Abstract
The budding yeast v-SNARE, Snc1, mediates fusion of exocytic vesicles to the plasma membrane (PM) and is subsequently recycled back to the Golgi. Postendocytic recycling of Snc1 requires a phospholipid flippase (Drs2-Cdc50), an F-box protein (Rcy1), a sorting nexin (Snx4-Atg20), and the COPI coat complex. A portion of the endocytic tracer FM4-64 is also recycled back to the PM after internalization. However, the relationship between Snx4, Drs2, Rcy1, and COPI in recycling Snc1 or FM4-64 is unclear. Here we show that rcy1∆ and drs2∆ single mutants, or a COPI mutant deficient in ubiquitin binding, display a defect in recycling FM4-64 while snx4∆ cells recycle FM4-64 normally. The addition of latrunculin A to acutely inhibit endocytosis shows that rcy1∆ and snx4∆ single mutants retain the ability to recycle Snc1, but a snx4∆rcy1∆ mutant substantially blocks export. Additional deletion of a retromer subunit completely eliminates recycling of Snc1 in the triple mutant (snx4∆rcy1∆vps35∆). A minor role for retromer in Snc1 recycling can also be observed in single and double mutants harboring vps35∆. These data support the existence of three distinct and parallel recycling pathways mediated by Drs2/Rcy1/COPI, Snx4-Atg20, and retromer that retrieve an exocytic v-SNARE from the endocytic pathway to the Golgi.
INTRODUCTION
Newly synthesized soluble and membrane proteins traveling the secretory pathway traffic from their origin in the endoplasmic reticulum (ER) to the trans-Golgi network (TGN) where they are recognized, sorted, and packaged into secretory vesicles that fuse with the plasma membrane (PM). In line with the SNARE hypothesis, principle components of these exocytic vesicles in Saccharomyces cerevisiae are Snc1 and Snc2, R-SNAREs homologous to mammalian synaptobrevin VAMP2 that confer specificity for fusion with the PM via interactions with complementary Q-SNAREs (Protopopov et al., 1993; Sollner et al., 1993). The “zippering” of the trans-SNARE complex will bring the two membranes into close proximity, ultimately driving the fusion of the vesicular membrane with the target membrane (Fiebig et al., 1999). At this point, the SNARE complex is now found in cis on the target membrane and is subsequently dissociated by the activity of the NSF AAA-ATPase, yeast Sec18, allowing recycling of SNAREs (Grote et al., 2000; Ishii et al., 2016).
After mediating fusion with the PM, Snc1 and Snc2 are endocytosed from the cell surface and trafficked back to the TGN, where they can then be repackaged into nascent secretory vesicles to facilitate additional rounds of transport (Lewis et al., 2000). Internalization from the PM is mediated by a conserved endocytosis signal within Snc1, which is recognized by the yeast homolog of the AP180/CALM clathrin adaptor protein (Lewis et al., 2000; Burston et al., 2009; Miller et al., 2011). Snc1 within endocytic vesicles mediates fusion with membranes containing the syntaxins (t-SNAREs) Tlg1 and Tlg2 (Holthuis et al., 1998; Gurunathan et al., 2002). Tlg1 labels an organelle that first receives bulk-phase endocytic tracers, like the lipophilic dye FM4-64, after its uptake by the cell (Vida and Emr, 1995; Wiederkehr et al., 2000; Day et al., 2018). As such, this Tlg1-positive organelle can be functionally described as an early endosome (EE). A recent study shows that Tlg1-positive structures eventually become labeled by the canonical TGN marker, Sec7, either via a fusion event or via maturation, suggesting Tlg1 marks a late Golgi compartment that also serves as the EE in yeast (Day et al., 2018). However, a portion of internalized FM4-64 recycles back to the PM from early endosomal membranes in a pathway that is independent of Sec7, but requires the F-box protein Rcy1 (Wiederkehr et al., 2000; MacDonald and Piper, 2017). In addition, rcy1∆ mutants possess enlarged compartments that contain Tlg1 and endocytosed cargo, including Snc1, Ste2, and FM4-64, and this mutant transports proteins through the secretory pathway with normal kinetics (Lewis et al., 2000; Wiederkehr et al., 2000). The latter observations support the existence of an EE (or recycling endosome) in yeast that functions independently of the Golgi.
The endosomal system is a dynamic, high-traffic system that is crucial in regulating the localization of cell-surface receptors, proteolytic degradation of unneeded proteins, and in recycling vesicular machinery back to TGN to facilitate exocytic trafficking. Transport through the endocytic pathway is thought to occur by maturation of EEs to late endosomes (LE) or prevacuolar endosomes (PVE), which fuse with vacuoles to deliver luminal content (Huotari and Helenius, 2011; Gautreau et al., 2014). Budding of intraluminal vesicles occurs during these maturation events to form multivesicular bodies (MVB) and proteins sorted into these vesicles are typically degraded on their arrival in lysosomes (Katzmann et al., 2001; Stahl and Barbieri, 2002). This fate is avoided, however, by certain resident Golgi proteins, cell surface receptors, SNAREs (Snc1 included), and endosomal proteins that are specifically retrieved from the endocytic pathway (Lewis et al., 2000). The retromer complex is composed of two subcomplexes: a cargo-selective subcomplex consisting of Vps26, Vps29, and Vps35 and a membrane-deformation subcomplex consisting of sorting nexin dimers (Seaman, 2012). Retromer plays a major role in retrieving proteins from PVEs back to the Golgi, although Snc1 (Lewis et al., 2000) and FM4-64 recycling (MacDonald and Piper, 2017) have been reported to be unperturbed in retromer mutants.
The recycling of Snc1 is facilitated by multiple factors including sorting nexin 4 (Snx4), Rcy1, the phosphatidylserine flippase Drs2-Cdc50, and COPI (Galan et al., 2001; Hua et al., 2002; Hettema et al., 2003; Chen et al., 2005; Furuta et al., 2007; Xu et al., 2017). The sorting nexin family of proteins shares a phosphoinositide-binding phox homology (PX) domain and is reported to play roles in various membrane trafficking and protein sorting functions within the endosomal system. Snx4 can form two distinct heterodimer complexes, either Snx4-Snx41 or Snx4-Atg20, which mediate trafficking from endosomes to the Golgi and also play a role in the initiation of the autophagic cytoplasm-to-vacuole targeting pathway (Bean et al., 2017; Ma et al., 2018). F-box proteins have been shown to confer substrate-selectivity to Skp1-Cullin-F-box (SCF) E3 ubiquitin (Ub) ligases (Skowyra et al., 1997); however, the Rcy1-Skp1 complex exerts an influence on Snc1 transport in a way that is both Cullin– and E2 Ub–conjugating enzyme (Cdc34) independent (Galan et al., 2001). Despite this, recent work reveals that K63 ubiquitylation of Snc1 is necessary for the efficient recycling of this v-SNARE via COPI-coated vesicles (Chen et al., 2011; Xu et al., 2017). In addition, Rcy1 binds to the regulatory domain of Drs2 and may activate Drs2-Cdc50’s flippase activity to support Snc1 recycling (Furuta et al., 2007; Hanamatsu et al., 2014). Inactivation of Drs2-Cdc50 causes accumulation of Snc1 in enlarged Tlg1-positive, Sec7-negative compartments (Furuta et al., 2007). However, it is not known whether all of these factors work together in a single pathway or define several different pathways for recycling Snc1. Here, we present data to suggest that Snc1 is retrieved to the TGN via distinct, parallel pathways mediated by Snx4, Rcy1/Drs2/COPI, and retromer.
RESULTS
Roles of Rcy1 and Snx4 in Snc1 recycling
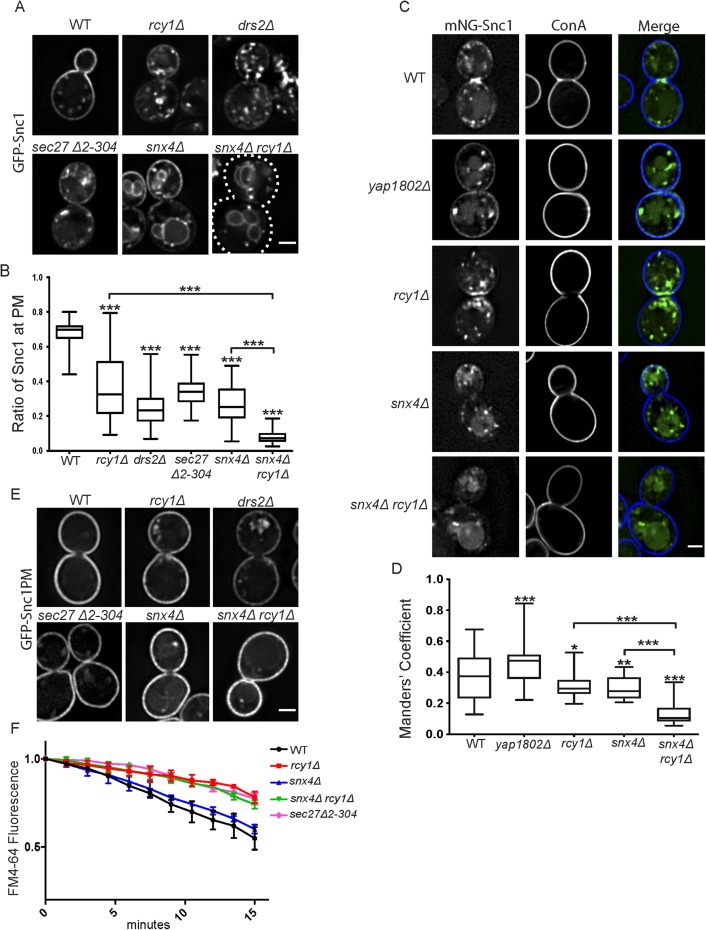
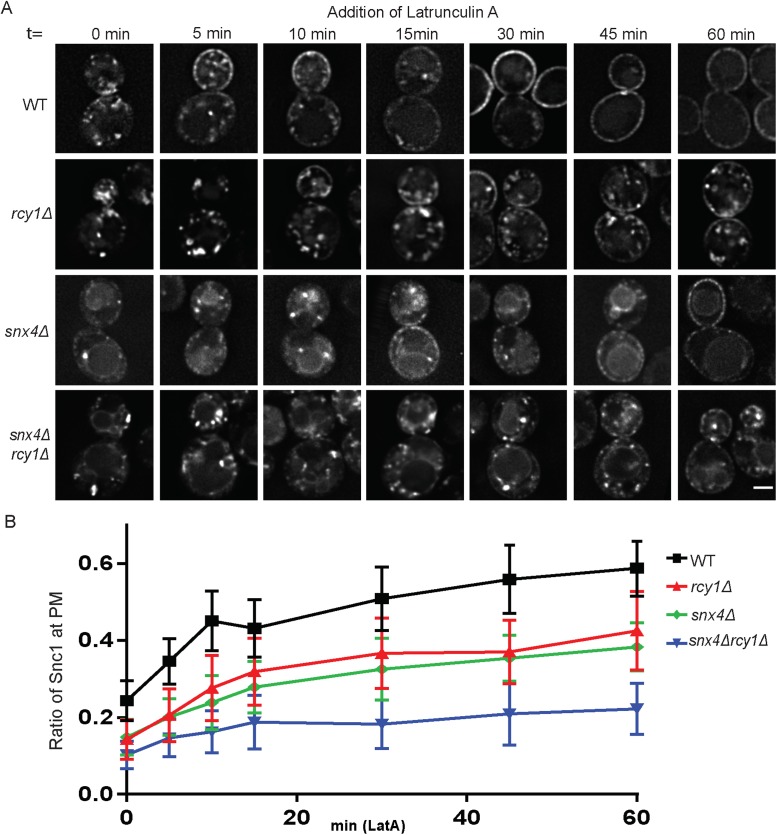
The GFP-Snc1 construct used to examine recycling of this protein in numerous studies is overexpressed from a strong promoter (Lewis et al., 2000). Under this condition, endocytosis from the PM appears to be a rate-limiting step in the recycling pathway and more than half of overexpressed GFP-Snc1 localized to the PM at steady state, typically enriched at the nascent bud site due to polarized secretion (Figure 1A and Lewis et al., 2000). If the trafficking of GFP-Snc1 is perturbed in such a way as to shift the rate-limiting step to another stop along the cargo’s itinerary, one would expect to observe a corresponding shift of steady-state localization toward that specific step. As previously observed, cells lacking the F-Box protein Rcy1 (rcy1Δ) displayed a substantial depletion of GFP-Snc1 from the cell surface and accumulation within internal punctae (Figure 1, A and B; Galan et al., 2001). A similar phenotype was observed in cells lacking the endosomal PE/PS flippase Drs2 (drs2Δ) (Hua et al., 2002; Furuta et al., 2007), as well as in mutants lacking the N-terminal propeller of β’-COP (sec27Δ2-304) important for Ub binding (Xu et al., 2017) (Figure 1, A and B), suggesting that they may be acting at a similar stage of GFP-Snc1 trafficking. This COPI mutant (sec27Δ2-304) retains the ability to recycle cargoes from the Golgi back to the ER and exhibits a relatively specific defect in Snc1 recycling with accumulation in Tlg1-marked compartments (Xu et al., 2017).
