Abstract
Technological advances in characterizing molecular heterogeneity at the single cell level have ushered in a deeper understanding of the biological diversity of cells present in tissues including atherosclerotic plaques. New subsets of cells have been discovered among cell types previously considered homogenous. The commercial availability of systems to obtain transcriptomes and matching surface phenotypes from thousands of single cells is rapidly changing our understanding of cell types and lineage identity. Emerging methods to infer cellular functions are beginning to shed new light on the interplay of components involved in multifaceted disease responses, like atherosclerosis. Here, we provide a technical guide for design, implementation, assembly, and interpretations of current single cell transcriptomics approaches from the perspective of employing these tools for advancing cardiovascular disease research.
Keywords: Atherosclerosis, methodology, immunology, single cell RNA sequencing
[1]. INTRODUCTION
Atherosclerosis is a major cause for many cardiovascular disease pathologies, such as coronary artery disease (CAD) leading to myocardial infarctions (MI), cerebrovascular disease (CVD) leading to stroke, and peripheral artery disease (PAD) leading to amputated limbs. Together, MI and stroke are the most common cause of death, ahead of cancer, accounting for 31% or deaths in the US and a similar percentage worldwide1. Atherosclerosis manifests as cell- and lipid-rich plaques in mid- and large-sized arteries. After initiation, infiltrating leukocytes promote vascular inflammation, plaque growth, calcification, instability, plaque rupture or erosion, ultimately resulting in catastrophic arterial thrombosis that fully or partially occludes the affected artery and leads to clinical manifestations2, 3.
It is now clear that inflammation is a key factor in human atherosclerosis. In the CANTOS clinical trial, more than 10,000 subjects with prior MI and high serum C-reactive protein were treated with the anti-IL-1β monoclonal antibody (mAb) canakinumab or placebo4. Canakinumab treatment significantly reduced the incidence of major adverse cardiovascular events (MACE). In the COLCOT trial, a cohort of patients with previous MI receiving low-dose colchicine as a broad anti-inflammatory treatment were protected against recurring ischemic cardiovascular events5. Inflammation is associated with the infiltration of various immune cells, most prominently macrophages, T cells, natural killer (NK) cells and B cells. Understanding the diversity of the cellular components of atherosclerotic plaques and unraveling the immune response in atherosclerosis holds the promise of enabling novel therapeutic strategies.
Traditionally, the composition of the immune cell infiltrate in atherosclerotic plaques was assessed by immunostaining6 or flow cytometry (FACS)7, 8. The number of parameters that can be studied simultaneously is limited to about 3 to 8 for immunostaining9 and about 8 to 16 for flow cytometry. New high-parametric flow cytometry allows to measure up to 28 fluorescent parameters10 and will soon increase to 40 (Cytek Aurora). Implementation of new approaches for expanded multi-dimensional analysis has expanded our understanding of cellular diversity present within atherosclerotic plaques. An intermediate step in broadening the number of antigens that could be assessed on a single cell was the development of mass cytometry11, which allows assessment of ~35–42 surface markers12 including intracellular cytokines or transcription factors13, but provides no information on mRNAs.
The entire repertoire of all mRNAs present in a cell is called the transcriptome. This review will provide a description of the current single-cell technologies for assessing transcriptomes and cell surface markers and outline emerging technologies that will continue to propel the field forward.
Primer to single cell approaches
Development of next-generation sequencing (NGS) approaches, along with the advent of microfluidics devices suitable for single cell encapsulation14 and advanced multi-well plates15, have enabled the interrogation of single cells for transcriptome analysis. NGS refers to DNA sequencing approaches that utilize a fluorescence- or ion-based multi-strand analysis approach, allowing for simultaneous measurements (sometimes referred to as “shotgun” sequencing) across the entire transcriptome at once16. Compared to traditional Sanger sequencing, NGS has dramatically reduced the need for large quantities of DNA template, increased read reliability and reduced cost and time associated with RNA sequencing. Complementing NGS technology was the development of microfluidics devices with capillary-like constraints able to enrich molecules or cells of a desired size, weight, or charge within a closed system17. Microfluidics platforms are widely used across many fields allowing for flow of reagents across a cell or substrate. They dramatically reduce sample and reagents needed for experiments. Single cell RNA sequencing (scRNA-seq) technologies typically use microfluidics platforms for separating cells of interest and performing the initial steps of sample preparation. Alternatively, multi-well plate systems are used for the same purpose with 250,000 (BD Rhapsody) to 1 million (Celsee) wells per plate. Together, NGS and modern microfluidics or multi-well approaches have enabled the single cell revolution and unveiled a new layer of complexity within biological systems.
Commercial scRNA-seq approaches have only been available since 2017 (Table 1), but experimental single cell transcriptomics have been attempted since the early 1990s18, 19. Early efforts were restricted by low cell numbers, as well as the number of transcripts that could be examined in a given experiment. Using modern approaches, transcriptome coverage with high sensitivity across tens of thousands of cells is readily achievable. The ability to identify rare cell populations and the implications of our new understanding of population heterogeneity have been immediately scientifically impactful. scRNA-seq approaches were used to identify the precursors of plasmacytoid dendritic cells (DC) in the bone marrow and for the first time unraveled detailed human thymus organogenesis and early T cell development 20, 21. Transcriptional signatures of murine aortic T cells or tumor-associated macrophages obtained from scRNA-seq were used to predict outcome in patients with recurrent ischemic events or prognosis in breast cancer patients, respectively12, 22. Combining transcriptomics with assessment of cell surface phenotype using oligonucleotide-labeled mAbs23 has allowed for the development of better identification markers for discrimination between cell populations and identified many new cell types. As an example, scRNA-seq was recently used to develop lineage-tracing networks within the zebrafish brain. These approaches allow for the identification of novel cell types and their cellular origins during embryonic development24, 25.
Table 1.
Comparison of scRNA-seq methodologies.
| Vendor | Method | # of Cells Loaded1 | Wet Lab Duration hrs | First Break hrs |
Bioinformatics Pipeline | Runtime | Cost per Cell2 | CITE-seq | WTA7 or Targeted | # of Genes | Sequencing Depth Required3 per cell |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10X Genomics | 3’ v3 | 16,000 | 22 | 4 | Cell Ranger | 1 Week | $0.684 | Yes | WTA | 1,600 | 50,000 |
| 5’ | 17,400 | 24 | 4 | Cell Ranger | 1 Week | $0.305 | Yes | WTA | 1,200 | 50,000 | |
| BD Biosciences | Rhapsody | 20,000 | 16 | 6 | Seven Bridges Genomics | 1 Day | $0.364 | Yes | Targeted | 500 | 20,000 |
| Rhapsody (WTA)7 | 10,000 | 16 | 6 | Seven Bridges Genomics | 1 Day | $0.754 | Yes | WTA | 800 | 50,000 | |
| Takara/Illumina | SMART-seq | 96 | 22 | 7 | Open source | 1 Week | $80 | No | WTA | 8,0006 | 2 million |
Based on manufacturer’s maximum loading capacity (input).
Cost excludes tax, shipping, labor, and sequencing.
Based on manufacturer’s maximum recommendations for transcriptome only.
Based on 50-plex oligonucleotide antibodies used.
Based on 20-plex oligonucleotide antibodies used.
Referencing62
Whole transcriptome analysis
Initial microarray approaches were utilized to determine bulk transcriptomes on tissues such as atherosclerotic plaques or mixtures of cells like peripheral blood mononuclear cells (PBMCs)26. Deeper transcriptomes of FACS-sorted cell populations such as aortic macrophages27 or newly identified atherosclerosis-relevant Treg subsets28 were obtained by bulk RNA-seq of sorted cells. Understanding gene expression derived from whole tissues or PBMCs is a complicated task, because measured gene expression is impacted by cellular composition. It was not until scRNA-seq data became available that the averaging problem of bulk RNA-seq became obvious. Even within one cell type, the transcriptome varies with activation, cell cycle, apoptosis, stress, or time of day (circadian)29. Deconvolution methods, such as Cibersort30, allow to dissect the cellular components found in a bulk transcriptome, but only for known cell types with known transcriptomes. Complete deconvolution methods of bulk RNA-seq data sets are being developed without the need for prior tissue composition knowledge. Most of such algorithms assume that a dataset obeys a linear mixing model, which refers to the linear relationship between the proportion of a cell-type specific gene in a bulk transcriptome to the abundance of this particular cell-type in the probed complex tissue. Multiple cell-type specific genes can show mutual expression linearity or collinearity, which is used to determine the varying cell types in bulk transcriptomes without the knowledge of its composition. For real-life datasets, an algorithm would need large enough input data to robustly determine the linear component of variability across multiple samples to faithfully determine the cellular composition e.g. in healthy and diseased tissue31. This requires a sizeable sample cohort and is limited by low abundance of a given cell type in a complex tissue. scRNA-seq allows us to identify the gene signatures required for accurate gene expression deconvolution within bulk RNA-seq datasets, so recently generated scRNA-seq datasets can be used to reanalyze bulk RNA-seq from previous studies32, 33. Using scRNA-seq will continue to help build our understanding of the relationship between cells within a given group and allow to predict differentiation trajectories within the bounds of a single snapshot in time.
Recently, several studies have introduced mass cytometry and scRNA-seq technologies into the cardiovascular field to study a diverse array of cell types, including stromal, endothelial, and immune cells34–42. In addition, fate-mapping approaches that insert artificial genes can also be used for tracking newly recruited cells or to map the differentiation of a heterogeneous population throughout disease kinetics42. Unbiased dimensionality reduction and clustering algorithms have resulted in the discovery of new cell types and better resolution of cell subsets. The results of such studies pave the way for identifying novel pathways for the regulation of atherosclerosis. Here, we review existing studies and aim to provide a guide for scRNA-seq experimental design, library preparation, processing, and bioinformatics approaches to developing reliable and interpretable results.
