Abstract
Topoisomerases are essential enzymes solving DNA topological problems such as supercoils, knots and catenanes that arise from replication, transcription, chromatin remodeling and other nucleic acid metabolic processes. They are also the targets of widely used anticancer drugs (e.g. topotecan, irinotecan, enhertu, etoposide, doxorubicin, mitoxantrone) and fluoroquinolone antibiotics (e.g. ciprofloxacin and levofloxacin). Topoisomerases manipulate DNA topology by cleaving one DNA strand (TOP1 and TOP3 enzymes) or both in concert (TOP2 enzymes) through the formation of transient enzyme-DNA cleavage complexes (TOPcc) with phosphotyrosyl linkages between DNA ends and the catalytic tyrosyl residue of the enzymes. Failure in the self-resealing of TOPcc results in persistent TOPcc that threaten genome integrity and lead to cancers and neurodegenerative diseases. The cell prevents the accumulation of topoisomerase-mediated DNA damage by excising the topoisomerase DNA-protein crosslinks (TOP-DPC) and ligating the associated breaks using multiple pathways conserved in eukaryotes. Tyrosyl-DNA phosphodiesterase (TDP1 and TDP2) cleave the tyrosyl-DNA bonds whereas structure-specific endonucleases such as Mre11 and XPF (Rad1) incise the DNA phosphodiester backbone to remove the TOP-DPC along with the adjacent DNA segment. The proteasome and metalloproteases of the WSS1/Spartan family typify proteolytic repair pathways that debulk TOP-DPC to make the peptide-DNA bonds accessible to the TDPs and endonucleases. The purpose of this review is to summarize our current understanding of how the cell excises TOP-DPC and why, when and where the cell recruits one specific mechanism for repairing topoisomerase-mediated DNA damage, acquiring resistance to therapeutic topoisomerase inhibitors and avoiding genomic instability, cancers and neurodegenerative diseases.
1. Topoisomerases, topoisomerase-DNA cleavage complexes (TOPcc) and topoisomerase-DNA protein crosslinks (TOP-DPC).
During transcription and replication, the intertwined DNA double helix (one right helical turn for 10.5 base pairs) needs to be separated to expose the DNA strands that serve as templates for the polymerases. In the flank of this local unwinding, the DNA duplex is overtwisted ahead of the DNA polymerase/helicase complex and undertwisted behind. This classical “twin helical model” explains the coordinated action of topoisomerases with DNA tracking processes [1]. DNA underwinding (undertwisting) can generate DNA superhelical twists (negative supercoiling) and the formation of abnormal DNA structures (palindromic hairpins, Z-DNA, R- and D-loops) while overwinding (overtwisting) can generate DNA positive supercoiling. Supercoils, to some extent, can absorb the abnormal DNA twisting. In the absence of topoisomerase activity, the torsional tension generated by unwinding and overwinding of DNA arrest DNA transactions, leading to genomic instability. Topoisomerases are employed by the cell to resolve DNA topological entanglements upon their formation, thereby securing DNA transactions and genome integrity [2].
Topoisomerases catalyze the topological transformations of DNA via reversible transesterification reactions in which the catalytic tyrosine in the enzymes attacks the phosphate group in DNA, acting as a nucleophile and coupling the breaks with a covalent linkage between the enzyme and DNA [2-4]. This protein-DNA covalent adduct generates a transient break in the DNA, which enables DNA topological transformations; hence resolution of the topological problems. By a reversed transesterification mechanism, topoisomerases catalyze the resealing of the DNA break with their release from DNA upon resolution of the DNA entanglements.
In humans, the six topoisomerases: TOP1, TOP1mt, TOP2α, TOP2β, TOP3α and TOP3β are distinct from each other in catalytic mechanisms, structures and functions [2-4]. Additionally, a seventh member of the family, Spo11 belongs to the type II topoisomerase family acting in meiotic recombination. Topoisomerases differ in the polarity with which they cleave the DNA phosphodiester bond via nucleophilic attack with their catalytic tyrosine residues. TOP1 enzymes attack and attach to the DNA 3′-end while TOP2 and TOP3 enzymes attack and attach to the 5′-end of DNA. TOP1 enzymes catalyze the resealing of the DNA breaks by promoting the nucleophilic attack of the phosphotyrosyl bond by the 5′-hydroxyl end of the DNA whereas TOP2 and TOP3 catalyze the resealing of the DNA 3′-hydroxyl ends. Hence, the reversal of TOPcc is coupled with DNA resealing.
TOP1 enzymes (TOP1 and its mitochondrial paralog TOP1mt [5]) resolve both DNA underwinding (undertwisting that can generate DNA superhelical negative supercoiling) and overwinding (overtwisting that can generate DNA positive supercoiling). The enzymes function as monomers by cutting one strand of the DNA duplex, controlling the rotation of the cleaved strand around the intact strand [6]. Eukaryotic TOP1 is associated with both transcription and replication complexes. It suppresses DNA overwinding ahead of RNA and DNA polymerases, enabling DNA translocation [7, 8]. It resolves the overwinding resulting from the convergence of replication forks and transcription bubbles as well as the DNA underwinding behind replication forks which lead to the formation of R-loops [9, 10]. TOP1 enzymes also prevent replication fork run-off and promote the advancement of replication forks [11].
Acting as a homodimers, TOP2 enzymes relax negative and positive supercoiling as well as DNA catenanes that are formed upon completion of replication. Unlike TOP1, TOP2 cuts both strands of a DNA duplex (the gate or G-segment) with a 4-base stagger in a Mg2+- and ATP-dependent manner, which allows a second duplex (the transport or T-segment) to pass through the enzyme and the broken DNA [2, 12]. TOP2α is highly expressed in dividing cells and in S-phase as it is essential for decatenating intertwined sister chromatids during mitosis. TOP2β performs a pivotal role in neural development by forming DNA double-strand breaks (DSB) at the promoter regions of neuronal genes to facilitate their transcription [13-16]. TOP2β also promotes the transcription of hormone (estrogen, androgen and insulins, etc.) responsive genes by generating DSB within their promoters [17-19].
Monomeric TOP3 enzymes handle underwound (negatively supercoiled) DNA by nicking one DNA strand thereby driving the passage of the intact strand though the nicked strand [4]. TOP3β is the sole topoisomerase that can act on RNA and relax RNA supercoiling (e.g. R-loops) using this strand passage mechanism [20].
Transient TOPcc are key steps for all topoisomerase reactions. Failure to reverse the TOPcc and their associated DNA break(s) results in DNA-protein crosslinks (TOP-DPC) that block DNA metabolic processes. Such irreversible TOPcc (which we refer to as TOP-DPC) may stem from various endogenous and exogenous sources. Endogenous DNA lesions, such as mismatches, abasic sites and SSB and DSB occur spontaneously and frequently [21-26]. Left unrepaired, they result in TOP-DPC that are genotoxic. Exogenous DNA damaging agents such as carcinogens, UV, IR and alkylating agents also irreversibly trap topoisomerases on DNA by inducing dimers and 6-4 photoproducts and cyclobutane pyrimidine dimers, SSB and DSB, O6-methylguanine, benzopyrene adducts (see Table 1 in [4]).
The prototypical exogenous sources that generate TOP-DPC are topoisomerase-targeting drugs, some of which are widely used in cancer therapy. These small molecules target topoisomerases by binding at the enzyme-DNA interface [27], thereby incapacitating topoisomerases for DNA rejoining and completion of their catalytic cycles. Camptothecin and its derivatives target TOP1 by preferentially binding at the interface of the enzyme-DNA complexes by forming preferential base stacking with guanine +1 and hydrogen bonds with key enzyme catalytic residues [28]. Cisplatin adducts have also been shown to trap TOP1cc [29]. Anticancer TOP2-targeting drugs etoposide and doxorubicin are also interfacial inhibitors that stabilize TOP2cc with a preference for cytosine −1 and adenine −1, respectively [30, 31]. Biochemical and structural studies have established a shared mechanism of action by which topoisomerase inhibitors bind the enzyme-DNA interface by contacting specific amino acids of the enzyme and stacking against the base pairs flanking the topoisomerase-mediated cleavage site, resulting in the drug-enzyme-DNA ternary complex due to “interfacial inhibition” [32-34]. TOP-DPC disrupt DNA transactions by generating collisions with traversing DNA or RNA polymerase. Consequently, TOP-DPC arrest cell cycle and eventually leads to cell death if left unrepaired.
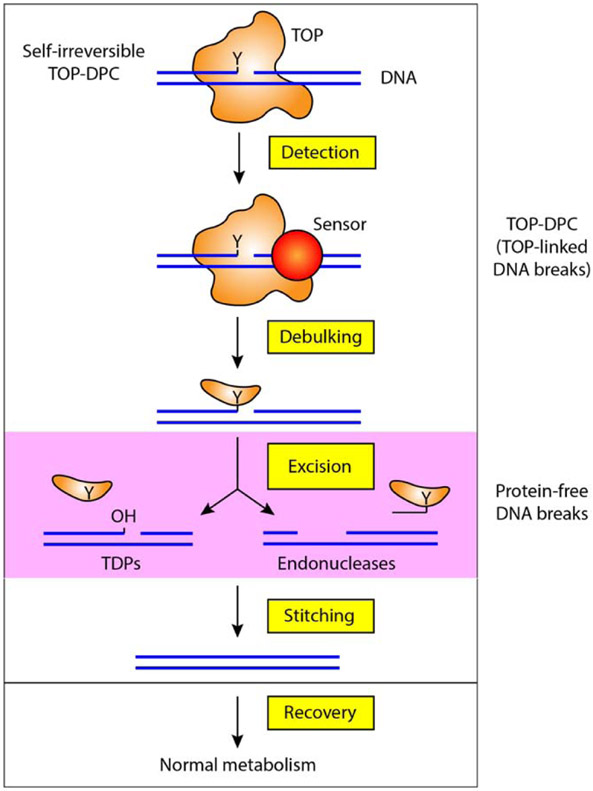
To remove TOP-DPC and repair the associated DNA breaks, the cell must follow a series of coordinated steps (Figure 1). First, it must detect the TOPcc as being potentially deleterious. Both replisome and transcription complexes carry DNA repair enzymes. As the DNA is translocated through these complexes, it is likely that irreversible and sterically blocking TOPcc (TOP-DPC) will elicit the detection mechanism such as activation of ubiquitylation pathways that target both the TOP-DPC and its surrounding chromatin environment. This mechanism is well-established for transcription-coupled nucleotide excision repair (TC-NER). Once detected, the TOP-DPC need to be “debulked” to enable access to the excision processes that remove the TOP-DPC and the repair of the associated DNA breaks. This debulking step is carried out by proteolytic enzymes that digest the TOP-DPC or denature the topoisomerase polypeptides. The proteasome and the involvement of DPC protease will be reviewed in more detail in an upcoming issue of DNA Repair. Following the excision of the TOP-DPC, the protein-free breaks need to be repaired by DNA polymerases, ligases, end-joining and recombination. Finally, once the DNA is restored, cells have to restart their metabolic processes. The detailed molecular elements of the DNA “stitching” and cell “Recovery” step are the least understood.
