Abstract
Background
In consideration of the difficulty in diagnosing high heterogeneous glioma, valuable prognostic markers are urgent to be investigated. This study aimed to verify that connective tissue growth factor (CTGF) is associated with the clinical prognosis of glioma, also to analyze the effect of CTGF on the biological function.
Methods
In this study, glioma and non-tumor tissue samples were obtained in 2012 to 2014 from the Department of Neurosurgery of Nanfang Hospital of Southern Medical University, Guangzhou, China. Based on messenger RNA (mRNA) data from the Cancer Genome Atlas (TCGA) and CCGA dataset, combined with related clinical information, we detected the expression of CTGF mRNA in glioma and assessed its effect on the prognosis of glioma patients. High expression of CTGF mRNA and protein in glioma were verified by reverse transcription-polymerase chain reaction, immunohistochemistry, and Western blotting. The role of CTGF in the proliferation, migration, and invasion of gliomas were respectively identified by methylthiazoletetrazolium assay, Transwell and Boyden assay in vitro. The effect on glioma cell circle was assessed by flow cytometry. For higher expression of CTGF in glioblastoma (GBM), the biological function of CTGF in GBM was investigated by gene ontology (GO) analysis.
Results
In depth analysis of TCGA data revealed that CTGF mRNA was highly expressed in glioma (GBM, n = 163; lowly proliferative glioma [LGG], n = 518; non-tumor brain tissue, n = 207; LGG, t = 2.410, GBM, t = 2.364, P < 0.05). CTGF mRNA and protein expression in glioma (86%) was significantly higher than that in non-tumor tissues (18%) verified by collected samples. Glioma patients with higher expression of CTGF showed an obviously poorer overall survival (35.4 and 27.0 months compared to 63.3 and 55.1 months in TCGA and Chinese Glioma Genome Atlas (CGGA) databases separately, CGGA: χ2 = 7.596, P = 0.0059; TCGA: χ2 = 10.46, P = 0.0012). Inhibiting CTGF expression could significantly suppress the proliferation, migration, and invasion of gliomas. CTGF higher expression had been observed in GBM, and GO analysis demonstrated that the function of CTGF in GBM was mainly associated with metabolism and energy pathways (P < 0.001).
Conclusions
CTGF is highly expressed in glioma, especially GBM, as an unfavorable and independent prognostic marker for glioma patients and facilitates the progress of glioma.
Keywords: Glioma, Connective tissue growth factor, Biological function, Prognosis, Proliferation, Migration, Invasion
Introduction
Glioma is the most common primary tumor of the central nervous system.[1–3] According to the traditional classification, gliomas are divided into non-invasive and lowly proliferative glioma (LGG), anaplastic astrocytoma (grade III), and glioblastoma (GBM).[2] GBM is the most malignant glioma, characterized by highly aggressive progression, increased proliferation, and robust angiogenesis.[4] The standard therapy of malignant glioma is surgical resection followed by chemo- and radiotherapy.[5] The poor median survival time of GBM is 14.6 months.[6–8] It is necessary to understand the molecular mechanisms of glioma as well as to develop novel therapeutic strategies.[9]
Connective tissue growth factor (CTGF/CCN2) is a 34,000 to 38,000 Da secreted protein, and the gene is located on human chromosome 6q23.1.[10] CTGF is reported to promote proliferation and differentiation of chondrocytes, mediate heparin- and divalent cation-dependent cell adhesion and so on.[11–13] CTGF is also commonly known for taking part in multifunctional signaling modulation in various pathological processes including tumor occurrence and development.[14,15]
CTGF is overexpressed in about 58% primary brain tumors and linked to 27 human cancers.[16] According to previous reports, the role of deregulated CTGF is complicated with a tissue-dependent manner, related to angiogenesis[17] and modulated signal pathways including Wnt.[18] The association and molecular mechanism between CTGF and prognosis of glioma remain to be clarified.
In this experiment, the data for investigation is obtained from public datasets (the Cancer Genome Atlas (TCGA), Chinese Glioma Genome Atlas [CGGA], Genotype-Tissue Expression (GTEx) and samples collected from glioma resection in Nanfang hospital, significantly overexpression of CTGF in glioma was clarified. Furthermore, the association of CTGF overexpression and poor prognosis of glioma was revealed by survival analysis. Downregulation of CTGF expression in glioma cells inhibited cell proliferation, migration and invasion in vitro. Our findings support the theory that overexpression of CTGF is potentially an unfavorable and independent prognostic marker for glioma patients.
Methods
Ethical approval
This study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committees of Nanfang Hospital of Guangdong Province (No. A3873-1), and each human tissue was obtained with prior consent from patients or their guardians before participation in the study.
Bioinformatic analysis
Data used in this study for bioinformatic analysis was obtained from public datasets, including messenger RNA (mRNA) chip data and RNA sequencing data. The gene expression data were downloaded from public datasets and normalized for analysis. Meanwhile, clinical case data in the database were extracted for clinical correlation analysis. Among them, TCGA (https://cancergenome.nih.gov/) contained 681 patients with glioma, while CGGA (http://www.cgga.org.cn/) contained 301 patients with glioma. The data was loaded into the corresponding statistical software to analyze the expression of CTGF in glioma, biological function, and its relationship with clinical prognosis.[19]
Clinical tissue sample collection
A total of 29 paraffin-embedded glioma and 11 non-tumor tissue samples (taken from epilepsy patients with cerebral lobectomy and cerebral hernia patients with intracranial decompression). The samples which were confirmed by hematoxylin-eosin staining and light microscopy were obtained in 2012 to 2014 from the Department of Neurosurgery of Nanfang Hospital of Southern Medical University, Guangzhou, China. Chemotherapy, radiotherapy, and other special treatments were not performed before surgery. All the gliomas had confirmed pathologic diagnosis and classification according to the World Health Organization criteria.
