Significance
Heparin is the most widely prescribed biopharmaceutical worldwide due to its potent anticoagulant activity. Therapeutic heparin is sourced primarily from porcine entrails and bovine lung. Mast cells appear to be the primary cell type that produces heparin, but all cells make a related polysaccharide, heparan sulfate. Understanding the factors that regulate the production of anticoagulant heparin would provide tools for bioengineering heparin. Here we provide evidence that key sulfotransferases in heparin/heparan sulfate production are under repression by the zinc finger protein ZNF263.
Keywords: heparin, zinc-finger transcription factor, heparan sulfate 3-O-sulfotransferases, anticoagulant, Factor X
Abstract
Heparin is the most widely prescribed biopharmaceutical in production globally. Its potent anticoagulant activity and safety makes it the drug of choice for preventing deep vein thrombosis and pulmonary embolism. In 2008, adulterated material was introduced into the heparin supply chain, resulting in several hundred deaths and demonstrating the need for alternate sources of heparin. Heparin is a fractionated form of heparan sulfate derived from animal sources, predominantly from connective tissue mast cells in pig mucosa. While the enzymes involved in heparin biosynthesis are identical to those for heparan sulfate, the factors regulating these enzymes are not understood. Examination of the promoter regions of all genes involved in heparin/heparan sulfate assembly uncovered a transcription factor-binding motif for ZNF263, a C2H2 zinc finger protein. CRISPR-mediated targeting and siRNA knockdown of ZNF263 in mammalian cell lines and human primary cells led to dramatically increased expression levels of HS3ST1, a key enzyme involved in imparting anticoagulant activity to heparin, and HS3ST3A1, another glucosaminyl 3-O-sulfotransferase expressed in cells. Enhanced 3-O-sulfation increased binding to antithrombin, which enhanced Factor Xa inhibition, and binding of neuropilin-1. Analysis of transcriptomics data showed distinctively low expression of ZNF263 in mast cells compared with other (non–heparin-producing) immune cells. These findings demonstrate a novel regulatory factor in heparan sulfate modification that could further advance the possibility of bioengineering anticoagulant heparin in cultured cells.
Heparin is one of the most frequently prescribed medications in the United States, administered to more than 12 million patients per year to prevent thrombosis during surgery and to treat thromboembolism (1, 2). A member of a family of linear polysaccharides known as glycosaminoglycans, heparin consists of alternating glucosamine and uronic acids that are heterogeneously N- and O-sulfated. The arrangement and orientation of the sulfated sugar residues specify distinct protein-binding sites that endow heparin with its various biological activities, including anticoagulant activity. Heparin shares a common biosynthetic pathway with heparan sulfate (HS), which is less sulfated than heparin, generally lacks anticoagulant activity, and is expressed on the cell surface and in the extracellular matrix of all mammalian cells (3).
The biosynthesis of heparin and HS is a nontemplated process, driven by the concerted activity of a large family of enzymes localized to the Golgi and endoplasmic reticulum (Fig. 1A). A key reaction in heparin biosynthesis is the addition of a sulfate group to the C3 position of an N-sulfoglucosamine residue within a sulfated pentasaccharide sequence, which confers high-affinity binding to antithrombin. Binding leads to a conformational change in antithrombin and ∼1,000-fold enhancement in its capacity to inactivate thrombin and Factor Xa, which accounts for its potent anticoagulant activity (4).
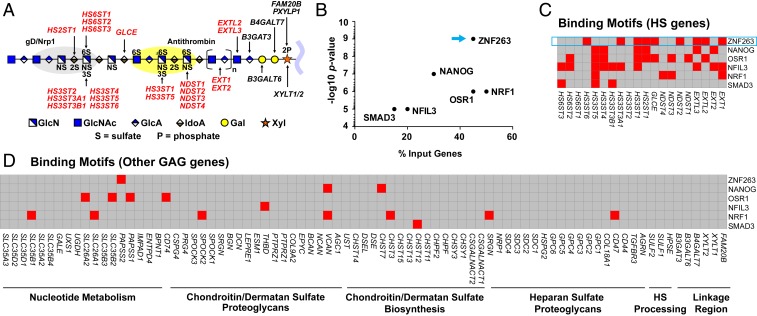
Fig. 1.
Heparin/HS structure, assembly, and regulation. (A) HS and heparin assemble while attached via a linkage tetrasaccharide to a core protein of a proteoglycan. NDSTs, HS2ST, HS3STs, and HS6STs install sulfate groups at specific sites along the HS/heparin chain, and an epimerase (GLCE) converts d-glucuronic acid to l-iduronic acid. The chain is rendered according the Symbol Nomenclature for Glycans (47). The gray and yellow oval shapes depict protein-binding sites for gD/NRP1 and antithrombin, respectively. Input genes for HOMER are indicated in bold red font. (B) HOMER motif enrichment analysis revealing TFs with predicted binding motifs in a set of HS biosynthesis genes. (C) Heatmap showing the presence (red) or absence (gray) of TF-binding motifs (y-axis) in regulatory regions of HS biosynthesis genes (x-axis), as predicted by HOMER. (D) Heatmap showing the presence (red) or absence (gray) of the same TF-binding motifs in a list of genes involved in nucleotide sugar and sulfate metabolism, CS/DS proteoglycans, CS/DS biosynthesis, HS proteoglycans, extracellular HS processing enzymes, and enzymes that generate the common linkage tetrasaccharide in heparin/HS and CS/DS.
