The envelope glycoprotein (Env) spike on the surface of human immunodeficiency virus type 1 (HIV-1) mediates the entry of the virus into host cells and is also the target for antibodies. During virus entry, Env needs to change shape. Env flexibility also contributes to the ability of HIV-1 to evade the host immune response; many shapes of Env raise antibodies that cannot recognize the functional Env and therefore do not block virus infection. We found that an HIV-1 entry inhibitor, BMS-806, stabilizes the functional shape of Env. We developed new variants of BMS-806 that stabilize Env in its natural state for long periods of time. The availability of such long-acting stabilizers of Env shape will allow the natural Env conformation to be characterized and tested for efficacy as a vaccine.
KEYWORDS: entry inhibitor, envelope, immunogen, lentivirus, retrovirus, structure, vaccine, virus
ABSTRACT
During human immunodeficiency virus type 1 (HIV-1) entry into cells, the viral envelope glycoprotein (Env) trimer [(gp120/gp41)3] binds the receptors CD4 and CCR5 and fuses the viral and cell membranes. CD4 binding changes Env from a pretriggered (state-1) conformation to more open downstream conformations. BMS-378806 (here called BMS-806) blocks CD4-induced conformational changes in Env important for entry and is hypothesized to stabilize a state-1-like Env conformation, a key vaccine target. Here, we evaluated the effects of BMS-806 on the conformation of Env on the surface of cells and virus-like particles. BMS-806 strengthened the labile, noncovalent interaction of gp120 with the Env trimer, enhanced or maintained the binding of most broadly neutralizing antibodies, and decreased the binding of poorly neutralizing antibodies. Thus, in the presence of BMS-806, the cleaved Env on the surface of cells and virus-like particles exhibits an antigenic profile consistent with a state-1 conformation. We designed novel BMS-806 analogues that stabilized the Env conformation for several weeks after a single application. These long-acting BMS-806 analogues may facilitate enrichment of the metastable state-1 Env conformation for structural characterization and presentation to the immune system.
IMPORTANCE The envelope glycoprotein (Env) spike on the surface of human immunodeficiency virus type 1 (HIV-1) mediates the entry of the virus into host cells and is also the target for antibodies. During virus entry, Env needs to change shape. Env flexibility also contributes to the ability of HIV-1 to evade the host immune response; many shapes of Env raise antibodies that cannot recognize the functional Env and therefore do not block virus infection. We found that an HIV-1 entry inhibitor, BMS-806, stabilizes the functional shape of Env. We developed new variants of BMS-806 that stabilize Env in its natural state for long periods of time. The availability of such long-acting stabilizers of Env shape will allow the natural Env conformation to be characterized and tested for efficacy as a vaccine.
INTRODUCTION
The entry of human immunodeficiency virus type 1 (HIV-1) into target cells is mediated by envelope glycoprotein (Env) spikes on the viral membrane (1). HIV-1 Env trimers consist of three gp120 exterior glycoproteins and three gp41 transmembrane glycoproteins. The gp120 subunits of Env bind the target cell receptors CD4 and either CCR5 or CXCR4 (2–5). CD4 binding drives Env from its pretriggered (state-1) conformation to an obligate intermediate (state 2) and then to the full CD4-bound (state-3) conformation (6–9). In the state-3 Env, the gp41 heptad repeat (HR1) region is formed and exposed (10). As a result of receptor-induced conformational changes in Env, the hydrophobic fusion peptide at the gp41 N terminus is thought to interact with the target cell membrane (11). Binding of gp120 to the CCR5 or CXCR4 chemokine receptor leads to the formation of a highly stable gp41 six-helix bundle, in which the HR2 helix near the viral membrane binds in an antiparallel manner to the HR1 coiled coil (12–14). The favorable energy change associated with six-helix bundle formation is used to fuse the viral and target cell membranes (15).
BMS-378806 (here called BMS-806) and its analogues are potent inhibitors of HIV-1 entry (16, 17). BMS-663068 (fostemsavir), the prodrug of the potent analogue BMS-626529 (temsavir; here called BMS-529), exhibits favorable antiviral and pharmacokinetic properties and is being evaluated as an anti-HIV-1 therapy in clinical trials (18, 19). BMS-806, BMS-529, and related HIV-1 entry inhibitors bind gp120 in a hydrophobic pocket located immediately adjacent to the CD4-binding site, between the β20-β21 loop and the α1 helix (20). At concentrations in the range where potent antiviral effects are seen, BMS-806 analogues block CD4-induced conformational changes in Env that lead to the formation/exposure of the gp41 HR1 coiled coil (10, 21, 22). At higher concentrations, BMS-806 can impede CD4 binding (16–18, 20, 21, 23). However, BMS-806 inhibits CD4-independent HIV-1 infection as efficiently as CD4-dependent infection (10, 24), indicating an antiviral mechanism that does not necessarily involve CD4 (25).
During natural HIV-1 infection, antibodies are elicited to many distinct conformations of Env. The majority of these antibodies recognize Env conformations other than state 1; most of these antibodies are sterically blocked from accessing their epitopes after Env engages CD4 on the target cell and thus have little or no ability to neutralize primary HIV-1 strains (26–28). We refer to these antibodies as poorly neutralizing antibodies. In a minority of individuals infected by HIV-1 for several years, antibodies capable of neutralizing a wide range of HIV-1 strains are elicited (29–32); we refer to these antibodies as broadly neutralizing antibodies (bNAbs). Most bNAbs recognize conserved epitopes on the surface of the state-1 Env conformation (6). Broadly neutralizing antibodies have not been elicited in animals or humans immunized with HIV-1 Env preparations, including stabilized soluble Env trimers (33–37). Conformational differences between these soluble trimers and state-1 membrane Envs have been documented (9, 38) and could explain the difficulty of eliciting broadly neutralizing antibodies capable of recognizing the state-1 Env on infectious virions.
Efforts to characterize the structure of the state-1 Env conformation on the cell or viral surface and to present this conformation to the immune system might benefit from small-molecule ligands like BMS-806 that block Env transitions from state 1. Here, we evaluate the effects of BMS-806 and BMS-529 treatment on the conformation of HIV-1 Env on the surface of cells and virus-like particles. We found that these effects are consistent with stabilization of a state-1-like conformation and are reversible upon removal of the compounds from Env. We also developed novel BMS-806 analogues that are long-acting and that stabilize a state-1-like conformation of membrane Env for at least 21 days after a single application. Such long-acting, state-1-stabilizing compounds should greatly assist efforts to characterize the metastable pretriggered conformation of Env and allow its presentation to the immune system.
RESULTS
Effects of BMS-806 on conformation of cell surface Env.
The full-length HIV-1AD8 Env was inducibly expressed in A549 cells (here designated A549-Env cells). The efficiency with which Env is proteolytically processed in these cells allows an evaluation of the conformation of both the mature (cleaved) Env and the uncleaved Env. The cell surface HIV-1AD8 Env was precipitated by a panel of Env ligands in the presence of BMS-806 or the dimethyl sulfoxide (DMSO) vehicle control (Fig. 1A and B). Consistent with previous observations (39–41), recognition of the mature HIV-1AD8 gp120 glycoprotein on the cell surface Env trimer was highly correlated with the ability of gp120 ligands to neutralize HIV-1AD8 (Fig. 1A, left); in the absence of BMS-806, broadly neutralizing antibodies exhibited significantly better gp120 recognition than poorly neutralizing antibodies (P < 0.0008, two-tailed unpaired t test). This observation suggests that the mature cell surface Env conformationally resembles the functional virion Env. In the absence of BMS-806, most broadly neutralizing antibodies (bNAbs) recognized the cleaved Env. Some bNAbs (PG9, PG16, PGT145, PGT151, and 35O22) directed against quaternary Env epitopes (42–45) recognized both uncleaved and mature Envs but exhibited preferential recognition of the cleaved Env (Fig. 1B). The 2F5, 4E10, and 10E8 bNAbs against the gp41 membrane-proximal external region (MPER) (46, 47) inefficiently precipitated the gp120 glycoprotein in the absence of BMS-806, although these MPER bNAbs recognized gp41 and the uncleaved gp160 Env efficiently. Some MPER-directed bNAbs have been shown to recognize the pretriggered Env, even though they exhibit a higher affinity for Env conformations induced by CD4 binding (48, 49). Poorly neutralizing antibodies preferentially precipitated the uncleaved Env, which samples non-state-1 conformations more readily than the mature Env (50, 51).
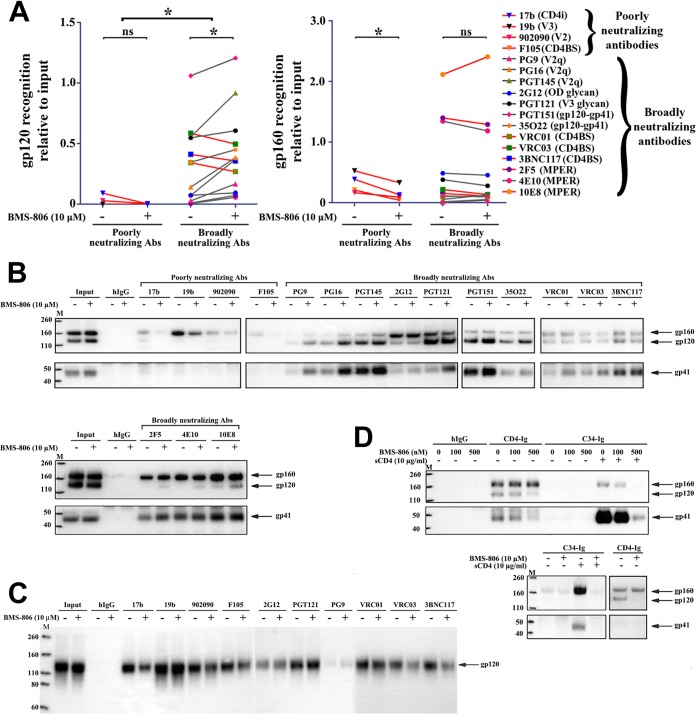
FIG 1.
Effect of BMS-806 on the conformation of cell surface Env. (A) A549-Env cells expressing HIV-1AD8 Env were incubated with the indicated antibodies in the presence of 10 μM BMS-806 (+) or the DMSO vehicle control (−). The cells were then washed and lysed, and the cell lysates were incubated with protein A-Sepharose beads. For the samples with BMS-806, the compound was also added to the cell lysates at a 10 μM concentration. Precipitates were analyzed by Western blotting with a rabbit anti-gp120 antiserum or the 4E10 anti-gp41 antibody. The precipitation of the gp120 and gp160 glycoproteins was quantified from two independent experiments like the experiment whose results are shown in panel B and is reported relative to the input level of each glycoprotein. Recognition of the indicated Env glycoprotein by poorly neutralizing antibodies and bNAbs was compared in the absence and presence of BMS-806, using a two-tailed paired Student's t test. *, P < 0.05; ns, not significant. The HIV-1 Env epitopes recognized by the antibodies are indicated in parentheses. CD4i, CD4-induced gp120 epitope; CD4BS, CD4-binding site gp120 epitope; V2q, V2 quaternary gp120 epitope; OD glycan, gp120 outer domain glycans; gp120-gp41, gp120-gp41 interface; MPER, membrane-proximal external region of gp41. (B) The blots from a representative experiment used to produce the results presented in panel A are shown. (Top) Western blotting analysis with a rabbit anti-gp120 antiserum; (bottom) Western blotting analysis with the 4E10 anti-gp41 antibody. hIgG, human immunoglobulin G. (C) Monomeric soluble HIV-1AD8 gp120 was precipitated by the indicated antibodies in the absence or presence of 10 μM BMS-806. The precipitated proteins were analyzed by Western blotting with a rabbit anti-gp120 antiserum. (D) The recognition of cell surface Env by C34-Ig was assessed in the absence or presence of sCD4 and BMS-806. In parallel, recognition of cell surface Env by CD4-Ig and by a negative control, human IgG, was studied. Lanes M, molecular mass markers. The numbers to the left are molecular masses (in kilodaltons).
