Significance
The light reactions of photosynthesis in algae and plants occur in thylakoid membranes within the chloroplast. Previous studies have implicated membrane contact sites, vesicle trafficking, and lipid-binding proteins in the formation of thylakoids at the chloroplast inner envelope, but the molecular basis of thylakoid biogenesis and maintenance remains enigmatic. Here we show that CPSFL1 is a chloroplast Sec14 domain protein that binds lipids and can transfer phosphoinositides between membranes in vitro. In the model plant Arabidopsis, CPSFL1 is necessary for the appearance of vesicles in chloroplasts. Our results provide molecular evidence supporting an essential role of chloroplast lipid and vesicle trafficking for photoautotrophic growth.
Keywords: chloroplast, thylakoid biogenesis, phosphoinositides, Sec14 domain, CRAL_TRIO domain
Abstract
In eukaryotic photosynthetic organisms, the conversion of solar into chemical energy occurs in thylakoid membranes in the chloroplast. How thylakoid membranes are formed and maintained is poorly understood. However, previous observations of vesicles adjacent to the stromal side of the inner envelope membrane of the chloroplast suggest a possible role of membrane transport via vesicle trafficking from the inner envelope to the thylakoids. Here we show that the model plant Arabidopsis thaliana has a chloroplast-localized Sec14-like protein (CPSFL1) that is necessary for photoautotrophic growth and vesicle formation at the inner envelope membrane of the chloroplast. The cpsfl1 mutants are seedling lethal, show a defect in thylakoid structure, and lack chloroplast vesicles. Sec14 domain proteins are found only in eukaryotes and have been well characterized in yeast, where they regulate vesicle budding at the trans-Golgi network. Like the yeast Sec14p, CPSFL1 binds phosphatidylinositol phosphates (PIPs) and phosphatidic acid (PA) and acts as a phosphatidylinositol transfer protein in vitro, and expression of Arabidopsis CPSFL1 can complement the yeast sec14 mutation. CPSFL1 can transfer PIP into PA-rich membrane bilayers in vitro, suggesting that CPSFL1 potentially facilitates vesicle formation by trafficking PA and/or PIP, known regulators of membrane trafficking between organellar subcompartments. These results underscore the role of vesicles in thylakoid biogenesis and/or maintenance. CPSFL1 appears to be an example of a eukaryotic cytosolic protein that has been coopted for a function in the chloroplast, an organelle derived from endosymbiosis of a cyanobacterium.
Photosynthetic organisms transform light energy into chemical potential, thereby providing the energy sources for nearly all life on Earth. The light reactions of oxygenic photosynthesis take place in thylakoids, a membrane system highly specialized for light energy capture, photosynthetic electron transfer reactions, and adenosine 5′-triphosphate (ATP) production (1). In plants, thylakoids are located in the chloroplast, an organelle of cyanobacterial origin. Two envelope membranes separate the chloroplast stroma, which is the aqueous phase surrounding the thylakoids, from the cytoplasm (2).
Photosynthetic electron transport generates highly reactive intermediates, which, in functional thylakoid membranes, are rapidly converted into more-stable intermediates to minimize production of unwanted byproducts, such as reactive oxygen species. Defects in thylakoid biogenesis have detrimental effects on plant viability (3, 4). The biogenesis of functional thylakoids requires the concerted interplay of lipid, protein, and pigment synthesis processes and assembly of these components (5–9). Although the synthesis of individual components necessary for thylakoid biogenesis is mostly understood, it still remains unclear how the resulting components are assembled to generate functional thylakoid membranes. In particular, the transport or transfer of lipids, which are synthesized exclusively at the envelope membranes (10), to the growing thylakoid membrane network during chloroplast biogenesis and growth is poorly understood (11–13).
During proplastid to chloroplast transition, thylakoids are synthesized de novo. For the initial establishment of thylakoid membranes, two mechanisms have been proposed based on microscopy: Primary thylakoid lamellae develop 1) by invagination of the inner envelope membrane (14, 15) or 2) by fusion of envelope membrane-derived vesicles (16–20). Once a set of initial chloroplasts has developed from proplastids in the differentiating cell, the chloroplasts divide and regrow in proliferating tissues (21, 22). Three routes for lipid deposition from the envelope to the growing thylakoid network during chloroplast growth have been postulated: 1) via envelope−thylakoid contact sites, 2) via envelope vesicles, and 3) via lipid-binding proteins (11, 12).
Several proteins, including VIPP1, have been proposed to play a role in chloroplast vesicle trafficking (23–28). Loss-of-function mutants of the chloroplast protein VIPP1 lack envelope-derived vesicles and do not form thylakoid membranes. Thus it was suggested that thylakoid biogenesis indeed involves vesicle trafficking (4). Still, the process of chloroplast vesicle trafficking remains enigmatic and is not fully understood on the molecular level. In addition, a consensus about whether any of the proposed proteins play a direct role in lipid transfer from the envelope membrane to the thylakoids is missing. Recently, it was suggested that the lack of vesicles in vipp1 mutants can be explained by a function of VIPP1 in envelope maintenance (29–31).
Here we provide evidence supporting a critical role for vesicle trafficking or protein-mediated lipid transport in chloroplasts from the inner envelope to the thylakoids. Arabidopsis thaliana has a chloroplast-localized Sec14 domain protein (CPSFL1) that is homologous to eukaryotic Sec14 proteins, which are found exclusively in eukaryotes and are essential for vesicle formation and trafficking at the trans-Golgi network (TGN) (32). Loss-of-function cpsfl1 mutants are seedling lethal and incapable of photoautotrophic growth (33). When grown heterotrophically on sucrose, they develop chloroplasts that are smaller in size and have reduced levels of thylakoid membranes. The chloroplasts of cpsfl1 mutants lack vesicles, and residual thylakoids occur in direct contact with the inner envelope. The CPSFL1 protein localizes to vesicle-like structures in the stroma or adjacent to the inner envelope. In vitro, CPSFL1 acts as a canonical Sec14 protein by binding and transferring phosphatidylinositol phosphate (PIP) between bilayers, and it can complement the yeast sec14 mutation. Altogether, our data show that CPSFL1 is essential for photoautotrophic growth and suggest a direct or indirect role in the regulation of thylakoid development by mediating vesicle trafficking and/or transfer of lipids or other hydrophobic molecules in the chloroplast.
Results
A Sec14 Domain Protein Is Localized in the Chloroplast of Arabidopsis.
All known mechanisms of membrane transport require the assistance of lipid-binding proteins, involved in either lipid transport or membrane curvature modulation (34–37). To identify candidate factors involved in membrane trafficking from the chloroplast inner envelope to thylakoids, we searched the Arabidopsis protein database for chloroplast-localized lipid-binding proteins. Out of 3,009 proteins with annotated lipid-binding domains, 304 had a predicted chloroplast transit peptide (cTP), and, out of this subset, we selected 34 candidates that had been experimentally found in the chloroplast by proteomics (http://ppdb.tc.cornell.edu/). Among the proteins without functional annotation, we found one protein (encoded by At5g63060) with homology to the yeast Sec14 protein. Proteins of the Sec14 superfamily are of eukaryotic origin (38, 39). They all contain a structural domain termed the CRAL_TRIO (cellular retinaldehyde binding-triple response) domain that allows binding and transfer of small hydrophobic molecules, such as lipids, sterols, and/or vitamin E (α-tocopherol). Yeast Sec14, the founding member of the superfamily, is known to regulate both lipid metabolism and vesicle transport, and it acts as a phosphatidylcholine (PC)/phosphatidylinositol transfer protein (32, 40).
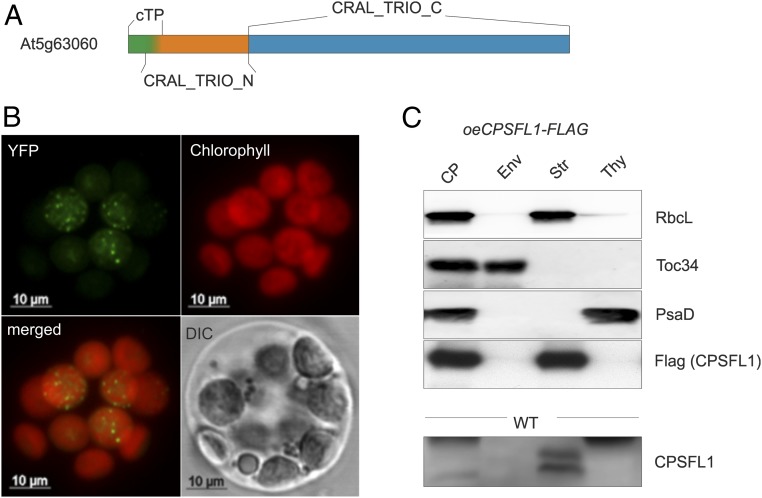
To verify the subcellular localization of the Sec14 homolog encoded by At5g63060, we expressed a full-length complementary DNA (cDNA) as a C-terminal YFP fusion in Arabidopsis protoplasts (Fig. 1B). The fusion protein was exclusively localized in plastids and showed a dual localization within the stroma and as distinct puncta with a higher local fluorescence intensity (Fig. 1B, YFP). Because of its homology to Sec14 and its plastid localization, we renamed At5g63060 as CPSFL1 for chloroplast-localized Sec14-like protein 1. We further analyzed the suborganelle localization of CPSFL1 by fractionating chloroplasts of wild-type (WT) and complemented cpsfl1 mutant lines overexpressing FLAG-tagged CPSFL1 (oeCPSFL1-FLAG/cpsfl1) into envelope, stroma, and thylakoids (Fig. 1C). In WT chloroplasts, CPSFL1 was detected within the stroma subfraction (Fig. 1C, CPSFL1 in WT). In oeCPSFL1-FLAG lines, CPSFL1-FLAG was detected almost exclusively in the plastid stroma and in traces within envelope and thylakoid fractions (Fig. 1C, FLAG [CPSFL1] in oeCPSFL1-FLAG/cpsfl1).
