Abstract
The Ehrlich pathway is a major route for the renewable production of higher alcohols. However, the product scope of the Ehrlich pathway is restricted, and the product selectivity is suboptimal. Here, we demonstrate that a Coenzyme A (CoA) detour, which involves conversion of the 2-keto acids into acyl-CoAs, expands the biological toolkit of reaction chemistries available in the Ehrlich pathway to include the gamut of CoA-dependent enzymes. As a proof-of-concept, we demonstrated the first biosynthesis of a tertiary branched-alcohol, pivalcohol, at a level of ~10 mg/L from glucose in Escherichia coli, using a pivalyl-CoA mutase from Xanthobacter autotrophicus. Furthermore, engineering an enzyme in the CoA detour, the Lactobacillus brevis CoA-acylating aldehyde dehydrogenase, allowed stringent product selectivity. Targeted production of 3-methyl-1-butanol (3-MB) in E. coli mediated by the CoA detour showed a 3-MB:side-product (isobutanol) ratio of >20, an increase over the ratios previously achieved using the conventional Ehrlich pathway.
Keywords: metabolic engineering, Ehrlich pathway, higher alcohol, tertiary branched-chemicals, pivalyl-CoA mutase
Graphical Abstract

The Ehrlich pathway, which converts 2-keto acids into alcohols via aldehydes (Figure 1), has been explored extensively to produce fuels, commodities, and value-added chemicals.1–7 However, the scope of the Ehrlich pathway is limited by the types of 2-keto acid precursors available. Biosynthetic pathways of amino acids provide a natural source of 2-keto acids, and recent efforts have substantially expanded the repertoire of products by elongating natural 2-keto acids with a synthetic “+1” cycle.3,4 Nevertheless, many product configurations are still inaccessible, mainly due to the limited chemistries catalyzed by 2-keto acid-utilizing enzymes. For example, the α,β-unsaturated and 3-functionalized alcohols have only been produced through the isoprenoid pathway or the reversed β-oxidation pathway.8,9 Furthermore, some configurations, such as the tertiary branched-alcohols, have not been produced in biomanufacturing, despite their important applications as fuel additives and solvents.10 Due to their bulky and hydrophobic nature, tertiary branched-compounds, such as tert-leucine, are essential reagents for molecular conformational control in chemical synthesis.11 However, de novo formation of tertiary branched-carbon chains has been elusive in biomanufacturing.
Figure 1.
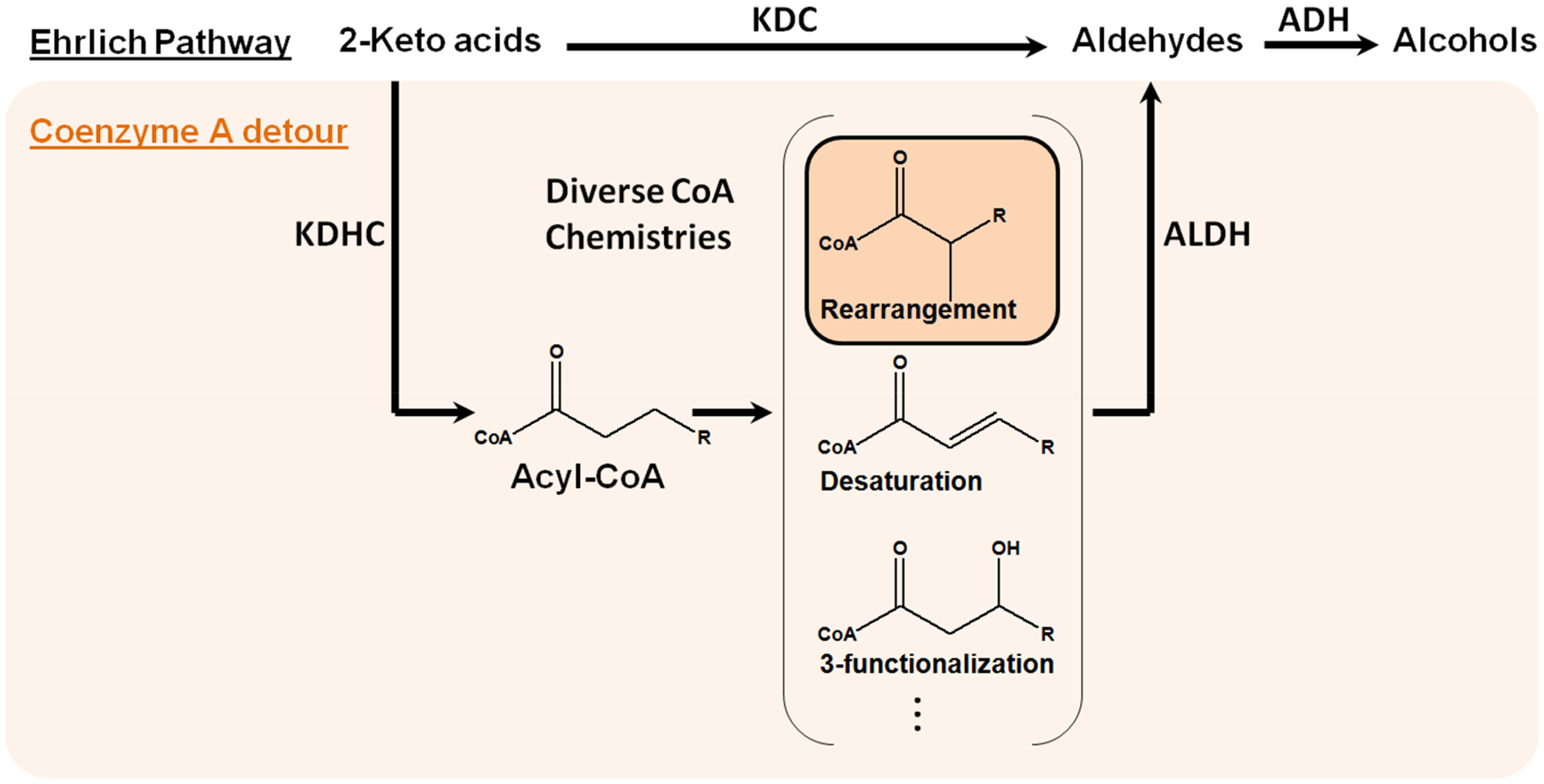
Coenzyme A (CoA) detour of the Ehrlich pathway. The conventional Ehrlich pathway converts 2-keto acids to alcohols via aldehydes. The CoA detour first converts 2-keto acids into acyl-CoAs, which allows the application of diverse CoA-dependent chemistries (The rearrangement chemistry explored in this study is highlighted). Finally, the CoA-derivatives are converted to aldehydes, re-entering the Ehrlich pathway. KDC, keto acid decarboxylase; KDHC, keto acid dehydrogenase complex; ALDH, CoA-acylating aldehyde dehydrogenase; ADH, alcohol dehydrogenase.
