Abstract
Retrograde membrane trafficking from plasma membrane to Golgi and endoplasmic reticulum typifies one of the key sorting steps emerging from the early endosome that affects cell surface and intracellular protein dynamics underlying cell function. While some cell surface proteins and lipids are known to sort retrograde, there are few effective methods to quantitatively measure the extent or kinetics of these events. Here we took advantage of the well-known retrograde trafficking of cholera toxin and newly defined split fluorescent protein technology to develop a quantitative, sensitive, and effectively real-time single-cell flow cytometry assay for retrograde membrane transport. The approach can be applied in high throughput to elucidate the underlying biology of membrane traffic and how endosomes adapt to the physiologic needs of different cell types and cell states.
INTRODUCTION
The functions of all eukaryotic cells depend on their ability to organize and distribute proteins, lipids, and nucleic acids to specific regions of the cell. This is largely achieved by dynamically sorting and sequestering specific molecules to membrane-bound subcellular organelles and the plasma membrane. The early sorting endosome and trans-Golgi network (TGN) play essential roles in these processes. One membrane trafficking pathway emerging from the sorting endosome leads retrograde in the secretory pathway to the TGN, connecting these key sorting stations for maintenance and regulation of cell structure and function. Some plasma membrane lipids are known to traffic retrograde all the way through the TGN into the endoplasmic reticulum (ER), an endogenous process co-opted by several bacterial toxins and viruses to cause disease. The retrograde pathways, however, have been difficult to measure with precision, sensitivity, and in real time. Biochemical assays for toxin transport into the Golgi and ER using protein sulfation and N-glycosylation to mark entry into TGN and ER, respectively, have proved highly informative, as has the use of ultrastructural electron and confocal light microscopy (Johannes et al., 1997; Torgersen et al., 2001; Saslowsky et al., 2010; Guimaraes et al., 2011). But these approaches are time consuming, technically challenging, and difficult to quantify or apply with high temporal resolution or in high throughput for gene or small molecule discovery. Here, we describe the development and validation of a novel, highly sensitive, quantitative, and robust single-cell assay for measuring retrograde trafficking from PM to TGN and ER, effectively in real time.
RESULTS AND DISCUSSION
Split-fluorescent protein toxin reporter
To model retrograde endosome sorting and transport, we applied the split fluorescent protein (split-FP) technologies (Feng et al., 2017) to Vibrio cholerae cholera toxin (CTx) and adapted the system to a flow cytometry assay. CTx typifies the family of AB5-subunit bacterial toxins that invade the ER of host cells to induce disease (Figure 1, a and b) (Fujinaga et al., 2003; Lencer and Tsai, 2003; Spooner et al., 2006). For CTx, the homopentameric B-subunit binds the glycosphingolipid GM1 at the plasma membrane and GM1 carries the toxin retrograde by vesicular transport into the TGN and ER. Once in the ER, a portion of the A-subunit, the enzymatically active A1-chain (Figure 1a, yellow), is dissociated from the B-subunit (red) and retro-translocated to the cytosol where it activates adenyl cyclase to cause disease. The other domain of the A-subunit, the A2-chain (cyan), remains assembled with the B-subunit in the ER lumen (Wernick et al., 2010).
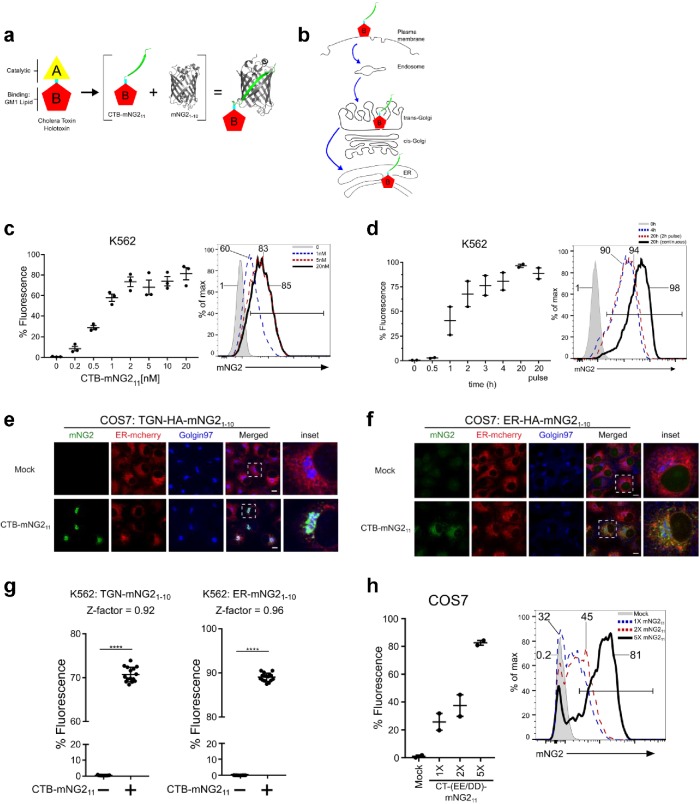
FIGURE 1:
Design, development, and validation of the split-fluorescence assay for retrograde trafficking. (a) Schematic of native holotoxin CTx structure and modifications to enable split-fluorescence complementation by fusing NG2 fragment mNG211 to the N-terminus of the CTx A2-chain and reassembly with CTx B-subunit to form CTB-mNG211. (b) Pathway of retrograde trafficking for CTB-mNG211 from the plasma membrane into the Golgi and ER. (c) Retrograde trafficking to ER in K562 cells stably expressing ER-HA-mNG21-10 as assessed by flow cytometry. Representative flow cytometry graph displayed to right. Cells were treated with CTB-mNG211 for 4 h at 37°C; n = 3 independent experiments. (d) Time course of retrograde trafficking into the ER in K562 cells with 10 nM CTB-mNG211—methods are as described above; n = 2 independent experiments. Representative flow cytometry graph shows timepoints at 0, 4, and 20 h for continuous CTB-mNG211 toxin uptake and in cells treated with a 2-h toxin pulse and chase for 20 h. (e, f) Confocal microscopy of CTB-mNG211 retrograde trafficking into TGN and ER of COS7 cells stably expressing TGN or ER mNG21-10 and ER-mCherry, 6 h incubation. Cells were stained with anti-Golgin97 to mark the TGN. (g) TGN and ER retrograde trafficking in cells with 15 biological replications per condition (treated or not treated). Z-factors = 0.92 for TGN and 0.96 for ER retrograde transport. (h) Addition of tandem mNG211 to CTx improves assay sensitivity. COS7 ER-HA-mNG21-10 cells were treated with 10 nM holotoxin CT-1XmNG211 or tandem CT-2XmNG211 or CT-5XmNG211 for 3.5 h at 37°C; n = 2 independent experiments. Error bars indicate ± SEM. ****p < 0.0001 (two-tailed Student’s t test). Scale bars are 10 μm.
