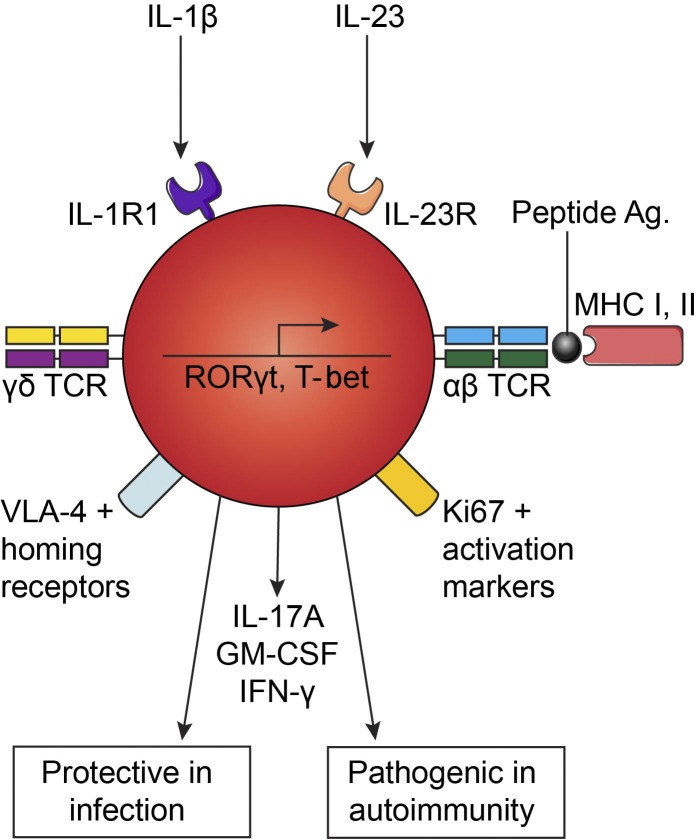
T cells classically express either αβ or γδ T cell receptors. We have identified T cells that express both pairs of receptors. These hybrid αβ-γδ T cells exhibit a hyperinflammatory and migratory phenotype and act as first responders in infection and CNS autoimmunity.
Abstract
T cells are classically recognized as distinct subsets that express αβ or γδ TCRs. We identify a novel population of T cells that coexpress αβ and γδ TCRs in mice and humans. These hybrid αβ-γδ T cells arose in the murine fetal thymus by day 16 of ontogeny, underwent αβ TCR–mediated positive selection into CD4+ or CD8+ thymocytes, and constituted up to 10% of TCRδ+ cells in lymphoid organs. They expressed high levels of IL-1R1 and IL-23R and secreted IFN-γ, IL-17, and GM-CSF in response to canonically restricted peptide antigens or stimulation with IL-1β and IL-23. Hybrid αβ-γδ T cells were transcriptomically distinct from conventional γδ T cells and displayed a hyperinflammatory phenotype enriched for chemokine receptors and homing molecules that facilitate migration to sites of inflammation. These proinflammatory T cells promoted bacterial clearance after infection with Staphylococcus aureus and, by licensing encephalitogenic Th17 cells, played a key role in the development of autoimmune disease in the central nervous system.
Graphical Abstract
Introduction
MHC-restricted CD4+ and CD8+ T cells typically mediate pathogen-specific adaptive immunity and express αβ TCRs. In contrast, γδ T cells play an important role in innate immunity at mucosal surfaces but can also display features of immunological memory, analogous to conventional αβ T cells (Misiak et al., 2017; Sutton et al., 2009). The accepted dogma is that common lymphoid progenitors develop into cells that express either αβ or γδ TCRs and that each population subsequently occupies a specific and highly conserved niche within the immune system.
γδ T cells are required for optimal innate and adaptive immune responses to infection and tumors (Murphy et al., 2014; Rei et al., 2014; Silva-Santos et al., 2015). They are the first lymphocytes to emerge in the fetus, and before full maturation of the immune system, they mediate protective functions in young animals (Shibata et al., 2007; Sinkora et al., 2005). A unique feature of murine γδ T cells is the preferential expression of different TCRγ variable region (Vγ) segments in different tissues. For example, Vγ5+ γδ T cells are present in skin, Vγ6+ γδ T cells localize to the reproductive mucosa, and Vγ1+ or Vγ4+ γδ T cells are found in secondary lymphoid organs (nomenclature of Heilig and Tonegawa, 1986). γδ T cells produce an array of cytokines, including IFN-γ, IL-4, IL-17A, IL-17F, IL-21, IL-22, GM-CSF, and TNF-α (Lockhart et al., 2006; Ribot et al., 2009; Sutton et al., 2012).
Although γδ T cells display characteristics of adaptive memory, they can also produce IL-17 upon stimulation with IL-1β and IL-23 in the absence of TCR engagement and provide an early source of innate proinflammatory cytokines that help amplify T helper type 17 (Th17) responses in certain autoimmune and infectious diseases (Conti et al., 2014; Crowley et al., 1997; Sutton et al., 2009). In humans with multiple sclerosis, increased frequencies of γδ T cells have been detected in acute brain lesions (Hvas et al., 1993; Wucherpfennig et al., 1992), and clonal expansions of γδ T cells have been observed in cerebrospinal fluid during the early stages of disease (Shimonkevitz et al., 1993). Similarly, IL-17–producing Vγ4+ T cells infiltrate the brain and spinal cord of mice with experimental autoimmune encephalomyelitis (EAE; Price et al., 2012; Sutton et al., 2009). Vγ4+ T cells also mediate inflammation via IL-17 production in the dermis of mice with psoriasis (Cai et al., 2011) and accumulate in the draining LNs and joints of mice with collagen-induced arthritis (Roark et al., 2007).
In this study, we identified a discrete population of T cells that coexpressed αβ and γδ TCRs. These hybrid αβ-γδ T cells were transcriptomically distinct from conventional γδ T cells, poised to migrate to sites of inflammation, and responsive to MHC class I (MHCI)–restricted or MHCII-restricted peptide antigens or stimulation with IL-1β and IL-23. In line with these findings, hybrid αβ-γδ T cells protected against infection with Staphylococcus aureus and, by licensing encephalitogenic Th17 cells, triggered autoimmune pathology in the central nervous system (CNS).
Results and discussion
Identification of hybrid αβ-γδ T cells
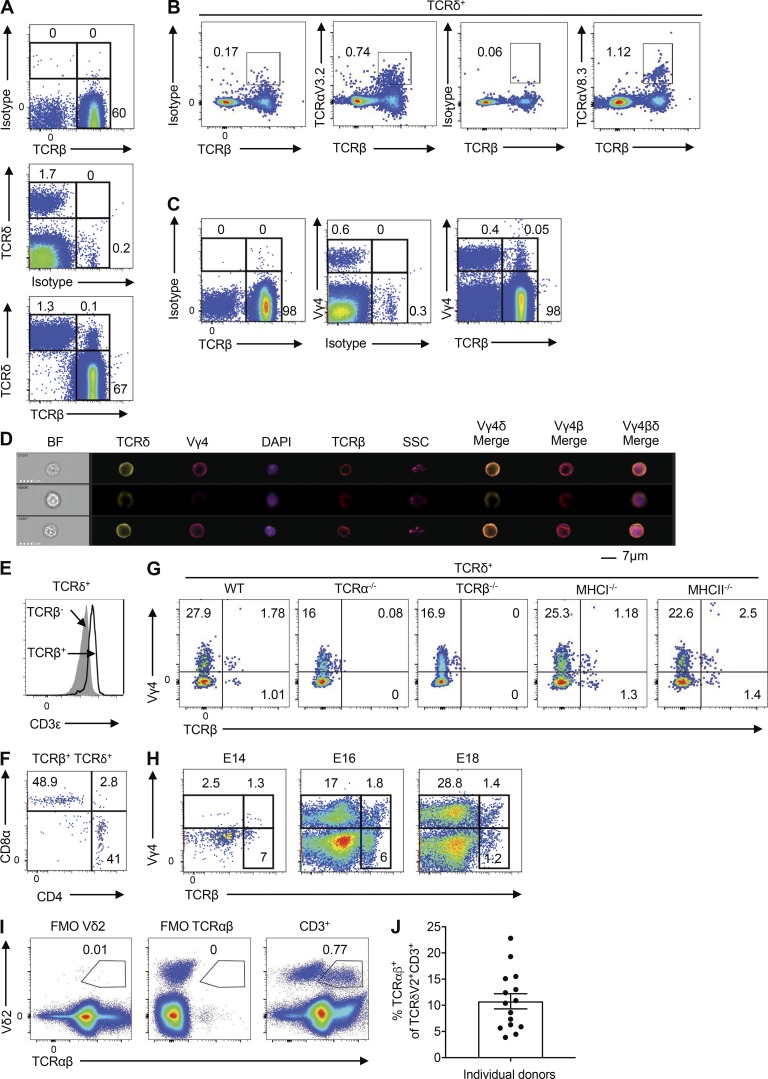
Initial flow cytometric analyses with antibodies specific for the constant regions of TCRδ and TCRβ unexpectedly revealed a rare population of TCRδ+TCRβ+ cells in the LNs of WT C57BL/6 mice (Fig. 1 A and Fig. S1 A). These findings were substantiated using confocal microscopy, which demonstrated surface expression of TCRβ on purified TCRδ+ cells (Fig. S1 B), and RT-PCR, which demonstrated the presence of transcripts encoding the joining region of TCRβ (Trbj) in purified TCRδ+ cells (Fig. S1 C). Moreover, TCRδ+ cells coexpressed TCRβ and various TCRα variable region (Vα) segments (Fig. 1 B), and some Vγ4+, Vγ1+, and Vγ4–Vγ1− T cells coexpressed TCRδ and TCRβ (Fig. 1 C and Fig. S1, D and E). ImageStream experiments confirmed coexpression of Vγ4 with TCRδ and TCRβ at the single-cell level (Fig. 1 D). In addition, flow cytometric analyses revealed that ∼6% of Vγ4+TCRδ+ cells expressed TCRβ (Fig. S1 F). These hybrid αβ-γδ T cells expressed higher levels of CD3ε than TCRδ+TCRβ− cells (Fig. 1 E), together with either CD4 or CD8α (Fig. 1 F). Coexpression of αβ and γδ TCRs was further validated using TCRα−/− and TCRβ−/− mice, which lacked hybrid αβ-γδ T cell populations, whereas ∼7% of Vγ4+TCRδ+ cells in WT mice expressed TCRβ (Fig. 1 G). Importantly, Vγ4+TCRδ+TCRβ+ cells were present in MHCI−/− and MHCII−/− mice (Fig. 1 G). CD4+Vγ4+TCRδ+TCRβ+ cells were absent in MHCII−/− mice, whereas CD8+Vγ4+TCRδ+TCRβ+ cells were absent in MHCI−/− mice (Fig. S1 G). Double-negative cells were the most predominant population in each strain (Fig. S1 G). An examination of the ontogeny of hybrid αβ-γδ T cells revealed that Vγ4+ cells emerged in the thymus between embryonic days (E) 14 and 16 (E14 and E16; Fig. 1 H). Although the majority of these Vγ4+ cells were TCRδ+TCRβ− (conventional γδ cells), we detected a clear population of Vγ4+ cells that coexpressed TCRβ. These findings suggest that hybrid αβ-γδ T cells develop in the embryonic thymus in synchrony with the early waves of conventional γδ T cells, following the same CD4 and CD8 expression patterns as γδ (double negative) or αβ T cells (MHC restricted).
Figure 1.
Identification of unconventional T cells that coexpress αβ and γδ TCRs. (A and B) Flow cytometric analysis of LN cells from WT mice, stained for TCRδ and TCRβ, gated on live CD3+ T cells (A), or stained for Vα2, Vα8.3, and TCRβ, gated on live CD3+TCRδ+ cells (B). (C) Flow cytometric analysis of LN cells from WT mice, stained for Vγ4 and TCRβ, gated on live CD3+ T cells. (D) ImageStream analysis of LN cells from WT mice, stained for Vγ4, TCRδ, and TCRβ, and costained with DAPI. (E and F) Flow cytometric analysis of LN cells from WT mice, showing mean fluorescence intensity (MFI) for CD3ε expression, gated on live CD3+TCRδ+TCRβ− or CD3+TCRδ+TCRβ+ cells (E), or stained for CD4 and CD8α, gated on live CD3+TCRδ+TCRβ+ cells (F). (G) Flow cytometric analysis of LN cells from WT, TCRα−/−, TCRβ−/−, MHCI−/−, or MHCII−/− mice, stained for Vγ4 and TCRβ, gated on live CD3+TCRδ+ cells. (H) Flow cytometric analysis of thymocytes isolated from WT mice on E14, E16, and E18, stained for Vγ4 and TCRβ, gated on live CD3+ cells. The gating strategy (A–H) is shown in Fig. S1 A. (I) Flow cytometric analysis of human PBMCs showing expression of TCRαβ and Vδ2, gated on live CD3+ cells. (J) Percent expression of TCRαβ among Vδ2+ cells in human PBMCs (n = 15 healthy donors), gated on live CD3+ cells. Data are representative of two independent experiments. Flow cytometry plots are representative of at least three independent experiments (n = 18 samples). BF, brightfield; FMO, fluorescence minus one; SSC, side scatter.
