Abstract
Bacterial products such as lipopolysaccharides (or endotoxin) cause systemic inflammation, resulting in a substantial global health burden. The onset, progression, and resolution of the inflammatory response to endotoxin are usually tightly controlled to avoid chronic inflammation. Members of the NF-κB family of transcription factors are key drivers of inflammation that activate sets of genes in response to inflammatory signals. Such responses are typically short-lived and can be suppressed by proteins that act post-translationally, such as the SOCS (suppressor of cytokine signaling) family. Less is known about direct transcriptional regulation of these responses, however. Here, using a combination of in vitro approaches and in vivo animal models, we show that endotoxin treatment induced expression of the well-characterized transcriptional repressor Krüppel-like factor 3 (KLF3), which, in turn, directly repressed the expression of the NF-κB family member RELA/p65. We also observed that KLF3-deficient mice were hypersensitive to endotoxin and exhibited elevated levels of circulating Ly6C+ monocytes and macrophage-derived inflammatory cytokines. These findings reveal that KLF3 is a fundamental suppressor that operates as a feedback inhibitor of RELA/p65 and may be important in facilitating the resolution of inflammation.
Keywords: inflammation, endotoxin, NF-kappa B (NF-KB), NF-kB transcription factor, gene regulation, macrophage, lipopolysaccharide (LPS), transcription, Krüppel-like factor 3 (KLF3)
Introduction
Inflammation is usually a tightly regulated physiological process that ensures appropriate onset, progression, and resolution. However, dysregulation of inflammation can lead to a host of inflammatory diseases, including macular degeneration, rheumatoid arthritis, inflammatory bowel disease, and multiple sclerosis (1–5). Understanding the checkpoints that control the activation and suppression of inflammation will inform the development of improved therapies for inflammatory disorders.
The nuclear factor κ–light-chain enhancer of activated B cells (NF-κB) family of transcription factors are central inflammatory mediators (6–8). Mammals have five NF-κB family proteins: RELA (p65), RELB, and REL, as well as NF-κB1 (p105; processed into p50) and NF-κB2 (p100; processed into p52). NF-κB functions in both innate and adaptive immune cells to direct proinflammatory gene expression programs, including activation of archetypal inflammatory cytokines such as tumor necrosis factor (TNF)2 (9), interleukin (IL) 1α (10), IL-1β (11), and IL-6 (12). NF-κB signaling has also been reported to perform anti-inflammatory roles (13–15). NF-κB function can be activated by bacterial products such as lipopolysaccharides (LPSs) that underlie the substantial global health burden represented by sepsis and endotoxemia (16, 17). Activation of NF-κB in response to stimuli such as LPS is well-characterized: prior to stimulation, inactive NF-κB dimers are retained in the cytosol by IκB proteins. Following stimulation, IκB kinase complexes disrupt this retention, allowing nuclear translocation of NF-κB, which can carry out induction of target genes. However, less is known about regulation of NF-κB family members at the transcriptional level.
Several members of the Krüppel-like factor (KLF) family of transcription factors have been implicated in immune cell function and inflammation, including KLF2 (18), KLF4 (19), and KLF6 (20). KLF3 has been well-established as a key transcription factor in red blood cell development (21–23); however, the Klf3−/− mouse lacks a severe erythropoietic phenotype. It has recently been shown that KLF3 regulates expression of the inflammatory modulator galectin-3 (24) and is also involved in B-cell maturation (25), suggesting a possible link between KLF3, immune cell function, and inflammation.
Here we report that the well-characterized transcriptional repressor KLF3 (21–24, 26–29) suppresses NF-κB–mediated inflammation through direct regulation of the p65-encoding gene Rela/RELA. In the absence of KLF3, mice exhibit pronounced systemic inflammation with elevated circulating inflammatory monocytes, constitutive proinflammatory cytokine production, and a heightened response to endotoxin treatment. KLF3 is induced in macrophages following LPS exposure and directly binds and represses the expression of the NF-κB subunit gene Rela in a simple feedback inhibition mechanism. These findings identify KLF3 as a fundamental suppressor of NF-κB–driven inflammation and, importantly, identify a novel repressor of RELA/p65 that acts at the transcriptional level.
Results
Pronounced systemic inflammation in the absence of KLF3
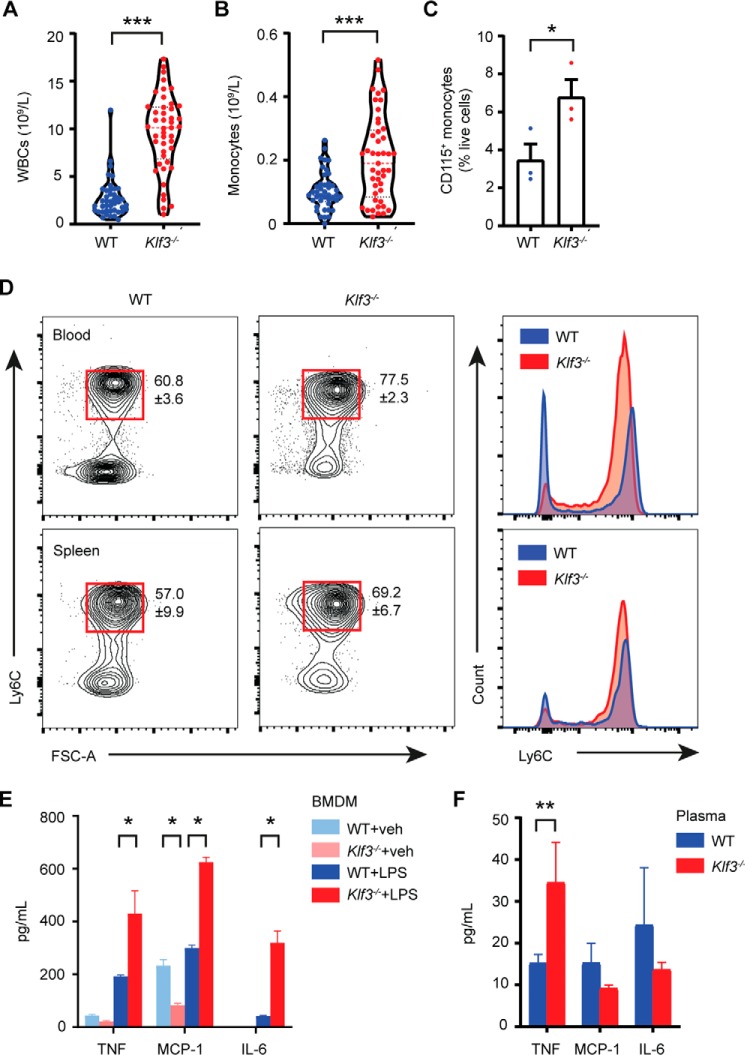
Given early findings that KLF3 is important for erythroid development, we performed blood counts to determine whether mice lacking KLF3 have altered numbers of circulating hematopoietic cells. Peripheral blood counts on samples from WT and Klf3−/− mice revealed that Klf3−/− mice have 3-fold more white blood cells (Fig. 1A) and, specifically, 2-fold more circulating monocytes (Fig. 1B). This result was confirmed by flow cytometry, in which we observed twice as many circulating CD115+ monocytes in Klf3−/− mice (Fig. 1C). More detailed analysis showed that Klf3−/− mice had a significantly higher proportion of Ly6C+ proinflammatory monocytes and a lower proportion of Ly6C− anti-inflammatory monocytes (Fig. 1D and Fig. S1A). This profile shift was also evident in spleen monocytes to a lesser extent. Interestingly, however, we did not observe increased abundances of monocytic precursors including macrophage/dendritic cell progenitors, common monocyte progenitors, or Ly6C+/− monocytes in the bone marrow of Klf3−/− mice (Fig. S1B). This suggests that circulating monocyte proliferation occurs downstream of myelopoiesis in response to endogenous and external cues.
Figure 1.
