Candida albicans is a major human fungal pathogen and the leading cause of systemic Candida infections. In recent years, Als3 and Ece1 were identified as important factors for fungal virulence. Transcription of both corresponding genes is closely associated with hyphal growth. Here, we describe how Tup1, normally a global repressor of gene expression as well as of filamentation, and the transcription factor Ahr1 contribute to full expression of ALS3 and ECE1 in C. albicans hyphae. Both regulators are required for high mRNA amounts of the two genes to ensure functional relevant protein synthesis and localization. These observations identified a new aspect of regulation in the complex transcriptional control of virulence-associated genes in C. albicans.
KEYWORDS: Candida albicans, Tup1, filamentation, fungal virulence, gene regulation
ABSTRACT
The capacity of Candida albicans to reversibly change its morphology between yeast and filamentous stages is crucial for its virulence. Formation of hyphae correlates with the upregulation of genes ALS3 and ECE1, which are involved in pathogenicity processes such as invasion, iron acquisition, and host cell damage. The global repressor Tup1 and its cofactor Nrg1 are considered to be the main antagonists of hyphal development in C. albicans. However, our experiments revealed that Tup1, but not Nrg1, was required for full expression of ALS3 and ECE1. In contrast to NRG1, overexpression of TUP1 was found to inhibit neither filamentous growth nor transcription of ALS3 and ECE1. In addition, we identified the transcription factor Ahr1 as being required for full expression of both genes. A hyperactive version of Ahr1 bound directly to the promoters of ALS3 and ECE1 and induced their transcription even in the absence of environmental stimuli. This regulation worked even in the absence of the crucial hyphal growth regulators Cph1 and Efg1 but was dependent on the presence of Tup1. Overall, our results show that Ahr1 and Tup1 are key contributors in the complex regulation of virulence-associated genes in the different C. albicans morphologies.
INTRODUCTION
The major human fungal pathogen Candida albicans can grow in different morphologies: unicellular yeast cells, pseudohyphae, and (true) hyphae (1). This morphological plasticity is crucial for fungal pathogenicity as formation of hyphae is involved in adhesion to and invasion of host cells and tissues, while yeast cells are required for dissemination within the bloodstream (2, 3). Within host cells, hyphae and their associated proteins are involved in the acquisition of trace elements such as iron and zinc (4, 5). The different growth forms of C. albicans exhibit distinct interaction patterns with host immune cells. Yeast cells are recognized and taken up by macrophages. However, some cells have the potential to survive and start to form hyphae. These hyphae are able to destroy macrophages by early induction of pyroptosis, host glucose consumption, and toxin production and later direct escape by physical forces (6–9). In contrast, neutrophils are activated only by hyphae and are crucial for fungal killing (10). Consequently, neutropenic patients have a significantly impaired outcome for infections with C. albicans (11). Fungal polymorphism is also important for the interaction with dendritic cells but not natural killer cells (12, 13). In recent years, genes ALS3 and ECE1 were found to encode key contributors to fungal virulence (14, 15). Together with DCK1, HGT2, HWP1, IHD1, RBT1, and orf19.2457, they are also part of the core filamentation response (CFR), since their expression is induced whenever C. albicans forms hyphae, regardless of the environmental stimulus (16). ALS3 encodes a multifunctional protein involved in adhesion, invasion, and iron acquisition (4, 14, 17). The ECE1 product is normally the transcript with the highest abundance in C. albicans hyphae, independently of the environmental stimulus that triggered filamentation (16). The encoded protein is processed intracellularly by the proteinases Kex1 and Kex2 into eight peptides (15, 18, 19). One of these peptides, candidalysin, is secreted from hyphae into the environment and mediates host cell cytolysis (15). Candidalysin is involved in the immunopathology of Candida vaginitis (20) and drives protective innate type 17 cell responses during oral candidiasis (15, 21).
Due to their importance for fungal virulence, knowledge about the regulation of ALS3 and ECE1 expression can contribute to a better understanding of fungal pathogenicity mechanisms. Both genes are downregulated in mutants lacking key activators of filamentation (16, 22). In contrast, they are both upregulated in mutants lacking the hyphal growth repressors Tup1 and Nrg1 (23, 24). Despite this, tup1Δ and nrg1Δ mutants exhibit attenuated virulence in several infection models (23, 25–27).
Here, we show that the transcriptional regulators Ahr1 and Tup1 play crucial roles in the activation of ALS3 and ECE1. While Tup1 contributes to repression and activation of the two genes, the presence of Ahr1 is important for high-level, functionally relevant expression of ALS3 and ECE1 in C. albicans hyphae.
RESULTS
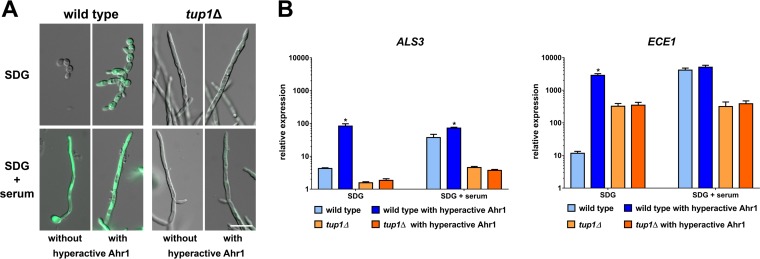
Tup1 is required for activation of ECE1 and ALS3.
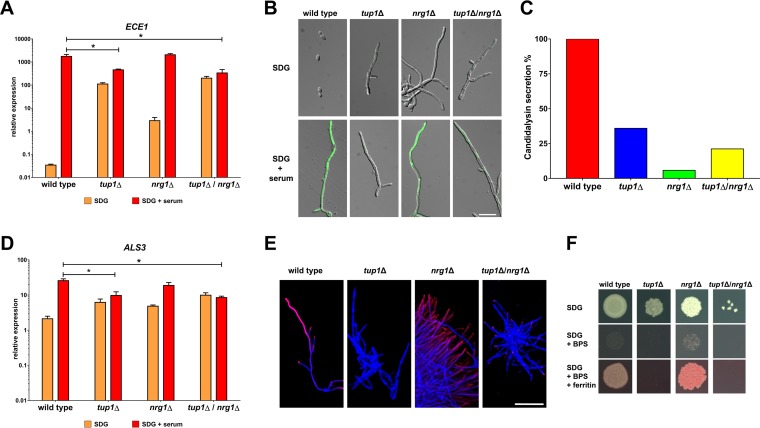
We used RT-qPCR (reverse transcription followed by qPCR) to quantify the transcription of ECE1 in the wild-type strain and in mutants lacking the hyphal growth repressors Nrg1 and Tup1 in minimal medium with or without 10% human serum. As expected, the gene was barely expressed in wild-type yeast cells but was highly upregulated in hyphae (Fig. 1A). It was expressed at higher levels in the filamentous mutants than in the wild type under yeast growth conditions, as expected (Fig. 1A). Under hyphal growth conditions, however, ECE1 was expressed at significantly lower levels in the tup1Δ mutant than in the wild type, which was not the case for the nrg1Δ mutant (Fig. 1A). In fact, ECE1 transcription in the tup1Δ mutant remained at an intermediate level under yeast and hyphal growth conditions. This unexpected low level of expression was not caused by Nrg1 activity as a tup1Δ/nrg1Δ double mutant showed levels of ECE1 transcription similar to those seen with the tup1Δ single mutant (Fig. 1A). We used an established green fluorescent protein (GFP) reporter system for ECE1 to visualize expression in these strains and observed that the level of upregulation of ECE1 in the mutants under yeast growth conditions was not sufficient for a visible GFP signal (Fig. 1B). In accordance with the RT-qPCR data, nrg1Δ filaments showed bright GFP signals under hyphal growth conditions, while only weak fluorescence or no fluorescence was observed in the tup1Δ and tup1Δ/nrg1Δ mutants (Fig. 1B). The decreased ECE1 expression in the tup1Δ mutant was not observed exclusively for serum-induced filamentation, as similar results were observed for hyphae which were induced by either pH or medium change (see Fig. S1 in the supplemental material). These results obtained with the GFP reporter system and RT-qPCR correlated with a significantly lower level of release of candidalysin by the tup1Δ mutant than by wild-type hyphae (Fig. 1C). However, we also noted that the nrg1Δ mutant was not able to release as much candidalysin as the wild type, indicating that some other defects in these mutants prevented the translation of high ECE1 mRNA amounts into released candidalysin (Fig. 1C). ECE1 is not the only core filamentation response gene which requires the presence of Tup1 for its high-level transcription in C. albicans hyphae. Similar expression patterns were observed for ALS3 (Fig. 1D), HWP1, and IHD1 but not for the other CFR genes (Fig. S2). As seen with ECE1 and candidalysin, the intermediate levels of expression of ALS3 in the tup1Δ and tup1Δ/nrg1Δ mutants were not sufficient for surface localization of the Als3 protein in these mutants (Fig. 1E). Both strains were also not able to use ferritin as an iron source; such use is known to be dependent on the presence of the Als3 protein on the cell surface (Fig. 1F) (3).
FIG 1.
