Abstract
Recently, a novel coronavirus (SARS-CoV-2; coronavirus disease 2019, COVID-19) has emerged, rapidly spreading and severely straining the capacity of the global health community. Many nations are employing combinations of containment and mitigation strategies, where early diagnosis of COVID-19 is vital in controlling illness progression and limiting viral spread within the population. Thus, rapid and accurate methods of early detection are vital to contain COVID-19 and prevent further spread and predicted subsequent infectious waves of viral recurrence in future. Immediately after its initial characterization, Chinese and American Centers for Disease Control and Prevention (CDCs) rapidly employed molecular assays for detection of COVID-19, mostly employing real-time polymerase chain reaction (RT-PCR) methods. However, such methods require specific expensive items of equipment and highly trained analysts, requiring upwards of 4–8 h to process. These requirements coupled with associated financial pressures may prevent effective deployment of such diagnostic tests. Loop mediated isothermal amplification (LAMP) is method of nucleic acid amplification which exhibits increased sensitivity and specificity are significantly rapid, and do not require expensive reagents or instruments, which aids in cost reduction for coronavirus detection. Studies have shown the successful application of LAMP assays in various forms to detect coronavirus RNA in patient samples, demonstrating that 1–10 copies of viral RNA template per reaction are sufficient for successful detection, ~100-fold more sensitive than conventional RT-PCR methods. Importantly, studies have also now demonstrated the effectiveness of LAMP methodology in the detection of SARS-CoV-2 RNA at significantly low levels, particularly following numerous improvements to LAMP assay protocols. We hypothesise that recent advancements in enhanced LAMP protocols assay perhaps represent the best chance for a rapid and robust assay for field diagnosis of COVID-19, without the requirement of specialized equipment and highly trained professionals to interpret results. Herein, we present our arguments with a view to disseminate such findings, to assist the combat of this virus that is proving so devastating. We hope that this strategy could be applied rapidly, and confirmed for viability with clinical samples, before being rolled out for mass-diagnostic testing in these current times.
Keywords: COVID-19, Polymerase chain reaction (PCR), Loop mediated isothermal amplification (LAMP), Diagnostic, Coronavirus
Introduction
Viral disease emergence represents a serious threat to global public health. Indeed, several viral epidemics over the past few decades have emerged with increasing frequency, including the severe acute respiratory syndrome coronavirus (SARS-CoV), H1N1 influenza, and more recently the Middle East respiratory syndrome coronavirus (MERS-CoV) [1]. Recently, a novel coronavirus (SARS-CoV-2; coronavirus disease 2019, COVID-19) emerged from its epicentre in Wuhan province in China, and its subsequent rapid spread has pushed the capacity of the global public health community to its sheer limits. As of 25 April 2020, there were over 2.8 million confirmed cases of COVID-19 and ~200,000 deaths recorded worldwide (https://www.worldometers.info/coronavirus/).
Many nations have employed combinations of containment and mitigation activities, intended to delay major surges of patients and levelling demand for care, while protecting the most vulnerable from infection [2]. However, early diagnosis of COVID-19 is vital in controlling illness progression and limiting viral spread within the population, employed as part of such mitigation strategies to stop onward transmission and decrease load on healthcare systems [3], [4].
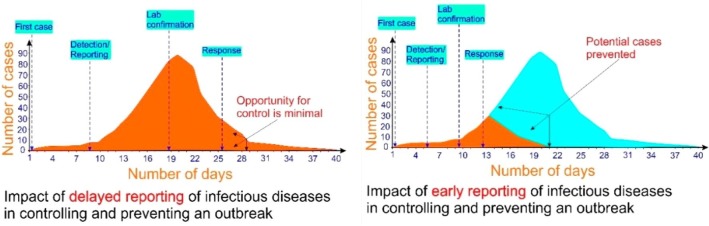
Thus, rapid and accurate methods of early detection are vital to contain COVID-19, and prevent further spread and predicted subsequent infectious waves of viral recurrence in future (Fig. 1 ). Although COVID-19 is generally thought to be milder compared to SARS and MERS-CoV, the full extent of the impact from COVID-19 remains pending. However, this outbreak represents an opportunity for multiple governmental and health sector stakeholders to partner and develop rapid diagnostic tests for infectious agents of global health concern [5].
Fig. 1.
