Abstract
Grapevine downy mildew, caused by oomycete fungus Plasmopara viticola, is one of the most devastating diseases of grapes across the major production regions of the world. Although many putative effector molecules have been identified from this pathogen, the functions of the majority of these are still unknown. In this study, we analyzed the potential function of 26 P. viticola effectors from the highly virulent strain YL. Using transient expression in leaf cells of the tobacco Nicotiana benthamiana, we found that the majority of the effectors could suppress cell death triggered by BAX and INF1, while seven could induce cell death. The subcellular localization of effectors in N. benthamiana was consistent with their localization in cells of Vitis vinifera. Those effectors that localized to the nucleus (17/26) showed a variety of subnuclear localization. Ten of the effectors localized predominantly to the nucleolus, whereas the remaining seven localized to nucleoplasm. Interestingly, five of the effectors were strongly related in sequence and showed identical subcellular localization, but had different functions in N. benthamiana leaves and expression patterns in grapevine in response to P. viticola. This study highlights the potential functional diversity of P. viticola effectors.
Keywords: grapevine, Plasmopara viticola, RxLR effectors, subnuclear localization, biological activities
Introduction
Grapevine downy mildew, caused by the oomycete Plasmopara viticola (Berk. & M. A. Curtis) Berl. & De Toni, is one of the most important diseases negatively impacting grape production worldwide (Panstruga and Dodds, 2009). As a biotrophic oomycete, P. viticola attacks almost all green tissues of the plant. In the asexual life cycle, an encysting zoospore forms a germinative tube which penetrates the leaf through a stoma, producing haustoria to obtain nutrients from the host (Panstruga and Dodds, 2009; Dodds and Rathjen, 2010).
Pathogenic oomycetes secrete effector molecules into the plant to disturb plant innate immunity, facilitating penetration and colonization (Judelson and Blanco, 2005; Kamoun, 2006; Tyler et al., 2006; Birch et al., 2009; Bozkurt et al., 2012; King et al., 2014). These effectors are divided into two important classes, apoplastic and cytoplasmic effectors. Cytoplasmic effectors include proteins of the RxLR and Crinkler (CRN) families (Mestre et al., 2016). The RxLR protein effectors are the largest class of effectors and have been the most extensively studied (Kamoun, 2006; Stassen and Van den Ackerveken, 2011). These comprise an amino- (N-)terminal signal peptide, followed by an RxLR motif (Arg-x-Leu-Arg, where x represents an any amino acid), followed by an EER motif (Kamoun, 2006; Birch et al., 2008). The signal peptide directs the secretion of the effector from the fungus, whereas the RxLR-EER motif participates in the delivery of the effector to the host cell (Whisson et al., 2007; Dou et al., 2008; Grouffaud et al., 2009). An exact RxLR-EER sequence is not always required for translocation to the host cell (Dou et al., 2008; Tian et al., 2011; Chen et al., 2013; Ye et al., 2015).
The battle between host and pathogen is multi-layered. Pathogen-associated molecular patterns (PAMPs) can be detected by pattern recognition receptor (PRR) proteins in the host cell membrane in a general mechanism referred to as PAMP-triggered immunity (PTI) (Jones and Dangl, 2006; Dodds and Rathjen, 2010). However, pathogens often secrete effectors inside the host cell to interfere with PTI, which the plant may respond to by effector-triggered immunity (ETI) (Dangl and Jones, 2001). For example, INF1, an elicitin secreted by Phytophthora infestans that acts as a PAMP, is recognized by the receptor-like protein ELICITIN RESPONSE (ELR), which then associates with the 1-ASSOCIATED KINASE1/SOMATIC EMBRYOGENESIS RECEPTOR KINASE 3 (BAK1/SERK3) protein kinase to trigger cell death (Du et al., 2015). This defense-related hypersensitive response (HR) has similirities with programmed cell death in induced by the pro-apoptotic mouse protein BAX (BCL2-Associated X) (Lacomme and Santa Cruz, 1999). However, oomycetes have evolved effectors that suppress PTI and ETI. For example, CRN70 and Avr1k, the two effectors of P. Infestan, have been showed to suppress a resistance (R) protein 3/AVR3a (gene to gene model)-induced HR (Kelley et al., 2010; Rajput et al., 2014; Fawke et al., 2015).
With recent advances in grapevine genomics and genomics technologies, rapid progress has been made in understanding resistance mechanisms to P. viticola. Numerous putative effectors have been identified from P. viticola based on their early expression during the infection process (Riemann et al., 2002; Sebastian et al., 2010; Mestre et al., 2012). Utilizing a cDNA-AFLP approach, 96 P. viticola sequences were obtained from infected grapevine leaves (Polesani et al., 2008). Additionally, many P. viticola cDNA sequences have been cataloged from expressed sequence tags (ESTs) from infected plant tissues (As-sadi et al., 2011; Cabral et al., 2011; Mestre et al., 2012). Fifty-four ESTs encoding potential secreted hydrolytic enzymes and effectors were identified from germinated zoospores of P. viticola (As-sadi et al., 2011; Cabral et al., 2011; Mestre et al., 2012). Based on the presence of a secretory signal sequence and an RxLR or CRN motif, 51 RxLR effectors and 10 CRN effectors were identified from P. viticola (Yin et al., 2015). RNA-based sequencing has been used to identify RxLR/CRN genes that are differentially expressed upon infection of grapevine (Brilli et al., 2018). The genome of P. viticola has been sequenced and hundreds of effectors including RxLR and CRN have been identified (Dussert et al., 2016, 2018; Yin et al., 2017). In recent years, there has been rapid progress in understanding P. viticola RxLR effectors. For example, the effector PvRxLR28, which can suppress cell death caused by some cell death elicitors, exhibits a burst of expression 6 h after infection (Xiang et al., 2016). This contributes to the pathogenicity of this strain. In contrast, the effector PvRxLR16 significantly enhances resistance, and can trigger cell death by itself in N. benthamiana cells (Xiang et al., 2017). A total of 83 putative RxLR effectors were identified from P. viticola “JL-7-2”; three of these were localized to chloroplasts while one was localized in both chloroplast and mitochondria (Liu et al., 2018). Effectors generally need to enter the host cell to carry out their function. The late blight resistance protein R1 and effector AVR1 require nuclear localization to activate the immune response, and PvRxLR16 and AVH241 need to localize to the nuclear and plasma membrane to trigger cell death, respectively (Yu et al., 2012; Du et al., 2015a; Xiang et al., 2017).
In this paper, we carried out genomic and RNA-based sequencing of the P. viticola strain “YL” (Yin et al., 2017). Based on nucleotide sequence, we identified a group of putative P. viticola RxLR effectors. Bioformatic surveys have revealed that a set of 25 P. viticola RxLR effectors were predicted in the genomic of the P. viticola (unpublished). One RxLR effector (PvAVH54804) was digged out during the infection of V. vinifera RNA-seq data (Liu et al., 2019). Here, we studied the virulence function of 26 of the RxLR effectors by transient expression in N. benthamiana cells. We examined both their ability to suppress cell death induced by INF1 and BAX, and their subcellular localization, both in N. benthamiana and V. vinifera. This provides a solid foundation for advanced study of the role of these and other effectors in grapevine downy mildew.
Materials and Methods
Bioinformatics and Sequence Analysis
The sequence of the putative P. viticola RxLR effectors in this paper can be found in GenBank date library of National Center for Biotechnology Information (NCBI), and all accession numbers were listed in Supplementary Table S2. Signal peptide cleavage sites were predicted using the SignalP 4.1 server1, and nuclear localization signals were identified using cNLS Mapper2. Sequences were compared with three published genome sequences (Dussert et al., 2016; Yin et al., 2017; Brilli et al., 2018). The phylogenetic tree was constructed with amino acid sequences using MEGA5 with the neighbor-joining method, 1,000 replicates, and the pairwise deletion option. Multiple alignment was carried out using BioEdit 7 software with ClustalW. Protein sequence alignment was performed using DNAMAN software.
