Here, Yang et al. sought to test whether p53 mediates the lethal consequences of autophagy deficiency using mice with conditionally deleted p53 and/or the essential autophagy gene Atg7 throughout adult mice. They show that Atg7 limits p53 activation and p53-mediated neurodegeneration, NRF2 mitigates lethal intestine degeneration upon autophagy loss, and their findings illustrate the tissue-specific roles for autophagy and functional dependencies on the p53 and NRF2 stress response mechanisms.
Keywords: autophagy, ATG7, p53, DNA damage, apoptosis, NRF2, oxidative stress
Abstract
Autophagy captures intracellular components and delivers them to lysosomes for degradation and recycling. Conditional autophagy deficiency in adult mice causes liver damage, shortens life span to 3 mo due to neurodegeneration, and is lethal upon fasting. As autophagy deficiency causes p53 induction and cell death in neurons, we sought to test whether p53 mediates the lethal consequences of autophagy deficiency. Here, we conditionally deleted Trp53 (p53 hereafter) and/or the essential autophagy gene Atg7 throughout adult mice. Compared with Atg7Δ/Δ mice, the life span of Atg7Δ/Δp53Δ/Δ mice was extended due to delayed neurodegeneration and resistance to death upon fasting. Atg7 also suppressed apoptosis induced by p53 activator Nutlin-3, suggesting that autophagy inhibited p53 activation. To test whether increased oxidative stress in Atg7Δ/Δ mice was responsible for p53 activation, Atg7 was deleted in the presence or absence of the master regulator of antioxidant defense nuclear factor erythroid 2-related factor 2 (Nrf2). Nrf2−/−Atg7Δ/Δ mice died rapidly due to small intestine damage, which was not rescued by p53 codeletion. Thus, Atg7 limits p53 activation and p53-mediated neurodegeneration. In turn, NRF2 mitigates lethal intestine degeneration upon autophagy loss. These findings illustrate the tissue-specific roles for autophagy and functional dependencies on the p53 and NRF2 stress response mechanisms.
Autophagy is the process by which cells direct their own intracellular proteins, lipids, and organelles to the lysosomal compartment for degradation (Mizushima 2010). Generally, the autophagy pathway involves the formation of double membrane-bound vesicles called autophagosomes that capture cargo such as cytoplasmic proteins, organelles, and bacteria. Autophagosomes with cargo then fuse with lysosomes to form autolysosomes where the cargo is degraded (Kaur and Debnath 2015). The breakdown products are then released into the cytoplasm where they are recycled and specifically used as substrates for central carbon metabolism to sustain survival (Rabinowitz and White 2010; Guo et al. 2016). These functions of autophagy are controlled by the autophagy-related genes (Atg) and other proteins that enable the formation of autophagosomes and recognition and capture of cargos (Mizushima and Komatsu 2011).
Autophagy maintains organelle function, prevents the accumulation of toxic cellular waste products, and sustains cell metabolism and survival during starvation (Poillet-Perez and White 2019). Autophagy is required to prevent the accumulation of damaged mitochondria, which is particularly important in the liver, muscle, and brain. In fact, the buildup of damaged mitochondria can lead to oxidative stress and perturbation of metabolism (Rabinowitz and White 2010). Autophagy is also important for removal of damaged proteins, functioning incoordination with proteasome degradation for protein quality control (Pohl and Dikic 2019). Autophagy defects lead to endoplasmic reticulum (ER) stress and accumulation of chaperone proteins due to loss of the ability to remove the unfolded protein and properly remodel the proteome in response to stress (Mathew et al. 2009, 2014). Under stress conditions such as nutrient starvation, autophagy is dramatically induced and essential for stress adaptation (Mizushima et al. 2004). Cargo-selective autophagy is also important, for example, to recycle iron from ferritin, which is critical for the iron homeostasis (Mancias et al. 2014).
Autophagy also has a critical role in mouse survival. Constitutively, Atg5- or Atg7-deficient mice are born developmentally normal but fail to survive the neonatal starvation period when the transplacental nutrient supply is interrupted but not yet restored by milk (Kuma et al. 2004; Komatsu et al. 2005). Force feeding only extends survival of autophagy-deficient newborn mice to neonatal starvation by 24 h. In contrast to newborn mice, adult mice have a greater tolerance to the loss of autophagy. Conditional whole-body ablation of the essential autophagy gene Atg7 in adult mice shortens life span to 2 to 3 mo due to susceptibility to infection and neurodegeneration (Karsli-Uzunbas et al. 2014). Autophagy also suppresses liver, brain, and muscle damage and prevents depletion of white adipose tissue (WAT). While adult mice tolerate autophagy deficiency in the short term in the fed state, fasting is lethal within 16 h due to hypoglycemia (Karsli-Uzunbas et al. 2014). These findings demonstrate that autophagy is required to maintain systemic mammalian metabolism and survival by mitigating metabolic stress during nutrient deprivation (Karsli-Uzunbas et al. 2014). Moreover, there are remarkable tissue-specific dependencies on autophagy, with brain, liver, muscle, and WAT being particularly autophagy-dependent (Karsli-Uzunbas et al. 2014).
Many major stress responses are controlled by p53, and there is mounting evidence for a functional interaction between the p53 and the autophagy pathways. p53 is a transcription factor and tumor suppressor that responds to diverse types of stresses including DNA damage, oncogene activation, oxidative stress, and hypoxia (Fischer 2017). In response to stress, p53 can induce apoptosis, senescence, and cell cycle arrest, and alter cell metabolism by regulating multiple p53 target genes (Toledo and Wahl 2006). It is generally thought that the p53 stress response can provide either protection and facilitate adaptation and recovery (e.g., cell cycle arrest) in the case of mild stress, or can eliminate cells (e.g., apoptosis) with excessive damage in the setting of high levels of stress (Kruiswijk et al. 2015). p53 thereby controls the nature of the stress response and its outcome.
p53 can also regulate autophagy. Under nutrient deprivation, a low ATP/AMP ratio activates 5′ AMP-activated protein kinase (AMPK), which then induces p53. Induction of p53 activates the transcription of genes in the AMPK pathway including tuberous sclerosis complex 2 (TSC2) and AMPK itself, and leads to the inhibition of mTOR and activation of autophagy (Feng et al. 2007). Some p53 target genes like BCL2-associated X protein (BAX) and p53-up-regulated modulator of apoptosis (PUMA) can directly activate autophagy in MEF cells (Yee et al. 2009). p53 can also directly turn on the expression of essential autophagy genes or induce autophagy via transcriptional activation of damage-regulated autophagy modulator (DRAM-1) in human and mouse cell lines (Crighton et al. 2006; Mah et al. 2012; Kenzelmann Broz et al. 2013). Autophagy deficiency can cause p53 induction in mouse models of lung, pancreatic, and breast cancer, and also in neurons, correlating with more apoptosis when p53 is intact, suggesting that autophagy may suppress p53 activation in some settings (Zhang et al. 2009; Yang et al. 2011; Guo et al. 2013; Huo et al. 2013; Rosenfeldt et al. 2013; Strohecker et al. 2013; Yang and Kimmelman 2014). As autophagy loss promotes p53 activation, and this p53 activation can be damaging, we sought to test the hypothesis that p53 was responsible for degenerative phenotypes induced by conditional autophagy loss in vivo.
To address how p53 and autophagy functionally interact in vivo and to determine the role that p53 plays in limiting the survival of mice without autophagy, we developed genetically engineered mouse models (GEMMs) to conditionally delete Atg7 and/or p53 systemically with tamoxifen (TAM). Whereas conditional, systemic Atg7 deletion (Atg7Δ/Δ) in adult mice limited their survival to 2–3 mo, codeletion of p53 and Atg7 (Atg7Δ/Δ p53Δ/Δ) remarkably extended life span to up to 6 mo and sustained survival during fasting. Atg7Δ/Δp53Δ/Δ mice showed decreased tissue damage, apoptosis, and DNA damage in the liver and brain in comparison with Atg7Δ/Δ mice. Activation of p53 by Nutlin-3 was inhibited by autophagy, which protected liver and brain from p53 hyperactivation and apoptosis, suggesting that autophagy may be a resistance mechanism to p53 activators. NRF2, in turn, is a resistance mechanism to loss of autophagy as conditional deletion of both Nrf2 and Atg7 in adult mice was synthetically lethal. Mice deficient for both Atg7 and Nrf2 (Nrf2−/−Atg7Δ/Δ) succumbed to damage to the small intestine, which was independent of p53 function. Thus, Atg7 protects against excessive p53 activation and damage in the liver and brain, whereas NRF-2 protects the intestine from damage upon loss of Atg7, demonstrating the functional interdependence and tissue specificity of stress response pathways.
