Abstract
Toxoplasma gondii is an obligate intracellular parasite which is capable of establishing life-long chronic infection in any mammalian host. During the intracellular life cycle, the parasite secretes an array of proteins into the parasitophorous vacuole (PV) where it resides. Specialized organelles called the dense granules secrete GRA proteins that are known to participate in nutrient acquisition, immune evasion, and host cell-cycle manipulation. Although many GRAs have been discovered which are expressed during the acute infection mediated by tachyzoites, little is known about those that participate in the chronic infection mediated by the bradyzoite form of the parasite. In this study, we sought to uncover novel bradyzoite-upregulated GRA proteins using proximity biotinylation, which we previously used to examine the secreted proteome of the tachyzoites. Using a fusion of the bradyzoite upregulated protein MAG1 to BirA* as bait and a strain with improved switch efficiency, we identified a number of novel GRA proteins which are expressed in bradyzoites. After using the CRISPR/Cas9 system to characterize these proteins by gene knockout, we focused on one of these GRAs (GRA55) and found it was important for the establishment or maintenance of cysts in the mouse brain. These findings highlight new components of the GRA proteome of the tissue-cyst life stage of T. gondii and identify potential targets that are important for maintenance of parasite persistence in vivo.
Introduction
Toxoplasma gondii is an apicomplexan parasite that chronically infects nearly every animal and approximately one-third of the world’s human population [1–3]. While the infection is typically asymptomatic in healthy persons, infection in immunocompromised patients (such as those with AIDS or patients taking immunosuppressive drugs) can result in life-threatening central nervous system disease [3,4]. While therapies exist that can combat the acute infection consisting of rapidly growing tachyzoites, there are no effective treatments that can clear the chronic infection which is mediated by slow-growing bradyzoite cysts. Thus, patients who are chronically infected with bradyzoites live under a life-long threat of reactivation of the parasite if a lapse in immune surveillance occurs [4]. A mechanistic understanding of how T. gondii bradyzoite cysts are formed and able to maintain lifelong persistence in the host is critical for the development of novel therapies that target this important intracellular pathogen.
T. gondii actively invades its host cells and replicates inside of a membrane-bound parasitophorous vacuole (PV) within the host cell cytoplasm [5]. Host cell invasion, PV formation and maintenance are mediated by a set of specialized secretory organelles known as micronemes, rhoptries, and dense granules [6–9]. While micronemes and rhoptries play roles in the initial stages of attachment and invasion, the dense granules secrete proteins called GRAs into the vacuolar space that participate in the remodeling and maintenance of the PV during intracellular replication [10–16]. While many GRAs function within the vacuole after secretion, some GRAs are able to cross the vacuolar membrane into the host cell and hijack cellular immune functions [17–22]. Most of the currently known GRAs have been characterized in the context of the acutely infectious tachyzoite life-cycle stage of the parasite. As expected, some of these have been found to have important roles during both stages of infection. GRA17, which operates as a small-molecule transporter at the PVM in tachyzoites, has also been shown to be important for growth and maintenance of bradyzoites [23,24]. Several other well-studied GRAs such as GRA1, GRA2, GRA4, GRA5, GRA6, GRA9 and GRA12 have been shown to express during the bradyzoite phase and localize to the cyst wall and matrix at different points of maturation [16,25–28]. Deletion of GRA2 results in defective formation of the cyst matrix and the failure of GRA4 and GRA6 to accumulate at the cyst periphery; this finding was in line with previous data which revealed the importance of GRA2, GRA4 and GRA6 in the formation of the intravacuolar network in tachyzoites [12,16,29].
While cyst formation and maintenance are likely to involve an array of bradyzoite-specific GRA proteins, only a few such proteins have been identified and little is known how they might enable the parasite to establish and maintain the chronic infection. These include CST1, bradyzoite pseudokinase 1 (BPK1) and the newly discovered CST2/GRA50, CST3/GRA51, CST4, CST5/GRA52, and CST6/GRA53 [30–34]. The deletion of the glycoprotein CST1 results in fragile cysts with thin cyst walls and lower cyst burden in a chronic-infection model [31]. BPK1 was found to be important in oral infectivity, and the disruption of this protein led to small in vivo cysts whose walls were more easily broken down by pepsin-acid treatment [34]. The purification of the cyst wall using Percoll gradient followed by immunoprecipitation with an anti-CST1 monoclonal antibody was used to identify cyst components. This study revealed 5 new bradyzoite-upregulated GRAs which localized to the tissue cyst wall and cyst matrix (named CST2 –CST6). Of these, CST2/GRA50 was important for parasite virulence and establishment of cyst burden [33]. Although this study revealed new proteins which are bradyzoite-specific cyst-wall components, it is likely that there are many more GRAs which participate in the formation and maintenance of bradyzoite tissue cysts during the chronic T. gondii infection.
To further bridge this gap in our knowledge, we have developed new approaches to rapidly identify and determine the function of novel GRAs. One important advance is our adaptation of the BioID technique to T. gondii for identifying proteins in specific subcellular compartments [35,36]. This approach utilizes a bait protein fused to a promiscuous biotin ligase (BirA*) that biotinylates interacting and proximal proteins in subcellular compartments in vivo. The resulting biotinylated proteins are isolated by streptavidin chromatography and are then identified by mass spectrometry. We have recently utilized this approach to identify a robust dataset of previously known and candidate vacuolar proteins in the tachyzoite stage of the parasite [36]. Our follow-up endogenous tagging of a subset of these candidate GRAs resulted in the verification of many new GRAs, demonstrating the effectiveness of this approach. This dataset thus provided an initial tachyzoite GRA proteome of T. gondii, many of which are also expressed in bradyzoites.
In this manuscript, we build on these results by extending the GRA BioID experiments to bradyzoites, utilizing a bradyzoite-upregulated protein as a bait and a background strain which enables improved switching to bradyzoites in vitro. Using this approach, we have identified five novel GRA proteins expressed in bradyzoite cysts. We subsequently used CRISPR/Cas9 to disrupt all five of the genes encoded these GRAs, demonstrating that they are not essential for growth in vitro. We then focused on one of these knockouts (GRA55) which is dispensable for growth in vitro and virulence of T. gondii during the acute phase of infection but is important for establishing or maintaining chronic infection in a mouse model. Together, this research identifies a group of novel bradyzoite-upregulated GRA proteins that are novel potential targets for the chronic T. gondii infection.
Results
Adaptation of BioID for Toxoplasma bradyzoite GRAs
Our previous success with using BioID to identify secreted GRAs in tachyzoites suggested that this approach would also work for bradyzoites. To improve our changes of obtaining bradyzoite GRAs, we made several modifications to our approach. First, we used as our background the PrugniaudΔku80 strain of T. gondii that contains GFP driven by the bradyzoite specific promoter LDH2, which has previously been shown to be effective at monitoring switching to bradyzoites [37]. Second, we sought to improve the efficiency of in vitro switching to bradyzoites, thus we deleted AP2IV-4 (S1 Fig), which encodes one of the nuclear Apetala-2 (AP2) proteins which has previously been shown to be important for the maintenance of the tachyzoite phenotype in vitro [38]. Lastly, we used the bradyzoite-upregulated GRA protein MAG1 [39,40] as our bait which we fused to the biotin ligase BirA* (Fig 1A). Deletion of AP2IV-4 and obtaining the PruΔku80::Δap2iv-4 and MAG1-BirA*::Δap2iv-4 parasites enables increased bradyzoite switching efficiency without disrupting the lytic cycle of the tachyzoites as previously described [38].
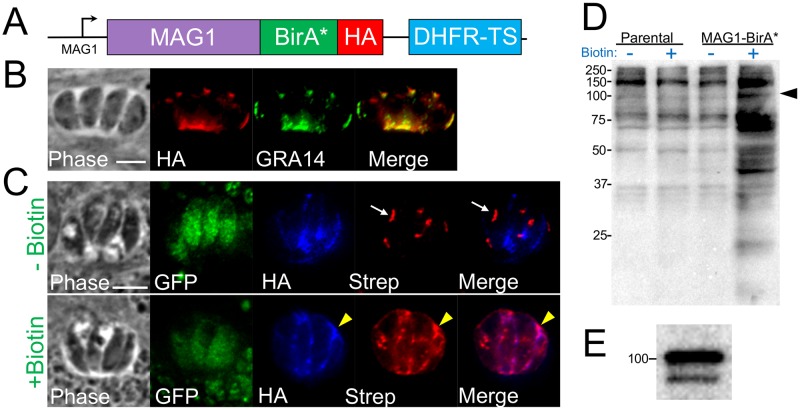
Fig 1. MAG1-BirA* localizes to the PV and biotinylates proteins in the cyst vacuole.
