Abstract
Colorectal cancer is a leading cause of cancer-related death internationally, with mounting evidence pointing to the role of the microbiome in adenoma and cancer development. This article aims to provide clinicians with a foundation for understanding the field of research into the microbiome. We also illustrate the various ways in which the microbiota have been linked to colorectal cancer, with a specific focus on microbiota with identified virulence factors, and also on the ways that byproducts of microbiota metabolism may result in oncogenesis. We also review strategies for manipulating the microbiome for therapeutic effects.
Keywords: Microbiome, colorectal cancer, colorectal adenomas
Introduction
Colorectal cancer (CRC) is a leading cause of cancer-related death worldwide [1]. Diet and bad habits, such as high consumption of alcohol and cigarette smoking, have been accepted as associative factors that place individuals at increased risk for the development of disease. However, the precise changes that occur as a result of these factors have not been clearly identified. Other malignancies have been linked to infectious etiologies, as in the case of human papillomavirus infection and cervical cancer [2], or Helicobacter pylori (H. pylori) and gastric cancer [3-5]. With more than 1014 bacteria residing in the colon [6], it is logical that we are now looking toward the microbiome as a potential driver of neoplastic transformation.
Defining the role of the microbiome
The human microbiome refers to the community of microorganisms sharing space within the human body; it consists of myriad bacteria, as well as fungi and viruses. Bacterial species greatly outnumber other constituents of the microbiome. The words “microbiome” and “microbiota” are often used interchangeably. However, the term “microbiome” encompasses the entire environment (microorganisms, genomes and microenvironment), while the term “microbiota” refers specifically to the microbes themselves. There are over 3.8 × 1013 bacterial cells in the human body (compared to 3.0 × 1013 human cells), with the majority residing in the colon (1011/mL bacteria) [6]. This constitutes a colonic microbiome of 3 million genes, approximately 100 times the number of genes encoded in the human genome [6,7]. Through the National Institutes of Health (NIH) Human Microbiome Project (HMP), our knowledge of the microbiome in times of health and disease has grown rapidly [8,9]. There are 3 dominant bacterial phyla within the human colon, Firmicutes, Bacteroidetes and Actinobacteria [10], and the distribution of microbiota varies along the gastrointestinal tract. Firmicutes are generally gram-positive cocci and form endospores. Familiar examples include Streptococcus spp., Enterococcus spp., Lactobacillus spp. and Bacillus spp. Bacteroidetes are gram-negative bacilli and are best known for the obligate anaerobic genus Bacteroides. Actinobacteria are gram-positive organisms that include Streptomyces spp. and Bifidobacterium spp. Through the study of germ-free animals, researchers have demonstrated the importance of the commensal bacteria, with roles in immune development and metabolic functions [11-16]. The microbiota appear to have a symbiotic relationship with humans.
Each individual’s microbiome develops in infancy and persists over time, with some variation through the different stages of life [17-19]. When there are significant pathological shifts in the microbiota, this is termed dysbiosis, and is associated with multiple disease states. For instance, dysbiosis has been associated with premature birth, type II diabetes and metabolic disease [8,20]. Additionally, the Inflammatory Bowel Disease Multi’omics Database (IBDMDB) has demonstrated that patients with Crohn’s disease have different microbial communities with different transcriptional and metabolic profiles than normal individuals, and that these populations may change cyclically with relation to disease flares, findings which have been supported by multiple groups [7,21,22].
Research foundations
Understanding the role of the microbiome requires that we broaden our lens when assessing human health. Incorporating the microbiome in this way, we need to look at the interconnected relationships of the human gut ecosystem. To study this complex system, research has become increasingly collaborative. Terminology and study methods from the field of ecology, adapt at studying communities and their connections, are being applied to human health. Additionally, computational biostatisticians adept at managing large data sets are working with clinicians to understand these populations and their impacts on health and disease. Certainly, the HMP has brought many experts in the field together to help coordinate efforts and develop new techniques [8].
Analysis of the microbiome may be performed by 16S ribosomal RNA (rRNA) amplification sequencing or via whole-genome shotgun (WGS) sequencing. 16S rRNA sequencing is most common, and takes advantage of the fact that bacterial species have highly conserved sequences flanking phylogenetically informative hypervariable regions that allow for the construction of nearly universal primer sets. This allows for a cost-effective mechanism for identification and quantification of the microbiome. WGS, however, allows for higher definition of the microbiota, though with less information on the taxonomic profile of communities and at a higher cost. Traditional methods, such as quantitative real time polymerase chain reaction (qPCR), have fallen out of favor given the new sequencing methodologies, as qPCR is unable to assess the full microbial diversity.
Understanding the research does require clinicians to re-familiarize themselves with taxonomic classification, as findings may be reported at any rank based on the details obtained in the sequencing and data processing (Table 1). Data reporting may occur at any taxa, which may be challenging to conceptualize if one is not familiar with microbiology. Microbiota may be reported as clades, or groups of microbes clustered based on a common ancestor. Frequently, findings are reported in operational taxonomic units (OTUs). An OTU refers to a group of genetically similar microbes, using this similarity to suggest a close relationship, and it is used in many current publications. However, the field is currently undergoing a transition toward the use of amplicon sequence variants or sub-OTUs, which use individual DNA sequences from high-throughput marker gene analysis and have the ability to account for sequence-based error. The use of these newer methods allows for more granular sequence resolution, though some experts remain concerned that reliance on this level of detail may complicate the understanding of larger trends.
Table 1.
Bacterial scientific classification by rank, with examples of commonly referenced bacterial species. Note that there are frequent deviations from the standardized suffix assigned to each rank
Currently, most data focus on the microbial populations at one given time, though some have proposed that relative changes over time are more indicative of disease. Questions regarding the most important metrics are still being debated. Is it more important to measure the absolute microbial populations or the relative population frequencies? Alternatively, are the specific microbiota more critical in driving human health, or is population diversity more important? Multiple groups have attempted to evaluate changes in bacterial networks as the drivers of disease [23,24]. These techniques remain controversial, though there are several techniques for evaluation of interconnected communities, with the enterotype model being the most prevalent [25,26]. Enterotypes refer to distinct symbiotic groupings of microbiota in the intestinal ecosystem. Currently there are data to suggest that, in the right context, changes in specific bacterial populations, changes in overall diversity and changes in networks all may contribute to diseased states.
While a thorough review of the statistical methods used to analyze the microbiome would be outside the scope of this review, it is important for the clinician to understand that the field of analytics used to assess this interrelated field continues to develop. Studies frequently measure alpha-diversity, or the diversity within a given environment, versus beta diversity, which is a comparison between the species from different environments. Alpha diversity is often assessed using the Shannon Index. Often data are found to lack a normal distribution, requiring nonparametric statistical methods such as the Wilcoxon rank sum or Kruskal-Wallis tests. Additionally, given the complexities of the data sets, multidimensional analyses are the foundation. There are different techniques to assess differences between groups, but the most common tests are fundamentally similar to an analysis of variance (ANOVA) [27], including permutational multivariate ANOVA, and analysis of group similarities. Additionally, comparisons between host factors and differences in the microbiome diversity are often performed using Spearman or Kendall rank correlation, among others. However, more nuanced methods are also commonly utilized, which generally require partnerships with individuals who have specialized training.
Finally, it is worth noting that the majority of studies rely on data from fecal samples, as these are more easily obtained and less invasive than biopsy specimens. There are some data to demonstrate that the fecal microbiome may be different than the mucosal microbiome [28-30]. This may be especially relevant when we attempt to understand the changes occurring at the mucosal level early on in the transition toward malignancy. The tumor microenvironment has been shown to be unique [31], and so it may prove that the global dysbiosis does not fully characterize the local impacts of certain microbiota.
Pathological impacts of the microbiota
A growing body of literature now points toward the association between the microbiota and the formation of adenomatous polyps (AP) and CRC. Studies have repeatedly demonstrated that alterations in overall microbial diversity are associated with diseased states across the gastrointestinal tract, including esophageal, gastric and colorectal neoplasms [32-37]. However, we have learned from the case of gastric cancer and H. pylori that the neoplastic driver may be more specific to an organism than just a state of general dysbiosis. Therefore, we have suspected that there is a specific genus or species associated with CRC and we are working to define the interactions between these microbes and the human host. Multiple microbial species have been consistently associated with CRC, though it is difficult to establish if the microbiota is driving tumorigenic changes in colonic cells, or if the pre-malignant changes are driving the microenvironment and microbiota residing there. Saus et al refer to bacterial “drivers”—those which may trigger epithelial cell DNA damage—and “passengers”—those which gain a competitive advantage in the altered microenvironment [38]. As most of the research to date does not demonstrate the changes over time, it may be difficult to ascertain which bacteria are “drivers” and which are “passengers,” but the observed differences in populations associated with AP and CRC are likely to be a mix of both.
