Abstract
Abnormal DUX4 expression in skeletal muscles plays a key role in facioscapulohumeral muscular dystrophy (FSHD) pathogenesis, although the molecular mechanisms regulating DUX4 expression are not fully defined. Using bioinformatic analysis of the genomic DUX4 locus, we have identified a number of putative G-quadruplexes (GQs) forming sequences. Their presence was confirmed in synthetic oligonucleotiode sequences derived from the enhancer, promoter and transcript of DUX4 through circular dichroism and nuclear magnetic resonance analysis. We further examined the binding affinity of a naturally occurring GQ stabilizing compound, berberine, to these non-canonical genetic structures using UV–Vis and fluorescence spectroscopy. Subsequent in vitro study in FSHD patient myoblasts indicated that berberine treatment reduced DUX4 expression and also expression of genes normally switched on by DUX4. Further investigation in a mouse model overexpressing exogenous DUX4 confirmed the therapeutic effects of berberine in downregulating DUX4 protein expression, inhibiting muscle fibrosis, and consequently rescuing muscle function. Our data demonstrate for the first time that GQs are present in the DUX4 locus and that the GQ interactive ligand reduces DUX4 expression suggesting potential role of GQs in FSHD pathogenesis. Our work provides the basis of a novel therapeutic strategy for the treatment of FSHD.
INTRODUCTION
Facioscapulohumeral dystrophy (FSHD) is one of the most prevalent muscular dystrophies (1:20 000) characterized by progressive weakness and destruction of the facial, shoulder and upper arm skeletal muscles (1–3). The molecular hallmark of the disease is the loss of epigenetic features from the D4Z4 macrosatellite repeat array positioned in the sub-telomeric end of chromosome 4 resulting in chromatin relaxation (4). The most common form of the disease, FSHD type 1 (FSHD1), is caused by a deletion of a large region of the D4Z4 repeat array (5, 6). In rare cases, mutations in Structural Maintenance of Chromosomes Hinge Domain Containing 1 gene (SMCHD1) that methylates the D4Z4 region leads to FSHD type 2 (FSHD2) (7,8). In both forms of the disease, the partial relaxation of the D4Z4 chromatin structure results in failure to repress expression of DUX4 gene within the D4Z4 unit on the permissive chromosome 4qA that encodes for a homeobox transcription factor (9). Overexpression of DUX4 de-regulates a number of cellular pathways causing skeletal muscle toxicity and this is recognized a major factor in the FSHD pathophysiology (3,10–12).
Despite recent advancements in our understanding of the genetic and epigenetic factors contributing to the development of FSHD, several questions remain unanswered regarding molecular mechanisms regulating DUX4 expression (13). Since the D4Z4 repeat arrays are very GC-rich (i.e. 73%), it has been suggested that these may contain biologically relevant epigenetic features (14). Indeed, it has been demonstrated that D4Z4 units adopt repressed chromatin structures in somatic cells through high levels of CpG methylation in association with repressive histone modifications (15, 16). Interestingly, the increased susceptibility to D4Z4 hypomethylation linked to a shorter D4Z4 repeat in FSHD1 and/or the SMCHD1 mutation in FSHD2 highly impacts the disease severity, indicating that the genetic and the epigenetic imbalances can influence development of FSHD (13,17). Another epigenetic modifier that has been identified within the D4Z4 array are secondary nucleic acid structures known as G-quadruplexes (GQs) (18–20). However, their role on DUX4 expression regulation has not been investigated.
GQs are formed within guanine-rich DNA and RNA sequences and consist of guanine-quartets held together by Hoogsten hydrogen bonding in a planar orientation (21), forming stacks of typically three or four quartets. Increasing in vitro and in vivo evidence has uncovered the presence of GQs in important regulatory regions (e.g., promoters (22–26), enhancers (27, 28), telomeres (29, 30), transcripts (31–33) and non-coding RNAs (34–36)). The data suggest an important role of these non-canonical structures in regulating key cellular functions linked to both DNA processes (e.g. telomere homeostasis, transcription and recombination) (37) and RNA post-transcriptional mechanisms (e.g. pre-mRNA processing and translation) (38). GQs have been found to be an important factor involved in regulating molecular mechanisms behind several human diseases, including cancer (39–41) and neurodegenerative disorders (42). As a result, a number of small-molecule ligands have been developed to target, modulate and/or visualize GQ structures (43–45).
In this study, we demonstrated the presence of novel GQ motifs within the enhancer, promoter and transcript of DUX4 by using bioinformatic and biophysical tools. We identified that berberine, a GQ stabilizing compound, could bind these DNA and RNA GQ structures with high affinity. Treatment of FSHD patient muscle cells and mice, injected with adeno-associated viral vectors (AAVs) overexpressing DUX4, with berberine resulted in downregulation of DUX4 expression and amelioration of DUX4-mediated pathological changes. These promising results indicate that GQ stabilizers offer a novel therapeutic strategy for targeting DUX4-related genetic elements as a potential treatment for FSHD.
MATERIALS AND METHODS
Reagents
Berberine chloride and calf thymus (CT) DNA were purchased from Sigma, UK. Berberine was dissolved in Milli-Q water and then further diluted to appropriate concentrations in either KP buffer (10 mM K2HPO4/KH2PO2, 100 mM KCl, pH 7.0) for circular dichroism (CD) and nuclear magnetic resonance (NMR) analyses, or in Tris buffer (10 mM Tris–HCl, 100 mM KCl, pH 7.0) for UV–Vis and fluorescence spectroscopy experiments. For the in vivo work, berberine was dissolved in DMSO at 50 mg/ml and further diluted in injectable saline prior to use. Ampicillin and PhenDC3 were purchased from Sigma, UK and dissolved in DMSO. DNA and RNA oligonucleotides listed in Table 2 were synthesized and HPLC purified by Integrated DNA Technologies (Belgium). All of the oligonucleotides and CT DNA were dissolved in KP buffer for CD and NMR analyses, or in Tris buffer for UV–Vis and fluorescence spectroscopy assessment.
Table 2.
Oligonucleotides used in secondary structure and berberine binding analysis
| Nucleic acid type | Construct name | Construct sequence | Structure |
|---|---|---|---|
| DNA | DME1 GQ | CAGGGGATGGTGGGGCTGGGGTTGAGTGATGGGC | G-quadruplex |
| D4P GQ | CGGGGTGGGGCGGGCTGTCCCAGGGGGGCT | G-quadruplex | |
| RNA | CSS_GQ | AGGGCCAGGCACCCGGGACAGGGUGGCAGGGC | Double stranded |
| E1_GQ | AGGGGAGUCCGUGGUGGGGCUGGGGCCGGGGU | G-quadruplex | |
| SS1_GQ | CGGGGUUGGGACGGGGUCGGGU | G-quadruplex |
Circular dichroism and nuclear magnetic resonance
For the CD and NMR analyses, annealed oligonucleotides in KP buffer were used at 2–4 μM. CD spectra were acquired using a Chirascan qCD spectrophotometer (Applied Photophysics Ltd, UK), equipped with a LTD6G circulating water bath (Grant Instruments, UK) and thermoelectric temperature controller (Melcor, USA). Oligonucleotides were annealed by heating to 95°C for 10 min and then cooled to room temperature for at least 4 h (i.e. ∼0.3°C/min). The samples were stored at –20°C. CD spectra were recorded over a wavelength range of 215–340 nm using a 1-cm path length strain-free quartz cuvette at room temperature. Data points were recorded at 1-nm intervals. A bandwidth of 3 nm was used, and 5000 counts acquired at each point with adaptive sampling enabled. Each trace is shown as the mean of three scans (± SD). NMR spectra (1H) were collected at 800 MHz using a Bruker Avance III spectrometer (US) with a triple resonance cryoprobe. Each sample contained 10% D2O. Standard Bruker acquisition parameters were used.
UV–visible and fluorescence spectroscopy
Annealed oligonucleotides were titrated in 0–10 μM range in both UV/Vis and fluorescent spectrometry readouts. The berberine concentration was kept constant at 10 and 5 μM in UV–Vis and fluorescent spectroscopic analyses, respectively. Tris buffer was used as a blank. The UV–Vis spectrophotometer was set to record spectra from 300 to 550 nm. Both types of spectra were recorded at room temperature. Fluorescent spectra were measured at λex/λem = 355/530 nm. To measure the binding constant (Ka), data was plotted into hyperbolic function using KaleidaGraph (Synergy Software, USA) following the equation below:
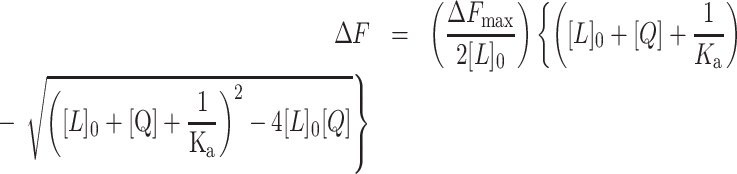 |
where:
ΔF = F – F0 and ΔFmax= Fmax – F0.
F 0 and F: initial and subsequent fluorescent intensities
[L]0: berberine concentration
[Q]: oligonucleotide concentration
K a: binding constant
This is a quadratic velocity equation for tight-binding substrates and was previously used to determine the affinity binding between berberine and human telomeric GQ (46).
DUX4 expressing AAV plasmid constructs
DUX4 expression was mediated through the use of adeno-associated viral vectors (AAVs). The AAV plasmid containing a D4Z4 construct driven by CMV or tMCK promoter as previously described (47) was kindly provided by Prof Scott Harper, Ohio State University School of Medicine, USA. To enhance muscle specificity, the Spc512 muscle-specific promoter was introduced into the plasmid to drive the gene expression. We also replace the downstream of the pLAM1 polyadenylation sequence (due to a lack in the cleavage site) which was subcloned from the eGFP.C1.ΔSV40.3′UTR DUX4 lentiviral vector plasmid that was kindly gifted by Dr Julie Dumonceaux, UCL, UK by Gibson assembly (NEB, UK). We named the newly generated plasmid as AAV.Spc512.DUX4.3′UTR. All plasmids were verified by restriction enzyme digestion confirmed by DNA sequencing (MWG, Germany).
Cell culture
Immortalized FSHD patient clones were kindly provided by Dr Vincent Mouly, Institute of Myology, Paris. Clone FSHD-A5 having three D4Z4 units was considered as being contracted whilst clone FSHD-6 having 13 D4Z4 units was considered as being non-contracted and used as a positive control. Cells were proliferated in culture medium composed of 4:1 (v/v) DMEM (Gibco, UK) to 199 medium (Gibco, UK), 20% (v/v) fetal bovine serum (FBS) (Gibco, UK), 50 μg/ml gentamycin, 5 μg/ml insulin, 0.2 μg/ml dexamethasone, 0.5 ng/ml β-FGF, 5 ng/ml hEGF and 25 μg/ml fetuin; reagents were purchased from Sigma, UK unless stated otherwise. Differentiation of the patient cells was induced by replacing the proliferation medium with high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with GlutaMAX (Gibco, UK) and 10 μg/ml insulin. On the second day of differentiation, cells were treated with berberine. RNA extraction (described below) was performed on the fourth day of cell differentiation.
