Abstract
Purpose
Preclinical data demonstrating androgen receptor (AR)-positive (AR+) TNBC cells are sensitive to AR antagonists and PI3K inhibition catalyzed an investigator-initiated, multi-institutional phase Ib/II study TBCRC032. The trial investigated the safety and efficacy of the AR-antagonist enzalutamide alone or in combination with the PI3K inhibitor taselisib in patients with metastatic AR+ (≥10%) breast cancer.
Patients and Methods
Phase Ib patients (ER+ or TNBC) with AR+ breast cancer received 160 mg enzalutamide in combination with taselisib to determine dose-limiting toxicities and the maximum tolerated dose (MTD). Phase II TNBC patients were randomized to receive either enzalutamide alone or in combination with 4 mg taselisib until disease progression. Primary endpoint was clinical benefit rate (CBR) at 16 weeks.
Results
The combination was tolerated and the MTD was not reached. The adverse events were hyperglycemia and skin rash. Overall, CBR for evaluable patients receiving the combination was 35.7%, and median progression-free survival (PFS) was 3.4 months. Luminal AR (LAR) TNBC subtype patients trended towards better response compared to non-LAR (75.0% vs. 12.5%, p=0.06), and increased PFS (4.6 vs. 2.0 months, p=0.082). Genomic analyses revealed subtype-specific treatment response; and novel FGFR2 fusions and AR splice variants.
Conclusions
The combination of enzalutamide and taselisib increased CBR in TNBC patients with AR+ tumors. Correlative analyses suggest AR protein expression alone is insufficient for identifying patients with AR-dependent tumors and knowledge of tumor LAR subtype and AR-splice variants may identify patients more or less likely to benefit from AR-antagonists.
Trial registration
ClinicalTrial.gov, Identifier: NCT02457910. Registered on May 29, 2015.
Keywords: TNBCtype, enzalutamide, PIK3CA, FGFR, FGFR2-TACC2, AR-V7/AR3, fusion, luminal AR
Introduction
Triple-negative breast cancers (TNBCs) are categorically defined by an absence of estrogen and progesterone receptor (ER and PR) expression, lack of amplification of the human epidermal growth factor receptor 2 (HER2) gene and a diverse transcriptional biology. TNBCs have been shown to be comprised of at least four distinct transcriptional subtypes: two basal subtypes (BL1 and BL2), a mesenchymal (M) subtype devoid of immune cells(1) and a luminal androgen receptor subtype (LAR) that is enriched in androgen receptor (AR) expression and downstream gene targets(2,3). Similar to ER and PR, AR is an intracellular steroid receptor that dimerizes and translocates to the nucleus after binding androgen ligands. In the nucleus, AR binds to androgen response elements to promote target gene transcription in a tissue-specific manner.
Approximately 70%–90% of all breast cancers express AR with the majority also expressing ER(4). AR is expressed in the absence of ER in a subset of TNBCs, ranging from 20% to 40%(4–7). Regardless of clinical subtype, AR expression has been associated with favorable prognosis, including improvements in overall survival(8) and lower risk of recurrence(9). Despite better prognosis, patients with AR-positive (AR+) tumors are far less sensitive to chemotherapy(10) and patients with AR+ TNBCs have a lower chance of achieving pathological complete response to neoadjuvant chemotherapy(11). Several retrospective studies have similarly demonstrated that patients with LAR subtype tumors are far less responsive to standard chemotherapy than other TNBC patients, highlighting the need for additional non-chemotherapy based therapeutic strategies in TNBC (11,12). The safety and efficacy of anti-androgens was first evaluated in metastatic AR+ TNBC patients (≥10% of tumor cells with nuclear protein expression by IHC), in which treatment with bicalutamide was well tolerated and patients had a 19% clinical benefit rate (CBR) at six months(13,14). A subsequent trial evaluating the second-generation AR-antagonist enzalutamide in women with advanced AR+ TNBC showed increased clinical activity with a 25% CBR at 16 weeks supporting additional evaluation of targeting AR in TNBC(13).
In contrast to other subtypes, LAR tumors are enriched (approximately 40–50%) in activating PIK3CA mutations(15,16). Further, LAR TNBC cell line models are sensitive to several androgen receptor (AR) antagonists(15,17,18), and all LAR TNBC cell line models contain PIK3CA mutations that confer sensitivity to PI3K inhibitors and synergy with AR antagonists(15). Given the dependency of pre-clinical LAR models on AR signaling and sensitivity to PI3K inhibitors and reciprocal feedback between the pathways in prostate cancer(19,20), we postulated that combined AR and PI3K inhibition would have greater efficacy than AR inhibition alone in AR+ TNBC patients. To translate our preclinical observations and identify new therapeutic options for patients with TNBC, we conducted an investigator-initiated, randomized phase Ib/II clinical trial evaluating orally administered enzalutamide with or without the PI3K inhibitor taselisib in patients with AR+ metastatic TNBC ( NCT02457910). The phase Ib portion included ER+ metastatic breast cancer patients as well. The primary objectives of the study were to determine: (i) the maximally tolerated dose (MTD) for the combination of enzalutamide and taselisib and (ii) the clinical benefit rate (CBR) at 16 weeks. Secondary objectives included progression-free survival (PFS), overall response rate (ORR) and if genomic and molecular correlates (PIK3CA mutation status, AR expression level and TNBCtype) predict sensitivity to enzalutamide alone or in combination with taselisib.
Materials and Methods
Study design
This multicenter, randomized, two-arm, open label, Simon two-stage phase I/II clinical trial evaluated the orally administered enzalutamide with or without taselisib in patients with AR+ metastatic breast cancer (ClinicalTrials.gov identifier NCT02457910). To facilitate accrual during dose escalation, both ER/PR+ and TNBC were eligible and enrolled in the phase 1b portion of the trial, whereas only TNBC patients were eligible for phase II. The study was conducted in accordance with Good Clinical Practice guidelines and the Declaration of Helsinki and approved by the Vanderbilt Institutional Review Board (IRB#150188). Written informed consent was obtained from all patients before enrollment, in agreement with approved protocols from respective ethics committees at every site. Eligible patients were randomized 3:1 according to a stratified permuted block scheme with two-thirds in the enzalutamide plus taselisib arm and one-third in the enzalutamide arm. Participants were randomized to receive:
Arm A
Patients received taselisib (4 mg) orally once daily on days 1–28 and enzalutamide (160 mg) PO QD on days 9–28 of cycle 1 and days 1–28 of subsequent cycles. Cycles repeated every 28 days in the absence of disease progression or unacceptable toxicity.
Arm B
Patients received enzalutamide (160 mg) orally once daily on days 1–28. Cycles repeated every 28 days in the absence of disease progression or unacceptable toxicity. Upon disease progression, patients could crossover to Arm A.
Efficacy endpoints
The primary endpoint for the Phase Ib portion was to determine the maximum tolerated dose (MTD), defined as the highest dose tested in which a dose limiting toxicity experienced by 0 out of 3 or 1 out of 6 patients among the dose levels over a four-week treatment. Toxicities were graded according to the National Cancer Institute CTCAE version 4.0 criteria. DLT criteria included events occurring during cycle 1 (first 4 weeks), that were possibly, probably, or definitively classified as drug-related. Key DLT criteria were defined as: rash or photosensitivity grade 3 for > 7 days despite skin toxicity treatment including oral steroids and antihistamines or photosensitivity ≥ grade 4; hyperglycemia grade 3 [Fasting Plasma Glucose (FPG) 250 ЈC 399 mg/dL; 13.9 – 22.2 mmol/L], confirmed with a repeat FPG within 24 hrs for > 7 consecutive days despite oral anti-diabetic treatment or hyperglycemia grade 4 (FPG ≥ 400 mg/dL; ≥ 22.3 mmol/L) or hyperglycemia leading to diabetic keto-acidosis and hospitalization for IV insulin infusion; diarrhea grade ≥ 3 for ≥ 48 hrs, despite the use of optimal anti-diarrhea therapy or nausea/vomiting grade ≥ 3 for ≥ 48 hrs, despite the use of optimal anti-emetic therapy or pancreatitis grade ≥ 3; febrile neutropenia grade ≥ 3 or ANC grade 3 for > 7 consecutive days or ANC grade 4 or platelet count grade 3 for > 7 consecutive days and/or with signs of excessive bleeding or platelet count grade 4 (G-CSF use was permitted during the DLT window).