Bacterial Strains and Plasmid Constructions
Escherichia coli strain Top10 (CWBIO) was used for cloning and amplification of recombinant plasmids, and was cultured at 37°C in LB (Luria-Bertani) medium. Agrobacterium tumefaciens strain GV3101 was used for transient expression in planta and was grown at 28°C in LB medium. The open reading frame (ORF) segments of putative P. viticola effectors, minus the predicted N-terminal signal sequence, were cloned from genomic DNA of P. viticola strain “YL” by PCR amplification and specific primers designed based on genomic sequence. PCR products were cloned into the plant expression PVX vector and pCAMBIA2300 vector. For PVX assay, the amplified segments were ligated into PVX vector pGR107 (containing a HA tag sequence) to form PVX-effectors construction. To identify the subcellular localization of those effectors in plant, effector gene sequences were ligated into pCAMBIA2300 vector containing green fluorescent protein (GFP) sequence. The primers in this paper were listed in Supplementary Table S1. The schematic diagrams of the constructs used for experiments were documented in Supplementary Material, Supplementary Figure S8.
Plant Materials, Transient Transformation Expression Assays
Recombinant plasmids were introduced into Agrobacterium by liquid nitrogen flash freezing, and transformants were selected on LB medium with kanamycin (50 μg/mL), gentamycin (50 μg/mL), and rifamycin (50 μg/mL). Agrobacterium containing effector-GFP constructs was cultured in liquid LB medium at 28°C with shaking at 200 rpm/min. After 20 h, cells were collected by centrifugation at 5,000 × g for 3 min, washed twice in 10 mM MgCl2, and then resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, pH 5.7, 200 μM acetosyringone) to a final OD600 of 0.4. Then, the solution was incubated for 3 h in the dark prior to infiltration as described (Van der Hoorn et al., 2000). Leaves of 4 to 5-week-old N. benthamiana and V. vinifera cv. Thompson seedless were subjected to infiltration using a 1-mL needless syringe. The healthy and fully expanded leaves of 4 to 5-week-old N. benthamiana leaves were used for protoplast preparation. The plant materials were grown in a greenhouse under 16-h light/8-h dark photoperiods at 25°C (light) and 18°C (dark). P. viticola isolate “YL” was maintain in grapevine leaves, and subcultured every 7 days at 22°C. To analyze expression levels of putative RxLR effectors, detached leaves of the susceptible V. vinifera cultivar “Pinot Noir” were inoculated with 40 μL P. viticola “YL” sporangial suspension (5 × 104 sporangia / mL) / water (as negative contral) on wet sterile filter papers, and then placed in growth champer at 22°C. Expression was evaluated by RT-PCR at 0 h, 6 h, 12 h, 24 h, 48 h, 72 h, 96 h, and 120 h after inoculation as described in Liu et al. (2019). These experiments were repeated at least three independent experimental repetitions with similar results.
Confocal Microscopy and Image Analysis
For characterizing the subcellular localization of putative P. viticola effectors, transient expression assays were conducted using leaves of 4 to 5-week-old N. benthamiana and V. vinifera plants with GFP-effector fusion proteins. Images were captured by confocal microscopy (Germany, LECIA) 3 days after infiltration. For transient expression in protoplasts, healthy and fully expanded N. benthamiana leaves were used for protoplast preparation. Protoplasts were subjected to transformation using a polyethylene glycol (PEG)-based method (Yoo et al., 2007). To unambiguously mark the nucleus, we also engineered and introduced an expression plasmid (pBI221-mCherry) fusing the NLS from SV40 T large antigen (amino acid sequence PKKKRKV) to a red fluorescent protein. Effector-GFP fusion proteins and GFP, together with NLS-mCherry which was considered as nuclear-localized marker were co-expressed in protoplasts following polyethylene glycol (PEG)-mediated transformation of the protoplasts. Protoplasts were incubated for 20 h under weak lighting at 25°C. Fluorescence of GFP and mCherry was excited at 488 nm and 561 nm, respectively.
Measurement of Ion Leakage
For measurement of ion leakage, five, 1-cm diameter leaf disks were incubated in 5 ml distilled water with gentle shaking (50 rpm/min) for 3 h at room temperature. Ion leakage was measured by a conductivity meter (DDS-307, LeiCi, ShangHai, China) as A. Total ion leakage was measured as B after incubation in a boiling water bath for 25 min. Results were expressed as percentage of total ion leakage A/B.
Protein Extraction and Western Blotting
Recombinant proteins were extracted from N. benthamiana leaves 2 days after infiltration using protein extraction buffer [50 mM HEPES-KOH, pH 7.5, 150 mM KCl, 1 mM EDTA, 0.5% (v/v) Triton X-100, 1 mM dithiothreitol, and 1 × protease inhibitor cocktail (ROCHE)]. Tissue was suspended in 1 × loading buffer, boiled in a water bath for 5 min, and subjected to centrifugation at 10,000 × g for 10 min. The supernatant containing solubilized protein was resolved on a 10% SDS-PAGE gel and transferred to a PVDF membrane. The membrane was blocked with 5% nonfat dry milk in TBST buffer (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.05% Tween-20) for 3 h. Mouse anti-GFP monoclonal antibody and anti-HA-monoclonal antibody (ABclonal) were added to the buffer at a 1:5000 dilution, and the membrane was shaken slowly at 4°C overnight. The membrane was then washed three times in TBST buffer, and goat anti-mouse IRDye 800CW in TBST was then added to the membrane at a dilution of 1:10000, and the membrane was shaken for an additional 1–2 h. The membrane was then washed 3–5 times for 5 min each in TBST and visualized using ChemiDocTM XRS+ Software with excitation at 700 and 800 nm.
RNA Extraction and Expression Pattern Analysis
For analysis of gene expression following P. viticola infection, RNA was purified from inoculated leaves using the Plant RNA Kit (OMEGA, United States). The quality of total RNA was assessed by electrophoresis on 1% agarose gels, and RNA concentration was determined by spectrophotometer (NanoPhotometer®; IMPLEN, CA, United States). Total RNA was treated with gDNA wiper mix (Vazyme) before complementary DNA (cDNA) synthesis. Total RNA (500 ng) was reverse transcribed to cDNA in 20 μL volume using the HiScript® Reverse Transcriptase Kit (Vazyme) according to instructions of the manufacturer. Oligonucleotide primers for RT-PCR assays were designed for specificity to each gene based on sequence information (Supplementary Table S2). A PvActin gene was used as an internal reference (Mestre et al., 2012).
Results
Identification and Phylogenetic Analysis of RxLR Effectors From P. viticola ‘YL’
Based on the presence of an N-terminal, signal-like sequence of 17–30 amino acids and conserved RxLR and EER motif, we identified ∼100 putative RxLR effector genes from genomic sequence of the P. viticola isolate “YL.” Of these, 26 were selected for analyses of function and subcellular localization (Table 1 and Supplementary Table S3). Compared with the P. viticola genome data, 3, 4, and 6 effectors were indentified the same as the published genomes from P. viticola isolate “JL-7-2,” “INRA-PV221” and “PvitFEM01,” respectively. In addition, most of these genes had the same length, and sequences query cover reached up to 96% (25/26), 100% (26/26), and 69% (18/26), but there were 8, 3, and 7 effectors had gaps compared with the three genomes, respectively (Supplementary Tables S4–S6). Two of these, PvAVH8 and PvAVH103, were previously reported as PvRxLR11 and PvRxLR9, respectively (Xiang et al., 2016). The RxLR-EER motifs were located in the N-terminal 20–80 amino acids of the putative effectors. All candidate effectors contained both RxLR and EER motifs except for PvAVH33, which had only an RxLR motif. The deduced amino acid sequences were aligned for 26 proteins, and a phylogenetic tree was constructed (Figure 1). This revealed that five proteins (PvAVH51, 52, 53, 71, and 102) had an especially close relationship, with up to ∼88% sequence homology (Figures 1A,B). The corresponding genes for these five were located on the same chromosome. Because the signal peptide is cleaved upon secretion and not present in the mature protein (Bozkurt et al., 2012; Fawke et al., 2015; Gascuel et al., 2016), we cloned the ORFs, minus the signal peptide sequence, from P. viticola genomic DNA for further experiments.
TABLE 1.
