Abstract
BEAF (Boundary Element-Associated Factor) was originally identified as a Drosophila melanogaster chromatin domain insulator-binding protein, suggesting a role in gene regulation through chromatin organization and dynamics. Genome-wide mapping found that BEAF usually binds near transcription start sites, often of housekeeping genes, suggesting a role in promoter function. This would be a nontraditional role for an insulator-binding protein. To gain insight into molecular mechanisms of BEAF function, we identified interacting proteins using yeast two-hybrid assays. Here, we focus on the transcription factor Serendipity δ (Sry-δ). Interactions were confirmed in pull-down experiments using bacterially expressed proteins, by bimolecular fluorescence complementation, and in a genetic assay in transgenic flies. Sry-δ interacted with promoter-proximal BEAF both when bound to DNA adjacent to BEAF or > 2-kb upstream to activate a reporter gene in transient transfection experiments. The interaction between BEAF and Sry-δ was detected using both a minimal developmental promoter (y) and a housekeeping promoter (RpS12), while BEAF alone strongly activated the housekeeping promoter. These two functions for BEAF implicate it in playing a direct role in gene regulation at hundreds of BEAF-associated promoters.
Keywords: BEAF, insulators, chromatin domains, gene regulation, enhancer–promoter looping, Drosophila
CHROMATIN domain insulator-binding proteins are thought to link nuclear architecture to gene regulation. There is evidence they can separate chromosomal topologically associating domains (TADs), and can either block or facilitate enhancer–promoter communication depending on context (Ali et al. 2016; Chetverina et al. 2017). The main known vertebrate insulator-binding protein is CTCF, often found with the protein complex cohesin (Bell et al. 1999; Vietri Rudan et al. 2015). CTCF usually binds in intergenic regions and introns (Kim et al. 2007). Pairwise mapping of chromatin interactions by Hi-C has found that many CTCF sites are found at TAD boundaries, with convergently oriented motifs at opposite boundaries interacting to form loop domains (Rao et al. 2014). While this contributes to nuclear architecture, it should be noted that CTCF also localizes within TADs, and not all TAD boundaries are associated with CTCF. Nonetheless, TADs play a role in gene regulation, and CTCF plays a role in establishing or maintaining many TAD boundaries (Guo et al. 2015; Lupiáñez et al. 2016).
In contrast, many DNA sequence-specific binding proteins have been identified as insulator proteins for the gene-dense genome of Drosophila melanogaster [Pauli et al. (2016) and references therein]. The Drosophila homolog of CTCF (dCTCF) does not pair to form loop domains and is not preferentially found at TAD boundaries (Rowley et al. 2017). In fact, many fly TADs appear to be separated by regions of active chromatin containing clustered housekeeping genes that form inter-TAD regions (Ulianov et al. 2016; Cubeñas-Potts et al. 2017; Hug et al. 2017). Multiple Drosophila insulator-binding and associated proteins often colocalize at inter-TADs (Van Bortle et al. 2014). The DNA-binding insulator protein with the strongest correlation with these regions is the Boundary Element-Associated Factor of 32 kDa, BEAF (Ulianov et al. 2016). Like their different associations with inter-TADs, the various insulator-binding proteins differ from each other with respect to their localization relative to genes. As examples, roughly 85% of BEAF peaks (Jiang et al. 2009), 35% of dCTCF peaks (Bushey et al. 2009), 30% of GAGA factor (GAF) peaks (Lee et al. 2008), 25% of Zw5 peaks (Model Organism ENCyclopedia Of DNA Elements 3303 and 3304), and 5% of Su(Hw) peaks (Bushey et al. 2009) are within 300 bp of a transcription start site (TSS). These differences suggest that there are differences in molecular mechanisms between vertebrate and insect insulator-binding proteins, as well as differences between the various Drosophila proteins.
Our focus is on BEAF, as a model insulator-binding protein. BEAF was discovered based on its binding to the Drosophila scs’ insulator (Zhao et al. 1995). Other BEAF-binding sites have subsequently been shown to be associated with insulator activity, supporting the idea that it plays a role in insulator function (Cuvier et al. 1998, 2002; Sultana et al. 2011). Consistent with the view that insulators play roles in nuclear architecture, a dominant-negative transgene and a null mutation in BEAF affect chromatin (Gilbert et al. 2006; Roy et al. 2007a). Both disrupt polytene chromosome structure and affect position effect variegation, in addition to affecting scs’ insulator function. Yet genome-wide mapping of BEAF binding found that it is normally found within a few hundred base pairs of TSSs (Bushey et al. 2009; Jiang et al. 2009; Nègre et al. 2010; Liang et al. 2014). It is unclear if BEAF is primarily an insulator protein or a promoter factor, or if these two functions are somehow linked.
Molecular mechanisms by which insulator-binding proteins function are generally unclear. To gain insight into BEAF function, we screened for physical interactions with other proteins. There are two 32-kDa BEAF isoforms encoded by one gene, BEAF-32A and BEAF-32B (Hart et al. 1997). These proteins differ by 80 aa at their N-termini, both of which contain a DNA-binding zinc finger. The remaining 200 aa are identical, and their C-termini have a BESS (BEAF, Suvar(3)7 and Stonewall) domain that mediates BEAF–BEAF interactions (Avva and Hart 2016). BEAF-32B is essential while BEAF-32A is not (Roy et al. 2007a), and genome-wide mapping found that the DNA binding of BEAF-32B is dominant (Jiang et al. 2009). Therefore, we focused on BEAF-32B or the portion of the protein common to both isoforms. We identified a transcription factor that interacts with BEAF: Serendipity δ (Sry-δ). This suggested that one function of promoter-proximal BEAF could be to facilitate communication, including enhancer–promoter looping, with specific transcription factors. Here, we characterize the interaction between BEAF and Sry-δ. We find synergistic activation when both proteins bind near the two promoters tested, and that gene activation by distantly bound Sry-δ is facilitated by promoter-proximal BEAF. There are differences between developmental and housekeeping promoters (Zabidi et al. 2015). We previously reported that BEAF is usually found near housekeeping promoters (Jiang et al. 2009; Shrestha et al. 2018). In the course of these experiments, we found that promoter-proximal BEAF can activate two housekeeping promoters on its own but does not have this effect on a developmental promoter. Our results provide insights into possible roles of BEAF at hundreds of housekeeping promoters in Drosophila.
Materials and Methods
Plasmid construction
Yeast two-hybrid screening:
All complementary DNAs (cDNAs) were PCR amplified using appropriate primers and fused in-frame as EcoRI-SalI restriction fragments on the 3′ side of sequences encoding the GAL4-activation domain (AD) in pOAD. Sources of the cDNAs are given in Table 1. The AbdB cDNA (Drosophila Genomics Resource Center clone RE47096) had a 1-bp deletion in the middle of the homeodomain-coding sequences, which was corrected by QuikChange mutagenesis (Stratagene, La Jolla, CA). Full-length and parts of BEAF-32B were similarly fused to the GAL4-binding domain (BD) in pOBD2, as previously described (Avva and Hart 2016). Gibson Assembly (New England Biolabs, Beverly, MA) was used to insert sequences encoding the N- or C-terminal half of Sry-δ into the EcoRI site of pOAD. All plasmids were confirmed by sequencing. The GAL4-AD yeast two-hybrid (Y2H) library was from Clontech, made in pGADT7 using equal quantities of D. melanogaster polyA RNA isolated from 20-hr embryos, larvae, and adults.
Table 1. Proteins tested in Y2H assays for interactions with BEAF.
| Protein | cDNA source | Y2H result |
|---|---|---|
| From Roy et al. (2007b) | ||
| Abd-A | RE04174 | — |
| Abd-B | RE47096 | — |
| Bcd | LD36304 | (+) |
| Dfd | A | — |
| Dll | IP14437 | — |
| Ftz | IP01266 | — |
| lab | RE63854 | — |
| MRTF | B | — |
| Pb | C | — |
| Scr | D | (+) |
| SpnE | IP03663 | — |
| Su(Hw) | LD15893 | — |
| Taf6 | LD24529 | — |
| zen | E | — |
| Zw5 | LD45751 | — |
| Other proteins | ||
| CP190 | LD02352 | — |
| dCTCF | GH14774 | — |
| D1 | RE39218 | — |
| DREF | CMH | — |
| GAF | F | — |
| NELF-A | F | — |
| NELF-B | F | — |
| NELF-D | F | — |
| NELF-E | F | — |
cDNA sources are Drosophila Genomics Resource Center clone identifiers except: A: (Kuziora and McGinnis 1988); B: (Han et al. 2004); C: (Benassayag et al. 1997); D: (Zeng et al. 1993); E: (Rushlow et al. 1987); F: (Lee et al. 2008); CMH: (Hart et al. 1999). (+) signifies an ambiguous Y2H result, as described in the Results. cDNA, complementary DNA; y2H, yeast two-hybrid.
Pull-down:
The Sry-δ gene lacks introns, so the coding sequence was directly PCR amplified from genomic DNA. Sequences encoding Sry-δ or its N- or C-terminal halves were PCR amplified such that each had an N-terminal Myc epitope tag. PCR products were cloned into a pET3 expression vector through Gibson Assembly, using a unique KpnI site in the plasmid. Construction of a pET plasmid encoding N-terminally FLAG epitope-tagged 32B was previously described (Avva and Hart 2016). All plasmids were confirmed by sequencing.
