Abstract
Liver plays essential roles in the metabolism of many endogenous chemicals and exogenous toxicants. Mechanistic studies in liver have been at the forefront of efforts to probe the roles of bioactivation and detoxication of environmental toxins and toxicants in hepatotoxicity. Moreover, idiosyncratic hepatoxicity remains a key barrier in the clinical development of drugs. The now vast NRF2 field emerged in part from biochemical and molecular studies on chemical inducers of hepatic detoxication enzymes and subsequent characterization of the modulation of drug/toxicant induced hepatotoxicities in mice through disruption of either Nrf2 or Keap1 genes. In general, loss of NRF2 increases the sensitivity to such toxic chemicals, highlighting a central role of this transcription factor and its downstream target genes as a modifier to chemical stress. In this review, we summarize the impact of NRF2 on the toxicology of multiple hepatotoxicants, and discuss efforts to utilize the NRF2 response in predictive toxicology.
Keywords: Nrf2, Keap1, hepatoxicity, hepatocarcinogenesis, xenobiotic metabolism
1. Nrf2: Foundations in Toxicology
The identification and initial functional characterization of the transcription factor Nrf2 (NF-E2-related factor 2) arose out of the convergence of studies in hematology and toxicology. Studies by the Talalay group beginning in the late 1970s established that induction of enzymes (e.g., glutathione S-transferases, GSTs) that conjugated electrophilic metabolites of toxins and carcinogens to promote their detoxication and elimination in response to treatment with food antioxidants and natural products such as flavones and isothiocyanates contributed to their protective actions against acute and chronic toxicities in rodents (Prestera et al. 1993). Rushmore and Pickett (1990) identified antioxidant responsive elements (ARE) in the GSTA1 gene using the protective inducer t-butyl hydroquinone (tBHQ), an antioxidative food additive. Simultaneously, Daniel’s group, also using tBHQ, named the regulatory sequence in the GSTA1 gene as an electrophile responsive element (EpRE) (Friling et al. 1990; Friling et al. 1992). The ARE was identified as 5’-puGTGACNNNGC-3’ (N: any nucleotide) and 5’-TGACATT-3’ and represented the core sequence of the ARE required for transcriptional activation (Rushmore et al. 1991). However, the identity of the transcription factors that trans-activate through the AREs remained unknown for several years thereafter.
Nrf2 was discovered as a NF-E2-like leucine zipper transcriptional activator that bound to the tandem NF-E2/AP1 repeat of the β-globin locus control region following cloning from a human erythroleukemic cell cDNA library (Moi et al. 1994). Kan’s group then generated a Nrf2 knockout (KO) mouse (Chan et al. 1996) that surprisingly demonstrated that Nrf2 was not essential for erythropoiesis. Moreover, these Nrf2 KO mice did not show any obvious phenotype, indicating that Nrf2 was dispensable for mouse development. At the same time, Yamamoto and colleagues cloned a chicken β-globin enhancer (ECH) that was very homologous to murine and human Nrf2 (Itoh et al. 1995) and also generated a Nrf2 KO mouse line (Itoh et al. 1997). Initial characterization of these mice provided the first in vivo evidence that inducible expression of xenobiotic metabolizing enzymes (GSTs and NAD(P)H:quinone oxidoreductase 1 (NQO1)) was regulated by a mechanism involving the ARE and Nrf2. Thus, toxicological and molecular biological aspects of Nrf2 merged. This Itoh et al. paper has become the most cited in the Nrf2 field. Initial follow-up toxicological studies using either of the original lines of Nrf2 KO mice demonstrated their increased sensitivity to acute hepatotoxicity from acetaminophen (Chan et al. 2001, Enomoto et al. 2001), acute pulmonary toxicity from butylated hydroxytoluene (BHT) (Chan and Kan 1999) and gastric carcinogenesis evoked by benzo[a]pyrene (Ramos-Gomez et al. 2001). These mice have now been used in dozens of models of chemically- and genetically-induced disease where pathological conditions are typically exacerbated by disruption of Nrf2 (Yamamoto et al. 2018). More recent development of a Nrf2 loxP mouse allows for isolation and interrogation of tissue-specific actions of the transcription factor in these pathologies (Xue et al. 2013). In addition, a Nrf2 KO rat has been generated by genome-editing technologies and applied to toxicological studies (Taguchi et al. 2016).
Prior to the cloning and functional characterization of Nrf2, Talalay et al. (1988) identified multiple classes of chemicals that induced detoxication enzymes and documented that for many their potencies paralleled their efficiency as electrophilic Michael addition acceptors and all shared a common chemical property of modifying sulfhydryl groups by oxidation, reduction or alkylation. A biosensor molecule coordinating the expression of detoxication genes by electrophilic and reactive oxygen-generating inducers was discovered in 1999 by Itoh et al. (1999). This thiol-rich, negative regulator of Nrf2 named Keap1 (Kelch-like ECH-associated protein 1) was isolated by yeast two-hybrid screening. Keap1 was subsequently identified as an adaptor protein for E3 ubiquitin ligase that collectively functions to maintain low steady-state levels of Nrf2 under quiescent conditions by marking it for proteasomal degradation by polyubiquitination (Sekhar et al. 2002, Kobayashi et al. 2004). However, perturbation of Keap1-Nrf2 interaction by modification of cysteine residues on Keap1 allow Nrf2 to escape degradation, translocate to the nucleus and activate its transcriptional program. A rat homologue of Keap1 was cloned in 2001 through the purification of Nrf2-interacting proteins from rat liver extracts (Dhakshinamoorthy and Jaiswal 2001). Recent reviews describe the emerging details of the structural mechanisms governing the interactions between Keap1, Nrf2 and E3 ubiquitin ligase and the chemical alterations imparted by inducers on the complex that affect the stability of Nrf2 (Tebay et al. 2015, Yamamoto et al. 2018).
A systemic Keap1 KO mouse was quickly developed and quite unexpectedly was shown to present with severe hyperkeratosis in the esophagus and forestomach, leading to death within weeks postnatally due to upregulation of Keratin 6 and occlusions in these tissues (Wakabayashi et al. 2003). To study physiological and pathological roles of constitutive, amplified Nrf2 signaling in vivo, in addition to the Keap1+/− mouse which is viable, single allele Keap1 loxP (Blake et al. 2010) and hypomorphic floxed knockdown mice (Okawa et al. 2006, Taguchi et al. 2010) have been created. In most cases studied, these lines of mice are more resistant to disease states than are wild-type or Nrf2 KO counterparts (Yamamoto et al. 2018).
2. Roles of Nrf2 in the Liver
Initial measures of enzyme specific activities in rodent liver indicated a coordinated expression of perhaps a half dozen xenobiotic metabolism genes by the enzyme inducers under study (typically phenolic antioxidants and dithiolethiones) (Benson et al. 1978, Benson et al. 1980, Kensler et al. 1985). Subtractive hybridization technology applied to identify dithiolethione-inducible genes in the liver tripled that number of likely gene battery members (Primiano et al. 1996). Nrf2 is now known to regulate the expression of about 250 genes through interactions with the ARE in combination with its heterodimeric binding partners, small MAFs. Studies across multiple tissues and cell lines indicate that these genes encode a network of enzymes involved in endo- and xeno-biotic metabolism, production of antioxidants and reducing equivalents, metabolism of carbohydrates and lipids, iron catabolism, protein degradation and regulators of inflammation (Yamamoto et al. 2018). Considerable analyses of gene and protein expression programs have been undertaken in rodent liver and primary human hepatocytes using microarray and proteomics in the Keap1 and Nrf2 genetic models and following siRNA targeting of these players, respectively (Kwak et al. 2001, Kwak et al. 2003, Yates et al. 2009, Kitteringham et al. 2010, Wu et al. 2012a, Walsh et al. 2014). These and other gene expression profiling studies have not only defined direct target genes of Nrf2, but have also presaged the cross-talk now recognized between Nrf2 and other signaling networks (Wakabayashi et al. 2010b).
