Abstract
Abstract: For many years it was thought that T lymphocytes recognized only peptide antigens presented by MHC class I or class II molecules. Recently, it has become clear that a wide variety of lipids and glycolipids are also targets of the T cell response. This novel form of cell‐mediated immune recognition is mediated by a family of lipid binding and presenting molecules known as CD1. The CD1 proteins represent a small to moderate sized family of β2‐microglobulin‐associated transmembrane proteins that are distantly related to MHC class I and class II molecules. They are conserved in most or all mammals, and control the development and function of T cell populations that participate in innate and adaptive immune responses through the recognition of self and foreign lipid antigens. Here we review the current state of our understanding of the structure and function of CD1 proteins, and the role of CD1‐restricted T cell responses in the immune system.
Keywords: adaptive immunity, CD1, glycolipid, innate immunity, lipid, NK T cell, T cell
The immune system of mammals is based on the coexistence of two strategies, now commonly referred to as innate and adaptive immunity. The innate immune response relies on the recognition of conserved pathogen‐associated molecular patterns by a limited number of germline‐encoded receptors with fixed structures, while the adaptive immune response allows the recognition of a potentially unlimited repertoire of antigens by specific receptors (i.e., immunoglobulins and T cell receptors) obtained by random recombination of genomic segments. One of the key mechanisms of this adaptive response is the T cell receptor‐mediated recognition of the complexes formed by the association of Major Histocompatibility Complex (MHC) class I or class II molecules with peptides.
During the past decade, it has been recognized that other mechanisms for specific antigen recognition by T lymphocytes also exist, such as that mediated by a third lineage of antigen presenting molecules that comprise the CD1 family. This novel pathway for antigen presentation represents an important advance in our understanding of the immune response because it allows the T cell system to recognize and respond to a class of antigens that is completely distinct from those that are presented by MHC class I and II. Instead of presenting peptides, the CD1 proteins bind and present lipid and glycolipid antigens of both self and foreign origin (1). Studies of this novel system for immune recognition now point to important roles in the processes of augmenting and regulating innate and adaptive immunity. Here we review recent developments in the study of CD1 molecules and the T cell populations that are controlled by them, and discuss their known and potential contributions to the immune response.
Diversification of the CD1 family
The CD1 molecules comprise a small to moderate sized family of β2‐microglobulin‐associated, transmembrane glycoproteins. They are structurally related to the MHC‐class I molecules, but are encoded by genes that are unlinked to the MHC locus and lack the extensive polymorphism of classical MHC encoded antigen presenting molecules. Variable numbers of CD1 genes have been found in all mammals studied to date, and these have been extensively characterized in several species including man, mouse, rat, rabbit and guinea pig (1, 2). It is currently not known whether CD1 genes exist in nonmammalian vertebrates such as birds or fish.
Three groups of CD1 molecules
In humans, the CD1 family has five members, designated as CD1a, CD1b, CD1c, CD1d and CD1e proteins. Among these, it has been proposed that two or perhaps three distinct groups of CD1 proteins can be discerned, based on sequence homologies, sites of expression and function. Thus, CD1a, CD1b and CD1c are typically classified as group 1 CD1 proteins, while the more divergent CD1d protein comprises group 2 (3). The CD1e protein has not been extensively studied, but it is intermediate in terms of its homology to group 1 and group 2 CD1 proteins, and appears to be unique in that its expression may be confined exclusively to intracellular compartments of myeloid lineage cells (4). Thus, we currently support the classification of CD1e as group 3 CD1. Most species studied to date show the presence of genes for multiple homologs of the human group 1 CD1 proteins, the exception being mice and rats, which only have genes encoding CD1d‐like proteins. The group 2 CD1 proteins appear to be even more highly conserved, and have been found in all mammals for which CD1 genes or proteins have been characterized in detail, with the exception of the guinea pig (Table 1).
Table 1.
Diversity of CD1 genes in mammals
| Total CD1 | Group 1 | Group 2 | Group 3 | |||
|---|---|---|---|---|---|---|
| Species | genes* | A | B | C | D | E |
| Man | 5 | 1 | 1 | 1 | 1 | 1 |
| Mouse | 2 | 0 | 0 | 0 | 2 | 0 |
| Rat | 1 | 0 | 0 | 0 | 1 | 0 |
| Guinea pig | ∼10 | ? | ≥ 4 | ≥ 3 | ? | ≥ 1 |
| Rabbit | ∼8 | ≥ 2 | ≥ 1 | ? | ≥ 1 | ≥ 1 |
| Sheep | ∼7 | ? | ≥ 3 | ? | ≥ 1 | ≥ 1 |
| Pig | ∼4 | ≥ 1 | ? | ? | ? | ? |
pseudogenes not included
? = presence or absence of isoform not established
Tissue distribution
Analysis of the tissue distribution of CD1 molecules provides further support for the classification of CD1 molecules into at least two distinct groups. The distribution of group 1 CD1 molecules is relatively restricted, with strong expression mostly limited to immature cortical thymocytes and a variety of specialized antigen‐presenting cells, most notably myeloid lineage dendritic cells and a subset of B lymphocytes (1). In contrast, human and murine CD1d molecules are widely expressed on hematopoietic cells (5, 6) and have also been detected on a variety of nonhematopoietic cell types, in particular epithelial cells (7, 8).
Cellular distribution and trafficking
An additional aspect of the diversity of CD1 molecules resides in their divergent cellular distribution (Fig. 1). The following is a summary of the differences in subcellular localization and trafficking that have been reported for the various CD1 isoforms.
Figure 1.
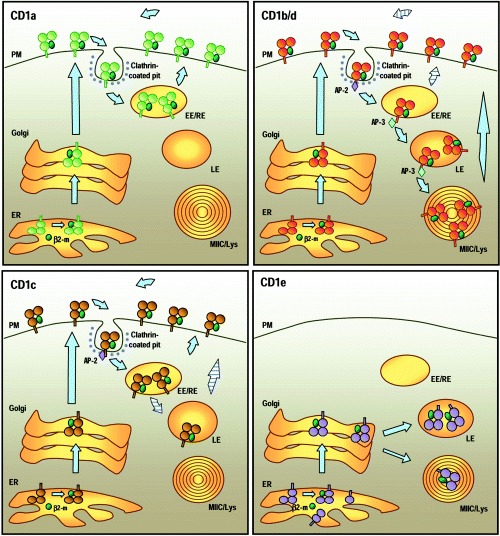
Cellular distribution and trafficking of human CD1 molecules. Top left: CD1a. After association with β2‐microglobulin in the endoplasmic reticulum (ER), CD1a molecules are transported to the plasma membrane (PM) through the secretory pathway. CD1a spontaneously internalizes via clathrin‐coated vesicles. It is not known whether this internalization requires association with other molecules. After internalization, CD1a recycles to the plasma membrane through early/sorting and early/recycling endosomes (EE/RE). Top right: CD1b and CD1d. The cellular distributions of CD1b and CD1d molecules are very similar. After expression at the cell surface, the interactions of the intracytoplasmic tails of CD1b and CD1d with the cytosolic adaptor complex AP‐2 mediate their internalization into clathrin‐coated pits and delivery to early endosomes. From there, they access the late endosomes (LE), the lysosomes and the MHC Class II compartments (MIIC/Lys) through their interaction with another cytosolic adaptor, AP‐3. CD1d molecules undergo several rounds of recycling to the cell surface (‘striped’ arrows) and tend to accumulate in the LE and MIIC/Lys over time. CD1b and CD1d that have been loaded with lipid antigens within the endocytic system escape to the cell surface and presumably accumulate there for antigen presentation. Bottom left: CD1c. CD1c molecules transit to the cell surface through the secretory pathway and are internalized via clathrin‐coated vesicles through their interaction with AP‐2. Most of the molecules recycle to the cell surface through the EE/RE, but a small fraction can be detected in the LE. Bottom right: CD1e. Alternative splicing potentially leads to the generation of various forms of CD1e in the ER (see text). Only CD1e molecules with three lumenal α domains can associate with β2‐microglobulin (β2‐m) and leave the ER. In immature dendritic cells (DC), most CD1e molecules are present in the Golgi. After activation and maturation of DC, CD1e molecules accumulate in the late endosomes and lysosomes where they are cleaved into a soluble form.
CD1a
At steady state, human CD1a molecules are prominently expressed at the plasma membrane of blood monocyte‐derived dendritic cells (DC) and epidermal Langerhans cells (LC). They are absent from the late endosomal and lysosomal compartments, but can be detected in the early/recycling endosomes (9). Analysis of CD1a trafficking in freshly extracted human LC suggests that CD1a molecules present at the cell surface spontaneously internalize via clathrin‐coated vesicles. Most likely, they are rapidly distributed to the early/sorting endosomes and delivered to early/recycling endosomes, and from this site they recycle back to the plasma membrane (10). It is still unclear which mechanisms drive the intracellular trafficking of CD1a, as its short intracytoplasmic tail does not contain any recognizable sorting motif, in contrast to most other CD1 molecules.
