Abstract
While a publicly available benchmark dataset provides a base for the development of new algorithms and comparison of results, hospital-based data collected from the real-world clinical setup is also very important in AI-based medical research for automated disease diagnosis, prediction or classifications as per standard protocol. Primary data must be constantly updated so that the developed algorithms achieve as much accuracy as possible in the regional context. This dataset would support research work related to image segmentation and final classification for a complete decision support system (https://doi.org/10.1016/j.tice.2020.101347) [1]. Liquid-based cytology (LBC) is one of the cervical screening tests. The repository consists of a total of 963 LBC images sub-divided into four sets representing the four classes: NILM, LSIL, HSIL, and SCC. It comprises pre-cancerous and cancerous lesions related to cervical cancer as per standards under The Bethesda System (TBS). The images were captured in 40x magnification using Leica ICC50 HD microscope collected with due consent from 460 patients visiting the O&G department of the public hospital with various gynaecological problems. The images were then viewed and categorized by experts of the pathology department.
Keywords: Cervical cancer, Pap smear, Liquid-based cytology, 40x, Cervical pre-cancerous lesions, Cervical cancerous lesions
Specifications table
| Subject | Computer Science, Computer Vision, and Pattern Recognition, |
| Specific subject area | Medical Image Processing, Cervical Cancer, Cell segmentation, Cell classification |
| Type of data | Images |
| How data were acquired | Images were captured using a Leica DM 750 microscope with camera model ICC50 HD, in 400x (40x objective lens × 10x eyepiece) magnifications (size 2048 × 1536pixels). |
| Data format | Raw JPG |
| Parameters for data collection | Images were captured in 400x (40x objective lens × 10x eyepiece) magnifications. The size of the images is 2048 × 1536 pixels. |
| Description of data collection | Liquid-based cytology provides more uniform fixation with a cleaner background and well-preserved samples for further HPV tests other than conventional Pap tests and hence it is preferred here. The LBC pap smear slides were collected from three distinguished medical diagnostic centers of the NER regions, India namely Babina Diagnostic Pvt. Ltd, Imphal, Gauhati Medical College and Hospital, Guwahati and Dr. B. Barooah Cancer Institute, Guwahati. All samples involve ethical clearance protocol from the three diagnostic centers along with patient consent from a total of 460 patients undergoing cervical screening tests. The images were captured in 400x magnifications using Leica DM 750 microscope, model ICC50 HD connected with the camera and a high-configured computer and software. The images represent the sub-categories of cervical lesions (malignant and pre-malignant) as NILM (Negative for Intraepithelial lesion or malignancy), LSIL (Low-grade intraepithelial lesions), HSIL (High-grade intraepithelial lesions), and SCC (Squamous Cell Carcinoma). |
| Data source location | 1. Babina Diagnostic Pvt. Ltd, Imphal, India |
| 2. Dr. B. Borooah Cancer Research Institute, Guwahati, Assam, India | |
| 3. Gauhati Medical College and Hospital, Guwahati, Assam, India | |
| Data accessibility | Hussain, Elima (2019), “Liquid-based cytology pap smear images for multi-class diagnosis of cervical cancer”, Mendeley Data, V4. |
| https://data.mendeley.com/datasets/zddtpgzv63/4 | |
| Related research article | E. Hussain, L.B. Mahanta, C. Ray, R. Kanta, Tissue and Cell A comprehensive study on the multi-class cervical cancer diagnostic prediction on pap smear images using a fusion-based decision from ensemble deep convolutional neural network, Tissue Cell. 65 (2020) 101347. |
Value of the data
-
•
This dataset can be used for a comparative assessment of one's experimental findings against publicly available conventional pap smear datasets such as the Sipakmed dataset by Plissiti et. al [2] and the Pap smear benchmark dataset by Jantzen et. Al [3]. Thin-Prep Liquid-based cytology pap smear datasets like Cervix93 by Phoulady et. al [4] also exists for experimental analysis.
-
•
Researchers can use this dataset for computer-assisted diagnosis of cervical cancer which necessitates interpretation of such images for different image segmentation algorithms, feature extraction or feature selection methodologies and in final classification step (both binary as well as multi-class classification). In the case of binary classification (normal vs. abnormal class), the NILM category can be grouped as normal whereas LSIL, HSIL, and SCC can be grouped as abnormal class.
-
•
Deep learning methodologies oriented classification or semantic segmentation tasks can also be incorporated with further data augmentation techniques using these images.
1. Data
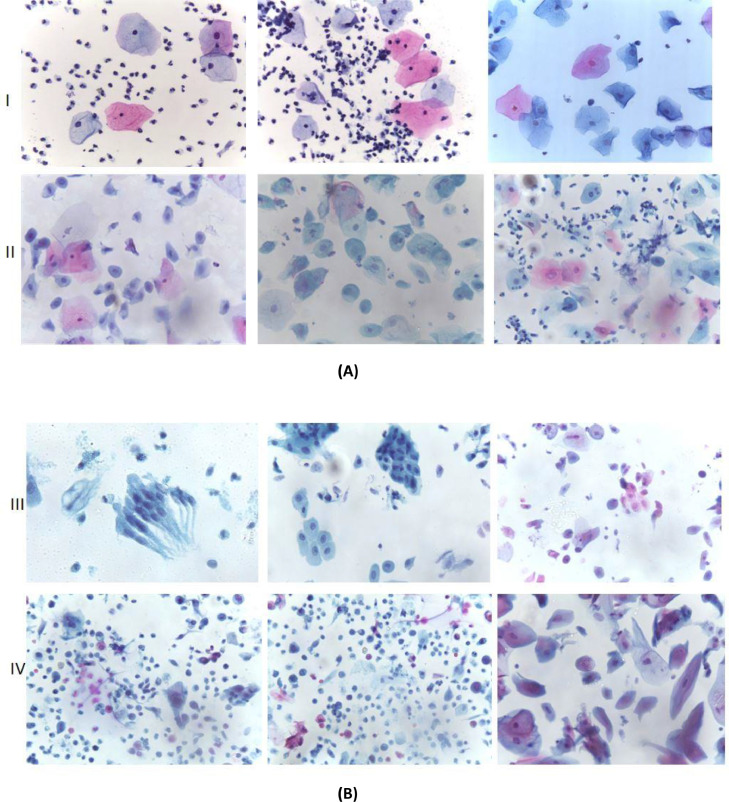
The dataset has been sub-divided into four categories each depicting the four classes of cervical cancer as per TBS standards. Table 1 quantifies the total images belonging to each category, a few samples of which are illustrated in Fig. 1. Total 963 images were captured from Pap smear slides at 400x magnification, out of which 613 images belong to NILM or normal category and 350 images belong to the abnormal category. The cytological description on cervical cell morphometry for distinguishing the following category is well explained by Gray et. al [5].
