Abstract
The dynamic organization of chromatin inside the cell nucleus plays a key role in gene regulation and genome replication, as well as maintaining genome integrity. Although the static folded state of the genome has been extensively studied, dynamical signatures of processes such as transcription or DNA repair remain an open question. Here, we investigate the interphase chromatin dynamics in human cells in response to local DNA damage, specifically, DNA double-strand breaks (DSBs). Using simultaneous two-color spinning-disk confocal microscopy, we monitor the DSB dynamics and the compaction of the surrounding chromatin, visualized by fluorescently labeled 53BP1 and histone H2B, respectively. Our study reveals a surprising difference between the mobility of DSBs located in the nuclear interior versus periphery (less than 1 μm from the nuclear envelope), with the interior DSBs being almost twice as mobile as the periphery DSBs. Remarkably, we find that the DSB sites possess a robust structural signature in a form of a unique chromatin compaction profile. Moreover, our data show that the DSB motion is subdiffusive and ATP-dependent and exhibits unique dynamical signatures, different from those of undamaged chromatin. Our findings reveal that the DSB mobility follows a universal relationship defined solely by the physical parameters describing the DSBs and their local environment, such as the DSB focus size (represented by the local accumulation of 53BP1), DSB density, and the local chromatin compaction. This suggests that the DSB-related repair processes are robust and likely deterministic because the observed dynamical signatures (DSB mobility) can be explained solely by their structural features (DSB focus size, local chromatin compaction). Such knowledge might help in detecting local DNA damage in live cells, as well as in aiding our biophysical understanding of genome integrity in health and disease.
Significance
The human genome undergoes dynamic rearrangements to which many processes such as transcription and DNA repair are thought to contribute, although specific dynamical signatures of these processes are unknown. We investigate chromatin dynamics in live human cells in response to local DNA damage, the DNA double-strand breaks (DSBs). Our data reveal that DSBs move differently from undamaged chromatin; moreover, we find that chromatin exhibits a unique compaction profile at the DSB sites. We identify a universal relationship between the physical parameters describing DSBs and their local environment, suggesting a deterministic nature of the DSB-related repair processes. Such knowledge might aid our biophysical understanding of genome integrity in health and disease.
Introduction
The proper function of interphase chromatin intimately depends on its structure, organization, and dynamics inside the cell nucleus. In higher eukaryotes, chromatin fiber consists of DNA wrapped around nucleosomes made of core histone proteins (1,2). The chromatin fiber is then spatially organized inside the nucleus into a hierarchy of loops; topologically associated domains; A and B compartments corresponding to transcriptionally active and inactive regions, respectively; and finally, at larger length scales, chromosome territories (3, 4, 5). Although the static folded state of the genome has been described in detail using chromosome conformation capture techniques, the mechanistic picture behind its dynamic nature remains elusive (4, 5, 6, 7, 8). In interphase nuclei, chromatin dynamics was found to be active, i.e., ATP-dependent, diffusive to subdiffusive with constraints, with occasional directed motion, and coherent over several seconds and micrometers (6,9, 10, 11, 12, 13). Although chromatin dynamics have been implied to play a major role in biological processes such as transcription, replication and DNA repair (6,14), their underlying mechanisms are largely unknown.
Chromatin structure and dynamics are strongly susceptible to DNA damage, which in general compromises genome integrity. DNA damage can range from chemical changes in the DNA molecule to a full DNA double-strand break (DSB) (15,16). Strikingly, the genome of a human cell suffers daily an estimated ∼103 DNA damage events (15,16), which, if left unrepaired, can have devastating consequences for the cell. For example, an unrepaired DSB can lead to apoptosis or cancer; therefore, robust DNA repair mechanisms are crucial for cellular health and survival (15, 16, 17). Illuminating chromatin dynamics upon DNA damage such as DSBs might provide insight into the dynamic organization of chromatin as well as the DNA repair processes.
The formation and repair of DSBs in higher eukaryotes have been previously investigated by inducing DSBs using endonucleases, radiation (ultraviolet, X-rays, γ-rays, α-particles) or chemicals (e.g., neocarzinostatin, zeocin, etoposide) (18,19). In these studies, DSBs were visualized by fluorescently labeled proteins that localize at the DSB site and associate with its repair (e.g., 53BP1, Rad52) (20). The motion of DSBs was largely described as subdiffusive (18,21), and overall chromatin dynamics, while remaining subdiffusive, was found to increase in response to DNA damage (12,18,21, 22, 23). Moreover, the micrometer-scale coherent motion of chromatin is eliminated upon DNA damage (12). Furthermore, the protein 53BP1 and the LINC complex, as well as microtubule activity, were shown to be required for DSB mobility, which is thought to be crucial for DSB repair (24). In contrast, DSB dynamics was mostly unaffected by depletion of single DNA remodelers ACF1 and PARP1 or tethering proteins MRE11 and cohesin, provided the ATM kinase was not compromised (25).
The physical picture behind the DSB dynamics in higher eukaryotes remains elusive. This is in contrast to similar studies in yeast (26, 27, 28, 29, 30, 31), the genome of which is smaller, more dilute, and below the entanglement threshold and thus can be well-approximated by an ideal Rouse chain (32, 33, 34). Moreover, the equilibrium Rouse model, which assumes an elastic coupling between the monomers, can describe the DSB motion, as well as local condensation changes in the yeast genome upon DNA damage (35,36). However, as suggested by earlier studies (9,12,25,37,38), more complex models may be required for the chromatin dynamics in higher eukaryotes, with increasing evidence that nonequilibrium effects, viscoelasticity and hydrodynamic interactions play important roles (12,13,39, 40, 41, 42, 43, 44, 45, 46).
The goal of this work is to identify physical parameters describing local DNA damage in human genome that would aid a mechanistic view of the impact of the DNA damage on the local chromatin structure and dynamics. Specifically, we investigate the physical relationship between the structural and dynamical signatures of DSBs in live human cells. We use neocarzinostatin (NCS) to introduce DSBs at different densities and measure their motion in the context of the surrounding chromatin and its packing density. Using HeLa cells expressing both histone H2B-GFP and 53BP1-mCherry, a key regulator in the cellular response to DSBs, we visualize chromatin compaction (H2B-GFP) and DSBs (53BP1-mCherry) and follow their behavior in time. Using simultaneous two-color spinning-disk confocal microscopy, we record concurrent streams of H2B-GFP and 53BP1-mCherry signals. We use single-particle tracking to obtain trajectories of single DSBs from the 53BP1-mCherry signal, which we correlate with the local chromatin packing obtained from the H2B-GFP signal. Our data reveal that DSB sites exhibit unique local chromatin compaction profile as well as dynamics. Moreover, we find a universal relationship between the structural and dynamical signatures of DSBs in live cells, which relies purely on physical parameters of the system and allows us to identify the DSB location in chromatin polymer network and predict the DSB mobility.
Materials and Methods
Cell culture
HeLa cells (CCL-2) were cultured according to American Type Culture Collection (Manassas, VA) recommendations. A stable HeLa cell line expressing both H2B-GFP and 53BP1-mCherry was cultured in a humidified 5% CO2 (vol/vol) atmosphere at 37°C in Gibco Dulbecco’s modified Eagle’s medium (Gaithersburg, MD) supplemented with 10% fetal bovine serum (vol/vol), 100 units per milliliter of penicillin, and 100 μg/mL streptomycin (Invitrogen, Carlsbad, CA) and 4.5 μg/mL of Plasmocin Prophylactic (Invitrogen). Before the experiment, cells were plated on 35-mm MatTek dishes with glass bottom no. 1.5 (MatTek, Ashland, MA) for 24 h. The medium was replaced by Gibco CO2-independent medium supplemented with L-glutamine (Invitrogen) before imaging. Cells were mounted on the microscope stage and kept at 37°C in a custom-built microscope incubator enclosure. For fixation experiments, cells were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) at room temperature for 20 min and then washed with PBS three times every 5 min. Coverslips were mounted on glass microscope slides using ProLong Diamond Antifade Reagent (Molecular Probes, Eugene, OR).
Biochemical perturbations
When indicated, 200, 500, or 800 ng/mL of NCS (Sigma-Aldrich, St. Louis, MO) dissolved in CO2-independent media supplemented with L-glutamine were added to cells and incubated for 60 min before imaging. We performed four independent experiments for every condition. To deplete ATP, 6 mM of 2-deoxyglucose (Sigma-Aldrich) and 1 μM trifluoromethoxy-carbonylcyanide phenylhydrazone (Sigma-Aldrich) dissolved in CO2-independent media supplemented with L-glutamine were added to cells and were incubated for 2 h before addition of 500 ng/mL of NCS. Cells were then incubated for another 60 min before imaging. We performed three independent repeats of the ATP depletion experiments. For the experiments investigating the time dependency of the DSB dynamics, cells were treated with 500 ng/mL of NCS (Sigma-Aldrich) dissolved in CO2-independent media supplemented with L-glutamine and imaged continuously from 10 to 100 min after the NCS addition. We performed three independent repeats of this experiment.