To develop a quantitative measure of retrograde trafficking, the N-terminus of the CTx A2-chain was fused to the split neon green2 (NG2) peptide mNG211 (Figure 1a, green). The fusion protein and CTx B-monomers were coexpressed in Escherichia coli and purified as an assembled mNG211-A2-chain associated with the CTx-B5 pentamer (Figure 1a and Supplemental Figure S1a). The modified toxin was called CTx B (CTB)-mNG211. It contains all structural features that underlie the retrograde trafficking of CTX: binding to GM1 by the B-subunit and binding to the ER-retention KDEL-receptor by the C-terminal KDEL-motif of the A2-chain (Figure 1b) (Lencer and Tsai, 2003; Spooner et al., 2006). To measure trafficking of CTB-mNG211 into the TGN or ER, we prepared several cell lines stably expressing the HA–epitope-tagged mNG21-10 protein, including both adherent epithelial cells (human embryonic kidney [HEK293T] and African green monkey kidney COS7) and human lymphocyte K562 cells that grow in suspension. HA-mNG21-10 was targeted to the TGN by fusing the molecule to the TGN targeting peptide from Gal-transferase (named TGN-HA-mNG21-10) and to the ER by encoding the signal sequence from ER resident chaperone BIP and the C-terminal ER-retention motif KDEL (named ER-HA-mNG21-10). In COS7 cells, TGN-HA-mNG21-10 localized principally to the TGN as assessed by immunofluorescence and colocalization with the TGN resident protein Golgin97 (Supplemental Figure S1b) (Barr, 1999). TGN-HA-mNG21-10 only marginally colocalized with the ER-mCherry marker when that was coexpressed (colocalization coefficients shown in Supplemental Figure S1c). Likewise, ER-HA-mNG21-10 localized in COS7 cells only to the ER (Supplemental Figure S1, c and d). When exposed to CTB-mNG211, all three cell lines expressing ER-HA-mNG21-10 became fluorescent. We found slightly different toxin dose profiles across K562, HEK293T, and COS7 cells as assessed by flow cytometry (Figure 1c and Supplemental Figure S1, e and f). Apparent ED50 was between 0.5 and 10 nM for retrograde trafficking, which corresponds closely to the ED50 for CTx-induced cell toxicity (Lencer et al., 1992, 1993). Time-course studies revealed temporal differences for the fluorescent signal induced by CTB-mNG211. K562 cells had a more rapid rise to maximum fluoresce when compared with HEK293T and COS7 cells (Figure 1d and Supplemental Figure S1, g and h). In pulse chase studies, both K562 and COS7 cells responded to CTB-mNG211 with nearly the same signal (Figure 1d and Supplemental Figure S1, g and h). Entry of CTB-mNG211 into the Golgi or ER was verified by colocalization with Golgin97 or ER-mCherry in COS7 cells expressing TGN-HA-mNG21-10 or ER-HA-mNG21-10, respectively (Figure 1, e and f). CTB-mNG211 retrograde trafficking and colocalization was also observed in COS7 cells stably expressing Sec61β-HA-mNG21-10, an ER-localized membrane-tethered mNG21-10 by fusion with Sec61β (Supplemental Figure S1, c, i, and j). The variability in kinetics of retrograde trafficking observed among cell types highlights how endosomes can adapt to the physiologic needs of different cells and cell states—as well appreciated for signal transduction events (Kholodenko, 2006).
To assess reproducibility and signal size, we studied 15 biologic replicates of K562 cells treated or not treated with CTB-mNG211 using both the TGN and the ER transport assays. We found high levels of reproducibility with calculated Z-factors for discerning retrograde transport of 0.92 and 0.96, respectfully (Figure 1g). Assays adapted for high-throughput screening are considered ideal if they have Z-factors >0.5 (Zhang et al., 1999). Signal sensitivity was enhanced by concatenating additional mNG211 strands. The addition of multiple (concatenated) mNG211 strands on CT-(EE/DD) progressively increased the PM to ER retrograde trafficking signal in COS7 cells (Figure 1h). We found the split-FP technology system can also be applied to red fluorescence, as evidenced by adaptation of split sfmCherry211 system (Feng et al., 2017) and generation of recombinant CTB-sfmCherry211 (Supplemental Figure S1k). Thus, the toxin-based split-FP assay enables robust, quantitative, effectively real-time, and high-throughput measurements of retrograde trafficking into TGN or ER.
General utility
The utility of the assay was first assessed in HEK293T and K562 cells acutely treated with small molecules implicated in affecting endosome traffic (Figure 2, a and b). Brefeldin A (BFA) causes collapse of the Golgi compartment into the ER and profoundly blocks the mechanics of vesicular transport into and out of the secretory pathway (Lencer et al., 1993). In both HEK293T and K562 cells, BFA completely blocked CTB–mNG211-induced fluorescence caused by trafficking into the ER (Figure 2, a and b). BFA had no effect on expression of ER-HA-mNG21-10 in either cell line (Supplemental Figure S2, a and b). Retrograde transport into the Golgi was also blocked by BFA (Figure 2c), as observed before using other measures of retrograde transport (Fujinaga et al., 2003). Also as expected (Johannes et al., 1997), we found that treatment with nocodazole to depolymerize microtubules resulted in inhibition of retrograde ER trafficking in both cell lines.
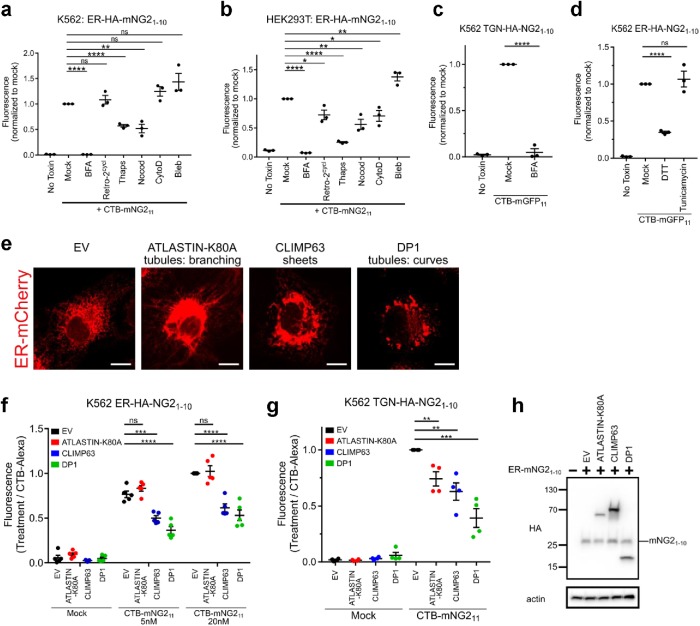
FIGURE 2:
Quantitative measures of retrograde traffic under different conditions. (a, b) Retrograde trafficking to ER as assessed in K562 (a) and HEK293T (b) treated with the indicated compounds for 30 min before addition of CTB-mNG211. Data normalized to Mock (untreated) cells; n = 3 independent experiments, each with three biologic replicates, and mean marked as one point for each condition. (c) Retrograde trafficking to TGN performed as in a; n = 3 independent experiments (BFA 10 µM). (d) Retrograde trafficking to ER in cells pretreated with DTT (4 mM) or tunicamycin (5 μg/ml); n = 3 independent experiments. (e) COS7 cells stably expressing ER-mCherry and ATLASTIN-K80A, CLIMP63, or DP1. (f) Retrograde trafficking to ER in cells overexpressing CLIMP63, DP1, or ATLASTIN1-K80A. Data normalized by CTB-Alexa Fluor 488 uptake; n = 5 independent experiments. (g) Retrograde trafficking to TGN as in f; n = 4 independent experiments. (h) Western blot for HA-epitope in HEK293T cells stably transfected with empty vector (EV), HA-tagged ATLASTIN-K80A, CLIMP63 or DP1, and HA-tagged TGN-mNG21-10. Mean ± SEM, ns not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (two-tailed Student’s t test). Scale bars are 10 μm.