Rhabdomyosarcoma (RD) TE671 cells (ATCC, UK) were cultivated in high glucose DMEM supplemented with GlutaMAX and 10% (v/v) FBS. The cells were treated with 0–120 μM of berberine dissolved in Milli-Q water and transfected with 1 μg of plasmid DNA using Lipofectamine 3000 (Fisher Scientific, UK) at the same time according to the manufacturer's instructions. Twenty-four hours after the transfection and treatment, RD TE671 cells were harvested for RNA extraction.
RT-PCR/qPCR quantification for DUX4 and its target genes
RNA from cultured human cells was extracted using RNeasy kit (QIAgen, UK). RNA from mouse tibialis anterior (TA) muscle (as described in the animal study below) was extracted using RNeasy Fibrous Tissue kit (QIAgen, UK) following tissue homogenization in lysis buffer at 25 Hz for 2–4 min on a TissueLyser II (QIAgen, UK). Purified total RNA was eluted in RNase-free water andquantification using a ND-1000 NanoDrop spectrophotometer (Thermo Scientific, UK). The cDNA was reversed transcribed using GoScript reverse transcription kit (Promega, UK), in a reaction containing 600 ng of total RNA and 500 ng of oligo(dT) primers (Fisher Scientific, UK). The PCR for all DUX4 mRNA isoforms and housekeeping gene B2M was performed using 100 ng of the cDNA and the Platinium Green Hot Start PCR 2× Master Mix kit (Fisher Scientific, UK). The PCR programme included a single denaturation step for 2 min at 92°C, followed by 30 cycles of amplification (denaturation for 1 min at 94°C, annealing for 1 min at 55°C, and extension for 45 se at 72°C), and one final cycle of extension for 5 min at 72°C. PCR products were verified on 2% (w/v) agarose TAE (40 mM Tris–acetate and 1 mM EDTA at pH 8.3) gel, run against HyperLadder IV (Bioline, UK) as a size marker. GeneTools Image Analysis software 4.02 (Syngene, UK) was used in densitometric analysis of gel images. Semi-quantitative analysis of DUX4 mRNA levels was subsequently calculated as the ratio of DUX4 PCR products to B2M PCR products. In qPCR, cDNA products were diluted at 1:20 in qPCR water (Roche, UK). Four-microlitre of diluted cDNA were then amplified using LightCycler480 SYBR Green Master I kit (Roche, UK). Reactions were run on LightCycler480 System, initialized at 95°C for 5 min, followed by 45 cycles at 95°C for 15 s, 60°C for 15 s, 72°C for 15 s. Relative quantification for DUX4 or its target genes (MBD3L2, TRIM43, ZSCAN4, Wfdc3) was performed against housekeeping genes,B2M or GAPDH. Primers used in RT-PCR and qPCR were purchased from Integrated DNA Technologies (Belgium) and are detailed in Table 1.
Table 1.
Primers list
| Target gene | Accession no. | Primer | Sequence (5′-3′) | Product size | |
|---|---|---|---|---|---|
| PCR primers | DUX4 | ENSG00000260596 | Forward | CCCAGGTACCAGCAGACC | 164 bp |
| Reverse | TCCAGGAGATGTAACTCTAATCCA | ||||
| B2M | NM_004048.2 | Forward | CTCTCTTTCTGGCCTGGAGG | 67 bp | |
| Reverse | TGCTGGATGACGTGAGTAAACC | ||||
| qPCR primers | DUX4 | ENSG00000260596 | Forward | CTCTGTGCCCTTGTTCTTC | 98 bp |
| Reverse | TCCAGGAGATGTAACTCTAATCCA | ||||
| Wfdc3 | ENSMUSG00000076434 | Forward | CTTCCATGTCAGGAGCTGTG | 134 bp | |
| Reverse | ACCAGGATTCTGGGACATTG | ||||
| Gapdh | ENSMUSG00000057666 | Forward | TCCATGACAACTTTGGCATTG | 103 bp | |
| Reverse | TCACGCCACAGCTTTCCA | ||||
| MBD3L2 | NM_144614.3 | Forward | CGTTCACCTCTTTTCCAAGC | 142 bp | |
| Reverse | AGTCTCATGGGGAGAGCAGA | ||||
| TRIM43 | NM_138800.1 | Forward | ACCCATCACTGGACTGGTGT | 100 bp | |
| Reverse | CACATCCTCAAAGAGCCTGA | ||||
| ZSCAN4 | NM_152677.2 | Forward | CTGGAGCAGTTTATGATTGG | 162 bp | |
| Reverse | AGCTTCCTGTCCCTGCATGT |
Immunocytochemistry for quantifying cell fusion index
FSHD immortalized myoblast cells were seeded and proliferated on six-well plates precoated with extracellular matrix gel from Engelbreth-Holm-Swarm murine sarcoma (Sigma, UK). The cell culture was immunostained on the fourth day of differentiation. The mouse anti-myosin heavy chain (MF20) primary antibody (DSHB, US) was used at 1:100 dilution. The goat anti-mouse AlexaFluor488 secondary antibody (Life Biotechnologies, UK) was diluted at1:200. Nuclei were stained with 1 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI) (Sigma, UK). Cell images were visualized on an inverted fluorescence Axio Observer D1 microscope (Zeiss, UK) and were captured at 10× magnification by an AxioCam MR3 combined with ZEN image software (Zeiss, UK). The cell fusion index was then calculated by counting the number of nuclei in MF20-positive myotubes containing three or more nuclei and expressed as the percentage of the total number of nuclei present in three random fields per image.
Animals and experimental design
Animal procedures were performed in accordance with the UK Animals (Scientific Procedures) Act, 1986. JAX™ C57BL/6J males were purchased from Charles River, UK. Mice were maintained in a standard 12-h light/dark cycle with free access to food and water. Eight-week old mice were randomized into three groups (n = 6 per group). Two groups were injected with AAV.Spc512.DUX4.3′UTR vectors into both TA muscles at 3e+10 vg/TA and 1 group received volume-matched saline into the same muscle type. Three days afterwards, mice in the AAV groups were injected intraperitoneally with either 10 mg/kg berberine (n = 6) or volume-matched saline (n = 6). Berberine or saline was reinjected every 3–4 days for a total of six injections. Three weeks after AAV administration, mice were put under terminal anaesthesia for in situ TA force measurement prior to collection of the muscle for subsequent analyses.
In situ muscle force measurement and tissue collection
Mice were anesthetized by intraperitoneal injection with a mixture of hypnorm (VetaPharma, UK) and hypnovel (Roche, UK). The distal tendon of TA muscle was dissected and attached to an isometric transducer, Dual-mode muscle lever (Aurora Scientific, Canada), through a loop made of braided silk suture (Harvard Apparatus, UK). The sciatic nerve was isolated and distally stimulated by a bipolar silver electrode using supramaximal square wave pulses at 0.1 ms duration. Data provided by the isometric transducer were recorded and analysed using Dynamic Muscle Control and Analysis Software (Aurora Scientific, Canada). All isometric measurements were obtained at an initial length at which a maximal tension was recorded during the tetanus. Responses to tetanic stimulations at increased pulse frequencies from 10 to 180 Hz were recorded and the maximal force was determined. The specific force was subsequently calculated based on a ratio of the maximal force and the muscle cross-sectional area, and was expressed as g force/cm2. All analyses were performed in a blind manner. Following contractile measurement, TA muscle from one side of the body was frozen immediately in liquid N2 for RNA extraction whilst muscle from the other side was embedded in optimal cutting temperature medium (VWR, UK) and subsequently frozen in liquid N2-cooled isopentane (Sigma, UK) for cryosectioning. All samples were kept at −80°C until use.
Muscle sectioning and immunostaining
Frozen TA muscle was cryosectioned on an OTF 5000 cryostat (Bright, UK) at 10 μm thickness for 12 serial levels through the muscle length. At each level, one transverse section was collected on a SuperFrost slide (VWR, UK) whilst 20 interval sections between the levels were collected into liquid N2-cooled 1.5 ml tubes and stored at −80°C for protein or DNA extraction. Serial section-containing slides were fixed in ice-cold acetone for 10 min and blocked in 1% (w/v) BSA, 1% (v/v) goat serum, 0.1% (v/v) Triton X-100, 1× PBS for 1 h. Subsequent incubation with rat anti-laminin (1:1000) and rabbit anti-collagen VI (1:300, Abcam, UK) antibodies was carried out at 4°C, overnight. Slides were washed three times in 1× PBS, 0.05% (v/v) Tween-20 prior to 1 h incubation with goat anti-rat AlexaFluor568 and goat anti-rabbit AlexaFluor488 (1:1000, Life Biotechnologies, UK). An additional incubation for 15 min with 1 μg/ml DAPI was performed before slides were mounted in Mowiol 4–88. Reagents were purchased from Sigma, UK unless stated otherwise. Mosaic images from each of mid-belly muscle sections were captured on Axio Observer D1 fluorescence microscope and generated by ZEN software (Zeiss, UK).
Histological analyses
Laminin immunostaining was used for identifying fibre perimeter. The minimal Ferret's diameter of an average of 3500 fibres per TA was semi-automatically measured by ZEN imaging analysis software (Zeiss, UK). Automatic analysis of the frequency distribution of the minimal Ferret's diameter was subsequently carried out using Prism5 software (GraphPad, USA). The mean intensity of collagen VI was also scored by ZEN image analysis software and shown as the percentage of the values of saline-injected group obtained in the same way. The number of fibres having internal nuclei was counted manually using ImageJ software (National Institutes of Health, Maryland, USA) and displayed as per mm2 of the muscle section.
Protein extraction and western blot quantification of DUX4 expression
Frozen TA sections collected from cryosectioning were homogenized in lysis buffer (0.15 M NaCl, 0.05 M HEPES, 1% (v/v) NP-40, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 0.01 M EDTA; (reagents were purchased from Sigma, UK) containing 1× protease inhibitors (Roche, UK) at 25 Hz for 1–2 min on a TissueLyser II (QIAgen, UK). Following centrifugation at 14 000 × g, 10 min, 4°C, the supernatant was transferred to fresh pre-chilled 1.5 ml tubes. The total protein was quantified by DC Protein Assay (Bio-Rad, UK). Fifty micrograms protein samples were resolved on 10% Bis–Tris NuPage gels (Life Technologies, UK). Novex Sharp pre-stained protein ladder (Life Technologies, UK) was used as a size standard. The gel was run at 150 V for 1.5 h, subsequently transferred to HyBond nitrocellulose membrane (GE Healthcare, UK) at 30 V for another 1.5 h. The membrane was quickly stained with Ponceau S to check transfer efficiency then horizontally cut at the 80-kDa position of the protein ladder. Two segments were incubated with blocking buffer (5% (w/v) skimmed milk, 1× PBS, 0.2% (v/v) Tween-20) for 1 h. The top segment was then incubated with mouse anti-vinculin antibody (1:10 000, Sigma, UK) whilst the bottom segment was incubated with mouse anti-V5 tag SV5-Pk1 antibody (1:1000, Abcam, UK) at 4°C overnight. Subsequent incubation with goat anti-mouse IRDye800CW (1:10 000, LI-COR Biosciences, UK) was carried out for 1 h at room temperature. The blots were visualized on an Odyssey Infrared Imaging System (LI-COR Biosciences, UK). Densitometric analysis of vinculin- and DUX4.V5 tag-positive bands was performed using Image Studio software (LI-COR Biosciences, UK). The values of DUX.V5 tag intensity were normalized to the values of corresponding vinculin intensity and expressed as percentage of the values of saline-injected group obtained in the same way.
