SUMMARY
RNA binding proteins (RBPs) comprise a large class of over 2000 proteins that interact with transcripts in all manner of RNA-driven processes. The structures and mechanisms that RBPs use to bind and regulate RNA are incredibly diverse. In this review we take a look at the components of protein-RNA interaction, from the molecular level to multi-component interaction. We first summarize what is known about protein-RNA molecular interactions based on analyses of solved structures. We additionally describe software currently available for predicting protein-RNA interaction and other resources useful for the study of RBPs. We then review the structure and function of seventeen known RNA binding domains and analyze the hydrogen bonds adopted by protein-RNA structures on a domain by domain basis. We conclude with a summary of the higher-level mechanisms that regulate protein-RNA interactions.
INTRODUCTION
RNA binding proteins (RBPs) potently and ubiquitously regulate transcripts throughout their life cycle (Lorković, 2012). RBP interactions with RNA range from single protein-RNA element interaction to the assembly of multiple RBPs and RNA molecules, such as the spliceosome. How RBPs selectively bind their targets is not always understood, although there are currently many techniques used to study these interactions. X-ray crystallography and nuclear magnetic resonance (NMR) experiments facilitate precise study of the amino acids and nucleotides that interact in protein-RNA complexes, and numerous such datasets have been generated for RBP domains in complex with RNA (Berman et al., 2000). Analyses of these data have inferred the number and types of intermolecular interactions and preferred amino acids that characterize specific protein-RNA binding (Han and Nepal, 2007; Perez-Cano and Fernandez-Recio, 2010). Furthermore, numerous studies have built on protein-RNA structural data to develop increasingly accurate software that predicts which residues in protein interact with RNA. We include a description of up-to-date software and data resources for the purpose of predicting and studying how RBPs interact with RNA.
RNA binding domains in protein are the functional units responsible for binding RNA. Multiple such domains often occur in a single RBP and these modular arrangements can coordinate and enhance binding to RNA (Cléry and Allain, 2011; Lunde et al., 2007). Additionally, RBPs tend to be enriched in intrinsically disordered regions, which themselves act as RNA-binding domains but limit the structural study of RBPs to ordered domains rather than full-length protein (Jarvelin et al., 2016). Several ordered domains have been studied for decades, although it is important to note that RNA binding domains are remarkably heterogeneous and can be difficult to classify (Gerstberger et al., 2014). Additionally, many domains remain to be characterized, where hundreds of RBPs lack known RNA-binding domains (Castello et al., 2016). Here we overview the strategies that seventeen well-characterized RNA binding domains use to achieve RNA binding. Furthermore, we present an analysis on the preferences in protein-RNA hydrogen bonds for eight of these domain types.
RBP binding ultimately achieves a range of cellular goals (Gerstberger et al., 2014; Glisovic et al., 2008), but many mechanisms—and many chances for regulation—lie in between binding and biological consequence. These mechanisms we categorize into several layers: protein-RNA assembly, combined action of the ribonucleoprotein, and modifications and interactions that regulate the previous two (Lovci et al., 2016; Lunde et al., 2007; Thapar, 2015). Here we describe these high-level processes and provide functional examples (Fiorini et al., 2015; Jackson et al., 2010; Sledz and Jinek, 2016), all of which are discovered by intense and detailed biochemical work, including by insights from protein-RNA structures. These summaries intersect the areas of study that enable a mechanistic understanding of RBP regulation and we hope serve as a useful and timely resource.
PROTEIN-RNA MOLECULAR INTERACTION
To understand RBP regulation of RNA targets one must understand the biochemical underpinnings that facilitate exact and specific interaction with these sites. RNA binding proteins (RBPs) bind their RNA targets through the molecular interactions of chemical moieties between protein residues and RNA nucleotides. At this resolution the distinction between RNA and protein begins to blend, as the same intermolecular forces that shape protein and RNA tertiary structures also stitch the two molecules together. These interactions occur dynamically, with sometimes quite large rearrangements in RNA and protein (Hainzl et al., 2005; Leulliot and Varani, 2001). In this section we will provide a detailed description of the molecular interactions that occur in protein-RNA structures and overview trends determined by previous research. We will also catalogue software that uses molecular level interaction data to predict protein-RNA binding.
Hydrogen bonds and Van der Waals
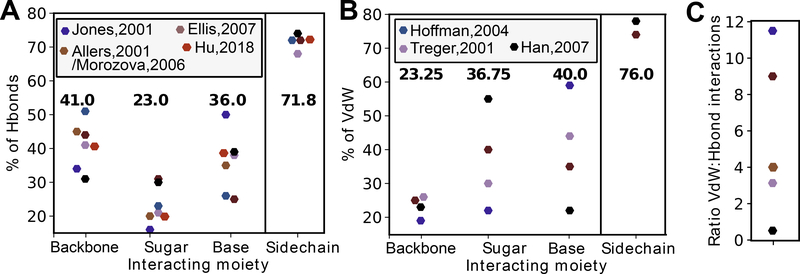
Hydrogen bonds and Van der Waals interactions have been extensively analyzed in protein-RNA interactions (Gupta and Gribskov, 2011; Han and Nepal, 2007; Hoffman et al., 2004; Perez-Cano and Fernandez-Recio, 2010; Treger and Westhof, 2001). Hydrogen bonds form between an electronegative atom bound to a hydrogen atom, whose partial positive charge attracts an electronegative partner. Hydrogen bonds can be formed by both neutral and ionic groups, and can be coordinated by water molecules (Figure 1B). They generally form at distances of 2.4–3.0 Å, contributing 0.5–4.5 kcal/mol per bond (Auweter et al., 2006). The weakest hydrogen bonds are considered to be Van der Waals (VdW) interactions, which are weak (0.5–1 kcal/mol) electrostatic interactions that occur above ~3.0 Å. All the studies that analyze VdW and hydrogen bonds in protein-RNA structures identify hydrogen bonds with HBPLUS (McDonald and Thornton, 1994) and identify VdW as the hydrogen bonds above a threshold donor-acceptor distance (Allers and Shamoo, 2001; Ellis et al., 2007; Han and Nepal, 2007; Hu et al., 2018; Jones et al., 2001; Morozova et al., 2006; Treger and Westhof, 2001). All RNA bases, the 2’-OH, and the phosphodiester backbone can form hydrogen bonds and VdW with protein (Figure 1A–C)(Teplova et al., 2011). Multiple analyses of hydrogen bond types in protein-RNA structures have found that hydrogen bonds with the base, 2’-OH (sugar), and the phosphate (RNA backbone) account for an average of 35.5%, 23.5%, and 41% of protein-RNA hydrogen bonds, respectively (Figure 2A) (Gupta and Gribskov, 2011; Han and Nepal, 2007; Hoffman et al., 2004; Treger and Westhof, 2001). Studies of VdW percentages with base, sugar, and phosphate are more variable (Figure 2B), perhaps reflecting inconsistent thresholds in categorizing VdW interactions.
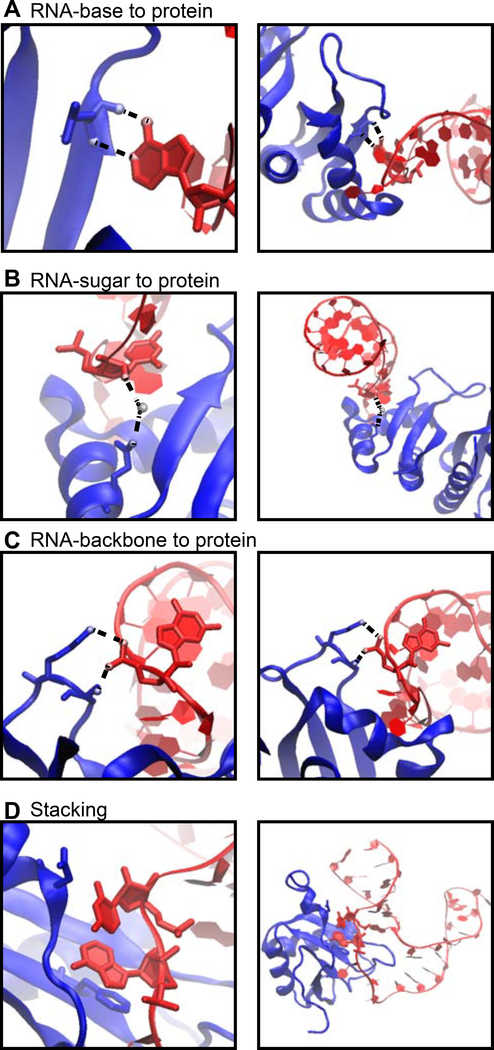
Figure 1.
Examples of protein-RNA hydrogen bonds and stacking interactions. The KH domain of human NOVA1 (PDB ID: 2ANR) (Teplova et al., 2011) and human U1A (PDB ID: 1AUD) (Oubridge et al., 1994) visualized with VMD (Humphrey et al., 1996) in detailed (left) and zoomed-out (right) perspectives. RNA in red, protein in blue. (A) Mainchain atoms of a Leu form hydrogen bonds with adenine. (B) Hydrogen bonds form between Gln and the 2’-OH of a cytosine, bridged by a water molecule. (C) Two hydrogen bonds form between the phosphate backbone atoms of guanine and Ser and Lys. (D) An adenine and cytosine in an unpaired loop stack between Asp and Phe.
Figure 2.
Meta-analysis of seven studies analyzing hydrogen bonds and Van der Waals interactions (VdW) in protein-RNA structures. (A) Reports across studies of the percent of hydrogen bonds in protein-RNA structures that occur with the RNA backbone (phosphate), sugar (2’-OH), or base. The percent of hydrogen bonds that occur with the protein sidechain (as opposed to the mainchain). Averages shown above each category. (B) Reports of the percent of VdWs in protein-RNA structures that occur with the RNA backbone (phosphate), sugar (2’-OH), or base. The percent of VdWs that occur with the protein sidechain (as opposed to the mainchain). Averages shown above each category. (C) Reports across studies of the average ratio of VdWs to hydrogen bonds per protein-RNA structure.
Proteins can interact with RNA using the mainchain of any residue and the sidechains of most residues. Studies have consistently found that the protein sidechain, versus mainchain, is employed in 71.5% of hydrogen bonds and 76% of VdW with RNA (Figure 2A–B). Polar amino acids Ser and Asn and positively charged amino acids Lys and Arg, which form strong ionic hydrogen bonds (salt bridges), predominate these interactions (Gupta and Gribskov, 2011; Han and Nepal, 2007; Hoffman et al., 2004; Perez-Cano and Fernandez-Recio, 2010; Treger and Westhof, 2001). VdW generally share the same preferences for amino acids that are observed for hydrogen bonds (Ellis et al., 2007; Han and Nepal, 2007; Jones et al., 2001; Treger and Westhof, 2001). In the overall set of interactions that occur at a protein-RNA interface, VdW are thought to predominate, although estimates of the ratio of VdW to hydrogen bond interactions per protein-RNA complex vary quite a bit (Figure 2C).
Hydrophobic, π-interactions, and stacking
Hydrophobic interactions occur at distances of 3.8–5.0 Å (Morozova et al., 2006; Onofrio et al., 2014) and contribute 1–2 kcal/mol per interaction (Dill et al., 2008). Hydrophobic interactions between RNA bases and hydrophobic sidechains can be important stabilizing factors at protein-RNA interfaces by sequestering hydrophobic residues and bases from solvent to form a “hydrophobic core” (Akopian et al., 2013; Allain et al., 1997; Yang et al., 2002; Yu et al., 2014). For example, the SRP54 “M” binding domain forms a methionine-rich hydrophobic surface with SRP RNA (Akopian et al., 2013). Hydrophobic interactions have been surveyed more sparsely in protein-RNA structures, but may account for up to 50% of the interactions at the protein-RNA interface, depending on the RBP (Hu et al., 2018).
π-interactions can form between any nitrogenous base ring and a π-containing amino acid, which includes the aromatic residues Trp, His, Phe, and Tyr, as well as the charged residues Arg, Glu, and Asp (Wilson et al., 2016). These interactions are relatively strong at ~2–6 kcal/mol per interaction, (Brylinski, 2018; Wilson et al., 2016) and often prefer to be stacked (referred to as π-stacking), occurring most frequently with an inter-atom distance of 2.7–4.3 Å (Auweter et al., 2006; Brylinski, 2018; Morozova et al., 2006; Wilson et al., 2016). Analyses of π-interactions occurring in protein-RNA crystal structures find multiple such interactions on average per structure (Hu et al., 2018; Wilson et al., 2016). These interactions can contribute considerable stability to protein-RNA binding, where some π-interactions are demonstrably crucial to binding function (Auweter et al., 2006; Liao et al., 2018; Oubridge et al., 1994). Such is the case with the extensive stacking interactions cementing human U1A spliceosomal protein with an RNA polyadenylation inhibition element, including two consecutive bases sandwiched between Phe and Asp residues (Figure 1D) (Oubridge et al., 1994). Stacking interactions also occur between bases and hydrophobic residues, and these may be mixed with π-π stacking interactions. For example, a single cytosine in a bulge in bacterial 4.5S SRP RNA stacks with both Phe and Leu in FtsY (Bifsha et al., 2007). More exotic π-stacking configurations include bases that stack on the protein mainchain between residues (Auweter et al., 2006) and perpendicular “T-stacks” between protein sidechains and bases. Stacking interactions with RNA are overall quite crucial and varied, possibly occurring at higher rates than in protein-DNA interactions (Wilson et al., 2016).
