Abstract
Adenosine deaminase-1 (ADA-1) plays both enzymatic and non-enzymatic roles in regulating immune cell function. Mutations in the ADA1 gene account for 15% of heritable severe-combined immunodeficiencies. We determined previously that ADA1 expression defines and is instrumental for the germinal center follicular helper T cell (TFH) phenotype using in vitro human assays. Herein, we tested whether ADA-1 can be used as an adjuvant to improve vaccine efficacy in vivo. In vitro, ADA-1 induced myeloid dendritic cell (mDC) maturation as measured by increased frequencies of CD40-, CD83-, CD86-, and HLA-DR-positive mDCs. ADA-1 treatment also promoted the secretion of the TFH-polarizing cytokine IL-6 from mDCs. In the context of an HIV-1 envelope (env) DNA vaccine, co-immunization with plasmid-encoded ADA-1 (pADA) enhanced humoral immunity. Animals co-immunized with env DNA and pADA had significantly increased frequencies of TFH cells in their draining lymph nodes and increased HIV-binding IgG in serum. Next, mice were co-immunized with subtype C env gp160 DNA and pADA along with simultaneous immunization with matched gp140 trimeric protein. Mice that received env gp160 DNA, pADA, and gp140 glycoprotein had significantly more heterologous HIV-specific binding IgG in their serum. Furthermore, only mice that received the combination ADA-1 adjuvanted vaccine had detectable neutralizing antibody responses. These studies support the use of ADA-1 as a vaccine adjuvant to qualitatively enhance germinal center responses and represent a novel application of an existing therapeutic agent that can be quickly translated for clinical use.
One Sentence Summary:
Adenosine deaminase-1 is a novel adjuvant that improves the quality of antibody response by augmenting germinal center function and its use in the context of vaccination represents a novel use of a clinically-approved therapeutic.
Introduction
Adenosine deaminase-1 (ADA-1) regulates intra- and extracellular adenosine levels by deaminating adenosine to produce inosine. ADA-1 functions both enzymatically and extra-enzymatically to regulate immune function. It is estimated that approximately 15% of heritable severe-combined immunodeficiencies (SCID) have their etiology in ADA1 loss-of-function mutations[1]. CD4+ T cells express CD26, an ADA-1 receptor, and it is proposed that ADA-1 may bridge the CD26 receptor on these cells and other ADA-1 receptors expressed on antigen-presenting cells, promoting the formation of an immunological synapse. We previously reported that ADA1 is critical to the TFH program, and that addition of exogenous ADA-1 enhanced the ability of less efficient pre-TFH to provide help to B cells[2].
Induction of broadly neutralizing antibodies (bnAbs) against HIV-1 is critical to the generation of a vaccine[3,4]. The development of nAbs occurs in some HIV patients however, these responses develop slowly, allowing for the evolution of viral escape mutants. No vaccine so far has been able to elicit broad nAbs in humans or animal models[5][6]. nAb development correlates with germinal center (GC) function[7]. Therefore, there is a critical need to develop immunogens which display neutralizing epitopes as well as vaccine modalities that can enhance GC function in the context of immunization with HIV-1 env vaccine antigens. The ability to encode molecular adjuvants targeting specific immune pathways makes DNA a useful platform for such vaccine modalities.
We hypothesized that the co-delivery of plasmid-encoded ADA-1 (pADA) as a molecular adjuvant would improve the quality of humoral immune responses to HIV-1 env DNA immunogens in a GC TFH-dependent manner. In vitro, we observed that treatment of human mDCs with ADA-1 induced maturation and resulted in a cytokine/chemokine secretion profile which likely supports TFH differentiation. In vivo, in the context of a consensus clade B HIV env DNA immunogen, we observed significant increases in the levels of HIV-binding IgG in the serum of mice co-immunized with pADA. These responses correlated with increased frequencies of GC TFH in the draining lymph nodes (DLNs) of vaccinated animals. We also evaluated the ability of pADA to enhance GC activities in the context of a DNA-protein co-immunization regimen. Again, addition of molecular ADA-1 to the DNA arm of this regimen resulted in superior HIV-binding antibody production and the production of heterologous tier-1 nAbs in vaccinated mice. These data indicate that pADA co-immunization enhances antigen-specific IgG in the serum of vaccinated animals in a GC TFH-dependent manner. Thus, ADA-1 is a promising adjuvant that targets vaccine-induced GC responses.
Results
ADA-1 induces the maturation of human mDCs
Whether ADA-1-matured mDCs influence the differentiation of CD4+ T cells towards a TFH phenotype has yet to be determined. To test this, monocytes from healthy PBMCs were treated with GM-CSF/IL-4 48h followed by maturation with either ADA-1 or LPS/IFNγ as a positive control for 24hrs. We assessed the frequency of surface costimulatory and HLA marker-expressing cells in ADA-1-treated mDCs compared to unstimulated cells after 24 hours. The gating strategy used for mDCs and the determination of their surface marker frequencies is shown in Supplemental Fig. 1A. We found that LPS/IFNγ and ADA-1 significantly up-regulated the percentage of CD40, CD83, CD86, and HLA-DR-positive human mDCs compared to unstimulated control. (Fig. 1A–D). The increase in ADA-1-mediated CD83 expression however, was noted to be significantly less than that induced by LPS/IFNγ but still significantly higher than immature unstimulated cells (Fig. 1B). Interestingly, neither LPS/IFNγ, nor ADA-1 treatments increased the frequency of mDCs positive for ligand of inducible co-stimulator (ICOSL) (Fig.1E). Both LPS/IFNγ and ADA-1 induced the expression of CD40 (Fig. 1A), which is known to be important for human TFH helper function via CD40L. These data confirm that ADA-1 induces the maturation of human mDCs in our in vitro culture system and suggest the possibility that ADA-1 may mediate its effect on TFH function at least in part, by altering the phenotype and maturation status of antigen-presenting cells.