AAV genome qPCR assay
DNA from TA transverse muscle sections were extracted using DNeasy Blood and Tissue kit (QIAgen, UK) following tissue homogenization at 25 Hz, for 1–2 min, on a TissueLyser II (QIAgen, UK). Extracted DNA samples were diluted 1:20 and the AAV.Spc512.DUX4.3′UTR plasmid was used for making the qPCR standard curve. The number of AAV copies was calculated based on the standard curve and expressed as AAV copies per ng of total DNA.
Statistical analysis
Data were analysed using Prism5 software (GraphPad, USA) and is shown as the means ± S.E.M. ‘N’ refers to the number of independent treatments or number of mice per group. Comparisons of statistical significance were assessed by one- or two-way ANOVA followed by Bonferroni's post-hoc test. Significant levels were set at *P < 0.05, **P < 0.01, ***P < 0.001.
RESULTS
Novel GQ motifs are predicted to form in the enhancer, promoter and transcript of DUX4
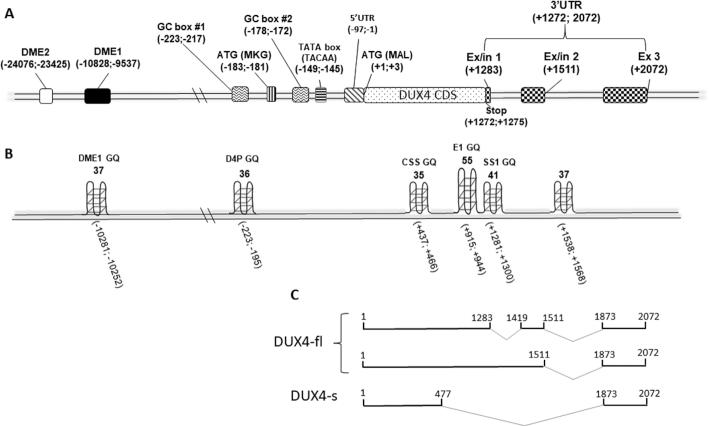
DUX4 enhancers, promoter and transcript-coding DNA regions have been previously described (14,48,49). The GQ predicting software, QGRS Mapper (50) was used to analyse for the presence of the putative motifs within these sequences. The analysed sequences with G-scores of ≥30 were considered to be strong candidates for GQ formation. Two myogenic enhancers are known to drive the DUX4 expression in skeletal muscle, namely DME1 and DME2 (48) (Figure 1A). However, a single putative GQ motif was predicted within the DME1 only (Figure 1B). In the promoter region, a single potential GQ motif was found 195 bp upstream of the DUX4 start codon (Figure 1A, B). The DUX4 transcript-coding sequence has been predicted to be particularly enriched in putative GQs. (Figure 1A, B). These mapped to the exon1 splice site of a DUX4 short isoform, to the exon1/intron1 boundary and to the 3′UTR of the DUX4 full length transcript (Figure 1C).
Figure 1.
Schematics of GQ motifs prediction scores. (A) Location of the genomic elements of DUX4 relative to the start codon (ATG) was mapped based on the data acquired from GenBank (accession number: AF117653) and previous publications: (48, 49). (B) Score prediction of the putative GQ structures was performed using QGRS Mapper software, aligned with the biologically relevant genetic signal elements of DUX4 genomic locus. (C) DUX4 transcript sequences were derived from Ensembl (ENSG00000260596). Abbreviations: DME1 and 2, DUX4 myogenic enhancers 1 and 2; DUX4-fl, DUX4 full length; DUX4-s, DUX4 short; Ex, exon; In, intron; CDS, coding DNA sequence; UTR, untranslated region; GQ, G-quadruplex.
Formation of DUX4 GQs
In order to establish whether the in silico predicted GQ-forming oligonucleotide sequences within the enhancer, promoter and transcript of DUX4 would form in solution, nuclear magnetic resonance (NMR) and circular dichroism (CD) spectroscopy were performed. The proton NMR spectrum demonstrated that the DNA oligonucleotides selected from the enhancer (DME1 GQ) and promoter (D4P GQ) show signals between 10 and 12 ppm, indicating the presence of Hoogsten hydrogen bonds that are characteristic of GQ structures (Figure 2A). The RNA oligonucleotides derived from the 5′-end of the exon 1 (E1 GQ) and exon 1/intron 1 boundary (SS1 GQ) also produced a broad GQ-specific signal within the 10–12 ppm range in the 1H NMR spectrum (Figure 2A). However, the RNA oligonucleotide sequences predicted to form a stable GQ structure within the cryptic splice site (CSS GQ) region of the DUX4 short isoform produced a clear set of signals >12 ppm, characteristic of the presence of Watson–Crick hydrogen bonding within the secondary structure (e.g. hairpin) of the annealed sequence indicating that the CSS-GQ was not a GQ motif (Figure 2A).
Figure 2.
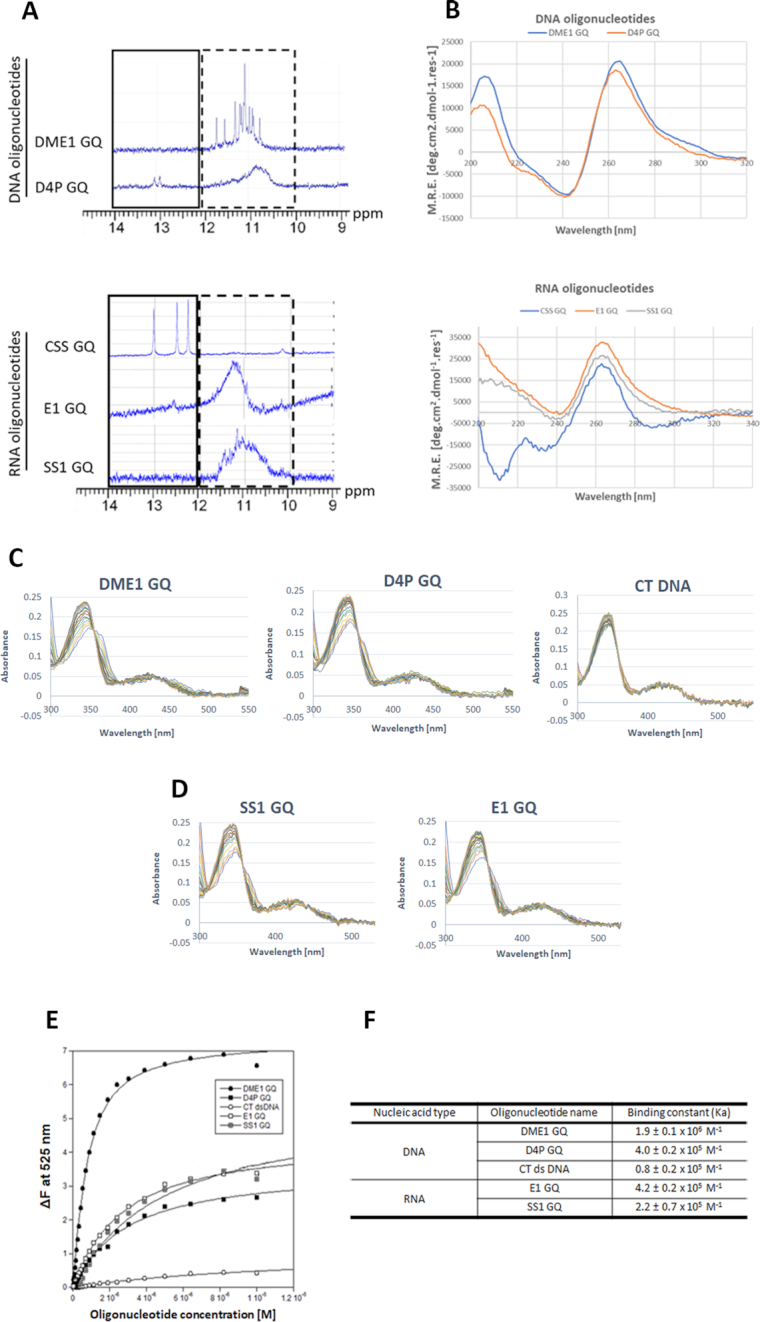
Biophysical characterization of DNA and RNA secondary structures and berberine binding of the predicted GQs in the DUX4 myogenic enhancer 1 (DME1), promoter (D4P) and transcript sequences. (A) 1H NMR spectrum of the oligonucleotides (4 μM) recorded at 800 MHz and 298 K showing groups of resonances characteristic for Watson-Crick (solid line box 12–14 ppm highlighting non-quadruplex structure signals) and Hoogsten H-bonded bases (dashed line box 10–12 ppm highlighting quadruplex structure). All of the analysed sequences produced signals characteristic of quadruplex conformations, except for the CSS GQ sequence which gave sharp signals (12–14 ppm) for a double stranded structure. (B) All of the analysed oligonucleotide sequences (except for the CSS GQ) at 4 μM using far-UV circular dichroism spectroscopy at 25°C reveal ellipticity maxima at 265 nm and ellipticity minima at 240 nm indicative of a parallel GQ structure formation by the oligonucleotides. Recorded ellipticity maximum and minimum for the CSS GQ oligonucleotide sequence was 260 nm and 210 nm, respectively, which is characteristic for A-form RNA secondary structure. UV–Vis absorbance spectra of 10 μM berberine in the presence of (C) DNA and (D) RNA oligonucleotides at a range of concentrations of 0–10 μM. The GQ-forming DNA oligonucleotide sequences, including: DME1 GQ and D4P GQ; E1 GQ and SS1 GQ were the RNA GQ-forming oligonucleotide sequences used in the analysis. The negative control used was CT DNA. (E) Plot of ΔF emission at 525 nm of (0.5 μM) berberine versus 0–10 μM DNA or RNA oligonucleotide concentrations was used to calculate binding constant (Ka) (F) of berberine to each of the oligonucleotide. Abbreviations: CT, calf thymus; DME1, DUX4 myogenic enhancer 1; D4P, DUX4 promoter; E1, exon 1; SS1, splice site 1; CSS, cryptic splice site, GQ, G-quadruplex.