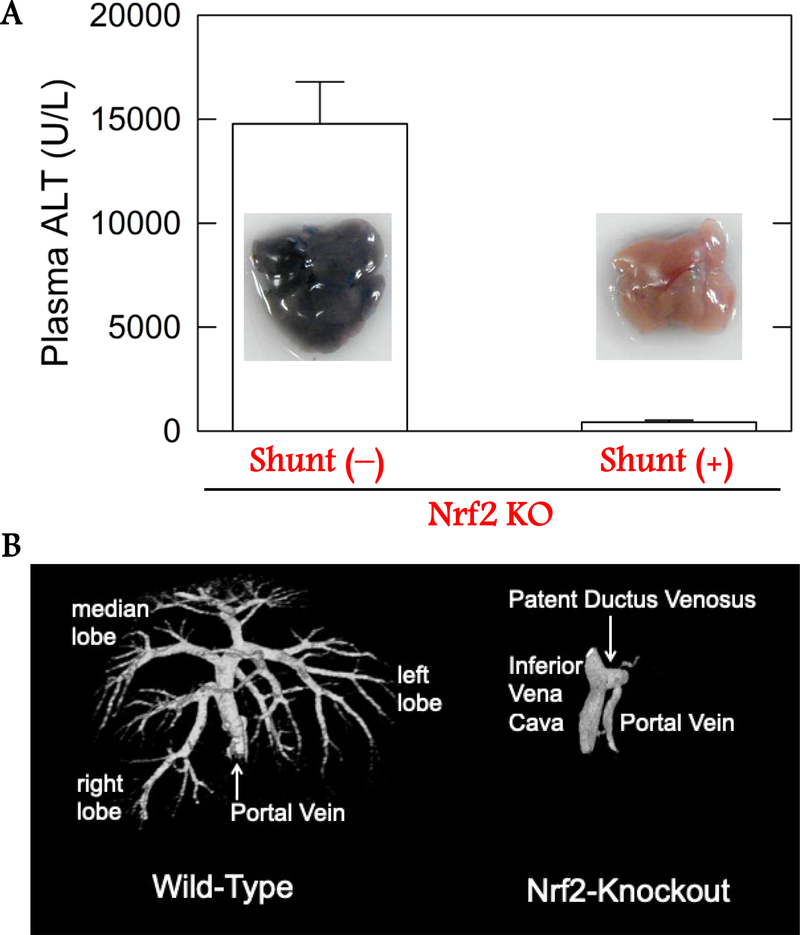
Nrf2 KO mice are now used extensively to study the etiopathogenesis and prevention of numerous diseases, often under the supposition of the original characterization of no intrinsic phenotype other than an altered adaptive response to stress (Chan et al. 1996, Itoh et al. 1997). One highly penetrant characteristic of these mice recognized early on is a lack of discolored incisor teeth (Yanagawa et al. 2004), reflecting a disorder in iron homeostasis. More recently, modulation of Nrf2 signaling has been found to affect liver size and growth. The liver to body weight ratio of Nrf2 KO mice is reduced when compared to wild-type (Beyer et al. 2008), while mice harboring liver-specific disruption of Keap1 display enhanced liver to body weight ratios (Wakabayashi et al. 2014). Morphological observations in Nrf2 KO mice have also indicated that bile ducts within the liver exhibit reduced micro-branching compared to wild-type and enhanced micro-branching with disruption of Keap1 (Wakabayashi et al. 2014). Disruption of Nrf2 signaling in the hepatic microenvironment could influence vascularization and physiological changes to the hepatic architecture could alter toxicological responses. Vascular corrosion casting of Nrf2 KO mice through the hepatic portal vein revealed a patent intrahepatic portacaval shunt (Skoko et al. 2014) (Figure 1), a vascular development defect also seen in aryl hydrocarbon receptor (AhR) KO mice (Lahvis et al. 2000). The congenital intrahepatic shunt in Nrf2 KO mice strongly influences hepatocyte oxygenation, gene expression and sensitivity to hepatotoxins in manners that may supersede the canonical effects of the loss of the Nrf2 transcription factor on xenobiotic and endobiotic metabolism through direct regulation of target gene expression. The penetrance of this phenotype in C57BL/6J is incomplete (~67%) and mice can be readily phenotyped by examining ketamine/xylazine sleeping times. However, few toxicological studies have accounted for a role of the shunt in their reported outcomes, so the impact of a portacaval shunt on the actions of toxins described below are largely unknown. The shunt does not appear to remain patent in Nrf2 KO rats (Taguchi et al. 2016).
Figure 1: Hepatic shunt in Nrf2 KO mice.
Altered acetaminophen toxicity and portal vasculature in wild-type versus Nrf2 KO C57BL/6J mice. Top. Plasma alanine aminotransferase (ALT) levels in 10-week old fasted wild-type and Nrf2 KO mice following acetaminophen challenge (250 mg/kg, i.p) after 6 hours. Means ± SE. Bottom. Micro computed tomography of livers perfused through the portal vein with radio-opaque Microfil in Nrf2 KO mice with intrahepatic shunt and wild-type mice. Figure adapted from Skoko et al. (2014).
3. Modulation of acute toxicity by NRF2
To evaluate the roles of Nrf2 in hepatotoxicity in vivo, a variety of hepatotoxicants have been studied in genetically modified mice and rats including Nrf2 KO, Nrf2 conditional KO, Keap1 knockdown and Keap1 conditional KO animals.
3–1. Acetaminophen
Acetaminophen, the primary ingredient of an antipyretic analgesic, is easily available due to marketing as an over-the-counter drug. As mentioned above, acetaminophen was the first toxic agent tested in Nrf2 KO mice to demonstrate that Nrf2 is a critical transcription factor that regulates the expression of cytoprotective genes. Nrf2 KO mice proved to be remarkably sensitive to acetaminophen hepatoxicity compared to wild-type mice (Chan et al. 2001, Enomoto et al. 2001). These early studies, which did not employ mice in the C57BL/6 background, were apparently not encumbered by the presence of patent shunts. C57BL/6 Nrf2 KO mice with the shunt are resistant to acetaminophen hepatotoxicity, because of the diminished blood flow to the liver. Keap1 knockdown (Keap1flox/−) mice, which manifest a systemic, partial activation of Nrf2, are able to survive at the lethal dose (Taguchi et al. 2010). Altered metabolism of acetaminophen likely accounts for these effects. Low doses of acetaminophen are conjugated with glucuronic acid by UDP-glucuronosyl transferases (UGTs) to facilitate excretion in urine. Overdosing with acetaminophen leads to acute hepatotoxicity through formation of the reactive metabolite, N-acetyl-p-benzoquinone imine (NAPQI), which is formed by Cyp isozymes (Laine et al. 2009). NQPQI binds to cellular proteins (Dahlin et al. 1984). Reisman et al. (2009) have reported decreased glucuronidation in Nrf2 KO mice and enhanced glucuronidation in Keap1 knockdown mice, highlighting a role for this detoxication pathway as a determinant of toxic responses. Acetaminophen also depletes hepatic glutathione (GSH) by as much as 90% (Mitchell et al. 1973). GSTs serve as detoxication enzymes that conjugate NAPQI with GSH. N-acetylcysteine (NAC), a precursor for GSH, prevents or decreases acetaminophen-induced hepatotoxicity. Nrf2 increases GSH synthesis through upregulation of glutamate-cysteine ligase modifier and catalytic subunits (Gclm and Gclc) to enhance excretion of the conjugate of NAPQI with GSH. Pretreatment with buthionine sulfoximine (BSO), a GSH depletor, cancels the resistance of these Keap1 knockdown mice. Transgenic mice that conditionally over-express Gcl subunits are protected from acetaminophen-induced liver injury (Botta et al. 2008). Small molecule inducers of Nrf2 signaling, such as dithiolethiones and the triterpenoid CDDO-Im, also protect against acetaminophen toxicity (Ansher et al. 1983, Reisman et al. 2009).
3–2. Carbon Tetrachloride (CCl4)
CCl4 is used in acute and chronic liver damage models to induce oxidative stress, necrosis, cholestasis and fibrosis. CCl4 is metabolized by Cyp enzymes to trichloromethyl-free radicals (CCl3•) that directly react with nucleic acids, proteins and lipids. In addition, CCl3• reacts with oxygen to form trichloromethyl peroxy radicals (CCl3•OO•) that subsequently cause lipid peroxidation. These reactive intermediate products are detoxified by enzymes such as GSTs and UGTs. Nrf2 protects the liver from acute and chronic toxin-mediated damage (Xu et al. 2008). Repair of hepatic injury that occurs after a single treatment with CCl4 was severely delayed in Nrf2 KO mice. The defect in repair was accompanied by an enhanced and prolonged inflammatory and profibrotic response. After long-term CCl4 treatment, the development of liver fibrosis was also enhanced in Nrf2 KO mice.
CCl4 has a potential to activate Nrf2 signaling, although the response is modest and insufficient to activate an early functional protective response to this and other hepatotoxins (Randle et al. 2008). Some plant extracts and chemicals protect against CCl4-induced hepatotoxicity; Curcuma longa Linn. extract (Lee et al. 2010), blackberry extract (Cho et al. 2011), isorhamnetin-3-O-galactoside - a flavonoid glycoside isolated from Artemisia capillaris Thunberg (Compositae) (Kim et al. 2012), ethanolic extract of A. paniculata (Chen et al. 2012), glycyrrhetinic acid - a derivative products of glycyrrhizic acid from liquorice root (Chen et al. 2013), and stevia (Ramos-Tovar et al. 2018).
3–3. Concanavalin A (ConA)
ConA is a mitogenic lectin extracted from the jack bean, Canavalia ensiformis. ConA is used as a model for T cell-mediated acute inflammatory liver injury. Activated inflammatory cells target hepatocytes, leading to apoptosis and necrosis (Tiegs et al. 1992). Amplified Nrf2 signaling, achieved either pharmacologically with CDDO-Im or genetically using liver-specific Keap1 KO mice inhibited ConA-mediated oxidative damage and hepatotoxicity. The protective effects of CDDO-Im were abrogated in Nrf2 KO mice (Osburn et al. 2008). Enhanced Nrf2 signaling had no effect on the initial increases in serum soluble proinflammatory proteins (e.g., IFN-γ, TNF-α, MCP-1 and IL-2), but suppressed late-stage inflammatory cell infiltration. ConA-induced hepatoxicity can also be inhibited by a variety of natural product inducers of Nrf2 signaling including dihydroquercetin (Zhao et al. 2015).
