Abstract
Background
Radiofrequency ultrasound data from the liver contain rich information about liver microstructure and composition. Deep learning might exploit such information to assess nonalcoholic fatty liver disease (NAFLD).
Purpose
To develop and evaluate deep learning algorithms that use radiofrequency data for NAFLD assessment, with MRI-derived proton density fat fraction (PDFF) as the reference.
Materials and Methods
A HIPAA-compliant secondary analysis of a single-center prospective study was performed for adult participants with NAFLD and control participants without liver disease. Participants in the parent study were recruited between February 2012 and March 2014 and underwent same-day US and MRI of the liver. Participants were randomly divided into an equal number of training and test groups. The training group was used to develop two algorithms via cross-validation: a classifier to diagnose NAFLD (MRI PDFF ≥ 5%) and a fat fraction estimator to predict MRI PDFF. Both algorithms used one-dimensional convolutional neural networks. The test group was used to evaluate the classifier for sensitivity, specificity, positive predictive value, negative predictive value, and accuracy and to evaluate the estimator for correlation, bias, limits of agreements, and linearity between predicted fat fraction and MRI PDFF.
Results
A total of 204 participants were analyzed, 140 had NAFLD (mean age, 52 years ± 14 [standard deviation]; 82 women) and 64 were control participants (mean age, 46 years ± 21; 42 women). In the test group, the classifier provided 96% (95% confidence interval [CI]: 90%, 99%) (98 of 102) accuracy for NAFLD diagnosis (sensitivity, 97% [95% CI: 90%, 100%], 68 of 70; specificity, 94% [95% CI: 79%, 99%], 30 of 32; positive predictive value, 97% [95% CI: 90%, 99%], 68 of 70; negative predictive value, 94% [95% CI: 79%, 98%], 30 of 32). The estimator-predicted fat fraction correlated with MRI PDFF (Pearson r = 0.85). The mean bias was 0.8% (P = .08), and 95% limits of agreement were -7.6% to 9.1%. The predicted fat fraction was linear with an MRI PDFF of 18% or less (r = 0.89, slope = 1.1, intercept = 1.3) and nonlinear with an MRI PDFF greater than 18%.
Conclusion
Deep learning algorithms using radiofrequency ultrasound data are accurate for diagnosis of nonalcoholic fatty liver disease and hepatic fat fraction quantification when other causes of steatosis are excluded.
© RSNA, 2020
Online supplemental material is available for this article.
See also the editorial by Lockhart and Smith in this issue.
Summary
When other causes of steatosis are excluded, de novo one-dimensional convolutional neural network algorithms can accurately identify nonalcoholic fatty liver disease and quantify hepatic fat fraction by using raw radiofrequency ultrasound data.
Key Results
■ Deep learning with raw ultrasound data provided hepatic fat fraction estimates correlated to proton density fat fraction measured with confounder-corrected chemical shift–encoded MRI (Pearson r = 0.85).
■ The proposed deep learning approach can diagnose nonalcoholic fatty liver disease (area under the receiver operating characteristic curve, 0.98) and is robust to changes in system settings, including transmit focal range and time gain compensation.
Introduction
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disease worldwide, affecting approximately 25% of the human population (1). NAFLD covers a spectrum of liver abnormalities ranging from simple steatosis to nonalcoholic steatohepatitis. Hepatic steatosis, characterized by the accumulation of fat droplets within hepatocytes, can progress to nonalcoholic steatohepatitis, fibrosis, cirrhosis, and even hepatocellular carcinoma (1,2). Early detection and treatment may halt or reverse NAFLD progression (2). Liver biopsy remains the reference standard for diagnosing NALFD and grading hepatic steatosis. However, biopsy is costly, invasive, and inappropriate for screening.
There is a critical need to develop noninvasive imaging methods to assess hepatic steatosis. Several modalities have been investigated (3–8), among which MRI and conventional (qualitative) US have the advantage of involving no ionizing radiation. Confounder-corrected chemical shift–encoded MRI can measure the proton density fat fraction (PDFF), a leading method for noninvasive quantification of hepatic steatosis (4,5). However, chemical shift–encoded MRI is not routinely accessible. Conventional US is widely available for NALFD assessment but is limited by its qualitative nature, operator dependency, and modest accuracy (3).
Quantitative analysis of raw radiofrequency (RF) ultrasound signals shows potential for objective and accurate disease assessment (9). This analysis is based on the premise that by altering tissue microstructures, disease processes can cause quantifiable RF signal changes. Of note, the RF signals contain more information than do gray-scale B-mode images because information is lost or altered when B-mode images are generated from the raw data (9). Thus, when compared with B-mode images, the rich RF data may allow more comprehensive characterization of pathophysiologic conditions. By using a well-characterized phantom for system calibration, quantitative ultrasound techniques analyze the fundamental RF signals to extract system-independent parameters, such as the attenuation and backscatter coefficients, with minimal operator dependency (10–14). These coefficients are correlated with hepatic fat fraction (15–17).
To take further advantage of RF signals and to eliminate the dependency on a calibration phantom, we propose a phantom-free deep-learning ultrasound approach for objective, accurate, and automated NAFLD diagnosis and liver fat quantification. Deep learning (18,19) based on convolutional neural networks (CNNs) can extract features from raw data and has been applied to ultrasound B-mode image analysis (20–25) but not RF analysis for steatosis assessment. The current study developed and evaluated one-dimensional CNNs for NAFLD diagnosis and liver fat quantification using contemporaneous MRI PDFF as the reference standard. MRI PDFF was used because it accurately quantifies liver fat (4,5) and can be safely and ethically acquired in asymptomatic control participants.
Materials and Methods
Study Participants
This study was a secondary analysis of 204 prospectively enrolled adult research participants with NAFLD and control participants without liver disease. The parent study was reported in a previous article (16) and used a different ultrasound analysis technique. For the current analysis, we developed and evaluated deep learning techniques in the same participants. The University of California, San Diego, approved this secondary analysis and the parent study, both of which complied with the Health Insurance Portability and Accountability Act. All participants provided written informed consent.
Participants were consecutively recruited by an expert hepatologist (R.L., >10 years of experience) from the University of California, San Diego, NAFLD Research Center between February 2012 and March 2014. Inclusion criteria were age of at least 18 years and willingness and ability to participate. Exclusion criteria were clinical, laboratory, or histologic evidence of a liver disease other than NAFLD; excessive alcohol consumption (>30 g per day within the past 10 years or >10 g per day in the previous year); and steatogenic or hepatoxic medication use. NAFLD in study participants was defined as MRI PDFF of 5% or greater, with other causes of steatosis excluded (16). Control participants (MRI PDFF <5%) had no liver disease based on comprehensive clinical and laboratory testing performed under the supervision of and interpreted by the hepatologist. All participants underwent same-day US and chemical shift–encoded MRI of the liver.