BMS-806 treatment altered Env recognition by several ligands (Fig. 1A and B). BMS-806 increased the recognition of the mature Env by some bNAbs, particularly those against V2 quaternary epitopes (PG9, PG16, and PGT145) and the gp120-gp41 interface (PGT151 and 35O22). The binding of bNAbs (2G12 and PGT121) to glycan-dependent gp120 outer domain epitopes (43, 52) was not affected by incubation with BMS-806. The binding of bNAbs (VRC01, VRC03, and 3BNC117) against the gp120 CD4-binding site (CD4BS) (53, 54) was either unchanged or slightly decreased by BMS-806. BMS-806 decreased the recognition of the uncleaved Env by the poorly neutralizing antibodies (17b, 19b, 902090, and F105) (55–59).
To help interpret the observed BMS-806 effects on ligand binding to cell surface Env trimers, monomeric soluble HIV-1AD8 gp120 was precipitated by Env ligands in the absence and presence of BMS-806. With the exception of the PG9, PG16, and PGT145 bNAbs, which recognize quaternary V2 epitopes at the Env trimer apex (42, 43), and the PGT151 and 35O22 bNAbs, which recognize epitopes at the gp120-gp41 interface (44, 45), all of the Env ligands tested efficiently precipitated the monomeric gp120 glycoprotein (Fig. 1C and data not shown). The very weak binding of the PG9 antibody against a V2 quaternary gp120 epitope to monomeric gp120 was slightly increased by BMS-806 (Fig. 1C). BMS-806 mildly decreased the gp120 binding of several CD4BS antibodies, the 17b antibody against a CD4-induced (CD4i) epitope (55), and a soluble CD4-immunoglobulin fusion protein (CD4-Ig) (Fig. 1C and data not shown). These observations indicate that some of the consequences of BMS-806 on cell surface Env recognition by CD4BS and CD4i antibodies involve local effects within a gp120 subunit; this interpretation is consistent with the proximity of the BMS-806 binding site to these gp120 epitopes (20). Many of the observed effects of BMS-806 on the binding of antibodies, particularly those (PG16, PGT145, PGT151, and 35O22) recognizing epitopes dependent on Env quaternary structure (42–45), likely involve interactions that occur in the context of the Env trimer.
To summarize, in the presence of BMS-806, the mature cell surface Env maintained a high level of recognition by bNAbs and a low level of recognition by poorly neutralizing antibodies, consistent with the pattern expected for a functional state-1 Env trimer (Fig. 1A). These results corroborate single-molecule fluorescence resonance energy transfer (smFRET) data indicating that HIV-1 membrane Envs incubated with BMS-806 analogues maintain a state-1-like conformation of gp120 (6, 9) (see below). Based on diminished recognition by poorly neutralizing antibodies, the sampling of non-state-1 conformations by the uncleaved Env on the cell surface appears to be decreased in the presence of BMS-806.
BMS-806 inhibits CD4-induced conformational changes in Env and, at higher concentrations, decreases Env-CD4 binding (10, 16–18, 20–23). We evaluated the effect of BMS-806 on the recognition of the cell surface HIV-1AD8 Env by CD4-Ig and C34-Ig. CD4-Ig is a soluble CD4 (sCD4)-immunoglobulin fusion protein, and C34-Ig contains the gp41 HR2 region fused to an immunoglobulin Fc (10). C34-Ig efficiently recognized the cell surface Env only after incubation with sCD4, as expected from the dependence of gp41 HR1 formation/exposure on CD4 binding (Fig. 1D) (10). BMS-806 blocked the C34-Ig recognition of Env in the presence of sCD4. At a high concentration (10 μM), BMS-806 inhibited CD4-Ig recognition of the gp120 glycoprotein on the mature Env but not the uncleaved gp160 Env. The titration of different concentrations of BMS-806 suggested that C34-Ig binding to Env was inhibited almost completely at 500 nM; at this concentration, CD4-Ig binding to the gp120 and gp160 Envs was still detected. These results verify that BMS-806 blocks CD4-induced conformational changes in Env; at higher concentrations, BMS-806 decreases CD4 binding, more so for the cleaved Env than the uncleaved Env.
Effects of BMS-806 on the conformation of Env on VLPs.
To produce virus-like particles (VLPs), the A549-Env cells expressing HIV-1AD8 Env were transduced with a lentivirus vector encoding an HIV-1 Gag-mCherry fusion protein. The Gag-mCherry protein produces noninfectious VLPs with an immature core morphology; filtration and low-speed centrifugation of the A549-Gag/Env cell supernatants minimized the levels of extracellular vesicles in the VLP preparations (H. Ding, S. Zhang, H. Nguyen, S. Zou, J. Sodroski, and J. D. Kappes, unpublished data). Both mature and uncleaved Envs were incorporated into VLPs (see Input in Fig. 2A and C).
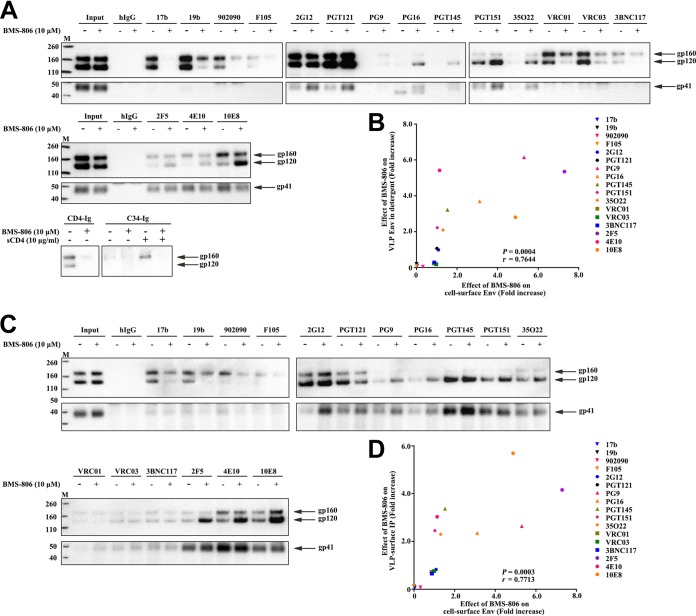
FIG 2.
Effect of BMS-806 on the conformation of VLP Env. (A) VLPs prepared from the supernatants of A549-Gag/Env cells, expressing HIV-1AD8 Env and Gag-mCherry, were solubilized in Triton X-100, and the VLP lysates were incubated with the indicated antibodies in the presence of BMS-806 (lanes +) or the DMSO vehicle control (lanes −). Precipitates were analyzed by Western blotting with a rabbit anti-gp120 antibody (top) or the 4E10 anti-gp41 antibody (bottom). (B) Correlation between the effect of BMS-806 on antibody recognition of gp120 from VLPs solubilized as described in the legend to panel A and the effect of BMS-806 on recognition of cell surface gp120 Env (as described in the legend to Fig. 1A and B). In each case, the effect of BMS-806 represents the level of gp120 precipitation by the antibody in the presence of BMS-806 divided by the level of gp120 precipitation in the absence of BMS-806. r, Pearson correlation coefficient. (C) VLPs from A549-Gag/Env cell supernatants were incubated with antibodies in the presence of BMS-806 (lanes +) or the DMSO vehicle control (lanes −) and then pelleted and washed. The repelleted VLPs were solubilized in Triton X-100, and the VLP lysates were incubated with protein A-Sepharose beads. The precipitates were analyzed by Western blotting with a rabbit anti-gp120 antibody (top) or the 4E10 anti-gp41 antibody (bottom). (D) Correlation between the effect of BMS-806 on antibody recognition of gp120 Env on intact, detergent-free VLPs (as shown in panel C) and the effect of BMS-806 on recognition of cell surface gp120 Env (as described in the legend to Fig. 1A). In each case, the effect of BMS-806 represents the level of precipitation of gp120 by the antibody in the presence of BMS-806 divided by the level of gp120 precipitation in the absence of BMS-806. IP, immunoprecipitation; r, Pearson correlation coefficient. Lanes M, molecular mass markers. The numbers to the left are molecular masses (in kilodaltons).
The recognition of the VLP HIV-1AD8 Env by a panel of Env ligands was evaluated in the presence of BMS-806 or the control, DMSO. In one assay format, the VLPs were solubilized in 1.5% Triton X-100 detergent before incubation with Env ligands (Fig. 2A and B). In a second assay format, the VLPs were preincubated with Env ligands in the absence of detergent; the VLPs were then pelleted and washed prior to solubilization in detergent and precipitation of the bound Envs with protein A-Sepharose beads (Fig. 2C and D). The results of the two assays were similar, except that Env recognition by the quaternary V2 bNAbs (PG9, PG16, and PGT145) was less efficient for the detergent-solubilized VLPs; moreover, without BMS-806 treatment, the recognition of the mature Env on the detergent-solubilized VLPs was slightly greater for the poorly neutralizing antibodies (the 19b anti-V3 antibody, the 17b antibody against a CD4i gp120 epitope, and the 902090 antibody against a linear V2 gp120 epitope). The VRC01 and VRC03 bNAbs against the CD4-binding site also precipitated the uncleaved and mature Envs more efficiently in the presence of Triton X-100. The recognition of the VLPs solubilized in 1.5% Cymal-5 detergent by the panel of antibodies was similar to that of the Triton X-100-solubilized VLPs (data not shown).
In the absence of BMS-806, all of the bNAbs recognized the mature Env on intact, detergent-free VLPs to various degrees (Fig. 2C). BMS-806 enhanced the recognition of the mature gp120 Env by bNAbs (PG9, PG16, PGT145, PGT151, and 35O22) directed against Env epitopes dependent on a quaternary conformation. In the presence of BMS-806, coprecipitation of gp120 by the bNAbs (2F5, 4E10, and 10E8) directed against the gp41 MPER was more efficient. BMS-806 reduced the recognition of the uncleaved Env by the poorly neutralizing antibodies and nearly eliminated the binding of the 19b and 17b antibodies to the mature gp120 Env. The effects of BMS-806 on antibody recognition of gp120 on both solubilized and intact VLPs directly correlated with the effects of BMS-806 on antibody recognition of cell surface gp120 Env (Fig. 2B and D). As was suggested by the analysis of cell surface Env, BMS-806 binding appears to be compatible with maintenance of a state-1-like conformation of the mature VLP-associated Env.
Effect of BMS-806 on gp120 association with Env complexes.
