Abstract
Recent years have witnessed the evolution of the cell biology of lipids into an extremely active area of investigation. Deciphering the involvement of lipid metabolism and lipid signaling in membrane trafficking pathways defines a major nexus of contemporary experimental activity on this front. Significant effort in that direction is invested in understanding the trans-Golgi network/endosomal system where unambiguous connections between membrane trafficking and inositol lipid and phosphatidylcholine metabolism were first discovered. However, powered by new advances in contemporary cell biology, the march of science is rapidly expanding that window of inquiry to include ever more diverse arms of the lipid metabolome, and to include other compartments of the secretory pathway as well.
Introduction
The endomembrane systems that define the organelles of the eukaryotic secretory pathway are remarkably dynamic structures. The landmark studies that unraveled the biochemical mechanism of membrane trafficking focused on the contributions of proteins in the membrane dynamics of the eukaryotic secretory pathway, and protein-centric views understandably dominated the early discussions [1–3]. In the nearly thirty years since the first convincing demonstrations that lipids and appropriately coordinated lipid metabolism are essential contributors to Golgi membrane trafficking and vesicle fusion were reported [4–9], our view of secretory organelle dynamics has evolved dramatically. Much attention has been, and continues to be, invested in the analysis of phosphoinositides as regulators of membrane trafficking and as determinants of compartmental identity. As structurally distinct molecules that are modified at specific positions on the inositol ring, phosphoinositides write chemical barcodes on membrane surfaces that help platform compartment-specific biochemical reactions (Figure 1). New advances highlight how phosphoinositides, and particularly how phosphatidylinositol-4-phosphate, integrate other aspects of lipid metabolism into the larger design for membrane traffic control.
Figure 1.

Phosphoinositides and their metabolism. Phosphoinositides are phosphorylated derivatives of phosphatidylinositol (PtdIns) and their production is catalyzed by distinct sets of lipid kinases that modify either the C3, C4 or C5 positions of the inositol headgroup. Reciprocally, phosphoinositides are dephosphorylated via the action of a cohort of phosphoinositide phosphatases. Whereas mammalian cells produce seven phosphoinositides [PtdIns 3-phosphate, PtdIns3P; PtdIns 4-phosphate, PtdIns(4)P; PtdIns-5-phosphate, PtdIns5P; PtdIns 3,5-bisphosphate, PtdIns(3,5)P2; PtdIns 4,5-bisphosphate PtdIns(4,5)P2; PtdIns 3,4-bisphosphate, PtdIns(3,4)P2; PtdIns 3,4,5-triphosphate, PtdIns(3,4,5)P3], most organisms produce fewer. For example, baker’s yeast (Saccharomyces cerevisiae) and plants produce the first five, but lack the capacity to produce PtdIns(3,4)P2 or PtdIns(3,4,5)P3. PtdIns(4)P metabolism is highlighted in this figure as this phosphoinositide is the focus of discussion.
Phosphoinositide distribution in organelles
With regard to phosphoinositides serving as membrane signaling platforms, early live cell lipid imaging studies were made possible by development of genetically encoded fluorescence biosensors with positional specificities for phosphorylated headgroups [10,11]. Initial studies with such biosensors reported compartment-specific enrichments for particular phosphoinositides. For example, the endocytic system typically scored as enriched in PtdIns(3)P, the TGN in PtdIns(4)P, the vacuole/lysosomal system in PtdIns(3,5)P2, and the plasma membrane in PtdIns(4,5)P2, PtdIns(3,4)P2 and PtdIns(3,4,5)P3. Those compartment-specific profiles gave birth to the view that phosphoinositides are primary determinants of compartmental identity [12].
While there is no doubt that specific phosphoinositides play important contributing roles as coincidence signals that help write a biochemical code for compartment identity, this concept is an oversimplification. That the phosphoinositide-binding biosensor domains register other factors in membranes than just the phosphoinositide ligands themselves was indicated by the results of cell phenotyping analyses where the biological consequences of biosensor overexpression were monitored [13,14•]. Phosphoinositide distributions are also a function of the model organism — suggesting a strict identity code is not a rigid principle. For example, plants do not follow the mammalian script in terms of the shape of organelle-specific enrichment of phosphoinositides. Biosensor imaging experiments suggest PtdIns(4)P is most highly represented in plant plasma membranes [14•] and enrichment of PtdIns(4,5)P2 is scored not only in plant plasma membranes but also in intracellular compartments (secretory or endocytic vesicles?) in the tip cytoplasm of growing root hairs [15,16].
Moreover, a clear demonstration that phosphoinositide distribution is less organelle specific than first advertised is highlighted by characterization of a PtdIns(4)P-specific probe (P4M) engineered from the SidM protein of the bacterial pathogen Legionella pneumophila. Consistent with the PtdIns(4)P distribution profiles previously described from immune-electron microscopy approaches [17], P4M-based biosensors detect PtdIns(4)P pools not only in mammalian late-Golgi compartments, but also in endosomes, lysosomes, and in the plasma membrane [18]. Those findings emphasize that phosphoinositide biosensor localization in cells cannot be confidently extrapolated to phosphoinositide mass. The biosensors only report phosphoinositide pools accessible to the specific biosensor being used, and we often do not understand the parameters that determine accessibility. This interpretive caveat applies to the use of biosensors for imaging the intracellular distribution and dynamics of any lipid.
Lipid signaling in the TGN/endosomal system
If one were tasked with monitoring the physiological status of a cell, one would be well-advised to place a stethoscope on the highly integrated system of trans-Golgi network (TGN) and endosomal compartments [19,20]. Collectively, this system consists of a continuum of maturing organelles that form a signaling node where membrane trafficking interfaces with multiple aspects of lipid metabolism. These aspects now include metabolism of glycerol-based phospholipids (e.g. phosphoinositides; phosphatidylcholine, PtdCho; phosphatidic acid, PtdOH; phosphatidylserine, PtdSer), neutral lipids (e.g. diacylglycerol, sterols), sphingolipids (ceramide, sphingomyelin), and lyso-phospholipids (lipids lacking one of the two acyl chains). The focus of this review will be on the interface between these various aspects of lipid metabolism and membrane trafficking in the TGN/endosomal system.
PtdIns(4)P production in the TGN/endosomal system
The available evidence identifies PtdIns(4)P signaling as a major nexus for integrating wider aspects of lipid metabolism with membrane trafficking through the TGN/endosomal system. This lipid is produced via the action of PtdIns 4-OH kinases (PI4K), and the pools of PtdIns(4)P that reside in different intracellular compartments are produced by distinct enzymes. Mammalian PI4Ks fall into two distinct structural classes categorized as type II enzymes (PI4KIIα, PI4KIIβ) and type III enzymes (PI4KIIIα, PI4KIIIβ) [21]. The yeast type II enzyme is Lsb6 whereas the yeast PI4KIIIα and PI4KIIIβ enzymes are designated Stt4 and Pik1, respectively [22–25]. The type II PI4Ks localize to the TGN/endosomal system, to post-Golgi vesicles and the plasma membrane [26–29], and perhaps to the endoplasmic reticulum [30,31]. The mammalian and yeast PI4KIIIα and PI4KIIIβ enzymes primarily localize to the plasma membrane and TGN/endosomal compartments, respectively [25,32].
The type II enzymes remain poorly characterized, but available evidence from mammalian, fly and yeast models indicates a role in the endosomal system [33–35]. Interestingly, the yeast type II PI4K (Lsb6) is neither essential for cell viability, nor does its recognized involvement in stimulating endosome dynamics require PI4K enzymatic activity [35]. In neither the mammalian nor the fly cases is it firmly established whether the endosomal dynamics phenotypes require type II PI4K lipid kinase activities, or not. By contrast, the type III proteins are each individually essential for cell viability, collectively account for >90% of the PtdIns(4)P synthesis in yeast cells, and these enzymes are non-redundant in terms of biological function [24]. This biological non-redundancy reflects the functional distinction of the respective PtdIns (4)P pools produced. The plasma membrane-localized PI4KIIIα generates plasma membrane pools of the phosphoinositide, whereas the TGN/endosome-localized PI4KIIIβ is responsible for producing TGN/endosomal pools of PtdIns(4)P [24,36,37].
In terms of membrane trafficking through the TGN/endosomal system, PtdIns(4)P pools produced by the type IIIβ otholog Pik1 are of primary importance. Direct evidence to that effect comes from demonstrations that impaired Pik1 PtdIns 4-OH kinase activity evokes a defect in protein export from late Golgi compartments in yeast [38,39]. Our current understanding for how PtdIns(4)P signaling regulates TGN/endosomal membrane trafficking functions rests on identification of PtdIns(4)P effectors that translate the PtdIns(4)P chemical code to action at the level of membrane trafficking. PtdIns(4)P effectors include clathrin adaptors [40,41], and other cargo adaptors such as the yeast Vps74 PtdIns(4)P binding protein and its mammalian ortholog GOLPH3 [42–45]. GOLPH3 is proposed to represent a core component of the TGN membrane trafficking machinery that interacts with the non-conventional actin-based myosin Myo18A to force membrane scission and release of TGN-derived transport vesicles [44].
The interface of PtdIns(4)P signaling with activities of membrane trafficking regulators such as Rab and Arf family GTPases is also well-established. It includes interactions with Rab nucleotide exchange factors and tethers [46–48], and with proteins that specifically bind GTP-bound Arf–GTP. Those include the Gga proteins [49–51], the ‘four phosphate adaptor proteins’ (FAPPs) that interact with both ARF-GTP and with PtdIns(4)P [52,53], and protein kinase D (PKD) that collectively contribute to the biogenesis of Golgi-derived transport carriers [54,55].
PtdIns(4)P and broader regulation of lipid metabolism in the TGN/endosomal system
Initial studies demonstrated that trafficking from the yeast TGN required a proper coordination of PtdCho biosynthesis with essential membrane trafficking pathways, and that the threshold requirement of PtdIns(4)P for trafficking from the TGN/endosomal system is powerfully modulated by metabolic flux through the CDP-choline pathway for PtdCho biosynthesis [5,6,56]. Those findings show that functional coordination of lipid metabolic pathways with phosphoinositide signaling is a foundational concept underlying control of TGN membrane trafficking. We now appreciate that PtdIns(4)P signaling coordinates the activities of other aspects of lipid metabolism in the TGN/endosomal system, and that it does so in a manner that modulates membrane trafficking functions as well. Significant focus in this regard is shifting to the various involvements of sphingolipids and cholesterol in traffic control (Figure 2).
