Abstract
Clostridioides difficile is a spore-forming bacterial pathogen that is the leading cause of hospital-acquired gastroenteritis. C. difficile infections begin when its spore form germinates in the gut upon sensing bile acids. These germinants induce a proteolytic signaling cascade controlled by three members of the subtilisin-like serine protease family, CspA, CspB, and CspC. Notably, even though CspC and CspA are both pseudoproteases, they are nevertheless required to sense germinants and activate the protease, CspB. Thus, CspC and CspA are part of a growing list of pseudoenzymes that play important roles in regulating cellular processes. However, despite their importance, the structural properties of pseudoenzymes that allow them to function as regulators remain poorly understood. Our recently solved crystal structure of CspC revealed that its pseudoactive site residues align closely with the catalytic triad of CspB, suggesting that it might be possible to ‘resurrect' the ancestral protease activity of the CspC and CspA pseudoproteases. Here, we demonstrate that restoring the catalytic triad to these pseudoproteases fails to resurrect their protease activity. We further show that the pseudoactive site substitutions differentially affect the stability and function of the CspC and CspA pseudoproteases: the substitutions destabilized CspC and impaired spore germination without affecting CspA stability or function. Thus, our results surprisingly reveal that the presence of a catalytic triad does not necessarily predict protease activity. Since homologs of C. difficile CspA occasionally carry an intact catalytic triad, our results indicate that bioinformatic predictions of enzyme activity may underestimate pseudoenzymes in rare cases.
Keywords: Clostridioides difficile, Clostridium difficile, pseudoenzyme, pseudoprotease, spore germination, subtilisin-like serine protease
Introduction
Catalytically inactive structural homologs of functional enzymes known as pseudoenzymes were first discovered more than 50 years ago, but they were generally dismissed as vestigial remnants of evolution because they lacked catalytic activity [1,2]. However, the prevalence of pseudoenzyme genes, estimated at 10–15% of a typical genome [3] across all domains of life [1,2], suggests that they have important biological functions. Indeed, recent work has established that pseudoenzymes perform diverse and crucial cellular functions [1,4,5], controlling metabolic and signaling pathways in processes ranging from cell cycle progression to protein trafficking. In acquiring these important functions, pseudoenzymes have in a way been ‘brought back to life’ [1,2,4,5] and have even been referenced as ‘zombie’ proteins [1,4].
While pseudoenzymes have been identified in over 20 different protein families, including pseudokinases, pseudophosphatases, and pseudoproteases [1,2], the mechanisms by which they modulate cellular processes are poorly understood, since relatively few predicted pseudoenzymes have been thoroughly studied in biological systems. Studies thus far indicate that pseudoenzymes can allosterically regulate the activity of cognate enzymes, nucleate protein complexes by acting as cellular scaffolds, control protein localization, and act as competitors for substrate binding or holoenzyme assembly [1,5].
Even less understood are the structural properties of pseudoenzymes that allow them to carry out these functions. Bioinformatic analyses imply that most pseudoenzymes have evolved from ancestral cognate enzymes due to loss of one or more residues required for catalysis or cofactor binding [6–8]. However, it is difficult to assess bioinformatically whether pseudoenzymes have acquired additional changes beyond these catalytic site substitutions that prevent their ancestral enzymatic function. Indeed, this question has only been experimentally addressed in a handful of studies [1]. Converting the pseudoactive glycine residue of the STYX pseudophosphatase to a catalytic cysteine restored its hydrolytic activity [9,10]. In contrast, substitutions that restore the catalytic site in pseudokinases have had differential effects depending on the pseudokinase. When residues required for binding ATP were restored to the human pseudokinase, RYK, it gained the ability to bind an ATP analog in thermal shift assays [11]. Conversely, when a catalytic cysteine was restored to the DivL histidine pseudokinase of the bacterium Caulobacter crescentus, it did not regain kinase activity [12]. Similarly, when the catalytic aspartate was restored to the ErbB3/HER3 pseudokinase, no increase in kinase activity was observed [13,14]. While this pseudokinase has vestigial kinase activity (∼1000-fold weaker than related kinases with intact catalytic motifs in vitro [14]), the ‘resurrection' mutation did not change ErbB3/HER3's ability to activate the neuregulin receptor in cells [15].
Beyond these relatively limited studies of pseudophosphatases and pseudokinases, the question of whether pseudoproteases can be converted back into active enzymes has not yet been tested. In this study, we attempted to resurrect the protease activity of two pseudoproteases, CspA and CspC, which play critical roles in the life cycle of Clostridioides difficile, a spore-forming bacterial pathogen that is the leading cause of nosocomial gastroenteritis worldwide [16,17]. C. difficile caused ∼225 000 infections and ∼13 000 deaths in 2017 in the United States alone [18] and has been designated by the Centers for Disease Control and Prevention as an urgent threat because of its intrinsic antibiotic resistance [19].
C. difficile’s resistant spore form is its major transmissive particle because C. difficile is an obligate anaerobe [20,21]. C. difficile infections begin when its metabolically dormant spore form germinates in the gut of vertebrate hosts in response to certain bile acids [22]. Notably, these bile acid germinants differ from the nutrient germinants sensed by almost all other spore-formers studied to date, and their signal transduction mechanism appears to be unique because C. difficile lacks the transmembrane germinant receptors found in all other spore formers [23–26]. Instead, the bile acid germinant signal is transduced by members of the clostridial serine protease family known as the Csps [27–30]. Csps are subtilisin-like serine protease family members [31,32] conserved in many clostridial species [33]. Three Csp proteins, CspA, CspB and CspC, participate in a signaling cascade that leads to the proteolytic activation of the SleC cortex lytic enzyme. Activated SleC then removes the protective cortex layer, which is essential for spores to exit dormancy [27,34,35].
Despite their conservation, the precise functions of the Csp family members differ between C. difficile and C. perfringens (and likely other members of the Clostridia). In C. perfringens, CspA, CspB, and CspC are all active proteases whose primary function is to proteolytically activate the SleC cortex lytic enzyme [31,36]. All three of these Csps carry the canonical catalytic triad of subtilisin-like serine protease (S8) family members, which consists of aspartate, histidine, and serine. They also all contain long N-terminal prodomains, which are characteristic of S8 family proteases [37]. The prodomains serve as intramolecular chaperones that induce folding of their subtilase domains [38]. Proper folding of the subtilase domain induces its protease activity, resulting in cleavage of the prodomain from the subtilase domain [38]. This autoprocessing event is essential for S8 family protease activity [32], and catalytic mutants of subtilisin-like serine proteases fail to undergo autoprocessing [32,39,40]. Accordingly, all three C. perfringens Csps autoproteolytically remove their prodomains [31].
In contrast, two of the three C. difficile Csps do not undergo autoprocessing, since they carry substitutions in their catalytic triad that render them pseudoproteases [27,28,41]. Unlike active Csps, the C. difficile CspC and CspA pseudoproteases cannot cleave the SleC cortex lytic enzyme. Instead, they regulate how C. difficile spores sense bile acid germinants as well as cation and amino acid co-germinant signals. C. difficile CspC is thought to directly sense bile acid germinants [28] and integrate signals from the two co-germinant classes [30], while C. difficile CspA may function as the co-germinant receptor [42] and is necessary for CspC to be packaged into mature spores [29]. Thus, C. difficile CspC and CspA both regulate the protease activity of CspB, whose intact catalytic triad is required for proteolytically activating SleC [27].
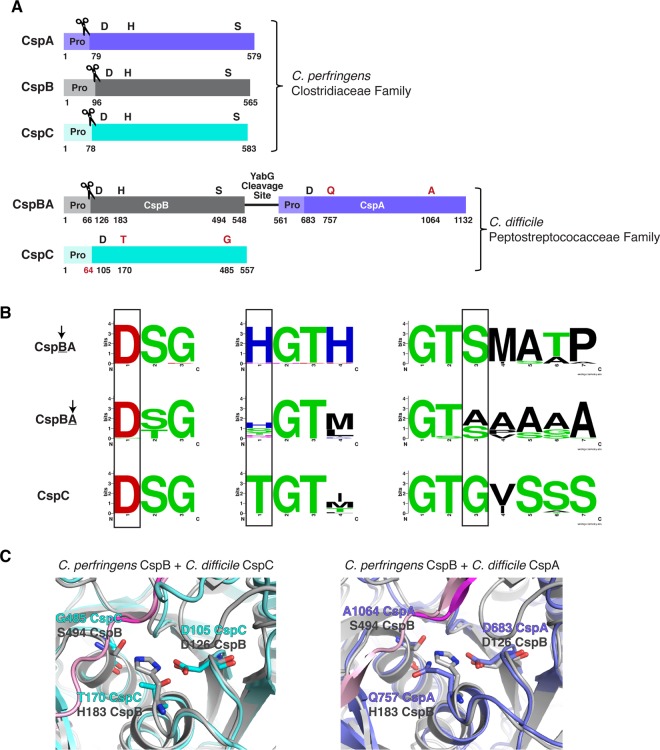
Interestingly, C. difficile cspA and cspB are encoded in a single open reading frame, cspBA, such that the resulting fusion protein physically links the active CspB protease to the inactive CspA pseudoprotease (Figure 1A). CspBA's protease-pseudoprotease arrangement is largely conserved in the Peptostreptococcaceae family to which C. difficile belongs [29], with the CspB domain carrying an intact catalytic triad in all sequences examined, and the CspA domain typically carrying at least one substitution in its catalytic triad ([29], Figure 1B). While the catalytic site substitutions present in the CspA pseudoprotease vary in the Peptostreptococcaceae family, the pseudoactive site residues of CspC are strictly conserved in this family ([29], Figure 1B). In contrast, members of the Lachnospiraceae and Clostridiaceae families all encode the three Csp proteins as individual proteases with intact catalytic triads, suggesting that Peptostreptococcaceae family CspA and CspC homologs specifically lost their catalytic activity.
Figure 1. Csp family subtilisin-like serine proteases in the Clostridia.
(A) Schematic of the active Csp proteases encoded by C. perfringens, CspA, CspB and CspC, compared with Clostridiodes difficile Csp proteins, where an active CspB protease is fused to an inactive CspA pseudoprotease domain, and CspC is also a pseudoprotease. ‘Pro' denotes the prodomain that functions as an intramolecular chaperone. The C-terminal residue of the prodomains that have been mapped are shown below the schematic [27,31]. The catalytic triad residues, aspartic acid (D), histidine (H) and serine (S), are shown in black; pseudoactive site residues are shown in red. The scissor icon marks the autoprocessing sites of the Csp family members that are catalytically active, which separates the prodomain from the subtilase domain. The predicted site that would be cleaved by autoprocessing of the CspC pseudoprotease based on the CspC structure [30] is also shown in red. The YabG cleavage site [42] between CspB and CspA is shown. (B) Sequence logos of the catalytic triad residue regions for CspBA and CspC of the Peptostreptococcaceae family. Regions shown correspond to the MEROPS protease database [53] definitions for the peptidase family S8A. Information regarding gene location and accession number for the proteins is included in the sequence logo analysis provided in Supplemental Table S3. (C) Cartoon model of the active/ pseudoactive site regions of either C. perfringens CspB (grey, PDB 4I0W) aligned to C. difficile CspC (cyan, PBD 6MW4) or CspB (grey) aligned to C. difficile CspA iTasser model (periwinkle), active site region. The cleaved prodomain of CspB is shown in dark magenta compared with the uncleaved prodomain of CspC (light pink) and CspA prodomain (light pink).