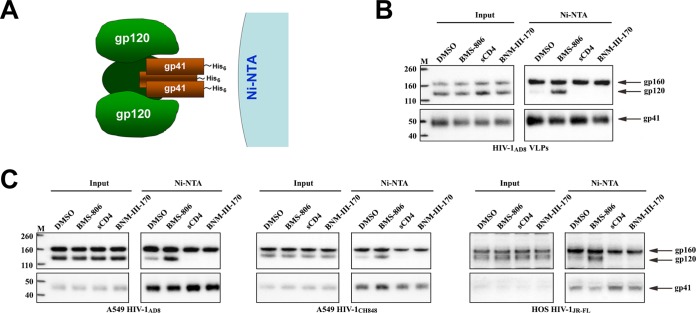
In the cell surface and VLP Env recognition assays, we noted several instances in which BMS-806 enhanced the coprecipitation of gp41 by an anti-gp120 antibody or the coprecipitation of gp120 by an anti-gp41 antibody. For example, the precipitation of cell surface Env by the PGT121, VRC01, and VRC03 bNAbs yielded more gp41 in the presence of BMS-806 than in its absence, even though the recognition of gp120 was not increased (Fig. 1B). The increased coprecipitation of gp41 by some anti-gp120 bNAbs in the cell surface Env immunoprecipitation assay was observed when BMS-806 was present throughout the procedure (as in the experiment whose results are presented in Fig. 1B) or only prior to cell lysis (data not shown). Conversely, more gp120 was precipitated by the 2F5, 4E10, and 10E8 bNAbs against the gp41 MPER in the presence of BMS-806 than in its absence, even though BMS-806 did not increase the amount of precipitated gp41 (Fig. 2A and C). These observations led us to hypothesize that BMS-806 stabilizes the noncovalent association of gp120 with the Env trimer. To test this hypothesis, we used the His6 tag on the gp41 C terminus to precipitate Env from detergent lysates of VLPs or cells expressing Env (Fig. 3A). The precipitates were then analyzed by Western blotting with antibodies against gp120 and gp41. When the DMSO control was added to the lysates of VLPs prepared from A549 cells expressing HIV-1AD8 Env and Gag-mCherry, both gp160 and gp41 were efficiently precipitated by the nickel-nitrilotriacetic acid (Ni-NTA) beads; however, little gp120 was coprecipitated with gp41 under these conditions (Fig. 3B). Addition of BMS-806 to the VLP lysates increased the amount of coprecipitated gp120. Addition of sCD4 or the small-molecule CD4-mimetic compound BNM-III-170 (60) decreased even the small amount of gp120 coprecipitated in the presence of DMSO, suggesting that these ligands induced shedding of gp120, as expected (61, 62). In assays examining Envs from different HIV-1 strains in cell lysates, BMS-806 increased the amount of coprecipitated gp120, whereas sCD4 and BNM-III-170 decreased the amount of coprecipitated gp120 (Fig. 3C). Consistent with the ability of BMS-806 to interfere with CD4 binding and CD4-induced Env conformational changes (see above), the sCD4-induced shedding of gp120 from the Env complexes could be blocked by the addition of 10 μM BMS-806 to the cell lysates (data not shown). We conclude that BMS-806 stabilizes the gp120 association with detergent-solubilized Env complexes.
FIG 3.
BMS-806 stabilization of gp120 association with Env complexes. (A) To evaluate the stability of the noncovalent association of gp120 with Env complexes, the detergent-solubilized, His6-tagged Envs were captured on Ni-NTA beads and analyzed by Western blotting with rabbit anti-gp120 antibody or the 4E10 anti-gp41 antibody. (B) VLPs were prepared from the supernatants of A549-Gag/Env cells expressing Gag-mCherry and HIV-1AD8 Env. Triton X-100 lysates of VLPs were incubated with DMSO or the indicated molecules. Env was then captured on Ni-NTA beads and analyzed by Western blotting, as described in the legend to panel A. (C) Triton X-100 lysates of A549 or HOS cells expressing Envs from different HIV-1 strains were incubated with Ni-NTA beads in the presence of the indicated molecules, and then the captured molecules were analyzed by Western blotting as described in the legend to panel A. Lanes M, molecular mass markers. The numbers to the left are molecular masses (in kilodaltons). The experiments for which the results are shown in panels B and C were performed at least twice, and the results of a representative experiment are shown.
BMS-806 analogues.
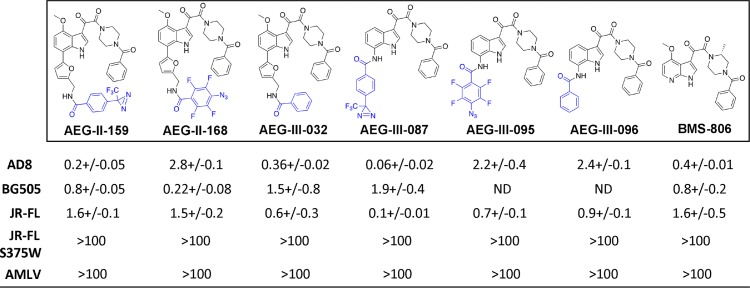
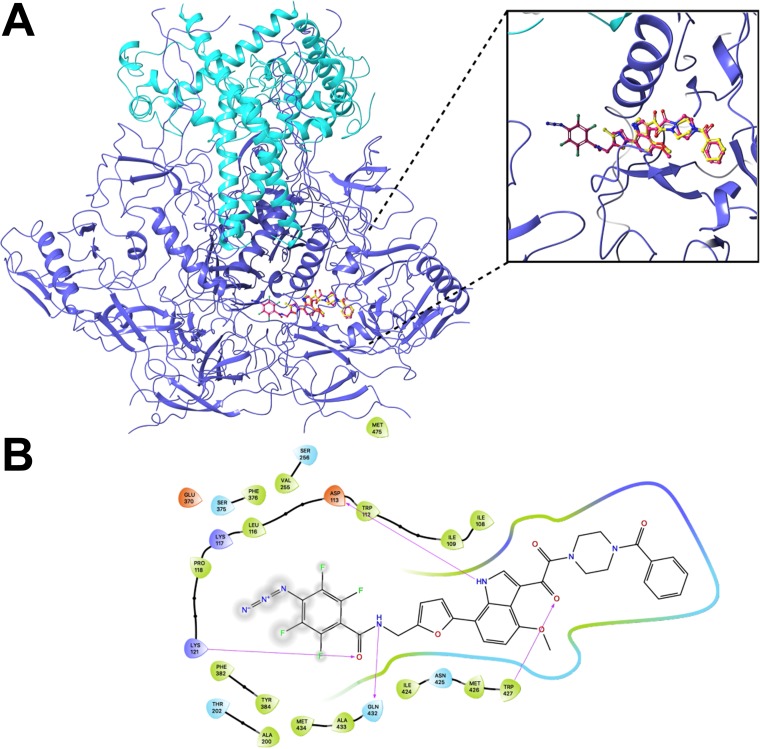
The available crystal structures of BMS-806 and BMS-529 complexed with a soluble gp140 SOSIP.664 Env trimer (PDB accession number 5U7O) indicate that the benzoyl ring of these compounds projects into a pocket on Env (20). A photoactivatable diazirine group was added to the benzoyl ring of BMS-529 to attempt to stabilize the compound-Env interaction. However, this analogue (MF463) failed to inhibit HIV-1 infection and was not evaluated further (data not shown). Instead, photoactivatable diazirine and azide groups were added to the other end of the compound (see AEG-II-159 and AEG-III-087 [with diazirine groups] and AEG-II-168 and AEG-III-095 [with azide groups] in Table 1) (see Supplemental Material S1 for details about the synthesis of these compounds.) Modeling studies suggested that these photoactivatable groups could be accommodated in complexes of the AEG compounds and soluble gp140 SOSIP.664 Env trimers (PDB accession number 5U7O) and could make additional favorable contacts. The binding energy scores for the best poses of AEG-II-159 and AEG-II-168 were more favorable than the score for the cognate docked pose of BMS-529: −13.3 and −14.5 kcal/mol for AEG-II-159 and AEG-II-168, respectively, versus −12.0 kcal/mol for BMS-529. A representative pose of AEG-II-168 is shown in Fig. 4A, and a corresponding interaction map is shown in Fig. 4B. All four AEG analogues inhibited HIV-1 infection with potencies comparable to the potency of BMS-806 (Table 1). The observed inhibition was completely abolished by the S375W change in Env, which fills the Phe 43 cavity that accommodates the benzoyl ring of the BMS-806 analogues (20, 63). Thus, the antiviral activity of AEG-II-159, AEG-II-168, AEG-III-087, and AEG-III-095 depends upon the availability of the Phe 43 cavity, a gp120 feature also required for BMS-806 and BMS-529 binding.
TABLE 1.
Inhibition of HIV-1 infection by BMS-806 analoguesa
Recombinant, luciferase-expressing single-round HIV-1 strains with the indicated HIV-1 Envs or the envelope glycoproteins of the amphotropic murine leukemia virus (AMLV) were incubated with different concentrations of the BMS-806 analogues for 30 min at 37°C. The virus-compound mixtures were then incubated with Cf2Th-CD4/CCR5 target cells for 48 h at 37°C in 5% CO2. Then, the cells were lysed and luciferase activity was measured. The 50% inhibitory concentrations (IC50; in nanomolars), presented here, were calculated from four independent experiments and are reported as means and standard errors. ND, not determined.
FIG 4.
Computational docking of BMS-806 analogues. The Maestro and Glide programs (Schrodinger) were used to dock BMS-529, AEG-II-159, and AEG-II-168 to the HIV-1BG505 soluble gp140 SOSIP.664 trimer complexed with BMS-529 (PDB accession number 5U7O) (20). Both AEG compounds docked in the existing BMS-529 pocket in the same orientation as that exhibited by BMS-529 in the crystal structure. (A) Rendering of the best docked pose of AEG-II-168 (pink sticks) with the crystallographic pose of BMS-529 (yellow sticks) superimposed. The gp120 subunits are depicted as dark blue ribbons, and the gp41 subunits are depicted as light blue ribbons. (B) Residue interaction map of the docked AEG-II-168 molecule. The gp120 residues are colored as follows: green, nonpolar; blue, polar; indigo, basic; red, acidic.
To determine if the photoactivatable azide or diazirine groups are required for the observed anti-HIV-1 activity of the AEG compounds, analogues lacking these groups (AEG-III-032 and AEG-III-096) were synthesized and tested. AEG-III-032 and AEG-III-096 inhibited HIV-1 infection specifically and had potencies comparable to the potency of BMS-806 (Table 1). Therefore, the photoactivatable azide or diazirine groups of AEG-II-159, AEG-II-168, AEG-III-087, and AEG-III-095 are not required for anti-HIV-1 activity.
Effects of AEG compounds on Env conformation.
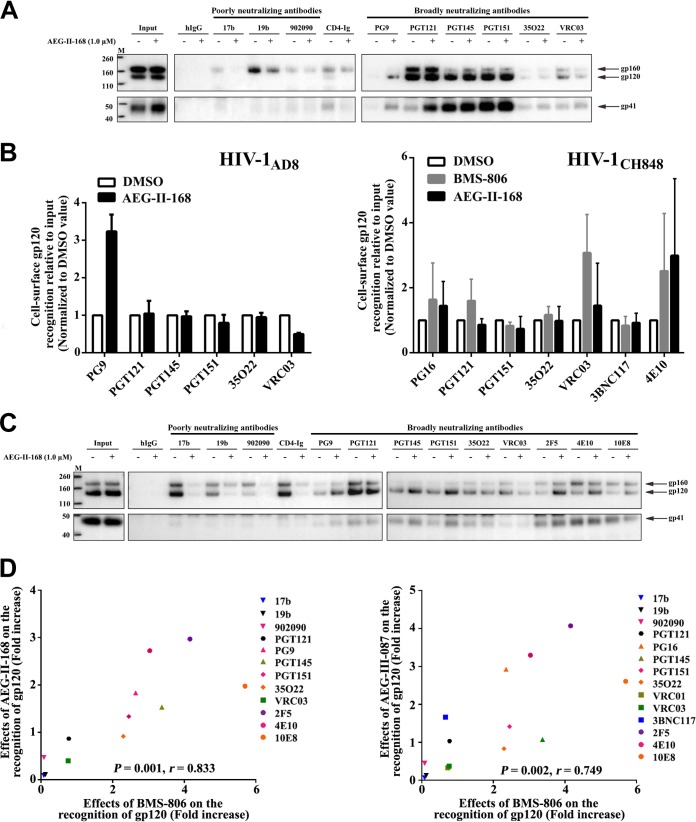
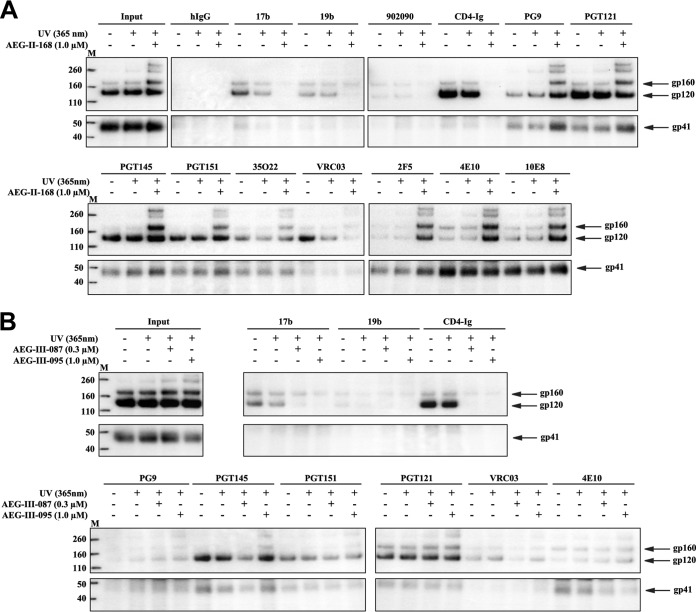
The effects of AEG-II-159, AEG-II-168, AEG-III-087, and AEG-III-095 on the HIV-1AD8 Env conformation were evaluated. The effects of the AEG compounds on the antigenic profile of the HIV-1AD8 Env on cell surfaces or VLPs were generally similar to those of BMS-806 (Fig. 5 and data not shown). Incubation with the AEG compounds resulted in decreased recognition by poorly neutralizing antibodies, whereas Env recognition by bNAbs was maintained or even increased. One exception was Env recognition by CD4BS bNAbs (VRC01, VRC03), whose binding was moderately decreased by the AEG compounds; although BMS-806 also exhibited a similar effect, the additional moieties of the AEG compounds presumably add to this competitive inhibition.