Fig. 1.
CPSFL1 is a plastid-localized Sec14-like protein. (A) Protein model of CPSFL1. Chloroplast targeting is encoded by an N-terminal cTP marked in green and overlapping with the first nine amino acids of the N-terminal CRAL_TRIO domain (orange), followed by the C-terminal CRAL_TRIO domain (blue) characteristic of all members of the Sec14 superfamily. The bipartite sequence motif is composed of CRAL_TRIO_C and N domains, with CRAL_TRIO_C forming the lipid-binding pocket. (B) Subcellular localization of CPSFL1-YFP. Protoplasts from WT Arabidopsis plants expressing CPSFL1 in-frame with a C-terminal YFP and under the control of the CaMV 35S promoter were analyzed by fluorescence confocal laser scanning microscopy. Differential interference contrast (DIC) was used to verify the intactness of protoplasts. Chlorophyll autofluorescence (red); CPSFL1-YFP (green). (C) Immunological localization of CPSFL1 and CPSFL1-FLAG in chloroplast (CP) subfractions from WT and oeCPSFL1-FLAG/cpsfl1 plants. Aliquots from the isolated thylakoid (Thy), stroma (Str), and envelope (Env) subfractions were probed using CPSFL1- and FLAG-specific antibodies. Subfraction purity was shown by probing with antibodies against thylakoid (PsaD), stromal (RbcL), or envelope (TOC34) proteins. PsaD, PSI subunit D; RbcL, large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase; TOC34, 34-kDa subunit of the translocon of the outer envelope membrane of chloroplasts.
CPSFL1 Is Essential for Photoautotrophic Growth.
To investigate the function of CPSFL1, we analyzed the phenotypes of an Arabidopsis cpsfl1 null mutant with a transfer DNA (T-DNA) insertion in the first exon of the CPSFL1 gene (SI Appendix, Fig. S2A). The cpsfl1 mutant does not accumulate detectable levels of CPSFL1 mRNA (SI Appendix, Fig. S2B) or CPSFL1 protein (Fig. 2B). When germinated on soil, homozygous cpsfl1 mutants were seedling lethal. Some homozygous cpsfl1 mutants that produce albino seedlings in the absence of sucrose can develop green leaves upon sucrose supplementation (33). Addition of 2% sucrose to the growth medium partially rescued the growth of cpsfl1 mutants (Fig. 2A). Under these specific growth conditions, cpsfl1 displayed a dwarf and pale-green phenotype (Fig. 2A). During growth, progressive bleaching of leaves was observed, resulting in senescence (SI Appendix, Fig. S3A, expanding and mature leaves, arrowheads). Photoautotrophic growth of cpsfl1 was completely restored by overexpression of a CPSFL1-FLAG fusion protein (line 38, used above for analysis of chloroplast subfractions). However, lines with very high CPSFL1 accumulation showed reduced growth, suggesting that very high levels of CPSFL1 have detrimental effects (SI Appendix, Fig. S3C, line 37).
Fig. 2.
The cpsfl1 mutants are seedling lethal and show a defect in photosynthesis. (A) Phenotype of WT, cpsfl1 mutants, and complemented lines. When grown on soil, homozygous cpsfl1 mutants were seedling lethal but could be partially rescued when grown on Murashige and Skoog medium supplemented with 2% sucrose (MS). Following transformation of cpsfl1 lines with a construct encoding the cDNA of CPSFL1 in-frame with a C-terminal FLAG tag and under the control of the 35S promoter, photoautotrophic growth could be fully restored as compared to WT (oeCPSFL1/cpsfl1). Fv/Fm measurements of WT, cpsfl1 mutants, and complemented lines indicated a moderate photosynthetic defect in cpsfl1. (B) Immunoblot analysis of CPSFL1 protein levels in total protein extracts of WT, cpsfl1 mutants, and complemented lines (oeCPSFL1/cpsfl1) using CPSFL1- and FLAG-specific antibodies. Ponceau Red stain (P.R.) served as loading control. (C) Quantification of chloroplast marker proteins. Total protein extracts of 4-wk-old WT, cpsfl1 mutants, and complemented lines (oeCPSFL1/cpsfl1) grown under identical conditions were analyzed for levels of respective chloroplast marker proteins. A dilution series of WT extracts was loaded for better estimation; 100% corresponds to 20 µg of protein in WT. Actin protein levels were analyzed as an internal standard. RbcL was used as stromal marker. PsaD, AtpB (subunit B of ATP synthase), Cytb6 (subunit cytochrome b6 of Cytb6f), and D2 (subunit of PSII) were used to estimate the abundance of respective thylakoid membrane protein complexes. TOC34 served as marker for envelope membranes. (D) HPLC pigment analysis of chlorophylls and carotenoids of WT and cpsfl1 mutant.
The seedling-lethal phenotype of cpsfl1 under photoautotrophic growth conditions and the localization of CPSFL1 to the chloroplast suggested that the cpsfl1 mutant might be impaired in photosynthesis. The maximum quantum efficiency of photosystem (PS) II (Fv/Fm) in heterotrophically grown cpsfl1 seedlings was decreased to 0.4 as compared to the WT value of 0.7, suggesting major defects in photosynthetic electron transfer (Fig. 2A, Fv/Fm). As shown by immunoblot analysis in Fig. 2C, the pale-green leaf phenotype of cpsfl1 was associated with an overall decreased abundance of components of the major photosynthetic complexes (i.e., PSII, cytochrome b6f complex, PSI, ATP synthase, and Rubisco) and the chloroplast envelope protein import machinery (Toc34). In cpsfl1, components of the photosynthetic electron transport chain accumulated to <25% of WT levels, suggesting a pleiotropic effect on photosynthetic membrane protein complexes or a defect in thylakoid development (Fig. 2C). The envelope protein Toc34 as well as the large subunit of Rubisco were reduced by ∼50% as compared to WT. To determine whether the decrease in photosynthetic membranes is associated with a defect in pigment synthesis, extracts of WT and cpsfl1 mutant plants were analyzed for changes in pigment composition by high-performance liquid chromatography HPLC (Fig. 2D). As expected from the reduced amount of plastids and thylakoids, the analysis of photosynthesis-relevant pigments showed an overall decrease of chlorophylls and carotenoid species, indicating a pleiotropic decrease in pigment accumulation. Whereas most carotenoids, including β-carotene-derived xanthophylls, accumulated to ∼40% of WT levels, α- and β-carotene showed a more pronounced reduction to ∼25% of WT levels. Altogether, our data suggest that cpsfl1 mutants contain fewer and/or smaller chloroplasts, with low amounts of thylakoid membranes and pigments.
The cpsfl1 Mutant Exhibits a Defect in Chloroplast Development and Thylakoid Ultrastructure.
To determine whether the photosynthetic phenotype of the cpsfl1 mutant was caused by a defect in chloroplast and/or thylakoid development, the ultrastructure of WT and cpsfl1 was analyzed by transmission electron microscopy (TEM). Leaf cells of cpsfl1 had fewer chloroplasts per cell as compared to WT (Fig. 3A, 2,000×). In addition, cpsfl1 plastids were highly irregular in shape and size and smaller in all cases than the uniformly shaped WT chloroplasts (Fig. 3A, 2,000×). Furthermore, the thylakoid network of cpsfl1 chloroplasts appeared to be underdeveloped, and the thylakoid area per plastid was reduced to 28.0 ± 7.7 in cpsfl1 compared to 49.2 ± 6.4 in WT (Fig. 3A, 5,000×; SI Appendix, Fig. S4B). Whereas WT thylakoids were clearly differentiated into grana stacks connected by stroma lamellae, thylakoids in cpsfl1 were much simpler, with grana stacks being interconnected only on the same plane (SI Appendix, Fig. S4B). These parallel thylakoid strands were in direct contact with the inner envelope membrane at two (or in some cases three) poles of the plastids (Fig. 3A, 5,000× and 12,000×). In contrast, no direct contact to inner envelope membranes was observed for WT thylakoids (Fig. 3A, 20,000×).
Fig. 3.