Since we recently showed that the pseudoactive site residues of C. difficile CspC closely align with the catalytic triad of an active CspB protease ([30], Figure 1C), we asked whether restoring an intact catalytic triad to CspC would be sufficient to convert it into an active protease capable of undergoing autoprocessing like other subtilisin-like serine proteases [32]. Since structural modeling of CspA also predicted close alignment of its pseudoactive site residues with CspB's catalytic triad residues (Figure 1C), we tested the effect of restoring the catalytic triad to C. difficile CspA to gain insight into the evolution of these ‘zombie' proteins from active proteases and the structural requirements for their function in C. difficile.
Experimental
Bacterial strains and growth conditions
C. difficile strain construction was performed using 630ΔermΔcspCΔpyrE [41] and 630ΔermΔpyrE-ΔcspBA as the parental strains via pyrE-based allele-coupled exchange (ACE [43]). This system allows for single-copy complementation of the ΔcspC and ΔcspBA parental mutants, respectively, from an ectopic locus. C. difficile strains are listed in Supplemental Table S1. They were grown on brain heart infusion media (BHIS) supplemented with taurocholate (TA, 0.1% w/v; 1.9 mM), thiamphenicol (10–15 µg/ml), kanamycin (50 µg/ml), cefoxitin (8 µg/ml), and l-cysteine (0.1% w/v; 8.25 mM) as needed. Cultures were grown under anaerobic conditions at 37°C using a gas mixture containing 85% N2, 5% CO2, and 10% H2.
Escherichia coli strains for BL21(DE3)-based protein production and for HB101/pRK24-based conjugations are listed in Supplemental Table S1. E. coli strains were grown shaking at 225 rpm in Luria-Bertani broth (LB) at 37°C. The media was supplemented with ampicillin (100 µg/ml), chloramphenicol (20 µg/ml) or kanamycin (30 µg/ml) as indicated.
E. coli strain construction
E. coli strains are listed in Supplemental Table S1 in the supplementary material. As previously described [30], the cspC complementation constructs were created using flanking primers, #2189 and 2242 (Supplemental Table S2), in combination with internal primers encoding a given point substitution, ΔcspBA genomic DNA was used as the template. This resulted in cspC complementation constructs carrying 282 bp of the cspBA upstream region in addition to the ΔcspBA sequence and the intergenic region between cspBA and cspC. This extended construct was required to produce wild-type levels of CspC when expressing the constructs in the pyrE locus [41,43]. For example, the T170H substitution was constructed using primer pair #2189 and 2355 to amplify a 5′ cspC complementation construct fragment encoding the T170H substitution at the 3′ end, while primer pair #2354 and 2242 were used to amplify a 3′ cspC complementation construct encoding the T170H substitution at the 5′ end. The individual 5′ and 3′ products were cloned into pMTL-YN1C digested with NotI/XhoI by Gibson assembly. In some cases, the two PCR products were used in a PCR SOE [44] prior to using Gibson assembly to clone the cspC construct into pMTL-YN1C digested with NotI and XhoI. The resulting plasmids were transformed into E. coli DH5α, confirmed by sequencing, and transformed into HB101/pRK24.
Similarly, for cspBA complementation constructs, each construct was designed with 126 bp of the ΔcspC sequence downstream of cspBA in order to fully complement the ΔcspBA mutant as previously described [41]. All primers used for strain construction are listed in Supplemental Table S2. For example, the Q757H point substitution was introduced into the complementation constructs by using primer pair #2189 and 3041 to amplify the 5′ end, and #3040 and 2242 to amplify the 3′ end. The A1064S mutant was designed in the same way, but with primer pair #3042 and 3043 to introduce the point substitution. The 5′ and 3′ products containing the various substitutions were cloned into pMTL-YN1C digested with NotI/XhoI and combined through Gibson assembly. Depending on the construct, some PCR products were combined by PCR SOE prior to using Gibson assembly. The resulting plasmids were transformed into E. coli DH5α, confirmed by sequencing, and transformed into HB101/pRK24.
To generate the construct encoding the cspBA prodomain trans-complementation construct, primer pair #2189 and 951 was used to amplify the 5′ fragment, and primer pair #950 and 2242 was used to amplify the 3′ fragment. In both cases, ΔcspC genomic DNA was used as a template as described previously [41]. The resulting two fragments were joined together using PCR SOE with primer pair #2189 and 2242, and the PCR SOE product was cloned into pMTL-YN1C digested with NotI and XhoI using Gibson assembly. A similar strategy was used to generate the cspC prodomain trans-complementation construct. Primers #2189 and #2553 were used to amplify the 5′ fragment, and primers #2552 and #2242 were used to amplify the 3′ fragment using ΔcspBA genomic DNA as a template. The fragments were joined together using SOE PCR with the primer pair #2189 and 2242, and the resulting SOE PCR product was cloned into pMTL-YN1C digested with NotI and XhoI using Gibson assembly.
To generate the recombinant protein expression constructs for producing CspC-His6 variants, primer pair #1128 and 1129 was used to amplify a codon-optimized version of cspC using pJS148 as the template (a kind gift from Joseph Sorg) as previously described [30]. The resulting PCR product was digested with NdeI and XhoI and ligated into pET22b cut with the same enzymes. The G171R variant was cloned using a similar procedure except that primer pair #1128 and 1342 and primer pair #1341 and 1129 were used to PCR the 5′ and 3′ fragments encoding the G171R mutation, respectively. The resulting PCR products were joined together using PCR SOE and flanking primer pair #1128 and 1129.
The remaining constructs encoding cspC codon-optimized variants for expression using pET22b were cloned using Gibson assembly. PCR SOE was used to introduce the G485S substitution: primer pairs #2311 and 2601 and #2600 and 2312 were used to generate the 5′ and 3′ fragments for the G485S substitution. The G485S-T170H construct was generated using the same procedure except that pET22b-cspCT170H was used as the template for the #2311 and #2601 PCR, and the resulting PCR product was purified following gel extraction. This 5′ fragment was then used in a PCR SOE reaction with the 3′ PCR fragment resulting from the primer pair #2600 and #2312 to assemble the double mutant construct. The resulting PCR products were cloned into pET22b digested with NdeI and XhoI using Gibson assembly.
To generate the recombinant protein expression constructs for producing CspC-CPD-His6, primer pair #1128 and 1166 were used to amplify the codon-optimized version of cspC from pJS148 (a kind gift from Joseph Sorg). The resulting PCR product was digested with NdeI and SacI and cloned into pET22b-CPDSacI digested with the same enzymes using DNA ligation. The T170H and G171R variants were cloned using a similar strategy as described above using PCR SOE, with the exception that primer pair #1372 and 1166 was used to clone a fragment encoding codon-optimized cspC carrying the relevant substitutions into pET22b-cspC-CPDSacI digested with BamHI and SacI. The 2xcat variant was cloned using the CspCT170H-CPD-His6 construct as the template for PCR with primer pair #2311 and 2602. The internal SOE primers used to introduce the G485S substitution were #2600 and 2601 as described above. The resulting PCR product (following gel extraction) was cloned into pET22b-CPDSacI digested with NdeI and SacI using Gibson assembly.
To generate the recombinant protein expression constructs for producing CspBA-His6 variants, primer pair #1505 and 1529 was used to amplify a codon-optimized version of cspB using a plasmid template (a kind gift from Joseph Sorg). The resulting PCR product was digested with NcoI and HindIII and ligated into pET28a digested with the same enzymes. Codon-optimized cspA was then amplified using primer pair #1507 and 1508 using another plasmid template from Joseph Sorg. The resulting PCR product was used as the template for a second PCR using primer pair #1530 and 1508. This PCR product was digested with HindIII and XhoI and then ligated into pET28a-cspB CO digested with the same enzymes. The CspBA-His6 recombinant protein expression constructs, were constructed using primers #3034 and 3035 to create a codon-optimized version of cspBA. The Q757H and A1064S point substitutions were introduced using primer pairs, #3036 and 3037, and #3038 and 3039, respectively. The resulting PCR products were digested with NcoI/XhoI, ligated into pET28a, and transformed into BL21.
Protein purification for recombinant E. coli analyses
E. coli BL21(DE3) strains listed in Supplemental Table S1 were used to produce and purify C-terminally His6-tagged codon-optimized cspC variants and cspBA variants as previously described [27]. Briefly, cultures were grown to mid-log phase in 2YT (5 g NaCl, 10 g yeast extract, and 15 g tryptone per liter) at 37°C. When cultures reached an OD600 ∼ 0.8, 250 µM isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added to induce expression of cspC. Cultures were then grown overnight at 18˚C. The cells were pelleted, resuspended in lysis buffer (500 mM NaCl, 50 mM Tris [pH 7.5], 15 mM imidazole, 10% [vol/vol] glycerol, 2 mM beta-mercaptoethanol), flash frozen in liquid nitrogen, thawed and finally sonicated. The insoluble material was pelleted, and the soluble fraction was incubated with Ni-NTA agarose beads (5 Prime) for 3 h, and eluted using high-imidazole buffer (500 mM NaCl, 50 mM Tris [pH 7.5], 200 mM imidazole, 10% [vol/vol] glycerol) after nutating the sample for 5–10 min.
Codon-optimized cspC from C. difficile was expressed in E. coli BL21(DE3) cells from a pET22b plasmid containing a C-terminal self-cleaving CPD tag [45], which is derived from the Vibrio cholerae MARTX toxin [46]. Starter cultures were grown in 20 ml LB broth with 100 μg/ml ampicillin. Terrific broth with 100 μg/ml ampicillin was inoculated with the starter culture (1 : 1000) and incubated for ∼60 h at 20°C with 225 rpm shaking.