Other DLT criteria included: total bilirubin grade 2 for > 7 consecutive days, if less normal-grade 1 at baseline or total bilirubin grade ≥ 3, if grade 2 at baseline or AST or ALT grade ≥ 2 in conjunction with blood bilirubin grade ≥ 2 of any duration or AST or ALT grade 3 for > 7 consecutive days or AST or ALT grade 4 or serum alkaline phosphatase grade 4 or serum lipase and/or serum amylase (asymptomatic) grade 3 for > 7 consecutive days or serum lipase and/or serum amylase (asymptomatic) grade 4 or ALT / AST of 3X ULN and concomitant bilirubin of >2 X ULN with no other explanation other than study drug; serum creatinine grade ≥ 3; fatigue grade 3 for > 7 consecutive days; persistent hypertension grade ≥ 3 requiring more than one drug or more intensive therapy than previously; cardiac toxicity grade ≥ 3 or cardiac event that is symptomatic or requires medical intervention or clinical signs of cardiac disease, such as unstable angina or myocardial infarction, or troponin grade 3 (confirmed with a repeat troponin within 24 hrs) or ECG QTc interval prolonged grade ≥ 3 (after repeat confirmation on at least 2 more ECGs at the same time point).
The primary outcome measurement for the phase II portion was CBR, as defined by the proportion of patients with a best response of complete response (CR), partial response (PR), or (SD) stable disease at 16 weeks. Secondary endpoints included overall progression-free survival (PFS) from cycle 1, day 1 until tumor progression and overall response rate (ORR) up to three years.
Eligibility criteria / Participants
Eligible patients were ≥18 years old, with metastatic invasive mammary carcinoma. All participants were required to: (i) provide informed written consent; (ii) have tumors demonstrating androgen receptor positivity (defined as ≥ 10% of tumor cell nuclei positive for AR by IHC) after central review at Vanderbilt University Medical Center; (iii) have an Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1; and (iv) an adequate hematologic, hepatic, and renal function. Patients were eligible for enrollment into the phase Ib portion regardless of tumor ER/PR status (negative or positive), while phase II patients were required to be triple-negative; defined as ER and PR negative (ER/PR expression <1% cells by IHC) and HER2 negative [acceptable methods of HER2 analysis included IHC (0, 1+), fluorescence in situ hybridization (FISH) with HER2/centromere on chromosome 17 (CEN17) ratio < 2, and/or chromogenic in situ hybridization with HER2/CEN-17 ratio<2 and average HER2 copy number < 4, and/or chromogenic in situ hybridization with HER2/CEN-17 ratio < 2 and average HER2 copy number < 4; by local assessment]. Patients with any number of prior therapies including prior anti-androgen therapy, other than enzalutamide, were allowed as long as they had adequate performance status and met all other eligibility criteria. Patients were also required to have measurable or bone-only evaluable disease; measurable disease was defined as at least one lesion that could be accurately measured in at least one dimension by Response Evaluation Criteria in Solid Tumors (RECIST) criteria 1.1, with radiology scans within 21 days of day 1, cycle 1.
Patients participating in the phase II portion were required to undergo biopsy of a metastatic lesion as long as a site for biopsy was safely accessible at baseline, day 14–21, and after progression of disease. If a metastatic lesion was not available, the patient could go on study provided that archived tissue was available. Patients crossing over from the enzalutamide-only arm to the combination arm were required to undergo a biopsy of metastatic lesion prior to cross-over.
Exclusion Criteria
Key exclusion criteria included concurrent anti-cancer treatment (chemotherapy, radiation therapy, surgery, immunotherapy, hormonal therapy or biological therapy) and clinically significant cardiac, pulmonary, or liver dysfunction, malabsorption symptoms, active autoimmune disease and immunocompromised status. Additional exclusion criteria included prior treatment with enzalutamide or prior use of PI3K or Akt inhibitors for more than four weeks in the metastatic setting for the treatment of cancer. Additional criteria for exclusion included: pregnant or lactating women, current or previously treated brain metastasis or active leptomeningeal disease, ongoing or active infection requiring parenteral antibiotics, psychiatric illness/social situations that would compromise patient safety or limit compliance with study requirements including maintenance of a compliance/pill diary and patients with type insulin-dependent type II diabetes not meeting inclusion criteria detailed in the protocol.
Prescreening evaluation of AR protein expression
Pre-screening consisted of consenting for archival tumor tissue analysis of AR by immunohistochemistry (IHC). AR IHC was performed on 5-μm formalin-fixed paraffin embedded (FFPE) sections from all potential clinical trial enrollees with Benchmark ULTRA (Ventana/Roche, Oro Valley, AZ) in the CLIA-certified clinical histology laboratory of Vanderbilt University Medical Center according to the manufacturer’s specifications. FFPE sections were deparaffinized with graded alcohols and xylenes, followed by antigen retrieval with cell conditioner #1 (Ventana/Roche, Oro Valley, AZ) at 95°C for 64 min. Tissue sections were incubated with 1.3 μl/ml rabbit anti-human AR antibody (SP107, Cell Marque, Rocklin, CA) recognizing the N-terminus of AR for 32 min at 37°C. Slides were washed with Ultrawash buffer (Ventana/Roche, Oro Valley, AZ) and counterstained with Hematox II (Ventana/Roche, Oro Valley, AZ). The percent of AR+ tumor cells was determined prior to enrollment by a certified surgical pathologist with expertise in breast pathology.
Imaging
Patients underwent baseline computed tomography scans of the chest, abdomen, and pelvis, along with bone scans. Scans were repeated every 2 cycles (every 8 weeks). Upon progression of disease by RECIST criteria by local assessment, patients were removed from study.
Treatment
In the phase Ib, taselisib (supplied by Genentech, Inc.) was administered orally at a starting dose of 2 mg and escalated to 4 mg, 6 mg and 8 mg in a 3+3 trial design. This was in combination with 160 mg enzalutamide (supplied by Astellas Pharma, US) taken daily orally without interruption for a 28-day cycle. Patients were treated until disease progression or unacceptable toxicity. In phase II, patients received either 160 mg enzalutamide alone or 160 mg enzalutamide in combination with 4 mg taselisib orally, as prior phase I single agent testing of taselisib showed activity at 4 mg(21).
Tissue collection and processing
Paired RNAlater/snap-frozen and FFPE biopsies were collected from metastatic sites at baseline (baseline tumor collection) prior to day 1, at day 14–21 (cycle 2 tumor collection), and after progression of disease (crossover arm) for correlative analyses.
Tumor RNA isolation and sequencing
RNA was extracted from frozen tumor biopsies preserved in RNAlater (ThermoFisher, AM7020) using RNAqueous-Micro Total RNA Isolation (Life Technologies/Ambion, catalog no. AM1931). Isolated RNA was quantified using the Qubit RNA HS Assay Kit (Life Technologies, catalog no. Q32852) and assessed with the Agilent 2100 bioanalyzer. RNA sequencing was performed with paired-end 2×75 bp fragments on the Illumina HiSeq 3000 using stranded mRNA (polyA-selected) sample prep kit, or with paired-end 2×150 bp on the Illumina NovaSeq6000 using the llumina Tru-seq total RNA sample prep kit. All samples were sequenced to a depth of 30e6 reads.
Tumor DNA isolation and sequencing
Additional sequencing was performed with the Tempus xO assay that combines a 1,711 targeted tumor and non-tumor (germline) DNA-sequencing with RNA sequencing to detect somatic alterations, germline variants and mRNA fusion events from chromosomal rearrangements. DNA was extracted from patient FFPE tumor tissue and peripheral blood mononuclear cells (germline), libraries constructed and sequencing performed on an Illumina HiSeq 4000 to a minimum depth of 300x. RNA was extracted from FFPE tumor tissue and after cDNA synthesis with random primers, a library was prepared from poly(A) selected transcripts and sequenced to obtain at least 500 million reads. Detailed methods on variant calling, copy number variation and fusion identification were performed as previously described(22).
RNA sequencing analysis
Reads were aligned to the hg38 genome using the STAR aligner 2-pass method(23) and gene-level read counts were quantified using subREAD(24). RNA expression transcripts per million (TPM) estimates were determined using default parameters of salmon(24,25). Variant calling was performed following the GATK Best Practices recommendations and used base quality score recalibration, indel realignment, duplicate removal, and variant/indel calling(26). Genetic variants were annotated with Annovar(27). Gene fusions were detected using EricScript(27,28) and Mapsplice 2.0(29). Differentially expressed genes were identified using DESeq2 correcting for batch and timepoint. Sequencing batch effects were removed with the combat function of the R package (sva)(30).