Differences from protein-DNA interactions
Similar analyses of protein-DNA structures allow comparison with protein-RNA interactions (Jones et al., 2001; Luscombe et al., 2001; Wilson et al., 2016). RBPs and DNA binding proteins show many of the same preferences for interacting residues, that is, positively charged and polar residues (Hoffman et al., 2004; Jones et al., 2001). However, the chemical and structural differences between DNA and RNA molecules results in observable differences in interactions. Approximately 20% of protein interactions with RNA occur with the 2’-OH while this is not available for protein-DNA interactions (Hoffman et al., 2004) (Figure 2). The base-pairing moieties in RNA bases are also much more extensively contacted by protein than in DNA as the Watson-Crick base face is normally base-paired in DNA (Allers and Shamoo, 2001; Luscombe et al., 2001). In this same vein, protein-DNA interactions more frequently use the phosphodiester backbone (Hoffman et al., 2004; Jones et al., 2001). DNA binding proteins tend to surround their target DNA helix, but this mode of binding is not always available to RBPs, which must accommodate a diverse range of stem-loops, bulges, and other complex structures (Jones et al., 2001). Additionally, double-stranded RNA (dsRNA) adopts a different helix than the standard DNA B-form helix (Bercy and Bockelmann, 2015), explaining why RBPs that interact with dsRNA are best suited to the RNA helix (Vukovic et al., 2014).
Despite these overall differences numerous proteins bind both DNA and RNA (Hudson and Ortlund, 2014). A canonical example is the CCHH zinc finger protein TFIIIA, which binds both the 5S rRNA gene and 5S rRNA with at least six tandem zinc finger (ZnF) domains (Hall, 2005). In binding 5S DNA, TFIIIA ZnFs 1, 3, and 5 interact with the major groove while ZnFs 4 and 6 serve as spacers. When binding RNA, ZnF 4 and 6 interact with unpaired 5S rRNA bases and ZnF 5 binds the RNA major groove, albeit by a different mode than it contacts the DNA major groove. Similarly, Human ADAR1 can bind both dsRNA and Z-form DNA, but uses separate domains for each (Barraud and Allain, 2012). Thus, even a dual DNA and RNA binding protein may still use unique strategies for contacting each molecule.
Binding dynamics
Protein-RNA interactions occur through dynamic rearrangements of both molecules (Leulliot and Varani, 2001). Nuclear magnetic resonance (NMR) and molecular dynamics (MD) simulations as well as crystal structures with and without ligand all shed light on the dynamic process of protein-RNA interaction (Loughlin et al., 2019; Tian et al., 2011; Yu et al., 2014). RNA and protein exhibit mostly local rearrangements during binding, which often entail backbone shifts and bases and residues that “flip out” (Hainzl et al., 2005; Leulliot and Varani, 2001; Matthews et al., 2016; Yang et al., 2002). Upon binding, the site of interaction becomes rigid, locking the molecules together, while adjacent elements in the two molecules loosen to balance the decrease in entropy. In this way, nucleotides or residues that do not directly interact can still be instrumental in binding if they direct the necessary compensatory changes for binding (Leulliot and Varani, 2001; Ravindranathan et al., 2010). Unstructured loops in both protein and RNA are common sites of rearrangement during binding, such as disordered linker regions between well-ordered binding domains in proteins. In fact, a large fraction of residues interacting with RNA tend to be in unstructured loops themselves (Barik et al., 2015; Cléry and Allain, 2011; Han and Nepal, 2007; Treger and Westhof, 2001), and these regions adopt structure upon binding RNA (Balcerak et al., 2019; Leulliot and Varani, 2001). Lastly, the large tertiary flexibility of RNA is a crucial functional feature in protein binding (Flores and Ataide, 2018; Leulliot and Varani, 2001). Computational modeling of protein-RNA binding found more success with simultaneous RNA folding and docking to a protein interface as opposed to RNA folding then docking (Kappel and Das, 2019), reflecting the importance of the tertiary rearrangements RNA requires to bind protein.
Protein-RNA prediction and resources
A great deal of interest lies in predicting RNA binding sites in proteins. For example, the above-mentioned analyses of protein-RNA structures indicated that protein-RNA interfaces prefer positively-charges residues, distinguishing them from protein-protein interfaces which prefer polar residues (Treger and Westhof, 2001). These sorts of metrics and the growing number of solved protein-RNA structures has enabled machine-learning based attempts at predicting protein-RNA interaction. A meta-analysis of these algorithms finds that the most successful features for the prediction of RNA binding sites in protein are residue composition, conservation, and solvent accessibility (Zhang et al., 2017). Additionally, improved thermodynamic models enable docking simulations helpful for understanding the tertiary dynamics of protein-RNA binding (Huang et al., 2013; Kappel and Das, 2019). We provide an up-to-date list and use description of these algorithms and how to access them, along with a few key RBP database resources (Table 1). Algorithms published for the purpose of RBP prediction whose source code or web server is no longer available were not included. We should note that these algorithms are biased by available structural data, which is dominated by the most abundant and readily-crystallized RNA binding domains such as the RNA recognition motif. Thus, predictive algorithms should greatly benefit in the future from the characterization of novel RNA-binding domains and data from alternative structural techniques such as cryogenic electron microscopy.
Table 1.
Summary of studies and software that catalogue or predict RBPs and their targets.
| Name | Link | Reference | Description |
|---|---|---|---|
| 3dRPC | http://biophy.hust.edu.cn/3dRPC.html | (Huang et al., 2013) | Command line software accepts PDB structures of a protein and an RNA and docks them. |
| aaRNA | http://sysimm.ifrec.osaka-u.ac.jp/aarna/ | (Li et al., 2014) | Web server accepts PDB structure or protein sequence to predict residues that bind RNA. Includes graphical output of the binding propensity of each residue. |
| Arpeggio | http://biosig.unimelb.edu.au/arpeggioweb/ | (Jubb et al., 2017) | Web server and command line software accepts PDB file and chain ID and returns all interactions that occur with the given chain. Ionic, polar, hydrogen bonds, aromatic ring stacking, etc. |
| ATtRACT | https://attract.cnic.es/index | (Giudice et al., 2016) | Database of RBPs with experimental data and their inferred bound sequence motif. |
| beRBP | http://bioinfo.vanderbilt.edu/beRBP/predict.html# | (Yu et al., 2019) | Web server and command line software, given protein and RNA sequence, predicts their interaction. |
| BioLiP | https://zhanglab.ccmb.med.umich.edu/BioLiP/ | (Yang et al., 2013) | Database of PDB protein structures with ligands (including RNA) that annotates atoms at the binding interface in a given structure. |
| catRAPID | http://s.tartaglialab.com/page/catrapid_group | (Bellucci et al., 2011) | Command line software (requires licensing) and web server. Accepts protein and RNA sequences and returns heatmap of interaction propensity at each residue/nucleotide pair. |
| DR_bind1 | http://drbind.limlab.ibms.sinica.edu.tw | (Chen et al., 2014) | Web server accepts single protein chain in PDB format and predicts which residues bind RNA. Also produces Jmol image of protein structure. |
| DRNApred | http://biomine.cs.vcu.edu/servers/DRNApred/ | (Yan and Kurgan, 2017) | Web server accepts (up to 100) protein sequence(s) and assesses each residue for its RNA (and DNA) interaction probability. |
| ENTANGLE | On request | (Morozova et al., 2006) | Software assesses hydrogen bonds, Van der Waals, stacking, and hydrophobic interactions between RNA and protein in given PDB structure. |
| HBPLUS | http://www.ebi.ac.uk/thornton-srv/software/HBPLUS/ | (McDonald and Thornton, 1994) | Command line software that lists hydrogen bonds in given PDB structure |
| hybridNAP | http://biomine.cs.vcu.edu/servers/hybridNAP/ | (Zhang et al., 2017) | Web server accepts (up to 10) protein sequence(s) and calculates each residue’s interaction probability with RNA, DNA, and/or protein. Also returns the feature values that determine this probability. |
| KYG | http://cib.cf.ocha.ac.jp/KYG/ | (Kim et al., 2006) | Web server accepts single chain PDB structure and predicts RNA interface propensity of each residue. Outputs graph, table, and downloadable PDB file with scores. |
| ndb | http://ndbserver.rutgers.edu | (Berman et al., 1992) (Coimbatore Narayanan et al., 2014) | Database of solved DNA and RNA structures. |
| NUCPLOT | https://www.ebi.ac.uk/thornton-srv/software/NUCPLOT/ | (Luscombe et al., 1997) | Command line software accepts a protein-RNA/DNA PDB structure and returns a graphic of protein interaction occurring at each nucleotide. |
| OPRA | https://life.bsc.es/pid/opra/default/index | (Perez-Cano and Fernandez-Recio, 2010) | Web server scores residues in PDB structure for interaction probability with RNA. Includes Jmol output of structure with residues colored by predicted value. |
| PDBsum | http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=index.html | (Laskowski et al., 2018) | Provides overview of given PDB structure, including protein sequence, defined structural regions, sequence of bound RNA/DNA, NUCPLOT depiction of bound DNA/RNA, etc. |
| PLIP | https://projects.biotec.tu-dresden.de/plip-web/plip/index | (Salentin et al., 2015) | Web server and command software accept PDB structure of protein-ligand and list each hydrogen bond, salt bridge, pi-interaction, and hydrophobic interaction with ligand. |
| PPRInt | https://webs.iiitd.edu.in/raghava/pprint/index.html | (Kumar et al., 2008) | Web server accepts protein sequence and predicts RNA-binding residues. |
| PredPRBA | http://PredPRBA.denglab.org/ | (Deng et al., 2019) | Web server accepts PDB file of protein-RNA structure and predicts the free energy of binding. |
| PRince | http://www.facweb.iitkgp.ac.in/~rbahadur/prince/home.html | (Barik et al., 2012) | Web server accepts PDB structure, given protein and RNA chain IDs will list atoms at the protein-RNA interface. |
| RAIDv2.0 | http://www.rna-society.org/raid2/index.html | (Yi et al., 2017) | Database of known RNA-RNA and protein-RNA interactions at the full transcript/protein level (not nucleotide/residue detail). |
| RBP prediction selection tool | www.iitm.ac.in/bioinfo/RNA-protein/ | (Nagarajan and Gromiha, 2014) | Online tool based on benchmark of various RBP prediction software. Shows the best software (limited selection) to use for a given RBP/RNA type. |
| RBPDB | http://rbpdb.ccbr.utoronto.ca | (Cook et al., 2011) | Database of RBPs with available experimental data, categorized by organism or RBP domain. |
| RBPmap | http://rbpma?.technion.a?.il/index.html | (Paz et al., 2014) | Web server and command line software, searches for given RBP binding motifs in given RNA sequence. |
| RCSB PDB | https://www.rcsb.org | (Berman et al., 2000) | Search parameters for RBPs with RNA: “Macromolecule Type: contains Protein AND contains RNA” |
| RNAbindPlus | http://ailab1.ist.psu.edu/RNABindRPlus/ | (Terribilini et al., 2007) | Web server predicts residues that bind RNA in given protein sequence. |
| RNAbindRv 2.0 | http://ailab1.ist.psu.edu/RNABindR/ | (Terribilini et al., 2007) | Web server predicts residues that bind RNA in given protein sequence. |
| RPISeq | http://pridb.gdcb.iastate.edu/RPISeq/ | (Muppirala et al., 2011) | Web server accepts protein and RNA sequence and predicts their interaction probability. |
| RsiteDB | http://bioinfo3d.cs.tau.ac.il/RsiteDB/ | (Shulman-Peleg et al., 2008) | Database searches for PDB structure (if published before 2008) and describes protein-RNA interactions: Jmol image, which base, etc. |
| SPOT-Seq-RNA | http://sparks-lab.org/server/SPOT-Seq-RNA/ | (Yang et al., 2014) | Web server and command line software, predicts whether given protein sequence is an RBP. |
| SPOT-Struct-RNA | http://sparks-lab.org/yueyang/server/SPOT-Struct-RNA/ | (Zhao et al., 2011) | Web server and command line software, predicts whether given PDB structure is an RBP. |
| TriPepSVM | https://github.com/marsicoLab/TriPepSVM | (Bressin et al., 2019) | Command line software accepts protein sequence and predicts whether it is an RBP. |
RNA BINDING DOMAINS
RBPs typically contain discrete domains for the purpose of binding RNA. Many RNA binding domains are quite small (<100 residues) and utilize only a handful of their residues to directly interact with RNA. This begs the question of how specific binding is achieved. The combination of multiple RNA binding regions engaging in hydrogen bonds, stacking interactions, and additional weaker interactions with all parts of the RNA nucleotide, as described above, cumulatively enables RBPs to bind specific regions in RNAs. Multiple binding domains often coexist in one RBP, enhancing specific RNA binding (Cléry and Allain, 2011; Lunde et al., 2007). Linkers between domains have been shown to mediate important RNA contacts as well, and the flexibility of linkers can determine whether adjacent RNA binding domains bind independently or cooperatively (Cléry and Allain, 2011; Lunde et al., 2007). Recognition of bipartite sequence motifs is a common occurrence, mediated by multiple RNA binding domains and their linkers and RNA structural arrangements that present bipartite sequences to the RBP (Dominguez et al., 2018; Loughlin et al., 2019; Lunde et al., 2007; Tan et al., 2014; Walden et al., 2012). Additionally, some RNA-binding domains are capable of mediating protein-protein interactions, including in concert with RNA binding (Cienikova et al., 2015; Yang et al., 2002). This multilayered approach allows almost limitless combinations, such that there exist hundreds of different RBPs that conduct a wide and diverse number of functions. Here we describe in detail seventeen structurally-characterized domains that have been described to bind RNA in multiple proteins (Table 2).
