Abstract
Studies comparing diverse groups have shown that many psychiatric diseases involve disruptions across distributed large-scale networks of the brain. There is hope that fMRI functional connectivity techniques will shed light on these disruptions, providing prognostic and diagnostic biomarkers as well as targets for therapeutic interventions. However, to date, progress on clinical translation of fMRI methods has been limited. Here, we argue that this limited translation is driven by a combination of inter-subject heterogeneity and the relatively low reliability of standard fMRI techniques at the individual level. We review a potential solution to these limitations: the use of new “precision” fMRI approaches that shift the focus of analysis from groups to single individuals through the use of extended data acquisition strategies. We begin by discussing the potential advantages of fMRI functional connectivity methods for improving our understanding of functional neuroanatomy and disruptions in psychiatric disorders. We then discuss the budding field of precision fMRI and findings garnered from this work. We demonstrate that precision fMRI can improve the reliability of functional connectivity measures, while showing high stability and sensitivity to individual differences. We close by discussing the application of these approaches to clinical settings.
Keywords: fMRI, functional connectivity, precision imaging, brain networks, individual differences, reliability
There is an enormous need to improve diagnosis and treatment of neuropsychiatric disorders given their personal and societal burden (1–3). A substantial body of neuroimaging work has documented variations in brain function that accompany psychiatric disorders with the hope of developing biomarkers and novel treatments. These studies generally observe that psychiatric disorders are associated not with focal brain pathology, but with widespread dysfunction of distributed brain networks (4–6). However, to date, few of these findings have been translated to clinical settings. At least two factors may contribute to this lack of translation. First, group comparisons do not capture the heterogeneity of phenomenological characteristics present across any given disorder. For example, individuals with schizophrenia can present with variable symptoms including delusions, hallucinations, disorganized behavior, and/or anhedonia. Individuals with depression may exhibit symptoms from anxiety to diminished reward sensitivity (7). Children with Tourette syndrome (TS) exhibit varied motor and vocal tics and frequently demonstrate comorbidity with ADHD and OCD (8). It is not surprising that findings from group studies do not adequately account for any individual’s unique characteristics. Second, many neuroimaging techniques are noisy and exhibit low reliability in single individuals, limiting the ability to capture neural characteristics related to individual heterogeneity. Thus, new approaches are needed to measure brain function reliably in individuals.
In this review, we highlight insights that neuroimaging methods can provide about systems-level brain function in health and disease. We then review recent efforts from our laboratories and others to develop individualized (“precision”) applications of fMRI using extended data acquisition strategies that can provide reliable and stable individual measures of brain organization. We close by discussing the practical application of precision fMRI in translational settings.
Measuring functional neuroanatomy with fMRI correlations
Human brain function is organized at many spatial scales, from local circuits to cortical columns, brain areas, and large-scale systems (9). The systems (10–14) and areal (15, 16) organization of human brains can be characterized noninvasively using functional connectivity MRI. Functional connectivity (FC) refers to temporal correlations in fMRI activity between different regions, which can be measured during experimental tasks or as spontaneous activity patterns during the resting-state (in the absence of task instructions). Resting-state methods have the advantage of not requiring patients to complete a task, which may prove challenging for patients and provide added burdens in administration for clinical centers. Despite the lack of experimental constraint, resting-state correlations demonstrate rich systematic patterns, with functionally-related regions showing high correlations to each other (“within system”), and lower correlations to regions in other systems. Indeed, resting-state fMRI has been used to define systems throughout the brain (11, 13, 17–20). These patterns are robust, with independent studies converging on similar descriptions of group-averaged system organization of the human brain (11, 13) (Figure 1A). Moreover, validation of FC patterns has been established through convergent evidence from other neural measures (21), lesion approaches (22, 23), and behavior (24). At a finer scale, FC can also be used to parcellate the brain into regions that approximate functional areas and map onto differences in task function (15, 16, 25).
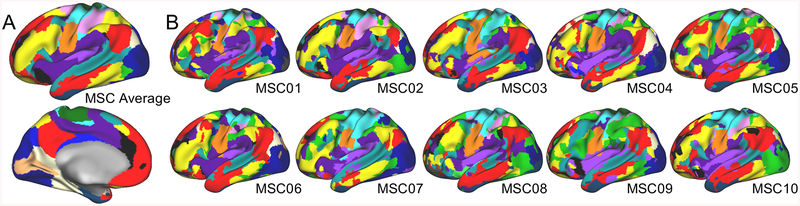
Figure 1: Group and individual functional brain networks.
(A) Robust group-average functional networks (different colors) are observed across multiple studies using different methods. This group-average map was created by averaging data from the MSC subjects at rest. (B) Functional networks in individuals exhibit similarities to the group-average, but also pronounced individual differences. For example, note variations in the network organization of the dorsolateral prefrontal cortex (colors are matched to group-average labeling in A). A and B are based on data from (69).
FC approaches can be used to measure differences in brain systems between neurotypical controls and diverse neuropsychiatric populations, such as in schizophrenia (5, 26), depression (27), or TS (28, 29), and to identify commonalities across diagnostic boundaries (e.g.,(30–32)). Additionally, functional network mapping provides context for interpreting brain activity during tasks. Task results are often described using large, poorly-defined anatomical locations (e.g., “lateral prefrontal cortex”). With coincident FC, task results can instead be ascribed to specific functional systems (e.g., frontoparietal or cinguloopercular), refining our understanding of task mechanisms in healthy individuals (17, 33, 34) and youths with diverse psychiatric symptoms (35). Furthermore, FC facilitates complex systems approaches to understanding brain function (36), which can be used to identify brain hubs (37–40) that support task function (41–45) and are vulnerable to damage (46–48). Network approaches hold promise for providing concise descriptions of how complex systems like the brain change across the lifespan (49–51) and are disrupted in psychiatric disorders (52), and may help develop mechanistic theories for these alterations (52–54).
Towards clinical utility of functional connectivity
Thus, FC has strong potential for clinical applications (55), including in the use of network mapping to enhance neurosurgical planning (56–60), providing systems-level biomarkers to identify at-risk individuals (61–63), sub-typing patients with diverse etiologies (32, 64), and identifying targets for treatment (e.g., with TMS) (55, 65, 66). However, despite a 20+ year history, FC techniques have yet to become widely used clinically. This observation raises the question of why clinical implementation has stalled.
For a measure to be clinically useful, it should be both reliable, such that repeated measurements produce the same result, and stable across contexts, such that it primarily reflects trait- rather than state-dependent effects. Additionally, it should show sensitivity to idiosyncratic features that may be clinically relevant. We suggest that new approaches are needed to reach clinically-relevant reliability, stability, and sensitivity of FC at the individual-level.
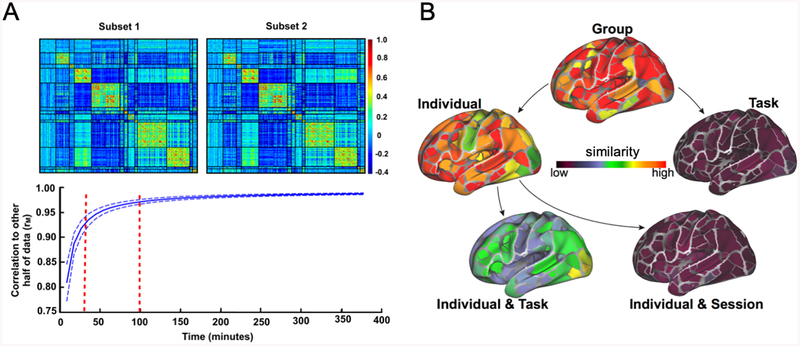
Achieving single-subject reliability is not trivial. While group descriptions of functional networks are robust, most studies collect insufficient data (5–10 min.) to obtain reliable FC estimates in single individuals (67–71). Recent estimates suggest that at least 40 min. of low-motion fMRI data are needed to achieve high reliability (test-retest r>0.9) across the full connectome (69, 70) (Figure 2A). While some networks may achieve reliability more quickly (e.g., the default-mode), most single connections show poor-to-fair reliability with < 40 min. of data (67, 71) (see Supplemental Discussion). Derived FC measures require even more data (e.g., areal parcellations require >50 min. (72); measures of lag structure require >200 min. (73, 74)). Moreover, reliability of fMRI data usually is worse in non-cortical regions that are implicated in many psychiatric disorders (75–77). Current estimates suggest that 90 min. of data are needed to achieve high reliability of cerebellar FC (78) and >100 min. are needed for the basal ganglia and thalamus (79). Notably, it may be possible to identify an individual within a large group – so-called “fingerprinting” approaches (80, 81)– using substantially less data (81). However, subject identification is not the same as individual-specific characterization of brain-based disease processes, which requires substantially more data and is the ultimate target for clinical utility.