Pronounced systemic inflammation in the absence of KLF3. A and B, full peripheral blood counts were performed to assess the number of circulating white blood cells (WBCs, A) and monocytes (B) in WT (n = 42) and Klf3−/− (n = 45) mice. C, flow cytometry was used to measure the proportion of CD115+ monocytes in WT and Klf3−/− blood as a percentage of live cells, following red blood cell lysis (n = 3). For A–C, the bars are representative of the mean, and for C, the error bars are means ± S.E. D, CD115+ Ly6C+ inflammatory monocytes were assessed in WT and Klf3−/− blood and spleen by flow cytometry. The means ± S.E. of four independent experiments are shown for the Ly6C+ populations indicated by red gates, with accompanying histograms showing the shift in Ly6C surface expression profile. E and F, levels of inflammatory cytokines TNF, MCP-1, and IL-6 were assessed in supernatant from WT and Klf3−/− BMDMs treated with vehicle (veh) or 100 ng/ml LPS for 24 h (n = 3 per genotype/treatment group, E) and plasma (WT: n = 5 and Klf3−/−: n = 7, F) with error bars representative of the mean ± S.E. Nonparametric Mann–Whitney U tests were performed to assess statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001. FSC-A, forward scatter area.
Given that Klf3−/− mice have more inflammatory monocytes in circulation, we hypothesized that they might also overexpress proinflammatory cytokines. We cultured WT and Klf3−/− bone marrow-derived macrophages (BMDMs; Fig. S1C), treated them with LPS (100 ng/ml) or vehicle for 24 h, and then harvested the supernatant. We measured secreted levels of several proinflammatory factors and found that upon LPS treatment, BMDMs lacking KLF3 produced significantly greater amounts of TNF, MCP-1/CCL2, and IL-6 (Fig. 1E and Fig. S1D). Interestingly, however, in vivo we only observed increased levels of TNF in Klf3−/− plasma, with no apparent difference in plasma levels of MCP-1, IL-6, or other cytokines between WT and Klf3−/− animals (Fig. 1F and Fig. S1E). Together, these results demonstrate that in the absence of the transcriptional repressor KLF3, mice exhibit systemic inflammation and that circulating monocytes are implicated in this phenotype.
Mice lacking KLF3 show a heightened and prolonged inflammatory response to endotoxin treatment
Given that Klf3−/− mice display increased Ly6C+ monocytes in circulation and higher levels of plasma TNF, both of which are symptoms of endotoxemia (30, 31), we tested to see whether they displayed a heightened sensitivity to inflammatory stimulants. We injected mice intraperitoneally with a low dose of the archetypal bacterial endotoxin LPS (0.167 mg/kg). Vehicle-treated mice maintained their body temperature over the 24-h period following injection, whereas LPS-treated mice experienced a drop in body temperature by up to 4 °C over the initial 2-h period postinjection, as expected following LPS treatment (32) (Fig. 2A). WT mice gradually recovered their body temperature, whereas Klf3−/− mice sustained a low body temperature for a further 4 h. This delayed recovery of body temperature was most evident at the 12-h postinjection time point, demonstrating the increased sensitivity of Klf3−/− mice to LPS. This observation also held, to a lesser extent, for mice given a very low dose of LPS (0.05 mg/kg), with LPS-treated Klf3−/− mice experiencing a delayed recovery of body temperature (Fig. S2A).
Figure 2.
Mice lacking KLF3 show a heightened and prolonged inflammatory response to endotoxin treatment. A, WT and Klf3−/− mice were intraperitoneally administered with vehicle (veh) or 0.167 mg/kg LPS, and their body temperatures were tracked by rectal probing over 24 h (vehicle: n = 4 per genotype and LPS: n = 5 per genotype). B–E, after 24 h, full blood counts were performed to assess the number of circulating white blood cells (WBCs, B), monocytes (C), lymphocytes (D), and neutrophils (E). F, TNF levels in plasma of WT and Klf3−/− mice treated with LPS (0.167 mg/kg) or vehicle were measured using ELISA. G and H, the proportion of CD115+ monocytes expressing Ly6C in the (G) blood and (H) spleen of WT and Klf3−/− mice treated for 24 h with vehicle or 0.167 mg/kg LPS was assessed by flow cytometry, with the mean values shown for each condition ± S.E. For G and H, representative median plots are shown from four or five mice per group (see Fig. S2, C and D for all plots). For A–F, error bars are means ± S.E., and nonparametric Mann–Whitney U tests were performed to assess statistical significance: *, p < 0.05 between genotypes; **, p < 0.01 between genotypes; #, p < 0.05 between conditions within the same genotype; ##, p < 0.01 between conditions within the same genotype. FSC-A, forward scatter area.
Peripheral blood was taken from mice given 0.167 mg/kg LPS (or vehicle) for total blood cell counts. Following LPS treatment, both WT and Klf3−/− mice displayed reduced circulating numbers of total white blood cells, monocytes, and lymphocytes (Fig. 2, B–D) but unchanged neutrophil abundance (Fig. 2E). We observed that vehicle-treated Klf3−/− mice had a higher abundance of total white blood cells, monocytes, and lymphocytes than vehicle-treated WT mice, as expected. Following LPS treatment, all white blood cell counts trended higher in Klf3−/− mice compared with WT mice, with monocyte abundance showing significant comparative elevation in KLF3-deficient mice. Alongside these findings, we confirmed that circulating TNF levels in vehicle-treated Klf3−/− mice were higher than in WT counterparts and likewise upon LPS treatment but with greater absolute amounts of TNF (Fig. 2F). The spleens of both WT and Klf3−/− were larger in mice treated with LPS for 24 h (Fig. S2B). We found an increased proportion of CD115+ Ly6C+ monocytes in vehicle-treated Klf3−/− blood when compared with WT, as expected, and this increase was retained in both the blood and spleen of LPS-treated Klf3−/− mice (with higher proportional abundance of Ly6C+ cells in Klf3−/− LPS-treated mice than WT LPS-treated mice) (Fig. 2, G and H, and Fig. S2, C and D). These results demonstrate that KLF3-deficient mice display an elevated sensitivity in response to treatment with endotoxin.
Macrophages lacking KLF3 exhibit enhanced inflammatory function
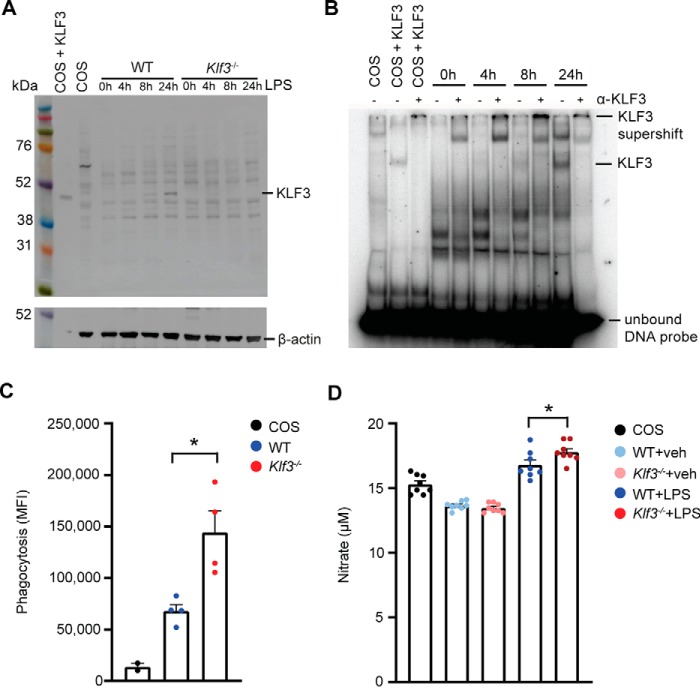
As a first step to exploring the molecular mechanism underlying these findings, we moved to experiments on cultured primary macrophages. We treated BMDMs with 100 ng/ml LPS for 0, 4, 8, and 24 h and immunoblotted for KLF3 protein (Fig. 3A). KLF3 levels increased by the 8-h mark, with very clear expression seen 24 h after LPS treatment. As expected, no KLF3 was detected in Klf3−/− samples. We also performed electrophoretic mobility shift assays (EMSAs) and found that KLF3–DNA-binding activity is most evident 24 h after LPS treatment (Fig. 3B). Klf3 is the third most highly expressed member of the Klf gene family in BMDMs, with levels only superseded by well-characterized macrophage transcription factors Klf2 and Klf6 (18, 20, 33, 34) (Fig. S3A). Given this, we sought to further characterize the effects of KLF3 on macrophage function. Phagocytosis of Escherichia coli bioparticles is enhanced 2-fold in Klf3−/− BMDMs compared with WT following 8 h of LPS treatment (Fig. 3C), whereas KLF3-deficient BMDMs treated with LPS for 24 h produce marginally but significantly more nitrate than LPS-induced WT cells (Fig. 3D). Using the reduction of resazurin blue as an indicator of cellular reduction environment, we found modest differences between WT and Klf3−/− BMDMs over a 24-h period of exposure, with the only significant difference observed at 2 h (Fig. S3B). BrdU incorporation assays on WT and Klf3−/− BMDMs demonstrated no significant differences in actively replicating cells (Fig. S3C), nor were any differences seen in BMDM cell counts over 8 days (Fig. S3D), suggesting no discernible proliferative differences between WT and Klf3−/− macrophages in vitro. Together, these results demonstrate that KLF3 protein levels increase in response to endotoxin and that macrophage inflammatory activity, including phagocytic capacity, nitrate production, and cellular reduction levels, are all enhanced in the absence of KLF3.
