Abstract
Influenza A viruses (IAVs) constitute a major threat to human health. The IAV genome consists of eight single-stranded viral RNA (vRNA) segments contained in separate viral ribonucleoprotein complexes (vRNPs) that are packaged together into a single virus particle. The structure of the vRNA is believed to play a role in assembling the different vRNPs into budding virions1–8 and in directing reassortment between IAVs9. Reassortment between established human IAVs and IAVs harboured in the animal reservoir can lead to the emergence of pandemic influenza strains to which there is little pre existing immunity in the human population10,11. While previous studies have revealed the overall organisation of the proteins within vRNPs, characterisation of vRNA structure using conventional structural methods is hampered by limited resolution and an inability to resolve dynamic components12,13. Here, we employ multiple high-throughput sequencing approaches to generate the first global high-resolution structure of the IAV genome. We show that different IAV genome segments acquire distinct RNA conformations and form both intra- and inter-segment RNA interactions inside influenza virions. We use our detailed map of IAV genome structure to provide the first direct evidence for how inter-segment RNA interactions drive vRNP co-segregation during reassortment between different IAV strains. The work presented here is a roadmap both for the development of improved vaccine strains and for the creation of a framework to ‘risk assess’ reassortment potential to better predict the emergence of new pandemic influenza strains.
In influenza virions, as well as in infected cells, RNA genome segments are assembled into vRNP complexes in which the termini of the vRNA associate with the viral RNA-dependent RNA polymerase while the rest of the vRNA is bound by an oligomeric nucleoprotein (NP)14,15. To examine IAV RNA structure at single-nucleotide resolution, we used SHAPE-MaP16–18, which probes the conformational flexibility (i.e. base pairing) of each nucleotide. We carried out our analysis both in virio and ex virio (Fig. 1a and Supplementary Fig. 1). For ex virio experiments, the eight vRNA segments of influenza A/WSN/1933 (H1N1) (WSN) were individually transcribed from PCR products using T7 RNA polymerase (ivtRNA) or “naked” vRNA was purified from deproteinated virus particles (nkvRNA). For in virio experiments, vRNA was probed in the context of vRNPs, directly inside purified WSN virions.
Figure 1 |. Analysis of the influenza A virus (IAV) genome structure using SHAPE-MaP.
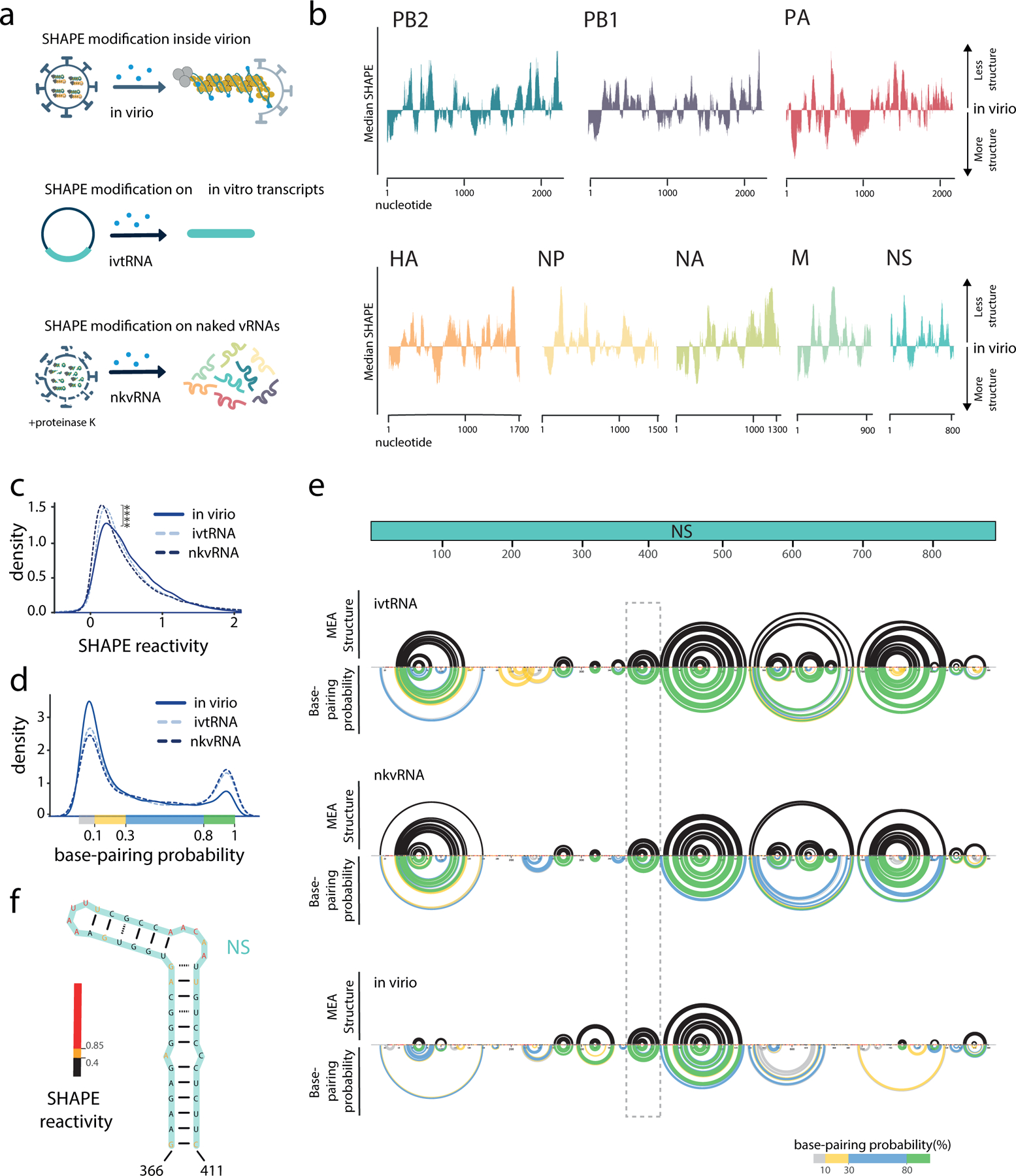
a, Schematic showing different samples used for SHAPE-MaP analysis. b, Median SHAPE reactivities of different WSN vRNA segments in virio. Medians were calculated over 50 nucleotide windows and plotted relative to the global median of a given segment. c, SHAPE reactivity distributions in different samples; **** P ≤ 2.2×10−16, two-sided Wilcox Rank-Sum Test, n = 13581 nucleotides per sample, in virio data is an average of 3 biologically independent samples, ivtRNA data is an average of 2 biologically independent samples, and nkvRNA data is from a single biological sample. d, Base-pairing probability distributions in different vRNA samples, calculated using SHAPE-informed partition function. e, Secondary RNA structure of the NS segment. Upper black arcs indicate the maximum expected accuracy RNA structure; only interactions associated with greater than 80% base pairing probabilities are shown. Lower coloured arcs indicate base-pairing probabilities. Dashed rectangle highlights the position of the hairpin shown in f. All sequence positions are annotated as 5′- 3′ in vRNA sense. nkvRNA, naked viral RNA; ivtRNA, in vitro transcribed RNA; MEA, maximum expected accuracy.