Figure. 1. ADA-1 treatment promotes DC maturation and the secretion of pro-TFH cytokines.
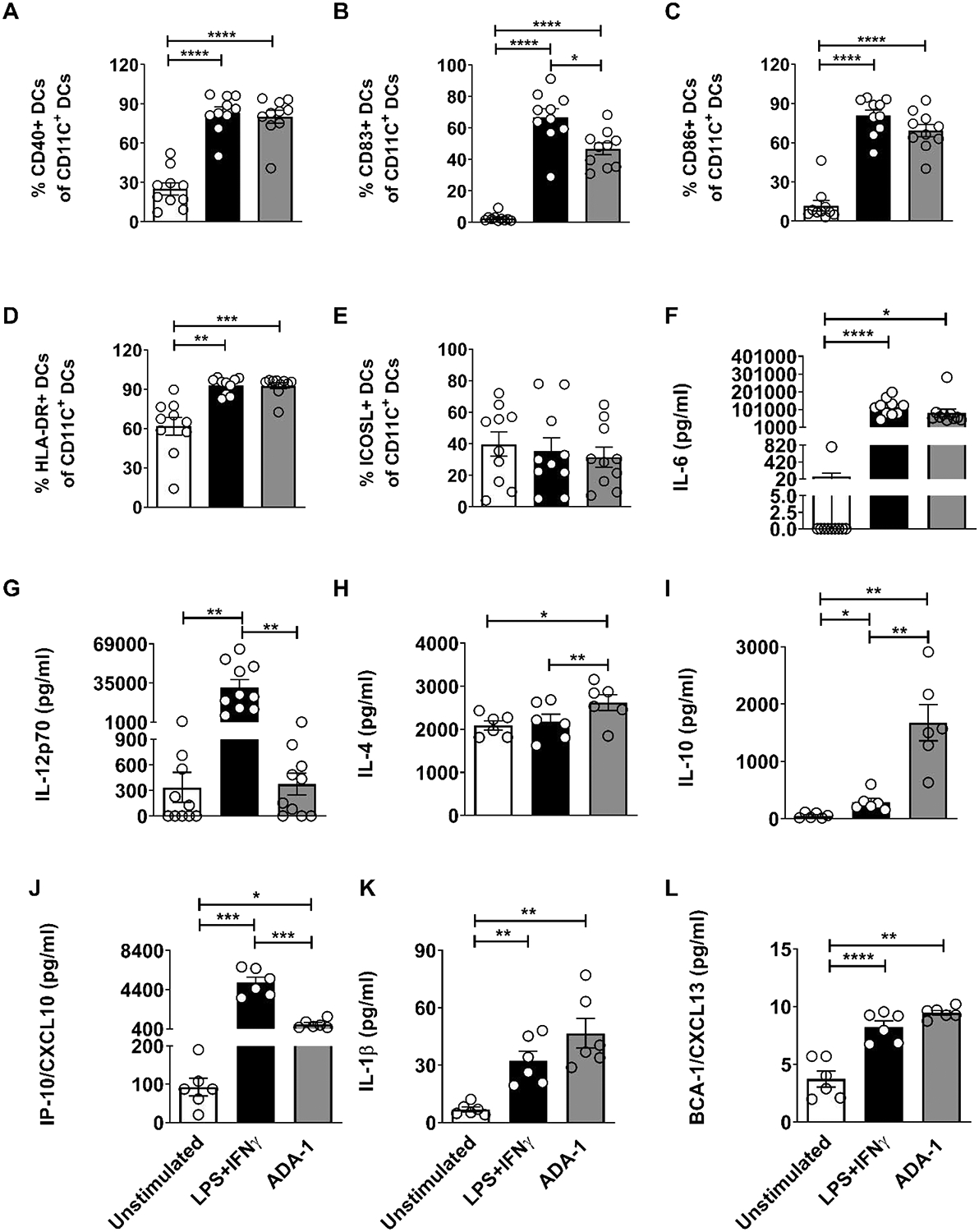
Myeloid dendritic cells (mDCs) differentiated in vitro from monocytes of healthy human PBMCs, were treated with 0.5μg/ml LPS plus 40ng/ml IFN-γ or 2.5μM ADA-1 for 24h, and compared to unstimulated cells as controls. mDCs were harvested and stained for surface HLA and costimulatory molecule expression and analyzed by flow cytometry. Values are shown as the frequencies of (A) CD40, (B) CD83, (C) CD86, (D) HLA-DR and (E) ICOS L expression in the live, CD3- CD19- CD11c+ mDC population. Results are expressed as mean ± SEM of 2 independent experiments (n=10). Culture supernatants were collected 24h post-stimulation and analyzed for the levels of pro-TFH cytokine/chemokine production. Levels of (F) IL-6 and (G) IL-12p70 were assessed by ELISA whereas levels of (H) IL-4, (I) IL-10, (J) IL-1ß and (K) BCA-1/CXCL13 were assessed by the Luminex assay. Results are expressed as mean ± SEM of 4 independent experiments (n=10) for (F) and (G), and as mean ± SEM of 2 independent experiments (n=6) for (H-K). Each individual circle represents one individual subject. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001 by one-way ANOVA
ADA-1 alters the cytokine and chemokine profile of human mDCs in vitro into a pro-TFH phenotype
We also evaluated the ADA-1 induced cytokine profile of mDCs in this system. LPS/IFNγ significantly induced the production of high levels of cytokines and chemokines consistent with a pro-inflammatory phenotype compared to unstimulated cells, namely IL-6, IL-12p70, IL-1ß, and CXCL13 (Fig. 1F, G, J, K). LPS plus IFNγ also induced minimal secretion of the anti-inflammatory cytokine IL-10 from mDCs (Fig. 1I). While ADA-1 administration significantly increased the secretion of the pro-inflammatory molecules IL-6, IL-1ß, and CXCL13 (Fig.1F, K, L) and the secretion of the anti-inflammatory molecules IL-4 and IL-10 (Fig.1H, I) from mDCs only in the case of IL-4 and IL-10 was this increase significantly more than that produced by LPS/IFNγ stimulation. IL-6 is a key regulator of the master TFH cell transcription factor Bcl6[8–11]. IL-1ß and CXCL13 have also been implicated in TFH development/function [12–14]. Interestingly, ADA-1-matured mDCs were not able to secrete IL-12p70 at significant levels compared to those treated with LPS/IFNγ (Fig. 1G). This observation suggests reduced inflammatory responses mediated by ADA-1 as compared to LPS/IFNγ. Thus, ADA-1-stimulated mDCs produce high levels of the key pro-TFH cytokine IL-6, high amounts of IL-1ß and CXCL13, which may play a role in TFH cell differentiation, function, and proliferation[13,14], alongside robust levels of the regulatory cytokines IL-4 and IL-10. Overall, our results demonstrate that ADA-1 can induce the maturation of dendritic cells and elicits an mDC cytokine/chemokine secretion phenotype that is distinct from TH1 and TH2 cytokine profiles, and which may support TFH cell differentiation.
pADA enhances TFH Cell frequency in vivo
To determine the effect that pADA co-immunization has on TFH cells in vivo, mice were vaccinated three times, separated by two weeks, with either empty plasmid (pVAX), DNA-encoded HIV-1 gp160 consensus envelop antigen from clade B viruses (pEnvB), or co-immunized with pEnvB and plasmid-encoded ADA-1 (+pADA) (Fig. 2A), and the frequency of germinal center B (GC B) and TFH cells in the spleens and DLNs of vaccinated animals was quantified at day 7 post-3rd immunization by flow cytometry (Supp. Fig 1B). We observed statistically significant increases in the frequency of GC TFH cells in the DLNs of pADA co-immunized animals as compared to animals receiving either empty plasmid vector or pEnvB alone (Fig. 2C, D). This increased frequency of TFH appears limited to the local DLNs (popliteal and inguinal), as such enhancements were not observed in the spleens of pADA co-immunized animals (Fig. 2B). These results indicate that molecular ADA-1 can enhance TFH cell frequency in vivo and importantly, demonstrate that the adjuvant effect of pADA is local and not systemic.
Figure 2. Co-immunization with pADA enhances GC formation.
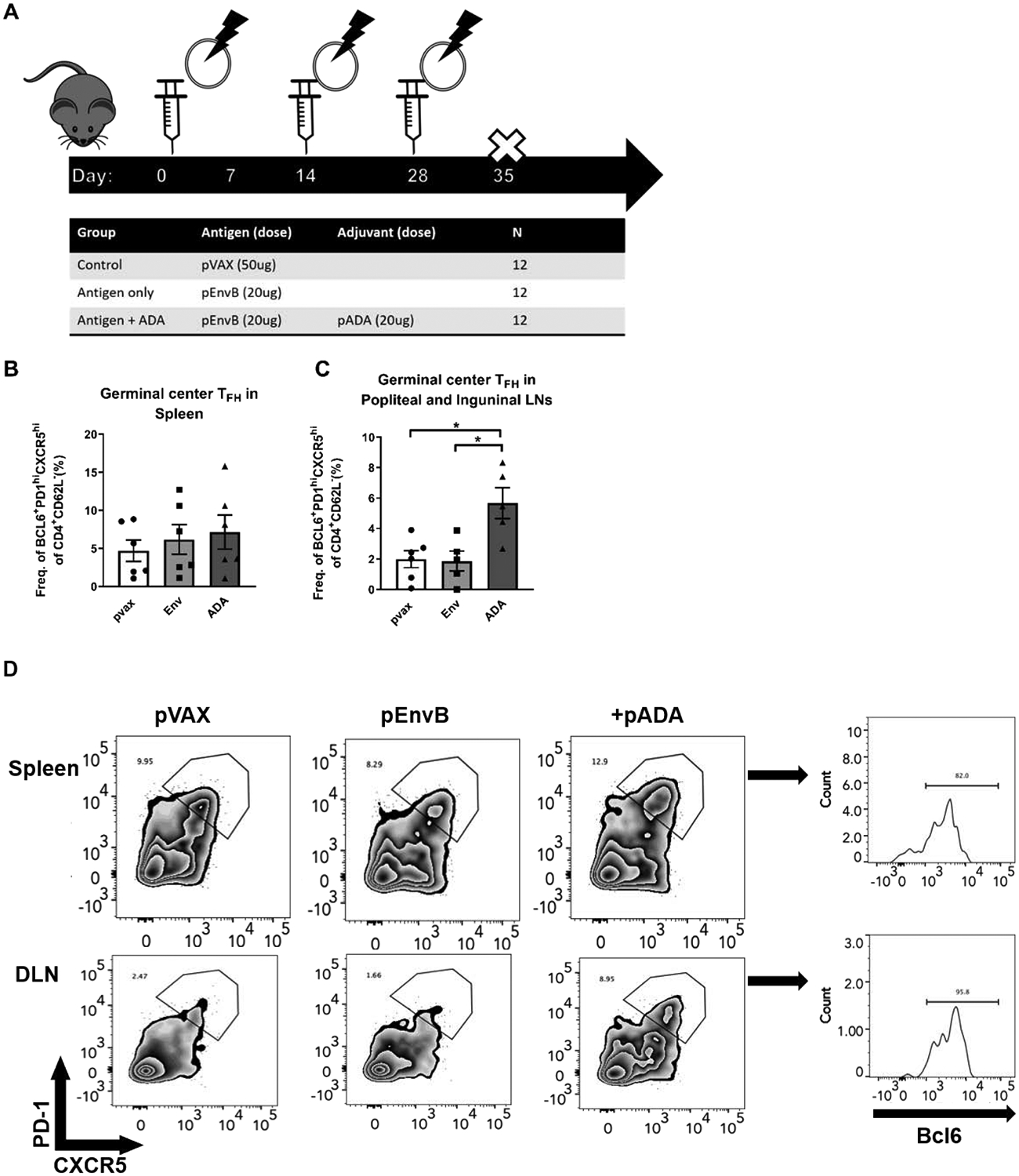
(A) female C57BL/6 mice were immunized thrice, separated by 2 weeks with either empty plasmid (pVAX), HIV envelope B consensus gp160-encoding plasmid (pEnvB), or co-immunized with pEnvB and pADA (+pADA). All immunizations was delivered via intramuscular injection, followed by electroporation using the CELLECTRA device (Inovio). (B) Frequency of TFH in spleens and (C) Popliteal and inguinal lymph nodes at day 7 post-3rd immunization. (D) Representative flow plots for the data presented in B and C. Each point represents an individual animal, bars represent the mean and SD. Each point represents the average of duplicate samples from an individual animal, bars represent the mean and error bars represent the SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 by one-way ANOVA. Data are representative of four independent experiments with n=3–6/group
pADA enhances HIV-specific IgG in vaccinated animals
We hypothesized that co-delivery of plasmid-encoded ADA-1 (pADA) in the context of our anti-HIV DNA vaccine would enhance HIV-specific IgG in vaccinated animals. Serum ELISA from vaccinated animals (Fig. 2A) revealed statistically significant increases in HIV-1 envB-binding IgG in pADA co-immunized animals compared to those receiving either empty plasmid or antigen alone (Fig. 3A). No differences in serum IgA (Fig. 3B) or IgM (not shown) were detected at this time point. These data indicate that pADA co-immunization enhances antigen-specific serum antibody responses. Further analysis of the IgG subclasses revealed that pADA co-immunization resulted in a statistically significant increase in HIV-specific IgG2b (Fig. 3D), which may be suggestive of TFH-type immunity. This was not true for IgG1 (Fig. 3C), IgG2c (Fig. 3E), or IgG3 (Fig. 3F). Importantly, IgG2 antibodies have been associated with long-term non-progression in HIV-infected human patients[15,16].