Cell culture
Human glioma U87MG and U251MG cells were purchased from the Chinese Academy of Sciences (Shanghai, China) and cultured in the Dulbecco modified Eagle medium (DMEM) (Hyclone, Logan, UT, USA) containing 10% fetal bovine serum (Hyclone). All cell lines were incubated in a humidified atmosphere of 5% CO2 at 37°C.
Immunohistochemistry
Paraffin sections were deparaffinized in 100% xylene for 1 h and rehydrated in descending ethanol series and phosphate buffer saline (PBS, Hyclone).[20] Heat-induced antigen retrieval was performed in 10 mmol/L citrate buffer for 15 min in a microwave oven. Endogenous peroxidase activity was inactivated with peroxidase blocking reagent (hydrogen peroxide 3%). After being washed with PBS for three times (5 min each time), the sections were incubated with primary antibodies overnight at 4°C. After washing three times with PBS to remove unbound antibody, the sections were incubated with a dedicated secondary antibody for 1 h at room temperature, and subsequently, the peroxidase reaction was developed using 3,3-diaminobenzidine (DAB) chromogen solution in DAB buffer substrate. Sections were counterstained with hematoxylin, mounted in neutral gum, and analyzed using a bright-field microscope equipped with a digital camera (Nikon, Japan).[21]
Immunohistochemistry results judgement
The immunohistochemically stained tissue sections were reviewed and scored separately by two pathologists blinded to the clinical parameters.[22] The score was evaluated according to the percentage of positive staining cells and the sum of staining intensity. The positive staining scores were defined 0 to 4 (0: negative, 1: low expression and <50%, 2: low expression and ≥50%, 3: high expression and <50%, and 4: high expression and ≥50%).
Western blotting
Western blotting was carried as described previously,[23] with rabbit polyclonal CTGF (1:1000; BOSTER Biological Technology, Pleasanton, CA, USA), β-actin (1:1000; Proteintech, Rosemont, IL, USA) antibodies. A horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse IgG antibody was used as the secondary antibody (1:2000; CoWin Bioscience, Beijing, China). Enhanced chemiluminescence reagents (Pierce, Rockford, IL, USA) were utilized to detect signals. All experiments were independently performed in triplicate.
Transient transfection with small-interfering RNAs (siRNAs)
siRNA for CTGF was designed and synthesized by Guangzhou RiboBio (RiboBio Inc, China). Three siRNAs targeting on CTGF gene were designed and synthesized, the most effective siRNA (siCTGF) identified by reverse transcription-polymerase chain reaction (RT-PCR) was applied for the further experiments. The target sequences: CTGF-seg1 was 5′-GCCTGCCATTACAACTGT-3′, CTGF-seg2 was 5′-GACCCAACTATGATTA-3′, CTGF-seg3 was 5′-GTGCATCCGTACTCCCAAA-3′.
RT-PCR
Total RNA from ten fresh non-tumor tissue samples and 12 glioma samples were isolated by using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and then treated with the TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific, Waltham, MA, USA), 0.3 μmol/L of each primer, and 0.2 μmol/L of probe. Reverse transcription synthesis of complementary DNA by reverse transcription kit according to the manufacturer's instructions. PCR cycling conditions were 95°C for 10 min to activate DNA polymerase, followed by 45 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 10 s.
Methylthiazoletetrazolium (MTT) assay
Glioma cells (1000/well) were seeded in 96-well plates with a volume of 200 μL medium, 20 μL of MTT solution was added to each well and incubated for 4 h. Supernate was abandoned, each well was filled with a volume of 100 μL DMEM. The absorbance value (A) was measured at 490 nm.
Transwell assay and Boyden assay
About 5 × 103 cells in 200 μL DMEM medium without fetal calf serum (FCS) were filled in the upper chamber of Transwell apparatus (Corning, NY, USA). DMEM with 10% FCS was seeded on the lower chamber. After putting Transwell apparatus in a 5% CO2 atmosphere at 37°C in 6 h, cells on the upper chamber were wiped by a cotton swab. The insert was washed with PBS twice. Cells on the lower chamber were fixed with methanol for 20 min, then stained with crystal violet for 20 min. The number of cells was counted under a microscope, averaged, and analyzed. Boyden assay was the same as Transwell assay, except that Transwell apparatus were pre-coated with Matrigel on the upper chamber.
Flow cytometry
The 5 × 106 cells were rinsed with cold PBS and filled with 70% ice-cold ethanol for 48 h at 4°C. Cold PBS was used to wash. Subsequently, cells were incubated with PBS containing 10 μg/mL propidium iodide and 0.5 mg/mL RNase A for 1 h at 37°C. A type of flow cytometer (EPICS XL, Beckman Co., USA) was accessed to analyze cell circle.
Survival analysis
Overall survival (OS) was identified by analyzing different data sets and clinical information of glioma. The partial case was abandoned. Survival curves were plotted using the Kaplan-Meier method in GraphPad Prism 7.0 (GraphPad Software, Inc., San Diego, CA, USA) and assessed using the log-rank test.
Gene ontology (GO) analysis
Among the upregulated expression of CTGF, 637 upregulated correlative genes in TCGA GBM data were obtained. Functional enrichment analysis tool was used to do GO analysis including molecular function, biological process and cellular component of CTGF (Ranked top six items were selected by P value).