The cpsfl1 mutants have a defect in chloroplast ultrastructure and lack chloroplast vesicles. (A) TEM micrographs of ultrathin sections of leaves from the WT and cpsfl1 mutant. To analyze the ultrastructure of cpsfl1 mutant leaves, plants were grown on MS medium supplemented with 1% sucrose under very low light intensities (10 µmol photons per m−2⋅s−1). Due to the growth differences between WT and cpsfl1 mutants, the first true leaves of cpsfl1 mutants were compared to WT leaves from the same developmental stage grown under identical conditions. (B) Chloroplast vesicles in WT, cpsfl1, and oeCPSFL1/cpsfl1. To analyze the capacity of chloroplast inner envelope vesicle formation, WT, cpsfl1 mutants, and oeCPSFL1/cpsfl1 plants were grown on MS medium supplemented with 1% sucrose in a growth chamber under very low light intensities (10 µmol photons per m−2⋅s−1). Due to the growth differences, the first true leaves of cpsfl1 mutants were compared to WT and oeCPSFL1/cpsfl1 leaves from the same developmental stage grown under identical conditions. In addition, developing proplastids within the same tissue were analyzed for vesicle abundance. To increase vesicle abundance, leaf samples were incubated for 1 h at 4 °C or kept at 25 °C (control) prior to fixation. (C) Quantification of chloroplast vesicles. The number of vesicles within the focal plane of 100 plastids was counted from cpsfl1, WT, and oeCPSFL1/cpsfl1 (line 38) leaf samples that were incubated for 1 h at 4 °C. Vesicles numbers were grouped into the following categories: 0, 1 to 2, 3 to 5, 6 to 10, and >10 vesicles per plastid section.
cpsfl1 Mutants Lack Inner Envelope Vesicles.
Sec14 proteins have been implicated in vesicle transport processes. Vesicle accumulation in eukaryotic cells can be induced by cold treatment, as shown for the Golgi apparatus (41) and also for the inner envelope membrane of the chloroplast (20). To investigate whether CPSFL1 is involved in formation of vesicles from the inner envelope, WT, cpsfl1, and oeCPSFL1-FLAG plants were incubated at 4 °C, and chloroplasts were analyzed by TEM (Fig. 3B).
Some mature WT chloroplasts showed vesicles budding at the inner envelope at room temperature with a vesicle diameter of ∼70 nm. The number of vesicles per chloroplast increased following incubation at 4 °C (Fig. 3B, WT 4 °C). Intriguingly, in cpsfl1 chloroplasts, almost no vesicles were observed, neither under ambient temperature nor following 4 °C treatment (Fig. 3 B and C, cpsfl1). In addition, under ambient temperature as well as following 4 °C treatment, large balloon-like structures with a diameter of up to 400 nm were observed in the stroma of most cpsfl1 plastids (SI Appendix, Fig. S4A). Analysis of serial sections showed a direct connection of these structures with envelope membranes (SI Appendix, Fig. S4B). Furthermore, some invaginations harbored mitochondria or cytoplasm, and thylakoid membranes showed a tight association with these envelope invaginations (SI Appendix, Fig. S4C). In oeCPSFL1-FLAG/cpsfl1 lines, WT-like vesicles were observed at both ambient temperature and 4 °C. The analysis of the ultrastructure of cpsfl1 mutants expressing FLAG-tagged CPSFL1 to WT levels showed the reoccurrence of stromal vesicle structures along with the development of WT-like thylakoids (Fig. 3 B and C, oeCPSFL1-FLAG/cpsfl1 [line 38]).
Vesicle-mediated membrane trafficking has been hypothesized to play an especially important role in developing plastids (16, 20). Therefore, developing chloroplasts of WT, cpsfl1, and oeCPSFL1-FLAG/cpsfl1 lines were analyzed by TEM (Fig. 3B, developing plastids). In comparison to mature chloroplasts, vesicle-like structures were present in nearly all plastids of WT and oeCPSFL1-FLAG/cpsfl1 even under ambient temperature (Fig. 3B, developing plastids of WT and oeCLSFL1-FLAG/cpsfl1). Again, no vesicles could be observed in developing plastids of the cpsfl1 mutant. Also, in contrast to mature chloroplasts, the balloon-like invaginations were absent in cpsfl1 (Fig. 3B, developing plastids of cpsfl1).
CPSFL1 Is Associated with Plastid Vesicles.
Fluorescence and immunolocalization of CPSFL1 showed localization in distinct spots (Fig. 1B). To analyze whether these spots could be attributed to the localization of CPSFL1 in vesicles, we performed immunogold TEM detection of CPSFL1 with anti-FLAG and anti-CPSFL1 antibodies (Fig. 4 A and B). In WT as well as in complemented lines, CPSFL1 was detected in nascent and pinched off vesicles emerging from the inner envelope membrane of chloroplasts. However, in WT chloroplasts probed with anti-CPSFL1 antibodies, the majority of gold particles marked the thylakoid membranes, due to a nonspecific binding of the antibodies (Figs. 2B and 4A). To rule out that the clustered arrangement of gold particles was caused by antibody aggregation, the PSII antenna protein Lhcb2 was immunolocalized in WT plastids as a control (Fig. 4C). In this case, no clustered localization could be observed, and the gold particles showed even labeling of Lhcb2 proteins throughout the thylakoid network. In addition, WT sections were probed with anti-FLAG primary and gold-labeled secondary antibodies to show background labeling when no FLAG tag was present (Fig. 4D). Only a few gold particles were found in this negative control. Altogether, these results strongly suggest that CPSFL1 localizes to vesicles, presumably originating from the inner envelope.
Fig. 4.
CPSFL1 is localized within vesicle-like structures. (A) Immunogold localization of CPSFL1-FLAG in oeCPSFL1/cpsfl1 leaves. Subcellular localization of CPSFL1 and CPSFL1-FLAG was shown by immunogold labeling and TEM of WT and oeCPSFL1/cpsfl1. Plants grown on soil in a growth chamber for 3 wk at 150 µmol photons per m−2⋅s−1 were used. In oeCPSFL1/cpsfl1 lines, CPSFL1-FLAG was detected with high specificity. In WT, the CPSFL1 antibody also recognized thylakoid proteins (according to a nonspecific signal shown in Fig. 2B). (B) Immunogold localization of CPSFL1 in WT leaves. White boxes in A and B mark the position of CPSFL1 clusters detected in plastids. Some of these are shown in higher magnification below. (C) Immunogold localization of Lhcb2 in WT leaves. As a control experiment, the thylakoid-specific Lhcb2 protein was detected by immunogold labeling and TEM. Subcellular localization of Lhcb2 was shown to be exclusively in the thylakoids. Inset in C shows a higher magnification of the plastid with gold particles bound to Lhcb2. (D) The use of secondary antibodies alone was included as an additional control and showed no labeling.
CPSFL1 Binds to Phosphatidic Acid and PIPs with High Specificity.
Sec14 domain proteins are known to transfer lipids, and many members of this superfamily specifically transfer phosphatidylinositides (42). Most members of the Sec14 superfamily catalyze phosphatidylinositol transfer activity (PITP) via their lipid-binding CRAL_TRIO domain. The bipartite CRAL_TRIO motif is composed of a large lobe encoding a hydrophobic lipid-binding pocket (CRAL_TRIO_C) that is closed off by a smaller N-terminal lobe (CRAL_TRIO_N) (43). To analyze the lipid-binding activity of CPSFL1, we used recombinant affinity-purified Sec14 proteins and performed protein−lipid blot overlay assays (Fig. 5A, CPSFL1). Full-length CPSFL1 and a version lacking the CRAL_TRIO_N domain (overlaps with the predicted cTP), as well as the positive controls yeast Sec14 and a Soybean Sec14p Homologue (SSH) Arabidopsis Ssh2p, were probed for their lipid binding specificity. CPSFL1 bound to phosphatidic acid (PA) and PIPs with high specificity (Fig. 5A, CPSFL1). Yeast Sec14 bound PC and PI, and Arabidopsis Ssh2p bound PA and PIP, as previously described (32, 44). However, yeast Sec14 preferentially bound to PA and PIPs in our assay (Fig. 5A). This was surprising, because PA has not been previously described as a Sec14 ligand. A mutant version of the CPSFL1 protein lacking the N terminus showed a reduction in PIP binding but still bound PA (Fig. 5A, CPSFL1ΔCRAL_TRIO_N), suggesting that the CPSFL1 CRAL_TRIO_N domain preferentially supports the binding of PIPs.
Fig. 5.
