Abstract
The aryl hydrocarbon receptor (AhR) is a cytoplasmic receptor and transcription factor, primarily activated through cognate ligand binding. It is an important factor in immunity and tissue homeostasis, and structurally diverse compounds from the environment, diet, microbiome, and host metabolism, can influence AhR activity. Emerging evidence suggests that AhR is a key sensor allowing immune cells to adapt to environmental conditions. Changes in AhR activity have been associated with autoimmune disorders and cancer, and AhR agonists or antagonists impacting immune disease outcomes have placed AhR as a potential actionable target for immunotherapy. We describe known ligands stimulating AhR activity, downstream pro-inflammatory and suppressive mechanisms potentiated by AhR, and how this understanding is applied to immunopathologies. Indeed, AhR-dependent immune responses might be modulated to help control pathologic outcomes.
An emerging link between environment and immune function
The aryl hydrocarbon receptor (AhR) is a cytoplasmic receptor that was initially discovered by virtue of its key role in mitigation of the toxic effects of environmental pollutants. However, recent evidence has shown that AhR responds to multiple exogenous and endogenous signals derived from diet, host metabolism, and the intestinal microbiome [1–3]. Upon ligand binding, AhR functions as a transcription factor impacting gene expression via promoter binding, recruitment of coactivators and corepressors at specific DNA regions, and interactions with signal transduction machinery in mouse and human cells [3–5]. Recent data have unexpectedly shown that AhR is critical for a wide range of immune functions including the maintenance of innate and adaptive cell populations at mucosal barrier sites, and the control of inflammation nodes (both at steady-state and in feedback loops during ongoing inflammatory reactions).
Exciting emerging evidence suggests that AhR activity can control immune disease severity and, perhaps most importantly, that AhR function can be targeted to improve disease outcomes in a variety of inflammatory pathologies (most notably certain autoimmune diseases) [3, 4, 6–8]. However, data suggest that AhR may be a double-edged sword that can drive both inflammatory and immune-suppressive phenotypes in T cells and myeloid cells depending on the AhR ligand and other unknown micro-environmental factors [3, 4, 9–13]. In this review, we explore mechanisms of ligand-dependent activation of AhR and its impact on cellular phenotypes. We also discuss the unexpected role AhR plays in controlling innate and adaptive immunity at multiple levels. Finally, we consider recent advances in our understanding of AhR as a systemic environmental sensor and modulator of inflammation during conditions of immune dysfunction, focusing especially on autoimmunity and cancer.
The Aryl Hydrocarbon Receptor (AhR): a sensor of the external and internal micro-environment.
In general, organisms are exposed to a multitude of environmental signals derived from internal physiologic processes and external sources via nutrients, air, and water. At the micro-level these cues may be products of metabolism providing feedback signals, while at the macro-level they may signal the presence of food sources or toxic material that should be avoided. Thus, the sensing of circuits responding to this everchanging chemical milieu includes key factors that can shape development, behavior, phenotype and survival.
The aryl hydrocarbon receptor is encoded by an ancient gene found in diverse animals such as nematodes (C. elegans), molluscs (e.g. M. edulis), fruit flies (D. melanogaster), and all chordates, indicating that the gene for the ancestral AhR protein was present at least 550 million years ago, prior to evolutionary divergence [14]. AhR was discovered via a search for the genetic locus responsible for 2,3,7,8-tetracholrodibenzo-p-dioxin (TCDD) inducible aryl hydrocarbon hydroxylase activity and expression of the cytochrome P450 monoxygenase encoded by CYP1A1 in mammalian cells [15].
AhR belongs to the basic helix-loop-helix (bHLH) family of transcription factors with a structure consisting of a ligand binding N-terminal bHLH domain, a C-terminal variable domain, and a DNA binding PER-ARNT-SIM (PAS) domain [16, 17]. AhR primarily functions as a transcription factor after ligand binding (Box 1). Multiple environmental pollutants bind AhR acting as ligands, including polycyclic (e.g. benzoflavones) and halogenated (e.g. dioxins) aromatic hydrocarbons [18] (Table 1); yet, other physiologic AhR ligands have been more difficult to identify. Of note, indole compound derivatives of the amino acid tryptophan appear to be a key family of AhR agonists (Table 1 and Figure 2) [2, 3, 19–23]. One of the most studied mechanisms of tryptophan-to-AhR ligand conversion concerns the metabolization of tryptophan to n-formylkynurenine (Kyn) by the enzymes indoleamine 2,3-dioxygenase (IDO) and tryptophan 2,3-dioxygenase (TDO) [24, 25]. Kyn is an intermediate affinity AhR ligand that has been implicated in regulatory T cell (Treg) functional maturation and suppression of inflammatory cytokine production in dendritic cells, modulating the course of inflammation in mouse models of immune disease [1, 24, 25]. However, one study recently reported that Kyn was significantly less potent than classic AhR ligand 6formylindolo[3,2-b]carbazole (FICZ) in activating AhR-dependent transcription, although condensation derivatives of Kyn have shown 100–1000 fold increases in AhR agonist potential [26]. In this vein, several downstream Kyn metabolites such as kynurenic acid, xanthurenic acid, and cinnabarinic acid are potent AhR ligands that have been implicated in modulating cancer cell migration and cytokine production by T cells in mice and humans [27, 28]. Tryptophan can also be altered by oxidative reactions to the AhR ligands 2-(1’H-indole-30-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE) and 6-formylindolo[3,2-b]carbazole (FICZ) in mouse and human cells [29–32] (Box 2).
Box 1. AhR controls cellular phenotype via transcriptionally dependent and independent mechanisms.
Upon Ligand binding, heterodimeric AhR binds to the DNA sequence motif 5’GCGTG-3’ referred as the xenobiotic response element (XRE) [79]. This results in transcriptional activation of genes encoding phase I and phase II detoxifying enzymes (Cyp1A1, Cyp1B1, etc.) and is the principal component of the xenobiotic detoxifying response [17] (Figure 1). At steady state conditions, AhR is retained in the cytosol in a complex with the chaperone HSP90, the co-chaperone p23, and AhR-interacting protein (AIP, also known as XAP2 or ARA9). HSP90 binds to both the bHLH and PAS domains preventing degradation of AhR and maintaining the receptor in a high ligand binding conformation [80]. P23 interacts with both AhR and HSP90, inhibiting nuclear translocation and interaction with another bHLH-PAS protein, the aryl hydrocarbon nuclear translocator (ARNT) [81]. Additionally, AIP and p23 prevent ubiquitination and degradation, thereby promoting maintenance of the cytosolic pool of AhR [82, 83].
Upon ligand binding, AhR undergoes a conformational change exposing a nuclear localization signal promoting release from the cytoplasmic moorings and nuclear accumulation (Figure 1) [17]. However, some data suggest that AhR release from HSP90 occurs in the nucleus, rather than the cytoplasm [84, 85]; nevertheless, HSP90 release is required for the formation of the AhR/ARNT heterodimer (the high affinity DNA binding form of AhR) and induction of AhR transcriptional activity [84].
In addition to direct transcriptional activity, TCDD treatment triggers interaction of the AhR/ARNT dimer with Brahma/SWI2-related gene 1 (Brg1), driving ATP-dependent chromatin remodeling [86]. Similarly, AhR can control chromatin remodeling by interacting with coactivators such as the steroid receptor coactivator-1 (SRC-1) complex or by displacing histone deacetylase complexes [87, 88]. Moreover, AhR can control the gene expression through other epigenetic mechanisms involving regulation of retrotransposons, micro-RNAs, and long non-coding RNAs [89–92].
Other mechanisms of AhR-dependent phenotypic responses include the regulation of E3 ubiquitin ligase activity driving selective protein degradation. In response to agonists such as 3-methylcholanthrene and β-naphthoflavone, AhR assembles into the CUL4B-based E3 ubiquitin ligase complex, promoting degradation of cytoplasmic receptors including the estrogen receptor (ER) α, ERβ and the androgen receptor; this has indicated that AhR might modulate hormonal responses by directly controlling receptor abundance [93].