TNBCtype
Normalized, batch-corrected, log2 transformed, gene-level RNA expression from variance stabilized transformed data was used as input to determine TNBCtype (http://cbc.mc.vanderbilt.edu/tnbc/) as previously described(31). The highest correlation coefficients were used to assign subtypes to either BL1, BL2, M or LAR subtype.
Statistics
The overall PFS data for the patients at the dose recommended for phase II were estimated using the Kaplan-Meier method with 95% confidence intervals. Logrank tests were performed to determine statistical significance between stratified populations in survival plots. Significance testing for CBR16 were performed with Fisher’s exact test.
Results
Patient screening, demographics, and baseline characteristics
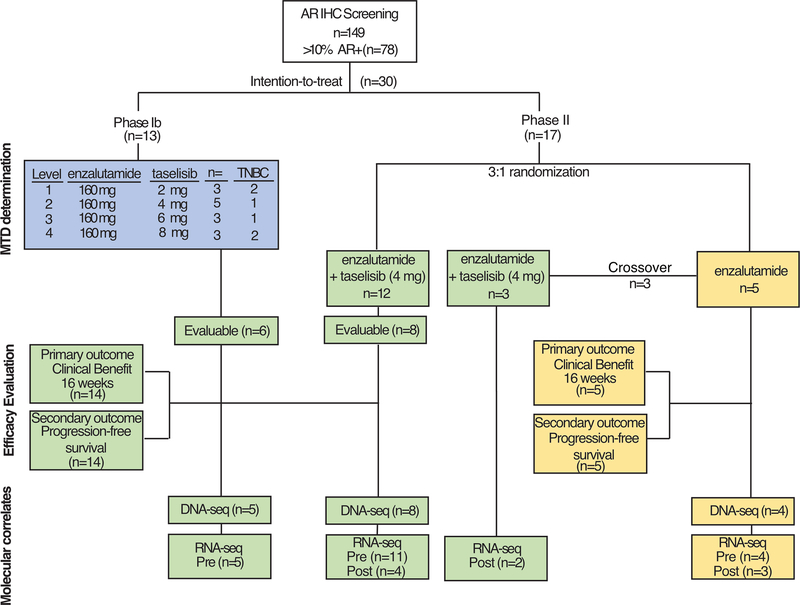
AR protein expression was assessed for 149 patients by IHC performed in central CLIA-approved laboratory and scored by a board-certified pathologist with breast pathology expertise. Patients could be prescreened for eligibility while receiving other treatment. Of the TNBC patients prescreened for phase II, one-half of the patients screened (63 of 127; 49.6%) were positive for AR (≥10% positive cells) and eligible for enrollment. Of these prescreened patients, 13 were enrolled in the phase I dose escalation and 17 in the phase II portion (Fig. 1). In the phase Ib dose escalation, 13 patients with AR+ tumors received 160 mg oral enzalutamide daily in combination with 2 mg (n=3), 4 mg (n=6), 6 mg (n=3) or 8 mg (n=2) oral taselisib (Fig. 1). Two patients receiving the 4 mg dose of taselisib were removed for toxicities. Seven of the patients were ER+/PR+ and six of the patients were TNBC.
Figure 1.
CONSORT diagram for enrollment onto TBCRC 032.
A total of 17 women were enrolled in the phase II portion at six cancer centers between May 2015 and August 2018 and randomized to either receive enzalutamide (n=5) or enzalutamide plus taselisib (n=12), representing the intention-to-treat (IIT) population in the phase II portion (Fig. 1 and Supplemental Table 1). Patients had previously received between 0 and 11 prior lines of therapy for metastatic disease with an average of 3.4 (+/− 2.6) prior therapies. In December 2018, the study was terminated due to the results of the phase III SANDPIPER trial showing the limited benefit of taselisib in metastatic breast cancer and a decision to halt development of the drug(32). Four patients were removed for drug-related toxicity. Table 1 lists the overall demographics and patient characteristics. The majority of patients were white (93%) followed by black/African American (3%) and Hispanic/Latino (3%). The average age at enrollment was 59.1 years (+/− 9.2) and number of days on treatment was 120+/− 116 days. The majority of women enrolled were post-menopausal (72%). Of the TNBC patient tumors sequenced (n=21 of the total 28 analyzed), 38% (n=8) carried a PIK3CA mutation. Mutations in the PIK3CA kinase domain (H1047R) were the most common (n=3), followed by mutations in the helical domain (E542K and E545K, n=2), then mutations in either the C2 domain (C420R, n=1) or Ras-binding domain (G118D, n=1). A summary of all patient clinical data is available in Supplemental Table S1.
Table 1.
Patient Demographics for all patients enrolled on TBRC032
| Age | 59.1 +/−9.2 | |
| Race | ||
| Black/AA | 3% ( 1) | |
| White | 93% (28) | |
| Unknown | 3% ( 1) | |
| Ethnicity | ||
| Hispanic/Latino | 3% ( 1) | |
| Non-Hispanic | 83% (25) | |
| Unknown | 13% ( 4) | |
| TNBCtype | ||
| Basal-like 1 | 25% (5) | |
| Basal-like 2 | 10% (2) | |
| Luminal AR | 40% (8) | |
| Mesenchymal | 25% (5) | |
| PIK3CA | ||
| Wild-type | 50% (14) | |
| Mutant | 50% (14) | |
| Menopausal Status | ||
| Pre-menopausal | 67% (20) | |
| Pre-menopausal | 17% ( 5) | |
| Hysterectomy | 13% ( 4) | |
| Male | 3% ( 1) |
Determination of MTD and safety assessment
To evaluate safety of the combination, 160 mg enzalutamide were administered in combination with taselisib starting dose of 2 mg and escalated to 4 mg, 6 mg and 8 mg in a traditional 3+3 trial design. In order to increase enrollment, AR+/ER+ patients were allowed to enrolled in the dose escalation, as these patients also had the potential to benefit from the treatment. We completed dose-escalation with 13 patients (7 ER+ and 9 TNBC) with none of the patients experiencing a DLT as per pre-defined criteria outlined above in Efficacy Endpoints. During dose-escalation, 50% of patients experienced a grade 3 treatment-related adverse event (AE) on any dose, with grade 3 hyperglycemia and skin rashes occurring in 25% and 33% of patients respectively, particularly at higher taselisib doses (Table 2). The hyperglycemia and rash were consistent with prior observations with taselisib and other PI3K inhibitors (21,31) and were controlled with anti-hyperglycemic medication and topical steroids or antihistamines within 7 days, thus not meeting criteria for DLT. Based on adverse events and discussions with personnel at Genentech and experience in other studies, the phase II dose was determined to be 4 mg taselisib in combination with 160 mg enzalutamide.
Table 2.