Figure S1.
A novel population of T cells that coexpresses αβ and γδ TCRs. (A) Gating strategy for the analysis of T cell subsets, including hybrid αβ-γδ T cells, conventional γδ T cells, and CD4+ T cells. (B) Confocal images of purified TCRδ+ cells costained for TCRβ. (C) Trbj expression in purified CD3+ or TCRδ+ cells quantified by RT-PCR. Control: heart cells. *, P < 0.05, ***, P < 0.001. (D) Confocal images of purified CD3+Vγ4+ cells costained for TCRβ. (E) Flow cytometry plots showing expression of Vγ1 versus Vγ4 on TCRδ+ cells, gated on live TCRβ+ cells. (F) Flow cytometry plots showing expression of TCRδ versus TCRβ on LN cells, gated on live CD3+Vγ4+TCRδ+ cells. (G) Flow cytometry plots showing CD4 versus CD8α on LN cells from MHCI−/− or MHCII−/− mice, gated on live TCRδ+ Vγ4+ TCRβ+ cells. Data are representative of at least three independent experiments. Results are shown as mean ± SEM. P values were calculated using a one-way ANOVA with Tukey's test for multiple comparisons (C). FSC, forward scatter.
Flow cytometric analyses of human peripheral blood mononuclear cells (PBMCs) revealed a rare population of cells (<1% of CD3+ T cells) that coexpressed TCRαβ and Vδ2 (Fig. 1 I) and further showed that ∼10% of all circulating Vδ2+ cells expressed TCRαβ (Fig. 1 J). These data provide preliminary evidence that hybrid αβ-γδ T cells are also present in humans.
Molecular analysis of TCR expression in Vγ4+TCRβ+ cells
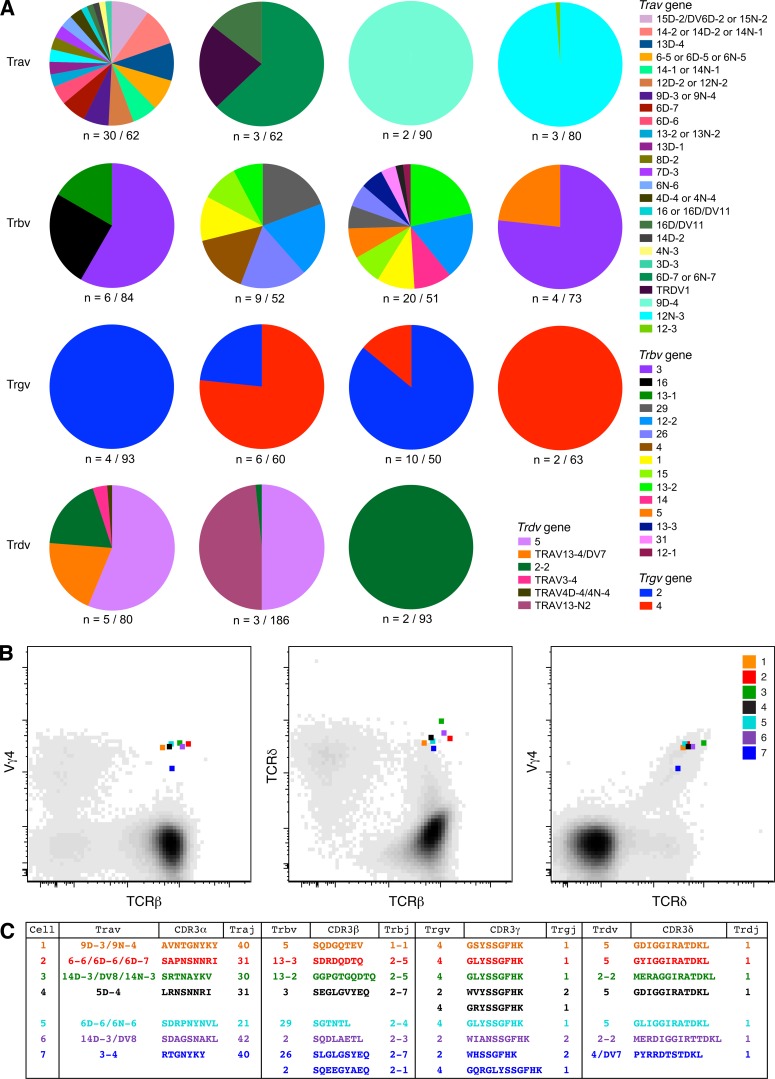
To examine the molecular basis of αβ and γδ TCR coexpression, we used an unbiased approach (Quigley et al., 2011) to amplify all expressed Tra, Trb, Trg, and Trd gene rearrangements in flow-purified Vγ4+TCRβ+ cells. Analysis of the Tra and Trb gene expression profiles revealed substantial heterogeneity (Fig. 2 A). We also detected Trgv2 gene transcripts, presumably reflecting antibody cross-recognition of Vγ2 and Vγ4. The associated Trd gene transcripts were heavily biased toward Trdv2-2 and Trdv5. Of note, disruption of a single chromatin loop facilitates “default” rearrangements with Trdv2-2 (Chen et al., 2015), which also occur in ∼42% of thymocytes in WT mice, and Trdv5 is located in a reversed transcriptional orientation 3′ of the Trdc gene (Glusman et al., 2001).
Figure 2.
Molecular analysis of TCR expression in flow-purified Vγ4+TCRβ+ cells. Viable CD3+Vγ4+TCRβ+ cells were flow purified from WT mice. (A) Pie charts display relative population-level frequencies for the Trav, Trbv, Trgv, and Trdv genes depicted in the key (IMGT nomenclature), with the total number of sequences indicated below. Concatenated data are shown (n = 4 mice). (B) Flow cytometry plots showing index-sorted Vγ4+TCRδ+TCRβ+ cells superimposed on cloud plots depicting the overall distribution of Vγ4 versus TCRβ (left panel), TCRδ versus TCRβ (middle panel), and Vγ4 versus TCRδ (right panel) in one mouse with EAE. (C) TCR sequences amplified from the single cells shown in B. Cells and sequences are color matched in B and C.
To confirm these findings, we flow-sorted single Vγ4+TCRδ+TCRβ+ cells from the LN cells of mice with EAE (Fig. 2 B). Functional Tra, Trb, Trg, and Trd gene rearrangements, consistent with the index sort parameters, were detected in ∼9% (7/79) of these cells (Fig. 2 C). In line with the population-level data, Trgv2 rearranged exclusively with Trgj2, Trgv4 rearranged exclusively with Trgj1, and Trdv2-2 and Trdv5 rearranged exclusively with Trdj1. Moreover, the CDR3γ and CDR3δ loop sequences were highly conserved, irrespective of gene use, and matched those identified in a recent study of CCR6+CD27− γδ T cells (Kashani et al., 2015). No clear patterns were observed among Tra and Trb gene transcripts amplified from single Vγ4+TCRδ+TCRβ+ cells. These data show that hybrid αβ-γδ T cells coexpress diverse TCRα and TCRβ chains and a limited array of TCRγ and TCRδ chains.
Hybrid αβ-γδ T cells can be activated innately or via αβ or γδ TCRs
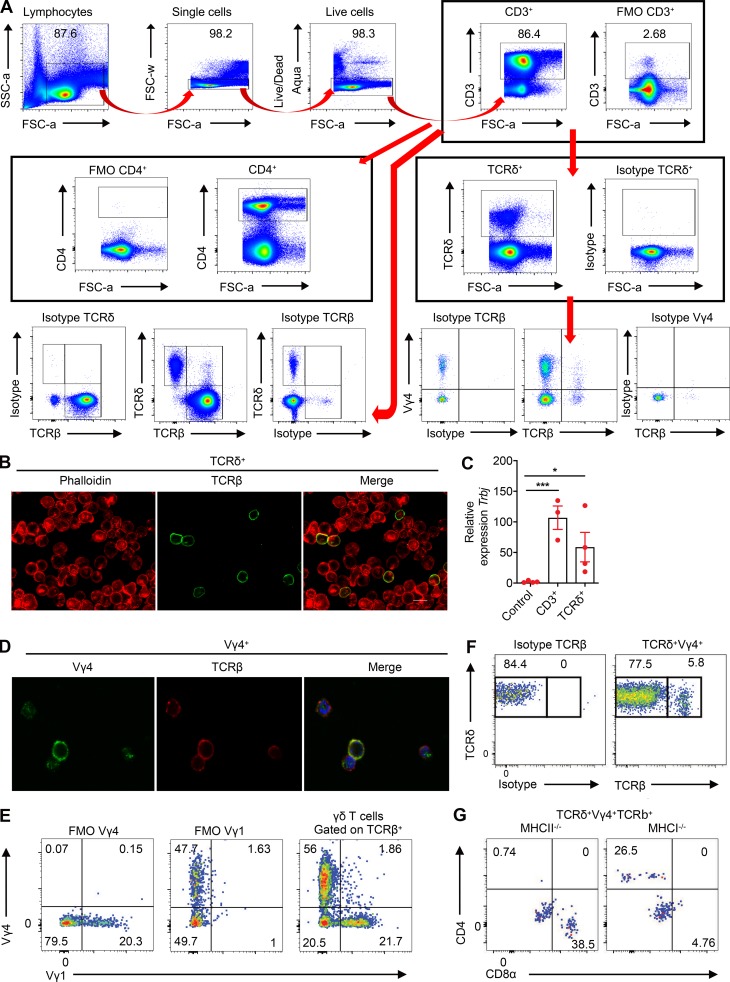
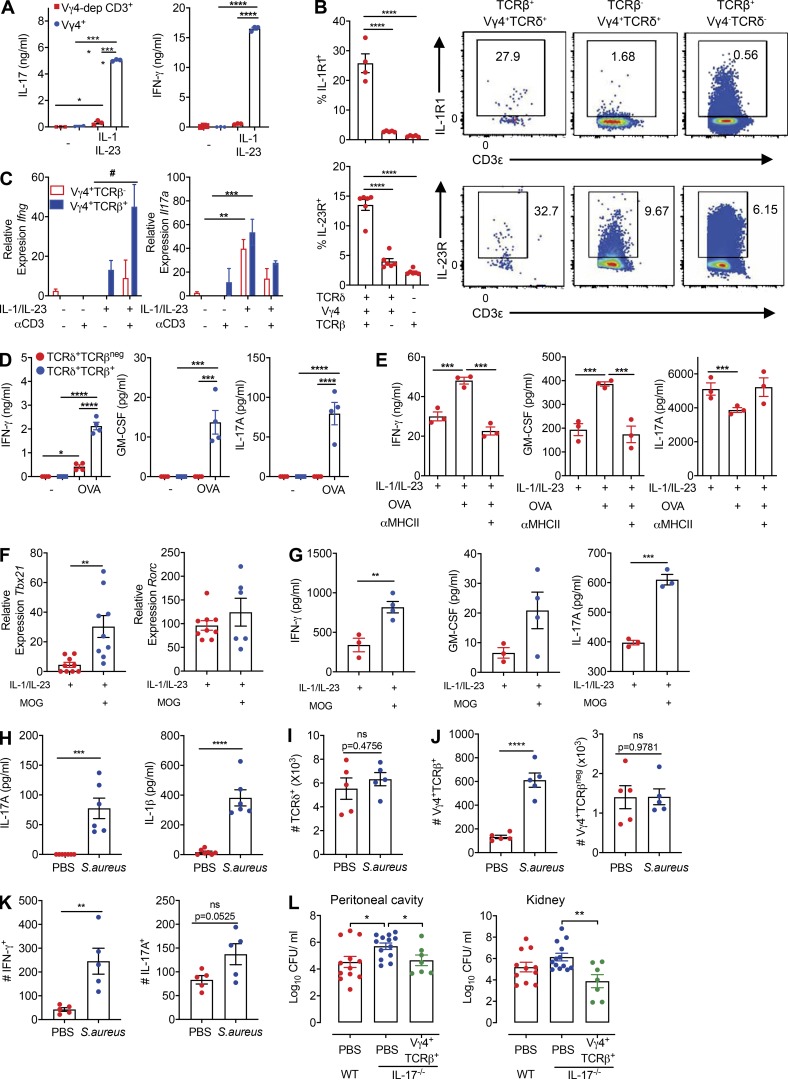
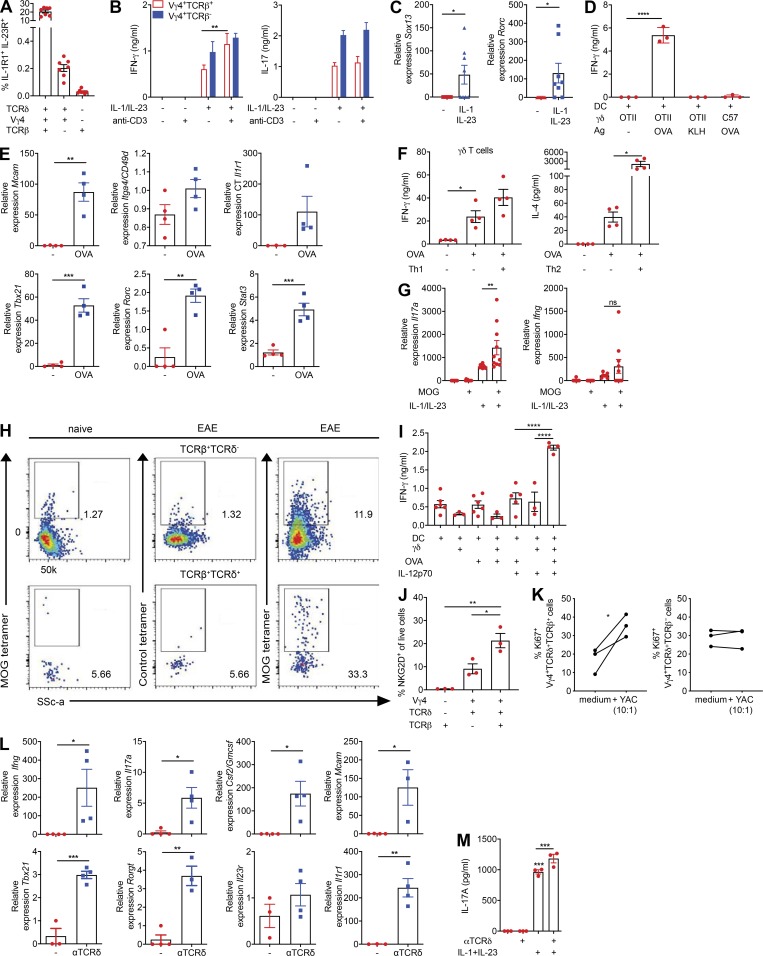
We then addressed the functionality of hybrid αβ-γδ T cells after stimulation with IL-1β and IL-23, which are known to promote the activation of innate γδ T cells (Sutton et al., 2009), or ligation of the expressed αβ or γδ TCRs. Vγ4+ T cells produced IFN-γ and IL-17 in response to IL-1β and IL-23 (Fig. 3 A). Moreover, depletion of Vγ4+ cells from the CD3+ T cell pool abrogated IFN-γ and IL-17 production elicited by IL-1β and IL-23 (Fig. 3 A), and Vγ4+ hybrid αβ-γδ T cells more frequently expressed IL-1R1 or IL-23R (Fig. 3 B) or coexpressed IL-1R1 and IL-23 (Fig. S2 A) compared with conventional Vγ4+ γδ or αβ T cells. Stimulation of hybrid αβ-γδ T cells with IL-1β and IL-23 induced the expression of Ifng and Il17a (Fig. 3 C), as well as the production of IFN-γ and IL-17 (Fig. S2 B). Although Ifng and Il17a expression was detected in the absence of TCR signaling, coactivation with anti-CD3 enhanced IFN-γ expression at the gene and protein levels. Hybrid αβ-γδ T cells also expressed Rorc, which encodes RORγt, the transcription factor associated with IL-17 production (Ivanov et al., 2006), and Sox13, which encodes the transcription factor associated with IL-17–producing Vγ4+ γδ T cells (Gray et al., 2013), after stimulation with IL-1β and IL-23 (Fig. S2 C).
