In this study, Ignatova et al. performed a systematic screen to uncover new RNA methyltransferases, and demonstrate that the methyltransferase-like 5 (METTL5) protein catalyzes m6A in 18S rRNA at position A1832. Their biochemical, molecular, and in vivo characterization of METTL5 highlights the importance of m6A in rRNA in stemness, differentiation, development, and diseases.
Keywords: m6A, methyltransferase, pluripotency
Abstract
Covalent chemical modifications of cellular RNAs directly impact all biological processes. However, our mechanistic understanding of the enzymes catalyzing these modifications, their substrates and biological functions, remains vague. Amongst RNA modifications N6-methyladenosine (m6A) is widespread and found in messenger (mRNA), ribosomal (rRNA), and noncoding RNAs. Here, we undertook a systematic screen to uncover new RNA methyltransferases. We demonstrate that the methyltransferase-like 5 (METTL5) protein catalyzes m6A in 18S rRNA at position A1832. We report that absence of Mettl5 in mouse embryonic stem cells (mESCs) results in a decrease in global translation rate, spontaneous loss of pluripotency, and compromised differentiation potential. METTL5-deficient mice are born at non-Mendelian rates and develop morphological and behavioral abnormalities. Importantly, mice lacking METTL5 recapitulate symptoms of patients with DNA variants in METTL5, thereby providing a new mouse disease model. Overall, our biochemical, molecular, and in vivo characterization highlights the importance of m6A in rRNA in stemness, differentiation, development, and diseases.
Almost all cellular RNAs are covalently modified and these modifications have emerged as a crucial layer in the regulation of RNA biogenesis and functions. Among the >170 distinct RNA modifications described, N6-methyladenosine (m6A) is currently one of the best studied. It is the most abundant internal mRNA modification, but it is also present in 18S and 28S rRNA, spliceosomal and noncoding RNAs (Roundtree et al. 2017).
m6A in RNA has been implicated in regulating RNA structure, stability, splicing, translation, localization, and interaction with distinct binding proteins (Liu et al. 2015). The precise location of m6A within transcripts can lead to different outcomes (Shi et al. 2019).
So far, several mammalian enzymes had been identified that catalyze m6A in RNAs. Among these are two members of the methyltransferase like (METTL) protein family that are characterized by the presence of a methyltransferase-like domain: METTL3, in a complex with METTL14 (Liu et al. 2014), catalyzes the formation of m6A in mRNAs, ncRNAs (Wang et al. 2014a, 2016), and pri-miRNAs (Alarcón et al. 2015), whereas METTL16 catalyzes m6A within a hairpin in the MAT2A mRNA and in spliceosomal U6 snRNA (Pendleton et al. 2017). An exception is ZCCHC4, which is a non-METTL family enzyme and has recently been identified as a 28S rRNA specific m6A methyltransferase (Ma et al. 2019). These m6A methyltransferases possess different substrate preferences and RNA recognition modes (Liu et al. 2014; Doxtader et al. 2018). Interestingly, METTL proteins, as well as ZCCHC4, have been implicated in different types of cancers (Lin et al. 2016; Barbieri et al. 2017; Deng et al. 2018).
In contrast to mRNAs, where multiple m6A sites per transcript exist, there are only two m6A sites in human rRNAs described: one in 18S rRNA at position A1832 (m6A1832) and one in 28S rRNA at position A4220a (m6A4220a) (Maden 1986, 1988). These modifications are located at functionally important regions within the ribosome (Natchiar et al. 2017) where they may tune translation and contribute to ribosome heterogeneity (Genuth and Barna 2018). Thus, they have the potential to be implicated in human diseases such as ribosomopathies that are often associated with cancer (Mills and Green 2017). In support of this, 28S rRNA m6A4220a has been linked to alterations in global translation rate, cell proliferation, and tumor growth (Ma et al. 2019). A very recent study showed that a METTL5 KO HCT116 cell line displays a loss of m6A in 18S rRNA (van Tran et al. 2019) and demonstrated that METTL5 is required for m6A formation in 18S rRNA. The authors further showed that METTL5 forms a complex with TRMT112, which was required for m6A formation in 18S rRNA in vivo. However, these studies did not demonstrate that METTL5 directly mediates methylation and the function of 18S rRNA m6A1832 remains largely unknown.
Due to the potential implications of RNA methyltransferases in human health, the characterization of novel enzymes and their function could not only uncover new mechanisms, but also new targets for the development of selective inhibitors and therapies. This prompted us to systematically search for uncharacterized methyltransferases in mammalian cells.
Here, we screened METTL proteins for RNA methyltransferase activity and show that METTL5 is an m6A-methyltransferase that methylates 18S rRNA. We demonstrate that METTL5 is required for efficient translation and has a profound impact on cell function and pluripotency. Phenotyping of Mettl5 KO mice revealed the importance of METTL5 for development.
Results
In vitro screening identifies METTL5 as an RNA methyltransferase
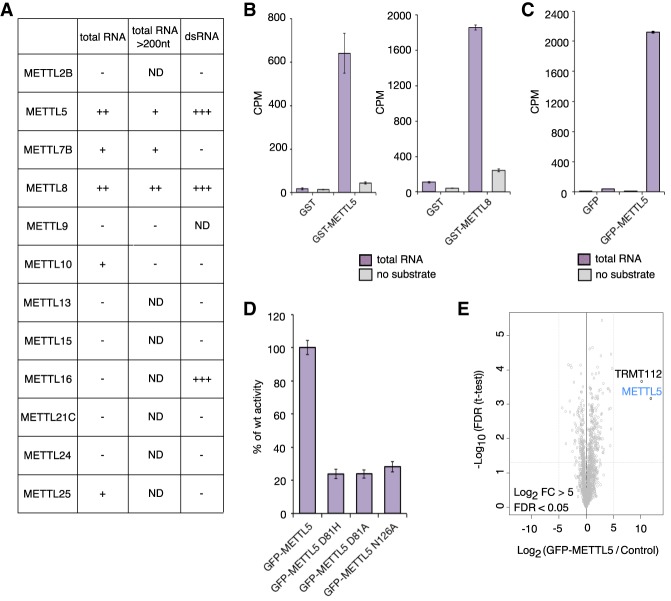
To identify new RNA methyltransferases, we cloned and expressed a collection of 12 METTL protein family members as GST fusions in E. coli and tested them in in vitro methyltransferase assays for activity. For this, we used different RNA species isolated from HeLa cells as substrates and radiolabeled S-adenosylmethionine (SAM) as methyl group donor. Figure 1A summarizes the results of this screen. By liquid scintillation counting, we detected strong and robust incorporation of methyl groups into total RNA by GST-METTL5 and GST-METTL8 (Fig. 1A,B), whereas for the other METTL proteins we observed lower or no activity (Fig. 1A). In our assays, METTL5 and METTL8 were not only active on total RNA, but also on RNAs >200 nt and double-stranded RNAs (Fig. 1A). Since METTL8 had been described recently as a new 3-methylcitidine (m3C) catalyzing enzyme (Xu et al. 2017), we focused our study on METTL5.
Figure 1.
METTL5 methylates RNA in vitro. (A) Results of screen of METTL proteins for RNA methyltransferase activity. In vitro RNA methyltransferases assays (MTA) with purified recombinant GST-METTL proteins on total RNA, RNA >200 nt, and double-stranded RNA as substrates and tritium-labeled SAM (S-adenosylmethionine) as methyl donor. RNA was purified and tritium incorporation quantified by liquid scintillation counting (LSC). Symbols were assigned based on counts per minute (CPM) of the in vitro reaction relative to the corresponding no-substrate control: (−) Activity less than threefold; (+) threefold to sevenfold; (++) sevenfold to 15-fold; (+++) >15-fold; (ND) substrate–protein pairs not tested. (B) In vitro RNA MTA with recombinant WT GST-METTL5 (left) and GST-METTL8 (right) on total HeLa RNA with tritium-labeled SAM. GST alone and reactions with no RNA substrate were used as controls. CPM for one representative experiment as averages of three technical replicates with standard deviation (SD) is shown. (C) In vitro RNA MTA with GFP-METTL5 purified from HeLa-FRT as in B. (D) In vitro RNA MTA with WT GFP-METTL5 and GFP-METTL5 D81-to-A, D81-to-H, and N126-to-A mutants as in C. Data are displayed as percentage of activity of WT enzyme calculated from an average of two biological and three technical replicates with SD. (E) TRMT122 interacts with METTL5. GFP-METTL5 was affinity-purified (in triplicates) from HeLa-FRT whole-cell extracts and enriched proteins identified by mass spectrometry. Lysates from cells expressing GFP alone were used as control. Data are visualized as a volcano plot. The log2 fold change (FC) of GFP-METTL5 to control in label-free quantification is plotted against the −log10 of the FDR calculated by a permutation-based FDR-adapted t-test as described before (Ignatova et al. 2019).
To exclude that the activity of GST-METTL5 is due to copurification of a bacterial methyltransferase we generated a HeLa-FRT cell line for inducible expression of GFP-METTL5 (van Nuland et al. 2013). We detected also a strong activity of GFP-METTL5 purified from these HeLa-FRT cells on total RNA (Fig. 1C). To further corroborate the methyltransferase activity of METTL5 we generated point mutations in the methyltransferase domain (at D81) and in the N6-adenosine-specific DNA methyltransferase signature (at N126) (Supplemental Fig. S1). These mutations abolished the activity of GFP-METTL5 (Fig. 1D). Together, these in vitro assays demonstrate for the first time that METTL5 is a bona fide RNA methyltransferase.
To identify METTL5-specific interactors, we performed label-free quantitative mass spectrometry. Due to the absence of immunoprecipitation (IP) grade antibodies, we used our GFP-METTL5 expressing cell line. We found that METTL5 has only one specific stable interaction partner (log2 fold change [FC] > 5 compared with the GFP expressing control cell line): the multifunctional methyltransferase subunit TRM112-like protein (TRMT112) (Fig. 1E; Supplemental Table S1), a recently described interactor of multiple different methyltransferases (Õunap et al. 2015) including METTL5 (van Tran et al. 2019). The presence of TRMT112 could explain why we observe higher activity of GFP-METTL5 purified from HeLa cells compared with recombinant GST-METTL5 purified from E. coli.