Summary of adverse effects to enzalutamide and taselisib combination
| Phase I | |||||
| Toxicity Category | Toxicity Code | Relationship | Taselisib (mg) | Grade 3 (n/%) | Grade 4 (n/%) |
| Metabolism and nutrition | Hyperglycemia | Definite | 4 | 3 (25.0) | |
| Skin and subcutaneous | Rash acneiform | Definite | 4 | 1 ( 8.3) | |
| Rash maculo-papular | Definite | 6,8 | 3 (25.0) | ||
| Investigation | Elevated alkaline phos | Possible | 4 | 1 ( 1.8) | |
| Phase II | |||||
| Toxicity Category | Toxicity Code | Relationship | Grade 3 (n/%) | Grade 4 (n/%) | |
| Blood and lymphatic | Febrile neutropenia | Possible | 1 ( 5.9) | ||
| Anemia | Possible | 1 ( 5.9) | |||
| Investigation | Elevated alkaline phos | Possible | 1 ( 5.9) | ||
| Gastrointestinal disorders | Nausea | Probable | 1 ( 5.9) | ||
| General disorders | Fatigue | Possible | 2 (11.8) | ||
| Fever | Possible | 1 ( 5.9) | |||
| Metabolism and nutrition | Dehydration | Possible | 1 ( 5.9) | ||
| Skin and subcutaneous | Rash maculo-papular | Probable | 3 (17.6) | 2 (11.8) | |
| Rash acneiform | Definite | 2 (11.8) | |||
| Pruritus | Definite | 1 ( 5.9) | |||
| Vascular disorders | Hypotension | Probable | 1 ( 5.9) | ||
Efficacy
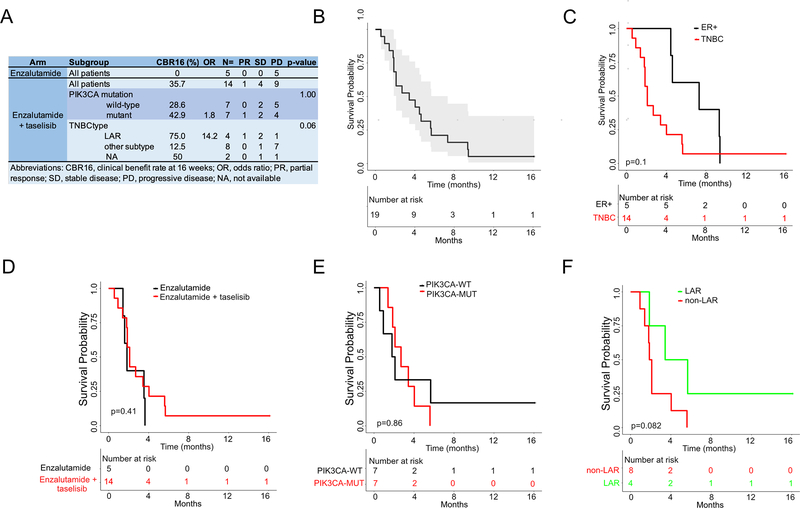
The primary outcome for the phase II portion of the trial was clinical benefit rate at 16 weeks (CBR16). Since the study was terminated prior to completion, we performed an analysis of evaluable TNBC patients who had at least one cycle of treatment and had their disease re-evaluated at 16 weeks or progressed prior to the 16-week assessment. Efficacy was evaluated in TNBC patients that received enzalutamide (n=5) or the combination of enzalutamide and taselisib (n=14). Patients that were removed from the trial due to toxicity (n=4), were not evaluated for efficacy. Evaluable patients receiving the combination included 6 TNBC patients from phase IB and 8 from phase II.
All of the patients on the enzalutamide alone arm had progression of disease at 16 weeks. Three patients in the enzalutamide-only arm crossed over to receive the combination, of which one had a partial response with the combination after crossing over. Evaluable patients receiving the combination had a clinical benefit rate of 35.7% with one patient achieving a partial response and four others displaying stable disease (Fig. 2A). There was not a significant difference in clinical benefit in the PIK3CA mutant TNBC population compared to PIK3CA wild-type (42.9% vs. 28.6% p=1.00). AR protein (% positive) by IHC displayed no trend according to response, but correlation strength to the LAR subtype appeared higher in responding patients (Fig. S1). TNBC patients with LAR subtype tumors trended towards better clinical benefit rate compared to all other subtypes (75% vs. 12.5%, p=0.06). Due to the limited sample size, these efficacy studies should be interpreted with caution. All patients progressed on the combination except for one LAR TNBC patient who continued to receive treatment without progression at 18 months when the study was terminated.
Figure 2. Efficacy of enzalutamide and taselisib in evaluable AR+ TNBC patients.
A, Clinical benefit rate (%) at 16 weeks for all TNBC patients treated with enzalutamide alone or in combination with taselisib stratified by PIK3CA mutation status and LAR subtype. Kaplan-Meier survival plots of progression-free survival (PFS) in B, all evaluable patients C, TNBC patients, or TNBC patients stratified by D, treatment arms, E, PIK3CA mutation status or F, TNBC subtype. Indicated p-values obtained by log-rank test.
The secondary outcome of median progression-free survival was 3.4 months for all evaluable patients receiving the combination (Fig. 2B). Median PFS was substantially lower for TNBC patients compared to ER+ patients (2.1 vs. 7.2 months, p=0.10) (Fig. 2C). There was no significant difference in PFS for evaluable TNBC patients receiving the combination compared to enzalutamide alone (2.7 vs. 2.0 months, p=0.41) (Fig. 2D). Patients with PIK3CA mutant TNBC did not have improved median PFS (2.7 vs. 2.0, p=0.83) compared to patients whose tumors had wild-type PIK3CA (Fig. 2E). However, patients with LAR subtype tumors exhibited a trend towards a better median PFS compared to all other subtypes (4.6 vs. 2.0 months, p=0.08), consistent with improved the clinical benefit rate observed at 16 weeks (Fig. 2F).
Adverse Events/Safety
The most common reason for treatment discontinuation was progressive disease; however, six patients came off study for toxicity reasons in the phase II. The phase Ib portion of the study was successfully completed without DLTs and a determination of a MTD of 8 mg of taselisib with enzalutamide. Based on other studies and discussions with the sponsor, taselisib 4 mg with enzalutamide was the recommended dose for phase II. The first six patients accrued to the phase II developed grade 3 or 4 rashes and the study was put on hold. After a thorough investigation, no cause was found, and this was thought to potentially be an issue with the batch of taselisib as not one of over 10 patients in the 4 mg or higher dose level in the phase I portion experienced such a severe reaction. The study re-opened with a new batch of taselisib, and two patients on the enzalutamide-only arm crossed over from enzalutamide alone to enzalutamide with taselisb and one new patient enrolled on the combination arm; none of these patients developed a rash. One patient was still receiving the combination of enzalutamide and taselisib at the end of the study, 616 days after cycle day 1. Adverse events of any grade or relationship occurred in 53% (9/17) of patients enrolled in the phase II portion and are listed in Supplemental Table S2. The most common adverse event was skin rash, which occurred in 29.4% (6/17) of patients (Table 2).
Correlative Analysis
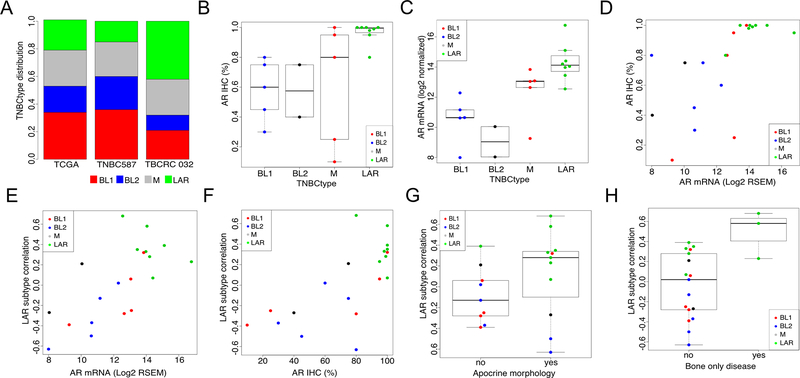
To determine baseline gene expression and subtype composition, RNA-seq was performed on RNA isolated from pre-treatment metastatic biopsies from 21 TNBC patients enrolled on the trial (5 patients from the phase Ib and 16 patients from the phase II portion of the trial). TNBC subtyping of pre-treatment biopsies resulted in a distribution of 38% LAR (n=8), 24% M (n=5), 24% BL1 (n=5) and 14% BL2 (n=3). As expected from the clinical trial enrollment criteria of >10% AR+ tumor cells, this distribution represents a near two-fold enrichment in LAR tumors compared to relative distribution of TNBC tumors in The Cancer Genome Atlas (TCGA) (21.6%, n=176)(3) and a meta-analysis of 587 TNBC tumors (15.0%)(2) (Fig. 3A); there was large range of AR positivity (10–100%) with a median of 77.5% AR+ (Fig. S2). However, when grouped by TNBC subtypes, AR+ cells were significantly greater in the LAR subtype (median = 99%, p=0.0023), compared to all other subtypes (median=75%) (Fig. 3B). AR mRNA levels showed a similar pattern across subtypes (Fig. 3C), and both AR protein and mRNA levels were highly correlated (spearman =0.753) between the cohort of individual tumors (Fig. 3D). Tumors with the highest AR mRNA and protein levels had higher correlations to the LAR subtype (Fig. 3 E and F). Tumors with apocrine histological features were enriched in the LAR subtype (Fig. 3G). Tumors characterized by bone-only metastatic disease were significantly (p=0.032) enriched in LAR subtype (3 of 8, 37.5%) of TNBC patients compared to patients with non-LAR TNBC subtypes (0 of 12, 0%) suggesting LAR tumors have patterns of metastasis similar to metastatic ER+ tumors (Fig. 3H).
Figure 3. Comparison of AR protein, mRNA and TNBCtype.