3–4. Ethanol
Alcohol drinking is a global health problem leading to a variety of medical conditions including cancers (Room et al. 2005). One-third of liver cirrhosis and one-fourth of liver cancers are thought to be attributable to alcohol consumption in humans (Room et al. 2005). Ethanol is metabolized by cytochrome P450s. Ethanol induces Cyp2e1 to generate acetaldehyde and resultant oxidative stress. When Cyp2e1 activity is elevated by ethanol, Nrf2 mRNA and protein levels are also observed to be increased in the liver (Gong and Cederbaum 2006). Several studies using Nrf2 KO mice demonstrate protective roles of Nrf2 in ethanol-induced hepatotoxicity. Loss of Nrf2 causes reduced detoxication of aldehyde and a marked steatosis in livers of ethanol-fed mice (Lamle et al. 2008). In ethanol-dosed Nrf2 KO mice, sterol regulatory element-binding protein 1, Srebp1, is elevated at mRNA and protein levels to induce the transcription of lipogenic enzymes. Nrf2 prevents ethanol-induced oxidative stress and accumulation of free fatty acids in liver by inducing expression of genes involved in antioxidant defense and decreasing expression of genes involved in lipogenesis (Wu et al. 2012b). Nrf2 KO mice exhibit increased mortality after ethanol exposure (Sun et al. 2018). Nrf2 plays an important protective role against acute (binge) induced hepatic and pancreatic damage by ethanol, which may be partially attributable to its primary regulatory role in antioxidant responses and its direct impact on ethanol metabolism. In this regard, sulforaphane has been shown to upregulate aldehyde dehydrogenase to metabolize acetaldehyde to nontoxic acetate, thereby reducing acetaldehyde toxicity (Ushida and Talalay 2013).
3–5. 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC)
While acetaminophen is used as a liver toxicant to evoke centrilobular damage, DDC is a hepatotoxicant that leads to injury around the portal vein. DDC-induced liver injury is ameliorated in Keap1 knockdown mice by promoting excretion of bilirubin (Taguchi et al. 2019). Fibrosis is enhanced in DDC-treated Nrf2 KO mice, but comprehensive liver injury is comparable to that observed in wild-type mice (Fragoulis et al. 2019). DDC is also used as an experimental model to expand oval cells that are liver progenitor cells. A mechanistic role for generation of oval cells by Nrf2 signaling has not yet been elucidated, but Nrf2 may improve hepatic regenerative capacity through enhanced proliferation of these cells.
3–6. Cadmium Chloride (CdCl2)
CdCl2 leads to kidney dysfunction, osteoporosis, and cancers in kidney, lung (Kolonel 1976) and prostate (Piscator 1981) and is causative of Itai-Itai disease that occurred in Japan in the 1900’s. Cadmium is also hepatotoxic. Acute treatment of mice with CdCl2 increased serum alanine aminotransferase and lactate dehydrogenase activities, and caused extensive hepatic hemorrhage and necrosis in Nrf2 KO mice. In contrast, Nrf2-enhanced mice (Keap1 knockdown and liver-specific Keap1 KO) had lower serum enzyme activities and less morphological alternations in the liver than wild-type mice. Studies in primary hepatocytes isolated from the four genotypes of mice indicated that oxidative stress was higher in Nrf2 KO cells, and lower in Nrf2-enhanced cells than in wild-type cells (Wu et al. 2012b). Wang et al. (Wang et al. 2015) have observed that cadmium selenide (CdSe) quantum dots used for imaging induce hepatic necrosis in mice. Protection was achieved by sub-chronic administration of sulforaphane, an inducer of Nrf2 signaling. Earlier studies indicated that exposure of CdCl2 upregulates Nrf2-target genes such as Nqo1 and Ho-1 (Alam et al. 1989, Beyersmann and Hechtenberg 1997, Stewart et al. 2003).
4. Effects of Nrf2 on chronic toxicity
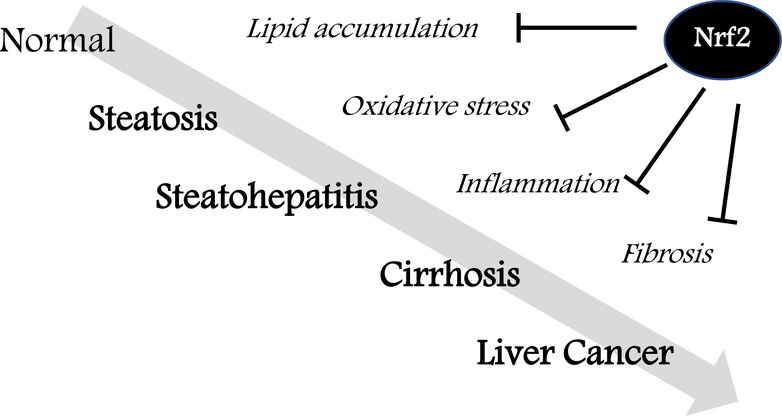
Many models of chronic hepatotoxicity have been established. Sequential alterations of steatosis with lipid accumulation, steatohepatitis with inflammation, cirrhosis and liver cancer are often seen and can be attenuated through enhancement of Nrf2 signaling (Figure 2).
Figure 2: Protective roles of Nrf2 in chronic liver diseases.
Sequential alterations of steatosis with lipid accumulation, steatohepatitis with inflammation, cirrhosis with fibrosis and liver cancer can be attenuated by Nrf2 activation.
4–1. Fatty liver disease
Fatty liver diseases can contribute to the onset of liver cancer. Methionine-choline deficient (MCD) diet is a classic dietary model that promotes the development of steatosis and then non-alcoholic steatohepatitis (NASH). This diet lacks methionine and choline, which are indispensable for hepatic mitochondrial ß-oxidation and very low-density lipoprotein (VLDL) synthesis (Anstee and Goldin 2006). The MCD diet induces oxidative stress and inflammation in the liver. Nrf2 is activated along with Nqo1 upregulation in MCD diet-fed wild-type mouse liver (Chowdhry et al. 2010, Sugimoto et al. 2010). In MCD diet-fed Nrf2 KO mice, NASH is aggravated in terms of fatty changes, inflammation, fibrosis, and iron accumulation. In the livers of the Nrf2 KO mice, oxidative stress is increased compared with that of wild-type mice based on increased levels of 4-hydroxy-2-nonenal and malondialdehyde. Contrary to Nrf2 KO mice, Keap1 knockdown mice are protected against MCD diet-induced NASH. Therefore, Nrf2 suppresses the rapid onset as well as the progression of NASH induced by MCD diet. Nrf2 inactivation reduces hepatic iron accumulation and counteracts oxidative stress-induced liver injury in NASH (Okada et al. 2012).
Use of the MCD diet is under debate as a model of NASH that reproduces human liver diseases (Stephenson et al. 2018). Unlike Western-like diets, the MCD diet decreases body weight, liver to body weight ratio, and serum glucose and leptin levels (Machado et al. 2015). Western-like diets high in saturated fat, trans fat and sugar better mimic the dietary patterns of humans. Effects of Nrf2 on diet-induced NASH has been examined in other models in addition to MCD diet. Using these models, several Nrf2 activators have been tested for prevention and treatment of NASH; the acetylenic tricyclic bis(cyano enone) TBE-31 in a high-fat plus fructose diet (Sharma et al. 2018), oltipraz and NK-252 (1-(5-(furan-2-yl)-1,3,4-oxadiazol-2-yl)-3-(pyridin-2-ylmethyl)urea) in rats fed a choline-deficient L-amino acid–defined diet (Shimozono et al. 2013), and curcumin in high-fat diet (Li et al. 2016). These chemicals attenuate the progression of NASH, although it remains to be elucidated whether the protective effects depend on Nrf2.
4–2. Aflatoxin B1 (AFB1)
AFB1, a mycotoxin produced by certain Aspergillus molds that contaminate foods, is a potent hepatocarcinogen in humans (Kensler et al. 2011). AFB1-induced hepatocarcinogenesis is well established in rats. Additionally, in rats the appearance of hepatic glutathione S-transferase P (GSTP)-positive foci predicts AFB1-induced hepatocarcinogenesis at an early stage before cancers develop. Co-treatment of rats with CDDO-Im, a potent Nrf2 inducer, with AFB1 completely prevents AFB1-induced hepatocarcinogenesis in a lifetime bioassay (Johnson et al. 2014) as well as formation of GSTP-positive foci (Yates et al, 2006). Diminished formation of aflatoxin-DNA adducts in the liver, driven by increased excretion of aflatoxin-mercapturic acid (a terminal product of GSH conjugation), are observed in the CDDO-Im treated animals; this altered metabolism of aflatoxin likely accounts for much of the protection and can be inferred to be mediated through Nrf2 signaling. In a more direct experimental approach, it is observed that Nrf2 KO F344 rats compared to wild-type are much more sensitive to the acute lethality from AFB1: an outcome associated with increased levels of DNA adducts and dampened detoxication (Taguchi et al. 2016). Whether Nrf2 KO rats are more or less sensitive to hepatocarcinogenesis remains an open question. Mice, where models of genetic manipulation of the Nrf2 pathway are abundant, are not an easy animal model to examine AFB1 toxicity because of intrinsic resistance to AFB1 hepatotoxicity due to a high constitutive expression of GSTs, particularly the GSTA3 subunit. In AFB1-dosed GSTA3 KO mice, DNA adducts are formed 100-fold more abundantly and excretion of aflatoxin-mercapturic acid is reduced greatly (Kensler et al. 2014). Interestingly, neither pharmacologic (CDDO-Im) nor genetic (Keap1 knockdown) strategies to elevate Nrf2 signaling offered protection in this hypersensitive mouse. Thus, GSTA3, which is a Nrf2-regulated gene, provided unilateral protection against AFB1. Interestingly, chronic treatment of the GSTA3 KO mice with AFB1 led to proliferation of oval cells, inhibition of hepatocyte proliferation and induction of cholangiocarcinoma rather than hepatocellular carcinoma (HCC) (Crawford et al. 2017).
