Graphical abstract

Highlights
-
•
Simian foamy viruses (SFV) are transmitted to humans after contact, mainly bites, with infected monkeys and apes.
-
•
Contexts of transmission include mainly hunting activities and monkeys’ sympatry.
-
•
In humans, active immune response probably explains SFV latency in blood and saliva.
-
•
It is a model of restriction of retroviral emergence after cross-species transmission.
Abstract
Most viral pathogens that have emerged in humans have originated from various animal species. Emergence is a multistep process involving an initial spill-over of the infectious agent into single individuals and its subsequent dissemination into the human population. Similar to simian immunodeficiency viruses and simian T lymphotropic viruses, simian foamy viruses (SFV) are retroviruses that are widespread among non-human primates and can be transmitted to humans, giving rise to a persistent infection, which seems to be controlled in the case of SFV. In this review, we present current data on the discovery, cross-species transmission, and molecular evolution of SFV in human populations initially infected and thus at risk for zoonotic emergence.
Current Opinion in Virology 2015, 10:47–55
This review comes from a themed issue on Emerging viruses: interspecies transmission
Edited by Antoine Gessain and Fernando Garcia-Arenal
For a complete overview see the Issue and the Editorial
Available online 17th February 2015
http://dx.doi.org/10.1016/j.coviro.2014.12.003
1879-6257/© 2014 Elsevier B.V. All rights reserved.
Most viral pathogens that have emerged in humans during the last century are thought to have originated from various animal species and are thus of zoonotic origin [1]. While the modes of viral dissemination within the human population are generally well characterized, the initial steps at the interspecies interface that lead to viral emergence remain poorly understood. Epidemiological field studies conducted in certain specific high-risk populations are thus crucial to obtain new insights into these early events of the emergence process.
Human infections by simian viruses represent an increasing public health concern. Indeed, non-human primates (NHPs) are considered to be the likely sources of viruses that infect humans and thus may pose a significant threat to human population [2]. This is well illustrated by some retroviruses, which have the ability to cross-species, possibly adapt to a new host and sometimes spread within this new species. It is now clear that the emergence of human immunodeficiency virus type 1 (HIV-1) and HIV-2 in humans has resulted from several independent interspecies transmissions. Different SIV types from chimpanzees and gorillas in the western part of Central Africa as well as sooty mangabeys in West Africa, gave rise to HIV-1 and HIV-2 respectively, probably during the first part of the last century [2]. Similarly, the origin of most Human T cell Lymphotropic virus type 1 (HTLV-1) subtypes appears to be linked to interspecies transmission between STLV-1-infected monkeys and humans, followed by variable periods of evolution in the human host [2]. In this brief review, we will present the current available data on the discovery, cross-species transmission and molecular evolution of the simian foamy viruses (SFV) present in different human populations at risk for zoonotic emergence.
Simian foamy viruses in humans
Since the initial description of foamy virus (FV) in rhesus monkey kidney cells in 1954 [3], such viruses have been isolated from several animal species, including numerous NHP species. The prevalence of FVs in naturally infected animals is generally high, but varies widely according to species [2]. Among NHP populations, SFV seroprevalence can reach up to 75–100% in adults [2]. In African green monkey and macaques, oral mucosa tissue is an important site for viral replication, explaining why foamy viral RNA is found at a high concentration in the saliva of such primates [4••]. Saliva-based means of transmission, such as bites, have been thus strongly suggested. Infection by SFV itself does not seem to cause any disease in infected NHPs, but studies have not been conducted to address this question specifically. However, when considering co-infections in macaques, SFV can increase the pathogenicity of simian immunodeficiency virus [5].
In 1971, the first human foamy virus was isolated from the cell culture of a Kenyan patient suffering of nasopharyngeal carcinoma [6]. Phylogenetic studies demonstrated that this virus originated from the East African chimpanzee subspecies (Pan troglodytes schweinfurtii). This strain is now considered to be the ‘prototype foamy virus’ (PFV). However, its exact origin remains unclear (in vivo cross-species transmission from a chimpanzee to the African patient, or cell culture contamination) [7]. In the 1970/80s, several papers showed conflicting results on the presence of SFV in human populations and in different patients [8, 9]. These findings reflected the high percentage of non-specific serological reactivity and the lack of specific confirmatory tests at that time.