Tup1 is required for high-level expression of ECE1 and ALS3. (A) Total RNA was isolated from the indicated strains after 6 h of growth in SDG with or without 10% human serum. ECE1 transcription was normalized against ACT1 and the control RNA (wild type, 5 h YPD, 37°C). Asterisks indicate significant transcription differences in a mutant compared to the wild type after growth in SDG with serum (P ≤ 0.05, two-tailed, unpaired Student's t test). (B) The indicated strains with integrated pECE1-GFP cassettes were grown for 6 h at 37°C in SDG with or without serum prior to microscopy. Shown are the overlays of the DIC channel and the GFP channel. Scale bar, 20 μm. (C) Candidalysin secretion was measured by LC-MS/MS after 18 h of growth in YNBS (pH 7.2). Candidalysin contents measured for wild-type hyphae were defined as 100%. (D) The total RNA isolated as described for panel A was used to determine the normalized relative expression levels of ALS3. Asterisks indicate significant transcription differences in a mutant compared to the wild type after growth in SDG with serum (P ≤ 0.05, two-tailed, unpaired Student's t test). (E) After 6 h of growth in SDG with 10% human serum at 37°C, cells of the indicated strains were stained first with a monoclonal anti-Als3 antibody (pink signal) and then with calcofluor white (blue signal). Overlays of the images taken in the Cy5 and DAPI (4′,6-diamidino-2-phenylindole) channel are shown. Scale bar, 50 μm. (F) From an overnight culture, 103 cells of the indicated strains were dropped on SDG medium with or without an iron chelator (bathophenanthroline disulfonate [BPS]) and with or without ferritin. The plates were grown for 3 days at 37°C in 5% CO2 before images were taken.
Tup1 is required for ECE1 expression after pH shift and medium change. The wild-type strain and tup1Δ strains with integrated pECE1-GFP were grown in the indicated media for 6 h at 37°C prior to fluorescence microscopy. Shown are the overlays of the DIC channel and the GFP channels. Scale bar, 20 μm. Download FIG S1, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Core filamentation gene expression response in filamentous mutants. The wild-type strain and the filamentous mutants were grown in SDG with or without 10% human serum for 6 h at 37°C prior to the isolation of total RNA, which was used for the determination of the relative levels of expression of the indicated genes. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student’s t test) from the results seen with the wild-type strain grown in SDG with serum. Download FIG S2, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overexpression of TUP1 does not inhibit hyphal morphology and expression of ALS3 and ECE1.
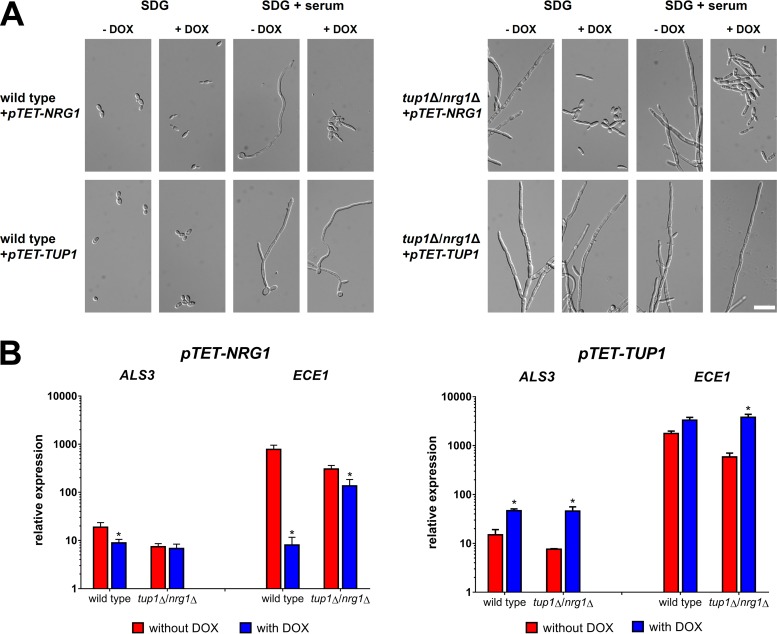
Due to the surprising finding that Tup1 was required for high-level ALS3 and ECE1 expression, we studied the effects of induced overexpression of TUP1 and NRG1 using pTET constructs. Overexpression of NRG1, but not TUP1, prevented filamentation in a wild-type background (Fig. 2A). Both pTET constructs were also integrated into the tup1Δ/nrg1Δ double mutant. Again, overexpression of NRG1 triggered filament-to-yeast reversion whereas overexpression of TUP1 did not affect filamentous growth (Fig. 2A). Induced overexpression of NRG1 in the wild type led to decreased ALS3 and ECE1 transcript levels (Fig. 2B). In contrast, TUP1 overexpression enhanced the expression of both genes and even restored high mRNA levels of ECE1 in the tup1Δ/nrg1Δ mutant (Fig. 2B). Nonetheless, the presence of Tup1 was important for the repression of ECE1 by NRG1 overexpression (Fig. 2B), illustrating that it contributes to repression and activation of this gene.
FIG 2.
Overexpression of TUP1 promotes ALS3 and ECE1 transcription. TET promoter-driven constructs of NRG1 and TUP1 were integrated into the ADH1 loci of wild-type strain SC5314 and the tup1Δ/nrg1Δ double mutant. (A) The morphology of the resulting strains was studied by microscopy after 6 h of incubation at 37°C in SDG or SDG with 10% human serum with or without the addition of 50 μg/ml doxycycline. Scale bar, 20 μm. (B) Total RNA of strains expressing either the pTET-TUP1 or the pTET-NRG1 construct was isolated after 6 h growth in SDG with 10% human serum. The pTET constructs were activated by the addition of 50 μg/ml doxycycline to the medium. The isolated RNA was used for RT-qPCR to determine the relative levels of gene expression of ALS3 and ECE1. A control RNA (wild type, 5 h YPD, 37°C) and the housekeeping gene ACT1 were used for normalization. Asterisks mark significant differences after overexpression of either NRG1 or TUP1 compared to the same strain without overexpression (P ≤ 0.05, two-tailed, unpaired Student's t test).
Ahr1 is required for the expression of ECE1 and ALS3.
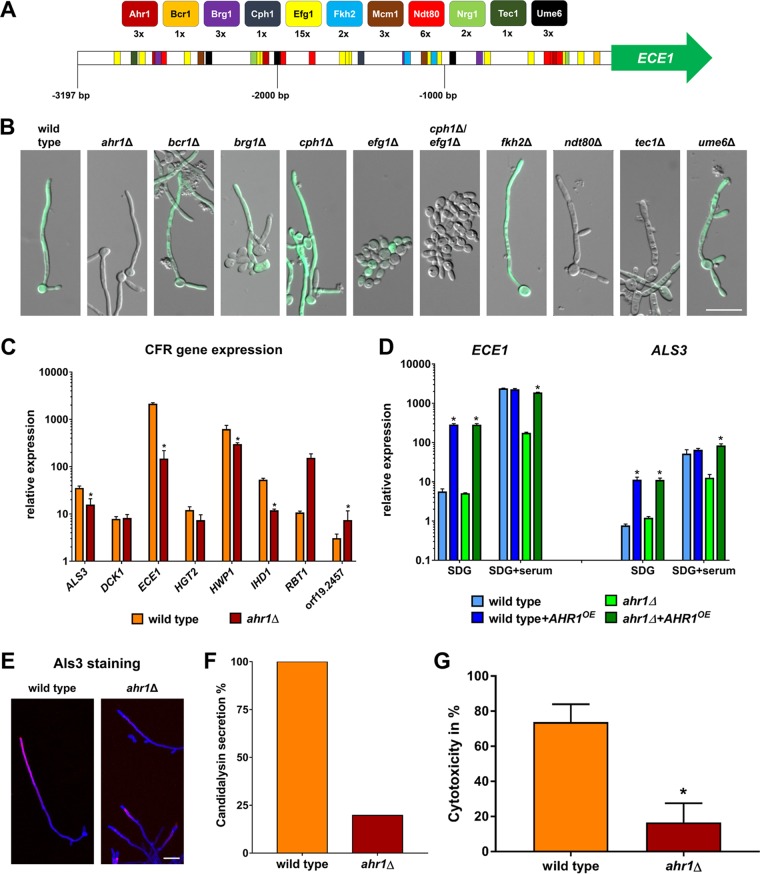
In addition to the influence of putative repressors, we also examined the influence of putative activators on the expression of ECE1. The 5′ intergenic region of the gene has a size of 3,197 bp and contains potential binding motifs for a variety of transcriptional activators which could at least in theory bind and regulate gene expression (Fig. 3A; see also Table S1). To visualize ECE1 expression, GFP was integrated into the ECE1 loci of mutants lacking these transcription factors to monitor ECE1 expression levels. The resulting strains were then examined for GFP signals under hyphal growth conditions. The efg1Δ, cph1Δ/efg1Δ, ndt80Δ, and tec1Δ mutants were not able to form hyphae, and in accordance with this phenotype, the GFP signal was mostly absent or weak (Fig. 3B). Strains lacking BCR1 or BRG1 partially formed hyphae with GFP signals present (Fig. 3B). The hyphae of the cph1Δ and fkh2Δ mutants showed bright GFP signals (Fig. 3B). The ume6Δ mutant initially formed hyphae with the expected GFP signal, although the hyphal growth quickly reverted to yeast growth (Fig. 3B). The only mutant which was able to form morphologically normal hyphae without a GFP signal was the ahr1Δ mutant (Fig. 3B). RT-qPCR results confirmed the microscopic observations and showed that the level of ECE1 transcription in ahr1Δ filaments was indeed significantly lower than in wild-type hyphae (Fig. 3C; see also Fig. S3). As seen with ECE1, the levels of transcription of ALS3, HWP1, and IHD1 were significantly reduced in ahr1Δ hyphae (Fig. 3C). Reintegration of AHR1 into the ADH1 locus, which also led to overexpression of the AHR1 gene, restored the high-level expression of ALS3 and ECE1 in ahr1Δ hyphae (Fig. 3D). Interestingly, pADH1-driven AHR1 induced upregulation of ALS3 and ECE1 in the wild type already under yeast growth conditions (Fig. 3D). As observed for the tup1Δ mutant, the intermediate expression levels of the ALS3 and ECE1 genes in the ahr1Δ mutant correlated with an absence of Als3 protein surface localization (Fig. 3E) and low secretion of candidalysin (Fig. 3F). In agreement with these findings, the level of virulence of the mutant in an oral epithelial cell infection model was found to be attenuated (Fig. 3G).