Summary of 26 putative Plasmopara viticola RxLR effectors description.
| Effectors | Peptide size | Signaip prediction | SignalP3-HMM | RxLR conserved motifs | EER motif | Effectors conserved prediction | Genomic location |
| PvAVH1 | 312 | 1–20 | 1 | RLLR | DEER | 46–61 | scaffold_4:1041171-1042109 |
| PvAVH33 | 559 | 1–21 | 0.999 | RLLR | 29–32 | scaffold_140:87996-89675 | |
| PvAVH36 | 726 | 1–19 | 1 | RVLP | DLKNKWAVHAGGEDR | 30–52 | scaffold_128:53655-55835 |
| PvAVH147 | 377 | 1–25 | 0.977 | RGLK | DEEAR | 21–43 | scaffold_59:284672-285805 |
| PvAVH3 | 642 | 1–21 | 0.999 | RFLR | DEER | 20–50 | scaffold_584:8728-10656 |
| PvAVH8(PvRxLR11a) | 202 | 1–23 | 0.992 | RSLD | DEER | 49–61 | scaffold_4:1177443-1178051 |
| PvAVH76 | 630 | 1–19 | 0.997 | RDSR | DEER | 31–49 | scaffold_19:447692-449584 |
| PvAVH21 | 620 | 1–23 | 0.905 | RFLR | DEER | 28–49 | scaffold_19:695886-697748 |
| PvAVH123 | 117 | 1–23 | 0.938 | RFLQ | DEER | 46–67 | scaffold_19:781377-781730 |
| PvAVH11 | 514 | 1–25 | 0.989 | RSLQ | DSEER | 53–70 | scaffold_76:145128-146672 |
| PvAVH133 | 724 | 1–21 | 1 | RILR | DEER | 30–51 | scaffold_128:156764-158938 |
| PvAVH135 | 722 | 1–23 | 0.999 | RILR | DEER | 28–49 | scaffold_128:171504-173672 |
| PvAVH67 | 414 | 1–19 | 0.998 | RFLL | DESR | 36–51 | scaffold_143:19374-20618 |
| PvAVH53 | 526 | 1–21 | 0.999 | RVIR | DAGNGER | 30–51 | scaffold_128:17264-18844 |
| PvAVH103(PvRxLR9b) | 116 | 1–17 | 0.987 | RILR | DGNVNREQER | 49–64 | scaffold_141:183645-183995 |
| PvAVH30 | 445 | 1–19 | 1 | KLLR | DNATNESR | 34–41 | scaffold_143:88038-89375 |
| PvAVH56 | 572 | 1–22 | 0.999 | RDPK | DLKLSAGNEER | 39–50 | scaffold_140:107427-109145 |
| PvAVH35 | 720 | 1–23 | 1 | RNLK | DEER | 37–52 | scaffold_140:56540-58702 |
| PvAVH61 | 729 | 1–19 | 0.999 | RVLR | DFTLSAGNEER | 30–52 | scaffold_140:44341-46530 |
| PvAVH51 | 502 | 1–21 | 0.999 | RVFR | DAGNGER | 30–51 | scaffold_128:4980-6488 |
| PvAVH52 | 526 | 1–19 | 0.999 | HVLR | DAGSEER | 31–53 | scaffold_128:92002-93582 |
| PvAVH71 | 527 | 1–19 | 0.988 | HVLR | DAGSEER | 31–53 | scaffold_128:45536-47119 |
| PvAVH102 | 520 | 1–19 | 0.998 | HVLR | DAGSEER | 31–53 | scaffold_128:32477-34039 |
| PvAVH47 | 399 | 1–25 | 0.999 | RILR | DEER | 30–57 | scaffold_128:247-1446 |
| PvAVH88 | 486 | 1–30 | 0.995 | RNLA | DDER | 36–73 | scaffold_59:287955-289415 |
| PvAVH54804 | 249 | 1–21 | 0.995 | RPLG | DDTNGEDR | 53–70 |
FIGURE 1.
Phylogeneticrelationship of 26 putative RxLR effectors from Plasmopara viticola. (A) A phylogenetic tree was constructed from alignment of predicted amino acid sequences using MEGA 5 with the neighbor-joining method, 1,000 replicates, and the pairwise deletion option. Five very closely related proteins are indicated with black diamonds. (B) Amino acid sequence alignment of the five very closely related RxLR proteins (PvAVH51, 52, 53, 71, and 102). The RxLR and EER motifs are indicated with black boxes. Positions where the amino acid is conserved in all five proteins is shown with black background.
A Subset of P. viticola RxLR Effectors Can Trigger Cell Death When Expressed in N. benthamiana
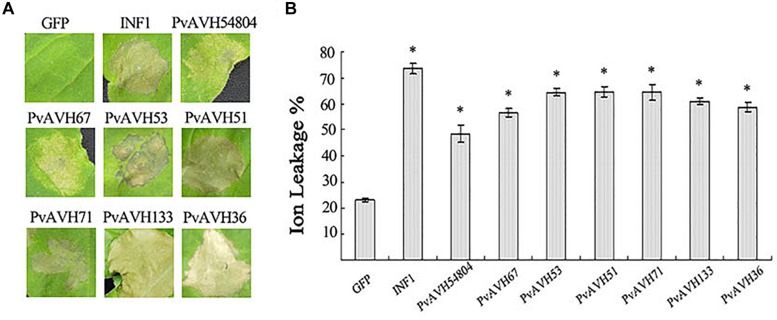
As a first step to assess function of the 26 effectors, we expressed the ORFs in N. benthamiana leaf cells using Agrobacterium infiltration, and assessed their potential to induce cell death. The ORF sequences of the 26 PvRxLR effectors were ligated into the PVX vector and pCAMBIA2300 vector as described in Yin et al. (2019). INF1, a known cell death inducer from P. infestans, and empty vector were used as positive and negative controls, respectively. We found that of the 26 effectors, expression of seven caused cell death by 4 days post infiltration (dpi) (Figure 2A). PvAVH51, 53, 71, 133, and 36 induced necrotic symptoms similar to those induced by INF1, although the effect of INF1 was more immediate (Figure 2A). Cells infiltrated with PvAVH54804 and PvAVH67 exhibited weaker cell death symptoms than the other five effectors. We also evaluated electrolyte leakage in the infiltrated N. benthamiana cells as an independent measure of cell death (Mittler et al., 1999). We found that ion leakage in leaves from plants transiently expressing the seven RxLR effectors was markedly higher than the empty vector control, but was less than the INF1-infiltrated plants (Figure 2B). These results showed that at least seven putative RxLR effectors from P. viticola could promote cell death when expressed heterologously in tobacco.
FIGURE 2.
P. viticola effectors triggered cell death in Nicotiana benthamiana. (A) Photographs of N. benthamiana leaves subjected to infiltration with agrobacterium carrying PVX-effectors at 4 days post-infiltration. Leaves infiltrated with the empty vector control (GFP) or positive control (INF1) is shown as reference. The experiment was repeated at least three times; representative results are shown. (B) Electrolyte (ion) leakage from infiltrated tissue at 4 days post-infiltration. Values were determined from three independent repetitions. Error bars indicate SE, with asterisk indicating significant difference from the empty vector control (GFP). Data are means ± SE based on three independent replicates, each including 10 individual leaves. (∗ indicate P < 0.05 in one-way ANOVA, using Turky HSDa test).
Suppression of INF1- and BAX -Triggered Cell Death by P. viticola Effectors
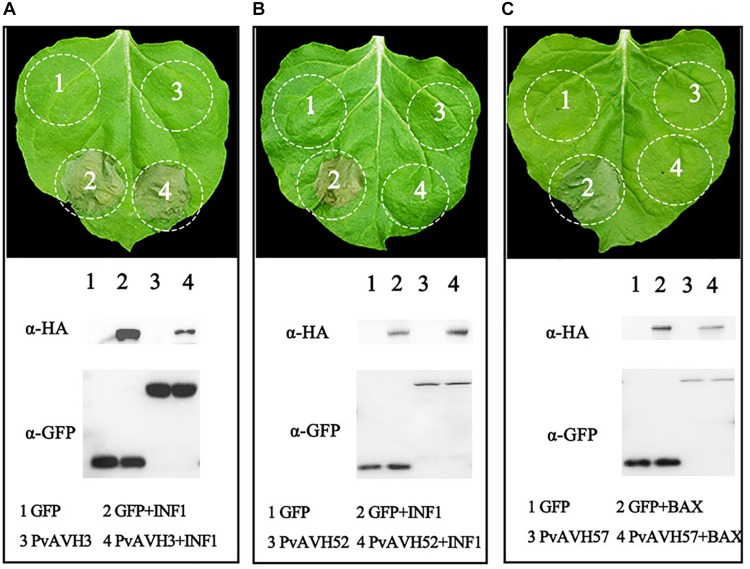
To further analyze the function of the 26 putative P. viticola effectors, we evaluated their potential to suppress cell death induced by INF1 and BAX when co-infiltrated into N. benthamiana (Table 2 and Supplementary Figure S1), an approach previously used to explore effector function (Wang et al., 2011). Agrobacterium carrying the recombinant effector plasmid was infiltrated into N. benthamiana leaves 24 h after infiltration of BAX or INF1, and cell death was examined after a further 4 days. Seven effectors were identified induced cell death themselves in N. benthamiana in above, remaining 19 effectors were analyzed. The results showed that six putative RxLR effectors failed to suppress the activity of INF1 like PvAVH3, 13 effectors could suppress the INF1-induced cell death completly like PvAVH52 and 19 could suppress BAX like PvAVH 47 (Figures 3A–C and Supplementary Figure S1). Western blotting detected the proteins expression and showed the constructs successfully transformed in N. benthamiana leaves spots. Among these, thirteen could suppress both INF1- and BAX-triggered cell deaths (Supplementary Figure S1).