Microbiota and human disease
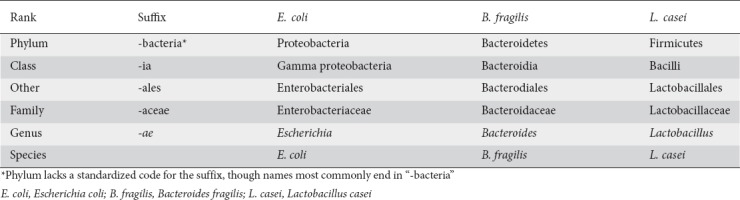
Human and animal studies using both fecal and tissue samples have demonstrated some consistent trends in the microbiota in relation to the development of colonic adenomas or neoplasms. The data have been reported at various taxa; however, there are overarching similarities in the findings. Fig. 1 shows the bacteria commonly associated with CRC and with adenoma formation, while maintaining the phylogenetic relationships of the microbes to each other. Microbiota commonly associated with CRC in human cohorts are primarily from 5 phyla. Additionally, there may be a unique profile of microbiota associated with adenomas [34,48,49], including the family Desulfovibrionaceae (Bilophila, Desulfovibrio) [49] and variants within the phylum Bacteroides (B. dorei, B. massiliensis, B. fragilis) [35,48,49].
Figure 1.
Microbiota associated with pre-cancerous and cancerous lesions. Microbiota associated with colorectal cancer and colorectal adenomas are often one of 5 phyla. Understanding these relationships helps consolidate the number of implicated microbiota, as studies frequently use different taxonomic levels in reporting results. In this Figure, the microbiota associated with colorectal cancer are in red, those associated with adenomas are in orange and those without specific comments in the literature but within the phylum are in grey [28-29,34-36,39-49]
Some of the microbes identified as being associated with adenocarcinoma formation do have close relatives associated with states of health, as discussed later. This does demonstrate that some microbes have specific characteristics that may be unique in relation to their clades. For instance, both A. parvulum and A. odontolyticus are oral microbiota associated with biofilm formation, and have been associated with CRC, whereas Bifidobacterium of the same phylum (Actinobacteria) is associated with health. Unlike its relatives, Bifidobacterium is native to the gastrointestinal tract, and Bifidobacterium performs carbohydrate fermentation.
Alterations in the microbiome have also been associated with distinct CRC characteristics. For instance, changes in the microbiome may also be associated with tumor grade [45]. Differences have also been identified in relation to different consensus molecular subtypes (CMSs), as originally defined by Guinney [50]. Using this framework, it would be most logical for CMS1 tumors to be associated with derangements in the microbiome, as they are most associated with immune activation. CMS1 tumors also have microsatellite instability (MSI), a high incidence of BRAF mutations, and are CpG island methylator phenotype (CIMP)-positive. Indeed, Purcell et al found that the microbiota did vary by tumor subtype [50], with CMS1 tumors being associated with increased populations of Fusobacteria and Bacteroidetes, including Fusobacterium (F.) hwasookii and Prophyromonas gingivalis [40]. Interestingly, both CMS1 tumors and the bacteria F. nucleatum are most commonly associated with the ascending colon [51,52]. Additionally, Koi et al have shown how F. nucleatum may contribute to the development of CIMP-positive and both MSI-H and MSI-L carcinogenesis [53]. Interestingly, Purcell et al also found CMS2 subtype tumors to have distinct microbial populations, with an increase in Selenomonas and Prevotella (also from the phylum Bacteroidetes). Burns et al demonstrated, using whole-exome sequencing, that specific communities were particularly associated with loss-of-function mutations in colonic tumors [44], including loss of function of the adenomatous polyposis coli (APC) gene, thus marrying changes in the microbiome with a CMS subtype, generally associated with a more traditional CRC pathway.
Virulence factors, inflammatory mediators and byproducts of metabolism
There are several specific bacteria that are consistently associated with CRC, and it is worth discussing these species in detail, as research is currently focused on these specific microbes and their potential roles as therapeutic targets.
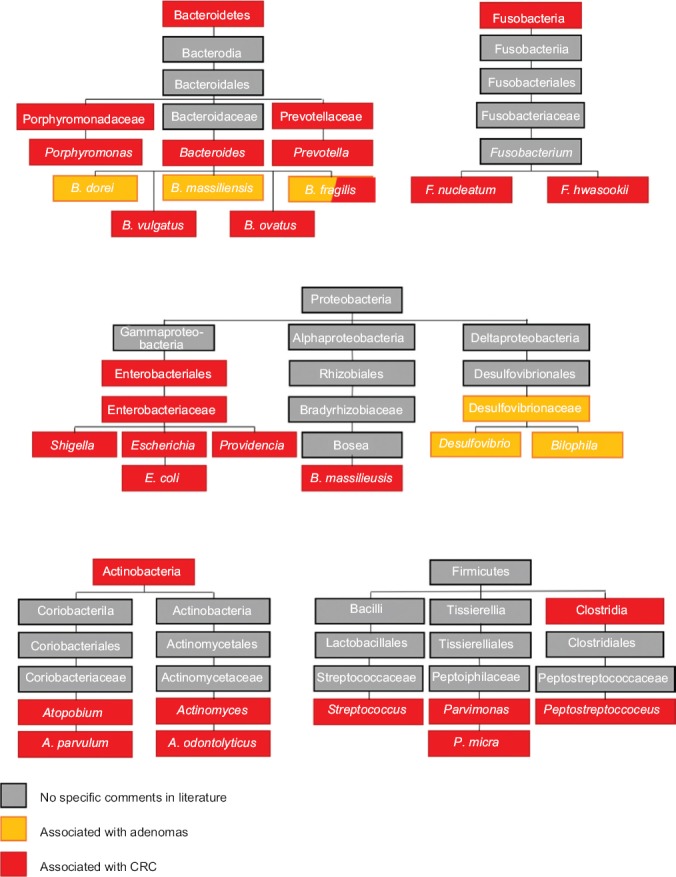
F. nucleatum is an oral anaerobe and is associated with biofilm formation [46,54,55], with some of the most consistent data associating its presence in the colon with CRC [56-59]. Multiple mechanisms by which F. nucleatum promotes and supports CRC have been delineated (Fig. 2A-D). The bacteria have been shown to selectively recruit tumor-infiltrating myeloid cells, promoting an inflammatory environment and tumorigenesis [57,59]. Its unique adhesin, FadA [60], is capable of binding E-cadherin and activating the Wnt/β-catenin signaling pathway, resulting in cellular proliferation and tumorigenesis [61]. It has recently been demonstrated that FadA, E-cadherin and Annexin A1, upregulated in CRC cells, form a complex to further accelerate tumor progression [62]. This pathway is further capable of activating NF-κB, resulting in an increase of proinflammatory cytokines as well as the expression of several oncogenes, such as c-Myc and Cyclin D1 [55,61]. In addition, F. nucleatum presents the protein Fap2 on its cellular outer membrane. Fap2 interacts with the receptor TIGIT, an inhibitory NK cell and T-cell receptor, therefore blocking immune cell cytotoxicity [63]. This same protein is capable of binding tumor cells via the Gal-GalNac polysaccharide [64], localizing the bacteria to tumor cells.
Figure 2.