(A) Diagram of the construct encoding the promoter and full genomic sequence of MAG1 fused to BirA* along with a 3xHA C-terminal epitope tag. (B) IFA of MAG1-BirA* showing that the fusion protein appropriately traffics to the tachyzoite PV and colocalizes with the known protein GRA14. Scale bar: 5 μm (applicable to all panels). (C) IFA of MAG1-BirA*-expressing parasites, showing the bradyzoite PV is labeled in a biotin-dependent manner (yellow arrowheads, +Biotin row). Endogenously biotinylated apicoplasts are observed with and without biotin (white arrows, -Biotin row). Scale bar: 5 μm (applicable to all panels in C). (D) Western blot of whole-cell lysates of parental (PrugniaudΔku80Δhxgprt) and MAG1-BirA*-expressing parasites -/+ biotin. Lysates were probed with streptavidin-HRP, revealing an increase in biotinylated proteins in MAG1-BirA*-expressing parasites upon addition of biotin. The MAG1-BirA* fusion protein is predicted to be ~105 kDa (arrowhead). (E) Western blot showing that MAG1-BirA* fusion migrates to ~105 kDa as detected by antibody against HA epitope tag.
When we expressed the MAG1-BirA* fusion driven from its endogenous promoter, the fusion was readily detectable in tachyzoites and trafficked appropriately to the PV when examined by immunofluorescence assay (IFA) (Fig 1B). While MAG1 is considered a bradyzoite GRA, this detection was expected as it is clearly expressed in tachyzoites and is one of the stronger hits in our tachyzoite GRA BioID experiments [36]. Upon in vitro switching to bradyzoites, the fusion expressed in bradyzoites and trafficked appropriately to cyst vacuole where it co-localized with the known GRA marker GRA14 (Fig 1B) [41]. The fusion protein migrated at a molecular weight of ~105 kDa as predicted [42]
In vivo biotinylation of the PV using MAG1-BirA* parasites
After producing switch-efficient MAG1-BirA* parasites, we assessed whether the fusion protein was active in the PV. To do this, we switched MAG1-BirA* parasites in vitro using high pH media for 5 days and supplemented biotin to the media on the final day [43]. Transition from the tachyzoite to bradyzoite stage was confirmed by the presence of GFP in the parasite cytosol (Fig 1C). As a control, MAG1-BirA* parasites were switched on a separate monolayer, without biotin supplementation. While the control parasites just showed the expected apicoplast background staining, we observed streptavidin staining overlapping with the fusion protein in the parasite vacuole when we supplemented biotin (Fig 1C). This demonstrated that the biotin was able to penetrate the PV compartment and the MAG1-BirA* was active during the induced bradyzoite stage. We additionally demonstrated by western blot that there was an increase in the number of biotinylated proteins in MAG1-BirA* lysates when excess biotin was supplemented to the media (Fig 1D). We then proceeded with a large-scale MAG1-BioID experiment by infecting human foreskin fibroblasts with a high multiplicity of infection, switching them in vitro to bradyzoites over the course of 5 days, supplementing biotin on day 4. We then harvested the cells, lysed the samples using stringent conditions, and purified the biotinylated protein fraction using streptavidin-affinity chromatography.
As with previous experiments, mass spectrometry analysis revealed a large number of tachyzoite and bradyzoite GRAs highly ranked by peptide spectrum matches when we examined the MAG1-BirA* lysate in comparison to the parental lysate (Table 1 and S1 Table) [36]. We found the bait protein (MAG1) as well as almost all numbered GRAs, MYR proteins, TgIST1, WNG1, WNG2, bradyzoite GRAs (such as CST proteins, MCP3, MCP4, and BPK1), cyclophilin, NTPaseI and NTPaseII. BPK1 did appear, although slightly lower ranking in abundance. There were several expected proteins that were missing from our dataset including the exported proteins GRA19, GRA20 and GRA21 [44]. In total, the MAG1-BioID experiment revealed 537 proteins that were not present in the control lysate, 675 proteins in total including some GRAs that were present in comparatively low levels in the control lysate (S1 Table). GRAs constituted many of the highest-ranking proteins in this experiment, however, as expected many other secreted proteins including rhoptry proteins (ROPs/RONs), and microneme proteins (MICs) were also found concentrated in the top hits of the dataset.
Table 1. Summary of known and novel GRAs found by MAG1-BioID.
| ToxoDB number | Protein Identification |
|---|---|
| TGME49_209755 | MAG2 |
| TGME49_264660 | SRS44/CST1 |
| TGME49_270240 | MAG1 |
| TGME49_217680 | GRA57 |
| TGME49_208730 | MCP4 |
| TGME49_204340 | GRA54 |
| TGME49_288650 | GRA12 |
| TGME49_254470 | MYR1 |
| TGME49_212300 | GRA32 |
| TGME49_228170 | IMC2a - GRA44 |
| TGME49_203310 | GRA7 |
| TGME49_289380 | GRA39 |
| TGME49_203290 | GRA34 |
| TGME49_231960 | GRA28 |
| TGME49_232600 | PLP1 |
| TGME49_240060 | TgIST1 |
| TGME49_222370 | SRS13 |
| TGME49_289540 | GRA29 homologue |
| TGME49_208740 | MCP3 |
| TGME49_269690 | GRA29 |
| TGME49_312420 | GRA38 |
| TGME49_230705 | CST3/GRA51 |
| TGME49_239740 | GRA14 |
| TGME49_251540 | GRA9 |
| TGME49_220240 | GRA31 |
| TGME49_226380 | GRA35 |
| TGME49_213067 | GRA36 |
| TGME49_253330 | BPK1 |
| TGME49_208830 | GRA16 |
| TGME49_261650 | CST4 |
| TGME49_232000 | GRA30 |
| TGME49_319340 | CST5/GRA52 |
| TGME49_227620 | GRA2 |
| TGME49_247440 | GRA33 |
| TGME49_309930 | GRA56 |
| TGME49_308093 | ROP32 |
| TGME49_310780 | GRA4 |
| TGME49_221210 | cyclophilin |
| TGME49_275440 | GRA6 |
| TGME49_290700 | GRA25 |
| TGME49_309760 | GRA55 |
| TGME49_215220 | GRA22 |
| TGME49_275470 | GRA15 |
| TGME49_304740 | ROP35/WNG1 |
| TGME49_227280 | GRA3 |
| TGME49_260520 | CST6/GRA53 |
| TGME49_203600 | CST2/GRA50 |
| TGME49_254720 | GRA8 |
| TGME49_277270 | NTPAse II |
| TGME49_201130 | ROP33 |
| TGME49_270250 | GRA1 |
| TGME49_237015 | GRA43 |
| TGME49_240090 | ROP34/WNG2 |
| TGME49_267740 | GRA48 |
| TGME49_277240 | NTPase I |
| TGME49_230180 | GRA24/TgBRADIN |
| TGME49_254000 | GRA47 |
| TGME49_279100 | MAF1a |
| TGME49_297880 | GRA23 |
| TGME49_237230 | MYR3 |
| TGME49_239010 | TEEGR/HCE1 |
| TGME49_219810 | GRA40 |
| TGME49_236890 | GRA37 |
| TGME49_268790 | GRA58 |
| TGME49_262050 | ROP39 |
| TGME49_316250 | GRA45 |
| TGME49_234950 | ROP48 |
| TGME49_237880 | GRA13 |
| TGME49_244530 | GRA49 |
| TGME49_286450 | GRA5 |
| TGME49_222170 | GRA17 |
Novel GRAs highlighted in bold and with grey shading.