Figure 2.

PtdIns(4)P and sphingolipid metabolism are coordinated in the TGN/endosomal system. Arf1-mediated recruitment of PKD to TGN membranes, and activation of the enzyme, requires diacylglcerol which itself is generated by sphingomyelin synthase (SMS) that consumes ceramide (chaperoned by CERT) and PtdCho to produce sphingomyelin (SM) and diacylglycerol (DAG). CERT and PI4KIIIβ are both PKD substrates. PKD downregulates the former and activates the latter, thereby providing a mechanism for both sensing and coordinating the activities of sphingolipid and PIP metabolic pathways in the TGN. Vesicle biogenesis can be generated by lateral segregation of sphingomyelin into microdomains that platform vesicle formation and perhaps clustering of cargo receptors such as those of the p24 family and Cab45 proteins, by the action of the PtdIns(4)P-binding protein FAPP2 which tubulates membranes and also regulates glucosylceramide (GlcCer) metabolism in the Golgi to promote complex GSL synthesis, and by the action of PtdIns(4)P in recruiting various cargo adaptors or ‘receptors’ that promote cargo packaging into forming vesicles.
One case in point involves the linkage of TGN membrane tubulation and vesicle biogenesis catalyzed by FAPP2 with integration of the metabolism of sphingolipids such as sphingomyelin, glucosylceramide and higher-order glyco-sphingolipids. FAPP2 is both a PtdIns(4)P-binding and a glucosylceramide-binding protein that regulates complex glycolipid synthesis in the Golgi system [52,57]. Ceramide transfer protein (CERT) is a PtdIns(4)P-binding protein required for sphingomyelin synthesis in Golgi membranes, and CERT is suggested to mediate non-vesicular transfer of ceramide from the endoplasmic reticulum to TGN membranes [58,59].
Sphingolipid metabolism and TGN PtdIns(4)P status are tightly coordinated [60••]. Specifically, the net efficiency by which the Sac1 PIP-phosphatase degrades PtdIns4(P) in the mammalian TGN is inversely proportional to metabolic flux through the SL-biosynthetic pathway in Golgi membranes. This metabolic coordination acts as a rheostat for maintaining TGN sphingolipid content in the face of sphingolipid flow into and from the organelle. This rheostat interfaces with membrane traffic because sphingomyelin (and other sphingolipids) can laterally organize themselves with cholesterol to form microdomains that provide a chemical environment that platforms formation of distinct classes of transport carriers into which transport cargo can be differentially sorted [61,62,63•]. In what might prove to be a related mechanism, the biological function of some cargo receptors, such as Cab45, is potentiated by the sphingolipid status of TGN membranes [64••].
Another example is provided by the yeast peripheral PtdIns(4)P-binding protein Vps74 whose function is responsive to both sphingolipid and PtdIns(4)P signals. Vps74 functions in retrograde transport of integral membrane glycosyltransferases from TGN to medial Golgi compartments, and the PtdIns(4)P pool involved in Vps74 recruitment to the TGN surface is produced by the yeast PI4KIIIβ enzyme Pik1 [42]. Yeast cells lacking Vps74 not only show deficits in specific Golgi sphingolipids, but show broadened PtdIns(4)P distribution throughout the Golgi system [65]. While Vps74-deficient cells are viable, these mutants require complex sphingolipid synthesis to maintain viability, and mutants compromised for complex sphingolipid production show deficits in glycosyl-transferase retrieval back to medial Golgi compartments. This is likely a conserved pathway. Rotini (Rti), the Drosophila Vps74 ortholog, also functions in retrieval of a special class of glycotransferases from the TGN to medial Golgi, and these particular glycosyltransferases are required for correct processing of heparan sulfate proteoglycans [66,67].
Lipid microdomains are but one mechanism by which sphingolipids can modulate trafficking processes. These can also function as de facto cofactors where a single sphingomyelin molecule of a specific molecular species interacts directly with a protein component of the membrane trafficking machinery and modulates its activity. The foundational example is provided by the single transmembrane p24 protein whose activity in recruiting cargo for packaging into COP1-coated vesicles requires its transmembrane domain to bind to a sphingomyelin molecule [68]. Interestingly, the SM-binding signature of the p24 transmembrane domain (VXXTLXXIY) is present in the transmembrane domains of a number of other proteins as well [69].
PtdIns(4)P and regulation of PtdSer membrane asymmetry in the TGN/endosomal system
Topological distribution of specific phospholipids is also responsive to PtdIns(4)P signaling. A phosphatidylserine (PtdSer) flippase of the P4-type ATPase family (Drs2) functionally interacts with Arf-dependent and clathrin-dependent pathways for TGN-derived vesicle biogenesis in yeast [70]. Drs2 retrieves PtdSer from the lumenal leaflet of TGN/endosomal membranes to the cytoplasmic leaflet and establishes a leaflet asymmetry so that PtdSer is relatively enriched on the TGN/endosome surface. The Arabidopsis Drs2 ortholog Ala3 functions similarly in coupling PtdSer flippase activity to membrane trafficking from TGN/endosomes [71].
In terms of mechanism, the ATPase and PtdSer flippase activities of Drs2 are stimulated by binding to both PtdIns (4)P generated by the PI4KIIIβ Pik1 and the Gea2 Arf guanine nucleotide exchange factor. Thus, PtdIns(4)P and the protein activator of Arf-GTP production cooperate in a coincidence detection mechanism to regulate Drs2 activity in TGN membranes [72]. Drs2-generated PtdSer asymmetry supports formation of TGN-derived clathrin coated vesicles by increasing local membrane curvature and negative charge so that the Arf GTPase activating protein Gcs1 can be recruited to TGN membranes to optimize vesicle formation and cargo loading [73]. Other evidence suggests that Drs2 activity controls lateral distribution of sterol in TGN membranes to regulate protein sorting into distinct classes of vesicles [74].
PITPs and biological specification of PtdIns(4)P signaling
How is PtdIns(4)P homeostasis controlled at the level of synthesis and at the level of degradation? With regard to the former, it is an underappreciated concept that both PI4KIIIα and PI4KIIIβ are intrinsically inefficient enzymes in biological contexts. That is, these enzymes, even as wild-type enzymes, are incapable of producing sufficient PtdIns(4)P to overcome the activities of erasers of PtdIns(4)P signaling to exert biological responses. Nature solves this problem for eukaryotic cells by the design of PtdIns transfer protein (PITPs) that potentiate PtdIns(4)P production by PI4Ks so that sufficient PtdIns (4)P is produced to support PtdIns(4)P signaling in the face of the opposing action of signaling antagonists [56,75,76]. Although the presently known PITPs fall into two ancient and distinct structural families [i.e. those with the Sec14 fold and those that adopt a StART (Steroidogenic Acute Regulatory Protein-related lipid transfer domain)-like fold], the available evidence indicates these PITPs operate on a similar mechanistic principle. That is, PITPs couple a heterotypic exchange cycle involving PtdIns and a second lipid ligand (classically PtdCho) to ‘present’ PtdIns to the PI4K. This heterotypic exchange cycle renders the engaged PtdIns molecule a superior substrate for the PI4K and thereby potentiates the catalytic activity of the enzyme [68,75–77].
The evolutionary design of PI4Ks as inefficient enzymes holds significant advantages. It not only allows cells to exert exquisite control over PI4K activity and PtdIns(4)P signaling, but it also allows the physical organization of PI4K signaling units into discrete nanocircuits (or signaling pixels) assembled around individual PI4K enzyme complexes [56,76]. High resolution ‘pixelation’ of membrane surfaces provides an attractive mechanism for functionally expanding what is a chemically homogenous PtdIns(4)P code into a high-resolution membrane screen capable of supporting a diverse set of biological outcomes for PtdIns(4)P signaling [4,75,78,79]. PITP-mediated instructive channeling of PtdIns(4)P synthesis and signaling to specific biological outcomes provides new opportunities for generating small molecule inhibitors that have the property of imposing PtdIns(4) P pool-specific interference on signaling [80–82].
PITP-mediated control of TGN/endosomal PtdIns(4)P in mammalian systems
The biological importance of PITPs is on display in vertebrate (zebrafish) and mammalian (mouse) contexts as well and, in all cases studied to date, the ability of the PITP to stimulate PtdIns(4)P production is required for biological activity [76,77]. More recent work with START-like PITPs connects Golgi PI4K signaling with establishment and/or maintenance of extreme cell polarity in embryonic neural stem cells (NSCs) of developing mammalian forebrain (Figure 3). A pair of START-like PtdIns/PtdCho-exchange PITPs (PITPα and PITPβ) support production of a TGN/endosomal PtdIns(4)P pool sufficient for recruitment of GOLPH3 and CERT in a cell-autonomous pathway essential for: (i) polarized distribution of the Golgi system and NSC polarity, and (ii) non-cell-autonomous maintenance of the neurogenic niche that supports development of the neocortex [83••]. Drosophila encodes one PITPα/PITPβ ortholog (Vib) and, similar to the case of embryonic mammalian NSCs, deficiencies in this activity impair fly neuroblast homeostasis. In flies, Vib supports production of a plasma membrane PtdIns(4)P pool (likely in cooperation with PI4KIIIα) to facilitate a non-muscle myosin and actin-based asymmetric partitioning of cell fate determination factors during cytokinesis [84••].
Figure 3.