Virulence factors of Fusobacterium (F.) nucleatum. (A) F. nucleatum has virulence factors, FadA, LPS and Fap2. (B) FadA binds E-cadherin, resulting in Wnt/β-catenin signaling and tumorigenesis, as well as activation of NF-κB and production of proinflammatory cytokines. Annexin A1, upregulated in colorectal cells, complexes with FadA and E-cadherin to further accelerate this pathway. (C) Fap2 interacts with the TIGIT receptor on NK and T-cells, inhibiting their ability to attack tumor cells. (D) Fap2 also binds to the Gal-GalNac polysaccharide expressed by tumor cells, which localized the F. nucleatum to tumor cells. (E) Binding of LPS to TRL-4 results in modulation of mi-RNA expression, in particular miR-18a and miR-4802, thereby disrupting autophagy Created with BioRender.com
LPS, Lipopolysaccharide; Fap2, Fusobacterium autotransporter protein 2; TIGIT, T-cell immunoreceptor with Ig and ITIM domains; Ga-GalNac, D-galactose-β (1-3)-N-acetyl-D-galactosamine; TRL-4, Toll-like receptor 4
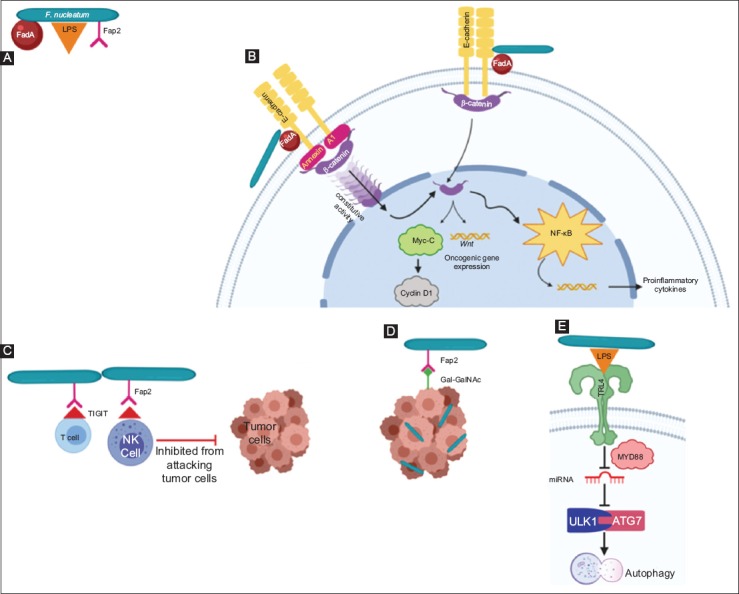
Bacteroides fragilis is an obligate anaerobe that has been associated with the development of CRC in both murine and human models [35,42,65-70]. The enterotoxigenic molecular subtype (ETBF) is associated with mutagenesis secondary to the bft gene, which encodes the BFT toxin, resulting in cellular proliferation and an inflammatory environment in multiple ways (Fig. 3). BFT causes barrier permeability and E-cadherin cleavage, which in turn activates the Wnt/β-catenin signal pathway, resulting in cellular proliferation [71,72]. It also incites NF-κB activation, resulting in the release of proinflammatory chemokines and cytokines [70]. Wu et al demonstrated in multiple intestinal neoplasia mice (heterozygous for the APC gene) that BFT activates STAT3, resulting in a selective TH17 response, inflammatory changes and development of CRC [69].
Figure 3.
Oncogenic properties of Bacteroides (B.) fragilis. Enterotoxigenic Bacteroides fragilis (ETBF) encodes the BFT toxin. BFT can penetrate the enterocyte. BFT can activate STAT3, resulting in Th17 cell response and inflammatory changes. It also activates E-cadherin, provoking the Wnt/β-catenin signaling pathway, which leads to cellular proliferation and NF-κB, resulting in production of proinflammatory cytokines BFT, Bacteroides fragilis toxin; STAT3, Signal transduce and activator of transcription 3
Escherichia coli (E. coli) is a gram-negative facultative anaerobe commensal to the gastrointestinal tract; however, the enteropathogenic E. coli (EPEC) strain has been associated with human neoplasia. EPEC is highly adherent to mucosal surfaces and has the ability to invade enterocytes [73,74]. In addition, EPEC can block mismatch repair (MMR) proteins MSH2 and MLH1 [75], linked with MSI. EPEC does this through secretion of the EspF protein, able to post-translationally deplete MMR proteins [76]. An increase in CRC has been demonstrated to occur by this pathway in cell lines [75,76]. Additionally, an increased level of reactive oxygen species (ROS) in host cells was seen independent of EspF [76], increasing the risk of DNA damage. Strains of E. coli also carry a genomic island called “pks”, which produces a polyketide-peptide genotoxin, Colibactin, leading to DNA double strand breaks and promoting CRC [77,78].
Enterococcus (E.) faecalis is a gram-positive facultative anaerobe primarily implicated in colitis-associated development of CRC, largely through the production of superoxide and hydroxyl radicals [79,80]. Superoxide results in upregulation of macrophage cyclooxygenase-2 promoting chromosomal instability, double-stranded DNA breaks and tetraploidy by oxidative damage via the bystander effect [81-83]. The aldehyde produced by COX-2 of infected macrophages is capable of diffusion to neighboring cells, resulting in DNA damage within these cells. In addition, polarized macrophages have been shown to increase tumor necrosis factor (TNF)-α and netrin-1, regulators of colonic cellular apoptosis [84]. The presence of E. faecalis has also been shown to arrest cells in G2 [82].
Streptococcus (S.) gallolyticus (previously S. bovis) is a gram-positive opportunistic bacterium first associated with CRC in the 1950s, when multiple patients with endocarditis were also found to have CRC [85]. It is best known for its ability to colonize collagen-rich surfaces, such as adenomatous tissue, by binding collagen (types I, IV and V) via the adhesin Acb [86]. While the exact mechanism has not been confirmed, it is believed that S. gallolyticus is able to penetrate beyond the epithelial barrier, inducing inflammation and tumorigenesis [87,88]. S. gallolyticus has several pilus surface proteins thought to perform key functions during infection [89]. The bacterium’s cell-associated protein isolates result in the release of proinflammatory markers (interleukin [IL]-8, prostaglandin E2) and result in pre-neoplastic changes in the colonic mucosa of rats, as well as the formation of aberrant crypt foci [90,91]. In human studies, the pilus proteins are associated with elevated inflammatory responses, including elevation of IL-1, COX-2 and IL-8, and these changes have been associated with colonic tumors [87,89]. In a case-controlled study, using antibodies to S. gallolyticus pilus proteins, Butt et al demonstrated an association between S. gallolyticus and an increased risk of CRC, consistent with the findings of others and suggests that S. gallolyticus plays a role in CRC [88,92].
Most of the bacteria reviewed have a specific virulence factor or protein identified, associated with the formation of pre-malignant or malignant lesions. The overarching mechanisms by which microbiota may induce neoplastic mutations within colonic enterocytes include direct actions of virulence factors, inflammatory mediators and the production of toxic or oncogenic byproducts (Fig. 4). More commonly, the effects of the microbiota in the colon are through metabolic byproducts and inflammatory mediation. Therefore, another way to evaluate the relationship of the microbiota and CRC is to evaluate the functional and metabolic roles they have in the colon and the interactions these products have with enterocytes. There are several byproducts of the colonic microbiota associated with negative impacts on the colon, many of which may be influenced by human diet.
Figure 4.
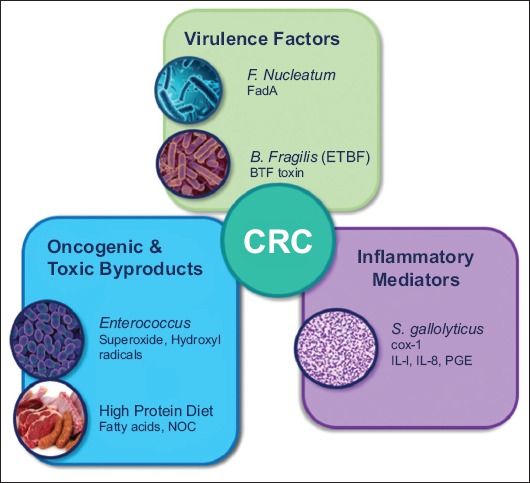
Mechanism by which microbiota promote colorectal cancer. Colorectal cancer may be promoted directly by bacterial virulence factors, oncogenic or toxic byproducts, or inflammatory mediators [28,33,38,42,45,47-48,120,131-133] F. Nucleatum, Fusobacterium nucleatum; ETBF, enterotoxigenic Bacteroides fragilis; BFT, Bacteroides fragilis toxin; NOC, N-nitroso compounds; S. gallolyticus, Streptococcus gallolyticus; COX-1, cylcooyxgenase-1; IL, interleukin; PGE, prostaglandin E
Fat metabolism
The colonic microbiota are altered with exposure to a diet rich in animal fats [93], with an increased incidence of microbial communities capable of metabolizing bile acids. Bile acids are a good energy source for multiple types of microbiota, though their hydrophobic nature may also result in cell membrane disruption for some microbial species, resulting in dynamic changes within the colonic lumen [94]. Intestinal microbiota metabolize primary bile acids that are not re-absorbed by the liver to secondary bile acids, most commonly deoxycholic acid and lithocholic acid [95], which have been demonstrated to result in an increased tumor burden in mice and DNA damage secondary to increased ROS and nitrogen species [96,97]. However, the ability of bile acids to behave as hormones and translocate to the nucleus can also have carcinogenic effects through alterations in signaling pathways [98-100]. Indeed, bile-tolerant microbiota have been associated with adenoma formation and CRC [41,49].