Bimolecular fluorescence complementation:
Plasmids using modified genomic BEAF sequences, so expression from endogenous BEAF promoters leads to the production of 32B-monomeric Red Fluorescent Protein (mRFP) or 32B-delBESS-mRFP (deletion of the BESS domain), have been described (Roy et al. 2007a; Avva and Hart 2016). Fluorescent protein-coding sequences were excised with KpnI and NotI, and replaced by Gibson assembly with PCR-amplified coding sequences of the Venus yellow fluorescent protein from the pTWV Drosophila gateway vector, incorporating a 7-aa spacer (GTRSAIT) between the BEAF and Venus sequences. Amino acids 1–173 were used for the N-terminal part of Venus (nV) and amino acids 155–239 were used for the C-terminal part (cV) (Hudry et al. 2011) to make plasmids capable of producing 32B-nV, 32B-cV, and 32B-delBESS-cV proteins in Drosophila cells. For Sry-δ, Gibson assembly was used to modify the Act5C promoter plasmid described below with cV sequences. The C-terminal cV fusion was done as described below for VP16 activation-domain tagging.
Luciferase:
Renilla luciferase (from pGL4.70; Promega, Madison, WI) and Sry-δ coding sequences were PCR amplified, and cloned by Gibson assembly into the BamHI site of pPac, which is located between a 2.6-kb Act5C promoter fragment and a 1.2-kb Act5C polyadenylation fragment in pUC18 (Krasnow et al. 1989). VP16-AD coding sequences were PCR amplified (from DD594; kind gift of D. Donze) and fused at the C-terminal end of Sry-δ by Gibson assembly using DraIII (four C-terminal amino acids of Sry-δ were removed). Looping test plasmids were built in pBSKS- (Stratagene). A PCR-amplified 225-bp simian virus 40 (SV40) polyadenylation region from pEGFP-N3 (Clontech) was inserted into the XbaI and SacI sites, followed by insertion of PCR-amplified firefly luciferase coding sequences from pGEM-luc (Promega) into the HindIII and BamHI sites. Gene blocks (IDT) with a 43-bp wild-type or mutant BEAF-binding site from scs’ (Zhao et al. 1995), connected to a minimal −69 to +71 y promoter (Morris et al. 2004; Melnikova et al. 2008) or −33 to +67 RpS12 promoter (Zabidi et al. 2015), were then inserted into the SalI and HindIII sites. Finally, a 2.3-kb λ phage HindIII fragment was PCR amplified with or without four tandem Sry-δ-binding sites on the 5′ or 3′ end, and inserted into the SalI site by Gibson assembly. The following sequence was used for the four tandem binding sites, with the binding sites underlined: 5′-AGATCTTCGCGCGTATTAGAGATGGAAACGATCGCGCGTATTAGAGATGGAAACGATCGCGCGTATTAGAGATGGAAACGATCGCGCGTATTAGAGATGGAAACCAAGATCT-3′ (Payre and Vincent 1991; Krystel and Ayyanathan 2013). The BEAF-binding site used is near the aurA TSS in scs’. To test the effects of the BEAF-binding site on aurA promoter function, a 215-bp scs’ fragment without or with the BEAF-binding site mutated (Cuvier et al. 1998) was inserted into the firefly luciferase-SV40 polyadenylation plasmid.
Y2H
Y2H assays were carried out using standard methods, as previously described (Avva and Hart 2016). Yeast strain Y2H-Gold (Clontech) or DDY2937 (MATα; trp1-901; leu2-3, 112; ura3-52; his3Δ200; gal4Δ; gal80Δ; LYS2::GAL1-HIS3; GAL2-ADE2; met2::GAL7-lacZ; kind gift of D. Donze) was transformed by the lithium acetate method with plasmids derived from pOAD and pOBD2, and plated on media lacking tryptophan and leucine (two-drop, selects for plasmids). After 3–5 days of growth at 30°, individual colonies were patched onto two- and four-drop (lacking tryptophan, leucine, adenine, and histidine; selects for reporter gene expression) plates. Colonies of interest were grown in liquid two-drop medium for 2 days and diluted to an OD600 of 0.1. Four fivefold serial dilutions were made in a 96-well plate, and 5 µl from each well was spotted onto two- and four-drop plates. Growth was compared after 2–3 days.
Library screening was done using the mate-and-plate method as described by the manufacturer (Clontech). The GAL4-AD plasmid library was in the Y187 yeast strain (MATα), and the GAL4-BD-BEAF-32B plasmid was in Y2H-Gold (MATa). Mated cells were plated on media containing X-α-Gal (α-galactosidase) and aureobasidin, and lacking Trp and Leu. Blue colonies were picked onto similar plates additionally lacking His and Ade. Blue colonies from these plates had their inserts PCR amplified and sequenced. Over 2.5e6 mated yeast were screened.
Pull-down assay
Proteins were expressed in Escherichia coli strain BL21, pLysS by growth at 25° for 24 hr in autoinduction medium ZYM-5052 (1% N-Z-amine, 0.5% yeast extract, 2 mM MgSO4, 25 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, 5 mM Na2SO4, 0.5% glycerol, 0.05% glucose, 0.2% lactose, 100 mg/liter ampicillin, and 34 mg/liter chloramphenicol), and protein extracts were prepared by standard methods (Studier et al. 1990; Studier 2005). Extracts containing Myc-tagged transcription factors and FLAG-tagged 32B were mixed and immunoprecipitated using anti-FLAG M2 beads (Sigma [Sigma Chemical], St. Louis, MO), followed by protein detection on western blots using anti-Myc (Santa Cruz Biotechnology) or anti-BEAF antibodies (Zhao et al. 1995), as previously described (Avva and Hart 2016).
Genetic interaction assay
Genetic interaction between BEAF and Sry-δ was tested using the rough-eye assay that was previously used to show genetic interactions between BEAF and other proteins (Roy et al. 2007b). This assay uses a GAL4-inducible, dominant-negative BEAF transgene called BID for BEAF self-Interaction Domain (Gilbert et al. 2006).The mutant Sry-δSF2, kindly provide by A. Vincent (Crozatier et al. 1992), and two UAS-RNAi (upstream activating sequence-RNA interference) stocks [Vienna Drosophila Resource Center (VDRC) 102786 and 41094] were tested. Briefly, ey-GAL4/CyO (Bloomington Drosophila Stock Center 5535) or ey-GAL4/CyO; UAS-BID flies were crossed to Sry-δSF2/TM3 and UAS-RNAi flies. Flies of the desired genotypes were collected, processed, and photographed using a JEOL JSM-6610LV scanning electron microscope at 10 kV under high vacuum, as previously described (Roy et al. 2007b).
Bimolecular fluorescence complementation assay
Drosophila S2 cells were grown at 25° in Shields and Sang M3 medium (M3, S8398; Sigma) with 10% fetal bovine serum (FBS; GIBCO [Grand Island Biological], Grand Island, NY), and antibiotic/antimycotic (anti/anti; 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 250 ng/ml amphotericin B; GIBCO) from 5 × 105 to 107 cells/ml. For transfection, 1.5 × 106 cells in 1 ml medium were grown per well in a 24-well plate for 24 hr. Cells were washed with serum-free medium and transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Briefly, 3 µl Lipofectamine 2000 was mixed with 500 µl M3 plus anti/anti and added to a mix of 250 ng N-Venus plasmid and 250 ng C-Venus plasmid. After 10 min, this was added to the washed cells and placed at 25° for 4.5 hr. The medium with DNA was removed and replaced by 1 ml M3 with 10% FBS and anti/anti. After 2 days, cells were resuspended in the medium plus 10 µg/ml Hoechst 33342 and placed on a slide with a Secure Seal Spacer (Invitrogen), covered with a coverslip, and a Leica DM6B fluorescence microscope was programed to scan and capture 50 images per slide. Venus-positive and total nuclei (Hoechst staining) were counted using CellProfiler (cellprofiler.org). Signal in the Venus channel had to overlap with signal in the Hoechst channel (i.e., had to be nuclear) to be counted. Values for the 50 images were added together to calculate the fraction of cells showing biomolecular fluorescence complementation (biFC). Three biological replicates were done.
Luciferase assay
Transfections were done as for the biFC assays. The plasmid DNAs used were a mix of 400 ng firefly luciferase (looping) plasmid, 5 ng pPac-Renilla luciferase (control) plasmid, and 100 ng pPac-transcription factor plasmid. After replacing the medium plus DNA by 1 ml M3 with 10% FBS and anti/anti, cells were grown for an additional 60 hr. Cells were lysed and assayed for luciferase activity using the dual-luciferase assay system (E1910; Promega) and a GloMax 20/20 luminometer (Promega). For each transfection, experimental firefly luciferase was divided by the control Renilla luciferase activity to control for transfection efficiency. For each plasmid set, values were then normalized to the BEAF-associated promoter without transcription factor-binding sites. Three biological replicates were done.
Data availability
Strains and plasmids are available upon request. Primer sequences are in Supplemental Material, Table S1. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.6084/m9.figshare.11988537.
Results
Identification of BEAF-interacting proteins
BEAF was originally identified as a chromatin domain insulator-binding protein, while subsequent genome-wide mapping found that it usually binds near TSSs (Zhao et al. 1995; Bushey et al. 2009; Jiang et al. 2009; Nègre et al. 2010). This raises questions about the function of promoter-proximal BEAF, as well as whether it is a traditional insulator protein. To gain insight into how BEAF works, we decided to identify proteins that physically interact with BEAF-32B using Y2H assays. BEAF-32B was used because it is essential, while BEAF-32A is not (Roy et al. 2007a), possibly because BEAF-32B has the dominant DNA-binding activity (Jiang et al. 2009). Both proteins are identical over ∼200 aa since they are produced from the same gene, differing only over their N-terminal 80 aa that encode DNA-BDs. So similar results should be obtained if BEAF-32A was used, unless interactions occur with the DNA-BD portion of the protein.