We then studied the poorly understood role and impact of intracellular calcium signaling on PM-ER retrograde trafficking using thapsigargin. Thapsigargin amplifies cytosolic Ca2+ levels by inhibition of the ER Ca2+-ATPase, thus also inducing ER stress. We found that thapsigargin inhibited retrograde transport from PM to ER (Figure 2, a and b). While the ER stress-inducing (but nonspecific) reducing agent dithiothreitol (DTT) caused inhibition of retrograde ER trafficking, thus phenocopying the cells treated with thapsigargin, we observed no such effect in cells treated with tunicamycin, a small molecule that causes ER stress by blockade of N-glycosylation (Figure 1d). Thus, thapsigargin likely inhibits retrograde membrane traffic via its effects on intracellular Ca2+ transients. Localized Ca2+ transients are well known to effect membrane traffic at nerve terminals, PM repair after injury, and exocytosis in secretory cell types (Bai and Chapman, 2004; Clapham, 2007; Andrews et al., 2014). We also note that our result with thapsigargin differs from two previous studies, which used indirect measures of retrograde trafficking and different experimental systems (Sandvig et al., 1996; Sorensen, 2017; Zhang et al., 2017), perhaps explaining the inconsistency.
In some cases, different cell lines exposed to the same treatments had different results. This was most readily evident in cells treated with cytochalasin D, which inhibits actin dynamics affecting endosome budding and movement (Lamaze et al., 1997; Radhakrishna and Donaldson, 1997) and in cells treated with Retro-2cycl, a small molecule recently found to block ER trafficking of the related AB5-subunit Shiga toxin (Figure 2, a and b) (Stechmann et al., 2010; Craig et al., 2017). In both cases, HEK293T epithelial cells responded to drug treatments, but K562 cells did not. We also note the enhancing effect of blebbistatin, a myosin II inhibitor (Straight et al., 2003), on retrograde trafficking. This result suggests the possibility, among others, that actin–myosin-based motility affects structure and function of some organelles or sorting steps in the pathway.
In all cases of small molecule treatment, there were no detectable effects of drug treatments on expression of the split-FP ER-HA-mNG21-10 as assessed by immunoblot (Supplemental Figure S2, a and b). Overall, our results highlight underappreciated, albeit in some cases expected, cell type-specific adaptations to endosome trafficking. They emphasize the value of the split FP-protein method for quantitative measurement of membrane transport with high degrees of temporal resolution.
Effects of ER structure: tubules and sheets
Retrograde vesicular trafficking to the ER necessarily entails factors intrinsic to ER structure and function. To test whether such functionality can be detected by the split-FP trafficking assay, we studied cells altered in ER morphology by overexpression of a mutant form of the dynamin-like GTPase ATLASTIN1 K80A, the coiled-coiled ER-luminal spacer protein CLIMP63, or DP1 (Figure 2e). Each of these ER intrinsic proteins affects the development and dynamics of ER tubules and sheets (Shibata et al., 2006)—amplifying ER tubules in cells overexpressing ATLASTIN1 K80A or DP1 and ER sheets in cells overexpressing CLIMP63 (Voeltz et al., 2006; Hu et al., 2009; Shibata et al., 2010). When retrograde trafficking of CTB-mNG211 was quantified in these cells, we found that overexpression of CLIMP63 and DP1 caused inhibition of transport into the ER compared with mock transfected cells (Figure 2f). Remarkably, retrograde trafficking from PM into the TGN was also affected (Figure 2g), implicating effects of ER structure on function of the closely related Golgi and TGN organelles. Overexpression of ATLASTIN1 K80A, in contrast, had reduced retrograde transport into the TGN but no detectable effect on trafficking into the ER. In all cases, expression levels of ER-HA-mNG21-10 in the different transfected cells were the same, as assessed by immunoblot (Figure 2h). The overall binding and uptake of Alexa-labeled CTxB was also assessed in each cell line under the same conditions—to control for this potentially confounding factor. There was no apparent effect on CTxB-Alexa uptake on overexpression of the various ER-altering proteins (Supplemental Figure S2c). Nevertheless, the results are shown normalized to the endocytic uptake of toxin among the different conditions. To test for the alternative possibility that the ER-shaping genes differentially affect membrane traffic by inducing an ER stress response, we used the same cell lines transfected with a luciferase reporter for IRE1α activation and XBP1 splicing (Yoshida et al., 2001; Iwawaki and Akai, 2006). Here, we found that CLIMP63 induced a mild ER stress response relative to thapsigargin treatment, but ATLASTIN1 K80A and DP1 did not (Supplemental Figure S2d). This also did not correlate with the observed phenotypes.
Thus, it appears that ER morphology can affect retrograde membrane transport into the ER. The different effects obtained by overexpression of ATLASTIN1 K80A and DP1 suggest that ER shape, on its own, cannot fully explain the mechanism, as both conditions are predicted to amplify ER tubules relative to sheets. And in the case of the ATLASTIN K80A mutant, the disparity in results between PM to TGN and PM to ER trafficking suggests an alternative pathway from PM to ER that can bypass the TGN, as implicated before (Llorente et al., 2003; Saslowsky et al., 2010).