Statistical analysis
IMB SPSS 22.0 (IBM Company, USA) and GraphPad Prism 7.0 software (GraphPad Software, USA) were mainly used to analyze statistical data and plot kinds of graphs. All values were expressed as mean ± standard deviation, and Student's t test was used for comparison of data between two groups, one-way analysis of variance was used for comparison of multiple groups. Different grade of malignant glioma was performed by Dunnett. A P value <0.05 was considered to indicate a statistically significant difference.
Results
TCGA databases revealed that CTGF mRNA was highly expressed in glioma
Compared with non-tumor tissues, the expression of CTGF was increased in glioma including LGG and GBM, analyzed with the web-server gene expression profiling interactive analysis (GBM, n = 163; LGG, n = 518; non-tumor brain tissue [NB], n = 207) [Figure 1A].
Figure 1.
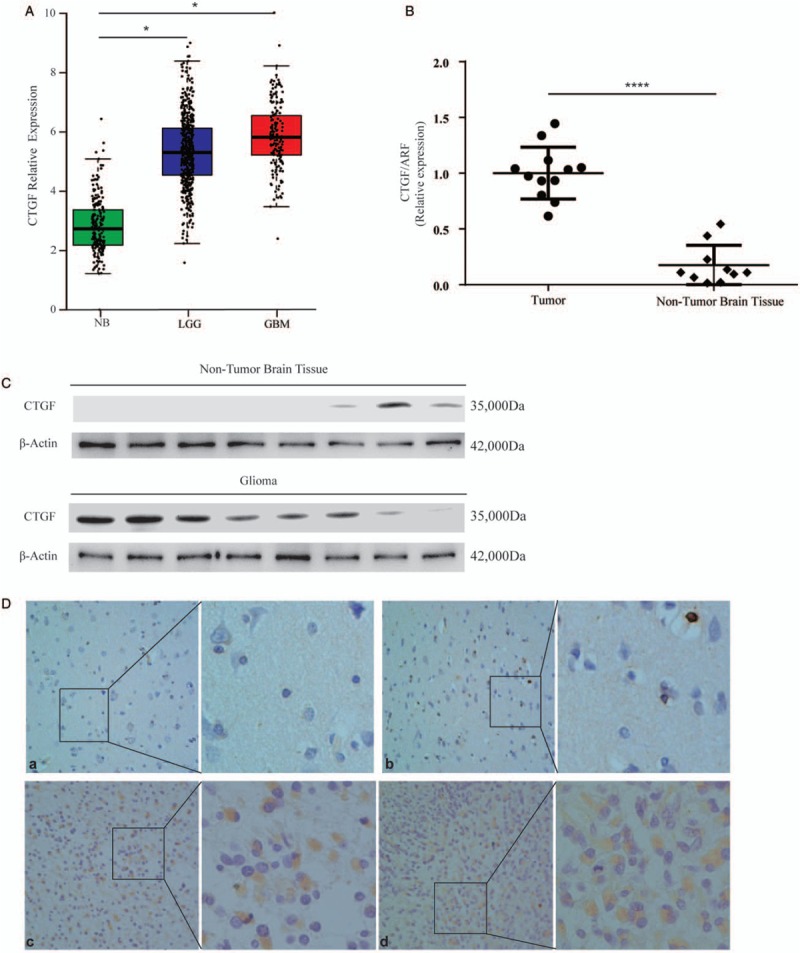
The expression of CTGF in glioma and non-tumor tissue. (A) The expression of CTGF mRNA was increased in glioma compared with non-tumor tissues (GBM, n = 163; LGG, n = 518; NB, n = 207), analyzed with the web server GEPIA and expression data presented at mean ± SD from TCGA and GTEx (LGG, t = 2.410, GBM, t = 2.364, P<0.05). (B) The expression of CTGF mRNA was evaluated in 12 glioma tissues compared with ten non-tumor tissues by RT-PCR (t = 9.117, P < 0.0001). (C) The expression of CTGF protein was higher in eight glioma samples than eight non-tumor tissues by Western blotting. (D) The expression and location of CTGF were examined by Immunohistochemical staining. (a and b) weak staining of CTGF in normal tissues. (c and d) strong staining of CTGF in glioma tissues. Original magnification× 400. CTGF: Connective tissue growth factor; GBM: Glioblastoma; LGG: Lowly proliferative glioma; mRNA: Messenger RNA; NB: Non-tumor brain; RT-PCR: Reverse transcription-polymerase chain reaction; SD: Standard deviation.
CTGF mRNA and protein expression in glioma was significantly higher than that in non-tumor tissues verified by collected samples
To determine whether CTGF played a role in glioma tumorigenesis, the expression of CTGF mRNA was evaluated in 12 glioma tissues compared with ten non-tumor tissues by RT-PCR [Figure 1B]. Similarly, it was higher expressed in glioma investigated in eight cases of glioma and eight NB tissues by Western blotting [Figure 1C]. Immunohistochemical staining showed the location and expression of CTGF in NB and glioma tissues, CTGF expression was evaluated in glioma in 29 archived paraffin-embedded glioma samples and 11 NB tissues using immunohistochemical staining [Figure 1D]. Thus, CTGF was highly expressed in 86% (25/29) of glioma samples compared to only 18% (2/11) of non-tumor tissue samples, which was a statistically significant difference [Table 1].
Table 1.
Expression of CTGF in glioma and non-tumor tissues.