Further CD spectroscopy analysis confirmed the NMR results. Except for the CSS GQ, all of the analysed oligonucleotide sequences showed a characteristic absorbance pattern for a parallel GQ structure with ellipticity maximum and minimum at 265 and 240 nm, respectively (51) (Figure 2B). In contrast, the CD measured absorbance of the CSS GQ RNA oligonucleotide had ellipticity maximum and minimum at 260 nm and 210 nm, respectively (Figure 2B), characteristic of a duplex structure.
Binding of DUX4 GQs by berberine
Berberine has been widely studied for its photochemical binding properties to different types of nucleic acid secondary structures, including: double stranded DNA, tRNA, triple helical nucleic acids and GQs (46, 52–54). The characteristic absorbance maxima for berberine have been reported to be 340 and 420 nm using UV–Vis spectroscopy (46, 55). In addition, berberine can act as a fluorophore when bound to GQs and emits light at 525 nm when excited with a wavelength of 355 nm (46, 55). Upon addition of nucleic acids to the berberine solution, the characteristic absorbance pattern changes and fluorescence increases (46). This can be utilized to gain an important insight into interaction between the ligand and DUX4 GQs. All of the analysed DNA and RNA GQ oligonucleotides investigated for binding interactions with berberine using UV-Vis and fluorescence spectroscopy were K+ stabilized.
UV–Vis analysis of the berberine solution demonstrated that addition of increasing amounts of DME1 GQ and D4P GQ DNA oligonucleotide resulted in λmax hypochromicity of 26% and 24%, respectively (Figure 2C). The E1 GQ and the SS1 GQ RNA oligonucleotides had comparable λmax hypochromicity of 28% and 30%, respectively (Figure 2D). Furthermore, a notable red shift of 6 and 8 nm for the two peaks at 340 and 420 nm was recorded for the DME1 GQ and D4P GQ, respectively. The E1 GQ shows a red shift of 6 nm at 340 nm peak only, whereas addition of the SS1 GQ resulted in 4 and 12 nm red shift at both 340 and 420 nm peaks, respectively (Figure 2D). The characteristic red shift of absorbance is typically associated with π–π interaction between the ligand and DNA/RNA bases, indicative of ends stacking (in case of GQ DNA/RNA) or intercalative (in case of double stranded DNA) binding mode (55). The CT DNA was used as a negative control and its addition to the berberine solution produced a considerable λmax hypochromicity of 14%, but no red shift was recorded at any of the peaks (Figure 2C). This suggests that berberine is a not a good duplex DNA intercalator, but rather a groove binder, as previously reported (46).
In order to study the binding event, fluorescence emission spectra for berberine were recorded in the absence and presence of increasing amounts of the DNA/RNA oligonucleotides (Figure 2E). Berberine alone in Tris buffer was non-fluorescent. Increasing concentration of the DME1 GQ and D4P GQ DNA oligonucleotide resulted in up to 7- and 2.5-fold increase of fluorescence intensity of berberine, respectively (Supplementary Figure S1). Addition of the E1 GQ and SS1 GQ RNA oligonucleotides increased the fluorescence emission of berberine by 3- and 4-fold, respectively (Supplementary Figure S1). The CT duplex DNA titration resulted in marginal increase of the fluoresce intensity of ∼0.5-fold (Supplementary Figure S1). The change in fluorescence emission of berberine at 525 nm recorded for each of the analysed oligonucleotides was used to produce the binding isotherm (Figure 2E). Analysis of the isotherm demonstrated that berberine has the highest binding affinity at 1.9 ± 0.1 × 106 M−1 towards the DME1 GQ (Figure 2F), which is similar to the previously reported binding affinity (i.e. 1.2 ± 0.2 × 106 M−1) of berberine to human telomeric quadruplex (46). Furthermore, binding affinity of berberine to D4P GQ DNA and E1 GQ oligonucleotides was measured at 4.0 ± 0.2 × 105 and 4.2 ± 0.2 × 105 M−1, respectively (Figure 2F). Lower binding affinity was recorded for the SS1 GQ RNA oligonucleotide at 2.2 ± 0.7 × 105 M−1 (Figure 2F). Binding to CT DNA showed the lowest affinity at 0.8 ± 0.2 × 105 M−1, indicating that berberine is a weak binder of the duplex DNA, as shown previously (56).
Berberine downregulates expression of DUX4 mRNA and its downstream genes in FSHD cells
To investigate the effects of berberine on DUX4 expression, FSHD immortalized myoblast cells were treated with berberine, one day after induction of cell differentiation. RNA was extracted three days later and RT-PCR quantification performed. Gel electrophoresis verification of PCR products from patient cells (FSHD-A5) and control cells (FSHD-6) is shown in Figure 3A, B. Subsequent semi-quantitative analysis of the DUX4 PCR products (Figure 3C) demonstrated downregulation of the DUX4 mRNA level in a dose-dependent manner with berberine treatment. At the lowest tested dose (i.e. 5 μM), the recorded DUX4 mRNA expression was not statistically significant (P > 0.05). A significant downregulation of the DUX4 was reached at the 10 μM dose of berberine (P < 0.01). At the highest berberine concentration tested (i.e. 25 μM), there was the most significant downregulation of the DUX4 PCR product at 78 ± 8% (P < 0.0001). No DUX4 expression was detected in the control cell line.
Figure 3.
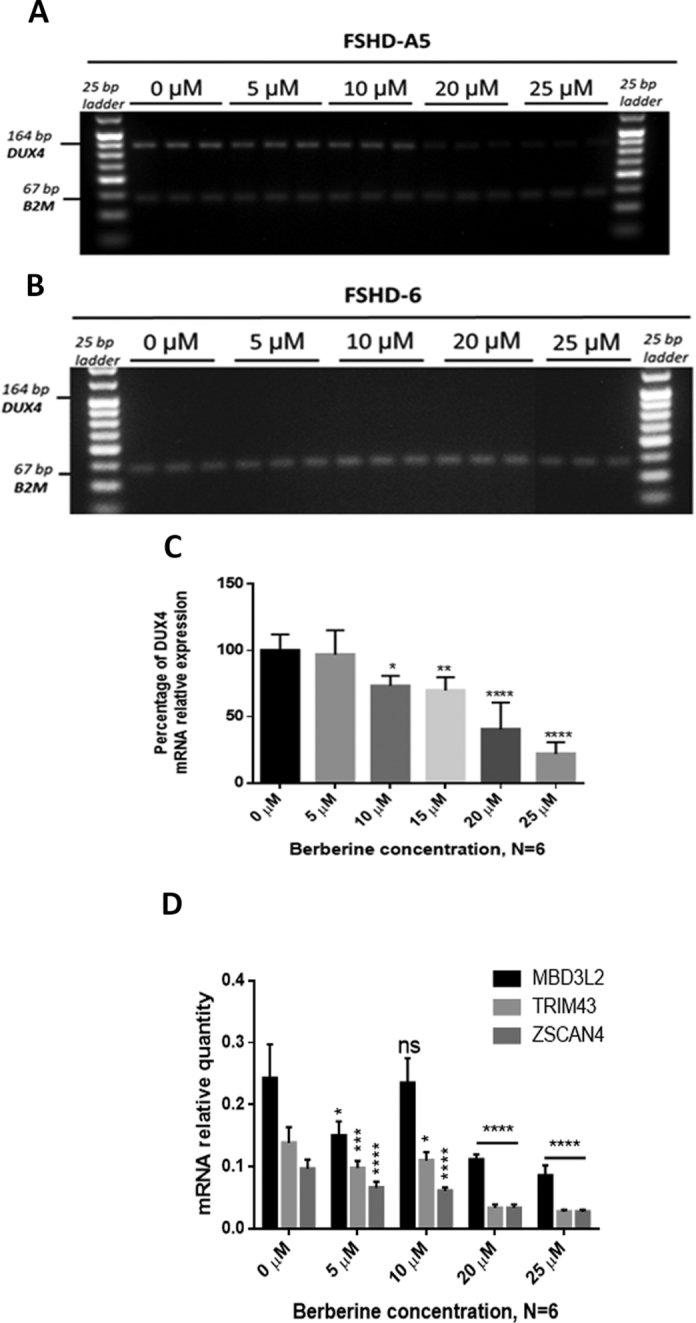
Expression of DUX4 and downstream genes in the presence of berberine. (A) Immortalized FSHD-A5 (DUX4 positive) and (B) FSHD-6 (DUX4 negative) myoblast cell cultures were treated with berberine in a dose-dependent manner on the second day of cell differentiation induction and incubated for an additional 48 h before harvesting the total RNA. Representative gel electrophoresis results display RT-PCR analysis using the primers detecting all DUX4 full-length isoforms. B2M was used as the housekeeping gene. (C) Semi-quantitative analysis of the DUX4 mRNA expression as a ratio of DUX4 band intensities normalized to their corresponding housekeeping band intensities shows a significant decrease of DUX4 mRNA level in a dose-dependent manner with berberine. Data are expressed as percentage of the level of control cells having no berberine added, N = 6 per concentration. (D) Expression levels of ZSCAN4, TRIM43 and MBD3L2 were measured by RT-qPCR in FSHD-A5 cell culture. Cells were treated with berberine on the second day of cell differentiation and RNA was harvested on the fourth day of differentiation. B2M was used as a housekeeping gene. Independent treatment at each concentration was performed, N = 6. Statistical analyses in (C, D) were by one-way ANOVA, *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001. Error bars represent S.E.M.
DUX4 is a transcription factor that affects expression of several downstream genes, including ZSCAN4, TRIM43 and MBD3L2 (57, 58). Since DUX4 expression levels in FSHD patient muscle tissue are very low and difficult to detect, the signature expression of these downstream genes can serve as a useful DUX4 biomarker (59). Hence, we performed additional RT-qPCR analysis of these genes to determine the effects of berberine on their expression. As shown in Figure 3D, mRNA levels of all of the analysed genes were significantly downregulated by berberine, in a dose-dependent manner compared to untreated samples. No detectable levels of expression in any of the analysed downstream genes was seen in the control cell line (data not shown).