Figure 2: Reliability and stability of functional brain networks.
(A) Recent studies using precision fMRI methods have demonstrated that more than 40 min. of high-quality fMRI data are necessary to achieve high test-retest reliability of functional networks in the cortex (top: functional networks from a single individual across two split-halves of the data; bottom: similarity of individual functional network measures across the connectome with increasing amounts of data). Even larger amounts of data may be needed to achieve high-reliability in non-cortical regions (78, 79). (B) Analysis of precision fMRI datasets allows for a decomposition of the sources of variance in functional networks. This work has shown that functional networks are dominated by stable factors, including common structure across groups and stable patterns of individual differences. Task-state makes modest contributions to FC that are largely individually specific, and daily variation is small with sufficient data. This is shown by examining how similar functional networks are from datasets that share group, individual, task, or session factors (or a combination). A is from (70), B is adapted from (93).
A precision fMRI approach
The findings above motivate an alternative strategy for data acquisition. The core feature of this approach is to collect larger quantities of fMRI data in single individuals as opposed to smaller quantities of data averaged over groups. For convenience, we call this acquisition strategy “precision” fMRI (pfMRI), though other terms, including “deep” or “high-sampling” have also been used. PfMRI data are frequently coupled with extensive phenotypic and behavioral measurements to aid in establishing validity. Moreover, pfMRI benefits from being combined with advanced analysis including dataset denoising, alignment, and network definition.
The MyConnectome dataset is one of the original demonstrations of the utility of pfMRI for functional network mapping*. For this dataset, Russ Poldrack scanned himself twice weekly for a year, and collected additional phenotypic variables regarding health, affect, genotype, gene expression, and metabolomics (90). This work established that even in a single person, classic network patterns are evident, but clear deviations from typical “group” organization are also observed (70). Importantly, despite modest day-to-day variability (perhaps associated with caffeine or arousal levels (70)), these network patterns were fundamentally stable over the course of the year (70, 91).
The MyConnectome study inspired several related pfMRI projects (for examples see Table 1, Supp. Table 1). Among these, the “Midnight Scan Club” (MSC) dataset collected >20 hrs. of fMRI data in each of 10 people (Figure 1B), including large amounts of task and resting-state data. Initial reports from this dataset established that extended data acquisitions can be used to generate reliable individual descriptions of functional brain organization (69). Robust individual differences in FC were observed and these network variations corresponded to brain activations during tasks (69).
Table 1:
A collection of precision fMRI datasets with resting-state data. ‘Precision’ datasets are defined here as datasets with >120 min. of resting state per individual collected in 2 or more sessions on the same scanner, and whole-brain data acquisition. These parameters allow for reliability and non-cortical analyses; additional “high-data” fMRI projects with 40–120 min. are listed in Supp. Table 1. Datasets were in part identified through Kong (149).
| Dataset name | Location of Dataset | Participants | Sessions & Collection Window | Resting-state data per session | Scan Parameters | Additional data collected |
|---|---|---|---|---|---|---|
| Anderson (68) | http://fcon_1000.projects.nitrc.org/indi/CoRR/html/utah_2.html | 1 M (age 39) | 5 sessions over 3 weeks | 50 min. |
|
|
| IPCAS – 3 Day [IPCAS 6] Zuo (143) |
http://fcon_1000.projects.nitrc.org/indi/CoRR/html/ipcas_6.html | 2 (1 F, ages 21 and 25) | 15 sessions over 3 days | 30 min. |
|
|
| MyConnectome Poldrack (90) |
https://openneuro.org/datasets/ds000031/versions/00001 | 1 M (age 45, author) | 89 sessions over 72 weeks | 10 min. |
|
|
| Kirby Weekly Dataset Choe (102) |
https://www.nitrc.org/projects/kirbyweekly | 1 M (age 40) | 158 sessions over 185 weeks | 7 min. |
|
|
| Midnight Scan Club (MSC) Gordon (69) |
https://openneuro.org/datasets/ds000224/versions/00001 | 10 (5 F, ages 24–34) | 10 sessions within 7 weeks | 30 min. |
|
|
| Buckner Lab Dataset Braga and Buckner (92) |
- | 4 (4 F, ages 21–26) | 24 sessions over ~16 weeks | 7 min. |
|
|
| Day2Day Dataset Filevich (144) |
e-mail authors | 8 (6 F, ages 24–32; N=6 with > 120 min.) | 11–50 sessions over 8–56 weeks | 5 min. |
|
|
| Healthy Brain Network Dataset (HBN-SSI) O’Connor (145) |
http://fcon_1000.projects.nitrc.org/indi/hbn_ssi/ | 13 (8 F, ages 21–42) | 13 sessions over 4–8 weeks | 10 min. |
|
|
| Yale Test-Retest Dataset Noble (71) |
http://fcon_1000.projects.nitrc.org/indi/retro/yale_trt.html | 12 (6 F, ages 27–56) | 4 sessions over ~1 week | 36 min. |
|
|
| Meszlénvi Dataset Meszlenyi (146) |
- | 1 F (age 28) | 10 sessions over 5 days | 20 min. |
|
|
| Gordon TBI Dataset Gordon (126) |
- | 26 (5 F, ages 37 +/− 11.8; 21 with TBI; N=24 with > 120 min.) | 2–5 sessions over less than 12 weeks | 5–44 min. |
|
|
| SIMON Dataset Duchesne (147) |
http://fcon_1000.projects.nitrc.org/indi/retro/SIMON.html | 1 M (age 29) | 15 sessions over 468 weeks | 10–11 min. |
|
|
| Donnelly-Kehoe Dataset Donnelly-Kehoe (148) |
- | 1 F (age 29) | 50 sessions over 26 weeks | 5 min. |
|
|
Additional pfMRI datasets have made important contributions to our understanding of functional systems. For example, Braga and Buckner (92) used a four person pfMRI dataset to describe substructure within several networks that is too individually variable to easily delineate in group data. Noble et al. (71) identified brain regions that are more or less reliable with a given quantity of data, which is critical for designing studies focused on specific regions. Given the public availability of many of these datasets (Table 1), their contribution to the field will likely increase in future years.
Precision approaches quantify stability and variability in functional brain networks
In addition to reliability, clinical utility requires that measurements show stability across contexts. That is, a diagnostic measurement would ideally be influenced only by the conditions of interest (e.g., long-term disease status, individual traits) rather than by day-to-day variation, ongoing states, or thoughts (e.g., whether the patient ate breakfast, if they were cold in the scanner room, if a technician was calm or abrupt, etc.). Thus, the stability of FC across different time-scales and contexts is important for its utility in psychiatric care.
The pfMRI design of the MSC is well-suited to examining this issue, as it contains data from multiple tasks spanning different cognitive domains collected in multiple individuals across multiple days (69). We used these data to identify cortical network patterns that were consistent across all measurements (static “group” effects) or that varied across individuals, days, or tasks (93). Our findings demonstrated that functional networks are largely stable, with shared group patterns as well as network features that were specific to individual subjects (Figure 2B).