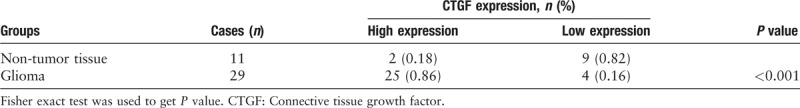
Glioma patients with higher expression of CTGF showed an obviously poorer OS
To investigate the prognostic of CTGF expression for glioma, clinical case data in CGGA and TCGA database were extracted for clinical correlation analysis. Kaplan-Meier survival analysis of OS was among 301 patients’ data in CGGA and 663 patients’ data in TCGA. P values were calculated by log-rank test (CGGA P = 0.0059; TCGA P = 0.0012) [Figure 2]. The curve graphs between the high and low expression of CTGF were observably different. The median OS among patients with higher CTGF expression was 35.4 months compared to 63.3 months among those with lower expression in TCGA databases (P < 0.01). The median OS among patients with higher CTGF expression was 26.97 months compared to 55.1 months among those with lower expression in CGGA databases (P < 0.01). We observed that the relation between CTGF protein expression and OS was inversely proportional. Glioma patients with low CTGF expression levels had better survival than those with high expression.
Figure 2.
Kaplan-Meier survival analysis of OS based on the data collected from 301 patients in CGGA databases and 663 patients in TCGA databases. The log-rank test was used to calculate P values (CGGA: χ2 = 7.596, P = 0.0059; TCGA: χ2 = 10.46, P = 0.0012). CGGA: Chinese Glioma Genome Atlas; OS: Overall survival; TCGA: The Cancer Genome Atlas.
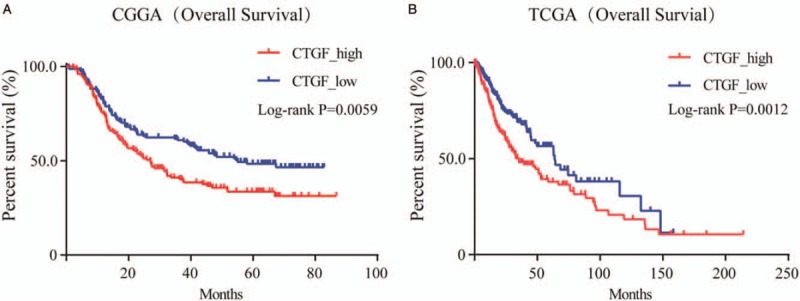
Inhibiting CTGF expression could significantly suppress glioma proliferation, migration, and invasion
To explore the role of CTGF in glioma, two human cell lines established from high-grade tumors (U251 and U87 cells) were utilized. After being assessed by RT-PCR, transcriptional levels of CTGF decreased up to 75.0% and 99.8% in siCTGF-seg1 transfected, 23.0% and 62.0% in siCTGF-seg2 transfected, 86.0% and 87.0% in siCTGF-seg3 transfected U251 cells and U87 cells respectively, compared to the negative control (NC) groups. The result indicated that siCTGF-seg1 and siCTGF-seg3 successfully suppressed gene translation, while siCTGF-seg2 did not. (P < 0.0001) [Figure 3A]. The effectivity of siCTGF-seg1 and siCTGF-seg3 in protein downregulation was shown by Western blotting [Figure 3B]. The growth curves determined by MTT assay showed that the downregulation of CTGF restrained cell proliferation of these two cell lines [Figure 3C]. To examine the influence of CTGF on cell migration, siCTGF transfected U251 and U87 cells were cultured on Transwell apparatus for 6 h incubation, and the percentage of migrated cells was less in both siCTGF transfected groups (for both P < 0.0001) [Figure 3D]. Change in cell invasiveness was verified with a Boyden chamber. After 6 h incubation, siCTGF transfected U251 and U87 cells both presented a significant decrease in invasiveness, compared with the NC groups (both P < 0.0001) [Figure 3E]. To further study the link between cell cycle control and CTGF in glioma cells, cell cycle distribution was assessed by Fluorescence Activated Cell Sorter (FACS) Caliber cytometry. After 48 h transfection, cells exhibited an increase in the cell fraction in G1 phase (U251-NC 34.68%, U251-siCTGF 44.87%, U87-NC 67.66%, U87-siCTGF 72.05%, P < 0.0001) with a corresponding reduction in the fraction of cells in S and G2/M phase. The outcome indicated inhibition of cell cycle progression from G1 to S phase, suggesting that CTGF may suppress the cell cycle by influencing DNA synthesis. But the escalation in the cell fraction in G1 phase was not remarkable enough, which illustrated inhibiting DNA synthesis may not be the only mechanism of suppressing cell cycle [Figure 3F].
Figure 3.
Downregulation of CTGF inhibited glioma cell proliferation, migration and invasion in vitro and induced cell cycle arrest at G1/S phase. (A) RT-PCR showed mRNA transcription levels of CTGF 48 h post-transfection, ADP-ribosyltransferase (ARF) used as loading control. The arbitrary units were plotted using mean ± SD of at least three individual repetitions. (B) Western blotting showed protein expression levels in NC and siCTGF treated U87 and U251 cell lines; β-actin served as a loading control. (C) Proliferation as measured by MTT assay. Absorbance was read at 490 nm with average from four repeated wells. (D and E) Downregulation of CTGF reduced U251 and U87 cells migration and invasion in vitro. Data were presented as mean ± SD for three independent experiments (D: U251, t = 3.251, U87, t = 3.206, P < 0.0001; E: U251, t = 3.302, U87, t = 3.293, P < 0.0001). Original magnification ×400. (F) The cell cycle distribution in siCTGF treated and NC groups of U251 and U87 cells was tested by FACS Caliber cytometry, three individual repetitions at least. CTGF: Connective tissue growth factor; LGG: Lowly proliferative glioma; mRNA: Messenger RNA; MTT: Methylthiazoletetrazolium; NB: Non-tumor brain; NC: Negative control; RT-PCR: Reverse transcription-polymerase chain reaction; SD: Standard deviation.