Protein metabolism
Bacterial protein metabolism results in a variety of byproducts, including branched chain fatty acids, N-nitroso compounds (NOCs), amines, ammonia, indoles, phenols, and hydrogen sulfide, many of which are harmful to colonocytes. Metabolomic studies demonstrate that the higher rates of CRC in individuals with increased protein intake are probably due to increased bacterial protein fermentation [41]. Increased dietary N-nitrosodimethylamine, a specific NOC, was demonstrated to lead to an increase in rectal cancer incidence in a large prospective cohort of 23,363 individuals in the UK [101]. However, intestinal microbiota are responsible for endogenous production of NOCs, accounting for approximately 45-75% of total NOC exposure [102]. In this way, an increased intake of heme iron via red meat results in the generation of NOCs [103,104] and is thought to promote CRC, primarily through DNA alkylation resulting in genetic mutations [105,106]. Ammonia is another product of protein fermentation, and has been well demonstrated to be carcinogenic at concentrations seen in western diets [107-109]. Bacterial fermentation of aromatic amino acids from proteins also leads to the production of bioactive products, or phenolic compounds. The bacterial cohorts most notable for production of these products are Bacteroides spp and some Firmicutes [110]. Increased phenol concentrations have been shown to induce cellular damage and break down the intestinal epithelial barrier [111-113].
Hydrogen sulfide
Hydrogen sulfide is a final bacterial product of protein fermentation. Sulfate reducing bacteria most commonly include Desulfovibrio spp. and Bilophila (B.) wadsworthia [114]. B. wadsorthia is an anaerobic gram-negative bacterium that produces hydrogen sulfide via taurine desulfonation and has been associated with inflammatory bowel disease and CRC [115]. Sulfide has been shown to be toxic to colonocytes, inducing mucosal hyperproliferation [116] and inhibiting butyrate oxidation, resulting in a state of energy deprivation and the breakdown of the epithelial barrier [117,118]. It has also been shown to be directly genotoxic, probably through the action of ROS [119,120]. For these reasons, it is not surprising that patients with higher levels of hydrogen sulfide have been associated with CRC [121].
Short-chain fatty acids (SCFA)
The outcomes associated with SCFA metabolism are more complicated, particularly in the case of butyrate. SCFAs are the primary fermentation products of dietary fiber. They include acetate, propionate and butyrate, and are often associated with Clostridium clusters IV and XIVa from the Firmicutes phylum and Bifidobacteria from the Actinobacteria phylum. Butyrate in particular is an important energy source for colonic enterocytes [122]. Overwhelmingly, data suggest that SCFA, including butyrate, are protective in relation to adenoma formation and CRC; however, there have been inconsistencies in these results. Belcheva et al demonstrated that low concentrations of butyrate resulted in hyperproliferation of colonic epithelial cells [123], though hyperproliferation is not seen at normal or increased levels of butyrate. Overall, SCFAs have been shown to decrease inflammation and carcinogenesis [124] and have several functions that support a protective effect. There are multiple extracellular G protein-coupled receptors on colonocytes, as well as immune cells, which interact with SCFAs [125,126]. The binding of butyrate with GPR109A in particular results in apoptosis of CRC cells via downregulation of Bcl-2, Bcl-xL and cyclin D1, as well as suppression of NF-κB [127]. However, the better described effects of butyrate and SCFA are intracellular, through inhibition of histone deacetylases leading to hyperacetylation of histones and transcription factors and resulting in downregulation of proinflammatory cytokines through signal transduction pathways [128]. Additionally, through this same mechanism, butyrate and propionate have been shown to induce the differentiation of regulatory colonic T-cells (cTreg) and increase expression of the transcription factor FOXP3, downregulating inflammation [129,130]. Although there are studies showing that butyrate may have a negative impact in low doses, studies overall suggest that, at physiologic levels, butyrate is associated with protective microbiota species and metabolites, while butyrate levels are inversely correlated with CRC and polyps [34,45,131].
Phytochemicals
Another group of bacterial products, phytochemicals, are derived most commonly from the breakdown of plant substrates and are therefore mediated by diet [132,133]. Phytochemicals, including flavonoids and benzoic acids, are associated with anti-inflammatory effects [110]. Their anti-inflammatory and anti-oxidative properties are probably secondary to downregulation of proinflammatory cytokines such as TNF and IL-6, and mediation of free radical effects and free radical scavenging [132,134,135].
A healthy microbial signature
Despite variation within the literature, there are specific microbiota that have been indicated as playing a protective role in regard to CRC development. The commonly cited microbiota are shown in Fig. 5 to allow for appreciation of similarities and relationships. The majority of studies have identified members of the Firmicutes phylum as associated with non-cancerous states.
Figure 5.
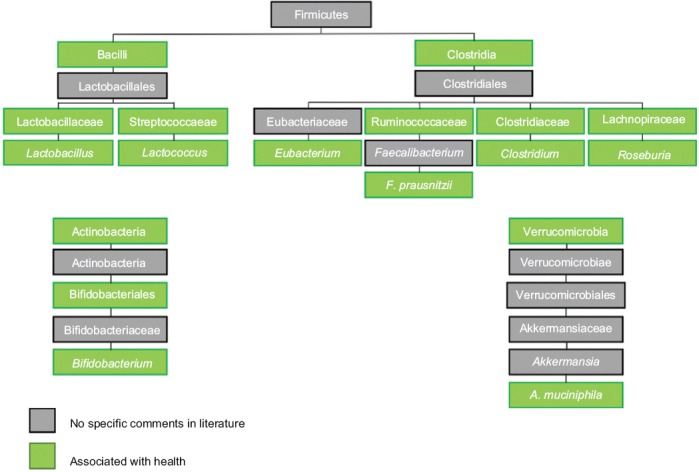
Microbiota associated with health. Microbiota associated with healthy individuals are often one of 3 phyla. Again, understanding the relationships between these microbiota helps to simplify the findings reported through the literature, as studies frequently use different taxonomic levels in reporting results. In this Figure, the microbiota associated with healthy individuals without cancerous or precancerous lesions are in green and those without specific comments in the literature but within the phylum are grey
The majority of these microbes are associated with butyrate production, and their repeated identification as protective microbes may be related to their metabolism as much as to the bacteria themselves. Identification of specific microbes and communities with anti-cancer effects is critical as we look to move toward therapeutic interventions. Understanding the protective microbiome may help guide modulation of the biome through supportive diet modification, probiotics and prebiotics. Given that the microbiome has been demonstrated to change in response to interventions, these relationships may lay the groundwork for future interventions.
Therapeutic directions
As we begin to appreciate the role of the microbiome in human health and disease, attention is shifting toward understanding ways that the microbiome can be altered to improve human health, including in relation to CRC. Currently, research is under way to evaluate ways in which the microbiome may help with disease diagnosis, prevention and therapy.
Detecting disease states
CRC is known to progress through the adenoma–adenocarcinoma sequence over many years, allowing a reasonable period to identify and intervene upon pre-malignant lesions prior to their progression to carcinoma. The US Preventative Services Task Force continues to recommend screening of individuals starting at age 50 (level A recommendation), though it does not specify the strategy for screening [139]. Colonoscopy remains the gold standard, though it does carry a small risk of harm, discomfort and inconvenience for patients. Stool-based tests, such as the guaiac-based fecal occult blood test and fecal immunochemical tests, lack sensitivity, especially if not used annually as directed. Therefore, the microbiome is being evaluated as a novel screening test for CRC.
Several authors have published on their models. Rezasoltani et al looked at 93 patients undergoing screening colonoscopy in Tehran, Iran, using qPCR to evaluate for F. nucleatum, E. faecalis, S. bovis (gallactolytica), ETBF, and Porphyromonas spp. [140]. Using a linear logistical analysis, they reported 91.4% sensitivity and 93.5% specificity in distinguishing those with adenomatous polyps or CRC from normal individuals. Flemer et al, from Ireland, investigated the use of oral and fecal microbiota in their ability to detect AP and CRC versus normal individuals in a cohort of 234 patients undergoing screening colonoscopy [46]. The oral microbiota were assessed using oral swabs. They found that with oral swabs alone, their sensitivity was 53% (CRC) and 67% (AP), with a specificity of 93.5%. With the addition of the fecal microbiota, their sensitivity improved to 76% (CRC) and 88% (AP). In this model, the investigators found that Peptostreoptocococcus, Parvimona and Fusobacterium were positivity associated with lesions, while the presence of Lachnospiraceae was negatively correlated. Both these studies demonstrate that the microbiota may serve as a useful screening tool for CRC, and this possibility supports the idea that the microbiota is an important aspect of CRC.
The microbiome as therapy
Given the interactions between the microbiome and the colonic environment, we are starting to look to the microbiome as a therapeutic tool. There are multiple ways to target the microbiome, to either increase or decrease specific microbes, or communities as a whole. The most obvious means of altering an individual’s microbiota is through changes in diet, supplementation with probiotics, antibiotics and even fecal microbiota transplantation (FMT).
