Abstract
Coronavirus infections have emerged as epidemic and pandemic threats in last two decades. After the H1N1 influenza pandemic in 2009, recently diagnosed novel betacoronavirus or severe acute respiratory syndrome coronavirus (SARS-CoV)-2 has spread across 203 countries and territories in all 5 major continents. World Health Organization (WHO) declared this as a public health emergency of international concern on January 30, 2020. Subsequently on February 11, 2020 a new name was given to this disease i.e. COVID-19 by an expert group from WHO. As of April 12, 2020, 10:00 CET, GMT+2:00, 1,696,588 confirmed cases and 105,952 confirmed deaths have been reported to the WHO. (Coronavirus disease 2019, situation report 83).
It possibly originated from a small animal market in Wuhan, China. A cluster of patients were admitted with unusual pneumonia not responding to treatment in various hospitals. Epidemiological, genomic analysis and correlation with other coronaviruses led to the isolation of new coronavirus, closely resembling the bat coronaviruses, from such patients in Wuhan. They were identified as the SARS-CoV-2. This virus infection presents as influenza like illness in the affected people. Fever, cough, respiratory distress with fatigue, diarrhea, nausea and vomiting are common symptoms seen in adults. This may progress on to severe respiratory distress, hypoxia, need for oxygen supplementation and ventilator support as seen in patients in the SARS-CoV-1 epidemic (2003) in Guangdong, China. The transmissibility of SARS-CoV-1 was less as compared to SARS-CoV-2 infection, and it was well controlled with good public health efforts. The present COVID-19 epidemic is still in the acceleration phase of 3 and 4 in various countries.
Without any effective antiviral agents available at present, the need of the hour is early case detection, isolation of cases, use of good preventive care measures by the household contacts and in the hospital set up. The results of ongoing clinical trials on hydroxychloroquine, azithromycin alone or in combination and a new antiviral agent remdesivir may help to treat some of the infections. A need for effective vaccine is being seen an as good preventive strategy in this pandemic. However the results of clinical trials and incorporation of vaccines in public health programs is a long way to go.
Keywords: Coronavirus, Betacoronavirus, COVID-19, SARS-CoV-1, SARS-CoV-2, Wuhan
1. Introduction
The world has seen the onset of a pandemic of a new infectious disease from December 2019. This has been formally named as the CoronaVirus Infectious Disease (COVID)-19 by a consensus group of WHO experts.1 Numerous clusters of patients started to surface in Wuhan, Hubei Province, China in mid December 2019. They presented with features of a viral respiratory illness with complaints of fever, cough, headache and breathlessness. Some of the patients had evidence of respiratory failure, shock, acute respiratory distress syndrome (ARDS) and sepsis.
A comprehensive evaluation for the etiological viral or bacterial organisms was conducted. Laboratory tests done were not diagnostic for various respiratory viruses, bacteria or fungal pathogens. A genome sequencing of the respiratory tract samples from these pneumonia patients ultimately led to the isolation of a new novel beta-coronavirus which was initially called “Wuhan virus”, novel coronavirus (nCoV-2019) and later severe acute respiratory syndrome (SARS)-2 coronavirus. Case reports of such patients were first published by various authors in Wuhan and Beijing.2 Subsequently other centers from Wuhan also reported the clinical, laboratory and epidemiological profile of such patients.
By January 7th, 2020, the nCoV-2019 had been isolated from the various specimens from admitted patients in Wuhan, and on January 10th, 2020 the viral genome data was shared by the virologists on various online databases for the viral genomes.
2. Epidemiology of COVID-19
Various centers for disease prevention and control all over the world like the Centers of Disease Control & Prevention (CDC, USA), WHO; Geneva, European Centre for Disease Prevention & Control (ECDPC) and Johns Hopkins University & Medicine; Baltimore have been providing daily update of the situation of the current pandemic from mid to late December onwards.
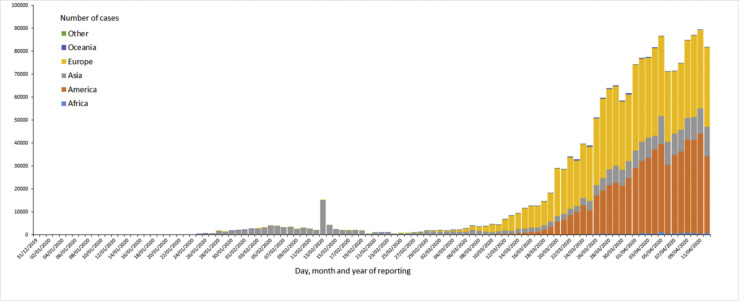
The ECDPC has a good data collection process, which is updated on daily basis between 6.00 and 10.00 Central European Time (CET). According to this centre there were 1,734,913 cases of the infection COVID-19 (in accordance to case definitions and testing strategies of various countries) as of April 12, 2020. About 108,192 deaths were reported for the same duration, since December 31, 2020, when this process of data collection was initiated. As on April 12, 2020, maximum numbers of cases have been reported in USA (529,951), Spain (161,852), Italy (152,271), followed by Germany (120,479), China (83,097), France (93,790), Iran (70,029), United Kingdom (78,991), Turkey (52,167), Canada (23,301) and Brazil (20,727) (Fig. 1 ).
Fig. 1.
The total no. of COVID-19 cases in the continents (China and other countries) as on April 12, 2020. Cases reported in accordance with the applied case definition and testing strategies in the affected countries. https://www.ecdc.europa.eu/geographical distribution-2019-ncov-cases.
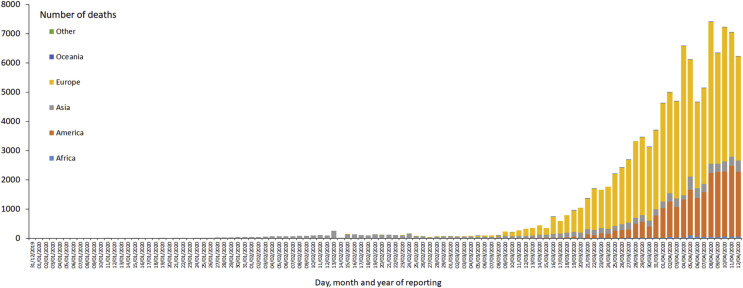
However the epidemic has led to more deaths in Italy (19,470), which surpassed the deaths which have occurred in China (3343). The deaths reported by other countries like USA (20,608), Spain (16,353), France (13,832), UK (9875), Iran (4357), Belgium (3342), Brazil (1124) and Turkey 1101 (Fig. 2).
Fig. 2.
Distribution of COVID-19 deaths in all continents as on 12 April 2020. Deaths reported in accordance with the applied case definition and testing strategies in the affected countries. https://www.ecdc.europa.eu/geographicaldistribution-2019-ncov-deaths.