Figure 3.
Hybrid αβ-γδ T cells can be activated innately or via αβ or γδ TCRs. (A) Purified Vγ4+ cells or Vγ4-depleted CD3+ cells were stimulated for 3 d with IL-1β and IL-23 or with medium alone. Data show cytokine production measured by ELISA. (B) Flow cytometric analysis of IL-1R1 and IL-23R expression on Vγ4+TCRδ+TCRβ+, Vγ4+TCRδ+TCRβ−, or Vγ4–TCRδ−TCRβ+ cells, gated on live CD3+ cells. (C) Relative Ifng and Il17a expression in purified Vγ4+TCRβ+ or Vγ4+TCRβ− cells stimulated for 3 d with IL-1β, IL-2, and IL-23 in the presence or absence of plate-bound anti-CD3. (D and E) IFN-γ, IL-17, and GM-CSF production by TCRδ+TCRβ+ or TCRδ+TCRβ− cells (D), or TCRδ+TCRβ+ cells (E), isolated from OT-II mice and stimulated for 3 d with or without OVA-pulsed DCs in the presence or absence of IL-1β and IL-23 and/or anti-MHCII. Cytokines were measured by ELISA. (F and G) Vγ4+TCRβ+ cells were isolated from MOG-immunized mice on day 7 and cultured for 3 d with DCs in the presence or absence of MOG and/or IL-1β and IL-23. Relative gene expression was measured by RT-PCR (F), and cytokines were measured by ELISA (G). PECs were isolated from naive mice or 3 h after i.p. challenge with S. aureus. (H) Cytokine levels in PEC culture supernatants measured by ELISA. (I) Flow cytometric quantification of TCRδ+ cells, gated on live CD3+ cells. (J) Flow cytometric quantification of Vγ4+TCRδ+TCRβ+ or Vγ4+TCRδ+TCRβ− cells, gated on live CD3+TCRδ+ cells. (K) Flow cytometric quantification of IFN-γ and IL-17 production by Vγ4+TCRδ+TCRβ+ cells. (L) Bacterial loads in the peritoneal cavity and kidneys of WT mice 3 d after S. aureus infection or IL-17−/− mice 3 d after adoptive transfer of Vγ4+TCRβ+ cells from WT mice or mock transfer (PBS). Data are representative of three independent experiments (n = 4–6 in A–K) or combined from two experiments (n = 8 or 12 in L). Flow cytometry plots in B are representative of six samples. Results are shown as mean ± SEM; *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001, #,P < 0.05; ns, not significant; two-way ANOVA with Tukey's test for multiple comparisons (A, D, and E), one-way ANOVA with Tukey's test for multiple comparisons (B and L), three-way ANOVA with Tukey's test for multiple comparisons (C), or unpaired t test (F–K).
Figure S2.
Hybrid αβ-γδ T cells can be activated innately or via αβ or γδ TCRs. (A) Coexpression of IL-1R1 and IL-23R on Vγ4+ hybrid αβ-γδ, Vγ4+ γδ T cells, or αβ T cells. (B) Production of IFN-γ and IL-17 by purified Vγ4+TCRβ+ or Vγ4+TCRβ− cells (3,000 cells/well) stimulated for 3 d with IL-1β and IL-23 in the presence or absence of plate-bound anti-CD3. (C) Expression of Sox13 and Rorc mRNA in purified Vγ4+TCRβ+ cells stimulated for 2 d with IL-1β and IL-23. (D) Production of IFN-γ by TCRδ+ cells isolated from OT-II or C57BL/6 mice and cultured for 2 d with DCs pulsed for 5 h with OVA peptide or KLH. (E) Gene expression in TCRδ+TCRβ+ cells isolated from OT-II mice and cultured for 3 d with DCs in the presence or absence of OVA peptide. (F) Cytokine production by γδ T cells isolated from OT-II mice stimulated for 3 d with or without OVA peptide in the presence or absence of IL-12p70 + IL-18 (Th1) or IL-4 + anti–IFN-γ + anti–IL-17 (Th2). (G) Gene expression in Vγ4+TCRβ+ cells isolated from MOG-immunized mice on day 7 and cultured for 3 d with DCs in the presence or absence of MOG and/or IL-1β and IL-23. (H) LN cells isolated from 7 d MOG + CFA immunized or naive mice incubated with MOG-tetramer-Pe or control tetramer-Pe, gated on CD3+CD4+CD44+ cells, examining TCRβ+TCRδ+ and TCRβ+TCRδ− populations. (I) γδ T cells from OT-I mice incubated for 3 d with DCs ± OVA peptide ± IL-12p70 and IFN-γ quantified in supernatants by ELISA. (J) Flow cytometry analysis of NKG2D expression on naive CD3+ T cells gating on TCRβ+ and TCRδ+ populations. (K) CD3+ T cells were incubated with and without YAC-1 cells (10:1) for 48 h. Proliferation was measured through expression of Ki67 by Vγ4+TCRδ+TCRβ+ versus Vγ4+TCRδ+TCRβ− cells, gated on live CD3+TCRδ+ cells. (L) Gene expression in TCRδ+TCRβ+ cells stimulated for 2 d with anti-TCRδ. (M) Gene expression in CD3+ cells cultured for 3 d with IL-1β and IL-23 in the presence or absence of anti-TCRδ. Data are representative of at least two independent experiments. Results are shown as mean ± SEM. P values were calculated using a one-way ANOVA with Tukey's test for multiple comparisons (B, D, F, G, I–K, and M) or an unpaired t test (C, E, and L). *, P < 0.05, **, P < 0.01, ***, P < 0.001, and ****, P < 0.0001. ns, not significant.
Next, we examined the ability of hybrid αβ-γδ T cells to respond to MHCII-restricted antigens, initially using OT-II transgenic mice, which exclusively express αβ TCRs that recognize an OVA peptide restricted by I-Ab. Purified TCRδ+ cells from OT-II mice, but not WT mice, produced IFN-γ in response to dendritic cells (DCs) pulsed with OVA, but not with KLH (Fig. S2 D). Moreover, purified TCRδ+TCRβ+ cells, but not TCRδ+TCRβ− cells, from OT-II mice produced IFN-γ, IL-17, and GM-CSF (Fig. 3 D) and significantly up-regulated Tbx21, Rorc, Stat3, and Mcam in response to OVA (Fig. S2 E). Co-stimulation of TCRδ+TCRβ+ cells from OT-II mice with IL-1β and IL-23 in the presence of OVA-pulsed DCs enhanced the production of IFN-γ and GM-CSF, which was reversed by the addition of anti-MHCII (Fig. 3 E). OVA stimulation reduced the production of IL-17 induced by IL-1β and IL-23 (Fig. 3 E), potentially reflecting enhanced expression of Tbx21, which encodes T-bet, a transcription factor that blocks Runx1-mediated transactivation of the Rorc promoter and subsequent expression of IL-17. Stimulation of OT-II–derived γδ T cells with OVA under Th1 and Th2 polarizing conditions also enhanced the production of IFN-γ and IL-4 (Fig. S2 F). Further evidence of MHCII-restricted peptide recognition by hybrid αβ-γδ T cells was obtained using Vγ4+TCRβ+ cells purified from WT mice immunized with myelin oligodendrocyte protein (MOG) and CFA. IL-1β and IL-23 induced the expression of Tbx21 and Rorc (Fig. 3 F), as well as the production of IFN-γ, IL-17, and GM-CSF, which was substantially enhanced in the presence of MOG and DCs (Fig. 3, F and G; and Fig. S2 G).
The antigen specificity of hybrid αβ-γδ T cells was confirmed using the corresponding MOG tetramer to stain LN cells 7 d after the induction of EAE. Approximately 33% of Vγ4+TCRβ+ cells stained with the MOG tetramer, in contrast to ∼12% of conventional αβ T cells (Fig. S2 H). Moreover, we found that purified γδ T cells from OT-1 mice, which harbor transgenic CD8+ T cells specific for OVA257–264 presented by H-2Kb, produced IFN-γ in the presence of OVA-pulsed DCs and IL-12 (Fig. S2 I). These data suggest that hybrid αβ-γδ T cells respond in an innate-like manner to IL-1β and IL-23, akin to conventional γδ T cells, but also recognize peptide antigens presented by MHCII or MHCI, akin to conventional αβ T cells.
The lack of well-defined antigens recognized by γδ T cells precluded analysis of activation via γδ TCRs. Nonetheless, we examined the expression of the natural killer group 2D (NKG2D) receptor, which is crucial for innate-like lymphocyte recognition of stressed or infected cells (Correia et al., 2013). Hybrid Vγ4+ αβ-γδ T cells expressed higher levels of NKG2D than conventional Vγ4+ γδ or αβ T cells (Fig. S2 J). Expression of Ki67, a marker of proliferation, was also significantly enhanced in hybrid Vγ4+ αβ-γδ T cells, but not in conventional Vγ4+ γδ T cells, after co-culture with YAC-1 cells, which constitutively express NKG2D ligands (Fig. S2 K). Moreover, stimulation of TCRδ+TCRβ+ cells with an activating anti-TCRδ antibody significantly increased the expression of Ifng, Il17a, Csf2, Mcam, Tbx21, Rorc, and Il1r1 (Fig. S2 L), as well as the production of IL-17 driven by IL-1β and IL-23 (Fig. S2 M).