The western diet has been identified as a risk factor for CRC, and it has been shown to modify the intestinal microbiota. Multiple groups have illustrated how diet modification may be a potential target [93,141-144]. Diet alterations may shift the microbiome away from a collection of microbes that are tumorigenic secondary to virulence factors, immunomodulation or metabolic products. Red meat consumption has been indicated as being uniquely supportive of tumorigenic microbiota, whereas plant-based diets support butyrate-producers and intestinal health. Donohoe et al demonstrated that dietary supplementation with fiber was enough to suppress tumor growth in a mouse model, and that the effects of the dietary fiber supplementation were mediated through increased butyrate production [144]. Diet, therefore, is the first line in inducing healthy adaptations in the colon and reducing the development of CRC.
Diet modification can be challenging for patients and works through large shifts in communities. An alternative strategy is to use probiotics to replace the specific microbiota that have been implicated in reducing the risk of CRC, or that have been specifically indicated as having anti-tumor effects. Probiotics have been demonstrated to change the microbiota, though their effects may be more nuanced than often presumed [145]. Appleyard et al demonstrated that, in proinflammatory rats, the progression from inflammation to dysplasia was mediated with treatment of a probiotic (VSL#3, containing strains of Bifidobacterium, Lactobacillus, and S. salivarus) [146]. Lactobacillus spp., in particular, have been demonstrated to inhibit the development of CRC via several mechanisms, including through mediation of the c-Jun N-terminal kinase pathway [137,138]. Additionally, Lactobacillus and Bifidobacterium have been shown to aid in restoring a normal balance of intestinal flora [147,148].
While these techniques cultivate a healthy microbiome through protective inputs, it is also possible to eradicate carcinogenic species when identified in order to prevent the development of cancerous or pre-cancerous lesions. Perhaps the best-known success story of preventative interventions would be that of H. pylori and gastric cancer. H. pylori, presumably through an inflammatory process, results in the accumulation of mitotic errors over time [5]. By treating H. pylori infection, we are able to modify the microbiota of the foregut in order to prevent the progression to malignancy. Similarly, researchers have looked at the use of antimicrobial treatment for prevention of CRC. In mice, antibiotic treatment was found to decrease the prevalence of Clostridium leptum and CRC tumor burden [149]. In mice infected with Fusobacterium spp., treatment with metronidazole slowed tumor growth and reduced cellular proliferation [52]. Zackular et al colonized germ-free mice with tumorigenic microbiota, resulting in tumor formation. They were able to partially reverse these effects with antibiotic treatment, demonstrating that the microbiome was a modifiable driver of tumorigenesis [150]. The risk of treatment with antibiotics is inducing a state of dysbiosis; however, treatment with antibiotics and co-treatment with probiotics may be a way to target high-risk microbiota while maintaining the global microbial balance.
Finally, FMT is another way to alter microbial communities. FMT has been successful in mouse models, though the transplant recipient is often germ-free at baseline. It has been validated as a tool for treatment of severe Clostridium difficile infections [151], and its use to restore dysbiotic microbiomes to normal is growing for treatment of other disorders [152,153]. However, enthusiasm was mildly tempered recently when a patient expired secondary to transfer of a drug-resistant bacterium. This unfortunate event prompted screening guidelines developed by the US Food and Drug Administration [154,155]. FMT remains a plausible treatment modality for management of dysbiosis, but will require further evaluation.
Modulation of existing therapies
There are therapies that target the microbiome directly, as well as others that use the microbiome as an adjunct to existing therapies. Lida et al used germ-free mice transplanted with lymphoma (EL4 cell line), colon carcinoma (MC38 cell line) and melanoma (B16 cell line) to demonstrate the importance of the microbiome in response to platinum-driven chemotherapy, finding that an intact microbiome was required for myeloid-derived cells to produce ROS and to result in tumor reduction [156]. Sivan et al demonstrated that Bifidobacterium was synergistic with PD-1 and PD-L1 inhibitors in epithelial derived tumors, improving tumor response to treatment in those with higher populations of Bifidobacterium [157]; similar findings also apply to Akkermansia muciniphila [136,158]. The colonic microbiota are also different in men with prostate cancer on androgen receptor axis-targeted therapies (ATT), and there is speculation that these changes may translate to variable responses to ATT [159]. To date, there are 2 currently enrolling NIH-funded trials looking at the role of FMT in patients with melanoma who failed PD-1 blockade. Further studies are needed to understand the role of the microbiota in relation to cancer treatment, including in CRC.
In the case of colorectal cancers, F. nucleatum has been consistently linked to neoplastic development. Yu et al demonstrated that F. nucleatum enrichment was associated with recurrent disease, and was also interestingly associated with chemotherapeutic resistance (Fig. 2E) [160,161]. Transfection with F. nucleatum in HCT116 and HT29 CRC cell lines was shown to decrease apoptosis secondary to oxaliplatin and 5-fluorouracil. As previously described, F. nucleatum has been implicated in the activation of the Wnt pathway. The Wnt signaling pathway has been associated with mutations in APC, key to the canonical CRC pathway [162]. This is clearly an important pathway in neoplasia development, and while therapeutics acting on it are not currently available, several are under investigation [163].
Moving forward, the microbiome is likely to play a role both in our understanding of cancer development, and as tool for modulating risk and treating disease. While we have seen time and time again the difficulties involved in diet modification, understanding the role of a likely intermediary, the microbiome, may help prevent development of disease through targeted therapies. As we solidify the connections between pathogenic species and neoplasms, it would be rational to anticipate that targeted antibiotic regimens may be developed, and probiotics and FMT may be developed to repopulate the gastrointestinal tract with health-provoking populations. Alternatively, the microbiome may be leveraged to help with treatment efficacy, again through repopulation of specific populations through probiotics or through transplantation of key microbiota.
Concluding remarks
We are just beginning to understand the role of the microbiome in human health and disease. Over the past decade, collaborative work has brought the role of the microbiome to the forefront of medical literature and is enriching our understanding of multiple diseases. The microbiome’s association with CRC is undeniable at this time. We are still working to understand the key microbiota, the impact of microbial communities and the mechanisms by which the microbiota interact with the human host. Ultimately, in any study of the microbiota, there are rarely bacteria that are clearly “good” or “bad.” However, multiple bacterial players, byproducts and interactions have already been identified as provoking or supporting neoplasia. Ultimately, our growing knowledge of the microbiota is anticipated to answer some of the questions regarding spontaneous cancer development, dietary links and treatment responses.
Acknowledgments
The authors would like to thank Christian P. Lanciault, MD, PhD, Department of Pathology, and Robert G. Martindale, MD, PhD, Department of Surgery, for their editorial contributions.
