Cues associated with intoxication can elicit cravings for alcohol, leading to increased consumption and relapse in people recovering from alcohol use disorder. Petruccelli et al. employed genetic tools in...
Keywords: alcohol, memory, RNA-sequencing, splicing, Drosophila
Abstract
Repeated alcohol experiences can produce long-lasting memories for sensory cues associated with intoxication. These memories can problematically trigger relapse in individuals recovering from alcohol use disorder (AUD). The molecular mechanisms by which ethanol changes memories to become long-lasting and inflexible remain unclear. New methods to analyze gene expression within precise neuronal cell types can provide further insight toward AUD prevention and treatment. Here, we used genetic tools in Drosophila melanogaster to investigate the lasting consequences of ethanol on transcription in memory-encoding neurons. Drosophila rely on mushroom body (MB) neurons to make associative memories, including memories of ethanol-associated sensory cues. Differential expression analyses revealed that distinct transcripts, but not genes, in the MB were associated with experiencing ethanol alone compared to forming a memory of an odor cue associated with ethanol. Adult MB-specific knockdown of spliceosome-associated proteins demonstrated the necessity of RNA-processing in ethanol memory formation. These findings highlight the dynamic, context-specific regulation of transcription in cue-encoding neurons, and the lasting effect of ethanol on transcript usage during memory formation.
ALCOHOL use disorder (AUD) affects millions of individuals and constitutes one of the most serious public health problems in the world today (Kendler et al. 2016; Grant et al. 2017; Cheng et al. 2018). This chronic, relapsing brain disorder can affect individuals for extremely long periods of time regardless of alcohol abstinence. Relapse is often triggered by exposure to cues that predict alcohol availability, as they evoke memories of the drug’s effects to result in cravings (Jasinska et al. 2014; Courtney et al. 2016; Groefsema et al. 2016; Valyear et al. 2017; Clemens and Holmes 2018; Logge et al. 2019). The complexity of alcohol’s molecular actions and our limited understanding of how these actions change distinct neurons in the brain’s reward memory circuitry has prevented our understanding of the mechanisms underlying these maladaptive memories.
Unlike the majority of other abused drugs, ethanol does not act on a particular molecular target, but instead affects a variety of molecules (Nestler 2013; Trudell et al. 2014). Many of the molecules implicated in ethanol-induced behaviors have broad roles in regulating diverse processes such as cell signaling, transcription, and neuronal plasticity (Ron and Barak 2016; Erickson et al. 2019). Furthermore, these roles are often dependent on both cell type and developmental stage. This complexity, as well as the diversity of experimental approaches in AUD research, often obscures ethanol’s context-specific molecular effects. However, recent advances in cell-type-specific isolation and sequencing technology can reveal the precise consequences of ethanol exposure on gene expression.
The genetic tools available in the fruit fly Drosophila melanogaster provide the ability to define where, when, and how alcohol may be acting in the nervous system, including within memory circuitry. Drosophila demonstrate ethanol-induced hyperactivity, tolerance (Wolf et al. 2002), and consummatory preference (Devineni and Heberlein 2009). They also remember and prefer the experience of intoxication, and this process requires mushroom body (MB) neuron activity (Kaun et al. 2011; Nunez et al. 2018). The MB integrates both sensory odor information from olfactory projection neurons and valence (aversive/rewarding) information from dopamine neurons. Downstream MB output neurons then drive avoidance of, or approach toward, an odor cue (Aso et al. 2014).
Long-term memory formation in many species requires both de novo transcription and protein synthesis (Bailey et al. 1996; Alberini and Kandel 2014; Sweatt 2016). Although long-term memory formation requires transcriptional changes (Alberini and Kandel 2014; Uchida and Shumyatsky 2018), it remains unclear how repeated ethanol experiences influence these processes. Similarly, whether the presentation of ethanol alone or ethanol paired with an odor induces the same transcriptional events is unknown. We hypothesized that ethanol exposures, alone or paired with an odor, would produce lasting transcriptomic changes within memory-associated MB neurons, thus altering memory formation.
Recent RNA-sequencing (RNA-seq) analysis in Drosophila (hereafter “flies”) has systematically identified the transcriptomic profiles of various cell types within MB circuitry (Shih et al. 2019), and examined gene expression changes in the context of associative memory formation (Crocker et al. 2016; Widmer et al. 2018). Here, we used nuclear, MB-specific RNA-seq to better understand transcriptional changes after the formation of odor-cue-induced ethanol memory. We found that ethanol exposures caused lasting expression changes at the transcript level, but not gene level, and that altered transcript usage was distinct between ethanol alone and ethanol cue memory groups. This suggests that differences in RNA-processing provide a distinct molecular landscape for encoding behavioral experiences.
Materials and Methods
Fly husbandry
Flies were raised on cornmeal agar food at 24°, 70% humidity, and with a 14:10 light:dark cycle. All flies used in this study were male. Male 4- to 5-day-old flies were isolated under CO2, given 1 day to recover, and then conditioned with specified paradigms detailed below. The fly lines used in this study include two pan-MB specific drivers, a MB010B split Gal4 line [#68293; Bloomington Drosophila Stock Center (BDSC)] (Aso et al. 2014) and R19B03 Gal4 line (#49830; BDSC) (Jenett et al. 2012), tubP-Gal80ts-7 (#7018; BDSC) (McGuire et al. 2004) recombined with R19B03-Gal4, a 5xUAS-unc84-2xGFP “INTACT” line (Henry et al. 2012), UAS-Cdc5-RNAi (#57425; BDSC) (Perkins et al. 2015), UAS-Rm62-RNAi (#34829; BDSC) (Perkins et al. 2015), UAS-Ref1-RNAi (#34626; BDSC) (Perkins et al. 2015), UAS-Pep-RNAi (#32944; BDSC) (Perkins et al. 2015), UAS-CG7971-RNAi (#52936; BDSC) (Perkins et al. 2015), GFP-RNAi (9330; BDSC), yw (Janelia Research Campus), and TRiP control (#36303; BDSC) (Perkins et al. 2015). elavC155-Gal4 (#458; BDSC) and Act5C-Gal4 (#25374; BDSC) were used to test efficacy of the RNA interference (RNAi) lines. Unless otherwise specified, experimental flies were heterozygous for transgenes. Control heterozygote crosses were performed with consideration to the respective genetic background of the trans-heterozygous Gal4 > UAS flies.
Odor-cue-induced ethanol memory
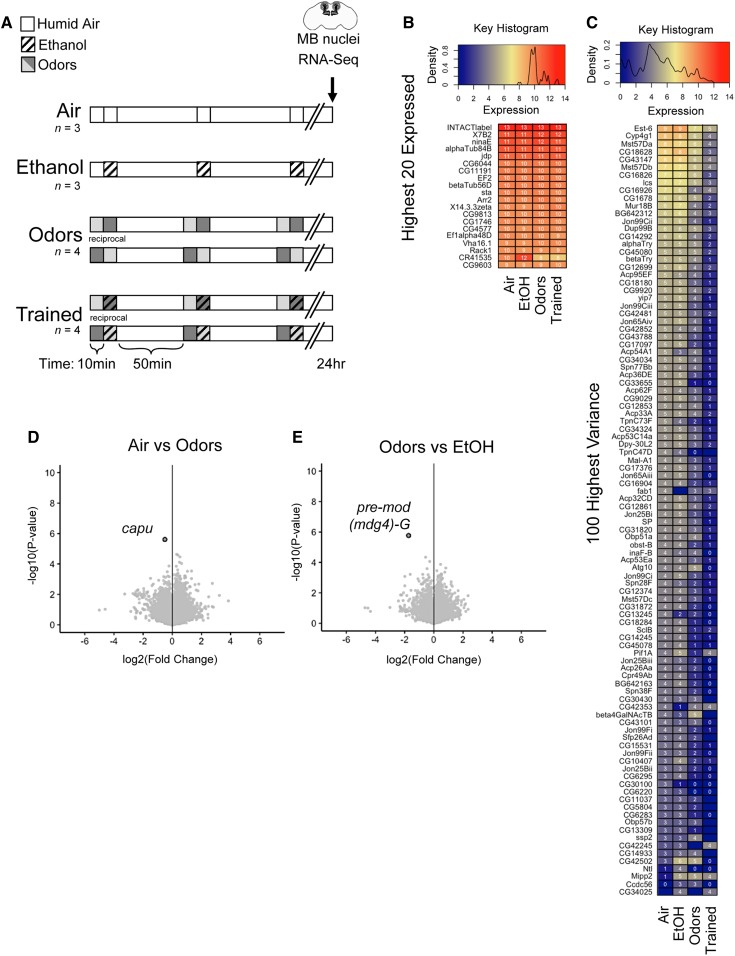
Odor-cue-induced ethanol memory was performed as published previously (Kaun et al. 2011; Nunez et al. 2018; Petruccelli et al. 2018). One minor difference for the RNA-seq data were the use of 50 flies per 14 ml training vial for the isolation of nuclei tagged in specific cell types (INTACT) experiment, whereas memory experiments were performed with 30 flies per vial. Humidified ethanol vapor (90:60 EtOH:air) for 10 min causes a moderate 13.8 ± 3 mM (0.01 g/dl) internal body ethanol concentration. Ethanol, odors (1:36 odorant:mineral oil with ethyl acetate or isoamyl alcohol), or both were delivered to flies in perforated vials for 10 min, followed by a 50 or 60 min rest before second and third trainings were performed (Figure 1A). Vapor treatments were delivered to flies on 1% agar and supplemented with yeast pellets overnight before being tested or killed the next day.