Results
Loss of p53 delays neurodegeneration and prolongs survival of Atg7-deficient mice
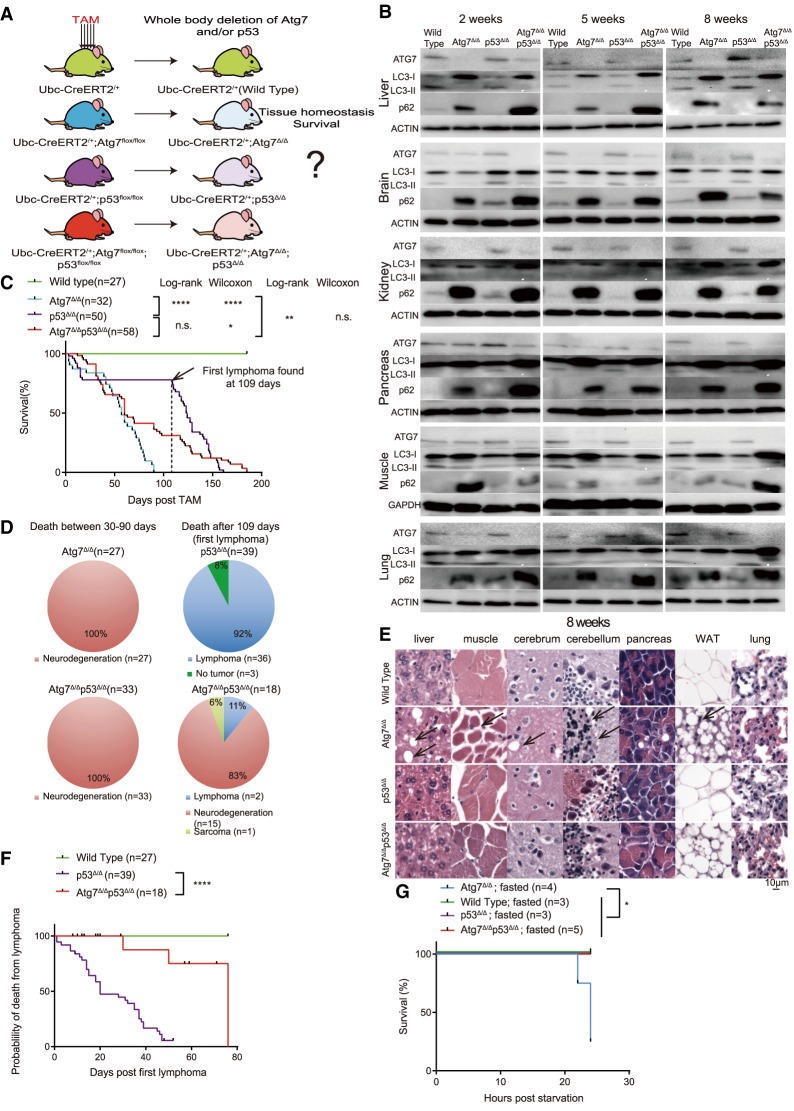
To test whether p53 plays a role in limiting the survival in mice without autophagy, adult mice were engineered with or without floxed alleles of Atg7 (Kuma et al. 2004), p53 (Marino et al. 2000), and a transgene expressing a TAM-regulated Cre recombinase under the control of ubiquitin C promoter that is ubiquitously expressed in the whole body (Ubc-CreERT2) (Ruzankina et al. 2007). Injecting TAM activates Cre throughout these mice and the floxed alleles of Atg7 and/or p53 are deleted separately or together (Fig. 1A). Mice with systemic loss of Atg7 or p53 or both in all tissues are thereby generated and gene deletion was confirmed by qRT-PCR at 2, 5, and 8 wk following the five consecutive days of TAM administration (Supplemental Fig. S1A). Loss of ATG7 protein expression was also associated with accumulation of an unprocessed form of microtubule-associated protein 1A/1B light chain 3 (LC3-I), decrease in or absence of the processed (active) form of LC3 (LC3-II), and accumulation of the autophagy substrate protein p62 in both Atg7Δ/Δ and Atg7Δ/Δp53Δ/Δ mice, indicating blockage of autophagy function (Fig. 1B). Atg7Δ/Δ mice had a life span of ∼2–3 mo, primarily due to susceptibility to infection early, and to neurodegeneration later, which is consistent with our previous findings (Fig. 1C,D; Karsli-Uzunbas et al. 2014). Similar to constitutively deficient p53−/− mice, p53Δ/Δ mice died from lymphoma, which limited life span to up to 6 mo (Fig. 1C,D; Donehower et al. 1995). In contrast to Atg7Δ/Δ mice, one-third of the Atg7Δ/Δp53Δ/Δ mice lived >3 mo and up to 6 mo after TAM, while all of the Atg7Δ/Δ mice died before 3 mo after TAM (Fig. 1C,D). Although Atg7Δ/Δ p53Δ/Δ lived longer than Atg7Δ/Δ mice, death was still predominantly from neurodegeneration (Fig. 1D). As loss of p53 did not alter survival to Atg7 deficiency early after deletion where death is due to susceptibility to infection (Karsli-Uzunbas et al. 2014), the role of p53 was specific to promoting death due to neurodegeneration (Fig. 1C,D). Therefore, p53 promotes neurodegeneration in mice deleted for Atg7.
Figure 1.
Atg7Δ/Δ, p53Δ/Δ mice have extended life span, delayed tissue damage and neurodegeneration compared with Atg7Δ/Δ mice. (A) Experimental design for generation of Atg7Δ/Δ mice, p53Δ/Δ mice, and Atg7Δ/Δ p53Δ/Δ mice. Ubc-CreERT2/+, Ubc-CreERT2/+; Atg7flox/flox mice, Ubc-CreERT2/+; p53flox/flox, Ubc-CreERT2/+; p53flox/flox; Atg7flox/flox mice were treated with TAM at 8–10 wk of age and analyzed at certain time points afterward. (B) Western blot for ATG7, p62, and LC3 at the indicated times of the indicated tissues from wild-type mice, Atg7Δ/Δ mice, p53Δ/Δ mice, and Atg7Δ/Δp53Δ/Δ mice. β-Actin was used as a loading control. (C) Kaplan-Meier survival curve of wild-type mice, Atg7Δ/Δ mice, p53Δ/Δ mice, and Atg7Δ/Δp53Δ/Δ mice. Dotted line indicates 109 d, when the first lymphoma was identified in p53Δ/Δ mice. (n.s,) Not significant; (*) P < 0.05; (**) P < 0.01; (****) P < 0.0001 (log-rank test and Gehan-Breslow-Wilcoxon test as indicated). (D) Percentage distribution for the cause of death of Atg7Δ/Δ, p53Δ/Δ, and Atg7Δ/Δp53Δ/Δ mice. The cause of death was analyzed at 30–90 d after TAM and 109–180 d after TAM. (E) Representative histology of liver, muscle, cerebrum, cerebellum, pancreas, white adipose tissue (WAT), and lung by hematoxylin and eosin stain (H&E) from wild-type, Atg7Δ/Δ, p53Δ/Δ, and Atg7Δ/Δp53Δ/Δ mice at the 8-wk time point. Black arrows indicate the damage site for these tissues. (F) Kaplan-Meier survival curve of wild-type mice, p53Δ/Δ mice, and Atg7Δ/Δp53Δ/Δ mice that died after 109 d. Black dots on the survival curve indicate the censoring times that mice died of no tumor development. (****) P < 0.0001 (log-rank test). (G) Kaplan-Meier survival curve of wild-type mice, Atg7Δ/Δ mice, p53Δ/Δ mice, and Atg7Δ/Δp53Δ/Δ mice during starvation at 10 d after TAM. (*) P < 0.05 (log-rank test). See also Supplemental Figure S1.