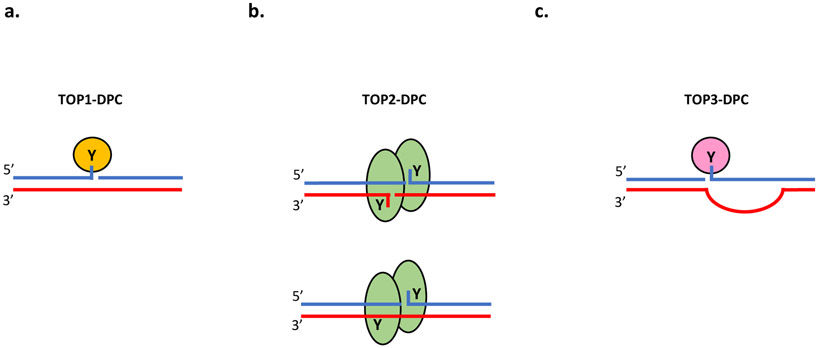
Figure 1. Architecture of TOP-DPC.
Topoisomerases cleave the DNA phosphodiester bond by carrying out nucleophilic attack of the hydroxyl end using their active tyrosine residues. This leads to formation of transient covalent linkages between the protein and the DNA, termed topoisomerase cleavage complexes (TOPcc). TOP1 enzymes attack the 3′-hydroxyl end of the DNA and cut one strand of the DNA duplex. TOP1cc can trapped by various endogenous and exogenous DNA damage and particularly by TOP1 inhibitors camptothecins. The trapped TOP1cc are referred to as TOP1 DNA-protein crosslinks (TOP1-DPC). b. Homodimeric TOP2 enzymes conduct nucleophilic attack of the 5′-hydroxyl end and cleave both strands of one DNA double helix with a 4-base stagger using Mg2+ and ATP The resulting transient TOP2cc can also be trapped by a wide range of DNA lesions and by TOP2 inhibitors such as etoposide and anthracyclines, leading to formation of TOP2-DPC. The cleavage by each subunit is not always concerted and single-strand breaks are readily produced by TOP2 enzymes, especially by their poisoning by etoposide (bottom). c. TOP3 enzymes nick one stand of single-stranded DNA by covalently linking its active tyrosine residue to the 5′ end of the DNA backbone using Mg2+ as a metal cofactor. Whether and how TOP3cc can be converted into TOP3-DPC require further investigation.
2. TOP-DPC excision by the tyrosyl-DNA Phosphodiesterases
The cell has specialized mechanisms to remove the covalently bound topoisomerases from 5′-ends (TOP2 and TOP3) or 3′-ends (TOP1) of the DNA. Tyrosyl-DNA Phosphodiesterases 1 and 2 represent a precise mechanism for excising TOP-DPC. They catalyze the hydrolysis of the phosphotyrosyl linkage between the enzyme and the DNA, thereby liberating the broken DNA termini, leaving a phosphate end which can be processed and restored (“stitched”; Figure 2) by end-joining through either homologous recombination (HR) or non-homologous end joining (NHEJ) (Figure 2).
Figure 2. Repair of TOP-DPC is a sequential multistep process.
It has been proposed that the molecular recognition mechanisms (or detection) of TOP-DPC involves encounter between the DPC and DNA transaction machineries such as DNA and RNA polymerases as they advance toward the TOP-DPC. Confrontation between TOP-DPC and ongoing DNA transactions elicits a signal for ubiquitylation and subsequent proteasomal degradation that removes the bulky protein component of the DPC (debulking) and expose the tyrosyl-DNA linkage of the TOP-DPC. The metalloprotease SPRTN, a reported component of the replisome, is also able to digest the DNA-bound topoisomerases likely upon the collision between replication fork and TOP-DPC. The debulking step appears to a prerequisite for the following excision that removes the DNA-linked topoisomerase remnant using nuclease activities, as it enables the nucleases to gain access to the protein-occluded broken ends. Tyrosyl-DNA phosphodiesterases (TDP) epitomize the nucleases that repair TOP-DPC. TDP1 and TDP2 carry out precise excision of TOP-DPC by hydrolyzing the phosphotyrosyl bond. On the other hand, more general nucleases such as the Mre11-Rad50-Nbs1 complex clear the remaining peptide likely by incising the DNA segment adjacent to the DPC. Complete removal of TOP-DPC by excision liberates the DNA termini, allowing repair of the strand breaks (stitching) by non-homologous end joining (NHEJ), homologous recombination (HR), single-strand annealing (SSA) or microhomology end-joining (MMEJ). Finally, efficient and accurate repair of the DNA breaks ensure the resumption of DNA metabolisms.
2.1. TDP1
Tyrosyl-DNA phosphodiesterase 1 (TDP1) was the first discovered eukaryotic enzyme that removes topoisomerase polypeptides [35]. It acts as DNA end-cleansing enzyme excising not only TOP1-DPC but also a variety of 3′-end blocking lesions formed by endogenous or by exogenous DNA damaging agents [36-42]. Its activity is restricted to DNA ends because TDP1 is devoid of exonuclease or endonuclease activity and processes only abnormal non-phosphorylated DNA ends with preference for 3′- vs. 5′-ends [35, 39, 43]. TDP1 deletion in combination with inactivating mutations affecting the DNA repair enzymes Rad9 or Rad1-Rad10 renders yeast cells hypersensitive to camptothecin [44-47], consistent with the important role of TDP1 in the repair of topoisomerase-mediated DNA damage. In addition, TDP1-deficient vertebrate cells are hypersensitive to oxidative and alkylation DNA damage [41-43, 48]. Human TDP1 can also serve as a backup excision pathway for TOP2-DPC [43]. Yeast cells do not express a specific TDP2, and their Tdp1 repairs both Top1- and Top2-DPC [49]. Vertebrate TDP1 exerts its activity against both TOP1 and TOP1mt, as TDP1 is active both in the nucleus and mitochondria [38, 50].
2.1.1. TDP1 structure, catalytic mechanism and substrates:
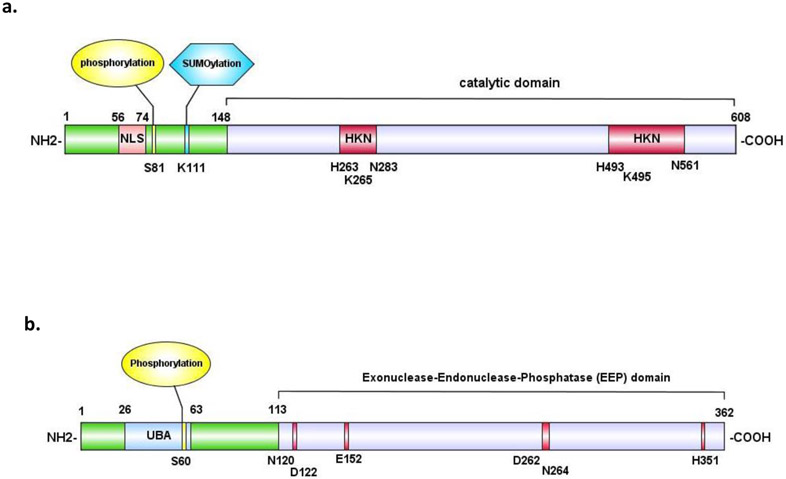
Following the discovery and characterization of TDP1 in yeast, human TDP1 was found by sequence analysis to be a highly conserved repair enzyme. It belongs to the phospholipase D (PLD) superfamily [51] characterized by the invariant sequence motif, H(X)K(X)4D. Human TDP1 is a 608 amino acid polypeptide with a molecular mass of 68.5 kDa (Figure 3a) consisting of two main domains. The N-terminal domain (NTD, residues 1-148) is not necessary for catalysis, and its structure has not yet been determined. It appears important for regulating the recruitment of TDP1 to DNA damage sites. The C-terminal catalytic domain (CTD, amino acid residues 149-608) bears the two active sites, H263 and H493 within the HKD1 and HKD2 motifs, respectively. Its crystallographic structure shows a wide DNA-peptide binding groove accommodating single-stranded DNA and the topoisomerase polypeptide [52-58].
Figure 3. Domain structures of human tyrosyl-DNA phosphodiesterases.
a. domain structures of human TDP1. The nuclear localization signal (NLS) is highlighted in pink, the known phosphorylation site in yellow and the known SUMOylation site in blue. The phosphodiesterase motifs (HKN) are highlighted in magenta. b. domain structure of human TDP2. The ubiquitin-associated domain (UBA) is highlighted in cyan. The known phosphorylation site is highlighted in yellow. Catalytic sites are highlighted in magenta. The UBA (ubiquitin binding domain) in the N-terminus is colored in blue.
TDP1 hydrolyzes tyrosyl-DNA phosphodiester bonds by forming an intermediate covalent complex, a TDP1cc, involving its active site residue H263 covalently linked to the 3′-phosphate DNA end as TDP1 cleaves the 3′-tyrosyl topoisomerase-DNA bond [48, 54, 59, 60]. In a second step, the phosphohistidine bond of the TDP1-DPC is hydrolyzed as the conserved H493 residue of TDP1 activates a water molecule. Mutation of H493 to arginine results in the neurodegenerative disease SCAN1 (spinocerebellar ataxia neuropathy) with TDP1 deficiency and accumulation of TDP1-DPC [56, 61-63]. Upon completion of its reaction, TDP1 releases the TOP1 polypeptide tyrosine and leaves a 3′-phosphate-end that needs to be further processed by polynucleotide kinase phosphatase (PNKP) and additional repair pathways [48, 59, 64].
TDP1 is also proficient at hydrolyzing a wide range of adducts from the 3′-ends of DNA, such as 3′-phosphoglycolate, 3′-abasic sites, TOP1-DPC 3′-phosphotyrosyl, and 3′-non-phosphorylated nucleotides. TDP1 does not require ATP or divalent ion for its activity and does not have a preference for single-stranded or blunt-ended substrates [43, 48, 59, 65]. As mentioned, TDP1 can also hydrolyze the 5′-phosphotyrosyl ends of TOP2-DPC [39, 43, 49, 66]. We recently showed that the conserved residue Y259 serves to position the nucleopeptide substrate in phase with the active site histidines in the DNA substrate groove [58, 66] and that residues S400, W590 and Y204, which are also in the DNA groove drive the selectivity for 3′- vs. 5′-tyrosyl substrates [66].
2.1.2. Post-translational modifications of TDP1 and recruitment to TOP1-DPC:
Post-translational modifications (PTM) of the NTD of TDP1 (Figure 3a) promote the recruitment of TDP1 to DNA lesion sites without exerting notable influence on the enzyme catalytic activity [67-70]. TDP1 phosphorylation at S81 by DNA-dependent protein kinase (DNA-PK) and Ataxia Telangiectasia Rad3 ortholog (ATR) increases the recruitment of TDP1 to DNA lesions as well as enhances the interaction of TDP1 with XRCC1 and ligase IIIα, which are crucial for PNKP recruitment [68, 69]. SUMOylation at lysine 111 of TDP1 enhances the accumulation of TDP1 at TOP1cc sites [70]. In addition, TDP1 forms a complex with poly (ADP-ribose) polymerase 1 (PARP1), which preexists DNA damage, and PARylation of TDP1 on its NTD stabilizes TDP1 and promotes the recruitment of XRCC1 to TOP1cc damage sites and PNKP participation in DNA repair [67]. Recently, TDP1 has been found in macromolecular complexes with the transcription-associated arginine methyltransferase (PRMT5) protein, which induces methylation of the CTD of TDP1 on positions R261 and R586. These modifications enhance TDP1 activity, the recruitment of TDP1 with XRCC1 to DNA and the survival of various cancer cell lines treated with CPT [71].