FIGURE 1:
Roles of Rcy1 and Snx4 in Snc1 recycling. (A) WT and mutant cells expressing GFP-Snc1 were imaged at 1000×. Images shown are single planes chosen from a z-stack series. (B) Quantification of images captured in A (n = 50); images were analyzed by determining the ratio of GFP signal at the PM as a function of total fluorescent signal. (C) WT and mutant cells expressing a Cu-induced mNG-Snc1 construct at low levels were stained with fluorescently labeled ConA and imaged at 1000×. Images shown are single planes. (D) Channels were separated and thresholded, then correlation was measured by finding the MCC between the channels (n = 50) (E) Wild-type and mutant cells expressing an endocytosis deficient GFP-Snc1(PM) were imaged at 1000×. Images shown are single planes. (F) Measurement of FM4-64 postendocytic recycling in WT and mutant cells. Fluorescence intensity was normalized to the initial value for each strain. Data represent three independent experiments. Scale bars represent 2 µm.
In addition, GFP-Snc1 was depleted from the PM in strains lacking the yeast sorting nexin 4 (snx4Δ). However, unlike the phenotypes observed in rcy1Δ, drs2Δ, or sec27Δ2-304, GFP-Snc1 mislocalized to the limiting membrane of the vacuole in snx4∆ cells (Figure 1, A and B, and Hettema et al., 2003; Obara et al., 2005). Snx4 is known to interact with additional sorting nexin proteins and associates with either Snx41 or Atg20 in order to perform distinct functions. The Snx4-Atg20 complex has previously been implicated in Snc1 recycling (Ma et al., 2017) and we similarly observed a mislocalization of GFP-Snc1 to the vacuole in atg20∆ cells relative to wild-type (WT) or snx41∆ cells (Supplemental Figure S1). The phenotypic difference between the sorting nexin mutants and Rcy1/Drs2/COPI mutants suggested that these proteins function at different steps along the Snc1 trafficking itinerary.
To address this hypothesis, we generated a double mutant deficient for Rcy1 and Snx4 (snx4Δrcy1Δ) in order to assay the effects on Snc1 trafficking. In this double mutant, GFP-Snc1 was almost completely absent from the PM, instead accumulating primarily on and within the vacuole, with the remainder of fluorescence in punctate structures (Figure 1, A and B). This additive phenotype, one in which the defects observed in the double-mutant is significantly worse than in either of the corresponding single mutants, is consistent with a model in which two parallel or partially redundant systems are both operating independently in the retrieval of a shared cargo. In contrast, we found that there was no difference between the defects observed in drs2Δ and rcy1Δdrs2Δ or between the defects observed in sec27Δ2-304 and rcy1Δsec27Δ2-304, suggesting that each of these factors acted within a singular pathway (Supplemental Figure S2, A and B).
As described above, an overexpressed GFP-Snc1 construct has been used to study these endosomal trafficking pathways. However, with the availability of more sensitive CCD cameras and brighter fluorescent proteins, like mNeonGreen (mNG) (Shaner et al., 2013), it was possible to assess these endosomal recycling pathways at low levels of mNG-Snc1 expression. We generated an mNG-SNARE construct under a copper-inducible promoter that allowed us to “tune” the expression levels to near-endogenous levels (Supplemental Figure S3; Ho et al., 2018). WT cells and mutants carrying a Cu-mNG-Snc1 plasmid were induced at 100 μM CuCl2 for 1 h and then treated with fluorescently labeled concanavalin A (ConA), a cell wall–binding lectin. The lower expression levels made it difficult to quantify the ratio of PM to cytoplasmic fluorescence in mutant cells using the same methods as in Figure 1B. Therefore, cells were imaged and analyzed for the degree of colocalization between mNG-Snc1 and ConA to determine the relative proportion of Snc1 at the cell surface in each of these strains. At the lower expression level, roughly one-third of mNG-Snc1 signal above the threshold was present at the PM in WT cells at steady state, while a significant fraction was found within small punctae. In cells lacking either Rcy1 or Snx4, PM localization of mNG-Snc1 was modestly but significantly reduced. mNG- Snc1 was substantially depleted from the PM in snx4Δrcy1Δ double mutants (Figure 1, C and D). Thus, GFP-Snc1 localized primarily to endosomes or Golgi cisternae when expressed at low levels, rather than the PM, but retained the same dependency on Rcy1 and Snx4 for recycling as previously shown with overexpressed GFP-Snc1.
Endocytosis of Snc1 from the PM requires the yeast AP180 homologues (Yap1801 and Yap1802) (Burston et al., 2009) and we suspected that overexpression of GFP-Snc1 saturated this interaction leading to a higher percentage at the PM of WT cells relative to mNG-Snc1. Consistent with this, we found that knocking out the more highly expressed YAP1802 gene increased mNG-Snc1 PM localization (Figure 1, C and D).
To address the possibility that the missorting of newly synthesized GFP-Snc1 trafficking from the Golgi caused its vacuolar localization in snx4Δrcy1Δ cells, we used a GFP-Snc1 variant that is unable to be internalized from the PM due to a mutation in the endocytic signal (Lewis et al., 2000). As such, any GFP-Snc1 molecules that are successfully secreted to the cell surface will then accumulate at the PM. In this experiment, GFP-Snc1(PM) robustly accumulated at the cell surface in the WT and in all tested mutants (Figure 1E), confirming that the observed defects in Figure 1, A and B, were primarily the result of perturbations in the recycling of GFP-Snc1 rather than a defect in exocytosis or the sorting of newly synthesized GFP-Snc1 at the level of the Golgi. A portion of the GFP-Snc1(PM) did accumulate internally in the mutants, which may reflect some degree of missorting into the endosomal system at the level of the Golgi.
Rcy1 was first identified for its role in postendocytic recycling of the lipophilic dye FM4-64 back to the PM (Wiederkehr et al., 2000) and so we examined this pathway in the snx4∆, snx4∆rcy1∆, and COPI mutants. After internalization of FM4-64 in WT cells, a fraction of the dye follows a recycling route and is secreted back into the media. Thus, WT cells displayed a gradual loss of fluorescence over 15 min while the rate of FM4-64 secretion was substantially slowed in rcy1∆ cells (Figure 1F and Wiederkehr et al., 2000). In this experiment, rcy1Δ and sec27Δ2-304 both displayed a nearly identical, reduced rate of dye recycling, consistent with a shared role in the same recycling pathway. In contrast, snx4Δ mutants were capable of recycling FM4-64 at a similar rate as WT cells (Figure 1F and MacDonald and Piper, 2017). Moreover, the rate of FM4-64 recycling in the snx4∆rcy1∆ double mutant was identical to rcy1∆. While both Snx4 and Rcy1 function in recycling GFP-Snc1 back to the PM (Figure 1, A–D), Snx4 does not significantly contribute to FM4-64 recycling (Figure 1E). These data support a model where Rcy1, Drs2, and COPI are acting in a recycling pathway traveled by both FM4-64 and Snc1, with Snx4 acting in a separate pathway traveled by Snc1, but not FM4-64. This set of experiments highlights the importance of these recycling activities under both overexpressed and more physiological levels of Snc1 expression and provides further evidence that Rcy1 and Snx4 operate at distinct steps along the Snc1 recycling itinerary. We use rcy1∆ to disrupt the Rcy1/Drs2/COPI pathway for the remainder of this study because Rcy1 appears to uniquely function in this pathway while drs2∆ also disrupts AP-1/clathrin function and COPI has roles earlier in the secretory pathway (Liu et al., 2008; Bethune and Wieland, 2018).
Combined loss of Rcy1 and Snx4 enhances GFP-Snc1 missorting to the vacuole
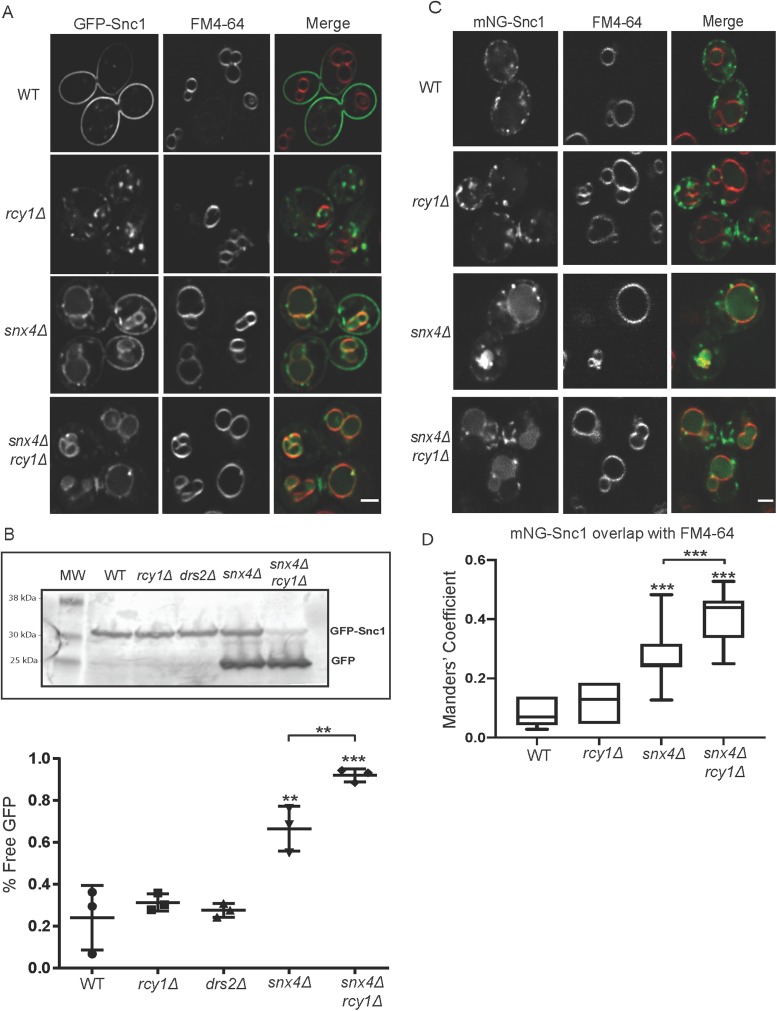
Here we sought to confirm and quantify the vacuolar mislocalization of Snc1 in snx4∆ and snx4∆rcy1∆ cells. To accomplish this, we first pulsed GFP–Snc1-overexpressing cells with FM4-64 for 10 min before washing out the dye and resuspending in fresh media at 30°C for 90 min. This “pulse/chase” treatment allowed the endocytosed dye to robustly and specifically accumulate at the vacuolar limiting membrane (Vida and Emr, 1995). GFP-Snc1 accumulated on and within the vacuole membrane in both snx4Δ and snx4Δrcy1Δ, but no colocalization between GFP-Snc1 and FM4-64 was observed in WT or rcy1∆ cells (Figure 2A). GFP is stable within the vacuolar lumen, while Snc1 is rapidly degraded; these properties lead to an accumulation of a smaller “free GFP” band and a depletion of the GFP-Snc1 fusion protein when this cargo is missorted to the vacuole. The ratio of free GFP to GFP-Snc1 on a Western blot probed with anti-GFP provides a method for quantifying mislocalization to the vacuole in a large population of cells. In line with the microscopy data, these Western blots indicated GFP-Snc1 was missorted to the vacuole in snx4Δ and a significantly enhanced sorting defect was observed in snx4Δrcy1Δ (Figure 2B). Similarly, we observed that fluorescently labeled Snc1 localized to the vacuole in snx4Δ and snx4Δrcy1Δ backgrounds even at lower expression levels, and this missorting phenotype was enhanced in the double mutant (Figure 2, C and D).
FIGURE 2:
Combined loss of Rcy1 and Snx4 enhances GFP-Snc1 missorting to the vacuole. (A) Cells expressing GFP-Snc1 were pulsed with FM4-64 and the dye was allowed to chase to the vacuole over 90 min. The cells were then imaged at 1000×; single planes are shown. (B) WT and mutant cells expressing GFP-Snc1 were cultured overnight, harvested, and lysed in buffer containing SDS. These lysates were used in a Western blot and blotted with mouse anti-GFP antibody and then a fluorescently labeled rabbit anti- mouse antibody and imaged. Lower molecular weight bands represent free GFP that has been proteolytically cleaved within the vacuole. The ratio of “free GFP” signal to total signal was used to quantify these blots. Data represent three independent experiments. (C) Cells expressing a Cu-induced mNG-Snc1 construct at endogenous level were pulsed with FM4-64, and the dye was allowed to chase to the vacuole. Micrographs were taken at 1000×; single planes are shown. (D) Channels were separated and thresholded and correlation was measured by finding the MCC between the channels (n = 50). Scale bars represent 2 µm.