Immunofluorescence staining
DNA DSBs were visualized using immunofluorescence staining of phosphorylated S139 γH2AX (#ab81299; Abcam, Cambridge, UK). First, cells were fixed for 20 min using 3.7% formaldehyde in PBS (Gibco) at room temperature, followed by three washes with PBS every 5 min. Cells were then permeabilized using 0.2% Triton X-100 in PBS for 10 min and washed two times with PBS. Next, the sample was incubated for 2 h with blocking reagent, 5% goat serum (LSPCN5000; Gibco) with 0.1% Tween20 in PBS. Cells were then treated with primary antibody (#ab81299; Abcam) diluted in the blocking reagent (1:2000) overnight in a humidified chamber at 4°C. After three washes with PBS, cells were incubated with secondary antibody (goat anti-rabbit Alexa 405, A31556; Invitrogen) diluted in PBS (1:200) for 1 h at room temperature. The cells were washed three final times with PBS before being mounted on a slide using ProLong Glass Antifade Reagent (Invitrogen).
Microscopy and image acquisition
Images were taken with a Yokogawa CSU-X1 spinning-disk confocal head (Yokogawa, Tokyo, Japan) with an internal motorized high-speed emission filter wheel and Spectral Applied Research Borealis modification (Spectral Applied Research, Richmond Hill, Ontario, Canada) for increased light throughput and illumination homogeneity on a Nikon Ti-E inverted microscope (Nikon, Tokyo, Japan) equipped with a 100× Plan Apo NA 1.4 objective lens and the Perfect Focus System. The microscope was mounted on a vibration-isolation air-table. To image H2B-GFP and 53BP1-mCherry at the same time, we illuminated the sample simultaneously with two excitation wavelengths, 488 and 561 nm, using two distinct solid-state lasers. The emission was collected with a 405/488/561/640 multibandpass dichroic mirror (Semrock, Rochester, NY) and separated by the W-View Gemini Image Splitter (Hamamatsu, Hamamatsu City, Japan) using the GFP/mCherry dichroic mirror (Chroma Technology, Bellows Falls, VT), and further passed through an ET525/30m emission filter (Chroma Technology) and an ET630/75m emission filter (Chroma Technology). The two fluorescent signals were allocated to the two halves of the image sensor, producing two distinct images. For three-color imaging, H2B-GFP, 53BP1-mCherry, and γH2AX (Alexa 405) were excited with 488, 561, and 405 nm solid-state lasers, respectively, and fluorescence was collected with a 405/488/561/640 multibandpass dichroic mirror (Semrock) and then ET525/50m, ET600/50m, and ET450/50m emission filters, respectively (Chroma Technology). Images were obtained with a Hamamatsu ORCA-R2 cooled CCD camera controlled with MetaMorph 7 (Molecular Devices, San Jose, CA) software. For two-color (red/green) signal registration, we imaged 4 μm TetraSpeck Fluorescent Microspheres (Molecular Probes) and obtained a two-dimensional local transformation matrix. The pixel size for the 100× objective was 0.065 μm. For two-color imaging, the observation duration was 25 s, with an exposure time of 250 ms. For three-color imaging, the H2B-GFP, 53BP1-mCherry, and γH2AX (Alexa 405) images were taken sequentially, each with an exposure time of 250 ms. The streams of 16-bit images were saved as multi-tiff stacks.
Image processing and data analysis
The nuclear contour was determined from the GFP signal using a previously published algorithm (47), and the DSB localization was determined from the 53BP1-mCherry signal, corrected for photobleaching using histogram matching (48). To obtain an accurate count of DSBs, we developed a machine-learning-assisted feature-finding algorithm. The 53BP1-mCherry signal across the first 10 frames was integrated, and a nuclear mask based on the nuclear contour was applied to remove the background signal. Using a local-maxima function, we found a large number of local maxima indicating possible features in the image, most of which correspond to noise. At each local maximum, feature descriptors such as the integrated intensity, gyration radius, and eccentricity were measured. Subsequently, 10 different filters were applied to the image, and 48 unique descriptor variables were obtained for each feature. We then manually sorted the DSBs from the noise features for 20 nuclei. The descriptor variables of these DSBs constituted a training data set, which we used to generate a binary-classification decision tree to sort features based on their descriptor variables. Next, we manually inspected a large sample of DSBs to confirm the goodness of the classification and performed manual corrections if needed and added these nuclei to the training data set. Our trained algorithm would correctly determine DSBs with ∼95% accuracy.
For the DSB tracking, we used previously published tracking algorithms (49,50) in combination with custom-made MATLAB (The MathWorks, Natick, MA) routines. Specifically, during the detection, we account for DSB foci of different sizes by searching for foci over a range of different radii. This way, some of the DSBs might be detected by different searches. To avoid multiple counting of a DSB, we perform watershed algorithm on the inverse of the 53BP1-mCherry signal, from which we define an approximate region of pixels for each DSB. Features located in the same region are deemed duplicates of the same DSB, and in such cases, the DSB centroid with the highest intensity is chosen. At this stage, each nucleus and all detected DSBs were manually inspected and approved before proceeding with their tracking.
To achieve high precision in tracking as well as to allow for tracking of all “trackable” features, we have developed an algorithm for finding of the window in which the feature is tracked. Such windows are obtained for every feature as follows: we filter the photobleach-corrected 53BP1-mCherry signal in each frame using a bandpass filter, then sum the filtered signal over all timeframes and perform watershed on the inverse of the summed signal. Regions too big to correspond to real features are removed, and trajectories estimated from the DSB centroid tracking are overlaid. If a trajectory is spanning neighboring windows, these windows are merged. The final window is held fixed for all frames, and the DSB centroid is found in every frame from the bandpass-filtered, photobleach-corrected signal within this window. This way, we can determine the trajectory of most DSBs with very high precision and a very low noise floor (∼13 nm). All DSB trajectories were corrected for potential nuclear motion by subtracting the nuclear centroid motion (51,52). The uncertainty of the size measurement of a 53BP1 focus is given by the uncertainty of the radial Gaussian fit and the variation of the size of the 53BP1 focus over the duration of the experiment, leading to an overall imprecision of ∼16 nm. The exposure of 250 ms leads to a size overestimate by less than 5% (53).
For the tracking of the mock DSBs, we analyzed the H2B-GFP signal of undamaged (control) cells. In each control cell, we find 150 random points. For the analysis of the interior random sites we remove those at the periphery and vice versa. For the obtained random site population, we measure Ich and Srel. We then remove all random sites whose Ich and Srel do not match those of the real interior or periphery DSBs. Lastly, we eliminate points that are too close to each other to avoid any signal overlap, choosing a minimal distance of 13 pixels between two mock DSBs. Next, the segmentation windows are obtained by the intersection of a circle (radius of 19 pixels) around the mock DSB center and the corresponding Voronoi cell. The segmentation window is held fixed for all timeframes, and the centroid of the mock DSB is found in every frame from the top-hat-filtered, photobleach-corrected H2B-GFP signal within this window. This way, the trajectories of the mock DSBs can be determined with high precision and a very low noise floor (∼19 nm). All mock DSB trajectories were corrected for a potential nuclear motion by subtracting the nuclear centroid motion. After the final mock DSB centroids were computed, Ich and Srel distributions of the interior/periphery mock DSBs were compared against the corresponding distributions of interior/periphery DSBs and the mock DSB Ich and Srel distributions, of which those not matching real DSBs were removed. The agreement between the Ich and Srel distributions for the mock and real DSBs was evaluated using the Kolmogorov-Smirnov test.
Results
Identification and quantification of DSBs
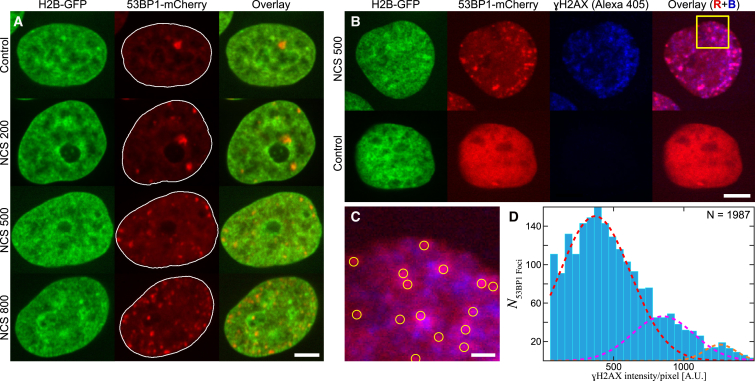
To investigate dynamics of DNA DSBs, we used HeLa cells expressing both histone H2B-GFP and 53BP1-mCherry (Fig. 1), allowing us to simultaneously visualize chromatin (H2B-GFP) and DNA damage foci (53BP1-mCherry). 53BP1 is a reliable indicator of the position of the DNA damage sites because it is a key molecular player in DNA damage repair, binding to DSBs shortly after they occur (54,55). We induce DSBs by adding NCS, whose mechanism of DSB generation is well-established (56), and observe 53BP1 foci 60 min after the NCS addition. At the 60 min mark, the 53BP1 foci correspond to the DSBs in the process of repair (57,58), which we confirmed by observing the colocalization with γH2AX, which marks DSBs in the repair (Fig. 1).
