Abstract
In skeletal muscle, proteins of the calcium release complex responsible for the excitation-contraction (EC) coupling are exclusively localized in specific reticulum–plasma membrane (ER-PM) contact points named triads. The CRC protein triadin (T95) is localized in the sarcoplasmic reticulum (SR) subdomain of triads where it forms large multimers. However, the mechanisms leading to the steady-state accumulation of T95 in these specific areas of SR are largely unknown. To visualize T95 dynamics, fluorescent chimeras were expressed in triadin knockout myotubes, and their mobility was compared with the mobility of Sec61β, a membrane protein of the SR unrelated to the EC coupling process. At all stages of skeletal muscle cells differentiation, we show a permanent flux of T95 diffusing in the SR membrane. Moreover, we find evidence that a longer residence time in the ER-PM contact point is due to the transmembrane domain of T95 resulting in an overall triad localization.
INTRODUCTION
Skeletal muscle contraction is triggered by a massive calcium release from intracellular stores upon plasma membrane depolarization. This phenomenon, known as the excitation-contraction (EC) coupling, occurs in specific sites of skeletal muscle, the triads. Each triad consists of two reticulum terminal cisternae, the junctional sarcoplasmic reticulum (jSR), flanking a single invagination of the plasma membrane, the transverse-tubule (T-tubule) (Flucher, 1992). From a structural point of view, triads are contact points between T-tubule membranes and the jSR, where proteins of the multimolecular calcium release complex (CRC) are located. The sharp organization of T-tubule and jSR membranes is of outmost importance because it allows a physical cross-talk between the two main components of the CRC that are each anchored in a different membrane compartment: the voltage-gated channel dihydropyridine receptor (DHPR) in the T-tubule and the intracellular calcium channel ryanodine receptor 1 (RyR1) in the jSR (Franzini-Armstrong and Jorgensen, 1994). As a consequence of this organization, the structural modification of DHPR induced by membrane depolarization can mechanically trigger the opening of RyR1 (Marty et al., 1994; Flucher and Franzini-Armstrong, 1996). In addition to both channels, the CRC includes several regulatory proteins such as calsequestrin, junctin, triadin, which can modulate RyR1 function or the organization of the molecular complex. Interestingly in skeletal muscles, all proteins of the CRC are exclusively localized in the triad membranes (Flucher, 1992), without a clear view on the underlying mechanisms.
Muscle fibers are long and multinucleated cells with a specific endomembrane system (Villa et al., 1993). The reticulum of a muscle cell is a continuum of membranes spread throughout the entire fiber and wrapping the actomyosin fibrils (Ogata and Yamasaki, 1997). It is specialized into subdomains either ubiquitously present in mammalian cells, such as rough endoplasmic reticulum for protein synthesis, or specific to the calcium cycle of the fiber, such as longitudinal SR and jSR (Rahkila et al., 1996; Kaisto and Metsikkö, 2003; Sorrentino, 2011). In the context of a highly differentiated membrane system, the traffic of CRC proteins to reach the jSR after their synthesis has so far remained controversial (Barone et al., 2015; Fu and Hong, 2016), with previous work suggesting either vesicular trafficking (Nori et al., 2004) or diffusion within the SR (McFarland et al., 2010). Moreover, once proteins of the CRC are localized in the jSR, the specific mechanisms that may confine them in this subcompartment are also unclear. Previous studies suggested that building of multiple protein–protein interactions within CRC and with external partners could contribute to this anchoring in jSR and that the localization of CRC proteins within jSR during differentiation was accompanied by a drastic reduction in their mobility (Cusimano et al., 2009). Despite these data, the mechanisms leading to the restricted localization of CRC components in membrane subcompartments such as the jSR are still elusive, as well as their specific dynamics within a mature SR network as compared with non jSR reticulum protein.
To better understand the molecular bases leading to the CRC formation and function, we have studied here the dynamic behavior of the triadin protein and compared it to another SR protein unrelated to EC coupling, Sec61β. Triadin (Trisk95 or T95), is the largest of the four isoforms obtained from alternative splicing of the TRDN gene (Marty et al., 2000; Vassilopoulos et al., 2005). It is a type II transmembrane (TM) protein composed of a short N-terminal domain, a single TM domain, and a long intraluminal domain that is able to form multimers and directly link calsequestrin and RyR1 (Knudson et al., 1993; Kobayashi et al., 2000; Lee et al., 2004; Marty et al., 2009). T95 is specifically expressed in the jSR side of skeletal muscle triads, where it plays a regulatory role on the RyR1 channel activity (Rezgui et al., 2005), and a structural role on jSR membranes (Fourest-Lieuvin et al., 2012). It has been proposed that T95 anchoring at the triad could represent a first event in the building of the CRC complex (Cusimano et al., 2009) and that some T95 domains may be responsible for its retention in the jSR (Rossi et al., 2014). To directly explore T95 dynamics and anchoring at the triad, fluorescent versions of T95 have been expressed in cultured myotubes and adult fibers from TRDN knockout (KO) animals. The accumulation of T95 in triads has been visualized by a photoactivatable form of the molecule. Our results demonstrate the existence of a constant flux of T95 toward and from jSR membranes, that is coupled to a retention mechanism driven by the TM domain of T95 in the jSR to create the steady-state localization of the molecule.
RESULTS
Triad organization and T95 behavior during cell differentiation
To study T95 dynamic inside a mature SR membrane system, fluorescent versions of T95 were expressed in primary myotubes cultured from TRDN KO mice, thus avoiding competition between the recombinant and endogenous proteins for localization in the jSR, a subcompartment of possibly limited size. The cultured myotubes were observed at two developmental stages, 3 d of differentiation (DIF3) when they are still immature, and 9 d (DIF9), when the overall organization of triads is close to the organization of an adult muscle fiber (Figure 1A). We had previously shown that in adult muscles of TRDN KO mice triad organization is unaffected at the macroscopic level (Oddoux et al., 2009), and first confirmed that the absence of endogenous T95 did not alter the presence nor the organization of triads in cultured myotubes, as shown by RyR1 staining, which displays double rows of dots intertwined with the Z-disk marker α-actinin at DIF9 (Figure 1A). Moreover, the localization of a GFP-tagged version of T95 in TRDN KO cultured myotubes was identical to the labeling of endogenous triadin in wild-type (WT) cells (Supplemental Figure S1). Video-microscopy experiments were next undertaken to follow T95-GFP dynamics in DIF3 and DIF9 myotubes (Figure 1B). The movies showed clusters of T95-GFP, similar to the clusters detected by immunolabeling on fixed cells. However, in DIF3 myotubes only a few T95-GFP clusters movements were detectable while no mobile clusters were visible at DIF9 during the 10 min recording (Figure 1B). To determine whether a fraction of the T95-GFP expressed in DIF9 myotubes was mobile but undetected by video-microscopy, we used FRAP experiments. After bleaching small areas, a partial recovery of the fluorescence was recorded, and the T95-GFP mobile fraction was estimated at 16% (Supplemental Figure S2). These results confirmed that only a small fraction of T95-GFP is under motion when expressed in DIF9 TRDN KO myotubes, and that simple video microscopy tracking of the T95-GFP is not sensitive enough to uncover its dynamics.
