Abstract
A large European multi-country Salmonella enterica serovar Enteritidis outbreak associated with Polish eggs was characterized by whole-genome sequencing (WGS)-based analysis, with various European institutes using different analysis workflows to identify isolates potentially related to the outbreak. The objective of our study was to compare the output of six of these different typing workflows (distance matrices of either SNP-based or allele-based workflows) in terms of cluster detection and concordance. To this end, we analysed a set of 180 isolates coming from confirmed and probable outbreak cases, which were representative of the genetic variation within the outbreak, supplemented with 22 unrelated contemporaneous S . enterica serovar Enteritidis isolates. Since the definition of a cluster cut-off based on genetic distance requires prior knowledge on the evolutionary processes that govern the bacterial populations in question, we used a variety of hierarchical clustering methods (single, average and complete) and selected the optimal number of clusters based on the consensus of the silhouette, Dunn2, and McClain–Rao internal validation indices. External validation was done by calculating the concordance with the WGS-based case definition (SNP-address) for this outbreak using the Fowlkes–Mallows index. Our analysis indicates that with complete-linkage hierarchical clustering combined with the optimal number of clusters, as defined by three internal validity indices, the six different allele- and SNP-based typing workflows generate clusters with similar compositions. Furthermore, we show that even in the absence of coordinated typing procedures, but by using an unsupervised machine learning methodology for cluster delineation, the various workflows that are currently in use by six European public-health authorities can identify concordant clusters of genetically related S . enterica serovar Enteritidis isolates; thus, providing public-health researchers with comparable tools for detection of infectious-disease outbreaks.
Keywords: whole-genome sequencing, epidemiology, surveillance, infectious disease, hierarchical clustering, unsupervised machine learning
Data Summary
The whole-genome sequencing data of the isolates used in this study are available in various public genomic databases, under the accession numbers indicated in Table S1 (available with the online version of this article). The distance matrices of these isolates, which were used in the statistical analysis, are included in Tables S2–S7.
Impact Statement.
Detection of closely related isolates is key in surveillance and control of pathogenic bacteria, as it allows tracing of potential outbreaks of infectious disease. While harmonization of workflows for genetic typing of the micro-organisms would facilitate the detection, in practice, the workflows are diverse in input and implementation. Therefore, it becomes relevant to assess their comparability. Our research shows that six distinct workflows, in use by several European institutions, can identify concordant clusters of genetically related Salmonella enterica serovar Enteritidis; thus, allowing identification of cross-border outbreaks. By using an unsupervised machine learning methodology and internal validation indices, we show that it is possible to detect an optimal number of clusters that separate outbreak from non-outbreak isolates. The clustering of the data can be done without a predefined distance for delineation of clusters and, thus, independently of knowledge from prior outbreaks.
Introduction
Foodborne pathogens are an important cause of morbidity and mortality, with non-typhoidal Salmonella enterica being one of the major foodborne disease agents. In 2017, 91 662 human cases of salmonellosis were reported in the European Union, representing an increase relative to recent previous years where the salmonellosis notification rates have shown a decreasing trend [1, 2].
Microbial typing is an essential tool for informing infectious-disease epidemiological investigation, providing a method for identification, surveillance, outbreak investigation and tracing of pathogenic micro-organisms. Microbial typing involves clustering – a form of unsupervised pattern recognition, which can be used to partition the input information (genetic information of the strains of interest) into clusters and has been applied to taxonomic challenges in biology for over 50 years [3]. Assigning isolates to genetic clusters composed of strains likely to be epidemiologically related often utilizes distance-based clustering approaches such as hierarchical agglomerative clustering [4–9], with average and single linkage, or modifications hereof, often being the preferred methods [4, 10, 11]. While more accurate and modern methods of inferring phylogenetic relationships among microbial isolates exist, such as maximum parsimony [12], maximum likelihood [13] and Bayesian [14], hierarchical clustering has the advantage of shorter computational time, so that it is even used in newly developed tools aiming to analyse large collections of microbial isolates [15]. In the process of assigning isolates to a certain cluster, a distance or similarity cut-off/threshold is typically used for cluster definition [16, 17]. Appropriate selection of similarity thresholds for clustering depends on interpreting variability in mutation rate, recombination rate and surveillance strategies across different microbial species and environmental settings. Furthermore, due to differences in the resolution of the various whole-genome sequencing (WGS) typing approaches and workflows, a direct comparison based on a fixed threshold is often not possible [18].
The increasing speed and decreasing operational and acquisition cost of WGS have made it a powerful high-resolution tool for epidemiological surveillance and outbreak investigations, and it is rapidly replacing traditional phenotyping and genotyping methods [19–23]. However, the methodological variety by which WGS data can be analysed may represent a significant hurdle in providing epidemiologists and decision makers with robust, interpretable information for action. A widely used analytical approach for WGS typing is based on protein-encoding alleles. Whole-genome multilocus sequence typing (wgMLST) generally takes an assembled genome as input and the alleles are subsequently identified based on nucleotide identity to a defined scheme [24, 25]. Downstream phylogenetic analysis is performed on the sequences of the shared loci or using distances computed based on the number of shared alleles, which are subsequently clustered. Another analytical approach is variant calling, which can be reference-free [26, 27] or reference-based, with the latter being more common in practice. Reference-based variant calling involves aligning the sequenced reads to a closely related reference genome (mapping) to identify SNPs [26–28]. The DNA sequences shared between the sequenced isolates and the reference genome can then be analysed based on differing SNPs. Currently, both SNP [17, 28, 29] and allele-based [30–34] workflows are routinely used for outbreak investigation and surveillance purposes. The pragmatics of the routine epidemiological surveillance of various potentially pathogenic micro-organisms is that, often, there is not a single, standardized workflow being used by different public-health institutions; instead, each institution develops its own workflow, according to the local resources and necessities. It is, however, unclear how robust some of these WGS-based typing workflows (i.e. the SNP- and gene-based workflows that are the subject of our analysis) are with respect to quantifying relatedness of microbial isolates and whether the clusters generated can be unambiguously used for epidemiological surveillance. This is particularly important since differences in cluster definition and similarity thresholds (i.e. at what point can isolates be considered part of the same cluster) can impact on case definitions and cluster composition for outbreak detection and outbreak investigation. Across the world, researchers are addressing this issue by comparing workflows commonly used in practice by various institutions for different micro-organisms [5, 7, 18, 35].
The objective of the present study was to compare the output of six SNP- or allele-based typing workflows currently in use by European public-health institutions in terms of cluster detection, and to establish the parameters and methodology that can facilitate the comparison of these workflows. To this end, we used a dataset selected from a recent large-scale multi-country outbreak of S . enterica serovar Enteritidis [8, 36] supplemented by a number of unrelated isolates for the genetic context, on which we used hierarchical agglomerative clustering with three different linkage methods: single, average and complete.
Methods
Strain set
The dataset analysed here is compiled from available whole-genome sequences from a large-scale European outbreak with S. enterica serovar Enteritidis sequence type (ST)11 spanning 14 countries [37]. A probable case was defined as a laboratory-confirmed S . enterica serovar Enteritidis infection with outbreak multiple loci variable-number tandem repeat analysis (MLVA) profiles 2-9-7-3-2 or 2-9-6-3-2 that occurred from 1 May 2016 through 1 October 2017. A confirmed case was considered to be an infection with a S . enterica serovar Enteritidis isolate sharing the same single-linkage t5-level SNP address based on WGS analysis using SNP workflow 1 described in the next section [38] (i.e. isolates that cluster together using single-linkage hierarchical clustering and a cut-off/threshold value of five SNPs for defining clusters) that occurred from 1 May 2016 through 1 October 2017; this definition was subsequently reinforced by epidemiological investigation [8]. As a result of using this ‘gold-standard’ definition, two major single-linkage clusters were defined as outbreak clusters – cluster 1 and cluster 2 [8, 36]. In total, isolates from 175 confirmed and 5 probable cases were selected for the current study, based on their representativeness for the genetic diversity observed during the outbreak. A number of other whole-genome sequences (n=22), not linked to the outbreak, were selected to reflect the genetic diversity of S . enterica serovar Enteritidis ST11 isolates with the MLVA profiles 2-9-7-3-2 or 2-9-6-3-2 circulating during the timespan of the outbreak in the representative countries [17]. Genetic diversity was assessed based on single-linkage clustering [4].