To overcome this limitation, a CoA-dependent detour is proposed (Figure 1), which is composed of three steps. First, 2-keto acids are converted to acyl-CoAs through the function of keto acid dehydrogenase complexes (KDHCs).12–16 Second, a range of acyl-CoA utilizing enzymes in natural product biosynthesis and various degradation pathways can be employed to diversify the carbon chains by conducting chemistries such as rearrangement,17 desaturation,18 and 3-functionalization.19 Third, the CoA-derivatives are converted to aldehydes by CoA-acylating aldehyde dehydrogenases (ALDHs), and re-enter the Ehrlich pathway. The aldehydes can then be turned into alcohols by alcohol dehydrogenases (ADHs). These three steps have been explored individually in different contexts;13–16,21 here we show that optimizing their compatibility is the key to successfully establishing the detour. Importantly, by using a recently discovered pivalyl-CoA mutase (PCM) from Xanthobacter autotrophicus as the rearrangement enzyme17 and matching it with an appropriate KDHC, ALDH, and ADH, we achieved the first bioproduction of a tertiary branched-alcohol, pivalcohol, at ~10 mg/L from glucose in Escherichia coli.
In addition, it has been a challenge to tune the product selectivity of the Ehrlich pathway.4,22 Since many 2-keto acids are present inside the cells, the Ehrlich pathway often produces a mixture of alcohols.2,4,22,23 Targeted production of long chain alcohols is especially difficult.22 Our hypothesis is that the attachment of the CoA handle can aid enzymes in substrate recognition, which may provide opportunities for engineering highly specific enzymes. The model system we studied here is the selective production of 3-methyl-1-butanol (3-MB) versus the side-product isobutanol, which only differ by one carbon in length. By employing the CoA detour and engineering the ALDH enzyme, we achieved a 3-MB:isobutanol ratio of >20 (~1250 mg/L 3-MB versus ~57 mg/L isobutanol produced from glucose by engineered E. coli), which exceeds the highest ratio (~12) achieved to date by the conventional Ehrlich pathway.23
In summary, we demonstrated that the engineered CoA detour can effectively expand the scope and enhance the selectivity of the Ehrlich pathway. We systematically compared a panel of KDHCs, ALDHs, and ADHs in higher alcohol production. We previously applied the vitamin B12-dependent carbon chain rearrangement chemistry to produce a secondary branched-carbon chain from a linear precursor.24 Here, we demonstrated that this method can be further extended to produce tertiary branched chemicals, which has not been achieved before.
RESULTS AND DISCUSSION
First, we sought to establish a functional CoA detour of the Ehrlich pathway in E. coli and demonstrate higher alcohol production (Figure 2A). The initial construction of the CoA detour was achieved by overexpressing the KDHC encoded by the bkdAA-bkdAB-bkdB-lpdV operon from Pseudomonas putida13 and the ALDH encoded by pduP from Klebsiella pneumoniae20 on a high-copy plasmid (pWB102). This plasmid was transformed into the ADH-defective E. coli strain AL70725 together with a second plasmid (pWB101) containing a wide substrate range ADH, YqhD from E. coli.26 The resulting strain was named WB101. Use of the AL707 strain as the host, which has all seven annotated ADHs disrupted,25 allowed us to study the effect of different ADHs on substrate selectivity, which will be discussed below. In rich medium, strain WB101 produced C3–C5 linear and C4–C5 branched alcohols, with titers ranging between 20 and 30 mg/L (Figure 2B), which can be used as gasoline substitutes. These results showed that a wide range of 2-keto acids naturally existing in E. coli can serve as inputs of the CoA detour. Upon further overexpressing the feedback-resistant LeuA and LeuBCD of E. coli23 on a third plasmid (pWB103), the resulting strain WB102 produced significantly increased titers of longer-chain higher alcohols such as 1-butanol, 1-pentanol, and 3-MB (Figure 2B), the production of which requires more cycles of chain elongation (Figure 2A). Interestingly, the titer of 1-propanol also increased in WB102. It might be possible that the LeuABCD were active in converting pyruvate to 2-ketobutryrate,27 the 2-keto acid precursor of 1-propanol. Alternatively, overexpression of the feedback-resistant LeuA may result in accumulation of branched chain amino acids, which have been shown to activate theP. putida KDHC complex.28
Figure 2.
