Abstract
Antibodies are widely used as cancer therapeutics, but their current use is limited by the low number of antigens restricted to cancer cells. A receptor tyrosine kinase, receptor tyrosine kinase-like orphan receptor 2 (ROR2), is normally expressed only during embryogenesis and is tightly down-regulated in postnatal healthy tissues. However, it is up-regulated in a diverse set of hematologic and solid malignancies, thus ROR2 represents a candidate antigen for antibody-based cancer therapy. Here we describe the affinity maturation and humanization of a rabbit mAb that binds human and mouse ROR2 but not human ROR1 or other human cell-surface antigens. Co-crystallization of the parental rabbit mAb in complex with the human ROR2 kringle domain (hROR2-Kr) guided affinity maturation by heavy-chain complementarity-determining region 3 (HCDR3)-focused mutagenesis and selection. The affinity-matured rabbit mAb was then humanized by complementarity-determining region (CDR) grafting and framework fine tuning and again co-crystallized with hROR2-Kr. We show that the affinity-matured and humanized mAb retains strong affinity and specificity to ROR2 and, following conversion to a T cell–engaging bispecific antibody, has potent cytotoxicity toward ROR2-expressing cells. We anticipate that this humanized affinity-matured mAb will find application for antibody-based cancer therapy of ROR2-expressing neoplasms.
Keywords: antibody engineering, antigen, epitope mapping, X-ray crystallography, cancer therapy, affinity maturation, kringle domain, receptor tyrosine kinase (RTK) ROR2
Introduction
mAbs are widely used as cancer therapeutics with ∼30 antibody-based cancer therapies FDA4-approved and marketed (1). Infrequent identification of new suitable cancer antigens restricts the indications and patients suited for mAb therapies. Due to their overexpression on cancer cells, receptor tyrosine kinases (RTKs) have proven their general suitability as cancer antigens. However, only a few out of more than 30 RTKs are currently targeted by FDA-approved mAbs (2). Two RTKs not yet targeted by FDA-approved mAbs are ROR1 and ROR2, which are expressed in embryogenesis and tightly down-regulated in postnatal tissues (3–6). A number of solid and hematologic malignancies have been shown to express ROR1 or ROR2, suggesting utility as targets for antibody-based cancer therapies (7). Multiple clinical trials for cancer therapies with ROR1-targeting antibodies are under way. Although ROR2-targeting campaigns were only recently translated from preclinical to clinical investigations (NCT03504488, NCT03393936, NCT03960060), they underscore the suitability and attraction of ROR2 as a candidate antigen for antibody-based cancer therapy.
ROR2 shares 58% amino acid sequence identity with ROR1 along with the same extracellular domain composed of an N-terminal Ig domain, a frizzled (Fz), and a kringle (Kr) domain (7, 8). ROR2 is involved in the WNT signaling pathway when associated with its ligand WNT5A and facilitates polarization of cells during embryonic development along with regulating migration and differentiation (9–12). While largely down-regulated after birth in mice (6) and humans (8, 13), ROR2 is overexpressed in several cancers (7, 9), including solid malignancies, such as renal cell adenocarcinoma and subsets of breast cancer, and hematologic malignancies, such as multiple myeloma (14–16). Among solid malignancies without FDA-approved and marketed antibody-based cancer therapies, a notable indication is sarcoma, where ROR2 overexpression was found in osteosarcoma, leiomyosarcoma, and gastrointestinal stromal tumor (13, 17). Numerous studies show that ROR2 increases invasiveness and takes part in tumorigenesis, making ROR2 a promising cancer target and biomarker.
We previously reported the generation of a diverse panel of ROR2-targeting mAbs derived from a large naive rabbit antibody library by phage display (18). Among these, mAb XBR2-401 (“401”) was shown to bind ROR2 with high affinity and exclusive specificity. Once converted into a chimeric antigen receptor T cell (CAR-T) format, 401 triggered selective killing in vitro (18). The epitope of 401 was mapped to the Kr domain. This plasma membrane–proximal location makes 401 a preferred candidate for T cell–engaging ROR2 × CD3 bispecific antibodies (biAbs). This is suggested by our ROR1 × CD3 biAb study, where ROR1-targeting mAb R11, with an epitope in the Kr domain, had superior in vitro and in vivo activity compared with ROR1 × CD3 biAbs with plasma membrane–distal epitopes (19). To map this epitope, we co-crystallized R11 in single-chain variable fragment (scFv) format with the Kr domain of ROR1 (Protein Data Bank (PDB): 6BA5 (55)). Notably, R11 does not cross-react with the Kr domain of ROR2, and 401 does not cross-react with the Kr domain of ROR1. Determination of the precise interaction of 401 and the Kr domain of ROR2 would allow for a full understanding of the mAb and aid its preclinical and clinical development.
mAb 401 was selected as a chimeric rabbit/human Fab with rabbit variable domains and human constant domains (18). An issue with utilizing chimeric mAbs for therapy is potential immunogenicity. Studies have shown that the risk of immunogenicity is lower with humanized mAbs versus chimeric mAbs (20). The chimeric mAbs in this comparison contained murine variable domains, and it is not known whether chimeric mAbs with rabbit variable domains have comparable immunogenicity. A well-established strategy of humanization is to graft murine complementarity-determining regions (CDRs) into human frameworks. This was later optimized by identifying human frameworks with the highest identity to the original murine antibody and using them as the backbone for the murine CDRs (21, 22). We and others showed that rabbit mAbs can also be humanized by CDR grafting (23). A number of humanized mAbs originating from the rabbit antibody repertoire are currently in clinical trials (24), with brolucizumab, an scFv-targeting VEGF in wet age-related macular degeneration, becoming the first to have received FDA approval on October 7, 2019. The United States Adopted Names Council and World Health Organization International Nonproprietary Names sector work together to update and determine classifications for the evolving world of nonhuman, chimeric, humanized, and human antibodies for diagnostic and therapeutic applications (25, 26). The current parameters that define a humanized antibody include the variable region identifying closer with human than any other species sequences. It also requires the identity of the final variable region sequences, including CDR regions, to be >85% human using IMGT (26).
Here we report the generation of a humanized mAb that binds to a membrane-proximal epitope of ROR2 with high affinity and specificity and as such can be utilized as an antibody-based cancer therapy. First, we co-crystallized 401 in scFv format with the hROR2-Kr domain and used this information for affinity maturation by phage display. The in vitro evolved mAb was humanized by CDR grafting and rational back-mutations, confirmed to have retained its exclusive specificity to ROR2, and was again co-crystallized with the hROR2-Kr domain, providing a detailed picture of the paratope and epitope. Finally, conversion to a T cell–engaging ROR2 × CD3 biAb demonstrated potent in vitro killing of ROR1−/ROR2+ but not ROR1+/ROR2− cancer cells. We anticipate broad therapeutic utility of this reagent as T cell–engaging biAb, CAR-T, and other antibody-based cancer therapies.
Results
Crystallization of mAb XBR2-401 in complex with the human ROR2 kringle domain
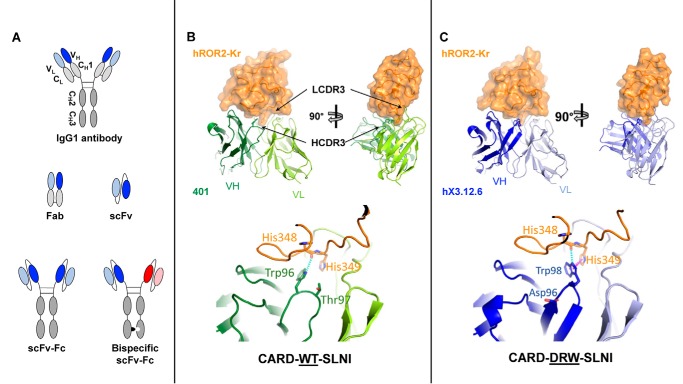
We previously reported a panel of 12 chimeric rabbit/human Fabs that were selected from a naive rabbit antibody library for binding to human ROR2 (18). Among these, mAb XBR2–401 (“401”) in Fab and IgG1 format (Fig. 1A) was shown to be specific for the kringle domain of ROR2 (hROR2-Kr) and to recognize both human and mouse orthologs but not its closest relative, ROR1 (18). To define the 401 paratope and epitope, we used X-ray crystallography to solve the structure of 401 in scFv (Fig. 1A) format in complex with hROR2-Kr at 1.2-Å resolution (Fig. 1B and Table S1) (Protein Data Bank ID (PDB): 6OSH). The crystal contained one complex in the asymmetric unit. All residues in the crystal were well-resolved except for the 15-amino acid scFv linker. The buried surface area between 401 and hROR2-Kr was 720 Å2, which comprised 7.0 and 15.9% of the total surface area of 401 and hROR2-Kr, respectively. The van der Waals contacts were dominated by HCDR2 and LCDR3 of 401. Notably, Ala-95 (Kabat numbering) from LCDR3 was nestled in a shallow hydrophobic pocket created by hROR2-Kr loop 3, 5, and 6 residues (27), Leu-350, Pro-368, Gln-371, Trp-376, Phe-378, and Met-386. On the other hand, His-349 of loop 3 from hROR2-Kr projected into the main pocket formed by the CDRs and made a salt bridge to Asp-32 of LCDR1 (Table 1). The interface also contained numerous direct and water-mediated hydrogen bond interactions dominated by residues from HCDR2 and LCDR1 (Table 1). Compared with the residues from HCDR2 and LCDR3 that are heavily involved in epitope recognition, Trp-96 (Kabat numbering) from HCDR3 provided limited interactions with hROR2-Kr through a suboptimal hydrogen bond with His-348 and van der Waals interactions with His-349 with the potential to form a π-π bond (Fig. 1B). This observation posed an opportunity for optimizing HCDR3 binding to the kringle domain.
