Significance
The ability to robustly control macromolecular shape on the nanometer length scale is important for a wide range of biomedical and materials applications. DNA nanotechnology has achieved considerable success in building up complex structures from a small number of types of building blocks. We describe a large library of protein building blocks and junctions between them that enable the design of proteins with a wide range of shapes through modular combination of blocks rather than traditional and more complex design at the level of amino acid residues.
Keywords: de novo protein design, biomaterials, modular protein design
Abstract
The ability to precisely design large proteins with diverse shapes would enable applications ranging from the design of protein binders that wrap around their target to the positioning of multiple functional sites in specified orientations. We describe a protein backbone design method for generating a wide range of rigid fusions between helix-containing proteins and use it to design 75,000 structurally unique junctions between monomeric and homo-oligomeric de novo designed and ankyrin repeat proteins (RPs). Of the junction designs that were experimentally characterized, 82% have circular dichroism and solution small-angle X-ray scattering profiles consistent with the design models and are stable at 95 °C. Crystal structures of four designed junctions were in close agreement with the design models with rmsds ranging from 0.9 to 1.6 Å. Electron microscopic images of extended tetrameric structures and ∼10-nm-diameter “L” and “V” shapes generated using the junctions are close to the design models, demonstrating the control the rigid junctions provide for protein shape sculpting over multiple nanometer length scales.
DNA nanotechnology has achieved considerable control over nanometer-length-scale structures using modular Watson–Crick base pairing as the fundamental design principle: the universality of base pairing makes it straightforward to build up complex structures by combining smaller modules (1). Such a modular combination of structured elements is more difficult with proteins because they can adopt a wide variety of folds that are not universally complementary. Fusing together multiple protein domains with flexible linkers is straightforward, but the rigid body orientation of domains in such constructs is not fixed, making it difficult to programmatically assemble larger structures using this approach. The design of complex structures would be considerably facilitated by general methods for rigidly fusing together preexisting modules. The SEWING method fuses proteins in helical regions and has been used to design a variety of protein shapes from modules extracted from native proteins (2); other methods have fused terminal helices (3–5). Crystal structures of designs generated using structure extension with native-fragment graphs (SEWING) and geometric assembly approaches (2–5) demonstrate that fusion of helical segments can generate quite rigid structures with well-defined geometries. Such approaches could potentially be even more powerful if applied to de novo designed proteins, which are much more modular and stable than native proteins.
Here we focus on the creation of a wide range of protein shapes using a diverse set of de novo designed protein building blocks with structural features that enable rigid fusion. Repeat proteins (RPs) are excellent building blocks for protein-based nanoscale materials as they can readily be shortened or lengthened by changing the number of repeats (6); hence each repeat protein generates a family of structures RPn, where n is the number of repeats. A rigid fusion of two different repeat proteins would provide access to the larger family of structures RP1mRP2n and fusion of three repeat proteins to the still larger family RP1mRP2nRP3l. The set of de novo designed helical repeat proteins (DHRs) is a particularly attractive starting point: DHRs are extremely stable with individual repeat units that, unlike the repeat proteins in nature, have favorable folding free energies (7) and are identical in each copy in the overall protein. Forty-four DHRs have been structurally validated: 15 by crystallography and the remainder by solution small-angle X-ray scattering (SAXS) (8). DHRs are quite versatile: they have been built into homo-oligomers (9), filaments (10), and lattices on inorganic crystals (11) and have been used as scaffolds for ligand-induced heterodimerization (12).
Here we describe a general approach for robustly joining together de novo designed repeat proteins to generate a wide range of shapes. We apply the method to rigidly combine DHRs, designed homo-oligomers, and DHR–ankyrin fusions (Fig. 1A) and demonstrate that the junctions enable the specification of protein shapes on the multiple nanometer length scale.
Fig. 1.