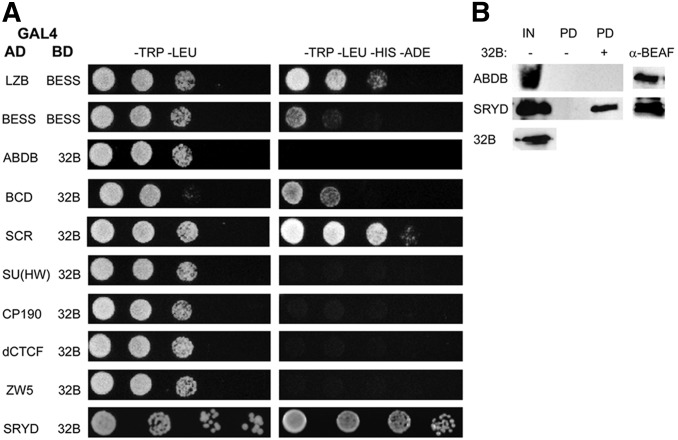
We had previously identified genetic interactions between BEAF and several proteins using a rough-eye assay (Roy et al. 2007b). To see if any genetic interactions reflected physical interactions, we started by testing these proteins. In addition, we tested a few other proteins of interest. Two proteins interacted with 32B in our Y2H assays (Figure 1A and Table 1), the homeodomain-containing transcription factors Bcd and Scr. Interestingly, we did not detect interactions with proteins that have previously been reported to interact with BEAF: Zw5 (Blanton et al. 2003), CP190 (Vogelmann et al. 2014), and D1 (Cuvier et al. 2002). Both Y2H interactions were atypical in the sense that only ∼30% of colonies containing both Y2H plasmids grew on four-drop plates selecting for HIS3 and ADE2 reporter gene expression. Growth on the four-drop plates was delayed and only a few colonies grew rather than the entire patch. When repatched onto a fresh four-drop plate, the entire patch would grow. Additionally, a third reporter gene was also activated (URA3::MEL1UAS-Mel1TATA encoding secreted α-GAL). GAL4-BD-BEAF-32B and GAL4-AD-transcription factor-coding sequences were PCR amplified from yeast growing on four-drop plates, and sequenced to look for mutations. No mutations were found, eliminating this as an explanation for the interactions. In contrast, using self-interactions of 32B, its leucine zipper plus BESS domain, or its BESS domain alone we found that 100% of colonies containing both Y2H plasmids grew on four-drop plates (Figure 1A) (Avva and Hart 2016).
Figure 1.
Y2H and pull-down tests for interactions between BEAF-32B and specific proteins. (A) BEAF-32B was fused to the C-terminal end of the GAL4 DNA-BD, and candidate proteins were fused to the C-terminal end of the GAL4-AD for use in Y2H assays. Interactions of the BEAF BESS domain with itself and the LZB domain were used as positive controls (see Figure 2A). As previously reported, the interaction of the BESS domain with itself was weaker than its interaction with the LZB (Avva and Hart 2016). Serial fivefold dilutions of OD600 0.1 yeast were spotted onto plates. Left panels (−TRP –LEU) show growth on plates selecting for plasmids. Right panels (−TRP –LEU –HIS –ADE) show growth on plates additionally selecting for reporter gene expression. Shown are proteins from Table 1 that interact with 32B, insulator proteins that do not interact with 32B as examples of negative results, and interaction with Sry-δ from the cDNA library screen. (B) Bacterial protein extracts containing N-terminal FLAG-tagged 32B and N-terminal Myc-tagged transcription factors were mixed and pulled down using anti-FLAG M2 beads. After SDS-PAGE, proteins were detected using anti-Myc or anti-BEAF antibodies. Sry-δ was pulled down only in the presence of FLAG-32B, while the negative control Myc-Abd-B was not pulled down. α-BEAF: detection of pulled down FLAG-32B from the (+) lanes (25% of pulldown); AD, activation domain; BD, binding domain; BEAF, Boundary Element-Associated Factor; cDNA, complementary DNA; IN, input proteins (20% of input); LZB, leucine zipper plus BESS; PD, proteins pulled down in the absence (−) or presence (+) of FLAG-32B (25% of pulldown); Sry-δ, Serendipity δ; Y2H, yeast two-hybrid.
Next, we screened a Drosophila cDNA library to identify additional proteins that interact with 32B. Over 2.5 million colonies were screened, resulting in 188 positive colonies that were sequenced and identified (Table 2). BEAF interacts with itself via a C-terminal BESS domain (Avva and Hart 2016), and 56 of the identified clones encoded BEAF. Of these, 16 had coding sequences only for the common part of BEAF, 32 also had sequences unique to 32B, and 8 also had sequences unique to 32A. The remaining cDNAs encoded 20 different proteins, with most identified once or twice. Annotations in FlyBase indicated that five of these proteins are nuclear. Like Bcd and Scr, one of these is a transcription factor, although Sry-δ has multiple zinc fingers rather than a homeodomain. Of the rest, eight proteins have unknown cellular locations and functions, six are found in the cytoplasm or nonnuclear organelles, and one is extracellular.
Table 2. Results of Y2H cDNA library screening for interactions with BEAF.
| Gene | FlyBase identifier | Hits | Location |
|---|---|---|---|
| BEAF-32 | FBgn0015602 | 16 | Nucleus |
| BEAF-32A | FBgn0015602 | 8 | Nucleus |
| BEAF-32B | FBgn0015602 | 32 | Nucleus |
| CG11164 | FBgn0030507 | 15 | Nucleus |
| Sry-δ | FBgn0003512 | 2 | Nucleus |
| Bin1, dSAP18 | FBgn0024491 | 1 | Nucleus |
| Polybromo, bap180 | FBgn0039227 | 1 | Nucleus |
| EAChm | FBgn0036470 | 1 | Nucleus |
| mRpL44 | FBgn0037330 | 48 | Mitochondria |
| CG32276 | FBgn0047135 | 7 | Endoplasmic reticulum |
| CG3625 | FBgn0031245 | 3 | Endomembrane system |
| Tango9 | FBgn0260744 | 1 | Golgi |
| Pfdn1 | FBgn0031776 | 1 | Cytoplasm |
| Tailor | FBgn0037470 | 1 | Cytoplasm |
| Lcp3 | FBgn0002534 | 1 | Extracellular |
| CkIIα-i3 | FBgn0025676 | 37 | Unknown |
| CG30424 | FBgn0050424 | 4 | Unknown |
| CG14317 | FBgn0038566 | 2 | Unknown |
| CG13285 | FBgn0035611 | 2 | Unknown |
| CG43088 | FBgn0262534 | 2 | Unknown |
| CG9947 | FBgn0030752 | 1 | Unknown |
| CG13083 | FBgn0032789 | 1 | Unknown |
| CG17162 | FBgn0039944 | 1 | Unknown |
We decided to focus our attention on the three transcription factors identified. Further Y2H testing of Sry-δ found that 100% of colonies containing the GAL4-BD-BEAF-32B and GAL4-AD-Sry-δ plasmids grew on four-drop plates. Because of the atypical Y2H results for Bcd and Scr, we will focus only on Sry-δ. To check its interaction with BEAF, we tested for copull-down after expression in E. coli. Protein extracts containing N-terminal Myc-tagged Sry-δ and FLAG-tagged 32B were mixed, and proteins were pulled down using anti-FLAG beads. Sry-δ was pulled down with 32B, while as a negative control Myc-tagged Abd-B was not (Figure 1B).
Mapping interaction regions
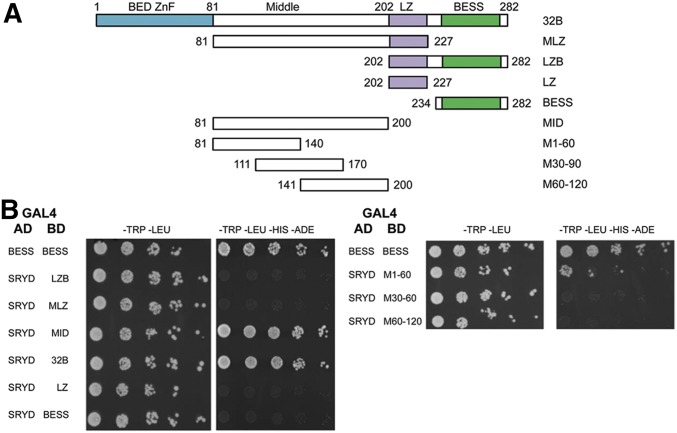
To further validate interactions between Sry-δ and BEAF, we mapped regions that interact by Y2H and pull-down assays. First, we tested parts of BEAF for interactions with Sry-δ, using BESS–BESS domain interactions and full-length 32B-Sry-δ interactions as positive controls (Figure 2). The parts of BEAF tested are present in both 32A and 32B. Sry-δ interacted with the middle (MID) region. For unknown reasons, possibly related to polypeptide folding or stability, the MID region with the putative leucine zipper (MLZ) did not interact. A point of interest is that roughly the first 75 aa of this 120-aa region are highly conserved among Drosophila species (Avva and Hart 2016). There is no reliable structural information for this region, so we split it into three overlapping 60-aa segments. M1-60 is sufficient for interactions with Sry-δ (Figure 2). The interaction is weaker than for the entire MID region, suggesting that additional sequences contribute to the interaction, or the proper folding or stability of M1-60.
Figure 2.