FIGURE 1:
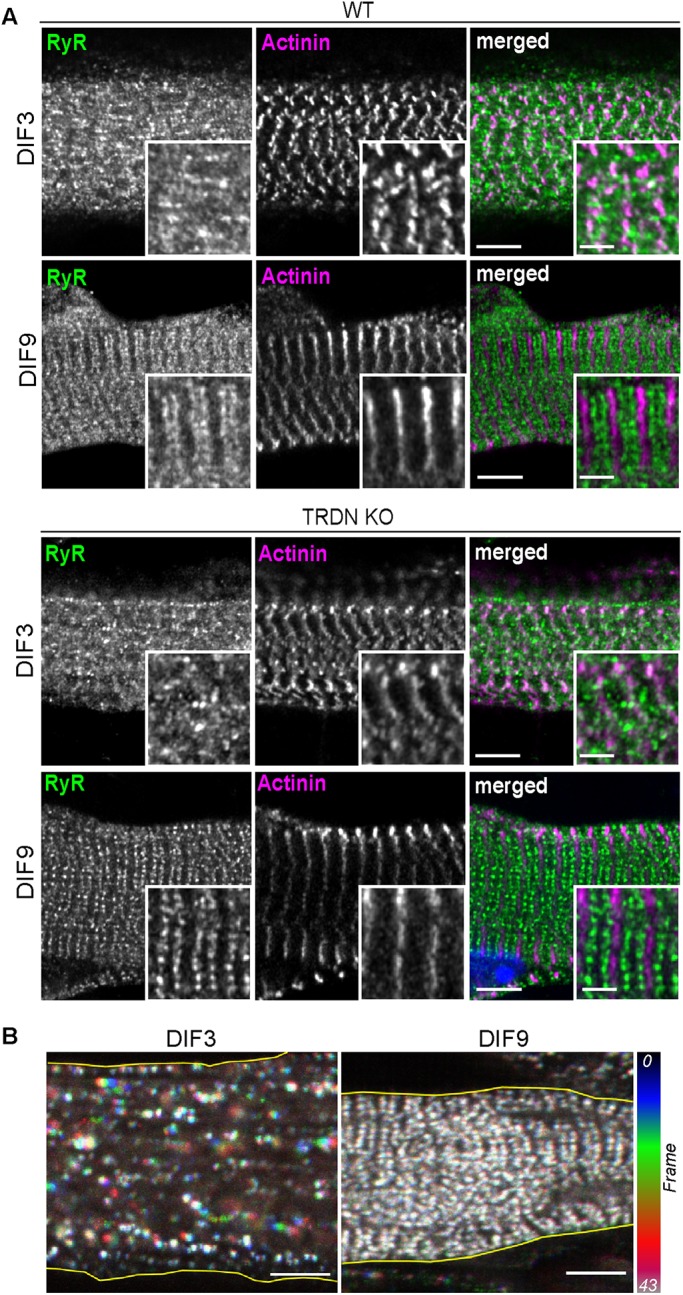
Triad organization and T95 behavior. (A) DIF3 and DIF9 WT (top panels) and TRDN KO (bottom panels) myotubes labeled with anti-RyR1 (green), and anti–α-actinin (magenta) antibodies. Single confocal planes, scale bars = 5 μm. For each image, insets of triad and of Z-disks for general sarcomere organization assessment are shown. Scale bars = 2 μm. (B) Color-coded representation of 10.6-min movies (43 frames) of DIF3 (left) and DIF9 (right) myotubes expressing T95-GFP. T95-GFP movements (represented by colored clusters) are only observed at DIF3. Scale bars = 5 μm.
T95 dynamics in SR membranes
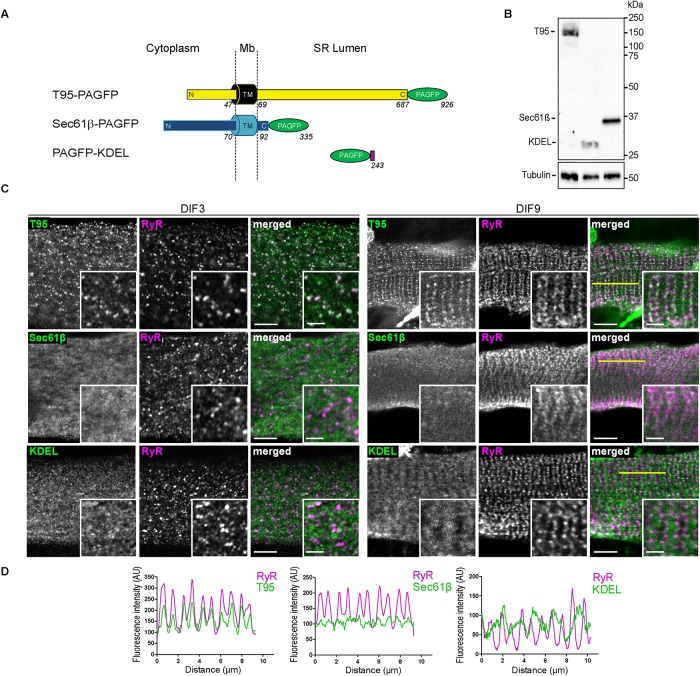
To observe T95 movements, we turned to a photoactivatable version of the protein (T95-PAGFP) (Patterson and Lippincott-Schwartz, 2002) and decided to compare its behavior to that of two recombinant proteins Sec61β-PAGFP and PAGFP-KDEL, which served as controls of SR proteins dynamics since they are unrelated to the EC coupling process and localized in different SR compartments. Sec61β is a subunit of the translocon, a type II TM protein of the reticulum with a single TM domain present in whole SR membrane (Rapoport et al., 1996; Greenfield and High, 1999). PAGFP-KDEL is localized in the lumen of the reticulum, thanks to its signal sequence and a KDEL retention motif (Munro and Pelham, 1987) (Figure 2A).
FIGURE 2:
T95, Sec61β, and KDEL localize, respectively, in SR membranes and lumen. (A) Schematic representation of T95, Sec61β, and KDEL constructs fused to PAGFP, with N-terminal (N), C-terminal (C), and transmembrane (TM) domains sequence length and localization regarding the SR membrane (cytoplasm, membrane [Mb] and SR lumen). T95-PAGFP: TM in black, N and C-terminal in yellow and PAGFP in green; Sec61β: TM, N-, and C-terminal in blue and PAGFP in green; PAGFP-KDEL: PAGFP in green and KDEL in purple. Amino acid numbers from N-terminal to C-terminal end are indicated. (B) Western blot analysis has been performed with antibody against GFP on 10 μg triadin KO cells lysates transduced with T95-PAGFP or PAGFP-KDEL or Sec61β-PAGFP. Three bands are detected at 150 kDa (T95-PAGFP), 37 kDa (Sec61β-PAGFP), and 28 kDa (PAGFP-KDEL). The β-tubulin has been used as a loading control (bottom panel). (C) DIF3 (left) and DIF9 (right) myotubes expressing T95, Sec61β, or KDEL immunolabeled with anti-GFP (green) and anti-RyR1 (magenta) antibodies. Scale bars = 5 μm. Insets of T95, Sec61β, or KDEL immunolabelings with that of RyR1 are shown. Scale bars = 2 μm. Percentages of colocalization are shown on Figure 6D and Supplemental Figure S7 for DIF9 and DIF3, respectively .(D) Plot profiles of constructs (green) vs. RyR1 (magenta) intensities as a function of distance for 10 μm (five successive triads as shown by the yellow line on C) in TRDN KO myotubes at DIF9.
All the photoactivatable constructs were expressed at a similar level, and displayed their expected localization in DIF3 and DIF9 myotubes (Figure 2, B and C). T95-PAGFP localizes with the jSR marker RyR1 in scattered clusters in the immature myotubes (DIF3) or in organized double rows of dots in mature myotubes (DIF9) (Figure 2, C and D). Sec61β-PAGFP and PAGFP-KDEL did not colocalize with RyR1 and showed diffuse labeling (Figure 2C). Moreover, PAGFP-KDEL showed denser striations in between RyR1 dots, therefore labeling the longitudinal SR (Figure 2, C and D). In the subsequent experiments, a red version of the KDEL construct (DsRed-KDEL) was used in combination with all photoactivatable proteins in order to label longitudinal SR and confirm that observed myotubes are mature.