Examination of in virio SHAPE-MaP profiles revealed that vRNA in the context of vRNPs is capable of accommodating secondary RNA structures with considerable base pairing, as evidenced by extensive regions of low SHAPE reactivities in virio (SHAPE values below the median, Fig. 1b and Supplementary Table 1). The eight different vRNPs in virio have unique vRNA conformations, and these structures are consistent and reproducible across three replicates (Supplementary Fig. 2). Comparison with ex virio SHAPE profiles and SHAPE-guided predictions of secondary RNA structures show that the presence of NP leads to less structurally constrained vRNA, as evidenced by a global increase in SHAPE reactivity (Fig. 1c) and decrease in the number of high-probability base-paired structures in virio (Fig. 1d–f). Some secondary structure remains resistant to NP, however: the 5′ end of each segment in virio tends to be more structured overall than the rest of the segment (Supplementary Fig. 3). We used the SHAPE data to predict local constrained RNA structures (<150 nt, see Methods), and found a substantial number present throughout each segment (e.g., Fig. 1f and Supplementary Figs. 4–5). In agreement, vRNA within regions reported to be enriched in NP19,20 has significantly higher SHAPE reactivity (Supplementary Fig. 6a). These observations are also consistent with early studies using enzymatic and chemical probing of naked and NP-bound short RNAs21 and recapitulate RNA structures that have been suggested using computational methods22,23 (Supplementary Fig. 5).
As a comparison, we carried out SHAPE-MaP on a closely-related IAV strain, A/Puerto Rico/8/34 (H1N1) (PR8), and the more distant A/Udorn/72 (H3N2) (Udorn) strain. We found that the SHAPE profiles of segments with high nucleotide sequence identity (>90%) preserve a similar RNA conformation to that of WSN, however segments with lower sequence identity (e.g. Udorn PB1, HA, and NA) tend to have different RNA conformations (Supplementary Fig. 6b–i), suggesting that the RNA sequence itself is the primary determinant of vRNA structure, even in the presence of NP.
Overall, our results suggest that parts of the vRNA could be exposed and accessible to form intermolecular RNA-RNA interactions between segments. We therefore proceeded to analyse such interactions occurring in virio using SPLASH24, which crosslinks base-paired RNAs and maps them using high-throughput sequencing (Supplementary Fig. 7). We performed two biological replicates of SPLASH using purified WSN virions to identify inter-segment RNA interactions (Fig. 2a) and were able to unambiguously identify discrete loci of interaction between segments (Supplementary Fig. 8). The method was highly reproducible between replicates with respect to both sequencing read coverage (Supplementary Fig. 7d–e) and the loci identified (Supplementary Fig. 8c). We also validated our results using qPCR (Supplementary Fig. 8d–f), and confirmed the presence of intra-segment 5’−3’ promoter interactions (Supplementary Fig. 8g).
Figure 2 |. Inter-segment RNA interactions in the IAV genome.
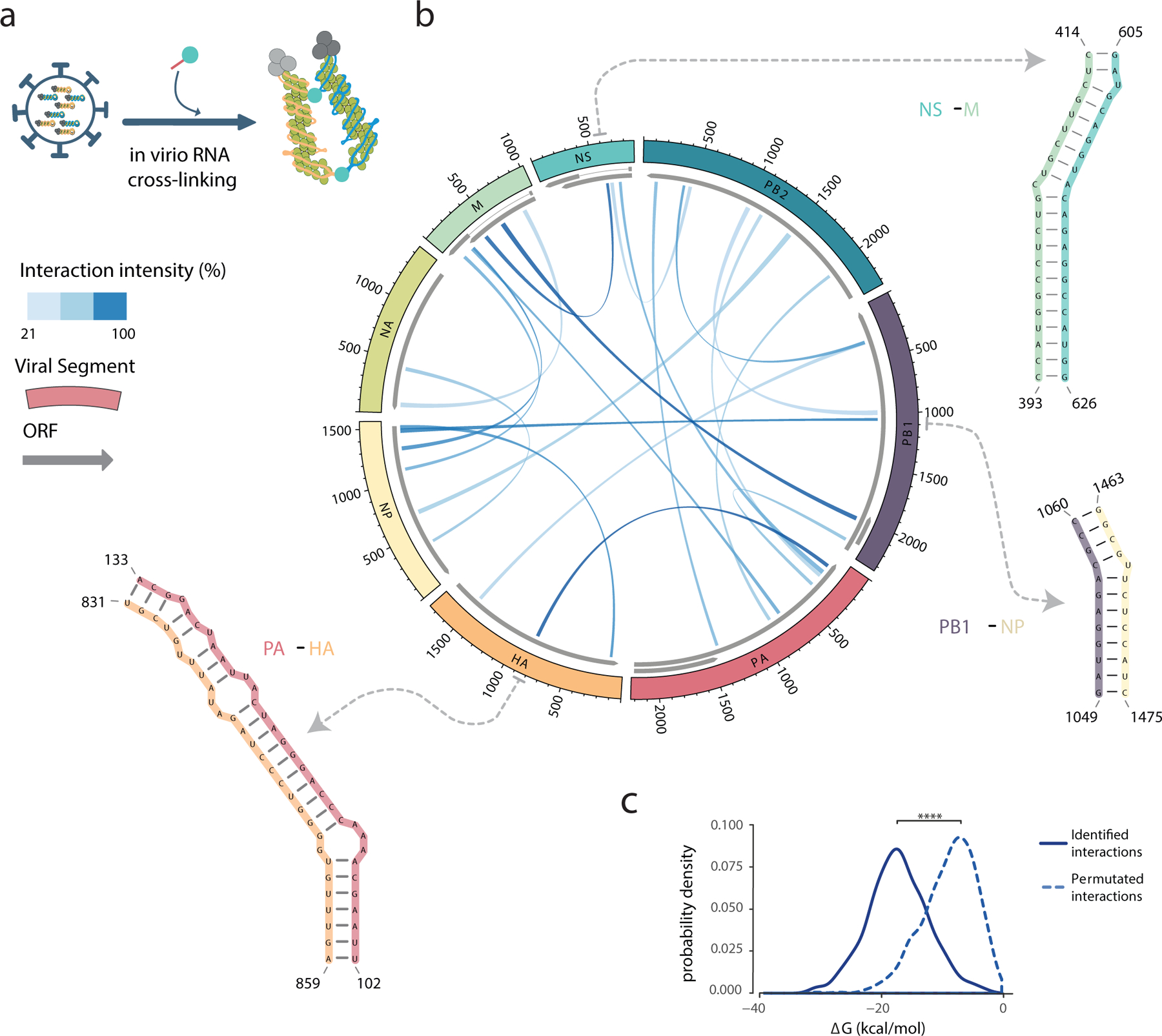
a, Schematic summarizing the SPLASH method. b, The most prevalent inter-segment RNA interactions identified using SPLASH for the WSN (H1N1) IAV strain. The eight vRNA segments are shown on the perimeter of the circle and the links indicate the regions involved in inter-segment base-pairing. The links are shaded by interaction frequencies based on the chimeric read contact matrix (see Methods). Dotted arrows point to SHAPE-guided RNA structure predictions of three representative interactions. c, Distribution of the ΔG energies associated with the interactions identified by SPLASH versus a permutated interaction dataset; **** P ≤ 1×10−16, two-sided Wilcox Rank-Sum Test, n = 611 interactions in common between two biologically independent experiments.