Mapping the region of BEAF that interacts with Sry-δ. (A) Schematic of the parts of BEAF that were fused to the GAL4 BD for Y2H assays. BED ZnF: 32B unique sequences, encompassing the DNA-binding BED finger (blue rectangle). LZ, purple rectangle; BESS domain, green rectangle. Numbers indicate the first and last amino acids present in the truncated proteins. (B) Results of Y2H assays, as in Figure 1A. BESS–BESS and Sry-δ-32B interactions were included as positive controls. Sry-δ interacts with M1-60. AD, activation domain; BD, binding domain; BEAF, Boundary Element-Associated Factor; LZ, putative leucine zipper; LZB, leucine zipper plus BESS; M/MID: middle region; MLZ, MID region with a putative leucine zipper; Sry-δ, Serendipity δ; Y2H, yeast two-hybrid; ZnF, zinc finger.
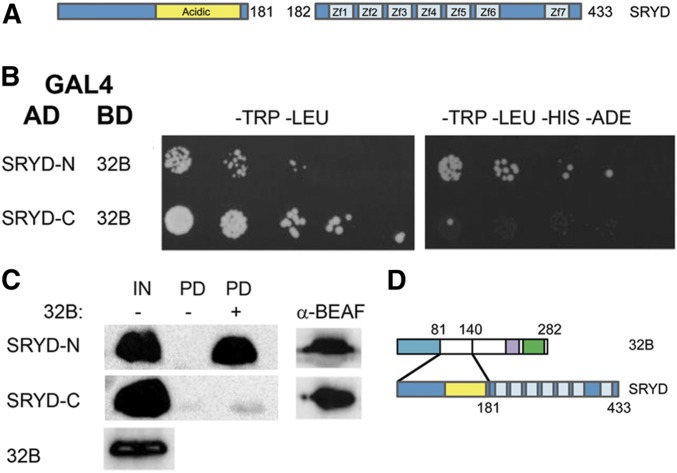
Next, we determined if 32B interacts with the N-terminal part of Sry-δ lacking zinc fingers (amino acids 1–181) or the C-terminal part with seven zinc fingers (amino acids 182–433). Sry-δ was split and fused either to an N-terminal GAL4-AD for Y2H assays, or to an N-terminal Myc tag for pull-down assays (Figure 3). The half of Sry-δ that has an acidic domain (amino acids 96–174) but lacks zinc fingers interacted with 32B in both Y2H and pull-down assays. These results are summarized in Figure 3D, and could be useful for future experiments designed to disrupt the interaction.
Figure 3.
Mapping the region of Sry-δ that interacts with 32B. (A) Schematic of the parts of Sry-δ that were fused to the GAL4 AD or a Myc tag. An acidic region and zinc fingers are indicated. (B) Results of Y2H assays, as in Figure 1A. The N-terminal half of Sry-δ, which lacks the zinc fingers, interacted with 32B. (C) Results of 32B pull-down assays, as in Figure 1B (IN: 20% of input; PD: 50% of the pulled down proteins; α-BEAF: 25% of the pulled down proteins). The half of Sry-δ lacking the zinc fingers was pulled down with 32B. (D) Summary of interactions between BEAF and Sry-δ. AD, activation domain; Sry-δ, Serendipity δ; Y2H, yeast two-hybrid.
Testing interactions by biFC
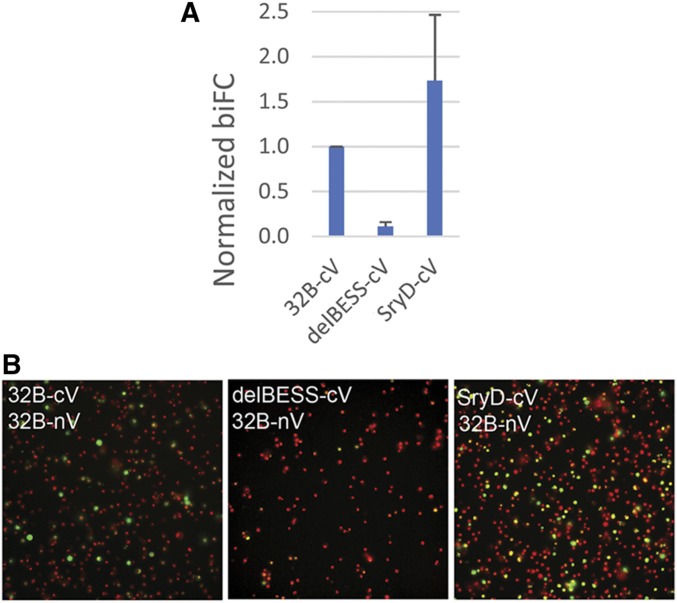
As a further test of interactions with BEAF, we used biFC (Figure 4). nV (amino acids 1–173) was fused to the C-terminus of 32B. As positive and negative controls, cV (amino acids 155–239) was fused to the C-terminus of 32B or 32B with the BESS domain deleted (32B-delBESS), respectively. The fraction of cells showing biFC of 32B-cV with 32B-nV was around nine times more than for 32B-delBESS-cV. A C-terminal cV fusion was made for Sry-δ and Abd-B. Abd-B-cV showed less interaction with 32B-nV than did delBESS-cV, indicating that little artifactual interaction with 32B-nV is driven by cV expression from the Act5C promoter (data not shown). A higher fraction of cells showed biFC of 32B-nV with Sry-δ-cV than with 32B-cV, clearly showing that Sry-δ and BEAF interact.
Figure 4.
Testing the interaction between Sry-δ and 32B using biFC. (A) Graph showing the fraction of cells showing biFC of indicated cV-tagged proteins with 32B-nV, normalized to 32B-cV with 32B-nV. A minimum of 50 images were counted per sample per experiment, and results are an average of three biological replicates with error bars showing the SD of the normalized replicates. Results for Sry-δ-cV were variable, but it clearly interacted with 32B-nV. (B) Representative micrographs for the indicated proteins. Nuclei were stained with Hoechst and false-colored red, while Venus is shown in green. All images were acquired using the same settings, but the image shown for delBESS-cV had the green channel enhanced to better show the Venus signal. 32B, Boundary Element-Associated Factor-32B; biFC, biomolecular fluorescence complementation; cV, C-terminal part of Venus; nV, N-terminal part of Venus; delBESS, 32B with the BESS domain deleted; Sry-δ, Serendipity δ.
Genetic interaction between BEAF and Sry-δ
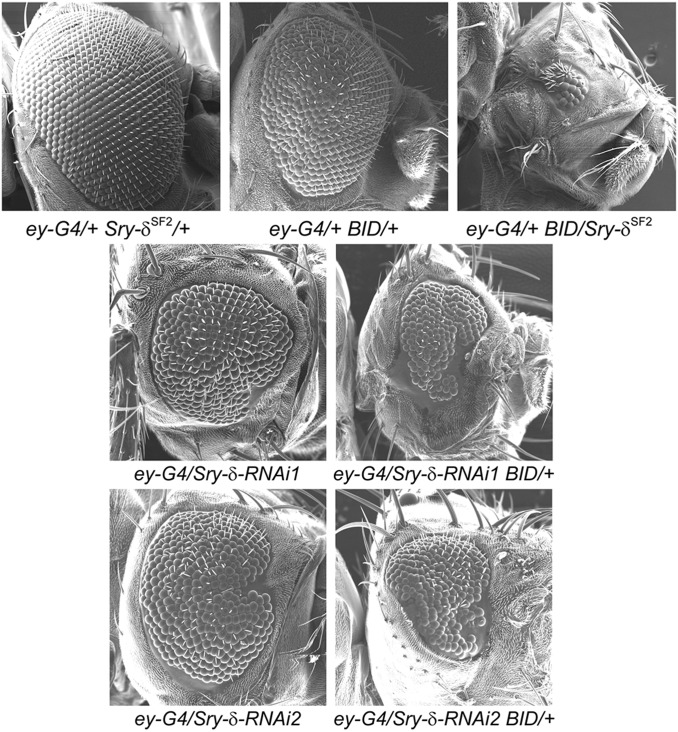
A genetic interaction between BEAF and other chromatin proteins was previously shown (Roy et al. 2007b), and guided our above selection of specific proteins to test for physical interactions. The assay utilized UAS-BID, a transgene encoding a dominant-negative form of BEAF lacking a DNA-BD. When produced under GAL4 control in eyes, it caused a rough-eye phenotype that was enhanced in the presence of heterozygous mutations in other proteins, including several transcription factors. We used the same assay to test for genetic interactions between BEAF and Sry-δ. Driving heterozygous UAS-BID expression using ey-GAL4 leads to a mild rough-eye phenotype, while combining ey-GAL4 and Sry-δSF2 does not affect eye development. The combination of heterozygous ey-GAL4, UAS-BID, and Sry-δSF2 has a dramatic effect on eye development: all flies have eyes with only a few ommatidia (Figure 5). We also tested two Sry-δ UAS-RNAi lines. Both gave a rough-eye phenotype with the ey-GAL4 driver, complicating the genetic interaction analysis. However, in both cases, the rough eye was clearly more extreme when a copy of UAS-BID was also present (Figure 5). We conclude that Sry-δ shows a genetic interaction with BEAF. While no mechanistic conclusions can be drawn from these results, the genetic interaction is consistent with our data showing a physical interaction between BEAF and Sry-δ.
Figure 5.
A rough-eye assay shows a strong genetic interaction between BEAF and Sry-δ. Shown are SEM images from eyes of 3–5-day-old females. Negative control ey-GAL4/+; Sry-δSF2/+ flies have normal eyes. A mild rough-eye phenotype is seen in ey-GAL4/+; UAS-BID/+ flies expressing a dominant-negative form of BEAF. The rough-eye phenotype is much stronger when the Sry-δSF2 mutation is also present (ey-GAL4/+; UAS-BID/ Sry-δSF2). This phenotype is 100% penetrant. Sry-δ UAS RNAi transgenes give a clear rough-eye phenotype when heterozygous with ey-GAL4 (ey-GAL4/Sry-δ-RNAi1 and ey-GAL4/Sry-δ-RNAi2). The phenotype is more extreme in combination with heterozygous UAS-BID (ey-GAL4/Sry-δ-RNAi1; BID/+ and ey-GAL4/Sry-δ-RNAi2; BID/+). RNAi1: VDRC 41094; RNAi2: VDRC 102786. BEAF, Boundary Element-Associated Factor; RNAi, RNA interference; Sry-δ, Serendipity δ; UAS, upstream activating sequence; VDRC, Vienna Drosophila Resource Center.