Variations in FC from day-to-day and task-to-task were also observed, but these effects were relatively small in magnitude. Interestingly, task effects on FC were largely individually-specific, rather than common across the group (93) (Figure 2B), consistent with previous studies showing subtle group-average task modulations of FC (45, 94) and added task-by-individual interactions (95, 96). The individual-specificity nature of task influences on brain networks suggests that their study may especially benefit from the use of pfMRI. In addition to characterizing task and daily variation, we also used pfMRI to show that FC is stable over shorter time-scales (i.e. minutes). In this work, apparent within-session variability in FC was primarily driven by sampling error, acquisition artifacts (such as motion), and subject arousal during scans (97).
The dominance of stable factors on FC was relatively consistent across spatial scales, from the full connectome, to single networks, regions, and even single connections (93). Subsequent work observed similar effects in subcortical and cerebellar regions (78, 79) with hints of increased individual variability (78) - an intriguing finding given evidence of variation within non-cortical structures in psychiatric disorders (75–77, 98–101).
Thus, FC techniques are well suited to measuring stable aspects of brain organization, including aberrant features that may underlie psychopathology. Moreover, these findings indicate a high sensitivity to individual differences in brain networks (see next section). Jointly, the strong reliability, stability over states, and sensitivity to individual differences seen with pfMRI methods make them strong candidates for clinical applications.
An emergent question is how FC varies across months and years. The MyConnectome dataset suggests that many aspects of FC are stable over a year (70, 91) (also seen for some networks over 3.5 years in the Kirby Weekly dataset (102)). However, new studies are needed to determine to what extent these patterns persist throughout the lifespan, from early infancy into aging, and whether they can be altered with prolonged or profound life experiences.
Characteristics of individual variation in brain networks
Individual FC differences (69, 80, 81, 103–106) have been identified at multiple scales, ranging from differences in brain-wide network organization (e.g., network “efficiency” (69)) to punctate regions that vary across individuals (70, 91, 103, 104). One open question is whether these disparate spatial scales measure related aspects of individual variation (e.g., variation in FC of a single region may cause apparent variation in brain-wide efficiency). Interestingly, in the MSC dataset different individuals are highlighted as ‘atypical’ depending on the scale of analysis: e.g., Gordon (69) found that brain-wide efficiency was significantly lower for subjects MSC02 and MSC06 relative to the group, but Seitzman (91) found that MSC02 and MSC06 sorted into different sub-groups based on regional variations, and Gratton (93) found that MSC01 was most different from the group in task and rest FC. An important question for future work is whether individual differences in FC reflect differences in the spatial organization of networks/areas or differences in the magnitude of functional correlations within a static spatial structure (e.g., (107, 108)).
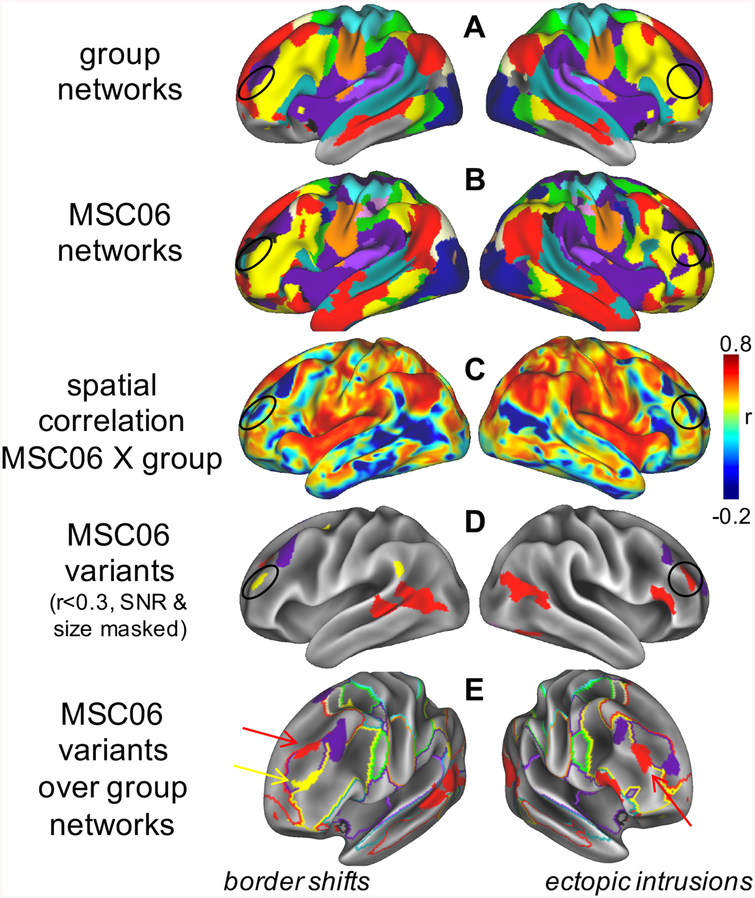
Recent reports have observed that individuals demonstrate localized regions of distinct FC relative to the group (70, 91). We term these “network variants” (Figure 3). Network variants occur most frequently in association cortex (e.g., parts of the frontoparietal, default mode, and cinguloopercular systems) and appear in two general forms: border shifts between networks (e.g., the default network enlarges, encroaching on classically frontoparietal regions) and ectopic intrusions (e.g., an isolated area in the frontoparietal network shows altered FC, such that it connects to the default network). Despite their low correspondence to the “average” architecture, network variants are common. Indeed, in preliminary investigations (70, 91) across datasets (109), we found network variants in every individual. These findings suggest that the average brain is not a veridical representation of any individual person.
Figure 3: Network variants.
Comparison of (A) group networks and (B) an individual from the MSC (MSC06, colors match Fig. 1). (C) Some locations exhibit low similarity to the group (a few examples are circled), which can be identified through vertex-wise spatial correlation; (D) we call these low-similarity locations network variants. (E) Variants may represent shifts of network borders (left) or isolated ectopic intrusions (right). We hypothesize that network variants are caused by stable factors that reprioritize the neural functions of cortical areas, causing shifts in the boundaries of cortical networks and ectopic intrusions. These altered prioritizations lead to changes in the dominant systems-level relationships of a region (e.g., increasing FC to relevant regions in alternate networks), causing these regions to appear as network variants. Thus, network variants may be related altered brain function during tasks and behavioral responses across individuals. See (91).
Network variants have a number of systematic trait-like properties (91) that suggest they may be good candidate clinical targets. They are stable across scans (reaching r>0.8 with >40 min. of data). Moreover, like other phenotypic traits such as eye-color, blood type, or personality, individuals can be grouped based on their characteristic patterns of network variants. Two sub-groups were identified across multiple datasets: one with variants more associated with the default-mode network and another with variants more associated with goal-directed control and sensorimotor processing systems (81). These findings suggest that network variants may relate to individual differences in complex goal-directed functions subserved by these networks (17, 110–113) - functions known to vary in the typical population (114) and implicated across a range of disorders including depression (27, 64, 115, 116), schizophrenia (103–105), and TS (28, 117).
Despite the nascent state of pfMRI studies, there is preliminary convergent evidence that validates the connection between these individual differences and differences in brain function (e.g., (80, 107, 108, 118–121)). For example, network variants re-assigned to the default-mode showed de-activations during tasks– much like canonical default-mode locations – even when found within cortical territories that are classically identified with task-activated networks (e.g. frontoparietal) (91). Thus, network variants may represent locations with shifted functions, leading to altered network correlations. Other findings have also suggested that individual-specific FC – defined at rest – overlaps well with task-related brain activations (118, 119, 121). Further preliminary validation of individualized FC measures comes from their successful guidance of TMS (e.g., (66, 122)) and by comparison with structural brain measures (e.g., overlap between FC variations and variations in myelin density in (69)).