p53 deficiency reduces tissue damage in Atg7Δ/Δ mice
Histological examination (H&E) of tissues from wild-type, Atg7Δ/Δ, p53Δ/Δ, and Atg7Δ/Δp53Δ/Δ mice revealed no differences 2 wk after TAM (Supplemental Fig. S1B). At 5 wk after TAM, Atg7Δ/Δ mice began to show early evidence of loss of hepatocytes in the liver, pyramidal neurons in the cerebrum, Purkinje cells in the cerebellum, and depletion of lipid in WAT as reported previously (Karsli-Uzunbas et al. 2014), which was not observed in the p53Δ/Δ or Atg7Δ/Δp53Δ/Δ mice (Supplemental Fig. S1C). Two months after TAM, Atg7Δ/Δ mice showed severe loss of hepatocytes, pyramidal neurons, Purkinje cells, and WAT, as well as muscle wasting, whereas the kidneys and lungs were not affected (Fig. 1E; Supplemental Fig. S1D; Karsli-Uzunbas et al. 2014). In contrast, these tissue damage phenotypes resulting from Atg7 deficiency were not observed in wild-type, p53Δ/Δ, and Atg7Δ/Δp53Δ/Δ mice (Fig. 1E; Supplemental Fig. S1D). These results suggest that tissue damage caused by autophagy deficiency is induced by p53. Atg7Δ/Δp53Δ/Δ mice did display the same phenotype as the Atg7Δ/Δ mice at 16 wk after deletion (Supplemental Fig. S1E), indicating that loss of p53 delays but does not prevent lethal neurodegeneration caused by autophagy deficiency.
ATG7 is required for tumorigenesis driven by p53 deletion
Constitutive p53 deficiency leads to the development of lethal thymic lymphomas, which limits life span to ∼6 mo (Donehower et al. 1995). Here, conditional p53 deficiency in adult mice (p53Δ/Δ) produced the same phenotype as 36 out of 39 mice died of thymic lymphoma between 3 and 6 mo after deletion (Fig. 1C,D). Atg7Δ/Δp53Δ/Δ mice showed similar life span limitation as the p53Δ/Δ mice; however, the vast majority of the mice died from neurodegeneration without tumor development. Fifteen out of 18 Atg7Δ/Δp53Δ/Δ mice died of neurodegeneration after the first lymphoma was identified in p53Δ/Δ mice at 109 d after TAM (Fig. 1C,D). Of the three Atg7Δ/Δp53Δ/Δ mice that died from cancer, two died from thymic lymphomas and one died from a sarcoma, in which all of these tumors were deleted for Atg7 and p53 (Fig. 1D; Supplemental Fig. S1F). Analysis of the Kaplan-Meier survival curve for p53Δ/Δ and Atg7Δ/Δp53Δ/Δ mice that died after 109 d revealed that the death probability from lymphoma in Atg7Δ/Δp53Δ/Δ mice is much lower compared with p53Δ/Δ mice (P-value < 0.0001, Log-rank test) (Fig. 1F). Thus, Atg7 promotes development of lethal thymic lymphomas driven by deletion of p53, consistent with the tumor-promoting role for autophagy reported in other settings (Poillet-Perez and White 2019).
p53 deficiency prevents fasting lethality in Atg7Δ/Δ mice
While Atg7Δ/Δ mice survive in the short term, in contrast to wild-type mice, fasting is lethal within 16 h due to hypoglycemia (Karsli-Uzunbas et al. 2014). Since p53 deficiency extended the life span and attenuated tissue damage in Atg7Δ/Δ mice, we sought to test whether p53 contributes to the death of Atg7Δ/Δ mice during fasting. In contrast to Atg7Δ/Δ mice where fasting was lethal, none of the Atg7Δ/Δp53Δ/Δ mice died upon fasting, suggesting that p53 was responsible for fasting-induced death of Atg7-deficient mice (Fig. 1F).
ATG7 is required to protect the liver and brain from p53-mediated damage
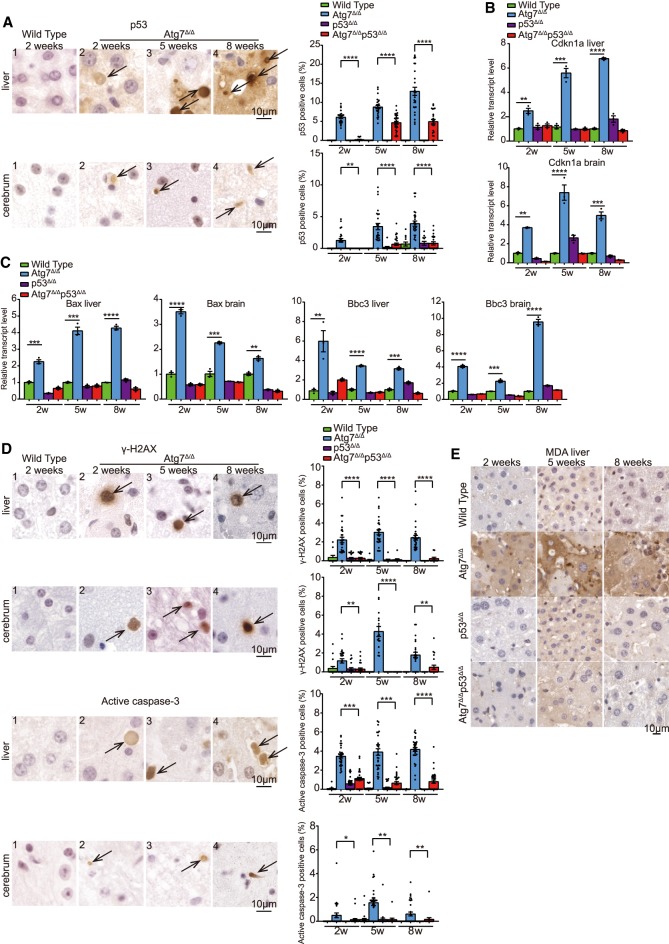
Autophagy deficiency causes p53 induction in neurons and in some cancer models and promotes cell death. Therefore, we tested whether p53 induction occurred in the whole body after Atg7 deletion. Immunohistochemistry (IHC) for p53 protein revealed that p53 accumulation was detectable at 2 wk after TAM administration, and was maintained at 5 and 8 wk after TAM in the livers and brains of Atg7Δ/Δ mice, while p53 activation was not apparent in wild-type, p53Δ/Δ, and Atg7Δ/Δp53Δ/Δ mice (Fig. 2A; Supplemental Fig. S2A,B). qRT-PCR for the p53 target genes cyclin-dependent kinase inhibitor 1A (Cdkn1a, or p21), BCL2-associated X (Bax), and BCL2-binding component 3 (Bbc3, or Puma) showed increased Cdkn1a, Bax, and Bbc3 expression in Atg7Δ/Δ mice at 2, 5, and 8 wk after TAM in the liver and brain compared with wild-type, p53Δ/Δ, and Atg7Δ/Δp53Δ/Δ mice (Fig. 2B,C). These data suggest that loss of Atg7 promotes activation of p53.
Figure 2.
Autophagy is required to protect liver and brain from p53 accumulation, DNA damage response activation, and apoptosis. (A) Representative liver and cerebrum IHC staining of p53 and quantification at the indicated times from wild-type and Atg7Δ/Δ mice. Black arrows indicate p53-positive cells. (2w) 2-wk time point; (5w) 5-wk time point; (8w) 8-wk time point. (B,C) Quantitative real-time PCR of Cdkn1a, Bax, and Bbc3 for liver and brain tissues from wild-type, Atg7Δ/Δ, p53Δ/Δ, and Atg7Δ/Δp53Δ/Δ mice at the indicated times. (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001 (unpaired t-test). (D) Representative liver and cerebrum IHC staining for γ-H2AX and active caspase-3 with quantification at the indicated times from wild-type and Atg7Δ/Δ mice. Black arrows indicate γ-H2AX or active caspase-3-positive cells. (2w) 2 wk; (5w) 5 wk; (8w) 8 wk. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 (****) P < 0.0001(unpaired t-test). (E) Representative liver IHC staining for MDA at the indicated times from wild-type, Atg7Δ/Δ, p53Δ/Δ, and Atg7Δ/Δp53Δ/Δ mice. See also Supplemental Figure S2.