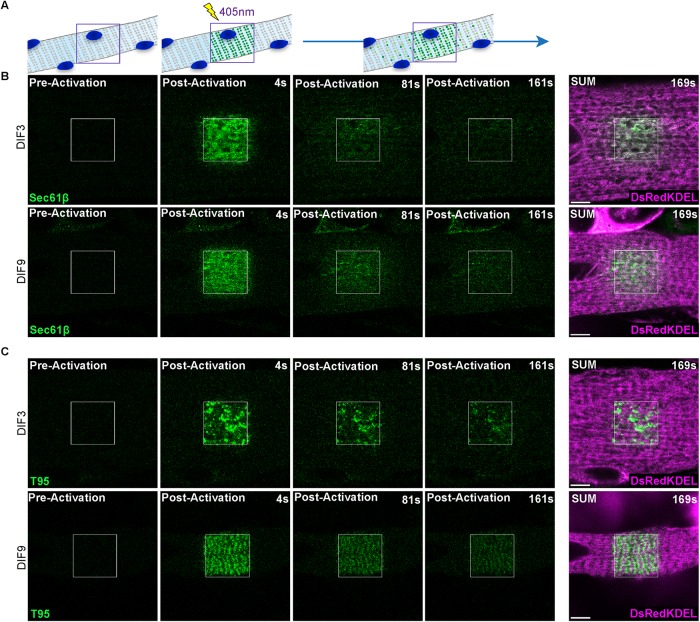
Photoactivation was achieved with a single 405 nm laser pulse in a large region of interest (ROI) in myotubes expressing PAGFP-tagged constructs and the fluorescent molecules were followed for 169 s (Figure 3A). The fluorescence of PAGFP-KDEL molecules initially detected in the ROI rapidly faded away after activation in both DIF3 and DIF9 myotubes (Supplemental Figure S3A and Supplemental Movies S1 and S2), as expected for a peptide freely diffusing in the SR lumen. Immediately after activation, Sec61β-PAGFP showed a diffuse fluorescence inside the ROI that decreased over time, and a wave of fluorescence was observed spreading outside the ROI in both DIF3 and DIF9 myotubes (Figure 3B and Supplemental Movies S3 and S4). In contrast, for T95-PAGFP the initial fluorescent patterns of activated proteins in the ROI were either scattered dots in DIF3 myotubes or double rows of dots in DIF9 myotubes and fluorescence spreading outside the ROI was too low to be quantified (Figure 3C and Supplemental Movies S5 and S6). For the three molecules, the fluorescence decrease in the activated ROI was fitted with an exponential decay curve, and the half time to decay (t1/2) was calculated (Table 1 and Supplemental Figure S3B). Values of t1/2 for T95-PAGFP and Sec61β-PAGFP were much higher than the ones recorded for PAGFP-KDEL, reflecting differences in the mobility of proteins localized within the lumen or in the membrane of SR. Comparison of t1/2 values suggested that T95-PAGFP mobility was more restricted than that of Sec61β-PAGFP. Although both molecules have a single TM domain, they differ in their topology and molecular weight, and we wondered whether the restricted diffusion of T95 could in fact arise from steric hindrance due to its much longer C-terminal luminal part (Figure 2A). To test this hypothesis, we investigated with the same protocol the mobility of a truncated version of T95 missing most of its luminal region (T95∆Cter, deletion of amino acid 113 to the C-terminal end, resulting in deletion of most functional domains already identified like the KEKE domain involved in molecular interaction with RyR1 and CSQ or the coiled-coil domain) (see scheme in Figure 6A later in the paper) (Fourest-Lieuvin et al., 2012). In DIF3 myotubes, T95∆Cter-PAGFP fluorescence in the ROI rapidly decreased over time, while in DIF9 myotubes a small decrease of fluorescence could be observed in the ROI together with spreading of a wave of fluorescence outside the ROI (Supplemental Figure S3C and Supplemental Movies S7 and S8). Calculated t1/2 for T95∆Cter-PAGFP were respectively close to that of Sec61β-PAGFP in DIF3 myotubes and no different from T95-PAGFP in DIF9 myotubes (Table 1 and Supplemental Figure S3B). Of note T95∆Cter shows the same pattern of triad localization than full length T95 at DIF9 (Supplemental Figure S3C, and see later T95-PAGFP in Figure 3C).
FIGURE 3:
T95 dynamics compared with control reticulum protein Sec61β in cultured myotubes. (A) Schematic representation of a photoactivation experiment: a myotube expressing a PAGFP-tagged protein of the triads is photoactivated in a specific ROI with a 405-nm laser, revealing the fluorescence of the proteins inside that region. The dynamic behavior of the activated proteins can be followed and recorded. (B) Single photoactivation of DIF3 (top panels) and DIF9 (bottom panels) myotubes coexpressing Sec61β-PAGFP (green) and DsRedKDEL (luminal reticulum marker, magenta). Shown are one image preactivation and three images postactivation (4, 81, and 161 s). SUM images are the sum intensity projections of all postactivation frames (169 s). Scale bars = 5 μm. (C) Single photoactivation of DIF3 (top panels) and DIF9 (bottom panels) myotubes coexpressing T95-PAGFP (green) and DsRedKDEL (luminal reticulum marker, magenta). Shown are one image preactivation and three images postactivation (4, 81, and 161 s). SUM images are the sum intensity projections of all postactivation frames (169 s). Scale bars = 5 μm.
TABLE 1:
Half-life time to decay values (t1/2) for each protein at DIF3 and DIF9.
| Localization | Proteins | DIF3 t1/2 (s) | DIF9 t1/2 (s) |
| Triads | T95 | 68.97 (64.86–73.64) | 104.00 (92.74–118.4) |
| SR membranes | Sec61β | 39.87 (38.05–41.88) | 57.33 (54.62–60.32) |
| SR lumen | KDEL | 7.73 (7.49–7.98) | 11.37 (11.01–11.75) |
| T95∆Cter | 34.54 (33.31–35.86) | 89.79 (79.76–102.7) |
Values are represented as means (c.i.), n = 32, 32, 36, and 34 cells for T95, Sec61β, T95∆Cter, and KDEL at DIF3, respectively, and n = 34, 40, 28, and 34 cells for T95, Sec61β, T95∆Cter, and KDEL at DIF9, respectively, from four independent experiments.
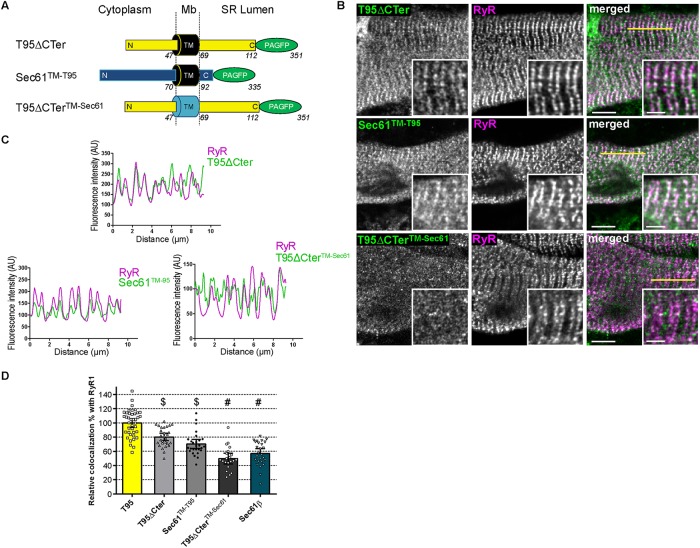
FIGURE 6:
Triadin TM domain is necessary for localization at triads. (A) Schematic representation of T95 ΔCter, Sec61 TM-T95, and T95ΔCTer TM-Sec61 constructs fused to PAGFP with N-terminal (N), C-terminal (C), and transmembrane (TM) domains sequence length and localization regarding the SR membrane (cytoplasm, membrane [Mb] and SR lumen). T95ΔCTer-PAGFP: TM in black, N, and C-terminal in yellow and PAGFP in green; Sec61TM-T95: TM in black, N- and C-terminal in blue, and PAGFP in green; T95ΔCTerTM-Sec61: TM in blue, N- and C-terminal in yellow, and PAGFP in green. Amino acid numbers from N-terminal to C-terminal end are indicated. (B) DIF9 myotubes expressing T95ΔCTer, Sec61TM-T95, and T95ΔCTer TM-Sec61 immunolabeled with anti-GFP (green) and anti-RyR1 (magenta) antibodies. Scale bars = 5 μm. Insets of T95 ΔCTer, Sec61TM-T95, or T95ΔCTerTM-Sec61 immunolabelings with that of RyR1 are shown. Scale bars = 2 μm. (C) Plot profiles of constructs (green) vs. RyR1 (magenta) intensities as a function of distance for 10 μm (five successive triads as shown by the yellow line in B) in TRDN KO myotubes at DIF9. (D) Percentage of PAGFP constructs colocalized with RyR1 labeling and normalized to that of DIF9 T95. Values are means ± c.i.; n = 40 cells from four independent experiments for T95; n = 26 cells from three independent experiments for Sec61β; n = 28 cells from three independent experiments for T 9 5 ΔCter; n = 21 cells from two independent experiments for T9 5 ΔCter TM-Sec61; n = 27 cells from two independent experiments for Sec61TM-T95. Using a parametric one-way analysis of variance followed by Tukey post-hoc comparisons: $, comparison with Sec61β: T95ΔCter (p < 0.0001) and Sec61TM-T95 (p < 0.05); #, comparison with T95ΔCter (p < 0.0001).
Movie S1.
Single photoactivation of DIF3 myotubes expressing PAGFP-KDEL. Scale bars = 5 μm. The fluorescence of PAGFP-KDEL molecules initially detected in the ROI rapidly faded away after activation.
Movie S2.
Single photoactivation of DIF9 myotubes expressing PAGFP-KDEL. Scale bars = 5 μm. The fluorescence of PAGFP-KDEL molecules initially detected in the ROI rapidly faded away after activation.
Movie S3.
Single photoactivation of DIF3 myotubes expressing Sec61β-PAGFP. Scale bars = 5 μm. The fluorescence of Sec61β-PAGFP spreads outside the ROI overtime.
Movie S4.
Single photoactivation of DIF9 myotubes expressing Sec61β-PAGFP. Scale bars = 5 μm. The fluorescence of Sec61β-PAGFP spreads outside the ROI overtime.
Movie S5.
Single photoactivation of DIF3 myotubes expressing T95-PAGFP. Scale bars = 5 μm. No spreading of T95-PAGFP fluorescence outside the ROI was observed.
Movie S6.
Single photoactivation of DIF9 myotubes expressing T95-PAGFP. Scale bars = 5 μm. No spreading of T95-PAGFP fluorescence outside the ROI was observed.
Movie S7.
