Abstract
To date, more than 200 monogenic, often devastating, skin diseases have been described. Because of unmet medical needs, development of long-lasting and curative therapies has been consistently attempted, with the aim of correcting the underlying molecular defect. In this review, we will specifically address the few combined cell and gene therapy strategies that made it to the clinics. Based on these studies, what can be envisioned for the future is a patient-oriented strategy, built on the specific features of the individual in need. Most likely, a combination of different strategies, approaches, and advanced therapies will be required to reach the finish line at the end of the long and winding road hampering the achievement of definitive treatments for genodermatoses.
More than 7000 monogenic diseases, caused by genetic mutations inherited in a dominant or recessive manner, affect millions of people worldwide. Gene therapy of recessively inherited disorders usually envisages the introduction of a correct copy of the affected gene (transgene) into the patient's cells by means of a vector, which could be either viral or nonviral, the former being either integrating (as retroviruses/lentiviruses) or nonintegrating (as adeno or herpes viruses) into the genome. Depending on the tissue, the transgene can be delivered ex vivo, as in cultured cells, or in vivo, either locally or systemically. Gene therapy of dominantly inherited disorders should instead focus on allele-specific silencing of the mutated gene or other gene-editing strategies (Pan et al. 2019; Sung and Kim 2019).
It soon became clear that long-term clinical success of gene therapy strictly depends on the type of target cell, as homeostasis of constantly renewing tissues relies on somatic stem cells holding self-renewal and proliferative potential (De Luca et al. 2019). Thus, to be successful, gene therapy requires a deep preliminary knowledge of cell and developmental biology of specific stem cells, alongside with the establishment of technologies allowing their maintenance in culture and in vivo after their transplantation. Such cultures can be used as advanced therapy medicinal products (ATMPs) either as they are (cell therapy) or through previous modification of their genome (gene therapy). Many stem-cell-based cell and gene therapies have entered clinical trials, covering a wide spectrum of diseases, but only a few reached official approval and registration as, for instance, Holoclar (Pellegrini et al. 2018) and Strimvelis (Aiuti et al. 2017).
Combined cell and gene therapy is thus becoming a central asset of regenerative medicine, which aims at replacing, engineering, or regenerating irredeemably damaged human tissues or organs, hence restoring their normal function. Among the tissues that might benefit from such combination, skin is one of the most attractive for several reasons. Epidermal keratinocytes can be easily collected on ordinary skin biopsy retrieval (Rheinwald and Green 1975). Keratinocyte cultures containing well-characterized stem cells, defined as holoclones (Barrandon and Green 1987; Hirsch et al. 2017), and have long been safely used for life-saving permanent regeneration of a functional epidermis in massive full-thickness skin burns (Gallico et al. 1984; Pellegrini et al. 1999b; Ronfard et al. 2000; De Luca et al. 2006). As with the hematopoietic system, and at variance with more complex tissues or organs, clinical translation of epidermal cultures into a clinical setting is thus doable. The transplanted cultured epidermis can be easily monitored throughout the lifetime of the patients and removed anytime, if any adverse event should occur. Finally, solid proof-of-principle has already been attained regarding the successful and safe use of transgenic epidermal cultures for the restoration of the human epidermis in a specific genetic skin disease, epidermolysis bullosa (EB) (Mavilio et al. 2006; De Rosa et al. 2014; Siprashvili et al. 2016; Bauer et al. 2017; Hirsch et al. 2017). The identification of a variety of severe monogenic skin diseases (Tadini et al. 2015) makes the epidermis an appealing candidate for a platform of combined cell and gene therapy approaches, which could become a clinical reality, a goal already achieved by transgenic hematopoietic stem cells for gene therapy of genetic immunodeficiencies and CAR-T cells (chimeric antigen receptor-genetically engineered T cells) for immunotherapy of B-cell acute lymphoblastic leukemia and diffuse large B-cell lymphoma (Kymriah) and primary mediastinal large B-cell lymphoma (Yescarta) (Aiuti et al. 2017; Sadelain et al. 2017; June and Sadelain 2018).
In this review, we will focus solely on combined cell and gene therapies tackling genetic skin diseases that made it to clinical trials (Fig. 1), leaving aside the many preclinical studies that still make up a large portion of genodermatoses panorama. In the final part, however, we will briefly address gene-editing strategies, which are deemed to offer promise for the treatment of many inherited genetic diseases. Although gene editing of genodermatoses is still at a preclinical stage, its fast development warrants an update.
Figure 1.
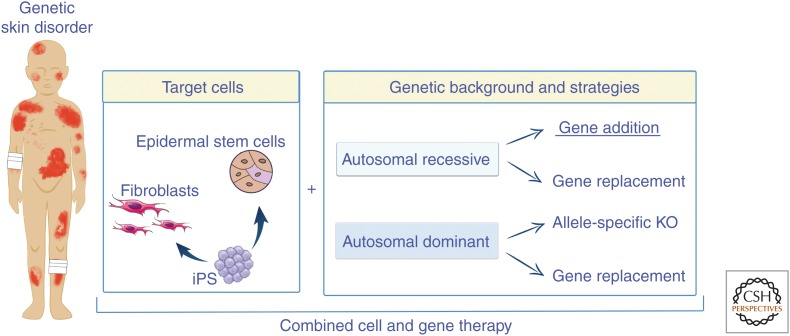
A view of current ex vivo gene therapy strategies for genodermatoses. Epidermal stem cells, fibroblasts, and induced pluripotent stem cells (iPS) can be genetically modified by means of gene addition (the most frequent approach), gene replacement, and allele-specific gene knockout (KO) for both recessively and dominantly inherited genetic skin diseases.
GENE CORRECTION OF SKIN CELLS
There are >200 monogenic skin diseases, featuring either dominant or recessive transmission patterns (Tadini et al. 2015). Most genodermatoses are caused by mutations affecting genes mainly expressed by epidermal keratinocytes, although some of them can also be expressed by dermal fibroblasts (Fig. 2). Some genetic skin disorders are marked by a mosaic pattern, often with alternating stripes of affected and unaffected skin that follow the lines of Blaschko, which can be caused by a postzygotic mutation during embryogenesis (Paller et al. 1994).
Figure 2.
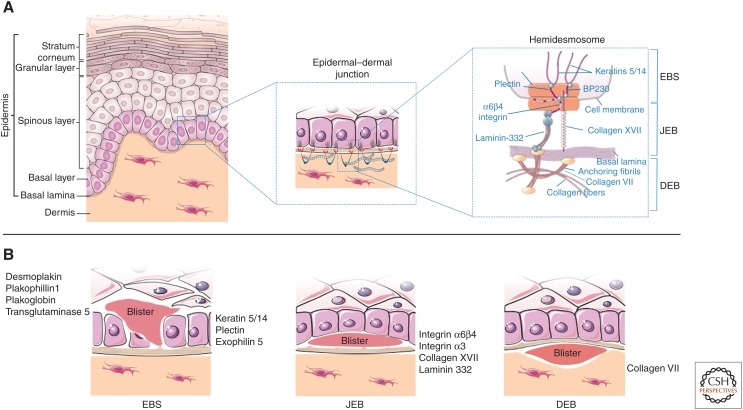
Schematic representation of human skin and epidermolysis bullosa (EB). (A) Epidermal layers (left) and the epidermal–dermal junction (middle) are designated. The right panel denotes the hemidesmosome and its components, in relation to EB simplex (EBS), junctional EB (JEB), and dystrophic EB (DEB). (B) Proteins involved in the pathogenesis of EBS, JEB, and DEB and site of blister formation.
Human epidermis constantly renews thanks to a small population of long-lived stem cells (Hirsch et al. 2017), which cannot be specifically targeted by local or systemic in vivo gene delivery. Therefore, permanent restoration of a functional epidermis requires ex vivo transduction of epidermal stem cells by means of integrating viral vectors and the subsequent transplantation of transgenic cultivated epidermal grafts, especially if large areas of the body are involved. For dominantly inherited genetic skin diseases, newly developed gene-editing methods are still at the preclinical stage and most likely will require an ex vivo correction of epidermal stem cells as well. Currently, retroviruses, lentiviruses, and herpes viruses dominate the scene of ex vivo gene therapy for genodermatoses.
Retroviral Vectors
To date, retroviral vectors (RVs) derived from Moloney murine leukemia virus (Markowitz et al. 1988; Riviere et al. 1995; Merten 2004), hereafter referred to as γRVs, are most frequently used in clinical trials for keratinocyte-based ex vivo gene therapy (Edelstein et al. 2007). They can accommodate up to ∼9 kb of genetic material and stably transduce nearly 100% of clonogenic keratinocytes (Dellambra et al. 1998; Hirsch et al. 2017). The construction of γRVs involves the generation of packaging cell lines carrying the transgene construct, the RNA packaging signals (Ψ), cis-acting viral sequences—such as 5′- and 3′-long terminal repeats (LTRs) used as a promoter—and gag, pol, and env viral sequences (Pear et al. 1993; Chang and Yee 2012). Packaging cells then need large-scale production in compliance with good manufacturing practices (GMPs), which are required to conform to the guidelines recommended by agencies that control the authorization of the manufacture and sale of pharmaceutical products, including ATMPs. Among others, complete absence of replication-competent viruses (RCVs) is a stringent GMP requirement (Cornetta et al. 2008).
A major concern about the use of γRVs is the risk of insertional mutagenesis. Indeed, these vectors show a preferential integration pattern in actively transcribed genes, which can interfere with physiological gene expression, leading to potential deregulation of cellular processes (Scherdin et al. 1990; Wu et al. 2003; Mitchell et al. 2004; Lewinski et al. 2006; Osborn et al. 2013). Lymphoproliferative or myelodysplastic disorders, resulting from proviral insertion into proto-oncogene promoters of hematopoietic stem cells, were observed in γRV-based clinical trials for gene therapy of X-linked severe combined immunodeficiency (SCID) (Hacein-Bey-Abina et al. 2003, 2008; Aiuti et al. 2009), Wiskott–Aldrich syndrome (WAS) (Aiuti et al. 2013; Braun et al. 2014; Hacein-Bey Abina et al. 2015), and X-linked chronic granulomatous disease (X-CGD) (Stein et al. 2010). Of note, none of these adverse events were reported when γRV-transduced hematopoietic stem cells were used to successfully treat adenosine deaminase (ADA)-deficient SCID (Aiuti et al. 2009). In fact, γRV-transduced CD34+ cells were approved in 2016 in the European Union for the treatment of ADA-SCID (Strimvelis). Similarly, no insertional oncogenesis has been detected in γRV-based ex vivo gene therapy of recessively inherited junctional EB (JEB) and dystrophic EB (RDEB) (see below). Thus, to trigger tumor formation, insertional mutagenesis might require other oncogenic factors, which could be related to cell type, patient's individual genetic background, age, disease, transgene, or other mutations (Howe et al. 2008; Cavazza et al. 2013).
Nevertheless, the development of safer γRVs is desirable and many efforts are going to achieve this. Self-inactivating-γRVs (SIN-γRVs) are characterized by a deletion of the U3 region of the 3′LTR, which eliminates a strong viral enhancer, hence reducing the tendency of γRV to activate genes. An internal, often tissue-specific, promoter can be used to drive the transcription of the transgene (Yu et al. 1986; Kraunus et al. 2004; Schambach et al. 2006). During these studies, it has been clearly shown that insertional gene activation is determined by the characteristics of transcriptional regulatory elements carried by the vector and is largely independent from the vector type or design (Maruggi et al. 2009). Indeed, the LTR has strong enhancer activity in both γRV and lentiviral vectors (LVs), whereas an internal cellular promoter induces deregulation of gene expression less frequently and to a lesser extent in both vectors (Maruggi et al. 2009).
An alternative vector system, derived from the Avian sarcoma-leukosis α-retrovirus (ASLV), is currently available. In preclinical models, αRVs showed favorable safety features as compared with both γRVs and LVs, at least in hematopoietic cells (Suerth et al. 2012). Small-scale surveys of integration sites of αRVs in different cell types pointed to a more haphazard integration pattern as compared with other RVs, with no apparent preference for promoters or transcription start sites (Mitchell et al. 2004; Hu et al. 2007, 2008; Suerth et al. 2010, 2012, 2014). This promising outcome led to the development of SIN-αRV being able to efficiently transduce mammalian cells, attain long-term transgene expression in murine and human hematopoietic progenitors, and redress X-CGD in a mouse model (Kaufmann et al. 2013).
Lentiviral Vectors
Lentiviruses are a subclass of retroviruses, the best-known member being human immunodeficiency virus (HIV), sharing with its fellow γRVs the gag, pol, and env genes. LVs own a few additional regulatory genes, namely tat and rev, and some other accessory genes depending on the subtype (Malim et al. 1988; Miller and Sarver 1997). LVs have many attractive properties. In contrast to γRVs, LVs infect both dividing and nondividing cells; hence, they could transduce a broader variety of human cells (Bukrinskaya et al. 1996; Kaushik and Ratner 2004). LVs can be pseudotyped with vesicular stomatitis virus glycoprotein (VSV-G), thus allowing the binding to a widely expressed receptor on the plasma membrane of target cells, with a consequent increase of transduction efficiency. Although LVs might have a slightly safer integration profile than γRVs (Cattoglio et al. 2010), LVs and γRVs have similar LTR-enhancer activity (Maruggi et al. 2009), potentially increasing the expression of the gene where they integrate. Of note, down-regulation of gene expression was observed only in the context of LVs, which might impact on tumor-suppressor genes (Maruggi et al. 2009).