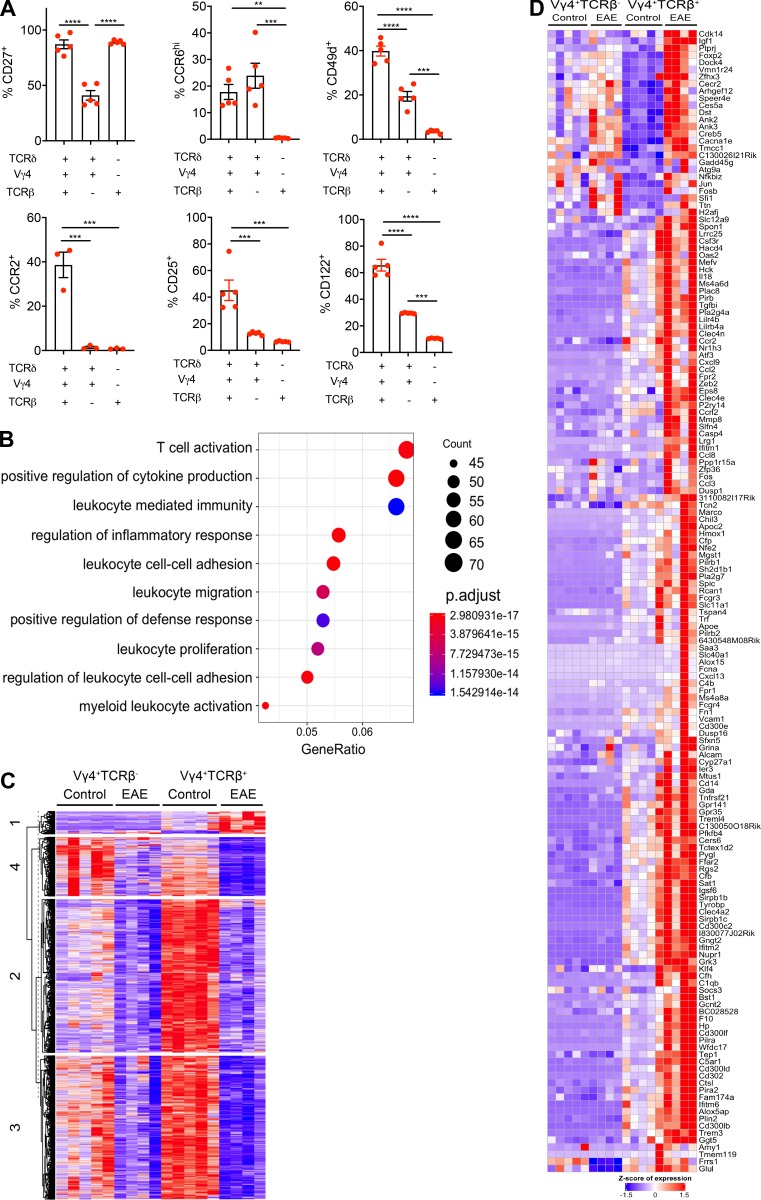
Further phenotypic analyses revealed that hybrid αβ-γδ T cells displayed unique properties and certain features associated with conventional γδ T cells. The majority of hybrid αβ-γδ T cells expressed CD27 (Fig. S3 A), whereas Vγ4+ γδ T cells were either CD27+ or CD27−, demarcating the production of IFN-γ or IL-17, respectively (Ribot et al., 2009). Moreover, CCR6, a chemokine receptor expressed on IL-17–secreting γδ T cells (Haas et al., 2009; Papotto et al., 2017) and pathogenic Th17 cells during EAE (Reboldi et al., 2009), was expressed by hybrid αβ-γδ and conventional Vγ4+ γδ T cells, but not by αβ T cells (Fig. S3 A). Similarly, CCR2, which allows Th17 cells to cross the blood–brain barrier and is associated with the development of EAE (Kara et al., 2015), was more commonly expressed by hybrid αβ-γδ T cells relative to conventional γδ or αβ T cells. The integrin CD49d, a subunit of the cell adhesion molecule α4β1, very late antigen-4(VLA-4), which is involved in the migration of encephalitogenic T cells into the CNS (Yednock et al., 1992), was expressed by ∼40% of hybrid αβ-γδ T cells but also by ∼20% of conventional Vγ4+ γδ T cells (Fig. S3 A). In addition, hybrid αβ-γδ T cells expressed IL-2R, CD25, and CD122 at higher frequencies than conventional Vγ4+ γδ T cells or αβ T cells (Fig. S3 A). These observations indicate that hybrid αβ-γδ T cells share some phenotypic features with γδ T cells but also exhibit characteristics typically associated with cell activation and migration.
Figure S3.
Hybrid αβ-γδ T cells are transcriptomically distinct from conventional γδ T cells and express Th17-associated markers. (A) Enriched T cells isolated from the spleens and LNs of WT mice were stained ex vivo for CCR2, CCR6, CD25, CD27, CD49d, and CD122. Expression was determined on live CD3+ cells coexpressing various combinations of Vγ4, TCRδ, and TCRβ. Data are representative of at least two independent experiments. Results are shown as mean ± SEM. P values were calculated using a one-way ANOVA with Tukey's test for multiple comparisons. (B) Dot plot of the top 15 significantly enriched biological processes inferred from differentially up-regulated genes in Vγ4+TCRβ+ versus Vγ4+TCRβ– cells. Dot color represents the P-adjusted enrichment value, and dot size represents the number of genes within each enriched ontology. (C) Heatmap of all protein-coding genes that are differentially expressed in Vγ4+TCRβ+ or Vγ4+TCRβ– cells from mice with EAE versus naive mice (n = 2,686). Genes are clustered using k-means clustering and a cluster size of 4. (D) Heatmap of all protein-coding genes that are up-regulated in either Vγ4+TCRβ+ or Vγ4+TCRβ− cells from naive mice or mice with EAE (cluster 1 from Fig. S3 C, n = 158 genes). **, P < 0.01, ***, P < 0.001, and ****, P < 0.0001.
Hybrid αβ-γδ T cells protect against S. aureus infection
To probe the biological relevance of these findings, we examined the ability of hybrid αβ-γδ T cells to protect against infection with S. aureus in a murine model, where a key role has been defined for IL-17–producing γδ T cells (Murphy et al., 2014). Mice infected with S. aureus displayed significantly elevated concentrations of IL-17 and IL-1β in the peritoneal fluid 3 h after infection (Fig. 3 H). At the same time point, there was a nonsignificant increase in the absolute number of TCRδ+ cells (Fig. 3 I) and a significant increase in the absolute number of Vγ4+TCRβ+ cells in the peritoneal cavity (Fig. 3 J). In addition, there was a significant increase in IFN-γ–producing and a nonsignificant increase in IL-17–producing Vγ4+TCRδ+TCRβ+ cells in infected versus naive mice (Fig. 3 K). Adoptive transfer of ∼10,000 flow-purified Vγ4+TCRβ+ cells from WT mice to IL-17A−/− mice before infection with S. aureus significantly reduced bacterial load in the peritoneal cavity and systemic dissemination to the kidney (Fig. 3 L). These data demonstrate that hybrid αβ-γδ T cells play a protective role in immunity to S. aureus infection.
Hybrid αβ-γδ T cells play a critical role in the development of EAE
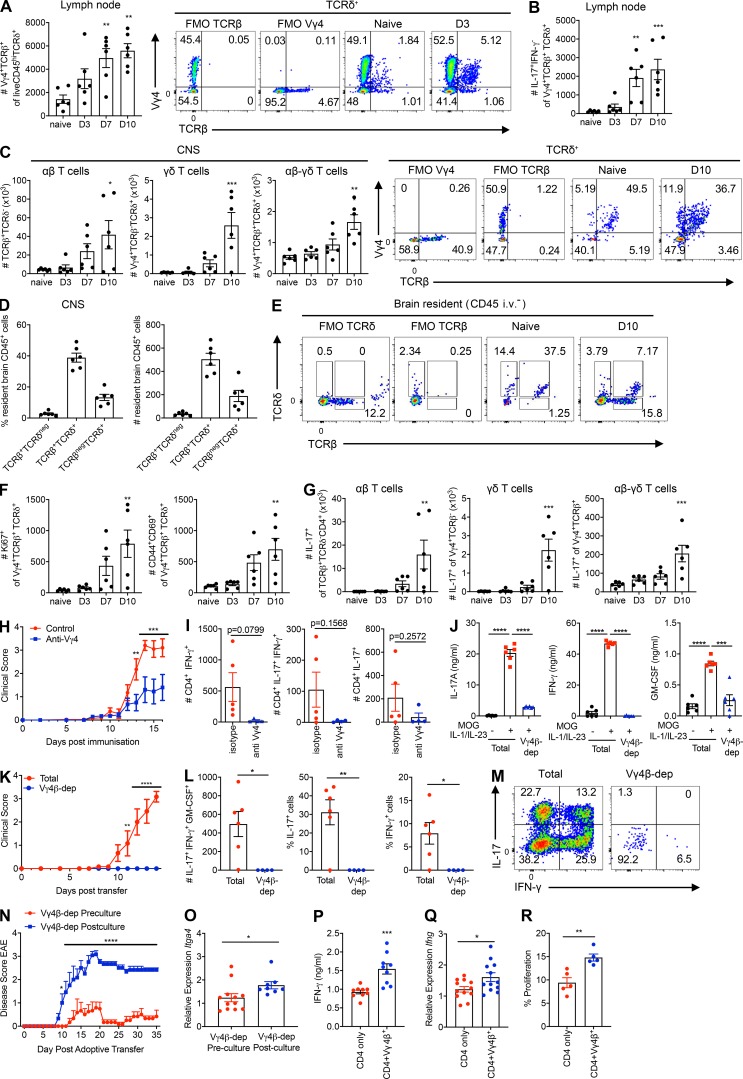
The role of γδ T cells in autoimmune diseases is well established (Sutton et al., 2009). We found that hybrid αβ-γδ T cells were significantly expanded and secreted IL-17 in the LNs of WT mice 3–10 d after the induction of EAE (Fig. 4, A and B). Hybrid αβ-γδ T cells also dominated the Vγ4+ compartment in the CNS of naive mice, and the number of hybrid αβ-γδ T cells in the CNS was significantly increased 10 d after the induction of EAE (Fig. 4 C). Conventional γδ T cells also infiltrated the brain 7–10 d after challenge, but CD4+ αβ T cells were numerically dominant by day 10 (Fig. 4 C). Using a validated in vivo staining technique with an anti-CD45 antibody administered i.v. immediately before sacrifice, we demonstrated that hybrid αβ-γδ T cells were the dominant CNS tissue-resident T cells in naive mice (Fig. 4 D) and that TCRδ+TCRβ+ cells outnumbered TCRδ+TCRβ– cells on day 10 of EAE (Fig. 4 E). These CNS-infiltrating hybrid αβ-γδ T cells displayed an activated, proliferative phenotype (Fig. 4 F) and secreted IL-17 (Fig. 4 G). Early in EAE (days 3–7), hybrid αβ-γδ T cells were the dominant IL-17–secreting subset in the CNS. However, conventional γδ T cells and CD4+ T cells were also important sources of IL-17 in the CNS, especially as the disease developed over time, and CD4+ αβ T cells formed the dominant IL-17–secreting population on day 10 of EAE (Fig. 4 G).
Figure 4.