FIG 5.
Effect of AEG compounds on the conformation of cell surface and VLP Envs. (A) Recognition of cell surface HIV-1AD8 Env by the indicated antibodies in the absence or presence of AEG-II-168 was assessed as described in the Fig. 1A legend. (B) The recognition of Env on the surface of A549 cells expressing HIV-1AD8 Env (left) or HIV-1CH848 Env (right) by the indicated antibodies was assessed as described in the Fig. 1A legend. The Env-expressing cells were incubated with 10 μM BMS-806, 1 μM AEG-II-168, or the DMSO vehicle control. The precipitation of the gp120 glycoprotein of the mature cell surface Env was quantified from at least two independent experiments, and the mean values were calculated. The values shown on the y axis represent these mean values relative to the amount of input gp120, normalized to the values obtained for DMSO. The level of recognition of the mature cell surface gp120 Env by the poorly neutralizing 17b, 19b, and 902090 antibodies was below the level of detection in both the absence and the presence of AEG-II-168 (data not shown). (C) The recognition of HIV-1AD8 Env on the surface of VLPs by the indicated antibodies in the absence or the presence of AEG-II-168 was assessed as described in the Fig. 2C legend. (D) Correlation between the effect of the indicated AEG compound on antibody recognition of gp120 Env on intact, detergent-free VLPs (as described in the legend to panel C above) and the effect of BMS-806 on antibody recognition of VLPs (as described in the Fig. 2C legend). In each case, the effect of the compound represents the level of precipitation of gp120 by the antibody in the presence of the compound divided by the level of gp120 precipitation in the absence of the compound. r, Pearson correlation coefficient. Lanes M, molecular mass markers. The numbers to the left are molecular masses (in kilodaltons).
The effects of BMS-806 and AEG-II-168 on antibody recognition of the cell surface Env from the transmitted/founder (T/F) HIV-1CH848 strain were evaluated. As was seen for the HIV-1AD8 Env (Fig. 1A and B and 5B, left), the bNAbs generally maintained their binding to the mature HIV-1CH848 Env in the presence of these compounds (Fig. 5B, right). The mature HIV-1CH848 Env was recognized inefficiently by the poorly neutralizing antibodies in both the presence and the absence of these compounds (data not shown). These results are consistent with the maintenance of a pretriggered (state-1) Env conformation in the presence of BMS-806 and AEG-II-168.
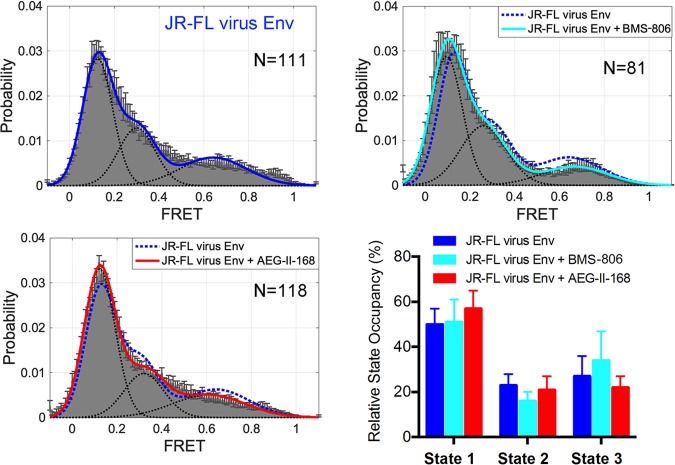
We also examined the effects of AEG-II-168 on the conformation of the HIV-1JR-FL Env on virions by smFRET. For these studies, smFRET probes were situated in the gp120 V1 and V4 variable regions, and the ratio of labeled to unlabeled Envs was kept low so that a single protomer of the Env trimer could be evaluated (6, 8, 9). In the presence of saturating concentrations of AEG-II-168, the HIV-1JR-FL virion Env maintained a state-1-dominant conformation (Fig. 6). These results are consistent with those of previous smFRET studies of the effect of BMS-806 and BMS-529 on the HIV-1 Env conformation (6, 9) and also are consistent with the above-described comparisons of Env antigenicity in the presence of the different BMS-806 analogues.
FIG 6.
Effect of AEG-II-168 on virion Env conformational states. The indicated numbers (N) of FRET traces were collected on HIV-1JR-FL Env labeled in the gp120 V1 and V4 regions in the absence and presence of a saturating concentration (100 μM) of BMS-806 or AEG-II-168. FRET histograms were compiled from the data and fitted for three Gaussian distributions centered at a low (0.1) FRET (state 1), intermediate (0.33) FRET (state 3), and high (0.65) FRET (state 2). Relative state occupancies are presented as means ± standard errors of the means.
Effect of AEG compounds on gp120 association with the Env trimer.
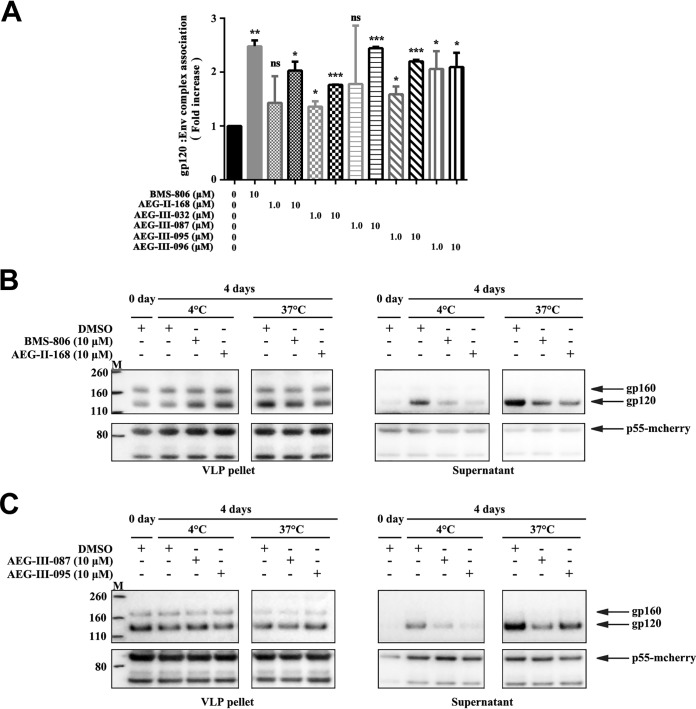
All four photoactivatable AEG compounds, as well as AEG-III-032 and AEG-III-096, lacking photoactivatable groups, stabilized the association of gp120 with detergent-solubilized HIV-1AD8 Env trimers (Fig. 7A and data not shown). AEG-II-168 also stabilized the gp120 association with HIV-1CH848 Env complexes solubilized in NP-40 lysis buffer (data not shown).
FIG 7.
Effect of BMS-806 and the AEG compounds on the gp120 association with solubilized Env complexes and with Env on VLPs. (A) Triton X-100 lysates of A549-Env cells expressing HIV-1AD8 Env were incubated with Ni-NTA beads in the presence of the indicated compounds at either a 1 or a 10 μM concentration. The precipitated Envs were analyzed by Western blotting with a rabbit anti-gp120 antibody and the 4E10 anti-gp41 antibody. The gp120 association with the Env complex was calculated as described in Materials and Methods for each compound and is shown relative to the value observed for the DMSO control. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. (B, C) VLPs with the HIV-1AD8 Env were incubated with DMSO, BMS-806, or the AEG compounds in physiological buffer for 4 days at 4°C, or 37°C. The VLPs were then pelleted and lysed; the lysed VLPs and supernatants were analyzed by Western blotting with a rabbit anti-gp120 antibody (top) and anti-Gag p55/p24/p17 antibody (bottom). Note that because the amount of gp120 shed was low compared with the amount of the VLP-associated gp120 shed, the level of gp120 in the VLP supernatants is a more accurate indicator of gp120 shedding. The results of typical experiments from two independent experiments are shown. Lanes M, molecular mass markers. The numbers to the left are molecular masses (in kilodaltons).
We evaluated the effect of BMS-806 and analogues on the spontaneous shedding of gp120 from HIV-1AD8 VLPs at 4°C and 37°C in the absence of detergent. BMS-806, AEG-II-168, AEG-III-087, and AEG-III-095 decreased the shedding of gp120 into the VLP supernatant at 4°C and 37°C (Fig. 7B and C). We conclude that these compounds stabilize the interaction of gp120 with the native membrane Env trimer under physiological conditions.
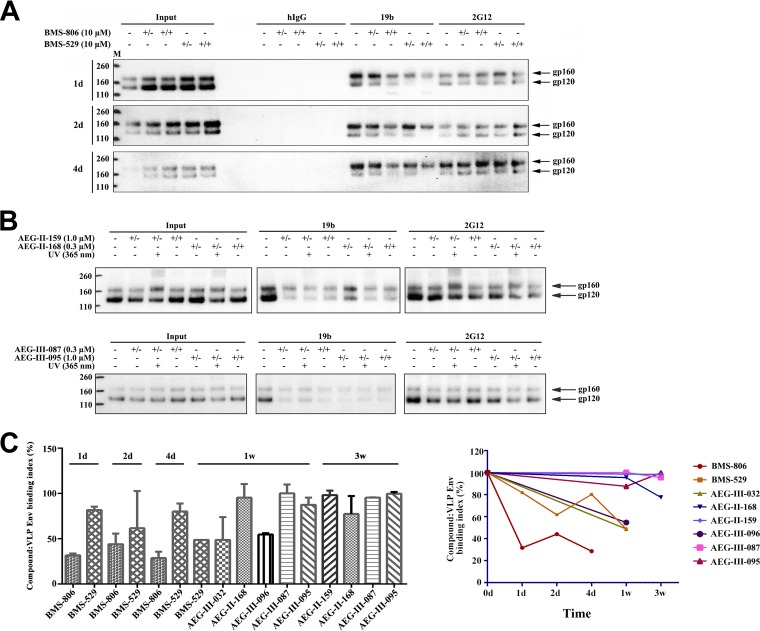
Reversibility of BMS-806 and BMS-529 binding to VLP Env.
The BMS-806-induced decrease in the binding of the 19b anti-V3 antibody can be used as an indicator of BMS-806–Env binding, allowing estimation of the stability of the Env-compound complex. BMS-806 and BMS-529 were incubated with the HIV-1AD8 Env on VLPs. The VLPs were then washed and incubated at room temperature for various lengths of time in a buffer with (+/+) or without (+/−) compound. After this incubation, the Env conformation was assessed using the 19b anti-V3 antibody, the 2G12 antibody (the binding of which is not affected by BMS-806), and a negative-control human immunoglobulin (hIgG) preparation. We compared the 19b recognition of Env in the washed samples (+/−) with that in the samples continuously incubated with the compound (+/+) and the untreated samples (−) (Fig. 8A). By 24 h, the conformational effects of BMS-806 on 19b binding were much less evident. The effects of the more potent analogue, BMS-529, were still apparent at 24 h but disappeared by 2 to 4 days. These results agree with those of a previous study suggesting that the binding of BMS-806 analogues to HIV-1 Env occurs with a low off rate (18); despite this low rate of dissociation of BMS-806 analogues from Env, the binding and the consequent effects of these compounds on Env conformation appear to be reversible.