FIG 3.
Ahr1 is required for high-level expression of ECE1 and ALS3. (A) Scheme of the intergenic region upstream of ECE1 with potential binding sites of transcriptional activators. (B) The wild-type strain and regulatory mutants expressing the pECE1-GFP construct were grown for 6 h at 37°C in SDG with 10% human serum prior to microscopy. Shown are the overlays of the DIC and the GFP channels. Scale bar, 20 μm. (C) The wild type and the ahr1Δ mutant were grown for 6 h at 37°C in SDG medium with serum prior to isolation of total RNA, which was used for the determination of relative gene expression levels. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student's t test) compared to the wild type. (D) Total RNA of the wild-type and ahr1Δ strains with or without AHR1 overexpression. The strains were grown for 6 h at 37°C in SDG with or without 10% human serum for the determination of relative gene expression levels. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student's t test) compared to the strain without AHR1 overexpression. (E) Cells of the indicated strains were grown for 6 h in SDG with 10% human serum at 37°C and were then stained with a monoclonal anti-Als3 antibody (pink signal), followed by a second staining with calcofluor white (blue signal). Shown are the overlays of the images taken in the Cy5 and DAPI channels. Scale bar, 20 μm. (F) Levels of candidalysin secretion of the wild type and the ahr1Δ mutant were measured by mass spectrometry after 18 h growth in YNBS (pH 7.2). Candidalysin contents measured for wild-type hyphae were defined as 100%. (G) Cytotoxicity of the indicated strains was determined by LDH release from the infected TR-146 cells after a 24 h coincubation. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student's t test) compared to the wild type.
ECE1 transcription in activator mutants. The wild-type strain and the indicated activator mutants were grown in SDG medium with 10% human serum for 4 h prior to the isolation of total RNA. The RNA was used to determine the relative levels of expression of ECE1. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student’s t test) compared to the wild type. Download FIG S3, TIF file, 0.9 MB (968KB, tif) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Putative transcription factor binding sites in the 5′ intergenic region of ECE1. Download Table S1, XLSX file, 0.01 MB (11.6KB, xlsx) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
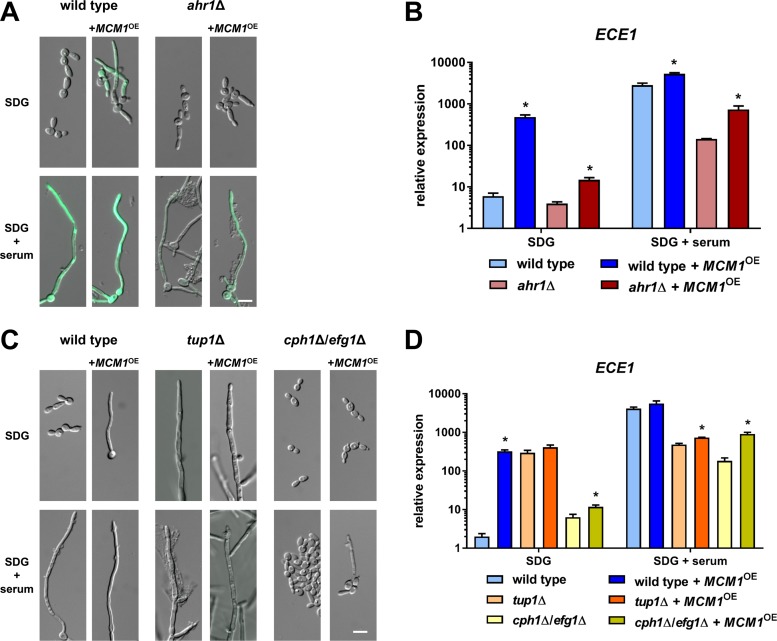
MCM1 overexpression can induce the expression of ECE1 and ALS3 in the presence of Ahr1 and Tup1.
Ahr1 is known for its interaction with Mcm1, a transcription factor which itself also binds to the ECE1 promoter (28, 29). As deletion of this essential transcriptional regulator is not feasible, we analyzed the effects of MCM1 overexpression under the control of the ADH1 promoter in the wild-type and ahr1Δ strains, each carrying the ECE1-GFP reporter system. This overexpression resulted in observations of GFP signals under yeast and hyphal growth conditions in the wild-type background (Fig. 4A). In the ahr1Δ mutant, however, the MCM1 overexpression led to a GFP signal in hyphae only (Fig. 4A). RT-qPCR showed that MCM1 overexpression triggered a strong upregulation of ECE1 in the wild-type background under both growth conditions (Fig. 4B). This effect of the overexpression was much lower in the absence of Ahr1 under yeast growth conditions (Fig. 4B). However, under hyphal growth conditions, MCM1 overexpression significantly increased ECE1 transcription in the ahr1Δ mutant, although not up to wild-type hyphal levels (Fig. 4B). Due to these observations, we studied the effects of MCM1 overexpression in the filamentous tup1Δ mutant and the nonfilamentous cph1Δ/efg1Δ double mutant. MCM1 overexpression partially rescued filamentation in the cph1Δ/efg1Δ double mutant (Fig. 4C), which was associated with upregulation of ECE1, although not to the level found in wild-type hyphae (Fig. 4D). In contrast, it was unable to compensate for the absence of Tup1 either morphologically or regarding ECE1 expression, which remained at intermediate levels (Fig. 4D). Similar gene expression patterns were observed for ALS3, HWP1, and IHD1 (Fig. S4).
FIG 4.
MCM1 overexpression activates transcription of ECE1. (A) The wild type and the ahr1Δ mutant with integrated pECE1-GFP and with or without pADH1-MCM1 were grown in SDG medium with or without 10% human serum for 6 h at 37°C prior to microscopy. Scale bar, 20 μm. (B and D) Total RNA from the wild-type and ahr1Δ mutant strains (B) and from the wild-type and tup1Δ and cph1Δ/efg1Δ mutant strains was isolated after 6 h growth and used to determine the relative ECE1 expression levels. Scale bars, 20 μm. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student's t test) compared to the corresponding strain without MCM1 overexpression. (C) The indicated strains were grown for 6 h at 37°C in SDG medium with or without 10% human serum prior to microscopy. Scale bar, 10 μm.
Core filamentation gene expression response during overexpression of MCM1. The indicated strains were grown for 4 h at 37°C in SDG medium with or without 10% human serum prior to the isolation of total RNA. The RNA was used to determine the relative levels of gene expression. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student’s t test) compared to the wild type. Download FIG S4, TIF file, 2.6 MB (2.7MB, tif) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
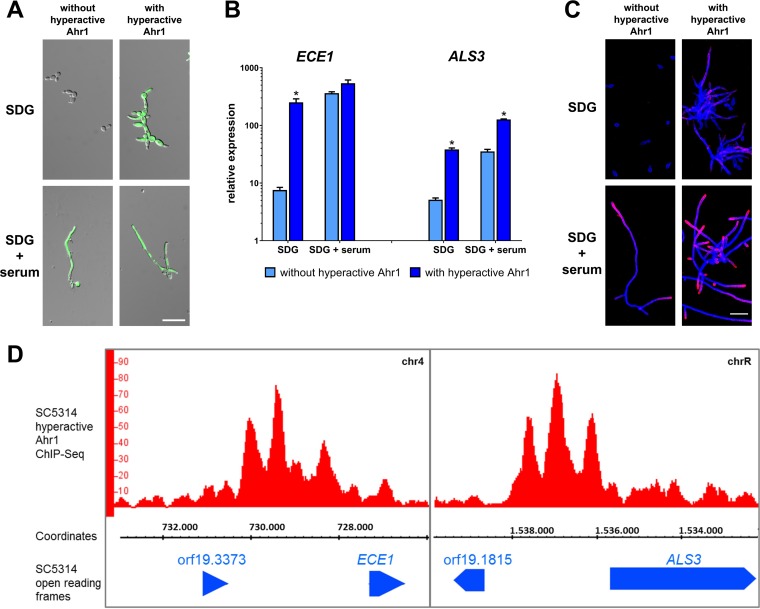
Hyperactive Ahr1 induces ALS3 and ECE1 expression without environmental stimuli by direct binding to the target promoters.