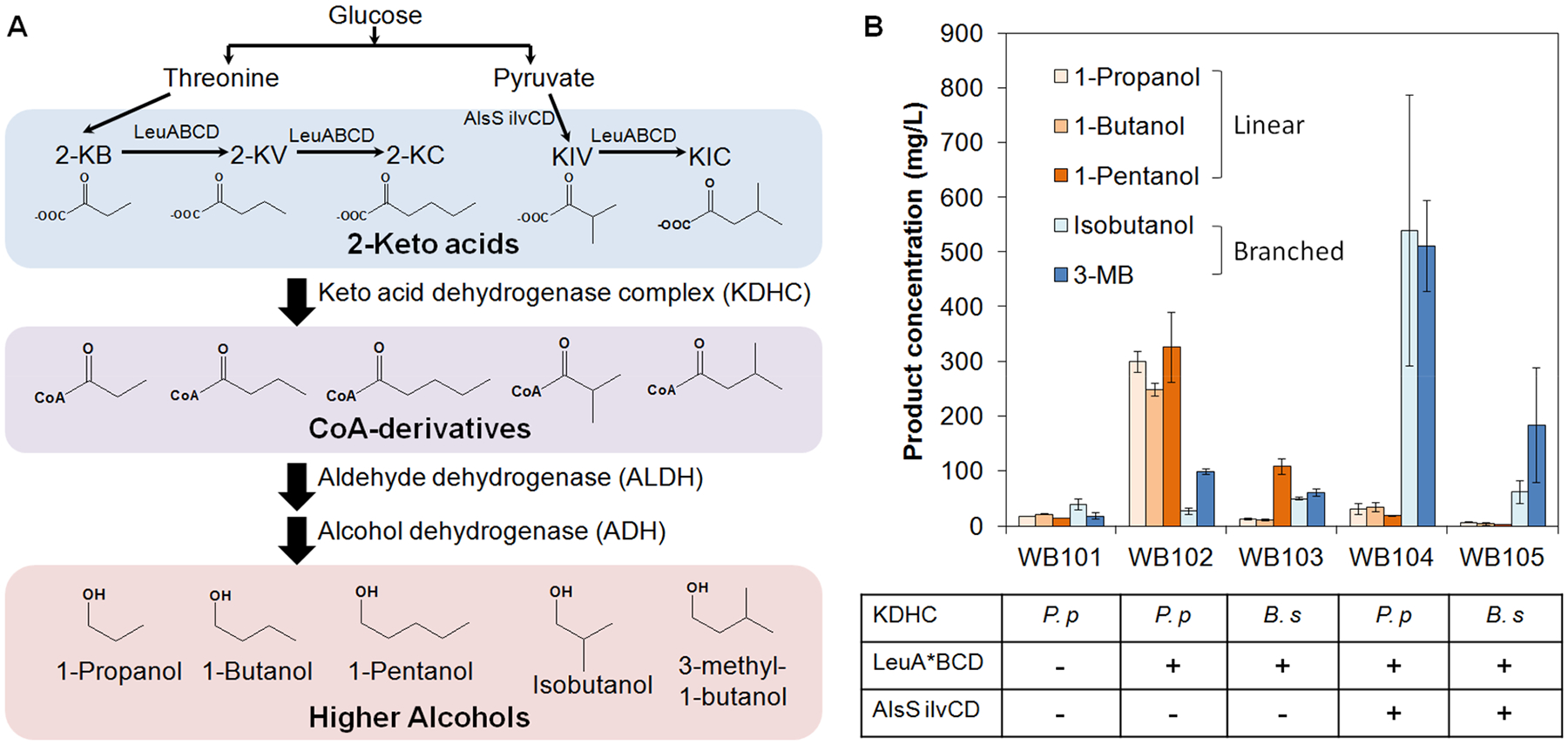
CoA detour of the Ehrlich pathway enables the production of various higher alcohols in Escherichia coli. (A) The production pathways for linear and branched alcohols. (B) Choice of KDHC and overexpression of upstream genes altered the product distribution and titers. 2-KB, 2-ketobutyrate; 2-KV, 2-ketovalerate; 2-KC, 2-ketoisocaproate; KIV, ketoisovalerate; KIC, ketoisocaproate; 3-MB, 3-methyl-1-butanol. KDHC, keto acid dehydrogenase complex; ALDH, CoA-acylating aldehyde dehydrogenase; ADH, alcohol dehydrogenase. LeuA*, the feedback-resistant variant (G462D) of E. coli LeuA. Pp, Pseudomonas putida; Bs, Bacillus subtilis. Alcohol titers were quantified at 72 h. Error bars represent one standard deviation of three replicates (n = 3).
When P. putida KDHC in WB102 was replaced with its homologue from B. subtilis12 (on plasmid pWB104), the resulting strain WB103 showed a distinct product spectrum. While the P. putida KDHC strongly favored the production of linear alcohols, the B. subtilis counterpart generally had higher preference toward the branched alcohols, as well as the long linear alcohol 1-pentanol (Figure 2B). These results suggested that B. subtilis KDHC might have a wider substrate binding pocket, and thus, it can better accommodate bulkier substrates (or wrapped long linear substrates). On the other hand, WB102 had a much higher total alcohol titer (~1000 mg/L) than WB103 (~224 mg/L), suggesting that P. putida KDHC has a higher activity in vivo in E. coli. KDHCs have been used to produce acyl-CoA precursors for ester and fatty acid production.12,13 Here, we report the direct comparison of the two commonly used KDHCs, which may guide future pathway construction involving these enzymes.
Interestingly, when AlsS from B. subtilis and IlvCD fromE. coli were overexpressed2 (on plasmid pWB105), the strains harboring P. putida or B. subtilis KDHC (WB104 and WB105, respectively) almost exclusively produced branched alcohols (Figure 2B). These results suggested that the overexpression of the upstream enzymes in branched-chain amino acid biosynthetic pathway (Figure 2A) substantially shifted the intracellular distribution of 2-keto acids. Despite different compositions, the total alcohol titers (~1136 mg/L for WB104 and ~260 mg/L for WB105) remained comparable to those before AlsS and IlvCD were overexpressed (strains WB102 and WB103), indicating that the pathway flux was mainly determined by the choice of KDHC. Given the high activity of P. putida KDHC, this enzyme was used for all further studies.