Table 1:
AhR agonists and antagonists
| Class | Compounds | Origin |
|---|---|---|
| AhR agonist | Endogenous | |
| • lndole-3-carbinol (I3C) [40] • 3,3’-diindolylmethane (DIM) [40] • Indolo [3,2-b]carbazole (ICZ) [40] • 2-(Indol-3-ylmethyl)-3,39-diindolylmethane (Ltr-1) [40] • lndole-3-acetonitrile (I3ACN) [40] • Curcumin [96] • Diosmin [97] |
Dietary | |
| • Indole [19, 37] • lndole-3-acetic acid (IAA) [19, 36] • lndole-3-aldehyde (IAld) [20] • Tryptamine [36] • lndoxyl-3-sulfate (I3S) [34] • 3-Methyl-indole (skatole) [38] |
Microbiome | |
| • Kynurenine (Kyn) [24, 25] • Kynurenic acid (KA) [98] • Xanthurenic acid [98] • Cinnabarinic acid (CA) [28] • 2-(19H-indole-39-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE) [31] • 6-Formylindolo [3,2-b] carbazole (FICZ) [32] • 5-hydroxy-tryptophan (5HTP) [99] • Bilirubin [100] • Biliverdin [100] • Lipoxin A4 [101] • Prostaglandin [102] |
Host metabolism | |
| • Indigo [103] • Indirubin [103] |
Plant / Mammalian enzymes | |
| • Trypthantrin [104] • Malassezin [104] |
Yeast / Fungi | |
| Exogenous | ||
| • 2,3,7,8-Tetrachlorodibenzop-dioxin (TCDD) [105] • 3-Methylcholanthrene [106] • Beta-naphthoflavone [107] |
HAH and PAH | |
| • Omeprazole [107] • VAF347 [108] • 4-hydroxy-tamoxifen (4OHT) [109] • 6-Methyl-1,3,8-trichlorodibenzofuran (6-MCDF) [110] |
Synthetic | |
| AhR antagonist | • Resveratrol [111] • Quercetin [112] |
Dietary |
| • CH223191 [113] • StemRegenin 1 (SR1) [114] • GNF351 [115] • MNF (3’-methoxy-4’-nitroflavone) [116] • 3′,4′-Dimethoxyflavone (DMF) [8] |
Synthetic | |
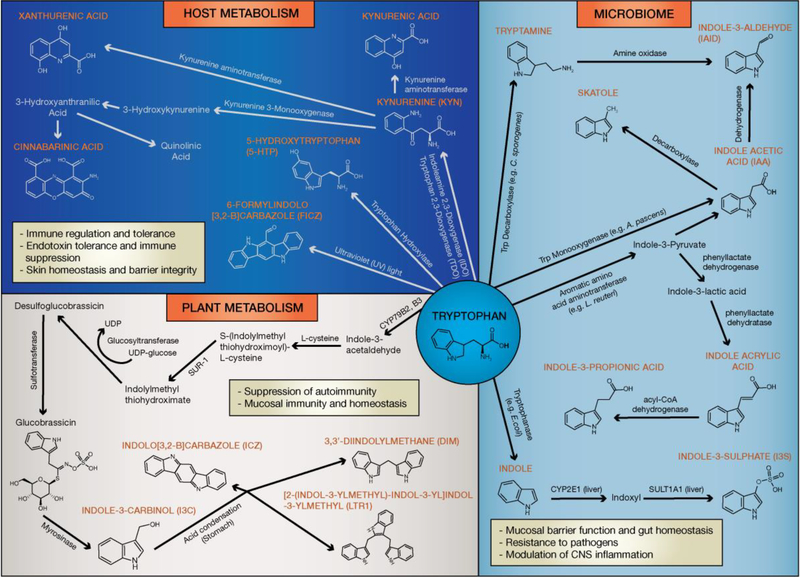
Figure 2: Biosynthesis of AhR ligands through the tryptophan metabolism.
Tryptophan is metabolized into a variety of AhR ligands affecting immunity, maintenance of epithelial barrier function and microbiome diversity. Host metabolites with AhR agonistic activity are primarily derived from tryptophan metabolism via the kynurenine pathway, with additional ligands produced by UV-exposure and oxidative reactions. In the GI tract multiple bacterial species (e.g. C. sporogenes, L. reuteri, E.coli, A. pascens) present in the microbiota metabolize tryptophan to products with potent AhR agonistic properties. Moreover, cruciferous vegetables contain the tryptophan metabolite glucosinolate, which undergoes a hydrolysis reaction forming the AhR protoagonist I3C. In the stomach I3C is metabolized via an acid-condensation reaction into the AhR ligands DIM, ICZ, and LTr1.
Box 2. ITE and FICZ, tryptophan-derived AhR ligands with opposing effects in immune function.
ITE was originally isolated from lung tissue as the product of a reaction between tryptophan and cysteine [31]. Competitive binding assays revealed ITE has an AhR binding affinity (Ki) of 3nM, which is slightly lower than TCDD (Ki of 0.5nM). FICZ, a regulator of skin homeostasis, is primarily produced via ultraviolet light driven photooxidative degradation of tryptophan, with an additional mechanism of generation via enzymatic reactions involving dehydrogenations and oxidative coupling [94, 95]. FICZ exhibits significant AhR agonistic activity with an EC50 of 36 pM, as opposed to Kyn which has a lower AhR-induction potency with an EC50 of 13 μM [26].
Commensal bacteria and fungi derived metabolites also show AhR agonistic properties. Tryptophanase (TnaA) produced by E. coli metabolizes dietary tryptophan to indole and its derivatives [33]. In the liver, bacterially-derived indole is further metabolized to indoxyl-3-sulfate (I3S) by the human sulfotransferase enzyme SULT1A1 [34, 35], while other bacterial species and their associated enzymes including A. pascens (tryptophan monooxygenase) and C. sporogenes (tryptophan decarboxylase) convert tryptophan into AhR ligands such as indole-3-acetic acid (IAA), and tryptamine (TA) respectively [36–38]. Lactobacilli interact with the host immune system in a variety of ways modulating homeostasis and gut immunity [20–23], and a study suggested that production of AhR ligands by L. reuteri could influence microbiome diversity by limiting the colonization of pathogenic microbes, primarily driven by IL-22 production from type 3 innate lymphoid cells (ILC3s) [20]. Similarly, deletion of CARD9 (glossary) rendered mice more susceptible to induced colitis due to an altered microbiome resulting in impaired tryptophan metabolism [22]. In contrast, CARD9−/− mice, gavaged with lactobacilli cultured from the gut microbiome of CARD9+/+ wildtype mice showed increased AhR function and reduced colitis; this suggested that restoration of tryptophan metabolism could reduce intestinal inflammation [22]. Moreover, the development of metabolic syndrome as a result of high fat diet in mice was associated with decreased fecal concentrationsof AhR ligands, while oral supplementation of lactobacilli, reduced hepatic lipid burden and serum triglycerides in mice on a high-fat western diet [21, 22]. Collectively, these studies suggested that the microbiome might play a key role in activating AhR signaling, which in turn can act as a key regulator of local and systemic inflammatory tone in the immune response.
Dietary sources such as vegetables, fruits, and tropical plants contain variety of AhR-modulating factors. In particular, cruciferous vegetables from the family Brassicaceae (e.g. cauliflower, cabbage, etc) are rich source of glucosinolate conjugates capable of activating AhR in mice and humans [39, 40]. After consumption, glucosinolates undergo hydrolysis generating indole-3-carbinol (I3C) and its downstream condensation derivatives 3,3’diindolylmethane (DIM), [2-(indol-3-ylmethyl)indol-3-yl] indol-3-ylmethane (LTr1), and indolo[3,4-b]carbazole (ICZ) which are AhR agonists [40]. These diet derived compounds are implicated in gut AhR activity required for the maintenance of intra epithelial lymphocytes and innate lymphoid cells, which in turn have been reported to promote epithelial cell proliferation, immune surveillance, and modulation of gut inflammation in mice [18, 41, 42]. This shows a critical link between AhR, dietary factors and intestinal immunity.