METTL5 catalyzes N6-methyladenosine formation in 18S rRNA at A1832
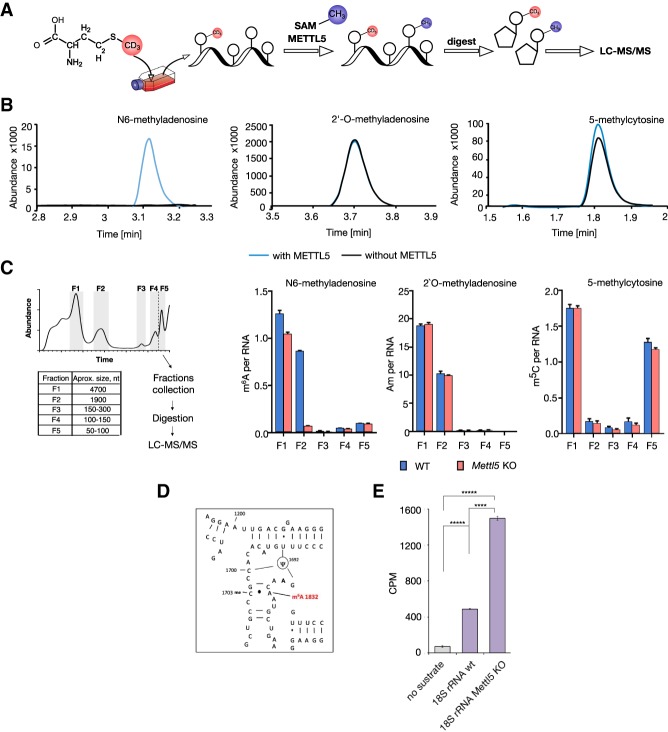
Next, we wanted to identify which type of methylation was catalyzed by METTL5. We therefore used methyl-NAIL-MS (nucleic acid isotope labeling-coupled mass spectrometry) (Fig. 2A; Supplemental Table S2; Reichle et al. 2019) as an unbiased method to identify RNA modifications. For this, we grew human WT and METTL5 KO HAP1 cells (Carette et al. 2011) in medium containing CD3-methionine for 7 d to ensure that all methyl groups in the cells contain deuterium. We then methylated the total RNA isolated from these cells in vitro with recombinant GST-METTL5 and unlabeled SAM as methyl group donor. This approach allowed us to exploit the mass difference of nucleosides methylated in cells and the nucleosides de novo methylated during the in vitro reaction. LC-MS/MS revealed a strong increase in N6-methyladenosine (m6A) abundance, but no major effects on the abundance of other modifications such as 2′-O-methyladenosine (Am) or 5-methylcytosine (m5C), suggesting that METTL5 catalyzes m6A deposition (Fig. 2B).
Figure 2.
METTL5 catalyzes m6A formation in RNA in vitro and in cells. (A) Principle of Methyl-NAIL-MS (Nucleic acid isotope labeling-coupled mass spectrometry) (Reichle et al. 2019). Cells were grown in the presence of CD3-methionine for metabolic D3-labeling of methylated nucleosides. Total RNA isolated from these cells was in vitro methylated by recombinant METTL5 with unlabeled SAM. RNA was digested and methylated nucleosides were analyzed by LC-MS/MS. (B) LC-MS/MS elution profiles of indicated modifications in in vitro methylated RNA. In vitro methylated nucleosides (blue) can be distinguished from endogenous methylations (black) by a mass shift of +3 Da. Overlaid MS/MS chromatograms of 6-methyladenosine (m6A), 2′-O-methyladenosine (Am), and 5-methylcytosine (m5C) are shown. (C, left) Scheme of RNA size exclusion chromatography followed by LC-MS/MS (Chionh et al. 2013). The collected and analyzed fractions are highlighted in gray with sizes of the predominant RNA species per fraction (F1: 28S rRNA; F2: 18S rRNA; F3: 5.8S rRNA and 5S rRNA; F4: 5S rRNA; F5: tRNAs and pri-miRNAs) in the table. (Right) Absolute quantifications of modified nucleosides per respective RNA for 6-methyladenosine (m6A), 2′-O-methyladenosine (Am), and 7-methylguanosine (m7G) (Borland et al. 2019). Average of three biological replicates and SD are plotted. See Supplemental Figure S2b for additional data. The small decrease in m6A abundance in fraction F1 could be due to a minor contamination with 18S rRNA as suggested by model from Piekna-Przybylska et al. (2008) (see also https://people.biochem.umass.edu/fournierlab/3dmodmap/humssu2dframes.php). (E) In vitro RNA MTA with recombinant GFP-METTL5 on 18S rRNA isolated from WT and Mettl5 KO mESC with tritium-labeled SAM. Tritium signal was quantified by liquid scintillation counting (LSC). CPMs for one representative experiment as averages of three technical replicates with SD are plotted. (****) P < 0.0001; (*****) P < 0.00001, calculated with Welch's test (unpaired t-test).
We then aimed to investigate the activity of METTL5 in cells. As HAP1 cells are an unstable haploid cell line that can spontaneously diploidize, we generated Mettl5 KO mESC lines (Supplemental Fig. S2a). We isolated RNA from Mettl5 KO and WT mESCs, size-fractionated (Fig. 2C, left) and analyzed abundance of RNA methylations in different factions. We observed a striking, >10-fold decrease in the amount of m6A per RNA in fraction F2 containing 18S rRNA as the major component (Fig. 2C). This effect is specific for m6A as we did not detect marked alterations in the levels of other types of methylations analyzed (Fig. 2C; Supplemental Fig. S2b), suggesting that in vivo METTL5 specifically catalyses m6A on 18S rRNA. The 18S rRNA has been shown to contain a single m6A site at position A1832 (m6A1832) with a high stoichiometry (Liu et al. 2013), located at the base of helix h44, in proximity to the decoding center of the ribosome (Fig. 2D). The drop from one molecule of m6A per RNA to <0.1 molecule of m6A per RNA suggests that loss of METTL5 strongly affects m6A at A1832, which is in line with a recent observation in human cells (van Tran et al. 2019). Of note, upon loss of METTL5, we did not observe any effect on m6A levels in the smaller RNA fractions that contain tRNAs and pri-miRNAs (Fig. 2C).
To demonstrate that METTL5 can indeed methylate 18S rRNA, we used equal amounts of 18S rRNA isolated from WT and Mettl5 KO mESCs (Supplemental Fig. S2c) as substrates in in vitro methyltransferase assays. We reasoned that since m6A1832 is strongly reduced in Mettl5 KO cells, the RNA from the KO cells would be a more efficient substrate for methylation by METTL5 in vitro. Consistent with this, we found that METTL5 displays a significantly stronger in vitro activity on 18S rRNA isolated from Mettl5 KO compared with 18S rRNA isolated from WT cells (Fig. 2E). Together, the combination of in vitro and in vivo assays establishes METTL5 as an enzyme that catalyzes m6A in 18S rRNA.
METTL5 is important for mESC pluripotency and correct differentiation
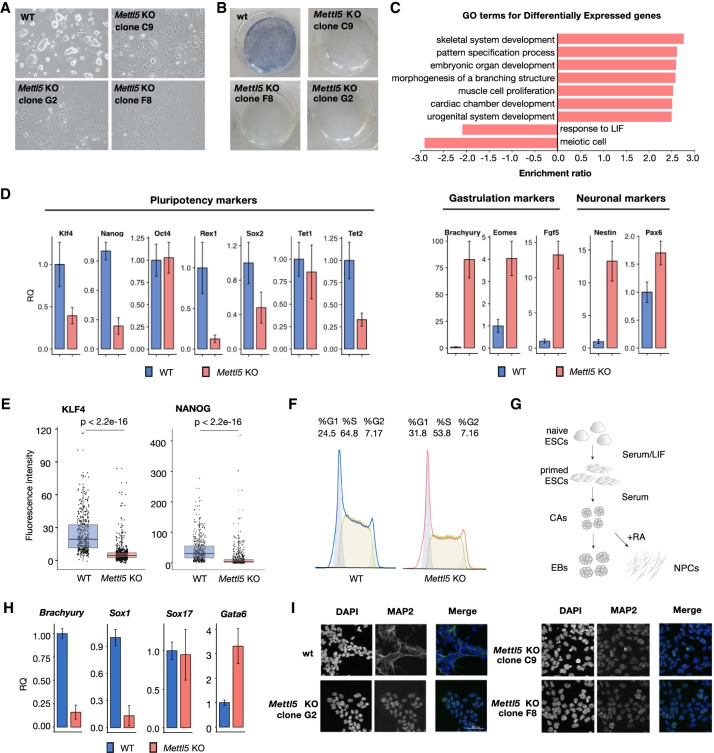
Next, we investigated whether loss of METTL5 affects mESC self-renewal and pluripotency. We observed changes in the morphology of mESC colonies grown under standard serum LIF conditions. Mettl5 KO cells displayed an increase in the number of flattened cells, as opposed to the typical dome-shaped, compact pluripotent WT mESC colonies, suggesting precocious differentiation (Fig. 3A). In line with these observations, there was a clear loss of alkaline phosphatase staining in all Mettl5 KO clones analyzed (Fig. 3B).
Figure 3.