Biography
Oregon Health & Science University, Portland, OR; Oregon Health & Science University, USA
Footnotes
Conflict of Interest: None
References
- 1.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 2.Bosch FX, Lorincz A, Muñoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55:244–265. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee YC, Chiang TH, Chou CK, et al. Association between Helicobacter pylori eradication and gastric cancer incidence:a systematic review and meta-analysis. Gastroenterology. 2016;150:1113–1124. doi: 10.1053/j.gastro.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 4.Mima K, Ogino S, Nakagawa S, et al. The role of intestinal bacteria in the development and progression of gastrointestinal tract neoplasms. Surg Oncol. 2017;26:368–376. doi: 10.1016/j.suronc.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson H, Chia N. Gut microbiome and colon cancer:a plausible explanation for dietary contributions to cancer. J Am Coll Surg. 2019;229:231–235. doi: 10.1016/j.jamcollsurg.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin J, Li R, Raes J, et al. Meta HIT Consortium. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Human Microbiome Project Consortium A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gagnière J, Raisch J, Veziant J, et al. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. 2016;22:501–518. doi: 10.3748/wjg.v22.i2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15:630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 13.Nagao-Kitamoto H, Kitamoto S, Kuffa P, Kamada N. Pathogenic role of the gut microbiota in gastrointestinal diseases. Intest Res. 2016;14:127–138. doi: 10.5217/ir.2016.14.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shanahan F. The host-microbe interface within the gut. Best Pract Res Clin Gastroenterol. 2002;16:915–931. doi: 10.1053/bega.2002.0342. [DOI] [PubMed] [Google Scholar]
- 16.Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol Immunol. 1995;39:555–562. doi: 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- 17.Faith JJ, Guruge JL, Charbonneau M, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tja Voigt AY, Costea PI, Kultima JR, et al. Temporal and technical variability of human gut metagenomes. Genome Biol. 2015;16:73. doi: 10.1186/s13059-015-0639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proctor LM, Creasy HH, Fettweis JM, et al. Integrative HMP (iHMP) Research Network Consortium. The integrative human microbiome project Nature. 2019;569:641–648. doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. IBDMDB Investigators. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colman RJ, Rubin DT. Fecal microbiota transplantation as therapy for inflammatory bowel disease:a systematic review and meta-analysis. J Crohns Colitis. 2014;8:1569–1581. doi: 10.1016/j.crohns.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knights D, Ward TL, McKinlay CE, et al. Rethinking “enterotypes”. Cell Host Microbe. 2014;16:433–437. doi: 10.1016/j.chom.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poudel R, Jumpponen A, Schlatter DC, et al. Microbiome networks:a systems framework for identifying candidate microbial assemblages for disease management. Phytopathology. 2016;106:1083–1096. doi: 10.1094/PHYTO-02-16-0058-FI. [DOI] [PubMed] [Google Scholar]
- 25.Costea PI, Hildebrand F, Arumugam M, et al. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol. 2018;3:8–16. doi: 10.1038/s41564-017-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arumugam M, Raes J, Pelletier E, et al. Meta HIT Consortium. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia Y, Sun J. Hypothesis testing and statistical analysis of microbiome. Genes Dis. 2017;4:138–148. doi: 10.1016/j.gendis.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu LC, Wei SC, Ni YH. Impact of microbiota in colorectal carcinogenesis:lessons from experimental models. Intest Res. 2018;16:346–357. doi: 10.5217/ir.2018.16.3.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flemer B, Lynch DB, Brown JM, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66:633–643. doi: 10.1136/gutjnl-2015-309595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Kempen LC, Ruiter DJ, van Muijen GN, Coussens LM. The tumor microenvironment:a critical determinant of neoplastic evolution. Eur J Cell Biol. 2003;82:539–548. doi: 10.1078/0171-9335-00346. [DOI] [PubMed] [Google Scholar]
- 32.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 33.Huipeng W, Lifeng G, Chuang G, Jiaying Z, Yuankun C. The differences in colonic mucosal microbiota between normal individual and colon cancer patients by polymerase chain reaction-denaturing gradient gel electrophoresis. J Clin Gastroenterol. 2014;48:138–144. doi: 10.1097/MCG.0b013e3182a26719. [DOI] [PubMed] [Google Scholar]
- 34.Peters BA, Dominianni C, Shapiro JA, et al. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome. 2016;4:69. doi: 10.1186/s40168-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 36.Burns MB, Lynch J, Starr TK, Knights D, Blekhman R. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med. 2015;7:55. doi: 10.1186/s13073-015-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanapareddy N, Legge RM, Jovov B, et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J. 2012;6:1858–1868. doi: 10.1038/ismej.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saus E, Iraola-Guzmán S, Willis JR, Brunet-Vega A, Gabaldón T. Microbiome and colorectal cancer:roles in carcinogenesis and clinical potential. Mol Aspects Med. 2019;69:93–106. doi: 10.1016/j.mam.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori G, Rampelli S, Orena BS, et al. Shifts of faecal microbiota during sporadic colorectal carcinogenesis. Sci Rep. 2018;8:10329. doi: 10.1038/s41598-018-28671-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep. 2017;7:11590. doi: 10.1038/s41598-017-11237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yachida S, Mizutani S, Shiroma H, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25:968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 42.Dai Z, Coker OO, Nakatsu G, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6:70. doi: 10.1186/s40168-018-0451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao R, Kong C, Huang L, et al. Mucosa-associated microbiota signature in colorectal cancer. Eur J Clin Microbiol Infect Dis. 2017;36:2073–2083. doi: 10.1007/s10096-017-3026-4. [DOI] [PubMed] [Google Scholar]
- 44.Burns MB, Montassier E, Abrahante J, et al. Colorectal cancer mutational profiles correlate with defined microbial communities in the tumor microenvironment. PLoS Genet. 2018;14:e1007376. doi: 10.1371/journal.pgen.1007376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu N, Yang X, Zhang R, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 46.Flemer B, Warren RD, Barrett MP, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018;67:1454–1463. doi: 10.1136/gutjnl-2017-314814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kasai C, Sugimoto K, Moritani I, et al. Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma:terminal restriction fragment length polymorphism and next-generation sequencing analyses. Oncol Rep. 2016;35:325–333. doi: 10.3892/or.2015.4398. [DOI] [PubMed] [Google Scholar]
- 48.Rezasoltani S, Asadzadeh Aghdaei H, Dabiri H, Akhavan Sepahi A, Modarressi MH, Nazemalhosseini Mojarad E. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microb Pathog. 2018;124:244–249. doi: 10.1016/j.micpath.2018.08.035. [DOI] [PubMed] [Google Scholar]
- 49.Hale VL, Chen J, Johnson S, et al. Shifts in the fecal microbiota associated with adenomatous polyps. Cancer Epidemiol Biomarkers Prev. 2017;26:85–94. doi: 10.1158/1055-9965.EPI-16-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mima K, Cao Y, Chan AT, et al. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clin Transl Gastroenterol. 2016;7:e200. doi: 10.1038/ctg.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bullman S, Pedamallu CS, Sicinska E, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koi M, Okita Y,M, Carethers J. Fusobacterium nucleatum infection in colorectal cancer:linking inflammation, DNA mismatch repair and genetic and epigenetic alterations. J Anus Rectum Colon. 2018;2:37–46. doi: 10.23922/jarc.2017-055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bachrach G, Ianculovici C, Naor R, Weiss EI. Fluorescence based measurements of Fusobacterium nucleatum coaggregation and of fusobacterial attachment to mammalian cells. FEMS Microbiol Lett. 2005;248:235–240. doi: 10.1016/j.femsle.2005.05.055. [DOI] [PubMed] [Google Scholar]
- 55.Proença MA, Biselli JM, Succi M, et al. Relationship between Fusobacterium nucleatum inflammatory mediators and microRNAs in colorectal carcinogenesis. World J Gastroenterol. 2018;24:5351–5365. doi: 10.3748/wjg.v24.i47.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komiya Y, Shimomura Y, Higurashi T, et al. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut. 2019;68:1335–1337. doi: 10.1136/gutjnl-2018-316661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han YW, Ikegami A, Rajanna C, et al. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol. 2005;187:5330–5340. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rubinstein MR, Baik JE, Lagana SM, et al. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β-catenin modulator Annexin A1. EMBO Rep. 2019;20 doi: 10.15252/embr.201847638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abed J, Emgård JE, Zamir G, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thiele Orberg E, Fan H, Tam AJ, et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 2017;10:421–433. doi: 10.1038/mi.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells:role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 67.Housseau F, Sears CL. Enterotoxigenic Bacteroides fragilis (ETBF)-mediated colitis in Min (Apc+/-) mice:a human commensal-based murine model of colon carcinogenesis. Cell Cycle. 2010;9:3–5. doi: 10.4161/cc.9.1.10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toprak NU, Yagci A, Gulluoglu BM, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12:782–786. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 69.Wu S, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chung L, Thiele Orberg E, Geis AL, et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe. 2018;23:203–214. doi: 10.1016/j.chom.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu S, Lim KC, Huang J, Saidi RF, Sears CL. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc Natl Acad Sci U S A. 1998;95:14979–14984. doi: 10.1073/pnas.95.25.14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology. 2003;124:392–400. doi: 10.1053/gast.2003.50047. [DOI] [PubMed] [Google Scholar]