[2]. SAMPLE PREPARATION
Obtaining a viable population of target cells in suspension and minimizing population bias or alterations in gene expression (Box: The promise of CITE-seq (or AB-seq) combined with scRNA-seq) is the first step in performing effective scRNA-seq. Cell liberation from dense tissue using enzymatic and mechanical isolation methods can often be a primary culprit in the introduction of variability within transcriptome analysis approaches. The investigator must determine whether tissue processing, digestion, or even cellular sorting are necessary, as each additional step introduces possible errors into the final dataset43.
Box. The promise of CITE-seq (or AB-seq) combined with scRNA-seq.
| Discovery | FACS panels are focused on known cell types. Unbiased approaches such as CITE-seq with 50 antibodies yield 1225 dot plots, many of which have never been seen before. This will lead to the discovery of new cell types in blood and tissues of humans and model animals in health and disease. The surface phenotype is superior to transcriptomes for cell identification, because it takes advantage of knowledge gained in 30 years of flow cytometry. |
| Complexity | Classical flow cytometry allows for detection of 16 markers. New analyzers promise up to 50 markers (Cytek Aurora, BD Symphony). Mass cytometry resolves up to 40 markers. Due to uniquely barcoded antibodies, CITE-seq is virtually unlimited. 100-plex panels are on the horizon. |
| Uncovering true heterogeneity | Machine learning algorithms perform dimensionality reduction (UMAP, tSNE) and group cells into clusters of similar surface antibody expression (Louvain). In a second analysis step, the single cell transcriptomes refine the clusters. |
| TCR, BCR | T and B cell receptor sequences can be assembled, for example using the 5’ solution by 10x Genomics, and combined with transcriptomes and surface phenotype. |
| Leveraging transcriptomes | Pathway analysis allows to determine the functionality of a given cell cluster, including activation, proliferation and apoptosis status. |
| Developmental cues | Single cell transcriptomes can be subjected to algorithms such as Monocle and RNAvelocity, which render data as pseudotime plots based on the expression status of a cell. This allows to infer developmental trajectories and lineage branching of cells in a complex environment. |
| Limitations | All scRNA-seq approaches require enzymatic and mechanical tissue dissociation, which induces artifacts. CITE-seq or AB-seq are not compatible with intracellular staining. The assays for CITE-seq or AB-seq are more elaborate and require a higher technical skill compared to sample preparation for flow cytometry. The workflooxw requires accessibility to multiple instruments including a FACS sorter, single cell platforms such as the 10x Chromium Controller and a sequencer. The biggest bottleneck today is the bioinformatic analysis of the generated data. |
For cell isolations from suspensions like blood, cells merely require a brief wash before being enrolled into a desired scRNA-seq platform. In some soft tissues like spleen or lymph nodes, lymphocytes can be isolated from single cell suspensions generated by only mechanical means (typically by a 70 μm nylon mesh) without enzymatic digestion. However, for the remainder of cells within tissues, typically some form of processing is required to liberate cells for single cell suspensions. Optimized protocols for liberating cells from mouse aortas8, 44 have been published and extensively tested. Enzymatic digestion with blends of collagenases and DNAses are the most common approach, particularly for cardiovascular disease-related tissues. For each tissue, the conditions (enzymes, concentrations, time, temperature, ion composition) need to be optimized and validated. Cold digestion protocols taking advantage of proteases with activity at 4–6°C are effective for tumor tissue dissection and revealed activation of stress response genes as a result of collagenase digestion at 37°C45. This stress response was particularly elevated when digestion persisted beyond 1 hour. Few genes have been associated with a cold-protease digest approach46. An alternative for cold-protease includes the use of a transcriptional inhibitor, such as actinomycin D, to prevent stress response elements from being activated during the isolation procedure47.
A reasonable validation step is using the enzyme cocktail on blood cells and comparing the phenotype of the cells exposed to the enzymes with that of untreated cells8. Protocols for large arteries and heart biopsies (unpublished) are being developed. Whether samples need to be digested in an enzyme cocktail and whether digestion must be performed at elevated temperatures (like 37°C), where RNA expression levels may be modified more rapidly and more profoundly, should be investigated on a case-by-case basis due to the potential to develop biased or non-reproducible data48. A recent study showed that a two-hour collagenase treatment of muscles to isolate muscle-resident satellite cells induced expression of immediate early genes including Fos, Jun, Socs3, and diverse heat shock proteins, marking a cellular stress response48. These genes were not detectable in cells derived from a one-hour collagenase digest. Thus, published data sets need to be carefully evaluated for the tissue dissociation protocols applied and might need to be reinterpreted in view of cell stress. A further source which may lead to decreased quality and yield of cells isolated from a complex tissue are lengthy manual processing times at the bench, such as removing perivascular adipose tissue or cutting samples for rapid digestion. Commercially available platforms including the Miltenyi gentleMACS allow for a more efficient and standardized tissue dissociation and homogenization.
In addition to potential processing artifacts from mechanical dissociation and enzymatic digestion, any given protocol may result in a misrepresentation of the actual cellular diversity present in the tissue. This is due to intrinsic predisposition of the preparation method to target some cell lineages over others. Often a historically established protocol was designed for the study of a specific cell type of interest, without consideration for other non-target cells that may be left to succumb to cell damage or death. An example would be that in our previous experiments to retrieve cholesterol-loaded foamy macrophages from within atherosclerotic plaque in the artery, an aggressive digest of the tissue with a relatively complex cocktail of enzymes was necessary36. This approach led to efficient recovery of the cells of interest but left other cells, particularly arterial endothelial and smooth muscle cells, almost uniformly unrecoverable. Using this same digestion approach for examining stromal cells within the arterial wall would result in poor quality transcriptomes that are difficult to interpret. With this possibility understood, the fraction of cells within a given sample may therefore be over- or under-represented. Validation may involve cell imaging in the tissue environment, but microscopy is by nature low-dimensional (few fluorochrome channels). Cell proportions obtained by scRNA-seq can be corrected by deconvolving bulk transcriptomes34, 49. When digesting tissues that will be subsequently stained for surface antigens, for cellular indexing of transcriptome and epitopes (CITE)-seq or FACS sorting, it is important to test for the loss of surface markers due to cleavage or shedding. For example, L-selectin (CD62L) and CD4 are lost after enzymatic treatment, but CD4 will recover after short-term (30 min) culture.
scRNA-seq approaches require a high viability (ideally >95%) of cellular input. Dead cells can be removed by bead-based approaches or FACS sorting, which is an approach favored by immunologists to isolate leukocytes or other cells of interest from tissues and organs. FACS sorting can also be used to narrow down the cell populations to be studied by scRNA-seq or bulk RNA-seq. Single cell suspensions are antibody-labeled on ice, undergo multiple buffer washes, and are pulled through a sorter. Narrowing down populations by pre-sorting can improve cell type resolution, but comes at the expense of potentially missing new cell types.
Different choices can be made for cells to be analyzed by scRNA-seq approaches. Broad populations from complex tissues have been chosen, for example arterial cells with no enrichment41, total hematopoietic cells expressing CD45-antigen from atherosclerotic aortas34, 35 or specific immune cell subpopulations such as regulatory T cells obtained from transgenic fluorescent reporter mice50. Advantages of using FACS sorting include the removal of dead cells and the ability to condense a viable, but relatively rare population or group of populations into a single sample. For example, purifying heart leukocytes is a laborious process and provides very few hematopoietic cells. Without sorting, there may not be sufficient cells to identify the diversity of macrophage populations within the tissue37. However, high pressure, shear stress and osmotic changes during the FACS sorting process can lead to cellular stress and altered gene expression51. To circumvent this problem, it is it is advisable to block the cells’ ability to produce new mRNAs by adding RNA polymerase inhibitors52, 53. Also, newer generations of low pressure-based cell sorters such as the MACSQuant Tyto may reduce these problems. In addition, faster and less invasive methods for cell enrichment include magnetic bead isolation approaches where cells of interest are negatively-selected by using antibody-magnetic bead labeling approaches to enrich for cells of interest by removing other cells from a mixed population. This procedure can be performed completely on ice in less than one hour and with low-budget equipment, making it a suitable replacement to sorting, which is time intensive and requires well-maintained and expensive equipment. Positive bead isolation strategies for cells of interest should be avoided as they can activate cells and lead to stress response signatures54. Approaches are being employed to limit cellular changes by shortening digestion periods, keeping cells on ice during digests, or even supplementing buffers with RNA polymerase inhibitors. Optimizing many of these methods appears to be ongoing in many laboratories and standards have not been established yet.
In some scenarios it is not feasible to isolate viable cells from tissues without substantial damage. For some clinical samples that are isolated following very long procedures like transplantation, were previously frozen or collected post mortem (such as brain aneurysm samples), it may be advantageous to take on an alternative isolation approach known as single-nuclei RNAseq (snRNA-seq)55. This approach takes advantage of the resistance of nuclei to degradation during freeze-thaw cycles. A typical protocol and work flow for snRNA-seq is highlighted here56. However, the quantity of mRNA available in the nucleus is limited. More unspliced mRNA may be recovered. Additionally, snRNA-seq is not compatible with CITE-seq, so no simultaneous protein expression information can be obtained. However, comparative analysis has shown that snRNA-seq faithfully replicates overall transcriptomes of whole-cell lysates57, 58.
[3]. scRNA-seq METHODOLOGY
scRNA-seq refers to a broad class of techniques and protocols that each contain their own inherent strengths and weaknesses. Depending on the readout being sought or the availability of resources, certain methods may be more desirable than others. The primary difference relates to the ability to process large (tens of thousands) or small (hundreds) numbers of cells. Higher cell numbers come at the expense of lower read depth within individual cells and impaired ability to decipher mRNA details such as splice variants or expression isoforms. However, all approaches share some universal steps. First is the ability to separate individual cells (Figure 1), either through microfluidic chambers, nano-droplet formation or plate-based systems. Generating the library entails lysing the cells, synthesizing the RNA into cDNA and amplifying the cDNA. Due to mRNA degradation after cell lysis and an inefficient reverse transcription reaction, only 10–20% of total transcripts are synthesized into cDNA14, 59, 60. Because amplification is not uniform, the number of reads mapping to a given transcript does not indicate the amount of transcript in the sample. This problem was solved by introducing unique molecular identifiers (UMIs)61. Counting UMIs is closely related to gene expression62, 63. Initially, the commercially available platform for scRNA-seq by 10x Genomics allowed to capture below 10% of a single cell’s transcripts14. This was significantly improved to 30–32% by new reagents and chemistry according to the manufacturer. All samples are processed for NGS. Illumina’s HiSeq or NovaSeq are the most commonly used platforms. In this section, we will reference three of the most popular approaches to get from single cells to a sequencing library. More approaches are reviewed here64. Single cell technologies continue to evolve and change.
