Keywords: adipose tissue, metabolism, metabolites, microbiome
Abstract
The intestinal commensal microbiome is an important component of host health, in part by contributing an abundance of metabolites that gain access to the systemic circulation. The microbiome thereby influences the physiology of numerous organ systems outside the gastrointestinal tract. The consequences of this signaling axis between the intestinal microbiome and host are profound, in particular for the modulation of organismal metabolism. Here, we review recent examples whereby the intestinal microbiome influences host metabolism by influencing the biology of adipose tissue. We place a special emphasis on metabolite-driven pathways by which adipose tissue responds to alterations in intestinal microbial colonization. Given its accessibility for therapeutic interventions, the gut microbiome is an attractive relay module for the remote control of systemic metabolism.
INTRODUCTION
Over the past decades, the prevalence of obesity has reached alarming levels. Despite increasing attention and substantial public health measures, more than half of the adult United States population is overweight, and the global trend is approaching similar proportions (49). Importantly, obesity is not only characterized by increased adiposity, but is also strongly associated with several metabolic conditions, including type 2 diabetes, fatty liver disease, and heart disease (29, 31). All together, this obesity pandemic has widespread implications on life span, quality of life, and health care spending. Consequently, there is an urgent need to understand the factors that regulate systemic metabolic homeostasis.
An emergent factor that plays a role in systemic metabolism is the intestinal microbiome (4, 5, 33). The intestinal metagenome consists of the combined genomes of the trillions of microbes that reside in the gastrointestinal tract, representing millions of microbial genes. Importantly, this rich diversity present in the microbiome substantially increases the range of biochemical and metabolic activities available to the colonized host. In turn, depending on the composition of the intestinal microbiome, distinct biochemical and metabolic activities will be occurring that can impact the development and function of the metabolic, immune, and nervous systems (15).
The study of the microbiome has been nucleated by technological advancements in next-generation sequencing to enable culture-independent analyses of microbiota composition (22, 24). Most notably, the decreasing cost of 16S ribosomal RNA (rRNA) sequencing has led to a remarkable quantity of observational reports finding changes in microbiome community composition in a number of diverse diseases compared with healthy controls (as reviewed in Refs. 15, 24). However, these reports have also elucidated two major limitations of observational studies of the microbiome. First, correlation does not mean causation; that is, correlative changes found in microbiome composition between disease states does not mean the microbiome plays a causative role in the pathophysiology of the disease. Second, microbiota taxonomy is highly diverse, yet, there exists considerable functional metabolic redundancy between different microbial species. This suggests that differences described in microbiome community composition by 16S rRNA sequencing may not accurately predict a difference in metabolic activity of the microbiome. Thus, the microbiome field will increasingly focus on moving our knowledge from correlation to causation and subsequently to the specific molecular mechanism whereby the microbiome influences host physiology and disease.
Here, we therefore aim to examine the mechanistic role of the microbiome in regulating systemic metabolism. A particular emphasis will be placed on the relationship between the microbiome and adipose tissue, given the central role of adipocytes in energy homeostasis and the development of obesity (16, 25, 45). We will provide structure to our discussion by organizing the review into three parts. First, we will examine the signals from the microbiome that have been found to modulate mammalian adipose tissue in the host. Second, we will consider the potential receivers of these microbiome-derived signals. Third, we will review the downstream response of this signaling on adipose tissue. This signal-receiver-response framework is a useful structure to analyze the microbiome-adipose tissue axis and its relationship to systemic metabolic homeostasis.
THE MICROBIOME IMPACT ON ADIPOSE TISSUE BIOLOGY
Before analyzing the microbiome-adipose tissue axis, we will evaluate the evidence suggesting the intestinal microbiome plays a role in regulating adipocyte metabolism.
Seminal studies more than a decade ago documented an altered microbiota in obesity (36). In the ob/ob mouse, which is characterized by excessive eating and adiposity due to recessive mutations in the leptin gene (60), it was found that feeding the exact same diet to genetically obese ob/ob mice and their lean siblings (heterozygous ob/+ or wild type) resulted in drastic differences in their microbiome regardless of kinship (36). Indeed, this altered gut microbiota has been reproduced independently in both obese mice and humans, showing that the microbiome has an increased capacity to harvest energy from the diet upon obesity (33, 44, 55). Several studies have furthermore addressed the question of causality for the microbiome in obesity. One of the earliest studies suggesting a causative role for the microbiome in regulating systemic metabolism involved the observation that germ-free mice, mice completely devoid of all microbial exposures from birth, have reduced body fat compared with mice with an unperturbed microbiome (4). Moreover, upon transplantation of the microbiota from conventionally raised mice into adult germ-free mice, there was approximately a 60% increase in body fat content and insulin resistance within 14 days (4, 5). Similar transplantation experiments have also been performed with the microbiota from obese mouse and human donors into germ-free mice, which resulted in enhanced adiposity compared with transplantation from a lean donor (44, 55). In one particular study of note, four female twin pairs discordant for obesity were recruited, and both the uncultured fecal microbiota and cultured collections of bacteria from these human donors were transferred into germ-free mice to show that the animals receiving the transplant from the obese twin developed increased adiposity compared with the transplant from the lean twin (44).