Higher expression of CTGF in GBM and GO analysis of CTGF-associated differentially expressed genes (DEGs)
Analysis of CTGF expression between LGG and GBM with the data from both CGGA database and TCGA database were conducted that GBM had a much higher expression quantity of CTGF [Figure 4A and 4B] (P < 0.0001). To further clarify the role of CTGF in GBM, we used the data of GBM genes from TCGA database and got DEGs through cases with different CTGF expression quantity. The GO enrichment analysis with DEGs indicated that in molecular function, 1.6% of high expressed genes involved in structural constituent of ribosome and 3.7% in catalytic activity [Figure 4C] (P < 0.001); in biological pathway, 6.3% of high expressed genes were associated with metabolism of proteins and 3.2% with translation [Figure 4C] (P < 0.001); in biological process, 11.7% of high expressed genes were relevant to metabolism and 11.3% to energy pathways [Figure 4C] (P < 0.001). These results provided more precise directions for us in future experiments.
Figure 4.
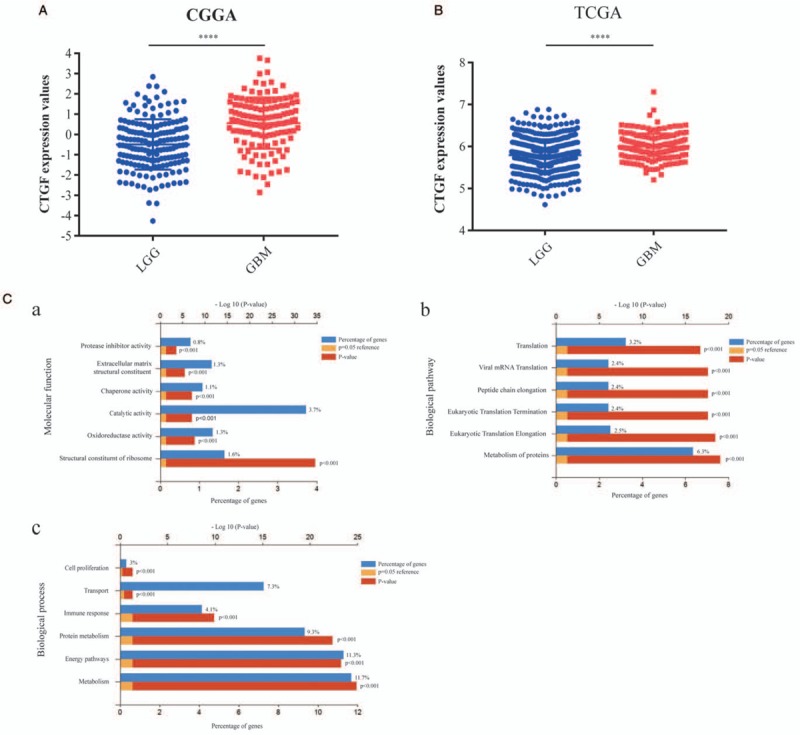
Higher expression and Go analysis of CTGF in GBM. (A) Comparison of CTGF expression values of LGG patients (n = 173) and GBM patients (n = 128) in CGGA databases (t = 7.91, P < 0.0001). (B) Comparison of CTGF expression values of LGG patients (n = 511) and GBM patients (n = 156) in TCGA databases (t = 6.106, P < 0.0001). (C) Dug differentially expressed genes (DEGs) with CTGF high expression in GBM were analyzed by GO functional enrichment analysis (first six bars, P < 0.001). (a) Molecular function for CTGF; (b) biological pathway for CTGF; (c) biological process for CTGF. CGGA: Chinese Glioma Genome Atlas; CTGF: Connective tissue growth factor; GBM: Glioblastoma; GO: Gene ontology; LGG: Lowly proliferative glioma; TCGA: The Cancer Genome Atlas.
Discussion
According to previous researches of CTGF, CTGF not only plays an indispensable role in embryonic development, endocrine regulation, wound repair, and angiogenesis but also is involved in several cancer occurrence and development.[24–28] As recent oncology researches revealed, overexpression of CTGF has been observed in a lot of types of tumors such as angiolipoma, vascular leiomyoma, and hepatocellular carcinoma and glioma.[29–32] CTGF is overexpressed in most tumors, while the expression is downregulated when it comes to colorectal cancer, lung cancer, and ovarian cancer.[33–36]
Though some articles mentioned that CTGF took part in glioma progression, the clinical value and mechanism of CTGF in human glioma have not been elucidated.[1,10,32] In our study, we presented comprehensive evidence that CTGF was upregulated in both mRNA and protein level, confirmed by RT-PCR, Western blotting, and immunohistochemistry. According to our data obtained from public datasets TCGA and CGGA, a higher level of CTGF was correlated to shorter OS in patients with glioma. Moreover, comparing the significant difference between GBM and LGG, we also observed that the level of CTGF expression was positively associated with pathology classification in human glioma.[37,38] All these results suggested that CTGF expression may serve as an unfavorable and independent prognostic factor for glioma patients.
The biological functions of CTGF explored in vitro in this study provided a mechanistic basis for the pathological and clinical observations. Interestingly, we found that knockdown of CTGF expression downregulated cell proliferation, migration, and invasion of glioma cells in vitro. To further explore the way of CTGF influencing cell proliferation, flow cytometry was utilized. The outcome indicated inhibition of cell cycle progression from G1 to S phase, suggesting that CTGF suppressed cell cycle by influencing DNA synthesis. However, DNA synthesis may not be the only mechanism of suppressing cell cycle.
