Abstract
Greater than 90% of significant genome-wide association study (GWAS) single-nucleotide polymorphisms (SNPs) are in noncoding regions of the genome, but only 25.6% are known expression quantitative trait loci (eQTLs). Therefore, the function of many significant GWAS SNPs remains unclear. We have identified a novel type of eQTL for which SNPs distant from ligand-activated transcription factor (TF) binding sites can alter target gene expression in a SNP genotype-by-ligand–dependent fashion that we refer to as pharmacogenomic eQTLs (PGx-eQTLs)—loci that may have important pharmacotherapeutic implications. In the present study, we integrated chromatin immunoprecipitation-seq with RNA-seq and SNP genotype data for a panel of lymphoblastoid cell lines to identify 10 novel cis PGx-eQTLs dependent on the ligand-activated TF aryl hydrocarbon receptor (AHR)—a critical environmental sensor for xenobiotic (drug) and immune response. Those 10 cis PGx-eQTLs were eQTLs only after AHR ligand treatment, even though the SNPs did not create/destroy an AHR response element—the DNA sequence motif recognized and bound by AHR. Additional functional studies in multiple cell lines demonstrated that some cis PGx-eQTLs are functional in multiple cell types, whereas others displayed SNP-by-ligand–dependent effects in just one cell type. Furthermore, four of those cis PGx-eQTLs had previously been associated with clinical phenotypes, indicating that those loci might have the potential to inform clinical decisions. Therefore, SNPs across the genome that are distant from TF binding sites for ligand-activated TFs might function as PGx-eQTLs and, as a result, might have important clinical implications for interindividual variation in drug response.
SIGNIFICANCE STATEMENT
More than 90% of single-nucleotide polymorphisms (SNPs) that are associated with clinical phenotypes are located in noncoding regions of the genome. However, the mechanisms of action of many of those SNPs have not been elucidated, and drugs may unmask functional expression quantitative trail loci (eQTLs). In the current study, we used drugs that bind to the ligand-activated transcription factor aryl hydrocarbon receptor (AHR) and identified SNPs that were associated with interindividual variation in gene expression following drug exposure—termed pharmacogenomic (PGx)-eQTLs. Possibly of greater significance, those PGx-eQTL SNPs were outside of AHR binding sites, indicating that they do not interrupt AHR DNA recognition. PGx-eQTLs such as those described in this work may have crucial implications for interindividual variation in drug.
Introduction
Expression quantitative trait loci (eQTLs) have identified important functions for noncoding single-nucleotide polymorphisms (SNPs) across the genome—some of which have been associated with drug response phenotypes identified during genome-wide association studies (GWAS) (Cornelis et al., 2011; Ji et al., 2014; Carithers et al., 2015; Gupta et al., 2016; Dudenkov et al., 2017). Furthermore, some of those SNPs are associated with variation in gene expression only in response to an environmental stimulus such as drug exposure, but are not associated with variation in gene expression under basal conditions (Knight et al., 1999; Bream et al., 2002; Ingle et al., 2010; Barreiro et al., 2012; Fairfax et al., 2014). However, molecular mechanisms by which SNPs are associated with variation in gene expression only after drug or xenobiotic exposure remain unclear. We have identified SNPs that are distant from ligand-activated transcription factor (TF) binding sites that can influence target gene expression in a SNP genotype-by-ligand–dependent fashion. We refer to those SNPs as pharmacogenomic eQTLs (PGx-eQTLs), DNA sequence variation that might have important clinical and pharmacotherapeutic implications. In this report, we will define a PGx-eQTL as a genetic locus that is associated with interindividual variation in transcription regulation that results in altered gene expression after drug or xenobiotic exposure (Liu et al., 2012, 2018a; Zhou, 2016; Ho et al., 2017).
Obviously, SNPs can create/disrupt canonical TF-binding motifs (Knight et al., 1999; Bream et al., 2002; Ingle et al., 2010), but our previous studies identified several examples of SNPs that map hundreds of base pairs (bp) distant from ligand-activated TF binding sites that could influence TF binding in a SNP genotype-by-drug exposure–dependent manner (Liu et al., 2012, 2018a; Zhou, 2016; Ho et al., 2017; Qin et al., 2017). One of those studies identified a PGx-eQTL for the aryl hydrocarbon receptor (AHR), a ligand-activated TF that is an important environmental modulator for several critical pathways and processes, including chemical toxicology, immune stimulation, cancer, and the kynurenine pathway (Stockinger et al., 2014; Liu et al., 2018b; Neavin et al., 2018). In that study, we identified a SNP (rs2470893) in the promoter of the cytochrome P450 1A1 (CYP1A1) gene that was associated with variation in differential expression of CYP1A1 after exposure to the AHR ligand 3-methylcholanthrene (3-MC) as compared with vehicle treatment, even though rs2470893 was not an eQTL for CYP1A1 prior to exposure to 3-MC. However, the rs2470893 SNP was distant from a canonical AHR response element (AHRE; also often referred to as a xenobiotic response element or a dioxin response element)—the DNA sequence recognized and bound by ligand-activated AHR (5′-CACGC-3′) (Liu et al., 2018a). CYP1A1 plays an important role in the metabolism of xenobiotics and drugs, so the rs2470893 SNP genotype could alter individual response to drugs that are metabolized by CYP1A1.
That observation raised the possibility that SNPs at a distance from AHREs might influence AHR ligand-dependent target gene expression and could have important implications for health, disease, and variation in drug response. However, it is unclear how prevalent PGx-eQTLs are—i.e., how many SNPs are not eQTLs in the absence of ligand exposure and do not create/destroy a canonical AHRE binding site but, nonetheless, demonstrate this SNP-by-drug interaction. In the present study, we queried SNPs at a distance from AHREs across the genome—SNPs beyond the previously described CYP1A1 SNP—in an effort to identify additional novel AHR ligand-dependent cis PGx-eQTLs using a panel of human lymphoblastoid cell lines (LCLs)—Epstein-Barr virus–transformed B cells. We have previously generated dense genomic data for this panel of cell lines derived from a large number of individual subjects. We identified 10 novel cis PGx-eQTLs and then further investigated those loci in additional cell types to identify possible SNP-by-ligand–dependent effects using luciferase reporter gene assays. Importantly, some of the cis PGx-eQTLs identified had previously been associated with clinical and pharmacotherapeutic response phenotypes, indicating that the findings reported in this work might provide insight into individual differences in patient response to drug therapy.
In summary, we systematically identified and tested SNPs across the genome that did not create/destroy AHREs and were associated with variation in AHR ligand–dependent expression but were not associated with variation in the absence of AHR ligand exposure. These observations suggest that knowledge of eQTLs under basal conditions is insufficient to fully understand the functional implications of many common polymorphisms, and that SNPs at a distance from ligand-activated TF binding sites may play an important role in interindividual variation in response to pharmacological and environmental agents that are ligands for ligand-activated TFs.
Materials and Methods
The Human Variation Panel.
The human variation panel of 300 LCLs—100 Han Chinese-American, 100 African-American, and 100 Caucasian-American—was generated from immortalized B cells purchased from the Coriell Institute (Camden, NJ). SNP genotype data and baseline microarray expression data were generated by Dr. L.W. and are available for all LCLs in the Human Variation Panel (Gene Expression Omnibus SuperSeries accession number GSE24277). This panel has proven extremely useful for generating and testing PGx hypotheses (Li et al., 2008; Niu et al., 2010; Ingle et al., 2013; Liu et al., 2013, 2014, 2017).
Cell Cultures.
LCLs were cultured in RPMI 1640 with 15% FBS and 1% penicillin/streptomycin. HMC3 (CRL-3304; American Type Culture Collection) and HepG2 (HB-8065; American Type Culture Collection) cells were cultured in EMEM with 10% FBS and 1% penicillin/streptomycin. All cells were cultured at 37°C with 5% CO2. For all treatment exposure experiments, cells were treated with 1 μM 3-methylcholanthrene (3-MC; Enzo Life Sciences, Farmingdale, NY), 1 μM N-[2-(3H-indol-3-yl)ethyl]-9-isopropyl-2-(5-methyl-3-pyridyl)purin-6-amine (GNF-351; MilliporeSigma, Burlington, MA), or 0.1% DMSO final concentration (vehicle) and were cultured for 24 hours. These conditions were previously optimized (Liu et al., 2018a).
RNA Sequencing.
GM17212, GM17223, GM17229, GM17240, GM17264, GM17276, GM17281, and GM17295 LCLs from the human variation panel were used to perform the RNA-seq studies. Metadata, including sex of the LCL donors, AHR expression levels in each LCL, and rs2470893 (CYP1A1 cis PGx-eQTL) SNP genotype status of each LCL studied, are provided in Supplemental Table 1. Cells were harvested by pelleting at 300 g for 5 minutes at 4°C and were washed with ice-cold phosphate-buffered saline before extracting RNA using the miRNeasy RNA extraction kit per manufacturer instructions (Qiagen, Germantown, MD). Sample RNA integrity numbers were between 7 and 10 (average = 9.45, median = 9.6) (Supplemental Table 2). RNA-seq libraries were prepared using the TruSeq RNA Library Prep Kit v2 (Illumina, San Diego, CA), and paired-end sequencing was performed on an Illumina HiSEq 4000. FastQC (Fairfax et al., 2014) was used to determine RNA-sequencing quality. RNA-sequencing data were aligned to the hg38 human genome using TopHat (2.0.14) (Kim et al., 2013). Raw counts were calculated using Subread (1.4.6) (Liao et al., 2013). RSeQC (2.4.6) was used for alignment quality control (Wang et al., 2012). RNA-seq quality control measures were within expectations (49%–50% GC content, sequencing quality score ≥32% and ≥80% reads concordantly mapped) (Supplemental Table 2). EdgeR was used to identify genes differentially expressed after 3-MC or GNF-351 treatment compared with vehicle (Robinson et al., 2010). EdgeR-normalized log of counts per million (cpm) demonstrated a high degree of correlation between duplicates (R2 > 0.992) (Supplemental Table 2).
Chromatin Immunoprecipitation Antibody Characterization.
