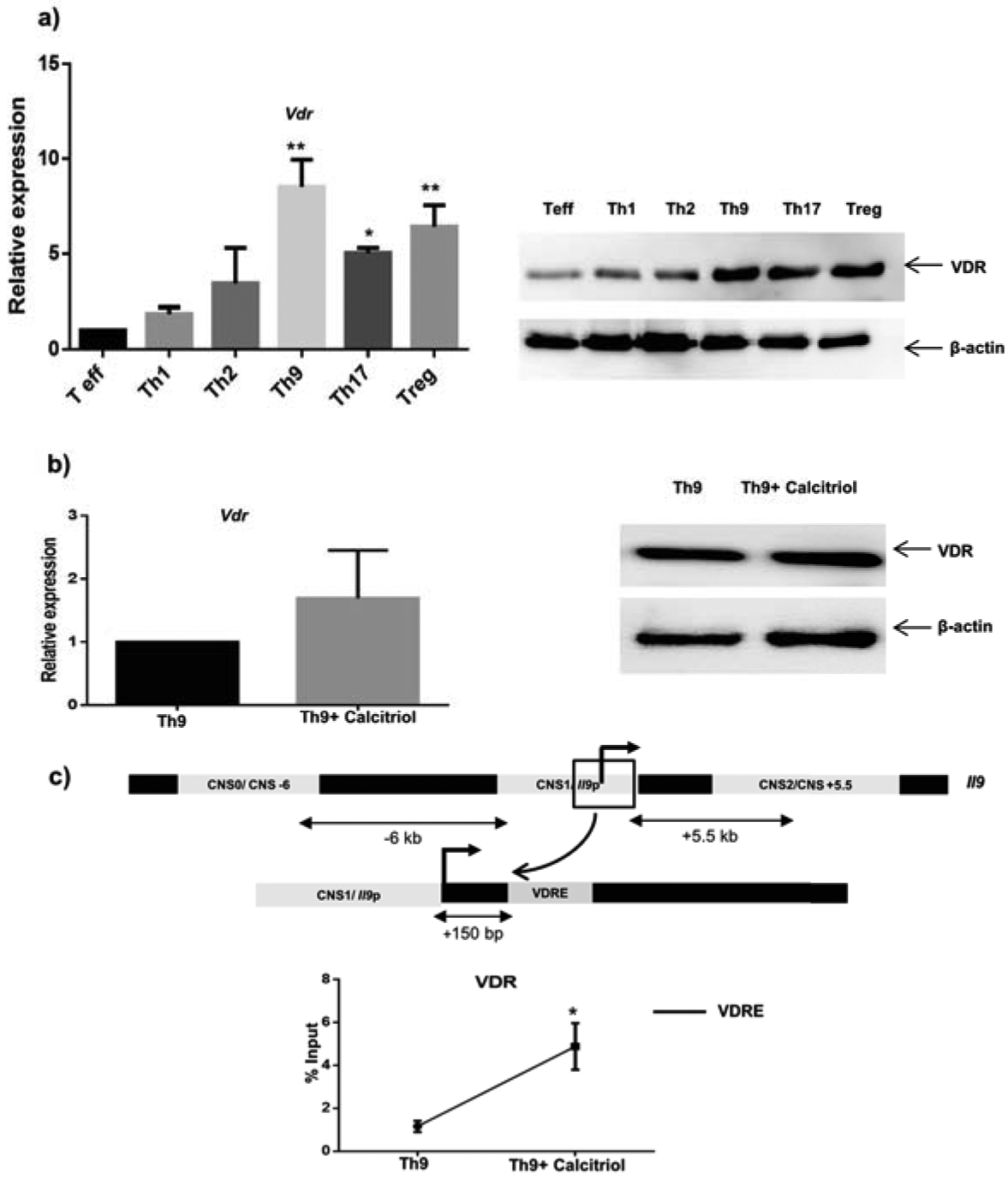
a) Naïve CD4+ T cells were isolated from C57BL/6 mice and activated under different T helper cell differentiating conditions (Th1, Th2, Th9, Th17 and Treg). VDR expression was determined by qPCR and immunoblot normalized against β-actin expression. b) Naive CD4+ T cells were activated in Th9 differentiating conditions with calcitriol (100 nM) and the expression by VDR by Th9 cells was investigated by qPCR and immunoblot. c) Binding of VDR at vitamin D response element upon treatment with calcitriol was tested using ChIP. Graphs depict mean± S.D of 3 independent experiments. *p<0.05 and **p<0.01 when compared to Teff (a) and Th9 (b and c).
Calcitriol impairs Th9 cell differentiation
Th cell subsets such as Th1, Th17 and Th9 cells are known to have both beneficial and detrimental functions. As mentioned previously, calcitriol levels and VDR expression are known to limit autoimmune responses. The immunomodulatory effect of calcitriol on Th1 and Th17 cell differentiation has been reported in several studies (18–21). Therefore, we wanted to define the role of calcitriol in the differentiation of Th9 cells. Naïve CD4+ T cells were polarized into Th9 cells in the presence of calcitriol (10, 50 and 100 nM) for 5 days and IL-9 production was assessed by intracellular cytokine staining. Calcitriol treatment resulted in a dose-dependent decrease in IL-9 production by Th9 cells with maximal reduction at 100 nM concentration with no change in cell viability (Fig S1a, 2a, b). These data correlate with the findings of Palmer and Keating (22, 23). IL-4 is the shared cytokine required for the differentiation of Th2 and Th9 cells. Th9 cells also secrete IL-4 at low levels. We observed that treatment with calcitriol did not impact IL-4 production by Th9 cells suggesting that calcitriol specifically dampens IL-9 secretion by Th9 cells and this effect is probably IL-4-independent (Fig 2a). We further confirmed this observation by determining the expression and secretion of IL-9 by Th9 cells in the presence of calcitriol using qPCR and ELISA (Fig 2b, c). Next we determined whether the effect of calcitriol on Th9 cell production was VDR-dependent. Developing Th9 cells were transduced with control plasmid (MIEG-hCD4) and VDR-expressing plasmid (MIEG-mVDR-hCD4). IL-9 secretion by Th9 cells was significantly reduced when VDR was ectopically expressed in Th9 cells, suggesting VDR-dependent function in the process (Fig 2d). Together, our findings confirm the inhibitory role of calcitriol on Th9 cell development in VDR-dependent manner.
Figure 2: Calcitriol downregulates Th9 cell development.
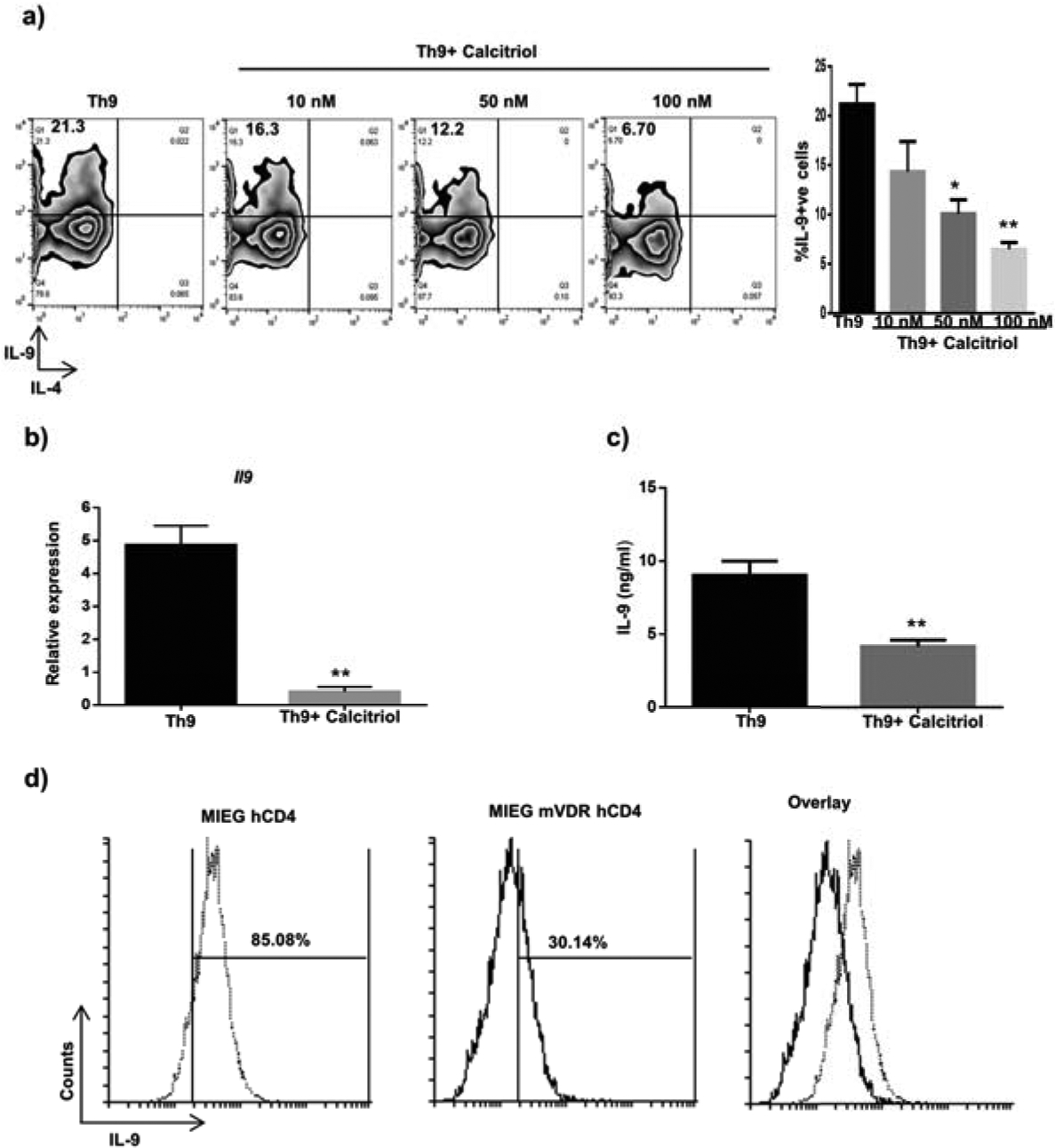
(a) Th9 cells were treated with different doses of calcitriol (10, 50 and 100 nM) and the frequency of IL-9+ cells was determined by flow cytometry. FACS plots depict the representative figure for the effect of different doses of calcitriol on the number of IL-9+ cells in Th9-differentiating condition. The graph represents number of IL-9 secreting Th9 cells when treated with different doses of calcitriol from at least three independent experiments. b) and c) Th9 cells were treated with calcitriol (100 nM) and the expression of Il9 as well as the amount of IL-9 secreted by Th9 cells was assessed by qPCR and ELISA. d) Murine vitamin D receptor was ectopically expressed in Th9 cells by retroviral transduction and the number of IL-9+ cells was detected by flow cytometry. Graphs represent mean± S.D of 3 independent experiments. *p<0.05 and **p<0.01 when compared to Th9 cells.