Figure 3. Co-immunization with pADA enhances serum IgG.
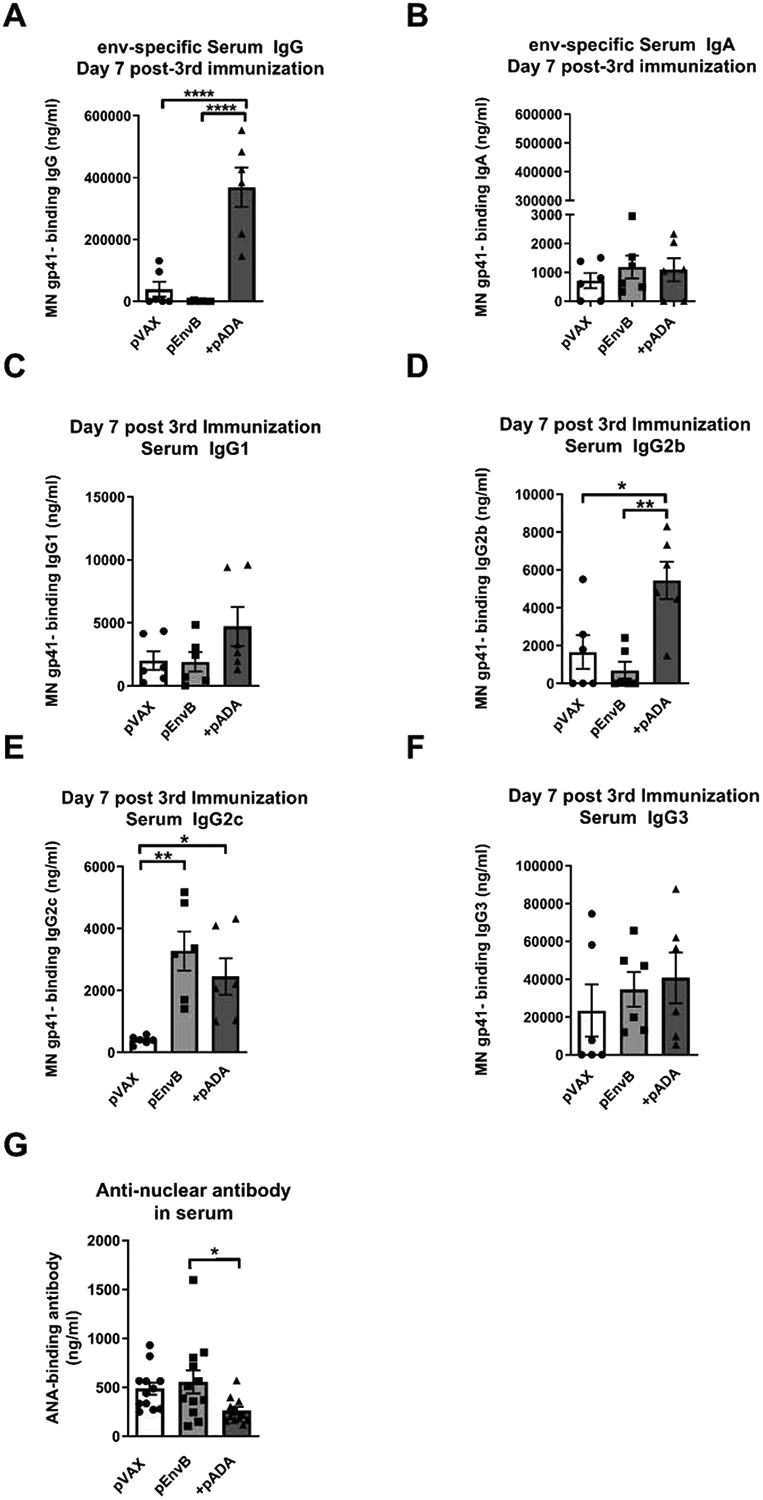
Mice were immunized as in figure 2. Total HIV-specific serum IgG (A), IgA (B), IgG1 (C), IgG2b (D), IgG2c (E), and IgG3 (F) was quantitated in serum via ELISA at day 7 post 3rd immunization. (G) Mice were immunized as in figure 2, and anti-nuclear antibody was quantified in serum at day 7 post-3rd immunization via ELISA. Each point represents the average of duplicate samples from an individual animal, bars represent the mean and error bars represent the SEM Each point represents the average of duplicate samples from an individual animal, bars represent the mean and SEM. *p<0.05, **p<0.01, ****p<0.0001 by one-way ANOVA. Data are representative of two independent experiments with n=6/group
As increased TFH frequency has been associated with autoimmunity [17–19], we quantified the levels of anti-nuclear antibody (ANA) present in the serum of vaccinated animals after three immunizations via ELISA. We detected no increase in the levels of ANA in the serum of mice co-immunized with pADA compared to mice receiving either empty plasmid vector or pEnvB alone (Fig. 3G). These data indicate that ADA-1 supports TFH expansion in an antigen-dependent manner which will be critical to translating the use of ADA-1 as a vaccine adjuvant to the clinic.
pADA enhances serum IgG in a DNA-protein co-immunization regimen
DNA-prime protein-boost vaccination regimens and other heterologous prime-boost regimens have demonstrated increased humoral immunity[20][21–23]. Recently we demonstrated that simultaneous delivery of DNA immunogens and protein immunogens together is superior to protein-only immunization with respect to the kinetics, magnitude, and breadth of antibody responses[24]. Such vaccination regimens can capitalize on the ability of the DNA platform to engender robust cell-mediated responses and the ability of the protein-adjuvant platform to induce robust humoral responses, while potentiating a shortened immunization regimen in the clinic. We again hypothesized that addition of pADA to the DNA arm of this regimen would enhance anti-HIV IgG in a GC TFH-dependent manner. The env immunogens used in this study were cloned from the plasma of an HIV subtype C-infected subject in the CAPRISA cohort who developed nAbs early in infection which continued to broaden subsequently over 4.5 years of study[25]. A single env sequence from 54 weeks post infection was motif optimized and used in this vaccine study as a DNA plasmid expressing gp160 native trimer (pCAP257–54wk_D), and as a modified truncated, non-cleaved trimeric glycoprotein gp140. We vaccinated mice with either clade C pCAP257–54wk_D env DNA alone (pEnvC), pEnvC DNA and gp140 CAP257–54wk_D protein (pEnvC+ rEnvC), pEnvC DNA and pADA without protein (pEnvC +pADA), or pEnvC DNA and pADA with simultaneous rEnvC protein immunization (pEnvC +rEnvC +pADA). All DNA immunogens and adjuvants were delivered to the left tibialis anterior followed by in vivo electroporation. Protein immunogens were delivered in aluminum phosphate adjuvant (AdjuPhos) in the right quadriceps femoris (Fig. 4A). While protein-adjuvant GC responses typically develop in 9–14 days, the kinetics of the GC response to DNA immunogens has not been well characterized, thus we vaccinated animals three times, separated by two weeks, and sacrificed them at days 3, 6, and 12 post-3rd immunization and quantified the frequencies of GC TFH and GC B in the DLNs and env-binding IgG in the serum. Interestingly, with this DNA antigen, in the absence of simultaneous protein immunization, co-immunization with pADA did not significantly enhance serum IgG responses compared to DNA antigen-only immunized animals. No differences were detected in serum IgG among any of the groups at days 3 and 6 post-3rd immunization (Fig. 4B, C). However, animals that received pADA with pEnvC and simultaneous protein immunization, had significantly more envC-binding IgG in serum than animals that received either DNA alone, DNA and protein, or DNA with pADA (Fig. 4D). These data confirm that pADA co-immunization can enhance antigen-specific serum antibody responses in the context of a DNA-protein co-immunization.
Figure 4. Co-immunization with pADA enhances serum IgG and promotes neutralizing antibody formation in a DNA-protein co-immunization regimen.