CD1b
Human monocyte‐derived DC express high levels of CD1b at their surface, but the molecule is also present in endosomes, lysosomes and the closely related MHC class II compartments (MIICs) of specialized antigen presenting cells (11, 12). After synthesis, CD1b molecules are transported rapidly to the cell surface via the secretory pathway, and from this site they are then internalized by clathrin‐mediated endocytosis. This internalization is dependent on a tyrosine‐based motif of the cytoplasmic tail (YXXΦ, where Y is tyrosine, X is any amino acid, and Φ is a bulky hydrophobic residue), probably through its interaction with cytosolic adaptor protein complex AP‐2 (11, 13). Subsequent sorting to the late endosomal compartment may be dependent on interaction of the CD1b tail with another adaptor protein AP‐3 (13, 14). It should be noted that these features of trafficking and localization have been described for human CD1b, but may not apply in all cases to orthologues of this protein in other species. For example, the guinea pig expresses several proteins that appear to be closely related in structure to human CD1b, but one of these (gpCD1b3) shows a pattern of cellular localization that is similar to that of human CD1a (15).
CD1c
The distribution of human CD1c partially overlaps those of CD1a and CD1b molecules. CD1c is expressed at the cell surface and most likely is internalized by clathrin‐mediated endocytosis, similar to CD1b. CD1c is abundantly present in the early/recycling compartment, but also colocalizes partially with CD1b in late endosomes (12). Despite the presence of a tyrosine‐based endosomal targeting motif very similar to the one present in CD1b, CD1c does not accumulate to any consistently appreciable extent in the MIIC compartment, probably because it cannot interact strongly with AP‐3 (13, 14).
CD1d
The steady state localizations and trafficking of human and murine CD1d molecules are very similar to those of human CD1b, with expression at the plasma membrane after synthesis and progressive accumulation in lysosomes and MIIC compartments, mediated by a tyrosine‐based internalization motif (16). However, two recent studies revealed the existence of alternative trafficking mechanisms for both murine and human CD1d, through their association, respectively, with the MHC class II‐associated invariant chain (Ii) and with MHC class II molecules themselves (17, 18).
Jayawardena‐Wolf et al. demonstrated that a fraction of murine CD1d coprecipitates with Ii, implying that the two molecules associate in vivo. The role of Ii on CD1d trafficking is supported by the observation that, in cells expressing murine CD1d from which the cytoplasmic tyrosine‐based targeting signal has been deleted, transfection of Ii results in redistribution of the mutant CD1d from the cell surface to late endosomes and lysosomes (17). Interestingly, Kang and Cresswell identified a very similar role of MHC Class II molecules in the trafficking of human CD1d (18). Taken together, these studies suggest the coexistence of two pathways of intracellular trafficking for CD1d (Fig. 2). Thus, one pathway allows newly synthesized CD1d molecules to move directly to the cell surface where they subsequently internalize to reach endosomes and MIICs. A second alternative pathway is followed by CD1d molecules that associate with the Ii (mice) or Ii/MHC class II complexes (human) in the ER. These would be predicted to enter the MIIC compartments directly as a result of intracellular sorting at the Golgi, and then subsequently be released to the cell surface after degradation of Ii and apparently still in association with MHC class II molecules. In agreement with this model, mouse CD1d trafficking is impaired in cathepsin S deficient cells, that are unable to normally degrade Ii, with accumulation of CD1d molecules in endosomal compartments where they colocalize with MHC II and Ii (19). While the currently published studies have examined Ii association only for CD1d, preliminary studies indicate that this can also be demonstrated for human CD1b (M. Sugita, V. Briken and S. A. Porcelli, unpublished data). Thus, it is possible that a similar alternative trafficking pathway resulting in direct Golgi to MIIC transport exists for at least some group 1 CD1 proteins.
Figure 2.
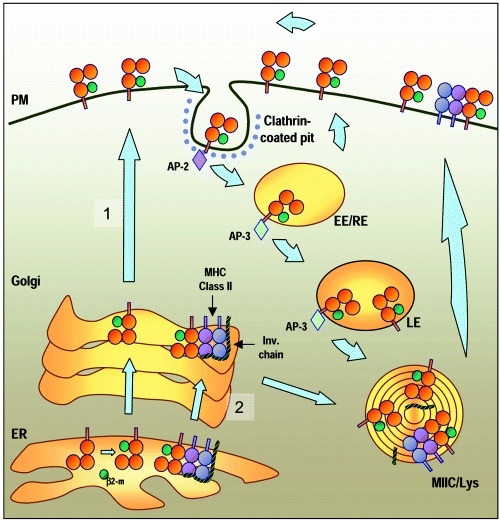
Alternative pathways for CD1d trafficking. Most CD1d molecules are transported directly to the plasma membrane (PM) through the secretory pathway (pathway 1). There, they are internalized via clathrin‐coated vesicles and access the endosomal compartments. After recycling several times between early endosomes (EE) and the cell surface, CD1d molecules accumulate over time in the lysosomes and MIIC compartments (MIIC/Lys). From there, they eventually return to the cell surface. A fraction of CD1d molecules form a complex in the ER with MHC class II molecules and the invariant chain (Ii). These most likely enter the MIIC compartment before being expressed at the cell surface as a result of sorting at the Golgi (pathway 2). After cleavage of Ii in the MIIC, the CD1d molecules are expressed at the cell surface. At least some of these CD1d molecules remain associated with MHC class II on the plasma membrane.
CD1e
Human CD1e has been identified only recently as a protein expressed by myeloid lineage DC (4). Analysis of CD1e mRNA from monocyte‐derived DC identified more than 15 mRNA species generated by alternative splicing. Some of these encode apparently full‐length transmembrane proteins with three extracellular domains (α1, α2 and α3) or membrane‐associated CD1e with only one (α3 only) or two α domains (α2α3, α1α3). Yet other transcripts lack transmembrane domain sequences and appear to encode putative soluble forms of the protein. Only the membrane‐associated isoforms with all three extracellular domains have been shown to leave the endoplasmic reticulum (ER), probably after they associate with β2‐microglobulin. These molecules are exported through the trans‐Golgi network to an acidic compartment, most likely the late endosomes or lysosomes, where they are cleaved into a soluble form and accumulate. This intracellular traffic is controlled by a short sequence in the C‐terminal end of the cytoplasmic tail containing two overlapping dilysine motifs (KKXK). In immature DC, CD1e molecules are found mainly in the Golgi compartments and trans‐Golgi network (TGN), whereas in LPS‐treated DC, CD1e molecules accumulate in late endosomes and lysosomes. Notably, CD1e molecules never seem to reach the cell surface, which appears to exclude their direct participation in antigen presentation. Homologues of human CD1e have been identified in guinea pigs and in several other mammals, but their expression and intracellular distributions have not been studied to date (Table 1). However, the marked conservation of many predicted structural features of CD1e in humans and other mammals suggests that the unusual intracellular localization may be conserved, and thus may be associated with some important function of this molecule.
T cell recognition of CD1‐presented antigens
Foreign and self antigens presented by CD1
Because of the resemblance between CD1 and MHC‐encoded antigen presenting molecules, early studies on the function of CD1 sought to demonstrate a role in specific recognition by T cells. A critical step was made with the isolation of a CD1b‐restricted human T cell line that was specific for a foreign antigen contained in Mycobacterium tuberculosis. The direct isolation and characterization of the mycobacterial substance presented to this T cell line led to the initial realization that CD1‐presented antigens are predominantly or exclusively lipid in nature (20).
The first CD1‐presented antigens to be precisely identified, all of which are presented by CD1b, were free mycolic acid, lipoarabinomannan (LAM) and glucose monomycolate (GMM), all major components of mycobacterial cell walls that do not have close structural homologs in mammalian cells (Fig. 3) (21). Additional studies revealed that other human group 1 CD1 molecules, CD1a and CD1c, could also mediate presentation of mycobacterial lipid antigens. While the identities of the foreign lipid antigens presented by CD1a are still not known, one prominent CD1c‐presented antigen from mycobacteria has been isolated and identified as a βl‐mannosyl‐phosphoisoprenoid (22). While these mycobacterial lipid antigens are in most cases structurally divergent from mammalian lipids, it has recently been demonstrated that a variety of mammalian glycosphingolipids are also presented by human group 1 CD1 molecules (Fig. 3) (23, 24). This fascinating finding suggests that CD1‐presented ligands are not solely, or even predominantly, molecular patterns expressed exclusively in microbial pathogens. This is important, because it indicates that in many situations the response to lipid antigens through CD1 presentation may represent a form of adaptive rather than innate immunity, requiring a fine distinction between self and foreign lipid antigens and a mechanism for maintaining tolerance to the former.
Figure 3.