A, Distribution of TNBCtype within TCGA (n=180), a microarray meta-analysis of TNBCs (n=587) and this study, TBCRC 032 (n=21). B, Percentage AR+ cells by TNBCtype. C, AR mRNA transcript levels (log2) by TNBCtype. D, Distribution of AR transcript and protein (IHC) levels by individual patients. Comparison of LAR subtype correlation strength and E, AR mRNA transcript levels or F, AR protein levels by individual patients. LAR subtype correlation stratified by G, apocrine morphology and H, presence of bone only clinical disease.
DNA Sequencing
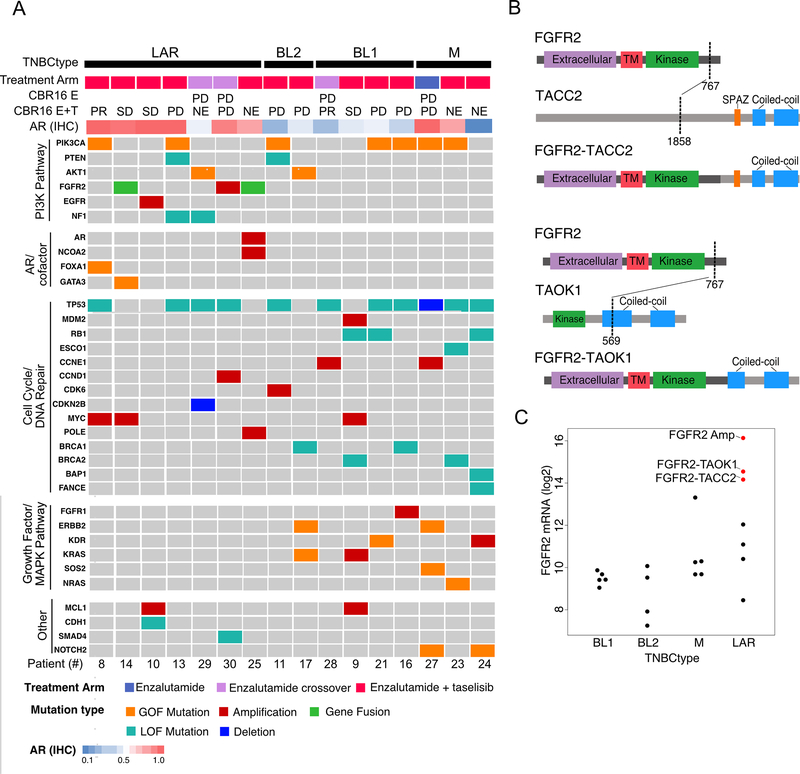
To determine the mutational landscape of patients’ tumors, we performed whole exome sequencing of DNA extracted from whole blood PBMCs and paired metastatic pre-treatment biopsies from 16 patients with evaluable specimens. The mutation burden ranged from 7 to 115 nonsynonymous mutations per tumor (average 30). We identified seven tumors distributed across all TNBC subtypes with activating PIK3CA mutations (H1047R, E545K, E542K, C420R and G118D) (Fig. 4A). Two tumors had activating AKT1 mutations (E17K) and two tumors had loss of function PTEN mutations (Fig. S3). FGFR amplifications and fusions were identified and exclusive to LAR tumors. Deleterious NF1 mutations were exclusive to LAR tumors and occurred in non-responding patients. One LAR tumor had co-amplification of AR and the nuclear coreceptor NCOA2, contributing to high levels of AR transcript (Fig. S4). LAR tumors were enriched in late frameshift mutations beyond the DNA binding domains in two chromatin modifying genes (GATA3 and FOXA1). Loss-of-function TP53 mutations were the most frequent alteration occurring across all subtypes (68.8%) as previously reported(33); and MDM2 amplification occurred in one patient with wild-type TP53. Non-LAR tumors were enriched in deleterious mutations in cell cycle (RB1; S82X and R467X), mitotic (ESCO1) and DNA-repair genes (BRCA1, BRCA2, BAP1 and FANCE) (Fig. 4A). Two tumors had deleterious mutations in BRCA1 (Q94X and S1617X) and one tumor had two deleterious BRCA2 mutations (W31X and E33X). Activating mutations in MAPK pathway (KRAS; G12D, NRAS; Q61K and SOS2; N426fs) as well as growth factor receptors (ERBB2, KDR and FGFR1) were identified in non-LAR tumors. All genetic DNA variants are available in Supplemental Table S3.
Figure 4. Genomic alterations in pre-treatment biopsies by TNBCtype.
A, Oncoplot shows the distribution of activating (gain-of-function (GOF) mutations, amplifications and gene fusions) and loss-of-function (LOF, mutations and deletions) alterations in genes organized by driver pathways and grouped by TNBC subtype. B, Diagrams show the protein domain structure of novel transcripts with FGFR2-fused to either TACC2 (upper) or TAOK1 (lower); abbreviation: TM, transmembrane. C, Beeswarm plot shows FGFR2 transcript levels by TNBCtype, with both FGFR2-amplified and FGFR2-fused tumors indicated in red. Abbreviations: CBR16, clinical benefit at 16 weeks; E, enzalutamide; T, taselisib; PD, progressive disease; SD, stable disease; PR, partial response; NE, not evaluated.
Gene Fusion Analysis
Since gene fusions frequently occur in AR-driven prostate cancer(34), we analyzed RNA-seq data generated from patients’ tumors for evidence of reads spanning two different genes. We identified two gene fusions (FGFR2-TACC2 and FGFR2-TAOK1) involving the kinase domain of fibroblast growth factor receptor 2 (FGFR2) in patient tumors of the LAR subtype. The breakpoints occurred after exon 17 of FGFR2 (chr10:121483697), fusing the FGFR2 kinase domain to the coiled-coil domains of either TACC2 or the serine/threonine-protein kinase TAOK1 (Fig. 4B and C, Supplemental Table S4). The FGFR2-TACC2 fusion is similar to oncogenic FGFR3-TACC3 fusions identified in glioblastomas(35) and likely activating, as we have previously shown for a FGFR3-TACC3 gene fusion in a LAR TNBC cell line(36). The tumor with the FGFR2-TACC2 fusion is also FGFR2 gene amplified; and, tumors with either amplifications or gene rearrangements expressed the highest levels of FGFR2 transcript (Fig. 4C). These fusions represent another mechanism by which LAR tumors activate the PI3K pathway.
Analysis of Gene Expression in Pre- and Post-treatment Tumor Biopsies
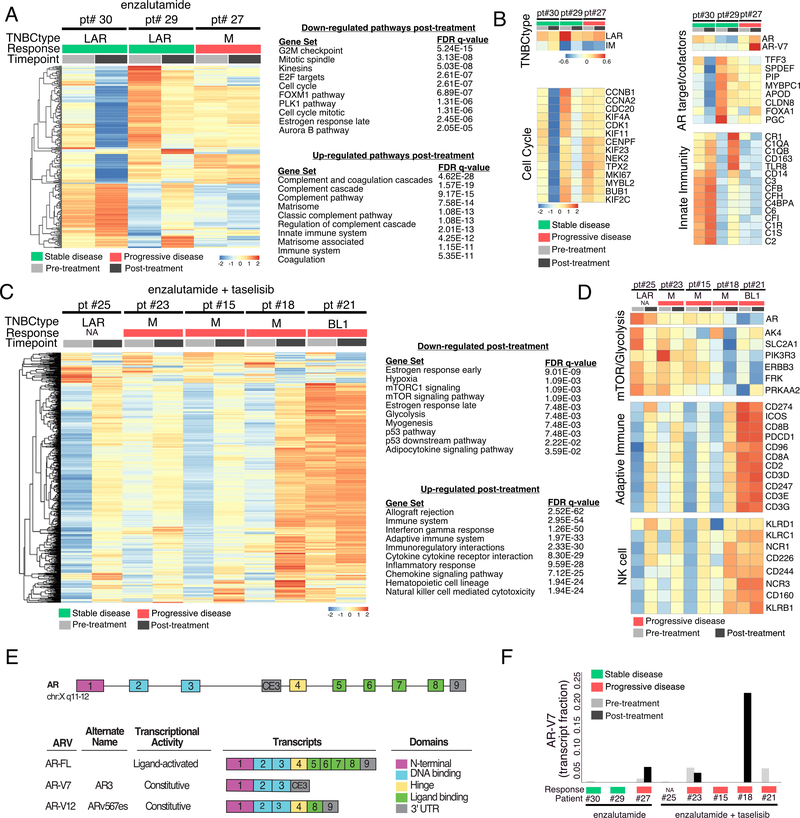
To assess changes in gene expression in tumors after enzalutamide treatment alone or in combination with taselisib, we performed RNA-seq on patient matched pre-treatment and cycle 2 (day 14–21) post-treatment metastatic site biopsies. Sequencing batch effects were removed as illustrated by principal component analysis (Fig. S5). For enzalutamide treatment alone, we identified differentially expressed genes from three available paired patient tumors before and after enzalutamide treatment (Fig. 5A). Gene ontology analysis of differentially expressed genes showed decreased enrichment in cell proliferation pathways (G2M checkpoint, mitotic spindle, kinesins, E2F targets, cell cycle, PLK1 pathway and Aurora B pathway) as well as estrogen receptor late genes after enzalutamide treatment (Fig. 5A). Pathways associated with innate immunity (complement and innate immune system) were increased in post-treatment tumors. Detailed analysis of individual tumors showed that gene expression differences were primarily driven by two LAR tumors (patient #30 and patient #29) that achieved stable disease, while the third M subtype tumor (Pt #27; who had progressive disease) had similar gene expression patterns before and after treatment (Fig. 5A). Consistent with these findings were the decreases in correlation strength to the LAR subtype and increased immunomodulatory (IM) subtype correlation observed in the responding LAR patients but not in the non-responding M patient (Fig. 5B). Cell cycle genes (MKI67, CCNB1, CCNA2, KIF11, BUB and KIF2C) were decreased after treatment in the two responding LAR patients (patient #30 and patient #29), suggesting enzalutamide inhibited proliferation. Additional evidence of enzalutamide activity is demonstrated by decreases in AR transcript and AR target genes (TFF3, SPDEF, PIP, MYBPC1, APOD, CLDN8, FOXA1 and PGC) in LAR tumors with stable disease and not in the M-subtype tumors of patients with progressive disease.