Figure 1.
Antibody formats used in this study and crystal structures of parental mAb 401 and affinity-matured and humanized mAb hX3.12.6 in complex with hROR2-Kr. A, from top to bottom, the IgG1 format is naturally found as a dimer containing constant regions (gray) and two N-terminal variable regions (darker and lighter shades of blue for VH and VL, respectively), which bind the antigen. Fab only consists of the VH, CH1, VL, and CL domains. The scFv-Fc format contains an Fc domain but without CL and CH1 domains. VH and VL are fused via a polypeptide linker. Both variable regions are identical on scFv-Fc. The bispecific scFv-Fc format contains an Fc with knobs-into-holes mutations to allow for a dimer to form between two different chains, enabling the combination of two different variable regions (blue and red). The monospecific scFv-Fc format serves as a control for the bispecific scFv-Fc format. B (top), the crystal structure of scFv 401 (green) in complex with hROR2-Kr (orange) was determined by X-ray crystallography at 1.2 Å resolution (PDB: 6OSH). The complex on the right is rotated 90° to make interactions of LCDR3 and HCDR3 with the epitope visible. Bottom, zoomed-in image of the 401:hROR2-Kr complex reveals HCDR3 residue Trp-96 weakly hydrogen-bonding (cyan) with the backbone of hROR2-Kr residue His-348 with a distance of 3.4 Å. C (top), crystal structure of affinity-matured and humanized scFv hX3.12.6 (blue) in complex with hROR2-Kr at 1.4 Å resolution (PDB: 6OSV). Bottom, zoomed-in image of the hX3.12.6:hROR2-Kr complex reveals increased hydrogen (2.8 Å) and π-π/π-cation (magenta) (4.2 Å) bonding of HCDR3 residue Trp-98 with the backbone of hROR2-Kr residue His-348 and His-349, respectively. VH is shown in a darker shade, whereas VL is shown in a lighter shade of green (B) or blue (C).
Table 1.
Residue interactions between hROR2-Kr and 401 or hX3.12.6
| scFv | Type of interaction | Kr domain | scFv presence |
|---|---|---|---|
| HCDR2 | |||
| Asn-51 (ND2) | Hydrogen bond | Pro-345 (O) | 401, hX3.12.6 |
| Asn-51 (ND2) | Hydrogen bond | Ser-347 (OG) | 401, hX3.12.6 |
| Ala-53 (O) | Hydrogen bond | Lys-382 (NZ) | 401, hX3.12.6 |
| Asn-55 (ND1) | Hydrogen bond | Pro-345 (O) | 401, hX3.12.6 |
| Asn-55 (ND2) | Hydrogen bond | Lys-382 (O) | 401, hX3.12.6 |
| Tyr-57 (OH) | Hydrogen bond | Ser-347 (N) | 401, hX3.12.6 |
| HCDR3 | |||
| Trp-98 (NE1) | Hydrogen bond | His-348 (O) | hX3.12.6 |
| Trp-98 | π-π/π-cation | His-349 | hX3.12.6 |
| LCDR1 | |||
| Ser-30 (OG) | Hydrogen bond | His-349 (O) | 401, hX3.12.6 |
| Ser-31 (N) | Hydrogen bond | Asp-354 (OD2) | 401, hX3.12.6 |
| Ser-31 (OG) | Hydrogen bond | Asp-354 (OD2) | 401, hX3.12.6 |
| Asp-32 (OD1/2) | Salt bridge | His-349 (ND1) | 401, hX3.12.6 |
| LCDR3 | |||
| Ala-95 (O) | Hydrogen bond | Arg-385 (NH2) | 401, hX3.12.6 |
| Thr-96 (OG) | Hydrogen bond | His-349 (NE2) | 401, hX3.12.6 |
hROR2-Kr and hROR1-Kr share 58% amino acid sequence identity (3). When the epitope residues of hROR2-Kr that are recognized by 401 were compared with those mediating R11:hROR1-Kr recognition (19), no residue overlap was found (Fig. S1). This observation explains why there is no cross-reactivity between 401 and R11 despite hROR2-Kr and hROR1-Kr's homologous amino acid sequences. When comparing the antibody-bound kringle domains from the 401:hROR2-Kr and the R11:hROR1-Kr complex, the root mean square deviation (RMSD) of Cα positions was found to be 0.695 Å, revealing a highly conserved tertiary structure of the two Kr domains.
We also crystallized and solved the structure of antibody-unbound hROR2-Kr at 1.1 Å resolution (PDB: 6OSN) (Fig. S1B (right) and Table S1). The RMSD of the unbound hROR2-Kr structure to 401-bound hROR2-Kr was 0.383 Å, revealing only minor differences between the coordinates. Notably, in the unbound hROR2-Kr structure, Arg-385 formed a mixed salt bridge/hydrogen bond interaction with an acetate from the crystallization solution (Fig. S1B, right). The binding site, which overlaps with the canonical lysine-binding sites (LBSs) in other kringle domains (28), was partially covered by 401 in the crystal structure of the 401:hROR2-Kr complex. Superposition of the unbound hROR2-Kr structure to the 401-bound hROR2-Kr showed a minor shift in loop 5 due to the bound acetate ion (Fig. S1B, right).
Overall, our findings of no overlap between ROR2 and ROR1 epitopes along with suboptimal binding of 401's HCDR3 to hROR2-Kr revealed opportunities for in vitro affinity maturation.
Affinity maturation via phage display
A phage display library was constructed to conduct focused mutagenesis on 401's HCDR3 residues 96 and 97 (Kabat numbering; Fig. 2) with 0, 1, or 2 additional randomized residues (Table 2). Additional randomized positions were investigated because the co-crystal structure of 401:hROR2-Kr depicted an open cavity between hROR2 and the HCDR3, which could be filled by a longer matured HCDR3 and improve mAb affinity. Selection for hROR2-Fc binding was performed three ways: surface, surface competition, and solution competition panning (see “Experimental procedures”). Both competition panning protocols applied selection pressure toward Fabs with lower dissociation rate constants (koff) and thus higher affinity. From the three combined libraries, 144 clones were selected and analyzed via ELISA for ROR2 binding and Fab expression from supernatants. The top 12 clones with the highest absorbance ratio (hROR2 binding to expression) were purified. Using surface plasmon resonance (SPR), thermodynamic (KD) and kinetic parameters (kon and koff) of the interaction with hROR2 were determined. Clone XBR2-401-X3.12 (X3.12), which was obtained from the X3 library, revealed the highest affinity (KD = 0.7 nm) (Table 3 and Fig. S2A), which is at least a 5-fold improvement from 401. The X3.12 HCDR3 sequence differs from 401 at two residue positions and is one residue longer, changing the HCDR3 sequence from DWTSLNI to DDRWSLNI (Table 3). To make the affinity-matured X3.12 more therapeutically relevant, the next step was to employ humanization.
Figure 2.
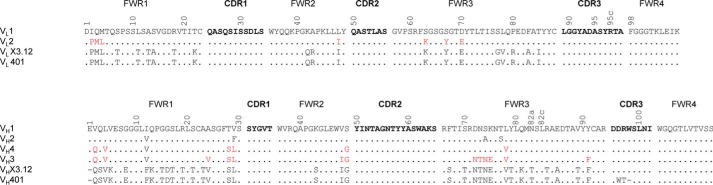
Amino acid sequence alignment of VL and VH of the affinity-matured and humanized variants. The location of the four framework regions (FWRs) and the three CDRs is indicated. The numbers refer to Kabat numbering of variable domain residues shown in single-letter code. Residues shown in red were back-mutated to the original rabbit residue. Dots, identical residues in the alignment to VL1 or VH1. CDR residues are shown in boldface type. Top, the VL1 FWRs are derived from human germline IGKV1-NL1*01; the VL1 CDRs are grafted from X3.12. The VL2 amino acid sequence is the same as VL1 but with seven back-mutations from the human germline to the original rabbit residues of X3.12. Bottom, the VH1 and VH2 FWRs are derived from human germlines IGHV3–66*03 and IGHV3–48*03, respectively, and their CDRs are grafted from X3.12. VH3 and VH4 vary from VH1 and VH2 by back-mutating FWR residues from the human germline to the original rabbit residues of X3.12.
Table 2.
Construction of focused randomized HCDR3 libraries
| ID | Randomized AAs | HCDR3 | Theoretical diversity |
|---|---|---|---|
| X2 | 2 | D XX SLNI | 103 |
| X3 | 2 + 1 insert | D XXX SLNI | 3.2 × 104 |
| X4 | 2 + 2 inserts | D XXXX SLNI | 106 |
Table 3.