CPSFL1 binds to PA and PIPs and has phosphatidylinositol phosphate transfer activity. (A) Lipid binding and specificity were verified by protein−lipid blot overlay assays shown on Left. CPSFL1, Yeast Sec14, Ssh2p, and CPSFL1ΔCRAL_TRIO_N specifically bind to PA and PIPs. A schematic representation of biological membrane lipids present on lipid strips used is shown on Right, including triglyceride (TG), diacylglycerol (DAG), PA, phosphatidylserine (PS), phosphatidylethanolamine (PE), PC, phosphatidylglycerol (PG), cardiolipin (CL), phosphatidylinositol (PtdIns), and phosphatidylinositol phosphates (PtdIns(4)P, PtdIns(4,5)P, and PtdIns(3,4,5)P) as well as cholesterol, sphingomyelin and sulfatides. (B) CPSFL1 specifically associates with PA- and SQDG-rich bilayers and extracts PI4P. Lipid binding or extraction/transport of recombinant FLAG-tagged CPSFL1 (final concentration 100 µg/mL) was tested by incubation with liposomes (final concentration 10 mg/mL) made from PC and containing 10 mol % MGDG, DGDG, SQDG, PI4P, or PA. Following ultracentrifugation, lipids in supernatant and pellet fractions were analyzed by TLC using 10% of the pellet fraction and 10% of the total liposomes used (with 2 mol % of the input) in comparison to the total amount of lipid extracted from the supernatant. Analysis of proteins in respective fractions shows CPSFL1-dependent extraction of PIP from liposomes but no liposome cosedimentation. Cosedimentation of CPSFL1 was observed only when PA was present in liposomes and, to a lesser extent, with SQDG. No extraction of respective lipids was observed. (C) PA-mediated PI4P transfer activity of CPSFL1 between donor and acceptor liposomes. PITP transfer activity of recombinant FLAG-tagged CPSFL1 (final concentration 100 µg/mL) was tested between light donor (without sucrose, with 2 mol % of fluorescent donor lipid in nonfluorescent PC moiety) (final 10 mg/mL) and heavy acceptor (with sucrose, without donor lipid or with fluorescent PA in nonfluorescent PC moiety) liposomes (10 mg/mL) by coincubation with or without recombinant CPSFL1-FLAG. Subsequently, liposomes were separated using sucrose density centrifugation, and lipids of 10% heavy liposomes were analyzed by TLC following extraction, in comparison to 10% of input donor liposomes using a fluorescence reader. Increasing PA levels in acceptor liposomes led to increased PI4P transfer mediated by CPSFL1. (D) Expression of CPSFL1 complements yeast sec14-1ts mutants. Overexpression of CPSFL1 lacking the cTP (-cTP) rescues the temperature-sensitive growth phenotype of the sec14-1ts mutant. Yeast dilution assays of sec14-1ts cells expressing full length CPSFL1 (Full), CPSFL1 without transit peptide (-cTP), or empty plasmid (pRS426) spotted onto uracil dropout synthetic media with 2% dextrose (Ura-Dextrose) or with 2% galactose (Ura-Galactose). Plates were incubated at the indicated temperatures and examined after 2 d to 6 d. (E) Superresolution immunolocalization of CPSFL1-FLAG (green) in oeCPSFL1-FLAG/cpsfl1 chloroplasts relative to PI4P (blue in Top; detected by PI4P-specific antibodies) and PA (blue in Bottom; detected using biotin-labeled TGD2) and thylakoids (chlorophyll autofluorescence [red]). Asterisk marks a plastid in the merged pictures that is shown in a higher magnification on the far right.
CPSFL1 Binds to PA and Extracts PIP from Membranes In Vitro.
Sec14 proteins bind to membranes and are able to transport lipids (45). To analyze whether PA or PIP facilitates the binding of CPSFL1 to membranes, we performed liposome/CPSFL1 cosedimentation experiments in which liposomes with bilayers containing PA or PIP (10% donor lipid/90% PC) were coincubated with recombinant CPSFL1 (Fig. 5B). Following coincubation, liposomes were collected by ultracentrifugation, and the resulting pellet and the supernatant fractions were analyzed for their lipid and protein content. Among the various PIPs, only the interaction of phosphatidylinositol 4-phosphate (PI4P) with CPSFL1 was tested in the subsequent analysis. Additionally, the interaction of CPSFL1 with the predominant glycolipids of envelope and thylakoid membranes (monogalactosyldiacylglycerol [MGDG], digalactosyldiacylglycerol [DGDG], and sulfoquinovosyldiacylglycerol [SQDG]), which were not present in the lipid blot overlay assays, were tested this way (Fig. 5B).
As expected based on the lipid overlay assay (Fig. 5A), CPSFL1 bound to liposomes containing PA. No interaction with MGDG and DGDG was observed (Fig. 5B, PA pellet); however, a moderate cosedimentation of CPSFL1 was observed when SQDG was present in membranes (Fig. 5B, pellet SQDG). No accumulation of PA or SQDG in the supernatant could be detected in this assay. This excludes a direct transport of these lipids by CPSFL1. In contrast, no cosedimentation of CPSFL1 was observed when PI4P was present in liposomes (Fig. 5B, pellet PIP and PC). Instead, PI4P accumulated in the supernatant fractions together with CPSFL1 (Fig. 5B).
CPSFL1 Acts as a Phosphatidylinositol Transfer Protein.
To clarify the functional relationship of CPSFL1 with respect to PA binding and PI4P extraction, we performed mixed liposome experiments. In particular, we wanted to know whether PA-rich membranes attract PI4P-loaded CPSFL1 or whether a higher preference of CPSFL1 for PA binding inhibits PI4P extraction. Therefore, the PI4P transfer protein activity of CPSFL1 was monitored between lipid bilayers of “light” donor liposomes that contained 10% PI4P and “heavy” acceptor liposomes that were filled with sucrose and lacked PI4P. Because acceptor liposomes were filled with sucrose, they could readily be separated from light donor liposomes by centrifugation. Subsequently, lipid extracts of both liposome fractions were analyzed by thin-layer chromatography (TLC). To highlight the role of CPSFL1 affinity to PA, PI4P transfer activity of CPSFL1 was monitored by incorporating variable amounts of PA into donor liposomes and acceptor liposomes (Fig. 5C). As an additional control, all liposome experiments were carried out with and without CPSFL1. As hypothesized, CPSFL1 was capable of transferring PI4P from donor to acceptor liposomes (Fig. 5C, PIP+CPSFL1). In addition, no inhibitory effect on PI4P transfer was observed when PA was present in donor liposomes. Instead, a marked increase of PI4P in acceptor liposomes was observed when PA levels in acceptor membranes were elevated as compared to donor liposomes. Thus, we could show that PA is required not only for membrane binding of CPSFL1, but this protein also promotes deposition of PI4P into PA-rich membrane bilayers (Fig. 5C).
Arabidopsis CPSFL1 Complements Yeast sec14.
The role of phosphatidylinositol transfer in yeast vesicle secretion is still unclear, in part because mutations affecting PC biosynthesis can suppress the growth defects of sec14 mutants (46), whereas not all Sec14 domain proteins with PITP activity can complement yeast sec14 mutations (47). However, the direct interaction of Sec14 with vesicle-releasing Golgi membranes and its essential role in TGN vesicle budding have been demonstrated extensively (32, 48, 49). Because both yeast Sec14 and CPSFL1 encode PITP activity and, like Sec14, CPSFL1 might also function in vesicle trafficking, we wanted to determine whether heterologous expression of Arabidopsis CPSFL1 can complement the yeast sec14 mutation. SEC14 is an essential gene in yeast, so we transformed a Saccharomyces cerevisiae temperature-sensitive sec14 mutant with the CPSFL1 cDNA under the control of a galactose-inducible promoter (Fig. 5D). The conditional lethal sec14-1ts mutant strain grew normally at 30 °C, but protein secretion and growth halted when cells were shifted to the nonpermissive temperature of 37 °C (48). Following expression of a full-length version of CPSFL1 in sec14-1ts, growth could not be restored. However, expression of CPSFL1 lacking the predicted N-terminal cTP fully restored growth of sec14-1ts on galactose-containing medium at the nonpermissive temperature (Fig. 5D), indicating that a plastid protein, CPSFL1, is able to compensate the function of Sec14 in TGN vesicle budding. The empty vector control showed no rescue of sec14-1ts.
CPSFL1 Partially Colocalizes with PA and PI4P.
The localization of CPSFL1 was analyzed by superresolution immunolocalization of FLAG-tagged CPSFL1 expressed in the cpsfl1 mutant using three-dimensional structured illumination microscopy (3D-SIM), which showed spots of anti-FLAG immunofluorescence at envelope membranes and also throughout the plastid and in the vicinity of thylakoids (Fig. 5E). We further localized CPSFL1 with respect to the distribution of PA and PI4P in chloroplasts by indirect immunofluorescence (Fig. 5E, PA [TGD2], PI4P). PA was detected by using a biotinylated version of the PA-binding protein TGD2 in combination with a fluorescence-labeled avidin, and PI4P was visualized by using PI4P-specific antibodies in combination with fluorescence-labeled secondary antibodies. A partial colocalization of PA and PI4P with CPSFL1 could be shown (Fig. 5E).
Discussion
A Role for Arabidopsis CPSFL1 in Chloroplast Vesicle Formation.
Our data show that CPSFL1 is involved in chloroplast development and is essential for photoautotrophic growth. Ultrastructural investigations of cpsfl1 chloroplasts showed a reduction in thylakoid membranes and a simplified membrane organization together with the loss of plastid vesicles (Fig. 3). Although it is difficult to prove the complete absence of vesicles, they were not observed under multiple conditions (varying temperature and developmental stage) in which they were readily apparent in WT chloroplasts. In addition, overexpression of CPSFL1 resulted in more vesicles (Fig. 3B); CPSFL1 appeared to be associated with vesicles in EM immunolocalization experiments (Fig. 4).