In 1995, based on specific serological and molecular assays, the first clear evidence of SFV in Humans was reported among 3 laboratory and monkey house personnel [10••]. Since then, other groups have reported similar findings in a series of workers occupationally exposed to NHPs in the USA and Canada and more recently in personnel from zoos or primate centers in Gabon and China [8]. Infection by SFV in a more natural setting was then demonstrated in villagers from Cameroon [11]. They were mostly hunters who reported direct contacts with blood and/or body fluids from wild NHPs. We extended such studies into different areas and populations of this Central African country and found the presence of SFV infection in at least 50 persons [12••]. The great majority of them (Bantus or Pygmies) were men who had been bitten by an ape (mostly gorillas but also chimpanzees) or a small monkey (mostly Cercopithecus nictitans) during hunting activities. A recent report from Gabon confirmed such frequent cross-species transmission in hunters after severe bites from mostly gorilla [13]. Infected women were also recently found in the Democratic Republic of Congo [14]. In South and Southeast Asian countries, SFV zoonotic infection has also been detected in various contexts of interspecies contact including ‘monkey temple’ workers, pet owners and people living around free-ranging macaques in South and Southeast Asia [15]. It is interesting to note that the Human/NHP interface in Asia differs greatly from that in Africa [16]. Human and macaques sympatry in Southeast Asia dates back as far as 25,000 years. Human-macaque commensalism is frequent in many monkey temples of these regions each year putting a very large number of persons, including tourists, at risk for macaques bites [17]. Indeed, human subjects in some South Asian countries came into contact with rhesus macaques mostly in the context of their daily lives, sharing a geographical area that is the ‘home range’ of both primate populations [16]. Such frequent interactions can lead to possible viral interspecies transmission. The situation is very different in central Africa where bush-meat hunting (including Apes such as gorillas and chimpanzees) is the major risk factor associated with SFV interspecies transmission [12••]. Furthermore, in Central Africa, the number of contacts between humans (mostly hunters and their wives and butchers) and potentially infected NHPs has probably greatly increased during the last century. This is believed to be due to increased hunting activities, resulting from a combination of urban demand for bush-meat, greater access to NHP habitats provided in part by logging roads, easier accessibility to fire arms, an increase in populations living in forest areas, and the associated increase in local food needs [2].
While nowadays, Humans are not considered to be a natural host of SFV, more than 100 cases of SFV infection have been reported so far in individuals in close contact with NHPs [8]. Among them, no specific pathology has been yet demonstrated. However, the selection bias inherent in the enrollment of healthy persons in the very few performed studies greatly limits the current ability to identify any potential associated disease.
SFV genomic organization and structural proteins
The SFV genome comprises the retroviral gag, pol and env genes, and two regulatory genes tas and bet. An internal promoter allows basal transcription of tas and bet (Figure 1 ). The transactivator Tas then activates a second promoter located in the long-terminal repeat, which induces the synthesis of the Gag, Pol and Env structural proteins. SFVs can also go through a late reverse transcription step, before the release of SFV particles. Thus, SFV particles can contain SFV RNA and SFV DNA genomes (Figure 2A) [18].
Figure 1.

Schematic representation of SFV genomic organization (chimpanzee strain). The SFV genome is flanked by two long terminal repeat (LTRs) which contain the unique 3′ (U3), repeated (R) and unique 5′ (U5) regions. gag encodes the full-length gag protein (74 kDa) and the shorter p70 protein. pol encodes the protease (PR)-reverse transcriptase (RT)-Rnase H protein and the integrase (INT). env encodes the leader peptide (LP), the surface glycoprotein (SU) and the transmembrane protein (TM). Two additional genes tas and bet encode proteins having regulatory functions. The transactivator Tas binds to the 5’LTR which activates the transcription of the structural genes gag, pol and env.
Figure 2.

Mechanisms of action of antiviral factors against FV. (A) SFV life cycle starts with infection of a new cell by SFV particles containing RNA and/or DNA, decapsidation of the viral genome, reverse transcription of RNA genomes and integration of viral DNA into cellular DNA. Cellular activation of the internal promoter induces tas and bet transcription. The transactivator Tas then activates a second promoter located in the long-terminal repeat, which induces the synthesis of the Gag, Pol and Env structural proteins. After assembling, viral particles bud essentially from the endoplasmic reticulum, with a late reverse-transcription step giving rise to both DNA and RNA particles. (B) SFV is detected by plasmacytoid dendritic cells (not exclusively) which triggers IFN-I production. Several antiviral factors that can be induced by IFN-I are active against SFV. TRIM5α are antiviral proteins that prevent SFV decapsidation [50]. Apolipoprotein B-editing catalytic polypeptide-like subunit (APOBEC) enzymes act on the negative strand DNA produced by the viral reverse transcriptase, resulting in G-to-A mutations in the viral genome, with potential deleterious consequences. FV Bet protein has been shown to partially prevent APOBEC action [40, 41, 42, 43]. N-Myc interactor (NMI) as well as a member of the interferon-induced protein (IFP) family, IFP35, can inhibit the replication of the prototype and the bovine FV respectively, by direct interaction with the viral transactivator Tas [46, 47]. Tetherin is known to block many different types of enveloped viruses by tethering the budding virus at the cell membrane and is also efficient to prevent FV release [48, 49].
SFV tropism
In vitro, SFV can infect most cell types. Heparan sulfates represent attachment factors [19]. However, an SFV receptor has not yet been discovered and is probably ubiquitous. Once productively infected, cells usually form syncitia with a ‘foam-like’ cytopathic effect, before cell death occurs (Figure 3 , A to D). However, some cell lines as well as primary cells of the myeloid or lymphoid lineage can remain chronically infected [20, 21]. In NHPs, SFV is latent in blood cells and replicates in the superficial epithelial layer of the oral mucosa [4••, 22], which explains the mode of transmission of SFV to humans, mainly through bites [12••]. Viral genomes in the buccal swab usually reflects those found in the blood [23]. In humans, SFV is also present in PBMCs and saliva. However, no active replication, as monitored by SFV RNA, has been detected in the PBMCs nor in the saliva [24].