2.1.3. TDP1-associated neurological diseases:
The persistence of TOP1-DPC or TDP1-DPC at DNA termini results in severe neurological disease, such as familial spinocerebellar ataxia with axonal neuropathy (SCAN1), which is caused by the homozygous H493R mutation on human TDP1 [61, 72, 73]. The cells derived from SCAN1 patients are hypersensitive to camptothecin (CPT) treatment due to 25-time reduction in TDP1 activity [61, 63]. Additionally, mice with TDP1 and ATM deficiency exhibit severe neurological defects [74]. Based on SCAN1-derived cell models, studies have shown that the TDP1 degradation on TDP1cc by ubiquitylation is negatively controlled by the deubiquitylating enzyme, UCHL3, which might be a promising focus for cancer and neurological research [75].
2.2. TDP2
Tyrosyl DNA phosphodiesterase (TDP2) was originally discovered as the TRAF and TNF receptor associated protein (TTRAP) promoting MAPK/JNK/p38 signaling as well as inhibiting the activation of Nuclear factor- kappa beta (NF-kB) [76]. An independent study discovered TDP2 as the ETS1-associated protein 2 (EAPII), which negatively regulates the transcription factor ETS1 and the synergistic action of ETS1 and AP-1 [77]. Subsequently, TTRAP/EAPII was discovered to display tyrosyl DNA phosphodiesterase activity by genetic screening of yeast S. cerevisiae for CPT hypersensitivity in strains lacking Tdp1 and Rad1. However, the 3′-tyrosyl DNA phosphodiesterase activity of TDP2 was weak in contrast to its strong 5′-tyrosyl DNA phosphodiesterase activity [78]. TTRAP/EAPII was renamed TDP2 [78] and its strong 5′-tyrosyl-DNA phosphodiesterase activity makes TDP2 pivotal for the repair of TOP2-DPC in higher eukaryotes, as demonstrated by the fact that TDP2 depletion sensitizes cells specifically to TOP2 poisons like etoposide [78-82]. Hence, while yeast has only one TDP (Tdp1), vertebrates have both TDP1 and TDP2 for the excision of TOP1-DPC and TOP2-DPC, respectively. Like TDP1, TDP2 is present both in the nucleus and mitochondria where it repairs TOP2-DPC that form in the mitochondrial genome [38, 83].
2.2.1. TDP2 structure, catalytic mechanism and substrates:
Human TDP2 is a small two-domain protein of molecular mass 41 kDa (362 amino acid residues) (Figure 3b). It contains an N-terminal ubiquitin-associated (UBA) domain (amino acids 26–63) that binds to ubiquitin (Ub) and a C-terminal exonuclease/endonuclease/phosphodiesterase (EEP) catalytic domain (amino acids 113–362) analogous to APE1 and DNase I [41, 59, 84-87]. Similar to other Mg2+/Mn2+-dependent nucleases, the catalytic domain of TDP2 contains four conserved catalytic motifs (TWN, LQE, GDXN and SDH), which are important for its phosphodiesterase activity [78, 88]. The non-canonical UBA domain of TDP2 has four short α-helices, lacks the highly conserved ‘MGF’ sequence of canonical UBA domains and instead has an ‘TA’ motif at the same position [85, 86]. Although the catalytic domain of TDP2 independently displays the 5′-tyrosyl DNA phosphodiesterase activity in vitro, the UBA domain of TDP2 is required for optimal activity of TDP2 and TOP2cc repair inside cells [86].
Crystal structures of the zebrafish, C. elegans, mouse and human TDP2 catalytic domains [84, 85, 87, 89] reveal that TDP2 proteins exhibit a monomeric organization with a N-terminal stretch without regular secondary structure elements, an α-helical bundle domain and highly ordered conserved C-terminal catalytic domain. TDP2 has a narrow DNA binding cleft, which can stabilize DNA substrates in a sequence independent manner. Oxygens present in phosphodiester groups of the three terminal nucleotides of the DNA phosphodiester backbone form H-bonds with the nitrogen residues of K240, R275, R303 and S320 and position the DNA substrate in the DNA binding channel. The metal coordination site of human TDP2 is formed by four residues N120, E152, D262 and H351 (N129, E161, D271 and H360 in zebrafish) and chelates one or two magnesium ion [59, 89-92].
The catalytic cycle of TDP2 is fundamentally different from TDP1. Indeed, TDP2 does not form a covalent intermediate with its DNA substrate but utilizes Mg2+ for its catalytic activity, which can be divided in the following steps: (a) a two-sided active site pocket recognizes both the DNA and protein components of a TOP2cc, (b) substrate binding allows conformational changes and activation of the catalytic site of TDP2; and (c) divalent cation (Mg2+)-mediated catalytic mechanism promotes hydrolysis of the phosphotyrosyl linkage [84, 93, 94]. Although biochemical metal-titration analysis suggested that TDP2 involves two metals for its catalytic cycle [88], crystallographic studies indicate that TDP2 may work via a one-metal ion mechanism [84, 85]. After cleaving off the tyrosine from the 5′-end of DNA, TDP2 leaves a 5′-phosphate group ready for repair and religation by the NHEJ pathway [78, 81].
The phosphodiesterase activity of TDP2 is specific for phosphotyrosyl bonds [59]. It optimally recognizes and cleaves 5′-phosphotyrosyl bonds linked to either DNA or RNA [87], but, as recombinant enzyme, TDP2 only displays weak 3′-phosphotyrosyl activity [78]. The lack of significant activity against 3′-phosphotyrosyl substrates might be different in cells as TDP2 inactivation sensitizes TDP1-deficient cells to CPT, indicating that TDP2 can serve as a backup for TDP1 [80]. Although TDP2 can bind apurinic site analogs (tetrahydrofuran) or 5′–alkylamine adducts (e.g. 6-amino hexanol), it cannot hydrolyze them [95]. TDP2 prefers 5′-tyrosyl bond at the end of single-stranded substrates (mimicking denatured TOP2-DPC or even TOP3-DPC) as well as double-stranded 5′-tyrosyl overhang substrates (mimicking TOP2-DPC) [84, 88]. TDP2 also can cleave a 5′-tyrosine covalently linked to a ribonucleotide or from a polyribonucleotide (mimics RNA-TOP3βcc) and co-crystal of TDP2 bound to a tyrosyl-RNA substrate further reinforces this biochemical data [87].
TDP2 also possesses a VPg unlinkase activity that hydrolyzes the covalent phosphotyrosyl bond between viral VPg protein and the 5′ end of viral RNA during the life cycle of viruses of the picornaviridae family [41, 59, 96].
2.2.2. Post-translational modifications and regulation of TDP2:
Phosphorylation of TDP2 at Serine 60 by extracellular signal-related kinase 3 (ERK3) enhances its phosphodiesterase activity, and S60A mutation renders A549 cells hypersensitive to etoposide [97]. Interaction with Ub (K48-linked and K63-linked di-Ub) via its N-terminal ubiquitin-associated (UBA) domain also regulates TDP2 activity [86]. Further studies are warranted to determine whether TDP2 is recruited by ubiquitylated histones or TOP-DPC and whether its Ub interactions induce a conformation change to remodel the TDP2 active site [41, 86, 94].
Ubiquitylation and proteasomal degradation of TOP2-DPC promotes TDP2-mediated repair [98] and purified TDP2 is unable to process native TOP2cc in vitro unless TOP2 is proteolyzed or denatured [87]. In addition, the UBA domain of TDP2 is required for optimal activity of TDP2 and TOP2-DPC repair inside cells [86]. Recently it was reported that TOP2-DPC are SUMOylated by the SUMO E3 ligase ZNF451/ZATT, which remodels the TOP2-DPC and allows TDP2 to access the 5′-phosphotyrosyl bond and process the SUMOylated TOP2-DPC in a proteasome-independent manner [95, 99].
2.2.3. TDP2-associated neurological diseases:
Biallelic mutations in TDP2 cause cerebellar ataxia, epilepsy, and intellectual disability and this condition is denoted as spinocerebellar ataxia autosomal recessive 23 (SCAR23). First identified in three siblings from a consanguineous Irish family, SCAR23 was recently found in a patient in the United States [100, 101]. Another recent study identified a girl patient of Italian origin having a different mutation (homozygous nonsense variant in exon 3 of TDP2) in TDP2 but displaying the main features SCAR23 [102]. An Egyptian known to have truncated mutant allele of TDP2 also suffered from intellectual disability, seizures and ataxia [100]. All these medical cases indicate that TDP2 is important for normal neural tissue function. The neurological diseases associated with lack of TDP2 activity are consistent with the normal occurrence of TOP2- DPC in neurons [14] and the requirement of both TDP2 and TDP1 to maintain genome integrity [103].
3. Mre11 and structure-specific nucleases: redundant excision pathways process TOP-DPC
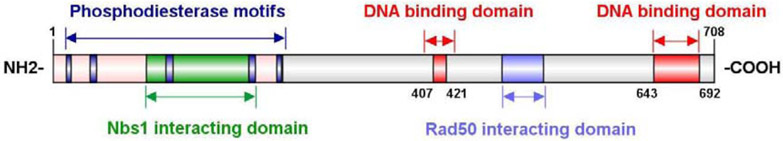
TOP-DPC can also be resolved by nucleases that cleave the DNA strand to which the topoisomerases are linked covalently [4, 104, 105] (Table 1). Meiotic recombination 11 (MRE11) is a versatile enzyme (Figure 4), as it can work both as double-strand-specific 3′–5′ DNA exonuclease and single-strand DNA endonuclease [106-108]. MRE11 forms a heterotrimeric complex, commonly referred to as the MRN complex, with RAD50 (an ATPase having DNA-binding activity) and Nijmegen breakage syndrome protein 1 (NBS1), a homologue of yeast Xrs2 [106, 109]. In coordination with CtIP, MRE11 mainly promotes the processing of DSB by generating 3′-single-stranded DNA tails for homologous recombination and single-strand annealing [107, 108, 110]. A study in yeast Saccharomyces cerevisiae provided direct evidence that the Mre11/Rad50/Xrs2 complex (MRN in humans) can excise TOP2-like-DPC (SPO11 adducts) from DNA ends [105, 111]. Consistently, in Schizosaccharomyces pombe, rad32 (S. pombe homolog of Mre11) in the MRX complex excises TOP1, TOP2 and SPO11 adducts from DNA ends [112, 113], with rad32 (S. pombe homolog of Mre11) deficiency causing hypersensitivity to topoisomerase poisons [114]. In mammalian cells, MRE11 is essential for the release and repair of TOP2-DPC [115, 116]. This process has been implicated for breast cancers with BRCA inactivation [19, 117].
