Abstract
The liver is the largest solid organ and a primary metabolic hub. In recent years, intact cell nuclei were used to perform single-nuclei RNA-seq (snRNA-seq) for tissues difficult to dissociate and for flash-frozen archived tissue samples to discover unknown and rare cell subpopulations. In this study, we performed snRNA-seq of a liver sample to identify subpopulations of cells based on nuclear transcriptomics. In 4282 single nuclei, we detected, on average, 1377 active genes and we identified seven major cell types. We integrated data from 94,286 distal interactions (p < 0.05) for 7682 promoters from a targeted chromosome conformation capture technique (HiCap) and mass spectrometry proteomics for the same liver sample. We observed a reasonable correlation between proteomics and in silico bulk snRNA-seq (r = 0.47) using tissue-independent gene-specific protein abundancy estimation factors. We specifically looked at genes of medical importance. The DPYD gene is involved in the pharmacogenetics of fluoropyrimidine toxicity and some of its variants are analyzed for clinical purposes. We identified a new putative polymorphic regulatory element, which may contribute to variation in toxicity. Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer and we investigated all known risk genes. We identified a complex regulatory landscape for the SLC2A2 gene with 16 candidate enhancers. Three of them harbor somatic motif breaking and other mutations in HCC in the Pan Cancer Analysis of Whole Genomes dataset and are candidates to contribute to malignancy. Our results highlight the potential of a multi-omics approach in the study of human diseases.
Keywords: snRNA-seq, proteomics, human liver, multi-omics data integration
Introduction
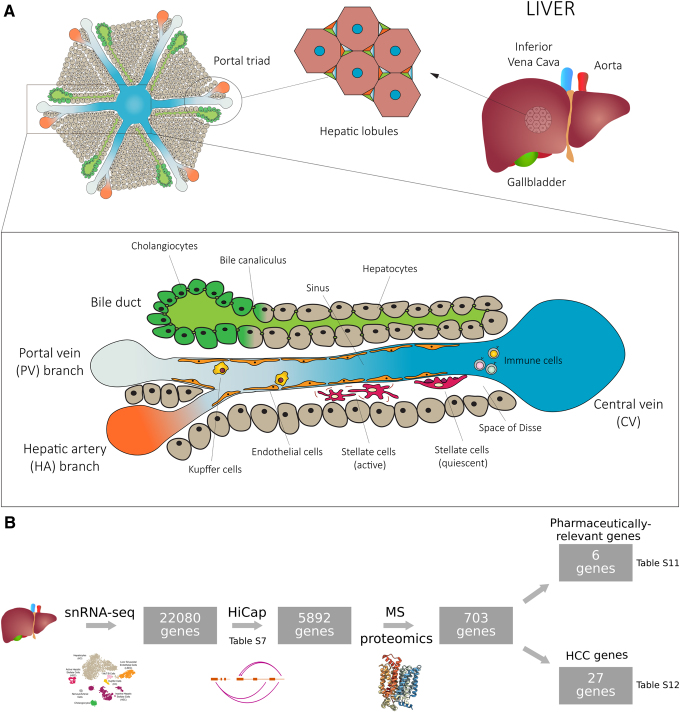
The liver is the largest solid organ of the human body and a primary metabolic hub. The parenchymal cells (PCs), that is, hepatocytes (HCs), constitute the biggest part of the liver and are involved in diverse physiological processes, for example, protein synthesis and storage of carbohydrates, lipid metabolism, urea and bile synthesis, drug metabolism, and detoxification processes for exogenous and endogenous compounds. HCs are arranged in hepatic lobules (Fig. 1), a microscopical hexagonal architecture with a central vein in the middle draining the blood coming from the distal hepatic artery (HA) and portal vein (PV) branches (Fig. 1).
FIG. 1.
Schematic cross-sectional view of the structural organization and cell populations of a liver lobule. (A) Each lobule presents a radial structure with a central vein (CV) in the middle from which HC cords radiate toward the so called portal triad consisting of branches of the portal vein (PV) and hepatic artery (HA) and bile ducts. The sinus is delimited by HCs that are arranged back to back in cords and it is lined by specialized sinusoidal endothelial cells. Kupffer and immune cells are located in the sinusoidal lumen, while hepatic stellate cells are localized in the space of Disse. (B) Workflow representing the design of the multi-omics study. The crystal structure of the glucose transporter was obtained from PDB (ID: 4ZWB, Deng et al., 2015). HC, hepatocyte.
Linear stretches of HCs (HC cords) define sinusoid capillaries where most of the nonparenchymal cells (NPCs) of the liver are located. NPCs release factors that regulate HCs both in physiological and pathological conditions (Kmiec, 2001). The best characterized NPCs include the liver sinusoidal endothelial cells (LSECs) lining the sinusoid capillaries, Kupffer cells (KCs), which are macrophages resident in the sinusoid capillary lumen, and hepatic stellate cells (HSCs) that act as storage for fat and vitamin A, and are located in the space of Disse defined by the HC cords and the sinus (Arii and Imamura, 2000).
Most knowledge of the function of liver cells is based on analysis of bulk tissue, but recent advances in next-generation sequencing (NGS) technologies are opening the field to the study of genomics and transcriptomics of single cells. Studying the gene expression profiles of single cells can potentially lead to the discovery of new and rare cell subpopulations or track cell lineages in development (Hwang et al., 2018). Single-cell RNA sequencing (scRNA-seq) has been successfully performed in several tissues in mice (Han et al., 2018); however, when it comes to human tissue, the scarcity of fresh tissue and the difficulty to obtain rare and difficult to isolate cell types constitute a major bottleneck (MacParland et al., 2018).
In recent years, it has been proposed that intact cell nuclei can be used to perform single-nuclei RNA-seq (snRNA-seq) for cell types difficult to dissociate, for flash-frozen archived tissue samples, and for large cells like fat and muscle (Nguyen et al., 2018). Several studies have reported high correlation between snRNA-seq and scRNA-seq gene expression, but at the same time revealed an enrichment bias for nuclear RNAs and lncRNAs in nuclei (Gao et al., 2017; Habib et al., 2017).
The differences in transcriptomics profile between defined liver cell populations stem from a precise genetic control of gene expression mostly driven by cell type-specific enhancers and other distal regulatory elements. Promoter-enhancer interactions has been studied on a genome-wide scale using HiC, a chromosome conformation capture technique that allows defining topologically associated domains and higher-order chromatin interactions (Lieberman-Aiden et al., 2009). Building on HiC, a novel technique known as targeted chromosome conformation capture (Capture-C, ChiC, and HiCap) has been developed to explore promoter-enhancer interactions to a much higher resolution (Dryden et al., 2014; Jäger et al., 2015; Sahlén et al., 2015). Today, it is possible to elucidate a genome-wide promoter-enhancer interaction network using as much as 50,000 capturing probes targeting gene promoters in a single HiCap experiment (Åkerborg et al., 2019).
Proteomics studies represent the ideal complementary analysis to evaluate the level of translation of an mRNA transcript into a protein. However, abundance in mRNA might not be proportionally correlated to the levels of proteins due to mRNA degradation, faulty translation mechanisms, or high rate of protein turnover (Maier et al., 2009). Moreover, transcriptomic analysis does not take into consideration crucial post-translational modifications that largely affect a protein's half-life and stability (Benjannet et al., 2006). Mass spectrometry (MS) has become the method of choice for analysis of proteins' primary structures, post-translational modifications, or protein-protein interactions in complex protein samples (Aebersold and Mann, 2003) both in bulk and single-cell environment (Specht and Slavov, 2018).
In this study, we performed snRNA-seq of a human liver sample to identify subpopulations of cells based on the nuclear transcriptomics. Using data from the same liver, we then integrated long-range HiCap interactions and proteomics data from bulk experiments with single nuclei transcriptomics data to “assign” specific enhancer-promoter interactions to specific liver cell populations.
Materials and Methods
Ethics statement
The study was approved by the Uppsala regional ethics committee (Dnr: 2009/028, 2011/037, 2014/433).
Preparation of single nuclei suspension
Human liver tissue was kindly provided by Prof. Per Artursson, Uppsala University. The liver sample was obtained from a partial hepatectomy of a patient with colon cancer and hepatic metastasis. The patient had provided written consent for the use of the biological sample for scientific research. Part of the resection, characterized as tumor free by a pathologist, was flash-frozen and subsequently employed for this study. The use of the sample for research purposes has been approved by the Uppsala ethics regional committee Dnr 2009/028 (updated 2011/037) for Prof. Artursson and Dnr 2014/433 for Prof. Wadelius.