4–2-2. Diethyl nitrosamine (DEN)
DEN, a constituent of tobacco smoke, is a widely used experimental hepatocarcinogen in rats and mice. DEN is metabolized by Cyp2e1 to a reactive α-hydroxydiethylnitrosamine, followed by the formation of an ethyl diazonium ion intermediate that gives rise to DNA adducts (Verna et al. 1996). Unexpectedly, deletion of Nrf2 confers resistance to DEN-induced hepatocarcinogenesis in mice (Ngo et al. 2017). Overactivated Nrf2 is required for tumor growth in DEN-induced HCC, perhaps through enhancement of the expression of metabolic enzymes required for cell proliferation.
Farber and colleagues established an experimental rat model of hepatocarcinogenesis that combined DEN, 2-acetylaminofluorene and partial hepatectomy (Solt et al. 1977). In this resistant hepatocyte model, Nrf2 or Keap1 mutations are found in 71% of GSTP-positive early preneoplastic lesions (Zavattari et al. 2015). The mutations are specifically concentrated in the Nrf2 Neh2 domain, which is the Keap1 binding region. Following this finding, a combination model of DEN and MCD diet was examined using Nrf2 KO rats (Orru et al. 2018). GSTP-positive foci with Nrf2 mutations appear in wild-type rats treated with DEN and MCD diet. Nrf2 disruption does not interfere with DEN-induced initiation but fully prevents the development of GSTP-positive preneoplastic lesions. Nrf2 appears to be a required for the clonal expansion of DEN-initiated hepatocytes.
4–4. Liver cancer
In human HCC, genetic analyses have been performed using whole-exome sequencing (Guichard et al. 2012). CTNNB1 encoding β-catenin, TP53, ARID1A and AXIN1 are altered in more than 10% of HCC. NFE2L2 encoding NRF2 is mutated in 6.4% of HCC. The NRF2 mutations are located within the DLG and ETGE motifs that are hotspots of somatic mutations. These mutations inhibit the binding with KEAP1 and stabilize NRF2. Six out of eight HCC with NRF2 mutations concurrently have CTNNB1 mutations. In another HCC analysis, KEAP1 mutations were identified by exome sequencing (Cleary et al. 2013). Somatic mutations of KEAP1 in HCC were reported at a frequency of 8.9% (Yoo et al. 2012).
In hepatitis C virus (HCV)-positive HCC, phosphorylated p62 accumulates and activates NRF2 by disrupting KEAP1-NRF2 binding (Saito et al. 2016). NRF2 activation through phosphorylated p62 directs glucose to the glucuronate pathway and glutamine towards GSH synthesis. A NRF2 inhibitor K67 (N-[2-acetonyl-4-(4-ethoxybenzenesulfonylamino)naphthalene-1-yl]-4-ethoxybenzenesulfonamide) disrupts the interaction between KEAP1 and p62 phosphorylated at S349. K67 suppresses the proliferation and chemoresistance of HCC. This finding suggests that NRF2 inhibitors might be effective especially in HCV-positive HCC patients. Limited numbers of NRF2 inhibitors have been reported to date. Use of current candidates are impeded due to their non-specific or toxic side effects.
Liver-specific Pten KO (Pten-Alb) mice are another model of liver cancer development driven through steatosis and NASH (Horie et al. 2004). In one-year old Pten-AlbCre mice, levels of well-known Nrf2 target gene products such as Gstp, Gstm and Gclc are increased along with p62 accumulation (Petersen et al. 2018). This suggests that Nrf2 signaling is activated in the liver cancers of Pten-AlbCre mice.
5. Interactions with other signaling pathways
Although complex in their own right, transcription factor signaling pathways such as Nrf2 do not operate unilaterally. There is extensive cross-talk with other signaling networks that fine tune tissue and organismal responses to stress and maintain cellular homeostasis (Wakabayashi et al. 2010b). Several examples are found in the liver.
5–1. Notch
Notch is a membrane-bound receptor-transcription factor complex. After accepting ligands such as Jags and Deltas, the receptor undergoes cleavage to yield the Notch intracellular domain (NICD) that translocates to the nucleus and functions as a transcription factor. Two of the four Notch receptors, Notch 1 and Notch 2 play important roles postnatally in liver development and maintenance of hepatic function. (Croquelois et al. 2005, Geisler et al. 2008). The binding core sequence, 5’-CGTGGGAG-3’, highly conserved in the Nrf2 promoter, interacts directly with the NICD to induce Nrf2 and Nrf2-target genes. Additionally, a functional ARE is located in upstream regulatory region of Notch1. Nrf2 to Notch signaling influences early phases of liver regeneration when hepatocyte renewal occurs following partial hepatectomy (Wakabayashi et al. 2010a). Notch to Nrf2 signaling leads to protection against acetaminophen toxicity (Wakabayashi et al. 2014). Morphological changes are also observed: hepatomegaly and enhanced micro-arborization of the biliary tree. These transcription factors likely affect the lineage fates of progenitor cells in the liver. Simultaneous activation of Notch and Nrf2 leads to substantial hepatomegaly in the liver in NICD::Keap1f/f-Albumin Cre mice (Wakabayashi et al. 2014).
5–2. Pten-PI3K-Akt signaling
As Nrf2 is polyubiquitinated and promptly degraded by the proteasome system, levels of Nrf2 protein expression are low in quiescent, unstressed cells. Nrf2 signaling is activated when Keap1 is inactivated as by electrophiles or oxidative stress. In addition to the Keap1-Cul3 complex (Kobayashi et al. 2004), βTrCP-Cul1 regulates the proteasomal degradation of Nrf2 (Chowdhry et al. 2013). Pten is a negative regulator of PI3K-Akt signaling. In the setting of inactivated Pten, Akt is phosphorylated and the downstream factor Gsk3 is phosphorylated and inactivated. Hepatic Nrf2 levels are markedly increased in liver-specific Pten::Keap1 double KO mice compared with liver-specific Keap1 single KO mice (Taguchi et al. 2014). This mouse model verifies the contributions of multiple degradation systems for Nrf2 in vivo.
5–3. AhR
AhR is a transcription factor that regulates genes encoding xenobiotic metabolizing oxygenases such as Cyp1a1 and Cyp1b1. When bound by polycyclic aromatic hydrocarbons such as dioxins, AhR translocates from the cytoplasm to the nucleus, heterodimerizes with the AhR nuclear translocator, and activates transcription through the xenobiotic response element (XRE). The XRE, 5’-T-GCGTG-3’, is distinct from the ARE sequence that binds Nrf2. In some instances, the response elements recognized by these transcription factors can be found in the regulatory domains of the same target genes. Dioxin induces Nqo1 in a Nrf2-dependent manner (Ma et al. 2004). AhR binds to the ARE in the Nrf2 promoter and modulates Nrf2 gene expression (Miao et al. 2005). Nrf2 also directly modulates AhR signaling, highlighting bidirectional interactions of these pathways (Shin et al. 2007).
Both AhR KO or Nrf2 KO mice are viable and fertile (Fernandez-Salguero et al. 1995, Itoh et al. 1997, Mimura et al. 1997). However, most (60%) AhR::Nrf2 double KO mice die within 1 week after birth due to unknown reasons (Noda et al. 2003). The surviving AhR::Nrf2 double KO mice show no apparent phenotype except for steatosis at the age of 1 week, which is also seen in AhR KO mice. AhR KO mice show portosystemic shunts in the liver and persistent abnormal vascular structures in the eyes and kidney (Lahvis et al. 2000). AhR plays an important role in developmental closure of the ductus venosus in the neonates (Lahvis et al. 2005). The liver sizes of AhR KO and Nrf2 KO mice are smaller than that of wild-type mice. As described at section 2, most of Nrf2 KO mice (67% in C57BL/6 strain) display a hepatic shunt (Skoko et al. 2014). Perhaps the lethality of the AhR::Nrf2 double KO mice may reflect problems in systemic circulation due to severe hepatic shunting. However, there is no report that examines this hypothesis.
5–4. Signal transducer and activator of transcription 5b (Stat5b)
Computational approaches were developed to identify factors that modulate Nrf2 target genes using an extensive compendium of gene expression outcomes in mouse liver from microarray analyses comparing Keap1 KO mice or Nrf2 KO mice treated with Nrf2 inducers CDDO-Im or oltipraz (Rooney et al. 2018). Activation of Nrf2 in the liver was consistently associated with suppression of the male-specific growth hormone-regulated transcription factor, Stat5b. One of 7 mammalian STAT transcription factors, Stat5b is regulated by estrogen or growth hormone and linked to sexual dimorphism and stress responses. Thus, feminization by chemical exposures were associated with Nrf2 activation. It is known that the livers of female mice normally exhibit higher Nrf2 activation than male mice. The enhanced basal and inducible levels of Nrf2 activation in females likely accounts for their greater resistance to age-dependent diseases and chemical-induced toxicity.