Figure 1.
Exposure paradigms and differentially expressed genes in MB nuclei. (A) Paradigm depicting three spaced 10 min exposures of “Air”, “Ethanol”, “Odors”, or “Trained” (odor + ethanol) and flies killed 24 hr later. RNA for each biological replicate (n) was extracted from mushroom body nuclei isolated from ∼2000 male heads [Air: n = 3, Ethanol: n = 3, Odors: n = 4 (two reciprocals of opposite odor order), Trained: n = 4 (two reciprocals of opposite odor order)]. (B and C) Volcano plots showing fold change of gene expression [log2(fc+1)] compared to the inverse of statistical significance [−log10(P-value)] (dark outline, FDR < 0.05). Both (B) Air vs. Odors and (C) Odors vs. EtOH had one statistically significant differentially expressed gene. (D and E) Density histogram and normalized FPKM expression (blue, yellow, and red color representing relative levels) of (D) the 100 most variable genes—the squared deviation from the mean—and (E) the 20 highest expressed genes postfiltering of RNA-associated genes. FDR, false discovery rate.
Memory testing was performed in a Y-Maze where flies were given a choice between the paired and unpaired odor. Conditioned preference index was calculated by counting the number of flies that moved toward the paired odor, subtracting the flies that moved toward the unpaired odor, and dividing that number by the total number of flies. The average conditioned preference index for pair of reciprocal tests was used as biological n = 1 for each genotype. Odor controls were performed similarly, but with preference indices corresponding to the number of flies facing a choice of either an odor (ethyl acetate or isoamyl alcohol) or no odor.
Isolation of MB nuclei (INTACT procedure)
INTACT was adapted from the method described in Pankova and Borst (2016) and pioneered in flies by Henry et al. (2012). The INTACT method of extracting nuclear RNA for sequencing provides several advantages compared to a general RNA-seq approach. First, the ability to use flash-frozen tissue, in contrast to FACS or dissected tissue samples, allows for an accurate examination of the current transcriptional profile of genetically targeted cells. Second, nuclear RNA from neurons contributes to the integrity of the active transcriptome snapshot by minimizing contamination from messenger RNA (mRNA) stored in the cytoplasm, along dendrites, and within axons, while allowing for the detection of experience-dependent differential expression (Lacar et al. 2016). Finally, given evidence that mRNA handling in subcellular compartments has been implicated in the formation and memory storage (Bramham and Wells 2007; Richter 2010; Shigeoka et al. 2016; Nakahata and Yasuda 2018; Biever et al. 2019), this approach provides a robust profile of the stable postexposure transcriptome unencumbered by the diversity of whole-cell RNA.
For each biological replicate used for INTACT RNA-seq, flies were pooled such that 2000 fly heads were collected via liquid nitrogen freezing and sieve-separated 24 hr after treatment conditioning. Flies were treated with one of four exposure paradigms (Figure 1A). Biological replicates were performed for air (n = 3), EtOH (n = 3), odors (n = 4), and trained (n = 4) conditions. Odors and trained conditions were performed with reciprocal odor groups (n = 2 ethyl acetate→isoamyl alcohol; n = 2 isoamyl alcohol→ethyl acetate).
Frozen heads were homogenized in a Kontes glass homogenizer (D9938-1SET; Sigma, St. Louis, MO) with 10 ml of Dounce buffer (10 mM β-glycerophosphate, 2 mM MgCl2, 0.5% IGEPAL buffer) and homogenized for ∼2 min with the loose A pestle. Homogenate was passed through a 190-µm nylon net filter (CMN-0185-C; Small Parts). The filter was subsequently rinsed with 2 ml of the same buffer before discarding. Homogenate was subsequently further homogenized gently ∼6–7 times with the tight B pestle, passed through a 20-µm filter (F020N-12-C; Small Parts), brought up to 50 ml by adding sucrose buffer (10 mM β-glycerophosphate, 2 mM MgCl2, 25 mM KCl, 250 mM sucrose). Next, 300 µl of Dynabead A magnetic beads (10001D; Thermo Fisher) were preincubated with rabbit α-GFP antibody (G10362; Thermo), and incubated with the homogenate for 30 min at 4° with gentle agitation. Beads were then captured with a magnet for 15 min and washed five times with 600 µl of sucrose buffer for 5 min at 4° with gentle agitation. Finally, RNA was extracted from bead-bound nuclei using TRIzol (Ambion, Life Technologies), resuspended in RNA-ase free water and DNA-ase treated according to manufacturer’s instructions (Ambion DNA-Free Kit).
RNA-seq
RNA libraries were polyA-enriched to identify mRNA transcripts ready for nuclear export and sequenced using a Hi-Seq 4000 (Illumina) machine at a depth of ∼30 million single-end 1 × 50 bp reads by GENEWIZ (South Plainfield, NJ). Read quality was assessed using FastQC 0.11.5 (Babraham Bioinformatics). Adapters were removed and trimmed using Trimmomatic 0.36 (Bolger et al. 2014). The “new tuxedo suite” was used to further process reads (Pertea et al. 2016). Reads were aligned with HISAT2 2.0.5 (Pertea et al. 2016) to the Ensembl BDGP6_transcriptome reference (Dm6), modified to include the 5xUAS-unc-84-2xGFP sequence present in our flies. Next, samtools 1.3.1 (Li et al. 2009) and Stringtie 1.3.3 (Pertea et al. 2016) were used to sort, merge, and quantify transcripts. Lastly, assembly and analysis of Ballgown objects (Pertea et al. 2016) and data visualization with ggplot2 was performed in RStudio 1.0.136 (Wickham 2009). No samples were discarded and default settings were used for the pipeline unless otherwise noted; command line code as follows:
#Trimmomatic:
TruSeq3-SE.fa:2:30:10:8:true LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36
#HISAT2:
hisat2 -q -x <reference index> -U <input fastq file> -S <output sam file> -p 50
#Samtools:
samtools view -Su alns.sam | samtools sort -o alns.sorted -@ 4
#StringTie:
stringtie <input.sorted.bam> -b <path to Ballgown tables> -o <output.gtf> -G <reference.gtf>
Immunohistochemistry
Fly brains were dissected in 1 × PBS solution and fixed in 2% paraformaldehyde S2 media overnight at 4°. Brains were continuously rocked on a nutator throughout the remainder of the protocol. Brains were washed four times for 15 min with PBS with 0.5% Triton X-100 (PBT), blocked at room temperature for 1 hr in PBT with 5% goat serum (PBT-GS). Primary antibodies (1:400 or 1:20) in PBT-GS were incubated overnight at 4°. Brains were then washed four times for 15 min with PBT, and secondary antibodies (1:500) in PBT-GS were incubated at room temperature for 2 hr. Again, brains were washed four times for 15 min with PBT, then mounted in DAPI Fluoromount-G (SouthernBiotech) or ProLong Diamond Antifade Mountant (Invitrogen, Carlsbad, CA).
Imaging
Images were obtained using a Nikon A1R Multiphoton Upright Confocal Microscope using NIS-Elements Software. Nonsaturating laser power and gain were determined for each channel and held constant throughout an experiment. We used 2-μm Z-sections with a ×20 air objective, and 0.5-μm Z-sections with a ×60 oil objective. Max stack images vary in depth shown. Z-series are available upon request. FIJI was used to apply contrast and brightness settings for visualization purposes.