The impact of rapid detection of infectious diseases in controlling and preventing an outbreak. Figure adapted from Nguyen et al 2020 [36].
Hypothesis: Loop mediated isothermal amplification (LAMP) assays are a robust and rapid diagnostic method to detect viral RNA
To enable effective identification and isolation strategies, a rapid and robust diagnostic test is essential, conductible in the field and at local point-of-care (POC) centres, without the requirement of specialized equipment and highly trained professionals to interpret results. Herein, we propose from a collection of recently published articles a potential protocol based on loop mediated isothermal amplification (LAMP).
Following its initial characterization, Chinese and American Centers for Disease Control and Prevention (CDCs) rapidly employed molecular assays for detection of COVID-19 in clinical samples [5], [6], [7]. These, and methods by other groups, mostly employed development of real-time polymerase chain reaction (RT-PCR) methods to diagnose COVID-19, mainly targeting various combinations of the open reading frame (ORF), envelope (E), nucleocapsid (N), and RNA-dependent RNA polymerase (RdRp) genes [5], [6], [7], [8], [9], [10]. Indeed, improved methods of quantitative RT-PCR characterized by rapid detection, high sensitivity and specificity, and are often prescribed as a gold standard for virus detection [3]. However, further novel PCR-based methods also present enhanced specificity and assay sensitivity.
PCR based methods of viral detection
PCR produces numerous copies (amplification) of a gene or series of genetic sequences by using a primer sequence and DNA polymerase enzymes to exponentially increase the amount of DNA required. PCR is widely used to amplify minute quantities of DNA to enable adequate requisite amounts for laboratory analysis of diagnosis. Owing to its simplicity, high sensitivity, and high sequence specificity, PCR-based methods are routinely and reliably capable of detecting coronavirus infection in patients [3], [11], [12]. In principle, such assays are employed following conversion of coronavirus RNA into complementary DNA by reverse transcription, following which PCR is performed and resultant amplification of DNA subject to specific detection or analytical methods such as electrophoresis or sequencing [3], [13], [14].
RT-PCR is significantly more sensitive than conventional methods [15], [16], and is routinely employed as the predominant method to detection most coronaviruses [17], [18], including COVID-19 [19]. A concern, however, particularly in the current demanding times, is that such analysis requires various specialist and expensive items of equipment, alongside highly trained analysts. Furthermore, current PCR-based methods of analysis require (realistically speaking, particularly when dealing with increasing volumes of potential infected patients) upwards of 4–8 h to process. These requirements, with the addition of financial pressures dictating that samples to be tested be sent potentially hundreds of miles away where requisite facilities and resources are present to perform such diagnostic analysis, may result in further inflation of the required time and finances to perform such diagnostic tests.
Evaluation of the Hypothesis: LAMP-based methods of viral detection
Loop mediated isothermal amplification (LAMP) is a novel isothermal nucleic acid amplification method. LAMP exhibits increased sensitivity and specificity due to an exponential amplification feature that utilises 6 different target sequences simultaneously identified by separate distinct primers in the same reaction [20]. LAMP assays are significantly rapid, and do not require expensive reagents or instruments, which aids in cost reduction for coronavirus detection [3].
Current improved LAMP assays employ a total of six primers, recognising eight distinct sites of the target sequence. A strand-displacing DNA polymerase is employed to initiate synthesis, while two primers form loop structures to facilitate and accelerate subsequent rounds of amplification. Specifically, LAMP employs two inner primers (FIP and BIP, which in turn consist of two parts each) and two outer primers (F3 and B3 which can recognize a total of six distinct regions within the target DNA). Two loop primers are employed (Forward loop primer; LF, and backward loop primer; LB) to accelerate amplification and detection efficiency [21], [22], [23].
First, FIP anneals to the template, and extension occurs using a polymerase with displacement activity (such as Bst polymerase), which will displace the product obtained from FIP by the extension reaction associated with the F3 primer. Subsequently, an extension reaction occurs using BIP on the product of FIP, and not on the template DNA due to displacement by DNA synthesis associated with the B3 primer. These reactions result in a product with a dumbbell-like structure which is essential for LAMP to establish isothermal amplification as the loop structures are always single stranded and can be annealed by FIP or BIP. This loop structure formation eliminates the denaturing step, which is otherwise essential in PCR for obtaining single-stranded DNA, and also establishes a cyclic reaction between the dumbbell-like structure and its complementary product, leading to elongated products with various copies of the target sequence produced (for reviews and detailed schematics, refer to [22], [23]).