Application to human disease
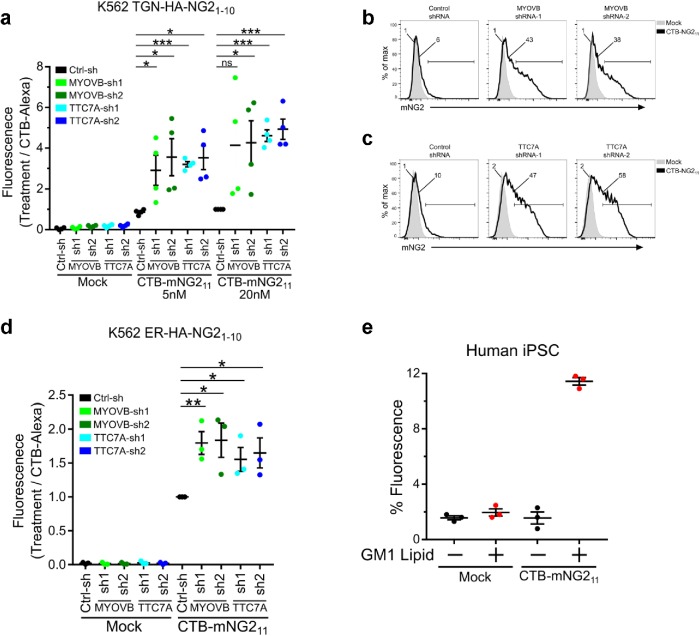
Alterations in cell polarity and trafficking genes underlie the pathophysiology of a number of poorly understood monogenic disorders (Li et al., 2005; Vilarino-Guell et al., 2011; Lo et al., 2015; Lesage et al., 2016). In the intestine, polarized trafficking is critical for normal transport and barrier function. Congenital diarrheas and enteropathies are a group of disorders resulting from monogenic mutations in intestinal epithelial genes (Thiagarajah et al., 2018), including a subset of disorders involving alterations in cell polarity and/or loss of apical membrane structure and function. These include loss of function mutations in MYO5B resulting in microvillus inclusion disease (Cutz et al., 1989; Muller et al., 2008) and in TTC7A (Chen et al., 2013; Samuels et al., 2013) resulting in infantile-onset enterocolitis. MYO5B function has been described to be important in the apical recycling pathway, but how MYO5B affects other trafficking pathways is incompletely understood. Loss of TTC7A causes altered determination of apical-basal polarity; however, there is currently no information about how the loss of this gene function impacts cellular trafficking. As all endomembrane structures of eukaryotic cells are essentially a network of interconnected pathways, we reasoned that measuring retrograde CTB-mNG211 trafficking in cells affected by these diseases may be informative of gene function and cellular phenotype. To test this, we used two unique shRNAs to knock down MYOVB or TTC7A in the human HEK293T cell line stably expressing TGN-HA-mNG21-10 and ER-HA- mNG21-10 (Supplemental Figure S3a). In both cases, and for both shRNAs against each gene, we found that trafficking from the PM into the TGN and ER was strikingly enhanced (Figure 3, a–d). Again, these results were controlled for total CTxB-Alexa uptake (Supplemental Figure SS3b). These results could not have been predicted. They raise interesting hypotheses for how trafficking networks are interconnected and for the subcellular mechanisms that underlie these gene-specific pathophysiologies.
FIGURE 3:
Retrograde trafficking in monogenic human disease. (a) Knockdown of MYOVB and TTC7a using two unique shRNAs in HEK293T cells stably expressing TGN-HA-mNG21-10 enhances retrograde trafficking to TGN. Data normalized to control shRNA at 20 nM CTB-mNG211/CTB Alexa Fluor 488 loading; n = 4 independent experiments. (b, c) Representative flow cytometry graphs from a. (d) Retrograde trafficking to ER as in a. n = 3 independent experiments. (e) Retrograde trafficking to TGN in human iPSCs stably expressing TGN-HA-mNG21-10 and treated or not with 2 μM GM1 C12:0. Error bars indicate ± SEM, ns not significant, *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed Student’s t test).
Another significant impediment in our understanding of intracellular trafficking in human diseases has been the lack of a quantitative assay to easily and quantitatively monitor membrane sorting in primary cells. To address this problem, we prepared human-induced pluripotent stem cells (iPSCs) lines stably expressing TGN-HA-mNG21-10. These undifferentiated cells, however, did not bind CTx in detectable amounts and gave no signal for retrograde transport (Figure 3e and Supplemental Figure S3, c and d). When the iPSCs cell plasma membranes were loaded with exogenous purified GM1 glycosphingolipids (in this case, GM1 containing short C12:0 acyl chains in the ceramide domain, termed GM1-C12:0), the iPSCs transported CTB-mNG211 into the TGN (Figure 3e, GM1 lipid-treated cells)—thus validating a primary human iPSCs model for assessing membrane dynamics.
Concluding remarks
Space and time are the two elements of membrane sorting that fundamentally underlie cell order. In large part, however, the timing (and rates) of intracellular membrane movement have been underexamined due to the challenges of quantifying membrane trafficking events with precision, sensitivity, and ease. Here, we define a method that overcomes these barriers for the retrograde pathways followed by GM1 and CTx. Importantly, we note the split-FP technology can be easily expanded to employ other toxins, lectins, or endogenous receptor ligands amenable to molecular modifications and recombinant expression—such as the bacterial Shiga (Sandvig et al., 1992), exotoxin A (Chaudhary et al., 1989), VacA (Lupetti et al., 1996; Zhang et al., 2019), and anthrax toxins (Petosa et al., 1997); the plant lectin ricin (Gonatas et al., 1975); and the extracellular signaling molecules epidermal growth factor or transferrin, which all follow different trafficking routes and engage other membrane processes underlying cellular function (O’Hare et al., 1987; Song et al., 2005; Tu et al., 2016; Zhang et al., 2019). In particular, the routes hijacked by VacA toxin from Helicobacter pylori (PM to mitochondria) (Cover and Blanke, 2005) and exotoxin A from Pseudomonas aeruginosa (PM to nucleus) (Chaumet et al., 2015) are good examples of well-observed but poorly understood pathways that may be discerned using the split-FP technology.
Furthermore, systems level understanding of trafficking networks and dynamics require large data sets of quantitative measurements on membrane movement over space and time and across different cell types and cell states. Similar to systemwide approaches on modeling signaling transduction networks (Kholodenko, 2006), the development of high throughput and quantitative trafficking technologies that assay for these factors will enable a fuller understanding of membrane traffic and sorting. We emphasize this approach can be applied in automation, in high throughput, and for gene or small molecule discovery, and the method has the power to compare endocytic networks across diverse immortalized cell lines or primary cells derived from normal or disease-affected individuals. These features, we argue, will enable systemwide analysis of membrane dynamics underlying cell function in health and disease with the potential for clinical translation.