Moreover, individual differences in network organization also appear to relate to behavioral variation (80, 107, 108). In the sub-groups described above, differences in network variants were associated with small differences in quality of life and drug use (91). Similarly, Smith (120) suggests that FC differences link to a “positive-negative mode” of behavioral variation. One important question to address in future research will be which variations in brain system organization have critical consequences on behavior and which reflect degenerate solutions to carrying out the same behavioral function.
Looking forward: pfMRI and psychiatry
Thus far, pfMRI approaches have primarily examined small, homogenous cohorts (Table 1). These datasets have highlighted the reliability of FC measurements, in the spirit of recent movements in psychology and psychiatry to increase reproducibility in research (123–125). This work has already provided important preliminary results regarding sources of variance and stability of FC techniques. Critically, the large amounts of data per subject in pfMRI approaches allow for observations to be verified reliably at the level of individuals (even in clinical samples (126)), which is the level most pertinent for clinical applications. It is this feature that makes pfMRI a compelling platform to address outstanding challenges in psychiatry.
One such challenge has been in understanding differences between the ‘normal’ and pathologic brain, which have so far been obscured in group format studies. Group studies typically find modest differences in FC across a range of comparisons: between tasks (45, 94), across states of consciousness (127), over development (128), and across major neurological and psychiatric disorders (30, 129, 130). One question is whether pfMRI approaches that take advantage of both group commonalities and individually specific features may be more sensitive to detecting heterogeneous differences, especially those relevant to clinical work. Early reports support this conjecture, as pfMRI approaches have demonstrated enhanced sensitivity not only to individual differences, but also to task-state effects (93), and clinical symptoms (126).
A second, related, challenge in psychiatry is to create tools for accurate diagnosis and prognosis of clinical features at the individual level. While pfMRI has yet to be used widely in clinical populations, initial accounts have suggested that high-data approaches can increase the association between fMRI measures and behavior in the neurotypical population (80, 107, 108, 120). We are aware of only one study to date to apply pfMRI to a clinical population (126). In that study, ~3.5 hrs. of MRI data were collected from 26 veterans with varying history of TBI and PTSD. Interestingly, FC mediated observed associations between TBI and PTSD symptoms. However, this relationship was only evident with large amounts of data; analyses using only 10 min. of data per subject were non-significant. Two other recent papers further highlight advantages of individualized analysis techniques in psychiatric datasets (although with lower amounts of per-subject data (131, 132)). In Wang (131), fMRI data were gathered from 158 participants diagnosed with schizophrenia, schizoaffective disorder, or bipolar disorder. FC derived from individually-specific regions significantly predicted symptom levels, while models using group regions performed consistently worse. Similarly, in participants with OCD, Brennan (132) found that brain networks modeled using individually-defined regions outperformed group-defined regions in predicting symptoms, and that individualized FC changes predicted treatment-based improvements. These findings highlight the added utility of individualized approaches to FC in psychiatry. Beyond post-hoc diagnosis, it is worth investigating whether pfMRI approaches will prove sensitive to risk factors for psychiatric disorders and disease progression, which would greatly enhance the utility of imaging in clinical management.
A third area of psychiatry that pfMRI can help to address are interventions that rely on subject-specific targeting of pathology. The spatially localized nature of many individual differences in FC (91, 103, 107, 108) means they may serve as patient-specific targets for stimulation-based interventions with TMS or DBS. While stimulation-based interventions can be effective, they suffer from variable patient responses (e.g. (133, 134)), which have been attributed to stimulation targeting procedures that do not respect individual variations in structural (135, 136) or functional (65, 137) neuroanatomy. Indeed, individual variability in FC of dorsolateral prefrontal cortex has been related to variation of TMS treatment efficacy in depression (66). Moreover, a recent pfMRI study of the sub-cortex showed that regions with consistent FC across individuals overlap with DBS stimulation sites that have shown more consistent treatment response, while DBS sites with known variability in response overlap with regions that exhibit variable FC (79). Use of pfMRI to identify individual-specific targets for brain stimulation thus has significant potential for improving treatment response rates (138).
A fourth challenge is to improve tracking of treatment efficacy and long-term remission. The multi-session nature of pfMRI allows researchers to determine how FC measurements vary over different time-scales. Thus far, evidence suggests that FC is stable across multiple sessions and even year-long periods, as long as sufficient data are collected per measurement to achieve high reliability (93). As such, pfMRI may be more sensitive to trait-like features that predict disease status or risk (e.g., whether a person has or will develop depression) than those that are associated with fluctuating behavior (e.g., current sad mood). However, pfMRI may also prove a more reliable baseline from which to expose rapid or profound changes in FC linked to treatment response to a pharmacologic or behavioral therapy. For example, lower-data but individualized FC approaches can predict changing OCD symptoms after intervention (132).
Feasibility of pfMRI in Clinical Samples
While pfMRI has many useful properties, the feasibility and cost of collecting extended datasets in patients are commonly cited as barriers to its use. Moreover, psychopathology is prevalent across the lifespan, and collecting pfMRI data from children and elderly adults may compound feasibility concerns. Such concerns can be addressed in a number of ways.
First, initial pfMRI studies have established that 30–45 minutes of low-artifact data may be sufficient to achieve good reliability for many cortical FC measures. In our initial investigations, we find that patient, pediatric, and elderly populations retain ~50–80% of data after motion denoising (29, 126, 129); thus, 45–90 min. of data collection would be needed to reach high reliability. While this is substantially more resting-state than is typically collected, this is not an unreasonable amount of scanning to ask of participants in general, as routine neurology assessments collect 2 hrs. of structural MRI. Clinicians may adjudicate whether the severity of psychiatric cases calls for similar scan investment (e.g., compare a relatively healthy patient with ADHD vs. a severely depressed patient with high suicide risk). Future methodological improvements may reduce the data needed to achieve reliable FC, though these may come at the cost of increased reliance on priors and decreased ability to observe divergent individual patterns (see Supplemental Discussion).
Second, since patient, pediatric, and elderly samples often exhibit increased head motion, additional strategies have been proposed to minimize motion and improve data quality (139). Our findings on FC stability suggest that data collection can effectively be split into several shorter runs within or across sessions to increase patient compliance. Indeed, breaking up data collection may even increase reliability levels (71), likely because of the autocorrelation structure of fMRI timeseries. Some have proposed combining task and rest to increase data quantities for FC (67), which could open up many current datasets for analysis. Care should be taken when mixing task and rest datasets, especially when effects of interest are small, but this may be an acceptable qualification for prediction/diagnosis (see Supplemental Discussion). Finally, data acquisition strategies such as movement feedback (140), on-line head motion estimates (141), and head cases to minimize motion (142) provide promising strategies to improve data quality during collection, rather than through post-hoc denoising.
Thus, while pfMRI studies require additional data per participant, for many applications the benefits of this investment (dramatically increased reliability and sensitivity to individual features) may be well worth the cost. Two or three hours of scanning may be of relatively small concern to patients considering having a DBS device implanted, suicidal individuals suffering from treatment-resistant depression, or parents seeking improved treatment for their child. We contend that it is not enormously useful to spend money on cheaper measures that do not replicate well within or across individuals – either for clinical practice or to forward research knowledge. Rather, direct translational application of neuroimaging results may be better afforded by pfMRI.
Supplementary Material
Acknowledgments:
This research was supported by a McDonnell Foundation Collaborative Activity Award (SEP); NIH grants R01MH118370 (CG), K01MH104592 (DJG), NS088590 (NUFD), and T32NS047987 (BTK); and a Career Development Award #1IK2CX001680 (EMG) from the US Department of Veterans Affairs Clinical Sciences Research and Development Service. The contents of this manuscript do not represent the views of the US Department of Veterans Affairs or the United States Government.