Table 2.
Summary of described RNA binding domains and example structures.
| Domain name | PDB search term | PDB structures | PDB structures with RNA | Size (a.a.) | Protein families found in | Ex. structure PDB | Example domain structure |
|---|---|---|---|---|---|---|---|
| Cold shock domain (CSD) | “Cold shock domain” | 46 | 13 | 70 | Cold shock proteins, Y-box proteins | 4A4I |  |
| Double-stranded RNA Binding Domain (dsRBD) | “dsRBD” | 59 | 27 | 65 | RNases, ADARs, Dicer | 3LLH |  |
| Helicase | “dead” OR “deah” OR “helicase domain” | 701 | 114 | 350–400 | DExH/D-box, Ski2-like, RIG-I-like, NS3, UPF1-like RNA binding helicases | 2I4I |  |
| Intrinsically disordered region (IDR) | “Intrinsically disordered region” | NA | NA | varies | Most RBPs | NA | NA |
| K homology (KH) | “KH domain” | 117 | 20 | 70 | hnRNPs, translation regulation proteins, very common | 1WH9 |  |
| La motif (LAM) | “La motif AND NOT rrm” | 54 | 5 | 90 | La proteins, La-related proteins (LARPs) | 1S29 |  |
| Piwi-Argonaute-Zwille (PAZ) | “PAZ domain” | 71 | 44 | 170 | Argonaute proteins, Dicer | 3O6E |  |
| P-element Induced Wimpy Testis (PIWI) | “PIWI domain” | 74 | 33 | 290 | Argonaute proteins | 1X4Q |  |
| Pentatricopeptide repeat (PPR) | “pentatricopeptide repeat” | 32 | 9 | 35 * n, 1 > n > 30 | RNA editing proteins | 4M59 |  |
| Pseudouridine synthase and archaeosine transglycoslyase (PUA) | “Pseudouridine synthase and archaeosine transglycoslyase” OR “PUA domain” | 119 | 28 | 66–98 | RNA modifying enzymes, metabolic enzymes | 1SQW |  |
| Pumillo-like repeat (PUM) | “Pumilio” OR “PUM domain” OR “Puf protein” | 60 | 49 | 334 | PUF proteins | 1M8W |  |
| RNA Recognition Motif (RRM) | “RNA Recognition Motif” OR “RRM” | 554 | 119 | 90 | hnRNPs, splicing factors, very common | 2MTG |  |
| Ribosomal S1-like (S1) | “S1 RNA binding domain” | 25 | 2 | 70 | Ribosomal proteins, Translation initiation factors, RNase II, PNPase | 2EQS |  |
| Sm and Like-Sm (Sm / Lsm) | “Sm RNA binding domain” | 31 | 23 | 80 | U1 spliceosomal proteins, Hfq | 2VC8 |  |
| thiouridine synthases, RNA methylases and pseudouridine synthases (THUMP) | “THUMP domain” | 12 | 3 | 100–110 | tRNA modifying enzymes | 2DIR |  |
| YT521-B homology (YTH) | “YTH domain” | 28 | 14 | 100–150 | YTH family m6A readers | 4RCI |  |
| Zinc Finger (ZnF) | “Zinc Finger” | 2677 | 63 | 30 | Transcription factors, METTL enzymes, Very common | 5ZC4 |  |
RNA recognition motif (RRM)
RNA Recognition Motifs (RRMs) are the most common and well-studied RNA binding domain. A search in the protein databank for “RRM” yields over 422 structures (Table 2), and RRMs are estimated to occur in 1% of all human proteins (Cléry and Allain, 2011). RRMs average 90 amino acids in size and adopt a β1α1β2β3α2β4 topology forming two α-helices against an antiparallel β-sheet, which houses the conserved RNA-binding RNP1 and RNP2 motifs in the central β1 and β3 strands (Cléry and Allain, 2011). RRMs interact with 2–8 nucleotides in single-stranded RNA (ssRNA) commonly through several sequential stacking interactions and hydrogen bonds with the RNP motifs, often with nanomolar affinities (Auweter et al., 2006; Clery et al., 2008). Each RRM has its own sequence preferences, often for degenerate sequences such as GU-rich tracts (Clery et al., 2008). The combination of consecutive RRMs in an RBP dramatically increases binding affinity and specificity (Maris et al., 2005). For example, the dual binding of both RRM domains in hnRNPA1 is crucial to its overall binding ability and function in repressing splicing (Beusch et al., 2017). RRMs have also been observed interacting with other protein domains, such as hnRNPC, whose single RRM domain drives multimerization with other hnRNPC molecules (Cienikova et al., 2015; Safaee et al., 2012).
K homology (KH)
The K homology (KH) domain was first discovered in heterogeneous nuclear ribonucleoprotein K (hnRNPK). At 70 amino acids, the KH domain is even smaller than the RRM domain, and typically recognizes 4 nucleotides in ssRNA or ssDNA (Cléry and Allain, 2011; Valverde et al., 2008). KH domains adopt either a type I β1α1α2β2β’α’ topology (in eukaryotes) or the reverse type II α’β’β1α1α2β2 topology (in prokaryotes), with a conserved “GXXG” RNA-binding motif located between the α1 and α2 helices (Valverde et al., 2008). RNA binding occurs in a hydrophobic pocket and includes several hydrogen bonds coordinated by the “GXXG” motif (Auweter et al., 2006). Stacking interactions between protein and RNA in KH domains are scarce, potentially explaining the domain’s weak micromolar affinities (Cléry and Allain, 2011; Valverde et al., 2008). As with RRMs, multiple KH domains (type I) in an RBP can independently or synergistically increase binding specificity (Valverde et al., 2008). For example, the two KH domains in MEX-3C increase its binding site to a 5 plus 4 nucleotide bipartite motif bound with 0.17 μM affinity (Yang et al., 2017). KH domains are also commonly found co-occuring with quaking (QUA) domains as part of the larger signal transduction and activation of RNA (STAR) domain, which greatly extends the binding surface and accommodates 7–8 nucleotides with 0.07 uM affinity (Sharma and Anirudh, 2017; Teplova et al., 2013).
Zinc finger (ZnF)
Zinc fingers (ZnFs) describe a large family of proteins that average 30 amino acids in size and form a simple ββα topology in which residues in the β-hairpin turn and α-helix are coordinated by a Zn2+ ion (Cléry and Allain, 2011). Most ZnFs bind DNA, but have been additionally shown to bind RNA, proteins, and small molecules (Lai et al., 2000). ZnF subtypes that interact with RNA include CCHC (zinc knuckle), CCCH, CCCC (RanBP2), and CCHH subtypes, where C and H refer to the interspersed cysteine (C) and histidine (H) residues that coordinate the zinc atom (Cléry and Allain, 2011). These subtypes display a range of sequence and structural specificities. Zinc knuckles (CCHC) recognize stem loop elements in RNA (or ssDNA) through contacts with bases in the loop and the phosphate backbone of the stem. CCCH and CCCC subtypes tend to recognize 3 nucleotide repeats through multiple such ZnFs in one RBP (Font and Mackay, 2010; Hall, 2005; Lai et al., 2000). These contacts are formed through hydrogen bonds with bases and the insertion of aromatic sidechains that stack between bases. The versatile and abundant CCHH ZnFs interact with both single-stranded and double-stranded RNA as well as DNA (Font and Mackay, 2010; Hall, 2005). Modular arrays of CCHH ZnFs have been successfully engineered to bind desired DNA sequences. Thus, designer ZnFs are thought to have potential for directed binding of RNA sequences, a goal that has been achieved with the much larger Pumilio homology domains (Font and Mackay, 2010).
Pumilio homology domain (PUM-HD)/(PUF)
The Pumilio and FBF (PUF) family of proteins occurs in most eukaryotes and is defined by the Pumilio homology domain (PUM-HD), or the PUF domain. The PUF domain is very large, consisting of eight α-helical repeats of a highly conserved 36 amino acid sequence that forms a concave RNA-binding surface (Wang et al., 2018). Each repeat recognizes one unpaired base through hydrogen bonds and a stabilizing stacking interaction, where the full domain recognizes up to 8 nucleotides in ssRNA with low nanomolar affinity (Zhao et al., 2018). Wild-type PUF repeats do not specifically recognize cytosine; however, protein engineering has produced repeats that do (Zhao et al., 2018). These advances combined with the PUF domain’s predictable base-recognition code allows modular design of pumilio proteins that recognize 8–10 nucleotide sequences containing all RNA bases (Zhao et al., 2018).
Pentatricopeptide repeat (PPR)
Very similar to PUF repeats, eukaryotic pentatricopeptide repeats (PPRs) are each ~35 residues in length and form two antiparallel α-helices. 2–30 repeats form a solenoid-shaped scaffold that binds specific ssRNA sequences with nanomolar affinity (Ke et al., 2013; Spahr et al., 2018) Two residues in each repeat determine base-specific binding through hydrogen bonds, enabling the development of designer PPRs that bind specified ssRNA or ssDNA sequences (Spahr et al., 2018).
Pseudouridine synthase and archaeosine transglycoslyase (PUA)
The pseudouridine synthase and archaeosine transglycoslyase (PUA) domain is found in the aforementioned enzymes as well several other RNA-modifying and metabolic enzymes (Perez-Arellano et al., 2007). PUA domains range from 67–94 amino acids in length, with a β1α1β2β3β4β5α2β6 architecture that forms a pseudobarrel encased by two α-helices. PUA domains have been characterized contacting dsRNA and its adjacent loops or overhangs through extensive hydrogen bonds with all parts of the RNA. These contact are typically formed by a glycine-rich loop between α1 and β2 or α2 and β6. Unlike many other domains, PUA domains are not found as tandem repeats (Perez-Arellano et al., 2007).
THUMP
Named for THioUridine synthase, Methyltransferase and Pseudouridine synthase, the THUMP domain is found in numerous tRNA-modifying enzymes. About 100 amino acids long, THUMP domains are always found in proximity to RNA-modifying domains and often in proximity to an N-terminal ferredoxin-like (NFLD) domain (Neumann et al., 2014). THUMP domains display a α1α2β1α3β2β2 topology that forms parallel α-helices flanking a β-sheet (Fislage et al., 2012). The first structure of a THUMP domain bound by RNA, bacterial 4-thiouridine synthetase in complex with tRNA, reveals a 3-dimensional fold that specifically recognizes the 3’-CCA tail and adjoining stem of tRNA. Several hydrogen bonds and VdWs contacts correctly position the tRNA for modification by the accompanying pyrophosphate domain (Neumann et al., 2014).
YT521-B homology (YTH)
The YT521-B homology (YTH) domain is found in the YTH family of proteins that “read” N6-methyladenosine (m6A) marks in RNA. The YTH domain ranges from 100–150 amino acids in length and forms a six-stranded β-barrel surrounded by four or five α-helices. Three residues in the hydrophobic core of the β-barrel trap the methyl group of m6A in an “aromatic cage” consisting of hydrogen bonds with the adenosine and π-interactions between tryptophan rings and the methyl group (Liao et al., 2018; Xu et al., 2014). The YTH domain specifically binds m6A over unmodified adenosines. Affinity of YTHDC1 for consensus DRm6(A)CH motifs was measured at 0.3 μM, while no binding was detected for the unmethylated sequence (Xu et al., 2014). Similarly, YTHDF2 affinity for methylated RNA was measured as 2.54 μM, with ten-fold lower affinity for the unmethylated target (Zhu et al., 2014).