Figure 5. Gene expression in patient tumors before and after treatment.
A, Heatmap shows differentially expressed genes and gene ontology pathways from three patient tumors, which were decreased or increased after cycle 2 (day 14–21) of enzalutamide treatment. B, Heatmaps highlight specific changes in gene expression from patient matched biopsies grouped by TNBC subtype, cell cycle genes, AR gene targets/cofactors, and innate immune genes. C, Gene expression changes and significantly enriched pathways in patient tumors before and after treatment with enzalutamide + taselisib. D, Heatmaps highlight specific changes in genes involved in mTOR signaling and adaptive immunity before and after treatment with the combination of enzalutamide and taselisib. E, Diagram depicts AR gene coding structure for full-length (FL) and alternatively spliced AR transcripts; numbers represent exons and cryptic exon 3 (CE3). F, Quantification of AR-V7 transcript (fraction relative to all AR transcripts) in pre-treatment and post-treatment biopsies of patients treated with enzalutamide or enzalutamide and taselisib. All colorbars representing response are derived from CBR16.
We performed a similar analysis of tumors from patients treated with the combination of enzalutamide and taselisib and found similar decreases in estrogen response genes, but also decreases in genes involved in mTOR signaling and glycolysis after treatment (Fig. 5C). The expression of genes involved in the immune system was increased after treatment, but in contrast to the enrichment in expression of genes involved in innate immunity and complement pathways that occurred after enzalutamide treatment alone, enzalutamide + taselisib treated tumors were enriched in adaptive immunity (Fig. 5C). Specifically, T-cell markers (CD8A, CD8B, CD3D, CD3E, CD3G, CD2, ICOS) and natural killer cell markers (KLRB1, KLRC1, KLRD1, NCR1, NCR3, CCD160, CD226 and CD244) were increased after treatment with the combination of enzalutamide and taselisib (Fig. 5D).
Since AR transcript levels increased and AR target genes did not change in the nonresponding patient #27 (Fig. 5B), we performed a detailed analysis of AR splice variants. We identified full-length AR transcript and two constitutively active splice variants (AR-V7 and AR-V12) lacking the ligand binding domain in four patients (Fig. 5E and Table S5). The AR-V7 transcript was present in three patients prior to treatment and increased in two patients post treatment, including patient #27 that did not respond to enzalutamide alone (Fig. 5F). The AR-V7 transcript was not present in the two patients that had stable disease after enzalutamide treatment alone, suggesting that this splice variant may drive resistance to AR antagonists in TNBC. Gene expression changes after enzalutamide treatment provide evidence that enzalutamide hit ‘target’; however, these molecular changes consistent with drug efficacy were limited to patients with the LAR tumor subtype.
Discussion
In the phase I portion we determined that 160 mg enzalutamide could be safely administered in combination with 4 mg taselisib with manageable toxicities, the most frequent being hyperglycemia and rash, particularly at higher 6 and 8 mg doses. In the phase II portion of the clinical trial, we examined the efficacy of enzalutamide in combination with the PI3K inhibitor taselisib in metastatic AR+ breast cancer. While the phase II portion was not completed due to the termination of the development of taselisib, we did enroll 17 of the proposed 49 patients. These patients did experience significant grade 3/4 rashes which may have been an effect of the batch of taselisib, as this was not observed in the phase Ib, however future studies could explore combinations with other PI3K inhibitors that have more tolerable side effect profiles such as apelisib (37). The correlatives from this biopsy-rich trial revealed interesting biology and potential mechanisms of resistance to anti-AR therapy in breast cancer, consistent with previous reports in prostate cancer(38,39). The primary endpoint was CBR at 16 weeks; 35.7% in evaluable patients treated with the enzalutamide and taselisib combination received clinical benefit at 16 weeks compared to none treated with enzalutamide alone. Stratification by PIK3CA mutation status showed no difference in clinical benefit for PIK3CA-mutated patients receiving the combination. However, when stratified by TNBC subtype, clinical benefit trended higher in the LAR subtype (p=0.06) with 75.0% of LAR patients receiving clinical benefit compared to 12.5% for all other subtypes. This increase in CBR was associated with increased in longer progression-free survival of LAR patients (4.6 vs. 2.0 months, p=0.08) compared to other subtypes. Thus, in addition to expressing AR protein, the presence of the LAR gene signature may identify those patients most likely to receive clinical benefit from AR antagonists. Interestingly, one patient achieved a partial response to the combination after progressing on enzalutamide alone, suggesting that combinatorial strategies will likely be needed for AR+ TNBC patients to achieve significant clinical benefit in the future. The evaluation of efficacy should be considered exploratory due to sample size limitation; and, observations should be confirmed in future trials.
Anti-androgen therapy was first evaluated in TNBC patients in a metastatic phase II Translational Breast Cancer Research Consortium (TBCRC 011) study that enrolled patients based on nuclear AR expression by IHC in ≥10% of tumor cells ( NCT00468715). In that study 12% of patients screened were AR+ and received 150 mg oral bicalutamide daily. While there were no complete or partial responses, bicalutamide was well-tolerated demonstrating some efficacy with a 19% CBR at six months(14). A recent phase II trial evaluated the second-generation AR-antagonist enzalutamide in women with advanced AR+ (AR IHC >0%) TNBC (NCT001889238)(13,40). In the 75 evaluable patients, the CBR was 33% at 16 weeks, meeting its primary objective and demonstrating clinical activity of enzalutamide in AR+ TNBC. The AR positive frequency in TNBC patients from that study (n=368) was 55% and similar to the current study (n=127) of 50% using the same cutoff of ≥10% AR+ tumor cells. However, an AR IHC score of ≥10% may not adequately identify AR-dependent tumors, since AR-positivity in tumors was not predictive of response in the prior study (NCT001889238) (40) and LAR subtype tumors were >80% AR+ in this study and associated more frequently with response.
RNA-seq analyses performed on pre- and post-treatment biopsies of enzalutamide-treated patients demonstrated that LAR subtype tumors displayed the greatest change in gene expression with decreases in the expression of cell cycle, AR and AR target genes, and increased expression of adaptive immune system related genes after treatment. These gene expression changes and decreased correlation to the LAR subtype after enzalutamide treatment could be used to identify patients in which AR-antagonists should be efficacious. Patients receiving the combination displayed decreased expression of genes involved in mTOR signaling and increased expression of genes related to adaptive immunity after treatment. These data are consistent with downstream PI3K inhibition, and the increase in expression of innate immune genes rather than adaptive immune genes, observed in patients treated with enzalutamide alone, suggests that the combination of PI3K inhibition and AR antagonism alters the immune response.