It was previously shown that a hyperactive version of Ahr1 can trigger filamentation in C. albicans (30). This construct, consisting of an AHR1 allele which was fused to a Gal4 activator domain (GAD) and a 3× hemagglutinin (HA3) tag, was integrated into the ADH1 locus of a wild-type derivative already carrying the pECE1-GFP construct. The resulting strain showed a bright GFP signal already under yeast growth conditions (Fig. 5A). Independently of the environment, the expression of ECE1 and ALS3 in the wild type with hyperactive Ahr1 was as high as in normal wild-type hyphae (Fig. 5B). Thus, addition of the Gal4 activator domain further enhanced the effects of AHR1 overexpression (Fig. 3D). We also observed slightly increased Als3 protein signals in the wild type with hyperactive Ahr1 under yeast growth conditions (Fig. 5C).
FIG 5.
Hyperactive Ahr1 induces high-level expression of ALS3 and ECE1. (A) The indicated strains with or without the hyperactive Ahr1 were grown in SDG or SDG with 10% human serum. Pictures were taken after 6 h of growth at 37°C. Shown are the overlays of the DIC and the GFP channels. Scale bar, 20 μm. (B) After 6 h of growth, total RNA of the strains grown as described for panel A was isolated and used for determination of relative gene expression levels. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student's t test) in mutants with hyperactive Ahr1 compared to their background strains without the hyperactive allele. (C) Cells of the indicated strains were grown for 6 h in SDG with 10% human serum at 37°C and were then stained with a monoclonal anti-Als3 antibody (pink signal), followed by a second staining with calcofluor white (blue signal). Shown are the overlays of the images taken in the Cy5 and DAPI channels. Scale bar, 20 μm. (D) ChIP-Seq shows direct binding of hyperactive Ahr1 to the promoters of ECE1 and ALS3. Genomic DNA used for ChIP-Seq was isolated from the wild-type strain with hyperactive Ahr1 after 6 h growth in SDG medium at 37°C. The binding peaks as shown in the IGB viewer are displayed.
To test whether induction of ECE1 was related to promoter binding of Ahr1, we used the wild-type derivative with the hyperactive Ahr1 to perform chromatin immunoprecipitation sequencing (ChIP-Seq) experiments under yeast growth conditions. ChIP-Seq provided clear evidence of physical binding of the hyperactive Ahr1 to the promoters of ECE1 and ALS3 (Fig. 5D) but also to those of other CFR genes (Table S2). The Ahr1 binding motif was found to be identical to one previously described (31). A nontagged version of the hyperactive Ahr1 was used to validate the ChIP-Seq results as shown for the ECE1 neighborhood (Fig. S5).
Validation of ChIP-Seq results for wild-type derivatives with hyperactive Ahr1. ChIP-Seq reads (red) mapped to the genome of the SC5314 wild type (ASM18296v3). An extract from chromosome 4 with the ORFs of ECE1, RBT1, and HWP1 (blue) is shown. (Top) ChIP-Seq reads of the control strain with hyperactive Ahr1 without a HA3 tag in the SC5314 background. (Middle) ChIP-Seq reads of the mutant with hyperactive Ahr1 with a HA3 tag in the SC5314 background. Identified peak summits are depicted in black. Download FIG S5, TIF file, 0.9 MB (994.1KB, tif) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Results of the ChIP-Seq analysis performed with hyperactive Ahr1. Download Table S2, XLSX file, 0.03 MB (34.5KB, xlsx) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
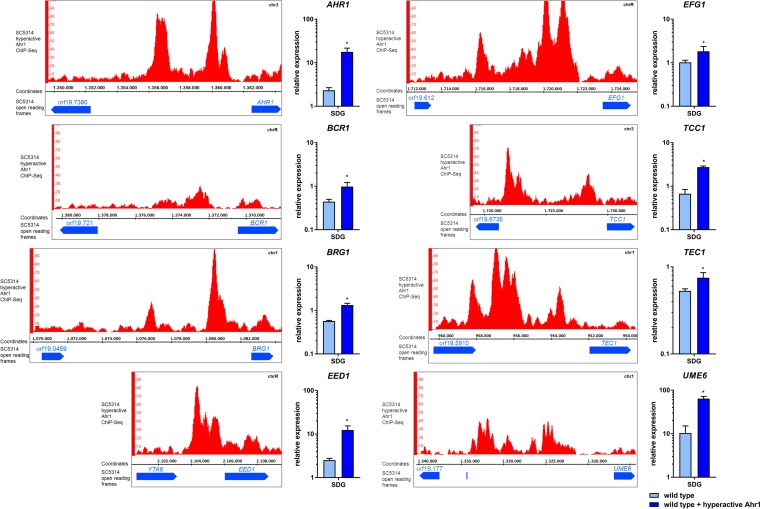
Hyperactive Ahr1 binds to genes encoding hypha-associated transcription factors and induces their expression.
Hyperactive Ahr1 also bound to the promoters of transcriptional regulator genes BCR1, BRG1, EFG1, TEC1, and UME6, and this binding correlated with their upregulation (Fig. 6; see also Table S2). We also detected binding to the promoter of EED1, which was associated with increased expression of the gene (Fig. 6; see also Table S2). In addition, hyperactive Ahr1 bound to the promoter of TCC1, which encodes a Tup1-interacting protein (Fig. 6; see also Table S2). However, physical attachment to the promoters of NRG1 and TUP1 was not detected. Binding of hyperactive Ahr1 to the promoter of the AHR1 gene itself might indicate a self-controlling feedback mechanism (Fig. 6; see also Table S2).
FIG 6.
Binding of hyperactive Ahr1 increases expression levels of regulatory genes. The wild-type strain with hyperactive Ahr1 was grown for 4 h in SDG medium at 37°C prior to the ChIP-Seq analyses. The binding of hyperactive Ahr1 to regulatory genes is displayed in IGB viewer images. After growth under the same conditions, total RNA of the wild type with or without hyperactive Ahr1 was isolated at the same time point and used for the determination of relative gene expression levels. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student's t test) in the presence of hyperactive Ahr1 compared to the wild type alone.
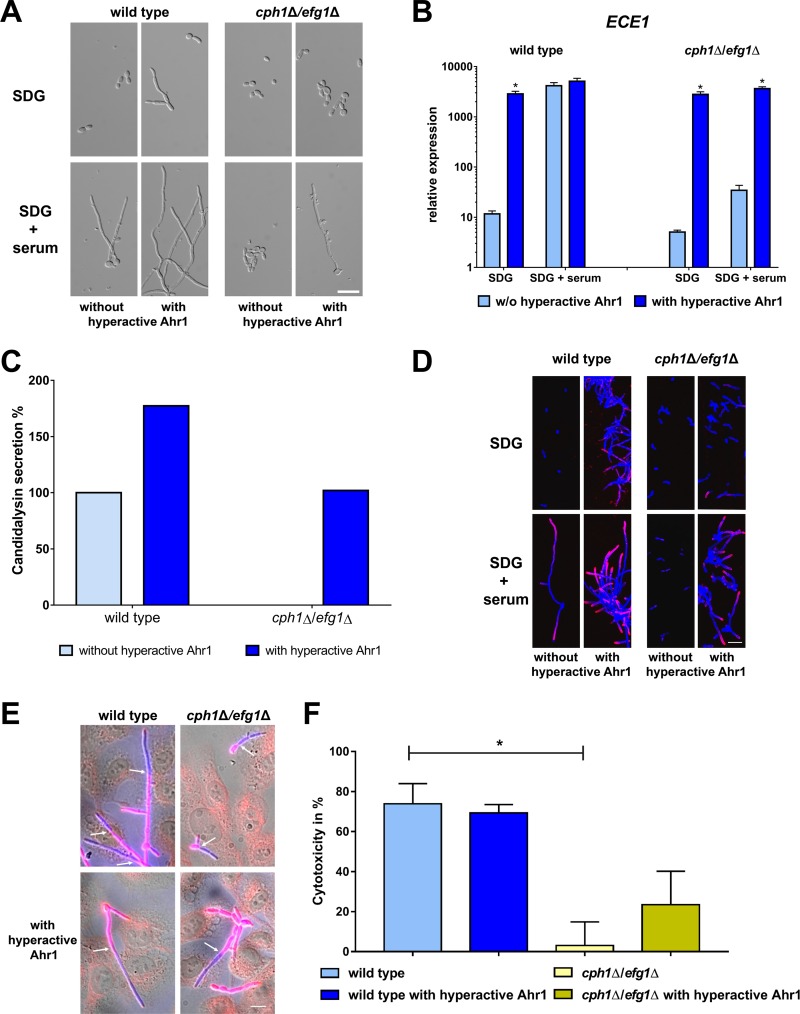
Hyperactive Ahr1 induces ALS3 and ECE1 expression in the absence of Cph1 and Efg1 but depends on Tup1.
