Abstract
Primary cilia in many cell types contain a periaxonemal subcompartment called the inversin compartment. Four proteins have been found to assemble within the inversin compartment: INVS, ANKS6, NEK8, and NPHP3. The function of the inversin compartment is unknown, but it appears to be critical for normal development, including left–right asymmetry and renal tissue homeostasis. Here we combine superresolution imaging of human RPE1 cells, a classic model for studying primary cilia in vitro, with a genetic dissection of the protein–protein binding relationships that organize compartment assembly to develop a new structural model. We observe that INVS is the core structural determinant of a compartment composed of novel fibril-like substructures, which we identify here by three-dimensional single-molecule superresolution imaging. We find that NEK8 and ANKS6 depend on INVS for localization to these fibrillar assemblies and that ANKS6-NEK8 density within the compartment is regulated by NEK8. Together, NEK8 and ANKS6 are required downstream of INVS to localize and concentrate NPHP3 within the compartment. In the absence of these upstream components, NPHP3 is redistributed within cilia. These results provide a more detailed structure for the inversin compartment and introduce a new example of a membraneless compartment organized by protein–protein interactions.
INTRODUCTION
The primary cilium is a signaling organelle that projects from the surfaces of specific cell types, including many human tissues. Each cilium is an elongated structure, 1–20 µm in length, consisting of a microtubule-based axoneme organized by the mother centriole. The cilium is bounded by a ciliary membrane that is continuous with the plasma membrane, but differs in composition and programming (Phua et al., 2019). Cells with primary cilia have just one cilium per cell, but multiciliated cells are present in some tissues including the brain and trachea. Generally, the function of the primary cilium is to establish a signaling compartment which coordinates events important in normal development and in metabolic or sensory functions specific to each tissue. Cilia-specific signaling pathways vary from tissue to tissue, and in many cases are not well understood. Mutations in proteins that localize to the primary cilium cause a spectrum of diseases and disorders, called ciliopathies (Waters and Beales, 2011). These range from lethal neonatal disorders to retinal degeneration and sensory differences such as anosmia.
Mutation or loss of the protein inversin (INVS) causes complete reversal of left–right body asymmetry (situs inversus, the condition for which the gene was originally named [ Mochizuki et al., 1998]) and also a recessive form of infantile cystic kidney disease called nephronophthisis (NPHP) (Otto et al., 2003; Okada et al., 2008; Tory et al., 2009; Bellavia et al., 2010). Both left–right asymmetry defects and NPHP are classic examples of ciliopathy. To date, 22 different NPHP genes have been identified, many of which encode proteins that localize to primary cilia. Four NPHP proteins, INVS (NPHP2), NPHP3, NEK8 (NPHP9), and ANKS6 (NPHP16), localize to a distinctive subcompartment within the primary cilium called the inversin compartment (INVc). The INVc is found at the proximal end of the cilium (the end closest to the basal body) and is defined by the localization of the INVS protein (Shiba et al., 2009). Many examples of primary cilia have been shown to contain an INVc including ductal epithelia from multiple organs in mice (Watanabe et al., 2003; Sohara et al., 2008; Nakajima et al., 2018), the left–right organizer of the embryonic node (Watanabe et al., 2003; Manning et al., 2013; Nakajima et al., 2018), renal epithelial cell lines (mIMCD3: Mahjoub et al., 2005; Otto et al., 2008; Trapp et al., 2008; Sang et al., 2011; MDCK: Nürnberger et al., 2004; Dai1: Shiba et al., 2009), and fibroblasts (derived from patients, Grampa et al., 2016, and 3T3 cell line, Hong et al., 2018). INVc proteins are well conserved in vertebrates, and an INVc-like region containing an INVS homologue has even been described in sensory neuronal cilia in Caenorhabditis elegans (Warburton-Pitt et al., 2014).
Based on phenotypes in patients and mouse models, the INVc has at least two physiological functions: maintaining tissue integrity in the kidney and left–right asymmetry determination in the early embryo. Control of these phenotypes may require the Wnt (Simons et al., 2005; Bellavia et al., 2010; Lienkamp et al., 2010; Sugiyama et al., 2011), Hippo (Grampa et al., 2016), or Akt (Suizu et al., 2016) signaling pathways, which have been linked to the INVc genes.
However, it remains unclear how the unique localization of INVS, NPHP3, NEK8, and ANKS6 to the INVc contributes to these signaling pathways, and so the function of the INVc is still unknown. Interactions have been detected between all four INVc proteins using a combination of affinity purification pull downs to identify interacting proteins (Sang et al., 2011; Hoff et al., 2013) and more directed coimmunoprecipitation experiments using tagged proteins expressed in cell culture (Hoff et al., 2013; Czarnecki et al., 2015). Here, we considered how the direct interactions between the proteins that localize to the INVc may organize or regulate the compartment and how the length of the INVc is determined. We were particularly intrigued by the mechanism of INVc length determination, as it had been reported to vary widely from cilium to cilium (Sang et al., 2011). Varying INVc length suggests the presence of an undiscovered mechanism for forming a compartment boundary within the primary cilium that is not bounded by a membrane or known structure.
Electron microscopy (EM) has been used widely to study cilia ultrastructure. However, EM works best for studying either the electron-dense components of the cilium, for example, the axoneme itself (Sun et al., 2019), or highly repeating structures such as intraflagellar (IFT) trains (Jordan et al., 2018) and microtubule inner proteins in motile cilia (Stoddard et al., 2018; Owa et al., 2019). In the absence of a robust EM-compatible INVc protein–labeling method, the inherent structural variability observed in the INVc would complicate EM reconstructions of the compartment, so we utilized the specificity of two fluorescence superresolution microscopy techniques, structured illumination microscopy (SIM) and single-molecule superresolution (SM SR) microscopy, both of which have recently been employed to study substructures at the ciliary base (Yang et al., 2015, 2018; Kanie et al., 2017; Shi et al., 2017; Bowler et al., 2019). Each of these techniques is compatible with immunofluorescence (IF) labeling. Both IF and fluorescently tagged proteins have been previously used to detect the INVc (Otto et al., 2008; Trapp et al., 2008; Shiba et al., 2009, 2010; Sang et al., 2011; Hoff et al., 2013; Czarnecki et al., 2015; Grampa et al., 2016; Nakajima et al., 2018). Using SIM to study colocalization of INVc proteins, 3D SM SR microscopy to determine the detailed three-dimensional (3D) structure of the compartment, and diffraction-limited microscopy (DLM) to measure the interdependence of the INVc proteins through pairwise correlations and quantitative studies in knockout lines, we present here a surprising new view of the INVc featuring improved detail on its structure and composition.
RESULTS
INVS, ANKS6, NEK8, and NPHP3 localize in the INVc
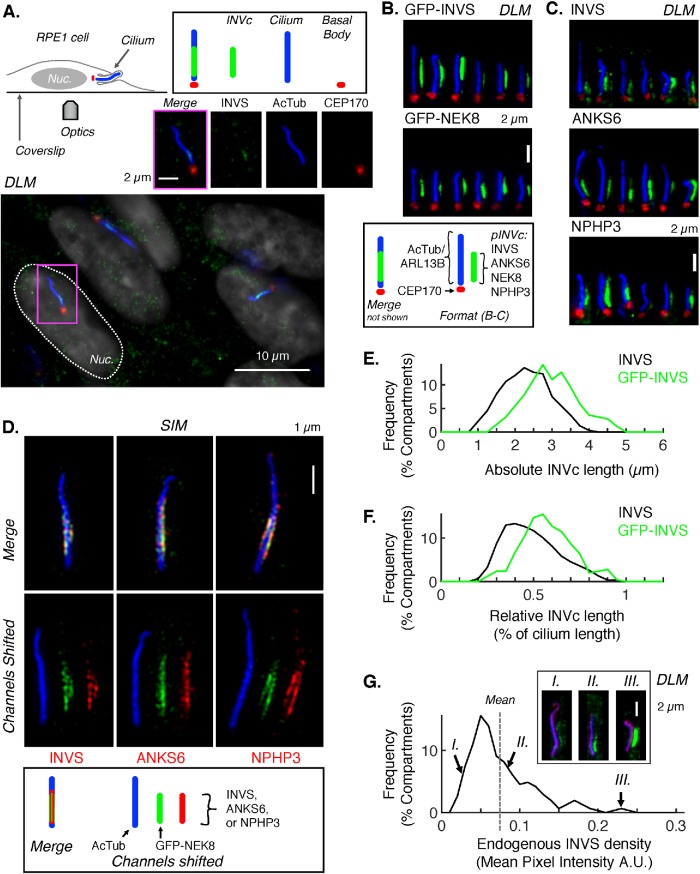
Confluent, serum-starved RPE1 cells (a retinal pigment epithelial cell line immortalized with telomerase hTERT) form primary cilia with 90% efficiency. Under these conditions, which were used throughout this study (Supplemental Figure S1A), we found that the INVc can be detected by either indirect IF or GFP-tagging of some INVc components in PFA-fixed cells. As expected, endogenous INVS, detected by anti-INVS, was localized in a distinct proximal ciliary subcompartment, approximately 300 nm above the basal body (inset Figure 1A, diagrammed in more detail in Supplemental Figure S2; 100 cilia from a representative experiment shown in Supplemental Figure S3). ANKS6 and NPHP3 were also detected in the INVc of RPE1 cilia by IF using commercially available antibodies to the endogenous proteins (Figure 1C). NEK8, for which we do not have an efficient antibody, localized to the INVc of RPE1 cells that stably express GFP-NEK8 (Figure 1B Middle). GFP-INVS also localizes to the INVc in RPE1 cells engineered to express GFP-INVS stably (Figure 1B, top). As GFP-ANKS6 and NPHP3-GFP did not localize efficiently to cilia in RPE1 cells, these constructs were not considered reliable INVc reporters (Supplemental Figure S4A). In addition to its localization in the INVc, we also noted that GFP-INVS localized to the ciliary base (likely to the outer distal appendages) and sometimes to the ciliary tip. However, these localizations were not detected for the endogenous INVS protein by staining with anti-INVS (Supplemental Figure S4, C–H), suggesting either that they are normally transient localizations or that these extracompartmental localizations result from tagging the protein.
FIGURE 1:
INVS, ANKS6, NEK8, and NPHP3 colocalize in the INVc. (A) Subcellular localization of the INVc in confluent, serum-starved WT RPE1 cells. Endogenous INVS (green) was detected by IF with a polyclonal anti-INVS primary and an AF488-conjugated secondary antibody measured by DLM. Cell nuclei (“Nuc.”) were labeled with DAPI (shown in gray). Cilia were detected by IF labeling of acetylated tubulin in the ciliary axoneme (AcTub, AF647 secondary, blue) and CEP170 at the basal body (AF568 secondary, red). The merge image of the inset (magenta border) contains only the INVS, AcTub, and CEP170 channels. The schematic of an RPE1 cell (top left, not drawn to scale) illustrates the positioning of the cilium relative to the nucleus in a typical RPE1 cell, as well as the positioning of the cilium relative to the coverslip and microscope objective (“Optics”). Most RPE1 cilia are positioned roughly parallel to the coverslip, as depicted. (B, C) DLM images of cilia with INVcs detected by fluorescence. For each INVc protein (pINVc), cilia from six individual cells have been oriented basal body down, cilium tip up, aligned side by side. (B, top two panels) DLM images of cilia in WT RPE1 cells stably expressing GFP-NEK8 or GFP-INVS. GFP was detected directly by fluorescence of the GFP protein. GFP signal is shown in green and shifted 600 nm to the right of the ciliary markers that were detected by IF: ARL13B (AF647 secondary, blue) and CEP170 (AF568 secondary, red). (B, bottom panel) Schematic illustrating the coloring and relative positioning of fluorescent markers shown in the images in B and C. (C) DLM of WT RPE1 cilia with inversin compartments detected by IF with primary antibodies to endogenous INVc proteins (pINVc): anti-INVS, anti-ANKS6, or anti-NPHP3, all detected with AF488 secondary antibodies and all shown here in green, shifted 600 nm to the right of the axoneme (AcTub, AF647 secondary, blue) and basal body (CEP170, AF568 secondary, red). (D) Pairwise colocalization of all four INVc proteins measured by SIM. GFP-NEK8 was detected by immunolabeling with anti-GFP primary and AF488-conjugated secondary antibodies (green). Endogenous ANKS6, NPHP3, and INVS were detected with specific primary antibodies and AF568-conjugated secondary antibodies (red). Samples were also stained with anti-acetylated tubulin (AcTub) primary detected with AF647-conjugated secondary (blue). Each of the three cilia is shown in a three-color merged image (top row of images) and with the red and blue channels shifted relative to the merge to show each channel individually (bottom row of images). (D, bottom panel) Schematic illustrating the coloring and relative positioning of fluorescent markers in the merged and shifted images. (E) Distribution of absolute INVc lengths measured in µm by DLM of endogenous INVS in WT cells (black, mean = 2.19, stdev = 0.61, n = 154) or GFP-INVS in WT cell lines stably expressing GFP fusion proteins (green, mean = 2.87, stdev = 0.70, n = 126) in one representative experiment. Only INVS-positive cilia are included. INVc lengths are significantly different between the two cell lines (p = 5.94 × 10–16). (F) Distribution of relative INVc lengths. INVc length was normalized relative to cilium length: [length INVc]/[length AcTub+ARL13B]. Endogenous INVS: mean = 0.46, stdev = 0.15, n = 154. GFP-INVS: mean = 0.55, stdev = 0.14, n = 126. Only INVS-positive cilia are included. Lengths are significantly different between the two cell lines (p = 4.00 × 10–7). (G) Distribution of INVS densities (see Supplemental Figure S2 for definition) within the INVc, measured as mean pixel intensity in arbitrary units (A.U.) for a manually masked INVS-positive region for the same 126 cilia as measured in E and F. Only INVS-positive cilia are included. Representative images of dim (low density, I), average (medium density, II), and bright (high density, III) compartments are shown.
The INVc varies in length and INVc length does not correlate with known proximal ciliary structures
In RPE1 cells, the mean INVc length of compartments detected by IF with anti-INVS is 2.2 µm (Figure 1E), or 46% of the cilium length (Figure 1F). As had previously been reported for mIMCD3 cells expressing GFP-INVS (Sang et al., 2011), the relative length of the compartment varies substantially from cilium to cilium (Figure 1, E and F). We found that most INVcs are 25–75% of the cilium length. Short compartments occupying just 19–25% of the total cilium length were found in 4% of cilia and long compartments extending from 75% of the cilium length to all the way to the ciliary tip in 3%. Stable expression of GFP-INVS increased both absolute INVc length (mean 2.9 µm, Figure 1E), and relative INVc length (55%, Figure 1F) compared with endogenous INVS compartments. We found no substantial correlation between INVc length and cilia length (Supplemental Figure S5A).
In an attempt to explain the variation in INVc length, we sought to determine whether the length of the INVc could be regulated by proximal structures in the primary cilium other than the known INVc components. We used SIM to measure colocalization between the INVc and the depth of the ciliary pocket, the polyglutamylated region of the axoneme, and the transition zone (TZ). We found that the ciliary pocket can be deeper or shallower than the height of the INVc (Supplemental Figures S5, B–D, and S6), indicating that the length of the INVc does not correlate directly with ciliary pocket depth. We also found that the INVc typically extends farther along the axoneme than the polyglutamylated region (Supplemental Figure S7A), suggesting that the INVc and tubulin polyglutamylation represent two overlapping yet independent ciliary subcompartments in RPE1 cells. Finally, we found that the INVc can begin well above the TZ (Supplemental Figure S8). The TZ and the start of INVc were separated by a median distance of 280 nm (SD 270 nm), which is a relatively large distance on a molecular scale, suggesting that the INVc and the TZ are not directly linked.