Three classes of known CD1‐presented lipid antigens. The sources of the structures illustrated are stated on the right, along with the CD1 isoform(s) known to present them. Top: mycolates. These are α‐branched, β‐hydroxy fatty acids produced by bacteria belonging to the order Actinomycetales (e.g., mycobacteria, nocardia, corynebacteria and rhodococci). CD1b is known to present both free and glycosylated forms of these, including a wide variety of different mycolate structures distinguished by variations in their chain lengths, the presence or absence of double bonds or cyclopropyl groups, and the presence or absence of oxygen containing substitutions (R group) near the distal end of the longer meromycolate branch. Illustrated are a dicyclopropanated C80 mycolate lacking an R group (α‐mycolic acid), and a typical methoxy‐mycolate of M. tuberculosis. Two of the many potential forms of glucose monomycolate are illustrated, both of which are known to be presented by human CD1b. Center: glycosphingolipids. The prototype for this class is the marine sponge‐derived glycolipid (2S,3S,4R)‐1‐O‐(α‐D‐galactopyranosyl)‐N‐hexacosanoyl‐2‐amino‐1,3,4‐octadecanetriol, generally referred to as α‐galactosyl ceramide. More recent studies have demonstrated CD1‐restricted presentation of several mammalian glycosphingolipids, two of which are illustrated. Bottom: glycophospholipids. Mycobacterial forms of this class were initially demonstrated to be presented by human CD1b, including the relatively simple phosphatidylinositol dimannoside structure shown and more heavily glycosylated versions of this basic structure such as lipoarabinomannans of M. tuberculosis and M. leprae. Mammalian phosphatidylinositol and more highly glycosylated versions of this structure (glycosylphosphatidylinositols (GPI)) have been shown to bind to mouse and human CD1d proteins, although evidence for their recognition by T cells is currently limited, The fully saturated β1‐mannosyl phosphoisoprenoids isolated from M. tuberculosis and M. avium are unique at present as the only CD1‐presented antigens bearing a single alkyl chain tail.
To date, no microbial pathogen‐associated antigen presented by CD1d has been isolated. The best described CD1d‐presented antigens are synthetic glycosylceramides, and in particular a form of α‐galactosylceramide containing a phytosphingosine base and a long chain (C26) fatty acid (αGalCer) (Fig. 3). This unusual glycolipid was initially identified as the active component of marine sponge extracts that possessed potent antitumor activity in mice. Subsequent studies showed that αGalCer was a potent ligand for human and murine CD1d‐restricted natural killer T cells (NK T cells) (25, 26). These NK T cells are a population of lymphocytes characterized by the coexpression of cell surface molecules usually observed in the natural killer lineage (NK1.1, Ly49, CD69 in mice; NKR‐P1A, CD94 and CD69 in human), and a TCR αβ with a limited and highly biased repertoire (27). In mice, the TCR of NK T cells are assembled from an invariant TCRα chain (Vα14Jα18; formerly designated Vα14Jα281) paired preferentially with TCRβ chains encoded by Vβ8.2, Vβ7 or Vβ2 combined with various Jβ segments. Human NK T cells express TCR chains that are homologous to the murine chains: an invariant Vα24Jα15 (formerly Vα24JαQ), paired predominantly with Vβ11. Attempts thus far to identify a natural homolog of αGalCer in mammalian tissues or in relevant microbial pathogens have been unsuccessful. It thus seems unlikely that this compound represents a true antigen for natural CD1d‐restricted immune responses, and efforts are still being made to identify the natural microbial or self antigens that are presented by CD1d. While it is generally accepted that the frequent CD1d‐restricted autoreactivity observed among murine NK T cells is likely to be due to recognition of self‐antigens presented on CD1d, the precise identity of these putative natural self ligands remains unknown. Some initial clues toward the identification of such self ligands may be provided by reports that CD1d restricted NK T cells can be activated by natural and synthetic glycosyl phosphatidylinositol (GPI) (28) and other common phospholipids including phosphatidylinositol (PI), phosphatidylethanolamine (PE) and phosphatidylglycerol (PG) (29).
These studies corroborate an earlier claim that GPI was the major natural ligand that could be identified in fractions of low molecular weight material extracted from murine CD1d proteins (30). Together, these findings on the binding of endogenous lipids by CD1 have suggested a model in which some endogenous lipids might play a chaperon‐like role during the biosynthesis of CD1 molecules (Fig. 4). As recently described for GPI and PI in the case of CD1d molecules, the binding of these endogenous lipids is likely to occur in the ER, shortly after association of the CD1 heavy chain with β2‐microglobulin, and would serve to stabilize the molecule and protect the binding site from inappropriate interactions until it reached the cell surface (31, 32). After internalization of CD1 molecules, the chaperon lipids would subsequently be exchanged for other endogenous glycolipids or exogenous lipids present in endocytic compartments.
Figure 4.
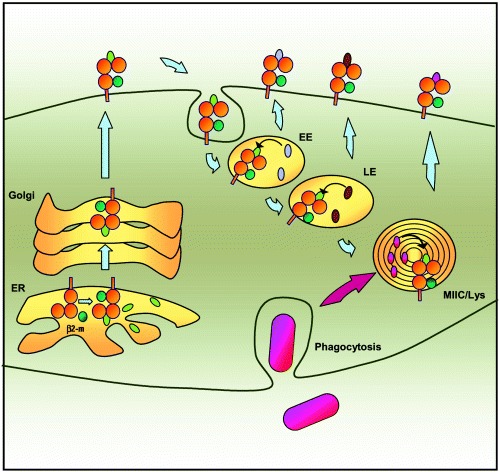
Binding of endogenous and exogenous lipids by CD1 molecules. CD1 molecules probably bind endogenous self lipids (green ovals) such as phosphatidylinositiol and glycosylphosphatidylinositol in the ER. The association may stabilize the molecule and protect the binding groove during migration to the cell surface. After internalization from the cell surface, the various CD1 isoforms follow similar pathways into the endocytic system. They accumulate at different sites (EE vs LE vs MIIC/Lys), which allows them to collectively sample the entire endocytic network. As they pass through the various compartments CD1 molecules may exchange the endogenous lipids loaded in the ER with other lipids accumulating in the cell via fluid phase or receptor‐mediated endocytosis (lavender and brown ovals) or phagocytosis (pink ovals). Most CD1 molecules recycle several times between the plasma membrane and the endosomes, thus enhancing their chances of encountering antigenic lipids at intracellular sites.
Molecular interactions involved in lipid antigen presentation
Interactions between CD1 proteins and their lipid ligands
As illustrated in Fig. 3, all identified CD1‐associated antigens are amphipathic lipid molecules, consisting of a hydrophilic head with polar or charged groups and a hydrophobic tail composed of one or two straight hydrocarbon chains. The molecular basis for TCR recognition of lipids in the context of CD1 presentation seems to parallel in many aspects the recognition of MHC/peptide complexes. Substantial insight in this area was provided by the crystal structure of the mouse CD1d protein (33). This revealed that CD1d molecules, like MHC class I proteins, possess an antigen‐binding groove formed by the αl and α2 domains. However, the binding groove in CD1d is narrower at its opening and deeper than the peptide‐binding groove in MHC molecules. In addition, it is formed mostly from two relatively large pockets (referred to as A′ and F′) that have a hydrophobic surface. Thus, the ligand‐binding groove of CD1d appears ideally suited to bind hydrophobic molecules, such as the alkyl tails of lipids and glycolipids. The general features of the MHC class I‐like fold with a large hydrophobic binding groove is likely to be conserved in all CD1 proteins, a view which is supported by recent structural data from the X‐ray crystallography of human CD1b molecules (34).
Based on the structure of CD1 molecules and the known ligands they present, a model has been proposed for CD1–lipid interactions. This predicts that the hydrophobic portion of the lipid ligand interacts with the A′ and F′ pockets, thus anchoring the lipid in the protein such that its polar head is positioned at or outside the opening of the groove, where it is available for direct contact with the TCR (35). This model is supported by several studies analyzing the structural requirements for glycolipid recognition by various CD1‐restricted T cells. For example, a CD1b‐restricted, GMM‐specific T cell line responds to several forms of GMM derived from different mycobacterial species that differ in mycolic acid composition, but do not respond to structural variants of the antigen that have even extremely subtle alterations in the hydrophilic head group (35). Similarly, αGalCer analogs with shortened acyl chains (as short as 2‐carbons) can stimulate murine and human CD1d‐restricted NK T cells, while analogs which lack hydroxyl groups on carbon 3 and 4 of the sphingosine base, are not antigenic (36). These studies suggest that while the nature of the hydrophobic tail certainly plays a role in the stability of the complex formed with CD1 molecules, it is unlikely to determine the specificity of the T cell response. In contrast, the fine structure of the hydrophilic head group is critical to TCR recognition, consistent with a model in which this portion of the antigen actually makes direct contact with the TCR.
TCR interactions with CD1 and CD1‐presented lipids
All of the CD1‐restricted T cell lines specific for mycobacterial antigens identified to date express αβ TCR. Analysis of clones restricted by human group 1 CD1 molecules showed heterogeneous usage of TCR Vα, Jα and Vβ segments, suggesting that, like peptide‐MHC specific T cells, the TCR repertoire of group 1 CD1‐restricted T cells is not strongly biased toward the usage of particular Vα/Vβ chains (37). The TCR of these cells also showed extensive junctional diversity of CDR3‐encoded residues. Analysis of site‐specific mutants of human CD1b and a CD1b‐specific TCR support the view that the TCR interacts both with the α‐helices of the CD1 molecule and with the lipid antigen protruding from its ligand binding groove (38, 39). In addition, as described previously for TCR interactions with MHC class I, the TCR of CD1‐restricted T cells are probably oriented diagonally to the horizontal axes of the αl and α2 helices of CD1. This orientation is predicted to place the highly variable CDR3 loops of the TCR over the opening of the binding groove where they could make direct interactions with the hydrophilic cap of the lipid antigen bound in the CD1 groove.