Sequencing and workflows
Six different post-sequencing bioinformatics workflows were used to analyse the dataset. These workflows were used for epidemiological surveillance during the outbreak where we drew our dataset from, and they are still in use by the respective institutions. The results of all workflows were expressed as Hamming distance matrices [39] among the bacterial isolates within the dataset. These distance matrices were used to test the concordance of the different workflows. For readability and consistency, we will use throughout the article the term 'workflow' to refer to the distance matrix it generated.
SNP-workflow 1 (SNP1)
Paired-end fastq files were quality trimmed and STs, based on the 7-loci multilocus sequence typing (MLST) scheme, determined using most software v 1.0 (github.com/phe-bioinformatics/MOST). vcf files were created using PHEnix software (github.com/phe-bioinformatics/PHEnix): short reads were mapped to an internal reference genome for S . enterica serovar Enteritidis, GenBank accession number AM933172, using bwa-mem [40]. sam files from bwa were sorted and indexed into bam files using SAMtools [41]. gatk v2.7 [42] was run in UnifiedGenotyper mode to create the vcf files with high-quality SNPs (>90 % consensus, minimum depth 10×, Mapping Quality (MQ)≥30). The total number of variant positions (1036) was used in the calculation of the Hamming distance matrix. Isolates Eng23, NL3, Scot3 and Scot28 were omitted from the distance matrix calculation as they appeared to be mixed sequences of more than one bacterial strain. SNP distances and clusters were determined and stored using SnapperDB software [4]. Single-linkage clustering was performed on the pairwise SNP difference between all isolates at 7 distance thresholds (250, 100, 50, 25, 10, 5 and 0 SNPs), resulting in a 7 digit ‘SNP address’ for each isolate, where each number confers membership in a cluster at each distance threshold [4].
SNP-workflow 2 (SNP2)
The fastq files were analysed as described elsewhere [43]. Briefly, the raw reads were trimmed using Trimmomatic v0.35 (the first two bases from every read, TruSeq adapters, reads <36 bp removed, reads clipped when the mean quality over 3 bp <22) and mapped to the reference (GenBank accession number AM933172) using bwa-mem v0.7.12–5 [40]. After marking duplicates with Picard v1.113–2, SNPs were called using gatk v3.60 [42] with default settings. The high-quality SNP profiles were converted to fasta (>90 % consensus, minimum depth 10×). Isolates Eng8, Eng59 and Scot63 were removed from further analyses as they yielded poor assemblies (>300 contigs). The differences between isolates were determined, ignoring mismatches between filtered SNPs. The total number of SNPs used in the calculation of the Hamming distance matrix was 1128.
cgMLST workflow 1 (MLSTcg1) and wgMLST workflow (MLSTwg)
The raw reads were trimmed to remove adaptor and barcode sequences (added during library generation) and low-quality reads using Trimmomatic v0.36 (min. Phred score 25). They were assembled with SPAdes v3.7.1 in BioNumerics version v7.6.2 (bioMérieux) including post-assembly optimization by mapping reads back onto the assembly and keeping the consensus. The cgMLST and wgMLST analyses were done based on the assembly as well as assembly-free calls using the schemes in BioNumerics including 3002 and 15 874 loci, respectively. Isolates Eng8, Eng51, Eng52, Eng63, Scot6, Scot7, Scot8, Scot9 and Scot14 were excluded from the analysis as <90 % of the core loci were detected in each of them. In addition, isolate NL3, with almost double the expected genome length and high number of loci with multiple alleles, was excluded. The Hamming distance matrix among the isolates was calculated with pairwise removal of the missing loci.
cgMLST workflow 2 (MLSTcg2)
fastq files were uploaded to EnteroBase (http://enterobase.warwick.ac.uk). Once received on the EnteroBase server, reads were parsed with metadata using MetaParser and then automatically processed using a versioned pipeline (V3). QAssembly, the assembly process, included read pre-processing, quality trimming for short reads using Sickle (keeping a maximum of 120× coverage for assembly), assembly using SPAdes and bwa, post-correction and filtering. Low-level contamination was removed and the quality checked using Kraken (v0.10.5-beta) [44]. Once the assembly had been carried out and passed the quality-control criteria (number of bases between 4 and 5.8Mbp, the sequence length of the shortest contig at 50% of the total genome length (N50) >20 kb, no. of contigs <600, proportion of Ns <3 %, correct species assignment in Kraken), cgMLST typing was performed using the 3002 loci EnteroBase cgMLST V2 [25]. The fasta files were used to carry out a blast search (version 2.2.31), a blastn search was carried out with the nucleotide reference sequences, and a usearch (version 8.0.1623) search was carried out with any available amino acid reference sequences. The search results were combined and parsed, and from this the alleles at the loci of interest in the target sequences were identified. Isolates Eng8, Eng51, Eng52, Eng63, Scot6, Scot7, Scot8, Scot9 and Scot14 were excluded from the analysis as <90 % of the core loci were detected in each of them. In addition, isolate NL3, with almost double the expected genome length and high number of loci with multiple alleles, was excluded. The Hamming distance matrix among the isolates was calculated with pairwise removal of the missing loci.
Ad hoc cgMLST workflow 3 (MLSTcg3)
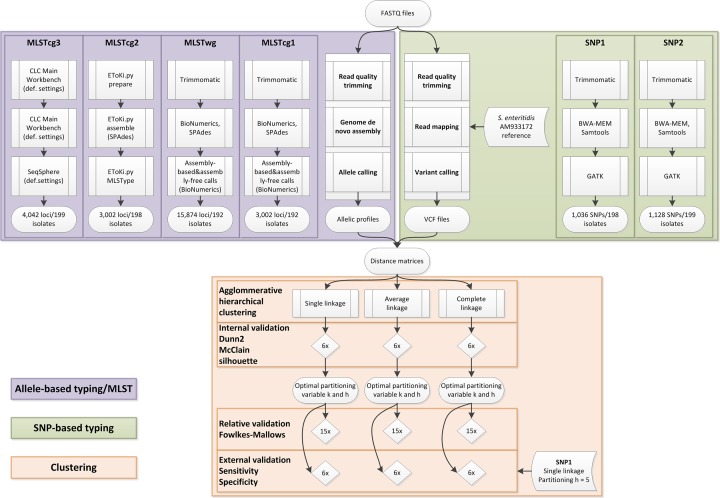
Paired-end fastq files were first quality trimmed and then de novo assembled using CLC Main Workbench (Qiagen; version 10.0) with default settings. Ridom SeqSphere software (Ridom; version 4.1.9) was used to define an ad hoc cgMLST set and to perform the gene-by-gene analysis. Based on 7-loci MLST, a closest finished reference genome was selected: strain P125109 (GenBank accession number NC_011294). To determine the cgMLST gene set, a genome-wide gene-by-gene comparison was performed using the MLST_target definer function of SeqSphere with default parameters [45]. Alleles for each locus were automatically assigned by the SeqSphere+ software to ensure a unique nomenclature. Isolates Eng8, Eng59 and Scot63 were removed as they showed less than 90 % core genome similarity with S . enterica serovar Enteritidis. The total number of loci used in the calculation of the distance matrix was 4042. The Hamming distance matrix among the isolates was calculated using the parameter ‘pairwise ignore missing values’. A summary representation of the workflows and statistical analysis can be found in Fig. 1.