Ultrasound Protocol
Nonenhanced US was performed by a research physician (E.H., 1 year of hands-on US training) using the 4C1 convex array (1–4 MHz) on a clinical ultrasound machine (Siemens S2000; Siemens, Issaquah, Wash) with an Ultrasound Research Interface option that allowed direct acquisition of RF data. Participants were positioned in the dorsal decubitus position, with the right arm at maximum abduction. The right liver lobe was visualized via a right intercostal approach, and a representative region of the parenchyma was identified, thereby avoiding major vasculature. The physician adjusted the transmit focal range and time gain compensation (TGC) (ie, a setting that reduces the effect of ultrasound attenuation on clinical images by increasing the received signal intensity with time [depth] [26]) for each participant, while fixing other settings. Ten consecutive RF frames were recorded at a rate of 10 frames per second when the participant was executing a breath hold in shallow expiration. Each frame had 560 lateral lines and was 10 cm deep. The machine automatically recorded the transmit focal range and the TGC settings (Appendix E1 [online]).
Ultrasound Data Preprocessing
A fixed region of interest with standard size and location (central 256 RF lines laterally; 1.8–9.7 cm axially) relative to the image frame was used for the one-dimensional CNN algorithms, yielding 2560 RF signals per participant (256 RF signals per frame × 10 frames per participant). The region of interest was intended to cover as much of the liver region below the liver capsule as possible while generally avoiding tissues outside the liver. A fixed region of interest rather than a hand-drawn one tailored to each participant’s liver anatomy was applied to minimize human intervention. No effort was made to completely exclude regions outside the liver, however, and the regions of interest contained variable amounts of extrahepatic tissue and structures.
Because TGC settings affect RF signals, quantitative analyses were performed before and after removal of the machine-recorded TGC settings.
The RF data of the last five frames were corrupted in two participants in the test group. The intact frames (frames 1–5) were duplicated for both participants to make 10 frames per participant for convenience of algorithm testing.
To reduce data size, the signals were downsampled by decimating the RF by four (ie, keeping every fourth sample) without filtering, to reduce the sampling frequency from 40 MHz to 10 MHz. According to the Nyquist-Shannon sampling theorem, the 10-MHz sampling frequency was sufficient to preserve useful information contained in the original signal acquired from the 4C1 transducer (bandwidth, approximately 2–4 MHz). The downsampled signal containing 1024 sample points was input to the one-dimensional CNNs.
CNN Algorithm Development
Two one-dimensional CNN algorithms were developed: a binary classifier and a fat fraction estimator. For each RF signal input, the classifier output an NAFLD classification score between 0 and 1, and the fat fraction estimator output the predicted fat fraction as a percentage.
The participants were equally divided into training (n = 102) and test (n = 102) groups by using stratified randomization (16). The algorithms were developed by using the training group via cross-validation and were evaluated by using the test group. Details are presented in Appendix E2 (online), and the code is available for research use at https://github.com/han51/nafld-1d-cnn.
Statistical Analysis
Algorithms were evaluated at the participant level. Because each algorithm generated one output per RF signal input and because there were 2560 signal inputs per participant, the classifier and fat fraction estimator generated 2560 NAFLD classification scores and 2560 fat fraction estimates per participant, respectively. The 2560 outputs were averaged for each algorithm to yield composite per-participant scores and estimates. A cutoff of 0.5 for the composite score was set a priori for the NAFLD classifier.
To evaluate classifier performance, we calculated sensitivity, specificity, positive predictive value, negative predictive value, and overall accuracy for NAFLD identification in the test group. We also generated the receiver operating characteristic curve of the composite NAFLD classification score for the test group and calculated the area under the receiver operating characteristic curve (AUC) and the 95% confidence interval (CI). The De Long test (27) was performed to compare the AUCs obtained by using signals without and with TGC.
To evaluate the fat fraction estimator performance, we calculated the correlation (Pearson r), bias, limits of agreement, and linearity between predicted fat fraction and MRI PDFF. Linearity was assessed by using sequential tests of polynomial fits to the plot of estimated fat fraction versus MRI PDFF (28). Linear range was identified if the linearity test over the entire MRI PDFF range failed. Linear regression slope, intercept, and R2 were evaluated. Analyses were performed by using MATLAB R2016a (Mathworks, Natick, Mass) and RStudio 1.2 (RStudio, Boston, Mass) software. A P value less than .05 indicated statistical significance.
Results
Participant Characteristics
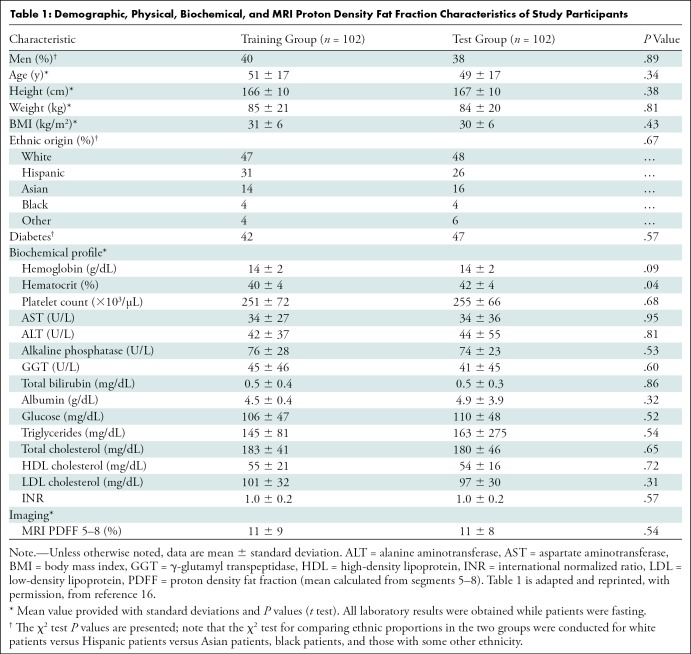
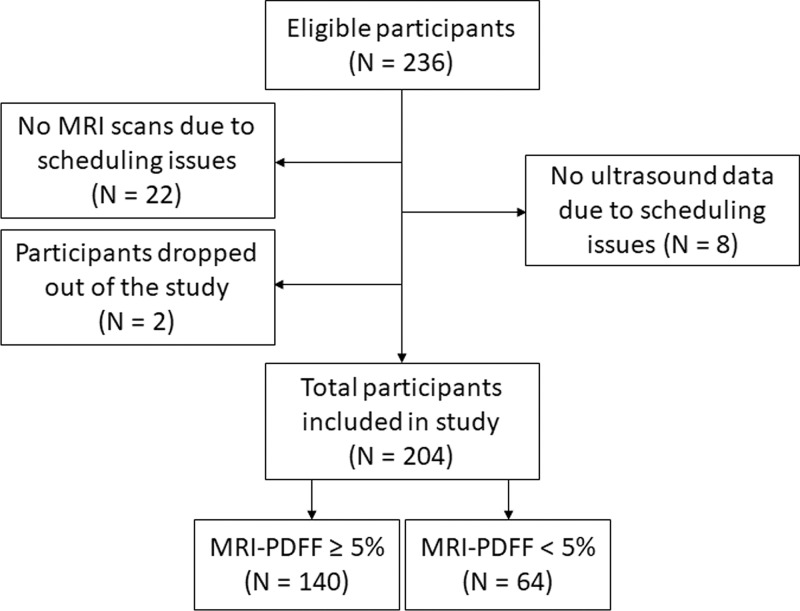
Participant characteristics are reported in Table 1. A total of 204 participants (Fig 1), 140 with NAFLD (mean age, 52 years ± 14 [standard deviation]; 82 women) and 64 control participants (mean age, 46 years ± 21; 42 women), were analyzed. Men made up 40% (41 of 102) of the training group and 38% (39 of 102) of the test group. The mean body mass index was 31 kg/m2 ± 6 in the training group and 30 kg/m2 ± 6 in the test group. The mean MRI PDFF (segments 5–8) was 11% ± 9 and 11% ± 8 in the training and test groups, respectively. In each group, MRI PDFF ranged from 1% to 35%, and 70 of 102 participants (69%) had NAFLD (MRI PDFF ≥ 5%).