Table 1:
Endonucleases excising TOP-DPC
| Nuclease | TOP1cc | TOP2cc | Notes | References |
|---|---|---|---|---|
| Rad1–Rad10 (yeast) (XPF–ERCC1 humans) | Yes | No | 3′-flap endonuclease complex involved in the nucleotide excision repair pathway (NER); functions in parallel pathway with TDP1; TDP1 and Rad1/Rad10 require a single-stranded gap between the 3-end to be processed and the 5′-end of the DNA. | [125, 129-132] |
| Slx4–Slx1 (yeast) SLX4–SLX1 (humans) | Yes | No | Endonuclease; works in parallel and redundant pathways with TDP1. | [125, 133] |
| Mus81–Mms4 (yeast) MUS81–EME1 (humans) | Yes | No | DNA endonuclease; preferentially cleaves broken replication forks and requires duplex DNA near the 3′-end; serves as an alternative pathway to TDP1 | [129, 130, 163] |
| XPG | No | Yes | 5′-flap endonuclease. | [114, 134] |
| FEN1 | No | Yes | 5′-flap endonuclease; works in vitro on ssDNA containing 5′-phosphotyrosyl substrate | [114, 134] |
| Mre11/Rad50/Xrs2 (yeast) MRE11/RAD50/NBS1 (humans) | Yes | Yes | DNA double-strand 3′–5′ exonuclease and single-strand endonuclease; requires a single-strand gap between the 3′-end to be processed and the 5′-end of the DNA. | [106, 112, 113, 123, 164] |
Figure 4. Domain structure of human Mre11.
The phosphodiesterase motifs are highlighted in indigo, the Nbs1 interacting domain is highlighted in green and the Rad50 interacting domain in purple. The DNA binding domains are highlighted in red.
The role of MRE11 and RAD50 in repairing TOP2-DPC is conserved in viruses and bacterial cells. In bacteriophage T4, gp46 (MRE11 in human) and gp47 (RAD50 in human) forms MR-like complex, which are important for the repair of m-AMSA induced T4-encoded Top2 cleavage complexes [118-120]. The Escherichia coli homolog of the MR complex consists in the heterodimeric complex of SbcC (RAD50 in human) and SbcD (MRE11 in human) proteins. The SbcCD complex can excise covalent DNA-bound proteins in vitro, and promotes repair of quinolone-induced gyrase-DNA complexes in E. coli cells [121, 122].
Apart from TOP2-DPC, the MRN complex also repairs TOP1-DPC in Xenopus laevis egg extracts [123] and yeast [45] and in vitro studies indicated that MRN can cleave and resect the 3′-phosphotyrosyl bonds between TOP1 and 3′ DNA end [106].
MRN-dependent TOP-DPC repair appears independent from the DSB resection activity of MRN during homologous repair (HR) and involves the nuclease activity of MRE11. Over-expression of nuclease-deficient mutants of MRE11 cannot complement MRE11-deficient cells, and those cells display increased accumulation of spontaneous TOP2-DPC [19, 116]. MRN complex dependent TOPcc repair starts with a nick in either the strand containing the covalently linked TOPcc or the complementary strand followed by strand resection in a 3′ to 5′ direction. A second nick in the complementary strand generates clean DSB which can be repaired by either mainly NHEJ or HR pathways [105, 116, 124]. This MRE11-dependent TOP2cc repair remains functional remains even during G1 phase (when HR is non-functional) in tissue cultured cells and in post-mitotic cells in the brains of embryonic mice [19, 116].
CtIP (ortholog of S. cerevisiae Sae2) physically and functionally interacts with the MRN/MRX complex [125, 126]. CtIP stimulates the endonuclease activity of the MRN complex in vitro and the removal of TOP2-DPs by MRN [105, 111-113, 124, 126, 127]. CtIP is tightly regulated and activated by cyclin-dependent kinase (CDK) during S-phase, which restricts the activity of the MRN/MRX complex to replicating cells (reviewed in [110]). Physical interaction between phosphorylated CtIP and BRCA1 further contributes to TOPcc repair [123, 128] and BRCA1 promotes the recruitment of MRE11 to TOPcc sites [19]. Surprisingly, CtIP-deleted cells are more proficient in removing TOP1-DPC than are wild-type cells, suggesting that CtIP might play an opposite role for TOP1-DPC by inhibiting their removal [112, 113].
Apart from MRE11, TOP-DPC are also repaired by other endonucleases. The 3′-flap endonuclease complex Rad1–Rad10 (XPF–ERCC1) has prominent role in the repair of TOP1-DPC in yeast, mice and humans [125, 129-132]. Another endonuclease SLX4–SLX1 works in a parallel pathway with TDP1 and can repair TOP1-DPC [125, 133]. Similarly, 5′-flap endonuclease XPG is involved in repair of TOP2cc [114, 134]. All the endonucleases having roles in repair of TOP-DPC have been reviewed in Table 1.
4. Synthetic lethality between the TDP1 and nuclease pathways
Synthetic lethality has been observed in yeast for the Tdp1 and the Rad1-Rad10 and the Mre11 exonuclease pathways [44, 130]. The ortholog of Rad1-Rad10 is XPF-ERCC1, which is known to be defective in a significant fraction of cancers, and particularly non-small cell lung cancers [135]. Mre11 is also frequently inactivated in cancers with mismatch repair deficiency, and particularly colon cancers [136-138].
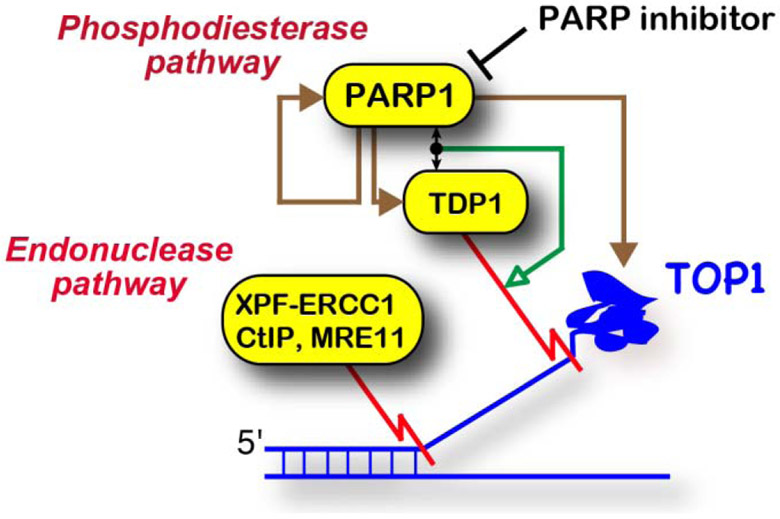
Due to the complementary actions of TDP1 and the nucleases XPF-ERCC1 and MRE11-RAD50-NBS1-CtIP, cancers with inactivation of either of these two nuclease pathways represent a potential indication for therapeutic strategies selectively enhancing the anticancer activity of TOP1 inhibitors by combining them with inhibitors of the TDP1 pathway (Figure 5). Because clinical TDP1 inhibitors are not presently available [139, 140], an alternative strategy to block TDP1 is to use PARP inhibitors [141, 142]. Indeed, TDP1 is coupled with PARP1 for the repair of TOP1-DPC [67] and inhibiting PARP effectively interferes with TDP1 action [141, 142], as PARylation of TDP1 by PARP1 recruits TDP1 to TOP1-DPC [67]. The synthetic lethal scheme illustrated in Figure 5 proposes that combination of PARP and TOP1 inhibitors, which is highly synergistic [142, 143] and often difficult to combine due to limiting bone marrow toxicity [144] would be most effective in cancer patients with XPF-ERCC1- or MRE11-deficient tumors.
Figure 5. Synthetic lethality between the TDP1 and nuclease pathways in human cancers.
PARP1 form a complex with TDP1, which promotes the recruitment of TDP1 to the TOP1-DPC and activates the TDP1 pathway (green arrow). PARP1 also PARylates TOP1, TDP1 and itself (autoPARylation) (brown arrows). Inactivation of the TDP1 pathway with PARP inhibitors renders ERCC1- or MRE11-deficient cells to TOP1 inhibitors.
5. Debulking of TOP-DPC by the proteasome and SPRTN
Because of the large size of the topoisomerases and the location of the DPC deep inside the topoisomerases, TDPs and endonucleases require a “debulking” step to gain access to the phosphotyrosyl bonds (for TDPs) and the DNA adjacent to the TOP-DPC (for the endonucleases) (Figure 2). Such debulking is primarily done by proteolytic digestion. Alternatively, topoisomerase protein denaturation and remodeling could be envisioned such as in the case of ZATT acting as a TOP2 denaturation chaperone [99].
The two main established pathways for proteolytic digestion of TOP-DPC are the proteasome and non-proteasomal proteases. They are briefly reviewed below and will be further developed in an upcoming DNA Repair review detailing the proteolytic “debulking” of TOP-DPC.
5.1. The ubiquitin-proteasome pathway:
Cells have the means to proteolytically digest the covalently DNA-bound topoisomerases, as exemplified by the ubiquitin-proteasome system (UPS). Ubiquitin is a post-translational modifier polypeptide consisting of 76 amino acids, whose polymeric conjugation to target proteins through its lysine 48 residue serves as a signal for proteasome recruitment to destroy the ubiquitylated proteins. The 26S proteasome acts as the endpoint of the UPS. It catalyzes the proteolytic degradation of the vast majority of cellular proteins. This large proteolytic machine comprises 31 subunits assembled into two main components, the 19S regulatory particle that recognizes and unfolds the polyubiquitylated proteins and the 20S proteolytic core that degrades the target proteins into oligopeptides [145-147].
The important role of UPS in degrading TOP-DPC is well-established. It was first reported in mouse mammary carcinoma cells that TOP1 inhibitor camptothecin rapidly (within 10 min) induce proteasome-mediated destruction of TOP1 in a ubiquitin activating enzyme (E1)-dependent manner [148]. Consequently, TOP2α was found to undergo proteasomal degradation in solid tumors upon exposure to the clinical TOP2 inhibitor etoposide [149]. Proteasome inhibitors were also shown to overcome resistance of solid tumor cell lines and of xenografts to etoposide and doxorubicin, another TOP2 inhibitor. Later studies in HeLa and various cancer cell lines treated with etoposide or its analog teniposide showed that the proteasome preferentially degrades TOP2β-DPC over TOP2α-DPC, and that the RNA polymerase II large subunit is also associated with the TOP2-DPC proteasomal destruction [150, 151]. Given the crucial role of TOP2β in transcriptional regulation, this finding suggested that obstruction of the progression of RNA polymerases by drug-trapped TOP2β-DPC elicits signal for ubiquitylation and subsequent proteasomal degradation of the TOP2β-DPC as well as the transcription machineries. Similarly, TOP1-DPC appear to transiently block the advancing the RNA polymerase elongation complexes, which arrests transcription and in turn evokes proteasome-mediated proteolysis of both TOP1-DPC and RNA polymerases [152].