Hybrid Vγ4+ αβ-γδ T cells promote CD4+ T cell homing to the CNS and drive the development of EAE. (A) Absolute numbers of Vγ4+TCRδ+TCRβ+ cells in the LNs of WT mice on days 0, 3, 7, and 10 of EAE. Right: representative flow cytometry plots. (B) Absolute numbers of IL-17-producing Vγ4+TCRδ+TCRβ+ cells in the LNs of WT mice on days 0, 3, 7, and 10 of EAE. (C) Absolute numbers of Vγ4+TCRδ+TCRβ–, Vγ4+TCRδ+TCRβ+, or Vγ4–TCRδ–TCRβ+ cells in the brains of WT mice on days 0, 3, 7, and 10 of EAE. Right: representative flow cytometry plots. (D) Absolute numbers and frequencies of brain-resident TCRδ−TCRβ+, TCRδ+TCRβ+, or TCRδ+TCRβ– cells after gating on CD45 i.v.– cells from naive mice injected with anti-CD45 10 min before euthanasia. (E) Representative flow cytometry showing expression of TCRβ versus TCRδ on tissue-resident (CD45 i.v.−) T cells from naive mice or mice with EAE (day 10). (F) Absolute numbers of Ki67+ or CD44+CD69+ Vγ4+TCRδ+TCRβ+ cells in the CNS of WT mice on days 0, 3, 7, and 10 of EAE. (G) Absolute numbers of IL-17–producing Vγ4+TCRδ+TCRβ+, Vγ4+TCRδ+TCRβ–, or CD4+ Vγ4–TCRδ–TCRβ+ cells in the CNS of WT mice on days 0, 3, 7, and 10 of EAE. (H and I) WT mice were treated on days –1, 2, 5, 7, 11, and 14 with Vγ4-depleting or isotype control antibodies. (H) Clinical scores for EAE. (I) Absolute numbers of cytokine-producing CD4+ T cells in the spinal cords on day 21 of EAE. (J–M) Total spleen and LN cells or Vγ4+TCRβ+ flow-depleted (Vγ4β-dep) spleen and LN cells from MOG-immunized mice (day 7) were cultured for 3 d with MOG, IL-1β, and IL-23. (J) IL-17, IFN-γ, and GM-CSF concentrations in culture supernatants quantified by ELISA. (K) Cultured T cells were transferred to naive mice and clinical scores were recorded for EAE. (L) Absolute numbers and frequencies of cytokine-producing cells in the brains of recipient mice on day 10 of EAE. (M) Representative flow cytometry plots showing production of IFN-γ versus IL-17, gated on live CD3+ cells. (N and O) Vγ4+ TCRβ+ T cells were depleted from a culture of LN and spleen cells from MOG-immunized mice before or after culture with MOG, IL-1β, and IL-23. (N) Cultured T cells were transferred to naive mice and clinical scores were recorded for EAE. (O) Itga4 mRNA was quantified by RT-PCR in cultured cells before transfer. (P–R) CD4+ T cells were isolated from the spleens and LNs of mice immunized with MOG and CFA (day 7) and cultured for 3 d with IL-1β, IL-23, and MOG, either in the presence or absence of Vγ4+TCRβ+ cells. (P) IFN-γ in culture supernatants quantified by ELISA. (Q) Ifng expression in purified CD4+ T cells. (R) Proliferation of CD4+ T cells measured by CFSE dilution. Data are representative of three independent experiments (n = 6 for A–N) or combined from two experiments (n = 6 for O–R). Results are shown as mean ± SEM; *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001; one-way ANOVA with Tukey's test for multiple comparisons (A–C, F, and G), two-way ANOVA with Tukey's test for multiple comparisons (J), repeated measures (H, K, and N), or unpaired t test (I, L, and O–R).
Depletion of Vγ4+ cells in vivo, which removes conventional Vγ4+ γδ and hybrid Vγ4+ αβ-γδ T cells, significantly impaired the development of EAE (Fig. 4 H) and limited the infiltration of cytokine-producing CD4+ T cells into the CNS (Fig. 4 I). To address the specific contribution of Vγ4+ hybrid αβ-γδ T cells in this model, we depleted Vγ4+TCRβ+ cells from the spleens and LNs of mice immunized with MOG and CFA. Adoptive transfer of unmanipulated T cells from MOG-immunized mice after expansion in vitro with MOG, IL-1β, and IL-23 induced EAE. Depletion of Vγ4+TCRβ+ cells significantly reduced the production of IFN-γ, IL-17, and GM-CSF in culture supernatants after incubation with MOG, IL-1β, and IL-23 (Fig. 4 J) and abrogated the induction of EAE (Fig. 4 K). This attenuated disease course was associated with significantly reduced frequencies and absolute numbers of various cytokine-producing T cells in the CNS (Fig. 4, L and M). To address the contribution of hybrid Vγ4+ αβ-γδ T cells in the priming of encephalitogenic CD4+ T cells, we depleted hybrid Vγ4+ αβ-γδ T cells from a culture of LN and spleen cells from MOG-immunized mice before or after culture with MOG, IL-1β, and IL-23. Depletion of hybrid αβ-γδ T cells before, but not after, culture significantly impaired the development of EAE following adoptive transfer to naive mice (Fig. 4 N). Cells depleted of hybrid αβ-γδ T cells before culture with MOG, IL-1β, and IL-23 also expressed significantly lower amounts of Itga4, which encodes the essential CD4+ T cell trafficking molecule VLA-4 (Fig. 4 O). Moreover, antigen-specific CD4+ T cells cultured with MOG, IL-1β, and IL-23 in the presence of hybrid αβ-γδ T cells expressed significantly more IFN-γ (Fig. 4 P) and Ifng (Fig. 4 Q) and proliferated to a greater extent (Fig. 4 R) than CD4+ T cells cultured in the absence of hybrid αβ-γδ T cells. These findings demonstrate that hybrid Vγ4+ αβ-γδ T cells play a nonredundant role in the immunopathogenesis of EAE via their ability to migrate to inflammatory sites and prime encephalitogenic CD4+ T cells.
Hybrid αβ-γδ T cells display a hyperactivated phenotype
Although hybrid αβ-γδ T cells are outnumbered by conventional γδ T cells, they are rapidly mobilized and play a key pathogenic role in development of CNS inflammation in the EAE model. To understand these proinflammatory effects in more detail, we performed a transcriptomic analysis of hybrid αβ-γδ T cells and conventional γδ T cells isolated from the LNs of WT mice under physiological conditions and during EAE.
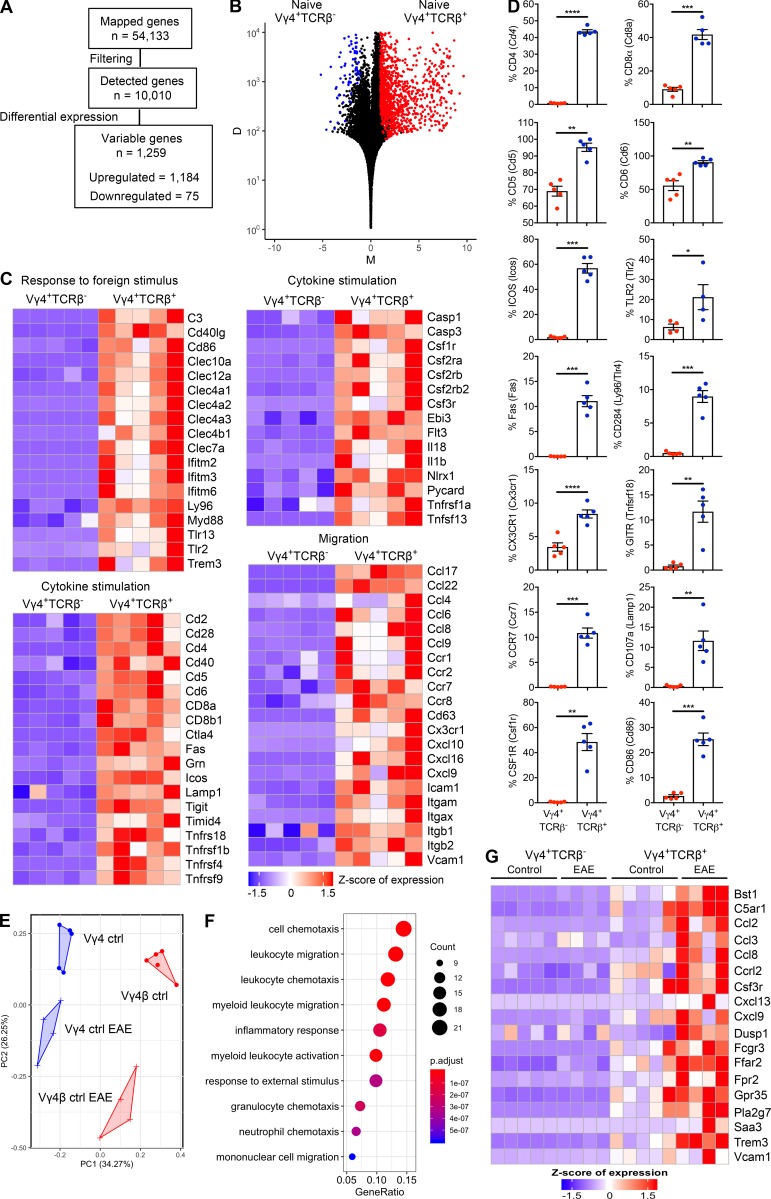
The data revealed that 1,259 genes were differentially expressed in hybrid αβ-γδ T cells (Vγ4+TCRβ+) relative to conventional γδ T cells (Vγ4+TCRβ−) under physiological conditions and that the majority (n = 1,184) of these differentially expressed genes (DEGs) were up-regulated in hybrid αβ-γδ T cells relative to conventional γδ T cells (Fig. 5, A and B). Functional enrichment analysis revealed that transcripts associated with T cell activation, cellular migration, cytokine stimulation, and immune responses to foreign stimuli were enriched in hybrid αβ-γδ T cells relative to conventional γδ T cells (Fig. 5 C and Fig. S3 B). Genes associated with the αβ T cell phenotype, including Cd4, Cd8, Cd6, and Cd28, were expressed at higher levels in hybrid αβ-γδ T cells relative to conventional γδ T cells. Hybrid αβ-γδ T cells also expressed higher levels of genes associated with cellular trafficking, including Ccr7, Cx3cr1, Ccl22, and Ccl17 (Fig. 5 C). In addition, hybrid αβ-γδ T cells expressed higher levels of Myd88, Ly96, Tlr2, and Tlr13, suggesting a capacity to respond to pathogen-associated molecular patterns, as well as genes associated with IL-1 signaling, including Casp3, Il1b, and Il18 (Fig. 5 C). A selection of identified DEGs were validated by flow cytometry (Fig. 5 D).
Figure 5.
Hybrid αβ-γδ T cells are transcriptionally distinct from conventional γδ T cells. RNA sequencing analysis of Vγ4+TCRδ+TCRβ+ (Vγ4+TCRβ+) or Vγ4+TCRδ+TCRβ− (Vγ4+TCRβ−) cells flow sorted from the spleens and LNs of naive WT mice or WT mice on day 3 of EAE. (A) Summary of the data preprocessing and filtering workflow. (B) NOISeq MD plot of genes passing the low-level filtering cutoff (n = 10,010), highlighting DEGs (n = 1,259) between naive Vγ4+TCRβ+ and Vγ4+TCRβ– cells. Red dots: genes up-regulated in Vγ4+TCRβ+ cells (n = 1,184). Blue dots: genes down-regulated genes in Vγ4+TCRβ+ cells (n = 75). M represents the log2-fold change in normalized expression values between Vγ4+TCRβ+ and Vγ4+TCRβ− cells. D represents the absolute value of the difference in expression between Vγ4+TCRβ+ and Vγ4+TCRβ− cells. D values are displayed on a log10 scale. Increasing D values represent increasing differences in expression levels between Vγ4+TCRβ+ and Vγ4+TCRβ− cells. (C) Heatmaps of selected genes from enriched biological processes derived using gene ontology enrichment analysis of up-regulated genes between naive Vγ4+TCRβ+ and Vγ4+TCRβ− cells. Expression values were z-transformed for visualization. (D) Flow cytometric analysis of purified CD3+ cells, comparing naive Vγ4+TCRβ+ and Vγ4+TCRβ− cells, gated on live CD3+TCRδ+ cells. Results are shown as mean ± SEM. (E) Reduced dimensionality representation of four cell populations via a principal-component analysis plot, where the Vγ4+TCRβ+ and Vγ4+TCRβ− populations separate along the first principal component (PC1) and the equivalent populations in naive mice or mice with EAE separate along the second principal component (PC2). (F) Dot plot of the top 10 significantly enriched biological processes inferred from differentially up-regulated genes in Vγ4+TCRβ+ or Vγ4+TCRβ− cells from mice with EAE versus naive mice (cluster 1 in Fig. S3 C; n = 158 genes). Dot color represents the P-adjusted enrichment value, and dot size represents the number of genes within each enriched gene ontology. (G) Heatmap of genes associated with chemotaxis/migration among all four populations derived using the gene ontology enrichment analysis in F. Expression values were z-transformed for visualization. Most of the data are shown for individual mice (n = 4 or 5 mice per group), except in D, where the data are representative of two experiments (n = 5 mice). *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001; unpaired t test.
Separation of hybrid αβ-γδ T cells from conventional γδ T cells in the principal component analysis plot indicated differences in the corresponding gene expression profiles under physiological conditions and during EAE (Fig. 5 E). Differential expression analysis was used to identify changes in gene expression levels for each cell type under each condition. DEGs were more common in hybrid Vγ4+ αβ-γδ T cells (total, n = 2,470; up-regulated, n = 157; down-regulated, n = 2,313) compared with conventional γδ T cells (total, n = 652; up-regulated, n = 3; down-regulated, n = 649), and most of these genes were similarly down-regulated in both cell types during EAE (Fig. S3 C). Enrichment analysis further revealed that these down-regulated genes were associated with T cell activation (cluster 2), cell cycle processes (cluster 3), and mRNA processing and transport (cluster 4; Fig. S3 C). In contrast, genes associated with cell migration and chemotaxis, including Vcam1, Ccl2, Ccl8, and Cxcl13, were preferentially up-regulated in hybrid Vγ4+ αβ-γδ T cells from mice with EAE (Fig. 5, F and G; and Fig. S3, C and D). These data suggest that hybrid αβ-γδ T cells are transcriptomically distinct from conventional γδ T cells, with a gene expression profile indicative of a proinflammatory and migratory phenotype.