Next, we tested the production capacity of the CoA detour. In the study by Connor et al.,23 3-MB production in E. coli using the conventional Ehrlich pathway was performed using strain JCL260 as the host, which is an engineered E. coli strain with major native fermentation pathways disrupted to conserve carbon and reducing equivalents in higher alcohol production. When we switched the host of the CoA detour-mediated 3-MB production pathway from AL707 to JCL260, the resulting strain WB106 produced ~2500 mg/L isobutanol and ~1900 mg/L 3-MB, roughly a 5-fold increase compared to WB104 (Figure 3). With the conventional Ehrlich pathway in JCL260, the final titer was ~120 mg/L isobutanol and ~1280 mg/L 3-MB.23 These data indicated that the CoA detour has a similar capacity to the conventional Ehrlich path way in higher alcohol production. Connor et al.29 also showed that two-phase extractive fermentation increased 3-MB production titer, presumably by removing toxic alcohol products from the medium. Using an organic layer of oleyl alcohol in the shake flasks, we also achieved higher production titers with WB104 (~2500 mg/L isobutanol and ~1880 mg/L 3MB) (Figure 3). To further increase the titers, extractive fermentation may be conducted with WB106.
Figure 3.
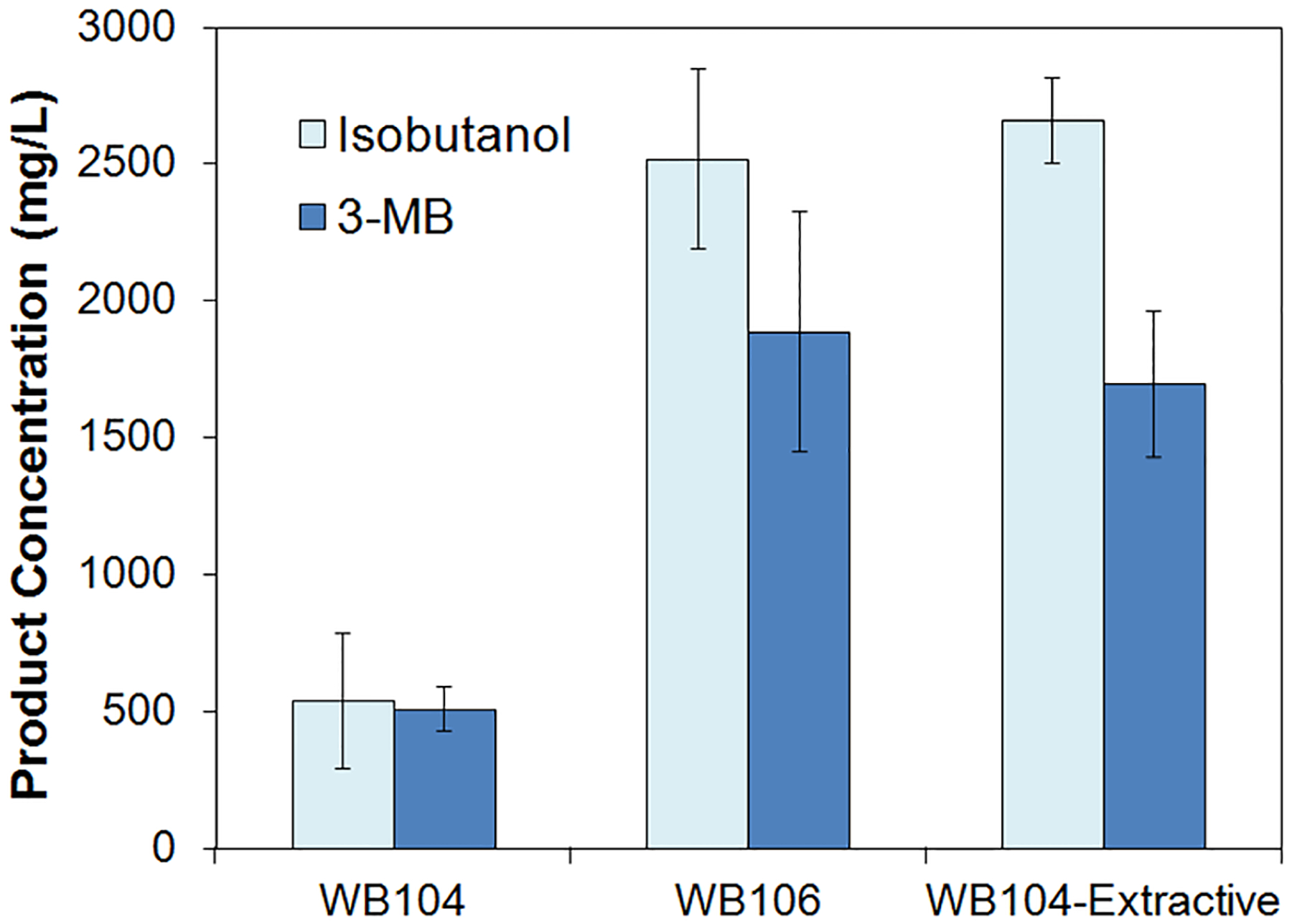
Optimization of isobutanol and 3-MB production through the CoA detour of the Ehrlich pathway. 3-MB, 3-methyl-1-butanol. Alcohol titers were quantified at 72 h. Error bars represent one standard deviation of three replicates (n = 3).