Double stranded RNA binding domain (dsRBD)
Double stranded RNA binding domains (dsRBDs), or motifs (dsRBMs), consist of 65–70 amino acids and are the third most common RNA binding domain (Masliah et al., 2013). dsRBDs specifically recognize and bind double-stranded RNA (dsRNA) and are found in proteins with roles in viral protection, RNAi, and cellular transport (Masliah et al., 2013). dsRBDs often appear as tandem repeats or in combination with other functional RNA binding domains, such as RNA editing or helicase domains (Cléry and Allain, 2011; Ranji et al., 2011). The domain is made up of an α1β1β2β3α2 fold that forms an antiparallel β-sheet flanked by α-helices on one face (Cléry and Allain, 2011; Masliah et al., 2013). dsRBDs specifically recognize the structure of an A-form RNA helix, spanning up to 16 base-pairs with hydrogen bond contacts to the phosphodiester back bone and 2’-OH (Cléry and Allain, 2011; Ramos et al., 2000). In some cases dsRBDs have demonstrated base-specific contacts, such as to bases in adjacent loops (Cléry and Allain, 2011; Masliah et al., 2013). Stacking interactions are rare, potentially explaining the low affinities (high nanomolar to micromolar range) of dsRBDs to RNA targets (Stefl et al., 2010; Wang et al., 2011).
Helicase
Helicase domains are found in all forms of life in helicase proteins, which unwind both DNA and dsRNA. Helicases comprise six superfamilies (SFs), of which SF1 and SF2 contain all the eukaryotic RNA and DNA helicases. RNA-binding helicases include the Upf1-like family in SF1 and the DEAD-box, DEAH, RIG-I-like, Ski2-like, NS3 families in SF2. The remaining SFs 3–6 contain bacterial and viral helicases that form multimeric rings (Jankowsky, 2011). Helicase domains are very large, containing 350–400 amino acids. In SF1 and SF2 the helicase domain is composed of two “recombinase A (recA)-like” subdomains, each of which contains an ATP-catalytic core, a nucleic acid binding region, and subdomains that coordinate the two. Within families of helicases these subdomains are quite conserved. Helicase monomers in the ring-forming superfamilies of helicases are similarly quite large and composed of multiple subdomains (Gai’ et al., 2004; Kainov et al., 2008). Bound RNA is surrounded by the recA-like domains; or in the case of multimeric helicases, RNA is pulled through the center of the ring. Contacts with RNA are dominated by hydrogen bonds to phosphate and sugar moieties, but contacts with bases are occasionally observed (Jankowsky, 2011; Kainov et al., 2008; Linder and Jankowsky, 2011; Weir et al., 2010). Multiple nucleotides are typically contacted simultaneously; DEAH/DEAD-box helicases, for example, tend to accommodate at least 5 single-stranded or base-paired nucleotides (Jiang et al., 2011; Linder and Jankowsky, 2011; Weir et al., 2010). Affinities to RNA are often in the nanomolar range, although they vary greatly by helicase and are modulated by other subdomains of the helicase. ATP binding generally promotes higher affinity to RNA by causing the helicase RNA-binding regions to “clamp”. ATP hydrolysis subsequently promotes conformational changes that cause the helicase to translocate 1 nucleotide and/or unwind its substrate (Iost et al., 1999; Jankowsky, 2011; Jiang et al., 2011; Kainov et al., 2003).
Cold shock domain (CSD)
The cold shock domain (CSD) is found in a large family of proteins associated with cold adaptation found in all domains of life. CSDs are composed of ~70 amino acids (more in eukaryotes) and five antiparallel β-strands that form a common β-barrel structure known as an oligosaccharide-/oligonucleotide-binding (OB) fold. CSDs contain the conserved RNP1 and RNP2 motifs common to RRMs, which bind ssRNA and ssDNA (Amir et al., 2018). CSDs contact 3–4 nucleotides through sequential stacking interactions and hydrogen bonds with bases, achieving nanomolar affinities (Kljashtorny et al., 2015; Sachs et al., 2012). CSD-containing proteins vary greatly in the types of sequences they recognize. Bacterial CspB is reported to bind pyrimidine-rich sequences and prefer ssDNA to ssRNA by up to ten-fold (Sachs et al., 2012). Y-box proteins contain the most well-studied eukaryotic CSDs, showing a preference for G-rich ssRNA sequences over ssDNA (Kljashtorny et al., 2015).
S1
The S1 RNA binding domain was originally discovered in S1 ribosomal protein, which binds both mRNAs and ribosomal RNA. The ~70 amino acid S1 domain forms a 5-stranded antiparallel β-barrel in the same OB-fold family as the CSD (Mihailovich et al., 2010). Despite sharing a common tertiary structure, the two domains show no sequence similarity, suggesting that their shared tertiary structure was achieved through convergent evolution (Mihailovich et al., 2010). S1 domains are additionally found in several exoribonucleases and eukaryotic translation initiation factors and in combination with other RNA binding domains such as the KH domain or CSDs (Amir et al., 2018; Chekanova et al., 2002; Hossain et al., 2016; Worbs et al., 2001). Despite their abundance, very little structural information is available for S1 domains in complex with RNA. S1 domains interact with both ssRNA and dsRNA in the context of the RNA binding channel of exoribonucleases (Hossain et al., 2016). Similarly, S1 domains of the ribosomal S1 protein likely interact with mRNA at the entry channel of the ribosome (Loveland and Korostelev, 2018).
Sm
The Sm RNA binding motif is found in Sm and Like-Sm (Lsm) proteins in eukaryotes and archaea and in Hfq protein in prokaryotes (Schumacher et al., 2002; Thore et al., 2003). The Sm motif consists of ~70 residues with an α1β1β2β3β4β5 topology that forms a curved antiparallel β-sheet. Sm-containing proteins readily multimerize through interactions between strands β4 and β5 in two Sm motifs. For example, Sm-Sm interactions link the seven human Sm proteins that make up the protein core of small nuclear ribonucleoproteins (snRNPs) in the spliceosome (Thore et al., 2003). The Sm multimers bind RNA with nanomolar affinity. Two Sm motifs form a 6-nucleotide binding surface that binds specific bases, often uridines, through hydrogen bonds and stacking interactions (Schumacher et al., 2002; Thore et al., 2003).
La motif (LAM)
The small ~90 residue La motif (LAM) is found in eukaryotic La and La-related proteins (LARPs). The LAM consists of five α-helices and three β-strands that form a small antiparallel β-sheet against a modified “winged-helix” fold (Bousquet-Antonelli and Deragon, 2009). The winged-helix structure itself is common to several other RNA and DNA binding proteins (Teichmann et al., 2012). LAMs are always found adjacent to at least one RRM, where the combination of these two domains likely evolve as a unit (Bousquet-Antonelli and Deragon, 2009). In La proteins, the dual LAM-RRM region tightly binds the UUU-OH elements at the 3’ ends of polymerase III-transcribed small RNAs. Binding occurs in a cleft between the LAM and RRM rather than the traditional RNA-binding surfaces of either the RRM or the LAM winged-helix fold. Several uracil bases stack with highly conserved aromatic residues in the LAM, and hydrogen bonds from both LAM and RRM coordinate bases, phosphates, and the terminating 2’-OH. These contacts result in low nanomolar affinities of the LAM for 3’-terminal UUU-OH elements (Teplova et al., 2006). The other LAM-containing proteins, LARPs, bind a diverse set of RNAs with as yet uncharacterized structural mechanisms (Schenk et al., 2012).
Piwi-Argonaute-Zwille (PAZ) and PIWI
Piwi-Argonaute-Zwille (PAZ) and PIWI RNA binding domains define the Argonaute family of proteins found in eukaryotes (Hock and Meister, 2008). Found on opposite sides of the Argonaute protein, both domains facilitate binding of small interfering RNA and microRNA guides to mRNA targets (Hock and Meister, 2008).
PAZ domains occur in Dicer proteins in addition to Argonaute proteins (Hock and Meister, 2008). Crystal structures of the PAZ domain display a six-stranded β-barrel topped with two α-helices and flanked on the opposite side by a special appendage containing a β-hairpin and short α-helix (Ma et al., 2004; Tian et al., 2011; Yan et al., 2003). A binding pocket formed between this appendage and the β-barrel binds the 2-nucleotide 3’ overhang in guide RNAs (gRNAs) with low micromolar affinity (Ma et al., 2004; Tian et al., 2011). Binding is coordinated mostly by conserved tyrosine residues that form hydrogen bonds with the phosphate backbone and sugar hydroxyls of the two terminal nucleotides (Ma et al., 2004; Tian et al., 2011).
The PIWI domain tertiary structure forms an RNase H-like fold consisting of a five-stranded β-sheet flanked by α-helices on both faces (Boland et al., 2011). The PIWI domain has endonucleolytic activity in some cases, but primarily stabilizes the gRNA-mRNA duplex seed region through hydrogen bonds with the gRNA backbone of nucleotides 3–5 and the 5’-overhang base (Boland et al., 2011; Ma et al., 2005; Miyoshi et al., 2016). The PIWI domain also contacts the PAZ domain in certain conformations, suggesting that its activity may be modulated by the conformational state of the PAZ domain (Boland et al., 2011).
Intrinsically Disordered Region (IDR)
Intrinsically disordered regions (IDRs) are unstructured and often consist of repeats of arginine/serine (RS repeat), arginine/glycine (RGG box), arginine or lysine-rich patches (R/K basic patches),or short linear motifs of amino acids (Balcerak et al., 2019; Jarvelin et al., 2016). Despite their lack of structure, IDRs have been found to dominate the composition in over 20% of RBPs (Jarvelin et al., 2016). It is increasingly observed that IDRs can be the sole RNA binding domain in an RBP and may actually drive the majority of protein-RNA interactions in the cell (Hentze et al., 2018). Like globular RNA binding domains, IDRs are conserved, often occur multiple times in one RBP, and can coordinate RNA binding in concert with other domains (Balcerak et al., 2019; Jarvelin et al., 2016; Loughlin et al., 2019). IDRs have been shown to drive higher affinity to RNA in RBPs that contain ordered RNA binding domains and can themselves transition to an ordered state once bound to RNA (Balcerak et al., 2019; Cruz-Gallardo et al., 2019; Jarvelin et al., 2016; Leulliot and Varani, 2001). IDRs show little RNA sequence dependence, however; suggesting that these regions’ high affinity for RNA is predominantly driven by electrostatic attraction to the phosphodiester backbone (Balcerak et al., 2019; Jarvelin et al., 2016).
Other RNA binding domains
The domains detailed above represent a mere fraction of the RNA-binding domains in existence, 21% of the 2,685 RNA-bound structures in PDB, while many hundreds more RNA-binding domains await characterization (Hentze et al., 2018). Several domains, such as the Brix domain, sterile alpha motif (SAM), and the SAF-A/B, Acinus and PIAS (SAP) domain are mostly known as protein- or DNA-binding domains but have one or two protein members shown to bind RNA. For example, the SAM domain is a well-known α-helical protein-protein interaction domain, but the SAM-containing proteins Smaug and its homologue VTS1p bind the pentaloop of an RNA hairpin element called the Smaug recognition element (SRE) with nanomolar affinity (Aviv et al., 2006; Ravindranathan et al., 2010). Conservation of the RNA-interacting residues among Smaug homologs suggests RNA-binding function exists in other SAM-containing proteins (Aviv et al., 2006). Many other RNA-binding domains are utterly unique, i.e. not yet found to resemble any other domain (Gerstberger et al., 2014; Hentze et al., 2018; Tan et al., 2013; Walden et al., 2012). The stem loop binding protein domain is found only in the protein of the same name (SLBP) that exclusively binds a conserved stem loop at the ends of histone mRNAs (Tan et al., 2013). IRP1, also known as ACO1, is an aconitase with a unique fold that tightly binds iron response elements in the UTRs of iron metabolism-related transcripts (Walden et al., 2012). Most ribosomal proteins each contain a unique RNA binding domain, the S1 domain excepted (Gerstberger et al., 2014). Structural data of viral RNA binding proteins reveals incredibly diverse and unique structures specialized for binding highly structured viral RNA elements. Recent large-scale studies have discovered hundreds of novel RNA binding regions in proteins, including in many well-characterized enzymatic proteins such as GAPDH, that surprisingly “moonlight” as RNA binding proteins (Hentze et al., 2018; Hudson and Ortlund, 2014). Overall, the diversity of RBPs is an astounding testament to their all-encompassing cellular roles in many domains of life.