In parallel, arrest of replication forks by TOP1cc also evokes the UPS to remove TOP1-DPC by proteolysis, allowing replication fork run-off on the leading strand, which leads to formation of DSB [153, 154]. A study in mouse carcinoma cells expressing thermolabile ubiquitin E1 and dominant-negative mutant ubiquitin has demonstrated that etoposide-induced TOP2β-DPC can still be targeted by the proteasome without being modified by ubiquitin, as opposed to TOP1-DPC, whose degradation requires proper polyubiquitylation [155]. These observations suggest TOP1-DPC and TOP2-DPC are recognized by the proteasome for destruction via different mechanisms, which warrant further investigations.
5.2. Spartan (SPRTN): a non-proteasomal replication-associated protease:
In recent years, mounting evidence demonstrates that SPRTN (SprT-like domain at the N-terminus), a DNA-binding metalloprotease, acts as an independent proteolytic mechanism to resolves TOP-DPC as well as other types of DPC in mammalian cells [105]. The initial study carried out in the yeast Saccharomyces cerevisiae identified Wss1 (Weak Suppressor of Smt3 Protein 1), the ortholog of SPRTN, as a novel protease processing TOP1-DPC in parallel to TDP1 [156]. Biochemical experiments showed that Wss1 cleaves TOP1 and other DPC substrates in a DNA-dependent manner. Subsequent reports in murine embryonic fibroblast cells (MEFs) and human cells revealed that SPRTN accumulates at ongoing replication forks where it proteolyzes TOP1-DPC via its SprT-like metalloprotease domain [157]. Although WSS1 and TDP1 function in distinct pathways to repair TOP1-DPC in yeast, SPRTN and TDP1 appear to act epistatically in mammalian cells [157, 158].
A role of SPRTN in repairing TOP2-DPC is evidenced by the findings that SPRTN knockout murine embryo fibroblast (MEFs) and MEFs expressing mutant SPRTN proteins compromised for their proteolytic activity exhibit increased accumulation and delayed clearance of TOP2α-DPC [159]. Also, depletion of the C-terminal DNA binding domain of SPRTN was found to retard TOP2αcc removal [159].
Proteolytic digestion by either the proteasome or SPRTN is unable to remove full-length covalently-linked topoisomerases from DNA due to the unproteolyzable phosphotyrosyl bond. Therefore, a succeeding nuclease pathway is necessary for clearing the covalent protein remnants to generate “clean” or “free” SSB or DSB for repair by homolog recombination or non-homologous end joining (Figure 2). The involvement of protease pathways for TOP-DPC repair also raises a question about whether proteolytic removal of the bulky protein adducts is required for nuclease action especially for TDPs, as the phosphotyrosyl bond is buried by the adducts hence presumably inaccessible.
Previous work has demonstrated that TDP1 is incapable of cleaving full-length native TOP1-DPC in vitro unless they are proteolyzed or denatured [160, 161]. Akin to processing of TOP1-DPC by TDP1, an earlier study shows that TDP2 can resolve only denatured or proteolyzed TOP2-DPC but not the native TOP2-DPC. As mentioned in section 2.2.2, a more recent report identified ZATT (ZNF451) as a SUMO ligase that catalyzes SUMO-2/3 modification of TOP2-DPC [99], which alters the conformation of TOP2, enabling TDP2 to process the DPC without proteolysis.
6. Conclusion and Perspectives
TOP-DPC as normal functional intermediates formed during the catalytic reaction cycles of topoisomerases can be stalled by multiple various sources including endogenous DNA lesions, carcinogens and exogenous agents particularly topoisomerase inhibitors, leading to a unique type of DNA damage often referred as irreversible or persistent TOP-DPC or protein-associated DNA breaks (PADB), which associate a DNA break (SSB or DSB) with each TOP-DPC. The repair of such frequently occurring DNA damage entails multiple evolutionarily conserved pathways. Among them, TDP1 and TPD2 constitute an emerging category of DNA repair enzymes discovered as excision enzyme for TOP-DPC. The additional participation of the MRN complex, SLX1-SLX4, MUS81 and XPF-ERCC1 in TOPcc repair [59] reveals the existence of crosstalk and redundancy between TDPs and more general nucleases to maintain genomic integrity and chromosomal stability in response to irreversible TOPcc.
Elucidating the mechanisms determining pathway choices and regulating the division of labor among such pathways will be of great value for basic cellular and molecular biology and for the potential development of inhibitors targeting these nucleases to enhance the activity of topoisomerase inhibitors as anticancer agents. For instance, PARP inhibitors are synthetic-lethal with TOP1 inhibitors in XPF-ERCC1-deficient cells due to the critical role of PARP in recruiting TDP1 and the redundancy of TDP1 and XPF-ERCC1 in TOP1-DPC removal [67, 141, 162]. Hence, PARP inhibitors act at least in part as TDP1 inhibitors in the case of TOP1-DPC [141]. Another important research area in need of further elucidation is how the different TOP-DPC excision repair pathways engage each of the different DNA repair pathways (HR vs. NHEJ vs. MMEJ or SSA) that must restore the DNA once the TOP-DPC is removed (see Figure 2).
Acknowledgements
Our studies are supported by the Center for Cancer Research, the Intramural Program of the National Cancer Institute, NIH (Z01 BC 006161 and Z01 BC 006150). We wish to thank all the present and past members of the Laboratory of Molecular Pharmacology and the Developmental Therapeutic Branch for their contribution.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no conflict of interest.
References
- [1].Wu HY, Shyy SH, Wang JC, Liu LF, Transcription generates positively and negatively supercoiled domains in the template, Cell, 53 (1988) 433–440. [DOI] [PubMed] [Google Scholar]
- [2].Vos SM, Tretter EM, Schmidt BH, Berger JM, All tangled up: how cells direct, manage and exploit topoisomerase function, Nat Rev Mol Cell Biol, 12 (2011) 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang JC, Cellular roles of DNA topoisomerases: a molecular perspective, Nat Rev Mol Cell Biol, 3 (2002) 430–440. [DOI] [PubMed] [Google Scholar]
- [4].Pommier Y, Sun Y, Huang SN, Nitiss JL, Roles of eukaryotic topoisomerases in transcription, replication and genomic stability, Nat Rev Mol Cell Biol, 17 (2016) 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang H, Barcelo JM, Lee B, Kohlhagen G, Zimonjic DB, Popescu NC, Pommier Y, Human mitochondrial topoisomerase I, Proc. Natl. Acad. Sci. U S A, 98 (2001) 10608–10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pommier Y, Topoisomerase I inhibitors: camptothecins and beyond, Nat Rev Cancer, 6 (2006) 789–802. [DOI] [PubMed] [Google Scholar]
- [7].Garg LC, DiAngelo S, Jacob ST, Role of DNA topoisomerase I in the transcription of supercoiled rRNA gene, Proc Natl Acad Sci U S A, 84 (1987) 3185–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Collins I, Weber A, Levens D, Transcriptional consequences of topoisomerase inhibition, Mol Cell Biol, 21 (2001) 8437–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Manzo SG, Hartono SR, Sanz LA, Marinello J, De Biasi S, Cossarizza A, Capranico G, Chedin F, DNA Topoisomerase I differentially modulates R-loops across the human genome, Genome biology, 19 (2018) 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cristini A, Ricci G, Britton S, Salimbeni S, Huang SN, Marinello J, Calsou P, Pommier Y, Favre G, Capranico G, Gromak N, Sordet O, Dual Processing of R-Loops and Topoisomerase I Induces Transcription-Dependent DNA Double-Strand Breaks, Cell Rep, 28 (2019) 3167–3181 e3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen SH, Chan NL, Hsieh TS, New mechanistic and functional insights into DNA topoisomerases, Annu Rev Biochem, 82 (2013) 139–170. [DOI] [PubMed] [Google Scholar]
- [12].Nitiss JL, DNA topoisomerase II and its growing repertoire of biological functions, Nat Rev Cancer, 9 (2009) 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].McKinnon PJ, Topoisomerases and the regulation of neural function, Nat Rev Neurosci, 17 (2016) 673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, Rueda R, Phan TX, Yamakawa H, Pao PC, Stott RT, Gjoneska E, Nott A, Cho S, Kellis M, Tsai LH, Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes, Cell, 161 (2015) 1592–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tiwari VK, Burger L, Nikoletopoulou V, Deogracias R, Thakurela S, Wirbelauer C, Kaut J, Terranova R, Hoerner L, Mielke C, Boege F, Murr R, Peters AH, Barde YA, Schubeler D, Target genes of Topoisomerase IIbeta regulate neuronal survival and are defined by their chromatin state, Proc Natl Acad Sci U S A, 109 (2012) E934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tsutsui K, Tsutsui K, Sano K, Kikuchi A, Tokunaga A, Involvement of DNA topoisomerase IIbeta in neuronal differentiation, J Biol Chem, 276 (2001) 5769–5778. [DOI] [PubMed] [Google Scholar]
- [17].Madabhushi R, The Roles of DNA Topoisomerase IIbeta in Transcription, International journal of molecular sciences, 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, Meeker AK, Netto G, De Marzo AM, Nelson WG, Yegnasubramanian S, Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements, Nat Genet, 42 (2010) 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sasanuma H, Tsuda M, Morimoto S, Saha LK, Rahman MM, Kiyooka Y, Fujiike H, Cherniack AD, Itou J, Callen Moreu E, Toi M, Nakada S, Tanaka H, Tsutsui K, Yamada S, Nussenzweig A, Takeda S, BRCA1 ensures genome integrity by eliminating estrogen-induced pathological topoisomerase II-DNA complexes, Proc Natl Acad Sci U S A, 115 (2018) E10642–E10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ahmad M, Shen W, Li W, Xue Y, Zou S, Xu D, Wang W, Topoisomerase 3beta is the major topoisomerase for mRNAs and linked to neurodevelopment and mental dysfunction, Nucleic Acids Res, 45 (2017) 2704–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pourquier P, Ueng LM, Kohlhagen G, Mazumder A, Gupta M, Kohn KW, Pommier Y, Effects of uracil incorporation, DNA mismatches, and abasic sites on cleavage and religation activities of mammalian topoisomerase I, J Biol Chem, 272 (1997) 7792–7796. [DOI] [PubMed] [Google Scholar]
- [22].Pourquier P, Pilon AA, Kohlhagen G, Mazumder A, Sharma A, Pommier Y, Trapping of mammalian topoisomerase I and recombinations induced by damaged DNA containing nicks or gaps. Importance of DNA end phosphorylation and camptothecin effects, J Biol Chem, 272 (1997) 26441–26447. [DOI] [PubMed] [Google Scholar]