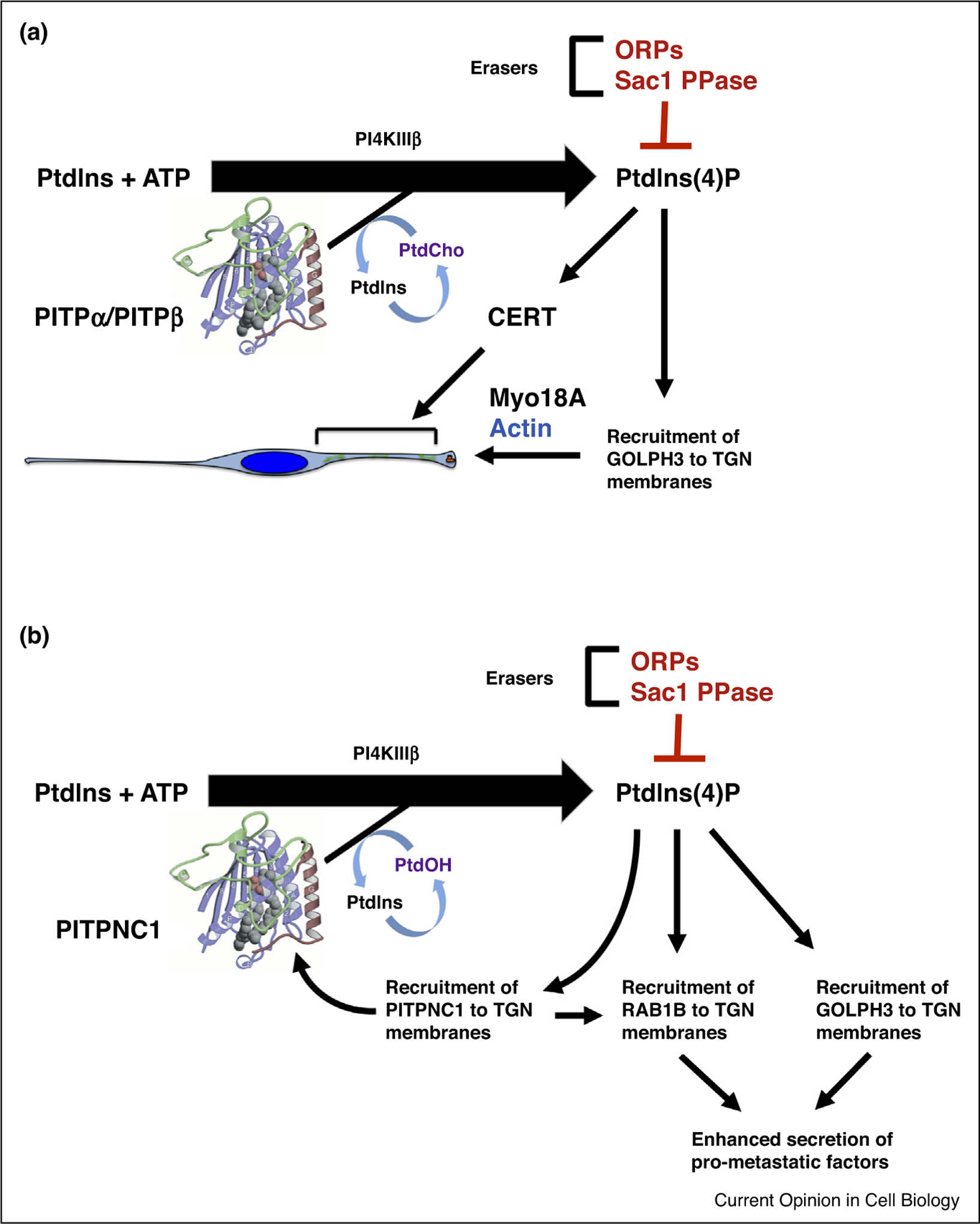
Class I START PITP function in apical loading of the Golgi system in neural stem cells. Class I START PITPs exchange PtdCho and PtdIns, thereby stimulating PtdIns 4-OH kinase activity on late Golgi membranes. The PtdIns(4)P pool recruits GOLPH3 and CERT to Golgi membranes with GOLPH3 subsequently engaging the apically directed actin machinery via the nonconventional myosin Myo18A. This interaction promotes loading of the Golgi system to the neural stem cell apical process, and in this fashion produces an asymmetry critical for establishment/maintenance of NSC polarity. CERT functions either downstream of GOLPH3 or in a parallel pathway to regulate Golgi positioning and NSC polarity. The underlying mechanism remains under investigation and might involve regulation of apically directed, rather than bulk, membrane trafficking.
GOLPH3, the START-like PITP Pitpnc1, and cancer
GOLPH3 not only binds PtdIns(4)P in the TGN/endosomes of mammalian NSCs, but there exists a compelling cancer link between GOLPH3 and yet another START-like PITP designated PITPnc1 [85,86••]. In that regard, PITPnc1 expression is downregulated by the ‘anti-metastatic’micro-RNA miR-126 whose silencing results in enhanced angiogenesis and aggressive tumor metastasis. PITPnc1 likely contributes to production of the TGN PtdIns(4)P pool required for membrane recruitment of GOLPH3 in cancer cells, and is itself reported to be a PtdIns(4)P binding protein — an activity that might help recruit this PITP to TGN membranes via a positive feedback loop which powers recruitment of GOLPH3 and the RAB1B GTPase to enhance secretory capacity of the TGN/endosomal system ([86••]; Figure 3). A GOLPH3/RAB1 connection is also evident in Drosophila where these components cooperate to support membrane incorporation at the point of cleavage furrow ingression during cytokinesis [87].
Erasers of PtdIns(4)P signaling — the Sac1 phosphatase
With regard to mechanisms of downregulation of PtdIns (4)P signaling, key players were first recognized in the ‘bypass Sec14’ genetic screen that identified loss-of-function mutations that restored cell viability (and efficient membrane trafficking through the TGN/endosomal system) to yeast cells completely lacking Sec14 PITP function. Those ‘bypass Sec14’ mutants fall into two general classes: (i) one where PtdIns(4)P levels are dramatically elevated, and (ii) another where mutants thrive in the face of low PtdIns(4)P levels normally insufficient to support either membrane trafficking from TGN/endosomes or cell viability [5,56,88,89].
The first class identifies the Sac1 lipid phosphatase that is the major enzyme for degrading PtdIns(4)P in the cell [90,91]. Its primary compartment of residence is the endoplasmic reticulum (ER; 88,92), and this localization gave rise to proposals that Sac1 acts in trans at ER-plasma membrane contact sites to hydrolyze plasma membrane PtdIns(4)P pools [93]. Structural studies contradict this notion. Sac1 is built for degrading PtdIns(4)P in its membrane of residence (i.e. in cis; 94,95). Second, in addition to first reports of Golgi pools of Sac1 [88], other data indicate this lipid phosphatase cycles from ER to Golgi membranes in response to nutrient-limiting or growth factor-limiting conditions [96,97]. Thus, Golgi PtdIns(4)P-dependent secretory activity can be controlled by regulating Golgi Sac1 pools.
In yet a third line of evidence, yeast Vps74 binds to both PtdIns(4)P and to Sac1, and has the capacity to locally control PtdIns(4)P pools [65]. In what is likely a constitutive process, Vps74 encounters Sac1 that has trafficked from the ER to the Golgi, and this complex maintains the increasing PtdIns(4)P gradient observed as one moves from early to medial to late Golgi compartments. Sac1 egress from ER is regulated in part by a Sac1 phosphorylation/14-3-3 protein-binding cascade that promotes Sac1 packaging into COPII vesicles [98].
Erasers of PtdIns(4)P signaling — the Kes1 PtdIns(4)P/sterol exchange protein
Included in the second class of ‘bypass Sec14’ mutants are loss-of-function mutations in a member of the oxysterol binding protein family Kes1/Osh4 [89]. Of the seven yeast oxysterol binding related proteins (ORPs or Osh proteins), Kes1/Osh4 is unique in its functional antagonism of Sec14-dependent PtdIns(4)P signaling [89,99••].Curiously, like the PtdIns(4)P signaling protagonist Sec14, Kes1 is also a lipid transfer protein in vitro — in this case one that executes a heterotypic PtdIns(4)P/ergosterol exchange reaction and functions as a negative regulator of PI4KIIIβ (Pik1)-dependent PtdIns(4)P signaling and membrane trafficking through the TGN/endosomal system [100,101]. Surprisingly, while Kes1 in vivo activity as PtdIns(4)P signaling antagonist in the TGN/endosomal trafficking program is fully ablated by defects in PtdIns(4)P binding, that same activity is enhanced by sterol binding deficits [102].
There are two competing ideas for how Kes1/Osh4 executes its biological function. One proposes an inter-organelle lipid transfer mechanism, while the second posits a sterol-based tuning mechanism that determines the amplitude of PtdIns(4)P sequestration by Kes1 from its pro-trafficking effectors (Figure 4). The first is the counter-current hypothesis where Kes1/Osh4 is envisioned to transfer sterol from ER to TGN membranes, and then reciprocally transfer PtdIns(4)P from the TGN to the ER for Sac1-mediated degradation [103,104]. This general idea has been expanded to ORPs in general, and to other inferred non-vesicular pathways for inter-organelle lipid transfer. Included in that extension is mobilization of PtdSer to the plasma membrane with a return itinerary for PtdIns(4)P back to the ER for Sac1-mediated hydrolysis — although there is some controversy on that point regarding which phosphoinositides and even which lipids are transported in counter-current fashion [105,106,107•,108••].
Figure 4.

Models for how Kes1/Osh4 downregulate PtdIns(4)P signaling in the TGN. Left panel: The sterol/PtdIns(4)P counter-current hypothesis. This model proposes the sterol/PtdIns(4)P exchange reaction occurs on TGN membranes to supply the system with sterol transported from the ER, and ferries PtdIns(4)P from the TGN to ER for its degradation in that compartment by the Sac1 lipid phosphatase. The degradation step is proposed to provide the energy to drive the cycle. In this scenario, both sterol transport to the TGN and PtdIns(4)P counter-transport to the ER are essential activities for Kes1/Osh4 biological function. Right panel: The sterol/sensing hypothesis. This model posits the operative sterol/PtdIns (4)P exchange reaction occurs on TGN membranes and does not involve inter-organelle lipid transport as functional prerequisite. The competition for Kes1/Osh4 binding to these lipids coordinates sterol status (scored by sterol binding) with the strength of Kes1/Osh4-mediated sequestration of PtdIns(4)P from its pro-trafficking effectors on TGN membranes (scored by PtdIns(4)P-binding). The basic principle is that Kes1/Osh sets a trafficking brake upon binding PtdIns(4)P, and lipid exchange with an available sterol molecule displaces a bound PtdIns(4)P molecule from Kes1/Osh4 – thereby releasing the signaling/trafficking brake by releasing sterol-bound Kes1/Osh4 from TGN membranes. Thus, the fraction of Kes1/Osh4 on TGN membranes reflects the strength of the signaling/trafficking brake, and this scalar is indirectly proportional to the availability of sterol for binding by Kes1/Osh4. This model predicts Kes1/Osh4 lacking sterol binding activity is a powerful signaling brake (enhanced function), while Kes1/Osh4 defective in PtdIns(4)P binding is bereft of biological function.