Quantitative PCR
Total RNA was extracted from ∼90 heads or 30 bodies using TRIzol (Ambion, Life Technologies). RNA samples were treated with DNase (Ambion DNA-Free Kit), and equal amounts of RNA (1 μg) were reverse-transcribed into complementary DNA (Applied Biosystems). Biological and technical replicates were then analyzed with SYBR Green Real-Time PCR (ABI PRISM 7700 Sequence Detection System; Bio-Rad) and performed using the following PCR conditions: 39 cycles (30 s at 95°, 30 s at 60°, 30 s at 72°). The Ct threshold was manually adjusted to 0.6 across all samples and targets. Then, each of the target genes was normalized to rp49 expression for the comparative ΔCt method analysis following subsequent comparison to the control genotype to assess fold enrichment (ΔΔCT method). Primers, listed below, were all tested for efficiency before using for experiments.
Stat92E-Exon1a_for TGCGCAACCAGTTGAATTCTT
Stat92E-Exon1a _rev CATTACACACACGACGCAGT
Stat92E-Exon1_for GGTAGTCGCGTTCGCAAAAA
Stat92E-Exon1_rev GCAGGTGTTGGGGGAAAAAC
Pep_for CGTCGTTCGGATCGGAG
Pep_rev GTCGAGAAGCGATGGTGG
Rm62_for GCGAGATCGTCATTGCCACAC
Rm62_rev GCGGAACTCACGGAGC
Ref1_for CGTCGGTAACCTGGACTAC
Ref1_rev CACGGCCACGTCTGAG
Rp49_for CATACAGGCCCAAGATCGTGAAG
Rp49_rev CACGTTGTGCACCAGGAACTTC
Statistical analysis
For RNA-seq, libraries were statistically analyzed in R to determine differential expression between treatment conditions. Raw P-values are shown, as well as corrected P-values (false discovery rate < 0.05). The freely available Ballgown package (Pertea et al. 2016) afforded generalized linear modeling statistics for differential expression analysis and data visualization tools. Libraries were normalized with Ballgown package default – sum of each library’s log expression measurements below the 75th percentile (Frazee et al. 2015). For intersectional analyses, hypergeometric statistics were performed on pairwise comparisons and considered different from chance at P-value < 0.05.
Memory and quantitative PCR experiment statistical analyses were performed using JMP Pro 14. Levene’s test was used to determine homogeneity of variance; Shapiro–Wilk goodness-of-fit was used to test normality. Data that did not violate assumptions of homogeneity of variance or normality were tested using an ANOVA test followed by Student’s t-test post hoc comparisons. Data that violated assumptions of normality and homogeneity of variance were tested with a rank-sums Wilcoxon test followed by Wilcoxon post hoc analysis for each pair. For all figures, the bar represents mean, error bars represent SE of the mean, and significance defined by *P < 0.05.
Data availability
Raw sequencing files and count data presented in this work are freely available on NCBI Gene Expression Omnibus via accession number GSE108525. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11929458.
Results
RNA-seq library characterization
We generated trans-heterozygous flies that express the nuclear-tagged GFP “INTACT” transgene (5xUAS-unc84-GFP) (Henry et al. 2012) in all MB neurons (MB010B-Gal4). Expression of INTACT was MB-specific (Supplemental Material, Figure S1, A and B) and did not disrupt odor-cue-induced ethanol memory (Figure S1C). MB-INTACT flies were subjected to one of four treatment paradigms—air, EtOH, odors, or trained—and flash frozen the following day (Figure 1A). MB nuclei were isolated and RNA-sequenced to a depth of ∼30 million reads per sample (Figure S2). Nearly all libraries showed >80% uniquely mapped reads and ∼15 million fragments per kilobase of transcript per million mapped reads (FPKM) per sample.
Across treatment conditions there was a similar frequency distribution of mean expression of the fly’s 17560 annotated genes (Figure S1D). Within the entire data set, 7984 genes (45%) showed little to no expression [arbitrarily defined as log2(FPKM + 1) < 2], whereas 9576 genes (55%) had robustly detected levels of gene expression [log2(FPKM + 1) ≥ 2]. Thus, adult MB nuclei were actively transcribing roughly 55% of annotated genes in the genome at the time of isolation.
The BDGP6 Dm6 annotation of the fly transcriptome is comprised of 17560 genes, with 34741 corresponding transcript isoforms, where 10010 genes are uniquely represented by a single transcript isoform. According to the current information (flybase.org), the Dm6 contains roughly 85% protein-coding isoforms, 2% noncoding RNAs, 1.5% microRNAs, 0.9% transfer RNAs, 0.8% small nucleolar RNAs, and 9.7% other/nonannotated biotype sequences. When pooling all expression counts across treatments, our data were comprised of 92% protein-coding genes/isoforms, 0.9% noncoding RNAs, 0.06% microRNAs, 0.06% transfer RNAs, 0.05% small nucleolar RNAs, and 6.3% other/nonannotated sequences (Figure S1E). The characterization of our RNA-seq libraries shows that adult MB neurons are actively transcribing detectable protein-coding genes to allow for interesting differential expression analyses.
Expression of a priori and novel gene lists across treatments
In light of the cell-type-specific RNA-seq and exposure conditions assayed (Figure 1A), we expected to see prominent expression of genes previously associated with MB function and or previously implicated ethanol-related behaviors. Using the “Vocabularies” search function on FlyBase (flybase.org), a list of genes was acquired representing “adult mushroom body” (FBbt:00003684) and ethanol-associated genes termed “behavioral response to ethanol” (FBbt:0048149). As expected, many of the MB-associated genes showed robust mean expression across our treatment libraries (Figure 1A, Figure S3A). Interestingly, many ethanol-associated genes were also detectable (Figure S3B).
We next determined the highest 20 expressed genes detected across treatments and found the “INTACTlabel” (manually added to the Dm6 transcriptome) was highly expressed across the libraries, demonstrating the robust nature of the GAL4/UAS binary expression system and further validating the specificity of the MB isolation technique. Most of the highly transcribed genes were RNA-associated genes (Figure S3Ci), which we posit to be a mix of transcripts from rough endoplasmic reticulum that was pulled down during nuclear isolation, as well as newly transcribed or nuclear-localized RNAs. We further curated the highest 20 mean expressed genes by excluding RNA-associated genes (Figure 1B). Together, these descriptive analyses indicate that the libraries represent comparable MB data sets in which lasting experience-dependent molecular effects could be explored.
Experience-dependent differential gene expression
To uncover experience-dependent effects on transcription, we examined genes that showed the greatest variance in expression (log-transformed counts per million means) across repeated air, ethanol, odors, and paired ethanol-odor (trained) treatments. Among the 100 most variable genes, many ribosomal proteins and previously uncharacterized (unnamed CG#) genes were identified (Figure S3D). Also included were Esterase-6 (Est-6), Cytochrome P450-4g1 (Cyp4g1), la costa (lcs), Mucin related 18B (Mur18B), alpha and beta Trypsin (αTry, βTry), yippee interacting protein 7 (yip7), fab1 kinase (fab1), Troponin C (TpnC73F and TpnC47D), Odorant binding protein 51a (Obp51a), obstructor-B (obst-B), inaF-B (inaF-B), Autophagy-related 10 (Atg-10), short spindle 2 (ssp2), neurotransmitter transporter-like (Ntl), Multiple inositol polyphosphate phosphatase 2 (Mipp2), and a number of Jonah peptidase transcripts (Figure 1C). Excluding RNA-associated genes further revealed Maltase Ai (Mal-A1), Sarcolamban B (SclB), Cuticular protein 49Ab (Cpr49Ab), and Coiled-coil domain containing 56 (Ccdc56) (Figure 1C).
To determine statistically significant differential expression between treatment conditions, we used “Ballgown” (Pertea et al. 2016), an R Bioconductor package. No genes or transcripts were removed from analyses and the adjusted P-value (a.k.a. q-value) was set to the standard false discovery rate < 0.05. All pairwise condition contrasts were examined at both the gene and transcript level. Only two genes were found to be differentially expressed 24 hr after the various treatments, the actin filament nucleating protein cappuccino (capu) (air vs. odors) (Figure 1D) and the nuclear DNA-binding protein pre-mod(mdg4)-G (odors vs. EtOH) (Figure 1E).
Intersectional analysis of differential transcript expression
In contrast to gene-level analysis, there were more statistically significant differences found at the transcript level, despite this data being subjected to more repeated measure corrections (Figure S4A). This implicates altered transcriptional start/stop site or RNA-processing events that influence transcript diversity. In light of this finding, subsequent analyses focused on the differential expression of transcripts between treatments.