In the next step, we integrated the allele coding for the hyperactive Ahr1 into the nonfilamentous cph1Δ/efg1Δ double mutant. This partially restored filamentation in the double mutant (Fig. 7A) and, more strikingly, induced a significant increase of ECE1 transcription to the levels in wild-type hyphae (Fig. 7B). In accordance with these observations, the cph1Δ/efg1Δ strain with hyperactive Ahr1 showed an increased secretion of candidalysin which was absent from the cph1Δ/efg1Δ supernatants (Fig. 7C). Hyperactive Ahr1 also triggered the upregulation of ALS3, HWP1, and IHD1 in cph1Δ/efg1Δ under both yeast and hyphal growth conditions (Fig. S6). This upregulation was sufficient to ensure a surface localization of Als3 protein on the cph1Δ/efg1Δ mutant cells under hyphal growth conditions (Fig. 7D). Cells of the double mutant with or without hyperactive Ahr1 were also able to invade human oral epithelial cells, although not as efficiently as the wild type (Fig. 7E). Despite a modestly increased level of lactate dehydrogenase (LDH) release of infected epithelial cells, hyperactive Ahr1 could restore virulence capacity only partially in the cph1Δ/efg1Δ mutant as the level of cell damage was still much lower than for the wild type (Fig. 7F). This suggests that secretion of candidalysin alone is not sufficient for full virulence in this mutant.
FIG 7.
Hyperactive Ahr1 activates ECE1 and ALS3 expression in the nonfilamentous cph1Δ/efg1Δ mutant. (A) Wild type SC5314 and cph1Δ/efg1Δ mutant with or without the hyperactive Ahr1 were grown for 6 h at 37°C in either SDG or SDG with 10% human serum prior to microscopy. Scale bar, 20 μm. (B) After 6 h growth under the same conditions as described for panel A, total RNA of these strains was isolated and used for determination of relative ECE1 expression levels. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student's t test) in mutants with hyperactive Ahr1 compared to their background strains without the hyperactive allele. (C) Candidalysin secretion of wild-type and cph1Δ/efg1Δ strains with or without the hyperactive Ahr1 was measured by LC-MS/MS after 18 h growth in YNBS (pH 7.2). Candidalysin secreted by wild-type hyphae was defined as 100%. (D) Cells of the indicated strains were grown for 6 h in SDG with 10% human serum at 37°C and then stained with the Als3 antibody (pink) and calcofluor white (blue signal). Shown are the overlays of the images taken in the Cy5 and DAPI channels. Scale bar, 20 μm. (E) TR-146 oral epithelial cells infected with wild-type and cph1Δ/efg1Δ strains with or without hyperactive Ahr1 after 4 h coincubation. C. albicans outside human cells is shown in pink; C. albicans inside human cells is shown in blue. Arrows mark the point of invasion. Scale bar, 20 μm. (F) Cytotoxicity of the indicated strains was determined by the release of LDH from infected TR-146 cells after a 24 h coincubation. Asterisks mark significant changes (P ≤ 0.05, two-tailed, unpaired Student's t test) to values of the wild-type strain.
Hyperactive Ahr1 induces expression of ALS3, HWP1, and IHD1 in the cph1Δ/efg1Δ double mutant. Wild-type and cph1Δ/efg1Δ strains with or without integrated hyperactive Ahr1 were grown for 6 h at 37°C in SDG medium with or without 10% human serum prior to the isolation of total RNA. The RNA was used to determine the relative levels of expression of genes ALS3, HWP1, and IHD1. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student’s t test) compared to the background strains without hyperactive Ahr1. Download FIG S6, TIF file, 0.9 MB (986.3KB, tif) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Interestingly, the hyperactive Ahr1 was able to induce a significant increase of ECE1 transcription in mutants lacking other hyphae regulators, such as ndt80Δ, tec1Δ, and ume6Δ mutants (Fig. S7). However, the hyperactive Ahr1 did not enhance the expression of ECE1 and ALS3 in the tup1Δ mutant, indicating that the presence of Tup1 is essential for the upregulation of both genes by the hyperactive Ahr1 (Fig. 8).
FIG 8.
Beneficial effects of the hyperactive Ahr1 for ALS3 and ECE1 expression depend on Tup1. (A) The wild-type strain and tup1Δ derivates with integrated pECE1-GFP and with or without hyperactive Ahr1 were grown for 4 h in SDG medium with or without 10% human serum at 37°C prior to microscopy. Shown are overlays of the DIC and the GFP channels. Scale bar, 10 μm. (B) After the same time of incubation in the indicated media, total RNA of the indicated strains was isolated and used for RT-qPCR to determine the relative levels of gene expression of ALS3 and ECE1. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student's t test) in mutants with AHR1-GAD compared to their background strains without the hyperactive allele.
Hyperactive Ahr1 restores ECE1 transcription in regulatory mutants. (A) The indicated strains with or without integrated hyperactive Ahr1 were grown for 4 h at 37°C in SDG medium with or without 10% human serum prior to microscopy. Scale bar, 10 μm. (B) Total RNA from the aforementioned strains was isolated after the same time of growth. The RNA was used to determine the relative levels of expression of the ECE1 gene. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student’s t test) compared to the background strains without hyperactive Ahr1. Download FIG S7, TIF file, 2.6 MB (2.7MB, tif) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
The results of our study revealed a complex picture of how the virulence-associated genes ALS3 and ECE1 of C. albicans are transcriptionally regulated. Both genes have long been known for high mRNA abundance in hyphae, and we found that these high transcript levels in hyphae are indeed required for function, as even a drop to a more intermediate level of ECE1 expression led to the absence of candidalysin secretion. A similar correlation was found between ALS3 expression and localization of its encoded protein on the cell surface. This striking disparity between intermediate transcript levels and a lack of functional protein (surface Als3 or secreted candidalysin) may indicate posttranscriptional regulation of mRNA and/or protein levels that warrant further studies. Our study results show that Ahr1 and Tup1 are key contributors in this complex regulation of the two genes in the different C. albicans morphologies. The presence of both regulators is required to reach high transcription levels, revealing new functions for both of them. Here, we showed that Ahr1 binds directly to the promoters of ALS3 and ECE1 and induces their transcription. These findings expand the previous knowledge about Ahr1, which was thus far known for its involvement in white-opaque switching and in fungal modulation of the environmental pH but also in regulation of fungal adhesion and filamentation (28, 30–32). This novel role of Ahr1 might explain why the deletion mutant not only is attenuated in an oral epithelium infection model but also fails to induce pyroptosis in infected phagocytes (7, 33, 34), for which candidalysin plays an important role (8, 35). According to the Candida Genome Database, orthologs of Ece1 can be found in the genomes of C. dubliniensis and C. tropicalis, which are the closest relatives of C. albicans (36). Ahr1 homologs can similarly be found in both fungi, hinting at the possibility that the regulatory mechanism described here is conserved in other Candida species as well.
In previous works, native Ahr1 was C-terminally tagged and used for ChIP-based identifications of the binding motifs in the promoters of target genes (28, 31). Although the hyperactive Ahr1 which we used displayed enhanced activity compared to a native version, we identified the same binding motifs, indicating that the C-terminal addition of the Gal4 activator domain did not lead to false-positive results.
In addition, a hyperactive version of Ahr1 was found to be bound to the promoters of several transcription factor genes, including BRG1, EFG1, TEC1, and UME6, indicating that it regulates a variety of processes which are linked with hyphal growth and virulence. Interestingly, the hyperactive Ahr1 can restore ALS3 and ECE1 expression in deletion mutants of these transcription factors, indicating that it has a central role in the regulation of the two genes. In this context, we also observed that hyperactive Ahr1 restored high-level ECE1 expression and candidalysin secretion in the nonfilamentous cph1Δ/efg1Δ mutant.
Hyperactive Ahr1 was not able to compensate for the absence of Tup1, in contrast to the aforementioned transcriptional activators. This illustrates the importance of the latter for high-level expression of ALS3 and ECE1. Our results also indicate that Tup1 not only participates in the repression of both genes in yeast cells but also is required for their full activation in hyphae. This activating role represents a new concept for C. albicans Tup1 but is in agreement with previous findings from Saccharomyces cerevisiae. There, it was shown that Tup1 and its corepressor Cyc8/Ssn6 can contribute to both the repression and activation of several genes, including GAL1 (37–41). In C. albicans, Tup1 might act as a backbone for the transcriptional control of ALS3 and ECE1 and change its interaction partners depending on whether repression or activation of the genes were required. We also observed that overexpression of MCM1 induced the transcription of these two genes, an effect which was strongly dependent on Tup1 but less so on Ahr1. Further experiments will be required to determine whether Ahr1 can act alone or requires Mcm1 for its function and how Tup1 is involved in the recruitment of Ahr1 or of both transcription factors to their target promoters.