Kinetic data of top 12 HDCR3 affinity-matured anti-ROR2 clones
| Clone Name | HCDR3 Seq. | KD (nM) | kon (104 m−1 s−1) | koff (10−3s−1) | χ2 |
|---|---|---|---|---|---|
| X3.12 | DDRWSLNI | 0.72 | 4.5 | 0.033 | 0.24 |
| X3.13 | DKGWSLNI | 1.1 | 2.6 | 0.027 | 0.095 |
| X2.12 | DTMSLNI | 1.2 | 3.4 | 0.040 | 0.41 |
| X4.3 | DWGNWSLNI | 1.3 | 3.0 | 0.038 | 0.47 |
| X2.23 | DYTSLNI | 1.4 | 3.3 | 0.047 | 0.36 |
| X2.7 | DSMSLNI | 1.5 | 6.9 | 0.10 | 0.47 |
| X3.21 | DGLTSLNI | 1.9 | 4.9 | 0.092 | 0.33 |
| X4.12 | DYMMNSLNI | 2.8 | 5.3 | 0.15 | 0.31 |
| X3.6 | DSRNSLNI | 3.2 | 6.0 | 0.19 | 0.53 |
| X4.21 | DSGVVSLNI | 3.4 | 2.4 | 0.081 | 0.12 |
| 401 | DWTSLNI | 7.0 | 3.1 | 0.22 | 0.43 |
| X4.9 | DNGSTSLNI | 10 | 3.3 | 0.34 | 0.22 |
| X4.35 | DSRRKSLNI | 31 | 5.4 | 1.7 | 1.5 |
Humanization by CDR grafting
Humanization of the affinity-matured chimeric rabbit/human X3.12 Fab was performed in three main steps. First, the human germlines with the closest identity to X3.12's variable light-chain (VL) and variable heavy-chain (VH) amino acid sequences were identified using IgBlast (see “Experimental Procedures” for online tools). The IMGT repertoire was then referenced to eliminate germlines with more than three polymorphisms. The human germlines that had the highest amino acid sequence identity to X3.12 with the least number of polymorphisms were heavy-chain germlines IGHV3-66*03 and IGHV3-48*03 and light chain germline IGKV1-NL1*01, which are 54.3, 54.8, and 65.6% identical to the X3.12 heavy and light chain, respectively. Second, CDRs from X3.12 determined using Kabat numbering were grafted into these three framework sequences (Fig. 2, VL1, VH1, and VH2). Third, residues determined to preserve affinity (29) were back-mutated from the human germline residues to the original rabbit residues. From these three steps comprised of CDR grafting and rational back-mutations, four heavy chain (VH1–VH4) and two light chain variants (VL1 and VL2) were formed (Fig. 2). These humanized chains were compared with the IMGT database of human germline antibody sequences using the IMGT/DomainGapAlign tool to determine the human identity percentage. The human identity of VL1, VL2, and VH1-VH4 was 88.6%*, 87.1%, 86.6%, 87.8%, 73.5%*, and 80.4%*, respectively, where the percentages with an asterisk indicate that the first “hit” on IMGT DomainGapAlign was not human. World Health Organization standards state that the first “hit” on IMGT DomainGapAlign tool must be human along with human identity being above 85% for the antibody to be considered humanized (26). All heavy- and light-chain Fab combinations were cloned into a pET11a variant (30) and expressed in the Escherichia coli Rosetta strain followed by quantification of Fab expression and hROR2 binding via ELISA to eliminate nonbinding clones. The remaining clones hX3.12.5, hX3.12.6, hX3.12.7, and hX3.12.8 had higher binding/expression ratios compared with X3.12. The overall percentage identity of these variants to human germlines was 87, 87, 80, and 84%, respectively, and all utilized VL2 (Table 4). Therefore, hX3.12.5 and hX3.12.6 are considered humanized, whereas hX3.12.7 and hX3.12.8 are considered chimeric mAbs by World Health Organization standards.
Table 4.
Kinetic data of humanized anti-ROR2 mAbs
| Clone namea | Heavy and light chains | Human identity (%) | KD (nm) | kon (104 m−1 s−1) | koff (10−3 s−1) | χ2 |
|---|---|---|---|---|---|---|
| hX3.12.5 | VH1,VL2 | 87 | 2.6 | 19 | 0.48 | 0.48 |
| hX3.12.6 | VH2,VL2 | 87 | 3.8 | 6.0 | 0.22 | 1.6 |
| hX3.12.7 | VH3,VL2 | 80b | 1.4 | 11 | 0.16 | 2.7 |
| hX3.12.8 | VH4,VL2 | 84b | 5.2 | 13 | 0.67 | 0.70 |
| h401.6 | VH2,VL2 | 87 | 16 | 1.4 | 0.23 | 0.09 |
a Clone names have been shortened for simplicity to exclude “XBR2-401” from the front.
b Closest reference gene on IMGT/DomainGapAlign is not human.
Characterization of affinity-matured and humanized Fabs
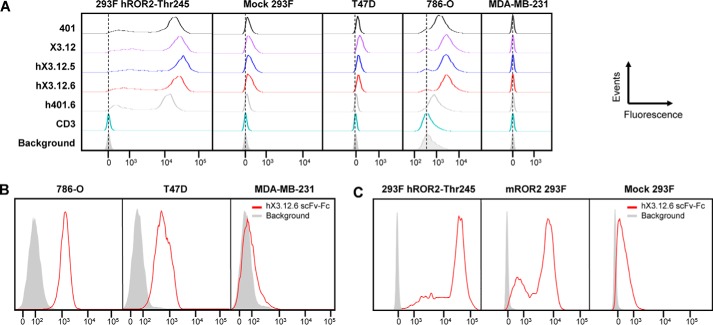
Following expression and purification (Fig. S3A), the affinities of hX3.12.5, hX3.12.6, hX3.12.7, and hX3.12.8 were determined to be 2.6, 3.8, 1.4, and 5.2 nm, respectively, by SPR (Table 4 and Fig. S2B). Moving forward, we focused on hX3.12.5 and hX3.12.6 Fabs, as they have the highest human identity while retaining nanomolar affinity for hROR2. Supporting flow cytometry data showed hX3.12.5- and hX3.12.6-bound HEK 293F cells stably overexpressing hROR2-Thr-245 allotype (18) while minimally binding the mock-transfected HEK 293F control cell line, which has some ROR2 expression (Fig. 3A). A Fab containing hX3.12.6 framework regions and parental 401 CDR sequences was generated and included for reference (h401.6, 16 nm affinity; Table 4). Clones hX3.12.5 and hX3.12.6 also bound to breast cancer cell line T47D (ROR2+, ROR1−) and renal cell adenocarcinoma cell line 786-O (ROR2+, ROR1+) but not to breast cancer cell line MDA-MB-231 (ROR2−, ROR1+) (Fig. 3A). The thermostability of humanized Fabs was addressed using LightCycler 480 to measure their melting temperatures. Chimeric rabbit/human Fabs X3.12 and 401 exhibited slightly higher thermostability than the humanized Fabs hX3.12.5 and hX3.12.6 along with h401.6 (Fig. S4). The melting temperatures of the affinity-matured and humanized Fabs are similar to previously reported Fab melting temperatures and suggest that they are stable (31, 32).
Figure 3.
Analysis of affinity-matured and humanized mAbs by flow cytometry. A, HEK 293F cells stably transfected with human ROR2 (allotype Thr-245) were stained with 5 μg/ml of the indicated parental, affinity-matured, and humanized Fabs followed by phycoerythrin-conjugated goat anti-human F(ab′)2 pAbs. Mock-transfected HEK 293F cells served as negative control. The Fabs were also tested against T47D (ROR2+, ROR1−), 786-O (ROR2+, ROR1+), and MDA-MB-231 (ROR2−, ROR1+) cell lines. Humanized anti-human CD3 Fab v9 and secondary antibody alone (Background; gray shade) served as additional negative controls. B, after its conversion from Fab to scFv-Fc, hX3.12.6 at 5 μg/ml followed by Alexa Fluor 647–conjugated donkey anti-human F(ab′)2 pAbs was used to stain 786-O, T47D, and MDA-MB-231 cell lines. Secondary antibody alone (gray shade) served as negative control. C, flow cytometry using hX3.12.6 scFv-Fc (5 μg/ml) followed by Alexa Fluor 647–conjugated donkey anti-human F(ab′)2 pAbs for staining HEK 293F cells stably transfected with human ROR2 (allotype Thr-245) or mouse ROR2. Mock-transfected HEK 293F cells and secondary antibody alone (Background; gray shade) served as negative controls. All events were normalized to mode.
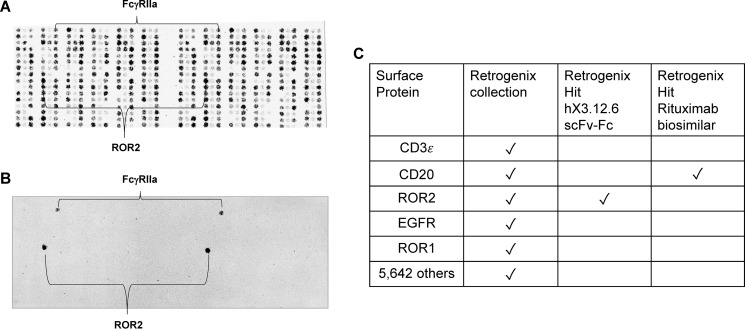
To test hX3.12.6's specificity to ROR2, the Fab was first converted to an IgG1-like format, scFv-Fc. This format contains two scFvs utilizing a (Gly4Ser)3 linker between VL and VH and fused to the human IgG1 Fc fragment (Fig. 1A). The hX3.12.6 scFv-Fc was screened against 786-O, T47D, and MDA-MB-231, confirming all to be ROR2+ (Fig. 3B) except MDA-MB-231, as described previously. We also confirmed hX3.12.6's cross-reactivity with mouse ROR2 (Fig. 3C), which was stably expressed on HEK 293 as described previously (18). As we did for the parental mAb XBR2–401 in chimeric rabbit/human IgG1 format (18), the hX3.12.6 scFv-Fc was screened against 5,647 human plasma membrane proteins (i.e. human cell surface antigens) expressed on the surface of human HEK 293 cells and arrayed on 16 slides in duplicate (33). The spotting pattern of ZsGreen1, which correlates with human cell surface antigen expression (Fig. 4A), is shown for the slide that contained human ROR2 (Fig. 4B). The only specific interaction identified in the hX3.12.6 scFv-Fc screen was ROR2 (Fig. 4, B and C), confirming that neither affinity maturation nor humanization diminished the high specificity of the parental mAb.
Figure 4.