Any potential colocalization between IFT and the INVc was also assessed by SIM (Supplemental Figure S9). As expected, IFT88 is most strongly enriched at the base of the cilium, with some IFT protein detected along the axoneme and at the ciliary tip. Axonemal IFT foci did not appear to be especially enriched or excluded either in the INVc or at the proximal and distal INV boundaries. No special relationship was observed between the INVc and axonemal IFT. We also evaluated the colocalization between GFP-INVS and GPR161, a ciliary GPCR that is readily detectable by IF in wild-type (WT) RPE1 cells, and similarly observed no special relationship between GFP-INVS and GPR161 (Supplemental Figure S10B). We also evaluated the colocalization of endogenous INVc proteins (ANKS6 and NPHP3) with another ciliary membrane–associated membrane protein, ARL13B, and again observed neither enrichment nor exclusion from the INVc (Supplemental Figure S10, C and D).
The INVc varies in INVc protein density
We noted that the fluorescence intensity of the INVc varies from cell to cell (Figure 1G) regardless of the marker used: antibodies to endogenous proteins or overexpressed GFP alleles of NEK8 or INVS. We interpret fluorescence intensity measured by microscopy as approximately reflecting INVc protein density in the cilium, as discussed in Materials and Methods and as diagrammed in Supplemental Figure S2E. Thus, in addition to varying in length, the INVc also varies in INVc protein density. We observe a modest correlation between INVc length and INVc protein density (Supplemental Figure S11, A and B), suggesting some relationship between the number of INVc proteins and the length of the INVc, but not excluding the possibility that INVc length and density are established by independent mechanisms that we explore in the Discussion.
SIM of the INVc in GFP-NEK8 cells supports colocalization with INVS, ANKS6, and NPHP3 and suggests an asymmetric periaxonemal structure
By DLM, all four INVc proteins (INVS, NEK8, ANKS6, and NPHP3) appear to colocalize in the INVc. To determine whether these proteins might be arranged within discrete subregions of the compartment that are not resolved by DLM, we used a superresolution-microscopy technique, SIM, to measure the colocalization of each INVc protein by staining for endogenous INVS, ANKS6, and NPHP3 in cells expressing GFP-NEK8. SIM achieves an XY resolution of approximately 100 nm and a Z resolution of approximately 300 nm (Schermelleh et al., 2010). By SIM, we found that INVc proteins still colocalized with each other in the INVc (Figure 1D; 20 additional cilia shown in Supplemental Figures S12–S14). We next considered what could be learned about the spatial arrangement of INVc proteins relative to the axoneme.
The diameter of the ciliary axoneme at the cilium base is 150–200 nm (Supplemental Figure S21, A and B), consistent with that estimated from EM images of RPE1 cells (Molla-Herman et al., 2010) and mIMCD3 cells (Gluenz et al., 2010; Sun et al., 2019), though the axonemes of motile cilia are larger, ∼200–250 nm (Bui et al., 2012; Oda et al., 2014). In SIM reconstructions of cilia oriented parallel to the imaging plane, as is the case for the majority of RPE1 cilia (Figure 1A), periaxonemal structures are expected to have a hollow signature: two parallel lines of signal (high-intensity pixels) separated by a lower-intensity center (Supplemental Figure S15). This dim center is the space where the axoneme excludes periaxonemal proteins. Consistent with this interpretation, SIM reconstructions of axonemes tend to lack the hollow signature, especially when imaged in the far red channel (AF647), which has a poorer resolution than either AF488 or AF568 (Supplemental Figure S15D and Schermelleh et al., 2010).
To establish a reference point for ciliary protein conformations, we measured the localization of ARL13B, an abundant ciliary protein that does not localize exclusively to the INVc, but instead localizes along the length of the axoneme (Supplemental Figure S10, C and D). ARL13B is associated with the inner leaflet of the ciliary membrane via palmitoylation near the N-terminus of the protein (Roy et al., 2017), and is therefore expected to exhibit some periaxonemal ciliary localization, but it also interacts with the axoneme (Revenkova et al., 2018). By SIM, ARL13B did appear as two parallel lines along the axoneme with signal symmetrically paired on either side of the axoneme (Supplemental Figures S10, C and D, and S15, B and D), which is consistent with enrichment of ARL13B in the periaxonemal space.
In the vast majority of SIM reconstructions of the INVc, INVc proteins are symmetrically paired, parallel lines (Figure 1D, INVS and NPHP3, and Supplemental Figures S12–S14), indicating that the INVc is a periaxonemal structure. This result is consistent with previously published TEM images of immuno-gold labeled INVS protein, which also showed INVS on either side of the axoneme in a longitudinal section of the cilium, as well as distributed around the axoneme in a cross-section (Shiba et al., 2009). However, in some SIM reconstructions, INVc proteins were notably arranged around the axoneme asymmetrically (Figure 1D; Supplemental Figures S16 and S17). In Figure 1D, the right side of the INVc, labeled with both INVS and GFP-NEK8, is shorter than the left side (300 nm compared with 700 nm). In the INVc labeled with GFP-NEK8 and ANKS6 (Figure 1D), the INVc signal is almost exclusively on the right side of the axoneme. We examined a total of 354 SIM reconstructions of GFP-NEK8-positive cilia and found that 36% of compartments contained asymmetric subregions ≥300 nm in length (Supplemental Figures S16 and S17). In comparison, 10% of ARL13B reconstructions (n = 75) contained such regions (Supplemental Figure S15). The prevalence of asymmetric INVcs observed by SIM suggested that the INVc could be composed of some variable structure that appeared to be evenly distributed about the axoneme in most cilia, but in some cilia could be enriched on one side only. A higher-resolution microscopy technique was needed to measure the spatial arrangement of the INVc structures around the axoneme, which we pursued next.
3D SM SR reveals that the INVc is composed of fibrilloid substructures
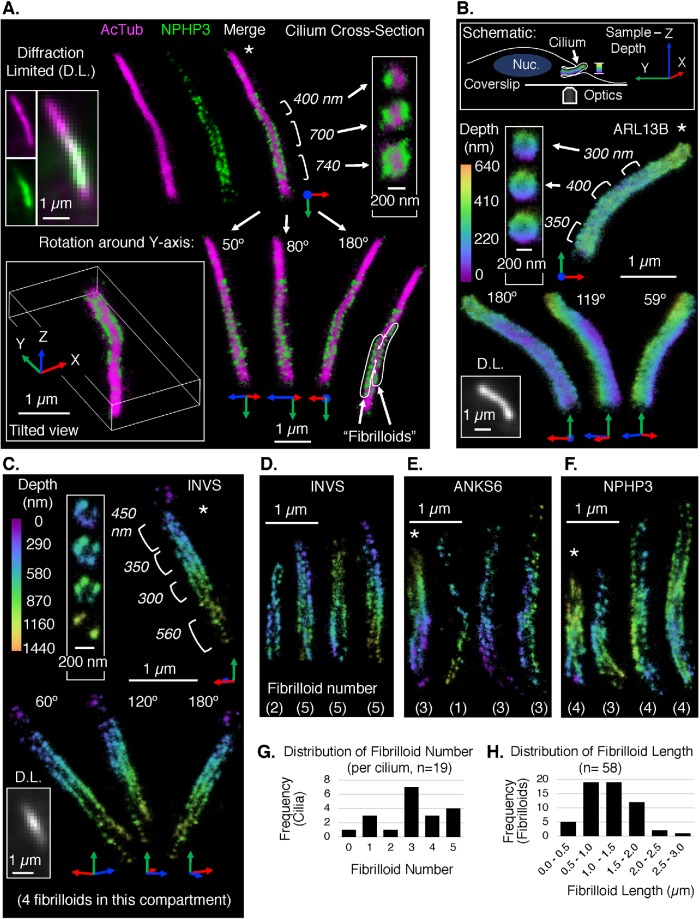
SM SR reconstructions of a whole dye-labeled structure can be created by localizing blinking dye molecules in many image frames. The effective resolution of the reconstruction exceeds the diffraction limit by computational localization of the position of each emitter with a precision that is finer than the diffraction-limited width of the point-spread function (PSF) of the emitter in the image. Conventional SM SR experiments result in 2D reconstructions containing localizations from a roughly 500 nm–deep axial section in the sample (approximately equal to the Z resolution of diffraction-limited microscopy). 3D SR methods provide far better resolution in the Z direction (von Diezmann et al., 2017). Here, we generated 3D SM SR reconstructions of cilia using a double-helix phase mask to transform the conventional PSF of each emitter into a bilobed PSF (Pavani et al., 2009; Yoon et al., 2019). The angle of the line between the lobes encodes the Z position of the emitter within the sample, allowing a reconstruction depth of approximately 2 µm, far thicker than the ∼500 nm axial range projected onto the plane in a conventional 2D image. Because RPE1 cilia tend to be oriented almost parallel to the coverslip (Figure 1A, top right) and the diameter of a cilium is at most 250 nm, the 2-µm region captured by our 3D SM SR approach was sufficient to measure whole cilia with high Z precision in the range ∼10–20 nm. Reconstruction statistics and labeling controls for our experiments are provided in Supplemental Figures S18–S20. This method was used to generate 3D SM SR reconstructions of the INVc and other cilia markers in IF-labeled RPE1 cells. Alexa Fluor (AF647) was used for labeling of all INVc proteins. For two-color 3D SM SR experiments, CF568 was used to label the axoneme with either anti-acetylated tubulin (Figure 2A and Supplemental Figure S21, A–C) or anti-polyglutamylated tubulin (Supplemental Figure S21D).
FIGURE 2:
3D SM SR reconstructions of the INVc and other ciliary markers. All localizations in the reconstructions are represented by 3D Gaussians with diameter 25 nm and an opacity set to show the localization density. Cilia are generally oriented tip up. Asterisks (*) indicate reconstructions for which supplemental movies and reconstruction statistics are provided (Supplemental Movies 1–4 and Supplemental Figures S18 and S19). (A) Two-color 3D SM SR reconstruction of a cilium immunolabeled with anti-NPHP3 to detect the INVc (AF647 secondary, green) and anti-acetylated tubulin to detect the axoneme (AcTub, CF568 secondary, magenta). Multiple views of the same cilium are shown, including diffraction-limited (D.L.) images of each channel separately and a merge (top left), SM SR reconstructions of each channel and a merge (top middle), rotated and tilted views of the merge (bottom), and cross-sections of three different regions of the INVc (top right). Cross-sections: the position and length of each region are annotated to the left of its corresponding cross-section. Rotated and tilted views: the XYZ-axes accompanying each reconstruction are defined by the position of the sample relative to the microscope, where the Z-axis represents sample depth as illustrated in the schematic in panel B. The “tilted view” is a tilted perspective viewing along the ciliary axis from ciliary tip (foremost) to base. The other three views (bottom right) are rotations of the reference image (connected by white arrows) around the Y-axis only. The degree of rotation is noted at the ciliary tip and is illustrated by the axes pinned to the ciliary base. The INVc (NPHP3) in this cilium appears to contain two linear substructures, “fibrilloids,” which are traced on a duplication of the 180° Y-rotation view (bottom right corner). (B–F) Single-color 3D SM SR reconstructions of ciliary proteins immunolabeled with specific primary antibodies and AF647-conjugated secondary antibodies. The color of each localization in the reconstruction maps to its Z-position in the sample. The depth range of the color scale is unique to each reconstruction. The 0 position (purple) is the edge of the reconstruction volume closest to the coverslip, not a fixed position relative to the sample stage. (B) Top: schematic defining the XYZ-axes of the sample, the coverage of the depth color scale used in single-color 3D SM SR reconstructions, and the relative orientations of the microscope objective (“Optics”), the coverslip, and an RPE1 cell with a cilium and nucleus (“Nuc.”) Objects are not drawn to scale. Bottom: 3D SM SR reconstruction of a cilium stained with anti-ARL13B to detect a ciliary protein independent of the INVc. Multiple views of the same cilium are shown (as in A): a D.L. image, cross-sections, and Y-axis rotations. (C) 3D SM SR reconstruction of a cilium stained with anti-INVS to detect the inversin compartment. Multiple views of the same cilium are shown (as in A and B): a D.L. image, cross-sections, and Y-axis rotations. The compartment in this reconstruction has four fibrilloids. (D–F) 3D SM SR reconstructions of multiple INVcs detected with either anti-INVS (D), anti-ANKS6 (E), or anti-NPHP3 (F). A single view of each cilium is shown (four cilia per panel, total 12 unique cilia in D–F). The number of fibrilloids counted in each reconstruction is noted at the bottom. (G) Distribution of the number of fibrilloids per cilium observed in 3D SM SR reconstructions of INVcs detected by immunolabeling for INVS, ANKS6, or NPHP3. Reconstructions of these INVcs are shown in panels C–F and Supplemental Figure S21, F–H. (H) Distribution of the lengths of individual fibrilloids measured in INVcs detected by INVS, ANKS6, or NPHP3 (as in G). From reconstructions of 18 compartments, a total of 58 fibrilloids were measured. The afibrilloid compartment in Supplemental Figure S21G is excluded from this graph.
Movie S1.
Two-color 3D SM SR reconstruction of a cilium where NPHP3 (green) and AcTub (magenta) are labeled.
Movie S2.
3D SM SR reconstruction of a cilium where ARL13B is labeled. Color corresponds to Z position in the sample.
Movie S3.
3D SM SR reconstruction of a cilium where INVS is labeled. Color corresponds to Z position in the sample.
Movie S4.
3D SM SR reconstruction of a cilium where ANKS6 is labeled. Color corresponds to Z position in the sample.
Movie S5.
3D SM SR reconstruction of a cilium where NPHP3 is labeled. Color corresponds to Z position in the sample.
Two-color 3D SM SR experiments labeling both the axoneme (AcTub, Figure 2A) and the INVc (NPHP3, Figure 2A) further confirmed that the INVc is a periaxonemal structure. Cross-sections of the reconstruction show that NPHP3 is excluded from the central acetylated tubulin-positive region (Figure 2A). NPHP3 was enriched in a subcompartment of the primary cilium, which, in the example shown (Figure 2A), occupied approximately 60% of the total ciliary length, starting approximately 300 nm from the base of the acetylated tubulin-positive axoneme and ending about 1 µm from the tip of the axoneme.
3D SM SR measurements of the axoneme demonstrate that the axoneme itself does not appear hollow for most of its length. A 3D SM SR reconstruction of an axoneme that is hollow at the ciliary base is provided in Supplemental Figure S21, A and B. While this is contrary to most conceptual models of the axoneme, which is frequently modeled as a hollow cylinder, these data are actually consistent with 3D ultrastructural studies using EM to measure the axonemes of mIMCD3 cells, in which a hollow center is only present at the base of the axoneme (the classic 9+0 arrangement reported for primary cilia) and most of the axoneme is composed of a more variable arrangement of the 9 doublets (9v arrangement), which eventually tapers into <9 singlets at the ciliary tip (Gluenz et al., 2010; Sun et al., 2019).