MATERIALS AND METHODS
C. albicans strains and media.
All C. albicans strains used in this study are listed in Table S3. Strains were routinely grown in YPD (20 g/liter glucose, 20 g/liter peptone, 10 g/liter yeast extract, with 20 g/liter agar if required) or SDG (synthetic defined glucose) minimal medium (20 g/liter glucose, 6.7 g/liter yeast nitrogen base [YNB] without amino acids; Sigma-Aldrich) at 37°C. For the induction of hyphal growth, strains were synchronized by two overnight incubations in SDG at 37°C. Cells (1 × 106/ml) were then transferred to prewarmed SDG with 10% human serum (Sigma-Aldrich) and incubated at 37°C for the indicated times. If necessary, 50 μg/ml doxycycline (DOX) was added to the medium. For the medium shift, cells grown overnight in YPD at 30°C were diluted to 1 × 106 cells/ml in prewarmed RPMI 1640 medium (Biochrom) or YPD and grown at 30°C (YPD) or 37°C (RPMI 1640). For the pH shift, overnight cultures of C. albicans strains grown in M199 medium (Sigma-Aldrich) (pH 4) were transferred to either M199 (pH 4) or M199 (pH 8) to reach a concentration of 1 × 106 cells/ml and grown at 37°C.
Strains, plasmids, and primers used in this study. Download Table S3, DOCX file, 0.02 MB (24.6KB, docx) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of plasmids.
All plasmids used in this study are listed in Table S3. The C. albicans SAT1 (caSAT1) marker of the previously described pSK-pECE1-GFP-SAT1 plasmid (15) was excised with NotI and replaced by C. albicans ARG4 from pFA-ARG4 (42) to generate plasmid pSK-pECE1-GFP-ARG4. For the construction of the pTET-TUP1 and pTET-NRG1 plasmids, TUP1 and NRG1 open reading frames (ORFs) were amplified from genomic C. albicans DNA with primers containing XhoI and BamHI restriction sites. The digested PCR product was then cloned into SalI-BglII-digested pNIM1 (43). To obtain an NRG1 deletion cassette, NRG1 upstream and downstream sequences were amplified with primers pairs NRG1P4/NRG1P5 and NRG1K/NRG1L, respectively, digested at the introduced restriction sites, and substituted for the OPT5 sequences flanking the SAT1 flipper cassette in the previously described pOPT5M2 plasmid (44) to generate pNRG1M2.
The previously described pECE1-GFP-SAT1 plasmid (45) was used for the creation of the MCM1 overexpression construct. First, a 5′ region for the integration into the ADH1 locus was amplified with primers 5′ADH1prom-KpnI-AscI-NarI and 3′-ADH1prom-XhoI and then cloned into NarI/XhoI-digested pECE1-GFP-SAT1, replacing the 5′ECE1 integration site. Second, a 3′ ADH1 homology region which was amplified with primers 5′ADH1term-SacII and 3′ADH1term-SacI was cloned via SacII/SacI into this plasmid which then contained only ADH1 homology regions GFP and caSAT1. In a final step, MCM1 was amplified with the primers 5′MCM1-XhoI and 3′MCM1-EcoRV and cloned via XhoI/EcoRV into the constructed plasmid, replacing the GFP to create plasmid pADH1-MCM1-SAT1. For the creation of the AHR1 overexpression construct, the AHR1 gene was amplified from genomic C. albicans DNA using the primers 5′CaAHR1-XhoI and 3′AHR1-PmlI. After restriction performed with XhoI/PmlI, the AHR1 ORF was cloned into the XhoI/EcoRV-restricted pADH1-GFP plasmid (45) plasmid, replacing the GFP and creating pADH1-AHR1-SAT1.
We used the already published pAHR1-GAD plasmid (30) to create a version without a HA3 tag. With the primers 5′XhoI-AHR1 and 3′AflII-GAD, the AHR1 ORF fused to the Gal4 activator domain was amplified from the pAHR1-GAD plasmid and restriction sites were introduced. Next, the DNA fragment was digested and ligated into the linearized vector (XhoI/AflII-digested pAHR1-GAD backbone).
Plasmids were validated by sequencing. Primers used for plasmid construction are listed in Table S3.
Construction of C. albicans strains.
To generate a tup1Δ/nrg1Δ double mutant, both NRG1 alleles were sequentially deleted in tup1Δ mutant BCa2-10 using the SAT1-flipping method (46) and the recyclable deletion cassette of plasmid pNRG1M2. Transformation cassettes were excised from the plasmids and cleaned up by gel extraction (QIAquick gel extraction kit; Qiagen). C. albicans strains were transformed by the use of the lithium acetate protocol (47) or by electroporation (48). Using the caSAT1 gene as a selection marker, cells were incubated for 4 h in YPD at 30°C after the heat shock or electroporation and then plated on YPD with 200 μg/ml nourseothricin. Otherwise, transformants were grown on SDG agar plates. Transformants were validated by Southern hybridization (for the tup1Δ/nrg1Δ double mutant) or colony PCR (all other strains). The primers used for strain verification are listed in Table S3.
Gene expression analysis.
Isolation of total fungal RNA was performed with a modified hot phenol chloroform method using zirconia beads to break up fungal cells. In short, cells were collected by centrifugation and resuspended in 500 μl AE-SDS buffer (including 10% SDS). A 500-μl volume of acid phenol chloroform-isoamyl alcohol and an amount of zirconia beads corresponding to a 500-μl volume were added. This mixture was then subjected to vortex mixing for 5 min and centrifuged. The supernatant was transferred to a new reaction tube, and further steps of isolation were performed as described previously (16). RT-qPCR and normalization against housekeeping gene ACT1 and a control RNA (from SC5314 cells, grown for 5 h in YPD at 37°C) were performed as previously described (13). Relative gene expression levels were calculated according to the threshold cycle (ΔCT) method (49). The primers used for RT-qPCR are listed in Table S3 in the format R1-gene name for the forward primer and R2-gene name for the reverse primer.
Fluorescence microscopy.
GFP fluorescence microscopy was performed with a Zeiss AxioObserver Z.1 microscope (Zeiss, Germany). The illumination time for the differential interference contrast (DIC) channel was 40 ms, and that for the GFP channel was 1,700 ms. The same illumination times were used for all samples to ensure that the GFP signals were comparable.
Immunofluorescence was performed with the anti-Als3 antibody and a Zeiss Z710 laser scanning microscope (Zeiss, Germany). Prior to microscopy, 1 × 106 cells/ml of C. albicans strains were grown in SDG or SDG plus 10% human serum for 6 h in petri dishes with a glass bottom (μDish; Mobitec, Germany) at 37°C. After 6 h, cells were washed with 1× phosphate-buffered saline (PBS) and stained with the anti-Als3 antibody (1:500 diluted in 1× PBS) (11) for 60 min at room temperature. After an additional washing with 1× PBS, cells were incubated with a secondary DyLight649 goat anti-rabbit antibody (Thermo Scientific, Germany) or goat anti-rabbit-488 IgG antibody (Jackson Immuno Research, USA). Afterwards, cells were fixed with Histofix (Carl Roth, Germany) for 5 min and washed three times with 1× PBS. Finally, cells were stained with calcofluor white for 15 min. Z-stack image series of hyphae were taken using Zen software (Zeiss, Germany), and images were merged using the extended depth of focus settings.
Ferritin growth assay.
The ferritin growth assay was performed as previously described (4).
Candidalysin measurement.
Detection of secreted candidalysin was performed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) with an Ultimate 3000 nano-RSLC system coupled to a QExactive Plus mass spectrometer (Thermo Fisher Scientific) as previously described (15). For hyphal growth conditions, strains were inoculated for 18 h in YNBS (75 mM MOPSO [3-morpholino-2-hydroxypropanesulfonic acid] buffer, 6.7 g/liter YNB, 20 g/liter sucrose, 1.1 g/liter N-acetyl-d-glucosamine, pH 7.2).
Chromatin immunoprecipitation sequencing (ChIP-Seq).
Two C. albicans strains with a hyperactive Ahr1 (with and without C-terminal HA3-tag) in the SC5314 wild-type background were used for ChIP-Seq analyses, which were performed by Active Motif (USA). Formaldehyde fixation of the strains was performed after 6 h of incubation in SDG medium at 37°C following the company’s protocol. The strain without the HA3 tag was used for antibody validation to determine false positives. ChIP-Seq reads were analyzed by Active Motif, resulting in 325 peaks after peak calling and filtering the results. Peak data were further analyzed by BioControl Jena GmbH. Using the chromosomal position of the peaks, neighboring genes on both strands were identified. A sequence of 500 nucleotides centering at the maximum of each peak region was used as input for the online motif analysis tool Meme-ChIP (v. 5.1.0, 50), resulting in a highly significant, centrally enriched motif. Further confirmation was done with MochiView version 1.46 (51) and the Integrated Genome Browser (IGB) (52).
Cytotoxicity and invasion assays.