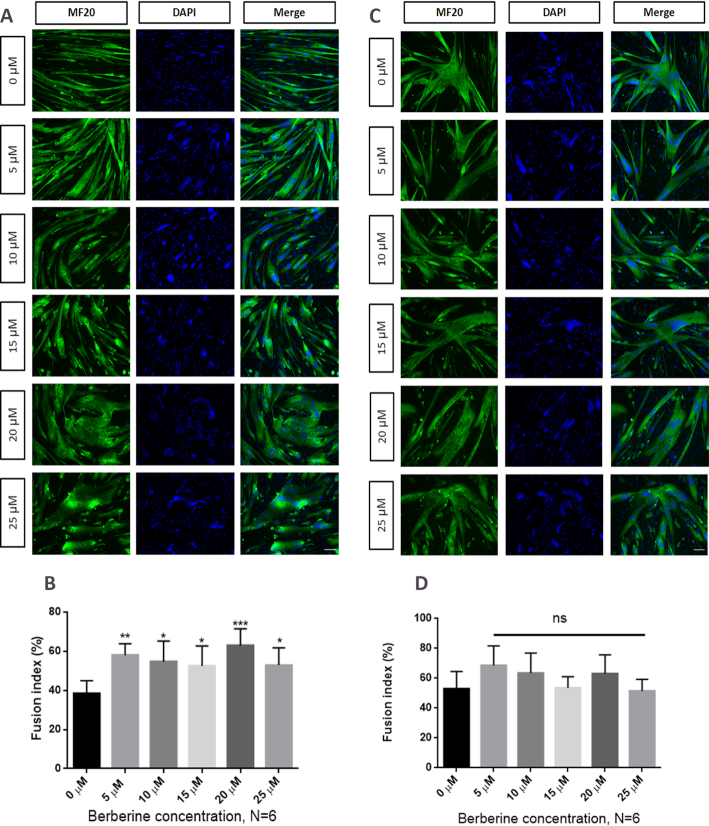
Berberine increases fusion index of FSHD myoblast cells
One of the mechanisms by which a rare protein such as DUX4 can cause myopathy is its ability to diffuse across the cytoplasm of myotubes to the neighbouring nuclei and lead to aberrant activation of many downstream genes (57).Therefore, in order to ensure that the berberine downregulation of the measured DUX4 downstream genes (i.e. ZSCAN4, TRIM43 and MBD3L2) was not a result of an indirect effect on myotube formation, the fusion index of the FSHD myotubes (clones A5 and 6) was assessed. Cells were immunostained with an antibody specific for the myosin heavy chain (MF20), and the nuclei were stained with DAPI (Figure 4A, B). The nuclei present in the MF20 positive myotubes were counted and divided by the total number of nuclei present in all cells in each image field. Quantification of the fusion index, expressed as the percentage of myotube-related nuclei versus total number of nuclei, is shown in Figure 4B, D. A statistically significant increase in the index of fusion was observed in FSHD-A5 cells at all tested concentrations of berberine (Figure 4B), suggesting that downregulation of DUX4 downstream genes was not caused by the impairment of the myotube formation. This idea was further supported by the fact that the control FSHD-6 cells did not show any statistically significant change (P > 0.05) in the fusion index with berberine treatment (Figure 4B, C).
Figure 4.
Fusion index of immortalized FSHD myotubes in the presence of berberine. Immortalized FSHD myotubes (A). FSHD-A5 or (C). FSHD-6 were treated with berberine at the second day of differentiation. Myosin heavy chain and nuclei were stained at the fourth day of differentiation with MF20 antibody and DAPI, respectively. Scale bar: 100 μm. Quantifying cell fusion index results for (B) FSHD-A5 and (D) FSHD-6, with nuclei counted in myotubes containing more than two nuclei and expressed as a percentage of the total nuclei present. Independent treatment at each concentration was performed, N = 6, *P < 0.05; **P < 0.01; ***P < 0.001, one -way ANOVA. Error bars represent S.E.M.
GQ ligands downregulate transiently expressed DUX4
RD cells, derived from human embryonal rhabdomyosarcoma, have been extensively used for studying muscle regulatory pathways and molecular responses to ectopic expression of DUX4 (49,58,60,61).
The levels of DUX4 driven by the tMCK promoter in the transfected RD cells were significantly downregulated by treatment with berberine as detected by the RT-qPCR (Figure 5A). To strengthen the idea that the berberine-mediated downregulation of DUX4 is mediated through targeting of DUX4-related GQs, another DNA/RNA GQ-specific ligand, PhenDC3 (60,61), was tested. As a result, the transfected cells expressing DUX4 driven by the tMCK promoter, showed significant downregulation of DUX4 expression when treated with PhenDC3 (Figure 5B). In contrast, the expression levels of DUX4 were not affected by the presence of ampicillin, a control molecule with no known GQ- binding properties (Figure 5B). These findings suggest that the downregulatory effect of the GQ ligands on DUX4 mRNA expression is in part related to targeting of the GQ motifs present in the coding (sense) strand of the genomic DNA sequence, and/or in the RNA transcript sequences.
Figure 5.
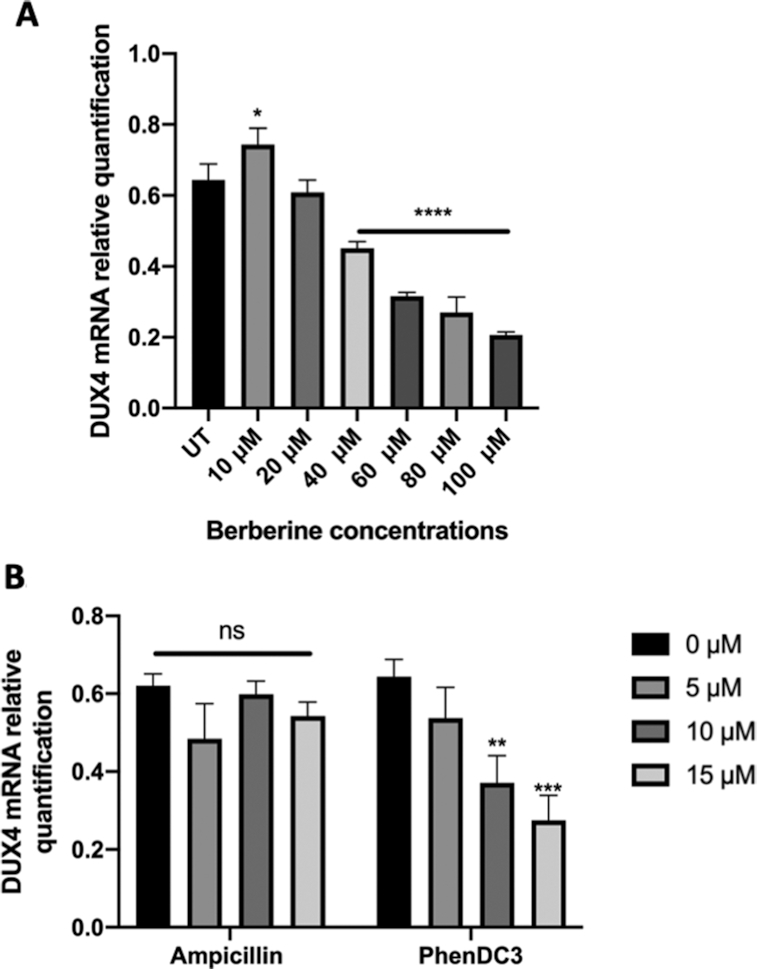
Expression levels of DUX4 transcript in RD TE671 cells treated with GQ ligands. RD TE671 cells were transfected with of DUX4-expressing plasmid and simultaneously treated with GQ ligands: berberine (A) and PhendDC3 (B). Ampicillin was used as control (B). Total RNA was harvested 24 hours after transfection and treatment. Expression levels of mRNA were measured using RT-qPCR, with GAPDH used as a housekeeping gene. Increasing concentrations of (A) berberine and (B) PhenDC3 downregulated DUX4 expression driven by the tMCK promoter. (B) Ampicillin control treatment had no effect on DUX4 expression driven by the tMCK promoter. Treatments were performed independently with N = 3, **P < 0.01; ****P < 0.0001, one-way ANOVA. Error bars represent S.E.M.
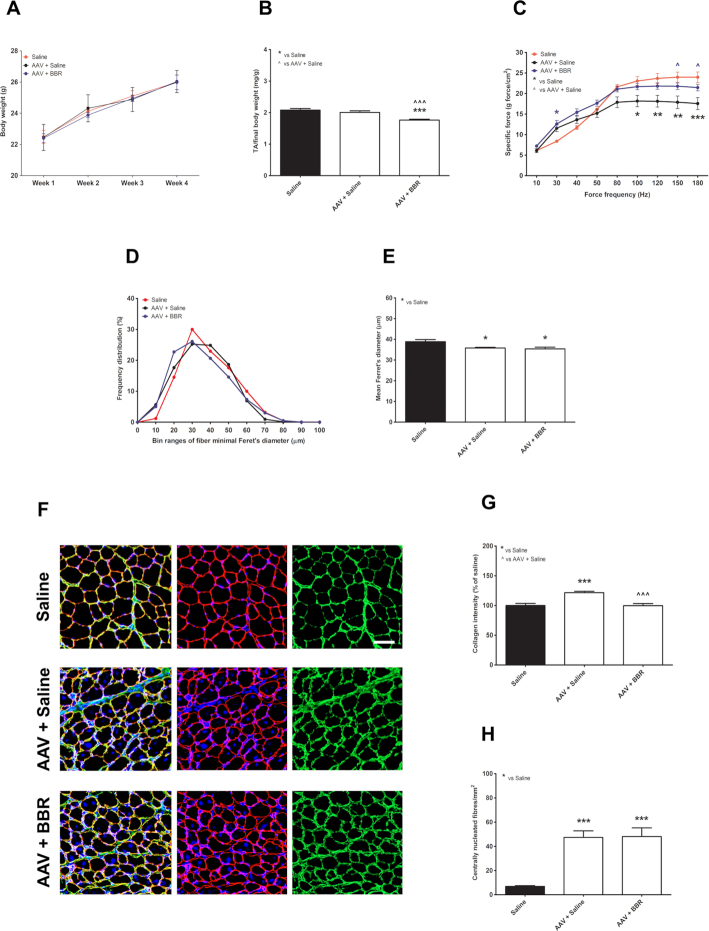
Berberine treatment improves muscle function and suppresses muscle fibrosis
Following promising results from cell culture work, we investigated the downregulatory effect of berberine in mice overexpressing DUX4. The body weight was scored weekly, showing a similar increase in weight among animal groups (P = 0.9995) (Figure 6A). However, in comparison to saline-injected group (considered as positive control), the TA mass normalized to the final body weight (Figure 6B) in mice injected with AAV.Spc512.DUX4.3′UTR and then treated with berberine (AAV+BBR) reduced 15.2 ± 1.4% (P = 0.0016), while mice injected with the AAVs and then saline (AAV+saline) showed 4.0 ± 2.1% TA mass reduction (P = 0.2429). Despite this, the muscle specific force of mice in AAV+BBR group was significantly stronger than the force of AAV+saline-treated muscle and was normalized towards the property of the positive control (Figure 6C). For instance, at the force frequency of 180 Hz, 26.7 ± 6.1% force drop was calculated in AAV+saline group as compared to positive control (P = 0.0015) whereas with berberine treatment the force was significantly improved (P = 0.0418) and remained at 89.5 ± 3.4% of the level of the positive control (P = 0.1129).
Figure 6.