Figure 3.
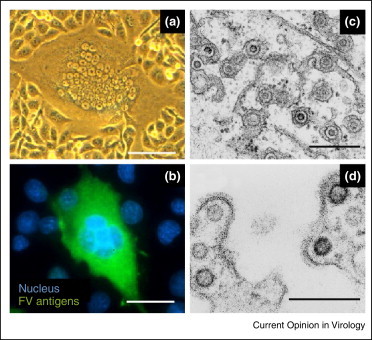
Observation of SFV and SFV-infected cells. BHK-21 cells were infected with a chimpanzee SFV strain isolated from an SFV-infected hunter [31]. A large syncytium with a ‘foamy’ aspect is visible using light microscopy (A). An immunofluorescent staining was performed as previously described [62]. SFV antigens are revealed in the green channel and nuclei are stained with DAPI (B). U87MG cells were infected by SFV-1 and used for electron microscopy (C,D). Scale bars represent 10 μm (A), 3 μm (B) or 300 nm (C,D).
In the blood, the cellular targets of SFV remain poorly studied. Preliminary studies, performed mostly in small series of chimpanzees, African green monkeys, and in few humans, have shown SFV DNA in lymphocytes, especially CD8+ T cells [25, 26]. By studying a larger cohort of humans infected with a gorilla strain of SFV in Cameroon, we demonstrated that CD8+, CD4+ T cells and CD19+ B lymphocytes were preferentially infected, compared to monocytes and NK cells (Table 1 ). SFV DNA was detected in both memory and naive CD4+ T lymphocytes and SFV DNA levels in CD4+ T cells were positively correlated with the duration of the infection [27].
Table 1.
SFV tropism and viral load in the blood of SFV-infected humans and NHPs. The proportion of SFV DNA positive samples among leukocyte populations in SFV-infected NHP and SFV-infected humans is indicated.
| Host | Infecting strain | Read-out for SFV DNA detection | Quantification method | SFV DNA detection and load in sorted white blood cell populations from SFV-infected individuals |
Refs | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CD8+ T lymphocytes | CD4+ T lymphocytes | CD19+ B lymphocytes | CD14+ monocytes | CD56+ NK lymphocytes | Granulocytes | |||||
| AGM | SFV-3 | Nb. of SFV DNA + samples/ind. tested | 9/9 | 1/3 | NTa | 2/7 | NTa | 1/7 | [26] | |
| SFV DNA copies/number of cells | Semi-quant. PCR (serial dilutions) | ≈1–100 SFV DNA/105 cells | ≈10 SFV DNA/105 cells | NTa | ≈1 SFV DNA/105 cells | NTa | ≈1 SFV DNA/105 cells | [26] | ||
| CPZ | SFV-6 | Nb. of SFV DNA + samples/ind. tested | 4/4 | 3/4 | NTa | 1/4 | NTa | 3/4 | [26] | |
| SFV DNA copies/number of cells | Semi-quant. PCR (serial dilutions) | ≈1 SFV DNA/105 cells | ≈1–10 SFV DNA/105 cells | NTa | ≈1 SFV DNA/105 cells | NTa | ≈1 SFV DNA/105 cells | [26] | ||
| Hum. | SFV-3 or HFV | Nb. of SFV DNA + samples/ind. tested | 2/2 | 0/2 | NTa | 0/2 | NTa | 0/2 | [26] | |
| SFV DNA copies/number of cells | Semi-quant. PCR (serial dilutions) | ≈1–10 SFV DNA/105 cells | NA | NTa | NA | NTa | NA | [26] | ||
| Hum. | SFV-HU1 (SFV-3) | Nb. of SFV DNA + samples/ind. tested | 0/1 | 0/1 | 1/1 | 1/1 | NT | NT | [25] | |
| SFV DNA copies/number of cells | Non-quant. PCR (one dilution) | NA | NA | >70 SFV DNA/105 cellsb | >70 SFV DNA/105 cellsb | NA | NA | [25] | ||
| Hum. | SFVgor | Nb of SFV DNA + samples/ind. tested | 10/11 | 9/11 | 7/11 | 2/11 | 1/11 | NT | [27] | |
| SFV DNA copies/number of cells | Quantitative PCR | 6–274 SFV DNA/105 cells | 6–61 SFV DNA/105 cells | 10–91 SFV DNA/105 cells | 4–5 SFV DNA/105 cells | 5 SFV DNA/105 cells | NA | [27] | ||
AGM: African green monkey; CPZ: chimpanzee; Hum.: Human; NT: Not tested; Nb.: Number; Ind.: Individuals; NA: Not applicable. SFV-HU1 is a strain close to SFV-3.
In the study from Von Laer et al. [26], the population tested is actually non-CD4+ non-CD8+ lymphocytes, and is thus likely enriched in both CD19+ B and CD56+ NK lymphocytes. The proportion of positive samples are 7/9 AGM, 3/4 CPZ and 0/2 humans. SFV DNA loads in this sorted cell population is ≈1/105 cells (in the 7/9 positive samples from AGM) and ≈1/105 cells (in the 3/4 samples from CPZ).