Whole-body ATG7 deficiency leads to DNA damage and apoptosis in the liver and cerebrum (Karsli-Uzunbas et al. 2014) and p53 is known to be activated by different stress signals including DNA damage and oxidative stress, and triggers cell cycle arrest and apoptosis (Fischer 2017). Therefore, we hypothesized that p53 induction in Atg7Δ/Δ mice may promote apoptosis. IHC for the DNA damage response activation marker γ-H2AX revealed accumulation of γ-H2AX in Atg7Δ/Δ liver hepatocytes, neurons, and nonneuronal cells in cerebrum starting at 2 wk that was apparent through 8 wk after TAM (Fig. 2D; Supplemental Fig. S2C,D). In contrast, γ-H2AX accumulation was not detected in wild-type, p53Δ/Δ, and Atg7Δ/Δ p53Δ/Δ mice (Supplemental Fig. S2C,D). As a likely consequence of p53 activation in Atg7Δ/Δ mice, they also showed more apoptosis marked by increased active caspase-3 in liver hepatocytes, neurons, and nonneuronal cells in cerebrum in comparison with Atg7Δ/Δp53Δ/Δ mice (Fig. 2D; Supplemental Fig. S2E,F). These data indicated that p53 induction caused apoptosis in Atg7Δ/Δ mice. Atg7Δ/Δ mice also displayed increased malondialdehyde (MDA) in the liver by IHC compared with wild-type, p53Δ/Δ, and Atg7Δ/Δp53Δ/Δ mice, indicating that p53 induction was associated with increased oxidative stress in Atg7Δ/Δ mice (Fig. 2E). As the phenotype of the Atg7Δ/Δp53Δ/Δ mice was similar to the Atg7Δ/Δ mice, just delayed, this suggests that other responses similar to p53 act later when p53 is absent. It is intriguing to speculate that other p53 family members, such as p63 and p73, may be responsible for the neurodegeneration in the Atg7Δ/Δp53Δ/Δ mice.
ATG7 limits p53 activation by Nutlin-3a
Regulation of p53 activity relies on the essential p53 antagonist MDM2, which is a direct transcriptional target of p53 and is up-regulated when p53 is activated by phosphorylation at specific serine and threonine residues (Bode and Dong 2004). MDM2 binds to p53, and the ubiquitin E3 ligase of MDM2 ubiquitinylates p53, which decreases its stability by targeting it to the proteasome for degradation (Honda et al. 1997; Kubbutat et al. 1997; Matsumine et al. 1997). Under stress conditions, p53 is phosphorylated at its transactivation domain (Ser15, Ser20, and Thr18), which disrupts its binding of MDM2, and is thereby stabilized and activated (Craig et al. 1999). In this way, p53 and MDM2 form a negative feedback loop resulting from p53-dependent induction of MDM2 and MDM2-dependent suppression of p53 activity, which helps the cell to deal with stress without hyperactivation of p53 (Montes de Oca Luna et al. 1995; Dotto 2009; Marine and Lozano 2010). Nutlin-3 works as an MDM2 antagonist and up-regulates the cellular p53 level by competing for the p53-binding site on MDM2, and is being assessed clinically to promote p53 activation for cancer therapy (Vassilev et al. 2004; Drost et al. 2015; Yee-Lin et al. 2018; Forte et al. 2019). Since Atg7 limits p53 accumulation and activation, we sought to test whether Atg7 also limited the ability of Nutlin-3 to activate p53 (Khoury and Domling 2012) as a potential resistance mechanism in normal tissues.
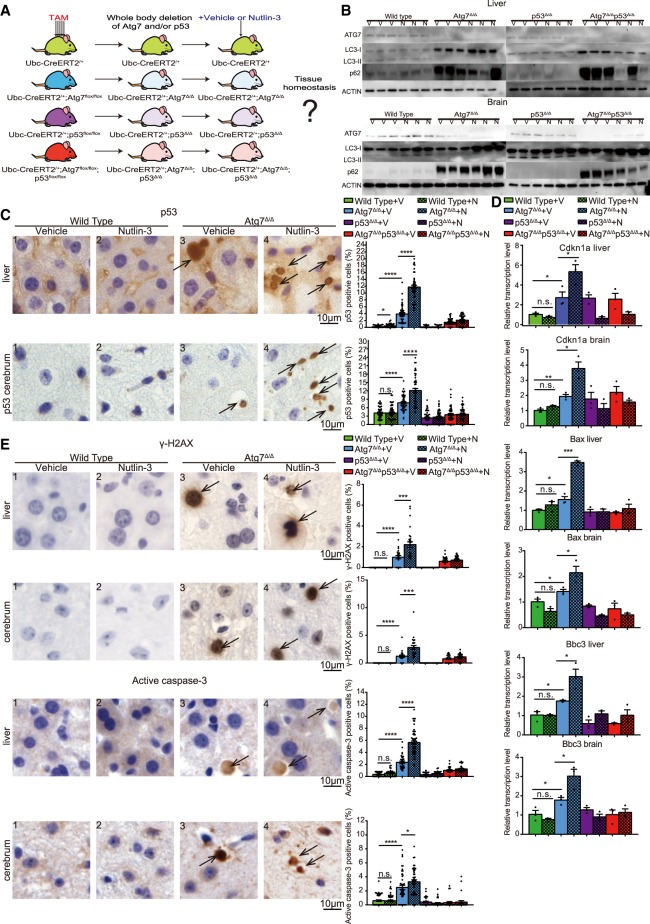
Following deletion of Atg7 and/or p53, mice were treated with either vehicle or Nutlin-3 (200 mg/kg) once per day for 1 wk (Fig. 3A). Deletion of Atg7 and p53 was confirmed by qRT-PCR (Supplemental Fig. S3A). Western blot for loss of ATG7 protein, accumulation of LC3-I and loss of LC3-II, and accumulation of p62 in the livers and brains from Atg7Δ/Δ and Atg7Δ/Δp53Δ/Δ mice indicated blockage of autophagy (Fig. 3B). As described previously, liver damage and neuron loss in Atg7Δ/Δ mice were confirmed by H&E, which was not affected by Nutlin-3 (Supplemental Fig. S3B). IHC of the livers and brains from Atg7Δ/Δ mice revealed increased p53 compared with wild-type mice, which was further increased in Nutlin-3-treated Atg7Δ/Δ mice, suggesting that Nutlin-3 induced p53 in the absence but not in the presence of autophagy. As expected, Nutlin-3 did not affect p53 levels in p53Δ/Δ mice and Atg7Δ/Δp53Δ/Δ mice (Fig. 3C; Supplemental Fig. S3C). qRT-PCR for the p53 target genes Cdkn1a, Bax, and Bbc3 showed increased Cdkn1a, Bax, and Bbc3 expression in untreated Atg7Δ/Δ mice, which was further increased in Nutlin-3-treated Atg7Δ/Δ mice in the liver and brain compared with vehicle or Nutlin-3-treated wild-type mice (Fig. 3D). This suggested that p53 was activated by Nutlin-3 only in the absence of Atg7 in normal tissues such as liver and brain. IHC of liver and brain revealed increased γ-H2AX and active caspase-3 in Atg7Δ/Δ mice compared with wild-type mice, and activation of p53 by Nutlin-3 greatly increased γ-H2AX and active caspase-3 levels. Induction of γ-H2AX and active caspase-3 were not observed in p53Δ/Δ and Atg7Δ/Δ p53Δ/Δ mice, suggesting that loss of autophagy induced apoptosis through p53 activation (Fig. 3E; Supplemental Fig. S3D,E). Therefore, autophagy is essential to protect tissues from apoptosis by limiting p53 activation.
Figure 3.