Identification of novel bradyzoite dense granule proteins from BioID datasets
Our dataset also contained a large number of hypothetical proteins. We narrowed this list of proteins as likely GRAs by selecting those proteins that were highly ranked in the dataset and lacked a C-terminal endoplasmic reticulum retention signal (K/HDEL) [11,45]. We prioritized those proteins with predicted signal peptides, however as many secreted proteins do not contain these sequences, we considered GRA candidates from the hypothetical proteins that also lacked this feature. Because we wished to focus on those proteins that were upregulated in bradyzoites, we also narrowed our candidate pool by focusing on proteins that were shown to be highly transcribed in bradyzoites. However, we also chose some candidates whose expression was similar between tachyzoites and bradyzoites [46]. We chose 9 candidates and used endogenous gene tagging to recombine a C-terminal 3xHA epitope tag for each gene product and examined their localization by IFA. We found that 5 of these trafficked to the PV in both tachyzoites and bradyzoites (Fig 2). We named these GRA55 (TgME49_309760), GRA56 (TgME49_309930), GRA57 (TgME49_217680), and GRA58 (TgME49_268790). In addition, while reviewing earlier datasets from a prior GRA-BioID experiments [36], we found another GRA–GRA59 (TgME49_313440)–which was also upregulated in bradyzoites. Of these confirmed GRA proteins, only GRA56 had an identifiable domain which has homology to melibiase, an alpha-galactosidase that catabolizes the disaccharide melibiose.
Fig 2. Identification of novel dense granule proteins, GRA55-GRA59 by MAG1-BioID.
(A) IFA of tachyzoites with rabbit anti-HA antibodies shows strong staining of the parasitophorous vacuole for each novel GRA that colocalizes with GRA14, demonstrating that these are novel dense granule proteins. Gene numbers and novel designations are as follows: GRA55 (TgME49_309760), GRA56 (TgME49_309930), GRA57 (TgME49_217680), GRA58 (TgME49_268790), and GRA59 (TgME49_313440). Scale bar: 5 μm. (B) IFA of bradyzoites with mouse anti-HA antibodies shows staining of each GRA at the cyst periphery and vacuolar space. Transgenic parasites expressed GFP, driven by LDH2 promoter, upon successful bradyzoite conversion. Images were taken three days after growth under alkaline-stressed conditions. Scale bar: 5 μm.
Targeted deletion of novel GRA proteins and functional characterization
Once we identified these novel GRAs, we assessed their function by gene deletion using CRISPR/Cas9 and homologous recombination [47–49]. Gene knockouts were assessed by loss of the epitope tag from the parental line and replacement of the DHFR selectable marker used in tagging for HXGPRT used in the knockout (e.g. by growth in mycophenolic acid and xanthine but failure to grow in pyrimethamine) (Fig 3A). Each of the knockouts was then confirmed by PCR (Fig 3B). We examined the deletion mutants by plaque assay and discovered none of the lines had impaired in vitro growth (Fig 4, S2 Fig). We then proceeded with a pilot study of each of the knockouts to identify severe changes in cyst burden. This pilot study suggested that Δgra55 parasites produced substantially lower cyst numbers per mouse brain, thus we focused on this strain for complementation and more detailed in vivo analyses.
Fig 3. Deletion of GRAs 55–59 using CRISPR/Cas9.
(A) IFA shows the absence of HA signal in the PV. GRA14 is a marker of the PV and is shown as a control. Scale bar: 5 μm. (B) PCR verification of the deletion of GRAs 55–59. Attempted amplification of genes in knockout parasite strains show an absence of the coding sequence in the knockout strain but present in the wild-type control (gene check). The insertion of HXGPRT into the correct locus is verified by PCR from upstream of the gene of interest into the selectable marker (KO check) which is present only in the knockouts (PruΔku80 genomic DNA used as positive control).
Fig 4. GRA55 is a novel GRA which is not important for in vitro growth or short-term virulence.
(A) Complementation strategy of GRA55 into the UPRT locus. The complemented gene contains a C-terminal 3xHA epitope tag and is driven by the native promoter. (B) IFA of GRA55-HA, Δgra55, and GRA55c parasites. Rabbit anti-HA shows strong staining in the vacuole of GRA55-HA and GRA55c parasites, colocalizing with GRA14. HA signal is absent in Δgra55 parasites. Scale bar: 5 μm. (C) Western blot demonstrating GRA55-HA migrating at the predicted size of 35.2 kDa (the size of GRA55-HA without the predicted signal peptide) and showing similar expression levels to the endogenously tagged strain. As expected, there is no HA signal in the Δgra55 strain. RON5C is used as a loading control [50]. (D) Quantitation of plaques formed by GRA55-HA, Δgra55, and GRA55c parasites. Results are shown as box-whisker plot with the middle line representing the median, the bottom and top of box representing the 25th and 75th percentile, and whiskers corresponding to smallest and largest plaques. (E, F, G) C57BL/6 female mice were infected with indicated doses of GRA55HA, Δgra55, and GRA55c parasites and Kaplan-Meier survival curves were generated. There was no statistical difference between the virulence of Δgra55 (F) vs. GRA55-HA (E) or GRA55c (G) parasites. (H) CBA/J mice were infected with 250 parasites each of the indicated strain. Mice were euthanized and mouse brains were examined by IFA after 30 days infection to enumerate T. gondii cyst burden. Δgra55 caused 10-fold lower burden of cysts when compared to GRA55-HA infection and GRA55c infection (p < 0.05 by two-tailed T-test). (I) Numerical counts of cysts per mouse brain (* = mouse dead prior to end of experiment).
GRA55 is not important for in vitro growth or virulence but affects chronic infection of Type II T. gondii
We engineered GRA55-complemented parasites by re-introducing the wild-type gene driven from its endogenous promoter into the uracil phosphoribosyltransferase (UPRT) locus of the Δgra55 parasite genome [51] (Fig 4A). We verified that GRA55 targeted properly to the PV by IFA and showed similar expression levels to wild type parasites by western blot and thus we named this strain GRA55c (Fig 4B and 4C). We verified that that Δgra55 parasites did not have any decreased growth by plaque assays (Fig 4D) compared to wild-type and complemented strains, confirming that this protein does not meaningfully participate in parasite growth in vitro. Upon examination of the role of GRA55 in acute infection in mice, we found that Δgra55 parasites killed mice in a dose-dependent manner, identical to that of mice infected with wild-type and GRA55c (Fig 4E, 4F and 4G). We then conducted an investigation into whether GRA55 was important for chronic toxoplasmosis in mice. We injected 3 separate groups of mice intraperitoneally with 250 parasites per mouse, allowed the infection to proceed to 30 days, and then euthanized the mice and examined tissue cyst burden in the brains. We found that mice infected with Δgra55 parasites suffered a consistent 10-fold decrease in cyst burden compared to the mice infected with wild-type and GRA55c parasites (Fig 4H and 4I). In summary, this data demonstrated that although GRA55 is not important for the acute infection and mortality, it is an important secreted protein for the establishment or maintenance of the chronic infection in mice.
Discussion
Toxoplasma bradyzoites are characterized by slow growing parasites encased in a cyst wall that enables transmission of the parasite to new hosts upon predation [25,52,53]. While the cysts are formed from secreted components from the parasite, the protein composition of the cyst has been challenging to determine as it is difficult to purify robust numbers of cysts and then separate the secreted components from the parasites. In addition, while switching from tachyzoites to bradyzoites can be accomplished in vitro, rapidly growing tachyzoites in the culture routinely overwhelm the slow growing bradyzoites and further complicate purifications. The use of the CST1 monoclonal antibody to purify fragments of the cyst wall and identify the purified components has been successful at identifying many cyst constituents [33]. However, purifying wall fragments requires relatively low stringency lysis conditions that may result in some contamination in these purifications. The adaptation of proximity biotinylation in T. gondii provides a new approach to label proteins prior to isolation and is ideal for compartments hard to access like the secreted GRAs of the cyst. We were additionally able to improve the in vitro switch efficiency using a strain which disrupted for the bradyzoite repressor AP2IV-4 [38]. While this approach helped us identify new GRAs, we found that challenges still remain with significant tachyzoite contamination, particularly at the high multiplicity of infection which we used in these experiments to minimize host biotinylated protein contamination in our streptavidin pulldowns. The recent identification of a master regulator of bradyzoite differentiation (BFD1) is likely to enable more robust cyst production without the stress of in vitro stage switching culture conditions and may yield a deeper insight into the cyst composition using proximity labeling [54].