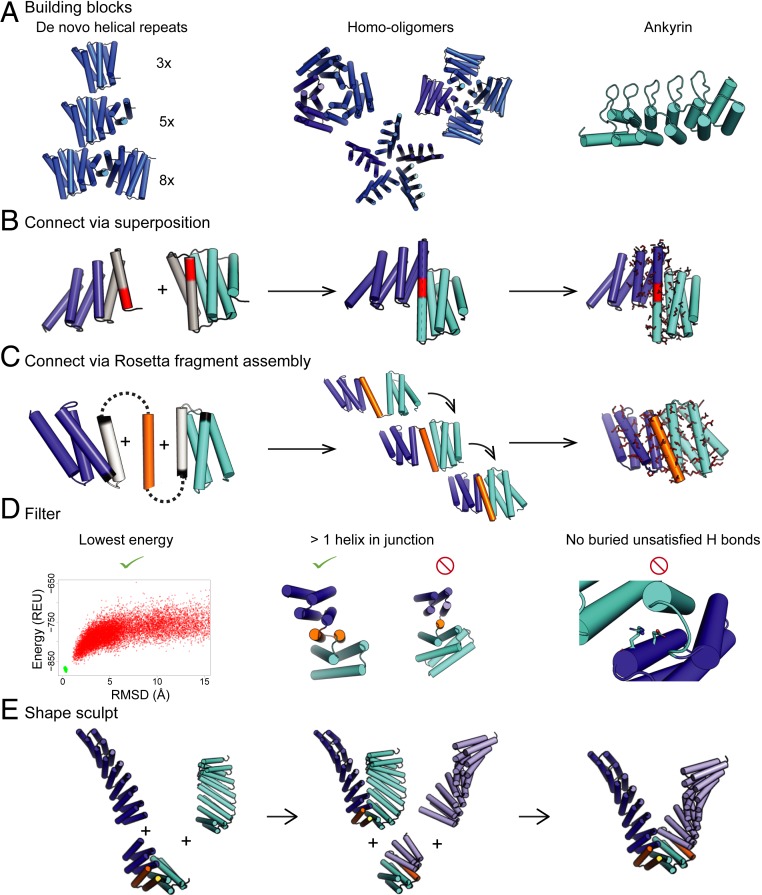
A general method to create arbitrary protein shapes using a library of designed junctions. (A) Building blocks: (Left, with different numbers of repeat units indicated in parentheses) DHRs, (Middle) homo-oligomers made from DHRs (9), and (Right) an ankyrin. (B) Junctions can be made by superimposing helices by overlapping six residues (red) in terminal repeats (gray). The nearby residues are then redesigned (red sticks). (C) Junctions can also be made by building additional protein backbone (gold) as a contiguous chain with Rosetta fragment assembly. Following removal of a helix (gray) and/or one to four terminal helix residues (black), the sequence near the interface is redesigned (red sticks). (D) Designs from both fusion methods are filtered to ensure they are lower in energy than other conformations in the energy landscape, contain two or more helices in contact throughout the junction, and there are no buried unsatisfied residues. To check that the design is the lowest energy we used either Rosetta@home to model the energy landscape or a machine-learning approximation to the Rosetta@home simulation (SI Appendix, Discussion S2). (E) The junction library is then used to sculpt proteins into various shapes. In this case, a repeat protein shown in dark blue is connected first to a repeat protein in cyan followed by a repeat protein in light blue. Junctions are shown in gold. REU, Rosetta energy units.
Results
Protein Fusion Approach.
We set out to develop methods for systematically generating large sets of rigid protein building blocks by combinatorially fusing DHRs. We explored two approaches, the first based on helical superposition and the second on Rosetta (https://www.rosettacommons.org/) fragment assembly. The helical superposition approach utilizes structure fusion through overlap of helical segments (as in ref. 2); in our approach, six-residue helical segments in a first DHR are superimposed onto a six-residue helical segment of a second DHR, and the sequences of residues adjacent to the junction are optimized using Rosetta design (Fig. 1B). We then select out the small fraction of the fusions in which the joined DHRs are in contact beyond the superimposed junction helix to reduce flexibility across the junction by requiring that at least two helices from each DHR make contact across the new interface. We also filtered out models with buried unsatisfied hydrogen bonds (13) and then used Rosetta de novo structure prediction calculations to identify sequences strongly specifying the designed structures in silico (Fig. 1D). With the helix fusion approach, we were able to generate an average of 2.7 junctions per DHR–DHR pair with sequences predicted to robustly fold into the designed shape in silico.
Rosetta Fragment Assembly Approach.
