SUMMARY
The recent Zika virus (ZIKV) and chikungunya virus (CHIKV) epidemics highlight the explosive nature of arthropod-borne (arbo)viruses transmitted by Aedes spp. mosquitoes1,2. Vector competence and the extrinsic incubation period (EIP) are two key entomological parameters used to assess the public health risk posed by arboviruses3. These are typically measured empirically by offering mosquitoes an infectious bloodmeal and temporally sampling mosquitoes to determine infection and transmission status. This approach has been used for the better part of a century; however, it does not accurately capture the biology and behavior of many mosquito vectors which refeed frequently (every 2–3 days)4. Here we demonstrate that acquisition of a second non-infectious bloodmeal significantly shortens the EIP of ZIKV-infected Ae. aegypti by enhancing virus dissemination from the mosquito midgut. Similarly, a second bloodmeal increases the competence of this species for dengue virus and CHIKV as well as Ae. albopictus for ZIKV, suggesting that this phenomenon may be common among other virus-vector pairings and that Ae. albopictus might be a more important vector than once thought. Bloodmeal-induced microperforations in the virus-impenetrable basal lamina which surrounds the midgut provide a mechanism for enhanced virus escape. Modeling of these findings reveals that a shortened EIP would result in a significant increase in the basic reproductive number, R0, estimated from experimental data. This helps explain how Ae. aegypti can sustain explosive epidemics like ZIKV despite relatively poor vector competence in single-feed laboratory trials. Together, these data demonstrate a direct and unrecognized link between mosquito feeding behavior, EIP, and vector competence.
Arthropod-borne (arbo)viruses represent an ongoing threat to human health as shown by the emergence and global spread of dengue virus (DENV; Flaviviridae), chikungunya virus (CHIKV; Togaviridae), and Zika virus (ZIKV; Flaviviridae)5,6. These three arboviruses are transmitted by mosquitoes of the genus Aedes, subgenus Stegomyia, which serve as epidemic vectors, and are known to cause disease outbreaks with high attack rates, necessitating research into the factors regulating virus transmission1. The urban-dwelling mosquito Aedes aegypti serves as a particularly efficient vector because it feeds predominately and frequently on human hosts (every 2–3 days) thereby increasing the frequency of host contact7–13. Nevertheless, in laboratory trials, Ae. aegypti populations from endemic regions often exhibit unexpectedly low vector competence values for their arboviruses as measured by the proportion of mosquitoes that become infected and transmit a pathogen after ingesting virus14–18. This could be explained, in part, by the techniques used to assess vector competence. In these studies, mosquitoes were offered an initial infectious bloodmeal and not allowed to refeed on blood again prior to assaying them for virus transmission, as is standard practice for assessing vector competence. Therefore, these studies do not recapitulate the natural biology of mosquitoes that refeed frequently. It is possible that differences in feeding history could help explain the seemingly paradoxical nature of Ae. aegypti-transmitted arboviruses.
Once a mosquito ingests an infected bloodmeal, arboviruses must overcome multiple barriers within the mosquito for transmission to occur19. The virus must infect the midgut, disseminate out of midgut cells, traverse the basal lamina layer to the hemolymph, and then infect the salivary glands before being transmitted to the next vertebrate host20. Failure of virus escape out of the mosquito midgut to the peripheral tissues has been identified as an important barrier to arbovirus transmission, but the underlying factors mediating this process are poorly understood14,15,21–23. Blood feeding triggers physiological changes within the mosquito—including mechanical distention of the midgut, apoptosis and regeneration of midgut epithelial cells, and altered permeability of the basal lamina—that could enhance or accelerate virus dissemination out of the midgut24–27. Based on these considerations, we evaluated the hypothesis that virus-infected mosquitoes fed an additional non-infectious bloodmeal will more effectively disseminate and transmit virus than mosquitoes fed only once.
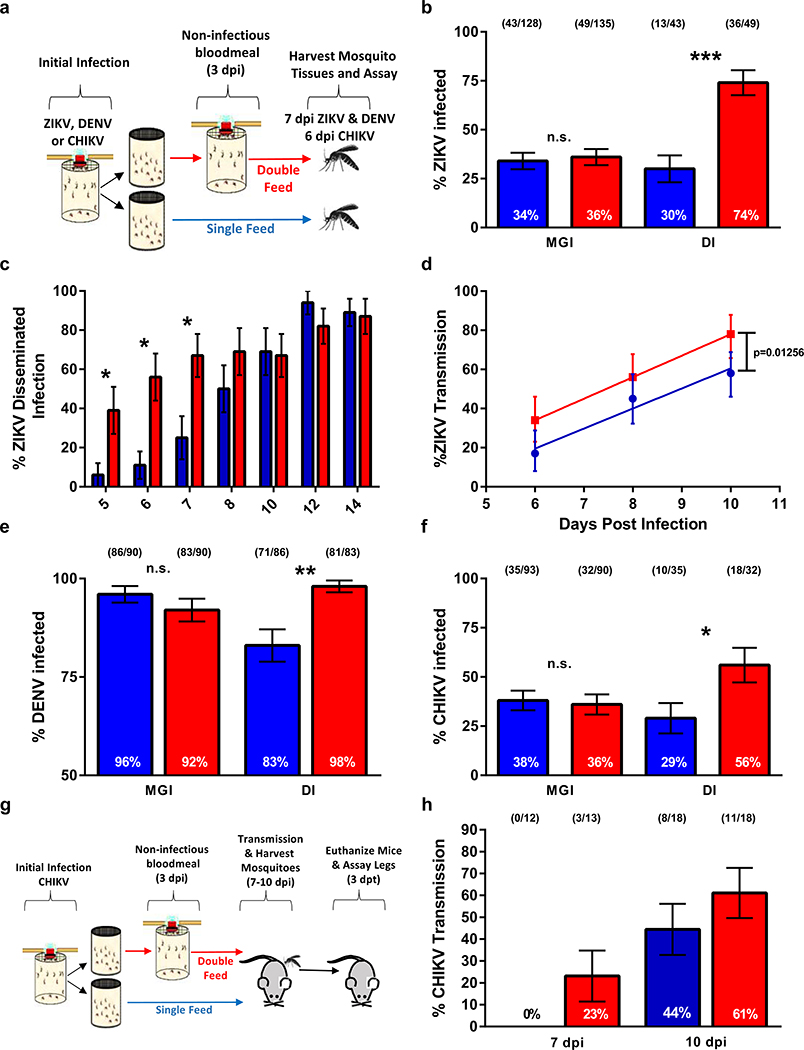
To test this hypothesis, we provided Ae. aegypti a second non-infectious bloodmeal 3 days after the infectious blood meal and compared virus infection and dissemination rates to those mosquitoes that received a single bloodmeal (Fig. 1a). Midgut infection prevalence of ZIKV was similar in the single- and double-feed groups, but the percentage of mosquitoes with disseminated ZIKV infection to leg tissues was significantly higher in mosquitoes receiving a second bloodmeal than those fed only once (Fig. 1b). Enhanced virus dissemination did not occur when mosquitoes were fed a non-infectious bloodmeal prior to receiving a ZIKV infectious bloodmeal (Extended Data 1). This suggests that an established midgut infection is a prerequisite for the observed enhanced rates of virus dissemination following a non-infectious bloodmeal. Temporal examination of this observation revealed that ZIKV disseminated more rapidly in the double-feed group than the single-feed group, but the difference in dissemination rates disappeared by day 10 post-infection (Fig. 1c). The increase in dissemination correlated with an increase of ZIKV positive saliva samples as the transmission rate regression line elevations (y-intercepts) were significantly different (p=0.01256) between the double-feed and single-feed cohorts, thus demonstrating enhanced early transmission potential (Fig. 1d and Extended Data 2). The dissemination and transmission studies were completed in different laboratories using different strains of Ae. aegypti further demonstrating the robustness of the double-feed observation.
Figure 1: Multiple feeding events increase dissemination and transmission rates of ZIKV, DENV and CHIKV in Aedes aegypti.
(a) Schematic of experimental design. (b) At 7 dpi paired bodies and legs were collected and assayed for the presence of ZIKV RNA by RT-qPCR (MGI (midgut infection rates) and DI (disseminated infection). (c) ZIKV dissemination rates of paired bodies and legs 5–14 dpi assayed for ZIKV RNA; only DI rates are presented. (d) 6, 8 and 10 dpi paired bodies and saliva were assayed for ZIKV RNA; only transmission rates (TR) are presented. TR data was analyzed by linear regression and analysis of covariance. (e,f) DENV-2 and CHIKV infection and dissemination rates, respectively. (g) Schematic of mouse transmission studies. (h) TR of CHIKV infected Ae. aegypti transmitting to suckling mice. (●) single-feed; (●) double-feed. Data were analyzed by two-sided Fisher’s exact test. (*) p<0.05, (**) p<0.01, (***) p<0.001. For b,c,e,f,h, center values represent the proportion and error bars represent the binomial SE of sample proportions. n for each experiment and precise p-values can be found in the linked source data.
To assess the impact of mosquito refeeding on vector competency for other arboviruses, Ae. aegypti were orally-exposed to dengue virus type 2 (DENV-2) and CHIKV and then given a second bloodmeal. As with ZIKV, the proportion of mosquitoes with disseminated infections for both DENV-2 (Fig. 1e) and CHIKV (Fig. 1f) were significantly increased compared to single-feed controls. The increased dissemination rates associated with double-feeding resulted in a higher proportion of mosquitoes transmitting CHIKV to mice (Fig. 1h). This demonstrates that the serial feeding behavior of Ae. aegypti enhances transmission of taxonomically diverse arboviruses and suggests that the mechanisms mediating this observation may be applicable to other virus-mosquito pairings. To evaluate whether our findings were unique to Ae. aegypti, we tested ZIKV rates of dissemination in a low-generation (F5) colony of Ae. albopictus which also refeeds within a single gonotrophic cycle28,29. Similar to Ae. aegypti, administration of a second non-infectious bloodmeal increased dissemination rates in Ae. albopictus (Extended Data 3). These findings suggest that under field conditions of frequent feeding, Ae. albopictus are more competent and could have contributed more to transmission during the ZIKV epidemic than previously thought.
To ascertain the basis of these findings, we determined whether an influx of energy-rich blood would promote viral replication and midgut escape. The number of virus genome equivalents was similar in mosquitoes regardless of feeding status (single- versus double-feed) or infection status (midgut restricted versus disseminated infection) (Extended Data 4). Our data indicates that once ZIKV has established an infection in the gut, its ability to escape is not conditioned by enhanced viral replication. These findings are consistent with studies that found no correlation between midgut titers and dissemination rates30,31 but contrast with the findings of Kramer et al. which showed higher midgut titers in mosquitoes with disseminated western equine encephalitis virus infection32.