Table 1:
Demographic, Physical, Biochemical, and MRI Proton Density Fat Fraction Characteristics of Study Participants
Figure 1:
Flowchart of study participants included and excluded in the study, adapted from reference 16. PDFF = proton density fat fraction.
RF Signals and B-Mode Images
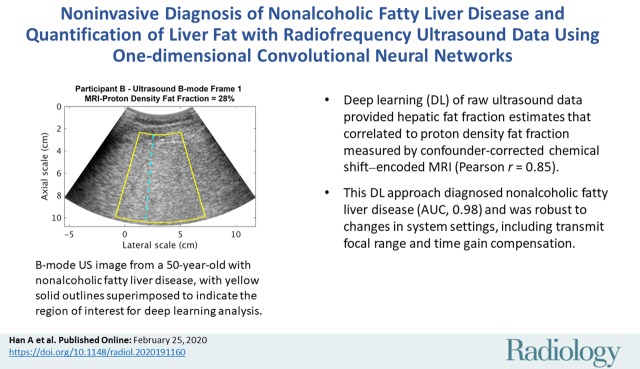
Representative B-mode images (with TGC) and RF signals are shown for two participants, referred to as participants A (MRI PDFF, 1%) (Fig 2) and B (MRI PDFF, 28%) (Fig 3). Figure 2a is a B-mode image reconstructed from frame 1 of the raw RF data (with TGC) acquired in participant A. The fixed region of interest is outlined by the superimposed yellow box. The blue dashed line is one of the 256 lines covered by the region of interest. The RF signals without and with TGC corresponding to the blue line in Figure 2a are shown in Figure 2b. Ultrasound attenuation with time (depth) was modest in the RF signal without TGC (Fig 2b), corresponding to the lower fat fraction. The TGC caused the deeper and weaker signals to be artificially more pronounced than the more superficial signals.
Figure 2a:
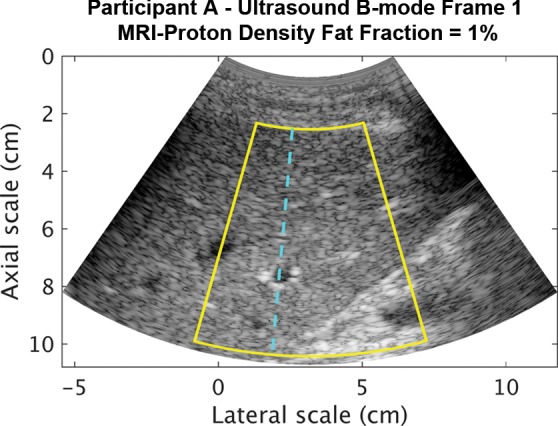
Data from 22-year-old woman with low proton density fat fraction (1%) (control participant, denoted participant A). Computer-reconstructed nonenhanced ultrasound B-mode images (sagittal plane with time gain compensation) and the underlying radiofrequency signals. (a) B-mode image frame 1 (with time gain compensation), with yellow outline superimposed to indicate the region of interest for deep learning analysis. (b) Radiofrequency signals corresponding to the blue line in a, without and with time gain compensation. (c) B-mode image frame 2 (with time gain compensation). (d) Radiofrequency signals corresponding to same location as indicated by the blue line in c but different frames (blue = frame 1, black = frame 2) without and with time gain compensation. Fixed region of interest includes signals from outside the liver.
Figure 3a:
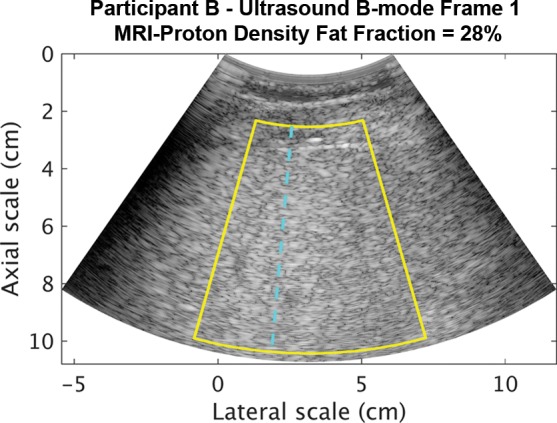
Data from 50-year-old man with high proton density fat fraction (28%) (participant with nonalcoholic fatty liver disease, denoted participant B). Computer-reconstructed nonenhanced ultrasound B-mode image (transverse plane with time gain compensation) and underlying radiofrequency signals. (a) B-mode image frame 1 for participant B, with yellow outline superimposed to indicate region of interest for deep learning analysis. (b) Radiofrequency signals corresponding to blue dashed line shown in a, without and with time gain compensation. Boundaries of the liver are not well delineated, and it is unclear whether the fixed region of interest includes signals from outside the liver.
Figure 2b:
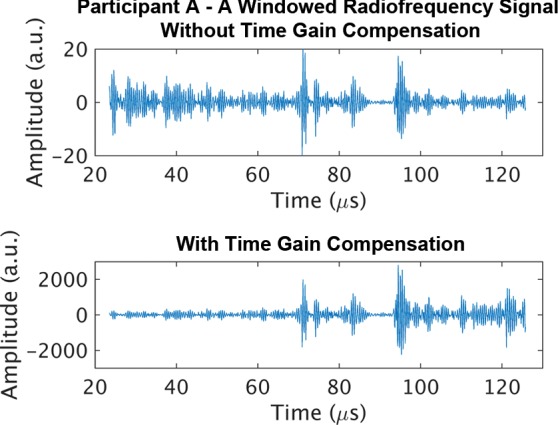
Data from 22-year-old woman with low proton density fat fraction (1%) (control participant, denoted participant A). Computer-reconstructed nonenhanced ultrasound B-mode images (sagittal plane with time gain compensation) and the underlying radiofrequency signals. (a) B-mode image frame 1 (with time gain compensation), with yellow outline superimposed to indicate the region of interest for deep learning analysis. (b) Radiofrequency signals corresponding to the blue line in a, without and with time gain compensation. (c) B-mode image frame 2 (with time gain compensation). (d) Radiofrequency signals corresponding to same location as indicated by the blue line in c but different frames (blue = frame 1, black = frame 2) without and with time gain compensation. Fixed region of interest includes signals from outside the liver.