The AHR antibody (D5S6H-lot1; Cell Signaling Technologies, Danvers, MA) that was used for chromatin immunoprecipitation (ChIP)-seq was characterized using primary and secondary characterization modes, as suggested by ENCODE guidelines (Landt et al., 2012). Immunoblot of whole-cell lysates was used as the primary characterization and identified a single crisp band that accounted for 81% (S.E. 3.9%) of the total immunoreactivity per lane at the expected molecular weight (∼100 kDa) for lysates from seven LCLs (Supplemental Fig. 1A). Both immunoblot and ChIP-quantitative polymerase chain reaction (qPCR) at the CYP1B1 promoter—a known and prototypic binding region for AHR (Shehin et al., 2000; Yang et al., 2008)—were decreased by ≥90% after AHR-pooled small interfering RNA knockdown (Dharmacon, Lafayette, CO) for secondary characterization of the antibody in HMC3 cells (Supplemental Fig. 1, B and C). An intergenic region on chromosome 19 was used as the negative locus for all ChIP-qPCR.
ChIP Sequencing.
Cells were incubated at room temperature for 10 minutes and cross-linked with 1% methanol-free formaldehyde for 10 minutes, and the reaction was quenched with 0.125 M glycine for 5 minutes. Twenty million cross-linked cells were pelleted, and the pellet was washed once with ice-cold Tris-buffered saline. Pelleted cells were frozen at −80°C, and ChIP-seq was carried out, as previously described (Zhong et al., 2017). The characterized AHR antibody (D5S6H-lot 1) was used to perform AHR ChIP-seq. FastQC was used to assess sequencing quality. Sequences were aligned to human genome version 19 (hg19) using Burrows–Wheeler Aligner, and duplicates were removed. Model-based analysis of ChIP-Seq (MACS2) was used to identify peaks (Zhang et al., 2008). The ChIP-seq peak detection threshold (q) was set to 0.01. The MEME suite (Bailey, 2011) was used to search for de novo motifs and compare those motifs with the motifs in the Homo sapiens Comprehensive Model Collection (HOCOMOCOv10) database (Kulakovskiy et al., 2018). The HOMER Motif Analysis Software (Heinz et al., 2010) was used to quantify the number of times a motif was identified in peaks. The ChromHMM (Kasowski et al., 2010; Ernst and Kellis, 2017) track for a LCL (GM12878) was used to identify the chromatin state of regions that contained AHR-binding peaks.
Quantitative Reverse-Transcription Polymerase Chain Reaction.
Total RNA was collected from LCLs using the Zymo Quick RNA Mini-prep kit (Zymo Research, Irvine, CA). RNA was quantified by NanoDrop, and 50–200 ng total RNA was used for quantitative reverse-transcription polymerase chain reaction (qRT-PCR) assays (based on baseline expression of the genes and the efficiency of the qRT-PCR primers). The Power SYBR Green RNA-to-Ct 1-Step Kit (Applied Biosystems, Foster City, CA) was used for all qRT-PCR assays, except for CYP1B1, in which a PrimeTime qPCR probe (Integrated DNA Technologies, San Diego, CA) was used. Primers and probes used for qRT-PCR are listed in Supplemental Table 3. Differential expression of a gene after 3-MC or GNF-351 treatment compared with vehicle was calculated using the comparative Ct method relative to β actin (Schmittgen and Livak, 2008). Experiments were carried out at least twice in duplicate or triplicate with a minimum of six different LCLs.
Data Analysis and Statistical Analysis.
Bedtools was used to compare the relative location of different genomic elements, including genes, AHR ChIP-seq–binding peaks, SNPs, ChromHMM-defined chomatin states (Kasowski et al., 2010; Ernst and Kellis, 2017), and chromatin states from the epigenome roadmap (Kundaje et al., 2015).
A Spearman correlation coeficient was used to test for correlation of SNPs with variation in differential gene expression following exposure to 3-MC or GNF-351 relative to vehicle and was tested for significance under the Student’s t distribution with an α of 0.1. An α of 0.1 allowed us to detect a correlation of 0.72 with 70% power. A two-sided Student’s t test was used to test the null hypothesis that the mean of differential gene expression after ligand treatment relative to vehicle was equal for two groups for qRT-PCR and luciferase assays. The null hypothesis was rejected when the calculated P value was less than 0.05. Statistical analyses were done in Excel and R, and plots were generated in R.
Luciferase Reporter Gene Assays.
Luciferase reporter gene constructs were prepared by cloning the region of interest into the pGL4 (E6651) backbone (Promega, Madison, WI). To investigate SNP function, regions were cloned from LCLs that contained SNPs of interest, but did not differ at any other SNP, unless otherwise noted, or were synthesized by GENEWIZ (South Plainfield, NJ). The FastStart Taq DNA polymerase kit (Roche, Indianapolis, IN) with or without the GC-RICH solution, the KAPA HiFi HotStart ReadyMixPCR Kit (Kapa Biosystems, Wilmington, MA), the Platinum Taq DNA Polymerase kit (Thermo Fisher Scientific, Waltham, MA), or the GC-RICH PCR system (Roche) was used to amplify the regions of interest.
Luciferase plasmids were cotransfected into cells with a 1:10 ratio of pRL renilla, which was used as a transfection control vector (E2231; Promega). Thermo Fisher Scientific (Morristown, NJ) Lipofectamine 3000 or Lipofectamine 2000 (HMC3 and HepG2) or the Lonza nucleofector kit V (LCLs) were used as transfection methods per manufacturer instructions. Twelve hours post-transfection, cells were exposed to 3-MC, GNF-351, or 0.1% DMSO (vehicle). Twenty-four hours post-treatment, medium was aspirated from wells and cells were washed with PBS (HMC3 and HepG2), or cells were pelleted at 2500 rpm and washed with PBS (LCLs) before resuspending in 1× passive lysis buffer (Promega). The Promega Dual-Luciferase Reporter Assay system was used for luciferase detection. Luciferase assays were carried out per manufacturer instructions. The ratio of luciferase to renilla (transfection control vector) was used to normalize transfection efficiency before calculating the fold change (Eggers et al., 2016). All luciferase assays were carried out at least twice with a minimum of six replicates.
Results
Experimental Strategy for the Identification and Validation of Putative AHR PGx-eQTLs
We reported previously that a SNP (rs2470893) at a distance from an AHRE in the promoter of the CYP1A1 gene was a cis PGx-eQTL for CYP1A1 after treatment with an exogenous AHR agonist, 3-MC, even though it was not an eQTL in the absence of 3-MC exposure (Liu et al., 2018a). In the present series of studies, to test the hypothesis that many other SNPs across the genome that are near AHR-regulated genes but that do not create/destroy AHREs—the DNA sequence recognized and bound by ligand-activated AHR—might influence variation in AHR ligand-dependent gene expression, we performed RNA-seq using eight LCLs—four homozygous wild-type and four homozygous variants for the rs2470893 SNP. We chose LCLs based on their rs2470893 SNP genotype so we could use that SNP as a cis PGx-eQTL positive control before using the RNA-seq data to test for novel cis PGx-eQTLs at additional genomic loci across the genome.
We understood that, with eight cell lines, we were underpowered to test possible associations of all SNPs across the genome that displayed differential gene expression following AHR ligand exposure as compared with vehicle. Therefore, we used a highly focused approach that allowed us to identify cis PGx-eQTLs by testing associations between SNPs surrounding known AHR binding sites near genes that were differentially expressed after exposure to the AHR agonist 3-MC or the AHR antagonist N-[2-(3H-indol-3-yl)ethyl]-9-isopropyl-2-(5-methyl-3-pyridyl)purin-6-amine (GNF-351) as compared with vehicle. Our approach to the identification of cis PGx-eQTLs included two phases: a hypothesis-generating phase that identified putatitve cis PGx-eQTLs, and a functional genomics phase designed to test the validity and generalizability of the putative cis PGx-eQTLs identified during the first phase.
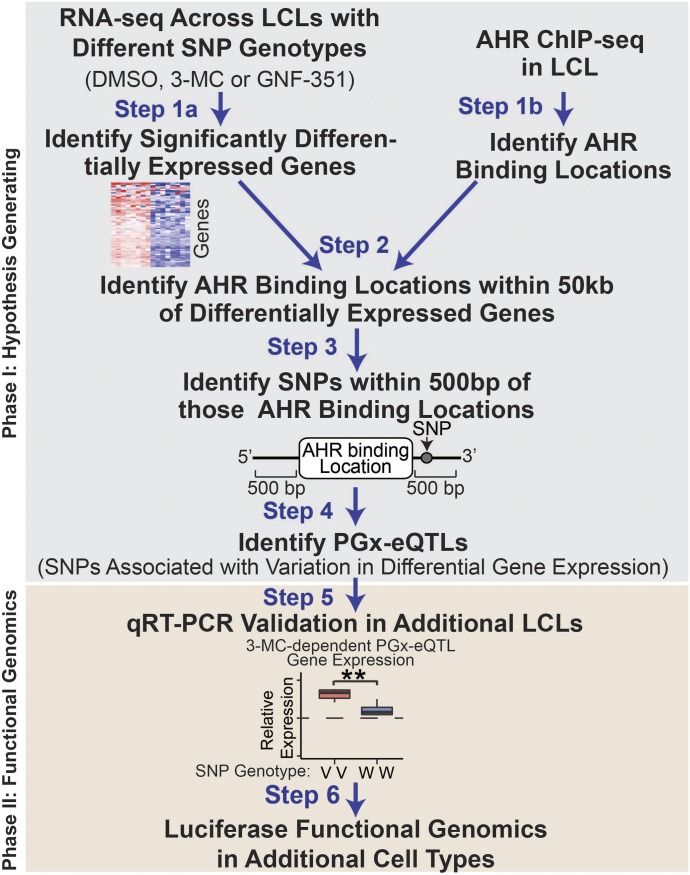
The first phase—the hypothesis-generating phase—included four steps that were designed to identify putative cis PGx-eQTLs. As an initial step, we identified genes that were highly differentially expressed [false discovery rate (FDR) ≤0.05, |fold change| ≥2] as determined by RNA-seq after treatment with the AHR agonist 3-MC or the AHR antagonist GNF-351 as compared with vehicle (Fig. 1, Step 1a). In parallel, we identified AHR binding sites throughout the genome by performing ChIP-seq using a single LCL (Fig. 1, Step 1b). We next identified which of the highly differentially expressed genes contained at least one AHR-binding peak within 50 kilobase (kb) of the gene (Fig. 1, Step 2) to identify genes that were most likely to be directly regulated by AHR. Next, SNPs within 500 bp of AHR-binding peaks that were within 50 kb of the differentially expressed genes (Fig. 1, Step 3) were tested for association (Spearman’s rank-order correlation) with variation in differential gene expression after treatment with 3-MC or GNF-351 (Fig. 1, Step 4). We selected a distance of 500 bp from a given AHR peak because all previously identified PGx-eQTLs have been within this distance and we were underpowered to investigate the association of all SNPs with variation in ligand-dependent gene expression. Only SNPs that had a minor allele frequency (MAF) >0.25 were considered for association because 0.25 was a MAF at which we were reasonably likely to include at least one homozygous variant LCL by chance among the eight LCLs based on Hardy–Weinberg equilibrium predictions. SNPs were excluded from consideration if they were eQTLs without treatment or if baseline eQTL information in LCLs was missing, as indicated by GTEx (Carithers et al., 2015). SNPs that were associated with variation in gene expression were tested for linkage disequilibrium, and all linked SNPs (R2 > 0.8) were considered part of a single locus. Therefore, one cis PGx-eQTL could include multiple SNPs and/or multiple AHR-binding peaks.