In the study from Callahan et al. [25], only one dilution of leucocytes DNA was shown, corresponding to a viral load of >70 SFV DNA copies/105 cells, assuming the usual sensitivity of 1 SFV DNA copies/105 cells.
SFV genetic stability
SFV genomes display a high evolutionary conservation among all the species infected and SFV genetic variability within one infected animal is very low over time (<1% variation over 13 years) [28]. This genetic stability might be explained by the long co-evolution with their host [29•] and their subsequent efficient adaptation. SFV genetic stability could also prevent or delay infection of new hosts as molecular changes may be required to infect cells from a different species and/or evade host-specific immune responses [30]. Genetic stability was also found in SFV-infected humans, with less than 1% divergence in the nucleotide sequence over several years [13, 24]. Several groups also reported genetic modifications including mutations in the Bet accessory gene [25, 31], deletion in the U3 sequence of the LTR which improves FV replication in vitro [31, 32] and deletions in the Tas sequence, which might be linked with the chronicity of the infection [31]. However, most of these modifications are not a unifying trait of zoonotic strains, and can also be found in NHP or experimentally infected animals [31]. This suggests that viral strains remain mainly stable, even decades after cross-species transmission. This is probably related to the apparent low level of SFV replication in infected humans, which could be linked with their efficient immune control.
In vivo super-infection and recombination of SFV
Some groups have reported cases of co-infection by two different SFVs in apes, monkeys, as well as in humans. Some chimpanzees infected by SFVcpz strains were co-infected with SFV from colobus monkeys [33], or cercopithecus monkeys [34]. More frequently, chimpanzees and macaques can harbor distinguishable SFVcpz and SFVmac strains respectively, likely the result of multiple independent infections that accumulate with age [16, 35, 36]. SFVcpz infection appears to first occur via vertical transmission but is followed in adult life by the acquisition of further infections, possibly stemming from aggressive interactions [35]. In some of these studies, as well as in a recent one, some SFV appears to be recombinant strains [34, 36, 37]. We, and others, have found different clones of SFV in single individuals [12••, 16, 24], suggesting a co-infection with multiple SFV strains. Whether they were acquired at a single time or through several contacts with NHP is not known.
Immune control of SFV
Innate restriction of SFV
Interferons are potent antiviral molecules and usually represent one of the first lines of defense to pathogens. In vitro, SFVs were first thought to be low interferon-inducers [38] but this actually depends on the cell type. Indeed, we found that SFVs are efficiently sensed by human hematopoietic cells and induce the production of high levels of IFN-I [39]. The main producers of IFN-I in the blood are the plasmacytoid dendritic cells, which detect SFV genome after uptake of SFV particles, through the endosomal toll-like receptor 7 [39]. In culture systems, the addition of type I IFN impairs SFV replication, suggesting that interferon-inducible genes might be involved in the control of SFV replication (Figure 2A and B):
-
-
Apolipoprotein B-editing catalytic polypeptide-like subunit (APOBEC) enzymes are a family of antiviral cytidine deaminases that act on the negative strand DNA produced by the viral reverse transcriptase, resulting in G-to-A mutations in the viral genome, with potential deleterious consequences. However, viruses often produce proteins that counteract cellular defense systems. FV Bet protein has been shown to prevent APOBEC action [40, 41, 42, 43]. Despite the action of Bet, it was reported that SFV genomes found in humans displayed some G-to-A mutations [24, 44]. Matsen et al. found that the frequence and the type of hypermutations were different in humans and macaques, with a majority of hypermutations leading to stop codons in humans, suggesting active restriction of the infecting strain in humans and not only passive acquisition of previously mutated strain [45•].
Additional studies have been performed in vitro to elucidate the mechanisms involved in the control of SFV replication, although their relevance in vivo has not been tested so far:
-
-
Two homologous proteins, N-Myc interactor (NMI) as well as a member of the interferon-induced protein (IFP) family, IFP35, can inhibit the replication of PFV and BFV, respectively, by direct interaction with the viral transactivator Tas [46, 47].
-
-
Tetherin is known to block many different types of enveloped viruses by tethering the budding virus at the cell membrane and is also efficient to prevent FV release [48, 49].
-
-
TRIM5α are antiviral proteins that prevent viral decapsidation [50]. They act in a species-specific manner: PFV and SFVmac are restricted by TRIM5α from most New World monkeys, but not from other primates including humans. This indicates that TRIM5α is probably not involved in the restriction of SFV after cross-species transmission from chimpanzees and macaques to humans, at least nowadays [51].
Of note, some known antiretroviral factors are not potent against SFV. For instance, as reverse transcription occurs predominantly before entry into target cell, these viruses are resistant to the dNTP levels reduction by SAMHD1 [52].
Adaptative restriction of SFV
Few studies have addressed the question of adaptative immune response to SFV.
First, neutralizing antibodies present in the serum of infected animals inhibit SFV transmission and infection in rhesus macaques [53], indicating a role of the adaptive immune response in the control of SFV in vivo. This is further supported by the fact that antibodies titers in feces of chimpanzees infected with SFV are inversely correlated with SFV viral load [34]. However, very few data are available concerning the adaptative immune response to SFV in humans. Antibodies against SFV are detected in the blood of infected individuals and were also found in saliva and urine samples in four SFV-infected individuals tested. However, antibody titers were lower than in infected chimpanzees, which could be linked to lower levels of SFV replication in humans [54]. Neutralization of SFV by sera from three SFVagm-infected individuals was reported in 1983 and 1997 [55, 56].