Though India has reported 8844 cases (including 856 cases have been cured or discharged, one migrated case) and 308 deaths according to Ministry of Health & Family Welfare, Govt. of India as on April 13, 2020 (8.00 A.M. GMT +5.30). The death rate may rise exponentially if the community transmission of the virus is not halted by various public health measures and epidemiological surveillance. Low prevalence of the infection is possibly due to very few people (160,000 approx.) have been actually tested according to Indian Council of Medical Research (ICMR) as on April 08, 2020.
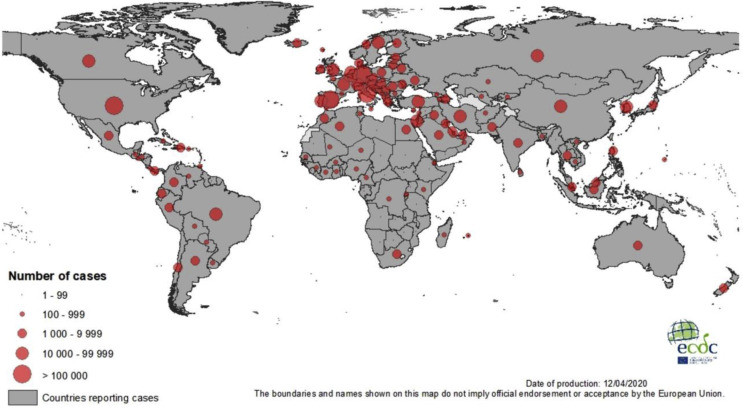
The number of cases substantially decreased in China as of first week of March 2020 to less than 500 per day, but they have exponentially increased in other countries like Italy, Iran, USA and Spain to 40,000 per day as in third week of March 2020 (ECDPC). The global number of cases was around 555 on Jan 22, 2020, which steadily increased to 12,000 cases on Feb 2, 2020. Within a span of one month the epidemic had quadrupled to 88,400 cases on Mar 1, 2020 and then it exponentially increased (doubled) to 167,500 cases on March 15, 2020. There was four-fold rise (4.67X) to 782,400 cases on March 30, 2020 in a span of only 15 days according to data from Coronavirus Resource Center, Johns Hopkins University (https://coronavirus.jhu.edu/map.html). This is similar to epidemiological data from other centers like ECDPC (Fig. 3 ).
Fig. 3.
Geographical distribution of COVID-19 cases world wide as of 12 April 2020. ECDC data.
Presuming that all the cases of COVID-19 have been reported and there is no sub-clinical transmission of the virus and there are no asymptomatic persons, which is unlikely, the crude fatality rate works out to be 5.87%, which is lower than seen in SARS epidemic of 2002–2003 (9.4%–15%) and MERS epidemic of 2012 (35.5%).
3. Coronaviruses
Coronaviruses (CoVs) are a group of enveloped positive-sense 5′-3′, single stranded RNA virus belonging to Coronaviridae family, present in various species of birds, snakes, bats and other mammals. In the zoonotic and avian population it primarily remains silent without any symptoms as they harbor the viruses. Avian species like birds and chicken may have respiratory tract infection or in cows and pigs may lead to enteritis. Humans however may be infected with various strains of previously known coronaviruses like 229E, OC43, NL63 and HKU1. They produce symptoms like rhinorrhea, mild cough (upper respiratory infection) or severe cough, tracheitis, bronchitis (lower respiratory tract infection).
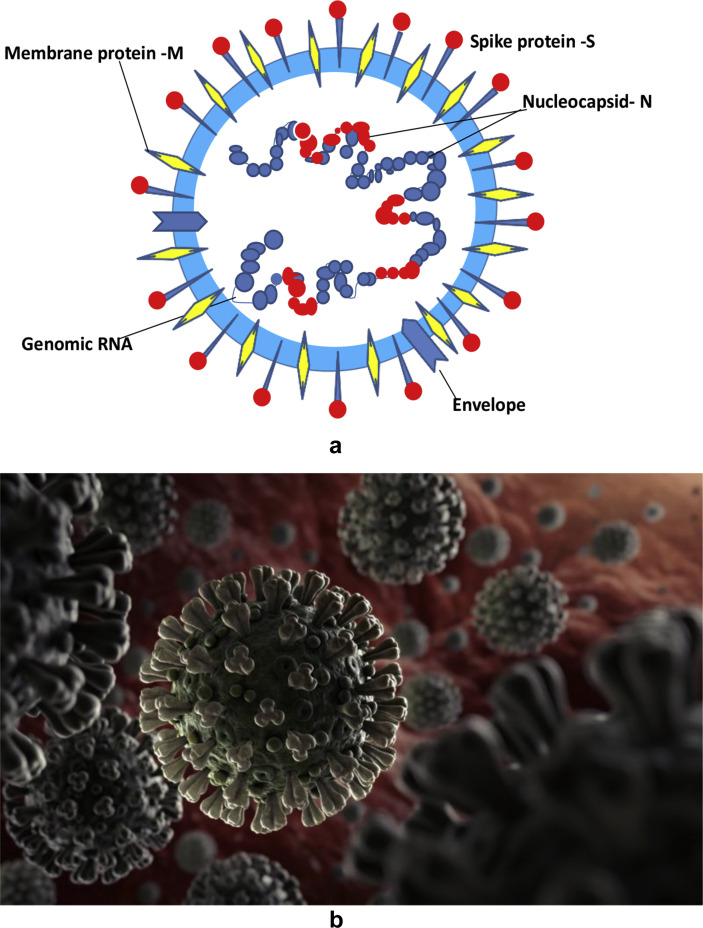
These viruses are 125 nm particles of spherical shape with club shaped spikes, the S protein bearing moiety which give the spiky appearance to the virions and resemble like the Sun's corona (crown) like pattern. This has been demonstrated with latest cryo-electron microscopy and tomographic techniques in various molecular biology research laboratories.3 These CoVs have been grouped as alpha, beta, gamma and delta based on the serological pattern. Recent genomic sequencing has classified them as various clades based on the phylogenetic analysis.
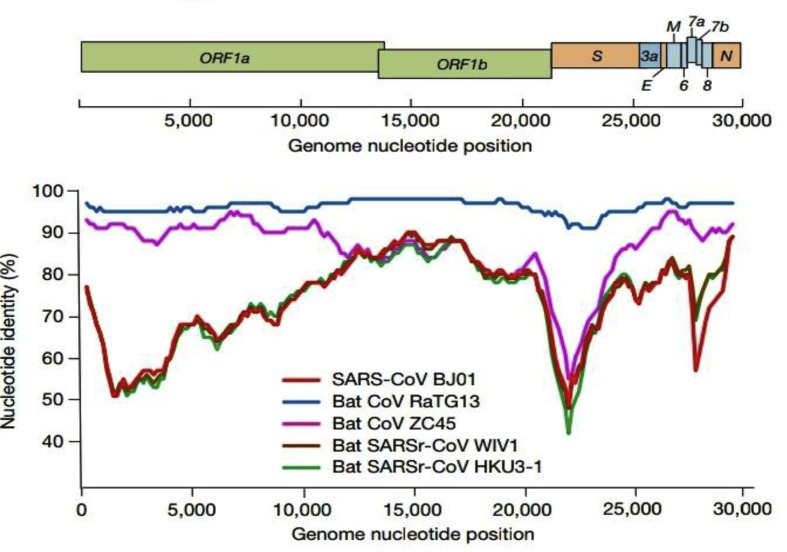
The viral genome is relatively large with approximately 30 (26–32) kb pairs.3 The virions have structural S-spike protein (outer spiky glycoprotein), M-membrane protein (a type III transmembrane glycoprotein), N-nucelocapsid protein (which is within the phospholipid bilayer) and non structural proteins, which are encoded by the various genetic loci on the RNA of the viruses. At the center of the virion lies a nucleocapsid composed of the genomic RNA and the nucleocapsid protein (Fig. 4, Fig. 5 ).