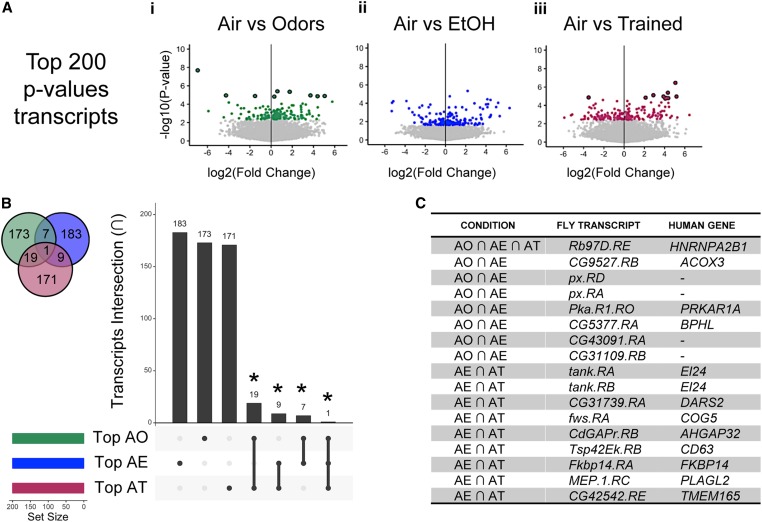
Using air treatment as a control, we compared the top 200 most significantly differentially expressed (lowest P-values) transcripts from air vs. odors (Figure 2Ai), air vs. EtOH (Figure 2Aii), and air vs. trained (Figure 2Aiii). There was surprisingly little intersectional overlap in specific transcripts between condition comparisons, including between air vs. EtOH and air vs. trained (Figure 2B). This indicated that experiencing odors, ethanol intoxication, or making ethanol-odor memories produced distinct, molecularly separable changes in the lasting transcriptional state of MB nuclei. Using hypergeometric statistics, the number of overlapping transcripts was found to be statistically greater than would be expected by chance alone (Figure S4B). The extent of intersection was visualized with an upgraded Venn diagram plot generated by an R package called “UpSetR” (Figure 2B, Figure S4B). Overlapping top P-value isoforms and their predicted human orthologs between condition comparisons are listed in Figure 2C.
Figure 2.
Comparison of differential transcripts in response to odor, ethanol, or trained (odor-ethanol) treatment. (A) Volcano plots showing fold change of transcript expression [log2(fc+1)] compared to the inverse of statistical significance [−log10(P-value)] (dark outline, FDR < 0.05). Plots for (i) Odors, (ii) EtOH, and (iii) Trained (ethanol-odor pairing) compared to Air (colors depict the top 200 P-value transcripts). (B) An upgraded Venn Diagram plot generated by an R package called “UpSetR” demonstrating the intersection in top 200 P-value transcripts (abbreviated by first letter in treatment). (C) A few of the transcripts from intersectional analysis are listed, along with the corresponding highest DIOPT-scored human genes. Hypergeometric statistics, *P < 0.05 (Figure S4C). FDR, false discovery rate.
We furthered compared the top 200 transcripts with the largest fold changes in expression (Figure S5). This approach demonstrated that specific transcripts have greater dynamic range in expression in response to experiencing odors, ethanol, or ethanol-odor training. Interestingly, there were more overlapping transcripts between pairwise comparisons than observed in the top P-value transcripts intersection analysis (Figure S5C), suggesting that particular transcripts in the MB neurons are more plastic than others. Hypergeometric statistics again showed that the number of overlapping transcripts was statistically greater than would be expected by chance alone (Figure S4C). A subset of the overlapping top fold-changing transcripts between treatment conditions are listed in Figure S5D.
Enrichment of genes associated with differential transcripts
To determine which types of transcripts are enriched in the top 200 P-value comparisons, a DAVID Gene Ontology (GO) analysis was performed on the associated gene IDs for each treatment: 167 air, 175 odor, 175, ethanol, and 188 trained genes. Using default DAVID settings, the top three GO annotations were identified (Figure S6). Compared to air controls, repeated ethanol exposures produced statistically significant GO terms: “Alternative splicing” (P-value = 0.0002), “Phosphoprotein” (P-value = 0.041), and “Coiled Coil” (P-value = 0.047). Interestingly, the response to trained treatment also produced “Alternative splicing” (P-value = 0.062) as a high-scoring enrichment category. It is important to note that the “Alternative splicing” GO term does not refer to spliceosome genes, but rather to genes that are “known to be alternatively spliced.” The enrichment analysis results further support the idea that there are experience-dependent changes in transcript usage, but not overall gene-level changes. Optimally, future ontology databases will include transcript-specific functional categorization.
Candidate transcripts underlying ethanol cue memories
Motivated by our interest in identifying lasting gene expression changes associated with formation of ethanol-cue-induced memories, we focused on the comparisons between odor vs. ethanol, ethanol vs. trained, and odor vs. trained flies (Figure 3).
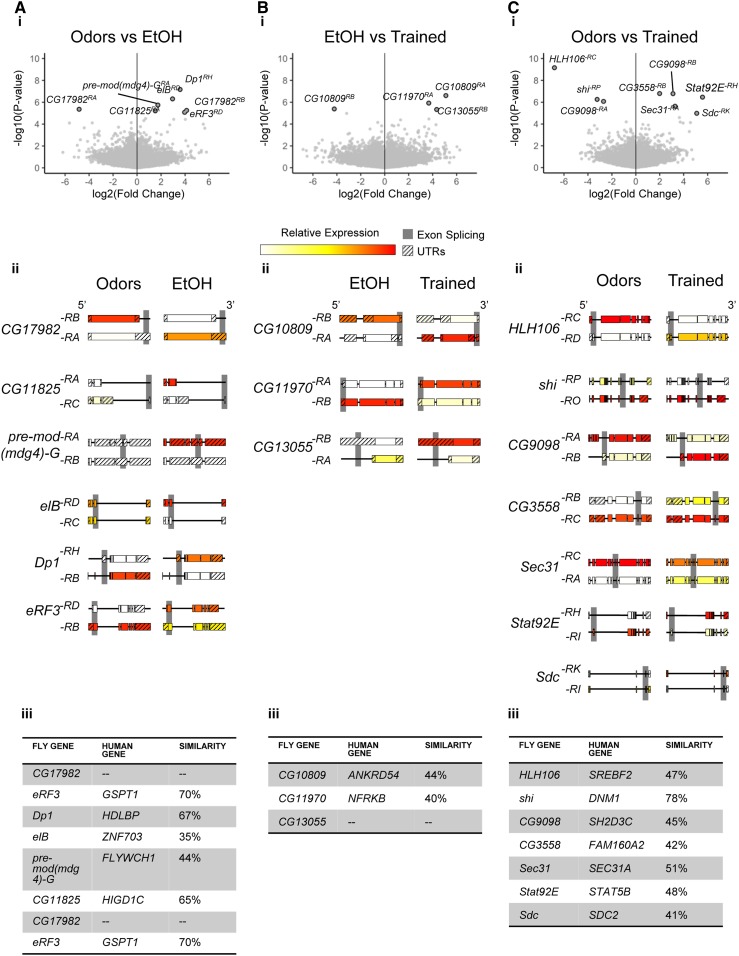
Figure 3.
Differential transcripts in particular pairwise treatment comparisons. (Ai–Ci) Volcano plots depicting (A) how EtOH differentially affects transcript expression compared to odors, (B) how Training (ethanol-odor pairing) differentially affects transcript expression compared to EtOH alone, and (C) how Training (ethanol-odor pairing) differentially affects transcript expression compared to Odors alone (dark outline, FDR < 0.05). (Aii–Cii) Significantly altered transcripts are depicted alongside the most highly expressed isoform of the same gene, gray shaded regions denote sequence differences between isoforms. (Aiii–Ciii) Transcripts are listed, along with the corresponding highest DIOPT-scored human genes. FDR, false discovery rate.
Seven transcripts (six genes) were found to be differentially expressed in response to ethanol treatment as compared to odors-only controls (Figure 3A). One transcript was downregulated (CG17982-RA) and six were upregulated {translational regulator Dodeca-satellite-binding protein 1 (Dp1-RH), transcription factor elbow B (elB-RD), transcription factor modifier of mdge 4 [pre-mod(mdg4)-G-RA], hypoxia-induced protein CG11825-RA, eukaryotic translation release factor 3 (eRF3), and a gene of unknown function CG17982-RB}.
Four transcripts (three genes, all of unknown function) were found to be differentially expressed in response to trained treatment as compared to ethanol alone (Figure 3B). One transcript was downregulated (CG10809-RB) and three were upregulated (CG11970-RA, CG13055-RB, and CG10809-RA).