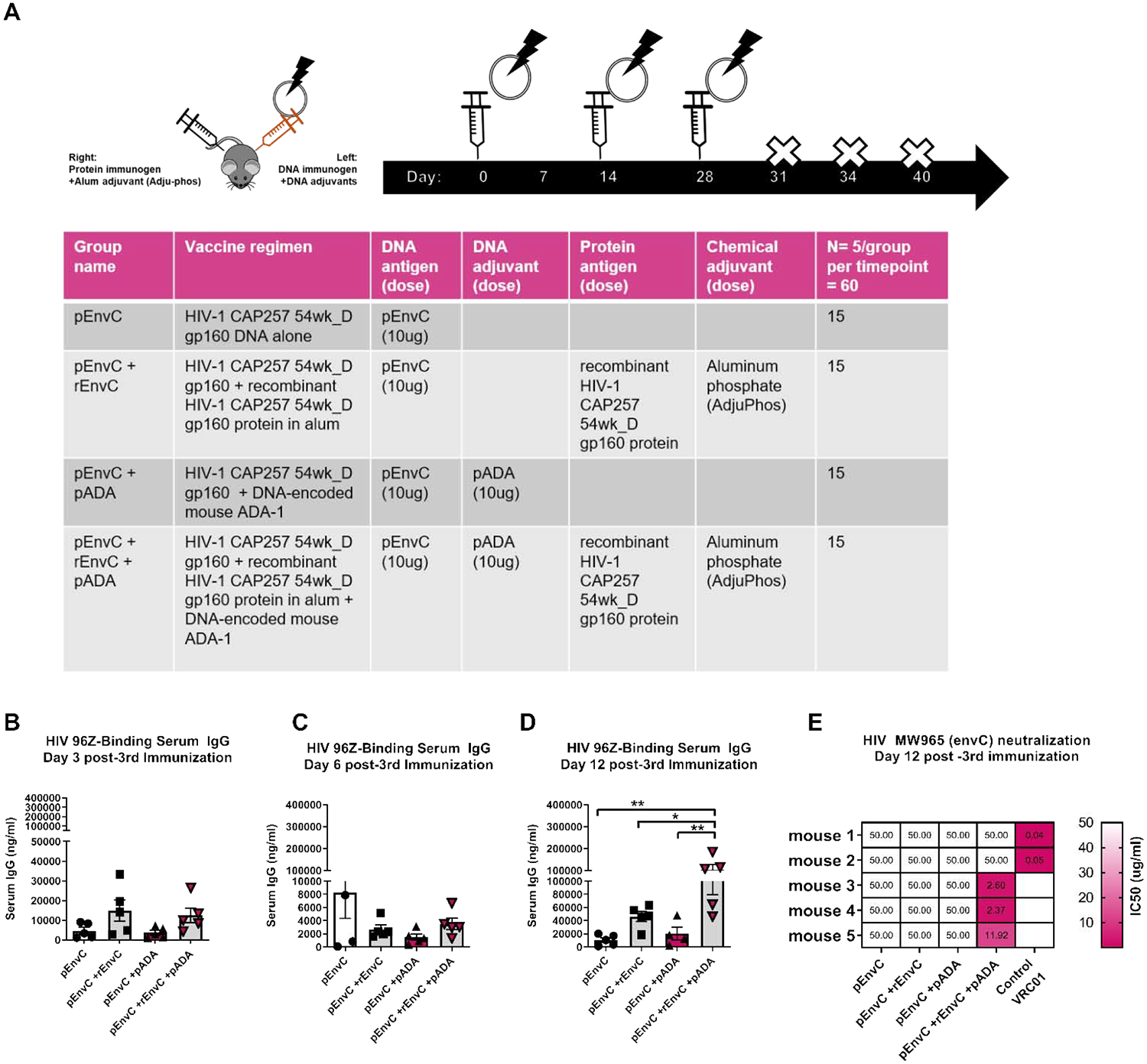
(A)Mice were immunized three times with envelope C DNA alone (pCapC), pCapC with simultaneous administration of matched recombinant envelope in alum adjuvant (+rCapC), DNA plus molecular ADA (+pADA), or DNA plus molecular ADA and simultaneous protein in alum (+pADA +rCapC). All DNA was delivered intramuscularly with in vivo electroporation in the left tibialis. All protein was delivered intramuscularly in the right quadriceps. env-binding IgG was quantified in the serum of vaccinated animals by ELISA at days 3 (B), 6 (C), and 12 (D) post-3rd immunization. Each point represents the average of duplicate samples from an individual animal, bars represent the mean and SEM. (E) IC50 values for purified mouse IgG were determined using the single-cycle TZM-bl neutralization assay against MW965 clade C HIV-1 viruses. 50ug/ml of mouse sera were added to test wells and values greater than 50ug/ml are represented as 50ug/ml. Each rectangle represents a single mouse within a group. 2ug/ml of VRC01 was used as a positive control for neutralization. Data are representative of one experiment with n=5/group. *p<0.05, **p<0.01 by one-way ANOVA. Data are representative of one experiment with n=5/group.
pADA induces nAbs during DNA-protein co-immunization
Only animals that received DNA and protein in the presence of pADA displayed increased envC-binding IgG at day 12 post-3rd immunization (Fig. 4D). This led us to hypothesize that administration of pADA supports enhanced GC function and thus alters the quality of humoral responses in this vaccination model. Therefore, we evaluated the quality of pADA-induced humoral responses in our mouse vaccination model using a TZM-bl neutralization assay using IgG purified from the serum of immunized mice. Remarkably, despite the similar frequencies of GC cells in animals which received simultaneous protein immunization, we were only able to detect neutralization of heterologous, tier-1 subtype C HIV-MW965 pseudovirus using IgG from mice that received DNA and protein along with pADA co-immunization (Fig. 4E). Three of the five immunized mice developed nAbs with potent activity. These data indicate that pADA enhances the function of GCs, independent of increases in frequencies of GC cell populations.
Immunization with pADA enhances TFH function in a DNA-protein co-immunization regimen
Analysis of HIV-specific IgG in the serum of animals receiving pEnvC and pADA with simultaneous rEnvC glycoprotein immunization indicated that pADA enhanced GC activities and antibody function. We collected the popliteal and inguinal lymph nodes from the left and right leg of each animal and quantified GC TFH and GC B cell frequencies by flow cytometry. Surprisingly, despite the observed increase in env-binding IgG in the serum of animals which received pEnvC, pADA, and simultaneous protein compared to those receiving pEnvC and simultaneous protein without pADA (Fig. 4D), all groups that received protein simultaneously displayed increases in the frequencies of GC TFH and GC B cells in the right DLNs irrespective of pADA co-immunization (Fig.5A–F). This data suggests that pADA enhances GC TFH function, but not necessarily GC TFH frequency, in this model. At all time points, there was no difference in the frequencies of GC TFH or GC B cells in the left DLNs (Supp. Fig. 2). As expected, at the later time points, GC B frequencies strongly correlated with TFH frequencies in the DLNs (Fig. 6A–F). These responses appeared to be specific to the DLNs, as we were only able to detect differences in the spleen at day 12 post-3rd immunization, when animals that received pADA co-immunization and simultaneous protein immunization exhibited increased GC B cell frequency compared to animals receiving pEnvC alone or pEnvC with pADA, but not those receiving pEnvC and protein (Supp. Fig. 3). The discrepancy between TFH frequencies induced by DNA and pADA alone in this experiment compared to our DNA only experiments (Fig. 2) may be due to differences in immunogenicity or kinetics of GC formation between the two env plasmids. It is also possible that the robust GC formation observed upon immunization with protein and alum adjuvant make it difficult to distinguish pADA-mediated changes in immune responses.
Figure 5. pADA supports protein-mediated GC formation.
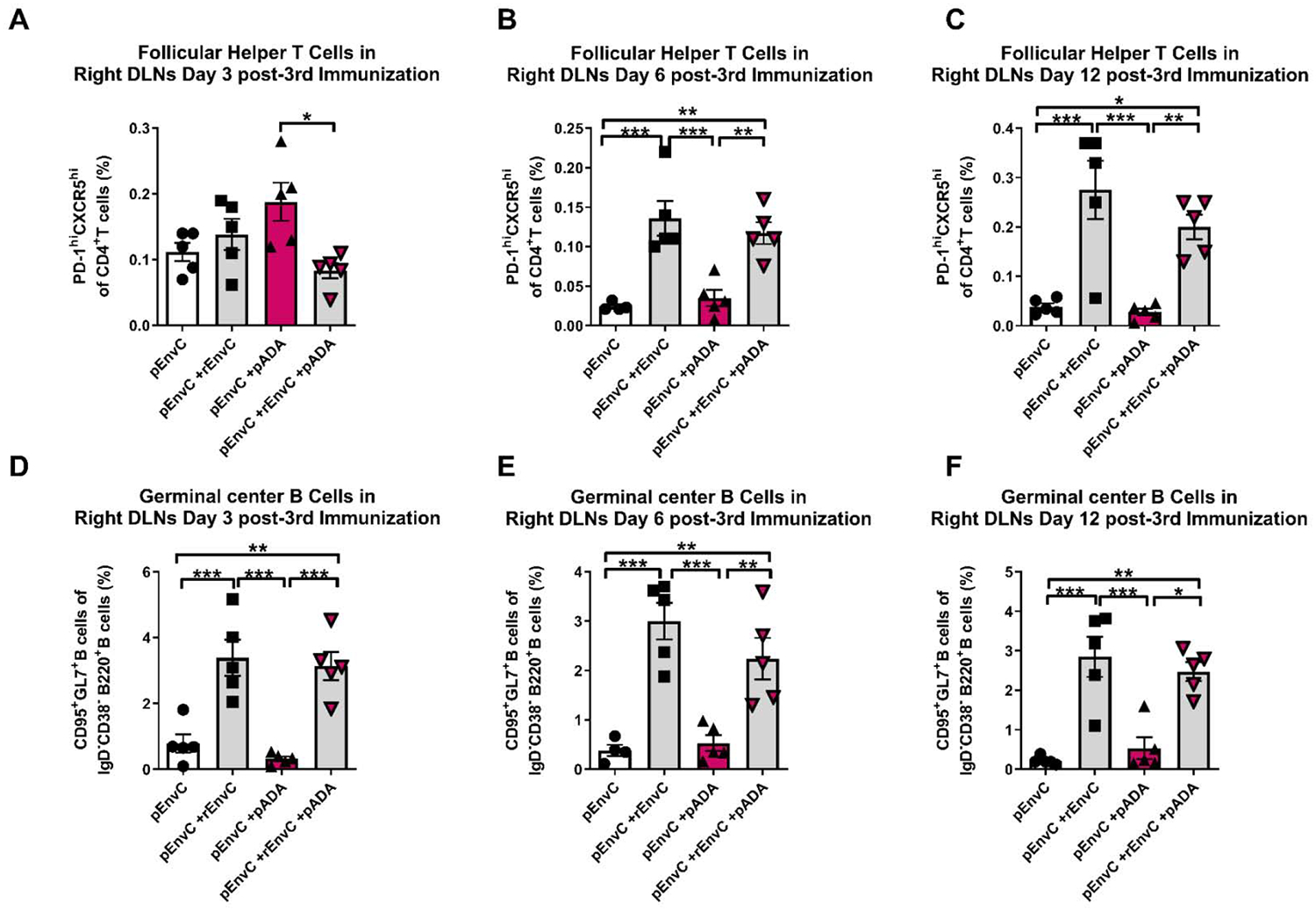
Mice were immunized as in fig. 5. TFH (A-C) and GC B (D-F) cell frequencies were determined in the right DLNs at days 3 (A,D), 6 (B,E), and 12 (C,F) post-3rd immunization. Each point represents an individual animal, bars represent the mean and SD. *p<0.05, **p<0.01, ***p<0.001 by one-way ANOVA. Data are representative of one experiment with n=5/group.
