Abstract
We introduce an R package and a web-based visualization tool for the representation, analysis and integration of epigenomic data in the context of 3D chromatin interaction networks. GARDEN-NET allows for the projection of user-submitted genomic features on pre-loaded chromatin interaction networks, exploiting the functionalities of the ChAseR package to explore the features in combination with chromatin network topology properties. We demonstrate the approach using published epigenomic and chromatin structure datasets in haematopoietic cells, including a collection of gene expression, DNA methylation and histone modifications data in primary healthy myeloid cells from hundreds of individuals. These datasets allow us to test the robustness of chromatin assortativity, which highlights which epigenomic features, alone or in combination, are more strongly associated with 3D genome architecture. We find evidence for genomic regions with specific histone modifications, DNA methylation, and gene expression levels to be forming preferential contacts in 3D nuclear space, to a different extent depending on the cell type and lineage. Finally, we examine replication timing data and find it to be the genomic feature most strongly associated with overall 3D chromatin organization at multiple scales, consistent with previous results from the literature.
INTRODUCTION
A decade after the first papers about Hi-C (1), the techniques to detect genomic structure experimentally at different scales have multiplied. We can now explore 3D chromatin contact dynamics across cell types (2), differentiation state (3), the cell-cycle (4) and even in single-cells (5). As the resolution of our datasets improves, the need for new genome visualization options is apparent. We have developed a set of tools for the integration and interpretation of genomics datasets in 3D, based on a network representation of chromatin contacts.
Despite the great advancement in our knowledge regarding chromatin conformation in the last few years, heatmaps are still the most widely used visualization frameworks. This works well, because contact matrices provide symmetric interaction profiles based on binning of contacts at a specific resolution. This mode of visualization is also ideal to spot two of the main sub-structures in the genome, namely topologically associated domains (TADs) (6,7), identified precisely as squares on the diagonal of Hi-C contact heatmaps, and compartments (1), directly related to the checkerboard patterns that are also easily observable in this representation.
Capture Hi-C is a variation of the Hi-C protocol that enables studying interactions involving a set of specific non-abundant regions of the genome, for example gene promoters (8). Promoter-capture Hi-C confirmed the existence and importance of long-range chromatin interactions and identified the role of Polycomb in creating a highly interconnected core of developmentally related genes in mouse embryonic stem cells (9).
Long-range interactions can also be detected in contact maps generated by identifying chromatin interactions that are mediated by specific proteins, for example RNA Polymerase II (RNAPII), with the ChIA-PET protocol (10,11). Hi-C datasets allow the detection of these long-range interactions only when extremely deep sequencing of the libraries is performed, due to the much higher abundance of short-range contacts that saturate sequencing libraries (12). One factor common to all chromosome conformation capture techniques is the reliance on hybridization and sequencing.
Alternative approaches that do not rely on sequencing, and even more so the ones that do not require the bias-prone ligation steps characterizing chromosome capture methods, provide us with an independent picture of nuclear organization. For example, different techniques exist for visualizing specific previously tagged chromatin regions by microscopy, leading to the inference of 3D interactions (DNA FISH Oligopaints (13), Hi-M (14)). Finally, hybrid methods that infer 3D proximity by segregating genomic fragments in different locations in the nucleus allow the inference of 3D contact maps independent of proximity ligation (GAM (15), SPRITE (16)).
There is a clear need to develop frameworks to represent, compare and integrate these different datasets in an intuitive and computationally efficient way. New tools to visualize chromatin will help us go beyond the identification of TADs and compartments, towards models of gene regulation. We suggest that interpreting chromatin structure as a network is a useful step in this direction.
A considerable number of papers have suggested interpreting chromatin structure as a network, either starting from Hi-Ccontact maps interpreted as distance matrices, see for example (17–23), or also increasingly using alternative chromosome capture techniques that detect longer-range specific contacts directly (24). Despite the processing required to obtain networks from 3C based datasets available at the time, network methods were shown to provide biological insight already as far back as 2010 (17). Importantly, thanks to recent improvements in experimental techniques, we are now able to generate meaningful chromatin networks directly from the contact datasets, not by filtering of contacts based on distance (25), and to provide an integration of epigenomic profiles reconstructing relationships between different epigenomic marks in 1D (26) or 3D (25). Network methods have also been used to detect common structural features such as TADs (27,28) and to identify the presence of different species in meta-genomic samples (29). Despite the presence of a considerable number of papers on network analysis of chromatin contacts and the clear potential of this analogy, most of the reviews on the subject still focus on different approaches for the analysis of chromosome capture datasets, with one exception to our knowledge (30).
The network representation is particularly useful when using PCHi-C networks, where we can subdivide the whole network into interactions involving only promoters (PP subnetwork) or those involving a promoter and an Other-end (PO subnetwork). This distinction can lead to interesting differences between these two types of genomic contacts, which are characterized by the presence of specific epigenomic features (25).
MATERIALS AND METHODS
ChAseR
The ChAseR package provides functions to read chromatin contact networks and many kinds of data available in bed file or other similar formats (gene lists, ChipSeq datasets, expression values) and perform the integration of the dataset onto a 3D chromatin structure network (Figure 1A). Once the data have been assigned to nodes in the network, ChAseR performs correlation analyses. Specifically, the ChAseR::ChAs function computes different forms of network correlations: assortativities, cross-assortativities, AND-assortativities, node by node correlation functions, assortativity for categorical features (see ChAseR vignette in Supplementary Material).
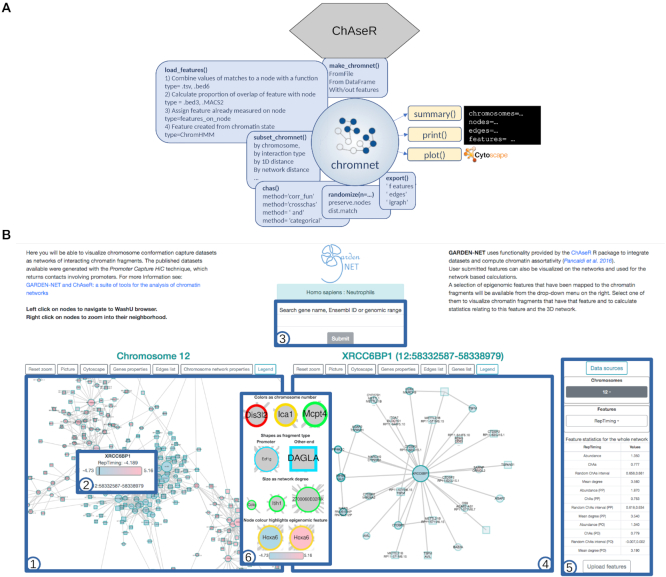
Figure 1.
Schematic overview of the two tools presented to perform chromatin network analysis. (A) Schematic view of the functionalities of the ChAseR R package. (B) A screen shot of the GARDEN-NET web-tool showing the main components: 1) the viewer on the left shows the network for the selected species and cell type; 2) the tool-tip shows properties of the node; 3) the search box allows to search for genes or genomic regions; 4) the viewer on the right shows the searched region or neighbours of selected genes; 5) the menu on the right allows for choosing chromosome (including the option PP to visualize the entire PP network for PCHi-C networks), selecting features to be displayed, and uploading user-defined features. The table under the menu displays calculated correlations and network properties of nodes with the selected feature. The legend at the center that can be opened from the option on the left-viewer shows the mapping between features and visual properties.
The value of chromatin assortativity is dependent on network topology, on features abundance (number of network nodes covered by the feature) and also on the relationship between the two (e.g. whether the feature tends to be found in nodes with high degree). It is therefore essential to perform randomizations to estimate the expected value of assortativity for a specific feature and compare it to its measured value.