Footnotes
Disclosures:
CG, BTK, DJG, EMG, SMN, and SEP have no biomedical financial disclosures or potential conflicts of interest. NUFD. has a financial interest in Nous Imaging Inc. and may financially benefit if the company is successful in marketing FIRMM software products. NUFD may receive royalty income based on FIRMM technology developed at Oregon Health and Sciences University and Washington University and licensed to Nous Imaging Inc. TOL is a patent holder on US Patent App. 16/136,996 ‘System and method for task-less mapping of brain activity’ and US Patent App. 16/141,605 ‘Supervised classifier for optimizing target for neuromodulation, implant localization, and ablation’.
While pfMRI has only recently been employed in the field of large-scale functional network mapping, the practice of extended data collection in single individuals has a long history in both behavioral psychophysics and visual field mapping with fMRI (82–84). Recently these methods have been extended to measuring responses to complex naturalistic stimuli (85–87). PfMRI also mirrors the tradition of monkey electrophysiology studies that typically collect large amounts of data from small samples of monkeys (often 1–3). While these samples are not large enough for statistical testing, their logic is that they serve as study replicates across samples: i.e., an effect can be more meaningful when seen within each individual in a small group than if it is only observable on average across a large population sample (88, 89). We argue that a similar logic applies to studies of brain network organization, especially in clinical settings where treatment is applied to individuals.
References
- 1.Knapp M, Mangalore R, Simon J (2004): The global costs of schizophrenia. Schizophr Bull. 30:279–293. [DOI] [PubMed] [Google Scholar]
- 2.Luppa M, Heinrich S, Angermeyer MC, Konig HH, Riedel-Heller SG (2007): Cost-of-illness studies of depression: a systematic review. J Affect Disord. 98:29–43. [DOI] [PubMed] [Google Scholar]
- 3.Vigo D, Thornicroft G, Atun R (2016): Estimating the true global burden of mental illness. Lancet Psychiatry. 3:171–178. [DOI] [PubMed] [Google Scholar]
- 4.Mayberg HS (2007): Defining the neural circuitry of depression: toward a new nosology with therapeutic implications. Biol Psychiatry. 61:729–730. [DOI] [PubMed] [Google Scholar]
- 5.Karbasforoushan H, Woodward ND (2012): Resting-state networks in schizophrenia. Curr Top Med Chem. 12:2404–2414. [DOI] [PubMed] [Google Scholar]
- 6.Barch DM (2017): Resting-State Functional Connectivity in the Human Connectome Project: Current Status and Relevance to Understanding Psychopathology. Harv Rev Psychiatry. 25:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotov R, Krueger RF, Watson D, Achenbach TM, Althoff RR, Bagby RM, et al. (2017): The Hierarchical Taxonomy of Psychopathology (HiTOP): A dimensional alternative to traditional nosologies. J Abnorm Psychol. 126:454–477. [DOI] [PubMed] [Google Scholar]
- 8.Leckman JF, Bloch MH, King RA, Scahill L (2006): Phenomenology of tics and natural history of tic disorders. Adv Neurol. 99:1–16. [PubMed] [Google Scholar]
- 9.Churchland PS, Sejnowski TJ (1988): Perspectives on cognitive neuroscience. Science. 242:741–745. [DOI] [PubMed] [Google Scholar]
- 10.Buckner RL, Krienen FM, Yeo BT (2013): Opportunities and limitations of intrinsic functional connectivity MRI. Nat Neurosci. 16:832–837. [DOI] [PubMed] [Google Scholar]
- 11.Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. (2011): The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of neurophysiology. 106:1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen SE, Sporns O (2015): Brain Networks and Cognitive Architectures. Neuron. 88:207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, et al. (2011): Functional network organization of the human brain. Neuron. 72:665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Power JD, Schlaggar BL, Petersen SE (2014): Studying brain organization via spontaneous fMRI signal. Neuron. 84:681–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eickhoff SB, Yeo BTT, Genon S (2018): Imaging-based parcellations of the human brain. Nature reviews Neuroscience. 19:672–686. [DOI] [PubMed] [Google Scholar]
- 16.Wig GS, Laumann TO, Petersen SE (2014): An approach for parcellating human cortical areas using resting-state correlations. Neuroimage. 93 Pt 2:276–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dosenbach NU, Fair DA, Miezin FM, Cohen AL, Wenger KK, Dosenbach RA, et al. (2007): Distinct brain networks for adaptive and stable task control in humans. Proceedings of the National Academy of Sciences of the United States of America. 104:11073–11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, et al. (2007): Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 27:2349–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corbetta M, Shulman GL (2002): Control of goal-directed and stimulus-driven attention in the brain. Nature reviews Neuroscience. 3:201–215. [DOI] [PubMed] [Google Scholar]
- 20.Greicius MD, Krasnow B, Reiss AL, Menon V (2003): Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proceedings of the National Academy of Sciences of the United States of America. 100:253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, et al. (2007): Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 447:83–86. [DOI] [PubMed] [Google Scholar]
- 22.Johnston JM, Vaishnavi SN, Smyth MD, Zhang D, He BJ, Zempel JM, et al. (2008): Loss of resting interhemispheric functional connectivity after complete section of the corpus callosum. J Neurosci. 28:6453–6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nomura EM, Gratton C, Visser RM, Kayser A, Perez F, D’Esposito M (2010): Double dissociation of two cognitive control networks in patients with focal brain lesions. Proceedings of the National Academy of Sciences of the United States of America. 107:12017–12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD (2012): Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cereb Cortex. 22:158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE (2016): Generation and Evaluation of a Cortical Area Parcellation from Resting-State Correlations. Cereb Cortex. 26:288–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheffield JM, Barch DM (2016): Cognition and resting-state functional connectivity in schizophrenia. Neurosci Biobehav Rev. 61:108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaiser RH, Andrews-Hanna JR, Wager TD, Pizzagalli DA (2015): Large-Scale Network Dysfunction in Major Depressive Disorder: A Meta-analysis of Resting-State Functional Connectivity. JAMA Psychiatry. 72:603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greene DJ, Church JA, Dosenbach NUF, Nielsen AN, Adeyemo B, Nardos B, et al. (2016): Multivariate pattern classification of pediatric Tourette syndrome using functional connectivity MRI. Developmental science. 19:581–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen AN, Gratton C, Church JA, Dosenbach NU, Black KJ, Petersen SE, et al. (in press): Atypical functional connectivity in Tourette syndrome differs between children and adults. Biol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker JT, Dillon DG, Patrick LM, Roffman JL, Brady RO Jr., Pizzagalli DA, et al. (2019): Functional connectomics of affective and psychotic pathology. Proceedings of the National Academy of Sciences of the United States of America. 116:9050–9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kebets V, Holmes AJ, Orban C, Tang S, Li J, Sun N, et al. (2019): Somatosensory-Motor Dysconnectivity Spans Multiple Transdiagnostic Dimensions of Psychopathology. BioRxiv.637827. [DOI] [PubMed]
- 32.Xia CH, Ma Z, Ciric R, Gu S, Betzel RF, Kaczkurkin AN, et al. (2018): Linked dimensions of psychopathology and connectivity in functional brain networks. Nat Commun. 9:3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gratton C, Neta M, Sun H, Ploran EJ, Schlaggar BL, Wheeler ME, et al. (2017): Distinct Stages of Moment-to-Moment Processing in the Cinguloopercular and Frontoparietal Networks. Cereb Cortex. 27:2403–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neta M, Miezin FM, Nelson SM, Dubis JW, Dosenbach NU, Schlaggar BL, et al. (2015): Spatial and temporal characteristics of error-related activity in the human brain. J Neurosci. 35:253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shanmugan S, Wolf DH, Calkins ME, Moore TM, Ruparel K, Hopson RD, et al. (2016): Common and Dissociable Mechanisms of Executive System Dysfunction Across Psychiatric Disorders in Youth. Am J Psychiatry. 173:517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sporns O (2010): Networks of the Brain. MIT press. [Google Scholar]
- 37.Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, et al. (2009): Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer’s disease. J Neurosci. 29:1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bullmore E, Sporns O (2009): Complex brain networks: graph theoretical analysis of structural and functional systems. Nature reviews Neuroscience. 10:186–198. [DOI] [PubMed] [Google Scholar]