Calcitriol downregulates the expression of Th9-specific transcription factors
Transcription factors differentiate effector T cells into Th cell subsets. Th9 cell differentiation is also regulated by a slew of transcription factors. Downstream of TGF-β signal, the transcription factor PU.1 and downstream of IL-4 signal the transcription factors IRF4 and Batf play important roles in Th9 cell differentiation (24). Additionally, the STAT6 signal is also indispensable for Th9 cell differentiation (25). Calcitriol modulates the differentiation of Th17 cells at the transcriptional level via reducing the expression of RORγt (22). Thus, we determined if calcitriol-mediated Th9 cell differentiation is controlled by modifying the expression of Th9-specific transcription factors. We observed that calcitriol significantly attenuated the expression of Sfpi1, Irf4, Batf and Stat6 mRNA in Th9 cells using qPCR (Fig 3a). This was further confirmed using immunoblot prepared from the nuclear extracts of Th9 cells treated with calcitriol. We demonstrated significantly attenuated expression of PU.1, IRF4 and Batf in the presence of calcitriol in Th9 cells (Fig 3b). Hence, calcitriol negatively regulates Th9 cell differentiation by downmodulating the transcriptional machinery required for the development of Th9 cells. Th9 cell differentiation requires the cytokines IL-4 and TGF-β, which are required for differentiating into Th2 and Th17 cells, respectively. This led us to speculate that the inhibitory effect of calcitriol on Th9 cells might be secondary to its effect in Th2 and Th17 cell differentiation. Therefore, we determined whether calcitriol attenuated IL-9 secretion indirectly by impairing the transcription machinery of Th2 and Th17 cells. Gata3 is lineage-specific transcription factor of Th2 cells and have been speculated to be regulating Th9 cell differentiation by suppressing the expression of the transcription factor, Foxp3 that negatively regulates Th9 cell differentiation (26). Th9 cells express Gata3; however, its expression was not altered in Th9 cells by calcitriol that correlated with unchanged expression of Il4 (Fig 3c). IL-9 has been suggested to be a mediator of Th17-mediated inflammatory diseases (27). Rorγt expression was unaltered when Th9 cells were treated with calcitriol (Fig 3c). This correlated with unchanged expression of Il17 (Fig 3c). Thus, our data collectively indicates that calcitriol inhibits IL-9-secreting Th9 cell development by attenuating the expression of Th9-differentiating transcription factors without impacting the expression of cytokines and transcription factors specific for the differentiation of Th2 and Th17 cells.
Figure 3: Calcitriol attenuates the expression of Th9-differentiating transcription factors.
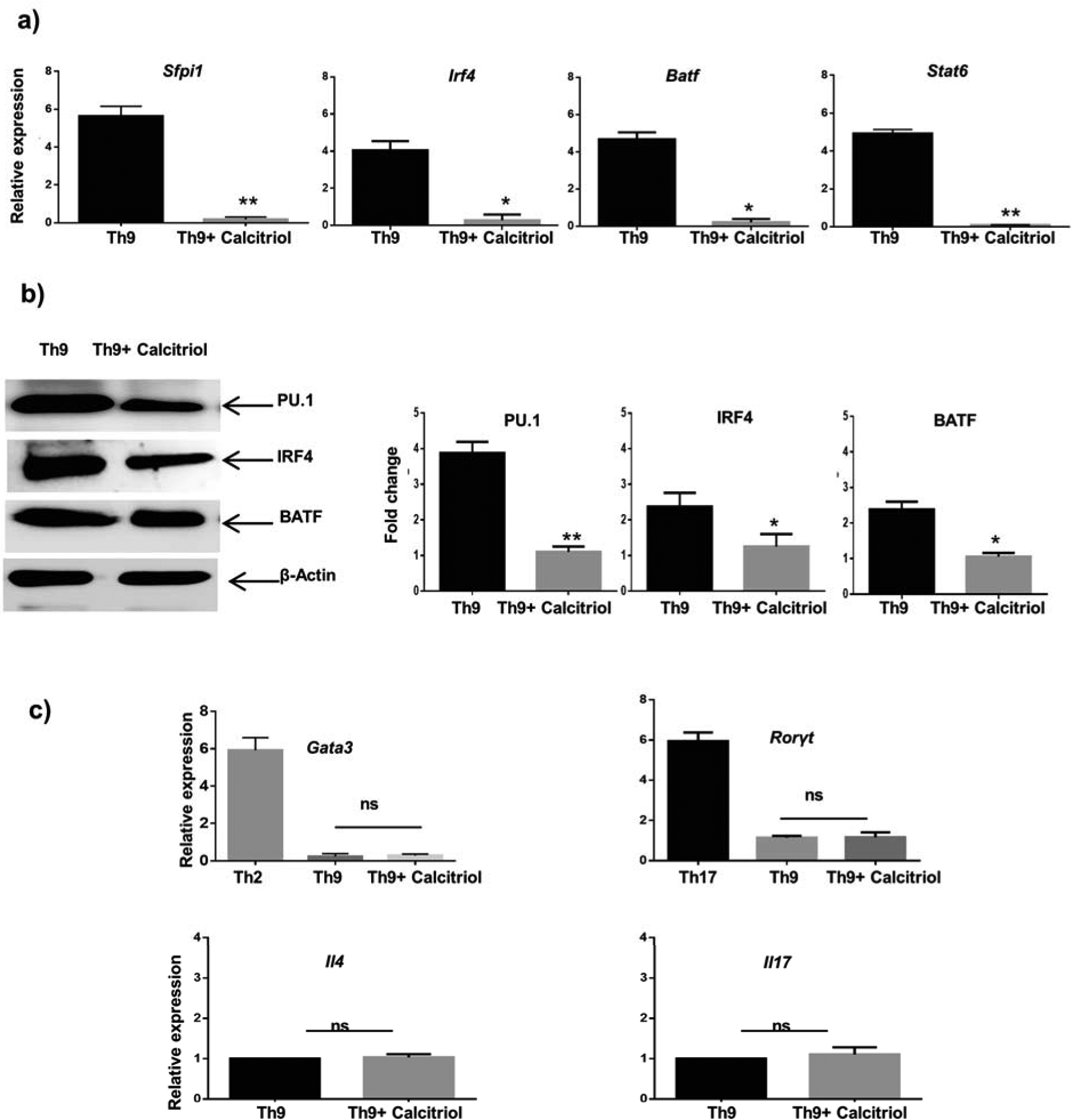
a) Th9 cells treated in the presence of calcitriol and the expression of various transcriptions factors Sfpi1, Irf4, Batf and Stat6 was determined by qPCR normalized against β-actin. b) The expression of PU.1, IRF4 and BATF by Th9 cells when treated with calcitriol was measured by western blot. c) Th9 cells were treated with calcitriol and the expression of Gata3, RORγt, Il4 and Il17 was investigated using qPCR. Gata3 and RORγt expression by Th2 and Th17 cells respectively, was used as standard. Graphs represent mean± S.D from 3 independent experiments. *p<0.05 and**p<0.01 when compared to Th9 cells. ns: not significant when compared to Th9 cells.
Vitamin D receptor interacts with PU.1 and impairs its binding to the Il9 gene locus
PU.1 is known to bind the Il9 gene locus and thereby regulate the development of Th9 cells (28). Our data suggested reduced expression of PU.1 by Th9 cells in the presence of calcitriol (Fig 3a, b). Hence, we wanted to determine whether decreased PU.1 expression is also associated with attenuated binding of PU.1 at Il9 gene locus thereby impeding Th9 cell development. We ascertained the effect of calcitriol on PU.1 binding at the Il9 gene using ChIP assay. We observed that treatment with calcitriol led to significantly decreased binding of PU.1 at CNS1 of the Il9 gene indicating that calcitriol impairs PU.1 binding at the Il9 gene culminating in attenuated Th9 cell development (Fig 4a). Additionally, we also identified the effect of calcitriol on the binding of PU.1 at putative VDRE on Il9 gene and observed significantly reduced PU.1 binding at VDRE in the presence of calcitriol (Fig S1b).
Figure 4: Vitamin D receptor associates with PU.1 and impairs the binding of PU.1 at Il9 gene.