There remains considerable debate regarding other experimental foundations of the counter-current model as well. A recent study in mammalian cells suggests as much as 50% of the cellular PtdIns(4)P is turned over in the Golgi system via the OSBP/Sac1 counter-current pathway as a function of the size of the sterol pool to be mobilized from the ER [109••]. In contradiction to that report, other evidence from mammalian cells shows regulation of PtdIns(4)P and cholesterol homeostasis by that very same mammalian OSBP is substantially (if not totally) independent of Sac1 activity [110••]. The base claim that yeast and mammalian ORPs are lipid carriers that promote non-vesicular sterol trafficking between intracellular membranes is also a controversial one. Some studies argue in favor [111,112], others against [113,114,115••]. However, the fact that neither ablation of all known membrane tethers that might support nonvesicular lipid transfer through contact sites, nor wholesale ablation of ORP function, has measurable effect on sterol transport in yeast makes a powerful case against the idea that ORPs are authentic sterol transporters – through contact sites or otherwise [115••].
Unlike the indirect assays typically used to infer non-vesicular lipid transport in living cells, the Sec14/Kes1 functional antagonism at the level of PtdIns(4)P signaling in the yeast TGN provides a physiologically relevant assay for Kes1/Osh4 function — one supported by an unambiguous biological phenotype. It is in this context that key predictions of the counter-current hypothesis can be examined with greater confidence. While the prediction of the model that PtdIns(4)P binding is an essential functional property of Kes1/Osh4 is fulfilled [99••,100,101], other key predictions are not supported by experiment. Contrary to the prediction that sterol binding is required for Kes1/Osh4 activity in vivo as a sterol carrier, Kes1 activity as an antagonist of PtdIns(4)P signaling at the TGN is enhanced upon loss of its ability to bind sterol [102]. Moreover, the role of the Sac1 PtdIns(4)P phosphatase as proposed by the counter-current model also does not easily conform to in vivo data. Whereas Kes1/Osh4 and Sac1 deficits have in common the ‘bypass Sec14’ phenotype, there is a curious PtdIns(4)P pool discrepancy with the basic foundation of the counter-current hypothesis. Kes1/Osh4 is responsive to the Pik1 (PI4KIIIβ)-generated pool of PtdIns(4)P in TGN/endosomes [100]. Yet, the cellular pool of PtdIns(4)P degraded by Sac1 in vivo is almost exclusively restricted to that produced by Stt4 (PI4KIIIα; 24,116). Arguments that these discrepancies can be accounted for by functional redundancy of the six other ORPs with Kes1/Osh4 is contra-indicated by experiment. None of the remaining ORPs – either individually or in combination – execute Kes1/Osh4 functions in the TGN [89,99••,102].
The alternative hypothesis proposes what is fundamentally a sterol sensing mechanism [102]. This model posits Kes1/Osh4 functions as a sensor which employs its differential lipid binding activities to run a heterotypic sterol/PtdIns(4)P exchange cycle on TGN membranes. This exchange cycle contributes to a rheostat mechanism that coordinates amplitude of the Kes1/Osh4 brake on PtdIns(4)P signaling and trafficking in response to ‘available’ sterol (Figure 4). This model is compatible with the existence of a TGN/endosomal Sac1 pool that degrades PtdIns(4)P locally, given the strength of any Kes1/Osh4 brake on PtdIns(4)P-signaling would be sensitive to Sac1 activity, and with the experimental result that loss of sterol binding capacity further activates Kes1/Osh4. How generally applicable this model is to the functional mechanisms of other ORPs remains to be determined.
New frontiers at the TGN/endosomal membrane trafficking/lipid signaling interface
That there is more to be discovered regarding PtdIns(4)P signaling in membrane traffic control is highlighted by a remarkable study that identifies a requirement for a Vps13/centrin complex on both TGN and late endosomal membranes for efficient membrane trafficking from the former compartment to the latter [117••]. The novelty here is that centrin is a core component of the spindle pole body that organizes the mitotic spindle in dividing cells. Further evidence to indicate a direct role for the Vps13/centrin complex in lipid signaling comes from studies of sporulating yeast cells that leverage PtdOH production via phospholipase D activity to build new membrane envelopes (prospore membranes) that encapsulate the newly replicated genomes produced by meiosis in the nutrient-deprived cells [118,119]. Prospore expansion is impaired in Vps13-deficient cells and this defect is associated with diminution of PtdOH and PtdIns(4)P pools. Congruent conclusions are reached in a mammalian cell model [120].
Taken together, the data identify the highly conserved Vps13 as yet another integrator for PtdIns(4)P, and perhaps PtdOH, signaling with TGN/endosomal membrane trafficking. The biological relevance of these findings is highlighted by the fact that inherited neurodegenerative diseases are associated with mutations in three of the four Vps13 isoforms expressed in humans [121]. On the basis of structural analyses and lipid transfer assays in vitro, it is suggested that Vps13 isoforms transfer lipids between organelles in vitro via a non-vesicular mechanism [122•].
Membrane trafficking and lyso-phospholipids
Active lipid metabolism in membranes both produces and consumes lipid precursors, or degradation products and modulates their ability to undergo dynamic processes such as vesiculation and tubulation. These intermediate metabolites do so in part by their disruption of membrane packing order and resultant enhancements of membrane malleability [123]. The Lands cycle of lipid deacylation/reacylation is one such a metabolic activity, and phospholipases A2 and the lyso-PtdOH acytransferase LPAAT3 cooperate to control Golgi dynamics and membrane trafficking [124,125].
New progress now elevates the rather sparsely investigated arena of acyl chain remodeling and lyso-phospholipid metabolism onto the main stage of lipid-mediated regulation of Golgi secretory function. With regard to the former, a protein complex that includes PI4KIIIβ catalyzes reacylation of lyso-PtdOH to produce of a pool of PtdOH that helps drive the membrane fission reaction that results in release of Golgi-derived vesicles [126•]. The demonstration that fatty-acid binding protein 5 (FABP5) modulates the kinetics of the Sar1 GTPase cycle, and promotes the biogenesis of COPII vesicles suitable for packaging of large cargo, might well reflect an involvement for the Lands cycle in that process [127•]. With regard to lyso-phospholipids, there is new evidence for a contribution of lyso-phosphatidylinositol to formation of COPII vesicles on the ER surface [128••]. While those findings were gleaned from a rather complex genetically sensitized yeast model, the general concept is likely to apply more broadly. A phospholipase A1 that converts PtdOH into lyso-PtdOH (i.e. by cleaving fatty acid from the sn-1 position) is a structural component of mammalian ER exit sites [129].
Global aspects of TGN/endosomal PtdIns(4)P signaling
An exciting new frontier is emerging — namely, the surprising linkage of TGN/endosomal lipid signaling with stage-specific regulation of cell cycle progression. One area in this new frontier involves PtdIns(4)P signaling and the mitotic spindle. This connection was alluded to above regarding the role of Vps13 and PtdIns(4)P in regulating membrane trafficking through the TGN/endosomal pathway in a mechanism that shows an obligate requirement for Vps13 binding to the spindle pole body component centrin to execute the process [117••]. The curious relationship between the Golgi system and the mitotic spindle is also on display in mammalian cells with diminished Sac1 PtdIns(4)-phosphatase activity. In that case, the Golgi is fragmented (but competent for bulk membrane trafficking) and the mitotic spindle is deranged with failure to progress through G2/M and resolve into a productive cell division [130].
Another direct linkage of chromosome metabolism to Golgi PtdIns(4)P signaling comes from the demonstration that the mammalian TGN/endosomal PtdIns(4)P effector GOLPH3 is phosphorylated by a DNA-damage-activated-protein-kinase. This modification results in fragmentation of the Golgi ribbon, as a result of enhanced interaction of GOLPH3 with Myo18A, and an accompanying membrane trafficking block [131]. Genetic studies in yeast further support the existence of a TGN/chromosome integrity checkpoint by showing that activity of the GOLPH3 ortholog Vps74 is required for prevention of telomere shortening, and that this level of control is likely imposed at the level of telomere capping [132].
The yin and yang of Sec14-mediated potentiated PI4KIIIβ signaling and Kes1/Osh4-mediated downregulation of PtdIns(4)P-signaling in the TGN/endosomal system provides another intriguing example where a membrane trafficking control circuit extends to cell cycle contexts [99••,102]. Kes1/Osh4 is not only a brake on membrane trafficking through the TGN/endosomal system, but it is also a physiologically relevant brake on PtdIns(4)P-dependent progression through the G1 stage of the cell cycle (Figure 4). The mechanistic basis for this activity is the reciprocal relationship between Kes1/Osh4 action and mTOR activity [102]. Moreover, a role for Kes1/Osh4 in promoting cell aging and shortening replicative lifespan is also indicated [99••].
The importance of Kes1/Osh4 as central integrator of TGN/endosomal PtdIns(4)P signaling with membrane trafficking and with the cell cycle is further emphasized by the demonstration that Kes1/Osh4 is a major non-histone target for the highly conserved NuA4 lysine acetyltransferase. NuA4 directly regulates Kes1/Osh4 PtdIns(4)P-binding by acetylating a key Lys residue involved in coordinating PtdIns(4)P headgroup binding [99••]. The base concept that cell cycle functions are regulated by lipid metabolism in the endosomal system is further reinforced by the demonstration that a late endosomal pool of PtdIns(3,4)P2 inhibits mTOR activation [133••].
Concluding remarks
We live in a fast pace era in the field of lipid cell biology — particularly as it relates to membrane traffic control. The field holds tremendous excitement as shown by the rapid progress with which fundamental advances are being made in the arena. However, a number of fundamental mechanistic questions remain under serious debate, and technical obstacles to further progress also exist and need to be overcome. Reliable functional assays that directly report on protein activity in a firm physiological context need to be developed. It is our opinion that a number of the ongoing controversies are the result of conclusions being based on inferences gleaned from indirect readouts of unclear physiological significance. We anticipate model organism systems will continue to play an ever larger role in clarifying direction. Biosensors that reliably report on membrane properties such as surface curvature, surface electrostatics, fluidity, and lateral tension also need to be either developed or improved. These biophysical parameters must be key contributors to the regulation of lipid signaling and we presently do not have a suitable handle on how to assess those contributions.