The drastic phenotype observed for cpsfl1 mutants indicates a significant role of vesicle transport in chloroplasts. However, because some thylakoid development can still be observed in cpsfl1 mutant chloroplasts, the formation of vesicles must not be essential for thylakoid biogenesis. In contrast to WT thylakoids, the thylakoids observed in cpsfl1 are in direct contact with the inner envelope (Fig. 3A). This suggests that, in the absence of CPSFL1 (and chloroplast vesicles), thylakoids develop exclusively from inner envelope thylakoid contact sites when plants are grown heterotrophically. In addition, CPSFL1 could also be involved in detachment of thylakoids from the envelope membranes. However, thylakoid development solely driven by envelope/thylakoid contact sites might not yield fully functional thylakoid membranes that are sufficient to sustain photoautotrophic growth during seedling development. Thus, our results indicate that both vesicle transport and inner envelope/thylakoid contact sites can contribute to thylakoid biogenesis. In fact, chloroplast vesicle transport has been shown to be particularly important in mature chloroplasts of expanding leaf tissue, whereas contact sites between the envelope and thylakoid membranes exist in the early stages of thylakoid development (14, 16, 21, 50).
Chloroplast vesicle transport could be involved in the trafficking of lipids from their site of synthesis in the envelope to thylakoids and other structures such as plastoglobules (11). It is possible that other hydrophobic molecules, including carotenoids, are also transported via this route. Indeed, we observed a preferential decrease in carotenes in the cpsfl1 mutant (Fig. 2D), similar to that seen in a cpsfl1 mutant of the unicellular green alga Chlamydomonas reinhardtii (51). Like the Arabidopsis mutant, Chlamydomonas cpsfl1 cannot grow photoautotrophically, and it is extremely light sensitive. These phenotypes might be explained by a defect in vesicle-mediated transfer of carotenoids from the envelope, which could impact early steps in carotenoid biosynthesis through feedback inhibition (51).
Putative components involved in chloroplast vesicle trafficking have been identified by previous genetic studies in Arabidopsis (52–54). Mutants such as fzl (25), thf1 (26), cprabA5e (23), and sco2 (24) were shown to accumulate chloroplast vesicles, whereas vipp1 mutants have been reported to lack chloroplast vesicles (4), similar to cpsfl1. The vipp1 and cpsfl1 mutants also share a similar pale-green phenotype and inability to grow photoautotrophically on soil, and both exhibit altered thylakoid morphology and high chlorophyll fluorescence. Like CPSFL1, VIPP1 is a lipid-binding protein that binds phosphatidylinositides in a protein−lipid blot overlay assay, and it binds PI4P in liposomes and promotes membrane tubulation in vitro (55, 56). These similarities suggest a possible functional relationship between CPSFL1 and VIPP1, which is an important subject for future work.
Lipid Binding and Transfer Activities of CPSFL1.
Sec14 domain proteins are also referred to as CRAL_TRIO proteins, and members of this family include proteins that bind lipids, vitamins, and retinoids (57–63). We have shown that CPSFL1 binds to PA and PIPs in vitro (Fig. 5 A and B), and we observed partial colocalization of PA (marked by recombinant TGD2) and CPSFL1-FLAG by superresolution microscopy of isolated chloroplasts (Fig. 5E). Binding of CPSFL1 to these lipids in the envelope could be involved in recruiting other chloroplast vesicle components. Indeed, PIPs have very important roles as signaling molecules and in mediating the recruitment of core components of the cytosolic vesicle trafficking machinery (64–67), such as GTPases from the Ypt/Rab and Ras family as well as Arf (ADP ribosylation factors) and GTPase-activating proteins. A variety of putative components involved in chloroplast vesicle transport, including GTPases, have been described by bioinformatic and proteomic studies of plastids (23, 54, 55, 68).
In chloroplasts, the understanding of the function of phosphatidylinositide metabolism is still at the beginning. Plastid phosphatidylinositides have mostly been analyzed in bulk as PI and found to represent between 1% and 5% of total lipids of envelope and thylakoid membranes (69). Only one study has detected PIP by radiolabeling experiments and found it, preferentially, in the outer leaflet of the chloroplast envelope (70). Mutants lacking enzymes for PI4P synthesis have more chloroplasts per cell, suggesting that PI4P negatively regulates chloroplast division (71). PI4P might also be a minor lipid in the chloroplast inner envelope membrane, where it could be bound by stromal proteins such as CPSFL1 and VIPP1 (56, 57).
Materials and Methods
Plant Material, Plant Generation, and Growth Conditions.
The cpsfl1 insertion line (SALK_116713C) was obtained from the Arabidopsis Biological Research Center. The T-DNA insertion into CPSFL1 is in the Col-0 background. Col-0 plants were used as WT. For plants expressing FLAG-tagged or YFP-tagged CPSFL1 proteins, the cDNA of CPSFL1 was cloned in frame with a C-terminal FLAG-tag into the pEARLEYGATE100 vector or into the pEARLEYGATE101 vector for C-terminal YFP fusion, both under control of the 35S promoter (72). For complementation experiments, heterozygous cpsfl1 mutant plants were transformed with a construct encoding FLAG-tagged CPSFL1 by floral dipping (73). For CPSFL1-YFP−expressing plants, WT plants were transformed. After transformation, plants were transferred to a growth chamber, and seeds were collected after 3 wk. Individual transgenic plants were selected by their resistance to ammonium glufosinate (Basta). Complementation and the presence and expression of the transgene were confirmed by PCR, RT-PCR, and protein immunoblot analyses for the FLAG epitope or fluorescence microscopy for YFP tag.
Plants were grown either on soil or on Murashige and Skoog (MS) medium supplemented with 1% sucrose in a growth chamber (day period of 12 h at 20 °C, 80 µmol photons per m−2⋅s−1 on leaf surfaces; night period of 12 h at 15 °C) or transferred once a week to fresh MS medium supplemented with 2% sucrose. For heterotrophic growth, light intensities were decreased to 10 µmol photons per m−2⋅s−1 for all genotypes.
Nucleic Acid Analysis.
For genotyping and transgene detection, the plant Phire direct PCR kit (ThermoFisher) was used. The cpsfl1 knockout lines were identified by using specific gene/gene and gene/T-DNA primer pairs (SI Appendix, Table S1). For transgene detection, a gene/FLAG tag or gene/YFP primer pair was used (SI Appendix, Table S1).
For cDNA synthesis, total RNA was extracted from leaf tissue with the RNeasy plant mini kit (Qiagen) according to the manufacturer’s instructions. The cDNA was prepared from 1 µg of total RNA applying the iScript cDNA synthesis kit (Bio-Rad) following the manufacturer's instructions. For RT-PCR, cDNA was diluted 100-fold in a final reaction mix and genes (ACTIN 9 [At2g42090] and CPSFL1 [At5g63060]) were amplified according to the manufacturer’s protocol either with Phire II Hot Start DNA polymerase (Qiagen) for genotyping or Phusion Polymerase (Finzymes) for cloning. Thermal cycling consisted of an initial step at 95 °C for 3 min, followed by 30 cycles of 10 s at 95 °C, 30 s at 58 °C, and 30 s at 72 °C.
Protoplast and Chloroplast Isolation, Chloroplast Fractionation, Protein Precipitation, and Total Protein Extracts.
Protoplasts were isolated according to ref. 29. Fully expanded leaves of Arabidopsis were excised and suspended in enzyme solution (1% [wt/vol] cellulase [Onozuka R10; Yakult], 0.5% [wt/vol] Pectolyase Y-23 [Kyowa Chemical], 400 mM mannitol, 10 mM CaCl2, 20 mM KCl, 5 mM ethylene glycol bis[β-aminoethyl ether]-N,N,N′,N′-tetraacetic acid [EGTA], and 20 mM MES, pH 5.7). Following incubation overnight in the dark and at room temperature (RT) with gentle agitation, protoplasts were collected by centrifugation at 800 × g for 1 min. Intact chloroplasts were obtained from leaves of 4-wk- to 5-wk-old plants. Following homogenization of leaf material in homogenization buffer (330 mM sorbitol, 20 mM Tricine/NaOH [pH 7.6], 5 mM EGTA, 5 mM [ethylenedinitrilo]tetraacetic acid [EDTA], 10 mM NaCO3, 0.1% [wt/vol] bovine serum albumin [BSA], 330 mg/L ascorbate), the sample was centrifuged (5 min, 2,000 × g), and the pellet was resuspended in resuspension buffer (300 mM sorbitol, 20 mM Hepes/KOH [pH 7.6], 5 mM MgCl2, 2.5 mM EDTA). Intact chloroplasts were purified by centrifugation (3,000 × g) through 80%/40% Percoll (GE Healthcare) step gradient and a single wash in resuspension buffer.
For chloroplast fractionation, plastids were subjected to hypotonic lysis in TE buffer (10 mM Tris, pH 7.5, and 2 mM EDTA). The lysate was loaded onto an 8.0-mL sucrose step gradient composed of 0.8 mL 1.2 M, 1.0 mL 1.0 M, and 1.0 mL of 0.46 M sucrose dissolved in TE buffer. The gradient was centrifuged at 200,000 × g for 1 h. The stroma, envelope membrane, and thylakoid fractions were collected from the supernatant (stroma), 0.46 M/1.0 M sucrose interface (envelope), and the pellet (thylakoids), respectively. Chloroplasts or chloroplast subfractions were precipitated using methanol/chloroform according to ref. 74 and resuspended in equal volumes of sample buffer containing 125 mM Tris-Cl, pH 6.8, 4% [wt/vol] sodium dodecyl sulfate (SDS), 10% [vol/vol] glycerol, 100 mM dithiothreitol (Sigma), and 0.05% [wt/vol] bromophenol blue.