Fig. 1.
PGx-eQTL analysis scheme. A PGx pipeline was developed to merge RNA-seq, ChIP-seq, and SNP-genotype data to identify putative PGx-eQTLs. As a first step, we identified the top differentially expressed genes [|Log2(fold change)| >1 and FDR <0.05] from our RNA-seq analyses for eight LCLs that were treated with 0.1% DMSO, 1 µM 3-MC, or 1 µM GNF-351 in duplicate (Step 1a). In parallel, we identified AHR-binding regions from AHR ChIP-seq analysis performed for one of the LCLs used in the RNA-seq studies (Step 1b). We next determined which of the top differentially expressed genes had AHR-binding peaks within 50 kb of the gene (Step 2) and identified those peaks that had SNPs with a MAF >0.25 that mapped within 500 bp of AHR ChIP-seq peaks (Step 3). We then identified putative PGx-eQTLs by testing the association of the SNPs from Step 3 with the differential expression of the nearby differentially expressed gene with the Spearman rank correlation and tested for significance under a two-tailed t-distribution (Step 4). We validated the putative PGx-eQTLs identified in additional LCLs (Step 5) and functionally validated them with luciferase reporter gene assays (Step 6).
The second phase—the functional genomics phase of the study—consisted of two steps designed to test the validity of the cis PGx-eQTLs and examine their possible function in additional cell types. Therefore, we first tested the putative cis PGx-eQTLs that had been identified during the hypothesis-generating phase by qRT-PCR in additional LCLs that were selected based on known SNP genotypes for each of the cis PGx-eQTLs (Fig. 1, Step 5). Finally, we examined the function of the validated cis PGx-eQTLs across three different cell types using luciferase reporter gene assays (Fig. 1, Step 6).
Phase I: Hypothesis-Generating Phase.
Phase I: Identification of differentially expressed genes after AHR ligand exposure.
RNA-seq performed in eight cell lines after exposure to the AHR agonist 3-MC or the AHR antagonist GNF-351 identified 1865 genes that were significantly differentially expressed (FDR ≤0.05) after 3-MC treatment and 3664 genes that were significantly differentially expressed (FDR ≤0.05) after GNF-351 treatment (Fig. 2, A and B). Sixty-nine genes were differentially expressed by at least 2-fold after either 3-MC or GNF-351 treatment (Fig. 2C). Importantly, there was wide variation in the degree of differential expression for these 69 genes among the eight cell lines after exposure to GNF-351 or 3-MC. That variation may be due, in part, to genomic variation. To test the validity of the RNA-seq data, 15 of these 69 genes were validated by qRT-PCR in independent experiments using all eight LCLs that had been used to perform RNA-seq (Supplemental Fig. 2). Those validation genes were selected from three groups: 1) prototypic AHR-regulated genes (CYP1A1, CYP1B1, AHRR, and TIPARP), 2) immune-relevant genes (RARRES2, IL2RA, IGF1, CXCL8, and CXCL12), and 3) other differentially expressed genes selected at random (SHF, WNT11, CRYM, GPR68, DMTN, and HES2).
Fig. 2.
LCL transcriptome after AHR activation (3-MC treatment) or inhibition (GNF-351 treatment). (A) A total of 1865 genes was differentially expressed after 3-MC treatment at a FDR <0.05, but only 16 of those genes were differentially expressed by at least 2-fold. More genes had expression that increased after 3-MC treatment than those that decreased. (B) A total of 3664 genes was differentially expressed after GNF-351 treatment at a FDR <0.05, but only 64 of those genes were differently expressed by at least 2-fold. (C) The figure shows a heatmap for the 69 genes that were differentially expressed by at least 2-fold with a FDR <0.05 after either 3-MC or GNF-351 treatment, but their degree of differential expression was variable across LCLs. (D) The SNP-dependent effect of rs2470893 after 3-MC treatment was determined by both qRT-PCR and RNA-seq in the samples that were used for RNA-seq and represented in violin plot. Each group contained four lymphoblastoid cell lines. P values were calculated using a two-tailed Student’s t test: *P < 0.05; ** P < 0.01; ***P < 0.001.
As a quality control measure, we next asked whether the previously identified rs2470893 SNP in the promoter of the CYP1A1 gene (Liu et al., 2018a) was a significant cis PGx-eQTL across the eight cell lines studied (four homozygous variant and four homozygous wild-type) based on our RNA-seq data. Consistent with our previous findings, the variant rs2470893 SNP genotype was associated with a greater increase in CYP1A1 expression than was the wild-type SNP genotype after 3-MC treatment compared with vehicle in these samples as tested by qRT-PCR (P = 0.001) and by RNA-seq (P = 0.006) (Fig. 2D). These results indicated that the RNA-seq data were consistent with the qRT-PCR findings and, as a result, that RNA-seq has adequate sensitivity to detect cis PGx-eQTLs.
In summary, we identified 69 genes that were highly differentially expressed after AHR ligand exposure compared with vehicle, with considerable variation across the eight cell lines studied. This variation might be due, at least in part, to genetic variation.
Phase I: Identification of AHR binding sites.
AHR ChIP-seq was performed using the GM17212 LCL to identify AHR binding regions across the genome in LCLs. Biologic replicates demonstrated a high degree of similarity based on the correlation coefficient for overlapping peak scores after either vehicle (R2 = 0.854, Supplemental Fig. 3A) or 3-MC treatment (R2 = 0.774, Supplemental Fig. 3B). A total of 17,535 peaks were in common across all of the samples studied, with just 904 peaks that were unique to the vehicle-treated samples and 810 peaks that were unique to the 3-MC–treated samples. Furthermore, the peaks unique to just 3-MC or vehicle treatment demonstrated less significant binding than did peaks that were in common across the LCLs (both P < 2.2E-16, one-sided Mann-Whitney U test). Because most AHR-binding peaks were shared across all samples, and because the AHR-binding peaks unique to either 3-MC or vehicle exposure demonstrated less significant binding than did peaks shared across all samples, the peaks that were common to all of the samples were used for the downstream analyses shown graphically in Fig. 1 (Steps 2–6). The majority of these AHR-binding peaks were located within active promoters, strong enhancers, weak enhancers, or weak promoters (Supplemental Fig. 3C), as defined by ChromHMM states developed in a LCL (GM12878) (Ernst et al., 2011; Ernst and Kellis, 2017).
The high degree of AHR-binding peak overlap between vehicle- and agonist-treated samples has been demonstrated previously in studies performed with breast cancer cell lines (Yang et al., 2018) and may result from the fact that endogenous compounds such as kynurenine and its metabolites are known ligands for AHR (Opitz et al., 2011). Furthermore, AHR is highly expressed in LCLs—even more highly expressed than in the liver (Supplemental Fig. 4A), where it plays an important role in xenobiotic and drug metabolism (Stockinger et al., 2014; Zhou, 2016). Furthermore, we observed a striking decrease in the expression of AHR-regulated genes from our panel of differentially expressed genes after AHR knockdown indicated that AHR regulated the expression of these genes at baseline (Supplemental Fig. 4B).
As the next step, to test the validity of the AHR ChIP-seq results, we applied the Discriminative Regular Expression Motif Elicitation (DREME-Tomtom) software from the MEME suite, which identified short de novo motifs that were highly represented in the ChIP-seq peaks, and then compared those de novo motifs with known transcription factor–binding motifs (Bailey, 2011). One of the top de novo motifs in our ChIP-seq peaks identified by DREME (E-value = 6.2E−65) was 5′-DCACGCA-3′, which was identified 1752 times in 1506 peaks. This 7-bp de novo motif was most similar to the 5-bp canonical AHR-binding motif (5′-CACGC-3′) (Li et al., 2016) when compared with all known motifs in the HOCOMOCO database (P value = 1E–05) (Supplemental Fig. 5). Some of the other highly represented DNA sequences included motifs for TFs that have previously been shown to interact with AHR such as SP1 (Kobayashi et al., 1996; Wang et al., 1999) and signal transducer and activator of transcription 5 (STAT5) (Kimura et al., 2008).
Phase I: Identification of putative cis PGx-eQTLs.
We next attempted to identify cis PGx-eQTLs from across the genome that did not create/destroy AHREs in ChIP-seq–identified AHR-binding peaks by testing SNPs for their association with variation in the 69 highly differentially expressed genes (FDR ≤0.05, |fold change| ≥2) that we had identified after exposure to either 3-MC or GNF-351 as compared with vehicle (Fig. 2C). Fifty-four of the 69 highly differentially expressed genes (78.3%) contained at least one AHR ChIP-seq–binding peak within 50 kb of the gene, which supported the possibility that those genes might be directly regulated by AHR. Thirty-eight of those AHR-binding peaks had at least one SNP (MAF ≥0.25) within 500 bp of the peak (measured from each end of the peak) that was not an eQTL for that gene under basal conditions, as reported by GTEx (Carithers et al., 2015). Of the 212 SNPs within 500 bp of the AHR-binding peaks, 36 SNPs at 19 loci were associated with variation in RNA-seq–based differential gene expression after exposure to 3-MC or GNF-351 relative to vehicle, thus raising the possibility that they might be cis PGx-eQTLs. A locus was defined as a single SNP or multiple SNPs that were in tight linkage disequilibrium (LD) (R2 > 0.8) that was associated with variation in differential gene expression after ligand exposure as compared with vehicle. Each of those 19 loci included between 1 and 10 SNPs in tight LD (R2 > 0.8). Most loci included just one AHR-binding peak, but one locus (in intron 4 of the AHRR gene) included three AHR-binding peaks because the tightly linked SNPs that were associated with variation in AHRR differential expression after exposure to 3-MC mapped across multiple peaks (Supplemental Table 4). Having identified a group of potential cis PGx-eQTLs by applying the strategy outlined in Fig. 1, we were ready to move to the functional genomics phase of the study.
Phase II: Functional Genomics Phase.
Phase II: Validation of putative cis PGx-eQTLs.