Randomization strategies
We implemented three randomization algorithms in the function ChAseR::randomize. The first consists in redistributing the features at random across nodes (Supplementary Figure S1A). This obviously preserves the average value of each feature in the network exactly.
A further option allows the user to split the nodes in the network in two non overlapping sets; features are randomly reshuffled but only among nodes belonging to the same set. For example, if the first set contains nodes (a, b, c) with feature values (1, 2, 3) and the second set contains nodes (d, f, g) with feature values (10, 11, 12), a possible randomization could associate nodes (a, b, c) with features (2, 3, 1) and nodes (d, e, f) with features (12, 11, 10); but the feature of a node from the first set can not be swapped with a feature from the second set. This might be useful, for example, when one wants to permute features distinguishing promoters from other-ends, thus preserving the mean abundance for each of these two specific node subsets.
Finally, the third algorithm rewires the network edges so that the distribution of edge distances (the genomic distance spanned by two nodes connected by an edge) after randomization is similar to the distribution of edge distances in the original network. Specifically, each logarithmic decade of edge distances (103−104, 104−105, …) contains approximately the same numbers of edges in the real and randomized networks (see Supplementary Figure S1B). This does not change the assignment of feature values to nodes, so that feature abundance is also preserved.
This last option is useful for the many features that have broad peaks, which are likely to overlap multiple fragments that are linearly close in the genome. In these cases, not preserving distance would produce a distortion of the genomic correlation and would lead to randomized ChAs values distributed around 0 for all features. On the contrary, using the distance-preserving randomizations leads to expected ChAs values that can be positive and high, so that only ChAs values outside of these random ranges can be considered significant (Supplementary Figure S1C).
GARDEN-NET
Generating chromatin contact networks
Chromatin networks are assembled starting from standard outputs of chromosome capture analysis software. We mostly focused on ‘targeted’ genome-wide techniques such as Capture Hi-C, ChIA-PET or Hi-ChIP, but also standard Hi-C datasets can be used to create networks, after filtering for significant interactions or detection of loops. To illustrate GARDEN-NET we will here consider data produced by Promoter-Capture Hi-C (PCHi-C).
This technique identifies contacts involving promoters, returning pairs of contacting fragments, stored for example as interaction files in .ibed format. The protocol is very similar to Hi-C, except the extra step of hybridization with a library of promoter baits. Similar to Hi-C, significant contacts have to be identified against a background of noise. The data in (2) is processed using the CHiCAGO pipeline, which uses a convolution noise model accounting for noise due to both distance-dependence of the signal (short genomic distance interactions are more likely) and ‘technical’ noise due to sequencing (37). An alternative pipeline presented in (38) was shown to better identify promoter–enhancer contacts but was not suited to identify promoter–promoter contacts, which are of interest in this study. We constructed networks from the CHiCAGO output, simply considering all contacts with CHiCAGO score >5 as edges, as suggested by the package guidelines. The networks are hence unweighted.
The other kind of contact datasets that we have included in GARDEN-NET are Hi-C derived networks (see Table 1). Hi-C data was generated and processed with the FitHiC pipeline (39), which allows us to retrieve a list of highly confident interactions associated with a P-value, generating chromatin contacts maps that are quite comparable to the PCHi-C networks.
Table 1.
Data sources for GARDEN-NET and for the present manuscript
| Data type | Cell types | Reference | Notes |
|---|---|---|---|
| Promoter Capture Hi-C | Human heamatopoietic primary cells | (2) | v.37 |
| Promoter Capture Hi-C | mESC | (8) | v.mm9 |
| Hi-C | mESC | (31) | v. mm9 |
| Hi-C | GM06990 | (1) | v.37 |
| ChIP-Seq (78 features) | mESC | (26) | v.mm9 |
| ChIP-Seq (4 hist. marks) | Human primary Monocytes, Neutrofils and Tcells | (32) | v.37 |
| ChIP-Seq (6 hist. marks) | Human primary B cells | (33) | v.38 |
| ChIP-Seq (2 hist. marks) | GM06990 | (34) | v.37 |
| ATAC-seq | Human primary B cells | (33) | v.38 |
| Repli-Chip | GM12978 | (35) | v.37 |
| Repli-Chip | GM06990 | (35) | v.37 |
| RNAseq | Human primary Monocytes, Neutrofils and Tcells | (32) | |
| RNAseq | Human primary B cells | (33) | |
| RNAseq | Human primary haematopoietic cells | (36) | |
| RNAseq | GM06990 | (34) | |
| DNA methylation | Human primary Monocytes, Neutrofils and Tcells | (32) | WGBS |
| DNA methylation | GM06990 | ENCODE (GSM999353) | 450K |
GARDEN-NET allows choosing specific species and cell types and returns a chromosome-wide visualization of the chromatin contact network on the left panel (Figure 1B). The user can then search for a specific region of interest by entering genomic coordinates, the gene name, or simply by right-clicking on one of the nodes in the left panel. After the search is activated, the panel on the right will display the neighbourhood of this region/gene while the zoomed-in regions are highlighted in the left panel. Alternatively, the user can select to visualize the entire network of only PP interactions and, in this case, connections between a selected promoter and other ends interacting with it will appear only on the right panel and after the search.
This visualization is based on a web implementation of Cytoscape (40,41) and hence visual properties of the graphics can be coupled to the represented dataset. In the case of PCHi-C, nodes can be either baits used in the promoter capture experiments (circles), or other ends (squares). All nodes are annotated with the names of genes that overlap the corresponding genomic fragment (tool-tip), and are linked to genome browser views of the regions (left-click). Bait nodes are additionally labelled with the name of the gene whose promoter was targeted by the capture system. Other ends often overlap with intronic regions, in which case the name of the gene harbouring the intron is shown in the tool-tip. The node border colour denotes the chromosome, to facilitate detection of inter-chromosomal interactions. A series of tabs above the left-most viewer window allow for resetting of the zoom, downloading of the data and images and loading of a table listing general network characteristics (number of nodes, number of promoter nodes etc.) and network statistics (average degree, size of connected component etc.) for the network represented on the left panel (either a single chromosome or the entire PP network).
Chromatin features
The second main function of GARDEN-NET is assigning the chromatin features to the nodes of the 3D structure network, which is performed by ChAseR. Depending on the organism and cell type chosen, a list of features is available in the drop-down menu on the right. In the case of mESCs, these are the features that were considered in a previous work (25) and include histone modifications, binding peaks of different transcription factors and cytosine modifications (see Table 1). For human haematopoietic cells, we currently provide expression, methylation, histone modifications and expression averaged over hundred of healthy individuals, as well as replication timing for cell lines (see Table 1).
Once one of these features is selected, it will be represented on the network visualization panel with a colour code mapping the node colour to the value. The tool-tip will show the exact value for the feature on the node and the range of feature values in the whole network for comparison. Upon selection of a feature, a table will also appear below the feature selection drop-down menu with a list of network statistics specific to the selected feature. This includes the proportion of nodes having the feature, their average degree and also the feature's chromatin assortativity (ChAs) value, calculated on the whole network or subnetworks (see ChAseR manual about the mapping procedure), with randomized values for reference. Users are invited to submit their own features through a menu which will require specifying the feature type and providing a feature file (multiple formats are accepted including bed3, bed6, chromhmm) and, once the feature is uploaded, users will be able to visualize their features on the chromatin network and calculate related statistics. More technical details about GARDEN-NET front and back-end can be found in Supplementary Material.