Eight transcripts (seven genes) were found to be differentially expressed in response to trained treatment as compared to odors-only controls (Figure 3C, Figure S7). Three of these transcripts were downregulated: the Sterol Regulatory Element Binding Protein SREBP (HLH106-RC), the Drosophila homolog of dynamin shibire (shi-RP), and a predicted Ras guanine-nucleotide exchange factor CG9098-RA. Four transcripts were upregulated: [a retinoic acid-like protein CG3558-RB, the same predicted Ras guanine-nucleotide exchange factor CG9098-RB, a Coat Protein Complex II factor Secretory 31 (Sec31-RA), and a transmembrane heparin sulfate proteoglycan Syndecan (Sdc-RK)], as well as a transcription factor that functions in the JAK/STAT pathway, Signal-transducer and activator of transcription protein at 92E (Stat92E-RH). Using the DRSC Integrative Ortholog Prediction Tool (DIOPT) (Hu et al. 2011), we identified, when possible, the human genes corresponding to the implicated fly genes (Figure 3Aiii, Figure 3Biii, Figure 3Ciii).
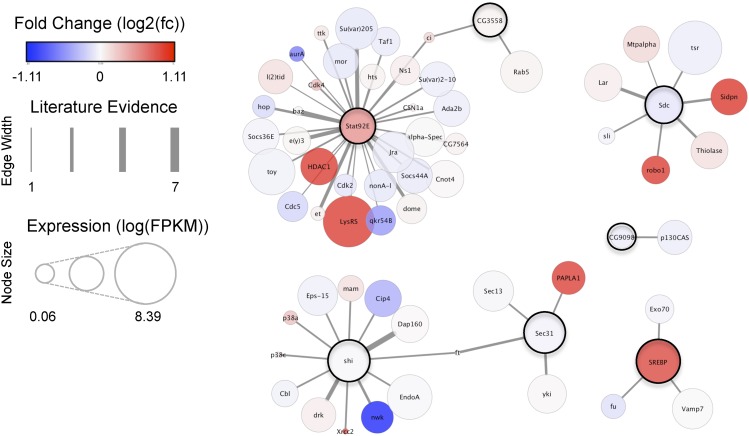
Molecular interactions with candidate ethanol-cue-induced memory genes
To visualize and probe deeper into the known functions of differentially expressed transcripts, we used the open source Cytoscape platform (Shannon et al. 2003) and MIST database (Hu et al. 2018) to identify proteins that interact with odor vs. trained transcripts of interest (Figure 4). By overlaying our data set information including expression level and fold change onto this network, we provided a foundation for prospective hypothesis-driven bioinformatic inquiries and experiments. Notably, Stat92E had the largest known network, including a number of proteins associated with nucleosome remodeling (HDAC1, brm, mor), transcriptional regulation (Taf1, Ada2b, kay, Jra, CG13510), and splicing (Cdc5, CG7564, tsu) (Figure 4).
Figure 4.
Protein interaction network analysis of alternatively expressed transcripts. Protein interaction network of proteins associated with “Odor vs. Trained” significant transcripts. Each node represents a protein, dark outlines denote proteins associated with significant “Odor vs. Trained” transcripts. Attributes of the nodes include fold change [log2(fc+1)] as color, expression level as size [log2(FKPM+1)] and edge thickness between nodes represents the extent of protein-protein interaction evidence from the MIST database.
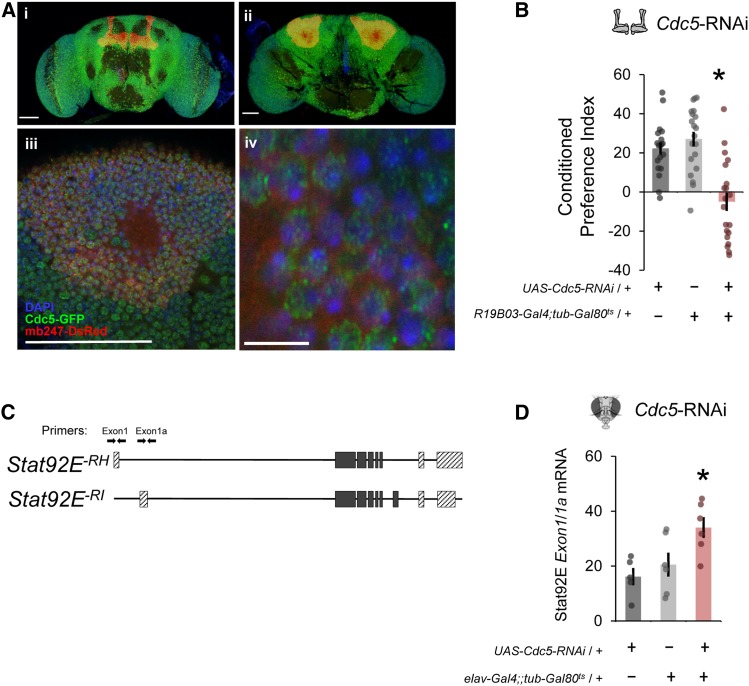
Considering our finding of differential transcript usage, we were intrigued to follow up on the possible requirement of spliceosomal proteins such as cell division cycle 5-like protein (Cdc5). A GFP-tagged version of Cdc5 was expectantly observed within nuclei throughout the adult central brain (Figure 5A, i and ii), including within MB neurons (Figure 5A, iii and iv). To test the necessity of Cdc5 in ethanol-cue-induced memory, Cdc5-RNAi was expressed during the adult stage using a pan-MB-specific Gal4 driver (R19B03-Gal4) (Figure S9A). Flies with adult Cdc5 knockdown had significant impairment in ethanol-cue-induced memory as compared to genetics controls (Figure 5B). To determine whether Cdc5 could affect individual Stat92E transcript expression, we performed whole-head quantitative RT-PCR with primers specific to each alternative first exon in the Stat-92E transcripts, where Exon1 includes Stat92E-RH and Exon1a includes Stat92E-RI (Henriksen et al. 2002) (Figure 5C, Figure S8A). Decreasing expression of Cdc5 in all adult neurons significantly altered the ratio of Exon1 to 1a (Stat92E-RH to -RI) transcripts (Figure 5D, Figure S8B).
Figure 5.
Spliceosome complex gene Cdc5 is required for cue-induced ethanol memory in adult MB neurons. (A) A Cdc5-GFP fusion shows that Cdc5 is expressed diffusely in the (i) anterior and (ii) posterior adult central brain, including (iii and iv) within MB nuclei (red). Bar in i, ii, and iii is 50 µM, Bar in iv is 5 µM). (B) Expression of Cdc5-RNAi in adult MB neurons significantly reduced conditioned preference for odor-cue-induced ethanol memory [F(2,56) = 21, *P < 0.0001]. Expression pattern of R19B03-Gal4 is shown in Figure S9A, and efficacy of the RNAi in Figure S10. (C) Primers designed to measure expression of transcripts that include the Stat92E–RH (Exon 1) or –RI (Exon 1a) isoforms (Henriksen et al. 2002), see Figure S8A for depiction of all Stat92E transcripts. (D) Decreasing expression of Cdc5 in adult neurons significantly alters the ratio of Exon1/1a transcripts in whole-head tissue [F(2,14) = 5.7, *P = 0.02].