MATERIALS AND METHODS
Plasmids
CTB subunit was cloned into pET28a vector (Clontech). mNG211 (and sfCherry211 strands) were fused to CTx A2 strand and cloned into pGEXTEV-SUMO vector. Single 1× and tandem 2× and 5× mNG211 were fused to the C-terminus of catalytically inactive (EE/DD) CTx A1 strand and cloned into pET24a (Clontech) with a His-SUMO tag. CTx A2 strand was cloned into pGEXTEV (a gift from Kim Orth, UT Southwestern). TGN-HA-mNG21-10 was cloned by fusion of GALT sequence from GAL-transferase and a HA epitope to the N-terminus of mNG21-10 into Ef1a-pHAGE-IRES-blasticidin vector derived from Ef1a-pHAGE-IRES-ZsGreen (obtained from the Harvard Plasmid Repository). The GFP was removed from the EF1a-pLVXGFP-N1 (Clontech) backbone and replaced with the human codon-optimized TGN-FLAG-sfmCherry21-10. CMV-pHAGE-IRES-Zsgreen ER- HA-mNG21-10 was fused with ER targeting signal sequence from BIP at the N-terminal and a C-terminal ER retention KDEL and cloned into Ef1a-pHAGE-IRES-blasticidin. ATLASITN1, CLIMP63, and DP1 cDNAs were obtained from the Harvard Plasmid Repository and cloned as HA-tagged fusion into CMV-pHAGE-IRES-ZsGreen (Harvard Plasmid Repository) or Ef1a-pHAGE-IRES-mCherry. ER-mCherry was cloned into pLVX-Puro (Clontech) using ER targeting signal sequence from BIP at the N-terminal and a C-terminal ER retention KDEL. Control short hairpin RNA (shRNA) targeting luciferase, MYOVB, and TTC7A were cloned into Ef1a-Puro-2a-mCherry-U6-shRNA vector. psPAX2 (Addgene plasmid 12260) was a gift from Didier Trono, EPFL. pCMV-VSVG was a gift from Bob Weinberg, MIT (Addgene 8454).
Protein purification
Recombinant CTB-mNG211 was expressed and purified by cotransformation of CTB pET28a and mNG211-CTA2 pGEXTEV-SUMO into Shuffle T7 Express (NEB). Transformed cells were grown overnight in a 30°C incubator and restreaked onto a new plate and regrown overnight at 30°C. Next day, cells were inoculated into 2× YT and shook for 3–5 h at 35°C. Starter culture was used to inoculate into larger culture until OD600 = 1.0–1.2. Cells were induced at 18°C for 16–20 h with 0.4 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cells were then collected and resuspended in 10 mM Tris, pH 8, 100 mM NaCl, 0.5% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride and passed through an Emulsiflex C5 homogenizer (Avestin). The lysate was spun at 15,000 × g for 10 min, and supernatant was collected and used for purification by glutathione resin at 4°C. Eluted protein was cleaved with ULP1 protease and dialyzed overnight at 4°C in 10 mM Tris, pH 8, 100 mM NaCl. Next day, protein was dialyzed to 5 mM Na2PO4, pH 7, and purified by HiTrap SP HP column (GE Healthcare) using SP column Buffer A: 5 mM Na2PO4, pH 7, and gradient elution with Buffer B: 5 mM Na2PO4, pH 7, 1 M NaCl. Bound SP fractions containing CTB-mNG211 were collected and run on SDS–PAGE gel and stained with Coomassie Blue reagent. Recombinant CTB-sfmCherry211 was expressed as above but dialyzed in 10 mM Tris, pH 8.5, 150 mM NaCl overnight with ULP1 protease and then to 10 mM Tris, pH 8.5, 5 mM NaCl before purification by HiTrap Q HP column. Separation was performed using Buffer A: 20 mM Tris, pH 8.5, and Buffer B: 20 mM Tris, pH 8.5, 1 M NaCl. Recombinant purified CTA2-CTB complex was generated using CTA2 pGEXTEV and CTB in pET28a. Recombinant CTA2-CTB was cotransformed, expressed, and purified as above but with TEV protease to remove GST tag. Holotoxin CT-1×mNG211, CT-2×mNG211, and CT-5×mNG211 were expressed and purified by expressing separately CTA1-1×mNG211, CTA1-2×mNG211, or CTA1-5×mNG211 subunits. CTA1 subunits were expressed and purified as above but with TEV protease to remove His6 tag cobalt resin. The CTA1 subunit was then incubated with purified CTA2-CTB complex overnight at 4°C in 10 mM Tris, pH 8, 150 mM NaCl to promote holotoxin assembly. Next day protein was dialyzed to S column A buffer. Holotoxin fractions were collected, and buffer was exchanged to Q column A buffer, 20 mM Tris, pH 8.5. Holotoxin fractions were collected using the HiTrap Q HP (GE Healthcare) separated by a salt gradient of Buffer B: 20 mM Tris, pH 8.5, 1 M NaCl. All recombinant protein were aliquoted and flash frozen by liquid nitrogen. For nucleotide sequence of split-FP toxin constructs, see Supplemental Table S1.
Cell culture
K562 cells were grown in RPMI + 10% fetal bovine serum (FBS) pen/strep. HEK293T and COS7 cells were grown in DMEM + 10% FBS pen/strep. COS7 cells stably expressing ER-mCherry and TGN-HA-mNG21-10 or ER-HA-mNG21-10 were generated by lentiviral transduction by using packaging plasmids psPAX2 and pVSVG. K562 and HEK293Tcells were generated by lentivirus transduction and drug selection for TGN-HA-mNG21-10, TGN-FLAG-sfmCherry21-10, or ER-HA-mNG21-10. Human iPSCs growth and maintenance were followed as described previously in Zhang et al. (2013). Human iPSCs were transduced and selected with blasticidin for transduced cells.TGN-HA-mNG21-10 lentivirus was concentrated by low-speed centrifugation before transduction to iPSCs (Jiang et al., 2015). Maintenance and incubation of cells were performed at 37°C + 5% CO2.
Western blotting
Cells were treated with respective treatments, harvested, and washed with 1× phosphate-buffered saline (PBS). Cells were lysed with 1× RIPA, 2 mM EDTA, and protease inhibitors. Lysates were spun at 20,000 × g for 5 min and supernatant collected. Sample buffer was added and heated for 10 min at 95°C. Samples were run on SDS–PAGE gels and immunoblotted with anti-HA for mNG21-10, anti-actin as loading control. Proteins were detected using SuperSignal West Femto (Fisher Scientific). Images were taken with the Azure c300 system.