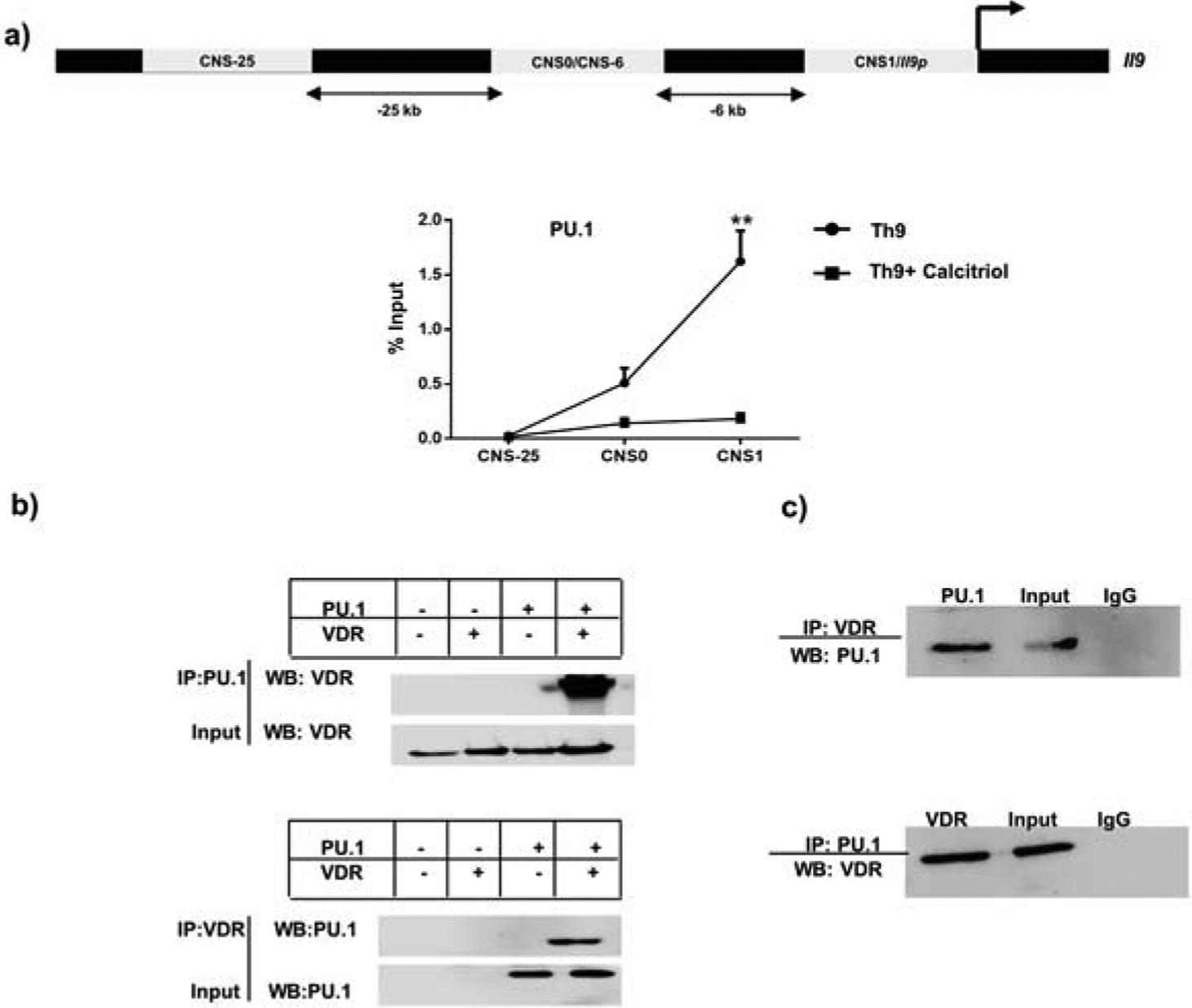
a) The three conserved non-coding sequences on Il9 gene have been depicted. The binding of PU.1 at the three CNS has been described. b) Top panel: HEK293T cells were transfected with VDR and PU.1-expressing plasmids, followed by immunoprecipitation with PU.1, probing against VDR. Bottom panel: Immunoprecipitation with VDR and blotting against PU.1 upon transfection of HEK293T cells with VDR and PU.1-expressing plasmids. c) Top panel: Th9 cells treated with calcitriol were immunoprecipitated with VDR and probed against PU.1. Bottom panel: Immunoprecipitation of nuclear lysate from calcitriol treated Th9 cells with PU.1, followed blotting against VDR.
We have observed previously that treatment with calcitriol leads to enhanced recruitment of VDR at putative VDRE on the Il9 gene (Fig 1c). Moreover, we also demonstrated decreased binding of PU.1 at Il9 gene locus (Fig 4a). Thus, we speculated that in the presence of calcitriol, VDR might bind to PU.1 binding sites on the Il9 gene and thereby attenuate PU.1 binding indirectly culminating in diminished Th9 cell development. However, PU.1 binding sites do not overlap with VDRE half-sites (Fig S1c) and thus we hypothesized that VDR in the presence of calcitriol might directly associate with PU.1 and thereby prevent PU.1 from binding at the Il9 gene to regulate Th9 cell development. In the presence of calcitriol, VDR is known to physically interact with Runx1 culminating in decreased binding of Runx1 at the Il17a gene (10). Moreover, VDR is also known to directly interact with PU.1 in alveolar macrophages suggesting a possible interaction between PU.1 and VDR in Th9 cells. We transfected human embryonic kidney (HEK293T) cells with PU.1 and VDR expressing plasmids as mentioned in the materials and methods section, followed by nuclear lysate preparation and co-immunoprecipitation to determine the association of VDR with PU.1. When immunoprecipitated with VDR, PU.1 was co-precipitated (Fig 4b). This was confirmed in reciprocal experiments that indicated VDR and PU.1 associate with each other (Fig 4b). We confirmed this observation by preparing nuclear lysates from Th9 cells cultured in the presence of calcitriol, immunoprecipitated with VDR followed by blotting with PU.1 (Fig 4c). This was confirmed in reciprocal experiments (Fig 4c). Consistent with our previous observation, we observed that VDR and PU.1 co-eluted in Th9 cells in the presence of calcitriol (Fig 4c). Hence, our results imply that VDR physically associates with PU.1 and thereby attenuates the binding of PU.1 at Il9 gene.
Next, we wanted to determine the domain(s) of VDR that interact with PU.1. Murine VDR is 422 amino acids long (29). It has a 76 amino acid-long DNA binding domain with a ligand binding domain that is 292 amino acids long (29). Thus, in order to map the region of VDR critical for association with PU.1, we generated deletion constructs of mouse VDR (mVDR) as described in the materials and methods section (Fig 5a). Briefly, we co-transfected HEK293T cells with full length mVDR and mutant mVDR lacking the DNA binding domain (ΔDBD), as well as distinct regions of ligand binding domain such as (Δ276, Δ304 and Δ403) along with PU.1-expressing plasmid. Next, we prepared nuclear lysates and immunoprecipitated with VDR followed by probe with PU.1 or vice-versa. We demonstrated that PU.1 co-precipitated with mVDR lacking regions of DNA binding domain as well as portions of ligand binding domain from 304–422 amino acids similar to the full length mVDR (Fig 5b and c; respectively). However, PU.1 precipitate was absent when mVDR lacked amino acids from 276–422 (Fig 5c). Thus, the region required for VDR interaction with PU.1 involves amino acids from 276–304. Overall, our results suggest that the ligand binding domain of VDR (276–304 amino acids) interacts with PU.1 and thereby impairs the binding of PU.1 at Il9 gene.
Figure 5: Domain-specific interaction of VDR with PU.1.
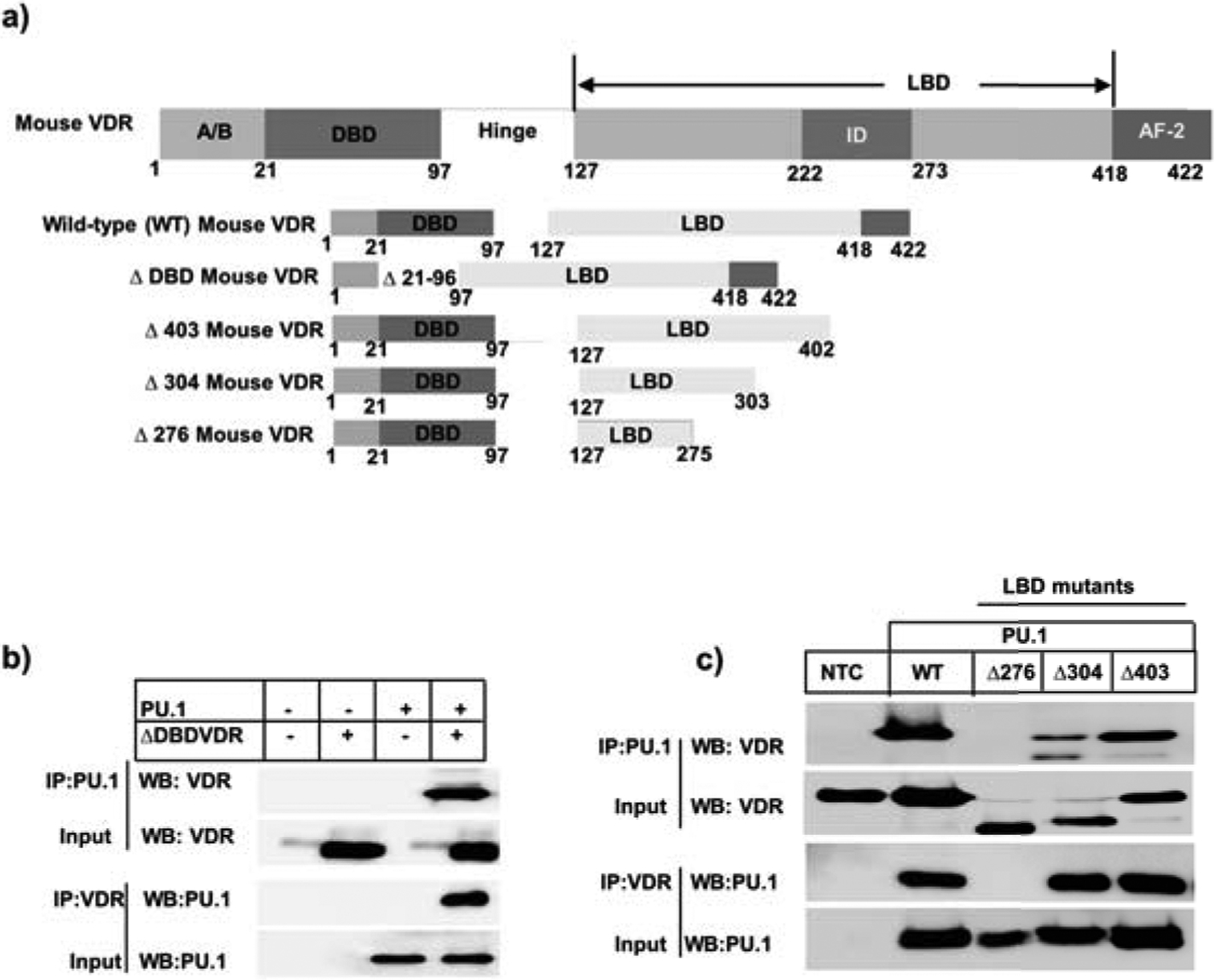
a) The wild-type VDR domain organization and the generation of DNA binding (DBD) and ligand binding (LBD) deletion mutants have been depicted. b) Top panel: Transfection of HEK293T cells with DBD deletion construct and PU.1-expressing plasmid and immunoprecipitation with PU.1 followed by probing against VDR. Bottom panel: Reciprocal immunoprecipitation of nuclear lysates prepared from calcitriol treated Th9 cells with VDR and probing against PU.1. c) Top panel: Co-transfection of HEK293T cells with wild-type mouse VDR or different ligand binding domain deletion mutants of mouse VDR with PU.1-expressing plasmid followed by immunoprecipitation with PU.1 and blotting against VDR. Bottom panel: Immunoprecipitation of nuclear lysates from the above mentioned co-transfected HEK293T cells with VDR and probing against PU.1.