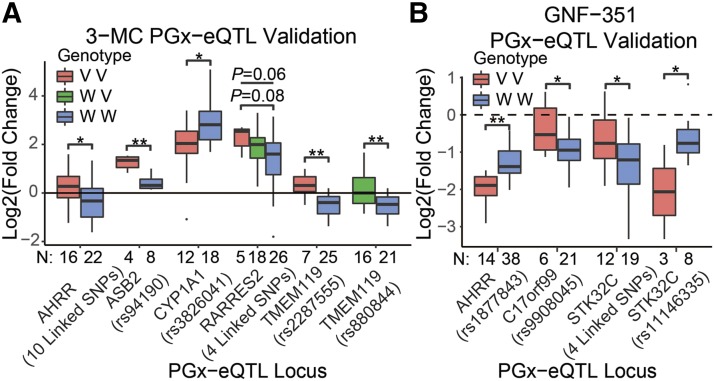
Of the 19 putative cis PGx-eQTLs that we had identified (Supplemental Table 4), four [AMZ1 (two separate linkage blocks), LOC151484, and TPRG1] were not included in the validation experiments because we were unable to identify specific primer sets to perform qRT-PCR (at least three primer sets were tested for each RNA). Of the remaining 15 putative cis PGx-eQTLs, 10 showed significant variation in SNP genotype-dependent gene expression after exposure to 3-MC or GNF-351 relative to vehicle in additional LCLs that were selected based on the SNP genotype for each independent cis PGx-eQTL (Fig. 3; Supplemental Table 4). Those 10 validated loci included 25 SNPs across 11 AHR-binding peaks that were associated with variation in differential gene expression after either 3-MC or GNF-351 exposure. Six of the 10 validated cis PGx-eQTLs were 3-MC dependent (Fig. 3A), and four were GNF-351 dependent (Fig. 3B). None of the cis PGx-eQTLs that we identified were both 3-MC dependent and GNF-351 dependent, which means that the SNPs that were associated with variation in gene expression after 3-MC exposure relative to vehicle were different from the SNPs that were associated with variation in gene expression after GNF-351 exposure. The variation in expression of three genes following ligand exposure were associated with two independent cis PGx-eQTLs for each of the genes—AHRR, TMEM119, and STK32C. One of the AHRR cis PGx-eQTLs was 3-MC dependent, whereas the other was GNF-351 dependent; both of the TMEM119 cis PGx-eQTLs were 3-MC dependent; and both of the STK32C cis PGx-eQTLs were GNF-351 dependent (Fig. 3). Whenever possible, for the genes that had two independent cis PGx-eQTLs, we selected cell lines that were homozygous wild-type at one locus while varying the SNP genotype at the other locus. Therefore, we identified 10 cis PGx-eQTLs, which included 25 SNPs across 11 AHR-binding peaks where SNP-by-treatment interactions altered the degree of AHR-regulated gene differential expression after exposure to either 3-MC or GNF-351 relative to vehicle. A canonical AHRE core motif (5′-CACGC-3′)—the DNA sequence that is recognized to be bound by AHR—was identified in 8 of the 11 AHR ChIP-seq–binding peaks that were present in 7 of the 10 cis PGx-eQTLs (Supplemental Table 4).
Fig. 3.
Validation of PGx-eQTL. Nine of the 15 testable putative PGx-eQTLs were validated by qRT-PCR, and one was suggestive. (A) Six validated PGx-eQTL loci were 3-MC dependent and (B) four validated PGx-eQTLs were GNF-351 treatment dependent. None of the loci were both 3-MC and GNF-351 treatment dependent. When possible, cell lines that were either homozygous variant (red) or homozygous wild-type (blue) were used to test differential expression. When unavailable (TMEM119-rs880844) or when a suggestive P value was detected (RARRES2-4–linked SNPs), cell lines heterozygous for those SNPs were used. All tests for significance used a two-tailed Student’s t test, and ANOVA was used across the three genotypes for the RARRES2 locus. The number of samples (N) for each group is at the bottom of the x-axis for each plot. *P < 0.05; **P < 0.01.
We also asked whether the 11 AHR binding sites across the 10 cis PGx-eQTLs (defined by SNP LD R2 > 0.8) might be functional in cell types other than LCLs by determining whether they were in genomic regions that were active or inactive across different cell types as defined by the epigenome roadmap (Kundaje et al., 2015). Active regions are those that are associated with expressed genes (i.e., enhancers and transcriptionally active sites), and inactive regions are those that are not associated with gene expression (i.e., heterochromatin and repressed polycomb states). Five of the 11 cis PGx-eQTL AHR-binding peaks were in genomic regions that were active across most cell types (Supplemental Fig. 6A). Five of the 11 cis PGx-eQTL AHR-binding peaks were in regions that were highly variable both across cell types and across cell lines within each cell type (Supplemental Fig. 6B). Only 1 of the 11 cis PGx-eQTL AHR-binding peaks was in an inactive genomic region across most cell types (Supplemental Fig. 6C). These observations indicate that many of these loci might be cis PGx-eQTLs in many tissues beyond just LCLs because the cis PGx-eQTL AHR-binding peaks fell within active regions across many cell types.
Therefore, we had identified 10 SNP loci that were not eQTLs under basal conditions, but that were cis PGx-eQTLs dependent on exposure to either the AHR agonist 3-MC or the AHR antagonist GNF-351. Those loci included 11 AHR-binding peaks and 25 SNPs that were associated with variation in differential gene expression of seven genes following 3-MC or GNF-351 exposure—none of which contained SNPs that interrupted a functional AHRE canonical motif. In addition, many of the cis PGx-eQTLs mapped to genomic regions that were active across many different cell types, raising the possibility that those loci may be cis PGx-eQTLs in additional cell types. All of these observations supported our original hypothesis that there might be additional SNPs near AHR-regulated genes that function as cis PGx-eQTLs in an AHR ligand-dependent fashion across the genome beyond the original SNP (rs2470893) that we had identified in the promoter of the CYP1A1 gene (Fig. 2D).
Phase II: Functional investigation of the identified AHR cis PGx-eQTLs.
We next set out to determine whether these 10 validated cis PGx-eQTLs were functional across different cell types by using luciferase reporter gene plasmids for all 10 of the cis PGx-eQTLs (Fig. 3). Those 10 loci included 25 SNPs across 11 peaks, which resulted in 12 SNP-peak regions. These 12 regions included one peak that was surrounded by two independent SNP LD blocks (R2 = 0.324) that were associated with variation in GNF-351–dependent STK32C gene expression and one SNP LD block that was associated with variation in 3-MC dependent AHRR gene expression that spanned three AHR-binding peaks over a 8944-bp genomic region (Supplemental Table 4). Variant and wild-type luciferase reporter gene constructs were generated for each cis PGx-eQTL peak region, and differential luciferase activity was determined relative to a pRL renilla transfection control vector. We tested the luciferase reporter gene constructs in three different cell types: LCL (GM17212), HMC3 microglia-derived cells, and HepG2 hepatic carcinoma-derived cells. We selected these cell lines because AHR is an important regulator of the immune response (LCL and HMC3) and because it plays an important role in xenobiotic metabolism in the liver (HepG2). For purposes of brevity, we only provide detailed results from the luciferase experiments in the main text for three of the cis PGx-eQTLs—one associated with 3-MC–dependent CYP1A1 expression and two associated with variation in ligand-dependent AHRR expression—because CYP1A1 and AHRR are prototypic targets of AHR. Results for the other cis PGx-eQTLs are described in a more concise manner in the main text with detailed results provided in the figure legends.
Functional investigation of CYP1A1 rs3826041 cis PGx-eQTL with luciferase reporter gene assays.
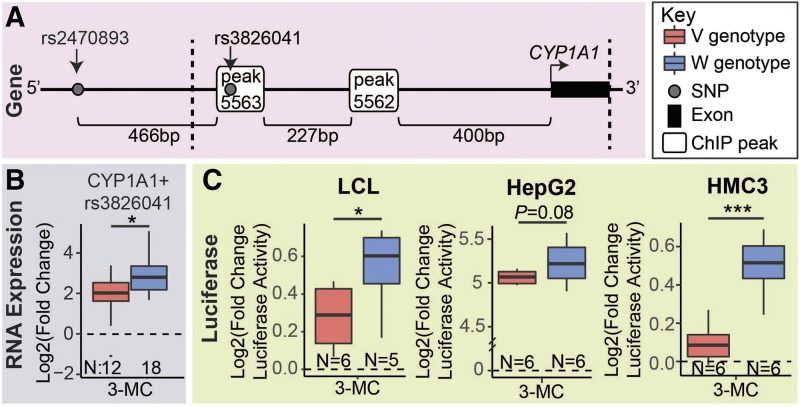
During the present studies, we identified a new cis PGx-eQTL SNP (rs3826041) in the promoter of the CYP1A1 gene. CYP1A1 is a prototypic target for AHR gene regulation and plays an important role in drug and xenobiotic metabolism. Binding profiles of AHR ChIP-seq–binding peaks within 50 kb of the CYP1A1 gene, including the peak near the cis PGx-eQTL SNP for this gene (indicated with an asterisk) are shown in Supplemental Fig. 7. The rs3826041 SNP in the promoter region of the CYP1A1 gene (Fig. 3A) was not in LD with the rs2470893 SNP (R2 = 0.079), which we had previously described as a cis PGx-eQTL (Liu et al., 2018a), and the rs3826041 SNP had a smaller effect on CYP1A1 3-MC–dependent differential expression (Cohen’s d = 1.0) than did the SNP that we had reported previously (rs2470893; Cohen’s d = 2.0). Furthermore, we previously reported that the rs2470893 SNP was 196 bp from a putative canonical AHRE motif (not shown in Fig. 4A) (Liu et al., 2018a), but the ChIP-seq data included in this manuscript showed that there was no AHR binding at that putative AHRE, indicating that it was not a functional AHRE in LCLs. Our AHR ChIP-seq data showed that the rs2470893 SNP was 466 bp from the nearest AHR-binding peak and 471 bp from the nearest AHRE located within that binding peak (Fig. 4A).
Fig. 4.
Functional validation of the CYP1A1 PGx-eQTL. (A) One SNP (rs3826041) in a ChIP-seq–identified AHR-binding peak in the promoter of the CYP1A1 gene was identified as a putative PGx-eQTL after 3-MC treatment. The rs3826041 SNP was different from the rs2470893 SNP previously described, and rs3826041 was within an AHR ChIP-seq peak, but was located 34 bp from an AHRE core motif (that was located within peak 5563). (B) The variant genotype for rs3826041 was associated with a smaller increase in CYP1A1 expression after 3-MC treatment as compared with the wild-type genotype. (C) Luciferase plasmids that contained the variant rs3826041 genotype and AHR-binding region, but not rs2470893 (indicated by dotted lines), demonstrated a smaller increase in luciferase activity after 3-MC treatment in LCLs, HepG2, and HMC3 cells when compared with luciferase plasmids containing the wild-type genotype for rs3826041. RNA expression significance was tested with a two-tailed Student’s t test, and luciferase significance was tested with a one-tailed Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.