Together, the data reveal that an extensive, redundant, complex network of RNA:RNA interactions exists between segments within influenza virions. Instead of a finite set of discrete interactions (e.g. what might be imagined from a single set of ‘packaging signals’), there are a large number of possible interactions within a virion population, with some interactions occurring much more frequently than the others. For WSN, we identified 611 interaction loci in common between the 2 replicates, with the top 3% of these loci (18/611 interactions, Fig. 2b) accounting for 25% of the sequencing reads in the dataset.
Importantly, we found that the distribution of the most prevalent interaction loci varies between the eight different vRNA segments, and that interaction sites are not restricted to certain regions (Fig. 2b). Most segments are able to interact with multiple other segments, and in some cases the same region can mediate interactions with multiple segments, suggesting that there is a level of redundancy in the network of intermolecular interactions and that multiple RNA conformations exist even within a genetically-identical population of virions.
We then predicted the single-nucleotide resolution structures of inter-segment interaction loci, and also incorporated our empirical SHAPE reactivity values to constrain these predictions. After benchmarking this approach against structured host RNAs (Supplementary Fig. 9), we calculated the intermolecular free energies (ΔG) of formation of predicted influenza intersegment RNA-RNA structures (e.g. in Fig. 2b, and Supplementary Table 2), and found they tended to be energetically highly favourable (median ΔG = −18 kcal/mol, solid line in Fig. 2c). In order to verify if the interaction loci were specific (vs. forming randomly due to confinement within a virion or higher GC content), we permutated the dataset (i.e. shuffled the partners of each interaction) and compared the intermolecular free energies (ΔG) of our real dataset to the permutated one: the permutated dataset was predicted to form much weaker interactions (median ΔG = −8 kcal/mol, dashed line in Fig. 2c). This difference in the distributions was highly significant (P < 1×10−16, Wilcox Rank-Sum test), confirming that the regions we identified by SPLASH were indeed specific. We also found that RNA within the most prominent interaction loci identified by SPLASH was overall more structured than RNA in the rest of the genome, according to our SHAPE-MaP data (P = 5.93×10−5, Supplementary Fig. 8h), though it is important to emphasise that substantial RNA structure is still present outside of SPLASH regions (e.g. hairpins and pseudoknots, Supplementary Figs. 4 and 5).
We then carried out SPLASH analysis on the closely-related PR8 strain (96% sequence identity to WSN) and compared the RNA-RNA interaction network of PR8 to that of WSN (Supplementary Fig. 10a): we found that the core of the network is broadly similar to that of WSN, but the prevalence of the interactions is prone to change, suggesting the structure is highly plastic in response to changes in nucleotide sequence. One-third of the top 2% of interaction loci in PR8 were also highly prevalent in WSN (in the top 10%), another third of the top PR8 loci were present in WSN as minor interactions (below the top 10%), while the remaining third of interactions were unique to PR8. We also carried out SPLASH analysis on the more-distant Udorn strain and found that it forms a much more distinct network of interactions (Supplementary Fig. 10a), with only 3/20 of the top interaction loci in common with WSN or PR8. To examine the effects of reassortment on the RNA-RNA interaction network, we then examined a reassortant virus containing the PB1 and NA gene segments from Udorn and the remaining 6 segments from PR8, PR8::Udorn(PB1+NA), as well as its specific parent Udorn and PR8 strains, which had >99% sequence identity to the Udorn and PR8 strains we examined earlier (Fig. 3 and Supplementary Table 2). We found that the reassortant inherited its interaction network from both parent strains (Supplementary Fig. 10b), with some previously-minor interaction loci rising substantially in prevalence to accommodate the reassorted segments. This suggests that the plasticity of the RNA interaction network allows influenza virus to assemble new gene constellations (as observed during antigenic shift) and to accommodate small sequence variations (as observed in antigenic drift, e.g. between WSN and PR8).
Figure 3 |. RNA interactions form a redundant, plastic network to accommodate variation and reassortment.
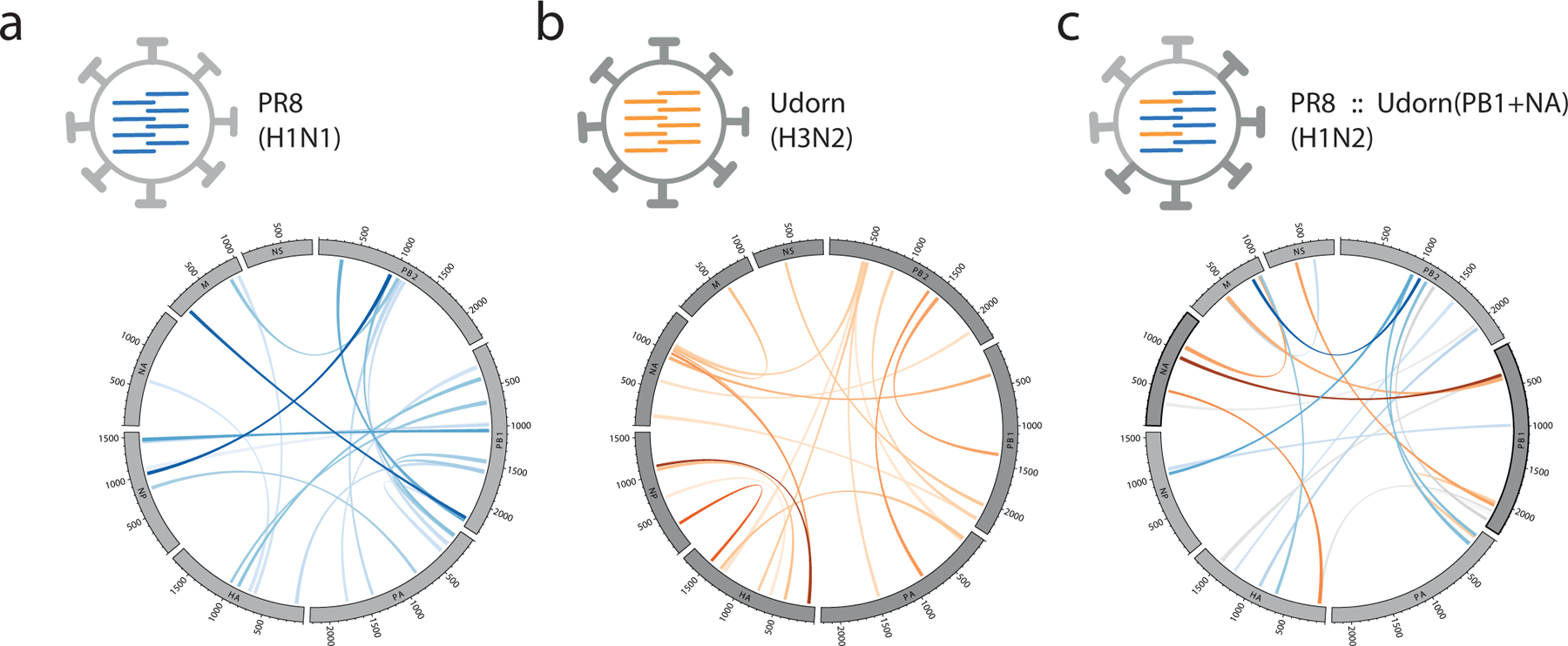
a-b, Inter-segment RNA interaction maps for two parent strains, PR8 (H1N1) and Udorn (H3N2), and c, a reassortant of PR8 that bears the Udorn PB1 and NA gene segments, PR8::Udorn(PB1+NA) (H1N2). Interactions are coloured according to the parent strain that donated the interaction and shaded according to their interaction frequency, as indicated above (see Figure 2). PR8, A/Puerto Rico/8/34; Udorn, A/Udorn/307/72.