Fig. 4.
a) Diagrammatic expression of a coronavirus virion: Various structural proteins which form the coronavirus particle and the genome, single stranded RNA are shown. (Adapted from Li G, et al. Journal of Medical Virology, 25 January 2020). b) Artistic impression of coronavirus virions developed from an electron micrograph.
Fig. 5.
Various structural and non-structural genes encoding for proteins of the 2019-nCoV and the genome nucleotide position. ORF = open reading frames, S = structural protein, N = nucleocapsid. Modified from original Fig. 1 showing genome characterization of 2019-nCoV WIV04 and the similarity plot based on the full length genome sequence of the isolate with sequences of other coronaviruses like the SARS-CoVBJ01, batSARSr-CoV and 3 other bat coronaviruses (batSARSr-CoV W1V1, bat coronavirus RaTG13 & ZC45). Nature, Vol. 579,12 March 2020.
Coronaviruses of pathogenic potential in humans are various α human coronavirus (HCoV) 223E, β severe acute respiratory syndrome (SARS) coronavirus (CoV), middle-east respiratory syndrome (MERS) coronavirus and the recently discovered SARS CoV-2. The gamma coronavirus is the infectious bronchitis virus (IBV) is an avian pathogen producing respiratory tract infection, kidney and gut related issues in chicken. The virus is excreted in the feces of chicken and tracheal secretions. This may produce infections in humans especially among those working in the poultry industry.4
3.1. Severe acute respiratory syndrome coronavirus (SARS-CoV)
From November 2002 to March 2003, an epidemic of unusual cases of pneumonia and acute respiratory syndrome cases was first identified in Guangdong province of southeastern China. About 8096 people were affected in 26 countries, with 774 deaths by the end of May 2004, when the epidemic was contained in China. The mortality rate was approximately 9.56%. This epidemic was contained within a few months and later few laboratories related accidental exposures were reported in China and Taipei. A new coronavirus subsequently identified and named as SARS coronavirus (SARS-CoV) was the incriminating pathogen responsible for the cluster of influenza like illness and respiratory distress cases in China and other countries like Hong Kong, Vietnam (Hanoi), Singapore, Canada (Toronto) etc. However with good epidemiological surveillance, monitoring of cases and their isolation led to timely containment and it did not become a pandemic.5
Genetic analysis has shown that the new SARS-CoV (RNA virus) resembles other coronavirues in about 50–60% nucleotide sequences only. It also has a high susceptibility for mutations, may be viable and able to culture after 24 h from various surfaces.6
Though the exact reservoir of the SARS-CoV is not known, bats have been found to harbor these viruses and transmit them to the human host.7 It is a highly contagious disease. The symptoms of this disease are influenza like, patients may present with fever, malaise, myalgia, headache, diarrhea and rigors. Fever may not be seen in the elderly population and immunocompromized patients. Later on in second week cough may develop, which may progress to dyspnea or respiratory distress. Such individuals may develop severe acute respiratory distress and may need intensive care and ventilator support. The radiological features on chest X-rays (CXR) and CT scans may be normal in early phase or have focal infiltrates progressing to generalized patchy infiltrates or areas of consolidation.
The laboratory diagnosis for confirmation of this infection includes immunofluorescent assay (IFA), enzyme-linked immunosorbent assay (ELISA) detecting antibodies to the virus after 10 days of illness (from serum or whole blood) and the polymerase chain reaction test (PCR). However the gold standard of diagnosis is the isolation of the virus by various tissue culture media techniques.8 This demonstrates the presence of live viable viruses, which have ability to replicate in various cell culture media like HeLa cell lines. The tissue or cell culture and PCR technique can be performed on various specimens like the nasopharyngeal swabs, sputum, bronchoalveolar lavage, blood and stool. The primers for the SARS-CoV were developed with onset of disease in 2003 and are available on the WHO website at www.who.int/csr/sars/primers. Presently ready to use PCR kits are available for detections of this virus from various specimens.9
The management is supportive and there are currently no approved antiviral agents for this virus to modify the clinical course of this disease.
3.2. Middle-East respiratory syndrome coronavirus (MERS)-CoV
A cluster of cases of respiratory tract infection (fever, cough, pneumonia and respiratory distress) started to surface in few middle-east countries (Saudi Arabia, Oman, UAE) in 2012 and subsequently spread to 24 other countries like Malaysia and USA. These cases were identified due to a new coronavirus originating from the camels, their genetic analysis revealed some homology with the SARS-CoV.10
This is a single stranded positive sense-RNA virus belonging to beta coronavirus genus of Coronaviridae family. The MERS-CoV genomes are phylogenetically classified into two clades; clade A (EMC/2012 & Jordan-N3/2012) and clade B. It has also been researched that these viruses have selective tropism for certain cell receptors (dipeptidyl peptidase 4, DPP4 or CD26) present on non-epithelial bronchial cells and thus evade the innate immune response and antagonize the IFN production in these cells.11 Human to human transmission is less likely and occurs in close contacts and health care personnel.
Between September 2012 and June 30, 2018, 2229 laboratory confirmed cases of Middle-East respiratory syndrome coronavirus (MERS-CoV) were reported to WHO. Most of the cases (83%) were from the Kingdom of Saudi Arabia. In this duration 791 individuals died due to other co-morbid illnesses (crude fatality rate 35.5%). Individuals with risk factors like diabetes, renal failure and hypertension had a higher risk of severe disease including death.12
No vaccines (preventive) or therapeutic treatment is available for this infection and the management lies in supportive care as per patient's clinical condition. General hygiene measures like regular hand washing, avoiding close contact with camels (touching or kissing them) should be advised. The consumption of raw or undercooked animal products (meat and milk) carries a high risk of infection from a variety of organisms. Animal products (including camel milk and meat) that are processed appropriately by cooking or pasteurization are safe for consumption.13
3.3. Severe acute respiratory syndrome coronavirus (SARS-CoV)-2
This novel beta-coronavirus has emerged as a threat to the human lives in last few months. This is a single stranded positive sense RNA virus which is akin to the previously known SARS-CoV. It bears about 80% genome homology with SARS-CoV and has about 96% identical genes with the bat coronavirus (BatCoV RaTG13) (see Table 1 ).