Snc1 passes through Tlg1 compartments where it accumulates in rcy1∆ cells
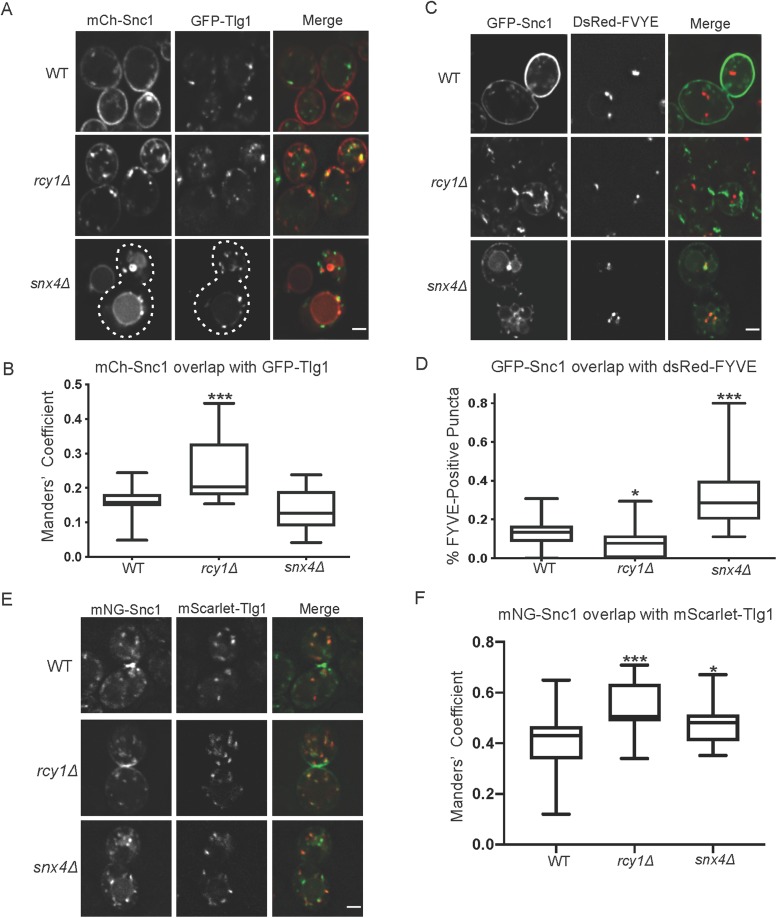
When overexpressed, Snc1 primarily localized to the PM of WT cells and internal punctae marked by Tlg1 (EE and/or TGN) in rcy1∆ cells (Figure 3, A and B). Even though Snc1 primarily localized to the vacuole of snx4∆ cells, most of these cells also displayed a few punctae with, in this case, mCherry(mCH)-Snc1 (Figure 3A). To ascertain the identity of these structures, we coexpressed fluorescent Snc1 along with a marker of the TGN and EE (GFP-Tlg1) (Siniossoglou and Pelham, 2001), or a LE marker (DsRed-FYVE) (Eugster et al., 2004) in WT and mutant strains, imaged them, and quantified the overlap of each marker via the Manders’ correlation coefficient (MCC) and by counting punctae, respectively. From these experiments, we observed a relatively low degree of overlap between Tlg1 and Snc1 in WT and snx4Δ cells. In contrast, the degree of colocalization between Snc1 and Tlg1 was increased nearly twofold in rcy1Δ (Figure 3, A and B). When comparing the colocalization between GFP-Snc1 and the LE marker, DsRed-FYVE, we observed little overlap in either the WT or the rcy1Δ strains (Figure 3, C and D). We did, however, see a significant increase in the proportion of Snc1 punctae that were also positive for DsRed-FYVE in snx4Δ, consistent with a transitory stop in the LE en route to the vacuole. Thus, rcy1∆ and snx4∆ are accumulating Snc1 at different steps along the endocytic pathway. This is consistent with published data showing Snx4 localizes to LEs while Rcy1 localizes to Tlg1-positive EE/TGN compartments (Chen et al., 2005; Arlt et al., 2015; Ma et al., 2017).
FIGURE 3:
Snc1 passes through Tlg1 compartments where it accumulates in rcy1∆. (A) WT and mutant cells expressing GFP-Tlg1 and mCh-Snc1 were imaged at 1000×; single planes are shown. (B) Channels were separated and thresholded and correlation was measured by finding the MCC between the channels (n = 50). (C) Cells expressing GFP-Snc1 and DsRed-FYVE were imaged at 1000×; single planes are shown. (D) Channels were split, and the total nonvacuolar GFP-positive structures were counted for each cell. Next, the number of GFP-positive structures that were also positive for dsRed- FYVE were counted and a ratio of these two values was calculated for each cell. (n = 40). (E) Cells expressing mCh-Tlg1 and mNG-Snc1 were imaged at 1000×; single plane is shown. (F) Channels were separated and thresholded and correlation was measured by finding the MCC between the two channels (n = 40). Scale bars represent 2 µm.
Similarly, we sought to confirm that the missorting phenotypes observed under overexpressed conditions were still applicable at low Snc1 expression levels, as it was possible that overexpression contributed to these phenotypes in the mutant cells. As noted, mNG-Snc1 expressed at low levels is observed both at the PM and within punctate structures in the cytoplasm. Colocalization analysis between mNG-Snc1 and mCH-Tlg1 reveals that these Snc1 punctae are also positive for mCh-Tlg1. This colocalization was significantly enhanced in rcy1Δ, suggesting that the efficiency of Snc1 retrieval from the Tlg1-positive compartment is impaired in this condition. The difference between WT and rcy1∆ was not as pronounced as with GFP-Snc1 because a larger percentage of mNG-Snc1 localized to Tlg1-positive compartments in WT cells at the lower expression level. The degree of colocalization between these two markers was slightly increased in snx4Δ, but not to the extent as observed in rcy1∆ cells (Figure 3, E and F).
Distinct sorting signals mediate Rcy1- and Snx4-dependent pathways
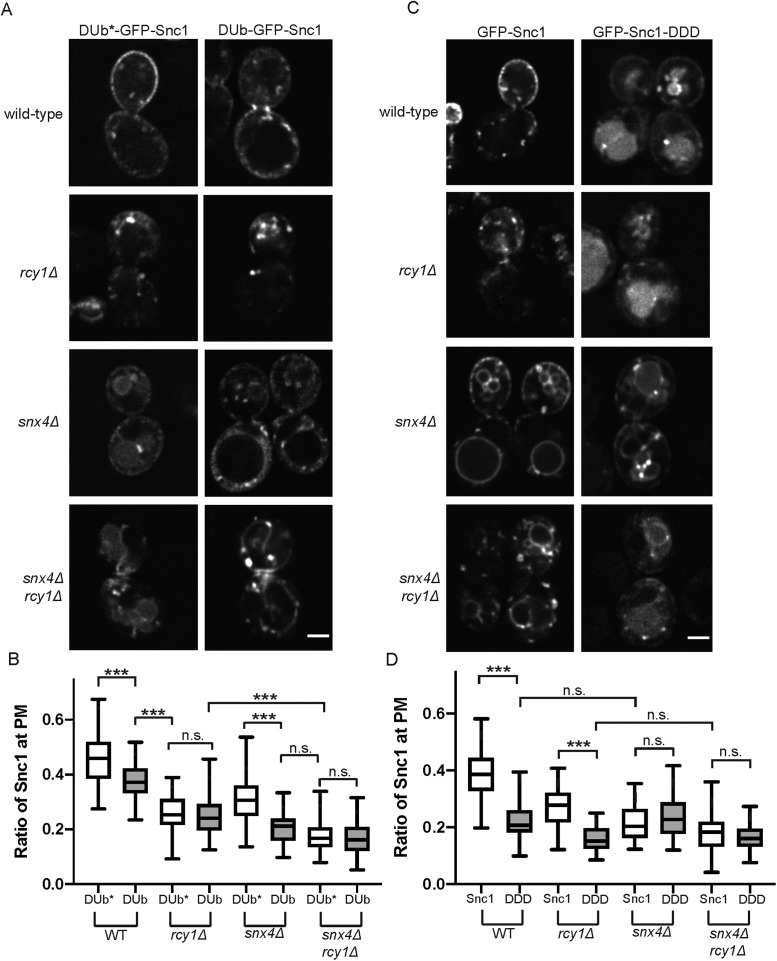
Earlier work from our group identified a requirement for the ubiquitination of Snc1 for interaction with the Rcy1/COPI recycling machinery. The UL36 deubiquitinase (DUb) from the herpes simplex virus, as well as a catalytically dead variant (DUb*), was fused to GFP-Snc1 and it was reported that DUb-GFP-Snc1 accumulated in Tlg1-positive membranes (Xu et al., 2017). Thus, we would predict an additive defect in Snc1 recycling when DUb-GFP-Snc1 is expressed in snx4∆ mutants (Figure 4A), comparable to what we observed for GFP-Snc1 in snx4∆rcy1∆. In WT cells, we observed a depletion of DUb-GFP-Snc1 from the PM, relative to DUb*-GFP-Snc1, and accumulation within cytoplasmic punctae and tubules. There was no significant difference in Snc1 recycling in rcy1Δ cells expressing either DUb-GFP-Snc1 or DUb*-GFP-Snc1. In contrast, we see a significantly enhanced missorting phenotype in snx4Δ cells expressing DUb-GFP-Snc1 when compared with the catalytically dead variant (Figure 4, A and B). Of note, rather than the primarily vacuolar localization of GFP-Snc1 observed in in snx4Δ mutants, DUb-GFP-Snc1 is completely excluded from the lumen of the vacuole in snx4Δ and snx4∆rcy1∆ mutants. Instead, the cargo accumulates within punctate structures, as well as on the vacuolar membrane and enriched in foci likely to represent PVEs (Figure 4A). As ubiquitination of membrane proteins is required for their entry into MVBs via the ESCRT machinery, this exclusion from the vacuolar lumen is expected, and indeed serves as a useful control to confirm that our DUb fusions are catalytically active. These results are consistent with a model in which there are two separate pathways capable of retrieval of Snc1 to the TGN, one dependent on ubiquitination of the cargo (Rcy1/COPI) and one independent of this modification (Snx4).
FIGURE 4:
Distinct sorting signals mediate Rcy1- and Snx4-dependent pathways. (A) WT and mutant cells expressing DUb-GFP-Snc1 and DUb*-GFP-Snc1 were imaged at 1000×; single planes are shown. (B) Quantification of images captured in A (n = 50); images were analyzed by determining the ratio of GFP signal at the PM as a function of total fluorescent signal. (C) WT and mutant cells expressing GFP-Snc1 and GFP-Snc1-DDD, both expressed from the PRC1 promoter, were imaged at 1000×; single planes are shown. The PRC1 promoter is comparable in strength to the CUP1 promoter and so approximately 40% of WT GFP-Snc1 localized to the PM in these conditions. (D) Quantification of images captured in C (n = 50); images were analyzed by determining the ratio of GFP signal at the PM as a function of total fluorescent signal.
Previous studies identified a series of charged residues (K88, K91, K93) within the transmembrane domain of Snc1 that are essential for recognition of the SNARE by the Snx4-mediated recycling machinery. While individual mutation of these residues did not affect Snc1 trafficking (Lewis et al., 2000), a charge reversal of all three residues (KKK to DDD) disrupted the ability of Snc1-DDD to be retrieved from the endosomal system by Snx4, resulting in an accumulation of the cargo within the vacuole in WT cells, recapitulating the defects seen in snx4Δ mutants (Ma and Burd, 2019). We predict that GFP-Snc1-DDD is still capable of engaging the distinct Rcy1/Drs2/COPI-dependent recycling module. Therefore, we would expect rcy1Δ cells to show a further diminished ability to recycle GFP-Snc1-DDD compared with GFP-Snc1, essentially phenocopying snx4Δrcy1Δ mutants described in Figure 1. WT cells expressing GFP-Snc1-DDD missorted the cargo to the vacuolar lumen and the fraction at the cell surface was diminished relative to GFP-Snc1. Consistent with our predictions, we observed a significantly enhanced defect in retrieval of GFP-Snc1-DDD in rcy1∆ cells relative to GFP-Snc1. Notably, there was no significant difference in PM localization between snx4Δ cells expressing either GFP-Snc1 or GFP-Snc1-DDD, indicating that, as the charge-reversal mutants are already unable to engage the Snx4 machinery, there is no further consequence to losing Snx4 itself. Last, we found that there is no measurable difference in PM localization of GFP-Snc1 or GFP-Snc1-DDD in snx4Δrcy1Δ double mutants (Figure 4, C and D). Taken together, these sets of experiments support a model of parallel retrieval mechanisms for Snc1, each reliant on distinct sorting signals to mediate interaction with the trafficking machineries.