The new rs3826041 CYP1A1 cis PGx-eQTL SNP was located in an AHR-binding peak, but was 34 bp distant from an AHR core-binding motif (5′-CACGC-3′), indicating that it did not disrupt the AHRE DNA motif recognized by AHR (Fig. 4A). The rs3826041 variant SNP genotype was associated with a smaller increase in CYP1A1 expression after 3-MC treatment (Fig. 4B), an observation that was confirmed by luciferase reporter gene assays in LCLs and HMC3 cells. The variation in luciferase activity was suggestive in HepG2 cells (P = 0.08) (Fig. 4, C). The luciferase reporter gene constructs that were used to investigate the rs3825041 SNP effect did not contain the previously identified rs2470893 SNP. Therefore, we identified a new CYP1A1 cis PGx-eQTL and had demonstrated that it was functional by luciferase reporter gene assays performed in three cell lines.
Functional investigation of AHRR cis PGx-eQTL SNPs with luciferase reporter gene assays.
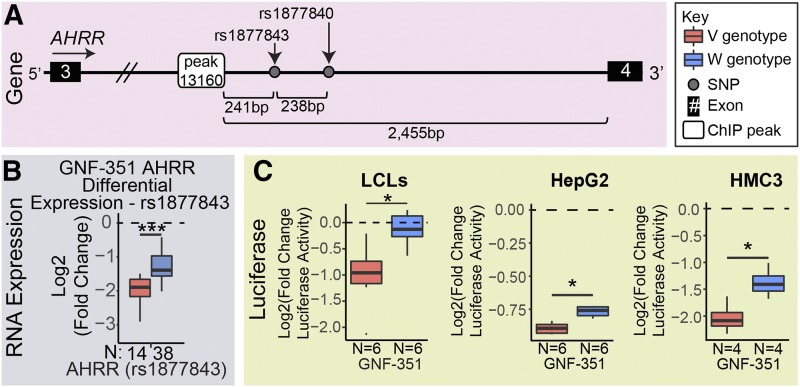
AHRR is another prototypic gene that is transcriptionally regulated by AHR. AHRR is the repressor of AHR and, therefore, provides crucial negative feedback to modulate AHR activation. The AHRR gene was the only gene to display both 3-MC–dependent and GNF-351–dependent cis PGx-eQTLs at two independent loci. AHR ChIP-seq–binding profiles within 50 kb of the AHRR gene, including those near cis PGx-eQTL SNPs (indicated with asterisks), are shown in Supplemental Fig. 8. The first AHRR cis PGx-eQTL—in intron 3 of the AHRR gene—included one SNP (rs1877843) that was 241 bp away from AHR-binding peak 13160 and was associated with variation in AHRR expression after GNF-351 exposure relative to vehicle (Fig. 5A). Therefore, we used luciferase reporter gene assays to study this locus in different cell types.
Fig. 5.
Functional investigation of the AHRR GNF-351–dependent PGx-eQTL. (A) The GNF-351–dependent PGx-eQTL for AHRR was in intron 3 of the AHRR gene and contained rs1877843, which was 241 bp from peak 13160. (B) The variant rs1877843 SNP genotype was associated with a larger decrease in AHRR expression after GNF-351 treatment than the wild-type SNP genotype. (C) Luciferase plasmids that contained the variant SNP genotypes for rs1877843 and rs1877840 demonstrated a larger decrease in luciferase activity after GNF-351 treatment compared with luciferase plasmids containing the wild-type SNP genotypes in all three cell lines. The number of samples (N) for each group is at the bottom of each plot. All tests for significance used a two-tailed Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.
Variant genotypes for the SNPs near AHR-binding peak 13160 (rs1877843 and rs1877840) in intron 3 of the AHRR gene (Fig. 5A) were associated with a larger decrease in AHRR expression after GNF-351 treatment than was observed for the wild-type genotype (Fig. 5B). This effect was consistent in all three cell lines—LCLs, HepG2, and HMC3 (Fig. 5C). These data indicate that this locus is a functional cis PGx-eQTL and that it has the potential to act functionally in cell types beyond just LCLs.
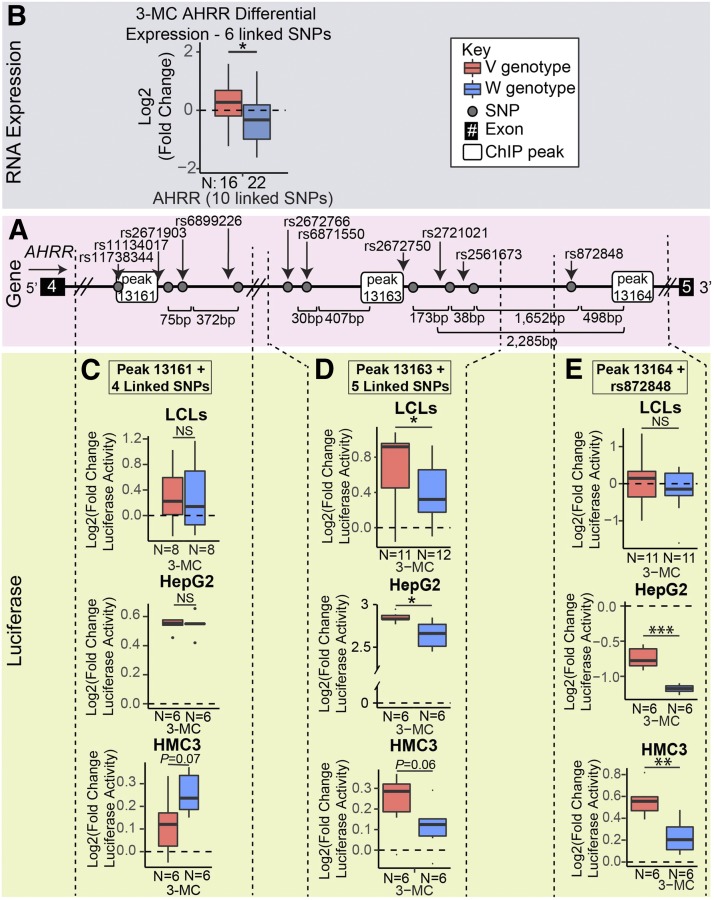
The other AHRR cis PGx-eQTL—which mapped to intron 4 of the AHRR gene—contained 10 tightly linked SNPs (R2 ≥ 0.88) across three AHR-binding peaks (Fig. 6A). Variant genotypes for these 10 SNPs were associated with higher AHRR expression relative to the wild-type genotypes after 3-MC exposure (Fig. 6B). The distances between the three AHR-binding peaks in this locus were large enough (6687 bp and 2284 bp) to permit the generation of separate luciferase plasmids for each AHR-binding peak together with the surrounding cis PGx-eQTL SNPs—thus making it possible to study and identify the three groupings of SNPs around each individual AHR-binding peak that comprised this cis PGx-eQTL, which included a large LD block.
Fig. 6.
Functional investigation of the AHRR 3-MC–dependent PGx-eQTL. (A) The 3-MC–dependent PGx-eQTL for AHRR included 10 SNPs across peaks 13161, 13163, and 13164 in intron 4 of the AHRR gene. (B) The variant SNP genotypes for the 10 linked SNPs that were across three AHR-binding peaks in intron 4 of the AHRR gene were associated with an increase after 3-MC treatment, and the wild-type was associated with a decrease after 3-MC treatment. (C) The luciferase plasmid that contained the variant SNP genotypes for the SNPs surrounding peak 13161 (as indicated by the dashed lines) demonstrated no difference in LCLs or HepG2 cells and a smaller increase than the wild-type SNP genotype in HMC3 cells, which was suggestive. (D) The luciferase plasmid that contained the variant genotypes for the five SNPs near peak 13163 in intron 4 of the AHRR gene (as indicated by the dashed lines) demonstrated a larger increase in luciferase activity after 3-MC treatment than the wild-type in LCLs, HepG2, and HMC3—the same direction observed for differential expression in LCLs. (E) The luciferase plasmid that contained the variant genotype for rs872848 and peak 13164 (indicated by the dashed lines) demonstrated no difference in luciferase activity in LCLs, but showed a smaller decrease than did the plasmid that contained the wild-type genotype for rs872848 with peak 13164 in HepG2 and displayed a larger increase in luciferase activity in HMC3. The number of samples (N) for each group is at the bottom of each plot. All tests for significance used a two-tailed Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.
The luciferase plasmids that contained AHR-binding peak 13161 did not demonstrate a SNP-by-treatment–dependent effect in any of the three cell lines studied (LCLs, HepG2 cells, or HMC3 cells) (Fig. 6C). This result indicates that the four SNPs surrounding AHR-binding peak 13161 may not contribute to the SNP-by-treatment–dependent variation in 3-MC–dependent AHRR expression observed by qRT-PCR (Fig. 6B) and that the SNPs that confer this SNP-by-treatment–dependent variation in gene expression may be those around AHR-binding peak 13163 or AHR-binding peak 13164 (Fig. 6A). Alternatively, the SNPs near AHR peak 13161 may operate over longer distances via peaks not included in this reporter construct or may require three-dimensional chromatin organization.
Indeed, the luciferase plasmids that contained AHR-binding peak 13163 with the surrounding SNPs (Fig. 6A) demonstrated SNP-by-treatment–dependent variation in luciferase activity following 3-MC treatment, and the responses were consistent in all three cells lines studied (LCLs, HepG2 cells, and HMC3 cells). The variant SNP genotypes demonstrated a larger increase in luciferase activity than the wild-type SNP genotypes after 3-MC treatment (Fig. 6D). These results indicate that AHR-binding peak 13163 together with the surrounding SNPs is functional and may contribute to the SNP-by-treatment–dependent variation in AHRR expression observed by qRT-PCR (Fig. 6B).
