Abstract
The vertebrate splicing factor RBM20 (RNA binding motif protein 20) regulates protein isoforms important for heart development and function, with mutations in the gene linked to cardiomyopathy. Previous studies have identified the four nucleotide RNA motif UCUU as a common element in pre-mRNA targeted by RBM20. Here, we have determined the structure of the RNA Recognition Motif (RRM) domain from mouse RBM20 bound to RNA containing a UCUU sequence. The atomic details show that the RRM domain spans a larger region than initially proposed in order to interact with the complete UCUU motif, with a well-folded C-terminal helix encoded by exon 8 critical for high affinity binding. This helix only forms upon binding RNA with the final uracil, and removing the helix reduces affinity as well as specificity. We therefore find that RBM20 uses a coupled folding-binding mechanism by the C-terminal helix to specifically recognize the UCUU RNA motif.
INTRODUCTION
Healthy cardiac development and function requires the regulated expression of many heart-specific genes. For several of these gene products, additional control through alternative splicing regulates a balance between cardiac protein isoforms that contain isoform-specific properties. A major example is the muscle protein titin, a giant protein that spans half the sarcomere and serves to modulate the elasticity of the stretched muscle (1–3). Titin exists as several isoforms and undergoes a notable shift in size from larger forms in early heart development towards shorter forms in the adult (reviews include (4–6)). The functional outcome of these alternative isoforms is to change titin from the longer compliant form toward the shorter stiffer form, thus affecting passive muscle tension during the cardiac cycle. Disruption of this splicing regulation leads to abnormal ratios of the compliant and stiff forms of titin, resulting in heart disease. For example, failure to produce sufficient levels of the shorter titin isoform can lead to abnormally compliant titin in dilated cardiomyopathy (DCM).
In a search for factors that regulate titin splicing, a chromosomal deletion in Rbm20 (RNA binding motif 20) was isolated as the genetic cause of titin mis-splicing from a mutant rat strain (7,8). RBM20 was itself first identified in a search for a familial genetic basis of DCM in human patients (9). Subsequent investigation has identified additional patients and mutations involving RBM20 related to DCM (10–19), as well as cardiac arrhythmia (20,21), pediatric restrictive cardiomyopathy (22) and left-ventricular non-compaction (23). Although representing only 3% of idiopathic cases, patients with DCM that have mutant RBM20 correlate with earlier disease onset, high penetrance and requirement for heart transplantation (9,12,18,24,25).
Biological characterization of RBM20 has incorporated rat, mouse and in vitro studies. Expression of Rbm20 is primarily localized to the heart, with lower expression in other striated muscle (7,26). Rats with homozygous or heterozygous loss of functional Rbm20 mimic human DCM symptoms as well as defects in the heart structure, age-related fibrosis and less capacity for exercise (7,27). Loss of RBM20 function in rats also coincides with extensive deregulation of titin splice isoforms towards an abnormal form of titin with all exons retained (7,8). Comparison to normal processing of titin pre-mRNA shows that RBM20 is required for several intron retention and exon skipping events, as well as the creation of circRNA (28). Mice with homozygous deletion of Rbm20 also reveal a large number of RBM20-dependent titin circRNA (29). Visualization of RBM20 in cardiomyocytes and muscle tissue show an organization into two nuclear clusters that colocalize with titin mRNA but not with other nuclear bodies, including paraspeckles or nuclear speckles (28). Purification of RBM20-bound titin RNA shows that introns remain, and thus it has been proposed that these clusters represent intermediate or co-transcriptional steps in titin pre-mRNA processing during which RBM20 remains bound to extended regions of titin pre-mRNA (28).
In addition to titin, RBM20 also regulates the splicing patterns of other transcripts that may contribute to the severity of DCM or promote separate cardiac pathologies. Human, rat and mouse studies identified additional splicing targets that encode proteins which bind and transport Ca2+ (7,26,30–33) such as the calcium channel ryanodine receptor 2 (RYR2), Ca2+/calmodulin-dependent protein kinase II delta (CAMK2δ), and the calcium channel voltage-gated L type alpha 1C subunit (CACNA1C/Cav1.2). Other characterized targets include formin homology 2 domain containing protein 3 (FHOD3), Z-band alternatively spliced PDZ-motif protein (ZASP/lDB3/CYPHER), and the PDZ and LIM domain protein 5 (PDLIM5/ENH) (7,32,34,35). Recent findings suggest that RBM20 processing clusters may connect to chromosomal regions for CACNA1C and CAMK2δ (36).
The RBM20 protein sequence is largely devoid of identifiable structured regions except for the central RNA Recognition Motif (RRM) domain as well as two small putative zinc finger domains (ZnF) (Figure 1A). To investigate a direct role in binding RNA, PAR-CLIP in HEK293 cells and HITS-CLIP in rat cardiomyocytes identified numerous transcripts that purified with tagged RBM20 (32). Analysis of the sequences revealed a prominently conserved RNA motif composed of the tetramer UCUU: this motif is enriched 50 nucleotides before and 100 nucleotides after exons regulated by RBM20, which largely fall into the categories of mutually exclusive or cassette exons (32). Furthermore, mutation of the UCUU motif to CGCG or CGUU sequences abolishes direct RBM20 binding to the Ryr2 transcript, with reduction in binding by a single base change to UCUG (32). Further analysis suggested that the RRM domain may indeed be responsible for UCUU recognition, since a construct from human RBM20 that covers the canonical RRM fold (residues 511–601) interacts only with titin-derived oligonucleotides that contain UCUU motifs (37). A minimal RRM region was also shown to bind to a titin transcript around titin exon 50 (7). Mouse strains with the RRM domain removed by deleting exons 6 and 7 of rbm20 result in mis-spliced titin, Camk2d, and Lbd3 (38), as well as increased titin stiffness in the diaphragm (39). However, cardiac pathology in this strain was less pronounced than for a larger size Rbm20 deletion (30). Although not corresponding to a folded domain, a region C-terminal to the RRM domain is enriched in arginine and serine residues (RS), and most of the disease mutations map to this small segment (9,18,40). Improper phosphorylation of key serine residues in this RS domain prevents proper nuclear localization of RBM20 mutants (41).
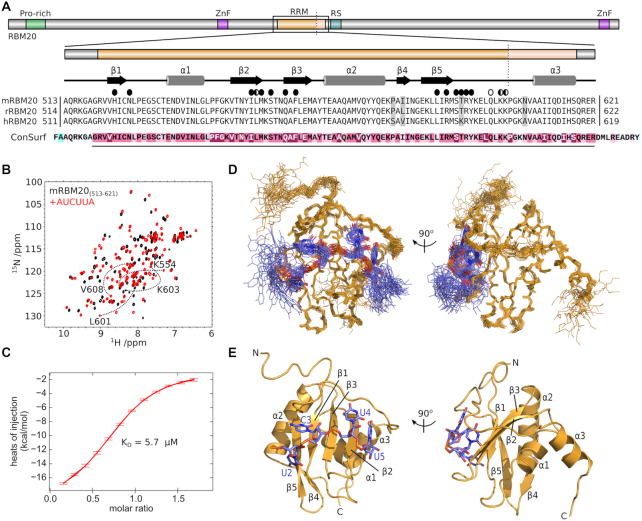
Figure 1.
Recognition of UCUU by the RBM20 RRM domain. (A) Domain composition of RBM20 with expanded details for the RRM domain. The alignment is composed of RBM20 sequences from mouse (mRBM20, UniProt Q3UQS8), rat (rRBM20; UniProt E9PT37) and human (hRBM20; UniProt Q5T481). The vertical dotted line shows the C-terminal end of previous RRM domain constructs. Secondary structure elements from the NMR structure are indicated above the alignment. Residues that contact the RNA by their sidechain and backbone atoms are indicated by full and open circles, respectively. Results from the ConSurf analysis is shown below the alignment, with the region of high conservation indicated with a line, and coloured purple. (B) 15N-HSQC overlay of [15N]mRBM20(513–621) in the absence (black) and presence (red) of 1.2 molar equivalents AUCUUA RNA. Annotated spectra can be found in Supplementary Figure S1. (C) Representative isothermal titration calorimetry (ITC) data for mRBM20(513–621) binding to AUCUUA RNA. (D) Ensemble of 25 lowest energy structures calculated for mRBM20(513–621) (backbone heavy atoms, orange lines) bound to AUCUUA RNA (all heavy atoms, purple lines). (E) Lowest energy structure model for the UCUU motif (all heavy atoms, purple sticks) and mRBM20 (residue 520–617, orange cartoon). RNA bases and protein secondary structure elements are labeled.
Removal of the RRM domain in mouse RBM20 leads to a phenotype consistent with, but less severe than, strains in which the complete RBM20 protein function is abolished (7,38). Nevertheless, few RBM20 disease mutations have so far been found within the RRM domain: there is a single example of a sporadic V535I mutation (V537I in mice) and the proximal I536T mutation that may be secondary to mRNA processing defects in Ldb3 (42). In contrast, the RS domain remains the hotspot of RBM20 disease mutations, and mutations such as S637A can act as a dominant negative toward titin splicing (41,43). It may be that functional mutants of RBM20 that can still bind to native pre-mRNA binding sites are more deleterious than RBM20 mutants that simply affect RNA-binding specificity. On the other hand, the modest effects observed for Rbm20ΔRRM mice help illustrate putative benefits of adjusting RBM20 function to help mediate certain heart pathologies. Mice strains that model abnormal titin stiffness show that crosses with Rbm20ΔRRM mice help promote a more compliant titin in the offspring with improvement of muscle function (38,44). Similarly, abnormal diastolic function in mouse models of heart failure with preserved ejection fraction (HFpEF) can be largely restored with heterozygous expression of Rbm20ΔRRM (45–47). In both cases, it is likely that effects on additional targets related to cardiac performance may contribute to phenotypic changes in the Rbm20ΔRRM mice, notably splicing and expression levels of proteins involved in calcium sensitivity and contraction regulation (31,48).
Given a biological role for the RRM domain and possible specificity for the UCUU motif exhibited by RBM20, we have used a structural approach to study RNA binding by the mouse RBM20 RRM domain. The atomic details confirm a role for the RRM domain in the direct interaction with the previously defined UCUU RNA motif. In addition to base recognition by the canonical RRM fold, we find that the specificity for the terminal uracil base in the motif is coupled with folding of an extra C-terminal α-helix encoded by exon 8 of the RBM20 gene.