FIG 8.
Duration of the effect of the BMS and AEG compounds on the VLP Env conformation. (A) VLPs prepared from A549-Gag/Env cells expressing HIV-1AD8 Env and Gag-mCherry were incubated with DMSO or 10 μM BMS-806 or BMS-529. The VLPs were pelleted and washed twice, resuspended in PBS with 2% DMSO or fresh compound, and incubated at room temperature for the indicated times. Then, the pelleted VLPs were lysed and incubated with antibodies and protein A-Sepharose beads. The precipitated proteins were analyzed by Western blotting with a rabbit anti-gp120 antiserum. The results are shown for the absence of the compound (−), in the continuous presence of the compound (+/+), or after initial exposure to the compound, washing, and incubation in PBS with 2% DMSO for the indicated times (+/−). (B) VLPs with the HIV-1AD8 Env were incubated with DMSO or the indicated AEG compound and, in some cases, irradiated with UV light. The VLPs were pelleted and washed twice and then resuspended in either PBS with 2% DMSO or fresh compound at room temperature for 3 weeks. The VLPs were then pelleted and lysed. The VLP lysates were directly analyzed by Western blotting (Input) or incubated with the 19b or 2G12 antibody and protein A-Sepharose beads. The precipitated proteins were analyzed by Western blotting with a rabbit anti-gp120 antiserum. Results are shown for the absence of the compound (−), in the continuous presence of the compound (+/+), or after initial exposure to the compound, washing, and incubation in PBS with 2% DMSO for 3 weeks (+/−). The results shown are typical of those obtained in two independent experiments. Lanes M, molecular mass markers. The numbers to the left are molecular masses (in kilodaltons). (C) The binding of the BMS and AEG compounds to the VLP Env is shown as a function of the time of incubation of the virus-compound complex in PBS with 2% DMSO at room temperature. The compound-VLP Env binding index was calculated as follows: compound-VLP Env binding index = [(−) – (+/−)]/[(−) – (+/+)] × 100%, where the symbols in parentheses represent the total amount of gp160 and gp120 recognized by the 19b antibody, as defined in the legend to panel B.
Long-term effects of AEG compounds on Env conformation.
We used the decrease in 19b antibody recognition to evaluate the stability of the association of the AEG compounds with HIV-1AD8 Env on VLPs. Pilot experiments established UV radiation doses that were compatible with our assay (data not shown). Surprisingly, even without UV irradiation, the effects of AEG-II-159, AEG-II-168, AEG-III-087, and AEG-III-095 on 19b recognition persisted for at least 3 weeks after washing (Fig. 8B and data not shown). The time-dependent association of the compounds with HIV-1AD8 Env on VLPs is shown in Fig. 8C. We note that the effects of AEG-III-032, which lacks a photoactivatable group, on the Env conformation were significantly reduced by 7 days following removal of the VLPs from the compound solution (Fig. 8C). Likewise, AEG-III-096, another analogue lacking a photoactivatable group, exhibited much faster reversibility than the corresponding compounds with diazirine and azide groups (AEG-III-087 and AEG-III-095, respectively) (Fig. 8C). Apparently, the durability of the effects of AEG-II-159, AEG-II-168, AEG-III-087, and AEG-III-095 on the HIV-1 Env conformation is enhanced by the presence of the photoactivatable groups but is not dependent on UV cross-linking.
Long-acting BMS-806 analogues potentially could be used to stabilize a state-1 conformation in Env immunogens. We compared the antigenic profile of HIV-1AD8 Env in VLPs 2 weeks after each of the following treatments: buffer/DMSO, UV irradiation, and AEG compounds (AEG-II-168, AEG-III-087, and AEG-III-095) plus UV irradiation. Following these treatments, the VLPs were pelleted, washed, and resuspended in buffer without compound at 4°C for 2 weeks, at which time a panel of antibodies was used to evaluate the Env conformation. All three AEG compounds lowered the level of Env recognition by poorly neutralizing antibodies and, with the exception of the CD4BS antibodies, maintained the integrity of the bNAb epitopes (Fig. 9 and data not shown). Treatment of the VLPs with AEG-II-168 and UV irradiation resulted in increased recognition by several bNAbs (PG9, PGT145, PGT151, 35O22, 2F5, 4E10, and 10E8); this increase was not seen for AEG-III-087 or AEG-III-095. UV irradiation of VLP Env in the presence of AEG-II-168 also led to an intensification of the gp160 band and the appearance of two new bands of approximately 240 and 280 kDa. We speculate that these represent gp120-gp41, gp120-gp120, and gp120-gp120-gp41 cross-linked products, respectively; the long extension (furan ring-amide linkage-tetrafluorobenzene ring) between the gp120-docking portion and the photoactivable azide of AEG-II-168 apparently allows these cross-links to form more readily than in AEG-III-087 and AEG-III-095, which have shorter spacers. Mass spectrometric analysis of UV-irradiated Env preparations incubated with AEG-II-168 failed to identify specific cross-links (data not shown). In summary, the results presented above indicate that several of the AEG compounds can exert long-term stabilizing effects on state-1-like Env conformations.
FIG 9.
Long-term effects of the AEG compounds on the HIV-1 Env conformation. (A, B) VLPs prepared from A549 cells expressing HIV-1AD8 Env were incubated with DMSO or the indicated AEG compound and, in some cases, irradiated with UV light. The pelleted VLPs were washed and then resuspended in PBS with 2% DMSO. The VLPs were incubated at room temperature for 2 weeks. The VLPs were pelleted and lysed, and the cell lysates were incubated with the indicated antibodies and protein A-Sepharose. The precipitated proteins were analyzed by Western blotting with a rabbit anti-gp120 antiserum (top) and with the 4E10 anti-gp41 antibody (bottom). The results of typical experiments of two independent experiments are shown. Lanes M, molecular mass markers. The numbers to the left are molecular masses (in kilodaltons).
DISCUSSION
In cells expressing HIV-1 Envs, both mature (cleaved) and uncleaved Envs are transported to the cell surface. The mature, functional Env, which largely resides in a state-1 conformation (6–9), is able to be recognized by most bNAbs but not by poorly neutralizing antibodies (38–41). The uncleaved Env has been suggested to assume a number of conformations and therefore binds both bNAbs and poorly neutralizing antibodies (50, 51). In general, we found that the binding of most bNAbs to the mature Env trimer is maintained or increased in the presence of BMS-806, whereas BMS-806 inhibits the binding of poorly neutralizing antibodies to the uncleaved Env. The effects of BMS-806 on the antigenicities of the cleaved cell surface Env and the cleaved VLP Env strongly correlated. The differential effects of BMS-806 on the binding of bNAbs and poorly neutralizing antibodies are consistent with the compound stabilizing a state-1-like, pretriggered conformation of the membrane Env trimer. Several smFRET studies of HIV-1 virion Env, including this study (Fig. 6), corroborate a model in which BMS-806 analogues bind and maintain a functional state-1 Env conformation (6, 9).
The gp120 exterior Env associates noncovalently with the mature Env trimer, creating the potential for the dissociation (shedding) of gp120 from the Env complex (61, 62). The shedding of gp120 occurs spontaneously but is enhanced by the binding of CD4 (61, 62); apparently, the CD4-induced opening of the Env trimer to the state-3 conformation destabilizes the association of the gp120 subunits with the membrane-anchored gp41 subunits. We found that BMS-806 and its analogues strengthened the gp120-Env association on the viral surface under physiological conditions, as well as in detergent lysates. The BMS-806-induced enrichment of a pretriggered, state-1-like Env conformation and the consequent decrease of open state-3 Env conformations likely relate to the observed stabilization of the gp120-trimer association. This assertion is supported by the observed BMS-806-induced increases in the binding of several bNAbs to quaternary epitopes that span the subunits of the state-1 Env trimer. As one example, BMS-806 enhanced the binding of the V2 quaternary bNAbs (PG9, PG16, PGT145), which recognize carbohydrate-dependent epitopes at the membrane-distal apex of the Env trimer formed by interactions of the gp120 protomers (42, 43, 64). HIV-1 mutants with Env changes that destabilize the state-1 conformation exhibit decreased sensitivity to the V2 quaternary antibodies, suggesting that these antibodies preferentially, although not exclusively, recognize a state-1 Env conformation (7, 65). BMS-806 induced smaller enhancements of the binding of the PGT151 and 35O22 bNAbs, which recognize quaternary structures spanning the gp120-gp41 interface (44, 45).
What is the mechanism by which the binding of a small molecule like BMS-806 stabilizes a state-1-like Env trimer structure? Recent data suggest that current high-resolution structures of HIV-1 Env trimers more closely represent state 2 than state 1 (9). In the absence of a detailed state-1 Env trimer structure, it is difficult to address the question asked above with precision. Nonetheless, the crystal structures of BMS-806 and its analogues have been solved in complexes with soluble gp140 SOSIP.664 trimers (20), which are in a state-2-like conformation (9). These observed BMS-806 and BMS-529 binding sites are consistent with data on HIV-1 escape mutants and are likely relevant to functional Envs (16, 20, 23, 66). In these structures, BMS-806 and its analogues bind in a hydrophobic pocket between the gp120 inner domain α1 helix and the β20-β21 loop (20). This pocket is adjacent to both the CD4-binding site and the trimer association domain, which governs the quaternary interactions among gp120 protomers. Thus, BMS-806 may be well-positioned to influence the adjacent trimer association domain, which includes the V1/V2 and V3 variable regions. The trimer association domains of gp120 are thought to open as state 1 makes transitions to downstream conformations, either in a spontaneous manner or as a result of CD4 binding (6–9, 63, 67, 68). BMS-806 may limit the flexibility of the trimer association domain, predisposing Env to maintain a state-1-like gp120 conformation. Such a model is consistent with our observation that BMS-806 enhances Env recognition by V2 quaternary antibodies. Indeed, some BMS-806-like HIV-1 entry inhibitors have been shown to impede the decrease in Env recognition by V2 quaternary antibodies that accompanies CD4 binding (22). Although BMS-806 did not exhibit this activity, BMS-806 can apparently decrease the spontaneous sampling of more open state-2/3 Env conformations (6, 9, 22); the blockade of spontaneous transitions is presumably more readily achieved by a small molecule than the blockade of the changes in the gp120 trimer association domain induced by a large protein like CD4.
A more universal activity observed for the BMS-806 class of HIV-1 entry inhibitors is the blockade of formation/exposure of the gp41 HR1 coiled coil (10, 21, 22). As the formation/exposure of the HR1 coiled coil is a natural consequence of CD4 binding to gp120 (10), conformational changes in gp120 that are impeded by BMS-806 may be prerequisites for these gp41 HR1 transitions. BMS-806 may also more directly influence gp41 in the as-yet-uncharacterized state-1 conformation. We observed that BMS-806 inhibited gp41 HR1 formation/exposure, as measured by C34-Ig binding, in response to sCD4 in different contexts, even in detergent lysates of VLPs.
Our assays measuring the reversibility of the Env conformational effects of the BMS-806 analogues indicate a remarkably low off rate of these compounds. Even the parental compounds, BMS-806 and BMS-529, as well as the control compounds, AEG-III-032 and AEG-III-096, which lack photoactivatable groups, required 1 to 4 days to reverse their conformational effects on Env. A previous study noted the low off rate of BMS-529 (18). Insertion of these compounds into a conserved, hydrophobic interdomain interface on Env may impose unfavorable enthalpic and entropic penalties on their extraction from Env and resolubilization. The addition of photoactivatable groups in AEG-II-159 and AEG-II-168 significantly decreased their reversibility, with maintenance of Env conformational effects being seen for at least 3 weeks. Surprisingly, even without UV irradiation and even when exposure to visible light was minimized, this prolongation of the Env conformational effect was observed. The antigenic profile of Env was not apparently altered by UV irradiation, leaving open the possibility of UV cross-linking of these or other BMS-806 analogues in the future.