Analysis of the specificity of affinity matured and humanized mAb hX3.12.6 by cell microarray technology. Using Retrogenix's custom cell microarray technology, mAb hX3.12.6 in scFv-Fc format was screened against 5,647 human plasma membrane proteins expressed on the surface of human HEK 293 cells. A, image of Retrogenix's custom cell microarray ZsGreen1 spotting pattern for ∼300 human plasma membrane proteins in duplicate on one of 16 slides. The array also included untransfected HEK 293 cells as a control. B, humanized scFv-Fc hX3.12.6 was screened at 20 μg/ml and was visualized using Alexa Fluor 647–conjugated goat anti-human IgG Fcγ pAbs. FcγRIIa is an expected nonspecific hit due to Fc binding. C, summary of the cell microarray screen where 20 μg/ml hX3.12.6 scFv-Fc against 5,647 human antigens arrayed in duplicate revealed ROR2 as the only specific hit. A rituximab biosimilar (1 μg/ml) was screened in parallel as control.
Next, hX3.12.6 scFv was co-crystallized with hROR2-Kr, and its structure was determined at 1.4 Å resolution (PDB: 6OSV) (Fig. 1C and Table S1). The structure of the complex of hX3.12.6 and hROR2-Kr allowed us to compare the affinity-matured HCDR3 with the parental HCDR3, particularly the π-π interaction formed with hROR2-Kr (Fig. 1C and Table 1). Similar to 401, in hX3.12.6, Ala-95 was buried in hROR2-Kr, and the salt bridge between light-chain Asp-32 and hROR2-Kr was also retained. All hydrogen bond interactions and van der Waals contacts present in 401:hROR2-Kr remained intact in the hX3.12.6:hROR2-Kr complex except for the changes in the HCDR3 due to affinity maturation, which include Asp-96 and Arg-97. Whereas these two residues do not directly interact with hROR2-Kr, they help to properly position Trp-98, which does contact hROR2-Kr. Trp-98 in hX3.12.6 further improves interactions with hROR2-Kr, having been optimized from 401's Trp-96, located at the tip of the HCDR3 loop (Fig. 1, B and C). Unlike in 401, the side chain of Trp-98 made an optimal hydrogen bond interaction with the backbone oxygen of hROR2-Kr's His-348. The side chain of Trp-98 also displayed geometric characteristics of π-π/π-cation interactions with hROR2-Kr His-349 (Fig. 1C (bottom) and Table 1). The RMSD between 401 and hX3.12.6 in their respective co-crystal structures was found to be 0.446 Å, suggesting subtle differences between the structures (Fig. S1A). The crystallized kringle domains in complex with either 401 or hX3.12.6 had an RMSD of 0.279 Å, indicating no relevant change between the two kringle domains. Collectively, these findings confirmed that our rational design of affinity maturation was critical in the improvement of ROR2 binding.
Generation and characterization of ROR2 × CD3 bispecific antibodies
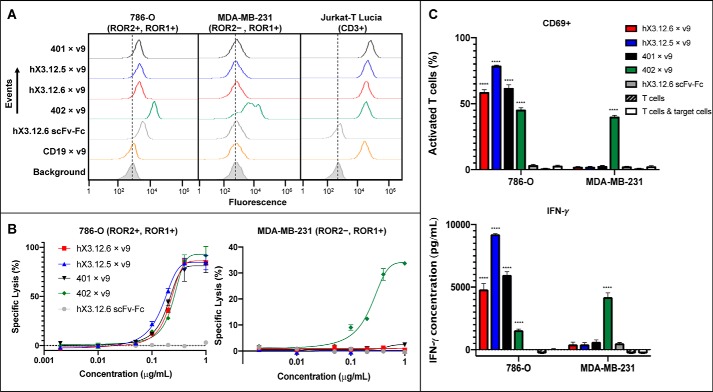
To determine the affinity-matured and humanized antibodies' functionality, hX3.12.6 was converted to a ROR2 × CD3 biAb and purified by Protein A and SEC (Fig. 1A and Fig. S3 (B and C)). We used the same heterodimeric and aglycosylated scFv-Fc format we previously reported for ROR1 × CD3 and CD19 × CD3 biAbs (19, 34). This was done by combining aglycosylation mutation N297A in the CH2 domain with knobs-into-holes mutations in the CH3 domain, specifically CH3 knob mutations S354C and T366W and CH3 hole mutations Y349C, T366S, L368A, and Y407V (35–37). As for the anti-CD3 arm, the well-defined affinity-matured and humanized anti-human CD3 mAb v9 was used (38). BiAbs were confirmed to bind 786-O and Jurkat-T Lucia (CD3+) while not binding MDA-MB-231 (Fig. 5A). ROR1-targeting XBR1-402 × v9 (402 × v9) biAb along with the monospecific hX3.12.6 scFv-Fc were used as controls. Primary T cells were expanded in vitro from two different healthy donor peripheral blood mononuclear cells (PBMCs) by the addition of anti-CD3/anti-CD28 beads and interleukin-2. Next, in vitro biAb-mediated target-dependent cytotoxicity by the expanded primary T cells was examined. Specific lysis of 786-O cells was seen for the three ROR2 × CD3 biAbs (401 × v9, hX3.12.6 × v9, and hX3.12.5 × v9) and positive control 402 × v9 with EC50 values of 0.19, 0.21, 0.15, and 0.25 μg/ml, respectively (1.9, 2.1, 1.5, and 2.5 nm) (Fig. 5B). Monospecific hX3.12.6 scFv-Fc was used as a negative control. To confirm that these biAbs are specifically killing via binding ROR2 on 786-O cells and CD3, we tested a ROR2−, ROR1+ cell line, MDA-MB-231, and found that all ROR2 × CD3 biAbs were inactive up to 1 μg/ml (Fig. 5B). The positive control ROR1 × CD3 biAb did show specific cell lysis as expected due to its binding to ROR1 on MDA-MB-231 cells. T-cell activation was quantified by flow cytometry using an anti-CD69 mAb, a known marker of early T-cell activation. Humanized biAbs hX3.12.6 × v9, hX3.12.5 × v9, and parental 401 × v9 incubated with T cells at 0.2 μg/ml up-regulated CD69 on over 50% of T cells in the presence of 786-O but not MDA-MB-231 cells (Fig. 5C). The negative control hX3.12.6 scFv-Fc did not reveal up-regulation of CD69. The release of type 1 cytokine IFN-γ was assessed by ELISA, where all ROR2 × CD3 biAbs caused cytokine release in the presence of ROR2+ but not ROR2− target cells (Fig. 5C). As shown previously (19), R11 × v9 caused comparable cytokine release in the presence of ROR1+ target cells.
Figure 5.
Activity of ROR2 × CD3 biAbs. A, the indicated ROR2 × CD3 biAbs along with control ROR1 × CD3 and CD19 × CD3 biAbs (all in heterodimeric scFv-Fc format) at 1 μg/ml in conjunction with Alexa Fluor 647–conjugated donkey anti-human F(ab′)2 pAbs were used to stain 786-O, MDA-MB-231, and Jurkat-T Lucia cells to confirm specific binding to ROR2 and CD3. Secondary antibody alone (Background; gray shade) served as negative control. B, a panel of ROR2 × CD3 biAbs based on the humanized and affinity-matured mAbs and the parental mAb was compared with a ROR1 × CD3 biAb (all in heterodimeric scFv-Fc format) and a monospecific negative control without T cell–engaging arm. Plotted is the specific lysis of cell lines 786-O (ROR2+, ROR1+) and MDA-MB-231 (ROR2+, ROR1−) after a 16-h incubation with the indicated range of biAb concentrations and ex vivo expanded T cells at an effector/target cell ratio of 10:1. T-cell activation was measured as percentage of CD69+ T cells determined by flow cytometry and cytokine IFN-γ release determined by ELISA (C). Based on independent triplicates shown as mean ± S.D. (error bars), one-way analysis of variance was used to analyze significant differences between ROR2 × CD3 (or ROR1 × CD3) biAbs and the monospecific scFv-Fc negative control (****, p < 0.0001).
Discussion
Obtaining a humanized antibody with high affinity for ROR2, a cancer target with relatively low cell surface density, facilitates investigations for therapeutic utility. A previous study (18) took the first step in selecting a well-defined anti-ROR2 mAb, 401, from a naive rabbit antibody library by phage display and confirming its high affinity and specificity. In the current study, we further characterized 401 by co-crystallization with its antigen, hROR2-Kr. The crystal structure revealed that the hROR2-Kr epitope residues bound by 401 did not overlap with the hROR1-Kr epitope residues bound by R11. This further defined 401 as specific to hROR2-Kr and not cross-reactive with ROR1. The dominant interactions between 401:hROR2-Kr were within the LCDR3 and HCDR2. Notably, LCDR3 dominance in rabbit antibody paratopes is not uncommon (39). Between HCDR3 and the hROR2-Kr there was weak hydrogen bonding and potential for a π-π interaction between Trp-96 and hROR2-Kr His-349. From these findings in the crystal structure, we hypothesized that elongating and mutating the HCDR3 would improve 401's affinity.
Affinity maturation of 401 was done by generating and selecting an HCDR3-targeted Fab-phage display library. This library allowed selection from variants that incorporated mutations in positions 96 and 97 of VH with the addition of up to two additional residues in the HCDR3. Selecting from this library produced 10 Fabs sustaining affinities below 10 nm as determined by SPR. The top 10 Fabs displayed varying lengths of HCDR3, with four containing one additional residue and three containing two additional residues. This suggests that seven of the 10 top clones would not have been selected from a strictly mutagenized HCDR3 library. Also, within the top four clones, three contained a Trp at position 98 or 99 compared with 401's Trp at 96, suggesting that improved π-π interactions were critical in the affinity maturation. The top Fab, X3.12, had a KD of 0.72 nm, a 5–10-fold improvement from parental mAb 401. The parental HCDR3 sequence starting at position 95 is DWTS, where the affinity-matured sequence is DDRWS. The affinity-matured HCDR3 sequence of X3.12 originated from the X3 library, as there is an extra residue in addition to the two randomized residues at positions 96 and 97. This selection emphasizes the value of structural guidance in affinity maturation as additional residues were incorporated in order to fill the cavity seen in the crystal structure between HCDR3 and hROR2-Kr.