Similar conclusions were obtained by analysis of antigen presentation by site‐specific mutants of murine CD1d, showing that activation of NK T hybridomas by αGalCer involves interaction of the TCR with both the galactose and specific residues of the α‐helices of CD1d (40). However, whereas the presence of the invariant Vα14Jα18 TCR α chain is generally required for αGalCer recognition, the observation that moderately diverse TCRβ chains containing several different Vβ regions (mainly Vβ8, Vβ7 and Vβ2) could be associated with αGalCer‐reactive NKT cells suggested that the TCRβ chains might not play a significant role in the recognition of the ligand (36). This hypothesis is supported by the finding that the CDR3 length of the Vβ chains of αGalCer‐reactive NKT cells seems to show little or no evidence for selection for particular CDR3β amino acid residues (41). The lack of stringent selection for CDR3β sequences suggests that the selection for Vβ8, Vβ7 and Vβ2 probably involves the CDR1 and CDR2 regions of these Vβ segments, most likely because of their ability to interact effectively with the α‐helical surface of CD1d rather than with the bound antigen.
In addition, T cells that show autoreactivity to CD1d but do not respond to αGalCer have been identified. These T cells have been called diverse CD1d‐restricted T cells because they do not express the invariant TCRα chain, and instead express a variety of Vα and Vβ gene segments. Although the diversity of the TCR of these T cells is not completely known yet, analysis of their repertoire has indicated preferential use of genes from the Vβ8 family, like NK T cells expressing the invariant TCRα chain (42, 43). Furthermore, it has also been suggested that their Vα repertoire is likely to be biased by the recurrent usage of a few Vα chains, such as Vα3.2 and Vα8 (43).
In summary, the data that have accumulated up to the present time emphasize that, in contrast to the apparent heterogeneity of the TCR of human group 1 CD1‐restricted T cells, the repertoire of murine CD1d‐restricted T cells is highly biased and shows much more limited diversity. This observation suggests that mouse CD1 might be involved in the recognition of a limited number of ligands with a relatively fixed structure. Possibilities for these ligands include either microbial lipids that function as pathogen associated molecular patterns, or possibly normal self or modified self lipids that are induced under conditions of stress. The recognition of such intrinsically foreign or stress‐induced ligands with a fixed structure would be consistent with the view that the CD1d system is more oriented toward innate rather than adaptive immune recognition. However, it is important to note that nearly all of the TCR repertoire analyses summarized above have focused on populations of CD1‐restricted T cells that show autoreactivity to the CD1d molecule. It remains possible that other populations of nonautoreactive CD1d‐restricted T cells may exist that use a more extensive repertoire of TCR to recognize a more diverse array of potential antigens. This possibility is strongly implied by a recent study demonstrating nonautoreactive human CD1d‐restricted T cells with highly diverse TCR that can be expanded by in vitro stimulation with αGalCer (44)
Lipid antigen loading and processing
Relatively little known is about the molecular details of antigen processing for lipid presentation, and the conditions that facilitate the loading of these ligands onto the CD1 proteins are mostly undetermined. Nonetheless, through combined analyzes of the cellular trafficking and of the requirements for presentation of various CD1‐presented antigens, some insight into these questions can be gathered for each CD1 isoform that is known to function in antigen presentation.
CD1a
Analysis of a human CD1a‐restricted T cell response to an unidentified mycobacterial lipid antigen suggests that antigen presentation requires endocytosis, but does not require endosomal acidification (9). Although the analysis of CD1a is limited to a single T cell line and a single unidentified lipid antigen at present, it appears most likely that this CD1 protein may use the early/recycling endosomes as its main loading compartment.
CD1b
Presentation of the long chain (∼C80) mycobacterial glycolipid GMM (Fig. 3) by human CD1b is completely inhibited by fixation of the antigen presenting cells (APC) with glutaraldehyde prior to antigen pulsing, or by treatment of APC with concanamycin A or chloroquine (9, 12). In AP‐3 deficient cells, the presentation of GMM is altered but not completely eliminated, suggesting that while lysosomes and MIIC compartments are probably the main sites of loading, some CD1b molecules might bind antigens earlier in the endocytic pathway (14).
Recent studies suggest that the mechanisms of antigen binding and presentation by CD1b may be more complex than originally thought and highly dependent on the chemical structure of a particular lipid antigen, specifically the length of its alkyl chains (45). Analysis of the cellular trafficking and antigenic potency of different molecular variants of the GMM antigen revealed that GMM with short alkyl chains (C32 GMM; Fig. 3) could rapidly load onto CD1b at the cell surface of dendritic cells (DC), whereas presentation of GMM with long alkyl chains (C80 GMM) by DC was a slower process that required delivery of the antigen to endosomal compartments. Subcellular fractionation revealed that DC sort the different forms of GMM antigen based on alkyl chain length, preferentially concentrating the long‐chain antigen in lysosomal compartments. Disruption of endosomal trafficking of CD1b by mutation of the targeting motif in its cytoplasmic tail increased the surface expression of CD1b as expected, and increased the presentation of C32 GMM while inhibiting the presentation of C80 GMM. Interestingly, presentation of GMM by nonprofessional APC gave opposite results with a preferential presentation of C32 GMM. These data demonstrate the coexistence of endosomal and nonendosomal pathways for antigen presentation by CD1b. It is thus suggested that specialized APC like immature DC, which are capable of phagocytosis, sort the antigens based on alkyl chain length and preferentially concentrate the long chain antigens in lysosomal compartments where they can be loaded on CD1b. In contrast, nonphagocytic CD1b+ cells such as thymocytes might preferentially present short chain self glycolipids, loaded during CD1b trafficking through the intracellular compartments to the cell surface or at the plasma membrane. In this way, they could provide self‐ligands for thymic selection of CD1‐restricted T cells.
CD1c
Antigen loading onto CD1c probably occurs in an endosomal compartment, as one study has found that the CD1c‐restricted T cell response is reduced if the APC are fixed before being pulsed with the antigen (46). However, presentation of the mycobacterial β1‐mannosyl isoprenoid glycolipid antigen by CD1c has rapid kinetics that are similar to those seen for the presentation of preprocessed peptide antigens that can bind to MHC class II on the cell surface. In addition, in marked contrast to CD1b, the presentation of glycolipid antigen by CD1c is not significantly reduced by deletion of the cytoplasmic tail endosomal targeting motif of the protein, and is also relatively resistant to the effects of inhibitors of endosomal acidification (chloroquine and concanannycin B) (12). Taken together, these results suggest that the loading of glycolipid antigens onto CD1c occurs efficiently in nonacidified or weakly acidified early endocytic compartments, and probably also to some extent at the cell surface.
CD1d
Studies using αGalCer as a model glycolipid antigen have created some controversy regarding the necessity of lipid antigen internalization for presentation by CD1d. The stimulation of NKT cells by αGalCer was initially reported to be inhibited by cell fixation before pulsing and by agents that block endosomal acidification (25, 26). However, Kronenberg and colleagues reported that immobilized recombinant murine and human CD1d molecules could be loaded at neutral pH with αGalCer for presentation to NKT cells (47). The study published by Gumperz et al. partially resolved this discrepancy by showing that while the binding and presentation of αGalCer was detectable at neutral pH, it was optimal at pH 4 with a 4‐fold increase in the NKT cell response (29). The absence of a strong or absolute requirement for acidification suggests that the loading of αGalCer could occur at various sites in the endosomal pathway, but probably more efficiently in acidified compartments.
Lipid antigen processing
Glycolipids with long alkyl chains or with large complex carbohydrate moieties may be too large to fit properly in the binding groove of CD1 molecules in a way that allows their recognition by T cell receptors. The loading of such antigens may require enzymatic cleavage of covalent bonds in the lipid antigen, or pH‐dependent modifications to the CD1 binding groove. Both of these mechanisms were originally suggested by studies of human CD1b (48). In addition, a recent study of the presentation of a disaccharide‐containing synthetic analog of αGalCer (Gal(αl→2)GalCer) revealed a clear requirement for removal of the terminal sugar by a lysosomal enzyme prior to effective presentation to CD1d‐restricted T cells (49). This provided a clear precedent for the requirement of enzymatic cleavage of the carbohydrate portion of a glycolipid antigen in a lysosomal compartment prior to its presentation to CD1‐restricted T cells. Based on these findings, it seems that enzymatic trimming of the carbohydrate portions of natural CD1‐presented antigens is likely to occur, although direct evidence for this is still lacking. It is currently not known whether enzymatic modification or cleavage of the alkyl tails of lipid antigens is possible in antigen presenting cells, or whether this may be required for presentation by CD1 in some cases.