Figure 1.
Induction of DSBs by NCS. (A) Micrographs of live HeLa cell nuclei expressing both H2B-GFP and 53BP1-mCherry are shown under the following conditions: control (under physiological conditions) and upon addition of 200, 500, and 800 ng/mL NCS. The first column shows the H2B-GFP signal, the second column the 53BP1-mCherry signal with the nuclear contour (white line), and the third column the overlay of H2B-GFP (green) and 53BP1-mCherry (red) signals. (B) A nucleus treated with 500 ng/mL NCS (top row) and a control nucleus (bottom row), both formaldehyde-fixed, are shown. H2B-GFP (green) signal visualizes chromatin. 53BP1-mCherry (red) and γH2AX (blue) signals visualize the DNA repair foci and DSBs, respectively. The overlay of the red and blue signals shows their colocalization (pink) in the NCS-treated nucleus, whereas no DNA damage is found in the control nucleus. (C) An enlarged view of the boxed-in region from (B) with yellow circles highlighting the position of 53BP1 foci is shown. (D) Distribution of γH2AX signal over the population of 53BP1 foci (N = 1987 over 36 cells) is shown. The histogram shows the presence of three distinct Gaussian peaks (red, pink, and orange dashed lines), with means at discrete values, where the means of the second and third peaks are about twofold and threefold the values of the first peak’s mean, respectively. This suggests that the first peak indicates the fraction of 53BP1 foci containing a single DSB, the second peak two DSBs, and the third peak three DSBs. Scale bars, (A)–(B) 5 μm, (C) 1 μm. To see this figure in color, go online.
Fig. 1 A shows a nucleus that was not exposed to NCS (control); its 53BP1-mCherry signal is mostly diffuse, with one large spot corresponding to a so-called “basal break,” i.e., a lesion that occurs in the cell spontaneously (19,59). Upon addition of NCS, 53BP1-mCherry localizes at the DNA damage sites, manifesting by a sharp punctate signal (Fig. 1 A). We collected data 60 min after the NCS addition for three different NCS concentrations: 200, 500, and 800 ng/mL. Our data show that increasing NCS concentration leads to a visible increase of the number of DSBs inside the cell nucleus (Fig. 1 A).
Earlier studies suggested that most 53BP1 foci contain exactly one DSB (60,61); thus, it is reasonable to assume that a 53BP1 focus corresponds to a DSB. To verify this assumption for our system, we performed an immunofluorescence staining visualizing the localization of γH2AX, which marks the position of a single DSB. Using machine learning algorithms, we evaluate a large population of 53BP1 foci (N = 2119) over 36 cells and find that ∼94% of 53BP1 foci show a colocalization with γH2AX (Fig. 1, B and C). We quantify the γH2AX intensity at every 53BP1 focus colocalizing with γH2AX (N = 1987) and find that the measured γH2AX intensity distribution exhibits exactly three distinct peaks (Fig. 1 D). We fit these peaks by three distinct Gaussians (red, pink, and orange dashed lines), with means at discrete intensity values: μ1 ∼ 400, μ2 ∼ 850, and μ3 ∼ 1250. Strikingly, μ2 and μ3 are about twofold and threefold the values of μ1, respectively. This suggests that the first, second, and third peaks indicate the fraction of 53BP1 foci containing a single DSB, two DSBs, and three DSBs, respectively. By counting the 53BP1 foci within these three subpopulations, we find that ∼80% of 53BP1 foci contain exactly one DSB, ∼15% two DSBs, and ∼5% three DSBs.
Different DSB dynamics in the nuclear interior versus periphery
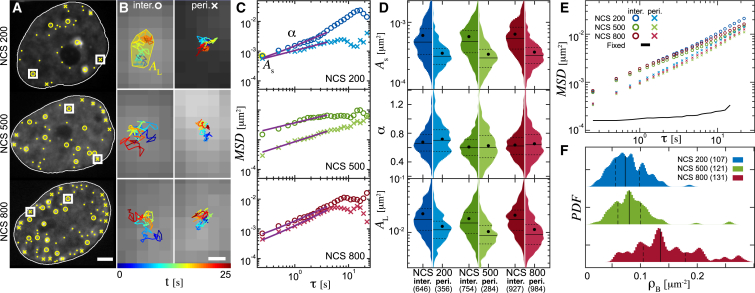
To obtain a comprehensive picture of the DSB dynamics, we examined the DSB dynamics based on their location in the cell nucleus. Specifically, we divided the DSBs into two categories: DSBs at the periphery (less than 1 μm from the nuclear envelope) and DSBs in the interior (more than 1 μm from the nuclear envelope). In addition, to explore a large variety of possible DSB positions in the cell nucleus, we vary the DSB density, i.e., the DSB number per unit area, by changing the NCS concentration in our experiments. We analyzed the motion of the DSBs at three different NCS concentrations, 200, 500, and 800 ng/mL (Fig. 2), and used machine learning algorithms (see Materials and Methods) to detect the position of the DSBs in the 53BP1-mCherry signal (Fig. 2 A, yellow markers). The trackable DSBs were divided into the periphery DSBs (yellow crosses) and the interior DSBs (yellow circles). The untrackable DSBs are marked by yellow dots. Such DSBs did not have a high enough signal/noise ratio and thus could not be tracked with high precision. In general, we found that DSBs with a low signal/noise ratio were out of focus. In our experiments, we tracked only DSBs that were in focus in the viewing plane and thus had a high signal/noise ratio.
Figure 2.
DSB dynamics as a function of the DSB position and NCS concentration. (A) DSBs are detected and tracked in 53BP1-mCherry signal (yellow markers). The trackable DSBs are divided into the periphery DSBs (less than 1 μm from the nuclear envelope, yellow crosses) and interior DSBs (yellow circles), and untrackable DSBs are marked by yellow dots (see Materials and Methods). (B) An enlargement of the boxed-in areas in (A) is given, showing examples of the interior and periphery DSB trajectories and their area AL given by a convex hull (yellow solid line). DSB trajectories are color-coded by their temporal evolution (blue to red). (C) Mean-square displacement (MSD) for trajectories of the periphery DSBs (crosses) and interior DSBs (circles) from (B) and a fit to MSD(τ) = C + Bτα are shown (purple line). (D) Distributions of the area explored at short timescales As, the exponents α, and area explored at long timescales AL obtained from individual DSB trajectories for populations of the interior and periphery DSBs are shown. Numbers in the x axis label indicate the number of DSBs analyzed. Dot, solid line, and dashed line in the violin plots correspond to the mean, median, and quartiles, respectively. (E) Average MSD for all trajectories for interior and periphery DSBs in a given experiment is shown. MSD measured for cells fixed in formaldehyde (black line, NDSB = 152, NCells = 42) defines the noise floor of our experiment. Error bars are shown in Fig. S1. (F) Distributions of the DSB density ρB are shown; numbers in legend indicate the number of nuclei analyzed. Scale bars, (A) 3 μm, (B) 100 nm. To see this figure in color, go online.
We focused on the fast, short-term DSB dynamics to elucidate the impact of material properties of the local chromatin network on the DSB dynamics. Thus, for all DSBs that were trackable, we followed their trajectories over 25 s with the temporal resolution of 250 ms (Fig. 2 B). First, we assess the area covered by an entire trajectory, AL (Fig. 2 B), which provides a measure for the mobility of a DSB over longer times. Further, from the obtained trajectories, we compute for each DSB the mean-square displacement (MSD), , where τ is the lag time (Fig. 2 C, markers), and fit the MSD to a power law f(τ) = C + Bτα (Fig. 2 C, solid lines). This simple model allows us to evaluate the type of motion that a DSB undergoes (e.g., diffusive, subdiffusive, and superdiffusive) at the timescale of our measurement while accounting for a possibility of an additional fast motion at timescales below our time resolution. Thus, for each DSB, we obtain exponent α as the fitting parameter. Next, we compare MSD values at the shortest time lag τ = 0.25 s, which we term As, informing on the DSB mobility over short times. Fig. 2 D shows the histograms of As, α, and AL for both the interior and periphery DSBs measured at three different NCS concentrations. The summary of the means and standard errors for As, α, and AL distributions is provided in Table S1 and the list of the corresponding p-values in Table S2.
Furthermore, we have calculated the average MSD over the entire ensembles of the interior and periphery DSBs for a given experiment: 200 ng/nL NCS (interior: NCell = 107, NDSB = 610; periphery: NCell = 99, NDSB = 336), 500 ng/nL NCS (interior: NCell = 121, NDSB = 682; periphery: NCell = 100, NDSB = 254), and 800 ng/nL NCS (interior: NCell = 128, NDSB =760; periphery: NCell = 131, NDSB = 862), showing the general trends under these conditions (Fig. 2 E; Fig. S1). As a negative control, we measured the dynamics of DSBs upon fixation of cells with formaldehyde, which shows elimination of the DSB dynamics and defines a noise floor for our measurements (black line, Fig. 2 E). To quantify the DSB density ρB (number of DSBs per unit area), we count all DSBs present in the first time frame (including those that could not be tracked) and find that ρB increases from 0.077 breaks/μm2 for 200 ng/nL NCS to 0.084 breaks/μm2 for 500 ng/nL NCS to 0.145 breaks/μm2 for 800 ng/nL NCS (Fig. 2 F). As displayed in Fig. 2 F, the ρB distributions at the 3 NCS concentrations are quite wide and exhibit partial overlaps with each other.