A suspension of single liver nuclei was prepared using the gentleMACS dissociator (Miltenyi). Five milligram of frozen liver tissue was transferred into a gentleMACS C-tube with 10 mL of 0.250 M sucrose solution. The tissue was homogenized using the gentleMACS dissociator program E. The homogenate was filtered using 100 μm MACS SmartStrainers and the volume was brought up to 15 mL with the sucrose solution. The homogenate was then centrifuged at 600 g at 4°C in an Eppendorf 5810R Centrifuge and the supernatant was discarded. The nuclei in the pellet were resuspended in 2 mL of phosphate-buffered saline (PBS) with 0.04% bovine serum albumin and RiboLock RNase Inhibitor (Thermo Scientific) and filtered using a 30 μm MACS SmartStrainers. The nuclei suspension was evaluated for purity and concentration using a hemocytometer (Fuchs-Rosenthal) for a final count of ∼1000 nuclei/μL.
Alternatively, the nuclei suspension was stained with 4′,6-diamidino-2-phenylindole (DAPI) and sorted using fluorescence-activated cell sorting (FACS) using the BD FACSMelody cell sorter with a 100-μm nozzle to achieve even a higher debris removal and precise determination of the nuclei concentration for a final count of ∼227 nuclei/μL.
snRNA-seq—cDNA library preparation and sequencing
After the determination of the nuclei suspension concentration, the suitable volume of nonsorted and sorted nuclei was calculated for a target cell recovery of 6000 and 3000 nuclei, respectively.
Samples were loaded on the 10 × Genomics single-cell-A chip (PN-no. 120236) and run on the Chromium System (Chromium Controller GCG-SR-1, 10 × Genomics).
After droplet generation and reverse transcription, the cDNA was recovered and amplified for 12 cycles. The samples where run on a High Sensitivity D1000 ScreenTape on the TapeStation (Agilent Technologies) and measured on the Qubit to determine the cDNA concentration.
The two sequencing libraries for FACS and notsorted nuclei were prepared using the Chromium Single Cell 3′ v2 Reagent kit (10 × Genomics, cat no. 120236/37/62) according to the manufacturer's protocol (CG00052 Single Cell 3′ Reagent Kit v2 User Guide). The adapter-ligated fragments were quantified by quantitative polymerase chain reaction using the Library quantification kit for Illumina (KAPA Biosystems) on a CFX384 Touch instrument (Bio-Rad Laboratories) before cluster generation and 26 + 8 + 0 + 91 cycles of sequencing of the two libraries in one S1 flowcell using the NovaSeq system and v1 sequencing chemistry (Illumina, Inc.).
snRNA-seq—data preprocessing, nuclei clustering, and differential expression
The raw base call files (bcl files) were demultiplexed using cellranger mkfastq tool and the resulting fastq files were analyzed with the cellranger count tool that performs alignment, filtering, barcode identification, and unique molecular identifier (UMI) counting.
We merged the unfiltered matrices from cellranger for the pre- and post-FACS replicates. We used the function emptyDrops of the R package DropletUtils (Griffiths et al., 2018; Lun et al., 2018) to filter out putative empty droplets at 1% false discovery rate (FDR) requiring nuclei to contain a minimum of 200 UMIs each. We used the function isOutlier of the R package scater3 to remove damaged or dying nuclei and removed nuclei with log-library sizes and log-transformed number of genes three median absolute deviations below the respective median. The same function was applied to filter out nuclei with percentage of counts for mitochondrial genes three median absolute deviations above the overall median.
Next, we removed all the genes that were expressed in <5 nuclei. We performed a rough clustering of the nuclei using the function quickCluster with min.mean = 0.1 and irlba.args = list(maxit = 1000) to control for potential heterogeneity between replicates and we computed the size factors using the function computerSumFactors with minimum library size-adjusted average counts 0.1 (McCarthy et al., 2017). We then applied the size factors to normalize the UMI counts using the function normalize (McCarthy et al., 2017). Lack of spike-in transcripts led us to model the technical noise after a Poisson distribution which serves as a lower bound for the variances of endogenous genes. For that, we used the scran (Lun et al., 2016) functions makeTechTrend with default arguments and trendVar with block set as replicates, a parametric curve fitted before smoothing, disabled spike-ins, and the degree of smoothing set to 0.05. This also provided a set of genes with positive biological components.
For batch effect correction between replicates, we used the function fastMNN from scran that uses a mutual nearest neighbors (MNN) approach (Haghverdi et al., 2018; Lun et al., 2016). In fastMNN, we assumed that every nucleus has ∼20 neighbors and we approximated the genes with positive biological components in all nuclei with the first 50 principal components, which are used for the batch effect correction. Next, we used the Poisson technical trend to compute the number of principal components that explain the majority of the variance in the data with the function denoisePCA from scran. We also applied the function runPCA from scater to detect and remove potential outliers. We computed the t-distributed stochastic neighbor embedding (t-SNE) dimensionality reduction for batch effect principal components from fastMNN and perplexity = 30.
Finally, we built a k-nearest neighbors graph with k = 50 using the function buildSNNGraph from scran to identify the clusters of cell types formed by the nuclei.
We next used Seurat v3.0 (Butler et al., 2018; Stuart et al., 2018) to explore the impact of a possible bias due to the cell cycle on our data and to regress out the impact of replicates from the data that would allow us to perform differential expression. Finally, we identified all the marker genes of each cluster using the function FindAllMarkers from Seurat filtering on 0.1% FDR and 1.5 log-fold change (Supplementary Table S1).
Based on the expression profile of marker genes in each cluster, we determined the identity of the different liver cell populations using a systematic literature search as reported in the results section. The random seed used throughout the whole study is 2019. R sessionInfo details can be found at Supplementary Table S2.
In silico bulk and bulk RNA-seq combined with mRNA-to-protein factors
In silico bulk snRNA-seq and scRNA-seq
To generate the in silico bulk RNA-seq sets, we used the filtered matrices from our snRNA-seq and publicly available scRNA-seq experiments (GSE115469) (MacParland et al., 2018). We used the function rowMeans of the R package Matrix to compute the average number of UMIs over all the single nuclei and cells, and we applied a log2 transformation to adjust their distributions and to make them comparable to other datasets.
Bulk RNA-seq
To generate the bulk RNA-seq set, we downloaded the publicly available GTEx V7 matrices of gene reads (Lonsdale et al., 2013). We extracted samples of liver origin, we used the function rowMeans of the R package Matrix to compute the average number of reads over all the samples, and we applied a log2 transformation to adjust their distributions and to make them comparable to other datasets.
mRNA-to-protein factors
We downloaded a list of tissue-independent and gene-specific protein abundance predictability factors (Moulana et al., 2018). We multiplied the log2-average in silico bulk and bulk RNA-seq gene expressions with their corresponding gene-specific factors for liver.
HiCap analysis
Mechanically fine-ground human liver tissue was used for studying the folding of the chromatin by high-throughput chromosome conformation capture coupled with subsequent targeted sequence capture (HiCap). Cells were crosslinked for 10 min in 1% formaldehyde solution and the reaction was quenched with the final concentration of 0.125 M of glycine. The samples were immediately lysed using ice-cold lysis buffer containing 10 mM Tris-HCl, pH 8.0, 10 mM NaCl, 0.2% Triton X-100, and protease inhibitor cocktail. Before incubating the lysis mixture for 10 min on ice, the cell agglomerates were dissociated by passing the cells in lysis buffer through a 27-gauge needle for up to eight times. Nuclei were then washed in ice-cold PBS with proteinase inhibitor blend and collected through mild centrifugation.
Sodium dodecyl sulfate (SDS) was used to partly solubilize the chromatin to aid the accessibility of fast digest MboI endonuclease (ThermoFisher Scientific) to the restriction sites. Triton X-100 was used to quench the SDS before introducing the enzyme into the mixture. After 4.5-h chromatin digestion at 37°C, the protruding 5′ends of the DNA left by the endonuclease were filled with a mixture of nucleosides containing biotin-14-dATP in a reaction catalyzed by Klenow fragment of DNA Polymerase I (ThermoFisher Scientific). The reaction was quenched by 10 mM ethylenediamine tetraacetic acid (EDTA) and brief incubation at 75°C. Blunt-end chromatin complex intramolecular ligation (proximity ligation) catalyzed by 12 Weiss Units of T4 DNA ligase (New England Biolabs) was then performed for 4.5 h at 16°C.