Internal pools of Snc1 can be recycled to the PM in snx4Δ and rcy1Δ mutants
Snc1 is mislocalized in snx4Δ and rcy1Δ mutants; however, these steady-state measurements fail to provide insight into the kinetics of postendocytic Snc1 recycling, or the fate of Snc1 molecules that appear to be “trapped” in these internal structures. If Rcy1 and Snx4 mediate two parallel pathways for recycling, each should be capable of recycling Snc1 in the absence of the other. To test this possibility, we acutely inhibited endocytosis with latrunculin A (LatA), a drug that inhibits the formation of F-actin (Ayscough, 2000), and measured the rate of Snc1 recycling as a function of its delivery to the PM over time from internal compartments. WT and mutant cells expressing mNG-Snc1 were induced for an hour and then washed and treated with both cycloheximide, a potent inhibitor of protein translation, and LatA. Cycloheximide was administered to eliminate any contribution from newly synthesized mNG-Snc1 in the secretory pathway so we would only measure recycling of pre-existing mNG-Snc1. These cells were then imaged over the course of an hour and the proportion of Snc1 at the PM was quantified for each of these time points.
In WT cells at t = 0, most of the mNG-Snc1 was visible within intracellular punctae, with a smaller portion visible at the cell surface. Between the 5- and 15-min time points, we saw a rapid increase in the accumulation of Snc1 at the PM, while internal pools began to dim or disappear. This trend continued, at a slower pace, throughout the entire hour, until Snc1 was almost exclusively localized to the PM and internal pools largely disappeared (Figure 5, A and B). At low expression levels, background fluorescence in the cytosol makes a significant contribution to the intracellular signal and so the absolute values likely underrepresent the percentage of Snc1 at the PM.
FIGURE 5:
Internal pools of Snc1 can be recycled to the PM in snx4Δ and rcy1Δ mutants. (A) Cells expressing a Cu-inducible mNG-Snc1 vector were induced for 1 h and then treated with LatA and cycloheximide and imaged at various time points over the course of an hour; images shown are of single planes. (B) For each time point, the fluorescence intensity at the PM was taken as a ratio as total cellular fluorescence and graphed (n = 30). Scale bar represents 2 µm.
Nonetheless, this result indicates that, in WT cells, the cytoplasmic Snc1-positive structures are readily accessible to the recycling machinery.
In rcy1Δ cells at t = 0, there was very little mNG-Snc1 visible at the cell surface; instead, the bulk of the cargo occupied both punctate and tubular structures within the cell. Compared to the WT, these structures were slower to deplete and delivery of Snc1 to the PM was incomplete. However, over the course of an hour, these internal structures showed a loss of signal intensity and mNG-Snc1 became localized to the PM to a greater extent, indicating that a portion of mNG-Snc1 was still accessible to recycling machinery in rcy1Δ cells. Even after an hour, a proportion of mNG-Snc1 was not recycled back to the PM and the measurable difference between WT and rcy1∆ cells at this stage is accounted for by a few tubular structures and punctae in the rcy1∆ cells where mNG-Snc1 appeared to be trapped (Figure 5, A and B).
Likewise, in the snx4Δ mutants, there was very little detectable Snc1 at the PM at t = 0. Instead, the majority of the protein was found on and within the vacuole, as well as within bright cytoplasmic punctate structures. These structures were persistent, and mNG-Snc1 was slow to appear at the PM, but most of the punctae disappeared by the 30-min time point, at which time mNG-Snc1 became more robustly localized to the PM. The vacuole, it seems, is a terminal destination for mNG-Snc1 in these cells, as the signal in this organelle did not deplete over time. These results suggest that nonvacuolar pools of Snc1 are still able to be recycled in snx4Δ mutants, albeit at a slower rate compared with WT cells (Figure 5, A and B).
We next asked whether recycling would be completely blocked in snx4Δrcy1Δ double mutants under these conditions. At t = 0, there is almost no mNG-Snc1 at the cell surface; instead, it was observed within the vacuole as well as in small internal structures. These structures largely persisted throughout the course of this experiment and mNG-Snc1 was very slow to appear at the cell surface. This apparent rate of recycling was slower than the measured rates for either the snx4Δ or the rcy1Δ single mutants, but showed that even in this double mutant condition, a small fraction of the internal pool of Snc1 was still competent for recycling. We hypothesize that, in this condition, a small amount of the cargo is capable of being recycled in a nonspecific manner by alternative means, perhaps the AP-1 or retromer pathways (Figure 5, A and B).
Loss of retromer in snx4Δrcy1Δ double mutants blocks Snc1 recycling
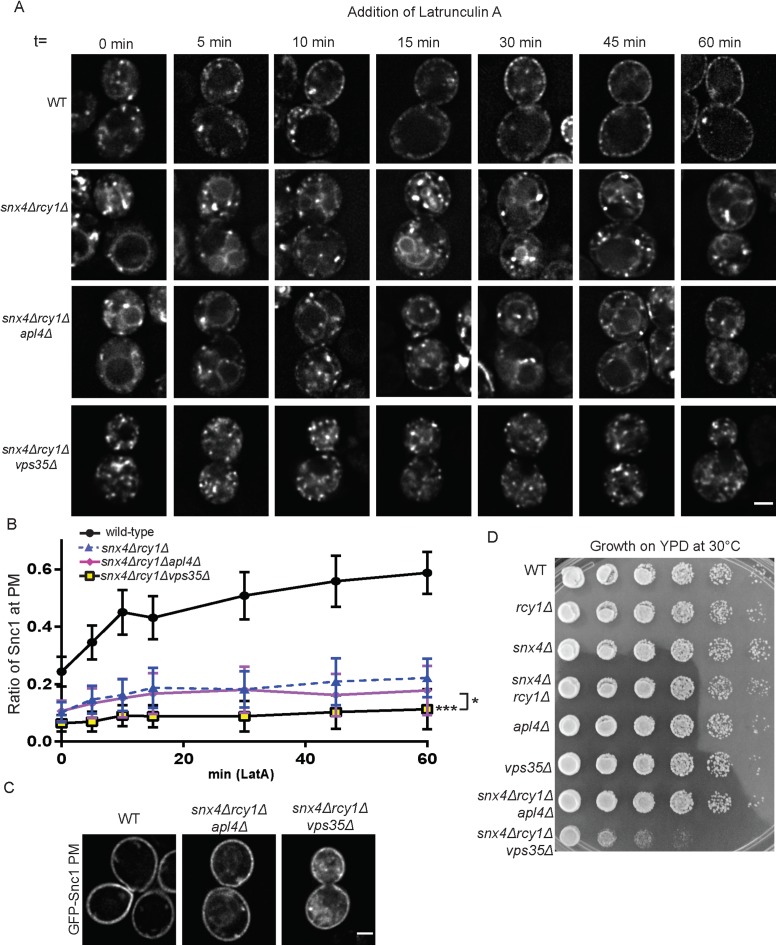
Despite previous reports finding that Snc1 trafficking was unaffected in retromer mutants (Hettema et al., 2003), we reasoned that a small contribution of retromer to recycling might be missed in cells with the Rcy1- and Snx4-dependent routes intact. To test this hypothesis, we generated a snx4Δrcy1Δvps35Δ triple mutant to ablate retromer function. We then tested these strains in a LatA experiment and visualized the localization of mNG-Snc1 over the course of an hour. As displayed in Figure 6, A and B, there was a very strong defect observed in mNG-Snc1 recycling and we saw very little accumulation of mNG-Snc1 at the cell surface in snx4Δrcy1Δvps35Δ triple mutants. mNG-Snc1 appeared to be completely retained within internal punctae that did not appreciably deplete even at later time points. In addition, the retromer mutation prevented mNG-Snc1 transport to the vacuole normally caused by snx4∆ (compare snx4∆rcy1∆ cells to snx4∆rcy1∆vps35∆ cells). This striking result suggested that, in conditions in which neither the Rcy1/Drs2/COPI- nor the Snx4-dependent pathways were available, small amounts of Snc1 are capable of recycling to the TGN from postendocytic structures via the retromer pathway.
FIGURE 6:
Loss of retromer in snx4Δrcy1Δ double mutants blocks Snc1 recycling. (A) WT and mutant cells expressing a Cu-inducible mNG-Snc1 vector were induced for 1 h and then treated with LatA and cycloheximide and imaged at various time points over the course of an hour; images shown are of single planes. (B) For each time point, the fluorescence intensity at the PM was taken as a ratio as total cellular fluorescence and graphed (n = 30). (C) Cells expressing an endocytosis deficient GFP-Snc1(PM) were imaged at 1000×. Images shown are single planes. (D) WT and mutant cells were cultured overnight in liquid culture and were transferred to a 96-well plate. These cultures then underwent serial 10-fold dilutions five times, were plated on YPD plates, and incubated at 30°C for 2 d before imaging. Scale bars represent 2 µm.
The role of the clathrin adaptor protein AP-1 in trafficking between the Golgi and endosomes has been unclear. Some studies proposed an anterograde function of AP-1 in delivering cargo from the TGN to endosomes (Le Borgne and Hoflack, 1997; Liu et al., 2008), other studies suggest a role in EE to TGN trafficking (Valdivia et al., 2002; Copic et al., 2007) (although Snc1 is unaffected in AP-1 single mutants [ Xu et al., 2017]), and a third possibility is that AP-1 mediates retrograde transport from the TGN to earlier (or younger) Golgi cisternae (Liu et al., 2008; Casler et al., 2019). In some cases, cargoes that are primarily retained in the Golgi/endosomal system are secreted to the PM in AP-1 mutants (Valdivia et al., 2002; Liu et al., 2008). The observation that a significant fraction of mNG-Snc1 appeared to be trapped internally in snx4∆rcy1∆ and was rather slowly exported in LatA-treated cells suggested it was trapped in a loop of retrograde and anterograde transport within the Golgi. Therefore, we disrupted the AP-1 subunit Apl4 to generate a snx4∆rcy1∆apl4∆ triple mutant and to assess the impact on Snc1 trafficking. Compared to snx4∆rcy1∆, the snx4∆rcy1∆apl4∆ triple mutant displayed a reduction in mNG-Snc1 trapped at the PM of LatA-treated cells, but the difference at the 60-min time point was not statistically significant (Figure 6, A and B). It was still possible to see a slight accumulation of Snc1 at the cell surface in snx4∆rcy1∆apl4∆ cells at later time points in this triple mutant. Thus, AP-1 is not involved in retaining mNG-Snc1 in the Golgi of snx4∆rcy1∆ cells. Compared to inactivating retromer, loss of AP-1 in this background also appears to impair mNG-Snc1 transport to the vacuole. These results suggest there is no significant role for AP-1 and clathrin in delivering Snc1 to an export competent site, although a minor role cannot be excluded.
The striking defects observed in these triple mutants led us to ask whether or not these phenotypes were indicative of a diminished ability to recycle internalized cargoes, or perhaps a more general disruption of secretory or exocytic function. To test this, we again used the GFP-Snc1(PM) mutant that lacks an AP180 endocytic signal to assess whether or not these strains were capable of secretion (Figure 6C). We observed that, in the snx4∆rcy1∆apl4∆ triple mutant, the vast majority of GFP-Snc1(PM) localized to the PM, though a small number of internal structures persisted. Similarly, GFP-Snc1(PM) was present at the PM in the snx4Δrcy1Δvps35Δ triple mutant, though there was a prominent cytoplasmic haze in these cells, as well as small number of punctate structures akin to those seen in snx4∆rcy1∆apl4∆. These results suggest that both of these triple mutants are secretion competent and that the lack of accumulation of GFP-Snc1 at the PM in these cells during the LatA experiment can be attributed to failure of recycling machinery, rather than a block in secretion. The internal pools of GFP-Snc1(PM) observed in snx4∆rcy1∆apl4∆ and snx4Δrcy1Δvps35Δ may arise from cargo internalized by AP180-independent bulk-phase endocytosis, a partial impairment of secretory function, or missorting at the level of the Golgi.
To measure the influence of perturbing endosome to Golgi trafficking on growth, we performed a serial dilution growth assay with WT, single mutant, and triple mutant strains deficient for recycling proteins (Figure 6D). After 2 d growth at 30°C, we saw no appreciable growth defects in any combination of our single or double mutants, relative to WT. Indeed, even the snx4Δrcy1Δapl4Δ grew at a relatively normal rate. In contrast, snx4Δrcy1Δvps35Δ showed a severe growth defect compared with all other strains. This synthetic growth defect among snx4∆, rcy1∆. and vps35∆ is consistent with these factors acting in three parallel pathways for endosome to Golgi transport.