Calcitriol increases the secretion of IL-10 by Th9 cells and enhances the binding of Foxp3 at Il9 gene locus
Foxp3 is the master regulator of Treg cells, which predominantly secrete anti-inflammatory cytokine IL-10 (30). Foxp3 expression is essential to maintain immune homeostasis and thereby limit autoimmunity (31). Induction of IL-10 also plays a critical role in preventing autoimmune diseases (32). Ectopic Foxp3 expression impairs IL-9 production in Th9 cells, thereby inhibiting Th9 cell development (33). Moreover, calcitriol deficiency has been correlated with dampened Foxp3+T cells in autoimmune patients (34). Calcitriol is also known to increase the ratio of Foxp3+T cells to IL-17+T cells associated with IL-10 induction in various autoimmune disease models (15). We therefore investigated whether there was concomitant increase in IL-10 production and Foxp3 expression upon treating Th9 cells with calcitriol. Calcitriol treatment led to a significant increase in Foxp3 expression and IL-10 production by Th9 cells (Fig 6a). This finding was confirmed by the significant increase in IL-10-secreting T cells that were inversely proportional to the percentage of IL-9-secreting T cells (Fig 6b). We then determined if the inhibitory effect of calcitriol on Th9 cells was mediated by IL-10. Th9 cells were treated with calcitriol along with IL-10 neutralizing antibody (anti-IL-10) to measure the frequency of IL-9+ cells in Th9 differentiating conditions. Calcitriol-treated Th9 cells in the presence of anti-IL-10 significantly increased IL-9 production (Fig 6b), as previously observed (22). However, this increase of IL-9 did not reach to the level achieved by Palmer et al. This could be due to the lack of complete blockade of IL-10. Weaver and colleagues blocked both IL-10 and its receptor, IL-10R. We also observed significant reduction of IL-10-secreting cells when anti-IL-10 was added to calcitriol-treated Th9 cells (Fig 6b). Thus, calcitriol decreased IL-9-producing T cells concomitant with an increase in IL-10-producing T cells.
Figure 6: Calcitriol induces IL-10 secretion by Th9 cells and promotes Foxp3 binding at Il9 gene.
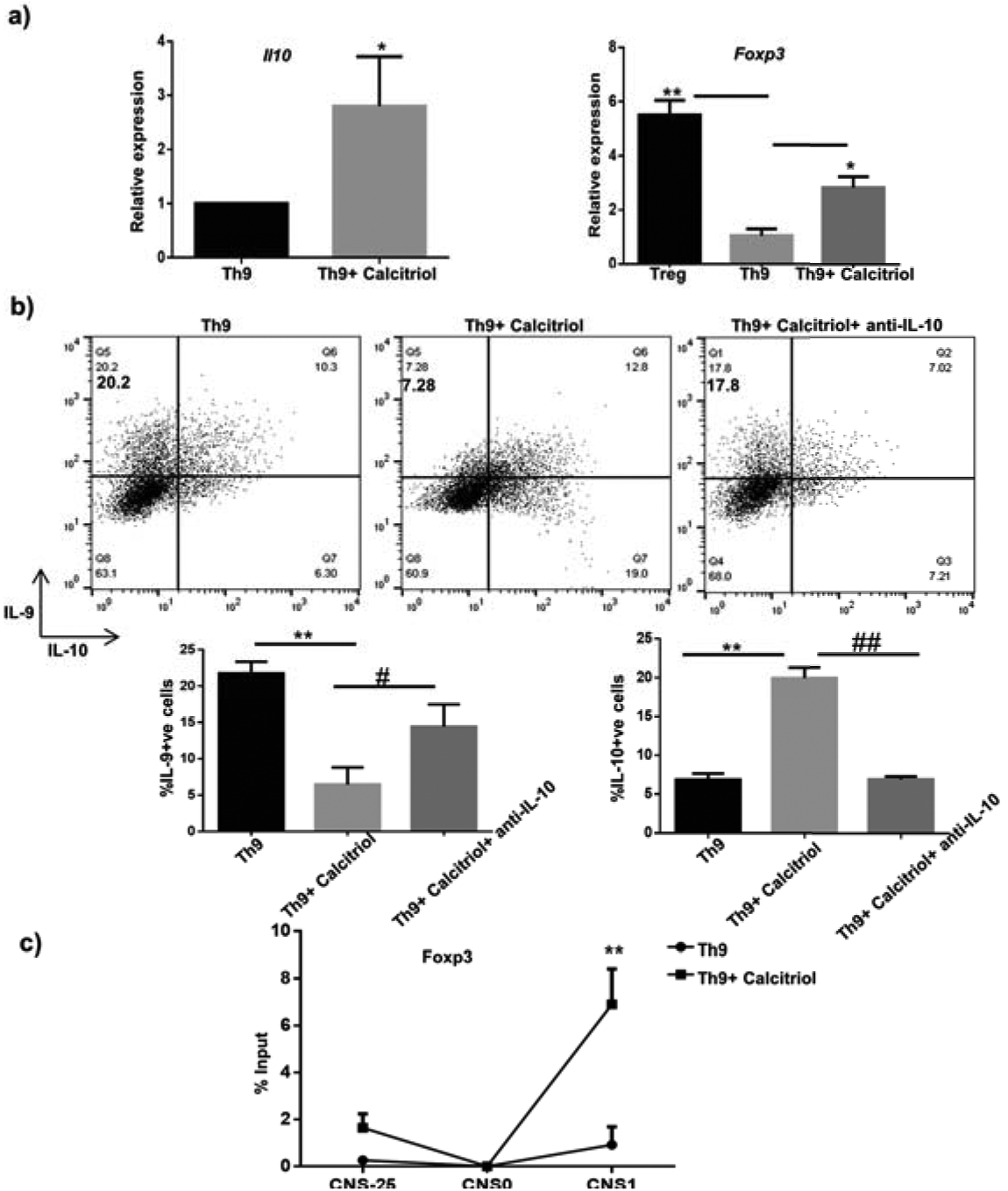
a) IL-10 and Foxp3 expression by Th9 cells treated with calcitriol is determined by q RT-PCR. b) The frequency of IL-10+ cells in the presence of calcitriol and anti-IL-10 (30 μg/mL) has been depicted in the representative FACS plots. The graphs represent the frequency of IL-9 and IL-10-positive cells in the presence of calcitriol and anti-IL-10 from 3 independent experiments. c) Foxp3 binding at three CNS of Il9 gene was determined by ChIP. Graphs depict mean± S.D of 3 independent experiments *p<0.05, **p<0.01 when compared to Th9 cells alone and #p<0.05, ##p<0.01 when compared to Th9 cells in the presence of calcitriol.
Even though Foxp3 expression was significantly increased in the presence of vitamin D in Th9 cells, it did not reach the level expressed by Treg cells (Fig 6a). We performed additional experiments to determine the function of Foxp3 in mediating the regulatory effect of calcitriol in Th9 cells. Foxp3 binding sites are present in the Il9 gene locus (33). Moreover, increased Foxp3 binding at the conserved non-coding sequences of Il9 gene prevents IL-9 expression resulting in impaired Th9 cell development (33). Thus, we performed ChIP to investigate whether calcitriol altered the Foxp3 binding at the CNSs of Il9 gene to regulate Th9 cell development. Interestingly, we observed that calcitriol treatment resulted in increased recruitment of Foxp3 at the Il9 promoter (Fig 6c). Collectively, our results imply that the immunomodulatory role of calcitriol in Th9 cell development is mediated by not affecting the expression of Foxp3, but by altering the recruitment of Foxp3 to the Il9 gene.