To access a larger number of junctions for a given repeat protein pair, we developed a Rosetta Monte Carlo fragment assembly approach that generates additional backbone structure to rigidly connect two DHRs. For each DHR pair, a new structural element was built to interface between the two domains, consisting of either a loop, a helix (with two loops), or two helices (with three loops). The lengths of the helices ranged from one less than the shortest of the helices in the DHRs being joined to one residue longer than the longest of the helices, and the lengths of the loops ranged from two to four residues. (The total length of the inserted structure ranged from 2 to 64 residues). For each junction, we exhaustively generated all secondary structure strings (“blueprints”) consistent with these rules and then built up backbone coordinates for each string through 3,200 Monte Carlo fragment assembly steps. Following each fragment insertion, the net rigid body transform was propagated to the downstream repeat protein domain (Fig. 1C, steps 1 and 2, and SI Appendix, Discussion S1); during this process the backbones in the flanking repeat proteins were kept rigid. Rosetta design was then used to design the amino acid sequence of the new residues and residues in the DHR that neighbor the new residues (Fig. 1C, step 3). The same filters used in the helix superposition approach were applied to eliminate implausible and flexible structures. With the fragment assembly approach we were able to design an average of 40 junctions per DHR–DHR pair and connect almost all pairs of DHRs (SI Appendix, Fig. S6).
To make the large-scale building of junction insertion regions between all pairs of repeat proteins computationally tractable, we increased the efficiency of the fragment assembly part of the second approach using several new algorithms, which resulted in designs more similar to native structures in their core sidechain packing and turn geometry. First, the centroid backbone stage was biased toward native-like hydrophobic packing arrangements using the residue-pair transform (RPX) (9) score, which favors residue–residue rigid body transforms observed between isoleucine, leucine, valine, and phenylalanine in the protein database (PDB). Incorporation of RPX motifs during low-resolution backbone sampling increases the downstream yield of well-packed designs 100-fold (SI Appendix, Fig. S3A). Second, we increased the quality of the local geometry in the junction regions, eliminating highly kinked helices and strained loops. Designs containing such structures fail the computationally expensive step of Rosetta de novo structure prediction, so it is advantageous to eliminate such local strain before structure prediction. To accomplish this, we developed techniques to filter out kinked helices and to connect secondary structure elements with unstrained loops within 0.4 rmsd of commonly occurring loops in the PDB (SI Appendix, Discussion S1, steps 4 and 5). Third, we biased sequence design with a sequence profile generated from protein fragments with a similar structure to the design (SI Appendix, Discussion S1, step 6). Together, the improvements in loop building and sequence design resulted in a 12% increase in the number of designs passing the final in silico validation by de novo structure prediction (SI Appendix, Fig. S3B). Finally, we improved the efficiency of this last evaluation step by developing a protocol that predicts the results of large numbers of de novo folding simulations (carried out on Rosetta@home) using features from a small number of de novo folding trajectories. These trajectories were biased by varying amounts toward the design model to sample both near the target structure and more broadly to allow more efficient estimation of the energy gap between the design and possible structurally divergent low-energy states. This method recapitulates the results obtained with unbiased folding trajectories with 100-fold lower computational cost (SI Appendix, Fig. S2 and Discussion S2).
Experimental Characterization.
Using the design and filtering methods described above, followed by clustering with a 1-Å-backbone rmsd threshold, we generated a set of 75,000 designs that pass the in silico filtering metrics as well or better than their component DHRs (SI Appendix, Discussion S5). Ninety-four percent of these designs were generated with the Rosetta fragment assembly approach, which explores more orientations between the DHRs and hence produces more solutions. Since the helix-fusion approach is similar to SEWING, which has been previously experimentally validated (2, 3), we focused our experimental characterization on designs made using the Rosetta fragment assembly approach.
We obtained synthetic genes encoding a diverse set of 34 designs, expressed the proteins in Escherichia coli, and purified them by nickel nitrilotriacetic acid (NTA) chromatography. Thirty-three of thirty-four of the designs were soluble and had the expected alpha-helical circular dichroism (CD) spectrum at 25 °C, and 28 of the 34 were folded at 95 °C. Thirty of these proteins were monomeric as measured by analytical size exclusion chromatography coupled to multiangle light scattering (Fig. 2A).
Fig. 2.