In addition to these studies involving crude microbiome transplantations into germ-free mice, there have also been multiple reports suggesting that targeted supplementations and alterations to the microbiome in adult animals can influence their metabolic parameters. In this context, much work has focused on the bacterium Akkermansia muciniphila. The presence of this bacterium inversely correlates with body weight in both humans and mice, and it has been found that administration of both live (23) and pasteurized (42) versions [but not heat killed (23)] can improve metabolic outcomes upon diet-induced obesity. Recently, a randomized, double-blind, placebo-controlled pilot study was performed in overweight and insulin-resistant human volunteers to test the benefits of A. muciniphila administration (21). This study demonstrated the safety of this intervention and preliminary evidence suggesting that it might restrain some aspects of metabolic syndrome in obese humans. Larger human cohort studies are required to validate these purported clinical benefits of A. muciniphila administration. Furthermore, another bacterium that has been associated with leanness is Lactobacillus plantarum (41), which has been suggested to ameliorate the effects of obesogenic diets through a mechanism that involves the bacteriocin plantaricin EF (30).
Thus, there is a growing consensus that the gut microbiome is an important environmental factor that regulates host metabolic homeostasis and may contribute to the development of obesity and metabolic disease. Moving forward, it will be crucial to gain a mechanistic understanding of these microbiome communications with host systemic metabolism.
On the basis of this rationale for the study of the microbiome in the context of systemic metabolism, we will now consider the microbiome-adipose tissue axis, using the signal-receiver-response framework.
THE SIGNALS
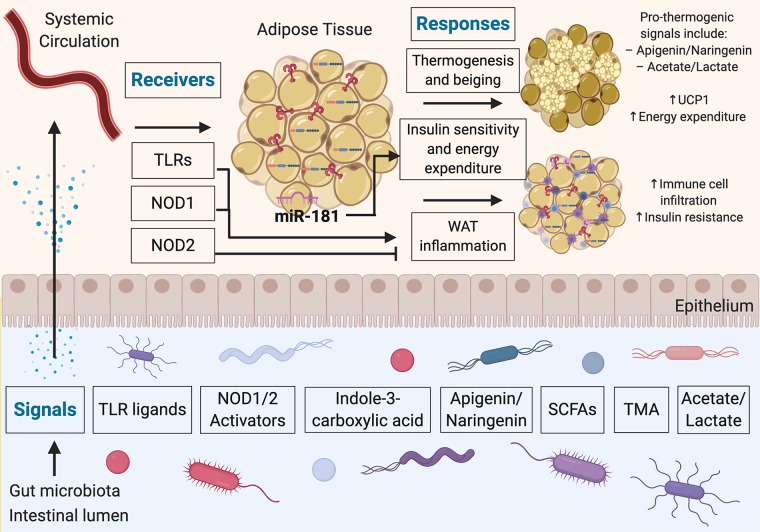
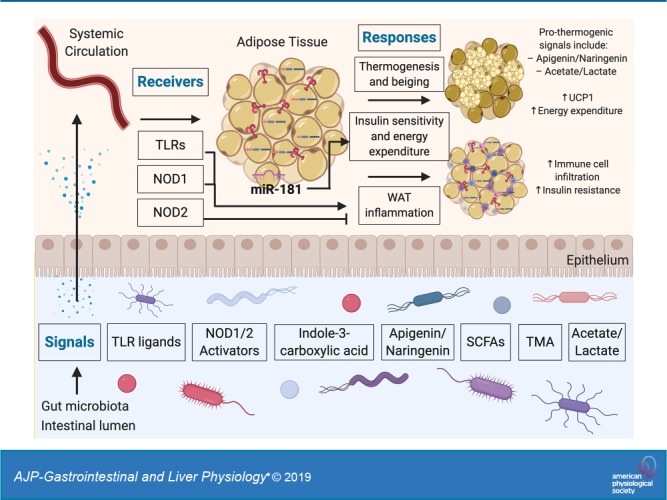
The biochemical and metabolic activities performed by the intestinal microbiome produces thousands of small, diffusible metabolites. Some of these metabolites have been found to regulate basic physiological processes both in the intestine and in distant tissues (40). Indeed, recent reports have found that a subset of these metabolites can regulate numerous aspects of adipose tissue biology, including the metabolic processes involved in the development of obesity (Fig. 1). Some recently reported examples of such metabolite signals include tryptophan-derived metabolites, flavonoids, and propionate. In addition, structural components of bacteria, such as Toll-like receptor (TLR) ligands, have been implicated in the microbiome-adipose tissue axis. Here, we consider each in turn.
Fig. 1.
Microbiome-adipose tissue axis in systemic metabolism through a signal-receiver-response framework. The gut microbiota produce molecules that act as signals to influence adipose tissue responses through various receptors. These secreted molecules can gain access to the systemic circulation and thereby reach adipocytes and other cells present in adipose tissue depots. The consequences of metabolite sensing by adipose tissue include modulation of thermogenic activity and inflammatory processes in adipose tissue. TLRs, Toll-like receptors; NOD1/2, nucleotide-binding oligomerization domain-containing protein-1/2; WAT, white adipose tissue; SCFAs, short-chain fatty acids; TMA, trimethylamine; miR, micro-RNA.