Numerous studies have now shown the successful application of LAMP assays in various forms to detect coronavirus RNA in patients samples [24], [25], [26], [27], demonstrating that 1–10 copies of viral RNA template per reaction was sufficient for successful detection, which were ~ 100-fold more sensitive than conventional RT-PCR methods [26], [27], [28], [29], [30], [31]. Of course, however, as with any emerging technology, there are some disadvantages associated with LAMP assays. Such methodology prevents inclusion of an internal PCR inhibition control, necessitating duplication of reactions while testing. Another disadvantage is the perceived complexity of the methodology, requiring a complex primer design system which can constrain target site selection and resolution or specificity. Furthermore, as the final product is a large fragment, downstream applications such as cloning are limited.
However, despite such drawbacks, LAMP is an ultrasensitive nucleic acid amplification method that can detect minute quantities of DNA or RNA templates within roughly an hour, far outstripping normally utilised RT-PCR methods, particularly with the current demands for rapid and sensitive testing. As the growing number of suspected COVID-19 cases exceeds the capacity of many hospitals, many patients remain untested impeding efforts to the control the disease. A rapid, point-of–care diagnostic for the COVID-19 is urgently needed, which we propose to be the LAMP method of detection (Table 1 ).
Table 1.
Comparison between PCR and LAMP based methods of viral RNA detection. Table adapted from Nguyen et al (2020).
| PCR | LAMP |
|---|---|
| Bulky and cumbersome | Smaller, simpler, portable. |
| Specialised thermal cyclers required | Only a heat block is required |
| 4–8 h until result | 1 h until result |
| Requires skilled technicians | Requires no specific skill |
| Requires an additional reverse transcription step | Can be performed directly on RNA |
| Unstable reactions prone to inhibitors requiring purification steps | Stable and inhibitors tolerated, and thus purification steps not required |
| Detects DNA | Detects DNA and RNA |
| Tested on patient samples | Less tested on patient samples |
Consequences of the hypothesis and Discussion: LAMP-based assays for rapid detection of COVID-19
El-Tholoth et al., [32] recently described design of a two stage LAMP (COVID-19 Penn-RAMP) strategy, which could be carried out in closed tubes with either fluorescence or colorimetric detection. Performance of such assays were not only comparable with conventional RT-PCR assays, but also exhibited ~ 10-fold higher sensitivity when testing purified targets. Similarly, Lamb et al., [33] also described successful and rapid detection of COVID-19 RNA within 30 min of experimentation. However, while significant advances, these assays and methods have not yet been applied to confirmed patient samples, with both these studies relying upon ‘simulated’ patient samples where swabs and blood samples were artificially ‘spiked’ with COVID-19 RNA.
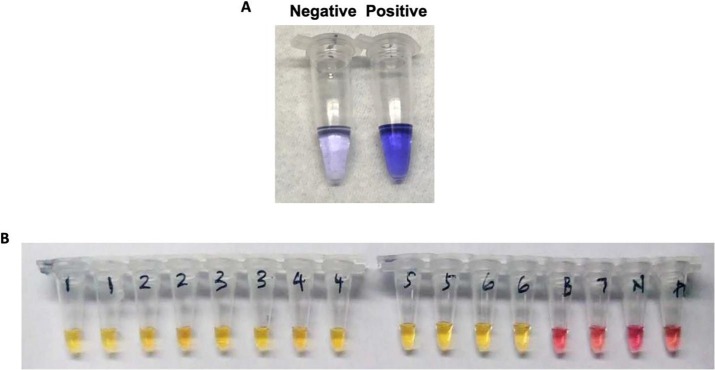
Importantly, however, Zhang et al., [34] have described the successful application of LAMP methodology to identify COVID-19 viral RNA from purified RNA or patient cell lysis using a visual, colorimetric detection. These results were further verified using RNA samples purified from respiratory swabs collected from COVID-19 patients in Wuhan, China with performance comparable to commercial RT-PCR tests, requiring only heating and visual inspection. Intriguingly, results obtained were also comparable between ‘spiked’ RNA samples and patient samples, suggesting that the improved methodology of El-Tholoth et al., [32] should yield satisfactory results with patient samples (Fig. 2 ).
Fig. 2.