Figure 3.
Macrophages lacking KLF3 exhibit enhanced inflammatory function. A, KLF3 protein levels in BMDMs following stimulation with 100 ng/ml LPS for 0–24 h were measured by Western blotting. 20 μg of nuclear extract was loaded per lane alongside a Rainbow molecular weight marker (GE Healthcare). The membranes were probed with a polyclonal anti-KLF3 antibody (raised in rabbit) followed by reprobing of the membrane with an anti–β-actin antibody (raised in mouse; A1978). A positive control (nuclear extract was from COS-7 cells overexpressing pMT3-Klf3) and negative control (nuclear extract was from untransfected COS-7 cells) were included, in addition to nuclear extracts from BMDMs lacking KLF3 to ensure antibody specificity. B, EMSA was used to detect functional KLF3 bound to a radiolabeled DNA probe in nuclear extracts from BMDMs stimulated with 100 ng/ml LPS for 0–24 h. Nuclear extracts from COS-7 cells overexpressing pMT3-Klf3 and untransfected COS-7 cells were used as positive and negative controls, respectively. Polyclonal anti-KLF3 antibody (α-KLF3) raised in rabbit was added to confirm the identity of bound protein as KLF3, labeled as a supershift band. Unbound radiolabeled DNA probe can be seen as an intense dark band at the bottom of the gel. C, phagocytosis assays were performed to compare the uptake of fluorescein-labeled K-12 E. coli bioparticles in WT and Klf3−/− BMDMs (n = 4), with the inclusion of COS-7 cells as a negative control (n = 2). The cells were stimulated with 100 ng/ml LPS for 8 h, and bioparticle uptake was read using 480-nm excitation/520-nm emission settings, represented as mean fluorescence intensity (MFI). D, production of nitrate was measured in the supernatant of BMDMs following exposure to 100 ng/ml LPS or vehicle (veh) for 24 h, with the inclusion of COS-7 cells as a reference point (n = 8). For C and D, error bars represent means ± S.E., and nonparametric Mann–Whitney U tests were carried out: *, p < 0.05.
Widespread gene deregulation in KLF3-deficient macrophages following endotoxin treatment
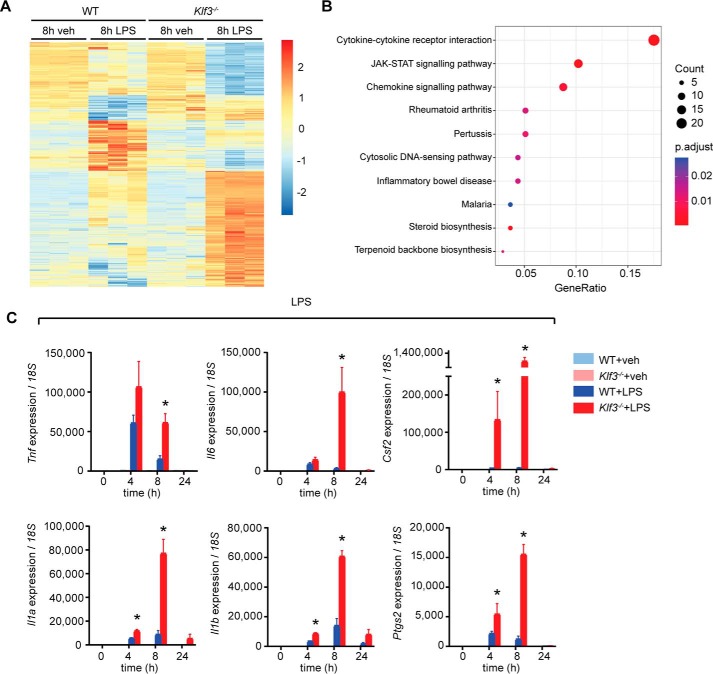
To study the role of KLF3 in genome-wide gene regulation in macrophages, we performed microarrays on BMDMs treated with LPS (100 ng/ml) or vehicle. We chose to compare WT and Klf3−/− cells treated for 8 h given that this time point coincides with optimal up-regulation of important proinflammatory cytokines and also with increases in KLF3 protein levels. The profiles of untreated WT and Klf3−/− BMDMs were broadly similar (Fig. 4A), in line with our finding that KLF3 expression levels are modest in unstimulated BMDMs. In contrast, Klf3−/− BMDMs treated with LPS for 8 h showed striking differences to WT cells. Motif discovery and enrichment analysis found that the promoters of the differentially expressed genes were enriched for motifs matching the binding sites for Sp/KLF transcription factor family members, suggesting that many of these are bona fide KLF3 target genes (Fig. S4). Cytokine–receptor interactions, JAK–STAT signaling, and chemokine pathways were the three most dysregulated biological pathways in Klf3−/− BMDMs (Fig. 4B), suggesting that in the absence of KLF3, macrophages undergo an enhanced inflammatory response. Using quantitative PCR, we validated deregulation of key cytokine genes (Fig. 4C). Proinflammatory genes such as Tnf, Il6, and Il1b are all activated in BMDMs following LPS treatment, with expression peaking between 4 and 8 h, before resolution of the inflammatory response. In the absence of KLF3, however, these genes are superactivated, and their up-regulation is prolonged. This suggests that KLF3 is directly or indirectly involved in the suppression of proinflammatory gene expression during the resolution of inflammation. We also observed elevated induction of Il1a and Il1b in Klf3−/− BMDMs treated with the synthetic dsRNA analog, poly(I·C), which activates signaling through TLR3 and NF-κB (Fig. S5A). Likewise, Il1a but not Stat1 was more highly induced in IFNγ-treated Klf3−/− cells (Fig. S5B). Upon treatment with IFNα (Fig. S5C) and IL-6 (Fig. S5D), which signal independently of NF-κB, no gene deregulation was observed in Klf3−/− BMDMs. These data implicate NF-κB signaling in the inflammatory effects mediated by KLF3.
Figure 4.
Widespread gene deregulation in macrophages lacking KLF3 following endotoxin treatment. A, heat map of significantly (p < 0.05) deregulated genes in Klf3−/− BMDMs following 8 h of treatment with 100 ng/ml LPS or vehicle (veh, n = 3 per condition and genotype). B, Kyoto Encyclopedia of Genes and Genomes analysis of the most highly deregulated pathways in LPS-treated Klf3−/− BMDMs. C, inflammatory cytokine genes showing deregulation in microarrays were validated using qPCR, with error bars representative of means ± S.E. of three independent biological replicates. Expression was normalized to 18S rRNA levels, and the 0-h time point for each gene was set to 1. Nonparametric Mann–Whitney U tests were carried out: *, p < 0.05.