Figure 3b:
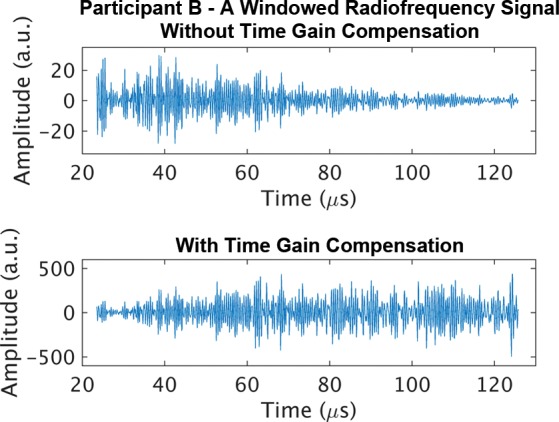
Data from 50-year-old man with high proton density fat fraction (28%) (participant with nonalcoholic fatty liver disease, denoted participant B). Computer-reconstructed nonenhanced ultrasound B-mode image (transverse plane with time gain compensation) and underlying radiofrequency signals. (a) B-mode image frame 1 for participant B, with yellow outline superimposed to indicate region of interest for deep learning analysis. (b) Radiofrequency signals corresponding to blue dashed line shown in a, without and with time gain compensation. Boundaries of the liver are not well delineated, and it is unclear whether the fixed region of interest includes signals from outside the liver.
The reconstructed B-mode images were visually similar between adjacent frames (Fig 2a, 2c), although careful examination revealed differences that were likely due to slight motion between frames. In contrast, the RF signals along the same scan line (Fig 2d) were noticeably different between adjacent frames (eg, different intensities around 40, 70, and 95 µsec), both without and with TGC, as might be expected due to random and structured effects, which contributed to speckle and noise.
Figure 2c:
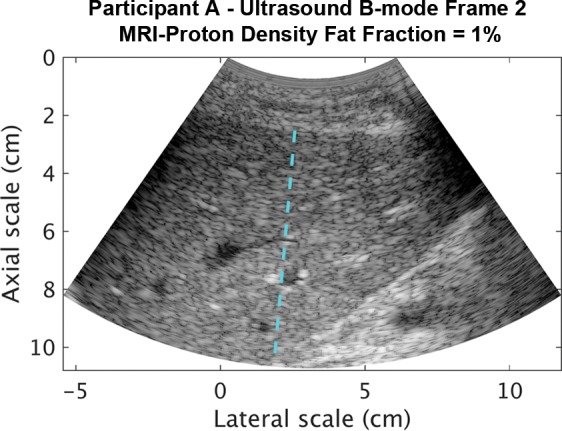
Data from 22-year-old woman with low proton density fat fraction (1%) (control participant, denoted participant A). Computer-reconstructed nonenhanced ultrasound B-mode images (sagittal plane with time gain compensation) and the underlying radiofrequency signals. (a) B-mode image frame 1 (with time gain compensation), with yellow outline superimposed to indicate the region of interest for deep learning analysis. (b) Radiofrequency signals corresponding to the blue line in a, without and with time gain compensation. (c) B-mode image frame 2 (with time gain compensation). (d) Radiofrequency signals corresponding to same location as indicated by the blue line in c but different frames (blue = frame 1, black = frame 2) without and with time gain compensation. Fixed region of interest includes signals from outside the liver.
Figure 2d:
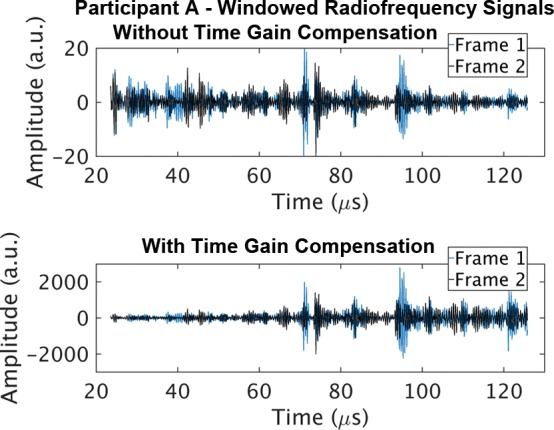
Data from 22-year-old woman with low proton density fat fraction (1%) (control participant, denoted participant A). Computer-reconstructed nonenhanced ultrasound B-mode images (sagittal plane with time gain compensation) and the underlying radiofrequency signals. (a) B-mode image frame 1 (with time gain compensation), with yellow outline superimposed to indicate the region of interest for deep learning analysis. (b) Radiofrequency signals corresponding to the blue line in a, without and with time gain compensation. (c) B-mode image frame 2 (with time gain compensation). (d) Radiofrequency signals corresponding to same location as indicated by the blue line in c but different frames (blue = frame 1, black = frame 2) without and with time gain compensation. Fixed region of interest includes signals from outside the liver.
The reconstructed B-mode image and RF signals from participant B (Fig 3) visually differed from those from participant A (Fig 2a, 2b). The B-mode image was more homogeneous for participant B than for participant A. Blood vessels were visible on the B-mode image for participant A but were obscured for participant B. For the RF signals from participant B, increased attenuation with time (depth) was evident without TGC, corresponding to the higher fat fraction. The TGC compensated for this attenuation by increasing signal amplification with time.
Classification
In the test group, the composite NAFLD classification score provided a high degree of discrimination between control participants without liver disease and participants with NALFD, as demonstrated by the receiver operating characteristic curves obtained by using RF signals without and with TGC (Fig 4). The AUCs were 0.98 (95% CI: 0.94, 1.00) and 0.95 (95% CI: 0.91, 0.99) for scores obtained by using RF signals without and with TGC, respectively. The two AUC estimates did not differ (P = .23).