Demonstrating the outcome of the LAMP assay using a colorimetric change to detect presence of COVID-19 viral DNA in A) simulated patient samples employed by El-Tholoth et al., 2020 (darker colour represents a positive assay, while lighter colour represents negative assay result), and B) actual patient samples (n = 7) from Wuhan province in China, analysed by Zhang et al., 2020 (yellow colour represents positive assays, while pink tubes represent negative assay results. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
An enhanced protocol for COVID-19 diagnosis
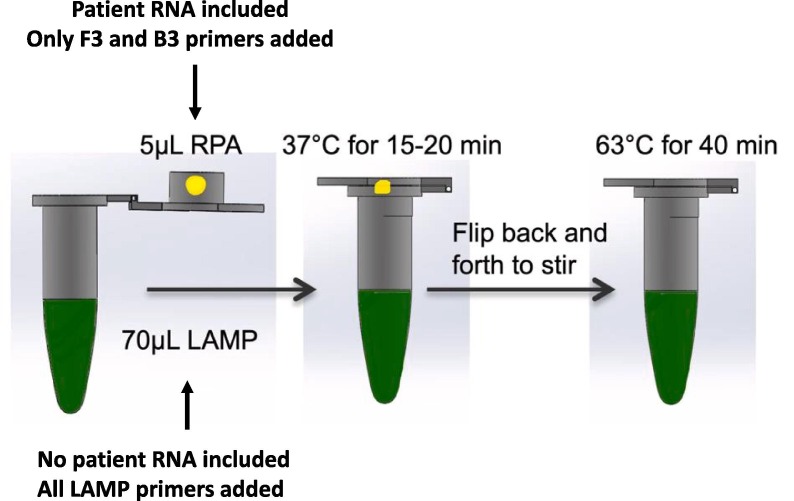
Considering the success obtained by Zhang et al., [34] using relatively less-sensitive LAMP, the enhanced sensitivity of the Penn-RAMP strategy employed by El-Tholoth et al., [32] attributable to a modified two-step LAMP protocol could prove significantly successful as a diagnostic. Penn-RAMP involves a preliminary reaction with outer LAMP primers to amplify all targets through recombinase polymerase amplification (RPA), in which all targets are amplified concurrently. After this, a second highly specific LAMP reaction is initiated. Specifically, the first stage uses outer LAMP primers F3 and B3, while the second stage further combines the other 4 RAMP primers. This ‘nested’ principle significantly enhanced LAMP sensitivity by ~ 10–100-fold compared to normal LAMP, particularly when operating with purified and crude samples [35]. Indeed, the Penn-RAMP methodology when applied on mock trials by El-Tholoth et al., [32] provided a success rate of 100% at 7–10 copies of viral RNA per reaction, compared to a 100% success rate at 700 viral RNA copies required for PCR methods [32], [35]. To this degree, we hypothesise that the modified LAMP assay proposed by El-Tholoth et al., [32] perhaps represent the best chance for a rapid and robust assay for field diagnosis of COVID-19 (Fig. 3 ), using the primer sequence strategies utilised (Table 2 )
Fig. 3.
Schematic representation of the experimental procedure of the Penn-RAMP procedure in the same tube. While the reactions could be performed in separate tubes and combined later, this envisaged procedure ensures rapid and simple flow-through and prevents potential for contamination. Figure adapted from El-Tholoth et al., (2020).
Table 2.
Sequences of COVID-19 Penn-RAMP primers used by El-Tholoth et al., (2020).
| Primer | Sequence (5′ – 3′) |
|---|---|
| F3 | TGCTTCAGTCAGCTGATG |
| B3 | TTAAATTGTCATCTTCGTCCTT |
| FIP | TCAGTACTAGTGCCTGTGCCCACAATCGTTTTTAAACGGGT |
| BIP | TCGTATACAGGGCTTTTGACATCTATCTTGGAAGCGACAACAA |
| Loop F | CTGCACTTACACCGCAA |
| Loop B | GTAGCTGGTTTTGCTAAATTCC |
We hypothesise that this assay is will be a rapid, cost-effective, and simple method that could be applied within the field at short-notice and utilised by users with even limited training. All the equipment that would be required would be a hot-block or heater capable of differential heating. Reagent-wise, the costs would be similar to that of real time RT-PCR, but the real advantage of this would be the rapidity of this assay, yielding results within an hour of testing, compared to 4–8 h taken with RT-PCR methods. The aim is not necessarily a quantitative measure of infection, but rather a simple positive/negative assay for rapid detection/confirmation. We consider that this strategy should be applied rapidly, and confirmed for viability with clinical samples, before being rolled out for mass-diagnostic testing in these current times.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The manuscript was conceived and written by both JK and AY, and submitted following approval of both authors.