After ligation, the samples were de-crosslinked by 8-h incubation at 65°C with the presence of Proteinase K (ThermoFisher Scientific) in the mixture. The samples were then purified using phenol-chloroform-isoamyl alcohol (25:24:1) blend and precipitated in ethanol with sodium acetate, pH 5.2, and glycogen. Any remaining intact RNA was removed by a 1-h incubation at 37°C with the presence of RNase A (ThermoFisher Scientific). Proximity ligation reaction artifacts (unligated biotinylated DNA ends) were then removed by T4 DNA Polymerase treatment. Resulting chimeric DNA constructs were subsequently sonicated to achieve fragments around 200 bp in length.
Covaris sonication system with SonoLab software was used for six cycles of 60 sec at 10% duty cycle, 200 cycles per burst, and intensity of 5. KAPA HTP Library preparation kit for Illumina platforms was used to prepare NGS-compatible libraries by following manufacturer's protocol for fragmented DNA end-repair, A-tailing, and TruSeq LT (Illumina, Inc.) adapter ligation. Biotin-avidin selection of fragments was performed before the adapter ligation step to further remove the technology artifacts and enrich the successful proximity ligation fragments.
Next, the targeted sequence capture was performed to further enrich the libraries of interest using SureSelect XT Target Enrichment System for Illumina Paired-End Multiplexed Sequencing libraries (Agilent Technologies). Hybridization and Capture protocol and reagents were used in this step following manufacturer's recommendations for custom RNA oligonucleotide probe hybridization to NGS-libraries for 24 h. After probe hybridization, the stringent washing of probe-captured libraries was performed to keep the target libraries and remove the mismatched hybridization artifacts. Enriched libraries were then sequenced in-house by Illumina single TruSeq LT index, paired-end sequencing on NextSeq 500 platform (Illumina, Inc.).
Throughout the chromosome conformation capture, library preparation, and target enrichment, the quality of the samples was checked by the 2100 Bioanalyzer system (Agilent Technologies) for automated electrophoresis on laboratory chip, and Qubit fluorometric quantitation system (Invitrogen) for nucleic acid quantitation.
HiCap analysis interaction calling
HiCap provides p-values relative to the null hypothesis that physical 3D distance is proportional to genomic distance (Anil et al., 2017). Contact occurrences among genomic segment pairs of interest are related to a carefully selected set of negative controls at corresponding genomic distance. Negative controls are regions with no known regulatory activity and far from promoters at a set distance, which in this study was 50 kb. Only interacting segments meeting a requirement of five p-value below 0.05 were taken forward. We merged DpnII fragments and liver-specific peaks from ChIP-Atlas database (Oki et al., 2018) (Supplementary Table S3) and called the interactions using the merged fragment dataset to increase the probability of calling interacting regions overlapping enhancer regions.
Distal elements interacting with promoters (probes) of genes were annotated with a ChromHMM model (Ernst and Kellis, 2012, 2017) that defines 25 chromatin states based on imputed data for 12 epigenetics marks including H3K4me1, H3K4me2, H3K4me3, H3K9ac, H3K27ac, H4K20me1, H3K79me2, H3K36me3, H3K9me3, H3K27me3, H2A.Z, and DNase. In this study, we utilized the ChromHMM annotation for healthy adult liver tissue (EID = E066) from the Roadmap Epigenomics Project (Kundaje et al., 2015).
Proteomics analysis
Liver sample was boiled in lysis buffer containing 10% Trifluoroethanol, 50 mM Tris pH 8 and 5 mM dithiothreitol (DTT) and 20 mM chloroacetamide. Sample was sonicated for 15 min on Bioruptor. Proteins were digested with LysC and Trypsin overnight at 37°. The peptides were purified on polystyrene divinyl benzene (SDB)-reverse phase sulphonate (RPS) material. The purified peptides were measured using liquid chromatography-mass spectrometry (LC-MS) instrumentation consisting of an EASY-nLC 1200 system coupled to a nanoelectrospray ion source and a Q Exactive HF Orbitrap (all Thermo Fischer Scientific). Purified peptides were separated on 50 cm high-performance liquid chromatography (HPLC) columns (in house packed into the tip with Reprosil-Pur C18-AQ 1.9 μm resin (Dr. Maisch GmbH). Purified peptides were loaded in buffer A (0.1% formic acid) and eluted with a linear 100-min gradient of 3–30% of buffer B (0.1% formic acid and 80% [v/v] acetonitrile).
The column temperature was kept at 60° by a Peltier element containing an in-house-developed oven. MS data were acquired with Top15 data-dependent MS/MS scan method (topN method). The target value for full scan MS spectra was set to 3e6 in the 300–1650 m/z range with a maximum injection time of 25 ms and a resolution of 60,000 at 200 m/z. Fragmentation of precursor ions was performed by high-energy C-trap dissociation with a normalized collision energy of 27 eV. MS/MS scans were performed at a resolution of 15,000 at m/z 200 with target ion values of 1e5 and maximum injection time of 25 ms.
MS raw file was analyzed using the MaxQuant software (Cox and Mann, 2008) and peptide list was searched against the human UniProt FASTA database with the Andromeda search engine (Cox et al., 2011). For the search, a contamination database was included, cysteine modification was set as a fixed modification, and N-terminal acetylation and methionine oxidation were set as variable modification. FDR was 0.01 for both the protein and peptide level with a minimum length of seven amino acids, and the FDR was determined by searching reverse database.
Enzyme specificity was set as C-terminal to arginine and lysine using trypsin protease, and maximum two missed cleavage were allowed for search. The peptides were identified with an initial precursor mass deviation of up to 7 ppm and fragment mass deviation of 20 ppm. In case of identified peptides that were shared between two or more proteins, these were combined and reported in protein groups. Contaminants and reverse identification were removed from further data analysis.
We log2 transformed the MS proteomics levels to approximate a normal distribution and to make the values better comparable to the in silico bulk or bulk RNA-seq experiments.
Data availability
All relevant data are presented within the article and its supporting information files. Access to the ENA (European Nucleotide Archive) database, which will host the raw sequencing data, and PRIDE (PRoteomics IDEntification Database), which will host the raw MS proteomics data, will be available at the EBI BioStudies database under the accession number S-BSST324. These data will be also available from the corresponding authors upon request.
Results
snRNA-seq identification of major liver cell types
Nuclei were isolated from a frozen sample of human liver tissue. The nuclei isolation was carried out in two independent replicates, with one replica undergoing an extra step of FACS. The two replicates were pooled together, corrected for batch effect (Supplementary Fig. S1), and underwent extensive quality control (cf. Methods; Supplementary Figs. S2 and S3).
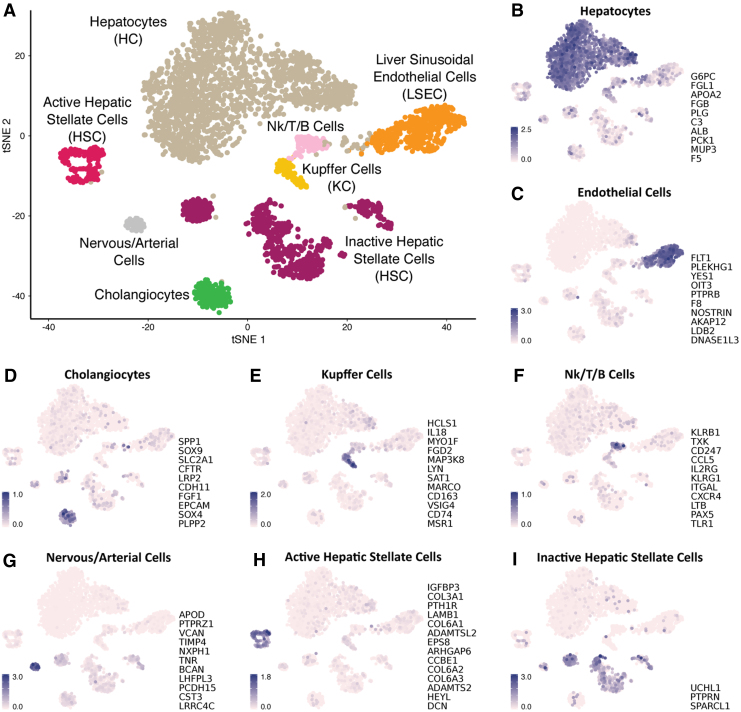
In total, we sequenced and analyzed 4282 nuclei from adult human liver tissue. On average, 1377 genes were detected per nucleus across the different cell types with HCs showing an average of 2103 transcribed genes per nucleus, suggesting a complex biology reflected in a more transcriptionally active liver parenchyma (Supplementary Table S4). We identified seven major clusters, which were assigned to different cell types based on expression of cell type-specific markers (Fig. 2): HCs, LSECs, KCs, active and several groups of inactive HSCs, cholangiocytes, intrahepatic immune cells (Nk-like/T/B cells), and a small fraction of cells that likely are of nervous/arterial origin.