TABLE 2.
Subcellular localization and suppression of cell deathinduced by BAX and INF1.
| Effectors | Subcellular localizationa |
Suppression of PCDb |
||
| INF1 | BAX | PCD | ||
| PvAVH1 | cp | − | + | |
| PvAVH33 | nc | + | + | |
| PvAVH36 | n | cd | ||
| PvAVH147 | nc | + | + | |
| PvAVH3 | n | − | + | |
| PvAVH8(PvRxLR11) | nc | + | + | |
| PvAVH76 | n | − | + | |
| PvAVH21 | n | + | + | |
| PvAVH123 | nc | + | + | |
| PvAVH11 | cp | + | + | |
| PvAVH133 | n | cd | ||
| PvAVH135 | n | + | + | |
| PvAVH67 | n | cd | ||
| PvAVH53 | n | cd | ||
| PvAVH103(PvRxLR9) | n | + | + | |
| PvAVH30 | n | + | + | |
| PvAVH56 | nc | + | + | |
| PvAVH35 | n | − | + | |
| PvAVH61 | n | − | + | |
| PvAVH51 | n | cd | ||
| PvAVH52 | n | + | + | |
| PvAVH71 | n | cd | ||
| PvAVH102 | n | − | + | |
| PvAVH47 | n | + | + | |
| PvAVH88 | n | + | + | |
| PvAVH54804 | nc | cd | ||
a n, nuclear; nc, nuclear and cytoplasm; cp, cytoplasm and plasma membrane. b +, consistent suppression; −, no suppression; cd, trigger cell death.
FIGURE 3.
Suppression of PCD by Plasmopara viticola RxLR effectors. Three phenotypes observed upon expression of effectors in N. benthamiana are shown. Effector-GFP recombinant constructs and GFP control were transiently expressed on opposite sides of N. benthamiana leaves. (A,B) Different phenotypes: no suppression of INF1-triggered cell death (example, PvAVH3) and suppression of INF1-triggered cell death (example, PvAVH52); (C) All effectors tested could supress BAX-triggered cell death (example, PvAVH47). Proteins were extracted from infiltrated areas to analyze expression. Each infiltration results from more than three leaves; each infiltration test was repeated at three times with similar results.
Subcellular Localization of 26 RxLR Effectors
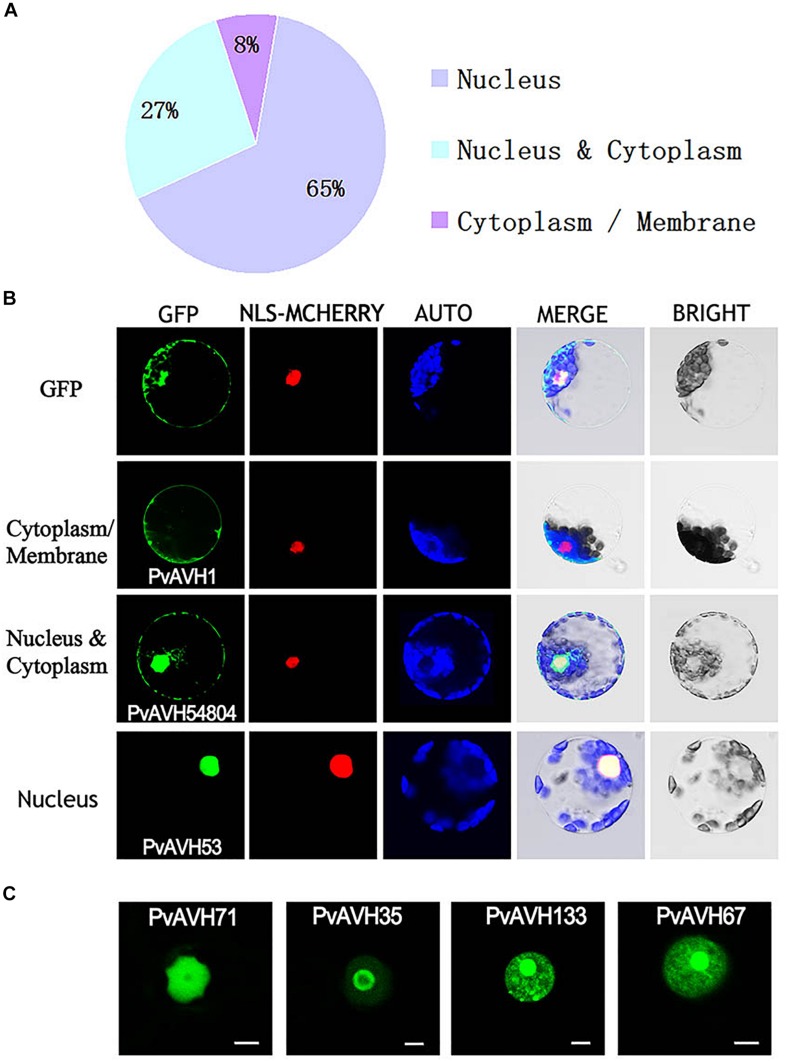
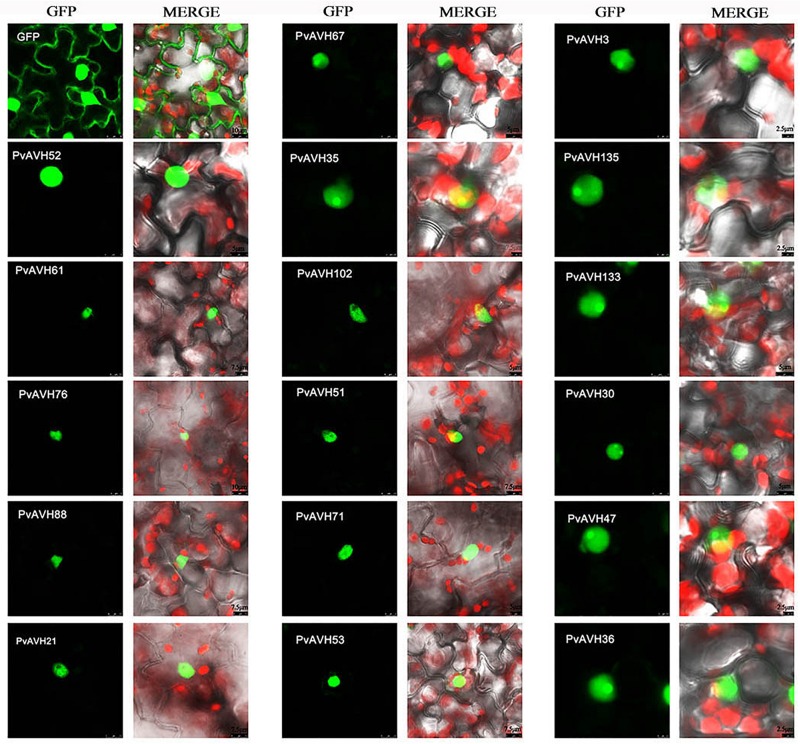
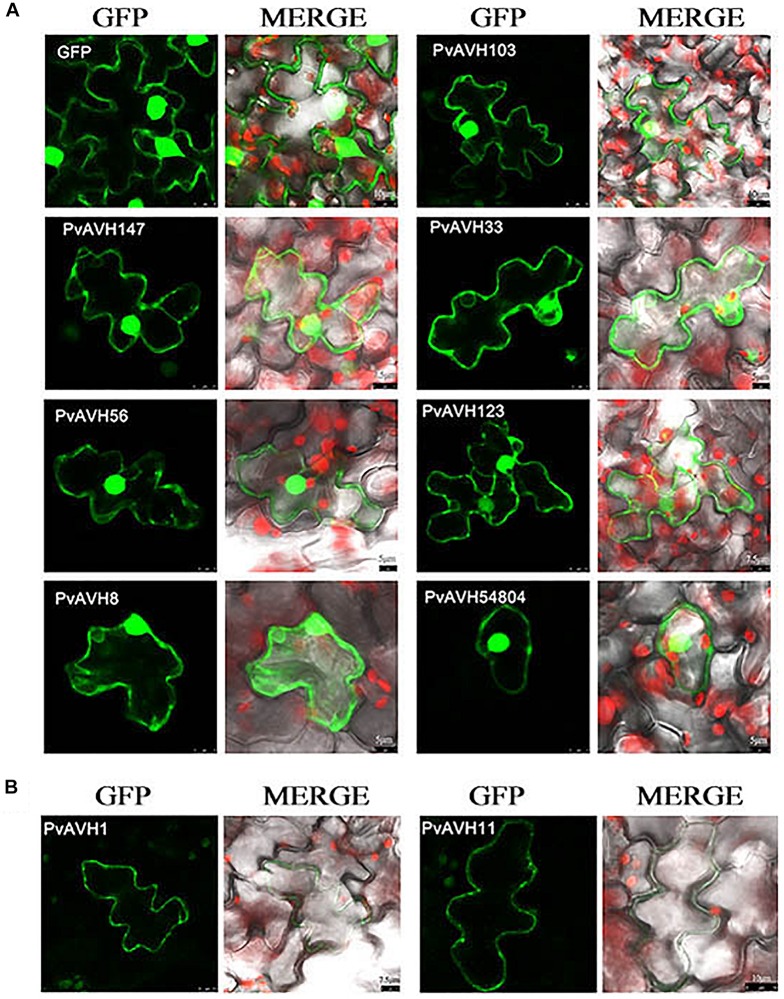
To further analyze the function of the 26 putative P. viticola RxLR effectors, the ORFs were ligated into the pCAMBIA2300 vector as fusions to GFP sequence. The plasmids were introduced into epidermal cells of N. benthamiana leaves through infiltration and agrobacterium-mediate transformation, and subcellular localization was visualized by confocal microscopy 3 days later (Table 2). Of the 26, 17 (65%) showed fluorescence concentrated in the nucleus, while 7 (27%) showed fluorescence in both the nucleus and cytoplasm. Two (8%) showed fluorescence in the cytoplasm and plasma membrane (Supplementary Figures S2–S4 and Figures 4A,B). Most of the 24 that were localized to the nucleus exhibited a classical nuclear localization signal (NLS) (Table 3). For each putative RxLR effector, the subcellular localizations of 26 effectors in N. benthamiana cells were the same as observed in V. vinifera (Figures 5, 6 and Supplementary Figures S5–S7).
FIGURE 4.
Subcellular localization of Plasmopara viticola RxLR effectors. Effector-GFP fusion proteins or GFP were expressed in Nicotiana benthamiana. Fluorescences accumulation of GFP and m-Cherry were analyzed by confocal microscopy, and excited at 488 nm and 561 nm, respectively. (A) Pie chart indicating the subcellular localization distribution for 26 effectors. (B) Subcellular localization of three representative effectors in N. benthamiana protoplasts: cytoplasm and plasma membrane (PvAVH1), the nucleus and cytoplasm (PvAVH54804), and nuclear specific (PvAVH53). Effectors-GFP fusion proteins and GFP were transiently expressed in N. benthamiana protoplasts following polyethylene glycol (PEG)-mediated transformation of the protoplasts. Photographs were taken 20–24 h after transformation. Scale bars = 25 μm. (C) Four sub-nuclear localization patterns observed for RxLR proteins (PvAVH71, 35, 133, and 67). Photographs were taken 72 h after infiltration. Scale bars = 5 μm.