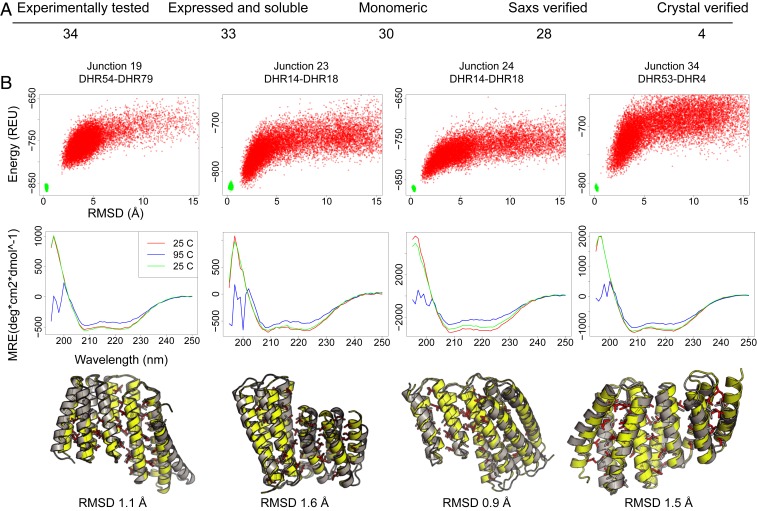
Experimental characterization of the designed junctions. (A) Numbers of designs at each characterization stage; the overall success rate through the SAXS stage is 82%. (B) Representative data for the four crystallized designs. Top row, junction names. Second row, the energy landscapes from Rosetta@home simulations. The y axis is energy as Rosetta energy units (REU; roughly 1 kcal/mol), and the x axis is the rmsd to the design. Third row, circular dichroism spectra collected at 25 °C (red), 95 °C (blue), and then cooled to 25 °C (green). All four proteins are stable to 95 °C. Bottom row, comparison of crystal structure to designed protein. In yellow and red are the backbone and side chains of the design; in gray is the crystal structure. Crystal structures have been deposited in the Protein data bank with the accession numbers 6W2R (Junction 19), 6W2V (Junction 23), 6W2W (Junction 24), and 6W2Q (Junction 34) (21–24). MRE, molar residue ellipticity.
We solved the crystal structures of four junctions with resolutions between 1.8 and 2.4 Å. The designs closely match the crystal structure with C-α rmsds ranging from 0.9 to 1.6 Å (Fig. 2B). These crystallized structures add two loops and a helix between two DHRs. The designs closely match the crystal structures in the junction region. Junction 19 has an rmsd of 1.2 Å and matches closely, 0.9 Å, over the middle 110 residues but deviates slightly (1.4 Å over the 76 residues of the N- and C-terminal repeats) due to movement in the terminal helices also observed in the crystal structures of the components DHR54 and DHR79. Junctions 23 and 24 are formed from the same building blocks (DHR14 and DHR18), but junction 24 takes a sharp turn at the connection while junction 23 is relatively straight; this difference is recapitulated in the crystal structures, showing that the junction method can assemble quite different geometries from the same building blocks. The crystal structure of the N-terminal DHR14 repeats in junction 24 better matches the original design (0.8 Å) than the crystal structures of DHR14 both in isolation (1.0 Å) and in junction 23 (0.9 Å); because of this the overall crystal structure of junction 24 is closer to the design model than that of junction 23 (0.9 vs. 1.6 Å). Junction 34 connects DHR53 to DHR4 with a slight twist at the junction; the crystal structure shows some deviation in the N- and C-terminal helices. See SI Appendix, Discussion S3 and Table S1 for further crystal structure analysis.
To characterize the overall shape of designs that did not crystallize we used SAXS (14, 15). For 28 of the 30 monomeric proteins the radius of gyration (RG) and maximum distance (dmax) estimates obtained from the scattering profiles were close to those computed from the design models. We further compared the experimentally observed SAXs profiles with simulated profiles calculated from the corresponding design models using the volatility ratio (Vr), which has been shown to be more robust to noise than the more traditional (16) (SI Appendix, Fig. S4, Table S2, and Discussion S4). The maximum value of Vr obtained for the design models of the four-junction crystal structures compared to the corresponding experimental SAXS spectra was 2.0, and among 15 previously determined crystal structures of DHRs that have similar size and aspect ratio as the junctions, the maximum value was 2.5 (8). Thus, designs with SAXS spectra matching spectra computed from the design models with Vr values less than 2.5 are likely to adopt structures close to the design models. Twenty-eight of the junction designs had Vr values below 2.5; the two proteins where the profiles did not match had dmax and RG approximately double that of the design, indicating likely aggregation (junctions 4 and 20).