Consistent with the INVc asymmetries first observed by SIM, 3D SM SR reconstructions demonstrated that NPHP3 is not evenly distributed around the axoneme. The asymmetry in NPHP3 distribution in Figure 2A is most apparent in the two cross-sections from the distal end of the compartment, in which NPHP3 was enriched either in one cluster (top cross-section) or two distinct clusters on opposite sides of the axoneme (middle cross-section). In the four views of the compartment where the reconstruction has been rotated around the Y-axis, we observe that the two opposed clusters in the cross-section actually represent top-down views of two linear fibril-like arrangements of NPHP3 protein that run most of the length of the INVc. From here on, we refer to these linear arrangements as “fibrilloids,” to reflect uncertainty as to whether these linear structures are true fibrils, which would be continuous polymers, or linear collections of INVc proteins arranged by an alternative mechanism. The reconstruction in Figure 2A demonstrates how a fibrilloid-based INVc could appear in SIM reconstructions to be either symmetrically or asymmetrically distributed about the axoneme: the 0°- and 180°-rotated views of the reconstruction would have nearly symmetric distributions of NPHP3 along the axoneme; the 80° reconstruction measured by SIM, however, would have an asymmetric periaxonemal configuration in which the NPHP3 signal in the distal compartment would be enriched on the left side.
The ciliary protein ARL13B does not form fibrilloids
ARL13B was again imaged as a reference for an INVc-independent ciliary protein. Cilia in WT RPE1 cells were indirectly fluorescently labeled using a polyclonal primary antibody against ARL13B and an AF647-conjugated secondary antibody, and then imaged by SM SR using the same conditions and setup as for the INVc proteins. Multiple views of one ARL13B reconstruction are shown in Figure 2B (additional cilia in Supplemental Figure S21E). For 3D display, all single-color reconstructions are rendered so that the color of each localization corresponds to its Z position in the reconstruction space. This is diagrammed showing the relative positions of the sample and optical setup in the schematic in Figure 2B. The 3D reconstructions of ARL13B demonstrated that this protein was mostly excluded from the center of the cilium where the axoneme is positioned (Figure 2B, cross-sections). Furthermore, the diameter of the base of the ARL13B reconstruction was 300 nm, whereas the diameter at the base of the acetylated tubulin reconstruction (Figure 2A) was 180 nm, supporting the interpretation that ARL13B primarily localizes to the space around the axoneme. Importantly, within the periaxonemal region where ARL13B was enriched, ARL13B proteins were evenly distributed both around the axoneme (Figure 2B cross-section) and along the axoneme (Figure 2B shows rotated Y-axis views). Thus ARL13B forms a continuous sheath that is distinct from the fibrilloids observed in the INVc. ARL13B reconstructions typically contained 6× more localizations than INVc protein reconstructions. To confirm that the fibrilloid arrangement observed in INVc protein reconstructions was not an artifact of sparse reconstruction, we resampled an ARL13B localization data set and generated new reconstructions using a number of localizations typically contained in an INVS 3D SM SR reconstruction (Supplemental Figure S22). As expected, sparse reconstructions of ARL13B did not have a fibrilloid arrangement, but showed even distribution of ARL13B protein throughout the ciliary volume. This continuous structure observed for ARL13B is completely different from the distinctive fibrilloid substructures observed in 3D SM SR reconstructions of the INVc proteins (Figure 2A, NPHP3), supporting the conclusion that the measured INVc substructure is not an artifact and appears to be unique to the INVc.
INVS, ANKS6, and NPHP3 share a fibrilloid substructure
To determine how widely the fibrilloid substructure of the INVc was observed, we generated single-color 3D SM SR reconstructions of multiple cilia from WT RPE1 cells by IF with anti-INVS, anti-ANKS6, or anti-NPHP3 using AF647-conjugated secondary antibodies. GFP-NEK8 was not included in these experiments, as it was our lowest-density INVc marker. Multiple views of the INVc of one cilium detected by labeling of INVS are shown in Figure 2C. Similarly to NPHP3, measured in Figure 2A, cross-sections of this cilium reveal that INVS was not evenly distributed around the axoneme. The clusters around the axoneme when viewed in the cross-section correspond to fibrilloids. These are best seen in the side views of the reconstruction and in Supplemental Movie 3. Rotation of this reconstruction around the Y-axis demonstrates that this particular compartment would appear from any angle to be symmetrically arranged around the axoneme if the same compartment were measured by SIM. This particular compartment is composed of four fibrilloids. Measurements of additional cilia also labeled with INVS (Figure 2D and Supplemental Figure S21F) demonstrate that the number of fibrilloids varies from cilium to cilium, though the compartment was essentially always composed of fibrilloids. Reconstructions of ANKS6 (Figure 2E and Supplemental Figure S21G) tended to be sparser (fewer localizations), but suggested that ANKS6 also localizes to multiple fibrilloid structures in the INVc. Single-color measurements of NPHP3 (Figure 2F and Supplemental Figure S21H) demonstrated that NPHP3 also localizes to multiple fibrilloid structures with variable fibrilloid number.
INVc fibrilloids vary in number and length
The number of fibrilloids counted for each 3D SM SR reconstruction of INVS, ANKS6, and NPHP3 varied from cilium to cilium (Figure 2, C–F); all but one of the compartments measured appeared to have a fibrilloid substructure (Supplemental Figure S21G). Fibrilloid number ranged from one to five fibrilloids, with a mean of three fibrilloids per INVc. Given the relatively small number of cilia explored with 3D SM SR reconstructions, measurements of fibrilloid number from all single-color 3D reconstructions of INVS, ANKS6, and NPHP3 (19 cilia total) were combined to present the distribution of fibrilloid number per cilium measured in this study (Figure 2G). The lengths of individual fibrilloids varied from 0.5 to 3 µm, with a median length of 1.2 µm (Figure 2H). The longest compartments tended to have more fibrilloids (Supplemental Figure S21I); however, individual fibrilloid length did not correlate with total compartment length (Supplemental Figure S21J). As both SIM and 3D SM SR microscopy require well labeled structures for interpretable reconstructions, the INVc-positive cilia, which were chosen for either 3D SM SR or SIM reconstructions, tended to be among the brightest or most densely labeled compartments present in the sample. We note that this bias may select for long compartments, compartments with high fibrilloid number, and symmetric compartments in SIM measurements.
On the internal features of fibrilloids
The diameter of the compartment measured by 3D SM SR imaging, which corresponds to the largest distance observed between two fibrilloids on opposite sides of the axoneme, was approximately 200 nm (Figure 2, A and C). The full-width half-maximum (FWHM) diameter of individual fibrilloids was measured to be 40 ± 7 nm (mean ± SD) derived from five fibrilloid measurements per cilium for three cilia for each protein measured. We estimate that labeling with primary and secondary antibodies could be responsible for up to 35 nm of this width. Each fibrilloid appeared to be composed of a series of clusters. A single cluster contained 50–100 distinct localizations (corrected for potential overcounting; see Materials and Methods). These clusters were spaced 0–120 nm apart (measured from centroid to centroid) with intervening dark regions in which there were almost no localizations. It is unclear whether the dark regions represent true gaps in the structure where there were truly no immunolabeled INVc proteins, or whether they are a result of a spatial limitation of indirect immunolabeling using polyclonal antibodies. In the indirect immunolabeling protocols used to prepare samples in this study, a single INVc protein molecule was bound by an unknown number of primary antibodies, which were also bound by an unknown number of secondary antibodies. A single protein of interest (INVS, ANKS6, or NPHP3) is thus likely represented by more than one localization. It is also possible that these gaps represent a fixation artifact that fragments what would otherwise be continuous INVc fibrils. Of note, the structural gaps detected in the 3D SM SR reconstructions of the INVc do not seem to correspond to the spacings observed in SIM reconstructions of the same proteins. Gaps in the 3D SM SR reconstructions were typically much smaller (40 nm) than what we expect to be resolved by SIM (100 nm). Additionally, no gaps were observed in 3D SM SR reconstructions of ARL13B or polyglutamylated tubulin (Supplemental Figure S21D), whereas SIM reconstructions of both ARL13B and polyglutamylated tubulin (Supplemental Figure S15B) featured a regular spacing, or granular appearance, similar to that observed for INVc proteins also imaged by SIM. On the basis of these observations, we conclude that the spacing observed in SIM reconstructions of INVc proteins (Figure 1D) may not represent true distributions of INVc proteins in the compartment, but might rather be a consequence of the SIM imaging and reconstruction technique.
Genetic dissection of INVc assembly by CRISPR editing of all four INVc genes
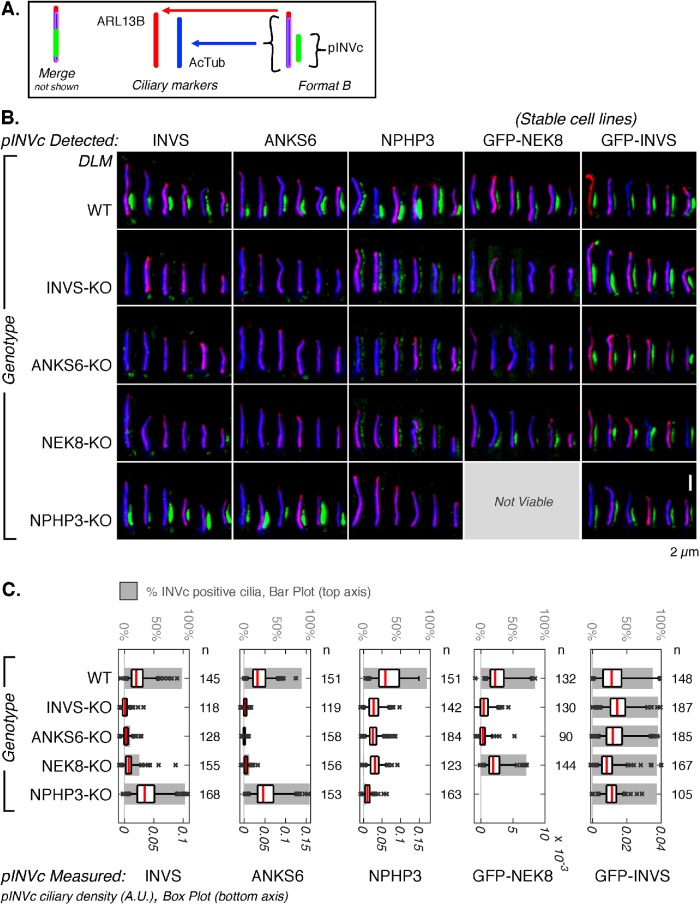
To determine the functional relationships between the four INVc proteins, we generated knockout (KO) cell lines of each INVc gene (gINVc) using guide RNA–directed CRISPR gene editing to introduce frame shift mutations in the INVS, NEK8, ANKS6, or NPHP3 genes in an RPE1 cell line stably expressing C-terminally BFP-tagged Cas9 nuclease (Kanie et al., 2017; Supplemental Figures S23–S26). We first determined that ciliation frequency and cilia length were not affected by loss of INVS, NEK8, ANKS6, or NPHP3 (Supplemental Figure S27). Next, we assessed the effect of loss of each gene on the formation and composition of the INVc. Ciliary localization of endogenous INVS, ANKS6, and NPHP3 detected by IF was measured by DLM in unedited WT RPE1 cell lines, and gINVc-KO cell lines (Figure 3B; quantified in Figure 3C and Supplemental Figure S28). Ciliary localization of NEK8 was measured by GFP fluorescence in WT and gINVc-KO cell lines stably expressing GFP-NEK8 (Figure 3B; expression of the GFP fusion protein is confirmed by immunoblot in Supplemental Figure S29B), which fully rescued NEK8-KO cells (Supplemental Figure S30). Ciliary localization of GFP-INVS was also measured by GFP fluorescence in WT and INVc-KO cell lines stably expressing GFP-INVS (Figure 3C; expression confirmed by immunoblot Supplemental Figure S29A), which fully rescued INVS-KO cells (Supplemental Figure S31).
FIGURE 3:
INVS is the primary organizer of the INVc. (A) Schematic illustrating the coloring and relative positioning of fluorescent markers in the images in B. (B) DLM images of cilia in which endogenous INVS, ANKS6, and NPHP3 were detected by IF (green, left three columns) in WT RPE1 cells and INVS-KO, ANKS6-KO, NEK8-KO, and NPHP3-KO cell lines. Each genotype is represented by a single row. GFP fluorescence measured by DLM in WT RPE1 cells and INVS-KO, ANKS6-KO, NEK8-KO, and NPHP3-KO cell lines stably expressing GFP-NEK8 or GFP-INVS (green, right two columns). In all samples, ARL13B (red) and AcTub (blue) were detected by IF to mark cilia. Six representative cilia are shown in each subpanel. Additional images are provided in Supplemental Figures S23–S25, S30, and S31. (C) Quantification of ciliary density (arbitrary units, A.U.) and INVc frequency for fluorescent INVc markers (anti-INVS, anti-ANKS6, anti-NPHP3, GFP-NEK8, and GFP-INVS, all shown in green in B) measured in WT and gINVc-KO cell lines. Ciliary area was masked using the ARL13B and AcTub channels. Ciliary densities measured for each sample are plotted as a box plot (bottom axis) on top of a gray bar representing the percentage of cilia that contained an INVc (top axis). The number of cilia (n) measured for each condition (sum of two replicates) is reported to the right of each box/bar. See Materials and Methods for a description of the statistics represented by these box plots. Supplemental Figure S28 reports the results of pairwise t tests performed for each sample represented in this panel.
ANKS6, NEK8, and NPHP3 fail to localize to the INVc in INVS-KO cells
Consistent with previously published results, we found that localization of ANKS6 (Hoff et al., 2013), NEK8 (Shiba et al., 2010), and NPHP3 (Shiba et al., 2010) all depend on INVS, as neither endogenous ANKS6, NPHP3, nor GFP-NEK8 was found in the proximal cilia of INVS-KO cells (Figure 3, B and C). Localization of ANKS6 and NPHP3 was restored in INVS-KO cells by stable expression of GFP-INVS (Figure 4, A, B, D, and E, and Supplemental Figure S31). The ciliary densities of endogenous ANKS6 (Figure 4C and Supplemental Figures S31, A, C, and D, and S32) or NPHP3 (Figure 4, D and E, and Supplemental Figures S31, B–D, S32F, S33, and S34B) were rescued to densities comparable to those observed in WT cells. Similarly to WT cells stably expressing GFP-INVS, INVS-KO cells rescued with GFP-INVS had compartments longer than endogenous INVcs (Supplemental Figure S31D). The expression of GFP-NEK8 had no effect on INVc lengths (Supplemental Figure S30F). In contrast to GFP-NEK8 and ANKS6, which were not detected in INVS-KO cilia, NPHP3 still localized to INVS-KO cilia, albeit with substantial deficit in NPHP3 protein density and subciliary localization. In INVS-KO cells, NPHP3 was present at significantly reduced levels compared with those in WT cilia (Figure 3C, Supplemental Figure S28C, Figure 4F, and Supplemental Figure S34B) and localized along the full length of the cilium rather than being concentrated in the proximal half of the cilium, as it is when localized to the INVc in WT cells. Overall, although INVS appears most important for INVc assembly, INVS, ANKS6, and NEK8 appear to cooperate to establish the INVc and to sequester NPHP3 within the compartment.