The purifications were carried out in three steps: nickel bead-based affinity purification, inositol hexakisphosphate (InsP6)-induced cleavage of the CPD tag, followed by size exclusion chromatography (SEC). Affinity purification was carried out as stated above: the cells were pelleted, resuspended in lysis buffer (500 mM NaCl, 50 mM Tris [pH 7.5], 15 mM imidazole, 10% [vol/vol] glycerol, 2 mM beta-mercaptoethanol), flash frozen in liquid nitrogen, thawed and then sonicated. The insoluble material was pelleted, and the soluble fraction was incubated with Ni-NTA agarose beads (Qiagen) for 4 h to capture the CspC-CPD-His6 fusion proteins. Washes were carried out three times with low imidazole buffer (500 mM NaCL, 10 mM Tris–HCl pH 7.5, 10% (v/v) glycerol, 15 mM imidazole, 2 mM β-mercaptoethanol) to decrease non-specific binding to the beads.
On-bead CPD cleavage was induced through the addition of 200 µM InsP6, which leaves behind a two amino acid Glu-Leu linker on the C-terminus of CspC variants. InsP6 was incubated with gentle shaking at 4°C overnight. The supernatant containing liberated CspC variants was collected, then the InsP6-induced cleavage was repeated for 2–4 h at 4°C. The second supernatant was collected, and both supernatants were pooled. Pooled protein elutions were concentrated to 20 mg/ml or less in a gel filtration buffer consisting of 200 mM NaCl, 10 mM Tris HCl pH 7.5, 5% glycerol and 1 mM DTT. SEC was carried out using a Superdex 200 Increase 10/300 GL (GE Healthcare) column. Protein fractions were assessed by Coomassie staining. The purified CspC variants were concentrated, aliquoted, flash frozen, and stored at −80°C for future analysis. Protein concentration was assessed using both A280 readings and Pierce 660 assay (ThermoFisher Scientific).
C. difficile strain construction
Complementation strains were constructed using CDDM to select for recombination of the complementation construct into the pyrE locus by restoring uracil prototrophy [43], as previously described [47]. At least two independent clones from each complementation strain were phenotypically characterized.
Sporulation
C. difficile strains were grown overnight on BHIS plates containing taurocholate (TA, 0.1% w/v, 1.9 mM). Liquid BHIS cultures were inoculated from the resulting colonies, which were grown to early stationary phase before being back-diluted 1 : 50 into BHIS. When the cultures reached an OD600 between 0.35 and 0.75, 120 µl of this culture were plated onto 70 : 30 agar plates and grown for 18–24 h as previously described [48]. Sporulating cells were harvested into phosphate-buffered saline (PBS), and cells were visualized by phase-contrast microscopy [49].
Spore purification
Sporulation was induced on 70 : 30 agar plates for 2–3 days as described above, and spores were purified as previously described [34]. Briefly, the samples were harvested into sterile water at 4°C. The samples were washed 6–7 times in 1 ml of ice-cold water per every two plates and incubated overnight in water at 4°C. The samples were then pelleted and incubated with DNase I (New England Biolabs) at 37°C for 60 min. Finally, samples were purified on a 20% : 50% HistoDenz (Sigma–Aldrich) gradient and washed 2–3 more times in water. Spore purity was assessed using phase-contrast microscopy (>95% pure). The optical density of the spore stock was measured at OD600, and spores were stored in water at 4°C.
Germination assay
As previously described [34], germination for each strain used the equivalent of 0.35 OD600 units, which corresponds to ∼1 × 107 spores. The proper number of spores was resuspended in 100 µl of water, 10 µl of this mixture was serially diluted in PBS, and the resulting dilutions were plated on BHIS-TA. Colonies arising from germinated spores were enumerated at 18–24 h. Germination efficiencies were calculated using mean CFUs produced by spores for a given strain relative to the mean CFUs produced by wild type. Analyses were based on at least three technical replicates performed on two independent spore preparations (i.e. two biological replicates). Statistical significance was determined by performing a one-way analysis of variance (ANOVA) on natural log-transformed data using Tukey's test.
OD600 kinetics assay
As previously described [50], ∼1.5 × 107 spores (0.48 OD600 unit) were resuspended in BHIS to a total volume of 1.1 ml. The sample was divided in two: 540 μl was added to a cuvette containing 60 µl of 10% taurocholate, while the other sample was added to a cuvette containing 60 μl of water, as a control. The samples were mixed, and the OD600 was measured every 3 min for the first 45 min and then every 15 min from 60 to 90 min. Statistical significance was determined by performing a two-way analysis of variance (ANOVA) using Tukey's test.
Western blot analysis
Samples for western blot analysis were prepared as previously described [51]. Briefly, sporulating cell pellets were resuspended in 100 µl of PBS, and 50 µl samples were removed and freeze-thawed for three cycles. The samples were resuspended in 100 µl EBB buffer (8 M urea, 2 M thiourea, 4% (w/v) SDS, 2% (v/v) β-mercaptoethanol), boiled for 20 min, pelleted, resuspended in the same volume. Subsequently, 7 µl of sample buffer was added to stain samples with bromophenol blue. C. difficile spores (∼1 × 107) were resuspended in 50 µl EBB buffer and processed similarly. The samples were resolved by 7.5% (for sporulating cell analyses of CspBA and CspC) or 12% SDS–PAGE gels. After, the gels were transferred to Millipore Immobilon-FL PVDF membranes and were blocked in Odyssey Blocking Buffer [47] for 30 min with 0.1% (v/v) Tween 20. Blots were incubated with rabbit polyclonal anti-CspB [27], anti-CspA (a generous gift from Joe Sorg, Texas A&M University, [42]) or anti-CotA antibodies and/or mouse polyclonal anti-SleC [27], anti-CspC [29], or anti-SpoIVA antibodies [34]. Additionally, western blotting for recombinant protein samples were blotted with mouse monoclonal anti-penta-His (ThermoScientific). The anti-CspB, anti-CspC, anti-SpoIVA antibodies were used at 1 : 2500 dilutions, the anti-SleC antibody was used at a 1 : 5000 dilution, and the anti-penta-His, anti-CotA, and anti-CspA antibodies were used at a 1 : 1000 dilution. IRDye 680CW and 800CW infrared dye-conjugated secondary antibodies were used at 1 : 20 000 dilutions. The Odyssey LiCor CLx was used to detect secondary antibody infrared fluorescence emissions. All blots shown are representative of analyses performed on two independent spore preparations.
Coomassie staining
Purified His-tagged CspC and CspBA were resolved on either 12 or 15% SDS–PAGE gels, and the gels were stained using GelCode Blue according to the manufacturer's instructions (ThermoFisher Scientific).
Protein modeling
Multiple protein model predictions were used to analyze the CspA structure. Specifically, we analyzed several predictions by I-TASSER (Iterative Threading ASSEmbly Refinement) [52]. All prediction models were downloaded as PDB files and were viewed using PyMol. The CspA sequence used was taken from the cspBA gene, starting at codon 560 which has been predicted to encode the YabG cleavage site [42]. CspA predictions were aligned with the RCSB PDB files of the CspB protease (PDB 4I0W, [27]) and CspC pseudoprotease (PDB 6MW4, [30]).
Protein sequence analysis
Protein sequences were obtained from NCBI protein by searching for homologous sequences to CspBA and CspC, respectively, in Clostridioides difficile strain 630 filtering only for species within the Peptostreptococcaceae family. The algorithm ‘PSI-BLAST' was used to identify distant relatives of the proteins of interest. Homologs that had >95% query cover were selected for analysis, and for CspC homologs only, sequences which additionally had >55% identity were selected to avoid redundancy with CspA or CspB individual homologs. For all analyses only the first three organisms from each species were selected, in order to avoid skewing of the data based on the most sequenced organisms. The selected sequences were analyzed using MacVector and aligned with the ClustalW algorithm. The regions surrounding the catalytic residues were selected based on previous analyses [29] using the MEROPS protease database [53] active site definitions for peptidase family S8A. Information regarding accession numbers for selected homologs is provided in Supplemental Tables S3 and S4.
Thermofluor assay
An amount of 1 µM of CspC protein variants was resuspended in gel filtration buffer (200 mM NaCl, 10 mM Tris HCl pH 7.5, 5% glycerol and 1 mM DTT) combined with 5X SYPRO orange dye. For the GuHCl control, 1 µM of protein was diluted into pH 2.5 phosphate buffer with 1.5 M GuHCl. Diluted protein in buffer was aliquoted into three wells at a final volume of 20 µl per well in 96-well white, low-profile PCR plate (ThermoFisher Scientific). The reaction was incubated with a 2 min hold at 25°C with a ramp rate of 1°C/min to a final temperature of 95°C with a 10 min hold using a Bio-Rad CFX Connect Real-Time PCR instrument.
Results
Restoring CspC's catalytic triad reduces CspC levels in C. difficile
To determine if protease activity could be resurrected in the CspC pseudoprotease, we restored the pseudoactive residues of CspC's catalytic triad (Figure 2A) individually and in combination. To this end, we generated strains producing CspC variants carrying the following amino acid substitutions: threonine 170 to histidine (CspCT170H), glycine 485 to serine (CspCG485S), and T170H-G485S (referred to as CspC2xcat). The constructs encoding these substitutions were integrated into the pyrE locus of a previously characterized in-frame cspC deletion mutant [41] using allele-coupled exchange [43]. These constructs, along with all other cspC constructs analyzed in this manuscript, were expressed from the native cspBA-cspC promoter as described previously [41].
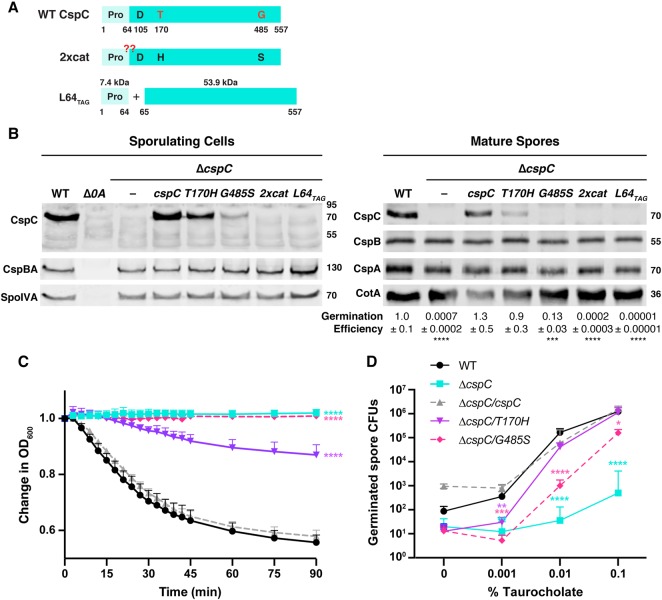
Figure 2. Restoring CspC's catalytic triad decreases CspC levels and germination efficiency.