As expected, our proximity labeling experiments with MAG1 as a bait protein identified many of the same proteins we found in the tachyzoite proteome. While MAG1 is one of the earliest described bradyzoite-enriched GRAs that is abundant in cysts, similar to CST1 there is also substantial MAG1 expression in tachyzoites (both proteins ranked high in our GRA-BioID of tachyzoites) [36]. Regardless, we were able to find many of the known bradyzoite upregulated proteins including CST1, BPK1, and the newly discovered CST proteins CST2, CST3, CST4, and CST5 [30,33,34]. We also found a group of candidate GRAs, five of which we verified by endogenous gene tagging. Interestingly, each of these new GRA proteins could be clearly detected in tachyzoites. This agrees with CST2, CST3, CST 4, and CST6 recently identified in the CST1 pulldown experiments which can also be observed in tachyzoites [33]. This may be due to the in vitro bradyzoite switching conditions used in these experiments and could indicate that these proteins are more important for transitioning to the bradyzoite stage rather than maintenance of mature cysts. In agreement with in vitro cysts not containing the full complement of proteins present in in vivo cysts, neither this study nor the CST1 pulldown identified BCP1, which has only been identified in the cyst wall of in vivo derived cysts [55]. It will be interesting to determine if cysts induced via BFD1 more faithfully replicate in vivo cysts, and thereby enable new categories of bradyzoite GRAs to be obtained, or if these are still lacking important characteristics of cysts isolated from mice.
In agreement with the T. gondii genome-wide CRISPR screen, we were able to disrupt all five of the new GRAs that we identified here [56]. While gene knockouts using CRISPR/Cas9 are generally efficient in Δku80 strains of Toxoplasma, we have found that replacing the Toxoplasma promoter driving HXGPRT with the Neospora GRA7 promoter improves the frequency of knockouts, frequently reaching 100% of the transfected population.
Most GRA proteins lack any identifiable homology to known eukaryotic proteins. GRA56, which we identified in this study, is a large protein with a domain that has homology to the melibiase family proteins. Melibiase proteins are alpha-galactosidases first discovered in plants, then found in several bacteria, animal tissues and fungi [57–65]. The enzyme acts to break apart melibiose, a disaccharide composed of galactose and glucose by a 1,6-alpha bond. In many fungi including Saccharomyces species, this enzyme is a large glycoprotein which contains an N-terminal signal sequence and is secreted outside of the cell, similar to GRA56 identified here [66–68]. We hypothesize that T. gondii may use this secreted enzyme to catabolize melibiose for energy utilization after uptake into the PV. While GRA56 does have a negative (-1.13) fitness score in the genome-wide CRISPR screen [56], it is possible that this knockout has compensated by upregulating other pathways as has previously been observed in Toxoplasma [69].
Our pilot in vivo assays suggested that disruption of GRA55 resulted in a defect in cyst counts, thus we complemented the strain with the wild-type gene. We found that the knockout showed no defect in plaque size or in acute virulence in vivo, but the chronic infection resulted in a ~10 fold decrease in cyst number in vivo, which was rescued by complementation. We did not observe any gross change is cyst size or morphology, and did not observe fragility of the cysts while manipulating the samples for counting as has been seen in the CST1 deletion mutants [31]. We performed an in vitro examination of switching efficiency between GRA55-HA, Δgra55, and GRA55c parasites. Although there was a general trend of increased efficiency from GRA55-HA to Δgra55 parasites, we found that there was no significantly increased switch efficiency between the three strains. Further, complementation of GRA55 did not reverse the trend (S3 Fig). This suggests that the defect may be in tissue tropism to the brain or in the formation or maintenance of cysts in vivo, although we cannot exclude the possibility that subtle changes in growth in vivo over the 30 day time period used for cyst development play a role in the decreased cyst count in GRA55 knockouts. Since GRA55 lacks homology to known proteins, the mechanism of action will be best be determined by mutagenesis experiments to identify functional domains and additional proximity biotinylation experiments to identify binding partners and regulators. Together, this research improves our understanding of the composition and function of GRA proteins in bradyzoites which will ultimately lead to new therapies to treat this poorly understood yet critical stage of infection.
Materials and methods
Host cell and parasite cultures
PruΔku80Δhxgprt (Type II, strain Prugniaud) and modified strains of T. gondii were used to infect human foreskin fibroblast (HFF) cells. HFFs were grown in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum and 2 mM glutamine, and maintained as previously described [70].
Antibodies
Primary antibodies used in immunofluorescence assays (IFAs) and Western blot: mouse polyclonal anti-GRA14 (1:1000), rabbit anti-HA (Covance, 1:2000), rabbit polyclonal anti-ROP13 (1:500).
IFA and Western blot
For IFA, HFFs were grown to confluence on coverslips and infected with T. gondii parasites. After 18 to 36 h, the coverslips were fixed and processed for indirect immunofluorescence as previously described [71]. Primary antibodies were detected by species-specific secondary antibodies conjugated to Alexa 594/488. The coverslips were mounted in ProLong Gold antifade reagent (ThermoFisher) and viewed with an Axio Imager Z1 fluorescence microscope (Zeiss) as previously described [41]. For Western blotting, parasites were lysed in Laemmli sample buffer (50 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 1% 2-mercaptoethanol, 0.1% bromophenol blue), and lysates were resolved by SDS-PAGE and transferred onto nitrocellulose membranes. Blots were probed with the indicated primary antibodies, followed by secondary antibodies conjugated to horseradish peroxidase (HRP). Target proteins were visualized by chemiluminescence.
Generation of Δap2IV-4 parasites using CRISPR/Cas9 and homologous recombination
An sgRNA sequence in the coding sequence of AP2IV-4 was chosen using the EuPaGDT tool (http://grna.ctegd.uga.edu/) and the gRNA scaffold forward and reverse sequences were engineered with BsaI ends (primers p1-p2 –S2 Table). This sequence was cloned into the pu6-Universal [48,49] plasmid at the same BsaI site. Separately, an HXGPRT cassette (driven by a Neospora GRA7 promoter) was amplified from the pJET1.2:NcGRA7pro:HXGPRT. The primers contained 39 bp homology to the 5’ UTR (forward) and 3’ UTR (reverse) regions of AP2IV-4 (primers: p3-p4 –S2 Table). PrugniaudΔku80 parasites were transfected with 30 μg of closed-circular pU6-universal/sgRNA with 60 μg of the HXGPRT/AP2IV-4-flanks double-stranded oligonucleotide. The transfected parasites were grown in medium containing 50 μg /ml MPA (mycophenolic acid) and 50 μg /ml xanthine and cloned by limiting dilution. Knockout clones were verified by PCR (primers: gene check: p44-p45, KO check: p43, p51 –S2 Table).
Generation of MAG1-BirA* fusion
To generate the MAG1-BirA* fusion, the MAG1 gene and promoter was PCR amplified from genomic DNA (primers: p5-p6 –S2 Table). This was cloned into the p.LIC.BirA*.3xHA.DHFR vector by ligation independent cloning as previously described [35,72]. 30 μg of the construct was linearized with HpaI and transfected into PruΔku80Δhxgprt::Δap2IV-4.hxgrprt parasites. The parasites were selected with medium containing 1 μM pyrimethamine, cloned by limiting dilution, and screened by IFA and Western blot against the HA tag. A clone expressing the fusion protein was selected and designated MAG1-BirA*:Δap2IV-4.hxgrprt.
Affinity capture of biotinylated proteins
HFF monolayers were infected with parasites expressing MAG1-BirA* fusions or the parental line (PruΔku80Δhxgprt:::Δap2IV-4.hxgrprt) for 3h. Media was changed to bradyzoite “switch” media (DMEM with 1%FBS, 50mM HEPES, 1%PSG, pH 8.1). [35,72]. Parasites were incubated at 37° C in ambient air (0.05% CO2) for 5 days to induce switch to tissue cysts, exchanging media every 2 days to maintain alkaline pH [33,43]. 150 μM biotin was supplemented in the media on the final 24 hrs. Cyst formation was verified by expression of LDH2-GFP. Intracellular parasites were collected, washed in PBS, and lysed in radioimmunoprecipitation assay (RIPA) buffer (50mM Tris pH 7.5, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40) supplemented with Complete Protease Inhibitor Cocktail (Roche) for 30 min on ice. Lysates were centrifuged for 15 min at 14,000 × g to pellet insoluble material and the supernatant was incubated with Streptavidin Plus UltraLink Resin (Pierce) at room temperature for 4 h under gentle agitation. Beads were collected and washed five times in RIPA buffer, followed by three washes in 8M urea buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl). Ten percent of each sample was boiled in Laemmli sample buffer, and eluted proteins were analyzed by Western blotting by streptavidin-HRP while the remainder was used for mass spectrometry.