Single photoactivation of DIF3 myotubes expressing T95ΔCter-PAGFP. Scale bars = 5 μm. The fluorescence of T95ΔCter-PAGFP spreads outside the ROI overtime.
Movie S8.
Single photoactivation of DIF9 myotubes expressing T95ΔCter-PAGFP. Scale bars = 5 μm. A small decrease of fluorescence of T95ΔCter-PAGFP could be observed in the ROI with a subtle spreading of a wave of fluorescence outside the ROI.
These experiments strongly suggested that although behaving like Sec61β in immature (DIF3) myotubes, T95∆Cter acquires some of the properties of the full length T95 as the myotube differentiates: a triad localization and a restricted mobility as compared with Sec61β. Hence, the restricted mobility of T95 compared with Sec61β cannot be entirely attributed to steric hindrance due to its long luminal domain but rather to specific molecular properties, at least in a mature SR.
T95 diffuses in SR membranes and is trapped in the triads
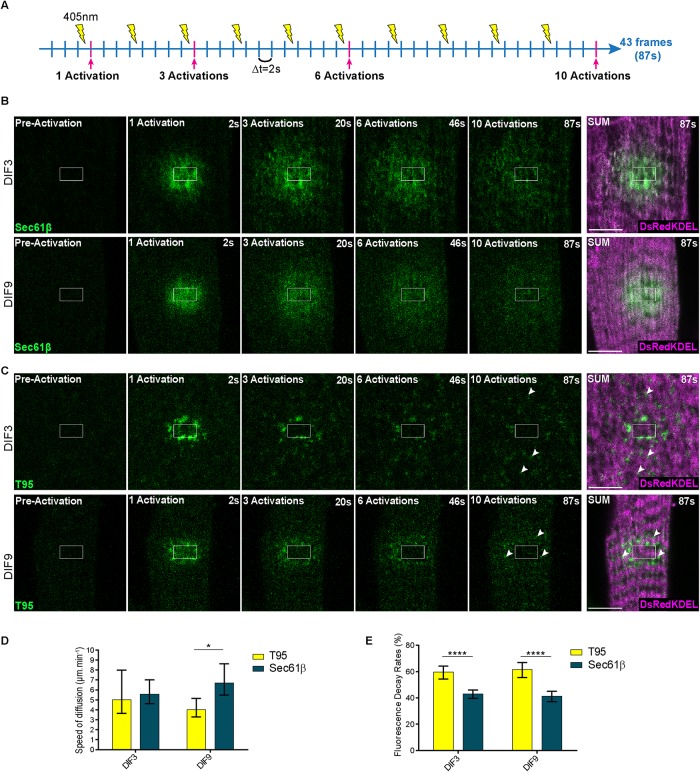
In the previous experiments, the absence of fluorescence detection outside the ROI after activation of T95-PAGFP correlated with the low mobile fraction previously calculated by FRAP (Supplemental Figure S2) and suggested that too few activated T95-PAGFP molecules had left the ROI to be detected. To directly observe T95 mobility we hypothesized that more T95-PAGFP needed to be activated and thus designed a protocol where photoactivation in a smaller ROI was performed iteratively during the video recording (one laser pulse every four movie frames), instead of using a single photoactivation (Figure 4).
FIGURE 4:
T95 diffuses in SR membranes of cultured myotubes. (A) Schematic representation of an iteration experiment: a myotube expressing a PAGFP-tagged protein of the triads is repeatedly photoactivated in a specific ROI with a 405-nm laser, revealing more fluorescence of the proteins inside that region. The dynamic behavior of the activated proteins can be followed and recorded. One frame recorded every 2 s and one activation every four frames. Frames preactivation and after 1, 3, 6, and 10 activations (pink lines) are shown on B and C. (B) Iterative photoactivation of DIF3 (top panels) and DIF9 (bottom panels) myotubes coexpressing Sec61β-PAGFP (green) and DsRedKDEL(magenta). Shown are one image preactivation and four images postactivation (2, 20, 46, and 87 s). SUM images are sum intensity projections of all postactivation frames (87 s). Scale bars = 5 μm. (C) Iterative photoactivation of DIF3 (top panels) and DIF9 (bottom panels) myotubes coexpressing T95-PAGFP (green) and DsRedKDEL (magenta). Shown are one image preactivation and four images postactivation (2, 20, 46, and 87 s). SUM images are sum intensity projections of all postactivation frames (87 s). T95 clusters appear at distant sites from the activation ROI (white arrowheads) and organize on each side of the DsRedKDEL striations at DIF9. Scale bars = 5 μm. (D) Speed of the fluorescence wave of Sec61β-PAGFP and T95-PAGFP at DIF3 and DIF9. Values are represented as means ± c.i. obtained from linear regressions as indicated on Supplemental Figures S4 and S5; *p = 0.01232 using model comparison of linear regressions, as performed by GraphPad Prism 6.0 (see Materials and Methods); n = 12 and 11 cells for Sec61β at DIF3 and DIF9, respectively, from three independent experiments; n = 10 and 7 cells for T95 at DIF3 and DIF9, respectively, from three independent experiments. (E) Fluorescence decay rates for Sec61β-PAGFP and T95-PAGFP at DIF3 and DIF9. Values are represented as means ± c.i. obtained from exponential decay modelization as indicated on Supplemental Figures S4 and S5; ****p < 0.0001 using model comparison of exponential decays, as performed by GraphPad Prism 6.0 (see Materials and Methods); n = 12 and 11 cells for Sec61β at DIF3 and DIF9, respectively, from three independent experiments; n = 10 and 7 cells for T95 at DIF3 and DIF9, respectively, from three independent experiments.
Repetitive activation of Sec61β-PAGFP in small ROIs resulted in diffuse fluorescence inside the ROI that progressively spread throughout the cell as a wave (Figure 4A and Supplemental Movies S9 and S10). This suggested a fast isotropic diffusion of Sec61β-PAGFP in all SR membranes, which was confirmed by colocalization with DsRed-KDEL on the average intensity projection images (Figure 4A). Two parameters were quantified to describe these movements: the speed of the wave of fluorescence, and the fluorescence decay rate as a function of the distance from the photoactivation zone (Supplemental Figures S4A and S5A).
Movie S9.
Iterative photoactivation of DIF3 myotubes expressing Sec61β-PAGFP. Scale bars = 5 μm. The fluorescence of Sec61β-PAGFP spreads throughout the cell overtime.
Movie S10.
Iterative photoactivation of DIF9 myotubes expressing Sec61β-PAGFP. Scale bars = 5 μm. The fluorescence of Sec61β-PAGFP spreads throughout the cell overtime.
For T95-PAGFP, repetitive activation of a small ROI led to more complex observations. First, after the initial fluorescence activation, a continuous decrease within the ROI was observed (Figure 4B). This was interpreted as photobleaching of the slowly mobile or motionless T95-PAGFP with repetitive laser pulses in the same ROI. Second, the initial fluorescence inside the ROI was followed by a fluorescence wave that diffused outside the ROI and became rapidly undetectable (Figure 4B and Supplemental Movies S11 and 12). When the speed of the wave of fluorescence and the fluorescence decay rate were calculated, it was obvious that this fluorescence was short-lived and did not spread more than 3 µm away from the ROI (Supplemental Figures S4B and S5B). The speed of the wave for T95-PAGFP was very similar to the speed for Sec61β-PAGFP in DIF3 myotubes whereas it seemed slower in DIF9 myotubes (Figures 4C; Supplemental Figure S4, Ac and Bc, for DIF3; Supplemental Figure S5, Ac and Bc, for DIF9). The fluorescence decay rates for T95-PAGFP in both DIF3 and DIF9 myotubes were significantly higher than the decay rates measured for Sec61β-PAGFP (Figure 4D; Supplemental Figure S4, Ad and Bd, for DIF3; and Supplemental Figure S5, Ad and Bd, for DIF9). Finally, clusters of more intense fluorescence were detected outside the ROI in both DIF3 and DIF9 myotubes (arrowheads on average intensity projections Figure 4B and Supplemental Movies S11 and 12). This showed that T95-PAGFP activated in the ROI reached specific spots located at distance from the activation zone. Comparison of T95-PAGFP with DsRed-KDEL localization on the average intensity projection showed no overlaps, but in DIF9 myotubes the clusters of T95-PAGFP were organized in rows on both sides of the DsRed-KDEL striations, which is characteristic of triad organization (Figure 4B, DIF9 Average image).
Movie S11.
Iterative photoactivation of DIF3 myotubes expressing T95-PAGFP. Scale bars = 5 μm. The fluorescence of T95-PAGFP diffuses outside the ROI and progressively accumulates in clusters far away from the activation zone
Movie S12.