With this experimental validation of the capability of building rigid junctions, we generated a library of 75,000 junctions between DHRs and 15 junctions between a DHR and a designed ankyrin (17) built with the fragment assembly strategy. Any pair of these single-junction proteins can be combined by matching a C- and N-terminal DHR (SI Appendix, Fig. S7A). There are 542 million two-junction combinations involving only DHRs and billions when also including individual repeat proteins, homo-oligomers, or ankyrin fusions (SI Appendix, Fig. S7B). To facilitate generation and exploration of such multiple-junction protein “sculpts,” we developed a parallelized python script that enumerates all DHR repeat lengths and junction combinations and writes a blueprint file that directs Rosetta to generate the three-dimensional structures and sequences.
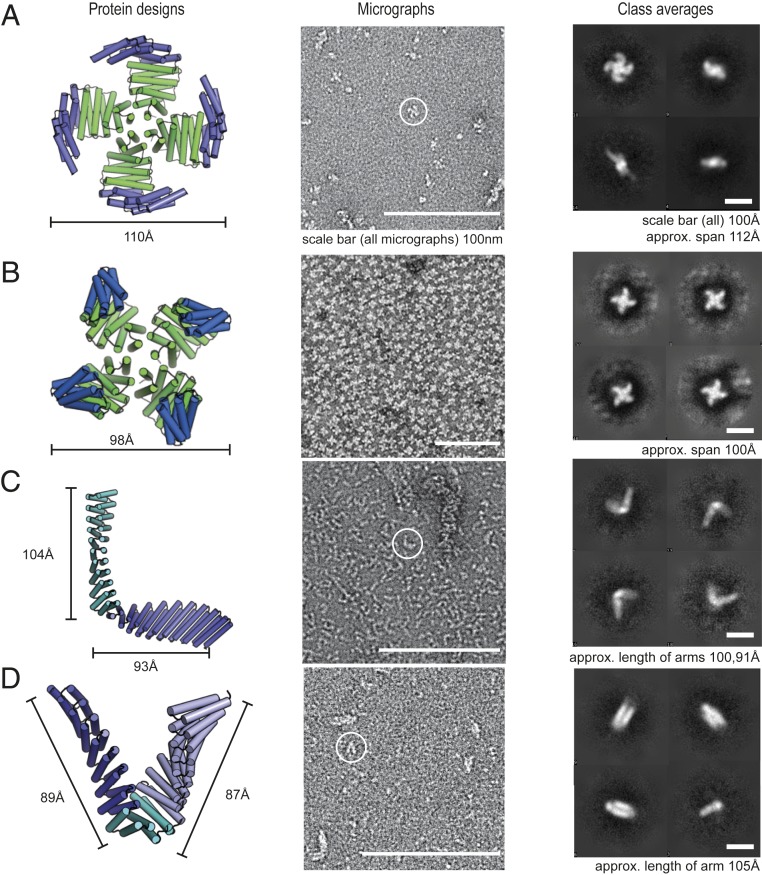
We used the enumerative method to generate large numbers of fused models and selected two designs for experimental testing with ∼10-nm arms flanking the junction site(s) likely to be visible in negative stain electron microscopy (EM). The 975-residue “L” shape design is composed of one junction, and the 853-residue “V” shape uses two junctions. To reduce possible recombination in synthetic genes encoding the designs, we introduced limited sequence variation in the surface helices of the structure. Both monomers expressed solubly in E. coli and their structures, as assessed by negative stain EM, are in agreement with design models (Fig. 3). The “L” shape design links together nine repeats of DHR14 and nine repeats of DHR76 via a DHR14–DHR76 junction that produces a roughly 90° angle between the two arms. The individual repeat units of DHR14 and DHR76 are built from different length helices, and the displacement along the repeat axis also differs; hence the longer arm, built from DHR14, is thinner than the shorter arm. The overall shape, the junction angle, and the differences in the thicknesses and lengths of the two arms are evident in the negative stain EM, both in the raw micrographs and in two-dimensional (2D) class averages (Fig. 3C; the shorter 93-Å arm is noticeably wider than the longer and thinner 104-Å arm). The “V” shape links together seven repeats of DHR14 to seven repeats of DHR54 via a DHR14–DHR79 and a DHR79–DHR54 junction. The negative stain 2D averages again are similar to the design model, with a close to “V” shape and with the two arms having similar widths and lengths. These results show that the junctions are sufficiently rigid to produce designs at the nanometer length scale.