With the matured affinity of X3.12, the next step was to make the mAb therapeutically relevant by humanization. Humanization of the rabbit variable domains is expected to lower the risk of possible immunogenicity after repeated administration (39). CDR grafting and rational back-mutations to X3.12 produced eight humanized variants. The optimal humanized anti-ROR2 mAb was hX3.12.6, which had a KD of 3.8 nm, a >5-fold improvement from h401.6, which contains the parental HCDR3 and the humanized frameworks from hX3.12.6, improving similarly to the affinity maturation. Thus, reversing the order of affinity maturation and humanization would have likely yielded similar clones. As discussed for parental 401, the light chain contained a large portion of the hydrogen bonds along with burial of the LCDR3 in the hROR2-Kr. These interactions are critical for retention of affinity and paratope structure, which supported the idea of conserving a higher percentage of the original rabbit residues in the light chain than the heavy chain when humanizing. All anti-ROR2 humanized variants presented (hX3.12.5, hX3.12.6, hX3.12.7, and hX3.12.8) maintained single-digit nanomolar affinity for hROR2-Kr. Humanization did not impair the mouse/human ROR2 cross-reactivity, which is an important characteristic for preclinical studies.
To distinguish the structural differences due to affinity maturation and humanization, the hX3.12.6:hROR2-Kr complex crystal structure was obtained. The percentage of buried surface area of each constituent in the 401:hROR2-Kr and hX3.12.6:hROR2-Kr complexes was 7.0%:15.9% and 6.8%:16%, respectively. The RMSD between the two co-crystal structures was 0.474 Å, with respect to the Cα positions, showing that they are essentially identical. The only noticeable difference between 401 and hX3.12.6 crystal structures is the HCDR3 loops due to affinity maturation changing the sequence, as discussed above, and a slight shift in HCDR1 due to changing the framework residue from Leu-29 to Phe-29 during humanization, located in the human framework region directly upstream of HCDR1. However, we mutated this residue back to Leu-29 and did not observe a notable difference in affinity (data not shown).
The kringle domain takes part in protein-protein interactions and is thought to be a binding mediator (27). In plasminogen, kringle domains bind to effector molecules such as fibrin via LBSs (28). Our crystal structures of ROR2-Kr in complex with 401 and hX3.12.6 revealed that it contains an LBS that may be involved in receptor-ligand or receptor-receptor (cis and trans) interactions. Whereas a functional role of the LBS of ROR2-Kr remains unknown, it is interesting to note that 401 and hX3.12.6 partially cover it and as such may sterically hinder it from binding a ligand or co-receptor.
Recent immunohistochemistry studies utilizing a mAb binding to an intracellular epitope found that ROR1 is more widely expressed than originally thought (40). There is currently limited knowledge of ROR2 compared with ROR1, mostly due to poor reagents and inconsistency. For example, one study found two of three commercially available anti-human ROR2 antibodies have off-target cross-reactivity (41). This issue is widespread, but unreliable studies can be reduced by having mAbs with defined and disclosed VH and VL sequences that are well-characterized (42). This is the case for the anti-ROR2 mAbs that we report here, laying the groundwork for consistency in the field. However, as in the case of ROR1, a mAb binding to an intracellular epitope of ROR2 may be a preferred reagent for immunohistochemistry and, as such, a suitable companion diagnostic. A recent study by Hellmann et al. (43) described the selection of fully human anti-human ROR2 mAbs and their conversion to antibody-drug conjugates. Although these mAbs are human, most are not cross-reactive with mouse ROR2. Further studies are needed to confirm the exclusive specificity of these mAbs for ROR2 and their epitopes.
Previous studies have shown that membrane-proximal epitopes of ROR1 can mediate more potent responses against cancer cells when targeted by CAR-Ts and T cell–engaging biAbs (18, 19, 44). These studies prompted us to convert hX3.12.6, which binds a membrane-proximal epitope on hROR2-Kr, into a T cell–engaging biAb. In the current study, we show that hX3.12.6 × v9 in an scFv-Fc aglycosylated format can cause T-cell activation specifically as seen in the in vitro cytotoxicity toward 786-O (ROR2+) but not MDA-MB-231 (ROR2−) cells. This suggests therapeutic utility of hX3.12.6 as the ROR2-targeting arm of T cell–engaging biAbs. In addition, as we reported previously, parental mAb 401 can be converted to potent and specific CAR-Ts (18). Its epitope mapping by co-crystallization, affinity maturation, and humanization in the current study affords a highly attractive molecule for investigating ROR2 targeting by T cell–engaging biAbs and CAR-Ts in preclinical models of ROR2-expressing malignancies.
Experimental procedures
Cell lines and primary cells
Breast cancer cell lines MDA-MB-231 and T47D and were purchased from ATCC and grown in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific), 100 units/ml penicillin/streptomycin (Thermo Fisher Scientific), and 10% (v/v) FBS (BioFluid Technologies). Renal cell adenocarcinoma cell line 786-O (an NCI-60 panel cell line obtained from The Scripps Research Institute's Cell-Based High-Throughput Screening Core) were grown in RPMI 1640 (Thermo Fisher Scientific), 100 units/ml penicillin/streptomycin (Thermo Fisher Scientific), and 10% (v/v) FBS (BioFluid Technologies). HEK 293F cells stably transfected with human ROR2 (allotype Thr-245), mouse ROR2, and mock vector were previously published (18). The same study showed that mAb 401 binds to both ROR2 allotypes, Thr-245 and Ala-245, which arise from an SNP (rs10820900) in the frizzled domain of human ROR2 (hROR2-Fz). Human PBMCs were purchased from AllCells and cultured in X-VIVO 20 medium (Lonza) with 5% (v/v) off-the-clot human AB serum (Gemini Bio-Products) and 100 units/ml interleukin-2 (Cell Sciences). Primary T cells were expanded from PBMCs as described previously (19) by using Dynabeads ClinExVivo CD3/CD28 (Thermo Fisher Scientific). The Jurkat-T Lucia NFAT reporter cell line was purchased from InvivoGen and cultured in RPMI 1640 (ATCC modification) medium with 100 units/ml penicillin/streptomycin and 10% (v/v) FBS (BioFluid Technologies); 100 μg/ml zeocin (InvivoGen) was added for every other passage.
Crystallization and structure determination of 401 and hX3.12.6 in complex with hROR2-Kr
Cloning, expression, and purification
DNA fragments encoding Fab 401, Fab hX3.12.6, and full-length human ROR2 (clone ID 40146553; GE Healthcare Dharmacon) were each amplified by PCR to create the corresponding scFv formats (VH-3x(GGGGS)-VL) and the kringle domain of human ROR2 (hROR2-Kr). The PCR product encoding hROR2-Kr was cloned into a pET15b expression vector (Novagen), which was modified to co-express E. coli chaperone/disulfide-isomerase (DsbC) (45), to create pET15b-hROR2-Kr-DsbC, which expresses hROR2-Kr with a thrombin-cleavable N-terminal hexahistidine tag. The scFv-encoding PCR products containing their own ribosome-binding sites were inserted between hROR2-Kr and DsbC, affording pET15b-hROR2-Kr-scFv401-DsbC and pET15b-hROR2-Kr-scFv hX3.12.6-DsbC. The three expression plasmids were each transformed into E. coli bacteria of the Rosetta-gami2(DE3) strain (Novagen). Transformed E. coli were grown in lysogeny broth medium containing ampicillin, tetracycline, and chloramphenicol, at 37 °C, with 230-rpm agitation. Protein expression was induced with 0.3 mm isopropyl β-d-thiogalactoside when the cell density reached A595 of 0.6. The cells were grown further for 18 h at 20 °C.
Protein purification
Bacterial pellets were resuspended in sonication buffer (20 mm HEPES, pH 8.0, 500 mm NaCl, 15 mm imidazole, 10% (v/v) glycerol), sonicated in an ice-water bath, and centrifuged for 25 min at 53,300 × g. The supernatants were loaded on a custom-packed 10-ml HIS-Select column (Sigma–Aldrich) and washed with sonication buffer. Bound proteins were eluted with a linear gradient of imidazole from 15 to 500 mm. The eluted proteins were treated overnight at 4 °C with thrombin (Sigma–Aldrich) to remove the N-terminal hexahistidine tags at hROR2-Kr. The cleaved proteins were purified further on a Superdex 200 26/60 column (GE Healthcare) equilibrated with 50 mm NaCl, 10 mm HEPES, pH 7.4.