Fig. 1.
Flowchart of the methodology used for the analysis of the bacterial isolates selected for this study.
Correlation of genetic distances
As a first indicator of the congruence of the six typing workflows, we calculated the linear correlations of the genetic distances in the corresponding distance matrices (lower half of the matrices, with diagonals excluded) as Spearman correlation coefficients. The higher values of this index indicate a better congruence of the compared typing workflows.
Clustering and internal validation
The comparison among the six distance matrices was performed using the hierarchical clustering with three of the most commonly used linkage criteria: single [46], average [47] and complete [48]. In order to assess the fit of the clustering to the distance matrix, we calculated the pairwise cophenetic correlation [3, 49] for each of the three clustering algorithms used. Cophenetic correlation is a linear correlation coefficient between the cophenetic distances obtained from the tree, and the original distances used to construct the tree. In other words, it is a measure of how faithfully a dendrogram preserves the original pairwise distances in the distance matrix. We compared the goodness of fit of the three clustering algorithms by means of pairwise Pearson correlation for dependent groups with overlapping variables, as implemented in the ‘cocor’ package v. 1.1–3 [50], with P<0.05 indicating a significant difference.
One of the common practices in clustering of microbial isolates for epidemiological purposes is using a pre-set distance threshold that has been derived from previous outbreaks [5, 8, 51]. While the comparison of two molecular typing workflows with the same resolution might be straightforward, allowing the use of identical thresholds for cluster delineation, it becomes less so when the units used in the typing process and the resolution are different. The concordance of any two workflows is then dependent on how a cluster is defined.
The selection of the optimal number of clusters (internal validation) was based on the consensus of three internal validity indices: silhouette [52], McClain–Rao [53], and Dunn2 index [54]. These were calculated using the functions NbClust and cluster.stats in packages NbClust v. 3.0 [55] and fpc v. 2.1–11.1 [56]. The silhouette index is a measure of how appropriately the data have been clustered. For each partition, the average silhouette is the average of the silhouettes for all objects in the dataset; that is in turn defined as the difference between the average distance of an object to any other object within the same cluster and the smallest average distance of the object to all objects in any other cluster [52]. The McClain–Rao index is defined as the quotient between the mean within-cluster and between-cluster distances [53]. The Dunn2 index measures the ratio between the minimum average dissimilarity between two clusters and the maximum average within-cluster dissimilarity [54]. We calculated these internal validity indices for a variable number of clusters, from k=3 to k=20, for all six workflows. The rationale behind these indices is to identify cluster sets that are compact and well separated. Thus, the number of clusters where the Dunn2 and silhouette indices were at maximum values, and the McClain–Rao at minimum values, were considered to be optimal and further analyses were based hereupon. It is, however, seldom that multiple validity indices validate the same number of clusters as the optimum, and it is often a consensus hereof that is used, with values approaching the maximum or the minimum ones, respectively, scoring a higher rank. The consensus of the three validity indices was calculated by aggregated ranking with cross-entropy Monte Carlo algorithm and Kendall distance, as implemented in package RankAggreg v. 0.6.5 [57].
Concordance
Concordance among the six typing workflows (relative validation)
For each of the clustering algorithms and the optimal number of clusters (as identified with the methodology described in the previous section), we performed a relative validation by assessing the concordance of each distance matrix with each of the other five. We represented the concordance between pairs of distance matrices as plots of dendrograms facing each other – also known as tanglegrams – using the function with the same name implemented in package dendextend v. 1.10.0 [58]. For the quantification of the concordance of the branches in the two facing dendrograms, we have calculated Baker’s gamma index [59], which is defined as the rank correlation between the stages at which pairs of objects combine in each of the two dendrograms. While Baker’s gamma gives a global measure of concordance, it cannot assess the concordance of the membership for each of the isolates in the various identified clusters. This second aspect we quantified by using the Fowlkes–Mallows index [60], as implemented in package profdpm v. 3.3 [61]. The visualization of the concordance among all six workflows was built with the function ‘alluvial’ in the package with the same name, v. 0.1–2 [62], and it was based on the optimal number of clusters for each of the clustering algorithms.
Concordance with the outbreak definition (external validation)
Normally, an external dataset is used for validation of the clusters, which in the case of an outbreak would consist of epidemiological data. For this outbreak, the case definition was given based on single linkage with a threshold of five, and identification of two major clusters as outbreak clusters. Therefore, we performed the external validation by comparing the two largest clusters in each of the workflows and clustering method with the outbreak clusters as defined above. We calculated the sensitivity of the WGS workflows as the TP/(TP +FN), and the specificity as the TN/(TN +FP), where TP=the number of true positives, isolates that were identified as coming from outbreak cases in each of the workflows as well as in the external validation dataset, FN=the number of false negatives, isolates that were assigned by each of the workflows as non-outbreak while in the external validation dataset they were defined as coming from outbreak cases, TN=the number of true negatives, isolates that were identified as non-outbreak in each of the workflows as well as in the external validation dataset, and FP=the number of false positives, isolates that were assigned by each of the workflows as coming from outbreak cases while in the external validation dataset they were assigned as non-outbreak. We assessed the concordance of the clusters corresponding to the outbreak clusters 1 and 2 for all workflows in our study by calculating the Fowlkes–Mallows index. All statistical analyses were performed in R version 3.4.3 [63].
Results
Sequence quality control
As part of the quality control, each institution decided to remove some of the isolates from the dataset as these did not correspond to their quality criteria. The resulting distance matrices had, therefore, variable sizes: SNP1 198×198 (Table S2), SNP2 199×199 (Table S3), MLSTcg1 192×192 (Table S4), MLSTcg2 198×198 (Table S5), MLSTcg3 199×199 (Table S6), MLSTwg 192×192 (Table S7). The precise subset of isolates retained by each workflow is indicated in Tables S2–S7 and a summary hereof in Table S1, where the presence of an isolate is indicated by 1 and absence by 0; 187 isolates of the initial dataset were shared by all workflows. We chose not to limit the analyses to the 187 isolates common to all workflows; instead, in order to recreate the commonly occurring situation in practice, where the number of isolates varies among the workflows (depending, among others, on the quality criteria used), we performed the analyses with a variable number of isolates. In pairwise comparisons between the workflows, the number of isolates used was the intersection of the two workflows. The number of SNPs and genes included in the various workflows were also variable and this was reflected in the distances among the isolates (Table 1).
Table 1.