Our findings reveal a striking difference between the dynamics of the periphery and interior DSBs (Fig. 2, C–E), with the periphery DSBs being much less mobile than the interior DSBs, as shown by the As and AL, while α remains similar. The mean values of As and AL are ∼80% and ∼70% larger, respectively, for the interior than the periphery DSBs. Interestingly, we find that for both the interior and the periphery DSBs, the average AL (mobility at longer times), As (mobility at short times), and α remain unchanged across different NCS concentrations. This suggests that the underlying physical mechanism driving the DSB dynamics is rather robust because it remains unchanged under all studied conditions. We hypothesize that the large difference in the mobility of the periphery and interior DSBs may occur because of a higher chromatin compaction present at the nuclear periphery, which could lead to an increased confinement for the dynamics of the periphery DSBs.
Unique chromatin compaction at DSB sites
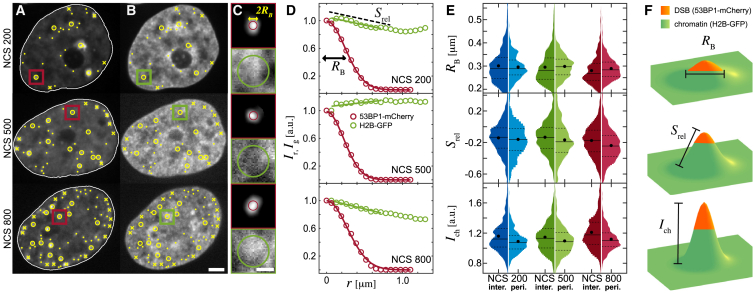
To evaluate the local compaction of chromatin at a DSB site, we have analyzed the H2B-GFP signal, which we have obtained simultaneously with the 53BP1-mCherry signal informing us on the DSB dynamics (Fig. 3). Histone H2B-GFP has been shown before to be a reliable reporter on chromatin position, and thus, changes in measured H2B-GFP intensity indicate also relative changes in the local compaction of the chromatin fiber (12,62). We identify positions of the DSBs in the 53BP1-mCherry signal (yellow markers, Fig. 3 A) and find corresponding sites in the H2B-GFP signal (yellow markers, Fig. 3 B). This allows us to evaluate chromatin compaction at and near each DSB by analyzing the H2B-GFP intensity (Fig. 3 C). First, we normalize the intensity in a small region of radius ∼1 μm around a DSB by the intensity at the DSB center for H2B-GFP and 53BP1-mCherry, respectively, and compute a radial average of the H2B-GFP and 53BP1-mCherry intensity profiles Ig(r) and Ir(r), respectively, for each DSB (Fig. 3 D). Then, we fit Ir(r) to a Gaussian function to determine the effective size of the 53BP1 focus, the radius of which we termed RB and defined as the halfwidth of the Gaussian curve (red curve, Fig. 3 D). At the same time, we fit Ig(r) for 0.75 μm from the DSB center with a linear function f(r) = B + Srelr, the slope (Srel) of which provides a measure of the change in chromatin compaction as a function of the radial distance r from the DSB (green line, Fig. 3 D). Finally, to compare the chromatin compaction at the DSB sites across different nuclei, we normalize the H2B-GFP signal of every nucleus by its mean H2B-GFP intensity. We then measure the chromatin compaction at the DSB center described by the normalized H2B-GFP intensity Ich.
Figure 3.
Local chromatin compaction at DSBs. (A) DSBs detected in 53BP1-mCherry signal are shown: interior DSBs (yellow circles), periphery DSBs (yellow crosses), and untrackable DSBs (yellow dots). (B) DSB positions in H2B-GFP signal are shown (yellow markers). (C) An enlargement of the boxed areas from (A) to (B) is shown. Red boxes show the 53BP1-mCherry signal with the DSB focus size estimated by the red circle. Green boxes show the H2B-GFP signal, and green circle marks an area of 0.75 μm radius around the DSB. (D) Radially averaged intensities of the 53BP1-mCherry (Ir, red markers) and the H2B-GFP (Ig, green markers) signals from (C), normalized by their value at the DSB center, are shown. To determine the 53BP1 focus size RB, a Gaussian was fitted to Ir (red line). To estimate chromatin compaction around a DSB, a line was fitted to Ig over 0.75 μm from the 53BP1 focus center (green line), and its slope Srel was measured. (E) Distributions of RB, Srel, and Ich (H2B-GFP intensity normalized by its mean in a nucleus) for all tracked periphery and interior DSBs are shown. (F) Schematics of RB, Srel, and Ich are shown. Green color corresponds to chromatin, and its height corresponds to the H2B-GFP intensity, which illustrates the chromatin compaction. The red color denotes the size and position of a 53BP1 focus. Scale bars, (A)–(B) 3 μm, (C) 1 μm. To see this figure in color, go online.
Fig. 3 E shows the histograms of RB, Srel, and Ich for both DSB populations, the interior and periphery DSBs, at different NCS concentrations. Across all samples (NDSB = 3504), we find an average size of 53BP1 focus RB ∼ 0.29 ± 0.06 μm (mean ± standard deviation), which is in an excellent agreement with an earlier study (53). No correlation was found between the 53BP1-mCherry expression level and the measured size of a 53BP1 focus (Fig. S2). Interestingly, we observe a slight decrease of the average RB with the increasing NCS concentration for the interior DSBs (Table S1). Strikingly, we find that the mean Srel value is ∼15–35% smaller for the periphery DSBs than for the interior DSBs. This suggests that the chromatin at the periphery DSBs tends to be more compact than at the interior DSBs.
Fig. 3 F provides a schematic illustration of RB, Ich, and Srel: the green color presents chromatin, its height corresponds to the H2B-GFP intensity, and a DSB is depicted in red. The height of the peak is given by Ich and its slope by Srel. Note that the smaller the value of Srel, the steeper the peak. As shown in the cartoon, an average DSB (red) is located at a local peak of H2B-GFP intensity (green), suggesting that chromatin has higher compaction at and around the DSB. Moreover, considering the trends shown by the ensemble averages obtained for RB, Srel, and Ich (Fig. 3 E), our data reveal that the DSB focus size RB for the interior DSBs decreases with an increasing NCS concentration; in other words, the more DSBs are present in the nucleus, the smaller the DSB foci. In addition, the local compaction as measured by Srel and Ich (slope and height of the green peak, respectively) slightly increases with the increasing DSB number (NCS concentration). On average, we find that the interior DSBs become smaller and the surrounding chromatin more compact with an increasing NCS concentration.
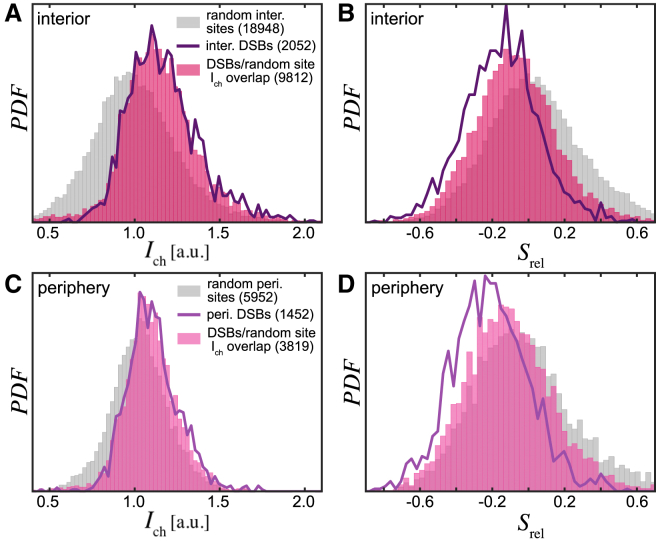
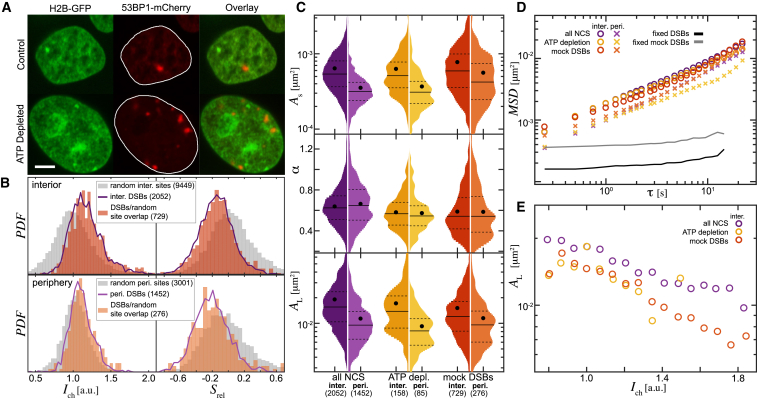
To examine the observed trends in chromatin compaction at DSB sites, we compare them against the chromatin compaction at random sites at the nuclear periphery and interior in nuclei under physiological conditions (Fig. 4). We measure Ich for 18,948 random interior sites and 5952 random periphery sites over 83 nuclei and find that their Ich distribution is centered around Ich ≈ 1, i.e., the mean intensity (Fig. 4, A and C, gray), whereas the Ich distributions for 2052 interior DSBs and 1452 periphery DSBs over 359 nuclei are clearly shifted toward Ich > 1 (Fig. 4, A and C, purple line), suggesting higher local chromatin compaction at DSBs. In fact, about half of the Ich values for random interior sites (9812) and random periphery sites (3819) overlap with those of DSBs (Fig. 4, A and C, pink). Moreover, when we compare the Srel distribution of these subpopulations of random sites (Fig. 4, B and D, pink) with that of the DSBs (Fig. 4, B and D, purple line), we find that Srel distribution of DSBs is clearly shifted toward negative Srel values. The Srel distribution for random sites (Fig. 4, B and D, gray) is centered around zero. We confirm these observations for all 53BP1 foci colocalizing with γH2AX (Fig. S3). This suggests that the higher chromatin compaction found at DSB sites is indeed specific to these damaged sites and likely caused by local DNA repair.