Another mechanism by which some CD1 proteins may accommodate the binding of very long lipids may involve the complex folding of alkyl chains within the groove. This possibility has been illustrated by recent functional studies on the presentation of very long chain mycolic acids by human CD1b (39). This demonstrated that the TCR of a CD1b restricted T cell clone specific for mycolic acids interacts with two hydrophilic components of this lipid antigen, the carboxylic acid in the polar head of the molecule and a hydrophilic keto or methoxy group on the long alkyl chain (Fig. 3). This observation suggests that a folding of the alkyl chain occurs during the binding to CD1b that brings these two distant hydrophilic components into close proximity to allow them to form a combinatorial epitope. Interestingly, the recent crystal structure data on CD1b supports the possibility that the groove of this molecule could potentially hold a very long chain mycolic acid in a folded configuration that might allow such positioning of the two hydrophilic components at the TCR binding interface (34).
Functions of CD1‐restricted T cells
Group 1 CD1‐restricted T cells reactive with bacterial antigens
The first group 1 CD1‐restricted T cell lines to be described were all derived from CD4–CD8– T cells, but subsequent studies demonstrated that CD1‐restricted T cells could also be found among CD8+ and CD4+ T cells. Most human group 1 CD1‐restricted T cells have been derived against mycobacterial antigens, and these invariably produce large amounts of IFNγ and extremely low or undetectable amounts of IL‐4 when cocultured with antigen‐pulsed dendritic cells. In addition, they generally show moderate to strong cytolytic activity against CD1+ target cells, such as dendritic cells that have been pulsed with mycobacterial antigens or infected with live mycobacteria (50). These functional characteristics of group 1 CD1‐restricted T cells defined by in vitro studies emphasize their potentially important role in vivo in microbial immunity. To date, this hypothesis has been examined in mycobacterial infections only, although limited data also suggest the capacity of CD1 molecules to present antigens derived from Gram negative bacteria. Perhaps the strongest evidence for an effective role of antigen‐specific CD1‐restricted T cells in host defense against mycobacteria is the discovery that CD1c‐restricted T cells specific for mycobacterial isoprenoid glycolipids are common in individuals previously infected with M. tuberculosis, but are not seen in naive control subjects (22). Also relevant is the observation that mycobacterial production of the CD1b‐presented antigen GMM absolutely requires a nonmycobacterial source of glucose (51). Therefore, a CD1b‐mediated T cell response to GMM would appear with pathogenic mycobacteria that have invaded host tissues and not with saprophytic mycobacteria that are unable to grow in animal tissues.
This implies that GMM could function essentially as a pathogen‐associated molecular pattern that signals the invasion of host tissues by actively growing mycobacteria, leading to an immune response that can be triggered through presentation by CD1b.
The role of CD1‐restricted T cell responses has also been studied in leprosy. CD4+ CD1b‐restricted and CD1c‐restricted T cell lines were derived from skin lesions of human subjects with M. leprae infection. These T cell lines produced IFNγ and GM‐CSF in response to activation with M. leprae antigens, suggesting their potential contribution in the immune response to M. leprae (50). Furthermore, analysis of skin biopsy specimens demonstrated that lesions of tuberculoid leprosy, in which a strong cell‐mediated immune response controls the growth of M. leprae, contain approximately 10 times more CD1+ dendritic cells than were observed in lepromatous leprosy lesions, where an inefficient Th1 immune response results in poorly organized cellular infiltrates and bacterial proliferation (52).
The investigation of group 1 CD1 in host immunity against infection has been hampered by the absence of an appropriate animal model. As mentioned previously, the muroid rodents (mice and rats) lack group 1 CD1 genes, as these were probably lost by deletion during the evolution of these species (2). The identification and characterization of guinea pig group 1 CD1 proteins has recently enabled studies demonstrating that a CD1‐restricted T cell response against M. tuberculosis lipid antigens can be induced by immunization in this animal. Therefore, the guinea pig may eventually provide a suitable model in which to study group 1 CD1‐restricted immune responses to mycobacterial pathogens in vivo (53).
Group 1 CD1‐recognition by γδ T cells
Like αβ T lymphocytes, γδ T cells express a TCR generated by somatic gene rearrangements, but with limited combinatorial diversity. Expansion of circulating γδ T cells during the course of various infectious diseases in humans and in animal models suggests a role in host defense against infections. They have been shown to respond to a variety of nonpeptide microbial ligands including mycobacterial isopentenyl pyrophosphate (IPP) and other prenyl pyrophosphate derivatives (54). While there is currently no evidence to demonstrate that the recognition of such microbial ligands is dependent on CD1 presentation, it has been clearly shown that some human γδ T cells recognize CD1 molecules, in particular CD1c (1, 55). The examples of CD1c‐specific γδ T cells described to date express the Vδ1 chain, which identifies them as tissue localizing rather than circulating γδ T cells. These cells are Th1‐like and cytolytic for CD1c expressing cells in the absence of foreign antigen (56).
Group 2 CD1‐restricted T cells
CD1d‐restricted NK T cells in the adjuvant cascade
The main population of CD1d‐restricted T cells is the αGalCer‐reactive Vα14J18+ NK T cells in mice, and the homologous Vα24Jα15+ T Cells in humans. One of the first identified functional properties of NK T cells was their ability to rapidly secrete both Th1 and Th2 cytokines in vitro in response to TCR stimulation without the need for priming, suggesting that they might have an important influence on the nature of the adaptive immune response through polarization of CD4+ T cells to Th1 or Th2 effectors (27).
A breakthrough in the analysis of NK T cells occurred with the discovery of the stimulatory activity of αGalCer, which created the possibility of specifically activating NK T cells in vivo (25, 26). In mice, injection of αGalCer induces a series of cellular activation events that have been referred to as the ‘adjuvant cascade’ (27). This begins with the rapid release of IL‐4 and IFNγ by NK T cells after CD1d‐mediated presentation of αGalCer, which directly stimulates NK cells and B cells, and potentially activates and influences the differentiation of conventional CD4+ and CD8+ T cells (Fig. 5).
Figure 5.
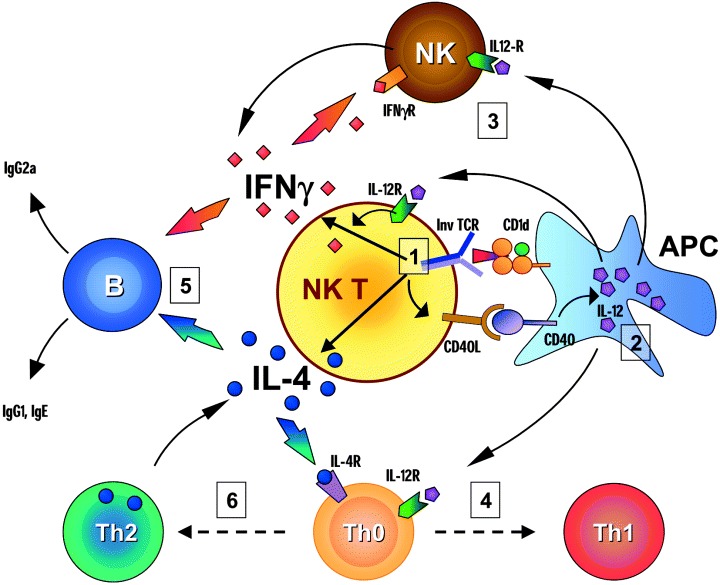
Cellular activation events induced by αGalCer. Administration of αGalCer in mice induces a complex series of cellular events referred to as an adjuvant cascade. (1) Recognition of αGalCer through the semi‐invariant TCR activates the NK T cells and induces rapid release of IL‐4 and IFNγ. The initial activation also leads to an up‐regulation of CD40L on the NK T cells. (2) The interaction of CD40L with CD40 activates the antigen‐presenting cell (APC) which produces IL‐12. (3) The release of IL‐12 amplifies the production of IFNγ by the NK T cells and acts together with IFNγ to activate NK cells. (4) IL‐12 also plays a major role in the polarization of Th0 naive T cells into Th1 helper and cytotoxic T cells. (5) IL‐4 production by NK T cells contributes to the activation of B cells. The immunoglobulin production by B cells is influenced by the balance between IL‐4 and IFNγ. While IFNγ promotes IgG2a production, IL‐4 stimulates IgG1 and lgE secretion. (6) In addition, IL‐4 can polarize Th0 T cells toward the Th2 phenotype. Under conditions that favor the preferential production of IL‐4 following initial activation of NK T cells, this effect on Th2 polarization may predominate.