Effects of berberine treatment in skeletal muscles injected with AAVs overexpressing DUX4. (A) Weekly-recorded body weight of experimental mice. (B) Normalized mass of tibialis anterior (TA) muscle to the final body weight. (C) In situ TA specific muscle force calculated as a ratio of the maximal force at each pulse frequency ranging from 10 to 180 Hz and the muscle cross-sectional area. (D) Frequency distribution of the minimal Feret's diameter of TA fibres is shown as percentage of the total fibre numbers. (E) Mean of the minimal Ferret's diameter of TA fibres. (F) Representative images of TA muscles following immunostaining for laminin (red), collagen VI (green), and nuclei staining with DAPI (blue); scale bar = 100 μm. (G) Evaluation of fibrosis in TA muscle, expressed as percentage of the intensity of collagen VI of saline-injected group. (H) Quantification of the number of centrally nucleated fibers per mm2 of TA muscle sections. All data are shown as means ± S.E.M.; error bars represent the S.E.M; N = 6 per group. Statistical comparison was by one-way ANOVA (in B, E, G, H) or two-way ANOVA (in C) followed by Bonferroni's post-hoc test; *P < 0.05, **P < 0.01, ***P < 0.001.
To further investigate the effect of AAV-mediated DUX4 overexpression and the potential therapeutic effect of berberine treatment on muscle histopathology, we performed co-immunostaining for laminin and collagen VI on TA transverse muscle sections. Laminin staining facilitated the identification of the myofibre sarcolemma for subsequent analysis of the minimal Ferret's diameter of myofibres and the frequency distribution of the fibre size (Figure 6D, E). Both AAV-injected groups displayed ∼10% reduction in the mean of the Ferret's diameter compared to the size of the positive control, P = 0.0109 for AAV+saline or P = 0.0236 for AAV+BBR. Staining for collagen VI was used to examine the level of excessive muscle fibrosis. DUX4 transgenic expression led to 21.7 ± 2.3% increase in the intensity of collagen VI as compared with the level of positive control (P = 0.0005). However, berberine treatment substantially prevented fibrosis deposition, with the level quantified at 99.7 ± 3.5% of the positive control, P = 0.9574 (Figure 6G). Muscle necrosis following DUX4 expression could be compensated for by innate muscle regeneration. Hence, in combination with DAPI staining for nuclei, we further counted the number of centrally nucleated fibres, which is an important hallmark of the muscle undergoing repeated degeneration-regeneration cycles (62). As presented in Figure 6H, we observed 47.3 ± 5.4% and 48.1 ± 7.2% myofibers having central nuclei in AAV-saline- and AAV-BBR-injected muscle groups, respectively, with no difference between the groups despite berberine treatment (P = 0.9361). Representative immunostained images are shown in Figure 6F.
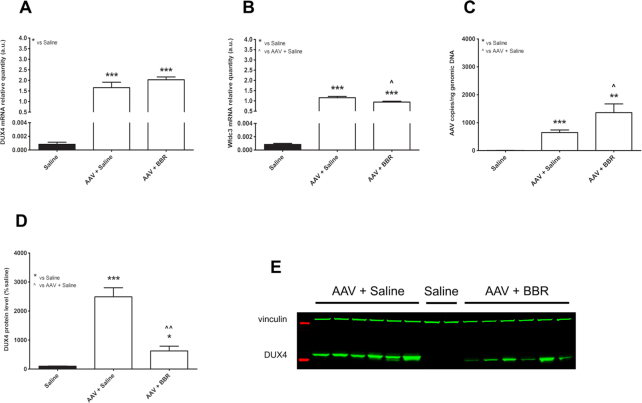
Expression of DUX4 and its target gene is reduced following berberine treatment
To explain the encouraging effects of berberine on muscle function and muscle histopathology, we carried out RT-qPCR quantification for DUX4 and one of its target genes, Wfdc3. As predicted, the mRNA levels of DUX4 and Wfdc3 in both AAV-injected groups were high, almost 2e+05-fold of the levels of saline group (Figure 7A, B). Berberine administration downregulated by 18.1 ± 3.4% the mRNA level of Wfdc3 seen in AAV+saline group (P = 0.0198). Although berberine had no effect on the DUX4 mRNA level (P = 0.2250), the treatment greatly reduced the level of DUX4 protein overexpressed in AAV+saline muscle by 74.9 ± 6.6% (P = 0.0019) (Figure 7D). Illustrative western blot analysis shown in Figure 7E confirms this. To assess the number of AAV copies remained, we performed additional qPCR quantification for DUX4 in DNA extracted from TA muscle. As shown in Figure 7C, the copies of AAVs in berberine-treated muscle was 2.1 ± 0.5-fold higher than in the other AAV-injected group (P = 0.0440).
Figure 7.
Effects of berberine treatment on DUX4 and its target gene. RT-qPCR results showing mRNA relative quantification of (A) DUX4 or (B) Wfdc3, against Gapdh. (C) qPCR assessment for AAV copies, expressed as the number of copies per ng of genomic DNA. (D) Western blot quantification for DUX4 protein levels, expressed as percentage of the value of saline-injected group. (E) Representative Western blot analysis showing AAV-mediated DUX4 expression. Each lane displays a sample from an individual muscle. Proteins from saline-injected muscle were loaded as controls. Vinculin was used as loading control for Western blot. Data are shown as means ± S.E.M.; error bars represent the S.E.M.; N = 6 per group. Statistical comparison was by one-way ANOVA followed by Bonferroni's post-hoc test; *P < 0.05, **P < 0.01, ***P < 0.001.
Taken together, systemic treatment with berberine, although was insufficient in downregulating DUX4 mRNA level, decreased AAV-mediated expression of DUX4 protein and its target gene Wfdc3. The effect consequently inhibited muscle fibrosis deposition and rescued muscle function in the treated mice.
DISCUSSION
The current consensus states that the postnatal expression of DUX4 is a key factor behind the pathophysiology of FSHD (63). Here, we describe the presence of novel GQ motifs within the enhancer, promoter and transcript of DUX4. Furthermore, we demonstrate that by using berberine, a GQ stabilizer, we could downregulate the expression of DUX4 and its target genes at the mRNA level in FSHD immortalized patient cells, without inhibition of myotube formation by the treated cells. The inhibitory effect of berberine on DUX4 expression was additionally demonstrated in mice overexpressing DUX4, with animals systemically treated with berberine displaying reduction in DUX4 protein expression and rescue of muscle function. Our data demonstrate for the first time that putative GQ forming sequences are present in the DUX4 locus. Targeting these structures, including those present within the DUX4 transcript, with a stabilizer such as berberine could potentially lead to downregulation of DUX4 expression and disease-related pathological changes, providing a novel therapeutic strategy for the treatment of FSHD.
The commonly set upper limit for GQ central loop length used in bioinformatic studies is seven nucleotides. However, in our study we found that all of the selected GQ-forming sequences within the enhancer, promoter and transcript (except for the SS1 GQ) exceeded that limit (Table 2). A body of accumulating experimental evidence has shown that extensive loop sizes did not prevent formation and stability of GQs. For example, a nine-nucleotide propeller loop was found within the stable GQ structure of the human CEB25 mini-satellite locus as determined by NMR studies (64). Furthermore, a very long 26-nucleotide loop, stabilized with a GC-based hairpin has been found to form within a promoter sequence of hTERT gene (65); or the human BCL-2 gene contained GQ-forming sequences containing a 13-nucleotide central loop (66). This may explain why in the study of Tsumagari et al. (18), when the GQ search algorithm was limited to GQ containing loops not exceeding seven nucleotides in length, no putative GQ structures were identified within the transcript of DUX4. In addition, we find that the presence of the C-tract within the loops of RNA GQ sequences has a strong inhibitory effect on the structure formation compared to the DNA counterparts. The CSS GQ RNA oligonucleotide despite a high G-score in QGRS Mapper analysis, was shown to form a stable hairpin structure in solution due to the presence of a run of C residues within its sequence. On the other hand, presence of the C-tract within the D4P GQ DNA sequence has not interfered with the GQ motif formation in solution. Together, our bioinformatic analysis support the idea that the consensus of GQ loops composed of maximum 7 bases should be revaluated, as has been previously suggested (67), and the scoring system, particularly in identifying the RNA GQs, should account for the presence of C-tracts present within the loop or flanking sequences of the motif (68).
Since RNA GQs are implicated in mRNA transcription and processing (38), it is possible that the berberine treatment mediates downregulation of DUX4 mRNA by interference with these processes. For example, during transcription, the newly produced pre-mRNA strand can fold into an intermolecular GQ structure with the coding (sense) DNA strand (69). The RNA transcript and the coding DNA strand require as little as two tandem G-tracks to form a stable GQ structure (70). Formation of such a hybrid RNA/DNA GQs has been predicted and confirmed in vitro using T7 RNA polymerase transcription, site specific mutagenesis and reporter-based transient transcription assays (71, 72). These studies conclude that formation of RNA/DNA hybrid GQ structures act as in-cis elements mediating transcription inhibition, providing a potential explanation behind the berberine-mediated DUX4 downregulation at the mRNA level.
RNA GQ motifs found in open reading frames have also been found to act as ‘roadblocks’, inhibiting progression of ribosomes along the mRNA, leading to significant decrease in protein synthesis efficiency (73). The RNA GQs downstream of the start codon have been found to be particularly effective in blocking translation, as demonstrated in the context of KMT2A and MLL proto-oncogenes, where the in-line probing combined with the G-A mutagenesis and the luciferase assay showed over 75% reduction in protein synthesis to be RNA GQ structure related (74). Presence of stable RNA GQ structures within the coding sequence of DUX4 could perhaps also negatively affect its translation, providing a potential explanation for the low levels of DUX4 protein present in the patient cells.
Berberine treatment has been demonstrated to lead to DUX4 mRNA downregulation in FSHD immortalized patient cells and in RD TE671 cells transiently expressing DUX4. Interestingly, two phenotypes of FSHD primary myotubes were previously reported: the atrophic and disorganized (75, 76). The atrophic phenotype was characteriszed by a narrow elongated myotubes with neatly aligned nuclei, whereas the disorganized myotubes were described as giant structures containing large clusters of nuclei. An antisense-mediated downregulation of DUX4 in FSHD primary cell culture has been demonstrated to lead to the prevention of the atrophic, but not the phenotype of disorganized myotube formation (76). We also show that berberine treatment leads to phenotypic switch from atrophic to disorganized state in FHSD immortalized myotubes. The clustering of nuclei in the berberine treated FSHD negative control cells was not as apparent, suggesting that the phenotypic switch could be DUX4 related (76). However, since berberine has been found to have a range of pharmacological activities, including anti-inflammatory, antimicrobial and anti-tumour effects, it is important to consider non-DUX4 specific effects (77). This is also further supported by the fact that berberine, apart from the DUX4 transcript, has a high affinity binding towards the GQ-forming sequences within the DUX4 enhancer and promoter regions. Although there is currently no feasible method to target specific GQs within the genome, new small molecule chemistries have begun to emerge that have high binding selectivity to GQs over duplex DNA or in some cases can even discriminate between RNA and DNA GQ structures. Two small-molecule ligands, carboxy pyridostatin and RGB1, have been recently developed to specifically target RNA GQs (78, 79). Since the DUX4 RNA GQs are demonstrated to be a likely target of berberine that leads to its mRNA downregulation, it would be interesting to evaluate these RNA GQ specific-ligands and their ability to suppress expression of the toxic transcription factor. The RBG1 is especially interesting candidate molecule, due to its ability to specifically bind RNA GQ not only over the DNA counterparts, but also other RNA secondary structures.