TLR Ligands
Metabolic endotoxemia is the systemic low-level elevation of gut-derived TLR ligands including lipopolysaccharide (LPS). Such metabolic endotoxemia has been shown to contribute to the onset and progression of insulin resistance and metabolic disease (10). Indeed, a recent example highlighting the importance of TLR signaling in adipose tissue involved the study of mice fed two different diets, lard and fish oil (9). The rationale behind this approach was that food rich in saturated dietary fat (e.g., lard) has been associated with enhanced white adipose tissue inflammation and metabolic dysfunction (32), whereas food rich in polyunsaturated fats (e.g., fish oil) results in reduced inflammation and promotes improved metabolic measures (8). The authors then asked whether these diets might in part be exerting their divergent effects on host metabolism through the microbiome. The study showed that mice fed a lard diet had increased TLR signaling and white adipose tissue inflammation compared with mice fed an isocaloric fish oil diet. In addition, mice genetically deficient in various components of the TLR signaling pathway (Trif−/− and Myd88−/−) were protected against this inflammation in white adipose tissue. The authors also showed that transplantation of the “fish oil microbiome” into mice pretreated with antibiotics were more protected from lard diet-induced adiposity and white adipose tissue inflammation than the mice transplanted with the “lard microbiome” (9). Taken together, this study suggests a link between the diet, the gut microbiome, systemically circulating gut-derived TLR ligands, and TLR signaling in white adipose tissue influencing systemic metabolic homeostasis.
Tryptophan-Derived Metabolites
Tryptophan is an essential amino acid in humans, meaning it must be obtained from the diet. However, tryptophan not only forms an integral component of many proteins as an amino acid, but the microbiota can also convert tryptophan to indole compounds that can accumulate to millimolar concentrations in the gut lumen (58). A recent report studied these tryptophan-derived metabolites in the context of a group of miRNAs that exhibit increased expression in obese white adipose tissue, called the miR-181 family. It was found that genetic ablation of the two most abundantly expressed miR-181 clusters protected mice from developing diet-induced obesity. Furthermore, it was found that germ-free mice had a greatly reduced expression of these miRNAs compared with conventionally reared mice, suggesting a role for signals from the microbiome. Likewise, antibiotic treatment of the conventional mice also decreased miR-181 expression in the white adipose tissue. A tryptophan-derived metabolite (indole-3-carboxylic acid) was identified as a metabolite reduced in high-fat diet-fed mice, which acts on adipocytes to inhibit miR-181 expression to regulate energy expenditure and insulin sensitivity (58).
Flavonoids
Polyphenols consumed by mammalian organisms through their diet are extensively modified by the metabolic activities of the intestinal microbiota (7, 56). Certain polyphenols, such as the flavonoids apigenin and naringenin, have been found to act on brown adipose tissue to increase the expression of uncoupling protein-1 (Ucp1), with the effect of enhancing energy expenditure (52, 53). Commensal bacteria express the enzymatic machinery to reduce the pool of bioavailable flavonoids, thereby limiting Ucp1 expression in brown adipose tissue and reducing energy expenditure. This mechanism has been implicated in postdieting weight regain, whereby flavonoid-degrading members of the commensal microbiome persist following a period of obesity. The reduced levels of flavonoids promote weight regain after successful weight loss (52).
Propionate
Propionate is a naturally occurring short-chain fatty acid (SCFA), useful as a potent inhibitor of molds that is extensively used as a food preservative, and is also endogenously produced by the intestinal microbiota (54). In a recent study, the metabolic effects of propionate consumption were investigated in both mice and humans. It was found that propionate can stimulate glycogenolysis and hyperglycemia in mice, but this effect was abrogated in mice deficient in fatty acid-binding protein-4 (FABP4) and liver glucagon receptor. Thus, propionate-induced glycogenolysis and hyperglycemia is dependent on FABP4 and glucagon. However, propionate did not directly promote glucagon or FABP4 secretion in ex vivo rodent pancreatic islets and adipose tissue, but instead seemed to act indirectly by activating the sympathetic nervous system, leading to the secretion of these hormones systemically to regulate glycogenolysis in adipose tissue and other metabolic organs. In humans, consumption of a meal containing propionate yielded increased plasma glucagon, FABP4, and norepinephrine postprandially (54). Thus, this study suggests that propionate and perhaps other gut-derived SCFAs play important roles in regulating adipose tissue and systemic metabolism. In addition, this study highlights the important notion that metabolite signals may regulate adipose tissue indirectly, for example through the nervous system, to influence systemic metabolic homeostasis. However, more work is required to further investigate the effect of 1) different routes of administration of SCFAs (e.g., oral propionate vs. colonic microbiome-derived propionate) as well as 2) different doses of SCFAs. Indeed, the unfavorable metabolic effects observed in this study directly contrast with previous reports attributing metabolic benefits to propionate, including suppressing food intake (3, 39) and reducing plasma fatty acid content (1). These differences likely stem from points 1 and 2 highlighted above, which are caveats that extend to many studies involving metabolite signals from the microbiome. Thus, more work is needed to understand the metabolic effects of propionate and other SCFAs, as well as metabolite signals from the microbiome more generally. Moreover, SCFAs have also been suggested as modulators of the transcription factor peroxisome proliferator-activated receptor-γ (PPARγ) (19, 34), which is a key regulator of adipocyte differentiation and metabolism and is also the target of the insulin-sensitizing drugs thiazolidinediones (TZDs) (35). Further research into the specific mechanisms between microbiome-derived SCFAs and the activity of PPARγ in adipose tissue will be an important avenue of future research.
Having reviewed some recent reports of metabolite signals from the microbiome influencing systemic metabolism, we will now move our discussion to the receivers of these signals.
