Abstract
Aims: Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) assay based on a TaqMan‐minor groove binder (MGB) probe was developed for the rapid detection of avian influenza virus subtype H5.
Methods and Results: Conserved regions in the haemagglutinin genes of avian influenza viruses subtype H5 served as targets for the primers and TaqMan‐MGB probe design. Concentrations of primers and probe were optimized to improve the sensitivity and specificity of the reactions. A plasmid containing the haemagglutinin gene was constructed and in vitro transcribed for a quantitative assay of copy numbers of the target gene. The results revealed that the optimal concentration of primers and probe was 640 and 480 nmol l−1, respectively. The threshold of 100 copies of target molecules could be detected. The linear range for detection was determined as 102 to 108 molecules in reaction.
Conclusions: It took less than 3 h to complete the detection from viral RNA extraction, with good sensitivity and repeatability.
Significance and Impact of the Study: Real‐time RT‐PCR assay with MGB probe was an effective means for quick and quantitative laboratory detection and monitoring of H5 avian influenza viruses.
Keywords: avian influenza virus, H5 subtype, laboratory detection, real‐time RT‐PCR, TaqMan‐MGB probe
Introduction
Highly pathogenic avian influenza (HPAI) is one of the most severe diseases communicable in birds. HPAI infection results in acute diseases with high morbidities and mortalities. Recent outbreaks and researches have revealed that HPAI could break through the species barrier to infect humans with high mortalities. The first outbreak was documented in Hong Kong in 1997 with 18 human cases including six fatalities (Yuen et al. 1998; Chan 2002). Since its reemergence from 2003 in Southeast Asia, 172 people have died from HPAI infection (WHO 2006). HPAI is becoming a serious threat to public health because of its fast genetic variation and increasing adaptation to humans. The establishment of rapid and reliable methods for H5 avian influenza virus detection is of great value for the control and prevention of HPAI.
Current methods for the diagnosis of H5N1 infection are virus isolation, serologic test and molecular detection of viral nucleic acids. Virus isolation is laborious, time‐consuming and has a high false‐negative rate. The serologic confirmation using serum samples collected from acute and recovery phases of infected human cases cannot give diagnostic results in the early phase of infection. The use of these two methods in detecting H5N1 virus has been limited because of these disadvantages (Gan 2004).
The real‐time RT‐PCR developed in recent years can give rapid, sensitive and specific results for the detection of virus infection (van Elden et al. 2001; Emery et al. 2004). Real‐time monitoring of amplified products cycle‐by‐cycle and direct virus quantification can be performed using this method. All the procedures were carried out in high containment conditions, and cross‐contaminations among samples and false‐positive results were therefore greatly avoided. The newly developed technique of minor groove binder (MGB) probe for diagnosis of virus infection has additional advantages over the traditional TaqMan probes (Fernandez et al. 2002; Zhao et al. 2005; Lole and Arankalle 2006). The MGB technique can enlarge the difference in T m values between the target template and other nonspecific nucleic acids, to obtain high specificity. The MGB probe can also improve hybridization stability and reduce the fluorescence background to obtain high sensitivity. In this report, TaqMan‐MGB real‐time quantitative RT‐PCR was used to detect H5‐specific nucleic acids.
Materials and methods
Materials
The RNeasy Kit from Qiagen (Hilden, Germany) was used for RNA extraction. DNA purification kit was purchased from Promega (Madison, WI). Premix Ex Taq (Perfect Real Time, Dalian, China) kit was from TaKaRa. SalI and T7 Riboprobe System were purchased from Promega. Primers and TaqMan‐MGB fluorescent probe were synthesized by Shanghai GeneCore BioTechnologies (Shanghai, China).