MATERIALS AND METHODS
Cloning
Constructs containing the RRM domain of murine RBM20 were created from a cDNA provided by Pamela Lorenzi (University of Verona, Italy), together with PCR primers containing an NcoI restriction site in the forward primer, with a stop codon and Acc65I restriction site in the reverse primers. The primer details are included in Supplementary Table S1. The PCR products, as well as a modified pET-9d vector, were digested with NcoI and Acc65I, followed by ligation, to produce plasmids that encode constructs with an N-terminal hexahistidine (His6) tag and cleavage site for tobacco etch virus (TEV) protease. The ligation products were transformed into Escherichia coli DH5α and resulting plasmids were verified by sequencing. Mutants were created by performing an initial PCR step with internal forward and reverse primers that harbour the mutant sequence.
Protein expression
Escherichia coli BL21(DE3) pLysY (New England Biolabs) were transformed with plasmids encoding the various mRBM20 constructs. Subsequent colonies were used for initial overnight culture growth at 37°C in lysogeny broth (LB) supplemented with 40 μg/ml kanamycin. Bacteria from the overnight cultures were used to start 500 ml cultures in LB for natural abundance protein, or in M9 minimal medium supplemented with isotopically-enriched compounds. For 15N-labelled protein, 1 g/l 15NH4Cl was added to the media, and additionally 2 g/l 13C-glucose was added for 13C,15N-labelled protein. For stereospecific assignment of methyl groups, the media was only enriched to 10% (w/w) 13C-glucose. Deuterated protein was grown in 99% (v/v) D2O with 2 g/l 2H,13C-glucose and 1 g/l 15NH4Cl. In all cases, initial growth of the 500 ml cultures at 37°C was followed by induction at an OD600nm of 0.6 with 0.25 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and protein expression continued for 16 h at 25°C. Cells were harvested by centrifugation at 4500 × g for 20 min at 4°C, resuspended in lysis buffer containing 5 mM imidazole, 50 mM Tris (pH 7.5), 500 mM NaCl, 5% (v/v) glycerol and stored at –80°C in the presence of added lysozyme.
Protein purification
Cells frozen in lysis buffer containing lysozyme were thawed on ice, with lysis aided by sonication. Soluble protein was separated from cellular debris by centrifugation at 20 000 × g for 30 min at 4°C. The supernatant was filtered through a GD/X 0.7 μm filter (GE Healthcare Life Sciences) and loaded onto 2 ml Nuvia IMAC Ni-charged resin (Bio-Rad Laboratories). The resin was then washed with 10 column volumes of buffer containing 5 mM imidazole, 50 mM Tris (pH 7.5), 500 mM NaCl, 5% (v/v) glycerol followed by 5 volumes of the same buffer but with 25 mM imidazole. Protein elution used the same buffer with 500 mM imidazole. Fractions containing protein were pooled and exchanged to the initial buffer containing 5 mM imidazole by using a PD10 desalting column (GE Healthcare Life Sciences). His-tagged TEV protease (0.1 mg/ml final concentration) was added for overnight cleavage at 4°C. The protease, hexahistidine tag and any uncleaved protein was removed by a second passage through the Nuvia IMAC Ni-charged resin. The purified samples were concentrated with 3 kDa MWCO Vivaspin centrifugal concentrators (Merck Millipore Corporation) followed by overnight dialysis in 20 mM sodium phosphate (pH 6.5) and 50 mM NaCl. Samples for NMR spectroscopy were supplemented with 2 mM dithiothreitol (DTT) and 10% (v/v) D2O. Samples for isothermal titration calorimetry (ITC) included 2.5 mM Tris(2-carboxyethyl)phosphine (TCEP) in the dialysis buffer. Protein purity was checked by SDS-PAGE and protein concentration was determined by absorbance at 280 nm with extinction coefficients obtained using ProtParam (http://web.expasy.org/protparam).
RNA synthesis
RNA oligonucleotides were synthesized on an Expedite 8909 (PerSeptive Biosystems) from phosphoramidite monomers, and purified from a mix of butanol and water. AUCUUA RNA was also purchased (Sigma-Aldrich). RNA concentrations were determined by absorbance at 260 nm with extinction coefficients obtained from OligoAnalzer (https://eu.idtdna.com/calc/analyzer).
NMR spectroscopy
NMR spectra were recorded at 298 K using a Bruker Neo spectrometer at 700 or 800 MHz, equipped with a standard triple resonance gradient probe or cryoprobe, respectively. Bruker TopSpin versions 4.0 (Bruker BioSpin) was used to collect data. NMR data were processed with NMR Pipe/Draw (49) and analysed with Sparky 3 (T.D. Goddard and D.G. Kneller, University of California).
Chemical shift assignment
Backbone 1HN, 1Hα, 13Cα, 13Cβ, 13C′ and 15NH chemical shifts for mRBM20(513–621) bound to AUCUUA were assigned based on a sample of 400 μM [13C,15N]mRBM20(513–621) bound to 480 μM unlabelled RNA, and used 2D 1H,15N-HSQC, 3D 15N-HNCO, 3D 15N-HNCACO, 3D 15N-HNCA, 3D 15N-HNCACB, 3D 15N-CBCACONH, and 3D 15N-HNHA spectra. Aliphatic side chain protons were assigned based on 2D 1H,13C-HSQC, 3D C(CO)NH-TOCSY, 3D H(C)CH-TOCSY and 3D (H)CCH-TOCSY spectra. Stereospecific assignment of leucine and valine methyl groups used a constant time 1H,13C-HSQC (50) on a 450 μM protein sample labelled with 10% (w/w) 13C-glucose in complex with 560 μM unlabelled RNA. Assignment of sidechain asparagine δ2 and glutamine ϵ2 amides used a 3D 15N-HSQC-NOESY (120 ms mixing time) on a sample of 400 μM [15N]mRBM20(513–621) with 480 μM unlabelled RNA. Aromatic 1H and 13C nuclei were assigned based on 2D 1H,13C-HSQC and 3D 13C-HSQC-NOESY (150 ms mixing time). Non-exchanging RNA 1H nuclei were assigned in 99% (v/v) D2O from 290 μM natural abundance AUCUUA RNA in complex with 300 μM [2H-99%]mRBM20(513–621) by using 2D 1H,1H-NOESY (120 ms mixing time) and 1H,1H-TOCSY spectra. Exchanging RNA 1H nuclei were assigned by using a x2-filtered 2D 1H,1H-NOESY (150 ms mixing time) at 278 K on a sample of 500 μM [13C,15N]mRBM20(513–621) bound to 600 μM AUCUUA.
For the unbound mRBM20(513–621) the backbone 1HN, 1Hα, 13Cα, 13Cβ, 13C′ and 15NH chemical shifts were assigned based on a sample of 400 μM [13C,15N]mRBM20(513–621) using 2D 1H,15N-HSQC, 3D 15N-HNCO, 3D 15N-HNCACO, 3D 15N-HNCA, 3D 15N-HNCACB and 3D 15N-CBCACONH spectra. Aliphatic side chain protons were assigned based on 2D 1H,13C-HSQC, 3D H(C)(CO)NH-TOCSY, 3D (H)C(CO)NH-TOCSY, 3D H(C)CH-TOCSY and 3D (H)CCH-TOCSY. Stereospecific assignment of leucine and valine methyl groups used a constant time 1H,13C-HSQC (50) on a 100 μM protein sample labelled with 10% (w/w) 13C-glucose. Assignment of sidechain asparagine δ2 amdies and glutamine ϵ2 amides used a 3D 15N-HSQC-NOESY (120 ms mixing time). Aromatic 1H and 13C nuclei were assigned based on 2D 1H,13C-HSQC (13C offset 120 ppm) and 3D 13C-HSQC-NOESY (120 ms mixing time, 13C offset 125 ppm).
Structure calculation
Structure ensembles were calculated by using Aria 2.3/CNS1.2 (51,52), with final ensembles refined in explicit water and consisting of the 25 lowest energy structures from a total of 100 calculated models. Complete refinement statistics are presented in Table 2.
Table 2.
NMR and refinement statistics for the RNA-bound and unbound RBM20(513–621)
| AUCUUA bound mRBM20 | Unbound mRBM20 | |
|---|---|---|
| Number of structures | 25 | 25 |
| Distance constraints | ||
| Total | 3693 | 2838 |
| Protein intraresidue | 929 | 823 |
| Protein interresidue | ||
| Sequential (|i – j| = 1) | 442 | 385 |
| Short range (1 < |i – j| < 4) | 199 | 177 |
| Medium range (3 < |i – j| < 6) | 79 | 76 |
| Long range (|i – j| > 5) | 398 | 336 |
| RNA | 104 | – |
| Intermolecular | 69 | – |
| Ambiguousa | 1445 | 1041 |
| Hydrogen bonds | 28 | – |
| Total dihedral angle constraints | ||
| Protein | 254 | 224 |
| RNA | 28 | – |
| Residual dipolar couplings | ||
| 1DHN | 82 | – |
| Structure statistics | ||
| Violations (mean and SD) | ||
| Distance constraints (Å)b | 0.018 ± 0.000 | 0.020 ± 0.001 |
| Dihedral angle constraints (°)c | 1.48 ± 0.06 | 1.40 ± 0.07 |
| Residual dipolar couplings, Qd | 0.24 ± 0.01 | – |
| Deviations from idealized geometry | ||
| Bond lengths (Å) | 0.004 ± 0.000 | 0.004 ± 0.000 |
| Bond angles (°) | 0.54 ± 0.01 | 0.51 ± 0.02 |
| Impropers (°) | 1.41 ± 0.04 | 1.5 ± 0.1 |
| Ramachandran plot (%) e | ||
| Most favoured | 93.5 (88.4) | 95.6 (82.1) |
| Additionally favoured | 6.3 (10.1) | 4.4 (14.5) |
| Generally allowed | 0.2 (0.6) | 0.0 (1.8) |
| Disallowed | 0.0 (0.8) | 0.0 (1.5) |
| Average pairwise rmsd (Å) | ||
| Protein backbone 2° structure | 0.25 ± 0.04 | 0.20 ± 0.04 |
| Protein heavy 2° structure | 0.56 ± 0.06 | 0.56 ± 0.07 |
| Protein backbone all | 1.5 ± 0.2 | 3.2 ± 0.8 |
| Protein heavy all | 1.9 ± 0.2 | 3.6 ± 0.8 |
aA standard class of distance restraints in Aria2.3, derived from cases in which the peak volume is contributed by two or more overlapping NOE crosspeaks
bNo violations >0.5 Å.
cNo violations >10°.
dCalculated for each structure in the ensemble using the method described by (73).
eDetermined by using PROCHECK-NMR (74). Main values display hetNOE > 0.5 and include residues 520–617 for the bound form, and 520–598 for the unbound form. Values that include all residues are in parentheses.