THE RECEIVERS
Receivers in biological signal cascades serve three main purposes: 1) specificity in signal reception, 2) signal propagation and amplification, and 3) signal translation into appropriate responses. Although it is still poorly characterized how adipose tissue senses signals derived from the gastrointestinal tract, recent examples have highlighted distinct mechanisms whereby receivers in the host recognize circulating factors originating in the gut to impact adipose tissue biology (Fig. 1). In the previous section we already considered the TLRs in the context of their signals, the TLR ligands; here, we therefore turn our attention to another type of pattern recognition receptors, nucleotide-binding oligomerization domain-containing (NOD) proteins NOD1 and NOD2.
NOD1 and NOD2, are intracellular pattern recognition receptors that recognize specific muropeptide sequences present in peptidoglycan-based bacterial cell walls (12). NOD1 specifically detects diaminopimelate-containing N-acetylglucosamine-N-acetylmuramic acid (GlcNAc-MurNAc) tripeptide motifs present in peptidoglycan (26), whereas NOD2 detects muramyl dipeptide (MDP) motifs, also present in peptidoglycan (27).
Recently, NOD1 and NOD2 have been associated with metabolic disease. Specifically, acute activation of NOD1 results in insulin resistance (46), and deletion of NOD1 protects against diet-induced insulin resistance in mice (2, 12). In addition, NOD1 activators are elevated in serum in response to high-fat diet in mice, and NOD1 is specifically required in the hematopoietic compartment for the accompanying metabolic inflammation and insulin resistance upon diet-induced obesity (13). In contrast, mice lacking functional NOD2 have increased adipose tissue inflammation and insulin resistance when fed a high-fat diet, independent of altered adiposity (20).
The mechanisms underlying these divergent effects of NOD1 and NOD2 are not fully understood; furthermore, how NOD1 or NOD2 detection of bacterial muropeptides influences glucose metabolism is largely unknown. To address some of these questions, one recent study (12) found that injecting MDP, recognized by NOD2, into obese mice lowered adipose inflammation and reduced glucose intolerance without affecting weight or intestinal microbiome composition. In contrast, and in line with previous observations, NOD1-activating muropeptides decreased glucose tolerance. Furthermore, the insulin-sensitizing effect of MDP was dependent on interferon regulatory factor 4 (IRF4), whereas the NOD1-induced glucose intolerance was unaffected by the genetic ablation of IRF4. Thus, IRF4 is suggested to play a key role in distinguishing these opposing glycemic responses to different types of peptidoglycan. It was also found that mifamurtide, a synthetic NOD2-activating adjuvant with orphan drug status, was an insulin sensitizer at clinically relevant doses in obese mice (12).
Collectively, these studies suggest that both NOD1 and NOD2 play important roles as receivers of microbiome-derived signals to influence adipose tissue biology and therefore systemic metabolism. However, many questions remain. What are the specific genera or species in the microbiota that lead to NOD1 versus NOD2 activation? It is known that the NOD1-binding motif is present in all Gram-negative bacteria and some Gram-positive bacteria (26), and the NOD2 motif is present in both Gram-negative and Gram-positive bacteria (27), but a more granular understanding of the microbes present in the gut that activate NOD1 and NOD2 has not yet been achieved. In addition, NOD1 and NOD2 are intracellular receptors, and understanding how gut-derived microbial cell products reach the intracellular milieu of distant cells remains unclear. Indeed, it has been suggested that some of the NOD1 or NOD2 activators may be endogenously produced and not necessarily derived only from the microbiome (13). Last, more work is required to identify the receivers of other microbiome-derived signals and how each of these receivers, both singly and integrated with the activity of other receivers, evokes a unique response.
THE RESPONSE
Several common themes have emerged over the past few years regarding the downstream response of the microbiome impact on adipose tissue. In the previous sections we have already broadly considered the effects of metabolic endotoxemia and white adipose tissue inflammation. Here, we will therefore highlight another specialized metabolic process in adipose tissue that appears to adapt to the state of intestinal microbial colonization: thermogenesis.
Thermogenesis refers to the generation of heat in organisms, and it has been suggested that this process can be leveraged to neutralize the hypercaloric state of obesity (17). Thermogenesis can be separated into shivering and nonshivering types. Nonshivering thermogenesis is facilitated by brown adipose tissue (BAT) and beige adipocytes to maintain host body temperature during cold exposure by a UCP1-dependent mechanism (11). The finding that adult humans have active depots of BAT, and that the activity of this tissue appears reduced in people with obesity (18, 57), has garnered significant interest to modulate nonshivering thermogenesis for potential therapeutic benefit in metabolic disease. Indeed, supporting this interest is the finding that BAT activity is a major determinant of plasma glucose and triglyceride levels (6, 48, 59).
Given the suggested relationship between the intestinal microbiome and systemic metabolism, several recent reports have studied the relationship between the microbiota and nonshivering thermogenesis in mice. The results to date are controversial. First, the composition of the gut microbiome has been found to be altered upon cold exposure, and the transfer of this “cold microbiota” increases white adipose tissue browning, energy expenditure, and cold tolerance (14). Second, the depletion of the microbiota both by antibiotics and in germ-free mice promoted upregulation of Ucp1 and a browning phenotype in white adipose tissue at room temperature (22°C) and at thermoneutrality (30°C) (50), thus suggesting that depleting the microbiota enhances thermogenic activity. In contrast, a more recent study reported that Ucp1 expression and whole body energy expenditure was impaired upon microbiota depletion (37). Indeed, most data in this study (37) were in conflict with the previous report (50), likely indicating that the microbiome influences thermogenesis in multiple different ways, which, depending on local microbiome composition in different animal facilities, determine the outcome of antibiotic treatment.