Virus strains and clinical specimen
The control antigens of H5N1, H7 and H9 avian influenza viruses were obtained from WHO. Influenza virus strains including A1/Bejiang/53/97, A3/Sydney/5/97 and B/Shenzhen/12/97 were provided by the National Influenza Center of China. Measles virus strain Shanghai191 and Respiratory syncytial virus strain Long were provided by the Shanghai Institute of Biological Products (China). Severe acute respiratory syndrome (SARS) coronavirus strain ZJ01 was isolated and conserved in our laboratory in 2003. Influenza virus H5N1 was isolated in our laboratory from gargle and tracheal aspirate specimens of a suspected human case in February 2006 and confirmed by the National Influenza Center of China.
Primers and probe for real‐time RT‐PCR
Sequence data of haemagglutinin (HA) genes of avian influenza H5 type isolated from birds and human beings were downloaded from GenBank. The program dnaman (Vaudreuil‐Dorion, Quebec, Canada) was used for homology analysis of the HA genes. The conserved regions were used for design of specific primers and TaqMan‐MGB fluorescent probe with Primer Express 2.0 Program (Foster City, CA). The probe was labelled with the fluorescent reporter FAM at the 5′ end; non‐fluorescent quencher and MGB were labelled at the 3′ end. The sequences of primers and probe were as follows: sense primer, 5′‐TGGATGGCTCCTCGGRAAC‐3′; antisense primer, 5′‐ARGACCATTCCGGCACATT‐3′; probe, 5′‐FAM‐TGTGTGACGAATTCAT‐3′MGB. The amplified fragment was 58 bp in length.
Viral RNA extraction
The nucleic acid extractions were carried out from clinical or standard virus samples under the containment conditions of a biosafety level 3 with additional safety precautions. RNeasy MiniKit (Qiagen) was used for viral RNA extraction following the manufacturer’s instructions.
Optimization of reaction conditions
Matrix titration method was carried out to optimize the concentration of primers and probe using fixed amount of template and reaction system. The annealing temperature was also optimized (45–65°C) by the amplification of positive samples. The conditions for optimal amplification was determined by the criteria that lower threshold cycle (C t) value and higher increase in fluorescence (ΔR n) were observed in the reaction.
Specificity evaluation of TaqMan‐MGB real‐time RT‐PCR
The nucleic acids from respiratory viruses, including avian influenza H5, H7 and H9 viruses, influenza viruses A1, A3 and B, SARS virus and RSV, were extracted for evaluating the specificity of TaqMan‐MGB real‐time RT‐PCR in the detection of H5 avian influenza.
Quantitative analyses
The coding region of HA gene of H5 avian influenza was amplified using specific primers as follows, HAF: 5′‐AGCAAAAGCAGGGGTCTAATCTG‐3′, HAR: 5′‐AGTCCAGACATCTAGGAAYCC‐3′. The amplified fragment was recovered from agarose gel and ligated with pGEM‐T vector. The DH5α competent cells were transformed with ligation mixture and spreaded on an LB plate containing ampicillin. Escherichia coli colonies were picked up and positive clones were identified using PCR amplification. Sequencing analysis was conducted to further confirm the insertion of HA gene. The concentration of recombinant plasmid DNA was determined by absorbance of UV light at 260 and 280 nm. The plasmid was then linearized at the SalI site and 5 μg DNA was used as template in an in vitro transcription reaction with the Riboprobe System using T7 RNA polymerase (Promega). After the in vitro transcription reaction at 37°C for 1 h, the DNA was digested by DNase I at 37°C for 15 min. The purified RNA was quantified with a spectrophotometer. The copy number of transcribed RNA was then determined following the method of Fronhoffs et al. (2002). The transcribed RNA was used for external standard in the amplification. Standard curve was created according to the reaction in which 10‐fold serial dilutions of transcribed RNA containing 108 to 101 molecules was employed as template. The amount of unknown sample could be extrapolated based on the standard curve. TaKaRa Premix Ex Taq (Perfect Real Time) kit was used for the quantitative real‐time RT‐PCR. The reaction was conducted in a total volume of 20 μl, consisting of 10 μl of Premix Ex Taq, 0·25 μl reverse transcriptase, 0·8 μl (20 μmol l−1) of sense and antisense primers, 0·6 μl (20 μmol l−1) of probe and 5 μl of diluted template. Fluorescence detection systems of MJ Opticon2 (Waltham, MA) or Roche LightCycler (Mannheim, Germany) were used for running the program containing a reverse transcription for 30 min, an initial denaturation for 2 min at 95°C and 40 cycles of amplification including 5 s at 95°C, 20 s at 50°C, and a step of single fluorescence detection at 50°C after each cycle. Three transcribed RNA samples with different concentrations were used to estimate the reproducibility of TaqMan‐MGB real‐time RT‐PCR in detection as external standard of avian influenza H5, each reaction contained five repeats.