Concluding remarks
Conventional αβ and γδ T cells originate from common thymocyte precursors, but the mechanisms that govern subsequent divergence and lineage fate are incompletely defined. The general consensus posits that a strong γδ TCR-mediated signal directs thymocytes to the γδ lineage, whereas a nonproductive signal permits TCRα rearrangements and commitment to the αβ lineage (Hayes et al., 2005). However, aberrant expression of TCR chains has been demonstrated among αβ and γδ T cell populations (Bowen et al., 2014; Hochstenbach and Brenner, 1989; Ishida et al., 1990). For example, in-frame TCRδ rearrangements have been identified in αβ T cells (Livak et al., 1995), and functional TCRβ rearrangements have been detected in γδ T cells (Bosco et al., 2008). The latter may even confer a proliferative advantage and selectively amplify certain subsets of murine TCRβ+ γδ thymocytes (Wilson and MacDonald, 1998). Similarly, up to 50% of all Vδ1+ cells in humans are natural killer T (NKT)–like cells that express TCRα and TCRδ segments with TCRβ (Pellicci et al., 2014). Of note, given the structure of the murine TCRα/δ locus, coexpression of αβ and γδ TCRs implies productive rearrangement of TCRα and TCRδ on different alleles. In addition, the murine TCRγ locus has been repeatedly duplicated on chromosome 13, potentially facilitating multiple rearrangements (Glusman et al., 2001). It is also notable that dual-lineage lymphocytes expressing a B cell receptor and an αβ TCR have recently been identified in humans and linked to the development of type 1 diabetes (Ahmed et al., 2019).
The collective data presented here identify a novel subset of hyperinflammatory T cells defined by the coexpression αβ and γδ TCRs. These intrathymically generated hybrid αβ-γδ T cells recognized MHC-restricted peptide antigens, like conventional αβ T cells, and produced IFN-γ, IL-17, and GM-CSF in response to IL-1β and IL-23, like conventional γδ T cells, defining a niche at the interface between adaptive and innate immunity. Moreover, hybrid αβ-γδ T cells expressed chemokine receptors and homing molecules, facilitating migration to sites of inflammation. Hybrid αβ-γδ T cells were expanded at the site of infection with S. aureus, and adoptive transfer of a very small number of purified hybrid Vγ4+ αβ-γδ T cells from WT mice conferred protection against S. aureus infection in IL-17−/− mice. In mice with EAE, IL-17–producing hybrid αβ-γδ T cells were found in the draining LNs before the appearance of IL-17–producing αβ T cells. Depletion of hybrid Vγ4+ αβ-γδ T cells suppressed the activation of Th17 cells and abrogated the development of EAE. These findings suggest that hybrid αβ-γδ T cells are equipped to act as highly proinflammatory “first responders,” illustrated here in the context of a bacterial infection and an autoimmune process in the CNS.
Materials and methods
Mice
C57BL/6 mice, IL-17−/− (C57BL/6 background) mice, OT-I mice (C57BL/6-Tg(TcraTcrb)1100Mjb/J), which exclusively express an αβ TCR specific for OVA257–264 restricted by H-2Kb, and OT-II mice (C57BL/6-Tg(TcraTcrb)425Cbn/J), which exclusively express an αβ TCR specific for OVA323–339 restricted by I-Ab, were sourced from the Jackson Laboratory. All mice were bred under specific pathogen–free conditions and maintained according to European Union Directives. Experiments were performed with sex-matched mice (aged 6–8 wk) under license BI00/2412 from the Irish Health Protection Regulatory Agency with approval from the Trinity College Dublin Comparative Medicine Ethics Committee. Embryonic thymus experiments were performed with C57BL/6 mice at the University of Birmingham, UK. For timed matings, the day of detection of a vaginal plug was designated as day 0. Experiments with TCRα−/−, TCRβ−/−, MHCI−/−, and MHCII−/− mice were performed at the Instituto de Medicina Molecular, Lisbon, Portugal.
Preparation of human PBMCs
PBMCs were isolated by Ficoll gradient centrifugation from leukocyte-enriched buffy coats obtained from anonymous healthy donors via the Irish Blood Transfusion Board, St. James’s Hospital, Dublin, Ireland. Ethical approval was granted by the School of Biochemistry and Immunology Research Ethics Committee, Trinity College Dublin, Ireland.
Antibodies
The following antibodies were used to characterize murine cells in flow cytometry experiments: anti-CCR2 (clone SA203G11, 0.1 µg/106 cells; BioLegend), anti-CCR6 (clone 140706, 0.1 µg/106 cells; BD Horizon), anti-CCR7 (clone 4B12, 0.1 µg/106 cells; eBioscience), anti-CD3 (clone 17A2, 0.05 µg/106 cells; BioLegend), anti-CD4 (clone RM4-5, 0.05 µg/106 cells; eBioscience), anti-CD5 (clone 53–7.3, 0.1 µg/106 cells; BD Biosciences), anti-CD6 (clone IM348, 0.1 µg/106 cells; eBioscience), anti-CD8α (clone 53–6.7, 0.05 µg/106 cells; eBioscience), anti-CD25 (clone PC61, 0.1 µg/106 cells; BioLegend), anti-CD27 (clone LG.7F9, 0.1 µg/106 cells; eBioscience), anti-CD49d (clone R1-2, 0.1 µg/106 cells; eBioscience), anti-CD86 (clone GL1, 0.1 µg/106 cells; BioLegend), anti-CD95 (clone 15A7, 0.1 µg/106 cells; eBioscience), anti-CD107a (clone 1D4B, 0.1 µg/106 cells; BD Biosciences), anti-CD115 (clone AFS98, 0.1 µg/106 cells, BioLegend), anti-CD122 (clone TM-b1, 0.1 µg/106 cells; eBioscience), anti-CD284 (clone MT5510, 0.1 µg/106 cells; BioLegend), anti-CX3CR1 (clone SA011F11, 0.1 µg/106 cells; BioLegend), anti-GITR (clone DTA1, 0.1 µg/106 cells; eBioscience), anti–GM-CSF (clone MP1-22E9, 0.1 µg/106 cells; eBioscience), anti-ICOS (clone 7E.17G9, 0.1 µg/106 cells; BioLegend), anti–IFN-γ (clone XMG1.2, 0.1 µg/106 cells; eBioscience), anti–IL-1R1 (clone 12A6, 0.1 µg/106 cells; BD Biosciences), anti–IL-17 (clone TC11-18H10.1, 0.1 µg/106 cells; BioLegend), anti–IL-23R (clone 078–1208, 0.1 µg/106 cells; BD Biosciences), anti-TCRβ (clone H57-597, 0.1 µg/106 cells; eBioscience), anti-TCRδ (clone GL3, 0.1 µg/106 cells; eBioscience), anti-TLR2 (clone T2.5, 0.1 µg/106 cells; BioLegend), anti-Vα2 (clone B20.1, 0.1 µg/106 cells; BioLegend), anti-Vα8.3 (clone B21.14, 0.1 µg/106 cells; BioLegend), anti-Vγ1 (clone 2.11, 0.1 µg/106 cells; BioLegend), and anti-Vγ4 (clone UC3-1OA6, 0.1 µg/106 cells; BioLegend). The following antibodies were used to characterize human cells in flow cytometry experiments: anti-TCRαβ (clone T10B9.1A-31, 5 µl/test; BD Horizon), anti-CD3 (clone SK7, 1 µl/test; BD Biosciences), and anti-Vδ2 (clone B6, 2.5 µl/test; BioLegend).
Immune cell purification
Purified cell populations were isolated from single-cell suspensions of leukocytes extracted from spleens and LNs. Briefly, WT cells were enriched by magnetic separation using a Pan T Cell Isolation Kit (Miltenyi Biotec), labeled with anti-Vγ4 (clone UC3-1OA6, 0.1 µg/106 cells; BioLegend) or anti-TCRδ (clone GL3, 0.1 µg/106 cells; eBioscience), sorted by flow cytometry, and incubated with anti-TCRβ (clone H57-597, 0.1 µg/106 cells; eBioscience). Alternatively, WT cells were enriched by magnetic separation using a γδ T Cell Isolation Kit (Miltenyi Biotec). In the EAE model, Vγ4+TCRβ+ cells were enriched from spleens and draining LNs on day 10 after immunization with MOG and CFA. Distinct cell populations were sorted by flow cytometry using a FACSAria Fusion (BD Biosciences) or a MoFlo Legacy (Beckman Coulter).
Cell culture
Cells were cultured in medium with or without various combinations of IL-1β, IL-4, IL-12p70, IL-18, and IL-23 (all 10 ng/ml), together with IL-2 (1 or 10 ng/ml) or anti–IFN-γ and anti–IL-17 (both 1 µg/ml), in the presence or absence of anti-CD3 (clone 145-2C11, 1 µg/ml; BD Biosciences), anti-CD28 (clone 37.51, 2 µg/ml; BD Biosciences), anti-MHCII (clone IA/IE, 10 µg/ml; eBioscience), and/or anti-TCRδ (clone UC7-13D5, 10 µg/ml; BioLegend). Culture supernatants were harvested 24–72 h after T cell activation, and concentrations of IFN-γ, IL-4, IL-17, and GM-CSF were quantified by ELISA (BD PharMingen or R&D Systems). For DC co-culture experiments, bone marrow from C57BL/6 mice was incubated with GM-CSF (20 ng/ml) for 6 d to generate mature DCs. Mature DCs were washed and incubated for 5 h at 20,000 cells/well (for culture with magnetically enriched γδ T cells) or 2,500 cells/well (for culture with flow-purified populations) with medium alone (negative control), OVA257–264 (4 µg/ml; Sigma-Aldrich), OVA323–339 (10 µg/ml; Sigma-Aldrich), KLH (10 µg/ml; Calbiochem), or MOG35–55 (10 µg/ml; GenScript). Magnetically enriched γδ T cells (105 cells/well from OT-I mice or 2 × 105 cells/well from OT-II mice), flow-purified TCRδ+TCRβ+ or TCRδ+TCRβ– cells (20,000 cells/well) from OT-II mice, or flow-purified Vγ4+TCRβ+ cells (20,000 cells/well) from mice immunized with MOG and CFA were then co-cultured in the presence or absence of various stimulants with mature DCs. Culture supernatants were harvested after 2 d and assayed by ELISA. Cells were harvested simultaneously into TRIzol Reagent (Thermo Fisher Scientific). For NKG2D ligand-induced activation, murine YAC-1 cells were cultured in RPMI 1640 medium (Gibco) supplemented with 10% FCS. WT cells were enriched by magnetic separation using a Pan T Cell Isolation Kit (Miltenyi Biotec) and incubated with or without YAC-1 cells at a 10:1 ratio for 48 h.
Ontogeny experiments
Thymuses from C57BL/6 embryos were isolated on E14, E16, and E18. Flow cytometric analyses were performed using either a FACSCantoII or an LSRFortessa (both BD Biosciences).
Flow cytometry
Flow cytometry was performed on LN cells, purified immune cell populations, and mononuclear cells enriched by Percoll density centrifugation from the brains and spinal cords of mice immunized with MOG and CFA. In all experiments, cells were washed, stained with LIVE/DEAD Fixable Aqua (Life Technologies), and blocked with anti-CD16/CD32 (1 µg/ml; BD PharMingen). For intracellular staining, mononuclear cells were incubated for 5 h with brefeldin A (5 µg/ml; Sigma-Aldrich) in the presence or absence of phorbol myristate acetate (10 ng/ml; Sigma-Aldrich) and ionomycin (1 µg/ml; Sigma-Aldrich). Intracellular staining for IFN-γ, IL-17, and GM-CSF was conducted after fixation and permeabilization using a FIX&PERM Cell Permeabilization Kit (Caltag). Data were acquired using either a FACSCantoII or an LSRFortessa (both BD Biosciences) and analyzed with FACSDiva (BD Biosciences) or FlowJo software (Tree Star). Gates were set on isotype or fluorescence minus one controls. The gating strategy is depicted in Fig. S1 A.
Imaging flow cytometry
T cells were enriched from LNs by magnetic separation using a Pan T Cell Isolation Kit (Miltenyi Biotec) and labeled with anti-TCRβ (clone H57-597, 0.1 µg/106 cells; eBioscience), anti-TCRδ (clone GL3, 0.1 µg/106 cells; eBioscience), and anti-Vγ4 (clone UC3-1OA6, 0.1 µg/106 cells; BioLegend). Cells were then fixed and permeabilized, stained with DAPI (30 mM), and analyzed using an ImageStreamX Mark II Imaging Flow Cytometer (Amnis).