RESULTS AND DISCUSSION
ChAseR and GARDEN-NET
Given the flexibility and efficiency of representing chromatin as a network, we have designed a pair of tools which can be used together or separately for the analysis of chromosome conformation capture data: GARDEN-NET and ChAseR (Figure 2). ChAseR is a stand-alone computational tool implemented as an R package, which provides functions to build and analyse chromatin interaction networks efficiently, integrate different epigenomic features on the network, and investigate the relation between chromatin structure and other genomic properties. For example, we have recently introduced the concept of chromatin assortativity (ChAs), which allows us to identify whether chromatin with specific features (chromatin marks, binding of transcription factors and replication timing, amongst others) tends to form preferential 3D contacts in the nucleus (25,42). ChAseR provides efficient calculations of ChAs and other related measures, including combined feature assortativity, local assortativity defined in linear or 3D space, and tools to explore these patterns (Figure 2B). GARDEN-NET is a web-based tool that exploits Cytoscape (40,41) functionality to allow exploration of pre-loaded chromatin contact networks (PCHi-C datasets in human haematopoietic cells (2) and mESCs (43)), in combination with different pre-loaded epigenomic datasets (histone modifications, expression, DNA methylation (32) and replication timing) also allowing users to submit features (.bed and other formats accepted). It provides user-friendly visualization and search of networks, calculations of network measures and ChAseR functionalities, including calculation of ChAs.
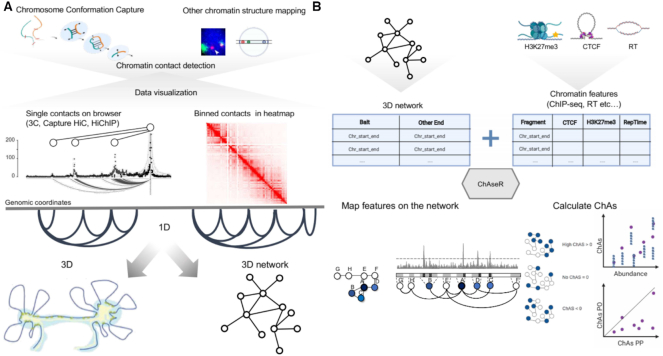
Figure 2.
Schematic representation of the chromatin assortativity approach to study genome organization. (A) Schema representing the different chromatin contact detection methods and different representations of these datasets. (B) Schema representing the different components that should be used as input in ChAseR and chromatin assortativity calculations and the two main ChAseR outputs: chromatin networks annotated with features and ChAs calculated on them. Mapping features on the networks involves assigning a value corresponding to a feature on the represented genomic fragment to each chromatin network node. Chromatin assortativity estimates whether there are preferential contacts connecting chromatin fragments that have a specific feature (blue nodes in the schematic). The ChAs calculation can be used to represent two types of plots: a ChAs versus abundance plot or, for networks that can be subdivided into PO and PP subnetworks, a plot showing ChAs in the PO vs PP subnetworks. These two plots suggest respectively whether the ChAs of the feature of interest (purple circle) is high compared to random expectation (blue smaller circles, see Materials and Methods) and whether it is most important for PP or PO contacts.
Assortativity analysis of histone modifications in primary neutrophils
We demonstrate the use of ChAseR and GARDEN-NET on epigenome datasets from the BLUEPRINT project, in which reference histone modifications, DNA methylation and gene expression were characterised for neutrophils, monocytes and T cells in 150 healthy individuals (32). We considered contacts identified using PCHi-C (2), taking all interactions called by CHiCAGO as unweighted (see Materials and Methods).
These networks include both contacts between promoter (P) nodes, denoted as PP edges, and contacts between a promoter (P) and another genomic region (O) denoted as PO edges. The total PCHi-C networks can hence be subdivided into PP and PO subnetworks. Comprehensive epigenomic data was collected for neutrophils, including four histone modifications, DNA methylation and expression levels across all individuals, allowing us to test the robustness of our findings in this cell type. For each of the following analyses we performed randomizations to test the significance of assortativity values, as described in Material and Methods.
We studied the assortativity of four histone marks in neutrophils from a single individual of the EPIVAR project (32). We first plotted ChAs of each histone mark on the PP subnetwork against each mark’s abundance on the same subnetwork (Figure 3A), also showing the ChAs resulting from distance-preserving randomizations (see Materials and Methods). We also investigated the relationship between ChAs calculated in the PO versus the PP network for each histone mark (Figure 3B).
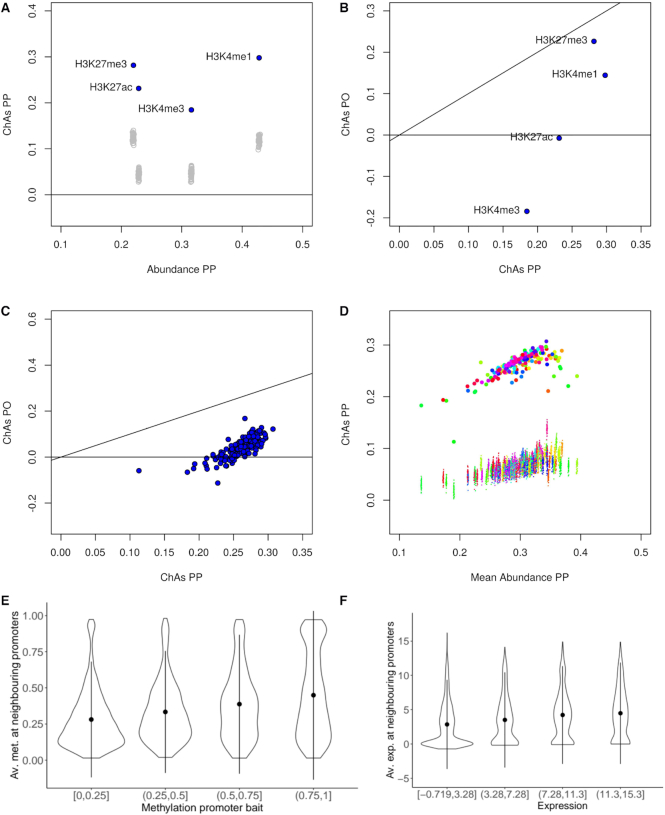
Figure 3.
Analysis of the assortativity of epigenomic features in primary neutrophils. (A) ChAs vs abundance on PCHi-C PP networks for 4 different histone modifications in a single individual of the EPIVAR project (32). Empty grey circles represent ChAs calculated on distance-preserving randomizations. (B) ChAs calculated for the same features as (A) in the PO and PP PCHi-C networks. The black line shows the diagonal (x = y). (C) ChAs for H3K27ac in the PO and PP PCHi-C networks for 150 different healthy individuals from EPIVAR. The black line shows the diagonal (x = y). (D) ChAs vs abundance for H3K27ac across all individuals from the same dataset as (C). Small dots correspond to ChAs values obtained for distance-preserving randomized networks and colours identify individuals. (E) Correlation between methylation quartiles at promoter baits and average methylation at neighbouring promoters in the PCHi-C PP neutrophil network. (F) Correlation between expression quantiles at promoter baits and average expression at neighbouring promoters in the PCHi-C PP neutrophil network.
The first feature that we examined was tri-methylation of Lysine 27 on histone 3 (H3K27me3), a chromatin mark which is strongly associated with Polycomb repressed chromatin (44) that was found to be assortative in mESCs (25). Confirming these results, H3K27me3 was found to have positive and significant ChAs in neutrophils in both PP and PO subnetworks. These results suggest the association of Polycomb with 3D chromatin contacts even in differentiated cells (Figure 3a,b). These results were confirmed for most of the individuals investigated (Supplementary Figure S2).
We then examined lysine 4 mono-methylation on histone 3 (H3K4me1), which has been associated to enhancers and promoters (44) and was found to be assortative in mESC (25). H3K4me1 was found to be assortative in neutrophils (Figure 3A and B). ChAs values were higher in the PP than in the PO subnetwork, suggesting that this mark is mostly associated to promoters that are in 3D contact with each other, but it can also be found in some regulatory regions in contact with promoters. These observations were strengthened by looking across individuals, which revelaed this mark to have very reproducible ChAs values (Supplementary Figure S2).