Figure 4a:
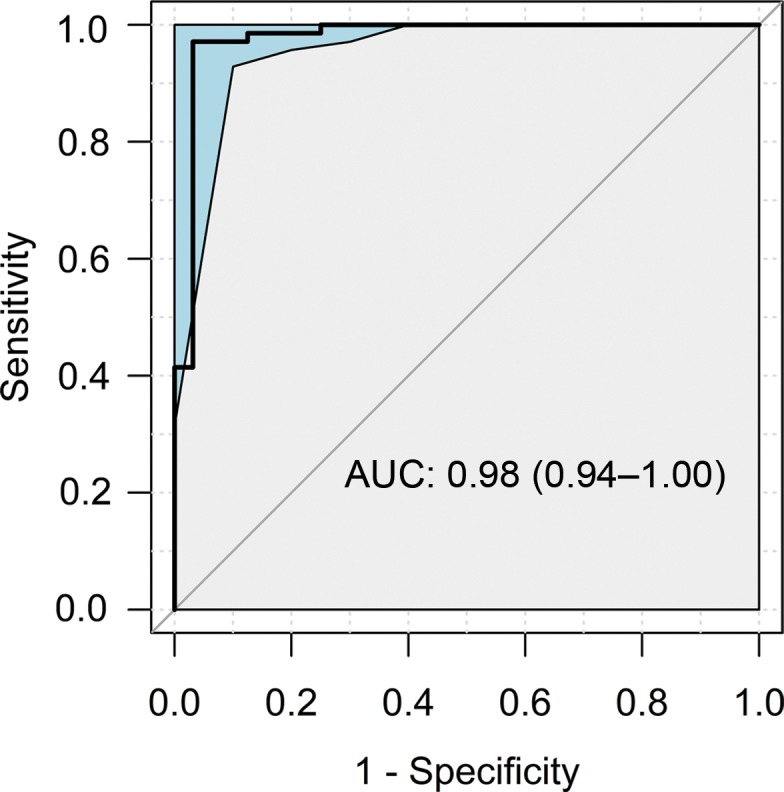
Receiver operating characteristic curves with 95% confidence bands of the composite nonalcoholic fatty liver disease classification scores yielded by the classifier for the test group using radiofrequency ultrasound signals (a) without and (b) with time gain compensation as the inputs. AUC = area under receiver operating characteristic curve.
Figure 4b:
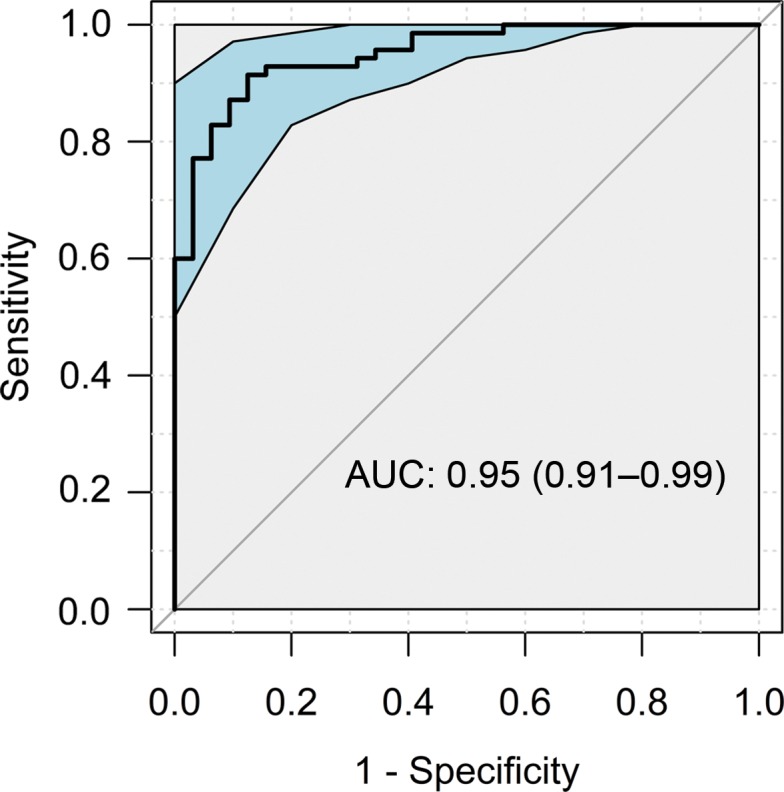
Receiver operating characteristic curves with 95% confidence bands of the composite nonalcoholic fatty liver disease classification scores yielded by the classifier for the test group using radiofrequency ultrasound signals (a) without and (b) with time gain compensation as the inputs. AUC = area under receiver operating characteristic curve.
Applying the predetermined threshold of 0.5 on the composite NAFLD classification score for NAFLD diagnosis in the test group yielded 68 true-positive results, two false-positive results, two false-negative results, and 30 true-negative results when RF signals without TGC were used and yielded 64 true-positive results, four false-positive results, six false-negative results, and 28 true-negative results when RF signals with TGC were used. These diagnostic results yielded a classification accuracy of 96% in the test group using RF signals without TGC, with 97% sensitivity, 94% specificity, 97% positive predictive value, and 94% negative predictive value (Table 2). They yielded a classification accuracy of 90% in the test group using RF signals with TGC, with 91% sensitivity, 88% specificity, 94% positive predictive value, and 82% negative predictive value (Table 2).
Table 2:
Performance Metrics for Nonalcoholic Fatty Liver Disease Diagnosis in Test Group
Fat Fraction Estimation
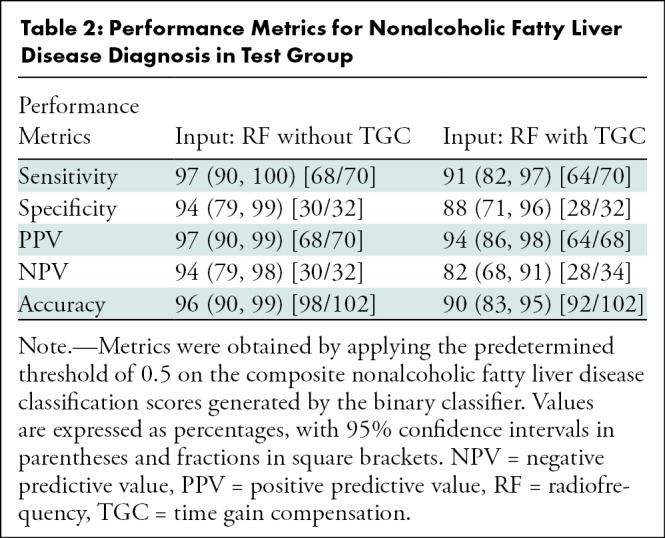
The predicted fat fraction values correlated with the MRI PDFF in the test group for RF signals without and those with TGC (Fig 5). The Pearson correlation coefficient was 0.85 (P < .001) and 0.80 (P < .001) for use of RF signals without and with TGC, respectively.
Figure 5a:
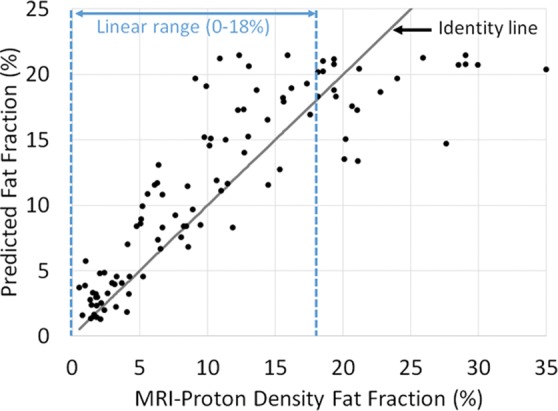
Predicted fat fraction versus MRI-derived proton density fat fraction obtained by using radiofrequency signals (a) without and (b) with time gain compensation. Blue lines represent the linear range. Gray line represents the identity line.