Similarly to γRV, the safety of LVs has been greatly improved by the generation of SIN-LVs (Zufferey et al. 1998). Many clinical trials have shown that SIN-LVs are quite safe for hematopoietic stem-cell-mediated gene therapy of X-SCID, WAS, and metachromatic leukodystrophy (Biffi et al. 2004, 2013; Galy et al. 2008; Zhou et al. 2010; De Ravin et al. 2016; Singh et al. 2017). Indeed, in the last few years, the use of LVs in gene therapy clinical trials has grown exponentially (Cavazzana et al. 2019; Naldini 2019) and will probably continue to do so in the future (The Journal of Gene Medicine, Gene Therapy Clinical Trials Worldwide, https://onlinelibrary.wiley.com/doi/full/10.1002/jgm.3015).
Despite the many advantages offered by LVs, RVs seem more manageable for ex vivo genetic modification of human keratinocytes. This is perhaps because Rev and the viral proteases of LVs are quite toxic for primary keratinocytes, leading to loss of a significant number of clonogenic cells during the transduction procedure (Haselhorst et al. 1998). Furthermore, although we have been able to limit the number of γRV integrations in keratinocytes to less than two per cell, we consistently detect more than 10 LV integrations in each clonogenic human keratinocyte, irrespective of which transduction protocol is used (Mavilio et al. 2006; Bauer et al. 2017; Hirsch et al. 2017; MC Latella et al., unpubl.). Of note, SIN-γRV showed a significantly lower bias for transcription start sites and promoters in human keratinocytes as compared with hematopoietic cells (Cavazza et al. 2013). Finally, preclinical studies aimed at combined cell and gene therapy of RDEB—a form of EB caused by mutations in COL7A1, a gene encoding for collagen VII (see below)—showed that virtually all clonogenic LV-transduced keratinocytes contained multiple rearranged, truncated, or fragmented collagen VII proteins (MC Latella et al., unpubl.). In contrast, only ∼25% of γRV-transduced keratinocytes, analyzed at the clonal level, contained a single aberrant collagen VII, always in the presence of full-length protein (JW Bauer, M De Luca et al., unpubl.). As a practical consideration, RVs allow the generation of stable GMP-certified packaging cell lines that can be used for years, hence fostering reproducibility of the procedure and lowering the cost of each transduction process. Instead, LV proved to be more efficient and less toxic in transducing human primary fibroblasts (Woodley et al. 2003; Georgiadis et al. 2016; Lwin et al. 2019).
Herpes Simplex Viruses
Herpes simplex viruses (HSVs) are DNA viruses extensively used for the generation of vectors. Their broad cellular tropism, high-packaging capacity (almost 150 kb), and ability to transduce both dividing and quiescent cells have quickly attracted the interest of researchers for their use in in vivo gene therapy approaches, particularly for the treatment of neurological disorders (Spaete and Frenkel 1982; Artusi et al. 2018).
In contrast to RVs and LVs, HSVs and their related vectors remain episomally recircularized in the nucleus and are thought to replicate as a concatemer via a rolling circle-like mechanism (Subak-Sharpe and Dargan 1998; Whitley and Roizman 2001). To date, different types of HSV-1-derived vectors exist: (1) replication-defective rHSV-1 lacking essential replication genes, which are provided in trans by the cell line, (2) attenuated rHSV-1, containing deletions in nonessential genes that hinder their replication in vivo but not in vitro, and (3) HSV-1-derived amplicon vectors, which are the safest by virtue of their carrying just an origin of viral replication, a packaging signal, and the cassette of interest (Manservigi et al. 2010). Thus, replication, structural, and packaging proteins must be supplied in trans by a helper/packaging cell line (lacking packaging signal), achieving a lower chance of RCV generation, and hence a better safety profile. A historical issue that concerned HSVs has been the presence of contaminating helper viruses in vector stocks, which could lead to unwanted immune responses (Bowers et al. 2003). However, the most important pitfall related to the use of HSVs in genodermatoses is their episomal nature, which hampers a long-lasting therapeutic effect (Marconi et al. 2008).
In conclusion, genodermatoses can best be tackled through combined cell and gene therapy by means of γRV, most preferably SIN forms. It would be of great interest to see whether SIN-αRVs could attain similar performances, considering their safer integration profile.
Epidermal Stem Cells as Target of Gene Therapy
Targeting stem cells is the cornerstone for successful combined cell and gene therapy of genetic diseases affecting any renewing tissues, including the skin (Fig. 3A). In fact, the remarkable clinical outcomes of regenerative medicine in blood and squamous epithelia accrue from a large body of studies on identification, characterization, and function of their specific stem cells (De Luca et al. 2006, 2019; Gonzales and Fuchs 2017; Belokhvostova et al. 2018; Li et al. 2019).
Figure 3.
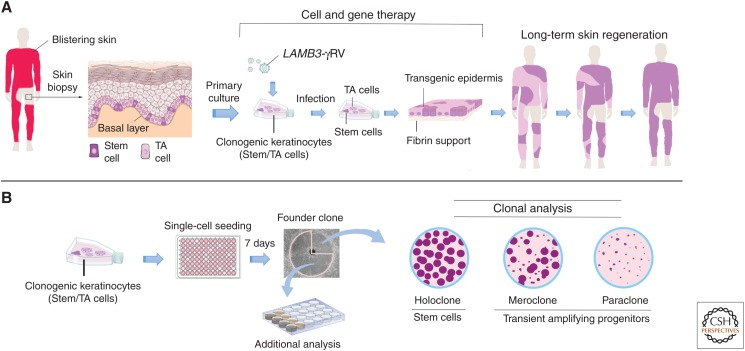
Schematic representation of ex vivo gene therapy and clonal analysis. (A) Scheme of combined cell and gene therapy for LAMB3-dependent junctional epidermolysis bullosa (JEB), as reported in Hirsch et al. (2017). Clonogenic keratinocytes, consisting of stem cells (rhodamine red) and transient amplyfing cells (light pink), were cultivated from a skin biopsy, transduced with γRVs, containing LAMB3 and used to prepare transgenic epidermal sheets, which were then transplanted on surgically prepared skin lesions. Of note, clonal tracing performed on cultures initiated from the restored skin (Hirsch et al. 2017) has shown that long-term skin regeneration (right part of the panel) is sustained only by long-lived, self-renewing stem cells detected as holoclone-forming cells (see panel B), as indicated by the rhodamine red color. (B) Clonal analysis. Keratinocytes are inoculated (0.5 cells per well) onto 96-multiwell plates containing irradiated 3T3-J2 cells. After 7 days of cultivation, single clones are transferred to two dishes and cultivated. One dish (one-quarter of the clone) is stained 12 days later for the classification of clonal type, which is determined by the percentage of aborted colonies formed by the progeny of the founding cell. The clone is scored as a holoclone when 0%–5% of colonies are terminal. When 95%–100% of colonies are terminal (or when no colonies formed), the clone is classified as a paraclone. When the number of terminal colonies is between 5% and 95%, the clone is classified as a meroclone. The second dish (three-quarters of the clone) is used for further analyses.
Keratinocyte stem cells, residing both in the epidermal basal layer and in the bulge of the hair follicle, underpin continuous renewal of the interfollicular epidermis, continuous cycles of hair growth, and degeneration and timely epidermal repair after injuries (Kretzschmar and Watt 2014; Fuchs 2016; Gonzales and Fuchs 2017; Belokhvostova et al. 2018). A crucial breakthrough fostering gene therapy of genodermatoses lies in the discovery that the features of human epithelial stem cells could be maintained out of the in vivo context, during serial cultivation in vitro (Rheinwald and Green 1975). This milestone in the history of human epidermal stem-cell research allowed the acquisition of crucial notions on the nature of these cells and paved the way for their successful life-saving clinical application in regenerative medicine (Gallico et al. 1984). Autologous keratinocyte cultures have been used worldwide for decades to regenerate the epidermis of thousands of patients with life-threatening full-thickness skin burns (De Luca et al. 2006; Green 2008). Similarly, the characterization and proper cultivation of human limbal stem cells has fostered their therapeutic use for the permanent restoration of a functional corneal epithelium in massive chemical burns of the eye (Pellegrini et al. 1997, 1999a, 2013; Rama et al. 2001, 2010). In 2015, autologous limbal cultures were the first stem-cell-based ATMP to receive marketing authorization from the European Medicine Agency (Holoclar).
Another crucial step was taken in 1987, when Barrandon and Green succeeded in cultivating human epidermal stem cells and transient amplifying progenitors (clonogenic cells with limited capacity for multiplication) at the clonal level (Barrandon and Green 1987). This led to the classification of proliferating keratinocytes into three groups, according to secondary clonal types produced by a founder clone generated by a single clonogenic cell (Fig. 3B). The founder clone was classified as paraclone if >95% of its secondary clones were aborted, terminal colonies. At the other side of the spectrum stood the holoclone, a founder clone generating <5% terminal clones. The meroclone lay in between, as 5%–95% of its secondary clones were terminal. These clonal types mark virtually all squamous epithelia (Barrandon and Green 1987; Pellegrini et al. 1999a). Clonal conversion, namely, the transition from holoclones to paraclones, is marked by reorganization of actin filament, being actin bundles distributed radially in holoclones and circumferentially in paraclones (Nanba et al. 2013).
The holoclone has all hallmarks of a stem cell, being the smallest colony-founding cell, uniquely endowed both with self-renewing capacity and tremendous proliferative potential (Rochat et al. 1994; Pellegrini et al. 1999a). Compelling, yet indirect, evidence of holoclone-forming cells being bona fide stem cells came from the notion that clinical success of epidermal cultures for full thickness skin burns and of limbal cultures for the treatment of massive ocular burns requires a defined number of holoclone-forming cells contained in the culture (Pellegrini et al. 1999a; Ronfard et al. 2000; Rama et al. 2010). It follows that the maintenance of a defined number of transgenic holoclones in keratinocyte cultures is mandatory to guarantee a successful outcome of combined cell and gene therapy for genodermatoses (Fig. 3A; Hirsch et al. 2017).
However, clonal analyses are rather laborious and time-consuming procedures. Efforts have thus been directed toward the identification of molecular markers that could specifically define each keratinocyte clonal type. There is evidence that the α isoform of the p63 transcription factor (Yang et al. 1998, 1999; Mills et al. 1999) is a human keratinocyte stem-cell marker, by virtue of its abundant expression in holoclones and its progressive decrease in expression during clonal conversion (Pellegrini et al. 2001; Di Iorio et al. 2005; De Rosa et al. 2019). In fact, p63 is a key, lineage-specific determinant of the proliferative capacity in stem cells of mammalian stratified epithelia (Yang et al. 1999; Pellegrini et al. 2001; Senoo et al. 2007). Thus, p63α—in particular the evaluation of the abundance of p63α in the nuclei of small keratinocytes (Di Iorio et al. 2005)—may prove a powerful tool for the prospective determination of the number of epidermal stem cells contained in transgenic epidermal grafts, as it is with cultured limbal/corneal epithelium (Rama et al. 2010).
Recently, Barrandon's group developed a strategy for ex vivo gene therapy that involves the expansion of a single holoclone-forming cell. Such clonal cultures regenerated a functional epidermis when transplanted onto athymic mice (Droz-Georget Lathion et al. 2015). In principle, this approach would minimize all safety burdens, as the entire epidermis would be generated from a single thoroughly characterized stem cell. However, its clinical applicability needs further investigation. On the one hand, this strategy would imply that all holoclone-forming cells are identical, which is far to be proven. On the other hand, long-lasting epidermal regeneration of large body sites (the body surface is 1–2 m2) would require a tremendous number of duplications of that single stem cell, which might raise other types of safety concerns and lead to a premature exhaustion of its proliferative potential.
Along this line, an alternative strategy for epidermal repair lies in induced pluripotent stem cells (iPS), which can be generated from any cell of the body (Takahashi et al. 2007). Their establishment involves a first step in which differentiated cells are brought back to an embryonic-like state by specific reprogramming factors, as Oct3/4, Sox2, c-Myc, and Klf4 (Takahashi et al. 2007). The resulting iPS can be expanded indefinitely in culture after gene correction. Selected clones of iPS can then be induced to differentiate into the desired cell types, such as fibroblasts or keratinocytes (Fig. 1), revealing themselves as a great tool for regenerative medicine protocols (Galach and Utikal 2011; Itoh et al. 2011; Tolar et al. 2011; Wenzel et al. 2014; Shinkuma et al. 2016; Abaci et al. 2017; Toyoshima et al. 2019). Although several phase I clinical trials, envisaging the use of iPS for other diseases, are ongoing (Guhr et al. 2018), this whole procedure bears potential risks regarding the safety of the cells, mainly related to possible aberration that might occur within their genome during such intense manipulation (Attwood and Edel 2019). In addition, fully reprogrammed iPS generated by current strategies might still display clone-to-clone variations in molecular properties and developmental potential. These considerations call for the setup of robust and reliable quality-control assays for the skin.
TACKLING GENETIC SKIN DISEASES
To date, clinical trials aimed at gene therapy of genodermatoses are focused on EB and Netherton syndrome (NS). These trials will pave the way for combined cell and gene therapy of other genetic skin diseases, as, for instance, lamellar ichthyosis.
Epidermolysis Bullosa
Inherited EB encompasses a heterogeneous group of rare, genetic disorders characterized by recurrent blistering of the skin and other stratified epithelia (Fig. 2). Blisters arise spontaneously or on minimal mechanical stress or trauma as a result of increased epithelial fragility, caused by molecular defects within genes encoding various structural proteins of the epidermal–dermal junction (Fig. 2; Fine et al. 2014). The incidence and prevalence of EB in the United States are ∼1/53,000 and 1/125,000, respectively, and similar estimates have been obtained in some European countries (Fine 2010, 2016). EB affects individuals from all ethnic origins without gender predilection and displays either dominant or recessive patterns of inheritance. More than 1000 mutations in at least 14 structural genes (Fig. 2) have been documented causing distinct clinical manifestations, ranging from mild to severe, with local or generalized involvement and significant morbidity and mortality. This variety depends on several factors, both phenotypic (distribution and severity of the lesions, involvement of mucosae, age of onset) and molecular (targeted protein, type of mutation and degree of function loss, mode of inheritance, and genetic background) (Fine 2010; Intong and Murrell 2012).