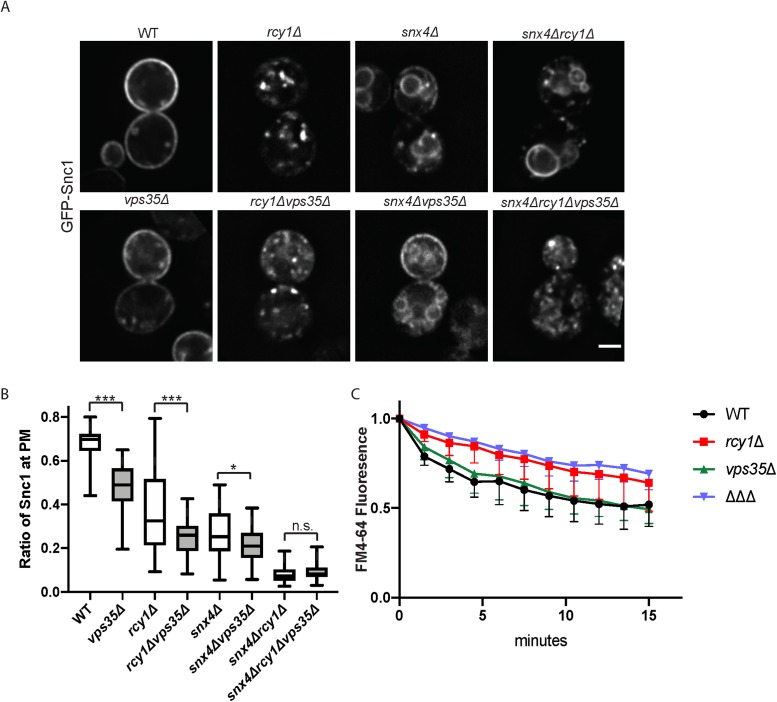
A previous, nonquantitative analysis suggested GFP-Snc1 recycling is independent of retromer (Hettema et al., 2003). In addition, it was possible that GFP-Snc1 only engages retromer when the primary routes are disrupted. To address these issues, we assayed our suite of retromer mutants for their ability to recycle GFP-Snc1. Our quantitative analysis reveals a modest, but statistically significant, defect in GFP-Snc1 localization the vps35Δ single mutant (Figure 7A). We also report enhanced defects in rcy1Δvps35Δ and snx4Δvps35Δ double mutants, as compared with rcy1Δ or snx4Δ single mutants, respectively, though these defects are not as strong as those observed in snx4Δrcy1Δ. There was no measurable difference in GFP-Snc1 recycling between snx4Δrcy1Δ and snx4Δrcy1Δvps35Δ mutants in this steady-state assay. We next measured the ability of WT and vps35∆ cells to recycle endocytosed FM4-64 back to the cell surface. The retromer single-mutant mutant did not display a defect in FM4-64 recycling (Figure 7C and MacDonald and Piper, 2017) and the defect in the snx4∆rcy1∆vps35∆ triple mutant (∆∆∆) mirrored that of rcy1∆ single mutants. Taken together, these results provide evidence that Rcy1/Drs2/COPI- and Snx4-dependent trafficking routes are the primary drivers of Snc1 recycling, and the retromer complex is able to retrieve modest amounts of this cargo back to the TGN but plays no role in FM4-64 recycling (Figure 7).
FIGURE 7:
Influence of Retromer on endosomal recycling. (A) WT and mutant cells expressing GFP-Snc1 were imaged at 1000×. Images shown are single planes chosen from a z-stack series. (B) Quantification of images captured in A (n = 50). (C) Measurement of FM4-64 postendocytic recycling in WT and mutant cells. Fluorescence intensity was normalized to the initial value for each strain. Data represent three independent experiments.
Localization of Snc1 in snx4Δ rcy1Δ vps35Δ mutants
Our initial observations with snx4Δrcy1Δvps35Δ revealed a pattern of GFP-Snc1 localization that was different from that presented in vps35Δ, rcy1Δ, or snx4Δ single mutants. Very little vacuolar localization was observed in the triple mutant and in addition to punctae, a dispersed, lacy fluorescence pattern was observed. To further characterize these compartments, we coexpressed a mCH-tagged Tlg1 and GFP-Snc1 in the triple mutant. The morphology of the mCh-Tlg1 compartments was greatly perturbed in snx4Δrcy1Δvps35Δ triple mutants and these structures colocalized with GFP-Snc1 primarily to the lacy network of membranes (Supplemental Figure S4A). Interestingly, the PI(3)P-binding marker of LEs, dsRED-FYVE, was less perturbed in this background and there was a small degree of overlap between dsRED-FYVE-decorated compartments and GFP-Snc1 punctae. These data suggested that Snc1 primarily accumulates in structures distinct from the LE in snx4Δrcy1Δvps35Δ mutants (Supplemental Figure S4A).
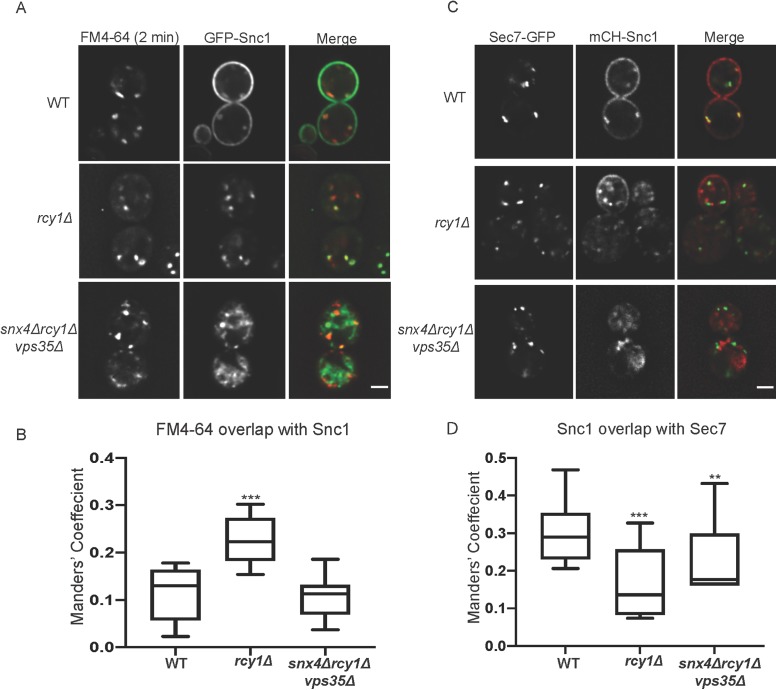
We also used a short pulse of FM4-64 to label endosomes at a very early stage of maturation to compare with GFP-Snc1 in WT, rcy1∆, and snx4Δrcy1Δvps35Δ cells (Figure 8A). To achieve this, cells were incubated on ice with FM4-64 for 30 min and were washed in ice-cold media. The samples were incubated at 30°C for 2 min and were placed back on ice and immediately imaged. While the majority of GFP-Snc1 was retained at the PM in WT cells, the internal Snc1-positive punctae colocalize tightly with FM4–64-labeled structures. Similar to the relationship between Tlg1 and Snc1 (Figure 3, A and B), GFP-Snc1 and FM4-64 primarily label the same structures in rcy1Δ mutants and indeed overlap to a greater extent than is measured in WT cells (Figure 8A). The FM4-64–decorated structures in snx4∆rcy1∆vps35∆ are morphologically similar to those seen in WT cells at early time points of FM4-64 internalization. Quantitatively, FM4-64 and GFP-Snc1 colocalize with one another to an equal extent in snx4Δrcy1Δvps35Δ mutants and WT cells; however, this measure overestimates their relationship, as GFP-Snc1 is dispersed over the majority of the cytoplasm in the triple mutants but is primarily at the PM of WT cells. The majority of the prominent GFP-Snc1 positive structures within snx4Δrcy1Δvps35Δ mutants remain distinct from membranes labeled by FM4-64 after a 2-min internalization (Figure 8, A and B). A longer pulse of FM4-64 (18 min) showed that the dye could transit through the endosomal system to the vacuolar membrane in snx4Δrcy1Δvps35Δ cells with similar kinetics as WT cells. Partial colocalization of these later endosomal structures with GFP-Snc1 was also observed (Supplemental Figure S4B). These results indicate that general traffic through the endosomes is not grossly disrupted in these triple mutants and GFP-Snc1 appears to be broadly distributed within this membrane system.
FIGURE 8:
Subcellular distribution of Snc1 in snx4Δrcy1Δvps35Δ mutants. (A) WT and mutant cells expressing GFP-Snc1 were incubated with FM4-64 and were allowed to internalize the dye for 2 min at 30°C before being put on ice and imaged at 1000×. (B) Channels were separated and thresholded and correlation was measured by finding the MCC between the channels (n = 20). (C) WT and mutant cells expressing Sec7-GFP and mCH-Snc1 were cultured and imaged at 1000×. (D) Channels were separated and thresholded and correlation was measured by finding the MCC between the channels (n = 30).
We also tested whether Snc1 accumulates within TGN compartments marked by Sec7. In WT cells, the cytoplasmic structures containing mCH-Snc1 were frequently colabeled with Sec7-GFP. Interestingly, while overlap between EE markers (both Tlg1 and short-pulsed FM4-64) and Snc1 was increased in rcy1Δ mutants, there was a significant decrease in the colocalization of mCH-Snc1 and Sec7-GFP in this background. Structures labeled by these markers appeared in close proximity to one another, though they were spatially distinct in rcy1Δ mutants. Similarly, we see a significant decrease in overlap between these two markers in snx4Δrcy1Δvps35Δ triple mutants compared with WT, and morphology of Sec7-labeled compartments appeared normal. The apparent increase in the MCC for the triple mutant relative to rcy1∆ is likely caused by the increased cytoplasmic haze exhibited by mCH-Snc1 in the triple mutant rather than an actual increase in colocalization. We conclude that the ability of Snc1 to enter Sec7-marked compartments is substantially perturbed in these mutants (Figure 8, C and D).
DISCUSSION
In this study, we have characterized recycling mechanisms that function in retrieving endocytosed cargoes from the endosomal system back to the PM. While the proteins Rcy1 and Snx4 have been implicated in Snc1 recycling in the past (Galan et al., 2001; Hettema et al., 2003), here we present data to suggest that these proteins act independently of one another, at different compartments within the endosomal system. Further, we provide evidence that a small amount of Snc1 (∼10%) within postendocytic structures is capable of using retromer as a means for retrieval to the TGN (Figures 6, A and B, and 7, A and B). Phenotypic analysis of snx4Δ and rcy1Δ indicates that both of these genes are important in the efficient recycling of Snc1, yet these defects manifest in contrasting ways. The snx4∆ mutant missorts Snc1 to the vacuole, while rcy1∆ cells retain Snc1 in an intracellular compartment marked by Tlg1 (Figures 2 and 3). However, Snc1 can be recycled to the PM in both single mutants where it can be trapped behind an endocytosis block, albeit with a lower efficiency than what is observed in WT cells (Figure 5). Analysis of snx4Δrcy1Δ double mutants reveals an additive phenotype, one in which Snc1 recycling is nearly ablated and most of Snc1 localizes to the vacuole (Figure 1, A and B). This phenotype suggests that we have disrupted the major avenues of Snc1 retrieval in these cells leading to its delivery to the vacuole by default. Additive defects are also observed in retromer double mutants (vps35∆rcy1∆ and vps35∆snx4∆) and the triple mutant is completely defective in Snc1 recycling (Figures 6 and 7). Thus, we think it is likely that Snc1 is recycled by all three routes in WT cells.
In contrast, FM4-64 recycling back to the cell surface from endosomes appears to be restricted to only the Rcy1/Drs2/COPI route. FM4-64 recycling is perturbed in rcy1∆ cells, but not snx4∆ or vps35∆, and snx4Δrcy1Δ or snx4Δrcy1Δvps35∆ strains are no more impaired than the rcy1Δ single mutant (Figure 7). Therefore, the Rcy1/Drs2/COPI pathway is required for recycling of FM4-64, while neither Snx4 nor retromer appear to participate in this activity. In addition, we find that rcy1∆ and a COPI mutant specifically defective in Snc1 recycling perturb FM4-64 recycling equivalently (Figure 1F). How FM4-64 is specifically incorporated into the early recycling pathway and not the later pathways is unclear, but may reflect differences in membrane lipid composition of carriers formed by each protein complex. The Rcy1/Drs2/COPI-dependent carriers may retain a lipid composition similar to the PM in which this lipophilic dye partitions. Conversely, the membranes of Snx4 or retromer carriers may be sufficiently remodeled to prevent this partitioning of FM4-64.