The availability of BMS analogues that can maintain state-1-like Env conformations for prolonged time periods may assist presentation of this conformation to the immune system. Given that state 1 is targeted by many bNAbs and is intrinsically metastable, limiting the conformations of Env immunogens to state 1 may facilitate the elicitation of bNAbs. The context in which Env immunogens are formulated could influence the success of this approach; for example, BMS analogues enrich state-1-like conformations in membrane-anchored Envs better than in soluble Env trimers (9; M. Lu, X. Ma, N. Reichard, D. S. Terry, J. Arthos, A. B. Smith III, J. G. Sodroski, S. C. Blanchard, and W. Mothes, submitted for publication). The influence of the BMS analogues on the immunogenicity of bNAb epitopes near their binding site will need to be determined empirically. BMS analogues with prolonged activity may also assist structural studies of state-1-like conformations, which could benefit from the availability of Env trimer preparations that are enriched in these pretriggered conformations.
MATERIALS AND METHODS
BMS-806 and BMS-529.
BMS-378806 (here called BMS-806) and BMS-626529 (temsavir; here called BMS-529) were purchased from Selleckchem and APExBIO, respectively.
Synthetic chemistry.
The synthesis of the AEG compounds is described in Supplemental Material S1.
Envelope glycoprotein constructs.
The HIV-1JR-FL Env and HIV-1AD8 mutant Envs were coexpressed in HOS cells with the Rev protein by the pSVIIIenv expression vector, using the natural HIV-1 env and rev sequences. The plasmids for the expression of HIV-1 virions with the HIV-1JR-FL Env used for single-molecule FRET have been previously described (6). The wild-type HIV-1AD8 and HIV-1CH848 Envs were expressed in A549 cells using a lentivirus vector, as described below.
Antibodies.
Antibodies against HIV-1 Env were kindly supplied by Dennis Burton (Scripps), Peter Kwong and John Mascola (Vaccine Research Center, NIH), Barton Haynes (Duke University), Michel Nussenzweig (Rockefeller University), Hermann Katinger (Polymun), James Robinson (Tulane University), and Marshall Posner (Mount Sinai Medical Center). In some cases, anti-Env antibodies were obtained through the NIH AIDS Reagent Program. Antibodies for Western blotting included goat anti-gp120 polyclonal antibody (Thermo Fisher), the 4E10 anti-gp41 antibody (Polymun), and anti-Gag p55/p24/p17 (Abcam). A horseradish peroxidase (HRP)-conjugated rabbit anti-goat IgG antibody (Thermo Fisher) or an HRP-conjugated goat anti-human IgG antibody (Santa Cruz) was used as the secondary antibody for Western blotting.
Cell lines.
293T cells (ATCC) were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 μg/ml of penicillin-streptomycin. Cf2Th-CD4/CCR5 cells stably expressing the human CD4 and CCR5 coreceptors for HIV-1 were grown in the same medium supplemented with 0.4 mg/ml of G418 and 0.2 mg/ml of hygromycin. HOS cells (ATCC) were grown in DMEM with 5% FBS and 100 μg/ml of penicillin-streptomycin. A549 lung epithelial cells (ATCC) were grown in DMEM–Ham’s F-12 medium with 10% FBS supplemented with l-glutamine and penicillin-streptomycin. All cell culture reagents were from Life Technologies.
Human A549 cells inducibly expressing Env or Env on virus-like particles (VLPs) were established. A549 cells constitutively expressing the reverse Tet transactivator were transduced with an HIV-1-based lentivirus vector expressing Rev and Env from HIV-1AD8, a primary HIV-1 strain. The vector transcribes a bicistronic mRNA comprising HIV-1AD8 rev and env and two selectable marker genes (puromycin-T2A-enhanced green fluorescent protein [EGFP]) fused in-frame with a T2A peptide-coding sequence. In the transduced cells, Env expression is controlled by the Tet-responsive element (TRE) promoter and Tet-on transcriptional regulatory elements. A similar strategy was used to express Rev and the transmitted/founder HIV-1CH848 Env in A549 cells. Env-expressing cells were enriched by doxycycline induction and fluorescence-activated cell sorting for the coexpressed EGFP marker. At approximately 72 h after treatment of these cells with 2-μg/ml doxycycline, the HIV-1AD8 and HIV-1CH848 gp160 Env precursor and the mature gp120 and gp41 glycoproteins were expressed. Here, we designate these cells A549-Env.
To produce cells expressing Env and VLPs, the A549-Env cells expressing the HIV-1AD8 Env were transduced with a lentivirus vector expressing the HIV-1 Gag precursor fused with mCherry. The doxycycline-regulated expression of the Gag-mCherry fusion protein resulted in the release of Env-containing VLPs into the medium. Here, we designate these cells A549-Gag/Env.
Immunoprecipitation of cell surface Env.
Doxycycline-induced A549-Env cells were washed twice with washing buffer (1× phosphate-buffered saline [PBS] plus 5% FBS) with or without 10 μM BMS-806 or an analogue. The cells were then incubated with 5-μg/ml antibody for 1 h at 4°C in the continued presence or absence of the BMS-806 analogue. After washing four times in washing buffer, the cells were lysed in NP-40 lysis buffer (0.5% NP-40, 0.5 M NaCl, 10 mM Tris, pH 7.5) for 5 min at 4°C with gentle agitation. For the samples with a BMS-806 analogue, the compound was added to the NP-40 lysis buffer at a 10 μM concentration. The lysates were cleared by centrifugation at 13,200 × g for 20 min at 4°C, and the clarified supernatants were incubated with protein A-Sepharose beads (50 μl of 25 mg/ml in PBS per sample) for 1 h at room temperature. The beads were pelleted (1,000 rpm for 1 min) and washed three times with final wash buffer [200 mM Tris-HCl, pH 8.0, 100 mM (NH4)2SO4, 1 M NaCl, 0.1% NP-40]. The beads were suspended in lithium dodecyl sulfate (LDS) sample buffer, boiled, and analyzed by Western blotting using 1:2,000-diluted goat anti-gp120 polyclonal antibody (Thermo Fisher) and 1:2,000-diluted HRP-conjugated rabbit anti-goat IgG (Thermo Fisher). The transmembrane Env was analyzed by Western blotting with the 4E10 anti-gp41 antibody and HRP-conjugated goat anti-human IgG (Santa Cruz).
For analysis of total Env expression in the cell, some of the clarified lysates were saved before the addition of protein A-Sepharose beads and analyzed by Western blotting as described above (these are referred to as the “input” samples). Detection of the antibody heavy and light chains in the gp41 Western blots provides an indication of the amount of antibody added, captured, and loaded in each experiment.
Antibody recognition of monomeric gp120.
To produce gp120 monomers, a stop codon was introduced into the HIV-1AD8 env gene sequence encoding the gp120-gp41 junction. Transfection of 293F cells with this plasmid DNA resulted in the transient expression of a secreted, monomeric gp120 glycoprotein into the cell supernatants. The supernatants were clarified by low-speed centrifugation and filtration through a 0.45-μm-pore-size filter and then used for precipitation by antibodies in the absence or presence of 10 μM BMS-806. The precipitates were analyzed by Western blotting with a goat anti-gp120 polyclonal antibody (Thermo Fisher) and 1:3,000-diluted HRP-conjugated rabbit anti-goat IgG antibody (Thermo Fisher).
Characterization of VLP-associated Env.
To prepare VLPs with HIV-1AD8 Env, 150-mm dishes of 30 to 40% confluent A549-Gag/Env cells were seeded and, on the following day, treated with 2 μg/ml doxycycline. At approximately 72 h after induction, cell supernatants were harvested and cleared by low-speed centrifugation (500 × g for 15 min at 4°C) and filtration through a 0.45-μm-pore-size filter. VLPs were pelleted by centrifugation at 100,000 × g for 1 h at 4°C. The resuspended VLP preparation was clarified by low-speed centrifugation. BMS-806 and its analogues were added to the clarified VLP-containing suspensions. In some cases, the VLP-compound mixtures were irradiated with a 100-watt, 365-nm UV lamp for 10 min at room temperature. For all UV studies, exposure of the samples to visible light was minimized.
For studies of the reversibility of BMS-806 analogue effects, the compounds were added to the VLPs and mixed for several seconds at room temperature. Then, the VLPs were pelleted (20,000 × g for 30 min at 4°C) twice and washed with 1.5 ml PBS with 2% DMSO. The pelleted VLPs were resuspended in 1 ml PBS with 2% DMSO and incubated at room temperature for various lengths of time (20 min to 3 weeks). Control VLPs were incubated in the continued presence of the BMS-806 analogues at room temperature for the same length of time. Then, the VLPs were pelleted (20,000 × g for 30 min at 4°C), and the pellet was incubated with 1.5% Triton X-100 for 30 min at 4°C. The VLP lysates were centrifuged (20,000 × g for 30 min at 4°C), and the supernatants were incubated with antibodies (10 μg/ml) in a 50-μl volume with 25 mg/ml of protein A-Sepharose beads for 1 h at 4°C. The beads were pelleted (1,000 rpm for 1 min at room temperature) and washed three times with 1 ml wash buffer with 1% Triton X-100. The beads were suspended in NuPAGE LDS sample buffer (Thermo Fisher), boiled, and analyzed by Western blotting, as described above.
For studies of the binding of ligands (antibodies, CD4-Ig, and C34-Ig) to detergent-solubilized VLP Env, VLPs prepared as described above were pelleted (20,000 × g for 30 min at 4°C). The pellet was incubated with 1.5% Triton X-100 for 30 min at 4°C. The VLP lysates were centrifuged (at 20,000 × g for 30 min at 4°C), and the supernatants were incubated with ligands (10 μg/ml) in a 50-μl volume with 25 mg/ml of protein A-Sepharose beads for 1 h at 4°C. The beads were pelleted (1,000 rpm for 1 min at room temperature), washed three times with 1 ml wash buffer with 1% Triton X-100, suspended in LDS sample buffer, and analyzed by Western blotting, as described above.
In a second assay format, we examined the effect of the BMS-806 analogues on the binding of antibodies to Env on intact VLPs in the absence of detergent. In this case, VLPs in A549-Gag/Env supernatants, clarified as described above, were pelleted by centrifugation at 100,000 × g for 1 h at 4°C. The pellet was resuspended in PBS, and the VLPs were pelleted by centrifugation at 20,000 × g for 30 min at 4°C. The VLPs were resuspended in PBS and incubated with antibodies (10 μg/ml) in a 100-μl volume for 1 h at 4°C. The VLPs were then pelleted (20,000 × g for 30 min at 4°C) and washed with PBS three times. The VLP pellet was then solubilized in 1.5% Triton X-100 for 30 min at 4°C, after which the VLP lysates were clarified by centrifugation at 20,000 × g for 30 min at 4°C. The supernatants were incubated with 50 μl of protein A-Sepharose beads for 1 h at 4°C. The beads were pelleted (1,000 rpm for 1 min at room temperature), washed three times with 1 ml wash buffer with 1% Triton X-100, suspended in LDS sample buffer, and analyzed by Western blotting, as described above.
Association of gp120 with Env complexes.