- [23].Lesher DT, Pommier Y, Stewart L, Redinbo MR, 8-Oxoguanine rearranges the active site of human topoisomerase I, Proc Natl Acad Sci U S A, 99 (2002) 12102–12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kingma PS, Corbett AH, Burcham PC, Marnett LJ, Osheroff N, Abasic sites stimulate double-stranded DNA cleavage mediated by topoisomerase II. DNA lesions as endogenous topoisomerase II poisons, J Biol Chem, 270 (1995) 21441–21444. [DOI] [PubMed] [Google Scholar]
- [25].Kingma PS, Osheroff N, Spontaneous DNA damage stimulates topoisomerase II-mediated DNA cleavage, J Biol Chem, 272 (1997) 7488–7493. [DOI] [PubMed] [Google Scholar]
- [26].Wilstermann AM, Osheroff N, Base excision repair intermediates as topoisomerase II poisons, J Biol Chem, 276 (2001) 46290–46296. [DOI] [PubMed] [Google Scholar]
- [27].Pommier Y, Drugging topoisomerases: lessons and challenges, ACS Chem Biol, 8 (2013) 82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jaxel C, Capranico G, Kerrigan D, Kohn KW, Pommier Y, Effect of local DNA sequence on topoisomerase I cleavage in the presence or absence of camptothecin, J Biol Chem, 266 (1991) 20418–20423. [PubMed] [Google Scholar]
- [29].van Waardenburg RC, de Jong LA, van Eijndhoven MA, Verseyden C, Pluim D, Jansen LE, Bjornsti MA, Schellens JH, Platinated DNA adducts enhance poisoning of DNA topoisomerase I by camptothecin, J Biol Chem, 279 (2004) 54502–54509. [DOI] [PubMed] [Google Scholar]
- [30].Capranico G, Kohn KW, Pommier Y, Local sequence requirements for DNA cleavage by mammalian topoisomerase II in the presence of doxorubicin, Nucleic Acids Res, 18 (1990) 6611–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pommier Y, Capranico G, Orr A, Kohn KW, Local base sequence preferences for DNA cleavage by mammalian topoisomerase II in the presence of amsacrine or teniposide, Nucleic Acids Res, 19 (1991) 5973–5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pommier Y, Marchand C, Interfacial inhibitors: targeting macromolecular complexes, Nat Rev Drug Discov, 11 (2011) 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB Jr., Stewart L, The mechanism of topoisomerase I poisoning by a camptothecin analog, Proc Natl Acad Sci U S A, 99 (2002) 15387–15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wu CC, Li TK, Farh L, Lin LY, Lin TS, Yu YJ, Yen TJ, Chiang CW, Chan NL, Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide, Science, 333 (2011) 459–462. [DOI] [PubMed] [Google Scholar]
- [35].Yang SW, Burgin AB, Huizenga BN, Robertson CA, Yao KC, Nash HA, A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases, P Natl Acad Sci USA, 93 (1996) 11534–11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pommier Y, Drugging topoisomerases: lessons and challenges, ACS chemical biology, 8 (2013) 82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pommier Y, DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition, Chem Rev, 109 (2009) 2894–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Huang SN, Pommier Y, Mammalian Tyrosyl-DNA Phosphodiesterases in the Context of Mitochondrial DNA Repair, International journal of molecular sciences, 20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Interthal H, Chen HJ, Champoux JJ, Human Tdp1 cleaves a broad spectrum of substrates including phosphoamide linkages, J Biol Chem, 280 (2005) 36518–36528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Brettrager EJ, Segura IA, van Waardenburg R, Tyrosyl-DNA Phosphodiesterase I N-Terminal Domain Modifications and Interactions Regulate Cellular Function, Genes (Basel), 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kawale AS, Povirk LF, Tyrosyl-DNA phosphodiesterases: rescuing the genome from the risks of relaxation, Nucleic Acids Res, 46 (2018) 520–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhou T, Akopiants K, Mohapatra S, Lin PS, Valerie K, Ramsden DA, Lees-Miller SP, Povirk LF, Tyrosyl-DNA phosphodiesterase and the repair of 3'-phosphoglycolate-terminated DNA double-strand breaks, DNA Repair (Amst), 8 (2009) 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Murai J, Huang SY, Das BB, Dexheimer TS, Takeda S, Pommier Y, Tyrosyl-DNA phosphodiesterase 1 (TDP1) repairs DNA damage induced by topoisomerases I and II and base alkylation in vertebrate cells, The Journal of biological chemistry, 287 (2012) 12848–12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pouliot JJ, Yao KC, Robertson CA, Nash HA, Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes, Science, 286 (1999) 552–555. [DOI] [PubMed] [Google Scholar]
- [45].Deng C, Brown JA, You D, Brown JM, Multiple Endonucleases Function to Repair Covalent Topoisomerase I Complexes in Saccharomyces cerevisiae, Genetics, 170 (2005) 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Karumbati AS, Deshpande RA, Jilani A, Vance JR, Ramotar D, Wilson TE, The role of yeast DNA 3'-phosphatase Tpp1 and rad1/Rad10 endonuclease in processing spontaneous and induced base lesions, J Biol Chem, 278 (2003) 31434–31443. [DOI] [PubMed] [Google Scholar]
- [47].Vance JR, Wilson TE, Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage, Proc. Natl. Acad. Sci. U S A, 99 (2002) 13669–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kawale AS, Akopiants K, Valerie K, Ruis B, Hendrickson EA, Huang SN, Pommier Y, Povirk LF, TDP1 suppresses mis-joining of radiomimetic DNA double-strand breaks and cooperates with Artemis to promote optimal nonhomologous end joining, Nucleic Acids Res, 46 (2018) 8926–8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nitiss KC, Malik M, He X, White SW, Nitiss JL, Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage, Proc Natl Acad Sci U S A, 103 (2006) 8953–8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Das BB, Dexheimer TS, Maddali K, Pommier Y, Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria, Proc Natl Acad Sci U S A, 107 (2010) 19790–19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Interthal H, Pouliot JJ, Champoux JJ, The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily, Proc Natl Acad Sci U S A, 98 (2001) 12009–12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Davies DR, Interthal H, Champoux JJ, Hol WG, The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1, Structure (Camb), 10 (2002) 237–248. [DOI] [PubMed] [Google Scholar]
- [53].Davies DR, Interthal H, Champoux JJ, Hol WG, Insights into substrate binding and catalytic mechanism of human tyrosyl-DNA phosphodiesterase (Tdp1) from vanadate and tungstate-inhibited structures, J Mol Biol, 324 (2002) 917–932. [DOI] [PubMed] [Google Scholar]
- [54].Davies DR, Interthal H, Champoux JJ, Hol WG, Crystal structure of a transition state mimic for tdp1 assembled from vanadate, DNA, and a topoisomerase I-derived Peptide, Chem Biol, 10 (2003) 139–147. [DOI] [PubMed] [Google Scholar]
- [55].Davies DR, Interthal H, Champoux JJ, Hol WG, Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes, J Med Chem, 47 (2004) 829–837. [DOI] [PubMed] [Google Scholar]
- [56].He X, van Waardenburg RC, Babaoglu K, Price AC, Nitiss KC, Nitiss JL, Bjornsti MA, White SW, Mutation of a Conserved Active Site Residue Converts Tyrosyl-DNA Phosphodiesterase I into a DNA Topoisomerase I-dependent Poison, J Mol Biol, 372 (2007) 1070–1081. [DOI] [PubMed] [Google Scholar]
- [57].Lountos GT, Zhao XZ, Kiselev E, Tropea JE, Needle D, Pommier Y, Burke TR, Waugh DS, Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening, Nucleic Acids Res, 47 (2019) 10134–10150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Flett FJ, Ruksenaite E, Armstrong LA, Bharati S, Carloni R, Morris ER, Mackay CL, Interthal H, Richardson JM, Structural basis for DNA 3'-end processing by human tyrosyl-DNA phosphodiesterase 1, Nat Commun, 9 (2018) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pommier Y, Huang SY, Gao R, Das BB, Murai J, Marchand C, Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2), DNA Repair (Amst), 19 (2014) 114–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Correction: Wnt5a Drives an Invasive Phenotype in Human Glioblastoma Stem-like Cells, Cancer Res, 77 (2017) 3962. [DOI] [PubMed] [Google Scholar]
- [61].Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, Champoux JJ, SCAN1 mutant Tdp1 accumulates the enzyme--DNA intermediate and causes camptothecin hypersensitivity, Embo J, 24 (2005) 2224–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, Stockton DW, Lupski JR, Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy, Nat. Genet, 32 (2002) 267–272. [DOI] [PubMed] [Google Scholar]
- [63].Miao ZH, Agama K, Sordet O, Povirk L, Kohn KW, Pommier Y, Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes, DNA Repair (Amst), 5 (2006) 1489–1494. [DOI] [PubMed] [Google Scholar]
- [64].Plo I, Liao ZY, Barcelo JM, Kohlhagen G, Caldecott KW, Weinfeld M, Pommier Y, Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions, DNA Repair (Amst), 2 (2003) 1087–1100. [DOI] [PubMed] [Google Scholar]
- [65].Raymond AC, Staker BL, Burgin AB Jr., Substrate specificity of tyrosyl-DNA phosphodiesterase I (Tdp1), J Biol Chem, 280 (2005) 22029–22035. [DOI] [PubMed] [Google Scholar]
- [66].Kiselev E, Dexheimer TS, Marchand C, Huang SN, Pommier Y, Probing the evolutionary conserved residues Y204, F259, S400 and W590 that shape the catalytic groove of human TDP1 for 3'- and 5'-phosphodiester-DNA bond cleavage, DNA Repair (Amst), 66–67 (2018) 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Das BB, Huang SY, Murai J, Rehman I, Ame JC, Sengupta S, Das SK, Majumdar P, Zhang H, Biard D, Majumder HK, Schreiber V, Pommier Y, PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage, Nucleic Acids Res, 42 (2014) 4435–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Das BB, Antony S, Gupta S, Dexheimer TS, Redon CE, Garfield S, Shiloh Y, Pommier Y, Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK, EMBO J, 28 (2009) 3667–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Chiang SC, Carroll J, El-Khamisy SF, TDP1 serine 81 promotes interaction with DNA ligase IIIalpha and facilitates cell survival following DNA damage, Cell Cycle, 9 (2010) 588–595. [DOI] [PubMed] [Google Scholar]
- [70].Hudson JJR, Chiang SC, Wells OS, Rookyard C, El-Khamisy SF, SUMO modification of the neuroprotective protein TDP1 facilitates chromosomal single-strand break repair, Nat Commun, 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Rehman I, Basu SM, Das SK, Bhattacharjee S, Ghosh A, Pommier Y, Das BB, PRMT5-mediated arginine methylation of TDP1 for the repair of topoisomerase I covalent complexes, Nucleic Acids Res, 46 (2018) 5601–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW, Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1, Nature, 434 (2005) 108–113. [DOI] [PubMed] [Google Scholar]
- [73].Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, Stockton DW, Lupski JR, Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy, Nat Genet, 32 (2002) 267–272. [DOI] [PubMed] [Google Scholar]