Domain differences in hydrogen bond formation with RNA
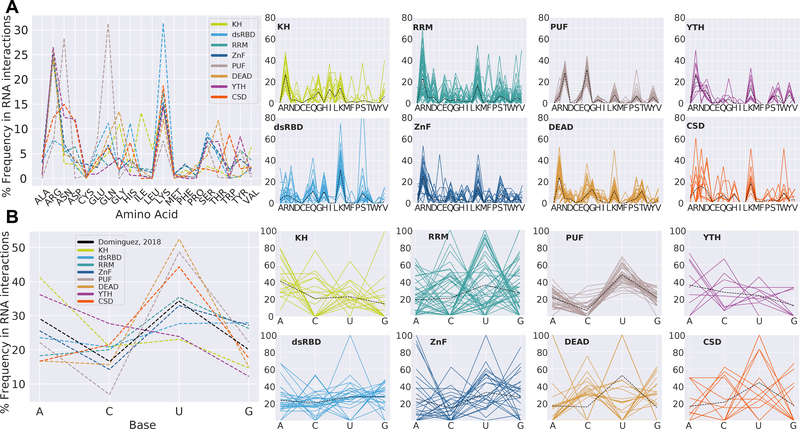
We additionally assessed the type and number of hydrogen bonds that proteins form with RNA in over 200 structures from eight common domain types (KH, dsRBD, RRM, ZnF, PUF/PUM-HD, DEAD helicase, YTH, and CSD). This includes the frequency of each amino acid used for protein-RNA hydrogen bonds, the frequency of each RNA base that contacts are formed with, and the moieties used to facilitate those bonds (Figures 3–4). This hydrogen bond analysis of protein-RNA structures has been conducted many times before (Figure 2) (Allers and Shamoo, 2001; Ellis et al., 2007; Han and Nepal, 2007; Hu et al., 2018; Jones et al., 2001; Morozova et al., 2006; Treger and Westhof, 2001), but without assessing domains separately. We immediately observe that across all domain types the positively charged amino acids Lys and Arg most frequently facilitate hydrogen bonds with RNA (Figure 3A), which directly agrees with previous analyses (Perez-Cano and Fernandez-Recio, 2010). Asp, Gln, His, and Ser are also frequently used, but are more dependent on domain type. Non-polar amino acids Ala, Cys, Met, and Pro are universally avoided. Trp is strongly avoided in hydrogen bonds with RNA by all domains except CSDs (p-val = 2.38e-5). Our analysis of amino acid frequencies considers the mainchain atoms of each residue as belonging to that particular amino acid. Thus we observe that Leu and Ile, whose sidechains are not capable of forming hydrogen bonds, are repeatedly involved in forming hydrogen bonds with RNA among KH domains, but no other domain (p-vals = 7.26e-6, 6.16e-9). This agrees with previous descriptions of salient RNA contacts in crystal structures of KH domains being formed via the mainchain of Ile residues (Cléry and Allain, 2011). Interestingly, a single point mutation replacing an Ile in the KH domain of FMRP is known to cause Fragile X Syndrome (De Boulle et al., 1993).
Figure 3.
Amino acid and base preferences in RNA-protein hydrogen bonds observed in over 200 structures, organized by domain. (A) The average frequency of each amino acid in forming hydrogen bonds with RNA across eight RNA binding domain types (left). The frequency of each amino acid (one-letter abbreviations) in forming hydrogen bonds with RNA in multiple structures, separated by domain type (right, smaller plots). (B) The average frequency of each RNA nucleotide in forming hydrogen bonds with protein across eight RNA binding domain types, as well as the average frequency of each base in sequence motifs from bind-N-seq data (Dominguez et al., 2018) (left). The frequency of each RNA nucleotide in forming hydrogen bonds with protein in multiple structures, separated by domain type (right, smaller plots).
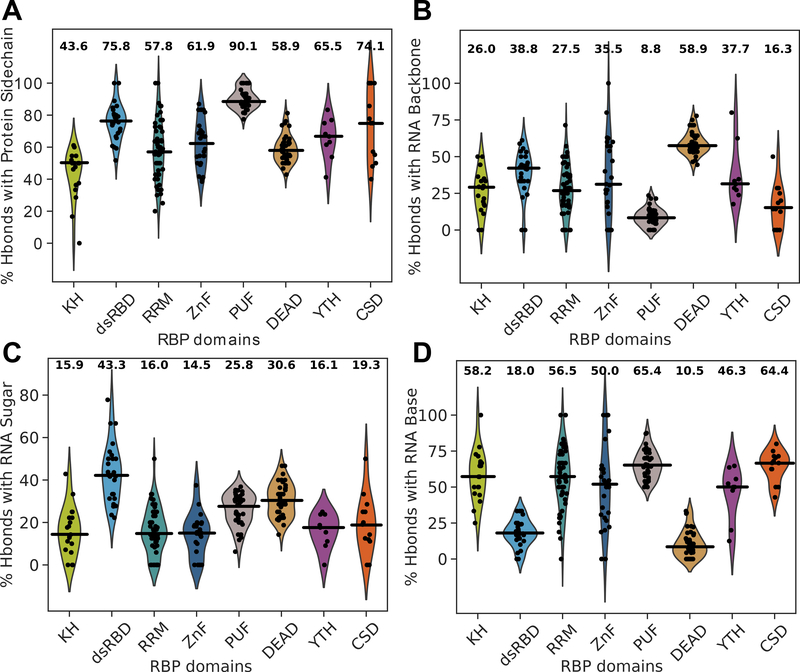
Figure 4.
Assessment of protein-RNA hydrogen bonds in over 200 structures, organized by RNA binding domain. Averages for each statistic are listed above each domain’s violin plot and medians are indicated with black horizontal bars. (A) The percent of protein-RNA hydrogen bonds that are formed using protein sidechains (as opposed to the mainchain). (B) The percent of protein-RNA hydrogen bonds that are formed with RNA backbone atoms. (C) The percent of protein-RNA hydrogen bonds that are formed with RNA sugar atoms. (D) The percent of protein-RNA hydrogen bonds that are formed with RNA base atoms.
Preferences for RNA nucleotides in protein-RNA hydrogen bonds were assessed as well. Note that even with small RNAs not necessarily all of the nucleotides interact with the RBP in a hydrogen bond. Previous analyses have varied in whether they report RBP preferences for interaction with specific nucleotides (Perez-Cano and Fernandez-Recio, 2010), and the types of RNA sequences that have been successfully co-crystallized with RBPs could be biased by technical reasons. Nevertheless, assessing our data we observe a preference for interactions with uracil, and an under-enrichment for cytosine (Figure 3B). Sequence motifs derived from RNA Bind-n-Seq experiments (Dominguez et al., 2018) also follow this pattern of base frequencies (r2 = 0.89, Pearson), providing orthogonal agreement for the observed base preferences. PUF domain structures exhibit the lowest percentage of hydrogen bonds with cytosine (p-val = 0.001), reflecting the lack of cytosine recognition in wild-type PUF repeats (Zhao et al., 2018). The KH domain re-asserts its status as an oddball as it forms hydrogen bonds with adenosines more frequently than any other domain (p-val = 0.002).
We were also interested in statistics summarizing the frequency of hydrogen bonds with the sidechain of amino acids and with the base, backbone, or sugar of the RNA (Figure 4). We observe pronounced domain differences with the percent usage of amino acid sidechains (versus the mainchain). For example, KH domains use sidechains in 43.6% of hydrogen bonds with RNA while PUF domains use sidechains in 90.1% of interactions (p-val of difference = 2.25e-13) (Figure 4A). In fact, sidechain rather than mainchain hydrogen bonds typically dominate protein-RNA interactions (Figure 2A), but KH domains are the only domain analyzed here that violate this trend (Figure 4A). The percentage of hydrogen bonds forming with either the backbone, sugar, or base of RNA nucleotides were also calculated for each protein structure (Figure 4B–D). PUF domains predictably hydrogen bond with the RNA backbone with the lowest frequency on average (p-val = 1.60e-13) (Figure 4B), instead interacting with the RNA base in 65.4% of its hydrogen bonds—the highest average of all domains (p-val = 1.31e-7) (Figure 4D). Also somewhat predictably, dsRBDs and DEAD helicase domains hydrogen bond least frequently with the RNA base (p-vals = 5.44e-8, p-val < 1e-307), and DEAD domains most frequently with the RNA backbone (p-val < 1e-307). This reflects the known ability of PUF domains to recognize sequences hyper-specifically, and of dsRBDs and DEAD domains to generally bind RNA without sequence preferences. dsRBDs also hydrogen bond most frequently—43.4% of the time—with the 2’-OH moiety (p-val = 1.90e-10) (Figure 4C), a testament to these domains’ specific recognition of dsRNA rather than DNA (Vukovic et al., 2014).
Overall our analysis highlights how domains differ and reinforces what is known about how certain domains form hydrogen bonds with RNA in service of their specific biology. In the future, assessing stacking, hydrophobic, and Van der Waals interactions by domain would additionally contribute to defining domains’ specific binding strategies. Quantifying binding strategies in this way will serve prediction and design efforts aimed at controlling the biology of RBPs.
REGULATION MECHANISMS
While RNA binding domains are directly responsible for interacting with RNA, we must consider the dynamic cellular context that regulates this interaction. In this section we will describe the factors that determine how RBPs find their targets, what combined function they perform with their targets, and how the first two are regulated.
Protein-RNA assembly
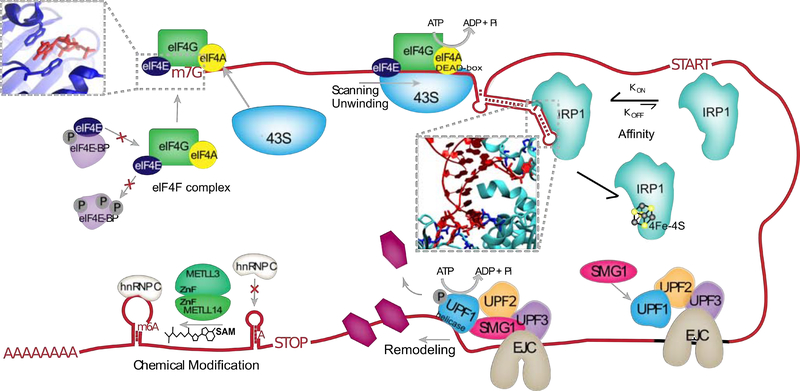
How does a given RBP find its target? The intermolecular interactions between protein and RNA are the raw starting material for determining their affinity, where interactions with specific moieties and/or binding pockets that “fit” an RNA substrate are common strategies for highly effective binding. In the case of IRP1 its “L-shaped” binding pocket and contacts with select bases yield an incredibly specific and strong picomolar affinity for the iron response element (Figure 5) (Walden et al., 2012). eIF4E, on the other hand, lacks specificity for select RNA elements like IRP1, instead generally binding all mRNAs through its recognition of their 5’-caps. This interaction is strong (nanomolar affinity), facilitated by stable stacking interactions with the m7Gppp structure (Figure 5) (Jackson et al., 2010; Niedzwiecka et al., 2002). Lastly, many RBPs exhibit neither specific nor high affinity binding for the purpose of functionally transient associations with RNA (Auweter et al., 2007; Linder and Jankowsky, 2011). RBP (or RNA) abundance also affects the free energy of binding, where high abundance pushes the equation in favor of association. Thus it is no wonder that IRP1 interaction with the iron response element in the highly abundant FTL transcript is one of the most reliable and well-studied protein-RNA interactions. Recruitment by other proteins is the primary mode by which many RBPs find their targets, especially in the assembly of multi-component ribonucleoprotein complexes. For example, eIF4E binds mRNA 5’-caps as a subunit of the eIF4F translation initiation complex, which delivers other members of the complex, such as helicase eIF4A, for action on the transcript (Figure 5). The eIF4F complex additionally recruits the 43S ribosomal complex to begin scanning the 5’UTR (Jackson et al., 2010). In the same way that RBPs are recruited, they may also be prevented from binding by sequestration by other proteins or modifications to their binding sites (described below).
Figure 5.