References
- 1.Cascella et al., StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2020 Jan. Available from: https://www.ncbi.nlm.nih.gov/books/NBK554776/.
- 2.Bedford J., Enria D., Giesecke J., Heymann D.L., Ihekweazu C., Kobinger G., Lane H.C., Memish Z., Oh M.D., Sall A.A., Schuchat A., Ungchusak K., Wieler L.H. WHO strategic and technical advisory group for infectious hazards. COVID-19: towards controlling of a pandemic. Lancet. 2020;395(10229):1015–1018. doi: 10.1016/S0140-6736(20)30673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen M, Zhou Y, Ye J, Abdullah Al-Maskri AA, Kang Y Zeng S, Cai S. Recent advances and perspectives of nucleic acid detection for coronavirus. Journal of pharmaceutical analytics. [Epub ahead of print; DOI: 10.1016/j.jpha.2020.02.010]. [DOI] [PMC free article] [PubMed]
- 4.WHO Coronavirus disease (COVID-2019) situation reports. Situation report—55. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200315-sitrep-55-covid-19.pdf?sfvrsn=33daa5cb_6. Date accessed: March 22, 2020.
- 5.Binnicker M.J. Emergence of a Novel Coronavirus Disease (COVID-19) and the Importance of Diagnostic Testing: Why Partnership between Clinical Laboratories, Public Health Agencies, and Industry Is Essential to Control the Outbreak. Clin Chem. 2020 doi: 10.1093/clinchem/hvaa071. pii: hvaa071 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J. novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2019;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 Mar;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020 Jan 31. pii: hvaa029. [DOI] [PMC free article] [PubMed]
- 11.Balboni A., Gallina L., Palladini A., Prosperi S., Battilani M. A real-time PCR assay for bat SARS-like coronavirus detection and its application to Italian greater horseshoe bat faecal sample surveys. ScientificWorldJournal. 2012;2012 doi: 10.1100/2012/989514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uhlenhaut C., Cohen J., Pavletic S., Illei G., Gea-Banacloche J.C., Abu-Asab M. Use of a novel virus detection assay to identify coronavirus HKU1 in the lungs of a hematopoietic stem cell transplant recipient with fatal pneumonia. Transpl Infect Dis. 2012 Feb;14(1):79–85. doi: 10.1111/j.1399-3062.2011.00657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adachi D., Johnson G., Draker R., Ayers M., Mazzulli T., Talbot P.J. Comprehensive detection and identification of human coronaviruses, including the SARS-associated coronavirus, with a single RT-PCR assay. J Virol Methods. 2004 Dec 1;122(1):29–36. doi: 10.1016/j.jviromet.2004.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Setianingsih T.Y., Wiyatno A., Hartono T.S., Hindawati E., Rosamarlina Dewantari AK, Myint K.S. Detection of multiple viral sequences in the respiratory tract samples of suspected Middle East respiratory syndrome coronavirus patients in Jakarta, Indonesia 2015–2016. Int J Infect Dis. 2019 Sep;86:102–107. doi: 10.1016/j.ijid.2019.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan Z, Zhang Y, He Z, Liu J, Lan K, Hu Y, Zhang C. A Melting Curve-Based Multiplex RT-qPCR Assay for Simultaneous Detection of Four Human Coronaviruses. Int J Mol Sci. 2016 Nov 23;17(11). pii: E1880. [DOI] [PMC free article] [PubMed]
- 16.Noh JY, Yoon SW, Kim DJ, Lee MS, Kim JH, Na W, Song D, Jeong DG, Kim HK. Simultaneous detection of severe acute respiratory syndrome, Middle East respiratory syndrome, and related bat coronaviruses by real-time reverse transcription PCR. Simultaneous detection of severe acute respiratory syndrArch Virol. 2017 Jun;162(6):1617-1623.ome, Middle East respiratory syndrome, and related bat coronaviruses by real-time reverse transcription PCR. [DOI] [PMC free article] [PubMed]