For the RNA-bound complex, the majority of protein 1H distances were obtained using NOE crosspeaks from 3D 15N-HSQC-NOESY (120 ms mixing time) on a sample of 400 μM [15N]mRBM20 with 480 μM unlabelled RNA, 3D 13C-HSQC-NOESY (150 ms mixing time) on a sample of 300 μM [15N]mRBM20 with 360 μM unlabelled RNA in 100% D2O, and 2D 1H,1H-NOESY (120 ms mixing time) on a sample of 450 μM [10%-13C,15N]mRBM20 with 480 μM unlabelled RNA. RNA–RNA 1H distances were derived from a 2D 1H,1H-NOESY (120 ms mixing time) spectrum using 290 μM natural abundance AUCUUA RNA in complex with fully deuterated 300 μM mRBM20(513–621) in 100% D2O. Intramolecular distances were mostly derived from a x2-filtered 2D 1H,1H-NOESY spectrum (240 ms mixing time, with a watergate suppression scheme) on a sample of 300 μM [13C,15N]mRBM20(513–621) bound to 360 μM AUCUUA. A similar spectrum was acquired at 278 K on a sample of 500 μM [13C,15N]mRBM20(513–621) bound to 600 μM AUCUUA to identify NOE crosspeaks involving exchangeable 1H RNA nuclei. Protein dihedral angles were obtained by using TALOS-N (53) and SideR (54,55). RNA dihedral angles were based on TOCSY and NOESY crosspeak patterns. The default parameters of Aria2.3 were used, except that the number of steps during the dynamics section were increased as follows: High-temp with 10 000 steps, Refine with 10 000 steps, Cool1 with 15 000 steps, Cool2 with 15 000 steps. All NOESY spectra peak lists were processed in the same manner, with spin diffusion correction used throughout all iterations (correlation time of 9.5 ns used for the spectra at 298 K, and 12 ns for the spectrum at 278 K). Following a preliminary structure calculation, residues within the centre of each α-helix were further restrained by hydrogen-bond restraints. Starting at iteration four, residual dipolar coupling (RDC) values were included based on alignment using the stretched-gel approach (56) by measuring interleaved spin state-selective TROSY experiments on RNA-bound [15N]-mRBM20(513–621). For the aligned sample, a solution of the complex was added to a 1 cm cylinder of dried 5% 19:1 acrylamide:bisacrylamide and stretched into an NMR tube by using the NE-373-A-6/4.2 kit (New Era Enterprises). RDC-based intervector projection angle restraints related to Da and R values of –6 and 0.35, respectively.
For unbound mRBM20(513–621) the 1H distances were obtained using NOE crosspeaks from a 3D 15N-HSQC-NOESY (120 ms mixing time) and 3D aliphatic 13C-HSQC-NOESY (120 ms mixing time) using a sample of 400 μM [13C,15N]mRBM20(513–621), a 3D aromatic 13C-HSQC-NOESY (120 ms mixing time) using a sample of 170 μM [13C]mRBM20(513–621) in 100% D2O, and a 2D 1H,1H-NOESY (120 ms mixing time) using a sample of 500 μM natural abundance mRBM20(513–621). Protein dihedral angles were obtained by using TALOS-N (53) and SideR (54,55). Spin diffusion correction used a correlation time of 13 ns.
Relaxation measurement
15N relaxation data were acquired at 298 K and a field strength of 700 MHz. {1H}–15N heteronuclear NOE spectra were recorded with and without 3 s of proton saturation. The resulting values represent the average and standard deviation of two independent measurements.
Isothermal titration calorimetry
Measurements were performed on an iTC200 microcalorimeter (Malvern Panalytical) at 298 K with a stir rate of 600 rpm and recorded with high sensitivity. The samples were dialysed overnight in 20 mM sodium phosphate (pH 6.5), 50 mM NaCl and 2.5 mM TCEP prior to ITC experiments, and the same dialysis buffer was used to dilute protein and RNA samples. The cell contained target concentrations of 40 or 80 μM RNA, with target syringe concentrations of 400 or 800 μM protein, respectively (details in Supplementary Figure S2). After an initial delay of 120 s, a first injection of 1 ul was followed by 12 injections of 3 ul, with a delay of 120 s between each injection. Data processing used NITPIC (57,58) to integrate the titration points, and SEDPHAT (59) to perform the curve fitting. The values in Table 2 present averages and standard deviations from at least two independent measurements. The graph in Figure 1C was prepared by using GUSSI (60).
RESULTS
RNA-binding by the RRM domain of RBM20
In order to first determine the boundaries of the construct to be produced for structural studies, we looked at sequence conservation around the RRM domain for RBM20 using ConSurf (61,62) within the PredictProtein server (63) (Figure 1A). The N-terminus of the RRM domain was clearly defined at the glycine preceding strand β1, Gly519 in mRBM20, whereas the C-terminal limit of conservation extended to Arg621 and therefore past the residues required for a typical RRM domain (Lys603 in mRBM20). To prevent unwanted truncation, we thus included six additional residues at the N-terminus and 18 additional residues at the C-terminus (Figure 1A). This construct derived from mouse RBM20 (hereafter mRBM20(513–621)) produced soluble protein and displayed NMR spectroscopy data consistent with a folded domain (Figure 1B, black spectrum; Supplementary Figure S1).
Our choice for RNA ligand was based on previous studies that have identified UCUU as an enriched RNA motif in RBM20-based PAR-CLIP of HEK293 cells and HITS-CLIP on rat cardiomyocytes (32). In particular, the UCUU sequence in Ryr2 is required for direct interaction by RBM20 (32), and therefore we have selected a ligand based on this sequence. To help with synthesis and purification of the RNA oligonucleotide, we have included an extra adenosine at the 5′ and 3′ ends of the motif derived from the Ryr2 sequence. ITC using this AUCUUA RNA with mRBM20(513–621) shows a 1:1 interaction with a KD of 5.7 μM (Figure 1C, Table 1). To provide additional details of binding specificity we performed a series of ITC measurements in which we made conservative mutations of the RNA ligand in which each base was mutated to the other purine or pyrimidine, respectively (Table 1). There were no significant changes in affinity upon mutation of the first or last base to guanine, in keeping with the primary importance of the central UCUU motif. Changing the first uracil of the motif to cytosine resulted in only a twofold increase in KD (13 μM versus 5.7 μM). In contrast, mutation of each base in the remaining CUU sequence resulted in larger KD increases of 16–36 times that of the original motif (KD values of 95–210 μM).
Table 1.
Isothermal titration calorimetry (ITC) measurements
| Protein | RNA | K d (μM) | ΔH (kcal/mol) | ΔS (cal/mol·K) |
|---|---|---|---|---|
| mRBM20(513–621) | AUCUUA | 5.7 ± 0.2 | −19.7 ± 0.5 | −42 ± 2 |
| mRBM20(513–621) | GUCUUA | 4.0 ± 0.1 | −21.0 ± 0.5 | −46 ± 2 |
| mRBM20(513–621) | ACCUUA | 13 ± 2 | −17 ± 1 | −36 ± 4 |
| mRBM20(513–621) | AUUUUA | 140 ± 30 | −10 ± 1 | −18 ± 4 |
| mRBM20(513–621) | AUCCUA | 210 ± 3 | −9 ± 2 | −14 ± 8 |
| mRBM20(513–621) | AUCUCA | 95 ± 1 | −27.0 ± 0.2 | −72 ± 1 |
| mRBM20(513–621) | AUCUUG | 4.8 ± 0.1 | −18.8 ± 0.4 | −39 ± 2 |
| mRBM20(513–621)H523A | AUCUUA | 67 ± 12 | −37 ± 4 | −104 ± 15 |
| mRBM20(513–621)N526A | AUCUUA | 9 ± 1 | −19 ± 1 | −40 ± 5 |
| mRBM20(513–621)V537I | AUCUUA | 10.3 ± 0.7 | −18.6 ± 0.4 | −40 ± 1 |
| mRBM20(513–621)Q558A | AUCUUA | 3.4 ± 0.7 | −17.4 ± 0.4 | −34 ± 2 |
| mRBM20(513–621)F560A | AUCUUA | 156 ± 6 | −29 ± 2 | −80 ± 5 |
| mRBM20(513–621)Q577A | AUCUUA | 6.6 ± 0.8 | −22 ± 1 | −50 ± 3 |
| mRBM20(513–621)R591M | AUCUUA | 12.4 ± 0.6 | −22.3 ± 0.34 | −52 ± 1 |
| mRBM20(513–621)R595M | AUCUUA | 29.3 ± 0.3 | −29.1 ± 0.8 | −77 ± 3 |
| mRBM20(513–621)Y596A | AUCUUA | 74 ± 13 | −6 ± 1 | 0 ± 5 |
| mRBM20(513–609)Δα3 | AUCUUA | 43 ± 1 | −16.2 ± 0.2 | −35 ± 1 |
| mRBM20(513–649)+RS | AUCUUA | 4.3 ± 0.4 | −17 ± 2 | −33 ± 7 |
| mRBM20(513–621) | vIRES-SL a | 8 ± 1 | −6.7 ± 0.6 | 1 ± 2 |
a GGGACCUGGUCUUUCCAGGUCCC (derived from PDB ID 2N3O in which the underlined sequence base-pairs to form the stem).
Despite the relatively modest affinity to AUCUUA RNA, we found that addition of this ligand to 15N-labelled mRBM20(513–621) resulted in clear backbone amide chemical shift perturbation for a majority of the residues (Figure 1B, red spectrum). This effect is consistent with the formation of a stable and intimate protein-RNA complex, and in fact involves a greater number of residues than would be expected for the simple binding of the RNA to one face of the domain. Given the high quality of the resulting spectrum we decided to proceed directly to determine the atomic structure of the mRBM20 RRM domain bound to AUCUUA RNA.
Molecular details of RNA binding to RBM20
Using a combination of distance, dihedral and residual dipolar coupling restraints, we determined an ensemble of 25 structures for mRBM20(513–621) bound to AUCUUA RNA (Figure 1D, Table 2). The most notable feature in the complex is the presence of an additional C-terminal helix (α3) that follows the canonical RRM fold (Figure 1E). The atomic details also illustrate specific interaction between all four bases in the UCUU motif with either sidechain or backbone atoms of mRBM20(513–621).