For total protein extracts, leaf samples were frozen in liquid nitrogen, homogenized to tissue powder, and thawed in sample buffer. Following 1 h incubation at RT, debris was removed by centrifugation, and the supernatant was used for further analysis.
Pigment Analysis.
Pigments were extracted and analyzed by HPLC as described previously (75).
SDS/Polyacrylamide Gel Electrophoresis, Immunoblotting, Antibodies, and Antibody Conjugation.
Protein samples were separated by SDS/polyacrylamide gel electrophoresis (PAGE) using 15% precast Gels (Biorad) according to the manufacturer’s instructions. For immunoblotting, the separated proteins were transferred from SDS/PAGE gels to polyvinylidene difluoride (PVDF) membranes (0.45 μm, Millipore) according to the manufacturer’s instructions. Protein transfer was assessed as uniform and complete by blot staining with Ponceau S Staining Solution (0.1% [wt/vol] Ponceau S in 5% [vol/vol] acetic acid). Following destaining with tris-buffered saline with Tween 20 (TBST) (50 mM Tris⋅HCl, pH 7.4, 200 mM NaCl, 0.2% [vol/vol] Tween 20), PVDF membranes were blocked for 60 min in blocking solution (TBST, containing 5% [wt/vol] nonfat dry milk [Safeway]), prior to overnight incubation with primary antibodies. Following three washes with TBST for 5 min each, membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies diluted 1:25,000 in blocking solution. Immunoreactivity was visualized by chemiluminescent detection using SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher) and an enhanced chemiluminescence (ECL) reader (Biorad).
Antibodies specific against the CSAETVRKEYFTEETLPSNFRS epitope of the CPSFL1 proteins were generated by YenZym Antibodies, LLC. Toc34 (Arabidopsis specific) and Tic40, Lhcb2, AtpB, PsaD, Cytb6, and D2 antibodies were obtained from Agrisera. FLAG and Avidin antibodies from rabbit and mouse were obtained from ThermoFisher. PI4P antibodies were obtained from abcam. HRP-labeled secondary antibodies from mouse were obtained from ThermoFisher. HRP-labeled secondary antibodies from rabbit were obtained from GE Healthcare. Fluorescent secondary antibodies (Alexa405 anti-mouse and Alexa488 anti-rabbit) were obtained from Invitrogen.
Generation and Purification of Recombinant Proteins.
For expression of recombinant proteins, N-terminal His6-tagged and C-terminal FLAG-tagged versions of CPSFL1 (AT5g63060), Ssh2P (AT5G47730), yeast SEC14 (GI_855103), a variant of CPSFL1 lacking the CRAL_TRIO_N domain (AT5g63060), and TGD2 (At3g20320) were inserted in pET151 (Invitrogen) and introduced into Escherichia coli BL21 (DB3.0 pLysS; Rosetta). The resulting strains were grown in 2xYT medium containing carbenicillin (50 mg/mL). Expression of recombinant proteins was induced at an OD 600 of 0.5 to 0.8 by the addition of 0.5 mM isopropyl-1-thio-β-D-thiogalactopyranoside (IPTG; Fisher). After induction, synthesis of recombinant proteins was allowed to proceed for 3 h at 30 °C. Bacteria were harvested by centrifugation (20 min, 4,000 × g, 4 °C), and the pellet was either used directly or stored at −80 °C. Recombinant proteins were purified using the N-terminal His-tag. After resuspension of cells in protein extraction buffer (50 mM NaH2PO4, 2% [vol/vol] Triton X-100, 300 mM NaCl, 10 mM imidazole, 1 mM phenylmethylsulfonylfluoride [pH 7.5] with NaOH), the samples were incubated at 4 °C for 30 min. The lysates were clarified by centrifugation (16,100 × g; 4 °C, 30 min). The supernatant including the recombinant proteins was loaded onto an immobilized metal-affinity chromatography column using a cobalt-nitriloacetic acid matrix (Co-NTA; ThermoFisher). The resin was washed with 50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole (pH 7.5), and bound proteins were eluted with 50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole (pH 7.5). Fractions containing recombinant proteins were identified using SDS/PAGE, pooled, and stored in 10% glycerol at −80 °C. Desalting, buffer exchange, and dialysis of protein solutions was performed prior to subsequent analysis using 10K Amicon Ultra-0.5 mL Centrifugal Filters (Millipore).
Lipid Blot Assay.
Lipid blots with equal quantities of lipids immobilized by spotting on a nylon membrane were purchased from Echelon. The membrane was blocked with 3% fatty acid-free bovine serum in TBST (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, and 0.1% vol/vol Tween-20) for 1 h at RT. Purified proteins were diluted in blocking solution to 0.2 mg/mL. The membrane was incubated with the proteins overnight at 4 °C on a rocking platform. After washing six times for 5 min with TBST, lipid blots were incubated with a 1:4,000 dilution of rabbit polyclonal FLAG antibody (ThermoFisher) in blocking reagent for 1 h. After washing six times for 5 min with TBST, the membrane was incubated with donkey anti-rabbit secondary antibody (GE Healthcare) diluted 1:25,000 into blocking reagent for 1 h. Following six additional washing steps with TBST for 5 min each, immunoreactivity was visualized using SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher) on an ECL reader (Biorad).
Liposome Preparation.
PC and indicated mol % of additional lipids (PA, PIP, MGDG, DGDG, and SQDG) were dissolved in methanol/chloroform (1:1) and dried into thin films in glass vials using nitrogen flow. Subsequent lipid films were dried for at least 16 h under vacuum at RT. Liposomes were formed by rehydrating the lipid films in rehydration buffer (10 mM Hepes pH 7.4, 250 mM NaCl with 1 M sucrose [heavy liposomes]) or without sucrose (light liposomes) to the total lipid concentration of 4 mM. Following 20 freeze/thaw cycles in liquid nitrogen, liposomes were filtered 21 times through a 400-nm carbonate filter (Millipore) and subsequently used for experiments.
In Vitro Lipid-Binding Assay.
Liposomes containing 10 mol % (0.20 mM) nitrobenzoxadiazole (NBD)-PA, boron-dipyrromethene (BODIPY)-PC, SQDG, MGDG, DGDG or BODIPY-PI4P were incubated with or without recombinant CPSFL1 (20 μM) in 200 μL of assay buffer (10 mM Hepes pH 7.4, 250 mM NaCl) at 25 °C for 30 min. After centrifugation at 400,000 × g for 30 min (SW60) to pellet the liposomes, the lipids in the supernatant were analyzed against 10% of the lipids in the pellet by TLC. CPSFL1 abundance in supernatant and pellet fractions was also analyzed by SDS/PAGE. Lipids were visualized by fluorescence (NBD-PA, BODIPY-PC, and BODIPY-PIP) or CuSO4 charring (MGDG, DGDG, SQDG) according to ref. 76. Therefore, TLC plates were submersed in CuSO4 solution (10% cupric sulfate [wt/vol] in 8% aqueous phosphoric acid [vol/vol]) and allowed to dry prior to charring at 180 °C for 8 min. Lipids were identified and quantified relative to the 10% of the individual lipids used for input liposomes (applied on the same TLC plates). Assays were performed at least in triplicates.
Biochemical Lipid Transfer Assay.
“Heavy” acceptor liposomes were prepared from PC (Avanti Polar Lipids) with and without 10 mol % NBD-PA (Avanti) in the assay buffer containing 1 M sucrose. The liposomes were pelleted at 16,100 × g and washed three times in the assay buffer. The “light” donor liposomes (200 nm) with 10 mol % PI4P and/or NBD-PA were prepared in assay buffer without sucrose and concentrated using discontinuous sucrose gradient between 0.8 M and 0.4 M sucrose in resuspension buffer. The donor and acceptor liposomes (corresponding to 2.0 mM and 1.0 mM total lipids, respectively) were incubated in 200 μL of assay buffer at 25 °C for 30 min with and without recombinant CPSFL1 at a sucrose concentration of 0.95 M and a protein concentration of 1.0 μM. The acceptor liposomes were then pelleted at 16,100 × g for 15 min and washed three times in 10 mM Hepes pH 7.4, 150 mM NaCl, and their lipid composition was analyzed by TLC (lipids). Ten percent of the donor liposomes were simultaneously separated to identify lipids and quantify the transfer. Lipids were visualized by using ultraviolet light in an ECL reader (Biorad).
TLC.