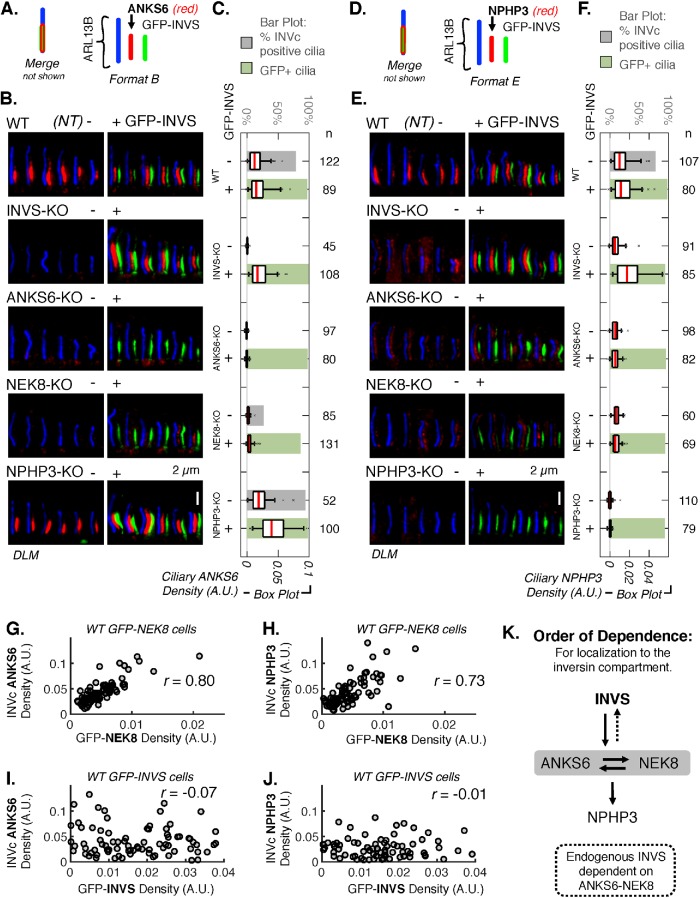
FIGURE 4:
ANKS6 and NEK8 link NPHP3 to INVS. (A) Schematic illustrating the coloring and relative positioning of fluorescent markers in the images in B. (B) DLM of ANKS6 detected by IF (AF568 secondary, red) in WT, INVS-KO, ANKS6-KO, NEK8-KO, and NPHP3-KO cell lines stably expressing GFP-INVS. GFP-INVS was detected by native GFP fluorescence (green). ARL13B (AF647 secondary, blue) marks the whole cilium. Additional images are provided in Supplemental Figure S32. (C) Mean ANKS6 density, measured as mean pixel intensity of the red channel in the ciliary region defined by ARL13B (arbitrary units, A.U.) in WT, INVS-KO, ANKS6-KO, NEK8-KO, and NPHP3-KO cell lines either not transduced (NT,-) or stably expressing GFP-INVS (+). Number of cilia (n) measured for each condition is noted to the right. See Supplemental Figure S34A for results of pairwise t tests performed for each sample represented in these plots. Supplemental Figure S32F provides INVc-specific density measurements. (D) Schematic illustrating the coloring and relative positioning of fluorescent markers in the images in E. (E) DLM of NPHP3 detected by IF (AF568 secondary, red) in WT, INVS-KO, ANKS6-KO, NEK8-KO, and NPHP3-KO cell lines stably expressing GFP-INVS. GFP-INVS detected by native GFP fluorescence (green). ARL13B (AF647 secondary, blue) marks the whole cilium. Additional images are provided in Supplemental Figure S33. (F) Mean ciliary intensity of NPHP3 in WT, INVS-KO, ANKS6-KO, NEK8-KO, and NPHP3-KO cell lines stably expressing GFP-INVS. Number of cilia (n) measured for each sample noted. See Supplemental Figure S34B for results of pairwise t tests performed for each sample represented in these plots. Supplemental Figure S32F provides INVc-specific density measurements. (G–J) Scatterplots and pairwise correlations of GFP-NEK8 or GFP-INVS (X-axis) with ANKS6 and NPHP3 (Y-axis) in WT RPE1 cells stably expressing GFP-NEK8 or GFP-INVS (see Supplemental Figure S36 for similar measurements for NEK8-KO cells rescued with GFP-NEK8 and INVS-KO cells rescued with GFP-INVS). GFP-NEK8 and GFP-INVS were detected directly by native GFP fluorescence and endogenous ANKS6 and NPHP3 were detected by indirect IF with AF568 secondary (as in B and E). The densities of the GFP-alleles and endogenous proteins within the INVc were measured as the mean pixel intensity in arbitrary units (A.U.) within a user-defined subcompartment mask drawn around the GFP and/or AF568-positive region of each cilium. (G) Correlation between ANKS6 and GFP-NEK8, n = 79, Pearson’s r value: r = 0.804 (p = 4.81 × 10–19). (H) Correlation between NPHP3 and GFP-NEK8, n = 79, Pearson’s r value: r = 0.726 (p = 2.62 × 10–15). (I) Correlation between ANKS6 and GFP-INVS, n = 86, Pearson’s r value: r = -0.0743 (p = 0.497). (J) Correlation between NPHP3 and GFP-INVS, n = 78, Pearson’s r value: r = -0.0139 (p = 0.904). (K) Diagram of hierarchical order of dependence for the localization of INVS, ANKS6, NEK8, and NPHP3 in the INVc, determined by IF of each INVc protein in gINVc-KO cells and gINVc-KO cells rescued with GFP-INVS.
ANKS6 and NEK8 are required for endogenous INVc assembly in RPE1 cells
It was previously shown that ANKS6 fails to localize to the INVc in NEK8-deficient cilia (Czarnecki et al., 2015; Grampa et al., 2016). Here we show that NEK8 localization to the INVc depends on ANKS6, as GFP-NEK8 failed to localize to the INVc in ANKS6-KO cell lines (Figure 3B). While it has previously been shown that INVS regulated the localization of ANKS6 (Hoff et al., 2013; Nakajima et al., 2018), NEK8 (Shiba et al., 2010; Nakajima et al., 2018), and NPHP3 (Shiba et al., 2010; Nakajima et al., 2018), we were surprised to find that INVcs detected by an antibody to the endogenous INVS protein were reduced in frequency and density in both NEK8-KO and ANKS6-KO cells (Figure 3B, far left). Thus, ANKS6 and NEK8 regulate endogenous INVS in RPE1 cells. Contrary to the behavior observed for endogenous INVS, we found that GFP-INVS formed INVcs efficiently when stably expressed in ANKS6-KO cells and NEK8-KO cells (Figure 3B, far right). Thus, ANKS6 and NEK8 do not regulate the localization of stably expressed GFP-tagged INVS.
Consistent with a reduction in ciliary localization of endogenous INVS, ANKS6-KO and NEK8-KO cells phenocopied INVS-KO cells, exhibiting the striking NPHP3 relocalization described above. Overall levels of ciliary NPHP3 were also reduced in both NEK8-KO and ANKS6-KO cells (quantified in Figure 3C). Localization of INVS, ANKS6, and NPHP3 was rescued by stable expression of GFP-NEK8 in NEK8-KO cells (Supplemental Figure S30, A and B). Stable overexpression of GFP-NEK8 did not appear to affect the range of protein densities of endogenous ANKS6, NPHP3, or INVS compared with those in WT cells (Figure 3C), nor did it affect INVc length (Supplemental Figure S30F).
INVS and ANKS6 still localize to the INVc in NPHP3-KO cells
Localization of endogenous INVS and ANKS6 were not affected in NPHP3-KO cells (Figure 3B). Localization of GFP-NEK8 in NPHP3-KO cells was not assessed, as we were unable to generate an NPHP3-KO cell line stably expressing GFP-NEK8, suggesting that NPHP3 may regulate NEK8 at some step preceding compartment assembly at which the constitutively expressed GFP-fusion protein is especially sensitive. On the basis of our observations that endogenous INVS and ANKS6 require NEK8 to assemble in the INVc, and that the localization of these two proteins are unaffected in NPHP3-KO cells, we expect that endogenous NEK8 also does not depend on NPHP3 for localization.
INVS localizes ANKS6 in the INVc, but ANKS6 density is regulated by NEK8
Although loss of ANKS6 or NEK8 by CRISPR gene editing nearly eliminated INVcs defined by the endogenous INVS protein (Figure 3B), forced expression of GFP-INVS in NEK8-KO cells led to the robust formation of GFP-INVS-positive compartments, referred to as GFP-INVSNEK8–KO compartments, in a high proportion of cells (>90%; Figures 3C and 4, C and F). To determine whether ANKS6 or NEK8 is more upstream in the INVc assembly pathway, we stained for endogenous ANKS6 protein in NEK8-KO cells expressing GFP-INVS. Some ANKS6 protein was detectable in GFP-INVSNEK8–KO compartments (Figure 4B); however, ANKS6 density was substantially decreased in GFP-INVSNEK8–KO cilia compared with cilia of WT cells (Figure 4C and Supplemental Figure S34A). To more closely examine ANKS6 localization in GFP-INVSNEK8–KO compartments, we measured the localization of GFP-INVS and ANKS6 by SIM (Supplemental Figure S35). In NEK8-KO cells, GFP-INVS does in fact effectively sequester ANKS6 in the compartment, as ANKS6 was found only in the region of the cilium that was also positive for GFP-INVS (Supplemental Figure S35D). The failure of GFP-INVS to concentrate ANKS6 in NEK8-KO cells is not a defect of the GFP-INVS allele, as GFP-INVS rescues ANKS6 to WT density levels when expressed in INVS-KO cells (Supplemental Figure S23E). This result suggests that ANKS6 physically associates with INVS independent of NEK8, and that NEK8 and INVS regulate distinct aspects of ANKS6 assembly in the INVc: INVS regulates ANKS6 compartmentalization (localization), whereas NEK8 regulates ANKS6 density within the INVc (concentration).
ANKS6 and NPHP3 protein densities in the INVc correlate with GFP-NEK8, but not with GFP-INVS
To further evaluate how combinations of INVc proteins might influence their respective distributions within the same compartment, we measured the correlation between INVc specific densities of endogenous ANKS6 and NPHP3 and either GFP-NEK8 or GFP-INVS (Figure 4, G–J, and Supplemental Figure S36). ANKS6 and GFP-NEK8 protein densities are strongly correlated with one another (Figure 4G and Supplemental Figure S36A), indicating that these two proteins assemble in the INVc as stoichiometric complexes. In contrast, ANKS6 density does not correlate with GFP-INVS density (Figure 4I and Supplemental Figure S36C), suggesting that, although INVS is required for ANKS6 localization to the INVc, the number of INVS proteins in an INVc is not a good predictor for the number of ANKS6 proteins. NPHP3 is moderately correlated with GFP-NEK8 (Figure 4H and Supplemental Figure S36B) and not correlated with GFP-INVS (Figure 4J and Supplemental Figure S36D), supporting a model in which NPHP3 interacts more directly with NEK8 than INVS and suggesting that the interaction between NEK8 and NPHP3 occurs with some predictive stoichiometry. We also assessed the localization of the first 200 amino acids of NPHP3 tagged with GFP (NPHP3(1-200)-GFP) relative to endogenous INVS by SIM. NPHP3(1-200)-GFP fails to be sequestered in the INVc (Supplemental Figure S37), suggesting that the C-terminus of NPHP3 is important for its interaction with the ANKS6-NEK8 complex.
Both ANKS6 and NEK8 are required downstream of INVS for localization and concentration of NPHP3 in the INVc
We were surprised to find that GFP-INVS does not rescue localization of endogenous NPHP3 to the INVc in either NEK8-KO cells or ANKS6-KO cells (Figure 4E). In both GFP-INVSNEK8–KO and GFP-INVSANKS6–KO compartments, NPHP3 protein was not only reduced in density (quantified in Figure 4F and Supplemental Figure S34), but was also redistributed from the INVc to the whole length of the cilium, as in INVS-KO cells (Figure 5; additional reconstructions provided in Supplemental Figures S38–S44). Ciliary localization of NPHP3 had previously been established to depend on a myristoylated protein trafficking pathway involving UNC119, ARL3, RP2,and ARL13B (Wright et al., 2011). The residual presence of some NPHP3 in either INVS-KO or otherwise INVc-deficient cilia suggests that the myristoylated protein trafficking pathway does not act downstream of the INVc, but rather first establishes a ciliary pool of NPHP3 protein, which is then sequestered and concentrated in the INVc by ANKS6 and NEK8. Because of these two regulatory pathways, NPHP3 is capable of two distinct ciliary localization patterns.
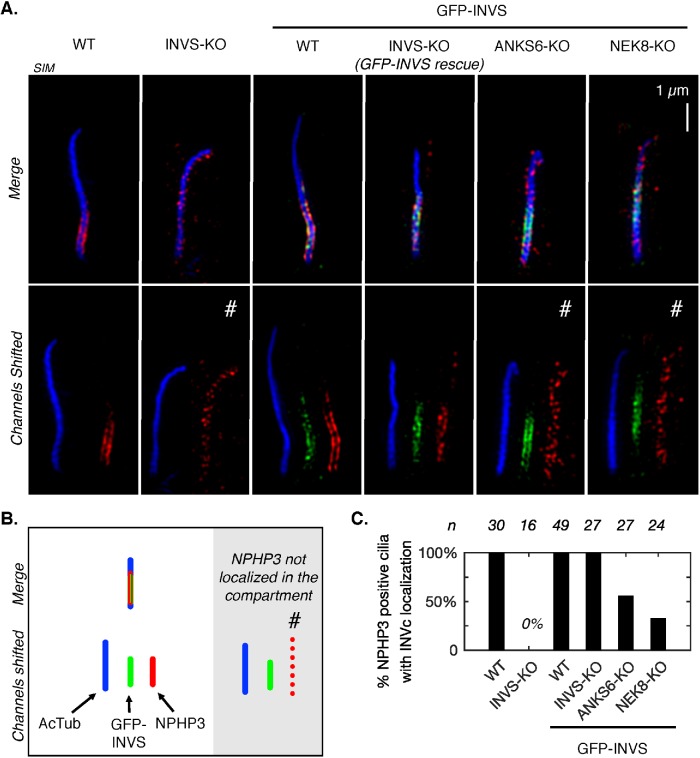
FIGURE 5:
SIM of two ciliary localization patterns of NPHP3. (A) SIM IF of NPHP3 (red, AF568 secondary) in a RPE1 cell lines of relevant genotypes. One cilium per genotype is shown in each column (left to right): WT and INVS-KO cell lines; WT, INVS-KO, ANKS6-KO, and NEK8-KO cell lines stably expressing GFP-INVS (green, detected with anti-GFP primary antibody, AF488 secondary). Acetylated tubulin (AcTub, AF647 secondary, blue) marks the axoneme. Top row: three-color merges of NPHP3, GFP-INVS, and AcTub. Bottom row: each color is shifted to show each channel individually, left to right: AcTub, GFP-INVS, and NPHP3. Pound symbols (#) mark cilia in which NPHP3 is not localized in the INVc. Additional reconstructions are provided in Supplemental Figures S38–S44. (B) Schematic illustrating the coloring and relative positioning of fluorescent markers in the images in A. (C) Frequency of cilia in which NPHP3 is localized within the INVc for each cell line, assessed by SIM reconstructions as shown in A and Supplemental Figures S38–S44. Only NPHP3-positive cilia are included. For GFP-INVS cell lines, only cilia which were positive for both NPHP3 and GFP are included.