Starting with the first uridine, U2 in the structure, the uracil base makes hydrophobic contacts to Leu589 and hydrogen bonds to the side chain of Asn526. Further contacts are made between Arg591 and the backbone 5′ phosphate (Figure 2A). Mutation of N526A results in only a small decrease in affinity (KD of 9 μM; Table 1) in keeping with the mild specificity for uracil. The base of the sole cytosine, C3, stacks onto His523 and the mutation H523A reduces affinity by a factor of 12 (KD of 67 μM; Table 1). The cytosine-specific amino group is recognized by a hydrogen bond from the sidechain of Thr594, with additional hydrogen bonds from Ser593 and the backbone amide of Thr594 (Figure 2B). The 5′ phosphate of C3 is in contact with the sidechains of Gln558 and Arg595, but only mutation of Arg affects binding affinity (Table 1, KD of 29.3 μM for R595M versus KD of 3.4 μM for Q558A). The U4 base stacks onto Phe560, with mutation F560A resulting in an increased KD of 156 μM (Table 1). In RBM20, there is notable binding affinity to the ribonucleotide granted by residues in the peptide linker that lies on top of the bound RNA. For the base of U4, a pair of hydrogen bonds from the backbone atoms of Gln600 aids in uracil specificity (Figure 2C). In addition, the aromatic ring of Tyr596, along with the sidechain of Phe560, defines a hydrophobic cleft which contacts hydrogens H5 and H6 of U4. Removal of the tyrosine aromatic ring in Y596A increases the KD to 74 μM (Table 1).
Figure 2.
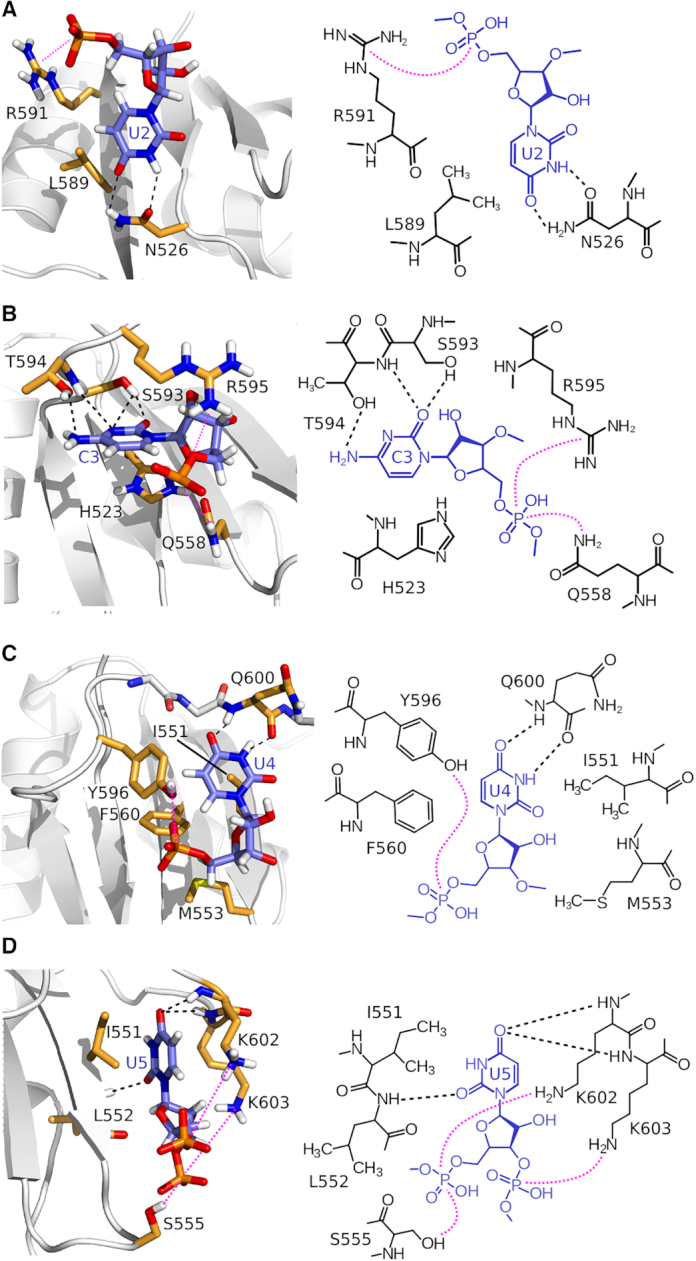
Molecular recognition of the UCUU motif by the RBM20 RRM domain. (A–D) Close up views of the intermolecular contacts between mRBM20(513–621) and (A) U2, (B) C3, (C) U4 and (D) U5 of the UCUU motif. On the left of each panel the contacts are indicated using the lowest energy structure, on the right is a schematic illustration showing the same contacts.
Compared to the canonical recognition of C3 and U4 bases across the strands β1 and β3, the final uracil in the motif, U5, binds in an atypical position between strand β2 and the loop before the extra C-terminal helix (Figure 2D). Hydrogen bonds from the backbone carbonyl of Leu552 and the backbone amides of Lys602 and Lys603 appear to guide specificity for the uracil base in this position. Due to the involvement of backbone atoms in this recognition, a simple site-directed mutation strategy can not be used to perturb binding of U5.
The C-terminal helix is required for RNA binding
Based on the structure of the RNA-bound complex, the extra C-terminal helix α3 in mRBM20(513–621) likely plays a role in the recognition of the 3′ uridine of the UCUU motif. By quantifying the change in 1H,15N amide crosspeak positions from the two spectra in Figure 1B, it is evident that addition of AUCUUA RNA causes widespread perturbation throughout mRBM20(513–621) (Figure 3A). The changes caused by ligand binding naturally include residues that lie below and above the bound RNA and are in direct contact with the ribonucleotides (Figure 3B). However, there is also significant chemical shift perturbation for all residues within the C-terminal helix, despite the fact that this helix lies distant to the bound RNA, on the opposite side of the RRM domain (Figure 3B).
Figure 3.
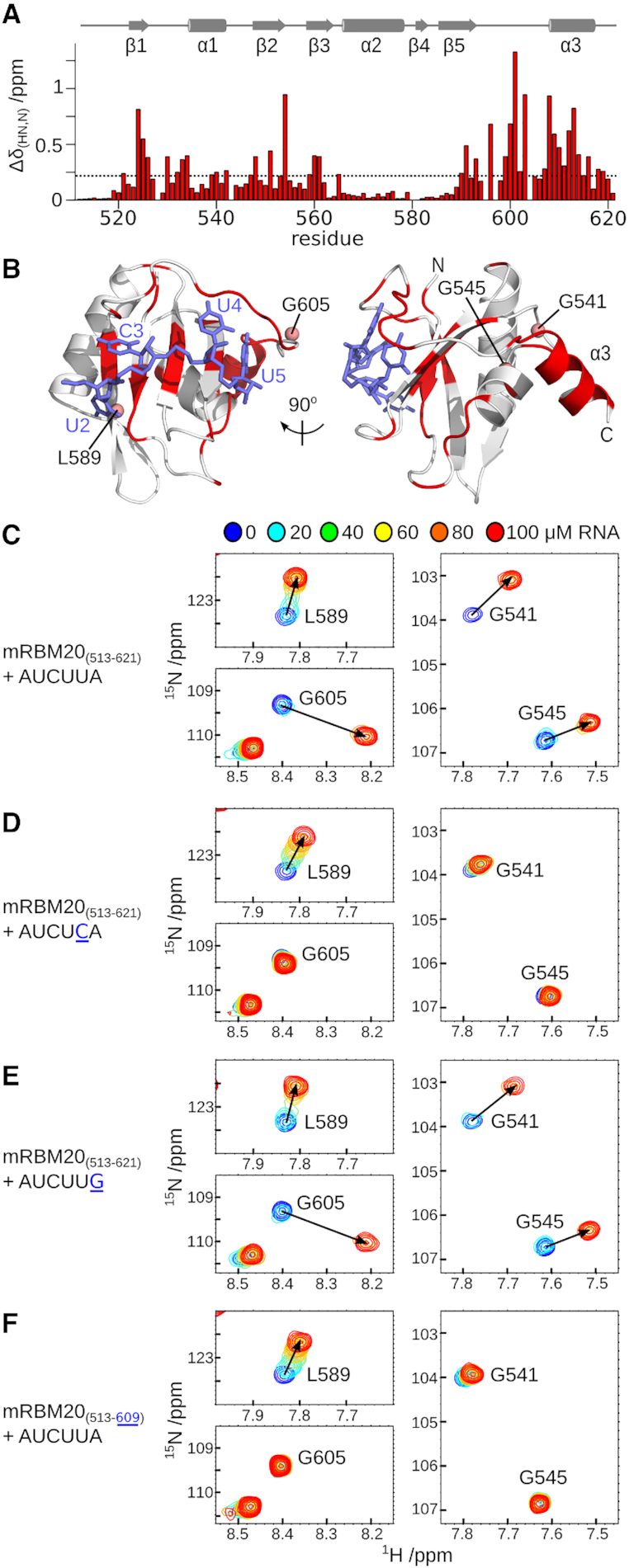
Helix α3 and nearby residues are affected by RNA binding. (A) Backbone amide chemical shift perturbation (ΔδHN,N) resulting from AUCUUA binding to mRBM20(513–621) from the spectra shown in Figure 1C, calculated as ((ΔδHN)2+(0.14*ΔδN)2)0.5. Secondary structure elements of RNA-bound mRBM20(513–621) are shown above the histogram. (B) Residues with ΔδHN,N greater than the average (0.23 ppm) are coloured red on the same views of RNA-bound mRBM20(513–621) as shown in Figure 1E. (C–F) Selected regions of the 1H,15N-HSQC spectra from 100 μM [15N]mRBM20(513–621) titrated with 0, 20, 40, 60, 80 and 100 μM RNA. The locations of Leu589, Gly605, Gly541 and Gly545 backbone amides in mRBM20(513–621) is indicated in (C). Complete spectra are shown in Supplementary Figure S3.
To gain further insight into the mechanism of RNA binding at the level of individual residues, we performed a series of titrations followed by NMR spectroscopy (Supplementary Figure S3). Starting with mRBM20(513–621), we followed the binding of the first and last uracil base within AUCUUA RNA by changes in 1H,15N-HSQC crosspeak positions for Leu589 and Gly605, respectively (Figure 3C). In addition, we find that residues distal to the RNA binding surface, but in contact with helix α3, are similarly affected by the ligand (Gly541 and Gly545; Figure 3C).