Inherited EB encompasses three major forms, primarily based on the level of skin cleavage (and subsequent blistering formation), that is intraepidermal in EB simplex (EBS), within the lamina lucida in JEB and beneath the lamina densa in DEB (Fig. 2; Intong and Murrell 2012; Fine et al. 2014). Kindler syndrome (KS) is a fourth, more rare EB form caused by mutations in FERMT-1, the gene encoding Kindlin-1. In KS, blisters occur at multiple levels, intraepidermal, and within or beneath the basement membrane (Intong and Murrell 2012; Fine et al. 2014).
EB, especially in its generalized intermediate/severe forms, greatly impairs the quality of life of people suffering from this painful, chronic and visible condition, representing a heavy health care burden (Horn and Tidman 2002; Fine et al. 2005; Dufresne et al. 2015). There is no cure for EB. Currently available therapies are largely palliative, only partially alleviating the devastating clinical manifestations (Prodinger et al. 2019). Such therapies, either systemic or topical (Prodinger et al. 2019), are not sufficient to provide decisive relief from pain, symptoms, and mental stress and achieve satisfactory living standards for these patients. Long-lasting, curative therapies are urgently needed and several attempts have been made in this respect. As of today, molecular-based therapeutic strategies aiming at gene correction in combination with cell-based approaches focus on individually designed treatments, holding promise for more effective results. We will now focus on JEB and RDEB, the only forms tackled by combined cell and gene therapy approaches that have made it to the clinic.
Junctional Epidermolysis Bullosa
Generalized JEB is a chronic, devastating, and life-threatening condition caused by mutations in LAMA3, LAMB3, or LAMC2—encoding, respectively, α3, β3, and γ2 chains of laminin-332 (also known as laminin 5)—and in genes encoding collagen XVII and α6β4 integrins (Fig. 2; Fine et al. 2008, 2014). Hemidesmosomes are rudimentary or absent, hence blisters occur within the lamina lucida of the basement membrane (Fig. 2) and predominate at sites exposed to friction, trauma, or heat. In addition, hair, nail, and enamel defects are common, along with the involvement of the ocular surface and gastrointestinal and renal systems (Laimer et al. 2010; Yancey and Hintner 2010). Deleterious mutations in genes encoding laminin 332 are usually lethal early in life (Hammersen et al. 2016). Approximately 40% of patients with intermediate to severe generalized JEB die before adolescence and adults are at high risk of developing squamous cell carcinoma (Fine et al. 2008, 2014).
LAMB3-deficient generalized intermediate JEB was the first genetic skin disease successfully tackled by ex vivo gene therapy (Mavilio et al. 2006). Combined cell and gene therapy was tested in a phase I/II clinical trial and eventually developed into a long-lasting curative treatment for skin lesions of this EB subtype. Preliminary preclinical studies showed that holoclones could be stably transduced by γRVs carrying different transgenes, including the LAMB3 full-length cDNA (Morgan et al. 1987; Mathor et al. 1996; Dellambra et al. 1998). Transgenic autologous epidermal cultures were then grafted on patients’ selected skin areas, on surgical removal of diseased epidermis and proper preparation of the wound bed. This approach has proven effective in restoring patches of normal epidermis on large nonhealing epidermal lesions in two adults LAMB3-dependent JEB patients (Mavilio et al. 2006; De Rosa et al. 2014; Bauer et al. 2017).
More recently, ex vivo gene therapy proved to be lifesaving, as it succeeded in regenerating virtually the entire epidermis of a 7-year-old child suffering from a devastating form of LAMB3-dependent JEB with very poor prognosis (Hirsch et al. 2017). Through the entire 4-year follow-up, the transgenic epidermis remained robust and resistant to mechanical stress, free from blisters or erosions. It expressed physiological levels of laminin-332, had normal thickness and continuity of the basement membrane, normal numbers and morphology of hemidesmosomes, and, notably, normal wound healing on injury (Hirsch et al. 2017). Strikingly, the regenerated epidermis was entirely transgenic, as LAMB3 messenger RNA (mRNA) and laminin 332 were uniformly detected in all the analyzed skin sections. No immune response or inflammation were observed. Of note, these studies not only attest to a most likely permanent (4- to 14-year follow-up) functional restoration of the epidermal–dermal junction but also reveal a good safety profile.
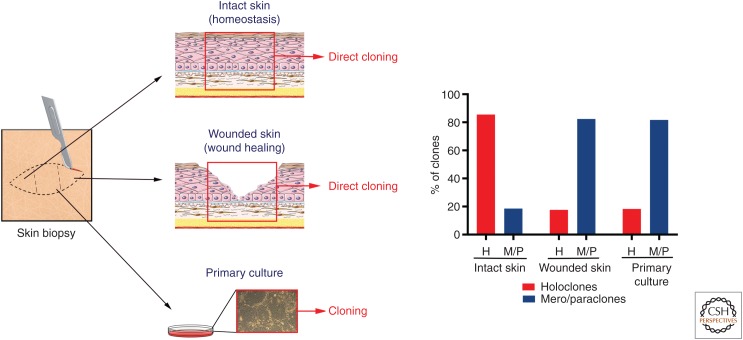
Targeting LAMB3 into the genome of long-lived stem cells underpins the stability of the transgenic epidermis. In fact, these studies provided formal, direct evidence that holoclone-forming cells identify authentic epidermal stem cells. Using γRV integrations as clonal genetic marks, clonal tracing has formally shown that, in the long-term, human epidermis is sustained uniquely by a defined number of self-renewing holoclones, which therefore have all the hallmarks of stem cells (Hirsch et al. 2017). Clonal tracing has also shown that holoclones continuously generate pools of meroclones and paraclones, which behave as short-lived progenitors, persist for various periods of time, replenish differentiated cells, and play a crucial role both in the engraftment of epidermal cultures and in epidermal regeneration during wound healing (Hirsch et al. 2017). This latter notion is consistent with the observation that single clonogenic keratinocytes derived directly from a biopsy taken from normal, unwounded skin generate mostly holoclones, whereas keratinocytes cloned from wounded skin generate predominantly mero- and paraclones (Fig. 4). Taken together, these data shed light on the issue, raised by lineage tracing experiments in mice, whether mammalian epidermis is sustained by specific long-lived, self-renewing stem cells (Mascré et al. 2012; Hirsch et al. 2017) or equipotent progenitors (Clayton et al. 2007; Rompolas et al. 2016), the latter being also suggested by the mode of proliferation of cultured clonogenic human keratinocytes (Roshan et al. 2016).
Figure 4.
Clonal analysis from skin biopsies and primary cultures. Clonal analysis was performed as described in Figure 3B. Human skin samples were obtained as anonymized surgical waste, typically from abdominoplasty or mammoplasty. Skin samples were cut in three parts for (1) direct cloning from intact skin, (2) direct cloning from wounded skin, (3) preparation of primary cultures. (1) Clonal analysis performed directly from the skin biopsy unveiled that the majority of clonogenic cells of intact skin generate holoclones (stem cells). (2) The epidermis was mechanically removed from the center of the biopsy to mimic a wound and the biopsy was kept in the incubator in keratinocyte growth medium (Hirsch et al. 2017). Clonal analysis, performed 24 hours later from a skin section taken in between the two edges of the wound, showed that the majority of clonogenic cells during the wound-healing process generates meroclones and paraclones (transient amplyfing progenitors). A similar distribution of clonogenic cells was detected in primary cultures (3), confirming that such cultures recapitulate a wound-healing process.
Altogether, the three JEB patients treated received ∼4 × 108 transgenic clonogenic keratinocytes, none of which developed into a transformed clone, within a time frame that entailed >150 epidermal renewal cycles (Mavilio et al. 2006; De Rosa et al. 2014; Bauer et al. 2017; Hirsch et al. 2017). The notion that long-term maintenance of transgenic epidermis is in fact sustained only by holoclones, a small percentage of such clonogenic cells (Mavilio et al. 2006; De Rosa et al. 2014; Bauer et al. 2017; Hirsch et al. 2017), indicates that the number of independent genomic hits stably introduced into the patient with the graft is finite, contributing to lower the risk of oncogene activation.
Taking all these results together, combined cell and gene therapy for LAMB3-dependent JEB is probably the most advanced therapeutic gene therapy approach for genodermatoses, and the closest to obtaining final approval by regulatory authorities as an advanced therapy for JEB skin lesions (De Luca et al. 2019).
Dystrophic Epidermolysis Bullosa
DEB is caused by mutations in COL7A1, the gene encoding collagen VII, the major component of dermal anchoring fibrils, whose absence (or alteration) induces blister formation just beneath the lamina densa of the basement membrane (Fig. 2). Dominant DEB (DDEB) generally displays a mild phenotype, with blisters occurring on areas of trauma (with a predilection for the extremities), often resulting in scars, milia (keratin-filled cysts of sweat glands), and loss of nails (Das and Sahoo 2004). Instead, generalized RDEB is more severe. Blisters develop at birth and progressively result in extensive scarring, particularly in joints, and there is highly disabling pseudosyndactyly—fusion of web spaces between the digits—and involvement of the mucous membranes (Nystrom and Bruckner-Tuderman 2016). The quality of life of RDEB patients is regarded as the poorest among all EB forms (Nystrom and Bruckner-Tuderman 2016) and patients’ life expectancy of usually highly reduced, because they almost invariably develop aggressive squamous cell carcinomas as the disease progresses (Fine et al. 2009; Montaudie et al. 2016; Kim et al. 2018).
A variety of preclinical studies attempted to deliver collagen VII to RDEB skin, underlining some crucial issues for future gene therapy in humans. Experiments included direct intradermal injection of LVs containing COL7A1 (Woodley et al. 2004b) and either local or systemic delivery of recombinant collagen VII protein (Woodley et al. 2004a; Remington et al. 2009). The outcome of the latter strategy pointed to its potential ability to safely reverse RDEB keratinocytes phenotype, at least temporarily because the half-life of collagen VII was estimated to be ∼1–2 months (Woodley et al. 2004a; Remington et al. 2009; Latella et al. 2017).
During the ex vivo gene therapy approaches pursued to tackle this ravaging disease, major hindrances were encountered in vector construction. The huge COL7A1 coding sequence (8833-nucleotides) makes its accommodation within γRVs a real challenge and negatively affects virus packaging, substantially reducing viral titer and limiting the efficiency of cell transduction (Christiano et al. 1994).
Marinkovich and colleagues recently reported early data from the first phase I/II ex vivo combined cell and gene therapy clinical trial for RDEB (Siprashvili et al. 2016). The trial involved transduction of cultured RDEB keratinocytes by means of γRVs bearing COL7A1, followed by generation of autologous transgenic epidermal grafts applied onto selected wounds of four patients. This approach appeared to be safe and well tolerated. In particular, no neutralizing immune response was observed to de novo synthesized collagen VII, no recombinant RV was detected in the bloodstream and no malignant transformation of transgenic keratinocytes occurred over a 12-month follow-up. To date, this study presented the most convincing evidence of deposition of functional collagen VII and restoration of anchoring fibril in the skin of RDEB patients. However, a variable, limited efficacy was reported, because at 12 months, ∼50% of the transplanted areas were covered by transgenic, functional epidermis. Three patients showed a decline of collagen VII expression and blistering at transplant sites after 6 months, whereas the fourth patient showed a decrease in collagen VII and anchoring fibrils at 12 months postgrafting, suggesting that the benefit was tapering off (Siprashvili et al. 2016) (ClinicalTrials.gov identifier: NCT01263379).
We have initiated a similar γRV-based phase I/II trial on RDEB (ClinicalTrials.gov identifier: NCT02984085) by means of autologous epidermal cultures transduced with the same type of γRVs used for gene therapy of JEB. This trial is still ongoing, but initial outcomes confirm the aforementioned results reported by Siprashvili et al. In contrast to gene therapy in JEB, our initial data point to mosaic epidermal regeneration in RDEB patients. Patches of transgenic epidermis expressing collagen VII are indeed interspersed with patches of untransduced, mutant epidermis (JW Bauer and M De Luca et al., unpubl.).
Marinkovich and colleagues speculate that the partial restoration of functional collagen VII might be caused by an age-related decline of the regenerative potential of epidermal cultures (Siprashvili et al. 2016). In our opinion, this is unlikely; autologous epidermal and limbal cultures permanently (more than a 35-year follow-up) regenerate the epidermis and the corneal epithelium in massive skin and ocular burns irrespective of the age of patients (Pellegrini et al. 1999b, 2013; De Luca et al. 2006; Rama et al. 2010). The first two JEB patients grafted with autologous transgenic epidermal sheets were 37 and 50 years old, respectively (Mavilio et al. 2006; Bauer et al. 2017). The first patient now had a 14-year follow-up and the transgenic epidermis is functioning well. In principle, the aforementioned findings could be ascribed to an insufficient number of epidermal stem cells retrieved from the skin biopsy and/or targeted by γRVs. Indeed, holoclone analysis was not performed in Siprashvili's study; therefore, stem cells were not quantified as they were in our JEB studies. Nevertheless, although we did evaluate the number of transduced stem cells by holoclone formation in our RDEB transgenic grafts, our clinical results are similar to those of Siprashvili and colleagues.