FIG. 2.
t-distributed stochastic neighbor embedding (t-SNE) plot for snRNA-seq analysis of 4282 liver nuclei. (A) Overview of different liver cell populations. (B–I) Expression of cell type-specific gene markers shown in the right-hand side of each panel. snRNA-seq, single-nuclei RNA-seq.
HCs are PCs that account for ∼70% of the liver's mass. They are involved in several biological processes from protein synthesis and lipid metabolism to exogenous and endogenous detoxification. We used established HC gene markers (Aizarani et al., 2019; Halpern et al., 2017, 2018; MacParland et al., 2018; Ramachandran et al., 2019) to identify 2155 nuclei (>50% of the total; Supplementary Table S4) as HCs, including G6PC, APOA2, FGB, ALB, PCK1, PLG, FGL1, MUP3, C3, and F5.
The remaining part of the liver is composed of NPCs that interact with HCs in a paracrine manner. The largest NPC population is LSECs, which line the sinusoidal capillary of the liver acting as a barrier, but are also involved in several physiological and immunological functions, particularly mediating the immune response upon liver injury (Shetty et al., 2018). We identified 488 (11.4%) LSECs based on the expression levels of gene markers (Aizarani et al., 2019; Halpern et al., 2017, 2018; MacParland et al., 2018; Ramachandran et al., 2019) such as FLT1, OIT3, PTPRB, F8, NOSTRIN, and PLEKHG1.
Between the HCs and the sinusoidal space lined by endothelial cells, in the so-called perisinusoidal space of the liver, is located the fourth major liver cell population: the HSCs. HSCs are lipid-storing cells that for the majority are maintained in an inactive/quiescent state. They can be activated in response to liver injury, inducing the production of collagen and extracellular matrix, making HSCs a major player involved in liver fibrosis.
Based on the expression profile of inactive and active-specific gene markers (Boers et al., 2006; MacParland et al., 2018; Mannaerts et al., 2013; Zhang et al., 2016), we identified 232 (5.4%) active HCSs: ADAMTS2, COL3A1, COL6A1/2/3, EPS2, LAMB1, IGFBP3, and DCN, and 868 (20.2%) inactive HSCs: PTPRN, UCHL1, and SPARCL1. Supplementary Table S5 reports the list of genes differentially expressed between inactive and active HSC. Interestingly, the inactive HSCs cluster in three major groups, indicating a phenotypic heterogeneity.
The transcriptome analysis of liver nuclei allowed us also to identify a distinct cluster of 194 cholangiocytes (4.5%), specialized epithelial cells of the bile duct that are involved in the bile secretion. Specific gene markers (Li et al., 2017; Sato et al., 2019) for this cluster were as follows: SOX4, SOX9, EPCAM, FGF1, CDH11, LRP2, CFTR, SLC2A1, and SPP1.
An immunity component of the NPCs is represented by KCs, which are liver specialized resident macrophages. They are located close to LSECs in the sinusoidal lumen and upon activation release cytokines and other signaling molecules affecting neighboring cells. We identified 135 (3.1%) KCs expressing the following gene markers (Aizarani et al., 2019; Halpern et al., 2017, 2018; MacParland et al., 2018; Ramachandran et al., 2019): IL18, MYO1F, FGD2, CD74, CD163, SAT1, MARCO, and MSR1.
We also identified a cluster of 131 (3%) liver nuclei expressing genes characteristic of immune cells, such as KLRB1, TXK, CD247, CCL5, IL2RG, KLRG1, ITGAL, CXCR4, LTB, PAX5, and TLR1. The expression of these gene markers has been reported characterizing B cells, T cells, and NK-like cells (Aizarani et al., 2019; Halpern et al., 2018; MacParland et al., 2018; Ramachandran et al., 2019) and in this study, defines a cluster of intrahepatic immune cells.
Finally, we identified a small cluster of 79 nuclei (1.8%). Analysis of gene markers characterizing this cluster suggested both a nervous (APOD, NXPH1, LHFPL3, and LRRC4C) and arterial (VCAN and TIMP4) origin. A hypothesis is that they could represent afferent and efferent autonomic nerves associated with branches of the PV or HA (Jensen et al., 2013; Yi et al., 2010), so we defined them as nervous/arterial cells.
Integrating bulk HiCap long-range interactions and single nuclei transcriptomics data
We performed a HiCap experiment on the same frozen liver sample using probes placed at the promoter regions of all genes to identify all the distal elements interacting with promoters. The rationale behind this experiment, performed in bulk tissue, was to identify cell population-specific long-range interactions by interpreting the HiCap results through the single nuclei transcriptome profiles. In total, we identified 97,709 significant (p < 0.05) long-range interactions between 7682 promoters and 94,286 distal regions (Fig. 1B; Supplementary Tables S6 and S7).
Using chromHMM annotations, 4.88% of distal elements were defined as enhancers, 0.68% as promoters for other genes, and 64.41% as silent regions, while 13.7% of the distal regions that overlapped at least two different annotation types were defined as mixed (Supplementary Fig. S4; Supplementary Table S8). The majority of the mixed set contained enhancer annotations, accounting for 70.4% of the whole group, increasing the overall portion of enhancers to ∼14% of the total regions.
To integrate information from the different datasets, we selected genes displaying expression profiles in snRNA-seq, reported HiCap interactions, and detected protein levels as discussed below. We focused on two groups of liver genes, fulfilling the above selection criteria, which are important for drug metabolism (pharmaceutically-relevant genes) and are used as prognostic biomarkers in patients with hepatocellular carcinoma (HCC), the most common type of primary liver cancer.
Overall, we identified a total of 52 distal interactions with putative active enhancers regulating pharmaceutically and HCC-related genes (highlighted in Supplementary Tables S9 and S10) that warrant deeper experimental validations to establish the molecular mechanism of regulation. In this study, we present two examples of integration of bulk and single nuclei data to identify putative genetic regulatory mechanisms mediating drug toxicity and HCC.
Integration with bulk proteomics
We also carried out proteomics analysis in bulk to identify the protein levels of the liver sample and explore correlations with the single nuclei transcriptomics data. We detected 2351 unique proteins. Genes expressing 703 of the proteins showed significant HiCap interactions with distal elements (Fig. 1B). The correlation between mRNA and protein levels has been reported to be highly affected by mRNA stability and protein degradation, making it difficult to predict the protein level from the mRNA.
Recently, it has been shown that considering a tissue-independent and gene-specific mRNA-to-protein (RTP) conversion factor may enhance the predictability of protein copy numbers from RNA levels (Edfors et al., 2016; Moulana et al., 2018). To compare transcriptomes from the single nuclei analysis of liver tissue and bulk liver proteomics, we first generated an in silico bulk dataset for snRNA-seq, averaging the expression data of all nuclei following a similar approach to a recent study integrating scRNA-seq and bulk proteomics in mice lung tissue (Angelidis et al., 2019). Finally, the bulk proteomics dataset was compared to the protein levels predicted by RTP ratios applied to the in silico bulk snRNA-seq, scRNA-seq, and bulk RNA-seq datasets (Supplementary Fig. S5).
As a benchmark, we compared the protein levels predicted by RTP ratios combined with the in silico bulk snRNA-seq for 4282 liver single nuclei, the in silico bulk scRNA-seq for 8444 single cells from 5 human liver samples (MacParland et al., 2018) that showed marked heterogeneity in the t-SNE plots, and the average RNA-seq expression in liver for 175 individuals from the GTEx project (Lonsdale et al., 2013). We observed a good correlation between in silico bulk snRNA-seq and scRNA-seq for the main liver cell types HC and LSEC (Pearson r = 0.57 and r = 0.45, respectively; Supplementary Fig. S6), and poorer correlation for other cell types.
As expected, the overall correlation of the in silico bulk snRNA-seq with the proteomics data was low (Pearson r = 0.19; Supplementary Fig. S5B). A better, yet moderate correlation with the proteomics data was observed for in silico bulk scRNA-seq and bulk RNA-seq (Pearson r = 0.4 and r = 0.46, respectively; Supplementary Fig. S5A, C), perhaps reflecting a quantitative “loss of information” when comparing protein abundancies with transcript levels from single nuclei, single cells, or bulk. The correlations improved largely when RNA-seq experiments were adjusted for RTP abundancy estimation factors (Pearson r = 0.47 for in silico bulk snRNA-seq, r = 0.61 for in silico bulk scRNA-seq, and r = 0.76 for bulk RNA-seq; Supplementary Fig. S5D–F), confirming once more that mRNA levels cannot predict protein levels on a general level, but must be verified in a gene-specific way.