In 2008, the US Food and Drug Administration issued a major recall of pharmaceutical heparin due to contamination of the raw heparin stock imported from China. This crisis prompted new guidelines for monitoring the purity of heparin, but the feedstock remains vulnerable to natural variation, susceptibility of the pig population to infectious agents, and potential contamination (5). Thus, finding an alternative source of heparin is important. While HS is produced by all animal cells, anticoagulant heparin is biosynthesized and stored primarily in connective-tissue type mast cells, a type of granulocyte derived from myeloid stem cells. The factors that regulate the enzymes and give rise to anticoagulant heparin are unknown. Defining these factors could facilitate the production of bioengineered heparin in typical cells used in the biopharmaceutical industry, such as Chinese hamster ovary cells (6).
In this study, we analyzed the expression and promoter sequences of the enzymes involved in HS and heparin assembly and identified ZNF263, a C2H2 zinc finger transcription factor (TF) with binding sites in 45% of the relevant genes. CRISPR-mediated targeting and siRNA-mediated knockdown of ZNF263 in human cell lines and primary cells revealed that ZNF263 is a transcriptional repressor, and its inactivation or silencing enhanced mRNA expression of HS3ST1 and HS3ST3A1, enzymes involved in the formation of binding sites for antithrombin and neuropilin-1 (NRP1) and glycoprotein D of herpes simplex virus, respectively. These findings uncover a well-characterized transcription regulatory factor in heparin and HS biosynthesis and provide insight into controlling this key step in heparin production.
Results
HS Biosynthetic Genes Exhibit the ZNF263 Consensus Motif.
To identify potential TFs that regulate heparin/HS biosynthesis, we analyzed the genomic regions circumscribing −400 to +100 bp around the transcription start sites of all of the biosynthetic genes involved in HS/heparin assembly. Specifically, we looked for TF-binding motifs using Hypergeometric Optimization of Motif Enrichment (HOMER). HOMER consists of an algorithm that identifies 8- to 20-bp motifs in large-scale genomics data (7). In its default settings, HOMER uses a hypergeometric model to score the enrichment of motifs in genomic sequences derived from chromatin immunoprecipitation sequencing (ChIP-seq) datasets compared with randomly selected sequences. However, here we used HOMER to analyze a set of gene promoter sequences in enzymes in a common biosynthetic pathway to determine whether there was enrichment of specific TF-binding sites. To validate this approach, we input genes involved in cholesterol biosynthesis and lysosome biogenesis, both of which are well-characterized metabolic pathways. HOMER correctly identified known master transcriptional regulators in these pathways, including SREBF2 and SOX17 in cholesterol biosynthesis (8, 9) and TFEB in lysosome biogenesis (10) (SI Appendix, Fig. S1).
Assuming that heparin/HS biosynthesis could also be controlled by master regulators, we analyzed an input list of 20 genes involved in its assembly (Fig. 1A). The enzymes responsible for the biosynthesis of the core tetrasaccharide linker region (XYLT1, XYLT2, FAM20B, PXYLP1, B4GALT7, B3GALT6, and B3GAT3) were excluded due to their involvement in both HS and chondroitin sulfate/dermatan sulfate (CS/DS) biosynthesis. Intriguingly, a motif associated with ZNF263 gained the greatest statistical significance, with predicted binding sites present on 45% of the input genes. Positive genes encoded enzymes involved in the initiation and elongation of HS (EXT1, EXTL2, and EXTL3), glucosamine N-sulfation (NDST2), uronic acid 2-O-sulfation (HS2ST1) and epimerization (GLCE), and 3-O-sulfation of glucosamine residues (HS3ST1, HS3ST3A1, and HS3ST6) (Fig. 1 B and C). Genes encoding the core proteins on which heparin and HS are assembled (HS proteoglycans), CS/DS biosynthesis, CS/DS proteoglycans, and nucleotide sugar transport and metabolism did not show enrichment for the ZNF263-binding motif, with the exception of PAPSS2, a synthase for the sulfate donor nucleotide 3′-phosphoadenyl-5′-phosphosulfate (Fig. 1D).
In this use of HOMER, the program found TF-binding motifs that were enriched in a list of genes provided by the user compared with a random list of genes. Specifically, HOMER searched a fixed, user-defined region in these genes, in this case 500 bp spanning the transcription start site, representing the putative regulatory region. In a complementary approach, we refined the identification of putative regulatory regions by using databases that link polymorphisms within a sequence window around a gene to its expression level (i.e., expression quantitative trait loci, cis-eQTL) (11), and leveraged additional evidence of TF and gene associations, such as coexpression and protein–protein interactions (12). Using this refined method, we also found ZNF263 to be significantly associated (P = 0.03) with the list of heparin/HS biosynthesis genes (Fig. 2A).