Activation of p53 by MDM2 antagonist Nutlin-3a in Atg7Δ/Δ mice leads to further increased DNA damage response and apoptosis in the liver and brain. (A) Experimental design for generation of Atg7Δ/Δ, p53Δ/Δ, and Atg7Δ/Δp53Δ/Δ mice and Nutlin-3 administration. Nutlin-3 was administered to mice by oral gavage 2 wk after TAM administration at a dosage of 200 mg/kg for 1 wk. (B) Western blot for ATG7, p62, and LC3 for liver and brain tissues from wild-type, Atg7Δ/Δ, p53Δ/Δ, and Atg7Δ/Δp53Δ/Δ mice treated with vehicle or Nutlin-3. β-Actin was used as a loading control. (V) Treated with vehicle; (N) treated with Nutlin-3. (C) Representative liver and cerebrum IHC staining for p53 and quantification from wild-type and Atg7Δ/Δ mice treated with vehicle or Nutlin-3. Black arrows indicate p53-positive cells. (V) Vehicle; (N) Nutlin-3. (*) P < 0.05; (****) P < 0.0001; (n.s.) not significant (unpaired t-test). (D) Quantitative real-time PCR of Cdkn1a, Bax, and Bbc3 for liver and brain tissues from wild-type, Atg7Δ/Δ, p53Δ/Δ, and Atg7Δ/Δp53Δ/Δ mice treated with vehicle or Nutlin-3. (V) Vehicle; (N) Nutlin-3. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (n.s.) not significant (unpaired t-test). (E) Representative liver and cerebrum IHC staining for γ-H2AX and active caspase-3 with quantification from wild-type and Atg7Δ/Δ mice treated with vehicle or Nutlin-3. Black arrows indicate γ-H2AX or active caspase-3-positive cells. (V) Vehicle; (N) Nutlin-3. (*) P < 0.05; (***) P < 0.001; (****) P < 0.0001; (n.s.) not significant (unpaired t-test).
Since p53 can induce a series of essential autophagy genes including Atg7 in MEF cells (Kenzelmann Broz et al. 2013), we hypothesized that up-regulation of p53 by Nutlin-3 can turn on essential autophagy genes and protect tissues from damage caused by p53 induction in wild-type mice. Real-time PCR on a series of autophagy essential genes indicated no significant difference in the autophagy gene mRNA levels, suggesting that the autophagy transcription program is not detectably induced by p53 at the times the tissues were collected (Supplemental Fig. S4).
Atg7 deficiency is synthetically lethal in the absence of Nrf2
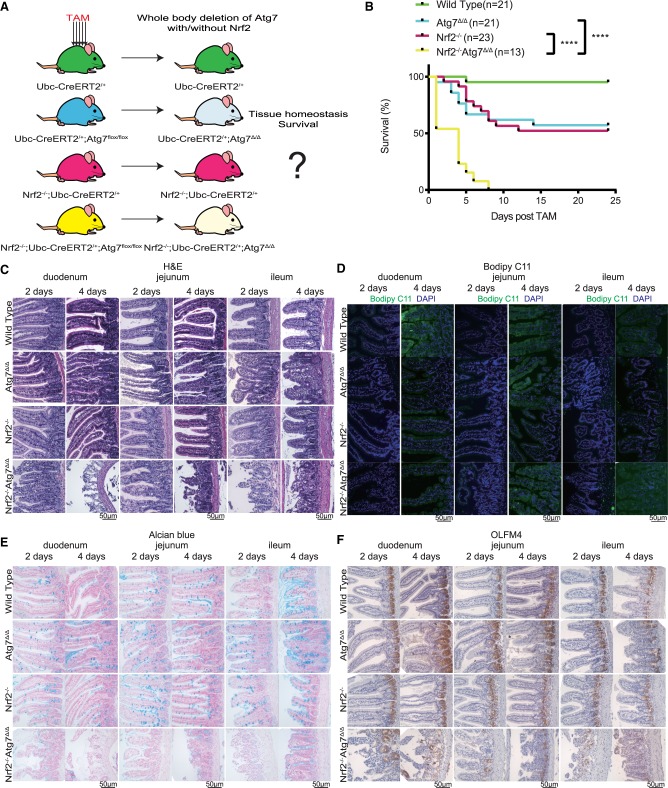
Autophagy can reduce reactive oxygen species (ROS) by removing damaged mitochondria and unfolded protein, and autophagy deficiency leads to increased ROS and accumulation of unfolded protein (Manjithaya et al. 2010; Mizushima 2010). Since we found induction of oxidative stress markers in Atg7Δ/Δ mice (Fig. 2E), we investigated whether the increased oxidative stress was responsible for tissue damage caused by p53 activation. NRF2 is the master regulator of the antioxidant defense and is ubiquitinated by an E3 ubiquitin ligase Kelch-like ECH-associated protein 1 (KEAP1) and degraded by the proteosome pathway under normal conditions (Kensler et al. 2007). With increased ROS, NRF2 is released from KEAP1 and triggers expression of a series of antioxidant genes, and NRF2 is induced by autophagy deficiency (Komatsu et al. 2010; Lau et al. 2010; Levonen et al. 2014). To examine the role of antioxidant defense in mice lacking autophagy, mice with constitutive deficiency in Nrf2 (Chan et al. 1996) were crossed with Ubc-CreERT2/+; Atg7flox/flox mice to generate Nrf2−/−; Ubc-CreERT2/+; Atg7flox/flox mice (Fig. 4A). TAM administration was then used to delete Atg7 in the presence and absence of Nrf2.
Figure 4.
Atg7 deficiency is synthetically lethal in the absence of Nrf2. (A) Experimental design for generation of Atg7Δ/Δ mice, Nrf2−/− mice, and Nrf2−/−Atg7Δ/Δ mice. (B) Kaplan-Meier survival curve of wild-type, Atg7Δ/Δ, Nrf2−/−, and Nrf2−/−Atg7Δ/Δ mice. (****) P < 0.0001(log-rank [Mantel-Cox] test). (C) Representative histology of duodenum, jejunum, and ileum by H&E at the indicated times from wild-type, Atg7Δ/Δ, Nrf2−/−, and Nrf2−/−Atg7Δ/Δ mice. (D) Representative Bodipy C11 stain of duodenum, jejunum, and ileum at the indicated times from wild-type, Atg7Δ/Δ, Nrf2−/−, and Nrf2−/−Atg7Δ/Δ mice. (E) Representative Alcian blue stain of duodenum, jejunum, and ileum at the indicated times from wild-type, Atg7Δ/Δ, Nrf2−/−, and Nrf2−/−Atg7Δ/Δ mice. (F) Representative duodenum, jejunum, and ileum IHC stain of OLFM4 at the indicated times from wild-type, Atg7Δ/Δ, Nrf2−/−, and Nrf2−/−Atg7Δ/Δ mice.
In contrast to Atg7Δ/Δ and Nrf2−/− mice, most of which survive, Nrf2−/−Atg7Δ/Δ mice had a life span of <7 d (Fig. 4B). Histological examination of tissues by H&E surprisingly showed no damage to the liver, brain, pancreas, lungs, and kidneys in Nrf2−/−Atg7Δ/Δ mice (Supplemental Fig. S5A). The only tissue with significant damage was the intestine (duodenum, jejunum, and ileum), which may be the cause of increased muscle wasting and loss of WAT (Fig. 4C; Supplemental Fig. S5B). Bodipy C11 staining of the whole small intestine was significantly increased in Nrf2−/−Atg7Δ/Δ intestine, indicating increased lipid peroxidation that results from ROS (Fig. 4D). IHC for active caspase-3 displayed increased staining in Nrf2−/−Atg7Δ/Δ intestine in both the crypt and villus, indicating increased apoptosis (Supplemental Fig. S5C–E). Thus, conditional deletion of the essential autophagy gene Atg7 in adult mice is synthetically lethal in the absence of Nrf2 due to damage to the intestine.
We then investigated which cell type in the intestine was most affected by deficiency in Atg7 in the absence of Nrf2. Alcian blue staining of paraffin sections from intestine tissues was significantly decreased in the Nrf2−/− Atg7Δ/Δ mouse intestine, suggesting loss of goblet cells (Fig. 4E). IHC for the stem cell marker OLFM4 revealed loss of OLFM4 staining in Nrf2−/− Atg7Δ/Δ but not in wild-type, Atg7Δ/Δ, and Nrf2−/− mouse intestine, suggesting loss of stem cells (Fig. 4F). IHC for the Paneth cell marker Lysozyme revealed diffuse staining in the Nrf2−/− Atg7Δ/Δ mouse intestines in comparison with wild-type, Atg7Δ/Δ, and Nrf2−/− intestines, which suggests that Paneth cell function is abnormal (Supplemental Fig. S5F; Cadwell et al. 2008). We then investigated whether this deleterious phenotype in the intestine was induced by p53 activation. p53flox/flox mice were crossed with Nrf2−/−; Ubc-CreERT2/+ mice and Nrf2−/−; Ubc-CreERT2/+; Atg7flox/flox mice to generate Nrf2−/−; Ubc-CreERT2/+; p53flox/flox mice and Nrf2−/−; Ubc-CreERT2/+; p53flox/flox; Atg7flox/flox mice. After TAM administration, loss of p53 in Nrf2−/−p53Δ/ΔAtg7Δ/Δ intestine was confirmed by qRT-PCR, and we found that Nrf2−/−p53Δ/ΔAtg7Δ/Δ mice did not survive longer than Nrf2−/−Atg7Δ/Δ mice, suggesting that the intestinal damage in the Nrf2−/−Atg7Δ/Δ mice was not caused by p53 (Supplemental Fig. S5G,H).