After αGalCer injection, NK T cells rapidly disappear from the liver and spleen within 3–12 h, most probably as the result of activation‐induced cell death (AICD). The NK T cells begin to reappear in the liver and the spleen after two or three days but their numbers remain lower than in control mice for several weeks (57, 58). A striking observation is that the NK T cells that repopulate the spleen have a Th2 phenotype (59). Several hypotheses have been raised to explain this phenomenon. One possibility is that αGalCer could polarize existing Th0 NK T cells, or NK T precursors, towards a Th2 phenotype. Alternatively, distinct populations of mature NK T cells may produce Th1 and Th2 cytokines, and αGalCer could selectively induce cell death of the Th1 NK T cells. Yet another possibility relates to the recently demonstrated developmentally regulated switch in NK T cell cytokine production. Two groups identified a thymic precursor of NK T cells that binds αGalCer‐loaded CD1d tetramers but does not express NK1.1, and showed that these cells after undergoing proliferation give rise to CD4+ and CD4–8– NK1.1+ NK T cells in the periphery (60, 61). Interestingly, these NK1.1 negative precursors of mature NK T cells appear to emigrate from the thymus, and produce predominantly IL‐4 upon stimulation with αGalCer. After one to several days in the extrathymic tissues, they appear to acquire NK1.1 expression, and then produce IFNγ almost exclusively upon stimulation (61). Thus, systemic injection of α GalCer may induce cell death in the majority of mature extrathymic NK T cells, leading to a subsequent repopulation of the tissues with recent thymic emigrants that will produce exclusively or mainly Th2 cytokines upon restimulation. Supporting this hypothesis is the observation that the Th2 polarization of NK T cells following α GalCer injection is relatively short lived, lasting no more than a few weeks (Dutronc Y, Porcelli SA, unpublished data).
Role of CD1d‐restricted NK T cells in antimicrobial immunity
As NK T cells can be studied in mouse models, it has been possible to assess their potential relevance in vivo in a wide variety of murine models of infectious disease. The following is a brief overview organized with respect to the broad categories of different pathogens that have been examined.
Bacteria
NK T cells have been implicated in the host response to several bacterial infections, but their role seems to vary according to the site or the nature of the pathogens (50). In Borrelia burdgoferi infection, CD1d deficiency in mice normally resistant to this pathogen results in arthritis associated with increased bacterial burden and enhanced production of specific IgG against Borrelia lipoprotein antigens (62). A protective role of CD1d‐restricted T cells has also been recently identified in a murine model of Pseudomonas aeruginosa pneumonia (63). Other models however, suggest that CD1d‐restricted T cells might play a regulatory role in immune response against bacterial pathogens by the secretion of Th2 cytokines (64).
Although, analysis of CD1d–/– mice does not support a critical role for CD1d‐restricted T cells in mycobacterial infection (65), granulomatous reaction in response to subcutaneous injection of mycobacterial extracts is impaired in NK T cell deficient mice. It was recently demonstrated that NK T cell accumulation at the site of injection in this experimental model is not dependent on TCR activation, and can be induced by a variety of glycolipids (66). The exact mechanisms of this phenomenon and their pertinence in bacterial infection remain to be determined.
Viruses
The role of NK T cells in immune responses against viruses has been particularly studied in the liver, where, at least in mice, NK T cells are the major resident T cell population. Activation of NK T cells by αGalCer inhibits the replication of hepatitis B virus (HBV) in HBV‐transgenic mice. This effect is associated with the production of IFNγ and IFNα/β and with activation of NK cells but not of conventional T cells (67). In a different model of hepatitis B infection, Baron et al. showed that a distinct population of CD1d‐restricted T cells that were not reactive to αGalCer was activated in response to hepatocytes expressing HBV antigens, and that this response produced hepatitis (68). Interestingly, a Th1‐biased CD1d‐restricted T cell population expressing diverse TCR was also identified recently in the livers of humans with chronic hepatitis C virus (HCV) infection. It is not known yet if these cells play a protective role against the infection, or if they contribute to the development of tissue damage (69).
Parasites and fungi
Several reports suggest a protective effect of NK T cells in mice during the course of infection with parasites or fungi including Plasmodium yoelii, P. berghei, Trypanosoma cruzi and Cryptococcus neoformans (50). However, most of these studies have focused on the effects of αGalCer mediated activation during the course of the infection rather than the natural role of NK T cells in these infections. These effects have generally been attributed to the induction of IFNγ secretion by αGalCer. Recently, it has also been reported that administration of αGalCer to mice with a suboptimal dose of irradiated P. yoelii sporozoites or suboptimal doses of recombinant viruses expressing malarial antigens enhances protective antimalarial immunity, thus indicating an adjuvant‐like property of this NK T cell activator (70).
NK T cells and tumor immunity
The antitumor activity of αGalCer, observed in several murine models depends on the activation of NK T cells as it is abolished in CD1‐deficient mice (reviewed in (71)). However, NK T cells are not believed to directly mediate the cytotoxicity against tumor cells that is induced by α GalCer in most tumor models, which is actually mediated by the NK cells (58). The critical elements of this activity of α GalCer are the production of IFNγ by NK T cells and of IL‐12 by dendritic cells, which amplifies IFNγ production. The cytotoxic activity of NK cells that is induced by αGalCer appears to be perforin‐independent and FasL‐independent. NK T cells can also be activated to produce IFNγ directly by IL‐12, and thus participate in the anti‐metastatic activities of this cytokine (71).
A natural role for NK T cells in tumor immunity has been identified more recently. NK T cells play a protective role against methylcholanthrene (MCA)‐induced sarcomas by the production of IFNγ, and the resulting antisarcoma responses involve NK and CD8+ T cells and are perforin‐dependent (72). Two different studies suggest that NK T could also have a suppressive role on tumor immunity. Terabe et al. observed, in a model where the tumor spontaneously regresses after initial growth and then recurs, that tumor recurrence was due to the incomplete elimination of tumor cells by CD8+ cytotoxic T lymphocytes (CTL), and that this is linked to a suppressive effect of NK T cells on CTL induction, mediated by the production of IL‐13 (73). Apparently similar findings were reported by Moodycliffe et al. who showed, by adoptive transfer, that NK T cells from UV‐irradiated mice have a strong suppressive activity on immune responses to a UV‐induced regressor tumor (74).
NK T cells and autoimmune diseases
Defects in NK T cell numbers and cytokine production have been reported in several animal models of autoimmune diseases and in human subjects with systemic sclerosis, type 1 diabetes, multiple sclerosis, and rheumatoid arthritis (reviewed in (75)). It has been shown that many mouse autoimmune disease models depend on pathogenic Th1 cells, and this is believed to be the case for their human counterparts also. The observation that αGalCer administration in vivo can give rise to Th2 polarized NK T cells suggests that this compound could be used to influence the course of these diseases. This idea has been evaluated in two experimental models to date, experimental autoimmune encephalomyelitis (EAE) and type 1 diabetes of NOD mice, and has shown encouraging results in both cases. In EAE, αGalCer injection was able to suppress the development of EAE (76). No protection was observed in IL‐4 KO and IL‐10 KC mice, highlighting the critical role of Th2 cytokines in this protective effect. Yamamura and colleagues also reported that an analog of αGalCer that preferentially induces IL‐4 production by NK T is able to attenuate the severity of EAE dramatically (77).
In NOD mice, repeated injections of αGalCer delay the onset and reduce the incidence of the diabetes 78, 79, 80). The protective effect of αGalCer treatment is associated with accumulation of NK T cells in the pancreas and the pancreatic lymph nodes, and the recruitment of tolerogenic myeloid DC in the pancreatic lymph nodes (80). After the onset of insulitis, αGalCer treatment does not protect against the development of diabetes if injected alone, but coadministration with IL‐7 restores the efficacy of treatment (79).
Concluding remarks
It is now clear from studies of the CD1 family that lipids and glycolipids represent relevant targets for T cell responses. As the missing pieces of information in our understanding of CD1 and the T cells that recognize it begin to be filled in, the appreciation of the potential role of lipid antigen recognition in the immune response continues to grow. Given the broad range of antigens in this chemical class, it seems likely that adaptive T cell mediated immune responses against foreign microbial lipids could represent important targets for generating resistance against certain pathogens. This may be particularly true for highly successful pathogens like M. tuberculosis or Plasmodium falciparum, which have clearly evolved multiple strategies to evade or subvert immune recognition. The same may be true for certain tumors, in which modified or aberrantly expressed glycolipids could represent a major basis for immune recognition. These aspects of the CD1‐restricted response are thus likely to have relevance to immunotherapies of infectious and neoplastic disease through novel approaches to vaccine development.
In addition to their role in the adaptive immune response, CD1‐restricted T cell responses also appear to be intimately connected to the generation of innate immunity. This is particularly evident from the many studies of CD1d‐restricted NK T cells, a highly conserved innate‐like lymphocyte in most or possibly all mammals that has the ability to trigger a powerful adjuvant cascade. While this can heighten inflammation and microbial resistance under some circumstances, the tremendous immunoregulatory potential of these T cells in other cases demonstrates their potential utility for controlling autoimmune and chronic inflammatory diseases. Understanding how to manipulate these T cells precisely through their recognition of CD1‐presented lipid ligands is likely to provide the key to exploiting their considerable potential as agents for promoting beneficial immunity and treating disease.
Acknowledgments:
Yves Dutronc is supported by a Marc Chaptal grant from Janssen‐Cilag Laboratoires. SAP is supported by grants from the NIH (AI45889 and AI48933) and a grant from the Irene Diamond Foundation.