Promoter-proximal BEAF facilitates Sry-δ action locally and from a distance
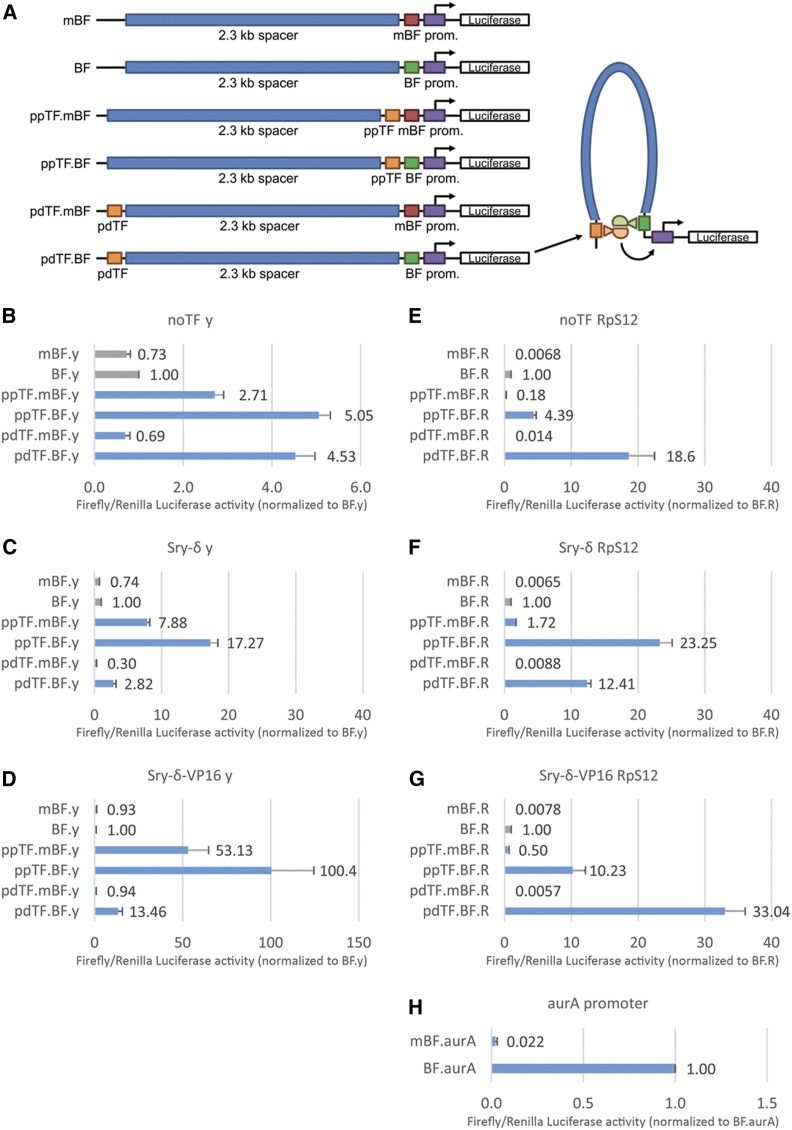
Genome-wide mapping has found that BEAF usually binds near TSSs (Bushey et al. 2009; Jiang et al. 2009; Nègre et al. 2010; Liang et al. 2014). This suggested to us that BEAF could facilitate long-distance enhancer–promoter communication with enhancers that utilize Sry-δ. We tested this using luciferase assays in transiently transfected S2 cells, similar to other studies (Nolis et al. 2009). As shown in Figure 6A, the high-affinity BEAF-binding site from the scs’ insulator (Hart et al. 1997) was placed next to a minimal promoter from the yellow (y) gene (Morris et al. 2004; Melnikova et al. 2008), with or without mutations that abrogate BEAF binding. Although this promoter is not normally active in S2 cells, a large body of evidence, including high-throughput studies (Arnold et al. 2017), shows that minimal promoters can be activated in any cell type by adjacent transcription factors. Upstream of this promoter was a 2.3-kb spacer sequence from a bacteriophage λ HindIII fragment. Four tandem Sry-δ transcription factor-binding sites were placed either in a promoter-proximal position adjacent to the BEAF-binding site or in a promoter-distal position upstream of the spacer sequence. Promoter-proximal Sry-δ-binding without and with BEAF will show if it can locally interact with BEAF to activate the reporter gene (normalized to cotransfected Renilla luciferase activity driven by an Act5C promoter, and then normalized to the BEAF-associated promoter without Sry-δ-binding sites). If BEAF facilitates activation by Sry-δ looping, this should be apparent by comparing the luciferase activity for the promoter-distal transcription factor-binding sites in the presence and absence of BEAF binding. Although Sry-δ is present in S2 cells (Gramates et al. 2017), we also made a plasmid to produce it from an Act5C promoter without and with a VP16 AD.
Figure 6.
Promoter-proximal BEAF facilitates local and long-range interactions between Sry-δ and promoters, and directly activates a housekeeping promoter. (A) Schematic of constructs used to drive firefly luciferase expression in transfected S2 cells. All transfections also had a plasmid with an Act5C promoter driving Renilla luciferase expression to normalize for transfection efficiency, with or without a plasmid with an Act5C promoter driving expression of Sry-δ or Sry-δ-VP16 (endogenous Sry-δ is expressed in S2 cells). Transfections with each set of plasmids were further normalized to firefly luciferase expression when only the BEAF-binding site was present. Error bars indicate the SD of three biological replicates. Test plasmids had either a minimal y (developmental) or RpS12 (housekeeping) promoter. Also shown is a model of Sry-δ interacting with BEAF to facilitate long-range activation of the promoter. The y promoter was tested (B) without an Sry-δ-expressing plasmid; (C) with an Sry-δ plasmid; and (D) with an Sry-δ-VP16 plasmid. Average luminometer readings for BF.y were 102,245 compared to a mock transfection background of 99. The RpS12 promoter was tested (E) without an Sry-δ-expressing plasmid; (F) with an Sry-δ plasmid; and (G) with an Sry-δ-VP16 plasmid. Average luminometer readings for BF.R were 1,145,492 compared to a mock transfection background of 102. (H) Testing the aurA promoter with and without mutations in the BEAF-binding site. Average luminometer readings for aurA were 924,577 compared to a mock transfection background of 122. In (B–G), comparison of the BF, ppTF.mBF, and ppTF.BF values show local interactions between Sry-δ and BEAF cooperatively activate the reporter gene, while comparison of the BF, pdTF.mBF, and pdTF.BF values indicate long-range interactions between Sry-δ and BEAF activate the reporter gene. In (B–D), comparisons of mBF and BF show that BEAF does not activate the y promoter. In (E) through (H), comparisons of mBF and BF show that BEAF activates the RpS12 and aurA promoters. BEAF, Boundary Element-Associated Factor; BF, promoter-proximal wild-type BEAF-binding site; mBF, promoter-proximal mutant BEAF-binding site; pdTF, promoter-distal four tandem Sry-δ TF-binding sites; ppTF, promoter-proximal four tandem Sry-δ TF-binding sites; prom., promoter; Sry-δ, Serendipity δ; TF, transcription factor.
Activation of the y promoter by promoter-proximal Sry-δ-binding sites increased from around three- to eightfold when ectopic Sry-δ was provided, and to 50-fold by ectopic Sry-δ-VP16 (Figure 6, B–D). In all cases, activation doubled when promoter-proximal BEAF also bound. Since BEAF binding alone did not activate, this demonstrates a synergistic interaction between BEAF and Sry-δ when they bind next to each other. In contrast, promoter-distal Sry-δ-binding sites did not activate when the promoter-proximal BEAF-binding site was mutated. However, promoter-proximal BEAF binding facilitated activation by promoter-distal Sry-δ-binding sites. Activation was three- to fivefold with and without ectopic Sry-δ, and increased to 13-fold with ectopic Sry-δ-VP16. This provides evidence for long-range communication between BEAF and distal Sry-δ.
Promoter-proximal BEAF activates a housekeeping promoter but not a developmental promoter
We expanded our analysis to include a minimal RpS12 housekeeping promoter (Zabidi et al. 2015). There are differences between promoters for developmental and housekeeping genes (Zabidi et al. 2015), and the y promoter is a developmental promoter with a TATA box, an initiator element, and a downstream promoter element (Morris et al. 2004; Melnikova et al. 2008). We previously found that BEAF usually localizes near promoters of housekeeping genes (Jiang et al. 2009). We extended this by compiling lists of genes with a TSS within 300 bp of the center of BEAF peaks from various additional sources (Bushey et al. 2009; Nègre et al. 2010; Liang et al. 2014) and compared them to lists of housekeeping genes, as defined by low variance in expression levels in various tissues, cell types, and developmental stages (Lam et al. 2012; Ulianov et al. 2016). We found that ∼85% of BEAF-associated genes are housekeeping genes (Shrestha et al. 2018). Note that the minimal RpS12 promoter has the DREF-binding site (Hirose et al. 1993) deleted, and presumably the sequences responsible for a BEAF peak near this promoter are as well.