AhR in innate immunity
Macrophages and Dendritic cells
Macrophages (Mϕ) possess enormous inflammatory potential that must be tightly controlled. Compelling evidence suggests AhR is a key factor restricting Mϕ inflammatory function in response to cell intrinsic regulatory feedback and extrinsic conditioning factors (for a list of AhR agonists and related changes in immune function see table 2). For example, Mϕ uptake and processing of apoptotic cells can drive immunosuppressive cytokine production (dominated by IL-10 and TGF-β) limiting the development of systemic autoimmunity in mice and humans [4]. Conversely, inhibition of AhR function has been found to completely abrogate apoptotic cell-induced Il10 gene expression in Mϕ, causing a shift to a proinflammatory state with increased production of IL-6, IL-12p40, and TNFα in vitro and in vivo, driving a loss of tolerance to apoptotic cell-associated antigens [4]. In co-culture experiments with apoptotic cells, Mϕ exhibited increased AhR binding to XREs in the promoters of Il10, Arg1, and Tgfb preferentially, when compared to Il6, Il12, and Tnfa promoters, suggesting direct transcriptional activation of regulatory cytokine gene expression [4]. In addition, another study reported that microbial metabolite-driven activation of AhR induced the expression of TGF-α in microglial cells limiting inflammatory pathology in a mouse model of MS (EAE) and in human multiple sclerosis (MS) [3]. These observations support a critical function of AhR in the induction of immuno-regulatory factors, further suggesting a putative dominant role of AhR in Mϕ polarization and induction of suppressive immune responses [3, 4].
Table 2:
Activation and function of AhR in different cell types
| Cell type | AhR activation and ligand type | Function |
|---|---|---|
| Macrophage (MΦ) | Apoptotic cells, LPS, CpG, Benzo[a]pyrene (BaP) | • IL-10 production [4, 45] • Suppression of inflammatory cytokines (IL-6, TNF-α, IL12, etc), activation markers (MHCII, CD86), and autoantibody responses [4, 45] • Vitamin D3 catabolism [117] |
| Dendritic cell (DC) | Apoptotic cells, LPS, CpG, ITE, Kyn, FICZ, I3C, VAF347, β-NF, TCDD | • Regulation of cytokine responses (IL-10, IL-6) [6, 43, 44] • Differentiation of DCs and suppression of co-stimulatory molecules (e.g. CD86, MHCII) [30, 44, 48, 49] • • Treg cell differentiation [25, 30, 43] |
| Treg | TCDD, ITE, Kyn, I3C, DIM, norisoboldine | • Treg cell differentiation and function [25, 30, 90, 118, 119] • Inhibition of inflammatory cytokines [59, 120] |
| Tr1 | IL-27 + TGFβ, TCDD, FICZ | • Tr1 cell differentiation and suppressive function [5, 59–61] • transcriptional regulation of IL-10, IL-21, and CD36 [5] |
| Th17 | IL-6 + TGFβ, FICZ | • Th17 cell differentiation [9, 10, 121] • IL-17, IL-22 production [9, 10, 121] |
| Th22 | TCDD, VAF347 | • IL-22 production [122] |
| Intraepithelial lymphocyte (IEL) | I3C, FICZ | • Development, IL-22 production [41, 123] • Gut homeostasis and barrier integrity [41, 123] |
| Innate lymphoid cell (ILC) | I3C, IAId | • Development and survival, IL-22 production [20, 42, 54] • Intestinal homeostasis and mucosal protection [20, 42] |
| CD8+ T cell | TCDD, viral infection | • Differentiation and proliferation [64, 65] • Maintenance of memory CD8 T cells [67] |
| NK cell | FICZ, I3C, DIM | • Development and maintenance [29] • IL-10, IFNγ production [51, 52] |
| B cell | TCDD, ITE, FICZ | • Suppression of plasmablast differentiation, antibody secretion, and class switching [124] • B cell proliferation [125] |
| Microglia, Astrocyte | Indole, I3S, IAId, Gut microbiome | • Suppression of CNS inflammation [2, 3] • modulation of TGFα and VEGFβ secretion [3] |
| Osteoblast/Osteoclast | TCDD, BaP | • Osteoblast and osteoclast differentiation [126] |
Similarly, AhR activity can significantly alter dendritic cell (DC) responses [4, 43]. Specifically, AhR deficient mouse bone marrow derived DCs (BMDC) display reduced ability to produce IL-10 in response to LPS and CpG relative to AhC competenr BMDCs [43]. Moreover, in monocyte-derived DCs from patients with Behçet’s disease, FICZ and ITE could suppress the expression of co-stimulatory molecules (HLA-DR, CD80, CD86) and reduce the production of proinflammatory cytokines after LPS stimulation (e.g. IL-1β, IL-6, and TNFα) in vitro relative to controls [44]. Thus, the data suggest that AhR can act as an important regulator of DC-driven immunosuppression and restriction of inflammatory potential.
AhR can also drive a feedback mechanism limiting ongoing innate proinflammatory responses. For example, Ahr expression is increased in vitro by LPS stimulation of Mϕ [45], and in vivo by type I interferons induced following infection with a number of different RNA and DNA viruses suggesting inflammatory stimulation can enhance AhR function via augmentation of protein amounts [1]. However, LPS (and interferons) can also increase expression and activity of IDO and TDO in mouse and human macrophages and dendritic cells, potentially increasing production of Kyn [46]. In this vein, endotoxin tolerance after secondary intra-peritoneal LPS administration in mice was reported to require TDO-dependent Kyn production in myeloid cells [6]. In this context, AhR activation drove Src-dependent phosphorylation of IDO1 that, in turn, induced production of TGF-β1, thus reducing inflammation and LPS-driven mortality, relative to controls [6]. Thus, AhR responses appear to be highly integrated in Mϕ and DC feedback pathways, reacting to inflammatory cues and enzymatic processes, as well as reinforcing AhR function and providing feedback that can limit the magnitude and duration of inflammation and associated pathology [47].
AhR may also influence innate immunity by controlling DC differentiation from monocyte precursors. A recent study reported that AhR-induced PDRM1 (Blimp1) expression could promote DC differentiation from blood monocytes in both mice and humans [48]. AhR activity could be augmented both by FICZ and I3C relative to controls, and AhR gene signatures were enriched in tuberculoid leprosy lesions, corresponding to increased monocyte-derived DC transcriptional signatures [48]. Similarly, another group found that AhR activation reduced the expression of the Mϕ transcription factor PU.1 and restricted the differentiation of human monocytes and Langerhans cells relative to controls [49]. This suggested that AhR sensing of micro-environmental indole compounds might potentially act as a key driver mechanism of the Mϕ/DC balance in inflamed tissues, controlling disease presentation and pathologic outcomes.
Innate Lymphoid Cells and AhR (ILCs)
ILCs are enriched in mucosal tissues and play a pivotal role in cytokine production and responses to symbiotic and pathogenic microbes [50]. Contact hypersensitivity responses to haptens or other small molecules can be driven by a subset of liver-resident CCR6+ NK (ILC1) cells in mice [29]. A report indicated that genetic deletion of AhR led to a dramatic reduction of this NK cell population due to increased cell turnover and reduced resistance to cytokine induced cell death [29]. Likewise, others reported that murine NK cell activation, IFNγ expression, and lytic function in response to lymphoma required AhR activity [51]. In contrast T. gondii infection required AhR-dependent IL-10 production in mouse NK cells to limit pathology and, ultimately, mortality [52]. This might argue that AhR is generally required for NK cell activation but does not drive a particular (i.e. tolerogenic) NK cell phenotype. However, this question requires further exploration.
ILC3s are primarily found in the gastrointestinal tract where they secrete IL-22, promoting intestinal homeostasis by induction of anti-microbial peptides and fucosylation of mouse epithelial cells [53]. ILC3s express AhR and its genetic disruption causes loss of a subpopulation of CCR6neg ILC3s and functional defects in CCR6+ ILC3s including reduced IL-22 production, a loss of cryptopatches (CP), and a failure to control C. rodentium infections relative to controls [54, 55]. The data suggest that the likely source of AhR ligands promoting ILC3 development and function is diet-derived, given that germ-free mice harbor ILC3s and preserved CP [54, 56]. Nevertheless, microbiome-driven AhR activity appears to play a key role in ILC3-dependent IL-22 production and regulation of the normal commensal flora in mice, providing protection from colonization with pathogenic microbes such as C. albicans [20]. It has not been determined why diet- versus microbiome-derived AhR ligands would preferentially impact ILC3 development and function, but the likely answer is that the many sources of AhR agonists might complement and substitute for each other. Thus, while singular sources of AhR ligands (e.g. dietary) can provide sufficient AhR activity for basal ILC3 population maintenance and function, the full spectrum of ILC3 functionality in control of microbial flora and restriction of inflammation and pathology may require multiple AhR ligands obtained from exogenous and endogenous sources. Collectively, the data suggest that microbiome and diet induced AhR activity may be critical in maintaining mucosal immune homenstasis, barrier function and protection from pathobiont colonization.