- 39.Power JD, Schlaggar BL, Lessov-Schlaggar CN, Petersen SE (2013): Evidence for hubs in human functional brain networks. Neuron. 79:798–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van den Heuvel MP, Sporns O (2013): Network hubs in the human brain. Trends in cognitive sciences. 17:683–696. [DOI] [PubMed] [Google Scholar]
- 41.Bertolero MA, Yeo BT, D’Esposito M (2015): The modular and integrative functional architecture of the human brain. Proceedings of the National Academy of Sciences of the United States of America. 112:E6798–6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menon V, Uddin LQ (2010): Saliency, switching, attention and control: a network model of insula function. Brain Struct Funct. 214:655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole MW, Repovs G, Anticevic A (2014): The frontoparietal control system: a central role in mental health. Neuroscientist. 20:652–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gordon EM, Lynch CJ, Gratton C, Laumann TO, Gilmore AW, Greene DJ, et al. (2018): Three Distinct Sets of Connector Hubs Integrate Human Brain Function. Cell Rep. 24:1687–1695 e1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gratton C, Laumann TO, Gordon EM, Adeyemo B, Petersen SE (2016): Evidence for Two Independent Factors that Modify Brain Networks to Meet Task Goals. Cell Rep. 17:1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crossley NA, Mechelli A, Scott J, Carletti F, Fox PT, McGuire P, et al. (2014): The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 137:2382–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gratton C, Nomura EM, Perez F, D’Esposito M (2012): Focal brain lesions to critical locations cause widespread disruption of the modular organization of the brain. J Cogn Neurosci. 24:1275–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warren DE, Power JD, Bruss J, Denburg NL, Waldron EJ, Sun H, et al. (2014): Network measures predict neuropsychological outcome after brain injury. Proceedings of the National Academy of Sciences of the United States of America. 111:14247–14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan MY, Park DC, Savalia NK, Petersen SE, Wig GS (2014): Decreased segregation of brain systems across the healthy adult lifespan. Proceedings of the National Academy of Sciences of the United States of America. 111:E4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fair DA, Cohen AL, Power JD, Dosenbach NU, Church JA, Miezin FM, et al. (2009): Functional brain networks develop from a “local to distributed” organization. PLoS Comput Biol. 5:e1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu S, Satterthwaite TD, Medaglia JD, Yang M, Gur RE, Gur RC, et al. (2015): Emergence of system roles in normative neurodevelopment. Proceedings of the National Academy of Sciences of the United States of America. 112:13681–13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bassett DS, Xia CH, Satterthwaite TD (2018): Understanding the Emergence of Neuropsychiatric Disorders With Network Neuroscience. Biol Psychiatry Cogn Neurosci Neuroimaging. 3:742–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bertolero MA, Yeo BTT, Bassett DS, D’Esposito M (2018): A mechanistic model of connector hubs, modularity and cognition. Nat Hum Behav. 2:765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu S, Pasqualetti F, Cieslak M, Telesford QK, Yu AB, Kahn AE, et al. (2015): Controllability of structural brain networks. Nat Commun. 6:8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fox MD, Greicius M (2010): Clinical applications of resting state functional connectivity. Front Syst Neurosci. 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee MH, Miller-Thomas MM, Benzinger TL, Marcus DS, Hacker CD, Leuthardt EC, et al. (2016): Clinical Resting-state fMRI in the Preoperative Setting: Are We Ready for Prime Time? Top Magn Reson Imaging. 25:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hacker CD, Laumann TO, Szrama NP, Baldassarre A, Snyder AZ, Leuthardt EC, et al. (2013): Resting state network estimation in individual subjects. Neuroimage. 82:616–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu H, Buckner RL, Talukdar T, Tanaka N, Madsen JR, Stufflebeam SM (2009): Task-free presurgical mapping using functional magnetic resonance imaging intrinsic activity. J Neurosurg. 111:746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mitchell TJ, Hacker CD, Breshears JD, Szrama NP, Sharma M, Bundy DT, et al. (2013): A novel data-driven approach to preoperative mapping of functional cortex using resting-state functional magnetic resonance imaging. Neurosurgery. 73:969–982; discussion 982–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang D, Buckner RL, Fox MD, Holt DJ, Holmes AJ, Stoecklein S, et al. (2015): Parcellating cortical functional networks in individuals. Nat Neurosci. 18:1853–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clark SV, Mittal VA, Bernard JA, Ahmadi A, King TZ, Turner JA (2018): Stronger default mode network connectivity is associated with poorer clinical insight in youth at ultra high-risk for psychotic disorders. Schizophr Res. 193:244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niznikiewicz MA (2019): Neurobiological approaches to the study of clinical and genetic high risk for developing psychosis. Psychiatry Res. [DOI] [PubMed] [Google Scholar]
- 63.Sorg C, Riedl V, Muhlau M, Calhoun VD, Eichele T, Laer L, et al. (2007): Selective changes of resting-state networks in individuals at risk for Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America. 104:18760–18765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, et al. (2017): Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med. 23:28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weigand A, Horn A, Caballero R, Cooke D, Stern AP, Taylor SF, et al. (2018): Prospective Validation That Subgenual Connectivity Predicts Antidepressant Efficacy of Transcranial Magnetic Stimulation Sites. Biol Psychiatry. 84:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fox MD, Liu H, Pascual-Leone A (2013): Identification of reproducible individualized targets for treatment of depression with TMS based on intrinsic connectivity. Neuroimage. 66:151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elliott ML, Knodt AR, Cooke M, Kim MJ, Melzer TR, Keenan R, et al. (2019): General functional connectivity: Shared features of resting-state and task fMRI drive reliable and heritable individual differences in functional brain networks. Neuroimage. 189:516–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson JS, Ferguson MA, Lopez-Larson M, Yurgelun-Todd D (2011): Reproducibility of single-subject functional connectivity measurements. AJNR Am J Neuroradiol. 32:548–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gordon EM, Laumann TO, Gilmore AW, Newbold DJ, Greene DJ, Berg JJ, et al. (2017): Precision Functional Mapping of Individual Human Brains. Neuron. 95:791–807 e797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen MY, et al. (2015): Functional System and Areal Organization of a Highly Sampled Individual Human Brain. Neuron. 87:657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Noble S, Spann MN, Tokoglu F, Shen X, Constable RT, Scheinost D (2017): Influences on the Test-Retest Reliability of Functional Connectivity MRI and its Relationship with Behavioral Utility. Cereb Cortex. 27:5415–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu T, Opitz A, Craddock RC, Wright MJ, Zuo XN, Milham MP (2016): Assessing Variations in Areal Organization for the Intrinsic Brain: From Fingerprints to Reliability. Cereb Cortex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Raut RV, Mitra A, Snyder AZ, Raichle ME (2019): On time delay estimation and sampling error in resting-state fMRI. Neuroimage. 194:211–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raut RV, Mitra A, Marek S, Ortega M, Snyder AZ, Tanenbaum A, et al. (2019): Organization of Propagated Intrinsic Brain Activity in Individual Humans. Cereb Cortex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bradshaw JL, Sheppard DM (2000): The neurodevelopmental frontostriatal disorders: evolutionary adaptiveness and anomalous lateralization. Brain Lang. 73:297–320. [DOI] [PubMed] [Google Scholar]
- 76.Hariri AR (2019): The Emerging Importance of the Cerebellum in Broad Risk for Psychopathology. Neuron. 102:17–20. [DOI] [PubMed] [Google Scholar]
- 77.Ramasubbu R, Konduru N, Cortese F, Bray S, Gaxiola-Valdez I, Goodyear B (2014): Reduced intrinsic connectivity of amygdala in adults with major depressive disorder. Front Psychiatry. 5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marek S, Siegel JS, Gordon EM, Raut RV, Gratton C, Newbold DJ, et al. (2018): Spatial and Temporal Organization of the Individual Human Cerebellum. Neuron. 100:977–993 e977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Greene DJ, Marek S, Siegel JS, Gordon EM, Gratton C, Newbold DJ, et al. (revise & resubmit): Individual specific and shared integrative zones of the human thalamus and basal ganglia.