Despite these seemingly inconsistent results with respect to microbiome depletion and thermogenic capacity, further studies have provided evidence to suggest specific mechanisms underlying the relationship between the microbiome and nonshivering thermogenesis in BAT and beige adipocytes. Here, we highlight three reported mechanisms relating to 1) bile acid synthesis, 2) the trimethylamine/flavin-containing monooxygenase-3/trimethylamine N-oxide (TMA/FMO3/TMAO) pathway, and 3) intermittent fasting. First, enhanced hepatic conversion of dietary cholesterol to bile acids during cold exposure has been found to play an important role in the response to cold temperatures in mice (59). In turn, this elevation of bile acids in the plasma and feces results in metabolically beneficial changes in the gut microbiome and adaptive thermogenesis (59). It remains to be seen whether these effects relating to bile acid synthesis will translate to humans. Second, the gut microbiota-initiated TMA/FMO3/TMAO pathway has been implicated as a potential modulator of metabolism. This pathway is an endocrine axis between the microbiome and host, whereby gut microbial metabolism of nutrients such as phosphatidylcholine, choline, and l-carnitine (abundant in Western diets) results in the production of TMA, which is metabolized into TMAO by the enzyme FMO3. TMAO levels are elevated in type 2 diabetic patients, and FMO3 levels in adipose tissue are correlated with obesity (47). Both knockdown and knockout of FMO3 protected mice from diet-induced obesity and stimulated the beiging and enhanced thermogenesis of white adipose tissue, thus suggesting a negative regulatory role for FMO3 in the beiging process (47). Third, intermittent fasting, or more precisely, an every-other-day fasting (EODF) regimen, has also been reported to stimulate beiging in white adipose tissue in mice through a gut microbiome-dependent mechanism (38). It was reported that EODF on a high-fat diet reduced obesity, insulin resistance, and hepatic steatosis and also resulted in an altered gut microbiota composition. The transplantation of the EODF-microbiota into microbiota-depleted mice stimulated white adipose tissue beiging, while EODF-induced beiging did not occur in mice treated with antibiotics. In addition, a systemic elevation of acetate and lactate was found, as well as the elevated expression in beige adipocytes of the monocarboxylate transporter 1, a lactate and acetate transporter. Previously, acetate and lactate have been implicated as beiging-inducers, suggesting a potential mechanism of EODF-induced beiging (38).
Taken together, the link between the microbiome and the thermogenic response in adipose tissue is a maturing field. Although there are some reports that have yielded conflicting results, as a whole, it appears that the microbiome can influence thermogenesis and other adipose tissue responses in multiple different ways. Moving forward, it will be crucial to gain a finer mechanistic understanding of these different responses in the adipose tissue and how these responses integrate with different gut-derived signals and different host receivers. Understanding these microbiome communications with host systemic metabolism has the potential to allow the therapeutic modulation or engineering of these signals, receivers, and responses, with the aim to improve outcomes in metabolic disease.
CONCLUSION
Although the study of gut microbiome-adipose tissue interactions is still a young field of research, several concepts have emerged from the recent literature. First, the microbiome can influence adipose tissue biology through secreted molecules that gain access to the systemic circulation and thereby reach adipocytes and other cells present in different adipose tissue depots. These secreted molecules can broadly be classified into metabolites (products of microbial metabolism) and microbial components (constituents of the microbial cell). Examples exist for how both types of molecules influence aspects of adipose tissue function, as discussed above.
The chemical nature of these molecules is of great diversity, ranging from SCFAs, cell wall components, flavonoids, and indoles as signals, to pattern recognition receptors, microRNAs, and transcriptional changes as receivers and responses. Future studies will show whether unifying patterns will emerge that provide a specific teleology behind the different signaling axes engaged by the microbiome to modulate adipose tissue biology.
Finally, given that most of the examples discussed in this review stem from animal models, it is important to consider the evidence for an impact of the intestinal microbiome on adipose tissue biology in humans. In a recent human trial, the impact of short-term oral antibiotic treatment in overweight individuals on insulin resistance was negligible (43). In contrast, long-term antibiotic exposure, done on human individuals several decades ago and commonly practiced in livestock until today, consistently leads to an overall increase in body fat mass (28, 51). Timing, duration, and specificity of the microbial impact on adipose tissue are therefore critical determinants of the metabolic outcome. Whether the same microbiome-derived molecules identified in mice are also active in humans remains to be determined. This young field of study is poised to experience several transformative discoveries in the near future.
GRANTS
C. A. Thaiss is supported by the National Institutes of Health Director’s New Innovator Award (DP2 AG-067492), the Edward Mallinckrodt, Jr. Foundation, the Global Probiotics Council, the Mouse Microbiome Metabolic Research Program of the National Mouse Metabolic Phenotyping Centers, and grants by the PennCHOP Microbiome Program, the Penn Institute for Immunology, the Penn Center for Molecular Studies in Digestive and Liver Diseases (P30-DK-050306), the Penn Skin Biology and Diseases Resource-based Center (P30 AR-069589), the Penn Diabetes Research Center (P30 DK-019525), and the Penn Institute on Aging. The figure was created using BioRender.com.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
C.A.T. conceived and designed research; P.L. analyzed data; P.L. interpreted results of experiments; P.L. prepared figures; P.L. and C.A.T. drafted manuscript; C.A.T. edited and revised manuscript; C.A.T. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank the members of the Thaiss laboratory for valuable input and apologize to those colleagues whose relevant work could not be cited owing to space constraints.