Mass spectrometry
The proteins bound to streptavidin beads were reduced and alkylated via sequential 20 minute incubations of 5mM TCEP 10mM iodoacetamide at room temperature in the dark while being mixed at 1200 rpm an Eppendorf thermomixer. Proteins were then digested by the addition of 0.1μg Lys-C (FUJIFILM Wako Pure Chemical Corporation, 125–05061) and 0.8μg Trypsin (Thermo Scientific, 90057) while shaking 37°C overnight. The digestions were quenched via addition of formic acid to a final concentration of 5% by volume. Each sample was desalted via C18 tips (Thermo Scientific, 87784) and then resuspended in 15μL of 5% formic acid before analysis by LC-MS/MS. Peptide samples were then separated on a 75μM ID, 25cm C18 column packed with 1.9μM C18 particles (Dr. Maisch GmbH) using on a 140-minute gradient of increasing acetonitrile and eluted directly into a Thermo Orbitrap Fusion Lumos instrument where MS/MS spectra were acquired by Data Dependent Acquisition (DDA). Data analysis was performed using the ProLuCID and DTASelect2 algorithms as implemented in the Integrated Proteomics Pipeline—IP2 (Integrated Proteomics Applications, Inc., San Diego, CA). Protein and peptide identifications were filtered using DTASelect and required a minimum of two unique peptides per protein and a peptide-level false positive rate of less than 1% as estimated by a decoy database strategy.
Generation of GRA-HA parasites (epitope tagging of BioID hits)
For GRA55, GRA56 and 59, LIC methodology was used: To epitope tag the novel GRA candidates, ~1500 bp of the 3’ genomic region of each gene was amplified using primers p7-p8 (GRA55), p15-16 (GRA66), p37-p38 (GRA59) (S2 Table) from PruΔku80Δhxgprt genomic DNA. The products were cloned into the p.LIC.3xHA.DHFR vector to add a 3xHA tag on the target gene as previously described [73–75]. The constructs were linearized and 50 μg of DNA was transfected by electroporation into PruΔku80Δhxgprt strain T. gondii.
For GRA57 and GRA58, CRISPR/Cas9 approach was used: An sgRNA sequence in the coding sequence of the intended GRA protein was chosen using the EuPaGDT tool (http://grna.ctegd.uga.edu/) and the gRNA scaffold forward and reverse sequences were engineered with BsaI ends. This sequence was cloned into the pU6-universal plasmid at the same BsaI site: p21-22 (GRA57), p29-p30 (GRA58) (S2 Table). Separately, a 3xHA:DHFR region was amplified from the p.LIC.3xHA.DHFR plasmid. The primers contained a 40 bp homology to a region just upstream of the gene-of-interest STOP codon (forward) and a 38 bp homology to a region in the gene-of-interest 3’ UTR (reverse) regions of the GRA gene: p23-24 (GRA57), p31-p32 (GRA58) (S2 Table). PrugniaudΔku80Δhxgprt parasites were transfected with 30 μg of closed-circular pU6-universal/sgRNA with 30 μg of the 3xHA-DHFR/GOI-C-term-flanks double-stranded oligonucleotide [49,72].
The transfected parasites were grown in media containing 1 μM pyrimethamine and selected parasites were cloned by limiting dilution. Clones were designated as GRA(X)-HA (e.g. GRA55-HA). All confirmed GRAs were examined in silico by BLAST to search for other T. gondii proteins with sequence similarity.
Deletion of GRAs 55–59 using CRISPR/Cas9
An sgRNA sequence in the coding sequence of the intended GRA protein was chosen using the EuPaGDT tool (http://grna.ctegd.uga.edu/) and the gRNA scaffold forward and reverse sequences were engineered with BsaI ends. This sequence was cloned into the pU6-universal plasmid at the same BsaI site [48,49]. Oligos: p9-p10 (GRA55), p17-18 (GRA46), p25-26 (GRA57), p33-p34 (GRA58), p39-p40 (GRA59) (S2 Table). Separately, an HXGPRT cassette (using the Neospora GRA7 promoter) was amplified from the pJET1.2:NcGRA7pro:HXGPRT. The primers contained 39 bp homology to the 5’ UTR (forward) and 3’ UTR (reverse) regions of the GRA gene. Primers: p11-p12 (GRA55), p19-20 (GRA56), p27-28 (GRA57), p35-p36 (GRA58), p41-p42 (GRA59) (S2 Table) PrugniaudΔku80Δhxgprt GRA-HA-tagged parasites were transfected with 30 μg of closed-circular pU6-universal/sgRNA with 30 μg of the HXGPRT/GRA-flanks double-stranded oligonucleotide. The transfected parasites were grown in medium containing 50 μg /ml MPA (mycophenolic acid) and 50 μg /ml xanthine and cloned by limiting dilution. Knockout clones were selected by loss of HA tag (IFA) and verified by PCR to examine for the absence of the gene of interest and presence of the HXGPRT cassette at the predicted recombination site (KO check primers: p46-p50, each paired with p51), (gene check: p7-p8 [GRA55], p15-16 [GRA56], p52-p53 [GRA57], p54-55 [GRA58] p37-p38 [GRA59]) (S2 Table).
Complementation of GRA55
GRA55-HA was re-introduced into the genome of the Δgra55 line in the uracil phosphoribosyltransferase (UPRT) locus [51]. This was done by amplifying the GRA55-3xHA gene as well as its native promoter from genomic DNA of the GRA55-HA parasite line (p13-p14) (S2 Table). The PCR product was cloned into the UPRT knockout vector with a tubulin promoter added using PacI and SpeI, forming a construct with the GRA55-3xHA cassette inserted between the UPRT 5’ and 3’ flanking regions. This plasmid was linearized using SacI-HF and transfected into Δgra55 parasites and selected with 5-Fluoro-5’-deoxyuridine (FUDR) to isolate parasites with the construct integrated at the UPRT locus [76]. Clones were selected by limiting dilution, examined by IFA (anti-HA antibody), and a positive clone was designated GRA55c.
Plaque assay
Serial dilutions of parasites were infected into separate wells of a 6-well plate with an HFF monolayer and allowed to form plaques. The dilutions were calibrated to infect 100 parasites/well, 200 parasites/well and 300 parasites/well. 6 days after infection, HFF monolayers were fixed in 3.7% formaldehyde for 15 minutes, washed with PBS, dried and stained with safranin. Plaques were then visualized with a Zeiss upright light microscope (Zeiss Axio Imager Z1). Plaque areas were measured using ZEN software (Zeiss) and plaque sizes were compared. 2 separate experiments of 30 plaques for each parasite line were measured to generate a mean plaque size along with interquartile ranges. Statistical significance comparing lines was calculated using two-sample two-tailed t-test.
Quantitation of in vitro bradyzoite differentiation efficiency in GRA55-HA, Δgra55, and GRA55c parasites
HFFs were grown on coverslips for seven days and infected with GRA55-HA, Δgra55 or GRA55c parasites for 3h. Media was changed to bradyzoite “switch” media (DMEM with 1%FBS, 50mM HEPES, 1%PSG, pH 8.1). [35,72] and parasites were incubated at 37°C in ambient air (0.05% CO2) for 3 days to induce switch to tissue cysts [33,43]. The Coverslips were then fixed for IFA analysis. To quantify bradyzoite switch efficiency, GFP positive and negative vacuoles were counted and percentages of GFP positive vacuoles were calculated. Two biological replicates were reported for each strain. Statistical significance was calculated comparing the means of GRA55-HA with Δgra55 or GRA55c with Δgra55 using a one-way ANOVA test (using p < 0.05 for significance).