Assuming that proper gene correction of epidermal stem cells has occurred, the partial restoration of collagen VII might relate to intrinsic differences between JEB and RDEB clonogenic keratinocytes and potential competition between COL7A1-transduced and -untransduced cells. JEB, but not RDEB, keratinocytes initiate colonies with low efficiency and show rapid clonal conversion leading to premature stem-cell depletion and replicative senescence (Mavilio et al. 2006; De Rosa et al. 2019). It has been recently shown that the epidermal stem-cell depletion and premature replicative senescence characteristic of JEB are caused by the perturbation of the YAP/TAZ signaling pathway (De Rosa et al. 2019). Such perturbation is not found in RDEB keratinocytes, whose clonogenicity and growth potential are similar to those of normal keratinocytes. Strikingly, gene therapy of JEB keratinocytes completely restores the YAP/TAZ pathway and, consequently, fully rescues epidermal stem-cell function and proliferative potential, both in vitro and in vivo after transplantation (De Rosa et al. 2019). It follows that, in contrast to RDEB, genetically corrected clonogenic JEB keratinocytes must hold a strong selective advantage over their gene corrected counterparts. Because the transduction efficiency of clonogenic RDEB keratinocytes was in the range of 50%–70% in both Siprashvili's and our study, it is tantalizing to speculate that cell competition between transduced and untransduced RDEB epidermal stem cells within the graft could explain the mosaic composition of the regenerated epidermis and the progressive decrease of collagen VII observed in RDEB patients treated with transgenic cultures.
It follows that a decisive improvement in combined ex vivo cell and gene therapy for RDEB could ensue from a precise control of the number of transduced holoclones contained in the graft, together with the development of new vectors and/or transduction procedures allowing genetic correction of virtually 100% of the entire stem-cell population of primary RDEB cultures. This should minimize the competition between transduced and untransduced RDEB cells and assure long-term regeneration of a fully functional transgenic epidermis. Such optimization procedures are underway. A hitherto unsolved issue might relate to potential competition between transgenic interfollicular epidermis and untransduced keratinocytes contained in the bulge or the isthmus of hair follicles (Snippert et al. 2010), which might contribute to interfollicular epidermis (Blanpain and Fuchs 2009). However, the notion that stem cells in the hair follicles contribute to wound repair but not to homeostasis of the epidermis (Claudinot et al. 2005; Ito et al. 2005), argues against this possibility, would the transgenic epidermis have a complete engraftment upon transplantation, as shown with JEB.
Last, but not least, hurdles also relate to intrinsic challenges in understanding the mechanisms underlying many rare diseases, which complicates the design of efficacious therapies. Particularly in RDEB, where skin cleavage is deeper than in JEB (Fig. 2), the clinical success of epidermal cultures strictly relies on proper selection of the patients including their immunological picture (Siprashvili et al. 2016). It also depends on control of inflammation and microenvironment of the receiving wound bed (which varies between JEB and RDEB and can hamper the engraftment of the culture), preparation of the wound bed, and postoperative procedures (De Luca et al. 2006), all of which are beyond the scope of this chapter.
Other gene therapy approaches for RDEB involve the use of LV- and HSV-derived vectors in autologous dermal fibroblasts, which can also produce to collagen VII (Marinkovich et al. 1993). A phase I clinical trial used SIN-LVs for correcting RDEB fibroblasts by an ex vivo strategy. This protocol (referred to as FCX-007) was used on five patients and envisaged local intradermal injection of transgenic fibroblasts (ClinicalTrials.gov identifier: NCT02810951) (Marinkovich et al. 2018). FCX-007 proved to be well-tolerated and safe in patients during the 52-week long follow-up, with no product-related adverse events or immunologic response being reported. However, data on the efficacy of FCX-007 are not yet available. A similar trial (ClinicalTrials.gov identifier: NCT0249381), but using codon-optimized COL7A1, reported comparable safety and early efficacy outcomes, pointing to a partial clinical synthesis and deposition of collagen VII, but without formation of mature anchoring fibrils (Lwin et al. 2019).
Another strategy to treat RDEB used KB103, an off-the-shelf topical gel delivering COL7A1 via an attenuated HSV-1 vector through an in vivo approach (ClinicalTrials.gov identifier: NCT03536143). KB103, targeting keratinocytes and fibroblasts, was developed for both severe RDEB and milder DDEB. However, its therapeutic effect is predicted to be temporary because of the episomal nature of HSV, which does not integrate within the patient's genome and is therefore eliminated on epidermal renewal. Finally, a proposed combined cell and gene therapy approach for RDEB envisages the use of iPS as these cells can be selected in culture after gene correction and induced to differentiate into keratinocytes. In principle, a single, very well-characterized iPS cell line can be used to produce large amounts of transgenic epidermis (Tolar et al. 2011; Sebastiano et al. 2014; Wenzel et al. 2014). This approach has not yet reached the stage of an approved clinical trial.
Recently, immunomyeloablation followed by allogeneic bone marrow (BM) transplantation has been proposed for the treatment of RDEB (Wagner et al. 2010). Variable clinical benefits were reported (improved healing of the wounds and reduced frequency of daily medication). But anchoring fibrils were not restored, despite increased collagen VII deposition at the epidermal–dermal junction (Wagner et al. 2010). Such an increased deposition might ensue from BM-derived subclinical inflammation resulting in heparin-binding EGF up-regulation and a consequent increase in mutated collagen VII synthesis in host cells, as observed with fibroblasts or mesenchymal stromal cells (Wong et al. 2008; Haniffa et al. 2009). In fact, it is not even clear which cell type contained in the BM aspirates and which mechanism could determine the limited reported clinical benefits, and whether such benefits could not rather be ascribed to a better management of the lesions attained during the long hospitalization of the patients. Indeed, the poorly defined trophic, anti-inflammatory or immune modulatory effects of BM-derived cells observed in other clinical settings is still a quite controversial issue (Bianco et al. 2013; De Luca et al. 2019). In summary, BM transplantation is not curative and in the absence of a curative effect, it is hard to accept 30% lethality (Wagner et al. 2010) in children whose life expectancy is of a few decades. Clinical trials envisaging less aggressive myeloablation are ongoing (NCT00881556).
Netherton Syndrome
NS is a rare, recessively inherited skin disease with a prevalence of 1–9/1,000,000 and an estimated incidence of 1/200,000. NS is caused by mutations in SPINK5, the gene encoding the serine protease inhibitor LEKTI. LEKTI deficiency results in an increase of trypsin-like hydrolytic activity in the stratum corneum, leading to premature desquamation and consequent severe defects of the skin barrier (Chavanas et al. 2000; Bitoun et al. 2003). NS is characterized by defective keratinization, hair shaft defects, recurrent infections, atopy (tendency to develop allergic diseases), and a predisposition to skin malignancies. Prognosis is often poor, with a high risk of life-threatening complications such as bronchopneumonia, sepsis, and hypernatremic (high concentration of serum sodium) dehydration secondary to severe water loss attributable to the defective skin barrier. Approximately 10% of infants die within the first year of life (Bitoun et al. 2003). There is no cure for NS currently.
Preclinical studies aimed at tackling NS have involved the use of SIN-LVs carrying SPINK5 for the transduction of murine and human keratinocytes (Di et al. 2011). Data pointed to high transduction efficiency of epidermal cells with subsequent correction of LEKTI expression in vitro as well as in a murine/human skin xenograft model (Di et al. 2011). Similarly, an adenoassociated vector 2 was used to successfully restore LEKTI expression in ∼75% of NS keratinocytes (Roedl et al. 2011). Both approaches have not yet been approved for clinical trials.
A phase I/II clinical trial, using combined ex vivo cell and gene therapy was recently attempted (ClinicalTrials.gov identifier: NCT01545323) (Di et al. 2013). The trial made use of SIN-LV bearing a codon-optimized SPINK5 gene under the control of the involucrin (an early marker of keratinocyte terminal differentiation) (Watt 1983) promoter, conferring compartment-specific LEKTI expression to NS keratinocytes. However, as of today, no data about the outcome of this trial have been published (Di et al. 2013).
In conclusion, all the outlined studies underscore the complex challenges of designing effective molecular therapies for EB, NS, and other genetic skin diseases. These strategies can be made effective only with a better understanding of cellular and molecular mechanisms underlying each condition, which necessitates additional basic research.
GENE EDITING FOR GENODERMATOSES
Gene editing aims at redressing a definite molecular defect through insertion, deletion, modification, or replacement of DNA at site-specific locations within the cell genome (Zhang et al. 2014). Gene-editing effectors engineered artificial nucleases combined with DNA-binding motifs, encompassing meganucleases, zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the CRISPR/CRISPR-associated (Cas) protein 9 system (CRISPR/Cas9) (Gupta and Shukla 2017). These protein-based molecular tools work by inducing double-strand breaks at user-specified sites on selected genomes, which are rapidly recognized by the DNA repair systems of the cell. Double-helix fixing can be performed mainly by nonhomologous end joining (NHEJ) and homologous direct repair (HDR) (Lieber 2010; Zhang et al. 2014; Chang et al. 2017; Komor et al. 2017; Zaboikin et al. 2017). NHEJ is a frequent outcome in response to DNA rupture. It may introduce small insertions and deletions (InDels) at the loci of interest, hence inducing frameshift mutations, destroying the open reading frame of target genes and abolishing their expression. Instead, whenever a template DNA is either present or provided, HDR can be triggered, although its occurrence is rather rare and variable among different cell types. Its prevalent form is known as homologous recombination and can perform error-free DNA repair (Lieber 2010; Zhang et al. 2014; Chang et al. 2017; Komor et al. 2017; Zaboikin et al. 2017).
The CRISPR/Cas9 system has proven to be the most fitting tool for accomplishing both NHEJ and HDR strategies (Jinek et al. 2012; Mali et al. 2013a,b; Pennisi 2013). Ex vivo, CRISPR-based gene-editing approaches have recently been investigated in genodermatoses, although they are still at a rather preclinical stage (Bonafont et al. 2019). Studies were carried out mainly on primary keratinocytes or cell lines derived by primary cells, whereas only few made use of iPS cells (Shinkuma et al. 2016; Bonafont et al. 2019).
Recessive Forms of EB
More than 650 different mutations causing RDEB have been identified within COL7A1. The majority of them cluster in distinct hotspots and are typical of certain ethnicities or groups of families (Intong and Murrell 2012; Fine et al. 2014). A proposed gene-editing strategy involves the eviction of a region carrying multiple genetic defects with the aim of simultaneously correcting a whole set of mutations (Escamez et al. 2010; van den Akker et al. 2011; Wertheim-Tysarowska et al. 2012). For instance, the c.6527insC mutation in COL7A1 was tackled by a combined delivery of template DNA and TALEN, by adenoassociated virus and adenoviral vectors, respectively. This strategy achieved correction of the mutation via HDR and additionally restored the reading frame via NHEJ (Chamorro et al. 2016).
An ex vivo exon-skipping approach was also developed to precisely excise COL7A1 exon 80 carrying the same c.6527insC mutation. This used a dual single-guide, RNA-guided Cas9 nuclease delivered as a ribonucleoprotein complex through electroporation and succeeded in deleting exon 80 and restoring COL7A1 reading frame in RDEB keratinocytes, which were able to produce functional collagen VII (Bonafont et al. 2019). Alternatively, the CRISPR/Cas9 system was delivered in RDEB-immortalized keratinocytes by plasmid transfection; however, genetic and functional HDR-based correction of the aforementioned mutation only occurred in a few selected clones (Hainzl et al. 2017).
CRISPR/Cas9-mediated homology-directed repair, delivered ex vivo in primary RDEB keratinocytes and fibroblasts through integration-deficient LVs, tackled a c.189delG (p.Leu64Trpfs*40) null mutation in exon 2 of COL7A1. Efficient COL7A1 editing was achieved, showing rescue of functional collagen VII and anchoring fibrils in vivo by means of transgenic skin xenografts (Izmiryan et al. 2018). An iPS-based approach was attempted to tackle a c.1837 C > T mutation on exon 14 of COL7A1. RDEB iPS cells were genetically corrected by means of TALEN-mediated HDR (Osborn et al. 2013). Finally, HDR-based gene editing restored laminin 332 expression in LAMB3-dependent JEB immortalized keratinocytes, but its efficiency was too low for clinical translation (Benati et al. 2018).
Dominant Forms of EB
Dominant forms of EB encompass most EBS and some DEB subtypes. Gene-editing techniques have been implemented for DDEB, covering TALEN nucleases and CRISPR/Cas9-mediated NHEJ approaches aimed at correcting c.8068_8084delinsGA mutation within COL7A1 (Shinkuma et al. 2016). A site-specific mutagenesis inducing NHEJ was performed on iPS generated from DDEB fibroblasts; gene-edited iPS cells produced keratinocytes and fibroblasts able to synthetize and secrete functional collagen VII (Shinkuma et al. 2016). A similar approach targeting RDEB was developed by combining CRISPR/Cas9 with iPS technology (Webber et al. 2016). A dominant hotspot mutation within the KRT14 gene, causing a generalized severe EBS was corrected using a double-nicking strategy (Kocher et al. 2017). This HDR-based approach involved CRISPR/Cas9 correction of a patient-derived immortalized EBS keratinocytes. This strategy resulted in a transfection efficiency of about 30% associated with a 16% pathogenic allele correction, assuming a wild-type/mutated allele ratio of 50/50 within treated cells (Kocher et al. 2017).
Other Genodermatoses
Epidermolytic ichthyosis (EI) is a rare autosomal genodermatosis, with an incidence of 1:100,000–400,000, characterized by blistering and subsequent reactive scaling of the skin (Müller et al. 2006). EI is inherited predominantly with a dominant pattern, although recessive forms have been reported. Cytoskeletal fragility of suprabasal epidermal cells is caused by dominant-negative mutations in either KRT1 or KRT10. As keratin 1 and keratin 10 are crucial cytoskeleton constituents, EI results in major impairment of intermediate filaments (Cheng et al. 1992; Chipev et al. 1992; Oji et al. 2010). To date, treating EI remains a challenge. Palliative treatments and medications aim at removing the excess of thickened skin layers. Recently, TALEN-based gene editing was performed with the aim of inactivating mutant KRT10 by introducing a frameshift mutation upstream of a premature termination codon in cultured EI keratinocytes. Despite phenotype restoration of selected clones, the therapeutic efficiency was low (3.7%) with high disruption rates on the wild-type allele (March et al. 2019).
In summary, proof-of-concept for the use of gene-editing technology in gene therapy of genodermatoses has been obtained, but several technical hurdles must be overcome before this technology can reach its full potential.