Novel insights on the pharmacogenetics of fluoropyrimidine toxicity
The individual response to a medication and the risk of adverse reaction have strong genetic component and several liver-expressed genes have been characterized as pharmacogenomics biomarkers. We explored the liver-related genes reported as “Pharmacogenomic biomarkers in Drug Labeling” by the FDA (https://www.fda.gov/drugs/scienceresearch/ucm572698.htm).
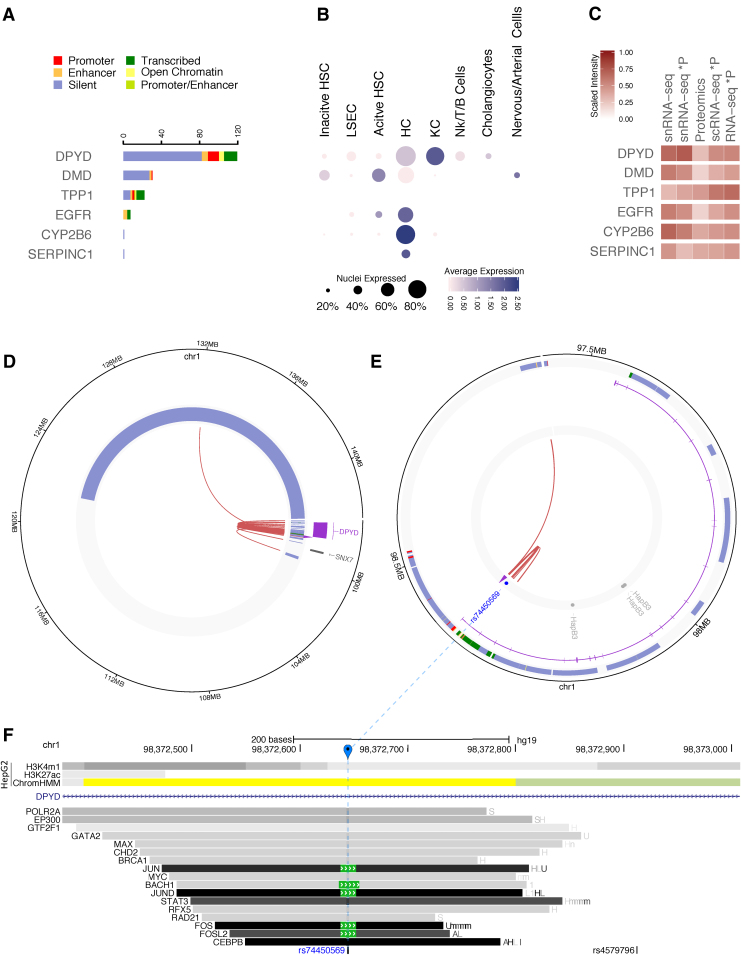
We selected pharmaceutically relevant genes that were expressed in snRNA-seq, and that had HiCap and proteomics signals (Fig. 1B; Supplementary Table S11). As an example, we investigated the highly polymorphic and clinically analyzed DPYD gene and checked its cell population-specific expression based on the snRNA-seq data, its HiCap interactions, and proteomics profile in bulk (Fig. 3; Supplementary Table S9).
FIG. 3.
Multi-omics overview of DPYD. (A) Number of probe-distal HiCap interactions associated to liver-specific chromHMM annotations. (B) Expression levels from snRNA-seq in different liver cell types. The size of the dot represents the number of nuclei that express the gene, while the color intensity the overall level of expression. (C) Heatmap comparing the expected and experimental levels of protein abundance from RNA-seq and MS proteomics experiments, respectively. The first column shows the log2-average expression of genes from the in silico bulk snRNA-seq, while the second one illustrates the estimated protein abundance calculated after calibrating the in silico bulk snRNA-seq levels for RTP abundancy estimation factors. The third column shows the experimental level of the protein abundance detected by MS. The last two columns show the estimated protein abundance calculated after calibrating the log2-average in silico bulk scRNA-seq levels and the log2-average number of reads of bulk RNA-seq. (D, E) Circos plots illustrating interactions of the probe for DPYD with distal elements. (D) The first track shows gene annotations overlapping probe-distal interactions. The second track shows manually curated chromHMM annotations overlapping interacting regions, while the inner one shows the experimental HiCap interactions. The purple arrow marks the probe location. (E) A zoom-in on the area of interest for the circos plot in (D). The first two tracks show chromHMM and gene annotations, respectively. The third track shows six enhancers interacting with the DPYD promoter. The inner track shows probe-distal (red) HiCap interactions. The purple arrow marks the probe location. (F) The genomic landscape for the SNP rs74450569 from (E) (marked in blue) located in a distal element overlapping an active enhancer and interacting with the DPYD probe. The UCSC genome browser tracks represent from the top: (1) the ChIP-seq signals for two active enhancer-specific histone modifications and ChromHMM annotations in HepG2 and (2) the transcription factors binding from ChIP-seq experiments from the ENCODE project with the coloring (light gray to dark gray) proportional to the signal strength observed in different cell lines (cell abbreviations can be found at: https://tinyurl.com/watv2v7). DPYD, dihydropyrimidine dehydrogenase; MS, mass spectrometry; RTP, mRNA-to-protein; scRNA-seq, single-cell RNA sequencing.
The enzyme dihydropyrimidine dehydrogenase (DPD) is encoded by the DPYD gene and represents the rate-limiting first step in the catabolism of pyrimidines in liver. Deficit in DPD is associated to missense alleles or alleles affecting splice sites, or sometimes alleles with unknown function. They increase the risk of toxicity in the form of severe bone marrow depression and neurotoxicity upon the use of the fluoropyrimidine chemotherapeutics fluorouracil (5-FU) or capecitabine, cancer drugs used in treatment of colon, rectum, stomach, breast, HCC, and other types of carcinomas. Analysis of these variants is now clinically implemented in several countries. In addition, over 30 single-nucleotide polymorphisms (SNPs), mainly missense or noncoding gene variants, have been identified in DPYD.
However, to date, the effect of these variants to the DPD enzymatic activity remains unclear. HiCap analysis identified 95 different distal regions interacting with probes at the DPYD promoter (Fig. 3D; Supplementary Table S9). Annotation of these distal regions using chromHMM data for liver tissue revealed six enhancer regions with different degrees of activity. The strongest one (Fig. 3E), located in the first intron of DPYD, shows transcription factors (TFs) binding sites for different TFs and the presence of the SNP rs74450569, a putative regulatory variant that may tune the gene expression and, in turn, contribute to the DPD enzyme levels and drug response.
The overall, yet not fully reached, goal is to identify the potential risk of toxicity upon fluoropyrimidine treatment based on the DPYD allelic makeup. Today, at least 14 different DPYD haplotypes have been reported from in vitro and clinical/ex vivo data (Whirl-Carrillo et al., 2012). rs74450569 has similar allele frequencies (D′ = 1) and is in linkage disequilibrium with the reported haplotype DPYD*HapB3 that contains three variants of unknown function associated to reduced DPD activity (rs75017182, rs56038477, and rs56276561 in Fig. 3E). Thus, it is possible that rs74450569 is the functional variant mediating the effect of DPYD*HapB3.
The regulatory element harboring rs74450569 contains several transcription factors binding sites (TFBSs) observed from ChIP-seq experiments in different cell lines in the ENCODE project (Dunham et al., 2012), including binding sites for FOSL2, CEBPB, JUN, and JUND in the HCC-derived cell line HepG2 (Fig. 3F).
Finally, the integration of snRNA-seq data allowed us “localizing” the expression of DPYD to both HCs and KCs (Fig. 3B), suggesting a novel nonparenchymal and macrophagic component in the pyrimidine catabolism.
Bulk proteomics data confirmed the expression of the DPD enzyme, although with a less predicted protein level than expected using RTP adjusted values of in silico bulk snRNA-seq (Fig. 3C). This trend was observed also for the other pharmacogenomics biomarkers and could suggest a high level of degradation of nuclear mRNA transcripts or rapid protein turnover. Further experimental validations with proteomics studies at single-cell level will be needed to precisely allocate the DPD protein level in HC and KC, and to verify the nonparenchymal contribution to the drug response.