Table 1.
Various clinical parameters seen in 191 enrolled patients. Adapted from Zhou F et al. Lancet. March 11, 2020. DOI: https://doi.org/10.1016/S0140-6736(20)30566-3.
| Total (n = 191) | Non-survivors (n = 54) | Survivors (n = 137) | P value | |
|---|---|---|---|---|
| Respiratory rate >24/min | 56 (29%) | 34 (63%) | 22 (16%) | <0.0001 |
| Pulse>125/min | 2 (1%) | 2 (4%) | 0 | 0.024 |
| Systolic BP < 90 mm Hg | 1 (1%) | 0 | 1(1%) | 0.53 |
| Fever (temp>37.3C) | 180 (94%) | 51 (94%) | 129 (94%) | 0.94 |
| Cough | 151 (79%) | 39 (72%) | 112 (82%) | 0.15 |
| Sputum production | 44 (23%) | 14 (26%) | 30 (22%) | 0.55 |
| Myalgia | 29 (15%) | 8 (15%) | 21 (15%) | 0.93 |
| Fatigue | 44 (23%) | 15 (28%) | 29 (21%) | 0.33 |
| Diarrhea | 9 (5%) | 2 (4%) | 7 (5%) | 0.67 |
| Nausea/vomiting | 7 (4%) | 3 (6%) | 4 (3%) | 0.40 |
Peng Zhou and others from the Wuhan Institute of Virology (CAS Key Laboratory of Special Pathogens) and other centers in Wuhan and Beijing reported the identification and characterization of a new coronavirus (nCoV-2019) which caused an epidemic of acute respiratory syndrome in humans in Wuhan, China. They analyzed the full length genome sequences from various samples from 5 patients, identified early in the epidemic. The genetic sequences of all isolates were identical and they shared 79.6% sequence identity to the SARS-CoV. It was also found that the isolate of the nCoV-2019, from the bronchoalveolar lavage (BAL) of one of the patients could be neutralized with the sera obtained from other several patients.14
A metagenomic RNA sequencing done on the first hospitalized patient from the BAL in mid December 2020 identified the novel coronavirus, with a viral genome of 29,903 nucleotides. This was named as WH human 1 coronavirus or 2019-nCoV by the authors.15
Other researchers had earlier found that nCoV-2019 utilizes a particular receptor (angiotensin converting enzyme II receptor; ACE2) to gain entry into the cells. The authors from the previous study also confirmed this finding. This was extrapolated from the molecular biological study from SARS-CoV experiments in a research laboratory in Hangzhou, China. They analyzed full length of the ACE2 receptor by a cryo-electron microscopic technique and the surface spike (S protein) of the SARS-CoV-2. They concluded that ACE2 is a dimeric assembly; this is docked on by the two S protein trimers and RBD of the S protein. This may facilitate the development of decoy ligands or neutralizing antibodies for the suppression of viral infection.16
In another remarkable report from Linlin Zhang and his colleagues (published recently in an online edition of Science on March 20, 2020) defined a particular drug target among coronaviruses. This viral protease (M,pro 3CLpro) is important in processing and translation of viral RNA. An X-ray structure of the non-ligand SARS-CoV-2 and it binding to an α-ketoamide inhibitor was studied. The researchers were able to develop this compound into a potent inhibitor of SARS-CoV-2 M.pro This inhibitor revealed tropism for lung tissue and could be administered by the inhalational route.17
3.3.1. Clinical trials for various therapeutic drugs in COVID-19
Various randomized clinical trials evaluating the efficacy of lopinavir/ritonavir (ChiCTR2000029308) and a new antiviral remdesivir (NCT04257656) in SARS-CoV-2 were initiated in China and USA. Lopinavir-ritonavir trial has been completed without any beneficial results. This was open-label, randomized control trial conducted on 199 sick patients with hypoxia (SaO2 < 94% & PaO2/FiO2 < 300 mm Hg) with laboratory confirmed SARS-CoV −2, who received the drug along with standard care for 14 days and other group received only the standard care. There was no difference in the time to improvement (hazard ratio 1.24: 95% confidence interval [CI], 0.90–1.72). There was no difference in mortality at 28 days of follow up.18
3.3.1.1. Clinical and radiological features of SARS-CoV-2
With the progression of the epidemic in China and USA, various papers were published online and in print with the collaborative efforts of various clinicians and researchers across the globe highlighting the transmission dynamics of the virus. Some authors studied the clinical course and risk factors for the mortality in adult patients with COVID-19 in Wuhan. Within a few months substantial information was available for the clinicians and researchers in open access articles.
3.4. Clinical course and risk factors
One of the early reported data about the epidemiological and clinical characteristics of patients with COVID-19 were published in February 2019. Zhou F et al. reported various clinical features and outcome in adult critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China.19 This retrospective study enrolled 191 confirmed patients suffering from the disease from a cohort of 813 patients admitted in two different hospitals in Wuhan, 54 patients died in the hospital and 137 were discharged. The median age of patients was 56 years (IQR 46–67), their age ranging from 18 to 87 years. Preponderance of males was seen, 119 (62%) males versus 72 (38%) females. The survival rate was significant in the age group of 45–58 years (mean 52 years) versus 63–76 years (mean 69 years).
Most of the patients had fever, seen in 51 (98%) out of 52 patients (35 males). Mean age was 59.7 years (SD ± 13.3). Various risk factors for death were older age >65 years, other co-morbidities like hypertension, diabetes and coronary artery disease. The death rate seems to higher with these underlying conditions as seen in the non survivor group (Table 2).
Table 2.
Various risk factors in 191 enrolled patients. Adapted from Zhou F et al. Lancet. March 11, 2020. DOI: https://doi.org/10.1016/S0140-6736(20)30566-3.
| Total (n = 191) | Non survivors (n = 54) | Survivors (n = 137) | P value | |
|---|---|---|---|---|
| Exposure history | 73 (38%) | 14 (26%) | 59 (43%) | 0.028 |
| Current smoker | 11 (6%) | 5 (9%) | 6 (4%) | 0.21 |
| Comorbidity | 91 (48%) | 36 (67%) | 55 (40%) | 0.001 |
| Hypertension | 58 (30%) | 26 (48%) | 32 (23%) | 0.0008 |
| Diabetes | 36 (19%) | 17 (31%) | 19 (14%) | 0.0051 |
| Coronary art. disease | 15 (8%) | 13 (24%) | 2 (1%) | <0.0001 |
| Chronic lung disease | 6 (3%) | 4 (7%) | 2 (1%) | 0.047 |
| Carcinoma | 2 (1%) | 0 | 2 (1%) | 0.37 |
| Chronic kidney disease | 2 (1%) | 2 (4%) | 0 | 0.024 |
| Others | 22 (12%) | 11 (20%) | 11 (8%) | 0.016 |
In addition to these clinical findings, the authors observed that various laboratory parameters like d-dimer greater than 1 μg/mL could help the clinicians to identify the patients who are likely to have poor prognosis. Poor outcome could also be predicted with high sequential organ failure assessment (SOFA) score (5.65, CI 2.61–12.23, p < 0.0001) in this study. The SOFA score is a marker for prognosis in patients with sepsis and shock and reflects the severity of multi-organ dysfunction.