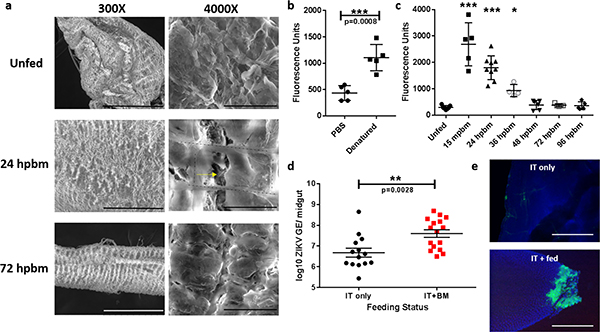
The midgut is encased in a proteoglycan extracellular matrix, termed the basal lamina, which provides protection and support to the epithelium. The pore size of the basal lamina is roughly 10 nm, yet the arboviruses tested in this study are 50–70 nm in size33. To determine if blood-feeding alters basal lamina integrity thereby accommodating transit of the larger virus particles, we performed scanning electron microscopy on Ae. aegypti midguts pre- and post-bloodmeal. The basal lamina of unfed midguts appeared intact and ruffled, while the basal lamina of midguts 24 hpbm (hours post-bloodmeal) were distended and had clear signs of damage (Fig. 2a). By 72 hpbm, the integrity of the basal lamina was reconstituted. Temporal quantification of basal lamina damage was performed using a collagen hybridizing peptide (CHP) binding assay. CHP specifically binds to damaged collagen IV, a major component of the basal lamina (Fig. 2b). Consistent with the SEM data, the CHP binding assay revealed high degrees of binding within 15 mpbm (minutes pbm) and statistically significant elevated levels of binding up to 36 hpbm (Fig. 2c). By 48 hpbm, CHP binding levels had returned to pre-bloodfed levels suggesting basal lamina repair. It has been proposed that bloodmeal-induced basal lamina damage is due to enzymatic degradation 25. However, our temporal data suggests that basal lamina damage is more likely the result of mechanical distention because it occurs immediately after blood engorgement. To evaluate how bloodmeal-induced microperforations in the midgut basal lamina might affect virus transit, we performed a retrograde infection assay. ZIKV was intrathoracically inoculated into a cohort of Ae. aegypti and half were provided a non-infectious bloodmeal 3 days post-inoculation. Nine days post-bloodmeal, mosquito midguts were removed, ZIKV antigen was detected by immunofluorescence, and ZIKV genome equivalents were quantified by RT-qPCR. ZIKV titers were significantly higher in those provided a bloodmeal (Fig. 2d) and clear signs of midgut epithelial infection could be detected in those provided a bloodmeal, whereas ZIKV antigen could not be detected in the epithelium of non-bloodfed individuals (Fig. 2e). Together these data provide a likely mechanism by which arboviruses are able to disseminate from the mosquito midgut and explain how virus already seeded in the midgut can easily escape upon acquisition of a second bloodmeal, while non-infectious bloodmeals provided prior to an infectious bloodmeal fail to enhance escape (Extended Data 1).
Figure 2: Bloodmeal acquisition induces microperforations in the midgut basal lamina.
(a) Representative SEM images of naïve and engorged Aedes aegypti midguts 24 hpbm and 72 hpbm at 300x (scale bars, 300 μm) and 4000x (scale bar, 20 μm) magnification. Two experimental replicates with 5 midguts/ replicate. Black dashed lines outline the visceral musculature and the yellow arrow denotes microperforations in the underlying basal lamina. (b) Pools of 5 unfed Ae. aegypti midguts per replicate were assayed for fluorescein labeled collagen hybridizing peptide (CHP) binding (n=5). Samples were analyzed by two-tailed t-test (p=0.0008). (c) Temporal binding profile of CHP binding to pools of 5 midguts/ replicate following a single bloodmeal. Differences in CHP binding were determined using a One-way ANOVA with a Dunnett’s multiple comparisons post-test. Precise p-values and samples sizes can be found in the linked source data. (d) Viral genome equivalents/ midgut of individuals intrathoracically inoculated with ZIKV (n=14) or inoculated and provided a bloodmeal 3 dpi (n=16). Midguts were assayed 12 dpi by RT-qPCR and statistically analyzed with a two-tailed t-test. (e) Midguts from ZIKV inoculated individuals with or without a bloodmeal assayed for ZIKV antigen 12 dpi. Representative images of two experimental replicates of 10 individual midguts/ per group. Images were acquired at 200X magnification (scale bar, 100 μm). For b-e, center values represent the means and error bars represent the SD.
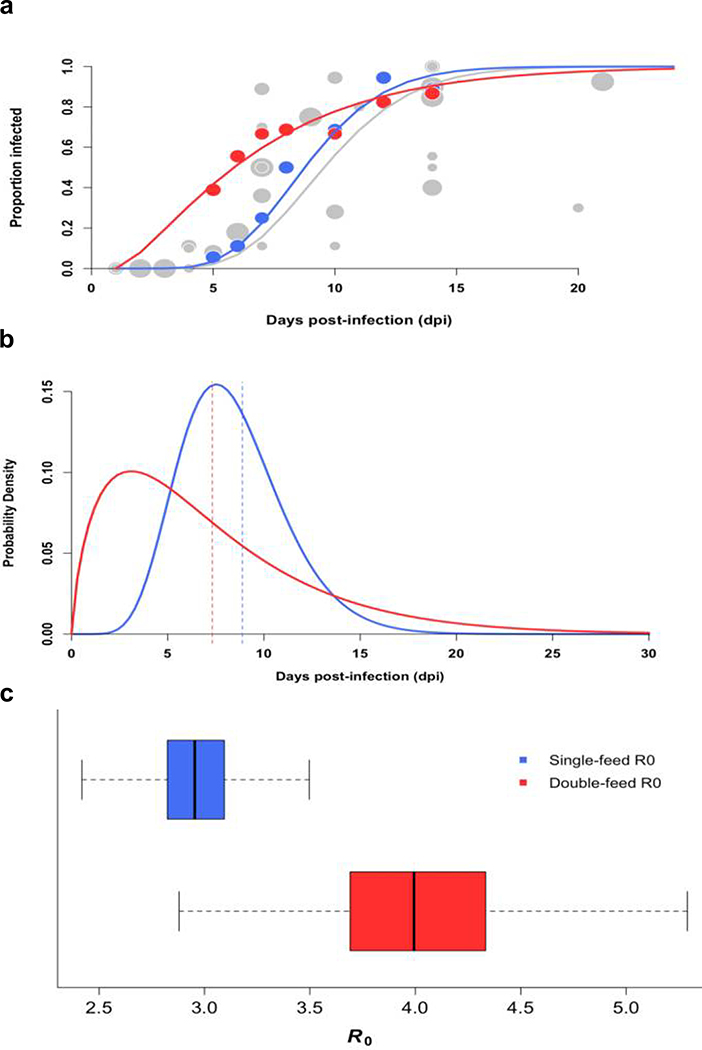
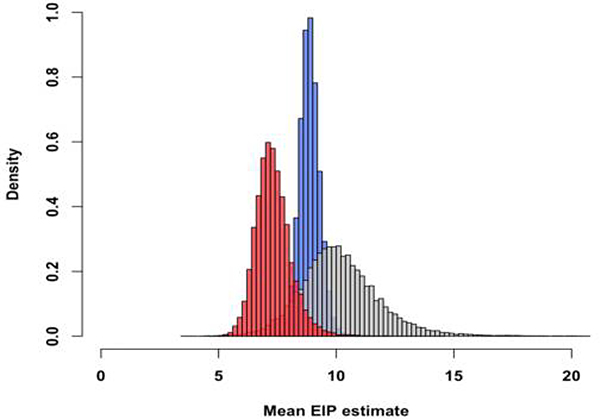
To quantify how a second bloodmeal would affect transmission of ZIKV by Ae. aegypti as measured by the basic reproductive number (R0), we first modeled the distributions of the ZIKV EIP when mosquitoes were fed only one bloodmeal and again when they were fed a second non-infectious bloodmeal, using our experimental data on disseminated infection (Fig. 3a). We estimated the mean EIP to be 8.88 days (posterior standard deviation (PSD) = 2.94 days) when mosquitoes were fed only one bloodmeal and 7.33 days (PSD = 5.96 days) when mosquitoes were fed a second non-infectious bloodmeal (Fig. 3b). The posterior probability that the mean for the single-feed EIP was larger than that of the double-feed EIP is 0.96 (Extended Data 5). The results were similar for the salivary gland infection data and when comparing our double-feed data to an estimate of the single-feed EIP derived from a meta-analysis of published studies that temporally assessed ZIKV infection rates (Fig. 3a).
Figure 3: Offering mosquitoes a second bloodmeal post-infection decreases mean EIP and increases transmission potential of ZIKV.
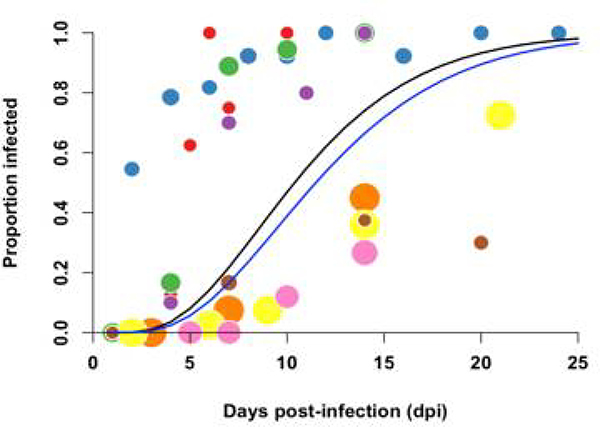
(a) Data from our single-feed disseminated infection experiments (●) and double-feed disseminated infection experiments (●) are shown with respective fitted gamma cumulative distribution function (CDF) ( (−) single-feed,; (−) double-feed). Each data point represents the proportion of infected mosquitoes at a single time point in a given experiment; the radii correspond proportionally to both the sample size and weight in estimating the parameters of the CDF. For reference, we also estimated ZIKV dissemination rates in Aedes aegypti mosquitoes offered only one infectious bloodmeal based on data from seven published studies along with our own single-feed results (●), and fitted a gamma CDF to the aggregated data (gray line). (b) Posterior densities for the single-feed EIP (−) and double-feed EIP (−) are plotted with corresponding means of the respective EIP shown by the dashed lines. (c) The boxplots display the distribution of mean R0 values for mosquitos offered a single bloodmeal (blue) versus a second bloodmeal (red). The boxes represent the interquartile range of the mean R0 values, the black line within the box marks the median, and the whiskers indicate the 10th and 90th percentiles of the mean R0 values.