Figure 4.
Local chromatin compaction at DSBs versus random chromatin sites. (A) Ich distribution measured for random interior sites (N = 18,948) over 83 control (undamaged) nuclei (gray) and for interior DSBs (N = 2052) over 356 NCS-treated nuclei (purple solid line) is shown. Note that the Ich distribution for random sites is centered around Ich ≈ 1, i.e., the mean intensity, whereas the Ich distribution for the DSBs is shifted toward Ich > 1. About half of the Ich values for random sites (pink, N = 9812) overlap with those of the DSBs (purple solid line). (B) Srel distribution for random sites (gray) and their subpopulation, whose Ich values overlap with those of the DSBs (pink from A), is shown. The Srel distribution for the DSBs is strongly shifted toward negative Srel values (purple solid line). This suggests that the higher chromatin compaction found at the DSB sites is indeed specific to the damaged sites. (C) and (D) present the same measurements for the periphery DSBs (N = 1452) and random sites (N = 5952). To see this figure in color, go online.
Unique dynamics of DSBs
Next, we investigate how the DSB dynamics compares to the dynamics of undamaged chromatin. Considering the unique chromatin compaction profile at the DSB sites (Fig. 4), undamaged chromatin sites of similar compaction must be used. To perform such comparisons, we have identified random chromatin sites in the H2B-GFP signal of 83 undamaged (control) nuclei, both in their interior (N = 9449) and periphery (N = 3001) (Fig. 5 A). Fig. 5 B shows distributions of Ich and Srel for random interior and periphery sites (gray) centered around 1 and 0, respectively. In comparison, the cumulative distributions of Ich and Srel for all DSBs (purple lines) are shifted toward higher values of Ich and lower values of Srel. From the random periphery and interior sites distributions (gray), we then select subpopulations (orange) that possess the same Ich and Srel characteristics as real DSBs (purple lines), which we term “mock DSBs.” Overall, we detected 729 interior mock DSBs and 276 periphery mock DSBs.
Figure 5.
Negative control for the DSB dynamics. (A) A control HeLa nucleus (top row) and an NCS-treated (500 ng/mL) nucleus upon ATP depletion (bottom row) are shown. 53BP1-mCherry (red) and H2B-GFP (green) signals visualize the DNA repair foci and chromatin, respectively. (B) Histograms of Ich and Srel for random chromatin sites (gray) at periphery and interior of control nuclei are given. From these distributions, we extract a subpopulation of random sites in undamaged nuclei, which has Ich and Srel distributions comparable to the ones measured for real DSBs (solid purple lines) in the interior and periphery (orange), respectively. (C) Distributions of As, α, and AL for DSBs measured at all 3 NCS concentrations (200, 500, and 800 ng/mL), an NCS (500 ng/mL)-treated nuclei upon ATP depletion, and the mock DSBs identified in (B) are shown. (D) MSDs for all samples from (C) are shown. MSD measured for DSBs in damaged cells fixed in formaldehyde (black line, NDSB = 152, NCells = 42) and mock DSBs in undamaged (control) cells fixed in formaldehyde (gray line, NDSB = 165, NCells = 12) defines the noise floor for DSBs and mock DSBs, respectively. (E) Relationship between Ich and AL for interior DSBs from (C) is shown. Error bars for (D) and (E) are shown in Fig. S4. Scale bars, 5 μm. To see this figure in color, go online.
Remarkably, the high chromatin compaction of the DSBs allows us to track the motion of the mock DSBs in the H2B-GFP signal and perform the same analysis as for the real DSBs. As shown in Fig. 5 C, the short-term mobility As of the mock DSBs is higher than that of the real DSBs for both interior and periphery. Conversely, the subdiffusive exponent α and the long-term mobility AL are strongly reduced for the mock DSBs. The summary of the means and standard errors for As, α, and AL distributions is provided in Table S3 and the list of the corresponding p-values in Table S4. The average MSD curves for both mock and real DSBs are displayed in Fig. 5 D (Fig. S4 A). Moreover, when we evaluate AL as a function of Ich, we find the real DSBs to be much more mobile than the mock DSBs (Fig. 5 E; Fig. S4 C). Our data show that DSBs exhibit unique dynamics, which is unlike that of an undamaged chromatin site of the same compaction in a live nucleus.
We hypothesize that the unique chromatin compaction, as well as dynamics of DSBs, might be caused by DNA-repair-related active processes conducted at DSB sites. To test this hypothesis, we induced DSBs using 500 ng/mL NCS and depleted ATP by blocking both the glycolysis and oxidative phosphorylation using 2-deoxyglucose and trifluoromethoxy-carbonylcyanide phenylhydrazone, respectively (Fig. 5 A). Then, we carried out the analysis of the DSB dynamics upon the ATP depletion. Our data show that α and AL of DSBs upon ATP depletion are reduced, whereas As remains unchanged (Fig. 5 C). The average MSD for DSBs upon ATP depletion is shown in Fig. 5 D. Strikingly, the ATP depletion causes a strong reduction of DSB mobility, as illustrated by AL as a function of Ich (Fig. 5 E). Thus, our findings suggest that the unique dynamics of DSBs is actively driven by ATP-consuming processes. This is further corroborated by the fact that the mock DSBs and the DSBs upon ATP depletion not only exhibit similar α, but also, their AL changes with Ich in the same fashion. These observations suggest that the unique DSB dynamics may be driven by active DNA-repair-related processes. Although it is known that ATPases participate on DNA repair (63), it is not obvious that their mechanistic action manifests as an effective translatory motion. Instead, they could (and many of them actually do) just change the local structure and organization of the chromatin fiber, possibly causing chromatin concentration fluctuations.
DSB dynamics follows a universal behavior
In light of the unique structural and dynamical signatures that we found for DSBs in live human cells (Figs. 4 and 5), we have reviewed the relationships between the measured physical parameters (RB, ρB, AL, As, α, Ich, and Srel) for all DSBs (Ninter = 2052, Nperi = 1452 over 359 cells) acquired across the three studied NCS concentrations. Their potential dependencies and/or correlations may provide insights into a general physical mechanism underlying the unique DSB dynamics.
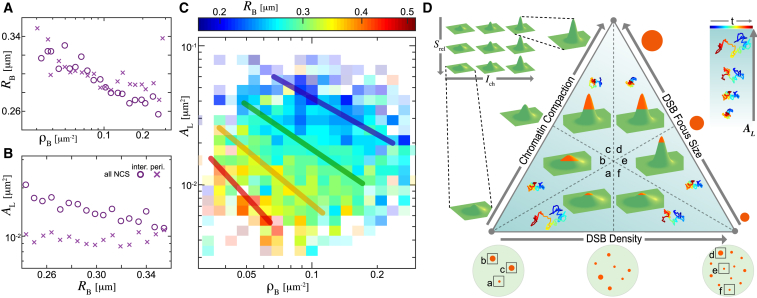
A close visual inspection reveals that RB systematically decreases with increasing ρB, suggesting that 53BP1 foci become smaller with their increasing number in the cell nucleus (Fig. 6 A; Fig. S5 A). This finding applies to both the interior and periphery DSBs. Furthermore, the mobility of the interior DSBs monotonously decreases with increasing RB, whereas no change is observed for the periphery DSBs (Fig. 6 B; Fig. S5 B). Next, we focus on the behavior of the interior DSBs because we anticipate that additional effects such as a possible chromatin tethering to the nuclear envelope might need to be investigated to illuminate the dynamics of the periphery DSBs. Strikingly, when we visualize simultaneously the dependencies of RB, AL, and ρB on each other by employing a three-variable heat map for the interior DSBs (Fig. 6 C; Fig. S5 C), we find that the DSB mobility strongly depends on the size and number of DSB foci. Specifically, the red, yellow, green, and blue lines indicate the areas in the phase diagram where RB is constant, with red corresponding to the largest and blue to the smallest RB. The diagonal position of these lines shows how large DSB foci (RB) occur in nuclei with fewer DSBs (ρB) and are less mobile (AL), whereas small 53BP1 foci (RB) are more mobile (AL) and found in nuclei with higher numbers of DSBs (ρB). A similar review of the three-variable heat map for AL, Srel, and Ich for the interior DSBs (Fig. S5, D–E) reveals that DSBs are less mobile (lower AL) at higher relative compaction states (lower Srel) and at higher compaction states (larger Ich).