Figure 6. GC B and TFH frequencies strongly correlate during DNA-protein co-immunization.
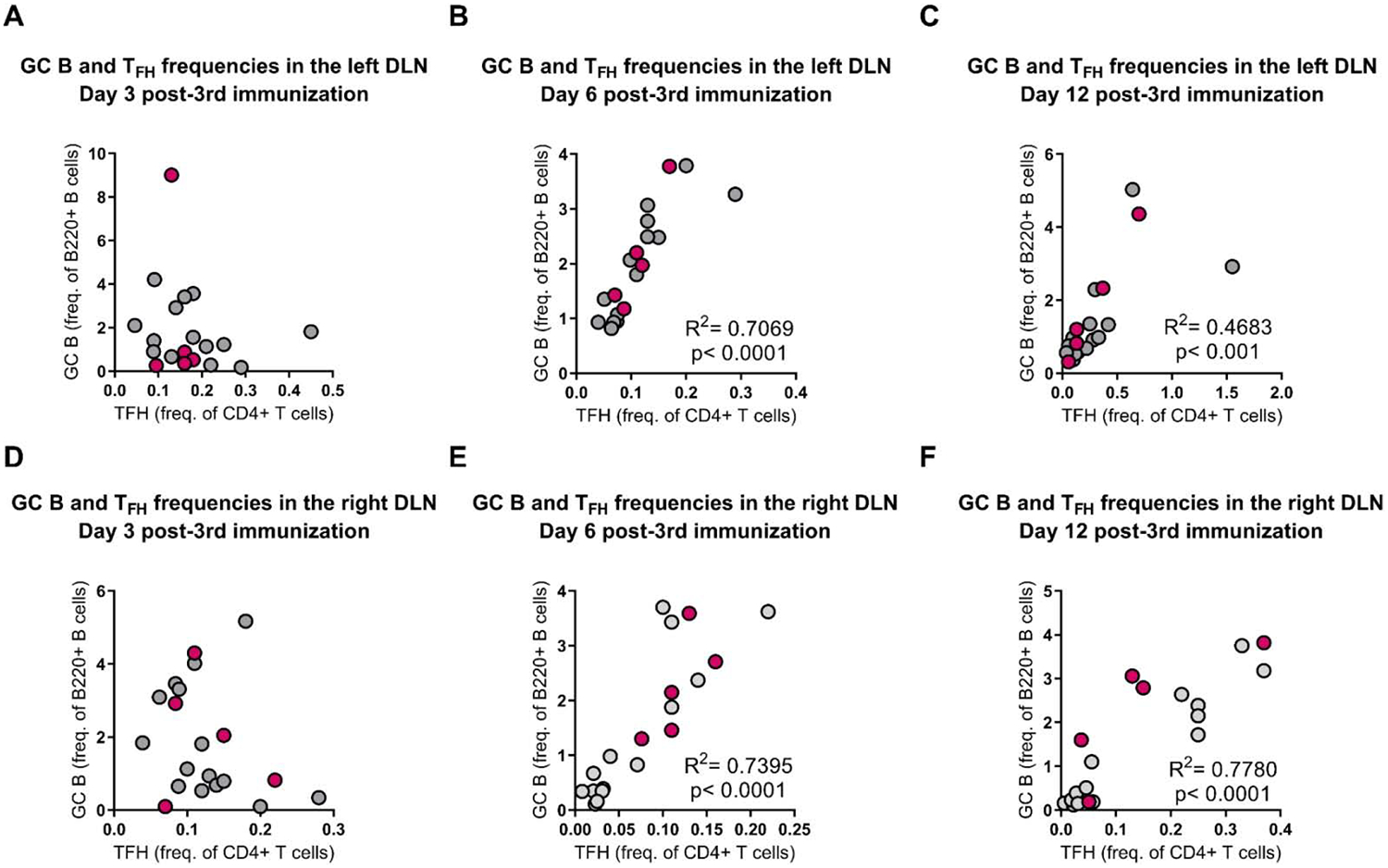
Mice were immunized as in figure 5. TFH and GC B frequencies were correlated in the left (top) and right (bottom) DLNs at days 3 (A,D), 6 (B,E), and 12 (C,F) post-3rd immunization. Each point represents an individual animal. Animals receiving DNA, protein and pADA are depicted in magenta. R2 and p values determined by Pearson correlation. Data are representative of one experiment with n=5/group
pADA promotes T cell responses in a DNA-protein co-immunization regimen
Next we investigated the effect of pADA co-immunization on cell mediated immunity. We detected increased frequencies of IFNγ spot-forming units (SFUs) in groups that were co-immunized with pADA and protein simultaneously at days 3 and 6 post 3rd immunization (Fig. 7A–B). At day 12 post-3rd immunization, groups which received simultaneous protein co-immunization with or without pADA had statistically significant increases in IFNγ-secreting cells in the spleen compared to groups which received pEnvC alone or pEnvC with pADA (Fig. 7C). However, at day 12 post 3rd immunization, animals which received pEnvC, pADA, and rEnvC had significantly more IFNγ SFUs compared to animals which received wither pEnvC alone, pEnvC with pADA, or pEnvC with protein. These data suggest that co-immunization with pADA in the context of a DNA-protein regimen promotes effector T cell function. Overall, our findings support the notion that pADA enhances TFH function which in turn, improves the quality of both humoral and cell mediated immune responses to vaccine antigens.
Figure 7. Molecular ADA enhances IFNy secretion during DNA-protein co-immunization.
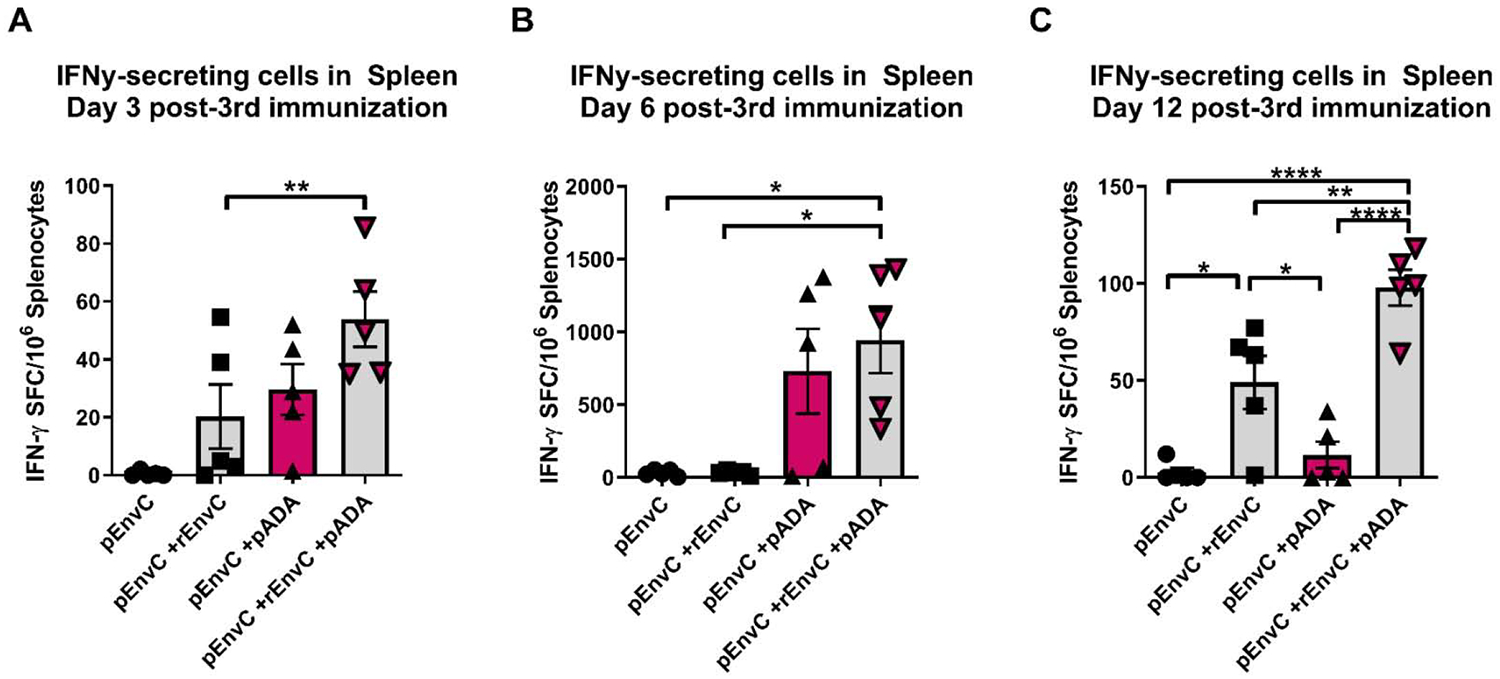
Mice were immunized as in Fig. 5. Spleenocytes were assayed for HIV-specific IFNy production at days 3(A), 6 (B), and 12 (C) post-3rd immunization. Each point represents the average of duplicate assays for an individual animal for days 3 and 6, and error bars represent the SEM. Each point represents a single assay for an individual animal for day 12 and error bars represent the SD. *p<0.05, **p<0.01, ****p<0.0001 by one-way ANOVA. Data are representative of one experiment with n=5/group.