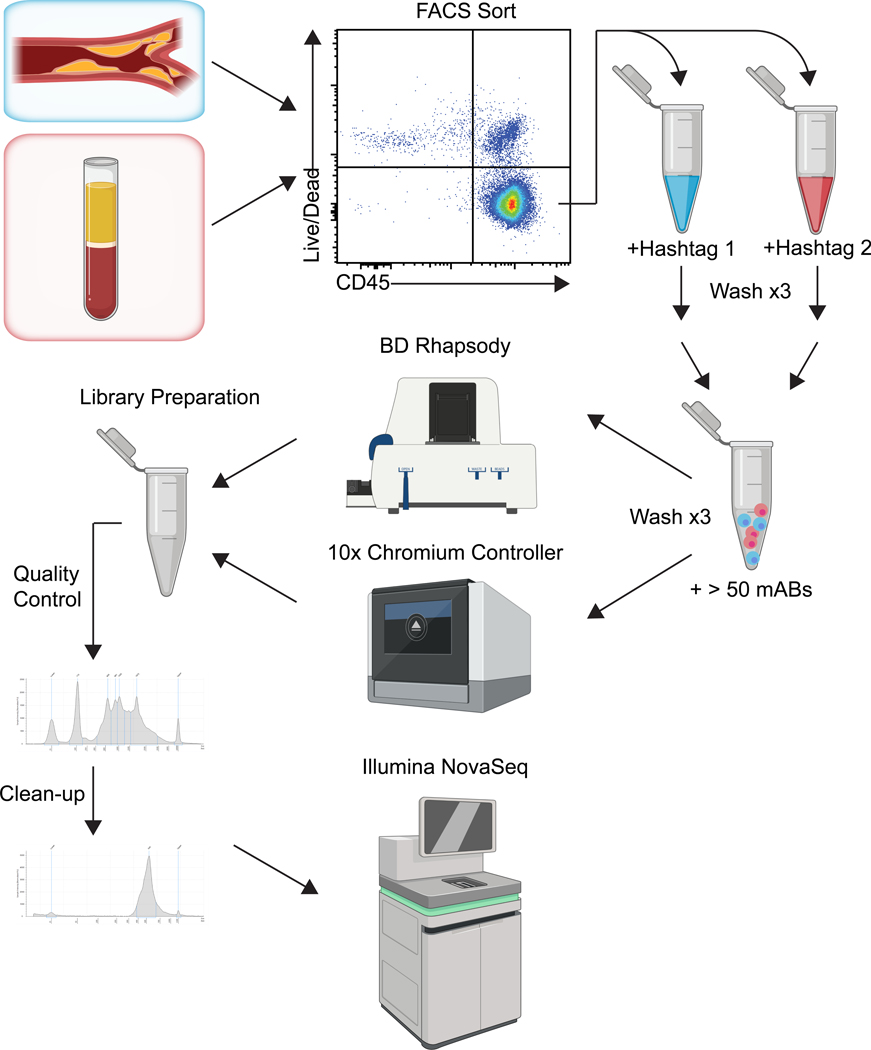
Figure 1.
Experimental workflow for CITE-seq and AB-seq experiments.
Single cell suspensions derived from artery wall (top) or PBMCs (bottom) are sorted for live cells and other desired markers (here: CD45 for leukocytes), hash-tagged, washed and incubated with the oligonucleotide-marked antibody panel (currently 50 mAbs are possible). After 3 more washes, the cells are loaded into the BD Rhapsody scanner or the 10x Chromium controller. Beads are retrieved and processed for library preparation. Pre-sequencing quality control (QC) is done by TapeStation. DNA fragments are removed, and the library is sequenced using NGS sequencers like the Illumina NovaSeq. The figure was created with BioRender.
Drop-seq
Droplet-based sequencing approaches, which include the popular 10X Genomics Chromium14 and Drop-seq65, 66 approaches and others take advantage of nanoliter droplet formation through microfluidics cartridges to place individual cells into an oil-based medium to separate cells for micro-reactions. These approaches are currently the most high-throughput with tens of thousands of cells being assessed in a single run (Table 1). They utilize UMIs that are associated with individual transcripts. UMIs are molecular barcodes that are attached on the surface of a bead to identify each transcript of a cell. Once cells are lysed, sample libraries can be combined after multiplexing. Multiplexing and sequencing multiple samples in one run reduces batch effects. Sequencing is limited to the cell barcode, multiplexing tags, UMI tags, and read-length size portions of transcripts that can be read. This limits the ability to detect many splice variants within Drop-seq approaches when compared to full-transcript sequencing approaches like SMART-seq. This makes it impossible to detect splice variants. Drop-seq provides high throughput, but low read depth. It is usually sufficient to obtain 40,000–50,000 reads per cell. This approach is useful for identifying rare cell populations and to map cellular diversity within whole organs.
SMART-seq
SMART-seq can be paired with index sorting or microfluidics platforms like the Fluidigm C1. Index sorting is a FACS-based approach of separating single cells into multi-well plates for subsequent single cell analysis. Fluidigm C1 is a fully automated system for separation of single cells that includes an automated platform for cell lysis, cDNA synthesis, and tagging procedures. The Fluidigm platform is suitable for analysis of surface protein expression by imaging, genomic DNA, epigenetic, or micro RNA abundance all within the microfluidics chamber. Whereas the first C1 generation allowed for the assessment of up to 96 cells, the C1 high-throughput integrated fluidics circuits platform allows to analyze mRNA of up to 800 individual cells The distribution of cells in the microfluidic chambers is gentler than flow cytometry sorting and may influence transcriptomes less. Optical assessment of the microchamber after cellular loading provides unparalleled confidence in the number of cells that are subjected to single cell RNA sequencing protocols. However, this approach comes at the cost of a high cellular input for loading.
Following separation of individual cells using cell sorting or microfluidics platforms, cells are lysed and RNA is hybridized to an oligo-dT containing primer after first strand cDNA synthesis to act as a barcode67. Current versions of this approach, termed SMART-seq2, have optimized the nucleic acid linking approach to improve kit performance68, 69. Full cDNA transcripts are then amplified and sequenced. This approach requires read depths beyond 1,000,000 reads per cell and can detect isoform variants and even single nucleotide polymorphisms (SNPs) in transcripts. Full sequence analysis improves mRNA sensitivity and provides great depth within individual cells. This comes at the cost of being low-throughput. The cost is high at >$50 per cell. Sophisticated new approaches based of SMART-seq2 protocols have been used to determine cellular heterogeneity in the developing mouse brain and an entire nematode using Split Pool Ligation-based Transcriptome sequencing (SPLiT-seq)70 and single-cell combinatorial indexing RNA sequencing (sci-seq)71, respectively. Both depend on multiple rounds of combinatorial indexing with oligonucleotide barcodes leading to uniquely barcoded individual cells and transcriptomes. For this, 100–1000 cells are sorted into 96- or 384 well-plates and kept intact. An in situ reverse transcription with barcoded primers will be performed, all cells will be collected, and randomly distributed into wells of another plate. This is followed by 2nd strand synthesis, tagmentation and a PCR to introduce a 2nd barcode. This technique allowed for the parallel analysis of 150,000 cells and drastically reduced cost per cell. Generation of SMART-seq3 is on the horizon; it detects transcripts in an allele-specific way and at increased sensitivity, which may lead to the detection of thousands of more genes per cell than are currently available with commercial products72.
BD Rhapsody
The BD Rhapsody approach allows for the targeted detection of a few hundred known targets. Several gene panels are available. Rhapsody performs paired barcoding in microwells with magnetic beads. Beads and attached mRNA are retrieved and synthesized into cDNA. cDNA is then amplified using targeted primers to specific genes of interest and labeled with library index barcodes. Samples are then sequenced and target gene expression is assessed. This approach leads to the ability to run more cells at a lower cost than other approaches. Rhapsody is compatible with the detection of surface antigens (by AB-seq) and hence provides protein information. A newly developed Rhapsody approach is designed to provide full (not targeted) transcriptomes.
CITE-seq, integrating transcriptome and proteomics
CITE-seq detects surface antigen abundance in tandem with scRNA-seq through the use of oligo-barcodes attached to antibody conjugates (instead of traditional flow cytometry or CyTOF, which use fluorochromes or rare metals, respectively). An analogous approach, REAP-seq (RNA expression and protein sequencing), has also been developed using an alternative method for conjugating oligonucleotide probes to antibodies73. Oligonucleotide barcodes can then be interrogated for enrichment within populations of cells to assist in identifying populations of interest. A typical workflow is shown in Figures 1 and 2. Matched transcriptome and cell surface phenotype information is retrieved, which allows for a more fine-grained analysis of cells. A first report in 2017 provided information of single cell transcriptomes and simultaneous assessment of 17 surfaces markers23.