Data used in the clustering analysis and the values corresponding to the optimal partition for each of the three hierarchical clustering methods used
Optimal_k, the optimal number of clusters as identified based on the silhouette, McClain–Rao and Dunn2 index; diameter, the maximum within-cluster distance; separation, minimum between-clusters distance.
|
Workflow |
Clustering |
Optimal_k |
Diameter |
Separation |
|---|---|---|---|---|
|
SNP1 |
Average |
k14 |
14 |
14 |
|
SNP2 |
Average |
k12 |
16 |
3 |
|
MLSTcg1 |
Average |
k12 |
13 |
10 |
|
MLSTcg2 |
Average |
k13 |
15 |
9 |
|
MLSTcg3 |
Average |
k13 |
13 |
12 |
|
MLSTwg |
Average |
k13 |
13 |
10 |
|
SNP1 |
Complete |
k14 |
14 |
14 |
|
SNP2 |
Complete |
k13 |
13 |
1 |
|
MLSTcg1 |
Complete |
k13 |
9 |
6 |
|
MLSTcg2 |
Complete |
k13 |
11 |
7 |
|
MLSTcg3 |
Complete |
k13 |
13 |
12 |
|
MLSTwg |
Complete |
k13 |
18 |
10 |
|
SNP1 |
Single |
k14 |
14 |
14 |
|
SNP2 |
Single |
k12 |
16 |
3 |
|
MLSTcg1 |
Single |
k11 |
17 |
10 |
|
MLSTcg2 |
Single |
k13 |
15 |
9 |
|
MLSTcg3 |
Single |
k13 |
13 |
12 |
|
MLSTwg |
Single |
k13 |
13 |
10 |
Correlation of genetic distances
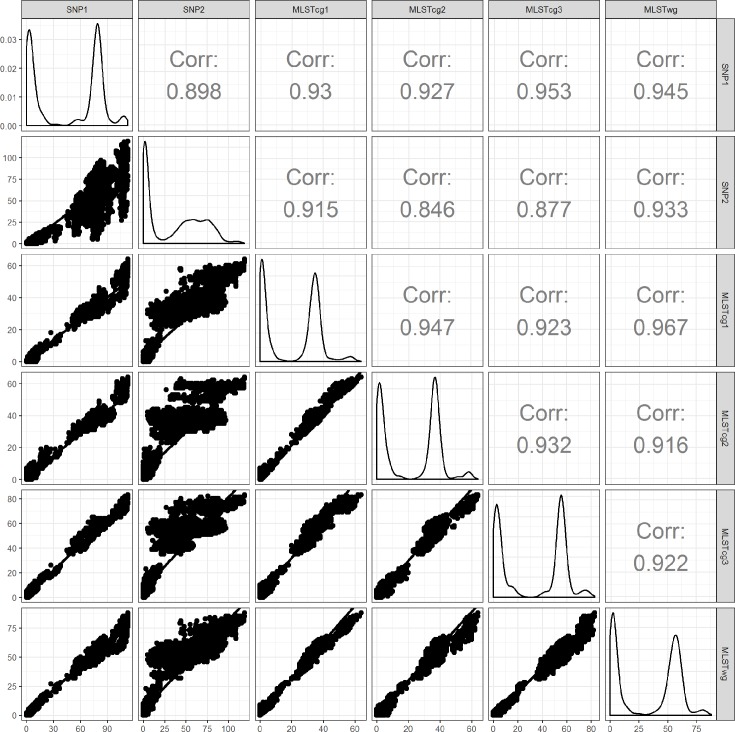
The genetic distances were highly correlated, even between the SNP- and allele-based workflows, with most of the Spearman coefficients of correlation >0.99. The correlation was weaker for the SNP2 distances, where Spearman correlation coefficients had values lower than 0.95, but still above 0.9 (Fig. 2).
Fig. 2.
Pairwise correlation between the genetic distances of the various workflows. The diagonal shows the density plots of the distances in each of the six workflows. The upper half of the plot indicates the Spearman coefficients of correlation for each combination of distance matrices. In the dotplots, the x-axis represents the genetic distance among isolates, as measured by the workflow indicated on the column label; the y-axis represents the genetic distance among isolates, as measured by the workflow indicated in the row label. Only the distances between isolates that are present in both workflows of a pairwise comparison are depicted in the figure.
Clustering
The cophenetic correlation was high for all three methods (Table S8), with the highest median value displayed by the average clustering (0.994 range: 0.925–0.997) and comparable median values for single (0.991 range: 0.607–0.995) and complete (0.991 range: 0.920–0.994) linkage. Average linkage showed significantly better fit to the distance matrices than either single or complete linkage (Table S8). Complete linkage had a better fit for the workflows incorporating the whole genome (wgMLST and SNP), while single linkage had a better fit for the workflows based on a subset of genes (cgMLST; Table S8); the differences between the two methods were, however, not statistically significant. Therefore, we assessed the optimal number of clusters for all three methods.
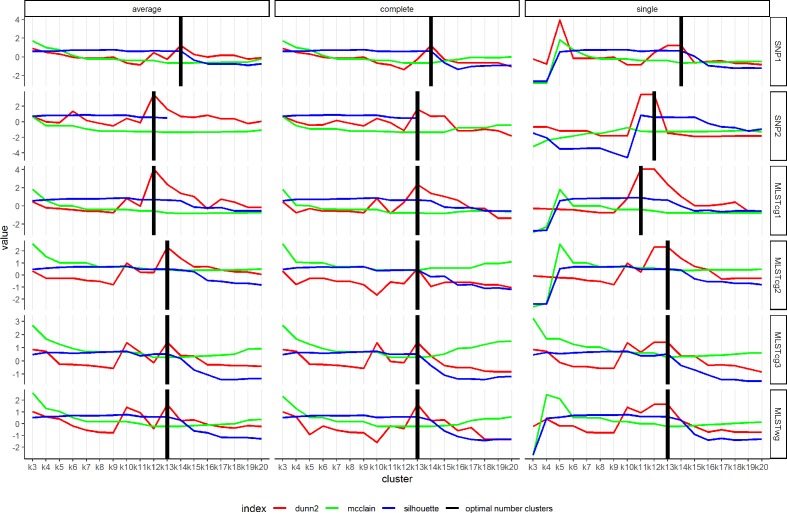
The optimal number of clusters for the various workflows showed higher variations for the average-linkage (12–14) and single-linkage algorithms (11–14), and it was almost unanimous for complete linkage (13 for all workflows and 14 for SNP1) (Table 1, Fig. 3). In the case of the SNP1, MLSTwg, MLSTcg2 and MLSTcg3 workflows, all three clustering algorithms yielded the same optimal number of clusters.
Fig. 3.
Internal validity indices for combinations of workflows and clustering algorithms, for k=3–20. The values of all indices are scaled and re-centred around 0 for better visualization. Maximum values of Dunn2 and silhouette, and minimum values of McClain–Rao indicate optimal number of clusters. The bold black vertical lines indicate the consensus optimal number of clusters as indicated in Table 1. The silhouette index is not defined for k>13 in average- and complete-linkage clustering of the SNP2 workflow.
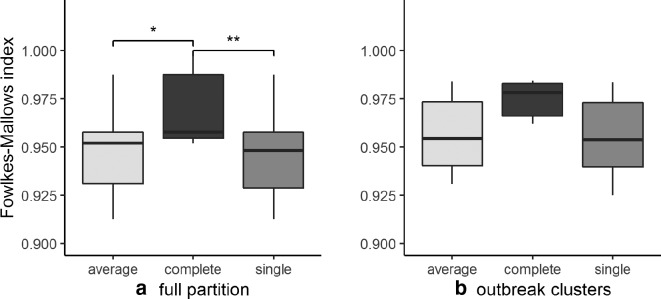
The concordance among the six workflows, at the optimal number of clusters as indicated above, was calculated using the Fowlkes–Mallows index (for reproducibility, all partitions are given in Table S10, for the six workflows, three clustering algorithms used and k=3–20). Complete linkage yielded the best concordance results when applied to the entire dataset, as it would also be intuitively expected from the consistent number of optimal clusters found for all workflows by this linkage algorithm.