Results
Optimizing of TaqMan‐MGB real‐time RT‐PCR reaction
To increase the efficiency and sensitivity of amplification, the concentration of primers and probe was optimized using a matrix titration method. Reactions with various annealing temperatures ranging from 45 to 65°C were also performed. The lowest C t value and highest ΔR n were observed in the amplification with an annealing temperature of 50°C; primer and probe concentration of 640 and 480 nmol l−1, respectively.
Specificity of TaqMan‐MGB real‐time RT‐PCR
The nucleic acids of H5, H7 and H9 avian influenza viruses and other respiratory viruses were extracted for TaqMan‐MGB real‐time RT‐PCR detection. Positive result was observed only in the detection of influenza virus with H5 type. No cross‐detection was observed among other influenza viruses or respiratory viruses. Fluorescence increasing was not detectable in the detection of negative control, H7, H9 avian influenza viruses and other respiratory viruses. The data are shown in Fig. 1. This result suggested that the TaqMan‐MGB real‐time RT‐PCR was an ideal technique for H5 avian influenza diagnosis.
Figure 1.
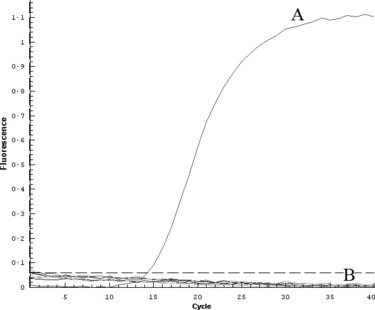
Specific evaluation of TaqMan‐MGB method for the detection of H5 avian influenza virus. The line of H5N1 amplification was indicated (A). The other lines represented the amplifications using nucleic acids from respiratory viruses (B), including avian influenza H7 and H9 viruses, influenza viruses A1 (H1N1), A3 (H3N2) and B, SARS‐CoV and RSV. The control reaction using DEPC‐treated water as template was also included.
Quantitative analysis
The in vitro transcribed RNA was used as a standard to determine the sensitivity of real‐time RT‐PCR. The 10‐fold serial dilutions (101–108 copies) of transcribed RNA were used to create the standard curve for quantitative analysis. The threshold for detection was determined as 102 molecules with the C t value under 36. Although the reaction containing 10 copies of target gene gave an amplification curve slightly higher than the threshold, it was uncertain to judge it as a positive amplification. The data of seven reactions with 108 to 102 molecules as template were used for plotting the standard curve related to C t value and copy number. The equation was obtained as Y = –3·551X + 43·75, R 2 = 0·996 (where X stands for log concentration and Y for cycle number). The amplification result and standard curve are shown in 2, 3.
Figure 2.
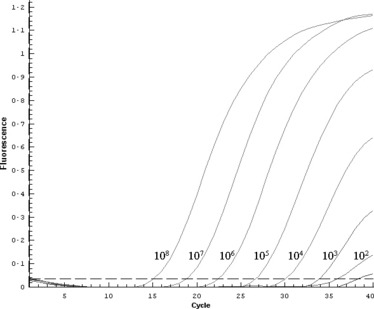
Amplification profile of detection for the HA nucleic acid of H5 avian influenza virus. From left to right, the amplification contained 108, 107, 106, 105, 104, 103, 102 and 101 copies of in vitro transcribed RNA, respectively.