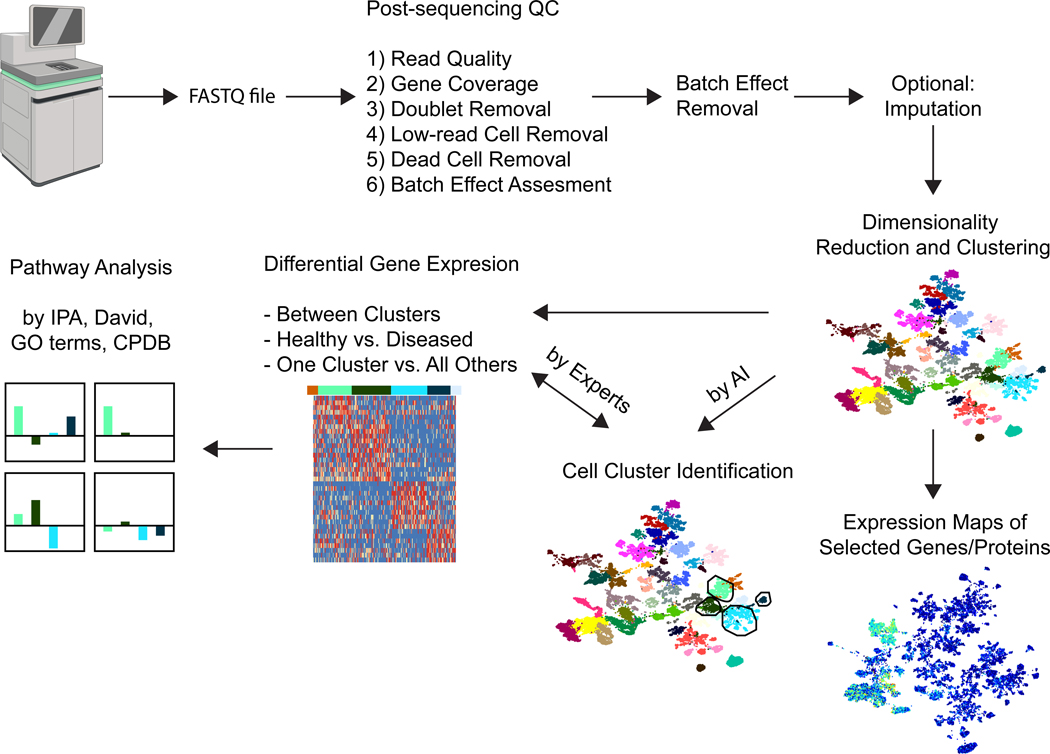
Figure 2.
Bioinformatic analysis workflow of CITE-seq and AB-seq experiments
The output of the sequencer is a FASTQ file with information about read quality. Further post-sequencing QC steps are indicated. If batch effects are detected, they can be removed if the experiment was designed to do so. Missing transcripts may be imputed. Dimensionality reduction (here by UMAP) and clustering (here by Louvain) result in 2 D maps of cells segregated by surface markers. Each antibody and transcript can be displayed as a heat map projected onto the UMAP. Differential gene expression is determined between clusters, or between samples from healthy and diseased individuals, or one cluster against all other cells. From the gene signature, pathway analysis is performed. The figure was created with BioRender.
The BioLegend brand of CITE-seq, called Total-seq, uses poly(dT) oligos and Drop-seq approaches utilizing 3’ or 5’ sequencing. Fernandez et al. recently used a 21-surface marker CITE-seq panel to describe leukocyte heterogeneity in human atherosclerotic plaques39, 74. Antibody numbers are limited only by the number of oligo barcodes developed by manufacturers, with custom options available. Therefore, expect antibody CITE-seq panels to dramatically expand in the near future.
[4]. PRACTICAL CONSIDERATIONS
scRNA-seq costs have dropped dramatically since 2017, but remain a limitation for researchers to access the technology. scRNA-seq is 10–200 times more expensive than bulk RNAseq approaches. Multiplexing scRNA-seq samples slightly reduced experimental costs, but most importantly helped to normalize between samples and control batch effects. These approaches utilize molecular barcoding at the sample level in addition to the cellular level. This allows for multiple samples to be combined and run on the same lanes, which becomes particularly important when collecting clinical samples with limited sample size, or when large numbers of samples need to be assessed. An intriguing new approach suitable for scRNA-seq or snRNA-seq approaches localized DNA-barcodes to lipid anchors on membranes termed MULTI-seq (Multiplexing using lipid-tagged indices) may be particularly helpful for sample barcoding approaches and discrimination of cell viability and endogenous gene expression patterns75. Antibody mediated multiplexing approaches have also been developed and modified for use with snRNA-seq analysis76.
Limitations of scRNA-seq include the limited depth of reads that can be detected per cell, often 10 times lower than what is obtained in bulk RNA-seq of sorted cells. Most bulk RNA-seq experiments are sequenced at millions of reads per sample. Shallow sequencing approaches are thought to underrepresent the transcriptional information. A greater sequencing depth increases the number of detectable transcripts. However, this is not linear and only marginally scalable, which means that sequencing depth needs to be increased tremendously to obtain information of additional transcripts. At a certain sequencing depth sequencing saturation occurs, which means that no additional unique transcripts will be detected.
Saturation is a function of both library complexity and sequencing depth. RNA content varies between cell types and their activation status, which will be represented by different numbers of transcripts in a library, called the complexity. The library complexity limits detection of transcripts even with increasing sequencing depths. As increasing sequencing depth comes with higher experimental costs, alternative strategies might be favored. A high-throughput scRNA-seq experiment can be used to uncover cellular heterogeneity in a complex tissue under homeostatic or pathologic conditions. Once an unusual cell subpopulations or transcript combinations within a cell population has been discovered, cell surface marker of this population can be identified. Subsequently, this population can be FACS-isolated and subjected to deep bulk RNA-seq to obtain a better picture. This was recently implemented for vascular macrophages in atherosclerotic arteries23.
In addition to sequencing depth, intrinsic bias between approaches has been reported. Specifically, full-length sequencing libraries are associated with increased sensitivity for longer mRNA fragments. By contrast, droplet approaches using UMI tagging acquire 3’ or 5’ ends for analysis, thus avoiding length biasing77.
[5]. POST-SEQUENCING PROCESSING
Post-sequencing processing for large data sets requires significant computational resources and experienced specialists, which remain limiting factors for many laboratories performing scRNA-seq (Figure 2). In droplet-based technologies, after paired-end sequencing is performed, two FASTQ files are obtained. The first contains cellular and UMI barcodes, which identifies the cellular source, the second contains sequence and quality scores of the sequenced cDNA. Subsequently, reads are mapped to the reference genome/transcriptome of the target species78, 79. Pseudoaligners instead of classical aligners can speed up the alignment process80. Droplet-based technologies, after alignment and quantification, yield a large matrix of detected gene expression in each cellular barcode.
A substantial proportion of droplets contain a bead but no cell. Other droplets contain one bead and one cell (desired). Yet other droplets contain one bead and two or more cells (multiplets). All multiplets must be identified and removed to prevent flawed results. Multiplets, unless removed, will look like unique cell types. There are three fundamental ways to remove multiplets: based on SNPs81, cell transcriptomes82, or hashtags83. Several methods to identify multiplets have been described84. Dead cells should also be removed. Overrepresentation of mitochondrial transcripts is commonly used to identify dead cells, but there is no consensus on the cutoff.
Regardless of the scRNA-seq strategy, the filtered count matrix must be normalized, scaled, and subjected to dimensionality reduction. Since scRNA-seq data contain extensive and highly variable data, visualization requires dimensionality reduction approaches for removal of noise and simplification of variation between samples. The variability between tens or hundreds of genes needs dimensionality reduction for an understandable display. Using algorithms such as tSNE, UMAP, or PCA, complex data can be generated into 2-dimensional models for simplified visualization. Additional downstream analyses include differential gene expression, followed by pathway/gene set enrichment, trajectory analysis, and others85.
Batch effects refer to preparation-dependent changes in how cells were isolated, RNA was processed, or any other unwanted experiment-to-experiment variations. Batch effects make it difficult to compare transcriptomes collected from different laboratories or at different times. Common batch effects in scRNA-seq datasets include protocol variations, variations among donors, and operator variations. Depending on the severity, batch effects can be so large as to completely obscure the biological information contained in the data. Principal Component Analysis (PCA) is often used to visually inspect whether cells cluster by some experimental co-variate. Recently, Büttner et al86 designed a test to quantify the extent of batch effects that is inherently more accurate than simple visual inspection. Although the optimal approach to prevent batch effects is following a balanced experimental design87, 88, balanced designs are rarely feasible or cost-effective. Hence, several batch correction algorithms have been proposed to correct for batch effects89–94, some of which have been initially designed for microarray and bulk RNA-seq89, whereas others have been specifically designed for scRNA-seq90–94. Generally, batch effect correction is applied to the normalized data before dimensionality reduction and clustering. These algorithms differ in the level at which they correct batch effects. Combat89 and MNNCorrect92 operate by directly correcting gene expression in each cell, whereas Seurat93 generates corrected low-dimensional reductions, similar to principal components, for clustering and visualization. BBKNN90 generates an adjusted neighbor graph taking into account the different batches for subsequent clustering and visualization. Other batch correction algorithms are embedded within frameworks specifically designed for scRNA-seq analysis95.
Dropouts and imputation of data
Dropouts are transcripts that are expected in a cell, but for which no reads were detected. The rate of dropouts is dramatically higher in scRNA-seq data than in bulk approaches, because the starting nucleotide concentration in a single cell is limiting. For a low-expressed gene, a transcript may be present in the cell or not at any given time. Even if present, transcripts might not be detected. This risk increases if the sequencing depth was insufficient, thus creating false negatives. The loss of “true” gene expression can lead to biased results and might necessitate the removal of cells or samples from further analysis. Conversely, similar dropouts in a subset of cells might aggregate them in the same cluster, producing false apparent cellular diversity. Multiple approaches to address this problem and fill the blanks in single cell data have been developed96–100. Each of these methods outperformed several others depending on the benchmarks and datasets used. Fundamentally, these approaches can be used to impute all gene expression values (e.g. MAGIC98) or only dropout genes (e.g. DrImpute97). Another approach adopted by SCRABBLE101 attempts to borrow information from a matched bulk RNA-seq sample in order to constrain imputation. This strategy is considered superior and outperforms five competitive platforms. The assumption underlying imputing all genes is that dropout is a broad phenomenon that affects all genes; albeit to varying degrees. These algorithms are not part of the necessary and routine scRNA-seq analysis workflow and should only be implemented when expected genes in cell populations are missing. On a cautionary note, Andrews et al102 recently suggested that imputation may be beneficial to visualization and clustering, but may introduce false positives when used for cell type markers and differential expression. New bioinformatic workflows allow to normalize and remove technical variation in molecular counts while preserving the true biological variation103. This procedure is independent of pseudocount addition or imputation and log transformation, thus improving the analysis of scRNA-seq data.