Figure 6.
Universal behavior of the DSB dynamics. (A) Relationship between the DSB focus size (RB) and nuclear DSB density (ρB) for the trajectories from all NCS experiments is shown, both in the interior and periphery. (B) Relationship between the long-term DSB mobility (AL) and RB for the trajectories from all NCS experiments is shown, both in the interior and periphery. (C) RB as a function of ρB and AL for the interior DSBs across all NCS concentrations (Fig. S5) is shown. Boxes with greater transparency present average over fewer DSBs. (D) A diagram illustrating the general relationship between DSB focus size (RB), DSB density (ρB), local chromatin compaction (Srel, Ich), and DSB mobility AL is given. (a)–(f) illustrate different situations: in a nucleus with low DSB density, smaller DSB foci located in a weakly compacted chromatin are most mobile (a), whereas larger DSB foci in a weakly compacted chromatin are less mobile (b), and large DSB foci located in a more condensed chromatin are least mobile (c). In a nucleus with high DSB density, large DSB foci located in a more compact chromatin have the lowest mobility (d), whereas small DSB foci in a more compact chromatin have a higher mobility (e), and small DSB foci located in less compact chromatin are the most mobile (f). This diagram hints at a close relationship between the physical properties of a DSB and its surrounding chromatin, both influencing DSB mobility. Error bars for (A) and (B) are shown in Fig. S5. To see this figure in color, go online.
Our mechanistic observations for the interior DSBs (Figs. 5 E, 6, A–C, and Document S1. Figs. S1–S7 and Tables S1–S4, Document S2. Article plus Supporting Material) suggest a universal relationship between the DSB focus size, DSB mobility, chromatin compaction at a DSB site, and the number of DSBs inside the cell nucleus. This relationship can be illustrated by a phase diagram in Fig. 6 D. Specifically, at low DSB density, small DSB foci exhibit high mobility, with chromatin only weakly condensed around them (Fig. 6 D, a). If, at low DSB density, the chromatin compaction or DSB focus size increases, the mobility of the DSB decreases (Fig. 6 D, b–c). At high DSB density, DSB foci are less mobile if they are large and located in more compact chromatin (Fig. 6 D, d) and more mobile if small and positioned in less compact chromatin (Fig. 6 D, e–f). Note that the periphery DSBs do not follow this universal behavior because their mobility seems to be much more restricted in general.
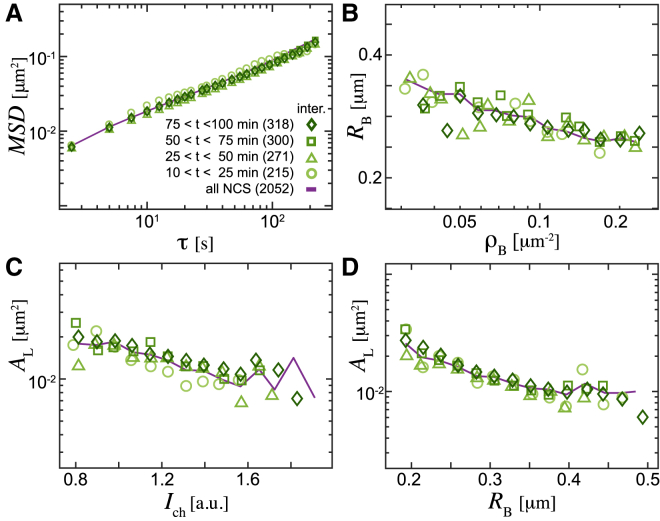
The above-described universal relationship for DSB dynamics was determined by investigating structural and dynamical signatures of DSBs at the 60 min mark upon their induction by NCS. However, DSB behavior is expected to vary with progress of their repair. To test the validity of this universal relationship at other times during DSB repair, we have performed additional experiments evaluating DSBs and their dynamics at 10–100 min after their induction using 500 ng/mL NCS. We chose the initial time of 10 min because earlier studies have shown that chromatin decondensation occurs at a DSB within 10–15 min from when the DNA damage was induced, followed by a chromatin condensation (64, 65, 66). The DNA repair process is anticipated to continue over 60–100 min (57,58). At longer times, additional effects of cell cycle progression would need to be decoupled. Fig. 7 A shows the average MSD curves for interior DSBs at 10–25, 25–50, 50–75, and 75–100 min after their induction (green markers) and the data from Fig. 5 analyzed at the 60 min mark (purple line). We find that MSD does not change during the first 100 min of DSB repair. Moreover, we find that for the interior DSBs, RB as a function of ρB (Fig. 7 B), AL as a function of Ich (Fig. 7 C), and AL as a function of RB (Fig. 7 D) exhibit the same type of dependencies at 10–25, 25–50, 50–75, and 75–100 min after DSB induction (green markers) as the data from Fig. 6 analyzed at the 60 min mark (purple line). Error bars are shown in Fig. S6.
Figure 7.
DSB dynamics as a function of time upon DNA damage. (A) Average MSD for the interior DSBs analyzed during 10–25 min (circles), 25–50 min (triangles), 50–75 min (squares), and 75–100 min (diamonds) after NCS 500 ng/mL addition is shown. The purple line represents data from Fig. 5C collected at the 60 min mark. (B) RB as a function of ρB for DSBs analyzed at different times is shown. (C) AL as a function of Ich for DSBs analyzed at different times is shown. (D) AL as a function of RB for DSBs analyzed at different times is shown. Error bars for (A)–(D) are shown in Fig. S6. To see this figure in color, go online.
Discussion
We investigate the dynamics of DNA DSBs in interphase chromatin of human cells. We find that the DSB sites possess a robust structural signature in a form of a unique chromatin compaction profile. Moreover, our data show that DSB motion is subdiffusive and ATP-dependent and exhibits unique dynamical signatures, different from those of undamaged chromatin. By systematically varying the DSB number per nucleus and analyzing the size and motion of each DSB focus as well as the compaction of the chromatin at each DSB site, we identify a universal relationship between the DSB focus size, DSB mobility, chromatin compaction at a DSB site, and the number of DSBs inside the cell nucleus, as illustrated by the phase diagram in Fig. 6 D. Moreover, we find this universal relationship persists for 10–100 min after the DSB induction.
Strikingly, based on the chromatin compaction, DSB focus size, and DSB density, we can predict the mobility of any DSB. This suggests that the physical properties of a DSB, as well as of its immediate environment, have strong influence on its mobility. Moreover, it hints that biological processes occurring at a DSB (likely DNA-repair-related processes (63)) are well-conserved across all DSBs. In other words, the molecular mechanism of the local DSB repair seems to be robust and likely deterministic, acting in the same fashion at all DSBs. Thus, the observed variation in DSB mobility might be explained by the physical resistance that these processes (and forces involved in them) feel in different local environments. Specifically, at an average 53BP1 focus size of RB ∼ 0.29 μm, viscous forces from the surrounding nucleoplasm dramatically impede the DSB motion. In fact, the Reynolds number (Re) describing the DSB motion is extremely low, Re = ρvRB/μnp ∼ 10−15, with characteristic quantities density ρ ∼ 103 kg m−3, velocity v ∼ 10−9 m s−1, and the nucleoplasm viscosity μnp ∼ 101–103 Pa s (39,40,42,67). A change in the DSB focus size leads to an effective change of its hydrodynamic radius and thus a different viscous friction, fv ∼ μnpRB. A smaller DSB focus thus experiences lower friction than a larger one and can move through the nucleoplasm faster than a larger particle with same forces applied to it. Indeed, this is not only consistent with our observations (Fig. 6 D, a–b) but also with an earlier study showing that large radiation-induced DSB foci are slower than the smaller ones (68).
Furthermore, we find that DSB foci become smaller with an increasing DSB density ρB. Previous studies found that a healthy nucleus contains a pool of free 53BP1 molecules, which are “ready” for an immediate response in case a DSB occurs (69). Taking this into account, if the available 53BP1 molecules respond to multiple DSBs at the same time, the number of 53BP1 molecules per DSB decreases with an increasing DSB number, leading to effectively smaller and thus more mobile DSB foci. This is in agreement with our experimental observations suggesting that the DSB focus size is indeed limited by the presence of the available 53BP1.