TABLE 3.
Canonical nuclear localization signalprediction of effectors.
| Effectors |
Prediction results |
|
| Monopartite NLS | Bipartite NLS | |
| PvAVH51 | LNNKRKRIDS | |
| PvAVH52 | LNSKRKRIDP | |
| PvAVH53 | LNNKRKRIDS | |
| PvAVH71 | LHSKRKRIDP/RNSKRQRIN | |
| PvAVH102 | LISKRKRIDP | |
| PvAVH67 | QVKTHHLSHQVVKT PDKMNTKASSRRFLL | |
| PvAVH135 | KNKKRQRID/QKLKRLKTL | RKAKNNDKNKRSDE VKPGNKKRQRIE |
| PvAVH47 | DMIRRKRQRID | EKNLKEAYSVKLLIMY ELFYDFCHGNKKLVG |
| PvAVH3 | RRRTKRPRAM/VLAKRRRTKR | |
| PvAVH76 | IVHRPKKMRLS | QGLKRWRLMYRDFF |
| PvAVH21 | KKKLRTK | |
| PvAVH133 | KKSKRQRIE/NIKKRQRID | |
| PvAVH30 | RPGKQHTDLSPYDL QTPVPEKNYFQHIMSND | |
| PvAVH88 | TILKAHFNEEELLGIA EEAEKVDSTKSIA | |
| PvAVH61 | KSRKRKSST | ENLSPKSWPNNWIL QPFHYNPSRYPRHKMLQ |
| PvAVH35 | RSNKRQRIV/GQVKRKRPNR | |
| PvAVH36 | NNRKRNRSD | KMLPKRVLPGSRDL KNKWAVHAGGEDRMLNRI |
FIGURE 5.
Nuclear localization of RxLRs in Vitis vinifera. Sub-nuclear localization of 17 RxLRs. Effectors-GFP fusion proteins were expressed in V. vinifera leaves using agroinfiltration. The accumulations of fluorescent protein-tagged RxLRs were captured by confocal microscopy at 72 h post-infiltration. The fluorescent of effectors-GFP fusing proteins was showed on the left panels (green channel) and the merged layer on the right panels. Scale bars = 5–10 μm.
FIGURE 6.
Localization of RxLRs to the nuclear and cytosolic, cytoplasm and plasma membrane in Vitis vinifera. (A,B) The nuclear and cytosolic, cytoplasm and plasma membrane localization of 9 RxLRs. GFP-effectors fusion protein were expressed in V. vinifera leaves using agroinfiltration. The accumulations of fluorescent protein-tagged RxLRs were captured by confocal microscopy at 72 h post-infiltration. The fluorescent of effectors-GFP fusing proteins was showed on the left panels (green channel) and the merged layer on the right panels. Scale bars = 5–10 μm.
Interestingly, for those 17 effectors that localized to the nucleus (Supplementary Table S3), we observed four, distinct subnuclear localization patterns (Figure 4C): (1) PvAVH53, 52, 51, 102, and 71 were distributed in the nucleoplasm; (2) PvAVH35 showed clear localization to the nucleoplasm and nucleolar cap-like structures; (3) PvAVH133, 76, and 21 showed clear accumulation in the nucleolus but also in foci within the nucleoplasm.; (4) PvAVH67, 135, 47, 3, 30, and 36 were concentrated in the nucleolus.
Analysis of Expression of P. viticola Effector Genes During Infection of V. vinifera
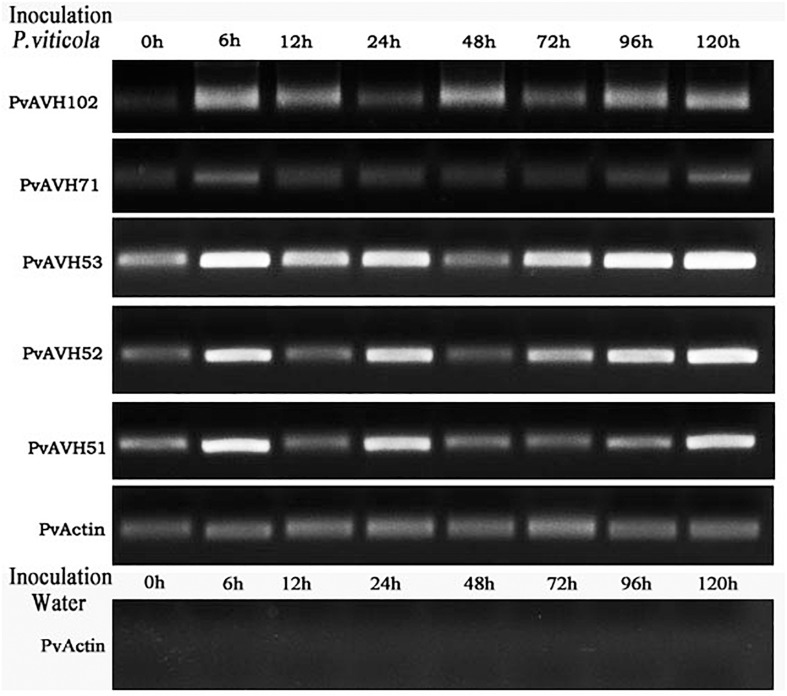
PvAVH51, 53, 71,102, and 52 have a close phylogenetic relationship, are located on the same chromosome, and exhibit the same subcellular localization. Three of these could induce plant cell death, while the remaining two suppressed INF1- and BAX-triggered cell death. To better understand the function of these genes in the host-pathogen interaction, we evaluated their temporal expression patterns during infection of V. vinifera (Figure 7). A PvActin gene was chosen to monitor the pathogen growth and normalize the measurement of the expression level of the effector genes. All five effector genes increased in expression by 6 hours post-inoculation (hpi) and then decreased by 12 hpi. PvAVH71 and PvAVH102 were not expressed or had a low expression level that could not be detected at 0 hpi. The relative expression of PvAVH53 was the highest at all time points, while that of PvAVH71 was the lowest. PvAVH51, PvAVH52, PvAVH71, and PvAVH53 showed a similar expression pattern that showed a low expression level at 48 hpi, and kept rising from 48 to 120 hpi. However, PvAVH102 showed a relatively high expression level at 48 hpi, and kept rising from 72 to 120 hpi. This apparent phenomenon in transcriptional response to infection suggests that each gene may be regulated to facilitate entry of the pathogen.