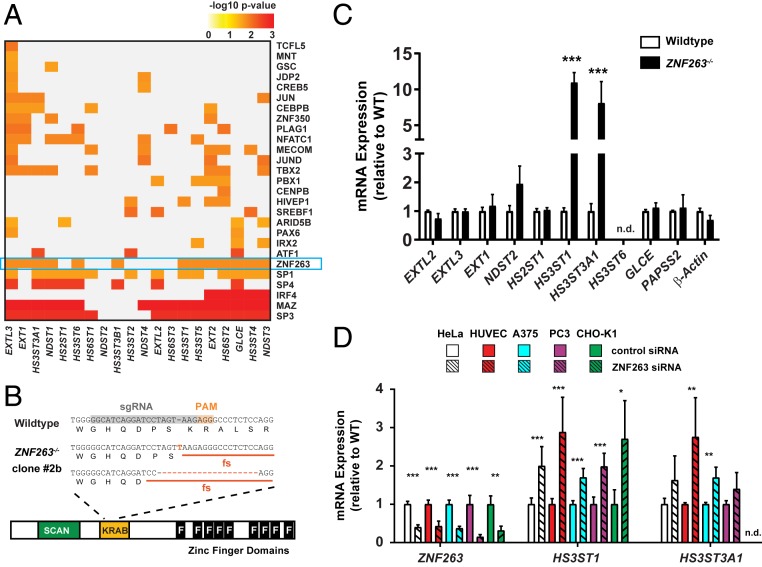
Fig. 2.
ZNF263 represses HS3ST1 and HS3ST3A1 expression. (A) TF-binding motifs (rows) significantly enriched in the set of HS biosynthesis genes (columns) based on cis-eQTL analysis. (B) CRISPR sgRNA targeting of human ZNF263 in HeLa cells. Frameshift mutations in the KRAB domain were confirmed in clone 2B and clone 5 (SI Appendix, Fig. S2A). (C) Fold change in mRNA expression of ZNF263 target genes in ZNF263−/− cells relative to wild-type cells. Only genes containing a ZNF263-binding motif are shown (Fig. 1C) (n = 4; t test after log2 transformation; Sidak–Holm correction for multiple tests). (D) mRNA expression of HS3ST1 and HS3ST3A1 in various primary, cancer, and CHO cell lines was measured after siRNA-mediated knockdown of ZNF263 mRNA (n = 4; t test after log2 transformation; Sidak–Holm correction for multiple tests). n.d., not detected.
ZNF263 Negatively Regulates the Expression of HS3ST1, the Key 3-O Sulfotransferase for Generating the Antithrombin-Binding Site on HS.
To uncover the potential regulatory role of ZNF263 in heparin/HS biosynthesis, we targeted an early constitutive exon of ZNF263 in HeLa cells using CRISPR/Cas9. Sequencing showed that clone 2B (ZNF263−/−) contained a single nucleotide insertion and a 19-bp deletion in the two alleles, respectively, resulting in loss-of-function alleles (Fig. 2B). HeLa cells were chosen because of ongoing studies of HS in this cell line and the compatibility of commercial gRNA libraries at the time that these studies were initiated. We then compared mRNA expression by qPCR of all HS input genes with predicted ZNF263-binding sites in wild-type and knockout cells (SI Appendix, Table S1). Interestingly, we found an ∼10-fold increase in HS3ST1 expression among the panel of HS target genes relative to expression in wild-type cells (Fig. 2C).
Previous studies have identified a ZNF263-binding site in the promoter of HS3ST1 based on ChIP-seq experiments (ENCODE Project Consortium). To confirm its functionality, we constructed a gene expression cassette with the HS3ST1 promoter controlling a luciferase reporter gene and found increased luciferase expression in the absence of ZNF263, corroborating negative regulation of HS3ST1 by ZNF263 through direct binding to its promoter (SI Appendix, Fig. S2A).
HS3ST1 encodes a sulfotransferase that catalyzes the addition of a 3-O-sulfate group on a specific glucosamine unit in heparin and HS that generates the antithrombin-binding site (Fig. 1A), and thus controls its anticoagulant activity (13, 14). We also found increased expression of HS3ST3A1, another 3-O sulfotransferase (Fig. 2C). Other putative ZNF263 targets in heparin/HS assembly identified by HOMER showed minimal changes in mRNA expression in HeLa ZNF263−/− cells relative to wild-type cells (Fig. 2C).
To corroborate these findings, we identified a second clone bearing different mutant alleles (SI Appendix, Fig. S2B). This strain also showed stimulation of HS3ST1 and HS3ST3A1 (SI Appendix, Fig. S2C). We also silenced expression of ZNF263 in a variety of other mammalian cell lines using an siRNA targeting ZNF263. Silencing ZNF263 resulted in significant increases in HS3ST1 and HS3ST3A1 expression in all the lines (Fig. 2D) but resulted in no major changes in expression levels of other HS genes (SI Appendix, Fig. S2D). These findings indicate that repression of 3-O-sulfation by ZNF263 is a specific and conserved process.
LC-MS Analysis of Lyase-Resistant 3-O-Sulfated Tetrasaccharides Derived from ZNF263−/− Cells.