- 73.Swidsinski A, Khilkin M, Kerjaschki D, et al. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 1998;115:281–286. doi: 10.1016/s0016-5085(98)70194-5. [DOI] [PubMed] [Google Scholar]
- 74.Martin HM, Campbell BJ, Hart CA, et al. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 75.Maddocks OD, Short AJ, Donnenberg MS, Bader S, Harrison DJ. Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans. PLoS One. 2009;4:e5517. doi: 10.1371/journal.pone.0005517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maddocks ODK, Scanlon KM, Donnenberg MS. An Escherichia coli effector protein promotes host mutation via depletion of DNA mismatch repair proteins. mBio. 2013;4:e00152–13. doi: 10.1128/mBio.00152-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A. 2010;107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23:529–536. doi: 10.1093/carcin/23.3.529. [DOI] [PubMed] [Google Scholar]
- 80.Huycke MM, Moore DR. In vivo production of hydroxyl radical by Enterococcus faecalis colonizing the intestinal tract using aromatic hydroxylation. Free Radic Biol Med. 2002;33:818–826. doi: 10.1016/s0891-5849(02)00977-2. [DOI] [PubMed] [Google Scholar]
- 81.Wang X, Huycke MM. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology. 2007;132:551–561. doi: 10.1053/j.gastro.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 82.Wang X, Allen TD, May RJ, Lightfoot S, Houchen CW, Huycke MM. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008;68:9909–9917. doi: 10.1158/0008-5472.CAN-08-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang X, Yang Y, Huycke MM. Commensal bacteria drive endogenous transformation and tumour stem cell marker expression through a bystander effect. Gut. 2015;64:459–468. doi: 10.1136/gutjnl-2014-307213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang Y, Wang X, Huycke T, Moore DR, Lightfoot SA, Huycke MM. Colon macrophages polarized by commensal bacteria cause colitis and cancer through the bystander effect. Transl Oncol. 2013;6:596–606. doi: 10.1593/tlo.13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McCoy W, Mason J. Enterococcal endocarditis associated with carcinoma of the sigmoid;report of a case. J Med Assoc State Ala. 1951;21:162–166. [PubMed] [Google Scholar]
- 86.Sillanpää J, Nallapareddy SR, Qin X, et al. A collagen-binding adhesin, Acb, and ten other putative MSCRAMM and pilus family proteins of Streptococcus gallolyticus subsp gallolyticus (Streptococcus bovis Group, biotype I) J Bacteriol. 2009;191:6643–6653. doi: 10.1128/JB.00909-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abdulamir AS, Hafidh RR, Bakar FA. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors:inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol Cancer. 2010;9:249. doi: 10.1186/1476-4598-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boleij A, van Gelder MM, Swinkels DW, Tjalsma H. Clinical importance of Streptococcus gallolyticus infection among colorectal cancer patients:systematic review and meta-analysis. Clin Infect Dis. 2011;53:870–878. doi: 10.1093/cid/cir609. [DOI] [PubMed] [Google Scholar]
- 89.Boleij A, Roelofs R, Danne C, et al. Selective antibody response to Streptococcus gallolyticus pilus proteins in colorectal cancer patients. Cancer Prev Res (Phila) 2012;5:260–265. doi: 10.1158/1940-6207.CAPR-11-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ellmerich S, Schöller M, Duranton B, et al. Promotion of intestinal carcinogenesis by Streptococcus bovis . Carcinogenesis. 2000;21:753–756. doi: 10.1093/carcin/21.4.753. [DOI] [PubMed] [Google Scholar]
- 91.Biarc J, Nguyen IS, Pini A, et al. Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S bovis) Carcinogenesis. 2004;25:1477–1484. doi: 10.1093/carcin/bgh091. [DOI] [PubMed] [Google Scholar]
- 92.Butt J, Romero-Hernández B, Pérez-Gómez B, et al. Association of Streptococcus gallolyticus subspecies gallolyticus with colorectal cancer:Serological evidence. Int J Cancer. 2016;138:1670–1679. doi: 10.1002/ijc.29914. [DOI] [PubMed] [Google Scholar]
- 93.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Islam KB, Fukiya S, Hagio M, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 95.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 96.Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res. 2005;589:47–65. doi: 10.1016/j.mrrev.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 97.Májer F, Sharma R, Mullins C, et al. New highly toxic bile acids derived from deoxycholic acid, chenodeoxycholic acid and lithocholic acid. Bioorg Med Chem. 2014;22:256–268. doi: 10.1016/j.bmc.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 98.Powell AA, Akare S, Qi W, et al. Resistance to ursodeoxycholic acid-induced growth arrest can also result in resistance to deoxycholic acid-induced apoptosis and increased tumorgenicity. BMC Cancer. 2006;6:219. doi: 10.1186/1471-2407-6-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sharma R, Prichard D, Majer F, et al. Ursodeoxycholic acid amides as novel glucocorticoid receptor modulators. J Med Chem. 2011;54:122–130. doi: 10.1021/jm100860s. [DOI] [PubMed] [Google Scholar]
- 100.Im E, Martinez J. Ursodeoxycholic acid (UDCA) can inhibit deoxycholic acid (DCA)-induced apoptosis via modulation of EGFR/Raf-1/ERK signaling in human colon cancer cells. J Nutr. 2004;134:483–486. doi: 10.1093/jn/134.2.483. [DOI] [PubMed] [Google Scholar]
- 101.Loh YH, Jakszyn P, Luben RN, Mulligan AA, Mitrou PN, Khaw KT. N-Nitroso compounds and cancer incidence:the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk Study. Am J Clin Nutr. 2011;93:1053–1061. doi: 10.3945/ajcn.111.012377. [DOI] [PubMed] [Google Scholar]
- 102.Tricker AR. N-nitroso compounds and man:sources of exposure, endogenous formation and occurrence in body fluids. Eur J Cancer Prev. 1997;6:226–268. [PubMed] [Google Scholar]
- 103.Constante M, Fragoso G, Calvé A, Samba-Mondonga M, Santos MM. Dietary heme induces gut dysbiosis, aggravates colitis, and potentiates the development of adenomas in mice. Front Microbiol. 2017;8:1809. doi: 10.3389/fmicb.2017.01809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuhnle GG, Story GW, Reda T, et al. Diet-induced endogenous formation of nitroso compounds in the GI tract. Free Radic Biol Med. 2007;43:1040–1047. doi: 10.1016/j.freeradbiomed.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 105.Lewin MH, Bailey N, Bandaletova T, et al. Red meat enhances the colonic formation of the DNA adduct O6-carboxymethyl guanine:implications for colorectal cancer risk. Cancer Res. 2006;66:1859–1865. doi: 10.1158/0008-5472.CAN-05-2237. [DOI] [PubMed] [Google Scholar]
- 106.Vanden Bussche J, Hemeryck LY, Van Hecke T, et al. O⁶-carboxymethylguanine DNA adduct formation and lipid peroxidation upon in vitro gastrointestinal digestion of haem-rich meat. Mol Nutr Food Res. 2014;58:1883–1896. doi: 10.1002/mnfr.201400078. [DOI] [PubMed] [Google Scholar]
- 107.Visek WJ. Diet and cell growth modulation by ammonia. Am J Clin Nutr. 1978;31:S216–S220. doi: 10.1093/ajcn/31.10.S216. [DOI] [PubMed] [Google Scholar]
- 108.Windey K, De Preter V, Verbeke K. Relevance of protein fermentation to gut health. Mol Nutr Food Res. 2012;56:184–196. doi: 10.1002/mnfr.201100542. [DOI] [PubMed] [Google Scholar]
- 109.Ichikawa H, Sakata T. Stimulation of epithelial cell proliferation of isolated distal colon of rats by continuous colonic infusion of ammonia or short-chain fatty acids is nonadditive. J Nutr. 1998;128:843–847. doi: 10.1093/jn/128.5.843. [DOI] [PubMed] [Google Scholar]
- 110.Russell WR, Duncan SH, Scobbie L, et al. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res. 2013;57:523–535. doi: 10.1002/mnfr.201200594. [DOI] [PubMed] [Google Scholar]
- 111.Pedersen G, Brynskov J, Saermark T. Phenol toxicity and conjugation in human colonic epithelial cells. Scand J Gastroenterol. 2002;37:74–79. doi: 10.1080/003655202753387392. [DOI] [PubMed] [Google Scholar]
- 112.Hughes R, Kurth MJ, McGilligan V, McGlynn H, Rowland I. Effect of colonic bacterial metabolites on Caco-2 cell paracellular permeability in vitro. Nutr Cancer. 2008;60:259–266. doi: 10.1080/01635580701649644. [DOI] [PubMed] [Google Scholar]
- 113.McCall IC, Betanzos A, Weber DA, Nava P, Miller GW, Parkos CA. Effects of phenol on barrier function of a human intestinal epithelial cell line correlate with altered tight junction protein localization. Toxicol Appl Pharmacol. 2009;241:61–70. doi: 10.1016/j.taap.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nava GM, Carbonero F, Croix JA, Greenberg E, Gaskins HR. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J. 2012;6:57–70. doi: 10.1038/ismej.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Peck SC, Denger K, Burrichter A, Irwin SM, Balskus EP, Schleheck D. A glycyl radical enzyme enables hydrogen sulfide production by the human intestinal bacterium Bilophila wadsworthia. Proc Natl Acad Sci U S A. 2019;116:3171–3176. doi: 10.1073/pnas.1815661116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Christl SU, Eisner HD, Dusel G, Kasper H, Scheppach W. Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa:a potential role for these agents in the pathogenesis of ulcerative colitis. Dig Dis Sci. 1996;41:2477–2481. doi: 10.1007/BF02100146. [DOI] [PubMed] [Google Scholar]