Various laboratory parameters like elevated interleukin (IL)-6, LDH, troponin I and lymphopenia were seen in patients with severe form of COVID-19 disease. Prolonged PT and APTT were observed in severe illness. These patients were more likely to need intensive care.
The outcome of these patients is like other critical care illness like dengue shock syndrome, who are admitted in the intensive care units may develop sepsis, respiratory failure, ARDS, heart failure, septic shock etc.
The disease severity of COVID-19 varied from a milder disease seen in 38% of the patients (who could be treated at home in isolation) to severe (35%) and critical category (28%). These patients needed to be admitted in the hospital for inpatient and critical care respectively.
The mean duration of illness from the onset of symptoms to hospital admission was 11 days (8–14 t). However the average intensive care unit duration of admission was 8 days (4–12 days) in the survivors.
3.5. Epidemiological factors for early transmission
After isolation of the virus in first week of January 2020 and as the number of cases started to rise exponentially in Wuhan and surrounding areas of Hubei, researchers looked at the dynamics of spread of infection in the community. Li Q and his colleagues analyzed the data of the first 425 confirmed cases, which had been reported till January 22, 2020.20 All of these were cases of human to human transmission occurring in close household and other contacts. The mean incubation period was 5.2 days (95% CI, 4.1–7.0). Maximum incubation period was 12.5 days. In the month of late December to last week of January 2020, the epidemic doubled in size every week (7.5 days, 95% CI, 5.3–19). One individual had the ability to transmit the infection to 2.2 persons (95% CI, 1.4–3.9).
The clinical characteristics of COVID-19 have been extensively published by various researchers in China and other countries like USA. An extensive retrospective data of 1099 patients from 552 hospitals in mainland China from the onset of epidemic till January 29, 2020 deals with the demographic and clinical features of the patients from Wuhan and other non residents.21 Only 1.9% of the patients had a direct contact with wildlife and seafood market in Wuhan, 72.3% had some contact with the residents of Wuhan. About 31.3% of the patients had visited the city. This epidemiological tracing is important for tracing the index case, which had been miles away, but still the virus was transmissible through various modes of air, road and rail traffic by person to person or fomite transmission.
The most common symptoms of illness were fever seen in 43.8% of the cases on admission. However during the course of hospital stay, 88.7% of the patients developed fever. Cough was the second commonest symptom (67.8%). Diarrhea was relatively uncommon (3.8%). Some of the salient clinical characteristics have been illustrated in Table 3 .
Table 3.
Clinical features of the study patients and according to disease severity. IQR = inter quartile range. Numerals in parenthesis represent percentages or IQR. Table modified from Table 1, illustrated in original article. Guan W et al. N Engl J Med. February 28, 2020. https://doi.org/10.1056/NEJMoa2002032.
| Characteristic | All patients | Non-severe disease | Severe disease |
|---|---|---|---|
| Age - median (IQR) years |
47 (35–58) |
45 (54–57) |
52 (40–65) |
|
Distribution |
no./total no. (%) |
||
| 0–14 yr | 9/1011 (0.9) | 8/848 (0.9) | 1/163 (0.6) |
| 15–49 yr | 557/1011 (55.1) | 490/848 (57.8) | 67/163 (41.1) |
| 50–64 yr | 292/1011 (28.9) | 241/848 (28.4) | 51/163 (31.3) |
| >65 yr | 153/1011 (15.1) | 109/848 (12.9) | 44/163 (27) |
| Females |
459/1096 (41.9) |
386/923 (41.8) |
73/173 (42.2) |
| Median incubation period days (IQR) |
4 (2–7) |
4 (2.8–7.0) |
4 (2.0–7.0) |
| Patients with fever | 473/1081 (43.8) | 391/910 (43) | 82/171 (48) |
| Symptom No. (%) | |||
| Conjunctiva cong. | 9 (0.8) | 5 (0.5) | 4 (2.3) |
| Nasal congestion | 53 (4.8) | 47 (5.1) | 6 (3.5) |
| Headache | 150 (13.6) | 124 (13.4) | 26 (15) |
| Cough | 745 (67.8) | 623 (67.3) | 122 (70.5) |
| Sore throat | 153 (13.9) | 130 (14) | 23 (13.3) |
| Sputum product | 370 (33.7) | 309 (33.4) | 61 (31.3) |
| Fatigue | 419 (38.1) | 350 (37.8) | 69 (39.9) |
| Hemoptysis | 10 (0.9) | 6 (0.6) | 4 (2.3) |
| Breathlessness | 205 (18.7) | 140 (15.1) | 65 (37.6) |
| Nausea/vomiting | 55 (5) | 43 (4.6) | 12 (6.9) |
| Diarrhea | 42 (3.8) | 32 (3.5) | 10 (5.8) |
| Myalgias/arthralgia | 164 (14.9) | 134 (14.5) | 30 (17.3) |
| Chills | 126 (11.5) | 100 (10.8) | 26 (15) |
| Signs of infection | |||
| Throat congestion | 19 (1.7) | 17 (1.8) | 2 (1.2) |
| Tonsils enlarged | 23 (2.1) | 17 (1.8) | 6 (3.5) |
| Lymphadenopathy | 2 (0.2) | 1 (0.1) | 1 (0.6) |
| Rash | 2 (0.2) | 0 | 2 (1.2) |
| Coexisting disorder | 261 (23.7) | 194 (21) | 67 (38.7) |
Chest X Rays and CT chest were essentially normal in 17.9% of the patients. About 162 (59.1%) of the patients had abnormal CXR. However CT chest revealed abnormalities in 840 (86.2%) of the total number (975), ground glass type of appearance was seen in 56.4% of the 975 patients screened. Other key findings were local patchy shadows (41.9%), bilateral patchy shadows (51.8%) and interstitial infiltrates in (14.7%) of the patients screened. The commonest hematological finding was lymphocytopenia (83.2%) of the patients on admission.
3.6. SARS-CoV-2 infection in children
Children under 18 years of age form a substantial group (29.3%) of the total world population of 7.75 billion people. However the epidemiological data from the initial studies is lacking. Chinese CDC estimates reveal that less than 1% of the children younger than 10 years of age were affected by the disease. In the study by Li and his colleagues regarding early transmission dynamics of CoV in Wuhan, it was quiet evident that there were no children less than 15 years of age.19 It may be possible that children had mild asymptomatic infection and were not detected, or they were less likely to be infected.