From our initial characterization of RNA binding to mRBM20(513–621) we noted that changing the terminal uracil to cytosine reduced binding affinity (Table 1). When we followed titration of mRBM20(513–621) with this AUCUCA RNA, the corresponding 1H,15N-HSQC shows that RNA still induced changes in Leu589 (Figure 3D). Therefore, the first uracil in AUCUCA is likely still recognized by RBM20. In contrast, the Gly605 crosspeak is unperturbed, implying that the introduced cytosine in position 5 is no longer able to interact with the protein (Figure 3D). At the same time, the crosspeaks of Gly541 and Gly545 are also unaffected by RNA binding (Figure 3D). Together, the data indicate that changing the terminal uracil to cytosine prevents interaction with mRBM20(513–621) and that this loss in binding also prevents chemical shift perturbation for residues in the C-terminal helix. As a control, the titration of mRBM20(513–621) with RNA ligand AUCUUG, in which the final adenine is replaced by guanine and affinity is maintained, does not affect the binding behaviour in the measured NMR spectra (Figure 3E).
We next designed a truncated form, mRBM20(513–609)Δα3, in which only the α3 helical region has been removed, but all RNA-binding residues are retained. From ITC measurements, the loss of α3 results in a decreased affinity by a factor of eight (KD of 43 μM; Table 1), supporting a key role for this helix in RNA recognition. When the mRBM20(513–609)Δα3 mutant is titrated with AUCUUA RNA, the corresponding 1H,15N-HSQC NMR data show that the overall domain fold is retained, and that Leu589 still reports on binding by the first uracil (Figure 3F). In contrast, the Gly605 crosspeak is unperturbed, implying that there is no interaction between the protein and the final nucleotide in the RNA motif even though the preferred uracil base is present (Figure 3F). This lack of perturbation once again extends to the crosspeaks of Gly541 and Gly545 (Figure 3F). The results indicate that although the C-terminal helix does not directly interact with the RNA, the helix is nonetheless required to form the binding site for the final uracil base in the motif. In addition, the RNA-induced chemical shift changes observed for Gly541 and Gly545 requires the presence of the C-terminal helix. We therefore hypothesize that helix α3 may not be a constant part of the RBM20 RRM domain, but may be selectively stabilized only when the final uridine is present in the RNA motif.
The C-terminus is disordered in the unbound state
To determine the nature of helix α3 in the unbound state, we calculated an ensemble of NMR-based structures for the free mRBM20(513–621) (Figure 4A, Table 2). The core RRM fold is nearly identical between the bound and free states with backbone rmsd of 0.01 Å for residues 520–598 calculated between the two ensembles using SuperPose (64). In contrast, the C-terminal region in unbound mRBM20(513–621) clearly lacks a stable helical fold (Figure 4B). This region instead displays increased disorder as compared to the RNA-bound state, and by NMR spectroscopy is more flexible (Figure 4C). A small plateau of similar backbone dynamics for a stretch of residues in the C-terminus might correspond to a transient helix for the unbound mRBM20(513–621). A very slight helicity in the α3 region in the free state is also indicated upon analysis of backbone 13Cα secondary chemical shifts (Figure 4D). Upon binding RNA, the helix α3 region and preceding loop shifts towards less dynamics to form the RNA-bound complex.
Figure 4.
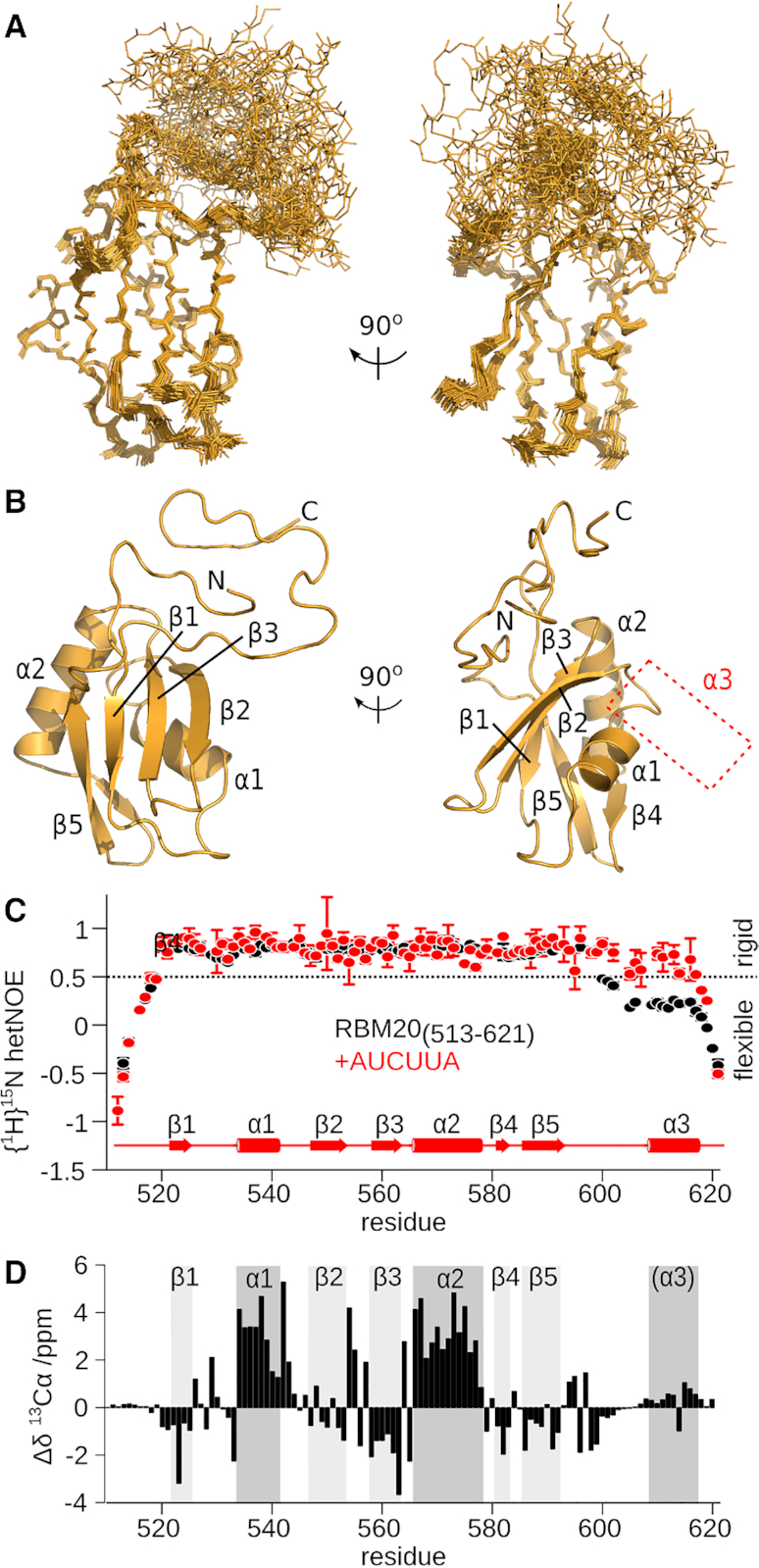
The helix α3 region is disordered in unbound mRBM20(513–621). (A) Ensemble of 25 lowest energy structures calculated for unbound mRBM20(513–621) (backbone heavy atoms, orange lines). (B) Lowest energy structure model for mRBM20 (orange cartoon). Protein secondary structure elements are labelled. The location of α3 in the RNA-bound complex is also indicated. (C) Flexibility of the backbone amides as measured by {1H}15N hetNOE of the unbound (black) and RNA-bound (red) [15N]mRBM20(513–621). The secondary structure elements of RNA-bound mRBM20(513–621) is also shown. Residues with {1H}15N hetNOE values <0.5 are considered to be disordered. (D) 13Cα secondary chemical shift (Δδ) of the unbound mRBM20(513–621) compared to a disordered peptide as predicted by using ncIDP (75). The secondary structure elements of RNA-bound mRBM20(513–621) are indicated by grey boxes.
Residues implicated in disease have little effect on RNA binding
Most of the RBM20 mutations implicated in cardiomyopathy are localized within the RS region, and these mutants mainly disrupt nuclear localization (41). To see if the addition of the RS region could in general affect RNA binding, we prepared a construct with the C-terminus extended to residue 649. This longer construct had the same affinity to AUCUUA RNA as mRBM20(531-621) (KD of 4.3 μM compared to 5.7 μM; Table 1). The similar KD values rule out a significant contribution to RNA binding by the RS region, at least for the unphosphorylated protein, and therefore mutations in this area would likely not act via simple changes in RNA binding. The only RRM domain mutant with possible link to disease, V537I, is located distant to the RNA-binding surface but is close to the area contacted by the C-terminal helix, as well as to Leu552 whose backbone amide is involved in U5 recognition (Supplementary Figure S4). By ITC, the affinity of V537I to AUCUUA RNA shows a modest two-fold reduction (KD of 10.3 μM; Table 1).
Structural similarity to polypyrimidine tract-binding protein
Given the importance of the added C-terminal helix in the RBM20 RRM domain, we searched for other RRM domains that may use the same RNA-binding mechanism. The only candidate we found was the first RRM domain of polypyrimidine tract-binding protein (PTBP1) bound to a viral internal ribosome entry site (vIRES) stemloop RNA (PDB ID 2N3O; (65). A sequence similarity was also previously identified in the RNP1 motifs of RBM20 and RRM1 of PTBP1 (66). In both RNA-bound structures, a UCUU sequence is involved in the interaction with a conserved hydrogen bond network to the CUU bases (Figure 5A, B). The 3′ uracil base in both cases is recognized by a hydrogen bond from the uracil O2 oxygen to a backbone amide within strand β2 (Leu552 in RBM20, and Lys92 in PTBP1; Figure 5B). Additional hydrogen bonds connect oxygen O4 to backbone amides in the loop preceding helix α3 (Figure 5B), although for PTBP1 these hydrogen bonds are present in only a subset of the ensemble. Despite the presence of α3 in both structures, the sequence conservation is low in the region C-terminal to the core RRM fold (Figure 5C, D). Most residues that lie atop the bound RNA differ between RBM20 and PTBP1 RRM1 (Figure 5D, left). More noticeable is that the C-terminal helix is significantly shifted between the two structures, although the overall path of the C-terminal residues is similar (Figure 5D, right). In terms of the RNA ligand, a difference between the two complexes is that RBM20 is bound to a short single-stranded RNA, whereas PTBP1 RRM1 binds UCUU within the loop of the vIRES stemloop. In the vIRES stemloop the first uridine is part of a U:U base pair within the stem structure, and thus can not be bound in the same orientation as with the ssRNA ligand. Using ITC we nevertheless find that mRBM20(513–621) can also interact with the vIRES stemloop UCUU with almost the same affinity as for ssRNA (KD of 8 μM verses 5.7 μM; Table 1).