Monolayers of TR146 buccal epithelial squamous cells, grown in Dulbecco’s modified Eagle’s medium/F12 (DMEM/F12) (Gibco) with 10% heat-inactivated fetal bovine serum, were infected with 4 × 104 fungal cells and incubated in serum-free DMEM/F12 for 24 h at 37°C in 5% CO2 in 96-well plates. To determine cytotoxicity, lactate dehydrogenase (LDH) release in culture supernatants was measured using a cytotoxicity detection kit (Roche). Supernatant from Triton-X-treated, uninfected TR146 cells served as a full-lysis control, corresponding to 100% cytotoxicity. The experiment was repeated three times with biological triplicates in each round. The invasion assay was performed with the same cell line as previously described (27).
ACKNOWLEDGMENTS
We thank Gerald Fink, Alexander Johnson, Susan Noble, Peter Sudbery, Arnold Bito, and Jürgen Wendland for providing strains and plasmids. Scott Filler provided the monoclonal Als3 antibody. We are also thankful to Stefanie Allert and Rita Müller for help with the infection assays.
This work was financed by funds from the Deutsche Forschungsgemeinschaft (DFG) within the Collaborative Research Center CRC 124 FungiNet (project C3 to O. Kurzai and project C2 to J. Morschhäuser and S. Vylkova, project C1 to B. Hube, project C5 to I. D. Jacobsen, project A1 to A. A. Brakhage, and project Z2 to O. Kniemeyer), DFG grant MO 846/7 to J. Morschhäuser, the International Leibniz Research School for Microbial and Biomolecular Interactions (to O. Kurzai), and the German Ministry for Education and Science (BMBF 03Z2JN21 to O. Kurzai and BMBF 03Z2JN11 to S. Vylkova).
Footnotes
Citation Ruben S, Garbe E, Mogavero S, Albrecht-Eckardt D, Hellwig D, Häder A, Krüger T, Gerth K, Jacobsen ID, Elshafee O, Brunke S, Hünniger K, Kniemeyer O, Brakhage AA, Morschhäuser J, Hube B, Vylkova S, Kurzai O, Martin R. 2020. Ahr1 and Tup1 contribute to the transcriptional control of virulence-associated genes in Candida albicans. mBio 11:e00206-20. https://doi.org/10.1128/mBio.00206-20.
Contributor Information
David Kadosh, University of Texas Health Science Center at San Antonio.
Judith Berman, Tel Aviv University.
REFERENCES
- 1.Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat Rev Microbiol 9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- 2.Zakikhany K, Naglik JR, Schmidt-Westhausen A, Holland G, Schaller M, Hube B. 2007. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol 9:2938–2954. doi: 10.1111/j.1462-5822.2007.01009.x. [DOI] [PubMed] [Google Scholar]
- 3.Mavor AL, Thewes S, Hube B. 2005. Systemic fungal infections caused by Candida species: epidemiology, infection process and virulence attributes. Curr Drug Targets 6:863–874. doi: 10.2174/138945005774912735. [DOI] [PubMed] [Google Scholar]
- 4.Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, Filler SG, Hube B. 2008. the hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog 4:e1000217. doi: 10.1371/journal.ppat.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Citiulo F, Jacobsen ID, Miramon P, Schild L, Brunke S, Zipfel P, Brock M, Hube B, Wilson D. 2012. Candida albicans scavenges host zinc via Pra1 during endothelial invasion. PLoS Pathog 8:e1002777. doi: 10.1371/journal.ppat.1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949. doi: 10.1016/S0092-8674(00)80358-X. [DOI] [PubMed] [Google Scholar]
- 7.Wellington M, Koselny K, Krysan DJ. 2012. Candida albicans morphogenesis is not required for macrophage interleukin 1beta production. mBio 4:e00433-12. doi: 10.1128/mBio.00433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasper L, König A, Koenig PA, Gresnigt MS, Westman J, Drummond RA, Lionakis MS, Groß O, Ruland J, Naglik JR, Hube B. 2018. The fungal peptide toxin Candidalysin activates the NLRP3 inflammasome and causes cytolysis in mononuclear phagocytes. Nat Commun 9:4260. doi: 10.1038/s41467-018-06607-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tucey TM, Verma J, Harrison PF, Snelgrove SL, Lo TL, Scherer AK, Barugahare AA, Powell DR, Wheeler RT, Hickey MJ, Beilharz TH, Naderer T, Traven A. 2018. Glucose homeostasis is important for immune cell viability during Candida challenge and host survival of systemic fungal infection. Cell Metab 27:988–1006. doi: 10.1016/j.cmet.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wozniok I, Hornbach A, Schmitt C, Frosch M, Einsele H, Hube B, Loffler J, Kurzai O. 2008. Induction of ERK-kinase signalling triggers morphotype-specific killing of Candida albicans filaments by human neutrophils. Cell Microbiol 10:807–820. doi: 10.1111/j.1462-5822.2007.01086.x. [DOI] [PubMed] [Google Scholar]
- 11.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurzai O, Schmitt C, Bröcker EB, Frosch M, Kolb-Mäurer A. 2005. Polymorphism of Candida albicans is a major factor in the interaction with human dendritic cells. Int J Med Microbiol 295:121–127. doi: 10.1016/j.ijmm.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Voigt J, Hunniger K, Bouzani M, Jacobsen ID, Barz D, Hube B, Loffler J, Kurzai O. 2014. Human natural killer cells acting as phagocytes against Candida albicans and mounting an inflammatory response that modulates neutrophil antifungal activity. J Infect Dis 209:616–626. doi: 10.1093/infdis/jit574. [DOI] [PubMed] [Google Scholar]
- 14.Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, Ibrahim AS, Edwards JE Jr, Filler SG. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol 5:e64. doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Hofs S, Gratacap RL, Robbins J, Runglall M, Murciano C, Blagojevic M, Thavaraj S, Forster TM, Hebecker B, Kasper L, Vizcay G, Iancu SI, Kichik N, Hader A, Kurzai O, Luo T, Kruger T, Kniemeyer O, Cota E, Bader O, Wheeler RT, Gutsmann T, Hube B, Naglik JR. 2016. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532:64–68. doi: 10.1038/nature17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin R, Albrecht-Eckardt D, Brunke S, Hube B, Hünniger K, Kurzai O. 2013. A core filamentation response network in Candida albicans is restricted to eight genes. PLoS One 8:e58613. doi: 10.1371/journal.pone.0058613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X, Oh SH, Cheng G, Green CB, Nuessen JA, Yeater K, Leng RP, Brown AJ, Hoyer LL. 2004. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 150:2415–2428. doi: 10.1099/mic.0.26943-0. [DOI] [PubMed] [Google Scholar]
- 18.Bader O, Krauke Y, Hube B. 2008. Processing of predicted substrates of fungal Kex2 proteinases from Candida albicans, C. glabrata, Saccharomyces cerevisiae and Pichia pastoris. BMC Microbiol 8:116. doi: 10.1186/1471-2180-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson JP, Mogavero S, Moyes DL, Blagojevic M, Kruger T, Verma AH, Coleman BM, De La Cruz Diaz J, Schulz D, Ponde NO, Carrano G, Kniemeyer O, Wilson D, Bader O, Enoiu SI, Ho J, Kichik N, Gaffen SL, Hube B, Naglik JR. 2018. Processing of Candida albicans Ece1p is critical for candidalysin maturation and fungal virulence. mBio 9:e02178-17. doi: 10.1128/mBio.02178-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richardson JP, Willems HME, Moyes DL, Shoaie S, Barker KS, Tan SL, Palmer GE, Hube B, Naglik JR, Peters BM. 22 January 2017, posting date Candidalysin drives epithelial signaling, neutrophil recruitment, and immunopathology at the vaginal mucosa. Infect Immun doi: 10.1128/IAI.00645-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verma AH, Richardson JP, Zhou C, Coleman BM, Moyes DL, Ho J, Huppler AR, Ramani K, McGeachy MJ, Mufazalov IA, Waisman A, Kane LP, Biswas PS, Hube B, Naglik JR, Gaffen SL. 3 November 2017, posting date Oral epithelial cells orchestrate innate type 17 responses to Candida albicans through the virulence factor candidalysin. Sci Immunol doi: 10.1126/sciimmunol.aam8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doedt T, Krishnamurthy S, Bockmuhl DP, Tebarth B, Stempel C, Russell CL, Brown AJ, Ernst JF. 2004. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol Biol Cell 15:3167–3180. doi: 10.1091/mbc.e03-11-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murad AM, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, Schnell N, Talibi D, Marechal D, Tekaia F, d'Enfert C, Gaillardin C, Odds FC, Brown AJ. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J 20:4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadosh D, Johnson AD. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell 16:2903–2912. doi: 10.1091/mbc.e05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun BR, Kadosh D, Johnson AD. 2001. NRG1, a repressor of filamentous growth in C.albicans, is down-regulated during filament induction. EMBO J 20:4753–4761. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moyes DL, Runglall M, Murciano C, Shen C, Nayar D, Thavaraj S, Kohli A, Islam A, Mora-Montes H, Challacombe SJ, Naglik JR. 2010. A biphasic innate immune MAPK response discriminates between the yeast and hyphal forms of Candida albicans in epithelial cells. Cell Host Microbe 8:225–235. doi: 10.1016/j.chom.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wachtler B, Wilson D, Haedicke K, Dalle F, Hube B. 2011. From attachment to damage: defined genes of Candida albicans mediate adhesion, invasion and damage during interaction with oral epithelial cells. PLoS One 6:e17046. doi: 10.1371/journal.pone.0017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Askew C, Sellam A, Epp E, Mallick J, Hogues H, Mullick A, Nantel A, Whiteway M. 2011. The zinc cluster transcription factor Ahr1p directs Mcm1p regulation of Candida albicans adhesion. Mol Microbiol 79:940–953. doi: 10.1111/j.1365-2958.2010.07504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavoie H, Sellam A, Askew C, Nantel A, Whiteway M. 2008. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics 9:578. doi: 10.1186/1471-2164-9-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schillig R, Morschhauser J. 2013. Analysis of a fungus-specific transcription factor family, the Candida albicans zinc cluster proteins, by artificial activation. Mol Microbiol 89:1003–1017. doi: 10.1111/mmi.12327. [DOI] [PubMed] [Google Scholar]