Crystallization and structure determination
Crystals of the scFv401:hROR2-Kr complex were grown by vapor diffusion at room temperature (RT) using 1.5 μl of 14 mg/ml protein and an equal volume of precipitant containing 0.1 m sodium citrate tribasic dihydrate, 15% (w/v) PEG 3350 and were fully grown within 2 days. hROR2-Kr produced clustered crystals in vapor diffusion at RT using 2 μl of 14 mg/ml protein with 1 μl of precipitant containing 0.2 m lithium acetate, 20% (w/v) PEG 3350. The crystal clusters were crushed and seeded to drops equilibrated with a protein/precipitant ratio of 3:1 to obtain single crystals. Crystals of the scFv hX3.12.6:hROR2-Kr complex were grown by vapor diffusion at RT using 1.5 μl of 3 mg/ml protein and an equal volume of precipitant containing 10 mm MgCl2 hexahydrate, 5 mm nickel (II) chloride hexahydrate, 0.1 m Na-HEPES, pH 7.0, 13% (w/v) PEG 4000. The crystals were flash-frozen in liquid nitrogen using nylon loops after removing excess mother liquor. Diffraction data sets with Bragg spacings set to 1.1 Å for both scFv401:hROR2-Kr and hROR2-Kr were collected on a Rayonix MX300 detector at the Advanced Photon Source (APS) beamline LS-CAT 21-ID-F synchrotron facility (Argonne National Laboratory). A diffraction data set with Bragg spacings set to 1.3 Å for scFv hX3.12.6:hROR2-Kr was collected on a PILATUS3 S 6M detector at the Advanced Light Source (ALS) beamline 5.0.2 synchrotron facility (Lawrence Berkeley National Laboratory). Data sets were processed with autoPROC using XDS as the processing engine (46). The structures were solved by the molecular replacement method using PHASER (47) with PDB entry 6BA5 (55) (scFvR11:hROR1-Kr) (19) as the search model. Crystallographic refinements were performed using PHENIX version 1.14 (48). Manual rebuilding and adjustment of the structures were done in Coot (49). Data processing and refinement statistics are shown in Table S1. Molecular images (Fig. 1 (B and C) and Fig. S1 (A and B)) were created using PyMOL (50). Interaction interfaces were analyzed using PDBePISA (51, 52). Structure validations were carried out with MolProbity (53).
Affinity maturation
Library generation
A stop mutant of 401 was generated using overlap extension PCR with sense primer STOP and antisense primer unirev to avoid contamination of the library with parental 401. Subsequently, three different libraries were generated to randomize and extend HCDR3 with NNK codons: two randomized codons (X2) located at residues 96 and 97 on HCDR3; an additional randomized codon immediately downstream of residue 97 (X3); and two additional randomized codons immediately downstream of residue 97 (X4). These variants were generated by overlap extension PCR utilizing the NNK-degenerated sense primer X2, X3, or X4 together with antisense primer unirev. The final constructs were cloned into phagemid pC3C as described previously along with flanking primers C-5′SFIVL and c-3′sfivh (18). Primer sequences were as follows: STOP, 5′-ACCTATTTCTGTGCGAGGATTGTAATCCCTTAACATCTGGGGACCA-3′; unirev, 5′-ATCTCTCGCACAGAAATAGGT-3′; X2, 5′-ACCTATTTCTGTGCGAGAGATNNKNNKTCCCTTAACATCTGGGGACCA-3′; X3, 5′-ACCTATTTCTGTGCGAGAGATNNKNNKNNKTCCCTTAACATCTGGGGACCA-3′; X4, 5′-ACCTATTTCTGTGCGAGAGATNNKNNKNNKNNKTCCCTTAACATCTGGGGACCA-3′.
Library selection
Following published protocols for the selection of chimeric rabbit/human Fab by phage display (54), three different panning approaches were investigated, where the first was conventional surface panning (3 rounds) using 1 μg of the hROR2-Fc (18) in 25 μl of PBS for immobilization on a 96-well ELISA plate (Costar 3690; Corning), 3% (w/v) BSA in PBS for blocking, and 10 wash steps using 0.05% (v/v) Tween 20 in PBS (TPBS). The second approach was surface competition panning (using the second round of the conventional surface panning, one round was conducted) using 100 ng of hROR2-Fc, immobilized and blocked as above, followed by a 2-h pre-incubation with a 10-fold molar excess of the parental Fab 401 and 15 wash steps with TPBS. The third approach was solution competition panning (5 rounds) using 2-fold decreasing amounts of biotinylated (54) hROR2-Fc (100 to 6.25 ng in PBS) and pre-incubation with a 10-fold molar excess of the parental Fab 401 to the hROR2-Fc at the various concentrations. At each step, capturing with streptavidin-coated magnetic beads (Dynabeads MyOne Streptavidin C1; Thermo Fisher Scientific), acid elution using 100 mm glycine-HCl (pH 2.2), coupling of panning rounds 2 and 3 and rounds 4 and 5 (i.e. without intermittent reamplification), and increasingly stringent wash steps (0.05% (v/v) to 0.5% (v/v) Tween 20 in PBS) were conducted. Using published protocols (54), final output colonies were screened by a Fab ELISA using immobilized hROR2-Fc, and the HCDR3 of positive clones was determined by DNA sequencing.
Humanization
Humanization of X3.12 was done by finding the closest human germline(s) using IgBlast (RRID:SCR_002873) with the least amount of polymorphisms, which was determined using IMGT's IGHV and IGKV mammalia human (Homo sapiens) links (RRID:SCR_018220). Rational mutations (29) were performed in varying severity to determine mutations that were necessary to retain ROR2 affinity.
Fab cloning, expression, and purification
All selected Fab variants were cloned as described with modifications (18). Briefly, Fab variants were cloned into bacterial expression vector pET11a (30), transformed into E. coli strain Rosetta (DE3) (EMD Millipore), and tandemly purified from culture supernatants using a 1-ml HiTrap Kappa Select HP column followed by a 1-ml HisTrap HP column in conjunction with an ÄKTA FPLC instrument (all from GE Healthcare). (The initial top 12 chimeric rabbit/human anti-human Fabs were purified using only the 1-ml HisTrap HP column.) Purity of protein was analyzed by SDS-PAGE and Coomassie Blue staining, and A280 absorbance was used to determine the concentration of purified Fab variants.
Surface plasmon resonance
Kinetic and thermodynamic parameters for the ROR2 binding of purified anti-ROR2 Fab variants were measured by the use of SPR as described previously (18), performed on a Biacore X100 instrument using Biacore reagents and software (GE Healthcare). Briefly, a CM5 sensor chip was immobilized with a mouse anti-human IgG CH2 mAb to capture the hROR2-Fc antigen. Each Fab variant was diluted to 100 nm 1× HBS-EP+ running buffer and further diluted 2-fold using 1× HBS-EP+ running buffer to make five dilutions in total with a replicate of the lowest concentration after measuring the highest concentration to confirm regeneration of the sensor chip.
Thermostability assay
Steps were followed as described in the LightCycler 480 Instrument Quick Guide (Roche Applied Science) for protein melting. Optimal conditions were determined using parental Fab 401 at 1 mg/ml and the suggested Optimization Table 1 in the Quick Guide. Roche Protein Melting software was used for analysis. The optimal conditions required 0.5 μl of 1 mg/ml Fab, 1.0 μl of SYPRO Orange Dye 100× stock, 8.50 μl of Dulbecco's PBS. All Fabs were tested under these conditions and in triplicates.
Retrogenix cell microarray
Custom prescreens, full screens, and postscreens were carried out by Retrogenix as described previously (18, 33).
Functionality studies
Production of ROR2 × CD3 bispecific antibody
Cloning, expression, and purification of ROR2 × CD3 biAbs in heterodimeric aglycosylated scFv-Fc format followed a previously described protocol with modifications (34). In short, the scFv-encoding sequences were synthesized as gBlocks containing a signal peptide–encoding sequence at the N terminus (Integrated DNA Technologies). Overlap extension PCR was used to include sequences encoding hinge and heavy-chain constant domains CH2 and CH3 of human IgG1. Previously described hole and knob mutations (37) were included in the CD3 scFv-hinge-CH2-CH3– and ROR2 scFv-hinge-CH2-CH3–encoding sequences, respectively. The aglycosylation mutation N297A in CH2 was included in both. These scFv-Fc–encoding sequences were then inserted into mammalian expression vector pCEP4 using KpnI and XhoI restriction sites. Following DNA sequencing (Eton Bioscience) for verification, the plasmids were transfected into HEK 293F cells (Thermo Fisher Scientific) using polyethyleneimine (Polysciences) at 3 × 106 cells/ml cultured in 150 ml of FreeStyle medium (Thermo Fisher Scientific) shaking at 37 °C in an atmosphere of 8% CO2 and 100% humidity. After 6–12 h, an additional 150 ml of FreeStyle medium was added. Supernatants were collected after 3 days followed by filtration and purification using a 1-ml HiTrap Protein A HP column (GE Healthcare) in conjunction with an ÄKTA FPLC instrument (GE Healthcare) followed by size-exclusion chromatography using a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with water followed by Dulbecco's PBS. Yields were typically ∼5–10 mg/liter. The purity of the biAbs was confirmed by SDS-PAGE followed by Coomassie Blue staining and quantified by A280 absorbance.
Flow cytometry
Similar to standard methods and as described previously (18, 19), staining of 1 × 105 target cells was done with 5 μg/ml Fab or biAb in 100 μl of cytometry buffer (PBS supplemented with 1% (w/v) BSA and 0.1% (w/v) sodium azide). After washing, the cells were incubated with a 1:1,000 dilution of phycoerythrin-conjugated goat anti-human IgG F(ab′)2 fragment–specific pAbs or Alexa Fluor 647–conjugated donkey anti-goat IgG (H+L) pAbs (both in F(ab′)2 format from Jackson ImmunoResearch) in 100 μl of flow cytometry buffer on ice for 1 h. Alexa Fluor 647–conjugated mouse anti-human CD69 mAb was purchased from BioLegend. Cells were analyzed using a FACSCanto instrument (BD Biosciences) and FlowJo analytical software (Tree Star).