KLF3 suppresses activation of NF-κB genes
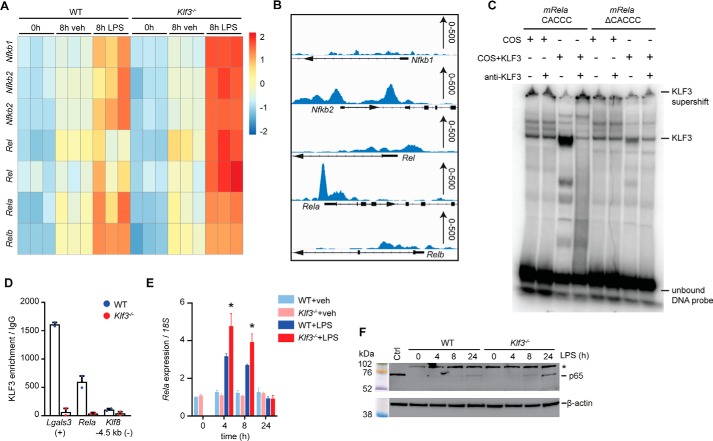
With the aim of identifying candidate genes that KLF3 binds and regulates directly, we consulted an existing KLF3 ChIP-Seq data set obtained from the analysis of murine embryonic fibroblasts (MEFs) (35). No significant KLF3 enrichment was observed at the promoter regions of Csf2, Tnf, Il6, Il1a, Il1b, or Ptgs2 (Fig. S6A), providing no evidence for direct regulation of these cytokine genes by KLF3 in this cell type. Instead, we hypothesized that KLF3 acts upon an upstream intermediary factor controlling expression of these genes during the inflammatory response. Interrogation of deregulated transcription factors from our BMDM microarray data revealed that genes encoding the NF-κB family, important regulators of the inflammatory response, are increased in the absence of KLF3 (Fig. 5A). In the MEF ChIP-Seq data, strong KLF3 binding was evident at the proximal promoter of Rela, encoding the NF-κB subunit RELA/p65 (Fig. 5B).
Figure 5.
KLF3 suppresses activation of NF-κB genes. A, a heat map was generated showing relative expression of genes encoding NF-κB subunits in WT and Klf3−/− BMDMs after 0 and 8 h of 100 ng/ml LPS or vehicle (veh) treatment (n = 3 per condition and genotype). B, KLF3 enrichment at the proximal promoter regions of genes encoding NF-κB from a KLF3-V5 ChIP-Seq in murine embryonic fibroblasts, with scale (0–500 read counts) provided on the right-hand side of each panel. C, EMSA was used to detect KLF3 binding to a radiolabeled DNA probe comprised of the WT (CACCC) or mutated (ΔCACCC) mouse Rela promoter consensus sequence. Nuclear extracts from COS-7 cells overexpressing pMT3-Klf3 and untransfected COS-7 cells were used as positive and negative controls, respectively. Polyclonal anti-KLF3 antibody raised in rabbit was added to confirm the identity of bound protein as KLF3, labeled as a supershift band. Unbound radiolabeled DNA probe can be seen as an intense dark band at the bottom of the gel. D, in vivo KLF3 binding at the Rela promoter in WT and Klf3−/− BMDMs treated with 100 ng/ml LPS for 4 h was performed using ChIP-qPCR (n = 2 mice per genotype). Lgals3 serves as a positive control region (+) where KLF3 is known to bind, and an unbound negative control locus −4.5 kb upstream of Klf8 (−) was also used (24). Error bars represent means ± S.E. E, expression of Rela was analyzed in WT and Klf3−/− BMDMs following treatment with LPS (100 ng/ml) or vehicle by qPCR, and mRNA levels were normalized to 18S. The 0-h WT vehicle treatment was set to 1, error bars represent the means ± S.E., and nonparametric Mann–Whitney U tests were performed: *, p < 0.05. F, protein levels of the NF-κB p65 subunit (RELA) were measured in BMDMs following stimulation with 100 ng/ml LPS for 0–24 h. 20 μg of nuclear extract was loaded per lane alongside a Rainbow molecular weight marker (GE Healthcare). The membranes were probed with a monoclonal anti-p65 antibody (raised in rabbit, 65 kDa) followed by reprobing of the membrane with an anti-β-actin antibody (raised in mouse; A1978). A control lane (nuclear extract from COS-7 cells, Ctrl) is also included, and the presence of a nonspecific background band at ∼102 kDa is demarcated by an asterisk.
Closer inspection of the DNA sequence corresponding to the KLF3 binding peak revealed a consensus KLF3 binding site, or CACCC box, in the Rela promoter: 5′-NCN CNC CCN-3′ (Fig. S7A). Using EMSA, we showed that KLF3 can bind the mouse Rela CACCC box, but binding is greatly reduced when this site is mutated (ΔCACCC) (Fig. 5C). Having shown that KLF3 binds the Rela promoter in vitro by EMSA and in vivo in MEFs by ChIP-Seq, we next investigated binding in a biologically relevant setting: primary BMDMs. KLF3 binding was evident at the Rela promoter following LPS stimulation, with negligible enrichment seen at a Klf8 negative control region where KLF3 is known to not bind (Fig. 5D). No binding was observed in Klf3−/− samples. This KLF3 enrichment at the Rela promoter in LPS-stimulated BMDMs correlates with that seen in unstimulated MEFs (Fig. 5B). We subsequently showed that KLF3 also binds the CACCC box in the human RELA promoter by EMSA (Fig. S6B) and, using CRISPR/Cas9 gene editing, generated human K562 cells lacking KLF3 (Fig. S7, B–D) to show that binding to RELA is equally evident in vivo (Fig. S6C). To confirm that KLF3 biologically binds and regulates transcription of Rela, we treated WT and Klf3−/− BMDMs with LPS in a 24-h time course and found that Rela expression levels were significantly more highly up-regulated in the absence of KLF3 at 4 and 8 h (Fig. 5E). Likewise, Klf3−/− BMDMs up-regulated Rela more highly than WT cells in response to poly(I·C) treatment but not with IL-6 treatment (Fig. S6, D and E). We next measured p65 levels by Western blotting in WT and Klf3−/− BMDMs treated with LPS for 0, 4, 8, or 24 h (Fig. 5F). Levels of nuclear p65 are significantly increased in the absence of KLF3 at 8 and 24 h, suggesting that KLF3 not only binds but also directly regulates the expression of the NF-κB subunit p65. The increased nuclear p65 levels were not due to genotype differences in mRNA (Fig. S6F) or protein (Fig. S6G) expression of total cellular IκB-α, which is responsible for sequestering inactive p65 in the cytoplasm. Additionally, we did not observe any differences in total cellular p65 levels between LPS-treated WT and Klf3−/− BMDMs (Fig. S6G). Together, these results demonstrate that NF-κB family genes are deregulated in the absence of KLF3 and that KLF3 directly binds and regulates expression of the gene encoding RELA/p65. This suggests that p65 acts as an intermediary factor that, in the absence of KLF3, is insufficiently repressed, permitting enhanced and prolonged activation of proinflammatory cytokines. These findings identify KLF3 as a potentially important regulator of endotoxin-induced inflammation through its suppression of the NF-κB subunit RELA/p65 (Fig. 6).
Figure 6.
Proposed model of KLF3-mediated suppression of inflammation following endotoxin exposure. Following the onset of an LPS-induced inflammatory response, KLF3 is responsible for regulating NF-κB signaling via repression of Rela/RELA (encoding the NF-κB subunit RELA/p65), suppressing the expression of inflammatory cytokines.
Discussion
NF-κB signaling plays a key role in driving the inflammatory response under both physiological and pathophysiological conditions. A diverse array of signals can elicit NF-κB activity, and the post-translational network governing NF-κB localization through the function of IκB and IκB kinase proteins is well-characterized. NF-κB can regulate its own activity through autoregulatory feedback loops (36), and it was recognized early on that κB subunits have the capacity to bind and autoregulate family members at the transcriptional level (37). Similarly, BCL3 has been shown to activate NF-κB expression, forming another autoregulatory loop (38). Beyond this autoregulation, however, remarkably little is known of how NF-κB gene expression is controlled at the level of transcription. Interestingly, it has been reported that KLF11 is able to physically inhibit the binding of NF-κB subunit p65 to its target genes, effectively suppressing NF-κB–induced proinflammatory signaling in endothelial cells (39).
In this study, using a combination of in vitro approaches and in vivo animal models, we have demonstrated that KLF3 is an important suppressor of inflammation, responsible for directly repressing RELA/p65 activity and thereby downstream cytokine expression. We showed that Klf3−/− mice display systemic inflammation and a distinct white blood cell phenotype, with increased sensitivity to endotoxin. Mice lacking another DNA-binding protein, NUPR1, also show increased sensitivity to endotoxin treatment, caused by an aberrant gene expression response (40), much like the widespread deregulation seen in the absence of KLF3. We propose that the physiological manifestations seen in LPS-treated Klf3−/− mice, including inflammatory monocyte abundance and overexpression of proinflammatory cytokines, are the result of a reduced capacity to suppress NF-κB signaling. It will be interesting to further pursue the downstream consequences of the absence of KLF3 on LPS tolerance and innate immune memory in future studies, given the clinical significance of these processes (41).