Limitations of CRISPR/Cas9
Unspecific cleavage of undesired sites, known as off-target activity, is the foremost concern of the CRISPR/Cas9 system (Zhang et al. 2017; Vakulskas and Behlke 2019). Classic SpCas9 could theoretically target a unique sequence of DNA in most genomes but it may tolerate a few mismatches between the gRNA and its complementary strand (Jinek et al. 2012; Fu et al. 2013; Hsu et al. 2013). Off-target effects are most likely to occur at sites with sequence similarity to the desired target site. However, to date, there are no fully reliable means to predict at which sites and with what frequency they will take place. This matter calls for special attention, as the introduction of a double-strand break or even a nick at the wrong off-target site can lead to unexpected mutations or rearrangements that may culminate in oncogenesis (Cullot et al. 2019; Thomas et al. 2019). Therefore, novel assays for genome-wide identification of off-targets have been proposed (Bae et al. 2014a,b; Park et al. 2015). The development of optimized mutant Cas9 nucleases aimed at overcoming off-target effects is also being actively pursued (Kleinstiver et al. 2016; Slaymaker et al. 2016).
A second major drawback of CRISPR/Cas9 concerns the extremely low efficiency of HDR-mediated DNA repair of double-strand breaks in many primary human cells (Ye et al. 2018; Cullot et al. 2019). Attempts to introduce specific alterations at a genomic site with HDR, which is active only in proliferating cells in S or G2 phase, can be compromised by NHEJ, which is active in all cells and in all phases of the cell cycle. This greatly impairs the generation of cells in which only one allele contains a desired HDR-mediated edit or cells in which both alleles contain a desired HDR-mediated edit, because NHEJ is a more frequent outcome on DNA cleavage. Currently, this pitfall poses a serious limitation to the clinical translation of such technology to human epidermis, because the highly efficient correction of epidermal stem cells is a fundamental prerequisite for long-lasting therapeutic effects of combined ex vivo cell and gene therapy. In principle, this hurdle could be overridden by the use of iPS cells, as a single iPS cell can theoretically be used to produce large amounts of cultured epidermis. Alternative approaches, aimed at increasing HDR efficiency (while suppressing NHEJ), include the use of small molecular inhibitors of NHEJ (Srivastava et al. 2012; Tomkinson and Sallmyr 2013; Robert et al. 2015; Vartak and Raghavan 2015; Yu et al. 2015), gene silencing (Chu et al. 2015), and cell-cycle synchronization (Lin et al. 2014).
Bringing all these considerations together, gene-editing technologies, in particular, CRISPR/Cas9, hold the potential to revolutionize combined cell and gene therapy for genodermatoses, provided that current drawbacks, such as low-HDR efficiency and off-target effects, can be overcome.
CONCLUSIONS
Major breakthroughs concerning advanced therapies of incurable inherited diseases or injuries have been achieved thanks to the considerable progress made within the three main fields of regenerative medicine, namely, cell therapy, gene therapy, and tissue engineering. Nevertheless, ATMPs must be granted authorization from regulatory authorities before these advances can be progressed into accessible therapies. Rigorous scientific research, as well as stringent assessments of quality, safety, and efficacy, are essential for these treatments to become a reality for the patients in need.
Regarding skin diseases, the early foundations of regenerative medicine approaches for treatment were laid more than three decades ago, on the first successful cell therapy achieved by means of cultured cells. Autologous keratinocyte cultures grafts were indeed capable of rescuing the lives of two brothers affected by virtually whole-body, full-thickness skin burns (Gallico et al. 1984). However, it was only at the end of the last century (Coulombe et al. 1991) that advances in molecular genetics enabled the detection and screening of mutations residing within known genes coding for structural proteins associated with the different inherited skin conditions (Has et al. 2018). The rapid pace of exome and genome sequencing led to the identification of novel disease-causing mutations, allowing the precise classification of the many diverse subtypes encompassed under EB and other disorders of the skin.
Meanwhile, gene therapy protocols began to enter the clinical panorama of rare inherited conditions, with the aim of correcting their underlying molecular defects (Aiuti et al. 2009). Success in tackling congenital immunodeficiencies or metabolic disorders by transgenic hematopoietic stem cells (Aiuti et al. 2009, 2013; Biffi et al. 2013) was crucial for the rational profiling of the feasibility of this regenerative medicine approach and the recent development of CAR-T-mediated cancer immunotherapy (Sadelain et al. 2017; June and Sadelain 2018). In fact, these studies paved the way for tackling diseases affecting other tissues and marked the starting point for the development of combined cell and gene therapy for genodermatoses.
From the various different methods grew a successful treatment for the skin lesions plaguing JEB patients (Mavilio et al. 2006; De Rosa et al. 2014; Bauer et al. 2017; Hirsch et al. 2017). However, it was soon clear that, by reason of the complexity and heterogeneity of inherited skin diseases, even within a single genodermatosis, such as EB, a one-size-fits-all solution for every clinical case is impractical. Instead, what can be envisioned is a more tailored approach, based on the specific features of the patient in need. A combination of successful strategies, approaches, and advanced therapies will be needed to attain improved living conditions or even a permanent treatment for the people suffering from these devastating diseases. Indeed, the long and winding road leading to this final ambition is challenging but must be pursued.
COMPETING INTEREST STATEMENT
Michele De Luca is a co-founder and member of the Board of Directors of Holostem Terapie Avanzate, s.r.l, Modena, Italy and consultant for J-TEC, Japan Tissue Engineering, Ltd.
ACKNOWLEDGMENTS
This work was supported by the Regione Emilia-Romagna, POR-FESR (E8IJ10000120007 and E92I16000220005).
Footnotes
Editors: Cristina Lo Celso, Kristy Red-Horse, and Fiona M. Watt
Additional Perspectives on Stem Cells: From Biological Principles to Regenerative Medicine available at www.cshperspectives.org
REFERENCES
- Abaci HE, Guo Z, Doucet Y, Jacków J, Christiano A. 2017. Next generation human skin constructs as advanced tools for drug development. Exp Biol Med (Maywood) 242: 1657–1668. 10.1177/1535370217712690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, Callegaro L, Scaramuzza S, Andolfi G, Mirolo M, Brigida I, et al. 2009. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med 360: 447–458. 10.1056/NEJMoa0805817 [DOI] [PubMed] [Google Scholar]
- Aiuti A, Biasco L, Scaramuzza S, Ferrua F, Cicalese MP, Baricordi C, Dionisio F, Calabria A, Giannelli S, Castiello MC, et al. 2013. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott–Aldrich syndrome. Science 341: 1233151 10.1126/science.1233151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiuti A, Roncarolo MG, Naldini L. 2017. Gene therapy for ADA-SCID, the first marketing approval of an ex vivo gene therapy in Europe: paving the road for the next generation of advanced therapy medicinal products. EMBO Mol Med 9: 737–740. 10.15252/emmm.201707573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artusi S, Miyagawa Y, Goins WF, Cohen JB, Glorioso JC. 2018. Herpes simplex virus vectors for gene transfer to the central nervous system. Diseases 6: 74 10.3390/diseases6030074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwood SW, Edel MJ. 2019. iPS-cell technology and the problem of genetic instability—can it ever be safe for clinical use? J Clin Med 8: 288 10.3390/jcm8030288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Kweon J, Kim HS, Kim JS. 2014a. Microhomology-based choice of Cas9 nuclease target sites. Nat Methods 11: 705–706. 10.1038/nmeth.3015 [DOI] [PubMed] [Google Scholar]
- Bae S, Park J, Kim JS. 2014b. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30: 1473–1475. 10.1093/bioinformatics/btu048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon Y, Green H. 1987. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci 84: 2302–2306. 10.1073/pnas.84.8.2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer JW, Koller J, Murauer EM, De Rosa L, Enzo E, Carulli S, Bondanza S, Recchia A, Muss W, Diem A, et al. 2017. Closure of a large chronic wound through transplantation of gene-corrected epidermal stem cells. J Invest Dermatol 137: 778–781. 10.1016/j.jid.2016.10.038 [DOI] [PubMed] [Google Scholar]
- Belokhvostova D, Berzanskyte I, Cujba AM, Jowett G, Marshall L, Prueller J, Watt FM. 2018. Homeostasis, regeneration and tumour formation in the mammalian epidermis. Int J Dev Biol 62: 571–582. 10.1387/ijdb.170341fw [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benati D, Miselli F, Cocchiarella F, Patrizi C, Carretero M, Baldassarri S, Ammendola V, Has C, Colloca S, Del Rio M, et al. 2018. CRISPR/Cas9-mediated in situ correction of LAMB3 gene in keratinocytes derived from a junctional epidermolysis bullosa patient. Mol Ther 26: 2592–2603. 10.1016/j.ymthe.2018.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P, Cao X, Frenette PS, Mao JJ, Robey PG, Simmons PJ, Wang CY. 2013. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat Med 19: 35–42. 10.1038/nm.3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A, De Palma M, Quattrini A, Del Carro U, Amadio S, Visigalli I, Sessa M, Fasano S, Brambilla R, Marchesini S, et al. 2004. Correction of metachromatic leukodystrophy in the mouse model by transplantation of genetically modified hematopoietic stem cells. J Clin Invest 113: 1118–1129. 10.1172/JCI200419205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A, Montini E, Lorioli L, Cesani M, Fumagalli F, Plati T, Baldoli C, Martino S, Calabria A, Canale S, et al. 2013. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 341: 1233158 10.1126/science.1233158 [DOI] [PubMed] [Google Scholar]
- Bitoun E, Micheloni A, Lamant L, Bonnart C, Tartaglia-Polcini A, Cobbold C, Al Saati T, Mariotti F, Mazereeuw-Hautier J, Boralevi F, et al. 2003. LEKTI proteolytic processing in human primary keratinocytes, tissue distribution and defective expression in Netherton syndrome. Hum Mol Genet 12: 2417–2430. 10.1093/hmg/ddg247 [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. 2009. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol 10: 207–217. 10.1038/nrm2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonafont J, Mencía Á, García M, Torres R, Rodríguez S, Carretero M, Chacón-Solano E, Modamio-Høybjør S, Marinas L, León C, et al. 2019. Clinically relevant correction of recessive dystrophic epidermolysis bullosa by dual sgRNA CRISPR/Cas9-mediated gene editing. Mol Ther 27: 986–998. 10.1016/j.ymthe.2019.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers WJ, Olschowka JA, Federoff HJ. 2003. Immune responses to replication-defective HSV-1 type vectors within the CNS: implications for gene therapy. Gene Ther 10: 941–945. 10.1038/sj.gt.3302047 [DOI] [PubMed] [Google Scholar]
- Braun CJ, Boztug K, Paruzynski A, Witzel M, Schwarzer A, Rothe M, Modlich U, Beier R, Gohring G, Steinemann D, et al. 2014. Gene therapy for Wiskott–Aldrich syndrome—long-term efficacy and genotoxicity. Sci Transl Med 6: 227ra33 10.1126/scitranslmed.3007280 [DOI] [PubMed] [Google Scholar]
- Bukrinskaya AG, Ghorpade A, Heinzinger NK, Smithgall TE, Lewis RE, Stevenson M. 1996. Phosphorylation-dependent human immunodeficiency virus type 1 infection and nuclear targeting of viral DNA. Proc Natl Acad Sci 93: 367–371. 10.1073/pnas.93.1.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoglio C, Maruggi G, Bartholomae C, Malani N, Pellin D, Cocchiarella F, Magnani Z, Ciceri F, Ambrosi A, von Kalle C, et al. 2010. High-definition mapping of retroviral integration sites defines the fate of allogeneic T cells after donor lymphocyte infusion. PLoS ONE 5: e15688 10.1371/journal.pone.0015688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazza A, Moiani A, Mavilio F. 2013. Mechanisms of retroviral integration and mutagenesis. Hum Gene Ther 24: 119–131. 10.1089/hum.2012.203 [DOI] [PubMed] [Google Scholar]
- Cavazzana M, Bushman FD, Miccio A, André-Schmutz I, Six E. 2019. Gene therapy targeting haematopoietic stem cells for inherited diseases: progress and challenges. Nat Rev Drug Discov 18: 447–462. 10.1038/s41573-019-0020-9 [DOI] [PubMed] [Google Scholar]
- Chamorro C, Mencía A, Almarza D, Duarte B, Büning H, Sallach J, Hausser I, Del Río M, Larcher F, Murillas R. 2016. Gene editing for the efficient correction of a recurrent COL7A1 mutation in recessive dystrophic epidermolysis bullosa keratinocytes. Mol Ther Nucleic Acids 5: e307 10.1038/mtna.2016.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T, Yee JK. 2012. General principles of retrovirus vector design. Methods Enzymol 507: 1–14. 10.1016/B978-0-12-386509-0.00001-6 [DOI] [PubMed] [Google Scholar]