Data deposition
Typically, most investigators deposit their raw data on the National Center for Biotechnology Information Gene Expression Omnibus database (NCBI GEO). Raw data from NCBI GEO can be downloaded using the freely available SRA toolkit. Some groups prefer to publish their data on their own websites, which usually have added features including the ability to browse individual genes and visualize clustering, e.g. Tabula Muris104 (https://tabula-muris.ds.czbiohub.org/) and the Human Cell Atlas (https://data.humancellatlas.org/). Other repositories are available, including the Single Cell Expression Atlas (https://www.ebi.ac.uk/gxa/sc) or the Pangloa database (https://panglaodb.se). When depositing scRNA-seq data, it is important to deposit at least the raw data and an accurate, detailed and complete metadata file. Although NCBI GEO provides a template for researchers to fill out the metadata information related to their data, it is up to the researcher to include as much detail as they want. This has not been standardized yet. Some GEO entries will have fastq files as raw files while others will have 10x bams files. Supplementary data is not standardized in GEO and can contain either count files, normalized expression values, h5 molecule matrices from the 10x cell ranger pipeline, or all of the above. Standardized and detailed data deposition is crucial for reproducibility, particularly due to the inherent variation observed in single cell studies. When using deposited data, it is very critical to ensure that data has been properly processed, especially when integrating multiple datasets. Processing the raw fastq data avoids many potential sources of error. The main challenges for using repositories is the ability to integrate the data in a way that retains as much biological information as possible while reducing the unwanted variation. A lot of progress has been made in developing tools for the integration of different datasets while accounting for potential batch effect and avoiding confounded experimental designs. An important consideration when integrating datasets is having the complete metadata, with as much detail as possible. Additionally, as more and more datasets are being generated, it is important to consider scaling both the hardware requirements, i.e. hard-disks and computing cores, and software requirements, i.e. tools and packages for the preprocessing and analysis of such data.
[6]. UNSUPERVISED CLUSTERING AND MACHINE LEARNING
Using unbiased computational approaches where gene expression similarities between individual cells leads to a clustering within the veil of a shared expression makeup has become the dominant approach for data analysis. The most popular algorithm is Louvain, which is a graph-based method for community detection. In-depth discussion of unsupervised clustering can be found elsewhere105. By allowing algorithms to sort the data, analytical bias can be avoided. These unbiased analysis approaches have proven that unique clusters of cells found in specimens can be reproduced in different labs using different implementations of this approach. Determining the boundaries between clusters in an unbiased way is still challenging. It is up to the researchers to identify or name clusters, based on prior knowledge or systematic naming conventions. Researchers often have to choose clustering resolution: leveraging this parameter will either merge similar clusters into one bigger cluster or split a bigger cluster into smaller, transcriptionally different clusters. While choosing this parameter is not straightforward, good practice needs confirmation of transcriptionally diverse cellular clusters with protein-based methods such as FACS. Also, in our experience, clusters are not always restricted to a single cell type. This is best exemplified by proliferating cells, which share a specific and dominant transcriptional profile. Thus, they cluster together although they are of different cell types. New bioinformatic tools including scmap106 and SingleR107 are aiding to call cell clusters. Both methods rely on reference data. They automatically choose sets of genes that identify a given cell type. However, both algorithms are prone to misclassifications if the transcriptome originating from a particular phenotype or cell of interest is not included in the reference. Particularly, aberrant cells or cells obtained from pathological conditions can harbor a heterogenous transcriptome which is not included in the reference. In case cells are not definable by the reference, scmap leaves these cells unclassified. A different algorithm, CHETAH108, also utilizes reference data, but infers classification on intermediate or unassigned cells by hierarchical clustering.
Analysis tools
Close to 500 scRNA-seq analysis packages are available by now, impossible to review in a short article. The expression of ligand-receptor pairs in single cell data obtained from complex tissue can be harnessed to predict how tissue cells interact and communicate with each other109. The cellular RNA content is a great indicator of the state of the cell at the time of analysis. However, dynamic processes such as embryogenesis cannot be addressed by a static method. Approaches to overcome the static nature of scRNA-seq include utilization of nucleotide analogs that can be pulsed prior to processing, allowing for temporal resolution of newly expressed RNA to be tracked within single cells, termed scSLAM-seq110. RNA velocity111 – which represents the time-resolved transcriptome state – can be predicted by analyzing unspliced and spliced mRNAs. In other words, RNA velocity allows to predict the future cell state given that unspliced mRNA has yet to be processed and expressed within the cell. Monocle112, 113 allows for unsupervised trajectory analysis of single cells by pseudo-time ordering. The second version of monocle68, a machine-learning technique based on reverse graph embedding (RGE) and parsimonious principal graphs, can be used on all scRNA-seq data sets and does not require input information about cell fates or branch points. Another algorithm available in the monocle2 package is Census59 . This algorithm converts fragments per kilobase per million reads/transcripts per million reads (FPKM/TPM) gene expression values in single cells to relative transcript counts without the need for spike-in standards or UMIs. For easier modeling of the data compared to normalized read counts, Census-obtained counts can be used to determine splice isoforms and provides allelic information which can be used to determine developmental regulation. An in-depth review of algorithms inferring trajectories of single cells can be found here114. Further useful analytical tools allow researchers to investigate not only the different transcriptional profiles of the cells but also their regulatory networks (SCENIC)115 and their potential interaction via ligand receptor relationships (CellPhoneDB116, NicheNet117). Multiple algorithms are particularly useful for studying adaptive immune responses in complex tissues. We find the most powerful tool in this regard to be TraCeR118, which can be used to reconstruct paired and full-length TCR chains from transcriptomes of single T cells. This allows to study the TCR V, D and J segment usage and clonality in homeostasis or under pathological conditions. It should be noted that TraCeR requires full-length transcriptomes. We have been able to assemble TCRα and β sequences from 10X Genomics 5’ sequencing.
Multi-omics approaches
The next step in single cell platform analysis includes broader multi-omics approaches like DNA methylation by bisulfite conversion sequencing or TAB-seq, non-coding RNAs, chromatin accessibility by ATAC-Seq, histone modification by CHIP-seq, and protein expression levels by CITE-seq (also called AB-seq). Integrating these data with ever-changing bioinformatics approaches enables more powerful analysis. For example, expressed quantitative trait loci (eQTLs) can help identify underlying regulatory networks associated with targets of interest.
All of today’s scRNA-seq methodologies require the preparation of single cell suspensions, which leads to complete loss of spatial information of cell types within a given tissue. There are some limited workarounds, like injecting an antibody intraveneously to label intravascular cells119. New approaches aim to fully retrieve spatial information. Recent publications introduced two new methodologies, seqFISH+120 and Slide-seq121, which allow for in situ spatial identification of transcripts at near-single cell resolution across a wide array of RNA probes. 10x Genomics offers a commercially available solution called Visium which allows for whole transcriptome detection, but not at single-cell resolution. Cartana provides a product based on in situ sequencing122. The technology allows to detect over 100 genes within tissue sections at single cell resolution123. In seqFISH+, tissue sections are incubated with transcript-specific probes containing a complementary sequence to an oligonucleotide attached to a fluorochrome. Key is the multiplexing with three different fluorochromes and multiple hybridization rounds, allowing to image multiple transcripts in every round. The images of every hybridization round are bioinformatically parsed together; detection of 10,000 transcripts in a complex tissue has been achieved. An alternative approach was used where barcoded-beads were spotted on a functionalized glass slide and sequenced subsequently to define areas of beads and establish a map of barcodes spotted on the slide121. Tissue sections were then incubated with the functionalized slide, RNA was released, hybridized with the spotted barcoded oligonucleotides, and subjected to sequencing. However, this approach does not provide true single cell resolution, because RNA from more than one cell may hybridize to each spot. Transcript detection at a resolution of 100 μm2 (10×10 μm) has been achieved.
The future of single cell methodologies
We are only in year 3 of commercially available scRNA-seq. Innovation continues at a rapid speed and new products are released weekly. CITE-seq and AB-seq are only the beginning of multi-omics approaches. Spatially resolved and high-throughput strategies are on the verge of becoming practical. ScRNA-seq approaches are being applied to high-throughput chemical screens to assess hundreds of thousands of cells in individual runs, allowing for molecular insights to be gained from large scale pharmacologic perturbations124. More than 100x cost reduction per cell has already been achieved compared to the pioneer days of sorting single cells into single wells. Sensitivity is improving, but still not satisfactory. Standard protocols for removing low quality transcriptomes, doublets and dead cells are sorely needed. Imputation and batch effect removal software programs exist, but there is no consensus in the field which ones should be used. Another important direction is the integrated and comparative analysis of single-cell data between different laboratories, donors, batches, single-cell platforms or even species. The goal of these methods is to identify common cell types/states that are present in different scRNA-seq samples while rendering dimensionality reduction and clustering robust to unwanted variation. While first techniques were only recently developed93, 125, enormous progress has already been made showing the possibility to extend these techniques to single-cell protein, chromatin and spatial data126. With more labs using scRNA-seq, the demand for unified processing strategies has increased. The biggest bottleneck in scRNA-seq today is the post-sequencing bioinformatic analysis, which is currently carried out by well-trained experts who are in high demand in academia and industry. Future strategies will aim to create user-friendly data analysis interfaces, databases and standard file formats that will streamline analysis and data sharing.
We expect methods and algorithms that will try to identify possible regulators, key transcription factors and epigenetic data. A single cell assay for transposase accessible chromatin (ATAC-Seq), a method to map open chromatin, is already available from 10x Genomics. Cell type calling and phenotypical associations based on scRNA-Seq data may use association rule mining or other rule-based machine learning techniques. These algorithms assist in generating hypotheses that can be tested in the lab.