After successfully establishing the CoA detour of the Ehrlich pathway in E. coli, we aimed to test if the detour would allow tuning of the product selectivity. We examined whether the enzymes in the CoA detour could serve as molecular sieves in chain length selection to specifically produce 3-MB (C5) while minimizing isobutanol (C4) formation. In the conventional Ehrlich pathway, the committing enzymes, keto acid decarboxylases (KDCs), have been engineered to select for longer chain lengths3,4,22 to prevent premature termination of the reiterative chain elongation cycle. However, KDCs are highly promiscuous, which makes narrowing their substrate range difficult. Our results (Figure 2B) indicate that similar challenges may exist in engineering the committing enzymes of the CoA detour, namely KDHCs. Therefore, we focused on the other enzyme in the detour, the ALDH. The broad substrate range ALDH that we used initially, PduP from K. pneumoniae, is highly active (Figure S1 and S2), but its specific activity ratio for 3-methylbutyraldehyde:isobutryaldehyde (3-MBA:IBA) is ~1 (Figure 4A), as assayed in vitro with purified proteins. Note, since the acyl-CoAs are not commercially available, we assayed the enzymes in the reverse direction using aldehydes as the substrates. We identified five additional ALDHs from the literature, which have been shown to use long or bulky substrates: Salmonella enterica PduP,20 Pseudomonas sp. DmpF,30 Lactobacillus brevis PduP,20 E. coli MphF,19 and Burkholderia xenovorans BphJ.31 However, the specific activity ratios (3-MBA:IBA) for these ALDHs also did not exceed 1 (Figure 4A). These findings necessitated protein engineering to create a 3-MBA favoring ALDH. We chose the L. brevis PduP to engineer for its high expression level. Through homology modeling of L. brevis PduP, we predicted that the methyl group of T428 extends into the active site, creating steric hindrance that potentially limits the maneuverability of larger substrates from aligning with catalytic residues to attain active geometry (Figure 4B). This finding is consistent with a recent study on an ALDH from Clostridium phytofermentans.32 After we changed the T428 to A, the resulting enzyme demonstrated a specific activity ratio (3-MBA:IBA) of ~3, which is higher than all native enzymes tested (Figure 4A). The catalytic efficiency (kcat/KM) of T428A L. brevis PduP toward 3-MBA was significantly increased compared to that of the wild type (Table S1). Computationally generated structure models also suggest that the engineered substrate binding pocket may better accommodate 3-methylbutyraldehyde (Figure 4C).
Figure 4.

Targeted production of 3-methyl-1-butanol (3-MB) by the CoA detour of the Ehrlich pathway. (A) The native and engineered CoA acylating aldehyde dehydrogenases (ALDHs) showed different ratios of specific activities toward 3-methylbutyraldehyde and isobutryaldehyde (3-MBA:IBA). The specific activities were measured using enzyme assays in vitro with purified enzymes. (B,C) Computational structure modeling revealed the role of the T428A mutation in accommodating 3-MBA. Contours of the binding pocket are shown in black. The T428A mutation allows the bulky 3-MBA (green) to orient in a catalytic geometry, with the carbonyl carbon positioned for nucleophilic attack by Cys275 (cyan) and hydride transfer to NAD (orange). (D) Isobutanol and 3-MB production titers by WB108 strain. (E) Comparison of the 3-MB/isobutanol production ratio among different strains harboring different ALDHs. Alcohol titers were quantified at 72 h. Error bars represent one standard deviation of three replicates (n = 3).
Following replacement of K. pneumoniae PduP with L. brevis PduP T428A, the resulting strain WB108 produced ~1110 mg/L 3-MB but only ~50 mg/L isobutanol (Figure 4D). The ratio of 3-MB to isobutanol was greater than 20 (Figure 4E), which was higher than those achieved using the conventional Ehrlich pathway in various organisms (Table S2. The highest ratio of ~12 was reported by Connor et al.23). In comparison, both the strains harboring K. pneumoniae PduP (WB104) and wild type L. brevis PduP (WB107) showed the 3-MB/isobutanol production ratio of ~1 (Figure 4E). These results highlight the effectiveness of chain length selection using the engineered CoA detour. Although the specificity was much higher, the 3-MB titer of WB108 was lower than that of WB104, presumably because the activity of L. brevis PduP T428A toward 3-MBA is relatively low (Figure S1), as assayed using purified proteins. Future work will focus on increasing the expression level of the engineered ALDH using stronger promoters, as well as engineering the highly active K. pneumoniae PduP to favor 3-MBA.
After characterizing the simplest CoA detour, which is only comprised of the entering enzyme KDHC and the exiting enzyme ALDH, we next sought to insert the diversifying step (Figure 1). We chose the rearrangement chemistry which can enable the formation of a tertiary branched-alcohol, pivalcohol (Figure 5A). The degree of branching has been shown to greatly affect the properties of fuels.12,16,33 Prior to this study, only linear and secondary branched-biofuels have been produced. Recently, a pivalyl-CoA mutase (PCM) was identified inX. autotrophicus that catalyzes the interconversion of isovaleryl CoA and pivalyl-CoA.17 We cloned the X. autotrophicus PCM as a fusion protein of the large and small subunits (PCM-F),17 and overexpressed it in E. coli. We detected significant activity in converting isovaleryl-CoA to pivalyl-CoA in crude cell lysates (Figure 5B), demonstrating functional overexpression of the enzyme. Since the vitamin B12-dependent acyl-CoA mutase family of proteins rely on chaperones to protect them from inactivation during catalysis,17 we next coexpressed the predicted G-protein chaperone with the PCM fusion protein. The G-protein chaperone, encoded by Xaut_5042 of X. autotrophicus, is in the same operon as the PCM catalytic subunits, but its function has not been characterized.17 Using crude lysate-based enzymatic assays, we showed that the chaperone significantly increased the PCM activity (Figure 5B).
Figure 5.