A complex regulatory landscape for a glucose transporter linked to HCC
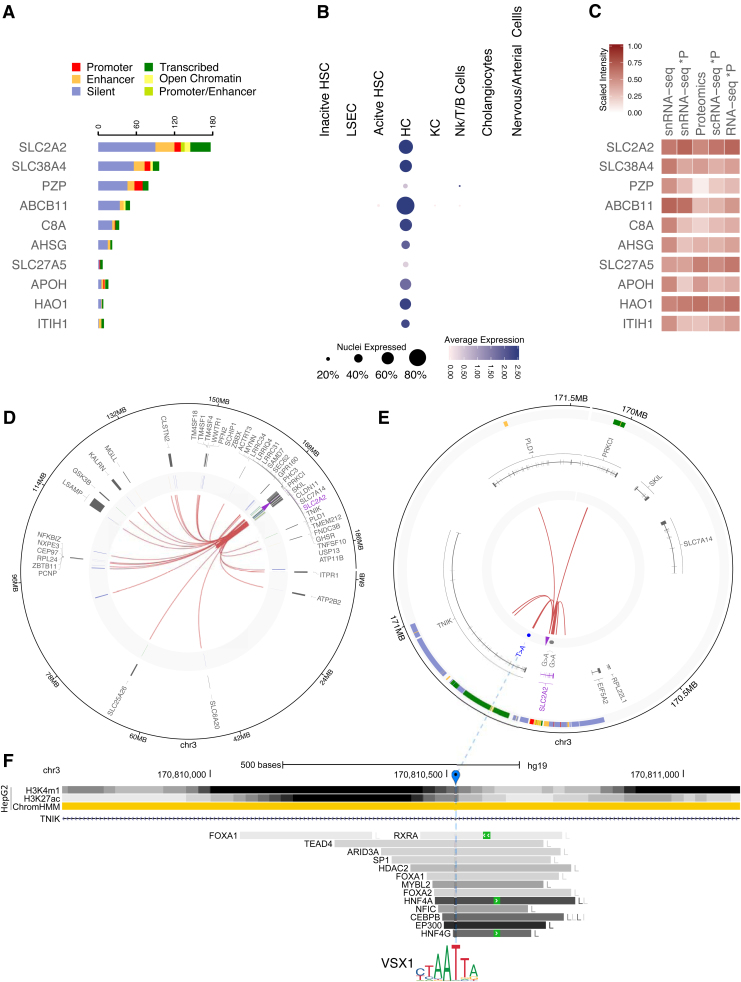
Collections of genes with biomarker characteristic are also available for HCC survival outcome. Li et al. (2018) curated a collection of 104 liver-enriched and prognostic genes in HCC. We focused on 27 of these 104 genes that we matched to the single nuclei transcriptomics data and for which we obtained also bulk HiCap and proteomics data (Figs. 1B and 4; Supplementary Table S12).
FIG. 4.
Overview of top 10 HCC genes with the largest number of significant probe-probe or probe-distal interactions detected in both proteomics and snRNA-seq. (A) Number of probe-distal (right panel) HiCap interactions associated to liver-specific chromHMM annotations. (B) Expression levels from snRNA-seq in different liver cell types. The size of the dot represents the number of nuclei that expresses the gene, while the color intensity the overall level of expression. (C) Heatmap comparing the expected and experimental levels of protein abundance from RNA-seq and MS proteomics experiments, respectively. The first column shows the log2-average expression of genes from the in silico bulk snRNA-seq, while the second one illustrates the estimated protein abundance calculated after calibrating the in silico bulk snRNA-seq levels for RTP abundancy estimation factors. The third column shows the experimental level of the protein abundance detected by MS. The last two columns show the estimated protein abundance calculated after calibrating the log2-average in silico bulk scRNA-seq levels and the log2-average number of reads of bulk RNA-seq. (D, E) Circos plots illustrating interactions of the probe for SLC2A2 with distal elements harboring active enhancer elements. (D) The first track shows gene annotations overlapping probe-distal interactions. The second track shows manually curated chromHMM annotations overlapping interacting regions, while the inner one shows the experimental HiCap interactions. The purple arrow marks the probe location. (E) A zoom-in on the area of interest for the circos plot in (D). The first two tracks show chromHMM and gene annotations, respectively. The third track shows HCC-specific mutations detected from the PanCancer consortium. The inner track shows 16 the probe-distal HiCap interactions overlapping active enhancer elements. The purple arrow marks the probe location. (F) The genomic landscape of the T > A motif-breaking mutation identified in (E) (marked in blue) located in one of the introns of the TNIK gene. The UCSC genome browser tracks represent from the top: (1) the ChIP-seq signals for two active enhancer-specific histone modifications and ChromHMM annotations in HepG2 and (2) the transcription factors binding from ChIP-seq experiments from the ENCODE project with the coloring (light gray to dark gray) proportional to the signal strength observed in different cell lines (cell abbreviations can be found at: https://tinyurl.com/watv2v7). HCC, hepatocellular carcinoma.
In total, we identified 469 distal HiCap interactions, of which 38 distal regions harbored active enhancers interacting with the promoters of 11 of the selected 27 genes (Supplementary Table S10). Six of these enhancers harbored at least one HCC-specific somatic noncoding mutation identified in the PanCancer Analysis of Whole Genomes consortium in 314 HCC cases (Supplementary Table S13) (Umer et al. article in preparation).
Not surprisingly, the vast majority of these essential genes associated to HCC survival, were highly expressed in HCs (Fig. 4B), in agreement with the established view that alterations of the balance of damage and regeneration of the liver parenchyma can result in fibrosis or cirrhosis, and eventually in the onset of HCC (Supplementary Fig. S7).
The top two HCC-related genes with most HiCap-reported interactions belong to the solute carrier (SLC) family of membrane transport proteins (Supplementary Fig. S7). One example is represented by novel putative distal regulatory elements identified for the SLC2A2 gene. SLC2A2 or GLUT2 is a glucose transporter historically associated to the glycogen storage disease type XI (also known as Fanconi-Bickel syndrome) where a nonfunctioning GLUT2 prevents the efflux of glucose from several tissues leading, among others, to glycogen accumulation, glucose and galactose intolerance, and fasting hypoglycemia (Santer et al., 1998).
In recent years, GLUT2 has been identified as a novel prognostic marker for HCC when it is overexpressed to meet the metabolic demands of proliferating cancer cells (Kim et al., 2017). HiCap probes designed at the TSS of SLC2A2 identified 143 interacting distal regions, including 16 chromHMM annotated enhancers, painting a rather complex regulatory landscape (Fig. 4D).
These distal elements (Fig. 4E; Supplementary Table S13) included seven enhancers located in introns 1, 3, 7, and 8 of the SLC2A2 gene and four enhancers in introns 9, 10/11, and 25 of the TNIK gene encoding a kinase involved in the Wnt signaling pathway. Nuclear expression of TNIK has been associated with poor HCC prognosis (Jin et al., 2014). HiCap data also revealed two intergenic enhancers and three enhancers located in introns of genes. One of the enhancers was located in intron 15 of the PLD1 gene, encoding for phospholipase D1 involved in cell proliferation and migration, and overexpressed in HCC (Xiao et al., 2016).
Another enhancer was harbored by intron six of PRKCI, a member of the protein kinase C family related to invasion and metastasis in HCC (Du et al., 2009). The third enhancer element was located in intron five of MGLL, a fatty acid metabolism enzyme downregulated in HCC, hence increasing HCC cell migration rate (Yang et al., 2018). Some of these enhancers showed liver-specific TFBS signals based on ENCODE data and three of them were defined by CTFC and RAD21 binding sites, suggesting a putative regulatory role through chromatin remodeling.
Not surprisingly, snRNA-seq analysis confirmed that the vast majority of HCC-related genes, including the ones harboring SLC2A2 enhancers, are primarily expressed in HCs (Tummala et al., 2017) (Fig. 4B). Bulk proteomics data confirmed the expression of SLC2A2 with a comparable RTP-predicted protein level than expected using RTP ratio-adjusted values of single nuclei transcriptomics (Fig. 4C). Predicted and observed protein levels for HCC genes did not follow a defined trend, with some genes showing lower (e.g., PZP, ACOT12, and ABCB11), similar (e.g., CYP8B1, FMO3, and APOB), or higher (e.g., SLC27A5, HAO1, and APOA1) protein levels than expected from adjustments of bulk RNA-seq data using RTP ratios (Supplementary Fig. S7). This suggests a cell- and gene-specific balance of mRNA degradation and protein turnover that calls for a validation at single-cell level.
Three SLC2A2 enhancer-harbored somatic mutations reported by the PanCancer consortium (Supplementary Table S13), in particular one somatic mutation in the enhancer located in the intron 25 of TNIK, have been reported as a motif breaker for the transcription factor VSX1, suggesting a putative noncoding, yet direct genetic role of this cancer mutation in the regulation of SLC2A2.
Further experimental validations editing and isolating these different putative regulatory elements are necessary to fully disentangle the genetic regulation of SLC2A2 in HCC.