Figure 3.
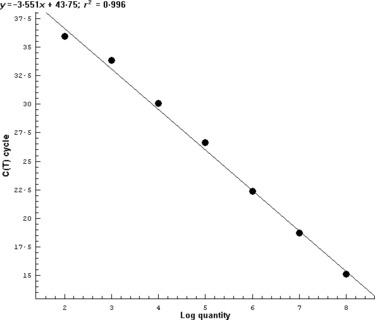
Standard curve created by the analysis of known copies of target nucleic acid. Reactions with copy numbers of target gene from 102 to 108 were used for creating the standard curve. The standard curve was plotted by the log concentration of copy numbers against C t values.
Repeatability of TaqMan‐MGB real‐time RT‐PCR
Reactions with the same concentration of in vitro transcribed RNA were used to evaluate the repeatability in the detection of the HA gene from H5 avian influenza; 107, 106 and 105 copies of HA fragments were used as template in 20 μl reactions. Amplifications from each concentration of target gene contained five repeats. The MGB probe‐based real‐time RT‐PCR was proved to be reliable because of the favourable repeatability (revealed by C t values) obtained in this assay. The amplification curve is shown in Fig. 4.
Figure 4.
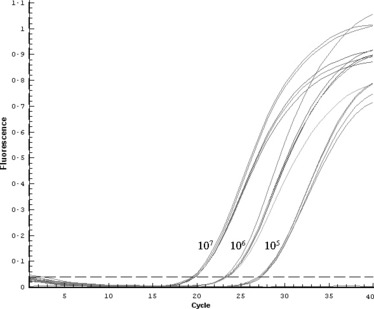
Reproducibility assay for the detection of HA gene using real‐time PCR. Each concentration of positive amplification contained five repeats. From left to right, 107, 106 and 105 copies of target molecules were used to estimate the reproducibility.
Detection of clinical samples from a suspected case of H5N1 infection
The diagnosis of a suspected case of HPAI (H5N1) infection was conducted using TaqMan‐MGB real‐time RT‐PCR. The tracheal aspirate specimen was collected in February 2006 from the 9‐year‐old girl who died on 6 March because of severe symptoms. The clinical sample was determined to be H5‐positive and this result was also confirmed by the National Influenza Center of China. The amplification profile was shown in Fig. 5.
Figure 5.
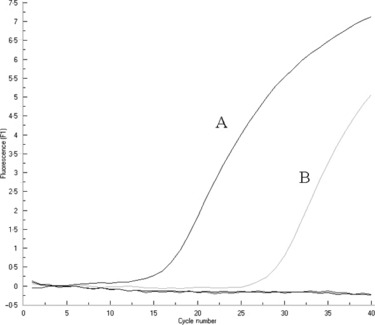
Detection of avian influenza virus H5N1 from a clinical sample using TaqMan‐MGB real‐time RT‐PCR. Sample A was positive control; sample B represented the amplification from the clinical specimen.
Discussion
The early laboratory detection of nucleic acid from suspected cases provided evidence for quarantine and medication in the control and prevention of HPAI outbreaks. There have been several papers describing the real‐time RT‐PCR detection of influenza viruses. Some have described the simultaneous detection of influenza A, including H5N1, while others described the subtyping of H5 influenza virus (Ng et al. 2005; Whiley and Sloots 2005; Payungporn et al. 2006; Wen et al. 2006). In this report, a rapid diagnosis of H5 avian influenza was implemented using TaqMan‐MGB real‐time RT‐PCR. The method combined a quick and sensitive detection of the nucleic acids of H5 avian influenza, and a threshold of 100 copies could be detected. Specificity was also improved by the fluorescent probe. Electrophoresis of amplified fragments was substituted by the direct fluorescence detection during the PCR process and the duration of detection was reduced. The newly developed TaqMan‐MGB probe is more stable and can improve signal‐to‐noise ratios due to the use of a non‐fluorescent quencher instead of the fluorescent quencher dye (Afonina et al. 2002). The combination of MGB stabilizes the hybridization of the probe and the T m value is about 10°C higher than conventional TaqMan probes. As a result, short oligonucleotides of about 15 nucleotides can be used for hybridization and therefore confer more options in the optimization of probe sequence. The MGB technique also has an advantage in the discrimination of mismatched nucleotide during hybridization (Lew et al. 2004). The specificity of detection using the MGB‐labelled probe was similar to that obtained by non‐MGB fluorescent probes, but more sensitivity was obtained when the MGB technique was employed in the detection of H5 avian influenza. One hundred molecules of external standard were detectable using the 16‐bp MGB probe specific for the H5 gene. The sensitivity was higher than using a conventional TaqMan probe which was obtained as 103–104 copies (Spackman et al. 2002).