- 117.Roediger WEW, Duncan A, Kapaniris O, Millard S. Sulphide impairment of substrate oxidation in rat colonocytes:A biochemical basis for ulcerative colitis? Clin Sci (Lond) 1993;85:623–627. doi: 10.1042/cs0850623. [DOI] [PubMed] [Google Scholar]
- 118.Leschelle X, Goubern M, Andriamihaja M, et al. Adaptative metabolic response of human colonic epithelial cells to the adverse effects of the luminal compound sulfide. Biochim Biophys Acta. 2005;1725:201–212. doi: 10.1016/j.bbagen.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 119.Attene-Ramos MS, Wagner ED, Gaskins HR, Plewa MJ. Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res. 2007;5:455–459. doi: 10.1158/1541-7786.MCR-06-0439. [DOI] [PubMed] [Google Scholar]
- 120.Attene-Ramos MS, Wagner ED, Plewa MJ, Gaskins HR. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res. 2006;4:9–14. doi: 10.1158/1541-7786.MCR-05-0126. [DOI] [PubMed] [Google Scholar]
- 121.Kanazawa K, Konishi F, Mitsuoka T, et al. Factors influencing the development of sigmoid colon cancer Bacteriologic and biochemical studies. Cancer. 1996;77:1701–1706. doi: 10.1002/(SICI)1097-0142(19960415)77:8<1701::AID-CNCR42>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 122.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article:the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 123.Belcheva A, Irrazabal T, Robertson SJ, et al. Gut microbial metabolism drives transformation of msh2-deficient colon epithelial cells [Erratum in Cell 2014159:456] Cell. 2014;158:288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 124.Chen ZF, Ai LY, Wang JL, et al. Probiotics Clostridium butyricum and Bacillus subtilis ameliorate intestinal tumorigenesis. Future Microbiol. 2015;10:1433–1445. doi: 10.2217/fmb.15.66. [DOI] [PubMed] [Google Scholar]
- 125.Brown AJ, Goldsworthy SM, Barnes AA, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 126.Singh N, Gurav A, Sivaprakasam S, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thangaraju M, Cresci GA, Liu K, et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009;69:2826–2832. doi: 10.1158/0008-5472.CAN-08-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wilson AJ, Chueh AC, Tögel L, et al. Apoptotic sensitivity of colon cancer cells to histone deacetylase inhibitors is mediated by an Sp1/Sp3-activated transcriptional program involving immediate-early gene induction. Cancer Res. 2010;70:609–620. doi: 10.1158/0008-5472.CAN-09-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 130.Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen HM, Yu YN, Wang JL, et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am J Clin Nutr. 2013;97:1044–1052. doi: 10.3945/ajcn.112.046607. [DOI] [PubMed] [Google Scholar]
- 132.Ramos S. Cancer chemoprevention and chemotherapy:dietary polyphenols and signalling pathways. Mol Nutr Food Res. 2008;52:507–526. doi: 10.1002/mnfr.200700326. [DOI] [PubMed] [Google Scholar]
- 133.Russell WR, Gratz SW, Duncan SH, et al. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am J Clin Nutr. 2011;93:1062–1072. doi: 10.3945/ajcn.110.002188. [DOI] [PubMed] [Google Scholar]
- 134.Huang WY, Cai YZ, Zhang Y. Natural phenolic compounds from medicinal herbs and dietary plants:potential use for cancer prevention. Nutr Cancer. 2010;62:1–20. doi: 10.1080/01635580903191585. [DOI] [PubMed] [Google Scholar]
- 135.Hsu CL, Yen GC. Phenolic compounds:evidence for inhibitory effects against obesity and their underlying molecular signaling mechanisms. Mol Nutr Food Res. 2008;52:53–61. doi: 10.1002/mnfr.200700393. [DOI] [PubMed] [Google Scholar]
- 136.Routy B, Gopalakrishnan V, Daillère R, Zitvogel L, Wargo JA, Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 2018;15:382–396. doi: 10.1038/s41571-018-0006-2. [DOI] [PubMed] [Google Scholar]
- 137.Konishi H, Fujiya M, Tanaka H, et al. Probiotic-derived ferrichrome inhibits colon cancer progression via JNK-mediated apoptosis. Nat Commun. 2016;7:12365. doi: 10.1038/ncomms12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chang CW, Liu CY, Lee HC, et al. Lactobacillus casei variety rhamnosus probiotic preventively attenuates 5-Fluorouracil/Oxaliplatin-induced intestinal injury in a syngeneic colorectal cancer model. Front Microbiol. 2018;9:983. doi: 10.3389/fmicb.2018.00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bibbins-Domingo K, Grossman DC, Curry SJ, et al. US Preventive Services Task Force. Screening for colorectal cancer:US Preventive Services Task Force Recommendation Statement. JAMA. 2016;315:2564–2575. doi: 10.1001/jama.2016.5989. [DOI] [PubMed] [Google Scholar]
- 140.Rezasoltani S, Sharafkhah M, Asadzadeh Aghdaei H, et al. Applying simple linear combination, multiple logistic and factor analysis methods for candidate fecal bacteria as novel biomarkers for early detection of adenomatous polyps and colon cancer. J Microbiol Methods. 2018;155:82–88. doi: 10.1016/j.mimet.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 141.Manzat-Saplacan RM, Mircea PA, Balacescu L, Chira RI, Berindan-Neagoe I, Balacescu O. Can we change our microbiome to prevent colorectal cancer development? Acta Oncol. 2015;54:1085–1095. doi: 10.3109/0284186X.2015.1054949. [DOI] [PubMed] [Google Scholar]
- 142.Uronis JM, Mühlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4:e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nelson H, Chia N. Gut microbiome and colon cancer:a plausible explanation for dietary contributions to cancer. J Am Coll Surg. 2019;229:231–235. doi: 10.1016/j.jamcollsurg.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 144.Donohoe DR, Holley D, Collins LB, et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014;4:1387–1397. doi: 10.1158/2159-8290.CD-14-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Suez J, Zmora N, Zilberman-Schapira G, et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018;174:1406–1423. doi: 10.1016/j.cell.2018.08.047. [DOI] [PubMed] [Google Scholar]
- 146.Appleyard CB, Cruz ML, Isidro AA, Arthur JC, Jobin C, De Simone C. Pretreatment with the probiotic VSL#3 delays transition from inflammation to dysplasia in a rat model of colitis-associated cancer. Am J Physiol Gastrointest Liver Physiol. 2011;301:G1004–G1013. doi: 10.1152/ajpgi.00167.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lenoir M, Del Carmen S, Cortes-Perez NG, et al. Lactobacillus casei BL23 regulates Treg and Th17 T-cell populations and reduces DMH-associated colorectal cancer. J Gastroenterol. 2016;51:862–873. doi: 10.1007/s00535-015-1158-9. [DOI] [PubMed] [Google Scholar]
- 148.Liang S, Xu L, Zhang D, Wu Z. Effect of probiotics on small intestinal bacterial overgrowth in patients with gastric and colorectal cancer. Turk J Gastroenterol. 2016;27:227–232. doi: 10.5152/tjg.2016.15375. [DOI] [PubMed] [Google Scholar]
- 149.Hattori N, Niwa T, Ishida T, et al. Antibiotics suppress colon tumorigenesis through inhibition of aberrant DNA methylation in an azoxymethane and dextran sulfate sodium colitis model. Cancer Sci. 2019;110:147–156. doi: 10.1111/cas.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zackular JP, Baxter NT, Iverson KD, et al. The gut microbiome modulates colon tumorigenesis. mBio. 2013;4:e00692–13. doi: 10.1128/mBio.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bakken JS, Borody T, Brandt LJ, et al. Fecal Microbiota Transplantation Workgroup. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol. 2011;9:1044–1049. doi: 10.1016/j.cgh.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Marotz CA, Zarrinpar A. Treating obesity and metabolic syndrome with fecal microbiota transplantation. Yale J Biol Med. 2016;89:383–388. [PMC free article] [PubMed] [Google Scholar]
- 153.Browne AS, Kelly CR. Fecal transplant in inflammatory bowel disease. Gastroenterol Clin North Am. 2017;46:825–837. doi: 10.1016/j.gtc.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 154.FDA. Information pertaining to additional safety protections regarding use of fecal microbiota for transplantation - screening and testing of stool donors for multi-drug resistant organisms. [[Accessed 7 February 2020]]. Available from: https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologics/information-pertaining-additional-safety-protections-regarding-use-fecal-microbiota-transplantation .
- 155.FDA Enforcement policy regarding investigational new drug requirements for use of fecal microbiota for transplantation to treat Clostridium difficile infection not responsive to standard therapies. U.S. Food and Drug Administration. 2016.
- 156.Lida N, Dzutsev A, Steward CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Routy B, Le C E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 159.Sfanos KS, Markowski MC, Peiffer LB, et al. Compositional differences in gastrointestinal microbiota in prostate cancer patients treated with androgen axis-targeted therapies. Prostate Cancer Prostatic Dis. 2018;21:539–548. doi: 10.1038/s41391-018-0061-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170:548–563. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang S, Yang Y, Weng W, et al. Fusobacterium nucleatum promotes chemoresistance to 5-fluorouracil by upregulation of BIRC3 expression in colorectal cancer. J Exp Clin Cancer Res. 2019;38:14. doi: 10.1186/s13046-018-0985-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schneikert J, Behrens J. The canonical Wnt signalling pathway and its APC partner in colon cancer development. Gut. 2007;56:417–425. doi: 10.1136/gut.2006.093310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer:Update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]