To determine whether augmented transcription of HS3ST1 and HS3ST3A1 translated into changes in the levels of HS 3-O-sulfation, HS was isolated from wild-type and ZNF263−/− HeLa cells using anion-exchange chromatography. Samples were subsequently depolymerized into disaccharides using heparin lyases, and commercial heparin was included as a reference. In addition to disaccharides, the lyase digestion generates a heterogeneous pool of resistant tetrasaccharides, carrying a single 3-O-sulfate group on the reducing end glucosamine unit (15, 16). This tetrasaccharide repertoire can be resolved by high-resolution LC-MS, with the limitation that quantitative values cannot be determined due to the lack of molecular standards (17, 18). Five tetrasaccharide components were detected in HeLa HS (Fig. 3 B and F) that differed in composition from the four tetrasaccharides present in commercial heparin (Fig. 3A). The presence of a 6-O-sulfated GlcNAc residue located on the nonreducing side of the 3-O-sulfated glucosamine unit (Tetra C and D) distinguishes heparin tetrasaccharides from the tetrasaccharides derived from HeLa HS, which contains N-sulfated glucosamine units in the comparable positions (Tetra B and E). Inactivation of ZNF263 in HeLa cells increased Tetra A significantly but decreased Tetra E (Fig. 3 C and D).
Fig. 3.
LC-MS analysis of HS in ZNF263−/− cells. (A and B) Extracted ion chromatograms of the lyase-resistant 3-O-sulfated tetrasaccharides found in unfractionated heparin (A) and wild-type HeLa HS (B) after digestion with heparin lyases. Resistant 3-O-sulfated tetrasaccharides are annotated. (C) Collision-induced dissociation MS/MS spectra of Tetra A. (D) 3-O-sulfated tetrasaccharide distribution based on % total peak intensity of the tetrasaccharides relative to the sum of all intensities for all resistant tetrasaccharides (n = 4; t test after arcsin transformation). (E) LC-MS quantification of disaccharides from HS in wild-type and ZNF263−/− cells (n = 3; z-test). The absolute values for the disaccharides and the different classes of disaccharides are shown in SI Appendix, Tables S2 and S3. The disaccharide structure code (48) is described in SI Appendix, Table S2. (Inset) LC-MS quantification of total HS showing no difference between wild-type and ZNF263−/− cells (n = 3; Mann–Whitney U test). (F) Assigned tetrasaccharide structures and masses (z = −2).
To determine whether any other changes in HS composition occurred, the lyase depolymerization products were subjected to aniline tagging under reductive amination conditions, which allows quantitation of recovered disaccharides (19). No major differences in the total amount of HS or the general disaccharide repertoire were observed, except for decreased levels of nonsulfated, N-acetylated disaccharides (D0A0) (Fig. 3E). Taken together, these data suggest that transcriptional regulation of HS3ST1 and HS3ST3A1 by ZNF263 induces a redistribution of 3-O-sulfation in HS.
Removal of ZNF263 Promotes Assembly of the Antithrombin-Binding Site in Cell Surface HS.
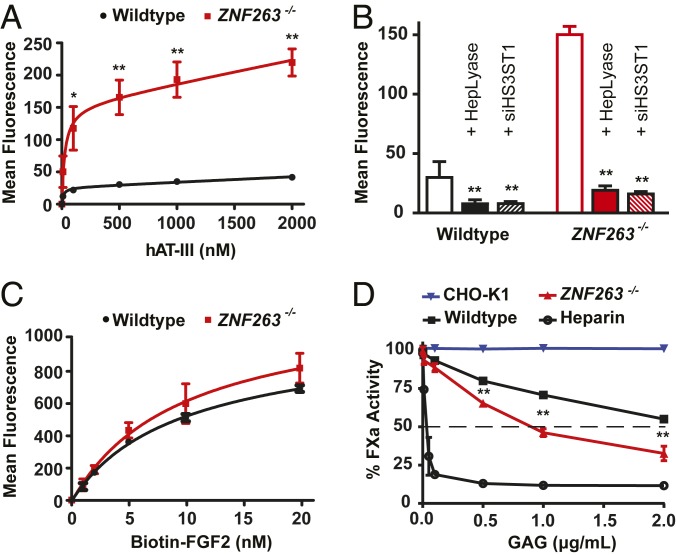
To elucidate whether the alteration of 3-O-sulfation in ZNF263−/− cells had an impact on anticoagulant activity, we first probed antithrombin binding to cell surface HS by flow cytometry. Incubation of HeLa cells with antithrombin revealed dose-dependent, saturable binding (Fig. 4A). Binding to ZNF263−/− cells was dramatically increased, with extrapolated Bmax values of 209 ± 14 for the mutant vs. 38 ± 2 for the wild type (Fig. 4A). A second ZNF263 knockout clone also showed a similar increase in antithrombin binding (SI Appendix, Fig. S2E). Treatment of the cells with heparin lyases abrogated antithrombin binding, indicating its dependence on cell surface HS (Fig. 4B). In addition, an siRNA targeting HS3ST1 yielded the same reduction in antithrombin binding as treatment with heparin lyase in both wild-type and ZNF263−/− cells, showing that the increased antithrombin binding in ZNF263−/− cells depended on 3-O-sulfation of HS mediated by HS3ST1 (Fig. 4B and SI Appendix, Fig. S2F) (20). In contrast, binding of fibroblast growth factor (FGF)-2, a protein that does not depend on 3-O-sulfation (21–23), showed no significant difference in binding to mutant and wild-type cells over a wide concentration range (Fig. 4C).
Fig. 4.