Calcitriol epigenetically regulates Th9 cell development
Global gene expression results from a balance between histone acetylation and deacetylation. We have previously observed that calcitriol recruited Foxp3 at Il9 gene locus culminating in impaired Th9 cell development (Fig 6c). Furthermore, Foxp3 is known to recruit histone deacetylase, HDAC1 at the Il9 gene locus resulting in repressive chromatin and suppression of Il9 gene transcription (33). Thus, we investigated whether calcitriol impacted Th9 cell development by regulating Il9 gene epigenetically. We speculated that recruitment of Foxp3 at the Il9 gene locus by calcitriol might in turn alter the recruitment of HDAC1 at Il9 gene locus and thereby regulate Il9 gene transcription. We assessed the binding of HDAC1 at all the three CNS of Il9 gene in the presence of calcitriol. We demonstrated that calcitriol treatment significantly increased the binding of HDAC1 at the three CNS (Fig 7a). Thus, it can be argued that calcitriol enhances Foxp3 binding, which in turn augments the binding of HDAC1 at Il9 gene locus. Consequently, this results in repressive chromatin at the Il9 gene leading to diminished Il9 gene transcription and Th9 cell development. Moreover, we also determined the binding of HDAC1 at putative VDRE using ChIP and observed increased binding of HDAC1 at VDRE as well in the presence of calcitriol (Fig S1d).
Figure 7: Calcitriol downmodulates Th9 cell development epigenetically.
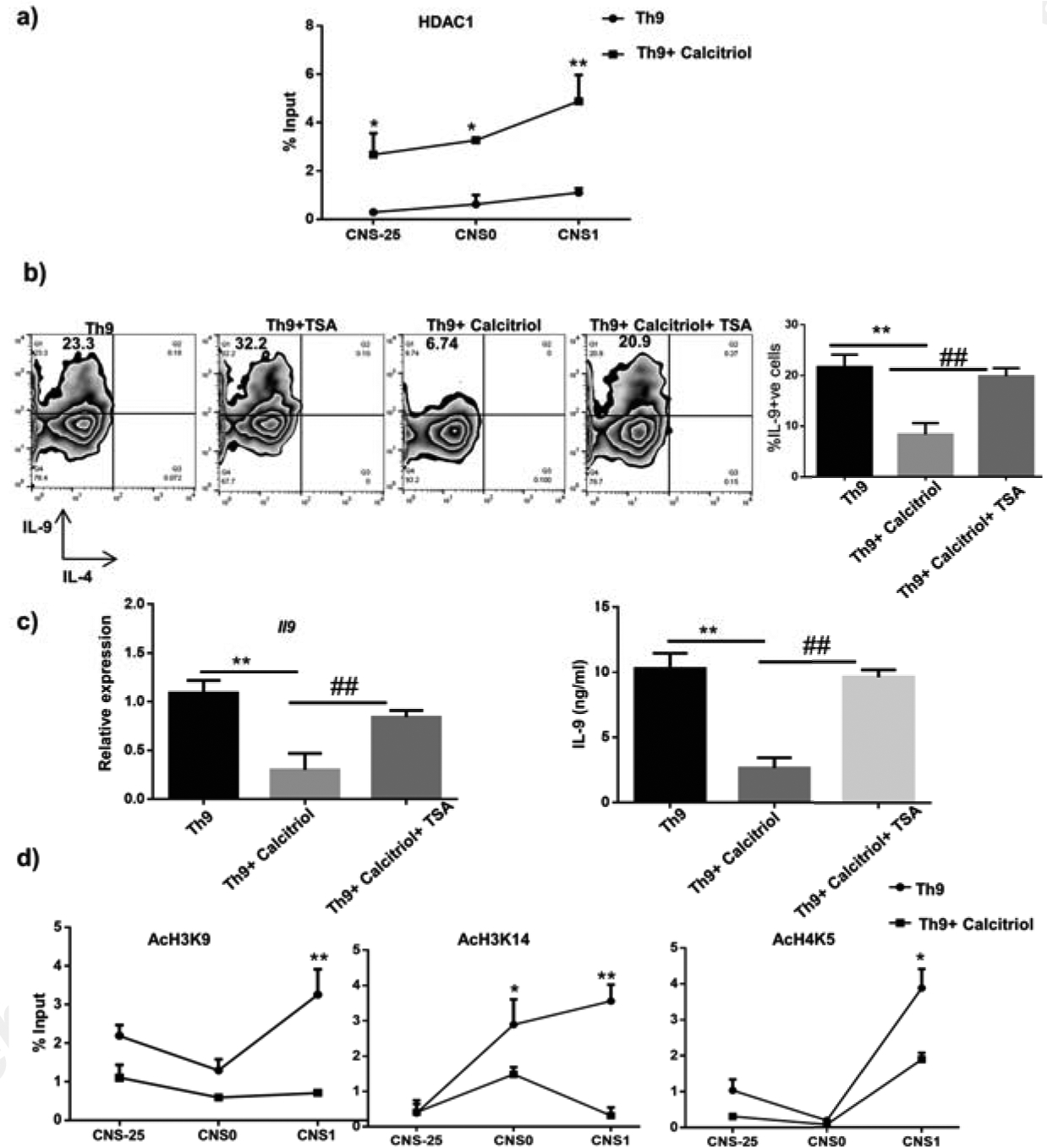
a) HDAC1 binding at the three CNS of Il9 gene in the presence of calcitriol was determined by ChIP. b) Th9 cells were cultured with calcitriol and trichostatin A (10 nM). The number of IL-9+ cells were analyzed by flow cytometry. Representative FACS plots indicate the frequency of IL-9+ cells upon treatment with calcitriol and TSA. The graphs represent IL-9+ cells from at least 3 independent experiments. c) The amount of IL-9 from Th9 cells treated with calcitriol and TSA was determined by qPCR and ELISA, respectively. d) Acetylation of specific histone residues including H3K9, H3K14 and H4K5 upon treatment of Th9 cells with calcitriol was determined using ChIP. *p<0.05, **p<0.01 when compared to Th9 cells and #p<0.05 and ##p<0.01 when compared to Th9 cells treated with calcitriol.
Several studies suggest that HDAC1/2 inhibitors such as trichostatin A (TSA) can reverse the repressive chromatin at Il9 gene locus and thereby rescue Foxp3-mediated suppression of Il9 gene transcription (33, 35). Thus, we tested whether TSA can rescue the inhibitory effect of calcitriol on Th9 cells. Differentiating Th9 cells were treated with calcitriol alone and or in combination with TSA and analysed the frequency of IL-9-positive cells. Upon treatment with TSA, an increase in IL-9 was observed, in accordance with our earlier publication (Fig 7b) (35). Consistent with our previous observation, calcitriol alone significantly inhibited the production of IL-9 (Fig 7b). Interestingly, the percentage of IL-9+ cells recovered to the level of control Th9 cells when Th9 cells were treated with both calcitriol and TSA (Fig 7b). Thus, our data implies that TSA can rescue the inhibitory effect of calcitriol on Th9 cells by reversing the repressive chromatin induced by calcitriol via promoting the recruitment of HDAC1 at the Il9 gene locus. We further confirmed this observation by determining the effect of TSA on calcitriol-mediated Il9 gene suppression using qPCR and ELISA. In the presence of TSA, the inhibitory effect of calcitriol was negated and associated with significant increase in the amount of IL-9 (Fig 7c).
Histone acetylation at lysine residues results in permissive chromatin leading to enhanced gene transcription (36). Increased histone acetylation at specific lysine residues leads to permissive Il9 gene locus and thereby increases Il9 gene transcription (35). Histone acetylation at Il9 gene is regulated by PU.1 and PU.1-deficiency results in attenuated histone acetylation at the Il9 promoter (35). We demonstrated decreased PU.1 levels as well as reduced PU.1 binding at Il9 gene locus in the presence of calcitriol. This prompted us to test whether decreased PU.1 binding at Il9 gene locus upon calcitriol treatment, impacts histone acetylation at Il9 gene as well. We initially tested the effect of calcitriol on total acetylation of histone 3 (H3) and 4 (H4) and demonstrated a significant decrease in the total acetylation of H3 but not total acetylation at H4 at Il9 promoter by ChIP (Fig S1e). Subsequently, we investigated the acetylation of specific histone residues at Il9 gene in the presence of calcitriol. We determined the acetylation at three specific lysine residues including H3K9, H3K14 and H4K5 at 3 CNS of Il9 gene. We observed a decrease in the acetylation of all the three histone acetylation marks in the presence of calcitriol at the Il9 gene promoter (Fig 7d). H3K14 acetylation was also significantly impaired at CNS0 region (Fig 7d). Even though a similar trend was observed at both CNS-25 and CNS0 for H3K9 acetylation, they did not reach statistical significance (Fig 7d). Overall, our data suggests that calcitriol attenuates Th9 cell development by increasing the recruitment of HDAC1 and by impairing histone acetylation at Il9 gene locus.
Discussion:
Multiple factors impact the function of immune system including micronutrients, such as vitamin D. A significant correlation exists between nutrition and immune system function as deficiency of single or multiple nutrients alter functional immune responses. The biologically active form vitamin D3, calcitriol is known to regulate the maturation, co-stimulation, cytokine production and migration, antigen presentation, and T cell skewing by dendritic cells (37–39). Function of calcitriol has been explored in adaptive immune cells including T helper cell subsets such as Th1, Th2, Th17, Treg cells. However, few studies have observed the impact of calcitriol in the differentiation of IL-9-secreting Th9 cells. The importance of IL-9 and Th9 cells has been reviewed extensively (13, 24). In this manuscript we have defined the molecular mechanisms of calcitriol-mediated regulation of Th9 cell differentiation.