From the polymer physics perspective, a DSB is associated with the chromatin fiber, which effectively confines the DSB motion. Equilibrium models of polymer dynamics describe an ideal chain comprised of beads connected by springs and account for different relaxation times (70,71). Although the Rouse model, which neglects the excluded volume and hydrodynamic interactions, predicts that the monomer motion scales as MSD(t) ∼ t1/2, the Zimm model, which accounts for both of these interactions, finds MSD(t) ∼ t2/3. The Rouse model has been successfully used to describe the yeast genome, both its physiological dynamics (32, 33, 34) as well as the DSB motion (35). In contrast, the power law we observe for the MSD of human DSBs (MSD ∼ tα with α = 0.6–0.68) suggests that the hydrodynamic interaction between the DSBs and the surrounding nucleoplasm might indeed play a role. However, it could also be given by the nonequilibrium nature of DSB motion, as demonstrated by the ATP-dependence of the measured DSB dynamics. Moreover, the local viscoelastic properties of the chromatin might also need to be considered, given an earlier observation that the coherent chromatin motion becomes eliminated upon the DSB induction, possibly preventing a long-range distance communication of forces (12). To compare the mobility of the human DSBs against the yeast DSBs, we estimated the length of the constraint of the motion (LC, used in (35)), which is roughly an equivalent to our (Fig. S7), and found that the yeast DSBs are ∼2–3 times more mobile than the human DSBs. This is consistent with the yeast genome being more dilute than the human genome. In summary, a more complex nonequilibrium description is required for the motion of human DSBs, which is consistent with the increasing evidence that the nonequilibrium nature, viscoelasticity, and hydrodynamic interactions play important roles in chromatin dynamics (12,13,39, 40, 41, 42, 43, 44, 45, 46).
Our data show that DSB sites exhibit a distinct structural signature of a unique local chromatin compaction profile, different from that of the undamaged chromatin. This is in agreement with previous studies (64, 65, 66), which observed a slow chromatin condensation around DSB sites. This local condensation of chromatin could stem from the DNA damage repair-related processes such as chromatin remodeling and/or histone modifications (72,73). In fact, it could be stabilized in heterochromatin-like fashion, considering recent findings of HP1α/β localization at DSBs (74,75), which could reduce transcription while facilitating repair. It remains unclear whether this structural feature of DSBs is a part or a consequence of the repair.
Remarkably, we find DSB motion to be strongly influenced by its position inside the cell nucleus. The DSBs located at the nuclear periphery are significantly less mobile than the ones in the nuclear interior. Interestingly, earlier studies found that undamaged chromatin sites also exhibit lower MSD at the nuclear periphery than those in the nuclear interior (76,77). The lower mobility of the periphery DSBs might be caused by the higher chromatin compaction of the perinuclear heterochromatin. An increased chromatin compaction could provide an additional resistance to DSB motion, which we can estimate by evaluating the local Young modulus E. Because E ∼ ξ, where ξ is the number of cross-links in the polymer network (chemical or topological) (70), it is conceivable that at higher chromatin compaction, there are more topological cross-links, and thus, E locally increases. However, the reduced mobility of the periphery DSBs could be also caused by a possible chromatin tethering to the nuclear envelope or steric effects influencing the DNA repair response in the proximity of the nuclear envelope.
In conclusion, our findings reveal that DSBs possess unique structural and dynamical signatures that clearly differ from the undamaged chromatin. This suggests that the underlying biophysical mechanism of DNA repair is a robust and likely a deterministic process, despite its complexity involving different types of nuclear ATPases at different stages of repair. Strikingly, the observed differences in DSB dynamics (i.e., dynamical signatures) can be explained solely by the differences in their physical properties such as focus size and local chromatin environment (i.e., structural features). Such knowledge might allow for detection of the local DNA damage in live human cells. Understanding the DSB dynamics might not only help to elucidate the molecular mechanisms of DNA repair but might also have important biomedical implications.
Author Contributions
A.Z. designed the research. J.A.E. and A.Z. performed the research, contributed new reagents/analytic tools, analyzed data, and wrote the manuscript.
Acknowledgments
This research was supported by the National Institutes of Health Grant R00-GM104152 and by the National Science Foundation Grants CAREER PHY-1554880 and CMMI-1762506.
Footnotes
Editor: Tamar Schlick.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.10.042.
Supporting Material
References
- 1.Alberts B., Johnson A., Walter P. Garland Science; New York: 2002. Molecular Biology of the Cell. [Google Scholar]
- 2.Van Holde K.E. Springer Science & Business Media; New York: 2012. Chromatin. [Google Scholar]
- 3.Bonev B., Cavalli G. Organization and function of the 3D genome. Nat. Rev. Genet. 2016;17:661–678. doi: 10.1038/nrg.2016.112. [DOI] [PubMed] [Google Scholar]
- 4.Dekker J., Marti-Renom M.A., Mirny L.A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibcus J.H., Dekker J. The hierarchy of the 3D genome. Mol. Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hübner M.R., Spector D.L. Chromatin dynamics. Annu. Rev. Biophys. 2010;39:471–489. doi: 10.1146/annurev.biophys.093008.131348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bickmore W.A., van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Sazer S., Schiessel H. The biology and polymer physics underlying large-scale chromosome organization. Traffic. 2018;19:87–104. doi: 10.1111/tra.12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levi V., Ruan Q., Gratton E. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys. J. 2005;89:4275–4285. doi: 10.1529/biophysj.105.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuang C.H., Carpenter A.E., Belmont A.S. Long-range directional movement of an interphase chromosome site. Curr. Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 11.Chen B., Gilbert L.A., Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zidovska A., Weitz D.A., Mitchison T.J. Micron-scale coherence in interphase chromatin dynamics. Proc. Natl. Acad. Sci. USA. 2013;110:15555–15560. doi: 10.1073/pnas.1220313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruinsma R., Grosberg A.Y., Zidovska A. Chromatin hydrodynamics. Biophys. J. 2014;106:1871–1881. doi: 10.1016/j.bpj.2014.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavalli G., Misteli T. Functional implications of genome topology. Nat. Struct. Mol. Biol. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misteli T. Higher-order genome organization in human disease. Cold Spring Harb. Perspect. Biol. 2010;2:a000794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burrell R.A., McGranahan N., Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 18.Miné-Hattab J., Rothstein R. DNA in motion during double-strand break repair. Trends Cell Biol. 2013;23:529–536. doi: 10.1016/j.tcb.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karanam K., Kafri R., Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol. Cell. 2012;47:320–329. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oshidari R., Mekhail K. Catch the live show: visualizing damaged DNA in vivo. Methods. 2018;142:24–29. doi: 10.1016/j.ymeth.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 21.Lebeaupin T., Sellou H., Huet S. Chromatin dynamics at DNA breaks: what, how and why. AIMS Biophys. 2015;2:458–475. [Google Scholar]
- 22.Roukos V., Voss T.C., Misteli T. Spatial dynamics of chromosome translocations in living cells. Science. 2013;341:660–664. doi: 10.1126/science.1237150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gothe H.J., Minneker V., Roukos V. Dynamics of double-strand breaks: Implications for the formation of chromosome translocations. Adv. Exp. Med. Biol. 2018;1044:27–38. doi: 10.1007/978-981-13-0593-1_3. [DOI] [PubMed] [Google Scholar]
- 24.Lottersberger F., Karssemeijer R.A., de Lange T. 53BP1 and the LINC complex promote microtubule-dependent DSB mobility and DNA repair. Cell. 2015;163:880–893. doi: 10.1016/j.cell.2015.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker A., Durante M., Jakob B. ATM alters the otherwise robust chromatin mobility at sites of DNA double-strand breaks (DSBs) in human cells. PLoS One. 2014;9:e92640. doi: 10.1371/journal.pone.0092640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dion V., Kalck V., Gasser S.M. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat. Cell Biol. 2012;14:502–509. doi: 10.1038/ncb2465. [DOI] [PubMed] [Google Scholar]
- 27.Miné-Hattab J., Rothstein R. Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 2012;14:510–517. doi: 10.1038/ncb2472. [DOI] [PubMed] [Google Scholar]
- 28.Neumann F.R., Dion V., Gasser S.M. Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination. Genes Dev. 2012;26:369–383. doi: 10.1101/gad.176156.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seeber A., Dion V., Gasser S.M. Checkpoint kinases and the INO80 nucleosome remodeling complex enhance global chromatin mobility in response to DNA damage. Genes Dev. 2013;27:1999–2008. doi: 10.1101/gad.222992.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hauer M.H., Seeber A., Gasser S.M. Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nat. Struct. Mol. Biol. 2017;24:99–107. doi: 10.1038/nsmb.3347. [DOI] [PubMed] [Google Scholar]