Mettl5 KO mESCs exhibit reduced pluripotency and differentiation defects. (A) Representative bright field images of WT and Mettl5 KO mESCs (clones C9, F8, and G2) after 6 d in serum LIF medium. (B) Representative alkaline phosphatase stainings of WT and Mettl5 KO mESCs (clones C9, F8, and G2). (C) Results of gene ontology (GO) analysis of differentially expressed genes in Mettl5 KO (clones C9) compared with WT mESCs. Normalized enrichment scores for the top GO terms with BH-adjusted P-values < 0.05 are plotted. (D) RT-qPCR analysis of the expression levels of pluripotency, gastrulation and lineage marker genes in WT (blue) and Mettl5 KO mESCs (red) after 6 d in Serum LIF media. Fold changes quantified relative (RQ) to WT are plotted. Error bars indicate the standard error on the average RQ-values of three independent KO clones (C9, F8, and G2) and three replicates of WT. (E) Boxplots showing quantification of immunostainings for pluripotency markers NANOG and KLF4 for two independent Mettl5 KO clones (F8 and G2) and WT mESCs. Each dot represents the mean fluorescence intensity of an individual cell. Welch two sample t-test P-values are shown. (F) Cell cycle distributions of Mettl5 KO and WT mESCs analyzed by flow cytometry after staining with propidium iodide (PI). The experiment was performed in three biological replicates. Representative distributions of PI intensities are shown. (G) Schematic representation of embryonic body formation assay (left) and neuronal differentiation protocol (right) via cellular aggregates (CAs). For details, see the Materials and Methods. (H) RT-qPCR analysis of the expression levels of lineage marker genes in WT (blue) and Mettl5 KO mESCs (red) at day 8 of the embryonic body (EB) formation assay. Fold changes quantified relative (RQ) to WT are plotted. Error bars indicate the standard error on the average RQ values of three independent KO clones (C9, F8, and G2) and three replicates of WT. (I) Immunostaining for the neuronal marker MAP2 in three Mettl5 KO clones (C9, F8, and G2) and WT control neuronal precursor cells (NPCs). Scale bar, 50 µM. DAPI staining (left) and merge (right) are shown.
To understand the molecular basis of the mESC phenotype, we performed RNAseq analysis of WT and Mettl5 KO cells. We detected global changes at the transcriptome level upon Mettl5 KO, with 2397 up-regulated and 1947 down-regulated genes (BH-adjusted P-values < 0.01, log2 FC > |1|) (Fig. 3C; Supplemental Fig. S3a; Supplemental Table S3). Gene ontology analysis of differentially expressed genes revealed an enrichment of genes related to development, differentiation, and morphogenesis, as well as response to LIF, in agreement with the observed precocious differentiation phenotype (Fig. 3A,B). To better characterize the Mettl5 KO mESCs, we next performed quantitative RT-PCR analysis of pluripotency and differentiation markers in three independently generated Mettl5 KO clones, compared with WT mESCs. We detected a decrease in the expression of almost all pluripotency markers analyzed, including Nanog, Klf4, Sox2, and Rex1/Zfp42, and an increase in the expression of differentiation markers (Fig. 3D). We confirmed the changes of Nanog, Klf4, and Sox2 expression at the protein level by quantitative immunofluorescence (Fig. 3E) and Western blot (Supplemental Fig. S3b). Cell cycle analysis of the Mettl5 KO cells indicated a decrease in the amount of cells in S-phase (Fig. 3F), which is known to occur during mESCs differentiation (Singh and Dalton 2009). Of note, we did not detect major changes in the expression of Mettl3 in the Mettl5 KO mESCs cells (Supplemental Fig. S3c). Additionally, we observed an increase of early apoptotic cells upon Mettl5 KO (Supplemental Fig. S4a) that could contribute to the effects on self-renewal and the precocious differentiation phenotype upon METTL5 depletion.
To investigate whether Mettl5 KO affects the capacity of mESCs to differentiate into all three germ layers, we assayed embryoid body (EB) formation (Fig. 3G, left). Mettl5 KO clones seem to aggregate into smaller EBs, compared with the WT mESCs (Supplemental Fig. S4b), in line with impaired self-renewal. We analyzed the expression of the lineage markers Brachyury (mesoderm), Sox17 (endoderm), Sox1 (neuroectoderm), and Gata6 (primitive endoderm) after 8 d of EB differentiation. Mettl5 KO EBs exhibited no major change in Sox17 expression, but showed a significant reduction in the mRNA levels of Brachyury and Sox1 and an increase in Gata6 (Fig. 3H). These data suggest that Mettl5 KO mESCs have a reduced differentiation potential, with impaired ability to activate neuroectodermal and mesodermal programs. To further investigate whether METTL5 loss can affect neuronal lineage specification, we applied a directed differentiation protocol into neuronal progenitor cells (NPCs) (Fig. 3G, right; Bibel et al. 2007). WT mESCs efficiently differentiated into NPCs as indicated by the homogenous expression and filamentous localization of the neuronal marker MAP2. In contrast, all Mettl5 KO clones examined failed to properly up-regulate MAP2 expression and instead exhibited an abnormal nuclear localization of the protein (Fig. 3I). Taken together, these findings indicate a reduction in the differentiation potential of Mettl5 KO mESCs toward neuronal lineages.
Mettl5 KO cells have reduced translation rates
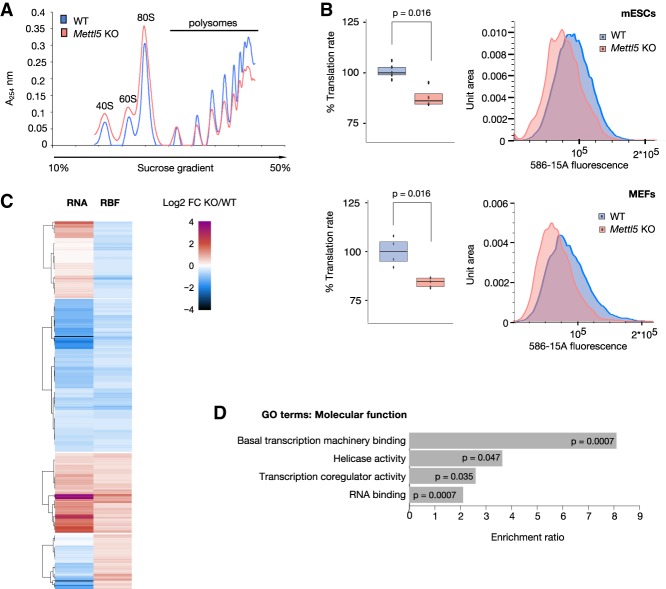
Next, we wanted to explore the molecular action of METTL5 and the consequences of loss of m6A methylation in the Mettl5 KO cells. We reasoned that the loss of an RNA modification located close to the decoding center of the ribosome, would lead to changes in translation. To address this, we first analyzed polysome profiles from WT and Mettl5 KO mESCs. Sucrose gradient profiles revealed a clear decrease in the abundance of polysomes, with the heavier polysomes being the most affected, and a concomitant increase in the 80S monosome peak in Mettl5 KO mESCs (Fig. 4A). Importantly, this overall shift in polysome abundance occurred in the absence of detectable changes in the levels or ratios of 18S and 28S rRNA (Supplemental Fig. S5a,b) and in the abundance of two ribosomal proteins (RPS2 and RPS4X) tested (Supplemental Fig. S5c). This, along with the increased levels of monosomes observed, indicates that the decrease in ribosome engagement in Mettl5 KO cells may not simply result from impaired rRNA biogenesis or ribosome amounts. To investigate whether the decrease in polysome abundance leads to reduced translation rates, we assessed global translation rates via measuring O-propargyl-puromycin (OPP) incorporation. This analysis revealed a global decrease in nascent protein synthesis rate of ∼15% in Mettl5 KO mESCs (Fig. 4B, top panel). Importantly, the global decrease in protein synthesis was not confined to ESCs, as we observed a similar decrease in OPP incorporation in Mettl5 KO MEFs, derived from Mettl5 KO mice (see below), compared with matching wild-type MEFs (Fig. 4B, bottom panel).
Figure 4.
Translation is altered in Mettl5 KO cells. (A) Polysome analysis of WT and Mettl5 KO (clone C9) mESCs. Mettl5 KO cells show increased monosome and decreased polysome profiles in sucrose gradients. The experiment was performed in three biological replicates. Data from one representative experiment is shown. (B) OPP-incorporation analysis to measure nascent translation rates in Mettl5 KO and WT mESCs (top panel) and Mettl5 KO and WT MEFs (passage 4) (bottom panel). OPP-Alexa 594 intensities of individual cells were measured by flow cytometry. Medians of OPP-Alexa 594 signal intensities from all replicates (normalized to WT) are displayed as box plots (left) and one representative OPP-Alexa 594 fluorescence intensity distribution per cell line is shown (right). Experiments were performed in two biological and at least two technical replicates. Welch two-sample t-test P-values are shown. (C) Genome-wide changes in transcript abundance and ribosome occupancy in Mettl5 KO (clone C9) and WT mESCs. Heat map displays changes in transcript abundance (RNA, left panel) or ribosome occupancy (RBF, right panel), expressed as log2 fold change (FC) relative to wild type in the range from −4 to 4, split into four groups: RBF log2 FC < −0.3 and RNA log2 FC > 0; RBF and RNA log2 FC < −0.3; RBF and RNA log2 FC > 0.3; RBF log2 FC > 0.3 and RNA log2 FC < 0. For fold changes, groups and transcript names see Supplemental Table S4. (D) Results of gene ontology (GO) by over-representation analysis of transcripts with decreased ribosomes occupancy (log2 FC < −0.3), but without reduction at the transcript level (log2 FC > 0) in Mettl5 KO compared with WT mESCs. Enrichment ratios and BH adjusted P-values are indicated.
Beyond the global decrease in overall translation, we next asked whether the translation of specific transcripts is affected. For this, we performed ribosome footprinting (Ingolia et al. 2009; Chou et al. 2017). Focusing on gene-level changes in transcript abundance and ribosome occupancy, we identified hundreds of transcripts that exhibited significant changes upon Mettl5 KO relative to wild-type mESCs (Fig. 4C; Supplemental Table S4). The majority of these transcripts exhibited changes in both mRNA abundance and ribosome occupancy and thus these changes reflect, most likely, altered mRNA abundance without changes in the translation rate. However, we identified 507 transcripts with altered ribosome occupancy without concordant changes in mRNA levels, reflecting transcript-specific changes in the ribosome loading and/or translation kinetics. Of these, 299 transcripts had decreased ribosome occupancy without reduction in mRNA abundance. Gene ontology analysis on these transcripts revealed an enrichment of genes involved in transcriptional regulation of various classes of RNAs (Fig. 4D). Altered translation of these transcripts could be implicated in the observed global effects on the transcriptome (Fig. 3C), including misregulation of pluripotency genes.