The noncovalent association of gp120 with HIV-1 Env complexes was studied using carboxy-terminally His6-tagged Envs from three different sources: (i) VLPs produced from A549-Gag/Env cells, (ii) A549 cells expressing HIV-1AD8 and HIV-1CH848 Envs, and (iii) HOS cells transiently expressing wild-type HIV-1JR-FL Env. VLPs and cell lysates from A549 cells were prepared in 1.5% Triton X-100. Cell lysates from HOS cells were prepared in 1.5% Cymal-5 (Anatrace). The VLP and cell lysates were clarified as described above. DMSO, a BMS-806 analogue, sCD4, or the CD4-mimetic compound BNM-III-170 (60) was added to the lysate. Aliquots of the lysates were saved for Western blotting to detect the gp160, gp120, and gp41 glycoproteins in the input sample. The bulk of the lysates was incubated with nickel-nitriloacetic acid (Ni-NTA) beads (Qiagen) for 1.5 h at 4°C. The beads were pelleted (1,000 rpm for 1 min at room temperature), washed 3 times at room temperature with wash buffer with 1% Triton X-100, boiled in LDS sample buffer, and analyzed by Western blotting as described above. The association of gp120 with the Env complex was calculated as follows: [(gp120/gp160)compound × (input gp120/input gp160)DMSO]/[(gp120/gp160)DMSO × (input gp120/input gp160)compound], where (gp120/gp160)compound is the ratio of gp120 to gp160 precipitated in the presence of the compound, (input gp120/input gp160)DMSO is the ratio of gp120 to gp160 in the input sample prepared in the presence of DMSO, (gp120/gp160)DMSO is the ratio of gp120 to gp160 precipitated in the presence of DMSO, and input gp120/input gp160)compound is the ratio of gp120 to gp160 in the input sample prepared in the presence of the compound.
Shedding of gp120 from VLP Env.
The effect of the BMS-806 analogues on the spontaneous shedding of gp120 from VLP Env was evaluated. VLPs with the wild-type HIV-1AD8 Env were prepared from the supernatants of A549-Gag/Env cells, as described above. The VLPs were suspended in PBS, to which was added either DMSO or a BMS-806 analogue. An aliquot of the VLP suspension in PBS-DMSO was processed as described below to serve as a day 0 control. The VLP suspensions were incubated for 4 days at 4°C or 37°C (with gentle rocking). Then, the VLPs were pelleted (100,000 × g for 30 min at 4°C). The pellets and supernatants were boiled in LDS sample buffer and analyzed by Western blotting either with 1:2,000-diluted goat anti-gp120 polyclonal antibody (Thermo Fisher) and HRP-conjugated rabbit anti-goat IgG antibody or with 1:5,000-diluted rabbit anti-Gag p55/p24/p17 antibody (Abcam) and HRP-conjugated goat anti-rabbit IgG antibody (Abcam).
Infection of single-round recombinant viruses.
To produce single-round HIV-1 expressing luciferase, 293T human embryonic kidney cells were cotransfected, using the Effectene transfection reagent (Qiagen), with plasmids expressing the pCMVΔP1Δenv HIV-1 Gag-Pol packaging construct, the HIV-1 envelope glycoproteins or the envelope glycoprotein of the control amphotropic murine leukemia virus (AMLV), and the firefly luciferase-expressing vector at a DNA ratio of 1:1:3 μg (66). The plasmids expressing the HIV-1 envelope glycoproteins and Rev protein were based on pSVIIIenv or pcDNA3.1 (Invitrogen Life Technologies, Carlsbad, CA). Cotransfection produced recombinant, luciferase-expressing viruses capable of a single round of infection. The virus-containing supernatants were harvested at between 36 and 40 h after transfection and cleared of debris by low-speed centrifugation. Aliquots of the virus preparations were frozen at −80°C until further use. The reverse transcriptase (RT) levels of all virus stocks were measured.
Cf2Th-CD4/CCR5 target cells were seeded at a density of 6 × 103 cells/well in 96-well luminometer-compatible tissue culture plates (PerkinElmer) at 24 h before infection. On the day of infection, BMS-806 analogues (0 to 100 nM) were incubated with recombinant viruses (10,000 RT units) at 37°C for 30 min. The virus-compound mixtures were added to the target cells and incubated for 48 h at 37°C. After this time, the medium was removed from each well and the cells were lysed by the addition of 30 μl passive lysis buffer (Promega) and three freeze-thaw cycles. An EG&G Berthold LB 96 V microplate luminometer was used to measure the luciferase activity in each well after the addition of 100 μl of luciferin buffer (15 mM MgSO4, 15 mM KPO4, pH 7.8, 1 mM ATP, 1 mM dithiothreitol) and 50 μl of 1 mM 99% firefly d-luciferin free acid (Prolume).
smFRET.
Viruses with HIV-1JR-FL Env that were double tagged in gp120 V1 (with the Q3 peptide) and V4 (with the A1 peptide) were prepared for single-molecule fluorescence resonance energy transfer (smFRET) imaging, as previously described (6). The high (40:1) ratio of wild-type Env to tagged Env ensures that, on average, only one tagged protomer is available for imaging on a single virus particle. The Q3 and A1 double-tagged viruses allowed the incorporation of Cy3B and Cy5 fluorescent labels, respectively. Thus, the relative movements of the V1 and V4 regions in one gp120 subunit of an individual Env trimer could be monitored in real time. The smFRET images were acquired on an in-house-built total internal reflection fluorescence microscope as described previously (6). HIV-1JR-FL incubated with saturating concentrations (100 μM) of AEG-II-168 or control HIV-1JR-FL not incubated with compound was used for smFRET imaging. Data were analyzed as described previously with the customized MATLAB (MathWorks) program SPARTAN (69). FRET trajectories meeting the criteria of quality (a sufficient signal/noise ratio, single-dye photobleaching, anticorrelated features between donor and acceptor intensity, and fluorescence lifetime) were compiled into FRET histograms. Hidden Markov modeling was used to fit the FRET histograms with the sum of three Gaussian distributions. The three-state model yielded the lowest log likelihood value in this case.
Modeling BMS-806 analogue interaction with Env.
The HIV-1BG505 soluble gp140 SOSIP.664 trimer complexed with BMS-529 (PDB accession number 5U7O) (20) was used as a docking target and prepared using the Maestro program (version 12.0.012; Schrodinger, 2019). The 20 best-ranked poses for BMS-529, AEG-II-168, and AEG-II-159 were determined with the Glide program (Schrodinger).
Statistical analysis.
Statistical analyses were performed using GraphPad Prism software (version 6; Graph Pad Software).
The Kolmogorov-Smirnov test was used to check the data distribution in Fig. 1. A two-tailed paired Student's t test or Wilcoxon matched-pairs signed-rank test was used to compare two groups and to determine if the data fitted a normal distribution, respectively.
Pearson’s correlation was used to analyze the correlation between groups in Fig. 2.
In all cases, we used a P value of less than 0.05 as a cutoff for statistical significance.
Data availability.
The customized MATLAB (MathWorks) program SPARTAN for smFRET analysis is publicly available at https://www.scottcblanchardlab.com/software.
Supplementary Material
ACKNOWLEDGMENTS
We thank Elizabeth Carpelan for manuscript preparation. Antibodies against HIV-1 Env were kindly supplied by Dennis Burton (Scripps), Peter Kwong and John Mascola (Vaccine Research Center, NIH), Barton Haynes (Duke University), Hermann Katinger (Polymun), James Robinson (Tulane University), and Marshall Posner (Mount Sinai Medical Center).
This work was supported by grants from the National Institutes of Health (grants AI100645, AI124982, AI145547, and GM56550/AI150471) and by a gift from the late William F. McCarty-Cooper.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 2.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 3.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 4.Feng Y, Broder CC, Kennedy PE, Berger EA. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 5.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 6.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB III, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herschhorn A, Ma X, Gu C, Ventura JD, Castillo-Menendez L, Melillo B, Terry DS, Smith AB III, Blanchard SC, Munro JB, Mothes W, Finzi A, Sodroski J. 2016. Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. mBio 7:e01598-16. doi: 10.1128/mBio.01598-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma X, Lu M, Gorman J, Terry DS, Hong X, Zhou Z, Zhao H, Altman RB, Arthos J, Blanchard SC, Kwong PD, Munro JB, Mothes W. 2018. HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. Elife 7:e34271. doi: 10.7554/eLife.34271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu M, Ma X, Castillo-Menendez LR, Gorman J, Alsahafi N, Ermel U, Terry DS, Chambers M, Peng D, Zhang B, Zhou T, Reichard N, Wang K, Grover JR, Carman BP, Gardner MR, Nikic-Spiegel I, Sugawara A, Arthos J, Lemke EA, Smith AB III, Farzan M, Abrams C, Munro JB, McDermott AB, Finzi A, Kwong PD, Blanchard SC, Sodroski JG, Mothes W. 2019. Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568:415–419. doi: 10.1038/s41586-019-1101-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Si Z, Madani N, Cox JM, Chruma JJ, Klein JC, Schon A, Phan N, Wang L, Biorn AC, Cocklin S, Chaiken I, Freire E, Smith AB III, Sodroski JG. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc Natl Acad Sci U S A 101:5036–5041. doi: 10.1073/pnas.0307953101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kowalski M, Potz J, Basiripour L, Dorfman T, Goh WC, Terwilliger E, Dayton A, Rosen C, Haseltine W, Sodroski J. 1987. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237:1351–1355. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 12.Chan DC, Fass D, Berger JM, Kim PS. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 13.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 14.Tan K, Liu J, Wang J, Shen S, Lu M. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci U S A 94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol 151:413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin PF, Blair W, Wang T, Spicer T, Guo Q, Zhou N, Gong YF, Wang HG, Rose R, Yamanaka G, Robinson B, Li CB, Fridell R, Deminie C, Demers G, Yang Z, Zadjura L, Meanwell N, Colonno R. 2003. A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc Natl Acad Sci U S A 100:11013–11018. doi: 10.1073/pnas.1832214100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T, Zhang Z, Wallace OB, Deshpande M, Fang H, Yang Z, Zadjura LM, Tweedie DL, Huang S, Zhao F, Ranadive S, Robinson BS, Gong YF, Ricarrdi K, Spicer TP, Deminie C, Rose R, Wang HG, Blair WS, Shi PY, Lin PF, Colonno RJ, Meanwell NA. 2003. Discovery of 4-benzoyl-1-[(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)oxoacetyl]-2-(R)-methylpiperazine (BMS-378806): a novel HIV-1 attachment inhibitor that interferes with CD4-gp120 interactions. J Med Chem 46:4236–4239. doi: 10.1021/jm034082o. [DOI] [PubMed] [Google Scholar]
- 18.Nowicka-Sans B, Gong YF, McAuliffe B, Dicker I, Ho HT, Zhou N, Eggers B, Lin PF, Ray N, Wind-Rotolo M, Zhu L, Majumdar A, Stock D, Lataillade M, Hanna GJ, Matiskella JD, Ueda Y, Wang T, Kadow JF, Meanwell NA, Krystal M. 2012. In vitro antiviral characteristics of HIV-1 attachment inhibitor BMS-626529, the active component of the prodrug BMS-663068. Antimicrob Agents Chemother 56:3498–3507. doi: 10.1128/AAC.00426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lalezari JP, Latiff GH, Brinson C, Echevarria J, Trevino-Perez S, Bogner JR, Thompson M, Fourie J, Sussmann Pena OA, Mendo Urbina FC, Martins M, Diaconescu IG, Stock DA, Joshi SR, Hanna GJ, Lataillade M, AI438011 Study Team. 2015. Safety and efficacy of the HIV-1 attachment inhibitor prodrug BMS-663068 in treatment-experienced individuals: 24 week results of AI438011, a phase 2b, randomised controlled trial. Lancet HIV 2:e427–e437. doi: 10.1016/S2352-3018(15)00177-0. [DOI] [PubMed] [Google Scholar]