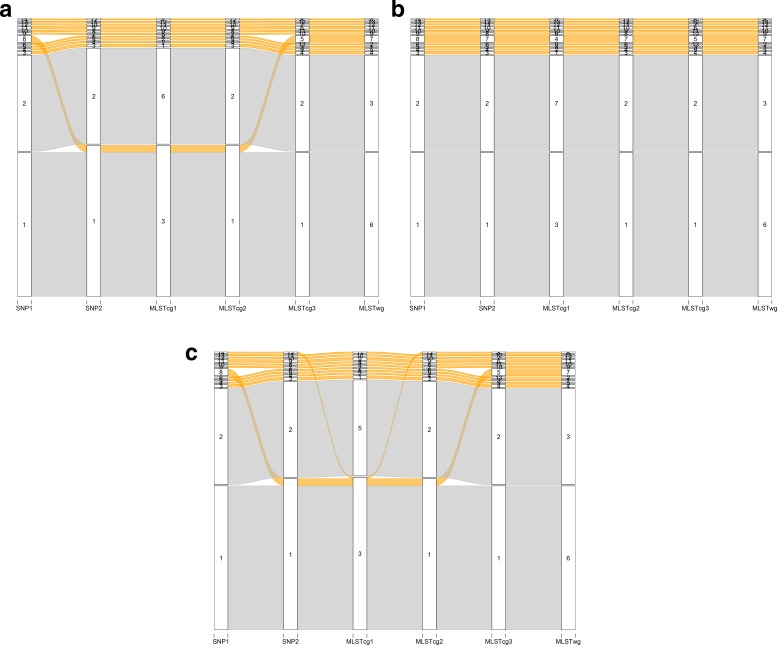
There was no significant difference between the three algorithms regarding the concordance of the outbreak isolates; the confirmed cases falling in clusters 1 and 2 of the outbreak were confirmed by all workflows using all clustering algorithms (Table S9). When average- or single-linkage clustering were employed, SNP1, MLSTcg3 and MLSTwg could distinguish the presence of three clusters, where SNP2, MLSTcg1 and MLSTcg2 could only identify two main ones (Fig. 4a, c). The highest Fowlkes–Mallows index was observed for the pair SNP1:MLSTwg (0.96).
Fig. 4.
Correspondence among the partitions of the six workflows following clustering with one of the following algorithms: (a) average linkage, (b) complete linkage, (c) single linkage. The grey alluvials stand for the outbreak-linked isolates, while the orange alluvials stand for the non-outbreak isolates.
With complete-linkage hierarchical clustering, all workflows placed the isolates in the same clusters, with values of Fowlkes–Mallows index between 0.95 and 1, as can be seen in Fig. 5. Absolute concordance (Fowlkes–Mallows index=1) for the entire partition was observed for the pairs SNP2/MLSTcg3 and MLSTcg1/MLSTwg. The lowest concordance (Fowlkes–Mallows index=0.952) was observed between the partitions MLSTcg1 and MLSTwg and those of MLSTcg3 and SNP2 (Table S11).
Fig. 5.
Summary of Fowlkes–Mallows indices of concordance between any two partitions: (a) for pairwise comparisons of the six partitions, (b) for pairwise comparisons of each of the six partitions with the reference outbreak clusters. The Fowlkes–Mallows index can take values on the interval 0–1, where values closer to 0 indicate absence of correlation, while values closer to 1 indicate close to perfect correlation. The asterisks indicate the P values for pairwise t -test: *difference with P<0.05; **difference with P<0.01.
In pairwise comparisons of the dendrograms corresponding to the six workflows, the concordance, as measured by Baker’s gamma, had mostly values above 0.9; with the average best concordances obtained with complete linkage (Table S11). We observed that for all the distance matrices, the optimal number of clusters in complete linkage was found at a similar threshold for the SNP-based workflows (14 SNP1, 13 SNP2) and at a threshold corresponding to less than 0.04 % allele distance (8–9/3002, 12/4042) – 99.6 % similarity [6] for the MLST-based workflows (Table 1).
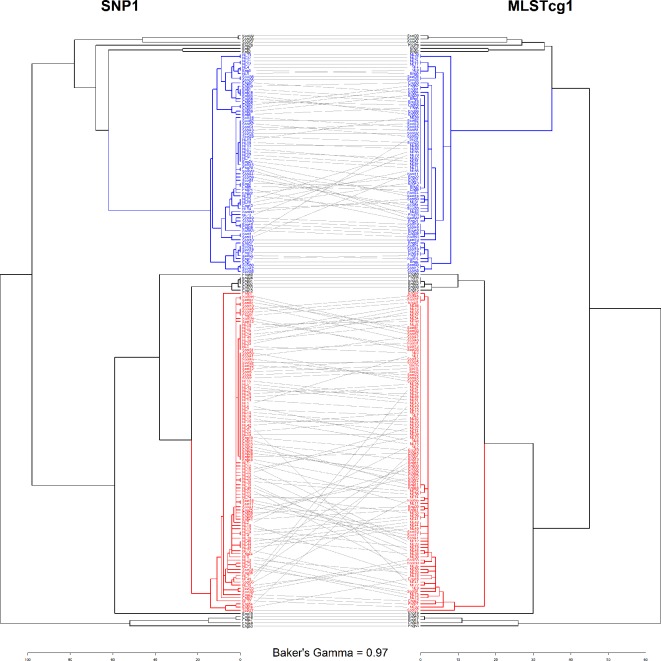
Our data indicate complete concordance in the clustering of the S . enterica serovar Enteritidis based on the genetic distances, when complete linkage is employed (Fig. 3). This is true for SNP-based workflows as well as for allele-based workflows (Figs 6 and S1–S14). The few isolates that change positions in the two dendrograms do so within the main clusters that were identified as optimal.
Fig. 6.
Tanglegram of SNP1 and MLSTcg1 clusterings with complete linkage. The outbreak clusters 1 and 2 are shown in red and blue, respectively.
The sensitivity of all workflows was 100 %. It is the specificity that was in all cases under 100 % (Table S9). When single or average linkage was employed, the specificity varied between 59.1 % for workflow SNP2 and 95.2 % for workflow SNP1. For complete linkage, the minimum specificity was observed for workflows SNP2 and MLSTcg3 (90.9 %), and the maximum for workflows SNP1 and MLSTcg2 (95.2 %).
Discussion
The speed of the molecular typing and the accuracy in identifying related microbial isolates are essential to outbreak detection and investigation. With the availability of WGS, a plethora of genomic typing workflows have emerged in the last years. In practice, different public-health institutes often use different, custom-made, analysis workflows, with small variations in the parameters used, that are not always straightforward to compare and the resulting differences in output difficult to reconcile. This causes, in turn, difficulties in setting single case definitions in multi-country outbreak settings. The question is then how can the outputs of the various workflows be reconciled? Should that not be the case, which workflows are best to use? Harmonization of the methodology to analyse the output data of these workflows is an important aspect in infectious-disease surveillance and consequent control of pathogenic micro-organisms spread. In this study, we attempted to use an objective method to compare the results of six different WGS-based typing workflows, used by different European public-health institutions, in order to see how these differ in terms of clustering results and whether there is a good-better-best hierarchy in the workflows at all.
Modern WGS typing techniques have a clear advantage over the classical genotyping workflows, such as MLST, in terms of discriminatory power. While, based on the 7-loci MLST [64], all 202 isolates belonged to ST11, based on WGS there were between 82, for MLSTcg1, and 132, for MLSTcg3, different genotypes identified. Thus, even among WGS typing techniques there are large differences in the discriminatory power. The intervals within which the distances can vary are smaller for the cgMLST workflows, which can be explained by the fact that cgMLST covers only part of the genome, and it is restricted to coding regions.