Surprisingly, BEAF alone activated the minimal RpS12 promoter over 100-fold (Figure 6, E–G). Aside from that, once again evidence of proximal and long-range communication between Sry-δ and BEAF was obtained. Promoter-proximal Sry-δ binding activated the RpS12 promoter in the absence of BEAF binding. As for the y promoter, there was synergistic activation together with BEAF binding, without or with ectopic Sry-δ or Sry-δ-VP16. Again, as for the y promoter, Sry-δ alone did not activate from promoter-distal-binding sites, but interacted with promoter-proximal BEAF to provide higher activation relative to BEAF alone. For some reason, the long-range communication gave three- to fourfold higher activation than local interactions between promoter-proximal Sry-δ and BEAF, for endogenous Sry-δ and ectopic Sry-δ-VP16 (Figure 6, E and G).
To summarize, these results show that local and long-range communication between Sry-δ and promoter-proximal BEAF facilitates gene activation. Unexpectedly, we also found that BEAF is a powerful activator of the housekeeping promoter that we used, but not the developmental promoter. To expand this analysis, we examined the ability of BEAF to activate another promoter. The BEAF-binding site we used comes from near the aurA TSS, which is in the scs’ insulator. Although aurA is not on the list of housekeeping genes, it must be expressed in all dividing cells because it encodes a protein essential for mitosis (Glover et al. 1995). Furthermore, our binding-site mutations are in the natural promoter context. Promoter activity dropped ∼50-fold when the BEAF-binding site was mutated (Figure 6H). This provides strong evidence that BEAF can directly participate in the activation of some promoters.
Discussion
BEAF was initially discovered as an insulator-binding protein, and transgenic assays have demonstrated that genomic sequences with BEAF-binding sites have insulator activity (Zhao et al. 1995; Cuvier et al. 1998, 2002; Sultana et al. 2011). Additionally, interfering with BEAF function with a dominant-negative protein or null mutation affects scs’ insulator activity (Gilbert et al. 2006; Roy et al. 2007a). Yet, genome-wide mapping found that BEAF is usually found near TSSs, suggesting it could play a role in promoter activity (Bushey et al. 2009; Jiang et al. 2009; Nègre et al. 2010). To gain insight into molecular mechanisms of BEAF function we conducted a Y2H screen for interacting proteins. We found a robust interaction between BEAF and the transcription factor Sry-δ. The interaction was confirmed by mapping interaction regions, pull-down experiments using bacterially expressed proteins, and biFC. A genetic interaction between BEAF and Sry-δ was shown using a previously described rough-eye assay (Roy et al. 2007b). Three other studies also found an interaction between BEAF and Sry-δ. One expressed 459 epitope-tagged chromatin proteins in S2 cells, immunoaffinity-purified the proteins, and did proteomic mass spectrometry to identify copurifying proteins (Rhee et al. 2014). BEAF co-immunoprecipitated with epitope-tagged Sry-δ and vice versa, finding multiple peptides for both proteins. We also detected Sry-δ by mass spectrometry of proteins that co-immunoprecipitated with BEAF from embryo nuclear protein extracts (M. Maharjan and C. M. Hart, personal communication). Second, an unpublished large-scale Y2H study found an interaction of BEAF with Sry-δ (http://flybi.hms.harvard.edu/results.php). Third, another large-scale Y2H study that focused on transcription factors also found an interaction between BEAF and Sry-δ (Shokri et al. 2019).
Sry-δ has seven zinc fingers, binds DNA as a dimer, and was shown to be a transcriptional activator in transient transfection experiments (Payre et al. 1997). It is closely related to, but functionally distinct from, Sry-β, which is encoded by a neighboring gene (Payre et al. 1994; Ruez et al. 1998). Like BEAF, Sry-δ is maternally provided and ubiquitous throughout development (Payre et al. 1990). Mutations are recessive embryonic lethal, although certain alleles allow the development of some adults when hemizygous over a deficiency (Crozatier et al. 1992). Almost all of these adults are small, sterile males, and some have phenotypes including rough eyes, extra humeral bristles, and missing thoracic macrochaetes. A dominant-negative form of BEAF is also embryonic lethal (Gilbert et al. 2006), and the few adults obtained from embryos lacking maternal and zygotic BEAF are nearly all males with rough eyes, although they are fertile (Roy et al. 2007a). Heterozygous mutations in sry-δ can suppress sterility caused by a piwi mutation, although Sry-δ does not appear to regulate piwi (Smulders-Srinivasan and Lin 2003). At this point, only the expression of bcd during oogenesis has been shown to require Sry-δ (Payre et al. 1994; Ruez et al. 1998; Schnorrer et al. 2000). However, the pleiotropic effects of sry-δ mutations during embryogenesis and later development indicate that many genes are regulated by Sry-δ.
The interaction with a transcription factor suggested that BEAF might be playing an activating role at BEAF-associated promoters, rather than insulating promoters. In support of this, we found higher activation when Sry-δ bound next to promoter-proximal BEAF than for either protein binding alone. We also tested the ability of promoter-proximal BEAF to facilitate gene activation by Sry-δ bound 2.3-kb upstream. We call this a looping assay because, although various models have been proposed (Furlong and Levine 2018), there is strong evidence that looping is a key component of enhancer–promoter communication (de Laat and Grosveld 2003; Deng et al. 2012; Weintraub et al. 2017). Evidence includes similar transient transfection experiments (Nolis et al. 2009). This has been confirmed at the genome-wide scale using methods such as Hi-C and chromatin interaction analysis by paired-end tag sequencing (Jin et al. 2013; Zhang et al. 2013). Promoter-distal Sry-δ binding alone did not activate the reporter gene, even with a VP16 AD. We obtained convincing evidence for looping between Sry-δ and BEAF leading to reporter gene activation.
There are prior demonstrations of a role for BEAF in activating BEAF-associated genes. Previous experiments found that many BEAF-associated genes are downregulated two- to fourfold after knockdown of BEAF in cultured S2 cells or in the absence of BEAF in embryos (Emberly et al. 2008; Jiang et al. 2009; Lhoumaud et al. 2014). In contrast, another study found that BEAF knockdown had minimal effects on gene expression in BG3 cells, with only six genes showing significant downregulation and none showing upregulation (Schwartz et al. 2012). These reports did not examine the effects of mutating BEAF-binding sites on gene expression. Further, they could not determine if the effects were direct or indirect, or if effects on gene regulation were due to activation by BEAF or insulation from repressive effects. By mutating a BEAF-binding site, we clearly show that BEAF can interact with the transcription factor Sry-δ to activate a promoter.
There are also earlier demonstrations that BEAF can participate in DNA looping interactions. It was shown that BEAF can interact with CP190 and chromator, and that homodimerization of either of these proteins can then act as a bridge between BEAF-binding sites, or BEAF and binding sites for other proteins these bridge proteins interact with, such as the insulator proteins dCTCF, Su(Hw), and GAGA factor (Vogelmann et al. 2014). In the case of CP190, it was shown that interactions with BEAF lead to looping interactions with genomic sites lacking BEAF-binding sites that are detected as indirect peaks by chromatin immunoprecipitation sequencing. These indirect peaks often have binding sites for dCTCF or GAGA factor. Mutating BEAF so that it does not interact with CP190 eliminated the indirect peaks, and also affected the expression of genes associated with BEAF and indirect peaks, suggesting that the CP190-mediated looping interactions are important for gene regulation (Liang et al. 2014). It is not known what effect the BEAF mutation has on interactions with other proteins such as chromator. We did not detect interactions between BEAF and CP190 by Y2H either by a direct test or in our cDNA library screen, although we more recently detected an interaction between BEAF and chromator (data not shown). The co-immunoprecipitation mass spectrometry study mentioned above also did not detect an interaction between BEAF and CP190, but did detect an interaction between BEAF and chromator (Rhee et al. 2014). We have similar co-immunoprecipitation mass spectrometry results (M. Maharjan and C. M. Hart, personal communication), and an earlier report also found that BEAF co-immunoprecipitated with chromator (Gan et al. 2011). Regardless of the contradictory CP190 results, chromator could be mediating long-range looping between BEAF and other chromatin proteins. However, neither CP190 nor chromator are typical transcription factors. They do not directly bind DNA (Vogelmann et al. 2014), and how they affect gene regulation is not clear. Here, we show DNA looping interactions between BEAF and Sry-δ, a typical transcription factor, leading to reporter gene activation without a need for bridging proteins.
An unexpected finding was that BEAF strongly activated the RpS12 housekeeping promoter and the aurA cell cycle-related promoter. It was previously found that sequences with BEAF-binding sites do not activate an hsp27 or hsp26 promoter after transient transfection (Zhao et al. 1995; Cuvier et al. 1998), or a w or hsp70 promoter in transgenic flies (Kellum and Schedl 1991, 1992; Cuvier et al. 1998). This led to the idea that BEAF is not a transcriptional activator. We obtained a similar result with the y promoter after transient transfection, supporting this idea. These are all regulated promoters. There are differences between regulated and housekeeping promoters (Zabidi et al. 2015), and we noticed that BEAF is usually found near the latter. Our results with the RpS12 promoter suggest that BEAF could be a transcriptional activator that is specific for housekeeping promoters, or a subset of these promoters. This could include the special class of ribosomal protein gene promoters (Wang et al. 2014), at least one-third of which (such as RpS12) are BEAF-associated. Although aurA was not on the list of housekeeping genes that we used, it has a BEAF-associated promoter (located in the scs’ insulator) and encodes an essential cell cycle protein (Glover et al. 1995). Thus, it must be expressed in all cycling cells and so could be considered a type of housekeeping gene. It will be interesting to expand the number of promoters tested, and to determine the mechanism behind the promoter-type specificity.