Figure 5.
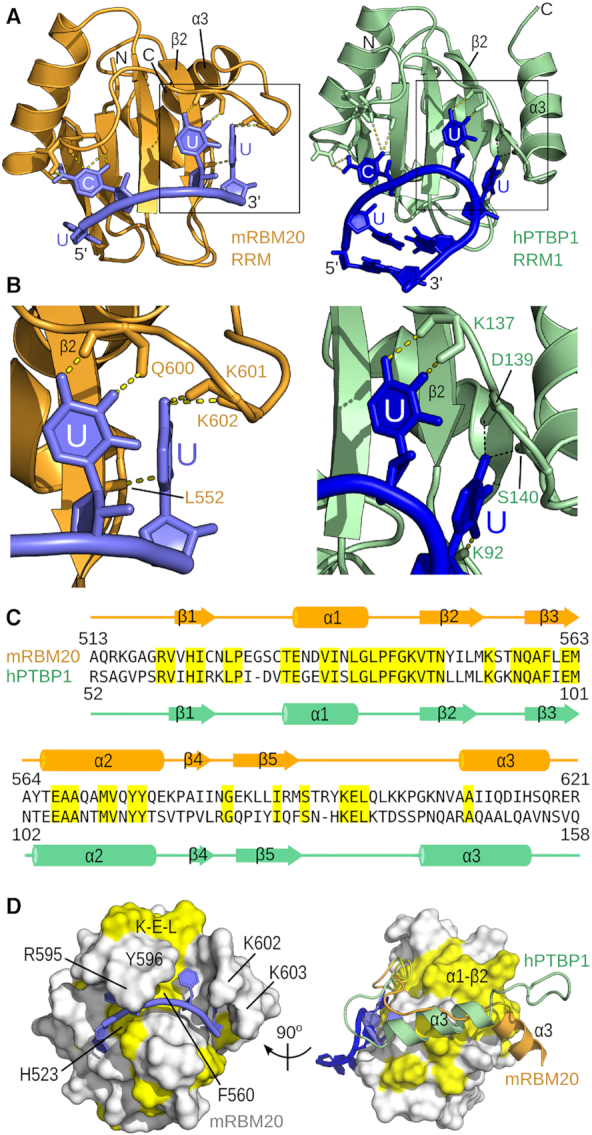
Structural similarity to RNA-bound PTBP1 RRM1. (A) Cartoon representation of protein-RNA complexes involving mRBM20 RRM domain (left) or hPTBP1 RRM1 (right, from PDB ID 2N30) bound to RNA containing a UCUU motif. Hydrogen bonds to the bases are shown as dashed lines. (B) Close-up views from (A) of hydrogen bonds to U4 and U5 in the RBM20 complex, and the corresponding interactions with PTBP1 RRM1. The hydrogen bonds from Asp139 and Ser140 to uracil in the PTBP1 structure, indicated by thin black dashed lines, are only present in a subset of structures within the ensemble. (C) Sequence alignment of mouse RBM20 RRM and human PTBP1 RRM1, with identical residues highlighted in yellow. Protein secondary structure elements are shown from each RNA-bound complex. (D) Conserved residues from (C) are coloured yellow on the model of RNA-bound mRBM20(513–621) using the same views as in Figure 1E. The conserved Lys-Glu-Leu region found in the β5-α3 loop of both proteins is indicated as K–E–L. On the right, the C-terminal residues of both mRBM20 (orange) and hPTBP1 (light green) are shown in cartoon representation, with the core RRM domain fold as a surface representation.
DISCUSSION
We have demonstrated in atomic detail the way in which the RRM domain from RBM20 is specific for the UCUU RNA sequence. Each base in the motif is recognized by the RRM domain, with an atypical binding of the final uridine nucleotide coupled to stabilization of a C-terminal helix (Figure 6A). These results establish the RRM domain as a major determinant of the RNA binding specificity of RBM20, and a basis of UCUU enrichment in target pre-mRNA.
Figure 6.
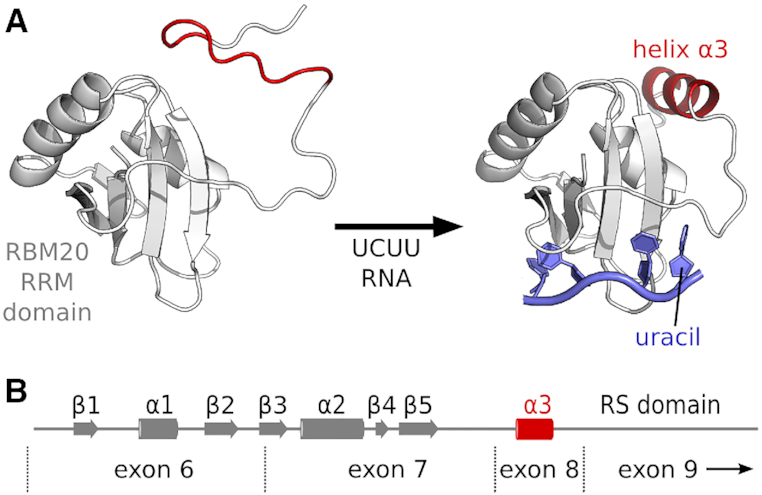
Model of RNA binding by RBM20. (A) In the unbound state, the RRM domain from RBM20 has a disordered C-terminus. Upon binding with an RNA ligand containing the sequence UCUU, the 3′ uracil combines with formation of a C-terminal helix to stabilize the protein-RNA complex. (B) The C-terminal α3 helix is encoded by exon 8 of the rbm20 gene, in between exons 6 and 7 that encode the canonical RRM domain fold, and the RS domain in exon 9.
The main finding from the structural studies is the unusual role of the C-terminal helix in RNA recognition. Despite its importance in RNA binding, helix α3 itself does not directly interact with the uridine nucleotide in the complex, but instead packs against β2 and helix α1 of the core RRM domain. This finding is notable since the low affinity binding by truncated construct mRBM20(513–609)Δα3, in which this helix is removed, nonetheless retains all of the residues and backbone atoms that directly contact RNA. In this context, the helix mainly represents an additional but key point of stabilization for the loop residues that directly contact RNA. This model explains the lack of sequence conservation between the C-terminal helices of RBM20 and PTBP1 RRM1 (Figure 5C).
An absolute need for residues C-terminal to the canonical RRM fold means that exon 8 of RBM20 is a required part of the functional RRM domain. The functional RRM domain from RBM20 therefore consists of the region stretching from exon 6 to exon 8, followed directly by the RS domain encoded by exon 9 (Figure 6B). This situation is shared with PTBP1 RRM1, in which the C-terminal helix is also encoded by the exon that follows the canonical RRM domain.
It is anticipated that additional domains in RBM20, as well as interaction with other proteins, help to further restrict RNA binding to specific motifs in target pre-mRNA. For example, the two zinc finger domains in RBM20 may contribute directly to RNA binding. The C-terminal zinc finger is essential for RBM20 function (37), however so far neither of the two zinc fingers have been shown to bind RNA. The RS domain is required for nuclear localization (41) and likely mediates protein-protein interactions (32). We have shown that inclusion of the unphosphorylated RS domain does not directly affect affinity to AUCUUA RNA (Table 1).
Determination of RBM20 binding sites may also be dictated by additional splicing factors that bind to the same pre-mRNA. The polypyrimidine tract-binding protein PTBP1 has been found to co-regulate splicing with RBM20 for FHOD3 pre-mRNA (34), and also towards a mini-gene reporter based on titin (37). This co-regulation could be explained by proximal binding of RBM20 and PTBP1 on the pre-mRNA. Both proteins display a similar preference of UC-rich motifs, and in this study we have found similarity in UCUU binding mechanisms between RBM20 and RRM1 of PTBP1 (Figure 5). RBM20 can equally recognize this RNA motif in short single-stranded RNA as well as in the context of a loop in stemloop RNA (Table 1), but it is possible that protein-protein interactions between RBM20 and PTBP1 could help define discrete high-affinity binding sites on the target pre-mRNA. However, a direct interaction between the two proteins has not yet been observed. Alternately, RBM20 and PTBP1 could independently contribute to splicing efficiency, and in this case additional structural features, other splicing factors, or surrounding sequence elements in the pre-mRNA may help refine RBM20 and PTBP1 binding sites.
RBM24 is another protein factor found to co-regulate splicing with RBM20, in this case towards. inclusion of exon 11 from Enh pre-mRNA (35). A direct interaction was found in vitro between the full-length RBM20 and RBM24 proteins, and a cooperative binding by both proteins might help define target specificity to the region upstream of exon 11. In support of functional cooperativity, single mouse knockouts of RBM20 or RBM24 have only minimal effect on exon 11, whereas simultaneously increasing or decreasing RBM20 and RBM24 shifts Enh towards the long or short isoform, respectively (35).
Our molecular study of RNA binding by the RBM20 RRM domain helps in the understanding of binding site specificity, but the long residence time of RBM20 on pre-mRNA targets such as titin (28) suggests the contribution of additional elements that stabilize the protein–RNA complexes. These additional contacts could include regions of RBM20 that mediate RBM20–RBM20 contacts, as well as possible contacts to proteins such as PTBP1 and RBM24.
The C-terminal helix in RBM20 RRM domain is critical to RNA selectivity, but the reverse is also true: binding of a high affinity UCUU motif is required to form the C-terminal helix. As a consequence, only by binding the complete motif would the residues following the canonical RRM domain condense into a stabilized helix. This triggered effect would reduce the distance to the following RS region in RBM20, and also alter the accessible surface features on the RRM domain that could either create or inhibit interaction with auxiliary binding partners.