- 80.Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, et al. (2015): Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat Neurosci. 18:1664–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miranda-Dominguez O, Mills BD, Carpenter SD, Grant KA, Kroenke CD, Nigg JT, et al. (2014): Connectotyping: model based fingerprinting of the functional connectome. PLoS One. 9:e111048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Engel SA, Glover GH, Wandell BA (1997): Retinotopic organization in human visual cortex and the spatial precision of functional MRI. Cerebral cortex (New York, NY: 1991). 7:181–192. [DOI] [PubMed] [Google Scholar]
- 83.Tootell RB, Hadjikhani NK, Mendola JD, Marrett S, Dale AM (1998): From retinotopy to recognition: fMRI in human visual cortex. Trends in cognitive sciences. 2:174–183. [DOI] [PubMed] [Google Scholar]
- 84.Tootell RB, Hadjikhani NK, Vanduffel W, Liu AK, Mendola JD, Sereno MI, et al. (1998): Functional analysis of primary visual cortex (V1) in humans. Proceedings of the National Academy of Sciences. 95:811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huth AG, de Heer WA, Griffiths TL, Theunissen FE, Gallant JL (2016): Natural speech reveals the semantic maps that tile human cerebral cortex. Nature. 532:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huth AG, Nishimoto S, Vu AT, Gallant JL (2012): A continuous semantic space describes the representation of thousands of object and action categories across the human brain. Neuron. 76:1210–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kay KN, Naselaris T, Prenger RJ, Gallant JL (2008): Identifying natural images from human brain activity. Nature. 452:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Normand MP (2016): Less is more: Psychologists can learn more by studying fewer people. Frontiers in psychology. 7:934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith PL, Little DR (2018): Small is beautiful: In defense of the small-N design. Psychonomic bulletin & review. 25:2083–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Poldrack RA, Laumann TO, Koyejo O, Gregory B, Hover A, Chen MY, et al. (2015): Long-term neural and physiological phenotyping of a single human. Nat Commun. 6:8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Seitzman BA, Gratton C, Laumann TO, Gordon EM, Adeyemo B, Gilmore AW, et al. (in press): Trait-like variants in human functional brain networks. Proceedings of the National Academy of Sciences of the United States of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Braga RM, Buckner RL (2017): Parallel Interdigitated Distributed Networks within the Individual Estimated by Intrinsic Functional Connectivity. Neuron. 95:457–471 e455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gratton C, Laumann TO, Nielsen AN, Greene DJ, Gordon EM, Gilmore AW, et al. (2018): Functional Brain Networks Are Dominated by Stable Group and Individual Factors, Not Cognitive or Daily Variation. Neuron. 98:439–452 e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cole MW, Bassett DS, Power JD, Braver TS, Petersen SE (2014): Intrinsic and task-evoked network architectures of the human brain. Neuron. 83:238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Geerligs L, Rubinov M, Cam C, Henson RN (2015): State and Trait Components of Functional Connectivity: Individual Differences Vary with Mental State. J Neurosci. 35:13949–13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xie H, Calhoun VD, Gonzalez-Castillo J, Damaraju E, Miller R, Bandettini PA, et al. (2017): Whole-brain connectivity dynamics reflect both task-specific and individual-specific modulation: A multitask study. Neuroimage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Laumann TO, Snyder AZ, Mitra A, Gordon EM, Gratton C, Adeyemo B, et al. (2017): On the Stability of BOLD fMRI Correlations. Cereb Cortex. 27:4719–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Greene DJ, Williams III AC, Koller JM, Schlaggar BL, Black KJ (2017): Brain structure in pediatric Tourette syndrome. Molecular psychiatry. 22:972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peterson BS, Thomas P, Kane MJ, Scahill L, Zhang H, Bronen R, et al. (2003): Basal Ganglia volumes in patients with Gilles de la Tourette syndrome. Arch Gen Psychiatry. 60:415–424. [DOI] [PubMed] [Google Scholar]
- 100.Giraldo-Chica M, Woodward ND (2017): Review of thalamocortical resting-state fMRI studies in schizophrenia. Schizophr Res. 180:58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McCutcheon RA, Abi-Dargham A, Howes OD (2019): Schizophrenia, Dopamine and the Striatum: From Biology to Symptoms. Trends Neurosci. 42:205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Choe AS, Jones CK, Joel SE, Muschelli J, Belegu V, Caffo BS, et al. (2015): Reproducibility and Temporal Structure in Weekly Resting-State fMRI over a Period of 3.5 Years. PLoS One. 10:e0140134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gordon EM, Laumann TO, Adeyemo B, Gilmore AW, Nelson SM, Dosenbach NUF, et al. (2017): Individual-specific features of brain systems identified with resting state functional correlations. Neuroimage. 146:918–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gordon EM, Laumann TO, Adeyemo B, Petersen SE (2017): Individual Variability of the System-Level Organization of the Human Brain. Cereb Cortex. 27:386–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mueller S, Wang D, Fox MD, Yeo BT, Sepulcre J, Sabuncu MR, et al. (2013): Individual variability in functional connectivity architecture of the human brain. Neuron. 77:586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Harrison SJ, Woolrich MW, Robinson EC, Glasser MF, Beckmann CF, Jenkinson M, et al. (2015): Large-scale probabilistic functional modes from resting state fMRI. Neuroimage. 109:217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bijsterbosch JD, Beckmann CF, Woolrich MW, Smith SM, Harrison SJ (2019): The relationship between spatial configuration and functional connectivity of brain regions revisited. Elife. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kong R, Li J, Orban C, Sabuncu MR, Liu H, Schaefer A, et al. (2019): Spatial Topography of Individual-Specific Cortical Networks Predicts Human Cognition, Personality, and Emotion. Cereb Cortex. 29:2533–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, et al. (2013): The WU-Minn Human Connectome Project: an overview. Neuroimage. 80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Braver TS (2012): The variable nature of cognitive control: a dual mechanisms framework. Trends in cognitive sciences. 16:106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Duncan J, Owen AM (2000): Common regions of the human frontal lobe recruited by diverse cognitive demands. Trends Neurosci. 23:475–483. [DOI] [PubMed] [Google Scholar]
- 112.Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL (2001): A default mode of brain function. Proceedings of the National Academy of Sciences of the United States of America. 98:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weissman DH, Roberts KC, Visscher KM, Woldorff MG (2006): The neural bases of momentary lapses in attention. Nat Neurosci. 9:971–978. [DOI] [PubMed] [Google Scholar]
- 114.Friedman NP, Miyake A, Young SE, Defries JC, Corley RP, Hewitt JK (2008): Individual differences in executive functions are almost entirely genetic in origin. J Exp Psychol Gen. 137:201–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Disner SG, Beevers CG, Haigh EA, Beck AT (2011): Neural mechanisms of the cognitive model of depression. Nature reviews Neuroscience. 12:467–477. [DOI] [PubMed] [Google Scholar]