In a previous study, the primers and MGB probe were designed based on the matrix gene sequences from animal and human influenza A viruses (Di Trani et al. 2006). In the present report, the HA sequences of various H5 type avian influenza viruses isolated from birds or humans were downloaded for the primer and probe design. To ensure the detection of various subtypes of H5 avian influenza viruses and to give evidence for pathogen tracing, degenerate primers were designed and proved to be sensitive and specific for laboratory detection.
In this study, the in vitro transcribed RNA from plasmid DNA was used to create a standard curve and subsequently extrapolate the quantity of unknown samples. Seven reactions were selected for plotting of the standard curve. The favourable reproducibility of TaqMan‐MGB real‐time RT‐PCR was evaluated by the detection of three in vitro transcribed RNA of different concentrations. The MGB technique developed was also used for laboratory confirmation of H5 avian influenza virus from a suspected human case in Zhejiang province. Less than 3 h was needed to obtain a positive result from receipt of the clinical sample. Sequencing analyses also confirmed that the H5N1 virus was avian‐specific. Sensitive and specific results were also obtained using the TaqMan‐MGB technique in the detection of 20 avian originated nucleic acid samples of H5N1 from the Zhejiang Academy of Agricultural Sciences (data not shown).
In this report, the TaqMan‐MGB real‐time RT‐PCR proved to be a sensitive and rapid method for the early diagnosis of H5 avian influenza virus infection. On the other hand, quantitative analysis using this technique was useful for viral load testing and evaluating the curative effects of medicines.
Acknowledgements
This work was funded by the Zhejiang Provincial Natural Sciences Foundation of China under grant no. Z303909.
References
- Afonina, I.A. , Reed, M.W. , Lusby, E. , Shishkina, I.G. and Belousov, Y.S. (2002) Minor groove binder‐conjugated DNA probes for quantitative DNA detection by hybridization‐triggered fluorescence. Biotechniques 32, 940–944, 946–949. [DOI] [PubMed] [Google Scholar]
- Chan, P.K. (2002) Outbreak of avian influenza A(H5N1) virus infection in Hong Kong in 1997. Clin Infect Dis 34, S58–S64. [DOI] [PubMed] [Google Scholar]
- Di Trani, L. , Bedini, B. , Donatelli, I. , Campitelli, L. , Chiappini, B. , De Marco, M.A. , Delogu, M. , Buonavoglia, C. et al. (2006) A sensitive one‐step real‐time PCR for detection of avian influenza viruses using a MGB probe and an internal positive control. BMC Infect Dis 6, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elden, L.J. , Nijhuis, M. , Schipper, P. , Schuurman, R. and Van Loon, A.M. (2001) Simultaneous detection of influenza viruses A and B using real‐time quantitative PCR. J Clin Microbiol 39, 196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, S.L. , Erdman, D.D. , Bowen, M.D. , Newton, B.R. , Winchell, J.M. , Meyer, R.F. , Tong, S. , Cook, B.T. et al. (2004) Real‐time reverse transcription‐polymerase chain reaction assay for SARS‐associated coronavirus. Emerg Infect Dis 10, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, C. , Boutolleau, D. , Manichanh, C. , Mangeney, N. , Agut, H. and Gautheret‐Dejean, A. (2002) Quantitation of HHV‐7 genome by real‐time polymerase chain reaction assay using MGB probe technology. J Virol Methods 106, 11–16. [DOI] [PubMed] [Google Scholar]