Calcitriol attenuated IL-9 production in murine Th9 cells that is consistent with previous studies (Fig 2) (22, 23). We also observed reduced IL-9 production and PU.1 expression when human Th9 cells were treated with calcitriol (unpublished data), suggesting the translatability of the findings to clinical settings. In Th2 cells, calcitriol enhanced the production of IL-4 and increased STAT6 expression in human (40). Unchanged IL-4-secreting cells in Th9 cells treated with calcitriol suggested that calcitriol-mediated modulation of IL-9 in Th9 cells is not directly dependent on IL-4 (Fig 2a). The cytokine TGF-β is required for differentiation of both Th9 and Th17 cells. Calcitriol did not alter the expression of Il17 gene in Th9 cells, suggesting attenuated IL-9 production in Th9 cells did not directly depend on IL-17 (Fig 3c). The expression of Il21, another cytokine produced by Th9 cells was not altered after calcitriol treatment (Fig S2a). The cytokine IL-10 has been reported to be secreted by Th9 cells; however low, compared to Th2 cells (28). Calcitriol impaired the differentiation of murine Th9 cells associated with increased IL-10 (Fig 6) (13). We also observed an increased level of Il10 gene expression that correlated with enhanced IL-10 secretion in Th9 cells treated with calcitriol (Fig 6b). Neutralizing IL-10 in Th9 cells significantly enhanced the secretion of IL-9 that affirms with the findings of Ulrich et al (Fig 6b) (41).
Previous studies have revealed that calcitriol altered the expression of the transcription factors specific to T helper subsets. We examined if calcitriol altered the expression of transcription factors essential for Th9 cell differentiation to regulate Il9 gene. In the presence of calcitriol, there was significant attenuation of the expression of PU.1, IRF4 and Batf, transcription factors that positively regulate Th9 cell differentiation (Fig 3a, b). Gata3, the lineage-specific transcription factor of Th2 cells, is also important for Th9 cell differentiation (13). We did not observe any difference of Gata3 expression when Th9 cells were treated with calcitriol (Fig 3c). Whether calcitriol regulates Il9 gene in Th9 cells indirectly by modifying ‘Th2-specific’ transcription factor will require further studies. Foxp3, is a negative regulator of Th9 cells (25). We observed an increased expression of Foxp3 in Th9 cells treated with calcitriol (Fig 6a). Calcitriol could additionally modulate IL-9 production by regulating IL-10. Loss of IL-9 is associated with enhanced level of IL-10 as well as augmented activated STAT3 levels (41). This data suggests that calcitriol not only attenuates the expression of transcription factors that positively regulate Th9 cells, but also enhances the expression of Foxp3, negative regulator of Th9 cells to modulate Il9 gene. Our results are in line with the study by Joshi et al that revealed the direct effect of Foxp3 induction by calcitriol to regulate pro-inflammatory Th17 responses (10).
VDR can bind to genomic DNA in a sequence-specific manner that is composed of two hexameric nucleotide half-sites. VDR-RXR-VDRE make up the core elements of calcitriol-mediated signaling. VDR can modulate the expression of a gene by interacting with other regulators. Studies have suggested context-dependent interaction of VDR with PU.1 (42, 43). Since PU.1, a ‘pioneer transcription factor’ can be regarded as the closest to a ‘master regulator’ of Th9 cells, we examined if VDR interacted with PU.1. We observed an interaction between VDR and PU.1 in cell lines and in primary Th9 cells (Fig 4b, c). Subsequently, we observed significantly reduced recruitment of PU.1 to Il9 gene promoter (Fig 4a). An enhancer 25 kb upstream of the Il9 gene (CNS-25) has been identified as the region to which most of the transcription factors required for Il9 gene expression bind (44). The significantly impaired recruitment of PU.1 was only observed at the Il9 promoter, but not observed at CNS-25 nor at CNS0, indicating that the Il9 promoter is probably the primary target of VDR-mediated altered recruitment of PU.1 (Fig 4a). Calcitriol could also regulate Il9 gene via IRF4, Batf and STAT6, transcription factors that are downstream of IL-4 signal in Th9 cells. There is increased representation of Batf in VDR binding peaks of DCs (45). VDR-IRF4 interaction could impact the regulation of IL-9. rs2853564, a SNP of VDR attenuated the binding of IRF4 (46). Batf has been shown to interact with IRF4 (47). It is possible that VDR can impact Batf-IRF4 interaction in Th9 cells to regulate IL-9. There could be additional transcriptional regulation of Il9 gene expression mediated by calcitriol. There are NFAT and AP-1 binding sites in the Il9 gene (48). The transcription factor NFATc1 eases the calcitriol-mediated suppression of transcription of human IL17A gene (10). Whether such regulation occurs at Il9 gene needs to be investigated.
We further dissected the VDR domain structure-function relationship involved in the regulation of Il9 gene. VDR is primarily composed of N-terminal variable A/B domain, core DNA binding domain (DBD) and structurally conserved C-terminal ligand binding domain (LBD) linked by a stretch of unstructured non-conserved hinge region (49). VDR LBD is able to recruit co-activators in its active conformation through the activation function-2 domain (50). To this end, we generated deletion mutation of the DBD (VDRΔDBD) and a series of mutations of LBD (VDRΔ276, VDRΔ304 and VDRΔ403) similar to the study conducted by Hsieh et al (51). The mutant VDRΔ403 lacking the AF-2 domain was used to determine its role in interaction of PU.1 with VDR. VDRΔ276 truncation having vitamin D interaction domain (insertion domain) intact was used to determine its importance. Another truncation in between the two above-mentioned regions led to VDRΔ304 mutant that was used to assess its importance in mediating interaction of VDR with PU.1. In the presence of vitamin D, the VDRΔ403 and VDRΔ304 were able to interact with PU.1 (Fig 5c). In contrast, VDRΔ276 abrogated this interaction, suggesting that a domain within aa 276–304 is required for VDR-PU.1 interaction. However, the absence of the DBD did not impair the binding of VDR with PU.1 as DBD is required to bind to its cognate DNA when activated (Fig 5b). These results are in line with the study by Hsieh et al where the VDRΔ134 truncation (lacking the entire LBD) abrogated the association of the Hairless (Hr)-corepressor with VDR. The same study showed that there was an insignificant difference of association between Hr and VDR when VDRΔ403 was used (51). However, VDRΔ304 mutation enhanced the association of Hr and VDR, suggesting the function of VDRΔ304 is context and partner-dependent. The DBD of VDR probably enhances the binding of VDR to the Il9 gene in the presence of the ligand. Additionally, DBD could also be responsible for the transregulation of a gene that is mediated by VDR. The domain-specific interaction between PU.1 and VDR in Th9 cells will require further studies.
Vitamin D can regulate the expression of a gene by orchestrating chromatin modification. Ligand-dependent impact of VDR with histone modifiers suggest the role calcitriol plays in modifying histone (52). Repression of IL17A gene involved the dissociation of histone acetylase activity and the recruitment of HDAC to the gene (10). We therefore wanted to examine if calcitriol modulated Il9 gene by modifying its chromatin in Th9 cells. When Th9 cells were treated with both calcitriol and TSA, the level of IL-9 was rescued to the level secreted by untreated Th9 cells (Fig 7b). This suggested that calcitriol would modulate Il9 gene epigenetically. While calcitriol alone increased IL-10 secretion in Th9 cells; calcitriol along with TSA did not alter IL-10 levels produced by Th9 cells (Fig S2b).
The VDR/RXR dimer has been shown to interact with histone modifiers to regulate gene transcription. Calcitriol enhances histone H3 and H4 acetylation in VDRE proximal regions of osteocalcin gene (53). We observed reduced AcH3K9, AcH3K14 and AcH4K5 marks in the presence of calcitriol at the Il9 promoter (Fig 7d). However, this difference was not observed at CNS0 region for any of the three specific histone acetylation marks in the presence of calcitriol, implying VDR works primarily through the Il9 promoter (Fig 7d). There could be additional histone acetylation marks of the Il9 gene that could be attenuated in the presence of calcitriol that could occur not only at the Il9 promoter, but also at other CNS. HDACs are also major regulators of gene expression. Not only immune pathologies are associated with HDACs, VDR-HDAC interaction has been observed in various diseases (54). Calcitriol resulted in the recruitment of HDAC2 to the IL17A promoter and suppressed its transcription (10). We therefore examined if calcitriol augmented the recruitment of HDAC1 to the Il9 gene. An enhanced recruitment of HDAC1 occurred at the Il9 promoter in Th9 cells (Fig 7a). This indicates that pharmacological modulation of histone acetylation could be used as therapeutic strategy for Th9 cell-mediated inflammatory diseases. Calcitriol also has been revealed to alter DNA methylation. Whether such regulation also modulates the Il9 gene is not known.
Calcitriol has been proposed to influence genomic VDR binding, affect the binding of ‘pioneer transcription factor’, and modify histones (55). In this study we have delineated the mechanisms of how calcitriol modulates the differentiation of IL-9-secreting Th9 cells by PU.1. Calcitriol attenuates the expression of PU.1 in Th9 cell differentiation but elevates the expression of Foxp3, a known negative regulator of Th9 cells. Ligand-activated VDR associates with PU.1 to prevent its binding to the Il9 gene. Additionally, calcitriol attenuates the positive histone modification marks of the Il9 gene, increases the recruitment of HDAC1 to the Il9 gene to suppress its expression. The model of calcitriol-mediated regulation of Il9 gene is shown in Figure 8. Further studies will feature how calcitriol regulates the functions of IRF4 and Batf, two other important Th9-specific transcription factors, to modulate the transcription of Il9 gene to reveal the control at the transcription factor network required for Th9 cell differentiation.
Figure 8: Mechanisms of calcitriol-mediated regulation of Th9 cells.