Mettl5 KO mice are subviable and have multiple developmental and behavioral phenotypes
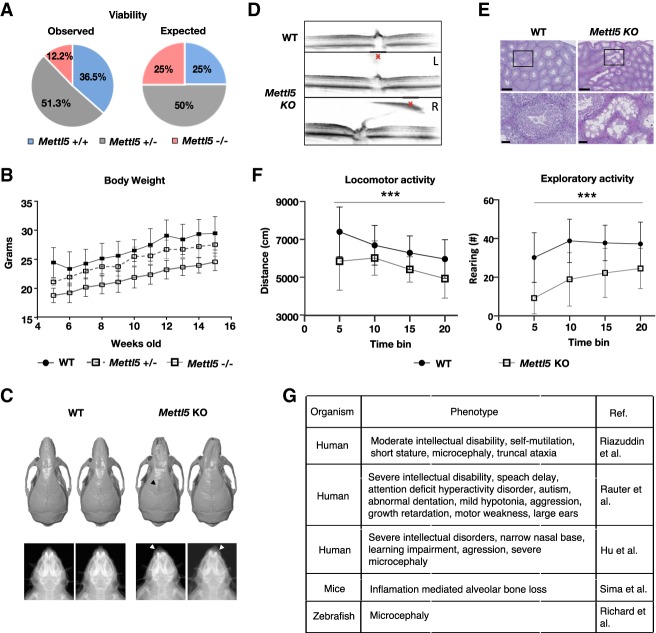
Having demonstrated effects of METTL5 loss in mESCs, we next wanted to investigate the importance of METTL5 in an animal model. Thus, we generated Mettl5 KO mice using CRISPR/Cas9 technology (Supplemental Fig. S6a) and comprehensively phenotyped them. Compared with WT control mice, we discovered several pronounced phenotypes in Mettl5 KO mice. Homozygous Mettl5 KO mice were subviable (the ratios of the genotypes differed significantly (P < 0.01) from Mendelian distribution, even if the seven mice that died between birth and weaning were considered to be all homozygous mutants) with <12.5% of born and weaned mice being knockouts (compared with 25% expected) (Fig. 5A). Between 5 and 15 wk of age, the weight of homozygote Mettl5 KO mice (but not of heterozygote) was significantly reduced (Fig. 5B). At 14 wk, microcomputed tomography and X-ray imaging revealed visible craniofacial abnormalities in about a half of the Mettl5 KO mice of both sexes. Image analyses demonstrated snout deviation due to altered nasal bone development and incomplete fusion of the frontal bone suture (Fig. 5C). At the same age, hearing thresholds were increased in Mettl5 KO mice in the auditory brainstem response testing (ABR) (Supplemental Fig. S6c). Optical coherence tomography (OCT) revealed abnormal retrolental tissue composed of melanocytes and endothelial cells, present in the vitreous body of 16-wk-old Mettl5 KO mice, likely due to alterations in the hyaloid vascular system regression. The eye phenotype appeared asymmetrically, with prevalence in the right eye (Fig. 5D). In fertility tests, none of the four females, mated with two Mettl5 KO males, showed signs of pregnancy and no litters were found at all. This observation is in agreement with the histological analysis in 16-wk-old males (Fig. 5E), where scattered degenerated seminiferous tubules of the testicles, as well as increased debris of spermatogonia in the epididymides were found with a penetrance of 50% (four out of eight Mettl5 KO males). At this age, none of seven WT control mice analyzed showed degenerative lesions. Remarkably, we also observed behavioral phenotypes. In an open field behavioral test that assesses spontaneous locomotor activity in a novel environment (Supplemental Fig. S6d), 8-wk-old Mettl5 KO mice were hypo-active and hypo-exploratory, as indicated by significantly reduced distance traveled and number of rearings (defined as the mouse standing on its hind legs to explore and to groom within the open field) (Fig. 5F).
Figure 5.
Mettl5 KO mice are subviable and display multiple phenotypic aberrations, including behavioral defects. (A) Mettl5 KO (−/−) mice are subviable; <12.5% of born and weaned mice were knockout animals. The total number of Mettl5 offspring born and weaned: 156 animals. Distribution of genotypes: 57 WT control animals (36.5%), 80 heterozygous (+/−) (51.3%), and 19 homozygous KO mutants (12.2%). From the 19 KO mice, three were females (16%) and 16 males (84%). (B) Weight monitoring of mice from the age of 5 to 15 wk showed significant differences (P = 0.001) between male Mettl5 KO (−/−) homozygous, heterozygous, and control animals (data are means ± SD, n = 23 WT+/+, n = 10 Mettl5+/−, n = 10 Mettl5−/−) (Supplemental Table S5a; Supplemental Fig. S6b for females). (C) Snout deviation was observed in 14-wk-old Mettl5 homozygous KO mice. Micro-CT (top panel) and X-ray (bottom panel) imaging analyses show nasal bones (indicated with white arrowheads in bottom panel) with abnormal growth pattern in seven out of 15 Mettl5 KO males and one out of three KO females. Also, frontal bone suture fusion was incomplete in three out of five Mettl5 KO males (black arrowhead in top panel). (D) Optical coherence tomography (OCT) images of retinas show abnormal retrolental (red X) tissue present in the eyes of 16-wk-old Mettl5 KO mice, due to alterations in hyaloid vascular system regression. This phenotype was observed in six out of eight Mettl5 KO males and three out of three KO females. The right eye was more affected (L- and R-labeled images). Normal hyaloid regression in WT mice is also displayed. (E) Hematoxylin and eosin staining of testis sections revealed scattered degeneration in the seminiferous tubules of four out of eight Mettl5 KO 16-wk-old males. Scale bars, 250 µm and 50 µm. (F) Eight-week-old Mettl5 KO mice were hypoactive (left) and hypoexploratory (right) compared with WT control mice during a 20-min open field test (Garrett et al. 2012). (***) P < 0.001 genotype effect with repeated measures ANOVA. Data are means ± SEM, males and females pooled, n = 45 WT, n = 13 Mettl5 KO (Supplemental Table S6). (G) Diseases and phenotypes described to be associated with METTL5 sequence variants in human and mice as well as in a zebrafish METTL5 morpholino model.
Altogether, these results highlight an important role of METTL5 in mouse development, in line with the impaired differentiation potential we observed in Mettl5 KO mESCs. Interestingly, the loss of the m6A methyltransferase METTL5 in mice reflects behavioral phenotypes and malformations of the skull also observed in human patients with METTL5 linked mutations (Fig. 5G; Hu et al. 2016; Reuter et al. 2017; Riazuddin et al. 2017).
Discussion
Here we have demonstrated that METTL5 is a bona fide RNA methyltransferase, which catalyzes m6A in 18S rRNA. We have shown that loss of Mettl5 impacts genomic output by affecting mRNA levels and translation. We found that KO of Mettl5 affects mESC pluripotency and differentiation. Furthermore, by generating Mettl5 KO mice, we identified the biological in vivo consequences of Mettl5 loss.
We systematically screened in in vitro assays recombinant METTL protein family members for novel RNA methyltransferase activities. Under our conditions, METTL5 and METTL8 were the most active RNA methyltransferases, with some activity for METTL7B, METTL10, and METTL25. However, for the latter three proteins we observed a significant batch-to-batch variation in enzymatic activities, indicating that further investigation is required to clarify whether they are indeed RNA methyltransferases.
Our combined in vitro and in vivo data demonstrate that METTL5 is an 18S rRNA-specific methyltransferase catalyzing m6A at A1832. This is in line with the recently reported changes in m6A abundance in 18S rRNA upon METTL5 depletion (van Tran et al. 2019). However, at this time we cannot exclude additional METTL5 targets, such as other types of RNA. For instance, it is possible that METTL5 methylates mRNAs that possess a structure and/or a motive similar to the region around A1832 of 18S RNA. Indeed, in our in vitro assays, METTL5 was able to methylate poly(A)-enriched RNA, although we detected less tritium incorporation than in total RNA (data not shown).
Modifications of rRNA can impact global ribosome structure and functions, including decoding, dwell time, peptidyl transfer, interactions with tRNAs, mRNAs, and translation factors (Liang et al. 2007; Polikanov et al. 2015). Thus, they would manifest itself as altered translation capacity, which is in line with our results of global translation defects observed upon Mettl5 deletion in mESCs. Further studies will be required to gain insights in the detailed mechanisms of how m6A1832 impacts on translation and to separate direct and indirect effects of loss of m6A1832. Since we did not detect changes in 18S rRNA levels in Mettl5 KO cells, it is unlikely that the loss of m6A from 18S rRNA has a substantial effect on the amount of the mature rRNAs themselves. Thus, our results point towards a role for m6A in 18S rRNA in translation efficiency, rather than through the regulation of rRNA or ribosome abundance.
The only described m6A site in human 18S rRNA is at A1832, located within the 3′ minor domain of the 18S rRNA, at the base of helix h44, in proximity to the decoding center (Fig. 2D), in a poorly accessible region. Thus, A1832 might only be accessible in 40S precursors and methylation of 18S rRNA by METTL5 might occur in nucleoli. This is in agreement with data from the Human Protein Atlas project, which describes METTL5 localization in the nucleoli. Because of the proximity to the mRNA it is possible that a modification of A1832 is involved in fine-tuning translation efficiency, which is in full agreement with our results.