Production of pivalcohol using the CoA detour of the Ehrlich pathway. (A) The pivalcohol production pathway in Escherichia coli.(B) Conversion of isovaleryl-CoA to pivalyl-CoA in vitro by crude cell lysates containing the pivalyl-CoA mutase fusion protein (PCM-F) from Xanthobacter autotrophicus, and with its G-protein chaperone. (C) Typical gas chromatography-flame ionization detector (GC-FID) traces showing the absence of pivalcohol in the production medium of WB109 (which has the E. coli YqhD), and the presence of pivalcohol in that of WB110 (which has Ralstonia sp. ADH). (D) Pivalcohol titers of WB109 and WB110. (E) Specific activities of various CoA-acylating aldehyde dehydrogenases (ALDHs) toward pivaldehyde, as measured in vitro using purified proteins. (F) Specific activities of various alcohol dehydrogenases (ADHs) toward pivaldehyde, as measured in vitro using purified proteins. N.D. not detected. See text for the sources of ALDHs and ADHs. Alcohol titers were quantified at 72 h. Error bars represent one standard deviation of three replicates (n = 3).
However, when the plasmid containing X. autotrophicus PCM-F and G-protein chaperone (pWB108) was added to the 3-MB production strain WB104, the resulting strain WB109 did not produce pivalcohol (Figure 5C, D). We hypothesized that the downstream enzymes, ALDH and ADH, might be incapable of accepting the sterically demanding tertiary branched carbon chain. Since we showed that the ALDH used in this strain, namely K. pneumoniae PduP, has positive activity with the tertiary branched-substrate (highest activity among all six tested ALDHs, Figure 5E), the ADH enzyme E. coli YqhD was likely the culprit. Indeed, YqhD showed virtually no activity in reducing pivaldehyde to pivalcohol (Figure 5F). We then characterized four ADHs from other organisms which were shown to have broad substrate-ranges or prefer bulky substrates. These ADHs include: ADH634 and Ypr1P35 from Saccharomyces cerevisiae, ADH from L. brevis,36 and ADH from Ralstonia sp.37 From in vitro enzyme assays with purified proteins, Ralstonia sp. ADH showed the highest activity toward pivaldehyde (Figure 5F). When YqhD was replaced with Ralstonia sp. ADH, the resulting strain WB110 was able to produce ~10 mg/L pivalcohol from glucose in rich medium in two-phase fermentation with oleyl alcohol as the extractive solvent23 (Figure 5C,D). The bioproduction of a tertiary-branched alcohol significantly expands the chemical inventory that can be accessed through metabolic engineering.
The pivalcohol production titer is still low. One of the major causes could be the relatively low activity of PCM, which is consistent with the observation that WB110 still produced significant amounts of 3-MB (data not shown). On the basis of sequence homology and literature,17 we tested another five PCMs from different organisms (Figure S3). Unfortunately, none of these PCMs performed better than the X. autotrophicus one, as assayed in vitro using crude cell lysates. Future work will focus on identifying highly active PCMs using bioinformatics tools17 from the ever-expanding genomic sequence database. In addition to the G-protein chaperone, the acyl-CoA mutase family of enzymes has been shown to require other auxiliary proteins, such as adenosyltransferases.38 Our ongoing work will explore the effects of these auxiliary proteins, as well as the effect of the coenzyme B12 biosynthesis system39 to better support PCM activity in vivo.
In conclusion, the CoA detour of the Ehrlich pathway shows promise to be a highly specific and versatile biosynthetic route. By leveraging both the capability of the 2-keto acid pathway to supply odd and even chain-length, linear, branched, and aromatic starter units and the extensive diversity of CoA-dependent carbon chain modifying chemistries, the CoA detour of the Ehrlich pathway may open up new compounds as biomanufacturing targets. Our results also suggest that KDHC, ALDH, and ADH all need to be carefully chosen to optimize the performance of the pathway, and their substrate specificities may be tuned simultaneously to offer a multidimensional control on product selectivity.
MATERIALS AND METHODS
The plasmids and strains used in this study are summarized in Table 1. The conditions used for higher alcohol production, enzyme assays, and computational modeling of protein structures are detailed in the Supporting Information.
Table 1.
Plasmids and Strainsa
| comments | reference | |
|---|---|---|
| Strains | ||
| JCL260 | BW25113/F′ [traD36, proAB+, lacIq ZΔM15, ΔadhE, ΔldhA, ΔfrdBC, Δfnr, Δpta, ΔpflB] | 2 |
| AL707 | JCL260: ΔadhP, ΔeutG, ΔyiaY, ΔyjgB, ΔbetA, ΔfucO, ΔeutE | 25 |
| WB101 | AL707 transformed with pWB101 and pWB102 | this study |
| WB102 | WB101 transformed with pWB103 | this study |
| WB103 | AL707 transformed with pWB101, pWB103, and pWB104 | this study |
| WB104 | AL707 transformed with pWB102, pWB103, and pWB105 | this study |
| WB105 | AL707 transformed with pWB103, pWB104, and pWB105 | this study |
| WB106 | JCL260 transformed with pWB102, pWB103, and pWB105 | this study |
| WB107 | AL707 transformed with pWB103, pWB105, and pWB106 | this study |
| WB108 | AL707 transformed with pWB103, pWB105, and pWB107 | this study |
| WB109 | WB104 transformed with pWB108 | this study |
| WB110 | AL707 transformed with pWB102, pWB103, and pWB108, pWB109 | this study |
| Plasmids | ||
| pWB101 | PLlacO1::Ec yqhD, SC101 ori, SpecR | this study |
| pWB102 | PLlacO1::Kp pdup-Pp KDHC, ColE1 ori, AmpR | this study |
| pWB103 | PLlacO1::Ec leuA G462D-leuBCD, p15A ori, KanR | this study |
| pWB104 | PLlacO1::Kp pdup-Bs_KDHC, ColE1 ori, AmpR | this study |
| pWB105 | PLlacO1::Bs alsS-Ec ilvCD-Ec yqhD, SC101 ori, SpecR | this study |
| pWB106 | PLlacO1::Lb pdup-Pp KDHC, ColE1 ori, AmpR | this study |
| pWB107 | PLlacO1::Lb pduP T428A-Pp KDHC, ColE1 ori, AmpR | this study |
| pWB108 | PBAD::-Xa PCM-F-Xaut 5042, RSF ori, CmR | this study |
| pWB109 | PLlacO1::Bs alsS-Ec ilvCD-Rs ADH, SC101 ori, SpecR | this study |
Abbreviations indicate source of the genes: Ec, Escherichia coli; Kp, Klebsiella pneumoniae; Bs, Bacillus subtilis; Pp, Pseudomonas putida; Lb, Lactobacillus brevis; Xa, Xanthobacter autotrophicus; Rs, Ralstonia sp.