FACS binding of HS-binding proteins in ZNF263 knockout cells. (A) ZNF263−/− cells show a large increase in binding of antithrombin (n = 2; t test). (B) Antithrombin binding (500 nM) is significantly reduced after treatment of cells with heparin lyases (I to III) or transfection of an siRNA targeting HS3ST1 (n = 4; t test). (C) Binding of FGF2 is unchanged in ZNF263−/− cells (t test; n = 2). (D) In vitro inhibition of Factor Xa activity by antithrombin in association with HS isolated from wild-type, ZNF263−/− cells, and CHO cells. (n = 2; t test after normalization).
The enhanced binding of antithrombin to cell surface HS in ZNF263−/− cells caused enhanced antithrombin-dependent anti-Factor Xa activity (IC50 ∼1 µg/mL) compared with HS from wild-type cells (IC50 >3 µg/mL) (Fig. 4D). HS isolated from CHO-K1 cells, which do not express any 3-O-sulfotransferases, did not exhibit anti-Xa activity when mixed with antithrombin (Fig. 4D) (6, 24). Heparin, in contrast, had very potent activity (IC50 <0.1 µg/mL). To rule out off-target effects of gene targeting in ZNF263−/− cells, we introduced the full-length human ZNF263 cDNA back into the genome of the knockout clone using a lentiviral expression vector. Transgenic expression of ZNF263 in the mutant reduced expression of HS3ST1 (Fig. 5A) and decreased antithrombin binding (Fig. 5B).
Fig. 5.
ZNF263 rescue reduces 3-O-sulfation. (A) Introduction of the ZNF263 cDNA through a lentiviral expression cassette restored repression of HS3ST1 and HS3STA1; note the change in scale on the y-axis (n ≥ 2; t test after log2 transformation). (B and C) Reintroduction of ZNF263 decreased binding of antithrombin (n = 2; t test) (B) and binding of another 3-O-sulfate–dependent ligand, NRP1 (C) (n = 4; t test).
NRP1 binding, which is enhanced by 3-O-sulfation catalyzed by HS3ST2 (23), was also enhanced in ZNF263−/− presumably through enhanced expression of HS3ST3A1 although the effect was not as dramatic as that observed for antithrombin dependent on higher HS3ST1 expression. Transfection of the mutant with ZNF263 cDNA reduced expression of HS3ST3A1 and diminished NRP1 binding (Fig. 5C).
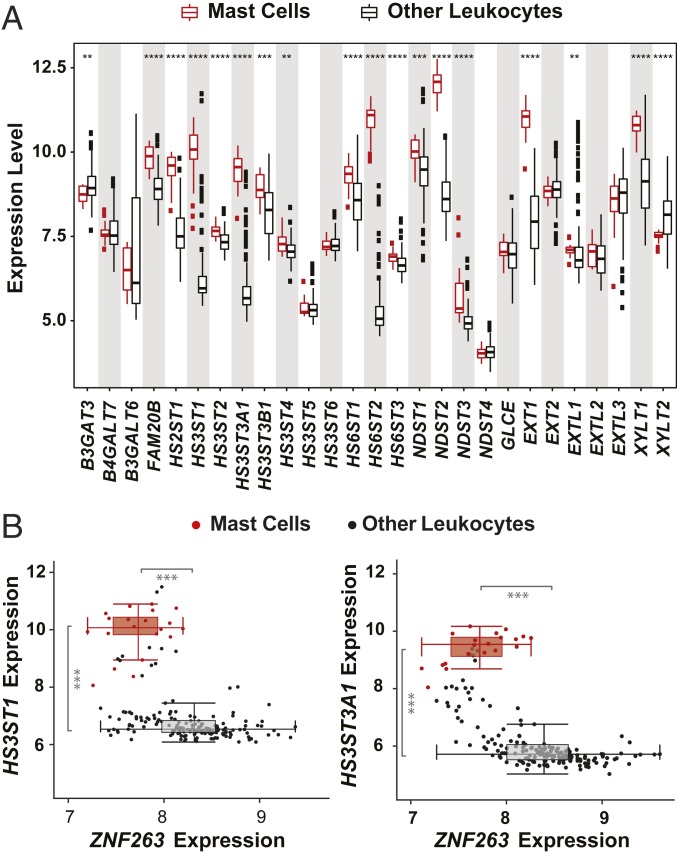
Mast Cells, Which Produce Anticoagulant Heparin, Show Low ZNF263 Expression.
To determine whether the expression of HS3ST isozymes and ZNF263 is relevant to cells that typically make heparin, we compared publicly available gene expression profiles from murine heparin-producing mast cells to other, non–heparin-producing white blood cells (25). Transcripts of several biosynthetic genes, including EXT1 (copolymerase), NDST2 (a GlcNAc N-deacetylase-N-sulfotransferase), HS6ST2 (a glucosamine 6-O-sulfotransferase), and enzymes mediating 3-O-sulfation (HS3ST1 and HS3ST3A1), were significantly elevated in mast cells (Fig. 6A). Interestingly, mast cells grouped into a distinctive cluster, largely separated from the bulk of other leukocytes, showing high expression levels of HS3ST1/HS3ST3A1 and low expression levels of ZNF263 (Fig. 6 B and C), in concordance with our findings of ZNF263 being a negative regulator of 3-O-sulfation.
Fig. 6.