While analysing the networks of the Udorn virus and its PR8 reassortant, we noticed that one of the most prominent interaction loci forms between the H3N2-origin PB1 and NA segments (Fig. 4a). Interestingly, twice in the past century, an avian PB1 segment has reassorted with an N2 NA segment during the generation of a pandemic influenza virus: first in 1957, leading to the Asian influenza pandemic (which arose from the reassortment of PB1, NA and HA segments from an avian H2N2 strain with other segments from the then-seasonal H1N1 virus), and again in 1968, leading to the H3N2 Hong Kong pandemic (when the seasonal H2N2 virus again acquired PB1 and HA segments from an avian source — perhaps as a result of an interaction between the seasonal N2 NA and the avian PB1)11. In addition, our previous studies examining seasonal H3N2 vaccine seed viruses produced using classical reassortment methods between egg-adapted PR8 and seasonal H3N2 viruses have noted that the PB1 and NA segments from H3N2 strains tend to co-segregate9,25,26. We showed that the region in the PB1 segment responsible for the co-segregation was within 272–566 nt (1776–2070 nt in 3’→5’ vRNA coordinates9) — precisely encompassing the prominent RNA-RNA interaction identified using SPLASH in this study (305–338 nt of PB1, Fig. 4a and Supplementary Fig. 11a–b) and suggesting this inter-segment interaction could drive the observed co-segregation patterns during reassortment.
Figure 4 |. Inter-segment RNA interactions drive IAV segment co-segregation during reassortment.
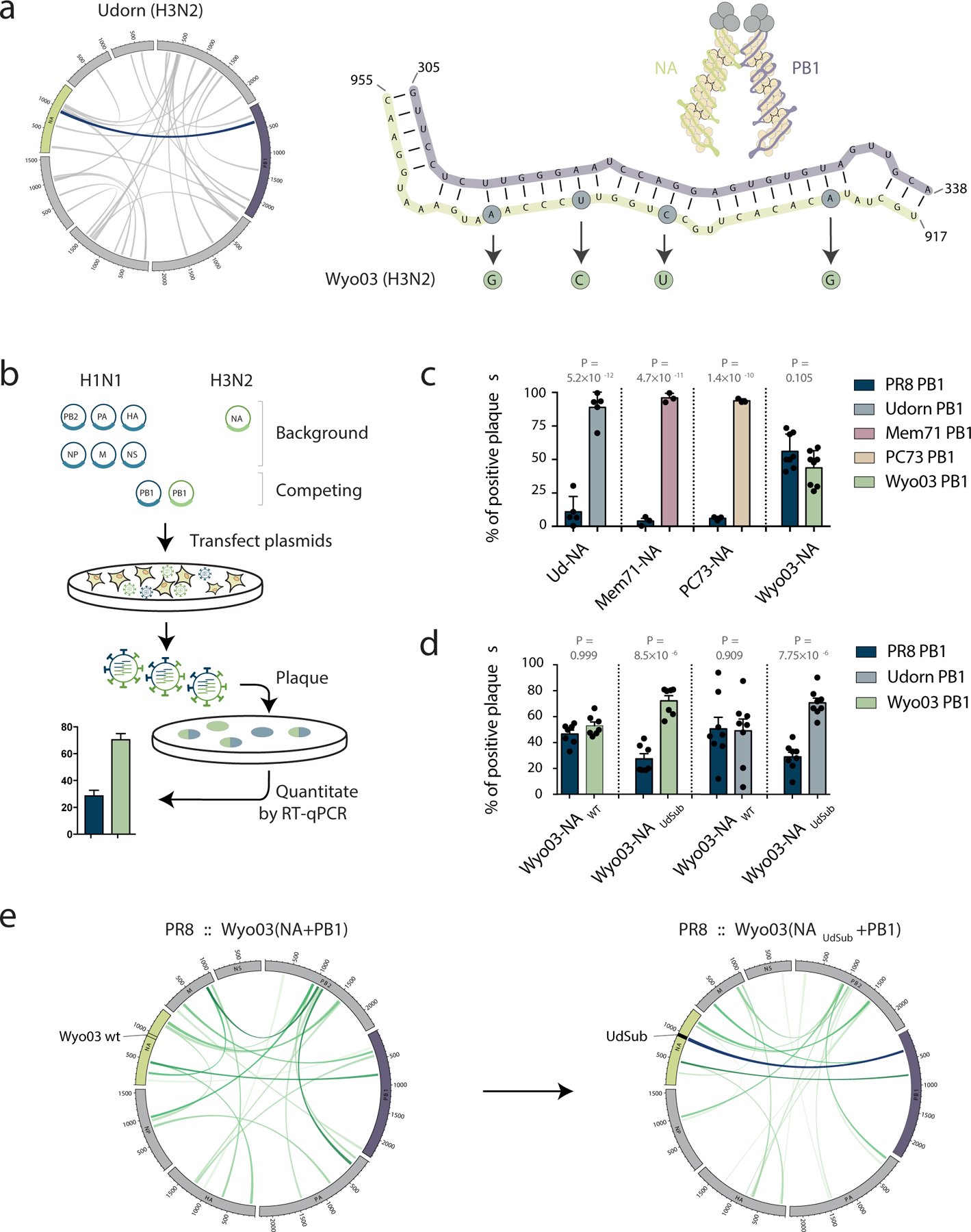
a, Inter-segment RNA interaction map for the Udorn virus, with the interaction between PB1 and NA segments highlighted in dark blue. Structure prediction for the highlighted PB1-NA interaction is shown on the right. Circled nucleotides highlight the bases that differ between the Udorn and Wyo03 strains. b, Competitive reverse-engineering of influenza viruses. Six plasmids encoding H1N1 background segments are transfected together with an H3N2 NA segment-encoding plasmid and PB1 segment-encoding plasmids from both H1N1 and H3N2 strains. The origin of the PB1 segment in the progeny viruses is determined using RT-qPCR. c, Co-segregation between H1N1 or H3N2 PB1 segments and H3N2 NA segments; P values as indicated, ANOVA with Sidak correction for multiple testing, n = 5 (Ud-NA), n = 3 (Mem71-NA and PC73-NA), and n = 8 (Wyo03-NA) biologically independent experiments; bar plot centre represents the mean, error bars indicate SEM. d, Preferential Wyo03 PB1 and NA segment co-segregation is recovered by substituting the 4 nucleotides that differ in Wyo03 NA from those in the PB1-interacting region of Udorn NA (highlighted in a) for a Udorn-like sequence; P values as indicated, ANOVA with Sidak correction for multiple testing, n = 7 (PR8 PB1 vs. Wyo03 PB1 competitions) and n = 8 (PR8 PB1 vs. Udorn PB1 competitions) biologically independent experiments; bar plot centre represents the mean, error bars indicate SEM. e, Substitution of the 4 Udorn-like nucleotides regenerates a strong inter-segment RNA interaction between the H3N2-origin PB1 and NA segments in reassortant viruses. Mem71, A/Memphis/1/71; Wyo03, A/Wyoming/3/03; PC73, A/Port Chalmers/73; Udorn, A/Udorn/307/72; PR8, A/Puerto Rico/8/34.