In prostate cancer, failure of androgen deprivation therapy is linked to AR gene amplifications(38) and increased expression of constitutively active, alternatively spliced AR transcripts such as AR-V7 that lack the C-terminal ligand-binding domain(39). Constitutively active AR variants are detected in breast cancer and have been shown to induce proliferation of a LAR cell line in the presence of enzalutamide(41). Herein, we identified one LAR patient with co-amplification of AR and the nuclear co-receptor NCOA2, both of which have previously been shown to promote castration-resistant prostate cancer(42,43). Further, the AR-V7 splice variant was present and increased post-treatment in two tumors from patients receiving enzalutamide alone or in combination with taselisib, suggesting that AR-driven cancers, regardless of tissue origin, evolve similar resistance mechanisms.
We identified RB1 mutations in 3 of 9 non-LAR and 0 of 5 LAR tumors. The lack of RB1 mutations and the preclinical efficacy of the cyclin-dependent kinase 4/6 (CDK4/CDK6) inhibitor palbociclib in all LAR cell line models(44) suggest an alternative enzalutamide combination therapy could include a CDK4/6 inhibitor. Furthermore, the gene encoding cyclin D1, CCND1, is a major transcriptional target of AR(45,46) and CDK4/6 inhibitors restore activity of anti-androgen treatment in castration resistant prostate cancer harboring an activating AR mutation(47). Currently, there is an ongoing phase I/II trial exploring the combination of palbociclib and bicalutamide for the treatment of metastatic AR+ TNBC ( NCT02605486) and the field awaits the results.
Genomic analysis of tumor biopsies in LAR tumors lacking PIK3CA mutations revealed novel gene fusions containing the kinase domain of FGFR2 fused 3’ to the coiled-coil domains of either TACC2 or TAOK1. The gene fusions were present in tumors from one patient with stable disease on the enzalutamide + taselisib combination, and from another patient that progressed on enzalutamide alone but achieved stable disease on the combination after crossover. These correlative genomic data suggest that the encoded FGFR2 fusion gene products may be drivers for the tumor cells harboring them and confer tumor sensitivity to PI3K inhibition, similar to the distinct reliance of FGFR2-amplified cell lines on PI3K signaling compared to other FGFR-driven cell lines(46). The transforming ability of FGFR3-TACC3 gene fusions was first described in glioblastomas(35), and subsequently in lung adenocarcinomas, lung squamous carcinomas, head and neck, bladder, cervical, and esophageal carcinomas. We previously identified FGFR3-TACC3 gene fusions in a LAR TNBC patient’s tumor and the LAR cell line SUM-185PE(36). The latter cell line displayed growth dependency on the gene fusion protein product and sensitivity to FGFR pharmacological inhibition(36). Recently, FGFR1-TACC1 fusions were identified as a recurrent alteration in extraventricular neurocytomas(36,48), demonstrating selective preference between the individual FGFR genes and TACC gene partners. This preference is the result of proximity of FGFR and TACC genes, as they retain the same orientation and close physical association, separated by only 255 kb (FGFR1 and TACC1), 386 kb (FGFR2 and TACC2) or 63 kb (FGFR3 and TACC3) on three different chromosomes (8p11, 10q26, 4p16, respectively). The FGFR2-TACC2 discovered in this study likely functions similarly to previously characterized FGFR1-TACC1 and FGFR3-TACC3 fusions. AR antagonists combined with FGFR inhibitors may be a reasonable combination for future clinical trials in AR-driven TNBCs.
In conclusion, this phase Ib/II study demonstrated enzalutamide can be given safely in combination with taselisib. Despite early trial termination, the combination appeared to increase clinical benefit in TNBC patients with AR+ tumors compared to enzalutamide alone, especially in those patients with LAR subtype tumors. Patients with LAR subtype tumors showed decreases in proliferation and AR-target gene expression after treatment with enzalutamide and had greater clinical benefit, suggesting AR IHC alone may not be sufficient to identify AR signaling-dependent tumors. Tumor genotyping/biomarker approaches involving RNA-seq may be necessary to identify patients most likely to benefit from AR antagonist-based therapies. Genomic analysis of patient tumors identified novel FGFR2 fusions that likely activate the PI3K pathway and AR splice variants that may contribute to enzalutamide resistance. Given the lack of effective non-chemotherapy based treatment options for metastatic TNBC, exploring subsets of TNBC that may respond to novel treatments is of the utmost importance; further studies are warranted to explore new combinations in AR+ TNBC.
Supplementary Material
Translational relevance.
Triple-negative breast cancer (TNBC) is a heterogeneous collection of biologically diverse cancers with a subset characterized by luminal gene expression, androgen receptor (AR) expression and enrichment of phosphatidylinositol-4, 5-bisphosphate 3-kinase (PIK3CA) activating mutations. Through the phase I/II trial described herein, we demonstrated the efficacy of the AR-antagonist enzalutamide alone or in combination with the PI3K inhibitor taselisib in AR-positive, metastatic TNBC. Pre-treatment and post-treatment biopsies were analyzed for molecular features associated with response. Patients with luminal AR (LAR) subtype tumors had increased clinical benefit, prolonged progression-free survival and gene expression changes consistent with AR signaling inhibition. FGFR2 amplifications and fusions were unique to the LAR subtype. The AR splice variant (AR-V7) which lacks ligand binding domain was expressed in a non-responding patient, suggesting a potential mechanism of enzalutamide resistance.
Acknowledgements
The authors thank all the patients who generously volunteered to participate in this study and the TBCRC investigators, research nurses, and study coordinators for their efforts on behalf of the patients. The authors are grateful to Thomas Stout, PhD for critical review of the manuscript. The authors also thank the Translational Breast Cancer Research Consortium and Susan G. Komen for the funding support.
Funding
This research was supported by: NCI/NCI grants CA098131 and CA068485 (JAP), and Susan G. Komen grants SAC110030 (JAP) and CCR13262005 (BDL). We would like to thank Vanderbilt Technologies for Advanced Genomics (VANTAGE) and the Translational Pathology Shared Resources for providing technologies supported by NIH S10 instrumentation grants RR028106, RR027764 and OD023475.
List of abbreviations
- AE
Adverse effect
- AR
androgen receptor
- BL1
basal-like 1
- BL2
basal-like 2
- CBR
clinical benefit rate
- CR
complete response
- ECOG
Eastern Cooperative Oncology Group
- FFPE
formalin-fixed paraffin embedded
- ITT
intention-to-treat
- LAR
luminal androgen receptor
- MTD
maximal tolerated dose
- M
mesenchymal
- ORR
overall response rate
- PR
partial response
- PIK3CA
phosphatidylinositol-4, 5-bisphosphate 3-kinase
- PFS
progression-free survival
- RECIST
Response evaluation criteria in solid tumors
- SD
stable disease
- TNBC
triple-negative breast cancer
- TPM
transcripts per million
Footnotes
Declarations
Ethics approval and consent to participate
The study was conducted in accordance with Good Clinical Practice guidelines and the Declaration of Helsinki. Written informed consent was obtained from all patients before enrollment, in agreement with approved protocols from respective ethics committees at every site.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the SRA repository (PRJNA562162) and processed data for the current study are available from the corresponding author on reasonable request.
Conflict of interest statement: BDL and JAP are inventors (PCT/US2012/065724) of intellectual property (TNBCtype) licensed by Insight Genetics Inc.