6. Monitoring Nrf2 for enzyme inducers and the detection of chemical toxicity
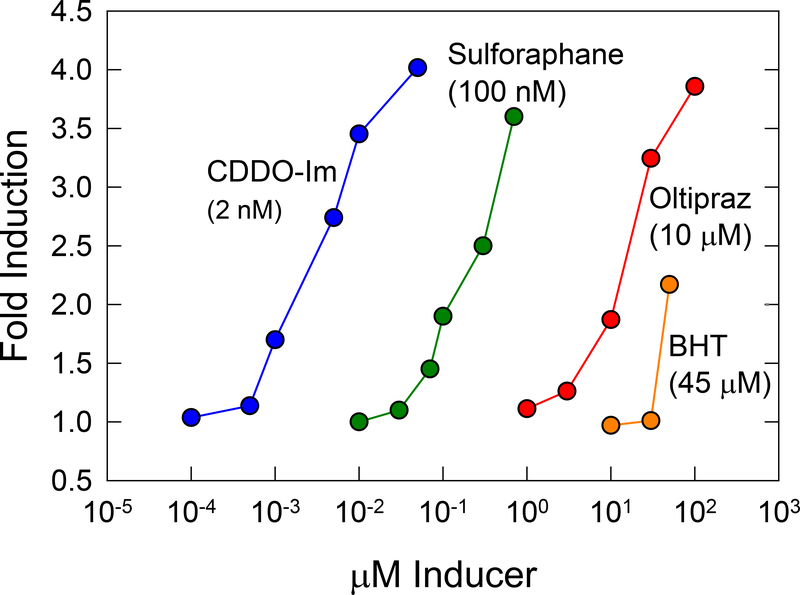
The measurement of enzyme induction in animals is laborious and expensive, and prompted Talalay and colleagues (Talalay et al. 1988) to developed a simple screening procedure in which the specific activity of NQO1, the product of a canonical Nrf2 target gene, is measured in Hepa1c1c7 murine hepatoma cells. They observed that these cells responded to many classes of inducers, and conversely induction of NQO1 in these cells was a reliable predictor of inducer activity in various rodent organs in vivo. The in vitro screening of extracts of fruits and vegetables lead to the discovery in 1992 of sulforaphane as a potent inducer of Nrf2 signaling in these hepatoma cells; intriguingly, sulforaphane, an isolate of broccoli, remains to date the most potent natural product inducer of the pathway (Dinkova-Kostova et al. 2017). Many dozens of inducers have been identified using high throughput screens of NQO1 activity. As depicted in Figure 3, concentration-dependent response curves indicate the progression of searches to find more potent inducers, beginning with the phenolic antioxidants (BHA) effective at mid-micromolar concentrations to more current efforts with CDDO-Im that are active a low nanomolar concentrations. Interestingly, not all inducers evoke identical programs of Nrf2-dependent gene expression in vivo. Wible et al. (2018) observed that while 1.2-dithiole-3-thione and CDDO-Im induced the Nrf2-regulated pathways of antioxidant defense and xenobiotic metabolism to comparable degrees in mouse liver, additional unique sets of Nrf2-dependent genes were regulated by each agent, e.g., cholesterol biosynthesis and ß-oxidation/lipid metabolism pathways, respectively.
Figure 3: Induction of Nrf2-regulated quinone reductase (NQO1) activity in Hepa1c1c7 mouse hepatoma cells.
There is a vast range in potency (>20,000-fold) of inducers of Nrf2 signaling used in cytoprotection studies. Optimal balance of potency, efficacy and specificity have not been defined in pre-clinical or clinical studies. Values in parentheses are the concentrations required to double NQO1 activity in Hepa1c1c7 cells. BHT, butylated hydroxytoluene; oltipraz, 5-(2-pyrazinyl)-4-methyl-1,2-dithiole-3-thione; sulforaphane, (−)-1-isothiocyanato-(4R)-(methylsulfinyl)butane; CDDO-Im, 1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole. Figure adapted from Kensler et al. (2010).
Given the critical role that Nrf2 may play as a modifier in the development and progression of chronic diseases, there has been a recent surge in interest by the pharmaceutical industry in the discovery and clinical development of small molecule inducers (Cuadrado et al. 2019). Issues of potency, specificity, mechanism of action, in vivo biodistribution and other factors including safety serve to define priority compounds. Hepatotoxicity accounts for a substantial number of drugs being withdrawn from the market. While direct signals for hepatotoxicity have not been described for Nrf2 activating compounds in current clinical trials (Dinkova-Kostova et al. 2017, Cuadrado et al. 2019), there is a broader role for monitoring activation of the Nrf2 pathway for in vivo toxicity prediction. The Tox21 program, a US inter-agency consortium, seeks to identify chemical structure-activity signatures derived through in vitro testing that could act as predictive surrogates for in vivo toxicity (Tice et al. 2013). A Tox21 10K library of compounds has been screened against 30 cell-based assays, including nuclear receptors and stress response pathways, at 15 concentrations leading to over 50 million data points (Huang et al. 2016). The stress response reporter assays include measures of mitochondrial toxicity, DNA damage, heat shock response, and activation of p53, AP-1, HIF-1α and ARE/Nrf2. For Nrf2, an ARE-β-lactamase reporter (ARE-bla) expressed in HepG2 cells, a human hepatocellular carcinoma line, is used as a measure of chemical induction of oxidative stress that indirectly leads to pathway activation (Shukla et al. 2012). From this study, it has been possible to cluster compounds by structural similarity and activity profile. The working assumption is that groups of similar cytotoxicity profiles may have similar mechanisms of action. Such data would then allow for prioritization of chemicals with little or no toxicological data for more in-depth toxicological evaluation. Follow-up studies have evaluated chemical in vitro-in vivo correlations between ARE pathway activation and hepatotoxicity (Kim et al. 2016) in which a liver damage data set of 1,314 chemicals provided the in vivo database. The authors examined 4 assays (agonists of PPARγ signaling, antagonists of thyroid receptor signaling, inhibitors of tyrosyl DNA phosphodiesterase, and activators of AREbla) thought to be relevant to ARE perturbations. None of the individual assay predictions were significantly associated with in vivo liver damage; however, combining all four assays resulted in a statistically significant association. Chemicals evoking mitochondrial dysfunction, linked to overproduction of reactive oxygen have also been linked to activation of the ARE/Nrf2 reporter (Xia et al. 2018). Copple et al. (2019) have characterized the NRF2 transcriptional network and its response to chemical insults in primary human hepatocytes as a tool for prediction of drug-induced liver injury. Microarray was used to identify 108 transcripts both downregulated by siRNA targeting NRF2 and upregulated by siRNA targeting KEAP1. Initial results indicate that activation of the expression of NRF2 target gene expression is a very good indicator of propensity to cause chemical stress; however, such activation had a lower sensitivity for the prediction of clinical drug induced liver injury in a test set of 158 compounds.
Electrophile and reactive oxygen-dependent protein modifications have emerged as cell signaling mechanisms that link adaptive responses to stress with cell function (Rudolph and Freeman 2009). Multiple transcription factors undergo posttranslational modification at reactive cysteine residues to mediate signal transduction. These highly conserved signaling pathways allow organisms to respond to electrophilic species that are produced as mediators of inflammation and metabolic stress, formed from a plethora of compounds in the diet and generated as consequences of toxin exposure. The chemical reactivity of endogenous and exogenous electrophiles (“soft” versus “hard”) towards nucleophilic targets determine proclivity towards proteomic versus genomic reactions and consequently, whether these events have therapeutic or toxicological implications. While induction of Nrf2 signaling is not a toxic response per se; it can be a harbinger of concern that requires detailed examination of the margin of safety of inducing compounds.
Acknowledgements
This work was supported by funding from MEXT/JSPS KAKENHI (16H01190 to K.T.); TERUMO LIFE SCIENCES FOUNDATION (18-III415 to K.T.) as well as NIH grant R35 CA197222 (T.W.K) and the Washington State Andy Hill CARE Fund (T.W.K.).