Conclusion
Single cell RNA-seq with CITE-seq or AB-seq is a powerful methodology for cell type discovery, lineage relationships, homeostasis, development and disease. Terabytes of data are generated every day. Single cell RNA sequencing is beginning to reveal the complex interactions between cell types in atherosclerosis initiation, progression, regression and plaque rupture as well as other fields of cardiovascular research. New insights will help to tailor new and innovative therapeutic strategies for atherosclerosis beyond controlling lipid levels.
Sources of funding
This work was supported by National Institutes of Health grant R01 HL121697, P01 HL136275, project 4 and core E as well as P01 HL088093 core to Dr. Ley. Dr. Winkels was supported by DFG award GZ WI 4811/1–1 from the German government. Dr. Williams was supported by NIH R00 HL138163. Konstantin Zaitsev was financially supported by the Government of the Russian Federation (Grant 08–08) and Systems Biology Program by Skoltech.
Nonstandard Abbreviations and Acronyms:
- ATAC-seq
Sequencing protocol which assesses open chromatin regions
- BCR
B cell receptor
- CHIP-seq
Chromatin precipitation sequencing, maps transcription factor binding
- CITE-seq
(also AB-seq) cellular indexing of transcriptome and epitopes, allows simultaneous assessment of antigens and transcriptomes
- CyTOF
Mass cytometry
- Deconvolution
Bioinformatic analysis of bulk RNA transcriptomes for cellular composition
- Drop-seq
Droplet-based sequencing
- Dropouts
Expected transcript for a given cell, but no read was detected
- eQTL
Expressed quantitative trait loci
- FACS
Flow cytometry
- FASTQ
File format obtained after sequencing
- FPKM
Fragments per kilobase per million reads
- Imputation
Algorithms that infer the expression of a dropout
- MULTI-seq
Multiplexing using lipid tagged indices-sequencing
- REAP-seq
RNA expression and protein sequencing
- RGE
Reversed graph embedding, used to resolve single-cell trajectories
- Saturation
Sequencing depth at which no additional transcripts will be detected
- Sci-seq
Single-cell combinatorial indexing RNA sequencing
- scRNA-seq
Single cell RNA sequencing
- scSLAM-seq
Single cell, thiol-(SH)-linked alkylation of RNA for metabolic labeling sequencing
- SMART-seq
Protocol to perform deep transcriptional profiling
- snRNA-seq
Single nuclei RNA sequencing
- SPLiT-seq
Split pool ligation-based transcriptome sequencing
- TAB-seq
Sequencing protocol which utilizes bisulfite sequencing and TET proteins to study 5-hydroxymethylcytosine DNA modifications
- Tagmentation
Sequencing library preparation with the Tn5 transposase
- TCR
T cell receptor
- TPM
Transcripts per million reads
- Trajectory analysis
Pseudotime projection obtained from single cell transcriptomes allowing to predict cell states and developmental relationships
- UMI
Unique molecular identifier; molecular tag used to detect and identify unique mRNA transcripts
Footnotes
Disclosure
The authors report no conflict of interest
References
- 1.Benjamin EJ, Muntner P, Alonso A, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Das SR, Delling FN, Djousse L, Elkind MSV, Ferguson JF, Fornage M, Jordan LC, Khan SS, Kissela BM, Knutson KL, Kwan TW, Lackland DT, Lewis TT, Lichtman JH, Longenecker CT, Loop MS, Lutsey PL, Martin SS, Matsushita K, Moran AE, Mussolino ME, O’Flaherty M, Pandey A, Perak AM, Rosamond WD, Roth GA, Sampson UKA, Satou GM, Schroeder EB, Shah SH, Spartano NL, Stokes A, Tirschwell DL, Tsao CW, Turakhia MP, VanWagner LB, Wilkins JT, Wong SS, Virani SS, American Heart Association Council on E, Prevention Statistics C and Stroke Statistics S. Heart Disease and Stroke Statistics-2019 Update: A Report From the American Heart Association. Circulation. 2019;139:e56–e528. [DOI] [PubMed] [Google Scholar]
- 2.Libby P, Pasterkamp G, Crea F and Jang IK. Reassessing the Mechanisms of Acute Coronary Syndromes. Circ Res. 2019;124:150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mori H, Torii S, Kutyna M, Sakamoto A, Finn AV and Virmani R. Coronary Artery Calcification and its Progression: What Does it Really Mean? JACC Cardiovasc Imaging. 2018;11:127–142. [DOI] [PubMed] [Google Scholar]
- 4.Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, Kastelein JJP, Cornel JH, Pais P, Pella D, Genest J, Cifkova R, Lorenzatti A, Forster T, Kobalava Z, Vida-Simiti L, Flather M, Shimokawa H, Ogawa H, Dellborg M, Rossi PRF, Troquay RPT, Libby P, Glynn RJ and Group CT. Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N Engl J Med. 2017;377:1119–1131. [DOI] [PubMed] [Google Scholar]
- 5.Tardif JC, Kouz S, Waters DD, Bertrand OF, Diaz R, Maggioni AP, Pinto FJ, Ibrahim R, Gamra H, Kiwan GS, Berry C, Lopez-Sendon J, Ostadal P, Koenig W, Angoulvant D, Gregoire JC, Lavoie MA, Dube MP, Rhainds D, Provencher M, Blondeau L, Orfanos A, L’Allier PL, Guertin MC and Roubille F. Efficacy and Safety of Low-Dose Colchicine after Myocardial Infarction. N Engl J Med. 2019. [DOI] [PubMed] [Google Scholar]
- 6.Jonasson L, Holm J, Skalli O, Bondjers G and Hansson GK. Regional accumulations of T cells, macrophages, and smooth muscle cells in the human atherosclerotic plaque. Arteriosclerosis. 1986;6:131–138. [DOI] [PubMed] [Google Scholar]
- 7.Koltsova EK, Garcia Z, Chodaczek G, Landau M, McArdle S, Scott SR, von Vietinghoff S, Galkina E, Miller YI, Acton ST and Ley K. Dynamic T cell-APC interactions sustain chronic inflammation in atherosclerosis. J Clin Invest. 2012;122:3114–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galkina E, Kadl A, Sanders J, Varughese D, Sarembock IJ and Ley K. Lymphocyte recruitment into the aortic wall before and during development of atherosclerosis is partially L-selectin dependent. J Exp Med. 2006;203:1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerner MY, Kastenmuller W, Ifrim I, Kabat J and Germain RN. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37:364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liechti T and Roederer M. OMIP-060: 30-Parameter Flow Cytometry Panel to Assess T Cell Effector Functions and Regulatory T Cells. Cytometry A. 2019;95:1129–1134. [DOI] [PubMed] [Google Scholar]
- 11.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE and Tanner SD. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. [DOI] [PubMed] [Google Scholar]
- 12.Winkels H, Ehinger E, Vassallo M, Buscher K, Dinh HQ, Kobiyama K, Hamers AAJ, Cochain C, Vafadarnejad E, Saliba AE, Zernecke A, Pramod AB, Ghosh AK, Anto Michel N, Hoppe N, Hilgendorf I, Zirlik A, Hedrick CC, Ley K and Wolf D. Atlas of the Immune Cell Repertoire in Mouse Atherosclerosis Defined by Single-Cell RNA-Sequencing and Mass Cytometry. Circulation research. 2018;122:1675–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe’er D, Tanner SD and Nolan GP. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ and Bielas JH. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gierahn TM, Wadsworth MH 2nd, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC and Shalek AK. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017;14:395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodwin S, McPherson JD and McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong JW, Studer V, Hang G, Anderson WF and Quake SR. A nanoliter-scale nucleic acid processor with parallel architecture. Nat Biotechnol. 2004;22:435–439. [DOI] [PubMed] [Google Scholar]
- 18.Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M and Coleman P. Analysis of gene expression in single live neurons. Proc Natl Acad Sci U S A. 1992;89:3010–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brady G, Barbara M and Iscove NN. Representative in Vitro cDNA Amplification From Individual Hemopoietic Cells and Colonies. Methods Mol Cell Biol. 1990:17–25. [Google Scholar]
- 20.Dress RJ, Dutertre CA, Giladi A, Schlitzer A, Low I, Shadan NB, Tay A, Lum J, Kairi M, Hwang YY, Becht E, Cheng Y, Chevrier M, Larbi A, Newell EW, Amit I, Chen J and Ginhoux F. Plasmacytoid dendritic cells develop from Ly6D(+) lymphoid progenitors distinct from the myeloid lineage. Nat Immunol. 2019;20:852–864. [DOI] [PubMed] [Google Scholar]
- 21.Zeng Y, Liu C, Gong Y, Bai Z, Hou S, He J, Bian Z, Li Z, Ni Y, Yan J, Huang T, Shi H, Ma C, Chen X, Wang J, Bian L, Lan Y, Liu B and Hu H. Single-Cell RNA Sequencing Resolves Spatiotemporal Development of Pre-thymic Lymphoid Progenitors and Thymus Organogenesis in Human Embryos. Immunity. 2019;51:930–948 e936. [DOI] [PubMed] [Google Scholar]
- 22.Tuit S, Salvagno C, Kapellos TS, Hau CS, Seep L, Oestreich M, Klee K, de Visser KE, Ulas T and Schultze JL. Transcriptional Signature Derived from Murine Tumor-Associated Macrophages Correlates with Poor Outcome in Breast Cancer Patients. Cell Rep. 2019;29:1221–1235 e1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R and Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spanjaard B, Hu B, Mitic N, Olivares-Chauvet P, Janjuha S, Ninov N and Junker JP. Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat Biotechnol. 2018;36:469–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raj B, Wagner DE, McKenna A, Pandey S, Klein AM, Shendure J, Gagnon JA and Schier AF. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat Biotechnol. 2018;36:442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perisic L, Aldi S, Sun Y, Folkersen L, Razuvaev A, Roy J, Lengquist M, Akesson S, Wheelock CE, Maegdefessel L, Gabrielsen A, Odeberg J, Hansson GK, Paulsson-Berne G and Hedin U. Gene expression signatures, pathways and networks in carotid atherosclerosis. J Intern Med. 2016;279:293–308. [DOI] [PubMed] [Google Scholar]