To further expound the biological functions of CTGF in glioma, we compared the expression levels in LGG and GBM. CTGF presented obviously higher expression in GBM, the most malignant glioma. Together with the effect of CTGF on the biological function of U87 and U251cells in vitro, we paid our attention to the mechanism of CTGF in GBM malignant phenotype succeedingly. Go analysis showed the CTGF function in GBM was mainly associated with metabolism and energy pathways.
CTGF has been reported to exert a range of effects in molecular pathways in tumors such as integrin-αvβ3–P-ERK1/2-dependent upregulation and CTGF-ITGB1-TrkA complex activation.[25,39,40] The previous CTGF-knockdown cells also suggested that CTGF promoted stem-like properties and upregulated pluripotency genes.[41,42] Furthermore, several findings suggested that CTGF overexpression was correlated with angiogenesis.[43–46]
Regrettably, the mechanism of CTGF in tumor metabolism and energy pathways has not been reported, not to mention in GBM. Existing knowledge still cannot explain how CTGF works on GBM malignant phenotype. The molecular biology study of CTGF in GBM remains for our team to complete in the future.
In summary, CTGF is highly expressed in glioma, especially GBM, and facilitates the progress of glioma. Combined with our research, present studies suggest that CTGF is a poor prognostic marker and a potential therapeutic target for human malignant tumor including glioma. Further exploration of the role of CTGF in cancers and other diseases remains to be done individually and conjointly.
Funding
This study was supported by a grant from the National Natural Science Foundation of China (No. 81872064).
Conflicts of interest
None.
Footnotes
How to cite this article: Song ZB, Yang HP, Xu AQ, Zhan ZM, Song Y, Li ZY. Connective tissue growth factor as an unfavorable prognostic marker promotes the proliferation, migration, and invasion of gliomas. Chin Med J 2020;133:670–678. doi: 10.1097/CM9.0000000000000683
Zi-Bin Song and Hui-Ping Yang contributed equally to this study.
References
- 1.Lee HK, Bier A, Cazacu S, Finniss S, Xiang C, Twito H, et al. MicroRNA-145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS One 2013; 8:e54652.doi: 10.1371/journal.pone.0054652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mendes FA, Coelho AJ, Kahn SA, Reis AH, Dubois LG, Romao LF, et al. Connective-tissue growth factor (CTGF/CCN2) induces astrogenesis and fibronectin expression of embryonic neural cells in vitro. PLoS One 2015; 10:e133689.doi: 10.1371/journal.pone.0133689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colombo MC, Giverso C, Faggiano E, Boffano C, Acerbi F, Ciarletta P. Correction: towards the personalized treatment of glioblastoma: integrating patient-specific clinical data in a continuous mechanical model. PLoS One 2015; 10:e143032.doi: 10.1371/journal.pone.0143032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song Y, Luo Q, Long H, Hu Z, Que T, Zhang X, et al. Alpha-enolase as a potential cancer prognostic marker promotes cell growth, migration, and invasion in glioma. Mol Cancer 2014; 13:65.doi: 10.1186/1476-4598-13-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Que T, Song Y, Liu Z, Zheng S, Long H, Li Z, et al. Decreased miRNA-637 is an unfavorable prognosis marker and promotes glioma cell growth, migration and invasion via direct targeting Akt1. Oncogene 2015; 34:4952–4963. doi: 10.1038/onc.2014.419. [DOI] [PubMed] [Google Scholar]
- 6.Romao LF, Mendes FA, Feitosa NM, Faria JC, Coelho-Aguiar JM, de Souza JM, et al. Connective tissue growth factor (CTGF/CCN2) is negatively regulated during neuron-glioblastoma interaction. PLoS One 2013; 8:e55605.doi: 10.1371/journal.pone.0055605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fayzullin A, Sandberg CJ, Spreadbury M, Saberniak BM, Grieg Z, Skaga E, et al. Phenotypic and expressional heterogeneity in the invasive glioma cells. Transl Oncol 2019; 12:122–133. doi: 10.1016/j.tranon.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng H, Yang Z, Xu N, Liu B, Fu Z, Lian C, et al. Connective tissue growth factor promotes temozolomide resistance in glioblastoma through TGF-beta1-dependent activation of Smad/ERK signaling. Cell Death Dis 2017; 8:e2885.doi: 10.1038/cddis.2017.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Li J, Chen L, Qi S, Yu S, Weng Z, et al. HERC3-mediated SMAD7 ubiquitination degradation promotes autophagy-induced EMT and chemoresistance in glioblastoma. Clin Cancer Res 2019; 25:3602–3616. doi: 10.1158/1078-0432.CCR-18-3791. [DOI] [PubMed] [Google Scholar]
- 10.Yin D, Chen W, O’Kelly J, Lu D, Ham M, Doan NB, et al. Connective tissue growth factor associated with oncogenic activities and drug resistance in glioblastoma multiforme. Int J Cancer 2010; 127:2257–2267. doi: 10.1002/ijc.25257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishida T, Kubota S, Kojima S, Kuboki T, Nakao K, Kushibiki T, et al. Regeneration of defects in articular cartilage in rat knee joints by CCN2 (connective tissue growth factor). J Bone Miner Res 2004; 19:1308–1319. doi: 10.1359/JBMR.040322. [DOI] [PubMed] [Google Scholar]