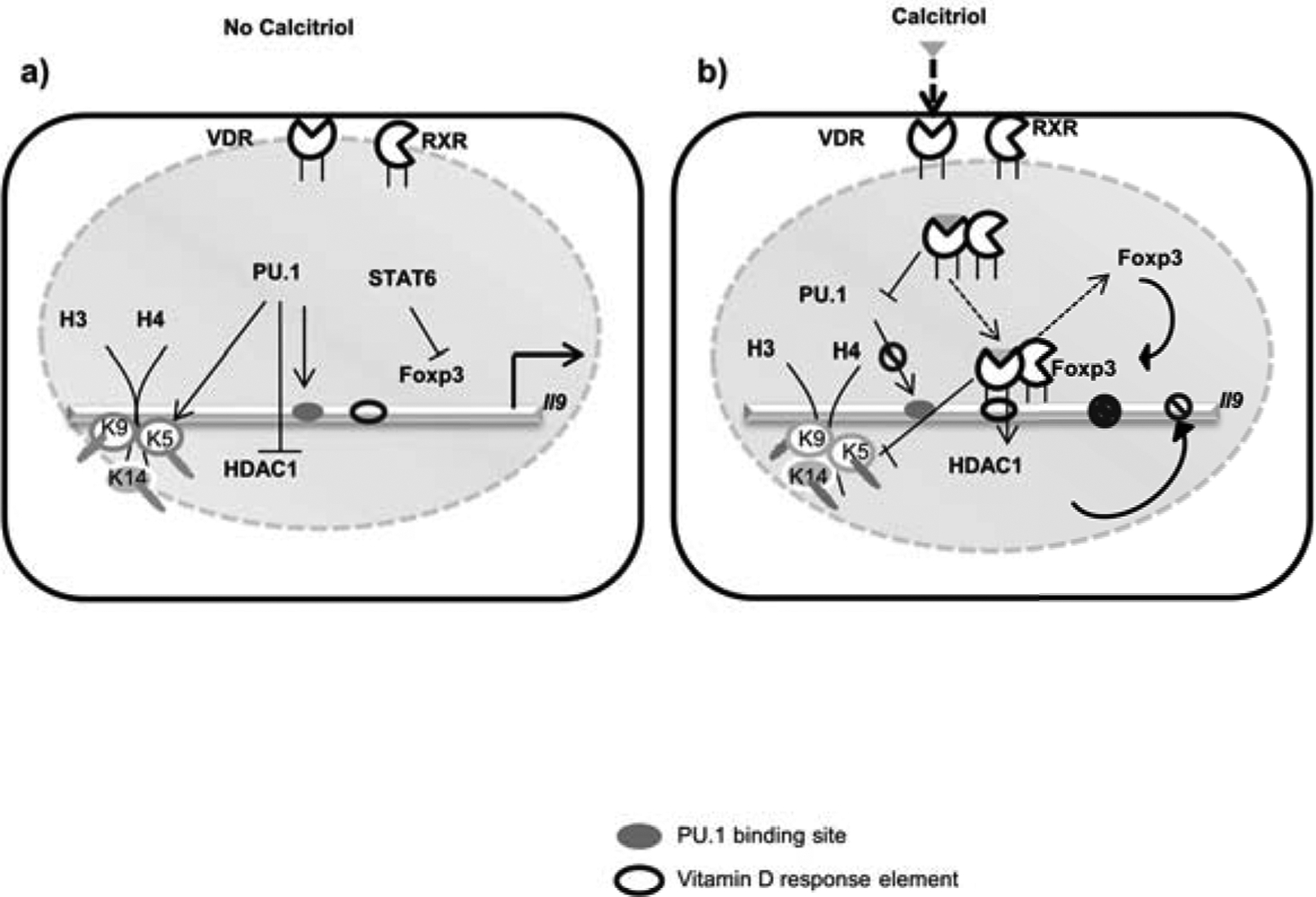
a) IL-9 production by Th9 cells is regulated at the transcriptional and epigenetic level by vitamin D. In the absence of calcitriol, PU.1 binds at Il9 gene locus and thereby inhibits the binding of HDAC1 at Il9 gene. PU.1 also promotes the acetylation of specific lysine residues of histone 3 and histone 4 thereby boosts Il9 gene transcription. Moreover, the binding of Foxp3 at Il9 gene (which attenuates Il9 gene transcription) is inhibited by STAT6. Thus, Il9 gene is controlled both at the transcriptional and at the chromatin level in Th9 cells. b) Calcitriol binds with VDR present in the nucleus, followed by heterodimerization with RXR and thereby binds to VDRE on the Il9 gene. Upon binding to the ligand (calcitriol), VDR impairs PU.1 expression, associates with PU.1 and thereby impairs the binding of PU.1 on Il9 gene locus. Moreover, calcitriol-VDR signaling recruits Foxp3 at Il9 gene. Additionally, calcitriol enhances the binding of HDAC1 at Il9 gene and by attenuating specific histone acetylation of Il9 gene.
Supplementary Material
Key Points:
Calcitriol impairs Th9 cell development.
Calcitriol downregulates PU.1 expression and its binding to the Il9 gene.
Calcitriol regulates chromatin remodelling at the Il9 gene locus.
Acknowledgment:
The authors acknowledge the Central Research Facility, IIT Kharagpur for providing access to the flow cytometer.
Funding: Shachi Pranjal Vyas acknowledges CSIR-UGC for providing fellowship. Arman Kunwar Hansda would like to acknowledge MHRD for providing fellowship. Dr Ritobrata Goswami has been supported by Young Scientist Scheme, SERB-DST, Government of India (YSS/2015/001147).
Footnotes
Conflict of interest: None to declare.
References
- 1.Singh P, Kumar M, and Al Khodor S. 2019. Vitamin D Deficiency in the Gulf Cooperation Council: Exploring the Triad of Genetic Predisposition, the Gut Microbiome and the Immune System. Frontiers in immunology 10: 1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang CY, Leung PS, Adamopoulos IE, and Gershwin ME. 2013. The implication of vitamin D and autoimmunity: a comprehensive review. Clinical reviews in allergy & immunology 45: 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haussler MR, Whitfield GK, Haussler CA, Hsieh JC, Thompson PD, Selznick SH, Dominguez CE, and Jurutka PW. 1998. The nuclear vitamin D receptor: biological and molecular regulatory properties revealed. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research 13: 325–349. [DOI] [PubMed] [Google Scholar]
- 4.Hayes CE, Hubler SL, Moore JR, Barta LE, Praska CE, and Nashold FE. 2015. Vitamin D Actions on CD4(+) T Cells in Autoimmune Disease. Frontiers in immunology 6: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Provvedini DM, Tsoukas CD, Deftos LJ, and Manolagas SC. 1983. 1,25-dihydroxyvitamin D3 receptors in human leukocytes. Science 221: 1181–1183. [DOI] [PubMed] [Google Scholar]
- 6.Mattner F, Smiroldo S, Galbiati F, Muller M, Di Lucia P, Poliani PL, Martino G, Panina-Bordignon P, and Adorini L. 2000. Inhibition of Th1 development and treatment of chronic-relapsing experimental allergic encephalomyelitis by a non-hypercalcemic analogue of 1,25-dihydroxyvitamin D(3). European journal of immunology 30: 498–508. [DOI] [PubMed] [Google Scholar]
- 7.Cippitelli M, and Santoni A. 1998. Vitamin D3: a transcriptional modulator of the interferon-gamma gene. European journal of immunology 28: 3017–3030. [DOI] [PubMed] [Google Scholar]
- 8.Blumberg RS, Dittel B, Hafler D, von Herrath M, and Nestle FO. 2012. Unraveling the autoimmune translational research process layer by layer. Nature medicine 18: 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordvik I, Myhr KM, Nyland H, and Bjerve KS. 2000. Effect of dietary advice and n-3 supplementation in newly diagnosed MS patients. Acta neurologica Scandinavica 102: 143–149. [DOI] [PubMed] [Google Scholar]
- 10.Joshi S, Pantalena LC, Liu XK, Gaffen SL, Liu H, Rohowsky-Kochan C, Ichiyama K, Yoshimura A, Steinman L, Christakos S, and Youssef S. 2011. 1,25-dihydroxyvitamin D(3) ameliorates Th17 autoimmunity via transcriptional modulation of interleukin-17A. Molecular and cellular biology 31: 3653–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamzaoui A, Berraies A, Hamdi B, Kaabachi W, Ammar J, and Hamzaoui K. 2014. Vitamin D reduces the differentiation and expansion of Th17 cells in young asthmatic children. Immunobiology 219: 873–879. [DOI] [PubMed] [Google Scholar]
- 12.Cantorna MT, Munsick C, Bemiss C, and Mahon BD. 2000. 1,25-Dihydroxycholecalciferol prevents and ameliorates symptoms of experimental murine inflammatory bowel disease. The Journal of nutrition 130: 2648–2652. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan MH, Hufford MM, and Olson MR. 2015. The development and in vivo function of T helper 9 cells. Nature reviews. Immunology 15: 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Chen S, Xiao X, Zhao Y, Ding W, and Li XC. 2017. IL-9 and Th9 cells in health and diseases-From tolerance to immunopathology. Cytokine & growth factor reviews 37: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantorna MT, Snyder L, Lin YD, and Yang L. 2015. Vitamin D and 1,25(OH)2D regulation of T cells. Nutrients 7: 3011–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veldman CM, Cantorna MT, and DeLuca HF. 2000. Expression of 1,25-dihydroxyvitamin D(3) receptor in the immune system. Archives of biochemistry and biophysics 374: 334–338. [DOI] [PubMed] [Google Scholar]
- 17.Pike JW, Meyer MB, and Bishop KA. 2012. Regulation of target gene expression by the vitamin D receptor - an update on mechanisms. Reviews in endocrine & metabolic disorders 13: 45–55. [DOI] [PubMed] [Google Scholar]
- 18.Kongsbak M, von Essen MR, Levring TB, Schjerling P, Woetmann A, Odum N, Bonefeld CM, and Geisler C. 2014. Vitamin D-binding protein controls T cell responses to vitamin D. BMC immunology 15: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemire JM 1992. Immunomodulatory role of 1,25-dihydroxyvitamin D3. Journal of cellular biochemistry 49: 26–31. [DOI] [PubMed] [Google Scholar]
- 20.Reichel H, Koeffler HP, Tobler A, and Norman AW. 1987. 1 alpha,25-Dihydroxyvitamin D3 inhibits gamma-interferon synthesis by normal human peripheral blood lymphocytes. Proceedings of the National Academy of Sciences of the United States of America 84: 3385–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang SH, Chung Y, and Dong C. 2010. Vitamin D suppresses Th17 cytokine production by inducing C/EBP homologous protein (CHOP) expression. The Journal of biological chemistry 285: 38751–38755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer MT, Lee YK, Maynard CL, Oliver JR, Bikle DD, Jetten AM, and Weaver CT. 2011. Lineage-specific effects of 1,25-dihydroxyvitamin D(3) on the development of effector CD4 T cells. The Journal of biological chemistry 286: 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keating P, Munim A, and Hartmann JX. 2014. Effect of vitamin D on T-helper type 9 polarized human memory cells in chronic persistent asthma. Annals of allergy, asthma & immunology : official publication of the American College of Allergy, Asthma, & Immunology 112: 154–162. [DOI] [PubMed] [Google Scholar]