References
- 1. Porcelli SA, Modlin RL. The CD1 system: antigen‐presenting molecules for T cell recognition of lipids and glycolipids. Annu Rev Immunol 1999: 17: 297–329. [DOI] [PubMed] [Google Scholar]
- 2. Calabi F, Milstein C. The molecular biology of CD1. Semin Immunol 2000: 12: 503–9. [DOI] [PubMed] [Google Scholar]
- 3. Calabi F, Jarvis JM, Martin L, Milstein C. Two classes of CD1 genes. Eur J Immunol 1989: 19: 285–92. [DOI] [PubMed] [Google Scholar]
- 4. Angenieux C, Salamero J, Fricker D et al. Characterization of CD1e, a third type of CD1 molecule expressed in dendritic cells. J Biol Chem 2000: 275: 37757–64. [DOI] [PubMed] [Google Scholar]
- 5. Brossay L, Jullien D, Cardell S et al. Mouse CD1 is mainly expressed on hemopoietic‐derived cells. J Immunol 1997: 159: 1216–24. [PubMed] [Google Scholar]
- 6. Exley M, Garcia J, Wilson SB et al. CD1d structure and regulation on human thymocytes, peripheral blood T cells, B cells and monocytes. Immunology 2000: 100: 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blumberg RS, Terhorst C, Bleicher P et al. Expression of a nonpolymorphic MHC class I‐like molecule, CD1D, by human intestinal epithelial cells. J Immunol 1991: 147: 2518–24. [PubMed] [Google Scholar]
- 8. Bonish B, Jullien D, Dutronc Y et al. Overexpression of CD1d by keratinocytes in psoriasis and CD1d‐dependent IFN‐gamma production by NK‐T cells. J Immunol 2000: 165: 4076–85. [DOI] [PubMed] [Google Scholar]
- 9. Sugita M, Grant EP, Van Donselaar E et al. Separate pathways for antigen presentation by CD1 molecules. Immunity 1999: 11: 743–52. [DOI] [PubMed] [Google Scholar]
- 10. Salamero J, Bausinger H, Mommaas AM et al. CD1a molecules traffic through the early recycling endosomal pathway in human Langerhans cells. J Invest Dermatol 2001: 116: 401–8. [DOI] [PubMed] [Google Scholar]
- 11. Sugita M, Jackman RM, Van Donselaar E et al. Cytoplasmic tail‐dependent localization of CD1b antigen‐presenting molecules to MIICs. Science 1996: 273: 349–52. [DOI] [PubMed] [Google Scholar]
- 12. Briken V, Jackman RM, Watts GFM, Rogers RA, Porcelli SA. Human CD1b and CD1c isoforms survey different intracellular compartments for the presentation of microbial lipid antigens. J Exp Med 2000: 192: 281–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Briken V, Jackman RM, Dasgupta S, Hoening IE, Porcelli SA. Intracellular trafficking pathway of newly synthesized CD1b molecules. EMBO J 2002: 21: 825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sugita M, Cao X, Watts GF, Rogers RA, Bonifacino JS, Brenner MB. Failure of trafficking and antigen presentation by CD1 in AP‐3‐deficient cells. Immunity 2002: 16: 697–706. [DOI] [PubMed] [Google Scholar]
- 15. Hiromatsu K, Dascher CC, Sugita M et al. Characterization of guinea‐pig group 1 CD1 proteins. Immunology 2002: 106: 159–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brossay L, Tangri S, Bix M, Cardell S, Locksley R, Kronenberg M. Mouse CD1‐autoreactive T cells have diverse patterns of reactivity to CD1+ targets. J Immunol 1998: 160: 3681–8. [PubMed] [Google Scholar]
- 17. Jayawardena‐Wolf J, Benlagha K, Chiu YH, Mahr R, Bendelac A. CD1d endosomal trafficking is independently regulated by an intrinsic CD1d‐encoded tyrosine motif and by the invariant chain. Immunity 2001: 15: 897–908. [DOI] [PubMed] [Google Scholar]
- 18. Kang SJ, Cresswell P. Regulation of intracellular trafficking of human CD1d by association with MHC class II molecules. EMBO J 2002: 21: 1650–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Riese RJ, Shi GP, Villadangos J et al. Regulation of CD1 function and NK1.1+ T cell selection and maturation by cathepsin S. Immunity 2001: 15: 909–19. [DOI] [PubMed] [Google Scholar]
- 20. Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1‐restricted αβ+ T cells. Nature 1994: 372: 691–4. [DOI] [PubMed] [Google Scholar]
- 21. Moody DB, Besra GS. Glycolipid targets of CD1‐mediated T‐cell responses. Immunology 2001: 104: 243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moody DB, Uirichs T, Muhlecker W et al. CD1c‐mediated T‐cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature 2000: 404: 884–8. [DOI] [PubMed] [Google Scholar]
- 23. Shamshiev A, Donda A, Carena I, Mori L, Kappos L, De Libero G. Self glycolipids as T‐cell autoantigens. Eur J Immunol 1999: 29: 1667–75. [DOI] [PubMed] [Google Scholar]
- 24. Shamshiev A, Gober HJ, Donda A, Mazorra Z, Mori L, De Libero G. Presentation of the same glycolipid by different CD1 molecules. J Exp Med 2002: 195: 1013–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kawano T, Cui J, Koezuka Y et al. CD1d‐restricted and TCR‐mediated activation of Vα14 NKT cells by glycosylceramides. Science 1997: 278: 1626–9. [DOI] [PubMed] [Google Scholar]
- 26. Spada FM, Koezuka Y, Porcelli SA. CD1d‐restricted recognition of synthetic glycolipid antigens by human natural killer T cells. J Exp Med 1998: 188: 1529–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benlagha K, Bendelac A. CD1d‐restricted mouse Vα14 and human Vα24 T cells: lymphocytes of innate immunity. Semin Immunol 2000: 12: 537–42. [DOI] [PubMed] [Google Scholar]
- 28. Schofield L, McConville MJ, Hansen D et al. CD1d‐restricted immunoglobulin G formation to GPI‐anchored antigens mediated by NKT cells. Science 1999: 283: 225–9. [DOI] [PubMed] [Google Scholar]
- 29. Gumperz JE, Roy C, Makowska A et al. Murine CD1d‐restricted T cell recognition of cellular lipids. Immunity 2000: 12: 211–21. [DOI] [PubMed] [Google Scholar]
- 30. Joyce S, Woods AS, Yewdell JW et al. Natural ligand of mouse CD1d1: cellular glycosylphosphatidylinositol. Science 1998: 279: 1541–4. [DOI] [PubMed] [Google Scholar]
- 31. De Silva AD, Park JJ, Matsuki N et al. Lipid protein interactions: the assembly of CD1d1 with cellular phospholipids occurs in the endoplasmic reticulum. J Immunol 2002: 168: 723–33. [DOI] [PubMed] [Google Scholar]
- 32. Roberts TJ, Sriram V, Spence PM et al. Recycling CD1d1 molecules present endogenous antigens processed in an endocytic compartment to NKT Cells. J Immunol 2002: 168: 5409–14. [DOI] [PubMed] [Google Scholar]
- 33. Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: an MHC‐like fold with a large hydrophobic binding groove. Science 1997: 277: 339–45. [DOI] [PubMed] [Google Scholar]
- 34. Gadola SD, Zaccai NR, Harlos K et al. Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nat Immunol 2002: 3: 721–6. [DOI] [PubMed] [Google Scholar]
- 35. Moody D, Besra GS, Wilson IA, Porcelli SA. The molecular basis of CD1‐mediated presentation of lipid antigens. Immunol Rev 1999: 172: 285–96. [DOI] [PubMed] [Google Scholar]
- 36. Brossay L, Naidenko O, Burdin N, Matsuda J, Sakai T, Kronenberg M. Structural requirements for galactosylceramide recognition by CD1‐restricted NK T cells. J Immunol 1998: 161: 5124–8. [PubMed] [Google Scholar]
- 37. Grant EP, Degano M, Rosat JP et al. Molecular recognition of lipid antigens by T cell receptors. J Exp Med 1999: 189: 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Melian A, Watts GF, Shamshiev A et al. Molecular recognition of human CD1b antigen complexes: evidence for a common pattern of interaction with αβ TCRs. J Immunol 2000: 165: 4494–504. [DOI] [PubMed] [Google Scholar]
- 39. Grant EP, Beckman EM, Behar SM et al. Fine specificity of TCR complementarity‐determining region residues and lipid antigen hydrophilic moieties in the recognition of a CD1‐lipid complex. J Immunol 2002: 168: 3933–40. [DOI] [PubMed] [Google Scholar]