- 12.Ramazani Y, Knops N, Elmonem MA, Nguyen TQ, Arcolino FO, van den Heuvel L, et al. Connective tissue growth factor (CTGF) from basics to clinics. Matrix Biol 2018; 68-69:44–66. doi: 10.1016/j.matbio.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Aguiar DP, de Farias GC, de Sousa EB, de Mattos CJ, Lobo JC, Casado PL, et al. New strategy to control cell migration and metastasis regulated by CCN2/CTGF. Cancer Cell Int 2014; 14:61.doi: 10.1186/1475-2867-14-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perbal B. CCN proteins: multifunctional signalling regulators. Lancet 2004; 363:62–64. doi: 10.1016/S0140-6736(03)15172-0. [DOI] [PubMed] [Google Scholar]
- 15.Battula VL, Chen Y, Cabreira MG, Ruvolo V, Wang Z, Ma W, et al. Connective tissue growth factor regulates adipocyte differentiation of mesenchymal stromal cells and facilitates leukemia bone marrow engraftment. Blood 2013; 122:357–366. doi: 10.1182/blood-2012-06-437988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox JL, Dews M, Minn AJ, Thomas-Tikhonenko A. Targeting of TGFbeta signature and its essential component CTGF by miR-18 correlates with improved survival in glioblastoma. RNA 2013; 19:177–190. doi: 10.1261/rna.036467.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rayego-Mateos S, Morgado-Pascual JL, Rodrigues-Diez RR, Rodrigues-Diez R, Falke LL, Mezzano S, et al. Connective tissue growth factor induces renal fibrosis via epidermal growth factor receptor activation. J Pathol 2018; 244:227–241. doi: 10.1002/path.5007. [DOI] [PubMed] [Google Scholar]
- 18.Zhang LX, Jin W, Zheng J, Dai YX, Song Y, Ni HB, et al. MicroRNA-375 regulates proliferation and apoptosis of glioma cancer cells by inhibiting CTGF-EGFR signaling pathway. Bratisl Lek Listy 2018; 119:17–21. doi: 10.4149/BLL_2018_004. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Xu G. Study of expressions of ING4 and CTGF in human gliomas and their meanings (in Chinese). Chin J Clin Neurosurg 2011; 16:224–227. doi: 10.3969/j.issn.1009-153X.2011.04.012. [Google Scholar]
- 20.Peng F, Li H, Yin B, Wang Y, Chen Y, Xu Z, et al. Effect of telmisartan on expression of metadherin in the kidney of mice with unilateral ureter obstruction (in Chinese). J South Med Univ 2019; 39:156–161. doi: 10.12122/j.issn.1673-4254.2019.09.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi RH, Yang SC, Wen ES, Hu Z, Long H, Zeng Y, et al. Negative nuclear expression of CDKL2 correlates with disease progression and poor prognosis of glioma. Int J Clin Exp Pathol 2018; 11:712–719. [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Zhang T, Chen Y, Song J, Meng Y, Liu S, et al. Expression of transcription factor SOX12 in lung adenocarcinoma and its clinical significance (in Chinese). J South Med Univ 2019; 39:186–191. doi: 10.12122/j.issn.1673-4254.2019.09.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi S, Song Y, Peng Y, Wang H, Long H, Yu X, et al. ZEB2 mediates multiple pathways regulating cell proliferation, migration, invasion, and apoptosis in glioma. PLoS One 2012; 7:e38842.doi: 10.1371/journal.pone.0038842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan T, Yu S, Tao H. The role of connective tissue growth factor (CTGF) in tumor and the research progress in CTGF and gastric cancer (in Chinese). J Chin Oncol 2013; 19:527–531. doi: 10.11735/j.issn.1671-170X.2013.07.B006. [Google Scholar]
- 25.Wells JE, Howlett M, Cole CH, Kees UR. Deregulated expression of connective tissue growth factor (CTGF/CCN2) is linked to poor outcome in human cancer. Int J Cancer 2015; 137:504–511. doi: 10.1002/ijc.28972. [DOI] [PubMed] [Google Scholar]
- 26.Eguchi D, Ikenaga N, Ohuchida K, Kozono S, Cui L, Fujiwara K, et al. Hypoxia enhances the interaction between pancreatic stellate cells and cancer cells via increased secretion of connective tissue growth factor. J Surg Res 2013; 181:225–233. doi: 10.1016/j.jss.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 27.Haque I, Banerjee S, Mehta S, De A, Majumder M, Mayo MS, et al. Cysteine-rich 61-connective tissue growth factor-nephroblastoma-overexpressed 5 (CCN5)/Wnt-1-induced signaling protein-2 (WISP-2) regulates microRNA-10b via hypoxia-inducible factor-1 alpha-TWIST signaling networks in human breast cancer cells. J Biol Chem 2011; 286:43475–43485. doi: 10.1074/jbc.M111.284158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang MY, Chen PS, Prakash E, Hsu HC, Huang HY, Lin MT, et al. Connective tissue growth factor confers drug resistance in breast cancer through concomitant up-regulation of Bcl-xL and cIAP1. Cancer Res 2009; 69:3482–3491. doi: 10.1158/0008-5472.CAN-08-2524. [DOI] [PubMed] [Google Scholar]
- 29.Chu CY, Chang CC, Prakash E, Kuo ML. Connective tissue growth factor (CTGF) and cancer progression. J Biomed Sci 2008; 15:675–685. doi: 10.1007/s11373-008-9264-9. [DOI] [PubMed] [Google Scholar]