(A) Schematic of WT CspC, CspC with a restored catalytic triad (2xcat), and the CspC prodomain (‘Pro') produced in trans with the subtilase domain to mimic the CspC products produced by autoprocessing (L64TAG). The L64TAG variant is generated from a construct where a stop codon was introduced after codon 64 and a ribosome binding site and start codon were added before codon 65. The red ‘??' indicates where the CspC2xcat variant would be expected to autoprocess if restoring the catalytic triad ‘resurrects' CspC protease activity. (B) Western blot analyses of CspB(A) and CspC in sporulating cells and purified spores from WT 630Δerm-p, ΔcspC, and ΔcspC complemented with either wild-type cspC or the L64TAG cspC trans-complementation variant. 2xcat refers to a complementation construct containing both the T170H and G485S point substitutions. Δspo0A (Δ0A) was used as a negative control for sporulating cells. CspBA was detected with anti-CspB and anti-CspA antibodies. SpoIVA was used as a loading control for sporulating cells, while CotA was used as a loading control for purified spores. The germination efficiency of spores from the indicated strains plated on BHIS medium containing 0.1% taurocholate is also shown relative to WT. The mean and standard deviations shown are based on multiple technical replicates performed on two independent spore purifications for purified spores. Statistical significance relative to WT was determined using a one-way ANOVA and Tukey's test. (C) Change in the OD600 in response to germinant of CspC catalytic mutant spores relative to WT spores. ΔcspC mutant spores serve as a negative control. 2xcat mutant spores are not shown, as they behaved similarly to the ΔcspC spores in the less sensitive plate-based germination assay. Purified spores were resuspended in BHIS, and germination was induced by adding taurocholate (1% final concentration). The ratio of the OD600 of each strain at a given time point relative to the OD600 at time zero is plotted. The mean of three assays from at least two independent spore preparations are shown. The error bars indicate the standard deviation for each time point measured. Statistical significance relative to WT was determined using a two-way ANOVA and Tukey's test. (D) Germinant sensitivity of CspC catalytic mutant spores compared with WT. Spores were plated on BHIS containing increasing concentrations of taurocholate. The number of colony forming units (CFUs) produced by germinating spores is shown. The mean and standard deviations shown are based on multiple replicates performed on two independent spore purifications. Statistical significance relative to WT was determined using a one-way ANOVA and Tukey's test. **** P < 0.0001, *** P < 0.001, ** P < 0.01, * P < 0.05.
To assess whether the T170H-G485S (2xcat) substitutions in CspC activated the autoprocessing activity characteristic of subtilisin-like serine proteases [32,38], we tested whether CspC2xcat gained the ability to cleave its prodomain by using western blotting to monitor changes in CspC size in sporulating cells. Rather than restoring autoprocessing activity, substitutions in the pseudoactive sites decreased CspC levels in sporulating cell lysates: no CspC was detectable in lysates of the double mutant (2xcat) strain (Figure 2B), and CspC levels were markedly diminished in the G485S mutant and slightly reduced in the T170H mutant. Thus, substitutions in CspC's pseudoactive residues would appear to reduce CspC production and/or stability in sporulating cells. In contrast, CspBA levels were unaffected in the mutant strains (Figure 2B), consistent with the observation that CspC does not affect CspBA levels [41].
C. difficile mutants expressing cspC encoding pseudoactive site substitutions exhibit decreased germination rates and germinant sensitivity
To evaluate the impact of the CspC pseudoactive site substitutions on CspC function during spore germination, we measured the ability of these mutant alleles to complement ΔcspC’s germination defect. Purified spores from WT, ΔcspC, ΔcspC/cspC, and the pseudoactive site mutant complementation strains were plated on a rich medium containing 0.1% taurocholate germinant, and the number of colony forming units (CFUs) that arose from germinating spores relative to WT was determined. The cspCT170H allele did not affect germination relative to WT even though CspCT170H protein levels were visibly decreased in western blot analyses of purified spores (Figure 2B). Surprisingly, the cspCG485S allele resulted in only a ∼10-fold defect in germination efficiency despite producing almost undetectable levels of CspCG485S protein. In contrast, the 2xcat double mutant exhibited a germination defect (∼5000-fold lower germination efficiency, Figure 2B) equivalent to that of the parental ΔcspC strain, consistent with CspC2xcat being undetectable in sporulating cells and purified spores by western blotting (Figure 2B). While low levels of ‘spontaneous' germination were observed in the ΔcspC spores, similar to previous analyses of other germinant receptor mutants [41,47,54,55], our results indicate that relatively little CspC is needed for C. difficile spores to germinate.
Although the G485S mutant exhibited only an ∼10-fold germination defect when spores were plated on a rich medium containing 0.1% taurocholate, G485S colonies arose more slowly than colonies derived from WT spores. To test whether the G485S and T170H pseudoactive site substitutions affected the rate of germination, we used an optical density-based germination assay. This assay measures the decrease in optical density of a population of germinating spores over time due to cortex degradation and core rehydration [50]. While the optical density of WT spores decreased by ∼40%, the optical density of the G485S mutant did not appreciably change over the assay period, similar to the ΔcspC strain (Figure 2C, P < 0.0001). Surprisingly, the T170H mutant germinated more slowly relative to WT (P < 0.0001) even though cspCT170H did not exhibit a germination defect in the plate-based CFU assay. Taken together, these results indicate that even single substitutions in CspC's pseudoactive active site impair folding and/or decrease CspC stability in sporulating cells. These decreased CspC levels correlate with slower spore germination rates. Restoring CspC's full catalytic triad reduces CspC levels even further and does not appear to restore autoprocessing to CspC.
We next wondered whether the reduced CspC levels in the T170H and G485S mutant spores would decrease their germinant sensitivity, since we previously showed that mutant spores with decreased CspB, CspA, and CspC levels are less responsive to germinant [47]. To measure germinant sensitivity, we plated cspC mutant spores on rich media containing varying concentrations of taurocholate germinant. On plates containing 0.001% taurocholate, T170H and G485S mutant spores germinated to a similar extent as ΔcspC spores (P ≤ 0.005, Figure 2D). However, on plates with 0.01% taurocholate, T170H mutant spores germinated to near wild-type levels, whereas G485S spores exhibited an ∼100-fold decrease relative to WT (P < 0.0001). At the highest concentration of germinant tested (0.1% taurocholate), T170H mutant spores were indistinguishable from WT spores, and the G485S mutant spores exhibited an ∼10-fold decrease in CFUs (Figure 2D, P < 0.0001) consistent with Figure 2B. Since the pseudoactive site mutants have wild-type levels of CspB and CspA in their spores (Figure 2B), the reduced sensitivity of their spores to germinant is caused by impaired CspC function and/or decreased protein levels.
The CspC prodomain cannot function in trans
Since our attempts to ‘resurrect' CspC's active site appeared to destabilize the protein, we wondered whether we could bypass the autoprocessing event by producing the CspC prodomain separately from the CspC subtilase domain. As mentioned earlier, subtilisin-like serine proteases use their long N-terminal prodomain as an intramolecular chaperone to promote folding of the subtilase domain into an active conformation [38]. The prodomains of other subtilisin-like serine proteases (including CspB in C. difficile) can perform this chaperone function in trans [27,32,38]. We thus tested whether C. difficile CspC's prodomain could function as a chaperone in trans, even though CspC normally does not undergo autoprocessing. To this end, we generated a complementation construct (L64TAG) that produces the prodomain (residues 1–64) separately from the remainder of the CspC protein (residues 65–557, Figure 2A). CspC was undetectable in the L64TAG mutant in western blot analyses of sporulating cells or purified spores (Figure 2B), suggesting that the CspC prodomain cannot function in trans unlike CspB [27] and other subtilisin-like serine proteases [32].
However, an important caveat to our prior finding that the CspB prodomain could function in trans was that these studies used plasmid overexpression [27]. Given that we recently determined that plasmid-based cspBA-cspC overexpression constructs can cause experimental artifacts [29,41], we tested whether chromosomally encoding the CspB prodomain in trans would allow for complementation of a cspBA deletion strain (Q66TAG, Supplemental Figure S1A). CspBA was detectable in sporulating cells of the Q66TAG complementation strain, albeit at reduced levels relative to WT and the wild-type cspBA complementation strain presumably because the chaperone activity of an intramolecular chaperone is more efficient than an intermolecular chaperone (Supplemental Figure S1B). Regardless, these results indicate that the CspB protease can still fold properly when its prodomain is supplied in trans even when Q66-TAG is expressed from the chromosome rather than a plasmid.
To assess how the decreased CspBA levels in the Q66TAG complementation strain would affect CspB, CspA, and CspC levels in mature Q66TAG spores (Supplemental Figure S1B), we analyzed the levels of these proteins in purified spores by western blotting. Consistent with our prior report that CspB and CspA are needed to incorporate and/or stabilize CspC in mature spores [29,41], reduced levels of CspB, CspA, and CspC were observed in Q66TAG spores. Importantly, the Q66TAG construct largely complemented the germination defect of the parental ΔcspBA strain, increasing the number of germinating spores by 1000-fold relative to ΔcspBA (P < 0.0001) and only 2-fold lower than WT spores (P < 0.01). Surprisingly, the greatly reduced levels of all three Csp proteins in Q66TAG spores did not strongly affect C. difficile spore germinant sensitivity, since only a ∼4-fold decrease in germination relative to WT was observed at the lowest concentration of taurocholate tested (0.001%, Supplemental Figure S1C, P < 0.005). This result suggests that Csp proteins are present in excess of what is needed to respond to germinant signals.
Restoring the catalytic triad to CspC reduces its stability
To test whether the pseudoactive site substitutions cause CspC to misfold, we determined the effect of these changes on recombinant CspC production, solubility, and autoprocessing in E. coli. We cloned the T170H, G485S, and 2xcat alleles into recombinant His-tagged protein expression vectors as well as a cspCG171R allele, a loss-of-function allele in C. difficile [28] that is thought to disrupt CspC folding due to steric hindrance (Supplemental Figure S2A, [30]). We then analyzed the resulting E. coli strains following IPTG induction and after affinity purification of the soluble fraction of E. coli lysates using Coomassie staining and western blotting. Notably, recombinant CspC2xcat was produced and purified at the same apparent molecular mass as wild-type CspC (Supplemental Figure S2B). Since an active CspC2xcat should generate a 7.5 kDa prodomain and ∼55 kDa subtilase domain through autoprocessing (Figure 2), these results indicate that restoring the catalytic triad does not reconstitute CspC protease activity.
Consistent with our finding that the pseudoactive site substitutions appear to destabilize CspC in sporulating C. difficile cells, markedly less recombinant CspC2xcat was purified from the soluble fraction of E. coli lysates relative to WT CspC-His6 (Elution fraction, Supplemental Figure S2B), even though CspC2xcat was observed at wild-type levels following IPTG induction (Induced fraction, Supplemental Figure S2B). The purification levels for the single pseudoactive site variants were also reduced relative to wild-type CspC, despite their wild-type induction levels in E. coli. Similarly, markedly reduced levels of the predicted protein folding mutant, CspCG171R, were purified despite the mutant protein being produced at wild-type levels in E. coli (Supplemental Figure S2B).