Confocal imaging
Confocal images were taken using a spinning disk confocal head coupled to an inverted Zeiss Axiovert 200 M microscope. To determine localization of stably expressing ER-mCherry and trans-Golgi and ER mNG21-10, we utilized COS7 cells, which are commonly used for membrane trafficking and ER imaging studies. Cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% saponin; anti-HA (Roche) was used to stain for mNG21-10 and anti-Golgin97. To determine ER shape morphology of ER shape genes, COS7 cells stably expressing ER-mCherry were transfected with ALASTIN1-K80A-IRES-ZsGreen, CLIMP63-IRES-ZsGreen, or DP1-IRES-ZsGreen. Cells were fixed and imaged as above. COS7 cells stably expressing ER-mCherry and Sec61β-HA-mNG21-10, TGN-HA-mNG21-10, or ER-HA-mNG21-10 were treated with recombinant CTB-mNG211 for 6 h. Cells were fixed, stained, and imaged as above. Pearson’s and Mander’s calculations were analyzed and calculated by Slidebook software (Intelligent 3i) using deconvolved images to generate masks by automatic threshold with the Otsu method.
Time-course retrograde assays
K562 cells stably expressing ER-HA-mNG21-10 were incubated with 10 nM CTB-mNG211 at indicated times. Cells were spun down, washed with 1× PBS and resuspended in 1× PBS 2% FBS for flow cytometry. HEK293T and COS7 cells expressing Sec61β-HA-mNG21-10 or ER-HA-mNG21-10 were incubated with 20 nM CTB-mNG211 at indicated times. Cells were collected and resuspended in 1× PBS 2% FBS for flow cytometry. Pulse-chase of CTB-mNG211 was performed by incubation of CTB-mNG211 for 2 h and incubated with cell culture media for 20 h before assayed by flow cytometry. Fluorescence positive gates were positioned relative to mock treated cells for all flow experiments.
Toxin dose retrograde assays
K562 cells stably expressing ER-HA-mNG21-10 were treated with increasing concentrations of CTB-mNG211 and incubated for 4 h at 37°C. Cells were collected and assayed by flow cytometry as above. COS7 and HEK293T cells stably expressing ER-HA-mNG21-10 were treated with increasing concentrations of CTB-mNG211 and incubated for 5 h at 37°C. Cells were collected and assayed by flow cytometry. K562 cells stably expressing TGN-FLAG-sfmCherry211 were incubated 20 nM CTB-sfmCherry211 for 5 h and assayed by flow cytometry.
Small molecular inhibitor retrograde assay
K562 cells stably expressing ER-HA-mNG21-10 were pretreated with each compound for 30 min before incubation of 10 nM CTB-mNG211 for an additional 4 h. HEK293T cells expressing ER-HA-mNG21-10 were treated as above but incubated with 20 nM CTB-mNG211 for 5 h. The final inhibitor concentrations used were DTT (4 mM), tunicamycin (5 μg/ml), BFA (5 μg/ml), Retro-2cycl (25 μM), thapsigargin (300 nM), nocodazole (1 μM), cytochalasin D (10 μM), and blebbistatin (50 μM). Figure 2, a–d, displays a positive percentage of fluorescence measurements pooled from independent experiments and normalized relative to CTB-mNG211 with no inhibitor treatment.
ER-shape retrograde assays
Lentivirus packaged with EF1a-IRES-mCherry empty vector, HA-ATLASTIN-K80A, HA-CLIMP63, and HA-DP1 were transduced into HEK293T stably expressing ER-HA-mNG21-10 cells. Retrograde assays were performed within 48–96 h after empty vector or ER shape gene transduction. Cells were incubated with 5 and 20 nM of CTB-mNG211 for 5 h. Cells were washed with 1× PBS and harvested for flow cytometry. CTB-Alexa Fluor 488 (2 nM) was treated for each cell line to act as a toxin loading control. An individual experiment was normalized using the ratio of CTB-mNG211/CTB-Alexa Fluor 488 to compare between cell lines and treatments. Data pooled from independent experiments were normalized relative to 20 nM CTB-mNG211 empty vector treatment.
Monogenic genes retrograde assay
Lentivirus packaged with shRNA against control targeting luciferase (Ctrl-sh) and two unique shRNAs each for MYOVB and TTC7A were produced. HEK293T cells stably expressing TGN-HA-mNG21-10 were transduced with shRNA packaged lentivirus. Forty-eight hours after transduction, cells were selected for transduction and then plated for PM to Golgi retrograde assay the next day. Cells were incubated with 20 nM CTB-mNG211 for 4 h, washed, and harvested to assay by flow cytometry. CTB-Alexa (2 nM) was used as loading control. Data were calculated as above but normalized relative to control shRNA treated with 20 nM CTB-mNG211. For oligo nucleotide sequences of shRNAs, see Supplemental Table S2.
Primary cells retrograde assay
The control iPSC line, GON0515-03, is derived from a 30-yr-old male (IRB# 10-02-0053) using Sendai virus reprogramming method, karyotypically normal at passage 5, and karyotyped after transducing with the Golgi split-mNG2 at passage 21. Human iPSCs were plated on Matrigel-coated plates with 10 μM rock inhibitor and iPSC-conditioned media. Cells were ± incubated with C12:0 GM1 lipid, which is known to traffic to the TGN. Lipid loading to cells was performed with 1:1 ratio of 2 μM lipid and 2 μM defatted bovine serum albumin resuspended in Advanced DMEM F12 for 10 min at 37°C. Cells were washed and then incubated with CTB-mNG211 for 5 h at 37°C. Cells were collected with Accutase and resuspended in 1× PBS + 2% FBS and analyzed by flow cytometry.
Z-factor determination
Fifteen biological replicates each for negative (Mock) and positive (CTB-mNG211) conditions were used to calculate Z-factor scores for the trans-Golgi and ER retrograde assay. For the trans-Golgi Z-factor experiment, K562 cells stably expressing TGN-HA-mNG21-10 were incubated with 10 nM CTB-mNG211 for 5 h. Cells were spun and washed before assay by flow cytometry. PM to ER retrograde Z-factor experiment was performed as above but with ER-HA-mNG21-10 at 4 h. Z-factor was calculated with the equation
: Z = 1 – 3 SD of sample ± 3 SD of control
│mean of sample – mean of control│
XBP1 luciferase assay
Firefly luciferase and renilla luciferase reagent was assayed with homemade luciferase reagent as stated previously (Baker and Boyce, 2014). HEK293T cells were transfected with empty vector or corresponding ER-shape genes and XBP1-luciferase reporter and renilla luciferase with Fugene HD (Promega). Cells treated for 4 h with thapsigargin (300 nM) served as positive control.
qRT-PCR
RNA was isolated by TRIzol (ThermoFisher) reverse transcribed with iScript cDNA Synthesis Kit (Bio-Rad) and qPCR by SYBR Green (Bio-Rad) on a CFX384 (Bio-Rad). Samples were normalized to actin. For qPCR primer sequences, see Supplemental Table S2.
Statistical analysis
Significance was assessed using two-tailed t test <0.05 was considered significant. Graphs were generated using GraphPad Prism.
Data availability
Data that support the findings of this study are available from the corresponding author on request. Relevant DNA sequences are listed in the Supplemental Information.