Supplementary Material
ACKNOWLEDGMENTS
The work is funded by the start-up fund of UC Irvine to H.L.
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssynbio.8b00358.
Plasmid and strain construction; Production conditions and analytic methods for higher alcohols; Enzyme assays for ALDHs, ADHs, and PCMs; Specific activity of ALDHs toward 3-MBA and isobutryaldehyde (Figure S1 and S2); Catalytic efficiency (kcat/KM) of the engineered CoA-acylating aldehyde dehydrogenase (Table S1); 3-MB to isobutanol ratios of previous efforts aimed at producing 3-MB using the conventional Ehrlich pathway (Table S2); Computational modeling of protein structures. Activity of various pivalyl-CoA mutases (PCMs) (Figure S3) (PDF)
REFERENCES
- (1).Hazelwood LA, Daran JM, van Maris AJ, Pronk JT, and Dickinson JR (2008) The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Applied and environmental microbiology 74, 2259–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Atsumi S, Hanai T, and Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89. [DOI] [PubMed] [Google Scholar]
- (3).Zhang K, Sawaya MR, Eisenberg DS, and Liao JC (2008) Expanding metabolism for biosynthesis of nonnatural alcohols. Proc. Natl. Acad. Sci. U. S. A 105, 20653–20658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Marcheschi RJ, Li H, Zhang K, Noey EL, Kim S, Chaubey A, Houk KN, and Liao JC (2012) A synthetic recursive ″+1″ pathway for carbon chain elongation. ACS Chem. Biol 7, 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Jambunathan P, and Zhang K (2014) Novel pathways and products from 2-keto acids. Curr. Opin. Biotechnol 29, 1–7. [DOI] [PubMed] [Google Scholar]
- (6).Tashiro Y, Rodriguez GM, and Atsumi S (2015) 2-Keto acids based biosynthesis pathways for renewable fuels and chemicals. J. Ind. Microbiol. Biotechnol 42, 361–373. [DOI] [PubMed] [Google Scholar]
- (7).Sarria S, Kruyer NS, and Peralta-Yahya P (2017) Microbial synthesis of medium-chain chemicals from renewables. Nat. Biotechnol 35, 1158–1166. [DOI] [PubMed] [Google Scholar]
- (8).George KW, Thompson MG, Kang A, Baidoo E, Wang G, Chan LJ, Adams PD, Petzold CJ, Keasling JD, and Lee TS (2015) Metabolic engineering for the high-yield production of isoprenoid-based C(5) alcohols in E. coli. Sci. Rep 5, 11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Gulevich AY, Skorokhodova AY, Stasenko AA, Shakulov RS, and Debabov VG (2016) Metabolic engineering of Escherichia coli for 1,3-butanediol biosynthesis through the inverted fatty acid β-oxidation cycle. Appl. Biochem. Microbiol 52, 15–22. [Google Scholar]
- (10).Global market of tert-butanol, http://www.europlat.org/globaltert-butanol-market.htm. Accessed in Aug 2018.
- (11).Bommarius AS, Schwarm M, Stingl K, Kottenhahn M, Huthmacher K, and Drauz K (1995) Synthesis and use of enantiomerically pure tert-leucine. Tetrahedron: Asymmetry 6, 2851–2888. [Google Scholar]
- (12).Haushalter RW, Kim W, Chavkin TA, The L, Garber ME, Nhan M, Adams PD, Petzold CJ, Katz L, and Keasling JD (2014) Production of anteiso-branched fatty acids in Escherichia coli; next generation biofuels with improved cold-flow properties. Metab. Eng 26, 111–118. [DOI] [PubMed] [Google Scholar]
- (13).Rodriguez GM, Tashiro Y, and Atsumi S (2014) Expanding ester biosynthesis in Escherichia coli. Nat. Chem. Biol 10, 259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Solomon KV, Ovadia E, Yu F, Mizunashi W, and O’Malley MA (2016) Mitochondrial targeting increases specific activity of a heterologous valine assimilation pathway in Saccharomyces cerevisiae. Metabolic engineering communications 3, 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jiang W, Qiao JB, Bentley GJ, Liu D, and Zhang F (2017) Modular pathway engineering for the microbial production of branched-chain fatty alcohols. Biotechnol. Biofuels 10, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Tao H, Guo D, Zhang Y, Deng Z, and Liu T (2015) Metabolic engineering of microbes for branched-chain biodiesel production with low-temperature property. Biotechnol. Biofuels 8, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kitanishi K, Cracan V, and Banerjee R (2015) Engineered and Native Coenzyme B12-dependent Isovaleryl-CoA/Pivalyl-CoA Mutase. J. Biol. Chem 290, 20466–20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Forster-Fromme K, and Jendrossek D (2008) Biochemical characterization of isovaleryl-CoA dehydrogenase (LiuA) of Pseudomonas aeruginosa and the importance of liu genes fora functional catabolic pathway of methyl-branched compounds. FEMS Microbiol. Lett 286, 78–84. [DOI] [PubMed] [Google Scholar]
- (19).Dellomonaco C, Clomburg JM, Miller EN, and Gonzalez R (2011) Engineered reversal of the beta-oxidation cycle for the synthesis of fuels and chemicals. Nature 476, 355–359. [DOI] [PubMed] [Google Scholar]