The questions and technical challenges that confront the field notwithstanding, there is no longer any doubt that lipid signaling is a crucial component of the membrane dynamics program of the eukaryotic cell. This point is further reinforced by progress in understanding the key involvements of lipids in cellular membrane trafficking systems not discussed here. These include: (i) the fusion/fission cycle of lysosomes [134], (ii) the maturation of the phosphoinositide content of clathrin-coated vesicles as these structures progress in their trafficking itinerary through the endocytic pathway [135], (iii) the conversion of PtdIns(3)P to PtdIns(4)P pools for egress of membrane vesicles from endosomes into the exocytic pathway [136], and the fission/fusion cycle of autophagosomes [137]. Indeed, the breadth of the lipid metabolism/membrane trafficking interface is of such magnitude that it is reasonable to ask whether there is any such thing as a lipid metabolic pathway that executes solely a house-keeping function. The time has come to consider lipids and lipid metabolism on an equal footing with proteins when considering the mechanisms that lay the foundation for membrane traffic design.
Acknowledgements
We apologize to our colleagues whose important contributions were not included due to space restrictions. Work in the authors’ laboratory is supported by N.I.H. grants GM44530 and GM112591, and an award from the Robert A. Welch Foundation (BE-0017), to VAB.
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Miller EA, Schekman R: COPII - a flexible vesicle formation system. Curr Opin Cell Biol 2013, 25:420–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothman JE: The future of Golgi research. Mol Biol Cell 2010, 21:3776–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ray K: From fission to fusion: a perspective on the research that won the Nobel Prize in Physiology or Medicine, 2013. J Biosci 2014, 39:3–12. [DOI] [PubMed] [Google Scholar]
- 4.Bankaitis VA, Aitken JR, Cleves AE, Dowhan W: An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 1990, 347:561–562. [DOI] [PubMed] [Google Scholar]
- 5.Cleves AE, McGee TP, Whitters EA, Champion KM, Aitken JR, Dowhan W, Goebl M, Bankaitis VA: Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell 1991, 64:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleves A, McGee T, Bankaitis V: Phospholipid transfer proteins: a biological debut. Trends Cell Biol 1991, 1:30–34. [DOI] [PubMed] [Google Scholar]
- 7.Eberhard DA, Cooper CL, Low MG, Holz RW: Evidence that the inositol phospholipids are necessary for exocytosis. Loss of inositol phospholipids and inhibition of secretion in permeabilized cells caused by a bacterial phospholipase C and removal of ATP. Biochem J 1990, 268:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hay JC, Martin TF: Phosphatidylinositol transfer protein required for ATP-dependent priming of Ca2+-activated secretion. Nature 1993, 366:572–575. [DOI] [PubMed] [Google Scholar]
- 9.Hay JC, Fisette PL, Jenkins GH, Fukami K, Takenawa T, Anderson RA, Martin TF: ATP-dependent inositide phosphorylation required for Ca2+-activated secretion. Nature 1995, 374:173–177. [DOI] [PubMed] [Google Scholar]
- 10.Cullen PJ, Cozier GE, Banting G, Mellor H: Modular phosphoinositide-binding domains—their role in signalling and membrane trafficking. Curr Biol 2001, 11:R882–R893. [DOI] [PubMed] [Google Scholar]
- 11.Lemmon MA: Phosphoinositide recognition domains. Traffic 2003, 4:201–213. [DOI] [PubMed] [Google Scholar]
- 12.Behnia R, Munro S: Organelle identity and the signposts for membrane traffic. Nature 2005, 438:597–604. [DOI] [PubMed] [Google Scholar]
- 13.Varnai P, Bondeva T, Tamas P, Toth B, Buday L, Hunyady L, Balla T: Selective cellular effects of overexpressed pleckstrinhomology domains that recognize PtdIns(3,4,5)P3 suggest their interaction with protein binding partners. J Cell Sci 2005, 118:4879–4888. [DOI] [PubMed] [Google Scholar]
- 14. •.Simon ML, Platre MP, Marques-Bueno MM, Armengot L, Stanislas T, Bayle V, Caillaud MC, Jaillais Y: A PtdIns(4)P-drivenelectrostatic field controls cell membrane identity and signalling in plants. Nat Plants 2016, 2:16089. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors use PtdIns(4)P biosensors that do not also bind the ArfGTPase to document a significant plant plasma membrane reservoir of PtdIns(4)P that is likely the largest pool of this phosphoinositide in the cell. This pool imparts high negative surface charge to the plasma membrane which is a defining feature of this membrane relative to other intracellular organelles.
- 15.Vincent P, Chua M, Nogue F, Fairbrother A, Mekeel H, Xu Y, Allen N, Bibikova TN, Gilroy S, Bankaitis VA: A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J Cell Biol 2005, 168:801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenzel I, Ischebeck T, Konig S, Holubowska A, Sporysz M, Hause B, Heilmann I: The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. Plant Cell 2008, 20:124–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond GR, Schiavo G, Irvine RF: Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns(4)P and PtdIns(4,5)P(2). Biochem J 2009, 422:23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammond GR, Machner MP, Balla T: A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J Cell Biol 2014, 205:113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y, Sirkis DW, Schekman R: Protein sorting at the trans-Golgi network. Annu Rev Cell Dev Biol 2014, 30:169–206. [DOI] [PubMed] [Google Scholar]
- 20.Kienzle C, von Blume J: Secretory cargo sorting at the trans-Golgi network. Trends Cell Biol 2014, 24:584–593. [DOI] [PubMed] [Google Scholar]
- 21.Clayton EL, Minogue S, Waugh MG: Mammalian phosphatidylinositol 4-kinases as modulators of membrane trafficking and lipid signaling networks. Prog Lipid Res 2013, 52:294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han GS, Audhya A, Markley DJ, Emr SD, Carman GM: The Saccharomyces cerevisiae LSB6 gene encodes phosphatidylinositol 4-kinase activity. J Biol Chem 2002, 277:47709–47718. [DOI] [PubMed] [Google Scholar]
- 23.Strahl T, Thorner J: Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta 2007, 1771:353–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Audhya A, Foti M, Emr SD: Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell 2000, 11:2673–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baird D, Stefan C, Audhya A, Weys S, Emr SD: Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J Cell Biol 2008, 183:1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clayton EL, Minogue S, Waugh MG: Phosphatidylinositol 4-kinases and PI4P metabolism in the nervous system: roles in psychiatric and neurological diseases. Mol Neurobiol 2013, 47:361–372. [DOI] [PubMed] [Google Scholar]
- 27.Salazar G, Craige B, Wainer BH, Guo J, De Camilli P, Faundez V: Phosphatidylinositol-4-kinase type II alpha is a component of adaptor protein-3-derived vesicles. Mol Biol Cell 2005, 16:3692–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balla A, Tuymetova G, Barshishat M, Geiszt M, Balla T: Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J Biol Chem 2002, 277:20041–20050. [DOI] [PubMed] [Google Scholar]
- 29.Wei YJ, Sun HQ, Yamamoto M, Wlodarski P, Kunii K, Martinez M, Barylko B, Albanesi JP, Yin HL: Type II phosphatidylinositol 4-kinase beta is a cytosolic and peripheral membrane protein that is recruited to the plasma membrane and activated by Rac-GTP. J Biol Chem 2002, 277:46586–46593. [DOI] [PubMed] [Google Scholar]
- 30.Waugh MG, Minogue S, Anderson JS, Balinger A, Blumenkrantz D, Calnan DP, Cramer R, Hsuan JJ: Localization of a highly active pool of type II phosphatidylinositol 4-kinase in a p97/valosincontaining-protein-rich fraction of the endoplasmic reticulum. Biochem J 2003, 373:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weixel KM, Blumental-Perry A, Watkins SC, Aridor M, Weisz OA: Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J Biol Chem 2005, 280:10501–10508. [DOI] [PubMed] [Google Scholar]
- 32.Boura E, Nencka R: Phosphatidylinositol 4-kinases: function, structure, and inhibition. Exp Cell Res 2015, 337:136–145. [DOI] [PubMed] [Google Scholar]
- 33.Balla A, Tuymetova G, Barshishat M, Geiszt M, Balla T: Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J Biol Chem 2002, 277:20041–20050. [DOI] [PubMed] [Google Scholar]
- 34.Burgess J, Del Bel LM, Ma CI, Barylko B, Polevoy G, Rollins J, Albanesi JP, Kramer H, Brill JA: Type II phosphatidylinositol 4-kinase regulates trafficking of secretory granule proteins in Drosophila. Development 2012, 139:3040–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang FS, Han GS, Carman GM, Blumer KJ: A WASP-binding type II phosphatidylinositol 4-kinase required for actin polymerization-driven endosome motility. J Cell Biol 2005, 171:133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakatsu F, Baskin JM, Chung J, Tanner LB, Shui G, Lee SY, Pirruccello M, Hao M, Ingolia NT, Wenk MR et al. : PtdIns(4)P synthesis by PI4KIIIalpha at the plasma membrane and its impact on plasma membrane identity. J Cell Biol 2012, 199:1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodgers MJ, Albanesi JP, Phillips MA: Phosphatidylinositol 4-kinase III-beta is required for Golgi maintenance and cytokinesis in Trypanosoma brucei. Eukaryot Cell 2007, 6:1108–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB: Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J Biol Chem 1999, 274:34294–34300. [DOI] [PubMed] [Google Scholar]
- 39.Walch-Solimena C, Novick P: The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat Cell Biol 1999, 1:523–525. [DOI] [PubMed] [Google Scholar]
- 40.Wang YJ, Wang J, Sun HQ, Martinez M, Sun YX, Macia E, Kirchhausen T, Albanesi JP, Roth MG, Yin HL: Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell 2003, 114:299–310. [DOI] [PubMed] [Google Scholar]
- 41.Daboussi L, Costaguta G, Payne GS: Phosphoinositide-mediated clathrin adaptor progression at the trans-Golgi network. Nat Cell Biol 2012, 14:239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wood CS, Schmitz KR, Bessman NJ, Setty TG, Ferguson KM, Burd CG: PtdIns(4)P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J Cell Biol 2009, 187:967–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu JW, Chang LC, Jang LT, Huang CF, Lee FJ: The N-terminus of Vps74p is essential for the retention of glycosyltransferases in the Golgi but not for the modulation of apical polarized growth in Saccharomyces cerevisiae. PLoS One 2013, 8:e74715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dippold HC, Ng MM, Farber-Katz SE, Lee SK, Kerr ML, Peterman MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu S et al. : GOLPH3 bridges phosphatidylinositol-4-phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell 2009, 139:337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergeron JJM, Au CE, Thomas DY, Hermo L: Proteomics identifies Golgi phosphoprotein 3 (GOLPH3) with a link between Golgi structure, cancer, DNA damage and protection from cell death. Mol Cell Proteomics 2017, 16:2048–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ortiz D, Medkova M, Walch-Solimena C, Novick P: Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J Cell Biol 2002, 157:1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benli M, Doring F, Robinson DG, Yang X, Gallwitz D: Two GTPase isoforms, Ypt31p and Ypt32p, are essential for Golgi function in yeast. EMBO J 1996, 15:6460–6475. [PMC free article] [PubMed] [Google Scholar]