Based on our single-feed empirical distribution of the EIP, we estimated the mean to be 2.96 (95% CI: 2.58–3.39), whereas when mosquitoes were fed a second bloodmeal following the initial infectious bloodmeal, the mean was 4.05 (95% CI: 3.22–5.17) (Fig. 3c). The distribution of R0 values was consistent with published estimates based on seroprevalence and incidence data from different locations (Extended Data 6). The median difference in was 1.03 (95% CI: 0.15, 2.25). The EIP was the first- or second-most influential parameter affecting the difference in R0 according to our two sensitivity analyses (Extended Data 7). The estimated increase in R0 following a second bloodmeal may help explain the magnitude of Ae. aegypti-vectored ZIKV epidemics despite the relatively low competence observed experimentally (after a single infectious bloodmeal) for this vector. Furthermore, our model predicted a greater epidemic potential of ZIKV (as indicated by R0>1) in regions where the daily probability of mosquito survival is lower (Extended Data 8). Thus, the potential range of ZIKV persistence may be slightly greater than previously estimated34.
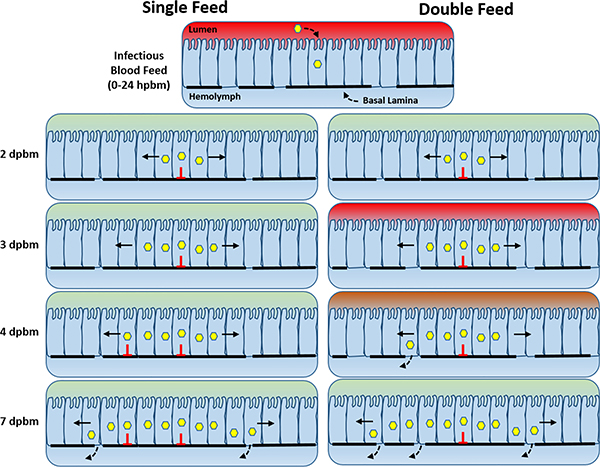
This study establishes a connection between mosquito feeding behavior and viral development within the vector that has direct impacts on the transmissibility and epidemic risk of arboviruses. We found that providing a second non-infectious bloodmeal to mosquitoes enhances viral dissemination from the midgut for a number of different virus-vector pairings. We propose that under field-relevant feeding regimens, viruses emerging from infected midgut epithelium cells can more readily traverse the basal lamina during subsequent feeding episodes as a result of bloodmeal-induced microperforations (Fig. 4). During infection of the mosquito midgut, only a handful of cells are initially infected but as infection progresses virus foci begin to expand covering more of the midgut tissue35. If the initial infectious blood meal resulted in sporadic basal lamina disruption followed by partial repair, then growing virus foci will eventually overlap with regions of discontinuous basal lamina that could serve as a conduit for virus escape. The addition of a second bloodmeal would only increase the number of microperforations and thus increase the likelihood that virus foci will chance upon a basal lamina break prior to repair. This proposed model helps explain how a second bloodmeal accelerates virus dissemination from an already established midgut infection.
Figure 4: Proposed model for double-feed enhanced early dissemination and transmission.
As Aedes spp. mosquitoes acquire an infectious bloodmeal, the basal lamina underlying the midgut becomes compromised due to mechanical distention. Initially, only a few midguts cells become infected, but with time, viruses spread cell-to-cell forming increasingly larger foci. Simultaneously, the basal lamina begins to repair, but not fully. Eventually the growing foci overlap with basal lamina microperforations allowing virus to bypass the basal lamina and enter the hemolymph. Acquisition of an additional bloodmeal would result in additional disruptions in the basal lamina thereby increasing the likelihood that the growing virus foci will chance upon a break resulting in earlier dissemination and transmission.
One limitation of the current study concerns the use of artificial blood meals to feed mosquitoes. This method is commonly used for colony maintenance and for vector competence studies, and has advantages over the use of laboratory animals for convenience and for arboviruses such as ZIKV, DENV, and CHIKV that do not produce infectious-level viremias in easy to maintain animal hosts. Despite these advantages, artificial feeding technique differs from natural blood feeding in ways that could potentially impact the results of this study. The technique requires the use of anticoagulated (defibrinated) blood that results in significantly lower arbovirus infections rates in mosquitoes than after feeding on viremic hosts36,37. To overcome this impediment, ZIKV and DENV were grown in mosquito cell culture rather than mammalian cells to generate virus titers that were greater than observed during human viremias. Viruses grown in insect cells express different N-linked glycan residues on the surface of envelope protein that could also affect virus infection of the mosquito midgut38. Nevertheless, our study shows the impact of sequential blood meals on arbovirus dissemination from the gut after initial infection. This occurs long after the initial infectious bloodmeal is ingested and therefore the source of virus, virus titer, or method of blood feeding is unlikely to alter our findings.
The volume of blood ingested during the second blood meal could potentially affect our results by impacting biophysical changes to the basal lamina layer surrounding the mosquito midgut. Most mosquitoes were fully engorged after blood feeding on a membrane feeder; however, in nature, blood feeding can often be interrupted resulting in partial engorgement which could influence virus dissemination associated with multiple feedings. A previous study found that collagen IV declined equally in the midguts of partially-fed vs fully engorged mosqtuioes25; however, it is unclear if this reduction in collagen IV results in loss of basal lamina integrity or influences rates of dissemination. Further research is needed to evaluate the impact of blood meal volume on basal lamina integrity and virus dissemination rates to better understand the epidemiological significance of this phenomenon.
Taken together, our findings emphasize the importance of considering feeding behavioral traits when performing vector competence studies. Past studies may underestimate the risks of arbovirus transmission by measuring vector competence after only a single infectious bloodmeal.
METHODS
Viruses, Cell Culture, Mosquitoes, and Mice
Viruses used in this study included ZIKV (PRVABC59; GenBank: KU501215), DENV-2 (125270/VENE93; GenBank: U91870), and CHIKV (R99659; GenBank: KX713902). C6/36 Ae. albopictus cells were used to amplify ZIKV (final passage history= Vero-3, C6/36–1) and DENV-2 (C6/36–3), CHIKV (Vero-2) were grown in Vero E6 cells, and BHK-21 (clone 15) cells were used to titrate infectious bloodmeals. Cell cultures were maintained in Minimal Essential Medium (MEM) with 10% fetal bovine serum, 100 U/ ml penicillin, 100 mg/ ml streptomycin, L-glutamine, 25 mg/ml amphotericin B, and sodium bicarbonate at 28°C for C6/36 cells or 37° C for Vero and BHK-21 cells with 5% CO2. C6/36 and BHK-21 cells were confirmed to be clear of mycoplasma. Colonies of Ae. aegypti (Orlando strain, collected from Orlando, FL in 1952 and Poza Rica strain, collected from Poza Rica Mexico in 2016) and Ae. albopictus (Stratford strain, generation F5, collected in Stratford, CT, 2015) were maintained on defibrinated sheep’s blood and reared under standard laboratory conditions39. Adult mosquitoes were housed at 27°C in environmental chambers with a 14:10 light: dark cycle. Litters of suckling mice (mixed sex) from pregnant CD-1 mice were obtained from Charles River Laboratories (Wilmington, MA). Statistical methods were not used to predetermine sample sizes. Litters of sucking mice were randomly assigned to cohorts of mosquitoes and individual suckling mice were randomly assigned to individual mosquitoes within the respective cohorts for the transmission studies. Similar to the mosquito samples, the right front leg of euthanized mice exposed to CHIKV infected mosquitoes were collected by one group and provided to a second group for processing and data analysis. Group 2 was unaware of the order in which samples were given to them. Only after processing and data analysis were completed for each experimental replicate were the treatments made aware to the sample order. Procedures for handling and care of animals were approved by and performed under the Animal Care and Use Committee at The Connecticut Agricultural Experiment Station (Protocol # P28–17).
Vector Competence Studies
Ae. aegypti and Ae. albopictus mosquitoes, 7–10 days post emergence, were offered an infectious bloodmeal containing a 1:1 mixture of defibrinated sheep’s blood and virus. After feeding, mosquitoes were cold-anesthetized and engorged females were transferred into two 32 oz. ice cream cartons containing a small cup with an egg-laying paper and housed in a 27°C environmental chamber. Mosquitoes had access to 10% sucrose sugar meals during the incubation period. Ingestion of sucrose was shown to be diverted to the mosquito crop rather than the midgut and therefore, will have no expected impact on midgut expansion and basal lamina permeability40.
Three to seven days after the initial infectious bloodmeal, one of the two cartons was provided a second non-infectious bloodmeal. Again, engorged females were collected and placed in a carton with a new egg-laying cup and provided access to 10% sucrose. Following variable extrinsic incubation periods, bodies, midguts, legs and salivary glands were harvested and macerated in 250 μl PBS-G [phosphate-buffered saline with 0.5% gelatin, 30% rabbit serum, and 1% 100x antibiotic-antimycotic (10,000 mg/ml of streptomycin, 10,000 U/ml penicillin, and 25 mg/ml of amphotericin B)] with a copper BB using a mixer mill. The inverse feeds (Extended Data 1) followed a similar experimental design; however, the double-feed group received a non-infectious bloodmeal prior to receiving a ZIKV-infectious bloodmeal. Either freshly grown virus or frozen virus stocks were used to complete the studies. Initially, all of the ZIKV studies were performed with frozen stocks of ZIKV (4.8 × 106 plaque forming units (pfu)/ml); however, in light of the poor midgut infection rates, all subsequent experiments were performed with freshly grown virus (1.0 × 106 –107 pfu/ml); C6/36 cells were infected at an multiplicity of infection of ~0.1 and harvested 4–5 days post infection. While this change in protocol did increase midgut infection rates, it did not alter the enhanced dissemination rate phenotype associated with multiple bloodmeals. For comparison, mean human viremia titers of ZIKV were estimated in the 104-105 genome equivalents/ ml range41,42. DENV-2 was grown fresh on C6/36 cells (5 × 106 – 3 × 107 pfu/ml) and frozen aliquots of CHIKV (4 × 106 pfu/ml) were used.
To evaluate ZIKV transmission by mosquitoes, saliva was collected in individual glass capillary tube containing immersion oil Type B. Salivation was allowed to occur for 25–30 min at room temperature. After salivation, the capillary tube was put into 100 μl of Dulbecco’s Modified Eagle Medium (DMEM) with 10% EquaFETAL (Atlas Biologicals, Fort Collins, CO) and 100 Units/mL of penicillin and 100 μg/mL of streptomycin, and the tube was centrifuged at maximum speed for 5 min. Outbred mice are susceptible to CHIKV infection43 and were used as host animals in CHIKV transmission experiments based on a previously published protocol44. Individual mosquitoes were allowed to feed on 5-day old mice. Mice were euthanized 3 days after exposure to mosquitoes. The front limb was removed from each mouse, homogenized in 500 ul of PBS-G, and tested for CHIKV infection by RT-qPCR.