Discussion
Our previous work demonstrates that ADA1 is significantly upregulated in GC TFH cells[2]. We observed increased frequencies of mDCs positive for surface costimulatory and antigen-presenting molecules following ADA-1 treatment in our in vitro culture system. This result is concordant with reports that stimulation with ADA-1 up-regulated the expression of surface markers of maturation on mDCs [26]. ADA-1 altered the cytokine and chemokine profile of mDCs to one that likely promotes the TFH program, including robust IL-6 production. While IL-6 is required to support the expression of the GC master regulator Bcl6 via STAT1[9,11], [27], the role of IL-12 in TFH differentiation is controversial. IL-12 can support the expression of Bcl6 via STAT4, however it also supports T-bet function, a master regulator which inhibits Bcl6[28],[29]. These responses have also been demonstrated to remain intact in STAT4-deficient animals, suggesting that IL-6 signaling through STAT1/STAT3 can compensate for IL-12 signaling. The balance between Bcl6 and T-bet may be regulated by expression of IL-2, where high expression of IL-2 in the context of IL-12 signaling promotes T-bet, and low expression of IL-2 in the context of IL-12 signaling promotes Bcl6 function[30,31]. CXCL13, a key TFH chemokine and ligand for the characteristic TFH marker CXCR5, was increased in the supernatant of ADA-1-treated mDCs. CXCL13 can promote CXCR5 expression in an autocrine fashion [32,33]. The ADA-1-mediated increase in IL-4 and IL-10 suggests a balance in the established mDC response. Significant increases in IL-4 observed upon ADA-1 treatment indicates a regulatory response which may dampen inflammation. Further experiments on the ability of ADA-1-stimulated mDCs to directly induce naïve CD4+ T cells to differentiate into TFH cells, in vitro as well as in vivo must be performed.
These data indicate that ADA-1 supports GC function by at least two distinct, but not mutually exclusive mechanisms. Firstly, expression and release of extracellular ADA-1 in the GC supports the GC TFH:B cell interaction. This is supported by our previous experiments which demonstrate that exogenous ADA-1 treatment improves the TFH phenotype and enhances the capacity of less-efficient CD4+ T cells to provide help to antibody secreting cells resulting in increased IgG secretion[2]. Secondly, ADA-1 promotes the maturation of APCs and the secretion of potentially TFH-polarizing cytokines from antigen presenting cells in vitro. In support of these mechanisms, we observed that co-immunization with pADA in the context of an HIV-1 env DNA vaccine increases DLN TFH cell frequencies and increases env-binding antibody in the serum of vaccinated mice. We were unable to detect increases in mouse anti-nuclear antibody (ANA) in the serum of animals that received pADA co-immunization, suggesting that increasing the frequencies of antigen-specific TFH cells is not detrimental. However, a complete characterization of the molecular mechanism of ADA-1 function in vivo is required in order to definitively demonstrate that ADA-1 administration in the context of an antigen-specific immune response does not promote autoimmunity.
Addition of pADA to the envelope DNA arm, and simultaneous co-delivery of protein-in-adjuvant, significantly increased HIV-specific IgG in the serum of vaccinated animals compared to those which received envelope DNA alone, DNA and protein, or DNA and pADA without protein. Interestingly in this experiment, while DNA, pADA, and protein together significantly enhanced serum IgG, we detected significant increases in TFH frequencies in the DLNs of the protein-vaccinated limb of all animals which received protein, irrespective of co-immunization with pADA. These data verify that protein and aluminum adjuvant formulations increase TFH cell frequency; however, the enhanced frequency of TFH cells induced by protein-in-alum without pADA did not result in significant enhancement of serum IgG in our experiments, suggesting that pADA enhanced the function of TFH cells as measured by antigen-binding antibody. This might be due to increased survivability of antigen-specific TFH cells and not necessarily increased proliferation of these cells which would explain the similar frequencies of TFH between groups that received protein with- and without pADA. It is also possible that ADA-1 may directly interact with B cells as ADA-1 deficiency is known to cause B cell defects, and this interaction may increase plasma cell fate-choice resulting in increased plasma cells and serum antibody without altering GC B cell frequency. However, we have previously reported that treatment of memory B cells with exogenous ADA-1 in the absence of TFH cells, does not enhance IgG production[2]. Recent elegant experiments indeed suggest that antigen availability is the primary driver for the plasma cell fate choice in the GC[34] and as DNA vaccine-transformed cells would likely be expressing both ADA-1 and env immunogens in the local DLN GCs, this could directly impact plasma cell differentiation. Interestingly, our initial experiments with DNA antigens and molecular ADA indicated that ADA enhanced frequencies of GC TFH however, in our DNA and protein experiments, addition of pADA did not enhance TFH frequencies. We hypothesize that the addition of aluminum adjuvant enhances GC cell frequency so much so, that and ADA-1 adjuvant effect on cell numbers could not be observed, however its effect on GC function as measured by antibody function, is readily observed. It is however possible that the ADA effect is highly antigen specific and thus future experiments to address this are necessary and ongoing.
The development of HIV neutralizing antibodies exclusively in the sera of mice immunized with the triple combination of pCAP257 (gp160), pADA, and CAP257 gp140 glycoprotein, but not in those mice receiving pCAP257 (gp160) and matched glycoprotein gp140 without pADA, further supports the notion that pADA improves GC TFH function in vivo. These data suggest that pADA enhances the functional output of the GC, namely the development of neutralizing antibodies. Further experiments to characterize the molecular mechanism by which pADA mediates its effects in vivo on antigen-presenting, T, and B cells are necessary to confirm this important finding. These results not only strongly support the use of pADA as a vaccine adjuvant to enhance humoral responses and T cell function, but also have implications for therapeutic interventions for SCID and autoimmune patients. Finally, as PEGylated ADA-1 is approved for the treatment of SCID patients and has an established safety and tolerability profile in the clinic[35,36], the use of ADA-1 to adjuvant vaccine-induced responses represents a novel application of an extant therapy with the potential to be fast-tracked for clinical application.
Materials and Methods
DNA plasmid preparation.
DNA vaccine constructs expressing complete HIV-1 consensus EnvB and EnvC gp160 proteins including gp120/gp41 fusion sites, were generated by Inovio Pharmaceuticals (Plymouth Meeting, PA) and have been previously reported [37–39]. Murine adenosine deaminase-1 (Gene Bank accession number NM001272052) was cloned into the pVAX vector (Invitrogen) and amplified by Genescript (Piscataway, NJ). Plasmid constructs expressing subtype C Env gp160 from subject CAP257 were kindly provided by Penny Moore and collaborators working with the CAPRISA cohort, as described[25]. The CAP257 54 week env gene sequence (CAP257 54wk_D) was motif-optimized using the Robins-Krasnitz methodology and cloned as gp160 into pEMC*. Plasmid DNA was purified using Endo-free reagents (Qiagen, Valencia, CA). Expression of full length gp160 in vitro was verified by immunofluorescence surface staining of transfected cells using the human mAb b12 and by complementation of env-negative HIV genomes to produce HIV pseudovirus expressing the Env-CAP257–54wk_D.
Protein production.
The gp140 DNA was derived from the gp160 envelope 54wk_D sequence by site-directed mutagenesis (QuickChange Multi Site-Directed Mutagenesis Kit, Stratagene, La Jolla, CA) to insert the previously described mutations[40] in the primary and secondary protease cleavage sites respectively: REKR was mutated to RSKS and KAKRR was mutated to KAISS. A large-scale endotoxin-free plasmid preparation (Qiagen, Valencia, CA) was used for stable expression in 293F cells for protein production[41]
Plasmid immunizations.
6- to 12-week-old male and female C57BL/6 (Charles River) mice were immunized thrice, 2 weeks apart, in the left tibialis anterior muscle with either 50 μg of pVAX, 20 μg of HIV-1 antigenic plasmid (pEnvB), or 20 μg of EnvB with 20 μg of ADA-1 (pADA) (n= 4–6 animals per group). The injection was followed by in vivo electroporation using the constant current CELLECTRA device (Inovio Pharmaceuticals, Plymouth Meeting, PA). In DNA-protein co-immunization experiments animals received 10ug of gp140 DNA (CAO25754wk_D) injected intramuscularly followed by in vivo electroporation.
Protein immunizations.
6- to 12-week-old female C57BL/6 mice were immunized thrice separated by two weeks in the right quadriceps femoris with 30 μl containing 10 μg of purified gp140 (CAP257–54wk_D). Protein immunogen was formulated in aluminum phosphate (Adju-phos) adjuvant (Invivogen, San Diego, CA Animal work was performed according to protocols approved by our Institutional Animal Care and Use Committee.
Mouse sacrifice, sample collection, and tissue harvest.
At time points designated in the immunization schedule, the animals were sacrificed and blood, spleens, popliteal, and inguinal lymph nodes were collected. Lymph nodes and spleens were processed to single cell suspensions. The resulting single cell suspensions from DLNs and spleens were washed and resuspended in R10 medium. All cells were counted, and cell viability was determined using a Countess Automated Cell Counter (Invitrogen, Life Technologies). Blood was collected in minicollect serum gel tubes (Grenier-Bio) and centrifuged at 13,000 RPM for 30 min at 4°C to separate serum.
Flow cytometry.
Single cell suspension of lymphocytes harvested from the DLNs, and spleens were suspended in cold RPMI 1640 containing 10% FBS and stained with the live/dead Fixable Aqua Dead Cell Stain Kit (Invitrogen) (dilution: 1/100; Cat. Number: L34957) to exclude dead cells. All cytometric analyses were performed using an LSR II flow cytometer (BD), and data were analyzed using FlowJo software (Treestar). All B-cell analyses were performed after gating out dead, IgD+, and CD3+F4/80+Gr1+ cells in the dump channel. TFH cells were stained with the following fluorochrome-conjugated anti-mouse antibodies: CD3 (17A2) (dilution: 1/100; Cat. Number: 100216), CD4 (GK1.5) (dilution: 1/200; Cat. Number: 100414), CD25 (PC61) (dilution: 1/100; Cat. Number: 102010), PD-1 (RMP1–30) (dilution: 1/100; Cat. Number: 109110), IgD (11–26c.2a) (dilution: 1/100; Cat. Number: 405742), B220 (RA3–6B2) (dilution: 1/100; Cat. Number: 103211), CD95 (SA367H8) (dilution: 1/100; Cat. Number: 152612), CD44 (IM7) (dilution: 1/100; Cat. Number: 103047), GL7 (GL7) (dilution 1/200; Cat. Number: 144610) were all from BioLegend. CXCR5 (2G8) (dilution: 1/20; Cat. Number: 560615) and CD62L (MEL-14) (dilution1/100; Cat. Number: 564108), were from BD Biosciences.