Snc1 recycling studies have typically been performed using a Snc1 construct driven from the strong promoter that substantially overexpresses Snc1 (Lewis et al., 2000; Galan et al., 2001; Hua et al., 2002; Hettema et al., 2003; Furuta et al., 2007; Xu et al., 2017). We have examined Snc1 trafficking at both high and low expression levels differing by approximately 100-fold and the results are generally congruent. Snc1 overexpression appears to saturate the AP180-dependent internalization step of endocytosis causing an aberrant accumulation at the PM of WT cells. By making endocytosis rate limiting, trafficking defects caused by rcy1∆ are easier to score as the steady-state Snc1 distribution shifts from PM to internal punctae. At low expression levels, the Snc1 mislocalization phenotype is more subtle in rcy1∆ cells, but we still observe a loss from the PM (Figure 1, C and D) and an increase in colocalization with Tlg1 (Figure 3, E and F). Partial mislocalization of Snc1 to the vacuole in snx4∆ cells is easily observed at low expression levels because the vacuole is morphologically distinct from Golgi and endosomes, which are not easily distinguishable from each other. Similarly, we see an additive effect of knocking out Rcy1 and Snx4 at both levels of expression, further suggesting these proteins mediate different routes for Snc1 recycling (Figure 2, C and D).
The localization of Snx4/Atg20 and Rcy1/Drs2/COPI to distinct compartments supports their action in separate pathways. Snx4 localizes to PI3P-positive PVEs that are also decorated with retromer, although it appears that retromer and Snx4 are operating independently to retrieve different cargoes back to the Golgi (Burda et al., 2002; Hettema et al., 2003; Ma et al., 2017). In addition, a significant amount of Snx4 localizes to the limiting membrane of the vacuole and may function at this site to retrieve Snc1 to either the PVE or the Golgi (Suzuki and Emr, 2018). While Snc1 is primarily routed to the vacuole in snx4Δ mutants, a smaller portion of Snc1 localizes to peri-vacuolar, DsRed–FYVE-positive punctae corresponding to LE/PVEs, suggesting this could be the site where Snc1 is normally packaged into Snx4 transport carriers. Drs2 and Rcy1 localize to compartments marked by Sec7 and Tlg1 and do not significantly colocalize with PVE markers (Liu et al., 2007; Ma and Burd, 2019). While most of COPI localizes to early Golgi compartments, about 18% colocalizes with Tlg1 and significantly less COPI colocalizes with the TGN marker Sec7 (Xu et al., 2017). Combined, these observations imply that Snc1 is retrieved from PVEs or vacuoles by Snx4 and is recycled from the Tlg1 compartment by Rcy1/Drs2/COPI.
What is the nature of the Tlg1 compartment, and are Snc1 and FM4-64 recycled by way of the Golgi or through a direct endosome-to-PM mechanism that bypasses the Golgi? In WT cells, Tlg1 punctae acquire Sec7 over a period of a few minutes, suggesting the Tlg1 compartment is potentially a trans-Golgi cisternae that matures into the TGN (Day et al., 2018). However, it is also possible that Tlg1-positive EEs recruit Sec7 as part of their maturation process, or merge with Sec7-decorated membranes. The observation that FM4-64 recycling is not perturbed when Sec7 is inactivated (MacDonald and Piper, 2017) suggests that fast recycling takes place from Tlg1 compartments prior to their acquisition of Sec7. This result also implies that the Rcy1/Drs2/COPI-dependent recycling of FM4-64 occurs independently of the Golgi, perhaps in a direct EE to PM pathway (Figure 9). Sec7 is an Arf-GEF and COPI requires Arf-GTP for vesicle formation, but Drs2 binds directly to another Arf-GEF, Gea2, therefore providing another means of activating Arf in the FM4-64 fast recycling pathway (Chantalat et al., 2004). In contrast, Snc1 recycling is perturbed by inactivation of Sec7 (MacDonald and Piper, 2017). This difference in Sec7 dependence for FM4-64 and Snc1 can be explained by our observation that the Snx4-dependent recycling pathway still operates in rcy1∆ cells, and that Snx4-dependent carriers deliver Snc1 to the Golgi where it can be trapped behind the Sec7 block. The GARP Golgi tether is also involved in Snc1 recycling (Conibear et al., 2003; Quenneville et al., 2006), supporting the idea that at least one of the recycling routes passes through the Golgi.
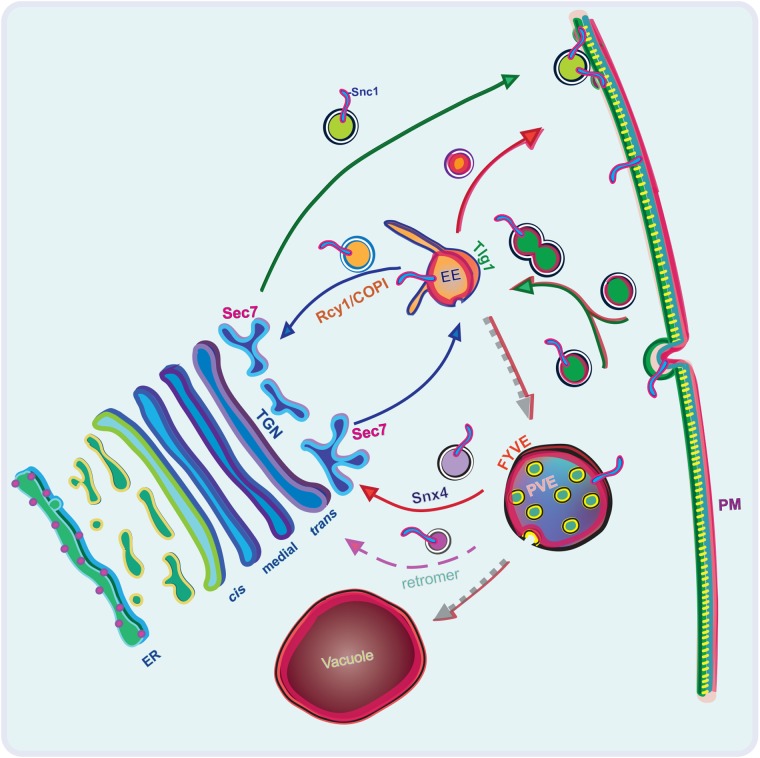
FIGURE 9:
Schematic of postendocytic recycling routes for Snc1. Proposed primary routes of transit for both Rcy1-/COPI- and Snx4-dependent Snc1 recycling after endocytosis, as well as a minor route mediated by retromer. The route of endocytosed FM4-64 is represented by a red outline on the outer leaflet of the PM, in the luminal leaflet of endosomal/vacuolar membranes, and along the arrows denoting the transit and maturation of these compartments. Endocytosed cargoes are first received by a Tlg1-positive EE, where they are either selectively transported by Drs2/Rcy1/COPI back to the Sec7-positive TGN, are directly recycled back to the PM, or remain in the EE as it matures into a PVE. This EE may acquire Sec7 by recruitment from the cytosol or through fusion with Sec7 compartments. As the EE matures into a PVE, specific cargoes are capable of retrieval by either retromer or Snx4-dependent mechanisms. The PVE will ultimately fuse with the vacuole.
Snc1 accumulates in Tlg1-positive compartments in Rcy1/Drs2/COPI-deficient cells, but are these Golgi or endosomal intermediates? An alternative model of endosomal organization in budding yeast has been proposed by Glick and colleagues (Day et al., 2018), one in which endocytosed material is directly trafficked to a Tlg1-positive Golgi cisternae, which later matures into a Sec7-positive TGN, as well as featuring a PVE distinct from these compartments. By this model, Rcy1/Drs2/COPI either would be required for maturation of the Tlg1 compartment (trans-Golgi) into a Sec7-positive TGN, or would promote the cargo-specific transport of Snc1 to the PM by inhibiting retrograde transport of the Snc1 within the Golgi. However, we do not favor these interpretations for the following reasons. Proteins are transported very quickly and efficiently through the yeast secretory pathway, requiring only ∼5 min to move from the ER, through the Golgi, and to the extracellular space (Graham and Emr, 1991). The rcy1∆ and drs2∆ mutants display no discernable defect in the kinetics of protein secretion (Chen et al., 1999; Wiederkehr et al., 2000; Hua et al., 2002; Furuta et al., 2007). COPI mutants show cargo-specific defects in ER to Golgi anterograde transport, but many cargoes are efficiently secreted after inactivating COPI (Gaynor and Emr, 1997). In addition, anterograde transport through the Golgi is thought to be driven by cisternal maturation, which implies that all secretory cargo in the Golgi should be delivered to the cell surface at the same pace, unless they were specifically packaged into retrograde vesicles (Losev et al., 2006; Matsuura-Tokita et al., 2006). Golgi enzymes, for example, are thought to be retained by constant retrograde retrieval in COPI vesicles or potentially AP-1/clathrin-coated vesicles (Tu et al., 2008; Ishii et al., 2016; Casler et al., 2019). Here we show that Snc1 can be secreted from Tlg1-positive compartments in rcy1∆ cells, but requires Snx4 and retromer for recycling. This implies Snc1 must pass through a PVE in rcy1∆ cells in order to be secreted, which is inconsistent with models representing the Tlg1 compartment as a Golgi compartment. However, it was possible that the pools of Tlg1-localized Snc1 we observe in rcy1Δ mutants are stuck in a “treadmilling” cycle of maturation and AP-1– or COPI-mediated retrieval within the Golgi. We found that, rather than accelerating exocytosis, COPI mutants cause the same retention of Snc1 in Tlg1 compartments seen in rcy1∆ or drs2∆ cells (Xu et al., 2017). We also tested whether an AP-1 mutation would suppress Snc1 mislocalization caused by deletion of Rcy1 and Snx4, but found no enhancement of Snc1 exocytosis. In contrast, exomer mutants that display a cargo-specific exocytosis defect for chitin synthase are efficiently suppressed by deletion of AP-1 subunit genes allowing chitin synthase secretion (Valdivia et al., 2002). We believe it is unlikely that Rcy1/Drs2/COPI deficiency induces Snc1 trafficking in an unknown intra-Golgi retrograde pathway to allow retention in the Golgi.
In contrast to the Day et al. (2018) model, which suggests that yeast lack a distinct EE organelle, our observations are more consistent with the view that Rcy1/Drs2/COPI is needed to recycle Snc1 from an EE or recycling endosome (Figure 9). Loss of Rcy1 would perturb the ability of Snc1 and FM4-64 to recycle out of this compartment and be delivered to the PM, but would not completely perturb movement of cargo to PVEs where Snc1 can be recycled by Snx4/Atg20 (primarily) or retromer. By shifting the rate-limiting step of GFP-Snc1 trafficking in rcy1Δ/drs2Δ/COPI mutants away from the saturated endocytic machinery and toward a recycling step at the EE, we force the cell to accumulate a greater-than-normal amount of GFP-Snc1 in these compartments, as the cargo will spend more time there. As these compartments mature toward a PVE, they are now accessible to the Snx4 recycling machinery, which is then able to shuttle Snc1 back to the TGN. For this reason, we do not observe GFP-Snc1 accumulation within the vacuole in rcy1Δ mutants, as GFP-Snc1 is specifically retrieved at a more intermediate stage of endosomal maturation. However, we did note that a portion of Snc1 failed to be exported in LatA-treated rcy1∆ cells over 60 min (Figure 5A), suggesting some accumulation in a dead-end compartment that cannot deliver content to either the Golgi or the vacuole. This effect was strongly pronounced in the triple mutant deficient for Rcy1, Snx4, and retromer. The retromer mutation in this background prevents transport of Snc1 to the vacuole and the SNARE is primarily retained in intracellular punctae after LatA treatment, indicating Snc1 cannot be secreted from these compartments (Figure 6A). In contrast, Snc1 bearing a mutation in its endocytosis signal accumulates at the PM in the snx4∆rcy1∆vps35∆ triple mutant, implying that newly synthesized Snc1 can be secreted in these cells, but cannot return to the secretory pathway if it is endocytosed (Figure 6C). We observe partial colocalization of Snc1 with early and LE markers in the triple mutant, including FM4-64 en route to the vacuole. Moreover, the kinetics of FM4-64 transport to the vacuole is not noticeably perturbed in the triple mutant. We speculate that Snc1 is primarily trapped in a dead-end compartment of endocytic origin in the triple mutant, but with some ability to exchange with functional endosomes mediating transport of FM4-64 through the endocytic pathway (resulting in the partial colocalizations we observe).