AhR in T cell adaptive immunity
One of the first indications that AhR directly influenced T cell activity was published in 2005 when TCDD was reported to induce regulatory function in CD4+ T cells suppressing murine graft versus host disease in an AhR-dependent mechanism [57]. A subsequent report found FICZ exacerbated disease in a mouse model of EAE by AhR-dependent CD4+ T cell acquisition of a Th17 cell phenotype [10]. Concurrently, one study reported similar results while also confirming the findings regarding TCDD induced Treg function [9]. These findings also highlight an interesting aspect of AhR biology, namely that different AhR agonists might have opposing effects on inflammatory versus regulatory T cell (Treg) functional maturation. The most often postulated reason for the divergence in ligand-dependent function might be related to ligand availability and affinity for AhR, given that TCDD shows 2–3-fold higher binding affinity relative to FICZ. However, molecular modeling suggests ITE has similar energetics of AhR binding compared to FICZ, yet is a potent driver of regulatory T cell function [58]. Moreover, ITE and FICZ appear to interact with the AhR binding pocket in a similar orientation suggesting the differences in binding may not be site-specific [58]. Given the divergent T cell functional outcomes of FICZ versus ITE exposure, it is clear that further work is warranted to fully understand how AhR perceives ligand interactions and how co-factors in the AhR transcriptional response interact to drive T cell maturation.
In addition to Th17 and Treg cells, AhR might be essential for differentiation and function of IL-10 producing type-1 regulatory (Tr1) T cells in mice and humans [5, 5961]. In particular, IL-27 induces Ahr expression, and AhR functionally cooperates with c-maf to induce promoter activity of Il10 and Il21 in CD4+ T cells [61]. Hif1α restricts function of Tr1 T cells by altering metabolism and antagonizing AhR via competitive binding to ARNT [5]. As reported, AhR can promote Tr1 suppressive function by restricting Hif1α activity via two distinct mechanisms. 1: extracellular ATP (eATP) suppresses Tr1 T cell activity by induction of Hif1α. However, AhR can induce expression of the exonuclease CD39, reducing eATP and limiting Hif1α protein in Tr1 T cells [5]. 2: AhR directly suppresses Hif1α protein levels by transcriptional induction of prolyl hydroxylase domain (PHD) proteins, promoting Hif1α proteasomal degradation [5].
Although less well understood mechanistically, AhR activity might also be critical for the maintenance and function of intra-epithelial lymphocytes [41]. IELs are TCRγδ and TCRαβ CD8αα+ T cells that populate the skin and GI tract prior to birth in mice [62]. AhR is highly expressed by Vγ3+ IELs in the skin and Vγ5+ IELs in the GI tract of mice [41]. Deletion of AhR did not reduce IEL precursor numbers, but maintenance of γδ T cell populations was compromised leading to a decrease in overall population numbers [41]. In the GI tract, IELs (along with ILCs) are major producers of IL-22, and reports suggest that IEL IL-22 production is dependent on AhR, limiting chronic inflammation and pathology of DSS-induced colitis in mice [63].
AhR activity is also crucial in the regulation of CD8+ T cell responses. TCDD was shown to inhibit the differentiation and proliferation of CD8+ T cells during influenza infection [64] and CTL-mediated pathology in acute graft versus host disease in mice [65]. When exposed to TCDD, CD8+ T cells exhibited methylation patterns similar to those of exhausted CD8+ T cells prior to infection, suggesting that AhR had a potent impact on basal functionality and initial responses to antigenic stimulation [64]. Similarly, IDO expression by tumor cells caused a Kyn- and AhR-dependent increase in expression of the exhaustion marker PD-1 on CD8+ intratumoral T cells in the B16 mouse model of melanoma relative to controls [66]. AhR has also been found to be highly expressed in tissue-resident memory (Trm) CD8+ T cells in the skin following herpes simplex virus infection in mice [67]. Specifically, Ahr did not influence CD8+ T cell homing as both Ahr−/− and Ahr+/+ CD8+ T cell migrated equally well to sites of skin inflammation, but AhR−/− Trm CD8+ T cells disappeared from the tissue at an increased rate over time compared to wild type T cells, suggesting that AhR was critical for Trm population maintenance in mice [67]. Taken together, the data argue that microenvironmental activation of AhR can play a crucial role balancing initial T cell responses and providing long-term protection, particularly at barrier surfaces.
AhR in autoimmunity
AhR activity is potently induced by inflammation in a variety of settings, thus it is perhaps not surprising that AhR can influence disease outcomes in a number of autoimmune conditions. Autoimmune inflammation in the central nervous system (CNS) is particularly impacted by AhR: genetic disruption of AhR in microglia, removal of commensal bacteria from the GI tract, or depletion of dietary tryptophan, can significantly increase CNS inflammation in mouse models of MS; conversely, CNS inflammation can be attenuated by supplementation with AhR activating metabolites such as indole or, in the case of antibiotic-treated mice, with the bacterial enzyme tryptophanase to mimic microbiome tryptophan metabolism [2, 3]. Recently, AhR was found to impact CNS autoimmune inflammation by controlling microglial communication with astrocytes [3]. In particular, relative to controls, microglial lineage deletion of AhR caused a loss of glial TGFα production driving an increase in expression of genes in astrocytes associated with inflammation and neurodegeneration, including Ccl2, Il1b and Nos2 [3]. While these results describe the importance of the microbiome in controlling pathologic CNS immunity, they beg a larger question regarding the role of AhR in environmental feedback regulation in inflammation. AhR is generally defined as a sensor of local environmental feedback, yet, in the context of CNS inflammation, the specific environmental conditions being sensed are systemically distributed metabolites. This illustrates the fact that AhR-mediated regulation is a dynamic process that depends on local and systemic availability of AhR ligands, and on the pattern of AhR expression in affected tissues.
While less is known about the role of AhR in systemic autoimmunity, recent evidence suggests that AhR is an important regulator of systemic autoimmune disease. TCDD administration to (NZB-NZW)F1 systemic lupus erythematosus (SLE)-prone mice had a protective effect, decreasing serum anti-DNA autoantibodies detectable by ELISA, relative to controls [68]. However, colonization of the pathobiont E. gallnarum in the liver induced AhR activity, enhancing autoimmune symptoms in SLE-prone (NZWBXSB)F1 mice [69]. Treatment with the AhR antagonist CH223191 reversed E. gallnarum effects, suggesting that AhR could be pathogenic in microbe-enhanced autoimmunity. While the AhR activating ligands were not determined in this study, the ability of AhR to drive both Th17 and FoxP3+ Treg differentiation indicated that AhR might be either pathogenic or protective in SLE, depending on the site of AhR activity and the ligands driving the response. In this vein, our laboratory recently reported that the dietary indole compounds (I3C and DIM) and tryptophan derivatives (ITE) showed protective effects in SLE reducing inflammatory autoimmunity and pathology in B6.Fcgr2b−/− and MRLlpr/lpr mice [4]. Notably, in advanced SLE-driven kidney disease, AhR induction was able to restore tissue function as assessed by reduced albuminuria, indicating that AhR could be relevant as a putative therapeutic target in established disease, at least in mice [4]. Furthermore, aged female C57BL/6 mice with a myeloid lineage AhR deletion developed an SLE-like phenotype with chronic immune system activation, increased serum autoantibody titers, and kidney pathology relative to wildtype mice [4]. Of clinical relevance, in human SLE patients, blood monocytes and DCs showed increased AhR transcriptional signatures, and furthermore, blocking AhR with CH223191 abrogated apoptotic cell-induced IL-10 production in human Mϕ, suggesting that AhR might serve a similar function in human SLE [4]. Collectively, these findings suggest that AhR represents an important regulatory node controlling inflammation, and which might be effectively targeted to potentially improve disease outcomes in certain autoimmune pathologies.