In the absence of NEK8, GFP-INVS and ANKS6 failed to sequester NPHP3 within the INVc in at least 77% of the cilia assessed by SIM (Figure 5, A and C, and Supplemental Figure S42). In ANKS6-KO cells expressing GFP INVS, NPHP3 was similarly mislocalized in at least 44% of cilia (Figure 5, A and C, and Supplemental Figure S43). These numbers are conservative estimates, as the substantial number of cilia still reported as having INVc-NPHP3 localization are likely explained by cases in which INVc extends all the way to the tip of the cilium (arrows, Supplemental Figures S42 and S43). In these cases, we interpret NPHP3 localization as in the compartment. Together, the deficit in NPHP3 concentration and frequent mislocalization in GFP-INVSANKS6–KO and GFP-INVSNEK8–KO compartments support a model in which NPHP3 is physically uncoupled from the INVc in the absence of either ANKS6 or NEK8.
Previous studies suggested that INVS and NPHP3 physically interact with one another (Bergmann et al., 2008; Sang et al., 2011) and that NPHP3 localization to the INVc is dependent on INVS (Shiba et al., 2010). Our data suggest that localization of NPHP3 to the INVc also requires both ANKS6 and NEK8. Interactions between NPHP3 and NEK8 and ANKS6 have also been shown by coimmunoprecipitation (Hoff et al., 2013; Czarnecki et al., 2015). Intriguingly, the interaction between NEK8 and NPHP3 was shown to be enhanced by coexpression of ANKS6 (Hoff et al., 2013). Our data showing that NPHP3 is mislocalized and reduced in density to a similar extent in both GFP-INVSANKS6–KO and GFP-INVSNEK8–KO compartments suggest either that NPHP3 interacts directly with NEK8, or that NEK8 is required in some way for ANKS6 to bind NPHP3 in the INVc. If NPHP3 interacts directly with NEK8, we can infer that NEK8 is not present in GFP-INVSANKS6–KO compartments where NPHP3 is not sequestered in the compartment. In fact, direct interaction between NPHP3 and ANKS6 would require or be enhanced by NEK8, as GFP-INVSNEK8–KO compartments contain a reduced but detectable amount of ANKS6 protein, which is insufficient to sequester NPHP3 in the compartment (SIM reconstructions of ANKS6 in gINVc-KO cell lines expressing GFP-INVS are provided in Supplemental Figures S45–S49 and summarized in Supplemental Figure S35).
While INVS is undoubtedly required for NPHP3 to localize in the compartment, it is unclear whether NPHP3 and INVS actually interact physically within the compartment. The requirement of ANKS6 and NEK8 suggests that these proteins could bridge an indirect interaction between INVS and NPHP3. This model is supported by the stronger correlation observed between GFP-NEK8 and NPHP3 (Figure 4H) than GFP-INVS and NPHP3 (Figure 4J), suggesting, as in the case of GFP-NEK8 and ANKS6, that NEK8 (or possibly NEK8 and ANKS6 together) stoichiometrically regulates the amount of NPHP3 concentrated in the INVc.
Kinase-inactivating and JCK mutations in NEK8 do not affect INVc assembly
An attractive mechanism by which NEK8 might facilitate interaction between ANKS6 and NPHP3 might involve NEK8 establishing activating phosphorylation on either protein. However, we find that NEK8 kinase activity is dispensable for NPHP3 concentration in the INVc, as the K33M (kinase-inactivating) mutation in NEK8 rescues both ANKS6 and NPHP3 INVc localization to WT-like densities in NEK8-KO cells (Supplemental Figures S50 and S51). An interaction between NEK8 and NPHP3, or a NEK8-dependent interaction between ANKS6 and NPHP3, is also not affected by the JCK (juvenile cystic kidney) mutation in the RCC1 domain of NEK8 (G448V), as JCK-NEK8 also completely rescues NPHP3 localization in the INVc in NEK8-KO cells (Supplemental Figure S51). GFP-NEK8-K33M and JCK INVcs were also measured by SIM to confirm that these mutations in NEK8 did not affect ANKS6 and NPHP3 sequestration (summarized in Supplemental Figure S52; additional reconstructions provided in Supplemental Figures S53–S58). On the basis of our measurements of the kinase-dead NEK8 allele, we conclude that kinase activity of NEK8 is not required to assemble the INVc, though it may be important for compartment function.
DISCUSSION
A new model for the structure and composition of the INVc
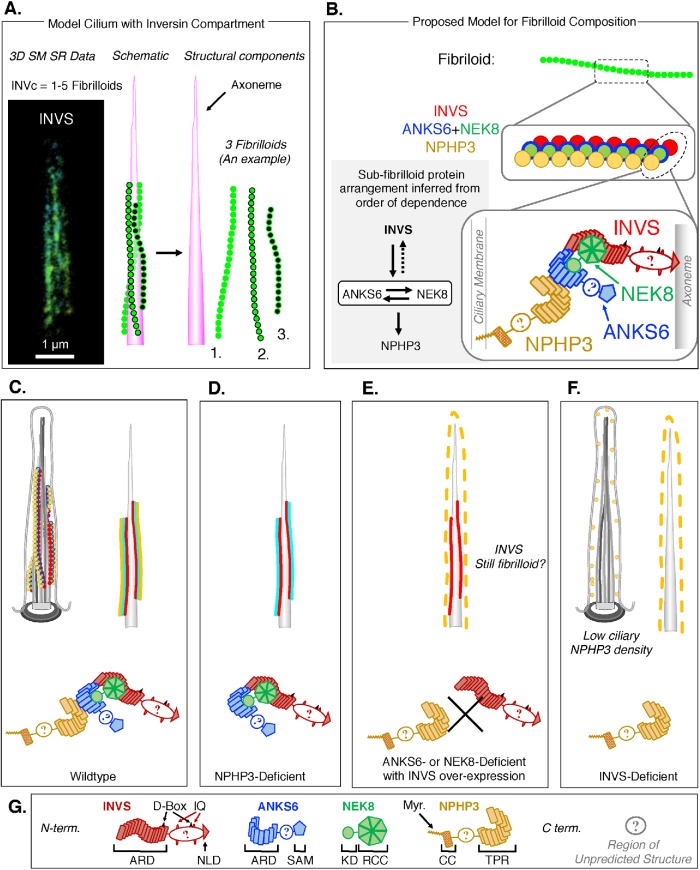
On the basis of the INVc measurements presented here, we propose a new model for the structure and composition of the INVc. 3D SM SR imaging revealed that the INVc is composed of a novel fibrilloid substructure (Figures 2 and 6A). From our genetic dissection of the compartment (Figures 3 and 4), we conclude that a complete INVc containing INVS, ANKS6, NEK8, and NPHP3, as observed in WT RPE1 cells, is assembled by three epistatic relationships: 1) INVS binds ANKS6-NEK8 complexes; 2) NEK8 plays an important role in establishing a high density of ANKS6-NEK8 complexes within the compartment (Figure 4), though it is dispensable for tethering ANKS6 to INVS (Supplemental Figure S35); and finally 3) high-density ANKS6-NEK8 complexes localize and concentrate NPHP3 in the INVc (Figures 4 and 5). These three relationships determine the final composition of the INVc and therefore also the composition of fibrilloids (Figure 6B).
FIGURE 6:
A new model for the structure and composition of the INVc. (A) 3D SM SR reconstructions of INVS (left) suggest that the INVc is composed of fibrilloid substructures. Based on these data, a schematic of a cilium with an INVc, which is composed in this example of three fibrilloids (green) that are aligned roughly parallel along the axoneme (magenta), is shown. (B–G) Illustrations of the structure and composition of the INVc at various levels of detail and in different genetic contexts (C–F). Each INVc protein is shown in following colors: INVS (red), ANKS6 (blue), NEK8 (green), and NPHP3 (yellow). In the highest-resolution illustrations, each INVc protein is illustrated with some structural details pertaining to predicted protein domains within each molecule (defined in G). (B) A possible model for fibrilloid composition based on the order of dependence for the assembly of the four INVc proteins in the compartment. The 1:1:1:1 stoichiometry for all INVc proteins shown here is symbolic, and the relative positions of the circular molecules in the middle-resolution illustration are intentionally ambiguous as to whether each fibrilloid is a heteropolymer of all or some INVc proteins or a homopolymer, consisting for example of INVS only, on which ANKS6-NEK8 and NPHP3 then assemble (see Supplemental Figure S59 for candidate polymer models). In the highest-resolution illustrations (bottom), NPHP3 associates directly with the ciliary membrane via its N-terminal myristoylation, and ANKS6–NEK8 complexes link NPHP3 to INVS. (C) Illustrations of a cilium in a WT cell in which the INVc is a fibrilloid structure composed of INVS, ANKS6, NEK8, and NPHP3. Three fibrilloids are shown in the more detailed illustration (top left). Two fibrilloids are shown in the simplified illustration (top right), where ANKS6 and NEK8 are together represented in cyan. INVS is the core organizer and is illustrated here as being more directly associated with the axoneme. (D) A cilium in an NPHP3-deficient cell. The INVc is still present and structurally unaffected by loss of NPHP3. (E) A cilium in an ANKS6- or NEK8-deficient cell in which GFP-INVS expression has rescued compartment formation. The INVc contains INVS, which we expect still forms fibrilloids. Ciliary NPHP3 density is reduced, and NPHP3 is not sequestered in the INVc, but rather relocalized throughout the ciliary volume. The failure of INVS to sequester NPHP3 is due to the loss of the ANKS6–NEK8 complex that is predicted to link these two proteins physically, based on our measurements of NPHP3 localization in ANKS6-KO or NEK8-KO cells stably expressing GFP-INVS. (F) A cilium in an INVS knockout cell. No INVc is present. NPHP3 is reduced and found throughout the cilium, albeit at a low density, similar to that observed for ANKS6- or NEK8-deficient cells. (G) Schematic of the protein domains and motifs present in the four INVc proteins as cartooned in panels B–F. All four proteins are oriented left to right from N- to C-terminus. The INVS protein contains an ankyrin repeat domain (ARD) consisting of 16 ankyrin repeats, two D-boxes (known to bind APC; Morgan et al., 2002), a bipartite NLS (Otto et al., 2003), two IQ domains (known to bind calmodulin; Yasuhiko et al., 2001), and a C-terminal ninein-like domain (NLD) that is important for centriolar targeting of INVS (Shiba et al., 2009) ANKS6 contains an N-terminal ARD with 11 ankyrin repeats, followed by a serine-rich patch and a C-terminal SAM domain. The ANKS6 ARD likely mediates an interaction with NEK8 through the N-terminal NEK8 kinase domain (KD; Czarnecki et al., 2015). NEK8 contains a C-terminal RCC (regulator of chromatin condensation) domain, which is predicted to fold into a propeller structure and is important for mediating an interaction of NEK8 and INVS (Zalli et al., 2012; Czarnecki et al., 2015). The N-terminus of NPHP3 is myristoylated (Myr.), which could insert in the ciliary membrane (Wright et al., 2011; Nakata et al., 2012; as illustrated in panel B). NPHP3 also contains a coiled-coil domain (amino acids 83–207), which may be important for targeting NPHP3 to the basal body (Nakata et al., 2012). The C-terminus of the NPHP3 contains at least 11 tetratricopeptide repeats (TPR). The first 200 amino acids of NPHP3 localize to the cilium (Wright et al., 2011) but fail to be sequestered in the INVc (Supplemental Figure S37), suggesting that the interaction between NPHP3 and ANKS6 or NEK8 is either through the TPR repeats or through some other domain present in the C-terminus of the protein (amino acids 201–1330).
The lengths of GFP-INVS–positive compartments in ANKS6-KO, NEK8-KO, and NPHP3-KO cells are similar to those in WT and INVS-KO cells expressing GFP-INVS (Figure 3B; quantified in Supplemental Figure S27C). As we have so far only detected fibrilloid compartment substructures, we expect that an INVc of WT length and density must be built of fibrilloids. Thus we conclude that INVS is likely competent to form fibrilloids independent of NEK8, ANKS6, or NPHP3, favoring a model in which INVS is the major determinant of INVc length. This model is further supported by the increase in compartment length driven by an increase in ciliary INVS in cells expressing GFP-INVS compared with endogenous INVS (Figure 1, E and F, and Supplemental Figure 11C).
The relationships governing INVc composition are illustrated in Figure 6, where an ANKS6-NEK8 complex physically links INVS to NPHP3 in WT cells (Figure 6, B and C). Loss of NPHP3 does not interfere with the binding of the ANKS6-NEK8 complex to INVS (Figure 6D). Loss of ANKS6 or NEK8 uncouples NPHP3 from INVS, though some NPHP3 is still found in the cilium (Figure 6E), similarly to its distribution in INVS-KO cells (Figure 6F). In this model the stoichiometry of the NEK8-ANKS6 complex is represented as 1:1, based on efficient copurification of these factors and mass spectrometry data (unpublished data), though the exact ratio has yet to be determined. INVS is shown associating more closely with the axoneme, as this protein has been shown previously to bind to microtubules (Nürnberger et al., 2004). NPHP3 is shown anchored to the ciliary membrane via myristoylation of the N-terminus of this protein which is important for its localization to cilia (Wright et al., 2011; Nakata et al., 2012; Nakajima et al., 2018).
New insights into the morphological variation of the INVc revealed by a fibrilloid model
At the onset of this study, the variation in compartment length measured here and previously reported (Shiba et al., 2009; Sang et al., 2011) and the variation in compartment density were intriguing morphological properties of the INVc. A fibrilloid substructure provides a structural basis for variation in both INVc length and density. In a fibrilloid model, a combination of individual fibrilloid lengths and how these structures are arranged in parallel (ends aligned in denser and shorter compartments or ends offset to establish extended compartments) could establish compartments of different lengths (Supplemental Figure S59A). The number of fibrilloids and how tightly they pack next to each other could build compartments of varying density (Supplemental Figure S59, B and C). Compartment density could also be influenced by the density of individual fibrilloids (Supplemental Figure S59D). These models provide a mechanism by which INVc length (fibrilloid length) and INVc protein density (fibrilloid number) are established independently. Owing to the structural (dye molecules displaced from the protein of interest by the length of up to two antibodies) and stoichiometric properties (INVc protein:1° antibody:2° antibody) of indirect immunolabeling, it is unclear how precisely our reconstructions capture fibrilloid substructure. Notably, we cannot determine whether the gapped clustered appearance of fibrilloids in 3D SM SR reconstructions (60-nm clusters separated by 40-nm gaps) truly represents underlying clusters of spaced proteins organized by an unidentified ciliary factor (Supplemental Figure S59D, model II), or rather a continuous structure (Supplemental Figure S59D, model I) that appears discontinuous only in the reconstructions.
We measured compartments in fixed cells only. Live-cell experiments recording INVS fluorescent fusion proteins suggest that, once assembled, the length of the INVc does not vary over a period of up to 90 min (Schaub and Stearns, 2013) and that GFP-INVS recovers from photobleaching of a 250–500–nm segment of the compartment within 60 s (Shiba et al., 2009). Together these results suggest that, while total INVc length is not a dynamic property on short (minute to hour) time scales in unperturbed cilia, individual INVS molecules do not occupy fixed positions, but are instead dynamically exchanging at a time scale on the order of seconds. The rate of recovery from photobleaching suggests an intracompartment mobility of 14 nm/s, which is much slower than either diffusion of proteins in the membrane or IFT. Intracompartmental INVS protein dynamics is therefore distinct from the other known modes of protein transport in the primary cilium. Notably, fast positional turnover for INVS suggests that the fibrilloids may not be stable, long-lived structures but either dynamic arrangements, which may assemble and disassemble within minutes, or an unidentified structure upon which INVS complexes dynamically assemble and disassemble.