Mouse virulence assays
Intracellular GRA55-HA, Δgra55 and GRA55c parasites mechanically liberated from infected HFF monolayers and resuspended in Opti-MEM Reduced Serum Medium (ThermoFisher) prior to intraperitoneal injection into groups of 4 female C57BL/6 mice each. Injected parasites were confirmed to be live and viable parasites by plaque assays with HFF monolayers. Mice were monitored for symptoms of infection, weight loss, and mortality for 21 days. Survival was plotted on a Kaplan-Meier curve. To confirm infection, surviving mice were sacrificed at 30 days after infection, and mouse brains were collected in aseptic fashion, homogenized, and examined by phase microscopy and fluorescence microscopy (for detection of GFP under LDH2 promoter for parasite encystation). In parallel, 1/3 of each brain was homogenized and incubated with HFF monolayers to qualitatively assess viability of parasites.
Mouse brain cyst quantitation
Intracellular GRA55-HA, Δgra55, and GRA55c parasites were mechanically liberated from infected HFF monolayers and resuspended in Opti-MEM prior to intraperitoneal injection into groups of 4 female CBA/J mice each. 250 parasites/mouse of GRA55-HA, Δgra55 and GRA55c were injected. The mice were monitored for 30 days after infection, and then sacrificed. Mouse brains were collected, homogenized and examined for T. gondii cysts (as above). Quantitation of cysts was performed by examining 25 μL aliquots of homogenate by fluorescence microscopy until approximately 25% (by volume) of each brain was examined. Total cyst burden was then extrapolated. Statistical significance of cyst burden between GRA55-HA, Δgra55, and GRA55c experiments was calculated by two-sample 2-tailed t-test [77].
Animal experimentation ethics statement
Specific details of our protocol were approved by the UCLA Institutional Animal Care and Use Committee, known as the Chancellor’s Animal Research Committee (protocol: 2004–005). Mice were euthanized for two purposes. In the acute infection model, they were euthanized when the animals reached a moribund state. For the chronic infection model, these mice were sacrificed at the conclusion of the experiment at 30 days postinfection. Euthanasia per AVMA guidelines—slow (20–30% per minute) displacement of chamber air with compressed CO2. This is followed by confirmatory method of cervical dislocation’.
Supporting information
(PDF)
(A) Diagram of the construct used to delete AP2IV-4. Cas9 induced double stranded break (DSB) is represented by the scissors. The AP2IV-4 locus was replaced with an HXGPRT selectable marker using homologous recombination simultaneously with a Cas9 induced DSB. (B) PCR verification of deletion of AP2IV-4 and confirmation of homologous recombination of the HXGPRT cassette in place of the deleted locus.
(TIF)
(A) GRA56, (B) GRA57, (C) GRA58, (D) GRA59. Results are shown as box-whisker plot with the middle line representing the median, the bottom and top of box representing the 25th and 75th percentile, and whiskers corresponding to smallest and largest plaques.
(TIF)
The mean ±SD was plotted for n = 2 biological replicates, where percentage of GFP positive vacuoles were calculated for each strain. No significance (p < 0.05) was reported comparing the means of GRA55-HA with Δgra55 or GRA55c with Δgra55 using a one-way ANOVA test.
(TIF)
There are two replicate experiments labeled MAG1-BioID-A and MAG1-BioID-B. Known GRAs shown in blue. Novel GRAs highlighted in Red. GRA-control = PruΔku80:Δap2iv-4. Proteins are ranked based on peptide spectrum matches (PSM). PSM is defined as the total number of theoretically generated peptide fragmentation spectra from the database matched to the experimental mass spectrometry fragmentation spectra. Columns are sortable using arrows next to each column heading.
(XLSX)
(DOCX)
Acknowledgments
Our thanks to Dr. Patricia Johnson, the Microbial Pathogenesis Training Grant T32, and supporting committee which provided professional and scientific mentorship to S.M.N. and A.C.T. throughout the grant period.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by NIH grants AI064616 and AI125106 to P.J.B., and no. GM089778 to J.A.W., the Ruth L. Kirschstein National Research Service Award (T32) AI007323 to S.M.N and A.T. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hill DE, Chirukandoth S, Dubey JP. Biology and epidemiology of Toxoplasma gondii in man and animals. Anim Health Res Rev 2005;6:41–61. 10.1079/AHR2005100. [DOI] [PubMed] [Google Scholar]
- 2.Dubey J, Jones J. Toxoplasma gondii infection in humans and animals in the United States. Int J Parasitol 2008;38:1257–78. 10.1016/j.ijpara.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Innes EA. Toxoplasmosis: Comparative species susceptibility and host immune response. Comp Immunol Microbiol Infect Dis 1997;20:131–8. 10.1016/s0147-9571(96)00038-0 [DOI] [PubMed] [Google Scholar]
- 4.Luft BJ, Remington JS. Toxoplasmic Encephalitis in AIDS. Clin Infect Dis 1992;15:211–22. 10.1093/clinids/15.2.211. [DOI] [PubMed] [Google Scholar]
- 5.Carruthers V, Boothroyd JC. Pulling together: an integrated model of Toxoplasma cell invasion. Curr Opin Microbiol 2007;10:83–9. 10.1016/j.mib.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 6.Carruthers VB, Sibley LD. Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur J Cell Biol 1997;73:114–23. [PubMed] [Google Scholar]
- 7.Opitz C, Soldati D. ‘The glideosome’: a dynamic complex powering gliding motion and host cell invasion by Toxoplasma gondii. Mol Microbiol 2002;45:597–604. 10.1046/j.1365-2958.2002.03056.x. [DOI] [PubMed] [Google Scholar]
- 8.Soldati D, Foth BJ, Cowman AF. Molecular and functional aspects of parasite invasion. Trends Parasitol 2004;20:567–74. 10.1016/j.pt.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Carruthers VB. Armed and dangerous: Toxoplasma gondii uses an arsenal of secretory proteins to infect host cells 1999:10. [DOI] [PubMed] [Google Scholar]
- 10.Mercier C, Adjogble KDZ, Däubener W, Delauw M-F-C. Dense granules: Are they key organelles to help understand the parasitophorous vacuole of all apicomplexa parasites? Int J Parasitol 2005;35:829–49. 10.1016/j.ijpara.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Nam H-W. GRA Proteins of Toxoplasma gondii: Maintenance of Host-Parasite Interactions across the Parasitophorous Vacuolar Membrane. Korean J Parasitol 2009;47:S29 10.3347/kjp.2009.47.S.S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mercier C, Dubremetz J-F, Rauscher B, Lecordier L, Sibley LD, Cesbron-Delauw M-F. Biogenesis of Nanotubular Network in Toxoplasma Parasitophorous Vacuole Induced by Parasite Proteins. Mol Biol Cell 2002;13:2397–409. 10.1091/mbc.e02-01-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sibley LD, Niesman IR, Parmley SF, Cesbron-Delauw M-F. Regulated secretion of multi-lamellar vesicles leads to formation of a tubulo- vesicular network in host-cell vacuoles occupied by Toxoplasma gondii n.d.:9. [DOI] [PubMed]
- 14.Beraki T, Hu X, Broncel M, Young JC, O’Shaughnessy WJ, Borek D, et al. Divergent kinase regulates membrane ultrastructure of the Toxoplasma parasitophorous vacuole. Proc Natl Acad Sci 2019;116:6361–70. 10.1073/pnas.1816161116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bittame A, Effantin G, Pètre G, Ruffiot P, Travier L, Schoehn G, et al. Toxoplasma gondii: Biochemical and biophysical characterization of recombinant soluble dense granule proteins GRA2 and GRA6. Biochem Biophys Res Commun 2015;459:107–12. 10.1016/j.bbrc.2015.02.078. [DOI] [PubMed] [Google Scholar]