- 20.Pancera M, Lai Y-T, Bylund T, Druz A, Narpala S, O'Dell S, Schön A, Bailer RT, Chuang G-Y, Geng H, Louder MK, Rawi R, Soumana DI, Finzi A, Herschhorn A, Madani N, Sodroski J, Freire E, Langley DR, Mascola JR, McDermott AB, Kwong PD. 2017. Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529. Nat Chem Biol 13:1115–1122. doi: 10.1038/nchembio.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho HT, Fan L, Nowicka-Sans B, McAuliffe B, Li CB, Yamanaka G, Zhou N, Fang H, Dicker I, Dalterio R, Gong YF, Wang T, Yin Z, Ueda Y, Matiskella J, Kadow J, Clapham P, Robinson J, Colonno R, Lin PF. 2006. Envelope conformational changes induced by human immunodeficiency virus type 1 attachment inhibitors prevent CD4 binding and downstream entry events. J Virol 80:4017–4025. doi: 10.1128/JVI.80.8.4017-4025.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herschhorn A, Gu C, Espy N, Richard J, Finzi A, Sodroski JG. 2014. A broad HIV-1 inhibitor blocks envelope glycoprotein transitions critical for entry. Nat Chem Biol 10:845–852. doi: 10.1038/nchembio.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Q, Ho HT, Dicker I, Fan L, Zhou N, Friborg J, Wang T, McAuliffe BV, Wang HG, Rose RE, Fang H, Scarnati HT, Langley DR, Meanwell NA, Abraham R, Colonno RJ, Lin PF. 2003. Biochemical and genetic characterizations of a novel human immunodeficiency virus type 1 inhibitor that blocks gp120-CD4 interactions. J Virol 77:10528–10536. doi: 10.1128/jvi.77.19.10528-10536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Zhou N, Sun Y, Ray N, Lataillade M, Hanna GJ, Krystal M. 2013. Activity of the HIV-1 attachment inhibitor BMS-626529, the active component of the prodrug BMS-663068, against CD4-independent viruses and HIV-1 envelopes resistant to other entry inhibitors. Antimicrob Agents Chemother 57:4172–4180. doi: 10.1128/AAC.00513-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langley DR, Kimura SR, Sivaprakasam P, Zhou N, Dicker I, McAuliffe B, Wang T, Kadow JF, Meanwell NA, Krystal M. 2015. Homology models of the HIV-1 attachment inhibitor BMS-626529 bound to gp120 suggest a unique mechanism of action. Proteins 83:331–350. doi: 10.1002/prot.24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 27.Moore PL, Ranchobe N, Lambson BE, Gray ES, Cave E, Abrahams MR, Bandawe G, Mlisana K, Abdool Karim SS, Williamson C, Morris L, CAPRISA 002 Study, NIAID Center for HIV/AIDS Vaccine Immunology (CHAVI). 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog 5:e1000598. doi: 10.1371/journal.ppat.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol 77:10557–10565. doi: 10.1128/jvi.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, Morris L, CAPRISA002 Study Team. 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol 85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray ES, Taylor N, Wycuff D, Moore PL, Tomaras GD, Wibmer CK, Puren A, DeCamp A, Gilbert PB, Wood B, Montefiori DC, Binley JM, Shaw GM, Haynes BF, Mascola JR, Morris L. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J Virol 83:8925–8937. doi: 10.1128/JVI.00758-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. 2014. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28:163–169. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. 2015. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torrents de la Pena A, de Taeye SW, Sliepen K, LaBranche CC, Burger JA, Schermer EE, Montefiori DC, Moore JP, Klasse PJ, Sanders RW. 2018. Immunogenicity in rabbits of HIV-1 SOSIP trimers from clades A, B, and C, given individually, sequentially, or in combination. J Virol 92:e01957-17. doi: 10.1128/JVI.01957-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klasse PJ, LaBranche CC, Ketas TJ, Ozorowski G, Cupo A, Pugach P, Ringe RP, Golabek M, van Gils MJ, Guttman M, Lee KK, Wilson IA, Butera ST, Ward AB, Montefiori DC, Sanders RW, Moore JP. 2016. Sequential and simultaneous immunization of rabbits with HIV-1 envelope glycoprotein SOSIP.664 trimers from clades A, B and C. PLoS Pathog 12:e1005864. doi: 10.1371/journal.ppat.1005864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pauthner MG, Nkolola JP, Havenar-Daughton C, Murrell B, Reiss SM, Bastidas R, Prevost J, Nedellec R, von Bredow B, Abbink P, Cottrell CA, Kulp DW, Tokatlian T, Nogal B, Bianchi M, Li H, Lee JH, Butera ST, Evans DT, Hangartner L, Finzi A, Wilson IA, Wyatt RT, Irvine DJ, Schief WR, Ward AB, Sanders RW, Crotty S, Shaw GM, Barouch DH, Burton DR. 2019. Vaccine-induced protection from homologous tier 2 SHIV challenge in nonhuman primates depends on serum-neutralizing antibody titers. Immunity 50:241–252.e6. doi: 10.1016/j.immuni.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montefiori DC, Karnasuta C, Huang Y, Ahmed H, Gilbert P, de Souza MS, McLinden R, Tovanabutra S, Laurence-Chenine A, Sanders-Buell E, Moody MA, Bonsignori M, Ochsenbauer C, Kappes J, Tang H, Greene K, Gao H, LaBranche CC, Andrews C, Polonis VR, Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Self SG, Berman PW, Francis D, Sinangil F, Lee C, Tartaglia J, Robb ML, Haynes BF, Michael NL, Kim JH. 2012. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J Infect Dis 206:431–441. doi: 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castillo-Menendez LR, Nguyen HT, Sodroski J. 2018. Conformational differences between functional human immunodeficiency virus envelope glycoprotein trimers and stabilized soluble trimers. J Virol 93:e01709-18. doi: 10.1128/JVI.01709-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pancera M, Wyatt R. 2005. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology 332:145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 40.Chakrabarti BK, Pancera M, Phogat S, O'Dell S, McKee K, Guenaga J, Robinson J, Mascola J, Wyatt RT. 2011. HIV type 1 Env precursor cleavage state affects recognition by both neutralizing and nonneutralizing gp41 antibodies. AIDS Res Hum Retroviruses 27:877–887. doi: 10.1089/AID.2010.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castillo-Menendez LR, Witt K, Espy N, Princiotto A, Madani N, Pacheco B, Finzi A, Sodroski J. 2018. Comparison of uncleaved and mature human immunodeficiency virus membrane envelope glycoprotein trimers. J Virol 92:e00277-18. doi: 10.1128/JVI.00277-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Protocol G Principal Investigators, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol G Principal Investigators, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Pena AT, Cupo A, Julien JP, van Gils M, Lee PS, Peng W, Paulson JC, Poignard P, Burton DR, Moore JP, Sanders RW, Wilson IA, Ward AB. 2014. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Pena AT, Derking R, Klasse PJ, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. 2014. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, Katinger H. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses 10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 47.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sattentau QJ, Moore JP. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med 174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakrabarti BK, Walker LM, Guenaga JF, Ghobbeh A, Poignard P, Burton DR, Wyatt RT. 2011. Direct antibody access to the HIV-1 membrane-proximal external region positively correlates with neutralization sensitivity. J Virol 85:8217–8226. doi: 10.1128/JVI.00756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haim H, Salas I, Sodroski J. 2013. Proteolytic processing of the human immunodeficiency virus envelope glycoprotein precursor decreases conformational flexibility. J Virol 87:1884–1889. doi: 10.1128/JVI.02765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alsahafi N, Bakouche N, Kazemi M, Richard J, Ding S, Bhattacharyya S, Das D, Anand SP, Prevost J, Tolbert WD, Lu H, Medjahed H, Gendron-Lepage G, Ortega Delgado GG, Kirk S, Melillo B, Mothes W, Sodroski J, Smith AB III, Kaufmann DE, Wu X, Pazgier M, Rouiller I, Finzi A, Munro JB. 2019. An asymmetric opening of HIV-1 envelope mediates antibody-dependent cellular cytotoxicity. Cell Host Microbe 25:578–587.e5. doi: 10.1016/j.chom.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol 70:1100–1108. doi: 10.1128/JVI.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67:3978–3988. doi: 10.1128/JVI.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boots LJ, McKenna PM, Arnold BA, Keller PM, Gorny MK, Zolla-Pazner S, Robinson JE, Conley AJ. 1997. Anti-human immunodeficiency virus type 1 human monoclonal antibodies that bind discontinuous epitopes in the viral glycoproteins can identify mimotopes from recombinant phage peptide display libraries. AIDS Res Hum Retroviruses 13:1549–1559. doi: 10.1089/aid.1997.13.1549. [DOI] [PubMed] [Google Scholar]
- 57.Madani N, Princiotto AM, Easterhoff D, Bradley T, Luo K, Williams WB, Liao HX, Moody MA, Phad GE, Vazquez Bernat N, Melillo B, Santra S, Smith AB III, Karlsson Hedestam GB, Haynes B, Sodroski J. 2016. Antibodies elicited by multiple envelope glycoprotein immunogens in primates neutralize primary human immunodeficiency viruses (HIV-1) sensitized by CD4-mimetic compounds. J Virol 90:5031–5046. doi: 10.1128/JVI.03211-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Posner MR, Hideshima T, Cannon T, Mukherjee M, Mayer KH, Byrn RA. 1991. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. J Immunol 146:4325–4332. [PubMed] [Google Scholar]
- 59.Thali M, Olshevsky U, Furman C, Gabuzda D, Posner M, Sodroski J. 1991. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J Virol 65:6188–6193. doi: 10.1128/JVI.65.11.6188-6193.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Melillo B, Liang S, Park J, Schon A, Courter JR, LaLonde JM, Wendler DJ, Princiotto AM, Seaman MS, Freire E, Sodroski J, Madani N, Hendrickson WA, Smith AB III. 2016. Small-molecule CD4-mimics: structure-based optimization of HIV-1 entry inhibition. ACS Med Chem Lett 7:330–334. doi: 10.1021/acsmedchemlett.5b00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKeating JA, McKnight A, Moore JP. 1991. Differential loss of envelope glycoprotein gp120 from virions of human immunodeficiency virus type 1 isolates: effects on infectivity and neutralization. J Virol 65:852–860. doi: 10.1128/JVI.65.2.852-860.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moore JP, McKeating JA, Weiss RA, Sattentau QJ. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 63.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andrabi R, Voss JE, Liang CH, Briney B, McCoy LE, Wu CY, Wong CH, Poignard P, Burton DR. 2015. Identification of common features in prototype broadly neutralizing antibodies to HIV envelope V2 apex to facilitate vaccine design. Immunity 43:959–973. doi: 10.1016/j.immuni.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Herschhorn A, Gu C, Moraca F, Ma X, Farrell M, Smith AB III, Pancera M, Kwong PD, Schon A, Freire E, Abrams C, Blanchard SC, Mothes W, Sodroski JG. 2017. The beta20-beta21 of gp120 is a regulatory switch for HIV-1 Env conformational transitions. Nat Commun 8:1049. doi: 10.1038/s41467-017-01119-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Madani N, Perdigoto AL, Srinivasan K, Cox JM, Chruma JJ, LaLonde J, Head M, Smith AB III, Sodroski JG. 2004. Localized changes in the gp120 envelope glycoprotein confer resistance to human immunodeficiency virus entry inhibitors BMS-806 and #155. J Virol 78:3742–3752. doi: 10.1128/JVI.78.7.3742-3752.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ozorowski G, Pallesen J, de Val N, Lyumkis D, Cottrell CA, Torres JL, Copps J, Stanfield RL, Cupo A, Pugach P, Moore JP, Wilson IA, Ward AB. 2017. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547:360–363. doi: 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang H, Cohen AA, Galimidi RP, Gristick HB, Jensen GJ, Bjorkman PJ. 2016. Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc Natl Acad Sci U S A 113:E7151–E7158. doi: 10.1073/pnas.1615939113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Juette MF, Terry DS, Wasserman MR, Altman RB, Zhou Z, Zhao H, Blanchard SC. 2016. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat Methods 13:341–344. doi: 10.1038/nmeth.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The customized MATLAB (MathWorks) program SPARTAN for smFRET analysis is publicly available at https://www.scottcblanchardlab.com/software.