AhR in the tumor immune-micro environment
Although AhR clearly influences tumorigenesis and metastasis, the immune cell-intrinsic impact of AhR in regulating immunity in the tumor microenvironment is not well understood. High AHR expression ahs been detected in several solid tumor types such as breast, prostate, and liver cancer [16], and increased nuclear AhR activity has correlated with poor prognosis in non-small cell lung cancer, urothelial cancer, and glioblastoma [24, 70, 71]. In contrast, AhR expression in breast cancer has been inversely correlated with histological grade, indicative of better prognosis and improved clinical outcomes relative to low AhR expression [72]. This suggests that perhaps, similar to certain autoimmune mechanisms, AhR might potentially function in a highly contextualized manner, acting as either a negative or positive factor in tumorigenesis, but this remains to be determined.
Nevertheless, several cancers exhibit constitutive expression of IDO and TDO with a significant presence of Kyn and downstream metabolites, suggesting that AhR might be actively engaged in the tumor microenvironment, although this has not been directly demonstrated. One study reported that IFNγ-induced IDO in chronic lymphocytic leukemia might limit the efficacy of chimeric antigen receptor (CAR)-T cell adoptive therapy [73]. While a definitive role for AhR was not established in this study, CAR-T cells presented increased CYP1A1 and reduced proliferation and persistence in vivo in response to tryptophan metabolites relative to controls, suggesting that AhR was able to impact CAR T-cell function [73]. Moreover, tumor repopulating cells (TRC) are a stem cell–like population of cancer cells that are important in tumorigenic processes, providing a pool of cells for tumor growth and metastasis [8]. In a mouse model of melanoma, the IDO-Kyn-AhR circuit was reported to protect TRC from IFNγ-mediated apoptosis by blocking STAT1 signaling, promoting tumor dormancy and persistence instead [66]. Furthermore, Kyn production by TRC was found to drive AhR-dependent CD8+ T cell exhaustion, inducing PD-1 expression and preventing CD8+ T cell-mediated killing of TRC [66]. Treatment of resting CD8+ T cells with Kyn was sufficient to induce PD-1 expression, an effect that was augmented by antigen-stimulation [8]. This suggested that Kyn could impact CD8+ T cell function in the tumor microenvironment in a promiscuous fashion, and capable of suppressing CD8+ T cell effector function [8].
There has been significant interest in the microbiome-tumor axis and data suggest that the composition of the microbiome influences tumor growth, responses to immune-oncology therapy (I/O), and chemotherapy [74]. In two recent studies, microbiome dysbiosis occurred as a result of antibiotic treatment reduced efficacy of I/O [75, 76]. This was associated with altered homeostasis and increased “leakiness” of the epithelial barrier in the GI tract, suggesting that alterations in host/microbiome interactions in the gut can drive effects at local and distant tumor sites. Evidence that microbial metabolites influence cancer is more limited; however, short chain fatty acids produced by microbial fiber fermentation in the GI tract can restrict colon cancer development, influencing composition of the microbiome and altering metabolism and function of resident immune cells in mice [77, 78]. Since AhR can influence immunity at sites both proximal and distal to the gut via microbial metabolites, it is likely that a functional relationship exists between the microbiome, AhR bioactive metabolites, and cancer outcomes (growth, resistance to therapy, etc); however, these potential interactions remain to be studied.
Concluding remarks
The initial discovery of this ancient AhR chemical sensing circuit was met with little interest outside of environmental toxicologist circles. However, it is now clear that AhR is a key environmental sensor and mechanistic regulator of immunity at epithelial barriers and systemically. Increased AhR activity appears to be a common feature of many inflammatory diseases and exciting new findings suggest that it may be a druggable target with wide applicability in diseases of immune dysfunction. However, it is important to note that AhR biology is complex, and the association of AhR with the toxicities of TCDD exposure remind us that caution must be exercised in attempting to manipulate this pathway. Thus, complex AhR biology and its effects in health and disease indicate that it is vital to properly explore the range of AhR ligand-receptor interactions, mechanisms and downstream biology (see Outstanding Questions). This increased knowledge may reveal general as well as disease-specific mechanistic iand functional nsight that will be critical for envisioning any possible and efficient AhR therapeutic manipulations in disease.
Figure 1: The AhR transcriptional circuit in mammalian cells.
The inactive form of the aryl hydrocarbon receptor (AhR) is present in the cytosol as a complex with the chaperone proteins HSP90, P23, and AIP. Multiple AhR ligands from the gut microbiome, host metabolism, diet, and environment induce a conformational change in AhR exposing the nuclear localization signal initiating nuclear shuttling. Recently an additional mechanism was shown to activate AhR via DNA exposed on apoptotic cells or apoptotic microparticles dependent on Toll-Like Receptor 9 (TLR9) [4]. Once in the nucleus, AhR forms a dimer with the AhR nuclear translocator (ARNT) binding to the XRE/DRE sequence motif 5’-GCGTG-3’. This induces expression of genes involved in AhR ligand metabolism and immune regulation. Furthermore, AhR drives expression of AhR repressor (AHRR) disrupting the AhR/ARNT heterodimer and suppressing its transcriptional activity. In addition, AhR functions as a CUL4B-based E3 ubiquitin ligase complex driving selective protein degradation. Regulation of AhR pathway is controlled through nuclear export and subsequent degradation of AhR via the ubiquitin-proteasome pathway.
Highlights.
AhR responds to diverse ligands in the environment; however, tryptophan metabolites have emerged as a key family of agonistic ligands.
AhR is a fundamental sensor and modulator of microbiome and immune cell homeostasis in the GI tract.
AhR can control differentiation and inflammatory potential in both innate and adaptive immune cells, locally at epithelial barriers, and systemically.
AhR responds to exogenously applied ligands by oral and injection routes, modulating inflammatory disease, and identifying it as an emerging druggable putative target for certain human diseases.
Outstanding questions.
Ligands that activate AhR can have dichotomous effects driving inflammation (FICZ) or regulatory immunity (ITE) despite having similar pharmacokinetics. However, the mechanisms driving differential immune outcomes are unknown. Further complicating advancement of AhR modulators as potential drugs are the well-known toxic effects of TCDD-induced AhR activity in humans. To advance AhR as a putative therapeutic target, can we gain further insight of the mechanisms underlying these contextual differences in AhR functionality?
Does AhR impact anti-tumor immunity? While AhR activity correlates with disease outcomes, the relative contribution of tumor cell-intrinsic versus immune impact of AhR needs further study. Similarly, whether targeted manipulation of AhR function can alter the immune microenvironment in tumors, or impact the cancer disease course, remains an open question.
Does a microbiome-AhR axis exist in cancer? If so, does it impact therapeutic responses? The microbiome profoundly alters therapeutic responses in chemo- and immunotherapy in cancer; however, it remains to be determined if microbial AhR agonistic metabolites significantly impact cancer prognosis via modulation of systemic or intratumoral immunity.
Acknowledgements
We thank the members of the McGaha lab for contributions to the research described in this manuscript, Dr. Marie Jo Halaby for critique of the article, and Kieran Manion for assistance with the figures. This work was supported by NIH grants AR067763 and CA190449 and grant #C1TPA-2016–20 from the Medicine by Design/Canada First Research Excellence Fund (TLM).
Glossary
- Behçet’s disease
inherited inflammatory disorder with highly inflamed blood vessels. Symptoms include mouth sores, eye inflammation, skin rashes, and genital sores.
- Caspase recruitment domain family member 9 (CARD9)
adaptor molecule operating downstream of pattern recognition receptors critical in regulating innate and adaptive immunity; highly expressed in DCs and Mϕ, representing an important regulator of immune responses against pathogens, driving downstream NF-κB activation.
- Cryptopatches (CP)
small clusters of closely packed lymphocytes located in the basal lamina propria of murine small and large intestine mucosae. Cells from CPs are positive for stem cell marker c-kit and mainly composed of lineage negative (CD3−CD4−CD8−B220−) cells.
- Glucosinolates
sulphur rich secondary metabolite found in plants from Brassica spp. (broccoli, cabbage, etc.); hydrolyzed into compounds such as isothiocynates; possess anti-inflammatory, anti-bacterial, anti-fungal, and anti-carcinogenic, etc. properties.