In addition, luciferase plasmids that contained peak 13164 and SNP rs872848 demonstrated a SNP-by-treatment–dependent variation in luciferase activity in HepG2 cells and HMC3 cells, but not in LCLs (Fig. 6E). However, the direction of this differential expression from baseline was not the same in all three cell lines for this region. Specifically, luciferase activity decreased in HepG2 cells following 3-MC exposure, but increased in HMC3 cells following 3-MC treatment. Even though the luciferase activity increased in HMC3 cells and decreased in HepG2 cells, the SNP-dependent effect was maintained, because the variant genotype demonstrated a larger increase after 3-MC treatment in HMC3 cells and a smaller decrease after 3-MC treatment in HepG2 cells (Fig. 6E). Therefore, peak 13164 and SNP rs872848 may contribute to the SNP-by-treatment–dependent variation observed for this locus, but only in specific cell types.
Therefore, this locus, which included 10 tightly linked SNPs across three AHR-binding peaks, demonstrated strong cell line–specific and SNP-by-treatment–dependent effects on gene expression. Specifically, the regions of this locus that included peak 13163 with the surrounding SNPs and peak 13164 with rs872848 both demonstrated SNP-by-treatment–dependent variation in luciferase activity, which suggests that they may both contribute to the overall SNP-by-treatment–dependent variation observed by qRT-PCR (Fig. 6B). The observed cell line–dependent effects may be due to differences in the cellular environment within each cell type, which could include differences in chromatin organization or differences in the expression of coregulator proteins that bind in a complex with AHR.
Functional investigation of additional cis PGx-eQTL SNPs with luciferase reporter gene assays.
The other seven cis PGx-eQTL genes functionally validated by qRT-PCR (see Fig. 3) were not prototypic AHR targets, but included several immune-related genes (RARRES2, TMEM119, and ASB2). We also studied generalizability across cell lines for these seven cis PGx-eQTLs by using luciferase reporter gene assays in LCLs, HepG2 cells, and HMC3 cells. Six of those seven cis PGx-eQTLs demonstrated SNP-by-treatment–dependent variation in luciferase activity following ligand exposure.
We first investigated the two 3-MC–dependent TMEM119 cis PGx-eQTLs that were 3′ of the TMEM119 gene. These SNPs were not in LD (R2 = 0.154) (Supplemental Fig. 9A). The cis PGx-eQTL that contained peak 3966 and SNP rs2287555 demonstrated a significant SNP-by-treatment–dependent variation in luciferase activity in HMC3 cells, where the variant SNP genotype was associated with decreased luciferase activity compared with the wild-type SNP genotype following 3-MC exposure—opposite to the LCL qRT-PCR results (Supplemental Fig. 9B). No significant variation was observed in LCLs or HepG2 cells (Supplemental Fig. 9C). The cis PGx-eQTL that included peak 3967 and SNP rs880844 demonstrated SNP-by-treatment–dependent variation in luciferase activity in all three cell lines (Supplemental Fig. 9D). However, consistent with the qRT-PCR results, the variant rs880844 SNP was associated with higher luciferase activity than the wild-type SNP genotype following 3-MC treatment in LCLs (Supplemental Fig. 9D). However, the variant rs880844 SNP genotype was associated with lower luciferase activity than observed for the wild-type SNP genotype in both HMC3 and HepG2 cells (Supplemental Fig. 9E).
The 3-MC–dependent RARRES2 cis PGx-eQTL mapped 3′ of the RARRES2 gene and included peak 15754 and four tightly linked SNPs (R2 = 1) (Supplemental Fig. 10A). The luciferase assays demonstrated SNP-by-treatment–dependent variation in luciferase activity for which variant SNP genotypes were associated with higher expression than were wild-type SNP genotypes following 3-MC exposure in HepG2 cells and LCLs (suggestive P = 0.06; Supplemental Fig. 10C). Those results were consistent with the LCL qRT-PCR results (Supplemental Fig. 10B). However, a similar response was not observed in HMC3 cells (Supplemental Fig. 10C).
The 3-MC–dependent ASB2 cis PGx-eQTL that mapped to intron 1 of the ASB2 gene (Supplemental Fig. 11A) demonstrated SNP-by-treatment–dependent variation in luciferase activity in all three cell lines studied. Consistent with the LCL qRT-PCR results (Supplemental Fig. 11B), the variant rs941490 SNP genotype was associated with higher luciferase activity compared with the wild-type SNP genoptype following 3-MC exposure in LCLs and HepG2 cells, but the variant SNP genotype was associated with lower luciferase activity compared with the wild-type SNP genotypes in HMC3 cells (Supplemental Fig. 11C).
We also identified SNP-by-treatment–dependent variation in luciferase activity for both of the GNF-351–dependent STK32C cis PGx-eQTLs in exon 1 of the STK32C gene (Supplemental Fig. 12A). The rs11146335 SNP was not in LD (R2 ≤ 0.32) with the other four SNPs, all of which were tightly linked (R2 ≥ 0.96). The cis PGx-eQTL that included peak 2391 and rs11146335 SNP, which was in the AHR-binding peak but was not in an AHRE, demonstrated SNP-by-treatment–dependent variation in luciferase activity following GNF-351 exposure in all three cell lines studied. Consistent with the LCL qRT-PCR results (Supplemental Fig. 12B), the variant rs11146335 SNP genotype was associated with lower luciferase activity compared with the wild-type SNP genotype in HepG2 cells, but the variant SNP genotype was associated with higher expression compared with the wild-type SNP genotype in LCLs and HMC3 cells (Supplemental Fig. 12C). The STK32C cis PGx-eQTL that included the four linked SNPs around peak 2391 demonstrated SNP-by-treatment–dependent variation in luciferase activity in LCLs in which variant SNP genotypes were associated with higher luciferase activity compared with wild-type SNP genotypes, which was consistent with the LCL qRT-PCR expression results (Supplemental Fig. 12D). However, a significant variation in luciferase activity was not observed in HMC3 or HepG2 cells (Supplemental Fig. 12E).
Finally, no SNP-by-treatment–dependent variation in luciferase activity was observed for the GNF-351 C17orf99 cis PGx-eQTL in any of the three cell lines studied (LCLs, HMC3, or HepG2; data not shown). It should be emphasized that this does not necessarily mean that this locus is not a cis PGx-eQTL, but rather that our in vitro assay, which lacks genomic context, was unable to detect a cis PGx-eQTL relationship. For example, the genomic context required to detect the effect might include the effects of three-dimensional chromatin organization or genomic regions that are required for the binding of coregulators.
In summary, three of the 10 validated cis PGx-eQTLs demonstrated SNP-by-treatment–dependent variation in luciferase activity that was consistent across all three different cell lines that we studied and was also consistent with the qRT-PCR results for LCLs. An additional three loci demonstrated SNP-by-treatment–dependent variation in luciferase activity across all three cell lines, but with opposite SNP-dependent effects in one of the cell lines studied. One of the 10 loci demonstrated a SNP-by-treatment–dependent variation in luciferase activity in two of the three cell lines, and two of the cis PGx-eQTLs demonstrated SNP-by-treatment–dependent variation in luciferase activity in only one of the three cell lines studied. Finally, only one locus failed to demonstrate SNP-by-treatment–dependent variation in luciferase activity in any of the cell lines studied. These results clearly demonstrate that cellular context is an important contriubutor to the functional implications of these cis PGx-eQTLs.
Discussion
We have described a novel type of eQTL in which SNPs at a distance from AHR binding sites are associated with interindividual variation in target gene expression in a ligand-by-genotype–dependent fashion. Our findings indicate that SNPs across the genome that do not create/destroy an AHRE can, nonetheless, demonstrate an AHR-dependent SNP-by-drug interaction that results in variation in the degree or direction of target gene expression following exposure to AHR ligands. However, those SNPs are not associated with variation in gene expression in the absence of AHR ligand treatment, so their functional impact is only detected in the presence of drug. We refer to these SNPs as PGx-eQTLs because they involve SNPs that are associated with variation in expression after drug or xenobiotic treatment. PGx-eQTLs include SNPs in transcription factor core–binding motifs as well as SNPs that are distant from transcription factor core motifs. However, in this study, we focused solely on SNPs that were outside of functional AHREs. These findings are potentially important because >90% of SNPs that are significantly associated with phenotypes (including PGx phenotypes) that have been identified during GWAS have mapped to noncoding regions of the genome (Hrdlickova et al., 2014; Farh et al., 2015; Tak and Farnham, 2015), but only 25.6% of GWAS SNPs are also eQTLs in the GTEx database (v7) (Carithers et al., 2015; MacArthur et al., 2017). Thus, the function of many clinically relevant SNPs remains unclear. Indeed, previous reports indicated that some SNPs only demonstrate associations with gene expression under specific conditions, such as exposure to drugs or other exogenous compounds (Li et al., 2008; Grundberg et al., 2011; Liu et al., 2012, 2018a; Fairfax et al., 2014; Moyerbrailean et al., 2016; Zhou, 2016; Ho et al., 2017; Knowles et al., 2017). The novelty of the results presented in this manuscript lies in 1) the genome-wide approach, which allowed identification of many novel cis PGx-eQTLs, and 2) our focus on SNPs that did not create/destroy functional AHREs, but still altered AHR ligand-dependent target gene expression.
Of interest is the fact that we identified different cis PGx-eQTLs after exposure to 3-MC (an AHR agonist) and GNF-351 (an AHR antagonist) (Fig. 3), possibly because different ligands may result in different AHR conformations that could potentially impact the effect of coregulatory protein complex interactions. Furthermore, many of those cis PGx-eQTLs were in active genomic regions across multiple cell types, and our luciferase assays demonstrated that some of the cis PGx-eQTLs were consistent across all cell types studied, whereas other loci were not.
We observed four general types of cis PGx-eQTLs: 1) cis PGx-eQTLs that were highly consistent across the three different cell types that we studied (LCLs, HMC3, and HepG2); 2) cis PGx-eQTLs that demonstrated opposing directions of differential expression in some cell lines regardless of SNP genotype; 3) cis PGx-eQTLs that demonstrated SNP-dependent effects in some, but not all of the cell lines studied; and 4) cis PGx-eQTLs that resulted in different directionality for some of the cell lines. The different directions for differential expression as well as the variation in SNP-dependent effects after the same treatment in different cell lines may be due to variation in the sensitivity of those cell lines to these exogenous compounds or to variation in the cellular environment such as the epigenetic landscape, including chromatin looping or variation in the expression of proteins, such as TF coregulators in different cell types.