Lipids were extracted according to ref. 77. Samples were mixed with 10-fold excess of extraction buffer containing methanol, chloroform, and formic acid (20:10:1). After 5-min incubation, a fivefold excess of 0.2 M phosphoric acid and 1 M KCl solution was added and mixed. Samples were centrifuged for 1 min at 13,000 × g at RT for phase separation. Lipids dissolved in the lower chloroform phase were dried under N2 stream and dissolved in chloroform prior to spotting on TLC plates. For general separation of lipids, a solvent mixture of acetone/toluene/water (91/31/7.5) was used. To separate phosphatidylinositides, a mixture of chloroform/methanol/water/concentrated NH4 (45/35/8.5/1.5 [vol/vol]) was used as solvent system. Following separation, lipids were visualized either by fluorescence or by charring at 180 °C for 8 min following dipping and drying of plates into 10% cupric sulfate in 8% aqueous phosphoric acid according to ref. 76.
Immunofluorescence, Confocal, and Super-Resolution Microscopy.
For immunofluorescence labeling, chloroplast suspensions were fixed in 4% paraformaldehyde (PFA) in PBS containing 1% dimethyl sulfoxide for 30 min at RT and subsequently rinsed three times for 10 min in 1× PBS. Unspecific binding sites were blocked by incubation of samples in 1× PBS with Tween 20 (PBST) with 2% dry milk for 1 h at RT. Subsequently, primary antibodies against CPSFL1-FLAG (anti-FLAG, Rabbit) and recombinant biotin-labeled TGD2 were added in a dilution of 1:100 to the samples and incubated for 1 h at RT. For PI4P detection, PI4P-specific antibody (from Mouse) were added in a 1:100 dilution. After three further rinses in PBST for 10 min each, AlexaFluor488 fluorescent anti-rabbit secondary antibodies and AlexaFluor405-labeled Avidin conjugates or Alexa Fluor405-labeled anti-mouse secondary antibodies were added in a 1:200 dilution, and samples were incubated for another hour at RT. Following three rinses with PBST and three rinses with PBS for 10 min each, chloroplasts were mounted on Poly-Lysine−coated coverslips using VectaShield mounting medium and observed by superresolution microscopy on a Zeiss Elyra PS.1 microscope (Zeiss).
For visualization of CPSFL1-YFP in protoplasts, protoplasts were mounted on coverslips in 400 mM mannitol, 10 mM CaCl2, 20 mM KCl, 5 mM EGTA, and 20 mM MES, pH 5.7, and imaged using an inverted Zeiss LSM710.
TEM.
Leaves of WT, cpsfl1 mutant plants, and CPSFL1 overexpressing lines were cut into 1-mm pieces prior to fixation overnight at 4 °C in 2% glutaraldehyde and 4% PFA in 0.1 M sodium phosphate buffer (pH 7.2). For temperature-dependent vesicle accumulation, according to ref. 20, leaves were preincubated for at least 1 h at 4 °C prior to fixation. The fixed leaf pieces were washed three times for 20 min in 0.1 M sodium phosphate buffer (pH 7.2) at 4 °C and then contrasted by incubation in 1% OsO4 overnight at 4 °C. After washing in 0.1 M sodium phosphate buffer (pH 7.2), samples were dehydrated in a graded ethanol series and embedded in LR White resin according to standard procedures. Ultrathin sections were cut with a diamond knife and put on formvar-coated single-hole grids. Sections were stained by incubation in 2% aqueous uranyl acetate for 10 min, followed by 5 min of incubation in lead citrate. Tomographic reconstruction was done with TrekEM2 software (Fiji) on 70-nm-thick serial sections.
For immunogold labeling, samples were fixed overnight at 4 °C in 4% PFA in PBS (vol/vol). Following three rinses with PBST, samples were embedded, sectioned, and mounted as described above. Unspecific antibody binding was decreased by incubation of samples in blocking buffer (PBST containing 2% BSA and 0.1% fish gelatin [Sigma]) for 1 h at RT. Antigens were detected by incubation in blocking buffer containing either anti-FLAG (1:50), anti-CPSFL1 (1:200), or anti-LHCB2 (1:100) for 1 h at RT. Excess antibodies were removed by six rinses with PBST. On section, primary antibodies were labeled at RT by incubation for 1 h in blocking buffer containing 20-nm gold particle-conjugated Protein A (1:10). Following six rinses with PBST and three rinses with ddH2O, samples were contrasted with 2% aqueous uranyl acetate for 10 min.
Yeast Strain and Growth.
The yeast strain used in this study, CTY1-1A (MATa ura3-52 lys2-801 Δhis3-200 s14-1ts), was kindly provided by Vytas A. Bankaitis, Texas A&M University, College Station, TX. CPSFL1 cDNA sequences encoding the full or the processed protein (i.e., lacking the cTP) were cloned by BamHI and EcoRI restriction enzyme digestion and ligation into galactose-inducible plasmid pRS426pGal-URA3, kindly provided by Randy Schekman, University of California, Berkeley, CA. Yeast transformation was performed as described in ref. 78. Yeast cells were grown on uracil drop-out synthetic media with 2% dextrose or with 2% galactose. Plates were incubated at the indicated temperatures and examined after 2 d to 6 d.
Data Availability.
All data presented in the paper are available in the main text and SI Appendix. Biological materials are available from the corresponding authors upon request.
Supplementary Material
Acknowledgments
We thank Vytas Bankaitis and Randy Schekman for providing materials, and Masakazu Iwai for critical reading of the manuscript. A.P.H. thanks Prof. Dr. Ralph Bock for use of equipment and consumables at the Max Planck Institute for Molecular Plant Physiology. This work was supported by the US Department of Energy, Office of Science, through the Photosynthetic Systems program in the Office of Basic Energy Sciences. K.K.N. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916946117/-/DCSupplemental.
References
- 1.Eberhard S., Finazzi G., Wollman F.-A., The dynamics of photosynthesis. Annu. Rev. Genet. 42, 463–515 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Wise R., Hoober J., The Structure and Function of Plastids (Springer, 2007). [Google Scholar]
- 3.Kobayashi K., Kondo M., Fukuda H., Nishimura M., Ohta H., Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 17216–17221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroll D., et al. , VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl. Acad. Sci. U.S.A. 98, 4238–4242 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer J., et al. , The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403, 203–207 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Dörmann P., Hoffmann-Benning S., Balbo I., Benning C., Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7, 1801–1810 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dörmann P., Balbo I., Benning C., Arabidopsis galactolipid biosynthesis and lipid trafficking mediated by DGD1. Science 284, 2181–2184 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Falbel T. G., Meehl J. B., Staehelin L. A., Severity of mutant phenotype in a series of chlorophyll-deficient wheat mutants depends on light intensity and the severity of the block in chlorophyll synthesis. Plant Physiol. 112, 821–832 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarvis P., et al. , Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc. Natl. Acad. Sci. U.S.A. 97, 8175–8179 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joyard J., et al. , Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol. Plant 2, 1154–1180 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Rast A., Heinz S., Nickelsen J., Biogenesis of thylakoid membranes. Biochim. Biophys. Acta 1847, 821–830 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Pribil M., Labs M., Leister D., Structure and dynamics of thylakoids in land plants. J. Exp. Bot. 65, 1955–1972 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Mechela A., Schwenkert S., Soll J., A brief history of thylakoid biogenesis. Open Biol. 9, 180237 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muehlethaler K., Frey-Wyssling A., Development and structure of proplastids [in German]. J. Biophys. Biochem. Cytol. 6, 507–512 (1959). [PMC free article] [PubMed] [Google Scholar]
- 15.Liang Z., et al. , Thylakoid-bound polysomes and a dynamin-related protein, FZL, mediate critical stages of the linear chloroplast biogenesis program in greening Arabidopsis cotyledons. Plant Cell 30, 1476–1495 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Wettstein D., The effect of genetic factors on the submicroscopic structures of the chloroplast. J. Ultrastruct. Res. 3, 234–240 (1959). [Google Scholar]
- 17.Carde J. P., Joyard J., Douce R., Electron microscopic studies of envelope membranes from spinach plastids. Biol. Cell. 44, 315–324 (1982). [Google Scholar]
- 18.Morré D. J., Morré J. T., Morré S. R., Sundqvist C., Sandelius A. S., Chloroplast biogenesis. Cell-free transfer of envelope monogalactosylglycerides to thylakoids. Biochim. Biophys. Acta 1070, 437–445 (1991). [DOI] [PubMed] [Google Scholar]
- 19.Morré D. J., Minnifield N., Paulik M., Identification of the 16 °C compartment of the endoplasmic reticulum in rat liver and cultured hamster kidney cells. Biol. Cell 67, 51–60 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westphal S., Soll J., Vothknecht U. C., A vesicle transport system inside chloroplasts. FEBS Lett. 506, 257–261 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Andersson M. X., Kjellberg J. M., Sandelius A. S., Chloroplast biogenesis. Regulation of lipid transport to the thylakoid in chloroplasts isolated from expanding and fully expanded leaves of pea. Plant Physiol. 127, 184–193 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersson M. X., Sandelius A. S., A chloroplast-localized vesicular transport system: A bio-informatics approach. BMC Genomics 5, 40 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karim S., et al. , A novel chloroplast localized Rab GTPase protein CPRabA5e is involved in stress, development, thylakoid biogenesis and vesicle transport in Arabidopsis. Plant Mol. Biol. 84, 675–692 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Tanz S. K., et al. , The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LHCB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings. Plant J. 69, 743–754 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Gao H., Sage T. L., Osteryoung K. W., FZL, an FZO-like protein in plants, is a determinant of thylakoid and chloroplast morphology. Proc. Natl. Acad. Sci. U.S.A. 103, 6759–6764 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q., et al. , Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 136, 3594–3604 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang S., Gao L., Blanchoin L., Staiger C. J., Heterodimeric capping protein from Arabidopsis is regulated by phosphatidic acid. Mol. Biol. Cell 17, 1946–1958 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia C., Khan N. Z., Nannmark U., Aronsson H., The chloroplast protein CPSAR1, dually localized in the stroma and the inner envelope membrane, is involved in thylakoid biogenesis. Plant J. 63, 73–85 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Zhang L., Kato Y., Otters S., Vothknecht U. C., Sakamoto W., Essential role of VIPP1 in chloroplast envelope maintenance in Arabidopsis. Plant Cell 24, 3695–3707 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S., Shen G., Li Z., Golbeck J. H., Bryant D. A., Vipp1 is essential for the biogenesis of Photosystem I but not thylakoid membranes in Synechococcus sp. PCC 7002. J. Biol. Chem. 289, 15904–15914 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L., Sakamoto W., Possible function of VIPP1 in maintaining chloroplast membranes. Biochim. Biophys. Acta 1847, 831–837 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Bankaitis V. A., Aitken J. R., Cleves A. E., Dowhan W., An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 347, 561–562 (1990). [DOI] [PubMed] [Google Scholar]