- Graft versus host disease (GvHD)
medical syndrome that can occurr post transplantation; a donor’s immune cells recognize the recipient’s body as a foreign and attack recipients’ normal cells. GvHD can be acute or chronic, depending on patient and donor variations in tissue type.
- Innate lymphoid cell (ILC)
derived from common lymphoid progenitors lacking expression of T cell or B cell receptors. Function is analogous to helper T cells and ILCs are primarily grouped based on transcription factor and cytokine expression.
- Type 3 innate lymphoid cells (ILC3)
predominantly located at mucosal surfaces in lungs and intestine. They express the transcription factor RORγt and produce effector cytokines including IL-22 and IL-17.
- Liver-resident CCR6+ NK (ILC1) cells
subset of group 1 ILCs (ILC1) expressing α1 integrin (CD49a) but not the classical NK cell marker α2 integrin (DX5). They require transcription factor T-bet for their development and produce robust amounts of IFNγ and TNF.
- CCR6-ve ILC3
do not express IL-17 or CD4, but are capable of making IL-22. They do not need RORγt for development, but require T-bet, differentiating them into NKp46+ ILC3.
- Intraepithelial lymphocytes (IEL)
heterogeneous population of T cells residing within the epithelium of the intestine and the skin. They express a αβ and γδ T cell receptor on their surface and are critical in maintaining the integrity of mucosal surfaces in the gut, protecting the epithelium from pathogen or immune-driven pathology.
- Regulatory T cell
population of Foxp3+CD4+ T cells that regulate or suppress other immune cells and maintain tolerance to self-antigens. They are implicated in cancer growth and prevention of autoimmune diseases.
- Xenobiotic response element (XRE) (or dioxin response element-DRE)
nucleotide sequence recognized by the ligand bound AhR-ARNT heterodimer; found at the promoter regions of AhR inducible genes e.g. CYP1A1, CYP1B1, etc.
- Th17 T cell
proinflammatory subclass of CD4+ T cells capable of secreting interleukin 17 (IL-17); implicated in the clearance of pathogens and in the pathogenesis of various inflammatory and autoimmune diseases.
- Type-1 regulatory (Tr1) T cells
population of Tregs with a Foxp3-ve phenotype exerting immunosuppressive function through high production of IL-10; implicated in the induction and maintenance of peripheral tolerance and suppression of inflammation in autoimmunity and GvHD.
- Tumor repopulating cells (TRCs) (or cancer stem cells)
subpopulation of tumor cells possessing stem cell-like properties; may serve as a reservoir for cancerous cells and tumor regrowth following tumor reductive therapy.
Footnotes
Disclaimer
The authors have no competing interests to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yamada T et al. (2016) Constitutive aryl hydrocarbon receptor signaling constrains type I interferon-mediated antiviral innate defense. Nat Immunol 17 (6), 687–94. [DOI] [PubMed] [Google Scholar]
- 2.Rothhammer V et al. (2016) Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med 22 (6), 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothhammer V et al. (2018) Microglial control of astrocytes in response to microbial metabolites. Nature 557 (7707), 724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinde R et al. (2018) Apoptotic cell-induced AhR activity is required for immunological tolerance and suppression of systemic lupus erythematosus in mice and humans. Nat Immunol 19 (6), 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mascanfroni ID et al. (2015) Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-alpha. Nat Med 21 (6), 638–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bessede A et al. (2014) Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature 511 (7508), 184–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moura-Alves P et al. (2014) AhR sensing of bacterial pigments regulates antibacterial defence. Nature 512 (7515), 387–92. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y et al. (2017) Blockade of IDO-kynurenine-AhR metabolic circuitry abrogates IFN-gamma-induced immunologic dormancy of tumor-repopulating cells. Nat Commun 8, 15207. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Quintana FJ et al. (2008) Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 453 (7191), 65–71. [DOI] [PubMed] [Google Scholar]
- 10.Veldhoen M et al. (2008) The aryl hydrocarbon receptor links TH17-cell-mediatedautoimmunity to environmental toxins. Nature 453 (7191), 106–9. [DOI] [PubMed] [Google Scholar]
- 11.Castaneda AR et al. (2018) Ambient particulate matter activates the aryl hydrocarbon receptor in dendritic cells and enhances Th17 polarization. Toxicol Lett 292, 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kado S et al. (2017) Aryl hydrocarbon receptor signaling modifies Toll-like receptor-regulated responses in human dendritic cells. Arch Toxicol 91 (5), 2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia M et al. (2015) Vehicular exhaust particles promote allergic airway inflammation through an aryl hydrocarbon receptor-notch signaling cascade. J Allergy Clin Immunol 136 (2), 441–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn ME (2002) Aryl hydrocarbon receptors: diversity and evolution. Chem Biol Interact 141 (1–2), 131–60. [DOI] [PubMed] [Google Scholar]
- 15.Legraverend C et al. (1982) Regulatory gene product of the Ah locus. Characterization of receptor mutants among mouse hepatoma clones. J Biol Chem 257 (11), 6402–7. [PubMed] [Google Scholar]
- 16.Murray IA et al. (2014) Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat Rev Cancer 14 (12), 801–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stockinger B et al. (2014) The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol 32, 403–32. [DOI] [PubMed] [Google Scholar]
- 18.Denison MS and Nagy SR (2003) Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol 43, 309–34. [DOI] [PubMed] [Google Scholar]
- 19.Miller CA 3rd (1997) Expression of the human aryl hydrocarbon receptor complex in yeast. Activation of transcription by indole compounds. J Biol Chem 272 (52), 328249. [DOI] [PubMed] [Google Scholar]
- 20.Zelante T et al. (2013) Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39 (2), 372–85. [DOI] [PubMed] [Google Scholar]
- 21.Natividad JM et al. (2018) Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metab. [DOI] [PubMed] [Google Scholar]
- 22.Lamas B et al. (2016) CARD9 impacts colitis by altering gut microbiota metabolismof tryptophan into aryl hydrocarbon receptor ligands. Nat Med 22 (6), 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cervantes-Barragan L et al. (2017) Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8alphaalpha(+) T cells. Science 357 (6353), 806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Opitz CA et al. (2011) An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478 (7368), 197–203. [DOI] [PubMed] [Google Scholar]
- 25.Mezrich JD et al. (2010) An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol 185 (6), 3190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seok SH et al. (2018) Trace derivatives of kynurenine potently activate the aryl hydrocarbon receptor (AHR). J Biol Chem 293 (6), 1994–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novikov O et al. (2016) An Aryl Hydrocarbon Receptor-Mediated Amplification Loop That Enforces Cell Migration in ER-/PR-/Her2- Human Breast Cancer Cells. Mol Pharmacol 90 (5), 674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe MM et al. (2014) Identification of cinnabarinic acid as a novel endogenous aryl hydrocarbon receptor ligand that drives IL-22 production. PLoS One 9 (2), e87877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang LH et al. (2016) The aryl hydrocarbon receptor is required for the maintenance of liver-resident natural killer cells. J Exp Med 213 (11), 2249–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quintana FJ et al. (2010) An endogenous aryl hydrocarbon receptor ligand acts on dendritic cells and T cells to suppress experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A 107 (48), 20768–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song J et al. (2002) A ligand for the aryl hydrocarbon receptor isolated from lung. Proc Natl Acad Sci U S A 99 (23), 14694–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rannug A et al. (1987) Certain photooxidized derivatives of tryptophan bind with very high affinity to the Ah receptor and are likely to be endogenous signal substances. J Biol Chem 262 (32), 15422–7. [PubMed] [Google Scholar]
- 33.Li G and Young KD (2013) Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology 159 (Pt 2), 402–10. [DOI] [PubMed] [Google Scholar]