Of interest, 4 of the 10 validated cis PGx-eQTLs that we identified were previously associated with drug and environment interaction phenotypes. The rs3826041 cis PGx-eQTL SNP in the promoter of the CYP1A1 gene (Fig. 6A) or SNPs in tight LD with that SNP (rs4886605, r2 = 0.87; rs1456432, r2 = 0.87) were previously associated with variability in coronary artery disease risk in Uygur men (Zou et al., 2014), variation in warfarin response (Luo et al., 2017), and in hepatocellular carcinoma risk (Li et al., 2009). In another study, the same SNPs were associated with risk for testicular germ cell tumors and variation in oxychlorodane concentration, a chemical that has been associated with AHR activation (Ng et al., 2010). Therefore, activation of AHR by oxychlorodane could alter CYP1A1 expression through this cis PGx-eQTL and could, hypothetically, lead to altered risk for testicular germ cell tumors.
In addition, the rs2671903 cis PGx-eQTL SNP in intron 4 of the AHRR gene near peak 13161 (Fig. 5A) has previously been associated with variation in CpG methylation in cigarette smokers versus nonsmokers, which indicated a gene-by-environment interaction that resulted in significant variation in AHRR expression (Tsaprouni et al., 2014). Cigarette smoke is known to contain polycyclic aromatic hydrocarbons that are AHR agonists, which could result in SNP-dependent AHRR variation in expression in response to cigarette smoke at this cis PGx-eQTL (Vu et al., 2015).
The rs880844 cis PGx-eQTL SNP 3′ of the TMEM119 (Supplemental Fig. 9) gene has been suggestively associated with rheumatoid arthritis risk. Of importance, AHR activity has been associated with rheumatoid arthritis as well as with increased risk for rheumatoid arthritis in smokers; and AHR activity has been demonstrated to contribute to rheumatoid arthritis severity in animal models through the modulation of environmental stimuli (Nakahama et al., 2011; Kazantseva et al., 2012; Vogel et al., 2014; Fu et al., 2018).
Finally, the SNPs that were in tight LD (r2 > 0.8) with the RARRES2 cis PGx-eQTL SNPs have previously been associated with variation in body mass index, metabolic, and inflammatory diseases and levels of chemerin (Tönjes et al., 2014) (the protein encoded by the RARRES2 gene). Chemerin is mainly expressed in adipose tissues and is an inflammatory modulator associated with metabolic and inflammatory diseases. AHR is known to modulate inflammatory response, and AHR is very highly expressed in both adipose tissue types reported by GTEx (Supplemental Fig. 4A). Therefore, regulation of RARRES2 by the AHR cis PGx-eQTL could have implications for metabolic and inflammatory diseases (Min et al., 2012; Tonjes et al., 2014; Er et al., 2018).
In summary, the AHR-based cis PGx-eQTLs studied in this work have demonstrated : 1) that cis PGx-eQTLs can be observed at multiple loci across the genome—at least for AHR; 2) that multiple cis PGx-eQTLs can regulate the expression of the same gene; 3) that there are cell line–dependent effects on differential expression after the same treatment; 4) that cis PGx-eQTLs can have different functional effects in different cell types; and 5) that cis PGx-eQTLs have potential clinical utility, including for drug response phenotypes. These results suggest that AHR cis PGx-eQTLs may play a role in disease pathophysiology and in response to drug therapy or xenobiotic exposure.
Obviously, this study has not resulted in an exhaustive list of AHR ligand–dependent PGx-eQTLs, but this series of experiments has provided evidence that SNPs across the genome that are at a distance from AHREs can influence AHR ligand–dependent expression. Based on the results of our previous studies, these SNPs may interact with coregulator(s) that binds to AHR to form a larger complex and may alter complex stability binding at that locus (Liu et al., 2012, 2018a; Zhou, 2016; Ho et al., 2017; Qin et al., 2017). Future studies aimed at understanding AHR PGx-eQTLs should focus on identifying mechanisms underlying the function of these SNPs and mechanisms responsible for their cell-specific effects.
Finally, there are many ligand-activated TFs other than AHR that have clinical relevance, including estrogen receptor α, androgen receptor, and glucocorticoid receptor, for which PGx-eQTLs might influence ligand-dependent expression and alter response to nuclear receptor–targeted drugs. Therefore, the present study represents only an initial step toward future studies of PGx-eQTLs designed to enhance our understanding of the molecular basis for this type of interindividual variation in gene expression, variation that might influence response after exposure to drugs or other xenobiotics. In summary, the data presented in this manuscript suggest that interindividual variation in response to drugs and xenobiotics that act as ligands for ligand-activated TFs may be due, in part, to SNP genotypes that are outside of ligand-activated TF binding sites. This functional SNP genotype information might potentially be used to predict and alter drug therapy response, i.e., for PGx purposes. The present results have clearly demonstrated that PGx-eQTLs differ from, but are complementary to, baseline eQTLs. Future studies should help us better understand how polymorphisms in noncoding regions of the genome might contribute to variation in gene expression after exposure to exogenous compounds and, as a result, how they might contribute to variation in drug response.
Accession Numbers
The accession numbers for the SNP genotype data for the lymphoblastoid cell lines reported in this paper are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo): GSE24277. The accession numbers for RNA-seq and ChIP-seq data reported in this paper are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo): GSE116638.
Abbreviations
- 3-MC
3-methylcholanthrene
- AHR
aryl hydrocarbon receptor
- AHRE
AHR response element
- bp
base pair
- ChIP
chromatin immunoprecipitation
- eQTL
expression quantitative trait locus
- FDR
false discovery rate
- GNF-351
N-[2-(3H-indol-3-yl)ethyl]-9-isopropyl-2-(5-methyl-3-pyridyl)purin-6-amine
- GWAS
genome-wide association study
- kb
kilobase
- LCL
lymphoblastoid cell line
- LD
linkage disequilibrium
- MAF
minor allele frequency
- PGx
pharmacogenomic
- qPCR
quantitative polymerase chain reaction
- qRT-PCR
quantitative reverse-transcription polymerase chain reaction
- SNP
single-nucleotide polymorphism
- TF
transcription factor
Authorship Contributions
Participated in research design: Neavin, Li, Wang, Ordog, Weinshilboum.
Conducted experiments: Neavin.
Performed data analysis: Neavin, Ye.
Wrote or contributed to the writing of the manuscript: Neavin, Lee, Liu, Li, Ordog, Weinshilboum.
Footnotes
L.W. and R.M.W. are cofounders of and stockholders in OneOme, a pharmacogenomic decision support company. The other authors have no conflicts to declare.
This work was supported, in part, by the National Institutes of Health National lnstitute of General Medical Sciences [Grants GM61388 (the Pharmacogenomics Research Network), GM28157, GM114838, GM072474], and in part by the National Science Foundation [Grant 1624615 (The Center for Computational Biology and Genomic Medicine)].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Bailey TL. (2011) DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27:1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro LB, Tailleux L, Pai AA, Gicquel B, Marioni JC, Gilad Y. (2012) Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc Natl Acad Sci USA 109:1204–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bream JH, Ping A, Zhang X, Winkler C, Young HA. (2002) A single nucleotide polymorphism in the proximal IFN-gamma promoter alters control of gene transcription. Genes Immun 3:165–169. [DOI] [PubMed] [Google Scholar]
- Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J, Gelfand ET, et al. GTEx Consortium (2015) A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv Biobank 13:311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis MC, Monda KL, Yu K, Paynter N, Azzato EM, Bennett SN, Berndt SI, Boerwinkle E, Chanock S, Chatterjee N, et al. (2011) Genome-wide meta-analysis identifies regions on 7p21 (AHR) and 15q24 (CYP1A2) as determinants of habitual caffeine consumption. PLoS Genet 7:e1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudenkov TM, Ingle JN, Buzdar AU, Robson ME, Kubo M, Ibrahim-Zada I, Batzler A, Jenkins GD, Pietrzak TL, Carlson EE, et al. (2017) SLCO1B1 polymorphisms and plasma estrone conjugates in postmenopausal women with ER+ breast cancer: genome-wide association studies of the estrone pathway. Breast Cancer Res Treat 164:189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CHB, Lewis S, Strayer C, Landreman A. (2016) Designing a Bioluminescent Reporter Assay: Normalization, Promega, Promega Corporation. [Google Scholar]
- Er LK, Wu S, Hsu LA, Teng MS, Sun YC, Ko YL. (2018) Pleiotropic associations of RARRES2 gene variants and circulating chemerin levels: potential roles of chemerin involved in the metabolic and inflammation-related diseases. Mediators Inflamm 2018:4670521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. (2017) Chromatin-state discovery and genome annotation with ChromHMM. Nat Protoc 12:2478–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, Jostins L, Plant K, Andrews R, McGee C, et al. (2014) Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343:1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, et al. (2015) Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Nogueira SV, Drongelen VV, Coit P, Ling S, Rosloniec EF, Sawalha AH, Holoshitz J. (2018) Shared epitope-aryl hydrocarbon receptor crosstalk underlies the mechanism of gene-environment interaction in autoimmune arthritis. Proc Natl Acad Sci USA 115:4755–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundberg E, Adoue V, Kwan T, Ge B, Duan QL, Lam KC, Koka V, Kindmark A, Weiss ST, Tantisira K, et al. (2011) Global analysis of the impact of environmental perturbation on cis-regulation of gene expression. PLoS Genet 7:e1001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Neavin D, Liu D, Biernacka J, Hall-Flavin D, Bobo WV, Frye MA, Skime M, Jenkins GD, Batzler A, et al. (2016) TSPAN5, ERICH3 and selective serotonin reuptake inhibitors in major depressive disorder: pharmacometabolomics-informed pharmacogenomics. Mol Psychiatry 21:1717–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho MF, Ingle JN, Bongartz T, Kalari KR, Goss PE, Shepherd LE, Mushiroda T, Kubo M, Wang L, Weinshilboum RM. (2017) TCL1A single-nucleotide polymorphisms and estrogen-mediated Toll-like receptor-MYD88-dependent nuclear factor-κB activation: single-nucleotide polymorphism- and selective estrogen receptor modulator-dependent modification of inflammation and immune response. Mol Pharmacol 92:175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdlickova B, de Almeida RC, Borek Z, Withoff S. (2014) Genetic variation in the non-coding genome: involvement of micro-RNAs and long non-coding RNAs in disease. Biochim Biophys Acta 1842:1910–1922. [DOI] [PubMed] [Google Scholar]