To test this hypothesis, we performed competitive reverse-engineering of viruses, in which plasmids encoding six viral genome segments of the PR8 virus were transfected together with a plasmid encoding the Udorn NA segment and plasmids encoding competing PB1 segments from the PR8 and Udorn viruses (Fig. 4b). The origin of the incorporated competing segment in the resulting reassortant virus was determined using RT-qPCR. Potential detrimental effects of protein incompatibility in the possible reassortant progeny were ruled out in experiments demonstrating their equivalent replication kinetics (Supplementary Fig. 11c). As previously shown25,26, a virus possessing the N2 NA gene segment originating from Udorn virus preferentially incorporates the Udorn PB1 segment, and we observed the same co-segregation pattern with other seasonal H3N2 strains (A/Memphis/1/71 and A/Port Chalmers/1/73) (Fig. 4c). In contrast, the NA segment from another H3N2 virus, A/Wyoming/3/03 (Wyo03), did not drive incorporation of the Wyo03 PB1 segment in the progeny viruses over the incorporation of the PR8 PB1 segment (Fig. 4c), in agreement with the gene constellations of vaccine seed viruses derived from this strain25. Intriguingly, the Udorn NA segment was able to drive co-selection of the Wyo03 PB1 segment, implying that the Wyo03 NA segment (vs. the Wyo03 PB1 segment) is responsible for the lack of NA-PB1 co-segregation in the Wyo03 strain (Supplementary Fig. 11d–e). Interestingly, the sequence of the PB1-interacting region of NA identified in our SPLASH data (917–955 nt) is conserved in Udorn, A/Memphis/1/71 and A/Port Chalmers/1/73, whereas there are 4 single nucleotide changes in Wyo03 (Fig. 4a and Supplementary Fig. 11f). We therefore generated a Udorn-like Wyo03 NA gene (Wyo03-NAUdSub) to restore complementarity and found that introducing these 4 nucleotide mutations into the Wyo03 NA segment was sufficient to bias the NA-PB1 interaction towards generation of PR8 reassortants with both Wyo03-derived NA and PB1 segments (Fig. 4d); SPLASH analysis of the reassortants confirmed the regeneration of the prominent ‘Udorn-like’ PB1-NA interaction at the site of mutation (Fig. 4e). Together, these results confirm the importance of RNA-RNA interactions in driving segment co-segregation during reassortment, and show that information on RNA structure can predict the ability of segments from different viral strains to reassort.
Overall, our study presents the first global high-resolution structure of the IAV genome. We show that different IAV genome segments in virions have distinct RNA conformations despite the presence of NP and form both intra- and inter-segment RNA interactions. This suggests that both nucleotide sequence changes of the RNA itself, as well as amino acid changes to the NP that alter its affinity to bind and restructure RNA (e.g. an “NP code”27) could affect this network, either directly at the interaction site, or indirectly by affecting which regions of vRNA along a vRNP are rendered accessible. It has been suggested that inter-segment interactions are mediated by the termini, supported by work showing that the minimal sequence for efficient incorporation of a given segment into virions includes ~50–150 nt of each end28, and that defective interfering RNAs (DI RNAs) maintain on average ~200nt of each end29. Nevertheless, studies examining inter-segment interactions directly have shown they can occur outside the termini3,9,30. Our work here supports the latter model, and also reveals that IAV has evolved a degree of flexibility in its genome structure, with multiple redundant loci involved in assembly of eight vRNPs into virions. Such a redundancy would explain the sufficiency of terminal sequences for segment incorporation and might act as an evolutionary strategy to accommodate changes in genome sequence caused by genetic drift or changing evolutionary pressures. Crucially, the redundancy in vRNP-vRNP interactions would accommodate the need for selective packaging of the eight vRNPs during infection while also allowing reassortment to occur in a co-infection event, thus providing influenza virus with a mechanism to escape established immunity in a particular host. Further exploration to generate a comprehensive understanding of the formation and dynamics of inter-segment RNA interactions in influenza viruses will enable us to better understand the likely gene constellations that may result from reassortment between a given set of strains, guiding vaccine design and risk assessment of potential pandemic influenza viruses.
Methods
Cell culture, virus amplification and purification for SHAPE and SPLASH experiments
Madin-Darby Bovine Kidney (MDBK) and Madin-Darby canine kidney (MDCK) cells were grown in Minimum Essential Medium (MEM; Merck), supplemented with 2 mM L-glutamine and 10% fetal calf serum. Stocks of A/WSN/33 (WSN) (H1N1) virus were produced by infecting MDBK cells with influenza virus at a multiplicity of infection (MOI) of 0.001. Viral stocks of A/Udorn/307/72 (H3N2) (Udorn) and A/Puerto Rico/8/1934 (H1N1) (PR8) viruses were produced by infecting MDCK cells at an MOI of 0.001 in the presence of 0.8 μg/ml of TPCK-trypsin (Merck). Viruses were harvested 2 days post infection. Viruses were purified by ultracentrifugation: firstly, the infected cell culture medium was clarified by centrifugation at 4000 rpm for 10 min at 4°C followed by centrifugation at 10,000 rpm for 15 min at 4°C. The virus was then purified by centrifugation through a 30% sucrose cushion at 25,000 rpm for 90 min at 4°C in a SW32 rotor (Beckman Coulter). The purified virus pellet was resuspended in a resuspension buffer (0.01 M Tris-HCl (pH 7.4), 0.1 M NaCl, 0.0001 M EDTA). We note that neither virion nor RNA tertiary structures are disrupted by ultracentrifugation31–33.
Selective 2′-hydroxyl acylation analysed by primer extension and mutational profiling (SHAPE-MaP)
SHAPE-MaP was performed according to published procotols17. 1-methyl-7-nitroisatoic anhydride (1M7) was synthesised from 4-nitroisatoic anhydride as described previously34. For the ivtRNA experiments each vRNA segment was synthesised from a linear DNA template using the HiScribe™ T7 High Yield RNA Synthesis Kit (NEB). The products were checked for size and purity on a 3.5% PAGE-urea gel. nkvRNA samples were prepared by purifying the WSN particles over sucrose cushion as described above. Purified viruses were treated with 250 μg/mL of Proteinase K (Roche) in PK buffer (10 mM Tris-HCl (pH 7.0), 100 mM NaCl, 1 mM EDTA, 0.5% SDS) for 40 min at 37°C. Before the modification ivtRNA and nkvRNA samples were folded at 37°C for 30 min in folding buffer (100 mM Hepes-NaOH (pH 8.0), 100 mM NaCl, 10 mM MgCl2). 1M7 (dissolved in anhydrous DMSO (Merck)) was added to a final concentration of 10 mM to the folded RNA and the samples were incubated for 75 s at 37°C. The in virio modifications were performed by adding 1M7 directly to purified virus. The ability of SHAPE reagents to penetrate viral particles was initially tested as described previously35 by performing 32P-labelled primer extensions on RNA extracted from SHAPE reagent-treated virus using an NA segment targeting primer (5’-AATTGGTTCCAAAGGAGACG-3’). In parallel to the 1M7-treated samples, control samples were treated with DMSO. N-methylisatoic anhydride (NMIA, Thermo Fisher) SHAPE reagent was also tested in virio. Experiments with NMIA were performed as described above for 1M7, except the purified virions were treated with NMIA for 45 min.