Infection rates were determined and reported as follows. Midgut infection (MGI) rate represents the total number of virus-positive bodies divided by the total number of mosquitoes tested. Similarly, disseminated infection (DI) rates were determined by dividing the total number of virus-positive legs by the total number of virus-positive bodies. Transmission rates were calculated by dividing the total number of virus-positive salivary secretions or virus-positive recipient mice by the total number of virus-positive bodies.
Viral RNA Detection by RT-qPCR
Total RNA was extracted from 50 μl of mosquito tissue and body homogenates using the Mag-Bind Viral DNA/RNA 96 Kit (Omega Bio-tek Inc., Norcross, GA) on a Kingfisher Flex automated nucleic acid extraction device (ThermoFisher Scientific, Waltham, MA) following the manufacturer’s instructions. Samples were eluted in 50 μl ddH2O. ZIKV RNA was detected in mosquito tissues using a previously described RT-qPCR primer-probe set (ZIKV 1087/1163c/1108 FAM)45. DENV-2 RNA was detected using a previously described primer-probe set spanning the 3’-UTR46 and CHIKV RNA was detected using the previously described 6856F/6981c/6919-FAM primer-probe set47. The same RT-qPCR protocol was used to detect all three viruses. In brief, 25 μl reactions containing 2.5 μl of total RNA were assayed with the TaqMan RNA-to-Ct 1-Step Kit (ThermoFisher Scientific) on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA) using the following parameters: RT – 50°C for 30 min, 95°C for 10 min, PCR – 95°C for 15 s., 60°C for 1 min followed by a plate read (50 cycles). Data were analyzed using the Bio-Rad CFX Manager 3.1 software. The cut-off value used for ZIKV exposed samples to be considered positive by RT-qPCR was Ct <37. This cut-off value was empirically determined by comparing paired serial ten-fold dilutions either inoculated on Vero cells or assayed by RT-qPCR (Table S5). Ct values <35 cycles were considered positive for CHIKV and <33 cycles for DENV-2. The use of RT-qPCR for scoring positives was validated by comparing RT-qPCR and cell culture isolation using salivary glands from ZIKV-exposed Ae. aegypti. While the percent positive rate was higher by RT-qPCR, the difference in infection rates between single-feed and double-feed samples was maintained (Table S6).
RNA standards were generated in order to quantify ZIKV RNA from mosquito midguts. Briefly, an ~ 680 bp fragment spanning the RT-qPCR primer set (positions 837–1520) was amplified with a forward primer containing a T7 promoter and a non-modified reverse primer. The amplicon was purified, sequenced, and used as template to generate RNA transcripts using the T7 Megascript Kit according to the manufacturer’s instructions (ThermoFisher Scientific). RNA was quantified on a Qubit Fluorometer (ThermoFisher Scientific) and diluted to achieve serial 10-fold genome equivalent (GE) dilutions. We detected 102–107 ZIKV GE/ reaction with a primer efficiency of 78.4% with an R2 value of 0.971, a slope of −3.977, and y-intercept = 46.965.
Scanning Electron Microscopy
Ae. aegypti mosquitoes, 7 days post emergence, were offered a non-infectious bloodmeal, sorted and housed as described in the main text. Ten midguts from unfed control and engorged mosquitoes were dissected at 24 and 72 hpbm and fixed in a 2% paraformaldehyde/ 2.5% glutaraldehyde solution containing 0.1% (w/v) CaCl2 and 1% (w/v) sucrose buffered in 100mM Na cacodylate (pH 7.3). Samples were fixed at 4°C for three days and postfixed in 1% (w/v) OsO4 in the same buffer at room temperature for one hour. Fixed specimens were dehydrated through a graded ethanol and acetone series and imaged on a Hitachi Tabletop SEM TM3030Plus.
Collagen Hybridizing Peptide Assay
Ae. aegypti mosquitoes, 7 days post emergence, were provided a non-infectious bloodmeal. Midguts were temporally dissected from mosquitoes 15 mpbm, 24 hpbm, 36 hpbm, 48 hpbm, 72 hpbm, and 96 hpbm and fixed in a 2.5% glutaraldehyde and 2% paraformaldehyde solution for 24 hrs. Extra care was taken to not rupture engorged midguts during dissection. Baseline levels of CHP binding were determined with unfed midguts. Positive control midguts included unfed midguts that were immediately heat denatured at 70°C for 2 min. prior to fixation. Midguts were grouped into five midguts per pool, each pool representing an experiential replicate of each time point. Upon fixation, midguts were washed three times in PBS and then incubated with fluorescein labelled CHP (3Helix) diluted in PBS at a final concentration of 5 μM and incubated overnight at 4°C. Subsequently, samples were washed three times in PBS and incubated with 1μg/ μl elastase in PBS for two hours at 27° C and agitated every 15 min. Samples were transferred to a black 96-well plate ensuring midguts were not transferred and diluted 1:1 with PBS. Fluorescence was determined on a BioTek SYNERGY H1 microplate reader using the area scan feature with and excitation/emission of 485/515.
Retrograde Infection Assay
Ae. aegypti mosquitoes were intrathoracically inoculated with approximately 70 ZIKV PFU’s. Three days post-inoculation (dpi), half of the individuals were provided a non-infectious bloodmeal. Mosquitoes were housed for nine additional days after which midguts were dissected. RNA was extracted and 15 samples from each group were assayed individually by RT-qPCR as described above. The remaining 10 midguts were fixed in 4% paraformaldehyde overnight. Midguts were washed twice in PBS and stained with a 1:200 dilution of the mouse anti-flavivirus E-glycoprotein clone FE1 in blocking buffer (PBS+5 % BSA+0.1% Tween 20) and incubated for 1 hr. at room temperature. Midguts were washed 3 times in PBS+0.1 Tween 20 and incubated with a donkey anti-mouse IgG conjugated to Alexafluor 488 diluted 1:200 in PBS+0.1% Tween 20. Samples were washed 3 times in PBS+0.1% Tween 20 and mounted on microscope slides with ProLong Gold Antifade with DAPI.
Statistical Analysis of Experimental Data
The data were pooled from two to four independent replicates for each experiment involving mosquitoes. Statistical methods were not used to predetermine sample size. Differences in the proportion of mosquitoes with midgut, disseminated, or salivary gland infection were analyzed using Fisher’s exact test. Standard error bars were determined by calculating the standard error of sample proportions. ZIKV transmission rate data was analyzed by linear regression and evaluated for differences in slope and y-intercept by analysis of covariance. Descriptive statistics are provided in the figure legends. All analyses were performed using GraphPad Prism Statistical software.
ZIKV Plaque Assay
In addition to RT-qPCR, salivary secretions from ZIKV-infected Ae. aegypti were assayed by plaque assay to confirm the presence of infectious virus particles. Briefly, saliva from individual mosquitoes were diluted in cell culture medium and plated on sub-confluent monolayers of Vero cells. Cells were incubated for 1 hr. at 37°C after which a semi-solid medium overlay (Dulbecco’s Modified Eagle Medium with 4% EquaFETAL, 2X Pen-Strep and 4 μg/mL Amphotericin B, mixed with an equal volume of 1.2% Tragacanth gum) (EquaFETAL, Atlas Biologicals, Fort Collins, USA) (Tragacanth, MP Biomedicals, Santa Ana, USA) was added. Cells were incubated for 4–5 days at 37oC in 5% CO2, stained with crystal violet, and plaques enumerated.
Estimates of EIP distributions for Zika virus
We developed models to estimate the distribution of the extrinsic incubation period (EIP) of Zika virus (ZIKV) under both single-feed and double-feed scenarios. Because the exact moment when a mosquito becomes infectious is not observable, observations of EIP for a particular vector-borne disease are generally reported as a range of days from the time when some proportion of mosquitoes first exhibit dissemination of the virus into the body or salivary glands following an infectious bloodmeal, to the time where a maximum proportion of mosquitoes exhibit disseminated infection or transmission 48. Whereas transmission studies often report the EIP as a range of days, we attempted to quantitatively describe the distribution of the EIP for ZIKV using our experimental results, and compare these results to published experimental data, in order to more accurately assess changes in the EIP when mosquitos are offered a second bloodmeal during the incubation period.
Published data on the EIP of ZIKV were collected by searching the relevant literature using Web of Science, PubMed and Google Scholar; search terms included “Aedes aegypti” AND “Zika” AND (“extrinsic incubation period” OR “dissemination” OR “salivary gland infection” OR “competence” OR “transmission”). We required that ZIKV infection data were collected on mosquitoes at three or more time points post-infection. Exclusions were not made based on language or date of publication. We attempted to account for sources of study variability (e.g. ZIKV strain, utilization of fresh or frozen virus stock, and geographic origin of mosquitos) in our models by incorporating a study-specific random effect. We identified nine papers in total that experimentally assessed ZIKV EIP in Aedes aegypti using traditional single-feed methodology (Table S1). We aggregated dissemination infection (DI) rate data for studies in which temporal infection data were collected on ZIKV dissemination to legs, wings or heads, including our own single-feed experimental results. Separately, we aggregated salivary gland infection (SGI) rate data for studies in which temporal infection data were collected on ZIKV dissemination to heads, salivary glands or in salivary secretions.
We assumed that the EIP for ZIKV was gamma distributed and that the data on the proportion of mosquitoes with disseminated infection sampled after t days could be modeled according to a gamma cumulative distribution function 48,49. We used a Bayesian framework to obtain estimates needed to parameterize the EIP distribution for ZIKV according to the formula below:
where the gamma CDF is defined by shape parameter αj and rate parameter βj for dataset j, with j=1 for the single-feed data and j=2 for the double-feed data; Γ(αj) refers to the incomplete gamma function. We included an additional parameter εi to account for between-study variability when fitting the model to the DI and SGI meta-analysis data composed of observations from multiple studies. The observed number of mosquitoes with disseminated infection at time t in experiment i (xt,i) was assumed to be binomially distributed with sample size (nt,i) and a success probability (pt,i):
We selected weakly informative priors for αj (Gamma(0.001,0.001)) and βj (Log-normal(0,0.001)), and assumed εi was normally distributed with mean 0 and variance τ; again, we specified a weakly informative Gamma(0.001,0.001) prior for τ. Posterior distributions were estimated via a Markov chain Monte Carlo (MCMC) sampling algorithm implemented using JAGS, run from the statistical program R with the rjags package 50,51. The algorithm was run for 100,000 iterations with 5,000 burn-in iterations for two chains. We fit the model to six datasets: the single-feed dissemination data (n=7 observations from our experimental results); the double-feed dissemination data (n=7 observations from our experimental results); the single-feed salivary gland data (n=3 observations from our experimental results); the double-feed salivary gland data (n=3 observations from our experimental results); the meta-analysis single-feed dissemination data (n=38 observations from 7 published studies aggregated with our single-feed dissemination results); and the meta-analysis single-feed salivary gland data (n=45 observations from 8 published studies). Convergence was assessed through visual inspection of trace plots and calculation of the Gelman–Rubin convergence diagnostic (97.5% quantile of <1.1) for all monitored parameters 52. Using a thinned subset (10%) of each model’s respective posterior shape and rate posterior distributions, we estimated the posterior distribution of mean EIP values as E[EIP] = α/β and Var(EIP)=α/β2 for each iteration.