- 31.Hernday AD, Lohse MB, Fordyce PM, Nobile CJ, DeRisi JL, Johnson AD. 2013. Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol Microbiol 90:22–35. doi: 10.1111/mmi.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vylkova S, Lorenz MC. 26 January 2017, posting date Phagosomal neutralization by the fungal pathogen Candida albicans induces macrophage pyroptosis. Infect Immun doi: 10.1128/IAI.00832-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wellington M, Koselny K, Sutterwala FS, Krysan DJ. 2014. Candida albicans triggers NLRP3-mediated pyroptosis in macrophages. Eukaryot Cell 13:329–340. doi: 10.1128/EC.00336-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allert S, Förster TM, Svensson CM, Richardson JP, Pawlik T, Hebecker B, Rudolphi S, Juraschitz M, Schaller M, Blagojevic M, Morschhäuser J, Figge MT, Jacobsen ID, Naglik JR, Kasper L, Mogavero S, Hube B. 2018. Candida albicans-induced epithelial damage mediates translocation through intestinal barriers. mBio 9:e00915-18. doi: 10.1128/mBio.00915-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swidergall M, Khalaji M, Solis NV, Moyes DL, Drummond RA, Hube B, Lionakis MS, Murdoch C, Filler SG, Naglik JR. 2019. Candidalysin is required for neutrophil recruitment and virulence during systemic Candida albicans infection. J Infect Dis 220:1477–1488. doi: 10.1093/infdis/jiz322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skrzypek MS, Binkley J, Binkley G, Miyasato SR, Simison M, Sherlock G. 2017. The Candida Genome Database (CGD): incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res 45:D592–D596. doi: 10.1093/nar/gkw924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Proft M, Struhl K. 2002. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell 9:1307–1317. doi: 10.1016/S1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- 38.Papamichos-Chronakis M, Petrakis T, Ktistaki E, Topalidou I, Tzamarias D. 2002. Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol Cell 9:1297–1305. doi: 10.1016/S1097-2765(02)00545-2. [DOI] [PubMed] [Google Scholar]
- 39.Desimone AM, Laney JD. 2010. Corepressor-directed preacetylation of histone H3 in promoter chromatin primes rapid transcriptional switching of cell-type-specific genes in yeast. Mol Cell Biol 30:3342–3356. doi: 10.1128/MCB.01450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han BK, Emr SD. 2011. Phosphoinositide [PI(3,5)P2] lipid-dependent regulation of the general transcriptional regulator Tup1. Genes Dev 25:984–995. doi: 10.1101/gad.1998611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han BK, Emr SD. 2013. The phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2)-dependent Tup1 conversion (PIPTC) regulates metabolic reprogramming from glycolysis to gluconeogenesis. J Biol Chem 288:20633–20645. doi: 10.1074/jbc.M113.452813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gola S, Martin R, Walther A, Dunkler A, Wendland J. 2003. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20:1339–1347. doi: 10.1002/yea.1044. [DOI] [PubMed] [Google Scholar]
- 43.Park YN, Morschhauser J. 2005. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot Cell 4:1328–1342. doi: 10.1128/EC.4.8.1328-1342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reuss O, Morschhauser J. 2006. A family of oligopeptide transporters is required for growth of Candida albicans on proteins. Mol Microbiol 60:795–812. doi: 10.1111/j.1365-2958.2006.05136.x. [DOI] [PubMed] [Google Scholar]
- 45.Hunniger K, Lehnert T, Bieber K, Martin R, Figge MT, Kurzai O. 2014. A virtual infection model quantifies innate effector mechanisms and Candida albicans immune escape in human blood. PLoS Comput Biol 10:e1003479. doi: 10.1371/journal.pcbi.1003479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reuss O, Vik A, Kolter R, Morschhauser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 47.Walther A, Wendland J. 2003. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr Genet 42:339–343. doi: 10.1007/s00294-002-0349-0. [DOI] [PubMed] [Google Scholar]
- 48.Kohler GA, White TC, Agabian N. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J Bacteriol 179:2331–2338. doi: 10.1128/jb.179.7.2331-2338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Homann OR, Johnson AD. 2010. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol 8:49. doi: 10.1186/1741-7007-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freese NH, Norris DC, Loraine AE. 2016. Integrated genome browser: visual analytics platform for genomics. Bioinformatics 32:2089–2095. doi: 10.1093/bioinformatics/btw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tup1 is required for ECE1 expression after pH shift and medium change. The wild-type strain and tup1Δ strains with integrated pECE1-GFP were grown in the indicated media for 6 h at 37°C prior to fluorescence microscopy. Shown are the overlays of the DIC channel and the GFP channels. Scale bar, 20 μm. Download FIG S1, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Core filamentation gene expression response in filamentous mutants. The wild-type strain and the filamentous mutants were grown in SDG with or without 10% human serum for 6 h at 37°C prior to the isolation of total RNA, which was used for the determination of the relative levels of expression of the indicated genes. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student’s t test) from the results seen with the wild-type strain grown in SDG with serum. Download FIG S2, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ECE1 transcription in activator mutants. The wild-type strain and the indicated activator mutants were grown in SDG medium with 10% human serum for 4 h prior to the isolation of total RNA. The RNA was used to determine the relative levels of expression of ECE1. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student’s t test) compared to the wild type. Download FIG S3, TIF file, 0.9 MB (968KB, tif) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Putative transcription factor binding sites in the 5′ intergenic region of ECE1. Download Table S1, XLSX file, 0.01 MB (11.6KB, xlsx) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Core filamentation gene expression response during overexpression of MCM1. The indicated strains were grown for 4 h at 37°C in SDG medium with or without 10% human serum prior to the isolation of total RNA. The RNA was used to determine the relative levels of gene expression. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student’s t test) compared to the wild type. Download FIG S4, TIF file, 2.6 MB (2.7MB, tif) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Validation of ChIP-Seq results for wild-type derivatives with hyperactive Ahr1. ChIP-Seq reads (red) mapped to the genome of the SC5314 wild type (ASM18296v3). An extract from chromosome 4 with the ORFs of ECE1, RBT1, and HWP1 (blue) is shown. (Top) ChIP-Seq reads of the control strain with hyperactive Ahr1 without a HA3 tag in the SC5314 background. (Middle) ChIP-Seq reads of the mutant with hyperactive Ahr1 with a HA3 tag in the SC5314 background. Identified peak summits are depicted in black. Download FIG S5, TIF file, 0.9 MB (994.1KB, tif) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Results of the ChIP-Seq analysis performed with hyperactive Ahr1. Download Table S2, XLSX file, 0.03 MB (34.5KB, xlsx) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Hyperactive Ahr1 induces expression of ALS3, HWP1, and IHD1 in the cph1Δ/efg1Δ double mutant. Wild-type and cph1Δ/efg1Δ strains with or without integrated hyperactive Ahr1 were grown for 6 h at 37°C in SDG medium with or without 10% human serum prior to the isolation of total RNA. The RNA was used to determine the relative levels of expression of genes ALS3, HWP1, and IHD1. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student’s t test) compared to the background strains without hyperactive Ahr1. Download FIG S6, TIF file, 0.9 MB (986.3KB, tif) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Hyperactive Ahr1 restores ECE1 transcription in regulatory mutants. (A) The indicated strains with or without integrated hyperactive Ahr1 were grown for 4 h at 37°C in SDG medium with or without 10% human serum prior to microscopy. Scale bar, 10 μm. (B) Total RNA from the aforementioned strains was isolated after the same time of growth. The RNA was used to determine the relative levels of expression of the ECE1 gene. Asterisks indicate significant changes (P ≤ 0.05, two-tailed, unpaired Student’s t test) compared to the background strains without hyperactive Ahr1. Download FIG S7, TIF file, 2.6 MB (2.7MB, tif) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains, plasmids, and primers used in this study. Download Table S3, DOCX file, 0.02 MB (24.6KB, docx) .
Copyright © 2020 Ruben et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.