Next, we considered, acetylation on Lysine 27 of histone 3 (H3K27ac), an activation mark which is often encountered on active gene promoters and active enhancers (44) and was also found to be assortative in mESCs (25). Strangely, in the single specific individual considered in Figure 3B, this mark was found to be neither assortative nor disassortative, with ChAs = 0 on the PO network, while having a significant ChAs on the PP network. Looking at multiple individuals, we observed variable levels of H3K27ac assortativity, especially in the PO subnetwork with ChAs values ranging from –0.1 to 0.1, compared to a range of 0.1 to 0.35 in PP (Figure 3C). This suggests that for all individuals there is a strong tendency for interactions between promoters that have similar levels of activation, but only in some individuals, regulatory regions in contact with active promoters also have this mark. The ChAs values on the PP networks are also variable, but all are found to be significantly higher than expected by distance-preserving randomizations (Figure 3D). This finding might suggest modulated expression levels of the genes across individuals depending on the presence of the H3K27ac activation mark on the corresponding enhancers. Neutrophils were previously found to display high levels of transcriptomic and epigenomic variability, even taking into account genetic differences between individuals, likely due to their role as the immune system’s first responders (45).
Finally, we looked at peaks of trimethylation on Lysine 4 of the same histone 3 (H3K4me3), a mark associated with active promoters (44). We observed strong assortativity in the promoter-promoter network and negative assortativity in the PO network (Figure 3B), consistent with what was found in mESC (25) and also with the expected behaviour for a promoter-specific mark. The ChAs values for this mark were found to be considerably less variable across individuals (Supplementary Figure S2). Results for H3K27ac and H3K4me1 could be largely confirmed in monocytes and T cells (Supplementary Figure S3).
Taken together, these results are consistent with recent reports of the existence of interactions between functional regulatory domains that can be evinced from correlation of histone marks (H3K4me3 and H3k27ac) across a large number of individuals (46). Moreover, we confirmed an association of chromatin marks to chromatin structure that leads to preferential contacts between chromatin regions with similar chromatin (47).
DNA methylation is assortative in primary neutrophil promoter-promoter networks
Given the strong association between de-methylation at gene promoters and active gene states, we went on to test assortativity of DNA methylation Beta values in the neutrophil PCHi-C PP subnetwork. We found positive values above 0.20 (Supplementary Figure S4). Using the corrfun option of the ChAseR::ChAs function, ChAseR allows for a more detailed investigation of the correlation of features in interacting genomic regions. We can plot the correlation between the value of methylation at one promoter and values of methylation of its 3D neighbouring promoters, separating methylation values in quartiles (Figure 3E). This correlation was clearly positive, indicating concordance of methylation states at interacting promoters.
Gene expression is assortative in primary neutrophil promoter–promoter networks
Having observed a strong relationship between chromatin marks related to gene expression and chromatin 3D structure, we predicted that gene expression levels of genes whose promoters establish contacts with each other should be correlated. We confirmed this hypothesis measuring a significant value of assortativity of gene expression on the cell-type specific PP subnetwork in all individuals and in all three cell types analysed (Supplementary Figure S5). This tendency was also evident as a correlation between expression quartiles on promoters and expression on their promoter neighbours on the PP subnetwork (Figure 3F).
Assortativity analysis of reference epigenome marks on B cells
To test assortativity of a larger number of epigenomic features, we considered the data from (33). We downloaded the six reference histone modifications (H3K9me3, H3K27me3, H3K4me1, H3K4me3, H3K36me3, H3K27ac) and ATAC-seq datasets (marking accessible chromatin) from B cells and projected them onto the B cell PCHi-C network (combining total and naive B cells). In addition to the previously discussed marks, this included methylation on Lysine 36 of histone 3, a modification, thought to be marking regions involved in active transcription and exons, and H3K9me3, a mark associated to constitutive heterochromatin.
As expected, we found all modifications to be significantly assortative with respect to random, as these marks are associated with chromatin states that are known to be attracting each other, such as active chromatin, making contacts within the active compartment, and heterochromatin and polycomb domains, also making preferential contacts with other regions with the same marks (Figure 4A).
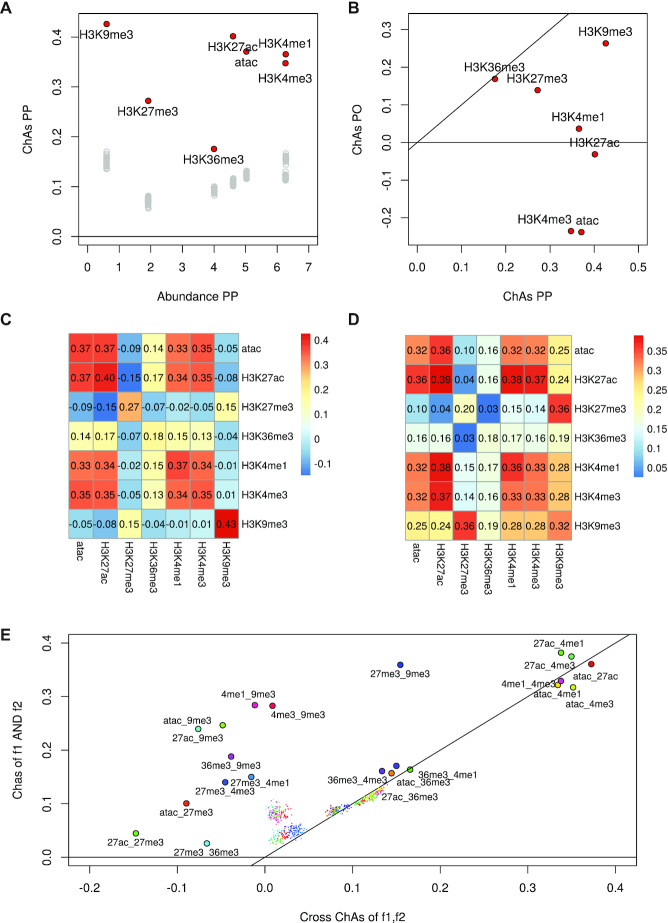
Figure 4.
Analysis of assortativity of six histone modifications and DNA accessibility (33) on the PCHi-C network for B cells. (A) ChAs versus Abundance calculated on the PCHi-C PP subnetwork for B cells (red circles) and values obtained by distance-preserving randomizations (grey circles). (B) ChAs in PO vs PP network for dataset in (A). (C) cross-ChAs of all possible feature combinations on the PCHi-C PP B cells network (the diagonal values correspond to single-feature ChAs). (D) AND-ChAs of all possible feature combinations on the PCHi-C PP B cells network. (E) Comparison between values of AND-ChAs and cross-ChAs. The insets show schematic representations of the two possible combined ChAs scenarios in the case of TF binding or histone modfications. cross-ChAs detects the presence of preferential contacts between regions with feature 1 and other regions with feature 2. AND-ChAs detects preferential contacts involving regions presenting both feature 1 and feature 2. Smaller coloured dots represent corresponding combined ChAs expected values according to distance preserving randomizations. All histone modifications are on histone 3 lysins (H3K removed for clarity from datapoint names).
H3K36me3 was the mark with the least increased ChAs in the PP subnetwork compared to distance preserving randomization, showing a moderate impact of 3D interactions in the preferential contacts established between regions with the mark. On the contrary, H3K9me3, followed by active chromatin marks, displayed strong assortativities. Separating the PP and PO networks we observe higher ChAs than expected for all features apart from H3K36me3 and H3K27me3 in the PP subnetwork, with ATAC-seq and H3K4me3 having strongly negative ChAs in the PO subnetwork (Figure 4B).