The models all showed clear indications of convergence, with the 97.5% quantile of <1.007 for all monitored parameters. We first compared the estimated EIP for our experimental results based on the dissemination data to those based on the salivary gland infection data. The estimated EIPs were similar: days, days, while days, = 9.7 days. However, the variance in the posterior distributions based on the salivary gland data was considerably greater due to the limited number of data points (n=3) (Table S2). We then compared model results for the meta-analysis DI and meta-analysis SGI datasets. We found the posterior means to be similar: α1dissem = 3.89 and β1,dissem = 0.34 and α1,SGI = 3.78 and β1,SGI = 0.37. However, the SGI data exhibited wider 95% credible intervals (CI) and posterior standard deviation of EIP (, (95% CI: 2.82–10.10)) compared to the CI and posterior standard deviation of EIP for the aggregated dissemination data (, . We also compared the aggregated single-feed dissemination data from the meta-analysis to our own single-feed dissemination data and found no significant difference in mean EIP estimates | data)= 0.22)).
We observed a large variability among studies in the literature on SGI rates over time (Extended Data 9) that did not appear to be attributable to the following study characteristics: mosquito strain, viral strain, viral titer, temperature or humidity during mosquito rearing, or fresh versus frozen viral stock. Notably, we observed a close similarity in posterior estimates of the mean EIP based on the dissemination and salivary gland data for both our experimental data and the meta-analysis (Table S2). Based on these observations, as well as the fact that published studies exhibited more consistency in estimating dissemination rates (Fig. 3a), and we had considerably more experimental data points to fit to the model for dissemination to legs/wings than to the salivary glands, we chose to focus our subsequent analysis exclusively on the dissemination data.
Summary estimates of the posterior distributions of the mean EIP for the single- and double-feed data from this study, along with the aggreated single-feed data from our meta-analysis, are listed in Table S2 and plotted in Fig. 3a–b for the dissemination data.
We assessed the difference in the distributions by calculating the posterior probability that the EIP for the double-feed data was greater than that of the single-feed data for the thinned subset of 20,000 random samples from the respective posterior distributions. The posterior probability that the aggregated mean single-feed EIP was greater than the mean double-feed EIP was 0.95; in other words, was less than or equal to 5.0% of the time. The posterior probability that the single-feed EIP was greater than the double-feed EIP was 0.96 based on our experimental data, i.e. was less than or equal to 4.2% of the time.
Estimation of the basic reproductive number (R0)
In order to determine how the effect of multiple feeding episodes on EIP translates into the overall transmission potential of ZIKV, we estimated R0 (defined as the average number of secondary human cases that a primary human case generates over the course of his or her infectious period in a fully susceptible population) under both single- and double-feed scenarios.
We estimated the basic reproductive number for the single feed scenario according to the Ross-Macdonald model 53,54:
where m is the ratio of mosquito to human population density, a is the mosquito human biting rate, p is the probability of daily survival for mosquitoes, b is the vector competence, N is the EIP, c is the probability that a mosquito is infected when biting an infectious human, and r is the human recovery rate. The first expression in the above equations represents the vectorial capacity (VC), whereas the ratio c/r represents the key human transmission parameters that dictate how VC relates to R0 54. The biting rate a is expressed intrinsically as the product of the time interval between bloodmeals and the proportion of mosquito bloodmeals on humans. Traditionally, and for our single-feed R0, a is squared to account for the necessity of two bites to transmit infection from an infected to susceptible human. To calculate R0 for the double-feed data we modified this equation to examine the impact of an additional bloodmeal—which we assumed could come from an infected or uninfected human or non-human vertebrate—by multiplying by an additional factor a/h, where h is the proportion of bloodmeals taken from humans:
We parameterized our model for R0 according to current understanding of ZIKV transmission dynamics. When parameters specific to ZIKV transmission were not available or as yet understood, we informed the ranges from dengue virus studies, as Zika and dengue are arboviruses of the same Flavivirus genus, and are both spread by Aedes genus mosquitoes. Parameters were as specific as possible to Aedes aegypti, although ranges often accounted for what may also be observed with Ae. albopictus. As a conservative assumption (and consistent with our experimental finding that the difference in dissemination rates disappeared by day 10), we used values of the vector competence from the literature and assumed b did not vary between the single- and double-feed models. Parameter values and ranges are summarized in Table S3. Parameter distributions are also specified, where triangular distributions signify the expectation that values close to the peak of the triangular distribution are more likely to occur (Table S3).
To estimate how R0 changes with feeding behavior, we used our posterior distributions for the EIP as input into the above equations, since we assert that the EIP is better represented by a gamma distribution than a single static value. For each of a thinned subset (10%) of MCMC posterior samples of the shape and rate parameters, and for each model (single- and double-feed), we generated 20,000 gamma random variables as input to calculate 20,000 respective R0 estimates. We then found the mean R0 for that particular parameter set, and repeated this process for each MCMC sample in the subset. We then compared the distributions of the single- and double-feed R0 defined by the respective EIP distributions.
Based on the single-feed distribution of the EIP for our experimental data, we estimated the mean to be 2.96 (95% CI: 2.58 −3.39), whereas when mosquitoes were fed a second bloodmeal following the initial infectious bloodmeal, the mean was 4.05 (95% CI: 3.22–5.17) (Extended Data 5). The mean was greater than the mean for 99.0% of the posterior samples . The median difference in R0 estimates was 1.03 (95% CI: 0.15–2.25). We also estimated to be 2.97 (95% CI: 1.84–4.29) based on the single-feed data from our meta-analysis, and found that the mean was greater than the mean meta-analysis for 92.0% of the posterior samples. Finally, we considered the threshold of R0=1, above which an epidemic can occur in a susceptible population, and found that on average, 94.1% of estimates, 94.8% of R0_SF_Meta estimates, and 85.1% of estimates were above this threshold.
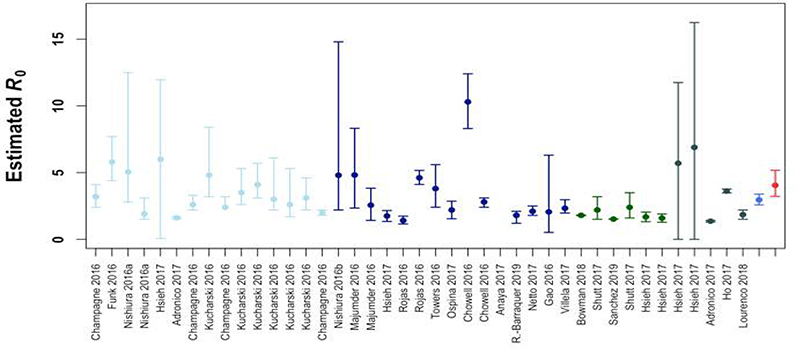
We compared our estimates of and to estimates of R0 from the literature. We identified 41 estimates of R0 for ZIKV from 22 studies. Studies were identified by searching Web of Science and PubMed; search terms included “Zika” AND (“reproductive OR reproduction number” OR “R naught” OR “R0” OR “transmission potential” OR “generation interval”). The estimates of R0 calculated in this study fell within the range of published estimates based on field data (Extended Data 6). We also noted a wide variation in estimates of the generation interval of ZIKV (Table S4), which is positively correlated with estimates of R0 and has been shown to be a major source of uncertainty in R0 estimates for ZIKV 55,56. Our results suggest that estimates of the generation interval of ZIKV based on natural history models may be upwardly biased, since the rely on estimates the EIP based on single-feed experimental data. Thus, estimates of R0 based on fitting to case report data may also be slightly upwardly biased. However, uncertainty in the generation interval used to estimate R0 in the literature is much greater than difference in the EIP that we estimated for the single-feed and double-feed experimental data.
Sensitivity analyses
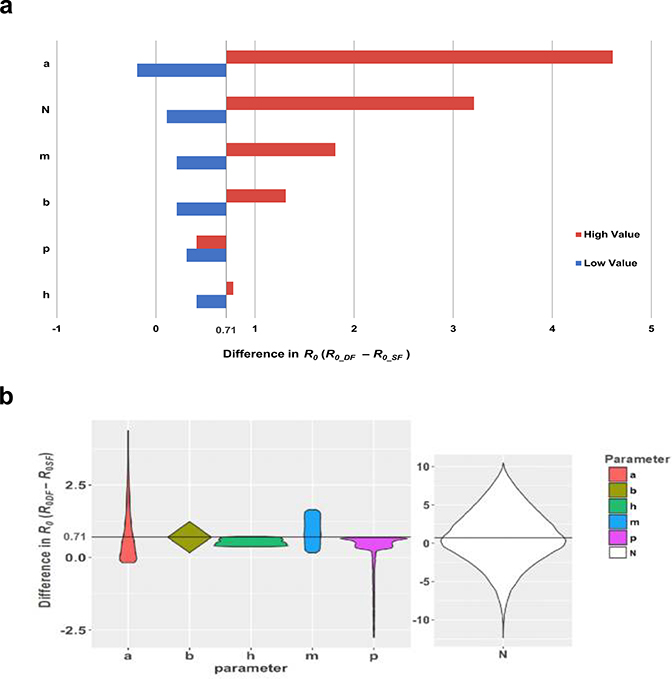
We assessed the sensitivity of the basic reproductive number estimates that resulted from variability in the input parameters N, a, m, b, p, and h using two analyses. First, using a best/worst-case scenario approach, we held all parameters at their given or mean value (Table S3) and calculated the difference in R0 between single feed and double feeds . We then varied each parameter individually according to the lowest and highest values in its specified range while holding all other parameters constant at their mean or given value (Table S3) and assessed how the difference in R0 between double- and single-feed estimates changed in magnitude from the initial difference. For the EIP estimates (N), we used our model results from the single-feed meta-analysis data because we felt they better represented the known variability in the distribution of EIP, as supported by numerous aggregated studies in addition to our own data. We found the most sensitive parameters to be the human biting rate (a), the EIP (N), and the mosquito density (m) according to this analysis (Extended Data 7a).