Sequencing library preparation was carried out as described previously17 following the randomer workflow. In brief, after 1M7 or control treatments, RNA was purified using the RNA Clean & Concentrator™−5 kit (Zymo Research). The RNA was reverse transcribed using Random Primer Mix (NEB) with Superscript II in MaP buffer (50 mM Tris-HCl (pH 8.0), 75 mM KCl, 6 mM MnCl2, 10 mM DTT and 0.5 mM dNTPs). Nextera XT DNA Library Prep Kit (Illumina) was used to prepare the DNA libraries. Final PCR amplification products were size selected using Agencourt AMPure XP beads (Beckman Coulter) and quality assessed using the Agilent DNA 1000 kit on a Bioanalyzer 2100 instrument (Agilent). For WSN, nkvRNA and ivtRNA the libraries were sequenced (2×150bp) on a HiSeq4000 instrument (Illumina), for PR8 and Udorn viruses the libraries were sequenced (1×150bp) on a NextSeq 500 instrument (Illumina).
Sequencing of psoralen crosslinked, ligated, and selected hybrids (SPLASH)
SPLASH samples were prepared as published previously24,36, with some modifications, for two replicates each of WSN, PR8, and Udorn viruses and a single replicate for each of the H3N2 reassortant viruses. Purified virus was incubated with 200 μM of EZ-Link™ Psoralen-PEG3-Biotin (Thermo Fisher) and 0.01% digitonin (Merck) for 5 min at 37°C. Virus was spread on a 6-well dish, covered with a glass plate, placed on ice, and irradiated for 45 min using a UVP Ultra Violet Product™ Handheld UV Lamp (Fisher). Crosslinked virus was treated with Proteinase K (Merck) and vRNA was extracted using TRIzol (Invitrogen). An aliquot of extracted vRNA was used to detect biotin incorporation using chemiluminescent nucleic acid detection module kit (Thermo Fisher) on Hybond-N+ nylon membrane (GE Healthcare Life Science). The rest of the extracted vRNA was fragmented using NEBNext® Magnesium RNA Fragmentation Module (NEB), and size selected for fragments shorter than 200 nt using RNA Clean & Concentrator™−5 (Zymo Research). The samples were enriched for biotinylated vRNA using Dynabeads MyOne Streptavidin C1 beads (Life Technologies) and on-bead proximity ligation and psoralen crosslink reversal were carried out as published previously. Sequencing libraries were prepared using the commercial SMARTer smRNA-Seq Kit (Clontech). Final size selection was done by running the PCR-amplified sequencing libraries on a 6% PAGE gel (Thermo Thermo Fisher Fisher) in TBE and selecting for 200–300bp DNA. Libraries were sequenced 1×150bp on a NextSeq 500 instrument (Illumina).
Processing of SHAPE-MaP sequencing reads
The sequencing reads were trimmed to remove adaptors using Skewer37. The SHAPE reactivity profiles were generated using the published ShapeMapper2 pipeline38, which aligns the reads to the reference genome using Bowtie2 and calculates mutation rates at each nucleotide position. The mutation rates are then converted to the SHAPE reactivity values defined as:
, where mutr1M7 is the nucleotide mutation rate in 1M7 treated sample, mutrDMSO is the mutation rate in the DMSO treated sample. All SHAPE reactivities are normalised to an approximate 0–2 scale by dividing the SHAPE reactivity values by the average reactivity of the 10% most highly reactive nucleotides after excluding outliers (defined as nucleotides with reactivity values that are greater than 1.5x the interquartile range). High SHAPE reactivities indicate more flexible (i.e. single-stranded) regions of RNA and low SHAPE reactivities indicate more structurally-constrained (i.e. base-paired) regions of RNA.
Processing of SPLASH sequencing reads
The sequencing reads were trimmed to remove adaptors using Skewer37. STAR39 was used to align the reads to the appropriate virus reference genome (Supplementary Table 2). Only the chimeric reads in which at least 20 nts aligned to the reference segments were used in further processing (STAR parameter -- chimSegmentMin 20). Chimeric reads were deduplicated using CIGAR strings and alignment positions. CIGAR strings in each read alignment were processed to find the read start and end coordinates. Chimeric read coordinates were used to produce a matrix of sequence interactions in R software. Discrete loci in the matrix were selected and individually fitted with a Gaussian curve based on read overlap intensity to define an interaction window; interaction windows of complex overlapping loci were separated into individual windows. The width of the interaction window was used to determine the start and end coordinates of each interaction and the number of reads that were within (or partially within) this region was used as a measure of the interaction frequency. For the generation of figures, the top 20 interactions in each virus were visualised using the circlize package40 in R. The full set of interaction loci is provided in Supplementary Table 2. For qPCR validation of interacting loci, psoralen-crosslinked samples were prepared and enriched as described above, but with an abbreviated fragmentation time (3 min vs. 4 min) to generate longer RNA fragments. RNA was polyadenylated with Poly(A) Polymerase (Takara) and cDNA was generated using smRNA dT Primer (Takara) and PrimeScript Reverse Transcriptase (Takara) according to the manufacturer’s instructions. A 50-cycle qPCR was carried out according to manufacturer’s instructions on a StepOnePlus instrument (ABI) using Brilliant II SYBR® Green QPCR Master Mix with ROX (Agilent) and primer pairs to test for inter-segment interactions. Primer sequences are given in Supplementary Table 3. Following 50 cycles of qPCR amplification, products were resolved by 8% PAGE (29:1 acrylamide/bis-acrylamide, 1x TBE buffer) and visualised using blue light transillumination.
RNA structure predictions
The IntaRNA (v2.3.1) algorithm41 was used to predict the ability of RNA-RNA interactions to occur in the regions identified during the SPLASH analysis, using the Exact mode (--mode E) and no seed constraint (--noSeed) options. SHAPE reactivities were included in the modeling of RNA-RNA interactions (--tShape and -- qShape). Permutated data sets were generated by randomly shuffling the specific interaction partners identified by SPLASH and assessing the interaction ΔG energies using IntaRNA. The significance of the difference between the probability distributions of the ΔG energies associated with the SPLASH-identified intermolecular RNA interactions versus the permutated datasets was calculated using Wilcox Rank-Sum Test in R software. The IntaRNA structure predictions were then used to trim the interaction regions to the nucleotides involved in the base pairing. Where SHAPE data was not available (PR8 reassortants with H3N2 viruses), pre-folding of each RNA strand (“accessibility”) was disabled in IntaRNA (--qAcc=N --tAcc=N). For validation against known structures, RNA secondary structure data was extracted from the RNA3dhub database42, based on cryoEM structures of the 80S ribosome43 (PDB: 6EK0) and from the U4/U6.U5 tri-snRNP spliceosomal complex44 (EMDB: EMD-2966). Reference RNA sequences were corrected to match bovine (MDBK) sequences for rRNA and U4/U6 snRNAs as previously described45. For intramolecular RNA structure predictions, the ViennaRNA Package 2.046 was used. The RNAfold command was used to predict secondary RNA structures and partition functions for each segment. SHAPE reactivities were included as pseudoenergy restraints. A 50 nt sliding median window correlation analysis between the ex virio and in virio SHAPE reactivity profiles was used to determine the extent of SHAPE correlation between the T7-transcribed RNA and vRNP-associated RNA. We found that no correlation existed >150 nt, and therefore set the maximum pairing distance constraint for structure and partition function predictions to 150 nt. For intramolecular structure predictions, we set the nucleotides within the promoter region to be single stranded.