In a broader sense, the RNA-binding mechanism of the RBM20 RRM domain further adds to the impressive diversity of structural features used by the family of RRM domains to interact with their ligands (reviewed in (67–69)). Such variety includes the region on the RRM domain involved in binding the ligand, as well as appended secondary structure elements key to specificity and affinity. For RBM20, the key aspect of helix formation upon binding RNA is uncommon. Some similarity can be seen in the linker region between the RRM and Leucine-Rich Repeat (LRR) domains of the export factor TAP which forms a helix upon binding the constitutive transport element (CTE) RNA (70). In the case of the La family protein p65, the C-terminal RRM domain also uses an appended helix α3 to interact with RNA. This C-terminal helix is already present in the absence of the ligand, but interaction with the telomerase stem IV RNA is coupled with extension of the helix by at least 14 residues (71). Outside of the RRM family, other RNA-binding proteins can be found for which ligand interaction helps to stabilize a helical fold. One such example is the RNA-binding domain of the transport factor PHAX, which undergoes substantial stabilization of its helical fold upon interaction with a range of RNA ligands (72).
Given the diversity of ligand binding strategies already observed within the family of RRM domains, it is likely that the structure determination of additional RRM domain complexes will reveal even more variation.
DATA AVAILABILITY
The structure ensemble of mRBM20(513–621) bound to AUCUUA RNA has been deposited at the Protein Data Bank (http://www.ebi.ac.uk/pdbe/) with accession ID 6SO9. The ensemble of structures for unbound mRBM20(513–621) has also been deposited, with accession ID 6SOE. Chemical shift assignments for RNA-bound and unbound mRBM20(513–621) have been deposited in the Biological Magnetic Resonance Data Bank (http://bmrb.wisc.edu/) under BMRB accession numbers 34428 and 34429, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
Plasmids encoding mouse RBM20 cDNA were provided by Pamela Lorenzi of the Department of Neurological, Biomedical and Movement Sciences, University of Verona. We thank Axelle Grélard, Estelle Morvan, and the structural biology platform at the Institut Europèen de Chimie et Biologie for access to the NMR spectrometers, equipment and technical assistance. SKU acknowledges Dr Souvik Maiti for support and assistance with the research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
DST INSPIRE Faculty Award [IFA-13 CH-102 to S.K.U.] from the Government of India, Department of Science and Technology; IR-RMN-THC Fr3050 CNRS; INSERM, CNRS and University of Bordeaux (to C.D.M.). Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1. Maruyama K., Natori R., Nonomura Y.. New elastic protein from muscle. Nature. 1976; 262:58–60. [DOI] [PubMed] [Google Scholar]
- 2. Wang K., McClure J., Tu A.. Titin: major myofibrillar components of striated muscle. Proc. Natl. Acad. Sci. U.S.A. 1979; 76:3698–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Azad A., Poloni G., Sontayananon N., Jiang H., Gehmlich K.. The giant titin: how to evaluate its role in cardiomyopathies. J. Muscle Res. Cell Motil. 2019; 40:159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Granzier H., Labeit S.. Cardiac titin: an adjustable multi-functional spring. J. Physiol. 2002; 541:335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guo W., Bharmal S.J., Esbona K., Greaser M.L.. Titin diversity–alternative splicing gone wild. J. Biomed. Biotechnol. 2010; 2010:753675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gigli M., Begay R.L., Morea G., Graw S.L., Sinagra G., Taylor M.R.G., Granzier H., Mestroni L.. A review of the giant protein titin in clinical molecular diagnostics of cardiomyopathies. Front. Cardiovasc. Med. 2016; 3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo W., Schafer S., Greaser M.L., Radke M.H., Liss M., Govindarajan T., Maatz H., Schulz H., Li S., Parrish A.M. et al.. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat. Med. 2012; 18:766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Greaser M.L., Krzesinski P.R., Warren C.M., Kirkpatrick B., Campbell K.S., Moss R.L.. Developmental changes in rat cardiac titin/connectin: transitions in normal animals and in mutants with a delayed pattern of isoform transition. J. Muscle Res. Cell Motil. 2005; 26:325–332. [DOI] [PubMed] [Google Scholar]
- 9. Brauch K.M., Karst M.L., Herron K.J., de Andrade M., Pellikka P.A., Rodeheffer R.J., Michels V.V., Olson T.M.. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J. Am. Coll. Cardiol. 2009; 54:930–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao Y., Feng Y., Zhang Y.-M., Ding X.-X., Song Y.-Z., Zhang A.-M., Liu L., Zhang H., Ding J.-H., Xia X.-S.. Targeted next-generation sequencing of candidate genes reveals novel mutations in patients with dilated cardiomyopathy. Int. J. Mol. Med. 2015; 36:1479–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Long P.A., Evans J.M., Olson T.M.. Diagnostic yield of whole exome sequencing in pediatric dilated cardiomyopathy. J. Cardiovasc. Dev. Dis. 2017; 4:E11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wells Q.S., Becker J.R., Su Y.R., Mosley J.D., Weeke P., D’Aoust L., Ausborn N.L., Ramirez A.H., Pfotenhauer J.P., Naftilan A.J. et al.. Whole exome sequencing identifies a causal RBM20 mutation in a large pedigree with familial dilated cardiomyopathy. Circ. Cardiovasc. Genet. 2013; 6:317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Waldmüller S., Schroeder C., Sturm M., Scheffold T., Imbrich K., Junker S., Frische C., Hofbeck M., Bauer P., Bonin M. et al.. Targeted 46-gene and clinical exome sequencing for mutations causing cardiomyopathies. Mol. Cell. Probes. 2015; 29:308–314. [DOI] [PubMed] [Google Scholar]
- 14. Chami N., Tadros R., Lemarbre F., Lo K.S., Beaudoin M., Robb L., Labuda D., Tardif J.-C., Racine N., Talajic M. et al.. Nonsense mutations in BAG3 are associated with early-onset dilated cardiomyopathy in French Canadians. Can. J. Cardiol. 2014; 30:1655–1661. [DOI] [PubMed] [Google Scholar]
- 15. Refaat M.M., Lubitz S.A., Makino S., Islam Z., Frangiskakis J.M., Mehdi H., Gutmann R., Zhang M.L., Bloom H.L., MacRae C.A. et al.. Genetic variation in the alternative splicing regulator RBM20 is associated with dilated cardiomyopathy. Heart Rhythm. 2012; 9:390–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rampersaud E., Siegfried J.D., Norton N., Li D., Martin E., Hershberger R.E.. Rare variant mutations identified in pediatric patients with dilated cardiomyopathy. Prog. Pediatr. Cardiol. 2011; 31:39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Millat G., Bouvagnet P., Chevalier P., Sebbag L., Dulac A., Dauphin C., Jouk P.-S., Delrue M.-A., Thambo J.-B., Le Metayer P. et al.. Clinical and mutational spectrum in a cohort of 105 unrelated patients with dilated cardiomyopathy. Eur. J. Med. Genet. 2011; 54:e570–e575. [DOI] [PubMed] [Google Scholar]
- 18. Li D., Morales A., Gonzalez-Quintana J., Norton N., Siegfried J.D., Hofmeyer M., Hershberger R.E.. Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin. Transl. Sci. 2010; 3:90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Robyns T., Willems R., Van Cleemput J., Jhangiani S., Muzny D., Gibbs R., Lupski J.R., Breckpot J., Devriendt K., Corveleyn A.. Whole exome sequencing in a large pedigree with DCM identifies a novel mutation in RBM20. Acta Cardiol. 2019; doi:10.1080/00015385.2019.1674490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nielsen J.B., Thorolfsdottir R.B., Fritsche L.G., Zhou W., Skov M.W., Graham S.E., Herron T.J., McCarthy S., Schmidt E.M., Sveinbjornsson G. et al.. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 2018; 50:1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parikh V.N., Caleshu C., Reuter C., Lazzeroni L.C., Ingles J., Garcia J., McCaleb K., Adesiyun T., Sedaghat-Hamedani F., Kumar S. et al.. Regional variation in RBM20 causes a highly penetrant arrhythmogenic cardiomyopathy. Circ. Heart Fail. 2019; 12:e005371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rindler T.N., Hinton R.B., Salomonis N., Ware S.M.. Molecular characterization of pediatric restrictive cardiomyopathy from integrative genomics. Sci. Rep. 2017; 7:39276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sedaghat-Hamedani F., Haas J., Zhu F., Geier C., Kayvanpour E., Liss M., Lai A., Frese K., Pribe-Wolferts R., Amr A. et al.. Clinical genetics and outcome of left ventricular non-compaction cardiomyopathy. Eur. Heart J. 2017; 38:3449–3460. [DOI] [PubMed] [Google Scholar]
- 24. Kayvanpour E., Sedaghat-Hamedani F., Amr A., Lai A., Haas J., Holzer D.B., Frese K.S., Keller A., Jensen K., Katus H.A. et al.. Genotype-phenotype associations in dilated cardiomyopathy: meta-analysis on more than 8000 individuals. Clin. Res. Cardiol. 2017; 106:127–139. [DOI] [PubMed] [Google Scholar]
- 25. Hey T.M., Rasmussen T.B., Madsen T., Aagaard M.M., Harbo M., Mølgaard H., Møller J.E., Eiskjær H., Mogensen J.. Pathogenic RBM20 -variants are associated with a severe disease expression in male patients with dilated cardiomyopathy. Circ. Heart Fail. 2019; 12:e005700. [DOI] [PubMed] [Google Scholar]
- 26. Beraldi R., Li X., Martinez Fernandez A., Reyes S., Secreto F., Terzic A., Olson T.M., Nelson T.J.. Rbm20-deficient cardiogenesis reveals early disruption of RNA processing and sarcomere remodeling establishing a developmental etiology for dilated cardiomyopathy. Hum. Mol. Genet. 2014; 23:3779–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo W., Pleitner J.M., Saupe K.W., Greaser M.L.. Pathophysiological defects and transcriptional profiling in the RBM20-/- rat model. PLoS One. 2013; 8:e84281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li S., Guo W., Dewey C.N., Greaser M.L.. Rbm20 regulates titin alternative splicing as a splicing repressor. Nucleic Acids Res. 2013; 41:2659–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khan M.A.F., Reckman Y.J., Aufiero S., van den Hoogenhof M.M.G., van der Made I., Beqqali A., Koolbergen D.R., Rasmussen T.B., van der Velden J., Creemers E.E. et al.. RBM20 regulates circular RNA production from the titin gene. Circ. Res. 2016; 119:996–1003. [DOI] [PubMed] [Google Scholar]
- 30. van den Hoogenhof M.M.G., Beqqali A., Amin A.S., van der Made I., Aufiero S., Khan M.A.F., Schumacher C.A., Jansweijer J.A., van Spaendonck-Zwarts K.Y., Remme C.A. et al.. RBM20 mutations induce an arrhythmogenic dilated cardiomyopathy related to disturbed calcium handling. Circulation. 2018; 138:1330–1342. [DOI] [PubMed] [Google Scholar]