ELISA assays.
ELISA was used to determine HIV-1 EnvB-specific IgG and IgA in mouse serum. Mouse blood samples were collected by cardiac puncture. Enzyme immunoassay/radioimmunoassay plates (Costar) were coated with HIV-1 EnvB (MNgp41), or EnvC (96Z) protein (NIH AIDS reagent repository) diluted in PBS at a concentration of 0.5 μg/ml in a final volume of 100 μl per well and incubated overnight at 4°C. Sera diluted in PBS/1% BSA containing protease inhibitor (Pierce) were added in triplicate incubated overnight at 4°C. Bound antibodies were detected with HRP-labeled goat anti-mouse IgG or IgA (KPL/Seracare, Gaithersburg, MD) and developed using TMB Ultra substrate (Thermo Fisher, Waltham, MA) according to the manufacturer’s instructions. The amount of total IgG (or subtypes) or IgA in sera was calculated by interpolating the optical densities on calibration curves created with known quantities of mouse immunoglobulin.
Detection of anti-nuclear antibody.
Mouse anti-nuclear antibody (ANA) was detected in the serum from vaccinated animals using a commercially available ELISA kit from Express Biotech (Frederick, MD), according to the manufacturer’s protocol. ANA in serum was also detected using a modified ANA/Hep-2 immunofluorescence assay (IFA) (Hemagen, Columbia, MD). Images were collected on a Leica DM500B microscope at 10x and 40x magnifications. Fluorescence intensity was graded blindly on a 0–4 scale according to the manufacturer’s protocol.
Neutralization assays.
The TZM-bl assay was performed as previously described[42] using purified mouse IgG (purified with Protein A). Recovery of IgG was determined by PAGE and Coomassie staining. All values were calculated with respect to virus only wells [(value for virus only minus cells only) minus (value for serum minus cells only)] divided by (value for virus minus cells only). The human monoclonal antibody VRC01, which is an IgG1, was used as a positive control. Viruses were grown as described as pseudoviruses and strains tested included HIV-SF162, HIV-MW965, and HIV-CAP257–54wk_D.
Human samples.
Blood samples from healthy donors were obtained from the Martin Memorial Health System (Stuart, Florida) after signed informed consent from all participants. All procedures were performed according to the Institutional Review Boards of the Martin Memorial Health System and Drexel University College of Medicine.
Generation of human monocyte-derived dendritic cells (mDCs).
Human PBMCs from healthy donors were obtained immediately after blood withdrawal using the Ficoll-Paque (GE Healthcare) gradient method and stored in liquid nitrogen until usage. The cells were thawed in RPMI 1640 (Corning) supplemented with 10% fetal bovine serum (Access Biologicals) and 1% penicillin/streptomycin (Gibco). CD14+ CD16+ monocytes were then enriched from total PBMCs by negative selection using the EasySep™ human monocyte enrichment kit without CD16 depletion (STEMCELL Technologies) according to the manufacturer’s protocol and counted. Cells were then resuspended in serum-free CellGenix® GMP dendritic cell medium (CellGenix) with 100ng/ml of recombinant human GM-CSF (BioLegend) and 20ng/ml of recombinant human IL-4 (Gemini Bio-Products) at a density of 2x×106 per ml in 24-well plates. After 48h of incubation, cells were stimulated with 0.5μg/ml of LPS (Invivogen) plus 40ng/ml of IFNγ (Gemini Bio-Products) or 2.5uM of ADA-1 (Sigma Aldrich) in 1ml medium and compared to the unstimulated control. Dendritic cells were harvested after 24h of stimulation and analyzed by flow cytometry.
Flow cytometric analysis of human mDCs.
Harvested mDCs were incubated with TruStain FcγR block (BioLegend) and fluorochrome-conjugated antibodies for 15–20 minutes on ice in the dark. The following BioLegend fluorochrome-conjugated anti-human antibodies were used: CD3 (clone HIT3α), CD19 (clone HIB19), CD14 (clone M5E2), CD11c (clone 3.9), HLA-DR (clone L243), CD86 (clone IT2.2), CD83 (clone HB15e), CD40 (clone 5C3) and ICOS L (clone 2D3). Dead cells were identified using both LIVE/DEAD fixable Aqua Dead Cell Stain Kit for flow cytometry (Vivid) (Life Technologies) and Annexin V (BD Biosciences). mDC samples were washed then resuspended in PBS plus 2% FBS then acquired on a BD LSR II and analyzed with FlowJo software (Treestar). The gating strategy excluded doublet cells and mDCs were gated on live (Vivid− Annexin V−) CD3− CD19− CD11c+ cells.
Analysis of cytokine level production by stimulated human mDCs using ELISA.
Cell culture supernatants were collected 24h post-stimulation and were analyzed for the levels of IL-6 and IL-12p70, using the Biolegend human IL-6 and human IL-12p70 ELISA kits respectively, according to the manufacturer’s protocol. Absorbance was read at 450nm using a Synergy HTX multi-mode (BioTek) spectrophotometer within 15 minutes of stopping the reaction. Standard curves were generated and sample concentrations were calculated in pg/ml.
Luminex mDC cytokine analysis.
The Bio-Plex Pro Human Chemokine Assay (40-Plex) (Bio-Rad) was used to determine the level of a 40 cytokine/chemokine panel produced by mDCs, 24h after stimulation. The following human chemokine/cytokine premixed panel was used according to the manufacturer’s protocol: 6Ckine (CCL21), MIG (CXCL9), GCP-2 (CXCL6), IL-6, I-309 (CCL1), IFNγ, SDF-1α+ß (CXCL12), I-TAC (CXCL11), MCP-3 (CCL7), IL-16, MCP-4 (CCL13), MDC (CCL22), Eotaxin-2 (CCL24), GM-CSF, MIF, TNF-α, MPIF-1 (CCL23), IL-2, IL-1ß, Eotaxin-1 (CCL11), TECK (CCL25), IL-4, MCP-1 (CCL2), IL-8, MIP-1α (CCL3), IL-10, MCP-2 (CCL8), Gro-α (CXCL1), MIP-3α (CCL20), SCYB16 (CXCL16), Eotaxin-3 (CCL26), MIP-1o (CCL15), TARC (CCL17), CTACK (CCL27), ENA-78 (CXCL5), BCA-1 (CXCL13), MIP-3ß (CCL19), Fractalkine (CXC3CL1), Gro-ß (CXCL2). Data was acquired on a Bio-Plex 200 System using beads regions defined in the protocol and analyzed with the Bio-Plex Manager 6.1 software (Bio-Rad). Standard curves were generated and sample concentrations were calculated in pg/ml.
Statistical analysis.
All statistical analysis was performed using GraphPad Prism software version 6. Flow cytometry data were analyzed using Flowjo versions 7 or 10 (Treestar). All data are representative of at least two independent experiments except where noted. Data are presented as the mean ± standard error of the mean or standard deviation as noted. Where appropriate, the statistical difference between immunization groups or treatment conditions was assessed using an ordinary one-way analysis of variance (ANOVA) (Tukey’s multiple comparison test). Correlations were assessed using a Pearson correlation. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Supplementary Material
Highlights.
We demonstrate using human mDCs, that ADA-1 can enhance antigen presentation and promote a TFH polarizing cytokine phenotype.
We have determined that ADA-1 selectively enhanced GC-TFH frequency in vivo.
We have shown that ADA-1 improves the quality of anti-HIV antibody responses, generating both binding and HIV neutralizing antibodies in a mouse model of vaccination.
Acknowledgements
We thank Penny Moore and Konstantinos Kurt Wibmer for the gift of the CAP257 env DNA sequences. Consensus envelope B DNA was purchased from Inovio pharmaceuticals (Plymouth Meeting, PA). We thank Laurent Humeau and Jean Boyer (Inovio Pharmaceuticals) for comments on the manuscript and thank Inovio pharmaceuticals for the consensus plasmids used in these studies as well as the use of the CELLECTRA device for electroporation.
Funding
This work was supported by funding to EKH from NIH 5RO1AI106482-01A and 1U19 AI128910-01
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
M. Kutzler receives grant funding, participates in industry collaborations, has received speaking honoraria, and honorarium for reviewing federal research grants at the NIH and DOD. Dr. Kutzler is currently funded through a sponsored research agreement with Pfizer, Inc. and has received royalties from patent licensure with Inovio Pharmaceuticals for molecular adjuvants and DNA vaccine antigens that are not being reported on in this manuscript. Thus, these financial relationships do not relate to the content of this manuscript and in no way pose a potential conflict of interest.