FIGURE 7.
Expression of selected effectors in infected “Pinot Noir” grapevine. Expression level of five effectors (PvAVH51, 52, 53, 71, and 102) was monitored by RT-PCR at 0 h, 6 h, 12 h, 24 h, 48 h, 72 h, 96 h, and 120 h after Plasmopara viticola inoculation and water (as negative control). PvActin gene was used as an internal reference.
Discussion
To date, knowledge of the pathology of P. viticola has been based on information from studies of P. infestans, which is a model for oomycete pathology (Bozkurt et al., 2012; King et al., 2014). RxLR-type effectors are the largest category and the most studied host-translocated proteins in oomycete. We identified effector-like proteins from P. viticola based on genomic and RNA-based sequence. We further identified 26 RxLR effectors for functional analysis. These sequences that blasted against the P. viticola the published genome “INRA-PV221” showed a stronger homology relationship. Phylogenetic analysis revealed that PvAVH51, 53, 71, 102, and 52 are closely related. Amino acid sequence alignment of these five proteins showed strong homology (up to 87.66% sequence similarity), and bioinformatics analysis revealed that the genes were located on the same chromosome. These five proteins also exhibited identical sub-nuclear localization. However, these putative effectors displayed distinct functions when expressed in N. benthamiana and distinct expression patterns at different stages of infection of V. vinifera. The presence of multiple, nearly identical gene copies has previously been reported in oomycete genomes (Mestre et al., 2016; Yin et al., 2017), and are thought to result from gene duplication during evolution, potentially facilitating infection of the host.
Characterization of the subcellular localizations ofP. viticola effectors will be critical for understanding theplant pathogenesis. P. viticola is an obligate biotrophicoomycete and therefore is unable to survive in vitro. Transient transformation of grapevine leaves is technically challenging and inefficient. Therefore, we used transient expression in the nonhost N. benthamiana to study the biological function and subcellular localization of the P. viticola effectors. This approach has been successfully applied for many bacterial and P. infestans effectors (Wang et al., 2011). However, we were able to use transient expression in leaves of V. vinifera to examine subcellular localization, and obtained results that were identical to those from the N. benthamiana system. In recent years, increasing evidences reveal that the plant localization for pathogen effectors is required for their function, which probably entre the host cell compartments to modulate plant innate immune responses and subsequently contribute to pathogenicity through plant-pathogen interaction (Boch and Bonas, 2010; Caillaud et al., 2012). For example, the RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death (Yu et al., 2012), and PvAvh74 and PvRxLR16 from P. viticola induced cell death depends on nuclear localization (Xiang et al., 2016; Yin et al., 2019). The effector AVR1 from P. infestans active the innate immune response by resistance protein R1 depending on the nuclear colocalization, and it also can target the exocyst to disturb the vesicle trafficking for potentially regulating plant immunity by interacting with exocyst subunit Sec5 protein (Du et al., 2015a, b). These results showed that the localization of the effectors were important to study the interaction between P. viticola and Vitis. Our functional analysis of the proteins was based on their ability to induce PCD or suppress the PCD triggered by INF and BAX in N. benthamiana. The number of pathogen effectors that can induce cell death is relatively small (Liu et al., 2018). Among the 26 RxLR effectors studied here, we found that seven could induce cell death, and that some of these could trigger a more rapid and stronger response than the others. Although effectors may often have redundant function, several individual effectors are keys to the full virulence of pathogen. For example, AVH241 and AVH238 could induce cell death and promote infection of P. sojae in N. benthamiana (Yu et al., 2012; Yang et al., 2017, 2019). Therefore, we hypothesized that 26 effectors might regulate the plant defense responses to contribute to virulence of P. viticola. Suppression of plant immunity responses is considered to be the primary activity of the P. viticola effectors (Liu et al., 2018). To evaluate this activity, we examined their ability to suppress cell death induced by INF1 and BAX. We found that most of the effectors tested could suppress cell death by BAX, while fewer could suppress cell death induced by INF1.
To assess the subcellular localization of the effectors, we carried out transient expression experiments in N. benthamiana leaves and protoplasts. Recently, increasing evidence suggests that while oomycete effectors can localize to a variety of plant cell compartments, the majority localize to the nucleus. For example, 82% of RxLR effectors from P. viticola and 66% of RxLRs from Hyaloperonospora arabidopsidis tested were found to be targeted to the nucleus (Caillaud et al., 2012; Liu et al., 2018). Our results are consistent with these reports, as 68% of the P. viticola effectors we studied were targeted to the nucleus and 24% were targeted to the nucleus and cytoplasm. Furthermore, we found that P. viticola effectors showed multiple sub-nuclear localizations. The majority mainly localized to nucleolus with additional weak localization to the nucleoplasm, but PvAVH35 strongly localized nucleolar cap-like structures. Others, such as PvAVH71, localized to the nucleoplasm. The significance of localization to discrete subcellular compartments is unknown. Previous studies revealed that effectors that localize to the nucleus may participate in reprogramming transcriptional mechanisms to suppress the plant innate immune response. For example, the P. infestans effector AVR2 suppresses plant innate immunity by up-regulating a brassinosteroid-responsive bHLH transcription factor (Boch and Bonas, 2010; Turnbull et al., 2017).
The nearly identical copies observed for some of the genes may be induced by LTR transposons, DNA transposons or other elements (Qutob et al., 2009; Mestre et al., 2016). PvAVH51, 53, 71, 102, and 52 are very closely related. These effectors (namely 51, 53, and 71) caused necrosis in Nicotiana leaves, but no cell death was observed in grape (V. vinifera) leaves. This phenomenon has been reported in three P. viticola RxLR effectors, PVITv1021061, PVITv1008294, and PVITv1008311 (Brilli et al., 2018). Meanwhile, a similar phenomenon was found that a P. infestans effector PITG_22798 induced cell death in tobacco but not in potato species (Wang et al., 2017). The cause of this result may be that these effectors can not be identified by the grapevine to trigger a immune response like cell death. Expression of these five effectors could be detected in infected grapevine leaves using RT-PCR, and were induced at an early stage of infection. All five effectors genes increased sharply at 6 hpi, but decreased at 12 hpi immediately. This similar expression patterns of RxLR effectors have been founded in H. arabidopsidis and P. sojae, namely “immediate-early, low” (Wang et al., 2011; Anderson et al., 2012). As previously reported, P. infestans haustoria appeared in the infected tissue at 3 hpi, and were abundant at 12 hpi (Wang et al., 2011). P. viticola encysted zoospores were observed on the stoma at 12 hpi (Yin et al., 2017). Moreover, PvRxLR18 and PvRxLR28 showed increased expression at 6 hpi, and this phenomenon was obvious in the highly virulent strains (Gomez-Zeledon and Spring, 2018). While PvAVH51, 53, 71, and PvAVH52 showed decreased expression levels after 24 h and then an increasing trend of up to 120 h. These similar expression patterns also have been reported for the effectors of P. viticola like PvRxLR28 and PvRxLR5 (Xiang et al., 2016). The P. viticola encysted zoospores were observed on the stoma at 12 hpi and the hyphae could be observed in spongy mesophyll at hpi (Yin et al., 2017). During the time (24–72 hpi), P. viticola may be busy in producing haustoria to obtain nutrients for growth and development at 48 hpi. Meanwhile, with the numbers of P. viticola increasing, the new infection will start, and the expression of these effectors will increase. Therefore, strong expression of effector genes early (prior to 12 h) during infection likely facilitates entry of the pathogen.