Library preparation and RNA sequencing
T cells were enriched from LNs by magnetic separation using a Pan T Cell Isolation Kit (Miltenyi Biotec), labeled as described above, and flow purified as Vγ4+TCRδ+TCRβ+ or Vγ4+TCRδ+TCRβ− populations. Total RNA was isolated using a NucleoSpin RNA XS Kit according to the manufacturer’s instructions (Clontech), omitting the carrier RNA and lysate filtration steps, and eluted in 10 µl of RNase-free water (Qiagen). RNA sequencing was performed on total RNA by normalizing the input mass to cell number according to cell type. For Vγ4+TCRδ+TCRβ− cells, input was normalized to 30,000 cells where possible (two samples fell below this input target, reaching totals of 12,687 and 21,772 cells). For Vγ4+TCRδ+TCRβ+ cells, input was normalized to 5,000 cells where possible (three samples fell below this input target, reaching totals of 2,036, 3,649, and 4,577 cells). cDNA was amplified using a SMARTer Ultra Low Input RNA Kit for Illumina Sequencing (Clontech) and sequenced at a target depth of 30 × 106 single-end (1 × 50 bp) reads per sample using a HiSeq 3000 System (Illumina).
Bioinformatic analysis of RNA sequencing data
Reads were mapped against the mouse reference genome (mm10, Ensembl release 76) using Kallisto (version 0.44; Bray et al., 2016). Transcript abundances were summarized to gene-level estimates in R (version 3.4.4) using the tximport Bioconductor package (version 1.8; Soneson et al., 2015) and the makeTxDbFromGFF package with a GTF annotation file from Ensembl (release 93). Differential expression analysis was performed using the NOISeq R Bioconductor package (version 2.22.1; Tarazona et al., 2015). Lowly expressed genes were removed on the basis of normalized read counts. Genes were kept if they reached a threshold of ≥100 read counts under any experimental condition. Data were normalized using the trimmed mean of log expression values approach in NOISeq. DEGs were detected using a probability value of 0.99, equivalent to a false discovery rate of 0.01, and an absolute fold-change cutoff of 2. A higher threshold of threefold was used for samples from mice with EAE, which exhibited higher levels of background noise. Heatmaps and clustering analyses, based on Euclidean distance with scaling by row, were generated using the ComplexHeatmap package in R (version 2.1.1; Gu, 2016). Principal-component analysis was performed using the R functions autoplot (ggfortify v0.4.8) and prcomp (stats v3.5.2) for all genes where the sum of the normalized read counts from all samples was at least 1,000 (n = 10,010). Gene ontology enrichment analysis was performed using the clusterProfiler package and the enrichGO tool in R (version 3.6.0; Yu et al., 2012).
Gene expression analysis
Total RNA was extracted from purified cell populations using the chloroform/isopropanol method. mRNA expression was evaluated by real-time PCR after reverse transcription using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR was performed for a variety of genes using predesigned TaqMan Gene Expression Assays (Applied Biosystems). 18s rRNA was used as an endogenous control. Samples were assayed using a 7500 Fast Real-Time PCR System (Applied Biosystems).
TCR sequencing
For population-level analyses, viable Vγ4+TCRβ+ cells from WT mice were flow purified directly into RNAlater (Applied Biosystems). Unbiased amplification of all expressed Tra, Trb, Trg, and Trd gene rearrangements was conducted using a template-switch anchored RT-PCR (Quigley et al., 2011). Amplicons were subcloned, sampled, sequenced, and analyzed as described previously (Price et al., 2005). For single-cell analyses, viable Vγ4+TCRβ+ cells from WT mice were index-sorted into a 96-well plate, and expressed Tra, Trb, Trg, and Trd gene rearrangements were amplified using a previously described protocol with minor modifications (Dash et al., 2011). Briefly, direct lysis and reverse transcription were performed using SuperScript Vilo (Invitrogen). The resultant cDNA was subjected to a first-round PCR incorporating a cocktail of validated Tra and Trb primers (Dash et al., 2011), together with newly designed Trg and Trd primers. The first-round products were subjected to a nested PCR using internal primer pools on separate 96-well plates for the α, β, γ, and δ chains, with each product assigned to an identical location for tracing back to the original cell. The final products were purified using Exonuclease I/Shrimp Alkaline Phosphatase. Sequencing was performed with the relevant constant region primers using an ABI Big Dye Sequencer (Applied Biosystems). Data were analyzed using a custom-built macro-enabled Microsoft Excel sheet to derive CDR3 nucleotide and amino acid sequences, and gene use was assigned by matching sequences against the IMGT database (Lefranc et al., 2009). If multiple species were observed, potentially reflecting biallelic rearrangements, the relevant transcripts were further resolved using additional PCRs. Briefly, the first-round products from the identified cell were subjected to another nested PCR using family-specific forward and reverse primers determined using a trace viewer. Resolution was further verified by adopting a “leave-one-out” amplification strategy, where the internal forward primer specific for the family determined in the previous experiment was withheld from the cocktail, and the resulting product was sequenced.
Immunofluorescence microscopy
γδ T cells or Vγ4+ cells were flow purified using a FACSAria Fusion (BD Biosciences). Purified cells were transferred onto poly-L-lysine–coated chamber slides, incubated for 2 h at 37°C, fixed in 4% paraformaldehyde for 15 min, and blocked with 20% FCS for 20 min γδ T cells were immunostained with anti-TCRβ–Alexa Fluor 647 and phalloidin, and Vγ4+ cells were immunostained in 5% bovine serum albumin with rabbit anti-TCRβ, washed, and labeled with a goat anti-mouse secondary antibody conjugated to Alexa Fluor 594. Slides were mounted using Mounting Medium with DAPI (DakoCytomation) and viewed on a point-scanning confocal microscope (FV1000; Olympus). Confocal images were selected to represent at least 20 captures (n = 3 independent experiments).
S. aureus infection model
Mice were inoculated i.p. with S. aureus (5 × 108 CFU in 100 µl) and sacrificed after 3 h or 3 d. Peritoneal exudate cells (PECs) were isolated from infected mice by lavage of the peritoneal cavity with 3 ml of sterile PBS. Lavage fluid was centrifuged and assayed for IL-1β, IL-17, and IFN-γ by ELISA. Kidneys were homogenized in 1 ml of sterile PBS. Total tissue bacterial load was established by plating serial dilutions of peritoneal lavage fluid or kidney homogenate on tryptic soy agar plates for 24 h at 37°C. Results were standardized to CFUs per milliliter.
EAE
Active EAE was induced by injecting mice s.c. with 100 µg of MOG35–55 peptide (GenScript) emulsified in CFA containing 4 mg/ml (0.4 mg/mouse) of heat-killed Mycobacterium tuberculosis (Chondrex). Mice were further injected i.p. with 250 ng of pertussis toxin (Kaketsuken) on days 0 and 2. In some experiments, mice were treated with anti-Vγ4 or an isotype control (250 µg/mouse; BioXCell), administered i.p. on days −1, 2, 5, 7, 11, 14, 17, and 20 of EAE. Passive EAE was induced by adoptive transfer of MOG-specific cells. C57BL/6 mice were immunized s.c. with 100 µg of MOG35–55 peptide (GenScript) emulsified in CFA containing 4 mg/ml (0.4 mg/mouse) of heat-killed M. tuberculosis (Chondrex). On day 10 after induction, the spleens and brachial, axillary, and inguinal LNs were removed from sacrificed mice and prepared as single-cell suspensions. For Vγ4β-depleted cultures, cells were labeled with anti-TCRβ (clone H57-597, 0.1 µg/106 cells; eBioscience) and anti-Vγ4 (clone UC3-1OA6, 0.1 µg/106 cells; BioLegend), and Vγ4+TCRβ+ cells were depleted by flow cytometry. Cells were stimulated with combinations of IL-1β (10 ng/ml), IL-23 (10 ng/ml), and/or MOG (100 µg/ml) in complete medium at 10 × 106 cells/ml. After 72 h, cells were washed, and cytokine production in the supernatants was measured by ELISA. A total dose of 5 × 106 viable cells was injected i.p. into each naive C57BL/6 recipient. Mice were monitored daily for signs of clinical disease. Clinical signs of EAE were assessed according to the following scores: no symptoms, 0; limp tail, 1; ataxic gait, 2; hindlimb weakness, 3; hindlimb paralysis, 4; tetraparalysis/moribund, 5.
MOG tetramer staining
Draining LNs were removed from mice 7 d after immunization with MOG and CFA and cultured at 20 × 106 cells/ml in the presence of IL-2 (5 ng/ml), 2.5% FCS, and MOG or control tetramer (National Institutes of Health Tetramer Facility) for 2.5 h at 37°C. Naive mice were processed similarly as controls. Cells were then washed twice and analyzed by flow cytometry as described above. Gates were set on fluorescence minus one controls among live CD3+CD4+CD44+ cells.
Data availability
RNA sequencing data for purified Vγ4+TCRδ+TCRβ+ or Vγ4+TCRδ+TCRβ− from LNs of naive mice or mice with EAE have been deposited to the Gene Expression Omnibus under accession no. GSE143500.
Statistical analysis
Statistical analyses were performed using one-way, two-way, and three-way ANOVAs and unpaired t tests in Prism (GraphPad). Error bars represent SD or SEM. Levels of significance are denoted as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Online supplemental material
Fig. S1 (related to Fig. 1) shows image and flow cytometry data from T cells that coexpress αβ and γδ TCRs. Fig. S2 (related to Fig. 4) shows activation data from hybrid αβ-γδ T cells. Fig. S3 (related to Fig. 5) shows RNA sequencing analysis of Vγ4+TCRβ+ and Vγ4+TCRβ− cells before or after activation.
Acknowledgments
The authors thank Owen Hughes and Barry Lewis (Merck) for assistance with ImageStream analysis, Andreea Petrasca and Jean Fletcher for isolation of human PBMCs, and Seth Coffelt for technical assistance and discussions.
This work was supported by grants from Science Foundation Ireland (15/IA/3041 to R.M. McLoughlin and 11/PI/1036, 12/RI/2340(7), 15/SPP/3212, and 16/IA/4468 to K.H.G. Mills), AbbVie (to K.H.G. Mills), Irish Higher Education Authority Program for Research in Third-Level Institutions (to S.C. Edwards and K.H.G. Mills), National Health and Medical Research Council (a CJ Martin ECR Fellowship to E.J. Grant), European Research Council (CoG_646701 to B. Silva-Santos), Medical Research Council (G1000213 to G. Anderson), Wellcome Trust (100326Z/12/Z to D.A. Price), National Institutes of Health (RO1AI107625 to P.G. Thomas), and American Lebanese Syrian Associated Charities (to P.G. Thomas).
Author contributions: S.C. Edwards and C.E. Sutton designed and performed most of the experiments; P. Dash and N. Apiwattanakul developed the single-cell method for sequencing murine γδ TCRs; K. Ladell, E.J. Grant, J.E. McLaren, P. Dash, N. Apiwattanakul, and W. Awad sequenced TCRs; F. Roche and K. Hokamp analyzed the RNA sequencing data; K. Ladell, K.L. Miners, and B. Moran designed flow cytometry panels and sorted cells; J.C. Ribot performed the studies with TCRα−/−, TCRβ−/−, MHCI−/−, and MHCII−/− mice; S. Baik performed the ontogeny studies; A. McGinley assisted with experimental work; S.J. Lalor and R.M. McLoughlin designed experiments and provided feedback on the manuscript; J. Paez-Cortez guided cell isolation and sorting methodologies for the single-cell RNA sequencing experiments; V. Pivorunas and L. Dowding designed and performed RNA extractions and designed the RNA sequencing input normalization strategy; M. Macoritto designed the sequencing strategy for single-cell RNA sequencing experiments; A. Slavin supervised and directed the RNA sequencing experiments with help from V. Pivorunas, L. Dowding, M. Macoritto, and J. Paez-Cortez; G. Anderson designed the ontogeny studies and provided feedback on the manuscript; B. Silva-Santos designed the studies with TCRα−/−, TCRβ−/−, MHCI−/−, and MHCII−/− mice and provided feedback on the manuscript; D.A. Price and P.G. Thomas designed the TCR sequencing studies and provided feedback on the manuscript; K.H.G. Mills directed the project, designed the studies, and wrote the manuscript with help from S.C. Edwards, C.E. Sutton, and D.A. Price. All authors edited and approved the final manuscript.