We propose a model for Snc1 recycling that is consistent with both our observations and those in the literature; Tlg-1 positive endosomes receive endocytosed cargoes and are capable of direct, rapid recycling of some Snc1 and FM4-64 back to the PM. In contrast to proposed models in which Tlg1 localizes solely to a trans-Golgi cisternae that mature into Sec7-labeled TGN, our findings are better represented by the idea that Tlg1-decorated EEs are capable of fusion or bidirectional exchange with the late Golgi cisternae that acquire Sec7. It is also possible that the majority of Tlg1 is in the Golgi of WT cells but Tlg1 cycles between the Golgi and EEs, thus accumulating within an enlarged and aberrant EE lacking Sec7 in rcy1∆ cells. This possibility is consistent with the observation that subcellular fractionation shows a pool of Tlg1 with Golgi membranes marked by Kex2 and a second pool of Tlg1 fractionating with membranes significantly denser than the Golgi, which likely represent EEs (Holthuis et al., 1998). Alternatively, it is possible that Sec7 is recruited to both maturing TGN and maturing recycling endosome compartments. These models are supported by our observation that the majority of Snc1 punctae in WT cells colabel with Sec7-mCH, while loss of Rcy1 results in a notable spatial separation of these markers (Figure 8, C and D). In contrast, Snc1 overlap with the Tlg1 compartments increases in rcy1Δ mutants, relative to WT (Figure 3, A and B). This series of results would be unexpected if Tlg1 compartments were simply maturing into Sec7-positive TGNs as part of Golgi maturation. The apparent uncoupling of Tlg1 and Sec7 compartments is especially difficult to reconcile with Tlg1-to-Sec7 models of maturation of the Golgi, given the fact that other cargoes progress through the secretory pathway in cells deficient for Rcy1 without kinetic delay (Wiederkehr et al., 2000). Instead, these data support a model in which Tlg1-positive EEs either acquire Sec7 or Sec7-positive TGN-derived vesicles fuse with Tlg1-EEs, each in a Rcy1-dependent manner. In each of these models, the LE or PVE is a distinct and stable compartment that receives carrier-mediated transport of materials from the Tlg1-positive endosome as well as the Sec7-positive TGN.
Taken together, our data support a model (Figure 9) in which endocytosed Snc1 is transported to the endosomal system before being recycled back to the TGN for additional rounds of secretion, or possibly via a direct route to the cell surface. Once Snc1 arrives in the EE, it is packed into COPI vesicles (Xu et al., 2017) in a Ub- and Rcy1-dependent manner and trafficked either back to the TGN or directly to the PM. As the contents of the Tlg1-positive endosome are transported to or mature into the PVE, Snx4/Atg20 is recruited to the membrane where it is capable of retrieving any remaining Snc1 back to the TGN. Snc1 that remains in the endosomal system and is not retrieved by either of these trafficking pathways then appears capable of using retromer to transit back to the TGN, though less efficiently. Recent work from Ma and Burd (2019) described similar observations on Snc1 recycling in snx4∆rcy1∆ double mutants and support the conclusion that these factors act in parallel recycling pathways. We hypothesize that both the Rcy1/Drs2/COPI and the Snx4 pathways are regularly employed to recycle secretory v-SNARES, and likely additional cargoes yet to be identified, in WT cells. Snc1 (∼10,000 copies per cell) and Snc2 (∼6000 copies per cell) are abundant proteins that constantly cycle through the secretory and endosomal systems in yeast. Thus, if only one means of retrieving these important cargoes existed, it could be easily saturated during particularly high-traffic periods (like polarized bud growth), leading to degradation of these proteins within the vacuole. In this type of system, it is logical that evolution would provide redundant means of recycling these important components. These studies serve to deepen our understanding of the dynamic endomembrane system and will pave the way for future studies into these distinct trafficking pathways.
MATERIALS AND METHODS
Strains and plasmids
Standard media and techniques for growing and transforming yeast were used. Plasmid constructions were performed using standard molecular manipulation. A list of strains used in this study can be found in Table 1. A list of plasmids used in this study can be found in Table 2.
TABLE 1:
Strains used in this study.
| Name | Genotype | Source | Notes |
|---|---|---|---|
| JBY001 | MATa his3∆1 leu2∆0 ura3∆0 met15∆0 | Invitrogen | BY4741 |
| JBY002 | MATα his3∆1 leu2∆0 ura3∆0 lys2∆0 | Invitrogen | BY4742 |
| JBY003 | MATa his3∆1 leu2∆0 ura3∆0 met15∆0 drs2∆::KanMX | This study | |
| JBY120 | MATa his3∆1 leu2∆0 ura3∆0 met15∆0 rcy1∆::KanMX | This study | |
| JBY122 | MATa his3∆1 leu2∆0 ura3∆0 met15∆0snx4∆::KanMX | This study | |
| JBY130 | MATa his3∆1 leu2∆0 ura3∆0 met15∆0snx4∆::KanMX rcy1∆::HIS3 | This study | |
| JBY150 | MATa his3∆1 leu2∆0 ura3∆0 met15∆0 yap1802∆::KanMX | Invitrogen | |
| JBY155 | MATa his3∆1 leu2∆0 ura3∆0 met15∆0 snx4∆::KanMX rcy1∆::HIS3 apl4∆::Hyg | This study | via cross to JBY130 |
| JBY157 | MATa his3∆1 leu2∆0 ura3∆0 met15∆0snx4∆::KAN rcy1∆::HIS3 vps35∆::Hyg | This study | via cross to JBY130 |
| PXY2175A | MATα his3∆1 leu2∆0 ura3∆0 lys2∆0sec27Δ::Hyg p315- sec27Δ2-304 | Xu et al., 2017 | |
| JBY203 | MATa his3∆1 leu2∆0 ura3∆0 met15∆0 snx41∆::KanMX | Invitrogen | |
| JBY204 | MATa his3∆1 leu2∆0 ura3∆0 met15∆0 Atg20∆::KanMX | Invitrogen | |
| JBY207 | MATa his3∆1 leu2∆0 ura3∆0 met15∆0 drs2∆::KanMX rcy1∆::HIS3 | This study | |
| JBY208 | MATα his3∆1 leu2∆0 ura3∆0 lys2∆0sec27Δ::Hyg p315- sec27Δ2-304 rcy1∆::KanMX | This study | |
| JBY210 | MATa his3∆1 leu2∆0 ura3∆0 met15∆0 vps35∆::KanMX | Invitrogen | |
| JBY211 | MATa his3∆1 leu2∆0 ura3∆0 met15∆0 vps35∆::KanMX rcy1∆::HIS3 | This study | |
| JBY212 | MATa his3∆1 leu2∆0 ura3∆0 met15∆0 vps35∆::KanMX snx4∆::HIS3 | This study |
TABLE 2:
lasmids used in this study.
| Name | Plasmid | Source |
|---|---|---|
| GFP-Snc1 (TPI) | pRS416-TPI-GFP-Snc1 | Lewis et al., 2000 |
| mCH-Snc1 | pRS416-ADH-mCH-Snc1 | This study |
| mCH-Tlg1 | pRS315-mCH-Tlg1 | Xu et al., 2013 |
| dsRed-FYVE | pRS425-dsRED-FYVE | Katzmann et al., 2003 |
| GFP-Snc1(PM) | pRS416-GFP-Snc1(PM) | Lewis et al., 2000 |
| mScarlet-Tlg1 | pRS315-mScarlet-Tlg1 | This study |
| mNG-Snc1 | pRS416-CUP1-mNG-Snc1 | This study |
| Kan KO | pfa6a::Kanamycin | Longtine et al., 1998 |
| Hyg KO | pfa6a::Hygromycin | Longtine et al., 1998 |
| His KO | pfa6a::HIS3 | Longtine et al., 1998 |
| Sec7-mEGFPx6 | pSEC7-mEGFPx6::URA3 | Papanikou et al., 2015 |
| GFP-Snc1 (PRC1) | pRS416-PRC1-GFP-Snc1 | Ma and Burd, 2019 |
| GFP-Snc1-DDD | pRS416-PRC1-GFP-Snc1(DDD) | Ma and Burd, 2019 |
| DUb-GFP-Snc1 | pRS416-DUb-GFP-Snc1 | Xu et al., 2017 |
| DUb*-GFP-Snc1 | pRS416-DUb*-GFP-Snc1 | Xu et al., 2017 |
Imaging and image analysis
To visualize GFP-, DsRed-, mScarlet, or mCH-tagged proteins, cells were grown to early- to-mid-logarithmic phase in BY4741 and BY4742 WT cells and mutants derived from this background, harvested, and resuspended in imaging buffer (10 mM Na2PHO4, 156 mM NaCl, 2 mM KH2PO4, and 2% glucose). Copper-inducible mNG strains were induced at 100 µM CuCl2 for 1 h before imaging. Cells were then mounted on glass slides and observed immediately at room temperature. Images were acquired using a DeltaVision Elite Imaging System (GE Healthcare Life Sciences, Pittsburgh, PA) equipped with a 100× objective lens followed by deconvolution using softWoRx software (GE Healthcare Life Sciences). Overlay images were created using the merge channels function of ImageJ software (National Institutes of Health [NIH]). GFP-Snc1 at the PM is quantified as previously described (Hankins et al., 2015). Briefly, concentric circles were drawn just inside and outside the PM using ImageJ to quantify the internal fluorescence and total fluorescence, respectively. The internal fluorescence was subtracted from the total to give the GFP intensity at the PM. This value was taken as a ratio of the total fluorescence to give the proportion of GFP intensity at the PM. At least 50 randomly chosen cells from three biological replicates (independently isolated strains with the same genotype) were used to calculate the mean and SD. To quantify GFP colocalization with mCH markers, a MCC for the two markers in each cell (n = 2, 20 cells each) was calculated using the ImageJ plugin Coloc 2 with Auto Local Thresholds applied in ImageJ (Manders et al., 1993; Bolte and Cordelieres, 2006). To assess the overlap of GFP-Snc1 with DsRed-FYVE, the two channels were split, the total nonvacuolar GFP-positive structures inside the cell were counted, and then the GFP-Snc1 punctae that also contained DsRed-FYVE were counted to determine the proportion of GFP-Snc1 structures that colocalized with DsRed-FYVE (n = 40 cells). To visualize the cell wall of cells, the cells were labeled with fluorescently labeled ConA at 50 µg/ml for 30 min and then spun down and washed in fresh media three times. Colocalization between ConA and mNG-Snc1 was conducted as described above. Statistical analysis consisted of one-way analyses of variance and Tukey post hoc tests for multiple comparisons. Probability values of less than 0.05, 0.01, and 0.001 were used to show statistically significant differences and are represented with *, **, or ***, respectively.
LatA endocytosis block
To assess the potential for intracellular pools of mNG-Snc1 to be recycled to the cell surface, cells expressing a Cu-inducible mNG-Snc1 plasmid were induced in 100 µM CuCl2 for 1 h, then washed with fresh Cu-free media, and treated with 10 µM of the F-actin polymerization inhibitor LatA and 250 µg/ml of the peptide translation inhibitor cycloheximide. Samples were taken at various time points after the addition of these inhibitors and were immediately placed on ice for the duration of the experiment. At this point, the cells were imaged and analyzed as described above. Previous experiments were conducted under these conditions, but translation was halted by the removal of CuCl2, rather than by the addition of cylcoheximide. The results of these experiments were consistent with the cycloheximide-treated cells, implying that the promoter shut-off was efficient under Cu-free conditions and that the cycloheximide translational block did not influence the trafficking phenotypes observed.
Immunoblotting
Cells were grown overnight until they reached mid-log phase. Their ODs were recorded and used to calculate the volume needed to harvest 2 OD units of cells. These cells were pelleted and the supernatant was aspirated off. Pellets were then resuspended in 100 µl of lysis buffer with SDS nonboiling sample buffer and were incubated at 95°C for 2 min before undergoing glass-bead lysis. The 0.5 equivalent OD600 of lysate was then loaded into a 4–20% gradient SDS–PAGE gel (Bio-Rad Cat. #4561095) and current was applied. The contents of these gels were transferred to PVDF membranes that were blocked with 10% milk in TBST for 1 h and then incubated with mouse anti-GFP antibody 1:1000 overnight at 4°C. IRDye 680LT Goat anti-Mouse secondary antibody was then applied at 1:10,000 for 1 h before the membranes were washed three times with TBST and one time with phosphate-buffered saline. Blots were visualized using an Odyssey CLx Licor imaging system. For mNG blotting, the samples were run in 4–20% gradient SDS–PAGE (Bio-Rad Cat. #4561095) and then transferred onto PVDF membrane (Bio-Rad Cat. #1620177). The PVDF membrane was blocked in blocking buffer (TBST + 10% nonfat milk) for 1 h at room temperature, then incubated with anti-mNG antibodies (1:1000) in blocking buffer overnight. After washing with TBST, the membrane was incubated with anti-mouse immunoglobulin G–horseradish peroxidase (1: 50,000 in TBST +10% nonfat milk) for 1 h at room temperature. The membrane was washed by TBST and then detected with chemiluminescent substrate (Thermo Scientific SuperSignal West Pico PLUS Cat. #34580). The membrane was imaged with ChemiDoc MP (Bio-Rad).
Antibodies
Rabbit mNG tag antibody (Cell Signaling Cat. #53061, 1:1000) was purchased from Cell Signaling Technology. Mouse anti-GFP (1C9A5, 1:1000) antibodies were purchased from the Vanderbilt Antibody and Protein Repository (Nashville, TN). IRDye 680LT Goat anti-mouse (LI-COR Biosciences Cat# 827–11080, RRID:AB_10795014, 1:20,000) was purchased from LI-COR Biosciences (Lincoln, NE).