Discussion
The analysis of transcriptome profiles from single nuclei today offers the possibility to identify cell types and states in biological samples difficult to dissociate or flash-frozen tissue samples with limited availability (Grindberg et al., 2013). Several studies have shown that a single nucleus displays an expression profile that is remarkably similar to one of the corresponding cells, with some expression bias toward transcripts of nuclear proteins or ncRNAs.
In this study, we performed snRNA-seq of a frozen human liver sample. The gene transcripts from liver nuclei were analyzed pre- and post-FACsorting to minimize the amount of cell debris from the tissue homogenization process. An accurate quality control did not reveal significant differences in the nuclei transcript quality, allowing us to pool the two preparations and obtain a collection of 4282 liver single nuclei transcriptomes (Supplementary Figs. S1–S3).
Nuclei clustering followed by differential expression analysis led to the identification of all the major liver cell types: HCs, endothelial cells, KCs, and HSCs both in quiescent and activated state. One of the advantages in the use of single cells/nuclei transcriptomics is the possibility to characterize and explore complex or rare cell populations.
Aside from the expected NPC types, we also identified a population of nuclei likely originated by resident immune cells, a distinct cluster of nuclei showing markers specific for cholangiocytes, and, perhaps more intriguingly, a small population of 79 nuclei exhibiting both nerve- and vascular-specific gene markers. While this finding must be validated in different liver samples, it is worth to remind that the liver innervation by the autonomic nervous system relies on the portal vasculature with both sympathetic and parasympathetic nerves wrapped around branches of the PV (posterior plexus) and HA (anterior plexus) (Jensen et al., 2013). The small cluster of nuclei we identified may represent a sample of this anatomical feature.
Two human liver cell atlases based on scRNA-seq have been recently released, revealing new subtypes of liver cells and providing a more detailed view of the organ design. (1) A study from Aizarani et al. (2019) on cryopreserved and freshly isolated human liver samples resulted in an atlas of about 10,000 single cells with a focus on epithelial liver cell progenitors. (2) A second study from Ramachandran et al. (2019) on liver cirrhosis at single-cell level performed on fresh liver biopsies, generated an atlas of liver-resident cells clustering more than 66,000 single cells into 21 populations with the spotlight on subpopulations of scar-associated macrophages and endothelial cells expanding in cirrhosis. Despite studying nuclei from a single liver sample, we were able to identify most of the major cell types reported in the aforementioned studies. However, as expected, the limited number of total nuclei and the confined view of the transcriptome coming from the analysis of nuclei and not whole cells prevented us from describing nuances such as zonation of cell types or rare subsets of immune cell populations.
The differential expression of genes in different cell populations stems from a precise genetic regulation with one or several distal elements affecting the transcriptome profile of each single nucleus. We used HiCap interaction data generated from the same liver tissue sample in bulk to identify putative regulatory elements modulating the expression of a set of liver-specific genes associated to drug metabolism and as prognostic biomarkers in patients with HCC. We presented two examples highlighting the potential of an integrative multi-omics approach in untangling some of the regulatory mechanisms behind the pharmacogenomics of fluoropyrimidine toxicity (DPYD) and the role of the glucose transporter GLUT2 (SLC2A2) in HCC.
HiCap data pinpointed a series of liver-specific enhancers that interact with the promoters of DPYD and SLC2A2.
SNPs located in DPYD-identified enhancers could affect the individual response to drug toxicity. For example, the regulatory element harboring rs74450569 presents several TF binding sites, among others, FOSL2 and JUN that are regulated by the ribosomal protein L34 (RPL34) through MAPK and p53 signaling. Silencing of RPL34 has been shown to inhibit tumor growth and proliferation, and simultaneously, to upregulate the expression of several TFs, including FOSL2 and JUN (Wei et al., 2016). A working hypothesis is that this may, in turn, increase the enhancer activity of the regulatory element harboring rs74450569 leading to an increase in DPYD activity and reduced toxicity.
At the same time, somatic mutations reported from the PanCancer consortium and harbored in SLC2A2-identified enhancers could shed more light in the HCC progression. While HiCap experiments offer a new level of resolution in the study of long-range 3D genomic interactions when compared to other Hi-C-derived techniques, they require further experimental validation to confirm the effect of distal element on the studied genes.
When it comes to the protein abundance in a cell, the mRNA translation and protein degradation processes are as important as the mRNA transcription and stability (Vogel et al., 2010). The nature of the mRNA also influences the correlation between mRNA and protein level: a relatively good correlation is expected for housekeeping genes that show stable mRNA and proteins opposed to TFs, signaling genes, or genes with cell cycle-specific functions that show unstable mRNA and proteins, and hence a poor correlation (Schwanhäusser et al., 2011).
An additional layer of complexity in trying to predict the protein level from the mRNA level is represented by post-translational modification with hundreds of ubiquitination enzymes, serine-threonine kinases, tyrosine kinases, and phosphatase dramatically altering the protein half-lives. Recently, it has been proposed that a better correlation could be achieved using a tissue-independent and gene-specific mRNA-to-protein conversion factor, which could enhance the predictability of protein copy numbers from RNA levels.
Conclusions
The integration of single nuclei transcripts with bulk HiCap and proteomics is a complex, heavily constrained multi-omics challenge. To overcome this, we focused on specific groups of genes that are of high importance to the liver. After exploring these gene sets in the single nuclei transcriptomics space, we focused on genes that showed interesting pharmacogenomics or HCC-related biomarker patterns. We explored these patterns through data generated in bulk, and we proposed potential genetic mechanisms that might affect the regulation of genes such as DPYD and SLC2A2, for instance by assigning specific promoter-distal interactions potentially affecting protein levels for genes expressed in distinct cell populations.
Supplementary Material
Acknowledgments
Sequencing was performed by the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX). We would like to thank Dr. Dirk Pacholsky at the BioVis Platform of Uppsala University for assistance in Flow Cytometry analysis.
Abbreviations Used
- CV
central vein
- DAPI
4′,6-diamidino-2-phenylindole
- DPYD
dihydropyrimidine dehydrogenase
- ENA
European Nucleotide Archive
- FACS
fluorescence-activated cell sorting
- FDR
false discovery rate
- HA
hepatic artery
- HC
hepatocyte
- HCC
hepatocellular carcinoma
- HSC
hepatic stellate cell
- KC
Kupffer cell
- LSEC
lined by specialized sinusoidal endothelial cell
- MNN
mutual nearest neighbors
- MS
mass spectrometry
- NGI
National Genomics Infrastructure
- NGS
next-generation sequencing
- NPC
nonparenchymal cell
- PBS
phosphate-buffered saline
- PC
parenchymal cell
- PV
portal vein
- PRIDE
PRoteomics IDEntification Database
- RPL34
ribosomal protein L34
- RTP
mRNA-to-protein
- SLC
solute carrier
- scRNA-seq
single-cell RNA sequencing
- SNP
single-nucleotide polymorphism
- SDS
sodium dodecyl sulfate
- snRNA-seq
single-nuclei RNA-seq
- TF
transcription factor
- TFBS
transcription factors binding site
- t-SNE
t-distributed stochastic neighbor embedding
- UMI
unique molecular identifier
- UPPMAX
Uppsala Multidisciplinary Center for Advanced Computational Science
Author Disclosure Statement
C.K. is an employee of AstraZeneca. The other authors declare no financial conflicts of interest.
Funding Information
The study was supported by grants from SciLifeLab (C.W.), AstraZeneca (C.W. and J.K.), the Swedish Diabetes Foundation (DIA 2017-269) (C.W.), EXODIAB (C.W.), the Swedish Cancer Foundation (CAN 2015/759, 2018/849) (C.W.), The National Science Centre (DEC-2015/16/W/NZ2/00314) (J.K.), Institute of Computer Science, Polish Academy of Sciences (J.K.), the eSSence program (J.K.), the Swedish Reseach Council (Vetenskapsrådet, grant no: 78081) (P.S.), and The Borgströms-Hedströms foundation (M.C.).