Table 1.
Dataset description.
 |
Fig. 1.
(A) Images belonging to class (I) NILM and (II) LSIL, and (B) Images belonging to class (III) HSIL and (IV) SCC.
The final classification step can be enhanced for better prediction accuracy with image pre-processing, image segmentation and feature extraction steps which will require quantitative analysis for identification of abnormal features based on cell-level morphometry like shape, color or texture analysis. Such an automated system based on artificial intelligence will enable computer-assisted diagnosis for early detection of pre-cancerous lesions to combat cervical cancer. This will contribute to rapid prognosis therapy in the end.
2. Experimental design, materials, and methods
Images in the datasets were collected using liquid-based cytology (LBC) (sure-path) technique in the Obstetric and Gynecology department of Gauhati Medical College and Hospital, the primary public healthcare center of the region. LBC technique involves a small brush to collect the sample with target from transformation zone (where a columnar epithelial cell changes into a squamous epithelial cell) in the same way as a conventional smear test, but instead of transferring the smear specifically to a microscopic slide, the samples are kept into a container with additive fluid. This fluid deals with evacuating different types of unwanted debris, like mucus, blood cells, etc., before setting a layer of cells on the slides. The vial containing cervical samples was finally placed at a vortex with 3000 rpm for 15-20 seconds to break mucotic and blood particles. After adding density reagent to the sample, it undergoes sedimentation and centrifugation at 2500 rpm for 5 minutes. This is mainly done so that particles having heavy molecular weight get settled down at the bottom of the slide. After one or two alcohol wash, the slides were stained using Haemotoxylin and Eosin (H&E) staining protocol.
These slides were then used to capture images using a Leica ICC50 HD microscope at 400x. The 400x magnification provides a better view of smear level image per slides than 100x and 200x with distinct cellular features as per the concerned categories. Ten best quality images per slides were acquired and maintained in a simple excel file along with medical reports per patient. While capturing these images, it is ensured that minimal overlap of image sections in a particular slide is happening. So images were essentially acquired by moving the microscope eyepiece over the slides in a sequential pattern. Although a subjective error is probable in this process, this sequence is repeated throughout to keep this error at a minimal percentage. The images were categorized as NILM, LSIL, HSIL and SCC based on the patient's report and finally confirmed with an expert pathologist's review from the pathology department. These images may now undergo different image processing tasks subjective to computer vision and machine learning fields.
Transparency document
Transparency documents associated with this article can be found in the online version at https://doi.org/10.1016/j.tice.2020.101347.
Acknowledgements
We acknowledge the Department of Biotechnology (DBT), Govt. of India for providing funds (grant no-DBTNER/Health/48/2016). Authors would like to thank the Department of Obstetrics & Gynaecology, Guwahati Medical College & Hospital, Bhangagarh, Guwahati, Assam (GMCH) for LBC set up. Authors would also like to acknowledge Dr. Anup K. Das, Senior Pathologists, Arya Wellness Centre, Guwahati, Assam for his valuable guidance mostly during the data acquisition phase and throughout. Lastly, authors would also like to thanks Dr. Dhabali Singh, Senior Pathologists, Babina Diagnostics, Imphal for contributing adequate LSIL, HSIL and SCC slides.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.105589.
Appendix. Supplementary materials
References
- 1.Hussain E., Mahanta L.B., Ray C., Kanta R. Tissue and Cell A comprehensive study on the multi-class cervical cancer diagnostic prediction on pap smear images using a fusion-based decision from ensemble deep convolutional neural network. Tissue Cell. 2020;65 doi: 10.1016/j.tice.2020.101347. [DOI] [PubMed] [Google Scholar]
- 2.Plissiti A.C.M.E., Dimitrakopoulos P., Sfikas G., Nikou C., Krikoni O. SIPAKMED: A new dataset for feature and image based classification of normal and pathological cervical cells in Pap smear images. IEEE Int. Conf. Image Process. 2018; Athens, Greece; 7-10 Oct. 2018. p. 2018. [Google Scholar]
- 3.Jantzen J., Dounias G. Proceeding NISIS-2006 Symp. 2006. The Pap Smear Benchmark.http://mde-lab.aegean.gr/index.php/downloads [Google Scholar]
- 4.Phoulady H.A., Moutan P.R. CVPR. 2018. A New Cervical Cytology Dataset for Nucleus Detection and Image Classification (Cervix93) and Methods for Cervical Nucleus Detection.https://github.com/parham-ap/cytology_dataset [Google Scholar]
- 5.Gray W., Kocjan G. Churchill Livingstone; 2010. Diagnostic cytopathology. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.