- Ingle JN, Liu M, Wickerham DL, Schaid DJ, Wang L, Mushiroda T, Kubo M, Costantino JP, Vogel VG, Paik S, et al. (2013) Selective estrogen receptor modulators and pharmacogenomic variation in ZNF423 regulation of BRCA1 expression: individualized breast cancer prevention. Cancer Discov 3:812–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle JN, Schaid DJ, Goss PE, Liu M, Mushiroda T, Chapman J-AW, Kubo M, Jenkins GD, Batzler A, Shepherd L, et al. (2010) Genome-wide associations and functional genomic studies of musculoskeletal adverse events in women receiving aromatase inhibitors. J Clin Oncol 28:4674–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Schaid DJ, Desta Z, Kubo M, Batzler AJ, Snyder K, Mushiroda T, Kamatani N, Ogburn E, Hall-Flavin D, et al. (2014) Citalopram and escitalopram plasma drug and metabolite concentrations: genome-wide associations. Br J Clin Pharmacol 78:373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowski M, Grubert F, Heffelfinger C, Hariharan M, Asabere A, Waszak SM, Habegger L, Rozowsky J, Shi M, Urban AE, et al. (2010) Variation in transcription factor binding among humans. Science 328:232–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantseva MG, Highton J, Stamp LK, Hessian PA. (2012) Dendritic cells provide a potential link between smoking and inflammation in rheumatoid arthritis. Arthritis Res Ther 14:R208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Naka T, Nohara K, Fujii-Kuriyama Y, Kishimoto T. (2008) Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Natl Acad Sci USA 105:9721–9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JC, Udalova I, Hill AV, Greenwood BM, Peshu N, Marsh K, Kwiatkowski D. (1999) A polymorphism that affects OCT-1 binding to the TNF promoter region is associated with severe malaria. Nat Genet 22:145–150. [DOI] [PubMed] [Google Scholar]
- Knowles DA, Davis JR, Edgington H, Raj A, Favé MJ, Zhu X, Potash JB, Weissman MM, Shi J, Levinson DF, et al. (2017) Allele-specific expression reveals interactions between genetic variation and environment. Nat Methods 14:699–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Sogawa K, Fujii-Kuriyama Y. (1996) Cooperative interaction between AhR.Arnt and Sp1 for the drug-inducible expression of CYP1A1 gene. J Biol Chem 271:12310–12316. [DOI] [PubMed] [Google Scholar]
- Kulakovskiy IV, Vorontsov IE, Yevshin IS, Sharipov RN, Fedorova AD, Rumynskiy EI, Medvedeva YA, Magana-Mora A, Bajic VB, Papatsenko DA, et al. (2018) HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res 46 (D1):D252–D259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al. Roadmap Epigenomics Consortium (2015) Integrative analysis of 111 reference human epigenomes. Nature 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. (2012) ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22:1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E-Y, Huang W-Y, Chang Y-C, Tsai M-H, Chuang EY, Kuok Q-Y, Bai S-T, Chao L-Y, Sher Y-P, Lai L-C. (2016) Aryl hydrocarbon receptor activates NDRG1 transcription under hypoxia in breast cancer cells. Sci Rep 6:20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Fridley B, Kalari K, Jenkins G, Batzler A, Safgren S, Hildebrandt M, Ames M, Schaid D, Wang L. (2008) Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res 68:7050–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Shugart YY, Zhou W, An Y, Yang Y, Zhou Y, Zhang B, Lu D, Wang H, Qian J, et al. (2009) Common genetic variations of the cytochrome P450 1A1 gene and risk of hepatocellular carcinoma in a Chinese population. Eur J Cancer 45:1239–1247. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Ho MF, Schaid DJ, Scherer SE, Kalari K, Liu M, Biernacka J, Yee V, Evans J, Carlson E, et al. (2017) Breast cancer chemoprevention pharmacogenomics: deep sequencing and functional genomics of the ZNF423 and CTSO genes. NPJ Breast Cancer 3:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Qin S, Ray B, Kalari KR, Wang L, Weinshilboum RM. (2018a) Single nucleotide polymorphisms (SNPs) distant from xenobiotic response elements can modulate aryl hydrocarbon receptor function: SNP-dependent CYP1A1 induction. Drug Metab Dispos 46:1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Ray B, Neavin DR, Zhang J, Athreya AP, Biernacka JM, Bobo WV, Hall-Flavin DK, Skime MK, Zhu H, et al. (2018b) Beta-defensin 1, aryl hydrocarbon receptor and plasma kynurenine in major depressive disorder: metabolomics-informed genomics. Transl Psychiatry 8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Goss PE, Ingle JN, Kubo M, Furukawa Y, Batzler A, Jenkins GD, Carlson EE, Nakamura Y, Schaid DJ, et al. (2014) Aromatase inhibitor-associated bone fractures: a case-cohort GWAS and functional genomics. Mol Endocrinol 28:1740–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Ingle JN, Fridley BL, Buzdar AU, Robson ME, Kubo M, Wang L, Batzler A, Jenkins GD, Pietrzak TL, et al. (2013) TSPYL5 SNPs: association with plasma estradiol concentrations and aromatase expression. Mol Endocrinol 27:657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wang L, Bongartz T, Hawse JR, Markovic SN, Schaid DJ, Mushiroda T, Kubo M, Nakamura Y, Kamatani N, et al. (2012) Aromatase inhibitors, estrogens and musculoskeletal pain: estrogen-dependent T-cell leukemia 1A (TCL1A) gene-mediated regulation of cytokine expression. Breast Cancer Res 14:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Li X, Zhu M, Tang J, Li Z, Zhou X, Song G, Liu Z, Zhou H, Zhang W. (2017) Identification of novel variants associated with warfarin stable dosage by use of a two-stage extreme phenotype strategy. J Thromb Haemost 15:28–37. [DOI] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, et al. (2017) The new NHGRI-EBI catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 45 (D1):D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JL, Nicholson G, Halgrimsdottir I, Almstrup K, Petri A, Barrett A, Travers M, Rayner NW, Mägi R, Pettersson FH, et al. GIANT Consortium. MolPAGE Consortium (2012) Coexpression network analysis in abdominal and gluteal adipose tissue reveals regulatory genetic loci for metabolic syndrome and related phenotypes. PLoS Genet 8:e1002505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyerbrailean GA, Richards AL, Kurtz D, Kalita CA, Davis GO, Harvey CT, Alazizi A, Watza D, Sorokin Y, Hauff N, et al. (2016) High-throughput allele-specific expression across 250 environmental conditions. Genome Res 26:1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahama T, Kimura A, Nguyen NT, Chinen I, Hanieh H, Nohara K, Fujii-Kuriyama Y, Kishimoto T. (2011) Aryl hydrocarbon receptor deficiency in T cells suppresses the development of collagen-induced arthritis. Proc Natl Acad Sci USA 108:14222–14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neavin DR, Liu D, Ray B, Weinshilboum RM. (2018) The role of the aryl hydrocarbon receptor (AHR) in immune and inflammatory diseases. Int J Mol Sci 19:3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CH, Janoo-Gilani R, Sipahimalani P, Gallagher RP, Gascoyne RD, Connors JM, Weber JP, Lai AS, Leach S, Le ND, et al. (2010) Interaction between organochlorines and the AHR gene, and risk of non-Hodgkin lymphoma. Cancer Causes Control 21:11–22. [DOI] [PubMed] [Google Scholar]
- Niu N, Qin Y, Fridley BL, Hou J, Kalari KR, Zhu M, Wu TY, Jenkins GD, Batzler A, Wang L. (2010) Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res 20:1482–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller M, et al. (2011) An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478:197–203. [DOI] [PubMed] [Google Scholar]
- Qin S, Ingle JN, Liu M, Yu J, Wickerham DL, Kubo M, Weinshilboum RM, Wang L. (2017) Calmodulin-like protein 3 is an estrogen receptor alpha coregulator for gene expression and drug response in a SNP, estrogen, and SERM-dependent fashion. Breast Cancer Res 19:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- Shehin SE, Stephenson RO, Greenlee WF. (2000) Transcriptional regulation of the human CYP1B1 gene: evidence for involvement of an aryl hydrocarbon receptor response element in constitutive expression. J Biol Chem 275:6770–6776. [DOI] [PubMed] [Google Scholar]
- Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. (2014) The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol 32:403–432. [DOI] [PubMed] [Google Scholar]
- Tak YG, Farnham PJ. (2015) Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin 8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tönjes A, Scholz M, Breitfeld J, Marzi C, Grallert H, Gross A, Ladenvall C, Schleinitz D, Krause K, Kirsten H, et al. (2014) Genome wide meta-analysis highlights the role of genetic variation in RARRES2 in the regulation of circulating serum chemerin. PLoS Genet 10:e1004854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaprouni LG, Yang TP, Bell J, Dick KJ, Kanoni S, Nisbet J, Viñuela A, Grundberg E, Nelson CP, Meduri E, et al. (2014) Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics 9:1382–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel CF, Khan EM, Leung PS, Gershwin ME, Chang WL, Wu D, Haarmann-Stemmann T, Hoffmann A, Denison MS. (2014) Cross-talk between aryl hydrocarbon receptor and the inflammatory response: a role for nuclear factor-κB. J Biol Chem 289:1866–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu AT, Taylor KM, Holman MR, Ding YS, Hearn B, Watson CH. (2015) Polycyclic aromatic hydrocarbons in the mainstream smoke of popular U.S. cigarettes. Chem Res Toxicol 28:1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang W, Safe S. (1999) Regulation of constitutive gene expression through interactions of Sp1 protein with the nuclear aryl hydrocarbon receptor complex. Biochemistry 38:11490–11500. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang S, Li W. (2012) RSeQC: quality control of RNA-seq experiments. Bioinformatics 28:2184–2185. [DOI] [PubMed] [Google Scholar]
- Yang SY, Ahmed S, Satheesh SV, Matthews J. (2018) Genome-wide mapping and analysis of aryl hydrocarbon receptor (AHR)- and aryl hydrocarbon receptor repressor (AHRR)-binding sites in human breast cancer cells. Arch Toxicol 92:225–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Solomon S, Fraser LR, Trombino AF, Liu D, Sonenshein GE, Hestermann EV, Sherr DH. (2008) Constitutive regulation of CYP1B1 by the aryl hydrocarbon receptor (AhR) in pre-malignant and malignant mammary tissue. J Cell Biochem 104:402–417. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-seq (MACS). Genome Biol 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Ye Z, Lenz SW, Clark CR, Bharucha A, Farrugia G, Robertson KD, Zhang Z, Ordog T, Lee JH. (2017) Purification of nanogram-range immunoprecipitated DNA in ChIP-seq application. BMC Genomics 18:985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L. (2016) AHR function in lymphocytes: emerging concepts. Trends Immunol 37:17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J-G, Ma Y-T, Xie X, Yang Y-N, Pan S, Adi D, Liu F, Chen B-D. (2014) The association between CYP1A1 genetic polymorphisms and coronary artery disease in the Uygur and Han of China. Lipids Health Dis 13:145. [DOI] [PMC free article] [PubMed] [Google Scholar]