In vitro cytotoxicity and T-cell activation assays
Cytotoxicity was measured by using CytoTox-Glo (Promega) following the manufacturer's protocol and a previous publication (34) with minor modifications. Primary T cells expanded from healthy donor PBMCs were used as effector cells, and 786-O or MDA-MB-231 cells were used as target cells at an effector/target cell ratio of 10:1. Cells were incubated in X-VIVO 20 medium (Lonza) with 5% (v/v) off-the-clot human AB serum. Target cells (2 × 104) were first incubated with the biAbs before adding the effector cells (2 × 105) in a final volume of 100 μl/well in a 96-well tissue culture plate followed by incubation at 37 °C for 16 h. A biAb concentration range from 2 ng/ml to 1 μg/ml was used. Plates were centrifuged, and 50 μl of the supernatants were transferred into a 96-well clear-bottom white-walled plate (Costar 3610, Corning) containing 25 μl/well CytoTox-Glo reagent. After a 15-min incubation at RT, a SpectraMax M5 instrument was used to read the plates with SoftMax Pro software set to luminescence. Following the ELISA Ready-SET-Go! Reagent protocols (eBioscience), additional supernatant from the previous study was diluted 20-fold and used in a human IFN-γ ELISA.
Data availability
Crystal structure files have been deposited to the Protein Data Bank and can be found by searching accession numbers 6OSH, 6OSV, and 6OSN, which correlate to 401:hROR2-Kr, hX3.12.6:hROR2-Kr, and hROR2-Kr crystal structures, respectively. All other data are contained within the article and supporting material.
Author contributions
R. S. G., J. W., J. S., J. F., and H. Park data curation; R. S. G., J. W., H. Peng, J. Q., J. S., J. F., H. Park, and C. R. formal analysis; R. S. G., J. W., H. Peng, J. S., J. F., H. Park, and C. R. investigation; R. S. G., J. S., J. F., and H. Park visualization; R. S. G., J. W., H. Peng, J. Q., J. S., J. F., and H. Park methodology; R. S. G., H. Park, and C. R. writing-original draft; R. S. G., J. W., H. Peng, J. Q., J. S., J. F., H. Park, and C. R. writing-review and editing; H. Peng, J. Q., and C. R. supervision; H. Peng and J. Q. validation; J. Q., J. S., J. F., and H. Park resources; C. R. conceptualization; C. R. funding acquisition.
Supplementary Material
Acknowledgments
We thank Drs. Meredith Gardner and Matthew Gardner (Farzan laboratory at The Scripps Research Institute) for helping to obtain the Fab melting data and Oluwarotimi Omorodion (Wilson laboratory at The Scripps Research Institute) for reading and editing the manuscript. We are grateful to the staff at Advanced Light Source synchrotron facility (Lawrence Berkeley National Laboratory) and LS-CAT Advanced Photon Source (Argonne National Laboratory) for kind support in synchrotron data collection.
This study was supported by National Institutes of Health Grants R01 CA174844, R01 CA181258, R01 CA204484, and R21 CA229961 (to C. R.). This is manuscript no. 29916 from The Scripps Research Institute. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Figs. S1–S4.
- FDA
- Food and Drug Administration
- RTK
- receptor tyrosine kinase
- Fz
- frizzled
- Kr
- kringle
- 401
- mAb XBR2-401
- CAR-T
- chimeric antigen receptor T cell
- biAb
- bispecific antibody
- PDB
- Protein Data Bank
- CDR
- complementarity-determining region
- ScFv
- single-chain variable fragment
- RMSD
- root mean square deviation
- LBS
- lysine-binding site
- SPR
- surface plasmon resonance
- X3.12
- clone XBR2-401-X3.12
- VL
- variable light-chain amino acid sequence
- VH
- variable heavy-chain amino acid sequence
- PBMC
- peripheral blood mononuclear cell
- IFN
- interferon
- FBS
- fetal bovine serum
- RT
- room temperature
- pAb
- polyclonal antibody
- FWR
- framework region
- hX3.12.6
- hXBR2-401-X3.12.6
- hX3.12.5
- hXBR2-401-X3.12.5.
References
- 1. Almagro J. C., Daniels-Wells T. R., Perez-Tapia S. M., and Penichet M. L. (2018) Progress and challenges in the design and clinical development of antibodies for cancer therapy. Front. Immunol. 8, 1751 10.3389/fimmu.2017.01751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Robinson D. R., Wu Y. M., and Lin S. F. (2000) The protein tyrosine kinase family of the human genome. Oncogene 19, 5548–5557 10.1038/sj.onc.1203957 [DOI] [PubMed] [Google Scholar]
- 3. Masiakowski P., and Carroll R. D. (1992) A novel family of cell surface receptors with tyrosine kinase-like domain. J. Biol. Chem. 267, 26181–26190 [PubMed] [Google Scholar]
- 4. Stricker S., Rauschenberger V., and Schambony A. (2017) ROR-family receptor tyrosine kinases. Curr. Top. Dev. Biol. 123, 105–142 10.1016/bs.ctdb.2016.09.003 [DOI] [PubMed] [Google Scholar]
- 5. Matsuda T., Nomi M., Ikeya M., Kani S., Oishi I., Terashima T., Takada S., and Minami Y. (2001) Expression of the receptor tyrosine kinase genes, Ror1 and Ror2, during mouse development. Mech. Dev. 105, 153–156 10.1016/S0925-4773(01)00383-5 [DOI] [PubMed] [Google Scholar]
- 6. Al-Shawi R., Ashton S. V., Underwood C., and Simons J. P. (2001) Expression of the Ror1 and Ror2 receptor tyrosine kinase genes during mouse development. Dev. Genes Evol. 211, 161–171 10.1007/s004270100140 [DOI] [PubMed] [Google Scholar]
- 7. Rebagay G., Yan S., Liu C., and Cheung N. K. (2012) ROR1 and ROR2 in human malignancies: potentials for targeted therapy. Front. Oncol. 2, 34 10.3389/fonc.2012.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Forrester W. C. (2002) The Ror receptor tyrosine kinase family. Cell. Mol. Life Sci. 59, 83–96 10.1007/s00018-002-8407-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Debebe Z., and Rathmell W. K. (2015) Ror2 as a therapeutic target in cancer. Pharmacol. Ther. 150, 143–148 10.1016/j.pharmthera.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Niehrs C. (2012) The complex world of WNT receptor signalling. Nat. Rev. Mol. Cell Biol. 13, 767–779 10.1038/nrm3470 [DOI] [PubMed] [Google Scholar]
- 11. DeChiara T. M., Kimble R. B., Poueymirou W. T., Rojas J., Masiakowski P., Valenzuela D. M., and Yancopoulos G. D. (2000) Ror2, encoding a receptor-like tyrosine kinase, is required for cartilage and growth plate development. Nat. Genet. 24, 271–274 10.1038/73488 [DOI] [PubMed] [Google Scholar]
- 12. Green J. L., Kuntz S. G., and Sternberg P. W. (2008) Ror receptor tyrosine kinases: orphans no more. Trends Cell Biol. 18, 536–544 10.1016/j.tcb.2008.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morioka K., Tanikawa C., Ochi K., Daigo Y., Katagiri T., Kawano H., Kawaguchi H., Myoui A., Yoshikawa H., Naka N., Araki N., Kudawara I., Ieguchi M., Nakamura K., Nakamura Y., and Matsuda K. (2009) Orphan receptor tyrosine kinase ROR2 as a potential therapeutic target for osteosarcoma. Cancer Sci. 100, 1227–1233 10.1111/j.1349-7006.2009.01165.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wright T. M., Brannon A. R., Gordan J. D., Mikels A. J., Mitchell C., Chen S., Espinosa I., van de Rijn M., Pruthi R., Wallen E., Edwards L., Nusse R., and Rathmell W. K. (2009) Ror2, a developmentally regulated kinase, promotes tumor growth potential in renal cell carcinoma. Oncogene 28, 2513–2523 10.1038/onc.2009.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Henry C., Quadir A., Hawkins N. J., Jary E., Llamosas E., Kumar D., Daniels B., Ward R. L., and Ford C. E. (2015) Expression of the novel Wnt receptor ROR2 is increased in breast cancer and may regulate both β-catenin dependent and independent Wnt signalling. J. Cancer Res. Clin. Oncol. 141, 243–254 10.1007/s00432-014-1824-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frenquelli M., Caridi N., Antonini E., Storti F., Viganò V., Gaviraghi M., Occhionorelli M., Bianchessi S., Bongiovanni L., Spinelli A., Marcatti M., Belloni D., Ferrero E., Karki S., Brambilla P., et al. (2020) The WNT receptor ROR2 drives the interaction of multiple myeloma cells with the microenvironment through AKT activation. Leukemia 34, 257–270 10.1038/s41375-019-0486-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edris B., Espinosa I., Mühlenberg T., Mikels A., Lee C. H., Steigen S. E., Zhu S., Montgomery K. D., Lazar A. J., Lev D., Fletcher J. A., Beck A. H., West R. B., Nusse R., and van de Rijn M. (2012) ROR2 is a novel prognostic biomarker and a potential therapeutic target in leiomyosarcoma and gastrointestinal stromal tumour. J. Pathol. 227, 223–233 10.1002/path.3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peng H., Nerreter T., Chang J., Qi J., Li X., Karunadharma P., Martinez G. J., Fallahi M., Soden J., Freeth J., Beerli R. R., Grawunder U., Hudecek M., and Rader C. (2017) Mining naive rabbit antibody repertoires by phage display for monoclonal antibodies of therapeutic utility. J. Mol. Biol. 429, 2954–2973 10.1016/j.jmb.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qi J., Li X., Peng H., Cook E. M., Dadashian E. L., Wiestner A., Park H., and Rader C. (2018) Potent and selective antitumor activity of a T cell-engaging bispecific antibody targeting a membrane-proximal epitope of ROR1. Proc. Natl. Acad. Sci. U.S.A. 115, E5467–E5476 10.1073/pnas.1719905115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davda J., Declerck P., Hu-Lieskovan S., Hickling T. P., Jacobs I. A., Chou J., Salek-Ardakani S., and Kraynov E. (2019) Immunogenicity of immunomodulatory, antibody-based, oncology therapeutics. J. Immunother. Cancer 7, 105 10.1186/s40425-019-0586-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones P. T., Dear P. H., Foote J., Neuberger M. S., and Winter G. (1986) Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature 321, 522–525 10.1038/321522a0 [DOI] [PubMed] [Google Scholar]