- 34.Schroeder JC et al. (2010) The uremic toxin 3-indoxyl sulfate is a potent endogenous agonist for the human aryl hydrocarbon receptor. Biochemistry 49 (2), 393400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banoglu E and King RS (2002) Sulfation of indoxyl by human and rat aryl(phenol) sulfotransferases to form indoxyl sulfate. Eur J Drug Metab Pharmacokinet 27 (2), 135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin UH et al. (2014) Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol Pharmacol 85 (5), 777–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hubbard TD et al. (2015) Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep 5, 12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weems JM and Yost GS (2010) 3-Methylindole metabolites induce lungCYP1A1 and CYP2F1 enzymes by AhR and non-AhR mechanisms, respectively. Chem Res Toxicol 23 (3), 696–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonderby IE et al. (2010) Biosynthesis of glucosinolates--gene discovery and beyond. Trends Plant Sci 15 (5), 283–90. [DOI] [PubMed] [Google Scholar]
- 40.Bjeldanes LF et al. (1991) Aromatic hydrocarbon responsiveness-receptor agonists generated from indole-3-carbinol in vitro and in vivo: comparisons with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Proc Natl Acad Sci U S A 88 (21), 9543–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y et al. (2011) Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 147 (3), 629–40. [DOI] [PubMed] [Google Scholar]
- 42.Schiering C et al. (2017) Feedback control of AHR signalling regulates intestinal immunity. Nature 542 (7640), 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen NT et al. (2010) Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc Natl Acad Sci U S A 107 (46), 19961–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C et al. (2014) Activation of the aryl hydrocarbon receptor affects activation and function of human monocyte-derived dendritic cells. Clin Exp Immunol 177 (2), 52130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura A et al. (2009) Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J Exp Med 206 (9), 2027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGaha TL et al. (2012) Amino acid catabolism: a pivotal regulator of innate and adaptive immunity. Immunol Rev 249 (1), 135–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simones T and Shepherd DM (2011) Consequences of AhR activation in steady state dendritic cells. Toxicol Sci 119 (2), 293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goudot C et al. (2017) Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages. Immunity 47 (3), 582–596 e6. [DOI] [PubMed] [Google Scholar]
- 49.Platzer B et al. (2009) Aryl hydrocarbon receptor activation inhibits in vitro differentiation of human monocytes and Langerhans dendritic cells. J Immunol 183 (1), 66–74. [DOI] [PubMed] [Google Scholar]
- 50.Klose CS and Artis D (2016) Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol 17 (7), 765–74. [DOI] [PubMed] [Google Scholar]
- 51.Shin JH et al. (2013) Modulation of natural killer cell antitumor activity by the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A 110 (30), 12391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagage S et al. (2014) The aryl hydrocarbon receptor promotes IL-10 production by NK cells. J Immunol 192 (4), 1661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goto Y et al. (2014) Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 345 (6202), 1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JS et al. (2011) AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol 13 (2), 144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiu J et al. (2012) The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36 (1), 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bouskra D et al. (2008) Lymphoid tissue genesis induced by commensals throughNOD1 regulates intestinal homeostasis. Nature 456 (7221), 507–10. [DOI] [PubMed] [Google Scholar]
- 57.Funatake CJ et al. (2005) Cutting edge: activation of the aryl hydrocarbon receptor by 2,3,7,8-tetrachlorodibenzo-p-dioxin generates a population of CD4+ CD25+ cells with characteristics of regulatory T cells. J Immunol 175 (7), 4184–8. [DOI] [PubMed] [Google Scholar]
- 58.Bisson WH et al. (2009) Modeling of the aryl hydrocarbon receptor (AhR) ligand binding domain and its utility in virtual ligand screening to predict new AhR ligands. J Med Chem 52 (18), 5635–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gandhi R et al. (2010) Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3(+) regulatory T cells. Nat Immunol 11 (9), 84653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu HY et al. (2011) In vivo induction of Tr1 cells via mucosal dendritic cells and AHR signaling. PLoS One 6 (8), e23618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Apetoh L et al. (2010) The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol 11 (9), 854–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carding SR et al. (1990) Developmentally regulated fetal thymic and extrathymic T-cell receptor gamma delta gene expression. Genes Dev 4 (8), 1304–15. [DOI] [PubMed] [Google Scholar]
- 63.Martin B et al. (2009) Interleukin-17-producing gammadelta T cells selectively expand in response to pathogen products and environmental signals. Immunity 31 (2), 321–30. [DOI] [PubMed] [Google Scholar]
- 64.Winans B et al. (2015) Linking the aryl hydrocarbon receptor with altered DNA methylation patterns and developmentally induced aberrant antiviral CD8+ T cell responses. J Immunol 194 (9), 4446–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rohlman D et al. (2013) Suppression of acute graft-versus-host response by TCDD is independent of the CTLA-4-IFN-gamma-IDO pathway. Toxicol Sci 135 (1), 8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y et al. (2018) Tumor-Repopulating Cells Induce PD-1 Expression in CD8(+) T Cells by Transferring Kynurenine and AhR Activation. Cancer Cell 33 (3), 480–494 e7. [DOI] [PubMed] [Google Scholar]
- 67.Zaid A et al. (2014) Persistence of skin-resident memory T cells within an epidermal niche. Proc Natl Acad Sci U S A 111 (14), 5307–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J and McMurray RW (2009) Effects of chronic exposure to DDT and TCDD on disease activity in murine systemic lupus erythematosus. Lupus 18 (11), 941–9. [DOI] [PubMed] [Google Scholar]
- 69.Manfredo Vieira S et al. (2018) Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 359 (6380), 1156–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Su JM et al. (2013) Prognostic value of nuclear translocation of aryl hydrocarbon receptor for non-small cell lung cancer. Anticancer Res 33 (9), 3953–61. [PubMed] [Google Scholar]
- 71.Ishida M et al. (2010) Activation of the aryl hydrocarbon receptor pathway enhances cancer cell invasion by upregulating the MMP expression and is associated with poor prognosis in upper urinary tract urothelial cancer. Carcinogenesis 31 (2), 287–95. [DOI] [PubMed] [Google Scholar]
- 72.Saito R et al. (2014) Aryl hydrocarbon receptor in breast cancer-a newly defined prognostic marker. Horm Cancer 5 (1), 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ninomiya S et al. (2015) Tumor indoleamine 2,3-dioxygenase (IDO) inhibits CD19CAR T cells and is downregulated by lymphodepleting drugs. Blood 125 (25), 3905–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gopalakrishnan V et al. (2018) The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 33 (4), 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gopalakrishnan V et al. (2018) Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359 (6371), 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Routy B et al. (2018) Gut microbiome influences efficacy of PD-1-basedimmunotherapy against epithelial tumors. Science 359 (6371), 91–97. [DOI] [PubMed] [Google Scholar]
- 77.Singh N et al. (2014) Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40 (1), 128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sivaprakasam S et al. (2016) An essential role of Ffar2 (Gpr43) in dietary fibre-mediated promotion of healthy composition of gut microbiota and suppression of intestinal carcinogenesis. Oncogenesis 5 (6), e238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fujisawa-Sehara A et al. (1987) Characterization of xenobiotic responsive elements upstream from the drug-metabolizing cytochrome P-450c gene: a similarity to glucocorticoid regulatory elements. Nucleic Acids Res 15 (10), 4179–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Antonsson C et al. (1995) Distinct roles of the molecular chaperone hsp90 in modulating dioxin receptor function via the basic helix-loop-helix and PAS domains. Mol Cell Biol 15 (2), 756–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kazlauskas A et al. (1999) Evidence that the co-chaperone p23 regulates ligand responsiveness of the dioxin (Aryl hydrocarbon) receptor. J Biol Chem 274 (19), 13519–24. [DOI] [PubMed] [Google Scholar]
- 82.Meyer BK and Perdew GH (1999) Characterization of the AhR-hsp90-XAP2 core complex and the role of the immunophilin-related protein XAP2 in AhR stabilization. Biochemistry 38 (28), 8907–17. [DOI] [PubMed] [Google Scholar]