To directly test whether the pseudoactive site substitutions and G171R mutation destabilized CspC, we measured the thermal stability of purified CspC variants carrying the following substitutions: T170H, T170H-G485S (2xcat), and G171R. Due to the low yields obtained with His-tagged CspCG171R and CspC2xcat variants, we generated constructs encoding C-terminal fusions to a self-cleaving CPD-His6 fusion tag. This purification system can enhance the solubility of some proteins and increase the purity of the preparation, since CspC is separated from the CPD-His6 fusion tag upon addition of inositol hexakisphosphate [45]. The fusion tag increased the yields of WT CspC by >2-fold (data not shown) and allowed us to purify sufficient quantities of the other CspC variants for biochemical analyses (Figure 3A,B).
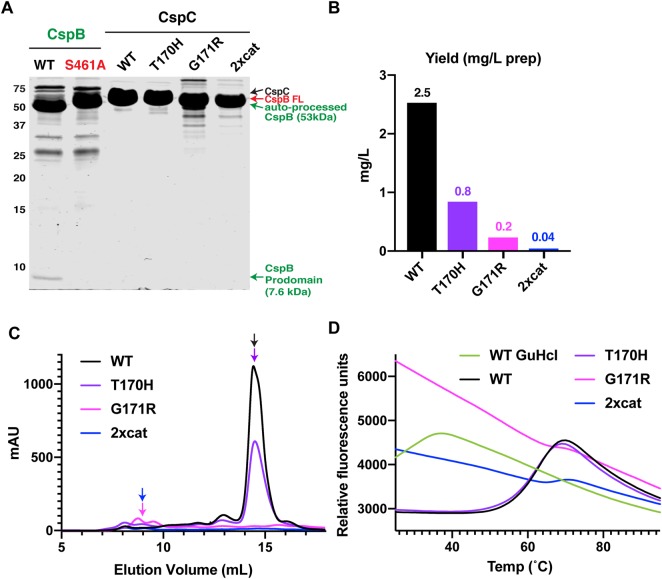
Figure 3. Changes to CspC's pseudoactive site region decrease its stability.
(A) Coomassie stain of purified C. difficile CspB and CspC. WT CspB and its catalytic site mutant, S461A, were affinity purified, while WT CspC and the CspC variants, T170H, G171R, and T170H-G485S (2xcat), were affinity purified using the self-cleaving CPD tag followed by gel filtration. WT CspB autoprocesses to release its 7.6 kDa N-terminal prodomain [27]; the catalytic mutant, CspBS461A, does not undergo autoprocessing (CspB-FL indicates full-length CspB). An active CspC2xcat would be expected to release a 7.5 kDa prodomain, but this is not observed. CspC2xcat and CspCG171R both run at slightly lower apparent MW than WT CspC, since these variants likely undergo CPD-induced trimming of their C-termini, and the C-terminal His-tagged variants run at the same apparent MW as WT CspC (Supplemental Figure S2). (B) Yields of CspC variants following CPD-mediated affinity purification and size exclusion chromatography per liter of culture. (C) Size exclusion chromatography elution profiles for CspC variants measured by mAU (A280). CspC variants purified using the CPD self-cleaving tag from 1 L of culture were concentrated and loaded onto a Superdex 200 Increase 10/300 GL column. (D) Thermofluor melt curves of the proteins purified in (C). GuHCl (guanidine hydrochloride) was used to denature WT CspC.
Similar to our findings with the His-tagged CspC variants, the yields for CspC2xcat and CspCG171R were 2% and 9% that of wild-type CspC, respectively, and the yield for CspCT170H was ∼30% that of WT CspC (Figure 3B). Notably, CspC2xcat and CspCG171R exhibited markedly different elution profiles from WT CspC and CspC2xcat during size exclusion chromatography (SEC). Whereas WT CspC and CspCT170H predominantly eluted as a monomer (estimated MW ∼60 kDa by SEC), the majority of CspC2xcat and CspCG171R appeared to aggregate, eluting at a MW ∼600 kDa by SEC (Figure 3C). They also ran at a slightly lower molecular mass than WT CspC in SDS–PAGE analyses (Figures 3 and Supplemental Figure S3), which likely reflects the self-cleaving CPD tag trimming the C-terminus of CspC2xcat and CspCG171R at Leucine 544. The CPD cleaves at leucine residues in unstructured regions [46], so partial unfolding of CspC2xcat and CspCG171R may have increased the accessibility of their C-terminal leucine residues to the CPD, especially since no difference in the apparent molecular mass of His-tagged CspC variants relative to WT CspC-His6 was observed (Supplemental Figure S2). Regardless, the identical mobilities of CspC2xcat and CspCG171R by SDS–PAGE indicate that CspC2xcat purified using the CPD tag does not undergo autoprocessing. Furthermore, the 7.4 kDa prodomain that would be liberated by an active CspC protease via autoprocessing was not detected by Coomassie staining, in contrast with the 7.6 kDa prodomain that was liberated from active CspB (Figure 3A).
When we measured the thermal stability of the purified CspC variants using a thermofluor assay, WT CspC and CspCT170H exhibited similar melting temperatures (∼68°C, Figure 3D). In contrast, high levels of SYPRO Orange binding to CspC2xcat and CspCG171R were observed even at room temperature, indicating that these variants have exposed hydrophobic regions. These results strongly suggest that purified CspC2xcat and CspCG171R are misfolded at room temperature, especially since their thermal denaturation profile resembled that of WT CspC treated with the denaturant, guanidium chloride (Figure 3D, [56]). Combined with their reduced purification yields and apparent aggregation in SEC analyses, the CspC2xcat and CspCG171R substitutions appear to destabilize CspC.
Resurrection of CspA's active site does not restore enzymatic function
Our finding that restoring the catalytic triad of CspC causes folding defects (Figures 2 and 3) is consistent with the strict conservation of the pseudoactive site residues, Thr170 and Gly485, in the Peptostreptococcaceae family ([29], Figure 1B). In contrast, the pseudoactive site substitutions in CspA's active site region are not strictly conserved, with Peptostreptococcaceae family members encoding CspA domains within CspBA fusion proteins that carry either one or two substitutions in residues of the catalytic triad and occasionally none at all (Figure 1). Since some Peptostreptococcaceae variants appear to encode active CspA domains, we hypothesized that C. difficile CspA might tolerate pseudoactive site substitutions more readily than CspC. Thus, we tested whether CspA could be converted back into an active protease by restoring its catalytic triad in C. difficile by cloning complementation constructs encoding amino acid substitutions of glutamine 757 to histidine (Q757H) and alanine 1064 to serine (A1064S) both individually and in combination. Notably, the CspA pseudoactive site substitutions did not affect CspBA levels in sporulating cells even in the mutant carrying an intact catalytic triad (BA2xcat, Figure 4B), in contrast with our analyses of the CspC pseudoactive site variants (Figure 2). The substitutions also did not affect the apparent molecular mass of CspBA2xcat relative to WT CspBA, indicating that restoring the catalytic triad to CspA is not sufficient to convert it into an active protease. If the pseudoactive site mutations had restored catalytic activity to the CspA domain, the resulting autoprocessing of the CspA2xcat prodomain would have generated a 73 kDa fragment (detectable by the anti-CspB antibody) and a 52 kDa fragment (detectable with the anti-CspA antibody, Figure 4A,B) in sporulating cells. Finally, the pseudoactive site substitutions did not affect CspBA function, since all three pseudoactive site mutants made functional, heat-resistant spores, which germinated at wild-type levels (Figure 4B).
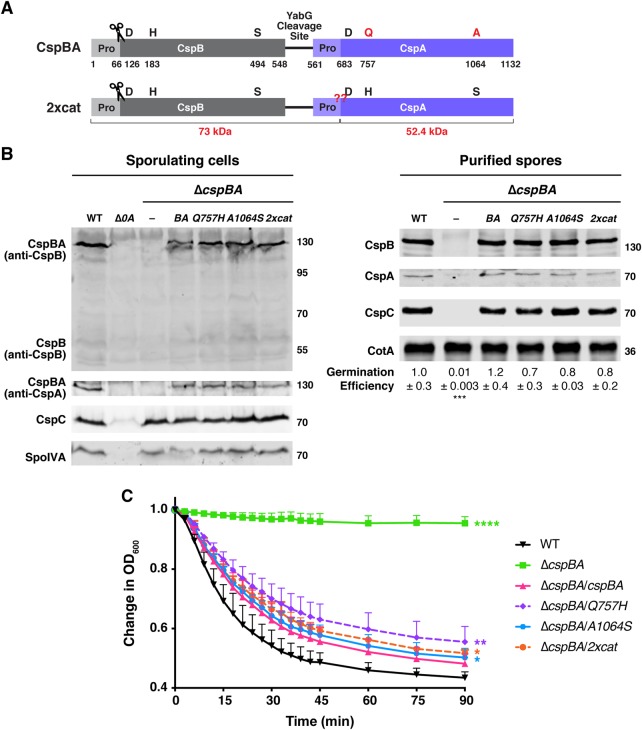
Figure 4. Restoring CspA's catalytic triad in C. difficile does not resurrect its protease activity or impact CspBA levels or function.
(A) Schematic of the CspBA fusion protein, where an active CspB protease is fused to an inactive CspA pseudoprotease domain. ‘Pro' denotes the prodomain. The C-terminal residue of the CspB prodomain following autoprocessing (denoted by the scissors) is shown [27]. Catalytic triad residues, aspartic acid (D), histidine (H) and serine (S), are shown in black; CspA pseudoactive site residues are shown in red. The CspA prodomain was identified using structural modeling [52]; the predicted cleavage site is shown in red below the schematic. The red ‘??' indicates the site where CspBA2xcat would be expected to undergo autoprocessing, if restoring the catalytic triad to CspA ‘resurrected' CspA protease activity. This would be expected to release the CspA subtilase domain (52.4 kDa) from the CspBA fusion protein, which also undergoes CspB-mediated autoprocessing. The YabG cleavage site [42] between CspB and CspA is shown. (B) Western blot analyses of CspB(A) and CspC in sporulating cells and purified spores from wild type 630Δerm-p, ΔcspBA, and ΔcspBA complemented with either wild-type cspBA or the cspBA catalytic variant (2xcat). CspBA was detected with anti-CspB and anti-CspA antibodies. Δspo0A (Δ0A) was used as a negative control for sporulating cells. SpoIVA was used as a loading control for sporulating cells. CotA was used as a loading control for purified spores. The germination efficiency of spores from the indicated strains plated on BHIS medium containing 0.1% taurocholate is shown relative to wild type. The mean and standard deviations shown are based on multiple technical replicates performed on two independent spore purifications. Statistical significance relative to wild type was determined using a one-way ANOVA and Tukey's test. (C) Change in the OD600 in response to germinant of CspBA catalytic mutant spores relative to wild-type spores. ΔcspBA mutant spores serve as a negative control. Purified spores were resuspended in BHIS broth, and germination was induced by adding taurocholate (1% final concentration). The ratio of the OD600 of each strain at a given time point relative to the OD600 at time zero is plotted. The mean of three independent assays from at least two independent spore preparations are shown. The error bars indicate the standard deviation for each time point measured. Statistical significance relative to wild type was determined using a two-way ANOVA and Tukey's test. **** P < 0.0001, *** P < 0.001, ** P < 0.01, * P < 0.05.