- 24.Goswami R, and Kaplan MH. 2011. A brief history of IL-9. Journal of immunology 186: 3283–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goswami R, Jabeen R, Yagi R, Pham D, Zhu J, Goenka S, and Kaplan MH. 2012. STAT6 dependent regulation of Th9 development. Journal of immunology 188: 968–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, Sobel RA, Mitsdoerffer M, Strom TB, Elyaman W, Ho IC, Khoury S, Oukka M, and Kuchroo VK. 2008. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(−) effector T cells. Nature immunology 9: 1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowak EC, Weaver CT, Turner H, Begum-Haque S, Becher B, Schreiner B, Coyle AJ, Kasper LH, and Noelle RJ. 2009. IL-9 as a mediator of Th17-driven inflammatory disease. The Journal of experimental medicine 206: 1653–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang HC, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, Jabeen R, McKinley C, Ahyi AN, Han L, Nguyen ET, Robertson MJ, Perumal NB, Tepper RS, Nutt SL, and Kaplan MH. 2010. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nature immunology 11: 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamei Y, Kawada T, Fukuwatari T, Ono T, Kato S, and Sugimoto E. 1995. Cloning and sequencing of the gene encoding the mouse vitamin D receptor. Gene 152: 281–282. [DOI] [PubMed] [Google Scholar]
- 30.Loser K, Hansen W, Apelt J, Balkow S, Buer J, and Beissert S. 2005. In vitro-generated regulatory T cells induced by Foxp3-retrovirus infection control murine contact allergy and systemic autoimmunity. Gene therapy 12: 1294–1304. [DOI] [PubMed] [Google Scholar]
- 31.Wing JB, Tanaka A, and Sakaguchi S. 2019. Human FOXP3(+) Regulatory T Cell Heterogeneity and Function in Autoimmunity and Cancer. Immunity 50: 302–316. [DOI] [PubMed] [Google Scholar]
- 32.Calcinaro F, Dionisi S, Marinaro M, Candeloro P, Bonato V, Marzotti S, Corneli RB, Ferretti E, Gulino A, Grasso F, De Simone C, Di Mario U, Falorni A, Boirivant M, and Dotta F. 2005. Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia 48: 1565–1575. [DOI] [PubMed] [Google Scholar]
- 33.Xiao X, Shi X, Fan Y, Zhang X, Wu M, Lan P, Minze L, Fu YX, Ghobrial RM, Liu W, and Li XC. 2015. GITR subverts Foxp3(+) Tregs to boost Th9 immunity through regulation of histone acetylation. Nature communications 6: 8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chambers ES, and Hawrylowicz CM. 2011. The impact of vitamin D on regulatory T cells. Current allergy and asthma reports 11: 29–36. [DOI] [PubMed] [Google Scholar]
- 35.Goswami R, and Kaplan MH. 2012. Gcn5 is required for PU.1-dependent IL-9 induction in Th9 cells. Journal of immunology 189: 3026–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, and Allis CD. 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383: 269–272. [DOI] [PubMed] [Google Scholar]
- 37.Barragan M, Good M, and Kolls JK. 2015. Regulation of Dendritic Cell Function by Vitamin D. Nutrients 7: 8127–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danova K, Grohova A, Strnadova P, Funda DP, Sumnik Z, Lebl J, Cinek O, Pruhova S, Kolouskova S, Obermannova B, Petruzelkova L, Sediva A, Fundova P, Buschard K, Spisek R, and Palova-Jelinkova L. 2017. Tolerogenic Dendritic Cells from Poorly Compensated Type 1 Diabetes Patients Have Decreased Ability To Induce Stable Antigen-Specific T Cell Hyporesponsiveness and Generation of Suppressive Regulatory T Cells. Journal of immunology 198: 729–740. [DOI] [PubMed] [Google Scholar]
- 39.Rampal R, Awasthi A, and Ahuja V. 2016. Retinoic acid-primed human dendritic cells inhibit Th9 cells and induce Th1/Th17 cell differentiation. Journal of leukocyte biology 100: 111–120. [DOI] [PubMed] [Google Scholar]
- 40.Mahon BD, Wittke A, Weaver V, and Cantorna MT. 2003. The targets of vitamin D depend on the differentiation and activation status of CD4 positive T cells. Journal of cellular biochemistry 89: 922–932. [DOI] [PubMed] [Google Scholar]
- 41.Ulrich BJ, Verdan FF, McKenzie AN, Kaplan MH, and Olson MR. 2017. STAT3 Activation Impairs the Stability of Th9 Cells. Journal of immunology 198: 2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nurminen V, Seuter S, and Carlberg C. 2019. Primary Vitamin D Target Genes of Human Monocytes. Frontiers in physiology 10: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seuter S, Neme A, and Carlberg C. 2017. Epigenomic PU.1-VDR crosstalk modulates vitamin D signaling. Biochimica et biophysica acta. Gene regulatory mechanisms 1860: 405–415. [DOI] [PubMed] [Google Scholar]
- 44.Koh B, Abdul Qayum A, Srivastava R, Fu Y, Ulrich BJ, Janga SC, and Kaplan MH. 2018. A conserved enhancer regulates Il9 expression in multiple lineages. Nature communications 9: 4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Booth DR, Ding N, Parnell GP, Shahijanian F, Coulter S, Schibeci SD, Atkins AR, Stewart GJ, Evans RM, Downes M, and Liddle C. 2016. Cistromic and genetic evidence that the vitamin D receptor mediates susceptibility to latitude-dependent autoimmune diseases. Genes and immunity 17: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Innocenti F, Owzar K, Jiang C, Etheridge AS, Gordan R, Sibley AB, Mulkey F, Niedzwiecki D, Glubb D, Neel N, Talamonti MS, Bentrem DJ, Seiser E, Yeh JJ, Van Loon K, McLeod H, Ratain MJ, Kindler HL, Venook AP, Nakamura Y, Kubo M, Petersen GM, Bamlet WR, and McWilliams RR. 2018. The vitamin D receptor gene as a determinant of survival in pancreatic cancer patients: Genomic analysis and experimental validation. PloS one 13: e0202272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huber M, and Lohoff M. 2014. IRF4 at the crossroads of effector T-cell fate decision. European journal of immunology 44: 1886–1895. [DOI] [PubMed] [Google Scholar]
- 48.Perumal NB, and Kaplan MH. 2011. Regulating Il9 transcription in T helper cells. Trends in immunology 32: 146–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pike JW, and Meyer MB. 2010. The vitamin D receptor: new paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D(3). Endocrinology and metabolism clinics of North America 39: 255–269, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molnar F, Perakyla M, and Carlberg C. 2006. Vitamin D receptor agonists specifically modulate the volume of the ligand-binding pocket. The Journal of biological chemistry 281: 10516–10526. [DOI] [PubMed] [Google Scholar]
- 51.Hsieh JC, Sisk JM, Jurutka PW, Haussler CA, Slater SA, Haussler MR, and Thompson CC. 2003. Physical and functional interaction between the vitamin D receptor and hairless corepressor, two proteins required for hair cycling. The Journal of biological chemistry 278: 38665–38674. [DOI] [PubMed] [Google Scholar]
- 52.Nurminen V, Neme A, Seuter S, and Carlberg C. 2019. Modulation of vitamin D signaling by the pioneer factor CEBPA. Biochimica et biophysica acta. Gene regulatory mechanisms 1862: 96–106. [DOI] [PubMed] [Google Scholar]
- 53.Shen J, Montecino M, Lian JB, Stein GS, Van Wijnen AJ, and Stein JL. 2002. Histone acetylation in vivo at the osteocalcin locus is functionally linked to vitamin D-dependent, bone tissue-specific transcription. The Journal of biological chemistry 277: 20284–20292. [DOI] [PubMed] [Google Scholar]
- 54.Milovanovic M, Heine G, Hallatschek W, Opitz B, Radbruch A, and Worm M. 2010. Vitamin D receptor binds to the epsilon germline gene promoter and exhibits transrepressive activity. The Journal of allergy and clinical immunology 126: 1016–1023, 1023 e1011–1014. [DOI] [PubMed] [Google Scholar]
- 55.Carlberg C 2019. Nutrigenomics of Vitamin D. Nutrients 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