Secondly, neutralization of IFN-gamma in activated PBMCs infected with SFV, increases viral expression [57] and SFV up-regulates MHC-I in vitro [58], suggesting that adaptative immunity might be elicited and efficient in the control of SFV in infected individuals. Although virus-specific T lymphocytes are key effectors that control the outcome of HTLV and HIV infection [59], no report has addressed their presence in SFV-infected NHPs or humans.
Conclusion
Human infection by SFV constitutes a unique natural model to study the various parameters involved in restriction of retroviral emergence. The future of SFV emergence in humans remains an open question. However, the current increase in NHP-to-human contacts (in part due to hunting activities and animal handling) may favor such cross-species transmission [2]. Furthermore, other factors such as SFV co-infection with HIV, already reported in Cameroon [60], may increase the risk of SFV pathogenicity and will require further investigation. Nevertheless, any prediction concerning viral emergence remains very difficult and hazardous, as recently exemplified by Ebola and MERS coronavirus outbreaks in West Africa and Saudi Arabia respectively [61]. In this regard, intensive research on mechanisms of the first steps of viral emergence should be further developed.
Conflict of interest
The authors declare no competing financial interests.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
RR was supported by the Bourse de l’Ecole Normale Supérieure from the Faculté Paris Diderot. This work was supported by the Institut Pasteur in Paris, France, by Programme Transversal de Recherche 437 from the Institut Pasteur, and by the French government program Investissement d’Avenir (grant ANR-10-LABX-62-IBEID). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wolfe N.D., Dunavan C.P., Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Locatelli S., Peeters M. Cross-species transmission of simian retroviruses: how and why they could lead to the emergence of new diseases in the human population. AIDS. 2012;26:659–673. doi: 10.1097/QAD.0b013e328350fb68. [DOI] [PubMed] [Google Scholar]
- 3.Enders J.F., Peebles T.C. Propagation in tissue cultures of cytopathogenic agents from patients with measles. Proc Soc Exp Biol Med. 1954;86:277–286. doi: 10.3181/00379727-86-21073. [DOI] [PubMed] [Google Scholar]
- 4••.Murray S.M., Picker L.J., Axthelm M.K., Hudkins K., Alpers C.E., Linial M.L. Replication in a superficial epithelial cell niche explains the lack of pathogenicity of primate foamy virus infections. J Virol. 2008;82:5981–5985. doi: 10.1128/JVI.00367-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides convincing evidence of SFV replication in the oral cavity of SFV-infected animals. This is a major argument in favor of the special mode of transmission of this retrovirus, mainly by bites, as supported by epidemiological data.
- 5.Choudhary A., Galvin T.A., Williams D.K., Beren J., Bryant M.A., Khan A.S. Influence of naturally occurring simian foamy viruses (SFVs) on SIV disease progression in the rhesus macaque (Macaca mulatta) model. Viruses. 2013;5:1414–1430. doi: 10.3390/v5061414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Achong B.G., Mansell P.W., Epstein M.A. A new human virus in cultures from a nasopharyngeal carcinoma. J Pathol. 1971;103:P18. [PubMed] [Google Scholar]
- 7.Murray S.M., Linial M.L. Foamy virus infection in primates. J Med Primatol. 2006;35:225–235. doi: 10.1111/j.1600-0684.2006.00171.x. [DOI] [PubMed] [Google Scholar]
- 8.Gessain A., Rua R., Betsem E., Turpin J., Mahieux R. HTLV-3/4 and simian foamy retroviruses in humans: discovery, epidemiology, cross-species transmission and molecular virology. Virology. 2013;435:187–199. doi: 10.1016/j.virol.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meiering C.D., Linial M.L. Historical perspective of foamy virus epidemiology and infection. Clin Microbiol Rev. 2001;14:165–176. doi: 10.1128/CMR.14.1.165-176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10••.Schweizer M., Turek R., Hahn H., Schliephake A., Netzer K.O., Eder G., Reinhardt M., Rethwilm A., Neumann-Haefelin D. Markers of foamy virus infections in monkeys, apes, and accidentally infected humans: appropriate testing fails to confirm suspected foamy virus prevalence in humans. AIDS Res Hum Retroviruses. 1995;11:161–170. doi: 10.1089/aid.1995.11.161. [DOI] [PubMed] [Google Scholar]; This is the first report of actual SFV infections in humans, after years of contradictory reports, mainly due to the lack of appropriate tests to detect SFV infection.
- 11.Wolfe N.D., Switzer W.M., Carr J.K., Bhullar V.B., Shanmugam V., Tamoufe U., Prosser A.T., Torimiro J.N., Wright A., Mpoudi-Ngole E. Naturally acquired simian retrovirus infections in central African hunters. Lancet. 2004;363:932–937. doi: 10.1016/S0140-6736(04)15787-5. [DOI] [PubMed] [Google Scholar]
- 12••.Betsem E., Rua R., Tortevoye P., Froment A., Gessain A. Frequent and recent human acquisition of simian foamy viruses through apes’ bites in central Africa. PLoS Pathog. 2011;7:e1002306. doi: 10.1371/journal.ppat.1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study elucidates modes of transmission of SFV from apes and monkeys to humans, mainly by bites.