- 40. Burdin N, Brossay L, Degano M et al. Structural requirements for antigen presentation by mouse CD1. Proc Natl Acad Sci U S A 2000: 97: 10156–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Matsuda JL, Gapin L, Fazilleau N, Warren K, Naidenko OV, Kronenberg M. Natural killer T cells reactive to a single glycolipid exhibit a highly diverse T cell receptor repertoire and small clone size. Proc Natl Acad Sci U S A 2001: 98: 12636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chiu YH, Jayawardena J, Weiss A et al. Distinct subsets of CD1d‐restricted T cells recognize self‐antigens loaded in different cellular compartments. J Exp Med 1999: 189: 103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Park SH, Weiss A, Benlagha K, Kyin T, Teyton L, Bendelac A. The mouse CD1d‐restricted repertoire is dominated by a few autoreactive T cell receptor families. J Exp Med 2001: 193: 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gadola SD, Dulphy N, Salio M, Cerundolo V. Vα24–JαQ–independent, CD1d‐restricted recognition of α–galactosylceramide by human CD4+ and CD8αβ+ T lymphocytes. J Immunol 2002: 168: 5514–20. [DOI] [PubMed] [Google Scholar]
- 45. Moody DB, Briken V, Cheng TY et al. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat Immunol 2002: 3: 435–42. [DOI] [PubMed] [Google Scholar]
- 46. Sugita M, Van Der WN, Rogers RA, Peters PJ, Brenner MB. CD1c molecules broadly survey the endocytic system. Proc Natl Acad Sci U S A 2000: 97: 8445–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Naidenko OV, Maher JK, Ernst WA, Sakai T, Modlin RL, Kronenberg M. Binding and antigen presentation of ceramide‐containing glycolipids by soluble mouse and human CD1d molecules. J Exp Med 1999: 190: 1069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Emst WA, Maher J, Cho S et al. Molecular interaction of CD1b with lipoglycan antigens. Immunity 1998: 8: 331–40. [DOI] [PubMed] [Google Scholar]
- 49. Prigozy TI, Naidenko 0, Qasba P et al. Glycolipid antigen processing for presentation by CD1d molecules. Science 2001: 291: 664–7. [DOI] [PubMed] [Google Scholar]
- 50. Gumperz JE, Brenner MB. CD1‐specific T cells in microbial immunity. Curr Opin Immunol 2001: 13: 471–8. [DOI] [PubMed] [Google Scholar]
- 51. Moody D, Guy MR, Grant E et al. CD1b‐mediated T cell recognition of a glycolipid antigen generated from mycobacterial lipid and host carbohydrate during infection. J Exp Med 2000: 192: 965–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sieling PA, Jullien D, Dahlem M et al. CD1 expression by dendritic cells in human leprosy lesions: correlation with effective host immunity. J Immunol 1999: 162: 1851–8. [PubMed] [Google Scholar]
- 53. Hiromatsu K, Dascher CC, LeClair KP et al. Induction of CD1‐restricted immune responses in guinea pigs by immunization with mycobacterial lipid antigens. J Immunol 2002: 169: 330–9. [DOI] [PubMed] [Google Scholar]
- 54. Tanaka Y, Morita CT, Tanaka Y, Nieves E, Benner MB, Bloom BR. Natural and synthetic non‐peptide antigens recognized by human γδ T cells. Nature 1995: 375: 155–8. [DOI] [PubMed] [Google Scholar]
- 55. Faure F, Jitsukawa S, Miossec C, Hercend T. CD1c as a target recognition structure for human T lymphocytes: analysis with peripheral blood γ/δ cells. Eur J Immunol 1990: 20: 703–6. [DOI] [PubMed] [Google Scholar]
- 56. Spada FM, Grant EP, Peters PJ et al. Self‐recognition of CD1 by γ/δ T cells: implications for innate immunity. J Exp Med 2000: 191: 937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Matsuda JL, Naidenko OV, Gapin L et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med 2000: 192: 741–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nakagawa R, Nagafune I, Tazunoki Y et al. Mechanisms of the antimetastatic effect in the liver and of the hepatocyte injury induced by α‐galactosylceramide in mice. J Immunol 2001: 166: 6578–84. [DOI] [PubMed] [Google Scholar]
- 59. Burdin N, Brossay IL, Kronenberg M. Immunization with α‐galactosylceramide polarizes CD1‐reactive NK T cells towards Th2 cytokine synthesis. Eur J Immunol 1999: 29: 2014–25. [DOI] [PubMed] [Google Scholar]
- 60. Pellicci DG, Hammond KJ, Uldrich AP, Baxter AG, Smyth MJ, Godfrey DL. A natural killer T (NKT) cell developmental pathway involving a thymus‐dependent NK1.1– CD4+ CD1d‐dependent precursor stage. J Exp Med 2002: 195: 835–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Benlagha K, Kyin T, Beavis A, Teyton IL, Bendelac A. A thymic precursor to the NK T cell lineage. Science 2002: 296: 553–5. [DOI] [PubMed] [Google Scholar]
- 62. Kumar H, Belperron A, Barthold SW, Bockenstedt LK. Cutting edge: CD1d deficiency impairs murine host defense against the spirochete, Borrelia burgdorferi . J Immunol 2000: 165: 4797–801. [DOI] [PubMed] [Google Scholar]
- 63. Nieuwenhuis EE, Matsumoto T, Exley M et al. CD1d‐dependent macrophage mediated clearance of Pseudomonas aeruginosa from lung. Nat Med 2002: 8: 588–93. [DOI] [PubMed] [Google Scholar]
- 64. Naiki Y, Nishimura H, Kawano T et al. Regulatory role of peritoneal NK1.1+αβ T cells in IL‐12 production during Salmonella infection. J Immunol 1999: 163: 2057–63. [PubMed] [Google Scholar]
- 65. Behar SM, Dascher CC, Grusby MJ, Wang CR, Brenner MB. Susceptibility of mice deficient in CD1D or TAP1 to infection with Mycobacterium tuberculosis . J Exp Med 1999: 189: 1973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mempel M, Ronet C, Suarez F et al. Natural killer T cells restricted by the monomorphic MHC class 1b, CD1d1 molecules behave like inflammatory cells. J Immunol 2002: 168: 365–71. [DOI] [PubMed] [Google Scholar]
- 67. Kakimi K, Guidotti LG, Koezulka Y, Chisari FV. Natural killer T cell activation inhibits hepatitis B virus replication in vivo. J Exp Med 2000: 192: 921–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Baron JL, Gardiner L, Nishimura S, Shinkai K, Locksley R, Ganem D. Activation of a nonclassical NKT cell subset in a transgenic mouse model of hepatitis B virus infection. Immunity 2002: 16: 583–94. [DOI] [PubMed] [Google Scholar]
- 69. Exley MA, He Q, Cheng O et al. Cutting edge: compartmentalization of Th1‐like noninvariant CD1d‐reactive T cells in hepatitis C virus‐infected liver. J Immunol 2002: 168: 1519–23. [DOI] [PubMed] [Google Scholar]
- 70. Gonzalez‐Aseguinolaza G, Van Kaer L, Bergmann CC et al. Natural killer T cell ligand α‐galactosylceramide enhances protective immunity induced by malaria vaccines. J Exp Med 2002: 195: 617–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Smyth MJ, Crowe NY, Hayakawa Y, Takeda K, Yagita H. Godfrey DI. NKT cells‐conductors of tumor immunity? Curr Opin Immunol 2002: 14: 165–71. [DOI] [PubMed] [Google Scholar]
- 72. Crowe NY, Smyth MJ, Godfrey DI. A critical role for natural killer T Cells in immunosurveillance of methylcholanthrene‐induced sarcomas. J Exp Med 2002: 196: 119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Terabe M, Matsui S, Noben‐Trauth N et al. NKT cell‐mediated repression of tumor immunosurveillance by IL‐13 and the IL‐4R‐STAT6 pathway. Nat Immunol 2000: 1: 515–20. [DOI] [PubMed] [Google Scholar]
- 74. Moodycliffe AM, Nghiern D, Clydesdale G, Ullrich SE. Immune suppression and skin cancer development: regulation by NKT cells. Nat Immunol 2000: 1: 521–5. [DOI] [PubMed] [Google Scholar]
- 75. Hammond KJ, Godfrey DL. NKT cells: potential targets for autoimmune disease therapy? Tissue Antigens 2002: 59: 353–63. [DOI] [PubMed] [Google Scholar]
- 76. Singh AK, Wilson MT, Hong S et al. Natural killer T cell activation protects mice against experimental autoimmune encephalomyelitis. J Exp Med 2001: 194: 1801–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Miyamoto K, Miyake S, Yamamura T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing TH2 bias of natural killer T cells. Nature 2001: 413: 531–4. [DOI] [PubMed] [Google Scholar]
- 78. Hong S, Wilson MT, Serizawa I et al. The natural killer T‐cell ligand α‐galactosylceramide prevents autoimmune diabetes in non‐obese diabetic mice. Nat Med 2001: 7: 1052–6. [DOI] [PubMed] [Google Scholar]
- 79. Sharif S, Arreaza GA, Zucker P et al. Activation of natural killer T cells α‐galactosylceramide treatment prevents the onset and recurrence of autoimmune Type 1 diabetes. Nat Med 2001: 7: 1057–62. [DOI] [PubMed] [Google Scholar]
- 80. Naumov YN, Bahjat KS, Gausling R et al. Activation of CD1d‐restricted T cells protects NOD mice from developing diabetes by regulating dendritic cell subsets. Proc Natl Acad Sci U S A 2001: 98: 13838–43. [DOI] [PMC free article] [PubMed] [Google Scholar]