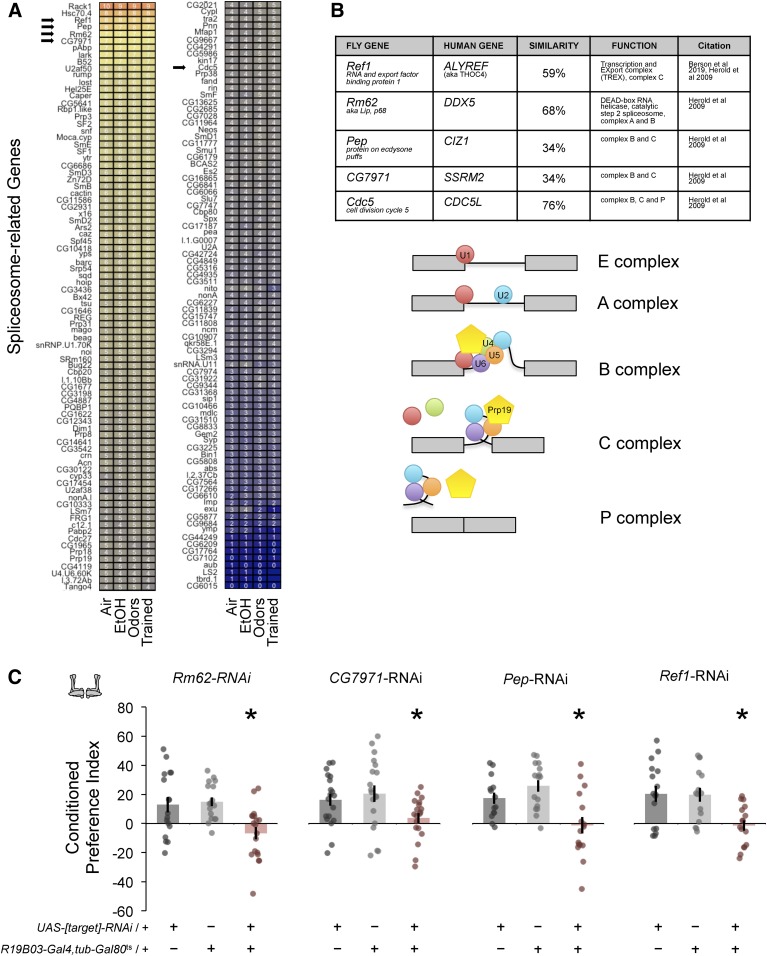
Knockdown of splice factors impairs ethanol-cue-induced memory
In light of differential transcript usage, GO analysis, and Cdc5 requirement, we assessed the detectable expression of FlyBase’s “spliceosome complex” gene list (GO:0005681) (Figure 6A). We predicted that decreasing expression of highly expressed splice factors in the MB neurons would affect alcohol associative memories. We choose candidate targets RNA export and export binding protein 1 (Ref1), Protein on ecdysone puffs (Pep), RNA helicase Rm62 (also referred to as p68), and CG7971, which is orthologous to serine/arginine repetitive matrix 2 (SRRM2) (Figure 6B). Adult MB-specific knockdown of each of these targets impaired ethanol-cue-induced memory without affecting odor sensitivity (Figure 6C, Figures S9 and S10).
Figure 6.
Knockdown of splice-associated targets in MB neurons reduces odor-cue-induced ethanol memory. (A) Density histogram and normalized FPKM expression (blue, yellow, and red color representing relative levels) of genes associated with the spliceosome; arrows mark selected candidate targets for testing. (B) Table listing four top-expressed spliceosome-associated genes and Cdc5, along with their corresponding highest DIOPT-scored human genes (Herold et al. 2009; Berson et al. 2019), and schematic demonstrating the complexes associated with these genes during splicing. (C) Expression of RNAi targeting Rm62 [F(2,49) = 5.72, P = 0.02], CG7971 [F(2,55) = 3.75, P = 0.03], Ref1 [F(2,44) = 6.96, P = 0.002], or Pep [F(2,43) = 9.30, P = 0.0004] in adult MB neurons significantly reduced conditioned preference for odor-cue-induced ethanol memory.
Discussion
The nuclear, cell-type-specific nature of our data provides a unique analysis of genes actively transcribed in memory-encoding neurons 24 hr after experience with ethanol or ethanol cue associative memory formation. The RNA-seq analysis highlighted the differential expression of particular transcripts. This work supports the essential role of RNA processing in the lasting transcriptional changes encoding ethanol-cue-induced memory.
Confirmation of known genes implicated with memory
Two of the genes implicated in this work have been previously implicated in memory and or ethanol-related behaviors. shi, the fly homolog of Dynamin (van der Bliek and Meyerowitz 1991), plays a role in endocytosis, which is critical for synaptic transmission in MB neurons during memory formation (McGuire et al. 2001; Kasuya et al. 2009) and in ethanol tolerance (Krishnan et al. 2012). We have also previously shown that expression of a temperature-sensitive form of shi in MB neurons disrupts ethanol cue memory at higher temperatures (Kaun et al. 2011). Stat92E, a transcription factor in the JAK/STAT signaling pathway, plays a role in long-term memory in Drosophila (Copf et al. 2011), and we have recently shown that reducing Stat92E in MB neurons similarly reduces ethanol cue memory (Petruccelli et al. 2018). Other targets identified in this work have been implicated in long-term memory in Drosophila (Figure S11). Similarly, a recent study showing the effects of acute response to alcohol shows some overlap in transcriptomic targets (Figure S11) (Signor and Nuzhdin 2018).
Context-specific transcript expression
An intriguing pattern that emerged from the data is that administration of repeated mild doses of ethanol, odors alone, or odors paired with ethanol all produced significantly altered transcript isoforms. Remarkably, the particular transcripts identified were specific to the behavioral paradigm the animals experienced. Even altered transcripts associated with the presentation of ethanol alone were very different from those when ethanol was presented concomitantly with an odor. This suggests that transcript expression is specific to the type of behavioral context flies experienced.
Alternative splicing associated with behavior in MB neurons
Alternative transcripts can be created through the use of alternative transcriptional start sites or alternative splicing. Alternative splicing of pre-mRNA transcribed from a finite genetic sequence enhances proteomic diversity. It is estimated that 95% of human genes undergo alternative splicing (Pan et al. 2008) and disruptions in the splicing status quo can lead to various human disease (Cieply and Carstens 2015). The most common cause of abnormal splicing is due to a mutation in the core splicing consensus sequences (Cieply and Carstens 2015). Our data, however, are derived from animals with the same genetic backgrounds and environmental conditions. This suggests that mutagenesis is not the cause of the potential transcript usage differences we observed.
Our data did not demonstrate differential expression in spliceosome factors with behavior condition, but indirectly implicated spliceosomal machinery. Specifically, known protein-protein interactions were revealed between Stat92E and spliceosome complex associated proteins, including Cdc5, CG7564, and tsu (Guruharsha et al. 2011) (Figure 4). The lack of differentially expressed splicing factors with treatment condition does not, however, rule out the possibility that spliceosome-associated proteins were acutely affected during treatment exposure. Consequently, our data may represent the aftermath of these, or post-translational response, effects.
A caveat to our data are that because we did not a priori expect to see splicing differences, our sequencing data were not performed with long-read sequencing techniques. Nor were statistical considerations, such as k-means clustering of transcripts or weighting exon-exon junction reads, used to compare splicing across genes. However, our data do show that reducing gene expression of five known spliceosome associated proteins in the adult MB disrupts formation of ethanol-cue-induced memory. Thus, we believe that the alternative transcript usage in our data, is in part, due to splicing that occurs during memory formation.
Splicing associated with memory formation
Splicing of particular targets in response to memory formation is not without precedent. For example, the splicing of the Orb2A transcript is required for both associative appetitive and courtship suppression memory in Drosophila (Gill et al. 2017). There is also evidence that this phenomenon may be conserved across species, since contextual fear conditioning induces differential splicing in the hippocampus of mice (Poplawski et al. 2016). It is, therefore, conceivable that while splicing may occur during all forms of synaptic plasticity, the isoform targets may be specific to the type of memory formed. The broad implication of this prediction is that the molecular engram of memory within relevant cells uses transcriptional diversity to provide the cellular plasticity associated with the type of memory being formed. This cellular encoding is bolstered by the diversity of epigenetic changes caused by neuronal activity associated with memory formation. For example, activity-induced histone modifications caused a late-onset shift in Neurexin-1 splicing reduced the stability of memories (Ding et al. 2017). Our data suggest that alcohol can affect transcriptional events, and thus shape how context is encoded during formation of memories.
Alcohol regulates splicing in cue-encoding neurons
Several recent studies have shown that alternative splicing is correlated to chronic alcohol consumption. Chronic self-administration of alcohol in cynomolgus macaques (Macaca fascicularis) is associated with alternative splicing of AMPA subunits in the prefrontal cortex (Acosta et al. 2012). Broader transcriptomic analysis demonstrated overrepresentation of genes associated with splicing across brain regions in chronic ethanol self-administrating rhesus macaques (Macaca mulatta) (Iancu et al. 2018). Similar alcohol-induced effects on splicing extend to humans (Farris and Mayfield 2014). Postmortem analysis of the brains of patients with AUD showed novel splicing in GABAB1 that decreased expression of the GABA binding site (Lee et al. 2014). Similarly, alcohol-induced splicing events also occurred in the developing human cortex in utero, potentially resulting in devastating neurodevelopmental consequences (Kawasawa et al. 2017).
Our data suggest that mild repeated alcohol exposures have lasting consequences on the transcript isoforms expressed in Drosophila. Importantly, altered isoform transcription occurred in a diversity of genes, including transcription factors like Stat92E, which could have long-lasting and broad effects on transcription within memory circuits (Copf et al. 2011). As alcohol alone did not induce expression of alternative transcripts of Stat92E, this suggests that splicing of this gene might occur due to memory formation. Since our analysis was restricted to neurons necessary for memories associated with punishment or reward, it is possible that even small effects could have widespread consequences for subsequent memory formation and vulnerability to dependence on drugs of abuse. Uncovering whether this is a broader phenomenon will require identifying lasting transcriptional events that occur in the same cell types across a diversity of memory and drug exposure paradigms.