Directly targeting transcription factors to therapeutically modulate cellular processes remains difficult, and this is no different with inflammation. Given that other KLF family members are involved in orchestrating the inflammatory response in various cell types, it has been proposed that targeting upstream mediators of KLFs may be the most viable option (42, 43).
Together, our findings identify KLF3 as a novel and direct suppressor of NF-κB signaling through regulation of Rela/RELA transcription. This advance fills a longstanding gap in our knowledge of the transcriptional regulation of the NF-κB family and provides a starting point for further studies into transcriptional control during the inflammatory response.
Experimental procedures
Animal husbandry
All animal work was carried out in accordance with approval from the University of New South Wales Animal Care and Ethics Committee (approval nos. 12/150A, 16/5B, and 18/34B). The animals were housed in a specific pathogen-free environment, at a constant ambient temperature of 22 °C, on a 12-h light–dark cycle with ad libitum access to standard chow food and water. Generation of global Klf3−/− mice on an FVB/NJ background has been previously described (29). Age-matched WT and Klf3−/− littermate mice from Klf3+/− × Klf3+/− crosses were used for all animal studies.
Animal procedures
For short-term endotoxin exposure experiments, WT and Klf3−/− mice were treated with 0.167 or 0.05 mg/kg LPS (Sigma) from E. coli or vehicle (0.1% saline) via intraperitoneal injection. Body temperature was measured over the first 6 h by rectal probing with a BAT-12 microprobe thermometer and then again at 12 and 24 h. After 24 h, the mice were euthanized, and blood and spleens were harvested for downstream analysis.
Blood analysis
Peripheral blood was obtained by cardiac bleed prior to euthanasia and stored in K2EDTA collection tubes (BD Biosciences). Whole blood was sent for blood count analysis at the University of Sydney Veterinary Pathology Diagnostic Services. Plasma was isolated by centrifugation of whole blood at 2,000 × g for 15 min (4 °C) and then stored for further analysis.
Cell culture and transfection
Bone marrow-derived macrophages were harvested from femora and tibiae as previously described (24). Equal numbers of WT and Klf3−/− BMDMs were seeded for all experiments. COS-7 cells were a gift from Stuart Orkin (Harvard) and were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin–glutamine. During passaging, adherent cells were lifted after a 5-min incubation at 37 °C with 2 mm PBS-EDTA. COS-7 cells were transfected with pMT3-Klf3 using FuGENE 6 (Promega) and harvested 48 h later for nuclear extraction. K562 cells were grown in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin–streptomycin–glutamine. Expression construct information can be found in Table S1.
Gene editing
For CRISPR/Cas9 genome editing, a plasmid encoding both the Cas9 protein and the single guide RNA (sgRNA) was used to delete KLF3 in K562 cells via double-strand breakage and nonhomologous end joining. pSpCas9(BB)-2A-GFP (PX458) was a gift from Feng Zhang (Addgene 48138) (44). We designed sgRNA sequences targeting KLF3 exon 3 using the optimized CRISPR/Cas9 design online tool provided by the Zhang laboratory from the Massachusetts Institute of Technology. All sgRNA and PCR primer sequences can be found in Table S2. K562 cells were transfected by nucleofection using a NeonTM transfection system (Life Technologies). The cells (5 × 105) were resuspended in nucleofection buffer R (NeonTM transfection kit) and given one pulse of 1,400–1,600 V for 20–30 ms. The cells were cultured for 72 h in RPMI 1640 medium with 10% FBS without antibiotics and then enriched by FACS, and clonal populations were established by sorting cells into 96-well culture plates. To screen clones for the desired KLF3 disruption, PCR was carried out on genomic DNA using Q5 polymerase (New England BioLabs), before confirmation using Sanger sequencing of PCR products and anti-KLF3 Western blotting. SnapGene version 4.2.6 was used to visualize sequencing traces.
Flow cytometry
Flow cytometry was performed using a BD LSRFortessa and BD LSRFortessa X-20. Sorting by FACS was performed using a BD Influx or BD FACSAria III. All cells were preblocked with anti-CD16/32 Fc block to reduce nonspecific binding. For identifying Ly6C+ monocytes in blood and spleen, red blood cell lysis was performed using double-distilled H2O prior to staining with the following antibodies: CD45-PE (BD Pharmingen), CD115-biotin (eBioscience), Streptavidin-BV711 (BD Horizon), CD11b-FITC (BD Pharmingen), Ly6C-BV421 (BD Horizon), Ly6G-APC (BD Pharmingen), CD19-APC (eBioscience), and CD3-APC (BD Pharmingen). ZombieNIR (Biolegend) was used as a viability dye. To analyze monocyte precursors in bone marrow, bone marrow was harvested and flushed, and then the cells were stained with the following antibodies: CD11b-FITC (BD Pharmingen), Ly6C-BV421 (BD Horizon), CD117-PE (BD Pharmingen), CD135-APC (BD Biosciences), CD115-BUV395 (BD Optibuild), Streptavidin-PE/CF594 (BD Biosciences) and a lineage mixture comprised of biotin-conjugated Ter119 (BD Biosciences), CD19 (BD Pharmingen), CD3 (eBioscience), NK1.1 (eBioscience), Ly6G (BD Pharmingen), and TCRγδ (eBioscience). 4′,6′-Diamino-2-phenylindole was used as a viability dye. For identifying BMDMs, the cells were stained with the following antibodies: CD11b-FITC (BD Pharmingen) and F4/80-PE/Cy5 (eBioscience) (gating strategy in Fig. S1C). Flow cytometry analysis was performed using FlowJo software, version 10. Detailed antibody information is available in Table S5.
Cytokine quantification
To measure the levels of inflammatory cytokines in mouse plasma and BMDM supernatant, we employed the LEGENDplexTM kit (Biolegend) according to the manufacturer's instructions. The assay was run using the BD LSRFortessa, and the output was analyzed with LEGENDplex version 8.0 software. To assess the levels of TNF in plasma from mice treated with LPS or vehicle, the mouse TNF QuantikineTM ELISA kit (R&D Systems) was used according to the manufacturer's instructions. A volume of 50 μl of undiluted plasma was run in duplicate for each sample for ELISA, and 25 μl of undiluted plasma or BMDM supernatant was used for LEGENDplex assays.
BMDM stimulations
BMDMs were differentiated in culture for 7–10 days and then treated with 100 ng/ml LPS (Sigma), 10 ng/ml IFNγ (Sigma), 10 μg/ml poly(I·C) (Sigma), 10 ng/ml IL-6 (Sigma), or 10 ng/ml IFNα (Thermo Fisher) for the stated lengths of time.
BMDM functional assays
To assess the phagocytic capacity of BMDMs, VybrantTM phagocytosis assays (Thermo Fisher) were performed according to the manufacturer's instructions. Briefly, the cells were seeded into 96-well plates at 5 × 104 cells/well and incubated at 37 °C for 6 h with 100 ng/ml LPS. After 6 h, supernatant was replaced with 100 μl of E. coli (K-12 strain) fluorescein-labeled bioparticles for 2 h in the presence of 100 ng/ml LPS. Bioparticles were removed entirely, and trypan blue was added for 1 min prior to reading at an excitation of 480 nm and emission of 520 nm to assess the fluorescence produced by phagocytosed bioparticles. To measure nitrate production, BMDMs were treated with 100 ng/ml LPS or vehicle for 24 h before collection of supernatant. The nitrate/nitrite colorimetric assay kit (Cayman Chemical) was employed to measure nitrate concentration in the supernatant and absorbance read at 540 nm, according to the manufacturer's instructions. To assess the cellular reduction environment in BMDMs, alamarBlueTM (Life Technologies) assays were performed according to the manufacturer's instructions. Briefly, the cells were seeded into 96-well plates at 5 × 104 cells/well overnight and then treated with 100 ng/ml LPS or vehicle the following day, in addition to a 0.1× volume of alamarBlue reagent. Fluorescence was read after 0, 2, 4, 6, 8, 10, 12, and 24 h with a peak excitation of 570 nm and peak emission at 585 nm.