Loss of Mettl3, an enzyme that catalyzes m6A in mRNA (Liu et al. 2014; Wang et al. 2014b), has been reported to promote self-renewal and to impair mESC exit from pluripotency, thereby reducing their capacity to differentiate into several lineages (Batista et al. 2014; Wu et al. 2016). In contrast to this, we found that the loss of Mettl5 can lead to spontaneous differentiation and reduced self-renewal of mESCs. This phenotype, together with our finding of a role for METTL5 in global translation, is in line with growing evidence suggesting that translational control, as well as a tight coordination of transcription and translation, are important for self-renewal, pluripotency, and differentiation of mESCs (Sampath et al. 2008; Tahmasebi et al. 2019). While Mettl3 and Mettl16 KOs are early embryonic lethal and lethal around implantation stage, respectively (Geula et al. 2015; Mendel et al. 2018), Mettl5 KO mice are viable, albeit with significant subviability and clear morphological and behavioral phenotypes. Of note, a recent study (van Tran et al. 2019) reported no strong phenotypes upon METTL5 KO in HCT116 human cancer cells, however, did not investigate specific effects on translation or development. The variations in the cellular phenotypes might be due to the cell type used (transformed human cancer cells vs. mouse ESCs) as well as an important role of METTL5 during mouse development.
rRNA modifications have been associated with altered translation in different diseases, such as cancer and infectious diseases (Sloan et al. 2017). DNA variants associated with METTL5 have been reported in human patients with learning impairment, intellectual disorders, motor-weakness, microcephaly, and nasal bone deformation (Hu et al. 2016; Reuter et al. 2017; Riazuddin et al. 2017). While our manuscript was under revision, a METTL5 morpholino zebrafish model was described that recapitulated the human microcephaly phenotype (Fig. 5G; Richard et al. 2019). In mice, alveolar bone loss upon Mettl5 mutations has been described (Sima et al. 2016). Our Mettl5 KO mice recapitulate these phenotypes. The partial impairment of differentiation of the mESCs in the neuroectodermal lineage could be a potential explanation behind the craniofacial and brain abnormalities in the Mettl5 KO mice. The shift in hearing sensitivity could be related to the skull abnormalities or ossicle malformations. The decreased locomotor activity of Mettl5 KO mice could represent altered novelty-induced anxiety or reflect motor dysfunction. Given that Mettl5 is expressed during brain development and also in the adult brain, it is tempting to speculate that maldevelopment and/or adult region-specific (e.g., hippocampal) synapse-dependent effects may account for the behavior abnormalities described here (Richard et al. 2019). However, understanding the full impact of Mettl5 loss on brain function will require additional investigation. Importantly, the reduced body weight, eye and craniofacial malformations, and a reduced locomotor activity we observed, together suggest that Mettl5 plays an important role during mouse development and that its loss can result in failure to thrive, as well as in morphological and behavioral abnormalities. In summary, our Mettl5 KO mice may be useful as a mouse model system in which to study the potential consequence of mutations affecting METTL5 in patients.
Overall, we demonstrate that METTL5 catalyzes m6A in 18S rRNA. Our work reveals the impact of METTL5 on translation, mESC pluripotency, and differentiation capacity. With our Mettl5 KO mice we have created a new mouse model system with human relevance.
Materials and methods
GST protein purification
BL21 Gold (DE3) E. coli cells were transformed with plasmids for recombinant protein expression (Supplemental Table S6) followed by induction with 0.5 M IPTG overnight at 18°C. Cell pellets were lysed in 25 mM Tris-HCl (pH 8), 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, and 0.2 mM PMSF with mild sonication. Lysates were centrifuged at 20,000g for 30 min at +4°C, and supernatants collected and incubated with glutathione-sepharose beads for 4 h at +4°C with rotation, followed by washes with lysis buffer (300 mM NaCl), elution with 10 mM reduced glutathione in 50 mM Tris-HCl (pH 8.0) and dialysis overnight against methyltransferase buffer assay.
In vitro methyltransferase assay (MTA)
Methyltransferase assays were performed in 6 mM HEPES-KOH (pH 8), 0.4 mM EDTA, 10 mM DTT, 80 mM KCl, 1.5 mM MgCl2, 0.2 U/mL RNasin, and 1.6% glycerol in the presence of 460 nM [3H]-SAM (1 mCi/mL; Perkin Elmer). Five micrograms of total RNA was used as a substrate. Assays were performed at +16°C overnight, followed by acid phenol-chloroform extraction. Tritium incorporation was analyzed by liquid scintillation counting (Triathler counter [HIDEX] in Ultima Gold LSC cocktail [Perkin Elmer]) as counts per minute (CPM). All data of in vitro methyltransferase assays are shown as mean ± SD (standard deviation) from three replicates. For nonradioactive assays, SAM from NEB was used as methyl group donor.
Cloning of METTL5 WT and catalytic mutants
Plasmid encoding GST-METTL5 was obtained from OriGene (EX-Z2099-B06; accession no. NM_014168) and subcloned into a HeLa-FRT vector as described before (Ignatova et al. 2019). GFP-METTL5 D81A, D81H, and N126A mutants were cloned by Q5 site-directed mutagenesis following the standard protocol from NEB and verified by Sanger sequencing. Mutagenesis primers are listed in Supplemental Table S7.
GFP-METTL5 affinity purification and mass spectrometry
GFP-METTL5 interactors in HeLa cells were identified using GFP affinity purifications from whole-cell extracts coupled to label-free mass spectrometry as described before (Ignatova et al. 2019).
Enzymatic digestion of RNA for mass spectrometry
Three-microgram to 100-ng portions of RNA were digested for 2 h at 37°C to single nucleosides with 0.2 U of alkaline phosphatase (Sigma-Aldrich), 0.02 U of phosphodiesterase I (VWR), and 0.2 U of benzonase in 5 mM Tris (pH 8) and 1 mM MgCl2 containing buffer. Tetrahydrouridine (THU; 0.5 µg; Merck), 1 µM butylated hydroxytoluene (BHT), and 0.1 µg of pentostatin were added to protect modification (Cai et al. 2015). Afterward, samples were filtered through 96-well filter plates (AcroPrep Advance 350 10K Omega, Pall Corporation) at 3000g for >10 min to remove digestive enzymes.
LC-MS/MS analysis of in vitro methylated RNA
Ribonucleosides were separated using a Luna Omega Polar, 150 × 2.1 mm, 2.5-µm particle size, 100 Å pore size from Phenomenex (Torrance), on an Agilent 1290 series HPLC system equipped with a diode array detector. Mobile phase A was 5 mM NH4OAc (≥99%, HiPerSolv Chromanorm, VWR) adjusted to pH 5.3 with glacial acetic acid (≥99%, HiPerSolv Chromanorm, VWR) and mobile phase B was pure acetonitrile (Roth, LC-MS grade, purity ≥99.95%). Gradient elution started with 98% A, increased to 10% B after 2 min, 30% B after 3 min, and 60% B after 3.5 min. Starting conditions are re-established at 4 min, followed by 2 min of equilibration. The flow rate was 0.4 mL/min and column temperature was 30°C. The effluent was directed through the DAD before entering the Agilent 6490 Triple Quadrupole mass spectrometer in dynamic multiple reaction monitoring (MRM) mode. The MS was operated in positive ion mode with the following parameters: electrospray ionization (ESI-MS, Agilent Jetstream), fragmentor voltage (set in tunefile to) 250 V, cell accelerator voltage 2 V, N2 gas temperature 150°C, N2 gas flow 15 L/min, nebulizer 30 psi, sheath gas (N2) temperature 275°C, sheath gas flow 11 l/min, capillary 2500 V, and nozzle voltage 500 V. The instrument was operated in dynamic MRM mode with the method listed in Supplemental Table S2a.
LC-MS/MS analysis of fractionated RNA
For quantification, an Agilent 1290 Infinity II equipped with a DAD combined with an Agilent Technologies G6470A Triple Quad system and electrospray ionization (ESI-MS, Agilent Jetstream) was used. Operating parameters were as follows: positive ion mode, skimmer voltage 15 V, cell accelerator voltage 5 V, N2 gas temperature 230°C and N2 gas flow 6 L/min, sheath gas (N2) temperature 400°C with a flow of 12 L/min, capillary voltage of 2500 V, nozzle voltage of 0 V, and the nebulizer at 40 psi. The instrument was operated in dynamic MRM mode with the method listed in Supplemental Table S2b. The mobile phases were A as 5 mM NH4OAc aqueous buffer, brought to pH 5.6 with glacial acetic acid and B as pure acetonitrile. A Synergi Fusion-RP column (Phenomenex, Synergi 2.5 µm Fusion-RP 100 Å, 150 × 2.0 mm) at 35°C and a flow rate of 0.35 mL/min was used. The gradient began with 100% A for 1 min, increased to 10% B by 5 min, and to 40% B by 7 min. The column was flushed with 40% B for 1 min and returned to starting conditions to 100% A by 8.5 min, followed by re-equilibration at 100% A for an additional 2.5 min.
Purification of RNA by size exclusion chromatography
For size exclusion chromatography (SEC), an Agilent 1100 HPLC system (Degasser G1279A, Quat Pump G1311A, ALS G1313A, COLCOM G1316A, VWD G1314A, and Analyt FC G1364C) with an AdvanceBio SEC-5 column, 1000 Å pore size, 5-µm particle size, and 7.8 × 300 mm (Agilent, Germany) was used. RNA was eluted with an isocratic gradient with a flow rate of 1 mL/min of 0.1 M ammonium acetate (≥98% purity). RNA was detected at 254 nm with a diode array detector. The fractions were collected and evaporated (GeneVac, EZ-2 Plus) to a volume of ∼30 µL before precipitation by a standard ammonium acetate protocol overnight at −20°C. Each RNA fraction was pelleted by centrifugation at 12,000g for 40 min at 4°C, washed once with 70% ethanol, and resuspended in 20 µL of water.
Cell culture
HAP1 WT and METTL5 KO (Horizon Discovery clones C631, HZGHC29081, and HZGHC005029c005) cells were cultured in high-glucose Dulbecco's modified Eagle medium (IMDM) supplemented with 10% FBS and 1% penicillin–streptomycin (Life Technologies, Inc.).
mESCs were maintained on 0.2% gelatin-coated dishes in Dulbecco's modified Eagle's medium (Sigma) supplemented with 16% FBS (Sigma), 0.1 mM β-mercaptoethanol (Invitrogen), 2 mM L-glutamine (Sigma), 1× MEM nonessential amino acids (Sigma), 100 U/mL penicillin, 100 μg/mL streptomycin (Sigma), homemade recombinant LIF tested for efficient self-renewal maintenance, and 2i (1 μM PD032591, 3 μM CHIR99021 [Axon Medchem, the Netherlands]).
For Methyl-NAIL-MS of in vitro reactions, cells were cultured in the RPMI1640 medium (Sigma Aldrich R7513) lacking methionine with the addition of L-methionine-methyl-D3 (98% atom) (Sigma-Aldrich), 10% FBS (Life Technologies, Inc.), and 2 mM L-glutamine (Sigma) for 5 d.