Fig. 3.
Characterization of long-armed junctions by negative stain EM. The designs shown match the EM averages at the resolution of the technique. Column 1: design model with each junction in a different shade of green or blue. Column 2: negative stain micrographs. Column 3: 2D class averages of the designs; the different views and orientations are consistent with the design models. A and B are symmetric oligomers composed of a homotrimer (9) and a repeat protein connected via a junction. C and D are monomeric designs composed of two repeat proteins connected by one junction, or three repeat protein connected by two junctions, respectively.
A potential application of the design methodology developed in this paper is to place receptor-binding domains in relative orientations appropriate for engaging with multiple cell surface receptor subunits. To test our repeat protein junctions in the context of homo-oligomers, we generated junctions to four previously verified DHR-based oligomers that ranged in symmetry from C2 to C5 (9). For each oligomer we generated fusions of two to three junctions that were at least 10 nm across to facilitate visualization in negative stain electron microscopy. Of the designs, two had negative stain EM images consistent with the design model. The spiral and X designs connect DHR53 to the HR04C4_1 oligomer via a junction between DHR53 and DHR4 (Fig. 3 A and B). The spiral design has two more DHR4 repeats than the X shape, which flip the arms of the spiral up and into a claw-like shape. A designed ankyrin–DHR–C2 fusion disassociated in negative stain, but the monomer has a distinctive shape recapitulated in negative stain 2D averages (SI Appendix, Fig. S8) with a DHR component wider and shorter than the ankyrin component. SAXs data suggest the ankyrin–C2 is a dimer at the concentrations used in the scattering experiments as the experimental radius of gyration of 55 is closer to the dimer RG of 49 than the monomer RG of 35. All three designs validated by EM had SAXS distance distributions (dmax) and RG consistent with the design (SI Appendix, Fig. S4C). Five of the designs that we were unable to validate by EM had SAXS, dmax, and RG values that differed from the design by more than 25%. The Vr values of the EM-validated designs ranged from 2.5 to 6.6, suggesting they are more flexible than the junction building blocks, which all had Vr < 2.5. (The Vr discrepancy also derives in part from the differences in sample size; the oligomer sculpt constructs are 10 nm across while the individual junctions span 4 nm or less).
Discussion
The design methods described in this paper enable the rapid and accurate design of new proteins by fusing de novo designed repeat proteins. Of the 34 experimentally characterized single-junction designs, 28 were close to the design model. The improvements in the efficiency and speed of the design protocol enabled the generation of 75,000 junctions strongly predicted to have the designed structure. The improvements in computational efficiency introduced here will enable more research groups to design de novo proteins without the need for extensive computational resources and facilitate the design of increasingly complex structures.
Modern manufacturing was revolutionized by parts that could be used interchangeably and easily connected to one another. Here we begin to apply this concept to de novo proteins. Fused proteins take seconds to computationally design, and the success rate is quite high (all three larger monomers described here had the correct shape by EM). The junction library is integrated in the modular design software Elfin (18) (https://github.com/Parmeggiani-Lab/elfin/). Similar to computer-assisted design (CAD) tools, Elfin allows users to trace out a desired geometry and identifies building blocks that assemble into that shape. More generally, the parts library developed here should enable rapid exploration of applications to imaging and cell signaling. In contrast to traditional approaches to joining domains with flexible linkers and bispecific antibodies, with the flexible hinge between the fragment crystallizable (FC) region and antigen-binding fragment (Fab) (19), our junction library enables precise control over the orientation of the fused domains. This is important for both design of higher-order protein assemblies and the arraying of receptor-binding domains in precise orientations to engage cell surface receptors in predefined geometries (20). Our junction library makes the exploration of these and other applications limited not by the design of the monomers and assemblies, but the creativity of the protein engineers deploying the methods.
Methods
Methods for protein expression, crystallization, SAXS, and negative stain electron microscopy are described in SI Appendix, Fig. S9.
Data Availability.
All data discussed in the paper has been deposited in the Protein Data Bank (PDB) (21–24). See SI Appendix for additional details.