A previously developed AAV-overexpressing mouse model (80) was used to study the potentially therapeutic effects of berberine on DUX4 expression. Although overexpression of DUX4 through a single intramuscular AAV delivery does not truly genocopy FSHD aspects or reflect physiological levels of DUX4, DUX4-induced muscle degeneration/regeneration, accompanied by abundant small myofibres with centrally located nuclei, inflammatory invasion, and fibrosis deposition has been reported (81). However, the reduction in muscle strength observed two weeks after AAV administration is resolved a week later by innate muscle regeneration. This clearly points out the complexity and the main drawback of AAV-overexpressing models. Once DUX4 is expressed, it causes damage to the muscle. Muscle deterioration, in turn, removes AAV particles, and as a result reduces the toxicity of DUX4. Since AAV-mediated expression is transient, regenerated fibres are not transduced with AAVs. Thereby, the repeated muscle degeneration-regeneration cycles eventually abolish AAV-based DUX4 and return the muscle to its ‘normal’ state. The presence of centrally nucleated fibres is an indicator of muscle regeneration, although after time the nuclei relocate to the myofibre periphery (82). As predicted, in the AAV-injected TA muscle at the end of the 4-week study, we observed normalization of the muscle mass to wild-type level, reduction in the fibre diameter, elevation in the percentage of fibres having centralized nuclei, and increase in muscle fibrosis. With berberine treatment, the specific muscle strength was significantly improved and normalized to the wild-type property and collagen VI deposition was completely prevented. In coherence with the reduction in DUX4 protein and downstream gene levels, these results clearly demonstrate the therapeutic effect of berberine in the mouse model used.
Despite these promising results, the muscle mass in berberine-treated TA surprisingly remained lower than the level in the wild-type or in the muscle receiving only AAVs. In FSHD, fatty infiltration, fibrosis, and atrophy of the muscle are proposed as the causes of reduction in the muscle cross-sectional area and contractile muscle function, contributing to muscle weakness (83). Although muscle regeneration can compensate for muscle loss, regenerative myofibres at the early stage of myogenesis (before nuclei repositioning to periphery) are not fully functional, hence, hampering normal muscle contraction. Given that the specific strength in berberine-treated muscle was comparable to the wild-type level, decrease in the muscle mass in association with higher percentage of small-diameter fibres (<30 μm), as compared to AAV-injected muscle, suggests a delay in muscle degeneration and regeneration. This consequently indicates suspension in DUX4 toxicity-mediated muscle damage. As mentioned above, muscle turnover dilutes AAV genome, ameliorating DUX4 pathogenesis. Therefore, myofibres rescued by berberine continue carrying the AAVs but may be destroyed over the time because the protective effect of berberine is not complete. Hence berberine treatment might potentially slow down DUX4-related damage to the muscle. This hypothesis is ascertained by the fact that the number of AAV copies remaining in the muscle of berberine-treated mice was higher than the level in animals without receiving berberine.
In summary, identification of GQ structures as potential regulators of DUX4 expression, provides a new platform for advancing our understanding of the molecular mechanisms underlining the FSHD pathogenesis. Furthermore, these structures serve as novel drug targets for development of future therapeutic strategies against the disease.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Prof. S. Harper, Ohio State University School of Medicine, USA for the AAV.CMV.DUX4 plasmid construct and Dr Julie Dumonceaux, UCL, UK for the eGFP.C1.ΔSV40.3′UTR DUX4 construct. We also acknowledge Dr Vincent Mouly, Institute of Myology, Paris for giving us the FSHD immortalized myoblast cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Rosetrees Trust (London, UK) [R10526]. Funding for open access charge: Royal Holloway REF 2021 OA fund.
Conflict of interest statement. None declared.
REFERENCES
- 1. Padberg G.W. Facioscapulohumeral muscular dystrophy: a clinician's experience. Facioscapulohumeral Muscular Dystrophy: Clinical Medicine and Molecular Cell Biology. 2004; Oxon: Garland Science/BIOS Scientific Publishers. [Google Scholar]
- 2. Mostacciuolo M.L., Pastorello E., Vazza G., Miorin M., Angelini C., Tomelleri G., Galluzzi G., Trevisan C.P.. Facioscapulohumeral muscular dystrophy: epidemiological and molecular study in a north-east Italian population sample. Clin. Genet. 2009; 75:550–555. [DOI] [PubMed] [Google Scholar]
- 3. Tawil R., van der Maarel S.M., Tapscott S.J.. Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skelet. Muscle. 2014; 4:12–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van der Maarel S.M., Miller D.G., Tawil R., Filippova G.N., Tapscott S.J.. Facioscapulohumeral muscular dystrophy: consequences of chromatin relaxation. Curr. Opin. Neurol. 2012; 25:614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R.. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992; 2:26–30. [DOI] [PubMed] [Google Scholar]
- 6. van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R.. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993; 2:2037–2042. [DOI] [PubMed] [Google Scholar]
- 7. Lemmers R.J.L.F., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W.E., Amell A.M., van der Vliet P.J., Almomani R., Straasheijm K.R. et al.. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 2012; 44:1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lemmers R.J.L.F., van den Boogaard M.L., van der Vliet P.J., Donlin-Smith C.M., Nations S.P., Ruivenkamp C.A.L., Heard P., Bakker B., Tapscott S., Cody J.D. et al.. Hemizygosity for SMCHD1 in facioscapulohumeral muscular dystrophy type 2: consequences for 18p deletion syndrome. Hum. Mutat. 2015; 36:679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lemmers R.J.L.F., van der Vliet P.J., Klooster R., Sacconi S., Camano P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W. et al.. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010; 329:1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kowaljow V., Marcowycz A., Ansseau E., Conde C.B., Sauvage S., Matteotti C., Arias C., Corona E.D., Nunez N.G., Leo O. et al.. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. NMD. 2007; 17:611–623. [DOI] [PubMed] [Google Scholar]
- 11. Wallace L.M., Garwick S.E., Mei W., Belayew A., Coppee F., Ladner K.J., Guttridge D., Yang J., Harper S.Q.. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol. 2011; 69:540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Matteotti C., van Acker A.M., Leo O. et al.. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:18157–18162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daxinger L., Tapscott S.J., van der Maarel S.M.. Genetic and epigenetic contributors to FSHD. Curr. Opin. Genet. Dev. 2015; 33:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hewitt J.E., Lyle R., Clark L.N., Valleley E.M., Wright T.J., Wijmenga C., van Deutekom J.C., Francis F., Sharpe P.T., Hofker M.. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum. Mol. Genet. 1994; 3:1287–1295. [DOI] [PubMed] [Google Scholar]
- 15. Zeng W., de Greef J.C., Chen Y.-Y., Chien R., Kong X., Gregson H.C., Winokur S.T., Pyle A., Robertson K.D., Schmiesing J.A. et al.. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet. 2009; 5:e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Balog J., Thijssen P.E., de Greef J.C., Shah B., van Engelen B.G.M., Yokomori K., Tapscott S.J., Tawil R., van der Maarel S.M.. Correlation analysis of clinical parameters with epigenetic modifications in the DUX4 promoter in FSHD. Epigenetics. 2012; 7:579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lemmers R.J.L.F., Goeman J.J., van der Vliet P.J., van Nieuwenhuizen M.P., Balog J., Vos-Versteeg M., Camano P., Ramos Arroyo M.A., Jerico I., Rogers M.T. et al.. Inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum. Mol. Genet. 2015; 24:659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsumagari K., Qi L., Jackson K., Shao C., Lacey M., Sowden J., Tawil R., Vedanarayanan V., Ehrlich M.. Epigenetics of a tandem DNA repeat: chromatin DNaseI sensitivity and opposite methylation changes in cancers. Nucleic Acids Res. 2008; 36:2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Falabella M., Kolesar J.E., Xiang I.M., Wang T., Horne W., Wallace C., Sun L., Taguchi Y., Wang C., Turek-Herman J. et al.. G-quadruplex dynamics contribute to epigenetic regulation of mitochondrial function. Sci Rep. 2019; 9:5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guilbaud G., Murat P., Recolin B., Campbell B.C., Maiter A., Sale J.E., Balasubramanian S.. Local epigenetic reprogramming induced by G-quadruplex ligands. Nat. Chem. 2017; 9:1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S.. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006; 34:5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hershman S.G., Chen Q., Lee J.Y., Kozak M.L., Yue P., Wang L.-S., Johnson F.B.. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008; 36:144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H.. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dexheimer T.S., Sun D., Hurley L.H.. Deconvoluting the Structural and Drug-Recognition Complexity of the G-Quadruplex-Forming Region Upstream of the bcl-2 P1 Promoter. J. Am. Chem. Soc. 2006; 128:5404–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun D., Liu W.-J., Guo K., Rusche J.J., Ebbinghaus S., Gokhale V., Hurley L.H.. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol. Cancer Ther. 2008; 7:880–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cogoi S., Paramasivam M., Spolaore B., Xodo L.E.. Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 2008; 36:3765–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bay D.H., Busch A., Lisdat F., Iida K., Ikebukuro K., Nagasawa K., Karube I., Yoshida W.. Identification of G-quadruplex structures that possess transcriptional regulating functions in the Dele and Cdc6 CpG islands. BMC Mol. Biol. 2017; 18:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. David A.P., Margarit E., Domizi P., Banchio C., Armas P., Calcaterra N.B.. G-quadruplexes as novel cis-elements controlling transcription during embryonic development. Nucleic Acids Res. 2016; 44:4163–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sen D., Gilbert W.. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364–366. [DOI] [PubMed] [Google Scholar]
- 30. Yang Q., Xiang J., Yang S., Zhou Q., Li Q., Tang Y., Xu G.. Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: I. recognizing mixed G-quadruplex in human telomeres. Chem. Commun. Camb. Engl. 2009; 9:1103–1105. [DOI] [PubMed] [Google Scholar]