- Fronhoffs, S. , Totzke, G. , Stier, S. , Wernert, N. , Rothe, M. , Bruning, T. , Koch, B. , Sachinidis, A. et al. (2002) A method for the rapid construction of cRNA standard curves in quantitative real‐time reverse transcription polymerase chain reaction. Mol Cell Probes 16, 99–110. [DOI] [PubMed] [Google Scholar]
- Gan, M.H. (2004) Avian Influenza. Beijing: China Agriculture Press. [Google Scholar]
- Lew, A.E. , Bock, R.E. , Molloy, J.B. , Minchin, C.M. , Robinson, S.J. and Steer, P. (2004) Sensitive and specific detection of proviral bovine leukemia virus by 5′ Taq nuclease PCR using a 3′ minor groove binder fluorogenic probe. J Virol Methods 115, 167–175. [DOI] [PubMed] [Google Scholar]
- Lole, K.S. and Arankalle, V.A. (2006) Quantitation of hepatitis B virus DNA by real‐time PCR using internal amplification control and dual TaqMan MGB probes. J Virol Methods 135, 83–90. [DOI] [PubMed] [Google Scholar]
- Ng, E.K. , Cheng, P.K. , Ng, A.Y. , Hoang, T.L. and Lim, W.W. (2005) Influenza A H5N1 detection. Emerg Infect Dis 11, 1303–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payungporn, S. , Chutinimitkul, S. , Chaisingh, A. , Damrongwantanapokin, S. , Buranathai, C. , Amonsin, A. , Theamboonlers, A. and Poovorawan, Y. (2006) Single step multiplex real‐time RT‐PCR for H5N1 influenza A virus detection. J Virol Methods 131, 143–147. [DOI] [PubMed] [Google Scholar]
- Spackman, E. , Senne, D.A. , Myers, T.J. , Bulaga, L.L. , Garber, L.P. , Perdue, M.L. , Lohman, K. , Daum, L.T. et al. (2002) Development of a real‐time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol 40, 3256–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, L.Y. , Xu, H. , Lan, Y. , Zhao, X. , Zhang, X.G. , Wang, D.Y. , Yao, L.H. , Dong, J. et al. (2006) Development of methods for detection of H5N1 from human clinical specimens. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 20, 24–26. [PubMed] [Google Scholar]
- Whiley, D.M. and Sloots, T.P. (2005) A 5′‐nuclease real‐time reverse transcriptase‐polymerase chain reaction assay for the detection of a broad range of influenza A subtypes, including H5N1. Diagn Microbiol Infect Dis 53, 335–337. [DOI] [PubMed] [Google Scholar]
- WHO (2006) Cumulative Number of Confirmed Human Cases of Avian Influenza A/(H5N1) Reported to WHO. http://www.who.int/csr/disease/avian_influenza/country/cases_table_2007_04_11/en/index.html (access date: 10 May 2007). [Google Scholar]
- Yuen, K.Y. , Chan, P.K. , Peiris, M. , Tsang, D.N. , Que, T.L. , Shortridge, K.F. , Cheung, P.T. , To, W.K. et al. (1998) Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351, 467–471. [DOI] [PubMed] [Google Scholar]
- Zhao, J.R. , Bai, Y.J. , Zhang, Q.H. , Wan, Y. , Li, D. and Yan, X.J. (2005) Detection of hepatitis B virus DNA by real‐time PCR using TaqMan‐MGB probe technology. World J Gastroenterol 11, 508–510. [DOI] [PMC free article] [PubMed] [Google Scholar]