Examples of mechanisms controlling RBP binding, interactions with RNA, and their regulation. eIF4E (dark blue) interacts with 7-methyl-guanosine cap (m7G), in part through stacking interactions (inset) and binds to RNA as part of the eIF4F which includes RBPs eIf4G and eIF4A. eIF4E association with eIF4F is prevented by sequestration to hypo-phosphorylated eIF4E-BP. The 43S ribosomal subunit is recruited to the eIF4F complex, and processively scans the 5’UTR, aided by ATP-driven helicase activity of the eIF4A DEAD-box domain. The RBP IRP1 specifically binds hairpin elements in the 5’UTR with high affinity through specific residues (inset, dark blue) that hydrogen bond with the bulge and apical loop of the RNA. RNA binding by IRP1 is prevented by 4Fe-4S ligand binding to IRP1. UPF1 is recruited to the exon junction complex (EJC), where its helicase activity is activated by interactions with SMG1 and UPF2. Driven by ATP, UPF1 removes both RNA structures and other bound RBPs in the 5’−3’ direction. A zinc-finger (ZnF) containing METTL3-METTL14 complex deposits methyl groups donated by S-adenosyl methionine (SAM) on targeted adenosines (m6A). M6A modifications reduce base-pairing in RNA, such that some locations become available for hnRNPC binding.
Combined function of protein and RNA
The individual components of protein and RNA have a different function once associated. This function can be guided by RNA binding domains themselves or cooperative enzymatic domains in the same RBP; or may require multiple protein/RNA components. We may separate protein-RNA functions into several broad categories: static binding, scanning/translocation, remodeling, and modification. Many RBPs statically bind specific RNA elements (often small hairpins) (Ravindranathan et al., 2010; Tan et al., 2013; Walden et al., 2006); that is to say, they do not alter the RNA further once bound. This mode of interaction often blocks access to the RNA, such as IRP1, which impedes translation initiation by blocking the scanning action of eIF4A (Figure 5)(Walden et al., 2006). Translocation along RNA substrates is common among RNA helicases, which includes eIF4A and UPF1 (Figure 5). UPF1, as well as a few other helicases, additionally remodels the RNP landscape of its substrate by processively removing other RBPs in its path (Fiorini et al., 2015; Jankowsky, 2011). Most frequently, helicases serve the purpose of removing RNA secondary structure, where scanning and structure unwinding can occur either together or independently (Fiorini et al., 2015; Jackson et al., 2010).
Performed by more than just helicases, manipulation of RNA structure can be considered a non-covalent form of RNA modification. Remodeling RNA includes chaperoning RNA structure formation in addition to removing structures. Chaperone activity on RNA has been observed among cold shock proteins and helicases, as well as the pseudouridine synthase TruB which modifies tRNA structure in addition to chemically modifying the RNA (Keffer-Wilkes et al., 2016; Rajkowitsch et al., 2007). RNA is chemically modified a number of ways by enzymes, known as writers, coupled with RNA binding domains. More than 100 chemical modifications have been identified in RNA involving all bases and the 2’-OH (Motorin and Helm, 2011). Among the most well-known writers are the methyltransferase like (METTL) proteins, such as the dual METTL3-METTL14 complex which uses a donor methyl group from the cosubstrate S-adenosyl methionine (SAM) to methylate the sixth carbon of adenine (m6A; Figure 5) (Sledz and Jinek, 2016). Some chemical modifications warp the local secondary structure of their resident RNAs, with reports of base-pair stabilization from pseudouridine modifications (Ge and Yu, 2013) and destabilization from inosine and m6A (Tanzer et al., 2019). Lastly, RNA is modified in a more extreme fashion by RBPs with nucleolytic activity, such as Argonaute proteins and the RNases Dicer and RNase II (Frazao et al., 2006; Jiang et al., 2011).
Regulation of binding and function
Protein-protein interactions (PPI) enable cooperative and competitive control over RBP binding. Such is the case for the translation initiation complex: eIf4E binding to the mRNA 5’-cap is enhanced by its association with subunit eIF4G; however, eIF4E interaction with eIF4E-BP prevents its binding to the eIF4F complex (Jackson et al., 2010). PPIs can also modulate RBP function or its efficiency thereof. Exon junction complex (EJC) interactors UPF2 and SMG1 induce conformational changes upon binding to UPF1 that de-repress its helicase activity (Figure 5) (Fiorini et al., 2015). m6A writers METTL3 and METTL14 are both able to modify RNA, but their activity is significantly enhanced by their mutual interaction (Sledz and Jinek, 2016). Non-protein ligands can also affect RBP binding, such as 4Fe-4S binding by IRP1 which activates its aconitase activity and mutually excludes iron response element binding (Figure 5) (Hentze et al., 2004). Similarly, ATP binding by helicases commonly induces ”clamping” of the helicase domain which increases its affinity for RNA (Gai’ et al., 2004; Linder and Jankowsky, 2011).
Post-translational modifications (PTMs) of residues in RBPs direct sophisticated regulation of their interaction sites and functions. RBPs often contain multiple sites for PTMs, the most common of which are phosphorylation (often serine), acetylation (often lysine) and arginine methylation (Hofweber and Dormann, 2019; Lovci et al., 2016). At the molecular level, PTMs introduce electrostatic charges that can affect the structural stability of binding regions or their ability to interact with RNA or other proteins (Drazic et al., 2016) (Law et al., 2003; Lovci et al., 2016). Phosphorylation and acetylation modifications introduce negative charge or neutralize positive charge, respectively. For example, acetylation of at least two lysine residues in the RRMs of PTBP1/2 disrupt their ability to hydrogen bond with the RNA backbone, likely by eliminating electrostatic attraction between the the positive lysine residues and the negatively charged RNA backbone (Pina et al., 2018). Arginine methylation on the other hand has been shown to decrease the favorability for cation-π interactions, such as the RGG-mediated interactions between IDRs that drive phase separation (Hofweber and Dormann, 2019). PTMs are most commonly observed modulating protein-protein interactions, such as the association between eIF4E and eIF4E-BP which is dependent on hypo-phosphorylation of eIF4-BP. Phosphorylation of at least two threonine residues in eIF4E-BP induces a structural change that buries its eIF4E binding site (Thapar, 2015), allowing eIF4E induction into the eIF4F complex instead (Figure 5).
Taking a less protein-centric view, RNA interactions and RNA modifications regulate RBP binding as well. M6A modifications were shown to ultimately determine whether binding sites are available to hnRNPC (Figure 5) (Liu et al., 2015). The small noncoding RNA BC1 inhibits eIF4A helicase activity, and long noncoding RNAs have been shown to act as RBP “sponges”, effectively reducing the abundance of an RBP for functional binding (HafezQorani et al., 2019; Linder and Jankowsky, 2011).
CONCLUSION
The current state of knowledge on RNA binding proteins is rapidly growing and includes many areas of study beyond the scope of this review. RNA binding site detection techniques, RBP synthetic design, and the role of RBPs in stress granules and neurodegenerative diseases, to name a few. All of these areas benefit from a solid mechanistic understanding of protein-RNA target interactions. In this review we took a mechanistic look at how RNA binding proteins interact with RNA, both at the molecular level and bird’s eye views. We repeatedly observe that detailed molecular structures explain the binding behavior and function of RBPs, such as helicase domains preferring interactions with the RNA backbone or specific residues in a YTH m6A reader protein “locking in” a methylated base. Similarly, our analysis of hydrogen bonds formed with RNA for different RNA binding domains reinforces the known structural function of several domains, but also shows how some domains are different from their peers. As the structures of more protein-RNA complexes are determined this analysis can be expanded to include more domain types, determining features that set domains apart or define their mechanism of binding. Additionally, studying RBP-substrate dynamics as well as larger multiprotein complexes are key for understanding the interactive process of protein-RNA regulation. We should note that the structural techniques used to study RNA binding domains are less capable of capturing the full RNA substrates that RBPs bind. Thus, while we are learning a great deal about what makes RBPs bind RNA, these techniques tell us much less about the reverse. Additional methods for the study of RNA molecules in complex with RBPs are needed. Finally, computational tools benefit greatly from our structural understanding of protein-RNA interaction mechanisms and in turn, enable rapid insights for any RBP of interest.
METHODS
Protein databank (PDB) format files for structures of RNA-interacting proteins were downloaded from RCSB (Berman et al., 2000) according to protein domain type, using search terms “KH domain”, “RRM”, “dsRBD”, “zinc finger”, “pumilio”, “DEAD”, “YTH domain”, and “cold shock domain”, and further narrowed by selecting x-ray crystallography structures and NMR structures consisting of both protein and RNA. PDB files were processed with HBPLUS (McDonald and Thornton, 1994), which infers hydrogen bond interactions between any two moieties, command:
echo file.pdb | clean #outputs to file.new
hbplus -d 3.35 -h 2.7 file.new file.pdb #outputs to file.hb2
All code used to assess protein-RNA hydrogen bond information in hb2 files is available on github.com/meracorley/hbplus_tools. Briefly, for each hb2 file, hydrogen bonds occurring between protein and RNA, including those coordinated by water molecules, were stored and organized by moiety type (base, sugar, backbone or sidechain, mainchain) as well as amino acid or base identity. To assess the frequency of each amino acid used in hydrogen bond interactions with RNA for a given structure, the number of interactions involving each amino acid was divided by the total number of potential protein-RNA interactions. To assess the frequency of each RNA nucleotide used in hydrogen bond interactions with protein, for a given structure the number of interactions involving each nucleotide was divided by the total number of potential protein-RNA interactions. The percentage of base|sugar|backbone|sidechain interactions was similarly calculated as the number of interactions of interest divided by the total number of potential interactions. The total number of hydrogen bonds per protein was counted per protein chain in a given structure and averaged over all chains. The total number of protein residues interacting with RNA was calculated as the count of unique interacting residues per protein chain in the given structure and averaged over all chains. P-values were calculated by two-sided t-tests.
ACKNOWLEDGMENTS
This work was supported by grants from the NIH (HG004659 and HD085902) to G.W.Y. M.C. was supported by the ALS Association Milton Safenowitz Post-doctoral Fellowship. The authors would like to thank Dr. Aaron Smargon for his input on protein-RNA structure analysis.