Since the YabG protease separates CspB from CspA coincident with spore maturation [29], we considered the possibility that CspA autoprocessing in the BA2xcat double mutant might occur after YabG-mediated cleavage. Thus, we analyzed the sizes and abundance of CspB and CspA in purified spores by western blotting. No change in the size or levels of CspA2xcat relative to WT CspA was observed in mature spores of cspBA2xcat (Figure 4B), further confirming that CspA does not undergo autoprocessing with an intact catalytic triad. No changes in CspB and CspC sizes or levels were observed in CspA pseudoactive site mutant spores (Figure 4B), which germinated with similar efficiency as WT on agar plates containing taurocholate germinant (Figure 4B). The mutant spores also germinated at similar rates in the optical density-based assay relative to the wild-type cspBA complementation strain (ΔcspBA/cspBA), albeit slightly slower than WT spores (Figure 4C). Taken together, these results indicate that factors beyond CspA's pseudoactive catalytic triad prevent the CspA pseudoprotease from functioning as an active protease and that CspA can tolerate substitutions in its pseudoactive site region more readily than CspC.
While the inability to resurrect CspA's protease activity likely reflects structural differences between the C. difficile CspA pseudoprotease and active Csp proteins in other clostridial organisms, it was formally possible that an unknown inhibitory factor in C. difficile prevented CspA2xcat from acquiring autoprocessing activity. To test this possibility, we cloned codon-optimized pseudoactive site mutant alleles of cspBA into vectors for IPTG-inducible recombinant protein production in E. coli. We then measured CspBA-His6 variant production and purification levels in E. coli. Constructs encoding full-length CspBA were generated, since the fusion protein reflects the form of CspA first produced in C. difficile sporulating cells, and CspB is needed to stabilize CspA in both sporulating C. difficile cells [41] and E. coli (data not shown).
While restoring the catalytic triad to CspC destabilized recombinant CspC2xcat, the equivalent mutations did not affect the production or purification of CspBA2xcat-His6 relative to WT CspBA based on Coomassie staining (Figure 5, top panel) and western blotting analyses (Figure 5, bottom panel). Although full-length CspBA was only ∼4% of the total protein purified based on quantifications of the Coomassie staining, no CspA autoprocessing was observed in CspBA2xcat in either the induced or purified fractions by western blotting (Figure 5). Autoprocessing would have led to CspA2xcat becoming separated from CspB by generating a 73 kDa fragment detectable with the anti-CspB antibody and a 52 kDa fragment detectable with the anti-His antibody.
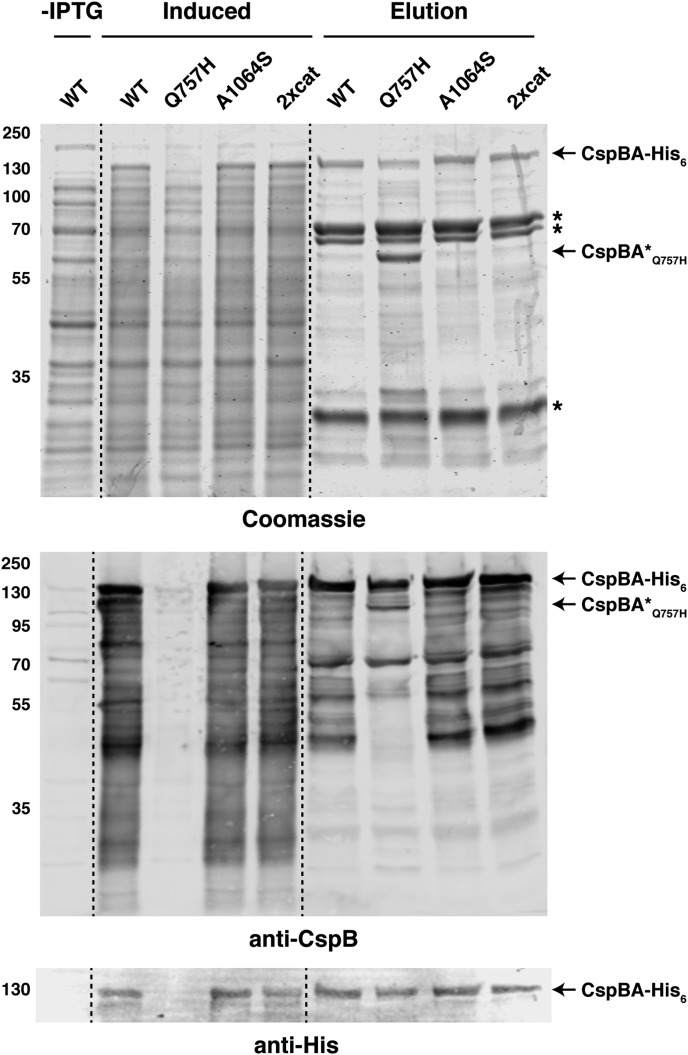
Figure 5. Resurrection of the CspA active site does not restore protease activity in E. coli.
Purification of CspBA variants from the soluble fraction. Cultures expressing the cspBA variants were induced with IPTG overnight at 18°C, and aliquots were removed for analysis of the ‘induced' fraction compared with the ‘uninduced' fraction prior to the addition of IPTG (-IPTG). Cultures were harvested, and cells were lysed using sonication. Following a high-speed centrifugation, the cleared lysate containing soluble proteins was incubated with Ni2+-NTA agarose beads. CspBA-His6 variants were eluted from the beads using imidazole (elution fraction). Samples were resolved by SDS–PAGE and analyzed by Coomassie staining (top) and western blotting (bottom). Non-specific proteins pulled-down with the Ni-NTA beads are marked with asterisks; a truncated CspBA Q757H variant is also marked. An anti-His6 antibody was used to detect full-length CspBA. An anti-CspB antibody was used to detect CspBA-His6 variants.
Unexpectedly, marked decreases in full-length CspBAQ757H were observed in the IPTG-induced fraction compared with WT and the other CspBA variants. CspBAQ757H appeared to be susceptible to protease cleavage in E. coli based on its altered banding pattern in the elution fraction relative to WT and the other CspBA variants (Figure 5), which could suggest that it has a folding defect. However, since the Q757H substitution did not affect CspBA size or function in C. difficile (Figure 4), if CspBAQ757H folding is impacted by the substitution, it does not appear to be physiologically relevant in C. difficile. Regardless, our data demonstrate that intrinsic structural features within the CspA pseudoprotease prevent it from being converted to a functional enzyme, even when its catalytic triad is restored.
Discussion
While pseudoenzymes most frequently arise from gene duplications [6,7], the extent to which a given pseudoenzyme's function has diverged from its ancestral enzymatic function varies for the limited number of pseudoenzymes that have been tested [1]. Our results indicate that protease activity is not restored to the CspA or CspC pseudoproteases of C. difficile when their catalytic triad is restored, since the autoprocessing that characterizes all S8 family proteases [32,38] was not observed in either natively or recombinantly produced variants. Thus, the C. difficile CspC and CspA pseudoproteases have evolved multiple mechanisms to ensure their loss of catalytic activity.
This loss of catalytic activity likely allowed the C. difficile CspA and CspC pseudoproteases to gain new functions separate from the role of active Csp family members in cleaving the SleC cortex lytic enzyme during germination [27]. In C. difficile, CspA and CspC are thought to bind small molecules, namely co-germinants and bile acid germinants, respectively [28,42]. Sensing of these small molecules somehow allows the CspA and CspC pseudoproteases to activate the CspB protease. Interestingly, this putative signaling architecture is similar to how many pseudoenzymes allosterically regulate the activity of their cognate enzymes [1,5], sometimes in response to small molecule binding [57].
Pseudoprotease-mediated allosteric regulation of a cognate protease has been best characterized for the c-FLIP-caspase-8 pseudoprotease-protease pair. c-FLIP has the same domain structure as caspase-8, but mutations in its active site dyad render it catalytically inactive. Heterodimerization of c-FLIP and procaspase-8 leads to low efficiency cleavage of the prodomain of c-FLIP by procaspase-8. Cleaved c-FLIP adopts a slightly different conformation that helps bring procaspase-8 into an active conformation, enhancing its catalytic activity [58,59]. By analogy, the structurally related C. difficile CspC and CspA pseudoproteases may allosterically regulate the CspB protease through heterodimerization, especially since some subtilisin-like serine proteases can function as dimers [60–62].
Regardless, another important finding of our studies is that the CspC and CspA pseudoproteases differentially tolerate changes to their pseudoactive site residues. Restoring CspC's catalytic triad destabilized CspC in C. difficile (Figure 2) and disrupted protein folding in E. coli (Figure 3), whereas the equivalent substitutions in CspA did not impact CspA folding in either organism or function in C. difficile (Figures 4 and 5). Given that these pseudoproteases tolerate changes in their pseudoactive sites to different degrees, our findings raise the question as to how CspC and CspA independently evolved to become pseudoproteases in C. difficile and other Peptostreptococcaceae family members.
In the case of CspC, loss of its catalytic site residues was likely critical to its evolution as a pseudoprotease. Although the crystal structure of CspC suggests that the substrate binding pocket can accommodate the catalytic triad residues (Figure 1, [30]), restoring these residues disrupts CspC folding (Figure 3D). Given that the specific identity of these pseudoactive site residues is strictly conserved in CspC homologs in the Peptostreptococcaceae family, the chemical properties of its specific pseudoactive site residues, namely threonine 170 and glycine 485 in C. difficile CspC (Figure 1, [29]), are likely crucial for the structural integrity of Peptostreptococcaceae family CspC homologs.