Cell growth and proliferation assays
For cell growth assays, BMDMs were grown for 7 days before seeding at 5 × 105 cells/ml. Cell counts were taken at days 0, 2, 4, and 6. Cell proliferation assays were performed using a BrdU cell proliferation assay kit, according to the manufacturer's instructions (Cell Signaling Technology). BMDMs were cultured for 7 days, and then 5 × 104 cells were plated into wells of a 96-well plate. The cells were then assayed at days 0, 2, 4, and 6. Briefly, 1× BrdU was added into each well and incubated for 4 h, then the cells were fixed and denatured for 30 min, and detection antibody solution was added for 1 h at room temperature. Next, HRP-conjugated secondary antibody solution was added and incubated at room temperature for 30 min. TMB substrate was then added and incubated for 30 min before adding stopping buffer, and then absorbance was read at 450 nm to detect BrdU incorporation.
Gene expression analysis
To assess mRNA expression, cells were lysed and homogenized in TRI reagent (Sigma) and then subjected to RNA extraction and cDNA synthesis as previously described (24). Quantitative real-time PCRs (qPCRs) were set up with Power SYBRTM Green PCR Master Mix and run with default cycle parameters on the Applied Biosystems 7500 fast real-time PCR system (for 96-well plate format) or the Applied Biosystems ViiA7 real-time PCR system (for 384-well plate format). Gene expression was quantified using the 2−ΔΔCT method, and relative mRNA expression was normalized to 18S rRNA levels, which have been shown to display stable expression across the cells and conditions studied. All qPCR primers were designed using primer3 and can be found in Table S4. For microarrays on BMDMs treated with 100 ng/ml LPS or vehicle, RNA was isolated using the RNeasyTM mini kit (Qiagen) and then subjected to quality control using the Agilent 2100 Bioanalyzer, following preparation with an Agilent RNA 6000 nano kit. An Affymetrix 3′ IVT PLUS kit was used for in vitro transcription prior to performing microarrays on an Affymetrix GeneChip HT MG-430PM array plate. Raw CEL files from the Affymetrix arrays were processed using the R package affy (45) (using ReadAffy) and the Robust Multi-array Average (RMA) method (46) (using rma also from the same package) was used to normalize the data and produce expression values. Differential expression analysis was done using the limma (47) R package. Heat maps were generated after hierarchical clustering and scaling of the row values using the RMA normalized expression values with R package pheatmap. Kyoto Encyclopedia of Genes and Genomes pathway analysis was done using the enrichKEGG function from clusterProfiler (48) with the dotplot function used to generate graphs. Microarray data sets are available from the Gene Expression Omnibus (accession no. GSE121646).
Motif discovery and enrichment analysis
To perform de novo motif discovery and known motif enrichment analysis, the transcription starts sites of all differentially expressed genes when comparing WT and Klf3−/− BMDMs treated with LPS for 8 h were first obtained from the GENCODE vM22 gtf file. Promoters were then defined as 1,000 bp upstream or downstream from the transcription start site (± 1,000 bp) and obtained using bedtools flank and slop functions (49) and HOMER findMotifsGenome.pl (to get sequences) (50). De novo motif analysis and known motif enrichment analysis using the JASPAR (51) and HOCOMOCO TF (52) databases was then performed using MEME-ChIP (53) (only the centered 500-bp region was considered for de novo motif discovery (using −ccut 500) with a maximum motif site of 25 bp (−meme−maxw 25)).
Protein extraction and Western blotting
Cytoplasmic, nuclear, and whole-cell protein extractions were performed as previously described (24, 27). For detection of KLF3 by Western blotting, 20 μg of nuclear extract was loaded onto Novex NuPAGETM 10% Bis-Tris gels, following denaturation and boiling. After blocking with 3.5% (w/v) skim milk, nitrocellulose membranes were incubated with anti-KLF3 antibody (Thermo Fisher). The membranes were then probed with HRP-linked anti-goat antibody (Santa Cruz Biotechnology) prior to imaging on the GE ImageQuant LAS 500 in the presence of Immobilon Western Chemiluminescent HRP substrate (Millipore). To detect RELA/p65 and IκB-α, 20 μg of protein was loaded per lane onto 10% Bis-Tris gels, following denaturation and boiling. After blocking with 5% (w/v) skim milk, nitrocellulose membranes were incubated overnight with anti-p65 antibody (Cell Signaling) or anti-IκB-α antibody (Abcam). The membranes were then probed with HRP-linked anti-rabbit antibody (GE Healthcare) prior to imaging. All membranes were stripped with 0.5 m NaOH and then reblocked with skim milk before probing with an anti-β-actin antibody (Sigma). Following this, HRP-linked anti-mouse antibody (GE Healthcare) was incubated on membranes prior to imaging. Detailed antibody information can be found in Table S5.
Electrophoretic mobility shift assays
EMSAs were performed as described previously (27). To assess KLF3 levels following LPS treatment, BMDMs were stimulated with 100 ng/ml LPS for 0, 4, 8 and, 24 h, and then nuclear extracts were obtained. The extracts were loaded onto 8% polyacrylamide gels along with prebleed immune serum or polyclonal anti-KLF3 antibody (raised in rabbit) and 32P-radiolabeled DNA probe. Nuclear extracts from untransfected COS-7 cells and COS-7 cells expressing pMT3-Klf3 were included as negative and positive controls, respectively. To assess binding of KLF3 to mouse and human RELA, probes were designed corresponding to the mouse and human CACCC boxes in the RELA promoter region and mutated versions of these sites (ΔCACCC). The gels were run for 1 h 45 min at 250 V and then imaged using a Typhoon FLA 9500 (GE Life Sciences). Probe sequences can be found in Table S3, and visual depiction of probes can be found in Fig. S7A.
Chromatin immunoprecipitation
ChIP experiments were performed as previously described (54). Briefly, 5–7 × 107 BMDM or K562 cells were used per immunoprecipitation. Prior to cross-linking, BMDMs were treated with 100 ng/ml LPS for 4 h. The cells were cross-linked with 1% formaldehyde for 10 min at room temperature, and the reaction was quenched with glycine at a final concentration of 125 mm. Cross-linked cells were then lysed and sonicated to obtain ∼200–400-bp fragments of chromatin. DNA bound to the protein of interest was pulled down at 4 °C overnight using anti-KLF3 antibody (Thermo Fisher) or normal goat IgG (Santa Cruz Biotechnology). qPCR was performed on ChIP material using an Applied Biosystems ViiA7 real-time PCR system. For analysis, immunoprecipitation was first normalized to the relative amount of input DNA and then to IgG controls. All qPCR primers for ChIP were designed using primer3 and can be found in Table S4. Detailed antibody information can be found in Table S5. The KLF3-V5 ChIP-Seq data set in MEFs is accessible at from the Gene Expression Omnibus (accession no. GSE44748). Data sets were visualized using Integrative Genomics Viewer software (Broad Institute).
Author contributions
A. J. K., L. Y., M. C., and K. G. R. Q. conceptualization; A. J. K. data curation; A. J. K., L. Y., M. S., L. J. N., G. S. G., E. S. S., E. J. V., and M. C. formal analysis; A. J. K., L. Y., M. S., L. J. N., G. S. G., E. S. S., E. J. V., M. C., and K. G. R. Q. investigation; A. J. K., L. Y., M. S., L. J. N., G. S. G., E. S. S., E. J. V., and M. C. methodology; A. J. K., L. Y., M. C., and K. G. R. Q. writing-original draft; A. J. K., M. C., and K. G. R. Q. project administration; A. J. K., L. Y., M. S., L. J. N., G. S. G., E. S. S., E. J. V., M. C., and K. G. R. Q. writing-review and editing; L. Y., M. C., and K. G. R. Q. supervision; L. Y. validation; L. Y., M. S., and L. J. N. visualization; M. C. funding acquisition.
Supplementary Material
Acknowledgments
We thank Chris Brownlee and Emma Johansson at the University of New South Wales Biological Resources Imaging Laboratory (BRIL) Flow Cytometry Facility for assistance with flow cytometry and sorting experiments. We also acknowledge the assistance of the Ramaciotti Centre for Genomics (University of New South Wales) for Sanger sequencing and microarray services.
This work was supported by Australian National Health and Medical Research Council (NHMRC) Grants APP1025873 and APP1025877 (to M. C.), Australian Postgraduate Awards (to A. J. K., L. J. N., M. S., and E. S. S.), a University of New South Wales Sydney Scientia Fellowship (to K. G. R. Q.), and a University of New South Wales Sydney Scientia Scholarship (to E. J. V.). The authors declare that they have no conflicts of interest with the contents of this article.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE121646.