In summary, our results provide insights into the biological activities and subcellular localization of 26 P. viticola RxLR effectors. P. viticola RxLR effectors showed similar localization in N. benthamiana and V. vinifera. The majority of the studied effectors localized to the nucleus, and may function to suppress plant innate immunity responses. Further study of the pathogenic molecular mechanisms of P. viticola will provide a theoretical foundation for understanding downy mildew disease in grapevine. This will enable the development of innovative approaches to control the disease as well as new, downy mildew-tolerant grapevine cultivars. To date, our understanding of how P. viticola effectors modulate plant immunity is still limited. Therefore, we will focus on the interaction between plant and pathogen, and the mechanism involved in the fight against infection.
Data Availability Statement
The datasets generated for this study can be found in the all datasets for this study are included in the Supplementary Material.
Author Contributions
TC and YX designed the research, and MD cloned all of the putative effectors genes and constructed the vectors in this study. TC and RL performed the analyses of subcellular localization and suppression. MYL and MJL prepared the plant materials. TC drafted the manuscript. XY, GL, and YW modified the language. All authors have read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff of the laboratory for their technical assistance.
Funding. This work was supported by the National Natural Science Foundation of China (Grant No. 31872054,31471844), the Ph.D Research Startup Foundation of Northwest A&F University (No. Z109021621), and the Central College Basic Scientific Research Business Expenses Special Funds (No. Z109021606).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00692/full#supplementary-material
References
- As-sadi F., Carrere S., Gascuel Q., Hourlier T., Rengel D., Le Paslier M. C., et al. (2011). Transcriptomic analysis of the interaction between Helianthus annuus and its obligate parasite Plasmopara halstedii shows single nucleotide polymorphisms in CRN sequences. BMC Genomics 12:498. 10.1186/1471-2164-12-498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. G., Casady M. S., Fee R. A., Vaughan M. M., Deb D., Fedkenheuer K., et al. (2012). Homologous RXLR effectors from Hyaloperonospora arabidopsidis and Phytophthora sojae suppress immunity in distantly relatedplants. Plant J. 72 882–893. 10.1111/j.1365-313X.2012.05079.x [DOI] [PubMed] [Google Scholar]
- Birch P. R. J., Armstrong M., Bos J., Boevink P., Gilroy E. M., Taylor R. M., et al. (2009). Towards understanding the virulence functions of RXLR effectors of the oomycete plant pathogen Phytophthora infestans. J. Exp. Bot. 60 1133–1140. 10.1093/jxb/ern353 [DOI] [PubMed] [Google Scholar]
- Birch P. R. J., Boevink P. C., Gilroy E. M., Hein I., Pritchard L., Whisson S. C. (2008). Oomycete RXLR effectors: delivery, functional redundancy and durable disease resistance. Curr. Opin. Plant Biol. 11 373–379. 10.1016/j.pbi.2008.04.005 [DOI] [PubMed] [Google Scholar]
- Boch J., Bonas U. (2010). Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 48:419. 10.1146/annurev-phyto-080508-081936 [DOI] [PubMed] [Google Scholar]
- Bozkurt T. O., Schornack S., Banfield M. J., Kamoun S. (2012). Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 15 483–492. 10.1016/j.pbi.2012.03.008 [DOI] [PubMed] [Google Scholar]
- Brilli M., Asquini E., Moser M., Bianchedi P. L., Perazzolli M., Si-Ammour A. (2018). A multi-omics study of the grapevine-downy mildew (Plasmopara viticola) pathosystem unveils a complex protein coding- and noncoding-based arms race during infection. Sci. Rep. 8:757. 10.1038/s41598-018-19158-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral A., Stassen J. H., Seidl M. F., Bautor J., Parker J. E., Van den Ackerveken G. (2011). Identification of Hyaloperonospora arabidopsidis transcript sequences expressed during infection reveals isolate-specific effectors. PLoS One 6:0019328. 10.1371/journal.pone.0019328.g001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud M. C., Piquerez S. J., Fabro G., Steinbrenner J., Ishaque N., Beynon J., et al. (2012). Subcellular localization of the Hpa RxLR effector repertoire identifies a tonoplast-associated protein HaRxL17 that confers enhanced plant susceptibility. Plant J. 69 252–265. 10.1111/j.1365-313X.2011.04787.x [DOI] [PubMed] [Google Scholar]
- Chen X. R., Xing Y. P., Li Y. P., Tong Y. H., Xu J. Y. (2013). RNA-Seq reveals infection-related gene expression changes in Phytophthora capsici. PLoS One 8:e74588. 10.1371/journal.pone.0074588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J. L., Jones J. D. G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. 10.1038/35081161 [DOI] [PubMed] [Google Scholar]
- Dou D., Kale S. D., Xia W., Jiang R. H. Y., Bruce N. A., Arredondo F. D., et al. (2008). RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 20 1930–1947. 10.1105/tpc.107.056093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P. N., Rathjen J. P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11:539. 10.1038/nrg2812 [DOI] [PubMed] [Google Scholar]
- Du J., Verzaux E., Chaparro-Garcia A., Bijsterbosch G., Keizer L. C. P., Zhou J., et al. (2015). Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat. Plants 1:15034. 10.1038/nplants.2015.34 [DOI] [PubMed] [Google Scholar]
- Du Y., Berg J., Govers F., Bouwmeester K. (2015a). Immune activation mediated by the late blight resistance protein R1 requires nuclear localization of R1 and the effector AVR1. New Phytol. 207 735–747. 10.1111/nph.13355 [DOI] [PubMed] [Google Scholar]
- Du Y., Mpina M., Birch P., Bouwmeester K., Govers F. (2015b). Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity. Plant Physiol. 169 1975–1990. 10.1104/pp.15.01169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussert Y., Gouzy J., Richart-Cervera S., Mazet I. D., Delière L., Couture C., et al. (2016). Draft genome sequence of plasmopara viticola, the grapevine downy mildew pathogen. Genome Announc. 4:e00987-16. 10.1128/genomeA.00987-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussert Y. D., Mazet I., Couture C., Gouzy J., Piron M.-C., Kuchly C., et al. (2018). A high-quality grapevine downy mildew genome assembly reveals rapidly evolving and lineage-specific putative host adaptation genes. bioRxiv [Preprint] 10.1101/350041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawke S., Doumane M., Schornack S. (2015). Oomycete interactions with plants: infection strategies and resistance principles. Microbiol. Mol. Biol. Rev. 79 263–280. 10.1128/mmbr.00010-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascuel Q., Buendia L., Pecrix Y., Blanchet N., Munos S., Vear F., et al. (2016). RXLR and CRN effectors from the sunflower downy mildew pathogen Plasmopara halstedii induce hypersensitive-like responses in resistant sunflower lines. Front. Plant Sci. 7:1887. 10.3389/fpls.2016.01887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Zeledon J., Spring O. (2018). Up-regulated RxLR effector genes of Plasmopara viticola in synchronized host-free stages and infected leaves of hosts with different susceptibility. Fungal Biol. 122 1125–1133. 10.1016/j.funbio.2018.07.006 [DOI] [PubMed] [Google Scholar]
- Grouffaud S., van West P., Avrova A., Birch P. R. J., Whisson S. C. (2009). Plasmodium falciparum and Hyaloperonospora parasitica effector translocation motifs are functional in Phytophthora infestans. Microbiology 154 3743–3751. 10.1099/mic.0.2008/021964-0 [DOI] [PubMed] [Google Scholar]
- Jones J. D. G., Dangl J. L. (2006). The plant immune system. Nature 444 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Judelson H. S., Blanco F. A. (2005). The spores of Phytophthora: weapons of the plant destroyer. Nat. Rev. Microbiol. 3 47–58. 10.1038/nrmicro1064 [DOI] [PubMed] [Google Scholar]
- Kamoun S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44 41–60. 10.1146/annurev.phyto.44.070505.143436 [DOI] [PubMed] [Google Scholar]
- Kelley B. S., Lee S. J., Damasceno C. M., Chakravarthy S., Kim B. D., Martin G. B., et al. (2010). A secreted effector protein (SNE1) from Phytophthora infestans is a broadly acting suppressor of programmed cell death. Plant J. 62 357–366. 10.1111/j.1365-313X.2010.04160.x [DOI] [PubMed] [Google Scholar]