- 33.Savage L. J., Imre K. M., Hall D. A., Last R. L., Analysis of essential Arabidopsis nuclear genes encoding plastid-targeted proteins. PLoS One 8, e73291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huijbregts R. P., Topalof L., Bankaitis V. A., Lipid metabolism and regulation of membrane trafficking. Traffic 1, 195–202 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Lee M. C. S., et al. , Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell 122, 605–617 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Wang C.-W., Hamamoto S., Orci L., Schekman R., Exomer: A coat complex for transport of select membrane proteins from the trans-Golgi network to the plasma membrane in yeast. J. Cell Biol. 174, 973–983 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lev S., Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat. Rev. Mol. Cell Biol. 11, 739–750 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Phillips S. E., et al. , The diverse biological functions of phosphatidylinositol transfer proteins in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 41, 21–49 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Mousley C. J., Tyeryar K. R., Vincent-Pope P., Bankaitis V. A., The Sec14-superfamily and the regulatory interface between phospholipid metabolism and membrane trafficking. Biochim. Biophys. Acta 1771, 727–736 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curwin A. J., Fairn G. D., McMaster C. R., Phospholipid transfer protein Sec14 is required for trafficking from endosomes and regulates distinct trans-Golgi export pathways. J. Biol. Chem. 284, 7364–7375 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martínez-Alonso E., Egea G., Ballesta J., Martínez-Menárguez J. A., Structure and dynamics of the Golgi complex at 15 °C: Low temperature induces the formation of Golgi-derived tubules. Traffic 6, 32–44 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Bankaitis V. A., Mousley C. J., Schaaf G., The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem. Sci. 35, 150–160 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sha B., Phillips S. E., Bankaitis V. A., Luo M., Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature 391, 506–510 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Kearns M. A., et al. , Novel developmentally regulated phosphoinositide binding proteins from soybean whose expression bypasses the requirement for an essential phosphatidylinositol transfer protein in yeast. EMBO J. 17, 4004–4017 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tripathi A., Nile A. H., Bankaitis V. A., Sec14-like phosphatidylinositol-transfer proteins and diversification of phosphoinositide signalling outcomes. Biochem. Soc. Trans. 42, 1383–1388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cleves A. E., Novick P. J., Bankaitis V. A., Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J. Cell Biol. 109, 2939–2950 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnabl M., et al. , Subcellular localization of yeast Sec14 homologues and their involvement in regulation of phospholipid turnover. Eur. J. Biochem. 270, 3133–3145 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Novick P., Field C., Schekman R., Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21, 205–215 (1980). [DOI] [PubMed] [Google Scholar]
- 49.Cleves A. E., et al. , Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell 64, 789–800 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strugger S., Über den Bau der Proplastiden und Chloroplasten. Naturwissenschaften 37, 166–167 (1950). [Google Scholar]
- 51.García-Cerdán J. G., et al. , Chloroplast Sec14-like 1 (CPSFL1) is essential for normal chloroplast development and affects carotenoid accumulation in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindquist E., Aronsson H., Chloroplast vesicle transport. Photosynth. Res. 138, 361–371 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karim S., Aronsson H., The puzzle of chloroplast vesicle transport—Involvement of GTPases. Front. Plant Sci. 5, 472 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khan N. Z., Lindquist E., Aronsson H., New putative chloroplast vesicle transport components and cargo proteins revealed using a bioinformatics approach: An Arabidopsis model. PLoS One 8, e59898 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohnishi N., Zhang L., Sakamoto W., VIPP1 involved in chloroplast membrane integrity has GTPase activity in vitro. Plant Physiol. 177, 328–338 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Theis J., et al. , VIPP1 rods engulf membranes containing phosphatidylinositol phosphates. Sci. Rep. 9, 8725 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kono N., et al. , Impaired α-TTP-PIPs interaction underlies familial vitamin E deficiency. Science 340, 1106–1110 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Panagabko C., et al. , Ligand specificity in the CRAL-TRIO protein family. Biochemistry 42, 6467–6474 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Phillips S. E., et al. , Yeast Sec14p deficient in phosphatidylinositol transfer activity is functional in vivo. Mol. Cell 4, 187–197 (1999). [DOI] [PubMed] [Google Scholar]
- 60.Min K. C., Kovall R. A., Hendrickson W. A., Crystal structure of human alpha-tocopherol transfer protein bound to its ligand: Implications for ataxia with vitamin E deficiency. Proc. Natl. Acad. Sci. U.S.A. 100, 14713–14718 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tahotna D., Holic R., Poloncova K., Simockova M., Griac P., Phosphatidylcholine transfer activity of yeast Sec14p is not essential for its function in vivo. Biochim. Biophys. Acta 1771, 83–92 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Intres R., Goldflam S., Cook J. R., Crabb J. W., Molecular cloning and structural analysis of the human gene encoding cellular retinaldehyde-binding protein. J. Biol. Chem. 269, 25411–25418 (1994). [PubMed] [Google Scholar]
- 63.Saari J. C., Nawrot M., Stenkamp R. E., Teller D. C., Garwin G. G., Release of 11-cis-retinal from cellular retinaldehyde-binding protein by acidic lipids. Mol. Vis. 15, 844–854 (2009). [PMC free article] [PubMed] [Google Scholar]
- 64.Ghosh R., Bankaitis V. A., Phosphatidylinositol transfer proteins: Negotiating the regulatory interface between lipid metabolism and lipid signaling in diverse cellular processes. Biofactors 37, 290–308 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Fang M., et al. , Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 15, 6447–6459 (1996). [PMC free article] [PubMed] [Google Scholar]
- 66.Li X., et al. , Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J. Cell Biol. 157, 63–77 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yanagisawa L. L., et al. , Activity of specific lipid-regulated ADP ribosylation factor-GTPase-activating proteins is required for Sec14p-dependent Golgi secretory function in yeast. Mol. Biol. Cell 13, 2193–2206 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kleffmann T., et al. , The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14, 354–362 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Block M. A., Douce R., Joyard J., Rolland N., Chloroplast envelope membranes: A dynamic interface between plastids and the cytosol. Photosynth. Res. 92, 225–244 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siegenthaler P. A., Müller M. O., Bovet L., Evidence for lipid kinase activities in spinach chloroplast envelope membranes. FEBS Lett. 416, 57–60 (1997). [DOI] [PubMed] [Google Scholar]
- 71.Okazaki K., Miyagishima S. Y., Wada H., Phosphatidylinositol 4-phosphate negatively regulates chloroplast division in Arabidopsis. Plant Cell 27, 663–674 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Earley K. W., et al. , Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Zhang X., Henriques R., Lin S.-S., Niu Q.-W., Chua N.-H., Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Wessel D., Flügge U. I., A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 (1984). [DOI] [PubMed] [Google Scholar]
- 75.Tran P. T., Sharifi M. N., Poddar S., Dent R. M., Niyogi K. K., Intragenic enhancers and suppressors of phytoene desaturase mutations in Chlamydomonas reinhardtii. PLoS One 7, e42196 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Churchward M. A., Brandman D. M., Rogasevskaia T., Coorssen J. R., Copper (II) sulfate charring for high sensitivity on-plate fluorescent detection of lipids and sterols: Quantitative analyses of the composition of functional secretory vesicles. J. Chem. Biol. 1, 79–87 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Z., Benning C., Arabidopsis thaliana polar glycerolipid profiling by thin layer chromatography (TLC) coupled with gas-liquid chromatography (GLC). J. Vis. Exp. 2518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gietz R. D., Schiestl R. H., Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 1–4 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in the paper are available in the main text and SI Appendix. Biological materials are available from the corresponding authors upon request.