- (20).Lan EI, Ro SY, and Liao JC (2013) Oxygen-tolerant coenzyme A-acylating aldehyde dehydrogenase facilitates efficient photosynthetic n-butanol biosynthesis in cyanobacteria. Energy Environ. Sci 6, 2672–2681. [Google Scholar]
- (21).Sheppard MJ, Kunjapur AM, Wenck SJ, and Prather KL (2014) Retro-biosynthetic screening of a modular pathway design achieves selective route for microbial synthesis of 4-methyl-pentanol. Nat. Commun 5, 5031. [DOI] [PubMed] [Google Scholar]
- (22).Mak WS, Tran S, Marcheschi R, Bertolani S, Thompson J, Baker D, Liao JC, and Siegel JB (2015) Integrative genomic mining for enzyme function to enable engineering of a non-natural biosynthetic pathway. Nat. Commun 6, 10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Connor MR, and Liao JC (2008) Engineering of an Escherichia coli strain for the production of 3-methyl-1-butanol. Applied and environmental microbiology 74, 5769–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Black WB, Zhang L, Kamoku C, Liao JC, and Li H (2018) Rearrangement of Coenzyme A-Acylated Carbon Chain Enables Synthesis of Isobutanol via a Novel Pathway in Ralstonia eutropha. ACS Synth. Biol 7, 794–800. [DOI] [PubMed] [Google Scholar]
- (25).Rodriguez GM, and Atsumi S (2012) Isobutyraldehyde production from Escherichia coli by removing aldehyde reductase activity. Microb. Cell Fact 11, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Jarboe LR (2011) YqhD: a broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl. Microbiol. Biotechnol 89, 249–257. [DOI] [PubMed] [Google Scholar]
- (27).Kohlhaw G, Leary TR, and Umbarger HE (1969) α-Isopropylmalate Synthase from Salmonella typhimurium: Purification and Properties. J. Biol. Chem 244, 2218–2225. [PubMed] [Google Scholar]
- (28).Sokatch JR, McCully V, and Roberts CM (1981) Purification of a branched-chain keto acid dehydrogenase from Pseudomonas putida. J. Bacteriol 148, 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Connor MR, Cann AF, and Liao JC (2010) 3-Methyl-1butanol production in Escherichia coli: random mutagenesis and two phase fermentation. Appl. Microbiol. Biotechnol 86, 1155–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Powlowski J, Sahlman L, and Shingler V (1993) Purification and properties of the physically associated meta-cleavage pathway enzymes 4-hydroxy-2-ketovalerate aldolase and aldehyde dehydrogenase (acylating) from Pseudomonas sp. strain CF600. J. Bacteriol 175, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Baker P, Carere J, and Seah SYK (2012) Substrate Specificity, Substrate Channeling, and Allostery in BphJ: An Acylating Aldehyde Dehydrogenase Associated with the Pyruvate Aldolase BphI. Biochemistry 51, 4558–4567. [DOI] [PubMed] [Google Scholar]
- (32).Tuck LR, Altenbach K, Ang TF, Crawshaw AD, Campopiano DJ, Clarke DJ, and Marles-Wright J (2016) Insight into Coenzyme A cofactor binding and the mechanism of acyl-transfer in an acylating aldehyde dehydrogenase from Clostridium phytofermentans. Sci. Rep 6, 22108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Li H, Cann AF, and Liao JC (2010) Biofuels: biomolecular engineering fundamentals and advances. Annu. Rev. Chem. Biomol. Eng 1, 19–36. [DOI] [PubMed] [Google Scholar]
- (34).Larroy C, Fernandez MR, Gonzalez E, Pares X, and Biosca JA (2002) Characterization Of The Saccharomyces Cerevisiae Ymr318c (Adh6) Gene Product As A Broad Specificity Nadph-Dependent Alcohol Dehydrogenase: Relevance In Aldehyde Reduction. Biochem. J 361, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Ford G, and Ellis EM (2002) Characterization of Ypr1p from Saccharomyces cerevisiae as a 2-methylbutyraldehyde reductase. Yeast 19, 1087–1096. [DOI] [PubMed] [Google Scholar]
- (36).Ernst M, Kaup B, Müller M, Bringer-Meyer S, and Sahm H (2005) Enantioselective reduction of carbonyl compounds by whole-cell biotransformation, combining a formate dehydrogenase and a (R)-specific alcohol dehydrogenase. Appl. Microbiol. Biotechnol 66, 629–634. [DOI] [PubMed] [Google Scholar]
- (37).Man H, Kędziora K, Kulig J, Frank A, Lavandera I, Gotor Fernandez V, Rother D, Hart S, Turkenburg JP, and Grogan G (2014) Structures of Alcohol Dehydrogenases from Ralstonia and Sphingobium spp. Reveal the Molecular Basis for Their Recognition of ‘Bulky–Bulky’ Ketones. Top. Catal 57, 356–365. [Google Scholar]
- (38).Li Z, Kitanishi K, Twahir UT, Cracan V, Chapman D, Warncke K, and Banerjee R (2017) Cofactor Editing by the G-protein Metallochaperone Domain Regulates the Radical B12 Enzyme IcmF. J. Biol. Chem 292, 3977–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Wang J, and Zhang K (2015) Production of mesaconate in Escherichia coli by engineered glutamate mutase pathway. Metab. Eng 30, 190–196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