Supplementary Material
Acknowledgments
We thank Michael J. Grey, Xiaomo Jiang, Stefanie S. Schmieder, and Jamie LeBarron for discussion and reagents. We thank Joseph Gonzalez-Heydrich and Catherine Brownstein for sharing the iPSC line and BCH IDDRC U54HD090255. This work is also supported by the American Liver Foundation and by National Institutes of Health (NIH) grants F32AA025271 (P.J.W.), R01CA205158 (S.H.H.), R01DK090311, R01DK105198, and R24OD017870; the Claudia Adams Barr Program in Innovative Basic Cancer Research and the Pew Scholar in the Biomedical Sciences (W.G.); K08DK113106, AGA Research Scholar Award, and BCH Faculty Career Development Award (J.R.T.); the Crohn’s and Colitis Foundation and a Rubin-Wolpow Fellowship; NIH grants T32HD007466 (P.L.), DK048106, DK084424, and the Clinical Translational Research Program of the Harvard Digestive Disease Center (S.B.S.); Harvard Digestive Diseases Center P30 DK034854 (Core C) (W.I.L.); and RC2DK118640 (J.R.T. and W.I.L.).
Abbreviations used:
- BFA
brefeldin A
- CTx
cholera toxin
- ER
endoplasmic reticulum
- EV
empty vector
- FBS
fetal bovine serum
- HEK
human embryonic kidney
- iPSC
induced pluripotent stem cell
- NG2
split neon green2
- PBS
phosphate-buffered saline
- split-FP
split fluorescent protein
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-07-0375) on November 27, 2019.
REFERENCES
- Andrews NW, Almeida PE, Corrotte M. (2014). Damage control: cellular mechanisms of plasma membrane repair. Trends Cell Biol , 734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Chapman ER. (2004). The C2 domains of synaptotagmin–partners in exocytosis. Trends Biochem Sci , 143–151. [DOI] [PubMed] [Google Scholar]
- Baker JM, Boyce FM. (2014). High-throughput functional screening using a homemade dual-glow luciferase assay. J Vis Exp , 50282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FA. (1999). A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr Biol , 381–384. [DOI] [PubMed] [Google Scholar]
- Chaudhary VK, Queen C, Junghans RP, Waldmann TA, FitzGerald DJ, Pastan I. (1989). A recombinant immunotoxin consisting of two antibody variable domains fused to Pseudomonas exotoxin. Nature , 394–397. [DOI] [PubMed] [Google Scholar]
- Chaumet A, Wright GD, Seet SH, Tham KM, Gounko NV, Bard F. (2015). Nuclear envelope-associated endosomes deliver surface proteins to the nucleus. Nat Commun , 8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Giliani S, Lanzi G, Mias GI, Lonardi S, Dobbs K, Manis J, Im H, Gallagher JE, Phanstiel DH, et al. (2013). Whole-exome sequencing identifies tetratricopeptide repeat domain 7A (TTC7A) mutations for combined immunodeficiency with intestinal atresias. J Allergy Clin Immunol , 656–664.e617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham DE. (2007). Calcium signaling. Cell , 1047–1058. [DOI] [PubMed] [Google Scholar]
- Cover TL, Blanke SR. (2005). Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat Rev Microbiol , 320–332. [DOI] [PubMed] [Google Scholar]
- Craig E, Huyghues-Despointes CE, Yu C, Handy EL, Sello JK, Kima PE. (2017). Structurally optimized analogs of the retrograde trafficking inhibitor Retro-2cycl limit Leishmania infections. PLoS Negl Trop Dis , e0005556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutz E, Rhoads JM, Drumm B, Sherman PM, Durie PR, Forstner GG. (1989). Microvillus inclusion disease: an inherited defect of brush-border assembly and differentiation. N Engl J Med , 646–651. [DOI] [PubMed] [Google Scholar]
- Feng S, Sekine S, Pessino V, Li H, Leonetti MD, Huang B. (2017). Improved split fluorescent proteins for endogenous protein labeling. Nat Commun , 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga Y, Wolf AA, Rodighiero C, Wheeler H, Tsai B, Allen L, Jobling MG, Rapoport T, Holmes RK, Lencer WI. (2003). Gangliosides that associate with lipid rafts mediate transport of cholera and related toxins from the plasma membrane to endoplasmic reticulm. Mol Biol Cell , 4783–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonatas NK, Steiber A, Kim SU, Graham DI, Avrameas S. (1975). Internalization of neuronal plasma membrane ricin receptors into the Golgi apparatus. Exp Cell Res , 426–431. [DOI] [PubMed] [Google Scholar]
- Guimaraes CP, Carette JE, Varadarajan M, Antos J, Popp MW, Spooner E, Brummelkamp TR, Ploegh HL. (2011). Identification of host cell factors required for intoxication through use of modified cholera toxin. J Cell Biol , 751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C. (2009). A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell , 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Akai R. (2006). Analysis of the XBP1 splicing mechanism using endoplasmic reticulum stress-indicators. Biochem Biophys Res Commun , 709–715. [DOI] [PubMed] [Google Scholar]
- Jiang W, Hua R, Wei M, Li C, Qiu Z, Yang X, Zhang C. (2015). An optimized method for high-titer lentivirus preparations without ultracentrifugation. Sci Rep , 13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L, Tenza D, Antony C, Goud B. (1997). Retrograde transport of KDEL-bearing B-fragment of Shiga toxin. J Biol Chem , 19554–19561. [DOI] [PubMed] [Google Scholar]
- Kholodenko BN. (2006). Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol , 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamaze C, Fujimoto LM, Yin HL, Schmid SL. (1997). The actin cytoskeleton is required for receptor-mediated endocytosis in mammalian cells. J Biol Chem , 20332–20335. [DOI] [PubMed] [Google Scholar]
- Lencer WI, de Almeida JB, Moe S, Stow JL, Ausiello DA, Madara JL. (1993). Entry of cholera toxin into polarized human intestinal epithelial cells. Identification of an early brefeldin A sensitive event required for A1-peptide generation. J Clin Invest , 2941–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencer WI, Delp C, Neutra MR, Madara JL. (1992). Mechanism of cholera toxin action on a polarized human intestinal epithelial cell line: role of vesicular traffic. J Cell Biol , 1197–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencer WI, Tsai B. (2003). The intracellular voyage of cholera toxin: going retro. Trends Biochem Sci , 639–645. [DOI] [PubMed] [Google Scholar]