Discussion
Both autophagy and p53 can protect tissues from stress such as DNA damage, oxidative stress, and hypoxia (Mizushima and Komatsu 2011; Fischer 2017), and their overlapping functions have suggested that these two pathways interact. p53 up-regulates the expression of essential autophagy genes and autophagy function in vitro (Crighton et al. 2006; Feng et al. 2007; Zhang et al. 2009; Mah et al. 2012; Kenzelmann Broz et al. 2013). In turn, autophagy inhibits p53 in some tumors providing a negative feedback loop (Guo et al. 2013; Rosenfeldt et al. 2013; Strohecker et al. 2013; Yang et al. 2014). Whether autophagy can regulate p53 in normal tissues in vivo, however, was not clear. We found that Atg7 suppresses p53 activation in the liver and brain, without which hyperactivation of p53 is responsible for damage to these tissues. Thus, essential autophagy gene Atg7 is a tissue-specific negative regulator of p53 and contributes to a negative feedback loop to limit p53 activation in vivo (Fig. 5A). Remarkably, eliminating p53 also rescued the survival of Atg7-deficient mice during fasting, suggesting that Atg7 restricts p53 activation in response to exogenous as well as endogenous stress (Fig. 5). Even when p53 activation is forced by Nutlin-3, Atg7 prevents these tissues from p53-mediated damage (Fig. 5A,B). However, how p53 is activated remains unclear, which could either be a direct effect of loss of Atg7, or an indirect effect caused by cellular microenvironment change after Atg7 deletion. These findings are consistent with the observation that Nutlin-3 does not activate p53 in normal tissues unless under stress conditions caused by Atg7 deficiency, but can efficiently activate p53 in different types of cancer cells without side effects on normal tissues (Vassilev et al. 2004; Drost et al. 2015; Yee-Lin et al. 2018; Forte et al. 2019). Therefore, autophagy may limit the effectiveness of MDM2 antagonists, and this should be tested in the cancer setting.
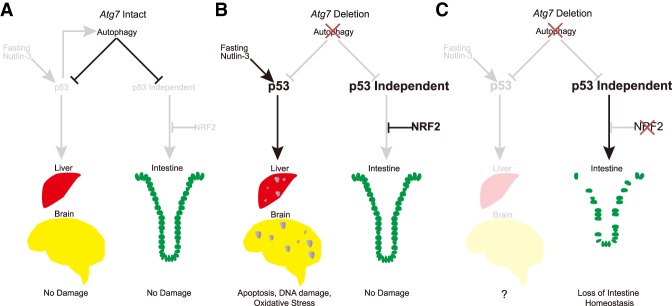
Figure 5.
Mechanism by which autophagy interacts with the p53 and NRF2 stress response mechanisms to protect tissues in a tissue specific manner. See the text for details.
The NRF2 and autophagy pathways both contribute to antioxidant defense. NRF2 is activated by autophagy deficiency in vitro and in tumors (Lau et al. 2010; Strohecker et al. 2013; Saito et al. 2016). The autophagy substrate p62, which accumulates when autophagy is blocked, interacts with KEAP1, thereby releasing and stabilizing NRF2 and promoting expression of its target genes (Komatsu et al. 2010; Lau et al. 2010; Ichimura et al. 2013; Levonen et al. 2014). We found that the protective function of NRF2 is essential for the survival of mice with loss of ATG7, as in stark contrast to Atg7Δ/Δ mice, Nrf2−/− Atg7Δ/Δ mice die rapidly, specifically from damage to the small intestine. Atg5 deficiency in intestine epithelia causes decreased numbers of intestinal stem cells, and these stem cells have a higher ROS level compared with wild-type mice, which can be rescued by treating mice with antioxidant N-acetyl cysteine (Asano et al. 2017). Atg16L1 is also required to protect the intestinal epithelium from necroptosis induction in response to virus-induced intestinal bowel disease by maintaining mitochondrial homeostasis (Matsuzawa-Ishimoto et al. 2017). We report here that knockout of NRF2 is synthetically lethal with loss of Atg7, which causes death within 1 wk before liver and brain damage can be observed, as NRF2 is specifically required to protect the survival of intestinal stem cells (Fig. 5C). The compensatory protective effect of NRF2 to loss of autophagy may be broad as recent cell-based screens identified NRF2 activation as a resistance mechanism selected for in cancer cells deleted for essential autophagy genes (Towers et al. 2019). In conclusion, autophagy limits p53 activation and damage in the liver and brain, while NRF2 limits intestinal stem cells damage due to loss of autophagy by a p53-independent mechanism. These findings demonstrate the functional interaction and tissue specificity of these stress regulated pathways (Fig. 5A–C).
ATG7 is an essential autophagy protein in the canonical autophagy pathway where it functions as an E1-like ubiquitin activating enzyme (Mizushima and Klionsky 2007). ATG7, however, also contributes to the other noncanonical LC3-dependent pathways LC3-associated phagocytosis (LAP) and LC3-associated endocytosis (LANDO). Macrophages use LAP to engulf and degrade particles and pathogens, which is immune-suppressive (Sanjuan et al. 2007; Florey et al. 2011; Martinez et al. 2011, 2015; Kim et al. 2013). LANDO is also immune suppressive and functions in microglia to remove the β-amyloid aggregates and protect the brain from neurodegeneration (Heckmann et al. 2019). Canonical autophagy, LAP, and LANDO all require autophagy machinery proteins including Beclin1, vacuolar protein sorting 34 (VPS34), ATG7, ATG5, and LC3, but activation of LAP or LANDO does not require some proteins in the autophagy initiation complexes such as RB1-inducible coiled-coil protein 1 (FIP200) (Martinez et al. 2011, 2015; Heckmann et al. 2019). Although LAP and LANDO have not been identified in hepatocytes, neurons, or intestinal stem cells as examined here, it would be interesting to test whether Fip200 deletion activates the p53 response in the liver and brain, or causes dependence on NRF2 in the intestine to distinguish roles of canonical and noncanonical autophagy pathways.
Material and methods
Mouse models
All animal care was carried out in compliance with Rutgers University Institutional Animal Care and Use Committee guidelines. Ubc-CreERT2/+ mice (The Jackson Laboratory) (Ruzankina et al. 2007) and Atg7flox/flox mice (provided by Dr. M. Komatsu, Tokyo Metropolitan Institute of Medical Science) (Komatsu et al. 2005) were cross-bred to generate the Ubc-CreERT2/+; Atg7flox/flox mice as described previously (Karsli-Uzunbas et al. 2014). To generate Ubc-CreERT2/+; Atg7flox/flox; p53flox/flox mice, p53flox/flox mice (The Jackson Laboratory) (Marino et al. 2000) were cross-bred with our previously created Ubc-CreERT2/+; Atg7flox/flox mice. To generate Nrf2−/−; Ubc-CreERT2/+ mice and Nrf2−/−; Ubc-CreERT2/+; Atg7flox/flox mice, Nrf2−/− mice (provided by YW Kan, University of California at San Francisco) (Chan et al. 1996) were cross-bred with Ubc-CreERT2/+ mice and our previously generated Ubc-CreERT2/+; Atg7flox/flox mice.