- Chang HHY, Pannunzio NR, Adachi N, Lieber MR. 2017. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 18: 495–506. 10.1038/nrm.2017.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavanas S, Bodemer C, Rochat A, Hamel-Teillac D, Ali M, Irvine AD, Bonafé JL, Wilkinson J, Taïeb A, Barrandon Y, et al. 2000. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat Genet 25: 141–142. 10.1038/75977 [DOI] [PubMed] [Google Scholar]
- Cheng J, Syder AJ, Yu QC, Letal A, Paller AS, Fuchs E. 1992. The genetic basis of epidermolytic hyperkeratosis: a disorder of differentiation-specific epidermal keratin genes. Cell 70: 811–819. 10.1016/0092-8674(92)90314-3 [DOI] [PubMed] [Google Scholar]
- Chipev CC, Korge BP, Markova N, Bale SJ, DiGiovanna JJ, Compton JG, Steinert PM. 1992. A leucine→proline mutation in the H1 subdomain of keratin 1 causes epidermolytic hyperkeratosis. Cell 70: 821–828. 10.1016/0092-8674(92)90315-4 [DOI] [PubMed] [Google Scholar]
- Christiano AM, Greenspan DS, Lee S, Uitto J. 1994. Cloning of human type VII collagen. Complete primary sequence of the α 1(VII) chain and identification of intragenic polymorphisms. J Biol Chem 269: 20256–20262. [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R. 2015. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33: 543–548. 10.1038/nbt.3198 [DOI] [PubMed] [Google Scholar]
- Claudinot S, Nicolas M, Oshima H, Rochat A, Barrandon Y. 2005. Long-term renewal of hair follicles from clonogenic multipotent stem cells. Proc Natl Acad Sci 102: 14677–14682. 10.1073/pnas.0507250102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E, Doupé DP, Klein AM, Winton DJ, Simons BD, Jones PH. 2007. A single type of progenitor cell maintains normal epidermis. Nature 446: 185–189. 10.1038/nature05574 [DOI] [PubMed] [Google Scholar]
- Cornetta K, Reeves L, Cross S. 2008. Production of retroviral vectors for clinical use. Methods Mol Biol 433: 17–32. 10.1007/978-1-59745-237-3_2 [DOI] [PubMed] [Google Scholar]
- Coulombe PA, Hutton ME, Letal A, Hebert A, Paller AS, Fuchs E. 1991. Point mutations in human keratin 14 genes of epidermolysis bullosa simplex patients: genetic and functional analyses. Cell 66: 1301–1311. 10.1016/0092-8674(91)90051-Y [DOI] [PubMed] [Google Scholar]
- Cullot G, Boutin J, Toutain J, Prat F, Pennamen P, Rooryck C, Teichmann M, Rousseau E, Lamrissi-Garcia I, Guyonnet-Duperat V, et al. 2019. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat Commun 10: 1136 10.1038/s41467-019-09006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das BB, Sahoo S. 2004. Dystrophic epidermolysis bullosa. J Perinatol 24: 41–47. 10.1038/sj.jp.7211019 [DOI] [PubMed] [Google Scholar]
- Dellambra E, Vailly J, Pellegrini G, Bondanza S, Golisano O, Macchia C, Zambruno G, Meneguzzi G, De Luca M. 1998. Corrective transduction of human epidermal stem cells in laminin-5-dependent junctional epidermolysis bullosa. Hum Gene Ther 9: 1359–1370. 10.1089/hum.1998.9.9-1359 [DOI] [PubMed] [Google Scholar]
- De Luca M, Pellegrini G, Green H. 2006. Regeneration of squamous epithelia from stem cells of cultured grafts. Regen Med 1: 45–57. 10.2217/17460751.1.1.45 [DOI] [PubMed] [Google Scholar]
- De Luca M, Aiuti A, Cossu G, Parmar M, Pellegrini G, Robey PG. 2019. Advances in stem cell research and therapeutic development. Nat Cell Biol 21: 801–811. 10.1038/s41556-019-0344-z [DOI] [PubMed] [Google Scholar]
- De Ravin SS, Wu X, Moir S, Anaya-O'Brien S, Kwatemaa N, Littel P, Theobald N, Choi U, Su L, Marquesen M, et al. 2016. Lentiviral hematopoietic stem cell gene therapy for X-linked severe combined immunodeficiency. Sci Transl Med 8: 335ra57 10.1126/scitranslmed.aad8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa L, Carulli S, Cocchiarella F, Quaglino D, Enzo E, Franchini E, Giannetti A, De Santis G, Recchia A, Pellegrini G, et al. 2014. Long-term stability and safety of transgenic cultured epidermal stem cells in gene therapy of junctional epidermolysis bullosa. Stem Cell Rep 2: 1–8. 10.1016/j.stemcr.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa L, Secone Seconetti A, De Santis G, Pellacani G, Hirsch T, Rothoeft T, Teig N, Pellegrini G, Bauer JW, De Luca M. 2019. Laminin 332–dependent YAP dysregulation depletes epidermal stem cells in junctional epidermolysis bullosa. Cell Rep 27: 2036–2049.e6. 10.1016/j.celrep.2019.04.055 [DOI] [PubMed] [Google Scholar]
- Di WL, Larcher F, Semenova E, Talbot GE, Harper JI, Del Rio M, Thrasher AJ, Qasim W. 2011. Ex-vivo gene therapy restores LEKTI activity and corrects the architecture of Netherton syndrome–derived skin grafts. Mol Ther 19: 408–416. 10.1038/mt.2010.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di WL, Mellerio JE, Bernadis C, Harper J, Abdul-Wahab A, Ghani S, Chan L, Martinez-Queipo M, Hara H, McNicol AM, et al. 2013. Phase I study protocol for ex vivo lentiviral gene therapy for the inherited skin disease, Netherton syndrome. Hum Gene Ther Clin Dev 24: 182–190. 10.1089/humc.2013.195 [DOI] [PubMed] [Google Scholar]
- Di Iorio E, Barbaro V, Ruzza A, Ponzin D, Pellegrini G, De Luca M. 2005. Isoforms of ΔNp63 and the migration of ocular limbal cells in human corneal regeneration. Proc Natl Acad Sci 102: 9523–9528. 10.1073/pnas.0503437102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droz-Georget Lathion S, Rochat A, Knott G, Recchia A, Martinet D, Benmohammed S, Grasset N, Zaffalon A, Besuchet Schmutz N, Savioz-Dayer E, et al. 2015. A single epidermal stem cell strategy for safe ex vivo gene therapy. EMBO Mol Med 7: 380–393. 10.15252/emmm.201404353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne H, Hadj-Rabia S, Taieb C, Bodemer C. 2015. Development and validation of an epidermolysis bullosa family/parental burden score. Br J Dermatol 173: 1405–1410. 10.1111/bjd.14072 [DOI] [PubMed] [Google Scholar]
- Edelstein ML, Abedi MR, Wixon J. 2007. Gene therapy clinical trials worldwide to 2007—an update. J Gene Med 9: 833–842. 10.1002/jgm.1100 [DOI] [PubMed] [Google Scholar]
- Escamez MJ, Cuadrado-Corrales N, Garcia M, Sanchez-Jimeno C, Illera N, Lopez-Martinez MA, Trujillo-Tiebas MJ, Ayuso C, Del Rio M. 2010. Novel human pathological mutations. Gene symbol: COL7A1. Disease: epidermolysis bullosa dystrophica. Hum Genet 127: 121. [PubMed] [Google Scholar]
- Fine JD. 2010. Inherited epidermolysis bullosa. Orphanet J Rare Dis 5: 12 10.1186/1750-1172-5-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine JD. 2016. Epidemiology of inherited epidermolysis bullosa based on incidence and prevalence estimates from the National Epidermolysis Bullosa Registry. JAMA Dermatol 152: 1231–1238. 10.1001/jamadermatol.2016.2473 [DOI] [PubMed] [Google Scholar]
- Fine JD, Johnson LB, Weiner M, Suchindran C. 2005. Impact of inherited epidermolysis bullosa on parental interpersonal relationships, marital status and family size. Br J Dermatol 152: 1009–1014. 10.1111/j.1365-2133.2004.06339.x [DOI] [PubMed] [Google Scholar]
- Fine JD, Eady RA, Bauer EA, Bauer JW, Bruckner-Tuderman L, Heagerty A, Hintner H, Hovnanian A, Jonkman MF, Leigh I, et al. 2008. The classification of inherited epidermolysis bullosa (EB): report of the Third International Consensus Meeting on Diagnosis and Classification of EB. J Am Acad Dermatol 58: 931–950. 10.1016/j.jaad.2008.02.004 [DOI] [PubMed] [Google Scholar]
- Fine JD, Johnson LB, Weiner M, Li KP, Suchindran C. 2009. Epidermolysis bullosa and the risk of life-threatening cancers: the National EB Registry experience, 1986–2006. J Am Acad Dermatol 60: 203–211. 10.1016/j.jaad.2008.09.035 [DOI] [PubMed] [Google Scholar]
- Fine JD, Bruckner-Tuderman L, Eady RA, Bauer EA, Bauer JW, Has C, Heagerty A, Hintner H, Hovnanian A, Jonkman MF, et al. 2014. Inherited epidermolysis bullosa: updated recommendations on diagnosis and classification. J Am Acad Dermatol 70: 1103–1126. 10.1016/j.jaad.2014.01.903 [DOI] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31: 822–826. 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. 2016. Epithelial skin biology: three decades of developmental biology, a hundred questions answered and a thousand new ones to address. Curr Top Dev Biol 116: 357–374. 10.1016/bs.ctdb.2015.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galach M, Utikal J. 2011. From skin to the treatment of diseases—the possibilities of iPS cell research in dermatology. Exp Dermatol 20: 523–528. 10.1111/j.1600-0625.2011.01282.x [DOI] [PubMed] [Google Scholar]
- Gallico GG III, O'Connor NE, Compton CC, Kehinde O, Green H. 1984. Permanent coverage of large burn wounds with autologous cultured human epithelium. N Engl J Med 311: 448–451. 10.1056/NEJM198408163110706 [DOI] [PubMed] [Google Scholar]
- Galy A, Roncarolo MG, Thrasher AJ. 2008. Development of lentiviral gene therapy for Wiskott–Aldrich syndrome. Expert Opin Biol Ther 8: 181–190. 10.1517/14712598.8.2.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis C, Syed F, Petrova A, Abdul-Wahab A, Lwin SM, Farzaneh F, Chan L, Ghani S, Fleck RA, Glover L, et al. 2016. Lentiviral engineered fibroblasts expressing codon-optimized COL7A1 restore anchoring fibrils in RDEB. J Invest Dermatol 136: 284–292. 10.1038/JID.2015.364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales KAU, Fuchs E. 2017. Skin and its regenerative powers: an alliance between stem cells and their niche. Dev Cell 43: 387–401. 10.1016/j.devcel.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H. 2008. The birth of therapy with cultured cells. Bioessays 30: 897–903. 10.1002/bies.20797 [DOI] [PubMed] [Google Scholar]
- Guhr A, Kobold S, Seltmann S, Seiler Wulczyn AEM, Kurtz A, Löser P. 2018. Recent trends in research with human pluripotent stem cells: impact of research and use of cell lines in experimental research and clinical trials. Stem Cell Rep 11: 485–496. 10.1016/j.stemcr.2018.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SK, Shukla P. 2017. Gene editing for cell engineering: trends and applications. Crit Rev Biotechnol 37: 672–684. 10.1080/07388551.2016.1214557 [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, Radford I, Villeval JL, Fraser CC, Cavazzana-Calvo M, et al. 2003. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med 348: 255–256. 10.1056/NEJM200301163480314 [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, Clappier E, Caccavelli L, Delabesse E, Beldjord K, et al. 2008. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest 118: 3132–3142. 10.1172/JCI35700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey Abina S, Gaspar HB, Blondeau J, Caccavelli L, Charrier S, Buckland K, Picard C, Six E, Himoudi N, Gilmour K, et al. 2015. Outcomes following gene therapy in patients with severe Wiskott–Aldrich syndrome. JAMA 313: 1550–1563. 10.1001/jama.2015.3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainzl S, Peking P, Kocher T, Murauer EM, Larcher F, Del Rio M, Duarte B, Steiner M, Klausegger A, Bauer JW, et al. 2017. COL7A1 editing via CRISPR/Cas9 in recessive dystrophic epidermolysis bullosa. Mol Ther 25: 2573–2584. 10.1016/j.ymthe.2017.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammersen J, Has C, Naumann-Bartsch N, Stachel D, Kiritsi D, Söder S, Tardieu M, Metzler M, Bruckner-Tuderman L, Schneider H. 2016. Genotype, clinical course, and therapeutic decision making in 76 infants with severe generalized junctional epidermolysis bullosa. J Invest Dermatol 136: 2150–2157. 10.1016/j.jid.2016.06.609 [DOI] [PubMed] [Google Scholar]
- Haniffa MA, Collin MP, Buckley CD, Dazzi F. 2009. Mesenchymal stem cells: the fibroblasts’ new clothes? Haematologica 94: 258–263. 10.3324/haematol.13699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Has C, Nyström A, Saeidian AH, Bruckner-Tuderman L, Uitto J. 2018. Epidermolysis bullosa: molecular pathology of connective tissue components in the cutaneous basement membrane zone. Matrix Biol 71-72: 313–329. 10.1016/j.matbio.2018.04.001 [DOI] [PubMed] [Google Scholar]
- Haselhorst D, Kaye JF, Lever AM. 1998. Development of cell lines stably expressing human immunodeficiency virus type 1 proteins for studies in encapsidation and gene transfer. J Gen Virol 79: 231–237. 10.1099/0022-1317-79-2-231 [DOI] [PubMed] [Google Scholar]
- Hirsch T, Rothoeft T, Teig N, Bauer JW, Pellegrini G, De Rosa L, Scaglione D, Reichelt J, Klausegger A, Kneisz D, et al. 2017. Regeneration of the entire human epidermis using transgenic stem cells. Nature 551: 327–332. 10.1038/nature24487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn HM, Tidman MJ. 2002. Quality of life in epidermolysis bullosa. Clin Exp Dermatol 27: 707–710. 10.1046/j.1365-2230.2002.01121.x [DOI] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, Brugman MH, Pike-Overzet K, Chatters SJ, de Ridder D, et al. 2008. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest 118: 3143–3150. 10.1172/JCI35798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31: 827–832. 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Ferris A, Larochelle A, Krouse AE, Metzger ME, Donahue RE, Hughes SH, Dunbar CE. 2007. Transduction of rhesus macaque hematopoietic stem and progenitor cells with avian sarcoma and leukosis virus vectors. Hum Gene Ther 18: 691–700. 10.1089/hum.2006.175 [DOI] [PubMed] [Google Scholar]