Expression of HS biosynthetic genes in mast cells. (A) Expression level (log2 mRNA abundance) of a set of 27 HS biosynthetic genes in murine mast cells and other leukocytes (t test). (B) Expression levels (log2 mRNA abundance) of HS3ST1 (Left) and HS3ST3A1 (Right) vs. ZNF263 expression in mast cells (red) and remaining leukocytes (black). Data analyzed from ref. 25.
Discussion
KRAB-containing C2H2 zinc finger proteins compose the largest family of TFs in the human genome (26). The high specificity and high affinity of these TFs mainly derives from multiple C-terminal zinc finger domains that bind to specific DNA motifs in the genome (27). ZNF263 is a C2H2 zinc finger TF that contains nine finger domains, a KRAB repression domain, and a SCAN domain (Fig. 2B). Overall, little is known about the function of this protein in the human genome, and no studies have shown a direct impact of ZNF263 on cell metabolism. A previous study identified more than 5,000 ZNF263-binding sites in human cancer cells and showed that this protein could act as either a transcriptional activator or repressor (28). Other recent reports have shown differential expression of ZNF263 target genes, along with other TFs, in hepatocellular carcinoma (29), cholangiocarcinoma (30), and gastric cancer (31). ZNF263 was predicted through computational methods to regulate TGFβ1 expression in atherosclerosis (32). Here we show that ZNF263 acts as a transcriptional repressor of specific HS biosynthetic enzymes (HS3ST1 and HS3ST3A1). Our findings provide evidence of transcriptional repression of genes involved in heparin/HS biosynthesis. Repression of these enzymes in tissues might reflect the need to restrict the production of specific forms of 3-O-sulfated HS due to its potential anticoagulant activity and effects on vascular biology and the nervous system (33).
In this report, ZNF263-binding sites were identified in genes encoding enzymes responsible for elongation/termination of HS (EXT1, EXTL2, and EXTL3), uronic acid 2-O-sulfation (HS2ST1) and epimerization (GLCE), gD-type 3-O-sulfation (HS3ST3A1 and HS3ST6), and heparin biosynthesis (HS3ST1 and NDST2). Despite having binding motifs in the promoters of these genes, only two genes (HS3ST1 and HS3ST3A1) were differentially expressed in ZNF263-targeted HeLa cells. However, this finding is not that surprising, because TFs typically regulate genes in complex with other TFs and coregulators, and thus differential effects on target genes are expected to occur in a cell type-specific manner (34, 35). Knockdown of ZNF263 in other cell lines resulted in increased expression of HS3ST1 and HS3ST3A1, suggesting a conserved, autonomous role of this TF at least in the context of these 3-O-sulfotransferases.
The addition of 3-O-sulfate groups to heparin and HS is thought to be a late biosynthetic event. Notably, some members of this enzyme family exhibit unique substrate preferences; for example, HS3ST1 is known to add a sulfate group to the C3 position of distinct N-sulfated glucosamine residues if the latter are located toward the reducing side of an adjacent 6-O-sulfated GlcNAc-GlcA unit. In fact, such a modification constitutes a key feature of the high-affinity pentasaccharide- binding site of antithrombin, an essential feature of anticoagulant heparin (36, 37). Binding of antithrombin to 3-O-sulfated heparin/HS results in dramatic conformational changes in antithrombin that accelerate the inhibition of coagulation factors such as Factor Xa and thrombin. In contrast to HS3ST1, HS3ST3A1 is one of five isozymes that catalyzes 3-O-sulfation of glucosamine residues to the reducing side of an iduronic acid moiety (Fig. 1A). HS3ST3A1 creates structural motifs that can be recognized by the envelope gD of herpes simplex virus type 1, impacting both virus attachment and cell–cell fusion events (38). Similar structural motifs can support binding to NRP1, a key factor in vasculogenesis and axonal guidance (23).
A comparison of 3-O-sulfated tetrasaccharides in wild-type HeLa and ZNF263−/− cells showed that 3-O-sulfation changed in the mutant, but not necessarily in ways predicted by the structure of the classic antithrombin-binding sequence in heparin. Typically, the high-affinity binding sites contain 6-O-sulfated GlcNAc residue located next to GlcA adjacent to the 3-O-sulfated unit. However, it should be noted that heparin contains other 3-O-sulfated sequences that might constitute lower-affinity sequences, which we suggest might have anticoagulant activity when present at high concentration. It is also notable that the nature of the antithrombin-binding sequence varies according to species and the organ from which the heparin was isolated (39–41).
Heparin production from pig mucosa carries considerable safety concerns, as well as regulatory challenges (42). To our knowledge, there is only one heparin-producing mast cell line, which was derived from the Furth murine mastocytoma, but the material lacks anticoagulant activity (43). Alternative biopharmaceutical production in animal cell cultures, such as CHO cells, presents a promising and safe alternative, but current strategies overexpressing certain HS biosynthetic enzymes have proven insufficient to produce heparin-like materials, primarily due to inadequate 3-O-sulfation (44–46). Our findings of ZNF263 as a negative regulator of 3-O-sulfation provides an attractive target for genetic engineering, and thus has a high potential for bioengineering efforts of heparin.