Whereas the negative PO subnetwork assortativity of H4K4me3 is to be expected for a promoter-specific mark, the similar ChAs value for ATAC-seq suggests a discordance between DNAse hypersensitivity of promoters with their connected regulatory regions. This might be related to differences in transcription factor occupancy at these regions. Again the H3K27ac is found to have a value close to 0 in the PO subnetworks, suggesting great variability across individuals, an aspect that could not be evaluated for this dataset.
Cross-ChAs and AND-ChAs, assortativity of combined features
Since ChAseR offers the possibility to calculate the combined correlation of two features, we decided to use the cross-ChAs and AND-ChAs options to evaluate ChAs of all the possible combinations of epigenomic marks included in this dataset. In this mode, ChAseR returns a matrix of values, which can be visualized as a heatmap highlighting which types of features are found to be in preferentially interacting chromatin fragments, either with one feature on each of the interacting fragments, or with both features on interacting fragments (Figure 4C, D).
We constructed a cross-ChAs matrix for the above mentioned features on B cells, as shown in Figure 4C. Clearly the diagonal of this matrix returns the traditional ChAs values (correlations of a feature with itself) but the off-diagonal elements can, in some cases, point to specific combinations of features present on interacting fragments or excluding each other.
This is the case, for example, for heterochromatin, which shows high ChAs only with itself, moderate cross-ChAs with H3K27me3, and negative cross-ChAs with all other features. It is in fact interesting to observe the difference between these cross-ChAs values in the two separate PP and PO subnetworks and to consider the values obtained by distance-preserving randomizations (Supplementary Figure S6). The tendencies observed for cross-ChAs for H3K9me3 are already present in the randomized networks, suggesting that 3D interactions might not have a strong effect on cross-ChAs estimations, apart from the cross-occurrence with H3K27me3, a mark heavily involved in 3D interactions. This is also confirmed looking at the cross-ChAsz-score (Supplementary Figure S6e).
The AND-ChAs matrix shows the strong tendency for fragments with both H3K4me1 and H3K4me3 to interact preferentially and also for fragments with any of these two features and H3K27ac to interact preferentially. The results show decreased tendency of regions with both H3K27me3 and H3K36me3 to establish preferential contacts but a strong likelihood for regions with both H3K27me3 and H3K9me3 to form contacts (Figure 4D). Comparing to the values obtained with distance-preserving randomizations (Supplementary Figure S6C and F) suggests that 3D interactions affect mostly the H3K27me3 mark AND-ChAs values when it is colocalized with H3K36me3. Both types of combined ChAs show an increased tendency for preferential contacts between active promoters (regions with H3K4me3 and H3K27ac especially (Figure 4C, D)).
Finally, we might want to compare the assortativity of combinations of features on the same or opposite nodes. In Figure 4E, we display the assortativity of all feature combinations where features are either on the same node (AND-ChAs) or on opposite interacting nodes (cross-ChAs). AND-ChAs values are generally higher than cross-ChAs and we also notice pairs including repressive marks (H3K27me3 or H3K9me3) to display lower ChAs than all other combinations. Results show combinations of 1 active and 1 repressive mark to be assortative in AND-ChAs but to have negative cross-ChAs. It must be mentioned that these results might be affected by the size of chromatin fragments considered (median: 5 kb) and the different average sizes of peaks of these epigenomic marks that might mean that features in different regions of a fragment appear to colocalize on our nodes.
Assortativity of chromatin states
Since specific histone marks are known to co-occur along the genome, denoting chromatin states, we investigated whether chromatin states are assortative on the PCHi-C networks. We downloaded chromatin states from (48) (11 states) for 15 haematopoietic primary samples (including T cells, monocytes, neutrophils and macrophages), assigned each PCHi-C network fragment the most abundant state along the fragment, and measured assortativity of the states using the categorical option of the ChAser::ChAs function.
We found categorical ChAs values of chromatin state to be larger than those obtained with distance-preserving randomization (Supplementary Figure S7), to a varying extent depending on the cell types. These findings suggest that the strong assortativity between regions with repressive marks, either constitutive heterochromatin or facultative poised Polycomb regions, drives a tendency for chromatin states assortativity. However, when analysed separately as single features, different chromatin states have varying levels of assortativity (data not shown).
Assortativity analysis of histone modifications, DNA methylation and expression in Hi-C derived contact networks
To confirm the applicability of our method to the widely available Hi-C datasets, we applied the same approach described above to measure assortativity of histone modifications on chromatin Hi-C networks derived from the lymphoblastoid cell line GM06990 (34). Hi-C data is inherently genome-wide and is thus not limited to interactions involving promoters. Interactions derived from Hi-C are normally binned at specific resolutions and can be associated to a score that determines whether the number of contacts joining two bins corresponding to genomic regions is significant compared to what would be expected based on the genomic distance between the regions, much like what is done for PCHi-C.
FitHiC is a method to estimate the significance of contacts based on modelling the random looping that is expected to occur in polymers (39). Thus FitHiC returns a P-value for each detected contact. We generated chromatin contact networks based on significant interactions detected in the GM06990 cell line by Hi-C processed with FitHiC at a 50 kb resolution.
Two replicates of ChipSeq for H3K27me3 and H3K36me3 were downloaded from (34). We confirmed that polycomb marked chromatin identified through H3K27me3 peaks was more assortative than expected at random (Supplementary Figure S8A). The ChAs values of H3K27me3 were comparable but lower than those obtained for PCHi-C neutrophil networks (median = 0.23 compared to 0.3 in neutrophils). We found H3K36me3 to be significantly assortative in both replicates on the FitHiC generated network of GM06990 cells (ChAs = 0.26), beyond what can be expected at random, possibly indicating preferential contacts between gene bodies occupied by RNAPII (Supplementary Figure S8). It should be noted that whereas there is variability in the two replicates of the ChIP experiments, as can be seen by differences in the abundance of the mark, the values of ChAs are much more comparable between replicates, pointing to the robustness of the ChAs statistics (Supplementary Figure S8A).
To consider a larger number of histone modifications and estimate consistency between results on Hi-Cand PCHi-C networks, we calculated Chas of the six histone marks and ATAC-seq from (33) on the FitHiC network for the lymphoblastoid cell line GM06990 (Supplementary Figure S8B). Despite the probable differences between primary B cells and the B cell derived cell line, we could see similar values of ChAs for all epigenomic marks on the Hi-C networks and the PP PCHi-C subnetwork (Figure 5A), with only the H3K27me3 and H3K9me3 marks having lower assortativity in Hi-C compared to PCHi-C (see Supplementary Figure S9 for other comparisons).
Figure 5.