- 16.Guevara RB, Fox BA, Falla A, Bzik DJ. Toxoplasma gondii Intravacuolar-Network-Associated Dense Granule Proteins Regulate Maturation of the Cyst Matrix and Cyst Wall. MSphere 2019;4:e00487–19, /msphere/4/5/mSphere487-19.atom. 10.1128/mSphere.00487-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bougdour A, Durandau E, Brenier-Pinchart M-P, Ortet P, Barakat M, Kieffer S, et al. Host Cell Subversion by Toxoplasma GRA16, an Exported Dense Granule Protein that Targets the Host Cell Nucleus and Alters Gene Expression. Cell Host Microbe 2013;13:489–500. 10.1016/j.chom.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Braun L, Brenier-Pinchart M-P, Yogavel M, Curt-Varesano A, Curt-Bertini R-L, Hussain T, et al. A Toxoplasma dense granule protein, GRA24, modulates the early immune response to infection by promoting a direct and sustained host p38 MAPK activation. J Exp Med 2013;210:2071–86. 10.1084/jem.20130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braun L, Brenier-Pinchart M-P, Hammoudi P-M, Cannella D, Kieffer-Jaquinod S, Vollaire J, et al. The Toxoplasma effector TEEGR promotes parasite persistence by modulating NF-κB signalling via EZH2. Nat Microbiol 2019;4:1208–20. 10.1038/s41564-019-0431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panas MW, Naor A, Cygan AM, Boothroyd JC. Toxoplasma Controls Host Cyclin E Expression through the Use of a Novel MYR1-Dependent Effector Protein, HCE 2019;10:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naor A, Panas MW, Marino N, Coffey MJ, Tonkin CJ, Boothroyd JC. MYR1-Dependent Effectors Are the Major Drivers of a Host Cell’s Early Response to Toxoplasma, Including Counteracting MYR1-Independent Effects. MBio 2018;9:e02401–17, /mbio/9/2/mBio.02401-17.atom. 10.1128/mBio.02401-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shastri AJ, Marino ND, Franco M, Lodoen MB, Boothroyd JC. GRA25 Is a Novel Virulence Factor of Toxoplasma gondii and Influences the Host Immune Response. Infect Immun 2014;82:2595–605. 10.1128/IAI.01339-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gold DA, Kaplan AD, Lis A, Bett GCL, Rosowski EE, Cirelli KM, et al. The Toxoplasma Dense Granule Proteins GRA17 and GRA23 Mediate the Movement of Small Molecules between the Host and the Parasitophorous Vacuole. Cell Host Microbe 2015;17:642–52. 10.1016/j.chom.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paredes-Santos T, Wang Y, Waldman B, Lourido S, Saeij JP. The GRA17 Parasitophorous Vacuole Membrane Permeability Pore Contributes to Bradyzoite Viability. Front Cell Infect Microbiol 2019;9:321 10.3389/fcimb.2019.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchholz KR, Fritz HM, Chen X, Durbin-Johnson B, Rocke DM, Ferguson DJ, et al. Identification of Tissue Cyst Wall Components by Transcriptome Analysis of In Vivo and In Vitro Toxoplasma gondii Bradyzoites. Eukaryot Cell 2011;10:1637–47. 10.1128/EC.05182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torpier G, Charif H, Darcy F, Liu JL, Dardé ML, Capron A. Toxoplasma gondii: Differential Location of Antigens Secreted from Encysted Bradyzoites. Exp Parasitol 1993;77:13–22. 10.1006/expr.1993.1056 [DOI] [PubMed] [Google Scholar]
- 27.Ferguson DJP, Jacobs D, Saman E, Dubremetz J-F, Wright SE. In vivo expression and distribution of dense granule protein 7 (GRA7) in the exoenteric (tachyzoite, bradyzoite) and enteric (coccidian) forms of Toxoplasma gondii. Parasitology 1999;119:259–65. 10.1017/S0031182099004692. [DOI] [PubMed] [Google Scholar]
- 28.Lemgruber L, Lupetti P, Martins‐Duarte ES, Souza WD, Vommaro RC. The organization of the wall filaments and characterization of the matrix structures of Toxoplasma gondii cyst form. Cell Microbiol 2011;13:1920–32. 10.1111/j.1462-5822.2011.01681.x. [DOI] [PubMed] [Google Scholar]
- 29.Labruyere et al. - 1999—Differential membrane targeting of the secretory p.pdf n.d. [DOI] [PubMed]
- 30.Zhang YW, Halonen SK, Ma YF, Wittner M, Weiss LM. Initial Characterization of CST1, a Toxoplasma gondii Cyst Wall Glycoprotein. Infect Immun 2001;69:501–7. 10.1128/IAI.69.1.501-507.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomita T, Bzik DJ, Ma YF, Fox BA, Markillie LM, Taylor RC, et al. The Toxoplasma gondii Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite Persistence. PLoS Pathog 2013;9:e1003823 10.1371/journal.ppat.1003823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomita T, Sugi T, Yakubu R, Tu V, Ma Y, Weiss LM. Making Home Sweet and Sturdy: Toxoplasma gondii ppGalNAc-Ts Glycosylate in Hierarchical Order and Confer Cyst Wall Rigidity. MBio 2017;8:e02048–16, /mbio/8/1/e02048-16.atom. 10.1128/mBio.02048-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu V, Mayoral J, Sugi T, Tomita T, Han B, Ma YF, et al. Enrichment and Proteomic Characterization of the Cyst Wall from In Vitro Toxoplasma gondii Cysts. MBio 2019;10:e00469–19, /mbio/10/2/mBio.00469-19.atom. 10.1128/mBio.00469-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buchholz KR, Bowyer PW, Boothroyd JC. Bradyzoite Pseudokinase 1 Is Crucial for Efficient Oral Infectivity of the Toxoplasma gondii Tissue Cyst. Eukaryot Cell 2013;12:399–410. 10.1128/EC.00343-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen AL, Kim EW, Toh JY, Vashisht AA, Rashoff AQ, Van C, et al. Novel Components of the Toxoplasma Inner Membrane Complex Revealed by BioID. MBio 2015;6:e02357–14. 10.1128/mBio.02357-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nadipuram SM, Kim EW, Vashisht AA, Lin AH, Bell HN, Coppens I, et al. In Vivo Biotinylation of the Toxoplasma Parasitophorous Vacuole Reveals Novel Dense Granule Proteins Important for Parasite Growth and Pathogenesis. MBio 2016;7:e00808–16, /mbio/7/4/e00808-16.atom. 10.1128/mBio.00808-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh U, Brewer JL, Boothroyd JC. Genetic analysis of tachyzoite to bradyzoite differentiation mutants in Toxoplasma gondii reveals a hierarchy of gene induction. Mol Microbiol 2002;44:721–33. 10.1046/j.1365-2958.2002.02903.x. [DOI] [PubMed] [Google Scholar]
- 38.Radke JB, Worth D, Hong D, Huang S, Sullivan WJ, Wilson EH, et al. Transcriptional repression by ApiAP2 factors is central to chronic toxoplasmosis. PLoS Pathog 2018;14:e1007035 10.1371/journal.ppat.1007035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferguson DJP, Parmley SF. Toxoplasma gondii MAG1 protein expression. Trends Parasitol 2002;18:482 10.1016/s1471-4922(02)02349-8 [DOI] [PubMed] [Google Scholar]
- 40.Di Cristina M, Del Porto P, Buffolano W, Beghetto E, Spadoni A, Guglietta S, et al. The Toxoplasma gondii bradyzoite antigens BAG1 and MAG1 induce early humoral and cell-mediated immune responses upon human infection. Microbes Infect 2004;6:164–71. 10.1016/j.micinf.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Rome ME, Beck JR, Turetzky JM, Webster P, Bradley PJ. Intervacuolar Transport and Unique Topology of GRA14, a Novel Dense Granule Protein in Toxoplasma gondii. Infect Immun 2008;76:4865–75. 10.1128/IAI.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parmley SF, Yang S, Harth G, David Sibley L, Sucharczuk A, Remington JS. Molecular characterization of a 65-kilodalton Toxoplasma gondii antigen expressed abundantly in the matrix of tissue cysts. Mol Biochem Parasitol 1994;66:283–96. 10.1016/0166-6851(94)90155-4 [DOI] [PubMed] [Google Scholar]