Our results indicate that there is a high correlation of the genetic distances among the various distance matrices, regardless of the measurement unit (gene in MLST-based and nucleotide in SNP-based workflows). However, the relations between the genetic distances as inferred from the various workflows are not always linear. It is generally the case that the differences among the output generated by various workflows stem from variations in the reference used, the values of filtering used for quality and coverage, and the inclusion or exclusion of high-density variants, or could be potentially generated by recombination processes (not analysed here). Thus, the choices made within the WGS typing workflow will lead to differences in distance matrices that are not amenable to using the same cut-off/threshold for defining clusters of potentially epidemiologically linked isolates. The shape and size of the clusters could be in these situations defined using internal and relative validation of the clustering.
No clustering method is applicable to all types of genetic data and all outbreak situations. An effective and pragmatic approach is to compute various partitions with different algorithms and to choose the one that best fits the data. While single linkage [46] is the method of choice for epidemiological studies and has been shown to be the best method for describing the genetic relationships between populations in a broad range of evolutionary histories [11], it is known to have a tendency to produce ‘long thin’ clusters in which nearby elements of the same cluster have small distances, but elements at opposite ends of a cluster may be much farther from each other than two elements of different clusters [47]. This behaviour arises since the similarity of two clusters is based on the minimum pairwise distance of their members. In complete linkage [48], the similarity of two clusters is based on the smallest maximum pairwise distance of their members, which means that the entire structure of the data is reflected in the clustering. Its tendency to reduce the intracluster distances will lead to well-structured, elliptical clusters. A compromise between the two methods is the average linkage [65], and it has been one of the most popular distance-based clustering methods for phylogenetic studies [10].
Our analysis aimed to verify the concordance of various WGS typing workflows, either SNP- or allele-based, using the example of a recent European S. enterica serovar Enteritidis outbreak, and applying the three clustering algorithms named above. On this dataset, average linkage had the best fit to the distance matrices, above single or complete linkage. One of the indications of concordance of two datasets is a comparable if not equal number of clusters. For the concordance of our six workflows, concordant results were obtained with average and single linkage. For both clustering algorithms, workflows making use of a smaller number of genetic loci – SNP2, MLSTcg1, MLSTcg2 – merged two of the clusters that were deemed well separated in the SNP1, MLSTcg3 and MLSTwg (workflows based on a higher number of genetic loci). This indicates that, even among the WGS typing workflows, variations in resolution will occur and they are relevant in the process of clustering. This is illustrated also in Fig. 3, where the silhouette index is no longer defined for cluster values higher than 13 for workflow SNP2, which had an overall lower resolution. Complete linkage yielded the best relative concordance among the workflows, as it allowed the identification of a similar number of clusters in all the datasets. This is due to the fact that complete linkage tends to produce well defined, elliptical clusters [47]. Thus, the chained clusters produced by single linkage, and to a lesser extent by average linkage, and responsible for the lower specificities observed with these algorithms (Table S9), were split in complete linkage.
As consequence of the aforementioned properties, complete-linkage clustering yielded the best results in the external validation process (Table S9), with lower specificity in single linkage due to the chaining property of this linkage algorithm. The small discordance (one isolate, resulting in a specificity of 94.7–95.2 %) of the complete-linkage partitions with the outbreak case definition is due to the fact that the latter used single-linkage clustering with a 5 SNPs distance threshold, originally selected as it could be shown empirically that isolates in the same 5 SNP cluster share a common source [66]; this threshold corresponds in our analysis to k=15. In our approach, the threshold was variable, dependent on the workflow, with up to 14 SNPs for the SNP-based workflows. The use of a threshold requires previous knowledge on the evolutionary processes that govern the bacterial populations in various environments, and knowledge on each particular outbreak; it is, therefore, cumbersome to infer for all the combinations micro-organism and source, requiring extensive epidemiological validations. The use of internal validation indices makes our approach more general, as it allows a transparent identification of the optimal number of clusters, independent of prior outbreaks. The clustering of the data becomes, thus, a dynamic process, and can accommodate the inclusion or removal of isolates, without the need of a predefined distance threshold for delineation of the clusters. The even lower specificity (90.9%, two isolates) for the SNP2 and MLSTcg3 when using complete-linkage clustering (Table S9) was due to the inclusion in one of the outbreak clusters of the isolate NL3; this isolate was excluded from the other distance matrices as it showed a mixed nucleotide sequence due to contamination with other species than S . enterica serovar Enteritidis. This situation indicates that even complete-linkage clustering is not sensitive enough to exclude such outliers from the outbreak clusters. We argue, however, that this situation could be easily avoided by a careful curation (preliminary screening) of the data. In this case, NL3 was already divergent from the other isolates based on the classical 7-loci MLST scheme. Furthermore, if the purpose of the clustering is identification of epidemiologically related isolates, the dataset can be filtered as to contain only isolates belonging to the same clonal complex. In order to identify mixes of bacteria belonging to the same species or even clonal complex, methods directed towards identification of heterozygous SNP positions in the genome assembly could be used in the future [67, 68].
The comparison of the six WGS typing workflows indicated a high concordance in clustering the S . enterica serovar Enteritidis isolates within ST11, regardless of whether they are reference-based (SNP workflows) or not (MLST workflows); in exemplification hereof, the tanglegram of the SNP1 and MLSTcg1 workflows clustered with complete linkage is shown in Fig. 6. The concordance between the SNP-based and MLST-based typing workflows has also been described for other micro-organisms [7, 18, 35, 69]. It is not surprising that the two approaches would agree with each other, as recombination is considered to play a minor role in the evolution of S . enterica serovar Enteritidis [70]; the only differences in clonal bacteria would stem from the point mutations in the non-coding regions. Indeed, this was reflected in the distances corresponding to the workflows in our analysis, with smaller distances in the cgMLST and wgMLST than in the SNP matrices (Fig. 2). There were, however, differences in the relative concordance attained with different clustering algorithms, and these stemmed primarily from the resolution of the workflows.
WGS-based typing has become a regular tool in infectious-disease epidemiology, complementary to classical epidemiological investigations. Harmonization of the typing schemes and workflows for pathogenic micro-organisms is, of course, desirable for reproducible results that would allow congruent and timely intervention measures for disease control. There are already international efforts underway in that direction [71]. However, the allele- and SNP-based typing methods offer, to a certain extent, complementary advantages and information, which is an argument for their parallel use. MLST is an internationally accepted standardized procedure, relatively easy to use on routine screenings by public-health laboratories, and with curated databases for the some important food-borne pathogens [6, 24, 25, 71], while SNP typing allows for identification of neutral mutations in the non-coding regions of the genome and has the advantage that it can be fully automated [4, 72]. Our analysis indicates that allele- and SNP-based typing workflows can generate clusters with similar compositions and, consequently, these specific typing workflows in use in Europe for S . enterica serovar Enteritidis have a high concordance. More importantly, we show that the methodology for comparing them can be built on unsupervised machine-learning tools, without much prior knowledge of the epidemiological data. We argue, though, that epidemiological information is crucial for outbreak investigation and that future studies can only benefit from incorporating it. Furthermore, validation of this approach for other datasets, preferably with available epidemiological data, and micro-organisms with different evolutionary strategies is advisable.
To conclude, our analysis shows that, even in the absence of coordinated typing procedures, but using a transparent and objective methodology for cluster delineation, the various workflows that are currently in use by the main European public-health authorities can identify highly concordant clusters of genetically related S. enterica serovar Enteritidis isolates; thus, providing researchers with comparable tools for detection of infectious-disease outbreaks.
Data Bibliography
The WGSs of the isolates used in this study are available in various public genomic databases, under the accession numbers indicated in Table S1 (2019).