TAM administration
For acute deletion of Atg7 and/or p53, detailed rationale and TAM preparation is described as published previously (Karsli-Uzunbas et al. 2014). For TAM delivery to the adult mice, 200 μL of the suspended solution per 20 g of body weight was injected intraperitoneally (IP) into 8- to 10-wk-old Ubc-CreERT2/+; Atg7flox/flox mice and Ubc-CreERT2/+; Atg7flox/flox; p53flox/flox mice once per day for five consecutive days to delete the floxed gene systematically. Additionally, the same dosages of TAM were given to Ubc-CreERT2/+ mice and Ubc-CreERT2/+; p53flox/flox mice, and these mice were examined as control groups.
Statistical methods
Survival curves were estimated using the Kaplan-Meier method. Comparisons of survival curves were made using the log-rank test and Gehan-Breslow-Wilcoxon test (Cox and Oakes 1984).
Fasting
Fasting was conducted as described previously (Karsli-Uzunbas et al. 2014).
Histology
Mouse tissues were collected and fixed in 10% formalin solution (Formaldehyde Fresh, Fisher Scientific SF94-4). Tissues were fixed overnight and then transferred to 70% ethanol for paraffin-embedded sections or 15% sucrose following by 30% sucrose for frozen sections. For Alcian blue staining, paraffin sections were processed with the Alcian blue stain kit (Abcam) following the manufacturer's protocol. For Bodipy C11 (Thermo Scientific D3861) stain, 5 µM Bodipy C11 dye were used to stain the frozen sections of intestine for 30 min and counterstained with DAPI. For IHC, paraffin sections were stained with antibodies against p53 (1:400; Novus Biologicals NB200-103), active caspase-3 (1:300; Cell Signaling 9661), γ-H2AX (1:480; Cell Signaling 9718), MDA (1:200; Cosmo Bio USA NOF-N213530-EX), OLFM4 (1:2000; Cell Signaling 39141), Lysozyme (1:2000; Agilent A0099). For quantification of IHC on p53, active caspase-3, and γ-H2AX, the liver and brain tissues were analyzed by quantifying at least 10 images at 60× magnification using ImageJ. A minimum of 100 cells in the liver and brain was scored for each image.
Nutlin-3a administration
TAM were injected via IP into 8- to 10-wk-old Ubc-CreERT2/+; Atg7flox/flox mice and Ubc-CreERT2/+; Atg7flox/flox; p53flox/flox mice once per day for five consecutive days. Additionally, same dosages of TAM were given to Ubc-CreERT2/+ mice and Ubc-CreERT2/+; p53flox/flox mice. After 2 wk, 200 mg/kg Nutlin-3 (Cayman Chemicals) resolved in 50% DMSO was delivered to the mice by oral gavage once per day for seven consecutive days. Mice were sacrificed 1 d after the last administration of Nutlin-3 and tissues were collected for histology and snap-frozen for Western blot and real-time PCR.
Real-time PCR
Total RNA were isolated from tissue by Trizol (Invitrogen). cDNA were then reverse-transcribed from the total RNA by MultiScribe RT kit (Thermo Fisher). Real-time PCR were performed on Applied Biosystems StepOne Plus machine. Atg7 and p53 were performed using SYBR Green for deletion detection (Atg7: forward 5′-ACTTGACCGGTCTTACCCTG-3′; reverse 5′-TACTCCTGAGCTGTGGTTGC-3′ p53: forward: 5′-CGACTACAGTTAGGGGGCAC-3′; reverse: 5′-GGAGGAAGTAGTTTCCATAAGCCT-3′; Actin: forward 5′-GAACCCTAAGGCCAACCGT GAAAAGATGAC-3′; reverse 5′ GCAGGATGGCGTGAGGGAGAGCA-3′). Besides that, all of the other genes were detected using predesigned commercial TaqMan primers for each gene accordingly (Cdkn1a: Mm00432448-m1; Bax: Mm00432050; Bbc3: Mm00519268; Actin: Mm00607939-s1; Uvrag: Mm00724370-m1; Ulk1: Mm0437238-m1; Ulk2: Mm03048846-m1; Vmp1: Mm00774656-m1; Atg2b: Mm00620760-m1; Atg4a: Mm04214755-s1; Atg4c: Mm00558175-m1; and Atg10: Mm00470550-m1). Results were calculated using the ΔΔCT method and then normalized to actin.
Western blot
Different tissues were ground in a Cryomill machine (Retsch) and then total protein extracts were isolated by Tris lysis buffer (1 mol/L Tris HCl, 1 mol/L NaCl, 0.1 mol/L EDTA, 10% NP40). Separated proteins were probed with antibodies against ATG7 (1:2000; Sigma A2856), LC3 (1:1500; Novus Biologicals NB600-1384), p62 (1:2000; American Research Products 03-GP62-C), GAPDH (1:1000; Santa Cruz Biotechnology sc-365062), and β-actin (1:5000; Sigma A1978).
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grants R01 CA130893, CA188096, and CA163591 (to E.W.). Services, results, and/or products in support of the research project were generated by the Rutgers Cancer Institute of New Jersey Biospecimen Repository and Histopathology Service Shared Resource, supported in part with funding from National Cancer Institute-Cancer Center Support Grant (NCI-CCSG) P30CA072720-5919; and the Biometrics Shared Resource, supported in part with funding from NCI-CCSG P30CA072720-5918.
Author contributions: Y.Y. performed the majority of the experimental work and wrote the manuscript. G.K.-U. performed some of the survival experiments and the fasting experiment and assisted with IHC tissue preparation from wild-type, Atg7Δ/Δ, p53Δ/Δ, and Atg7Δ/Δp53Δ/Δ mice. L.P.-P. and A.S. assisted with the Western blot and quantification of IHC. Z.S.H. assisted with the mouse husbandry. Y.Z. and W.H. assisted with the qRT-PCR. D.M. assisted with the statistics. E.W. was the leading principal investigator who conceived and supervised on the project and edited the paper.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.335570.119.
References
- Asano J, Sato T, Ichinose S, Kajita M, Onai N, Shimizu S, Ohteki T. 2017. Intrinsic autophagy is required for the maintenance of intestinal stem cells and for irradiation-induced intestinal regeneration. Cell Rep 20: 1050–1060. 10.1016/j.celrep.2017.07.019 [DOI] [PubMed] [Google Scholar]
- Bode AM, Dong Z. 2004. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer 4: 793–805. 10.1038/nrc1455 [DOI] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. 2008. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456: 259–263. 10.1038/nature07416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Lu R, Chang JC, Kan YW. 1996. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci 93: 13943–13948. 10.1073/pnas.93.24.13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR, Oakes D. 1984. Analysis of survival data. Chapman and Hall, London; New York. [Google Scholar]
- Craig AL, Burch L, Vojtesek B, Mikutowska J, Thompson A, Hupp TR. 1999. Novel phosphorylation sites of human tumour suppressor protein p53 at Ser20 and Thr18 that disrupt the binding of mdm2 (mouse double minute 2) protein are modified in human cancers. Biochem J 342: 133–141. 10.1042/bj3420133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. 2006. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126: 121–134. 10.1016/j.cell.2006.05.034 [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Vogel H, McArthur MJ, Montgomery CA Jr, Park SH, Thompson T, Ford RJ, Bradley A. 1995. Effects of genetic background on tumorigenesis in p53-deficient mice. Mol Carcinog 14: 16–22. 10.1002/mc.2940140105 [DOI] [PubMed] [Google Scholar]
- Dotto GP. 2009. Crosstalk of Notch with p53 and p63 in cancer growth control. Nat Rev Cancer 9: 587–595. 10.1038/nrc2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, et al. 2015. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521: 43–47. 10.1038/nature14415 [DOI] [PubMed] [Google Scholar]
- Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ. 2007. The regulation of AMPKβ1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res 67: 3043–3053. 10.1158/0008-5472.CAN-06-4149 [DOI] [PubMed] [Google Scholar]
- Fischer M. 2017. Census and evaluation of p53 target genes. Oncogene 36: 3943–3956. 10.1038/onc.2016.502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. 2011. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol 13: 1335–1343. 10.1038/ncb2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forte IM, Indovina P, Iannuzzi CA, Cirillo D, Di Marzo D, Barone D, Capone F, Pentimalli F, Giordano A. 2019. Targeted therapy based on p53 reactivation reduces both glioblastoma cell growth and resistance to temozolomide. Int J Oncol 54: 2189–2199. 10.3892/ijo.2019.4788 [DOI] [PubMed] [Google Scholar]
- Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, et al. 2013. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev 27: 1447–1461. 10.1101/gad.219642.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Teng X, Laddha SV, Ma S, Van Nostrand SC, Yang Y, Khor S, Chan CS, Rabinowitz JD, White E. 2016. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev 30: 1704–1717. 10.1101/gad.283416.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann BL, Teubner BJW, Tummers B, Boada-Romero E, Harris L, Yang M, Guy CS, Zakharenko SS, Green DR. 2019. LC3-associated endocytosis facilitates β-amyloid clearance and mitigates neurodegeneration in murine Alzheimer's disease. Cell 178: 536–551 e514. 10.1016/j.cell.2019.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 420: 25–27. 10.1016/S0014-5793(97)01480-4 [DOI] [PubMed] [Google Scholar]