HS isolated from ZNF263−/− HeLa cells inhibited Factor Xa activity when mixed with antithrombin in vitro; however, the activity was still much lower than that of pharmaceutical heparin. Several factors may explain this result. First, the HS produced by HeLa cells and other cell types is typically poorly sulfated compared with heparin, suggesting that other factors besides ZNF263 must drive the high sulfation of the chains. Second, therapeutic heparin is a fractionated form of heparin/HS isolated from porcine mucosa, using a process that enriches for highly sulfated chains. Third, the extent of 3-O-sulfation in HeLa cells lacking ZNF263 might be lower than that observed in transfected cells or in commercial heparin, but measuring 3-O-sulfation is difficult owing to the limited availability of only a few standards and the instability of the resistant tetrasaccharides (18).
Additional experiments are needed to examine whether other cell types might prove more amenable for engineering a highly anticoagulant form of HS through suppression of ZNF263 (45). While further work is needed to produce HS with the structural features and anticoagulant activity of pharmaceutical heparin, the identification of ZNF263 as an important regulatory factor in the pathway of heparin biosynthesis may enable the development of new strategies to achieve this ultimate goal.
Materials and Methods
The application of HOMER and eQTL analysis for binding motif prediction and statistical methods is described in SI Appendix, Materials and Methods. CRISPR/Cas9 targeting of the KRAB domain of ZNF263 is described in Fig. 2B. Rescue of the ZNF263−/− mutant was achieved by lentiviral transduction. Antithrombin binding was performed by flow cytometry with an antibody directed to antithrombin. Factor Xa activity was measured by standard protocols. qPCR probes are described in SI Appendix, Table S1. LC-MS analysis of HS was performed as described previously (19). Additional information on cell culture, purification of glycosaminoglycans, and analysis of heparinase-resistant tetrasaccharides is provided in SI Appendix, Materials and Methods.
Data Availability Statement.
Readers can access the data, associated protocols, and materials within the manuscript, the SI Appendix, or in public databases as indicated.
Supplementary Material
Acknowledgments
We thank Craig van der Kooi, University of Kentucky, for providing the B1B2 form of NRP1. We also thank the GlycoAnalytics Core Facility at University of California San Diego for help with analytical experiments. Support was provided by NIH grants (R01 GM33063, to J.D.E.; R21 CA199292, to J.D.E. and N.E.L.; R35 GM119850, to N.E.L; and R21 HD089067, to P.L.S.M.G.), the Fondation Leducq (to C.K.G and P.L.S.M.G.), a fellowship award to R.J.W. from the NIH (K12HL141956), and the Novo Nordisk Foundation Center for Biosustainability at the Technical University of Denmark (N.E.L.).
Footnotes
Competing interest statement: The University of California San Diego and J.D.E. have a financial interest in TEGA Therapeutics, Inc. The terms of this arrangement have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920880117/-/DCSupplemental.
References
- 1.Lindahl U., What else can “heparin” do? Haemostasis 29 (suppl. S1), 38–47 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Oduah E. I., Linhardt R. J., Sharfstein S. T., Heparin: Past, present, and future. Pharmaceuticals (Basel) 9, E38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu D., Esko J. D., Demystifying heparan sulfate-protein interactions. Annu. Rev. Biochem. 83, 129–157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damus P. S., Hicks M., Rosenberg R. D., Anticoagulant action of heparin. Nature 246, 355–357 (1973). [DOI] [PubMed] [Google Scholar]
- 5.Vilanova E., Tovar A. M. F., Mourão P. A. S., Imminent risk of a global shortage of heparin caused by the African Swine Fever afflicting the Chinese pig herd. J. Thromb. Haemost. 17, 254–256 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Baik J. Y., et al. , Metabolic engineering of Chinese hamster ovary cells: Towards a bioengineered heparin. Metab. Eng. 14, 81–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madison B. B., Srebp2: A master regulator of sterol and fatty acid synthesis. J. Lipid Res. 57, 333–335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rommelaere S., et al. , Sox17 regulates liver lipid metabolism and adaptation to fasting. PLoS One 9, e104925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sardiello M., et al. , A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Miles C. M., Wayne M., Quantitative trait locus (QTL) analysis. Nat. Educ. 1, 208 (2008). [Google Scholar]
- 12.Sonawane A. R., et al. , Understanding tissue-specific gene regulation. Cell Rep. 21, 1077–1088 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsberg E., et al. , Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature 400, 773–776 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Deligny A., et al. , NDST2 (N-deacetylase/N-sulfotransferase-2) enzyme regulates heparan sulfate chain length. J. Biol. Chem. 291, 18600–18607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada S., et al. , Structural studies on the tri- and tetrasaccharides isolated from porcine intestinal heparin and characterization of heparinase/heparitinases using them as substrates. Glycobiology 4, 69–78 (1994). [DOI] [PubMed] [Google Scholar]
- 16.Lawrence R., Kuberan B., Lech M., Beeler D. L., Rosenberg R. D., Mapping critical biological motifs and biosynthetic pathways of heparan sulfate. Glycobiology 14, 467–479 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Huang Y., et al. , Discovery of a heparan sulfate 3-O-sulfation specific peeling reaction. Anal. Chem. 87, 592–600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhurandhare V. M., Pagadala V., Ferreira A., Muynck L., Liu J., Synthesis of 3-O-sulfated disaccharide and tetrasaccharide standards for compositional analysis of heparan sulfate. Biochemistry, 10.1021/acs.biochem.9b00838 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence R., et al. , Evolutionary differences in glycosaminoglycan fine structure detected by quantitative glycan reductive isotope labeling. J. Biol. Chem. 283, 33674–33684 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L., et al. , The retinoic acid and cAMP-dependent up-regulation of 3-O-sulfotransferase-1 leads to a dramatic augmentation of anticoagulantly active heparan sulfate biosynthesis in F9 embryonal carcinoma cells. J. Biol. Chem. 273, 27998–28003 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Yayon A., Klagsbrun M., Esko J. D., Leder P., Ornitz D. M., Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 64, 841–848 (1991). [DOI] [PubMed] [Google Scholar]