This article contains Tables S1–S5 and Figs. S1–S7.
- TNF
- tumor necrosis factor
- IL
- interleukin
- LPS
- lipopolysaccharide
- KLF
- Krüppel-like factor
- BMDM
- bone marrow-derived macrophage
- EMSA
- electrophoretic mobility shift assay
- BrdU
- bromodeoxyuridine
- IFN
- interferon
- MEF
- murine embryonic fibroblast
- FBS
- fetal bovine serum
- HRP
- horseradish peroxidase
- qPCR
- quantitative real-time PCR
- sgRNA
- single guide RNA.
References
- 1. Kauppinen A., Paterno J. J., Blasiak J., Salminen A., and Kaarniranta K. (2016) Inflammation and its role in age-related macular degeneration. Cell. Mol. Life Sci. 73, 1765–1786 10.1007/s00018-016-2147-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pérez-Cerdá F., Sánchez-Gómez M. V., and Matute C. (2016) The link of inflammation and neurodegeneration in progressive multiple sclerosis. Multiple Sclerosis Demyelinating Disorders 1, 9 10.1186/s40893-016-0012-0 [DOI] [Google Scholar]
- 3. Smolen J. S., Aletaha D., and McInnes I. B. (2016) Rheumatoid arthritis. Lancet 388, 2023–2038 10.1016/S0140-6736(16)30173-8 [DOI] [PubMed] [Google Scholar]
- 4. Gajendran M., Loganathan P., Catinella A. P., and Hashash J. G. (2018) A comprehensive review and update on Crohn's disease. Dis. Mon. 64, 20–57 10.1016/j.disamonth.2017.07.001 [DOI] [PubMed] [Google Scholar]
- 5. Straub R. H., and Schradin C. (2016) Chronic inflammatory systemic diseases: an evolutionary trade-off between acutely beneficial but chronically harmful programs. Evol. Med. Public Health 2016, 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tak P. P., and Firestein G. S. (2001) NF-κB: a key role in inflammatory diseases. J. Clin. Invest. 107, 7–11 10.1172/JCI11830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mao L., Nie B., Nie T., Hui X., Gao X., Lin X., Liu X., Xu Y., Tang X., Yuan R., Li K., Li P., Ding K., Wang Y., Xu A., et al. (2017) Visualization and quantification of browning using a Ucp1–2A-Luciferase knock-in mouse model. Diabetes 66, 407–417 10.2337/db16-0343 [DOI] [PubMed] [Google Scholar]
- 8. Bonizzi G., and Karin M. (2004) The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 25, 280–288 10.1016/j.it.2004.03.008 [DOI] [PubMed] [Google Scholar]
- 9. Shakhov A. N., Collart M. A., Vassalli P., Nedospasov S. A., and Jongeneel C. V. (1990) κB-type enhancers are involved in lipopolysaccharide-mediated transcriptional activation of the tumor necrosis factor α gene in primary macrophages. J. Exp. Med. 171, 35–47 10.1084/jem.171.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mori N., and Prager D. (1996) Transactivation of the interleukin-1α promoter by human T-cell leukemia virus type I and type II Tax proteins. Blood 87, 3410–3417 10.1182/blood.V87.8.3410.bloodjournal8783410 [DOI] [PubMed] [Google Scholar]
- 11. Hiscott J., Marois J., Garoufalis J., D'Addario M., Roulston A., Kwan I., Pepin N., Lacoste J., Nguyen H., and Bensi G. (1993) Characterization of a functional NF-κB site in the human interleukin 1β promoter: evidence for a positive autoregulatory loop. Mol. Cell. Biol. 13, 6231–6240 10.1128/MCB.13.10.6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Libermann T. A., and Baltimore D. (1990) Activation of interleukin-6 gene expression through the NF-κB transcription factor. Mol. Cell. Biol. 10, 2327–2334 10.1128/MCB.10.5.2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gadjeva M., Tomczak M. F., Zhang M., Wang Y. Y., Dull K., Rogers A. B., Erdman S. E., Fox J. G., Carroll M., and Horwitz B. H. (2004) A role for NF-κB subunits p50 and p65 in the inhibition of lipopolysaccharide-induced shock. J. Immunol. 173, 5786–5793 10.4049/jimmunol.173.9.5786 [DOI] [PubMed] [Google Scholar]
- 14. Saccani A., Schioppa T., Porta C., Biswas S. K., Nebuloni M., Vago L., Bottazzi B., Colombo M. P., Mantovani A., and Sica A. (2006) p50 nuclear factor-κB overexpression in tumor-associated macrophages inhibits M1 inflammatory responses and antitumor resistance. Cancer Res. 66, 11432–11440 10.1158/0008-5472.CAN-06-1867 [DOI] [PubMed] [Google Scholar]
- 15. Lawrence T., Gilroy D. W., Colville-Nash P. R., and Willoughby D. A. (2001) Possible new role for NF-κB in the resolution of inflammation. Nat. Med. 7, 1291–1297 10.1038/nm1201-1291 [DOI] [PubMed] [Google Scholar]
- 16. Martin G. S., Mannino D. M., Eaton S., and Moss M. (2003) The epidemiology of sepsis in the United States from 1979 through 2000. N. Engl. J. Med. 348, 1546–1554 10.1056/NEJMoa022139 [DOI] [PubMed] [Google Scholar]
- 17. Andreakos E., Sacre S. M., Smith C., Lundberg A., Kiriakidis S., Stonehouse T., Monaco C., Feldmann M., and Foxwell B. M. (2004) Distinct pathways of LPS-induced NF-κB activation and cytokine production in human myeloid and nonmyeloid cells defined by selective utilization of MyD88 and Mal/TIRAP. Blood 103, 2229–2237 10.1182/blood-2003-04-1356 [DOI] [PubMed] [Google Scholar]
- 18. Das H., Kumar A., Lin Z., Patino W. D., Hwang P. M., Feinberg M. W., Majumder P. K., and Jain M. K. (2006) Kruppel-like factor 2 (KLF2) regulates proinflammatory activation of monocytes. Proc. Natl. Acad. Sci. U.S.A. 103, 6653–6658 10.1073/pnas.0508235103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alder J. K., Georgantas R. W. 3rd, Hildreth R. L., Kaplan I. M., Morisot S., Yu X., McDevitt M., and Civin C. I. (2008) Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J. Immunol. 180, 5645–5652 10.4049/jimmunol.180.8.5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Date D., Das R., Narla G., Simon D. I., Jain M. K., and Mahabeleshwar G. H. (2014) Kruppel-like transcription factor 6 regulates inflammatory macrophage polarization. J. Biol. Chem. 289, 10318–10329 10.1074/jbc.M113.526749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eaton S. A., Funnell A. P., Sue N., Nicholas H., Pearson R. C., and Crossley M. (2008) A network of Kruppel-like factors (Klfs): Klf8 is repressed by Klf3 and activated by Klf1 in vivo. J. Biol. Chem. 283, 26937–26947 10.1074/jbc.M804831200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Funnell A. P., Norton L. J., Mak K. S., Burdach J., Artuz C. M., Twine N. A., Wilkins M. R., Power C. A., Hung T. T., Perdomo J., Koh P., Bell-Anderson K. S., Orkin S. H., Fraser S. T., Perkins A. C., et al. (2012) The CACCC-binding protein KLF3/BKLF represses a subset of KLF1/EKLF target genes and is required for proper erythroid maturation in vivo. Mol. Cell. Biol. 32, 3281–3292 10.1128/MCB.00173-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Funnell A. P., Mak K. S., Twine N. A., Pelka G. J., Norton L. J., Radziewic T., Power M., Wilkins M. R., Bell-Anderson K. S., Fraser S. T., Perkins A. C., Tam P. P., Pearson R. C., and Crossley M. (2013) Generation of mice deficient in both KLF3/BKLF and KLF8 reveals a genetic interaction and a role for these factors in embryonic globin gene silencing. Mol. Cell. Biol. 33, 2976–2987 10.1128/MCB.00074-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Knights A. J., Yik J. J., Mat Jusoh H., Norton L. J., Funnell A. P., Pearson R. C., Bell-Anderson K. S., Crossley M., and Quinlan K. G. (2016) Kruppel-like factor 3 (KLF3/BKLF) is required for widespread repression of the inflammatory modulator galectin-3 (Lgals3). J. Biol. Chem. 291, 16048–16058 10.1074/jbc.M116.715748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vu T. T., Gatto D., Turner V., Funnell A. P., Mak K. S., Norton L. J., Kaplan W., Cowley M. J., Agenès F., Kirberg J., Brink R., Pearson R. C., and Crossley M. (2011) Impaired B cell development in the absence of Kruppel-like factor 3. J. Immunol. 187, 5032–5042 10.4049/jimmunol.1101450 [DOI] [PubMed] [Google Scholar]