Iterative photoactivation of DIF9 myotubes expressing T95-PAGFP. Scale bars = 5 μm. The fluorescence of T95-PAGFP diffuses outside the ROI and progressively accumulates in clusters far away from the activation zone.
Overall, these experiments allowed direct visualization of a diffusion and accumulation of T95 in the jSR: 1) the detection of clusters of T95-PAGFP corresponding to triads outside the ROI showed that molecules of T95-PAGFP activated in the ROI had reached distant jSR membranes and have been trapped there and 2) the detection of the spreading of an isotropic wave of T95-PAGFP fluorescence similar to what is observed during diffusion of Sec61β-PAGFP strongly suggested that T95 diffuses in SR membranes before reaching the jSR.
T95 diffuses and is trapped in the triads of adult muscle fibers
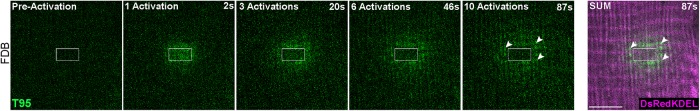
Although mature cultured myotubes represent a good model to observe CRC proteins dynamics, they may not recapitulate all phenomena taking place in an adult muscle. We next wondered whether the dynamic behavior of T95 so far described by our experiments could also be observed in a mature muscle fiber. To this end, T95-PAGFP and DsRed-KDEL were expressed in flexor digitorum brevis (FDB) muscles of adult TRDN-KO mice using an electroporation protocol (DiFranco et al., 2009). Iterative activations of T95-PAGFP in a small ROI induced a fluorescence wave detected outside the ROI, together with photobleaching in the ROI, reminiscent of what was observed in DIF9 myotubes (Figure 5 and Supplemental Movie 13). Analysis of time-lapse images also showed the accumulation of fluorescence in clusters outside the ROI. These clusters were easily detected on the average intensity projection image as well-aligned dots, typical for triad localization in muscle fibers (Figure 5, arrowheads). This experiment strongly suggested that a fraction of the T95-PAGFP is also permanently diffusing in the SR of adult muscle fibers, as observed in DIF9 myotubes, with an accumulation in triad membranes.
FIGURE 5:
T95 is dynamic in adult muscle fibers. Iterative photoactivation of a representative FDB fiber coexpressing T95-PAGFP (green) and DsRedKDEL (luminal reticulum marker, magenta). Shown are one image preactivation and four images postactivation (2, 20, 46, and 87 s). SUM images are sum intensity projections of all postactivation frames (87 s). White arrowheads indicate clusters of T95 organized in double rows of dots around DsRedKDEL striations. Scale bars = 5 μm.
Movie S13.
Iterative photoactivation of an adult muscle fiber (FDB) expressing T95-PAGFP. Scale bars = 5 μm. In an adult muscle fiber, the fluorescence of T95-PAGFP also diffuses outside the ROI and progressively accumulates in clusters far away from the activation zone.
T95 resides at the triads thanks to its TM domain
Previous experiments allowed the recording of a permanent flux of T95 within endomembranes of muscle cells, together with a progressive accumulation of T95 in the jSR. We next investigated the molecular motifs that could account for the acquisition of a longer residence time for T95 when it reaches the jSR membranes. The C-terminal luminal domain of T95 is involved in oligomerization and binding to other CRC proteins, and could therefore play a role in T95 targeting to the jSR (Fourest-Lieuvin et al., 2012; Rossi et al., 2014). Our previous experiment suggested that removing most of the C-terminal luminal part of T95 (T95∆Cter-PAGFP) did not affect its apparent steady-state localization in triads, as observed on images of photoactivation (Supplemental Figure S3C). The quantification performed by immunolocalization showed that the relative amount of T95∆Cter colocalized with RyR1 reached 80% of the T95/RyR1 colocalization (Figure 6D). In comparison, due to Sec61β diffuse pattern of expression, Sec61β and RyR1 colocalization was quantified at 55% of the T95/RyR1 colocalization (Figure 6D). These experiments suggested that motifs present in the first 113 amino acids of T95 were sufficient to drive the localization of T95∆Cter at the triad. As Sec61β and T95 have the same TM domain length, we decided to swap them in order to test whether T95 TM may confer triad localization to a Sec61 hybrid molecule, and whether its removal induces a delocalization of T95∆Cter (Figure 6A). Sec61TM-T95 and T95∆CterTM-Sec61 hybrids molecules were produced and expressed in cultured DIF9 myotubes (Supplemental Figure S6). The T95∆CterTM-Sec61 construct accumulated in small clusters clearly different from RyR1-labeled triads (Figures 6, B–D). Strikingly, Sec61TM-T95 formed double rows of clusters that were colocalized with RyR1 (Figures 6, B and C). Moreover, the respective levels of colocalization of Sec61TM-T95 and T95∆Cter with RyR1 were not different (Figure 6D). We concluded that exchanging of the TM domain of Sec61β by the one of T95 induced the localization of the Sec61TM-T95 hybrid in the triads demonstrating that TM domain of T95 is the main molecular motif responsible for its anchoring in the jSR.
DISCUSSION
In skeletal muscle, the EC coupling process is mediated by the CRC localized in triad membranes (Flucher, 1992; Flucher et al., 1992; Villa et al., 1993). Reticulum proteins of the CRC are exclusively localized in specific subdomains, the jSR, but the mechanisms by which they are confined in this restricted region are elusive, as well as their dynamic within the SR membranes. One major difficulty to address these questions is that upon observation, a majority of CRC proteins are localized in triads where they show a low mobility (Cusimano et al., 2009). Nevertheless, the visualization of the small fraction of mobile CRC proteins is important to understanding how they can reach the triad and be retained there. To explore the dynamic behavior of triadin (T95), we expressed T95 fluorescent chimeras in TRDN KO myotubes or adult muscles. We confirm in our experimental system that only a small proportion of T95 is mobile, which explains why video-microscopy cannot be used for this study. We therefore followed small pools of a photoactivated fluorescent T95 chimera (T95-PAGFP), and compared their dynamic behaviors to other proteins of the SR, whose traffic is unrelated to EC coupling.
T95 can diffuse in SR membranes
Our first goal was to characterize the dynamic behavior of T95. Single photoactivation experiments could theoretically uncover the dynamic of PAGFP chimeras expressed in myotubes in two ways: the direct observation of protein movement outside the ROI and the decrease of fluorescence inside the ROI. T95-PAGFP movement was hardly detectable outside the activated ROI, in contrast to movement of PAGFP-KDEL or Sec61β-PAGFP. However, T95-PAGFP dynamics could be evaluated with the half time to fluorescence decay (t1/2) within the ROI, which was comparable to that of Sec61β-PAGFP, a protein known to diffuse within the reticulum membrane (Cui et al., 2015). This comparison suggested that mobile T95 molecules were undergoing diffusion within the SR membranes. When iterative photoactivation in a ROI were next performed, the movement of T95-PAGFP outside the ROI became detectable, and its analysis confirmed a diffusion mechanism. In young (DIF3) and mature (DIF9) myotubes, the movement of Sec61β-PAGFP was characterized by a wave of fluorescence spreading isotropically. The same pattern of isotropic diffusion was found for T95-PAGFP fluorescence, although it could be followed on a smaller distance. The speed of diffusion of these waves was comparable for Sec61β-PAGFP and T95-PAGFP in young myotubes, but seemed slower for T95-PAGFP in mature myotubes. Importantly during these experiments, ROI were selected in random locations showing that diffusion of T95-PAGFP is not restricted to specific areas of the myotubes and suggesting the existence of a permanent flux of T95 molecules in SR.
T95 acquisition of a long residence time in jSR membranes
The comparison of Sec61β-PAGFP and T95-PAGFP dynamics after photoactivation also demonstrated that in addition to diffusion, T95-PAGFP acquires a long residence time whenever reaching the jSR. During iterative photoactivation, the calculated fluorescence decay rate was larger for T95-PAGFP at both stages of myotubes differentiation. Because the speed of diffusion of the wave of Sec61β-PAGFP and T95-PAGFP fluorescence was similar, one can assume that the enhanced fluorescence decay rate observed for T95-PAGFP is not due to an accelerated diffusion. More likely, this decay rate reflects a restriction in the movement of T95-PAGFP. Accordingly T95-PAGFP fluorescence was locally strongly enhanced in spots outside the ROI, whose localization is typical for triads: at DIF3 these clusters are spread in the whole cell, whereas at DIF9 they were aligned on each side of the Z-disks. Therefore, our experiments suggest that even if T95-PAGFP can constantly diffuse in the SR membranes, it acquires a longer residence time in the jSR membranes leading to a progressive accumulation of T95-PAGFP in the jSR. The fact that the same phenomenon was observed in adult muscles indicates that this hypothesis is not restricted to developing myotubes. Our experiments thus suggest that the constitution of the T95 pool inside the jSR membranes in skeletal muscle results from an equilibrium between constant fluxes of T95 molecules diffusing toward and from the triads, where a specific mechanism confines T95 molecules.