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- Alam J, Shibahara S, Smith A (1989) Transcriptional activation of the heme oxygenase gene by heme and cadmium in mouse hepatoma cells. J Biol Chem 264:6371–6375. [PubMed] [Google Scholar]
- Ansher SS, Dolan P, Bueding E (1983) Chemoprotective effects of two dithiolthiones and of butylhydroxyanisole against carbon tetrachloride and acetaminophen toxicity. Hepatology 3:932–935. [DOI] [PubMed] [Google Scholar]
- Anstee QM, Goldin RD (2006) Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int J Exp Pathol 87:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson AM, Batzinger RP, Ou SY, Bueding E, Cha YN, Talalay P (1978) Elevation of hepatic glutathione S-transferase activities and protection against mutagenic metabolites of benzo(a)pyrene by dietary antioxidants. Cancer Res 38:4486–4495. [PubMed] [Google Scholar]
- Benson AM, Hunkeler MJ, Talalay P (1980) Increase of NAD(P)H:quinone reductase by dietary antioxidants: possible role in protection against carcinogenesis and toxicity. Proc Natl Acad Sci U S A 77:5216–5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer TA, Xu W, Teupser D, auf dem Keller U, Bugnon P, Hildt E, Thiery J, Kan YW, Werner S (2008) Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. EMBO J 27:212–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyersmann D, Hechtenberg S (1997) Cadmium, gene regulation, and cellular signalling in mammalian cells. Toxicol Appl Pharmacol 144:247–261. [DOI] [PubMed] [Google Scholar]
- Blake DJ, Singh A, Kombairaju P, Malhotra D, Mariani TJ, Tuder RM, Gabrielson E, Biswal S (2010) Deletion of Keap1 in the lung attenuates acute cigarette smoke-induced oxidative stress and inflammation. Am J Respir Cell Mol Biol 42:524–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta D, White CC, Vliet-Gregg P, Mohar I, Shi S, McGrath MB, McConnachie LA, Kavanagh TJ (2008) Modulating GSH synthesis using glutamate cysteine ligase transgenic and gene-targeted mice. Drug Metab Rev 40:465–477. [DOI] [PubMed] [Google Scholar]
- Chan K, Han XD, Kan YW (2001) An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc Natl Acad Sci U S A 98:4611–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Kan YW (1999) Nrf2 is essential for protection against acute pulmonary injury in mice. Proc Natl Acad Sci U S A 96:12731–12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Lu R, Chang JC, Kan YW (1996) NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A 93:13943–13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HW, Huang YJ, Yao HT, Lii CK (2012) Induction of Nrf2-dependent antioxidation and protection against carbon tetrachloride-induced liver damage by Andrographis Herba (chuan xin lian) ethanolic extract. J Tradit Complement Med 2:211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zou L, Li L, Wu T (2013) The protective effect of glycyrrhetinic acid on carbon tetrachloride-induced chronic liver fibrosis in mice via upregulation of Nrf2. PLoS One 8:e53662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BO, Ryu HW, Jin CH, Choi DS, Kang SY, Kim DS, Byun MW, Jeong IY (2011) Blackberry extract attenuates oxidative stress through up-regulation of Nrf2-dependent antioxidant enzymes in carbon tetrachloride-treated rats. J Agric Food Chem 59:11442–11448. [DOI] [PubMed] [Google Scholar]
- Chowdhry S, Nazmy MH, Meakin PJ, Dinkova-Kostova AT, Walsh SV, Tsujita T, Dillon JF, Ashford ML, Hayes JD (2010) Loss of Nrf2 markedly exacerbates nonalcoholic steatohepatitis. Free Radic Biol Med 48:357–371. [DOI] [PubMed] [Google Scholar]
- Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD (2013) Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 32:3765–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary SP, Jeck WR, Zhao X, Chen K, Selitsky SR, Savich GL, Tan TX, Wu MC, Getz G, Lawrence MS, Parker JS, Li J, Powers S, Kim H, Fischer S, Guindi M, Ghanekar A, Chiang DY (2013) Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 58:1693–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copple IM, den Hollander W, Callegaro G, Mutter FE, Maggs JL, Schofield AL, Rainbow L, Fang Y, Sutherland JJ, Ellis EC, Ingelman-Sundberg M, Fenwick SW, Goldring CE, van de Water B, Stevens JL, Park BK (2019) Characterisation of the NRF2 transcriptional network and its response to chemical insult in primary human hepatocytes: implications for prediction of drug-induced liver injury. Arch Toxicol 93:385–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DR, Ilic Z, Guest I, Milne GL, Hayes JD, Sell S (2017) Characterization of liver injury, oval cell proliferation and cholangiocarcinogenesis in glutathione S-transferase A3 knockout mice. Carcinogenesis 38:717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croquelois A, Blindenbacher A, Terracciano L, Wang X, Langer I, Radtke F, Heim MH (2005) Inducible inactivation of Notch1 causes nodular regenerative hyperplasia in mice. Hepatology 41:487–496. [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Rojo AI, Wells G, Hayes JD, Cousin SP, Rumsey WL, Attucks OC, Franklin S, Levonen AL, Kensler TW, Dinkova-Kostova AT (2019) Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat Rev Drug Discov 18:295–317. [DOI] [PubMed] [Google Scholar]
- Dahlin DC, Miwa GT, Lu AY, Nelson SD (1984) N-acetyl-p-benzoquinone imine: a cytochrome P-450-mediated oxidation product of acetaminophen. Proc Natl Acad Sci U S A 81:1327–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakshinamoorthy S, Jaiswal AK (2001) Functional characterization and role of INrf2 in antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene 20:3906–3917. [DOI] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Fahey JW, Kostov RV, Kensler TW (2017) KEAP1 and done? Targeting the NRF2 pathway with sulforaphane. Trends Food Sci Technol 69:257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto A, Itoh K, Nagayoshi E, Haruta J, Kimura T, O’Connor T, Harada T, Yamamoto M (2001) High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicological Sciences 59:169–177. [DOI] [PubMed] [Google Scholar]
- Fernandez-Salguero P, Pineau T, Hilbert DM, McPhail T, Lee SS, Kimura S, Nebert DW, Rudikoff S, Ward JM, Gonzalez FJ (1995) Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science 268:722–726. [DOI] [PubMed] [Google Scholar]
- Fragoulis A, Schenkel J, Herzog M, Schellenberg T, Jahr H, Pufe T, Trautwein C, Kensler TW, Streetz KL, Wruck CJ (2019) Nrf2 ameliorates DDC-induced sclerosing cholangitis and biliary fibrosis and improves the regenerative capacity of the liver. Toxicol Sci doi: 10.1093/toxsci/kfz055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friling RS, Bensimon A, Tichauer Y, Daniel V (1990) Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc Natl Acad Sci U S A 87:6258–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friling RS, Bergelson S, Daniel V (1992) Two adjacent AP-1-like binding sites form the electrophile-responsive element of the murine glutathione S-transferase Ya subunit gene. Proc Natl Acad Sci U S A 89:668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler F, Nagl F, Mazur PK, Lee M, Zimber-Strobl U, Strobl LJ, Radtke F, Schmid RM, Siveke JT (2008) Liver-specific inactivation of Notch2, but not Notch1, compromises intrahepatic bile duct development in mice. Hepatology 48:607–616. [DOI] [PubMed] [Google Scholar]
- Gong P, Cederbaum AI (2006) Nrf2 is increased by CYP2E1 in rodent liver and HepG2 cells and protects against oxidative stress caused by CYP2E1. Hepatology 43:144–153. [DOI] [PubMed] [Google Scholar]
- Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, Clement B, Balabaud C, Chevet E, Laurent A, Couchy G, Letouze E, Calvo F, Zucman-Rossi J (2012) Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 44:694–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie Y, Suzuki A, Kataoka E, Sasaki T, Hamada K, Sasaki J, Mizuno K, Hasegawa G, Kishimoto H, Iizuka M, Naito M, Enomoto K, Watanabe S, Mak TW, Nakano T (2004) Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest 113:1774–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Xia M, Sakamuru S, Zhao J, Shahane SA, Attene-Ramos M, Zhao T, Austin CP, Simeonov A (2016) Modelling the Tox21 10 K chemical profiles for in vivo toxicity prediction and mechanism characterization. Nat Commun 7:10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y (1997) An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236:313–322. [DOI] [PubMed] [Google Scholar]
- Itoh K, Igarashi K, Hayashi N, Nishizawa M, Yamamoto M (1995) Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol Cell Biol 15:4184–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M (1999) Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13:76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NM, Egner PA, Baxter VK, Sporn MB, Wible RS, Sutter TR, Groopman JD, Kensler TW, Roebuck BD (2014) Complete protection against aflatoxin B(1)-induced liver cancer with a triterpenoid: DNA adduct dosimetry, molecular signature, and genotoxicity threshold. Cancer Prev Res (Phila) 7:658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler KH, Slocum SL, Chartoumpekis DV, Dolan PM, Johnson NM, Ilic Z, Crawford DR, Sell S, Groopman JD, Kensler TW, Egner PA (2014) Genetic or pharmacologic activation of Nrf2 signaling fails to protect against aflatoxin genotoxicity in hypersensitive GSTA3 knockout mice. Toxicol Sci 139:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Egner PA, Trush MA, Bueding E, Groopman JD (1985) Modification of aflatoxin B1 binding to DNA in vivo in rats fed phenolic antioxidants, ethoxyquin and a dithiothione. Carcinogenesis 6:759–763. [DOI] [PubMed] [Google Scholar]