Supplementary Data
Funding information
This study was financed by the Dutch Ministry of Health, Welfare and Sports.
Author contributions
E.F. and C.E.C. conceived and designed the analysis. E.F., T.J.D., D.B., M.V. and S.K. have provided resources for the study, in the form of WGSs of the bacterial isolates. H.H., D.B., R.R.V., D.P. and T.V. performed the WGS typing. C.E.C. performed the statistical analysis and wrote the manuscript. All authors reviewed and edited the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: cgMLST, core-genome multilocus sequence typing; MLST, multilocus sequence typing; SNP, single nucleotide polymorphism; ST, sequence type; wgMLST, whole-genome multilocus sequence typing; WGS, whole-genome sequencing.
Eleven supplementary tables and fourteen supplementary figures are available with the online version of this article.
References
- 1.EFSA. ECDC The European Union summary report on trends and sources of zoonoses, zoonotic agents and food‐borne outbreaks in 2016. EFSA J. 2017;15:e05077. doi: 10.2903/j.efsa.2017.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.EFSA. ECDC The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018;16:e05500. doi: 10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sokal RR, Sneath PHA. Principles of Numerical Taxonomy. San Francisco, CA and London: W. H. Freeman; 1963. [Google Scholar]
- 4.Dallman T, Ashton P, Schafer U, Jironkin A, Painset A, et al. SnapperDB: a database solution for routine sequencing analysis of bacterial isolates. Bioinformatics. 2018;34:3028–3029. doi: 10.1093/bioinformatics/bty212. [DOI] [PubMed] [Google Scholar]
- 5.Katz LS, Griswold T, Williams-Newkirk AJ, Wagner D, Petkau A, et al. A comparative analysis of the Lyve-SET phylogenomics pipeline for genomic epidemiology of foodborne pathogens. Front Microbiol. 2017;8:375. doi: 10.3389/fmicb.2017.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moura A, Criscuolo A, Pouseele H, Maury MM, Leclercq A, et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes . Nat Microbiol. 2017;2:16185. doi: 10.1038/nmicrobiol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearce ME, Alikhan N-F, Dallman TJ, Zhou Z, Grant K, et al. Comparative analysis of core genome MLST and SNP typing within a European Salmonella serovar Enteritidis outbreak. Int J Food Microbiol. 2018;274:1–11. doi: 10.1016/j.ijfoodmicro.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pijnacker R, Dallman TJ, Tijsma ASL, Hawkins G, Larkin L, et al. An international outbreak of Salmonella enterica serotype Enteritidis linked to eggs from Poland: a microbiological and epidemiological study. Lancet Infect Dis. 2019;19:778–786. doi: 10.1016/S1473-3099(19)30047-7. [DOI] [PubMed] [Google Scholar]
- 9.Leekitcharoenphon P, Nielsen EM, Kaas RS, Lund O, Aarestrup FM. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica . PLoS One. 2014;9:e87991. doi: 10.1371/journal.pone.0087991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenstein J. Inferring Phylogenies. Sunderland, MA: Sinauer; 2003. [Google Scholar]
- 11.Kalinowski ST. How well do evolutionary trees describe genetic relationships among populations? Heredity. 2009;102:506–513. doi: 10.1038/hdy.2008.136. [DOI] [PubMed] [Google Scholar]
- 12.Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool. 1971;20:406–416. doi: 10.2307/2412116. [DOI] [Google Scholar]
- 13.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 14.Rannala B, Yang Z. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol. 1996;43:304–311. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Z, Alikhan N-F, Sergeant MJ, Luhmann N, Vaz C, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kluytmans-van den Bergh MFQ, Rossen JWA, Bruijning-Verhagen PCJ, Bonten MJM, Friedrich AW, et al. Whole-genome multilocus sequence typing of extended-spectrum-beta-lactamase-producing Enterobacteriaceae . J Clin Microbiol. 2016;54:2919–2927. doi: 10.1128/JCM.01648-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashton P, Nair S, Peters T, Tewolde R, Day M, et al. Revolutionising public health reference microbiology using whole genome sequencing: Salmonella as an exemplar. bioRxiv. 2015 doi: 10.1101/033225. [DOI] [Google Scholar]
- 18.Saltykova A, Wuyts V, Mattheus W, Bertrand S, Roosens NHC, et al. Comparison of SNP-based subtyping workflows for bacterial isolates using WGS data, applied to Salmonella enterica serotype Typhimurium and serotype 1,4,[5],12:i:- PLoS One. 2018;13:e0192504. doi: 10.1371/journal.pone.0192504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dallman TJ, Byrne L, Ashton PM, Cowley LA, Perry NT, et al. Whole-genome sequencing for national surveillance of Shiga toxin-producing Escherichia coli O157. Clin Infect Dis. 2015;61:305–312. doi: 10.1093/cid/civ318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong JC, Mercoulia K, Tomita T, Easton M, Li HY, et al. Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes . J Clin Microbiol. 2016;54:333–342. doi: 10.1128/JCM.02344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franz E, Gras LM, Dallman T. Significance of whole genome sequencing for surveillance, source attribution and microbial risk assessment of foodborne pathogens. Curr Opin Food Sci. 2016;8:74–79. doi: 10.1016/j.cofs.2016.04.004. [DOI] [Google Scholar]
- 22.ECDC Expert Opinion on Whole Genome Sequencing for Public Health Surveillance. Stockholm: European Centre for Disease Prevention and Control; 2016. [Google Scholar]
- 23.WHO Whole Genome Sequencing for Foodborne Disease Surveillance: Landscape Paper. Geneva: World Health Organization; 2018. [Google Scholar]
- 24.Maiden MCJ, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol. 2013;11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alikhan N-F, Zhou Z, Sergeant MJ, Achtman M. A genomic overview of the population structure of Salmonella . PLoS Genet. 2018;14:e1007261. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pajuste F-D, Kaplinski L, Möls M, Puurand T, Lepamets M, et al. FastGT: an alignment-free method for calling common SNVs directly from raw sequencing reads. Sci Rep. 2017;7:2537. doi: 10.1038/s41598-017-02487-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Standage DS, Brown CT, Hormozdiari F. Kevlar: a mapping-free framework for accurate discovery of de novo variants. iScience. 2019;18:28–36. doi: 10.1016/j.isci.2019.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leekitcharoenphon P, Lukjancenko O, Friis C, Aarestrup FM, Ussery DW. Genomic variation in Salmonella enterica core genes for epidemiological typing. BMC Genomics. 2012;13:88. doi: 10.1186/1471-2164-13-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashton PM, Baker KS, Gentle A, Wooldridge DJ, Thomson NR, et al. Draft genome sequences of the type strains of Shigella flexneri held at Public Health England: comparison of classical phenotypic and novel molecular assays with whole genome sequence. Gut Pathog. 2014;6:7. doi: 10.1186/1757-4749-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inns T, Ashton PM, Herrera-Leon S, Lighthill J, Foulkes S, et al. Prospective use of whole genome sequencing (WGS) detected a multi-country outbreak of Salmonella Enteritidis. Epidemiol Infect. 2017;145:289–298. doi: 10.1017/S0950268816001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mair-Jenkins J, Borges-Stewart R, Harbour C, Cox-Rogers J, Dallman T, et al. Investigation using whole genome sequencing of a prolonged restaurant outbreak of Salmonella Typhimurium linked to the building drainage system, England, February 2015 to March 2016. Euro Surveill. 