Supplementary Material
References
- Aebersold R, and Mann M (2003). Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- Aizarani N, Saviano A, Sagar, et al. (2019). A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572, 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åkerborg Ö, Spalinskas R, Pradhananga S, et al. (2019). High-resolution regulatory maps connect vascular risk variants to disease-related pathways. Circ Genom Precis Med 12, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelidis I, Simon LM, Fernandez IE, et al. (2019). An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat Commun 10, 963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anil A, Spalinskas R, Åkerborg Ö, and Sahlén P (2017). HiCapTools: A software suite for probe design and proximity detection for targeted chromosome conformation capture applications. Bioinformatics 34, 675–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arii S, and Imamura M (2000). Physiological role of sinusoidal endothelial cells and Kupffer cells and their implication in the pathogenesis of liver injury. J Hepatobiliary Pancreat Surg 7, 40–48 [DOI] [PubMed] [Google Scholar]
- Benjannet S, Rhainds D, Hamelin J, Nassoury N, and Seidah NG (2006). The proprotein convertase (PC) PCSK9 is inactivated by furin and/or PC5/6A: Functional consequences of natural mutations and post-translational modifications. J Biol Chem 281, 30561–30572 [DOI] [PubMed] [Google Scholar]
- Boers W, Aarrass S, Linthorst C, Pinzani M, Elferink RO, and Bosma P (2006). Transcriptional profiling reveals novel markers of liver fibrogenesis: Gremlin and insulin-like growth factor-binding proteins. J Biol Chem 281, 16289–16295 [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, and Mann M (2011). Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- Deng D, Sun PC, Yan CY, et al. (2015). Molecular basis of ligand recognition and transport by glucose transporters. Nature 526, 391–396 [DOI] [PubMed] [Google Scholar]
- Dryden NH, Broome LR, Dudbridge F, et al. (2014). Unbiased analysis of potential targets of breast cancer susceptibility loci by Capture Hi-C. Genome Res 24, 1854–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G-S, Wang J-M, Lu J-X, et al. (2009). Expression of P-aPKC-ι, E-cadherin, and β-catenin related to invasion and metastasis in hepatocellular carcinoma. Ann Surg Oncol 16, 1578–1586 [DOI] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, et al. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edfors F, Danielsson F, Hallström BM, et al. (2016). Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol Syst Biol 12, 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, and Kellis M (2012). ChromHMM: Automating chromatin-state discovery and characterization. Nat Methods 9, 215–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, and Kellis M (2017). Chromatin-state discovery and genome annotation with ChromHMM. Nat Protoc 12, 2478–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Kim C, Sei E, et al. (2017). Nanogrid single-nucleus RNA sequencing reveals phenotypic diversity in breast cancer. Nat Commun 8, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths JA, Richard AC, Bach K, Lun ATL, and Marioni JC (2018). Detection and removal of barcode swapping in single-cell RNA-seq data. Nat Commun 9, 2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindberg RV, Yee-Greenbaum JL, McConnell MJ, et al. (2013). RNA-sequencing from single nuclei. Proc Natl Acad Sci 110, 19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Avraham-Davidi I, Basu A, et al. (2017). Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 14, 955–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Lun ATL, Morgan MD, and Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 36, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern KB, Shenhav R, Massalha H, et al. (2018). Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat Biotechnol 36, 962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern KB, Shenhav R, Matcovitch-Natan O, et al. (2017). Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang R, Zhou Y, et al. (2018). Mapping the mouse cell atlas by Microwell-seq. Cell 172, 1091.e17–1107.e17 [DOI] [PubMed] [Google Scholar]
- Hwang B, Lee JH, and Bang D (2018). Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med 50, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger R, Migliorini G, Henrion M, et al. (2015). Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci. Nat Commun 6, 6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KJ, Alpini G, and Glaser S (2013). Hepatic nervous system and neurobiology of the liver. Compr Physiol 3, 655–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Jung HY, Wang Y, et al. (2014). Nuclear expression of phosphorylated TRAF2- and NCK-interacting kinase in hepatocellular carcinoma is associated with poor prognosis. Pathol Res Pract 210, 621–627 [DOI] [PubMed] [Google Scholar]
- Kim YH, Jeong DC, Pak K, et al. (2017). SLC2A2 (GLUT2) as a novel prognostic factor for hepatocellular carcinoma. Oncotarget 8, 68381–68392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec Z. (2001). Cooperation of Liver Cells in Health and Disease: With 18 Tables. Heidelberg, Germany: Springer Berlin Heidelberg; [DOI] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dorrell C, Canaday PS, et al. (2017). Adult mouse liver contains two distinct populations of cholangiocytes. Stem Cell Rep 9, 478–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Xu T, Liu C, et al. (2018). Liver-enriched genes are associated with the prognosis of patients with hepatocellular carcinoma. Sci Rep 8, 11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, et al. (2013). The genotype-tissue expression (GTEx) project. Nat Genet 45, 580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL, McCarthy DJ, and Marioni JC (2016). A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5, 2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL, Riesenfeld S, Andrews T, Dao TP, Gomes T, and Marioni JC (2018). Distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. bioRxiv, 234872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacParland SA, Liu JC, Ma X-Z, et al. (2018). Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 9, 4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T, Güell M, and Serrano L (2009). Correlation of mRNA and protein in complex biological samples. FEBS Lett 583, 3966–3973 [DOI] [PubMed] [Google Scholar]
- Mannaerts I, Schroyen B, Verhulst S, et al. (2013). Gene expression profiling of early hepatic stellate cell activation reveals a role for Igfbp3 in cell migration. PLoS One 8, e84071-e [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Campbell KR, Lun ATL, and Wills QF (2017). Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics (Oxford, England) 33, 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulana A, Scanteianu A, Jones D, Stern AD, Bouhaddou M, and Birtwistle MR (2018). Gene-specific predictability of protein levels from mRNA data in humans. bioRxiv, 399816 [Google Scholar]
- Nguyen QH, Pervolarakis N, Nee K, and Kessenbrock K (2018). Experimental considerations for single-cell RNA sequencing approaches. Front Cell Dev Biol 6, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki S, Ohta T, Shioi G, et al. (2018). ChIP-Atlas: A data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep 19, e46255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, Dobie R, Wilson-Kanamori JR, et al. (2019). Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlén P, Abdullayev I, Ramsköld D, et al. (2015). Genome-wide mapping of promoter-anchored interactions with close to single-enhancer resolution. Genome Biol 16, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santer R, Schneppenheim R, Suter D, Schaub J, and Steinmann B (1998). Fanconi-Bickel syndrome—The original patient and his natural history, historical steps leading to the primary defect, and a review of the literature. Eur J Pediatr 157, 783–797 [DOI] [PubMed] [Google Scholar]
- Sato K, Marzioni M, Meng F, Francis H, Glaser S, and Alpini G (2019). Ductular reaction in liver diseases: Pathological mechanisms and translational significances. Hepatology 69, 420–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, et al. (2011). Global quantification of mammalian gene expression control. Nature 473, 337. [DOI] [PubMed] [Google Scholar]
- Shetty S, Lalor PF, and Adams DH (2018). Liver sinusoidal endothelial cells—Gatekeepers of hepatic immunity. Nat Rev Gastroenterol Hepatol 15, 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht H, and Slavov N (2018). Transformative opportunities for single-cell proteomics. J Proteome Res 17, 2565–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, et al. (2018). Comprehensive integration of single cell data. bioRxiv, 460147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummala KS, Brandt M, Teijeiro A, et al. (2017). Hepatocellular carcinomas originate predominantly from hepatocytes and benign lesions from hepatic progenitor cells. Cell Rep 19, 584–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Abreu RdS, Ko D, et al. (2010). Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 6, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Ding L, Wei Z, et al. (2016). Ribosomal protein L34 promotes the proliferation, invasion and metastasis of pancreatic cancer cells. Oncotarget 7, 85259–85272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. (2012). Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 92, 414–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Sun Q, Bei Y, et al. (2016). Therapeutic inhibition of phospholipase D1 suppresses hepatocellular carcinoma. Clin Sci 130, 1125. [DOI] [PubMed] [Google Scholar]
- Yang X, Zhang D, Liu S, Li X, Hu W, and Han C (2018). KLF4 suppresses the migration of hepatocellular carcinoma by transcriptionally upregulating monoglyceride lipase. Am J Cancer Res 8, 1019–1029 [PMC free article] [PubMed] [Google Scholar]
- Yi C-X, la Fleur SE, Fliers E, and Kalsbeek A (2010). The role of the autonomic nervous liver innervation in the control of energy metabolism. Biochim Biophys Acta 1802, 416–431 [DOI] [PubMed] [Google Scholar]
- Zhang DY, Goossens N, Guo J, et al. (2016). A hepatic stellate cell gene expression signature associated with outcomes in hepatitis C cirrhosis and hepatocellular carcinoma after curative resection. Gut 65, 1754–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are presented within the article and its supporting information files. Access to the ENA (European Nucleotide Archive) database, which will host the raw sequencing data, and PRIDE (PRoteomics IDEntification Database), which will host the raw MS proteomics data, will be available at the EBI BioStudies database under the accession number S-BSST324. These data will be also available from the corresponding authors upon request.