- Kensler TW, Roebuck BD, Wogan GN, Groopman JD (2011) Aflatoxin: a 50-year odyssey of mechanistic and translational toxicology. Toxicol Sci 120 Suppl 1:S28–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N. Nrf2: friend or foe for chemoprevention? (2010) Carcinogenesis. 31:90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Cho HI, Kim KM, Kim SJ, Choi JS, Kim YS, Lee SM (2012) Isorhamnetin-3-O-galactoside protects against CCl4-induced hepatic injury in mice. Biomol Ther (Seoul) 20:406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MT, Huang R, Sedykh A, Wang W, Xia M, Zhu H (2016) Mechanism profiling of hepatotoxicity caused by oxidative stress using antioxidant response element reporter gene assay models and big data. Environ Health Perspect 124:634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitteringham NR, Abdullah A, Walsh J, Randle L, Jenkins RE, Sison R, Goldring CE, Powell H, Sanderson C, Williams S, Higgins L, Yamamoto M, Hayes J, Park BK (2010) Proteomic analysis of Nrf2 deficient transgenic mice reveals cellular defence and lipid metabolism as primary Nrf2-dependent pathways in the liver. J Proteomics 73:1612–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M (2004) Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24:7130–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolonel LN (1976) Association of cadmium with renal cancer. Cancer 37:1782–1787. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Itoh K, Yamamoto M, Sutter TR, Kensler TW (2001) Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1, 2-dimethiole-3-thione. Mol Med 7:135–145. [PMC free article] [PubMed] [Google Scholar]
- Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW (2003) Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem 278:8135–8145. [DOI] [PubMed] [Google Scholar]
- Lahvis GP, Lindell SL, Thomas RS, McCuskey RS, Murphy C, Glover E, Bentz M, Southard J, Bradfield CA (2000) Portosystemic shunting and persistent fetal vascular structures in aryl hydrocarbon receptor-deficient mice. Proc Natl Acad Sci U S A 97:10442–10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahvis GP, Pyzalski RW, Glover E, Pitot HC, McElwee MK, Bradfield CA (2005) The aryl hydrocarbon receptor is required for developmental closure of the ductus venosus in the neonatal mouse. Mol Pharmacol 67:714–720. [DOI] [PubMed] [Google Scholar]
- Laine JE, Auriola S, Pasanen M, Juvonen RO (2009) Acetaminophen bioactivation by human cytochrome P450 enzymes and animal microsomes. Xenobiotica 39:11–21. [DOI] [PubMed] [Google Scholar]
- Lamle J, Marhenke S, Borlak J, von Wasielewski R, Eriksson CJ, Geffers R, Manns MP, Yamamoto M, Vogel A (2008) Nuclear factor-eythroid 2-related factor 2 prevents alcohol-induced fulminant liver injury. Gastroenterology 134:1159–1168. [DOI] [PubMed] [Google Scholar]
- Lee HS, Li L, Kim HK, Bilehal D, Li W, Lee DS, Kim YH (2010) The protective effects of Curcuma longa Linn. extract on carbon tetrachloride-induced hepatotoxicity in rats via upregulation of Nrf2. J Microbiol Biotechnol 20:1331–1338. [DOI] [PubMed] [Google Scholar]
- Li B, Wang L, Lu Q, Da W (2016) Liver injury attenuation by curcumin in a rat NASH model: an Nrf2 activation-mediated effect? Ir J Med Sci 185:93–100. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kinneer K, Bi Y, Chan JY, Kan YW (2004) Induction of murine NAD(P)H:quinone oxidoreductase by 2,3,7,8-tetrachlorodibenzo-p-dioxin requires the CNC (cap ‘n’ collar) basic leucine zipper transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2): cross-interaction between AhR (aryl hydrocarbon receptor) and Nrf2 signal transduction. Biochem J 377:205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado MV, Michelotti GA, Xie G, Almeida Pereira T, Boursier J, Bohnic B, Guy CD, Diehl AM (2015) Mouse models of diet-induced nonalcoholic steatohepatitis reproduce the heterogeneity of the human disease. PLoS One 10:e0127991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao W, Hu L, Scrivens PJ, Batist G (2005) Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem 280:20340–20348. [DOI] [PubMed] [Google Scholar]
- Mimura J, Yamashita K, Nakamura K, Morita M, Takagi TN, Nakao K, Ema M, Sogawa K, Yasuda M, Katsuki M, Fujii-Kuriyama Y (1997) Loss of teratogenic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in mice lacking the Ah (dioxin) receptor. Genes Cells 2:645–654. [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Jollow DJ, Potter WZ, Davis DC, Gillette JR, Brodie BB (1973) Acetaminophen-induced hepatic necrosis. I. Role of drug metabolism. J Pharmacol Exp Ther 187:185–194. [PubMed] [Google Scholar]
- Moi P, Chan K, Asunis I, Cao A, Kan YW (1994) Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A 91:9926–9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo HKC, Kim DH, Cha YN, Na HK, Surh YJ (2017) Nrf2 mutagenic activation drives hepatocarcinogenesis. Cancer Res 77:4797–4808. [DOI] [PubMed] [Google Scholar]
- Noda S, Harada N, Hida A, Fujii-Kuriyama Y, Motohashi H, Yamamoto M (2003) Gene expression of detoxifying enzymes in AhR and Nrf2 compound null mutant mouse. Biochem Biophys Res Commun 303:105–111. [DOI] [PubMed] [Google Scholar]
- Okada K, Warabi E, Sugimoto H, Horie M, Tokushige K, Ueda T, Harada N, Taguchi K, Hashimoto E, Itoh K, Ishii T, Utsunomiya H, Yamamoto M, Shoda J (2012) Nrf2 inhibits hepatic iron accumulation and counteracts oxidative stress-induced liver injury in nutritional steatohepatitis. J Gastroenterol 47:924–935. [DOI] [PubMed] [Google Scholar]
- Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M (2006) Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun 339:79–88. [DOI] [PubMed] [Google Scholar]
- Orru C, Szydlowska M, Taguchi K, Zavattari P, Perra A, Yamamoto M, Columbano A (2018) Genetic inactivation of Nrf2 prevents clonal expansion of initiated cells in a nutritional model of rat hepatocarcinogenesis. J Hepatol 69:635–643. [DOI] [PubMed] [Google Scholar]
- Osburn WO, Yates MS, Dolan PD, Chen S, Liby KT, Sporn MB, Taguchi K, Yamamoto M, Kensler TW (2008) Genetic or pharmacologic amplification of nrf2 signaling inhibits acute inflammatory liver injury in mice. Toxicol Sci 104:218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen DR, Saba LM, Sayin VI, Papagiannakopoulos T, Schmidt EE, Merrill GF, Orlicky DJ, Shearn CT (2018) Elevated Nrf-2 responses are insufficient to mitigate protein carbonylation in hepatospecific PTEN deletion mice. PLoS One 13:e0198139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscator M (1981) Role of cadmium in carcinogenesis with special reference to cancer of the prostate. Environ Health Perspect 40:107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestera T, Zhang Y, Spencer SR, Wilczak CA, Talalay P (1993) The electrophile counterattack response: protection against neoplasia and toxicity. Adv Enzyme Regul 33:281–296. [DOI] [PubMed] [Google Scholar]
- Primiano T, Gastel JA, Kensler TW, Sutter TR (1996) Isolation of cDNAs representing dithiolethione-responsive genes. Carcinogenesis 17:2297–2303. [DOI] [PubMed] [Google Scholar]
- Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW (2001) Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A 98:3410–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Tovar E, Hernandez-Aquino E, Casas-Grajales S, Buendia-Montano LD, Galindo-Gomez S, Camacho J, Tsutsumi V, Muriel P (2018) Stevia prevents acute and chronic liver injury induced by carbon tetrachloride by blocking oxidative stress through Nrf2 upregulation. Oxid Med Cell Longev 2018:3823426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randle LE, Goldring CE, Benson CA, Metcalfe PN, Kitteringham NR, Park BK, Williams DP (2008) Investigation of the effect of a panel of model hepatotoxins on the Nrf2-Keap1 defence response pathway in CD-1 mice. Toxicology 243:249–260. [DOI] [PubMed] [Google Scholar]
- Reisman SA, Buckley DB, Tanaka Y, Klaassen CD (2009) CDDO-Im protects from acetaminophen hepatotoxicity through induction of Nrf2-dependent genes. Toxicol Appl Pharmacol 236:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Room R, Babor T, Rehm J (2005) Alcohol and public health. Lancet 365:519–530. [DOI] [PubMed] [Google Scholar]
- Rooney J, Oshida K, Vasani N, Vallanat B, Ryan N, Chorley BN, Wang X, Bell DA, Wu KC, Aleksunes LM, Klaassen CD, Kensler TW, Corton JC (2018) Activation of Nrf2 in the liver is associated with stress resistance mediated by suppression of the growth hormone-regulated STAT5b transcription factor. PLoS One 13:e0200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph TK, Freeman BA (2009) Transduction of redox signaling by electrophile-protein reactions. Sci Signal 2:re7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushmore TH, Morton MR, Pickett CB (1991) The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem 266:11632–11639. [PubMed] [Google Scholar]
- Rushmore TH, Pickett CB (1990) Transcriptional regulation of the rat glutathione S-transferase Ya subunit gene. Characterization of a xenobiotic-responsive element controlling inducible expression by phenolic antioxidants. J Biol Chem 265:14648–14653. [PubMed] [Google Scholar]
- Saito T, Ichimura Y, Taguchi K, Suzuki T, Mizushima T, Takagi K, Hirose Y, Nagahashi M, Iso T, Fukutomi T, Ohishi M, Endo K, Uemura T, Nishito Y, Okuda S, Obata M, Kouno T, Imamura R, Tada Y, Obata R, Yasuda D, Takahashi K, Fujimura T, Pi J, Lee MS, Ueno T, Ohe T, Mashino T, Wakai T, Kojima H, Okabe T, Nagano T, Motohashi H, Waguri S, Soga T, Yamamoto M, Tanaka K, Komatsu M (2016) p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat Commun 7:12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhar KR, Yan XX, Freeman ML (2002) Nrf2 degradation by the ubiquitin proteasome pathway is inhibited by KIAA0132, the human homolog to INrf2. Oncogene 21:6829–6834. [DOI] [PubMed] [Google Scholar]