T95 domains involved in jSR localization
Previous studies have highlighted the molecular properties of T95 and several domains could be involved in the mechanisms leading to the confinement of T95 in the jSR membranes. Multimerization of T95 via disulfide bridges was suggested to contribute to triad formation (Froemming et al., 1999; Marty, 2015) and may represent a mechanism by which T95 residence time in the jSR is enhanced. However, we show that deletion of the majority of the luminal sequence of T95, which contains the reactive cysteines of the protein, does not change the localization of T95ΔCter. Of note, the T95 C-terminal domain also contains the motifs responsible for interaction with other proteins such as calsequestrin and RyR1 (Knudson et al., 1993; Marty et al., 1994, 2009; Kobayashi et al., 2000; Lee et al., 2004), or the structural protein Climp-63 (Osseni et al., 2016). Neither multimerization via disulfide-bridges nor binding to known interactors seems required for T95 localization at the triad. However these motifs contribute to the mobility of T95 in the SR because 1) it has previously been shown that a version of T95 deleted from its C-terminus interaction motif and mutated on its cysteines has an enhanced mobility in muscle fibers (Fourest-Lieuvin et al., 2012; Rossi et al., 2014) and 2) in this study the removal of the luminal domain of T95 confers to T95ΔCter-PAGFP a rate of diffusion similar to the rate of Sec61β in immature (DIF3) myotubes.
Nevertheless we show that removing the TM domain of T95∆Cter strongly reduces its localization at the jSR, while replacing the TM domain of Sec61β by the T95’s one confers to Sec61TM-T95 hybrid a jSR membrane localization. This directly demonstrates that the TM domain of T95 is the main driver for the protein localization. The intimate mechanisms leading to retention in the jSR through such a hydrophobic domain has to be deciphered but may involve direct molecular interactions with other proteins, a mechanism described for two proteins of the longitudinal part of skeletal muscle SR, phospholamban and SERCA (Martin et al., 2018), or interaction with the jSR membrane lipids. Indeed, the specific lipid composition of the jSR membrane could affect the dynamic of its protein components, and the lipid bilayer thickness of intracellular organelles is known to contribute to segregation of proteins according to their TM domain size (Sharpe et al., 2010). Of note, triads represent endoplasmic reticulum-plasma membrane (ER-PM) contact sites that are specific to striated muscle. In many cell types, ER-PM contact site have a specific lipid composition (Scorrano et al., 2019). The localization of TM proteins of the ER in the contact sites can rely on their cytosolic domain that contains specific domain binding to lipids present in the PM, which allows it to bridge between the two membranes, like junctophilin whose isoforms fulfill this role in muscle triads and in neurons (Takeshima et al., 2015). Our data suggest that lipids inside ER membrane microdomains may also contribute to the localization of some ER resident protein of the ER-PM contact sites.
Model for T95 localization during SR differentiation
Overall, our set of experiments supports a model in which T95 diffuses in SR membranes before being trapped in the jSR, a mechanism contributing to the dynamics of T95 in jSR in adult muscle. Interestingly, specific features were observed in developing myotubes during this study. First, when T95-GFP was followed by video microscopy experiments, a few vesicular movements were detected in young (DIF3) myotubes that were absent in mature (DIF9) myotubes. One cannot exclude that the mobility of T95 proteins may rely on vesicular trafficking as well as diffusion inside membranes of the SR, albeit in variable proportions as the muscle cells differentiate. Such a switch in trafficking mechanisms between different stages of muscle cells differentiation was already proposed for phospholamban, a modulator of the muscle calcium ATPase (Stenoien et al., 2007). Second, the comparison of PAGFP-chimera’s dynamics between young and mature myotubes also revealed that the dynamics of T95, Sec61β, and KDEL evaluated with t1/2 (Table 1) were all slowed down at the same rate (67%) in mature myotubes. This shows that a phenomenon affecting whole SR properties or morphology is responsible for lowering the mobility of the three molecules. During myoblast differentiation, the SR becomes tethered to the contractile acto-myosin apparatus (Bagnato et al., 2003), which fills an important part of the cell cytoplasm. A reduction in the space occupied by the SR may explain the reduction in mobility of SR proteins. The apparition within the SR of subcompartments, such as the jSR, with specific structure or composition that would limit diffusion of molecules, could also explain this drop in mobility during differentiation. Such a mechanism was suggested to explain the local retention of cargoes destined to secretion in the endoplasmic reticulum of neurons (Cui-Wang et al., 2012). Cui-Wang et al. (2012) pointed out that the reticulum molecule Climp-63 played a pivotal role in the building of reticulum zone whose molecular complexity could hinder a cargo’s mobility. Interestingly we recently showed that Climp-63 interacts with T95 (Osseni et al., 2016), and that removing the interaction motif of T95 with Climp-63 enhanced T95 mobility (Fourest-Lieuvin et al., 2012). The modulation of SR properties or composition during muscle cell differentiation could therefore alter T95 properties regarding the jSR membrane. Along these lines, we show here that the dynamic behavior of T95ΔCter that is close to the Sec61β dynamics in DIF3 myotubes shifts to a behavior close to full-length T95’s one in DIF9 myotubes showing that the acquisition of jSR retention due the TM domain requires an underlying mechanism related to myotube differentiation.
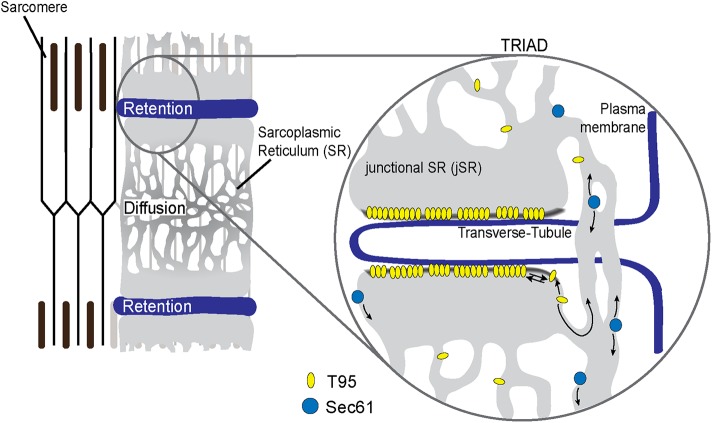
Overall, the present study demonstrates in a model of mature myotubes, as well as in adult muscle fibers, the dynamic behavior of T95. It allowed a direct visualization of T95 movement in SR and suggests that T95 can diffuse in SR membranes before being trapped in the triads (Figure 7). The model we propose also implies that a permanent flux of T95 may be responsible for the steady-state composition of the CRC. The molecular mechanisms leading to the exclusive localization of T95 in the jSR involve the presence of its TM segment, suggesting that the composition of the jSR may be important in this process. The question of whether other components of the CRC follow the same traffic and retention mechanisms remains open, especially for the giant RyR1 tetrameric calcium channel.
FIGURE 7:
Model of T95 dynamic behavior in adult muscle fibers. Schematic representation of T95 (yellow ovals) and Sec61β (blue disks) dynamics inside an adult muscle fiber. As Sec61β, T95 constantly diffuses inside the SR. However, once in the jSR T95 accumulates in this subcompartment thanks to its TM domain. The residence time of T95 inside the jSR could be due to a specific membrane composition (gray line) of this compartment and/or to the multimerization of T95.
MATERIALs AND METHODS
Antibodies and immunolabeling
Polyclonal antibodies against triadin and RyR1 were described previously (Marty et al., 1994; Vassilopoulos et al., 2005). Monoclonal antibodies against RyR1, GFP, α-actinin, and β-tubulin were purchased from the Developmental Studies Hybridoma Bank (DSHB, #34C), Abcam (#ab290), and Sigma (#A7811 and #T5201), respectively.
After fixation with 4% paraformaldehyde (PFA), the differentiated cells were permeabilized for 15 min in phosphate-buffered saline (PBS) supplemented with 1% Triton X-100 and saturated for 30 min in PBS with 0.1% Triton X-100, 0.5% bovine serum albumin, and 2% goat serum. Incubations with primary and secondary antibodies were done in the saturation buffer overnight at 4°C and for 2 h at room temperature, respectively.