- 116.Fales CL, Barch DM, Rundle MM, Mintun MA, Snyder AZ, Cohen JD, et al. (2008): Altered emotional interference processing in affective and cognitive-control brain circuitry in major depression. Biol Psychiatry. 63:377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Church JA, Wenger KK, Dosenbach NU, Miezin FM, Petersen SE, Schlaggar BL (2009): Task control signals in pediatric tourette syndrome show evidence of immature and anomalous functional activity. Front Hum Neurosci. 3:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tobyne SM, Somers DC, Brissenden JA, Michalka SW, Noyce AL, Osher DE (2018): Prediction of individualized task activation in sensory modality-selective frontal cortex with ‘connectome fingerprinting’. Neuroimage. 183:173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li M, Wang D, Ren J, Langs G, Stoecklein S, Brennan BP, et al. (2019): Performing group-level functional image analyses based on homologous functional regions mapped in individuals. PLoS Biol. 17:e2007032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Smith SM, Nichols TE, Vidaurre D, Winkler AM, Behrens TE, Glasser MF, et al. (2015): A positive-negative mode of population covariation links brain connectivity, demographics and behavior. Nat Neurosci. 18:1565–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tavor I, Parker Jones O, Mars RB, Smith SM, Behrens TE, Jbabdi S (2016): Task-free MRI predicts individual differences in brain activity during task performance. Science. 352:216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lynch CJ, Breeden A, Gordon EM, Cherry JB, Turkeltaub PE, Vaidya CJ (2019): Precision inhibitory stimulation of individual-specific cortical hubs disrupts information processing in humans. Cerebral cortex (New York, NY: 1991). 29:3912–3921. [DOI] [PubMed] [Google Scholar]
- 123.Collaboration OS (2015): Estimating the reproducibility of psychological science. Science. 349:aac4716. [DOI] [PubMed] [Google Scholar]
- 124.Insel TR (2015): The NIMH experimental medicine initiative. World Psychiatry. 14:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Poldrack RA, Baker CI, Durnez J, Gorgolewski KJ, Matthews PM, Munafò MR, et al. (2017): Scanning the horizon: towards transparent and reproducible neuroimaging research. Nature Reviews Neuroscience. 18:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gordon EM, Scheibel RS, Zambrano-Vazquez L, Jia-Richards M, May GJ, Meyer EC, et al. (2018): High-Fidelity Measures of Whole-Brain Functional Connectivity and White Matter Integrity Mediate Relationships between Traumatic Brain Injury and Post-Traumatic Stress Disorder Symptoms. J Neurotrauma. 35:767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Larson-Prior LJ, Zempel JM, Nolan TS, Prior FW, Snyder AZ, Raichle ME (2009): Cortical network functional connectivity in the descent to sleep. Proceedings of the National Academy of Sciences of the United States of America. 106:4489–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nielsen AN, Greene DJ, Gratton C, Dosenbach NUF, Petersen SE, Schlaggar BL (2019): Evaluating the Prediction of Brain Maturity From Functional Connectivity After Motion Artifact Denoising. Cereb Cortex. 29:2455–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gratton C, Koller JM, Shannon W, Greene DJ, Maiti B, Snyder AZ, et al. (2019): Emergent Functional Network Effects in Parkinson Disease. Cereb Cortex. 29:1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Repovs G, Csernansky JG, Barch DM (2011): Brain network connectivity in individuals with schizophrenia and their siblings. Biol Psychiatry. 69:967–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang D, Li M, Wang M, Schoeppe F, Ren J, Chen H, et al. (2018): Individual-specific functional connectivity markers track dimensional and categorical features of psychotic illness. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brennan BP, Wang D, Li M, Perriello C, Ren J, Elias JA, et al. (2019): Use of an Individual-Level Approach to Identify Cortical Connectivity Biomarkers in Obsessive-Compulsive Disorder. Biol Psychiatry Cogn Neurosci Neuroimaging. 4:27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gaynes BN, Lloyd SW, Lux L, Gartlehner G, Hansen RA, Brode S, et al. (2014): Repetitive transcranial magnetic stimulation for treatment-resistant depression: a systematic review and meta-analysis. J Clin Psychiatry. 75:477–489; quiz 489. [DOI] [PubMed] [Google Scholar]
- 134.Nestor KA, Jones JD, Butson CR, Morishita T, Jacobson CEt, Peace DA, et al. (2014): Coordinate-based lead location does not predict Parkinson’s disease deep brain stimulation outcome. PLoS One. 9:e93524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fitzgerald PB, Hoy K, McQueen S, Maller JJ, Herring S, Segrave R, et al. (2009): A randomized trial of rTMS targeted with MRI based neuro-navigation in treatment-resistant depression. Neuropsychopharmacology. 34:1255–1262. [DOI] [PubMed] [Google Scholar]
- 136.Tolleson C, Pallavaram S, Li C, Fang J, Phibbs F, Konrad P, et al. (2015): The optimal pallidal target in deep brain stimulation for dystonia: a study using a functional atlas based on nonlinear image registration. Stereotact Funct Neurosurg. 93:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sack AT, Cohen Kadosh R, Schuhmann T, Moerel M, Walsh V, Goebel R (2009): Optimizing functional accuracy of TMS in cognitive studies: a comparison of methods. J Cogn Neurosci. 21:207–221. [DOI] [PubMed] [Google Scholar]
- 138.Luber BM, Davis S, Bernhardt E, Neacsiu A, Kwapil L, Lisanby SH, et al. (2017): Using neuroimaging to individualize TMS treatment for depression: Toward a new paradigm for imaging-guided intervention. Neuroimage. 148:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Greene DJ, Black KJ, Schlaggar BL (2016): Considerations for MRI study design and implementation in pediatric and clinical populations. Developmental cognitive neuroscience. 18:101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Greene DJ, Koller JM, Hampton JM, Wesevich V, Van AN, Nguyen AL, et al. (2018): Behavioral interventions for reducing head motion during MRI scans in children. Neuroimage. 171:234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dosenbach NUF, Koller JM, Earl EA, Miranda-Dominguez O, Klein RL, Van AN, et al. (2017): Real-time motion analytics during brain MRI improve data quality and reduce costs. Neuroimage. 161:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Power JD, Silver BM, Silverman MR, Ajodan EL, Bos DJ, Jones RM (2019): Customized head molds reduce motion during resting state fMRI scans. Neuroimage. 189:141–149. [DOI] [PubMed] [Google Scholar]
- 143.Zuo XN, Anderson JS, Bellec P, Birn RM, Biswal BB, Blautzik J, et al. (2014): An open science resource for establishing reliability and reproducibility in functional connectomics. Sci Data. 1:140049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Filevich E, Lisofsky N, Becker M, Butler O, Lochstet M, Martensson J, et al. (2017): Day2day: investigating daily variability of magnetic resonance imaging measures over half a year. BMC Neurosci. 18:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.O’Connor D, Potler NV, Kovacs M, Xu T, Ai L, Pellman J, et al. (2017): The Healthy Brain Network Serial Scanning Initiative: a resource for evaluating inter-individual differences and their reliabilities across scan conditions and sessions. Gigascience. 6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Meszlenyi RJ, Hermann P, Buza K, Gal V, Vidnyanszky Z (2017): Resting State fMRI Functional Connectivity Analysis Using Dynamic Time Warping. Front Neurosci. 11:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Duchesne S, Chouinard I, Potvin O, Fonov VS, Khademi A, Bartha R, et al. (2019): The Canadian Dementia Imaging Protocol: Harmonizing National Cohorts. J Magn Reson Imaging. 49:456–465. [DOI] [PubMed] [Google Scholar]
- 148.Donnelly-Kehoe P, Saenger VM, Lisofsky N, Kuhn S, Kringelbach ML, Schwarzbach J, et al. (2019): Reliable local dynamics in the brain across sessions are revealed by whole-brain modeling of resting state activity. Hum Brain Mapp. 40:2967–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kong X multiBrain. https://github.com/Conxz/multiBrain.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.