Figure 5b:
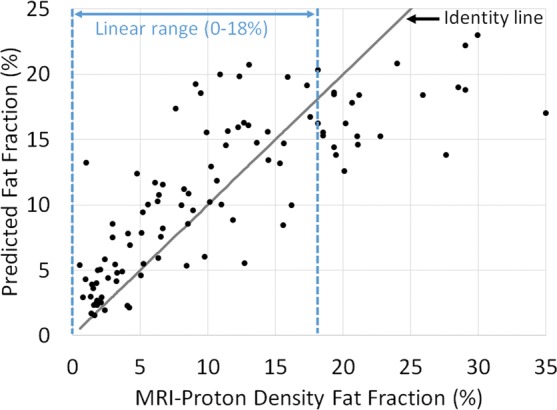
Predicted fat fraction versus MRI-derived proton density fat fraction obtained by using radiofrequency signals (a) without and (b) with time gain compensation. Blue lines represent the linear range. Gray line represents the identity line.
Graphically, the predicted fat fraction versus MRI PDFF scatterplots (Fig 5) track the identity line. A linearity test (28) showed no nonlinearity between predicted fat fraction and MRI PDFF for MRI PDFF of 18% or less, regardless of whether TGC was removed. Linear regression of the predicted fat fraction against MRI PDFF within the linear range (MRI PDFF ≤ 18%) yielded a slope of 1.1, an intercept of 1.3, and R2 of 0.79 (Pearson r = 0.89) when signals without TGC were used and a slope of 0.9, an intercept of 3.1, and R2 of 0.59 (Pearson r = 0.77) when signals with TGC were used. The fat fraction estimator underestimated the fat fraction for MRI PDFF greater than 18%, suggesting a saturation effect outside the linear range. Linear regression of the predicted fat fraction against MRI PDFF over the entire MRI PDFF range (MRI PDFF < 35%) yielded a slope of 0.7, an intercept of 3.8, and R2 of 0.73 (Pearson r = 0.85) when signals without TGC were used and a slope of 0.6, an intercept of 4.8, and R2 of 0.64 (Pearson r = 0.80) when signals with TGC were used; the R2 values were equal to the squared values of the Pearson correlation coefficients, as expected.
The mean bias of the predicted fat fraction over the entire MRI PDFF range was 0.8% (P = .08), and 95% limits of agreement were -7.6% to 9.1% when signals without TGC were used (Fig 6). When signals with TGC were used, the mean bias became 0.34% (P = .49), and the 95% limits of agreement were -9.4% to 10.0%.
Figure 6a:
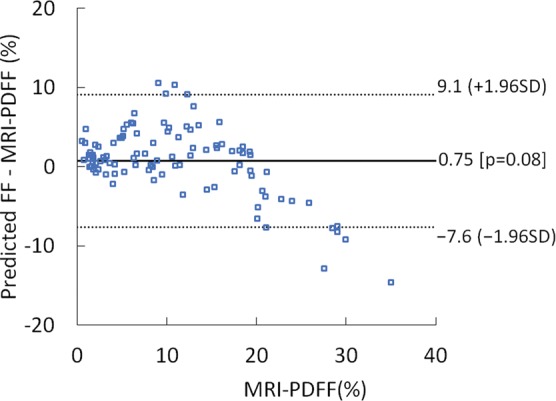
Difference between predicted fat fraction (FF) and MRI-derived proton density fat fraction (PDFF) versus the MRI-derived PDFF plots obtained by using radiofrequency signals (a) without and (b) with time gain compensation. SD = standard deviation.
Figure 6b:
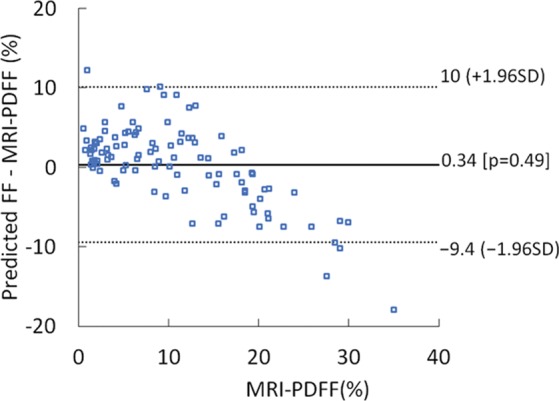
Difference between predicted fat fraction (FF) and MRI-derived proton density fat fraction (PDFF) versus the MRI-derived PDFF plots obtained by using radiofrequency signals (a) without and (b) with time gain compensation. SD = standard deviation.
Discussion
We developed one-dimensional convolutional neural network (CNN) algorithms for nonalcoholic fatty liver disease (NAFLD) diagnosis and fat fraction estimation using ultrasound radiofrequency (RF) signals as the input and MRI proton density fat fraction (PDFF) as the reference standard. The algorithms showed promising performance in a test group of 102 participants. The classifier yielded high classification accuracy (96%) and an area under the receiver operating characteristic curve of 0.98. The fat fraction estimator predicted fat fraction values that correlated with MRI PDFF (r = 0.85; P < .001) and that were linear with MRI PDFF over a broad range of clinically relevant MRI PDFF values. However, we also observed a possible saturation effect at MRI PDFF greater than 18%, the exact cause of which is not yet well understood. A potential explanation was insufficient training data for MRI PDFF greater than 18%. Another potential explanation was that ultrasonic signals could be insensitive to fat fraction changes at high MRI PDFF values. We also demonstrated the feasibility to develop and train one-dimensional CNNs de novo using RF signals, without using techniques, such as transfer learning (ie, reuse of a model pretrained on a different problem) and data augmentation (ie, artificial expansion of the input data through various transformations). We showed algorithm robustness under varying transmit focal range and time gain compensation (TGC) settings, although better performance was achieved by using signals without TGC. Other settings (eg, transmit frequency, line density) potentially critical to the algorithm performance were fixed. However, the model robustness with focal range and TGC suggested the one-dimensional CNN algorithms could be robust to more settings, possibly providing a phantom-free approach for ultrasound diagnosis using RF signals.
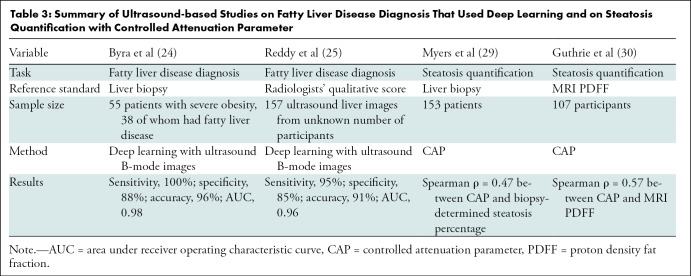
Several studies have used deep learning with B-mode images for steatosis classification (Table 3). Byra et al (24) proposed a transfer learning approach to diagnose fatty liver disease using ultrasound B-mode images with a deep CNN pretrained with nonmedical images. They evaluated the approach in 55 patients with severe obesity, 38 of whom had fatty liver (with biopsy used as the reference standard), yielding 100% sensitivity, 88% specificity, 96% overall accuracy, and an AUC of 0.98. Reddy et al (25) used a similar transfer learning approach to diagnose fatty liver disease on 157 ultrasound liver images, with radiologists’ qualitative score used as the ground truth, yielding 95% sensitivity, 85% specificity, 91% accuracy, and an AUC of 0.96. Although our classifier achieved performances nominally similar to those of Byra et al (24) and better than those of Reddy et al (25), it is difficult to directly compare the various studies because of differences in the reference and participant samples.