Footnotes
DECLARATION OF INTERESTS
G.W.Y. is co-founder, member of the Board of Directors, equity holder, and paid consultant for Locana and Eclipse BioInnovations. G.W.Y is a Distinguished Visiting Professor at the National University of Singapore. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. The authors declare no other competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akopian D, Shen K, Zhang X, and Shan SO (2013). Signal Recognition Particle: An Essential Protein-Targeting Machine. Annual Review of Biochemistry, Vol 82 82, 693–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain FH, Howe PW, Neuhaus D, and Varani G (1997). Structural basis of the RNA-binding specificity of human U1A protein. EMBO J 16, 5764–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers J, and Shamoo Y (2001). Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J Mol Biol 311, 75–86. [DOI] [PubMed] [Google Scholar]
- Amir M, Kumar V, Dohare R, Islam A, Ahmad F, and Hassan MI (2018). Sequence, structure and evolutionary analysis of cold shock domain proteins, a member of OB fold family. J Evolution Biol 31, 1903–1917. [DOI] [PubMed] [Google Scholar]
- Auweter SD, Oberstrass FC, and Allain FH (2006). Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res 34, 4943–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Oberstrass FC, and Allain FHT (2007). Solving the structure of PTB in complex with pyrimidine tracts: An NMR study of protein-RNA complexes of weak affinities. Journal of Molecular Biology 367, 174–186. [DOI] [PubMed] [Google Scholar]
- Aviv T, Lin Z, Ben-Ari G, Smibert CA, and Sicheri F (2006). Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p. Nat Struct Mol Biol 13, 168–176. [DOI] [PubMed] [Google Scholar]
- Balcerak A, Trebinska-Stryjewska A, Konopinski R, Wakula M, and Grzybowska EA (2019). RNA-protein interactions: disorder, moonlighting and junk contribute to eukaryotic complexity. Open Biol 9 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik A, C, N., Pilla SP, and Bahadur RP (2015). Molecular architecture of protein-RNA recognition sites. J Biomol Struct Dyn 33, 2738–2751. [DOI] [PubMed] [Google Scholar]
- Barik A, Mishra A, and Bahadur RP (2012). PRince: a web server for structural and physicochemical analysis of protein-RNA interface. Nucleic Acids Res 40, W440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud P, and Allain FHT (2012). ADAR Proteins: Double-stranded RNA and Z-DNA Binding Domains. Curr Top Microbiol 353, 35–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellucci M, Agostini F, Masin M, and Tartaglia GG (2011). Predicting protein associations with long noncoding RNAs. Nature Methods 8, 444–445. [DOI] [PubMed] [Google Scholar]
- Bercy M, and Bockelmann U (2015). Hairpins under tension: RNA versus DNA. Nucleic Acids Research 43, 9928–9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh SH, Srinivasan AR, and Schneider B (1992). The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J 63, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, and Bourne PE (2000). The Protein Data Bank. Nucleic Acids Research 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beusch I, Barraud P, Moursy A, Clery A, and Allain FH (2017). Tandem hnRNP A1 RNA recognition motifs act in concert to repress the splicing of survival motor neuron exon 7. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bifsha P, Landry K, Ashmarina L, Durand S, Seyrantepe V, Trudel S, Quiniou C, Chemtob S, Xu Y, Gravel RA, et al. (2007). Altered gene expression in cells from patients with lysosomal storage disorders suggests impairment of the ubiquitin pathway. Cell death and differentiation 14, 511–523. [DOI] [PubMed] [Google Scholar]
- Boland A, Huntzinger E, Schmidt S, Izaurralde E, and Weichenrieder O (2011). Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc Natl Acad Sci U S A 108, 10466–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, and Deragon JM (2009). A comprehensive analysis of the La-motif protein superfamily. RNA 15, 750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressin A, Schulte-Sasse R, Figini D, Urdaneta EC, Beckmann BM, and Marsico A (2019). TriPepSVM: de novo prediction of RNA-binding proteins based on short amino acid motifs. Nucleic Acids Res 47, 4406–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brylinski M (2018). Aromatic interactions at the ligand-protein interface: Implications for the development of docking scoring functions. Chem Biol Drug Des 91, 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, Curk T, Krijgsveld J, and Hentze MW (2016). Comprehensive Identification of RNA-Binding Domains in Human Cells. Mol Cell 63, 696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Dutko JA, Mian IS, and Belostotsky DA (2002). Arabidopsis thaliana exosome subunit AtRrp4p is a hydrolytic 3 ‘-> 5 ‘ exonuclease containing S1 and KH RNA-binding domains. Nucleic Acids Research 30, 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Sargsyan K, Wright JD, Huang YS, and Lim C (2014). Identifying RNA-binding residues based on evolutionary conserved structural and energetic features. Nucleic Acids Research 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cienikova Z, Jayne S, Damberger FF, Allain FHT, and Maris C (2015). Evidence for cooperative tandem binding of hnRNP C RRMs in mRNA processing. Rna 21, 1931–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cléry A, and Allain FHT (2011). From structure to function of RNA binding domains In RNA Binding Proteins, Zdravko L, ed. (Landes Bioscience Springer Science+Business Media; ). [Google Scholar]
- Clery A, Blatter M, and Allain FH (2008). RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol 18, 290–298. [DOI] [PubMed] [Google Scholar]
- Coimbatore Narayanan B, Westbrook J, Ghosh S, Petrov AI, Sweeney B, Zirbel CL, Leontis NB, and Berman HM (2014). The Nucleic Acid Database: new features and capabilities. Nucleic Acids Res 42, D114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook KB, Kazan H, Zuberi K, Morris Q, and Hughes TR (2011). RBPDB: a database of RNA-binding specificities. Nucleic Acids Res 39, D301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Gallardo I, Martino L, Kelly G, Atkinson RA, Trotta R, De Tito S, Coleman P, Ahdash Z, Gu Y, Bui TTT, et al. (2019). LARP4A recognizes polyA RNA via a novel binding mechanism mediated by disordered regions and involving the PAM2w motif, revealing interplay between PABP, LARP4A and mRNA. Nucleic Acids Res 47, 4272–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boulle K, Verkerk AJ, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra BA, and Willems PJ (1993). A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet 3, 31–35. [DOI] [PubMed] [Google Scholar]
- Deng L, Yang WY, and Liu H (2019). PredPRBA: Prediction of Protein-RNA Binding Affinity Using Gradient Boosted Regression Trees. Front Genet 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill KA, Ozkan SB, Shell MS, and Weikl TR (2008). The protein folding problem. Annual Review of Biophysics 37, 289–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D, Freese P, Alexis MS, Su A, Hochman M, Palden T, Bazile C, Lambert NJ, Van Nostrand EL, Pratt GA, et al. (2018). Sequence, Structure, and Context Preferences of Human RNA Binding Proteins. Mol Cell 70, 854–867 e859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazic A, Myklebust LM, Ree R, and Arnesen T (2016). The world of protein acetylation. Bba-Proteins Proteom 1864, 1372–1401. [DOI] [PubMed] [Google Scholar]
- Ellis JJ, Broom M, and Jones S (2007). Protein-RNA interactions: Structural analysis and functional classes. Proteins-Structure Function and Bioinformatics 66, 903–911. [DOI] [PubMed] [Google Scholar]
- Fiorini F, Bagchi D, Le Hir H, and Croquette V (2015). Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nature communications 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fislage M, Roovers M, Tuszynska I, Bujnicki JM, Droogmans L, and Versees W (2012). Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life. Nucleic Acids Res 40, 5149–5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores JK, and Ataide SF (2018). Structural Changes of RNA in Complex with Proteins in the SRP. Front Mol Biosci 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font J, and Mackay JP (2010). Beyond DNA: zinc finger domains as RNA-binding modules. Methods Mol Biol 649, 479–491. [DOI] [PubMed] [Google Scholar]
- Frazao C, Mcvey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, and Carrondo MA (2006). Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature 443, 110–114. [DOI] [PubMed] [Google Scholar]
- Gai’ DH, Zhao R, Li DW, Finkielstein CV, and Chen XS (2004). Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119, 47–60. [DOI] [PubMed] [Google Scholar]
- Ge J, and Yu YT (2013). RNA pseudouridylation: new insights into an old modification. Trends Biochem Sci 38, 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, and Tuschl T (2014). A census of human RNA-binding proteins. Nature Reviews Genetics 15, 829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice G, Sanchez-Cabo F, Torroja C, and Lara-Pezzi E (2016). ATtRACT-a database of RNA-binding proteins and associated motifs. Database : the journal of biological databases and curation 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, and Dreyfuss G (2008). RNA-binding proteins and post-transcriptional gene regulation. Febs Letters 582, 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, and Gribskov M (2011). The role of RNA sequence and structure in RNA--protein interactions. J Mol Biol 409, 574–587. [DOI] [PubMed] [Google Scholar]
- HafezQorani S, Houdjedj A, Arici M, Said A, and Kazan H (2019). RBPSponge: genomewide identification of lncRNAs that sponge RBPs. Bioinformatics 35, 4760–4763. [DOI] [PubMed] [Google Scholar]
- Hainzl T, Huang S, and Sauer-Eriksson AE (2005). Structural insights into SRP RNA: an induced fit mechanism for SRP assembly. RNA 11, 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TM (2005). Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol 15, 367–373. [DOI] [PubMed] [Google Scholar]
- Han K, and Nepal C (2007). PRI-Modeler: extracting RNA structural elements from PDB files of protein-RNA complexes. FEBS Lett 581, 1881–1890. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Castello A, Schwarzl T, and Preiss T (2018). A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol 19, 327–341. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, and Andrews NC (2004). Balancing acts: Molecular control of mammalian iron metabolism. Cell 117, 285–297. [DOI] [PubMed] [Google Scholar]
- Hock J, and Meister G (2008). The Argonaute protein family. Genome Biol 9, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MM, Khrapov MA, Cox JC, Yao J, Tong L, and Ellington AD (2004). AANT: the Amino Acid-Nucleotide Interaction Database. Nucleic Acids Res 32, D174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofweber M, and Dormann D (2019). Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J Biol Chem 294, 7137–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain ST, Malhotra A, and Deutscher MP (2016). How RNase R Degrades Structured RNA: ROLE OF THE HELICASE ACTIVITY AND THE S1 DOMAIN. Journal of Biological Chemistry 291, 7877–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Qin L, Li ML, Pu XM, and Guo YZ (2018). A structural dissection of protein-RNA interactions based on different RNA base areas of interfaces. Rsc Adv 8, 10582–10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YY, Liu SY, Guo DC, Li L, and Xiao Y (2013). A novel protocol for three-dimensional structure prediction of RNA-protein complexes. Scientific reports 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson WH, and Ortlund EA (2014). The structure, function and evolution of proteins that bind DNA and RNA. Nat Rev Mol Cell Bio 15, 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, and Schulten K (1996). VMD: visual molecular dynamics. J Mol Graph 14, 33–38, 27–38. [DOI] [PubMed] [Google Scholar]
- Iost I, Dreyfus M, and Linder P (1999). Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. Journal of Biological Chemistry 274, 17677–17683. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CUT, and Pestova TV (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Bio 11, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E (2011). RNA helicases at work: binding and rearranging. Trends Biochem Sci 36, 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvelin AI, Noerenberg M, Davis I, and Castello A (2016). The new (dis)order in RNA regulation. Cell Commun Signal 14, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang FG, Ramanathan A, Miller MT, Tang GQ, Gale M, Patel SS, and Marcotrigiano J (2011). Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479, 423–U184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Daley DTA, Luscombe NM, Berman HM, and Thornton JM (2001). Protein-RNA interactions: a structural analysis. Nucleic Acids Research 29, 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubb HC, Higueruelo AP, Ochoa-Montano B, Pitt WR, Ascher DB, and Blundell TL (2017). Arpeggio: A Web Server for Calculating and Visualising Interatomic Interactions in Protein Structures. Journal of Molecular Biology 429, 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainov DE, Mancini EJ, Telenius J, Lisai J, Grimes JM, Bamford DH, Stuart DI, and Tuma R (2008). Structural basis of mechanochemical coupling in a hexameric molecular motor. Journal of Biological Chemistry 283, 3607–3617. [DOI] [PubMed] [Google Scholar]
- Kainov DE, Pirttimaa M, Tuma R, Butcher SJ, Thomas GJ, Bamford DH, and Makeyev EV (2003). RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein. Journal of Biological Chemistry 278, 48084–48091. [DOI] [PubMed] [Google Scholar]
- Kappel K, and Das R (2019). Sampling Native-like Structures of RNA-Protein Complexes through Rosetta Folding and Docking. Structure 27, 140−+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J, Chen RZ, Ban T, Zhou XE, Gu X, Tan MH, Chen C, Kang Y, Brunzelle JS, Zhu JK, et al. (2013). Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat Struct Mol Biol 20, 1377–1382. [DOI] [PubMed] [Google Scholar]
- Keffer-Wilkes LC, Veerareddygari GR, and Kothe U (2016). RNA modification enzyme TruB is a tRNA chaperone. Proc Natl Acad Sci U S A 113, 14306–14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim OTP, Yura K, and Go N (2006). Amino acid residue doublet propensity in the protein-RNA interface and its application to RNA interface prediction. Nucleic Acids Research 34, 6450–6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kljashtorny V, Nikonov S, Ovchinnikov L, Lyabin D, Vodovar N, Curmi P, and Manivet P (2015). The Cold Shock Domain of YB-1 Segregates RNA from DNA by Non-Bonded Interactions. PLoS One 10, e0130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Gromiha AM, and Raghava GPS (2008). Prediction of RNA binding sites in a protein using SVM and PSSM profile. Proteins-Structure Function and Bioinformatics 71, 189–194. [DOI] [PubMed] [Google Scholar]
- Lai WS, Carballo E, Thorn JM, Kennington EA, and Blackshear PJ (2000). Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem 275, 17827–17837. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Jablonska J, Pravda L, Varekova RS, and Thornton JM (2018). PDBsum: Structural summaries of PDB entries. Protein Science 27, 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law LMJ, Everitt JC, Beatch MD, Holmes CFB, and Hobman TC (2003). Phosphorylation of rubella virus capsid regulates its RNA binding activity and virus replication. Journal of Virology 77, 1764–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulliot N, and Varani G (2001). Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry 40, 7947–7956. [DOI] [PubMed] [Google Scholar]