- 26. Bell-Anderson K. S., Funnell A. P., Williams H., Mat Jusoh H., Scully T., Lim W. F., Burdach J. G., Mak K. S., Knights A. J., Hoy A. J., Nicholas H. R., Sainsbury A., Turner N., Pearson R. C., and Crossley M. (2013) Loss of Kruppel-like factor 3 (KLF3/BKLF) leads to upregulation of the insulin-sensitizing factor adipolin (FAM132A/CTRP12/C1qdc2). Diabetes 62, 2728–2737 10.2337/db12-1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crossley M., Whitelaw E., Perkins A., Williams G., Fujiwara Y., and Orkin S. H. (1996) Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol. Cell. Biol. 16, 1695–1705 10.1128/MCB.16.4.1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dewi V., Kwok A., Lee S., Lee M. M., Tan Y. M., Nicholas H. R., Isono K., Wienert B., Mak K. S., Knights A. J., Quinlan K. G., Cordwell S. J., Funnell A. P., Pearson R. C., and Crossley M. (2015) Phosphorylation of Kruppel-like factor 3 (KLF3/BKLF) and C-terminal binding protein 2 (CtBP2) by homeodomain-interacting protein kinase 2 (HIPK2) modulates KLF3 DNA binding and activity. J. Biol. Chem. 290, 8591–8605 10.1074/jbc.M115.638338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sue N., Jack B. H., Eaton S. A., Pearson R. C., Funnell A. P., Turner J., Czolij R., Denyer G., Bao S., Molero-Navajas J. C., Perkins A., Fujiwara Y., Orkin S. H., Bell-Anderson K., and Crossley M. (2008) Targeted disruption of the basic Kruppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol. Cell. Biol. 28, 3967–3978 10.1128/MCB.01942-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oettgen H. F., Carswell E. A., Kassel R. L., Fiore N., Williamson B., Hoffmann M. K., Haranaka K., and Old L. J. (1980) Endotoxin-induced tumor necrosis factor. Recent Results Cancer Res. 75, 207–212 10.1007/978-3-642-81491-4_32 [DOI] [PubMed] [Google Scholar]
- 31. Geng S., Chen K., Yuan R., Peng L., Maitra U., Diao N., Chen C., Zhang Y., Hu Y., Qi C.-F., Pierce S., Ling W., Xiong H., and Li L. (2016) The persistence of low-grade inflammatory monocytes contributes to aggravated atherosclerosis. Nat. Commun. 7, 13436 10.1038/ncomms13436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Steiner A. A., Molchanova A. Y., Dogan M. D., Patel S., Pétervári E., Balaskó M., Wanner S. P., Eales J., Oliveira D. L., Gavva N. R., Almeida M. C., Székely M., and Romanovsky A. A. (2011) The hypothermic response to bacterial lipopolysaccharide critically depends on brain CB1, but not CB2 or TRPV1, receptors. J. Physiol. 589, 2415–2431 10.1113/jphysiol.2010.202465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lingrel J. B., Pilcher-Roberts R., Basford J. E., Manoharan P., Neumann J., Konaniah E. S., Srinivasan R., Bogdanov V. Y., and Hui D. Y. (2012) Myeloid-specific Kruppel-like factor 2 inactivation increases macrophage and neutrophil adhesion and promotes atherosclerosis. Circ. Res. 110, 1294–1302 10.1161/CIRCRESAHA.112.267310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim G. D., Das R., Goduni L., McClellan S., Hazlett L. D., and Mahabeleshwar G. H. (2016) Kruppel-like factor 6 promotes macrophage-mediated inflammation by suppressing B cell leukemia/lymphoma 6 expression. J. Biol. Chem. 291, 21271–21282 10.1074/jbc.M116.738617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burdach J., Funnell A. P., Mak K. S., Artuz C. M., Wienert B., Lim W. F., Tan L. Y., Pearson R. C., and Crossley M. (2014) Regions outside the DNA-binding domain are critical for proper in vivo specificity of an archetypal zinc finger transcription factor. Nucleic Acids Res. 42, 276–289 10.1093/nar/gkt895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Renner F., and Schmitz M. L. (2009) Autoregulatory feedback loops terminating the NF-κB response. Trends Biochem. Sci. 34, 128–135 10.1016/j.tibs.2008.12.003 [DOI] [PubMed] [Google Scholar]
- 37. Liptay S., Schmid R. M., Nabel E. G., and Nabel G. J. (1994) Transcriptional regulation of NF-κB2: evidence for κB-mediated positive and negative autoregulation. Mol. Cell. Biol. 14, 7695–7703 10.1128/MCB.14.12.7695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fujita T., Nolan G. P., Liou H. C., Scott M. L., and Baltimore D. (1993) The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-κB p50 homodimers. Genes Dev. 7, 1354–1363 10.1101/gad.7.7b.1354 [DOI] [PubMed] [Google Scholar]
- 39. Fan Y., Guo Y., Zhang J., Subramaniam M., Song C. Z., Urrutia R., and Chen Y. E. (2012) Kruppel-like factor-11, a transcription factor involved in diabetes mellitus, suppresses endothelial cell activation via the nuclear factor-κB signaling pathway. Arterioscler. Thromb. Vasc. Biol. 32, 2981–2988 10.1161/ATVBAHA.112.300349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vasseur S., Hoffmeister A., Garcia-Montero A., Barthet M., Saint-Michel L., Berthézène P., Fiedler F., Closa D., Dagorn J. C., and Iovanna J. L. (2003) Mice with targeted disruption of p8 gene show increased sensitivity to lipopolysaccharide and DNA microarray analysis of livers reveals an aberrant gene expression response. BMC Gastroenterol. 3, 25 10.1186/1471-230X-3-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Biswas S. K., and Lopez-Collazo E. (2009) Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 30, 475–487 10.1016/j.it.2009.07.009 [DOI] [PubMed] [Google Scholar]
- 42. Jain M. K., Sangwung P., and Hamik A. (2014) Regulation of an inflammatory disease: Krüppel-like factors and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 34, 499–508 10.1161/ATVBAHA.113.301925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sweet D. R., Fan L., Hsieh P. N., and Jain M. K. (2018) Krüppel-like factors in vascular inflammation: mechanistic insights and therapeutic potential. Front. Cardiovasc. Med. 5, 6–6 10.3389/fcvm.2018.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gautier L., Cope L., Bolstad B. M., and Irizarry R. A. (2004) affy: analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 10.1093/bioinformatics/btg405 [DOI] [PubMed] [Google Scholar]
- 46. Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., and Speed T. P. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15 10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., and Smyth G. K. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu G., Wang L. G., Han Y., and He Q. Y. (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quinlan A. R., and Hall I. M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., and Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fornes O., Castro-Mondragon J. A., Khan A., van der Lee R., Zhang X., Richmond P. A., Modi B. P., Correard S., Gheorghe M., Baranašić D., Santana-Garcia W., Tan G., Chèneby J., Ballester B., Parcy F., et al. (2020) JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kulakovskiy I. V., Vorontsov I. E., Yevshin I. S., Sharipov R. N., Fedorova A. D., Rumynskiy E. I., Medvedeva Y. A., Magana-Mora A., Bajic V. B., Papatsenko D. A., Kolpakov F. A., and Makeev V. J. (2018) HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 46, D252–D259 10.1093/nar/gkx1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Machanick P., and Bailey T. L. (2011) MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697 10.1093/bioinformatics/btr189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schmidt D., Wilson M. D., Spyrou C., Brown G. D., Hadfield J., and Odom D. T. (2009) ChIP-seq: using high-throughput sequencing to discover protein–DNA interactions. Methods 48, 240–248 10.1016/j.ymeth.2009.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.