- 31. Guo W., Zhu C., Yin Z., Wang Q., Sun M., Cao H., Greaser M.L.. Splicing factor RBM20 regulates transcriptional network of titin associated and calcium handling genes in the heart. Int. J. Biol. Sci. 2018; 14:369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maatz H., Jens M., Liss M., Schafer S., Heinig M., Kirchner M., Adami E., Rintisch C., Dauksaite V., Radke M.H. et al.. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J. Clin. Invest. 2014; 124:3419–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wyles S.P., Li X., Hrstka S.C., Reyes S., Oommen S., Beraldi R., Edwards J., Terzic A., Olson T.M., Nelson T.J.. Modeling structural and functional deficiencies of RBM20 familial dilated cardiomyopathy using human induced pluripotent stem cells. Hum. Mol. Genet. 2016; 25:254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lorenzi P., Sangalli A., Fochi S., Dal Molin A., Malerba G., Zipeto D., Romanelli M.G.. RNA-binding proteins RBM20 and PTBP1 regulate the alternative splicing of FHOD3. Int. J. Biochem. Cell Biol. 2019; 106:74–83. [DOI] [PubMed] [Google Scholar]
- 35. Ito J., Iijima M., Yoshimoto N., Niimi T., Kuroda S.’ichi, Maturana A.D.. RBM20 and RBM24 cooperatively promote the expression of short enh splice variants. FEBS Lett. 2016; 590:2262–2274. [DOI] [PubMed] [Google Scholar]
- 36. Bertero A., Fields P.A., Ramani V., Bonora G., Yardimci G.G., Reinecke H., Pabon L., Noble W.S., Shendure J., Murry C.E.. Dynamics of genome reorganization during human cardiogenesis reveal an RBM20-dependent splicing factory. Nat. Commun. 2019; 10:1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dauksaite V., Gotthardt M.. Molecular basis of titin exon exclusion by RBM20 and the novel titin splice regulator PTB4. Nucleic Acids Res. 2018; 46:5227–5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Methawasin M., Hutchinson K.R., Lee E.-J., Smith J.E. 3rd, Saripalli C., Hidalgo C.G., Ottenheijm C.A.C., Granzier H.. Experimentally increasing titin compliance in a novel mouse model attenuates the Frank-Starling mechanism but has a beneficial effect on diastole. Circulation. 2014; 129:1924–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van der Pijl R., Strom J., Conijn S., Lindqvist J., Labeit S., Granzier H., Ottenheijm C.. Titin-based mechanosensing modulates muscle hypertrophy. J. Cachexia Sarcopenia Muscle. 2018; 9:947–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Watanabe T., Kimura A., Kuroyanagi H.. Alternative splicing regulator RBM20 and cardiomyopathy. Front Mol Biosci. 2018; 5:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murayama R., Kimura-Asami M., Togo-Ohno M., Yamasaki-Kato Y., Naruse T.K., Yamamoto T., Hayashi T., Ai T., Spoonamore K.G., Kovacs R.J. et al.. Phosphorylation of the RSRSP stretch is critical for splicing regulation by RNA-Binding Motif Protein 20 (RBM20) through nuclear localization. Sci. Rep. 2018; 8:8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamamoto T., Miura A., Itoh K., Takeshima Y., Nishio H.. RNA sequencing reveals abnormal LDB3 splicing in sudden cardiac death. Forensic Sci. Int. 2019; 302:109906. [DOI] [PubMed] [Google Scholar]
- 43. Kimura A. Molecular genetics and pathogenesis of cardiomyopathy. J. Hum. Genet. 2016; 61:41–50. [DOI] [PubMed] [Google Scholar]
- 44. Buck D., Smith J.E. 3rd, Chung C.S., Ono Y., Sorimachi H., Labeit S., Granzier H.L.. Removal of immunoglobulin-like domains from titin's spring segment alters titin splicing in mouse skeletal muscle and causes myopathy. J. Gen. Physiol. 2014; 143:215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hinze F., Dieterich C., Radke M.H., Granzier H., Gotthardt M.. Reducing RBM20 activity improves diastolic dysfunction and cardiac atrophy. J. Mol. Med. 2016; 94:1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bull M., Methawasin M., Strom J., Nair P., Hutchinson K., Granzier H.. Alternative splicing of titin restores diastolic function in an HFpEF-Like genetic murine model (TtnΔIAjxn). Circ. Res. 2016; 119:764–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Methawasin M., Strom J.G., Slater R.E., Fernandez V., Saripalli C., Granzier H.. Experimentally increasing the compliance of titin through RNA binding motif-20 (RBM20) inhibition improves diastolic function in a mouse model of heart failure with preserved ejection fraction. Circulation. 2016; 134:1085–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pulcastro H.C., Awinda P.O., Methawasin M., Granzier H., Dong W., Tanner B.C.W.. Increased titin compliance reduced length-dependent contraction and slowed cross-bridge kinetics in skinned myocardial strips from rbm (20ΔRRM) mice. Front. Physiol. 2016; 7:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 50. Senn H., Werner B., Messerle B.A., Weber C., Traber R., Wüthrich K.. Stereospecific assignment of the methyl 1H NMR lines of valine and leucine in polypeptides by nonrandom 13C labelling. FEBS Lett. 1989; 249:113–118. [Google Scholar]
- 51. Rieping W., Habeck M., Bardiaux B., Bernard A., Malliavin T.E., Nilges M.. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007; 23:381–382. [DOI] [PubMed] [Google Scholar]
- 52. Brunger A.T. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2007; 2:2728–2733. [DOI] [PubMed] [Google Scholar]
- 53. Shen Y., Bax A.. Protein structural information derived from NMR chemical shift with the neural network program TALOS-N. Methods Mol. Biol. 2015; 1260:17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hansen D.F., Neudecker P., Kay L.E.. Determination of isoleucine side-chain conformations in ground and excited states of proteins from chemical shifts. J. Am. Chem. Soc. 2010; 132:7589–7591. [DOI] [PubMed] [Google Scholar]
- 55. Hansen D.F., Neudecker P., Vallurupalli P., Mulder F.A.A., Kay L.E.. Determination of Leu side-chain conformations in excited protein states by NMR relaxation dispersion. J. Am. Chem. Soc. 2010; 132:42–43. [DOI] [PubMed] [Google Scholar]
- 56. Liu Y., Prestegard J.H.. A device for the measurement of residual chemical shift anisotropy and residual dipolar coupling in soluble and membrane-associated proteins. J. Biomol. NMR. 2010; 47:249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Keller S., Vargas C., Zhao H., Piszczek G., Brautigam C.A., Schuck P.. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 2012; 84:5066–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Scheuermann T.H., Brautigam C.A.. High-precision, automated integration of multiple isothermal titration calorimetric thermograms: new features of NITPIC. Methods. 2015; 76:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhao H., Piszczek G., Schuck P.. SEDPHAT – a platform for global ITC analysis and global multi-method analysis of molecular interactions. Methods. 2015; 76:137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brautigam C.A. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 2015; 562:109–133. [DOI] [PubMed] [Google Scholar]
- 61. Berezin C., Glaser F., Rosenberg J., Paz I., Pupko T., Fariselli P., Casadio R., Ben-Tal N.. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004; 20:1322–1324. [DOI] [PubMed] [Google Scholar]
- 62. Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N.. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016; 44:W344–W350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yachdav G., Kloppmann E., Kajan L., Hecht M., Goldberg T., Hamp T., Hönigschmid P., Schafferhans A., Roos M., Bernhofer M. et al.. PredictProtein—an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014; 42:W337–W343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Maiti R., Van Domselaar G.H., Zhang H., Wishart D.S.. SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 2004; 32:W590–W594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dorn G., Leitner A., Boudet J., Campagne S., von Schroetter C., Moursy A., Aebersold R., Allain F.H.-T.. Structural modeling of protein-RNA complexes using crosslinking of segmentally isotope-labeled RNA and MS/MS. Nat. Methods. 2017; 14:487–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Filippello A., Lorenzi P., Bergamo E., Romanelli M.G.. Identification of nuclear retention domains in the RBM20 protein. FEBS Lett. 2013; 587:2989–2995. [DOI] [PubMed] [Google Scholar]
- 67. Maris C., Dominguez C., Allain F.H.-T.. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005; 272:2118–2131. [DOI] [PubMed] [Google Scholar]
- 68. Cléry A., Blatter M., Allain F.H.-T.. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008; 18:290–298. [DOI] [PubMed] [Google Scholar]
- 69. Daubner G.M., Cléry A., Allain F.H.-T.. RRM-RNA recognition: NMR or crystallography…and new findings. Curr. Opin. Struct. Biol. 2013; 23:100–108. [DOI] [PubMed] [Google Scholar]
- 70. Teplova M., Wohlbold L., Khin N.W., Izaurralde E., Patel D.J.. Structure-function studies of nucleocytoplasmic transport of retroviral genomic RNA by mRNA export factor TAP. Nat. Struct. Mol. Biol. 2011; 18:990–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Singh M., Wang Z., Koo B.-K., Patel A., Cascio D., Collins K., Feigon J.. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol. Cell. 2012; 47:16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mourao A., Varrot A., Mackereth C.D., Cusack S., Sattler M.. Structure and RNA recognition by the snRNA and snoRNA transport factor PHAX. RNA. 2010; 16:1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cornilescu G., Marquardt J.L., Ottiger M., Bax A.. Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J. Am. Chem. Soc. 1998; 120:6836–6837. [Google Scholar]
- 74. Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M.. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996; 8:477–486. [DOI] [PubMed] [Google Scholar]
- 75. Tamiola K., Acar B., Mulder F.A.A.. Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J. Am. Chem. Soc. 2010; 132:18000–18003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure ensemble of mRBM20(513–621) bound to AUCUUA RNA has been deposited at the Protein Data Bank (http://www.ebi.ac.uk/pdbe/) with accession ID 6SO9. The ensemble of structures for unbound mRBM20(513–621) has also been deposited, with accession ID 6SOE. Chemical shift assignments for RNA-bound and unbound mRBM20(513–621) have been deposited in the Biological Magnetic Resonance Data Bank (http://bmrb.wisc.edu/) under BMRB accession numbers 34428 and 34429, respectively.