- 31. Kumari S., Bugaut A., Huppert J.L., Balasubramanian S.. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007; 3:218–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arora A., Dutkiewicz M., Scaria V., Hariharan M., Maiti S., Kurreck J.. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008; 14:1290–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chambers V.S., Marsico G., Boutell J.M., Antonio Di, Smith M., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 34. Jayaraj G.G., Pandey S., Scaria V., Maiti S.. Potential G-quadruplexes in the human long non-coding transcriptome. RNA Biol. 2012; 9:81–86. [DOI] [PubMed] [Google Scholar]
- 35. Arachchilage Mirihana, Dassanayake G., Basu S.. A potassium ion-dependent RNA structural switch regulates human pre-miRNA 92b maturation. Chem. Biol. 2015; 22:262–272. [DOI] [PubMed] [Google Scholar]
- 36. Xu Y., Komiyama M.. Structure, function and targeting of human telomere RNA. Methods San Diego Calif. 2012; 57:100–105. [DOI] [PubMed] [Google Scholar]
- 37. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fay M.M., Lyons S.M., Ivanov P.. RNA G-quadruplexes in biology: principles and molecular mechanisms. J. Mol. Biol. 2017; 429:2127–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cimino-Reale G., Zaffaroni N., Folini M.. Emerging role of G-quadruplex DNA as target in anticancer therapy. Curr. Pharm. Des. 2016; 22:6612–6624. [DOI] [PubMed] [Google Scholar]
- 40. Cammas A., Millevoi S.. RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acids Res. 2017; 45:1584–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu Y., Brosh R.M.. G-quadruplex nucleic acids and human disease. FEBS J. 2010; 277:3470–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Simone R., Fratta P., Neidle S., Parkinson G.N., Isaacs A.M.. G-quadruplexes: emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett. 2015; 589:1653–1668. [DOI] [PubMed] [Google Scholar]
- 43. Che T., Wang Y.-Q., Huang Z.-L., Tan J.-H., Huang Z.-S., Chen S.-B.. Natural alkaloids and heterocycles as G-quadruplex ligands and potential anticancer agents. Mol. Basel Switz. 2018; 23:doi:10.3390/molecules23020493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Le Vy Thi, Han T., Chae S., Park H.-J.. G-quadruplex binding ligands: from naturally occurring to rationally designed molecules. Curr. Pharm. Des. 2012; 18:1948–1972. [DOI] [PubMed] [Google Scholar]
- 45. Duarte A.R., Cadoni E., Ressurreição A.S., Moreira R., Paulo A.. Design of modular G-quadruplex ligands. ChemMedChem. 2018; 13:869–893. [DOI] [PubMed] [Google Scholar]
- 46. Arora A., Balasubramanian C., Kumar N., Agrawal S., Ojha R.P., Maiti S.. Binding of berberine to human telomeric quadruplex - spectroscopic, calorimetric and molecular modeling studies. FEBS J. 2008; 275:3971–3983. [DOI] [PubMed] [Google Scholar]
- 47. Ansseau E., Domire J.S., Wallace L.M., Eidahl J.O., Guckes S.M., Giesige C.R., Pyne N.K., Belayew A., Harper S.Q.. Aberrant splicing in transgenes containing introns, exons, and V5 epitopes: lessons from developing an FSHD mouse model expressing a D4Z4 repeat with flanking genomic sequences. PLoS One. 2015; 10:e0118813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Himeda C.L., Debarnot C., Homma S., Beermann M.L., Miller J.B., Jones P.L., Jones T.I.. Myogenic enhancers regulate expression of the facioscapulohumeral muscular dystrophy-associated DUX4 gene. Mol. Cell. Biol. 2014; 34:1942–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Mattéotti C., van Acker A.M., Leo O. et al.. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:18157–18162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kikin O., D’Antonio L., Bagga P.S.. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006; 34:W676–W682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kypr J., Kejnovská I., Renčiuk D., Vorlíčková M.. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009; 37:1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Basu A., Jaisankar P., Kumar Suresh. Binding of the 9-O-N-aryl/arylalkyl amino carbonyl methyl substituted berberine analogs to tRNA(phe.). PLoS One. 2013; 8:e58279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mazzini S., Bellucci M.C., Mondelli R.. Mode of binding of the cytotoxic alkaloid berberine with the double helix oligonucleotide d(AAGAATTCTT)(2). Bioorg. Med. Chem. 2003; 11:505–514. [DOI] [PubMed] [Google Scholar]
- 54. Nandi R., Debnath D., Maiti M.. Interactions of berberine with poly(A) and tRNA. Biochim. Biophys. Acta. 1990; 1049:339–342. [DOI] [PubMed] [Google Scholar]
- 55. Li Z.-Q., Liao T.-C., Dong C., Yang J.-W., Chen X.-J., Liu L., Luo Y., Liang Y.-Y., Chen W.-H., Zhou C.-Q.. Specifically targeting mixed-type dimeric G-quadruplexes using berberine dimers. Org. Biomol. Chem. 2017; 15:10221–10229. [DOI] [PubMed] [Google Scholar]
- 56. Bhadra K., Maiti M., Kumar G.S.. Berberine-DNA complexation: new insights into the cooperative binding and energetic aspects. Biochim. Biophys. Acta. 2008; 1780:1054–1061. [DOI] [PubMed] [Google Scholar]
- 57. Ferreboeuf M., Mariot V., Bessières B., Vasiljevic A., Attié-Bitach T., Collardeau S., Morere J., Roche S., Magdinier F., Robin-Ducellier J. et al.. DUX4 and DUX4 downstream target genes are expressed in fetal FSHD muscles. Hum. Mol. Genet. 2014; 23:171–181. [DOI] [PubMed] [Google Scholar]
- 58. Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R. et al.. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell. 2012; 22:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Marsollier A.-C., Ciszewski L., Mariot V., Popplewell L., Voit T., Dickson G., Dumonceaux J.. Antisense targeting of 3′ end elements involved in DUX4 mRNA processing is an efficient therapeutic strategy for facioscapulohumeral dystrophy: a new gene-silencing approach. Hum. Mol. Genet. 2016; 25:1468–1478. [DOI] [PubMed] [Google Scholar]
- 60. Lista M.J., Martins R.P., Billant O., Contesse M.-A., Findakly S., Pochard P., Daskalogianni C., Beauvineau C., Guetta C., Jamin C. et al.. Nucleolin directly mediates Epstein-Barr virus immune evasion through binding to G-quadruplexes of EBNA1 mRNA. Nat. Commun. 2017; 8:16043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lombardi Puig, Holmes E., Verga A., Teulade-Fichou D., M.-P. Nicolas, Londoño-Vallejo A.. Thermodynamically stable and genetically unstable G-quadruplexes are depleted in genomes across species. Nucleic Acids Res. 2019; 47:6098–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Blaeser A., Awano H., Wu B., Lu Q.-L.. Progressive dystrophic pathology in diaphragm and impairment of cardiac function in FKRP P448L mutant mice. PLoS ONE. 2016; 11:e0164187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tawil R., van der Maarel S.M., Tapscott S.J.. Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skelet. Muscle. 2014; 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Amrane S., Adrian M., Heddi B., Serero A., Nicolas A., Mergny J.-L., Phan A.T.. Formation of pearl-necklace monomorphic G-quadruplexes in the human CEB25 minisatellite. J. Am. Chem. Soc. 2012; 134:5807–5816. [DOI] [PubMed] [Google Scholar]
- 65. Palumbo S.L., Ebbinghaus S.W., Hurley L.H.. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J. Am. Chem. Soc. 2009; 131:10878–10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Agrawal P., Lin C., Mathad R.I., Carver M., Yang D.. The major G-quadruplex formed in the human BCL-2 proximal promoter adopts a parallel structure with a 13-nt loop in K+ solution. J. Am. Chem. Soc. 2014; 136:1750–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Guédin A., Gros J., Alberti P., Mergny J.-L.. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010; 38:7858–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Beaudoin J.-D., Jodoin R., Perreault J.-P.. New scoring system to identify RNA G-quadruplex folding. Nucleic Acids Res. 2014; 42:1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang J., Zheng K., Xiao S., Hao Y., Tan Z.. Mechanism and manipulation of DNA:RNA hybrid G-quadruplex formation in transcription of G-Rich DNA. J. Am. Chem. Soc. 2014; 136:1381–1390. [DOI] [PubMed] [Google Scholar]
- 70. Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N.. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004; 18:1618–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wanrooij P.H., Uhler J.P., Shi Y., Westerlund F., Falkenberg M., Gustafsson C.M.. A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 2012; 40:10334–10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zheng K., Xiao S., Liu J., Zhang J., Hao Y., Tan Z.. Co-transcriptional formation of DNA:RNA hybrid G-quadruplex and potential function as constitutional cis element for transcription control. Nucleic Acids Res. 2013; 41:5533–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Endoh T., Sugimoto N.. Mechanical insights into ribosomal progression overcoming RNA G-quadruplex from periodical translation suppression in cells. Sci. Rep. 2016; 6:22719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Thandapani P., Song J., Gandin V., Cai Y., Rouleau S.G., Garant J.-M., Boisvert F.-M., Yu Z., Perreault J.-P., Topisirovic I. et al.. Aven recognition of RNA G-quadruplexes regulates translation of the mixed lineage leukemia protooncogenes. eLife. 2015; 4:e06234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tassin A., Laoudj-Chenivesse D., Vanderplanck C., Barro M., Charron S., Ansseau E., Chen Y.-W., Mercier J., Coppée F., Belayew A.. DUX4 expression in FSHD muscle cells: how could such a rare protein cause a myopathy. J. Cell. Mol. Med. 2013; 17:76–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ansseau E., Vanderplanck C., Wauters A., Harper S.Q., Coppée F., Belayew A.. Antisense oligonucleotides used to target the DUX4 mRNA as therapeutic approaches in facios scapulo humeral muscular dystrophy (FSHD). Genes. 2017; 8:doi:10.3390/genes8030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ganesan K., Xu B.. Telomerase inhibitors from natural products and their anticancer potential. Int. J. Mol. Sci. 2017; 19:doi:10.3390/ijms19010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Antonio Di, Biffi M., Mariani G., Raiber A., E.-A. Rodriguez, Balasubramanian S.. Selective RNA versus DNA G-quadruplex targeting by in situ click chemistry. Angew. Chem. Int. Ed Engl. 2012; 51:11073–11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Katsuda Y., Sato S.-I., Asano L., Morimura Y., Furuta T., Sugiyama H., Hagihara M., Uesugi M.. A small molecule that represses translation of G-quadruplex-containing mRNA. J. Am. Chem. Soc. 2016; 138:9037–9040. [DOI] [PubMed] [Google Scholar]
- 80. Wallace L.M., Garwick S.E., Mei W., Belayew A., Coppee F., Ladner K.J., Guttridge D., Yang J., Harper S.Q.. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol. 2011; 69:540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wallace L.M., Liu J., Domire J.S., Garwick-Coppens S.E., Guckes S.M., Mendell J.R., Flanigan K.M., Harper S.Q.. RNA interference inhibits DUX4-induced muscle toxicity in vivo: implications for a targeted FSHD therapy. Mol. Ther. J. Am. Soc. Gene Ther. 2012; 20:1417–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Roman W., Gomes E.R.. Nuclear positioning in skeletal muscle. Semin. Cell Dev. Biol. 2018; 82:51–56. [DOI] [PubMed] [Google Scholar]
- 83. Janssen B.H., Voet N.B.M., Nabuurs C.I., Kan H.E., de Rooy J.W.J., Geurts A.C., Padberg G.W., van Engelen B.G.M., Heerschap A.. Distinct disease phases in muscles of facioscapulohumeral dystrophy patients identified by MR detected fat infiltration. PLoS One. 2014; 9:e85416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.