- Huo Y, Cai H, Teplova I, Bowman-Colin C, Chen G, Price S, Barnard N, Ganesan S, Karantza V, White E, et al. 2013. Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer Discov 3: 894–907. 10.1158/2159-8290.CD-13-0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, et al. 2013. Phosphorylation of p62 activates the Keap1–Nrf2 pathway during selective autophagy. Mol Cell 51: 618–631. 10.1016/j.molcel.2013.08.003 [DOI] [PubMed] [Google Scholar]
- Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, et al. 2014. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov 4: 914–927. 10.1158/2159-8290.CD-14-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J, Debnath J. 2015. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol 16: 461–472. 10.1038/nrm4024 [DOI] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. 2007. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47: 89–116. 10.1146/annurev.pharmtox.46.120604.141046 [DOI] [PubMed] [Google Scholar]
- Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, Sidow A, Attardi LD. 2013. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev 27: 1016–1031. 10.1101/gad.212282.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury K, Domling A. 2012. P53 mdm2 inhibitors. Curr Pharm Des 18: 4668–4678. 10.2174/138161212802651580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Zhao H, Martinez J, Doggett TA, Kolesnikov AV, Tang PH, Ablonczy Z, Chan CC, Zhou Z, Green DR, et al. 2013. Noncanonical autophagy promotes the visual cycle. Cell 154: 365–376. 10.1016/j.cell.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434. 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12: 213–223. 10.1038/ncb2021 [DOI] [PubMed] [Google Scholar]
- Kruiswijk F, Labuschagne CF, Vousden KH. 2015. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol 16: 393–405. 10.1038/nrm4007 [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. 1997. Regulation of p53 stability by Mdm2. Nature 387: 299–303. 10.1038/387299a0 [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. 2004. The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036. 10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. 2010. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol 30: 3275–3285. 10.1128/MCB.00248-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levonen AL, Hill BG, Kansanen E, Zhang J, Darley-Usmar VM. 2014. Redox regulation of antioxidants, autophagy, and the response to stress: implications for electrophile therapeutics. Free Radic Biol Med 71: 196–207. 10.1016/j.freeradbiomed.2014.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah LY, O'Prey J, Baudot AD, Hoekstra A, Ryan KM. 2012. DRAM-1 encodes multiple isoforms that regulate autophagy. Autophagy 8: 18–28. 10.4161/auto.8.1.18077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. 2014. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509: 105–109. 10.1038/nature13148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R, Nazarko TY, Farre JC, Subramani S. 2010. Molecular mechanism and physiological role of pexophagy. FEBS Lett 584: 1367–1373. 10.1016/j.febslet.2010.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Lozano G. 2010. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ 17: 93–102. 10.1038/cdd.2009.68 [DOI] [PubMed] [Google Scholar]
- Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. 2000. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev 14: 994–1004. [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. 2011. Microtubule-associated protein 1 light chain 3α (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci 108: 17396–17401. 10.1073/pnas.1113421108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, Orchard R, Guan JL, Tan H, Peng J, et al. 2015. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol 17: 893–906. 10.1038/ncb3192 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell 137: 1062–1075. 10.1016/j.cell.2009.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Khor S, Hackett SR, Rabinowitz JD, Perlman DH, White E. 2014. Functional role of autophagy-mediated proteome remodeling in cell survival signaling and innate immunity. Mol Cell 55: 916–930. 10.1016/j.molcel.2014.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumine H, Saito M, Shimoda-Matsubayashi S, Tanaka H, Ishikawa A, Nakagawa-Hattori Y, Yokochi M, Kobayashi T, Igarashi S, Takano H, et al. 1997. Localization of a gene for an autosomal recessive form of juvenile Parkinsonism to chromosome 6q25.2-27. Am J Hum Genet 60: 588–596. [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa-Ishimoto Y, Shono Y, Gomez LE, Hubbard-Lucey VM, Cammer M, Neil J, Dewan MZ, Lieberman SR, Lazrak A, Marinis JM, et al. 2017. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med 214: 3687–3705. 10.1084/jem.20170558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. 2010. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol 22: 132–139. 10.1016/j.ceb.2009.12.004 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Klionsky DJ. 2007. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr 27: 19–40. 10.1146/annurev.nutr.27.061406.093749 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. 2011. Autophagy: renovation of cells and tissues. Cell 147: 728–741. 10.1016/j.cell.2011.10.026 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15: 1101–1111. 10.1091/mbc.e03-09-0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378: 203–206. 10.1038/378203a0 [DOI] [PubMed] [Google Scholar]
- Pohl C, Dikic I. 2019. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 366: 818–822. 10.1126/science.aax3769 [DOI] [PubMed] [Google Scholar]
- Poillet-Perez L, White E. 2019. Role of tumor and host autophagy in cancer metabolism. Genes Dev 33: 610–619. 10.1101/gad.325514.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E. 2010. Autophagy and metabolism. Science 330: 1344–1348. 10.1126/science.1193497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway R, et al. 2013. p53 status determines the role of autophagy in pancreatic tumour development. Nature 504: 296–300. 10.1038/nature12865 [DOI] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. 2007. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1: 113–126. 10.1016/j.stem.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Ichimura Y, Taguchi K, Suzuki T, Mizushima T, Takagi K, Hirose Y, Nagahashi M, Iso T, Fukutomi T, et al. 2016. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat Commun 7: 12030 10.1038/ncomms12030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al. 2007. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450: 1253–1257. 10.1038/nature06421 [DOI] [PubMed] [Google Scholar]
- Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, McMahon M, White E. 2013. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov 3: 1272–1285. 10.1158/2159-8290.CD-13-0397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. 2006. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 6: 909–923. 10.1038/nrc2012 [DOI] [PubMed] [Google Scholar]
- Towers CG, Fitzwalter BE, Regan D, Goodspeed A, Morgan MJ, Liu CW, Gustafson DL, Thorburn A. 2019. Cancer cells upregulate NRF2 signaling to adapt to autophagy inhibition. Dev Cell 50: 690–703 e696. 10.1016/j.devcel.2019.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303: 844–848. 10.1126/science.1092472 [DOI] [PubMed] [Google Scholar]
- Yang A, Kimmelman AC. 2014. Inhibition of autophagy attenuates pancreatic cancer growth independent of TP53/TRP53 status. Autophagy 10: 1683–1684. 10.4161/auto.29961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell'antonio G, et al. 2011. Pancreatic cancers require autophagy for tumor growth. Genes Dev 25: 717–729. 10.1101/gad.2016111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, Von Hoff DD, Maitra A, Kimmelman AC. 2014. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov 4: 905–913. 10.1158/2159-8290.CD-14-0362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee KS, Wilkinson S, James J, Ryan KM, Vousden KH. 2009. PUMA- and Bax-induced autophagy contributes to apoptosis. Cell Death Differ 16: 1135–1145. 10.1038/cdd.2009.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee-Lin V, Pooi-Fong W, Soo-Beng AK. 2018. Nutlin-3, A p53-Mdm2 antagonist for nasopharyngeal carcinoma treatment. Mini Rev Med Chem 18: 173–183. 10.2174/1389557517666170717125821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD, Wang Y, Wang Y, Zhang X, Han R, Wu JC, Liang ZQ, Gu ZL, Han F, Fukunaga K, et al. 2009. p53 mediates mitochondria dysfunction-triggered autophagy activation and cell death in rat striatum. Autophagy 5: 339–350. 10.4161/auto.5.3.8174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.