- 31.Seeber A., Hauer M.H., Gasser S.M. Chromosome dynamics in response to DNA damage. Annu. Rev. Genet. 2018;52:295–319. doi: 10.1146/annurev-genet-120417-031334. [DOI] [PubMed] [Google Scholar]
- 32.Zimmer C., Fabre E. Principles of chromosomal organization: lessons from yeast. J. Cell Biol. 2011;192:723–733. doi: 10.1083/jcb.201010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amitai A., Toulouze M., Holcman D. Analysis of single locus trajectories for extracting in vivo chromatin tethering interactions. PLoS Comput. Biol. 2015;11:e1004433. doi: 10.1371/journal.pcbi.1004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimmer C., Fabre E. Chromatin mobility upon DNA damage: state of the art and remaining questions. Curr. Genet. 2019;65:1–9. doi: 10.1007/s00294-018-0852-6. [DOI] [PubMed] [Google Scholar]
- 35.Amitai A., Seeber A., Holcman D. Visualization of chromatin decompaction and break site extrusion as predicted by statistical polymer modeling of single-locus trajectories. Cell Rep. 2017;18:1200–1214. doi: 10.1016/j.celrep.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 36.Amitai A., Holcman D. Encounter times of chromatin loci influenced by polymer decondensation. Phys. Rev. E. 2018;97:032417. doi: 10.1103/PhysRevE.97.032417. [DOI] [PubMed] [Google Scholar]
- 37.Platani M., Goldberg I., Swedlow J.R. Cajal body dynamics and association with chromatin are ATP-dependent. Nat. Cell Biol. 2002;4:502–508. doi: 10.1038/ncb809. [DOI] [PubMed] [Google Scholar]
- 38.Sinha D.K., Banerjee B., Shivashankar G.V. Probing the dynamic organization of transcription compartments and gene loci within the nucleus of living cells. Biophys. J. 2008;95:5432–5438. doi: 10.1529/biophysj.108.135921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tseng Y., Lee J.S., Wirtz D. Micro-organization and visco-elasticity of the interphase nucleus revealed by particle nanotracking. J. Cell Sci. 2004;117:2159–2167. doi: 10.1242/jcs.01073. [DOI] [PubMed] [Google Scholar]
- 40.de Vries A.H., Krenn B.E., Kanger J.S. Direct observation of nanomechanical properties of chromatin in living cells. Nano Lett. 2007;7:1424–1427. doi: 10.1021/nl070603+. [DOI] [PubMed] [Google Scholar]
- 41.Pajerowski J.D., Dahl K.N., Discher D.E. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. USA. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Celedon A., Hale C.M., Wirtz D. Magnetic manipulation of nanorods in the nucleus of living cells. Biophys. J. 2011;101:1880–1886. doi: 10.1016/j.bpj.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erdel F., Baum M., Rippe K. The viscoelastic properties of chromatin and the nucleoplasm revealed by scale-dependent protein mobility. J. Phys. Condens. Matter. 2015;27:064115. doi: 10.1088/0953-8984/27/6/064115. [DOI] [PubMed] [Google Scholar]
- 44.Stephens A.D., Banigan E.J., Marko J.F. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell. 2017;28:1984–1996. doi: 10.1091/mbc.E16-09-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Pierro M., Potoyan D.A., Onuchic J.N. Anomalous diffusion, spatial coherence, and viscoelasticity from the energy landscape of human chromosomes. Proc. Natl. Acad. Sci. USA. 2018;115:7753–7758. doi: 10.1073/pnas.1806297115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saintillan D., Shelley M.J., Zidovska A. Extensile motor activity drives coherent motions in a model of interphase chromatin. Proc. Natl. Acad. Sci. USA. 2018;115:11442–11447. doi: 10.1073/pnas.1807073115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu F.Y., Haley S.C., Zidovska A. On the origin of shape fluctuations of the cell nucleus. Proc. Natl. Acad. Sci. USA. 2017;114:10338–10343. doi: 10.1073/pnas.1702226114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cox I.J., Roy S., Hingorani S.L. Proc. IEEE Image Process. Vol. 2. 1995. Dynamic histogram warping of image pairs for constant image brightness; pp. 366–369. [Google Scholar]
- 49.Crocker J.C., Grier D.G. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 1996;179:298–310. [Google Scholar]
- 50.Pelletier V., Fournier P., Kilfoil M. 2005. 2D feature finding and tracking algorithms.https://people.umass.edu/kilfoil/tools.php [Google Scholar]
- 51.Heun P., Laroche T., Gasser S.M. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–2186. doi: 10.1126/science.1065366. [DOI] [PubMed] [Google Scholar]
- 52.Crocker J.C., Hoffman B.D. Multiple-particle tracking and two-point microrheology in cells. Methods Cell Biol. 2007;83:141–178. doi: 10.1016/S0091-679X(07)83007-X. [DOI] [PubMed] [Google Scholar]
- 53.Reindl J., Girst S., Dollinger G. Chromatin organization revealed by nanostructure of irradiation induced γH2AX, 53BP1 and Rad51 foci. Sci. Rep. 2017;7:40616. doi: 10.1038/srep40616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Panier S., Boulton S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 55.Schultz L.B., Chehab N.H., Halazonetis T.D. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shiloh Y., van der Schans G.P., Becker Y. Induction and repair of DNA damage in normal and ataxia-telangiectasia skin fibroblasts treated with neocarzinostatin. Carcinogenesis. 1983;4:917–921. doi: 10.1093/carcin/4.7.917. [DOI] [PubMed] [Google Scholar]
- 57.Hamilton C., Hayward R.L., Gilbert N. Global chromatin fibre compaction in response to DNA damage. Biochem. Biophys. Res. Commun. 2011;414:820–825. doi: 10.1016/j.bbrc.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andegeko Y., Moyal L., Rotman G. Nuclear retention of ATM at sites of DNA double strand breaks. J. Biol. Chem. 2001;276:38224–38230. doi: 10.1074/jbc.M102986200. [DOI] [PubMed] [Google Scholar]
- 59.Suchánková J., Kozubek S., Bártová E. Distinct kinetics of DNA repair protein accumulation at DNA lesions and cell cycle-dependent formation of γH2AX- and NBS1-positive repair foci. Biol. Cell. 2015;107:440–454. doi: 10.1111/boc.201500050. [DOI] [PubMed] [Google Scholar]
- 60.Sedelnikova O.A., Rogakou E.P., Bonner W.M. Quantitative detection of (125)IdU-induced DNA double-strand breaks with γ-H2AX antibody. Radiat. Res. 2002;158:486–492. doi: 10.1667/0033-7587(2002)158[0486:qdoiid]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 61.Lopez Perez R., Best G., Huber P.E. Superresolution light microscopy shows nanostructure of carbon ion radiation-induced DNA double-strand break repair foci. FASEB J. 2016;30:2767–2776. doi: 10.1096/fj.201500106R. [DOI] [PubMed] [Google Scholar]
- 62.Kimura H., Cook P.R. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ceccaldi R., Rondinelli B., D’Andrea A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burgess R.C., Burman B., Misteli T. Activation of DNA damage response signaling by condensed chromatin. Cell Rep. 2014;9:1703–1717. doi: 10.1016/j.celrep.2014.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiolo I., Tang J., Costes S.V. Nuclear dynamics of radiation-induced foci in euchromatin and heterochromatin. Mutat. Res. 2013;750:56–66. doi: 10.1016/j.mrfmmm.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hinde E., Kong X., Gratton E. Chromatin dynamics during DNA repair revealed by pair correlation analysis of molecular flow in the nucleus. Biophys. J. 2014;107:55–65. doi: 10.1016/j.bpj.2014.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caragine C.M., Haley S.C., Zidovska A. Surface fluctuations and coalescence of nucleolar droplets in the human cell nucleus. Phys. Rev. Lett. 2018;121:148101. doi: 10.1103/PhysRevLett.121.148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Falk M., Lukasova E., Kozubek S. Chromatin dynamics during DSB repair. Biochim. Biophys. Acta. 2007;1773:1534–1545. doi: 10.1016/j.bbamcr.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 69.Rübe C.E., Lorat Y., Rübe C. DNA repair in the context of chromatin: new molecular insights by the nanoscale detection of DNA repair complexes using transmission electron microscopy. DNA Repair (Amst.) 2011;10:427–437. doi: 10.1016/j.dnarep.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 70.De Gennes P.-G. Cornell University Press; Ithaca, New York: 1979. Scaling Concepts in Polymer Physics. [Google Scholar]
- 71.Rubinstein M., Colby R.H. Oxford University Press; New York: 2003. Polymer Physics, volume 23. [Google Scholar]
- 72.Hendzel M.J., Strickfaden H. The Functional Nucleus. Springer; 2016. DNA repair foci formation and function at DNA double-strand breaks; pp. 219–237. [Google Scholar]
- 73.Nair N., Shoaib M., Sørensen C.S. Chromatin dynamics in genome stability: roles in suppressing endogenous DNA damage and facilitating DNA repair. Int. J. Mol. Sci. 2017;18:1486. doi: 10.3390/ijms18071486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baldeyron C., Soria G., Almouzni G. HP1α recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J. Cell Biol. 2011;193:81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ayoub N., Jeyasekharan A.D., Venkitaraman A.R. HP1-β mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 76.Shinkai S., Nozaki T., Togashi Y. Dynamic nucleosome movement provides structural information of topological chromatin domains in living human cells. PLoS Comput. Biol. 2016;12:e1005136. doi: 10.1371/journal.pcbi.1005136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chubb J.R., Boyle S., Bickmore W.A. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 2002;12:439–445. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.