References
- Ahmed R., Omidian Z., Giwa A., Cornwell B., Majety N., Bell D.R., Lee S., Zhang H., Michels A., Desiderio S., et al. 2019. A Public BCR Present in a Unique Dual-Receptor-Expressing Lymphocyte from Type 1 Diabetes Patients Encodes a Potent T Cell Autoantigen. Cell. 177:1583–1599.e16. 10.1016/j.cell.2019.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco N., Engdahl C., Bénard A., Rolink J., Ceredig R., and Rolink A.G.. 2008. TCR-beta chains derived from peripheral gammadelta T cells can take part in alphabeta T-cell development. Eur. J. Immunol. 38:3520–3529. 10.1002/eji.200838668 [DOI] [PubMed] [Google Scholar]
- Bowen S., Sun P., Livak F., Sharrow S., and Hodes R.J.. 2014. A novel T cell subset with trans-rearranged Vγ-Cβ TCRs shows Vβ expression is dispensable for lineage choice and MHC restriction. J. Immunol. 192:169–177. 10.4049/jimmunol.1302398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., and Pachter L.. 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34:525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Cai Y., Shen X., Ding C., Qi C., Li K., Li X., Jala V.R., Zhang H.G., Wang T., Zheng J., and Yan J.. 2011. Pivotal role of dermal IL-17-producing γδ T cells in skin inflammation. Immunity. 35:596–610. 10.1016/j.immuni.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Carico Z., Shih H.-Y., and Krangel M.S.. 2015. A discrete chromatin loop in the mouse Tcra-Tcrd locus shapes the TCRδ and TCRα repertoires. Nat. Immunol. 16:1085–1093. 10.1038/ni.3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti H.R., Peterson A.C., Brane L., Huppler A.R., Hernández-Santos N., Whibley N., Garg A.V., Simpson-Abelson M.R., Gibson G.A., Mamo A.J., et al. 2014. Oral-resident natural Th17 cells and γδ T cells control opportunistic Candida albicans infections. J. Exp. Med. 211:2075–2084. 10.1084/jem.20130877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia D.V., Lopes A., and Silva-Santos B.. 2013. Tumor cell recognition by γδ T lymphocytes: T-cell receptor vs. NK-cell receptors. OncoImmunology. 2:e22892 10.4161/onci.22892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley M.P., Reich Z., Mavaddat N., Altman J.D., and Chien Y.. 1997. The recognition of the nonclassical major histocompatibility complex (MHC) class I molecule, T10, by the gammadelta T cell, G8. J. Exp. Med. 185:1223–1230. 10.1084/jem.185.7.1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash P., McClaren J.L., Oguin T.H. III, Rothwell W., Todd B., Morris M.Y., Becksfort J., Reynolds C., Brown S.A., Doherty P.C., and Thomas P.G.. 2011. Paired analysis of TCRα and TCRβ chains at the single-cell level in mice. J. Clin. Invest. 121:288–295. 10.1172/JCI44752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G., Rowen L., Lee I., Boysen C., Roach J.C., Smit A.F.A., Wang K., Koop B.F., and Hood L.. 2001. Comparative genomics of the human and mouse T cell receptor loci. Immunity. 15:337–349. 10.1016/S1074-7613(01)00200-X [DOI] [PubMed] [Google Scholar]
- Gray E.E., Ramírez-Valle F., Xu Y., Wu S., Wu Z., Karjalainen K.E., and Cyster J.G.. 2013. Deficiency in IL-17-committed Vγ4(+) γδ T cells in a spontaneous Sox13-mutant CD45.1(+) congenic mouse substrain provides protection from dermatitis. Nat. Immunol. 14:584–592. 10.1038/ni.2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z. 2016. ComplexHeatmap. R package version 2.1.1. https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html (accessed October 2019).
- Haas J.D., González F.H., Schmitz S., Chennupati V., Föhse L., Kremmer E., Förster R., and Prinz I.. 2009. CCR6 and NK1.1 distinguish between IL-17A and IFN-gamma-producing gammadelta effector T cells. Eur. J. Immunol. 39:3488–3497. 10.1002/eji.200939922 [DOI] [PubMed] [Google Scholar]
- Hayes S.M., Li L., and Love P.E.. 2005. TCR signal strength influences alphabeta/gammadelta lineage fate. Immunity. 22:583–593. 10.1016/j.immuni.2005.03.014 [DOI] [PubMed] [Google Scholar]
- Heilig J.S., and Tonegawa S.. 1986. Diversity of murine gamma genes and expression in fetal and adult T lymphocytes. Nature. 322:836–840. 10.1038/322836a0 [DOI] [PubMed] [Google Scholar]
- Hochstenbach F., and Brenner M.B.. 1989. T-cell receptor delta-chain can substitute for alpha to form a beta delta heterodimer. Nature. 340:562–565. 10.1038/340562a0 [DOI] [PubMed] [Google Scholar]
- Hvas J., Oksenberg J.R., Fernando R., Steinman L., and Bernard C.C.A.. 1993. γ δ T cell receptor repertoire in brain lesions of patients with multiple sclerosis. J. Neuroimmunol. 46:225–234. 10.1016/0165-5728(93)90253-U [DOI] [PubMed] [Google Scholar]
- Ishida I., Verbeek S., Bonneville M., Itohara S., Berns A., and Tonegawa S.. 1990. T-cell receptor gamma delta and gamma transgenic mice suggest a role of a gamma gene silencer in the generation of alpha beta T cells. Proc. Natl. Acad. Sci. USA. 87:3067–3071. 10.1073/pnas.87.8.3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., and Littman D.R.. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133. 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Kara E.E., McKenzie D.R., Bastow C.R., Gregor C.E., Fenix K.A., Ogunniyi A.D., Paton J.C., Mack M., Pombal D.R., Seillet C., et al. 2015. CCR2 defines in vivo development and homing of IL-23-driven GM-CSF-producing Th17 cells. Nat. Commun. 6:8644 10.1038/ncomms9644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashani E., Föhse L., Raha S., Sandrock I., Oberdörfer L., Koenecke C., Suerbaum S., Weiss S., and Prinz I.. 2015. A clonotypic Vγ4Jγ1/Vδ5Dδ2Jδ1 innate γδ T-cell population restricted to the CCR6+CD27− subset. Nat. Commun. 6:6477 10.1038/ncomms7477 [DOI] [PubMed] [Google Scholar]
- Lefranc M.-P., Giudicelli V., Ginestoux C., Jabado-Michaloud J., Folch G., Bellahcene F., Wu Y., Gemrot E., Brochet X., Lane J., et al. 2009. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 37(Database):D1006–D1012. 10.1093/nar/gkn838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak F., Petrie H.T., Crispe I.N., and Schatz D.G.. 1995. In-frame TCR delta gene rearrangements play a critical role in the alpha beta/gamma delta T cell lineage decision. Immunity. 2:617–627. 10.1016/1074-7613(95)90006-3 [DOI] [PubMed] [Google Scholar]
- Lockhart E., Green A.M., and Flynn J.L.. 2006. IL-17 production is dominated by gammadelta T cells rather than CD4 T cells during Mycobacterium tuberculosis infection. J. Immunol. 177:4662–4669. 10.4049/jimmunol.177.7.4662 [DOI] [PubMed] [Google Scholar]
- Misiak A., Wilk M.M., Raverdeau M., and Mills K.H.. 2017. IL-17-Producing Innate and Pathogen-Specific Tissue Resident Memory γδ T Cells Expand in the Lungs of Bordetella pertussis-Infected Mice. J. Immunol. 198:363–374. 10.4049/jimmunol.1601024 [DOI] [PubMed] [Google Scholar]
- Murphy A.G., O’Keeffe K.M., Lalor S.J., Maher B.M., Mills K.H., and McLoughlin R.M.. 2014. Staphylococcus aureus infection of mice expands a population of memory γδ T cells that are protective against subsequent infection. J. Immunol. 192:3697–3708. 10.4049/jimmunol.1303420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papotto P.H., Ribot J.C., and Silva-Santos B.. 2017. IL-17+ γδ T cells as kick-starters of inflammation. Nat. Immunol. 18:604–611. 10.1038/ni.3726 [DOI] [PubMed] [Google Scholar]
- Pellicci D.G., Uldrich A.P., Le Nours J., Ross F., Chabrol E., Eckle S.B.G., de Boer R., Lim R.T., McPherson K., Besra G., et al. 2014. The molecular bases of δ/αβ T cell-mediated antigen recognition. J. Exp. Med. 211:2599–2615. 10.1084/jem.20141764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D.A., Brenchley J.M., Ruff L.E., Betts M.R., Hill B.J., Roederer M., Koup R.A., Migueles S.A., Gostick E., Wooldridge L., et al. 2005. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J. Exp. Med. 202:1349–1361. 10.1084/jem.20051357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.E., Reinhardt R.L., Liang H.E., and Locksley R.M.. 2012. Marking and quantifying IL-17A-producing cells in vivo. PLoS One. 7:e39750 10.1371/journal.pone.0039750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley M.F., Almeida J.R., Price D.A., and Douek D.C.. 2011. Unbiased molecular analysis of T cell receptor expression using template-switch anchored RT-PCR. Curr. Protoc. Immunol. Chapter 10:Unit10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboldi A., Coisne C., Baumjohann D., Benvenuto F., Bottinelli D., Lira S., Uccelli A., Lanzavecchia A., Engelhardt B., and Sallusto F.. 2009. C-C chemokine receptor 6-regulated entry of TH-17 cells into the CNS through the choroid plexus is required for the initiation of EAE. Nat. Immunol. 10:514–523. 10.1038/ni.1716 [DOI] [PubMed] [Google Scholar]
- Rei M., Gonçalves-Sousa N., Lança T., Thompson R.G., Mensurado S., Balkwill F.R., Kulbe H., Pennington D.J., and Silva-Santos B.. 2014. Murine CD27(-) Vγ6(+) γδ T cells producing IL-17A promote ovarian cancer growth via mobilization of protumor small peritoneal macrophages. Proc. Natl. Acad. Sci. USA. 111:E3562–E3570. 10.1073/pnas.1403424111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot J.C., deBarros A., Pang D.J., Neves J.F., Peperzak V., Roberts S.J., Girardi M., Borst J., Hayday A.C., Pennington D.J., and Silva-Santos B.. 2009. CD27 is a thymic determinant of the balance between interferon-gamma- and interleukin 17-producing gammadelta T cell subsets. Nat. Immunol. 10:427–436. 10.1038/ni.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roark C.L., French J.D., Taylor M.A., Bendele A.M., Born W.K., and O’Brien R.L.. 2007. Exacerbation of collagen-induced arthritis by oligoclonal, IL-17-producing gamma delta T cells. J. Immunol. 179:5576–5583. 10.4049/jimmunol.179.8.5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata K., Yamada H., Hara H., Kishihara K., and Yoshikai Y.. 2007. Resident Vdelta1+ gammadelta T cells control early infiltration of neutrophils after Escherichia coli infection via IL-17 production. J. Immunol. 178:4466–4472. 10.4049/jimmunol.178.7.4466 [DOI] [PubMed] [Google Scholar]
- Shimonkevitz R., Colburn C., Burnham J.A., Murray R.S., and Kotzin B.L.. 1993. Clonal expansions of activated gamma/delta T cells in recent-onset multiple sclerosis. Proc. Natl. Acad. Sci. USA. 90:923–927. 10.1073/pnas.90.3.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Santos B., Serre K., and Norell H.. 2015. γδ T cells in cancer. Nat. Rev. Immunol. 15:683–691. 10.1038/nri3904 [DOI] [PubMed] [Google Scholar]
- Sinkora M., Sinkorová J., and Holtmeier W.. 2005. Development of gammadelta thymocyte subsets during prenatal and postnatal ontogeny. Immunology. 115:544–555. 10.1111/j.1365-2567.2005.02194.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C., Love M.I., and Robinson M.D.. 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000 Res. 4:1521 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton C.E., Lalor S.J., Sweeney C.M., Brereton C.F., Lavelle E.C., and Mills K.H.. 2009. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity. 31:331–341. 10.1016/j.immuni.2009.08.001 [DOI] [PubMed] [Google Scholar]
- Sutton C.E., Mielke L.A., and Mills K.H.. 2012. IL-17-producing γδ T cells and innate lymphoid cells. Eur. J. Immunol. 42:2221–2231. 10.1002/eji.201242569 [DOI] [PubMed] [Google Scholar]
- Tarazona S., Furió-Tarí P., Turrà D., Pietro A.D., Nueda M.J., Ferrer A., and Conesa A.. 2015. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 43:e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., and MacDonald H.R.. 1998. A limited role for beta-selection during gamma delta T cell development. J. Immunol. 161:5851–5854. [PubMed] [Google Scholar]
- Wucherpfennig K.W., Newcombe J., Li H., Keddy C., Cuzner M.L., and Hafler D.A.. 1992. Gamma delta T-cell receptor repertoire in acute multiple sclerosis lesions. Proc. Natl. Acad. Sci. USA. 89:4588–4592. 10.1073/pnas.89.10.4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yednock T.A., Cannon C., Fritz L.C., Sanchez-Madrid F., Steinman L., and Karin N.. 1992. Prevention of experimental autoimmune encephalomyelitis by antibodies against alpha 4 beta 1 integrin. Nature. 356:63–66. 10.1038/356063a0 [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., and He Q.Y.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA sequencing data for purified Vγ4+TCRδ+TCRβ+ or Vγ4+TCRδ+TCRβ− from LNs of naive mice or mice with EAE have been deposited to the Gene Expression Omnibus under accession no. GSE143500.