- 30.Shimo T, Kubota S, Yoshioka N, Ibaragi S, Isowa S, Eguchi T, et al. Pathogenic role of connective tissue growth factor (CTGF/CCN2) in osteolytic metastasis of breast cancer. J Bone Miner Res 2006; 21:1045–1059. doi: 10.1359/JBMR.060416. [DOI] [PubMed] [Google Scholar]
- 31.Aikawa T, Gunn J, Spong SM, Klaus SJ, Korc M. Connective tissue growth factor-specific antibody attenuates tumor growth, metastasis, and angiogenesis in an orthotopic mouse model of pancreatic cancer. Mol Cancer Ther 2006; 5:1108–1116. doi: 10.1158/1535-7163.MCT-05-0516. [DOI] [PubMed] [Google Scholar]
- 32.Xie D, Yin D, Wang HJ, Liu GT, Elashoff R, Black K, et al. Levels of expression of CYR61 and CTGF are prognostic for tumor progression and survival of individuals with gliomas. Clin Cancer Res 2004; 10:2072–2081. doi: 10.1158/1078-0432.Ccr-0659-03. [DOI] [PubMed] [Google Scholar]
- 33.Kato S, Yokoyama S, Hayakawa Y, Li LH, Iwakami Y, Sakurai H, et al. P38 pathway as a key downstream signal of connective tissue growth factor to regulate metastatic potential in non-small-cell lung cancer. Cancer Sci 2016; 107:1416–1421. doi: 10.1111/cas.13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim T, Lee I, Kim J, Kang WK. Synergistic effect of simvastatin plus radiation in gastric cancer and colorectal cancer: implications of BIRC5 and connective tissue growth factor. Int J Radiat Oncol Biol Phys 2015; 93:316–325. doi: 10.1016/J.Ijrobp.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 35.Guo YH, Li XR, Lin CW, Zhang Y, Hu G, Zhou JY, et al. MicroRNA-133b inhibits connective tissue growth factor in colorectal cancer and correlates with the clinical stage of the disease. Mol Med Rep 2015; 11:2805–2812. doi: 10.3892/mmr.2014.3075. [DOI] [PubMed] [Google Scholar]
- 36.Ren W, Sun XX, Wang K, Feng HL, Liu YH, Fei C, et al. BMP9 inhibits the bone metastasis of breast cancer cells by downregulating CCN2 (connective tissue growth factor, CTGF) expression. Mol Biol Rep 2014; 41:1373–1383. doi: 10.1007/s11033-013-2982-8. [DOI] [PubMed] [Google Scholar]
- 37.Yang F, Tuxhorn JA, Ressler SJ, McAlhany SJ, Dang TD, Rowley DR. Stromal expression of connective tissue growth factor promotes angiogenesis and prostate cancer tumorigenesis. Cancer Res 2005; 65:8887–8895. doi: 10.1158/0008-5472.CAN-05-1702. [DOI] [PubMed] [Google Scholar]
- 38.Lin BR, Chang CC, Che TF, Chen ST, Chen R, Yang CY, et al. Connective tissue growth factor inhibits metastasis and acts as an independent prognostic marker in colorectal cancer. Gastroenterology 2005; 128:9–23. doi: 10.1053/j.gastro.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 39.Chen PS, Wang MY, Wu SN, Su JL, Hong CC, Chuang SE, et al. CTGF enhances the motility of breast cancer cells via an integrin-alpha v beta 3-ERK1/2-dependent S100A4-upregulated pathway. J Cell Sci 2007; 120:2053–2065. doi: 10.1242/jcs.03460. [DOI] [PubMed] [Google Scholar]
- 40.Edwards LA, Woolard K, Son MJ, Li A, Lee J, Ene C, et al. Effect of brain- and tumor-derived connective tissue growth factor on glioma invasion. J Natl Cancer Inst 2011; 103:1162–1178. doi: 10.1093/jnci/djr224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang CC, Hsu WH, Wang CC, Chou CH, Kuo M, Lin BR, et al. Connective tissue growth factor activates pluripotency genes and mesenchymal-epithelial transition in head and neck cancer cells. Cancer Res 2013; 73:4147–4157. doi: 10.1158/0008-5472.CAN-12-4085. [DOI] [PubMed] [Google Scholar]
- 42.Mao ZF, Ma XY, Rong YF, Cui L, Wang XQ, Wu WC, et al. Connective tissue growth factor enhances the migration of gastric cancer through downregulation of E-cadherin via the NF-kappa B pathway. Cancer Sci 2011; 102:104–110. doi: 10.1111/j.1349-7006.2010.01746.x. [DOI] [PubMed] [Google Scholar]
- 43.Edwards LA, Woolard K, Son MJ, Li AG, Lee JW, Ene C, et al. Effect of brain- and tumor-derived connective tissue growth factor on glioma invasion. J Natl Cancer Inst 2011; 103:1162–1178. doi: 10.1093/jnci/djr224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen CN, Chang CC, Lai HS, Jeng YM, Chen CI, Chang KJ, et al. Connective tissue growth factor inhibits gastric cancer peritoneal metastasis by blocking integrin alpha 3 beta 1-dependent adhesion. Gastric Cancer 2015; 18:504–515. doi: 10.1007/s10120-014-0400-0. [DOI] [PubMed] [Google Scholar]
- 45.Chien WW, O’Kelly J, Lu DN, Leiter A, Sohn J, Yin D, et al. Expression of connective tissue growth factor (CTGF/CCN2) in breast cancer cells is associated with increased migration and angiogenesis. Int J Oncol 2011; 38:1741–1747. doi: 10.3892/ijo.2011.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin BR, Chang CC, Chen R, Jeng YM, Liang JT, Lee PH, et al. Connective tissue growth factor acts as a therapeutic agent and predictor for peritoneal carcinomatosis of colorectal cancer. Clin Cancer Res 2011; 17:3077–3088. doi: 10.1158/1078-0432.CCR-09-3256. [DOI] [PubMed] [Google Scholar]