- 48.Mizuno-Yamasaki E, Medkova M, Coleman J, Novick P: Phosphatidylinositol 4-phosphate controls both membrane recruitment and a regulatory switch of the Rab GEF Sec2p. Dev Cell 2010, 18:828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heldwein EE, Macia E, Wang J, Yin HL, Kirchhausen T, Harrison SC: Crystal structure of the clathrin adaptor protein 1core. Proc Natl Acad Sci U S A 2004, 101:14108–14113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Sun HQ, Macia E, Kirchhausen T, Watson H, Bonifacino JS, Yin HL: PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol Biol Cell 2007, 18:2646–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Demmel L, Gravert M, Ercan E, Habermann B, Muller-Reichert T, Kukhtina V, Haucke V, Baust T, Sohrmann M, Kalaidzidis Y et al. : The clathrin adaptor Gga2p is a phosphatidylinositol 4-phosphate effector at the Golgi exit. Mol Biol Cell 2008, 19:1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA: FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol 2004, 6:393–404. [DOI] [PubMed] [Google Scholar]
- 53.Cao X, Coskun U, Rossle M, Buschhorn SB, Grzybek M, Dafforn TR, Lenoir M, Overduin M, Simons K: Golgi protein FAPP2 tubulates membranes. Proc Natl Acad Sci U S A 2009, 106:21121–21125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liljedahl M, Maeda Y, Colanzi A, Ayala I, Van Lint J, Malhotra V: Protein kinase D regulates the fission of cell surface destined transport carriers from the trans-Golgi network. Cell 2001, 104:409–420. [DOI] [PubMed] [Google Scholar]
- 55.Baron CL, Malhotra V: Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science 2002, 295:325–328. [DOI] [PubMed] [Google Scholar]
- 56.Bankaitis VA, Mousley CJ, Schaaf G: The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem Sci 2010, 35:150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.D’Angelo G, Polishchuk E, Di Tullio G, Santoro M, Di Campli A, Godi A, West G, Bielawski J, Chuang CC, van der Spoel AC et al. : Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature 2007, 449:62–67. [DOI] [PubMed] [Google Scholar]
- 58.Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M: Molecular machinery for non-vesicular trafficking of ceramide. Nature 2003, 426:803–809. [DOI] [PubMed] [Google Scholar]
- 59.Kumagai K, Kawano M, Shinkai-Ouchi F, Nishijima M, Hanada K: Interorganelle trafficking of ceramide is regulated by phosphorylation-dependent cooperativity between the PH and START domains of CERT. J Biol Chem 2007, 282:17758–17766. [DOI] [PubMed] [Google Scholar]
- 60. ••.Capasso S, Sticco L, Rizzo R, Pirozzi M, Russo D, Dathan NA, Campelo F, van Galen J, Holtta-Vuori M, Turacchio G et al. : Sphingolipid metabolic flow controls phosphoinositide turnover at the trans-Golgi network. EMBO J 2017, 36:1736–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]; By acutely manipulating sphingolipid metabolism in TGN membranes by offering cells short-chain ceramide, the authors provide evidence that ceramide consumption into sphingomyelin production via sphingomyelin synthase lowers PtdIns(4)P in the TGN system. This reduction in PtdIns(4) P reduces ceramide import into the organelle and thereby sets up a homeostatic circuit that coordinates TGN PtdIns(4)P levels with flux through the sphingolipid biosynthetic pathway.
- 61.Klemm RW, Ejsing CS, Surma MA, Kaiser HJ, Gerl MJ, Sampaio JL, de Robillard Q, Ferguson C, Proszynski TJ, Shevchenko A et al. : Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J Cell Biol 2009, 185:601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duran JM, Campelo F, van Galen J, Sachsenheimer T, Sot J, Egorov MV, Rentero C, Enrich C, Polishchuk RS, Goni FM et al. : Sphingomyelin organization is required for vesicle biogenesis at the Golgi complex. EMBO J 2012, 31:4535–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. •.Deng Y, Rivera-Molina FE, Toomre DK, Burd CG: Sphingomyelin is sorted at the trans Golgi network into a distinct class of secretory vesicle. Proc Natl Acad Sci U S A 2016, 113:6677–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using a novel genetically encoded sphingomyelin biosensor coupled with sophisticated imaging approaches, the authors identify a distinct sphingomyelin-enriched vesicle population that buds from the TGN and exhibits specificity regarding what cargo is packaged for delivery to the PM.
- 64. ••.Deng Y, Pakdel M, Blank B, Sundberg EL, Burd CG, von Blume J: Activity of the SPCA1 calcium pump couples sphingomyelin synthesis to sorting of secretory proteins in the trans-Golgi network. Dev Cell 2018, 47:464–478 e468. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cab45 is a Ca2+-binding cargo receptor protein Cab45 whose oligomerization is stimulated by the action of a sphingomyelin-activated SPCA1 calcium pump of the TGN. Cab45 oligomers are functional cargo receptors competent for cargo packaging into a class of TGN-derived vesicles enriched in sphingomyelin — a signature that the vesicles are derived from sphingomyelin-rich TGN membrane domains.
- 65.Wood CS, Hung CS, Huoh YS, Mousley CJ, Stefan CJ, Bankaitis V, Ferguson KM, Burd CG: Local control of phosphatidylinositol 4-phosphate signaling in the Golgi apparatus by Vps74 and Sac1 phosphoinositide phosphatase. Mol Biol Cell 2012, 23:2527–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang WL, Chang CW, Chang YY, Sung HH, Lin MD, Chang SC, Chen CH, Huang CW, Tung KS, Chou TB: The Drosophila GOLPH3 homolog regulates the biosynthesis of heparan sulfate proteoglycans by modulating the retrograde trafficking of exostosins. Development 2013, 140:2798–2807. [DOI] [PubMed] [Google Scholar]
- 67.Chang W-L, Chang C-W, Chang Y-Y, Sung H-H, Lin M-D, Chang S-C, Chen C-H, Huang C-W, Tung K-S, Chou T-B: The Drosophila GOLPH3 homolog regulates the biosynthesis of heparan sulfate proteoglycans by modulating the retrograde trafficking of exostosins. Development 2013, 140:2798–2807. [DOI] [PubMed] [Google Scholar]
- 68.Contreras FX, Ernst AM, Haberkant P, Bjorkholm P, Lindahl E, Gonen B, Tischer C, Elofsson A, von Heijne G, Thiele C et al. : Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature 2012, 481:525–529. [DOI] [PubMed] [Google Scholar]
- 69.Bjorkholm P, Ernst AM, Hacke M, Wieland F, Brügger B, von Heijne G: Identification of novel sphingolipid-binding motifs in mammalian membrane proteins. Biochim Biophys Acta 2014, 1838:2066–2070. [DOI] [PubMed] [Google Scholar]
- 70.Bjorkholm P, Ernst AM, Hacke M, Wieland F, Brügger B, von Heijne G: Identification of novel sphingolipid-binding motifs in mammalian membrane proteins. Biochim Biophys Acta 2014, 1838:2066–2070. [DOI] [PubMed] [Google Scholar]
- 71.Poulsen LR, Lopez-Marques RL, McDowell SC, Okkeri J, Licht D, Schulz A, Pomorski T, Harper JF, Palmgren MG: The Arabidopsis P4-ATPase ALA3 localizes to the Golgi and requires a beta-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell 2008, 20:658–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Natarajan P, Liu K, Patil DV, Sciorra VA, Jackson CL, Graham TR: Regulation of a Golgi flippase by phosphoinositides and an ArfGEF. Nat Cell Biol 2009, 11:1421–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu P, Baldridge RD, Chi RJ, Burd CG, Graham TR: Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J Cell Biol 2013, 202:875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hankins HM, Sere YY, Diab NS, Menon AK, Graham TR: Phosphatidylserine translocation at the yeast trans-Golgi network regulates protein sorting into exocytic vesicles. Mol Biol Cell 2015, 26:4674–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schaaf G, Ortlund EA, Tyeryar KR, Mousley CJ, Ile KE, Garrett TA, Ren J, Woolls MJ, Raetz CR, Redinbo MR et al. : Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol Cell 2008, 29:191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grabon A, Khan D, Bankaitis VA: Phosphatidylinositol transfer proteins and instructive regulation of lipid kinase biology. Biochim Biophys Acta 2015, 1851:724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grabon A, Bankaitis VA, McDermott MI: The interface between phosphatidylinositol transfer protein function and phosphoinositide signaling in higher eukaryotes. J Lipid Res 2019, 60:242–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ren J, Pei-Chen Lin C, Pathak MC, Temple BR, Nile AH, Mousley CJ, Duncan MC, Eckert DM, Leiker TJ, Ivanova PT et al. : A phosphatidylinositol transfer protein integrates phosphoinositide signaling with lipid droplet metabolism to regulate a developmental program of nutrient stress-induced membrane biogenesis. Mol Biol Cell 2014, 25:712–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu WI, Routt S, Bankaitis VA, Voelker DR: A new gene involved in the transport-dependent metabolism of phosphatidylserine, PSTB2/PDR17, shares sequence similarity with the gene encoding the phosphatidylinositol/phosphatidylcholine transfer protein, SEC14. J Biol Chem 2000, 275:14446–14456. [DOI] [PubMed] [Google Scholar]
- 80.Nile AH, Tripathi A, Yuan P, Mousley CJ, Suresh S, Wallace IM, Shah SD, Pohlhaus DT, Temple B, Nislow C et al. : PITPs as targets for selectively interfering with phosphoinositide signaling in cells. Nat Chem Biol 2014, 10:76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khan D, McGrath KR, Dorosheva O, Bankaitis VA, Tripathi A: Structural elements that govern Sec14-like PITP sensitivities to potent small molecule inhibitors. J Lipid Res 2016, 57:650–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pries V, Nocker C, Khan D, Johnen P, Hong Z, Tripathi A, Keller AL, Fitz M, Perruccio F, Filipuzzi I et al. : Target Identification and mechanism of action of picolinamide and benzamide chemotypes with antifungal properties. Cell Chem Biol 2018, 25:279–290 e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. ••.Xie Z, Hur SK, Zhao L, Abrams CS, Bankaitis VA: A Golgi lipid signaling pathway controls apical Golgi distribution and cell polarity during neurogenesis. Dev Cell 2018, 44:725–740 e724. [DOI] [PMC free article] [PubMed] [Google Scholar]; Functional interference with PITPa/b activity results in cell-autonomous loss of GOLPH3 localization from TGN membranes, and redistribution of the Golgi system from a ribbon network in the NSC apical process to a compact perinuclear location with accompanying loss of NSC polarity. Notably, the PITP-deficient and GOLPH3-deficient NSCs are viable cells competent for directional migration over large, and for bulk secretion. Those behaviors are surprising given previous reports re GOLPH3 function in cell line models and suggest more specialized TGN secretory functions for the PITP/PtdIns(4)P/GOLPH3 axis.