- 27.McArdle S, Buscher K, Ghosheh Y, Pramod AB, Miller J, Winkels H, Wolf D and Ley K. Migratory and Dancing Macrophage Subsets in Atherosclerotic Lesions. Circ Res. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, McArdle S, Gholami A, Kimura T, Wolf D, Gerhardt T, Miller J, Weber C and Ley K. CCR5+T-bet+FoxP3+ Effector CD4 T Cells Drive Atherosclerosis. Circ Res. 2016;118:1540–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raj A and van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaitsev K, Bambouskova M, Swain A and Artyomov MN. Complete deconvolution of cellular mixtures based on linearity of transcriptional signatures. Nat Commun. 2019;10:2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Park J, Susztak K, Zhang NR and Li M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat Commun. 2019;10:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baron M, Veres A, Wolock SL, Faust AL, Gaujoux R, Vetere A, Ryu JH, Wagner BK, Shen-Orr SS, Klein AM, Melton DA and Yanai I. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Syst. 2016;3:346–360 e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winkels H, Ehinger E, Vassallo M, Buscher K, Dinh H, Kobiyama K, Hamers A, Cochain C, Vafadarnejad E, Saliba AE, Zernecke A, Pramod A, Ghosh A, Anto Michel N, Hoppe N, Hilgendorf I, Zirlik A, Hedrick C, Ley K and Wolf D. Atlas of the Immune Cell Repertoire in Mouse Atherosclerosis Defined by Single-Cell RNA-Sequencing and Mass Cytometry. Circ Res. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cochain C, Vafadarnejad E, Arampatzi P, Jaroslav P, Winkels H, Ley K, Wolf D, Saliba AE and Zernecke A. Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis. Circ Res. 2018. [DOI] [PubMed] [Google Scholar]
- 36.Kim K, Shim D, Lee JS, Zaitsev K, Williams JW, Kim KW, Jang MY, Seok Jang H, Yun TJ, Lee SH, Yoon WK, Prat A, Seidah NG, Choi J, Lee SP, Yoon SH, Nam JW, Seong JK, Oh GT, Randolph GJ, Artyomov MN, Cheong C and Choi JH. Transcriptome Analysis Reveals Nonfoamy Rather Than Foamy Plaque Macrophages Are Proinflammatory in Atherosclerotic Murine Models. Circ Res. 2018;123:1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bajpai G, Schneider C, Wong N, Bredemeyer A, Hulsmans M, Nahrendorf M, Epelman S, Kreisel D, Liu Y, Itoh A, Shankar TS, Selzman CH, Drakos SG and Lavine KJ. The human heart contains distinct macrophage subsets with divergent origins and functions. Nat Med. 2018;24:1234–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin JD, Nishi H, Poles J, Niu X, McCauley C, Rahman K, Brown EJ, Yeung ST, Vozhilla N, Weinstock A, Ramsey SA, Fisher EA and Loke P. Single-cell analysis of fate-mapped macrophages reveals heterogeneity, including stem-like properties, during atherosclerosis progression and regression. JCI Insight. 2019;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez DM, Rahman AH, Fernandez NF, Chudnovskiy A, Amir ED, Amadori L, Khan NS, Wong CK, Shamailova R, Hill CA, Wang Z, Remark R, Li JR, Pina C, Faries C, Awad AJ, Moss N, Bjorkegren JLM, Kim-Schulze S, Gnjatic S, Ma’ayan A, Mocco J, Faries P, Merad M and Giannarelli C. Single-cell immune landscape of human atherosclerotic plaques. Nat Med. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cole JE, Park I, Ahern D, Kassiteridi C, Danso Abeam D, Goddard M, Green P, Maffia P and Monaco C. Immune cell census in murine atherosclerosis: cytometry by time of flight illuminates vascular myeloid cell diversity. Cardiovasc Res. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalluri AS, Vellarikkal SK, Edelman ER, Nguyen L, Subramanian A, Ellinor PT, Regev A, Kathiresan S and Gupta RM. Single-Cell Analysis of the Normal Mouse Aorta Reveals Functionally Distinct Endothelial Cell Populations. Circulation. 2019;140:147–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wirka RC, Wagh D, Paik DT, Pjanic M, Nguyen T, Miller CL, Kundu R, Nagao M, Coller J, Koyano TK, Fong R, Woo YJ, Liu B, Montgomery SB, Wu JC, Zhu K, Chang R, Alamprese M, Tallquist MD, Kim JB and Quertermous T. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med. 2019;25:1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen QH, Pervolarakis N, Nee K and Kessenbrock K. Experimental Considerations for Single-Cell RNA Sequencing Approaches. Front Cell Dev Biol. 2018;6:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butcher MJ, Herre M, Ley K and Galkina E. Flow cytometry analysis of immune cells within murine aortas. J Vis Exp. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Flanagan CH, Campbell KR, Zhang AW, Kabeer F, Lim JLP, Biele J, Eirew P, Lai D, McPherson A, Kong E, Bates C, Borkowski K, Wiens M, Hewitson B, Hopkins J, Pham J, Ceglia N, Moore R, Mungall AJ, McAlpine JN, Team CIGC, Shah SP and Aparicio S. Dissociation of solid tumor tissues with cold active protease for single-cell RNA-seq minimizes conserved collagenase-associated stress responses. Genome Biol. 2019;20:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denisenko E, Guo BB, Jones M, Hou R, de Kock L, Lassmann T, Poppe D, Clement O, Simmons RK, Lister R and Forrest ARR. Systematic bias assessment in solid tissue 10x scRNA-seq workflows. BioRxiv. [Preprint] November 06, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Hove H, Martens L, Scheyltjens I, De Vlaminck K, Pombo Antunes AR, De Prijck S, Vandamme N, De Schepper S, Van Isterdael G, Scott CL, Aerts J, Berx G, Boeckxstaens GE, Vandenbroucke RE, Vereecke L, Moechars D, Guilliams M, Van Ginderachter JA, Saeys Y and Movahedi K. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci. 2019;22:1021–1035. [DOI] [PubMed] [Google Scholar]
- 48.van den Brink SC, Sage F, Vertesy A, Spanjaard B, Peterson-Maduro J, Baron CS, Robin C and van Oudenaarden A. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods. 2017;14:935–936. [DOI] [PubMed] [Google Scholar]
- 49.Winkels H, Ehinger E, Ghosheh Y, Wolf D and Ley K. Atherosclerosis in the single-cell era. Curr Opin Lipidol. 2018;29:389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zemmour D, Zilionis R, Kiner E, Klein AM, Mathis D and Benoist C. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat Immunol. 2018;19:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romero-Santacreu L, Moreno J, Perez-Ortin JE and Alepuz P. Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA. 2009;15:1110–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakai M, Troutman TD, Seidman JS, Ouyang Z, Spann NJ, Abe Y, Ego KM, Bruni CM, Deng Z, Schlachetzki JCM, Nott A, Bennett H, Chang J, Vu BT, Pasillas MP, Link VM, Texari L, Heinz S, Thompson BM, McDonald JG, Geissmann F and Glass CK. Liver-Derived Signals Sequentially Reprogram Myeloid Enhancers to Initiate and Maintain Kupffer Cell Identity. Immunity. 2019;51:655–670 e658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu YE, Pan L, Zuo Y, Li X and Hong W. Detecting Activated Cell Populations Using Single-Cell RNA-Seq. Neuron. 2017;96:313–329 e316. [DOI] [PubMed] [Google Scholar]
- 54.Beliakova-Bethell N, Massanella M, White C, Lada S, Du P, Vaida F, Blanco J, Spina CA and Woelk CH. The effect of cell subset isolation method on gene expression in leukocytes. Cytometry A. 2014;85:94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grindberg RV, Yee-Greenbaum JL, McConnell MJ, Novotny M, O’Shaughnessy AL, Lambert GM, Arauzo-Bravo MJ, Lee J, Fishman M, Robbins GE, Lin X, Venepally P, Badger JH, Galbraith DW, Gage FH and Lasken RS. RNA-sequencing from single nuclei. Proc Natl Acad Sci U S A. 2013;110:19802–19807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krishnaswami SR, Grindberg RV, Novotny M, Venepally P, Lacar B, Bhutani K, Linker SB, Pham S, Erwin JA, Miller JA, Hodge R, McCarthy JK, Kelder M, McCorrison J, Aevermann BD, Fuertes FD, Scheuermann RH, Lee J, Lein ES, Schork N, McConnell MJ, Gage FH and Lasken RS. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc. 2016;11:499–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lacar B, Linker SB, Jaeger BN, Krishnaswami SR, Barron JJ, Kelder MJE, Parylak SL, Paquola ACM, Venepally P, Novotny M, O’Connor C, Fitzpatrick C, Erwin JA, Hsu JY, Husband D, McConnell MJ, Lasken R and Gage FH. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016;7:11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bakken TE, Hodge RD, Miller JA, Yao Z, Nguyen TN, Aevermann B, Barkan E, Bertagnolli D, Casper T, Dee N, Garren E, Goldy J, Graybuck LT, Kroll M, Lasken RS, Lathia K, Parry S, Rimorin C, Scheuermann RH, Schork NJ, Shehata SI, Tieu M, Phillips JW, Bernard A, Smith KA, Zeng H, Lein ES and Tasic B. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One. 2018;13:e0209648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiu X, Hill A, Packer J, Lin D, Ma YA and Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14:309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Svensson V, Natarajan KN, Ly LH, Miragaia RJ, Labalette C, Macaulay IC, Cvejic A and Teichmann SA. Power analysis of single-cell RNA-sequencing experiments. Nat Methods. 2017;14:381–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lonnerberg P and Linnarsson S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11:163–166. [DOI] [PubMed] [Google Scholar]
- 62.Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I and Enard W. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell. 2017;65:631–643 e634. [DOI] [PubMed] [Google Scholar]
- 63.Chen W, Li Y, Easton J, Finkelstein D, Wu G and Chen X. UMI-count modeling and differential expression analysis for single-cell RNA sequencing. Genome Biol. 2018;19:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valihrach L, Androvic P and Kubista M. Platforms for Single-Cell Collection and Analysis. Int J Mol Sci 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A and McCarroll SA. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA and Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP and Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S and Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. [DOI] [PubMed] [Google Scholar]
- 69.Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G and Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 2013;10:1096–1098. [DOI] [PubMed] [Google Scholar]
- 70.Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, Graybuck LT, Peeler DJ, Mukherjee S, Chen W, Pun SH, Sellers DL, Tasic B and Seelig G. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360:176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, Adey A, Waterston RH, Trapnell C and Shendure J. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hagemann-Jensen M, Ziegenhain C, Chen P, Ramsköld D, Hendriks G-J, Larsson AJM, Faridani OR and Sandberg R. Single-cell RNA counting at allele- and isoform-resolution using Smart-seq3. BioRxiv. [Preprint] October 25, 2019. [DOI] [PubMed] [Google Scholar]