- Lesage S, Drouet V, Majounie E, Deramecourt V, Jacoupy M, Nicolas A, Cormier-Dequaire F, Hassoun SM, Pujol C, Ciura S, et al. (2016). Loss of VPS13C Function in autosomal-recessive Parkinsonism causes mitochondrial dysfunction and increases PINK1/Parkin-dependent mitophagy. Am J Hum Genet , 500–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Tiab L, Jiao X, Munier FL, Zografos L, Frueh BE, Sergeev Y, Smith J, Rubin B, Meallet MA, et al. (2005). Mutations in PIP5K3 are associated with Francois-Neetens mouchetee fleck corneal dystrophy. Am J Hum Genet , 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente A, Lauvrak SU, van Deurs B, Sandvig K. (2003). Induction of direct endosome to endoplasmic reticulum transport in Chinese hamster ovary (CHO) cells (LdlF) with a temperature-sensitive defect in epsilon-coatomer protein (epsilon-COP). J Biol Chem , 35850–35855. [DOI] [PubMed] [Google Scholar]
- Lo B, Zhang K, Lu W, Zheng L, Zhang Q, Kanellopoulou C, Zhang Y, Liu Z, Fritz JM, Marsh R, et al. (2015). AUTOIMMUNE DISEASE. Patients with LRBA deficiency show CTLA4 loss and immune dysregulation responsive to abatacept therapy. Science , 436–440. [DOI] [PubMed] [Google Scholar]
- Lupetti P, Heuser JE, Manetti R, Massari P, Lanzavecchia S, Bellon PL, Dallai R, Rappuoli R, Telford JL. (1996). Oligomeric and subunit structure of the Helicobacter pylori vacuolating cytotoxin. J Cell Biol , 801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller T, Hess MW, Schiefermeier N, Pfaller K, Ebner HL, Heinz-Erian P, Ponstingl H, Partsch J, Rollinghoff B, Kohler H, et al. (2008). MYO5B mutations cause microvillus inclusion disease and disrupt epithelial cell polarity. Nat Genet , 1163–1165. [DOI] [PubMed] [Google Scholar]
- O’Hare M, Roberts LM, Thorpe PE, Watson GJ, Prior B, Lord JM. (1987). Expression of ricin A chain in Escherichia coli. FEBS Lett , 73–78. [DOI] [PubMed] [Google Scholar]
- Petosa C, Collier RJ, Klimpel KR, Leppla SH, Liddington RC. (1997). Crystal structure of the anthrax toxin protective antigen. Nature , 833–838. [DOI] [PubMed] [Google Scholar]
- Radhakrishna H, Donaldson JG. (1997). ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J Cell Biol , 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels ME, Majewski J, Alirezaie N, Fernandez I, Casals F, Patey N, Decaluwe H, Gosselin I, Haddad E, Hodgkinson A, et al. (2013). Exome sequencing identifies mutations in the gene TTC7A in French-Canadian cases with hereditary multiple intestinal atresia. J Med Genet , 324–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig K, Garred O, Prydz K, Kozlov JV, Hansen SH, van Deurs B. (1992). Retrograde transport of endocytosed Shiga toxin to the endoplasmic reticulum. Nature , 510–512. [DOI] [PubMed] [Google Scholar]
- Sandvig K, Garred O, van Deurs B. (1996). Thapsigargin-induced transport of cholera toxin to the endoplasmic reticulum. Proc Natl Acad Sci USA , 12339–12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saslowsky DE, Cho JA, Chinnapen H, Massol RH, Chinnapen DJ, Wagner JS, De Luca HE, Kam W, Paw BH, Lencer WI. (2010). Intoxication of zebrafish and mammalian cells by cholera toxin depends on the flotillin/reggie proteins but not Derlin-1 or -2. J Clin Invest , 4399–4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. (2010). Mechanisms determining the morphology of the peripheral ER. Cell , 774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Voeltz GK, Rapoport TA. (2006). Rough sheets and smooth tubules. Cell , 435–439. [DOI] [PubMed] [Google Scholar]
- Song S, Xue J, Fan K, Kou G, Zhou Q, Wang H, Guo Y. (2005). Preparation and characterization of fusion protein truncated Pseudomonas exotoxin A (PE38KDEL) in Escherichia coli. Protein Expr Purif , 52–57. [DOI] [PubMed] [Google Scholar]
- Sorensen JB. (2017). Ride the wave: retrograde trafficking becomes Ca(2+) dependent with BAIAP3. J Cell Biol , 1887–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner RA, Smith DC, Easton AJ, Roberts LM, Lord JM. (2006). Retrograde transport pathways utilised by viruses and protein toxins. Virol J , 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stechmann B, Bai SK, Gobbo E, Lopez R, Merer G, Pinchard S, Panigai L, Tenza D, Raposo G, Beaumelle B, et al. (2010). Inhibition of retrograde transport protects mice from lethal ricin challenge. Cell , 231–242. [DOI] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. (2003). Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science , 1743–1747. [DOI] [PubMed] [Google Scholar]
- Thiagarajah JR, Kamin DS, Acra S, Goldsmith JD, Roland JT, Lencer WI, Muise AM, Goldenring JR, Avitzur Y, Martin MG, et al. (2018). Advances in evaluation of chronic diarrhea in infants. Gastroenterology , 2045–2059.e2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgersen ML, Skretting G, van Deurs B, Sandvig K. (2001). Internalization of cholera toxin by different endocytic mechanisms. J Cell Sci , 3737–3747. [DOI] [PubMed] [Google Scholar]
- Tu W, Li T, Wang Q, Cai K, Gao X, Wang H. (2016). A simple method for expression and purification of Shiga toxin 1 (Stx1) with biological activities by using a single-promoter vector and native signal peptide. Biotechnol Appl Biochem , 539–545. [DOI] [PubMed] [Google Scholar]
- Vilarino-Guell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, Soto-Ortolaza AI, Cobb SA, Wilhoite GJ, Bacon JA, et al. (2011). VPS35 mutations in Parkinson disease. Am J Hum Genet , 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell , 573–586. [DOI] [PubMed] [Google Scholar]
- Wernick NL, Chinnapen DJ, Cho JA, Lencer WI. (2010). Cholera toxin: an intracellular journey into the cytosol by way of the endoplasmic reticulum. Toxins (Basel) , 310–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell , 881–891. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. (1999). A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen , 67–73. [DOI] [PubMed] [Google Scholar]
- Zhang X, Jiang S, Mitok KA, Li L, Attie AD, Martin TFJ. (2017). BAIAP3, a C2 domain-containing Munc13 protein, controls the fate of dense-core vesicles in neuroendocrine cells. J Cell Biol , 2151–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, et al. (2013). Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron , 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Zhang H, Li S, Pintilie GD, Mou TC, Gao Y, Zhang Q, van den Bedem H, Schmid MF, Au SWN, et al. (2019). Cryo-EM structures of Helicobacter pylori vacuolating cytotoxin A oligomeric assemblies at near-atomic resolution. Proc Natl Acad Sci USA , 6800–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study are available from the corresponding author on request. Relevant DNA sequences are listed in the Supplemental Information.