Next, for each parameter, we randomly sampled from its specified distribution (Table S3) while holding all other parameters constant at their mean or given value. For example, we took 10,000 random samples of m from the Unif(1,10) distribution as input for both the single-feed and double-feed transmission models and then examined the distribution of differences in R0 for each of the 10,000 iterations (Extended Data 7b). According to this analysis, we found the three most sensitive parameters to be the EIP (N), the human biting rate (a), and the probability of daily survival for mosquitoes (p), in that order. This analysis also sheds light on the anomalous behavior of p; although random sampling generally resulted in , the long tail of the distribution of differences suggests that a minority (14.4%) of random samples of p resulted in , and the majority of samples resulted in a difference less than the constant difference of 0.71. The largest values of resulted from samples of p in the middle of its range.
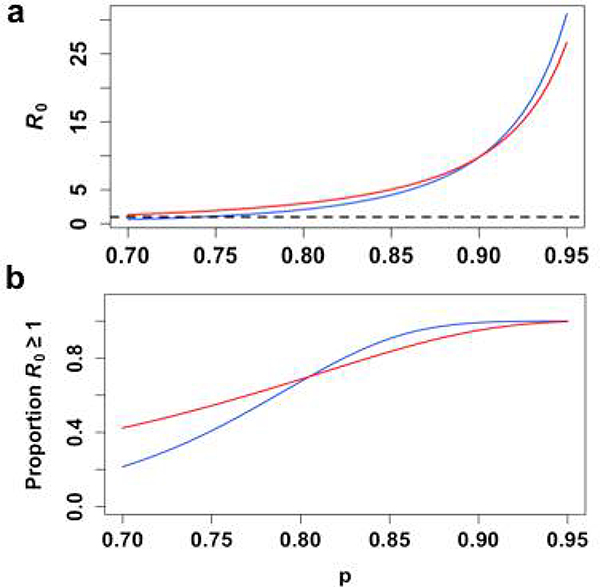
We also plotted the relationship between R0 and p (i.e. the probability of daily survival of mosquitoes), as this parameter plays an important role in the variability of R0 across different climates (Extended Data 8). For each value of p specified in Table S3, we generated 10,000 gamma-distributed estimates of the EIP as input to calculate 10,000 respective R0 estimates, as previously described. At high values of p, R0 is slightly larger for the single-feed model, although the overall magnitude of R0 for either model is large and unlikely to be observed in a natural setting. At lower values of p, within the range where ZIKV epidemics have been observed, R0 is slightly higher under the double-feed model (Extended Data 8a) and is more likely to be greater than 1 (Extended Data 8b). Our results therefore suggest that multiple bloodmeals in a natural setting may lead to greater persistence of ZIKV on the edges of observed transmission zones.
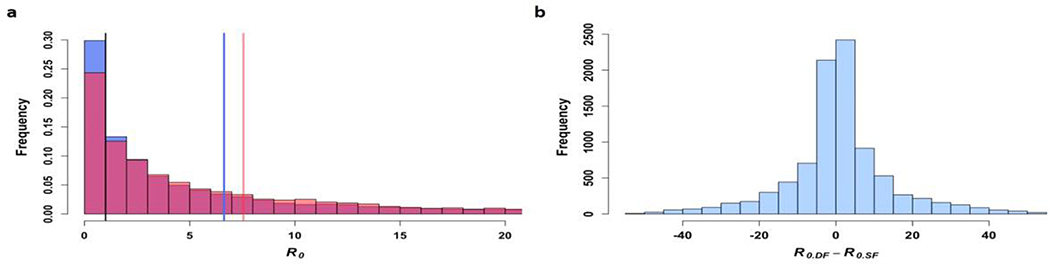
Finally, we performed a full probabilistic uncertainty analysis where we estimated the total uncertainty of the mean difference in R0 between single- and double-feeds using Latin Hypercube Sampling. With this approach, we created a matrix of uniform random output of 10,000 samples using the R software randomLHS function as input to the quantile function for each parameter’s specified distribution (Table S3) 57. We then determined the distribution of R0 values and removed those outside of the 95% CI to maintain realistic estimates. The and histograms are shown in Extended Data 10a. The two R0 distributions overlap significantly, but the mean value for is larger than that of . The distribution of ( for each random sample is shown in Extended Data 10b. The distribution is skewed slightly to the right with a mean difference of 0.87. Note that in this analysis, we are implicitly assuming that all of the parameters—including the EIP for the single- and double-feed models—are independent of one another, which is unlikely to be the case, but covariance of the different parameters is unknown.
Extended Data
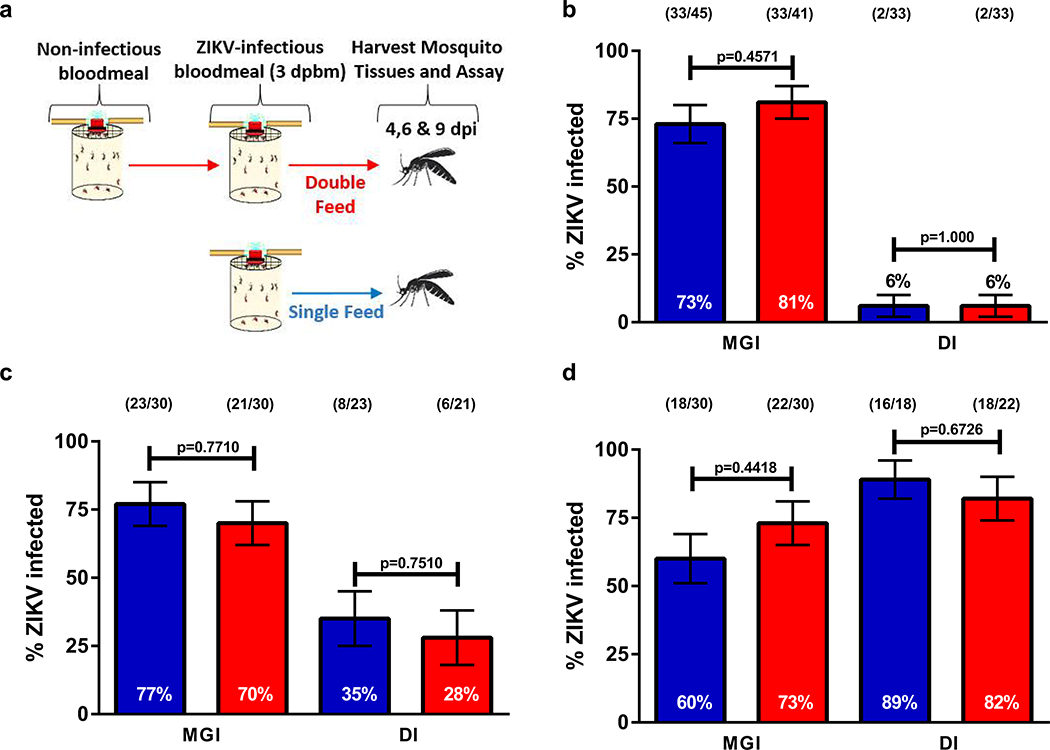
Extended Data Fig. 1. Non-infectious bloodmeals prior to infectious bloodmeals do not increase dissemination rates.
(a) Schematic of experimental design. At (b) 4, (c) 6, and (d) 9 dpi, midguts and legs were dissected from both cohorts and tested for the presence of ZIKV RNA by RT-q-PCR. (●) single feed; (●) double feed. Data were analyzed by Two-sided Fisher’s exact test. Sample sizes (represented as a fraction of positive samples/ total samples) for each treatment/ timepoint are embedded in the figures above each experimental group. Center values represent the proportion and error bars represent the binomial SE of sample proportions.
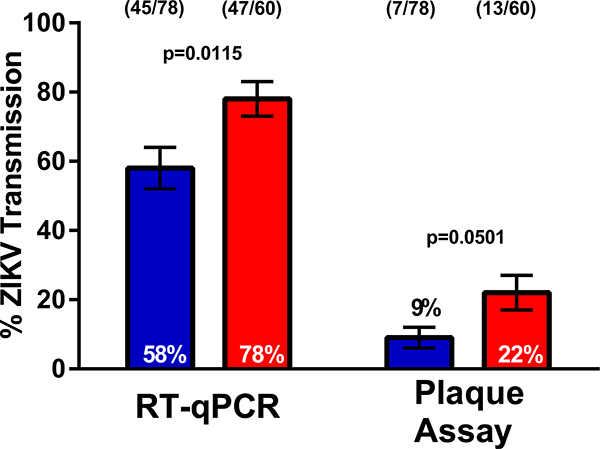
Extended Data Fig. 2. Increased transmission of ZIKV RNA and infectious virions associated with acquisition of an additional non-infectious bloodmeal.
Aedes aegypti mosquitoes were offered a ZIKV infectious bloodmeal and at 3 dpi individuals in the double-feed groups were fed a second, non-infectious bloodmeal. 10 dpi mosquito saliva was collected and assayed for ZIKV RNA and infectious virions by RT-qPCR and plaque assay respectively. (●) single feed; (●) double feed. Data were analyzed by Two-sided Fisher’s exact test. (*) p<0.05. Sample sizes (represented as a fraction of positive samples/ total samples) for each treatment/ timepoint are embedded in the figures above each experimental group. Center values represent the proportion and error bars represent the binomial SE of sample proportions.
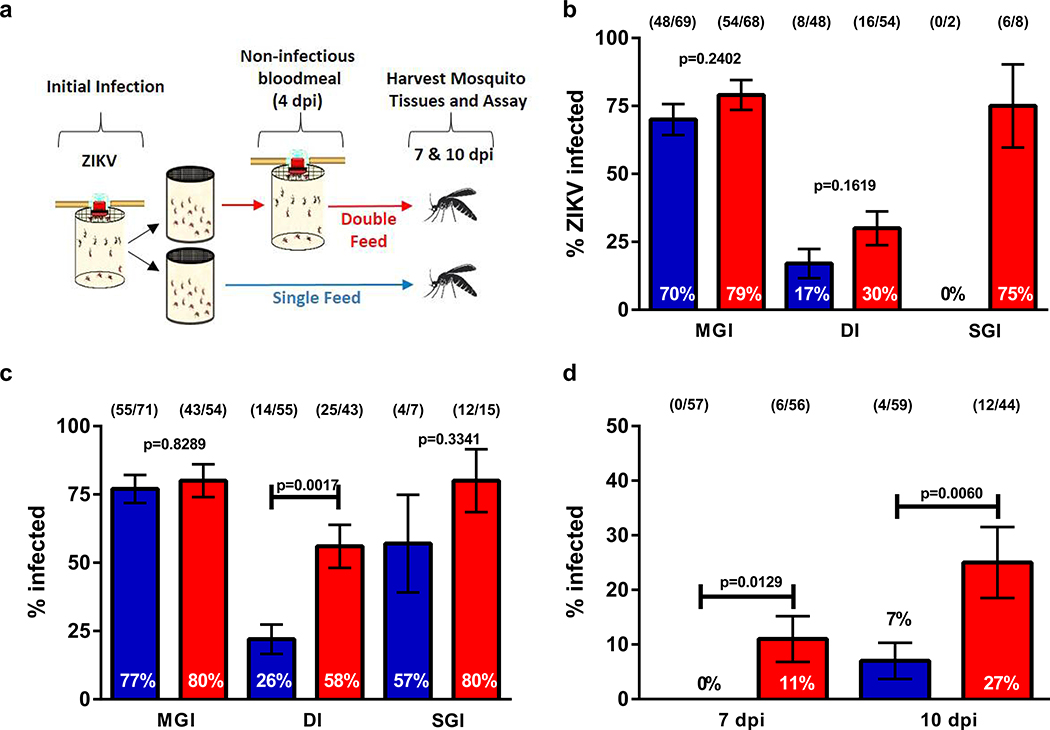
Extended Data Fig. 3. Multiple feeding events increase the potential of Aedes albopictus to transmit ZIKV.