Growth assay
Cells were grown overnight to saturation and then diluted 1:10 in sterile water into wells of the first column of a 96-well plate. These wells then underwent five 10-fold dilutions into adjacent columns. The 96-well plates were replica plated onto yeast extract peptone with 2% dextrose (YPD) plates and grown at 30°C for 2 d before being photographed.
Molecular cloning
Plasmid constructions were performed using standard molecular manipulation with Gibson Assembly (Xu et al., 2017). Mutations were introduced using a Q5 Site-Directed Mutagenesis Kit. All plasmids were confirmed by sequencing. Table 2 contains a list of plasmids used in this study.
FM4-64 recycling assays
Cells were grown overnight to mid-log phase in 10-ml cultures of YPD. These cultures were then pelleted and resuspended in 1 ml YPD and incubated with 40 µM FM4-64 solubilized in dimethyl sulfoxide for 8 min at 30°C. Cells were then placed on ice for 10 min before being pelleted and washed three times in fresh, ice-cold media. While still on ice, these cultures were resuspended in 4 ml of SM media and then 12 250-µl aliquots were loaded into a 96-well plate. The plates were taken off of the ice for 4 min to allow them to heat to room temperature and then read using a BD Accuri C6 flow cytometer, taking 10,000 events per reading with roughly 90 s elapsing between each time point. The relative fluorescence of each well was recorded and set relative to the first recorded time point to measure the decrease in fluorescence over the course of the experiment. The data displayed are the mean of three replicated experiments. Additional recycling assays were performed in a similar manner, but using a BD FACSAria III (BD Biosciences) running FACSDiva 6.1.3. Forward and side scatter were used to isolate single-cell populations. Error bars are SEM.
Supplementary Material
Acknowledgments
We are grateful to our colleague Jason MacGurn for his assistance with microscopy and input on the manuscript. We also thank Hugh Pelham, Kathy Gould, David Katzmann, Chris Burd, and Scott Emr for providing plasmids used in these studies. Flow cytometry experiments were performed in the Vanderbilt Medical Center (VMC) Flow Cytometry Shared Resource. The VMC Flow Cytometry Shared Resource is supported by NIH grants to the Vanderbilt Ingram Cancer Center (P30-CA68485) and the Vanderbilt Digestive Disease Research Center (P30-DK058404). The research reported in this publication was made possible by support from the National Institute of General Medical Sciences of the NIH under award number R01 GM118452 to T.R.G. and in part by the National Institute of General Medical Sciences of the NIH under award number T32 GM008554.
Abbreviations used:
- ConA
concanavalin A
- DUb
deubiquitinase
- EE
early endosome
- ER
endoplasmic reticulum
- LatA
latrunculin A
- LE
late endosome
- MCC
Manders’ correlation coefficient
- mCH
mCherry
- mNG
mNeonGreen
- MVB
multivesicular bodies
- PM
plasma membrane
- PVE
prevacuolar endosome
- PX
phox homology
- SCF
Skp1-Cullin-F-box
- TGN
trans-Golgi network
- Ub
ubiquitin
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-05-0290) on February 19, 2020.
REFERENCES
- Arlt H, Auffarth K, Kurre R, Lisse D, Piehler J, Ungermann C. (2015). Spatiotemporal dynamics of membrane remodeling and fusion proteins during endocytic transport. Mol Biol Cell , 1357–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR. (2000). Endocytosis and the development of cell polarity in yeast require a dynamic F-actin cytoskeleton. Curr Biol , 1587–1590. [DOI] [PubMed] [Google Scholar]
- Bean BD, Davey M, Conibear E. (2017). Cargo selectivity of yeast sorting nexins. Traffic , 110–122. [DOI] [PubMed] [Google Scholar]
- Bethune J, Wieland FT. (2018). Assembly of COPI and COPII vesicular coat proteins on membranes. Annu Rev Biophys , 63–83. [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J Microsc-Oxford , 213–232. [DOI] [PubMed] [Google Scholar]
- Burda P, Padilla SM, Sarkar S, Emr SD. (2002). Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J Cell Sci , 3889–3900. [DOI] [PubMed] [Google Scholar]
- Burston HE, Maldonado-Baez L, Davey M, Montpetit B, Schluter C, Wendland B, Conibear E. (2009). Regulators of yeast endocytosis identified by systematic quantitative analysis. J Cell Biol , 1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casler JC, Papanikou E, Barrero JJ, Glick BS. (2019). Maturation-driven transport and AP-1-dependent recycling of a secretory cargo in the Golgi. J Cell Biol , 1582–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantalat S, Park SK, Hua Z, Liu K, Gobin R, Peyroche A, Rambourg A, Graham TR, Jackson CL. (2004). The Arf activator Gea2p and the P-type ATPase Drs2p interact at the Golgi in Saccharomyces cerevisiae. J Cell Sci , 711–722. [DOI] [PubMed] [Google Scholar]
- Chen SH, Chen S, Tokarev AA, Liu F, Jedd G, Segev N. (2005). Ypt31/32 GTPases and their novel F-box effector protein Rcy1 regulate protein recycling. Mol Biol Cell , 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Ingram MF, Rosal PH, Graham TR. (1999). Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J Cell Biol , 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Shah AH, Segev N. (2011). Ypt31/32 GTPases and their F-Box effector Rcy1 regulate ubiquitination of recycling proteins. Cell Logist , 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Cleck JN, Stevens TH. (2003). Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol Biol Cell , 1610–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copic A, Starr TL, Schekman R. (2007). Ent3p and Ent5p exhibit cargo-specific functions in trafficking proteins between the trans-Golgi network and the endosomes in yeast. Mol Biol Cell , 1803–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day KJ, Casler JC, Glick BS. (2018). Budding Yeast Has a Minimal Endomembrane System. Dev Cell , 56–72.e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster A, Pecheur EI, Michel F, Winsor B, Letourneur F, Friant S. (2004). Ent5p is required with Ent3p and Vps27p for ubiquitin-dependent protein sorting into the multivesicular body. Mol Biol Cell , 3031–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig KM, Rice LM, Pollock E, Brunger AT. (1999). Folding intermediates of SNARE complex assembly. Nat Struct Biol , 117–123. [DOI] [PubMed] [Google Scholar]
- Furuta N, Fujimura-Kamada K, Saito K, Yamamoto T, Tanaka K. (2007). Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol Biol Cell , 295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JM, Wiederkehr A, Seol JH, Haguenauer-Tsapis R, Deshaies RJ, Riezman H, Peter M. (2001). Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol Cell Biol , 3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau A, Oguievetskaia K, Ungermann C. (2014). Function and regulation of the endosomal fusion and fission machineries. Cold Spring Harb Perspect Biol , a016832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Emr SD. (1997). COPI-independent anterograde transport: cargo-selective ER to Golgi protein transport in yeast COPI mutants. J Cell Biol , 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Emr SD. (1991). Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J Cell Biol , 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote E, Carr CM, Novick PJ. (2000). Ordering the final events in yeast exocytosis. J Cell Biol , 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan S, Marash M, Weinberger A, Gerst JE. (2002). t-SNARE phosphorylation regulates endocytosis in yeast. Mol Biol Cell , 1594–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamatsu H, Fujimura-Kamada K, Yamamoto T, Furuta N, Tanaka K. (2014). Interaction of the phospholipid flippase Drs2p with the F-box protein Rcy1p plays an important role in early endosome to trans-Golgi network vesicle transport in yeast. J Biochem , 51–62. [DOI] [PubMed] [Google Scholar]
- Hankins HM, Sere YY, Diab NS, Menon AK, Graham TR. (2015). Phosphatidylserine translocation at the yeast trans-Golgi network regulates protein sorting into exocytic vesicles. Mol Biol Cell , 4674–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Lewis MJ, Black MW, Pelham HR. (2003). Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J , 548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B, Baryshnikova A, Brown GW. (2018). Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst , 192–205.e193. [DOI] [PubMed] [Google Scholar]
- Holthuis JC, Nichols BJ, Dhruvakumar S, Pelham HR. (1998). Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J , 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Fatheddin P, Graham TR. (2002). An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol Biol Cell , 3162–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari J, Helenius A. (2011). Endosome maturation. EMBO J , 3481–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M, Suda Y, Kurokawa K, Nakano A. (2016). COPI is essential for Golgi cisternal maturation and dynamics. J Cell Sci , 3251–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. (2001). Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell , 145–155. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Stefan CJ, Babst M, Emr SD. (2003). Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol , 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R, Hoflack B. (1997). Mannose 6-phosphate receptors regulate the formation of clathrin-coated vesicles in the TGN. J Cell Biol , 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HR. (2000). Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell , 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Hua Z, Nepute JA, Graham TR. (2007). Yeast P4-ATPases Drs2p and Dnf1p are essential cargos of the NPFXD/Sla1p endocytic pathway. Mol Biol Cell , 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Surendhran K, Nothwehr SF, Graham TR. (2008). P4-ATPase requirement for AP- 1/clathrin function in protein transport from the trans-Golgi network and early endosomes. Mol Biol Cell , 3526–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast , 953–961. [DOI] [PubMed] [Google Scholar]
- Losev E, Reinke CA, Jellen J, Strongin DE, Bevis BJ, Glick BS. (2006). Golgi maturation visualized in living yeast. Nature , 1002–1006. [DOI] [PubMed] [Google Scholar]
- Ma M, Burd CG. (2019). Retrograde trafficking and quality control of yeast synaptobrevin, Snc1, are conferred by its transmembrane domain. Mol Biol Cell , 1729–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Burd CG, Chi RJ. (2017). Distinct complexes of yeast Snx4 family SNX-BARs mediate retrograde trafficking of Snc1 and Atg27. Traffic , 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Kumar S, Purushothaman L, Babst M, Ungermann C, Chi RJ, Burd CG. (2018). Lipid trafficking by yeast Snx4 family SNX-BAR proteins promotes autophagy and vacuole membrane fusion. Mol Biol Cell , 2190–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C, Piper RC. (2017). Genetic dissection of early endosomal recycling highlights a TORC1-independent role for Rag GTPases. J Cell Biol , 3275–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders EMM, Verbeek FJ, Aten JA. (1993). Measurement of colocalization of objects in dual-color confocal images. J Microsc-Oxford , 375–382. [DOI] [PubMed] [Google Scholar]
- Matsuura-Tokita K, Takeuchi M, Ichihara A, Mikuriya K, Nakano A. (2006). Live imaging of yeast Golgi cisternal maturation. Nature , 1007–1010. [DOI] [PubMed] [Google Scholar]
- Miller SE, Sahlender DA, Graham SC, Honing S, Robinson MS, Peden AA, Owen DJ. (2011). The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell , 1118–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Miyashita N, Xu C, Toyoshima I, Sugita Y, Inesi G, Toyoshima C. (2005). Structural role of countertransport revealed in Ca(2+) pump crystal structure in the absence of Ca(2+). Proc Natl Acad Sci USA , 14489–14496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikou E, Day KJ, Austin J, II, Glick BS. (2015). COPI selectively drives maturation of the early Golgi. eLife , e13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protopopov V, Govindan B, Novick P, Gerst JE. (1993). Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell , 855–861. [DOI] [PubMed] [Google Scholar]
- Quenneville NR, Chao TY, McCaffery JM, Conibear E. (2006). Domains within the GARP subunit Vps54 confer separate functions in complex assembly and early endosome recognition. Mol Biol Cell , 1859–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN. (2012). The retromer complex—endosomal protein recycling and beyond. J Cell Sci , 4693–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al. (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods , 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Pelham HR. (2001). An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J , 5991–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell , 209–219. [DOI] [PubMed] [Google Scholar]
- Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. (1993). A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell , 409–418. [DOI] [PubMed] [Google Scholar]
- Stahl PD, Barbieri MA. (2002). Multivesicular bodies and multivesicular endosomes: the “ins and outs” of endosomal traffic. Sci STKE , e32. [DOI] [PubMed] [Google Scholar]
- Suzuki SW, Emr SD. (2018). Membrane protein recycling from the vacuole/lysosome membrane. J Cell Biol , 1623–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L, Tai WC, Chen L, Banfield DK. (2008). Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science , 404–407. [DOI] [PubMed] [Google Scholar]
- Valdivia RH, Baggott D, Chuang JS, Schekman RW. (2002). The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell , 283–294. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol , 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A, Avaro S, Prescianotto-Baschong C, Haguenauer-Tsapis R, Riezman H. (2000). The F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J Cell Biol , 397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Baldridge RD, Chi RJ, Burd CG, Graham TR. (2013). Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J Cell Biol , 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Hankins HM, MacDonald C, Erlinger SJ, Frazier MN, Diab NS, Piper RC, Jackson LP, MacGurn JA, Graham TR. (2017). COPI mediates recycling of an exocytic SNARE by recognition of a ubiquitin sorting signal. eLife , e28342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.