- 84. ••.Koe CT, Tan YS, Lonnfors M, Hur SK, Low CSL, Zhang Y, Kanchanawong P, Bankaitis VA, Wang H: Vibrator and PI4KIIIalpha govern neuroblast polarity by anchoring non-muscle myosin II. eLife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study describes a PITP-dependent and PtdIns(4)P-dependent pathway for regulating the asymmetric cell division program and self-renewal of Drosophila neuroblasts. This pathway involves establishment of a PM pool of PtdIns(4)P that positions a non-muscle myosin appropriately to organize the asymmetric cell division.
- 85.Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K: The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer 2008, 47:939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. ••.Halberg N, Sengelaub CA, Navrazhina K, Molina H, Uryu K, Tavazoie SF: PITPNC1 recruits RAB1B to the Golgi network to drive malignant secretion. Cancer Cell 2016, 29:339–353. [DOI] [PMC free article] [PubMed] [Google Scholar]; PITPnc1 is identified as a highly cancer-relevant gene and a mechanism is described where PITPnc1 potentiates the secretion of pro-metastatic factors, metalloproteases and pro-angiogenic growth factor. The enhanced secretory phenotypes is indicated to result from the ability of PITPnc1 to bind to PtdIns(4)P and promote GOLPH3 and RAB1B recruitment to the TGN.
- 87.Sechi S, Frappaolo A, Belloni G, Giansanti MG: The roles of the oncoprotein GOLPH3 in contractile ring assembly and membrane trafficking during cytokinesis. Biochem Soc Trans 2015, 43:117–121. [DOI] [PubMed] [Google Scholar]
- 88.Cleves AE, Novick PJ, Bankaitis VA: Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J Cell Biol 1989, 109:2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fang M, Kearns BG, Gedvilaite A, Kagiwada S, Kearns M, Fung MK, Bankaitis VA: Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J 1996, 15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- 90.Guo S, Stolz LE, Lemrow SM, York JD: SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem 1999, 274:12990–12995. [DOI] [PubMed] [Google Scholar]
- 91.Rivas MP, Kearns BG, Xie Z, Guo S, Sekar MC, Hosaka K, Kagiwada S, York JD, Bankaitis VA: Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to “bypass Sec14p” and inositol auxotrophy. Mol Biol Cell 1999, 10:2235–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Whitters EA, Cleves AE, McGee TP, Skinner HB, Bankaitis VA: SAC1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J Cell Biol 1993, 122:79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD: Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 2011, 144:389–401. [DOI] [PubMed] [Google Scholar]
- 94.Cai Y, Deng Y, Horenkamp F, Reinisch KM, Burd CG: Sac1-Vps74 structure reveals a mechanism to terminate phosphoinositide signaling in the Golgi apparatus. J Cell Biol 2014, 206:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zewe JP, Wills RC, Sangappa S, Goulden BD, Hammond GR: SAC1 degrades its lipid substrate PtdIns(4)P in the endoplasmic reticulum to maintain a steep chemical gradient with donor membranes. eLife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Blagoveshchenskaya A, Cheong FY, Rohde HM, Glover G, Knodler A, Nicolson T, Boehmelt G, Mayinger P: Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. J Cell Biol 2008, 180:803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Piao H, MacLean Freed J, Mayinger P: Metabolic activation of the HOG MAP kinase pathway by Snf1/AMPK regulates lipid signaling at the Golgi. Traffic 2012, 13:1522–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bajaj Pahuja K, Wang J, Blagoveshchenskaya A, Lim L, Madhusudhan MS, Mayinger P, Schekman R: Phosphoregulatory protein 14-3-3 facilitates SAC1 transport from the endoplasmic reticulum. Proc Natl Acad Sci U S A 2015, 112:E3199–E3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. ••.Huang J, Mousley CJ, Dacquay L, Maitra N, Drin G, He C, Ridgway ND, Tripathi A, Kennedy M, Kennedy BK et al. : A lipid transfer protein signaling axis exerts dual control of cellcycle and membrane trafficking systems. Dev Cell 2018, 44:378–391 e375. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study describes an interface between PtdIns(4)P signaling, membrane trafficking and cell cycle control. Kes1/Osh4 is shown to be a negative regulator of progression through G1 when cells are nutrient deprived, and that Kes1/Osh4 and Sec14 play opposing roles in regulating the initiation of cell division. The specificity of Kes1/Osh4 interaction with Sec14 is again on display, and the physiological importance of Kes1/Osh4 is further emphasized by the demonstration that Kes1/Osh4 is a significant non-histone target for the NuA4 lysine acetyltransferase which targets a key lysine involved in coordination of PtdIns(4)P binding. Similar, but functionally non-redundant, activities in cell cycle control and regulation of lifespan are suggested for other yeast ORPs.
- 100.Li X, Rivas MP, Fang M, Marchena J, Mehrotra B, Chaudhary A, Feng L, Prestwich GD, Bankaitis VA: Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J Cell Biol 2002, 157:63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G: Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J Cell Biol 2011, 195:965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mousley CJ, Yuan P, Gaur NA, Trettin KD, Nile AH, Deminoff SJ, Dewar BJ, Wolpert M, Macdonald JM, Herman PK et al. : A sterolbinding protein integrates endosomal lipid metabolism with TOR signaling and nitrogen sensing. Cell 2012, 148:702–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B: A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell 2013, 155:830–843. [DOI] [PubMed] [Google Scholar]
- 104.Antonny B, Bigay J, Mesmin B: The oxysterol-binding protein cycle: burning off PI(4)P to transport cholesterol. Annu Rev Biochem 2018, 87:809–837. [DOI] [PubMed] [Google Scholar]
- 105.Moser von Filseck J, Copic A, Delfosse V, Vanni S, Jackson CL, Bourguet W, Drin G: INTRACELLULAR TRANSPORT. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science 2015, 349:432–436. [DOI] [PubMed] [Google Scholar]
- 106.Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, Narayanaswamy P, Wenk MR, Nakatsu F, De Camilli P: INTRACELLULAR TRANSPORT. PI4P/phosphatidylserine counter transport at ORP5-and ORP8-mediated ER-plasma membrane contacts. Science 2015, 349:428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. •.Ghai R, Du X, Wang H, Dong J, Ferguson C, Brown AJ, Parton RG, Wu JW, Yang H: ORP5 and ORP8 bind phosphatidylinositol-4,5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane. Nat Commun 2017, 8:757. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work challenges the conclusions of ref 106 that ORP5 and ORP8 are PtdSer/PtdIns(4)P counter-transporters and that PtdIns(4)P binding is required for ORP5/8 recruitment to ER-PM contact sites for PtdSer/PtdIns(4)P counter-current transport. Structural and biochemical of the PH domains of these proteins are concluded to identify these as PtdIns2-binding modules, that recruitment of ORP5/8 to ER-PM contact sites requires PtdIns(4,5)P2-binding, and that ORP5/8 catalyze PtdSer/PtdIns (4,5)P2 counter-current transport between the ER and PM.
- 108. ••.Sohn M, Korzeniowski M, Zewe JP, Wills RC, Hammond GRV, Humpolickova J, Vrzal L, Chalupska D, Veverka V, Fairn GD et al. : PI(4,5)P2 controls plasma membrane PI4P and PS levels via ORP5/8 recruitment to ER-PM contact sites. J Cell Biol 2018, 217:1797–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study is experimentally the most comprehensive of the group on the ORP5/8 topic and it occupies a middle ground between the contrary positions of Refs. [106] and [107]. Structural, cell biological and biochemical data report that the OrRP5/8 PH-domains bind both PtdIns(4,5)P2 and PtdIns(4)P, and that ORP5 recruitment to ER-PM contact sites is efficient is resting cells whereas elevated PtdIns(4,5)P2 is required for ORP8 recruitment to such sites to support PtdSer/PtdIns(4)P counter-current transport. The base conclusion is that ORP5/8 cooperatively regulate PM PtdIns(4,5)P2 by controlling flux through the PtdSer/PtdIns(4)P counter-current pathway.
- 109. ••.Mesmin B, Bigay J, Polidori J, Jamecna D, Lacas-Gervais S, Antonny B: Sterol transfer, PI4P consumption, and control of membrane lipid order by endogenous OSBP. EMBO J 2017, 36:3156–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]; OSBP and Sac1 co-localize to ER-Golgi contact sites and this colocalization is part of the argument for these two proteins running a reciprocal ER to Golgi and return sterol/PtdIns(4)P counter-current to promote sterol egress from ER which is obligately coupled to PtdIns(4)P transfer to ER for degradation. Refs. [109] and [110] test the same key foundation of the model by assessing whether the Sac1 intracellular localization or activity have any bearing on known activities of OSBP. This study presents a body of experimental data from which the authors conclude that OSBP and Sac1 function is closely coupled and that the OSBP/Sac1 circuit is responsible for turnover of the major fraction of the cellular PtdIns(4)P generated. This conclusion is taken as evidence in support of the counter-current hypothesis.