- 13.Mouinga-Ondeme A., Caron M., Nkoghe D., Telfer P., Marx P., Saib A., Leroy E., Gonzalez J.P., Gessain A., Kazanji M. Cross-species transmission of simian foamy virus to humans in rural Gabon, Central Africa. J Virol. 2012;86:1255–1260. doi: 10.1128/JVI.06016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Switzer W.M., Tang S., Ahuka-Mundeke S., Shankar A., Hanson D.L., Zheng H., Ayouba A., Wolfe N.D., LeBreton M., Djoko C.F. Novel simian foamy virus infections from multiple monkey species in women from the Democratic Republic of Congo. Retrovirology. 2012;9:100. doi: 10.1186/1742-4690-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones-Engel L., May C.C., Engel G.A., Steinkraus K.A., Schillaci M.A., Fuentes A., Rompis A., Chalise M.K., Aggimarangsee N., Feeroz M.M. Diverse contexts of zoonotic transmission of simian foamy viruses in Asia. Emerg Infect Dis. 2008;14:1200–1208. doi: 10.3201/eid1408.071430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engel G.A., Small C.T., Soliven K., Feeroz M.M., Wang X.X., Hasan M.K., Oh G., Alam S.M.R., Craig K.L., Jackson D.L. Zoonotic simian foamy virus in Bangladesh reflects diverse patterns of transmission and co-infection. Emerg Microbes Infect. 2013:2. doi: 10.1038/emi.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones-Engel L., Engel G.A., Schillaci M.A., Rompis A., Putra A., Suaryana K.G., Fuentes A., Beer B., Hicks S., White R. Primate-to-human retroviral transmission in Asia. Emerg Infect Dis. 2005;11:1028–1035. doi: 10.3201/eid1107.040957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rethwilm A. Molecular biology of foamy viruses. Med Microbiol Immunol. 2010;199:197–207. doi: 10.1007/s00430-010-0158-x. [DOI] [PubMed] [Google Scholar]
- 19.Plochmann K., Horn A., Gschmack E., Armbruster N., Krieg J., Wiktorowicz T., Weber C., Stirnnagel K., Lindemann D., Rethwilm A. Heparan sulfate is an attachment factor for foamy virus entry. J Virol. 2012;86:10028–10035. doi: 10.1128/JVI.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikovits J.A., Hoffman P.M., Rethwilm A., Ruscetti F.W. In vitro infection of primary and retrovirus-infected human leukocytes by human foamy virus. J Virol. 1996;70:2774–2780. doi: 10.1128/jvi.70.5.2774-2780.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu S.F., Stone J., Linial M.L. Productive persistent infection of hematopoietic cells by human foamy virus. J Virol. 1996;70:1250–1254. doi: 10.1128/jvi.70.2.1250-1254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray S.M., Picker L.J., Axthelm M.K., Linial M.L. Expanded tissue targets for foamy virus replication with simian immunodeficiency virus-induced immunosuppression. J Virol. 2006;80:663–670. doi: 10.1128/JVI.80.2.663-670.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soliven K., Wang X., Small C.T., Feeroz M.M., Lee E.G., Craig K.L., Hasan K., Engel G.A., Jones-Engel L., Matsen FAt. Simian foamy virus infection of rhesus macaques in Bangladesh: relationship of latent proviruses and transcriptionally active viruses. J Virol. 2013;87:13628–13639. doi: 10.1128/JVI.01989-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rua R., Betsem E., Gessain A. Viral latency in blood and saliva of simian foamy virus-infected humans. PLoS One. 2013;8:e77072. doi: 10.1371/journal.pone.0077072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Callahan M.E., Switzer W.M., Matthews A.L., Roberts B.D., Heneine W., Folks T.M., Sandstrom P.A. Persistent zoonotic infection of a human with simian foamy virus in the absence of an intact orf-2 accessory gene. J Virol. 1999;73:9619–9624. doi: 10.1128/jvi.73.11.9619-9624.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Laer D., Neumann-Haefelin D., Heeney J.L., Schweizer M. Lymphocytes are the major reservoir for foamy viruses in peripheral blood. Virology. 1996;221:240–244. doi: 10.1006/viro.1996.0371. [DOI] [PubMed] [Google Scholar]
- 27.Rua R., Betsem E., Montange T., Buseyne F., Gessain A. In vivo cellular tropism of gorilla simian foamy virus in blood of infected humans. J Virol. 2014;88:13429–13435. doi: 10.1128/JVI.01801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schweizer M., Schleer H., Pietrek M., Liegibel J., Falcone V., Neumann-Haefelin D. Genetic stability of foamy viruses: long-term study in an African green monkey population. J Virol. 1999;73:9256–9265. doi: 10.1128/jvi.73.11.9256-9265.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Switzer W.M., Salemi M., Shanmugam V., Gao F., Cong M.E., Kuiken C., Bhullar V., Beer B.E., Vallet D., Gautier-Hion A. Ancient co-speciation of simian foamy viruses and primates. Nature. 2005;434:376–380. doi: 10.1038/nature03341. [DOI] [PubMed] [Google Scholar]; This important study sets the date of cospeciation between foamy viruses and primates as far as 30 MY. The recent discovery of endogenous viruses, and FV in fishes, pushed the cospeciation date to even more than 400 MY.