- 73.Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, Moore R, McClanahan TK, Sadekova S and Klappenbach JA. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol. 2017;35:936–939. [DOI] [PubMed] [Google Scholar]
- 74.Huynh K Distinct immune microenvironments in atherosclerotic plaques. Nat Rev Cardiol. 2019. [DOI] [PubMed] [Google Scholar]
- 75.McGinnis CS, Patterson DM, Winkler J, Conrad DN, Hein MY, Srivastava V, Hu JL, Murrow LM, Weissman JS, Werb Z, Chow ED and Gartner ZJ. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat Methods. 2019;16:619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gaublomme JT, Li B, McCabe C, Knecht A, Yang Y, Drokhlyansky E, Van Wittenberghe N, Waldman J, Dionne D, Nguyen L, De Jager PL, Yeung B, Zhao X, Habib N, Rozenblatt-Rosen O and Regev A. Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. Nat Commun. 2019;10:2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Phipson B, Zappia L and Oshlack A. Gene length and detection bias in single cell RNA sequencing protocols. F1000Res. 2017;6:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R and Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Melsted P, Booeshaghi AS, Gao F, Beltrame E, Lu L, Hjorleifsson KE, Gehring j and Pachter L. Modular and efficient pre-processing of single-cell RNA-seq. BioRxiv [Preprint] July 26, 2019. [DOI] [PubMed] [Google Scholar]
- 81.Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, Gate RE, Mostafavi S, Marson A, Zaitlen N, Criswell LA and Ye CJ. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nature biotechnology. 2018;36:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McGinnis CS, Murrow LM and Gartner ZJ. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019;8:329–337 e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck WM 3rd, Smibert P and Satija R. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018;19:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lun ATL, Riesenfeld S, Andrews T, Dao TP, Gomes T, participants in the 1st Human Cell Atlas J and Marioni JC. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 2019;20:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luecken MD and Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019;15:e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buttner M, Miao Z, Wolf FA, Teichmann SA and Theis FJ. A test metric for assessing single-cell RNA-seq batch correction. Nat Methods. 2019;16:43–49. [DOI] [PubMed] [Google Scholar]
- 87.Tung PY, Blischak JD, Hsiao CJ, Knowles DA, Burnett JE, Pritchard JK and Gilad Y. Batch effects and the effective design of single-cell gene expression studies. Sci Rep. 2017;7:39921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goh WWB, Wang W and Wong L. Why Batch Effects Matter in Omics Data, and How to Avoid Them. Trends Biotechnol. 2017;35:498–507. [DOI] [PubMed] [Google Scholar]
- 89.Leek JT, Johnson WE, Parker HS, Jaffe AE and Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Polanski K, Park JE, Young MD, Miao Z, Meyer KB and Teichmann SA. BBKNN: Fast Batch Alignment of Single Cell Transcriptomes. Bioinformatics. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang F, Wu Y and Tian W. A novel approach to remove the batch effect of single-cell data. Cell Discov. 2019;5:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haghverdi L, Lun ATL, Morgan MD and Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nature biotechnology. 2018;36:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Butler A, Hoffman P, Smibert P, Papalexi E and Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Risso D, Perraudeau F, Gribkova S, Dudoit S and Vert JP. Publisher Correction: A general and flexible method for signal extraction from single-cell RNA-seq data. Nature communications. 2019;10:646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang T, Johnson TS, Shao W, Lu Z, Helm BR, Zhang J and Huang K. BERMUDA: a novel deep transfer learning method for single-cell RNA sequencing batch correction reveals hidden high-resolution cellular subtypes. Genome Biol. 2019;20:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li WV and Li JJ. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nature communications. 2018;9:997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gong W, Kwak IY, Pota P, Koyano-Nakagawa N and Garry DJ. DrImpute: imputing dropout events in single cell RNA sequencing data. BMC bioinformatics. 2018;19:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, Bierie B, Mazutis L, Wolf G, Krishnaswamy S and Pe’er D. Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell. 2018;174:716–729 e727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang M, Wang J, Torre E, Dueck H, Shaffer S, Bonasio R, Murray JI, Raj A, Li M and Zhang NR. SAVER: gene expression recovery for single-cell RNA sequencing. Nature methods. 2018;15:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eraslan G, Simon LM, Mircea M, Mueller NS and Theis FJ. Single-cell RNA-seq denoising using a deep count autoencoder. Nature communications. 2019;10:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Peng T, Zhu Q, Yin P and Tan K. SCRABBLE: single-cell RNA-seq imputation constrained by bulk RNA-seq data. Genome Biol. 2019;20:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Andrews TS and Hemberg M. False signals induced by single-cell imputation. F1000Res. 2018;7:1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hafemeister C and Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial expression. BioRxiv [Preprint] March 14, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tabula Muris C, Overall c, Logistical c, Organ c, processing, Library p, sequencing, Computational data a, Cell type a, Writing g, Supplemental text writing g and Principal i. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kiselev VY, Andrews TS and Hemberg M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat Rev Genet. 2019;20:273–282. [DOI] [PubMed] [Google Scholar]
- 106.Kiselev VY, Yiu A and Hemberg M. scmap: projection of single-cell RNA-seq data across data sets. Nat Methods. 2018;15:359–362. [DOI] [PubMed] [Google Scholar]
- 107.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, Butte AJ and Bhattacharya M. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.de Kanter JK, Lijnzaad P, Candelli T, Margaritis T and Holstege FCP. CHETAH: a selective, hierarchical cell type identification method for single-cell RNA sequencing. Nucleic Acids Res. 2019;47:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cohen M, Giladi A, Gorki AD, Solodkin DG, Zada M, Hladik A, Miklosi A, Salame TM, Halpern KB, David E, Itzkovitz S, Harkany T, Knapp S and Amit I. Lung Single-Cell Signaling Interaction Map Reveals Basophil Role in Macrophage Imprinting. Cell. 2018;175:1031–1044 e1018. [DOI] [PubMed] [Google Scholar]
- 110.Erhard F, Baptista MAP, Krammer T, Hennig T, Lange M, Arampatzi P, Jurges CS, Theis FJ, Saliba AE and Dolken L. scSLAM-seq reveals core features of transcription dynamics in single cells. Nature. 2019;571:419–423. [DOI] [PubMed] [Google Scholar]
- 111.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lonnerberg P, Furlan A, Fan J, Borm LE, Liu Z, van Bruggen D, Guo J, He X, Barker R, Sundstrom E, Castelo-Branco G, Cramer P, Adameyko I, Linnarsson S and Kharchenko PV. RNA velocity of single cells. Nature. 2018;560:494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS and Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA and Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14:979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Saelens W, Cannoodt R, Todorov H and Saeys Y. A comparison of single-cell trajectory inference methods. Nat Biotechnol. 2019;37:547–554. [DOI] [PubMed] [Google Scholar]
- 115.Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, van den Oord J, Atak ZK, Wouters J and Aerts S. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Efremova M, Vento-Tormo M, Teichmann S and Vento-Tormo R. CellPhoneDB v2.0: Inferring cell-cell communication from combined expression of multi-subunit receptor-ligand complexes. BioRxiv. [Preprint] June 24, 2019. [DOI] [PubMed] [Google Scholar]
- 117.Browaeys R, Saelens W and Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2019. [DOI] [PubMed] [Google Scholar]
- 118.Stubbington MJT, Lonnberg T, Proserpio V, Clare S, Speak AO, Dougan G and Teichmann SA. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods. 2016;13:329–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Galkina E, Thatte J, Dabak V, Williams MB, Ley K and Braciale TJ. Preferential migration of effector CD8+ T cells into the interstitium of the normal lung. J Clin Invest. 2005;115:3473–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Eng CL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, Cronin C, Karp C, Yuan GC and Cai L. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature. 2019;568:235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F and Macosko EZ. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wahlby C and Nilsson M. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 2013;10:857–860. [DOI] [PubMed] [Google Scholar]
- 123.Soldatov R, Kaucka M, Kastriti ME, Petersen J, Chontorotzea T, Englmaier L, Akkuratova N, Yang Y, Haring M, Dyachuk V, Bock C, Farlik M, Piacentino ML, Boismoreau F, Hilscher MM, Yokota C, Qian X, Nilsson M, Bronner ME, Croci L, Hsiao WY, Guertin DA, Brunet JF, Consalez GG, Ernfors P, Fried K, Kharchenko PV and Adameyko I. Spatiotemporal structure of cell fate decisions in murine neural crest. Science. 2019;364. [DOI] [PubMed] [Google Scholar]
- 124.Srivatsan SR, McFaline-Figueroa JL, Ramani V, Saunders L, Cao J, Packer J, Pliner HA, Jackson DL, Daza RM, Christiansen L, Zhang F, Steemers F, Shendure J and Trapnell C. Massively multiplex chemical transcriptomics at single-cell resolution. Science. 2020;367:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Barkas N, Petukhov V, Nikolaeva D, Lozinsky Y, Demharter S, Khodosevich K and Kharchenko PV. Joint analysis of heterogeneous single-cell RNA-seq dataset collections. Nat Methods. 2019;16:695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P and Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]