Diverse splicing mechanisms across addictions
Alternative splicing has been recently implicated in cocaine addiction (Cates et al. 2018). Transcription factor E2F3a regulates cocaine-induced alternative splicing in the mouse nucleus accumbens, and E2F3b mediates cocaine responses in the prefrontal cortex (Cates et al. 2019). Drosophila E2F1 was expressed in our MB nuclei data set (Figure 1Cii), although it was not alternatively spliced in response to alcohol or alcohol-cue training (see transcript count data in Gene Expression Omnibus). We speculate that cocaine utilizes the E2F transcription factor family more than alcohol does to drive drug-cue memory formation and retention. Future investigations will likely identify both conserved drug-specific and convergent molecular mechanisms that influence transcriptional activity in reward circuitry.
Acknowledgments
We thank Lee Henry (Cold Spring Harbor Laboratory, Southern Illinois University Edwardsville, National Institute of General Medical Sciences) for advice on INTACT, and members of the Kaun Laboratory, especially Reza Azanchi, Michael Feyder, and Yanabah Jaques, for assistance with Drosophila husbandry and laboratory tasks. We thank Dr. Faith Liebl (SIUE), Dr. Kate O’Connor-Giles (Brown University), and members of the Kaun lab for helpful feedback on the manuscript. This work was supported by the Richard and Susan Smith Family Foundation (Newton, MA), the Carney Institute for Brain Science Center of Biomedical Research Excellence “Center for Nervous System Function” (NIGMS grant P20-GM103645), and the National Institute on Alcohol Abuse and Alcoholism (grant R01-AA024434).
Author contributions: E.P. and K.R.K. designed the research and wrote the manuscript. E.P. and N.L. performed INTACT nuclear extraction. N.L. developed the pipeline to align RNA-seq reads. K.R.K. acquired alcohol memory data and performed associated data analysis. T.B. performed immunohistochemistry and acquired brain images. A.W. performed quantitative PCR experiments. E.P. performed RNA-seq statistical analyses and produced figures in consultation with K.R.K. Revision of the manuscript was performed by E.P. and K.R.K.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.11929458.
Communicating editor: M. Wolfner
Literature Cited
- Acosta G., Freidman D. P., Grant K. A., and Hemby S. E., 2012. Alternative splicing of AMPA subunits in prefrontal cortical fields of cynomolgus monkeys following chronic ethanol self-administration. Front. Psychiatry 2: 72 10.3389/fpsyt.2011.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberini C. M., and Kandel E. R., 2014. The regulation of transcription in memory consolidation. Cold Spring Harb. Perspect. Biol. 7: a021741 10.1101/cshperspect.a021741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y., Hattori D., Yu Y., Johnston R. M., Iyer N. A. et al. , 2014. The neuronal architecture of the mushroom body provides a logic for associative learning. eLife 3: e04577 10.7554/eLife.04577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C. H., Bartsch D., and Kandel E. R., 1996. Toward a molecular definition of long-term memory storage. Proc. Natl. Acad. Sci. USA 93: 13445–13452. 10.1073/pnas.93.24.13445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson A., Goodman L. D., Sartoris A. N., Otte C. G., Aykit J. A. et al. , 2019. Drosophila Ref1/ALYREF regulates transcription and toxicity associated with ALS/FTD disease etiologies. Acta Neuropathol. Commun. 7: 65 10.1186/s40478-019-0710-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biever A., Donlin-Asp P. G., and Schuman E. M., 2019. Local translation in neuronal processes. Curr. Opin. Neurobiol. 57: 141–148. 10.1016/j.conb.2019.02.008 [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramham C. R., and Wells D. G., 2007. Dendritic mRNA: transport, translation and function. Nat. Rev. Neurosci. 8: 776–789. 10.1038/nrn2150 [DOI] [PubMed] [Google Scholar]
- Cates H. M., Heller E. A., Lardner C. K., Purushothaman I., Pena C. J. et al. , 2018. Transcription factor E2F3a in nucleus accumbens affects cocaine action via transcription and alternative splicing. Biol. Psychiatry 84: 167–179. 10.1016/j.biopsych.2017.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cates H. M., Bagot R. C., Heller E. A., Purushothaman I., Lardner C. K. et al. , 2019. A novel role for E2F3b in regulating cocaine action in the prefrontal cortex. Neuropsychopharmacology 44: 776–784. 10.1038/s41386-018-0296-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. G., Kaakarli H., Breslau J., and Anthony J. C., 2018. Assessing changes in alcohol use and alcohol use disorder prevalence in the United States: evidence from national surveys from 2002 through 2014. JAMA Psychiatry 75: 211–213. 10.1001/jamapsychiatry.2017.4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieply B., and Carstens R. P., 2015. Functional roles of alternative splicing factors in human disease. Wiley Interdiscip. Rev. RNA 6: 311–326. 10.1002/wrna.1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens K. J., and Holmes N. M., 2018. An extended history of drug self-administration results in multiple sources of control over drug seeking behavior. Prog. Neuropsychopharmacol. Biol. Psychiatry 87: 48–55. 10.1016/j.pnpbp.2017.11.011 [DOI] [PubMed] [Google Scholar]
- Copf T., Goguel V., Lampin-Saint-Amaux A., Scaplehorn N., and Preat T., 2011. Cytokine signaling through the JAK/STAT pathway is required for long-term memory in Drosophila. Proc. Natl. Acad. Sci. USA 108: 8059–8064. 10.1073/pnas.1012919108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney K. E., Schacht J. P., Hutchison K., Roche D. J., and Ray L. A., 2016. Neural substrates of cue reactivity: association with treatment outcomes and relapse. Addict. Biol. 21: 3–22. 10.1111/adb.12314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker A., Guan X. J., Murphy C. T., and Murthy M., 2016. Cell-type-specific transcriptome analysis in the Drosophila mushroom body reveals memory-related changes in gene expression. Cell Rep. 15: 1580–1596. 10.1016/j.celrep.2016.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devineni A. V., and Heberlein U., 2009. Preferential ethanol consumption in Drosophila models features of addiction. Curr. Biol. 19: 2126–2132. 10.1016/j.cub.2009.10.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Liu S., Tian M., Zhang W., Zhu T. et al. , 2017. Activity-induced histone modifications govern Neurexin-1 mRNA splicing and memory preservation. Nat. Neurosci. 20: 690–699. 10.1038/nn.4536 [DOI] [PubMed] [Google Scholar]
- Erickson E. K., Grantham E. K., Warden A. S., and Harris R. A., 2019. Neuroimmune signaling in alcohol use disorder. Pharmacol. Biochem. Behav. 177: 34–60. 10.1016/j.pbb.2018.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris S. P., and Mayfield R. D., 2014. RNA-Seq reveals novel transcriptional reorganization in human alcoholic brain. Int. Rev. Neurobiol. 116: 275–300. 10.1016/B978-0-12-801105-8.00011-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazee A. C., Pertea G., Jaffe A. E., Langmead B., Salzberg S. L. et al. , 2015. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 33: 243–246. 10.1038/nbt.3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill J., Park Y., McGinnis J. P., Perez-Sanchez C., Blanchette M. et al. , 2017. Regulated intron removal integrates motivational state and experience. Cell 169: 836–848.e15. 10.1016/j.cell.2017.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B. F., Chou S. P., Saha T. D., Pickering R. P., Kerridge B. T. et al. , 2017. Prevalence of 12-month alcohol use, high-risk drinking, and DSM-IV alcohol use disorder in the United States, 2001–2002 to 2012–2013: results from the national epidemiologic survey on alcohol and related conditions. JAMA Psychiatry 74: 911–923. 10.1001/jamapsychiatry.2017.2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groefsema M., Engels R., and Luijten M., 2016. The role of social stimuli content in neuroimaging studies investigating alcohol cue-reactivity. Addict. Behav. 58: 123–128. 10.1016/j.addbeh.2016.02.033 [DOI] [PubMed] [Google Scholar]