- 30.Cleaveland S., Haydon D.T., Taylor L. Overviews of pathogen emergence: which pathogens emerge, when and why? Curr Top Microbiol Immunol. 2007;5(3):85–111. doi: 10.1007/978-3-540-70962-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rua R., Betsem E., Calattini S., Saib A., Gessain A. Genetic characterization of simian foamy viruses infecting humans. J Virol. 2012;86:13350–13359. doi: 10.1128/JVI.01715-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt M., Herchenroder O., Heeney J., Rethwilm A. Long terminal repeat U3 length polymorphism of human foamy virus. Virology. 1997;230:167–178. doi: 10.1006/viro.1997.8463. [DOI] [PubMed] [Google Scholar]
- 33.Leendertz F.H., Zirkel F., Couacy-Hymann E., Ellerbrok H., Morozov V.A., Pauli G., Hedemann C., Formenty P., Jensen S.A., Boesch C. Interspecies transmission of simian foamy virus in a natural predator-prey system. J Virol. 2008;82:7741–7744. doi: 10.1128/JVI.00549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W., Worobey M., Li Y., Keele B.F., Bibollet-Ruche F., Guo Y., Goepfert P.A., Santiago M.L., Ndjango J.B., Neel C. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathog. 2008;4:e1000097. doi: 10.1371/journal.ppat.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blasse A., Calvignac-Spencer S., Merkel K., Goffe A.S., Boesch C., Mundry R., Leendertz F.H. Mother-offspring transmission and age-dependent accumulation of simian foamy virus in wild chimpanzees. J Virol. 2013;87:5193–5204. doi: 10.1128/JVI.02743-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feeroz M.M., Soliven K., Small C.T., Engel G.A., Pacheco A., Yee J., Wang X.X., Hasan M.K., Oh G., Levine K. Population dynamics of rhesus macaques and associated foamy virus in Bangladesh. Emerg Microbes Infect. 2013;2:e29. doi: 10.1038/emi.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galvin T.A., Ahmed I.A., Shahabuddin M., Bryan T., Khan A.S. Identification of recombination in the envelope gene of simian foamy virus serotype 2 isolated from Macaca cyclopis. J Virol. 2013;87:8792–8797. doi: 10.1128/JVI.03555-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabile A., Rhodes-Feuillette A., Jaoui F.Z., Tobaly-Tapiero J., Giron M.L., Lasneret J., Peries J., Canivet M. In vitro studies on interferon-inducing capacity and sensitivity to IFN of human foamy virus. Res Virol. 1996;147:29–37. doi: 10.1016/0923-2516(96)80237-8. [DOI] [PubMed] [Google Scholar]
- 39.Rua R., Lepelley A., Gessain A., Schwartz O. Innate sensing of foamy viruses by human hematopoietic cells. J Virol. 2012;86:909–918. doi: 10.1128/JVI.06235-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell R.A., Wiegand H.L., Moore M.D., Schafer A., McClure M.O., Cullen B.R. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J Virol. 2005;79:8724–8731. doi: 10.1128/JVI.79.14.8724-8731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lochelt M., Romen F., Bastone P., Muckenfuss H., Kirchner N., Kim Y.B., Truyen U., Rosler U., Battenberg M., Saib A. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc Natl Acad Sci U S A. 2005;102:7982–7987. doi: 10.1073/pnas.0501445102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perkovic M., Schmidt S., Marino D., Russell R.A., Stauch B., Hofmann H., Kopietz F., Kloke B.P., Zielonka J., Strover H. Species-specific inhibition of APOBEC3C by the prototype foamy virus protein bet. J Biol Chem. 2009;284:5819–5826. doi: 10.1074/jbc.M808853200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaguva Vasudevan A.A., Perkovic M., Bulliard Y., Cichutek K., Trono D., Haussinger D., Munk C. Prototype foamy virus Bet impairs the dimerization and cytosolic solubility of human APOBEC3 G. J Virol. 2013;87:9030–9040. doi: 10.1128/JVI.03385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delebecque F., Suspene R., Calattini S., Casartelli N., Saib A., Froment A., Wain-Hobson S., Gessain A., Vartanian J.P., Schwartz O. Restriction of foamy viruses by APOBEC cytidine deaminases. J Virol. 2006;80:605–614. doi: 10.1128/JVI.80.2.605-614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Matsen FAt, Small C.T., Soliven K., Engel G.A., Feeroz M.M., Wang X., Craig K.L., Hasan M.K., Emerman M., Linial M.L. A novel Bayesian method for detection of APOBEC3-mediated hypermutation and its application to zoonotic transmission of simian foamy viruses. PLoS Comput Biol. 2014;10:e1003493. doi: 10.1371/journal.pcbi.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]; This interesting study provides evidence of distinct effects of APOBEC in humans and monkeys, suggesting that mutated strains found in humans are not passively acquired from macaques but are efficiently targeted by anti-viral mechanisms in humans.