(a) Schematic of experimental design. Paired bodies (MGI; midgut infection; % of mosquitoes with viral RNA in their bodies), legs (DI; disseminated infection; % of body positive mosquitoes with viral RNA in their legs) and salivary glands (SGI; salivary gland infection; % of leg positive mosquitoes with viral RNA in their salivary glands) were collected and assayed for the presence of viral RNA (b) 7 dpi and (c) 10 dpi. (d) SGI data from (b) and (c) analyzed as the % of ZIKV-exposed mosquitoes with a salivary gland infection. The data presented represents at least three experimental replicates. (●) single-feed; (●) double-feed. Data were analyzed by Two-sided Fisher’s exact test. (*) p<0.05, (**) p<0.01, (***) p<0.001. Sample sizes (represented as a fraction of positive samples/ total samples) for each treatment/ timepoint are embedded in the figures above each experimental group. Center values represent the proportion and error bars represent the binomial SE of sample proportions.
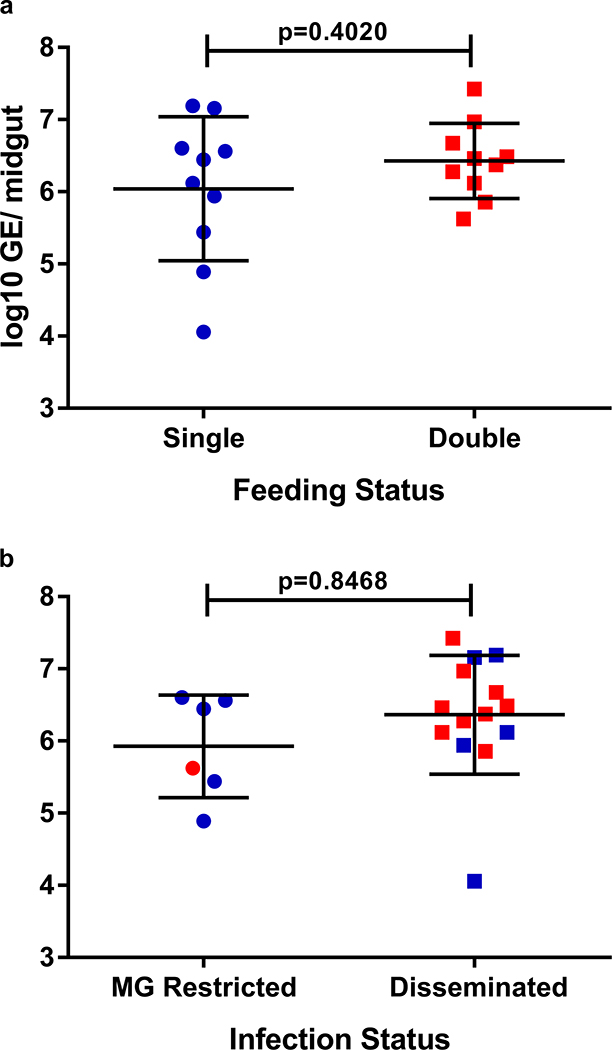
Extended Data Fig. 4. Increased dissemination rates associated with multiple feeding are not due to increased midgut replication.
Aedes aegypti mosquitoes were offered a ZIKV-infectious bloodmeal and 4 dpi individuals in the double feed groups were fed a second, non-infectious bloodmeal. At 7 dpi, mosquito midguts and legs were dissected and viral genomic equivalents were determined. (a) Midgut genome equivalents based on feeding status regardless if the infection was restricted to the midgut or disseminated (n=10/ group). (b) Midgut genome equivalents based on infection status (disseminated (n=14) vs. not disseminated (n=6) (i.e. midgut restricted)) regardless of feeding status. (●) single feed; (●) double feed. Data were analyzed by Two-sided T-test. Center values represent means and error bars represent SD.
Extended Data Fig. 5. Distributions of mean EIP estimates.
(●) experimental single-feed; (●) meta-analysis single-feed; (●) double feed, from a thinned subset (10,000 iterations) of each model’s respective posterior shape and rate estimates.
Extended Data Fig. 6. Comparison of R0 estimates from experimental data to published estimates from field studies.
Circles correspond to point estimates of R0 derived from an individual study and setting and/or methodology, while the lines show the corresponding 95% CI (n=41 estimates from 22 studies). Colors are used to show estimates from different regions (light blue: Oceania, dark blue: South America, dark green: Central America, gray: other) and our own experimental results (blue: single-feed model, red: double-feed model).
Extended Data Fig. 7. Results of a best/worst case scenario sensitivity analysis and a one-way random sampling sensitivity analysis.
(a) Scenario sensitivity analysis assessing the effect of varying each parameter according to its lowest and highest bounds on the difference in R0 ( (blue=lowest value; red=highest value for each parameter). The initial difference of 0.71 was obtained by holding all parameters constant at their mean or given value (Table S4). (b) Violin plots show the probably density of the difference in R0 when we randomly sampled each parameter 10,000 times from its specified distribution (Table S4), while holding all other parameters constant. Each violin spans the 98% quantile of the distribution of R0 differences with the width proportional to the probability of observing a particular value of difference. The horizontal line at 0.71 represents the difference in R0 when all parameters are held constant at their mean or given value.
Extended Data Fig. 8. Comparison of R0 and p.
(a) The mean value for 10,000 simulations of R0 is plotted for every value of p (blue= single-feed model, red= double-feed model, dashed black= reference line at R0=1), along with (b) the proportion of those simulations for which R0 >1.
Extended Data Fig. 9. Salivary Gland Infection Meta-Analysis.
Salivary Gland Infection (SGI) data aggregated from 8 published studies are plotted, with each observation (n=45) weighted by sample size and color-coded by study. The black line shows the best-fit gamma CDF model for salivary gland data and the blue line for combined meta-analysis single-feed dissemination data (data points not shown).
Extended Data Fig. 10. Results of an uncertainty analysis assessing the distribution of R0 under different feeding assumptions.
(a) We compared histograms of the single-feed R0 distribution specified by the uncertainty analysis (blue) and double-feed R0 distribution (red) and the overlap (purple) generated from 10,000 iterations of Latin Hypercube Sampling. The mean R0 for each respective distribution is also shown as a horizontal line . (b) The distribution of the difference in R0 is plotted for each random sample (n=10,000).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Michael C. Thomas, Michael Olson, Tanya Petruff, and Maria Correa who provided technical assistance with rearing, processing, and virus testing of mosquitoes. This publication was supported by the Cooperative Agreement Number U01CK000509, funded by the Centers for Disease Control and Prevention. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention or the Department of Health and Human Services. This work was further supported by the US Department of Agriculture Hatch Funds and Multistate Research Project (CONH00773 and NE1443), the National Center for Advancing Translational Sciences of the National Institutes of Health (CTSA grant number UL1 TR001863 and KL2 TR001862) (JLW), and the National Institutes of Health/National Institute of Allergy and Infectious Diseases (R01 AI112970) (VEP) and R21 AI129464 (BDF).
Footnotes
COMPETING INTERESTS STATEMENT:
The authors declare no competing interests.