Cell culture for reverse-engineering of influenza viruses
MDCK cells and human embryonic kidney (293T) cells were sourced from an existing collection in the Department of Microbiology and Immunology, The University of Melbourne. MDCK cells were grown in RPMI-1640 (Sigma Aldrich, NSW, Australia) and 293T cells were grown in Dulbecco’s modified Eagles medium (Life Technologies, Victoria, Australia). Both media were supplemented with 10% heat-inactivated fetal calf serum (FCS), 2 mM L-glutamine, 2mM sodium pyruvate, 24 μg/ml gentamicin, 50 μg/ml streptomycin and 50 IU/ml penicillin. Co-cultures of MDCK cells and 293T cells for transfection were established in Opti-MEM (Life Technologies) with 50 μg/ml streptomycin and 50 IU/ml penicillin.
Construction of reverse-engineered influenza viruses
Individual gene segments from PR8, Udorn, Memphis/1/71 (H3N2) (Mem71), Port Chalmers/1/73 (H3N2) (PC73) and Wyoming/3/03 (H3N2) (Wyo03) viruses were reverse transcribed and cloned into pHW2000 plasmids47. The reverse genetics-derived viruses contained either a PR8, Udorn, Mem71, PC73 or Wyo03 PB1 gene with either a wildtype or modified NA gene in a genetic background comprising 6 segments from PR8 virus (PB2, PA, HA, NP, M, NS). The modified NA gene (Wyo03UdSub) was derived from Wyo03 and carried 4 substitutions towards the Udorn NA sequence: nt G943A, C938U, U933C, and G923A. Three of these four nucleotide changes were silent, with the fourth (A534G) resulting in a conservative lysine to arginine change (K172R). Complementary fragments incorporating these changes were generated by PCR and joined together by another round of PCR using segment-specific primers containing BsmBI restriction sites48 and the product was cloned into the pHW2000 expression vector for virus rescue. Primer sequences are given in Supplementary Table 3. Rescued viruses were amplified in 10-day embryonated hen’s eggs. Infectious allantoic fluid was titrated for virus content by plaque formation in MDCK cells49 and stored at −80°C.
Determination of viral replication kinetics of reverse-engineered viruses
The replication characteristics of the reverse-engineered viruses were determined by infecting MDCK cells at an MOI of 0.01. Following 1 hr absorption (at t=0 hr) the inoculum was removed and cells were washed and incubated at 37°C, 5% CO2 in RPMI-1640 supplemented with 2mM L-glutamine, 2mM sodium pyruvate, 24 μg/ml gentamicin, 50 μg/ml streptomycin and 50 IU/ml penicillin and 1 μg/ml of TPCK-trypsin (Worthington Biochemical Corporation, NJ, USA). Cell culture supernatants were harvested at various time points post infection and stored at −80°C for analysis. Viral titres were determined by plaque formation on confluent monolayers of MDCK cells.
Nine-plasmid competitive reverse-engineering of influenza viruses
Competitive reverse-engineering of influenza viruses was undertaken using a modified version of the eight-plasmid reverse genetics system26 as described previously25. Briefly, plasmids encoding the PB2, PB1, PA, HA, NP, M and NS vRNA segments of PR8 were co-transfected with plasmids encoding NA vRNA and competing PB1 vRNA of either Udorn, Mem71, PC73 or wildtype or modified Wyo03. Each plasmid (1 μg) was mixed with FuGENE6 transfection reagent (Promega, NSW, Australia) in Opti-MEM and added to a co-culture of 239T and MDCK cells. Six hrs post-transfection, media was replaced with Opti-MEM supplemented with 50 μg/ml streptomycin and 50 IU/ml penicillin. Twenty-four hrs later, TPCK-trypsin (1 μg/ml) was added and the supernatant harvested after a further 42 hrs and stored at −80°C. To determine the incorporation frequencies of the competing PB1 genes, the progeny viruses in the transfection supernatant were subjected to a plaque assay in MDCK cells. Randomly chosen plaques (~36/experiment) were picked by sampling through the agarose and resuspended in 0.05% Triton-X. The source of the competing gene segments was identified by gene-specific RT-qPCR using the SensiFast probe no-ROX one-step RT-PCR Kit (Bioline, NSW, Australia). Each 20 μl reaction was performed using 5 μl of plaque-picked virus suspension, 10 μl of 2x SensiFast No-ROX one-step mix, 0.2 μl of reverse transcriptase, 0.4 μl of Ribosafe RNase inhibitor, 0.8 μl of each 10 μM gene-specific forward and reverse primer and 0.08 μl of each 25 μM gene-specific probe. Primer and probe sequences are given in Supplementary Table 3. The RT reaction was incubated for 10 min at 45°C, before proceeding with qPCR amplification, per manufacturer’s instructions. The reported values are combined data from three to eight independent transfection experiments for each competition.
Data availability statement
All sequencing data have been deposited to Sequence Read Archive (accession numbers: SRP127020, SRP126994, SRP150669, SRP150677, and SUB5763755) and the processed SHAPE reactivities are available in SNRNASM format as Supplementary Table 1 and processed SPLASH data as Supplementary Table 2.
Supplementary Material
Acknowledgements:
We thank G.G. Brownlee and J. Kenyon for helpful discussions, J. Kenyon and J.G. Aw sharing protocols, J. Robertson for making 1M7 reagent and St Jude Children’s Research Hospital, Memphis, TN, USA, for provision of the pHW2000 plasmids. This work was supported by a Wellcome Trust studentship [105399/Z/14/Z] (to B.D.), a UK Biotechnology and Biological Sciences Research Council (BBSRC) studentship [BB/M011224/1] (to M.K.), grants from the US National Institutes of Health [HL111527, GM101237 and HG008133] (to A.L.), a National Health and Medical Research Council of Australia programme grant [ID1071916] (to L.E.B.), UK Medical Research Council programme grants [MR/K000241/1 and MR/R009945/1] (to E.F.), and an EPA Cephalosporin Junior Research Fellowship (to D.L.V.B.).
Footnotes
Competing Interests
The authors declare no competing financial interests.