2017;22:17-00037. doi: 10.2807/1560-7917.ES.2017.22.49.17-00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One. 2011;6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Luo Y, Carleton H, Timme R, Melka D, et al. Whole genome and core genome multilocus sequence typing and single nucleotide polymorphism analyses of Listeria monocytogenes isolates associated with an outbreak linked to cheese, United States, 2013. Appl Environ Microbiol. 2017;83:e00633-17. doi: 10.1128/AEM.00633-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmid D, Allerberger F, Huhulescu S, Pietzka A, Amar C, et al. Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011-2013. Clin Microbiol Infect. 2014;20:431–436. doi: 10.1111/1469-0691.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandwagt D, van den Wijngaard C, Tulen AD, Mulder AC, Hofhuis A, et al. Outbreak of Salmonella Bovismorbificans associated with the consumption of uncooked ham products, the Netherlands, 2016 to 2017. Euro Surveill. 2018;23:17-00335. doi: 10.2807/1560-7917.ES.2018.23.1.17-00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Revez J, Espinosa L, Albiger B, Leitmeyer KC, Struelens MJ, et al. Survey on the use of whole-genome sequencing for infectious diseases surveillance: rapid expansion of European national capacities, 2015-2016. Front Public Health. 2017;5:347. doi: 10.3389/fpubh.2017.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.EFSA. ECDC Multi-country Outbreak of Salmonella Enteritidis Infections Linked to Polish Eggs. Stockholm and Parma: European Food Safety Authority, European Centre for Disease Prevention and Control; 2017. [Google Scholar]
- 38.Dallman TJ, Crook PD, Godbole G, Mook P, Chattaway MA, et al. Use of whole-genome sequencing for the public health surveillance of Shigella sonnei in England and Wales, 2015. J Med Microbiol. 2016;65:882–884. doi: 10.1099/jmm.0.000296. [DOI] [PubMed] [Google Scholar]
- 39.Hamming RW. Error detecting and error correcting codes. Bell Syst Technic J. 1950;29:147–160. doi: 10.1002/j.1538-7305.1950.tb00463.x. [DOI] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van den Berg RR, Dissel S, Rapallini MLBA, van der Weijden CC, Wit B, et al. Characterization and whole genome sequencing of closely related multidrug-resistant Salmonella enterica serovar Heidelberg isolates from imported poultry meat in the Netherlands. PLoS One. 2019;14:e0219795. doi: 10.1371/journal.pone.0219795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ridom . Ridom SeqSphere+ User Guide; 2019. https://www.ridom.de/u/User_Guide.html
- 46.Sneath PHA. The application of computers to taxonomy. Microbiology. 1957;17:201–226. doi: 10.1099/00221287-17-1-201. [DOI] [PubMed] [Google Scholar]
- 47.Carlsson G, Memoli F. Characterization, stability and convergence of hierarchical clustering methods. J Mach Learn Res. 2010;11:1425–1470. [Google Scholar]
- 48.Sørensen T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons. Biol Skr. 1948;5:1–34. [Google Scholar]
- 49.Henri C, Leekitcharoenphon P, Carleton HA, Radomski N, Kaas RS, et al. An assessment of different genomic approaches for inferring phylogeny of Listeria monocytogenes . Front Microbiol. 2017;8:2351. doi: 10.3389/fmicb.2017.02351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diedenhofen B, Musch J. cocor: a comprehensive solution for the statistical comparison of correlations. PLoS One. 2015;10:e0121945. doi: 10.1371/journal.pone.0121945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McLauchlin J, Aird H, Andrews N, Chattaway M, de Pinna E, et al. Public health risks associated with Salmonella contamination of imported edible betel leaves: analysis of results from England, 2011–2017. Int J Food Microbiol. 2019;298:1–10. doi: 10.1016/j.ijfoodmicro.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- 53.McClain JO, Rao VR. CLUSTISZ: a program to test for the quality of clustering of a set of objects. J Mark Res. 1975;12:456–460. [Google Scholar]
- 54.Halkidi M, Batistakis Y, Vazirgiannis M. On clustering validation techniques. J Intell Inf Syst. 2001;17:107–145. doi: 10.1023/A:1012801612483. [DOI] [Google Scholar]
- 55.Charrad M, Ghazzali N, Boiteau V, Niknafs A. NbClust : an R package for determining the relevant number of clusters in a data set. J Stat Softw. 2014;61:6. doi: 10.18637/jss.v061.i06. [DOI] [Google Scholar]
- 56.Hennig C . fpc: Flexible Procedures for Clustering; 2018. https://CRAN.R-project.org/package=fpc
- 57.Pihur V, Datta S, Datta S. RankAggreg, an R package for weighted RANK aggregation. BMC Bioinformatics. 2009;10:62. doi: 10.1186/1471-2105-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galili T. dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics. 2015;31:3718–3720. doi: 10.1093/bioinformatics/btv428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baker FB. Stability of two hierarchical grouping techniques case 1: sensitivity to data errors. J Am Stat Assoc. 1974;69:440–445. [Google Scholar]
- 60.Fowlkes EB, Mallows CL. A method for comparing two hierarchical clusterings. J Am Stat Assoc. 1983;78:553–569. doi: 10.1080/01621459.1983.10478008. [DOI] [Google Scholar]
- 61.Shotwell MS. profdpm: an R package for MAP estimation in a class of conjugate product partition models. J Stat Softw. 2013;53:8. doi: 10.18637/jss.v053.i08. [DOI] [Google Scholar]
- 62.Bojanowski M, Edwards R. alluvial: R Package for Creating Alluvial Diagrams. 2016
- 63.R Core Team R: a Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 64.Achtman M, Wain J, Weill F-X, Nair S, Zhou Z, et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica . PLoS Pathog. 2012;8:e1002776. doi: 10.1371/journal.ppat.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sokal RR, Michener CD. A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull. 1958;28:1409–1438. [Google Scholar]
- 66.Waldram A, Dolan G, Ashton PM, Jenkins C, Dallman TJ. Epidemiological analysis of Salmonella clusters identified by whole genome sequencing, England and Wales 2014. Food Microbiol. 2018;71:39–45. doi: 10.1016/j.fm.2017.02.012. doi: 10.1016/j.fm.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 67.Sobkowiak B, Glynn JR, Houben RMGJ, Mallard K, Phelan JE, et al. Identifying mixed Mycobacterium tuberculosis infections from whole genome sequence data. BMC Genomics. 2018;19:613. doi: 10.1186/s12864-018-4988-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Low AJ, Koziol AG, Manninger PA, Blais B, Carrillo CD. ConFindr: rapid detection of intraspecies and cross-species contamination in bacterial whole-genome sequence data. PeerJ. 2019;7:e6995. doi: 10.7717/peerj.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cunningham SA, Chia N, Jeraldo PR, Quest DJ, Johnson JA, et al. Comparison of whole-genome sequencing methods for analysis of three methicillin-resistant Staphylococcus aureus outbreaks. J Clin Microbiol. 2017;55:1946–1953. doi: 10.1128/JCM.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Didelot X, Bowden R, Street T, Golubchik T, Spencer C, et al. Recombination and population structure in Salmonella enterica . PLoS Genet. 2011;7:e1002191. doi: 10.1371/journal.pgen.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nadon C, Van Walle I, Gerner-Smidt P, Campos J, Chinen I, et al. Pulsenet international: vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Euro Surveill. 2017;22:30544. doi: 10.2807/1560-7917.ES.2017.22.23.30544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davis S, Pettengill JB, Luo Y, Payne J, Shpuntoff A, et al. CFSAN SNP pipeline: an automated method for constructing SNP matrices from next-generation sequence data. PeerJ Computer Science. 2015;1:e20. doi: 10.7717/peerj-cs.20. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.