References
- 1.Mayer SV, Tesh RB & Vasilakis N The emergence of arthropod-borne viral diseases: A global prospective on dengue, chikungunya and zika fevers. Acta Trop 166, 155–163, doi: 10.1016/j.actatropica.2016.11.020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faria NR et al. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 546, 406–410, doi: 10.1038/nature22401nature22401 [pii] (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kramer LD & Ciota AT Dissecting vectorial capacity for mosquito-borne viruses. Current opinion in virology 15, 112–118, doi: 10.1016/j.coviro.2015.10.003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott TW & Takken W Feeding strategies of anthropophilic mosquitoes result in increased risk of pathogen transmission. Trends Parasitol 28, 114–121, doi: 10.1016/j.pt.2012.01.001S1471-4922(12)00004-9 [pii] (2012). [DOI] [PubMed] [Google Scholar]
- 5.Weaver SC & Reisen WK Present and future arboviral threats. Antiviral Res 85, 328–345, doi: 10.1016/j.antiviral.2009.10.008S0166-3542(09)00495-1 [pii] (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Musso D & Gubler DJ Zika Virus. Clin Microbiol Rev 29, 487–524, doi: 10.1128/CMR.00072-1529/3/487 [pii] (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott TW et al. Blood-feeding patterns of Aedes aegypti (Diptera: Culicidae) collected in a rural Thai village. J Med Entomol 30, 922–927 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Scott TW et al. Longitudinal studies of Aedes aegypti (Diptera: Culicidae) in Thailand and Puerto Rico: blood feeding frequency. J Med Entomol 37, 89–101 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Ponlawat A & Harrington LC Blood feeding patterns of Aedes aegypti and Aedes albopictus in Thailand. J Med Entomol 42, 844–849 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Baak-Baak CM et al. Blood Feeding Status, Gonotrophic Cycle and Survivorship of Aedes (Stegomyia) aegypti (L.) (Diptera: Culicidae) Caught in Churches from Merida, Yucatan, Mexico. Neotrop Entomol 46, 622–630, doi: 10.1007/s13744-017-0499-x [pii] (2017). [DOI] [PubMed] [Google Scholar]
- 11.Sivan A, Shriram AN, Sunish IP & Vidhya PT Host-feeding pattern of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) in heterogeneous landscapes of South Andaman, Andaman and Nicobar Islands, India. Parasitol Res 114, 3539–3546, doi: 10.1007/s00436-015-4634-5 (2015). [DOI] [PubMed] [Google Scholar]
- 12.De Benedictis J et al. Identification of the people from whom engorged Aedes aegypti took blood meals in Florida, Puerto Rico, using polymerase chain reaction-based DNA profiling. Am J Trop Med Hyg 68, 437–446 (2003). [PubMed] [Google Scholar]
- 13.Barrera R et al. Vertebrate hosts of Aedes aegypti and Aedes mediovittatus (Diptera: Culicidae) in rural Puerto Rico. J Med Entomol 49, 917–921 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diagne CT et al. Potential of selected Senegalese Aedes spp. mosquitoes (Diptera: Culicidae) to transmit Zika virus. BMC Infect Dis 15, 492, doi: 10.1186/s12879-015-1231-2[pii] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chouin-Carneiro T et al. Differential Susceptibilities of Aedes aegypti and Aedes albopictus from the Americas to Zika Virus. PLoS Negl Trop Dis 10, e0004543, doi: 10.1371/journal.pntd.0004543 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diagne CT et al. Vector competence of Aedes aegypti and Aedes vittatus (Diptera: Culicidae) from Senegal and Cape Verde archipelago for West African lineages of chikungunya virus. Am J Trop Med Hyg 91, 635–641, doi: 10.4269/ajtmh.13-0627 [pii] (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calvez E et al. Dengue-1 virus and vector competence of Aedes aegypti (Diptera: Culicidae) populations from New Caledonia. Parasit Vectors 10, 381, doi: 10.1186/s13071-017-2319-x [pii] (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diallo M et al. Vector competence of Aedes aegypti populations from Senegal for sylvatic and epidemic dengue 2 virus isolated in West Africa. Trans R Soc Trop Med Hyg 102, 493–498, doi: 10.1016/j.trstmh.2008.02.010S0035-9203(08)00063-1 [pii] (2008). [DOI] [PubMed] [Google Scholar]
- 19.Black W. C. t. et al. Flavivirus susceptibility in Aedes aegypti. Arch Med Res 33, 379–388, doi:S0188-4409(02)00373-9 [pii] (2002). [DOI] [PubMed] [Google Scholar]
- 20.Hardy JL in The Arboviruses: Epidemiology and Ecology Vol. 1 (ed Monath TP) 87–126 (CRC Press, 1988). [Google Scholar]
- 21.Franz AW, Kantor AM, Passarelli AL & Clem RJ Tissue Barriers to Arbovirus Infection in Mosquitoes. Viruses 7, 3741–3767, doi: 10.3390/v7072795 [pii] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vazeille M, Dehecq JS & Failloux AB Vectorial status of the Asian tiger mosquito Aedes albopictus of La Reunion Island for Zika virus. Med Vet Entomol, doi: 10.1111/mve.12284 (2017). [DOI] [PubMed] [Google Scholar]
- 23.O’Donnell KL, Bixby MA, Morin KJ, Bradley DS & Vaughan JA Potential of a Northern Population of Aedes vexans (Diptera: Culicidae) to Transmit Zika Virus. J Med Entomol 54, 1354–1359, doi: 10.1093/jme/tjx0873797264 [pii] (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okuda K et al. Cell death and regeneration in the midgut of the mosquito, Culex quinquefasciatus. J Insect Physiol 53, 1307–1315, doi: 10.1016/j.jinsphys.2007.07.005 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Dong S et al. Chikungunya virus dissemination from the midgut of Aedes aegypti is associated with temporal basal lamina degradation during bloodmeal digestion. PLoS Negl Trop Dis 11, e0005976, doi: 10.1371/journal.pntd.0005976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kantor AM, Grant DG, Balaraman V, White TA & Franz AWE Ultrastructural Analysis of Chikungunya Virus Dissemination from the Midgut of the Yellow Fever Mosquito, Aedes aegypti. Viruses 10, doi:E571 [pii] 10.3390/v10100571 [pii] (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weaver SC, Scott TW, Lorenz LH, Lerdthusnee K & Romoser WS Togavirus-associated pathologic changes in the midgut of a natural mosquito vector. J Virol 62, 2083–2090 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delatte H et al. Blood-feeding behavior of Aedes albopictus, a vector of Chikungunya on La Reunion. Vector Borne Zoonotic Dis 10, 249–258, doi: 10.1089/vbz.2009.0026 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Egizi A, Healy SP & Fonseca DM Rapid blood meal scoring in anthropophilic Aedes albopictus and application of PCR blocking to avoid pseudogenes. Infect Genet Evol 16C, 122–128, doi:S1567–1348(13)00025–7 [pii] 10.1016/j.meegid.2013.01.008 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Dickson LB, Sanchez-Vargas I, Sylla M, Fleming K & Black W. C. t. Vector competence in West African Aedes aegypti Is Flavivirus species and genotype dependent. PLoS Negl Trop Dis 8, e3153, doi: 10.1371/journal.pntd.0003153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosio CF, Beaty BJ & Black W. C. t. Quantitative genetics of vector competence for dengue-2 virus in Aedes aegypti. Am J Trop Med Hyg 59, 965–970 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Kramer LD, Hardy JL, Presser SB & Houk EJ Dissemination barriers for western equine encephalomyelitis virus in Culex tarsalis infected after ingestion of low viral doses. Am J Trop Med Hyg 30, 190–197 (1981). [DOI] [PubMed] [Google Scholar]
- 33.Houk EJ, Hardy JL & Chiles RE Permeability of the midgut basal lamina in the mosquito, Culex tarsalis Coquillett (Insecta, Diptera). Acta Trop 38, 163–171 (1981). [PubMed] [Google Scholar]
- 34.Perkins TA, Siraj AS, Ruktanonchai CW, Kraemer MUG & Tatem AJ Model-based projections of Zika virus infections in childbearing women in the Americas. Nature Microbiology 1, 16126, doi:10.1038/nmicrobiol.2016.12610.1038/nmicrobiol.2016.126https://www.nature.com/articles/nmicrobiol2016126#supplementary-informationhttps://www.nature.com/articles/nmicrobiol2016126#supplementary-information (2016). [DOI] [PubMed] [Google Scholar]
- 35.Salazar MI, Richardson JH, Sanchez-Vargas I, Olson KE & Beaty BJ Dengue virus type 2: replication and tropisms in orally infected Aedes aegypti mosquitoes. BMC Microbiol 7, 9, doi: 10.1186/1471-2180-7-9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer RP, Hardy JL & Presser SB Comparative vector competence of Culex tarsalis and Culex quinquefasciatus from the coachella, imperial, and San Joaquin Valleys of California for St. Louis encephalitis virus. Am J Trop Med Hyg 32, 305–311, doi: 10.4269/ajtmh.1983.32.305 (1983). [DOI] [PubMed] [Google Scholar]
- 37.Turell MJ Reduced Rift Valley fever virus infection rates in mosquitoes associated with pledget feedings. Am J Trop Med Hyg 39, 597–602, doi: 10.4269/ajtmh.1988.39.597 (1988). [DOI] [PubMed] [Google Scholar]
- 38.Hsieh P & Robbins PW Regulation of asparagine-linked oligosaccharide processing. Oligosaccharide processing in Aedes albopictus mosquito cells. J Biol Chem 259, 2375–2382 (1984). [PubMed] [Google Scholar]
- 39.Armstrong PM & Rico-Hesse R Differential susceptibility of Aedes aegypti to infection by the American and Southeast Asian genotypes of dengue type 2 virus. Vector Borne Zoonotic Dis 1, 159–168, doi: 10.1089/153036601316977769 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Airs PM, Kudrna KE & Bartholomay LC Impact of sugar composition on meal distribution, longevity, and insecticide toxicity in Aedes aegypti. Acta Trop 191, 221–227, doi: 10.1016/j.actatropica.2019.01.005 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Waggoner JJ et al. Viremia and Clinical Presentation in Nicaraguan Patients Infected With Zika Virus, Chikungunya Virus, and Dengue Virus. Clin Infect Dis 63, 1584–1590, doi:ciw589 [pii] 10.1093/cid/ciw589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Musso D et al. Molecular detection of Zika virus in blood and RNA load determination during the French Polynesian outbreak. J Med Virol 89, 1505–1510, doi: 10.1002/jmv.24735 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haese NN et al. Animal Models of Chikungunya Virus Infection and Disease. J Infect Dis 214, S482–S487, doi:jiw284 [pii] 10.1093/infdis/jiw284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson JF, Main AJ, Delroux K & Fikrig E Extrinsic incubation periods for horizontal and vertical transmission of West Nile virus by Culex pipiens pipiens (Diptera: Culicidae). J Med Entomol 45, 445–451 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Lanciotti RS et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14, 1232–1239, doi: 10.3201/eid1408.080287 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alm E et al. Universal single-probe RT-PCR assay for diagnosis of dengue virus infections. PLoS Negl Trop Dis 8, e3416, doi: 10.1371/journal.pntd.0003416PNTD-D-14-01106 [pii] (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lanciotti RS et al. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis 13, 764–767, doi: 10.3201/eid1305.070015 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan M & Johansson MA The incubation periods of Dengue viruses. PLoS One 7, e50972, doi: 10.1371/journal.pone.0050972 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferguson NM et al. Countering the Zika epidemic in Latin America. Science 353, 353–354, doi: 10.1126/science.aag0219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.rjags: Bayesian Graphical Models using MCMC v. R package version 4–6 (2016). [Google Scholar]
- 51.R: A language and environment for statistical computing. (http://www.R-project.org/. R Foundation for Statistical Computing, Vienna, Austria, 2014). [Google Scholar]
- 52.Brooks SP & Gelman A General Methods for Monitoring Convergence of Iterative Simulations. Journal of Computational and Graphical Statistics 7, 434–455, doi: 10.1080/10618600.1998.10474787 (1998). [DOI] [Google Scholar]
- 53.Christofferson RC & Mores CN Estimating the magnitude and direction of altered arbovirus transmission due to viral phenotype. PLoS One 6, e16298, doi: 10.1371/journal.pone.0016298 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith DL et al. Ross, Macdonald, and a Theory for the Dynamics and Control of Mosquito-Transmitted Pathogens. PLoS Pathog. 8, e1002588, doi: 10.1371/journal.ppat.1002588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chowell G et al. Using Phenomenological Models to Characterize Transmissibility and Forecast Patterns and Final Burden of Zika Epidemics. PLoS Curr 8, doi: 10.1371/currents.outbreaks.f14b2217c902f453d9320a43a35b9583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Funk S et al. Comparative Analysis of Dengue and Zika Outbreaks Reveals Differences by Setting and Virus. PLOS Neglected Tropical Diseases 10, e0005173, doi: 10.1371/journal.pntd.0005173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.lhs: Latin Hypercube Samples. v. R package. version 0.14. (2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.