References
- 1.Gerber M, Isel C, Moules V, Marquet R. Selective packaging of the influenza A genome and consequences for genetic reassortment. Trends Microbiol. 2014;22(8):446–455. [DOI] [PubMed] [Google Scholar]
- 2.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. Genome packaging in influenza A virus. J Gen Virol. 2010;91(2):313–328. [DOI] [PubMed] [Google Scholar]
- 3.Gavazzi C, Yver M, Isel C, et al. A functional sequence-specific interaction between influenza A virus genomic RNA segments. Proc Natl Acad Sci U S A. 2013;110(41):16604–16609. doi: 10.1073/pnas.1314419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fournier E, Moules V, Essere B, et al. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res. 2011;40(5):2197–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marsh GA, Rabadan R, Levine AJ, Palese P. Highly conserved regions of influenza a virus polymerase gene segments are critical for efficient viral RNA packaging. J Virol. 2008;82(5):2295–2304. doi: JVI.02267-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gog JR, Afonso EDS, Dalton RM, et al. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res. 2007;35(6):1897–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutchinson EC, Curran MD, Read EK, Gog JR, Digard P. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J Virol. 2008;82(23):11869–11879. doi: 10.1128/JVI.01634-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutchinson EC, Wise HM, Kudryavtseva K, Curran MD, Digard P. Characterisation of influenza A viruses with mutations in segment 5 packaging signals. Vaccine. 2009;27(45):6270–6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbertson B, Zheng T, Gerber M, et al. Influenza NA and PB1 gene segments interact during the formation of viral progeny: Localization of the binding region within the PB1 gene. Viruses. 2016;8(8):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowen AC. Constraints, drivers, and implications of influenza A virus reassortment. Annual Review of Virology. 2017;4(1). [DOI] [PubMed] [Google Scholar]
- 11.Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic formation. Cell host & microbe. 2010;7(6):440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arranz R, Coloma R, Chichon FJ, et al. The structure of native influenza virion ribonucleoproteins. Science. 2012;338(6114):1634–1637. doi: 10.1126/science.1228172. [DOI] [PubMed] [Google Scholar]
- 13.Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. Organization of the influenza virus replication machinery. Science. 2012;338(6114):1631–1634. doi: 10.1126/science.1227270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.te Velthuis AJ, Fodor E. Influenza virus RNA polymerase: Insights into the mechanisms of viral RNA synthesis. Nature Reviews Microbiology. 2016;14(8):479–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pflug A, Lukarska M, Resa-Infante P, Reich S, Cusack S. Structural insights into RNA synthesis by the influenza virus transcription-replication machine. Virus Res. 2017. [DOI] [PubMed] [Google Scholar]
- 16.Wilkinson KA, Gorelick RJ, Vasa SM, et al. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS biology. 2008;6(4):e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM. Selective 2 [prime]-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nature protocols. 2015;10(11):1643–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watts JM, Dang KK, Gorelick RJ, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460(7256):711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee N, Le Sage V, Nanni AV, Snyder DJ, Cooper VS, Lakdawala SS. Genome-wide analysis of influenza viral RNA and nucleoprotein association. Nucleic Acids Res. 2017;45(15):8968–8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams GD, Townsend D, Wylie KM, et al. Nucleotide resolution mapping of influenza A virus nucleoprotein-RNA interactions reveals RNA features required for replication. Nature communications. 2018;9(1):465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baudin F, Bach C, Cusack S, Ruigrok RW. Structure of influenza virus RNP. I. influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 1994;13(13):3158–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi Y, Dadonaite B, van Doremalen N, Suzuki Y, Barclay WS, Pybus OG. Computational and molecular analysis of conserved influenza A virus RNA secondary structures involved in infectious virion production. RNA biology. 2016;13(9):883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gultyaev AP, Tsyganov-Bodounov A, Spronken MI, Van Der Kooij S, Fouchier RA, Olsthoorn RC. RNA structural constraints in the evolution of the influenza A virus genome NP segment. RNA biology. 2014;11(7):942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aw JGA, Shen Y, Wilm A, et al. In vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Mol Cell. 2016;62(4):603–617. [DOI] [PubMed] [Google Scholar]
- 25.Cobbin JC, Verity EE, Gilbertson BP, Rockman SP, Brown LE. The source of the PB1 gene in influenza vaccine reassortants selectively alters the hemagglutinin content of the resulting seed virus. J Virol. 2013;87(10):5577–5585. doi: 10.1128/JVI.02856-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cobbin JC, Ong C, Verity E, Gilbertson BP, Rockman SP, Brown LE. Influenza virus PB1 and neuraminidase gene segments can cosegregate during vaccine reassortment driven by interactions in the PB1 coding region. J Virol. 2014;88(16):8971–8980. doi: 10.1128/JVI.01022-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreira ÉA et al. A conserved influenza A virus nucleoprotein code controls specific viral genome packaging. Nat Commun 7, 12861 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutchinson EC, Kirchbach, von JC, Gog JR & Digard P Genome packaging in influenza A virus. J. Gen. Virol 91, 313–328 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Saira K. et al. Sequence analysis of in vivo defective interfering-like RNA of influenza A H1N1 pandemic virus. J. Virol 87, 8064–8074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noda T. et al. Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nat Commun 3, 639 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutchinson EC et al. Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry. PLoS Pathog. 8, e1002993 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hutchinson EC et al. Conserved and host-specific features of influenza virion architecture. Nat Commun 5, 4816 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitra S. Detecting RNA tertiary folding by sedimentation velocity analytical ultracentrifugation. Methods Mol Biol 1086, 265–288 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Turner R, Shefer K, Ares M Jr. Safer one-pot synthesis of the ‘SHAPE’ reagent 1-methyl-7-nitroisatoic anhydride (1m7). RNA. 2013;19(12):1857–1863. doi: 10.1261/rna.042374.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkinson KA, Merino EJ, Weeks KM. Selective 2 [prime]-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nature protocols. 2006;1(3):1610–1616. [DOI] [PubMed] [Google Scholar]
- 36.Aw JGA, Shen Y, Nagarajan N, Wan Y. Mapping RNA-RNA interactions globally using biotinylated psoralen. JoVE (Journal of Visualized Experiments). 2017(123):e55255–e55255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang H, Lei R, Ding S, Zhu S. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Busan S, Weeks KM. Accurate detection of chemical modifications in RNA by mutational profiling (MaP) with ShapeMapper 2. RNA. 2018;24(2):143–148. doi: 10.1261/rna.061945.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobin A, Davis CA, Schlesinger F, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu Z, Gu L, Eils R, Schlesner M & Brors B circlize Implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Busch A, Richter AS, Backofen R. IntaRNA: Efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008;24(24):2849–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leontis NB & Zirbel CL in RNA 3D Structure Analysis and Prediction (eds. Leontis N & Westhof E) 281–298 (Springer Berlin; Heidelberg, 2012). [Google Scholar]
- 43.Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I & Klaholz BP Visualization of chemical modifications in the human 80S ribosome structure. Nature. 2017;551:472–477. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen THD et al. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature. 2015;523:47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.te Velthuis AJW et al. Mini viral RNAs act as innate immune agonists during influenza virus infection. Nature Microbiology. 2018;3:1234–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorenz R. et al. ViennaRNA Package 2.0. Algorithms Mol Biol 6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A. 2000;97(11):6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffmann E, Stech J, Guan Y, Webster R, Perez D. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146(12):2275–2289. [DOI] [PubMed] [Google Scholar]
- 49.Tannock GA, Paul JA, Barry RD. Relative immunogenicity of the cold-adapted influenza virus A/ann arbor/6/60 (A/AA/6/60-ca), recombinants of A/AA/6/60-ca, and parental strains with similar surface antigens. Infect Immun. 1984;43(2):457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited to Sequence Read Archive (accession numbers: SRP127020, SRP126994, SRP150669, SRP150677, and SUB5763755) and the processed SHAPE reactivities are available in SNRNASM format as Supplementary Table 1 and processed SPLASH data as Supplementary Table 2.