Plasmids and lentivirus production
Full-length rat T95 (accession number #AJ243304) and the mutant T95ΔCTer were cloned in a pcDNA3.1 plasmid backbone as previously described (Marty et al., 2000; Fourest-Lieuvin et al., 2012). The pPAGFP-N1 plasmid was a gift from Jennifer Lippincott-Schwartz (National Institutes of Health, Bethesda, MD) (Addgene plasmid # 11909; http://n2t.net/addgene:11909; RRID:Addgene_11909), and pDsRed2ER was purchased from Clontech (#632409). pWPXLd was a gift from Didier Trono (EPFL, Lausanne, Switzerland) (Addgene plasmid #12258; http://n2t.net/addgene:12258; RRID:Addgene_12258) and tdTomato-Sec61-N-18 was a gift from Michael Davidson (Florida State University) (Addgene plasmid #58130; http://n2t.net/addgene:58130; RRID:Addgene_58130). The plasmids were amplified by transformation in Stbl3 bacteria and purified with Macherey-Nagel maxi kit (#740424).
PAGFP-T95 in the lentiviral vector pWPXLd (PAGFP-T95pW) was produced by digestion of T95 by EcoRI, ligation in pPAGFP-N1, and insertion in pWPXLd with In-Fusion cloning kit (Clontech) after removal of GFP. The Sec61β and the mutant T95∆Cter cDNA sequences were produced by PCR amplification and inserted by recombination in place of T95 in PAGFP-T95pW. Hybrid sequences were obtained by gene synthesis (Genscript) and inserted in place of T95 in PAGFP-T95pW after EcoRI digestion.
The lentiviral vectors were produced by a triple transfection of pWPXLd, psPAX2, and pCMV-VSV-G in HEK293T, as described previously (Osseni et al., 2016). Titrations were done with an accuri6 cell counter for constitutively fluorescent constructs and with a confocal microscope equipped with a FRAP module and a 405-nm laser for photo-activatable constructs.
Animals
The TRDN KO mice described previously (Oddoux et al., 2009) were used to produce WT and TRDN KO littermates. All procedures using animals were approved by the Institutional Ethics Committee and followed the guidelines of the National Research Council Guide for the care and use of laboratory animals.
Western blot analysis
The presence of T95, Sec61β, KDEL, T95∆Cter, T95∆CterTM-Sec61, and Sec61TM-T95 in triadin KO cells was assayed by Western blot analysis using a chemiluminescent reagent after electrophoretic separation of the protein on a 12% acrylamide gel and electrotransfer on Immobilon P (Millipore) (Oddoux et al., 2009). The signal was compared with the amount of β-tubulin. The secondary antibody were labeled with horseradish peroxidase and supplied by Jackson ImmunoResearch Laboratories.
Cell culture and transduction
Satellite cell preparations were performed as previously described from the hind limbs of newborn pups (Marty et al., 2000). Cells (50,000 cells/dish) were seeded on Matrigel (BD; 1:100 dilution)-coated ibidi dishes (#81156 Biovalley–Clinisciences) in HamF10 (Life Technologies) supplemented with 20% fetal bovine serum (Life Technologies) and 1% penicillin/streptomycin. The cells were transduced with lentivirus before induction of differentiation (DMEM supplemented with 2% horse serum and 1% penicillin/streptomycin). After 2 d of differentiation, a second layer of Matrigel (dilution 1:3) was added on the cells and the differentiation medium was supplemented with 50 ng/ml-1 of agrin (R&D Systems) as described previously (Falcone et al., 2014; Pimentel et al., 2017)
Electroporation and fiber dissociation
Mice FDB muscles were electroporated as described before (DiFranco et al., 2009) by injection of 100 µg pPAGFP-T95 and 56 µg pDsRed-KDEL. FDB muscles were dissected 5 d after electroporation, dissociated enzymatically for 45 min at 37°C in 1.5 mg/ml-1 collagenase (Sigma), then mechanically, and the fibers were plated on ibidi dishes (#81156 Biovalley–Clinisciences) for analysis.
Photoactivation and video-microscopy experiments
For photoactivation and video-microscopy experiments, cells differentiated for 3 d (DIF3, young myotubes) or 9 d (DIF9, mature myotubes) were placed in fresh medium devoid of phenol red at least 1 h prior to observations. Cell monitoring was performed on a 37°C heating plate, with a Zeiss LSM710 confocal microscope. A 63 × 1.40 N.A. Plan-Apochromat oil immersion objective and a pinhole aperture of 1 Airy unit were used.
For video-microscopy experiments, cells were monitored with an image every 15 s for 10.6 min.
For single photoactivation experiments, three images were acquired before PAGFP activation in the ROI with 50% power of the 405-nm laser. Laser scan speed and gain parameters were the same for the acquisition of all photo-activated constructs. All images were acquired without any averaging. Single activated cells were monitored with an image every 4 s for 169 s. Each time lapse has been corrected for photobleaching and normalized between 1 and 0 (0 being the baseline before photoactivation and 1 the intensity right after activation). The obtained curves were fitted to an exponential decay from which half-time values (t1/2) were extracted.
For iterative photoactivation experiments, activated cells were acquired every 2 s for 87 s with an activation every four images. Fluorescence intensities (Supplemental Figures S4 and S5) were measured within concentric squares drawn around the initially photoactivated ROI and plotted against time. For each colored curve—corresponding to each concentric square—a temporal maximum was detected and associated to the distance vis-à-vis the initial ROI. After plotting mean delays to maximum fluorescence for each concentric square, a linear regression performed with GraphPad Prism 6.0 software enabled for the calculation of a slope and thus the speed of fluorescence wave, following the equation indicated.
Alternatively, fluorescence values at the time of maximal fluorescence, observed in each concentric square and normalized to that observed in the first square, gave rise to a plot that could be fitted with an exponential decay using GraphPad Prism 6.0 software. The k parameter of the plot was estimated, and thus a decay rate, following the equation indicated.
FRAP on differentiated myotubes
Live-cell video recordings were performed using an inverted microscope (Axio Observer, Zeiss) coupled to a spinning-disk confocal system (CSU-W1-T3, Yokogawa, pinhole aperture of 1 Airy unit) connected to wide-field electron-multiplying CCD camera (ProEM+ 1024, Princeton Instruments) and maintained at 37°C and 5% CO2. Images were taken at 1 Hz for 60 s using a 63× oil-immersion objective (1.46 NA). The raw fluorescence values from the unbleached area were subtracted from the values obtained in the photobleached areas. The values were then normalized to 0–100%, with 100% corresponding to the fluorescence value recorded just before photobleaching and 0% just after. A nonlinear fit with an unconstrained model “One site binding–Specific binding” from GraphPad Prism 6.0 was applied and indicated Bmax values for each experiment. To obtain a more accurate mean value, square roots of Bmax values were then represented.
Colocalization measure
Colocalization percentages were obtained as described before (Osseni et al., 2016) by generation of binary masks of the overlap between the two channels.
Statistical analysis
All statistical analyses were performed with Microsoft Office Excel and GraphPad Prism 6.0. Data are presented as means ± confidence intervals (c.i.). The number of independent experiments (n) is indicated in the figure legends.
Supplementary Material
Acknowledgments
We thank Eric Denarier and Sami Chaaban for fruitful discussions and critical review of the manuscript and the imaging facility platform (PIC-GIN) for help with image acquisitions. This work was supported by grants from the Institut National de la Santé et de la Recherche Médicale (INSERM), from the Fondation Ducoin to M.S., and from the Association Française contre les Myopathies (AFM-Téléthon) to M.S., P.A., and I.M.
Abbreviations used:
- c.i.
confidence intervals
- CRC
calcium release complex
- DHPR
dihydropyridine receptor
- EC
excitation-contraction
- FDB
flexor digitorum brevis
- jSR
junctional sarcoplasmic reticulum
- PBS
phosphate-buffered saline
- ROI
region of interest
- RyR
ryanodine receptor
- SR
sarcoplasmic reticulum
- TM
transmembrane
- T-tubule
transverse-tubule
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-07-0399) on December 26, 2019.