- 46.Hu X., Yang W., Liu R., Geng Y., Qiao W., Tan J. N-Myc interactor inhibits prototype foamy virus by sequestering viral Tas protein in the cytoplasm. J Virol. 2014;88:7036–7044. doi: 10.1128/JVI.00799-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan J., Qiao W., Wang J., Xu F., Li Y., Zhou J., Chen Q., Geng Y. IFP35 is involved in the antiviral function of interferon by association with the viral tas transactivator of bovine foamy virus. J Virol. 2008;82:4275–4283. doi: 10.1128/JVI.02249-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu F., Tan J., Liu R., Xu D., Li Y., Geng Y., Liang C., Qiao W. Tetherin inhibits prototypic foamy virus release. Virol J. 2011;8:198. doi: 10.1186/1743-422X-8-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dietrich I., McMonagle E.L., Petit S.J., Vijayakrishnan S., Logan N., Chan C.N., Towers G.J., Hosie M.J., Willett B.J. Feline tetherin efficiently restricts release of feline immunodeficiency virus but not spreading of infection. J Virol. 2011;85:5840–5852. doi: 10.1128/JVI.00071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pacheco B., Finzi A., McGee-Estrada K., Sodroski J. Species-specific inhibition of foamy viruses from South American monkeys by New World Monkey TRIM5{alpha} proteins. J Virol. 2010;84:4095–4099. doi: 10.1128/JVI.02631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yap M.W., Lindemann D., Stanke N., Reh J., Westphal D., Hanenberg H., Ohkura S., Stoye J.P. Restriction of foamy viruses by primate Trim5 alpha. J Virol. 2008;82:5429–5439. doi: 10.1128/JVI.02462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gramberg T., Kahle T., Bloch N., Wittmann S., Mullers E., Daddacha W., Hofmann H., Kim B., Lindemann D., Landau N.R. Restriction of diverse retroviruses by SAMHD1. Retrovirology. 2013;10:26. doi: 10.1186/1742-4690-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams D.K., Khan A.S. Role of neutralizing antibodies in controlling simian foamy virus transmission and infection. Transfusion. 2010;50:200–207. doi: 10.1111/j.1537-2995.2009.02372.x. [DOI] [PubMed] [Google Scholar]
- 54.Cummins J.E., Jr., Boneva R.S., Switzer W.M., Christensen L.L., Sandstrom P., Heneine W., Chapman L.E., Dezzutti C.S. Mucosal and systemic antibody responses in humans infected with simian foamy virus. J Virol. 2005;79:13186–13189. doi: 10.1128/JVI.79.20.13186-13189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neumann-Haefelin D., Rethwilm A., Bauer G., Gudat F., zur Hausen H. Characterization of a foamy virus isolated from Cercopithecus aethiops lymphoblastoid cells. Med Microbiol Immunol. 1983;172:75–86. doi: 10.1007/BF02124508. [DOI] [PubMed] [Google Scholar]
- 56.Schweizer M., Falcone V., Gange J., Turek R., Neumann-Haefelin D. Simian foamy virus isolated from an accidentally infected human individual. J Virol. 1997;71:4821–4824. doi: 10.1128/jvi.71.6.4821-4824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Falcone V., Schweizer M., Toniolo A., Neumann-Haefelin D., Meyerhans A. Gamma interferon is a major suppressive factor produced by activated human peripheral blood lymphocytes that is able to inhibit foamy virus-induced cytopathic effects. J Virol. 1999;73:1724–1728. doi: 10.1128/jvi.73.2.1724-1728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colas S., Bourge J.F., Wybier J., Chelbi-Alix M.K., Paul P., Emanoil-Ravier R. Human foamy virus infection activates class I major histocompatibility complex antigen expression. J Gen Virol. 1995;76(Pt 3):661–667. doi: 10.1099/0022-1317-76-3-661. [DOI] [PubMed] [Google Scholar]
- 59.Bangham C.R. CTL quality and the control of human retroviral infections. Eur J Immunol. 2009;39:1700–1712. doi: 10.1002/eji.200939451. [DOI] [PubMed] [Google Scholar]
- 60.Switzer W.M., Garcia A.D., Yang C., Wright A., Kalish M.L., Folks T.M., Heneine W. Coinfection with HIV-1 and simian foamy virus in West Central Africans. J Infect Dis. 2008;197:1389–1393. doi: 10.1086/587493. [DOI] [PubMed] [Google Scholar]
- 61.McCloskey B., Dar O., Zumla A., Heymann D.L. Emerging infectious diseases and pandemic potential: status quo and reducing risk of global spread. Lancet Infect Dis. 2014;14:1001–1010. doi: 10.1016/S1473-3099(14)70846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Calattini S., Betsem E.B., Froment A., Mauclere P., Tortevoye P., Schmitt C., Njouom R., Saib A., Gessain A. Simian foamy virus transmission from apes to humans, rural Cameroon. Emerg Infect Dis. 2007;13:1314–1320. doi: 10.3201/eid1309.061162. [DOI] [PMC free article] [PubMed] [Google Scholar]