- King S. R., McLellan H., Boevink P. C., Armstrong M. R., Bukharova T., Sukarta O., et al. (2014). Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKK epsilon to suppress plant immune signaling. Plant Cell 26 1345–1359. 10.1105/tpc.113.120055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme C., Santa Cruz S. (1999). Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. U.S.A. 96 7956–7961. 10.1073/pnas.96.14.7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Lan X., Song S., Yin L., Dry I. B., Qu J., et al. (2018). In planta functional analysis and subcellular localization of the oomycete pathogen Plasmopara viticola candidate RXLR effector repertoire. Front. Plant Sci. 9:286. 10.3389/fpls.2018.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Weng K., Dou M., Chen T., Yin X., Li Z., et al. (2019). Transcriptomic analysis of Chinese wild Vitis pseudoreticulata in response to Plasmopara viticola. Protoplasma 256 1409–1424. 10.1007/s00709-019-01387-x [DOI] [PubMed] [Google Scholar]
- Mestre P., Carrere S., Gouzy J., Piron M.-C., Tourvieille de Labrouhe D., Vincourt P., et al. (2016). Comparative analysis of expressed CRN and RXLR effectors from two Plasmopara species causing grapevine and sunflower downy mildew. Plant Pathol. 65 767–781. 10.1111/ppa.12469 [DOI] [Google Scholar]
- Mestre P., Piron M.-C., Merdinoglu D. (2012). Identification of effector genes from the phytopathogenic oomycete Plasmopara viticola through the analysis of gene expression in germinated zoospores. Fungal Biol. 116 825–835. 10.1016/j.funbio.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Mittler R., Herr E. H., Orvar B. L., van Camp W., Willekens H., Inze D., et al. (1999). Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyperresponsive to pathogen infection. Proc. Natl. Acad. Sci. U.S.A. 96 14165–14170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panstruga R., Dodds P. N. (2009). Terrific protein traffic: the mystery of effector protein delivery by filamentous plant pathogens. Science 324 748–750. 10.1126/science.1171652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesani M., Desario F., Ferrarini A., Zamboni A., Pezzotti M., Kortekamp A., et al. (2008). cDNA-AFLP analysis of plant and pathogen genes expressed in grapevine infected with Plasmopara viticola. BMC Genomics 9:142. 10.1186/1471-2164-9-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qutob D., Tedman-Jones J., Dong S., Kuflu K., Pham H., Wang Y., et al. (2009). Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a. PLoS One 4:3. 10.1371/journal.pone.0005066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput N. A., Zhang M., Ru Y., Liu T., Xu J., Liu L., et al. (2014). Phytophthora sojae effector PsCRN70 suppresses plant defenses in Nicotiana benthamiana. PLoS One 9:e98114. 10.1371/journal.pone.0098114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemann M., Büche C., Kassemeyer H. H., Nick P. (2002). Cytoskeletal responses during early development of the downy mildew of grapevine (Plasmopara viticola). Protoplasma 219 13–22. 10.1007/s007090200001 [DOI] [PubMed] [Google Scholar]
- Sebastian S., Mireille V. D., Bozkurt T. O., Cano L. M., Matthew S., Marco T., et al. (2010). Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. U.S.A. 107 17421–17426. 10.1073/pnas.1008491107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassen J. H., Van den Ackerveken G. (2011). How do oomycete effectors interfere with plant life? Curr. Opin. Plant Biol. 14 407–414. 10.1016/j.pbi.2011.05.002 [DOI] [PubMed] [Google Scholar]
- Tian M., Win J., Savory E., Burkhardt A., Held M., Brandizzi F., et al. (2011). 454 genome sequencing of Pseudoperonospora cubensis reveals effector proteins with a QXLR translocation motif. Mol. Plant Microbe Interact. 24 543–553. 10.1094/mpmi-08-10-0185 [DOI] [PubMed] [Google Scholar]
- Turnbull D., Yang L., Naqvi S., Breen S., Welsh L., Stephens J., et al. (2017). RXLR effector AVR2 up-regulates a brassinosteroid-responsive bHLH transcription factor to suppress immunity. Plant Physiol. 174 356–369. 10.1104/pp.16.01804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler B. M., Tripathy S., Zhang X., Dehal P., Jiang R. H., Aerts A., et al. (2006). Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313 1261–1266. 10.1126/science.1128796 [DOI] [PubMed] [Google Scholar]
- Van der Hoorn R. A., Laurent F., Roth R., De Wit P. J. (2000). Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant Microbe Interact. 13 439–446. 10.1094/MPMI.2000.13.4.439 [DOI] [PubMed] [Google Scholar]
- Wang Q., Han C., Ferreira A. O., Yu X., Ye W., Tripathy S., et al. (2011). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. The Plant Cell 23 2064–2086. 10.1105/tpc.111.086082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ren Y., Zhou J., Du J., Hou J., Jiang R., et al. (2017). The cell death triggered by the nuclear localized RxLR effector PITG_22798 from Phytophthora infestans is suppressed by the effector AVR3b. Int. J. Mol. Sci. 18:409. 10.3390/ijms18020409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson S. C., Boevink P. C., Lucy M., Avrova A. O., Morales J. G., Gilroy E. M., et al. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450 115–118. 10.1038/nature06203 [DOI] [PubMed] [Google Scholar]
- Xiang J., Li X., Wu J., Yin L., Zhang Y., Lu J. (2016). Studying the mechanism of Plasmopara viticola RxLR effectors on suppressing plant immunity. Front. Microbiol. 7:709. 10.3389/fmicb.2016.00709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., Li X., Yin L., Liu Y., Zhang Y., Qu J., et al. (2017). A candidate RxLR effector from Plasmopara viticola can elicit immune responses in Nicotiana benthamiana. BMC Plant Biol. 17:75. 10.1186/s12870-017-1016-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Junli T., Qunqing W., Wenwu Y., Kai T., Shuyi D., et al. (2012). The RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death. New Phytol. 196 247–260. 10.1111/j.1469-8137.2012.04241.x [DOI] [PubMed] [Google Scholar]
- Yang B., Wang Q., Jing M., Guo B., Wu J., Wang H., et al. (2017). Distinct regions of the Phytophthora essential effector Avh238 determine its function in cell death activation and plant immunity suppression. New Phytol. 214 361–375. 10.1111/nph.14430 [DOI] [PubMed] [Google Scholar]
- Yang B., Wang Y., Guo B., Jing M., Zhou H., Li Y., et al. (2019). The Phytophthora sojae RXLR effector Avh238 destabilizes soybean Type2 GmACSs to suppress ethylene biosynthesis and promote infection. New Phytol. 222 425–437. 10.1111/nph.15581 [DOI] [PubMed] [Google Scholar]
- Ye W., Wang Y., Wang Y. (2015). Bioinformatics analysis reveals abundant short alpha-helices as a common structural feature of oomycete RxLR effector proteins. PLoS One 10:e0135240. 10.1371/journal.pone.0135240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., An Y., Qu J., Li X., Zhang Y., Dry I., et al. (2017). Genome sequence of Plasmopara viticola and insight into the pathogenic mechanism. Sci. Rep. 7:46553. 10.1038/srep46553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Li X., Xiang J., Qu J., Zhang Y., Dry I. B., et al. (2015). Characterization of the secretome of Plasmopara viticola by de novo transcriptome analysis. Physiol. Mol. Plant Pathol. 91 1–10. 10.1016/j.pmpp.2015.05.002 [DOI] [Google Scholar]
- Yin X., Liu R.-Q., Su H., Su L., Guo Y.-R., Wang Z.-J., et al. (2017). Pathogen development and host responses to Plasmopara viticola in resistant and susceptible grapevines: an ultrastructural study. Horticult. Res. 4 17033–17033. 10.1038/hortres.2017.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Shang B., Dou M., Liu R., Chen T., Xiang G., et al. (2019). The nuclear-localized RxLR effector PvAvh74 from Plasmopara viticola induces cell death and immunity responses in Nicotiana benthamiana. Front. Microbiol. 10:1531. 10.3389/fmicb.2019.01531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. D., Cho Y. H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2 1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the all datasets for this study are included in the Supplementary Material.