The stable GFP-METTL5 doxycycline (DOX)-inducible HeLa-FRT cell line was created by transfecting HeLa-FRT cells (van Nuland et al. 2013) with modified pcDNA5/FRT/TO and pOG44 plasmids. Cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 1% penicillin−streptomycin (Life Technologies, Inc.). At 80% confluency, cells were treated with doxycycline at a final concentration of 1 μg/mL for 16 h.
CRISPR/Cas9 gene editing in mESCs
For the generation of the Mettl5 KO mutants, Mettl5-specific gRNAs were cloned into a modified version of the SpCas9-T2A-Puromycin/gRNA vector (Addgene 62988) (Ran et al. 2013), where we fused truncated human Geminin to SpCas9 for increasing homology-directed repair efficiency. A pUC57 minivector harboring a mNeonGreen poly(A) cassette of 1068 bp and ∼300 bp of homology to the genomic locus was synthesized (GenScript). For targeting in WT J1 ESCs, cells were transfected with a 4:1 ratio of donor oligo and Cas9/gRNA construct. After 2 d, cells were subjected to puromycin selection (1 µg/mL) for 48 h and enriched by flow cytometry. Colonies were allowed to grow for 6 d, then picked into 96-well plates, and screened using restriction fragment length polymorphism analysis as described previously (Mulholland et al. 2015). Mettl5 mutation was confirmed by Sanger sequencing.
AP staining
Five-thousand serum LIF cultured cells were seeded, fixed after 5 d (4% formaldehyde), and stained with alkaline phosphatase blue membrane substrate solution (Sigma-Aldrich AB0300), according to the manufacturer's protocol.
Embryonic body formation
For embryonic body (EB) formation, mESCs were cultured for 8 d in serum LIF medium. mESCs (4 × 106) were seeded on bacteriological Petri dishes and cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% FBS (Sigma), 0.1 mM β-mercaptoethanol (Invitrogen), 2 mM L-glutamine (Sigma), 1× MEM nonessential amino acids (Sigma), 100 U/mL penicillin, and 100 μg/mL streptomycin (Sigma). Every second day, the EBs were transferred to a new Petri dish.
Neural progenitor differentiation
Neural progenitor differentiation was performed as described in Bibel et al. (2007). Briefly, mESCs were cultured in serum LIF for 5 d. Every second day, the formed cellular aggregates (CAs) were transferred to a new Petri dish. After 4 d, retinoic acid (RA) was added with a final concentration of 5 µM. After 4 d, CAs were dissociated using 0.05% trypsin and seeded at 2 × 105 per cm2 in DMEM/F-12 supplemented with 1× N-2 supplement (Gibco), 0.5× L-glutamine (Sigma), 1× Pen/Strep (Sigma), and 50 µg/mL BSA on PORN/laminin-coated plates.
Immunofluorescence
For immunostaining, mESCs were grown on coverslips as described (Bibel et al. 2007). For the quantification of the NANOG and KLF4 immunostaining, the mESCs were cultured for 1 wk in serum LIF and seeded in 5 × 106 densities on geltrex-treated (Gibco A1569601) coverslips. All of the following steps were performed at room temperature. The cells were fixed for 10 min with 4% paraformaldehyde (pH 7.0), washed three times for 10 min with PBST (PBS, 0.01% Tween-20), permeabilized for 5 min in PBS supplemented with 0.5% Triton X-100, and washed twice for 10 min with PBS. Cells were then incubated in blocking solution (PBST, 4% BSA) for 1 h. Coverslips were incubated with primary and secondary antibodies (diluted in blocking solution) in dark humid chambers for 1 h and washed three times for 10 min with PBST. For DNA counterstaining, coverslips were incubated 10 min in PBST containing a final concentration of 2 μg/mL DAPI (Sigma-Aldrich) and washed three times for 10 min with PBS-T. Coverslips were mounted in antifade medium (VectaShield, Vector Laboratories) and sealed with colorless nail polish.
The following primary antibodies were used: polyclonal rabbit anti-Klf4 (1:300; Proteintech 11880-1-AP), polyclonal rabbit anti-Nanog (1:300; Abcam ab80892), polyclonal chicken anti-MAP2 (1:500; Acris TA336617), polyclonal goat antirabbit conjugated to Alexa 488 (1:500; Thermo Fisher Scientific A-11034), polyclonal donkey antichicken conjugated to Alexa 488 (1:500; Jackson ImmunoResearch 711-547-003).
Immunofluorescence intensity analysis
For immunofluorescence, images were collected on a Nikon TiE microscope equipped with a Yokogawa CSU-W1 spinning disk confocal unit (50-μm pinhole size), an Andor Borealis illumination unit, Andor ALC600 laser beam combiner (405 nm/488 nm), Andor IXON 888 Ultra EMCCD camera, and a Nikon 100×/1.45 NA oil immersion objective. The microscope was controlled by software from Nikon (NIS Elements, ver. 5.02.00). DAPI or Alexa 488 were excited with 405-nm and 488-nm lasers. Within each experiment, cells were imaged using the same settings on the microscope to compare signal intensities between cell lines. Fiji software was used to analyze images. The region of interest of the entire nucleus was manually selected or thresholded using the DAPI signal. Fluorescence intensities of these regions of interest were measured in the 488-nm channel. The mean fluorescence intensities were extracted for each cell line.
Polysome profiling and analysis
mESCs were grown in serum LIF for 5 d. At 80% confluency, cyclohexamide was added to the medium (final concentration 100 µg/mL) and incubated for 10 min at 37°C. Cells were washed twice in PBS/cyclohexamide and harvested by scraping. Cells were pelleted at 500g for 5 min at 4°C. Pellets were resuspended in lysis buffer (10 mM Tris-HCL at pH 7.5, 5 mM MgCl2, 100 mM KCl, 1% Triton X-100, 2 mM DTT, 100 µg/mL cyclohexamide, protease inhibitor) and incubated for 10 min on ice. Lysate was spun at 1300g for 10 min at 4°C and supernatant was collected. Equal concentrations of samples were carefully pipetted on top of 10%–50% (w/v) sucrose gradients (20 mM HEPES-KOH at pH 7.4, 5 mM MgCl2, 100 mM KCL, 2 mM DTT, 100 µg/mL cyclohexamide). Gradients were centrifuged in a SW 41 Ti rotor at 35,000 rpm for 160 min at 4°C and fractions were measured in a UV/VIS absorbance detector. Experiments were performed in biological triplicates.
Total RNA sequencing according to RiboZero protocol was performed to determine RNA abundance. For ribosome footprinting we used RNase A + T1 instead of RNase 1, which has been found to be detrimental to mammalian ribosome integrity (Gerashchenko and Gladyshev 2017). Cell lysates were incubated with 60 U of RNase T1 and 30 U for RNase A per A260 absorbance unit of cell extract at room temperature, then loaded onto a 10%–50% (w/v) sucrose gradient (20 mM HEPES-KOH at pH 7.4, 5 mM MgCl2, 100 mM KCl, 2 mM DTT, 100 µg/mL cycloheximide) and spun at 35,000 rpm for 2 h, 40 min in a SW40Ti rotor. The 80S fraction was collected. Total RNA was isolated using an equal volume of Trizol followed by isopropanol precipitation. RNAs 26–32 nt in size were isolated from the total RNA by running the samples on a 15% denaturing acrylamide gel.
Single-end libraries were demultiplexed using in-line barcodes with Novobarcode (Novocraft V3.02.08), and adapters removed keeping only reads >26 nt using Fastx toolkit (V0.0.14). Reads were mapped to the transcriptome (Gencode VM20) using the splice-aware STAR-aligner (V2.5.3a), and uniquely mapped reads were extracted using samtools (V1.3). RPKM per gene (NCBI Refseq) for both ribosome footprint density and RNA abundance was counted by RSEM (V1.3). Differential expression was calculated using DESeq2 in R (V3.6). Replicates with other KO clones gave similar results.
Gene ontology (GO) by overrepresentation analysis was performed with WebGestalt (Liao et al. 2019) on transcripts (identified by > 10 reads) with decreased ribosome occupancy (log2 FC < −0.3) but without down-regulation at the transcript level (log2 FC > 0) in Mettl5 KO (clones C9) compared with WT mESCs using BH P-values < 0.05.
Flow cytometric analysis
For apoptosis and cell cycle assays, mESCs grown in serum LIF for 5 d were stained with Annexin V/propidium iodide according to the manufacturer's protocol (Annexin V apoptosis detection kit FITC, eBioscience), fixed in ethanol, and stained with PI followed by flow cytometry analysis.
For translation assays mESCs grown in Serum LIF for 8 d and primary MEFs (passage 4) were stained with Click-iT Plus OPP Alexa fluor 594 protein synthesis assay kit (Life Technologies). Cells were incubated with OPP (20 μM final concentration) for 30 min, fixed with ethanol, and processed according to the manufacturer's instructions.
Events were recorded by the FACSAriaIII sorter and analyzed with the FlowJo 10.6.1 software. Sequential electronic gating was performed to obtain single cells. Cell cycle analysis was performed in FlowJo 10.6.1, statistical analysis for translation assay was performed in R.
RNA-seq and analysis
mESCs were lysed in Trizol, total RNA was isolated by Trizol extraction followed by purification with a ZymoReseach RNA mini-prep kit, two rounds of poly(A) selection (with oligo dT beads) and RNA fragmentation to 100 nt with NEB RNA fragmentation solution. Illumina TruSeq stranded mRNA protocol was used with 0.1 µg of poly(A) RNA for the sequencing libraries preparation. Libraries were pooled and sequenced on Illumina HiSeq 4000. Reads were mapped to the complete transcriptome (mm10) and quantified using Kallisto (V0.45.0) (Bray et al. 2016). Differential expression was calculated using Sleuth (V0.30.0) (Pimentel et al. 2017) in R (V3.5.3). Gene Ontology (GO) analysis was performed with WebGestalt (Liao et al. 2019) on misregulated (Benjamin-Hochberg adjusted P-values < 0.01, log2 FC > |1|) genes using gene set enrichment analysis (GSEA) method.