The changes beyond the active site substitutions in C. difficile CspC likely include residues involved in binding its prodomain. Our data indicate that the CspC prodomain cannot act as a chaperone when supplied in trans (Figure 2B), unlike CspB and many other subtilisin-like serine proteases (Supplemental Figure S1, [27,32,38]), suggesting that the requirements for C. difficile CspC folding differ from those of other S8 family proteases. Notably, C. difficile CspC's prodomain is bound more tightly to its subtilase domain than the prodomain of C. perfringens CspB [27,30], in part due to a ‘clamp' region in C. difficile CspC that holds the prodomain in place but is absent from other subtilisin-like proteases [30]. In addition, CspC is highly sensitive to changes in its pseudoactive site region, since the loss-of-function cspC alleles identified in a prior genetic screen by Francis et al. [28] all cluster to this region and likely prevent prodomain binding to the pseudoactive site region via steric occlusion (Supplemental Figure S2). Consistent with this interpretation, we showed that one of these alleles, cspCG171R, caused misfolding of recombinant CspC (Figure 3), strongly suggesting that the loss-of-function mutations identified in the germination mutant screen [28] destabilize CspC in sporulating cells.
While CspC structure and function rely critically on the identity of its pseudoactive site residues, CspA structure and function were unaffected by substitutions that restore the catalytic triad (Figure 4). These results suggest that loss of C. difficile CspA's catalytic residues was not critical for its evolution into a pseudoprotease, unlike C. difficile CspC. While substitution of catalytic site residues is thought to be the most frequent driving force behind the evolution of new functions for pseudoenzymes, substitutions that prevent substrate binding or catalysis through other mechanisms have also been observed in some pseudoenzymes [6,7]. It is likely that C. difficile’s CspA pseudoprotease domain has evolved analogous substitutions that render it catalytically inactive, but the nature of these substitutions remains unclear in the absence of a CspA crystal structure. Regardless, our results raise the important possibility that CspA domains in Peptostreptococcaeae family CspBA homologs with intact catalytic triads (Figure 1B) may include substitutions that occlude substrate binding and prevent protease activity.
A similar case of pseudoenzymes lacking catalytic activity despite retaining their active site residues has been observed in the iRhom family of proteins, which are widely conserved, inactive homologs of rhomboid proteases [6]. While most iRhom pseudoproteases lack either one or both catalytic dyad residues required for rhomboid protease activity, at least one iRhom family member carries an intact catalytic dyad yet lacks protease activity [63]. Notably, a distinguishing feature of iRhoms is the presence of a proline residue adjacent to the catalytic serine (or pseudoactive site residue) that is sufficient to prevent proteolytic activity in rhomboid proteases [63]. Whether a similar type of inactivating residue occurs in the CspA (or CspC) pseudoproteases in the Peptostreptococcaceae family remains to be determined, but it is worth noting that iRhom pseudoproteases carry additional structural features that distinguish iRhoms from rhomboid proteases beyond the catalytic site substitutions [6] including an extended cytoplasmic amino terminus and conserved cysteine-rich luminal loop domain.
However, unlike iRhoms and rhomboid proteases, the CspC pseudoprotease exhibits a high degree of structural similarity to the CspB protease, with the structures almost being superimposable (rmsd of ∼1 Å, [30]). A similar degree of structural similarity to CspB is predicted for the CspA pseudoprotease [52], so the elements of CspA structure that prevent its catalytic activity remain unclear. Regardless, our data suggest that the bioinformatics-based analyses used to predict catalytic activity based on ‘the absence of one or more catalytic residues' [2] could under-estimate the prevalence of inactive enzymes in rare cases [63]. As more pseudoenzymes are directly studied rather than bioinformatically predicted, it is likely that additional pseudoenzymes with intact catalytic triads will be discovered.
Indeed, studying the mechanism and function of pseudoenzymes has revealed that these proteins can be excellent targets for drug discovery, since small molecules can differentially target their remodeled active sites without impacting their active enzyme counterparts [1,5]. Further elucidating the critical properties and mechanisms of action of Csp pseudoproteases could represent a promising avenue for therapeutic intervention, since C. difficile spore germination is required to initiate infection [28,64,65].
Acknowledgements
We would like to thank J. Sorg for generously sharing codon-optimized versions of cspA, cspB, and cspC and the anti-CspA antibody; N. Minton (U. Nottingham) for providing us with access to the 630ΔermΔpyrE strain and pMTL-YN1C and pMTL-YN3 plasmids for allele-coupled exchange (ACE); and Marcin Dembek for directly providing these materials to us and sharing his specific protocols on ACE with us.
Abbreviations
- Csp
clostridial serine protease
- InsP6
inositol hexakisphosphate
- SEC
size exclusion chromatography
- TA
taurocholate
- WT
wild-type
Competing Interests
A.S. has a paid consultancy for BioVector, Inc., a diagnostic start-up.
Funding
Research in this manuscript was funded by Award Number K12 GM133314 to A.E.R., who is a Tufts IRACDA fellow, and T32 GM007310 to E.R.F., and Award Number R01GM108684 to A.S from the National Institutes of General Medical Sciences and R21AI26067 to A.S. from the National Institutes of Allergy and Infectious Disease. A.S. is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease supported by the Burroughs Wellcome Fund. M.L.D was supported in part by the Alpha Omega Alpha Carolyn L. Kuckein Student Research Fellowship. The content is solely the responsibility of the author(s) and does not necessarily reflect the views of the Burroughs Wellcome Fund, or the National Institutes of Health. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author Contribution
A.S. conceived the hypothesis and supervised the project with help from A.E.R. M.L.D, E.R.F, A.E.R. and A.S designed the experiments. A.S. constructed the single pseudoactive site mutants for CspC in C. difficile, codon-optimized cspC mutants carrying single pseudoactive site mutants, and codon-optimized cspBA expression construct. M.L.D. constructed the double pseudoactive site mutant of CspC, all the CspA pseudoactive site mutants, and cspBA codon-optimized expression constructs encoding pseudoactive site substitutions. E.R.F. cloned the G171R and double pseudoactive site mutant expression constructs for codon-optimized CspC. M.L.D. performed the phenotypic characterization of C. difficile strains (heat-resistance, plate-based and optical density-based germination assays, and western blot analyses) unless otherwise indicated, as well as the E. coli protein purification analyses of CspBA, purification and analysis of CspC-CPD-His6 variants using size exclusion chromatography and thermofluor assays. E.R.F. performed the E. coli protein purification analyses of CspC-His6. A.S. performed the phenotypic analyses of the cspBA prodomain trans-complementation analyses. A.E.R. optimized the thermofluor assay. M.L.D and A.S. wrote the manuscript with help from A.E.R. and E.R.F.
Supplementary Material
References
- 1.Murphy J.M., Farhan H. and Eyers P.A. (2017) Bio-Zombie: the rise of pseudoenzymes in biology. Biochem. Soc. Trans. 45, 537–544 10.1042/BST20160400 [DOI] [PubMed] [Google Scholar]
- 2.Ribeiro A.J.M., Das S., Dawson N., Zaru R., Orchard S., Thornton J.M. et al. (2019) Emerging concepts in pseudoenzyme classification, evolution, and signaling. Sci. Signal. 12, eaat9797 10.1126/scisignal.aat9797 [DOI] [PubMed] [Google Scholar]
- 3.Pils B. and Schultz J. (2004) Inactive enzyme-homologues find new function in regulatory processes. J. Mol. Biol. 340, 399–404 10.1016/j.jmb.2004.04.063 [DOI] [PubMed] [Google Scholar]
- 4.Eyers P.A. and Murphy J.M. (2016) The evolving world of pseudoenzymes: proteins, prejudice and zombies. BMC Biol. 14, 98 10.1186/s12915-016-0322-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy J.M., Mace P.D. and Eyers P.A. (2017) Live and let die: insights into pseudoenzyme mechanisms from structure. Curr. Opin. Struct. Biol. 47, 95–104 10.1016/j.sbi.2017.07.004 [DOI] [PubMed] [Google Scholar]
- 6.Adrain C. and Freeman M. (2012) New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat. Rev. Mol. Cell Biol. 13, 489–498 10.1038/nrm3392 [DOI] [PubMed] [Google Scholar]
- 7.Todd A.E., Orengo C.A. and Thornton J.M. (2002) Sequence and structural differences between enzyme and nonenzyme homologs. Structure 10, 1435–1451 10.1016/S0969-2126(02)00861-4 [DOI] [PubMed] [Google Scholar]
- 8.Zaru R., Magrane M., Orchard S. and UniProt C. (2019) Challenges in the annotation of pseudoenzymes in databases: the UniProtKB approach. FEBS J. 10.1111/febs.15100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wishart M.J., Denu J.M., Williams J.A. and Dixon J.E. (1995) A single mutation converts a novel phosphotyrosine binding domain into a dual-specificity phosphatase. J. Biol. Chem. 270, 26782–26785 10.1074/jbc.270.45.26782 [DOI] [PubMed] [Google Scholar]
- 10.Wishart M.J. and Dixon J.E. (1998) Gathering STYX: phosphatase-like form predicts functions for unique protein-interaction domains. Trends Biochem. Sci. 23, 301–306 10.1016/S0968-0004(98)01241-9 [DOI] [PubMed] [Google Scholar]
- 11.Murphy J.M., Zhang Q., Young S.N., Reese M.L., Bailey F.P., Eyers P.A. et al. (2014) A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem. J. 457, 323–334 10.1042/BJ20131174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Childers W.S., Xu Q., Mann T.H., Mathews I.I., Blair J.A., Deacon A.M. et al. (2014) Cell fate regulation governed by a repurposed bacterial histidine kinase. PLoS Biol. 12, e1001979 10.1371/journal.pbio.1001979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prigent S.A. and Gullick W.J. (1994) Identification of c-erbB-3 binding sites for phosphatidylinositol 3′-kinase and SHC using an EGF receptor/c-erbB-3 chimera. EMBO J. 13, 2831–2841 10.1002/j.1460-2075.1994.tb06577.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi F., Telesco S.E., Liu Y., Radhakrishnan R. and Lemmon M.A. (2010) Erbb3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc. Natl Acad. Sci. U.S.A. 107, 7692–7697 10.1073/pnas.1002753107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendrola J.M., Shi F., Park J.H. and Lemmon M.A. (2013) Receptor tyrosine kinases with intracellular pseudokinase domains. Biochem. Soc. Trans. 41, 1029–1036 10.1042/BST20130104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abt M.C., McKenney P.T. and Pamer E.G. (2016) Clostridium difficile colitis: pathogenesis and host defence. Nat. Rev. Microbiol. 14, 609–620 10.1038/nrmicro.2016.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rupnik M., Wilcox M.H. and Gerding D.N. (2009) Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7, 526–536 10.1038/nrmicro2164 [DOI] [PubMed] [Google Scholar]