References
- 1.Harano K, Wang Y, Lim B, Seitz RS, Morris SW, Bailey DB, et al. Rates of immune cell infiltration in patients with triple-negative breast cancer by molecular subtype. PLoS One. 2018;13:e0204513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One. 2016;11:e0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niemeier LA, Dabbs DJ, Beriwal S, Striebel JM, Bhargava R. Androgen receptor in breast cancer: expression in estrogen receptor-positive tumors and in estrogen receptor-negative tumors with apocrine differentiation. Mod Pathol. 2010;23:205–12. [DOI] [PubMed] [Google Scholar]
- 5.Collins LC, Cole KS, Marotti JD, Hu R, Schnitt SJ, Tamimi RM. Androgen receptor expression in breast cancer in relation to molecular phenotype: results from the Nurses’ Health Study. Mod Pathol 2011;24:924–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park S, Koo J, Park HS, Kim J-H, Choi S-Y, Lee JH, et al. Expression of androgen receptors in primary breast cancer. Ann Oncol. 2010;21:488–92. [DOI] [PubMed] [Google Scholar]
- 7.McGhan LJ, McCullough AE, Protheroe CA, Dueck AC, Lee JJ, Nunez-Nateras R, et al. Androgen receptor-positive triple negative breast cancer: a unique breast cancer subtype. Ann Surg Oncol 2014;21:361–7. [DOI] [PubMed] [Google Scholar]
- 8.Vera-Badillo FE, Templeton AJ, de Gouveia P, Diaz-Padilla I, Bedard PL, Al-Mubarak M, et al. Androgen receptor expression and outcomes in early breast cancer: a systematic review and meta-analysis. J Natl Cancer Inst. 2014;106:djt319. [DOI] [PubMed] [Google Scholar]
- 9.Qu Q, Mao Y, Fei X-C, Shen K-W. The impact of androgen receptor expression on breast cancer survival: a retrospective study and meta-analysis. PLoS One. 2013;8:e82650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loibl S, Müller BM, von Minckwitz G, Schwabe M, Roller M, Darb-Esfahani S, et al. Androgen receptor expression in primary breast cancer and its predictive and prognostic value in patients treated with neoadjuvant chemotherapy. Breast Cancer Res Treat. 2011;130:477–87. [DOI] [PubMed] [Google Scholar]
- 11.Santonja A, Sánchez-Muñoz A, Lluch A, Chica-Parrado MR, Albanell J, Chacón JI, et al. Triple negative breast cancer subtypes and pathologic complete response rate to neoadjuvant chemotherapy. Oncotarget. 2018;9:26406–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Echavarria I, López-Tarruella S, Picornell A, García-Saenz JÁ, Jerez Y, Hoadley K, et al. Pathological Response in a Triple-Negative Breast Cancer Cohort Treated with Neoadjuvant Carboplatin and Docetaxel According to Lehmann’s Refined Classification. Clin Cancer Res. 2018;24:1845–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Traina TA, Miller K, Yardley DA, Eakle J, Schwartzberg LS, O’Shaughnessy J, et al. Enzalutamide for the Treatment of Androgen Receptor-Expressing Triple-Negative Breast Cancer. J Clin Oncol. 2018;36:884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gucalp A, Tolaney S, Isakoff SJ, Ingle JN, Liu MC, Carey LA, et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic Breast Cancer. Clin Cancer Res. 2013;19:5505–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmann BD, Bauer JA, Schafer JM, Pendleton CS, Tang L, Johnson KC, et al. PIK3CA mutations in androgen receptor-positive triple negative breast cancer confer sensitivity to the combination of PI3K and androgen receptor inhibitors. Breast Cancer Res. 2014;16:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Millis SZ, Gatalica Z, Winkler J, Vranic S, Kimbrough J, Reddy S, et al. Predictive Biomarker Profiling of > 6000 Breast Cancer Patients Shows Heterogeneity in TNBC, With Treatment Implications. Clin Breast Cancer. 2015;15:473–81.e3. [DOI] [PubMed] [Google Scholar]
- 17.Gasparini P, Fassan M, Cascione L, Guler G, Balci S, Irkkan C, et al. Androgen receptor status is a prognostic marker in non-basal triple negative breast cancers and determines novel therapeutic options. PLoS One. 2014;9:e88525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barton VN, D’Amato NC, Gordon MA, Lind HT, Spoelstra NS, Babbs BL, et al. Multiple molecular subtypes of triple-negative breast cancer critically rely on androgen receptor and respond to enzalutamide in vivo. Mol Cancer Ther. 2015;14:769–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas C, Lamoureux F, Crafter C, Davies BR, Beraldi E, Fazli L, et al. Synergistic targeting of PI3K/AKT pathway and androgen receptor axis significantly delays castration-resistant prostate cancer progression in vivo. Mol Cancer Ther. 2013;12:2342–55. [DOI] [PubMed] [Google Scholar]
- 21.Juric D, Krop I, Ramanathan RK, Wilson TR, Ware JA, Sanabria Bohorquez SM, et al. Phase I Dose-Escalation Study of Taselisib, an Oral PI3K Inhibitor, in Patients with Advanced Solid Tumors. Cancer Discov. 2017;7:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beaubier N, Tell R, Huether R, Bontrager M, Bush S, Parsons J, et al. Clinical validation of the Tempus xO assay. Oncotarget. 2018;9:25826–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benelli M, Pescucci C, Marseglia G, Severgnini M, Torricelli F, Magi A. Discovering chimeric transcripts in paired-end RNA-seq data by using EricScript. Bioinformatics. 2012;28:3232–9. [DOI] [PubMed] [Google Scholar]
- 29.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Li J, Gray WH, Lehmann BD, Bauer JA, Shyr Y, et al. TNBCtype: A Subtyping Tool for Triple-Negative Breast Cancer. Cancer Inform. 2012;11:147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baselga J, Dent SF, Cortés J, Im Y-H, Diéras V, Harbeck N, et al. Phase III study of taselisib (GDC-0032) fulvestrant (FULV) v FULV in patients (pts) with estrogen receptor (ER)-positive, PIK3CA-mutant (MUT), locally advanced or metastatic breast cancer (MBC): Primary analysis from SANDPIPER. Journal of Clinical Oncology. 2018. 36:18_suppl, LBA1006–LBA1006. [Google Scholar]
- 33.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaver TM, Lehmann BD, Beeler JS, Li C-I, Li Z, Jin H, et al. Diverse, Biologically Relevant, and Targetable Gene Rearrangements in Triple-Negative Breast Cancer and Other Malignancies. Cancer Res. 2016;76:4850–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N Engl J Med. 2019. May 16;380(20):1929–1940. [DOI] [PubMed] [Google Scholar]
- 38.Friedlander TW, Roy R, Tomlins SA, Ngo VT, Kobayashi Y, Azameera A, et al. Common structural and epigenetic changes in the genome of castration-resistant prostate cancer. Cancer Res. 2012;72:616–25. [DOI] [PubMed] [Google Scholar]
- 39.Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A, et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A. 2010;107:16759–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker JS, Peterson AC, Tudor IC, Hoffman J, Uppal H. A novel biomarker to predict sensitivity to enzalutamide (ENZA) in TNBC. Journal of Clinical Oncology. 2015. page 1083–1083. [Google Scholar]
- 41.Hickey TE, Irvine CM, Dvinge H, Tarulli GA, Hanson AR, Ryan NK, et al. Expression of androgen receptor splice variants in clinical breast cancers. Oncotarget. 2015;6:44728–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin J, Lee H-J, Wu S-P, Lin S-C, Lanz RB, Creighton CJ, et al. Androgen deprivation-induced NCoA2 promotes metastatic and castration-resistant prostate cancer. J Clin Invest. 2014;124:5013–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bubendorf L, Kononen J, Koivisto P, Schraml P, Moch H, Gasser TC, et al. Survey of gene amplifications during prostate cancer progression by high-throughout fluorescence in situ hybridization on tissue microarrays. Cancer Res. 1999;59:803–6. [PubMed] [Google Scholar]
- 44.Asghar US, Barr AR, Cutts R, Beaney M, Babina I, Sampath D, et al. Single-Cell Dynamics Determines Response to CDK4/6 Inhibition in Triple-Negative Breast Cancer. Clin Cancer Res. 2017;23:5561–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearson A, Smyth E, Babina IS, Herrera-Abreu MT, Tarazona N, Peckitt C, et al. High-Level Clonal FGFR Amplification and Response to FGFR Inhibition in a Translational Clinical Trial. Cancer Discov. 2016;6:838–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gong X, Litchfield LM, Webster Y, Chio L-C, Wong SS, Stewart TR, et al. Genomic Aberrations that Activate D-type Cyclins Are Associated with Enhanced Sensitivity to the CDK4 and CDK6 Inhibitor Abemaciclib. Cancer Cell. 2017;32:761–76.e6. [DOI] [PubMed] [Google Scholar]
- 47.Korpal M, Korn JM, Gao X, Rakiec DP, Ruddy DA, Doshi S, et al. An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide). Cancer Discov. 2013;3:1030–43. [DOI] [PubMed] [Google Scholar]
- 48.Sievers P, Stichel D, Schrimpf D, Sahm F, Koelsche C, Reuss DE, et al. FGFR1:TACC1 fusion is a frequent event in molecularly defined extraventricular neurocytoma. Acta Neuropathol. 2018;136:293–302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.