- 17.Corman V.M., Müller M.A., Costabel U., Timm J., Binger T., Meyer B. Eurosurveillance. 2012;17:20334. doi: 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- 18.Lu X., Whitaker B., Sakthivel S.K., Kamili S., Rose L.E., Lowe L. Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J Clin Microbiol. 2014 Jan;52(1):67–75. doi: 10.1128/JCM.02533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DGJC, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020 Jan;25(3). [DOI] [PMC free article] [PubMed]
- 20.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000 Jun 15;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002 Jun;16(3):223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 22.Gallas-Lindemann C., Sureshkumar P., Noack M.J., Sotiriadou I. Loop-Mediated Isothermal Amplification: An Advanced Method for the Detection of Giardia, Current Topics in Giardiasis, Alfonso. J. Rodriguez-Morales, IntechOpen, 2017 doi: 10.5772/intechopen.70804. [DOI] [Google Scholar]
- 23.Mori Y., Tomita N., Kanda H., Notomi T. Novel Molecular Diagnostic Platform for Tropical Infectious Diseases, Current Topics in Tropical Medicine, Alfonso. J Rodriguez-Morales, IntechOpen, 2012 doi: 10.5772/29561. [DOI] [Google Scholar]
- 24.Poon L.L., Leung C.S., Tashiro M., Chan K.H., Wong B.W., Yuen K.Y. Rapid detection of the severe acute respiratory syndrome (SARS) coronavirus by a loop-mediated isothermal amplification assay. Clin Chem. 2004 Jun;50(6):1050–1052. doi: 10.1373/clinchem.2004.032011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pyrc K., Milewska A., Potempa J. Development of loop-mediated isothermal amplification assay for detection of human coronavirus-NL63. J Virol Methods. 2011 Jul;175(1):133–136. doi: 10.1016/j.jviromet.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001 Nov 23;289(1):150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 27.Shirato K., Yano T., Senba S., Akachi S., Kobayashi T., Nishinaka T. Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP) Virol J. 2014 Aug;8(11):139. doi: 10.1186/1743-422X-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong T.C., Mai Q.L., Cuong D.V., Parida M., Minekawa H., Notomi T. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004 May;42(5):1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Njiru Z.X. Loop-mediated isothermal amplification technology: towards point of care diagnostics. PLoS Negl Trop Dis. 2012;6(6) doi: 10.1371/journal.pntd.0001572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirato K., Semba S., El-Kafrawy S.A., Hassan A.M., Tolah A.M., Takayama I. Development of fluorescent reverse transcription loop-mediated isothermal amplification (RT-LAMP) using quenching probes for the detection of the Middle East respiratory syndrome coronavirus. J Virol Methods. 2018 Aug;258:41–48. doi: 10.1016/j.jviromet.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang P., Wang H., Cao Z., Jin H., Chi H., Zhao J. A Rapid and Specific Assay for the Detection of MERS-CoV. Front Microbiol. 2018 May;29(9):1101. doi: 10.3389/fmicb.2018.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Tholoth et al., 2020. Available at: https://chemrxiv.org/articles/A_Single_and_Two-Stage_Closed-Tube_Molecular_Test_for_the_2019_Novel_Coronavirus_COVID-19_at_Home_Clinic_and_Points_of_Entry/11860137/1. Accessed March 22, 2020.
- 33.Lamb et al., 2020. medRxiv 2020.02.19.20025155; Available at: https://www.medrxiv.org/content/10.1101/2020.02.19.20025155v1. Accessed March 22, 2020.
- 34.Zhang et al., 2020. medRxiv 2020.02.26.20028373; Available at: https://www.medrxiv.org/content/10.1101/2020.02.26.20028373v1. Accessed March 22, 2020.
- 35.Song J., Liu C., Mauk M.G., Rankin S.C., Lok J.B., Greenberg R.M. Two-Stage Isothermal Enzymatic Amplification for Concurrent Multiplex Molecular Detection. Clin Chem. 2017 Mar;63(3):714–722. doi: 10.1373/clinchem.2016.263665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen T., Bang D.D., Wolff A. 2019 Novel Coronavirus Disease (COVID-19): Paving the Road for Rapid Detection and Point-of-Care Diagnostics. Micromachines. 2020;11(3):306. doi: 10.3390/mi11030306. [DOI] [PMC free article] [PubMed] [Google Scholar]