- 22.Ye S., et al. , Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs. Biochemistry 40, 14429–14439 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Thacker B. E., et al. , Expanding the 3-O-sulfate proteome-enhanced binding of neuropilin-1 to 3-O-sulfated heparan sulfate modulates its activity. ACS Chem. Biol. 11, 971–980 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baik J. Y., et al. , Optimization of bioprocess conditions improves production of a CHO cell-derived, bioengineered heparin. Biotechnol. J. 10, 1067–1081 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dwyer D. F., Barrett N. A., Austen K. F.; Immunological Genome Project Consortium , Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat. Immunol. 17, 878–887 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tupler R., Perini G., Green M. R., Expressing the human genome. Nature 409, 832–833 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Ecco G., Imbeault M., Trono D., KRAB zinc finger proteins. Development 144, 2719–2729 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frietze S., Lan X., Jin V. X., Farnham P. J., Genomic targets of the KRAB and SCAN domain-containing zinc finger protein 263. J. Biol. Chem. 285, 1393–1403 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J., Qian Z., Li F., Li J., Lu Y., Integrative analysis of microarray data to reveal regulation patterns in the pathogenesis of hepatocellular carcinoma. Gut Liver 11, 112–120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L., Feng S., Yang Y., Identification of transcription factors (TFs) and targets involved in the cholangiocarcinoma (CCA) by integrated analysis. Cancer Gene Ther. 23, 439–445 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Xu G., Li K., Zhang N., Zhu B., Feng G., Screening driving transcription factors in the processing of gastric cancer. Gastroenterol. Res. Pract. 2016, 8431480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dhaouadi N., et al. , Computational identification of potential transcriptional regulators of TGF-ß1 in human atherosclerotic arteries. Genomics 103, 357–370 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Thacker B. E., Xu D., Lawrence R., Esko J. D., Heparan sulfate 3-O-sulfation: A rare modification in search of a function. Matrix Biol. 35, 60–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suhovskih A. V., et al. , Tissue-specificity of heparan sulfate biosynthetic machinery in cancer. Cell Adhes. Migr. 9, 452–459 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreuger J., Kjellén L., Heparan sulfate biosynthesis: Regulation and variability. J. Histochem. Cytochem. 60, 898–907 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindahl U., Bäckström G., Thunberg L., Leder I. G., Evidence for a 3-O-sulfated D-glucosamine residue in the antithrombin-binding sequence of heparin. Proc. Natl. Acad. Sci. U.S.A. 77, 6551–6555 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atha D. H., Stephens A. W., Rosenberg R. D., Evaluation of critical groups required for the binding of heparin to antithrombin. Proc. Natl. Acad. Sci. U.S.A. 81, 1030–1034 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shukla D., et al. , A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99, 13–22 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Loganathan D., Wang H. M., Mallis L. M., Linhardt R. J., Structural variation in the antithrombin III binding site region and its occurrence in heparin from different sources. Biochemistry 29, 4362–4368 (1990). [DOI] [PubMed] [Google Scholar]
- 40.Fu L., et al. , Structural characterization of pharmaceutical heparins prepared from different animal tissues. J. Pharm. Sci. 102, 1447–1457 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kouta A., et al. , Comparative pharmacological profiles of various bovine, ovine, and porcine heparins. Clin. Appl. Thromb. Hemost. 25, 10.1177/1076029619889406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lord M. S., Whitelock J. M., Bioengineered heparin: Is there a future for this form of the successful therapeutic? Bioengineered 5, 222–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montgomery R. I., et al. , Stable heparin-producing cell lines derived from the Furth murine mastocytoma. Proc. Natl. Acad. Sci. U.S.A. 89, 11327–11331 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Datta P., et al. , Bioengineered Chinese hamster ovary cells with Golgi-targeted 3-O-sulfotransferase-1 biosynthesize heparan sulfate with an antithrombin-binding site. J. Biol. Chem. 288, 37308–37318 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gasimli L., et al. , Bioengineering murine mastocytoma cells to produce anticoagulant heparin. Glycobiology 24, 272–280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhaskar U., et al. , Engineering of routes to heparin and related polysaccharides. Appl. Microbiol. Biotechnol. 93, 1–16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varki A., et al. , Symbol nomenclature for graphical representations of Glycans. Glycobiology 25, 1323–1324 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrence R., Lu H., Rosenberg R. D., Esko J. D., Zhang L., Disaccharide structure code for the easy representation of constituent oligosaccharides from glycosaminoglycans. Nat. Methods 5, 291–292 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Readers can access the data, associated protocols, and materials within the manuscript, the SI Appendix, or in public databases as indicated.