A report by Xiaoxia Lu and his colleagues at Wuhan Children Hospital, Wuhan and other centers in Huazhong, Hong Kong, Beijing and California describe varied spectrum of illness seen in children infected with SARS-CoV-2.22 It was seen that children have a less serious form of disease and asymptomatic infections are not uncommon. This may of public health importance as they may spread the infection to elderly population with or without underlying co-morbidity.
A review of 72,314 cases from the hospital database was analyzed, 1391 children were assessed and tested over a span of one month (January 28 to February 26, 2020). A total of 171(12.3%) of the children were infected. The median age of infected children with SARS-CoV-2 was 6.7 years. Fever was the most prominent symptom (41.5%) at any time during the illness. Cough and pharyngeal congestion was seen in 48.5% and 46.2% of the infected children respectively. Other manifestations were diarrhea (8.8%), fatigue (7.6%), rhinorrhea (7.6%), vomiting (6.4%), nasal congestion (5.3%), tachypnea (28.7%) and tachycardia (42.1). Abnormal CECT chest findings (ground glass opacities, local patchy infiltrates, bilateral infiltrates & interstitial abnormalities) were seen in 63% of the total CTs done. Very few children (2.3%) had evidence of hypoxia i.e. (SaO2<92%).
In a correspondence to the NEJM (March 12, 2020), Liu and his colleagues extracted data of six children < 15 years of age from 366 children admitted with respiratory tract infection in 3 Tongji hospitals around Wuhan in a span of one week. Among the viral pathogens isolated, SARS-CoV-2 was detected in 6 (1.6%) isolates only. The duration of fever varied from 3 to 11 days, only one child needed intensive care unit admission.24
The relatively asymptomatic nature of this infection in children has been studied in another small retrospective observational study in 36 children (age group 0–16 years) in Zhejiang. China. All the children were RT-PCR positive for SARS-CoV-2. The primary route of transmission was history of close contact and exposure in affected areas (89%). 19 (53%) had moderate clinical type with evidence of pneumonia, fever was seen in 13 (36%) children. A comparative prevalence of clinical features with adult patients has been evaluated in this study. Fever, cough, sore throat, dypnea and pneumonia seem to be less common in pediatric patients.25 The prevalence of pneumonia in COVID-19 children was 53%, similar to pneumonia seen in adults (SARS-1), higher than as seen with H1N1 influenza patients (15%).
3.7. Aerosol and surface transmission of coronaviruses, SARS-CoV-1 and SARS-CoV-2 and their viability
One of the important questions asked is about kinetics of transmission of new coronaviruses. How this infection spreads in a community in close contact scenarios (households, work places, public places like closed air conditioned restaurants, metros, buses etc.)? The likely spread by fomites, touching contaminated objects and fine steel surfaces is also of interest to the clinicians, researchers and general public alike.
Respiratory infections can be transmitted thorough respiratory droplets (5–10 μm in diameter) or via respiratory nuclei (<5 μm) in diameter. The transmission of infection from person to person in COVID-19 occurs through respiratory droplet and surface contact routes. The transmission of infection by air-borne route has not been reported as per data of 75,465 cases of the infection in China.26 , 27
Doremalen, Bushmaker and their colleagues conducted in-vitro experiments to study the viability of the virions (SARS)-CoV and SARS-CoV-2 in aerosols, plastic, stainless steel, copper and cardboard.28 In this experiment aerosols were generated by using a three-jet Collison nebulizer and fed in Goldberg drum in a laboratory. It was observed that the SARS-CoV−2 was detected on these surfaces even after 72 h after initial application over these surfaces. The viral titers decreased after 24 h. On plastic surfaces the viruses were viable for 72 h, however the viral load decreased exponentially from 103.7 to 100.6 TCID50. In case of stainless surfaces the viral load decreased from 103.6 to 100.6 after 48 h. However on copper surfaces no viable SARS-CoV-2 were detected after 4 h and no viable SARS-CoV-1 were measured after 8 h. On cardboard surfaces no viable SARS-CoV-1 and CoV-2 after 24 and 8 h respectively. In aerosol generating experiments the researchers found the viability of SARS-CoV-1 and CoV-2 in 3 h, though the titers decreased from 104.3 to 103.5 TCID50 and 103.5 to 102.5 TCID50 per liter of air respectively.
However there is lot to speculate and translate this experimental setting in actual transmissibility of infection in the human host and what is the minimum infective dose of the virus to cause disease symptoms after a period of latency. It is important to note if intact virus particles can be detected in the air of the patient's rooms or in aerosol generating medical procedures. The host's initial mucosal interaction with the virus will also determine the development of infection.
4. Laboratory testing for the COVID-19
The confirmation of clinical diagnosis for COVID-19 can be done by certain laboratory tests. WHO released its interim guidelines for laboratory biosafety and prioritized laboratory testing strategy on 19 and 22 March 2020.29 In the first week of March 2020, about 16 selected labs all over the world had been designated as WHO reference laboratories for confirmatory testing for COVID-19.
In India, National Institute of Virology (NIV) ICMR, Pune is the apex laboratory for testing facility. At present 123 Govt. laboratories and few private labs have been accredited for the testing procedure as on March 31, 2020.
4.1. Real-Time Reverse transcriptase (RT)-Polymerase chain reaction (PCR)
Various biological samples like nasopharyngeal swabs/washes/aspirates, sputum, BAL etc. can be tested for the presence of virus. Initially in house PCR technique was used by various research laboratories using the primers developed after analyzing the genome of this novel coronavirus. Gene targets in N gene, ORF1ab, nucleoprotein, RdRP, spike protein have been used for detection of the virus.30
CDC has developed a test kit for testing of patient specimens for SARS-CoV-2. The test kit is called CDC 2019-nCoV Real-Time Reverse transcriptase (RT)-PCR Diagnostic panel. It is designed to be used with certain systems of PCR instruments with specific software. The test is intended to be used with upper and lower respiratory specimens collected from persons who meet the CDC criteria for COVID-19 testing.31
Presently various commercial kits for detection of the virus have been validated. They employ these targets for detection of the virus by real time reverse transcription polymerase chain reaction (RT-PCR). They detect the nucleic acids of the SARS-CoV-2 in various specimens as specified earlier. The tests are run with positive and negative controls.
4.2. Rapid diagnostic tests
In 2nd or 3rd week of illness, various antibodies have been detected in the convalescent serum of the donors. Serological assays to detection of SARS-CoV-2 are in the process of development. They need to undergo clinical trials and the regulatory review process. These assays will help to understand the epidemiology of the disease and detection of asymptomatic infection in a given population.32
Rapid diagnostic test kits have been recently developed in 4–6 weeks time. They can detect the SARS-CoV-2 in few hours and can be used as screening tests in certain hot-spot areas of the epidemic. Various commercial and research labs like Bosch and Abbott have launched the rapid diagnostic tests. These molecular cartridge based assays have an accuracy of 95% for detection of infection (nasal/throat swabs) and meet various quality control standards as per WHO. They can be used at various points of patient care and no transportation of samples is required. However the test results by these rapid diagnostic kits need to be confirmed by RT-PCR for SARS-CoV-2.