Known structural features of INVc proteins and possible contributions to INVc composition
Generally, the structural features present in the four INVc proteins (diagrammed in Figure 6G) do not currently inform a mechanism by which these proteins would assemble a fibril-like structure. The model we propose for INVc composition at the subfibrilloid level (Figure 6B) relies on several INVc protein interaction features published by other groups.
The SAM domain of ANKS6 is the only established oligomerization domain present in an INVc protein. Although some SAM domains have been shown to form extensive oligomers in vitro (Leettola et al., 2014), it is unclear whether an ANKS6-based SAM polymer provides the structural basis of the INVc because the ANKS6 SAM domain fails to polymerize in vitro (Leettola et al., 2014), ANKS6 proteins deleted in the SAM domain still localize to the INVc (Czarnecki et al., 2015), and we show here that GFP-INVS still forms an INVc in ANKS6-KO cells.
Reciprocal pull downs of two different exogenously expressed INVS fusion proteins suggest that INVS proteins can self-associate (Suizu et al., 2016). This raises the intriguing possibility that INVS may form oligomers. Future structural and biochemical studies of INVS may determine whether INVS can polymerize outside the axoneme, although the fibrilloid structures that we observed by 3D SM SR microscopy may require both the axoneme and the ciliary membrane, and possibly some additional unidentified ciliary factor.
Like INVS, NEK8 has been shown to self-associate by reciprocal pull downs of NEK8 fusion proteins (Choi et al., 2013). While NEK8 homooligomerization is likely dispensable for fibrilloid formation, as GFP-INVS forms compartments efficiently in NEK8-KO cells, it may provide a mechanism by which NEK8 raises the concentration of ANKS6 and NPHP3 in the INVc. The domain that mediates NEK8 self-association, and therefore potentially also the concentrator function of NEK8, has yet to be identified.
The kinase activity of NEK8 has been established previously by in vitro kinase assays (Zalli et al., 2012; Choi et al., 2013; Czarnecki et al., 2015). NEK8 is the only INVc protein with demonstrable enzymatic activity, and, as kinases play important regulatory roles in many signaling pathways, the concept of a NEK8-driven signaling compartment has been an attractive model for the molecular function of the INVc. We show here that, although NEK8 is required for ANKS6 concentration and NPHP3 tethering, NEK8 kinase activity is dispensable for either of these functions (Supplemental Figures S50–S52). NEK8 therefore plays an important structural role in INVc assembly, independent of its kinase activity. This structural role of NEK8 in regulating INVc composition may be upstream of an important signaling event that does require NEK8 kinase activity. Two ciliary NEK8 substrates have been identified: PC-2 and ANKS6. PC-2, an ion channel and major genetic determinant in polycystic kidney disease, has been shown to be phosphorylated in a NEK8-dependent manner (Sohara et al., 2008) and is an intriguing candidate for an INVc effector in the kidney. ANKS6 is not only a NEK8 interactor and activator, but also very likely a substrate that could have an unidentified role in an important cell signaling pathway (Czarnecki et al., 2015; Nakajima et al., 2018). Recently, the G2A-NPHP3 mouse model for cystic kidney disease demonstrated that phosphorylation of ANKS6 by NEK8 was impaired in the G2A mouse, suggesting that NPHP3, while dispensable for INVc formation, is critical for INVc function and also that colocalization of ANKS6, NEK8, and NPHP3 in the INVc is necessary for this signaling event (Nakajima et al., 2018).
Results from the G2A mouse are consistent with our structural model, which suggests that the primary function of the INVc could be to establish a scaffold that both sequesters and concentrates ciliary NPHP3 (Figure 6, C–F). We are intrigued by the possibility that NPHP3, as the most downstream protein to assemble in the compartment, might be a potential effector of the compartment. Importantly, persistent cytogenesis in the G2A-NPHP3 mouse (Nakajima et al., 2018) confirms that a functionally competent INVc contains myristoylated NPHP3. Future studies aiming to identify the molecular function of the INVc should consider how NPHP3-dependent phosphorylation of ANKS6 (Nakajima et al., 2018) might prevent kidney cytogenesis, and whether NPHP3 could also regulate an unidentified ciliary protein that is important for disease-relevant ciliary signaling.
Conclusion
Our results provide a new model of the INVc featuring unprecedented molecular detail on both INVc substructure and how the four INVc proteins are arranged within the compartment. The structural functions that we have detailed for the four INVc proteins have the potential to inform future cell-biological studies of the regulation of the INVc: for example, 1) how might calcium signaling and APC (Morgan et al., 2002) or Akt (Suizu et al., 2016) activity signal through INVS to affect fibrilloid structure and association with the ANKS6-NEK8 complex? 2) What signals might affect the concentrator function of NEK8 and ANKS6? 3) Are INVc fibrilloids organized and assembled by an unidentified ciliary or axonemal structure? 4) Finally, how might these fibrilloids interact with other ciliary molecules and pathways? More broadly, the structure we report here may also provide insights into other subcellular structures built from proteins with structural similarities to INVc proteins, or possibly INVc proteins in other subcellular compartments. How the distinctive compartmentalization of these four proteins achieved by this unique fibrilloid structure contributes to the functional significance of the INVc, for example, in establishing left–right asymmetry during development or protecting healthy kidneys from cyst formation, remains an open question for the field of ciliopathy research.
MATERIALS AND METHODS
Cell culture
RPE1 cells were maintained in DMEM/F12 (12400024, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (100-106, Gemini), 1X GlutaMAX (35050-079, Thermo Fisher Scientific), and 100 U/ml penicillin–streptomycin (15140163, Thermo Fisher Scientific) at 37°C in 5% CO2. Cells were consistently passaged every 2–3 d to prevent their reaching confluency during maintenance. Cell lines used in this study tested negative for mycoplasma by PCR (Praetorius, 2017). For microscopy experiments, cells were grown to confluency and serum-starved in DMEM/F12 supplemented only with penicillin–streptomycin and GlutaMAX for 24 h to induce ciliation.
Cloning GFP expression vectors
Expression vectors for lentiviral transduction were generated by Gateway cloning (Invitrogen) using the BP (11789-100, Invitrogen) and LR (11791-100, Invitrogen) clonase reactions according to the manufacturer’s instructions. The K33M and G448V mutations in NEK8 were introduced by Quickchange site-directed mutagenesis reactions (200521, Agilent) of the entry clone containing mNEK8 cDNA. The vectors and primers used in this study are listed in Supplemental Tables S1 and S2.
Generation of GFP stable cell lines
Stable cell lines expressing GFP-tagged proteins were generated using lentivirus. Lentivirus carrying genes of interest were produced by cotransfecting 293T cells in a six-well dish (24 h after seeding 2 × 105 cells/well) with 150 ng of pCMV-VSV-G, 350 ng of pCMV-dR8.2 dvpr, and 500 ng of pWPXLd expression vector containing the gene of interest (1 µg DNA total) and 3 μl of Fugene 6 (E2692, Promega). Media were replaced 6–24 h after transfection to omit transfection reagent, and virus was harvested 48–72 h posttransfection. RPE1 cells were infected 24 h after seeding 5 × 104 cells per well of a six-well dish with fresh viral supernatant filtered with a 0.45-μm PVDF filter (SLHV013SL, Millipore) and mixed with a fourfold volume of fresh medium containing 12.5 μg/ml polybrene (TR-1003-G, Millipore). After infection for 72 h, cells were selected with 10 µg/ml blasticidin, sorted for GFP-positive cells on a BD Digital Vantage sorter with 488-nm laser excitation, and subsequently maintained in 10 μg/ml blasticidin. The vectors used in this study are listed in Supplemental Table S1.
Generation of knockout cell lines using CRISPR-Cas9
To generate knockout cell lines, RPE1-Cas9-BFP cells (Kanie et al., 2017) were infected with lentivirus carrying a single guide RNA (sgRNA) targeted to an exon of the target gene in the pMCB306 vector. After selection with 10 μg/ml puromycin, the sgRNA–puromycin resistance–EGFP cassette was removed by infecting cells with adenovirus carrying Cre-recombinase (1045, Vector BioLabs) at a multiplicity of infection of 50. At 10 d after adenovirus infection, GFP-negative single cells were sorted using FACSaria (BD). Single-cell clones were expanded and subjected to genomic DNA PCR combined with TIDE analysis (Brinkman et al., 2014) as well as screening by IF. Clones with loss of ciliary protein were considered effective knockouts. It should be noted that by these methods, the cell lines generated might express truncated forms of the targeted proteins. ANKS6-KO clones were screened by IF with anti-ANKS6. NPHP3-KO clones were screened by IF with anti-NPHP3. INVS-KO clones were screened both by TIDE to confirm biallelic frame shift mutations and also for loss of ciliary INVS by IF with anti-INVS. NEK8-KO clones were screened by TIDE and also by loss of ciliary ANKS6 that could be rescued by expression of GFP-NEK8.
To reduce the possibility that phenotypes observed in the knockout cells were due to off-target Cas9 editing or other clonal effects, mixed pools of knockout clones were created by combining single-cell clones as follows: the ANKS6-KO cell line was a combination of 10 single-cell clones, consisting of 4 clones edited by guide #1 and 6 clones by guide #4; INVS-KO was four single-cell clones all edited by guide #1; NEK8-KO was seven single-cell clones, four clones edited by guide #1 and three clones by guide #2; the NPHP3-KO was five single-cell clones, one clone edited by guide #1, three clones by guide #2, and one clone guide #3. The targeting sequences of guide RNAs and primers used in this study are provided in Supplemental Table S2. The vectors used in this study are listed in Supplemental Table S1.
Immunofluorescence for diffraction-limited microscopy
For conventional IF experiments using DLM, cells were grown to confluency on glass coverslips, serum-starved for 24 h to induce ciliation, and then fixed at room temperature using a 4% PFA solution prepared in 1X phosphate-buffered saline (PBS; Supplemental Figure S1A). After fixation, coverslips were stored up to 1 wk in PBS at 4°C before staining, though typically prepared within 1 d. For IF, samples were washed in PBS, blocked in 5% normal serum prepared in IF buffer (IFB; composition 3% BSA, 0.1% NP40, 0.0125% sodium azide in PBS) for 30–60 min. Samples were rinsed in IFB and then stained with primary antibodies diluted in IFB for 2–4 h (see Supplemental Table S3 for specifications and dilutions of the antibodies used in this study). Samples were washed five times in IFB (5-min incubations for the middle two washes) and then stained with fluorescent secondary antibodies (1 µg/ml each, Supplemental Table S4) and DAPI (0.1 µg/ml) for 30–60 min. Samples were washed twice with IFB and four times with PBS, and then briefly dried by wicking away residual PBS and mounted on a glass microscope slide with FluoromountG (Southern Biotech). All incubations of fixed samples were performed at room temperature and protected from light. Prepared slides were stored at 4°C, protected from the light, and imaged within 1 wk.
Antibodies
The specifications and concentrations of primary and secondary antibodies used in this study are provided in Supplemental Tables 3 and 4, respectively.
Image acquisition for diffraction-limited microscopy
Diffraction-limited images were acquired on an Everest workstation (Intelligent Imaging Innovations) equipped with a Zeiss AxioImager Z1 microscope, a 63× NA1.4 Plan-Apochromat objective lens (420780-9900, Zeiss), and a CoolSnapHQ cooled CCD camera (Roper Scientific; Supplemental Figure S1B). Replicate coverslips were prepared for each sample, and at least five images were captured for each coverslip. Within an experiment, each channel was imaged with consistent exposure times and instrument settings. To avoid camera saturation, exposure times were chosen so that the pixel intensities for Alexa Fluor–labeled structures were typically around 30% of the camera’s dynamic range. To reduce photobleaching, exposure times for structures that were detected by native GFP fluorescence were limited such that they typically yielded pixel intensities around 5–10% of the dynamic range.
Quantitative diffraction-limited microscopy workflow for measuring cilia and INVc length, density, and frequency
Owing to the observed variation in INVc frequency and INVc length in RPE1 cells, we decided to minimize stochastic sampling error by using a medium-throughput quantitative microscopy approach to compare INVc measurements for many cilia in each sample. Throughout this study, cilia and INVcs were measured by analyzing images of fluorescently labeled samples acquired by DLM using a semiautomated workflow in MATLAB. As outlined in Supplemental Figure S1, C and D, our custom MATLAB scripts first store indexed images of cilia to measure cilium length and ciliary protein density and then, based on user input, also measure INVc length and density. Supplemental Figures S1–S2 provide diagrams and definitions for each of the parameters measured. This approach allows quantitative comparisons of gross ciliary and INVc morphologies between different samples by plotting stitched images of many cilia (cilia plots) and by box and bar plot representations of the distribution of INVc protein densities (boxplot) and INVc frequency (bars) in that sample (Supplemental Figure S1F). For DLM images presented within a single figure panel (cilia plots), each marker (IF-labeled protein or GFP fusion protein) was imaged with the same illumination settings, and the image intensities (contrast) were adjusted to the same range for the final rendering of the image. MATLAB codes used for image processing and measurements of DLM images will be provided upon request.
Diffraction-limited microscopy image processing
Images were exported from SlideBook (Intelligent Imaging Innovations) as separate tiff files for each channel. For each experiment, images were then imported into MATLAB and indexed and stored for each sample in a structural array. Images were converted to a grayscale format with pixel intensities ranging from 0 to 1. The upper bound input for this conversion was defined by the bit depth of the original image to preserve the raw intensity ranges of each image; for 12-bit images, converted image = mat2gray(original image, [0 4096]). Subsequent image measurements were performed on these normalized grayscale images. Multichannel images were registered in MATLAB image by image to correct for random shifts between channels introduced by our DLM imaging system.
Cilia identification
Image contrast settings both for identifying and for masking cilia were determined in MATLAB. Contrast adjustments ensure more accurate threshold-based masking by setting a lower bound that would eliminate (set pixel intensity to 0) weakly stained background structures and an upper bound that would make the cilia readily visible (brightest pixels to 1). The same contrast settings were then applied to all images to be compared within the experiment. Contrast-adjusted images, typically containing only the ciliary markers to prevent INVc selection bias, were exported as TIFFs for cilia annotation. Cilia were annotated with the rectangular selection tool in FIJI, which was preferred over MATLAB for this step for accessibility and ease of use. The bounding box, or coordinates, for each rectangular selection in an image was exported as a csv file, which was then imported into MATLAB.
Cilia masking
Cilia masks were generated automatically by thresholding the contrast-adjusted images of the ciliary marker (AcTub or ARL13B). In experiments where two ciliary markers were used, ciliary masks were made from the sum of the two channels. The optimal masking threshold was initially determined by trial and error; however, because the pixel intensities of each image were normalized, and the contrasts were adjusted consistently to best represent cilia according to an experienced user’s eye, the same threshold (0.3) was used for most of the experiments presented in this study. Mask accuracy was assessed in each experiment by generating cilia plots showing the mask (Supplemental Figure S3D).
Subcompartment (INVc) masking
The INVc was masked manually for all cilia of a specified sample, using the cursor to draw around the region of interest with the imfreehand and createMask MATLAB functions. In samples that were detected by two INVc proteins, the mask was drawn around the region defined by at least one of the INVc protein channels.