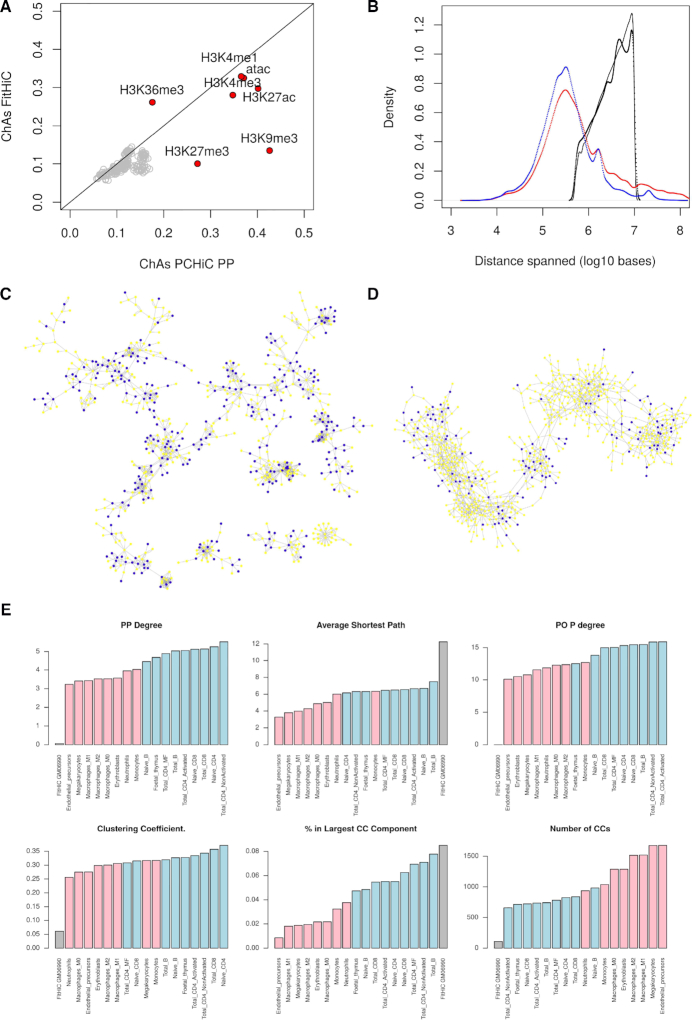
Comparison of PCHi-C and Hi-C networks. (A) Comparison of ChAs values calculated for histone modifications on the Hi-C network compared to the PCHi-C PP subnetwork. Grey dots indicate distance preserving randomizations. (B) Distribution of distances spanned by different networks. PCHi-C PP networks for neutrophils (blue) and B cells (red), GM06990 Hi-C network with FitHiC P-value < 0.05 (black), GM06990 Hi-C network with all contacts identified by FitHiC (grey). (C) A portion of the PCHi-C PP subnetwork for neutrophils. Expression is denoted by node color (yellow RPKM < 1, blue RPKM > 1, (32)). (D) A portion chromosome 1 on the Hi-C network for GM06690 (35) showing contacts with FitHiC P-value < 0.05. Expression is denoted by node color (yellow RPKM < 1, blue RPKM > 1 (34)). (E) Topological network properties of haematopoietic networks. PO P degree = degree of promoter nodes in the PO subnetwork. CC = connected component.
We then considered DNA methylation data for the GM06690 cell line. Contrary to our finding in neutrophil (ChAs in PP > 0.2, cf. Figure 3E), DNA methylation was not found to be assortative in these Hi-C networks (Supplementary Figure S10), as could be expected given that Hi-C data does not capture exclusively promoter interacting regions. However, gene expression as estimated by FPKM values for GM06990 (34) had significant ChAs (0.17, distance-preserving randomization between 0.07 and 0.9) (Supplementary Figure S10B), higher than the value found in neutrophils (ChAs PCHi-C PP = 0.12).
The lower assortativity of both heterochromatic marks and methylation in the FitHiC network might be explained by differences in the coverage of interactions spanning particular ranges of chromosomic distances between the two experimental techniques.
We compared the frequency distribution of distances spanned in the interactions of PCHi-C PP and FitHiC networks (Figure 5B). The FitHiC network displays a different distribution of interactions distances, possibly due to inherent limitation of sequencing depth in the original Hi-C experiments which limit the detection of long-range (>10MB) interactions. This difference in coverage of long-range interactions between Hi-C and PCHi-C might explain the observed differences in ChAs for repressive marks (facultative and constitutive heterochromatin), which are known to involve long-range contacts. The FitHiC and PCHi-C PP networks are topologically very different from each other (Figure 5C, D and Supplementary Figure S11). In addition, Hi-C cannot easily assess the significance of inter-chromosomal interactions. These differences in topology are reflected also by the network measures reported in Figure 5E (Supplementary Figure S11). Interestingly, considering the PCHi-C PP networks, we observe a strong difference between myeloid and lymphoid lineages: lymphoid networks have higher degrees and they show fewer and larger connected components, a stronger similarity to undifferentiated pluripotent cells, suggesting their possibly enhanced plasticity compared to myeloid ones.
Replication timing is the most assortative feature on promoter-promoter chromatin networks
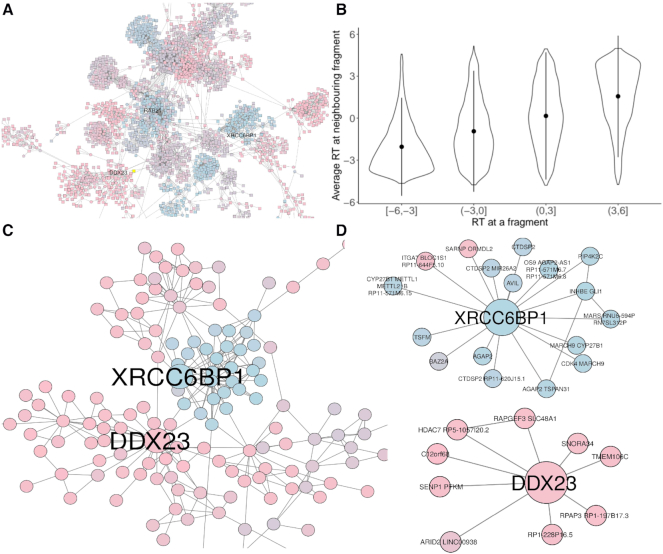
The general applicability of ChAseR and GARDEN-NET allows us to investigate assortativity of any feature defined along the genome. For example, we found replication timing (RT) of genes to be the most assortative genomic feature of the ones analysed, with a value ranging from 0.6 in mESCs to 0.77 in neutrophils (Figure 6). Looking at a specific neighbourhood on the neutrophil network, we observe the separation of early and late RT domains (Figure 6A), which is reflected in a strong correlation between RT values at neighbouring fragments (Figure 6B). This separation between early and late RT regions is maintained when looking at the PP subnetwork (Figure 6C).
Figure 6.
Replication timing on the neutrophil PCHi-C network. (A) A portion of the PCHi-C network for neutrophils (2). (B) Correlation between replication timing (RT) at one promoter node with average RT on interacting promoters. (C) A portion of the PCHi-C PP subnetwork extracted from (a) highlighting interactors of the DDX23 and XRCC6BP1 genes, showing a clear separation between early and late RT neighbourhoods. (D) PP interactions of the DDX23 and XRCC6BP1 genes. DDX23 is an early RT gene involved in splicing which interacts with other early RT genes involved in other basic cellular processes (chromatin remodelling, protein modifications etc. see Supplementary Text). XRCC6BP1 (ATP23) is a gene involved in DNA break repair, often amplified in glioblastoma. Its neighbouring promoters include other cancer-related genes with functions in development, cell cycle control and apoptosis (see Supplementary Text andSupplementary Figure S13).
It should be noted that distance preserving randomizations return very high values of expected ChAs in the neutrophils (0.66), due to a strong linear correlation in RT values. These correlations have been observed in the past by Pope et al. (35) amongst others, who proposed that TADs would correspond to replication domains. Our values of ChAs, which are higher than expected based on the linear correlation of RT values (Supplementary Figure S12), suggest an important role for 3D long-range contacts in bringing together regions and genes with similar replication timing across large distances. Similar observations regarding the 3D organization of RT have been made on Hi-C based datasets (39) mostly considering contacts within TADs. We think that targeted capture methods, very deep Hi-C libraries or non-sequencing based methods offer the potential to observe chromatin interactions at longer distances, reinforcing evidence for a strong 3D organization of replication and other biological processes across scales (49).