- Sharma RS, Harrison DJ, Kisielewski D, Cassidy DM, McNeilly AD, Gallagher JR, Walsh SV, Honda T, McCrimmon RJ, Dinkova-Kostova AT, Ashford MLJ, Dillon JF, Hayes JD (2018) Experimental Nonalcoholic Steatohepatitis and Liver Fibrosis Are Ameliorated by Pharmacologic Activation of Nrf2 (NF-E2 p45-Related Factor 2). Cell Mol Gastroenterol Hepatol 5:367–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimozono R, Asaoka Y, Yoshizawa Y, Aoki T, Noda H, Yamada M, Kaino M, Mochizuki H (2013) Nrf2 activators attenuate the progression of nonalcoholic steatohepatitis-related fibrosis in a dietary rat model. Mol Pharmacol 84:62–70. [DOI] [PubMed] [Google Scholar]
- Shin S, Wakabayashi N, Misra V, Biswal S, Lee GH, Agoston ES, Yamamoto M, Kensler TW (2007) NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol Cell Biol 27:7188–7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SJ, Huang R, Simmons SO, Tice RR, Witt KL, Vanleer D, Ramabhadran R, Austin CP, Xia M (2012) Profiling environmental chemicals for activity in the antioxidant response element signaling pathway using a high throughput screening approach. Environ Health Perspect 120:1150–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoko JJ, Wakabayashi N, Noda K, Kimura S, Tobita K, Shigemura N, Tsujita T, Yamamoto M, Kensler TW (2014) Loss of Nrf2 in mice evokes a congenital intrahepatic shunt that alters hepatic oxygen and protein expression gradients and toxicity. Toxicol Sci 141:112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt DB, Medline A, Farber E (1977) Rapid emergence of carcinogen-induced hyperplastic lesions in a new model for the sequential analysis of liver carcinogenesis. Am J Pathol 88:595–618. [PMC free article] [PubMed] [Google Scholar]
- Stephenson K, Kennedy L, Hargrove L, Demieville J, Thomson J, Alpini G, Francis H (2018) Updates on dietary models of nonalcoholic fatty liver disease: Current studies and insights. Gene Expr 18:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart D, Killeen E, Naquin R, Alam S, Alam J (2003) Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J Biol Chem 278:2396–2402. [DOI] [PubMed] [Google Scholar]
- Sugimoto H, Okada K, Shoda J, Warabi E, Ishige K, Ueda T, Taguchi K, Yanagawa T, Nakahara A, Hyodo I, Ishii T, Yamamoto M (2010) Deletion of nuclear factor-E2-related factor-2 leads to rapid onset and progression of nutritional steatohepatitis in mice. Am J Physiol Gastrointest Liver Physiol 298:G283–294. [DOI] [PubMed] [Google Scholar]
- Sun J, Fu J, Zhong Y, Li L, Chen C, Wang X, Wang L, Hou Y, Wang H, Zhao R, Zhang X, Yamamoto M, Xu Y, Pi J (2018) NRF2 mitigates acute alcohol-induced hepatic and pancreatic injury in mice. Food Chem Toxicol 121:495–503. [DOI] [PubMed] [Google Scholar]
- Taguchi K, Hirano I, Itoh T, Tanaka M, Miyajima A, Suzuki A, Motohashi H, Yamamoto M (2014) Nrf2 enhances cholangiocyte expansion in Pten-deficient livers. Mol Cell Biol 34:900–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi K, Maher JM, Suzuki T, Kawatani Y, Motohashi H, Yamamoto M (2010) Genetic analysis of cytoprotective functions supported by graded expression of Keap1. Mol Cell Biol 30:3016–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi K, Masui S, Itoh T, Miyajima A, Yamamoto M (2019) Nrf2 activation ameliorates hepatotoxicity induced by a heme synthesis inhibitor. Toxicol Sci 167:227–238 [DOI] [PubMed] [Google Scholar]
- Taguchi K, Takaku M, Egner PA, Morita M, Kaneko T, Mashimo T, Kensler TW, Yamamoto M (2016) Generation of a new model rat: Nrf2 knockout rats are sensitive to aflatoxin B1 toxicity. Toxicol Sci 152:40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talalay P, De Long MJ, Prochaska HJ (1988) Identification of a common chemical signal regulating the induction of enzymes that protect against chemical carcinogenesis. Proc Natl Acad Sci U S A 85:8261–8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebay LE, Robertson H, Durant ST, Vitale SR, Penning TM, Dinkova-Kostova AT, Hayes JD (2015) Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic Biol Med 88:108–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice RR, Austin CP, Kavlock RJ, Bucher JR (2013) Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect 121:756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiegs G, Hentschel J, Wendel A (1992) A T cell-dependent experimental liver injury in mice inducible by concanavalin A. J Clin Invest 90:196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushida Y, Talalay P (2013) Sulforaphane accelerates acetaldehyde metabolism by inducing aldehyde dehydrogenases: relevance to ethanol intolerance. Alcohol Alcohol 48:526–534. [DOI] [PubMed] [Google Scholar]
- Verna L, Whysner J, Williams GM (1996) N-nitrosodiethylamine mechanistic data and risk assessment: bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacol Ther 71:57–81. [DOI] [PubMed] [Google Scholar]
- Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, Harada T, Engel JD, Yamamoto M (2003) Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet 35:238–245. [DOI] [PubMed] [Google Scholar]
- Wakabayashi N, Shin S, Slocum SL, Agoston ES, Wakabayashi J, Kwak MK, Misra V, Biswal S, Yamamoto M, Kensler TW (2010a) Regulation of notch1 signaling by nrf2: implications for tissue regeneration. Sci Signal 3:ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N, Skoko JJ, Chartoumpekis DV, Kimura S, Slocum SL, Noda K, Palliyaguru DL, Fujimuro M, Boley PA, Tanaka Y, Shigemura N, Biswal S, Yamamoto M, Kensler TW (2014) Notch-Nrf2 axis: regulation of Nrf2 gene expression and cytoprotection by notch signaling. Mol Cell Biol 34:653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N, Slocum SL, Skoko JJ, Shin S, Kensler TW (2010b) When NRF2 talks, who’s listening? Antioxid Redox Signal 13:1649–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J, Jenkins RE, Wong M, Olayanju A, Powell H, Copple I, O’Neill PM, Goldring CE, Kitteringham NR, Park BK (2014) Identification and quantification of the basal and inducible Nrf2-dependent proteomes in mouse liver: biochemical, pharmacological and toxicological implications. J Proteomics 108:171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, He Y, Yu G, Li B, Sexton DW, Wileman T, Roberts AA, Hamilton CJ, Liu R, Chao Y, Shan Y, Bao Y (2015) Sulforaphane protects the liver against CdSe quantum dot-induced cytotoxicity. PLoS One 10:e0138771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wible RS, Tran QT, Fathima S, Sutter CH, Kensler TW, Sutter TR (2018) Pharmacogenomics of chemically distinct classes of Keap1-Nrf2 activators identify common and unique gene, protein, and pathway responses in v ivo. Mol Pharmacol 93:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KC, Cui JY, Klaassen CD (2012a) Effect of graded Nrf2 activation on phase-I and -II drug metabolizing enzymes and transporters in mouse liver. PLoS One 7:e39006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KC, Liu J, Klaassen CD (2012b) Role of Nrf2 in preventing ethanol-induced oxidative stress and lipid accumulation. Toxicol Appl Pharmacol 262:321–329. [DOI] [PubMed] [Google Scholar]
- Xia M, Huang R, Shi Q, Boyd WA, Zhao J, Sun N, Rice JR, Dunlap PE, Hackstadt AJ, Bridge MF, Smith MV, Dai S, Zheng W, Chu PH, Gerhold D, Witt KL, DeVito M, Freedman JH, Austin CP, Houck KA, Thomas RS, Paules RS, Tice RR, Simeonov A (2018) Comprehensive analyses and prioritization of Tox21 10K chemicals affecting mitochondrial function by in-depth mechanistic studies. Environ Health Perspect 126:077010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Hellerbrand C, Kohler UA, Bugnon P, Kan YW, Werner S, Beyer TA (2008) The Nrf2 transcription factor protects from toxin-induced liver injury and fibrosis. Lab Invest 88:1068–1078. [DOI] [PubMed] [Google Scholar]
- Xue P, Hou Y, Chen Y, Yang B, Fu J, Zheng H, Yarborough K, Woods CG, Liu D, Yamamoto M, Zhang Q, Andersen ME, Pi J (2013) Adipose deficiency of Nrf2 in ob/ob mice results in severe metabolic syndrome. Diabetes 62:845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Kensler TW, Motohashi H (2018) The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol Rev 98:1169–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa T, Itoh K, Uwayama J, Shibata Y, Yamaguchi A, Sano T, Ishii T, Yoshida H, Yamamoto M (2004) Nrf2 deficiency causes tooth decolourization due to iron transport disorder in enamel organ. Genes Cells 9:641–651. [DOI] [PubMed] [Google Scholar]
- Yates MS, Kwak M-K, Egner PA, Groopman JD, Bodreddigari S, Sutter TR, Baumgartner KJ, Roebuck BD, Yore MM. Honda T, Gribble GW, Sporn MB and Kensler TW (2006) Potent protection against aflatoxin-induced tumorigenesis through induction of Nrf2-regulated pathways by the triterpenoid, 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole. Cancer Res. 66:2488–2494. [DOI] [PubMed] [Google Scholar]
- Yates MS, Tran QT, Dolan PM, Osburn WO, Shin S, McCulloch CC, Silkworth JB, Taguchi K, Yamamoto M, Williams CR, Liby KT, Sporn MB, Sutter TR, Kensler TW (2009) Genetic versus chemoprotective activation of Nrf2 signaling: overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis 30:1024–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo NJ, Kim HR, Kim YR, An CH, Lee SH (2012) Somatic mutations of the KEAP1 gene in common solid cancers. Histopathology 60:943–952. [DOI] [PubMed] [Google Scholar]
- Zavattari P, Perra A, Menegon S, Kowalik MA, Petrelli A, Angioni MM, Follenzi A, Quagliata L, Ledda-Columbano GM, Terracciano L, Giordano S, Columbano A (2015) Nrf2, but not beta-catenin, mutation represents an early event in rat hepatocarcinogenesis. Hepatology 62:851–862. [DOI] [PubMed] [Google Scholar]
- Zhao M, Chen J, Zhu P, Fujino M, Takahara T, Toyama S, Tomita A, Zhao L, Yang Z, Hei M, Zhong L, Zhuang J, Kimura S, Li XK (2015) Dihydroquercetin (DHQ) ameliorated concanavalin A-induced mouse experimental fulminant hepatitis and enhanced HO-1 expression through MAPK/Nrf2 antioxidant pathway in RAW cells. Int Immunopharmacol 28:938–944. [DOI] [PubMed] [Google Scholar]