- 110. ••.Charman M, Goto A, Ridgway ND: Oxysterol-binding protein recruitment and activity at the endoplasmic reticulum-Golgi interface are independent of Sac1. Traffic 2017, 18:519–529. [DOI] [PubMed] [Google Scholar]; The authors test the same foundation of the counter-current model addressed in Ref. [109] as it relates to OSBP and Sac1. The study presents several experimental results indicating OSBP and Sac1 function independently. This conclusion is contrary to the one reached in Ref. 109.
- 111.Raychaudhuri S, Im YJ, Hurley JH, Prinz WA: Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J Cell Biol 2006, 173:107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Prinz WA: Lipid trafficking sans vesicles: where, why, how? Cell 2010, 143:870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dittman JS, Menon AK: Speed limits for nonvesicular intracellular sterol transport. Trends Biochem Sci 2017, 42:90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Georgiev AG, Sullivan DP, Kersting MC, Dittman JS, Beh CT, Menon AK: Osh proteins regulate membrane sterol organization but are not required for sterol movement between the ER and PM. Traffic 2011, 12:1341–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. ••.Quon E, Sere YY, Chauhan N, Johansen J, Sullivan DP, Dittman JS, Rice WJ, Chan RB, Di Paolo G, Beh CT et al. : Endoplasmic reticulum-plasma membrane contact sites integrate sterol and phospholipid regulation. PLoS Biol 2018, 16:e2003864. [DOI] [PMC free article] [PubMed] [Google Scholar]; As test of whether Kes1/Osh4 and other ORPs are sterol transporters that operate via inter-membrane contact sites, the authors combinatorially deleted the known membrane tethers and found bidirectional transport of sterols between the ER and PM to be unaffected in tetherless cells. Those cells show significant derangements of the lipidome and sterol organization, including accumulation of PtdIns(4)P in the PM. These derangements can be rescued by synthetic tethers or by simple feeding of cells with precursors of lipid synthesis such as choline. The authors propose that neither ORPs nor inter-membrane contact sites function directly in lipid exchange. Rather, they infer that such structures are integrators that control lipid synthesis. These conclusions challenge a number of inferences regarding the function of contact sites, and are relevant to discussions of the counter-current hypothesis for ORP function in cells.
- 116.Nemoto Y, Kearns BG, Wenk MR, Chen H, Mori K, Alb JG Jr, De Camilli P, Bankaitis VA: Functional characterization of a mammalian Sac1 and mutants exhibiting substrate-specific defects in phosphoinositide phosphatase activity. J Biol Chem 2000, 275:34293–34305. [DOI] [PubMed] [Google Scholar]
- 117. ••.De M, Oleskie AN, Ayyash M, Dutta S, Mancour L, Abazeed ME, Brace EJ, Skiniotis G, Fuller RS: The Vps13p-Cdc31p complex is directly required for TGN late endosome transport and TGN homotypic fusion. J Cell Biol 2017, 216:425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using a permeabilized cell reconstitution assay, this study identifies Vps13 as a factor required for docking and/or homotypic fusion of TGN compartments. This activity is demonstrated to require a tight physical interaction between Vps13 and centrin, and the role of phosphoinositides in this process is also discussed. A structural description of the large Vps13/centrin complex using single particle electron microscopy imaging suggests how this complex may tether two distinct membrane system in close proximity. This work provides an outstanding demonstration of the power of resolution/reconstitution approaches.
- 118.Park JS, Neiman AM: VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J Cell Sci 2012, 125:3004–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Park JS, Okumura Y, Tachikawa H, Neiman AM: SPO71 encodes a developmental stage-specific partner for Vps13 in Saccharomyces cerevisiae. Eukaryot Cell 2013, 12:1530–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Park JS, Halegoua S, Kishida S, Neiman AM: A conserved function in phosphatidylinositol metabolism for mammalian Vps13 family proteins. PLoS One 2015, 10:e0124836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hariri H, Ugrankar R, Liu Y, Henne WM: Inter-organelle ER-endolysosomal contact sites in metabolism and disease across evolution. Commun Integr Biol 2016, 9:e1156278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. •.Kumar N, Leonzino M, Hancock-Cerutti W, Horenkamp FA, Li P, Lees JA, Wheeler H, Reinisch KM, De Camilli P: VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J Cell Biol 2018, 217:3625–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]; An N-terminal domain of Vps13 is shown to have the ability to mobilize glycerophospholipids between membrane bilayers in vitro in a rather non-specific manner. X-ray structure determination of a fungal Vps13 show that domain adopts a tubular conformation with a hydrophobic cavity with sufficient dimension to potentially accommodate several lipid molecules. Different isoforms of the human proteins localize differentially to contact sites between the ER and other organelles that include mitochondria, lipid droplets, late endosomes, and to lysosomes. The authors propose Vps13 transfers lipids between the ER and these organelles.
- 123.Henriksen JR, Andresen TL, Feldborg LN, Duelund L, Ipsen JH: Understanding detergent effects on lipid membranes: a model study of lysolipids. Biophys J 2010, 98:2199–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brown WJ, Chambers K, Doody A: Phospholipase A2 (PLA2) enzymes in membrane trafficking: mediators of membrane shape and function. Traffic 2003, 4:214–221. [DOI] [PubMed] [Google Scholar]
- 125.Schmidt JA, Brown WJ: Lysophosphatidic acid acyltransferase 3 regulates Golgi complex structure and function. J Cell Biol 2009, 186:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. •.Pagliuso A, Valente C, Giordano LL, Filograna A, Li G, Circolo D, Turacchio G, Marzullo VM, Mandrich L, Zhukovsky MA et al. : Golgi membrane fission requires the CtBP1-S/BARS-induced activation of lysophosphatidic acid acyltransferase delta. Nat Commun 2016, 7:12148. [DOI] [PMC free article] [PubMed] [Google Scholar]; A protein complex consisting of PI4KIIIβ, Arf, PKD, the PAK kinase, and the BARS protein scaffolded around a 14-3-3γ dimer is assembled on the TGN surface. This complex promotes a spatially restricted burst of PtdOH acyl chain remodeling by stimulating activity of a lyso-PtdOH acyltransferase (LPAATδ). This activity, in turn, promotes the scission of forming vesicular carriers from the TGN surface.
- 127. •.Melville D, Gorur A, Schekman R: Fatty-acid binding protein 5 modulates the SAR1 GTPase cycle and enhances budding of large COPII cargoes. Mol Biol Cell 2019, 30:387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]; The rules that govern formation of large COPII vesicles capable of incorporating very large cargos such as chylomicrons and procollagen for delivery from the ER to the Golgi system is poorly understood. Using a cell-free assay as primary instrument, this study identifies fatty acid binding protein (FABP5) as modulator of the Sar1 GTPase cycle. This modulation involves enhancement of Sar1 GTP loading and, to a lesser extent, stimulation of GTPase activity. Such altered kinetics of Sar1 activity potentiates biogenesis of such large COPII-coated vesicles. In vivo data support the in vitro results as elevated FABP5 expression enhances secretion of large cargo from cells.
- 128. ••.Melero A, Chiaruttini N, Karashima T, Riezman I, Funato K, Barlowe C, Riezman H, Roux A: Lysophospholipids facilitate COPII vesicle formation. Curr Biol 2018, 28:1950–1958 e1956. [DOI] [PMC free article] [PubMed] [Google Scholar]; Several independent experimental approaches converge on the conclusion that accumulation of lyso-phosphatidylinositol in ER membranes reduces their rigidity and helps promote formation of ER exit sites (i.e. sites where COPII vesicles form), and, therefore, of COPII vesicles, under conditions where the activation of the Arf-like Sar1 GTPase is suboptimal.
- 129.Ong YS, Tang BL, Loo LS, Hong W: p125A exists as part of the mammalian Sec13/Sec31 COPII subcomplex to facilitate ER-Golgi transport. J Cell Biol 2010, 190:331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu Y, Boukhelifa M, Tribble E, Bankaitis VA: Functional studies of the mammalian Sac1 phosphoinositide phosphatase. Adv Enzyme Regul 2009, 49:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Farber-Katz SE, Dippold HC, Buschman MD, Peterman MC, Xing M, Noakes CJ, Tat J, Ng MM, Rahajeng J, Cowan DM et al. : DNA damage triggers Golgi dispersal via DNA-PK and GOLPH3. Cell 2014, 156:413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rodrigues J, Banks P, Lydall D: Vps74 connects the Golgi apparatus and telomeres in Saccharomyces cerevisiae. G3 (Bethesda) 2018, 8:1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. ••.Marat AL, Wallroth A, Lo WT, Muller R, Norata GD, Falasca M, Schultz C, Haucke V: mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science 2017, 356:968–972. [DOI] [PubMed] [Google Scholar]; This work demonstrates a late endosomal pool of PtdIns(3,4)P2 produced by class II PI3KC2b kinase represses mTORC1 activity in mammalian cells upon growth factor deprivation. The repression involves an induced interaction of the lipid kinase with the Raptor subunit of mTORC1. It is proposed that the resultant local synthesis of PtdIns(3,4)P2 recruits 14-3-3proteins that in turn inhibit Raptor.
- 134.Wickner W, Rizo J: A cascade of multiple proteins and lipids catalyzes membrane fusion. Mol Biol Cell 2017, 28:707–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.He K, Marsland R III, Upadhyayula S, Song E, Dang S, Capraro BR, Wang W, Skillern W, Gaudin R, Ma M et al. : Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature 2017, 552:410–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ketel K, Krauss M, Nicot AS, Puchkov D, Wieffer M, Muller R, Subramanian D, Schultz C, Laporte J, Haucke V: A phosphoinositide conversion mechanism for exit from endosomes. Nature 2016, 529:408–412. [DOI] [PubMed] [Google Scholar]
- 137.Yu S, Melia TJ: The coordination of membrane fission and fusion at the end of autophagosome maturation. Curr Opin Cell Biol 2017, 47:92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]