- Li SL, Yamashita K, Amada KM, and Standley DM (2014). Quantifying sequence and structural features of protein-RNA interactions. Nucleic Acids Research 42, 10086–10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Sun H, and Xu C (2018). YTH Domain: A Family of N(6)-methyladenosine (m(6)A) Readers. Genomics Proteomics Bioinformatics 16, 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, and Jankowsky E (2011). From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Bio 12, 505–516. [DOI] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, and Pan T (2015). N(6)-methyladenos-inedependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković ZJ (2012). RNA binding proteins (Austin, Tex.: Landes Bioscience; ). [Google Scholar]
- Loughlin FE, Lukavsky PJ, Kazeeva T, Reber S, Hock EM, Colombo M, Von Schroetter C, Pauli P, Clery A, Muhlemann O, et al. (2019). The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity. Molecular Cell 73, 490−+. [DOI] [PubMed] [Google Scholar]
- Lovci MT, Bengtson MH, and Massirer KB (2016). Post-Translational Modifications and RNA-Binding Proteins. Rna Processing: Disease and Genome-Wide Probing 907, 297–317. [DOI] [PubMed] [Google Scholar]
- Loveland AB, and Korostelev AA (2018). Structural dynamics of protein S1 on the 70S ribosome visualized by ensemble cryo-EM. Methods 137, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, and Varani G (2007). RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8, 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Laskowski RA, and Thornton JM (1997). NUCPLOT: a program to generate schematic diagrams of protein-nucleic acid interactions. Nucleic Acids Res 25, 4940–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Laskowski RA, and Thornton JM (2001). Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Research 29, 2860–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Ye K, and Patel DJ (2004). Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429, 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, and Patel DJ (2005). Structural basis for 5 ‘end-specific recognition of guide RNA by the A-fulgidus Piwi protein. Nature 434, 666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, and Allain FH (2005). The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272, 2118–2131. [DOI] [PubMed] [Google Scholar]
- Masliah G, Barraud P, and Allain FH (2013). RNA recognition by double-stranded RNA binding domains: a matter of shape and sequence. Cell Mol Life Sci 70, 1875–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews MM, Thomas JM, Zheng Y, Tran K, Phelps KJ, Scott AI, Havel J, Fisher AJ, and Beal PA (2016). Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat Struct Mol Biol 23, 426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald IK, and Thornton JM (1994). Satisfying hydrogen bonding potential in proteins. J Mol Biol 238, 777–793. [DOI] [PubMed] [Google Scholar]
- Mihailovich M, Militti C, Gabaldon T, and Gebauer F (2010). Eukaryotic cold shock domain proteins: highly versatile regulators of gene expression. Bioessays 32, 109–118. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Ito K, Murakami R, and Uchiumi T (2016). Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute. Nature communications 7, 11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova N, Allers J, Myers J, and Shamoo Y (2006). Protein-RNA interactions: exploring binding patterns with a three-dimensional superposition analysis of high resolution structures. Bioinformatics 22, 2746–2752. [DOI] [PubMed] [Google Scholar]
- Motorin Y, and Helm M (2011). RNA nucleotide methylation. Wiley interdisciplinary reviews. RNA 2, 611–631. [DOI] [PubMed] [Google Scholar]
- Muppirala UK, Honavar VG, and Dobbs D (2011). Predicting RNA-Protein Interactions Using Only Sequence Information. Bmc Bioinformatics 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R, and Gromiha MM (2014). Prediction of RNA Binding Residues: An Extensive Analysis Based on Structure and Function to Select the Best Predictor. Plos One 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Lakomek K, Naumann PT, Erwin WM, Lauhon CT, and Ficner R (2014). Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification. Nucleic Acids Res 42, 6673–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras AC, Mak P, Darzynkiewicz E, Sonenberg N, et al. (2002). Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5 ‘ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. Journal of Molecular Biology 319, 615–635. [DOI] [PubMed] [Google Scholar]
- Onofrio A, Parisi G, Punzi G, Todisco S, Di Noia MA, Bossis F, Turi A, De Grassi A, and Pierri CL (2014). Distance-dependent hydrophobic-hydrophobic contacts in protein folding simulations. Phys Chem Chem Phys 16, 18907–18917. [DOI] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo CH, and Nagai K (1994). Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372, 432–438. [DOI] [PubMed] [Google Scholar]
- Paz I, Kosti I, Ares M, Cline M, and Mandel-Gutfreund Y (2014). RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Research 42, W361–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Arellano I, Gallego J, and Cervera J (2007). The PUA domain - a structural and functional overview. FEBS J 274, 4972–4984. [DOI] [PubMed] [Google Scholar]
- Perez-Cano L, and Fernandez-Recio J (2010). Optimal Protein-RNA Area, OPRA: A propensity-based method to identify RNA-binding sites on proteins. Proteins-Structure Function and Bioinformatics 78, 25–35. [DOI] [PubMed] [Google Scholar]
- Pina JM, Reynaga JM, Truong AAM, and Keppetipola NM (2018). Post-Translational Modifications in Polypyrimidine Tract Binding Proteins PTBP1 and PTBP2. Biochemistry 57, 3873–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, and Schroeder R (2007). RNA chaperones, RNA annealers and RNA helicases. RNA Biol 4, 118–130. [DOI] [PubMed] [Google Scholar]
- Ramos A, Grunert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St Johnston D, and Varani G (2000). RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J 19, 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranji A, Shkriabai N, Kvaratskhelia M, Musier-Forsyth K, and Boris-Lawrie K (2011). Features of double-stranded RNA-binding domains of RNA helicase A are necessary for selective recognition and translation of complex mRNAs. J Biol Chem 286, 5328–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranathan S, Oberstrass FC, and Allain FHT (2010). Increase in Backbone Mobility of the VTS1p-SAM Domain on Binding to SRE-RNA. Journal of Molecular Biology 396, 732–746. [DOI] [PubMed] [Google Scholar]
- Sachs R, Max KEA, Heinemann U, and Balbach J (2012). RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution. Rna 18, 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaee N, Kozlov G, Noronha AM, Xie J, Wilds CJ, and Gehring K (2012). Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol Cell 48, 375–386. [DOI] [PubMed] [Google Scholar]
- Salentin S, Schreiber S, Haupt VJ, Adasme MF, and Schroeder M (2015). PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res 43, W443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk L, Meinel DM, Strasser K, and Gerber AP (2012). La-motif-dependent mRNA association with Slf1 promotes copper detoxification in yeast. RNA 18, 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, and Brennan RG (2002). Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. Embo Journal 21, 3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, and Anirudh CR (2017). Mechanism of mRNA-STAR domain interaction: Molecular dynamics simulations of Mammalian Quaking STAR protein. Scientific reports 7, 12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman-Peleg A, Shatsky M, Nussinov R, and Wolfson HJ (2008). Prediction of interacting single-stranded RNA bases by protein-binding patterns. Journal of Molecular Biology 379, 299–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz P, and Jinek M (2016). Structural insights into the molecular mechanism of the m(6)A writer complex. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahr H, Chia T, Lingford JP, Siira SJ, Cohen SB, Filipovska A, and Rackham O (2018). Modular ssDNA binding and inhibition of telomerase activity by designer PPR proteins. Nature communications 9, 2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl R, Oberstrass FC, Hood JL, Jourdan M, Zimmermann M, Skrisovska L, Maris C, Peng L, Hofr C, Emeson RB, et al. (2010). The Solution Structure of the ADAR2 dsRBM-RNA Complex Reveals a Sequence-Specific Readout of the Minor Groove. Cell 143, 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Marzluff WF, Dominski Z, and Tong L (2013). Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3’hExo ternary complex. Science 339, 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan DZ, Zhou M, Kiledjian M, and Tong L (2014). The ROQ domain of Roquin recognizes mRNA constitutive-decay element and double-stranded RNA. Nature Structural & Molecular Biology 21, 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer A, Hofacker IL, and Lorenz R (2019). RNA modifications in structure prediction - Status quo and future challenges. Methods 156, 32–39. [DOI] [PubMed] [Google Scholar]
- Teichmann M, Dumay-Odelot H, and Fribourg S (2012). Structural and functional aspects of winged-helix domains at the core of transcription initiation complexes. Transcription 3, 2–7. [DOI] [PubMed] [Google Scholar]
- Teplova M, Hafner M, Teplov D, Essig K, Tuschl T, and Patel DJ (2013). Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites. Gene Dev 27, 928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Malinina L, Darnell JC, Song J, Lu M, Abagyan R, Musunuru K, Teplov A, Burley SK, Darnell RB, et al. (2011). Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1. Structure 19, 930–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Yuan YR, Phan AT, Malinina L, Ilin S, Teplov A, and Patel DJ (2006). Structural basis for recognition and sequestration of UUUOH 3 ‘ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Molecular Cell 21, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terribilini M, Sander JD, Lee JH, Zaback P, Jernigan RL, Honavar V, and Dobbs D (2007). RNABindR: a server for analyzing and predicting RNA-binding sites in proteins. Nucleic Acids Research 35, W578–W584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapar R (2015). Structural Basis for Regulation of RNA-Binding Proteins by Phosphorylation. ACS chemical biology 10, 652–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore S, Mayer C, Sauter C, Weeks S, and Suck D (2003). Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA - Common features of RNA binding in Archaea and Eukarya. Journal of Biological Chemistry 278, 1239–1247. [DOI] [PubMed] [Google Scholar]
- Tian Y, Simanshu DK, Ma JB, and Patel DJ (2011). Structural basis for piRNA 2’-Omethylated 3’-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains. Proc Natl Acad Sci U S A 108, 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treger M, and Westhof E (2001). Statistical analysis of atomic contacts at RNA-protein interfaces. J Mol Recognit 14, 199–214. [DOI] [PubMed] [Google Scholar]
- Valverde R, Edwards L, and Regan L (2008). Structure and function of KH domains. FEBS J 275, 2712–2726. [DOI] [PubMed] [Google Scholar]
- Vukovic L, Koh HR, Myong S, and Schulten K (2014). Substrate Recognition and Specificity of Double-Stranded RNA Binding Proteins. Biochemistry 53, 3457–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden WE, Selezneva A, and Volz K (2012). Accommodating variety in iron-responsive elements: Crystal structure of transferrin receptor 1 B IRE bound to iron regulatory protein 1. FEBS Lett 586, 32–35. [DOI] [PubMed] [Google Scholar]
- Walden WE, Selezneva AI, Dupuy J, Volbeda A, Fontecilla-Camps JC, Theil EC, and Volz K (2006). Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science 314, 1903–1908. [DOI] [PubMed] [Google Scholar]
- Wang M, Oge L, Perez-Garcia MD, Hamama L, and Sakr S (2018). The PUF Protein Family: Overview on PUF RNA Targets, Biological Functions, and Post Transcriptional Regulation. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZH, Hartman E, Roy K, Chanfreau G, and Feigon J (2011). Structure of a Yeast RNase III dsRBD Complex with a Noncanonical RNA Substrate Provides New Insights into Binding Specificity of dsRBDs. Structure 19, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir JR, Bonneau F, Hentschel J, and Conti E (2010). Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. P Natl Acad Sci USA 107, 12139–12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KA, Holland DJ, and Wetmore SD (2016). Topology of RNA-protein nucleobase-amino acid pi-pi interactions and comparison to analogous DNA-protein pi-pi contacts. RNA 22, 696–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worbs M, Bourenkov GP, Bartunik HD, Huber R, and Wahl MC (2001). An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Molecular Cell 7, 1177–1189. [DOI] [PubMed] [Google Scholar]
- Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, and Min J (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol 10, 927–929. [DOI] [PubMed] [Google Scholar]
- Yan J, and Kurgan L (2017). DRNApred, fast sequence-based method that accurately predicts and discriminates DNA- and RNA-binding residues. Nucleic Acids Res 45, e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, and Zhou MM (2003). Structure and conserved RNA binding of the PAZ domain. Nature 426, 468–474. [DOI] [PubMed] [Google Scholar]
- Yang JY, Roy A, and Zhang Y (2013). BioLiP: a semi-manually curated database for biologically relevant ligand-protein interactions. Nucleic Acids Research 41, D1096–D1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wang C, Li F, Zhang J, Nayab A, Wu J, Shi Y, and Gong Q (2017). The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity. J Biol Chem 292, 16221–16234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YD, Zhao HY, Wang JH, and Zhou YQ (2014). SPOT-Seq-RNA: Predicting Protein-RNA Complex Structure and RNA-Binding Function by Fold Recognition and Binding Affinity Prediction. Protein Structure Prediction, 3rd Edition 1137, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YS, Declerck N, Manival X, Aymerich S, and Kochoyan M (2002). Solution structure of the LicT-RNA antitermination complex: CAT clamping RAT. Embo Journal 21, 1987–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y, Zhao Y, Li C, Zhang L, Huang H, Li Y, Liu L, Hou P, Cui T, Tan P, et al. (2017). RAID v2.0: an updated resource of RNA-associated interactions across organisms. Nucleic Acids Res 45, D115–D118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Wang J, Sheng QH, Liu Q, and Shyr Y (2019). beRBP: binding estimation for human RNA-binding proteins. Nucleic Acids Research 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QF, Ye W, Jiang C, Luo R, and Chen HF (2014). Specific Recognition Mechanism between RNA and the KH3 Domain of Nova-2 Protein. Journal of Physical Chemistry B 118, 12426–12434. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ma Z, and Kurgan L (2017). Comprehensive review and empirical analysis of hallmarks of DNA-, RNA- and protein-binding residues in protein chains. Brief Bioinform. [DOI] [PubMed] [Google Scholar]
- Zhao HY, Yang YD, and Zhou YQ (2011). Structure-based prediction of RNA-binding domains and RNA-binding sites and application to structural genomics targets. Nucleic Acids Research 39, 3017–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YY, Mao MW, Zhang WJ, Wang J, Li HT, Yang Y, Wang Z, and Wu JW (2018). Expanding RNA binding specificity and affinity of engineered PUF domains. Nucleic Acids Res 46, 4771–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu TT, Roundtree IA, Wang P, Wang X, Wang L, Sun C, Tian Y, Li J, He C, and Xu YH (2014). Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell research 24, 1493–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]