Supplementary Material
Acknowledgments
The SAXS work was conducted at the Advanced Light Source (ALS) supported by Department of Energy Office of Biological and Environmental Research. Additional support comes from the NIH project ALS Efficiently Networking Advanced BeamLine Experiments (P30 GM124169), a High-End Instrumentation Grant S10OD018483 from the Open Philanthropy Project at the Institute for Protein Design. This work was also supported by National Institute of General Medical Sciences (Grants R01GM12764 and R01GM118396) (to J.M.K.). Additional acknowledgments are in SI Appendix, Fig. S10.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. B.K. is a guest editor invited by the Editorial Board.
Data deposition: The crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank (PDB), http://www.rcsb.org/ (accession nos. 6W2R, 6W2V, 6W2W, and 6W2Q).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908768117/-/DCSupplemental.
References
- 1.Hong F., Zhang F., Liu Y., Yan H., DNA origami: Scaffolds for creating higher order structures. Chem. Rev. 117, 12584–12640 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Jacobs T. M., et al. , Design of structurally distinct proteins using strategies inspired by evolution. Science 352, 687–690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glover D. J., Giger L., Kim S. S., Naik R. R., Clark D. S., Geometrical assembly of ultrastable protein templates for nanomaterials. Nat. Commun. 7, 1–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai Y.-T., et al. , Designing and defining dynamic protein cage nanoassemblies in solution. Sci. Adv. 2, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Youn S.-J., et al. , Construction of novel repeat proteins with rigid and predictable structures using a shared helix method. Sci. Rep. 7, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parmeggiani F., Huang P.-S., Designing repeat proteins: A modular approach to protein design. Curr. Opin. Struct. Biol. 45, 116–123 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Geiger-Schuller K., et al. , Extreme stability in de novo-designed repeat arrays is determined by unusually stable short-range interactions. Proc. Natl. Acad. Sci. U.S.A. 115, 7539–7544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunette T. J., et al. , Exploring the repeat protein universe through computational protein design. Nature 528, 580–584 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fallas J. A., et al. , Computational design of self-assembling cyclic protein homo-oligomers. Nat. Chem. 9, 353–360 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen H., et al. , De novo design of self-assembling helical protein filaments. Science 362, 705–709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pyles H., Zhang S., De Yoreo J. J., Baker D., Controlling protein assembly on inorganic crystals through designed protein interfaces. Nature 571, 251–256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foight G. W., et al. , Multi-input chemical control of protein dimerization for programming graded cellular responses. Nat. Biotechnol. 37, 1209–1216 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maguire J. B., Boyken S. E., Baker D., Kuhlman B., Rapid sampling of hydrogen bond networks for computational protein design. J. Chem. Theory Comput. 14, 2751–2760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hura G. L., et al. , Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat. Methods 6, 606–612 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rambo R. P., Tainer J. A., Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annu. Rev. Biophys. 42, 415–441 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Hura G. L., et al. , Comprehensive macromolecular conformations mapped by quantitative SAXS analyses. Nat. Methods 10, 453–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parmeggiani F., et al. , A general computational approach for repeat protein design. J. Mol. Biol. 427, 563–575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeh C.-T., Brunette T. J., Baker D., McIntosh-Smith S., Parmeggiani F., Elfin: An algorithm for the computational design of custom three-dimensional structures from modular repeat protein building blocks. J. Struct. Biol. 201, 100–107 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Labrijn A. F., Janmaat M. L., Reichert J. M., Parren P. W. H. I., Bispecific antibodies: A mechanistic review of the pipeline. Nat. Rev. Drug Discov. 18, 585–608 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Mohan K., et al. , Topological control of cytokine receptor signaling induces differential effects in hematopoiesis. Science 364, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunette T., Bick M., Baker D., Junction 19, DHR54-DHR79. PDB, http://www.rcsb.org/structure/6W2R. Deposited 7 March 2020.
- 22.Brunette T., Bick M., Baker D., Junction 23, DHR14-DHR18. PDB, http://www.rcsb.org/structure/6W2V. Deposited 7 March 2020.
- 23.Brunette T., Bick M., Baker D., Junction 24, DHR14-DHR18. PDB, http://www.rcsb.org/structure/6W2W. Deposited 7 March 2020.
- 24.Brunette T., Bick M., Baker D., Junction 34, DHR53-DHR4. PDB, http://www.rcsb.org/structure/6W2Q. Deposited 7 March 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper has been deposited in the Protein Data Bank (PDB) (21–24). See SI Appendix for additional details.