REFERENCES
- 1.Al-Lahham S, Roelofsen H, Rezaee F, Weening D, Hoek A, Vonk R, Venema K. Propionic acid affects immune status and metabolism in adipose tissue from overweight subjects. Eur J Clin Invest 42: 357–364, 2012. doi: 10.1111/j.1365-2362.2011.02590.x. [DOI] [PubMed] [Google Scholar]
- 2.Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermúdez-Humarán LG, Smirnova N, Bergé M, Sulpice T, Lahtinen S, Ouwehand A, Langella P, Rautonen N, Sansonetti PJ, Burcelin R. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med 3: 559–572, 2011. doi: 10.1002/emmm.201100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora T, Sharma R, Frost G. Propionate. Anti-obesity and satiety enhancing factor? Appetite 56: 511–515, 2011. doi: 10.1016/j.appet.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101: 15718–15723, 2004. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA 104: 979–984, 2007. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Weller H, Waurisch C, Eychmüller A, Gordts PLSM, Rinninger F, Bruegelmann K, Freund B, Nielsen P, Merkel M, Heeren J. Brown adipose tissue activity controls triglyceride clearance. Nat Med 17: 200–205, 2011. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- 7.Braune A, Blaut M. Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes 7: 216–234, 2016. doi: 10.1080/19490976.2016.1158395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckley JD, Howe PRC. Anti-obesity effects of long-chain omega-3 polyunsaturated fatty acids. Obes Rev 10: 648–659, 2009. doi: 10.1111/j.1467-789X.2009.00584.x. [DOI] [PubMed] [Google Scholar]
- 9.Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Bäckhed F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab 22: 658–668, 2015. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56: 1761–1772, 2007. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 11.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev 84: 277–359, 2004. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 12.Cavallari JF, Fullerton MD, Duggan BM, Foley KP, Denou E, Smith BK, Desjardins EM, Henriksbo BD, Kim KJ, Tuinema BR, Stearns JC, Prescott D, Rosenstiel P, Coombes BK, Steinberg GR, Schertzer JD. Muramyl dipeptide-based postbiotics mitigate obesity-induced insulin resistance via IRF4. Cell Metab 25: 1063–1074.e3, 2017. doi: 10.1016/j.cmet.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 13.Chan KL, Tam TH, Boroumand P, Prescott D, Costford SR, Escalante NK, Fine N, Tu Y, Robertson SJ, Prabaharan D, Liu Z, Bilan PJ, Salter MW, Glogauer M, Girardin SE, Philpott DJ, Klip A. Circulating NOD1 activators and hematopoietic NOD1 contribute to metabolic inflammation and insulin resistance. Cell Reports 18: 2415–2426, 2017. doi: 10.1016/j.celrep.2017.02.027. [DOI] [PubMed] [Google Scholar]
- 14.Chevalier C, Stojanović O, Colin DJ, Suarez-Zamorano N, Tarallo V, Veyrat-Durebex C, Rigo D, Fabbiano S, Stevanović A, Hagemann S, Montet X, Seimbille Y, Zamboni N, Hapfelmeier S, Trajkovski M. Gut microbiota orchestrates energy homeostasis during cold. Cell 163: 1360–1374, 2015. doi: 10.1016/j.cell.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet 13: 260–270, 2012. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chouchani ET, Kajimura S. Metabolic adaptation and maladaptation in adipose tissue. Nat Metab 1: 189–200, 2019. doi: 10.1038/s42255-018-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chouchani ET, Kazak L, Spiegelman BM. New advances in adaptive thermogenesis: UCP1 and beyond. Cell Metab 29: 27–37, 2019. doi: 10.1016/j.cmet.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng Y-H, Doria A, Kolodny GM, Kahn CR. Identification and importance of brown adipose tissue in adult humans. N Engl J Med 360: 1509–1517, 2009. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.den Besten G, Bleeker A, Gerding A, van Eunen K, Havinga R, van Dijk TH, Oosterveer MH, Jonker JW, Groen AK, Reijngoud D-J, Bakker BM. Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes 64: 2398–2408, 2015. doi: 10.2337/db14-1213. [DOI] [PubMed] [Google Scholar]
- 20.Denou E, Lolmède K, Garidou L, Pomie C, Chabo C, Lau TC, Fullerton MD, Nigro G, Zakaroff-Girard A, Luche E, Garret C, Serino M, Amar J, Courtney M, Cavallari JF, Henriksbo BD, Barra NG, Foley KP, McPhee JB, Duggan BM, O’Neill HM, Lee AJ, Sansonetti P, Ashkar AA, Khan WI, Surette MG, Bouloumié A, Steinberg GR, Burcelin R, Schertzer JD. Defective NOD2 peptidoglycan sensing promotes diet-induced inflammation, dysbiosis, and insulin resistance. EMBO Mol Med 7: 259–274, 2015. doi: 10.15252/emmm.201404169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, Falony G, Raes J, Maiter D, Delzenne NM, de Barsy M, Loumaye A, Hermans MP, Thissen J-P, de Vos WM, Cani PD. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med 25: 1096–1103, 2019. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science 308: 1635–1638, 2005. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA 110: 9066–9071, 2013. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franzosa EA, Hsu T, Sirota-Madi A, Shafquat A, Abu-Ali G, Morgan XC, Huttenhower C. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat Rev Microbiol 13: 360–372, 2015. doi: 10.1038/nrmicro3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gesta S, Tseng Y-H, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell 131: 242–256, 2007. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Girardin SE, Boneca IG, Carneiro LAM, Antignac A, Jéhanno M, Viala J, Tedin K, Taha MK, Labigne A, Zähringer U, Coyle AJ, DiStefano PS, Bertin J, Sansonetti PJ, Philpott DJ. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300: 1584–1587, 2003. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 27.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 278: 8869–8872, 2003. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 28.Haight TH, Pierce WE. Effect of prolonged antibiotic administration of the weight of healthy young males. J Nutr 56: 151–161, 1955. doi: 10.1093/jn/56.1.151. [DOI] [PubMed] [Google Scholar]
- 29.Hajer GR, van Haeften TW, Visseren FLJ. Adipose tissue dysfunction in obesity, diabetes, and vascular diseases. Eur Heart J 29: 2959–2971, 2008. doi: 10.1093/eurheartj/ehn387. [DOI] [PubMed] [Google Scholar]
- 30.Heeney DD, Zhai Z, Bendiks Z, Barouei J, Martinic A, Slupsky C, Marco ML. Lactobacillus plantarum bacteriocin is associated with intestinal and systemic improvements in diet-induced obese mice and maintains epithelial barrier integrity in vitro. Gut Microbes 10: 382–397, 2019. doi: 10.1080/19490976.2018.1534513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hruby A, Hu FB. The epidemiology of obesity: a big picture. Pharmacoeconomics 33: 673–689, 2015. doi: 10.1007/s40273-014-0243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy A, Martinez K, Chuang C-C, LaPoint K, McIntosh M. Saturated fatty acid-mediated inflammation and insulin resistance in adipose tissue: mechanisms of action and implications. J Nutr 139: 1–4, 2009. doi: 10.3945/jn.108.098269. [DOI] [PubMed] [Google Scholar]
- 33.Khan MT, Nieuwdorp M, Bäckhed F. Microbial modulation of insulin sensitivity. Cell Metab 20: 753–760, 2014. doi: 10.1016/j.cmet.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, Keller MP, Attie AD, Rey FE, Denu JM. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell 64: 982–992, 2016. doi: 10.1016/j.molcel.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazar MA. PPAR γ, 10 years later. Biochimie 87: 9–13, 2005. doi: 10.1016/j.biochi.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 36.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102: 11070–11075, 2005. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B, Li L, Li M, Lam SM, Wang G, Wu Y, Zhang H, Niu C, Zhang X, Liu X, Hambly C, Jin W, Shui G, Speakman JR. Microbiota depletion impairs thermogenesis of brown adipose tissue and browning of white adipose tissue. Cell Reports 26: 2720–2737.e5, 2019. doi: 10.1016/j.celrep.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Li G, Xie C, Lu S, Nichols RG, Tian Y, Li L, Patel D, Ma Y, Brocker CN, Yan T, Krausz KW, Xiang R, Gavrilova O, Patterson AD, Gonzalez FJ. Intermittent fasting promotes white adipose browning and decreases obesity by shaping the gut microbiota. Cell Metab 26: 672–685.e4, 2017. [Erratum in: Cell Metab 26: 801, 2017.] doi: 10.1016/j.cmet.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin HV, Frassetto A, Kowalik EJ Jr, Nawrocki AR, Lu MM, Kosinski JR, Hubert JA, Szeto D, Yao X, Forrest G, Marsh DJ. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms [Online]. PLoS One 7: e35240, 2012. doi: 10.1371/journal.pone.0035240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin F-PJ, Dumas M-E, Wang Y, Legido-Quigley C, Yap IKS, Tang H, Zirah S, Murphy GM, Cloarec O, Lindon JC, Sprenger N, Fay LB, Kochhar S, van Bladeren P, Holmes E, Nicholson JK. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol 3: 112, 2007. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Million M, Angelakis E, Paul M, Armougom F, Leibovici L, Raoult D. Comparative meta-analysis of the effect of Lactobacillus species on weight gain in humans and animals. Microb Pathog 53: 100–108, 2012. doi: 10.1016/j.micpath.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, Myridakis A, Delzenne NM, Klievink J, Bhattacharjee A, van der Ark KCH, Aalvink S, Martinez LO, Dumas M-E, Maiter D, Loumaye A, Hermans MP, Thissen JP, Belzer C, de Vos WM, Cani PD. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 23: 107–113, 2017. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 43.Reijnders D, Goossens GH, Hermes GD, Neis EPJG, van der Beek CM, Most J, Holst JJ, Lenaerts K, Kootte RS, Nieuwdorp M, Groen AK, Olde Damink SW, Boekschoten MV, Smidt H, Zoetendal EG, Dejong CHC, Blaak EE. Effects of gut microbiota manipulation by antibiotics on host metabolism in obese humans: a randomized double-blind placebo-controlled trial. Cell Metab 24: 63–74, 2016. doi: 10.1016/j.cmet.