- 18.Lessa F.C., Mu Y., Bamberg W.M., Beldavs Z.G., Dumyati G.K., Dunn J.R. et al. (2015) Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 372, 825–834 10.1056/NEJMoa1408913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CDC, (2019) Antibiotic Resistance Threats in the United States https://www.cdc.gov/drugresistance/biggest-threats.html
- 20.Giordano N., Hastie J.L. and Carlson P.E. (2018) Transcriptomic profiling of Clostridium difficile grown under microaerophillic conditions. Pathog Dis 76, fty010 10.1093/femspd/fty010 [DOI] [PubMed] [Google Scholar]
- 21.Paredes-Sabja D., Shen A. and Sorg J.A. (2014) Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol. 22, 406–416 10.1016/j.tim.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen A. (2015) A Gut Odyssey: the impact of the microbiota on Clostridium difficile spore formation and germination. PLoS Pathog. 11, e1005157 10.1371/journal.ppat.1005157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharjee D., McAllister K.N. and Sorg J.A. (2016) Germinants and their receptors in Clostridia. J. Bacteriol. 198, 2767–2775 10.1128/JB.00405-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kochan T.J., Foley M.H., Shoshiev M.S., Somers M.J., Carlson P.E. and Hanna P.C. (2018) Updates to Clostridium difficile spore germination. J. Bacteriol 200, e00218-18 10.1128/JB.00218-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen A., Edwards A.N., Sarker M.R. and Paredes-Sabja D. (2019) Sporulation and germination in Clostridial pathogens. Microbiol. Spectr 7 10.1128/microbiolspec.GPP3-0017-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu D., Sorg J.A. and Sun X. (2018) Clostridioides difficile biology: sporulation, germination, and corresponding therapies for C. difficile infection. Front Cell Infect Microbiol. 8, 29 10.3389/fcimb.2018.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams C.M., Eckenroth B.E., Putnam E.E., Doublie S. and Shen A. (2013) Structural and functional analysis of the CspB protease required for Clostridium spore germination. PLoS Pathog. 9, e1003165 10.1371/journal.ppat.1003165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francis M.B., Allen C.A., Shrestha R. and Sorg J.A. (2013) Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog. 9, e1003356 10.1371/journal.ppat.1003356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kevorkian Y., Shirley D.J. and Shen A. (2016) Regulation of Clostridium difficile spore germination by the CspA pseudoprotease domain. Biochimie 122, 243–254 10.1016/j.biochi.2015.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohlfing A.E., Eckenroth B.E., Forster E.R., Kevorkian Y., Donnelly M.L., Benito de la Puebla H. et al. (2019) The CspC pseudoprotease regulates germination of Clostridioides difficile spores in response to multiple environmental signals. PLoS Genet. 15, e1008224 10.1371/journal.pgen.1008224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimamoto S., Moriyama R., Sugimoto K., Miyata S. and Makino S. (2001) Partial characterization of an enzyme fraction with protease activity which converts the spore peptidoglycan hydrolase (SleC) precursor to an active enzyme during germination of Clostridium perfringens S40 spores and analysis of a gene cluster involved in the activity. J. Bacteriol. 183, 3742–3751 10.1128/JB.183.12.3742-3751.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shinde U. and Thomas G. (2011) Insights from bacterial subtilases into the mechanisms of intramolecular chaperone-mediated activation of furin. Methods Mol. Biol. 768, 59–106 10.1007/978-1-61779-204-5_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paredes-Sabja D., Setlow P. and Sarker M.R. (2011) Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 19, 85–94 10.1016/j.tim.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 34.Fimlaid K.A., Jensen O., Donnelly M.L., Francis M.B., Sorg J.A. and Shen A. (2015) Identification of a novel lipoprotein regulator of Clostridium difficile spore germination. PLoS Pathog. 11, e1005239 10.1371/journal.ppat.1005239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francis M.B., Allen C.A. and Sorg J.A. (2015) Spore cortex hydrolysis precedes dipicolinic acid release during Clostridium difficile spore germination. J Bacteriol. 197, 2276–2283 10.1128/JB.02575-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urakami K., Miyata S., Moriyama R., Sugimoto K. and Makino S. (1999) Germination-specific cortex-lytic enzymes from Clostridium perfringens S40 spores: time of synthesis, precursor structure and regulation of enzymatic activity. FEMS Microbiol Lett. 173, 467–473 10.1111/j.1574-6968.1999.tb13540.x [DOI] [PubMed] [Google Scholar]
- 37.Siezen R.J. and Leunissen J.A. (1997) Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 6, 501–523 10.1002/pro.5560060301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shinde U. and Inouye M. (2000) Intramolecular chaperones: polypeptide extensions that modulate protein folding. Semin. Cell Dev. Biol. 11, 35–44 10.1006/scdb.1999.0349 [DOI] [PubMed] [Google Scholar]
- 39.Comellas-Bigler M., Maskos K., Huber R., Oyama H., Oda K. and Bode W. (2004) 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin; structure of an intact pro-subtilase. Structure 12, 1313–1323 10.1016/j.str.2004.04.013 [DOI] [PubMed] [Google Scholar]
- 40.Tanaka S.I., Matsumura H., Koga Y., Takano K. and Kanaya S. (2007) Four new crystal structures of Tk-subtilisin in unautoprocessed, autoprocessed and mature forms: insight into structural changes during maturation. J. Mol. Biol. 372, 1055–1069 10.1016/j.jmb.2007.07.027 [DOI] [PubMed] [Google Scholar]
- 41.Kevorkian Y. and Shen A. (2017) Revisiting the role of Csp family proteins in regulating Clostridium difficile spore germination. J. Bacteriol 199, e0026-17 10.1128/JB.00266-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shrestha R., Cochran A.M. and Sorg J.A. (2019) The requirement for co-germinants during Clostridium difficile spore germination is influenced by mutations in yabG and cspA. PLoS Pathog. 15, e1007681 10.1371/journal.ppat.1007681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng Y.K., Ehsaan M., Philip S., Collery M.M., Janoir C., Collignon A. et al. (2013) Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS ONE 8, e56051 10.1371/journal.pone.0056051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horton R.M., Hunt H.D., Ho S.N., Pullen J.K. and Pease L.R. (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68 10.1016/0378-1119(89)90359-4 [DOI] [PubMed] [Google Scholar]
- 45.Shen A., Lupardus P.J., Morell M., Ponder E.L., Sadaghiani A.M., Garcia K.C. et al. (2009) Simplified, enhanced protein purification using an inducible, autoprocessing enzyme tag. PLoS ONE 4, e8119 10.1371/journal.pone.0008119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen A., Lupardus P.J., Albrow V.E., Guzzetta A., Powers J.C., Garcia K.C. et al. (2009) Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin. Nat. Chem. Biol. 5, 469–478 10.1038/nchembio.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donnelly M.L., Li W., Li Y.Q., Hinkel L., Setlow P. and Shen A. (2017) A Clostridium difficile-specific, gel-forming protein required for optimal spore germination. mBio 8, e02085-16 10.1128/mBio.02085-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ribis J.W., Ravichandran P., Putnam E.E., Pishdadian K. and Shen A. (2017) The conserved spore coat protein SpoVM is largely dispensable in clostridium difficile spore formation. mSphere 2, e00315-17 10.1128/mSphere.00315-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen A., Fimlaid K.A. and Pishdadian K. (2016) Inducing and quantifying Clostridium difficile spore formation. Methods Mol. Biol. 1476, 129–142 10.1007/978-1-4939-6361-4_10 [DOI] [PubMed] [Google Scholar]
- 50.Donnelly M.L., Fimlaid K.A. and Shen A. (2016) Characterization of Clostridium difficile spores lacking either SpoVAC or dipicolinic acid synthetase. J. Bacteriol. 198, 1694–1707 10.1128/JB.00986-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Putnam E.E., Nock A.M., Lawley T.D. and Shen A. (2013) SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J. Bacteriol. 195, 1214–1225 10.1128/JB.02181-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang J. and Zhang Y. (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181 10.1093/nar/gkv342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rawlings N.D., Barrett A.J., Thomas P.D., Huang X., Bateman A. and Finn R.D. (2018) The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 46, D624–D632 10.1093/nar/gkx1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paidhungat M. and Setlow P. (2000) Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182, 2513–2519 10.1128/JB.182.9.2513-2519.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sturm A. and Dworkin J. (2015) Phenotypic diversity as a mechanism to exit cellular dormancy. Curr. Biol. 25, 2272–2277 10.1016/j.cub.2015.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sviben D., Bertosa B., Hlousek-Kasun A., Forcic D., Halassy B. and Brgles M. (2018) Investigation of the thermal shift assay and its power to predict protein and virus stabilizing conditions. J. Pharm. Biomed. Anal. 161, 73–82 10.1016/j.jpba.2018.08.017 [DOI] [PubMed] [Google Scholar]
- 57.Huang H., Zeqiraj E., Dong B., Jha B.K., Duffy N.M., Orlicky S. et al. (2014) Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity. Mol. Cell 53, 221–234 10.1016/j.molcel.2013.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang D.W., Xing Z., Pan Y., Algeciras-Schimnich A., Barnhart B.C., Yaish-Ohad S. et al. (2002) c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J. 21, 3704–3714 10.1093/emboj/cdf356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu J.W., Jeffrey P.D. and Shi Y. (2009) Mechanism of procaspase-8 activation by c-FLIPL. Proc. Natl Acad. Sci. U.S.A. 106, 8169–8174 10.1073/pnas.0812453106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gamble M., Kunze G., Brancale A., Wilson K.S. and Jones D.D. (2012) The role of substrate specificity and metal binding in defining the activity and structure of an intracellular subtilisin. FEBS Open Bio. 2, 209–215 10.1016/j.fob.2012.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ottmann C., Rose R., Huttenlocher F., Cedzich A., Hauske P., Kaiser M. et al. (2009) Structural basis for Ca2+-independence and activation by homodimerization of tomato subtilase 3. Proc. Natl Acad. Sci. U.S.A. 106, 17223–17228 10.1073/pnas.0907587106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vevodova J., Gamble M., Kunze G., Ariza A., Dodson E., Jones D.D. et al. (2010) Crystal structure of an intracellular subtilisin reveals novel structural features unique to this subtilisin family. Structure 18, 744–755 10.1016/j.str.2010.03.008 [DOI] [PubMed] [Google Scholar]
- 63.Lemberg M.K. and Freeman M. (2007) Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 17, 1634–1646 10.1101/gr.6425307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Howerton A., Patra M. and Abel-Santos E. (2013) Fate of ingested Clostridium difficile spores in mice. PLoS ONE 8, e72620 10.1371/journal.pone.0072620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Howerton A., Seymour C.O., Murugapiran S.K., Liao Z., Phan J.R., Estrada A. et al. (2018) Effect of the synthetic bile salt analog CamSA on the Hamster model of Clostridium difficile infection. Antimicrob. Agents Chemother 62, e02551-17 10.1128/AAC.02251-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.