One question is whether the transcription factor DREF (Hirose et al. 1993; Tue et al. 2017) rather than BEAF might account for the effects we observed. The consensus motif for DREF (TATCGATA) is related to that for BEAF (clustered CGATA motifs); however, their binding sites do not always overlap. We previously found that DREF does not bind to the BEAF-binding site used here, and that BEAF and DREF compete rather than cooperate for binding when their binding sites overlap (Hart et al. 1999). We did not detect an interaction between BEAF and DREF in our Y2H screen. As mentioned in the Results, the minimal RpS12 promoter lacks the DREF motif present at the endogenous promoter. It is unlikely that DREF influenced our results.
Metazoan chromosomes are organized into TADs. Vertebrate TAD boundaries often have convergent CTCF sites that interact to form TAD loops. In contrast, fly TADs appear to be separated by regions of active chromatin containing clustered housekeeping genes that form inter-TAD regions (Ulianov et al. 2016; Cubeñas-Potts et al. 2017; Hug et al. 2017). BEAF is found near the TSSs of hundreds of housekeeping genes. By contributing to the activation of these promoters, BEAF could contribute to nuclear organization by helping to establish and maintain active genes that form inter-TAD regions. This could explain why BEAF is found at TAD boundaries and inter-TADs. The interaction with Sry-δ could be important at a subset of sites.
Here, we demonstrate two functions for the BEAF insulator protein: activating a gene through local or long-range communication with a transcription factor, and directly activating a housekeeping promoter. It should be noted that nucleosomes form on nonreplicating transfected DNA, although with irregular density and positioning on most plasmid copies (Reeves et al. 1985; Archer et al. 1992; Jeong and Stein 1994). Future experiments testing chromosomally integrated reporter genes would be informative to determine if normal chromatin affects these functions. This provides insight into BEAF, although it is currently unclear how these functions relate to insulator activity. It will be interesting to determine if BEAF can mediate long-range interactions with additional transcription factors, and what characteristics allow direct activation of a promoter by BEAF. Integrating this information with understanding of insulator activity, and the potential role of BEAF in helping to establish or maintain genomic TAD organization, remain challenges for the future.
Acknowledgments
The authors would like to thank the Drosophila Genomics Resource Center [National Institutes of Health (NIH) grant 2P40 OD-010949] for cDNA clones; the Bloomington Drosophila Stock Center (NIH grant P40 OD-018537) and Alain Vincent for fly stocks; FlyBase as an essential Drosophila resource; David Donze for plasmids, help with Y2H, and discussions; and Jamie Wood for advice on luciferase assays. This work was supported by National Science Foundation grant 1244100 from the Division of Molecular and Cellular Biosciences (www.nsf.gov).
Footnotes
Supplemental material available at figshare: https://doi.org/10.6084/m9.figshare.11988537.
Communicating editor: P. Geyer
Literature Cited
- Ali T., Renkawitz R., and Bartkuhn M., 2016. Insulators and domains of gene expression. Curr. Opin. Genet. Dev. 37: 17–26. 10.1016/j.gde.2015.11.009 [DOI] [PubMed] [Google Scholar]
- Archer T. K., Lefebvre P., Wolford R. G., and Hager G. L., 1992. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science 255: 1573–1576. 10.1126/science.1347958 [DOI] [PubMed] [Google Scholar]
- Arnold C. D., Zabidi M. A., Pagani M., Rath M., Schernhuber K. et al. , 2017. Genome-wide assessment of sequence-intrinsic enhancer responsiveness at single-base-pair resolution. Nat. Biotechnol. 35: 136–144. 10.1038/nbt.3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avva S. V., and Hart C. M., 2016. Characterization of the Drosophila BEAF-32A and BEAF-32B insulator proteins. PLoS One 11: e0162906 10.1371/journal.pone.0162906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A. C., West A. G., and Felsenfeld G., 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396. 10.1016/S0092-8674(00)81967-4 [DOI] [PubMed] [Google Scholar]
- Benassayag C., Seroude L., Boube M., Erard M., and Cribbs D. L., 1997. A homeodomain point mutation of the Drosophila proboscipedia protein provokes eye loss independently of homeotic function. Mech Dev 63: 187–198. [DOI] [PubMed] [Google Scholar]
- Blanton J., Gaszner M., and Schedl P., 2003. Protein:protein interactions and the pairing of boundary elements in vivo of boundary elements in vivo. Genes Dev. 17: 664–675. 10.1101/gad.1052003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey A. M., Ramos E., and Corces V. G., 2009. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 23: 1338–1350. 10.1101/gad.1798209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverina D., Fujioka M., Erokhin M., Georgiev P., Jaynes J. B. et al. , 2017. Boundaries of loop domains (insulators): determinants of chromosome form and function in multicellular eukaryotes. Bioessays 39: 1600233. 10.1002/bies.201600233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozatier M., Kongsuwan K., Ferrer P., Merriam J. R., Lengyel J. A. et al. , 1992. Single amino acid exchanges in separate domains of the Drosophila serendipity delta zinc finger protein cause embryonic and sex biased lethality. Genetics 131: 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeñas-Potts C., Rowley M. J., Lyu X., Li G., Lei E. P. et al. , 2017. Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture. Nucleic Acids Res. 45: 1714–1730. 10.1093/nar/gkw1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvier O., Hart C. M., and Laemmli U. K., 1998. Identification of a class of chromatin boundary elements. Mol. Cell. Biol. 18: 7478–7486. 10.1128/MCB.18.12.7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvier O., Hart C. M., Kas E., and Laemmli U. K., 2002. Identification of a multicopy chromatin boundary element at the borders of silenced chromosomal domains. Chromosoma 110: 519–531. 10.1007/s00412-001-0181-1 [DOI] [PubMed] [Google Scholar]
- de Laat W., and Grosveld F., 2003. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 11: 447–459. 10.1023/A:1024922626726 [DOI] [PubMed] [Google Scholar]
- Deng W., Lee J., Wang H., Miller J., Reik A. et al. , 2012. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149: 1233–1244. 10.1016/j.cell.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberly E., Blattes R., Schuettengruber B., Hennion M., Jiang N. et al. , 2008. BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol. 6: 2896–2910 [corrigenda: PLoS Biol. 7 (2009)]. 10.1371/journal.pbio.0060327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong E. E. M., and Levine M., 2018. Developmental enhancers and chromosome topology. Science 361: 1341–1345. 10.1126/science.aau0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan M., Moebus S., Eggert H., and Saumweber H., 2011. The Chriz-Z4 complex recruits JIL-1 to polytene chromosomes, a requirement for interband-specific phosphorylation of H3S10. J. Biosci. 36: 425–438. 10.1007/s12038-011-9089-y [DOI] [PubMed] [Google Scholar]
- Gilbert M. K., Tan Y. Y., and Hart C. M., 2006. The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics 173: 1365–1375. 10.1534/genetics.106.056002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D. M., Leibowitz M. H., McLean D. A., and Parry H., 1995. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 81: 95–105. 10.1016/0092-8674(95)90374-7 [DOI] [PubMed] [Google Scholar]
- Gramates L. S., Marygold S. J., Santos G. D., Urbano J. M., Antonazzo G. et al. , 2017. FlyBase at 25: looking to the future. Nucleic Acids Res. 45: D663–D671. 10.1093/nar/gkw1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Xu Q., Canzio D., Shou J., Li J. et al. , 2015. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162: 900–910. 10.1016/j.cell.2015.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Li X., Wu J., and Olson E. N., 2004. A myocardin-related transcription factor regulates activity of serum response factor in Drosophila. Proc. Natl. Acad. Sci. 101: 12567–12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart C. M., Zhao K., and Laemmli U. K., 1997. The scs’ boundary element: characterization of boundary element-associated factors. Mol. Cell. Biol. 17: 999–1009. 10.1128/MCB.17.2.999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart C. M., Cuvier O., and Laemmli U. K., 1999. Evidence for an antagonistic relationship between the boundary element-associated factor BEAF and the transcription factor DREF. Chromosoma 108: 375–383. 10.1007/s004120050389 [DOI] [PubMed] [Google Scholar]
- Hirose F., Yamaguchi M., Handa H., Inomata Y., and Matsukage A., 1993. Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase alpha and proliferating cell nuclear antigen. J. Biol. Chem. 268: 2092–2099. [PubMed] [Google Scholar]
- Hudry B., Viala S., Graba Y., and Merabet S., 2011. Visualization of protein interactions in living Drosophila embryos by the bimolecular fluorescence complementation assay. BMC Biol. 9: 5 10.1186/1741-7007-9-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug C. B., Grimaldi A. G., Kruse K. and Vaquerizas J. M., 2017. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169: 216–228.e19. 10.1016/j.cell.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Jeong S., and Stein A., 1994. Micrococcal nuclease digestion of nuclei reveals extended nucleosome ladders having anomalous DNA lengths for chromatin assembled on non-replicating plasmids in transfected cells. Nucleic Acids Res. 22: 370–375. 10.1093/nar/22.3.370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Emberly E., Cuvier O., and Hart C. M., 2009. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol. Cell. Biol. 29: 3556–3568. 10.1128/MCB.01748-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F., Li Y., Dixon J. R., Selvaraj S., Ye Z. et al. , 2013. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503: 290–294. 10.1038/nature12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R., and Schedl P., 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64: 941–950. 10.1016/0092-8674(91)90318-S [DOI] [PubMed] [Google Scholar]