ICMR-NIV had recently approved 20 kits developed by various labs from across the world. About 12 rapid diagnostic kits have undergone strict validation by ICMR. They have been approved as on March 27, 2020. However all of these kits are not US-FDA approved. https://www.icmr.nic.in/contents/covid-19.
5. Therapeutic drugs for the COVID-19
There are no approved drugs for treatment or prophylaxis for this infection. The key to management is isolation of the patient, hand hygiene measures, use of face masks and other sanitary measures for prevention of respiratory aerosol/droplet infection generated by the patients. This will reduce the risk of transmission to care givers and close contacts. A clinical trial for the efficacy of a new antiviral drug, remdesivir (Gileads Sciences Inc.) has been initiated in hospitalized adults in Nebraska, USA (NCT04257656). This investigational drug has earlier been tried in animal models infected with Ebola virus, SARS and MERS virus.23 This drug is an inhibitor of RNA polymerase which are essential for viral replication in the host cells. Remdesivir reduced the severity of disease, virus replication and damage to the lungs when administered as pre-exposure prophylaxis and as therapeutic treatment in rhesus macaques.
There has been lot of interest generated in use of various antimicrobial/immunomodulatory drugs like chloroquine and hydroxychloroquine (HCQ) in SARS or SARS-2 infected patients. A recent controlled clinical study conducted by Raoult D et al. in France revealed that 100% of the patients who received a combination of HCQ plus azithromycin tested negative after 6 days of treatment (unpublished data).
HCQ had shown higher in-vitro activity against SARS-CoV-2 and has been used in admitted COVID-19 patients on uncontrolled basis in many countries including USA. A small non-randomized, open label study of the use HCQ with or without azithromycin has reported reduced detection of SARS-CoV-2 on repeat samples in the test group of patients as compared to control group.33
An open-data clinical trial to evaluate the efficacy of HCQ for prevention of COVID-19 infection in a cohort of healthy medical professionals and healthcare workers is ongoing in France. Their presumption for the use of HCQ is based on the in-vitro experiments showing the efficacy of HCQ in preventing the novel coronavirus infection in various primate cells.34 , 35 Other countries affected by this pandemic like South Korea and China have drafted guidelines for the use of HCQ and chloroquine for therapeutic treatment of COVID-19.
Due to lack of an effective alternative drugs for treating sick patients in intensive care set up, especially with co-morbidities like diabetes, hypertension, coronary artery disease and immunocompromized status, various drug regulatory authorities have allowed the use of HCQ. However they are to be used as ‘off the label indications’ on compassionate grounds. Health care professionals taking care of sick patients may also receive HCQ with or without azithromycin on case to case basis for prophylaxis. There are no clinical trials available to define the exact dosing schedule for HCQ alone or in combination with azithromycin for treatment in COVID-19 patients. Both drugs may lead to prolonged QT interval. ECG should be done (baseline, on treatment) and used with caution in patients with chronic medical conditions or who are on other medications to prevent occurrence of arrhythmias.36
6. Vaccines for coronaviruses
The epidemics of SARS (2003), MERS (2012) and the recent SARS-2 (2019) has generated a lot of interest in the preventive strategies like the drug chemoprophylaxis for health care individuals and close contacts of index patients of the COVID-19. The role of drugs has been discussed in a separate section.
After the SARS epidemic in 2003, various researchers all over world got in the effort of developing a candidate vaccine for the same. There are about 43 vaccines that have been developed. However most of them are in the pre-clinical phase. They are yet to undergo phase 1 randomized trials in humans. These vaccines are DNA, inactivated, live attenuated vaccines (LAV), viral vector based (non-replicating) protein sub-unit vaccines and other 6 unknown vaccine types.37
One of the RNA based vaccines (LNP-encapsulated mRNA) has been made available for the phase 1 clinical trial in humans. This vaccine has been developed by collaborative effort of scientists at NIAID/NIH and a biotechnology company (Moderna Therapeutics) in USA. The clinical trial began in mid March 2020 and has already enrolled few patients in Seattle, USA (NCT04283461). It will study the efficacy and adverse reactions of the candidate vaccine in 45 adult volunteers from 18 to 55 years of age.
Another vaccine that has gone in phase 1 clinical trial is a non-replicating viral vector vaccine (ChiCTR2000030906). It has been bioengineered by using an adenovirus vector (type 5) at Beijing Institute of Biotechnology and CanSino Biological Inc. It uses the same platform as was used for developing a vaccine for Ebola virus. However this is just the beginning and it will take 12–18 months for the trial to complete and have the data to interpret for antibody response and long term efficacy.
7. Host Immune responses
An understanding of the various host cell immune responses evoked by the coronaviruses may help us to develop effective drugs and vaccines for this infection. Various researchers have studied the T cell immune response to SARS-CoV in the past. Recently with the evolution of the SARS-CoV-2 or COVID-19 epidemic in China a resurgence of interest in the immune mechanisms has been generated.
An innate immune response evoked by the macrophage activation leads to a T cell mediated response. The macrophages present CoV antigens to the T cell subsets (Th 17) which lead to a massive release of various cytokines like IL-1, IL-6, IL-8, IL-21, TNF-b and MCP-1. This is responsible for the immune amplification. These cytokines and chemokines are responsible for recruitment of lymphocytes and other leukocytes to the site of inflammation. This immune amplification is partly responsible for the tissue damage in the respiratory alveoli, bronchioles, pulmonary interstitial walls etc. The increased expression of the inflammatory mediators has a down regulatory effect on NK cells and CD8 cells, which are important for the lymphocytes to clear the virus. Binding of the S-protein to various host cell receptors like ACE2 and DPP4R enables the viral RNA to gain entry in the host cell cytoplasm. Various toll like receptors (TLR) like TLR-3, TLR-4 are important to either evade the immune response or recognition of S-protein. The S-protein recognition leads to activation of pro-inflammatory cytokines.38
8. Conclusion
Coronavirus infections have lead to few epidemics and a new pandemic in last 2 decades. The infections vary in clinical manifestations from self limiting viral respiratory tract infections or gastroenteritis to severe form like the SARS-CoV-1, MERS and the recent SARS-CoV-2 infections. These have led to a significant morbidity and mortality and a global economic crisis. Newer developments in therapeutics, preventive therapy in the form of chemoprophylaxis and vaccines are underway. Newer information about the molecular mechanisms, clinical manifestations, epidemiological pattern and preventive public measures is available each week in all the scientific or medical journals. In addition lot of input is being provided by electronic and print media for public awareness. Only few articles published in the initial phase of COVID-19 epidemic have been cited to maintain brevity of the article. It will not be appropriate if a little note is not made about the unsung heroes of this pandemic. The world’s ‘new heroes’ are the medical workers. The doctors, nurses, paramedical staff and other health care workers who are directly or indirectly involved with patient care in isolation wards and critical care areas. They risk their own lives, knowing that there are no effective drugs or vaccines available at present. They are on the frontline of a new kind of war according to Stephen Collinson, CNN new analyst.39 At least 23 physicians treating such patients have died in Italy. A number of doctors have died in France and China, including Dr. Li Wenliang, who was the first to alert the world. Health care professionals face lot of psychological stress as many patients die in this type of scenario. Prioritizing scarce health care resources is a challenge for them. Lack of masks, gloves, scrub-suits, equipment like ventilators, ECMO machines, dialysis machines etc. is a problem in developed and developing countries alike. However, they still head on to confront this new war of emerging infections like Ebola virus and SARS-CoV 1 and 2.