- 83.Nguyen PM et al. (2012) p23 co-chaperone protects the aryl hydrocarbon receptor from degradation in mouse and human cell lines. Biochem Pharmacol 84 (6), 838–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tsuji N et al. (2014) The activation mechanism of the aryl hydrocarbon receptor (AhR) by molecular chaperone HSP90. FEBS Open Bio 4, 796–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Heid SE et al. (2000) Role of heat shock protein 90 dissociation in mediating agonist-induced activation of the aryl hydrocarbon receptor. Mol Pharmacol 57 (1), 82–92. [PubMed] [Google Scholar]
- 86.Wang S and Hankinson O (2002) Functional involvement of the Brahma/SWI2related gene 1 protein in cytochrome P4501A1 transcription mediated by the aryl hydrocarbon receptor complex. J Biol Chem 277 (14), 11821–7. [DOI] [PubMed] [Google Scholar]
- 87.Beischlag TV et al. (2002) Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol Cell Biol 22 (12), 4319–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schnekenburger M et al. (2007) HDAC1 bound to the Cyp1a1 promoter blocks histone acetylation associated with Ah receptor-mediated trans-activation. Biochim Biophys Acta 1769 (9–10), 569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morales-Hernandez A et al. (2016) Alu retrotransposons promote differentiation of human carcinoma cells through the aryl hydrocarbon receptor. Nucleic Acids Res 44 (10), 4665–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Singh NP et al. (2016) Dietary Indoles Suppress Delayed-Type Hypersensitivity by Inducing a Switch from Proinflammatory Th17 Cells to Anti-Inflammatory Regulatory T Cells through Regulation of MicroRNA. J Immunol 196 (3), 1108–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hanieh H and Alzahrani A (2013) MicroRNA-132 suppresses autoimmune encephalomyelitis by inducing cholinergic anti-inflammation: a new Ahr-based exploration. Eur J Immunol 43 (10), 2771–82. [DOI] [PubMed] [Google Scholar]
- 92.Garcia GR et al. (2017) In Vivo Characterization of an AHR-Dependent Long Noncoding RNA Required for Proper Sox9b Expression. Mol Pharmacol 91 (6), 609619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ohtake F et al. (2009) AhR acts as an E3 ubiquitin ligase to modulate steroid receptor functions. Biochem Pharmacol 77 (4), 474–84. [DOI] [PubMed] [Google Scholar]
- 94.Wincent E et al. (2009) The suggested physiologic aryl hydrocarbon receptor activator and cytochrome P4501 substrate 6-formylindolo[3,2-b]carbazole is present in humans. J Biol Chem 284 (5), 2690–6. [DOI] [PubMed] [Google Scholar]
- 95.Smirnova A et al. (2016) Evidence for New Light-Independent Pathways for Generation of the Endogenous Aryl Hydrocarbon Receptor Agonist FICZ. Chem Res Toxicol 29 (1), 75–86. [DOI] [PubMed] [Google Scholar]
- 96.Ciolino HP et al. (1998) Effect of curcumin on the aryl hydrocarbon receptor and cytochrome P450 1A1 in MCF-7 human breast carcinoma cells. Biochem Pharmacol 56 (2), 197–206. [DOI] [PubMed] [Google Scholar]
- 97.Ciolino HP et al. (1998) Diosmin and diosmetin are agonists of the aryl hydrocarbon receptor that differentially affect cytochrome P450 1A1 activity. Cancer Res 58 (13), 2754–60. [PubMed] [Google Scholar]
- 98.DiNatale BC et al. (2010) Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicol Sci 115 (1), 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bittinger MA et al. (2003) Aspartate aminotransferase generates proagonists of the aryl hydrocarbon receptor. Mol Pharmacol 64 (3), 550–6. [DOI] [PubMed] [Google Scholar]
- 100.Phelan D et al. (1998) Activation of the Ah receptor signal transduction pathway by bilirubin and biliverdin. Arch Biochem Biophys 357 (1), 155–63. [DOI] [PubMed] [Google Scholar]
- 101.Schaldach CM et al. (1999) Lipoxin A4: a new class of ligand for the Ah receptor. Biochemistry 38 (23), 7594–600. [DOI] [PubMed] [Google Scholar]
- 102.Seidel SD et al. (2001) Activation of the Ah receptor signaling pathway by prostaglandins. J Biochem Mol Toxicol 15 (4), 187–96. [DOI] [PubMed] [Google Scholar]
- 103.Adachi J et al. (2001) Indirubin and indigo are potent aryl hydrocarbon receptor ligands present in human urine. J Biol Chem 276 (34), 31475–8. [DOI] [PubMed] [Google Scholar]
- 104.Magiatis P et al. (2013) Malassezia yeasts produce a collection of exceptionally potent activators of the Ah (dioxin) receptor detected in diseased human skin. J Invest Dermatol 133 (8), 2023–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Poland A et al. (1976) Stereospecific, high affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J Biol Chem 251 (16), 4936–46. [PubMed] [Google Scholar]
- 106.Kim H et al. (1995) 3-Methylcholanthrene and pyridine effects on CYP1A1 and CYP1A2 expression in rat renal tissue. Drug Metab Dispos 23 (8), 818–24. [PubMed] [Google Scholar]
- 107.Daujat M et al. (1996) Induction of CYP1A1 gene by benzimidazole derivatives during Caco-2 cell differentiation. Evidence for an aryl-hydrocarbon receptor-mediated mechanism. Eur J Biochem 237 (3), 642–52. [DOI] [PubMed] [Google Scholar]
- 108.Lawrence BP et al. (2008) Activation of the aryl hydrocarbon receptor is essential for mediating the anti-inflammatory effects of a novel low-molecular-weight compound. Blood 112 (4), 1158–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.DuSell CD et al. (2010) Regulation of aryl hydrocarbon receptor function by selective estrogen receptor modulators. Mol Endocrinol 24 (1), 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McDougal A et al. (2001) Tamoxifen-induced antitumorigenic/antiestrogenic action synergized by a selective aryl hydrocarbon receptor modulator. Cancer Res 61 (10), 3902–7. [PubMed] [Google Scholar]
- 111.Beedanagari SR et al. (2009) Resveratrol inhibits dioxin-induced expression of human CYP1A1 and CYP1B1 by inhibiting recruitment of the aryl hydrocarbon receptor complex and RNA polymerase II to the regulatory regions of the corresponding genes. Toxicol Sci 110 (1), 61–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ciolino HP et al. (1999) Dietary flavonols quercetin and kaempferol are ligands of the aryl hydrocarbon receptor that affect CYP1A1 transcription differentially. Biochem J 340 ( Pt 3), 715–22. [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao B et al. (2010) CH223191 is a ligand-selective antagonist of the Ah (Dioxin) receptor. Toxicol Sci 117 (2), 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Boitano AE et al. (2010) Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 329 (5997), 1345–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fang ZZ et al. (2014) In vivo effects of the pure aryl hydrocarbon receptor antagonist GNF-351 after oral administration are limited to the gastrointestinal tract. Br J Pharmacol 171 (7), 1735–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Henry EC et al. (1999) Flavone antagonists bind competitively with 2,3,7, 8tetrachlorodibenzo-p-dioxin (TCDD) to the aryl hydrocarbon receptor but inhibit nuclear uptake and transformation. Mol Pharmacol 55 (4), 716–25. [PubMed] [Google Scholar]
- 117.Matsunawa M et al. (2009) The aryl hydrocarbon receptor activator benzo[a]pyrene enhances vitamin D3 catabolism in macrophages. Toxicol Sci 109 (1), 50–8. [DOI] [PubMed] [Google Scholar]
- 118.Kaye J et al. (2016) Laquinimod arrests experimental autoimmune encephalomyelitis by activating the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A 113 (41), E6145–E6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lv Q et al. (2018) Norisoboldine, a natural AhR agonist, promotes Treg differentiation and attenuates colitis via targeting glycolysis and subsequent NAD(+)/SIRT1/SUV39H1/H3K9me3 signaling pathway. Cell Death Dis 9 (3), 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ye J et al. (2017) The Aryl Hydrocarbon Receptor Preferentially Marks and Promotes Gut Regulatory T Cells. Cell Rep 21 (8), 2277–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Veldhoen M et al. (2009) Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med 206 (1), 43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Basu R et al. (2012) Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity 37 (6), 1061–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kadow S et al. (2011) Aryl hydrocarbon receptor is critical for homeostasis of invariant gammadelta T cells in the murine epidermis. J Immunol 187 (6), 3104–10. [DOI] [PubMed] [Google Scholar]
- 124.Vaidyanathan B et al. (2017) The aryl hydrocarbon receptor controls cell-fate decisions in B cells. J Exp Med 214 (1), 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Villa M et al. (2017) Aryl hydrocarbon receptor is required for optimal B-cell proliferation. EMBO J 36 (1), 116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Izawa T et al. (2016) The Nuclear Receptor AhR Controls Bone Homeostasis by Regulating Osteoclast Differentiation via the RANK/c-Fos Signaling Axis. J Immunol 197 (12), 4639–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]