- [74].Katyal S, Lee Y, Nitiss KC, Downing SM, Li Y, Shimada M, Zhao J, Russell HR, Petrini JH, Nitiss JL, McKinnon PJ, Aberrant topoisomerase-1 DNA lesions are pathogenic in neurodegenerative genome instability syndromes, Nat Neurosci, Advance Online Publication (2014). [DOI] [PMC free article] [PubMed]
- [75].Liao C, Beveridge R, Hudson JJR, Parker JD, Chiang SC, Ray S, Ashour ME, Sudbery I, Dickman MJ, El-Khamisy SF, UCHL3 Regulates Topoisomerase-Induced Chromosomal Break Repair by Controlling TDP1 Proteostasis, Cell Rep, 23 (2018) 3352–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Pype S, Declercq W, Ibrahimi A, Michiels C, Van Rietschoten JG, Dewulf N, de Boer M, Vandenabeele P, Huylebroeck D, Remacle JE, TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs), and that inhibits nuclear factor-kappa B activation, J Biol Chem, 275 (2000) 18586–18593. [DOI] [PubMed] [Google Scholar]
- [77].Pei H, Yordy JS, Leng Q, Zhao Q, Watson DK, Li R, EAPII interacts with ETS1 and modulates its transcriptional function, Oncogene, 22 (2003) 2699–2709. [DOI] [PubMed] [Google Scholar]
- [78].Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW, A human 5'-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage, Nature, 461 (2009) 674–678. [DOI] [PubMed] [Google Scholar]
- [79].Zeng Z, Cortes-Ledesma F, El Khamisy SF, Caldecott KW, TDP2/TTRAP is the major 5'-tyrosyl DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase II-induced DNA damage, J Biol Chem, 286 (2011) 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zeng Z, Sharma A, Ju L, Murai J, Umans L, Vermeire L, Pommier Y, Takeda S, Huylebroeck D, Caldecott KW, El-Khamisy SF, TDP2 promotes repair of topoisomerase I-mediated DNA damage in the absence of TDP1, Nucleic Acids Res, 40 (2012) 8371–8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Gomez-Herreros F, Romero-Granados R, Zeng Z, Alvarez-Quilon A, Quintero C, Ju L, Umans L, Vermeire L, Huylebroeck D, Caldecott KW, Cortes-Ledesma F, TDP2-dependent non-homologous end-joining protects against topoisomerase II-induced DNA breaks and genome instability in cells and in vivo, PLoS Genet, 9 (2013) e1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Maede Y, Shimizu H, Fukushima T, Kogame T, Nakamura T, Miki T, Takeda S, Pommier Y, Murai J, Differential and Common DNA Repair Pathways for Topoisomerase I- and II-Targeted Drugs in a Genetic DT40 Repair Cell Screen Panel, Mol Cancer Ther, 13 (2014) 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Huang SN, Dalla Rosa I, Michaels SA, Tulumello DV, Agama K, Khiati S, Jean SR, Baechler SA, Factor VM, Varma S, Murai J, Miller Jenkins LM, Kelley SO, Pommier Y, Mitochondrial tyrosyl-DNA phosphodiesterase 2 and its TDP2(S) short isoform, EMBO Rep, 19 (2018) e42139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Schellenberg MJ, Appel CD, Adhikari S, Robertson PD, Ramsden DA, Williams RS, Mechanism of repair of 5'-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2, Nat Struct Mol Biol, 19 (2012) 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Shi K, Kurahashi K, Gao R, Tsutakawa SE, Tainer JA, Pommier Y, Aihara H, Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2, Nat Struct Mol Biol, 19 (2012) 1372–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Rao T, Gao R, Takada S, Al Abo M, Chen X, Walters KJ, Pommier Y, Aihara H, Novel TDP2-ubiquitin interactions and their importance for the repair of topoisomerase II-mediated DNA damage, Nucleic Acids Res, 44 (2016) 10201–10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Gao R, Schellenberg MJ, Huang SY, Abdelmalak M, Marchand C, Nitiss KC, Nitiss JL, Williams RS, Pommier Y, Proteolytic degradation of topoisomerase II (Top2) enables the processing of Top2.DNA and Top2.RNA covalent complexes by tyrosyl-DNA-phosphodiesterase 2 (TDP2), J Biol Chem, 289 (2014) 17960–17969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Gao R, Huang SY, Marchand C, Pommier Y, Biochemical characterization of human tyrosyl-DNA phosphodiesterase 2 (TDP2/TTRAP): a Mg(2+)/Mn(2+)-dependent phosphodiesterase specific for the repair of topoisomerase cleavage complexes, J Biol Chem, 287 (2012) 30842–30852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Hornyak P, Askwith T, Walker S, Komulainen E, Paradowski M, Pennicott LE, Bartlett EJ, Brissett NC, Raoof A, Watson M, Jordan AM, Ogilvie DJ, Ward SE, Atack JR, Pearl LH, Caldecott KW, Oliver AW, Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2, Biochem J, 473 (2016) 1869–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Shi K, Kurahashi K, Gao R, Tsutakawa SE, Tainer JA, Pommier Y, Aihara H, Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2, Nature structural & molecular biology, 19 (2012) 1372–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Schellenberg MJ, Appel CD, Adhikari S, Robertson PD, Ramsden DA, Williams RS, Mechanism of repair of 5'-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2, Nature structural & molecular biology, 19 (2012) 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Gao R, Huang SY, Marchand C, Pommier Y, Biochemical Characterization of Human Tyrosyl-DNA Phosphodiesterase 2 (TDP2/TTRAP): a Mg2+/Mn2+-dependent phosphodiesterase specific for the repair of topoisomerase cleavage complexes, J. Biol. Chem, 287 (2012) 30842–30852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Schellenberg MJ, Perera L, Strom CN, Waters CA, Monian B, Appel CD, Vilas CK, Williams JG, Ramsden DA, Williams RS, Reversal of DNA damage induced Topoisomerase 2 DNA-protein crosslinks by Tdp2, Nucleic Acids Res, 44 (2016) 3829–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Riccio AA, Schellenberg MJ, Williams RS, Molecular mechanisms of topoisomerase 2 DNA-protein crosslink resolution, Cell Mol Life Sci, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Riccio AA, Schellenberg MJ, Williams RS, Molecular mechanisms of topoisomerase 2 DNA-protein crosslink resolution, Cell Mol Life Sci, 77 (2020) 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Virgen-Slane R, Rozovics JM, Fitzgerald KD, Ngo T, Chou W, van der Heden van Noort GJ, Filippov DV, Gershon PD, Semler BL, An RNA virus hijacks an incognito function of a DNA repair enzyme, Proc Natl Acad Sci U S A, 109 (2012) 14634–14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Bian K, Muppani NR, Elkhadragy L, Wang W, Zhang C, Chen T, Jung S, Seternes OM, Long W, ERK3 regulates TDP2-mediated DNA damage response and chemoresistance in lung cancer cells, Oncotarget, 7 (2016) 6665–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Lee KC, Swan RL, Sondka Z, Padget K, Cowell IG, Austin CA, Effect of TDP2 on the Level of TOP2-DNA Complexes and SUMOylated TOP2-DNA Complexes, International journal of molecular sciences, 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Schellenberg MJ, Lieberman JA, Herrero-Ruiz A, Butler LR, Williams JG, Munoz-Cabello AM, Mueller GA, London RE, Cortes-Ledesma F, Williams RS, ZATT (ZNF451)-mediated resolution of topoisomerase 2 DNA-protein cross-links, Science, 357 (2017) 1412–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Gomez-Herreros F, Schuurs-Hoeijmakers JH, McCormack M, Greally MT, Rulten S, Romero-Granados R, Counihan TJ, Chaila E, Conroy J, Ennis S, Delanty N, Cortes-Ledesma F, de Brouwer AP, Cavalleri GL, El-Khamisy SF, de Vries BB, Caldecott KW, TDP2 protects transcription from abortive topoisomerase activity and is required for normal neural function, Nat Genet, 46 (2014) 516–521. [DOI] [PubMed] [Google Scholar]
- [101].Zagnoli-Vieira G, Bruni F, Thompson K, He L, Walker S, de Brouwer APM, Taylor RW, Niyazov D, Caldecott KW, Confirming TDP2 mutation in spinocerebellar ataxia autosomal recessive 23 (SCAR23), Neurol Genet, 4 (2018) e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Ciaccio C, Castello R, Esposito S, Pinelli M, Nigro V, Casari G, Chiapparini L, Pantaleoni C, Group TS, D'Arrigo S, Consolidating the Role of TDP2 Mutations in Recessive Spinocerebellar Ataxia Associated with Pediatric Onset Drug Resistant Epilepsy and Intellectual Disability (SCAR23), Cerebellum, 18 (2019) 972–975. [DOI] [PubMed] [Google Scholar]
- [103].McKinnon PJ, Genome integrity and disease prevention in the nervous system, Genes Dev, 31 (2017) 1180–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Menon V, Povirk LF, End-processing nucleases and phosphodiesterases: An elite supporting cast for the non-homologous end joining pathway of DNA double-strand break repair, DNA Repair (Amst), 43 (2016) 57–68. [DOI] [PubMed] [Google Scholar]
- [105].Stingele J, Bellelli R, Boulton SJ, Mechanisms of DNA-protein crosslink repair, Nat Rev Mol Cell Biol, 18 (2017) 563–573. [DOI] [PubMed] [Google Scholar]
- [106].Sacho EJ, Maizels N, DNA Repair Factor MRE11/RAD50 Cleaves 3'-Phosphotyrosyl Bonds and Resects DNA to Repair Damage Caused by Topoisomerase 1 Poisons, J Biol Chem, 286 (2011) 44945–44951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Oh J, Symington LS, Role of the Mre11 Complex in Preserving Genome Integrity, Genes (Basel), 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Paull TT, 20 Years of Mre11 Biology: No End in Sight, Mol Cell, 71 (2018) 419–427. [DOI] [PubMed] [Google Scholar]
- [109].Larson ED, Cummings WJ, Bednarski DW, Maizels N, MRE11/RAD50 cleaves DNA in the AID/UNG-dependent pathway of immunoglobulin gene diversification, Mol Cell, 20 (2005) 367–375. [DOI] [PubMed] [Google Scholar]
- [110].Kass EM, Jasin M, Collaboration and competition between DNA double-strand break repair pathways, FEBS Lett, 584 (2010) 3703–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Neale MJ, Pan J, Keeney S, Endonucleolytic processing of covalent protein-linked DNA double-strand breaks, Nature, 436 (2005) 1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Hartsuiker E, Mizuno K, Molnar M, Kohli J, Ohta K, Carr AM, Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions, Mol Cell Biol, 29 (2009) 1671–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Hartsuiker E, Neale MJ, Carr AM, Distinct Requirements for the Rad32(Mre11) Nuclease and Ctp1(CtIP) in the Removal of Covalently Bound Topoisomerase I and II from DNA, Mol Cell, 33 (2009) 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Malik M, Nitiss JL, DNA repair functions that control sensitivity to topoisomerase-targeting drugs, Eukaryot Cell, 3 (2004) 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Lee KC, Padget K, Curtis H, Cowell IG, Moiani D, Sondka Z, Morris NJ, Jackson GH, Cockell SJ, Tainer JA, Austin CA, MRE11 facilitates the removal of human topoisomerase II complexes from genomic DNA, Biology open, 1 (2012) 863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Hoa NN, Shimizu T, Zhou ZW, Wang ZQ, Deshpande RA, Paull TT, Akter S, Tsuda M, Furuta R, Tsutsui K, Takeda S, Sasanuma H, Mre11 Is Essential for the Removal of Lethal Topoisomerase 2 Covalent Cleavage Complexes, Mol Cell, 64 (2016) 1010. [DOI] [PubMed] [Google Scholar]