Measurement of cilium and INVc length
For DLM images, both ciliary length and INVc length were calculated as half the mask perimeter (regionprops, “Perimeter” property) and converted to µm or nm.
Measurement of cilia and INVc density
The mean pixel intensity values within cilia or INVc masks, measured using the (regionprops, “meanintensity” property) function in MATLAB, were interpreted in this study to represent ciliary or compartment protein densities for the protein detected in the channel measured within the masked region (illustrated in Supplemental Figure S2). To correct for nonuniform background signal, the mean pixel intensity of the local background is subtracted from each density measurement (Supplemental Figures S1D and S2C). We note that for GFP alleles detected by native GFP fluorescence, intensity measurements were direct readings of GFP protein density. For endogenous proteins detected by indirect IF with polyclonal antibodies, the measured fluorescence intensities may not correspond to the number of target proteins for several reasons: epitopes on the target protein may be variably accessible to the primary antibodies throughout the sample; the number of primary antibodies that bind to a single target proteins is unknown and may vary within the sample; similarly, the number of secondary antibodies that bind to each primary antibody is unknown and may vary within the sample; the number of fluorophores per secondary antibody is variable; immunoglobulin G (IgG) molecules are bivalent, but the spatial arrangement of epitopes in the sample may variably promote or prevent the binding of a single IgG molecule to more than one epitope-bearing protein; and finally the number of antibodies that can bind to a single protein of interest in the sample may be sterically hindered by the sample or limited by the sizes of the antibodies.
Additional image processing for diffraction-limited microscopy cilia plots
For making cilia plots, images were rescaled by a factor of 4 with bicubic interpolation to reduce pixilated appearance when cilia were rotated. Cilia were oriented vertically with the angle of rotation determined by the mask orientation (regionprops, “orientation” property) and cropped. Some cilia were then flipped automatically to attempt to orient cilia tip up based on which end of the image contained the highest pixel intensities for a specified “bottom-enriched” channel. This method worked well only when basal body marker was included as the bottom-enriched channel. Cilia orientation was corrected for all cilia shown in the main figures, but not in the supplemental figures. In most multichannel IF micrographs showing more than one cilium, each channel is shown with some X-axis displacement so that intensities and relative sizes of individual channels can be compared.
Immunofluorescence for structured illumination microscopy
Samples for structured illuminated microscopy (SIM) were prepared in a manner similar to that for those for DLM ,with the following modifications: cells were grown on high-performance 18 × 18 mm 0.170 ± 0.005–mm glass coverslips (Zeiss). Incubations in primary antibody were 4–6 h, and samples were stained with secondary antibodies (2 µg/ml) for 2 h. Coverslips were mounted with Slowfade gold, dried for 20 min, and then sealed with clear nail varnish and stored at -20°C until imaging.
Image acquisition and processing for structured illumination microscopy
SIM experiments on samples labeled with AF488-, AF568-, and AF647-conjugated secondary antibodies were acquired on a DeltaVision OMX V4 system (488 nm [100 mW], 561 nm [100 mW], and 642 nm [300 mW] coherent sapphire solid state lasers; Evolve 512 EMCCD cameras [Photometrics]). SIM images were acquired with a 100× objective and stacks of 2.00-μm Z-steps were taken in 0.125-μm increments. Images were reconstructed and registered using SoftWorx 6.5.1 software, and registration settings were calibrated by the Cell Science Imaging Facility (Stanford). Reconstructions were then SUM projected in FIJI. Images were rotated to orient the cilia tips up. The contrast for each channel was adjusted to crop out low-intensity reconstruction artifacts such as ghosting and best display the protein of interest. The pixel intensities represented in SIM reconstructions should therefore not be compared between cilia. Images of individual cilia were rescaled to reduce the pixelated appearance when converting to different image types. Images of individual cilia were imported into MATLAB to make cilia plots.
Immunofluorescence for three-dimensional single-molecule superresolution imaging
Samples for 3D SM SR imaging were prepared in a manner similar to that for samples for DLM with the following modifications: cells were grown in four-well chambered coverslip µ-Slides (Ibidi, 80426). At 24 h before serum starvation, fluorescent beads (TetraSpeck, T7280, 0.2 µm, Invitrogen, diluted 1:5000) were added to each well to allow the cells to internalize them. Cells were blocked for 1 h and incubated in primary antibody staining solution for 4 h and secondary staining solution for 2 h. Following secondary staining and IFB and PBS washes, cells were postfixed in 4% PFA for 10 min to immobilize antibodies and then washed and stored in PBS at 4°C until imaging (usually the next day). Primary and secondary antibodies were used at a final concentration of 2 µg/ml, with the exception of some ANKS6 reconstructions (primary: 0.2 µg/ml). These labeling conditions resulted in high labeling specificity for all antibodies used, as demonstrated in the diffraction-limited images of labeling controls shown in Supplemental Figure S20.
Optical setup for 3D SM SR imaging
The optical setup was built around a conventional inverted microscope (IX71, Olympus) as described previously (Gustavsson et al., 2018). Illumination lasers (560 and 647 nm, both 1000 mW CW, MPB Communications) were spectrally filtered (560 nm: ff01-554/23-25 excitation filter; 647 nm: ff01-631/36-25 excitation filter, both Semrock), circularly polarized (LPVISB050-MP2 polarizers, WPQ05M-561 and WPQ05M-633 quarter-wave plates, all Thorlabs), and expanded and collimated using lens telescopes. The toggling of the lasers was controlled with shutters (VS14S2T1 with VMM-D3 three-channel driver, Vincent Associates Uniblitz) and synchronized with the detection optics via MATLAB. Both lasers were introduced into the back port of the microscope through a Köhler lens to allow wide field epi-illumination.
The sample was mounted on a coarse xy translation stage (M26821LOJ, Physik Instrumente) and a precision xyz piezoelectric stage (P-545.3C7, Physik Instrumente). The light emitted from the fluorophores was detected by a high NA detection objective (UPLSAPO100XO, 100×, NA 1.4, Olympus) and spectrally filtered (Di01-R405/488/561/635 dichroic, Semrock; ZET561NF notch filter, Chroma; ZET647NF notch filter, Chroma; 561 EdgeBasic long-pass filter, Semrock; and ET605/70 bandpass filter, Chroma, for red detection and ET700/75m bandpass filter, Chroma, for far red detection). The emission produced an intermediate image by the microscope tube lens before entering a 4f imaging system. The first lens of the 4f system (f = 90 mm, G322389000, Qioptiq) was positioned one focal length from the intermediate image plane. To create a two color–channel 4f system, a dichroic mirror (T660lpxrxt, Chroma) was inserted after the first 4f lens to transmit far red light into the first light path and reflect light with wavelengths shorter than 660 nm into the second light path. In the planes one focal length after the first 4f lens (the Fourier planes of the imaging paths), the phases of the emitted light were modulated to reshape the point spread function (PSF) to encode the axial position of the emitter using transmissive dielectric double-helix (DH) phase masks (615 nm for the green channel, 686 nm for the red channel, 2.7 mm diameter, both Double-Helix Optics). After phase modulation, the light was focused by second 4f lenses and imaged using two EM-CCD cameras (iXon DU-897E and iXon DU-897U, Andor).
3D SM SR imaging
For diffraction-limited imaging, cells were imaged in PBS using low-power 560-nm or 647-nm excitation (∼1 W/cm2). For 3D SM SR imaging, the PBS was replaced by a reducing and oxygen-scavenging buffer (Halpern et al., 2015) optimized for dSTORM blinking comprising 100 mM Tris-HCl (Invitrogen), 10% (wt/vol) glucose (BD Difco), 2 µl/ml catalase (C100, Sigma-Aldrich), 560 µg/ml glucose oxidase (Sigma-Aldrich), and 10 mM cysteamine (Sigma-Aldrich). For two-color imaging, the buffer was also supplemented with 143 mM β-mercaptoethanol (Sigma-Aldrich). For all single-molecule measurements, the sample was excited either by ∼10 kW/cm2 647-nm light or by ∼15 kW/cm2 560-nm light and imaged using an exposure time of 35 ms and a calibrated EM gain of 184. Sample drift was measured using fiducial beads (TetraSpeck, 0.2 µm, Invitrogen) in the sample and removed during postprocessing. After each imaging experiment, at least 300 dark frames were acquired when the shutter of the camera was closed. The mean of these dark frames was subtracted from the images before analysis. To facilitate calibration of the PSFs and registration between the two channels, beads (TetraSpeck, T7280, 0.2 µm, Invitrogen) on a coverslip were imaged when scanning over a 2-µm axial range using the piezoelectric stage. For registration measurements, the beads were also translated in xy between scans to more densely sample the entire field of view. Custom scripts were written in MATLAB to synchronize the laser shutters, image acquisition on the camera, and translation by the piezoelectric stage for these calibration and registration scans.
Analysis of 3D SM SR data
Calibration, registration, and fitting of single-molecule and fiducial bead images acquired using the DH-PSF was performed using the open-source Easy-DHPSF software version 2.0 (Lew et al., 2013; Bayas et al., 2019; https://sourceforge.net/projects/easy-dhpsf/). When images of the DH-PSF were analyzed, the lobes of each PSF were fitted using nonlinear least-squares functions in MATLAB with a pair of identical radially symmetric two-dimensional Gaussians as the objective function. The localization precision was calculated using the calibration described previously (Gahlmann et al., 2013). To obtain an estimate of the number of dye molecules per cluster in the fibrilloids, a correction for overcounting was applied as follows: localizations that were found within a three-frame sequence and within a distance of 27.3 nm were treated as one dye molecule.
The data used for image reconstruction were filtered to remove localizations with too large or small distance between lobes, dlobe (5.3 px < dlobe(green) < 7.5 px and 5.8 px < dlobe(red) < 7.2 px, 1 px = 160 nm), and x or y localization precision larger than 30 nm. This resulted in median localization precisions in the ranges 6–13 nm and 9–19 nm in xy and z, respectively (see Supplemental Figures S18 and S19). The localizations were rendered using the Vutara SRX software from Bruker, where each localization was represented by a 3D Gaussian with a diameter of 25 nm and an opacity set to show the localization density.
Reconstructions from the two-color 3D SM SR experiments in this study were manually overlaid to correct for nanoscale residual shifts after registration of the two channels. This was acceptable for two-color experiments measuring the axoneme and INVc, where small misalignments (<50 nm) would not change the interpretation of the data (Figure 2A; Supplemental Figure S21, C and D).
The total INVc length, the length of individual fibrilloids (Supplemental Figure S21J), and the number of fibrilloids per cilium (Supplemental Figure S21I) were estimated manually from the 3D reconstructions. Ambiguous cases were not counted as fibrilloids, and the fibrilloid length estimation was stopped at gaps in a fibrilloid. Not counting ambiguous cases might have caused an underestimation of the total number of fibrils due to insufficient dye label density. The requirement of continuity of localizations for fibrilloid estimation might, on the other hand, have caused both an overestimation of the number of fibrilloids and an underestimation of the fibrilloid length.
Software used for image analysis: DLM epifluorescence microscopy—FIJI, MATLAB; SIM experiments—SoftWorx, FIJI, MATLAB; and 3D SM SR microscopy—Easy-DHPSF, FIJI, MATLAB.
Statistical analyses
Box plots.
Box-and-whisker plots were used to represent the distribution of ciliary protein densities measured in a ciliary mask (Figures 3 and 4). Box-and-whisker features represent the following statistics (outlined in Supplemental Figure S1F): red line, median; box edges, 25th and 75th percentiles; whiskers, 5th and 95th percentiles; “outlying” data points in the bottom 5% and top 95% of the data were plotted as “x.” In some figures the scale of the axis on which the box was plotted did not include all outliers.
Box–bar plots.
Box plots (described above) were plotted on top of a horizontal bar plot. Bars were plotted on an independent axis from the box plot, ranging from 0 to 100%. Bar length represented the percentage of cilia that contained visible INVcs. INVc-positive cilia were determined subjectively (user-defined). Cilia were considered INVc-positive if they contained INVS, ANKS6, or NEK8, irrespective of subcompartment length or density. NPHP3-positive cilia, in samples where NPHP3 was the only INVc protein measured, were scored as containing an INVc only if the NPHP3-positive region was relatively high in density (bright) and localized in the proximal cilium. These criteria likely underestimate the presence of NPHP3-positive INVcs in NEK8-KO cells where INVS and ANKS6 staining confirmed that 10–20% of cilia contained low-density compartments, but also potentially ignores NPHP3 inversin-like compartments in INVS-KO and ANKS6-KO cells.
Pearson’s correlation.
The correlation between the densities of two INVc proteins measured in a user-defined compartment mask (as in Figure 4) was calculated using the corr function in MATLAB: [r, p] = corr(X,Y). The variable X is a list of the INVc protein density measurements for one protein, and Y lists the INVc protein–density measurements in the same compartments for the second protein. The resulting r value is the Pearson linear correlation coefficient between X and Y, and p is a probability value for testing the null hypothesis that X and Y are not correlated. Pearson’s r values are interpreted as follows: r = –1, X and Y are anti-correlated; r = 0, no correlation; and r = 1, perfect correlation. Here we considered p < 0.05 to indicate confident rejection of the null hypothesis, supporting the result that the densities of the two INVc proteins with positive r values were significantly correlated, and r > 0.5 to represent a biologically meaningful positive correlation.
t test of statistical significance.
For comparisons between two different samples, the probability value p was calculated for a two-tailed t test using the ttest2 function in MATLAB: [h, p] = ttest2(X,Y). The variable X is a list of the measurements for one sample, and Y lists measurements of the same parameter for a second sample. We considered p < 0.05 to indicate confident rejection of the null hypothesis (h = 0).
Supplementary Material
Acknowledgments
We thank Tomoharu Kanie and Keren Hilgendorf of the Jackson Lab for scientific guidance and establishing the cloning vectors and parental cell lines used in this study, Karlene Cimprich for providing the mNEK8 cDNA, Mike Bassik (Stanford) for providing a genome-wide reference for CRISPR guide sequences, and Galo Garcia III and Jeremy Reiter for providing antibodies and advice for IF detection of the ciliary pocket. We acknowledge the Beckman Cell Science Imaging Facility (Stanford) for hosting the Deltavision OMX Blaze microscope: the project described was supported, in part, by Award Number 1S10OD01227601 from the National Center for Research Resources (NCRR). H.W.B. was supported by a Stanford Graduate Fellowship and the Stanford Training Program in Aging Research. Research reported in this publication was supported in part by the National Institute on Aging of the National Institutes of Health under Award T32 AG0047126. A.K.G. acknowledges partial financial support from National Institute of General Medical Sciences of the National Institutes of Health Grant 1K99GM134187-01, from Swedish Research Council Grant 2016-00130, and from the Foundation BLANCEFLOR Boncompagni-Ludovisi, née Bildt. This project was also supported in part by U.S. National Institute of General Medical Sciences Grant R35GM118067 (W.E.M.), by Grant R01GM114276 (P.K.J.), and by Goldfinch Bio. We thank Carl G. Ebeling, worldwide application scientist for Bruker Fluorescence Microscopy, for his support and for the use of the Bruker SRX visualization and analysis software for rendering localization data. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the NCRR.