- Kellum R., and Schedl P., 1992. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 12: 2424–2431. 10.1128/MCB.12.5.2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H., Abdullaev Z. K., Smith A. D., Ching K. A., Loukinov D. I. et al. , 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245. 10.1016/j.cell.2006.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnow M. A., Saffman E. E., Kornfeld K., and Hogness D. S., 1989. Transcriptional activation and repression by Ultrabithorax proteins in cultured Drosophila cells. Cell 57: 1031–1043. 10.1016/0092-8674(89)90341-3 [DOI] [PubMed] [Google Scholar]
- Krystel J., and Ayyanathan K., 2013. Global analysis of target genes of 21 members of the ZAD transcription factor family in Drosophila melanogaster. Gene 512: 373–382. 10.1016/j.gene.2012.09.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuziora M. A., and McGinnis W., 1988. Autoregulation of a Drosophila homeotic selector gene. Cell 55: 477–485. [DOI] [PubMed] [Google Scholar]
- Lam K. C., Muhlpfordt F., Vaquerizas J. M., Raja S. J., Holz H. et al. , 2012. The NSL complex regulates housekeeping genes in Drosophila. PLoS Genet. 8: e1002736 10.1371/journal.pgen.1002736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Li X., Hechmer A., Eisen M., Biggin M. D. et al. , 2008. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol. Cell. Biol. 28: 3290–3300. 10.1128/MCB.02224-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhoumaud P., Hennion M., Gamot A., Cuddapah S., Queille S. et al. , 2014. Insulators recruit histone methyltransferase dMes4 to regulate chromatin of flanking genes. EMBO J. 33: 1599–1613. 10.15252/embj.201385965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Lacroix L., Gamot A., Cuddapah S., Queille S. et al. , 2014. Chromatin immunoprecipitation indirect peaks highlight long-range interactions of insulator proteins and Pol II pausing. Mol. Cell 53: 672–681. 10.1016/j.molcel.2013.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez D. G., Spielmann M., and Mundlos S., 2016. Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet. 32: 225–237. 10.1016/j.tig.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Melnikova L., Kostuchenko M., Silicheva M., and Georgiev P., 2008. Drosophila gypsy insulator and yellow enhancers regulate activity of yellow promoter through the same regulatory element. Chromosoma 117: 137–145. 10.1007/s00412-007-0132-6 [DOI] [PubMed] [Google Scholar]
- Morris J. R., Petrov D. A., Lee A. M., and Wu C. T., 2004. Enhancer choice in cis and in trans in Drosophila melanogaster: role of the promoter. Genetics 167: 1739–1747. 10.1534/genetics.104.026955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N., Brown C. D., Shah P. K., Kheradpour P., Morrison C. A. et al. , 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6: e1000814 10.1371/journal.pgen.1000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolis I. K., McKay D. J., Mantouvalou E., Lomvardas S., Merika M. et al. , 2009. Transcription factors mediate long-range enhancer-promoter interactions. Proc. Natl. Acad. Sci. USA 106: 20222–20227. 10.1073/pnas.0902454106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli T., Vedder L., Dowling D., Petersen M., Meusemann K. et al. , 2016. Transcriptomic data from panarthropods shed new light on the evolution of insulator binding proteins in insects: insect insulator proteins. BMC Genomics 17: 861 10.1186/s12864-016-3205-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payre F., and Vincent A., 1991. Genomic targets of the serendipity beta and delta zinc finger proteins and their respective DNA recognition sites. EMBO J. 10: 2533–2541. 10.1002/j.1460-2075.1991.tb07793.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payre F., Noselli S., Lefrere V., and Vincent A., 1990. The closely related Drosophila sry beta and sry delta zinc finger proteins show differential embryonic expression and distinct patterns of binding sites on polytene chromosomes. Development 110: 141–149. [DOI] [PubMed] [Google Scholar]
- Payre F., Crozatier M., and Vincent A., 1994. Direct control of transcription of the Drosophila morphogen bicoid by the serendipity delta zinc finger protein, as revealed by in vivo analysis of a finger swap. Genes Dev. 8: 2718–2728. 10.1101/gad.8.22.2718 [DOI] [PubMed] [Google Scholar]
- Payre F., Buono P., Vanzo N., and Vincent A., 1997. Two types of zinc fingers are required for dimerization of the serendipity delta transcriptional activator. Mol. Cell. Biol. 17: 3137–3145. 10.1128/MCB.17.6.3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. S., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D. et al. , 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680 [corrigenda: Cell 162: 687–688 (2015)]. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R., Gorman C. M., and Howard B., 1985. Minichromosome assembly of non-integrated plasmid DNA transfected into mammalian cells. Nucleic Acids Res. 13: 3599–3615. 10.1093/nar/13.10.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee D. Y., Cho D. Y., Zhai B., Slattery M., Ma L. et al. , 2014. Transcription factor networks in Drosophila melanogaster. Cell Rep. 8: 2031–2043. 10.1016/j.celrep.2014.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley M. J., Nichols M. H., Lyu X., Ando-Kuri M., Rivera I. S. M. et al. , 2017. Evolutionarily conserved principles predict 3D chromatin organization. Mol. Cell 67: 837–852.e7. 10.1016/j.molcel.2017.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Gilbert M. K., and Hart C. M., 2007a Characterization of BEAF mutations isolated by homologous recombination in Drosophila. Genetics 176: 801–813. 10.1534/genetics.106.068056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Tan Y. Y., and Hart C. M., 2007b A genetic screen supports a broad role for the Drosophila insulator proteins BEAF-32A and BEAF-32B in maintaining patterns of gene expression. Mol. Genet. Genomics 277: 273–286. 10.1007/s00438-006-0187-8 [DOI] [PubMed] [Google Scholar]
- Ruez C., Payre F., and Vincent A., 1998. Transcriptional control of Drosophila bicoid by Serendipity delta: cooperative binding sites, promoter context, and co-evolution. Mech. Dev. 78: 125–134. 10.1016/S0925-4773(98)00159-2 [DOI] [PubMed] [Google Scholar]
- Rushlow C., Doyle H., Hoey T., and Levine M., 1987. Molecular characterization of the zerknullt region of the Antennapedia gene complex in Drosophila. Genes Dev. 1: 1268–1279. [DOI] [PubMed] [Google Scholar]
- Schnorrer F., Bohmann K., and Nusslein-Volhard C., 2000. The molecular motor dynein is involved in targeting swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat. Cell Biol. 2: 185–190. 10.1038/35008601 [DOI] [PubMed] [Google Scholar]
- Schwartz Y. B., Linder-Basso D., Kharchenko P. V., Tolstorukov M. Y., Kim M. et al. , 2012. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 22: 2188–2198. 10.1101/gr.138156.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokri L., Inukai S., Hafner A., Weinand K., Hens K. et al. , 2019. A comprehensive Drosophila melanogaster transcription factor interactome. Cell Rep. 27: 955–970.e7. 10.1016/j.celrep.2019.03.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha S., Oh D. H., McKowen J. K., Dassanayake M., and Hart C. M., 2018. 4C-seq characterization of Drosophila BEAF binding regions provides evidence for highly variable long-distance interactions between active chromatin. PLoS One 13: e0203843 10.1371/journal.pone.0203843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smulders-Srinivasan T. K., and Lin H., 2003. Screens for piwi suppressors in Drosophila identify dosage-dependent regulators of germline stem cell division. Genetics 165: 1971–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41: 207–234. 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., and Dubendorff J. W., 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185: 60–89. 10.1016/0076-6879(90)85008-C [DOI] [PubMed] [Google Scholar]
- Sultana H., Verma S., and Mishra R. K., 2011. A BEAF dependent chromatin domain boundary separates myoglianin and eyeless genes of Drosophila melanogaster. Nucleic Acids Res. 39: 3543–3557. 10.1093/nar/gkq1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tue N. T., Yoshioka Y., Mizoguchi M., Yoshida H., Zurita M. et al. , 2017. DREF plays multiple roles during Drosophila development. Biochim. Biophys. Acta 1860: 705–712. 10.1016/j.bbagrm.2017.03.004 [DOI] [PubMed] [Google Scholar]
- Ulianov S. V., Khrameeva E. E., Gavrilov A. A., Flyamer I. M., Kos P. et al. , 2016. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 26: 70–84. 10.1101/gr.196006.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K., Nichols M. H., Li L., Ong C. T., Takenaka N. et al. , 2014. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 15: R82 10.1186/gb-2014-15-5-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M., Barrington C., Henderson S., Ernst C., Odom D. T. et al. , 2015. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 10: 1297–1309. 10.1016/j.celrep.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelmann J., Le Gall A., Dejardin S., Allemand F., Gamot A. et al. , 2014. Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the Drosophila genome. PLoS Genet. 10: e1004544 10.1371/journal.pgen.1004544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. L., Duttke S. H., Chen K., Johnston J., Kassavetis G. A. et al. , 2014. TRF2, but not TBP, mediates the transcription of ribosomal protein genes. Genes Dev. 28: 1550–1555. 10.1101/gad.245662.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub A. S., Li C. H., Zamudio A. V., Sigova A. A., Hannett N. M. et al. , 2017. YY1 is a structural regulator of enhancer-promoter loops. Cell 171: 1573–1588.e28. 10.1016/j.cell.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi M. A., Arnold C. D., Schernhuber K., Pagani M., Rath M. et al. , 2015. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518: 556–559. 10.1038/nature13994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Andrew D. J., Mathies L. D., Horner M. A., and Scott M. P., 1993. Ectopic expression and function of the Antp and Scr homeotic genes: the N terminus of the homeodomain is critical to functional specificity. Development 118: 339–352. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wong C. H., Birnbaum R. Y., Li G., Favaro R. et al. , 2013. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature 504: 306–310. 10.1038/nature12716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Hart C. M., and Laemmli U. K., 1995. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81: 879–889. 10.1016/0092-8674(95)90008-X [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. Primer sequences are in Supplemental Material, Table S1. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.6084/m9.figshare.11988537.