RT-PCR analysis
Total RNA was isolated using the NucleoSpin RNA miniprep kit (Macherey-Nagel). Five-hundred nanograms of RNA was reverse-transcribed using high-capacity cDNA reverse transcription kit (Applied Biosystems). Quantitative PCR analysis was performed in triplicate for each sample by using 5 ng of the reverse transcription reaction in a Light Cycler 480 (Roche) with Fast SYBR Master Mix (Roche). Error bars indicate standard deviation of pooled triplicate measurements.
Western blot
Cells growing in Serum LIF for 5 d were harvested, washed with PBS, and lysed by adding five cell pellet volumes of RIPA buffer (1% NP-40, 400 mM NaCl, 50 mM Tris at pH 8.0, 1% SDS, 0.5% sodium deoxicholate). Cells were sonicated twice for 0.5 sec (10% amplitude), incubated for 30 min at 4°C on a rotation wheel, and centrifuged at 16,000g for 30 min, after which soluble whole-cell extracts (WCEs) were collected. Total protein concentration was measured with a Pierce BCA protein assay kit (Thermo Fisher Scientific). Twenty micrograms to 40 µg of whole-cell extract was used per lane.
Antibodies used for immunoblotting were as follows: anti-SOX2 (1:1000; Abcam ab92494), anti-RPS2 clone 3G6 (1:1000; Abnova H00006187-M01), and anti-RPS4X (1:1000; Abnova PAB17574) primary antibodies followed by horseradish peroxidase (HRP)-conjugated secondary antibodies goat antimouse (1:1500, Dako P0447) and antirabbit (1:100,000, Jackson Laboratory 111-035-003).
CRISPR/Cas9 gene editing in mice
The Mettl5 KO mouse model was derived using CRISPR/Cas technology and systematically characterized in the German Mouse Clinic phenotyping screen as described previously (Gailus-Durner et al. 2005; Fuchs et al. 2018). For the single-guide RNA (sgRNA) selection in vitro, two 20-bp sgRNA sequences targeting Mettl5 exon1 (gene ID 75422) were screened on the Web site http://crispor.tefor.net and synthesized by using the in vitro transcription EnGen Kit (NEB E3322S). The primers were generated with the NEB tool (http://nebiocalculator.neb.com/#!/sgrna) from Metabion. Following in vitro transcription, RNA was purified using the RNA Clean & Concentrator-25 from Zymo (R1017). Cas9 protein was purchased from IDT (1081059). The deletion allele injection mixes consisted of 50 ng/μL Cas9 mRNA, 40 ng/µL Cas9 protein, and 25 ng/µL sgRNA (each) in a final volume of 20 μL of 1× modified TE (10 mM Tris, 0.1 EDTA; RNase-free).
For microinjection C57BL/6NCrl female mice (32 d old) were injected with 5 IU/mouse of pregnant mare serum, followed 48 h later with 5 IU/mouse of human chorionic gonadotropin. The females were then mated to C57BL/6NCrl males, and fertilized oocytes were collected at 0.5 dpc. The Cryo Unit of the Institute of Experimental Genetics microinjected the sgRNA/Cas9 mixture into the pronucleus of pronuclear stage zygotes. Microinjections were performed using a microinjection needle (1 mm outer and 0.75 mm inner) with a tip diameter of 0.4–1.6 µm, an Eppendorf Femto Jet 4i to set pressure, and time to control injection volume (0.5–1 pL per embryo). Injections were performed under a 400× magnification with Hoffman modulation contrast for visualizations. Injected zygotes were cultured overnight and developed two-cell embryos were transferred into pseudopregnant (day 0.5pc) CD-1-females on the day after the injection, on average, 16 embryos per recipient female.
Mice phenotyping
For testing fertility of Mettl5 mutant mice, two homozygous Mettl5 males (9 wk old) were mated with WT females of the same age (two females per male). Breedings were kept for 2 wk and separated afterward. Female cages were inspected daily to report litters born dead or alive.
For the analysis of body weight, a linear mixed-effects model was applied with random intercept and slope as well as the fixed effects genotype, age, and the interaction of these two.
The open field (OF) analysis was carried out as described previously (Garrett et al. 2012). The arena consisted of a transparent and infrared light-permeable acrylic test arena with a smooth floor (internal measurements: 45.5 × 45.5 × 39.5 cm). Illumination levels were set at ∼150 lux in the corners and 200 lux in the middle of the test arena. Data were recorded and analyzed using the ActiMot system (TSE).
Morphological alterations of the eye posterior segment, eye fundus, and retina were analyzed with a Spectralis OCT (Heidelberg Engineering) modified with a 78-diopter double-aspheric lens (Volk Optical, Inc.) fixed directly to the outlet of the device. A contact lens with a focal length of 10 mm (Roland Consult) was applied to the eye of the mouse with a drop of methylcellulose (2% methocel; OmniVision). For measurements, anaesthetized mice were placed on a platform in front of the Spectralis OCT such that the eye was directly facing the lens of the recording unit. The parameters evaluated were number of main blood vessels (fundus), retinal thickness, and morphology of retinal layers. Images were taken as described previously (Puk et al. 2013). X-ray imaging was performed in an UltraFocus DXA system (Faxitron Bioptics, LLC) with automatic exposure control.
For the skull analysis, data sets were acquired at 36-µm³ voxel resolution using a SkyScan 1176 in vivo micro-CT system (Bruker). All scans were acquired with the following parameters: 50 kV, 45 ms exposure time, 500-µA source current, 0.7° rotation step, 0.5-mm aluminum filter. Image reconstruction was performed using InstaRecon (InstaRecon, Inc.), 3D model generation was done with CTAn (Bruker), and GOM Inspect software (GOM GmbH) was used for 3D rendering and visualization.
For pathological analyses, hematoxylin and eosin (H&E) staining was performed on formalin-fixed paraffin-embedded sections (4 µm) of testis and epididymis. The slides were analyzed by two independent pathologists.
Mice
Mice were maintained in IVC cages with water and standard mouse chow according to the directive 2010/63/EU, German laws, and GMC housing conditions. All tests were approved by the responsible authority of the district government of Upper Bavaria.
Mouse embryonic fibroblast (MEF) generation
MEFs were isolated from postcoitus day E13.5–E14.5 embryos. Each embryo was dissected into 10 mL of cold sterile PBS, voided of its internal organs, and head and blade minced. The resulted material was cultured in 25 mM DMEM high glucose without L-glutamine supplemented with 100 µg/mL puromycin, glutamax (alanine + glutamine) 200 mM, and 10% FCS. MEFs were kept in liquid nitrogen tanks.
Genotype analysis
Genomic DNA was extracted from tissue samples collected from mice during ear labeling at weaning age (3-wk-old) and PCR reaction was performed with Mettl5 specific primers (Mettl5-1 for AGAACTTCCTTATATCCAAATCGCT and Mettl5-1 rev CTGGGGAGTTTCGGGTTAGGTAGAC). After purification, PCR products were sequenced.
Data accessibility
Proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al. 2016), data set identifier PXD015582. RNA-seq data (Fig. 3C) have been deposited to GEO under accession number GSE144346. Ribosome footprinting and RNA-seq data (Fig. 4C) have been deposited to GEO under accession number GSE144313.
Supplementary Material
Acknowledgments
We thank Marat Yusupov and Lasse Bohl Jenner for helpful discussions. S.M.K. thanks Thomas Carell and his group for instrument time (LC-MS/MS) and advice. Work in R.S.’s laboratory was supported by the Deutsche Forschungsgemeinschaft (DFG) through SFB 1064 and SFB 1309 (project ID 325871075), the EpiTrio Consortium, the AmPro Program (ZT0026), and the Helmholtz Gesellschaft. The S.B. laboratory was supported by the DFG through SFB 1064. S.M.K. is grateful for funding by the DFG (SFB1309 and KE1943/3-1). The M.V. laboratory is part of the Oncode institute, partly funded by the Dutch Cancer Society (KWF). The GMC was funded by the German Federal Ministry of Education and Research (Infrafrontier grant 01KX1012) and the German Center for Diabetes Research (DZD).
Author contributions: V.V.I., S.B., S.M.K., and R.S. designed the study and analyzed the data. V.V.I. and R.S. wrote the manuscript with the feedback from all authors. V.V.I. performed and analyzed in vitro, cell culture (with help from PRL), flow cytometry, and RNAseq experiments. S.K., K.B., E.V.D.L., and S.M.K. performed LC/MS-MS experiments. T.H.G. and O.J.R. performed polysome and ribosome profiling. T.H.G., O.J.R., and V.V.I. analyzed ribosome profiling. P.S. and S.B. generated KO mESCs and studied pluripotency, differentiation, and EB formation with help from M.V. P.W.T.C.J., M.P.B., and M.V. performed and analyzed GFP-METTL5 affinity purifications. A.S.-M., Y.-L.C., O.V.A., A.A.-P., T.K.-R., J.C.-W., L.B., M.K., L.G., and S.M.H. performed mouse phenotyping tests and analyzed data. C.S. and S.M. generated the mouse line. S.M. analyzed viability data. W.W., H.F., and V.G.-D. supervised the mouse phenotyping. M.H.d.A. conceived and designed the mouse phenotyping study.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.333369.119.