- [117].Morimoto S, Tsuda M, Bunch H, Sasanuma H, Austin C, Takeda S, Type II DNA Topoisomerases Cause Spontaneous Double-Strand Breaks in Genomic DNA, Genes (Basel), 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Neece SH, Carles-Kinch K, Tomso DJ, Kreuzer KN, Role of recombinational repair in sensitivity to an antitumour agent that inhibits bacteriophage T4 type II DNA topoisomerase, Mol Microbiol, 20 (1996) 1145–1154. [DOI] [PubMed] [Google Scholar]
- [119].Woodworth DL, Kreuzer KN, Bacteriophage T4 mutants hypersensitive to an antitumor agent that induces topoisomerase-DNA cleavage complexes, Genetics, 143 (1996) 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Stohr BA, Kreuzer KN, Repair of topoisomerase-mediated DNA damage in bacteriophage T4, Genetics, 158 (2001) 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Connelly JC, de Leau ES, Leach DR, Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex, DNA Repair (Amst), 2 (2003) 795–807. [DOI] [PubMed] [Google Scholar]
- [122].Aedo S, Tse-Dinh YC, SbcCD-mediated processing of covalent gyrase-DNA complex in Escherichia coli, Antimicrob Agents Chemother, 57 (2013) 5116–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Aparicio T, Baer R, Gottesman M, Gautier J, MRN, CtIP, and BRCA1 mediate repair of topoisomerase II-DNA adducts, J Cell Biol, 212 (2016) 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Deshpande RA, Lee JH, Arora S, Paull TT, Nbs1 Converts the Human Mre11/Rad50 Nuclease Complex into an Endo/Exonuclease Machine Specific for Protein-DNA Adducts, Mol Cell, 64 (2016) 593–606. [DOI] [PubMed] [Google Scholar]
- [125].Deng C, Brown JA, You D, Brown JM, Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae, Genetics, 170 (2005) 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP, Human CtIP promotes DNA end resection, Nature, 450 (2007) 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Cannavo E, Cejka P, Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks, Nature, 514 (2014) 122–125. [DOI] [PubMed] [Google Scholar]
- [128].Nakamura K, Kogame T, Oshiumi H, Shinohara A, Sumitomo Y, Agama K, Pommier Y, Tsutsui KM, Tsutsui K, Hartsuiker E, Ogi T, Takeda S, Taniguchi Y, Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair, PLoS Genet, 6 (2010) e1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Liu C, Pouliot JJ, Nash HA, Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1, Proc Natl Acad Sci U S A, 99 (2002) 14970–14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Vance JR, Wilson TE, Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage, Proc Natl Acad Sci U S A, 99 (2002) 13669–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Zhang C, Liu Y, Hu Z, An L, He Y, Hang H, Targeted deletion of mouse Rad1 leads to deficient cellular DNA damage responses, Protein Cell, 2 (2011) 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Takahata C, Masuda Y, Takedachi A, Tanaka K, Iwai S, Kuraoka I, Repair synthesis step involving ERCC1-XPF participates in DNA repair of the Top1-DNA damage complex, Carcinogenesis, 36 (2015) 841–851. [DOI] [PubMed] [Google Scholar]
- [133].Kim Y, Spitz GS, Veturi U, Lach FP, Auerbach AD, Smogorzewska A, Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4, Blood, 121 (2013) 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Kametani Y, Takahata C, Narita T, Tanaka K, Iwai S, Kuraoka I, FEN1 participates in repair of the 5'-phosphotyrosyl terminus of DNA single-strand breaks, Carcinogenesis, 37 (2016) 56–62. [DOI] [PubMed] [Google Scholar]
- [135].Postel-Vinay S, Soria JC, ERCC1 as Predictor of Platinum Benefit in Non-Small-Cell Lung Cancer, J Clin Oncol, 35 (2017) 384–386. [DOI] [PubMed] [Google Scholar]
- [136].Giannini G, Ristori E, Cerignoli F, Rinaldi C, Zani M, Viel A, Ottini L, Crescenzi M, Martinotti S, Bignami M, Frati L, Screpanti I, Gulino A, Human MRE11 is inactivated in mismatch repair-deficient cancers, EMBO Rep, 15 (2002) 248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Wen Q, Scorah J, Phear G, Rodgers G, Rodgers S, Meuth M, A mutant allele of MRE11 found in mismatch repair-deficient tumor cells suppresses the cellular response to DNA replication fork stress in a dominant negative manner, Mol Biol Cell, 19 (2008) 1693–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Takemura H, Rao VA, Sordet O, Furuta T, Miao ZH, Meng L, Zhang H, Pommier Y, Defective Mre11-dependent Activation of Chk2 by Ataxia Telangiectasia Mutated in Colorectal Carcinoma Cells in Response to Replication-dependent DNA Double Strand Breaks, J. Biol. Chem, 281 (2006) 30814–30823. [DOI] [PubMed] [Google Scholar]
- [139].Marchand C, Huang SY, Dexheimer TS, Lea WA, Mott BT, Chergui A, Naumova A, Stephen AG, Rosenthal AS, Rai G, Murai J, Gao R, Maloney DJ, Jadhav A, Jorgensen WL, Simeonov A, Pommier Y, Biochemical Assays for the Discovery of TDP1 Inhibitors, Mol Cancer Ther, 13 (2014) 1535–7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Huang SY, Pommier Y, Marchand C, Tyrosyl-DNA Phosphodiesterase 1 (Tdp1) inhibitors, Expert Opin Ther Pat, 21 (2011) 1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Murai J, Marchand C, Shahane SA, Sun H, Huang R, Zhang Y, Chergui A, Ji J, Doroshow JH, Jadhav A, Takeda S, Xia M, Pommier Y, Identification of novel PARP inhibitors using a cell-based TDP1 inhibitory assay in a quantitative high-throughput screening platform, DNA Repair (Amst), 21 (2014) 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Pommier Y, O'Connor MJ, de Bono J, Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action, Sci Transl Med, 8 (2016) 362ps317. [DOI] [PubMed] [Google Scholar]
- [143].Murai J, Zhang Y, Morris J, Ji J, Takeda S, Doroshow JH, Pommier YG, Rationale for PARP inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition, J Pharmacol Exp Ther, 349 (2014) 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Thomas A, Pommier Y, Targeting Topoisomerase I in the Era of Precision Medicine, Clin Cancer Res, 25 (2019) 6581–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Kleiger G, Mayor T, Perilous journey: a tour of the ubiquitin-proteasome system, Trends Cell Biol, 24 (2014) 352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A, Structure and Function of the 26S Proteasome, Annu Rev Biochem, 87 (2018) 697–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Oh E, Akopian D, Rape M, Principles of Ubiquitin-Dependent Signaling, Annu Rev Cell Dev Biol, 34 (2018) 137–162. [DOI] [PubMed] [Google Scholar]
- [148].Desai SD, Liu LF, Vazquez-Abad D, D'Arpa P, Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin, J Biol Chem, 272 (1997) 24159–24164. [DOI] [PubMed] [Google Scholar]
- [149].Ogiso Y, Tomida A, Lei S, Omura S, Tsuruo T, Proteasome inhibition circumvents solid tumor resistance to topoisomerase II-directed drugs, Cancer Res, 60 (2000) 2429–2434. [PubMed] [Google Scholar]
- [150].Mao Y, Desai SD, Ting CY, Hwang J, Liu LF, 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes, J Biol Chem, 276 (2001) 40652–40658. [DOI] [PubMed] [Google Scholar]
- [151].Xiao H, Mao Y, Desai SD, Zhou N, Ting CY, Hwang J, Liu LF, The topoisomerase II beta circular clamp arrests transcription and signals a 26S proteasome pathway, Proc Natl Acad Sci U S A, 100 (2003) 3239–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Desai SD, Zhang H, Rodriguez-Bauman A, Yang JM, Wu X, Gounder MK, Rubin EH, Liu LF, Transcription-dependent degradation of topoisomerase I-DNA covalent complexes, Mol Cell Biol, 23 (2003) 2341–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Lin CP, Ban Y, Lyu YL, Liu LF, Proteasome-dependent processing of topoisomerase I-DNA adducts into DNA double strand breaks at arrested replication forks, J Biol Chem, 284 (2009) 28084–28092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y, Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5'-phosphorylated DNA double-strand breaks by replication runoff, Mol. Cell. Biol, 20 (2000) 3977–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Ban Y, Ho CW, Lin RK, Lyu YL, Liu LF, Activation of a novel ubiquitin-independent proteasome pathway when RNA polymerase II encounters a protein roadblock, Mol Cell Biol, 33 (2013) 4008–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Stingele J, Schwarz MS, Bloemeke N, Wolf PG, Jentsch S, A DNA-dependent protease involved in DNA-protein crosslink repair, Cell, 158 (2014) 327–338. [DOI] [PubMed] [Google Scholar]
- [157].Maskey RS, Flatten KS, Sieben CJ, Peterson KL, Baker DJ, Nam HJ, Kim MS, Smyrk TC, Kojima Y, Machida Y, Santiago A, van Deursen JM, Kaufmann SH, Machida YJ, Spartan deficiency causes accumulation of Topoisomerase 1 cleavage complexes and tumorigenesis, Nucleic Acids Res, 45 (2017) 4564–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Vaz B, Popovic M, Newman JA, Fielden J, Aitkenhead H, Halder S, Singh AN, Vendrell I, Fischer R, Torrecilla I, Drobnitzky N, Freire R, Amor DJ, Lockhart PJ, Kessler BM, McKenna GW, Gileadi O, Ramadan K, Metalloprotease SPRTN/DVC1 Orchestrates Replication-Coupled DNA-Protein Crosslink Repair, Mol Cell, 64 (2016) 704–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Lopez-Mosqueda J, Maddi K, Prgomet S, Kalayil S, Marinovic-Terzic I, Terzic J, Dikic I, SPRTN is a mammalian DNA-binding metalloprotease that resolves DNA-protein crosslinks, eLife, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Debethune L, Kohlhagen G, Grandas A, Pommier Y, Processing of nucleopeptides mimicking the topoisomerase I-DNA covalent complex by tyrosyl-DNA phosphodiesterase, Nucleic Acids Res, 30 (2002) 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Interthal H, Chen HJ, Champoux JJ, Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages, J Biol Chem, 280 (2005) 36518–36528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Zhang YW, Regairaz M, Seiler JA, Agama KK, Doroshow JH, Pommier Y, Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells, Nucleic Acids Res, 39 (2011) 3607–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Bastin-Shanower SA, Fricke WM, Mullen JR, Brill SJ, The mechanism of mus81-mms4 cleavage site selection distinguishes it from the homologous endonuclease rad1-rad10, Mol Cell Biol, 23 (2003) 3487–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Hamilton NK, Maizels N, MRE11 function in response to topoisomerase poisons is independent of its function in double-strand break repair in Saccharomyces cerevisiae, PLoS ONE, 5 (2010) e15387. [DOI] [PMC free article] [PubMed] [Google Scholar]