CONCLUSION
We have presented two complementary approaches to investigate genome-wide datasets in the context of the organization of chromatin inside the nucleus. GARDEN-NET is a chromatin network visualization tool, which allows to upload user-defined feature files and project these features on the available 3D chromatin contact networks. The underlying functionalities of GARDEN-NET for the production and investigation of chromatin networks and for the mapping of genome-wide features on them are provided by the ChAseR package, which can also be used on its own. We believe these two tools will empower researchers interested in epigenomics, enabling them to perform both genome-wide and region-specific investigations of their data within the context of 3D genome architecture evinced by chromosome capture experiments, especially when separating different subnetworks is of interest. It should also be noted that contact networks generated by any other method, including non-sequencing based techniques, can be just as easily used in ChAseR, once the experimentally detected contacts are expressed as a list of pairs of nodes corresponding to chromatin fragments.
Even if assortativity can summarise relationships between 3D structure and specific features with a single correlation coefficient, the complexity of genome architecture will rarely be fully resolved by this approach. For this reason, we have provided additional tools to more carefully dissect each feature or combination of features in the context of 3D contacts. This can involve using extensions of the concept of chromatin assortativity to local and global measures specific for the feature types, or exploring scatter plots and violin plots to study the robustness of the correlation at a more granular level. We have provided extensive descriptions of the randomization procedures employed to assess significance of ChAs values obtained. Further work will be required to provide a randomization strategy that correctly preserves distance-relationships and works on asymmetric networks (for example PO subnetworks).
We have shown how users can easily perform integrated analyses of (epi)genomic data in the context of 3D chromatin structure networks, efficiently gaining a quantitative insight on which genome properties potentially display spatial organization patterns in 3D nuclear space.
We find histone modifications to have similar assortativity patterns in haematopoietic human cells to the ones described in mESCs (25). We also noticed a highly variable abundance and assortativity of H3K27ac, especially in the PO subnetworks and especially in neutrophils, which might be related to the previously documented high inter-individual variability in methylation and expression in this cell type (45), related to the plasticity required by these cells. We were also able to estimate assortativity of feature pairs (histone modifications) either appearing on the same chromatin fragment or each on one of two interacting fragments. Interestingly, higher assortativity was observed for feature pairs involving combinations of active marks (H3K27ac, ATAC-seq and H3K36me3), with an important role played by 3D contacts (as shown by distance-preserving randomizations). Two-mark combinations involving repressive marks (H3K27me3 and H3K9me3) on the same fragment showed high assortativity (AND-ChAs), while the same marks were not found preferentially on opposite ends of contacts (low cross-ChAs). Further work on higher resolution capture Hi-C networks could shed further light on the biological meaning of these results.
Finally, we compared our results between two network types: PCHi-C datasets and Hi-Cdata processed with the FitHiC pipeline. We observed very consistent results of assortativity of expression, histone modifications and replication timing between these two types of networks, despite clear differences in their topology. DNA methylation was found to be assortative in the PCHi-C PP subnetwork of neutrophils but not in the Hi-C networks for a lymphoblastoid cell line. These results can be due to the different coverage of interactions spanning defined genomic distances in the two networks, to the different coverage of promoter regions in the two networks (no ChAs of DNA methylation on PO networks), and to the different roles methylation plays outside of promoters.
Our recent findings of assortativity of efficiency of replication origins strengthen reported results that replication could have a major association with 3D chromatin structure across scales (49) and be a major force in shaping genome architecture, both in terms of replication timing and of single origin activation.
Beyond the examples given in this paper, the tools provided will allow researchers with or without advanced bioinformatics skills to test hypotheses on 3D organization of any genomic property that can be measured at specific positions along the genome.
DATA AVAILABILITY
GARDEN-NET is available at https://pancaldi.bsc.es/garden-net/ and on GitHub https://github.com/VeraPancaldiLab/GARDEN-NET while ChAseR can be downloaded at https://bitbucket.org/eraineri/ChAseR/ Code to generate all examples is available at https://github.com/VeraPancaldiLab/ChAseR_demo. Published data used is listed in Table 1.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the Barcelona Supercomputing Science for hosting the GARDEN-NET website, Daniel Rico, Biola Javierre, José María Fernández González and Alfonso Valencia for discussions related to the manuscript.
Notes
Present address: Tran Bich Ngoc Cao, Master in Life Sciences, Département de Biologie, École Normale Supérieure, Paris 75005, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
INSERM; Fondation Toulouse Cancer Santé and Pierre Fabre Research Institute as part of the Chair of Bioinformatics in Oncology of the CRCT [to M.M., V.P.]; BioInfo4Women programme at the Barcelona Supercomputing Center [to V.P.]; N.C. acknowledges an Internship scholarship from University of Science and Technology of Hanoi, Vietnam; Spanish Ministry of Economy, Industry and Competitiveness (MEIC) through the Instituto de Salud Carlos III and the 2014–2020 Smart Growth Operating Program [to E.R.]; EMBL partnership and co-financing with the European Regional Development Fund (MINECO/FEDER) [BIO2015-71792-P to E.R.]; Centro de Excelencia Severo Ochoa, and the Generalitat de Catalunya through the Departament de Salut, Departament d’Empresa i Coneixement and the CERCA Programme [to E.R.]; Plan Nacional [PGC2018-099640-B-I00 to E.R.]. Funding for open access charge: Chair of Bioinformatics in Oncology of the CRCT [to V.P.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O. et al.. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (New York, N.Y.). 2009; 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Javierre B.M., Burren O.S., Wilder S.P., Kreuzhuber R., Hill S.M., Sewitz S., Cairns J., Wingett S.W., Várnai C., Thiecke M.J. et al.. Lineage-Specific genome architecture links enhancers and Non-coding disease variants to target gene promoters. Cell. 2016; 167:1369–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G.L., Lubling Y., Xu X., Lv X., Hugnot J.-P., Tanay A. et al.. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017; 171:557–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giorgetti L., Servant N., Heard E.. Changes in the organization of the genome during the mammalian cell cycle. Genome Biol. 2013; 14:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nagano T., Lubling Y., Várnai C., Dudley C., Leung W., Baran Y., Mendelson Cohen N., Wingett S., Fraser P., Tanay A.. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature. 2017; 547:61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J. et al.. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012; 485:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G.. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012; 148:458–472. [DOI] [PubMed] [Google Scholar]
- 8. Schoenfelder S., Furlan-Magaril M., Mifsud B., Tavares-Cadete F., Sugar R., Javierre B.-M., Nagano T., Katsman Y., Sakthidevi M., Wingett S.W. et al.. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015; 25:582–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schoenfelder S., Sugar R., Dimond A., Javierre B.-M., Armstrong H., Mifsud B., Dimitrova E., Matheson L., Tavares-Cadete F., Furlan-Magaril M. et al.. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat. Genet. 2015; 47:1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fullwood M.J., Liu M.H., Pan Y.F., Liu J., Xu H., Mohamed Y.B., Orlov Y.L., Velkov S., Ho A., Mei P.H. et al.. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009; 462:58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng M., Tian S.Z., Capurso D., Kim M., Maurya R., Lee B., Piecuch E., Gong L., Zhu J.J., Li Z. et al.. Multiplex chromatin interactions with single-molecule precision. Nature. 2019; 566:558–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G.L., Lubling Y., Xu X., Lv X., Hugnot J.-P., Tanay A. et al.. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017; 171:557–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beliveau B.J., Apostolopoulos N., Wu C.-T.. Visualizing genomes with Oligopaint FISH probes. Curr. Protoc. Mol. Biol. 2014; 105:14.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cardozo Gizzi A.M., Cattoni D.I., Fiche J.-B., Espinola S.M., Gurgo J., Messina O., Houbron C., Ogiyama Y., Papadopoulos G.L., Cavalli G. et al.. Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms. Mol. Cell. 2019; 74:212–222. [DOI] [PubMed] [Google Scholar]