Abbreviations used:
- 3D SM SR
three-dimensional single-molecule superresolution
- ANKS6
ankyrin repeat and SAM domain containing protein 6
- DLM
diffraction-limited microscopy
- dSTORM
direct stochastic optical reconstruction microscopy
- gINVc
genes encoding pINVc
- IF
immunofluorescence
- IFT
intraflagellar transport
- IMCD
inner medullary collecting duct
- INVc
inversin compartment
- INVS
inversin
- NEK8
NIMA-related kinase 8
- NPHP
nephronophthisis
- pINVc or INVc protein
known proteins that localize to the inversin compartment NVS, ANKS6, NEK8, NPHP3
- RPE
retinal pigmented epithelium
- SIM
structured illumination microscopy
- TZ
transition zone.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-09-0499) on January 2, 2020.
REFERENCES
- Bayas C, von Diezmann A, Gustavsson A-K, Moerner WE. (2019). Easy-DHPSF 2.0: open-source software for three-dimensional localization and two-color registration of single molecules with nanoscale accuracy. Nat Res Protocol Exchange. 10.21203/rs.2.9151/v1 (accessed 10 June 2019). [DOI] [Google Scholar]
- Bellavia S, Dahan K, Terryn S, Cosyns J-P, Devuyst O, Pirson Y. (2010). A homozygous mutation in INVS causing juvenile nephronophthisis with abnormal reactivity of the Wnt/beta-catenin pathway. Nephrol Dial Transplant , 4097–4102. [DOI] [PubMed] [Google Scholar]
- Bergmann C, Fliegauf M, Bruchle NO, Frank V, Olbrich H, Kirschner J, Schermer B, Schmedding I, Kispert A, Kranzlin B, et al. (2008). Loss of nephrocystin-3 function can cause embryonic lethality, Meckel-Gruber-like syndrome, situs inversus, and renal–hepatic–pancreatic dysplasia. Am J Hum Genet , 959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler M, Kong D, Sun S, Nanjundappa R, Evans L, Farmer V, Holland A, Mahjoub MR, Sui H, Loncarek J. (2019). High-resolution characterization of centriole distal appendage morphology and dynamics by correlative STORM and electron microscopy. Nat Commun , 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, Amendola M, van Steensel B. (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res , e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T. (2012). Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J Cell Biol , 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HJC, Lin J-R, Vannier J-B, Slaats GG, Kile AC, Paulsen RD, Manning DK, Beier DR, Giles RH, Boulton SJ, et al. (2013). NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol Cell , 423–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki PG, Gabriel GC, Manning DK, Sergeev M, Lemke K, Klena NT, Liu X, Chen Y, Li Y, San Augustin JT, et al. (2015). ANKS6 is the critical activator of NEK8 kinase in embryonic situs determination and organ patterning. Nat Commun , 6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlmann A, Ptacin JL, Grover G, Quirin S, von Diezmann ARS, Lee MK, Backlund MP, Shapiro L, Piestun R, Moerner WE. (2013). Quantitative multicolor subdiffraction imaging of bacterial protein ultrastructures in three dimensions. Nano Lett , 987–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluenz E, Höög JL, Smith AE, Dawe HR, Shaw MK, Gull K. (2010). Beyond 9+0: noncanonical axoneme structures characterize sensory cilia from protists to humans. FASEB J , 3117–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grampa V, Delous M, Zaidan M, Odye G, Thomas S, Elkhartoufi N, Filhol E, Niel O, Silbermann F, Lebreton C, et al. (2016). Novel NEK8 mutations cause severe syndromic renal cystic dysplasia through YAP dysregulation. PLos Genet , e1005894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson A-K, Petrov PN, Lee MY, Shechtman Y, Moerner WE. (2018). 3D Single-molecule super-resolution microscopy with a tilted light sheet. Nat Commun , 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern AR, Howard MD, Vaughan JC. (2015). Point by point: an introductory guide to sample preparation for single-molecule, super-resolution fluorescence microscopy. Curr Protocols Chem Biol , 103–120. [DOI] [PubMed] [Google Scholar]
- Hoff S, Halbritter J, Epting D, Frank V, Nguyen T-MT, van Reeuwijk J, Boehlke C, Schell C, Yasunaga T, Helmstädter M, et al. (2013). ANKS6 is a central component of a nephronophthisis module linking NEK8 to INVS and NPHP3. Nat Genet , 951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-R, Wang C-L, Huang Y-S, Chang Yu-C, Chang Ya-C, Pusapati GV, Lin C-Y, Hsu N, Cheng H-C, Chiang Y-C, et al. (2018). Spatiotemporal manipulation of ciliary glutamylation reveals its roles in intraciliary trafficking and hedgehog signaling. Nat Commun , 1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan MA, Diener DR, Stepanek L, Pigino G. (2018). The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat Cell Biol , 1250–1255. [DOI] [PubMed] [Google Scholar]
- Kanie T, Abbott KL, Mooney NA, Plowey ED, Demeter J, Jackson PK. (2017). The CEP19-RABL2 GTPase complex binds IFT-B to initiate intraflagellar transport at the ciliary base. Dev Cell , 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leettola CN, Knight M, Cascio D, Hoffman S, Bowie JU, Wilson PD, Chapin HC, Caplan MJ, Torres VE, Harris PC, et al. (2014). Characterization of the SAM domain of the PKD-related protein ANKS6 and its interaction with ANKS3. BMC Struct Biol , 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew MD, von Diezmann ARS, Moerner W. 2013. Easy-DHPSF open-source sofware for three-dimensional localization of single molecules with precision beyond the optical difraction limit. Protocol Exchange, 10.1038/protex.2013.026. [DOI] [Google Scholar]
- Lienkamp S, Ganner A, Boehlke C, Schmidt T, Arnold SJ, Schafer T, Romaker D, Schuler J, Hoff S, Powelske C, et al. (2010). Inversin relays Frizzled-8 signals to promote proximal Pronephros development. Proc Natl Acad Sci USA , 20388–20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Trapp ML, Quarmby LM. (2005). NIMA-related kinases defective in murine models of polycystic kidney diseases localize to primary cilia and centrosomes. J Am Soc Nephrol , 3485–3489. [DOI] [PubMed] [Google Scholar]
- Manning DK, Sergeev M, van Heesbeen RG, Wong MD, Oh J-H, Liu Y, Henkelman RM, Drummond I, Shah JV, Beier DR. (2013). Loss of the ciliary kinase Nek8 causes left–right asymmetry defects. J Am Soc Nephrol , 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki T, Saijoh Y, Tsuchiya K, Shirayoshi Y, Takai S, Taya C, Yonekawa H, Yamada K, Nihei H, Nakatsuji N, et al. (1998). Cloning of Inv, a gene that controls left/right asymmetry and kidney development. Nature , 177–181. [DOI] [PubMed] [Google Scholar]
- Molla-Herman A, Ghossoub R, Blisnick T, Meunier A, Serres C, Silbermann F, Emmerson C, Romeo K, Bourdoncle P, Schmitt A, et al. (2010). The ciliary pocket: an endocytic membrane domain at the base of primary and motile cilia. J Cell Sci (Pt 10), 1785–1795. [DOI] [PubMed] [Google Scholar]
- Morgan D, Eley L, Sayer J, Strachan T, Yates LM, Craighead AS, Goodship JA. (2002). Expression analyses and interaction with the anaphase promoting complex protein Apc2 suggest a role for inversin in primary cilia and involvement in the cell cycle. Hum Mol Genet , 3345–3350. [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Kiyonari H, Mukumoto Y, Yokoyama T. (2018). The Inv compartment of renal cilia is an intraciliary signal-activating center to phosphorylate ANKS6. Kidney Int , 1108–1117. [DOI] [PubMed] [Google Scholar]
- Nakata K, Shiba D, Kobayashi D, Yokoyama T. (2012). Targeting of Nphp3 to the primary cilia is controlled by an N-terminal myristoylation site and coiled-coil domains. Cytoskeleton , 221–234. [DOI] [PubMed] [Google Scholar]
- Nürnberger J, Kribben A, Saez AO, Heusch G, Philipp T, Phillips CL. (2004). The invs gene encodes a microtubule-associated protein. J Am Soc Nephrol , 1700–1710. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Yagi T, Kikkawa M. (2014). Mechanosignaling between central apparatus and radial spokes controls axonemal dynein activity. J Cell Biol , 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Sugimoto K, Shimada Y, Fujita S, Yanagida H, Yagi K, Takemura T. (2008). Association of INVS (NPHP2) mutation in an adolescent exhibiting nephronophthisis (NPH) and complete situs inversus. Clin Nephrol , 135–141. [DOI] [PubMed] [Google Scholar]
- Otto EA, Schermer B, Obara T, O’Toole JF, Hiller KS, Mueller AM, Ruf RG, Hoefele J, Beekmann F, Landau D, et al. (2003). Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left–right axis determination. Nat Genet , 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto EA, Trapp ML, Schultheiss UT, Helou J, Quarmby LM, Hildebrandt F. (2008). NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. J Am Soc Nephrol , 587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owa M, Uchihashi T, Yanagisawa H-A, Yamano T, Iguchi H, Fukuzawa H, Wakabayashi K-I, Ando T, Kikkawa M. (2019). Inner lumen proteins stabilize doublet microtubules in cilia and flagella. Nat Commun , 1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavani SRP, Thompson MA, Biteen JS, Lord SJ, Liu Na, Twieg RJ, Piestun R, Moerner WE. (2009). Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc Natl Acad Sci USA , 2995LP–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua SC, Chiba S, Suzuki M, Su E, Roberson EC, Pusapati GV, Schurmans S, Setou M, Rohatgi R, Reiter JF, et al. (2019). Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell , 261. [DOI] [PubMed] [Google Scholar]
- Praetorius C. (2017). Homemade PCR test for mycoplasma contamination. BiteSize Bio. https://bitesizebio.com/23682/homemade-pcr-test-for-mycoplasma-contamination/. [Google Scholar]
- Revenkova E, Liu Q, Gusella GL, Iomini C. (2018). The Joubert syndrome protein ARL13B binds tubulin to maintain uniform distribution of proteins along the ciliary membrane. J Cell Sci , jcs212324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K, Jerman S, Jozsef L, McNamara T, Onyekaba G, Sun Z, Marin EP. (2017). Palmitoylation of the ciliary GTPase ARL13b is necessary for its stability and its role in cilia formation. J Biol Chem , 17703–17717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, Baye LM, Wen X, Scales SJ, Kwong M, et al. (2011). Mapping the NPHP–JBTS–MKS protein network reveals ciliopathy disease genes and pathways. Cell , 513–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub JR, Stearns T. (2013). The Rilp-like proteins Rilpl1 and Rilpl2 regulate ciliary membrane content. Mol Biol Cell , 453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Heintzmann R, Leonhardt H. (2010). A guide to super-resolution fluorescence microscopy. J Cell Biol , 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Garcia G, Van De Weghe JC, McGorty R, Pazour GJ, Doherty D, Huang Bo, Reiter JF. (2017). Super-resolution microscopy reveals that disruption of ciliary transition-zone architecture causes Joubert syndrome. Nat Cell Biol , 1178–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba D, Manning DK, Koga H, Beier DR, Yokoyama T. (2010). Inv acts as a molecular anchor for Nphp3 and Nek8 in the proximal segment of primary cilia. Cytoskeleton , 112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba D, Yamaoka Y, Hagiwara H, Takamatsu T, Hamada H, Yokoyama T. (2009). Localization of Inv in a distinctive intraciliary compartment requires the C-terminal ninein-homolog-containing region. J Cell Sci , 44–54. [DOI] [PubMed] [Google Scholar]
- Simons M, Gloy J, Ganner A, Bullerkotte A, Bashkurov M, Kronig C, Schermer B, Benzing T, Cabello OA, Jenny A, et al. (2005). Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat Genet , 537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohara E, Luo Y, Zhang J, Manning DK, Beier DR, Zhou J. (2008). Nek8 regulates the expression and localization of polycystin-1 and polycystin-2. J Am SocNephrol , 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard D, Zhao Y, Bayless BA, Gui L, Louka P, Dave D, Suryawanshi S, Tomasi RF-X, Dupuis-Williams P, Baroud CN, et al. (2018). Tetrahymena RIB72A and RIB72B are microtubule inner proteins in the ciliary doublet microtubules. Mol Biol Cell , 2566–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Tsukiyama T, Yamaguchi TP, Yokoyama T. (2011). The canonical Wnt signaling pathway is not involved in renal cyst development in the kidneys of Inv mutant mice. Kidney Int , 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suizu F, Hirata N, Kimura K, Edamura T, Tanaka T, Ishigaki S, Donia T, Noguchi H, Iwanaga T, Noguchi M. (2016). Phosphorylation-dependent Akt-inversin interaction at the basal body of primary cilia. EMBO J , 1346–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Fisher RL, Bowser SS, Pentecost BT, Sui H. (2019). Three-dimensional architecture of epithelial primary cilia. Proc Natl Acad Sci USA , 9370 LP–9379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tory K, Rousset-Rouviere C, Gubler M-C, Moriniere V, Pawtowski A, Becker C, Guyot C, Gie S, Frishberg Y, Nivet H, et al. (2009). Mutations of NPHP2 and NPHP3 in infantile nephronophthisis. Kidney Int , 839–847. [DOI] [PubMed] [Google Scholar]
- Trapp ML, Galtseva A, Manning DK, Beier DR, Rosenblum ND, Quarmby LM. (2008). Defects in ciliary localization of Nek8 is associated with cystogenesis. Pediatr Nephrol , 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Diezmann A, Shechtman Y, Moerner WE. (2017). Three-dimensional localization of single molecules for super-resolution imaging and single-particle tracking. Chem Rev , 7244–7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton-Pitt SRF, Silva M, Nguyen KCQ, Hall DH, Barr MM. (2014). The Nphp-2 and Arl-13 genetic modules interact to regulate ciliogenesis and ciliary microtubule patterning in C. elegan. PLoS Genet , e1004866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe D, Saijoh Y, Nonaka S, Sasaki G, Ikawa Y, Yokoyama T, Hamada H. (2003). The left–right determinant inversin is a component of node monocilia and other 9+0 cilia. Development , 1725LP–1734. [DOI] [PubMed] [Google Scholar]
- Waters AM, Beales PL. (2011). Ciliopathies: an expanding disease spectrum. Pediatr Nephrol , 1039–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KJ, Baye LM, Olivier-Mason A, Mukhopadhyay S, Sang L, Kwong M, Wang W, Pretorius PR, Sheffield VC, Sengupta P, et al. (2011). An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev , 2347–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Chong WM, Wang W-J, Mazo G, Tanos B, Chen Z, Tran TMN, Chen Y-D, Weng RR, Huang C-E, et al. (2018). Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat Commun , 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Su J, Wang W-J, Craige B, Witman GB, Tsou M-FuB, Liao J-C. (2015). Superresolution pattern recognition reveals the architectural map of the ciliary transition zone. Sci Rep , 14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhiko Y, Imai F, Ookubo K, Takakuwa Y, Shiokawa K, Yokoyama T. (2001). Calmodulin binds to Inv protein: implication for the regulation of Inv function. Dev Growth Differ , 671–681. [DOI] [PubMed] [Google Scholar]
- Yoon J, Comerci CJ, Weiss LE, Milenkovic L, Stearns T, Moerner WE. (2019). Revealing nanoscale morphology of the primary cilium using super-resolution fluorescence microscopy. Biophys J , 319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalli D, Bayliss R, Fry AM. (2012). The Nek8 protein kinase, mutated in the human cystic kidney disease nephronophthisis, is both activated and degraded during ciliogenesis. Hum Mol Genet , 1155–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.