- Hu J, Renaud G, Golmes T, Ferris A, Hendrie PC, Donahue RE, Hughes SH, Wolfsberg TG, Russell DW, Dunbar CE. 2008. Reduced genotoxicity of avian sarcoma leukosis virus vectors in rhesus long-term repopulating cells compared to standard murine retrovirus vectors. Mol Ther 16: 1617–1623. 10.1038/mt.2008.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intong LR, Murrell DF. 2012. Inherited epidermolysis bullosa: new diagnostic criteria and classification. Clin Dermatol 30: 70–77. 10.1016/j.clindermatol.2011.03.012 [DOI] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G. 2005. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med 11: 1351–1354. 10.1038/nm1328 [DOI] [PubMed] [Google Scholar]
- Itoh M, Kiuru M, Cairo MS, Christiano AM. 2011. Generation of keratinocytes from normal and recessive dystrophic epidermolysis bullosa-induced pluripotent stem cells. Proc Natl Acad Sci 108: 8797–8802. 10.1073/pnas.1100332108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izmiryan A, Ganier C, Bovolenta M, Schmitt A, Mavilio F, Hovnanian A. 2018. Ex vivo COL7A1 correction for recessive dystrophic epidermolysis bullosa using CRISPR/Cas9 and homology-directed repair. Mol Ther Nucleic Acids 12: 554–567. 10.1016/j.omtn.2018.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- June CH, Sadelain M. 2018. Chimeric antigen receptor therapy. N Engl J Med 379: 64–73. 10.1056/NEJMra1706169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann KB, Brendel C, Suerth JD, Mueller-Kuller U, Chen-Wichmann L, Schwäble J, Pahujani S, Kunkel H, Schambach A, Baum C, et al. 2013. α Retroviral vector-mediated gene therapy for X-CGD: functional correction and lack of aberrant splicing. Mol Ther 21: 648–661. 10.1038/mt.2012.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik R, Ratner L. 2004. Role of human immunodeficiency virus type 1 matrix phosphorylation in an early postentry step of virus replication. J Virol 78: 2319–2326. 10.1128/JVI.78.5.2319-2326.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Li M, Intong-Wheeler LRA, Tran K, Marucci D, Murrell DF. 2018. Epidemiology and outcome of squamous cell carcinoma in epidermolysis bullosa in Australia and New Zealand. Acta Derm Venereol 98: 70–76. 10.2340/00015555-2781 [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. 2016. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529: 490–495. 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher T, Peking P, Klausegger A, Murauer EM, Hofbauer JP, Wally V, Lettner T, Hainzl S, Ablinger M, Bauer JW, et al. 2017. Cut and paste: efficient homology-directed repair of a dominant negative KRT14 mutation via CRISPR/Cas9 nickases. Mol Ther 25: 2585–2598. 10.1016/j.ymthe.2017.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Badran AH, Liu DR. 2017. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168: 20–36. 10.1016/j.cell.2016.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraunus J, Schaumann DH, Meyer J, Modlich U, Fehse B, Brandenburg G, von Laer D, Klump H, Schambach A, Bohne J, et al. 2004. Self-inactivating retroviral vectors with improved RNA processing. Gene Ther 11: 1568–1578. 10.1038/sj.gt.3302309 [DOI] [PubMed] [Google Scholar]
- Kretzschmar K, Watt FM. 2014. Markers of epidermal stem cell subpopulations in adult mammalian skin. Cold Spring Harb Perspect Med 4: a013631 10.1101/cshperspect.a013631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimer M, Lanschuetzer CM, Diem A, Bauer JW. 2010. Herlitz junctional epidermolysis bullosa. Dermatol Clin 28: 55–60. 10.1016/j.det.2009.10.006 [DOI] [PubMed] [Google Scholar]
- Latella MC, Cocchiarella F, De Rosa L, Turchiano G, Gonçalves M, Larcher F, De Luca M, Recchia A. 2017. Correction of recessive dystrophic epidermolysis bullosa by transposon-mediated integration of COL7A1 in transplantable patient-derived primary keratinocytes. J Invest Dermatol 137: 836–844. 10.1016/j.jid.2016.11.038 [DOI] [PubMed] [Google Scholar]
- Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, Collins F, Shinn P, Leipzig J, Hannenhalli S, et al. 2006. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog 2: e60 10.1371/journal.ppat.0020060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, He XC, Li L. 2019. Hematopoietic stem cells: self-renewal and expansion. Curr Opin Hematol 26: 258–265. 10.1097/MOH.0000000000000506 [DOI] [PubMed] [Google Scholar]
- Lieber MR. 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79: 181–211. 10.1146/annurev.biochem.052308.093131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, Doudna JA. 2014. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3: e04766 10.7554/eLife.04766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lwin SM, Syed F, Di WL, Kadiyirire T, Liu L, Guy A, Petrova A, Abdul-Wahab A, Reid F, Phillips R, et al. 2019. Safety and early efficacy outcomes for lentiviral fibroblast gene therapy in recessive dystrophic epidermolysis bullosa. JCI Insight 4: 126243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. 2013a. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31: 833–838. 10.1038/nbt.2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013b. RNA-guided human genome engineering via Cas9. Science 339: 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Hauber J, Fenrick R, Cullen BR. 1988. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature 335: 181–183. 10.1038/335181a0 [DOI] [PubMed] [Google Scholar]
- Manservigi R, Argnani R, Marconi P. 2010. HSV recombinant vectors for gene therapy. Open Virol J 4: 123–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March OP, Lettner T, Klausegger A, Ablinger M, Kocher T, Hainzl S, Peking P, Lackner N, Rajan N, Hofbauer JP, et al. 2019. Gene editing-mediated disruption of epidermolytic ichthyosis-associated KRT10 alleles restores filament stability in keratinocytes. J Invest Dermatol 139: 1699–1710. [DOI] [PubMed] [Google Scholar]
- Marconi P, Argnani R, Berto E, Epstein AL, Manservigi R. 2008. HSV as a vector in vaccine development and gene therapy. Hum Vaccin 4: 91–105. 10.4161/hv.4.2.6212 [DOI] [PubMed] [Google Scholar]
- Marinkovich MP, Keene DR, Rimberg CS, Burgeson RE. 1993. Cellular origin of the dermal–epidermal basement membrane. Dev Dyn 197: 255–267. 10.1002/aja.1001970404 [DOI] [PubMed] [Google Scholar]
- Marinkovich M, Lane A, Sridhar K, Keene D, Malyala A, Maslowski J. 2018. A phase 1/2 study of genetically corrected, collagen VII expressing autologous human dermal fibroblasts injected into the skin of patients with recessive dystrophic epidermolysis bullosa (RDEB). J Invest Dermatology 138: S100 10.1016/j.jid.2018.03.599 [DOI] [Google Scholar]
- Markowitz D, Goff S, Bank A. 1988. A safe packaging line for gene transfer: separating viral genes on two different plasmids. J Virol 62: 1120–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruggi G, Porcellini S, Facchini G, Perna SK, Cattoglio C, Sartori D, Ambrosi A, Schambach A, Baum C, Bonini C, et al. 2009. Transcriptional enhancers induce insertional gene deregulation independently from the vector type and design. Mol Ther 17: 851–856. 10.1038/mt.2009.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascré G, Dekoninck S, Drogat B, Youssef KK, Brohée S, Sotiropoulou PA, Simons BD, Blanpain C. 2012. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 489: 257–262. 10.1038/nature11393 [DOI] [PubMed] [Google Scholar]
- Mathor MB, Ferrari G, Dellambra E, Cilli M, Mavilio F, Cancedda R, De Luca M. 1996. Clonal analysis of stably transduced human epidermal stem cells in culture. Proc Natl Acad Sci 93: 10371–10376. 10.1073/pnas.93.19.10371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavilio F, Pellegrini G, Ferrari S, Di Nunzio F, Di Iorio E, Recchia A, Maruggi G, Ferrari G, Provasi E, Bonini C, et al. 2006. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat Med 12: 1397–1402. 10.1038/nm1504 [DOI] [PubMed] [Google Scholar]
- Merten O-W. 2004. State-of-the-art of the production of retroviral vectors. J Gene Med 6: S105–S124. 10.1002/jgm.499 [DOI] [PubMed] [Google Scholar]
- Miller RH, Sarver N. 1997. HIV accessory proteins as therapeutic targets. Nat Med 3: 389–394. 10.1038/nm0497-389 [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398: 708–713. 10.1038/19531 [DOI] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol 2: E234 10.1371/journal.pbio.0020234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaudié H, Chiaverini C, Sbidian E, Charlesworth A, Lacour JP. 2016. Inherited epidermolysis bullosa and squamous cell carcinoma: a systematic review of 117 cases. Orphanet J Rare Dis 11: 117 10.1186/s13023-016-0489-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JR, Barrandon Y, Green H, Mulligan RC. 1987. Expression of an exogenous growth hormone gene by transplantable human epidermal cells. Science 237: 1476–1479. 10.1126/science.3629250 [DOI] [PubMed] [Google Scholar]
- Müller FB, Huber M, Kinaciyan T, Hausser I, Schaffrath C, Krieg T, Hohl D, Korge BP, Arin MJ. 2006. A human keratin 10 knockout causes recessive epidermolytic hyperkeratosis. Hum Mol Genet 15: 1133–1141. 10.1093/hmg/ddl028 [DOI] [PubMed] [Google Scholar]
- Naldini L. 2019. Genetic engineering of hematopoiesis: current stage of clinical translation and future perspectives. EMBO Mol Med 11: e9958 10.15252/emmm.201809958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanba D, Toki F, Matsushita N, Matsushita S, Higashiyama S, Barrandon Y. 2013. Actin filament dynamics impacts keratinocyte stem cell maintenance. EMBO Mol Med 5: 640–653. 10.1002/emmm.201201839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyström A, Bruckner-Tuderman L. 2016. Gene therapy for epidermolysis bullosa: sticky business. Mol Ther 24: 2035–2036. 10.1038/mt.2016.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oji V, Tadini G, Akiyama M, Blanchet Bardon C, Bodemer C, Bourrat E, Coudiere P, DiGiovanna JJ, Elias P, Fischer J, et al. 2010. Revised nomenclature and classification of inherited ichthyoses: results of the first Ichthyosis Consensus Conference in Sorèze 2009. J Am Acad Dermatol 63: 607–641. 10.1016/j.jaad.2009.11.020 [DOI] [PubMed] [Google Scholar]
- Osborn MJ, Starker CG, McElroy AN, Webber BR, Riddle MJ, Xia L, DeFeo AP, Gabriel R, Schmidt M, von Kalle C, et al. 2013. TALEN-based gene correction for epidermolysis bullosa. Mol Ther 21: 1151–1159. 10.1038/mt.2013.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paller AS, Syder AJ, Chan YM, Yu QC, Hutton E, Tadini G, Fuchs E. 1994. Genetic and clinical mosaicism in a type of epidermal nevus. N Engl J Med 331: 1408–1415. 10.1056/NEJM199411243312103 [DOI] [PubMed] [Google Scholar]
- Pan D, Büning H, Ling C. 2019. Rational design of gene therapy vectors. Mol Ther Methods Clin Dev 12: 246–247. 10.1016/j.omtm.2019.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Bae S, Kim JS. 2015. Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31: 4014–4016. 10.1093/bioinformatics/btv537 [DOI] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci 90: 8392–8396. 10.1073/pnas.90.18.8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini G, Traverso CE, Franzi AT, Zingirian M, Cancedda R, De Luca M. 1997. Long-term restoration of damaged corneal surfaces with autologous cultivated corneal epithelium. Lancet 349: 990–993. 10.1016/S0140-6736(96)11188-0 [DOI] [PubMed] [Google Scholar]
- Pellegrini G, Golisano O, Paterna P, Lambiase A, Bonini S, Rama P, De Luca M. 1999a. Location and clonal analysis of stem cells and their differentiated progeny in the human ocular surface. J Cell Biol 145: 769–782. 10.1083/jcb.145.4.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini G, Ranno R, Stracuzzi G, Bondanza S, Guerra L, Zambruno G, Micali G, De Luca M. 1999b. The control of epidermal stem cells (holoclones) in the treatment of massive full-thickness burns with autologous keratinocytes cultured on fibrin. Transplantation 68: 868–879. 10.1097/00007890-199909270-00021 [DOI] [PubMed] [Google Scholar]
- Pellegrini G, Dellambra E, Golisano O, Martinelli E, Fantozzi I, Bondanza S, Ponzin D, McKeon F, De Luca M. 2001. p63 identifies keratinocyte stem cells. Proc Natl Acad Sci 98: 3156–3161. 10.1073/pnas.061032098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini G, Rama P, Matuska S, Lambiase A, Bonini S, Pocobelli A, Colabelli RG, Spadea L, Fasciani R, Balestrazzi E, et al. 2013. Biological parameters determining the clinical outcome of autologous cultures of limbal stem cells. Regen Med 8: 553–567. 10.2217/rme.13.43 [DOI] [PubMed] [Google Scholar]
- Pellegrini G, Ardigò D, Milazzo G, Iotti G, Guatelli P, Pelosi D, De Luca M. 2018. Navigating market authorization: The path Holoclar took to become the first stem cell product approved in the European Union. Stem Cells Transl Med 7: 146–154. 10.1002/sctm.17-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. 2013. The CRISPR craze. Science 341: 833–836. 10.1126/science.341.6148.833 [DOI] [PubMed] [Google Scholar]
- Prodinger C, Reichelt J, Bauer JW, Laimer M. 2019. Epidermolysis bullosa: advances in research and treatment. Exp Dermatol 34: 976–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rama P, Bonini S, Lambiase A, Golisano O, Paterna P, De Luca M, Pellegrini G. 2001. Autologous fibrin-cultured limbal stem cells permanently restore the corneal surface of patients with total limbal stem cell deficiency. Transplantation 72: 1478–1485. 10.1097/00007890-200111150-00002 [DOI] [PubMed] [Google Scholar]