- 15. Beagrie R.A., Scialdone A., Schueler M., Kraemer D.C.A., Chotalia M., Xie S.Q., Barbieri M., de Santiago I., Lavitas L.-M., Branco M.R. et al.. Complex multi-enhancer contacts captured by genome architecture mapping. Nature. 2017; 543:519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quinodoz S.A., Ollikainen N., Tabak B., Palla A., Schmidt J.M., Detmar E., Lai M.M., Shishkin A.A., Bhat P., Takei Y. et al.. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell. 2018; 174:744–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Botta M., Haider S., Leung I.X., Lio P., Mozziconacci J.. Intra-and inter-chromosomal interactions correlate with CTCF binding genome wide. Mol. Syst. Biol. 2010; 6:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sandhu K.S., Li G., Poh H.M., Quek Y.L.K., Sia Y.Y., Peh S.Q., Mulawadi F.H., Lim J., Sikic M., Menghi F. et al.. Large-scale functional organization of long-range chromatin interaction networks. Cell Rep. 2012; 2:1207–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boulos R.E., Arneodo A., Jensen P., Audit B.. Revealing long-range interconnected hubs in human chromatin interaction data using graph theory. Phys. Rev. Lett. 2013; 111:118102. [DOI] [PubMed] [Google Scholar]
- 20. MacKay K., Kusalik A., Eskiw C.H.. GrapHi-C: graph-based visualization of Hi-C datasets. BMC Res. Notes. 2018; 11:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan Z.W., Guarnera E., Berezovsky I.N.. Exploring chromatin hierarchical organization via Markov state modelling. PLOS Comput. Biol. 2018; 14:e1006686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paulsen J., Liyakat Ali T.M., Nekrasov M., Delbarre E., Baudement M.-O., Kurscheid S., Tremethick D., Collas P.. Long-range interactions between topologically associating domains shape the four-dimensional genome during differentiation. Nat. Genet. 2019; 51:835–843. [DOI] [PubMed] [Google Scholar]
- 23. Lesne A., Riposo J., Roger P., Cournac A., Mozziconacci J.. 3D genome reconstruction from chromosomal contacts. Nat. Methods. 2014; 11:1141–1143. [DOI] [PubMed] [Google Scholar]
- 24. Thibodeau A., Márquez E.J., Shin D.-G., Vera-Licona P., Ucar D.. Chromatin interaction networks revealed unique connectivity patterns of broad H3K4me3 domains and super enhancers in 3D chromatin. Sci. Rep. 2017; 7:14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pancaldi V., Carrillo-de Santa-Pau E., Javierre B.M., Juan D., Fraser P., Spivakov M., Valencia A., Rico D.. Integrating epigenomic data and 3D genomic structure with a new measure of chromatin assortativity. Genome Biol. 2016; 17:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Juan D., Perner J., Carrillo de Santa Pau E., Marsili S., Ochoa D., Chung H.-R., Vingron M., Rico D., Valencia A.. Epigenomic Co-localization and Co-evolution reveal a key role for 5hmC as a communication Hub in the chromatin network of ESCs. Cell Rep. 2016; 14:1246–1257. [DOI] [PubMed] [Google Scholar]
- 27. Norton H.K., Emerson D.J., Huang H., Kim J., Titus K.R., Gu S., Bassett D.S., Phillips-Cremins J.E.. Detecting hierarchical genome folding with network modularity. Nat. Methods. 2018; 15:119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J., Hero A.O., Rajapakse I.. Spectral identification of topological domains. Bioinformatics. 2016; 32:2151–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marbouty M., Cournac A., Flot J.F., Marie-Nelly H., Mozziconacci J., Koszul R.. Metagenomic chromosome conformation capture (meta3C) unveils the diversity of chromosome organization in microorganisms. eLife. 2014; 3:e03318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morlot J.-B., Mozziconacci J., Lesne A.. Network concepts for analyzing 3D genome structure from chromosomal contact maps. EPJ Nonlinear Biomed. Phys. 2016; 4:2. [Google Scholar]
- 31. Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B.. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012; 485:376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen L., Ge B., Casale F., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yan Y., Kundu K., Ecker S. et al.. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016; 167:1398–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beekman R., Chapaprieta V., Russiñol N., Vilarrasa-Blasi R., Verdaguer-Dot N., Martens J.H., Duran-Ferrer M., Kulis M., Serra F., Javierre B.M. et al.. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat. Med. 2018; 24:868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang X., Zhang Y., Zhu X., Purmann C., Haney M.S., Ward T., Khechaduri A., Yao J., Weissman S.M., Urban A.E.. Local and global chromatin interactions are altered by large genomic deletions associated with human brain development. Nat. Commun. 2018; 9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pope B.D., Ryba T., Dileep V., Yue F., Wu W., Denas O., Vera D.L., Wang Y., Hansen R.S., Canfield T.K. et al.. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014; 515:402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Finotello F., Trajanoski Z.. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol. Immunother.: CII. 2018; 67:1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cairns J., Freire-Pritchett P., Wingett S.W., Várnai C., Dimond A., Plagnol V., Zerbino D., Schoenfelder S., Javierre B.-M., Osborne C. et al.. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 2016; 17:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zouari Y.B., Molitor A.M., Sikorska N., Pancaldi V., Sexton T.. ChiCMaxima: a robust and simple pipeline for detection and visualization of chromatin looping in Capture Hi-C. Genome Biology. 2018; 20:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ay F., Bailey T.L., Noble W.S.. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res. 2014; 24:999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Franz M., Lopes C.T., Huck G., Dong Y., Sumer O., Bader G.D.. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics. 2015; 32:309–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Newman M. E.J. Networks. An Introduction. 2010; Oxford: Oxford University Press. [Google Scholar]
- 43. Mifsud B., Tavares-Cadete F., Young A.N., Sugar R., Schoenfelder S., Ferreira L., Wingett S.W., Andrews S., Grey W., Ewels P.A. et al.. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015; 47:598–606. [DOI] [PubMed] [Google Scholar]
- 44. Zhou V.W., Goren A., Bernstein B.E.. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011; 12:7–18. [DOI] [PubMed] [Google Scholar]
- 45. Ecker S., Pancaldi V., Valencia A., Beck S., Paul D.. Epigenetic and transcriptional variability shape phenotypic plasticity. BioEssays. 2018; 40:1700148. [DOI] [PubMed] [Google Scholar]
- 46. Delaneau O., Zazhytska M., Borel C., Giannuzzi G., Rey G., Howald C., Kumar S., Ongen H., Popadin K., Marbach D. et al.. Chromatin three-dimensional interactions mediate genetic effects on gene expression. Science (New York, N.Y.). 2019; 364:eaat8266. [DOI] [PubMed] [Google Scholar]
- 47. Robson M.I., Ringel A.R., Mundlos S.. Regulatory landscaping: how enhancer-promoter communication is sculpted in 3D. Mol. cell. 2019; 74:1110–1122. [DOI] [PubMed] [Google Scholar]
- 48. Carrillo-de Santa-Pau E., Juan D., Pancaldi V., Were F., Martin-Subero I., Rico D., Valencia A.. Automatic identification of informative regions with epigenomic changes associated to hematopoiesis. Nucleic Acids Res. 2017; 45:9244–9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jodkowska K., Pancaldi V., Almeida R., Rigau M., Graña-Castro O., Fernández-Justel J.M., Rodríguez-Acebes S., Rubio-Camarillo M., Pau E.C.-D.S., Pisano D. et al.. Three-dimensional connectivity and chromatin environment mediate the activation efficiency of mammalian DNA replication origins. 2019; bioRxiv doi:29 May 2019, preprint: not peer reviewed 10.1101/644971. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GARDEN-NET is available at https://pancaldi.bsc.es/garden-net/ and on GitHub https://github.com/VeraPancaldiLab/GARDEN-NET while ChAseR can be downloaded at https://bitbucket.org/eraineri/ChAseR/ Code to generate all examples is available at https://github.com/VeraPancaldiLab/ChAseR_demo. Published data used is listed in Table 1.