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 44.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341: 1241214, 2013. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell 156: 20–44, 2014. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schertzer JD, Tamrakar AK, Magalhães JG, Pereira S, Bilan PJ, Fullerton MD, Liu Z, Steinberg GR, Giacca A, Philpott DJ, Klip A. NOD1 activators link innate immunity to insulin resistance. Diabetes 60: 2206–2215, 2011. doi: 10.2337/db11-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schugar RC, Shih DM, Warrier M, Helsley RN, Burrows A, Ferguson D, Brown AL, Gromovsky AD, Heine M, Chatterjee A, Li L, Li XS, Wang Z, Willard B, Meng Y, Kim H, Che N, Pan C, Lee RG, Crooke RM, Graham MJ, Morton RE, Langefeld CD, Das SK, Rudel LL, Zein N, McCullough AJ, Dasarathy S, Tang WHW, Erokwu BO, Flask CA, Laakso M, Civelek M, Naga Prasad SV, Heeren J, Lusis AJ, Hazen SL, Brown JM. The TMAO-producing enzyme flavin-containing monooxygenase 3 regulates obesity and the beiging of white adipose tissue. Cell Reports 19: 2451–2461, 2017. [Erratum in: Cell Rep 20: 279, 2017.] doi: 10.1016/j.celrep.2017.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stanford KI, Middelbeek RJW, Townsend KL, An D, Nygaard EB, Hitchcox KM, Markan KR, Nakano K, Hirshman MF, Tseng Y-H, Goodyear LJ. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J Clin Invest 123: 215–223, 2013. doi: 10.1172/JCI62308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevens J, Oakkar EE, Cui Z, Cai J, Truesdale KP. US adults recommended for weight reduction by 1998 and 2013 obesity guidelines, NHANES 2007–2012. Obesity (Silver Spring) 23: 527–531, 2015. doi: 10.1002/oby.20985. [DOI] [PubMed] [Google Scholar]
- 50.Suárez-Zamorano N, Fabbiano S, Chevalier C, Stojanović O, Colin DJ, Stevanović A, Veyrat-Durebex C, Tarallo V, Rigo D, Germain S, Ilievska M, Montet X, Seimbille Y, Hapfelmeier S, Trajkovski M. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat Med 21: 1497–1501, 2015. doi: 10.1038/nm.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thaiss CA, Elinav E. The remedy within: will the microbiome fulfill its therapeutic promise? J Mol Med (Berl) 95: 1021–1027, 2017. doi: 10.1007/s00109-017-1563-z. [DOI] [PubMed] [Google Scholar]
- 52.Thaiss CA, Itav S, Rothschild D, Meijer MT, Levy M, Moresi C, Dohnalová L, Braverman S, Rozin S, Malitsky S, Dori-Bachash M, Kuperman Y, Biton I, Gertler A, Harmelin A, Shapiro H, Halpern Z, Aharoni A, Segal E, Elinav E. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature 540: 544–551, 2016. doi: 10.1038/nature20796. [DOI] [PubMed] [Google Scholar]
- 53.Thaiss CA, Shapiro H, Elinav E. Post-dieting weight gain: the role of persistent microbiome changes. Future Microbiol 12: 555–559, 2017. doi: 10.2217/fmb-2017-0045. [DOI] [PubMed] [Google Scholar]
- 54.Tirosh A, Calay ES, Tuncman G, Claiborn KC, Inouye KE, Eguchi K, Alcala M, Rathaus M, Hollander KS, Ron I, Livne R, Heianza Y, Qi L, Shai I, Garg R, Hotamisligil GS. The short-chain fatty acid propionate increases glucagon and FABP4 production, impairing insulin action in mice and humans. Sci Transl Med 11: eaav0120, 2019. doi: 10.1126/scitranslmed.aav0120. [DOI] [PubMed] [Google Scholar]
- 55.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031, 2006. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 56.van Duynhoven J, Vaughan EE, Jacobs DM, Kemperman RA, van Velzen EJ, Gross G, Roger LC, Possemiers S, Smilde AK, Doré J, Westerhuis JA, Van de Wiele T. Metabolic fate of polyphenols in the human superorganism. Proc Natl Acad Sci USA 108, Suppl 1: 4531–4538, 2011. doi: 10.1073/pnas.1000098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JMAFL, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJJ. Cold-activated brown adipose tissue in healthy men. N Engl J Med 360: 1500–1508, 2009. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 58.Virtue AT, McCright SJ, Wright JM, Jimenez MT, Mowel WK, Kotzin JJ, Joannas L, Basavappa MG, Spencer SP, Clark ML, Eisennagel SH, Williams A, Levy M, Manne S, Henrickson SE, Wherry EJ, Thaiss CA, Elinav E, Henao-Mejia J. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci Transl Med 11: eaav1892, 2019. doi: 10.1126/scitranslmed.aav1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Worthmann A, John C, Rühlemann MC, Baguhl M, Heinsen FA, Schaltenberg N, Heine M, Schlein C, Evangelakos I, Mineo C, Fischer M, Dandri M, Kremoser C, Scheja L, Franke A, Shaul PW, Heeren J. Cold-induced conversion of cholesterol to bile acids in mice shapes the gut microbiome and promotes adaptive thermogenesis. Nat Med 23: 839–849, 2017. doi: 10.1038/nm.4357. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature 372: 425–432, 1994. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]