- Rama P, Matuska S, Paganoni G, Spinelli A, De Luca M, Pellegrini G. 2010. Limbal stem-cell therapy and long-term corneal regeneration. N Engl J Med 363: 147–155. 10.1056/NEJMoa0905955 [DOI] [PubMed] [Google Scholar]
- Remington J, Wang X, Hou Y, Zhou H, Burnett J, Muirhead T, Uitto J, Keene DR, Woodley DT, Chen M. 2009. Injection of recombinant human type VII collagen corrects the disease phenotype in a murine model of dystrophic epidermolysis bullosa. Mol Ther 17: 26–33. 10.1038/mt.2008.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinwald JG, Green H. 1975. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 6: 331–343. 10.1016/S0092-8674(75)80001-8 [DOI] [PubMed] [Google Scholar]
- Riviere I, Brose K, Mulligan RC. 1995. Effects of retroviral vector design on expression of human adenosine deaminase in murine bone marrow transplant recipients engrafted with genetically modified cells. Proc Natl Acad Sci 92: 6733–6737. 10.1073/pnas.92.15.6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Barbeau M, Éthier S, Dostie J, Pelletier J. 2015. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med 7: 93 10.1186/s13073-015-0215-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat A, Kobayashi K, Barrandon Y. 1994. Location of stem cells of human hair follicles by clonal analysis. Cell 76: 1063–1073. 10.1016/0092-8674(94)90383-2 [DOI] [PubMed] [Google Scholar]
- Roedl D, Oji V, Buters JT, Behrendt H, Braun-Falco M. 2011. rAAV2-Mediated restoration of LEKTI in LEKTI-deficient cells from Netherton patients. J Dermatol Sci 61: 194–198. 10.1016/j.jdermsci.2010.12.004 [DOI] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR, Kawaguchi K, Park S, Gonzalez D, Brown S, Boucher J, Klein AM, Greco V. 2016. Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 352: 1471–1474. 10.1126/science.aaf7012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronfard V, Rives JM, Neveux Y, Carsin H, Barrandon Y. 2000. Long-term regeneration of human epidermis on third degree burns transplanted with autologous cultured epithelium grown on a fibrin matrix. Transplantation 70: 1588–1598. 10.1097/00007890-200012150-00009 [DOI] [PubMed] [Google Scholar]
- Roshan A, Murai K, Fowler J, Simons BD, Nikolaidou-Neokosmidou V, Jones PH. 2016. Human keratinocytes have two interconvertible modes of proliferation. Nat Cell Biol 18: 145–156. 10.1038/ncb3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadelain M, Rivière I, Riddell S. 2017. Therapeutic T cell engineering. Nature 545: 423–431. 10.1038/nature22395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambach A, Mueller D, Galla M, Verstegen MM, Wagemaker G, Loew R, Baum C, Bohne J. 2006. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther 13: 1524–1533. 10.1038/sj.gt.3302807 [DOI] [PubMed] [Google Scholar]
- Scherdin U, Rhodes K, Breindl M. 1990. Transcriptionally active genome regions are preferred targets for retrovirus integration. J Virol 64: 907–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiano V, Zhen HH, Haddad B, Bashkirova E, Melo SP, Wang P, Leung TL, Siprashvili Z, Tichy A, Li J, et al. 2014. Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Sci Transl Med 6: 264ra163 10.1126/scitranslmed.3009540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. 2007. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129: 523–536. 10.1016/j.cell.2007.02.045 [DOI] [PubMed] [Google Scholar]
- Shinkuma S, Guo Z, Christiano AM. 2016. Site-specific genome editing for correction of induced pluripotent stem cells derived from dominant dystrophic epidermolysis bullosa. Proc Natl Acad Sci 113: 5676–5681. 10.1073/pnas.1512028113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Khan I, Khim S, Seymour B, Sommer K, Wielgosz M, Norgaard Z, Kiem HP, Adair J, Liggitt D, et al. 2017. Safe and effective gene therapy for murine Wiskott–Aldrich syndrome using an insulated lentiviral vector. Mol Ther Methods Clin Dev 4: 1–16. 10.1016/j.omtm.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siprashvili Z, Nguyen NT, Gorell ES, Loutit K, Khuu P, Furukawa LK, Lorenz HP, Leung TH, Keene DR, Rieger KE, et al. 2016. Safety and wound outcomes following genetically corrected autologous epidermal grafts in patients with recessive dystrophic epidermolysis bullosa. JAMA 316: 1808–1817. 10.1001/jama.2016.15588 [DOI] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. 2016. Rationally engineered Cas9 nucleases with improved specificity. Science 351: 84–88. 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, van de Wetering M, van den Born M, Begthel H, Vries RG, et al. 2010. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327: 1385–1389. 10.1126/science.1184733 [DOI] [PubMed] [Google Scholar]
- Spaete RR, Frenkel N. 1982. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell 30: 295–304. 10.1016/0092-8674(82)90035-6 [DOI] [PubMed] [Google Scholar]
- Srivastava M, Nambiar M, Sharma S, Karki SS, Goldsmith G, Hegde M, Kumar S, Pandey M, Singh RK, Ray P, et al. 2012. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell 151: 1474–1487. 10.1016/j.cell.2012.11.054 [DOI] [PubMed] [Google Scholar]
- Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, Schmidt M, Krämer A, Schwäble J, Glimm H, et al. 2010. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med 16: 198–204. 10.1038/nm.2088 [DOI] [PubMed] [Google Scholar]
- Subak-Sharpe JH, Dargan DJ. 1998. HSV molecular biology: general aspects of herpes simplex virus molecular biology. Virus Genes 16: 239–251. 10.1023/A:1008068902673 [DOI] [PubMed] [Google Scholar]
- Suerth JD, Maetzig T, Galla M, Baum C, Schambach A. 2010. Self-inactivating α retroviral vectors with a split-packaging design. J Virol 84: 6626–6635. 10.1128/JVI.00182-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerth JD, Maetzig T, Brugman MH, Heinz N, Appelt JU, Kaufmann KB, Schmidt M, Grez M, Modlich U, Baum C, et al. 2012. α Retroviral self-inactivating vectors: long-term transgene expression in murine hematopoietic cells and low genotoxicity. Mol Ther 20: 1022–1032. 10.1038/mt.2011.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerth JD, Labenski V, Schambach A. 2014. α Retroviral vectors: from a cancer-causing agent to a useful tool for human gene therapy. Viruses 6: 4811–4838. 10.3390/v6124811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YK, Kim SW. 2019. Recent advances in the development of gene delivery systems. Biomater Res 23: 8 10.1186/s40824-019-0156-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadini G, Brena M, Gelmetti C, Pezzani L. 2015. Atlas of genodermatoses, 2nd ed. CRC, Boca Raton, FL. [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Thomas M, Burgio G, Adams DJ, Iyer V. 2019. Collateral damage and CRISPR genome editing. PLoS Genet 15: e1007994 10.1371/journal.pgen.1007994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar J, Xia L, Riddle MJ, Lees CJ, Eide CR, McElmurry RT, Titeux M, Osborn MJ, Lund TC, Hovnanian A, et al. 2011. Induced pluripotent stem cells from individuals with recessive dystrophic epidermolysis bullosa. J Invest Dermatol 131: 848–856. 10.1038/jid.2010.346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkinson AE, Sallmyr A. 2013. Structure and function of the DNA ligases encoded by the mammalian LIG3 gene. Gene 531: 150–157. 10.1016/j.gene.2013.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima KE, Ogawa M, Tsuji T. 2019. Regeneration of a bioengineered 3D integumentary organ system from iPS cells. Nat Protoc 14: 1323–1338. 10.1038/s41596-019-0124-z [DOI] [PubMed] [Google Scholar]
- Vakulskas CA, Behlke MA. 2019. Evaluation and reduction of CRISPR off-target cleavage events. Nucleic Acid Ther 29: 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Akker PC, Jonkman MF, Rengaw T, Bruckner-Tuderman L, Has C, Bauer JW, Klausegger A, Zambruno G, Castiglia D, Mellerio JE, et al. 2011. The international dystrophic epidermolysis bullosa patient registry: an online database of dystrophic epidermolysis bullosa patients and their COL7A1 mutations. Hum Mutat 32: 1100–1107. 10.1002/humu.21551 [DOI] [PubMed] [Google Scholar]
- Vartak SV, Raghavan SC. 2015. Inhibition of nonhomologous end joining to increase the specificity of CRISPR/Cas9 genome editing. FEBS J 282: 4289–4294. 10.1111/febs.13416 [DOI] [PubMed] [Google Scholar]
- Wagner JE, Ishida-Yamamoto A, McGrath JA, Hordinsky M, Keene DR, Woodley DT, Chen M, Riddle MJ, Osborn MJ, Lund T, et al. 2010. Bone marrow transplantation for recessive dystrophic epidermolysis bullosa. N Engl J Med 363: 629–639. 10.1056/NEJMoa0910501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM. 1983. Involucrin and other markers of keratinocyte terminal differentiation. J Invest Dermatol 81: 100s–103s. 10.1111/1523-1747.ep12540786 [DOI] [PubMed] [Google Scholar]
- Webber BR, Osborn MJ, McElroy AN, Twaroski K, Lonetree CL, DeFeo AP, Xia L, Eide C, Lees CJ, McElmurry RT, et al. 2016. CRISPR/Cas9-based genetic correction for recessive dystrophic epidermolysis bullosa. NPJ Regen Med 1: 16014 10.1038/npjregenmed.2016.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel D, Bayerl J, Nystrom A, Bruckner-Tuderman L, Meixner A, Penninger JM. 2014. Genetically corrected iPSCs as cell therapy for recessive dystrophic epidermolysis bullosa. Sci Transl Med 6: 264ra165 10.1126/scitranslmed.3010083 [DOI] [PubMed] [Google Scholar]
- Wertheim-Tysarowska K, Sobczyńska-Tomaszewska A, Kowalewski C, Skroński M, Święćkowski G, Kutkowska-Kaźmierczak A, Woźniak K, Bal J. 2012. The COL7A1 mutation database. Hum Mutat 33: 327–331. 10.1002/humu.21651 [DOI] [PubMed] [Google Scholar]
- Whitley RJ, Roizman B. 2001. Herpes simplex virus infections. Lancet 357: 1513–1518. 10.1016/S0140-6736(00)04638-9 [DOI] [PubMed] [Google Scholar]
- Wong T, Gammon L, Liu L, Mellerio JE, Dopping-Hepenstal PJ, Pacy J, Elia G, Jeffery R, Leigh IM, Navsaria H, et al. 2008. Potential of fibroblast cell therapy for recessive dystrophic epidermolysis bullosa. J Invest Dermatol 128: 2179–2189. 10.1038/jid.2008.78 [DOI] [PubMed] [Google Scholar]
- Woodley DT, Krueger GG, Jorgensen CM, Fairley JA, Atha T, Huang Y, Chan L, Keene DR, Chen M. 2003. Normal and gene-corrected dystrophic epidermolysis bullosa fibroblasts alone can produce type VII collagen at the basement membrane zone. J Invest Dermatol 121: 1021–1028. 10.1046/j.1523-1747.2003.12571.x [DOI] [PubMed] [Google Scholar]
- Woodley DT, Keene DR, Atha T, Huang Y, Lipman K, Li W, Chen M. 2004a. Injection of recombinant human type VII collagen restores collagen function in dystrophic epidermolysis bullosa. Nat Med 10: 693–695. 10.1038/nm1063 [DOI] [PubMed] [Google Scholar]
- Woodley DT, Keene DR, Atha T, Huang Y, Ram R, Kasahara N, Chen M. 2004b. Intradermal injection of lentiviral vectors corrects regenerated human dystrophic epidermolysis bullosa skin tissue in vivo. Mol Ther 10: 318–326. 10.1016/j.ymthe.2004.05.016 [DOI] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300: 1749–1751. 10.1126/science.1083413 [DOI] [PubMed] [Google Scholar]
- Yancey KB, Hintner H. 2010. Non-herlitz junctional epidermolysis bullosa. Dermatol Clin 28: 67–77. 10.1016/j.det.2009.10.008 [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dötsch V, Andrews NC, Caput D, McKeon F. 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 2: 305–316. 10.1016/S1097-2765(00)80275-0 [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398: 714–718. 10.1038/19539 [DOI] [PubMed] [Google Scholar]
- Ye L, Wang C, Hong L, Sun N, Chen D, Chen S, Han F. 2018. Programmable DNA repair with CRISPRa/i enhanced homology-directed repair efficiency with a single Cas9. Cell Discov 4: 46 10.1038/s41421-018-0049-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SF, von Ruden T, Kantoff PW, Garber C, Seiberg M, Ruther U, Anderson WF, Wagner EF, Gilboa E. 1986. Self-inactivating retroviral vectors designed for transfer of whole genes into mammalian cells. Proc Natl Acad Sci 83: 3194–3198. 10.1073/pnas.83.10.3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, Liu H, La Russa M, Xie M, Ding S, et al. 2015. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16: 142–147. 10.1016/j.stem.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaboikin M, Zaboikina T, Freter C, Srinivasakumar N. 2017. Non-homologous end joining and homology directed DNA repair frequency of double-stranded breaks introduced by genome editing reagents. PLoS ONE 12: e0169931 10.1371/journal.pone.0169931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wen Y, Guo X. 2014. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet 23: R40–R46. 10.1093/hmg/ddu125 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Long C, Li H, McAnally JR, Baskin KK, Shelton JM, Bassel-Duby R, Olson EN. 2017. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci Adv 3: e1602814 10.1126/sciadv.1602814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Mody D, DeRavin SS, Hauer J, Lu T, Ma Z, Hacein-Bey Abina S, Gray JT, Greene MR, Cavazzana-Calvo M, et al. 2010. A self-inactivating lentiviral vector for SCID-X1 gene therapy that does not activate LMO2 expression in human T cells. Blood 116: 900–908. 10.1182/blood-2009-10-250209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, Trono D. 1998. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol 72: 9873–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]