References
- 1.Buckley RH: Molecular defects in human severe combined immunodeficiency and approaches to immune reconstitution. Annu. Rev. Immunol 2004, 22:625–655. [DOI] [PubMed] [Google Scholar]
- 2.Tardif V, Muir R, Cubas R, Chakhtoura M, Wilkinson P, Metcalf T, Herro R, Haddad EK: Adenosine deaminase-1 delineates human follicular helper T cell function and is altered with HIV. Nature communications 2019, 10:823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caskey M, Schoofs T, Gruell H, Settler A, Karagounis T, Kreider EF, Murrell B, Pfeifer N, Nogueira L, Oliveira TY: Antibody 10–1074 suppresses viremia in HIV-1-infected individuals. Nature medicine 2017, 23:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheid JF, Horwitz JA, Bar-On Y, Kreider EF, Lu C-L, Lorenzi JC, Feldmann A, Braunschweig M, Nogueira L, Oliveira T: HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 2016, 535:556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L: HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 2015, 349:aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA: Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity 2017, 46:1073–1088. e1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havenar-Daughton C, Carnathan DG, de la Peña AT, Pauthner M, Briney B, Reiss SM, Wood JS, Kaushik K, van Gils MJ, Rosales SL: Direct probing of germinal center responses reveals immunological features and bottlenecks for neutralizing antibody responses to HIV Env trimer. Cell reports 2016, 17:2195–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chavele K-M, Merry E, Ehrenstein MR: Cutting edge: circulating plasmablasts induce the differentiation of human T follicular helper cells via IL-6 production. The Journal of Immunology 2015, 194:2482–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R, Yusuf I, Crotty S: IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PloS one 2011, 6:e17739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papillion AM, Bachus H, Fuller M, León B, Ballesteros-Tato A: IL-6 counteracts IL-2-dependent suppression of T follicular helper cell responses. Edited by: Am Assoc Immnol; 2018. [Google Scholar]
- 11.Choi YS, Eto D, Yang JA, Lao C, Crotty S: Cutting edge: STAT1 is required for IL-6–mediated Bcl6 induction for early follicular helper cell differentiation. The Journal of Immunology 2013, 190:3049–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu X-B, Cao D-L, Zhang X, Jiang B-C, Zhao L-X, Qian B, Gao Y-J: CXCL13/CXCR5 enhances sodium channel Nav1. 8 current density via p38 MAP kinase in primary sensory neurons following inflammatory pain. Scientific reports 2016, 6:34836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cucak H, Yrlid U, Reizis B, Kalinke U, Johansson-Lindbom B: Type I interferon signaling in dendritic cells stimulates the development of lymph-node-resident T follicular helper cells. Immunity 2009, 31:491–501. [DOI] [PubMed] [Google Scholar]
- 14.Ritvo P-G, Klatzmann D: Interleukin-1 in the response of follicular helper and follicular regulatory T cells. Frontiers in immunology 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez V, Costagliola D, Bonduelle O, N’go N, Schnuriger A, Théodorou I, Clauvel J-P, Sicard D, Agut H, Debré P: Combination of HIV-1-specific CD4 Th1 cell responses and IgG2 antibodies is the best predictor for persistence of long-term nonprogression. Journal of Infectious Diseases 2005, 191:2053–2063. [DOI] [PubMed] [Google Scholar]
- 16.Ngo-Giang-Huong N, Candotti D, Goubar A, Autran B, Maynart M, Sicard D, Clauvel J-P, Agut H, Costagliola D, Rouzioux C: HIV type 1-specific IgG2 antibodies: markers of helper T cell type 1 response and prognostic marker of long-term nonprogression. AIDS research and human retroviruses 2001, 17:1435–1446. [DOI] [PubMed] [Google Scholar]
- 17.Zhu C, Ma J, Liu Y, Tong J, Tian J, Chen J, Tang X, Xu H, Lu L, Wang S: Increased frequency of follicular helper T cells in patients with autoimmune thyroid disease. The Journal of Clinical Endocrinology & Metabolism 2012, 97:943–950. [DOI] [PubMed] [Google Scholar]
- 18.Linterman MA, Rigby RJ, Wong RK, Yu D, Brink R, Cannons JL, Schwartzberg PL, Cook MC, Walters GD, Vinuesa CG: Follicular helper T cells are required for systemic autoimmunity. Journal of Experimental Medicine 2009, 206:561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morita R, Schmitt N, Bentebibel S-E, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, Sabzghabaei N: Human blood CXCR5+ CD4+ T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 2011, 34:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu S: Heterologous prime–boost vaccination. Current opinion in immunology 2009, 21:346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Kennedy JS, West K, Montefiori DC, Coley S, Lawrence J, Shen S, Green S, Rothman AL, Ennis FA: Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime–protein boost HIV-1 vaccine in healthy human volunteers. Vaccine 2008, 26:3947–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letvin NL, Montefiori DC, Yasutomi Y, Perry HC, Davies M-E, Lekutis C, Alroy M, Freed DC, Lord CI, Handt LK: Potent, protective anti-HIV immune responses generated by bimodal HIV envelope DNA plus protein vaccination. Proceedings of the National Academy of Sciences 1997, 94:9378–9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Diepen MT, Chapman R, Douglass N, Galant S, Moore PL, Margolin E, Ximba P, Morris L, Rybicki EP, Williamson A-L: Prime-boost immunizations with DNA, modified vaccinia virus Ankara, and protein-based vaccines elicit robust HIV-1 tier 2 neutralizing antibodies against the CAP256 superinfecting virus. Journal of Virology 2019, 93:e02155–02118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pissani F, Malherbe DC, Schuman JT, Robins H, Park BS, Krebs SJ, Barnett SW, Haigwood NL: Improvement of antibody responses by HIV envelope DNA and protein co-immunization. Vaccine 2014, 32:507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wibmer CK, Bhiman JN, Gray ES, Tumba N, Karim SSA, Williamson C, Morris L, Moore PL: Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS pathogens 2013, 9:e1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casanova V, Naval-Macabuhay I, Massanella M, Rodríguez-García M, Blanco J, Gatell JM, García F, Gallart T, Lluis C, Mallol J: Adenosine deaminase enhances the immunogenicity of human dendritic cells from healthy and HIV-infected individuals. PloS one 2012, 7:e51287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ballesteros-Tato A, Randall TD: Priming of T follicular helper cells by dendritic cells. Immunology and cell biology 2014, 92:22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma CS, Suryani S, Avery DT, Chan A, Nanan R, Santner Nanan B, Deenick EK, Tangye SG: Early commitment of naïve human CD4+ T cells to the T follicular helper (TFH) cell lineage is induced by IL-12. Immunology and cell biology 2009, 87:590–600. [DOI] [PubMed] [Google Scholar]
- 29.León B, Ballesteros-Tato A, Browning JL, Dunn R, Randall TD, Lund FE: Regulation of T H 2 development by CXCR5+ dendritic cells and lymphotoxin-expressing B cells. Nature immunology 2012, 13:681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oestreich KJ, Mohn SE, Weinmann AS: Molecular mechanisms that control the expression and activity of Bcl-6 in T H 1 cells to regulate flexibility with a T FH-like gene profile. Nature immunology 2012, 13:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi YS, Yang JA, Crotty S: Dynamic regulation of Bcl6 in follicular helper CD4 T (Tfh) cells. Current opinion in immunology 2013, 25:366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandruvada SN, Yuvaraj S, Liu X, Sundaram K, Shanmugarajan S, Ries WL, Norris JS, London SD, Reddy SV: Role of CXC chemokine ligand 13 in oral squamous cell carcinoma associated osteolysis in athymic mice. International journal of cancer 2010, 126:2319–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan P, Shi M, Lai L, Tang Z, Xie N, Xu H, Wei Q, Zhang X, Yang L, Wu L: Regulative role of the CXCL13-CXCR5 axis in the tumor microenvironment. Precision Clinical Medicine 2018, 1:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kräutler NJ, Suan D, Butt D, Bourne K, Hermes JR, Chan TD, Sundling C, Kaplan W, Schofield P, Jackson J: Differentiation of germinal center B cells into plasma cells is initiated by high-affinity antigen and completed by Tfh cells. Journal of Experimental Medicine 2017, 214:1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Booth C, Gaspar HB: Pegademase bovine (PEG-ADA) for the treatment of infants and children with severe combined immunodeficiency (SCID). Biologics: targets & therapy 2009, 3:349. [PMC free article] [PubMed] [Google Scholar]
- 36.Lee CR, McKenzie CA, Webster KD, Whaley R: Pegademase bovine: replacement therapy for severe combined immunodeficiency disease. DICP 1991, 25:1092–1095. [DOI] [PubMed] [Google Scholar]
- 37.Yan J, Corbitt N, Pankhong P, Shin T, Khan A, Sardesai NY, Weiner DB: Immunogenicity of a novel engineered HIV-1 clade C synthetic consensus-based envelope DNA vaccine. Vaccine 2011, 29:7173–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muthumani K, Zhang D, Dayes NS, Hwang DS, Calarota SA, Choo AY, Boyer JD, Weiner DB: Novel engineered HIV-1 East African Clade-A gp160 plasmid construct induces strong humoral and cell-mediated immune responses in vivo. Virology 2003, 314:134–146. [DOI] [PubMed] [Google Scholar]
- 39.Desrosiers MD, Cembrola KM, Fakir MJ, Stephens LA, Jama FM, Shameli A, Mehal WZ, Santamaria P, Shi Y: Adenosine deamination sustains dendritic cell activation in inflammation. The Journal of Immunology 2007, 179:1884–1892. [DOI] [PubMed] [Google Scholar]
- 40.Malherbe DC, Doria-Rose NA, Misher L, Beckett T, Puryear WB, Schuman JT, Kraft Z, O’Malley J, Mori M, Srivastava I: Sequential immunization with a subtype B HIV-1 envelope quasispecies partially mimics the in vivo development of neutralizing antibodies. Journal of virology 2011, 85:5262–5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sellhorn G, Caldwell Z, Mineart C, Stamatatos L: Improving the expression of recombinant soluble HIV Envelope glycoproteins using pseudo-stable transient transfection. Vaccine 2009, 28:430–436. [DOI] [PubMed] [Google Scholar]
- 42.Montefiori DC: Measuring HIV neutralization in a luciferase reporter gene assay. In HIV protocols. Edited by: Springer; 2009:395–405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