Declaration of Competing Interest
None.
Acknowledgements
This article wouldn't have been possible without the technical assistance from various persons. My special thanks to P.L. Ogra MD, Emeritus Professor of Pediatrics and Microbiology, Children's Hospital of Buffalo and UTMB, Galveston, USA. Mr Amit Jain, Journal Co-ordinator, CMRP for providing editorial manager assistance. Mr Gaurav Gund and Ms Manju Sayal, Department of Pediatrics for providing secretarial assistance.
References
- 1.Naming the Coronavirus Disease (COVID-19) and the Virus that Causes it. WHO; 11 February 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it [Google Scholar]
- 2.Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ijnjatovic J., Sapats S. Avian infectious bronchitis virus. Rev Sci Tech. 2000;19:493–508. doi: 10.20506/rst.19.2.1228. [DOI] [PubMed] [Google Scholar]
- 5.Severe acute respiratory syndrome. International Travel and Health. WHO. https://www.who.int/ith/diseases/sars/en/ (Accessed on 22 March 2020.)
- 6.Drosten C., Gunther S., Preiser W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 7.Li W., Shi Z., Yu M. Bats are natural reservoirs of SARS-like coronavirues. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 8.Cooke F.J., Shapiro Global outbreak of severe acute respiratory syndrome. Int J Infect Dis. 2003;7:80–85. doi: 10.1016/S1201-9712(03)90001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emery S.L., Erdman D.D., Bowen M.D. Real-time reverse transcription-PCR assay for SARS-associated coronavirus. Emerg Infect Dis. 2004;10:311–316. doi: 10.3201/eid1002.030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu K.W., Poon L.M., GomaaMM MERS in coronavirus dromedary camels, Egypt. Emerg Infect Dis. 2014;20:1049–1053. doi: 10.3201/eid2006.140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raj V.S., Mou H., Smits S.L. Dipeptidyl dipeptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Global Summary. WHO; Geneva, Switzerland: 2018 August. WHO MERS Global Summary and Assessment of Risk.https://who.int/publications-detail/who-mers-global-summary-and-assessment-of-risk---august-2018 [Google Scholar]
- 13.Middle East respiratory syndrome coronavirus (MERS-CoV) Key Facts. 11 March 2019 https://www.who.int/news-room/fact-sheets/detail/middle-eastcoronavirus WHO. [Google Scholar]
- 14.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu F., Zhao S., Yu B. Complete genome characterization of a novel coronavirus associated with severe human respiratory disease in Wuhan, China. BioRxiv. 2020 doi: 10.1101/2020.01.24.919183. [DOI] [Google Scholar]
- 16.Yan R., Zhang Y., Li Y., Lu Xia, Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full length human ACE2. Science. 2020;10 doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L., Lin D., Sun X. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science. 20 Mar 2020 doi: 10.1126/SCIENCE.abb3405. eabb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao B., Wang Y., Wen D. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. March 18, 2020 doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou F., Yu T., Du R. Clinical course and risk factors for mortality of adult in patients with COVID-19 in Wuhan, China: a retrospective study. Lancet. March 11, 2020 doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q., Med M., Guan X. Early transmission dynamics in Wuhan, China, of a novel coronavirus-infected pneumonia. N Engl J Med. January 29, 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan W., Ni Z., Hu Y. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. February 28, 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu X., Zhang J., Qu J. SARS-CoV-2 infection in children. N Engl J Med. March 18, 2020 doi: 10.1056/NEJMc2005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wit E, Feldmann F, Cronin J. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. PNAS. 2020;117(12):6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu W., Zhang Q., Chen J. Detection of COVID-19 in children in early January 2020 in Wuhan, China. N Engl J Med. March 12, 2020 doi: 10.1056/NEJMc2003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Health Organization Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19) 16-24. Feb 2020. https://www.who.int/docs/default-source/coronaviruses/who-china-joint mission on covid-19-final report.pdf Geneva. Available from:
- 26.Ong S.W., Tan Y.K., Chia P.Y. Air, surface, environmental and personal protective contamination by SARS-CoV-2 from asymptomatic patient. JAMA. Mar 4 2020 doi: 10.1001/jama.2020.3227. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qui H., Wu J., Hong L., Luo Y., Song Q., Chen D. Clinical and epidemiological features of 36 children with coronavirus disease (COVID-19) in Zhejang, China, an observational cohort study. Lancet. 2020 doi: 10.1016/S1473-3099(20)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorenmalen N., Bushmaker T., Morris D.H. Aerosol and surface stability of SARS-CoV-2 as compared to SARS-CoV-1. N Engl Med. March 17, 2020 doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Use of laboratory methods for SARS diagnosis. https://www.who.int/csr/sars/labmethods/en/ Accessed at:
- 30.COVID-19 testing-Wikipedia. https://en.m.wikepedia.org/wiki/COVID-19_testing Accessed at:
- 31.CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic Panel. March 30, 2020. https://www.fda.gov/media/134922/download Accessed at: [DOI] [PMC free article] [PubMed]
- 32.Loeffelholz M.J., Tang Y. Laboratory diagnosis of emerging human coronavirus infections – the state of the art. Emerg Microb Infect. 2020;9:747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gautret P., Lagier J.C., Parola P. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020 Mar 20:105949. doi: 10.1016/j.ijantimicag.2020.105949. [Epub ahead of print] PubMed PMID: 32205204; PubMed Central PMCID: PMC7102549. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Liu J., Cao R., Wang M. Hydroxychloroquine is less toxic derivative of chloroquine is effective in inhibiting SARS-CoV-2 infection in vito. Cell Discovery. 2020;6 doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An Open Data clinical Trial for COVID-19 Prevention. https://www.covidtrial.io/
- 36.Therapeutic Options for Patients with COVID-19. Centers for Disease Control and Prevention; 2019. https://www.cdc.gov/coronavirus/2019-ncov/hcp/therapeutic-options.html [Google Scholar]
- 37.World Health Organization; 20 March 2020. Draft Landscape of COVID-19 Candidate Vaccines.https://www.who.in [Google Scholar]
- 38.Li G., Fan Y., Lai Y. Coronavirus infections and immune responses. J Med Virol. 2020;92:424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collinson S. From cnn.com; Tue March 24, 2020. The world's new heroes are medical workers. Analysis by Stephen Collinson, CNN. Updated 4.55 PM EDT. [Google Scholar]