Table 3:
Summary of Ultrasound-based Studies on Fatty Liver Disease Diagnosis That Used Deep Learning and on Steatosis Quantification with Controlled Attenuation Parameter
Several studies quantified liver steatosis by using MRI PDFF or liver biopsy as the reference standard (Table 3). For example, a study of 153 patients (29) showed that controlled attenuation parameter was correlated with the percentage of steatosis (Spearman ρ = 0.47), with biopsy used as the reference standard, and a study (30) in 107 participants showed a 0.57 Spearman correlation coefficient between the controlled attenuation parameter and MRI PDFF.
Use of RF signals has several potential advantages. Not only do RF signals contain more information than B-mode images (9) or the envelope data (Appendix E3 [online]), they are also less dependent on system settings and postprocessing operations or can be corrected for these, which can reduce variability. For instance, RF signals are not influenced by the dynamic range setting and filtering operations that affect the appearance of B-mode images. Additionally, diagnostic techniques based on RF signals are potentially more suitable for devices that do not easily produce B-mode images (eg, emerging wearable ultrasound devices [31]). Although training the one-dimensional CNN algorithms takes a considerable amount of time, the trained algorithms can be run in real time to analyze new data.
Our study had several limitations. First, the ultrasound data were acquired from a single scanner platform by one physician. The cross-platform and cross-operator generalizability of the algorithms remains to be tested. Second, the RF data are not yet readily available on all commercial ultrasound systems. However, more manufacturers are starting to provide RF capabilities. Third, this study did not address whether deep learning algorithms using RF data could outperform those using B-mode images. Finally, despite our efforts to provide methodologic details, other investigators might still have difficulty reproducing this deep learning study. To facilitate reproducibility, we have made our code available for research use.
A possible direction for future studies is to optimize the regions of interest to include only signals in the liver. The fixed region of interest used in this study was easy to implement and required no human intervention but included signals in a variable and uncontrolled manner from structures outside the liver, which probably reduced the algorithm performance. Another direction could be to assess the role of our algorithms in providing a cost-effective solution for quantifying longitudinal changes of liver fat in response to treatment.
In conclusion, one-dimensional convolutional neural network algorithms can be developed and trained de novo to accurately identify nonalcoholic fatty liver disease and quantify hepatic fat fraction using raw radiofrequency ultrasound data in the appropriate clinical context. The algorithms are robust to changes in several machine settings, including transmit focal range and time gain compensation.
APPENDIX
SUPPLEMENTAL FIGURES
Current address: Department of Radiology, SUNY Upstate Medical University, Syracuse, NY.
Supported by the National Institutes of Health (R01DK106419).
Disclosures of Conflicts of Interest: A.H. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: institution received funding from Siemens Healthineers for the prior study. Other relationships: disclosed no relevant relationships. M.B. disclosed no relevant relationships. E.H. disclosed no relevant relationships. M.P.A. disclosed no relevant relationships. J.W.E. disclosed no relevant relationships. R.L. Activities related to the present article: received funding from NIEHS (5P42ES010337), NCATS (5UL1TR001442), NIDDK (R01DK106419, P30DK120515), and DOD PRCRP (CA170674P2); received an investigator-initiated study grant from Siemens. Activities not related to the present article: disclosed no relevant relationships. Other relationships: is a consultant or advisory board member for Arrowhead Pharmaceuticals, AstraZeneca, Bird Rock Bio, Boehringer Ingelheim, Bristol-Myer Squibb, Celgene, Cirius, CohBar, Conatus, Eli Lilly, Galmed, Gemphire, Gilead, Glympse bio, GNI, GRI Bio, Intercept, Ionis, Janssen Inc., Merck, Metacrine, NGM Biopharmaceuticals, Novartis, Novo Nordisk, Pfizer, Prometheus, Sanofi, Siemens, and Viking Therapeutics; institution has received grant support from Allergan, Boehringer-Ingelheim, Bristol-Myers Squibb, Cirius, Eli Lilly, Galectin Therapeutics, Galmed Pharmaceuticals, GE, Genfit, Gilead, Intercept, Grail, Janssen, Madrigal Pharmaceuticals, Merck, NGM Biopharmaceuticals, NuSirt, Pfizer, pH Pharma, Prometheus, and Siemens; is the co-founder of Liponexus. C.B.S. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: is on the board of the Society of Abdominal Radiology, AMRA, Guerbet, and Bristol Myers Squibb; is a consultant for GE Healthcare, Bayer, AMRA, Fulcrum Therapeutics, and IBM/Watson Health; institution received grants from Gilead, GE Healthcare, Siemens, GE MRI, Bayer, GE Digital, GE US, ACR Innovation, Philips, and Celgene; is a speaker for GE Healthcare and Bayer; institution receives royalties from Wolters Kluwer Health (UpToDate Publishing); developed educational presentations for Medscape and Resoundant; institution has lab service agreements with Enanta, ICON Medical Imaging, Gilead, Shire, Virtualscopics, Intercept, Synageva, Takeda, Genzyme, Janssen, NuSirt, Celgene-Parexel, and Organovo; has independent consulting contracts with Epigenomics and Blade Therapeutics; developed educational presentations or articles for Medscape. Other relationships: disclosed no relevant relationships. W.D.O. disclosed no relevant relationships.