REFERENCES
- Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V. (2003). Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J Cell Biol , 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone V, Randazzo D, Del Re V, Sorrentino V, Rossi D. (2015). Organization of junctional sarcoplasmic reticulum proteins in skeletal muscle fibers. J Muscle Res Cell Motil , 501–515. [DOI] [PubMed] [Google Scholar]
- Cui XA, Zhang H, Ilan L, Liu AX, Kharchuk I, Palazzo AF. (2015). mRNA encoding Sec61β, a tail-anchored protein, is localized on the endoplasmic reticulum. J Cell Sci , 3398–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui-Wang T, Hanus C, Cui T, Helton T, Bourne J, Watson D, Harris KM, Ehlers MD. (2012). Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell , 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusimano V, Pampinella F, Giacomello E, Sorrentino V. (2009). Assembly and dynamics of proteins of the longitudinal and junctional sarcoplasmic reticulum in skeletal muscle cells. Proc Natl Acad Sci USA , 4695–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFranco M, Quinonez M, Capote J, Vergara J. (2009). DNA transfection of mammalian skeletal muscles using in vivo electroporation. J Vis Exp , 1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone S, Roman W, Hnia K, Gache V, Didier N, Lainé J, Auradé F, Marty I, Nishino I, Charlet-Berguerand N, et al (2014). N-WASP is required for Amphiphysin-2/BIN1-dependent nuclear positioning and triad organization in skeletal muscle and is involved in the pathophysiology of centronuclear myopathy. EMBO Mol Med , 1455–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flucher BE. (1992). Structural analysis of muscle development: transverse tubules, sarcoplasmic reticulum, and the triad. Dev Biol , 245–260. [DOI] [PubMed] [Google Scholar]
- Flucher BE, Franzini-Armstrong C. (1996). Formation of junctions involved in excitation-contraction coupling in skeletal and cardiac muscle. Proc Natl Acad Sci USA , 8101–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flucher BE, Phillips JL, Powell JA, Andrews SB, Daniels MP. (1992). Coordinated development of myofibrils, sarcoplasmic reticulum and transverse tubules in normal and dysgenic mouse skeletal muscle, in vivo and in vitro. Dev Biol , 266–280. [DOI] [PubMed] [Google Scholar]
- Fourest-Lieuvin A, Rendu J, Osseni A, Pernet-Gallay K, Rossi D, Oddoux S, Brocard J, Sorrentino V, Marty I, Fauré J. (2012). Role of triadin in the organization of reticulum membrane at the muscle triad. J Cell Sci , 3443–3453. [DOI] [PubMed] [Google Scholar]
- Franzini-Armstrong C, Jorgensen AO. (1994). Structure and development of E-C coupling units in skeletal muscle. Annu Rev Physiol , 509–534. [DOI] [PubMed] [Google Scholar]
- Froemming GR, Murray BE, Ohlendieck K. (1999). Self-aggregation of triadin in the sarcoplasmic reticulum of rabbit skeletal muscle. Biochim Biophys Acta , 197–205. [DOI] [PubMed] [Google Scholar]
- Fu Y, Hong T. (2016). BIN1 regulates dynamic t-tubule membrane. Biochim Biophys Acta , 1839–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield JJ, High S. (1999). The Sec61 complex is located in both the ER and the ER-Golgi intermediate compartment. J Cell Sci (Pt 10), 1477–1486. [DOI] [PubMed] [Google Scholar]
- Kaisto T, Metsikkö K. (2003). Distribution of the endoplasmic reticulum and its relationship with the sarcoplasmic reticulum in skeletal myofibers. Exp Cell Res , 47–57. [DOI] [PubMed] [Google Scholar]
- Knudson CM, Stang KK, Jorgensen AO, Campbell KP. (1993). Biochemical characterization of ultrastructural localization of a major junctional sarcoplasmic reticulum glycoprotein (triadin). J Biol Chem , 12637–12645. [PubMed] [Google Scholar]
- Kobayashi YM, Alseikhan BA, Jones LR. (2000). Localization and characterization of the calsequestrin-binding domain of triadin 1. Evidence for a charged beta-strand in mediating the protein-protein interaction. J Biol Chem , 17639–17646. [DOI] [PubMed] [Google Scholar]
- Lee JM, Rho S-H, Shin DW, Cho C, Park WJ, Eom SH, Ma J, Kim DH. (2004). Negatively charged amino acids within the intraluminal loop of ryanodine receptor are involved in the interaction with triadin. J Biol Chem , 6994–7000. [DOI] [PubMed] [Google Scholar]
- Martin PD, James ZM, Thomas DD. (2018). Effect of Phosphorylation on Interactions between Transmembrane Domains of SERCA and Phospholamban. Biophys J , 2573–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty I. (2015). Triadin regulation of the ryanodine receptor complex. J Physiol (Lond) , 3261–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty I, Fauré J, Fourest-Lieuvin A, Vassilopoulos S, Oddoux S, Brocard J. (2009). Triadin: what possible function 20 years later? J Physiol (Lond) , 3117–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty I, Robert M, Villaz M, De Jongh K, Lai Y, Catterall WA, Ronjat M. (1994). Biochemical evidence for a complex involving dihydropyridine receptor and ryanodine receptor in triad junctions of skeletal muscle. Proc Natl Acad Sci USA , 2270–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty I, Thevenon D, Scotto C, Groh S, Sainnier S, Robert M, Grunwald D, Villaz M. (2000). Cloning and characterization of a new isoform of skeletal muscle triadin. J Biol Chem , 8206–8212. [DOI] [PubMed] [Google Scholar]
- McFarland TP, Milstein ML, Cala SE. (2010). Rough endoplasmic reticulum to junctional sarcoplasmic reticulum trafficking of calsequestrin in adult cardiomyocytes. J Mol Cell Cardiol , 556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S, Pelham HR. (1987). A C-terminal signal prevents secretion of luminal ER proteins. Cell , 899–907. [DOI] [PubMed] [Google Scholar]
- Nori A, Bortoloso E, Frasson F, Valle G, Volpe P. (2004). Vesicle budding from endoplasmic reticulum is involved in calsequestrin routing to sarcoplasmic reticulum of skeletal muscles. Biochem J , 505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddoux S, Brocard J, Schweitzer A, Szentesi P, Giannesini B, Brocard J, Fauré J, Pernet-Gallay K, Bendahan D, Lunardi J, et al (2009). Triadin deletion induces impaired skeletal muscle function. J Biol Chem , 34918–34929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata T, Yamasaki Y. (1997). Ultra-high-resolution scanning electron microscopy of mitochondria and sarcoplasmic reticulum arrangement in human red, white, and intermediate muscle fibers. Anat Rec , 214–223. [DOI] [PubMed] [Google Scholar]
- Osseni A, Sébastien M, Sarrault O, Baudet M, Couté Y, Fauré J, Fourest-Lieuvin A, Marty I. (2016). Triadin and CLIMP-63 form a link between triads and microtubules in muscle cells. J Cell Sci , 3744–3755. [DOI] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. (2002). A photoactivatable GFP for selective photolabeling of proteins and cells. Science , 1873–1877. [DOI] [PubMed] [Google Scholar]
- Pimentel MR, Falcone S, Cadot B, Gomes ER. (2017). In vitro differentiation of mature myofibers for live imaging. J Vis Exp , 10.3791/55141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahkila P, Alakangas A, Väänänen K, Metsikkö K. (1996). Transport pathway, maturation, and targetting of the vesicular stomatitis virus glycoprotein in skeletal muscle fibers. J Cell Sci (Pt 6), 1585–1596. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. (1996). Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem , 271–303. [DOI] [PubMed] [Google Scholar]
- Rezgui SS, Vassilopoulos S, Brocard J, Platel JC, Bouron A, Arnoult C, Oddoux S, Garcia L, De Waard M, Marty I. (2005). Triadin (Trisk 95) overexpression blocks excitation-contraction coupling in rat skeletal myotubes. J Biol Chem , 39302–39308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Bencini C, Maritati M, Benini F, Lorenzini S, Pierantozzi E, Scarcella AM, Paolini C, Protasi F, Sorrentino V. (2014). Distinct regions of triadin are required for targeting and retention at the junctional domain of the sarcoplasmic reticulum. Biochem J , 407–417. [DOI] [PubMed] [Google Scholar]
- Scorrano L, De Matteis MA, Emr S, Giordano F, Hajnóczky G, Kornmann B, Lackner LL, Levine TP, Pellegrini L, Reinisch K, et al (2019). Coming together to define membrane contact sites. Nat Commun , 1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe HJ, Stevens TJ, Munro S. (2010). A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell , 158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino V. (2011). Sarcoplasmic reticulum: structural determinants and protein dynamics. Int J Biochem Cell Biol , 1075–1078. [DOI] [PubMed] [Google Scholar]
- Stenoien DL, Knyushko TV, Londono MP, Opresko LK, Mayer MU, Brady ST, Squier TC, Bigelow DJ. (2007). Cellular trafficking of phospholamban and formation of functional sarcoplasmic reticulum during myocyte differentiation. Am J Physiol Cell Physiol , C2084–C2094. [DOI] [PubMed] [Google Scholar]
- Takeshima H, Hoshijima M, Song LS. (2015). Ca(2)(+) microdomains organized by junctophilins. Cell Calcium , 349–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilopoulos S, Thevenon D, Rezgui SS, Brocard J, Chapel A, Lacampagne A, Lunardi J, De Waard M, Marty I. (2005). Triadins are not triad-specific proteins: two new skeletal muscle triadins possibly involved in the architecture of sarcoplasmic reticulum. J Biol Chem , 28601–28609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa A, Podini P, Nori A, Panzeri MC, Martini A, Meldolesi J, Volpe P. (1993). The endoplasmic reticulum-sarcoplasmic reticulum connection. II. Postnatal differentiation of the sarcoplasmic reticulum in skeletal muscle fibers. Exp Cell Res , 140–148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.