- Guruharsha K. G., Rual J. F., Zhai B., Mintseris J., Vaidya P. et al. , 2011. A protein complex network of Drosophila melanogaster. Cell 147: 690–703. 10.1016/j.cell.2011.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen M. A., Betz A., Fuccillo M. V., and Darnell J. E. Jr, 2002. Negative regulation of STAT92E by an N-terminally truncated STAT protein derived from an alternative promoter site. Genes Dev. 16: 2379–2389 (erratum: Genes Dev. 16: 2729). 10.1101/gad.1020702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry G. L., Davis F. P., Picard S., and Eddy S. R., 2012. Cell type-specific genomics of Drosophila neurons. Nucleic Acids Res. 40: 9691–9704. 10.1093/nar/gks671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold N., Will C. L., Wolf E., Kastner B., Urlaub H. et al. , 2009. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol. Cell. Biol. 29: 281–301. 10.1128/MCB.01415-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B. et al. , 2011. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12: 357 10.1186/1471-2105-12-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Vinayagam A., Nand A., Comjean A., Chung V. et al. , 2018. Molecular Interaction Search Tool (MIST): an integrated resource for mining gene and protein interaction data. Nucleic Acids Res. 46: D567–D574. 10.1093/nar/gkx1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu O. D., Colville A., Walter N. A. R., Darakjian P., Oberbeck D. L. et al. , 2018. On the relationships in rhesus macaques between chronic ethanol consumption and the brain transcriptome. Addict. Biol. 23: 196–205. 10.1111/adb.12501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinska A. J., Stein E. A., Kaiser J., Naumer M. J., and Yalachkov Y., 2014. Factors modulating neural reactivity to drug cues in addiction: a survey of human neuroimaging studies. Neurosci. Biobehav. Rev. 38: 1–16. 10.1016/j.neubiorev.2013.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A., Rubin G. M., Ngo T. T., Shepherd D., Murphy C. et al. , 2012. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2: 991–1001. 10.1016/j.celrep.2012.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuya J., Ishimoto H., and Kitamoto T., 2009. Neuronal mechanisms of learning and memory revealed by spatial and temporal suppression of neurotransmission using shibire, a temperature-sensitive dynamin mutant gene in Drosophila melanogaster. Front. Mol. Neurosci. 2: 11 10.3389/neuro.02.011.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaun K. R., Azanchi R., Maung Z., Hirsh J., and Heberlein U., 2011. A Drosophila model for alcohol reward. Nat. Neurosci. 14: 612–619. 10.1038/nn.2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasawa Y. I., Mohammad S., Son A. I., Morizono H., Basha A. et al. , 2017. Genome-wide profiling of differentially spliced mRNAs in human fetal cortical tissue exposed to alcohol. Alcohol 62: 1–9. 10.1016/j.alcohol.2017.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler K. S., Ohlsson H., Sundquist J., and Sundquist K., 2016. Alcohol use disorder and mortality across the lifespan: a longitudinal cohort and Co-relative analysis. JAMA Psychiatry 73: 575–581. 10.1001/jamapsychiatry.2016.0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan H. R., Al-Hasan Y. M., Pohl J. B., Ghezzi A., and Atkinson N. S., 2012. A role for dynamin in triggering ethanol tolerance. Alcohol. Clin. Exp. Res. 36: 24–34. 10.1111/j.1530-0277.2011.01587.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacar B., Linker S. B., Jaeger B. N., Krishnaswami S. R., Barron J. J. et al. , 2016. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun. 7: 11022 [corrigenda: Nat. Commun. 8: 15047 (2017)]. 10.1038/ncomms11022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Mayfield R. D., and Harris R. A., 2014. Altered gamma-aminobutyric acid type B receptor subunit 1 splicing in alcoholics. Biol. Psychiatry 75: 765–773. 10.1016/j.biopsych.2013.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logge W. B., Morley K. C., Haber P. S., and Baillie A. J., 2019. Executive functioning moderates responses to appetitive cues: a study in severe alcohol use disorder and alcoholic liver disease. Alcohol Alcohol. 54: 38–46. 10.1093/alcalc/agy083 [DOI] [PubMed] [Google Scholar]
- McGuire S. E., Le P. T., and Davis R. L., 2001. The role of Drosophila mushroom body signaling in olfactory memory. Science 293: 1330–1333. 10.1126/science.1062622 [DOI] [PubMed] [Google Scholar]
- McGuire S. E., Mao Z., and Davis R. L., 2004. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE 2004: pl6 10.1126/stke.2202004pl6 [DOI] [PubMed] [Google Scholar]
- Nakahata Y., and Yasuda R., 2018. Plasticity of spine structure: local signaling, translation and cytoskeletal reorganization. Front. Synaptic Neurosci. 10: 29 10.3389/fnsyn.2018.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler E. J., 2013. Cellular basis of memory for addiction. Dialogues Clin. Neurosci. 15: 431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez K. M., Azanchi R., and Kaun K. R., 2018. Cue-induced ethanol seeking in Drosophila melanogaster is dose-dependent. Front. Physiol. 9: 438 10.3389/fphys.2018.00438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankova K., and Borst A., 2016. RNA-seq transcriptome analysis of direction-selective T4/T5 neurons in Drosophila. PLoS One 11: e0163986 10.1371/journal.pone.0163986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L. J., Frey B. J., and Blencowe B. J., 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40: 1413–1415 [corrigenda: Nat. Genet. 41: 762 (2009)]. 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- Perkins L. A., Holderbaum L., Tao R., Hu Y., Sopko R. et al. , 2015. The transgenic RNAi project at harvard medical school: resources and validation. Genetics 201: 843–852. 10.1534/genetics.115.180208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Kim D., Pertea G. M., Leek J. T., and Salzberg S. L., 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11: 1650–1667. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruccelli E., Feyder M., Ledru N., Jaques Y., Anderson E. et al. , 2018. Alcohol activates scabrous-notch to influence associated memories. Neuron 100: 1209–1223.e2. 10.1016/j.neuron.2018.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplawski S. G., Peixoto L., Porcari G. S., Wimmer M. E., McNally A. G. et al. , 2016. Contextual fear conditioning induces differential alternative splicing. Neurobiol Learn Mem 134 Pt B: 221–235. 10.1016/j.nlm.2016.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J. D., 2010. Translational control of synaptic plasticity. Biochem. Soc. Trans. 38: 1527–1530. 10.1042/BST0381527 [DOI] [PubMed] [Google Scholar]
- Ron D., and Barak S., 2016. Molecular mechanisms underlying alcohol-drinking behaviours. Nat. Rev. Neurosci. 17: 576–591. 10.1038/nrn.2016.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T. et al. , 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T., Jung H., Jung J., Turner-Bridger B., Ohk J. et al. , 2016. Dynamic axonal translation in developing and mature visual circuits. Cell 166: 181–192. 10.1016/j.cell.2016.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih M. M., Davis F. P., Henry G. L., and Dubnau J., 2019. Nuclear transcriptomes of the seven neuronal cell types that constitute the Drosophila mushroom bodies. G3 (Bethesda) 9: 81–94. 10.1534/g3.118.200726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signor S., and Nuzhdin S., 2018. Dynamic changes in gene expression and alternative splicing mediate the response to acute alcohol exposure in Drosophila melanogaster. Heredity 121: 342–360. 10.1038/s41437-018-0136-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt J. D., 2016. Neural plasticity and behavior - sixty years of conceptual advances. J. Neurochem. 139: 179–199. 10.1111/jnc.13580 [DOI] [PubMed] [Google Scholar]
- Trudell J. R., Messing R. O., Mayfield J., and Harris R. A., 2014. Alcohol dependence: molecular and behavioral evidence. Trends Pharmacol. Sci. 35: 317–323. 10.1016/j.tips.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S., and Shumyatsky G. P., 2018. Synaptically localized transcriptional regulators in memory formation. Neuroscience 370: 4–13. 10.1016/j.neuroscience.2017.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valyear M. D., Villaruel F. R., and Chaudhri N., 2017. Alcohol-seeking and relapse: a focus on incentive salience and contextual conditioning. Behav. Processes 141: 26–32. 10.1016/j.beproc.2017.04.019 [DOI] [PubMed] [Google Scholar]
- van der Bliek A. M., and Meyerowitz E. M., 1991. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature 351: 411–414. 10.1038/351411a0 [DOI] [PubMed] [Google Scholar]
- Wickham H., 2009. Ggplot2: Elegant Graphics for Data Analysis, Springer, New York: 10.1007/978-0-387-98141-3 [DOI] [Google Scholar]
- Widmer Y. F., Bilican A., Bruggmann R., and Sprecher S. G., 2018. Regulators of long-term memory revealed by mushroom body-specific gene expression profiling in Drosophila melanogaster. Genetics 209: 1167–1181. 10.1534/genetics.118.301106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F. W., Rodan A. R., Tsai L. T., and Heberlein U., 2002. High-resolution analysis of ethanol-induced locomotor stimulation in Drosophila. J. Neurosci. 22: 11035–11044. 10.1523/JNEUROSCI.22-24-11035.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequencing files and count data presented in this work are freely available on NCBI Gene Expression Omnibus via accession number GSE108525. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11929458.