Abbreviations:
- AUC
- area under receiver operating characteristic curve
- CI
- confidence interval
- CNN
- convolutional neural network
- NAFLD
- nonalcoholic fatty liver disease
- PDFF
- proton density fat fraction
- RF
- radiofrequency
- TGC
- time gain compensation
References
- 1. Loomba R, Sanyal AJ. . The global NAFLD epidemic . Nat Rev Gastroenterol Hepatol 2013. ; 10 ( 11 ): 686 – 690 . [DOI] [PubMed] [Google Scholar]
- 2. Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. . Mechanisms of NAFLD development and therapeutic strategies . Nat Med 2018. ; 24 ( 7 ): 908 – 922 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Machado MV, Cortez-Pinto H. . Non-invasive diagnosis of non-alcoholic fatty liver disease. A critical appraisal . J Hepatol 2013. ; 58 ( 5 ): 1007 – 1019 . [DOI] [PubMed] [Google Scholar]
- 4. Le TA, Chen J, Changchien C, et al . Effect of colesevelam on liver fat quantified by magnetic resonance in nonalcoholic steatohepatitis: a randomized controlled trial . Hepatology 2012. ; 56 ( 3 ): 922 – 932 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Noureddin M, Lam J, Peterson MR, et al . Utility of magnetic resonance imaging versus histology for quantifying changes in liver fat in nonalcoholic fatty liver disease trials . Hepatology 2013. ; 58 ( 6 ): 1930 – 1940 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Park CC, Nguyen P, Hernandez C, et al . Magnetic resonance elastography vs transient elastography in detection of fibrosis and noninvasive measurement of steatosis in patients with biopsy-proven nonalcoholic fatty liver disease . Gastroenterology 2017. ; 152 ( 3 ): 598 – 607.e2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reeder SB, Cruite I, Hamilton G, Sirlin CB. . Quantitative assessment of liver fat with magnetic resonance imaging and spectroscopy . J Magn Reson Imaging 2011. ; 34 ( 4 ): 729 – 749 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Artz NS, Hines CDG, Brunner ST, et al . Quantification of hepatic steatosis with dual-energy computed tomography: comparison with tissue reference standards and quantitative magnetic resonance imaging in the ob/ob mouse . Invest Radiol 2012. ; 47 ( 10 ): 603 – 610 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oelze ML, Mamou J. . Review of quantitative ultrasound: envelope statistics and backscatter coefficient imaging and contributions to diagnostic ultrasound . IEEE Trans Ultrason Ferroelectr Freq Control 2016. ; 63 ( 2 ): 336 – 351 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han A, Andre MP, Erdman JW, Jr, Loomba R, Sirlin CB, O’Brien WD., Jr . Repeatability and reproducibility of a clinically based QUS phantom study and methodologies . IEEE Trans Ultrason Ferroelectr Freq Control 2017. ; 64 ( 1 ): 218 – 231 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Han A, Andre MP, Deiranieh L, et al . Repeatability and reproducibility of the ultrasonic attenuation coefficient and backscatter coefficient measured in the right lobe of the liver in adults with known or suspected nonalcoholic fatty liver disease . J Ultrasound Med 2018. ; 37 ( 8 ): 1913 – 1927 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han A, Labyed Y, Sy EZ, et al . Inter-sonographer reproducibility of quantitative ultrasound outcomes and shear wave speed measured in the right lobe of the liver in adults with known or suspected non-alcoholic fatty liver disease . Eur Radiol 2018. ; 28 ( 12 ): 4992 – 5000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han A, Zhang YN, Boehringer AS, et al . Inter-platform reproducibility of ultrasonic attenuation and backscatter coefficients in assessing NAFLD . Eur Radiol 2019. ; 29 ( 9 ): 4699 – 4708 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yao LX, Zagzebski JA, Madsen EL. . Backscatter coefficient measurements using a reference phantom to extract depth-dependent instrumentation factors . Ultrason Imaging 1990. ; 12 ( 1 ): 58 – 70 . [DOI] [PubMed] [Google Scholar]
- 15. Andre MP, Han A, Heba E, et al . Accurate diagnosis of nonalcoholic fatty liver disease in human participants via quantitative ultrasound . 2014 IEEE International Ultrasonics Symposium, Chicago, September 3–6, 2014 . Piscataway, NJ: : IEEE; , 2014. ; 2375 – 2377 . [Google Scholar]
- 16. Lin SC, Heba E, Wolfson T, et al . Noninvasive diagnosis of nonalcoholic fatty liver disease and quantification of liver fat using a new quantitative ultrasound technique . Clin Gastroenterol Hepatol 2015. ; 13 ( 7 ): 1337 – 1345.e6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paige JS, Bernstein GS, Heba E, et al . A pilot comparative study of quantitative ultrasound, conventional ultrasound, and MRI for predicting histology-determined steatosis grade in adult nonalcoholic fatty liver disease . AJR Am J Roentgenol 2017. ; 208 ( 5 ): W168 – W177 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. LeCun Y, Bengio Y, Hinton G. . Deep learning . Nature 2015. ; 521 ( 7553 ): 436 – 444 . [DOI] [PubMed] [Google Scholar]
- 19.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542(7639):115–118 [Published correction appears in Nature 2017;546(7660):686.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Han S, Kang HK, Jeong JY, et al . A deep learning framework for supporting the classification of breast lesions in ultrasound images . Phys Med Biol 2017. ; 62 ( 19 ): 7714 – 7728 . [DOI] [PubMed] [Google Scholar]
- 21. Xu Y, Wang Y, Yuan J, Cheng Q, Wang X, Carson PL. . Medical breast ultrasound image segmentation by machine learning . Ultrasonics 2019. ; 91 : 1 – 9 . [DOI] [PubMed] [Google Scholar]
- 22. Yap MH, Pons G, Martí J, et al . Automated breast ultrasound lesions detection using convolutional neural networks . IEEE J Biomed Health Inform 2018. ; 22 ( 4 ): 1218 – 1226 . [DOI] [PubMed] [Google Scholar]
- 23. Byra M, Galperin M, Ojeda-Fournier H, et al . Breast mass classification in sonography with transfer learning using a deep convolutional neural network and color conversion . Med Phys 2019. ; 46 ( 2 ): 746 – 755 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Byra M, Styczynski G, Szmigielski C, et al . Transfer learning with deep convolutional neural network for liver steatosis assessment in ultrasound images . Int J CARS 2018. ; 13 ( 12 ): 1895 – 1903 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reddy DS, Bharath R, Rajalakshmi P. . A novel computer-aided diagnosis framework using deep learning for classification of fatty liver disease in ultrasound imaging . 2018 IEEE 20th International Conference on e-Health Networking, Applications and Services (Healthcom), Ostrava, Czech Republic, September 17–20, 2018 . Piscataway, NJ: : IEEE; , 2018. . [Google Scholar]
- 26. Reid JM, Wild JJ. . Ultrasonic ranging for cancer diagnosis . Electronics (Basel) 1952. ; 25 ( 5 ): 136 – 138 . [Google Scholar]
- 27. DeLong ER, DeLong DM, Clarke-Pearson DL. . Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach . Biometrics 1988. ; 44 ( 3 ): 837 – 845 . [PubMed] [Google Scholar]
- 28. Raunig DL, McShane LM, Pennello G, et al . Quantitative imaging biomarkers: a review of statistical methods for technical performance assessment . Stat Methods Med Res 2015. ; 24 ( 1 ): 27 – 67 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Myers RP, Pollett A, Kirsch R, et al . Controlled attenuation parameter (CAP): a noninvasive method for the detection of hepatic steatosis based on transient elastography . Liver Int 2012. ; 32 ( 6 ): 902 – 910 . [DOI] [PubMed] [Google Scholar]
- 30. Guthrie H, Castro N, Beysen C, Morrow L, Hompesch M. . Relationship between controlled attenuation parameter (CAP) and magnetic resonance imaging-derived proton density fat fraction (MRI-PDFF) in subjects at high risk for nonalcoholic fatty liver disease (NAFLD) , ProSciento, Inc. https://prosciento.com/wp-content/uploads/2019/02/NASHTAG2019-Poster_Hompesch_FINAL-CB12292018L.pdf. Accessed April 23, 2019 .
- 31. Wang C, Li X, Hu H, et al . Monitoring of the central blood pressure waveform via a conformal ultrasonic device . Nat Biomed Eng 2018. ; 2 ( 9 ): 687 – 695 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.