References
- Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. 2015. N6-methyladenosine marks primary microRNAs for processing. Nature 519: 482–485. 10.1038/nature14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millán-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, et al. 2017. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 552: 126–131. 10.1038/nature24678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. 2014. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15: 707–719. 10.1016/j.stem.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibel M, Richter J, Lacroix E, Barde Y-A. 2007. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat Protoc 2: 1034–1043. 10.1038/nprot.2007.147 [DOI] [PubMed] [Google Scholar]
- Borland K, Diesend J, Ito-Kureha T, Heissmeyer V, Hammann C, Buck AH, Michalakis S, Kellner S. 2019. Production and application of stable isotope-labeled internal standards for RNA modification analysis. Genes (Basel) 10: 26 10.3390/genes10010026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Cai WM, Chionh YH, Hia F, Gu C, Kellner S, McBee ME, Ng CS, Pang YLJ, Prestwich EG, Lim KS, et al. 2015. A platform for discovery and quantification of modified ribonucleosides in RNA: application to stress-induced reprogramming of tRNA modifications. Methods Enzymol 560: 29–71. 10.1016/bs.mie.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, et al. 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477: 340–343. 10.1038/nature10348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chionh YH, Ho C-H, Pruksakorn D, Ramesh Babu I, Ng CS, Hia F, McBee ME, Su D, Pang YLJ, Gu C, et al. 2013. A multidimensional platform for the purification of non-coding RNA species. Nucleic Acids Res 41: e168 10.1093/nar/gkt668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HJ, Donnard E, Gustafsson HT, Garber M, Rando OJ. 2017. Transcriptome-wide analysis of roles for tRNA modifications in translational regulation. Mol Cell 68: 978–992.e4. 10.1016/j.molcel.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Su R, Weng H, Huang H, Li Z, Chen J. 2018. RNA N6-methyladenosine modification in cancers: current status and perspectives. Cell Res 28: 507–517. 10.1038/s41422-018-0034-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxtader KA, Wang P, Scarborough AM, Seo D, Conrad NK, Nam Y. 2018. Structural basis for regulation of METTL16, an S-Adenosylmethionine homeostasis factor. Mol Cell 71: 1001–1011.e4. 10.1016/j.molcel.2018.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs H, Aguilar-Pimentel JA, Amarie OV, Becker L, Calzada-Wack J, Cho Y-L, Garrett L, Hölter SM, Irmler M, Kistler M, et al. 2018. Understanding gene functions and disease mechanisms: phenotyping pipelines in the German Mouse Clinic. Behav Brain Res 352: 187–196. 10.1016/j.bbr.2017.09.048 [DOI] [PubMed] [Google Scholar]
- Gailus-Durner V, Fuchs H, Becker L, Bolle I, Brielmeier M, Calzada-Wack J, Elvert R, Ehrhardt N, Dalke C, Franz TJ, et al. 2005. Introducing the German Mouse Clinic: open access platform for standardized phenotyping. Nat Methods 2: 403–404. 10.1038/nmeth0605-403 [DOI] [PubMed] [Google Scholar]
- Garrett L, Lie DC, Hrabé de Angelis M, Wurst W, Hölter SM. 2012. Voluntary wheel running in mice increases the rate of neurogenesis without affecting anxiety-related behaviour in single tests. BMC Neurosci 13: 61 10.1186/1471-2202-13-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genuth NR, Barna M. 2018. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol Cell 71: 364–374. 10.1016/j.molcel.2018.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Gladyshev VN. 2017. Ribonuclease selection for ribosome profiling. Nucleic Acids Res 45: e6 10.1093/nar/gkw822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. 2015. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347: 1002–1006. 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- Hu H, Haas SA, Chelly J, Van Esch H, Raynaud M, de Brouwer APM, Weinert S, Froyen G, Frints SGM, Laumonnier F, et al. 2016. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry 21: 133–148. 10.1038/mp.2014.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatova VV, Jansen PWTC, Baltissen MP, Vermeulen M, Schneider R. 2019. The interactome of a family of potential methyltransferases in HeLa cells. Sci Rep 9: 6584 10.1038/s41598-019-43010-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Liu Q, Fournier MJ. 2007. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell 28: 965–977. 10.1016/j.molcel.2007.10.012 [DOI] [PubMed] [Google Scholar]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. 2019. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47: W199–W205. 10.1093/nar/gkz401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Choe J, Du P, Triboulet R, Gregory RI. 2016. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell 62: 335–345. 10.1016/j.molcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. 2013. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19: 1848–1856. 10.1261/rna.041178.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. 2014a. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10: 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. 2015. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518: 560–564. 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Wang X, Cai J, Dai Q, Natchiar SK, Lv R, Chen K, Lu Z, Chen H, Shi YG, et al. 2019. N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol 15: 88–94. 10.1038/s41589-018-0184-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden BEH. 1986. Identification of the locations of the methyl groups in 18S ribosomal RNA from Xenopus laevis and man. J Mol Biol 189: 681–699. 10.1016/0022-2836(86)90498-5 [DOI] [PubMed] [Google Scholar]
- Maden BEH. 1988. Locations of methyl groups in 28 S rRNA of Xenopus laevis and man: clustering in the conserved core of molecule. J Mol Biol 201: 289–314. 10.1016/0022-2836(88)90139-8 [DOI] [PubMed] [Google Scholar]
- Mendel M, Chen K-M, Homolka D, Gos P, Raman Pandey R, McCarthy AA, Pillai RS. 2018. Methylation of structured RNA by the m6A writer METTL16 is essential for mouse embryonic development. Mol Cell 71: 986–1000.e11. 10.1016/j.molcel.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EW, Green R. 2017. Ribosomopathies: there's strength in numbers. Science 358: eaan2755 10.1126/science.aan2755 [DOI] [PubMed] [Google Scholar]
- Mulholland CB, Smets M, Schmidtmann E, Leidescher S, Markaki Y, Hofweber M, Qin W, Manzo M, Kremmer E, Thanisch K, et al. 2015. A modular open platform for systematic functional studies under physiological conditions. Nucleic Acids Res 43: e112 10.1093/nar/gkv550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I, Klaholz BP. 2017. Visualization of chemical modifications in the human 80S ribosome structure. Nature 551: 472–477. 10.1038/nature24482 [DOI] [PubMed] [Google Scholar]
- Õunap K, Leetsi L, Matsoo M, Kurg R. 2015. The stability of ribosome biogenesis factor WBSCR22 is regulated by interaction with TRMT112 via ubiquitin-proteasome pathway. PLoS One 10: e0133841 10.1371/journal.pone.0133841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, Conrad NK. 2017. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169: 824–835.e14. 10.1016/j.cell.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna-Przybylska D, Decatur WA, Fournier MJ. 2008. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res 36: D178–D183. 10.1093/nar/gkm855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. 2017. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14: 687–690. 10.1038/nmeth.4324 [DOI] [PubMed] [Google Scholar]
- Polikanov YS, Melnikov SV, Söll D, Steitz TA. 2015. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat Struct Mol Biol 22: 342–344. 10.1038/nsmb.2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puk O, de Angelis MH, Graw J. 2013. Longitudinal fundus and retinal studies with SD-OCT: a comparison of five mouse inbred strains. Mamm Genome 24: 198–205. 10.1007/s00335-013-9457-z [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. 2013. Double nicking by RNA-guided CRISPR cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichle VF, Kaiser S, Heiss M, Hagelskamp F, Borland K, Kellner S. 2019. Surpassing limits of static RNA modification analysis with dynamic NAIL-MS. Methods 156: 91–101. 10.1016/j.ymeth.2018.10.025 [DOI] [PubMed] [Google Scholar]
- Reuter MS, Tawamie H, Buchert R, Hosny Gebril O, Froukh T, Thiel C, Uebe S, Ekici AB, Krumbiegel M, Zweier C, et al. 2017. Diagnostic yield and novel candidate genes by exome sequencing in 152 consanguineous families with neurodevelopmental disorders. JAMA Psychiatry 74: 293 10.1001/jamapsychiatry.2016.3798 [DOI] [PubMed] [Google Scholar]
- Riazuddin S, Hussain M, Razzaq A, Iqbal Z, Shahzad M, Polla DL, Song Y, van Beusekom E, Khan AA, Tomas-Roca L, et al. 2017. Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol Psychiatry 22: 1604–1614. 10.1038/mp.2016.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard EM, Polla DL, Assir MZ, Contreras M, Shahzad M, Khan AA, Razzaq A, Akram J, Tarar MN, Blanpied TA, et al. 2019. Bi-allelic variants in METTL5 cause autosomal-recessive intellectual disability and microcephaly. Am J Hum Genet 105: 869–878. 10.1016/j.ajhg.2019.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C. 2017. Dynamic RNA modifications in gene expression regulation. Cell 169: 1187–1200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, Murry CE. 2008. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2: 448–460. 10.1016/j.stem.2008.03.013 [DOI] [PubMed] [Google Scholar]
- Shi H, Wei J, He C. 2019. Molecular cell review: where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell 74: 640–650. 10.1016/j.molcel.2019.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sima C, Cheng Q, Rautava J, Levesque C, Sherman P, Glogauer M. 2016. Identification of quantitative trait loci influencing inflammation-mediated alveolar bone loss: insights into polygenic inheritance of host-biofilm disequilibria in periodontitis. J Periodontal Res 51: 237–249. 10.1111/jre.12303 [DOI] [PubMed] [Google Scholar]
- Singh AM, Dalton S. 2009. The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell 5: 141–149. 10.1016/j.stem.2009.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Warda AS, Sharma S, Entian K-D, Lafontaine DLJ, Bohnsack MT. 2017. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol 14: 1138–1152. 10.1080/15476286.2016.1259781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi S, Amiri M, Sonenberg N. 2019. Translational control in stem cells. Front Genet 9: 709 10.3389/fgene.2018.00709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nuland R, Smits AH, Pallaki P, Jansen PWTC, Vermeulen M, Timmers HTM. 2013. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol Cell Biol 33: 2067–2077. http://mcb.asm.org/cgi/doi/10.1128/MCB.01742-12 10.1128/MCB.01742-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, Bohnsack KE, Bohnsack MT, Jaffrey SR, Graille M, et al. 2019. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res 47: 7719–7733. 10.1093/nar/gkz619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44: D447–D456. 10.1093/nar/gkv1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. 2014a. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. 2014b. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 16: 191–198. 10.1038/ncb2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al. 2016. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 534: 575–578. 10.1038/nature18298 [DOI] [PubMed] [Google Scholar]
- Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, Liu Y, Byrum SD, Mackintosh SG, Zhong M, et al. 2016. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature 532: 329–333. 10.1038/nature17640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Liu X, Sheng N, Oo KS, Liang J, Chionh YH, Xu J, Ye F, Gao YG, Dedon PC, et al. 2017. Three distinct 3-methylcytidine (m3C) methyltransferases modify tRNA and mRNA in mice and humans. J Biol Chem 292: 14695–14703. 10.1074/jbc.M117.798298 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al. 2016), data set identifier PXD015582. RNA-seq data (Fig. 3C) have been deposited to GEO under accession number GSE144346. Ribosome footprinting and RNA-seq data (Fig. 4C) have been deposited to GEO under accession number GSE144313.