- 22. Queen C., Schneider W. P., Selick H. E., Payne P. W., Landolfi N. F., Duncan J. F., Avdalovic N. M., Levitt M., Junghans R. P., and Waldmann T. A. (1989) A humanized antibody that binds to the interleukin-2 receptor. Proc. Natl. Acad. Sci. U.S.A. 86, 10029–10033 10.1073/pnas.86.24.10029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rader C., Ritter G., Nathan S., Elia M., Gout I., Jungbluth A. A., Cohen L. S., Welt S., Old L. J., and Barbas C. F. 3rd (2000) The rabbit antibody repertoire as a novel source for the generation of therapeutic human antibodies. J. Biol. Chem. 275, 13668–13676 10.1074/jbc.275.18.13668 [DOI] [PubMed] [Google Scholar]
- 24. Mage R. G., Esteves P. J., and Rader C. (2019) Rabbit models of human diseases for diagnostics and therapeutics development. Dev. Comp. Immunol. 92, 99–104 10.1016/j.dci.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hwang W. Y., and Foote J. (2005) Immunogenicity of engineered antibodies. Methods 36, 3–10 10.1016/j.ymeth.2005.01.001 [DOI] [PubMed] [Google Scholar]
- 26. Jones T. D., Carter P. J., Plückthun A., Vásquez M., Holgate R. G., Hötzel I., Popplewell A. G., Parren P. W., Enzelberger M., Rademaker H. J., Clark M. R., Lowe D. C., Dahiyat B. I., Smith V., Lambert J. M., et al. (2016) The INNs and outs of antibody nonproprietary names. mAbs 8, 1–9 10.1080/19420862.2015.1114320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee C. H., Park K. J., Sung E. S., Kim A., Choi J. D., Kim J. S., Kim S. H., Kwon M. H., and Kim Y. S. (2010) Engineering of a human kringle domain into agonistic and antagonistic binding proteins functioning in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 107, 9567–9571 10.1073/pnas.1001541107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang M., Zajicek J., Geiger J. H., Prorok M., and Castellino F. J. (2010) Solution structure of the complex of VEK-30 and plasminogen kringle 2. J. Struct. Biol. 169, 349–359 10.1016/j.jsb.2009.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y. F., and Ho M. (2017) Humanization of rabbit monoclonal antibodies via grafting combined Kabat/IMGT/Paratome complementarity-determining regions: rationale and examples. mAbs 9, 419–429 10.1080/19420862.2017.1289302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stahl S. J., Watts N. R., Rader C., DiMattia M. A., Mage R. G., Palmer I., Kaufman J. D., Grimes J. M., Stuart D. I., Steven A. C., and Wingfield P. T. (2010) Generation and characterization of a chimeric rabbit/human Fab for co-crystallization of HIV-1 Rev. J. Mol. Biol. 397, 697–708 10.1016/j.jmb.2010.01.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Menzen T., and Friess W. (2014) Temperature-ramped studies on the aggregation, unfolding, and interaction of a therapeutic monoclonal antibody. J. Pharm. Sci. 103, 445–455 10.1002/jps.23827 [DOI] [PubMed] [Google Scholar]
- 32. McConnell A. D., Zhang X., Macomber J. L., Chau B., Sheffer J. C., Rahmanian S., Hare E., Spasojevic V., Horlick R. A., King D. J., and Bowers P. M. (2014) A general approach to antibody thermostabilization. mAbs 6, 1274–1282 10.4161/mabs.29680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Freeth J., and Soden J. (2020) New advances in cell microarray technology to expand applications in target deconvolution and off-target screening. SLAS Discov. 25, 223–230 10.1177/2472555219897567 [DOI] [PubMed] [Google Scholar]
- 34. Robinson H. R., Qi J., Cook E. M., Nichols C., Dadashian E. L., Underbayev C., Herman S. E. M., Saba N. S., Keyvanfar K., Sun C., Ahn I. E., Baskar S., Rader C., and Wiestner A. (2018) A CD19/CD3 bispecific antibody for effective immunotherapy of chronic lymphocytic leukemia in the ibrutinib era. Blood 132, 521–532 10.1182/blood-2018-02-830992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hristodorov D., Fischer R., Joerissen H., Müller-Tiemann B., Apeler H., and Linden L. (2013) Generation and comparative characterization of glycosylated and aglycosylated human IgG1 antibodies. Mol. Biotechnol. 53, 326–335 10.1007/s12033-012-9531-x [DOI] [PubMed] [Google Scholar]
- 36. Atwell S., Ridgway J. B. B., Wells J. A., and Carter P. (1997) Stable heterodimers from remodeling the domain interface of a homodimer using a phage display library. J. Mol. Biol. 270, 26–35 10.1006/jmbi.1997.1116 [DOI] [PubMed] [Google Scholar]
- 37. Merchant A. M., Zhu Z., Yuan J. Q., Goddard A., Adams C. W., Presta L. G., and Carter P. (1998) An efficient route to human bispecific IgG. Nat. Biotechnol. 16, 677–681 10.1038/nbt0798-677 [DOI] [PubMed] [Google Scholar]
- 38. Zhu Z., Lewis G. D., and Carter P. (1995) Engineering high affinity humanized anti-p185HER2/anti-CD3 bispecific F(ab′)2 for efficient lysis of p185HER2 overexpressing tumor cells. Int. J. Cancer 62, 319–324 10.1002/ijc.2910620315 [DOI] [PubMed] [Google Scholar]
- 39. Weber J., Peng H., and Rader C. (2017) From rabbit antibody repertoires to rabbit monoclonal antibodies. Exp. Mol. Med. 49, e305 10.1038/emm.2017.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Balakrishnan A., Goodpaster T., Randolph-Habecker J., Hoffstrom B. G., Jalikis F. G., Koch L. K., Berger C., Kosasih P. L., Rajan A., Sommermeyer D., Porter P. L., and Riddell S. R. (2017) Analysis of ROR1 protein expression in human cancer and normal tissues. Clin. Cancer Res. 23, 3061–3071 10.1158/1078-0432.CCR-16-2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ma S. S., Henry C. E., Llamosas E., Higgins R., Daniels B., Hesson L. B., Hawkins N. J., Ward R. L., and Ford C. E. (2017) Validation of specificity of antibodies for immunohistochemistry: the case of ROR2. Virchows Arch. 470, 99–108 10.1007/s00428-016-2019-5 [DOI] [PubMed] [Google Scholar]
- 42. Bradbury A., and Plückthun A. (2015) Standardize antibodies used in research. Nature 518, 27–29 10.1038/518027a [DOI] [PubMed] [Google Scholar]
- 43. Hellmann I., Waldmeier L., Bannwarth-Escher M. C., Maslova K., Wolter F. I., Grawunder U., and Beerli R. R. (2018) Novel antibody drug conjugates targeting tumor-associated receptor tyrosine kinase ROR2 by functional screening of fully human antibody libraries using Transpo-mAb display on progenitor B cells. Front. Immunol. 9, 2490 10.3389/fimmu.2018.02490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hudecek M., Lupo-Stanghellini M. T., Kosasih P. L., Sommermeyer D., Jensen M. C., Rader C., and Riddell S. R. (2013) Receptor affinity and extracellular domain modifications affect tumor recognition by ROR1-specific chimeric antigen receptor T cells. Clin. Cancer Res. 19, 3153–3164 10.1158/1078-0432.CCR-13-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park H., Adsit F. G., and Boyington J. C. (2005) The 1.4 Å crystal structure of the human oxidized low density lipoprotein receptor lox-1. J. Biol. Chem. 280, 13593–13599 10.1074/jbc.M500768200 [DOI] [PubMed] [Google Scholar]
- 46. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 47. Vagin A., and Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 10.1107/S0021889897006766 [DOI] [Google Scholar]
- 48. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 10.1107/S0907444902016657 [DOI] [PubMed] [Google Scholar]
- 49. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 50. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 51. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., and Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 10.1107/S0907444998003254 [DOI] [PubMed] [Google Scholar]
- 52. Brunger A. T. (2007) Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2, 2728–2733 10.1038/nprot.2007.406 [DOI] [PubMed] [Google Scholar]
- 53. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rader C. (2009) Generation and selection of rabbit antibody libraries by phage display. Methods Mol. Biol. 525, 101–128, xiv 10.1007/978-1-59745-554-1_5 [DOI] [PubMed] [Google Scholar]
- 55. doi: 10.1073/pnas.1719905115. https://www.rcsb.org/search?q=audit_author.name:Park,%20H. Park, H., https://www.rcsb.org/search?q=audit_author.name:Rader,%20C. Rader, C. (2018) Potent and Selective Antitumor Activity of a T-Cell Engaging Bispecific Antibody Targeting a Membrane-Proximal Epitope of ROR1. Protein Data Bank. 6BA5. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystal structure files have been deposited to the Protein Data Bank and can be found by searching accession numbers 6OSH, 6OSV, and 6OSN, which correlate to 401:hROR2-Kr, hX3.12.6:hROR2-Kr, and hROR2-Kr crystal structures, respectively. All other data are contained within the article and supporting material.