- 43.Mayoral J, Di Cristina M, Carruthers VB, Weiss LM. Toxoplasma gondii: Bradyzoite Differentiation In Vitro and In Vivo. Methods Mol Biol Clifton NJ 2020;2071:269–82. 10.1007/978-1-4939-9857-9_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsiao C-HC, Luisa Hiller N, Haldar K, Knoll LJ. A HT/PEXEL Motif in Toxoplasma Dense Granule Proteins is a Signal for Protein Cleavage but not Export into the Host Cell: T. gondii HT/PEXEL Motif for Protein Processing. Traffic 2013;14:519–31. 10.1111/tra.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mercier C, Cesbron-Delauw M-F. Toxoplasma secretory granules: one population or more? Trends Parasitol 2015;31:60–71. 10.1016/j.pt.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Pittman KJ, Aliota MT, Knoll LJ. Dual transcriptional profiling of mice and Toxoplasma gondii during acute and chronic infection. BMC Genomics 2014;15:806 10.1186/1471-2164-15-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen B, Brown K, Long S, Sibley LD. Development of CRISPR/Cas9 for Efficient Genome Editing in Toxoplasma gondii. Methods Mol Biol Clifton NJ 2017;1498:79–103. 10.1007/978-1-4939-6472-7_6. [DOI] [PubMed] [Google Scholar]
- 48.Shen B, Brown KM, Lee TD, Sibley LD. Efficient Gene Disruption in Diverse Strains of Toxoplasma gondii Using CRISPR/CAS9. MBio 2014;5:e01114–14. 10.1128/mBio.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sidik SM, Hackett CG, Tran F, Westwood NJ, Lourido S. Efficient Genome Engineering of Toxoplasma gondii Using CRISPR/Cas9. PLoS ONE 2014;9:e100450 10.1371/journal.pone.0100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beck JR, Chen AL, Kim EW, Bradley PJ. RON5 Is Critical for Organization and Function of the Toxoplasma Moving Junction Complex. PLOS Pathog 2014;10:e1004025 10.1371/journal.ppat.1004025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reese ML, Zeiner GM, Saeij JPJ, Boothroyd JC, Boyle JP. Polymorphic family of injected pseudokinases is paramount in Toxoplasma virulence. Proc Natl Acad Sci 2011;108:9625–30. 10.1073/pnas.1015980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dubey JP, Lindsay DS, Speer CA. Structures of Toxoplasma gondiiTachyzoites, Bradyzoites, and Sporozoites and Biology and Development of Tissue Cysts. Clin Microbiol Rev 1998;11:267–99. 10.1128/CMR.11.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watts E, Zhao Y, Dhara A, Eller B, Patwardhan A, Sinai AP. Novel Approaches Reveal that Toxoplasma gondii Bradyzoites within Tissue Cysts Are Dynamic and Replicating Entities In Vivo. MBio 2015;6 10.1128/mBio.01155-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waldman BS, Schwarz D, Wadsworth MH, Saeij JP, Shalek AK, Lourido S. Identification of a master regulator of differentiation in Toxoplasma. BioRxiv 2019:660753 10.1101/660753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milligan-Myhre K, Wilson SK, Knoll LJ. Developmental change in translation initiation alters the localization of a common microbial protein necessary for Toxoplasma chronic infection. Mol Microbiol 2016;102:1086–98. 10.1111/mmi.13538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sidik SM, Huet D, Ganesan SM, Huynh M-H, Wang T, Nasamu AS, et al. A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell 2016;166:1423–1435.e12. 10.1016/j.cell.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Friis J, Ottolenghi P. Localization of melibiase in a strain of veast. Comptes-Rendus Trav Lab Carlsberg 1959;31:272–81. [PubMed] [Google Scholar]
- 58.Bailey RW. alpha-Galactosidase activity of rumen bacteria. Nature 1962;195:79–80. 10.1038/195079b0. [DOI] [PubMed] [Google Scholar]
- 59.Ahmed MU, Cook FS. SEPARATION OF AN ALPHA-GALACTOSIDASE FROM WATERMELON SEEDS. Can J Biochem 1964;42:605–12. 10.1139/o64-073. [DOI] [PubMed] [Google Scholar]
- 60.Schmitt R, Rotman B. Alpha-galactosidase activity in cell-free extracts of Escherichia coli. Biochem Biophys Res Commun 1966;22:473–9. 10.1016/0006-291x(66)90297-x [DOI] [PubMed] [Google Scholar]
- 61.Mital BK, Shallenberger RS, Steinkraus KH. Alpha-glactosidase activity of lactobacilli. Appl Microbiol 1973;26:783–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Malhotra OP, Dey PM. Purification and physical properties of sweet-almond alpha-galactosidase. Biochem J 1967;103:508–13. 10.1042/bj1030508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coleman RL. Separation of an alpha-galactosidase from rat uterus. Biochim Biophys Acta 1968;159:192–3. 10.1016/0005-2744(68)90261-1 [DOI] [PubMed] [Google Scholar]
- 64.Rao KS, Pieringer RA. The presence of an alpha-galactosidase in brain and its role in the degradation of the digalactosyl diglyceride of brain. J Neurochem 1970;17:483–8. 10.1111/j.1471-4159.1970.tb00525.x. [DOI] [PubMed] [Google Scholar]
- 65.Harpaz N, Flowers HM, Sharon N. Purification of coffee bean alpha-galactosidase by affinity chromatography. Biochim Biophys Acta 1974;341:213–21. 10.1016/0005-2744(74)90082-5 [DOI] [PubMed] [Google Scholar]
- 66.Herrero P, Moreno F, Gascón S. Role of vesicles on the transport and secretion of exocellular enzymes by yeast. Cell Mol Biol Cyto-Enzymol 1980;26:485–92. [PubMed] [Google Scholar]
- 67.Lazo PS, Ochoa AG, Gascón S. alpha-Galactosidase (melibiase) from Saccharomyces carlsbergensis: structrual and kinetic properties. Arch Biochem Biophys 1978;191:316–24. 10.1016/0003-9861(78)90094-2 [DOI] [PubMed] [Google Scholar]
- 68.Lazo PS, Ochoa AG, Gascón S. alpha-Galactosidase from Saccharomyces carlsbergensis. Cellular localization, and purification of the external enzyme. Eur J Biochem 1977;77:375–82. 10.1111/j.1432-1033.1977.tb11677.x. [DOI] [PubMed] [Google Scholar]
- 69.Lamarque MH, Roques M, Kong-Hap M, Tonkin ML, Rugarabamu G, Marq J-B, et al. Plasticity and redundancy among AMA–RON pairs ensure host cell entry of Toxoplasma parasites. Nat Commun 2014;5:1–13. 10.1038/ncomms5098. [DOI] [PubMed] [Google Scholar]
- 70.Roos DS, Donald RG, Morrissette NS, moulton LC. Molecular Tools for Genetic Dissection of the Protozoan Parasite Toxoplasma gondii Methods Cell Biol., Academic Press; 1994, p. 27–63. 10.1016/s0091-679x(08)61845-2 [DOI] [PubMed] [Google Scholar]
- 71.Bradley PJ, Ward C, Cheng SJ, Alexander DL, Coller S, Coombs GH, et al. Proteomic Analysis of Rhoptry Organelles Reveals Many Novel Constituents for Host-Parasite Interactions in Toxoplasma gondii. J Biol Chem 2005;280:34245–58. 10.1074/jbc.M504158200. [DOI] [PubMed] [Google Scholar]
- 72.Bradley PJ, Rayatpisheh S, Wohlschlegel JA, Nadipuram SM. Using BioID for the Identification of Interacting and Proximal Proteins in Subcellular Compartments in Toxoplasma gondii. Methods Mol Biol Clifton NJ 2020;2071:323–46. 10.1007/978-1-4939-9857-9_18. [DOI] [PubMed] [Google Scholar]
- 73.Huynh M-H, Carruthers VB. Tagging of Endogenous Genes in a Toxoplasma gondii Strain Lacking Ku80. Eukaryot Cell 2009;8:530–9. 10.1128/EC.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fox BA, Ristuccia JG, Gigley JP, Bzik DJ. Efficient Gene Replacements in Toxoplasma gondii Strains Deficient for Nonhomologous End Joining. Eukaryot Cell 2009;8:520–9. 10.1128/EC.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fox BA, Falla A, Rommereim LM, Tomita T, Gigley JP, Mercier C, et al. Type II Toxoplasma gondii KU80 Knockout Strains Enable Functional Analysis of Genes Required for Cyst Development and Latent Infection. Eukaryot Cell 2011;10:1193–206. 10.1128/EC.00297-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Donald RG, Roos DS. Insertional mutagenesis and marker rescue in a protozoan parasite: cloning of the uracil phosphoribosyltransferase locus from Toxoplasma gondii. Proc Natl Acad Sci 1995;92:5749–53. 10.1073/pnas.92.12.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Q, Sibley LD. Assays for Monitoring Toxoplasma gondii Infectivity in the Laboratory Mouse. Methods Mol Biol Clifton NJ 2020;2071:99–116. 10.1007/978-1-4939-9857-9_5. [DOI] [PubMed] [Google Scholar]